Abstract
Bark protects the tree against environmental insults. Here, we analyzed whether this defensive strategy could be utilized to broadly enhance protection against colitis. As a proof of concept, we show that exosome‐like nanoparticles (MBELNs) derived from edible mulberry bark confer protection against colitis in a mouse model by promoting heat shock protein family A (Hsp70) member 8 (HSPA8)‐mediated activation of the AhR signaling pathway. Activation of this pathway in intestinal epithelial cells leads to the induction of COP9 Constitutive Photomorphogenic Homolog Subunit 8 (COPS8). Utilizing a gut epithelium‐specific knockout of COPS8, we demonstrate that COPS8 acts downstream of the AhR pathway and is required for the protective effect of MBELNs by inducing an array of anti‐microbial peptides. Our results indicate that MBELNs represent an undescribed mode of inter‐kingdom communication in the mammalian intestine through an AhR‐COPS8‐mediated anti‐inflammatory pathway. These data suggest that inflammatory pathways in a microbiota‐enriched intestinal environment are regulated by COPS8 and that edible plant‐derived ELNs may hold the potential as new agents for the prevention and treatment of gut‐related inflammatory disease.
Keywords: AhR, anti‐microbial peptides, COP9/COPS8, heat shock protein family A (Hsp70) member 8, inflammation, Mulberry bark derived exosome‐like nanoparticles
Subject Categories: Immunology; Microbiology, Virology & Host Pathogen Interaction; Signal Transduction
Mulberry bark derived exosome‐like nanoparticles (MBELNs) prevent gut inflammation via plant heat shock protein HSPA8‐mediated activation of AhR/COPS8 pathways. Treatment with MBELNs promotes the restoration of gut microbiome homeostasis, ameliorating intestinal inflammatory pathologies.
Introduction
Developing a variety of avenues to deal with or to adapt to stresses such as changing environment, climate, lifestyle, or social structure is central to maintaining human health. Deficiency in or loss of strategies to deal with new or previous unmet changes or stresses can result in disease. For plants to remain healthy, they must develop more comprehensive strategies to deal with a higher frequency of changes and a more diversified variety of stresses in order to survive. Plant bark is considered an accumulation of several different outer layers of a woody plant and constitutes 10–20% of the weight of most woody plants (Loub et al, 1975; Vane et al, 2006) that protects the plant against numerous insults coming from desiccation, agents causing disease, and extreme temperatures. Whether these features can be utilized for improving human healthy has not been studied in detail.
Recently, we (Mu et al, 2014) and others (Xiao et al, 2018) have identified exosome‐like nanoparticles (ELNs) from tissues of edible plants. ELNs are naturally released, and they are taken up by host cells and subsequently communicate with ELN recipient cells, which is a biological hallmark of mammalian cell‐derived exosomes. Unlike animal exosomes which are difficult to produce in large quantities, ELNs can easily be isolated and purified in large quantities. Therefore, in this study, we sought to identify the therapeutic factor(s) in ELNs, which may provide ELN recipient cells a strategy to deal with stress analogous to plant cells.
Mulberry, a deciduous plant from the genus Morus (family Moraceae) that includes several species (most commonly Morus notabillis, Morus alba, and Morus rubra) is grown for several beneficial roles including fruit production, leaf production for silkworm feeding (Sanchez, 1999), its clinical efficacy in glucose metabolism (Asai et al, 2011), against untreated type 2 diabetes (Nakamura et al, 2011), and mild dyslipidemia (Aramwit et al, 2013), to yield bark for paper production and because of its multiple usage in traditional medicines (Asano et al, 1994; Kim et al, 1999; Chan et al, 2016). We selected the plant Morus notabillis for this study because of its widespread presence, therapeutic application, and the availability of the complete genome sequence (He et al, 2013).
The aryl hydrocarbon receptor (AhR)‐mediated signaling pathway plays a role in response to stress and a number of chemicals isolated from plants (Rothhammer & Quintana, 2019). There are a number of natural ligands from plant sources that have been shown to be effective in neurological disease, inhibiting tumor growth, lowering cholesterol, and having positive effects in other chronic illnesses (Hirano et al, 1995; Diplock et al, 1998; Datla et al, 2001; Kurowska & Manthey, 2004). These AhR agonists exert their function by AhR‐mediated CYP1A1 activation, downregulation of nuclear factor kappa B (NF‐kB)‐mediated inflammatory signaling, and activation of inflammatory molecules (Singh et al, 2007; Sarkar et al, 2008; Choi et al, 2010; Potapovich et al, 2011). Whether edible plant bark‐derived factors have an effect on AhR signaling is not known. Further, the molecular mediators and mechanisms governing the association between molecules from plants and the AhR signaling pathway in mammalian cells in general are still elusive.
The purpose of this study was to determine whether edible plant bark‐derived ELNs could be utilized to protect against colitis in a mouse model that results in numerous insults to the gut microbiome and the resulting metabolites. We thus tested whether a plant defensive strategy could be used to enhance protection against colitis in a mammalian model. We used mulberry bark‐derived exosome‐like nanoparticles (MBELNs) as a proof of concept to study the effect of MBELNs on gut epithelial AhR‐mediated signaling in mice. MBELN‐fed mice showed enhanced mucus barrier function. Moreover, we found that MBELN‐derived HSPA8 is required for MBELN‐mediated induction of the expression of AhR as well as activation of AhR. Further, activation of the AhR signaling pathway leads to induction of the expression of COP9/COPS8. The role of induced COPS8 was further demonstrated using intestinal specific COPS8 knockout (KO) mice, showing that MBELN‐mediated protection of mice from dextran sulfate sodium (DSS)‐induced colitis was abolished in COPS8 KO mice. Mechanistically, we show that MBELN treatment results in deneddylation of cullin 1 (CUL1). The COP9 signalosome inhibits the E3 ubiquitin ligase activity of Cullin‐RING ubiquitin ligases (CRLs) by promoting the cleavage of neural precursor cell expressed developmentally downregulated protein 8 (NEDD8)‐CUL1 conjugates (also conceptualized as deneddylation) (Lyapina et al, 2001). Our findings thus link plant communication with the mammalian kingdom via plant‐derived exosome‐like nanoparticle‐mediated regulation of CUL1 deneddylation and the mammalian proteasome degradation machinery.
Results
MBELNs enhance the expression and activate the aryl hydrocarbon receptor (AhR)‐mediated pathway
We isolated exosome‐like nanoparticles from mulberry bark (MBELNs) by differential centrifugation (Appendix Fig S1A). The sucrose‐purified MBELNs had an average diameter of 151.3 ± 45.4 nm (Appendix Fig S1B and C) and showed exosome‐like characteristics based on a bilayer structure and size revealed by electron microscopy (Appendix Fig S1D). Like other liposomes, the presence of a number of explicit lipid molecules (Appendix Fig S1E and F) and proteins (Appendix Fig S1G and Dataset EV1) was demonstrated by mass spectrometry (MS) analysis. The presence of RNA was confirmed by agarose gel electrophoresis with and without digestion by RNase. Isolated RNA was further analyzed using mRNA sequencing that confirmed the presence of several peculiar RNAs with critical roles in metabolic and anti‐pathogenicity as well (Appendix Fig S1H and I and Dataset EV2).
To evaluate the potential efficacy and use of MBELNs as an agent to improve gut healthiness, the toxicity and tropism were evaluated. MBELNs were orally administered to C57BL/6J mice (once daily for 15 days) at two different doses (2 × 109 and 1 × 1010 MBELNs/100 µl/dose/mouse/day) and mice rested for the next 15 days (no treatment) and were then humanely sacrificed on day 30. Mice showed no adverse effects including no significant changes in body weight (Appendix Fig S2A), skin rash, or abnormal fecal discharge during MBELN administration or follow‐up. However, at the dose of 1 × 1010 MBELNs/mouse, mice had a slightly reduced body weight that was not significant; this group of mice did not show any other adverse changes. Mice did not show any abnormal effects regarding morphology of internal organs, gut tissue microscopic structure, blood cholesterol, triglycerides, or liver enzyme alanine transaminase (ALT), while liver enzyme aspartate transaminase (AST) was lower in the MBELN group, but still in a normal range (Appendix Fig S2B–D). The in vivo bio‐distribution and trafficking of DiR‐labeled MBELNs after oral administration was evaluated in mice using an Odyssey imaging system. Three hours (h) after a single oral administration of DiR‐labeled MBELNs (10 × 109 particles/mouse), DiR fluorescent signals were predominantly detected in the duodenum–ileum of the small intestine, colon, and cecum. A small fraction of fluorescent signal was also observed in spleen, liver, lung, kidney, heart, and blood. The presence and intensity of the imaging signal indicated that DiR‐labeled MBELNs were detectable up to 6 h after a single dose in blood, heart, liver, lung, kidney, and brain, while in the small intestine and colon tissue, it was detectable up to 12 h later. The signal was reduced over time and was not detectable at 24 h (Appendix Fig S2E and F). Confocal microscopy analysis demonstrated that 3 h after orally administering MBELNs, it was predominantly taken up by gut epithelial cells, Paneth cells (yellow arrow), and colon tissue. Additionally, in the spleen and liver, the MBELNs were predominantly present in F4/80+ macrophages (Appendix Fig S2G).
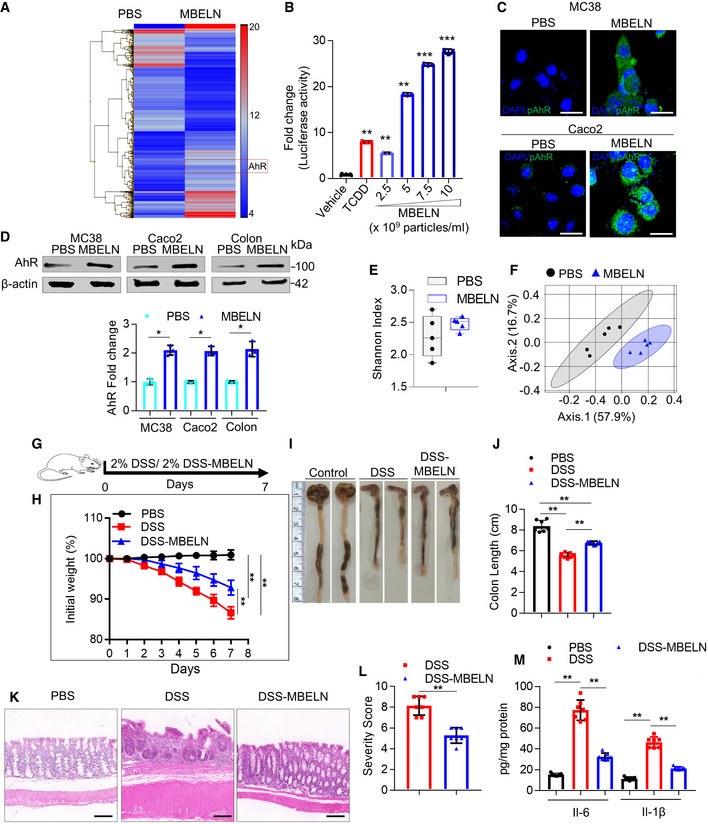
Next, the effect of MBELNs on the genes expressed in colon tissue was further determined. MBELNs were orally administered to wild‐type C57BL/6J mice for seven days and the expression of genes was profiled using cDNA array analysis. The results suggested that MBELN treatment has an effect on the genes expressed in colon tissue (Fig 1A). We noticed that the expression of AhR was increased due to MBELN treatment. Our recent work indicated that edible plant‐derived exosomes regulate AhR‐mediated signaling pathways in the intestinal epithelium (Teng et al, 2018). AhR is an important ligand‐activated receptor molecule responding to endogenous or exogenous ligands, including plant‐derived ligands, and plays a significant role in the host defense mechanism (Moura‐Alves et al, 2014, 2019). To determine whether MBELNs can activate the AhR‐mediated pathway, mouse Hepa1.1 cells, stably harboring an AhR responsive luciferase reporter construct, were incubated with vehicle, 2,3,7,8‐tetrachlorodibenzo‐p‐dioxin (TCDD; 1 nM) as a positive control, or increasing concentrations of MBELNs (2.5 × 109, 5 × 109, 7.5 × 109, 10 × 109 particles/ml). Exposure to MBELNs resulted in a dose‐dependent increase in luciferase expression. A significant 2‐fold induction over vehicle treatment was observed at the lowest concentration of 2.5 × 109 particles/ml and a maximal 27‐fold expression increase at 10 × 109 particles/ml. The data indicate that MBELNs are a more potent activator of the AhR pathway than the prototypical AhR agonist TCDD (Fig 1B). Intestinal epithelial cells are one of the major cell types targeted by MBELNs. Therefore, we further explored whether MBELNs affect activation of the AhR‐mediated pathway in intestinal epithelial cells. C57BL/6 murine colon MC38 cells and human colon Caco2 cells were exposed to vehicle or MBELNs for 3 h. First, we found that MBELN treatment (5 × 109 particles/ml) increases levels of phosphorylated‐AhR as well as total AhR in both mouse MC38 cells and human intestinal epithelial Caco2 cells (Fig 1C and Appendix Fig S2H). The induction of AhR expression was further demonstrated in colon epithelial tissue of C57BL/6J mice following 3 h of a single dose of oral administration of MBELNs (10 × 109 particles/mouse) (Fig 1D). Induction of AhR target genes, including Cytochrome P450, family 1, subfamily A, polypeptide 1 (CYP1A1), and Indoleamine 2,3‐dioxygenase (IDO1), and corresponding proteins, was also observed in MBELN‐treated MC38 cells, Caco2 cells and in C57BL/6J mouse colon epithelial tissue in vivo (Appendix Fig S2I and J).
Figure 1. MBELNs activate AhR signaling.
-
AMice were orally administered with mulberry bark‐derived exosome‐like nanoparticles (MBELNs) (10 × 109 particles/100 µl/mouse) or phosphate‐buffered saline (PBS) for 7 days. Heat map showing influence of MBELNs on colonic gene expression from three biological replicates.
-
BIn vitro assessment of MBELN‐dependent induction of aryl hydrocarbon receptor (AhR) promoter using HEPA1.1 cells (contain AhR responsive luciferase reporter construct). Data are mean ± SEM from three biological replicates. **P < 0.01, ***P < 0.001 using one‐way ANOVA.
-
CRepresentative images showing expression of pAhR in MC38 cells and Caco2 cells treated with MBELNs from three biological replicates. Scale bar 20 μm.
-
DWestern blot (top) and graphical representation of fold changes (bottom) for AhR in MC38 cells, Caco2 cells, and colon epithelial cells after treatment with MBELNs. Data are mean ± SEM of three biological replicates. *P < 0.05 using Student’s t‐test.
-
EFecal microbiota were analyzed following administration of MBELNs (10 × 109 particle/100 µl/dose/day/mouse) for 7 days to C57BL/6 mice. Alpha diversity (Shannon index) was calculated at the family level and is displayed as a bar‐and‐whiskers plot for each individual combination of control and MBELN treatment. The center line represents the median and the box encloses the 1st and 3rd quartiles (“hinges”). The upper and lower whiskers represent the furthermost points from the respective hinges, which are no more than 1.5 interquartile ranges from the hinge. The individual points are overlaid. Data are presented from five mice or biological replicates.
-
FBeta diversity analysis, biplots samples (points), and bacterial taxa at family level (axes) are simultaneously projected on the two‐dimensional canonical axes (CA1 and CA2). Data are presented from five mice or biological replicates.
-
GPictorial representation of dextran sulfate sodium (DSS; 2%)‐induced colitis development in mice.
-
HGraph showing loss of weight following DSS‐induced colitis with and without MBELN treatment. Data are mean ± SEM of seven biological replicates, **P < 0.01 using Mann–Whitney test.
-
IRepresentative image showing changes in colon morphology and length following DSS‐induced colitis with concurrent treatment with MBELNs.
-
JColumn graph showing changes in colon length. Data are mean ± SEM from seven biological replicates, **P < 0.01 using one‐way ANOVA.
-
K, LHematoxylin and eosin (HE) staining to show histological changes in colon tissue (left) and a graph showing severity score (right) based on histological data. Scale bar 200 μm. Data are mean ± SEM of seven biological replicates, **P < 0.01 using Student’s t‐test.
-
MEnzyme‐linked immunosorbent assay (ELISA) for interleukin (IL)‐6 and IL‐1β from colon tissue. Data are mean ± SEM from seven biological replicates, **P < 0.01 using one‐way ANOVA.
Source data are available online for this figure.
