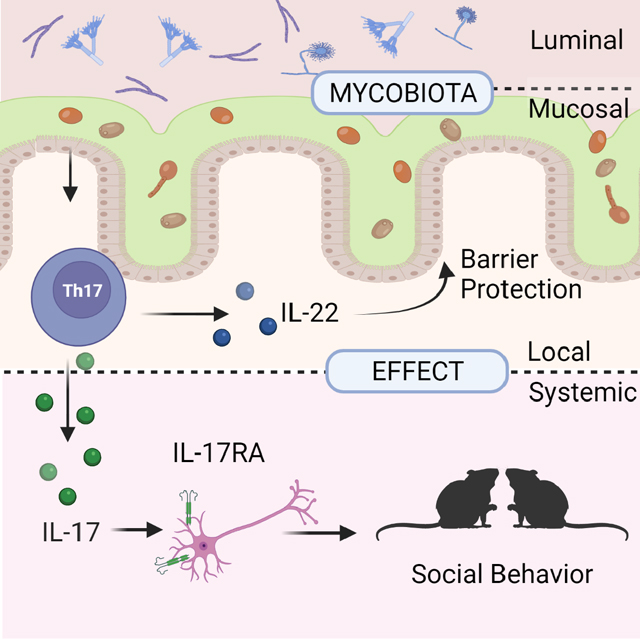
Summary
Fungal communities (the mycobiota) are an integral part of the gut microbiota, and the disruption of their integrity contributes to local and gut-distal pathologies. Yet, the mechanisms by which intestinal fungi promote homeostasis remain unclear. We characterized the mycobiota biogeography along the gastrointestinal tract and identified a subset of fungi associated with the intestinal mucosa of mice and humans. Mucosa-associated fungi reinforced intestinal epithelial function and protected mice against intestinal injury and bacterial infection. Notably, intestinal colonization with a defined consortium of mucosa-associated fungi promoted social behavior in mice. The gut-local effects on barrier function were dependent on IL-22 production by CD4+ T helper cells while the effects on social behavior were mediated through IL-17R-dependent signaling in neurons. Thus, spatial organization of the gut mycobiota is associated with host-protective immunity, epithelial barrier function and might be a driver of neuroimmune modulation of mouse behavior through complementary type 17 immune mechanisms.
Keywords: Mycobiota, Gut-brain axis, Social Behavior, Th17, Fungal consortia, Microbiota Biogeography, Intestinal barrier
In Brief:
Mucosa-associated fungi protect against intestinal injury and infection through the induction of IL-22 by Th cells, and foster social behavior via IL-17 signaling in neurons
Graphical Abstract
Introduction
A diverse community of bacteria, viruses, protozoa, and fungi coexists within the mammalian gastrointestinal (GI) tract (Hall et al., 2017). The physiology of the intestine varies dramatically along its length forming unique environmental niches and creating a distinct microbiota biogeography that shapes the dynamics of microbial interactions with the host (Costello et al., 2019; Donaldson et al., 2016; Johansson et al., 2008; Martinez-Guryn et al., 2019; Pelaseyed et al., 2014; Tropini et al., 2017). The distance from the intestinal mucosa adds another dimension allowing further differentiation between luminal and mucosa-associated microbiota. Mucosa-associated microbiota have the ability to induce host-protective immunity through the direct interaction with the underlying epithelium and mucosal immune cells (Atarashi et al., 2017; Ivanov et al., 2009; Ladinsky et al., 2019; Sonnenberg et al., 2012). The spatial organization of the microbiota, including altered longitudinal distribution or translocation of luminal species into the mucosa, might represent an important factor contributing to several inflammatory, metabolic and cognitive diseases (Atarashi et al., 2017; Hooper et al., 2012; Rowan et al., 2010; Bajaj et al., 2012; Everard et al., 2013; Gevers et al., 2014). Recently, pathophysiological changes in the gut have been shown to mediate effects of microbiota on host behavior and neurological disorders (Buffington et al., 2016; Chu et al., 2019; Hall et al., 2017; Muller et al., 2020; Veiga-Fernandes and Mucida, 2016).
In addition to bacteria that can influence a wide range of physiological processes ranging from immune development to behavior, a growing body of research suggests an involvement of intestinal fungi in the modulation of immune-homeostasis (Doron et al., 2021 a, b; Jiang et al., 2017; Ost et al., 2021; Yang et al., 2017). While often investigated in the context of infectious disease, fungi are common inhabitants of a healthy mammalian GI tract (Chiaro et al., 2017; Hoarau et al., 2016; Iliev and Leonardi, 2017; Jiang et al., 2017; Sokol et al., 2017). Consistently, changes in the composition of gut fungal populations (termed “fungal dysbiosis”) can contribute to local and gut-distal pathologies in colitis, alcoholic liver disease, and allergic lung disease (Jiang et al., 2017; Leonardi et al., 2018; Limon et al., 2019; Leonardi et al., 2020; Jain et al., 2021; Sokol et al., 2017; Wheeler et al., 2016; Yang et al., 2017), altogether suggesting a possible beneficial role of a balanced intestinal fungal community in maintaining host immune homeostasis and human health.
Here, we initially characterized the biogeography of the fungal communities along the gastrointestinal tract and identified a subset of fungi associated with the intestinal mucosa in both laboratory mice and humans. In contrast to bacterial communities, mycobiota showed a stronger clustering based on the luminal vs. mucosal sampling site rather than on the longitudinal location. A defined consortium of mucosa-associated fungi, but not a luminal fungal consortium, exerted an immuno-protective effect in the gut by increasing barrier function and transcription of epithelial genes involved in JAK/STAT signaling and DNA repair. Intestinal mucosa associated fungi were potent inducers of CD4+ T helper cell-derived IL-22 and IL-17. Gut-local production of IL-22 promoted barrier function, and protected mice from intestinal injury during both antibiotic treatment and bacterial infection. Consistent with recent findings suggesting the existence of crosstalk between the gut microbiota and the brain (Buffington et al., 2016; Hall et al., 2017; Veiga-Fernandes and Mucida, 2016), intestinal colonization with mucosa-associated fungi increased the responsiveness of mice to social stimuli, highlighting the ability of gut fungi to modulate host behavior. We found that mucosal fungi induced a systemic release of IL-17A and that IL-17 sensing in neurons was necessary for the fungal-modulation of mouse social behavior. Altogether, our findings suggest that mucosa associated fungi have the unique ability to drive protective mucosal immunity with effects on intestinal epithelial cells, gut barrier function, intestinal disease, and neuroimmune-mediated host behavior.
Results
Specific bacterial communities are associated with different GI sites with strong compositional differences along the length of the digestive tract (Hsiao et al., 2013; Tropini et al., 2017). Fecal composition cannot fully grasp the complexity of the intestinal microbiota as feces solely provide a “representative picture” of the mycobiota throughout the entire GI tract. To explore the biogeography of the fungal microbiota along the GI tract, we collected luminal and mucosal samples from several GI regions including the stomach, jejunum, ileum, cecum, and colon of C57BL/6 mice and performed ITS sequencing of fungal ribosomal DNA (rDNA) (Figure 1A). Non-metric dimensional scaling of fungal communities revealed a robust clustering based on the luminal vs. mucosal sampling site rather than on the longitudinal location (Figure 1B). Further analysis revealed a high diversity of luminal mycobiota irrespective of the longitudinal sampling location, while mucosa-associated fungal communities were significantly less diverse (Figure 1C). Since the large intestine harbored the highest fungal loads (Iliev et al., 2012), we focused our further analysis on this body site. Differential abundance analysis of samples from the large intestinal mucosa versus luminal content identified several fungal genera that appeared to be preferentially associated with either location (Figure 1D). The luminal-associated mycobiota included genera that were likely transient-environmental organisms such as Cladosporium and Aspergillus (Hallen-Adams and Suhr, 2016), while the mucosa-enriched operational taxonomic units (OTUs) belonged to several “immuno-reactive” fungal genera (Bacher et al., 2019; Chiaro et al., 2017; Iliev et al., 2012; Leonardi et al., 2018), such as Candida, Saccharomyces, and Saccharomycopsis (Figure 1D). A similar mycobiota composition was observed in human subjects where Candida spp. and Saccharomyces spp. were the dominant genera associated with the intestinal mucosa of surgical specimens (Figure 1 E, F, Table S1). The presence of distinct communities, together with the reduced alpha diversity observed in the mucosal mycobiota, suggests that only a subset of specialized fungi can colonize the unique environment of the intestinal mucosa.
Figure 1. A distinct fungal community is associated with the murine intestinal mucosa.
A. Luminal and mucosal samples were collected from the stomach, jejunum, ileum, cecum, and colon of 5 female C57Bl/6J mice for DNA isolation and ITS sequencing. Relative abundance at the genus level of the 14 most abundant genera in the mouse intestine. B. Non-metric multidimensional scaling (NMDS) plot of the fungal community structure from the sampled location. C. Shannon diversity index representing the alpha diversity of the fungal community at the OTU level in the sampled location; filled circles: luminal mycobiota (L); empty circles: mucosal associated mycobiota (M). Mean +/− SEM, dots represent individual samples. D. Differentially abundant OTUs between the luminal mycobiota and the mucosal associated mycobiota. Dots represents individual OTUs with a FDR < 0.01 and abundance > 1%. E-F. Characterization of the human mucosa-associated mycobiota, mean +/− SEM, dots represent individual subjects. E. Relative abundance of the major Phyla. F. Relative abundance of the major fungal genera in individual samples (left) and as boxplots (right) in the intestinal mucosa of 7 individuals. G. Mice were colonized with luminal fungi (LUM) or mucosal fungi (MUC) every other day for 10 days prior to DSS treatment. Control groups were orally gavaged with PBS at the same time points. Body weight and the presence of occult blood were assessed daily. Mice were administered sterile 3% dextran sodium sulphate (DSS) in an antibiotic cocktail (ABX: ampicillin 0.4g/l, vancomycin 0.4g/l, metronidazole 0.3g/l, cefoperazone 0.4g/l, 10% sucrose) for 5 days followed by a recovery period of 7 days with ABX and a second 5 days DSS+ABX and ABX cycle until achievement of end point. H. Survival following the first day of DSS administration in the PBS group and LUM colonized group (left). Survival following the first day of DSS administration in the PBS group and MUC colonized group (right). Log-rank (Mantel-Cox) test I. Intestinal permeability was measured by quantification of FITC-dextran serum levels following intragastric gavage of the LUM or MUC consortia in SPF mice. Dots represent individual mice, Mann-Whitney T Test.
