Abstract
Plastidial acetyl-coenzyme A carboxylase from most plants is a multi-enzyme complex comprised of four different subunits. One of these subunits, the biotin carboxyl carrier protein (BCCP), was previously proposed to be encoded by a single gene in Arabidopsis. We report and characterize here a second Arabidopsis BCCP (AtBCCP2) cDNA with 42% amino acid identity to AtBCCP1 and 75% identity to a class of oilseed rape (Brassica napus) BCCPs. Both Arabidopsis BCCP isoforms were expressed in Escherichia coli and found to be biotinylated and supported carboxylation activity when reconstituted with purified, recombinant Arabidopsis biotin carboxylase. In vitro translated AtBCCP2 was competent for import into pea (Pisum sativum) chloroplasts and processed to a 25-kD polypeptide. Extracts of Arabidopsis seeds contained biotinylated polypeptides of 35 and 25 kD, in agreement with the masses of recombinant AtBCCP1 and 2, respectively. AtBCCP1 protein was present in developing tissues from roots, leaves, flowers, siliques, and seeds, whereas AtBCCP2 protein was primarily expressed in 7 to 10 d-after-flowering seeds at levels approximately 2-fold less abundant than AtBCCP1. AtBCCP1 transcript reflected these protein expression profiles present in all developing organs and highest in 14-d leaves and siliques, whereas AtBCCP2 transcript was present in flowers and siliques. In protein blots, four different BCCP isoforms were detected in developing seeds from oilseed rape. Of these, a 35-kD BCCP was detected in immature leaves and developing seeds, whereas developing seeds also contained 22-, 25-, and 37-kD isoforms highly expressed 21 d after flowering. These data indicate that oilseed plants in the family Brassicaceae contain at least one to three seed-up-regulated BCCP isoforms, depending upon genome complexity.
The committed step for de novo fatty acid biosynthesis is the formation of malonyl-coenzyme A (CoA), catalyzed by acetyl-CoA carboxylase (ACCase, EC 6.4.1.2). ACCase catalyzes the ATP-dependent carboxylation of acetyl-CoA through a carboxylation and carboxyltransferase two-step reaction (Guchait et al., 1974). The number and organization of ACCase isoforms is variable in plants (Sasaki et al., 1995). All plants contain a homomeric cytosolic isoform that is proposed to be involved in flavonoid and long-chain fatty acid biosynthesis (Shorrosh et al., 1994). A second isoform is present within the plastid stroma of plant cells and is involved in de novo fatty acid biosynthesis (Kannangara and Stumpf, 1972; Alban et al., 1994). In graminae plants, plastidial ACCase is a homomeric enzyme, whereas in dicots and non-graminae monocots it is composed of at least four polypeptides assembled as a complex of approximately 600 kD (Sasaki et al., 1995; Ohlrogge and Jaworski, 1997). Because de novo fatty acid biosynthesis occurs predominantly in plastids (Ohlrogge et al., 1979) and malonyl-CoA is impermeable to the plastid envelope, it is widely accepted that plastidial ACCase is the key initial enzyme in this pathway. In addition to a heteromeric isoform, plastids from Brassicaceae plants also contain a homomeric ACCase, the role of which remains unknown (Schulte et al., 1997).
A wealth of direct and indirect evidence suggests plastidial ACCase is one major control point for light-induced fatty acid biosynthesis in leaves and likely one rate-limiting step for this pathway in other organs. Evidence in support of this premise is summarized below. Measurement of in vivo acyl-acyl carrier protein (ACP) pools from isolated pea (Pisum sativum) chloroplasts indicated malonyl-ACP was shifted to acetyl-ACP after the transition to darkness (Post-Beittenmiller et al., 1991, 1992). Biochemical properties of plastidial ACCase indicate optimal activity during light-adapted stromal conditions (Nikolau and Hawke, 1984; Hunter and Ohlrogge, 1998). Regulation of ACCase via reversible redox activation (Sasaki et al., 1997) or reversible protein phosphorylation (Savage and Ohlrogge, 1999) is mediated by light. In graminae monocots, inhibitor studies coupled with in vivo labeling indicated strong flux control coefficients at this step (Page et al., 1994). In suspension cells, feedback inhibition of fatty acid biosynthesis occurred at the ACCase step (Shintani and Ohlrogge, 1995). Targeting of a homomeric ACCase to the plastids of rapeseed resulted in a 5% increase in total seed fatty acid content (Roesler et al., 1997).
Besides being present in dicotyledon plant and green algal plastids, multisubunit ACCases are also found in gram-negative and gram-positive prokaryotes (Li and Cronan, 1992a, 1992b; Marini et al., 1995). Multisubunit ACCases are comprised of biotin carboxyl carrier protein (BCCP), biotin carboxylase (BC), α-carboxyltransferase, and β-carboxyltransferase subunits. Though the organization of the plant complex is unknown, the prokaryotic counterpart is a homodimer of BC polypeptides assembled with a homodimer of BCCP, which is loosely associated to a heterotetramer of α- and β-carboxyltransferase subunits (Guchait et al., 1974). In higher plants the β-carboxyltransferase subunit is plastid-encoded (Li and Cronan, 1992c; Sasaki et al., 1993), whereas the remaining three subunits are nuclear-encoded. Genes or cDNAs for the nuclear-encoded subunits have been characterized from Arabidopsis (Choi et al., 1995; Bao et al., 1997; Sun et al., 1997), oilseed rape (Brassica napus; Elborough et al., 1996), soybean (Reverdatto et al., 1999), and pea (Shorrosh et al., 1996). The BCCP subunit from Arabidopsis was previously reported to be a single gene copy (CAC1, Choi et al., 1995). In contrast, the allotetraploid relative oilseed rape contains at least six BCCP gene copies (Elborough et al., 1996). The oilseed rape isoforms group into two distinct classes based upon amino acid and nucleotide sequence comparisons. The previously identified Arabidopsis BCCP (AtBCCP1; accession no. AF236873) is most similar to class one oilseed rape BCCP subunits. In this report we present evidence for a second BCCP isoform from Arabidopsis, which is a homolog to class two BCCPs from oilseed rape. Based upon semiquantitative immunoblot analyses we conclude that class one BCCPs are strongly expressed in most plant organs, whereas class two BCCPs accumulate in developing seed and might have an evolved role in fatty acid biosynthesis for lipid deposition.
RESULTS
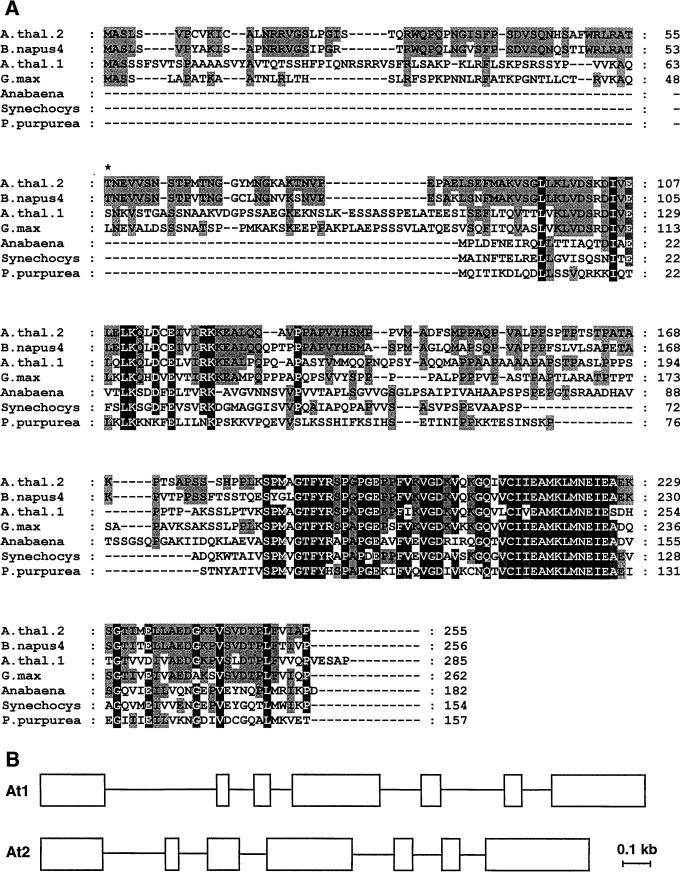
In a search of the Arabidopsis expressed sequence tag (EST) database using the AtBCCP1 polypeptide as a search query, a total of 13 putative BCCP ESTs were found. Two of these 13 were partial cDNAs that correspond to the previously described AtBCCP1 transcripts. The 11 remaining cDNAs encoded a protein related to AtBCCP1, primarily in the C-terminal one-half of the polypeptide. Eight of these 11 encoded the entire open reading frame (ORF) and four also contained the entire 5′-untranslated region (UTR; GenBank accession nos. H37386, H37396, AI992947, and N38652). cDNA clone H37386 was completely sequenced and is reported here as AtBCCP2. The deduced polypeptide exhibits 42% amino acid identity with AtBCCP1, 40% with oilseed rape class one isoforms, 75% with oilseed rape class two isoforms, and 50% with soybean BCCP. The gene for AtBCCP2 (accession no. AF223948) was recently sequenced and mapped to chromosome V. Based upon the intron/exon structure of these two genes (Fig. 1B) and their location on chromosome V it appears the second gene arose from an ancestral gene duplication event.
