Abstract
Biliary tract cancers, including intra- and extra-hepatic cholangiocarcinoma as well as gallbladder cancer, are associated with poor prognosis and the majority of patients present with advanced-stage, non-resectable disease at diagnosis. Biliary tract cancer may develop through an accumulation of genetic and epigenetic alterations and can be influenced by microbial exposure. Furthermore, the liver and biliary tract are exposed to the gastrointestinal microbiome through the gut–liver axis. The availability of next-generation sequencing technology has led to an increase in studies investigating the relationship between microbiota and human disease. In particular, the interplay between the microbiome, the tumour micro-environment and response to systemic therapy is a prospering area of interest. Given the poor outcomes for patients with biliary tract cancer, this emerging field of research, through which new biomarkers may be identified, offers potential as a tool for early diagnosis, prognostication or even as a future therapeutic target. This review summarises the available evidence on the microbiome environment in patients with biliary tract cancer, including a discussion around confounding factors, implications for therapy and proposed future directions.
Subject terms: Biliary tract cancer, Cancer microenvironment
Background
Biliary tract cancers (BTC) are sub-classified into intra- and extra-hepatic cholangiocarcinoma (the latter being further subdivided into perihilar cholangiocarcinoma and distal cholangiocarcinoma [1]) and gallbladder cancer. Although cholangiocarcinoma is considered relatively rare in the Western world, 0.35–2 cases per 100,000 population [2], both incidence and mortality are rising, particularly of intrahepatic cholangiocarcinoma [3, 4]. Furthermore, estimated cancer-related deaths for the United States suggest that liver and intrahepatic bile duct cancer will surpass colorectal cancer to become the third most common cause of cancer-related death by 2040 [5].
Patients with BTC have a poor prognosis and the majority present with advanced-stage, non-resectable disease at diagnosis [6]. The Advanced Biliary tract Cancer (ABC)-02 study reported a median progression-free survival (PFS) and overall survival (OS) in the advanced setting of 8 and 11.7 months, respectively, with first-line standard-of-care cisplatin/gemcitabine chemotherapy [7]. Additionally, the ABC-06 study determined benefit from second-line FOLFOX (5-fluorouracil, folinic acid and oxaliplatin) chemotherapy compared with active symptom control alone, median OS of 6.2 months versus 5.3 months [8]. Given the modest survival gains observed thus far, new therapeutic options are required.
Risk factors for cholangiocarcinoma vary globally and collectively account for a small number of cases [9]. In East Asia, liver fluke infections with Opisthorchis viverrini or Clonorchis sinensis, due to the consumption of uncooked river fish, are the driving risk factor for cholangiocarcinoma [10]. A recent meta-analysis evaluated risk factors for cholangiocarcinoma in Eastern and Western world populations, excluding the established risk factors of primary sclerosing cholangitis (PSC) and liver fluke infection, and reported that the strongest risk factors for both intra- and extra-hepatic cholangiocarcinoma were biliary cysts and stones, cirrhosis, hepatitis B and hepatitis C [11]. Gallbladder carcinoma has a different range of established risk factors, including, but not limited to, cholelithiasis [12], PSC [13], structural biliary tree abnormalities [14] and obesity [15].
Biliary tract cancer may therefore develop through the accumulation of genetic and epigenetic alterations, and can be influenced by host immunity, diet, environmental and microbial exposure [16]. The ‘microbiome’ refers to the collective genomes of microorganisms within a particular environment, and the human intestinal microbiome comprises ~1014 microorganisms that have a crucial role in host functions, including modulating immunity, protecting the host against pathogenic microbes and regulating metabolic processes [17–20]. Microbial dysbiosis (an imbalance in the gut microbial community) has been associated with multiple diseases, including cancer [21, 22], obesity and insulin resistance [23, 24], inflammatory bowel disease [25] and neurodegenerative disease [26, 27].
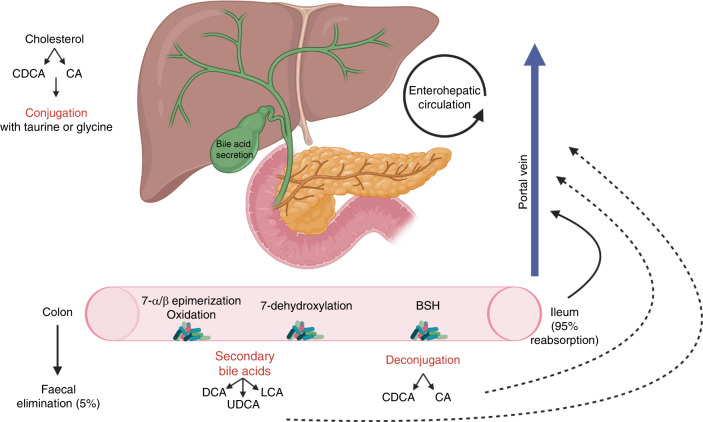
The liver and biliary tract are exposed to the gastrointestinal microbiome through the gut–liver axis, which refers to the bidirectional communication between the gastrointestinal tract and the liver via the portal vein, biliary tract and systemic circulation [28]. Primary bile acids, cholic acid (CA) and chenodeoxycholic acid (CDCA) are conjugated with glycine or taurine in the liver and released into the duodenum via the biliary tract. Approximately 95% are reabsorbed in the terminal ileum, recirculated to the liver via the portal circulation and secreted again in a process known as enterohepatic circulation [29]. The remaining primary bile acids are deconjugated by bacteria with bile salt hydrolase (BSH) activity such as Clostridium, Enterococcus, Bifidobacterium and Lactobacillus, before being absorbed or metabolised into secondary bile acids via a further gut microbiota-mediated process involving dehydroxylation (Fig. 1) [30, 31]. Secondary bile acids either return to the liver via passive absorption or are excreted in the faeces. Bile acids and the gut microbiota closely interact, and in addition to microbiota affecting bile acid metabolism, bile acids also affect microbiota composition [29–31].
Fig. 1. Microbial metabolism of bile acids and the enterohepatic circulation [28–31].
(CDCA: chenodeoxycholic acid, CA: cholic acid, LCA: lithocholic acid, DCA: deoxycholic acid, UDCA: ursodeoxycholic acid, BSH: bile salt hydrolase). Created with BioRender.com.
There is growing evidence that disruptions in the gut–liver axis contribute to the pathogenesis of many liver diseases, including cancer [32]. A recent study by Ma et al. demonstrated that gut microbiota-mediated bile acid metabolism regulates liver cancer (hepatocellular carcinoma (HCC) and liver metastases) via accumulation of hepatic natural killer T (NKT) cells. Expression of CXCL16, a ligand on liver sinusoidal endothelial cells responsible for NKT-cell accumulation, was increased by primary bile acids and enhanced tumour inhibition, whereas increasing secondary bile acids had opposing effects [33].
Additionally, patients with BTC are at risk of developing biliary obstruction and may require interventions, including endoscopic retrograde cholangiopancreatography (ERCP) or percutaneous transhepatic cholangiography (PTC). These necessary interventions may inadvertently introduce gut microbiota into the biliary tract. Biliary stenting has been shown to induce significant bile microbiota shifts and is associated with a higher risk of post-operative surgical-site infection [34, 35], however, the effects on oncological outcomes are less clear. A recent study by Shrader et al. treated human pancreatic cancer cells in vitro with bile samples collected during pancreaticoduodenectomy from patients (stented and non-stented) in order to observe if bacterial contamination had any effect on pancreatic cell survival. They first demonstrated that bile from patients with stents had less effect on reducing pancreatic cancer-cell survival than non-stented bile. Secondly, they found that pre-incubation of non-stented bile samples with live Enterococcus faecalis or Streptococcus oralis reduced the inhibitory effect of the bile on pancreatic cancer-cell survival [36]. These findings support the concept that introduction of gut bacteria into the biliary system through biliary stenting may alter the bile composition and its biological behaviour towards cancer cells. Further preclinical experiments and clinical studies are required to confirm this theory.
The human biliary system has often been considered to be a sterile environment in individuals without biliary pathology or prior biliary intervention, although the technical and ethical challenges of bile sample collection in ‘healthy’ individuals limit the evidence within this field. Molinero et al. aimed to address this limitation by collecting bile samples from the gallbladders of liver-transplant donors with no biliary pathology (control group), and comparing them with bile samples from the gallbladders of patients undergoing surgery for cholelithiasis. The results demonstrated that bacterial communities were in fact present in the bile of the control group, and the microbiota composition differed significantly from the cholelithiasis group (p < 0.05) [37]. Further studies are needed to improve understanding of the physiologically ‘normal’ bile microbiota, which will be crucial in unravelling their potential role in human disease.
Furthermore, the development of cost-effective, high-throughput next-generation sequencing (NGS) technology, which obtains 16S ribosomal ribonucleic acid (rRNA) hypervariable region or whole-genome shotgun (WGS) sequence reads for analysis of the microbiome, has resulted in a substantial increase in studies investigating the relationship between the human microbiome and cancer. This review focuses on the available evidence on the microbiome environment in patients with BTC and potential implications this may have for therapy, including a discussion on potential directions for future research.