Close proximity of bacterial strains to the intestinal epithelium has been associated with the ability to induce specific immune responses. To assess whether luminal and mucosa-associated fungi have immuno-protective potential, we used a model of antibiotics-induced ecological perturbation coupled with prolonged DSS-mediated injury previously used by others to study how specific microbiota constituents impact barrier function (Kernbauer et al., 2014; Schieber et al., 2015). Based on our analysis of luminal and mucosa-associated communities in mice and humans, we generated two defined model consortia consisting of three representative species from each niche: mucosa-associated (MUC) consisting of Candida albicans, Saccharomyces cerevisiae, and Saccharomycopsis fibuligera, and luminal (LUM) consisting of Aspergillus amstellodamii, Cladosporium cladosporioides, and Wallemia sebi (Figure 1D). To assess whether our model consortia had a distinct effect on the disease progression, we colonized antibiotic-treated mice for three weeks with either LUM or MUC before DSS administration (Figure 1G). These experiments revealed that colonization with the mucosal consortium was protective against intestinal injury and reduced mortality in mice. In contrast, colonization with the luminal consortium did not confer protection in this model (Figure 1G-H).
The intestinal epithelium forms the first cellular barrier between the microbiota and the mucosal immune system in the gut. We thus assessed the effect of the consortium of mucosa-associated fungi on intestinal epithelial cells (IECs). To minimize noise and confounding effects from the pre-existing intestinal mycobiota, we colonized fungi-free altered Schaedler’s flora (ASF) mice (Li et al., 2018; Schaedler et al., 1965) with the mucosal fungal consortium and performed RNA-seq on colonic epithelial cells (IEC, Figure 2A-B, Figure S1A). Intestinal colonization with the MUC consortium induced a distinct transcriptional profile in IECs with upregulation of genes involved in immune response and cell proliferation (Figure 2B). Specifically, factors involved in oxidative phosphorylation, IFN responses, JAK/STAT signaling, and DNA repair were upregulated, while genes involved in TGFβ signaling, protein secretion, and G2M cell-cycle checkpoint were downregulated (Figure 2B). Given these data, we further evaluated the intestinal barrier function upon colonization with MUC and LUM consortia. Barrier permeability in SPF mice colonized with either consortium revealed a markedly reduced intestinal permeability upon MUC colonization when compared to LUM colonization (Figure 1I), consistent with the finding that the latter did not confer protection in our intestinal injury model (Figure 1H).
Figure 2. A defined mucosal fungal consortium promotes barrier integrity and induces the production of IL-22 by CD4+ T cells.
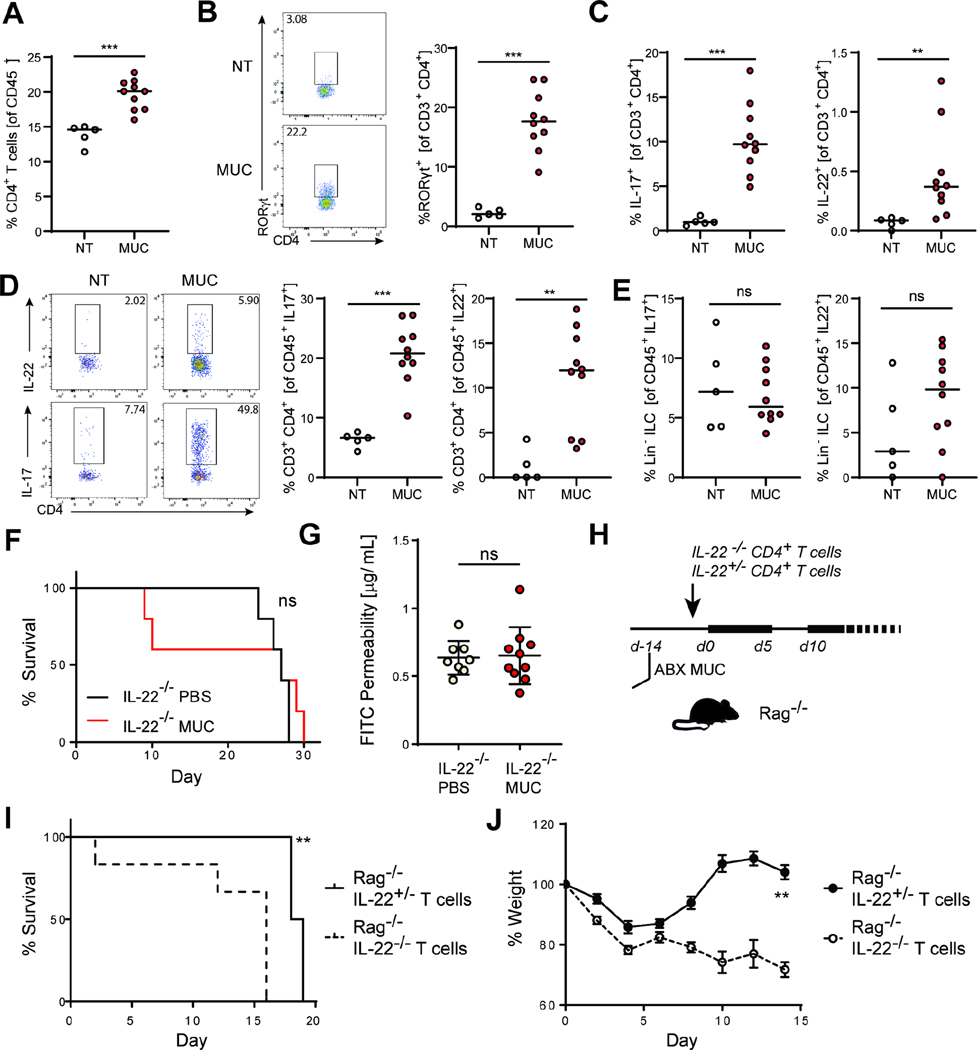
A-B: ASF mice were colonized with mucosal fungi (MUC) every other 4 days for 10 days. Single reads RNAseq was performed on colonic epithelial cells (n=4 mice per group, see also Figure S1A, Table S2). A. Volcano plot of RNA-seq on epithelial cells from MUC colonized ASF mice and untreated ASF controls. Red dots represent genes with FDR < 0.1 B. Gene set enrichment analysis (GSEA) of the MUC and ASF RNA-seq results. Bars show normalized enrichment scores (NESs) of the gene set enrichment analysis (GSEA) on the hallmark gene sets. C-I: male and female SPF C57Bl/6J mice were treated with antibiotics and colonized with luminal fungi (LUM) or mucosal fungi (MUC) for 10 days. C. Quantification of myeloid cell populations and in the colonic lamina propria (cLP). D-I. Representative graphs and characterization of the lymphoid populations in the cLP. D. Frequency of type 3 innate lymphoid cells (ILC3) in the cLP. E. Quantification of T helper cells (Th), cytotoxic T cells (Tc), and γδ T cells (γδT) among the CD45+ hematopoietic compartment in the cLP. F. Quantification and representative flow cytometry plots of Th cells subsets in the cLP. G. Quantification and representative flow cytometry plots of IL-17A and IL-17F production by T helper cells in the cLP. H. Quantification and representative flow cytometry plots of IL-22 production by T helper cells in the cLP. I. Quantification and representative flow cytometry plots of IFNγ production by T helper cells in the cLP. Gating strategy for C-H shown in Figure S1 B-C. Immunophenotyping of the mLN is shown in Figure S2. Experiments in C-I were repeated at least three times with n>4 per group. *P<0.05, **P<0.01, ***P<0.001 Mann-Whitney T-test. Data are shown as Tukey box and whiskers plots.
The intestinal epithelium is in constant cross-talk with the immune cells of the underlying intestinal mucosa. Distinct populations of intestinal phagocytes (Coombes and Powrie, 2008; Pull et al., 2005; Sun et al., 2007; Denning et al., 2007; Spadoni et al., 2015; Chikina et al., 2020; Leonardi et al., 2018), innate lymphoid cells (ILCs) (Fung et al., 2016; Sonnenberg et al., 2011), and T cells (Asseman et al., 2003; Chen et al., 2002; Fontenot et al., 2003; Li et al., 2007; Maloy et al., 2003) play a vital role in the protection and maintenance of the intestinal barrier. As fungi can induce strong immune responses, we hypothesized that the protective function of the MUC consortium could be mediated via the immune system. Immunophenotyping of mice colonized with either fungal consortium showed no differences in the composition of the myeloid, ILCs, FoxP3+ regulatory T cells (Tregs), T-bet+ T helper (Th1) cells, and cytotoxic T cells (Figure 2 C-F, Figure S1 B-C, Figure S2, Figure S3). In contrast, we observed a significant increase in the frequency of RORγt+ type 17 T helper cells (Th17) in the colonic lamina propria (cLP) and mesenteric lymph nodes (mLN) of MUC mice (Figure 2 F & G, Figure 3B, Figure S2A). Th cells in MUC colonized mice produced high levels of the effector cytokines IL-22, IL-17A, and IL-17F (Figure 2 G & H, 3A-D, Figure S2 B-D, Figure S3). Colonization of ASF mice with MUC recapitulated the phenotype observed in antibiotics-treated mice, suggesting that MUC induces Type 17 responses independently from antibiotic-induced dysbiosis (Figure S4 A-C). When assessed individually, two of the members of the MUC consortium –C. albicans and S. cerevisiae –induced an increase in the frequency of colonic Th17 cells. However, the compound effect of the MUC consortium appeared to be stronger (Figure S3 A-B). Given these data and the known role of IL-22 and IL-17 in the maintenance of intestinal barrier function (Nagalakshmi et al., 2004; Aujla et al., 2008; Backert et al., 2014; Pham et al., 2014; Ahlfors et al., 2014; Lee et al., 2015; Gronke et al., 2019; Neil et al., 2019; Aggor et al., 2020) we next assessed whether IL-17A and or IL-22 mediates the protective effect of the MUC consortium. We induced intestinal injury in IL-17A (Il17a−/−) and IL-22 (Il22−/−) deficient mice in the presence or absence of MUC colonization. As expected, Il17a−/− mice were more susceptible to intestinal injury (Gaffen et al., 2014; Yang et al., 2008; Zhou et al., 2021). In these mice MUC colonization led to a slightly increased survival (Figure S4 D-E). In contrast, the absence of IL-22 abrogated the protective effect of MUC consortia on both survival and barrier function (Figure 3F-G). Type 3 Innate lymphoid cells (ILC3) are a major source of IL-22 in the colon (Ahlfors et al., 2014; Fung et al., 2016) but their frequency and production of IL-17 and IL-22 was not affected by MUC colonization (Figure 2E, 3E, Figure S3 D-F). After determining that CD4+ T cells are the major source of MUC-induced IL-22, we next assessed whether IL-22 producing CD4+ T cells were sufficient for the protective effect of the MUC consortia. To specifically explore the role of IL-22 production by T helper cells, T and B cells deficient Rag1−/− mice (Fung et al., 2016) were adoptively transferred with CD4+ T cells derived from either Il22+/− (able to produce IL-22) or Il22−/− mice before the induction of intestinal injury (Figure 3H). Notably, transfer of Il22+/− CD4+ T cells significantly prolonged the survival of Rag1−/− mice colonized with the MUC consortium when compared to mice adoptively-transferred with IL-22 deficient CD4+ T cells (Figure 3 H-J), suggesting a key role of T cells as a source of IL-22 in response to mucosa-associated fungi colonization.