Diet can alter the composition of gut microbiota. Analysis of MBELN‐related alterations in microbial community composition following a 7‐day treatment at a family level showed an increase in bacterial species richness in the feces (Fig 1E). Beta diversity analysis demonstrated significant dissimilarities between microbiota populations (Fig 1F). MBELN treatment enhanced the bacterial communities of Firmicutes (Dehalobacteriaceae, Lachnospiraceae, Ruminococcaceae, Erysipelotrichaceae, and Mogibacteriaceae), Porphyromonadaceae, Prevotellaceae, Rikenellaceae, Paraprevotellaceae from Bacteroidetes, Proteobacteria (Pseudomonadaceae, Desulfovibrionaceae), and Verrucomicrobia (Verrucomicrobiaceae), while it reduced the community richness for Actinobacteria (Corynebacterium), Bacteroidetes (Bacteroidaceae, S24.7 and Odoribacteraceae), Firmicutes (Lactobacillaceae, Turicibacteraceae, and Clostridiaceae), Proteobacteria (Alcaligenaceae and Helicobacteraceae) and Tenericutes (Mycoplasmataceae) (Appendix Fig S3A). Next, we tested whether MBELNs could inhibit species‐specific growth of bacteria. We found that MBELNs had species‐specific growth inhibitory effects on virulent Listeria (L.) monocytogenes‐EGD (Lis‐EGD), while they did not affect the growth of Escherichia coli (E. coli), Porphyromonas gingivalis (PG), Streptococcus gordonii (SG), and non‐virulent Listeria monocytogenes (Lis) (Appendix Fig S3B and C). We also noticed that MBELNs were able to inhibit the expression of bacterial mRNAs (Appendix Fig S3D; Dataset EV3) that are crucial for the pathogenesis of virulence and growth (Chatterjee et al, 2006; Toledo‐Arana et al, 2009; Hamon et al, 2012). Next, we determined whether the protein complexes formed by a virulent strain of L. monocytogenes‐EGD with MBELNs are different from the non‐virulent strain of L. monocytogenes. We identified a unique protein profile of virulent L. monocytogenes‐EGD that interacted with biotin‐labeled MBELNs when compared to non‐virulent L. monocytogenes (Appendix Fig S3E and F; Dataset EV4). The interacting proteins may contribute to the efficiency in uptake of virulent versus non‐virulent strains (12.6 vs. 30.8% in non‐virulent vs. virulent L. monocytogenes; Appendix Fig S3G). The phenotypes, including AhR activation and bacterial species‐specific growth inhibition, were reproduced with MBELNs from different lots of mulberry bark harvested at different time points (summer, fall, winter, and spring). In addition, there was no difference on AhR induction in MC38 cells, suggesting that the therapeutic factors from MBELNs are not affected by seasonal variation or another factor (Fig EV1A).
Figure EV1. MBELN‐induced induction of AhR.
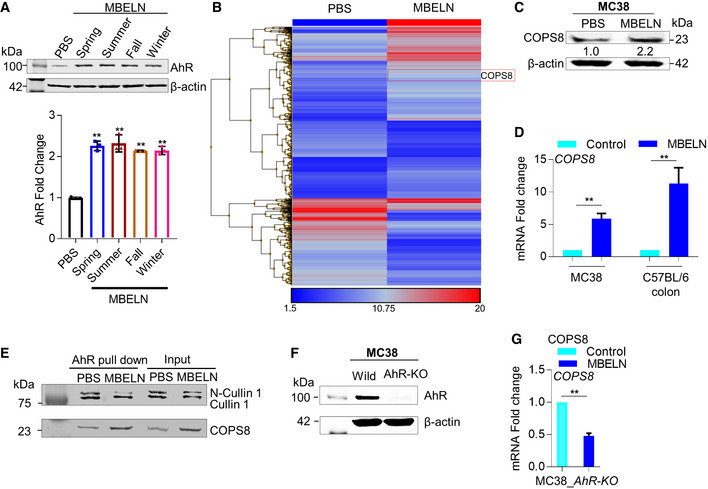
- To test the reproducibility of mulberry bark‐derived exosome‐like nanoparticles (MBELN) collected during different seasons of the year, MBELNs were extracted in spring, summer, fall, and winter and used to treat MC38 cells to evaluate AhR expression. Data are mean ± SEM from three biological replicates per group. **P < 0.01 using one‐way ANOVA.
- Heat map showing gene influences in crypts of villi in C57BL/6J mice. Data shown are from three technical replicates.
- Western blot for COP9/COP9 Constitutive Photomorphogenic Homolog Subunit 8 (COPS8) in MC38 cells following MBELN treatment. Data shown are from three biological replicates.
- mRNA expression of COPS8 in MC38 and after MBELN administration and C57BL/6J colon epithelial cells following MBELN administration for 7 days. Data are mean ± SEM from three biological replicates per group. **P < 0.01 using Student’s t‐test.
- MC38 cells treated with MBELN and protein extract were immunoprecipitated using anti‐aryl hydrocarbon receptors (AhR) antibody. The precipitate was subjected to Western blot with antibody against Cullin 1 (CUL1) and COPS8. Data shown are from three biological replicates.
- Western blot to confirm knockout of AhR in MC38 cells. Data shown are from three biological replicates.
- COPS8 mRNA expression in AhR‐knockout (AhR‐KO) MC38 cells following treatment with MBELNs. Data are mean ± SEM from three biological replicates per group. **P < 0.01 using Student’s t‐test.
Source data are available online for this figure.
To further determine whether the data generated above can be translated to a mammalian setting under physiological conditions for the prevention of gut‐related disease, we tested our central hypothesis as to whether ELNs from an edible plant (Mulberry; Morus notabillis) can be utilized for enhancing intestinal barrier function, which would play a critical role in the prevention of inflammatory bowel disease (Antoni et al, 2014; Lechuga & Ivanov, 2017). To assess the influence of MBELNs on DSS‐induced colitis development, mice were orally administered MBELNs (10 × 109 particles/dose/day/mouse) for 7 days in addition to 2% DSS in the drinking water (Fig 1G). Mice treated with MBELNs showed reduced colitis severity, in particular a reduction in loss of body weight, colon shortening, and inflammatory severity (Fig 1H–J). MBELN treatment in conjunction with DSS administration resulted in attenuated crypt damage, reduction of the infiltration of inflammatory cells (Fig 1K and L), and reduction in the levels of inflammatory cytokines interleukin (IL)‐6 and IL‐1β (Fig 1M).
MBELNs induce the expression of specific genes via the AhR pathway
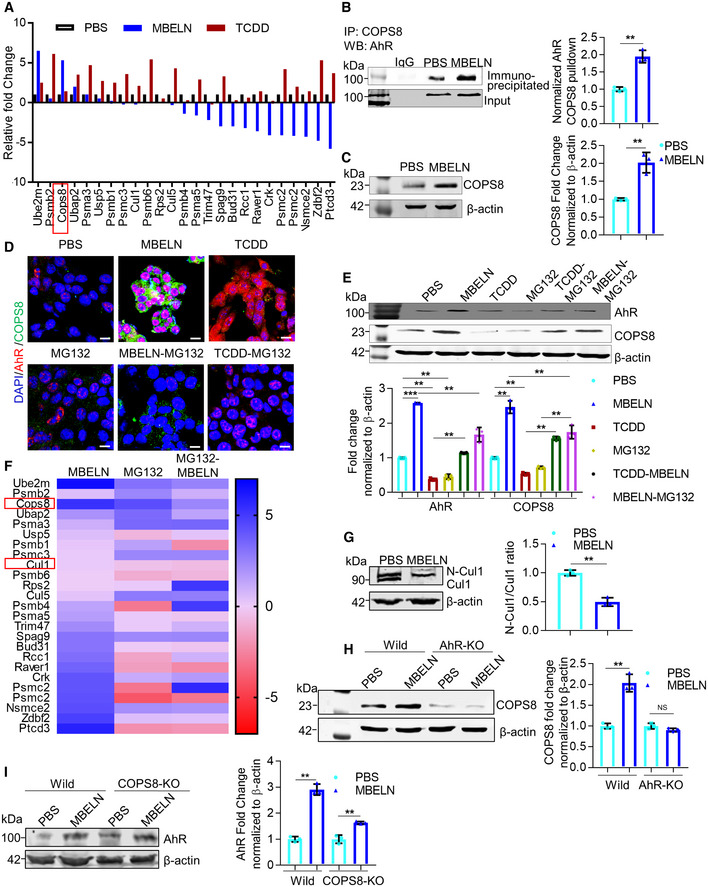
AhR is a ligand‐activated transcription factor that integrates environmental, dietary, microbial, and metabolic cues to control complex transcriptional programs in a ligand‐specific, cell type‐specific, and context‐specific manner. AhR interacting intracellular proteins in MC38 cells were investigated while being treated with MBELNs and compared to cells treated with TCDD and PBS as controls. Interaction between AhR and MBELN‐induced proteins in the AhR complex was identified by co‐immunoprecipitation using an AhR antibody followed by MS (Fig 2A). Interestingly, the MBELN‐treated group had several proteins interacting with AhR specially involved in proteasome degradation and ubiquitin pathways (Fig 2A). COPS8 was selected for further investigation, as COPS8 plays a crucial role in the COP9 signalosome complex and is important for the crosstalk with proteasome‐mediated pathways (Schwechheimer et al, 2001; Perazzo et al, 2003). To confirm the MS data, in a reciprocal immunoprecipitation assay using a COPS8 antibody, we demonstrated that MBELN treatment recruited more AhR into the COPS8 complex (Fig 2B). Furthermore, MBELN treatment also increased COPS8 at the protein and mRNA levels in ileum crypts and colon epithelial cells (Figs 2C and EV1B–D).
Figure 2. MBELN treatment leads to inducing the expression of COPS8 via the AhR signaling pathway.
-
AMass spectrometry (MS) analysis of aryl hydrocarbon receptor (AhR) pulldown protein from mulberry bark‐derived exosome‐like nanoparticles (MBELNs) and 2,3,7,8‐tetrachlorodibenzo‐p‐dioxin (TCDD)‐treated MC38 cell lysate. Data from three technical replicates are shown.
-
BCo‐immunoprecipitation of COP9/COP9 Constitutive Photomorphogenic Homolog Subunit 8 (COPS8) bound AhR from MC38 cell lysate treated with MBELNs. Data are mean ± SEM from three biological replicates, **P < 0.01 using Student’s t‐test.
-
CWestern blot analysis showing expression of COPS8 in mice ileum crypts. Data are mean ± SEM of three biological replicates, **P < 0.01 using Student’s t‐test.
-
DRepresentative images showing effect of MBELNs with and without TCDD (1 nm)/ carbobenzoxy‐Leu‐Leu‐leucinal (MG132; 20 µM) on co‐expression of AhR and COPS8 using immunofluorescence. Scale bar 10 μm, images are from three biological replicates.
-
EWestern blot showing MBELNs, TCDD, and MG132 effect on co‐expression of AhR and COPS8. The data represent as mean ± SEM from three biological replicates **P < 0.01, ***P < 0.001 using one‐way ANOVA.
-
FHeat map showing MS analysis of AhR pulldown protein from MC38 cells treated with MBELNs in combination with proteasome inhibitor, MG132. Data are represented from three technical replicates.
-
GExpression of Cullin 1 (CUL1) and neddylated CUL1 (N‐CUL1) following MBELN treatment using Western blot. Data are mean ± SEM from three biological replicates. **P < 0.01 using Student’s t‐test.
-
H, IWestern blot showing expression of COPS8 in wild‐type (Wild) and AhR‐KO MC38 cells and expression of AhR in wild and COPS8‐KO MC38 cells. MC38 cells were KO using CRISPR/ CAS9 KO plasmid transfection.
Data information: Data represent mean ± SEM from three biological replicates, **P < 0.01, NS—non‐significant, using one‐way ANOVA.
Source data are available online for this figure.
Degradation of AhR via the ubiquitin–proteasome pathway takes place upon AhR ligand stimulation (Ma & Baldwin, 2000) as a feedback mechanism for maintaining AhR homeostasis. We tested the stability of AhR upon MBELN stimulation. Interestingly, in contrast to the results generated using TCDD stimulation, MBELN treatment resulted in an increase of AhR expression. MC38 cells treated with proteasome inhibitor MG132 showed attenuated MBELN‐mediated induction of cytoplasmic AhR, AhR translocation, and COPS8 expression (Fig 2D). These results suggest that the pathway causing MBELN‐mediated activation of AhR and subsequently inducing expression of COPS8 is promoted by the proteasome degradation pathway. Data further indicate that MBELNs enhanced the expression of COPS8 protein in contrast to the fact that the prototypical AhR agonist, TCDD, decreased the expression of COPS8 protein (Fig 2E). MS analysis further suggests that MG132 treatment altered the composition of the AhR complex of MC38 cells treated with MBELNs (Fig 2F). The presence of COPS8 and cullin 1 was confirmed by co‐immunoprecipitation using AhR pulldown (Fig EV1E). To examine the effect of MBELNs on the proteosome pathway through COPS8‐CUL1 (neddylation and deneddylation), CUL1 was examined, as CUL1 is known to be involved in the maintenance of F‐box stability and dynamic SCF (Skp1–CUL1–F‐box protein) assembly and is collectively controlled by the activity of the COP9 signalosome (Lyapina et al, 2001). The deneddylation of CUL1 increased following treatment with MBELNs which is in agreement with the fact that COP9 has deneddylation activity (Fig 2G) (Lyapina et al, 2001).
To test whether AhR is an upstream factor to regulate the expression of COPS8, KO for AhR in MC38 cells was accomplished using CRISPR/CAS9 and confirmed by Western blot (Fig EV1F). AhR KO inhibited the MBELN‐mediated COPS8 expression at the protein and transcriptional level when compared to wild‐type cells (Figs 2H and EV1G), while COPS8 KO did not affect MBELN‐dependent AhR induction (Fig 2I), which demonstrates MBELNs mediate induction of COPS8 through AhR.
Deletion of COPS8 in intestinal epithelial cells leads to higher susceptibility to DSS‐induced intestinal inflammation
Next, the significance of the induction of COP9/COPS8 was further investigated. Although COP9 regulates the cell cycle and proliferation (Yang et al, 2002), it is not known whether COPS8 plays a role in regulating gut barrier function. To address the in vivo role of COPS8, we generated mice expressing Villin‐Cre and COPS8‐lox alleles (COPS8fl/fl ) to KO COPS8 in intestinal epithelial cells (IEC) (COPS8ΔIEC ). To verify the cell‐type specificity of COPS8 KO, we performed immunofluorescent staining (IF) for COPS8 along the anterior–posterior axis of the mouse small and large intestine. In control COPS8fl/fl intestine, COPS8 is expressed in both intestinal epithelial cells and interstitial cells (Fig EV2A). In COPS8ΔIEC intestine, however, the majority of the crypt/villus units are negative for COPS8, while all the interstitial cells remain positive for COPS8 (Fig EV2A). The efficiency of COPS8 deletion in isolated IECs was confirmed by Western blotting (Fig EV2B). Additionally, we examined COPS8 protein levels in other organs and found no changes in liver, kidney, and lung of COPS8ΔIEC mice when compared to the controls (Fig EV2B). These results show that the deletion of COPS8 is specific to IECs. COPS8 deletion or knockdown has been reported to cause instability of other COPS subunits (Menon et al, 2007; Lei et al, 2011; Su et al, 2011; Liu et al, 2013). We found COPS5, COPS6, and COPS7 protein amounts were also decreased to various degrees (Fig EV2C), indicating the essential role of COPS8 for COP9 complex integrity in IECs. The best‐known function of the COP9 complex is the regulation of cullin‐Ring‐E3 ubiquitin ligase activity through deneddylation (Su et al, 2011). Consistent with the loss‐of‐function mutants of individual COPS subunits, COPS8‐deficient IECs displayed a marked increase of neddylated cullins (Fig EV2C), indicating that the cullin deneddylation activity was compromised (Wei & Deng, 2003). In addition, COPS8ΔIEC mice displayed a significant reduction in body weight (Fig EV2D) and small intestinal length, while colon length remained the same (Fig EV2E).
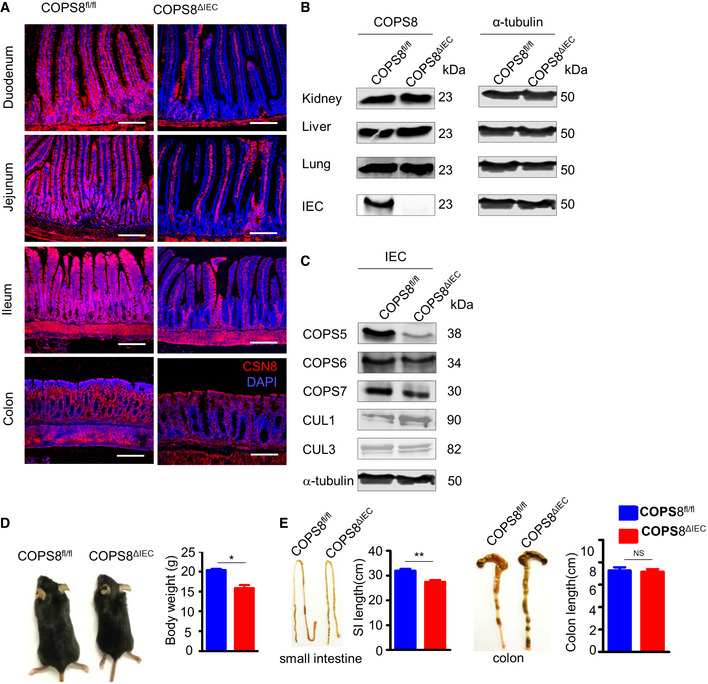
Figure EV2. Deletion of COP9/COP9 Constitutive Photomorphogenic Homolog Subunit 8 (COPS8) in intestinal epithelial cells (IEC) affects COP9 complex integrity.
