Abstract
Microbial communities in wastewater treatment plants (WWTPs) play a key role in water purification. Microbial communities of activated sludge (AS) vary extensively based on plant operating technology, influent characteristics and WWTP capacity. In this study we performed 16S rRNA gene profiling of AS at nine large-scale WWTPs responsible for the treatment of municipal sewage from the city of Moscow, Russia. Two plants employed conventional aerobic process, one plant—nitrification/denitrification technology, and six plants were operated with the University of Cape Town (UCT) anaerobic/anoxic/oxic process. Microbial communities were impacted by the technology and dominated by the Proteobacteria, Bacteroidota and Actinobacteriota. WWTPs employing the UCT process enabled efficient removal of not only organic matter, but also nitrogen and phosphorus, consistently with the high content of ammonia-oxidizing Nitrosomonas sp. and phosphate-accumulating bacteria. The latter group was represented by Candidatus Accumulibacter, Tetrasphaera sp. and denitrifiers. Co-occurrence network analysis provided information on key hub microorganisms in AS, which may be targeted for manipulating the AS stability and performance. Comparison of AS communities from WWTPs in Moscow and worldwide revealed that Moscow samples clustered together indicating that influent characteristics, related to social, cultural and environmental factors, could be more important than a plant operating technology.
Subject terms: Environmental biotechnology, Microbial communities, Microbial ecology, Microbiome, Water microbiology, Environmental microbiology
Introduction
The removal of numerous pollutants produced by agriculture, industry, and households is important for the protection of natural ecosystems and human health. Wastewater treatment plants (WWTPs) employ a series of mechanical and biological processes that convert contaminated water into a sufficiently clean state through a series of steps removing different types of organic and inorganic pollutants1,2. Typically, wastewater treatment in large facilities takes place in three stages. The first stage includes physical methods of water purification, the second stage—chemical and/or biological treatment in bioreactors with suspended or attached activated sludge (AS). The third stage is the final treatment of water and its disinfection.
At the second stage, a consortium of microorganisms of AS transforms pollutants into harmless products or into products less hazardous to the environment and humans than the original components3. AS is a taxonomically and metabolically diverse microbial community with complex trophic relationships between its members4. It is the largest managed artificial ecosystem, continuously functioning in many cases for decades. The composition of the microbial community, which is shaped by both operating conditions and influent characteristics5,6, determines the main biochemical processes of wastewater treatment, and its change, for example, the massive development of filamentous forms of bacteria, can lead to a decrease in the efficiency of treatment and the occurrence of emergency situations7.
In municipal wastewater treatment plants, microbial consortia of AS are often developed under similar conditions, since the content of the main components of wastewater is limited to a rather narrow range of concentrations (except in some extreme cases of highly diluted or concentrated wastewater), pH is usually 7–8, temperatures vary from 10 to 30 °C8. In addition, in modern technologies certain biologically determined rules are followed: maintaining the minimum aerobic age of sludge necessary for the development of nitrification (the retention time of solid matter), ensuring the optimal retention time of the sludge in the anaerobic zone for effective enhanced biological phosphorus removal, optimizing the ratio of biochemical oxygen demand, nitrogen and phosphorus, etc.3. Therefore, it can be expected that the composition of microbial communities of activated sludge will contain a common component, which was confirmed by the results of studies comparing the composition of AS communities of various WWTPs6,9,10. At the same time a high diversity and differences of microbial communities of AS were noted, which was associated with climatic factors and the specificity of certain treatment plants: the share of the industrial component in the total influent, the temperature regime, the peculiarity of the used technologies and the exploitation of plants9. It has been shown that there is a relationship between the diversity and composition of the microbial community and the performance of treatment facilities2, although the authors noted that the real effect is not the performance itself, but the variation of indicators such as chemical oxygen demand, the retention time of suspended matter etc.
Despite the extensive application of traditional and modern molecular methods for studying microbial consortia of AS, their ecophysiology, population dynamics and diversity are far from being comprehensively understood. Most microorganisms of activated sludge are not cultivated, and the role of many typical inhabitants is not clearly known11,12.
Recently, using a systematic worldwide sampling, a Global Water Microbiome Consortium (GWMC) analysed the 16S ribosomal RNA gene sequences from ~ 1,200 AS samples taken from 269 WWTPs10. This study revealed that although the global AS bacterial communities contain ~ 1 billion phylotypes, ASs has a small, global core bacterial community of 28 phylotypes that is strongly linked to WWTP performance10. This study showed that although the type of treatment process exerted significant effects on microbial community structures, it was overwhelmed by geographical separation, and the compositions of AS microbial communities were significantly different between any two continents10. Although the GWMC study included WWTPs from 23 countries on 6 continents, the distribution of samples was geographically biased and covered mostly North America, Western and Central Europe, Eastern Asia (mostly China), Australasia, and several cities in South America and South Africa10.
Another large-scale initiative, the MiDAS project, analyzed samples (mostly AS) from 740 WWTPs using different types of treatment technologies, and represents the largest global sampling of WWTPs to date13,14. The resulting full-length 16S rRNA gene reference database, MiDAS 4, represent a comprehensive catalogue of bacteria in wastewater treatment systems and taxonomy from the domain to species level14. Although this study targeted WWTPs located in 425 cities, 31 countries on 6 continents, like the GWMC project, MiDAS mostly covered the same geographic regions.
In order to expand the geographical coverage and our knowledge about microbiomes of AS in general, we analyzed the composition of microbial communities of AS at large-scale WWTPs in the city of Moscow (Russia). Although the Moscow WWTPs are among the largest in the world, they (as well as other WWTPs from Russia) were not studied in the framework of GWMC and MiDAS projects. There are only a few studies on the analysis of the composition of microbial communities of AS from Moscow WWTPs using modern high throughput molecular genetic methods. Kallistova et al.15 using FISH method analyzed AS samples from four Moscow WWTPs and characterized the abundance of the major technologically important microbial groups (ammonium- and nitrite-oxidizing, phosphate-accumulating, foam-inducing, anammox bacteria, and methanogens) in the aeration tanks. Later Shchegolkova et al.16 performed 16S rRNA gene profiling of AS communities in three WWTPs responsible for processing sewage with different origins: municipal wastewater, slaughterhouse wastewater, and refinery sewage. The taxonomic structures of AS microbiomes were found to become stable in time, and each WWTP demonstrated a distinct pattern16. Several studies were devoted to 16S rRNA profiling, metagenomics and FISH studies of nitritation/ anammox wastewater treatment bioreactors applied for the treatment of NH4-rich wastewater17–20.
In this study, we present the results of an analysis of the composition of AS microbial communities at nine large-scale WWTPs in Moscow employing three different technologies.
Methods
Characteristics of WWTPs in the city of Moscow
Wastewater treatment plants of JSC "Mosvodokanal" carry out the treatment of sewage in the city of Moscow. Household and industrial wastewater entering the city sewerage system undergoes a full purification process, including biological treatment with AS. The largest treatment facilities, the Lyuberetskiy and Kuryanovskiy WWTP complexes (hereafter referred to as LOS and KOS, respectively), each with a capacity of about 2 million m3 per day, consist of several wastewater treatment units in which a number of modern technologies for biological wastewater treatment are implemented, including biological nutrient removal (carbon, nitrogen, phosphorus)21–24. These treatment units (hereafter referred to as WWTPs) at each WWTP complex are fed by the same inflow water but otherwise are independent installations between which there is no transfer and mixing of AS.
After the primary treatment, the wastewater is subjected to purification in bioreactors with suspended activated sludge. The characteristics of the wastewater entering bioreactors at the two WWTP complexes are somehow different. The waters at the Kuryanovskiy WWTP complex contain approximately 1.5 times lower concentrations of organic matter and phosphorus compared to the Lyuberetskiy WWTPs, while the ammonium content do not differ significantly (Table 1).
Table 1.