Figure 3. The protective effect of a defined mucosal fungal consortium is mediated by IL-22 producing CD4+ T cells.
Male and female SPF C57Bl/6J mice were treated with antibiotics and colonized with luminal fungi (LUM) or mucosal fungi (MUC) for 10 days. A. Frequency of CD3+ CD4+ T cells among CD45+ cells. B. Representative flow cytometry plot and quantification of RORγ+ expressing Th17 cells/. C. IL-17A+, and IL-22+ expression among CD3+ CD4+ T cells in the colonic lamina propria (cLP). D. Representative flow cytometry plot of IL-22 and IL-17 expression by CD4+ T cells and frequency of T cells among IL-17 and IL-22 expressing CD45+ cells E. Frequency of ILC3s among IL-17 and IL-22 expressing CD45+ cells. Gating strategy: CD4+ T cells: Lin+ CD3+CD4+; ILC3s: Lin- (lineage: CD11c, CD11b, NK1.1, Gr1, TCRβ, CD5, CD19), CD3-, CD90.2+, CD127+ RORγ+. See also Figure S3-4. F-J. Il22−/− mice were colonized with mucosal fungi (MUC) every 3 days for 10 days prior to DSS treatment. Control groups were orally gavaged with PBS at the same time points. Mice were administered with sterile 3% dextran sodium sulphate (DSS) in an antibiotic cocktail (ABX: ampicillin 0.4g/l, vancomycin 0.4g/l, metronidazole 0.3g/l, cefoperazone 0.4g/l, 10% Sucrose) for 5 days followed by a recovery period of 7 days with ABX and a second 5 days DSS+ABX and ABX cycle until achievement of end point. F. Survival following the first day of DSS administration in the PBS group and MUC colonized group. G. Intestinal permeability was measured by quantification of FITC-dextran serum levels following intragastric gavage. H. Rag1−/− mice were colonized with mucosal fungi (MUC) and gavaged PBS every 3 days for 10 days followed by adoptive transfer with 4.106 CD4+ T cells from either Il22−/− or Il22+/− mice one day prior to DSS+ABX administration. I. Percent survival. J. Percent change in weight in Rag−/− mice. Dots represent individual mice, mean +/− SEM. *P<0.05, **P<0.01, ***P<0.001. Log-rank (Mantel-Cox) test (C, F); Mann-Whitney T Test (B, D); Experiments were repeated twice with n=4–7 per group.
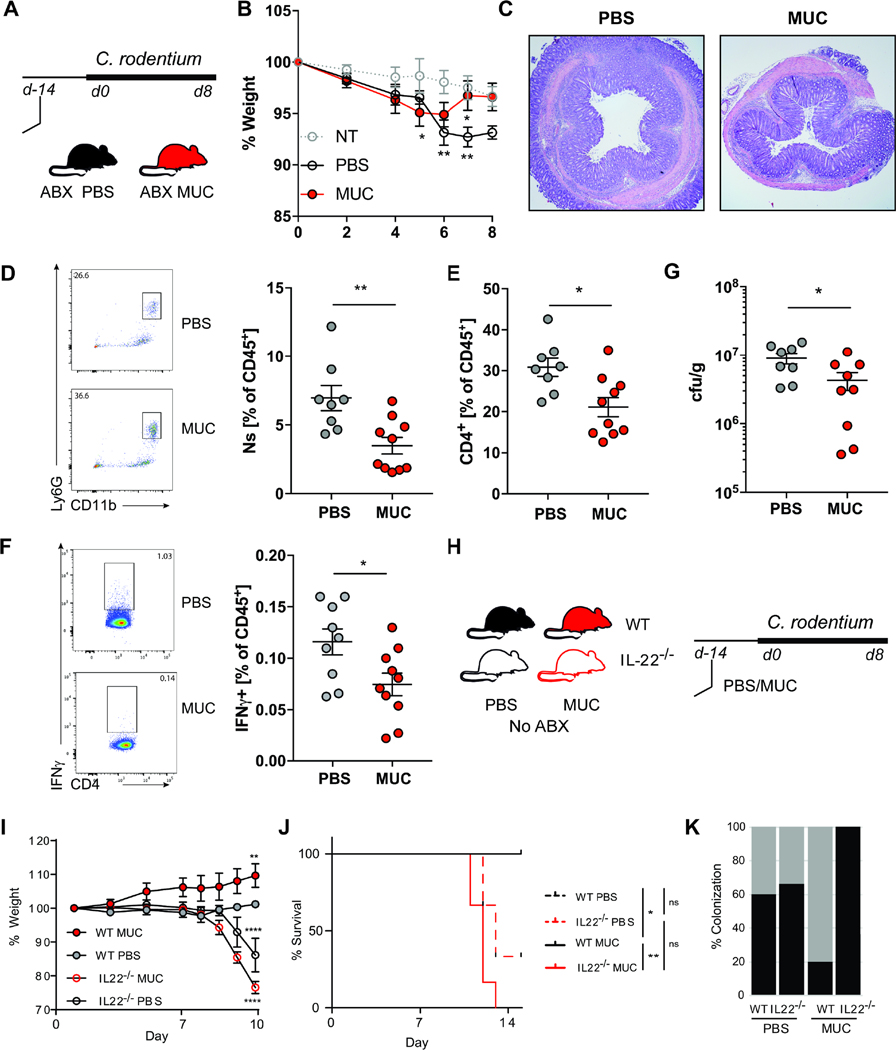
We next assessed whether the MUC consortium would provide protection during infection with intestinally attaching and effacing (A/E) pathogens (Collins et al., 2014; Mundy et al., 2005; Wong et al., 2011). The A/E bacterium Citrobacter rodentium adheres to colonic IEC causing mucosal hyperplasia and wasting disease (Collins et al., 2014). Consistent with its protective effect on the intestinal epithelium, colonization with the MUC consortium reduced the severity of C. rodentium infection and pathogen burden in the intestines in antibiotics treated mice (Figure 4A-G). To determine whether the protective effect was mediated by IL-22, we next infected IL-22 deficient mice in the presence or the absence of MUC. Since Il22−/− mice are highly susceptible to C. rodentium (Ahlfors et al., 2014; Sonnenberg et al., 2011; Zheng et al., 2008) inoculation was performed in absence of antibiotic treatment (Figure 4H). The MUC consortia did not provide any benefit to Il22−/− mice but rather aggravated disease severity (Figure 4 I-J) and resulted in increased C. rodentium colonization (Figure 4K). These data suggest that the protective effect of the MUC consortia in infectious colitis is IL-22-associated and is not due to direct competition with C. rodentium.
Figure 4. A defined mucosal fungal consortium is protective against C. rodentium induced colitis.
A. C57BL/6J mice were colonized with a fungal consortium (MUC) or administered PBS (PBS) every 3 days for 2 weeks. Mice were administered with an antibiotic cocktail (ampicillin 0.4g/l, vancomycin 0.4g/l, metronidazole 0.3g/l, cefoperazone 0.4g/l, 10% sucrose) or not treated (NT) for 3 days before the inoculation with 109 C. rodentium. Mice were sacrificed at day 8 post C. rodentium infection B. Weight change following C. rodentium infection. #: 2 PBS mice reached the humane end-point and had to be sacrificed on day 6. C. Representative histological H&E section of the colon at day 8. D. Representative flow cytometry plot and quantification of CD11b+ Gr-1+ neutrophils (shown as percent among CD45+ lymphocytes) infiltration in the colonic lamina propria (cLP). E. Quantification of CD4+ T cells infiltration in the cLP. F. Representative flow cytometry plot and quantification of IFNγ+ CD4+ T cells (shown as percent among CD45+ lymphocytes) in the mesenteric lymph nodes (mLN). G. C. rodentium colony forming units (cfu/g) in the feces of at day 7 post infection. Il22−/− or control littermates (WT) were colonized with a fungal consortium (MUC) or administered PBS (PBS) every 3 days for 2 weeks prior to the inoculation with 109 C. rodentium. H. WT and Il22−/− mice were colonized with MUC or administered PBS every 3 days for 2 weeks before the inoculation with 109 C. rodentium. I. weight loss and J. percent survival. K. Percent C. rodentium colonization following colonization with a fungal consortium (MUC) or in control mice fed with PBS. Dots represent individual mice, mean +/− SEM. Experiments were repeated twice with n=6–10 per group. *P<0.05, **P<0.01, ***P<0.001; Mann-Whitney T Test (D-G); two-way ANOVA (B, I); Log-rank (Mantel-Cox) test (J).