Methods
A search to identify eligible studies and conference abstracts was undertaken using the Ovid Medline and Embase databases. Keywords used to identify eligible publications included (((microbio* OR microflor* OR ‘gastrointestinal microbiome’/OR microbiome/ or ‘bacterial microbiome’/) AND ((‘biliary tract’ OR gallbladder OR ‘ampulla of Vater’ OR klatskin) ADJ3 (cancer* OR tumor* OR tumour* OR carcinoma* OR neoplas* OR malignanc*)) OR BTC OR ((perihilar OR klatskin OR hilar OR “distal bile duct” OR intrahepatic) AND cholangiocarcinoma)) OR ‘biliary tract neoplasms’/OR ‘biliary tract cancer’/OR exp ‘bile duct carcinoma’/OR ((‘bile ducts, intrahepatic’/) AND (cancer* OR tumor* OR tumour* OR carcinoma* OR neoplas* OR malignanc*))). No dates of publication or language limits were applied.
Eligible studies were required to include results on microbes associated with BTC in human participants. Meta-analyses, conference abstracts, prospective studies and retrospective series were included. The references of eligible studies and relevant review articles (identified from the database search) were examined to detect other studies of interest. Exclusion due to non-relevance consisted of studies reporting on benign disease only, other malignancies, biliary tract infections, complications of biliary tract procedures or interventions, and studies reporting on antibiotic excretion, susceptibility or resistance. Eligible studies that were duplicated in a meta-analysis were also excluded.
Data collected from each of the eligible studies included author’s name, country, year of publication, type of study, number of patients, disease site, sample type and handling, method of analysis and key microbiota results.
Results
The database search identified 413 studies (last updated 11th March 2021): 360 ineligible studies were excluded after review of the abstracts and the remaining 53 studies were assessed firstly for results reporting microbes associated with BTC, and secondly for references detailing other studies of interest. A total of 20 eligible studies were identified: four meta-analyses, three conference abstracts and thirteen observational studies (2013–2021) (Fig. 2). One further study [38] was added following peer review, resulting in a total of 21 eligible studies.
Fig. 2. Flow diagram describing identification of studies reporting on the microbiome environment in biliary tract cancer.
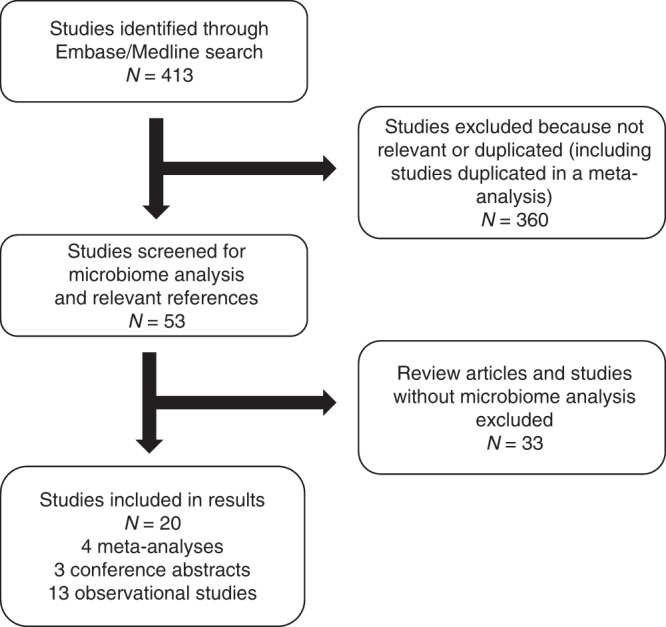
Reasons for inclusion and exclusion are stated.
The potential association of Helicobacter species and Salmonella typhi with BTC
Three prospective studies and two meta-analyses investigating the association of Helicobacter species with BTC were identified (Table 1). The meta-analyses both included ten case–control studies and six studies were duplicated across the two analyses. Xiao et al. restricted inclusion criteria to case–control studies in which control participants had no known diagnosis of cholelithiasis [39], whereas Zhou et al. included case–control studies in which inter-study control participants were with and without benign biliary pathology [40]. Both meta-analyses found a significant association between Helicobacter species and the presence of BTC compared with control subjects without biliary pathology, and Zhou et al. also reported significantly higher Helicobacter infection rates in patients with BTC compared to those with benign biliary pathology. Zhou et al. also performed a species subgroup analysis and found significantly higher rates of H. pylori and H. bilis in BTC compared with benign disease, and no significant differences in H. hepaticus or H. ganmani between groups.
Table 1.
Selected studies reporting on microbes associated with benign biliary disease and biliary tract cancer in human studies.
| Author | Country | Type of study | No. of patients | Disease site | Sample type | Method of analysis | Microbiota results |
|---|---|---|---|---|---|---|---|
| Helicobacter species | |||||||
| Murphy et al. [43] | Finland | Prospective | 410 |
Biliary tract carcinoma (BTC) (89) Hepatocellular carcinoma (HCC) (97) Age-matched controls (224) |
Serum | Helicobacter spp. Multiplex serology assay |
Helicobacter pylori (H. pylori) seropositivity in 100% gallbladder cancer, 97% of extra-hepatic bile duct cancer, 96% of intrahepatic bile duct cancer and 91% of ampulla of Vater cancer. OR 5.47 (95% CI 1.17–25.65) for H.pylori seropositivity and risk of developing BTC. |
| Segura-López et al. [41] | Mexico | Prospective | 194 |
Extra-hepatic cholangiocarcinoma (ECCA) (103) Control—benign biliary pathology (91) |
aBiliary brushing | Polymerase chain reaction (PCR) |
Helicobacter bilis (H. bilis) infection was significantly associated with ECCA (43% of cases) compared to benign biliary pathology (21% of cases) (p = 0.002). Helicobacter hepaticus (H. hepaticus) infection not significantly different between the two groups (p = 0.82). |
| Avilés-Jiménez et al. [42] | Mexico | Prospective | 200 |
Extra-hepatic cholangiocarcinoma (100) Control—benign biliary pathology (100) |
aBiliary brushing |
PCR (all samples) 16S ribosomal ribonucleic acid (rRNA) gene sequencing (20 samples) |
H. pylori significantly more abundant in ECCA than in control group (p = 0.035) Significant difference in microbiota composition between ECCA and benign biliary pathology group (p = 0.01). Methylophilaceae, Fusobacterium, Prevotella, Actinomyces, Novosphingobium and H. pylori increased in ECCA. |
| Zhou et al. [40] |
Japan Thailand India Pakistan Germany |
Meta-analysis |
10 studies (726 patients) |
Biliary tract cancer (190) Benign biliary pathology (434) Control—no biliary pathology (102) |
Serum Bile Tissue |
PCR Enzyme linked immunosorbent assay (ELISA) Culture Immunohistochemistry |
Infection rate of Helicobacter spp significantly higher in BTC group compared to: -benign biliary pathology group (OR 3.2, 95% CI 2.15–4.77, p = 0.0001) -control group (OR 6.5, 95% CI 3.14–13.63, p = 0.0001) Higher rate of H. pylori (49.5% vs 33.3%, p = 0.003) and H. bilis (52.2% vs 23.7%, p < 0.0001) in BTC group vs benign biliary pathology group. No significant difference in rate of H. hepaticus and Helicobacter ganmani between groups. |
| Xiao et al. [39] |
Sweden Germany Japan Thailand China |
Meta-analysis |
10 studies (418 patients) |
Cholangiocarcinoma (CCA) Control—without cholelithiasis |
Serum Bile Tissue |
PCR ELISA Culture Immunohistochemistry |
Significant association between Helicobacter spp and CCA (cumulative OR 8.88, 95% CI 3.67–21.49). Subgroup analysis based on geographic distribution: -Asia (OR 6.68, 95% CI 2.29–19.49) -Europe (OR 14.9, 95% CI 4.79–46.35) |
| Salmonella typhi | |||||||
| Author | Country | Type of study | No. of patients | Disease site | Sample type | Method of analysis | Microbiota results |
| Nagaraja et al. [44] |
UK India Chile USA China Japan Bolivia Mexico |
Meta-analysis | 17 studies | Gallbladder carcinoma (GBC) |
Serum Bile Tissue |
Antibody levels Culture PCR |
Subgroup analysis of studies from South-East Asia showed significant association between chronic Salmonella typhi (S. typhi) status and GBC (OR 4.13, 95% CI 2.87–5.94, p < 0.01). Chronic S. typhi carrier state significantly associated with GBC based on S. typhi antibody levels (OR 3.52, 95% CI: 2.48–5.00, p < 0.01) and culture (OR: 4.14, 95% CI 2.41–7.12, p < 0.01). |
| Koshiol et al. [45] |
Chile India USA Denmark China Japan Bolivia Mexico |
Meta-analysis | 16 studies | Gallbladder carcinoma |
Serum Bile Stool Tissue |
Antibody levels Culture PCR |
Elevated S. typhi Vi antibody seropositivity associated with GBC risk (RR 4.6, 95% CI 3.1–6.8). Positive S. typhi bile or stool culture associated with GBC risk (RR 5.0, 95% CI 2.7–9.3) |
| Other identified microbiota | |||||||
| Author | Country | Type of study | No. of patients | Disease site | Sample type | Method of analysis | Microbiota results |
| Lenz et al. [55] | Germany | Prospective | 58 |
Biliary tract carcinoma (24%) Chronic pancreatitis (14%) Choledocholithiasis (14%) Unclear stenosis of common bile duct (9%) Remaining 39% not specified |
aBile | 16S rRNA gene sequencing |
In all patients, the 9 most common bacterial phyla were Pseudomonadales (11.8%), Enterobacteriales (10.0 %), Sphingomonadales (8.3 %), Burkholderiales (8.1%), Lactobacillales (7.6%), Caulobacterales (6.7%), Alteromonadales (6.3%), Rhizobiales (6.0%) and Clostridiales (5.8%). No significant correlation between phyla and primary diagnosis (p = 0.803), CCA (p = 0.664) or malignant biliary stenosis (p = 0.529). |
| Tsuchiya et al. [48] 2018 |
Bolivia Chile |
Prospective | 37 |
Gallbladder carcinoma (7) Cholelithiasis (30) |
bBile | 16S rRNA gene sequencing |
Fusobacterium nucleatum, Escherichia coli (E. coli) and Enterobacter sp. were the predominant species in patients with GBC. E. coli, Enterococcus gallinarum and Salmonella sp. were the predominant species in patients with cholelithiasis. |