Characteristics of analyzed WWTPs.
| WWTP ID | WWTP1 | WWTP2 | WWTP3 | WWTP4 | WWTP5 | WWTP6 | WWTP7 | WWTP8 | WWTP9 |
|---|---|---|---|---|---|---|---|---|---|
| WWTPs complex | LOS | LOS | LOS | LOS | LOS | LOS | KOS | KOS | KOS |
| Technology | SF | N-DN | UCT | UCT | UCT | UCT | CAS | UCT | UCT |
| WWTP capacity (103 m3/day) | 1000 | 500 | 80 | 80 | 80 | 500 | 1000 | 600 | 600 |
| Bioreactors hydraulic regime | Plug-flow | Plug-flow | Carrousel | Plug-flow | Carrousel | Carrousel | Plug-flow | Plug-flow | Plug-flow |
| Hydraulic retention time (h) | 12 | 12 | 9 | 9 | 9 | 12 | 8 | 10 | 10 |
| BOD –inflow (mg/L) | 180–190 | 180–190 | 180–190 | 180–190 | 180–190 | 170–190 | 90–130 | 80–120 | 90–130 |
| BOD –outflow (mg/L) | 3–4 | 2–3 | < 3 | < 3 | < 3 | 1–2 | 5–6 | 1–3 | 1–3 |
| COD –inflow (mg/L) | 500–550 | 500–550 | 500–550 | 500–550 | 500–550 | 450–500 | 310–350 | 300–340 | 310–350 |
| COD –outflow (mg/L)` | 30–40 | 30–40 | nd | nd | nd | 30–40 | 30–40 | 30–40 | 30–40 |
| N-NH4- inflow (mg/L) | 40–50 | 40–55 | 40–50 | 40–50 | 40–50 | 35–45 | 40–50 | 35–45 | 40–50 |
| N-NH4- outflow (mg/L) | 5–6 | 0.4–0,5 | < 0,4 | < 0,4 | < 0,4 | 0,3–0,4 | 12–16 | 0,3–0,5 | 0,3–0,5 |
| N-NO3 – outflow (mg/L) | 10–11 | 10–11 | < 9 | < 9 | < 9 | 7–8 | 5–9 | 8–9 | 8–9 |
| P-PO4 – inflow (mg/L) | 4–5 | 4–5 | 4–5 | 4–5 | 4–5 | 4–5 | 2–4 | 2–4 | 2–4 |
| P-PO4 – outflow (mg/L) | 2,5–3,5 | 3–4 | < 1 | < 1 | < 1 | 0,2–0,3 | 0,3–0,6 | 0,2–0,4 | 0,2–0,4 |
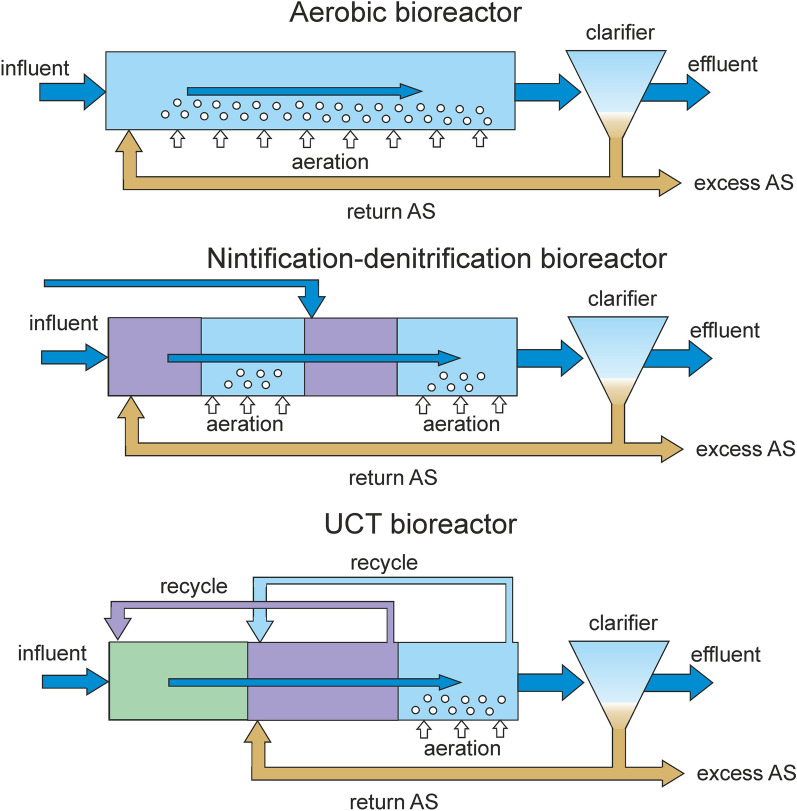
All bioreactors are continuous: the inflowing wastewater and the returned AS are continuously fed to the inlet (or to a certain point) of the bioreactor, where they are mixed. Then the mixed liquor passes the bioreactor and enters the clarifier where the sludge is separated from the outflowing purified water by sedimentation. Chemical coagulants are not used. Three main technologies are used in the investigated WWTPs (Fig. 1).
Figure 1.
Schemes of three types of bioreactors showing the different compartments and flow directions. Anaerobic, anoxic and oxic zones are colored in green, violet and blue, respectively.
In the simplest process, implemented at WWTPs 1 and 7, the oxidation of organic matter by heterotrophic aerobic microorganisms and the oxidation of ammonium by nitrifiers with the formation of nitrate are carried out in plug-flow bioreactors under aerobic conditions. The sludge mixture moves continuously along an elongated corridor from the point of mixing of the return AS with incoming wastewater to the clarifier. Wastewater is supplied either only at the beginning of the bioreactor (Conventional Activated Sludge process, CAS) (WWTP7), or along the entire aeration tank corridor (Step Feed Process, SF) (WWTP1). The entire volume of the bioreactor is aerated by blowing air.
The nitrification/ denitrification technology (N-DN) is realized at WWTP2. Wastewater is fed into the plug-flow bioreactor at the beginning and in the middle of the bioreactor and passes through zones without aeration, each followed by an aerobic zone. In these two anoxic zones, denitrification occurs due to the organic matter contained in the wastewater and nitrates contained in the return AS mixture (in the first anoxic zone) or arriving from the first aerobic zone (in the second anoxic zone). In two aerated zones organics and ammonium are oxidized.
Six other WWTPs are operated by anaerobic/anoxic/oxic process, also known as the University of Cape Town (UCT) technology. At the first stage, the sludge mixture enters the anaerobic zone, where phosphate-accumulating microorganisms (PAO) consume easily degradable organic matter, then to the anoxic zone, where denitrification and accumulation of phosphates by denitrifying PAO occur, and finally to the aerobic zone, where organic matter and ammonium are oxidized. Recycling from the aerobic zone ensures the inflow of nitrates into the anoxic zone, and the recycle of the AS mixture from the end of anoxic zone into the anaerobic zone minimizes the ingress of nitrates. Plug-flow bioreactors are used at WWTPs 4, 8 and 9, and carousel bioreactors at WWTPs 3, 5 and 6 (Supplementary Fig. S1).
The bulk dissolved oxygen concentration in aerobic zones of all bioreactors was 2–3 mg/L, the solid retention time was 15–25 days, and the hydraulic retention time was 8–12 h. The temperature in the time of AS sampling was 18–20 °C, and did not fall below 16 °C even in winter. The concentrations of AS were similar (2–4 g/L) in all WWTPs, and volatile suspended solids accounted for 60–65%.
Sampling and chemical analysis
The AS samples were obtained from nine wastewater treatment facilities at at Luberetsky and Kuryanovsky WWTP complexes in the Moscow region between March and May 2021. Samples of return activated sludge at 9 WWTPs (each in three replicas, 27 samples in total) were taken in BD Falcon tubes and immediately transferred to the laboratory at + 4 °C.
Samples of inflow and outflow water were collected between December 2020 and May 2021 and kindly provided by “Mosvodokanal” JSC. Chemical analysis was carried out according to standard methods. Water quality values (biochemical oxygen demand (BOD), chemical oxygen demand (COD), ammonium nitrogen (NH4-N) and phosphorus (P-PO4) of influent and effluent, as well as nitrate nitrogen (NO3-N) in the effluent were measured using environmental standard methods twice a week for 6 months. Monthly average values are shown in Table 1.
DNA isolation, amplification and sequencing of the 16S rRNA gene fragments
Total genomic DNA from each AS sample was extracted using a Power Soil DNA isolation kit (MO BIO Laboratories, Inc., Carlsbad, CA, USA) and stored at − 20 °C. PCR amplification of 16S rRNA gene fragments comprising the V3–V4 variable regions was carried out using the universal prokaryotic primers PRK 341F (5′-CCTAYG GGDBGCWSCAG) and PRK 806R (5′-GGA CTA CNVGGG THTCTAAT)25. The PCR fragments were bar-coded using the Nextera XT Index Kit v.2 (Illumina, San Diego, CA, USA) and purified using Agencourt AMPure beads (Beckman Coulter, Brea, CA, USA). The concentrations of PCR products were determined using the Qubit dsDNA HS Assay Kit (Invitrogen, Carlsbad, CA, USA). All PCR fragments were then mixed and sequenced on Illumina MiSeq (2 × 300 nt from both ends). Pairwise overlapping reads were merged using FLASH v.1.2.1126. The final dataset consisted of 2,173,862 16S rRNA gene reads (Supplementary Table S1).
Bioinformatics analysis of microbial community composition and diversity
All sequences were clustered into operational taxonomic units (OTUs) at 97% identity using the USEARCH v. 11 program27. Low quality reads, chimeric sequences, and singletons were removed by the USEARCH algorithm. To calculate OTU abundances, all reads obtained for a given sample (including singletons and low-quality reads) were mapped to OTU sequences at a 97% global identity threshold by USEARCH. The taxonomic assignment of OTUs was performed by searching against the SILVA v.138 rRNA sequence database using the VSEARCH v. 2.14.1 algorithm28. The recently developed MiDAS 414, a full-length 16S rRNA gene reference database for wastewater treatment systems, was used to taxonomically identify OTUs up to the species level in the same way.
The diversity indices at a 97% OTU cut-off level were calculated using Usearch v.1127. To avoid sequencing depth bias, the number of reads generated for each sample were randomly sub-sampled to the size of the smallest dataset (94,942 reads) using the «single_rarefaction.py» script of QIIME29.