Recent studies have shown that the composition of gut fungal communities is altered in neuropsychiatric conditions that are associated with behavioral changes in humans (Strati et al., 2017; Zou et al., 2021). A behavior-modulatory role for bacteria has been established in mice (Buffington et al., 2016; Clarke et al., 2013; Desbonnet et al., 2014; Hsiao et al., 2013; Mueller et al., 2019; Sudo et al., 2004; Veiga-Fernandes and Mucida, 2016; Wu et al., 2022). In particular, disruption of the gut bacterial communities with antibiotic treatment can lead to behavioral changes in rodents (Bercik et al., 2011; Desbonnet et al., 2015; Kayyal et al., 2020). We thus used a model of antibiotics-induced bacterial dysbiosis (Shao et al., 2019) and examined social and repetitive behaviors in mice colonized with the MUC consortium and age and sex-matched untreated controls (Albelda and Joel, 2012; Desbonnet et al., 2014). MUC colonization did not affect repetitive, anxiety, and compulsion-related behaviors (Albelda and Joel, 2012; Hoeffer et al., 2008) (Figure S5). When subjected to the three-chamber sociability test mice colonized with the MUC consortium demonstrated increased social preference and spent significantly more time in the chamber with a social stimulus and less time in the empty chamber (Figure 5A-E). Further, MUC colonized mice spent more time interacting with the social stimulus as indicated by the direct interaction index (Figure 5F). These results demonstrate that mucosa-associated fungi can promote social behavior, but not anxiety-related or repetitive behaviors. We next wondered whether the changes could be driven by a compensatory effect of gut fungi following the antibiotic-mediated depletion of the bacterial communities. Importantly, MUC colonization promoted social behavior in presence of a stable community of bacteria (the ASF bacterial consortium) suggesting that the observed effect can occur in absence of antibiotics-induced bacterial dysbiosis (Figure 5 G-J).
Figure 5. The mucosal consortium promotes mouse social behavior in the three-chamber social test.
A. 3 weeks old male C57Bl/6J mice were administered with an antibiotic cocktail (ABX: ampicillin 0.4g/l, vancomycin 0.4g/l, metronidazole 0.3g/l, cefoperazone 0.4g/l, 10% sucrose) and were colonized with a fungal consortium (MUC) or administered PBS (PBS) every 4 days until 8 weeks of age. Mice were tested using a three-chamber social task setting. Mice were allowed to explore the empty three-chamber arena for 10 minutes (Habituation Phase). Then, mice were allowed to approach age and sex-matched individuals confined in a cage (S) on one side of the arena or an empty cage (NS) on the other side of the arena (Social Phase). C. center chamber. B. Position heat maps for a single representative mouse of the PBS and MUC group for the habituation (left) and social phase (right). C. Cumulative time spent in the non-social (NS), center (C), and social (S) chamber during the habituation phase. D. Cumulative time spent in the non-social (NS), center (C), and social (S) chamber during the social phase. E. The social preference index for the social phase was calculated based on the time spent in the social (S) and non-social chamber (NS) as (S-NS)/(S+NS). F. Direct interaction index for the social phase. Data shown are mean values, dots represent individual mice, n=10 mice per group. See also Figure S5. G-J. Altered Schadler’s Flora (ASF) colonized mice (males and females) were colonized with a fungal consortium (ASF-MUC) or administered PBS (ASF-PBS) and subjected to the three-chamber social task as described above. Results are pooled from two separate experiments with n>10 per group. G. Position heat maps for a single representative mouse of the ASF-PBS and ASF-MUC group during the social phase. H. Cumulative time spent in the non-social (NS), center (C), and social (S) chamber during the habituation phase. I. Cumulative time spent in the non-social (NS), center (C). C, D, H, I: Data are shown as Tukey box and whiskers plot, Two way ANOVA. E,F,J: Data shown are mean+/−SEM, dots represent individual mice.; E, F: Mann-Whitney T Test. *P<0.05, **P<0.01, ***P<0.001.
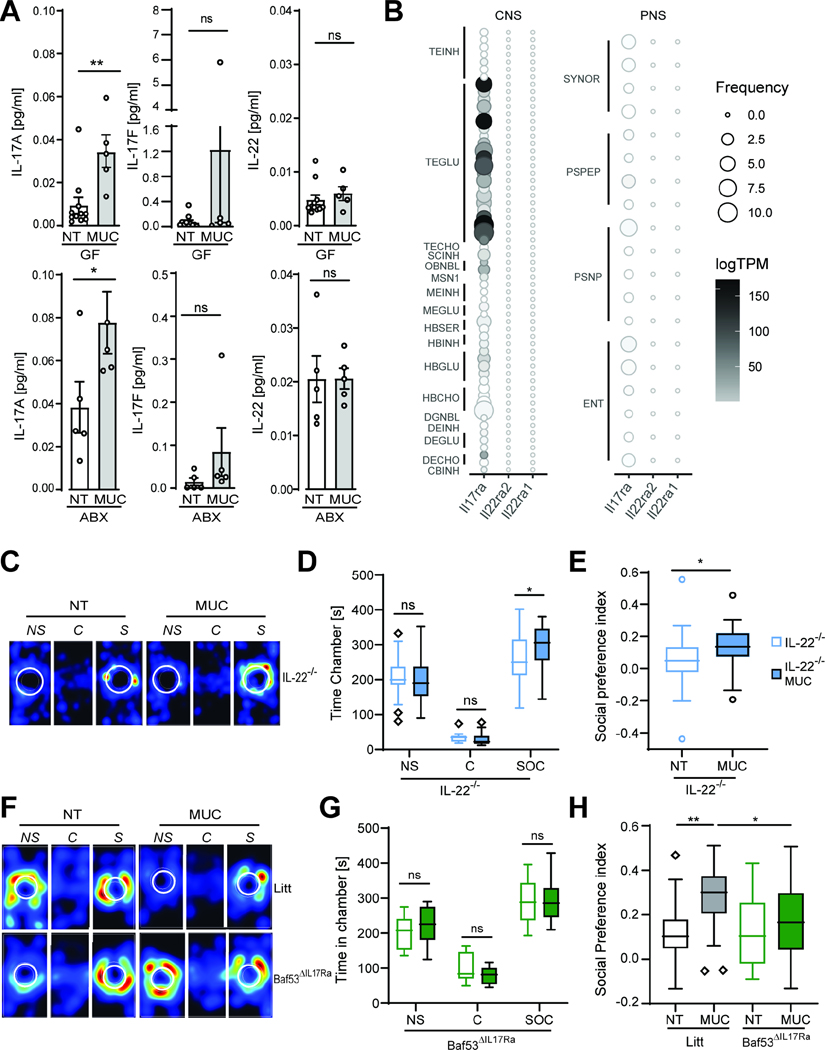
Growing evidence suggests that the interaction between the nervous and immune system play a critical role in the development of social behavior (Kopec et al., 2018). Since various cytokines possess a direct neuro-modulatory role in the central and peripheral neural system (CNS and PNS) (Salvador et al., 2021) we reasoned that the MUC-induced production of IL-22 and or IL-17 observed in both antibiotics treated and ASF mice could be an important driver of the observed behavioral changes. Despite the increase of type 17 cytokines induced by MUC in the intestine, only IL-17A was systemically increased in the serum and spleens of MUC colonized mice (Figure 6A, Figure S6 A). Through the analysis of previously published single-cell RNA Seq data (Zeisel et al., 2018), we confirmed that both enteric and CNS neurons express high levels of the IL-17 receptor gene Il17ra but not the IL-22 receptors genes Il22ra1 and Il22ra2 (Figure 6 B, Table S3). As expected, Il22 deletion did not affect the ability of the MUC consortium to increase social behavior (Figure 6C-E).
Figure 6. IL-17Ra signaling on neurons mediates the MUC-induced increase in social behavior.
A. Mice treated with a cocktail of antibiotics (ABX, 0.3g/l Metronidazole, 0.4 g/l Cefoperazone, 0.4g/l Ampicillin, 0.4 g/l Vancomycin) or germ-free mice (GF) were colonized with the MUC consortium (MUC) or not treated (NT). Circulating levels of cytokines in the serum were measured using a bead-based immune assay. Data shown are mean+/− SEM, dots represent individual mice. See also Figure S6 B. B. Expression pattern of the IL-22 receptors genes Il22ra1 and Il22ra2, and the IL-17A receptor gene Il17ra in a previously published single-cell RNA-seq dataset (Zeisel et al., 2018) of the mouse adult central nervous system (CNS) and peripheral nervous system (PNS). TPM: Transcript per million. Neuron cluster names as assigned in Zeisel et al., 2018 are reported in Table S3. C-E. Male and female Il22−/− mice treated with antibiotics and colonized (MUC) or administered PBS (NT) with the mucosal fungi consortium and subjected to the three-chamber social task. C. Representative position heat maps for individual mice of each group during the social phase. D Cumulative time spent by NT or MUC in the non-social (NS), center (C), and social (S) chamber. E. Social preference index for the social phase. F-H. BAF53bCre/+ mice were crossed with Il17rafl/fl mice to selectively abrogate IL-17ra signaling on neurons (Baf53bΔIL17Ra) littermates were used as controls (Litt). Male and female mice were used in the experiment. Mice were treated with antibiotics and colonized with a fungal consortium (MUC) or not (NT). F. Representative position heat maps for individual mice of each group during the social phase. G. Total time spent in the respective chambers during the social phase. Data is representative of three independent experiments. H. Percent time spent in the chamber center or border of the open field test chamber. Data are pooled from three independent experiments. See also Figure S6 C-F. *P<0.05, **P<0.01, ***P<0.001; A, E: Mann-Whitney T Test. D-G: Two ways ANOVA. H: One-way ANOVA. Tukey box and whiskers plot.
IL-17A has been shown to possess direct neuro-modulatory properties and to influence social behavior in mice (Alves de Lima et al., 2020; Chen et al., 2017; Choi et al., 2016; Reed et al., 2020). To assess whether the direct sensing of IL-17A by neurons contributes to the observed behavioral phenotype we crossed pan neuronal Baf53b-Cre mice (Morarach et al., 2021; Zhan et al., 2015) with Il17ra fl/fl mice to specifically abrogate IL-17 signaling in neurons (Baf53Δ17ra). Following antibiotics induced dysbiosis, MUC colonized Baf53Δ17ra mice had no overt differences in terms of distance traveled, mobility, and anxiety-related behavior when compared to non-treated Baf53Δ17ra controls (Figure S6 C-D). As expected, MUC colonization promoted social behavior in the IL-17R competent cre-littermates (Litt, Figure 6F-H). In contrast, the social behavior of Baf53Δ17ra did not improve upon MUC colonization and was significantly reduced when compared to MUC colonized cre-littermates (Figure 6F-H). Further, MUC colonization did not increase the direct interaction with the age and sex-matched test mice in Baf53Δ17ra mice (Figure S6 E-F). Collectively, our results suggest that fungal colonization of the intestinal mucosa promotes social behavior in mice through a direct action of IL-17A on neurons.