-
AImmunofluorescence analysis of COPS8 protein expression with anti‐COPS8 antibody (red) and with DAPI (blue) in the duodenum, jejunum, ileum, and colon from Villin‐Cre and COPS8‐lox alleles expressing (COPS8fl / fl ) and knockout COPS8 in IEC (COPS8ΔIEC ) mice. Scale bar, 100 μm, data are represented from seven biological replicates per genotype.
-
B, CImmunoblot analysis of COPS8 protein expression in the liver, kidney, lung, and IECs isolated from ileum of COPS8fl / fl and COPS8ΔIEC mice and analysis of the associated proteins (COPS5, COPS6, COPS7, Cullin (CUL) 1 and 3) in lysates prepared from isolated IECs from COPS8fl / fl and COPS8ΔIEC mice. Data are represented from seven biological replicates per genotype.
-
DBody size (representative pictures, left) and weight (right) of COPS8fl / fl and COPS8ΔIEC mice at 4 weeks of age. Data are represented as mean ± SEM from seven biological replicates per genotype. *P < 0.05, NS—non‐significant using Student’s t‐test.
-
ELength of small intestine and colon from COPS8fl / fl and COPS8ΔIEC mice. Data are represented as mean ± SEM from seven biological replicates per genotype. **P < 0.01, NS—non‐significant using Student’s t‐test.
Source data are available online for this figure.
Histological analysis revealed a sharp reduction of Paneth cells throughout the length of the small intestine (Figs 3A and EV3A). Paneth cell granules store lysozyme, which were barely detectable in COPS8ΔIEC crypts (Figs 3B and EV3B). Electron microscopy (EM) showed the presence of degenerating organelle membranes and the loss of granules in Paneth cells of COPS8ΔIEC mice (Black arrows in Fig 3C). Although numerous secretory granules are apically located and secreted into the cryptic lumen of COPS8fl/fl mice, complete absence of secretory granules was observed in the lumen of COPS8ΔIEC mice (Blue arrows in Figs 3C and EV5C). EM analysis also revealed microvilli were abundant in the inner surfaces of cryptic lumina in COPS8fl/fl mice, but markedly reduced in COPS8ΔIEC mice (yellow arrows in Fig EV3C). These data indicate that COPS8 quantitatively regulates the secretory granules. EM revealed a contracted endoplasmic reticulum (ER) in COPS8ΔIEC goblet cells (white arrow in Fig 3C). The number of alcian blue+ goblet cells was mildly reduced in duodenum and jejunum (Fig 3D) but not in ileum (Fig EV3D). Though the number of entero‐endocrine cells remained unaffected, chromogranin A+ (ChgA+) staining revealed a clear mislocalization of ChgA+ cells (Figs 3E and EV3E). Paneth cells are one of the major cell types that release lysozyme and anti‐microbial peptides (AMPs) that play a protective in IBD (Ayabe et al, 2000; Wehkamp et al, 2005; Salzman et al, 2010). To test whether COPS8 deficiency triggers dysbiosis via a defect in the production of AMPs, global gene expression profiles were examined in crypt IECs using the Clariom S sDNA array. We analyzed mRNA panels of AMPs produced in COPS8ΔIEC and COPS8fl/fl mice. Consistent with the marked reduction of Paneth cells, the expression of defensins was barely detectable and the expression of lysozyme was also dramatically reduced in IECs of distal ileum of COPS8ΔIEC mice (Fig 3F). Real‐time quantitative reverse transcription polymerase chain reaction (qRT–PCR) confirmed that the expressions of AMPs (viz. Defensin Alpha (Defa)‐1, Defa‐21, Defa‐b1, Defa‐rs1, and Angiopoietin (Ang)‐4) were reduced significantly in ileum and colon (Fig 3G and H).
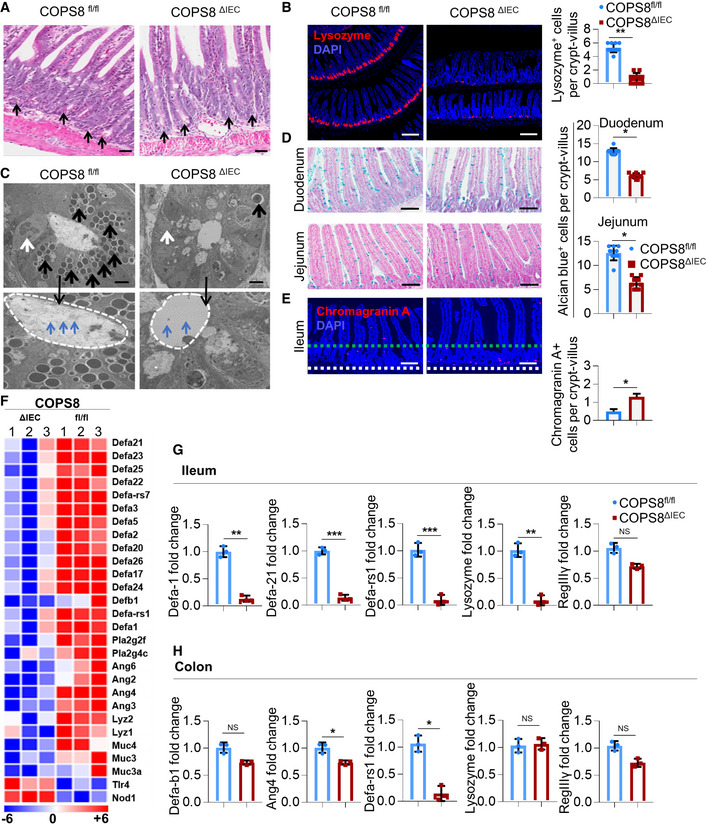
Figure 3. Deletion of COP9/COP9 constitutive photomorphogenic homolog subunit 8 (COPS8) in IECs leads to the loss of the Paneth cell lineage in distal ileum crypts.
-
ARepresentative hematoxylin and eosin (HE) staining of ileum. Paneth cells with typical eosinophilic granules (black arrows) on HE‐stained sections at the base of crypts in Villin‐Cre and COPS8‐lox alleles expressing (COPS8fl / fl ), but not COPS8 knockout (KO) (COPS8ΔIEC ) epithelium. Scale bar 50 μm, n = 7 biological replicate per genotype.
-
BThe granule protein, lysozyme, was examined by immunofluorescence (Red) and counted in the ileum of COPS8fl/fl and COPS8ΔIEC mice (scale bar 100 μm). The column graph represents number of lysozyme+ cells presented as mean ± SEM from seven biological replicates. **P < 0.01 using Student’s t‐test.
-
CTransmission electron microscopy (TEM) of crypts of COPS8fl/fl and COPS8ΔIEC mice. The base of the crypt in COPS8ΔIEC mice is occupied by poorly differentiated columnar epithelial cells that lack secretory granules, rudimentary electron‐dense granules (black arrows), granules in the lumen (blue arrows) and have a contracted endoplasmic reticulum (ER) (white arrows). Scale bar 5 μm, data are from seven biological replicates per genotype.
-
DGoblet cells were stained by Alcian blue and counted in COPS8fl/fl and COPS8ΔIEC epithelia of duodenum and jejunum (scale bar 100 μm). The column graph represents number of Alcian blue+ cells presented as mean ± SEM from seven biological replicates. *P < 0.05 using Student’s t‐test.
-
EThe marker for enteroendocrine cells (between two dashed line), chromogranin, was detected by immunofluorescence and counted in the ileum of COPS8fl/fl and COPS8ΔIEC mice (scale bar 100 μm). The column graph represents number of chromogranin A+ cells per crypt‐villus presented as mean ± SEM from seven biological replicates. *P < 0.05 using Student’s t‐test.
-
FRNA sequencing‐based measurements of transcripts comprising antimicrobial peptides (AMPs)‐related genes in crypts isolated from COPS8fl/fl and COPS8ΔIEC mice. Representative data from three biological replicates per genotype.
-
G, HReal‐time quantitative reverse transcription polymerase chain reaction (PCR) (qRT–PCR) analysis of the expression of genes encoding AMPs in the ileum (G) and colon (H) in COPS8fl/fl and COPS8ΔIEC mice. Error bars indicate the mean ± SD from three biological replicates. *P < 0.05, **P < 0.01, ***P < 0.001 using Student’s t‐test.
Figure EV3. Paneth cell loss in COPS8ΔIEC mice.
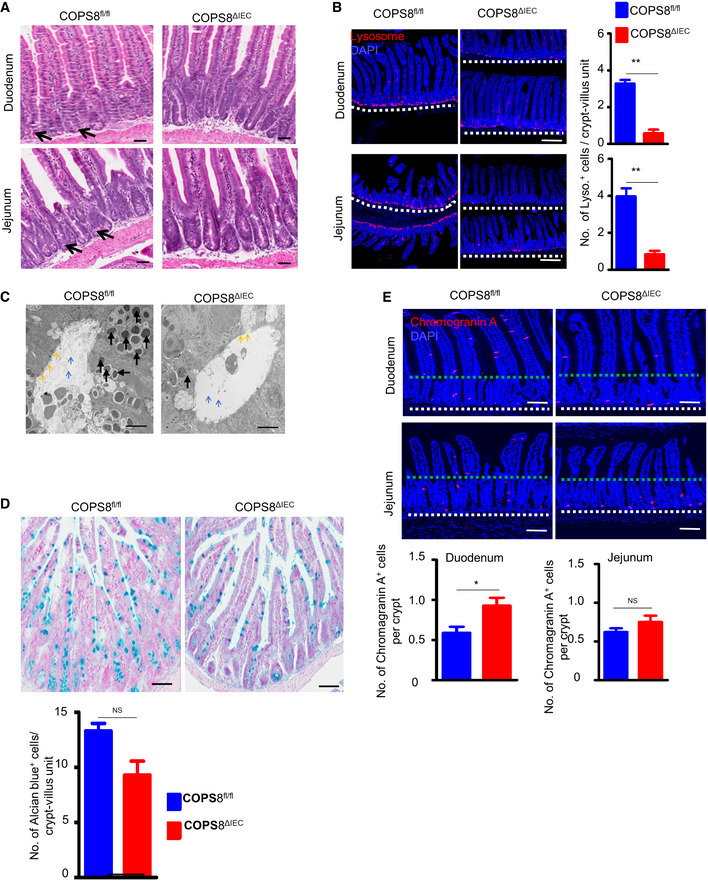
- Representative hematoxylin and eosin (HE) staining of duodenum and jejunum showing loss of Paneth cells (black arrow) in COPS8ΔIEC mice. Scale bar 50 μm, data are represented from seven biological replicates per genotype.
- The granule protein, lysozyme, was examined by immunofluorescence (Red) and counted in the duodenum and jejunum (located above dashed line) of Villin‐Cre and COPS8‐lox alleles expressing (COPS8fl / fl ) and COPS8ΔIEC mice. Scale bar 200 μm. Data are represented as mean ± SEM from seven biological replicates per genotype. **P < 0.01 using Student’s t‐test.
- Transmission electron microscopy (TEM) of crypts of COPS8fl / fl and COPS8ΔIEC mice. The base of the crypt in COPS8ΔIEC mice is occupied by poorly differentiated columnar epithelial cells that lack secretory granules, rudimentary electron‐dense granules (black arrows), microvilli (yellow arrows), and granules in the lumen (blue arrows). Scale bar 5 μm.
- Goblet cells from COPS8fl/fl and COPS8ΔIEC mice were stained with Alcian blue and counted in the ileum. Scale bar 50 μm. Data are represented as mean ± SEM from seven biological replicates per genotype. NS—statistically non‐significant using Student’s t‐test.
- The marker for enteroendocrine cells, chromogranin, was detected by immunofluorescence and counted in the duodenum and jejunum of COPS8fl / fl and COPS8ΔIEC mice. Scale bar 100 μm. Data are represented as mean ± SEM from seven biological replicates per genotype. **P < 0.01 using, NS—non‐significant using Student’s t‐test.
Figure EV5. Analysis of the MBELN factor (protein, lipid, and RNA) liable for AhR induction.
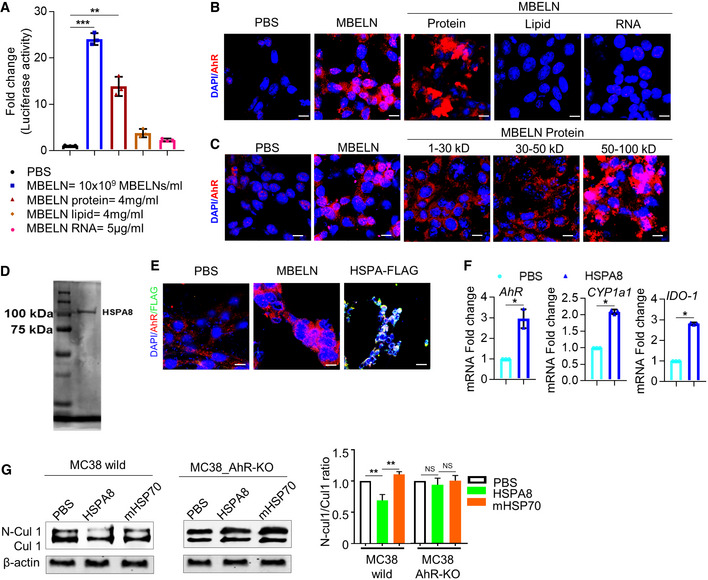
- Effect of different fractions of mulberry bark‐derived exosome‐like nanoparticles (MBELNs) on the expression of aryl hydrocarbon receptor (AhR) promoter using HEPA1.1 cells (AhR responsive luciferase reporter construct). Data are represented as mean ± SEM from three biological replicates. *P < 0.05, NS—non‐significant using one‐way ANOVA.
- Confocal image showing the effect of MBELN fractions on localization of AhR. Scale bar 10 μm, Data are represented from three biological replicates.
- The effect of different molecular weight fractions of MBELN‐derived proteins on AhR localization in MC38 cells. Scale bar 10 μm, Data are represented from three biological replicates.
- Cloning, expression, and purification of recombinant MBELN‐HSPA8.
- Confocal microscopy showing FLAG‐tagged HSPA8 protein treatment to MC38 colocalized with AhR. Scale bar 10 μm, Data are represented from three biological replicates.
- Real‐time quantitative reverse transcription polymerase chain reaction (PCR) for expression of AhR and its targeted genes Cytochrome P450, family 1, subfamily A, polypeptide 1 (CYP1a1), and Indoleamine 2,3‐dioxygenase (IDO‐1) in MC38 cells following HSPA8 treatment. Data are represented as mean ± SEM from three biological replicates. *P < 0.05 using Student’s t‐test.
- Western blot showing the ratio of CUL1 and neddylated cullin 1 (N‐CUL1) expression in wild and AhR knockout (KO) MC38 cells. Data are represented as mean ± SEM from three biological replicates. *P < 0.05, NS—non‐significant using Student’s t‐test.
Source data are available online for this figure.
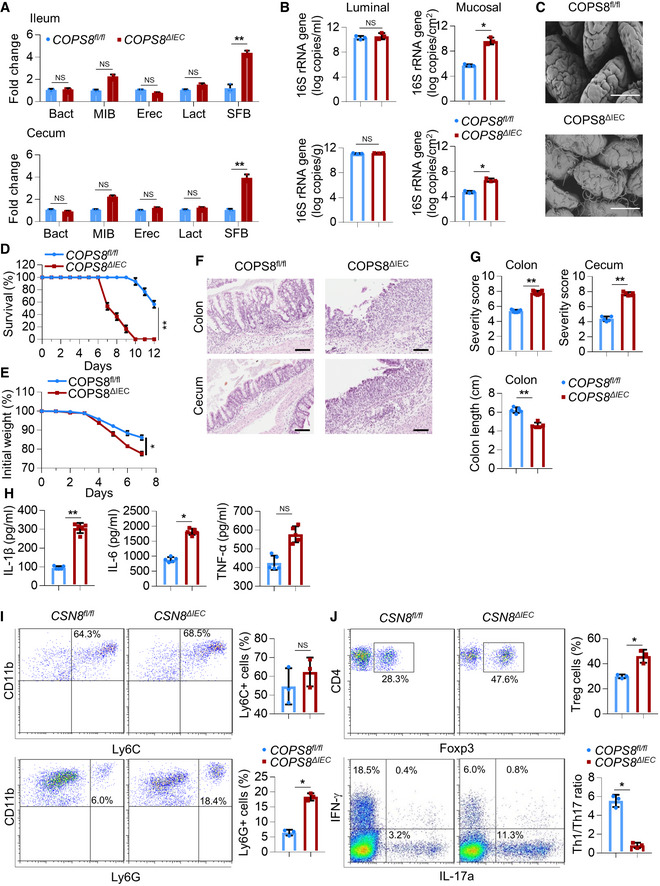
Susceptibility to intestinal inflammation is often associated with alterations in commensal bacterial populations (Alenghat et al, 2013; Peterson & Artis, 2014). Using real‐time PCR with 16S rRNA primers targeting the dominant bacterial populations in the mouse intestinal tract (Salzman et al, 2002), we compared the relative abundance of individual bacterial groups in distal ileum and colon of COPS8ΔIEC mice and their single‐housed controls. These included members of the gram‐positive Firmicutes phylum (Eubacteriumrectale, Lactobacillus, and Segmented Filamentous Bacteria (SFB) groups) and the gram‐negative Bacteroidetes phylum (Bacteroides and Mouse Intestinal Bacteroides (MIB) groups). Surprisingly, although the relative number of gram‐negative Bacteroides and MIB and gram‐positive Eubacteriumrectale and Lactobacillus remained similar, there was a dramatic increase of SFB (Fig 4A). Increase of the mucus‐associated bacteria plays a role in gut inflammation (Matsuoka & Kanai, 2015). The altered composition of bacterial species as a result of KO of COPS8 in gut epithelial cells may be due to their different spatial distribution, luminal vs. anchoring the apical surface of epithelia (Vaishnava et al, 2008, 2011). We quantitatively analyzed bacteria in the mucus versus lumen using real‐time PCR. Although the luminal bacterial loads did not show a significant difference, the bacteria recovered from the intestinal surface showed a consistent higher trend in COPS8ΔIEC mice as compared to COPS8fl/fl mice (Fig 4B). Indeed, transmission EM also revealed a thicker network of bacteria in the distal ileum of COPS8ΔIEC mice comparing to that of COPS8fl/fl mice (Fig 4C). These results suggest that KO of COPS8 in gut epithelial cells leads to more bacteria accessing the gut mucus, and more dense mucus‐associated bacteria due to KO of COPS8 is in accordance with the observed reduction of expression of genes encoding for AMPs secreted by Paneth cells.