| Poudel et al. [49] | USA | Prospective | 10 |
Cholangiocarcinoma (3) Ampullary carcinoma (1) Pancreatic ductal adenocarcinoma (PDAC) (5) Gallstone pancreatitis (1) |
aBile | 16S rRNA gene sequencing |
Most reads were from phyla Firmicutes (57.9%) and Proteobacteria (14.9%). Benign specimen (pancreatitis) separated clearly from the rest showing 98.9% of reads from Clostridium sensu stricto (phylum Firmicutes). Beta-diversity analysis: a cluster including 3 samples (2 with CCA, 1 with PDAC) had higher abundance of phyla Fusobacteria (90.6%) and Verrucomicrobia (92.9%) |
| Jia et al. [50] | China | Prospective | 84 |
Intrahepatic cholangiocarcinoma (ICCA) (28) Hepatocellular carcinoma (28) Liver cirrhosis (16) Control—no biliary pathology (12) |
Faecal | 16S rRNA gene sequencing |
ICCA patients showed higher prevalence rates of Lactobacillus, Actinomyces, Peptostreptococcaceae, and Alloscardovia than the other groups The family Ruminococcaceae was more abundant in patients with ICCA with vascular invasion (compared to those without vascular invasion) |
| Chng et al. [51] |
Singapore Thailand Romania |
Retrospective | 60 |
Cholangiocarcinoma Opisthorchis.viverrini associated (28) Non O. viverrini associated (32) |
cTumour | 16S rRNA gene sequencing |
Dietziaceae, Pseudomonadaceae and Oxalobacteraceae were the major inhabitants of bile duct tissues in CCA patients. Bifidobacteriaceae, Enterobacteriaceae and Enterococcaceae enrichment in O. viverrini versus non - O. viverrini groups. Stenotrophomonas significantly enriched in tumour vs adjacent normal tissue in non - O. viverrini group (p = 0.039) |
| Lee et al. [57] | South Korea | Prospective | 155 |
Biliary tract cancer (24) Cholecystitis/cholangitis (43) Control—no biliary pathology (88) |
dPlasma | 16S rRNA gene sequencing | Compositional differences of Bifidobacteriaceae family and Oxalobacteraceae Ralstonia found to be a significant positive marker, and the Pseudomonadaceae family, Corynebacteriaceae Corynebacterium and Comamonadaceae Comamonas were significant negative markers to differentiate BTC patients from control group. |
| Chen et al. [52] | China | Prospective | 68 |
Distal cholangiocarcinoma (dCCA) (8) Common bile duct stones (44) Recurrent choledocholithiasis (16) |
aBile | 16S rRNA gene sequencing |
Proteobacteria, Firmicutes, Bacteroidetes, and Actinobacteria are the most dominant phyla in the bile of patients with dCCA and common bile duct stones. Significant increase in the phyla Gemmatimonadetes, Nitrospirae, Chloroflexi, Latescibacteria, and Planctomycetes in dCCA patients compared to common bile duct stones. |
| Dangtakot et al. [53] | Thailand | Prospective | 60 |
Cholangiocarcinoma (30) Choledocholithiasis (CDL) (30) |
aBile | 16S rRNA gene sequencing |
Enterobacter, Stenotrophomonas and Pseudomonas significantly more abundant in CCA compared to CDL (p < 0.05). Cetobacterium, Pyramidobacter and Streptococcus species significantly less abundant in CCA compared to CDL (p < 0.05). |
| Katsuyuki et al. [56] | USA | Prospective |
95 bile (b) samples 58 faecal (f) samples |
Cholangiocarcinoma (11f, 26b) Primary sclerosing cholangitis (31f, 35b) PSC co-existing with CCA (16f, 17b) Control—cholelithiasis or choledocholithiasis (17b) |
eBile Faecal |
16S rRNA gene sequencing |
CCA bile samples had significant difference of alpha diversity compared to control group, with less Streptococcaceae and Desulfovibrionaceae. An increased abundance of Fusobacteria was found after biliary stent placement and increased with the number of stents in the bile duct. |
| Zhang et al. [54] | China | Prospective | 71 |
Cholangiocarcinoma (8) Hepatocellular carcinoma (10) Mixed-type liver cancer (6) Liver cirrhosis (24) Control—healthy (23) |
Faecal | 16S rRNA gene sequencing |
Enterobacter and Escherichia-Shigella most significantly represented in patients with primary liver cancer (CCA, HCC, mixed-type). Relative abundance of Enterobacter ludwigii highest in primary liver cancer group and 100× greater than liver cirrhosis and healthy controls. Significantly decreased Firmicutes/Bacteroidetes ratio in primary liver cancer group compared to healthy controls (p < 0.05). |
| Song et al. [58] | China | Prospective | 14 |
Gallbladder carcinoma (7) Chronic calculous cholecystitis (7) |
bTissue | Metagenomic sequencing | Peptostreptococcus stomatis, Fusobacterium mortiferum, Acinetobacter junii and Enterococcus faecium positively correlated and significantly contributed to GBC group. |
| Saab et al. [38] | Iran | Prospective | 75 |
Extra-hepatic cholangiocarcinoma (28) Control—benign biliary pathology (47) |
aBile | 16S rRNA gene sequencing |
The most abundant genera in ECCA group were Enterococcus, Streptococcus, Bacteroides, Klebsiella and Pyramidobacter. In a subgroup analysis of patients without comorbidities (19 ECCA and 37 controls), relative abundance of Bacteroides, Geobacillus, Meiothermus and Anoxybacillus significantly higher in ECCA compared with controls (p < 0.05). |
| Serra et al. [46] | Italy | Prospective | 53 |
Cholangiocarcinoma (20) Gallbladder carcinoma (2) Carcinoma of the head of the pancreas (31) |
bBile | Phoenix Automated Microbiology System |
E. coli and Pseudomonas spp were significant positive predictors for presence of CCA (p < 0.0001). Pseudomonas spp only significant positive predictor for presence of GBC (p = 0.0001). |
| Di Carlo et al. [47] 2019 | Italy | Retrospective | 152 |
Cholangiocarcinoma (42) Gallbladder carcinoma (5) Ampullary carcinoma (4) Carcinoma of the head of the pancreas (72) Cholelithiasis (27) Cholangitis (1) Chronic pancreatitis (1) |
cBile | Phoenix Automated Microbiology System or Vitek-2 System | E. coli, Pseudomonas aeruginosa, and Klebsiella pneumoniae were identified in the cancer population and their presence was associated with reduced survival time. |
aSamples obtained at time of scheduled endoscopic retrograde cholangiopancreatography (ERCP) or percutaneous transhepatic biliary drainage (PTBD).
bSamples obtained at time of surgery.
cArchived samples (previous biopsy or biliary tract procedure).
dBacteria-derived extracellular vesicles isolated in the plasma.
eTiming of sample collection not specified.
Three prospective studies carried out since the meta-analyses support an association between Helicobacter species and BTC. Segura-López et al. [41] and Avilés-Jiménez et al. [42] identified significant associations between extra-hepatic cholangiocarcinoma and H. bilis or H. pylori, respectively, detected by polymerase chain reaction (PCR) analysis of biliary brushings at the time of scheduled ERCP. Alternatively, Murphy et al. used a Helicobacter species multiplex serology assay, and prospectively evaluated associations between seropositivity and BTC. The study concluded that seropositivity to H. pylori proteins was associated with an increased risk of developing BTC [43], however, the participants comprised entirely of Finnish male smokers and the results require validation in other populations.
Two meta-analyses investigating the association of Salmonella typhi and gallbladder carcinoma were identified (Table 1). Nagaraja et al. included seventeen studies [44], Koshiol et al. included 22 studies [45] and fifteen studies were duplicated across the two analyses. Both analyses included case–control and cohort studies. A variety of sample types, including bile, stool, tissue and serum from patients with and without gallbladder carcinoma, were analysed, and both meta-analyses found an association between chronic S. typhi carrier state and gallbladder carcinoma, based on S. typhi antibody levels and culture-detection methods. Nagaraja et al. also conducted a subgroup analysis of studies from South-East Asia and reported a significant association between chronic S. typhi carrier state and gallbladder carcinoma in this geographical distribution.
The potential association of other microbiota with BTC
The following section will highlight the reported associations of other microbiota identified with BTC.
Two studies used automated microbiology systems (Phoenix or Vitek-2) to analyse bile microbiota in patients with BTC (stage not specified). Serra et al. conducted a cross-sectional study on bile samples from females undergoing surgery for confirmed biliary tract or pancreatic cancer, and found Pseudomonas species to be a significant positive predictor for the presence of cholangiocarcinoma and gallbladder carcinoma [46]. Di Carlo et al. retrospectively evaluated bile samples taken at the time of scheduled ERCP in a cohort of patients with confirmed BTC, carcinoma of the head of the pancreas and benign biliary pathology, and found that the presence of E. coli, P. aeruginosa, and K. pneumoniae was associated with reduced survival in the cancer population (death within 6 months of diagnosis) [47].