Discussion
The close interaction between the gut microbiota and its host is critical in maintaining mammalian health (Atarashi et al., 2017; Everard et al., 2013; Fung et al., 2014; Hooper et al., 2012). Gut fungi are increasingly recognized as a key component of the gut microbiota with the ability to influence host immune responses (Iliev et al., 2012; Jiang et al., 2017, 2017; Leonardi et al., 2018; Li et al., 2018; Sokol et al., 2017; Yang et al., 2017). However, far less attention has been focused on the role of gut-native fungal communities in homeostasis, despite data suggesting that fungi are important players in the diversification of the host immune repertoire and a key component of a balanced intestinal ecosystem (Iliev and Leonardi, 2017; Wheeler et al., 2017). Akin to bacteria, the outcomes of fungal colonization with select fungi are likely to depend not only on the characteristic of the colonizing species but also on their site-specific ability to interact with the host (Atarashi et al., 2017; Everard et al., 2013; Hooper et al., 2012). Using next-generation sequencing of the fungal rDNA ITS region we characterized the biogeography of the intestinal mycobiota and revealed a site-specific clustering of the gut fungal communities. We identified a subset of fungal genera that are closely associated with the intestinal mucosa. In both humans and mice, these mucosa-associated fungi include genera such as Candida and Saccharomyces, which have been previously shown to modulate host-immunity (Bacher et al., 2019; Chiaro et al., 2017; Leonardi et al., 2018). In contrast, many of the lumen-associated genera that we identified are unlikely to represent true colonizers and might rather be transient environmental fungi lacking the ability to adhere to the mucosa or interact with the intestinal epithelium (Hallen-Adams and Suhr, 2016).
By establishing a defined consortium of fungi that can colonize the intestinal mucosa and comparing their immunomodulatory properties to a consortium of luminal fungi, we unearthed compelling evidence that these mucosa-associated fungi promoted barrier function and protected mice from intestinal injury upon antibiotic treatment and during bacterial infection. In particular, we found that mucosal-associated fungi drove a distinct immunological response which reduced intestinal permeability, and increased mouse social behavior.
Th cells have been shown to play distinct roles during fungal infection and colonization (Speakman et al., 2020). We revealed that the potent induction of IL-22 and IL-17A production in CD4+ T cells differentially mediate the effect of intestinal mucosal fungi on the mammalian host. Complex gene-environment interactions contribute to the etiology of autism spectrum disorders (ASD). While gut bacteria have been implicated in the development of neurological disorders and are known to influence mouse behavior (Chu et al., 2019; Desbonnet et al., 2014), key questions about the involvement of the mycobiota are just beginning to be investigated. We showed that mucosal associated fungi promote murine social behavior both in presence and absence of bacterial communities suggesting a direct effect of fungi along the gut-brain axis. Although previous work by multiple investigators and our study demonstrate that intestinal microbes can modulate host behavior in mice the functional role of the gut microbiota in the etiology of ASD and other neurological manifestations remains controversial (Yap et al., 2021). Greater insight into the mechanism of microbial-mediated behavioral modulation might help shed light into their role in the development of this and other conditions. Cytokine aberrations are common in ASD (Zhao et al., 2021) and might play role in modulating neural function. Cytokines have been shown to regulate the neuro-immune cross-talk via receptors expressed in both neural and immune cells (Veiga-Fernandes and Mucida, 2016; Chen et al., 2017; Salvador et al., 2021; Reed et al., 2020). In this study, we show that mucosal-associated fungi induce unique local and systemic cytokine signatures. In particular, mucosal fungi increase the levels of IL-17A both enterically and in the systemic circulation. IL-17A possesses direct neuro-modulatory properties (Alves de Lima et al., 2020; Chen et al., 2017; Choi et al., 2016; Reed et al., 2020). In mice, IL-17A has been shown to act on neurons of the somatosensory cortex and promote social behavior (Reed et al., 2020). By specifically abrogating IL-17RA on neurons we have shown that IL-17 signaling is an important mediator of the effect of fungi along the gut-brain axis. Our work suggests that direct neuromodulation by fungi-induced IL-17A mediates an increase in social behavior in mice.
In conclusion, our findings suggest that the spatial organization of the mycobiota within the intestinal niche is a key factor in driving protective mucosal immunity. This interaction has a profound local and systemic impact on the intestinal epithelium, neurons, barrier function, intestinal disease, and host behavior.
Limitations of the Study
Although our study demonstrates that mucosal associated fungi can modulate both the host susceptibility to intestinal disease and its behavior, there are a few outstanding issues:
Recent studies have highlighted strong strain-specific differences in the immune-modulatory properties of gut fungi (Li et al., 2022). The experiments described in the current study were performed with a limited number of fungal strains and can thus not be generalized to every member of each species. Further studies are needed to elucidate the functional effects of individual strains.
Neural circuits are subject to sensitive temporal windows of increased plasticity (Lupien et al., 2009). In our behavioral experiments, mice were colonized with fungi immediately after weaning (3 weeks of age). Our study did not identify the critical temporal window for the effect of fungi on mouse behavior.
Our study suggests that IL-17RA on neurons is essential to mediate the effects of fungi along the gut-brain axis. However, we did not thoroughly delineate which neuron populations are involved.
Our work demonstrates that mucosa-associated fungi can promote social behavior in mice. Whether this phenomenon can be translated to humans with the existence of a complex commensal microbial community remains to be determined.
STAR Methods
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. Iliyan D. Iliev, iliev@med.cornell.edu.
Materials availability
This study did not generate new unique reagents.
Data and code availability
The ITS sequencing data are deposited in NCBI Sequence Read Archive (SRA, http://www.ncbi.nlm.nih.gov/Traces/sra, PRJNA594055). The RNA sequencing data are deposited in NCBI Gene expression omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/ GSE141622). This paper does not report original code. All other data needed to evaluate the conclusions in the manuscript are available within the main text or supplementary materials.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
C57BL/6J (Stock #: 000664), Rag1−/− (B6.129S7-Rag1tm1M°m/J, Jackson Stock# 002216), and Il17a−/− (Il17atm1.1(icre)Stck/J, #016879) mice were purchased from the Jackson Laboratories (Bar Harbor, ME) and bred at WCMC for at least 3 generations. IL-22 reporter mice (C57BL/6-Tg (Il22-EGFP)1Gson/J, Jackson Stock# 035005) were generated in the laboratory of Dr. Sonnenberg (Teng et al., 2019). C57BL/6-Il22tm1.1 (icre)Stck/J (Jackson Stock# 027524) x R26R-EYFP (Jackson Stock# 006148), the specific cross was annotated as Il22cre/+ YFP mice allowing for tracking of Il22 expression and was provided by Dr. Gregory Sonnenberg. The Il22cre/cre YFP mice lack Il22 expression and are annotated throughout the study as Il22-/−. BAF53b-Cre mice (Tg (Actl6b-Cre)4092Jiwu/J, #027826) and Il17ra fl/fl mice (B6.Cg-Il17ratm2.1K°ll/J, # 031000) were bred to obtain F1: Actl6b cre+/− Il17ra fl/+ mice. F1 was bred with Il17ra fl/fl mice. Actl6b cre+/− (BAF53bΔIL17ra) or Actl6 cre −/− littermates (Litt) were used as control and co-house with cre+ mice (separated by sex) for the whole length of the experiment. For chemically induced and infectious colitis, littermates were randomly assigned to experimental groups. Animals were used between 8 and 12 weeks of age unless otherwise specified. Males and females were used in approximately equal ratios or as noted in figure legend. All animals were housed under specific pathogen-free (SPF) conditions unless otherwise described at Weill Cornell Medicine. Experiments were performed after prior approval by the Institutional Animal Care and Use Committee of Weill Cornell Medicine.
Human samples
Human colonic mucosa of deidentified individuals according to the Institutional Review Board approved protocol from the Weill Cornell Medicine in accordance with the Helsinki Declaration. Luminal contents were removed by gentle washing with saline and mucosal associated tissues was collected by gentle scraping with a disposable sterile inoculation loop. Samples were snap frozen upon collection and stored at −80C. Upon collection of all specimens, samples were processed for ITS sequencing as described below.
Fungal strains
Fungal cultures are started from stocks in 30% glycerol in Sabouraud dextrose broth (SDB; EMD Chemicals). Candida albicans (SC5314) and Saccharomyces cerevisiae Hansen (ATCC MYA796™) were obtained from the American Type Culture Collection (Manassas, VA). Saccharomycopsis fibuligera (Ili7) was previously isolated from the mouse gut (Iliev et al., 2012). hMUC C. albicans and S. cerevisiae were previously isolated from a mucosal wash. C. albicans, S. cerevisiae and S. fibuligera were cultured overnight in aerobic conditions on SBD agar plates at 37°C. For mice inoculations, cultures were started from single colonies (agar-plates) in liquid SDB at 37°C. For mice inoculation, fungi were centrifuged 10 min at 500 rcf, washed once with sterile PBS and counted. Aspergillus amstelodami (ATCC 46362) was cultured on SDB at 30°C. Wallemia sebi (FRR 1471, ATCC 42964) and Cladosporium cladosporioides (ATCC 38810) were grown on Sabouraud dextrose agar (SDA; EMD Chemicals) at room temperature (20–22°C). For collection of W. sebi and C. cladosporioides plates were flooded with 7 ml 0.05% Tween 80 (in H2O) and carefully scraped with sterile inoculation loop. Liquid was filtered through a 40μm cell strainer, centrifuged 5 min at 450 rcf, washed once in sterile PBS and counted for mice inoculation.
METHOD DETAILS
Experimental replication, randomization, and blinding
Age matched groups of mice were randomly allocated to the experimental groups. To ensure reproducibility, each experiment was performed independently at least twice.
Assessment of Fungal induced immune response
Cefoperazone (0.4 g/l; Sigma-Aldrich, St. Louis, MO) was provided to mice ad libitum in drinking water for the whole length of the experiments. 3 days after starting the cefoperazone treatment, mice were fed with fungal consortia every 4 days. Mice were sacrificed at day 14 and sampled as described below. To assess colonization fecal pellets were collected 3 days after the first fungal gavage. For the assessment of intestinal mucosa colonization, after mouse euthanasia, intestines were removed at day 10 post colonization (or in one set of experiments at day 3 and day 7), rinsed thoroughly with ice cold PBS until removal of all luminal contents. The mucosa was then collected by gentle scraping with a disposable, sterile inoculation loop. Feces or mucosal scrapings were weighted and plated as serial dilutions in PBS on SD agar. Fungal load is quantified as colony forming units (cfu) per gram of feces or assessed by qPCR upon DNA isolated as described below.