Figure 4. COP9/COP9 Constitutive Photomorphogenic Homolog Subunit 8 (COPS8) deficiency increases susceptibility to DSS‐induced colitis.
-
AReal‐time quantitative reverse transcription polymerase chain reaction (PCR) (qRT–PCR) analysis of the differential abundance of selected bacterial taxa in stool samples of ileum and colon of Villin‐Cre and COPS8‐lox alleles expressing (COPS8fl / fl ) and COPS8 knockout (KO) (COPS8ΔIEC ) mice. Error bars indicate the mean ± SD from three biological replicates per genotype. **P < 0.01, NS—non‐significant using Student’s t‐test.
-
BqRT–PCR analysis of the abundance of luminal and mucosal bacteria in ileum (top) and cecum (bottom) of COPS8fl / fl and COPS8ΔIEC mice. Error bars indicate the mean ± SD from three biological replicates per genotype. *P < 0.05, NS—non‐significant using Student’s t‐test.
-
CScanning electron microscopy of ileum of COPS8fl / fl and COPS8ΔIEC mice. Scale bar 50 μm, representative data from three biological replicates per genotype.
-
D–JMice were treated by 2% dextran sodium sulfate (DSS) in drinking water for 7 days. (D) Survival curves of COPS8fl / fl and COPS8ΔIEC mice during DSS‐induced colitis. Difference in survival was determined with Kaplan–Meier analysis, **P < 0.01. Representative data from 20 biological replicates per genotype. (E) Changes in body weight are presented as percent of initial weight was determined using Mann–Whitney test, *P < 0.05. Representative data from 20 biological replicates per genotype. (F) Representative hematoxylin and eosin (HE)‐stained sections of colon and cecum from COPS8fl / fl and COPS8ΔIEC mice after DSS‐induced colitis (day 9). Scale bar 100 μm, representative data from five biological replicates per genotype. (G) Histopathologically scored sections of distal colon; cecum and colon length were analyzed from COPS8fl / fl and COPS8ΔIEC mice after DSS‐indcued colitis (day 7). Data are represented as mean ± SEM from five biological replicates per genotype. **P < 0.01 using Student’s t‐test. (H) Cytokine levels (interleukin (IL)‐1β, IL‐6, and tumor necrosis factor‐α (TNF‐α)) in the colon collected on day 7 in colitis induced COPS8fl / fl and COPS8ΔIEC mice. Data are represented as mean ± SEM from five biological replicates per genotype. *P < 0.05, **P < 0.01, NS—non‐significant using Student’s t‐test. (I) The frequency of CD11b+Ly6C+ and CD11b+Ly6G+ cells in colonic lamina propria (cLP) of COPS8fl / fl and or COPS8ΔIEC mice with DSS‐induced colitis. The column graph represents percentage of Ly6C+ and Ly6G+ cells, presented as mean ± SEM from three biological replicates per genotype. *P < 0.05, NS—non‐significant using Student’s t‐test. (J) Representative FACS plots and percentage of intracellular staining of Forkhead box protein P3 (FOXP3), Interferon gamma (IFN‐γ) and IL‐17A in CD3+CD4+ T cells from colonic lamina propria of COPS8fl / fl or COPS8ΔIEC mice with DSS‐induced colitis. The column graph represents percentage of Treg+ and ratio of Th1/Th17 cells, represented as mean ± SEM from three biological replicates per genotype. *P < 0.05 using Student’s t‐test.
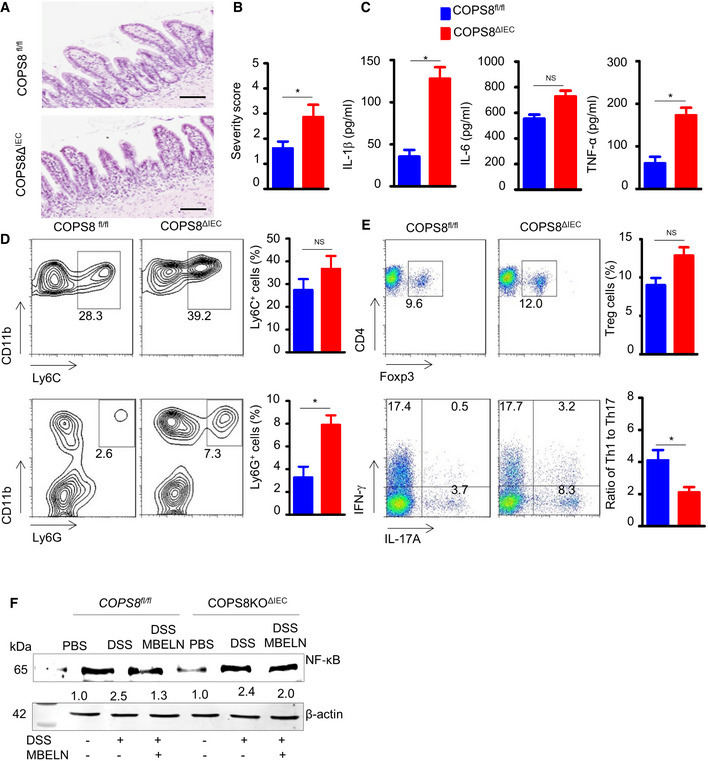
To assess the significance of IEC‐intrinsic COPS8 expression in the context of intestinal damage and inflammation, a DSS‐induced mouse colitis model was used. After the administration of DSS (2%) for 7 days, COPS8ΔIEC mice showed significantly increased mortality and severe weight loss compared to COPS8fl/fl mice (Fig 4D and E). Histological examination of COPS8ΔIEC large intestine revealed severe inflammation with massive cellular infiltration into the mucosa, and loss of crypts and surface epithelium, particularly in the cecum (Fig 4F), which is in sharp contrast to the minimally affected large intestine of COPS8fl/fl mice. The severity of the colitis in COPS8ΔIEC mice is also reflected by the markedly increased colitis score and the shortened length of the colon (Fig 4G). Consistent with increased inflammation, ELISA of the COPS8ΔIEC colon showed significantly increased secretion of the pro‐inflammatory cytokines IL‐1β and IL‐6 compared to control mice (Fig 4H). There were significantly more CD11b+Ly6G+ neutrophils in the colon of COPS8ΔIEC mice (Fig 4I). Correlated to severe inflammation, a higher Treg frequency was observed in colonic lamina propria (cLP) of COPS8ΔIEC mice than that of control mice (Fig 4J). The ratio of Th1 cells compared toTh17 cells, as indicated by the respective production of interferon gamma (IFN‐γ) and IL‐17 in CD4+ T cells, was very low in COPS8ΔIEC mice (Fig 4J). Moreover, histological analysis of the distal ileum demonstrated a notably larger area with the loss of crypts and increased cellularity in the lamina propria (Fig EV4A and B). The increased susceptibility to ileitis in the absence of COPS8 in IECs was further supported by increased production of pro‐inflammatory cytokines IL‐1β, IL‐6, and tumor necrosis factor‐α (TNF‐α) (Fig EV4C), by elevated infiltration of the lamina propria with neutrophils (Fig EV4D) and an increased Th1/Th17 cell ratio (Fig EV4E). These data suggested that COPS8ΔIEC mice were more susceptible than COPS8fl/fl mice to intestinal inflammation, which is accompanied by reduced AMP secretion by Paneth cells.
Figure EV4. Deficiency of COP9/COP9 Constitutive Photomorphogenic Homolog Subunit 8 (COPS8) increases susceptibility to dextran sodium sulfate (DSS)‐induced colitis.
-
A, BRepresentative image of histological analysis of the distal ileum of COPS8fl / fl and COPS8ΔIEC mice by hematoxylin and eosin (HE) staining (A) and column graph (B) showing cellularity (severity score) in the lamina propria. Scale bar 100 μm. Data are represented as mean ± SEM from seven biological replicates per genotype. *P < 0.05 using Student’s t‐test.
-
CProduction of proinflammatory cytokines interleukin (IL)‐1β, IL‐6 and tumor necrosis factor‐α (TNF‐α) in intestinal epithelial cells (IEC) of COPS8fl / fl and COPS8ΔIEC mice. Data are represented as mean ± SEM from seven biological replicates per genotype. *P < 0.05 using Student’s t‐test.
-
DNeutrophils, CD11b+‐Ly6C+ (upper panel) and CD11b+‐Ly6G+ (lower panel), infiltration in colonic lamina propria of COPS8fl / fl and COPS8ΔIEC mice. Column graph showing percentage of neutrophils CD11b+‐Ly6C+ and CD11b+‐Ly6G+ presented as mean ± SEM from seven biological replicates per genotype. *P < 0.05, NS—non‐significant using Student’s t‐test.
-
EPercentage of Treg cells (upper panel) and ratio of Th1/Th17 cells (lower panel) colonic lamina propria of COPS8fl / fl and COPS8ΔIEC mice. Data are represented as mean ± SEM from three biological replicates per genotype. *P < 0.05, NS—non‐significant using Student’s t‐test.
-
FWestern blot for nuclear factor kappa B (NF‐κB) in Villin‐Cre and COPS8‐lox alleles expressing (COPS8fl / fl ) and IEC COPS8 knockout (COPS8ΔIEC ) mice colon tissue following treatment with phosphate‐buffered saline (PBS), DSS and DSS with mulberry bark‐derived exosome‐like nanoparticles (MBELNs). Data are mean ± SEM. *P < 0.05, using Student’s t‐test and one‐way ANOVA.
Source data are available online for this figure.
Induced intestinal inflammation is accompanied by altered fecal gut microbiota composition in COPS8ΔIEC mice
Paneth cells are a primary source for releasing AMPs and preventing dysbiosis (Peterson & Artis, 2014). COPS8 KO leads to Paneth cell dysfunction. A 16S rRNA gene‐based microbiota sequencing analysis showed that the fecal microbiota derived from COPS8ΔIEC mice clustered apart from the COPS8fl/fl control (Fig 5A). The microbial composition was strikingly different between COPS8fl/fl and COPS8ΔIEC mice and significantly altered in COPS8ΔIEC mice (Fig 5B). Out of 11,160 operational taxonomic units, the top 30 genera were differentially represented in feces between COPS8fl/fl and COPS8ΔIEC mice (Fig 5C). Intriguingly, metagenomic analysis of gut microbiota revealed that the abundance of Bacteriodetesis was similar in COPS8fl/fl and COPS8ΔIEC mice. However, COPS8 deficiency led to a high abundance of Gram(−) bacteria related to Proteobacteria and a to lower abundance of Gram(+) bacteria related to Firmicutes in feces (Fig 5C). Notably, linear discriminant analysis effect size (LEfSe) analysis identified Beta‐Proteobacteria mostly from Gram(−) Proteobacteria as specific for the microbiota in COPS8ΔIEC mice (Fig 5C), whereas levels of Clostridiales related to Gram(+) Clostridia were relatively higher in COPS8fl/fl mice (Fig 5D ).
Figure 5. COP9/COP9 Constitutive Photomorphogenic Homolog Subunit 8 (COPS8) deficiency modifies gut microbiota composition and bacterial diversity that contributes to intestinal inflammation.
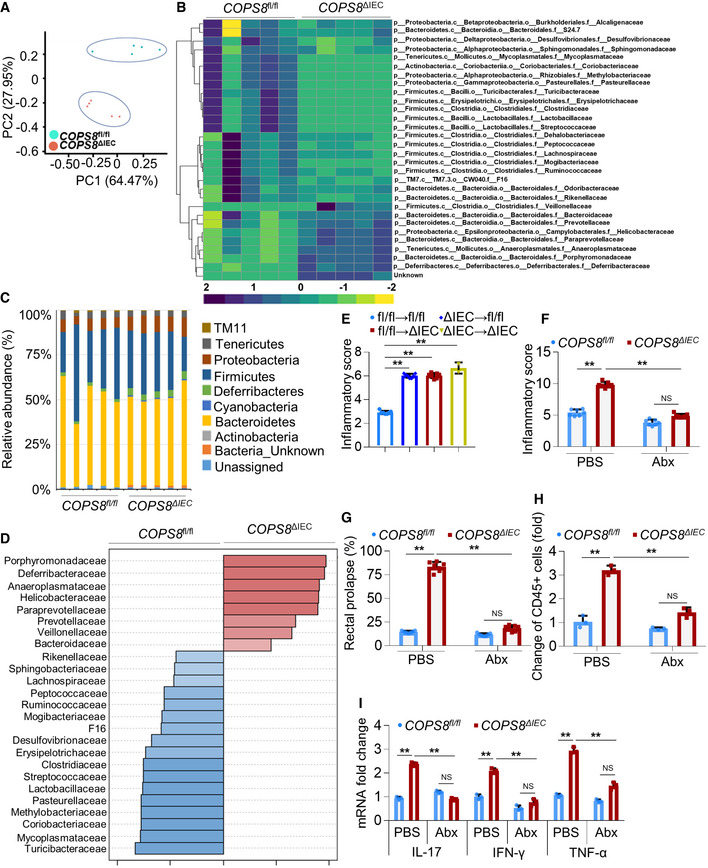
- Principal components analysis of 16S rRNA gene‐sequencing analysis of gut microbes obtained from Villin‐Cre and COPS8‐lox alleles expressing (COPS8fl / fl ) and COPS8 knockout (KO) (COPS8ΔIEC ) mice. PC1 and PC2 explain the 64.47 and 27.95% variation, respectively. Representative data from five biological replicates per genotype.
- Heatmap of differentially represented bacterial species in feces between COPS8fl / fl and COPS8ΔIEC mice. The top 30 genera that are shared by all of the samples shown. The scale was the percentage composition (log2) based on the 16S rRNA gene sequences analyzed using the Greengene database (gg_otus2013). The hierarchical clustering based on Spearman rank correlation (average linkage method) was performed using Gene Cluster 3.0 and images generated on Java Treeview. Representative data from five biological replicates per genotype.
- The relative abundance of bacteria phylum in the feces of COPS8fl / fl and COPS8ΔIEC mice. Representative data from five biological replicates per genotype.
- LEfSe analysis was applied to identify high‐dimensional biomarkers that discriminate between feces from COPS8fl / fl and COPS8ΔIEC mice. Representative data from five biological replicates per genotype.
- Histologic severity scores of COPS8fl / fl mice and COPS8ΔIEC after fecal transplantation. Data represents the histological score of mice analyzed on day 160. Data are represented as mean ± SEM from seven biological replicates per genotype. **P < 0.01 using one‐way ANOVA.
- Inflammation scores in the colon of 5‐month‐old COPS8fl / fl and COPS8ΔIEC mice. Data are represented as mean ± SEM from seven biological replicates per genotype. **P < 0.01, NS—non‐significant using one‐way ANOVA.
- Percentage of rectal prolapse of 5‐month‐old mice COPS8fl / fl and COPS8ΔIEC . Data are represented as mean ± SEM from seven biological replicates per genotype. **P < 0.01, NS—non‐significant using one‐way ANOVA.
- Flow cytometric analysis of CD45+ infiltrating immune cells in the lamina propria of the ileum. Data are represented as mean ± SEM from three biological replicates per genotype. **P < 0.01, NS—non‐significant using one‐way ANOVA.
- Real‐time quantitative reverse transcription polymerase chain reaction (qRT–PCR) analysis of the expression of genes encoding inflammatory cytokines in the ileum. COPS8fl / fl and COPS8ΔIEC mice were either untreated (phosphate‐buffered saline (PBS)) or treated with the antibiotic cocktail (Abx) for 3 months. Data are mean ± SEM from three biological replicates. **P < 0.01, NS—non‐significant using one‐way ANOVA.