Figure 1.
Amino acid sequence comparison of plant and cyanobacterial BCCPs. Alignment was performed with Clustal X software using default parameters. Shading reflects the degree of amino acid conservation; black shading indicates amino acid identity with all polypeptides. Plastid processing site for soybean BCCP occurs between residues 48 and 49. By comparison, the mature AtBCCP2 was postulated to start with Thr-56, indicated by an asterisk. GenBank accession numbers are X90730, oilseed rape4; U23155, Arabidopsis1; U40666, soybean; L14863, Anabaena; D64001, Synechocys; and U38804, P. purpurea. B, Gene organization of Arabidopsis BCCP isoforms. A scale model of intron and exon (white boxes) organization for Arabidopsis BCCP isoforms one and two are illustrated. Scale bar at the lower right is 0.1 kb in length. GenBank accession numbers are as follows: At1, AB005242 and At2, AP002074.
The AtBCCP2 cDNA is 1,235 bp in length with a 765-bp ORF starting with an initiating codon at bp 94 and a stop codon at bp 859. A consensus translation start motif (90aacaatggc) containing the initiating Met is present at the 5′ end and a poly(A) initiation signal (1190aaataaa) at the 3′ end. The ORF encodes a 255-amino acid polypeptide with a predicted Mr of 27,248. The highest similarity between AtBCCP1 and AtBCCP2 is concentrated at the C terminus, in the proximity of the biotinylation motif 217EAMKLMNEIE226, which contains the putative biotinyl Lys (Samols et al., 1998; Fig. 1). The amino terminus (approximately 50 amino acids) resembles a plastid-targeting peptide as revealed by organellar sorting algorithms. This putative plastid targeting peptide is only 30% identical to AtBCCP1 and soybean BCCP-targeting peptides, but is 85% identical to those in class two oilseed rape BCCP isoforms. The low similarity to soybean BCCP for which information about plastid processing is known (Reverdatto et al., 1999) precludes an accurate prediction for AtBCCP2. Therefore, to verify this protein is plastidial and estimate the apparent Mr of the mature protein, in vitro translated protein was incubated with intact pea chloroplasts and was assayed for protein uptake.
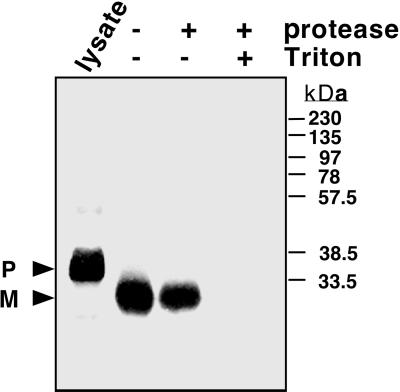
In vitro translation of AtBCCP2 transcript resulted in a 34-kD radiolabeled protein, approximately 7 kD larger than the predicted molecular mass (Fig. 2). When incubated with intact pea chloroplasts, radiolabeled AtBCCP2 was imported with greater than 50% efficiency. Chloroplast import was verified by processing of the precursor protein to a 25-kD mature polypeptide and protection of this protein to exogenous thermolysin protease. As a control for protease activity, detergent was added to permeabolize the envelope, which rendered the radiolabeled protein susceptible to proteolysis. These data demonstrated that the cDNA-encoded protein contained a transit peptide of approximately 9 kD, which was capable of directing it to plastids.
Figure 2.
Chloroplast import of Arabidopsis BCCP2. Intact pea chloroplasts were incubated with in vitro translated AtBCCP2 lysate (lane 1) for protein uptake. The chloroplast suspension was then aliquoted, washed, and re-isolated (lane 2), or treated with thermolysin protease after import (lane 3) or protease treated in the presence of Triton X-100 detergent (lane 4) prior to resolving by SDS-PAGE. AtBCCP2 precursor (P) and mature (M) polypeptides were visualized by autoradiography. Mr markers are indicated.
Recombinant Expression of BC and BCCPs and Reconstitution of BC-BCCP Half-Reaction of ACCase
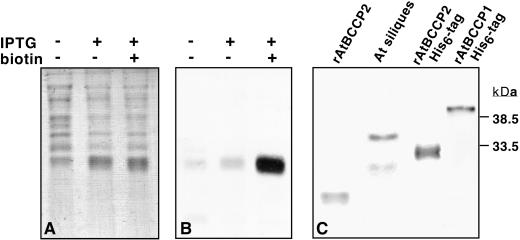
To confirm the molecular masses of AtBCCP1 and 2 and to verify these were biotin-containing proteins, the coding regions for the predicted mature proteins were PCR-amplified and subcloned into expression vectors. The N-terminal amino acids of mature AtBCCP1 and 2 were predicted to be Ser64 and Thr56, respectively, based upon loose conservation with the experimentally determined processing site for soybean BCCP (Fig. 1). Two AtBCCP2 fusion proteins were expressed in Escherichia coli. Fusion one contained a (His)6-tag within a 41-residue amino-terminal fusion to the mature AtBCCP2 polypeptide and fusion two contained only a MG-dipeptide at the amino-terminus to initiate translation for the mature AtBCCP2 polypeptide. Protein expression in the protease-deficient E. coli host BL21(DE3) resulted in highly induced polypeptides of 32 and 22 kD for the two respective constructs, as observed by Coomassie Blue staining and immunoblot analyses with anti-biotin antibodies. For comparison, when fused to a 38-residue tag, AtBCCP1 migrated as a 42-kD polypeptide during SDS-PAGE (Fig. 3), 10 kD larger than a similar fusion for AtBCCP2. Because the size of AtBCCP2 polypeptide without a (His)6-tag did not agree with the chloroplast imported mature AtBCCP2, it was reasoned that the predicted mature start site (Thr56) was invalid. We inevitably conclude that recombinant expressed AtBCCP2 was not complete mature protein. When expressed under standard growth conditions, detection of recombinant AtBCCP2 with anti-biotin antibodies was poor, an indication of insufficient biotinylation. It was previously noted that recombinant expression of the dihydrolipoamide acetyltransferase component to the mitochondrial pyruvate dehydrogenase complex required exogenous lipoic acid for complete lipoylation of apoprotein (Thelen et al., 1999). It was, therefore, hypothesized that augmenting AtBCCP expression cultures with free biotin might ameliorate the low biotinylation frequency. The addition of biotin during growth and induction phases significantly improved the biotinylation efficiency, which demonstrated the E. coli biotin ligase was capable of biotinylating this plant-derived BCCP (Fig. 3B).
Figure 3.
Expression of Arabidopsis BCCP isoforms in E. coli. A and B, Recombinant BL21(DE3) cells were grown for 6 h in Luria broth (plus antibiotic) at 37°C with shaking. Cells were aliquoted (0.2 mL) into three culture tubes containing 3 mL of Luria broth and nothing (control), 2 mm isopropylthio-β-galactoside, or 2 mm isopropylthio-β-galactoside plus 0.1 mm biotin and grown for another 6 h. Cells were then collected by centrifugation, resuspended in 0.4 mL of SDS-PAGE sample buffer, and heated at 95°C for 15 min prior to SDS-PAGE. A, Coomassie Blue-stained gel containing 5 μL of poly-(His) AtBCCP2-induced cell lysates. B, Anti-biotin stained immunoblot of A containing 0.5 μL of each sample. C, Anti-biotin stained immunoblot of recombinant AtBCCP2, minus fusion tag (approximately 5 μg of cell lysate; lane 1), poly-(His) AtBCCP2 (50 ng of purified protein; lane 3); and poly-(His) AtBCCP1 (approximately 5 μg of cell lysate; lane 4) resolved next to Arabidopsis total silique protein (10 μg) for comparison.
Mature AtBC (starting with Cys71), expressed as a 36-amino acid fusion, migrated as a 55-kD polypeptide, which is close to the 51 kD size of native Arabidopsis BC. Recombinant (His)6-tagged AtBCCPs and BC were purified to greater than 90% homogeneity by Ni-chelate chromatography (data not shown). The yield of rAtBC was 10 mg L−1 culture and subsequent to purification was concentrated to 37 mg mL−1 without loss of solubility. From three preparations the yield of rAtBCCP1 was 1.6, 4.4, and 3.8 mg L−1, whereas rAtBCCP2 was 7.2, 22, and 24 mg L−1 culture media. Purified rAtBC had a specific activity of 0.64 ± 0.04 (sd) nmol CO2 min−1 mg−1 protein when assayed under the conditions described here. This value for purified enzyme is modest when compared with the specific activities observed from lysed pea chloroplasts (0.2–0.7 nmol min−1 mg−1) assayed under the same conditions. The low value for rAtBC is possibly due to the amino-terminal tag present on the recombinant form and the lack of BCCP partner protein, which together might hinder correct folding. Because activity was completely abolished by boiling rAtBC prior to assaying, this is clearly the result of an enzymatic activity. As shown in Table I, both rAtBCCP isoforms were capable of accepting 14CO2 from rAtBC, confirming their activity as cofactors. Free biotin at the same concentration gave a 23- to 100-fold lower rate. When specific activities are compared, rAtBCCP2 appears to be the superior carboxy acceptor.