Calculation of Jaccard and weighted Unifrac distance metrics and trees was performed applying “beta_div” command in USEARCH. For the UniFrac analyses, a tree for the OTUs based on the sequence identity was constructed in USEARCH using “cluster_agg” command.
Integration of data from this study with data from GWMC
All 16S rRNA gene sequences assigned to OTUs obtained in the present work were mapped to OTU sequences from GWMC (http://gwmc.ou.edu/). About 95% of sequences obtained in our experiments were mapped to GWMC OTUs at a 97% global identity threshold. OTU tables of both sets of samples were merged using command of “otutab_merge” of USEARCH. The obtained combined OTU table served as the input data for constructing a neighbor-joining tree generated from the Bray–Curtis dissimilarity matrix, which was calculated using the “beta_div” command of the Usearch program. The obtained tree was annotated and visualized in R package using ggtree method30,31.
Network analysis
Co-occurrence networks were inferred based on a Spearman correlation matrix32 and constructed using only significant correlation33. The cutoff for correlation coefficients was determined to be 0.6 and the cutoff for adjusted p-values was 0.00134. Only OTUs, the relative abundance of which was at least 0.5% in at least one sample, were included in the analysis. Visualization of co-occurrence network was performed using Cytoscape v.3.8.2 platform35,36.
Data availability
The raw data generated from 16S rRNA gene sequencing have been deposited in the NCBI Sequence Read Archive (SRA) and are available via the BioProject PRJNA764866.
Results and discussion
Performance characteristics of WWTPs
Three main technologies are used in the investigated WWTPs. The simplest aeration process is implemented at WWTPs 1 and 7, where wastewater is supplied either at the beginning of the bioreactor corridor (CAS process, WWTP7), or along the entire aeration tank (SF process, WWTP1). The WWTP1 removed more than 98% of organics (according to the BOD data) and about 90% of ammonium, while the purification efficiency of WWTP7 was lower (95% removal of organics and 70% of ammonium) (Table 1). Interestingly, although these units were not designed to remove phosphorus, the WWTP7 removed more than 70% of phosphorus, while in WWTP1 its removal was inefficient. The nitrification/ denitrification technology, realized at WWTP2, enabled removal of more than 98% of organics and more than 99% of ammonium, while phosphorous was not removed (Table 1). Six WWTPs operated by the UCT technology enabled efficient purification of the wastewater from both organics (> 98%), ammonium (> 99%) and phosphorous (> 90%) (Table 1). The concentrations of N-NO3 in the effluent were much lower than N-NH4 in the influent indicating efficient denitrification in all WWTPs (Table 1).
Diversity of microbial communities of AS
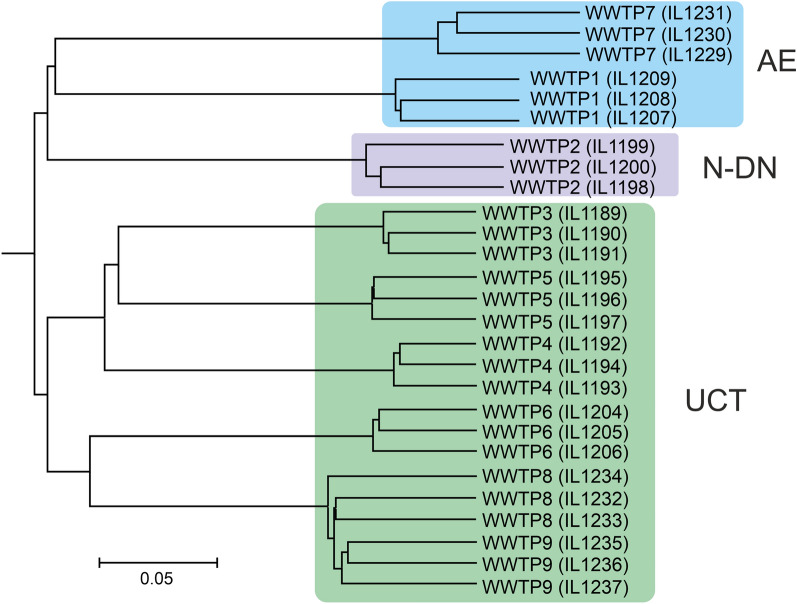
Between 23,167 and 84,634 16S rRNA gene sequences were obtained for 27 analysed AS samples (9 WWTPs, 3 replicas) and clustered into 14,690 OTUs at the level of 97% identity. Neighbour-joining trees based of the UniFrac analysis (Fig. 2) and Jaccard similarity (Supplementary Fig. S1) of OTU datasets revealed that replicate samples formed distinct branches for all WWTPs except for WWTP8 and WWTP9 which use identical bioreactors and treatment technologies. Therefore, for subsequent analysis, for each of the 9 WWTPs, three replicates were combined into one dataset. Both UniFrac and Jaccard trees revealed clustering of samples according to the technology used (Fig. 2 and Supplementary Fig. S2).
Figure 2.
Neighbor joining tree illustrating weighted UniFrac distances between microbial communities of AS samples from 9 WWTPs (three replications). Sample IDs are shown in brackets after the WWTP number.
The number of species-level OTUs present in AS at individual WWTPs ranged between 3860 and 4868, these values are typical for large-scale wastewater treatment plants10. Overall, the diversity of microbial communities did not significantly vary between WWTPs employing different technologies, while the evenness of microbial communities in plants operated by the UCT process was slightly lower than in the others (Supplementary Table S2).
Microbial community patterns at the phylum level
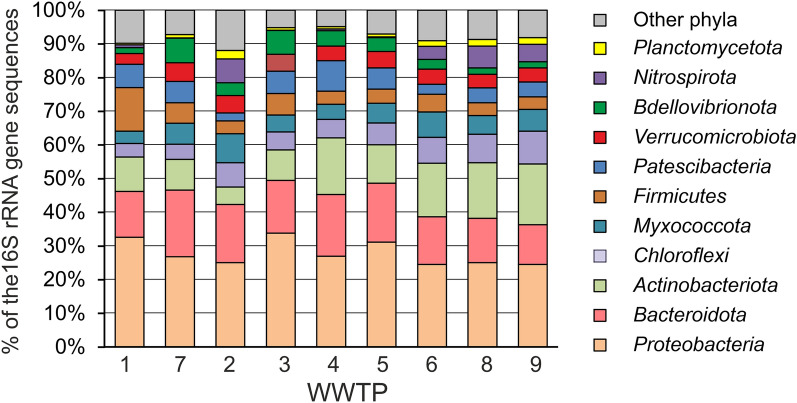
Taxonomic assignment of OTUs revealed the presence of 53 phylum-level lineages of Bacteria and Archaea, recognized in the Genome Taxonomy Database (GTDB)37. However, top 11 phyla comprising on average more than 1% of all the 16S rRNA gene sequences together accounted for more than 90% of the community (Fig. 3 and Supplementary Table S3).
Figure 3.
Bacterial and archaeal community composition in AS samples according to the results of 16S rRNA gene sequencing. The composition is displayed at the phylum level. Average values for three replicas are shown for each WWTP.
Archaea represented less than 2% of sequences in all samples and were assigned to the phyla Nanoarchaeota, Halobacterota, Euryarchaeota and Thermoplasmatota; each was detected in all samples. Besides members of the phylum Nanoarchaeota, known to comprise partner-dependent parasites or symbionts with small genome size and limited metabolic capacities38, most of other Archaea represented known methanogenic lineages of the families Methanobacteriaceae and Methanosaetaceae.
Bacterial communities were dominated by the Proteobacteria (on average 27.8% of 16S rRNA gene reads), mostly of classes gamma (23.5%) and alpha (4.3%). Other major groups were the Bacteroidota (15.7%), Actinobacteriota (12.5%), Chloroflexi (6.6%), Myxococcota (5.9%), Firmicutes (5.6%), Patescibacteria (5.5%), Verrucomicrobiota (4.5%), Bdellovibrionota (3.9%), Nitrospirota (2.7%), and Planctomycetota (1.3%). The relative abundances of these lineages in different samples differ by no more than several times, with the exception of Nitrospirota, the share of which is minimal (0.07%) in WWTP3 and maximal (7.1%) in WWTP2. All other bacterial phyla accounted for less than 2% of 16S rRNA gene reads in all samples except for the Campylobacterota representing about 3.5% of the community in WWTP1.
The main microbial drivers of wastewaters treatment
The biological wastewater treatment includes several microbial-driven processes, such as mineralization of organics by heterotrophs, oxidation of ammonia to nitrite and finally to nitrate, denitrificartion with the production of N2 gas, enhanced biological phosphorus removal etc. In this section we analyze the presence of the microbial groups which could be involved in these processes. The activities of microbial processes depend on the absolute concentrations of microorganisms, but given that the concentration of AS in all bioreactors was similar, we can consider the efficiency of processes in relation to the relative abundance of the corresponding functional group in the community. Data on the shares of the discussed groups of microorganisms are shown in Table 2.