To determine whether bacterial dysbiosis plays a causal role in the exacerbated intestinal inflammation in COPS8ΔIEC mice, we performed fecal transfer experiments with COPS8ΔIEC and COPS8fl/fl mice to study alterations in inflammation. COPS8fl/fl mice that received microbiota from COPS8ΔIEC mice showed more intestinal inflammation when compared to mice only receiving COPS8fl/fl microbiota, as indicated by a higher histological score (Fig 5E). To test whether microbiota directly affects intestinal inflammation in COPS8ΔIEC mice, mice were treated with antibiotics. Antibiotic treatment led to a large decrease in both inflammation scores and rectal prolapse in the colon of COPS8ΔIEC mice (Fig 5F and G). Strikingly, antibiotics suppressed both immune cell infiltration (Fig 5H) and pro‐inflammatory cytokine production (Fig 5I) in the ileum of COPS8ΔIEC mice. These data indicate that the more severe intestinal inflammation in COPS8ΔIEC mice is likely due to dysbiosis of gut microbiota‐induced intestinal inflammation.
Deletion of IEC COPS8 does not lead to protection of COPS8 KO mice treated with MBELNs against DSS‐induced intestinal inflammation
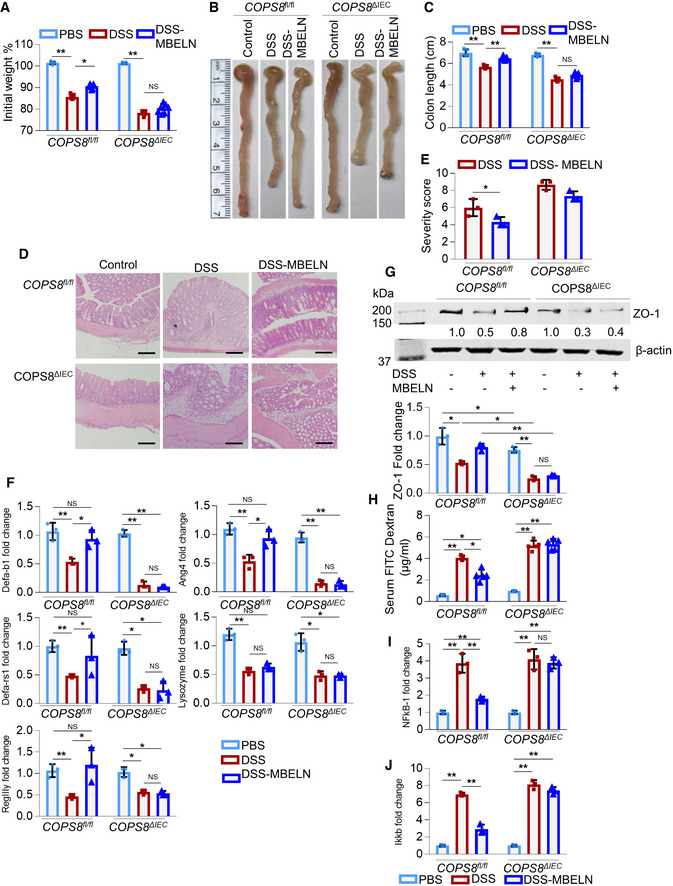
To further test whether COPS8 plays a role in MBELN‐mediated protection of mice against DSS‐induced colitis, we treated COPS8fl/fl mice and COPS8ΔIE mice with 2% DSS with and without MBELNs for seven consecutive days to induce colitis. In COPS8fl/fl mice, treatment with MBELNs prevented weight loss, shortening of the colon, and inflammation upon DSS induction of colitis. However, in COPS8ΔIE mice treated with MBELNs, none of these beneficial effects were evident (Fig 6A–E). COPS8fl/fl mice treated with MBELNs showed enhanced expression of AMPs, whereas there was no induction of AMPs in the colon of COPS8ΔIE mice (Fig 6F). Collectively, the results suggest that COPS8 is essential for MBELN‐mediated induction of AMPs and prevents DSS‐induced colitis. Western blot analysis showed that tight junction proteins ZO‐1 (Zonula occludens‐1 ZO‐1) that play an important role in maintaining gut barrier function are upregulated in the small intestine epithelium of MBELN‐treated COPS8fl/fl mice, while there was no evidence of upregulation in COPS8ΔIE mice (Fig 6G). This result also is in agreement with the data generated using serum FITC‐dextran assessment (Fig 6H), suggesting that gut epithelial COPS8 induced by MBELNs plays a role in enhancing gut barrier function. Analysis of ZO‐1 protein expression in COPS8fl/fl and COPS8ΔIE mice colon tissue demonstrated that COPS8ΔIE mice showed lower but not significantly different ZO‐1 expression compared to COPS8fl/fl mice, while DSS‐induced gut injury significantly reduced ZO‐1 expression in COPS8ΔIE mice compared to COPS8fl/fl mice. Similarly, gut permeability increased in COPS8ΔIE mice following DSS‐induced injury. This suggests that COPS8ΔIE mice are more vulnerable to DSS‐induced gut injury. MBELN treatment resulted in inhibition of expression of NF‐κB and IKK‐β in the colon of COPS8fl/fl mice; however, COPS8 KO attenuated MBELN‐mediated reduction of NF‐κB and IKK‐β expression (Figs 6I and J, and EV4F). Collectively, the cumulative inhibitory effect of MBELNs on gut inflammation via COPS8‐mediated release of AMP and inhibition of the inflammatory pathway could play a pivotal role in the prevention of colitis.
Figure 6. Intestinal epithelial COPS8 is required for MBELN‐mediated protection against mouse colitis.
-
AGraph showing loss of weight following oral dextran sodium sulfate (DSS) (2%) treatment with and without mulberry bark‐derived exosome‐like nanoparticles (MBELNs) for 7 days (10 × 109 particles/dose/day/mouse) once daily for 7 days in Villin‐Cre and COP9/COP9 constitutive photomorphogenic homolog subunit 8 (COPS8)‐lox alleles expressing (COPS8fl / fl ) and COPS8 knockout (KO) (COPS8ΔIEC ) mice. Changes in body weight are presented as percent of initial weight determined using Mann–Whitney test, *P < 0.05, **P < 0.01, NS—non‐significant. Representative data from seven biological replicates per genotype.
-
BRepresentative image showing changes in colon morphology and length following DSS‐induced colitis with concurrent treatment with MBELNs COPS8fl / fl and COPS8ΔIEC mice. Representative data from seven biological replicates per genotype.
-
CColumn graph showing changes in colon length. Data are mean ± SEM from seven biological replicates. **P < 0.01, NS—non‐significant using one‐way ANOVA.
-
D, EHematoxylin and eosin staining (HE) to show histological changes (D) and graph showing severity scores based on histological data (E). Scale bar 200 μm, column graph represents colon severity score, presented as mean ± SEM from three biological replicates per genotype. *P < 0.05, NS—non‐significant using Student’s t‐test.
-
FExpression of anti‐microbial peptide (AMPs) in the colon of COPS8fl / fl and COPS8ΔIEC mice while being treated with MBELNs by oral administration. Data are mean ± SEM from three biological replicates per group. *P < 0.05, **P < 0.01, NS—non‐significant using one‐way ANOVA. (G) Protein expression tight junction protein Zonula occludens‐1 (ZO‐1) using Western blotting in colonic intestine epithelial cells. Data are mean ± SEM from three biological replicates per group. *P < 0.05, **P < 0.01, NS—non‐significant using one‐way ANOVA.
-
HMeasurement of serum fluorescein isothiocyanate (FITC)‐dextran to evaluate the permeability of the intestines. Data are mean ± SEM from five biological replicates per group. *P < 0.05, **P < 0.01 using one‐way ANOVA.
-
I, JTranscriptional expression of nuclear factor kappa B (NF‐κB) (I) and inhibitor of nuclear factor kappa‐β kinase (IKK‐β) (J) mRNA using real‐time quantitative reverse transcription polymerase chain reaction (qRT–PCR) in colonic epithelial cells upon treatment with MBELNs in addition to DSS. Data are mean ± SEM from three biological replicates per group. **P < 0.01, NS—non‐significant using one‐way ANOVA.
Source data are available online for this figure.
MBELN‐derived protein HSPA8 binds to and subsequently activates AhR
To further investigate the factors from MBELNs responsible for the activation of AhR, we screened proteins, lipids, and RNA extracted from MBELNs. Treatment with MBELN‐derived protein extracts led to the activation of AhR (Figs 7A and EV5A and B). Upon treating with different MBELN protein fractions based on molecular weight, we found that proteins ranging from 50 to 100 kDa were responsible for its activity (Figs 7B and EV5C). Surface Plasmon resonance (SPR) analysis indicated that proteins derived from MBELNs bind to His‐tagged recombinant human AhR (His‐hAhR) captured on an NTA chip, demonstrating that AhR directly binds to MBELN‐derived proteins (Fig 7C).
Figure 7. MBELN‐derived HSPA8 protein binds to hAhR and activates AhR.
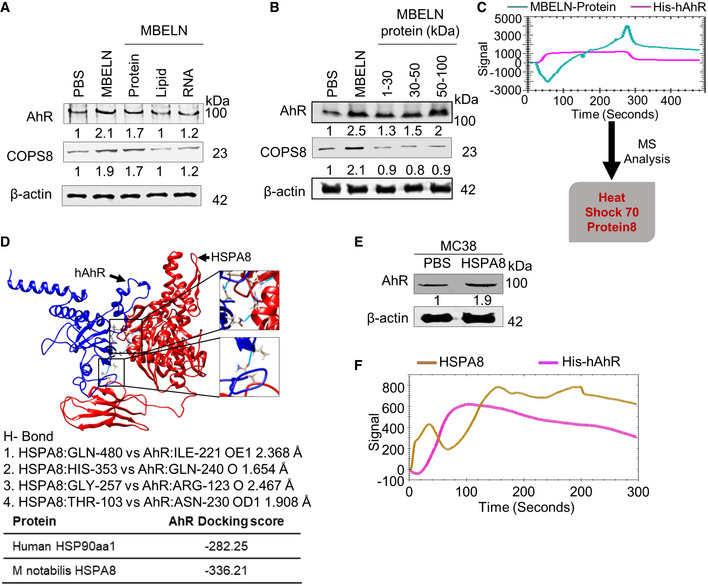
- Western blots showing expression of AhR in MC38 cells induced by different fractions (protein, lipid, and RNA) derived from mulberry bark‐derived exosome‐like nanoparticles (MBELNs). Data shown are from three biological replicates.
- The effect of different size protein fractions of mulberry bark‐derived exosome‐like nanoparticles (MBELNs) on the expression of aryl hydrocarbon receptor (AhR) demonstrated in Western blots. Data shown are from three biological replicates.
- Surface Plasmon resonance (SPR) experiment showing MBELN‐derived protein binding to recombinant human His‐AhR (hAhR) and mass spectrometry (MS) analysis of proteins binding to hAhR on a SPR chip. Data shown are from three biological replicates.
- Computational analysis showing docking of Heat Shock Protein Family A (Hsp70) Member 8 (HSPA8; represented in red color) to hAhR (represented in blue color). Data shown are from three biological replicates.
- MC38 cells treated with MBELN‐HSPA8 showing activation of AhR by Western blot. Data shown are from three biological replicates.
- SPR experiment showing recombinant HSPA8 protein interaction with recombinant hAhR on a SPR chip. Data shown are from three biological replicates.
Source data are available online for this figure.
The MBELN‐derived protein fraction binding to recombinant hAhR on the NTA chip was eluted and analyzed using MS to characterize the proteins contained in MBELNs that bind to AhR. MS analysis revealed that there are several proteins (Dataset EV5) present in the fraction that binds to AhR. Based on a homology analysis with human HSP90aa1, an AhR binding protein that provides stability to AhR (Rothhammer & Quintana, 2019), MBELN‐derived heat shock protein family A (Hsp70) member 8 (HSPA8) had the highest similarity (71.2%) to human HSP90aa1 (Appendix Fig S4A). Subsequently, we performed docking analyses for AhR with HSPA8 and AhR with HSP90 which demonstrated that HSPA8 has a higher binding affinity than homologous protein HSP90, with HSPA8 having a docking score of −336.21 when compared to HSP90 with a docking score of −282.25. Collectively, this analysis demonstrated HSPA8 could be an AhR activating ligand and could be one of the contributors to AhR activation (Fig 7C). As full gene and protein sequences of HSPA8 were available, we computationally analyzed the docking of HSPA8 to AhR. Docking analysis demonstrated that HSPA8 derived from MBELNs has a stronger binding affinity compared to endogenous human HSPA8 (Fig 7D). The amino acid sequence of MBELN‐derived HSPA8 is 35% identical to human HSPA8 (Appendix Fig S4B).
To confirm our result that MBELN‐HSPA8 binds to AhR and induces its activity, we cloned His‐, MBP‐, and FLAG‐tagged mulberry HSPA8 gene into E. coli and expressed a His‐MBP‐FLAG‐tagged mulberry HSPA8 protein (~106.75 kDa) (Fig EV5D). The SPR experiment demonstrated that purified MBELN‐HSPA8 (TEV protease cleaved and purified to eliminate His and MBP tagging) binds to AhR (Fig 7F). The binding of MBELN‐HSPA8 to AhR was confirmed by treating MC38 cells with FLAG‐tagged MBELN‐HSPA8 for 3h and then staining the cells with anti‐FLAG and anti‐pAhR (Fig EV5E). MC38 cells treated with purified HSPA8 confirmed the inducible effect of MBELN‐HSPA8 on AhR and its target genes at a similar level as with MBELNs (Figs 7E and EV5F). Collectively, we showed that MBELN‐derived HSPA8 binds to and subsequently activates AhR. To further determine whether HSPA8 plays a role in MBELN‐mediated deneddylation of CUL1 via the AhR pathway, MC38 cells with and without AhR KO were treated with HSPA8 packed in liposomes made from the total lipids extracted from MBELNs or mouse Hsp70 as a control. The Western blot analysis indicates that cells treated with liposome‐containing mulberry HSPA8 show deneddylation of CUL1, while those treated with liposomes‐containing mouse Hsp70 as a control did not (Fig EV5G).
Discussion
In this study, as proof of concept, edible mulberry plant bark‐derived exosome‐like nanoparticles were used to demonstrate beneficial effects on the mammalian intestinal tract. Four lines of experimentation demonstrated the critical roles of MBELNs in AhR/COPS8‐mediated intestinal epithelial cell protection in a DSS‐induced mouse colitis model. First, the expression as well as activation of AhR is MBELN‐HSPA8 specific, as HSPA8 cannot be replaced with mouse HSP70. Further, we show that MBELN‐HSPA8 is only 35% identical to human HSP70 and it is thus unlikely that human HSP70 has the same biological function as MBELNs, although data published by others indicate that host‐derived HSP70 is constitutively highly expressed in intestinal epithelial cells (Musch et al, 1999; Yang et al, 2002; Kojima et al, 2003; Petrof et al, 2004). Second, our data indicated that HSPA8 is incorporated into mulberry bark‐derived exosomes for targeted delivery to intestinal epithelial cells. Third, impacting on the AhR pathway, MBELNs selectively activate COP9/COPS8 in intestinal epithelial cells. This was further supported by the observation that AhR KO abolished COPS8 induction. Fourth, induced COP9/COPS8 is essential for MBELN‐mediated prevention of DSS‐induced mouse colitis via the induction of an array of AMPs. Based on the data presented in this study, we propose a model in which MBELNs prevent mouse colitis via AhR/COP9/COPS8‐mediated induction of AMPs. Binding of MBELN‐HSPA8 to AhR is required for activation of AhR signaling, and the activity of proteasome degradation pathway cross‐talking with COP9/COPS8 regulates the homeostasis of MBELN‐mediated AhR signaling.
The finding that mulberry bark, that is, MBELN‐derived heat shock protein HSPA8 binding to AhR leads to the induction of COPS8 and the promotion of dennydylation of CUL1 opens up a new avenue for studying the role of plant heat shock proteins in regulating mammalian protein stability. It is well known that Hsp70‐containing molecular chaperone machines are one of the major contributors to prevent stress‐induced protein misfolding, restoring the normal function of aggregated proteins or promoting the degradation of misfolded and aggregated proteins by the proteasome (Wong & Cuervo, 2010; Chen et al, 2011; Hipp et al, 2014; Balchin et al, 2016) if the aggregated proteins cannot be functionally restored. Defects in any of these mechanisms can cause severe proteotoxicity, which in turn can lead to cancer and neurodegenerative disorders (Balch et al, 2008; Chen et al, 2011; Hipp et al, 2014; Schmidt & Finley, 2014; Labbadia & Morimoto, 2015). It is conceivable that many other identified and unidentified factors including HSP70 are involved in these complicated mechanisms. Our findings provide a rationale for further studying whether HSPA8 treatment can prevent certain diseases as described above via regulating the COP9/AhR/proteasome axis.