Thirteen studies were identified, which used 16S rRNA gene sequencing [42, 48–57] or shotgun metagenomics [58] to analyse the microbiome in patients with BTC (Table 1). A variety of sample types from patients with BTC were analysed: seven studies used bile, three used faecal samples, three used tissue and one study used plasma. Seven studies stated that samples were frozen at −80 °C prior to DNA extraction, one study stated that samples were stored in a freezer but did not specify the temperature and another study stated that samples were thawed on ice prior to placing them in lysing tubes. Three studies were identified from conference abstracts, which did not include details on sample storage. Eight different DNA-extraction kits were used across the studies, the most commonly used kits were the PowerSoil DNA Isolation Kit and the QIAamp DNA Easy Kit (used in two studies each). The most common variable regions of the 16S gene targeted for sequencing were V3 and V4. An overview of sample storage, DNA extraction and DNA sequencing is provided in Table 2.
Table 2.
Methodology of studies using 16S rRNA gene sequencing or shotgun metagenomics to analyse the microbiome in patients with BTC.
| Author | Sample storage | DNA extraction | Selected region of 16 S gene | Sequencing platform |
|---|---|---|---|---|
| Lenz et al. [55] | Not stated | Not stated | V3–V4 | Illumina |
| Tsuchiya et al. [48] | Stored at −80 °C | NucleoSpin Soil | V3–V4 | Illumina Miseq |
| Jia et al. [50] | Stored at −80 °C | PowerSoil DNA Isolation kit | V4 | Illumina Miseq |
| Lee et al. [57] | Stored in a freezer | PowerSoil DNA Isolation kit | V3–V4 | Illumina Miseq |
| Chen et al. [52] | Stored at −80 °C | OMEGA DNA kit | V3–V4 | Illumina Miseq |
| Chng et al. [51] | Thawed on ice and transferred into lysing tubes | EZ1 DNA Tissue kit | V3–V6 | Illumina Hiseq 2000 |
| Poudel et al. [49] | Not stated | PowerViral DNA Isolation kit | Not stated | Illumina |
| Avilés-Jiménez et al. [42] | Stored at −80 °C | QIAamp DNA easy kit | V4 | Illumina Miseq |
| Dangtakot et al. [53] | Stored at −80 °C | QIAamp DNA easy kit | V3–V4 | Illumina Hiseq 2500 |
| Katsuyuki et al. [56] | Not stated | Not stated | V3–V5 | Illumina Miseq |
| Zhang et al. [54] | Placed on ice and transferred to laboratory | QIAamp DNA mini kit | V3–V4 | Illumina Hiseq |
| Song et al. [58] | Stored at −80 °C | QIAamp DNA mini kit | N/A (metagenomic study) | Illumina Hiseq ×10 |
| Saab et al. [38] | Stored at −80 °C | QIAsymphony kit | V3-4 | Illumina Miseq |
Differentially abundant taxa described across the thirteen studies covered all taxonomic levels, from phylum to species. In particular, an increase in Fusobacteria (four studies), Enterobacteriaceae (three studies) and Pseudomonadaceae (three studies) was consistently reported in patients with BTC. The stage of BTC was not specified in nine out of ten of these studies. Of the four studies reporting on Fusobacteria, two studies found it to be a predominant species in different sample types (bile and tissue), specifically in patients with gallbladder carcinoma. Enterobacteriaceae was reported at an increased abundance in three different study populations and in two different patient sample types: cholangiocarcinoma (bile sample), gallbladder carcinoma (bile sample) and primary liver cancer, including cholangiocarcinoma, HCC and mixed-type HCC/cholangiocarcinoma (faecal sample). In two studies, Pseudomonadaceae was reported at an increased abundance in patients with cholangiocarcinoma in two different sample types (tissue and bile). No other bacterial taxa were consistently reported in more than two studies.
Establishing consistent relationships between specific taxa and a disease is challenging, and some taxon have been both positively and negatively associated with BTC. For example, Serra et al. reported Pseudomonas in bile samples as a significant positive predictor for the presence of cholangiocarcinoma [46]; however, Lee et al. analysed plasma samples and found it to be a significant negative marker for the presence of BTC [57]. Notably, the studies had similar sample sizes (22 versus 24 patients with BTC), however, they were conducted in two different geographical regions (Italy compared with South Korea), and used different methods of analysis (automated microbiology system versus 16S rRNA gene sequencing).
Discussion
To some degree, the heterogeneity of the microbiota results can be attributed to the inherent intra- and inter-person variability of microbiome composition [59, 60], however, the methodological disparities between studies should also be considered. Differences in study population, sample type, sample handling and method of analysis of all of the included studies will now be discussed and assessed for their potential to confound the results.
Study population
Large differences in sample size existed between the studies, ranging from four to 103 participants with BTC and one to 224 control participants. The inability to replicate the results could in part be due to the low sample size of some studies. The control-group population is another variable to consider, and eleven studies included control participants with benign biliary pathology. This is largely due to the majority of studies reporting on the bile or biliary tissue microbiome, necessitating the choice of control group to include individuals that have a clinical indication for an invasive procedure, such as an ERCP, as opposed to including healthy subjects.
Nineteen different countries are represented across the results dataset, including the meta-analyses. Geographical location has been shown to have an effect on human gut microbiome variations [61], and may contribute to intra- or inter-study differences. Chng et al demonstrated compositional differences in the biliary tract tissue microbiota of liver fluke-related and non-related cholangiocarcinoma [51]. This supports the role of O.viverrini in enabling an altered microbiome, but the intra-study differences observed could in part be due to the multiple origins of the samples (Thailand, Singapore and Romania). Additionally, the two meta-analyses investigating an association between S. typhi and gallbladder carcinoma include studies that are mostly conducted in regions with a high incidence of typhoid fever, although Koshiol et al. found that the association remained even when stratified by geographical region [45].
Sample type, collection and storage
Inter-subject variation is dependent on sampling site [62], and beta-diversity analysis has demonstrated that the overall structure of microbiota is significantly different between different samples types [63]. The variety of sample types analysed in BTC studies may therefore contribute to the inter-study variability of microbiota results. The majority of the included studies collected and analysed bile or biliary tract tissue, whilst other studies analysed blood or faecal samples. Additionally, description of the biliary tract tissue micro-environment has typically been generalised from bile fluid analysis; however, Chng et al. found a significantly different composition between the two sample types in patients with cholangiocarcinoma (stage not specified) [51].
A selection of studies to date have identified similarities between bile or biliary tract tissue microbiota and faecal microbiota in patients with BTC. For example, Jia et al. reported higher abundances of four genera, including Actinomyces, in the gut microbiota of patients with intrahepatic cholangiocarcinoma [50], whilst Avilés-Jiménez et al. previously described an increased abundance of bile duct Actinomyces in patients with extra-hepatic cholangiocarcinoma [42]. Furthermore, Saab et al., who took methodological precautions to avoid contamination of collected bile with the duodenal mucosa, reported levels of Proteobacteria that were close to values previously described in the small intestine [38]. Factors influencing bacterial colonisation within the biliary tract may include gastric or duodenal contamination, altered sphincter of Oddi function or transportation through the process of enterohepatic circulation [38]. Further studies are needed to assess the comparability of microbiota results from studies reporting on different sample types, particularly comparisons between biliary and intestinal microbiota.
Additionally, studies comparing the methodology used for microbiome analysis have demonstrated that sample collection, transportation and storage all have an impact on sample quality [64, 65]. Optimising faecal microbiome studies is of particular interest, as participants often collect the sample at home, presenting logistical challenges. Ideally, collected samples should be transported as soon as possible and stored at −20 °C to −80 °C to prevent microbial composition alteration. Where this is impractical, a DNA stabiliser can be used as a preservation tool, and multiple commercial kits have been validated [66]. In this review, of the thirteen studies that used NGS techniques (Table 2), seven stored the samples at −80 °C, whereas others transported the samples on ice to the laboratory, or stored them in a freezer at an unspecified temperature. The three studies presented as conference abstracts did not report transportation or storage conditions [49, 55, 56]. Microbial shifts due to inconsistencies in sample transportation and storage conditions should therefore be considered as a potential source of inter-study differences.
Method of analysis
The traditional approach of using culture methods to identify bacteria involved in human disease has many limitations, including a bias towards bacteria that proliferate under laboratory conditions, and an underestimation of the diverse microbial population in question [67, 68]. In this review, culture methods were used in studies investigating an individual species (Helicobacter or Salmonella). Other included studies used PCR, enzyme-linked immunosorbent assay (ELISA), immunohistochemistry or multiplex serology assays, as a means of microbial detection. Arguably of more importance is the spectrum of methods used between studies, and the impact that this will have on differences in the results, for example, Zhou et al. reported detection rates of Helicobacter species varying from 0 to 83% in the bile, serum and biliary tissue of patients with BTC and benign biliary pathology across ten case–control studies using a range of detection methods [40].
In the following sections, important considerations regarding sample processing and analysis for studies using NGS techniques will be discussed. Firstly, the process of DNA extraction can introduce fundamental bias, especially in relation to extraction kits and the inclusion of a mechanical, as well as a chemical method for cell lysis [69]. The addition of a mechanical lysis step has been linked to a higher DNA yield, higher bacterial diversity and more efficient extraction of DNA from Gram-positive bacteria [70, 71]. Of the thirteen studies using NGS techniques to evaluate the microbiome in patients with BTC, eight different DNA-extraction kits were used, comprising different methods of chemical and/or mechanical lysis, and therefore may have contributed to the heterogeneity of the results between studies.