Table 2.
Relative abundancies (% of total 16S rRNA gene sequences) of microbial genera involved in nitrogen and phosphorous removal.
| WWTP ID | WWTP1 | WWTP2 | WWTP3 | WWTP4 | WWTP5 | WWTP6 | WWTP7 | WWTP8 | WWTP9 |
|---|---|---|---|---|---|---|---|---|---|
| Technology | SF | N-DN | UCT | UCT | UCT | UCT | CAS | UCT | UCT |
| Ammonia oxidizers | |||||||||
| Nitrosomonas | 0.15 | 1.81 | 0.61 | 1.24 | 0.87 | 0.85 | 0.25 | 2.62 | 2.37 |
| Nitrosospira | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.02 | 0.02 |
| Nitrite oxidizers | |||||||||
| Nitrospira | 0.78 | 7.06 | 0.07 | 0.48 | 0.20 | 3.92 | 0.08 | 6.49 | 5.20 |
| Ca. Nitrotoga | 0.00 | 0.00 | 0.26 | 0.46 | 0.38 | 0.01 | 0.01 | 0.00 | 0.00 |
| PAO (including potential) | |||||||||
| Ca. Accumulibacter | 0.59 | 0.12 | 1.01 | 0.43 | 0.76 | 0.48 | 0.90 | 0.73 | 1.05 |
| Tetrasphaera | 1.35 | 0.36 | 1.32 | 0.80 | 1.08 | 0.42 | 0.81 | 0.19 | 0.23 |
| Zoogloea | 0.66 | 0.19 | 0.35 | 0.09 | 0.23 | 0.48 | 1.41 | 0.15 | 0.13 |
| Thiothrix | 0.93 | 0.04 | 0.20 | 0.03 | 0.01 | 0.03 | 1.22 | 0.00 | 0.01 |
| Dechloromonas | 5.03 | 0.58 | 5.84 | 3.07 | 3.85 | 1.45 | 1.48 | 1.60 | 1.87 |
| Ca. Obscuribacter | 0.37 | 0.88 | 0.11 | 0.56 | 0.17 | 1.85 | 0.00 | 0.07 | 0.04 |
| Gemmatimonas | 0.01 | 0.03 | 0.01 | 0.00 | 0.00 | 0.03 | 0.06 | 0.03 | 0.03 |
| Thauera | 0.98 | 1.42 | 0.24 | 0.44 | 0.25 | 1.74 | 0.69 | 2.24 | 1.35 |
| Ca. Microthrix | 3.92 | 1.23 | 2.65 | 11.80 | 3.10 | 11.10 | 3.37 | 11.00 | 12.81 |
| GAO | |||||||||
| Ca. Competibacter | 3.54 | 0.98 | 8.50 | 7.13 | 7.33 | 2.51 | 0.95 | 2.53 | 3.66 |
| Defluviicoccus | 0.05 | 0.02 | 0.03 | 0.06 | 0.05 | 0.03 | 0.02 | 0.04 | 0.05 |
Oxidation of ammonia and denitrification
Oxidation of ammonia to nitrate via nitrite is usually accomplished by two groups of microorganisms, although complete oxidation of ammonia to nitrate by Nitrospira sp. (comammox process) has been also reported39. Among known ammonia oxidizers (AOM) only members of the family Nitrosomonadaceae, mostly of the genus Nitrosomonas, were detected. The relative abundance of Nitrosomonadaceae clearly correlated with efficiency of ammonia removal: it ranged from 0.8% to 2.9% in all WWTPs enabling good removal of ammonia (> 99%) and was below 0.4% in WWTPs 1 and 7 where ammonia removal was less efficient.
Two genera of nitrite-oxidizing bacteria (NOB) were identified, Nitrospira and Candidatus Nitrotoga. The relative abundance of Nitrospirae sp. was the highest (7.06%) in WWTP2, which uses nitrification–denitrification process and much lower in WWTPs 1 and 7 (0.78% and 0.08%). WWTPs using the UCT process in terms of Nitrospirae content were clearly divided into two groups. AS samples of WWTPs 6, 8 and 9 harbored between 3.9% and 6.5% of Nitrospirae sp., while its relative abundance was less than 0.5% in WWTPs 3, 4 and 5. However, microbial communities of these three bioreactors contained 0.26 to 0.46% of Ca. Nitrotoga, nearly absent in other WWTPs. This recently described Ca. Nitrotoga species can be functionally and sometimes dominant important nitrite oxidizers in WWTPs due to their relatively higher resistance to free nitrous acid and free ammonia than other NOB and the presence of complete pathways for hydrogen and sulfite oxidation, suggesting that alternative energy metabolisms enable these bacteria to survive nitrite depletion40–42.
Regardless of the used purification technology and the efficiency of ammonia removal, the concentration of N-NO3 in the effluent was in the range from 7 to 10.5 mg/L, which corresponds to 15–20% of N–NH4 in the influent and indicates effective denitrification and removal of nitrogen in the gaseous form. The presence of numerous heterotrophic denitrifying bacterial is consistent with this observation.
Biological phosphorus removal
The next main issue of wastewater treatment is the removal of phosphorus. This process is carried out by microorganisms of the group of phosphate-accumulating organisms (PAO) capable of intracellular accumulation of polyphosphates under cyclic growth conditions, with alternated presence and absence of electron acceptors43–46.
Typical PAO, most often found in WWTPs, are members of the candidate species Candidatus Accumulibacter phosphatis (family Rhodocyclaceae, Gammaproteobacteria)47–50. During the anaerobic phase, Ca. Accumulibacter phosphatis takes up volatile fatty acids present in the wastewater and stores the carbon from these substrates intracellularly as polyhydroxyalkanoates51. At the same time, intracellular polyphosphate is degraded to form ATP, releasing phosphate into the medium. During the aerobic phase, stored polyhydroxyalkanoates are used for energy production while phosphate is taken up from the medium and accumulated as polyphosphate. Ca. Accumulibacter phosphatis were found in all AS samples. Its relative abundance was the lowest (0.12%) in WWTP2 employing the nitrification/ denitrification process and poorly removing phosphorus. In other plants Ca. Accumulibacter phosphatis accounted for 0.43 to 1.05% of the communities and its share does not correlate with the phosphorus removal efficiency or the type of treatment process.
Several other bacteria, despite the difference in their metabolism from Ca. Accumulibacter phosphatis, are considered as likely PAO46, including members of Proteobacteria (Dechloromonas52,53, Zooglea54,55, Thauera56, Thiothrix57, Ca. Accumulimonas58, Malikia59), Actinobacteria (Tetrasphaera60,61, Microlunatus62, Candidatus Microthrix63), Gemmatimonadetes (Gemmatimonas64) and Melainabacteria (Candidatus Obscuribacter phosphatis65). Among these bacteria, members of the genera Tetrasphaera, Zoogloea, Thiothrix, Dechloromonas, Ca. Obscuribacter, Gemmatimonas, and Ca. Microthrix were detected.
The most abundant potential PAO, Ca. Microthrix sp., (average share 6.8%) was presented in all WWTPs and its share was higher in WWTPs employing the UCT process than in other types of bioreactors. Earlier it has been shown that Candidatus Microthrix parvicella contained of large polyphosphate granules and this microbial group might have been responsible for phosphorus removal during the sludge bulking period when Ca. Accumulibacter phosphatis was excluded from the system63. Dechloromonas sp., denitrifying bacteria capable of acetate uptake and polyphosphate storage, accounted for 1.5% to 5.8% of AS microbiomes in bioreactors operating under aerobic and UCT processes, while in WWTP2 it accounted for only 0.6%. Members of the genera Zoogloea and Thiothrix, aerobic bacteria capable of denitrification, often occur in ASs57,66,67. Both genera were more abundant in WWTPs employing aerobic process. Ca. Obscuribacter was most frequent in the WWTP6 (about 1.9%) and accounted for less than 1% in other samples. Members of the genus Tetrasphaera, known to be capable of aerobic polyphosphate accumulation under condition of assimilating glucose and/or amino acids anaerobically in advance68 were found in all WWTPs. Their relative abundance was minimal in WWTPs 8 and 9 (about 0.2%) and reached 1.35% in WWTP1. Although nitrate-reducing bacteria of the genus Thauera are primary known for their ability to perform anaerobic degradation of aromatic and other refractory compounds69, recently Thauera sp. strain SND5 was found to be a phosphate-accumulating organism56. The relative abundance of Thauera sp. ranged from 0.2 to 2.2% and was maximal in WWTP8.
Glycogen accumulating organisms (GAOs) can accumulate glycogen and polyhydroxyalkanoates inside cells, but do not have ability to intracellular accumulation of polyphosphates. They are commonly found together with PAO in EBPR bioreactors where their predominance may lead to EBPR failure due to the competition for carbon source with PAO70. Known GAO belongs to the Gammaproteobacteria (Candidatus Competibacter phosphatis) and Alphaproteobacteria (Defluvicoccus sp.)71,72. Ca. Competibacter accounted for up to 8.5% of the communities in UCT WWTPs, but were less numerous (< 1%) in WWTPs 2 and 7. The relative abundance of Defluvicoccus sp. was less than 0.1% in all samples. Thus, in WWTPs using the UCT process, a high relative abundance of GAO did not adversely affected the efficiency of phosphorus removal.