Table I.
Activity assays of purified recombinant BC and BCCP isoforms
| Biotin Carboxylase Assay
|
BC-BCCP
Reconstitution Assay
|
||
|---|---|---|---|
| Sample | Specific activity | Sample | Specific activity |
| pmol CO2 min−1 mg−1 | pmol CO2 min−1 mg−1 | ||
| Minus rAtBC (background) | 12 (1.7) | 4.2 μM biotin | 0.0 (0.0) |
| Boiled rAtBC | 14 (1.3) | 22 μM biotin | 0.2 (0.05) |
| rAtBC | 640 (4.0) | 22 μM rAtBCCP1 | 4.5 (1.0) |
| 22 μM rAtBCCP2 | 21.0 (10) | ||
Biotin carboxylase activity was quantitated as the amount of 14C transferred from NaH14CO3 to free biotin as determined by stability to bubbling with unlabeled CO2. Biotin carboxylase assay was performed with 50 mm d-biotin, 72 μg of purified rAtBC, and 5 mm NaH14CO3 (approximately 3,000 dpm nmol−1). Reconstitution assay of purified recombinant BC and BCCP isoforms was performed with equimolar amounts of BC and BCCP (or free biotin as a substitute for BCCP) as indicated below. All values are the mean of at least three samples and sd is noted in parentheses.
Differential Expression of Arabidopsis BCCP Isoforms
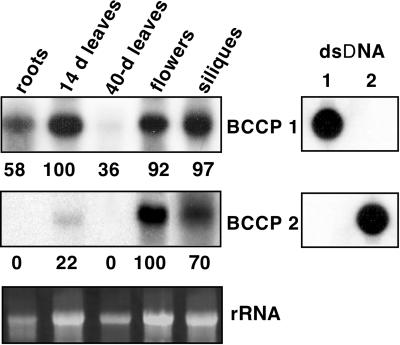
The presence of two BCCP genes in Arabidopsis might be an indication of expression or biochemical differences that are important for ACCase function. To determine the expression patterns for these isoforms, northern-blot analyses were performed. Using a probe containing the 3′ one-half of AtBCCP1 cDNA, transcript abundance from various organs was analyzed. Though amino acid identity between the two BCCP isoforms is high near the carboxyl terminus, the nucleotide identity is sufficiently low to prevent cross-hybridization under the conditions employed here (Fig. 4). The transcript for AtBCCP1 was detected in all organs analyzed, which included roots, leaves, flowers, and siliques. AtBCCP1 transcript abundance was similar in 14-d leaves to developing siliques and was estimated to be 0.001% to 0.005% of the total RNA, based upon standard DNA controls. Estimating the rRNA to be in 100-fold excess over mRNA, AtBCCP1 transcript represented about 0.1% to 0.5% of mRNA in developing leaves and siliques. In agreement with Choi et al. (1995) we observed that AtBCCP1 transcript is down-regulated during leaf development since 14-d rosette leaves contain approximately 3-fold more transcript than 40-d rosette leaves. Although developing leaves and siliques contained the highest amount of BCCP1 transcript, flowers were also rich in this transcript, containing 92% as much. Of all the organs analyzed, roots contained the least amount of transcript, 58% as much as developing leaves.
Figure 4.
Transcript expression analysis of Arabidopsis BCCP isoforms. Total RNA (30 μg) isolated from total roots, 14-d rosette leaves, 40-d rosette leaves, flowers, and developing siliques was analyzed. As a loading control, 16 S rRNA amounts are shown. DNA probes were quality-controlled and assayed for cross-hybridization using 15 ng of double-stranded DNA (dsDNA). Cross-hybridization between the two isoforms was negligible. Intensity and area of the hybridization signals were quantitated with Image Quant software and expressed as relative percentages.
An early indication of AtBCCP2 transcript expression came from microarray analyses using cDNAs derived from a developing seed EST library (Girke et al., 2000). Comparing total mRNA derived from developing seeds and leaves the ratio of expression using four different AtBCCP2 ESTs averaged 5.1. These data indicated developing seeds contained several-fold more transcript than developing leaves. Northern analyses showed AtBCCP2 transcript was mostly expressed in flowers and developing siliques and minimally expressed in roots and leaves. The overall abundance of AtBCCP2 transcript was approximately 4-fold lower than AtBCCP1, when specific activity of the hybridization probes and exposure times were considered. Taken together, the expression profiles indicate isoform one is present in all organs and more abundant overall compared with isoform two.
To confirm differential expression of these BCCP isoforms, protein levels were monitored with anti-biotin antibodies. Anti-biotin antibodies were used instead of isoform-specific antibodies so the abundance of each isoform could be directly compared. Arabidopsis organs display predominantly three biotinylated polypeptides, 25 ± 3 (sd), 35 ± 3, and 88 ± 2 kD in size. The 88-kD polypeptide was previously characterized as the 3-methylcrotonoyl-CoA carboxylase (Song et al., 1994) and the 35-kD polypeptide, a putative BCCP to ACCase (Choi et al., 1995). The 25-kD biotin-containing protein has never been reported from Arabidopsis, but has an apparent Mr identical to the mature size of chloroplast imported AtBCCP2 (Fig. 2) and more similar to recombinant AtBCCP2 than AtBCCP1 (Fig. 3C).
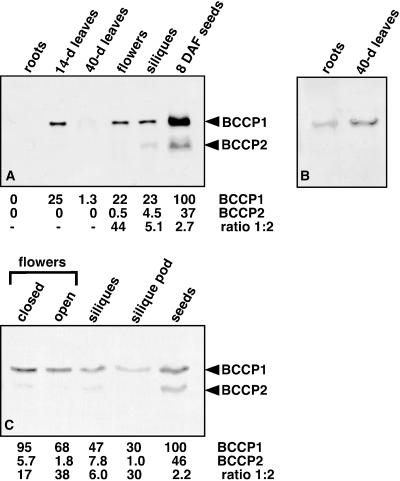
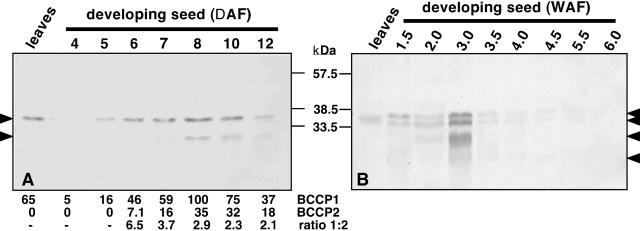
The expression pattern for the 35-kD polypeptide resembles the profile for AtBCCP1 transcript; most abundant in 14-d leaves, flowers, and developing siliques, but lower in roots and 40-d leaves (Fig. 5A). Of all the organs analyzed, 8 d-after-flowering seeds contained the greatest amount of 35-kD BCCP, approximately 4-fold more than in 14-d leaves. Using purified recombinant AtBCCPs as standards, the 35-kD BCCP was estimated to be approximately 0.05% of the total protein in 14-d leaves, flowers, and siliques. The 25-kD polypeptide is expressed predominantly in siliques, though a faint band was observed in flowers (Fig. 5, A and B). Closer inspection of dissected siliques revealed the 25-kD polypeptide is not present in the silique pod, but rather specifically expressed in the developing seeds (Fig. 5C). Throughout Arabidopsis seed development, the 35-kD biotin-containing polypeptide was generally 2-fold more abundant than the 25-kD form (Fig. 6A). Both polypeptides were temporally regulated during seed development and expression of AtBCCP2 was highest in 7- to 10-d-after-flowering seeds and was difficult to detect during earlier stages.
Figure 5.
Protein expression analysis of Arabidopsis BCCP isoforms. BCCPs were detected with anti-biotin, alkaline phosphatase-conjugated antibodies. Relative molecular weights for BCCPs are 35 ± 3 (sd) and 25 ± 3 kD for isoforms one and two, respectively. A, Total Arabidopsis protein (10 μg) was extracted from roots, 14-d rosette leaves, 40-d rosette leaves, flowers, developing siliques, and developing seeds 8 d after flowering for immunoblot analyses. Arrows indicate BCCP isoforms. B, Higher amount (40 μg) of root and 40-d leaf protein was immunoblotted to determine if AtBCCP2 was present in these organs. Only the 35-kD form (BCCP1) was detected in these organs. C, Immunoblot analysis of total Arabidopsis protein (20 μg) from closed or open flowers, developing siliques, developing silique pods, and developing seeds (mixed stages). Amount of each BCCP polypeptide was quantitated with Image Quant software and is expressed as relative percentages of the maximum value. Ratios of BCCP levels are also noted.