The COP9 signalosome (COP9) is highly conserved in all eukaryotes, from fungi to humans (Lingaraju et al, 2014). It is composed of eight canonical subunits named COPS1 through COPS8, according to their descending molecular weights (Kwok et al, 1998). Depletion of any of the subunits impairs Cullin deneddylation, demonstrating that the deneddylase activity of COP9 requires the formation of the COP9 holocomplex by the eight subunits. The deneddylase activity of COP9 resides in COPS5, but COPS5 exerts Cullin deneddylation activity only when it is situated in the fully assembled COP9 holocomplex consisting of all eight subunits, which is why the loss of any of the eight COPS subunits abolishes Cullin deneddylation (Wei & Deng, 2003). In this study, we have shown that cullin‐1 deneddylation is increased as a result of MBELNs treatment. MBELNs activate the AhR pathway which induces the expression of COPS8, and KO of COPS8 leads to decreasing cullin‐1 deneddylation. However, we cannot exclude the possibility of MBELNs regulating the cullin‐1 deneddylation activity of COP9 holocomplex also via other COPS subunits.
AMPs have key roles in plant defense via their microbicidal activity. COP9 interacts with AMP in plant cells (Hind et al, 2011; Damon et al, 2012). COPS is required for proper plant development and has a profound effect on plant defense responses. Silencing of genes for COPS subunits in tomato plants resulted in a mild morphological phenotype and reduced expression of wound‐responsive genes in response to mechanical wounding, attack by Manduca sexta larvae, and Prosystemin over‐expression (Hind et al, 2011; Damon et al, 2012). Whether mammalian COP9 plays a role in regulating production of antimicrobial peptides is not known.
Paneth cells are specialized intestinal epithelial cells that are located at the base of small intestinal crypts and play a vital role in preserving gut epithelium homeostasis. The dense granules produced by Paneth cells contain an abundance of antimicrobial peptides and immunomodulating proteins that function to regulate the composition of the intestinal flora. Paneth cells act as a safeguard from bacterial translocation across the epithelium and constitute the niche for intestinal stem cells in the small intestine by providing multiple niche signals. Recently, Paneth cells have become the focal point of investigations defining the mechanisms underlying epithelium–microbiome interactions, the pathogenesis of chronic gut mucosal inflammation and bacterial infection (Bevins & Salzman, 2011; Farin et al, 2014; Lueschow & McElroy, 2020; Yu et al, 2020). In this study, we found that knockout of intestinal epithelial‐specific COP9/COPS8 causes a sharp reduction of Paneth cells, suggesting that COPS8 is essential for the survival of Paneth cells. Since COP9/COPS8 has multiple functions including regulating cell proliferation, apoptosis, and differentiation, this finding opens a new avenue for further studying COP9 holocomplex‐mediated molecular pathways that contribute to regulation of Paneth cell biology in general, and moreover, how MBELNs mediate the AhR signaling pathway to promote the expression of Paneth COPS8 is warranted.
Any conclusion related to the role of HSPA8 in the context of MBELNs must be cautiously drawn. We cannot exclude other MBELN factors that may work with HSPA8 to regulate the intracellular fate of MBELNs. In general, the intracellular fate of MBELNs after internalization may follow the typical endosomal pathway, from early endosomes as sorting compartments to acidic vesicles, that is, late endosomes and MVBs, which fuse with lysosomes, eventually undergoing degradation. However, ELN cargoes likely bypass degradation, as various studies have demonstrated ELN‐mediated functional changes in recipient cells. The function of several potential intracellular sites could be temporarily altered due to MBELN internalization. For instance, the endoplasmic reticulum (ER), which is a nucleation site for translation, could be a route for lysosomal escape enabling cargo release, as ER scanning can occur after ELN sorting into the endosomal trafficking circuit. This would be a route of choice for MBELNs to release their cargoes in the ER for rapid translation and mediation of altered gene expression. ELNs could also be able to use pathways similar to viruses to avoid lysosomal degradation. Other possible routes that allow ELNs to escape lysosomal degradation include redirection of ELN cargoes from the endosomal pathway to the trans‐Golgi network through retrograde trafficking. ELNs can also be redirected back to the plasma membrane of early endosomes via recycling the endosomes. Hence, MBELN cargoes besides HSPA8 could play a role in bypassing direct lysosomal degradation to fulfill HSPA8‐mediated activation of the AhR signaling pathway. Here, we show that HSPA8 can activate the AhR signaling pathway, which occurs after MBELNs escape the degradation of HSPA8. This finding provides a foundation for further studies regarding whether other MBELN‐derived factors work together with HSPA8 and contribute to the escape mechanism.
Our finding that COP9/COPS8 regulates the induction of an array of AMPs will have broad preventive and therapeutic applications. Bacterial drug resistance leads to serious health problems worldwide due to the long‐term use and abuse of traditional antibiotics, leading to drug‐resistant bacteria. AMPs are essential components of the immune defenses of multicellular organisms and are widely distributed throughout the animal and plant kingdoms (Zasloff, 2002). AMPs have been demonstrated to have their own advantages over traditional antibiotics with a broad spectrum of anti‐microbial activities including anti‐bacterial, anti‐fungal, anti‐viral, and anti‐cancer activities, and even overcoming bacterial drug resistance (Zhang & Gallo, 2016). Our findings demonstrate that oral administration of MBELNs leads to the induction of an array of AMPs and the AhR/COP9/COPS8 pathways opens a way to a range of clinical applications that could protect against a broad array of inflammatory signals, infection‐causing agents, such as bacteria, fungi, parasites and viruses and also exhibits immunomodulatory activity.
Our findings also demonstrate that MBELNs selectively interact with L. monocytogenes‐EGD‐derived proteins compared to non‐virulent L. monocytogenes. This set of MS data provides a foundation to further determine which pathogenic L. monocytogenes protein(s) is/are targeted by MBELNs to inhibit L. monocytogenes growth.
Materials and Methods
Isolation and purification of exosome‐like nanoparticles from the mulberry bark, leaf, and fruit
Bark, that is, the tissues outside the vascular cambium, from mulberry stems were manually removed using a chisel, washed with running water, and soak into phosphate‐buffered saline (PBS). The bark was cut into small pieces followed by electronic blending at high speed for 5 min. The large particulates were filtered out using a strainer which resulted in a slurry. The slurry from the bark was differentially centrifuged to obtain ELNs from bark (MBELNs) as mentioned earlier (Wang et al, 2013). In brief, the slurry was serially centrifuged at 1,000 g for 10 min, 3,000 g for 20 min, and 10,000 g for 40 min to remove large particulates. The supernatant was then centrifuged at 150,000 g for 2 h. The pellet was resuspended in sterile PBS. The purification of ELNs was performed using a sucrose density gradient (8, 30, 45, and 60% sucrose in 20 mM Tris–Cl, pH 7.2) followed by centrifugation at 150,000 g for 2 h at 4°C. The band between the 30 and 45% layers was collected. To confirm the bilayer structure of the ELNs, they were imaged and analyzed using electron microscopy (Zeiss EM 900). ELN size distribution and concentration were determined using a NanoSight NS300 (Malvern Instrument).
MBELN lipid analysis
Lipid was isolated from resuspended MBELNs using the chloroform–methanol method (Wang et al, 2013). One volume of resuspended MBELNs was mixed with three volumes of chloroform: methanol (2:1 v/v) by manual shaking until the MBELNs were dispersed in the solvent. Additionally, one volume of chloroform was added and mixed. Finally, one volume of water was added, mixed, centrifuged at 2,000 g for 10 min at room temperature and the bottom layer carefully collected. Lipids were dried on a heating block under nitrogen gas. Lipids were analyzed by thin‐layer chromatography using HPTLC‐plates (silica gel 60 with concentrating zone, 20 cm × 10 cm; Merck) with a mobile phase of chloroform: methanol: H2O (65:25:4) and detected under an iodine vapor. To characterize the lipid composition, the lipid samples were analyzed using a triple quadrupole mass spectrometer (Applied Biosystems Q‐TRAP, Applied Biosystems, Foster City, CA) at the Lipidomics Research Center, Kansas State University (Manhattan, KS) as described earlier (Wang et al, 2013). The data are presented as nmol/mg dry weight of total lipid.
MBELN protein analysis
The MBELN pellet was mixed with lysis buffer (100 mM Tris–Cl (pH 6.8), 4% (w/v) sodium dodecyl sulfate (SDS); 200 mM dithiothreitol (DTT)) containing protease inhibitors at 4°C and vortexed for 15 s and kept on ice for 15 min. The mixture was centrifuged at 12,000 g for 15 min at 4°C. The supernatant was discarded, and the pellet was retained, stored at −80°C for further downstream processing. For SDS–PAGE analysis, 40 µg of sample were mixed with Laemmli sample buffer, boiled for 5 min, and loaded on 10% SDS–PAGE gel. The gels were stained with Coomassie blue.
Bacterial culture, treatment, and MBELN uptake by bacterial cells
Listeria (L.) monocytogenes (non‐virulent), L. monocytogenes‐EGD (virulent strain), and Streptococcus (S) gordonii were cultured in Brain‐Heart Infusion (BHI) broth containing 0.5% yeast extract, Escherichia coli was cultured in LB Media (Luria–Bertani) broth, and Porphyromonas (P) gingivalis 33277 was cultured in tryptic soy broth supplemented with yeast extract (1 mg/ml), hemin (5 μg/ml) and menadione (1 μg/ml). Both strains of L. monocytogenes were cultured in an aerobic chamber while S. gordonii and P. gingivalis were cultured in anaerobic chamber at 37°C.
MBELNs were labeled with PKH26 Red Fluorescent Cell Linker Kit (SIGMA) as per the manufacturer’s instructions. L. monocytogenes and L. monocytogenes‐EGD bacteria were incubated with PKH26 labeled MBELNs (1 × 109 particles/ml) for 3 h at 37°C and cells were harvested by centrifugation at 5,000 g for 15 min at 4°C. The uptake of MBELNs by L. monocytogenes and L. monocytogenes‐EGD was determined by flow cytometry (BDFACSCanto™, Becton Dickinson).
Listeria proteomic analysis
Protein from L. monocytogenes and L. monocytogenes‐EGD was isolated by suspending bacterial cells in lysis buffer (2% SDS, 100 mM DTT, 20 mM Tris–HCl, pH 8.8) followed by sonication at 200 W. Protein lysate was isolated after centrifugation at 12,000 g for 20 min at 4°C. MBELNs were labeled with biotin using an EZ‐Link™ Sulfo‐NHS‐Biotinylation Kit according to the manufacturer’s protocol (Thermo Fisher Scientific). The biotinylated MBELNs were incubated with bacterial protein lysate overnight at 4°C on a rotary shaker. Then, the MBELN‐biotin‐protein lysate mixture was incubated with streptavidin magnetic beads for 1 h at 4°C. Biotin‐bound magnetic beads were washed and cleaned thrice with lysis buffer to remove unbounded protein. Streptavidin magnetic beads were re‐suspended in fresh lysis buffer (2% SDS, 100 mM DTT, 20 mM Tris–HCl pH‐8.8) at 95°C for 20 min. Eluted proteins were run on a PAGE gel and analyzed by MS.
mRNA Seq
RNA was extracted from purified MBELNs using a Qiagen RNeasy Plant mini kit (Qiagen, 74904) and from bacteria using a Rneasy Mini Kit (Qiagen, 74104). An RNA‐Seq library was prepared using a QiaSEQ RNA library kit per the manufacturer’s instructions. The mRNA‐seq sequencing was performed on an Illumina instrument using high output NextSeq sequencing kits (FC‐404‐2005).
MBELN uptake by bacterial cells
MBELNs were labeled with PKH26 Red Fluorescent Cell Linker Kit (SIGMA) as per the manufacturer’s instructions. L. monocytogenes and L. monocytogenes‐EGD bacteria were incubated with PKH26 labeled MBELNs (1 × 109 particles/ml) for 3 h at 37°C and cells were harvested by centrifugation at 5,000 g for 15 min at 4°C. The uptake of MBELNs by L. monocytogenes and L. monocytogenes‐EGD was determined by flow cytometry (BDFACSCanto™, Becton Dickinson).
Animals
All animal procedures were approved by the University of Louisville Institutional Animal Care and Use Committee. All the mice were housed in a pathogen‐free facility on a 12‐h light/dark cycle, unless otherwise specified. C57BL/6J were obtained from Jackson Laboratories. The COPS8‐floxed mouse model (COPS8flox/flox) was originally created as described previously (Menon et al, 2007). To delete COPS8 in IECs, Villin‐Cre+/COPS8flox/flox mice (termed COPS8ΔIEC ) were generated by crossing COPS8flox/flox mice with Villin‐Cre transgenic mice. The Villin‐Cre mice were in a C57BL/6 background. Littermate Villin‐Cre‐ / COPS8flox/flox mice (termed COPS8fl/fl ) were obtained and used as controls.
Animals and treatments
All animal procedures were approved by the University of Louisville Institutional Animal Care and Use Committee. All the mice were housed in a pathogen‐free facility on a 12‐h light/dark cycle.
The mouse model of colitis was chemically induced in age‐matched (8–12 weeks) mice using 2% (w/v) dextran sulfate sodium salt (DSS; molecular weight: 36–50 kDa; MP Biomedicals) in the drinking water for 7 days. Water was changed every third day. The MBELN group mice were administered MBELNs orally (10 × 109 particles/100 µl/ mouse) once every day in addition to DSS in the drinking water. Mice were sacrificed on day 7 and tissue samples were snap‐frozen and stored at −80°C for downstream processing. Mice were checked for body weight, stool softness, diarrhea, and blood in stools on a daily basis. A disease severity score was calculated as described previously (La et al, 1997). For the survival assay, mice were followed up to 12 days.
At the endpoint of the DSS‐induced colitis experiment, intestinal permeability was evaluated by oral administration of FITC‐dextran (44 mg/100 g body weight). After 4 h, mice were anesthetized by isoflurane inhalation and 1 ml blood was collected by cardiac puncture then the mice were sacrificed. Serum was collected by centrifugation, diluted with an equal volume, and 100 µl diluted serum in duplicates was used to determine the concentration of FITC in serum by fluorometry spectrophotometer with an excitation of 485 nm and an emission wavelength of 528 nm.
In vivo trafficking of MBELNs
In vivo trafficking of MBELNs was demonstrated using DiR‐labeled MBELNs. The Dir‐labeled MBELNs were orally administered as a single dose of 10 × 109 particle/100 µl/mouse. Mice were examined in vivo at 3, 6, 12, and 24 h after administration. The mice were humanly sacrificed at designated times and organs were excised for ex vivo imaging of respective organs using an Odyssey CLx (LI‐COR Biosciences) imaging system.
To confirm the localization of MBELNs at the cellular level in different organs, mice were orally gavaged with 10 × 109 PKH26 labeled MBELNs and were sacrificed 3 h later. All of the internal organs were excised and fixed overnight in periodate‐lysine paraformaldehyde (PLP), transferred to a 30% sucrose solution for 6 h, and frozen in OCT at −80°C. Tissue samples were cut at 5 µm thickness and immuno‐stained with appropriate primary antibodies (1:200). Primary antibodies were detected by Alexa Fluor 488‐ and 594‐conjugated goat anti‐mouse, anti‐rabbit IgG (1:600, Invitrogen). Sections were counterstained with 4,6‐diamidino‐2‐phenylindole (DAPI), and images were captured on a Nikon confocal microscope equipped with a digital image analysis system (Nikon).
Cell culture and treatment
Cell lines used in this study were the mouse colon cancer MC38 cell line and the human Caco2 colon cancer cell line that were originally obtained from ATCC and maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 100 U/ml penicillin/streptomycin. The Hepa1.1, mouse hepatoma cell line harboring the stably integrated pAhRDtkLuc3 reporter gene construct was cultured in α‐modified essential media (Sigma) supplemented with 10% FBS. All cells were grown in a humidified atmosphere of 5% CO2 at 37°C. In vitro cells were treated with MBELNs at a concentration of 5 × 109 particles/ml for 3 h unless otherwise stated.
AhR KO in MC38 cells
MC38 cells (2 × 105 cells/well) in a six‐well plate were transfected with AhR CRISPR/Cas9 KO Plasmid (sc‐419054, Santa Cruz Biotechnology Inc. Dallas, TX, USA) using Lipofectamine™ 3000 Transfection Reagent (Invitrogen, L3000001). Following 72 h of transfection, cells were confirmed as eGFP positive cells by confocal fluorescence microscopy (Nikon). The eGFP‐positive cells were sorted and cultured in complete DMEM supplemented with 10% FBS and 100 U/ml penicillin/streptomycin. AhR knockout was confirmed by Western blot analysis.
Immunocytochemistry and immunohistochemistry
For immunocytochemistry MC38 and Caco2 cells (6 × 104 cells/well) were grown on four‐chamber slides and cultured for 24 h at 37°C. Upon 70–80% cell confluency, chambers had spent media replaced with fresh complete media and treated with MBELNs (5 × 109 particles/ml) for 6 h in a 5% CO2 incubator at 37°C. Cells were then fixed with 2% paraformaldehyde in PBS for 20 min at room temperature, followed by permeabilization using 0.2% Triton X‐100 for 15 min.