Microbial community composition and efficiency of wastewater purification
All five WWTPs using the UCT process ensured effective removal of not only organic matter, but also nitrogen and phosphorus, which is consistent with the high content of nitrifying and phosphate accumulating bacteria.
WWTP2, using the nitrification–denitrification process, effectively removed nitrogen, and the relative abundance of nitrifiers in this AS community was the highest. However, there was almost no phosphorus removal, and the share of PAO was the lowest. The low efficiency of phosphorus removal and the low abundance of PAO in WWTP2 are associated with the absence of anaerobic zones in bioreactors, as well as the consumption of easily degradable organic matter by denitrifying microorganisms.
Contrary to expectations, the two WWTPs, using a simple aeration process, provided the poorest removal of both organics and ammonia, while the nitrate content in the effluent was approximately the same as in other installations. However, the total nitrogen content in the form of nitrate and ammonium in the effluent was much lower than in the influent, indicating that nitrogen was removed during denitrification. Probably, under conditions of insufficient aeration, formation of local anoxic zones within microbial flocs facilitates the denitrification performed by heterotrophic bacteria73, while part of the ammonia and organic matter remain non-oxidized and enters the effluent water.
Interestingly, relatively efficient phosphorus removal was observed in WWTP7 but not in WWTP1. These two bioreactors differ in that in WWTP7 the organic-rich influent is fed to the beginning of the bioreactor, while in WWTP1 it is distributed along the entire length. It is likely that in WWTP7 local organic-rich anaerobic zones are formed within AS flocks near the point of influent supply, while in WWTP1 the concentration of organic substances is everywhere lower. In such organic-rich zones denitrifying PAO of the genera Zoogloea and Thiothrix can proliferate and their shares were maximal in WWTP7.
Other abundant community members
Analysis of the compositions of microbial communities revealed 16 genus-level lineages, the average share of which across nine WWTPs exceeds 1% (Supplementary Table S4). In total, these 16 genera accounted for 28 to 51% (on average about 36%) of analyzed microbiomes. The most abundant genus, Ca. Microthrix, was the most numerous in AS samples of WWTPs 4, 6, 8 and 9 (11—13%) using the UCT process, while in other samples its relative abundance was much lower (1.2—3.4%). Besides its potential role in phosphorus removal, Ca. Microthrix is known as a slowly growing filamentous bacterium causing bulking and foaming in activated sludge systems thus seriously affecting the stable operation of WWTPs74. The growth of cultured species of this genus, M. parvicella, is susceptible to both environmental and operational parameters, such as temperature, oxygen concentration, type of substrate, electron acceptor, organic loading, and sludge retention time (reviewed in75). Probably, enrichment of Ca. Microthrix in the AS of the UCT WWTPs was associated with low oxygen concentrations in the anaerobic and anoxic zones of these bioreactors, favorable for the growth of these bacteria7. The dominant Ca. Microthrix phylotype was identified as Ca Microthrix subdominans, a species typically present along with Ca. M. parvicella, although usually in lower abundances76.
Besides Ca. Microthrix and above mentioned Ca. Competibacter, Dechloromonas, Nitrospira, Nitrosomonas and Thauera, the list of most abundant genera includes 3 cultured (Trichococcus, Haliangium, and Chitinivorax) and 7 uncultured lineages, defined in the Silva database as OM27_clade (Bdellovibrionota, Bdellovibrionaceae), Candidatus Nomurabacteria (Patescibacteria), AKYH767 (Bacteroidota, Sphingobacteriales), a member of the family Moraxellaceae (Gammaproteobacteria) defined as “Agitococcus lubricus group’, mle1-27 (Myxococcota), env.OPS_17 (Bacteroidota, Sphingobacteriales), and OLB12 (Bacteroidota, Microscillaceae).
Members of the genera Trichococcus, Haliangium, and Chitinivorax are often found in AS of wastewater treatment facilities. Trichococcus sp. are filamentous bacteria that can degrade a wide range of carbohydrates77, while Chitinivorax sp. are devoted to the hydrolysis of chitin78. Similarly to Dechloromonas sp., members of the Haliangium are active denitrifiers in the AS communities79.
All uncultured genus-level lineages were previously detected in bioreactors treating wastewater80–83, but their functional roles remain elusive. The most abundant uncultured OM27_clade, with an average share of about 3%, was hypothesized to be predatory bacteria84. Members of the candidate genus OLB12 are aerobic heterotrophs abundant in the anammox granules collected from partial-nitritation anammox reactor85. The only cultured member of “Agitococcus lubricus group’, A. lubricus, is a lipolytic aerobic heterotroph found in freshwater bodies86. The relative abundance of this lineage reached 4% in WWTP2, while it was below 0.2% in all AS samples currently described in the MiDAS database14.
Network analysis
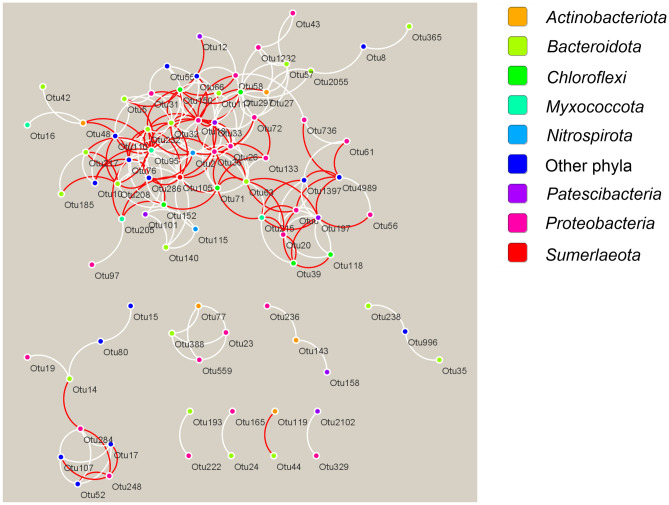
Various functional characteristics of microorganisms are closely related to the differentiation of ecological niches, and network analysis allows one to deduce patterns of interactions that develop within the WWTP ecosystem. Network analysis indicated possible cooperation (co-occurrence) and mutual exclusion among diverse microorganisms in WWTPs (Fig. 4).
Figure 4.
Network of co-occurring abundant microbial OTUs of AS based on correlation analysis. A connection stands for a strong (Spearman’s rho > 0.6) and significant (adjusted p value < 0.001) correlation. OTUs are colored according to phylum. Co-presence and mutual exclusion of OTUs are shown in white and red lines, respectively.
Of 104 OTUs, with abundances more than 0.5% in at least one of the AS samples, 83 had statistically significant connections with other OTUs the maximum number of which reached 18 (Supplementary Table S5). OTUs with a high number of connections (“degree”) are considered as hub members, supposed to be key players in sustaining the overall community87. The largest number of connections (18) was observed for OTU assigned to the genus Haliangium, whose members, together with Dechloromonas sp., are active denitrifiers in WWTPs communities79. Notably, bacteria participating in ammonia and phosphorous removal were among the hub members, namely, Nitrosomonas, Nitrospira, Ca. Competibacter, Dechloromonas, and Thauera. Positive connection was detected between denitrifiers (Thauera Otu26 and Haliangium Otu95) and bacteria involved in the oxidation of ammonia (Nitrosomonas Otu18 and Nitrospira Otu2). This link is consistent with the fact that the source of nitrate is mainly microbial oxidation of ammonium, rather than influent sewage water. Interestingly, we did not detected mutual exclusion between Ca. Accumulibacter and Ca. Competibacter.
The nest largest number of connections was detected for Otu105, assigned to the candidate genus Sumerlaea (candidate phylum Sumerlaeota, previously known as BRC1). Members of this genus were predicted to be metabolically versatile bacteria capable to utilize various carbohydrates, including chitin, performing fermentation as well as complete oxidation of organic substrates through aerobic and anaerobic (with nitrate and sulfur compounds) respiration88. Co-presence of Sumerlaea and denitrifiers (Thauera and Haliangium) suggest that Sumerlaea sp. could be involved in degradation of organic substrates linked to dissimilatory nitrate reduction. Interestingly, no connection to other community members was found for the most abundant species, Ca. Microthrix (Otu1). Ca. Microthrix has been considered a specialized consumer of long-chain fatty acids that have been confirmed to be the key carbon and energy sources for its growth both under aerobic, anoxic and anaerobic conditions75,89. Probably due to such metabolic specialization Ca. Microthrix has limited interactions with other community members.
Microbial communities of Moscow WWTPs on a global scale
Variations in community composition are key points for understanding community functioning. To understand how the composition of microbial communities of AS varied across different geographic locations, we compared taxonomic structures of our microbiomes with that reported in a worldwide study performed by a Global Water Microbiome Consortium10. That study showed that although the taxonomic community structures observed on different continents were not clearly separated at the OTU level in two dimensional ordinations, it was significantly different between any two continents10, and although the type of treatment process exerted significant effects on microbial community structures, it was overwhelmed by continental geographical separation10.