A variable region of the 16S rRNA gene must be selected for PCR amplification and sequencing. Importantly, the choice of hypervariable region and the design of PCR primers have an effect on phylogenetic resolution [72, 73]. All of the included studies that reported on the choice of variable region incorporated the V4 region for sequencing, therefore reducing the likelihood of bias. To a lesser extent, choice of sequencing platform can also explain inter-study differences [73], however, all of the included studies used a version of the Illumina sequencing platform, making this an unlikely contribution to variability observed.
Bioinformatic analysis involves the translation of bacterial sequences (generated using NGS techniques) into taxonomic profiles and relative abundance estimations. A study comparing three different 16S rRNA pipelines used for taxonomic assignment (QIIME1, MALT and DADA2) with the outputs of whole- metagenomic sequencing (WMS) reported that two of the pipelines (QIIME1 and DADA2) yielded results that were more consistent with WMS taxonomic assignments in comparison with the third choice of pipeline (MALT). Furthermore, the lower the abundance of a bacterial genus (<0.5%, as detected by WMS), the lower the probability of it being correctly identified by any of the three 16S rRNA pipelines [74]. These results indicate that the bioinformatic-processing pipeline should be considered as a source of analytical bias, and may have contributed to the heterogeneity of results observed between studies.
Studies of the microbiome in other cancer types
Key lessons from studies of the microbiome in other cancer populations will now be explored, and future directions for studies in BTC proposed.
Predictive and prognostic role of the microbiome
Recent studies show significant microbial contributions in select cancer types, primarily of the faecal microbiome. Significant differences in the diversity and composition of the gut microbiome have been demonstrated in patients with melanoma, non-small-cell lung cancer (NSCLC) and renal-cell carcinoma (RCC) who respond to anti-programmed cell death-1 (PD-1) immunotherapy (versus non-responders) [75–77]. Routy et al. analysed faecal samples from patients with NSCLC and RCC and found that Akkermansia muciniphila was significantly associated with patients who responded to immunotherapy (p = 0.004) [75]. Both Gopalakrishnan et al. and Matson et al. analysed faecal samples from patients with metastatic melanoma, and reported a significantly higher abundance of Ruminococcaceae (p < 0.01) and Bifidobacteriaceae (p < 0.05), respectively, in responders compared with non-responders [76, 77]. Direct comparison of differentially abundant microbiota across these studies is limited by differences in methods of analysis, as well as differences in the method of discriminating between responders and non-responders. Nonetheless, there is agreement that there is an association between the composition and diversity of the gut microbiome and anti-PD-1 efficacy, and further studies are needed to determine the composition of a ‘favourable’ gut microbiome.
The tumour-associated microbiome is another area of interest, and a distinctive signature (Pseudoxanthomonas–Streptomyces–Saccharopolyspora–Bacillus clausii) was predictive of long-term survival (>5 years after surgery) in a study analysing the tumour specimens of 43 patients with resected pancreatic ductal adenocarcinoma (PDAC) [78]. Microbiota identified in the tumour may also have a role in tumour response to chemotherapy, for example, Gammaproteobacteria (a common class of bacteria identified in PDAC tumours) is able to metabolise gemcitabine into its inactive form and could account for gemcitabine resistance in this patient group [79]. Additionally, an enrichment of Fusobacterium nucleatum has been observed in multiple patient cohorts with colon cancer across the world [80–82], and tumour Fusobacterium load has been significantly associated with poorer survival outcomes among patients with caecum and ascending colon tumours [83].
Furthermore, studies assessing precancerous conditions have identified associations between the microbiome and disease progression. Pereira et al. compared the bile microbiota between control subjects undergoing routine ERCP with patients diagnosed with early-stage PSC, advanced-stage PSC or biliary dysplasia/cholangiocarcinoma. The results demonstrated that the bile microbiota composition of control subjects and subjects with early-stage PSC was similar, however, the presence of members of the Streptococcus genus in bile was positively correlated with PSC disease progression [84]. These results underline the need to further explore the role of Streptococcus in PSC progression and development of cholangiocarcinoma.
Potential effects of antibiotic use on the microbiome and response to systemic treatment in patients with malignancy
Biliary obstruction arises as a result of direct tumour compression in patients with BTC, and is frequently complicated by superadded infection and a requirement for antibiotics. Antibiotics can influence the gut microbiome and may affect response to cancer therapy. Specifically, administration of antibiotics within 2 months before, or 1 month after, initiation of immunotherapy, is associated with significantly shorter PFS and OS in patients with advanced NSCLC, RCC and urothelial carcinoma [75]. The link between antibiotic use, immunotherapy efficacy and reduced OS is supported by a number of recent meta-analyses [85–87]. Furthermore, the effect of antibiotics on OS was greater, depending on time of exposure in relation to immunotherapy initiation (Lurienne et al. report −60 to +60 days [85]); however, the heterogeneity of included studies in the meta-analyses remains a limiting factor.
Alternatively, the use of antibiotics to target key constituents of the cancer microbiota has been a point of interest, and Bullman et al. demonstrated that treatment of mice, bearing a colon-cancer xenograft, with metronidazole, reduced Fusobacterium load, cancer-cell proliferation and overall tumour growth [83]. Further studies looking into this targeted microbiota approach are needed.
The microbiome as a therapeutic target
The role of the reinstatement of the microbiome on therapeutic response in solid tumours is also currently being investigated. Modulation of the gut microbiota by means of faecal microbial transplantation (FMT) has demonstrated promising results in preclinical cancer models in mice [75, 76], and more recently in a phase-I clinical trial [88]. Using donors who achieved complete response to PD-1 blockade, FMT and re-induction of anti-PD-1 therapy in patients with refractory metastatic melanoma is safe, feasible and potentially effective (clinical response in 3/10 patients) [88]. However, the characteristics of optimal microbiota compositions of FMT donors and recipients remain elusive and future studies in larger cohorts are needed.
There are currently seven studies listed on clinicaltrials.gov (last updated 1st April 2021) recruiting patients with cancer for FMT interventions. The majority of eligible patients are those with immunotherapy-resistant disease and the cancer types being investigated are RCC, melanoma, prostate and gastrointestinal, and one study is recruiting patients with advanced solid tumours who are being treated with immunotherapy (NCT4116775, NCT04130763, NCT04264975, NCT03353402, NCT03341143, NCT04758507 and NCT037772899). Additionally, four studies are recruiting patients to investigate the effectiveness of FMT in treating immunotherapy-induced colitis (NCT04038619, NCT03819296, NCT04721041 and NCT04163289). There are currently no recorded studies recruiting patients with BTC in this specific research area.
Conclusion
This review highlights accumulating evidence for an association between the microbiome and BTC; however, studies to date have not yet distinguished whether changes in the composition of microbiota have a causative role, or whether they are solely an effect of the cancer. Small sample size, a lack of methodological standardisation and non-availability of information about confounding factors limit the comparability of the obtained results. Many of the studies also lack information on characteristics such as the stage of BTC, therefore limiting the applicability of associations between specific microbes and survival outcomes. Standardisation of study protocols, as well as collection and publication of information on other confounding factors, including medication history, diet and geography, must be considered in the design of future studies.
In this review, Fusobacteria, Enterobacteriaceae and Pseudomonadaceae were the most consistently reported taxa at an increased abundance in patients with BTC. Fusobacteria, and more specifically F. nucleatum, has been frequently associated with colorectal carcinogenesis [89, 90], however, it has also been reported in oral [91, 92], oesophageal [93, 94], cervical [95] and gastric cancer [96, 97]. Enterobacteriaceae may also promote colon cancer [89, 98]. The biliary tract is exposed to the gastrointestinal microbiome via the gut–liver axis, and therefore microbes involved in colorectal carcinogenesis and progression, such as Fusobacteria and Enterobacteriaceae, may play a similar role in BTC. Further studies are needed to confirm this relationship.
Future prospective studies in BTC should aim to explore the prognostic and predictive ability of the microbiome, and establish if differences in the diversity and composition of the microbiome are correlated with response to treatment and survival. Furthermore, studies to investigate the dynamic nature of the microbiome in BTC should assess differences between patients with resectable versus advanced disease, as well as identifying any longitudinal changes in the microbiome, for example, pre- compared with post treatment. Additionally, emerging evidence suggests a link between antibiotic use and immunotherapy efficacy; future studies need to establish whether this link exists in patients with BTC receiving systemic treatment.
Formal diagnosis of BTC, in particular cholangiocarcinoma, is made difficult by numerous factors, including a lack of screening programmes, non-specificity and late presentation of symptoms, and technical difficulties in obtaining tissue for cytology or histopathology. The microbiome is an emerging field through which new biomarkers may be identified, and offers potential as a tool for early diagnosis of BTC, prognostication or even as a future therapeutic target. As a result, this evolving field of research warrants further investigation in both preclinical and clinical BTC studies.
Acknowledgements
Not applicable
Author contributions
RW drafted and revised the paper. EK reviewed and approved the final version of the paper. TJ performed the literature search and reviewed and approved the final version of the paper. AL reviewed and approved the final version of the paper. RAH reviewed and approved the final version of the paper. JWV reviewed and approved the final version of the paper. MMN concept initialisation and review and approval of draft and final version of the paper.
Funding information
Dr. Roseanna C Wheatley is studying for an MD, funded by the Timpson Fellowship. The salary of Dr. Angela Lamarca is in part funded by The Christie Charity and the European Union’s Horizon 2020 Research and Innovation Programme [grant number 825510, ESCALON].
Ethics approval and consent to participate
Not applicable.