DSS-induced epithelial injury.
Mice were orally supplemented with the mucosal fungal consortium every 3 days for 10 days prior to the DSS treatment. Control groups were orally gavaged with PBS at the same time points. Body weight and the presence of occult blood were assessed daily. Mice were administered with sterile 3% dextran sodium sulphate (DSS) in an antibiotic cocktail (ABX: ampicillin 0.4g/l, vancomycin 0.4g/l, metronidazole 0.3g/l, cefoperazone 0.4g/l, 10% sucrose) for 5 days followed by a recovery period of 7 days with ABX and a second 5 days DSS+ABX and ABX cycle until achievement of end point. For the adaptive transfer of CD4+ T cells in Rag1 −/− mice spleens and mesenteric lymph nodes of Il22−/− or Il22+/− donor mice were removed by dissection. Tissues were mashed into a nylon screen, and the cells obtained were pooled, washed twice in Hanks’ balanced salt solution (HBSS; Sigma-Aldrich), and resuspended at 108 cells/ml. CD4+ T cells were isolated by negative selection, using MagniSort™ Mouse CD4+ T cell Enrichment Kit (eBioscience), according to the manufacturer’s protocol. Isolated CD4+ T cells were transferred into recipient Rag1 −/− mice by retro-orbital injection 1 day prior to the administration of DSS.
Citrobacter rodentium infection
C. rodentium was grown on a Luria-Bertani (LB) plate 2 days before infection. A liquid culture from a single colony was started in LB and incubated overnight (37°C at 200 rpm) and then regrown on 50ml of LB for 3–4 hours until growth to 0.6 at OD600. Mice were orally inoculated with 109 bacteria per mouse (in 100 μl PBS). Body weights was assessed daily. Feces were collected, weighted, and plated in serial dilutions on MacConkey agar No 1 (Sigma) for C. rodentium quantification.
Gnotobiotic mice
Altered Schaedler flora (ASF) mice were generated from germ-free (GF) C57BL/6 mice inoculated with ASF and bred for at least 5 generations to obtain fully immunocompetent progeny. ASF mice were maintained within sterile vinyl isolators at Weill Cornell Medical College Gnotobiotic Mouse Facility. For the generation of gnotobiotic mice colonized with the fungal mucosal consortium, ASF mice were colonized at 5 weeks of age and housed in isolated cages.
Isolation of large intestine lamina propria cells
Intestinal lamina propria (LP) cells were isolated as previously described (Leonardi et al., 2018) with some modifications. Briefly, colons and ceca were isolated, opened longitudinally, washed of fecal contents, and then cut into 1 cm pieces. Intestinal pieces were transferred into HBSS medium (Sigma), supplemented with 2 mM EDTA, and were shaken for 8 min at 37°C. The remaining tissue was washed, cut in small pieces, and subsequently incubated in digestion medium consisting of RPMI 1640, 5% fetal bovine serum (FBS), 0.5 mg/ml collagenase type VIII (Sigma), 5 U/ml DNase (Roche Diagnostics), 100 IU/ml penicillin and 100 μg/ml streptomycin for 25 min at 37°C by gentle shaking. The cell suspensions were washed with PBS, filtered through a mesh, and used directly for immune-phenotyping of LP cells.
Antibodies and flow cytometry
Cell suspensions were prepared as described above, blocked with CD16/CD32 (Mouse BD Fc Block™, 2.4G2, BD Biosciences) and stained with Fixable Viability Dye (eFluor® 506, eBioscience, 1:1000 concentration) and with surface antibodies at a 1:250 concentration in PBS 0.5% Bovine Serum Albumine (antibodies are listed in the Key Resource Table). For intracellular staining of transcription factors, cells were stained with surface markers, fixed permeabilized over night at 4°C with eBioscience™ Fixation/Permeabilization buffer. Cells were stained with anti-TF antibodies at a 1:200 concentration in eBioscience™ Permeabilization buffer. For intracellular cytokine detection, cells were incubated with 50 ng/ml phorbol 12-myristate 13-acetate (PMA; Sigma-Aldrich), 500 ng/ml ionomycin (Sigma-Aldrich) and 10 μg/ml Brefeldin A (BFA; Sigma-Aldrich) in complete RPMI media at 37° C for 6 h. After surface staining cells were fixed, permeabilized overnight at 4 °C with BD Cytofix/Cytoperm™ buffer. Staining was performed using the relevant anti-cytokine antibodies for 1h 4 °C at a 1:100 concentration in BD Perm/Wash™ Buffer. Flow cytometry was performed using a LSRFortessa (BD Biosciences) and data were analyzed with FlowJo software (TreeStar Inc.). See supplementary figure 2 for gating procedure.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD16/CD32 monoclonal antibody (93), eBioscience™ | Thermo Fisher Scientific (eBioscience™) | Cat # 14-0161-81; RRID: AB_467132 |
| Anti-mouse I-A/I-E a | BioLegend | Cat # 107639; RRID: AB_2565894 |
| Anti-mouse CD11b monoclonal antibody (M1/70), PE-Cyanine7 | Thermo Fisher Scientific (eBioscience™) | Cat # 25-0112-82; RRID: AB_469588 |
| Anti-mouse Ly-6C monoclonal antibody (HK1.4), eFluor 450 | Thermo Fisher Scientific (eBioscience™) | Cat # 48-5932-82; RRID: AB_10805519 |
| Anti-mouse Siglec-F (E50-2440), PE | BD Biosciences | Cat # 552126; RRID: N/A |
| Anti-mouse F4/80 antibody (BM8), Alexa Fluor 488 | BioLegend | Cat # 123120; RRID: AB_893479 |
| Anti-mouse Syk antibody (5F5), PE | BioLegend | Cat # 646004; RRID: AB_2565306 |
| Anti-mouse Ly-6G monoclonal antibody (1A8-Ly6g), APC | Thermo Fisher Scientific (eBioscience™) | Cat # 17-9668-80; RRID: AB_2573306 |
| Anti-mouse CX3CR1 antibody (SA011F11), BV421 | BioLegend | Cat # 149023; RRID: AB_2565706 |
| Anti-mouse CD11c antibody (N418), Alexa Fluor 700 | BioLegend | Cat # 117319; RRID: AB_528735 |
| Anti-mouse CD45 monoclonal antibody (30-F11), APC-eFluor 780 | Thermo Fisher Scientific (eBioscience™) | Cat # 47-0451-82; RRID: AB_1548781 |
| Anti-mouse CD4 Antibody (RM4-5), Brilliant Violet 605™ | BioLegend | Cat # 100548; RRID: AB_2563054 |
| Anti-mouse Gata-3 monoclonal antibody (TWAJ), PerCP-eFluor 710 | Thermo Fisher Scientific (eBioscience™) | Cat # 46-9966-42; RRID: AB_10804487 |
| Anti-mouse FOXP3 monoclonal antibody (FJK-16s), APC | Thermo Fisher Scientific (eBioscience™) | Cat # 17-5773-82; RRID: AB_469457 |
| Anti-mouse ROR gamma (t) Monoclonal antibody (B2D), PE | Thermo Fisher Scientific (eBioscience™) | Cat # 12-6981-82; RRID: AB_10807092 |
| Anti-mouse IFN gamma monoclonal antibody (XMG1.2), FITC | Thermo Fisher Scientific (eBioscience™) | Cat # 11-7311-82; RRID: AB_465412 |
| Anti-mouse IL-17A Antibody (TC11-18H10.1) Pacific Blue™ | BioLegend | Cat # 506918; RRID: AB_893544 |
| Anti-mouse CD127/IL-7Ra Antibody (A7R34) | Thermo Fisher Scientific (eBioscience™) | Cat# 135014, RRID: AB_1937265 |
| Anti-mouse CD45 antibody (30-F11), Brilliant Violet 650™ | Thermo Fisher Scientific (eBioscience™) | Cat # 103151; RRID: AB_2565884 |
| Anti-mouse CD3ε antibody (145-2C11), APC-Cy7 | BioLegend | Cat#100330;RRID:AB_1877170 |
| Anti-mouse TCR β antibody (H57-597), FITC | Tonbo | Cat#:35-5961-U100; RRID: AB_1851905 |
| Anti-mouse NK-1.1/CD161 (PK136) antibody, PerCP-Cy5.5 | BioLegend | Cat#:108728 RRID: AB_2132705 |
| Anti-mouse CD5 (53-7.3) antibody, FITC | Thermo Fisher Scientific (eBioscience™) | Cat#:11-0051-82 RRID: AB_464908 |
| Anti-mouse IL-22 (IL-22JOP) antibody, PE | Thermo Fisher Scientific (eBioscience™) | Cat#: 17-7222-82, RRID: AB_10597583 |
| Anti-mouse IL-17F (9D3.1C8) antibody, PE | BioLegend | Cat#:517007 RRID: AB_10661730 |
| eBioscience™ Fixable Viability Dye eFluor™ 506 | Thermo Fisher Scientific (eBioscience™) | Cat # 65-0866-14 |
| Bacterial and fungal Strains | ||
| Altered Schaedler Flora (ASF) | N/A | (Gomes-Neto et al., 2017) |
| Citrobacter rodentium | ATCC | ATCC 51459 |
| Candida albicans SC5314 | ATCC | ATCC MYA-2876) |
| Saccharomyces cerevisiae Meyen ex E.C. Hansen | ATCC | ATCC® MYA-796 |
| Aspergillus amstelodami | ATCC | ATCC 46362 |
| Wallemia sebi (FRR 1471) | ATCC | ATCC 42964 |
| Cladosporium cladosporioides | ATCC | ATCC 38810 |
| Candida albicans human mucosal isolate | This study | N/A |
| Saccharomyces cerevisiae human mucosal isolate | This study | N/A |
| Saccharomycopsis fibuligera ili7 mouse gut isolate | This study | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Collagenase type VIII | Sigma-Aldrich | Cat# C2139 |
| DNase I | Sigma-Aldrich | Cat# D5025 |
| Brefeldin A | Thermo Fisher Scientific (eBioscience™) | Cat# 00-4506-51 |
| Ionomycin | Sigma-Aldrich | Cat# I0634 |
| Phorbol 12-myristate 13-acetate (PMA) | Sigma-Aldrich | Cat# P8139 |
| Fetal Bovine Serum | Corning | Cat# MT35016CV |
| Hanks’ balanced salt solution (HBSS) | Sigma | Cat# H4385 |
| 0.5M EDTA, pH 8.0 | Corning | Cat# 46-034-CI |
| Probumin® Bovine Serum Albumin Diagnostic Grade, Powder | VWR | Cat# IC820451 |
| Sabouraud dextrose broth | VWR | Cat# 89406-400 |
| Sabouraud 4% dextrose agar | VWR | Cat# EM1.05438.0500 |
| MacConkey agar Agar No 1 (Sigma) | Sigma-Aldrich | Cat# 70143 |
| LB Agar Miller Dehydrated Culture Media | Millipore Sigma | Cat# 110283 |
| Penicillin-Streptomycin (10,000 U/mL) | Thermo Fisher Scientific | Cat# 15140122 |
| Tween® 80 (Polysorbate) | VWR | Cat# 97061-674 |
| Dextran sodium sulphate | MP Biomedicals | Cat# 216011090 |
| Ampicillin (Sodium), USP Grade | Gold Biotechnology, Inc. | Cat# A-301-5 |
| Vancomycin | Gold Biotechnology, Inc. | Cat# V-200-5 |