For tissue immunofluorescence analysis, tissue was excised and fixed in periodate‐lysine paraformaldehyde (PLP) and frozen in OCT at −80°C. Tissue samples were cut at 5 µm thickness, immuno‐stained, and the slides incubated with designated primary antibody (AhR (Invitrogen), Phospho‐AhR (Invitrogen), COPS8 (Abcam), Mucin‐2 (MUC2, Abcam), Lysozyme C (Abcam), and F4/80 (Santacruz)) overnight at 4°C in a humidified chamber. Tissues were washed and stained with anti‐mouse/rabbit fluorescein isothiocyanate–Alexa Fluor 488/594 secondary antibody and DAPI for confocal microscope examination.
For histology, tissue specimens were fixed in 10% formalin, dehydrated, and then embedded in paraffin. Tissue samples were cut at 5 µm thicknesses and stained with hematoxylin and eosin. Histologic grades and inflammation were assessed with a modified scoring system (Read et al, 2000). Each sample was graded semi‐quantitatively from 0 to 3 for the four following criteria: degree of goblet cell depletion and epithelial hyperplasia; leukocyte infiltration in the lamina propria; area of tissue affected and the presence of markers of severe inflammation such as crypt abscesses, submucosal inflammation, and ulcers. Scores for each criterion were added to give an overall inflammation score for each sample of 0–12.
Electron microscopy
For tissue samples, two‐millimeter tissue pieces were fixed in 2.5% glutaraldehyde in 0.1 M Sorenson’s phosphate buffer and kept at 4°C overnight or for a specific period. After several washes in 0.1 M Sorenson’s phosphate buffer, the blocks after embedded in epoxy resin, 300–400 nm semi‐thin sections were cut with good orientation of the samples; then, 80–90 nm sections were cut on a Reichert Ultracut E ultramicrotome. The thin sections were transferred onto 150‐mesh copper grids before staining with uranyl acetate and lead citrate. The sections were viewed under a JEOL 1011 transmission electron microscope with a GATAN Erlangshen CCD (charge‐coupled device) camera.
RNA extraction and PCR
Total RNA was isolated from the cells, tissue, or exosomes using a RNeasy Mini Kit (Qiagen, 74104) according to the manufacturer’s instructions. cDNA was synthesized from RNA (1μg) and reverse‐transcribed using SuperScript III reverse transcriptase and oligo dT primers (Invitrogen). For quantitation of genes of interest, cDNA samples were amplified in a CFX96 Realtime System (Bio‐Rad) using SYBR Green Master Mix (Invitrogen) and specific primers (Appendix Table S1) according to the manufacturer's instructions. Fold changes in mRNA expression between treatments and controls were determined by the δCT method. Results for each sample were normalized to the concentration of GAPDH mRNA measured in the same samples and expressed as fold increase over control/baseline levels, which are set at a value of 1. Differences between groups were determined using a two‐sided Student's t‐test and one‐way ANOVA. Error bars on plots represent ± SEM, unless otherwise noted.
16S rDNA sequencing
Mice were sacrificed and feces were collected from both small intestine and colon. A small sample of feces was snap‐frozen and stored at −80°C. Total genomic DNA was isolated using a QIAamp DNA Microbiome Kit (Qiagen, 19091) as per the manufacturer’s instructions. 15 ng of DNA was used as template to amplify 16S rRNA gene using High Fidelity PCR system kit (Roche). The v3‐v4 regions of 16S ribosomal RNA gene were amplified using primer 319F (ACTCCTACGGGAGGCAGCAG) and 806R (GGACTACHVGGGTWTCTAAT) (1 μM). The primers were anchored with adaptor (adopter A: 5′ CCATCTCATCCCTGCGTGTCTCCGACTCAG 3′ and adopter B: 5′ CCTATCCCCTGTGTGCCTTGGCAGTCTCAG 3′) and Multiplex Identifiers (MIDs; 10 bp long). The following PCR protocol was used for amplifying fragments: 95°C for 3 min, followed by 27 cycles of 95°C for 15 s, 58°C for 15 s, 72°C for 15 s, and a final extension at 72°C for 5 min. PCR products were pooled and purified on a 1% TAE ultrapure agarose gel using purification columns (Qiagen) for the generation of Illumina libraries. The amplicon sequence was conducted using the 454 Jr. Sequencing platform. The 16S rRNA gene sequences were analyzed using QIIME platform scripts (www.qiime.org). The sequences were verified at randomly selected 1,500 sequences/sample and downstream analysis was performed. The microbial classification was performed using the GreenGenes reference data base (gg_otus‐13_8) and QIIME tools. The sequences reference picked into Operational Taxonomic Units (OTUs) by clustering 97% sequence similarity (uclust) and classified at various taxonomic ranks (phylum, order, class, family, genus, and species). The beta diversity principle co‐ordinate plots were generated using phylogenetic metrics of UniFrac distances (Lozupone & Knight, 2005). The phylogenetic analysis was performed using Figtree with default parameter. The evolutional tree and percentage for each bacterial species were virtualized by Interactive Tree of Life (iTOL) software.
Immunoblotting
Cells and tissue samples were snap‐frozen and stored at −80°C for later downstream processing. Samples were thawed on ice and proteins were extracted using a modified radioimmunoprecipitation assay (RIPA) buffer (Sigma) in the presence of protease and phosphatase inhibitors (Roche). Protein was separated using 10% SDS–PAGE and transferred to a nitrocellulose membrane. The nitrocellulose membrane was blocked with 5% non‐fat milk and incubated overnight at 4°C with primary antibodies, 1:1,000 dilution in PBST (COPS8‐Abcam; COPS5‐ BIOMOL, COPS6‐Santa Cruz, COPS7‐Santa Cruz, a‐tubulin‐Santa Cruz, β‐actin‐Santa Cruz, AhR‐Invitrogen, ZO‐1‐Invitrogen, CYP1a1‐ Invitrogen, IDO‐1‐Proteintech, CUL1‐Epitomics, and CUL3‐Epitomics) (Appendix Table S2) followed by incubation with goat anti‐mouse/rabbit secondary antibodies conjugated to Alexa Fluor® 790 (Invitrogen) for 1 h at room temperature. The bands were visualized using an Odyssey Imager (LiCor Inc).
For co‐immunoprecipitation, the MC38 cells were treated with the respective agent or vehicle for 3 h at 37°C. The cells were washed with cold PBS three times and lysed in TNE buffer (Tris–HCl 10 mM pH 7.8, NaCl 0.15 M, Nonidet P‐40 1%, EDTA 1 mM and protease inhibitor cocktail complete (Millipore Sigma)). The lysate was centrifuged at 12,000 g for 15 min at 4°C and the supernatant was collected. The supernatant was mixed with AhR antibody (Invitrogen; 1 µg/100 µg protein lysate) and incubated for 2 h at 4°C. 100 µl of Protein A agarose beads were washed three times with TNE buffer and added to the lysate‐antibody mixture, incubated for 1 h at 4°C on a rotary shaker. The lysate mixture was washed three times with TNE buffer. The protein was eluted using 2X sample buffer for Western blotting, and MS analysis proteins were eluted using 0.1 M glycine‐pH 2.7 and neutralized by 0.1 M Tris‐pH 8.
ELISA
The quantity of IL‐17A, IL‐6, TNF‐a, IL‐10, IL‐1b, and IFN‐γ (eBioscience) (Appendix Table S2) was determined in culture supernatants, serum, and tissue using ELISA kits according to the manufacturer’s instructions. The sensitivity of the assay was < 20 pg/ml.
Microarray analysis
Total RNA from colon tissue and crypts was isolated using the RNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol (n = 3 RNA samples per genotype). For microarray whole transcript expression analysis, 100 ng of total RNA was amplified and labeled following the GeneChip® WT PLUS Reagent kit protocol from Affymetrix (Thermo Fisher, Waltham, MA), followed by hybridization to Affymetrix Mouse ClariomTM S arrays. RNA samples were submitted to Invitrogen/Thermo Fisher Scientific Affymetrix facility, Santa Carla, CA, USA, for analysis. Transcriptome Analysis Console (TAC) 4.0 from Thermo Fisher Scientific was used to analyze the data.
Surface plasmon resonance (SPR)
SPR experiments were conducted on an OpenSPR (NicoyaLife) as described earlier (Sundaram et al, 2019). The experiment was conducted using a NTA sensor chip (NicoyaLife). Tests were run at a flow rate of 20 µl/min using HBS running buffer (20 mM HEPES, 150 mM NaCl, pH 7.4). Purified His‐Tag hAhR protein was first captured on a NTA sensor chip subsequently rinsed thoroughly with running buffer to remove unattached proteins. After a stable signal was obtained, MBELN total cell lysates (5 µg/ml of protein) were run over the immobilized His‐tag hAhR. After 5 min of interaction time, the bound proteins were eluted using a 200 mM imidazole solution. The eluted protein was characterized by MS proteomic analysis. The sensograms were analyzed using TraceDrawer kinetic analysis software.
Sequence alignment and docking analysis
Protein sequence alignment was done using online webtool (https://www.expasy.org/). Docking of proteins (human AhR and Mulberry HSPA8) was performed using ROSIE Rosetta Online Server (https://rosie.graylab.jhu.edu/docking2) (Lyskov & Gray, 2008; Chaudhury et al, 2011). The output pdb files were analyzed using UCSF Chimera windows software (https://www.cgl.ucsf.edu/chimera/).
Cloning and expression
The PAS‐A and PAS‐B domain of the human AhR gene (121–400 bp) from Caco2 cells and Mulberry HSPA8 full‐length gene from mulberry bark cloned into pET28‐MBP‐His‐TEV‐FLAG vector and protein was expressed into a bacterial expression system. Briefly, a cDNA library was prepared from Caco2 cells and mulberry bark mRNA using SuperScript III reverse transcriptase (Invitrogen) and oligo dT primers (Eurofins Genomics). The gene insert of the human AhR gene and Mulberry HSPA8 was amplified using primer (Appendix Table S1). The assembly of gene insert and vector DNA (2:1; insert: vector) was done using NEBuilder® HiFi DNA Assembly Master Mix as per the manufacturer's instructions. The assembled vector with gene insert was transformed into competent DH5a (NEB), plated on a LB agar‐kanamycin plate (50 mg/l) and incubated overnight at 37°C. Positive colonies were confirmed by colony PCR and Sanger sequencing. Plasmid DNA was extracted using a plasmid extraction kit (QIAprep Spin Miniprep Kit; 27106) and transformed into BL21 (DE3). Protein expression was induced by isopropyl β‐D‐1‐thiogalactopyranoside (IPTG; 1 mM) when an OD 0.8 at 600 nm wavelength was achieved and then incubated overnight at 18°C. Cells were harvested and protein was extracted and purified using Ni‐NTA beads (Qiagen) as per the manufacturer’s instructions and protein was confirmed on SDS–PAGE and following dialysis, stored at −80°C in storage buffer (50 mM Tris–HCl, 150 mM NaCl, 1 mM DTT, 10% glycerol, pH 7.6).
Reagents, antibodies, and flow cytometry
For analysis of surface markers, cells were stained in PBS containing 2% (wt/vol) BSA. Intracellular staining of the transcription factors Foxp3 was performed using the Foxp3 Fix/Perm Buffer Set (eBioscience). For detection of intracellular cytokines, cells were first stimulated for 4 h with 50 ng/ml PMA and 1 µg/ml ionomycin in the presence of Brefeldin A (5 µg/ml; all obtained from Sigma), followed by staining for surface markers. Cells were then fixed and permeabilized using the Foxp3 Fix/Perm Buffer Set (eBioscience) and stained for intracellular cytokines. The following antibodies were used at a dilution of 1/200–1/600: PerCP‐Cy5.5, PE‐, FITC‐, or APC‐labeled anti‐IL‐17A (TC11–18H10.1), APC‐ or PE‐Cy7‐labeled anti‐IFN‐γ (XMG1.2), PE‐labeled anti‐Foxp3 (FJK‐16s, eBioscience), PE‐, FITC‐, or APC‐labeled anti‐CD11b (M1/70), PE‐, FITC‐, or APC‐labeled anti‐CD4 (RM4‐5), PE‐Cy7‐labeled anti‐CD3 (145‐2C11), PE‐anti‐Gr‐1 (RB6‐8C5), PE‐, or FITC‐labeled anti‐mouse Ly6G (1A8). All antibodies were obtained from Biolegend unless otherwise noted (Appendix Table S2). Flow cytometry data were acquired on BDFACSCanto™ (Becton Dickinson) and analyzed using FlowJo software (Treestar).
Plasma lipid and liver enzyme analysis
Peripheral blood (200 µl) was collected in sodium citrate vials by retro‐orbital bleeding under anesthesia. Blood plasma was extracted after centrifugation at 1,500 g for 15 min. Level of blood plasma lipids (cholesterol, triglycerides) and liver enzyme (alanine transaminase, aspartate transaminase) was analyzed from 100 µl of blood plasma using a Piccolo® Lipid Panel and analyzed using the Piccolo Xpress analyzer.
Statistical analysis
All the data are shown as mean ± SEM for three or more biological replicates, and all the statistical analyses were performed using GraphPad Prism 7. A t‐test was used to compare the means of two groups and one‐way analysis of variance test to compare multiple experimental groups. The significance is showed as P ≤ 0.05*, P ≤ 0.01**, and P < 0.001*** were considered to be statistically significant.
Author contributions
Mukesh Sriwastva: Data curation; Formal analysis; Validation; Investigation; Methodology; Writing – original draft; Writing – review and editing. Zhongbin Deng: Data curation; Formal analysis; Investigation; Methodology. Bomei Wang: Investigation; Methodology. Yun Teng: Conceptualization; Resources; Software; Validation. Anil Kumar: Formal analysis; Investigation. Kumaran Sundaram: Formal analysis; Investigation. Jingyao Mu: Data curation; Formal analysis; Investigation; Methodology. Chao Lei: Formal analysis; Investigation; Methodology. Gerald W Dryden: Resources; Funding acquisition; Project administration. Fangyi Xu: Investigation; Methodology. Lifeng Zhang: Resources; Data curation; Funding acquisition; Project administration. Jun Yan: Resources; Data curation; Software. Xiang Zhang: Data curation; Software. Juw Won Park: Software; Formal analysis. Michael Merchant: Data curation; Formal analysis. Nejat, K Egilmez: Data curation; Funding acquisition; Project administration. Huang‐Ge Zhang: Conceptualization; Supervision; Funding acquisition; Validation; Investigation; Writing – original draft; Project administration; Writing – review and editing.
In addition to the CRediT author contributions listed above, the contributions in detail are
MKS and HGZ designed the study, analyzed and interpreted the data, and prepared the manuscript; MKS, ZBD, and BW performed the experiments and interpreted the data; YT and JWP analyzed the 16S data; AK performed flow cytometry; KS performed tissue processing and analysis; JM provided histological analysis; CL provided technical support for cloning, MM, FX, and XZ performed protein analysis; LZ provided technical support; GWD, NE, and JY interpreted the findings.
Disclosure statement and competing interests
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Dataset EV1
Dataset EV2
Dataset EV3
Dataset EV4
Dataset EV5
Source Data for Appendix
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 6
Source Data for Figure 7
Acknowledgments
We thank Dr. Ning Wei for providing anti‐CSN8 antibody, Dr. D. Wilkey for mass spectrometry analysis; Ms. Mary Roth; Dr. Monsen Robert Chandos for docking analysis, and Dr. Ruth Welt for lipid analysis; and Dr. J. Ainsworth for editorial assistance. This work was supported by a grant from the National Institutes of Health (NIH) (R01AT008617), the Robley Rex VA Medical Center Merit Review Grants (H‐GZ), and COBRE Pilot Project, NIH (P20GM125504, YT). Huang‐Ge Zhang is supported by a Research Career Scientist (RCS, IK6 BX004199) Award and P20GM125504. J. M. is partly supported by R01AA023190. L.Z. is partly supported by R01AA028435 and R01DK115406. Zhong‐bin Deng is supported by R01DK115406. X.Z. and M.M. are supported by P50AA024337 and P20GM113226. J.P. is supported by the NIH National Institute of General Medical Sciences (P20GM103436) and NIH National Institute of Environmental Health Sciences grant (P30ES030283).
EMBO reports (2022) 23: e53365.