Despite its high diversity, AS microbiome has a small global core bacterial community (28 OTUs) that is strongly linked to WWTP performance and accounted for an average 12.4% of the 16S rRNA gene sequences in AS samples10. 26 of these OTUs were present in Moscow WWTPs and accumulated from 11.3 to 23.5% of 16S rRNA gene reads (Supplementary Table S6).
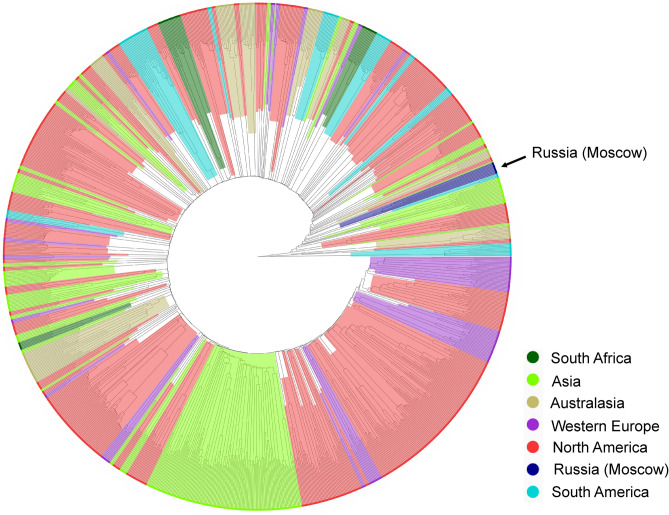
Clustering based on the Bray–Curtis dissimilarity showed that all samples from Moscow WWTPs clustered together and formed a distinct branch not embedded into European or any other large groups, but forming a sister lineage to a subsets of Asian, South American and North American clusters (Fig. 5). These data indicate that influent characteristics, related to cultural, social and environmental factors in each region, could be more important than a plant operating conditions. Similar observations have been previously reported for the microbial communities of AS from WWTPs in Korea and Vietnam2. For example, due to the relatively low cost of water for household consumption, wastewater in Moscow has a relatively low content of organic matter compared to the values typical for most large cities (COD of 600–900 mg/L). It should be noted that the primers used in our work for the 16S rRNA gene profiling differed from those used by the GWMC consortium (515F/806R), which may affect the observed shares of particular microbial taxa. However, both primer pairs (515F/806R and 341F/806R) enabled good coverage of overall microbial diversity and provided similar estimates of the relative abundancies of most species in various environments90,91.
Figure 5.
Neighbor joining tree illustrating clustering based on the Bray–Curtis dissimilarity between microbial communities of AS samples from 9 WWTPs obtained in this study and ones reported by a Global Water Microbiome Consortium (http://gwmc.ou.edu/). Samples are colored according to geographic location.
Conclusions
This study provides the data on the composition of microbial communities of AS from large-scale WWTPs of the Moscow city and the evidences of the impact of purification technology. Although the taxonomic structure of communities at high ranks was similar and the most numerous groups were found in all WWTPs, UniFrac and Jaccard trees revealed clustering of samples according to the employed technology. WWTPs employing the UCT process enabled efficient removal of organics, nitrogen and phosphorus, while WWTPs operated under simple aeration or nitrification–denitrification process removed these components less efficiently. Co-occurrence network analysis provided information on key hub microorganisms in AS, which may be targeted for manipulating the AS stability and performance. Comparison of AS communities from Moscow WWTPs with ones analyzed worldwide by GWMC revealed that all Moscow samples clustered together, indicating that influent characteristics, related to cultural, social and environmental factors in each region, could be more important than a plant operating conditions.
Supplementary Information
Acknowledgements
This work was partly supported by the Russian Science Foundation (Project 21-64-00019 to A.V.M.).
Author contributions
A.V.M. and N.V.R. designed and supervised the research project; A.G.D. collected the samples and analysed chemical composition of wastewater; V.V.K. and A.V.M. performed 16S rRNA gene profiling; S.B., A.V.B., N.V.P., and N.V.R. analysed the sequencing data; S.B. and N.V.R. wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Nikolai V. Ravin, Email: nravin@biengi.ac.ru
Andrey V. Mardanov, Email: mardanov@biengi.ac.ru
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-07132-4.
References
- 1.Ahkola H, et al. A preliminary study on the ecotoxic potency of wastewater treatment plant sludge combining passive sampling and bioassays. Sci. Total. Environ. 2021;758:143700. doi: 10.1016/j.scitotenv.2020.143700. [DOI] [PubMed] [Google Scholar]
- 2.Kim YK, et al. The capacity of wastewater treatment plants drives bacterial community structure and its assembly. Sci. Rep. 2019;9:14809. doi: 10.1038/s41598-019-50952-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tchobanoglous, G., Burton, F.L. & Stensel, H.D. Wastewater Engineering: Treatment and Reuse. 1819 (McGraw-Hill Education, 2003).
- 4.Wagner M, Loy A. Bacterial community composition and function in sewage treatment systems. Curr. Opin. Biotechnol. 2002;3:218–227. doi: 10.1016/s0958-1669(02)00315-4. [DOI] [PubMed] [Google Scholar]
- 5.Ibarbalz FM, Figuerola EL, Erijman L. Industrial activated sludge exhibit unique bacterial community composition at high taxonomic ranks. Water Res. 2013;11:3854–3864. doi: 10.1016/j.watres.2013.04.010. [DOI] [PubMed] [Google Scholar]
- 6.Saunders AM, Albertsen M, Vollertsen J, Nielsen PH. The activated sludge ecosystem contains a core community of abundant organisms. ISME J. 2015;10:11–20. doi: 10.1038/ismej.2015.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jenkins, D., Richard, M.G., & Daigger, G.T. (ed. Jenkins, D.). Manual on the causes and control of activated sludge bulking, foaming, and other solids separation problems. 224 (CRC Press, 2003).
- 8.Henze, M., Harremoës, P., Jansen, L. C. & Arvin E. (eds. Henze, M., Harremoës, P., Jansen, L. C. & Arvin E) Wastewater treatment. Biological and chemical processes. 383 (Springer, 1997).
- 9.Zhang T, Shao MF, Ye L. 454 Pyrosequencing reveals bacterial diversity of activated sludge from 14 sewage treatment plants. ISME J. 2012;6:1137–1147. doi: 10.1038/ismej.2011.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu L, et al. Global diversity and biogeography of bacterial communities in wastewater treatment plants. Nat. Microbiol. 2019;7:1183–1195. doi: 10.1038/s41564-019-0426-5. [DOI] [PubMed] [Google Scholar]
- 11.Seviour, R.J. & Nielsen, P.H. (eds. Seviour, R.J. & Nielsen, P.H.) Microbial Ecology of Activated Sludge. 688 (IWA Publishing, 2010).
- 12.Nielsen, P. H., Daims, H., & Lemmer, H. (eds. Nielsen, P. H., Daims, H., & Lemmer, H.) FISH Handbook for Biological Wastewater Treatment. 200 (IWA Publishing, 2009).