Figure 6.
Expression of BCCP isoforms during Arabidopsis and rapeseed development. BCCP polypeptides were detected with anti-biotin, alkaline phosphatase-conjugated antibodies. Arrows indicate the major biotinylated polypeptides. A, Arabidopsis developing seed harvested 4 to 12 d after flowering (5 μg of protein) was resolved next to 14-d developing leaves (15 μg) for comparison. Top and bottom biotinylated polypeptides correspond to BCCP1 and 2, respectively. BCCPs were quantitated and expressed as relative percentages. Ratio of absolute BCCP levels are also noted. B, Developing rapeseed harvested 1.5 to 6.0 weeks after flowering (100 μg) was resolved next to developing leaf protein (100 μg).
In the related oilseed plant rapeseed, four BCCP polypeptides were observed in developing seeds, 22, 25, 35, and 37 kD in size (Fig. 6B). The 35-kD BCCP was also present in developing leaves, however, the 22-, 25-, and 37-kD BCCPs were not observed in developing leaves from equally loaded gels. Expression of these three BCCPs peaked around 21 d after flowering and unlike developing seeds from Arabidopsis, the 25-kD BCCP was nearly equal in abundance to the 35- and 37-kD forms during seed development.
Ion-Exchange Chromatography of Arabidopsis Silique Extracts
It was previously observed that pea chloroplast BC and BCCP remain associated during salt elution from diethylaminoethyl- (DEAE) sepharose chromatography, whereas the carboxyltransferase subunits easily dissociate from the BC-BCCP partner proteins (Shorrosh et al., 1996). To determine if the two biotinylated polypeptides from Arabidopsis seeds were assembled with BC, a total protein extract from developing siliques was applied to a DEAE-sepharose matrix, washed with a low ionic strength wash buffer, and was subsequently eluted with a step gradient of increasing sodium chloride in wash buffer (Fig. 7). Approximately 80% of BC and BCCPs from the soluble extract bound to the DEAE matrix as determined by immunoblot analysis. A single Arabidopsis polypeptide of 51 kD cross-reacted with anti-castor BC antibodies, which is similar in size to various other plant BCs (Roesler et al., 1996). Most of the 51-kD polypeptide eluted between 80 and 140 mm sodium chloride, which coincided with the elution of the two biotin-containing polypeptides. Co-elution with BC is supportive evidence that both of these biotinylated proteins are assembled with BC and represent the two BCCP isoforms.
Figure 7.
Association of Arabidopsis BC-BCCP polypeptides by co-elution from DEAE-sepharose. Arabidopsis developing silique proteins were applied to an ion-exchange matrix, washed with five column volumes of low ionic strength wash buffer, and eluted with a sodium chloride step gradient of 1.0-mL fractions. Equal volumes of each fraction were resolved by SDS-PAGE for immunoblotting with anti-castor BC or anti-biotin antibodies.
DISCUSSION
Our interest in control of fatty acid biosynthesis in oilseeds has led to the investigation of the multisubunit plastidial ACCase and its regulation at the biochemical and molecular levels. With the completion of the Arabidopsis genome, the genes that comprise a multisubunit plastidial ACCase have all been identified. The BC and α- carboxyltransferase subunits are apparently encoded by a single nuclear gene, whereas β-carboxyltransferase is encoded by a single plastome gene (Mekhedov et al., 2000). It was suggested that the BCCP subunit was also encoded by a single nuclear gene based upon a Southern blot probed with AtBCCP1 (Choi et al., 1995). In this report we characterize the presence of a second gene by identifying a cDNA encoding a second isoform for this ACCase subunit.
Even though the amino acid identity is only 42% to the previously characterized isoform, the evidence summarized below conclusively demonstrates the cDNA described in this report encodes a second BCCP isoform to Arabidopsis plastid ACCase. The hallmark BCCP biotinylation motif is present in the deduced amino acid sequence (Fig. 1). In vitro translated protein is efficiently imported and processed in chloroplasts (Fig. 2). Recombinant protein is biotinylated and is an active acceptor of carboxy groups from BC (Fig. 3; Table I). A biotin-containing protein of a similar molecular mass is present in Arabidopsis seeds and co-elutes with BC during ion-exchange chromatography (Figs. 5–7).
The reason two BCCP isoforms are present in Arabidopsis is not clear, though it is intriguing since the other ACCase subunits are encoded by single genes. To help understand this, the expression patterns of these isoforms was investigated. An earlier investigation showed that AtBCCP1 transcript is highly expressed in developing leaves (Choi et al., 1995). Here we show this transcript is also present in roots, and is highly expressed in flowers and siliques. This transcript expression profile is similar to that observed for AtBC transcript that was quantitated by northern-blot and promoter-β-glucuronidase analysis (Bao et al., 1997). In that investigation siliques were the primary organ where β-glucuronidase protein accumulated, containing 25% more activity than flowers. It has been observed previously that in oilseed plants, developing leaves and seeds are two organs that exhibit high rates of fatty acid biosynthesis (Ohlrogge and Jaworski, 1997). As expected, developing leaves were rich in AtBCCP1 transcript, accumulating to approximately 0.1% of mRNA. Flowers and siliques contained similar amounts of transcript, which probably reflects the demand for lipids during pollination and reserve deposition (Evans et al., 1992). It is striking that mature leaves are nearly devoid of this transcript, suggesting flux through ACCase, and consequently fatty acid biosynthesis, is minimal at this point in development. In an alternate manner, this might indicate a low rate of protein turnover. However, since BCCP protein was abundant in 14-d leaves yet barely detectable in 40-d leaves, the protein must be degraded during leaf development.
Unlike AtBCCP1, AtBCCP2 transcript was not abundant in roots or developing leaves, instead flowers and siliques contained most of the message, albeit at approximately 4-fold lower amounts than AtBCCP1. These data were in agreement with microarray analyses, revealing lower expression in developing leaves compared with developing seeds (Girke et al., 2000). Based upon EST abundance it might be argued that AtBCCP2 is more abundant than AtBCCP1. In general, this is an approximate method to compare transcript abundance (Mekhedov et al., 2000); cDNA construction and library amplification steps can alter the representation of transcripts from RNA preparations. Because eight out of 11 AtBCCP2 ESTs contain the entire ORF, whereas the two AtBCCP1 ESTs contain less than 50% of the ORF, perhaps the AtBCCP1 transcript has secondary structure hindering reverse transcription. This is one possible explanation for the under-representation of AtBCCP1 transcript in Arabidopsis cDNA libraries. A different library of 10,500 ESTs derived from Arabidopsis developing seed mRNA contained eight AtBCCP2 versus three AtBCCP1 cDNAs (White et al., 2000). The abundance of AtBCCP2 ESTs from this library correlates with our prediction (based upon transcript abundance), but AtBCCP1 EST abundance is approximately 10-fold lower.
Analysis of these isoforms at the protein level required evidence for a difference in apparent Mr. The low sequence homology between the two Arabidopsis isoforms in conjunction with their low Mr led to the speculation that these isoforms could be resolved by conventional SDS-PAGE. We predicted that AtBCCP2 would have a lower apparent mass than AtBCCP1 since it contained fewer amino acids and a lower percentage of Pro and Ala residues than AtBCCP1 in the hinge region upstream of the biotinylation domain. It was necessary to confirm this experimentally, since it is well documented that these proteins migrate anomalously during SDS-PAGE (Li and Cronan, 1992a) and processing site predictions were based upon a poorly conserved homolog. In vitro synthesized AtBCCP2 migrated at an apparent molecular mass of 34 kD, and after chloroplast import was processed to 25 kD. Both of these masses are higher than those predicted for the precursor and mature polypeptides, which is attributed to the hypervariable hinge domain (Li and Cronan, 1992a). The hinge region is rich in Pro and Ala residues to increase the flexibility for active-site coupling (Brocklehurst and Perham, 1993). Pro- and Ala-rich regions alter the migration of proteins during SDS-PAGE, resulting in a larger apparent size. Anomalous migration during SDS-PAGE has also been observed with dihydrolipoamide acetyltransferases, which also contain Pro, Ala-rich flexible-hinge domains necessary for active-site coupling (Thelen et al., 1999). Since recombinant AtBCCP2 without a poly-(His) fusion migrated at a molecular mass of 22 kD, the predicted start site of Thr56 must be incorrect. Based upon the 32-kD apparent size of the 41-amino acid (His)6-tag fusion, mature AtBCCP2 likely begins near Val42.