Consent to publish
Not applicable.
Competing interests
RCW: no competing interests to declare. EK: no competing interests to declare. TJ: no competing interests to declare. AL: received travel and educational support from Ipsen, Pfizer, Bayer, AAA, SirtEx, Novartis, Mylan and Delcath; speaker honoraria from Merck, Pfizer, Ipsen, Incyte and AAA; advisory honoraria from EISAI, Nutricia Ipsen, QED and Roche; she is a member of the Knowledge Network and NETConnect Initiatives funded by Ipsen; all outside scope of this work. RAH has served on the advisory board for Roche, BMS, Eisai, Celgene, Beigene, Ipsen and BTG. He has received speaker fees from Eisai, Ipsen, Mylan and PrimeOncology, and has received travel and educational support from Bayer, BMS and Roche; all outside of the scope of this work. JWV received honoraria from Agios, AstraZeneca, Baxter, Genoscience Pharma, Hutchison Medipharma, Imaging Equipment Ltd (AAA), Incyte, Ipsen, Mundipharma EDO, Mylan, QED, Servier, Sirtex and Zymeworks; and grants, honoraria and non-financial support from NuCana, all outside of the scope of this work. MMN received research grant support from Servier, Ipsen and NuCana. She has received travel and accommodation support from Bayer and Ipsen and speaker honoraria from Pfizer, Ipsen, NuCana and Mylan. She has served on advisory boards for Celgene, Ipsen, Sirtex, Baxalta and Incyte; all outside of the scope of this work.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Nakeeb A, Pitt HA, Sohn TA, Coleman JA, Abrams RA, Piantadosi S, et al. Cholangiocarcinoma: a spectrum of intrahepatic, perihilar, and distal tumors. Ann Surg. 1996;224:463–75. doi: 10.1097/00000658-199610000-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banales JM, Cardinale V, Carpino G, Marzioni M, Andersen JB, Invernizzi P, et al. Expert consensus document: cholangiocarcinoma: current knowledge and future perspectives consensus statement from the European Network for the Study of Cholangiocarcinoma (ENS-CCA) Nat Rev Gastroenterol Hepatol. 2016;13:261–80. doi: 10.1038/nrgastro.2016.51. [DOI] [PubMed] [Google Scholar]
- 3.Patel T. Increasing incidence and mortality of primary intrahepatic cholangiocarcinoma in the United States. Hepatology. 2001;33:1353–7. doi: 10.1053/jhep.2001.25087. [DOI] [PubMed] [Google Scholar]
- 4.Taylor-Robinson SD, Toledano MB, Arora S, Keegan TJ, Hargreaves S, Beck A, et al. Increase in mortality rates from intrahepatic cholangiocarcinoma in England and Wales 1968–1998. Gut. 2001;48:816–20. doi: 10.1136/gut.48.6.816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rahib L, Wehner MR, Matrisian LM, Nead KT. Estimated projection of US cancer incidence and death to 2040. JAMA Netw Open. 2021;4:e214708. doi: 10.1001/jamanetworkopen.2021.4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forner A, Vidili G, Rengo M, Bujanda L, Ponz-Sarvisé M, Lamarca A. Clinical presentation, diagnosis and staging of cholangiocarcinoma. Liver Int. 2019;39:98–107. doi: 10.1111/liv.14086. [DOI] [PubMed] [Google Scholar]
- 7.Valle J, Wasan H, Palmer DH, Cunningham D, Anthoney A, Maraveyas A, et al. Cisplatin plus gemcitabine versus gemcitabine for biliary tract cancer. N Engl J Med. 2010;362:1273–81. doi: 10.1056/NEJMoa0908721. [DOI] [PubMed] [Google Scholar]
- 8.Lamarca A, Palmer DH, Wasan HS, Ross PJ, Ma YT, Arora A, et al. Second-line FOLFOX chemotherapy versus active symptom control for advanced biliary tract cancer (ABC-06): a phase 3, open-label, randomised, controlled trial. Lancet Oncol. 2021;22:690–701. doi: 10.1016/S1470-2045(21)00027-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valle JW, Kelley RK, Nervi B, Oh DY, Zhu AX. Biliary tract cancer. Lancet. 2021;397:428–44. doi: 10.1016/S0140-6736(21)00153-7. [DOI] [PubMed] [Google Scholar]
- 10.Sithithaworn P, Yongvanit P, Duenngai K, Kiatsopit N, Pairojkul C. Roles of liver fluke infection as risk factor for cholangiocarcinoma. J Hepatobiliary Pancreat Sci. 2014;21:301–8. doi: 10.1002/jhbp.62. [DOI] [PubMed] [Google Scholar]
- 11.Clements O, Eliahoo J, Kim JU, Taylor-Robinson SD, Khan SA. Risk factors for intrahepatic and extrahepatic cholangiocarcinoma: A systematic review and meta-analysis. J Hepatol. 2019;72:95–103. doi: 10.1016/j.jhep.2019.09.007. [DOI] [PubMed] [Google Scholar]
- 12.Hsing AW, Bai Y, Andreotti G, Rashid A, Deng J, Chen J, et al. Family history of gallstones and the risk of biliary tract cancer and gallstones: A population-based study in Shanghai, China. Int J Cancer. 2007;121:832–8. doi: 10.1002/ijc.22756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewis JT, Talwalkar JA, Rosen CB, Smyrk TC, Abraham SC. Prevalence and risk factors for gallbladder neoplasia in patients with primary sclerosing cholangitis: Evidence for a metaplasia-dysplasia-carcinoma sequence. Am J Surg Pathol. 2007;31:907–13. doi: 10.1097/01.pas.0000213435.99492.8a. [DOI] [PubMed] [Google Scholar]
- 14.Kamisawa T, Kaneko K, Itoi T, Ando H. Pancreaticobiliary maljunction and congenital biliary dilatation. Lancet Gastroenterol Hepatol. 2017;2:610–8. doi: 10.1016/S2468-1253(17)30002-X. [DOI] [PubMed] [Google Scholar]
- 15.Li ZM, Wu ZX, Han B, Mao YQ, Chen HL, Han SF, et al. The association between BMI and gallbladder cancer risk: a meta-analysis. Oncotarget. 2016;7:43669–79. doi: 10.18632/oncotarget.9664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakamura H, Arai Y, Totoki Y, Shirota T, Elzawahry A, Kato M, et al. Genomic spectra of biliary tract cancer. Nat Genet. 2015;47:1003–10. doi: 10.1038/ng.3375. [DOI] [PubMed] [Google Scholar]
- 17.Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016;14:e1002533. [DOI] [PMC free article] [PubMed]
- 18.Honda K, Littman DR. The microbiota in adaptive immune homeostasis and disease. Nature. 2016;535:75–84. doi: 10.1038/nature18848. [DOI] [PubMed] [Google Scholar]
- 19.Fukuda S, Toh H, Hase K, Oshima K, Nakanishi Y, Yoshimura K, et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469:543–9. doi: 10.1038/nature09646. [DOI] [PubMed] [Google Scholar]
- 20.Tremaroli V, Bäckhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489:242–9. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- 21.Gao R, Wang Z, Li H, Cao Z, Gao Z, Chen H, et al. Gut microbiota dysbiosis signature is associated with the colorectal carcinogenesis sequence and improves the diagnosis of colorectal lesions. J Gastroenterol Hepatol. 2020;35:2109–21. doi: 10.1111/jgh.15077. [DOI] [PubMed] [Google Scholar]
- 22.Chen J, Domingue JC, Sears CL. Microbiota dysbiosis in select human cancers: evidence of association and causality. Semin Immunol. 2017;32:25–34. doi: 10.1016/j.smim.2017.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541–6. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- 24.Bäckhed F, Ding H, Wang T, Hooper LV, Gou YK, Nagy A, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA. 2004;101:15718–23. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma HQ, Yu TT, Zhao XJ, Zhang Y, Zhang HJ. Fecal microbial dysbiosis in Chinese patients with inflammatory bowel disease. World J Gastroenterol. 2018;24:1464–77. doi: 10.3748/wjg.v24.i13.1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu SC, Cao ZS, Chang KM, Juang JL. Intestinal microbial dysbiosis aggravates the progression of Alzheimer’s disease in Drosophila. Nat Commun. 2017;8:24. [DOI] [PMC free article] [PubMed]
- 27.Keshavarzian A, Green SJ, Engen PA, Voigt RM, Naqib A, Forsyth CB, et al. Colonic bacterial composition in Parkinson’s disease. Mov Disord. 2015;30:1351–60. doi: 10.1002/mds.26307. [DOI] [PubMed] [Google Scholar]
- 28.Tripathi A, Debelius J, Brenner DA, Karin M, Loomba R, Schnabl B, et al. The gut-liver axis and the intersection with the microbiome. Nat Rev Gastroenterol Hepatol. 2018;15:397–411. doi: 10.1038/s41575-018-0011-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Aguiar Vallim TQ, Tarling EJ, Edwards PA. Pleiotropic roles of bile acids in metabolism. Cell Metab. 2013;17:657–69. doi: 10.1016/j.cmet.2013.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ridlon JM, Harris SC, Bhowmik S, Kang DJ, Hylemon PB. Consequences of bile salt biotransformations by intestinal bacteria. Gut Microbes. 2016;7:22–39. doi: 10.1080/19490976.2015.1127483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wahlström A, Sayin SI, Marschall H-U, Bäckhed F. Intestinal Crosstalk between Bile Acids and Microbiota and Its Impact on Host Metabolism. Cell Metab. 2016;24:41–50. doi: 10.1016/j.cmet.2016.05.005. [DOI] [PubMed] [Google Scholar]