- 13.Nierychlo M, et al. MiDAS 3: an ecosystem-specific reference database, taxonomy and knowledge platform for activated sludge and anaerobic digesters reveals species-level microbiome composition of activated sludge. Water Res. 2020;182:115955. doi: 10.1016/j.watres.2020.115955. [DOI] [PubMed] [Google Scholar]
- 14.Dueholm MS, et al. MiDAS 4: A global catalogue of full-length 16S rRNA gene sequences and taxonomy for studies of bacterial communities in wastewater treatment plants. bioRxiv. 2021 doi: 10.1101/2021.07.06.451231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kallistova AIu, et al. Microbial composition of the activated sludges of the Moscow wastewater treatment plants. Microbiology. 2014;83:699–708. [PubMed] [Google Scholar]
- 16.Shchegolkova NM, et al. Microbial community structure of activated sludge in treatment plants with different wastewater compositions. Front. Microbiol. 2016;7:90. doi: 10.3389/fmicb.2016.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mardanov AV, et al. Metagenome of the microbial community of anammox granules in a nitritation/anammox wastewater treatment system. Genome Announc. 2017;42:e01115–e1117. doi: 10.1128/genomeA.01115-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mardanov AV, et al. Dynamics of the composition of a microbial consortium during start-up of a single-stage constant flow laboratory nitritation/anammox setup. Microbiology. 2016;85:681–692. [Google Scholar]
- 19.Nikolaev YuA, et al. Novel design and optimisation of a nitritation/anammox set-up for ammonium removal from filtrate of digested sludge. Environ. Technol. 2018;5:593–606. doi: 10.1080/09593330.2017.1308442. [DOI] [PubMed] [Google Scholar]
- 20.Mardanov AV, et al. Genome of a novel bacterium “Candidatus Jettenia ecosi” reconstructed from the metagenome of an anammox bioreactor. Front. Microbiol. 2019;10:2442. doi: 10.3389/fmicb.2019.02442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Danilovich DA. Blok udaleniya biogennyh elementov Lyubereckih ochistnyh sooruzhenij g. Moskvy–etapy vnedreniya sovremennyh tekhnologij. Nailuchshie dostupnye tekhnologii vodosnabzheniya i vodootvedeniya. 2014;2:20–37. [Google Scholar]
- 22.Danilovich DA, Kozlov MN. 25 let promyshlennogo vnedreniya tekhnologij udaleniya azota i fosfora na moskovskih ochistnyh sooruzheniyah: 20 aprobirovannyh tekhnologicheskih reshenij. Nailuchshie dostupnye tekhnologii vodosnabzheniya i vodootvedeniya. 2019;1:42–55. [Google Scholar]
- 23.Kevbrina MV, et al. Anammoks–perspektivnaya tekhnologiya udaleniya azota iz stochnyh vod. Vodosnabzhenie i sanitarnaya tekhnika. 2019;5:28–35. [Google Scholar]
- 24.Kevbrina MV, Gavrilin AM, Dorofeev AG, Kozlov MN, Aseeva VG. Nailuchshie dostupnye tekhnologii ochistki stochnyh vod: opyt vnedreniya AO «Mosvodokanal». Vodosnabzhenie i sanitarnaya tekhnika. 2019;6:40–49. [Google Scholar]
- 25.Frey, B. et al. Microbial diversity in European alpine permafrost and active layers. FEMS Microbiol Ecol. 3, fiw018 (2016). [DOI] [PubMed]
- 26.Magoc T, Salzberg S. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 28.Rognes T, Flouri T, Nichols B, Quince C, Mahé F. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4:e2584. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caporaso J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu G, Lam TTY, Zhu H, Guan Y. Two methods for mapping and visualizing associated data on phylogeny using ggtree. Mol. Biol. Evol. 2018;2:3041–3043. doi: 10.1093/molbev/msy194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu G, Smith DK, Zhu H, Guan Y, Lam TTY. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 2017;1:28–36. doi: 10.1111/2041-210X.12628. [DOI] [Google Scholar]
- 32.Langfelder P, Horvath S. Fast R functions for robust correlations and hierarchical clustering. J. Stat. Softw. 2012;46:i11. [PMC free article] [PubMed] [Google Scholar]
- 33.Barberan A, et al. Why are some microbes more ubiquitous than others? Predicting the habitat breadth of soil bacteria. Ecol. Lett. 2014;17:794–802. doi: 10.1111/ele.12282. [DOI] [PubMed] [Google Scholar]
- 34.Luo X, Bhattacharya CB. Corporate social responsibility, customer satisfaction, and market value. J. Mark. 2006;70:1–18. [Google Scholar]
- 35.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;11:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Faust K, Raes J. CoNet app: inference of biological association networks using Cytoscape. F1000Res. 2016;5:1519. doi: 10.12688/f1000research.9050.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parks DH, et al. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 2018;36:996–1004. doi: 10.1038/nbt.4229. [DOI] [PubMed] [Google Scholar]
- 38.Dombrowski N, Lee JH, Williams TA, Offre P, Spang A. Genomic diversity, lifestyles and evolutionary origins of DPANN archaea. FEMS Microbiol Lett. 2019;366:fnz008. doi: 10.1093/femsle/fnz008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Kessel MA, et al. Complete nitrification by a single microorganism. Nature. 2015;528:555–559. doi: 10.1038/nature16459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lücker S, et al. Nitrotoga-like bacteria are previously unrecognized key nitrite oxidizers in full-scale wastewater treatment plants. ISME J. 2015;9:708–720. doi: 10.1038/ismej.2014.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kitzinger K, et al. Characterization of the first “Candidatus nitrotoga” isolate reveals metabolic versatility and separate evolution of widespread nitrite-oxidizing bacteria. mBio. 2018;9:e01186-18. doi: 10.1128/mBio.01186-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng M, et al. Critical factors facilitating Candidatus nitrotoga to be prevalent nitrite-oxidizing bacteria in activated sludge. Environ Sci Technol. 2020;54:15414–15423. doi: 10.1021/acs.est.0c04192. [DOI] [PubMed] [Google Scholar]
- 43.Van Loosdrecht MCM, Hooijmans CM, Brdjanovic D, Heijnen JJ. Biological phosphate removal processes. Appl. Microbiol. Biotechnol. 1997;48:289–296. [Google Scholar]
- 44.Seviour RJ, Mino T, Onuki M. The microbiology of biological phosphorus removal in activated sludge systems. FEMS Microbiol. Rev. 2003;27:99–127. doi: 10.1016/S0168-6445(03)00021-4. [DOI] [PubMed] [Google Scholar]
- 45.Dorofeev AG, Nikolaev YA, Mardanov AV, Pimenov NV. Cyclic metabolism as a mode of microbial existence. Microbiology. 2019;88:402–415. [Google Scholar]
- 46.Akbari A, et al. Unrevealed roles of polyphosphate-accumulating microorganisms. Microb. Biotechnol. 2021;14:82–87. doi: 10.1111/1751-7915.13730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hesselmann RPX, Werlen C, Hahn D, van der Meer JR, Zehnder AJB. Enrichment, phylogenetic analysis and detection of a bacterium that performs enhanced biological phosphate removal in activated sludge. Syst. Appl. Microbiol. 1999;22:454–465. doi: 10.1016/S0723-2020(99)80055-1. [DOI] [PubMed] [Google Scholar]
- 48.Stockholm-Bjerregaard M, et al. Critical assessment of the microorganisms proposed to be important to enhanced biological phosphorus removal in full-scale wastewater treatment systems. Front. Microbiol. 2017;8:718. doi: 10.3389/fmicb.2017.00718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zeng W, Bai X, Guo Y, Li N, Peng Y. Interaction of “Candidatus Accumulibacter” and nitrifying bacteria to achieve energy-efficient denitrifying phosphorus removal via nitrite pathway from sewage. Enzyme. Microb. Technol. 2017;105:1–8. doi: 10.1016/j.enzmictec.2017.06.005. [DOI] [PubMed] [Google Scholar]
- 50.Qiu G, et al. Polyphosphate-accumulating organisms in full-scale tropical wastewater treatment plants use diverse carbon sources. Water Res. 2019;149:496–510. doi: 10.1016/j.watres.2018.11.011. [DOI] [PubMed] [Google Scholar]
- 51.Mino T, van Loosdrecht MCM, Heijnen JJ. Microbiology and biochemistry of the enhanced biological phosphate removal process. Water Res. 1998;32:3193–3207. [Google Scholar]
- 52.Kong Y, Xia Y, Nielsen JL, Nielsen PH. Structure and function of the microbial community in a full-scale enhanced biological phosphorus removal plant. Microbiology (SGM). 2007;153:4061–4073. doi: 10.1099/mic.0.2007/007245-0. [DOI] [PubMed] [Google Scholar]
- 53.Terashima M, et al. Culture-dependent and -independent identification of polyphosphate-accumulating Dechloromonas spp. Predominating in a full-scale oxidation ditch wastewater treatment plant. Microbes. Environ. 2016;31:449–455. doi: 10.1264/jsme2.ME16097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shao Y, et al. Zoogloea caeni sp. nov., a floc-forming bacterium isolated from activated sludge. Int. J. Syst. Evol. Microbiol. 2009;59:526–30. doi: 10.1099/ijs.0.65670-0. [DOI] [PubMed] [Google Scholar]
- 55.Pelevina AV, et al. A microbial consortium removing phosphates under conditions of cyclic aerobic-anaerobic cultivation. Microbiology. 2021;90:66–77. [Google Scholar]
- 56.Wang Q, He J. Complete nitrogen removal via simultaneous nitrification and denitrification by a novel phosphate accumulating Thauera sp. strain SND5. Water Res. 2020;185:116300. doi: 10.1016/j.watres.2020.116300. [DOI] [PubMed] [Google Scholar]