| Cefoperazone sodium salt | Sigma | Cat# C4292-5G |
| Sucrose | Sigma | Cat# S0389-500G |
| Metronidazole | Sigma | Cat# M3761-5G |
| Critical Commercial Assays | ||
| FoxP3 /Transcription Factor Staining Buffer set | Thermo Fisher Scientific (eBioscience™) | Cat# 00-5523-00 |
| BD Fixation/Permeabilization Solution Kit | BD Biosciences | Cat# 554714 |
| LEGENDplex™ Custom Mouse 11plex Panel | BioLegend | Cat# 900001231 |
| MagniSort™ Mouse CD4 Naïve T cell Enrichment Kit | ThermoFisher | Cat# 8804-6824-74 |
| Direct-zol RNA Miniprep | Zymo Research | Cat# R2051 |
| Deposited Data | ||
| RNA sequencing | NCBI |
https://www.ncbi.nlm.nih.gov/geo/ GSE141622 |
| ITS Sequencing | NCBI |
https://www.ncbi.nlm.nih.gov/sra/docs/ GenBank: PRJNA594055 |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6 | The Jackson Laboratory | JAX# 000664 |
| Mouse: C57BL/6-Il22tm1.1(icre)Stck/J | The Jackson Laboratory | JAX# 027524 |
| Mouse: B6.129X1-Gt(ROSA)26Sortm1(EYFP)Cos/J | The Jackson Laboratory | JAX# 006148 |
| Mouse: C57BL/6-Tg(Il22-EGFP)1Gson/J | The Jackson Laboratory | JAX# 035005 |
| Mouse: R26R-EYFP | The Jackson Laboratory | JAX# 006148 |
| Mouse: Tg(Actl6b-Cre)4092Jiwu/J | The Jackson Laboratory | JAX# 027826 |
| Mouse: B6.Cg-Il17ratm2.1Koll/J | The Jackson Laboratory | JAX# 031000 |
| Mouse: Il17atm1.1(icre)Stck/J | The Jackson Laboratory | JAX #016879 |
| Mouse: Rag1−/− B6.129S7-Rag1tm1Mom/J | The Jackson Laboratory | JAX# 002216 |
| Software and Algorithms | ||
| FlowJo LLC 10.1. | Becton Dickinson | N/A |
| EthoVision 12.0.1138 | Noldus Information Technology | |
| GraphPad Prim 8.3.0 (538) | GraphPad Software | N/A |
| RStudio Desktop 1.2.1335 | https://rstudio.com/ | N/A |
| R version 3.5.0 (2018-04-23) | www.r-project.org | N/A |
Behavioral testing
Age and sex matched C57BL/6J mice were purchased from Jackson Laboratories (Bar Harbor, ME) and housed at WCMC in temperature-controlled rooms with a 12-hour light/dark cycle. Mice were tested during the light phase of the light cycle under dim light conditions (18–20 LUX); to control for time-of-day effects, cages of different groups were alternated. To minimize animal stress, cages were changed weekly by the investigator performing the behavioral testing; care was taken not to test animals within three days of cage changes. To further minimize confounding results due to stress, mice were handled for three consecutive days prior to the start of testing. Mice were permitted a minimum 60-minute habituation period to the testing room, and testing was performed at the same time of day. All comparisons were made between littermates. Behaviors were tested in the following order: social behavior, marble burying, elevated plus maze.
Three chamber sociability test
During a ten-minute habituation period, a test mouse was placed in the center chamber and given free access to the entire arena (Figure 5A). A clean pair of gloves was used to handle each mouse. The two lateral chambers contained an inverted empty metal cylinder. Immediately following the habituation phase, an unfamiliar, age and sex-matched C57BL/6J mouse was placed in one of the two cylinders for a total of 10 additional min. Between subjects, the placement of the social stimulus mouse was successively alternated between the two lateral chambers (Moy et al., 2004). The entire arena setting was cleaned with water and Clidox-S (Pharmacal, Naugatuck, CT) after each trial. EthoVision software (EthoVision, Noldus Information Technology, Netherlands) was linked via an overhead camera and tracked the movement of each subject in every compartment. The distance traveled (cm) and the duration of stay (s) in each compartment were measured automatically by EthoVision. Nose, center, and tail points were tracked. The direct interaction index for the social phase was calculated based on the time spent in the social (S) and non-social chamber (NS) as (S-NS)/(S+NS).
Elevated plus maze test
The elevated plus maze consisted of two opposite open arms (25 × 5 cm, with 3-mm-high ledges) and two perpendicular closed arms (25 × 5 cm, with 15-cm-high walls) of the same size. The maze was made of opaque plastic plates and was elevated to a height of 50 cm. Each mouse was placed in the central square of the maze (5 × 5 cm), facing one of the open arms, and was recorded for 10 min. The distance traveled (cm) and the duration of stay (s) in each compartment were measured automatically by EthoVision.
Marble burying
Mice were placed in a cage bottom with floor area of 483 cm2 filled with 4 cm of fresh, autoclaved wood chip bedding. Each cage contained 20 homogenously distributed glass marbles (4 × 5) on top of the bedding. Mice were returned to their cage after 10 min. The number of buried marbles (50% or more covered) was recorded. Bedding and cage were replaced for each mouse, marbles were washed with Clidox-S (Pharmacal, Naugatuck, CT), rinsed with water and dried between tests.
DNA isolation, fungal and bacterial rDNA gene quantitative analysis
Mouse intestinal content or mucosal scrapings were collected. DNA for fungal sequencing and validation RT-qPCR was isolated from mouse intestinal content or mucosal scrapings, or from human colonic mucosa of deidentified individuals according to the Institutional Review Board approved protocol from the Weill Cornell Medicine, following lyticase treatment, bead beating with 0.1mm and 0.5 mm beads, and processing using QIAmp DNA mini kit (Qiagen) as in (Tang et al., 2015).
Microbiome sequencing analysis, Illumina library generation and sequencing
Fungal ITS1–2 regions were amplified by PCR for analysis of mouse and human fungal microbiomes that were sequenced using the Illumina MiSeq platforms. The following PCR primers: ITS1F CTTGGTCATTTAGAGGAAGTAA; ITS2R GCTGCGTTCTTCATCGATGC, were modified to include forward (5’ TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG‐[locus-specific sequence]) and reverse (5’ GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAG‐[locus-specific sequence]) sequencing adaptors. ITS amplicons were generated with 35 cycles, using Invitrogen AccuPrime PCR reagents (Carlsbad, CA). Amplicons were then used in the second PCR reaction, using Illumina Nextera XT v2 (San Diego, CA) barcoded primers to uniquely index each sample and 2×300 paired end sequencing was performed on the Illumina MiSeq (Illumina, CA). DNA was amplified using the following PCR protocol: Initial denaturation at 94°C for 10 min, followed by 40 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and elongation at 72°C for 2 min, followed by an elongation step at 72°C for 30 min. All libraries were subjected to quality control using qPCR, DNA 1000 Bioanalyzer (Agilent), and Qubit (Life Technologies) to validate and quantitate library construction prior to preparing a Paired End flow cell. The sequencing data are deposited in NCBI Sequence Read Archive (SRA, http://www.ncbi.nlm.nih.gov/Traces/sra, SUB6653651).
Data Analysis
Raw FASTQ ITS1 sequencing data were filtered to enrich for high quality reads, removing the adapter sequence by cutadapt v1.4.1 or any reads that do not contain the proximal primer sequence (Tang et al., 2015). Sequence reads were then quality-trimmed by truncating reads not having an average quality score of 20 (Q20) over a 3 base pair sliding window and removing reads shorter than 100 bp (57). These high quality reads were then aligned to Targeted Host Fungi (THF) ITS1 database, using BLAST v2.2.22 and the pick_otus.py pipeline in the QIIME v1.6 wrapper with an identity percentage ≥97% for operational taxonomic unit (OTU) picking (Altschul et al., 1990). The alignment results were then tabulated across all reads, using the accession identifier of the ITS reference sequences as surrogate OTUs and using a Perl script (Tang et al., 2015). For Illumina bacterial analysis, because of the abundance of Illumina reads and higher overall sequence quality of the reads, we used the QIIME package with minimal customization (Edgar, 2010). Shannon diversity index (H) was calculated at the OTUs levels as (1)
| (1) |
where pi is the proportional abundance of OTUs i.
Simpson Diversity Index (1-D) (60) was calculated as (2)
| (2) |
Where N= total number of individuals of all species, ni = total number of individuals for each species i.
Gene expression profiling by RNA-seq and bioinformatics analyses
For RNA-sequencing (RNA-seq), ASF mice were colonized with the mucosal consortium by oral gavage. A single litter of 9-week-old animals was used for the experiment. Two weeks after gavage of the mucosal consortium, mice were sacrificed, and the colons were removed and opened longitudinally. The colons were rinsed thoroughly with ice cold PBS until removal of all luminal contents. Intestinal epithelial cells (IEC) were collected by gentle scraping with a disposable, sterile inoculation loop. IEC were collected in Trizol and lysed by repeated passing through a 27G syringe needle. RNA was extracted by chloroform phase separation followed by RNA extraction with in-column DNase treatment using the Direct-zol RNA Kit (Zymo Research). Agilent Bioanalyzer Sample QC – Nanogel was used to determine the Total RNA concentration and quality (Table S2).