- 32.Albillos A, de Gottardi A, Rescigno M. The gut-liver axis in liver disease: pathophysiological basis for therapy. J Hepatol. 2020;72:558–77.. doi: 10.1016/j.jhep.2019.10.003. [DOI] [PubMed] [Google Scholar]
- 33.Ma C, Han M, Heinrich B, Fu Q, Zhang Q, Sandhu M, et al. Gut microbiome–mediated bile acid metabolism regulates liver cancer via NKT cells. Science. 2018;360:eaan5931. [DOI] [PMC free article] [PubMed]
- 34.Scheufele F, Aichinger L, Jäger C, Demir IE, Schorn S, Sargut M, et al. Effect of preoperative biliary drainage on bacterial flora in bile of patients with periampullary cancer. Br J Surg. 2017;104:e182–e188. doi: 10.1002/bjs.10450. [DOI] [PubMed] [Google Scholar]
- 35.Scheufele F, Schorn S, Demir IE, Sargut M, Tieftrunk E, Calavrezos L, et al. Preoperative biliary stenting versus operation first in jaundiced patients due to malignant lesions in the pancreatic head: a meta-analysis of current literature. Surg (U S) 2017;161:939–50. doi: 10.1016/j.surg.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 36.Shrader HR, Miller AM, Tomanek-Chalkley A, McCarthy A, Coleman KL, Ear PH, et al. Effect of bacterial contamination in bile on pancreatic cancer cell survival. Surg (U S) 2021;169:617–22. doi: 10.1016/j.surg.2020.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Molinero N, Ruiz L, Milani C, Gutiérrez-Díaz I, Sánchez B, Mangifesta M, et al. The human gallbladder microbiome is related to the physiological state and the biliary metabolic profile. Microbiome. 2019;7:1–17. doi: 10.1186/s40168-019-0712-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saab M, Mestivier D, Sohrabi M, Rodriguez C, Khonsari MR, Faraji A, et al. Characterization of biliary microbiota dysbiosis in extrahepatic cholangiocarcinoma. PLoS ONE. 2021;16:e0247798. [DOI] [PMC free article] [PubMed]
- 39.Xiao M, Gao Y, Wang Y. Helicobacter species infection may be associated with cholangiocarcinoma: a meta‐analysis. Int J Clin Pr. 2014;68:262–70. doi: 10.1111/ijcp.12264. [DOI] [PubMed] [Google Scholar]
- 40.Zhou D, Wang JD, Weng MZ, Zhang Y, Wang XF, Gong W, et al. Infections of Helicobacter spp. in the biliary system are associated with biliary tract cancer: A meta-analysis. Eur J Gastroenterol Hepatol. 2013;25:447–54. doi: 10.1097/MEG.0b013e32835c0362. [DOI] [PubMed] [Google Scholar]
- 41.Segura-López FK, Avilés-Jiménez F, Güitrón-Cantú A, Valdéz-Salazar HA, León-Carballo S, Guerrero-Pérez L, et al. Infection with Helicobacter bilis but not Helicobacter hepaticus was Associated with Extrahepatic Cholangiocarcinoma. Helicobacter. 2015;20:223–30. doi: 10.1111/hel.12195. [DOI] [PubMed] [Google Scholar]
- 42.Avilés-Jiménez F, Guitron A, Segura-López F, Méndez-Tenorio A, Iwai S, Hernández-Guerrero A, et al. Microbiota studies in the bile duct strongly suggest a role for Helicobacter pylori in extrahepatic cholangiocarcinoma. Clin Microbiol Infect. 2016;22:178. doi: 10.1016/j.cmi.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 43.Murphy G, Michel A, Taylor PR, Albanes D, Weinstein SJ, Virtamo J, et al. Association of seropositivity to Helicobacter species and biliary tract cancer in the ATBC study. Hepatology. 2014;60:1963–71. doi: 10.1002/hep.27193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nagaraja V, Eslick GD. Systematic review with meta-analysis: the relationship between chronic Salmonella typhi carrier status and gall‐bladder cancer. Aliment Pharm Ther. 2014;39:745–50. doi: 10.1111/apt.12655. [DOI] [PubMed] [Google Scholar]
- 45.Koshiol J, Wozniak A, Cook P, Adaniel C, Acevedo J, Azócar L, et al. Salmonella enterica serovar Typhi and gallbladder cancer: a case-control study and meta-analysis. Cancer Med. 2016;5:3310–25.. doi: 10.1002/cam4.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Serra N, Di Carlo P, Gulotta G, d’Arpa F, Giammanco A, Colomba C, et al. Bactibilia in women affected with diseases of the biliary tract and pancreas. A STROBE guidelines-adherent cross-sectional study in Southern Italy. J Med Microbiol. 2018;67:1090–5. doi: 10.1099/jmm.0.000787. [DOI] [PubMed] [Google Scholar]
- 47.Di Carlo P, Serra N, D’arpa F, Agrusa A, Gulotta G, Fasciana T, et al. The microbiota of the bilio-pancreatic system: a cohort, STROBE-compliant study. Infect Drug Resist. 2019;12:1513–27. doi: 10.2147/IDR.S200378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsuchiya Y, Loza E, Villa-Gomez G, Trujillo CC, Baez S, Asai T, et al. Metagenomics of microbial communities in gallbladder bile from patients with gallbladder cancer or cholelithiasis. Asian Pacific. J Cancer Prev. 2018;19:961–7. doi: 10.22034/APJCP.2018.19.4.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Poudel SK, Padmanabhan R, Chahal P, Sanaka M, Stevens T, Guinta K, et al. Microbiome signature of bile from pancreatic and biliary tract cancer patients: A pilot study. J Clin Oncol. 2019;37:e15744–e15744. [Google Scholar]
- 50.Jia X, Lu S, Zeng Z, Liu Q, Dong Z, Chen Y, et al. Characterization of gut microbiota, bile acid metabolism, and cytokines in intrahepatic cholangiocarcinoma. Hepatology. 2020;71:893–906. doi: 10.1002/hep.30852. [DOI] [PubMed] [Google Scholar]
- 51.Chng KR, Chan SH, Ng AHQ, Li C, Jusakul A, Bertrand D, et al. Tissue microbiome profiling identifies an enrichment of specific enteric bacteria in opisthorchis viverrini associated cholangiocarcinoma. EBioMedicine. 2016;8:195–202. doi: 10.1016/j.ebiom.2016.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen B, Fu SW, Lu L, Zhao H. A preliminary study of biliary microbiota in patients with bile duct stones or distal cholangiocarcinoma. Biomed Res Int. 2019;1092563. [DOI] [PMC free article] [PubMed]
- 53.Dangtakot R, Intuyod K, Ahooja A, Wongwiwatchai J, Hanpanich P, Lulitanond A, et al. Profiling of bile microbiome identifies district microbial population between choledocholithiasis and cholangiocarcinoma patients. Asian Pac J Cancer Prev. 2021;22:233–40. doi: 10.31557/APJCP.2021.22.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang L, Wu YN, Chen T, Ren CH, Li X, Liu GX. Relationship between intestinal microbial dysbiosis and primary liver cancer. Hepatobiliary Pancreat Dis Int. 2019;18:149–57. doi: 10.1016/j.hbpd.2019.01.002. [DOI] [PubMed] [Google Scholar]
- 55.Lenz P, Steidl L, Cordes F, Kahl BC, Karch H, Dobrindt U, et al. Sa1328 analysis of the human biliary microbiome and its alterations in biliary tract diseases. Gastroenterology. 2015;148:S–293. [Google Scholar]
- 56.Katsuyuki M, Chandrasekhara V, Wongjarupong N, Chen J, Johnson S, Chia N, et al. The Role of the Biliary and Gut Microbiome in the Progression of Cholangiocarcinoma and Primary Sclerosing Cholangitis. Am J Gastroenterol. 2019;114:S25–6. [Google Scholar]
- 57.Lee H, Lee HK, Min SK, Lee WH. 16S rDNA microbiome composition pattern analysis as a diagnostic biomarker for biliary tract cancer. World J Surg Oncol. 2020;18:19. doi: 10.1186/s12957-020-1793-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Song X, Wang X, Hu Y, Li H, Ren T, Li Y, et al. A metagenomic study of biliary microbiome change along the cholecystitis‐carcinoma sequence. Clin Transl Med. 2020;10:e97. doi: 10.1002/ctm2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Voigt AY, Costea PI, Kultima JR, Li SS, Zeller G, Sunagawa S, et al. Temporal and technical variability of human gut metagenomes. Genome Biol. 2015;16:73. doi: 10.1186/s13059-015-0639-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Galloway-Peña JR, Smith DP, Sahasrabhojane P, Wadsworth WD, Fellman BM, Ajami NJ, et al. Characterization of oral and gut microbiome temporal variability in hospitalized cancer patients. Genome Med. 2017;9:21. doi: 10.1186/s13073-017-0409-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.He Y, Wu W, Zheng HM, Li P, McDonald D, Sheng HF, et al. Regional variation limits applications of healthy gut microbiome reference ranges and disease models. Nat Med. 2018;24:1532–5. doi: 10.1038/s41591-018-0164-x. [DOI] [PubMed] [Google Scholar]