There are six presently known biotinylated enzymes in plants, namely homomeric ACCase (plastidial and cytosolic), 3-methylcrotonoyl-CoA carboxylase, propionyl-CoA carboxylase, pyruvate carboxylase, and BCCPs. In addition, a biotin-containing protein of unknown function is abundant in pea seeds (Duval et al., 1994). Of these, only BCCPs are low molecular mass proteins that are abundant in developing tissues. Based upon these criteria, the biotinylated polypeptides of 25 and 35 kD are likely BCCP subunits. The 25-kD isoform is identical in size to chloroplast-imported AtBCCP2 and similar to recombinantly expressed protein. The 35-kD isoform is most likely AtBCCP1 since recombinant protein for this isoform is 10 kD larger than AtBCCP2 (Fig. 3). Further support of this nomenclature is observed in the expression patterns for these isoforms and their relative abundance. AtBCCP1 transcript is observed in all organs, much like the 35-kD polypeptide. AtBCCP2 transcript is primarily present in reproductive organs, in agreement with the 25-kD isoform. The transcript for isoform two is approximately 4-fold less abundant than isoform one, which is roughly true for the 25-kD protein in comparison with the 35-kD protein. Although it is possible the smaller isoform is actually a proteolytic derivative of the larger, more abundant form we argue against this for two reasons. The protein extracts in this investigation were prepared under denaturing conditions. The 25-kD form is only observed in flowers and seeds, whereas the 35-kD form is present in all organs.
The presence of two BCCP genes suggests at least two different types of ACCase complexes exist in Arabidopsis plastids. Although the significance of this is unclear, some possibilities are discussed here. Expression analyses indicate AtBCCP1 could be considered a housekeeping isoform since it is present in all Arabidopsis organs. In agreement with this proposed function, down-regulation of AtBCCP1 through antisense technology resulted in chlorotic, curled, and variegated leaves, demonstrating the constitutively expressed isoform is critical for maintenance of cellular lipids (Fatland et al., 1999). AtBCCP2, on the other hand, appears to have a more specific role since it is predominantly expressed in reproductive organs. Expression patterns such as these have been observed for other multigenic families of fatty acid biosynthetic proteins and one particularly well-characterized gene family is the ACP. At least three Arabidopsis ACPs are constitutively expressed, whereas three more appear to be specific to seed, leaf, or root organs (Hlousek-Radojcic et al., 1992). This level of complexity within the simple Arabidopsis genome was enigmatic for a number of years.
It was recently discovered that optimal desaturase activity for the production of unusual monoenic fatty acids required a seed-specific coriander ACP (Suh et al., 1999). Similar ACPs from spinach and E. coli did not support the high rates observed in coriander seed extracts. Furthermore, mitochondrial and Cuphea lanceolata ACPs promote short-chain termination when compared with a plastidial and a related C. lanceolata ACP isoform, respectively (Shintani and Ohlrogge, 1994; Schutt et al., 1998). The reason for this difference is uncertain, but indicates ACPs may have specific interactions with desaturase and perhaps thioesterase function and have co-evolved with unusual fatty acid synthesis in oilseeds. Like ACPs, BCCPs have no catalytic activity, but instead function more like cofactors since free biotin can substitute for BCCP with E. coli and plastidial ACCases (Guchait et al., 1974; Sun et al., 1997). However, the catalytic efficiency (Vmax/Km ratio) of biotin versus BCCP is 8,000- and 2,000-fold lower in carboxylase and carboxyltransferase assays, respectively (Blanchard et al., 1999). Activity differences between free and BCCP-conjugated biotin indicate association of the BCCP apoprotein with biotin carboxylase and carboxyltransferase subunits is critical for optimal ACCase activity. In an alternate manner, ordered presentation or increased solubility of the biotin cofactor through a biotin-protein conjugate might also explain these activity differences. Further evidence for the involvement of apo-BCCP in active-site coupling is observed here. Both purified rAtBCCP isoforms were preferred to free biotin as carboxy acceptors from rAtBC (Table I). Although rAtBCCP2 appears to be a superior carboxy-acceptor than rAtBCCP1, this might be explained by differences in biotinylation or native conformation and further experimentation will be necessary to determine which isoform is optimal for ACCase activity. These findings further define the role of BCCP in ACCase function and suggests that amino acid differences between the Arabidopsis BCCPs, in combination with differential expression profiles, might be one mechanism for regulating ACCase activity.
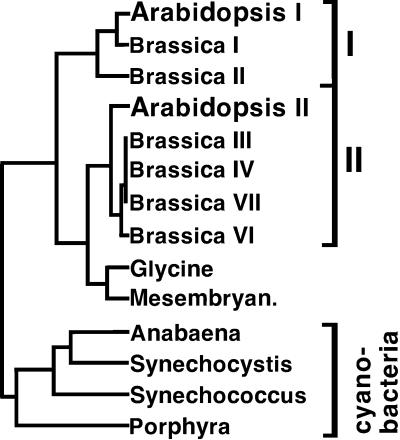
The possibility that BCCP isoforms might influence overall ACCase activity led to the speculation that these genes have diverged to perform specific roles in oilseed plants. If so, conserved homologs should exist for the two Arabidopsis BCCPs. Dendrogram analysis with all known BCCPs revealed at least two different subclasses within the plant kingdom (Fig. 8). The BCCPs from Arabidopsis are split between these two phylogenetic classes as follows. AtBCCP1 and oilseed rape isoforms 1 and 2 define group one, whereas AtBCCP2 and oilseed rape isoforms 3, 4, 6, and 7 belong to the second group. It is interesting that other plant BCCPs from soybean and Mesembryanthemum crystallinum sources formed a group distinct from class one and two Brassicaceae BCCPs. Relatedness of AtBCCP2 with class two oilseed rape BCCPs prompted the investigation of BCCPs from developing seed. Oilseed rape developing seed contained at least four BCCP polypeptides ranging in size from 22 to 37 kD. Analysis of developing oilseed rape seed from other laboratories also revealed multiple biotin-containing proteins in this size range, though the lower Mr forms were attributed to proteolysis (Elborough et al., 1996). Since the sizes of these low molecular mass biotin-containing polypeptides (22 and 25 kD) agree favorably with the apparent size of AtBCCP2, we suggest they represent the class two BCCPs rather than proteolytic derivatives of the 35- and 37-kD putative class one isoforms. This is supported by direct evidence that oilseed rape contains at least four genes that encode class two BCCP isoforms (Elborough et al., 1996) and like AtBCCP2, the oilseed rape genes appear to have evolved for expression in reproductive organs (Fig. 6B). The higher BCCP copy number in oilseed rape compared with Arabidopsis is likely due to its allotetraploid genome. The potential importance of BCCPs in ACCase function and fatty acid biosynthesis is reflected not only in gene complexity, but by the observation that all BCCP isoforms from Arabidopsis and oilseed rape are up-regulated midway through seed development, coinciding with the period of maximal oil accumulation (Turnham and Northcote, 1983; Mansfield and Briarty, 1992).
Figure 8.
Dendrogram analysis of BCCPs. Clustal analysis was performed with Genetics Computer Group DNA analysis software (Madison, WI) using default parameters. Length of horizontal lines indicates inverse degree of relatedness. Cyanobacterial and plant sequences are indicated. GenBank accession numbers not mentioned in Figure 1 are X90727, X90728, X90729, X90731, and X90732, for oilseed rape isoforms 1, 2, 3, 6, and 7; AI822996, Mesembryanthemum; and U59235, Synechococcus.
In conclusion, two classes of BCCP genes exist in Brassicaceae plants. The data presented here demonstrate class one BCCPs are expressed in all tissues examined, whereas class two are expressed in reproductive organs, particularly developing seeds. An understanding of class two BCCP isoforms and their possible specialized function during seed development awaits further biochemical and molecular genetic studies.
MATERIALS AND METHODS
Plant Materials
All seeds were sown in a 1:1 mixture of water-saturated vermiculite and peat moss-enriched soil. Arabidopsis (Columbia ecotype) plants were grown at 18°C under low light fluence (49 μmol photon m−2 s−1). Oilseed rape (Brassica napus) was grown at 20°C during a 16-h photoperiod (294 μmol photon m−2 s−1), and 15°C during the dark. Pea (Pisum sativum L. cv Little Marvel) seedlings were grown at a constant 22°C with an 8-h photoperiod (210 μmol photon m−2 s−1). For seed development analyses, flowers were tagged at the point when petals first appeared from the flower whorl.
Identification and Sequencing of AtBCCP2 cDNA
The AtBCCP1 cDNA (GenBank accession no. U23155) was used as a query of the Arabidopsis EST database (Newman et al., 1994) using the BLAST (BLAST, Altschul et al., 1990). A different class of BCCP cDNAs was observed as a result of this query. The cDNA that appeared to be the longest at the 5′ end (GenBank accession no. H37386) was ordered from the Arabidopsis Biological Research Center (Columbus, OH). Both strands of the cDNA were sequenced by primer walking using 20-mer oligonucleotides. All other steps were performed according to the dye-deoxynucleotide chain termination procedure (Applied Biosystems, Foster City, CA). Reaction products were analyzed on an ABI automated sequencer at the Michigan State University DNA core facility.