Data availability
The gene expression microarray dataset generated in this study is available in following database:
Gene Expression Omnibus GSE185349: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE185349
Gene Expression Omnibus GSE185351: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE185351
References
- Alenghat T, Osborne LC, Saenz SA, Kobuley D, Ziegler CGK, Mullican SE, Choi I, Grunberg S, Sinha R, Wynosky‐Dolfi M et al (2013) Histone deacetylase 3 coordinates commensal‐bacteria‐dependent intestinal homeostasis. Nature 504: 153–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoni L, Nuding S, Wehkamp J, Stange EF (2014) Intestinal barrier in inflammatory bowel disease. World J Gastroenterol 20: 1165–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aramwit P, Supasyndh O, Siritienthong T, Bang N (2013) Mulberry leaf reduces oxidation and C‐reactive protein level in patients with mild dyslipidemia. Biomed Res Int 2013: 787981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai A, Nakagawa K, Higuchi O, Kimura T, Kojima Y, Kariya J, Miyazawa T, Oikawa S (2011) Effect of mulberry leaf extract with enriched 1‐deoxynojirimycin content on postprandial glycemic control in subjects with impaired glucose metabolism. J Diabetes Invest 2: 318–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano N, Tomioka E, Kizu H, Matsui K (1994) Sugars with nitrogen in the ring isolated from the leaves of Morus bombycis. Carbohydr Res 253: 235–245 [DOI] [PubMed] [Google Scholar]
- Ayabe T, Satchell DP, Wilson CL, Parks WC, Selsted ME, Ouellette AJ (2000) Secretion of microbicidal alpha‐defensins by intestinal Paneth cells in response to bacteria. Nat Immunol 1: 113–118 [DOI] [PubMed] [Google Scholar]
- Balch WE, Morimoto RI, Dillin A, Kelly JW (2008) Adapting proteostasis for disease intervention. Science 319: 916–919 [DOI] [PubMed] [Google Scholar]
- Balchin D, Hayer‐Hartl M, Hartl FU (2016) In vivo aspects of protein folding and quality control. Science 353: aac4354 [DOI] [PubMed] [Google Scholar]
- Bevins CL, Salzman NH (2011) Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat Rev Microbiol 9: 356–368 [DOI] [PubMed] [Google Scholar]
- Chan EWC, Lye PY, Wong SK (2016) Phytochemistry, pharmacology, and clinical trials of Morus alba. Chin J Nat Med 14: 17–30 [DOI] [PubMed] [Google Scholar]
- Chatterjee SS, Hossain H, Otten S, Kuenne C, Kuchmina K, Machata S, Domann E, Chakraborty T, Hain T (2006) Intracellular gene expression profile of Listeria monocytogenes. Infect Immun 74: 1323–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury S, Berrondo M, Weitzner BD, Muthu P, Bergman H, Gray JJ (2011) Benchmarking and analysis of protein docking performance in Rosetta v3.2. PLoS One 6: e22477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Retzlaff M, Roos T, Frydman J (2011) Cellular strategies of protein quality control. Cold Spring Harbor Perspect Biol 3: a004374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YJ, Arzuaga X, Kluemper CT, Caraballo A, Toborek M, Hennig B (2010) Quercetin blocks caveolae‐dependent pro‐inflammatory responses induced by co‐planar PCBs. Environ Int 36: 931–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damon C, Dmitrieva J, Muhovski Y, Francis F, Lins L, Ledoux Q, Luwaert W, Markó IE, Mauro S, Ongena M et al (2012) Interaction network of antimicrobial peptides of Arabidopsis thaliana, based on high‐throughput yeast two‐hybrid screening. Plant Physiol Biochem 58: 245–252 [DOI] [PubMed] [Google Scholar]
- Datla KP, Christidou M, Widmer WW, Rooprai HK, Dexter DT (2001) Tissue distribution and neuroprotective effects of citrus flavonoid tangeretin in a rat model of Parkinson's disease. NeuroReport 12: 3871–3875 [DOI] [PubMed] [Google Scholar]
- Diplock AT, Charleux JL, Crozier‐Willi G, Kok FJ, Rice‐Evans C, Roberfroid M, Stahl W, Vina‐Ribes J (1998) Functional food science and defence against reactive oxidative species. Br J Nutr 80(Suppl 1): S77–S112 [DOI] [PubMed] [Google Scholar]
- Farin HF, Karthaus WR, Kujala P, Rakhshandehroo M, Schwank G, Vries RG, Kalkhoven E, Nieuwenhuis EE, Clevers H (2014) Paneth cell extrusion and release of antimicrobial products is directly controlled by immune cell‐derived IFN‐gamma. J Exp Med 211: 1393–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamon MA, Ribet D, Stavru F, Cossart P (2012) Listeriolysin O: the Swiss army knife of Listeria. Trends Microbiol 20: 360–368 [DOI] [PubMed] [Google Scholar]
- He N, Zhang C, Qi X, Zhao S, Tao Y, Yang G, Lee T‐H, Wang X, Cai Q, Li D et al (2013) Draft genome sequence of the mulberry tree Morus notabilis. Nat Commun 4: 2445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hind SR, Pulliam SE, Veronese P, Shantharaj D, Nazir A, Jacobs NS, Stratmann JW (2011) The COP9 signalosome controls jasmonic acid synthesis and plant responses to herbivory and pathogens. Plant J 65: 480–491 [DOI] [PubMed] [Google Scholar]
- Hipp MS, Park S‐H, Hartl FU (2014) Proteostasis impairment in protein‐misfolding and ‐aggregation diseases. Trends Cell Biol 24: 506–514 [DOI] [PubMed] [Google Scholar]
- Hirano T, Abe K, Gotoh M, Oka K (1995) Citrus flavone tangeretin inhibits leukaemic HL‐60 cell growth partially through induction of apoptosis with less cytotoxicity on normal lymphocytes. Br J Cancer 72: 1380–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Gao JJ, Lee WC, Ryu KS, Lee KR, Kim YC (1999) Antioxidative flavonoids from the leaves of Morus alba. Arch Pharm Res 22: 81–85 [DOI] [PubMed] [Google Scholar]
- Kojima K, Musch MW, Ren H, Boone DL, Hendrickson BA, Ma A, Chang EB (2003) Enteric flora and lymphocyte‐derived cytokines determine expression of heat shock proteins in mouse colonic epithelial cells. Gastroenterology 124: 1395–1407 [DOI] [PubMed] [Google Scholar]
- Kurowska EM, Manthey JA (2004) Hypolipidemic effects and absorption of citrus polymethoxylated flavones in hamsters with diet‐induced hypercholesterolemia. J Agric Food Chem 52: 2879–2886 [DOI] [PubMed] [Google Scholar]
- Kwok SF, Solano R, Tsuge T, Chamovitz DA, Ecker JR, Matsui M, Deng XW (1998) Arabidopsis homologs of a c‐Jun coactivator are present both in monomeric form and in the COP9 complex, and their abundance is differentially affected by the pleiotropic cop/det/fus mutations. Plant Cell 10: 1779–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La D, As P, Sg M, van Rees EP (1997) Role of animal models for the pathogenesis and treatment of inflammatory bowel disease. Scand J Gastroenterol Suppl 223: 99–104 [PubMed] [Google Scholar]
- Labbadia J, Morimoto RI (2015) The biology of proteostasis in aging and disease. Annu Rev Biochem 84: 435–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechuga S, Ivanov AI (2017) Disruption of the epithelial barrier during intestinal inflammation: quest for new molecules and mechanisms. Biochim Biophys Acta Mol Cell Res 1864: 1183–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei D, Li F, Su H, Tian Z, Ye B, Wei N, Wang X (2011) COP9 signalosome subunit 8 is required for postnatal hepatocyte survival and effective proliferation. Cell Death Differ 18: 259–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingaraju GM, Bunker RD, Cavadini S, Hess D, Hassiepen U, Renatus M, Fischer ES, Thomä NH (2014) Crystal structure of the human COP9 signalosome. Nature 512: 161–165 [DOI] [PubMed] [Google Scholar]
- Liu C, Guo L‐Q, Menon S, Jin D, Pick E, Wang X, Deng XW, Wei N (2013) COP9 signalosome subunit Csn8 is involved in maintaining proper duration of the G1 phase. J Biol Chem 288: 20443–20452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loub WD, Wattenberg LW, Davis DW (1975) Aryl hydrocarbon hydroxylase induction in rat tissues by naturally occurring indoles of cruciferous plants. J Natl Cancer Inst 54: 985–988 [PubMed] [Google Scholar]
- Lozupone C, Knight R (2005) UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71: 8228–8235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lueschow SR, McElroy SJ (2020) The paneth cell: the curator and defender of the immature small intestine. Front Immunol 11: 587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyapina S, Cope G, Shevchenko A, Serino G, Tsuge T, Zhou C, Wolf DA, Wei N, Shevchenko A, Deshaies RJ (2001) Promotion of NEDD‐CUL1 conjugate cleavage by COP9 signalosome. Science 292: 1382–1385 [DOI] [PubMed] [Google Scholar]
- Lyskov S, Gray JJ (2008) The RosettaDock server for local protein‐protein docking. Nucleic Acids Res 36: W233–W238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, Baldwin KT (2000) 2,3,7,8‐tetrachlorodibenzo‐p‐dioxin‐induced degradation of aryl hydrocarbon receptor (AhR) by the ubiquitin‐proteasome pathway. Role of the transcription activaton and DNA binding of AhR. J Biol Chem 275: 8432–8438 [DOI] [PubMed] [Google Scholar]
- Matsuoka K, Kanai T (2015) The gut microbiota and inflammatory bowel disease. Semin Immunopathol 37: 47–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon S, Chi H, Zhang H, Deng XW, Flavell RA, Wei N (2007) COP9 signalosome subunit 8 is essential for peripheral T cell homeostasis and antigen receptor‐induced entry into the cell cycle from quiescence. Nat Immunol 8: 1236–1245 [DOI] [PubMed] [Google Scholar]
- Moura‐Alves P, Faé K, Houthuys E, Dorhoi A, Kreuchwig A, Furkert J, Barison N, Diehl A, Munder A, Constant P et al (2014) AhR sensing of bacterial pigments regulates antibacterial defence. Nature 512: 387–392 [DOI] [PubMed] [Google Scholar]
- Moura‐Alves P, Puyskens A, Stinn A, Klemm M, Guhlich‐Bornhof U, Dorhoi A, Furkert J, Kreuchwig A, Protze J, Lozza L et al (2019) Host monitoring of quorum sensing during Pseudomonas aeruginosa infection. Science 366: eaaw1629 [DOI] [PubMed] [Google Scholar]
- Mu J, Zhuang X, Wang Q, Jiang H, Deng Z‐B, Wang B, Zhang L, Kakar S, Jun Y, Miller D et al (2014) Interspecies communication between plant and mouse gut host cells through edible plant derived exosome‐like nanoparticles. Mol Nutr Food Res 58: 1561–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musch MW, Sugi K, Straus D, Chang EB (1999) Heat‐shock protein 72 protects against oxidant‐induced injury of barrier function of human colonic epithelial Caco2/bbe cells. Gastroenterology 117: 115–122 [DOI] [PubMed] [Google Scholar]
- Nakamura S, Hashiguchi M, Yamaguchi Y, Oku T (2011) Hypoglycemic effects of Morus alba leaf extract on postprandial glucose and insulin levels in patients with type 2 diabetes treated with sulfonylurea hypoglycemic agents. J Diabetes Metab 2: 1–5 [Google Scholar]
- Perazzo A, Peng Z, Young YN, Feng Z, Wood DK, Higgins JM, Stone HA (2003) Evidence for a physical association of the COP9 signalosome, the proteasome, and specific SCF E3 ligases in vivo . Curr Biol 13: R504–R505 [DOI] [PubMed] [Google Scholar]
- Peterson LW, Artis D (2014) Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat Rev Immunol 14: 141–153 [DOI] [PubMed] [Google Scholar]
- Petrof EO, Ciancio MJ, Chang EB (2004) Role and regulation of intestinal epithelial heat shock proteins in health and disease. Chin J Dig Dis 5: 45–50 [DOI] [PubMed] [Google Scholar]
- Potapovich AI, Lulli D, Fidanza P, Kostyuk VA, De Luca C, Pastore S, Korkina LG (2011) Plant polyphenols differentially modulate inflammatory responses of human keratinocytes by interfering with activation of transcription factors NFkappaB and AhR and EGFR‐ERK pathway. Toxicol Appl Pharmacol 255: 138–149 [DOI] [PubMed] [Google Scholar]
- Read S, Malmström V, Powrie F (2000) Cytotoxic T lymphocyte‐associated antigen 4 plays an essential role in the function of CD25(+)CD4(+) regulatory cells that control intestinal inflammation. J Exp Med 192: 295–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothhammer V, Quintana FJ (2019) The aryl hydrocarbon receptor: an environmental sensor integrating immune responses in health and disease. Nat Rev Immunol 19: 184–197 [DOI] [PubMed] [Google Scholar]
- Salzman NH, Hung K, Haribhai D, Chu H, Karlsson‐Sjöberg J, Amir E, Teggatz P, Barman M, Hayward M, Eastwood D et al (2010) Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol 11: 76–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman NH, de Jong H, Paterson Y, Harmsen HJM, Welling GW, Bos NA (2002) Analysis of 16S libraries of mouse gastrointestinal microflora reveals a large new group of mouse intestinal bacteria. Microbiology 148: 3651–3660 [DOI] [PubMed] [Google Scholar]
- Sanchez MD (1999) Mulberry: an exceptional forage available almost worldwide. World Anim Rev 93: 36–46 [Google Scholar]
- Sarkar FH, Li Y, Wang Z, Kong D (2008) NF‐kappaB signaling pathway and its therapeutic implications in human diseases. Int Rev Immunol 27: 293–319 [DOI] [PubMed] [Google Scholar]
- Schmidt M, Finley D (2014) Regulation of proteasome activity in health and disease. Biochem Biophys Acta 1843: 13–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer C, Serino G, Callis J, Crosby WL, Lyapina S, Deshaies RJ, Gray WM, Estelle M, Deng XW (2001) Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCFTIRI in mediating auxin response. Science 292: 1379–1382 [DOI] [PubMed] [Google Scholar]
- Singh NP, Hegde VL, Hofseth LJ, Nagarkatti M, Nagarkatti P (2007) Resveratrol (trans‐3,5,4'‐trihydroxystilbene) ameliorates experimental allergic encephalomyelitis, primarily via induction of apoptosis in T cells involving activation of aryl hydrocarbon receptor and estrogen receptor. Mol Pharmacol 72: 1508–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su H, Li J, Menon S, Liu J, Kumarapeli AR, Wei N, Wang X (2011) Perturbation of cullin deneddylation via conditional Csn8 ablation impairs the ubiquitin‐proteasome system and causes cardiomyocyte necrosis and dilated cardiomyopathy in mice. Circ Res 108: 40–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram K, Miller DP, Kumar A, Teng Y, Sayed M, Mu J, Lei C, Sriwastva MK, Zhang L, Yan J et al (2019) Plant‐derived exosomal nanoparticles inhibit pathogenicity of Porphyromonas gingivalis. iScience 21: 308–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng Y, Ren YI, Sayed M, Hu X, Lei C, Kumar A, Hutchins E, Mu J, Deng Z, Luo C et al (2018) Plant‐derived exosomal microRNAs shape the gut microbiota. Cell Host Microbe 24: 637–652.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo‐Arana A, Dussurget O, Nikitas G, Sesto N, Guet‐Revillet H, Balestrino D, Loh E, Gripenland J, Tiensuu T, Vaitkevicius K et al (2009) The Listeria transcriptional landscape from saprophytism to virulence. Nature 459: 950–956 [DOI] [PubMed] [Google Scholar]
- Vaishnava S, Behrendt CL, Ismail AS, Eckmann L, Hooper LV (2008) Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host‐microbial interface. Proc Natl Acad Sci USA 105: 20858–20863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, Ley R, Wakeland EK, Hooper LV (2011) The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science 334: 255–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vane CH, Drage TC, Snape CE (2006) Bark decay by the white‐rot fungus Lentinula edodes: Polysaccharide loss, lignin resistance and the unmasking of suberin. Int Biodeterior Biodegradation 57: 14–23 [Google Scholar]
- Wang Q, Zhuang X, Mu J, Deng Z‐B, Jiang H, Zhang L, Xiang X, Wang B, Yan J, Miller D et al (2013) Delivery of therapeutic agents by nanoparticles made of grapefruit‐derived lipids. Nat Commun 4: 1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehkamp J, Fellermann K, Herrlinger KR, Bevins CL, Stange EF (2005) Mechanisms of disease: defensins in gastrointestinal diseases. Nat Clin Pract Gastroenterol Hepatol 2: 406–415 [DOI] [PubMed] [Google Scholar]
- Wei N, Deng XW (2003) The COP9 signalosome. Annu Rev Cell Dev Biol 19: 261–286 [DOI] [PubMed] [Google Scholar]
- Wong E, Cuervo AM (2010) Integration of clearance mechanisms: the proteasome and autophagy. Cold Spring Harbor Perspect Biol 2: a006734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J, Feng S, Wang X, Long K, Luo YI, Wang Y, Ma J, Tang Q, Jin L, Li X et al (2018) Identification of exosome‐like nanoparticle‐derived microRNAs from 11 edible fruits and vegetables. PeerJ 6: e5186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Menon S, Lykke‐Andersen K, Tsuge T, Di Xiao , Wang X, Rodriguez‐Suarez RJ, Zhang H, Wei N (2002) The COP9 signalosome inhibits p27(kip1) degradation and impedes G1‐S phase progression via deneddylation of SCF Cul1. Curr Biol 12: 667–672 [DOI] [PubMed] [Google Scholar]
- Yu S, Balasubramanian I, Laubitz D, Tong K, Bandyopadhyay S, Lin X, Flores J, Singh R, Liu Y, Macazana C et al (2020) Paneth cell‐derived lysozyme defines the composition of mucolytic microbiota and the inflammatory tone of the intestine. Immunity 53: 398–416.e398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zasloff M (2002) Antimicrobial peptides of multicellular organisms. Nature 415: 389–395 [DOI] [PubMed] [Google Scholar]
- Zhang L‐J, Gallo RL (2016) Antimicrobial peptides. Curr Biol 26: R14–R19 [DOI] [PubMed] [Google Scholar]