- 57.Rubio-Rincón FJ, et al. Long-term effects of sulphide on the enhanced biological removal of phosphorus: The symbiotic role of Thiothrix caldifontis. Water Res. 2017;116:53–64. doi: 10.1016/j.watres.2017.03.017. [DOI] [PubMed] [Google Scholar]
- 58.Nguyen HT, Nielsen JL, Nielsen PH. 'Candidatus Halomonas phosphatis', a novel polyphosphate-accumulating organism in full-scale enhanced biological phosphorus removal plants. Environ Microbiol. 2012;10:2826–2837. doi: 10.1111/j.1462-2920.2012.02826.x. [DOI] [PubMed] [Google Scholar]
- 59.Spring S, Wagner M, Schumann P, Kampfer P. Malikia granosa gen. nov., sp. nov., a novel polyhydroxyalkanoate- and polyphosphate-accumulating bacterium isolated from activated sludge, and reclassification of Pseudomonas spinosa as Malikia spinosa comb. nov. Int. J. Syst. Evol. Microbiol. 2005;55:621–629. doi: 10.1099/ijs.0.63356-0. [DOI] [PubMed] [Google Scholar]
- 60.Maszenan AM, et al. Three isolates of novel polyphosphate-accumulating Gram-positive cocci, obtained from activated sludge, belong to a new genus, Tetrasphaera gen. nov., and description of two new species, Tetrasphaera japonica sp. nov. and Tetrasphaera australiensis sp. nov. Int. J. Syst. Evol. Microbiol. 2000;50:593–603. doi: 10.1099/00207713-50-2-593. [DOI] [PubMed] [Google Scholar]
- 61.Nielsen JL, Nguyen H, Meyer RL, Nielsen PH. Identification of glucose-fermenting bacteria in a full-scale enhanced biological phosphorus removal plant by stable isotope probing. Microbiology (SGM). 2012;158:1818–1825. doi: 10.1099/mic.0.058818-0. [DOI] [PubMed] [Google Scholar]
- 62.Nakamura K, et al. Microlunatus phosphovorus gen. nov., sp. nov., a new gram-positive polyphosphate-accumulating bacterium isolated from activated sludge. Int. J. Syst. Bacteriol. 1995;45:17–22. doi: 10.1099/00207713-45-1-17. [DOI] [PubMed] [Google Scholar]
- 63.Wang J, et al. The potential role of 'Candidatus Microthrix parvicella' in phosphorus removal during sludge bulking in two full-scale enhanced biological phosphorus removal plants. Water Sci. Technol. 2014;70:367–375. doi: 10.2166/wst.2014.216. [DOI] [PubMed] [Google Scholar]
- 64.Zhang H, et al. Gemmatimonas aurantiaca gen. nov., sp. nov., a Gram-negative, aerobic, polyphosphate-accumulating microorganism, the first cultured representative of the new bacterial phylum Gemmatimonadetes phyl. nov. Int. J. Syst. Evol. Microbiol. 2003;53:1155–1163. doi: 10.1099/ijs.0.02520-0. [DOI] [PubMed] [Google Scholar]
- 65.Soo RM, et al. An expanded genomic representation of the phylum Cyanobacteria. Genome Biol. Evol. 2014;6:1031–1045. doi: 10.1093/gbe/evu073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen H, Wang M, Chang S. Disentangling community structure of ecological system in activated sludge: core communities, functionality, and functional redundancy. Microb. Ecol. 2020;80:296–308. doi: 10.1007/s00248-020-01492-y. [DOI] [PubMed] [Google Scholar]
- 67.Mardanov AV, et al. Genomic and metabolic insights into two novel Thiothrix species from enhanced biological phosphorus removal systems. Microorganisms. 2020;8(12):2030. doi: 10.3390/microorganisms8122030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu R, Hao X, Chen Q, Li J. Research advances of Tetrasphaera in enhanced biological phosphorus removal: a review. Water Res. 2019;166:115003. doi: 10.1016/j.watres.2019.115003. [DOI] [PubMed] [Google Scholar]
- 69.Rabus R. Functional genomics of an anaerobic aromatic-degrading denitrifying bacterium, strain EbN1. Appl. Microbiol. Biotechnol. 2005;68:580–587. doi: 10.1007/s00253-005-0030-x. [DOI] [PubMed] [Google Scholar]
- 70.Oehmen A, et al. Advances in enhanced biological phosphorus removal: from micro to macro scale. Water Res. 2007;41:2271–2300. doi: 10.1016/j.watres.2007.02.030. [DOI] [PubMed] [Google Scholar]
- 71.Crocetti GR, Banfield JF, Keller J, Bond PL, Blackall LL. Glycogen-accumulating organisms in laboratory-scale and full-scale wastewater treatment processes. Microbiology (SGM). 2002;148:3353–3364. doi: 10.1099/00221287-148-11-3353. [DOI] [PubMed] [Google Scholar]
- 72.Wong MT, Tan FM, Ng WJ, Liu WT. Identification and occurrence of tetrad-forming Alphaproteobacteria in anaerobic-aerobic activated sludge processes. Microbiology. 2004;150:3741–3748. doi: 10.1099/mic.0.27291-0. [DOI] [PubMed] [Google Scholar]
- 73.Schramm A, et al. On the occurrence of anoxic microniches, denitrification, and sulfate reduction in aerated activated sludge. Appl. Environ. Microbiol. 1999;65:4189–4196. doi: 10.1128/aem.65.9.4189-4196.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rossetti S, Tomei MC, Nielsen PH, Tandoi V. "Microthrix parvicella", a filamentous bacterium causing bulking and foaming in activated sludge systems: a review of current knowledge. FEMS Microbiol. Rev. 2005;29:49–64. doi: 10.1016/j.femsre.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 75.Fan NS, Qi R, Huang BC, Jin RC, Yang M. Factors influencing Candidatus Microthrix parvicella growth and specific filamentous bulking control: a review. Chemosphere. 2020;244:125371. doi: 10.1016/j.chemosphere.2019.125371. [DOI] [PubMed] [Google Scholar]
- 76.Nierychlo M, et al. Low global diversity of Candidatus Microthrix, a troublesome filamentous organism in full-scale WWTPs. Front. Microbiol. 2021;12:690251. doi: 10.3389/fmicb.2021.690251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Strepis N, et al. Genome-guided analysis allows the identification of novel physiological traits in Trichococcus species. BMC Genom. 2020;21:24. doi: 10.1186/s12864-019-6410-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen WM, Yang SH, Huang WC, Cheng CY, Sheu SY. Chitinivorax tropicus gen. nov., sp. nov., a chitinolytic bacterium isolated from a freshwater lake. Int. J. Syst. Evol. Microbiol. 2012;62:1086–1091. doi: 10.1099/ijs.0.031310-0. [DOI] [PubMed] [Google Scholar]
- 79.McIlroy SJ, et al. Identification of active denitrifiers in full-scale nutrient removal wastewater treatment systems. Environ. Microbiol. 2016;18:50–64. doi: 10.1111/1462-2920.12614. [DOI] [PubMed] [Google Scholar]
- 80.Lu HP, Shao YH, Wu JH, Hsieh CH. System performance corresponding to bacterial community succession after a disturbance in an autotrophic nitrogen removal bioreactor. mSystems. 2020;5:e00398-20. doi: 10.1128/mSystems.00398-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Iannacone F, Di CF, Granata F, Gargano R, Esposito G. Simultaneous nitrification, denitrification and phosphorus removal in a continuous-flow moving bed biofilm reactor alternating microaerobic and aerobic conditions. Bioresour. Technol. 2020;310:123453. doi: 10.1016/j.biortech.2020.123453. [DOI] [PubMed] [Google Scholar]
- 82.LaPara TM, Nakatsu CH, Pantea L, Alleman JE. Phylogenetic analysis of bacterial communities in mesophilic and thermophilic bioreactors treating pharmaceutical wastewater. Appl. Environ. Microbiol. 2000;66:3951–3959. doi: 10.1128/aem.66.9.3951-3959.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Huang X, et al. Sludge alkaline fermentation enhanced anaerobic- multistage anaerobic/oxic (A-MAO) process to treat low C/N municipal wastewater: nutrients removal and microbial metabolic characteristics. Bioresour. Technol. 2020;302:122583. doi: 10.1016/j.biortech.2019.122583. [DOI] [PubMed] [Google Scholar]
- 84.Orsi WD, et al. Diverse, uncultivated bacteria and archaea underlying the cycling of dissolved protein in the ocean. ISME J. 2016;10:2158–2173. doi: 10.1038/ismej.2016.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Speth DR, In ‘t Zandt MH, Guerrero-Cruz S, Dutilh BE, Jetten MS. Genome-based microbial ecology of anammox granules in a full-scale wastewater treatment system. Nat. Commun. 2016;7:11172. doi: 10.1038/ncomms11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Franzmann PD, Skerman VBD. Agitococcus lubricus gen. nov., sp. nov., a lipolytic, twitching coccus from freshwater. Int. J. Syst. Bacteriol. 1981;31:177–183. [Google Scholar]
- 87.Berry D, Widder S. Deciphering microbial interactions and detecting keystone species with co-occurrence networks. Front. Microbiol. 2014;5:219. doi: 10.3389/fmicb.2014.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kadnikov VV, et al. Phylogeny and physiology of candidate phylum BRC1 inferred from the first complete metagenome-assembled genome obtained from deep subsurface aquifer. Syst. Appl. Microbiol. 2019;42:67–76. doi: 10.1016/j.syapm.2018.08.013. [DOI] [PubMed] [Google Scholar]
- 89.Nielsen PH, Roslev P, Dueholm TE, Nielsen JL. Microthrix parvicella, a specialized lipid consumer in anaerobic-aerobic activated sludge plants. Water Sci. Technol. 2002;46:73–80. [PubMed] [Google Scholar]
- 90.Klindworth A, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2012;41:e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wasimuddin, et al. Evaluation of primer pairs for microbiome profiling from soils to humans within the One Health framework. Mol. Ecol. Resour. 2020;6:1558–1571. doi: 10.1111/1755-0998.13215. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data generated from 16S rRNA gene sequencing have been deposited in the NCBI Sequence Read Archive (SRA) and are available via the BioProject PRJNA764866.