Northern-Blot Analyses
Total RNA was isolated from Arabidopsis organs using a guanidinium thiocyanate extraction procedure (Sambrook et al., 1989). RNA was quantitated by A260. RNA samples were brought up to 15 μL with sterile water and equal volumes of formamide and formaldehyde were added prior to heating for 15 min at 50°C. Electrophoresis of RNA was performed in a 1% (w/v) agarose gel containing 2.2 m formaldehyde and subsequently blotted to Hybond nylon membrane (Amersham, Buckinghamshire, UK) by capillary action for approximately 16 h. After transfer, membranes were rinsed briefly with 0.2× SSC, 0.1% (w/v) SDS, and were then dried at 80°C for 3 h. Immobilized rRNA was visualized by rehydrating the membrane in 0.03% (w/v) methylene blue, 0.3 m sodium acetate, pH 6.0 for 1 min. Excess stain was removed by iterative washes with sterile water. Membranes were prehybridized for 6 h at 42°C in 15 mL of 50% (v/v) deionized formamide, 5× SSPE, 0.5% (w/v) SDS, 5× Denhardt's (50× = 1% [w/v] of the following: Ficoll, polyvinylpyrrolidone, bovine serum albumin), 5% (w/v) dextran sulfate, and 100 μg mL−1 sheared salmon sperm DNA. Hybridization was performed at 42°C for 16 h in the same solution using the 3′ one-half of BCCP1 (GenBank accession no. Z25714, 627–1,089 bp) or the 3′-UTR of BCCP2 (BamHI, 871–1,235 bp) labeled by random hexamer extension. Subsequent to hybridization, the membranes were washed once at 28°C with 2× SSPE, 0.5% (w/v) SDS for 5 min then twice at 28°C for 15 min. Final two washes were performed at 50°C with 0.1× SSPE, 0.5% (w/v) SDS for 15 min each. Membranes were covered with cellophane and exposed to autoradiography film. Hybridization signals were quantitated by Image Quant software (Molecular Dynamics, Sunnyvale, CA) accounting for area and signal intensity. Signals were expressed as values relative to the highest signal on the blot.
Extraction of Arabidopsis Protein for SDS-PAGE and Detection of Biotin-Containing Polypeptides
Extraction of protein from roots, leaves, flowers, and siliques was performed as follows. Up to 0.1 g of plant material was harvested and placed into a 1.5-mL plastic microfuge tube. Plant material was pulverized with a microcentrifuge pestle in the presence of liquid nitrogen. Powder was immediately reconstituted in 0.2 to 0.5 mL of extraction buffer (0.2% [w/v] SDS, 8 m urea, and 2% [v/v] 2-mercaptoethanol) by vortexing. Samples were then heated at 95°C for 15 min. Insoluble debris was collected by centrifugation for 15 min at 12,000g. Supernatant was removed and placed into a fresh microfuge tube. Seed protein was extracted by placing the dissected seeds directly into extraction buffer and homogenizing with a plastic microfuge pestle. Insoluble debris was collected by centrifugation and the supernatant was saved for analysis. Protein concentration was determined by dye-binding protein assay using bovine serum albumin as the standard (Bradford, 1976). Equal amounts of total protein were resolved by SDS-PAGE for immunoblot analyses.
SDS-PAGE and protein transfer to nitrocellulose was performed using standard conditions (Towbin et al., 1979). Nitrocellulose membranes were blocked for at least 1 h with 10 mm Tris-HCl, pH 8.0, 0.15 m sodium chloride, 0.3% (v/v) Tween 20 (TBS-T), and 2% (w/v) nonfat dry milk. Anti-biotin antibodies conjugated to alkaline phosphatase (Kirkegaard and Perry Laboratories, Gaithersburg, MD) were directly added at a 1:2,000 dilution. The sensitivity of these antibodies (2 ng of purified recombinant BCCP) was over 100-fold higher than similar products from various manufacturers. Probing proceeded for 16 h at 25°C after which the membranes were briefly rinsed with TBS-T. The membranes were then washed 6 times for 10 min each with TBS-T. Blots were then washed 5 min with developing solution (0.1 m Tris-HCl, pH 9.5, 0.1 m sodium chloride, and 5 mm magnesium chloride) before colorimetric detection in developing solution containing 0.33 mg mL−1 p-nitro blue tetrazolium chloride and 0.17 mg mL−1 5-bromo-4-chloro-3-indoyl phosphate p-toluidine salt. Immunoblot signals were quantitated using Image Quant software, which accounted for band area plus intensity. Signals were corrected for background and expressed as a value relative to the most abundant signal on the blot.
Chloroplast Import of AtBCCP2
Chloroplasts were isolated from 10-d-old pea leaves according to a previous procedure (Bruce et al., 1994). In vitro transcription and translation was performed with the quick-coupled T7 polymerase rabbit reticulocyte lysate kit available from Promega (Madison, WI). Lysate (5 μL) containing radiolabeled AtBCCP2 was added directly to 6 μL of 100 mm MgATP and 139 μL of import buffer (50 mm HEPES- [4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid] KOH, pH 8.0, and 330 mm sorbitol). Isolated pea chloroplasts were diluted to 1 mg chlorophyll mL−1 with import buffer and 50 μL was added to diluted lysate mixture for a final volume of 200 μL. Import proceeded for 30 min at 25°C after which the chloroplasts were diluted 5-fold with import buffer and centrifuged at 1,500g for 3 min. The supernatant was discarded and the chloroplast pellet was gently washed in 1 mL of import buffer. The chloroplasts were collected by centrifugation and washed a second time. The washed chloroplast pellet was resuspended in 160 μL of import buffer and 50-μL aliquots were placed into three microfuge tubes. All tubes contained 1 mm calcium chloride, tube 2 and 3 contained 0.4 mg mL−1 thermolysin protease (Sigma, St. Louis), and tube 3 also contained 0.05% (v/v) Triton X-100, all final concentrations. Protease digestion proceeded for 30 min at 4°C and was terminated by the addition of 1 mL of import buffer containing 5 mm EDTA and 5 mm EGTA. Intact chloroplasts were collected by centrifugation and washed with 1 mL of import buffer, 5 mm EDTA, and 5 mm EGTA. The final chloroplast pellet was resuspended in 50 μL of SDS-PAGE sample buffer (2% [w/v] SDS, 8 m urea, and 2% [v/v] 2-mercaptoethanol) and heated at 95°C for 15 min prior to loading onto SDS-PAGE.
Recombinant Expression of Arabidopsis BCCPs and BC
Oligonucleotide primers were designed to PCR amplify the mature ORF of AtBCCP1 and AtBCCP2 for subcloning into pET28b expression vector (Novagen, Madison, WI). Based upon the experimentally determined processing site for soybean BCCP, primers were designed to start with Ser-64 (AtBCCP1) or Thr56 (AtBCCP2). Primers JO625, gcgtgaGAATTCcatgggctctaacaaggttagtactggtgca (5′-sense), and JO634, gcgaagCTCGAGtacggttgaaccacaaacagagg (3′-antisense), were used to amplify AtBCCP1 mature ORF (nucleotides 245–898). Primers JO627, ccctaTCTAGAGAATTCagaaggagatatgggcactaatgaggttgtttctaac (5′-sense), and JO628, gcgaagCTCGAGtcaaggtgcgatgacaaaaag (3′-antisense), were used to PCR amplify the AtBCCP2 mature ORF (nucleotides 258–861). Primer JO627 contained an artificial ribosomal binding site upstream of an ATG start codon (noted in italics) to initiate translation for the mature AtBCCP2 polypeptide without a poly (His) tag. Primers JO685, gcgtgaGAATTCtgcagtggtggtgataagatt (5′-sense), and JO686, gggtcaCTCGAGctaaaccgttgcgtttgtcagatc (3′-antisense), were used to amplify the AtBC ORF (nucleotides 329–1732). PCR was performed under standard conditions with plasmid DNA as template (Thelen et al., 1999). The AtBCCP1 amplicon was subcloned into the EcoRI and XhoI sites of pET28b to obtain an amino-terminal poly-(His) fusion-tagged fusion protein. AtBCCP2 amplicon was subcloned into the XbaI, XhoI ,and EcoRI, XhoI sites of pET28b to obtain mature protein without and with an amino-terminal poly-(His) fusion tag, respectively. AtBC amplicon was subcloned into the EcoRI and XhoI sites of pET28a to obtain an amino-terminal poly (His) fusion.
To amplify the mature ORF of AtBCCP1, a 3′ primer was initially designed according to the defined ORF of GenBank sequence U23155 (CAC1, Choi et al., 1995). When used in combination with the 5′ primer, no amplicon was obtained by RT-PCR or PCR using total RNA and plasmid DNA as template, respectively. As a consequence, the nucleotide sequence of U23155 was verified against two EST sequences (AI999240 and Z25714) and chromosome V genomic sequence (AB005242). A frameshift at nucleotide 895, due to an extra cytosine at this position, was revealed. This resulted in a GTA read-through, instead of a TAG stop codon, and a subsequent -VESAP carboxy-terminal tail in the deduced amino acid sequence (Fig. 1). The gene sequence for AtBCCP1 also contains a frameshift mutation at this same position (U62029, Ke et al., 1997). To check for any additional differences, the full-length AtBCCP1 EST (AI999240) was completely sequenced. Besides the frameshift mutation, clone AI999240 contained 37 and 124 bp longer 5′- and 3′-UTRs, respectively. Due to the significant differences between these two AtBCCP1 cDNAs, the full sequence for clone AI999240 was submitted to GenBank (assigned accession no. AF236873). To proceed with PCR amplification of AtBCCP1, the antisense primer was redesigned to account for the aforementioned frameshift.