- 62.Flores GE, Caporaso JG, Henley JB, Rideout JR, Domogala D, Chase J, et al. Temporal variability is a personalized feature of the human microbiome. Genome Biol. 2014;15:531–531. doi: 10.1186/s13059-014-0531-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Langheinrich M, Wirtz S, Kneis B, Gittler MM, Tyc O, Schierwagen R, et al. Microbiome Patterns in Matched Bile, Duodenal, Pancreatic Tumor Tissue, Drainage, and Stool Samples: Association with Preoperative Stenting and Postoperative Pancreatic Fistula Development. J Clin Med. 2020;9:2785. doi: 10.3390/jcm9092785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang Z, Zolnik CP, Qiu Y, Usyk M, Wang T, Strickler HD, et al. Comparison of fecal collection methods for microbiome and metabolomics studies. Front Cell Infect Microbiol. 2018;8:301. doi: 10.3389/fcimb.2018.00301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Song SJ, Amir A, Metcalf JL, Amato KR, Xu ZZ, Humphrey G, et al. Preservation methods differ in fecal microbiome stability, affecting suitability for field studies. mSystems. 2016;1:21–37. doi: 10.1128/mSystems.00021-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu WK, Chen CC, Panyod S, Chen RA, Wu MS, Sheen LY, et al. Optimization of fecal sample processing for microbiome study—the journey from bathroom to bench. J Formos Med Assoc. 2019;118:545–55.. doi: 10.1016/j.jfma.2018.02.005. [DOI] [PubMed] [Google Scholar]
- 67.Gupta S, Mortensen MS, Schjørring S, Trivedi U, Vestergaard G, Stokholm J, et al. Amplicon sequencing provides more accurate microbiome information in healthy children compared to culturing. Commun Biol. 2019;2:1–7. doi: 10.1038/s42003-019-0540-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Woo PCY, Lau SKP, Teng JLL, Tse H, Yuen KY. Then and now: Use of 16S rDNA gene sequencing for bacterial identification and discovery of novel bacteria in clinical microbiology laboratories. Clin Microbiol Infect. 2008;14:908–34. doi: 10.1111/j.1469-0691.2008.02070.x. [DOI] [PubMed] [Google Scholar]
- 69.Claassen S, du Toit E, Kaba M, Moodley C, Zar HJ, Nicol MP. A comparison of the efficiency of five different commercial DNA extraction kits for extraction of DNA from faecal samples. J Microbiol Methods. 2013;94:103–10.. doi: 10.1016/j.mimet.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pollock J, Glendinning L, Wisedchanwet T, Watson M. The madness of microbiome: attempting to find consensus “best practice” for 16S microbiome studies. Appl Environ Microbiol. 2018;84:e02627–17. doi: 10.1128/AEM.02627-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Guo F, Zhang T. Biases during DNA extraction of activated sludge samples revealed by high throughput sequencing. Appl Microbiol Biotechnol. 2013;97:4607–16. doi: 10.1007/s00253-012-4244-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang B, Wang Y, Qian PY. Sensitivity and correlation of hypervariable regions in 16S rRNA genes in phylogenetic analysis. BMC Bioinforma. 2016;17:135. doi: 10.1186/s12859-016-0992-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tremblay J, Singh K, Fern A, Kirton ES, He S, Woyke T, et al. Primer and platform effects on 16S rRNA tag sequencing. Front Microbiol. 2015;6:771. doi: 10.3389/fmicb.2015.00771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bergsten E, Mestivier D, Sobhani I. The limits and avoidance of biases in metagenomic analyses of human fecal microbiota. Microorganisms. 2020;8:1954. doi: 10.3390/microorganisms8121954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillère R, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359:91–7. doi: 10.1126/science.aan3706. [DOI] [PubMed] [Google Scholar]
- 76.Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018;359:97–103. doi: 10.1126/science.aan4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre M-L, et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science. 2018;359:104–8. doi: 10.1126/science.aao3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Riquelme E, Zhang Y, Zhang L, Montiel M, Zoltan M, Dong W, et al. Tumor microbiome diversity and composition influence pancreatic cancer outcomes. Cell. 2019;178:795–806. doi: 10.1016/j.cell.2019.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Geller LT, Barzily-Rokni M, Danino T, Jonas OH, Shental N, Nejman D, et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science. 2017;357:1156–60. doi: 10.1126/science.aah5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McCoy AN, Araújo-Pérez F, Azcárate-Peril A, Yeh JJ, Sandler RS, Keku TO. Fusobacterium is associated with colorectal adenomas. PLoS ONE. 2013;8:e53653. doi: 10.1371/journal.pone.0053653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li YY, Ge QX, Cao J, Zhou YJ, Du YL, Shen B, et al. Association of Fusobacterium nucleatum infection with colorectal cancer in Chinese patients. World J Gastroenterol. 2016;22:3227–33. doi: 10.3748/wjg.v22.i11.3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ito M, Kanno S, Nosho K, Sukawa Y, Mitsuhashi K, Kurihara H, et al. Association of Fusobacterium nucleatum with clinical and molecular features in colorectal serrated pathway. Int J Cancer. 2015;137:1258–68. doi: 10.1002/ijc.29488. [DOI] [PubMed] [Google Scholar]
- 83.Bullman S, Pedamallu CS, Sicinska E, Clancy TE, Zhang X, Cai D, et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science. 2017;358:1443–8. doi: 10.1126/science.aal5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pereira P, Aho V, Arola J, Boyd S, Jokelainen K, Paulin L, et al. Bile microbiota in primary sclerosing cholangitis: Impact on disease progression and development of biliary dysplasia. PLoS ONE. 2017;12:e0182924. [DOI] [PMC free article] [PubMed]
- 85.Lurienne L, Cervesi J, Duhalde L, de Gunzburg J, Andremont A, Zalcman G, et al. NSCLC immunotherapy efficacy and antibiotic use: a systematic review and meta-analysis. J Thorac Oncol. 2020;15:1147–59.. doi: 10.1016/j.jtho.2020.03.002. [DOI] [PubMed] [Google Scholar]
- 86.Wilson BE, Routy B, Nagrial A, Chin VT. The effect of antibiotics on clinical outcomes in immune-checkpoint blockade: a systematic review and meta-analysis of observational studies. Cancer Immunol Immunother. 2020;69:343–54. doi: 10.1007/s00262-019-02453-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Petrelli F, Iaculli A, Signorelli D, Ghidini A, Dottorini L, Perego G, et al. Survival of patients treated with antibiotics and immunotherapy for cancer: a systematic review and meta-analysis. J Clin Med. 2020;9:1458. doi: 10.3390/jcm9051458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Baruch EN, Youngster I, Ben-Betzalel G, Ortenberg R, Lahat A, Katz L, et al. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science. 2021;371:602–9. doi: 10.1126/science.abb5920. [DOI] [PubMed] [Google Scholar]
- 89.Gagnaire A, Nadel B, Raoult D, Neefjes J, Gorvel JP. Collateral damage: Insights into bacterial mechanisms that predispose host cells to cancer. Nat Rev Microbiol. 2017;15:1–20. doi: 10.1038/nrmicro.2016.171. [DOI] [PubMed] [Google Scholar]
- 90.Brennan CA, Garrett WS. Fusobacterium nucleatum—symbiont, opportunist and oncobacterium. Nat Rev Microbiol. 2019;17:156–66. doi: 10.1038/s41579-018-0129-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhao H, Chu M, Huang Z, Yang X, Ran S, Hu B, et al. Variations in oral microbiota associated with oral cancer. Sci Rep. 2017;7:11773. [DOI] [PMC free article] [PubMed]
- 92.Harrandah AM, Chukkapalli SS, Bhattacharyya I, Progulske-Fox A, Chan EKL. Fusobacteria modulate oral carcinogenesis and promote cancer progression. J Oral Microbiol. 2021;13:1849493. doi: 10.1080/20002297.2020.1849493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yamamura K, Baba Y, Nakagawa S, Mima K, Miyake K, Nakamura K, et al. Human microbiome Fusobacterium nucleatum in esophageal cancer tissue is associated with prognosis. Clin Cancer Res. 2016;22:5574–81. doi: 10.1158/1078-0432.CCR-16-1786. [DOI] [PubMed] [Google Scholar]
- 94.Liu Y, Baba Y, Ishimoto T, Tsutsuki H, Zhang T, Nomoto D, et al. Fusobacterium nucleatum confers chemoresistance by modulating autophagy in oesophageal squamous cell carcinoma. Br J Cancer. 2021;124:963–74. doi: 10.1038/s41416-020-01198-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Audirac-Chalifour A, Torres-Poveda K, Bahena-Román M, Téllez-Sosa J, Martínez-Barnetche J, Cortina-Ceballos B, et al. Cervical microbiome and cytokine profile at various stages of cervical cancer: a pilot study. PLoS ONE. 2016;11:e0153274. doi: 10.1371/journal.pone.0153274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hsieh YY, Tung SY, Pan HY, Yen CW, Xu HW, Lin YJ, et al. Increased Abundance of Clostridium and Fusobacterium in gastric microbiota of patients with gastric cancer in Taiwan. Sci Rep. 2018;8:158. [DOI] [PMC free article] [PubMed]
- 97.Boehm ET, Thon C, Kupcinskas J, Steponaitiene R, Skieceviciene J, Canbay A, et al. Fusobacterium nucleatum is associated with worse prognosis in Lauren’s diffuse type gastric cancer patients. Sci Rep. 2020;10:16240. doi: 10.1038/s41598-020-73448-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yurdakul D, Yazgan-Karataş A, Şahin F. Enterobacter strains might promote colon cancer. Curr Microbiol. 2015;71:403–11. doi: 10.1007/s00284-015-0867-x. [DOI] [PubMed] [Google Scholar]