Poly (His)-tagged recombinant protein was expressed and purified from the protease-deficient BL21(DE3) E. coli cell line according to standard conditions (Thelen et al., 1998), except for the inclusion of 0.2 mm biotin throughout the growth and induction phases for BCCPs. Protein was dialyzed in 2 L of 10 mM TES-KOH, pH 7.4, 10% (v/v) glycerol, and 1 mm dithiothreitol for 16 h at 4°C and was subsequently concentrated on ultrafiltration membranes with a 5-kD cutoff.
Reconstitution of BC Half-Reaction with Recombinant BC and BCCPs
Biotin carboxylase activity was assayed as the enzymatic transfer of 14CO2 from NaH14CO3 to free d-biotin (Sigma) or purified, recombinant BCCPs to form carboxybiotin, which is stable to unlabeled CO2 bubbling. Reaction conditions were similar to those described previously (Guchait et al., 1974) except for the use of 100 mm HEPES-KOH, pH 8.0, 4 mm MgCl2, and 3 mm dithiothreitol in reaction medium. Reactions (150 μL) were initiated with rBC enzyme, proceeded for 30 min at 25°C, and were terminated by pipetting the reaction into 2 mL of ice-cold water containing 200 μL of 1-octanol. Unlabeled CO2 was then bubbled into the reaction vial for 30 min at 4°C. After gasing, 0.2 mL of 0.1 n NaOH was added prior to liquid scintillation spectroscopy. Minus enzyme background rates were determined for each experiment and were typically less than 5% of the actual rate. The specific activity of NaH14CO3 in reaction was approximately 3,000 dpm nmol−1 NaH14CO3 and was verified for each experiment. All values are the mean of at least three reactions. Use of BCCPs as carboxy acceptors was assayed in triplicate using independent protein preparations.
DEAE Chromatography of Arabidopsis Silique Extract
Arabidopsis siliques were homogenized in a mortar and pestle with liquid nitrogen and reconstituted in homogenization buffer, 50 mm HEPES-KOH, pH 8.0, 10% (v/v) glycerol, 1% (v/v) Triton X-100, 2 mm phenylmethanesulfonyl fluoride, 2 mm benzamidine, 2 mm ε-amino-n-caproic acid, 5 mm EDTA, and 14 mm 2-mercaptoethanol by vortexing for 30 s. Particulate matter was removed by centrifugation, 14 K g for 1 min and supernatant was applied to a DEAE-sepharose matrix (0.6-mL bed volume) pre-equilibrated with homogenization buffer. Column was washed with three volumes of homogenization buffer (minus glycerol and Triton X-100) and eluted with a sodium chloride gradient in wash medium. Elution fractions were heated in sample buffer and resolved by SDS-PAGE for immunoblotting with anti-castor BC (Roesler et al., 1997) and anti-biotin antibodies.
ACKNOWLEDGMENT
The authors thank the Arabidopsis Biological Resource Center for providing the cDNA clones used in this investigation.
Footnotes
This work was supported by the National Science Foundation (grant no. MCB94–06466) and by the Michigan Agricultural Experiment Station.
LITERATURE CITED
- Alban C, Baldet P, Douce R. Localization and characterization of two structurally different forms of acetyl-CoA carboxylase in young pea leaves, of which one is sensitive to aryloxyphenoxyproprionate herbicides. Biochem J. 1994;300:557–565. doi: 10.1042/bj3000557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bao X, Shorrosh BS, Ohlrogge J. Isolation and characterization of an Arabidopsis biotin carboxylase gene and its promoter. Plant Mol Biol. 1997;35:539–550. doi: 10.1023/a:1005881006620. [DOI] [PubMed] [Google Scholar]
- Blanchard CZ, Chapman-Smith A, Wallace JC, Waldrop GL. The biotin domain peptide from the biotin carboxyl carrier protein of Escherichia coli acetyl-CoA carboxylase causes a marked increase in the catalytic efficiency of biotin carboxylase and carboxyltransferase relative to free biotin. J Biol Chem. 1999;274:31767–31769. doi: 10.1074/jbc.274.45.31767. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brocklehurst SM, Perham RN. Prediction of the three-dimensional structure of the biotinylated domain from yeast pyruvate carboxylase and of the lipoylated H-protein from the pea leaf glycine cleavage system: a new automated method for the prediction of protein tertiary structure. Protein Sci. 1993;2:626–639. doi: 10.1002/pro.5560020413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce BD, Perry S, Froehlich J, Keegstra K. In vitro import of proteins into chloroplasts. Plant Mol Biol Man. 1994;J1:1–15. [Google Scholar]
- Choi JK, Yu F, Wurtele E, Nikolau BJ. Molecular cloning and characterization of the cDNA coding for the biotin-containing subunit of the chloroplastic acetyl-coenzyme A carboxylase. Plant Physiol. 1995;109:619–625. doi: 10.1104/pp.109.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duval M, Job C, Alban C, Douce R, Job D. Developmental patterns of free and protein-bound biotin during maturation and germination of seeds of Pisum sativum: characterization of a novel seed-specific biotinylated protein. Biochem J. 1994;299:141–150. doi: 10.1042/bj2990141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elborough KM, Winz R, Deka RK, Markham JE, White AJ, Rawsthorne S, Slabas AR. Biotin carboxyl carrier protein and carboxyltransferase subunits of the multi-subunit form of acetyl-CoA carboxylase from Brassica napus: cloning and analysis of expression during oilseed rape embryogenesis. Biochem J. 1996;315:103–112. doi: 10.1042/bj3150103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans DE, Taylor PE, Singh MB, Knox RB. The interrelationship between the accumulation of lipids, protein and the level of acyl carrier protein during the development of Brassica napus L. pollen. Planta. 1992;186:343–354. doi: 10.1007/BF00195314. [DOI] [PubMed] [Google Scholar]
- Fatland BL, Qian H, Nikolau BJ, Wurtele E (1999) Antisense expression of the CAC1 subunit of ACCase in Arabidopsis results in significant modifications in leaf and cell organization (abstract no. 276). Plant Physiol Plant Biol Suppl 78
- Girke T, Todd J, Ruuska S, White J, Benning C, Ohlrogge J. Microarray analysis of developing Arabidopsis seeds. Plant Physiol. 2000;124:1570–1581. doi: 10.1104/pp.124.4.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guchait RB, Polakis SE, Dimroth P, Stoll E, Moss J, Lane MD. Acetyl coenzyme A carboxylase system of Escherichia coli: purification and properties of the biotin carboxylase, carboxyltransferase, and carboxyl carrier protein components. J Biol Chem. 1974;249:6633–6645. [PubMed] [Google Scholar]
- Hlousek-Radojcic A, Post-Beittenmiller D, Ohlrogge JB. Expression of constitutive and tissue-specific acyl carrier protein isoforms in Arabidopsis. Plant Physiol. 1992;98:206–214. doi: 10.1104/pp.98.1.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter SC, Ohlrogge JB. Regulation of spinach chloroplast acetyl-CoA carboxylase. Arch Biochem Biophys. 1998;359:170–178. doi: 10.1006/abbi.1998.0900. [DOI] [PubMed] [Google Scholar]
- Kannangara CG, Stumpf PK. Fat metabolism in higher plants: a prokaryotic type acetyl-CoA carboxylase in spinach chloroplasts. Arch Biochem Biophys. 1972;152:83–91. doi: 10.1016/0003-9861(72)90196-8. [DOI] [PubMed] [Google Scholar]
- Ke J, Choi JK, Smith M, Horner HT, Nikolau BJ, Wurtele ES. Structure of the CAC1 gene and in situ characterization of its expression: the Arabidopsis thaliana gene coding for the biotin-containing subunit of the plastidic acetyl-coenzyme A carboxylase. Plant Physiol. 1997;113:357–365. doi: 10.1104/pp.113.2.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S-J, Cronan JE., Jr The gene encoding the biotin carboxylase subunit of Escherichia coli acetyl-CoA carboxylase. J Biol Chem. 1992a;267:855–863. [PubMed] [Google Scholar]
- Li S-J, Cronan JE., Jr The genes encoding the two carboxylase subunits of Escherichia coli acetyl-CoA carboxylase. J Biol Chem. 1992b;267:16841–16847. [PubMed] [Google Scholar]
- Li S-J, Cronan JE., Jr Putative zinc finger protein encoded by a conserved chloroplast gene is very likely a subunit of a biotin-dependent carboxylase. Plant Mol Biol. 1992c;20:759–761. doi: 10.1007/BF00027147. [DOI] [PubMed] [Google Scholar]
- Mansfield SG, Briarty LG. Cotyledon cell development in Arabidopsis thaliana during reserve deposition. Can J Bot. 1992;70:151–164. [Google Scholar]
- Marini P, Li SJ, Gardiol D, Cronan JE, de Mendoza D. The genes encoding the biotin carboxyl carrier protein and biotin carboxylase subunits of Bacillus subtilis acetyl coenzyme A carboxylase, the first enzyme of fatty acid synthesis. J Bacteriol. 1995;177:7003–7006. doi: 10.1128/jb.177.23.7003-7006.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekhedov S, Martinez de Ilarduya O, Ohlrogge JB. Toward a functional catalog of the plant genome: a survey of genes for lipid biosynthesis. Plant Physiol. 2000;122:389–401. doi: 10.1104/pp.122.2.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman T, de Bruijn FJ, Green P, Keegstra K, Kende H, McIntosh L, Ohlrogge J, Raikhel N, Somerville S, Thomashow M. Genes galore: a summary of methods for accessing results from large-scale partial sequencing of anonymous Arabidopsis cDNA clones. Plant Physiol. 1994;106:1241–1255. doi: 10.1104/pp.106.4.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolau BJ, Hawke C. Purification and characterization of maize leaf acetyl-coenzyme A carboxylase. Arch Biochem Biophys. 1984;228:85–96. doi: 10.1016/0003-9861(84)90049-3. [DOI] [PubMed] [Google Scholar]
- Ohlrogge JB, Jaworski JG. Regulation of fatty acid biosynthesis. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:109–136. doi: 10.1146/annurev.arplant.48.1.109. [DOI] [PubMed] [Google Scholar]
- Ohlrogge JB, Kuhn DN, Stumpf PK. Subcellular localization of acyl carrier protein in leaf protoplasts of Spinacia oleracea. Proc Natl Acad Sci USA. 1979;78:1194–1198. doi: 10.1073/pnas.76.3.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page RA, Okada S, Harwood JL. Acetyl-CoA carboxylase exerts strong flux control over lipid synthesis in plants. Biochim Biophys Acta. 1994;1210:369–372. doi: 10.1016/0005-2760(94)90242-9. [DOI] [PubMed] [Google Scholar]
- Post-Beittenmiller MA, Jaworski JG, Ohlrogge JB. In vivo pools of free and acylated acyl carrier proteins in spinach: evidence for sites of regulation of fatty acid biosynthesis. J Biol Chem. 1991;266:1858–1865. [PubMed] [Google Scholar]
- Post-Beittenmiller MA, Roughan G, Ohlrogge JB. Regulation of plant fatty acid biosynthesis: analysis of acyl-CoA and acyl-acyl carrier protein substrate pools in spinach and pea chloroplast. Plant Physiol. 1992;100:923–930. doi: 10.1104/pp.100.2.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reverdatto S, Beilinson V, Nielsen NC. A multisubunit acetyl-coenzyme A carboxylase from soybean. Plant Physiol. 1999;119:961–978. doi: 10.1104/pp.119.3.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesler KR, Savage LJ, Shintani DK, Shorrosh BS, Ohlrogge JB. Co-purification, co-immunoprecipitation, and coordinate expression of acetyl-coenzyme A carboxylase activity, biotin carboxylase, and biotin carboxyl carrier protein of higher plants. Planta. 1996;198:517–525. doi: 10.1007/BF00262637. [DOI] [PubMed] [Google Scholar]
- Roesler KR, Shintani D, Savage L, Boddupalli S, Ohlrogge JB. Targeting of the Arabidopsis homomeric acetyl-coenzyme A carboxylase to Brassica napus seed plastids. Plant Physiol. 1997;113:75–81. doi: 10.1104/pp.113.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Samols D, Thornton CG, Murtif VL, Kumar GK, Haase FC, Wood HG. Evolutionary conservation among biotin enzymes. J Biol Chem. 1988;263:6461–6464. [PubMed] [Google Scholar]
- Sasaki Y, Hakamada K, Suama Y, Nagano Y, Furusawa I, Matsuno R. Chloroplast-encoded protein as a subunit of acetyl-CoA carboxylase in pea plant. J Biol Chem. 1993;268:25118–25123. [PubMed] [Google Scholar]
- Sasaki Y, Kozaki A, Hatano M. Link between light and fatty acid synthesis: thioredoxin-linked reductive activation of plastidic acetyl-CoA carboxylase. Proc Natl Acad Sci USA. 1997;94:11096–11101. doi: 10.1073/pnas.94.20.11096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y, Konishi T, Nagano Y. The compartmentation of acetyl-coenzyme A carboxylase in plants. Plant Physiol. 1995;108:445–449. doi: 10.1104/pp.108.2.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage LJ, Ohlrogge JB. Phosphorylation of pea chloroplast acetyl-CoA carboxylase. Plant J. 1999;18:521–527. doi: 10.1046/j.1365-313x.1999.00478.x. [DOI] [PubMed] [Google Scholar]
- Schulte W, Topfer R, Stracke R, Schell J, Martini N. Multi-functional acetyl-CoA carboxylase from Brassica napus is encoded by a multi-gene family: indication for plastidic localization of at least one isoform. Proc Natl Acad Sci USA. 1997;94:3465–3470. doi: 10.1073/pnas.94.7.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutt BS, Brummel M, Schuch R, Spener F. The role of acyl carrier protein isoforms from Cuphea lanceolata seeds in the de novo biosynthesis of medium-chain fatty acids. Planta. 1998;205:263–268. doi: 10.1007/s004250050320. [DOI] [PubMed] [Google Scholar]
- Shintani DK, Ohlrogge JB. The characterization of a mitochondrial acyl carrier protein isoform isolated from Arabidopsis thaliana. Plant Physiol. 1994;104:1221–1229. doi: 10.1104/pp.104.4.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani DK, Ohlrogge JB. Feedback inhibition of fatty acid synthesis in tobacco suspension cells. Plant J. 1995;7:577–587. [Google Scholar]
- Shorrosh BS, Dixon RA, Ohlrogge JB. Molecular cloning, characterization, and elicitation of acetyl-CoA carboxylase from alfalfa. Proc Natl Acad Sci USA. 1994;91:4323–4327. doi: 10.1073/pnas.91.10.4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorrosh BS, Savage LJ, Soll J, Ohlrogge JB. The pea chloroplast membrane-associated protein, IEP96, is a subunit of acetyl-CoA carboxylase. Plant J. 1996;10:261–268. doi: 10.1046/j.1365-313x.1996.10020261.x. [DOI] [PubMed] [Google Scholar]
- Song J, Wurtele ES, Nikolau BJ. Molecular cloning and characterization of the cDNA coding for the biotin-containing subunit of 3-methylcrotonoyl-CoA carboxylase: identification of the biotin carboxylase and biotin-carrier domains. Proc Natl Acad Sci USA. 1994;91:5779–5783. doi: 10.1073/pnas.91.13.5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh MC, Schultz DJ, Ohlrogge JB. Isoforms of acyl carrier protein involved in seed-specific fatty acid synthesis. Plant J. 1999;17:679–688. doi: 10.1046/j.1365-313x.1999.00418.x. [DOI] [PubMed] [Google Scholar]
- Sun J, Ke J, Johnson JL, Nikolau BJ, Wurtele ES. Biochemical and molecular biological characterization of CAC2, the Arabidopsis thaliana gene coding for the biotin carboxylase subunit of the plastidic acetyl-coenzyme A carboxylase. Plant Physiol. 1997;115:1371–1383. doi: 10.1104/pp.115.4.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelen JJ, Muszynski MG, David NR, Luethy MH, Elthon TE, Miernyk JA, Randall DD. The dihydrolipoamide S-acetyltransferase subunit of the mitochondrial pyruvate dehydrogenase complex from maize contains a single lipoyl domain. J Biol Chem. 1999;274:21769–21775. doi: 10.1074/jbc.274.31.21769. [DOI] [PubMed] [Google Scholar]
- Thelen JJ, Muszynski MG, Miernyk JA, Randall DD. Molecular analysis of two pyruvate dehydrogenase kinases from maize. J Biol Chem. 1998;273:26618–26623. doi: 10.1074/jbc.273.41.26618. [DOI] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnham E, Northcote DH. Changes in the activity of acetyl-CoA carboxylase during rape-seed formation. Biochem J. 1983;212:223–229. doi: 10.1042/bj2120223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JA, Todd J, Newman T, Focks N, Girke T, Martinez de Ilarduya O, Jaworski JG, Ohlrogge J, Benning C. A new set of Arabidopsis ESTs from developing seeds: the metabolic pathway from carbohydrates to seed oil. Plant Physiol. 2000;124:1582–1594. doi: 10.1104/pp.124.4.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]