Poly-A pull-down was performed to enrich mRNAs from total RNA samples, followed by library construction using Illumina TruSeq chemistry. Cluster generation and 50–base pair single-end 50 cycled sequencing was performed on the Illumina HiSeq4000 system. Raw sequence reads were mapped to the mouse genome mm10/GRCm38 using the STAR aligner (Dobin et al., 2013) version 2.5.2b. Mapped reads were counted at the gene level using the Rsubread R package (Liao et al., 2019). Lowly expressed genes were prefiltered, keeping only genes with 50 or more raw counts in at least 2 samples. Library size normalization and further analysis was performed using the R package DESeq2 version 1.22.2 (Love et al., 2014). Principal components analysis (PCA) was performed based on the 500 genes with highest variance after applying the DESeq2 variance stabilizing transformation. Differential expression was assessed using the Wald test as implemented in DESeq2, with a False Discovery Rate (FDR) threshold of 0.1 being used as a significance threshold. Gene set enrichment analysis vas performed using the R package fgsea version 1.8.0 (Korotkevich et al., 2021) and the Molecular Signatures Database (MSigDB) H hallmark gene sets for mouse (http://bioinf.wehi.edu.au/software/MSigDB).
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis was calculated by GraphPad Prism (GraphPad Software) and R. Statistical details of experiments can be found in the figures’ legend. Unless otherwise indicated individual dots represent each individual mouse and data are presented as the mean ± SEM. Significance is defined as: *p < 0.05, **p < 0.01, **p < 0.00. Mann-Whitney Test or one-way analysis of variance (ANOVA) were performed to data comparison as specified in the figures’ legend.
Supplementary Material
A. rt-qPCR of mucosal scraping of mice colonized with the mucosal fungal consortium 10 days post-colonization. Data shown are mean values, dots represent individual mice. B. Gating strategy of myeloid cells in the colonic lamina propria. Lineage: CD3, NK1.1, Gr1, TCRβ, CD5, CD19). B. Gating strategy of lymphocytes in the colonic lamina propria. CD4+ T cells are gated as Live (Fixable Viability Dye negative), CD45+ CD3+ CD90.2+ CD4+. Type 3 innate lymphoid cells are gated as Live, CD45+ Lin- CD3- CD90.2+ CD127+ CD127+ RORγt+. Lin: CD11c, CD11b, NK1.1, Gr1, TCRβ, CD5, CD19).
SPF mice were treated with antibiotics were colonized with consortia of mucosal (MUC) or luminal (LUM) fungi. A. Representative flow cytometry plot and quantification of the numbers of FoxP3+ RORγt-, FoxP3+ RORγt+, and FoxP3- RORγt+ CD4+ T cells among CD45+ hematopoietic cells in the mLN. B. Representative flow cytometry plot and quantification of the total numbers of IL-17A and/or IL-22 producing CD4+ Th cells. B. Representative flow cytometry plot and quantification of the total numbers of IL-17A and IL-17F producing CD4+ Th cells. C. Representative flow cytometry plot and quantification of the total numbers of IFNγ producing CD4+ Th cells. Experiment was repeated three times with n>4 per group. *P<0.05, **P<0.01, ***P<0.001. Data are shown as Tukey Box Whiskers plots. Mann-Whitney T Test (F, G).
A-B: C57Bl/6J mice were treated with antibiotics and inoculated with the Mucosal consortium (MUC), the individual members of the consortium (Sc: S. cerevisiae, Ca: C. albicans, Sf: S. fibuligera), or left untreated. A. Frequency of IL-17A+ CD3+ CD4+ T cells among the CD45+ cLP population. B. Frequency of IL-22A+ CD3+ CD4+ T cells among the CD45+ cLP population. C-D: IL-22 reporter mice (expressing eGFP under the control of the IL-22 promoter) were treated with antibiotics and inoculated with the Mucosal consortium (MUC) or left untreated. cLP were harvested at day 3 and 7. C. Frequency of eGFP+ CD3+ CD4+ T cells among the CD45+ cLP population. D. Frequency of eGFP+ Lin-CD90.2+ CD127+ ILC3 among the CD45+ cLP population. E-F. Fate mapper mice were colonized with the MUC consortium. Expression of IL-22 by CD45+ cells in the colonic lamina propria (cLP, E) and mesenteric lymph nodes (F). G-H. Fate mapper mice were colonized with a MUC consortium (MUC) composed of mucosa-associated C. albicans and S. cerevisiae, isolated from healthy individuals. Control mice were administered PBS (PBS). G. Expression of IL-22 by CD4+ T cells in the cLP. H. Expression of IL-22 by CD4+ T cells in the mLN. Data shown are mean values +/− SEM, dots represent individual mice. *P<0.05, **P<0.01, ***P<0.001; Mann-Whitney T est.
A-B. Representative flow cytometry plots and quantification of IL-22, IL17A, IL-17F, and IFNγ production by CD3+ CD4+ T cells in the cLP. C. Quantification of IL-22, IL-17A, IL-17F, and IFNγ production by CD3+ CD4+ T cells in the mesenteric lymph nodes (mLN). Data shown are mean values, dots represent individual mice. *P<0.05, **P<0.01, ***P<0.001; Mann Whitney Test. E. WT and Il17a−/− mice were treated with broad spectrum antibiotics prior to fungal colonization (MUC) or not (NT) and administration of Dextran Sodium Sulphate as described in Figure 2 G. F. Percent survival. Log-rank (Mantel-Cox) test.
A-E. 3 weeks old male C57Bl/6J mice (n=10 per group) were administered with an antibiotic cocktail (ABX: ampicillin 0.4g/l, vancomycin 0.4g/l, metronidazole 0.3g/l, cefoperazone 0.4g/l, 10% sucrose) or not treated (NT). ABX mice were colonized with a fungal consortium (MUC) or administered PBS (PBS) every 4 days until testing at 8 weeks of age. A. Marble burying behavior in the marble burying test. Graph shows the total number of buried marbles (out of 20) during a 10-minute trial. B. Experimental layout of the elevated plus-maze test and total distance travelled (cm) by individual mice. C. Frequency of visits to the closed arms of the elevated plus-maze. D. Percent time spent in the closed arms. E. Percent time spent in the open arms of the elevated plus maze. Data shown are mean values, dots represent individual mice. *P<0.05, **P<0.01, ***P<0.001; Mann-Whitney T Test.
A. Representative flow cytometry plot and quantification of the production of cytokines by CD4+ T helper cells in the mouse spleen following colonization with a luminal consortium (LUM) or mucosal consortium (MUC). B. Levels of Cytokines in the serum of mice following colonization with the MUC consortium. Mice treated with a cocktail of antibiotics (ABX, 0.3g/l Metronidazole, 0.4 g/l Cefoperazone, 0.4g/l Ampicillin, 0.4 g/l Vancomycin) or germ-free mice (GF) were colonized with the MUC consortium (MUC) or not treated (NT). Circulating levels of cytokines in the serum were measured using a bead-based immune assay. Data shown are mean values, dots represent individual mice. *P<0.05, **P<0.01, ***P<0.001; Mann Whitney Test. C-F. Baf53bCre/+ mice were crossed with Il17rafl/fl mice to selectively abrogate IL-17ra signaling on neurons (Baf53bΔIl17ra) littermates were used as controls (Litt). Mice were treated with antibiotics and colonized with a fungal consortium (MUC) or not (NT). C. Total fungal loads expressed as colony forming unit (cfu) per gram of feces in Litt and Baf53bΔIl17ra mice. D. Direct interaction index during the social phase. E. Total distance travelled and moving time in the habituation phase. F. Percent time spent in the chamber center or border of the open field test chamber. Dots represent individual mice (triangles: females, diamonds males). Mann-Whitney T test.
Supplemental Table 1. Related to Figure 1. Metadata for human subjects’ samples.
Supplemental Table 2. Related to Figure 2. RNA quality
Supplemental Table 3. Related to Figure 6. Publicly available RNA-Seq data obtained from Zeisel, 2018 (Zeisel et al., 2018)
Highlights.
A specific community of fungi is present in the intestinal mucosa of humans and mice
Mucosa-associated fungi (MAF) induce Type 17 immunity by T helper cells
MAF protect mice against intestinal injury and infection via IL-22-dependet mechanisms
MAF promote social behavior in mice through IL-17-mediated signaling in neurons
Acknowledgments
We thank the members of the Iliev lab for their critical feedback and for their help in designing the graphical abstract using Biorender. We thank Gregory Sonnenberg for discussions and for providing key reagents. Irina Leonardi is supported though CCF 568319 Research Fellowship Award; Research in the Iliev laboratory is supported by US National Institutes of Health (R01DK113136, R01DK121977, and R01AI163007), the Leona M. and Harry B. Helmsley Charitable Trust, the Irma T. Hirschl Career Scientist Award, Crohn’s and Colitis Foundation, and the Burrough Welcome Trust PATH Award.
Footnotes
Declaration of Interests
The authors declare that they have no conflicts of interest with the contents of this article.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aggor FEY, Break TJ, Trevejo-Nuñez G, Whibley N, Coleman BM, Bailey RD, Kaplan DH, Naglik JR, Shan W, Shetty AC, McCracken C, Durum SK, Biswas PS, Bruno VM, Kolls JK, Lionakis MS, Gaffen SL, 2020. Oral epithelial IL-22/STAT3 signaling licenses IL-17–mediated immunity to oral mucosal candidiasis. Science Immunology 5, eaba0570. 10.1126/sciimmunol.aba0570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlfors H, Morrison PJ, Duarte JH, Li Y, Biro J, Tolaini M, Meglio PD, Potocnik AJ, Stockinger B, 2014. IL-22 Fate Reporter Reveals Origin and Control of IL-22 Production in Homeostasis and Infection. The Journal of Immunology 193, 4602–4613. 10.4049/jimmunol.1401244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albelda N, Joel D, 2012. Animal models of obsessive-compulsive disorder: Exploring pharmacology and neural substrates. Neuroscience & Biobehavioral Reviews 36, 47–63. 10.1016/j.neubiorev.2011.04.006 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ, 1990. Basic local alignment search tool. Journal of Molecular Biology 215, 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Alves de Lima K, Rustenhoven J, Da Mesquita S, Wall M, Salvador AF, Smirnov I, Martelossi Cebinelli G, Mamuladze T, Baker W, Papadopoulos Z, Lopes MB, Cao WS, Xie XS, Herz J, Kipnis J, 2020. Meningeal γδ T cells regulate anxiety-like behavior via IL-17a signaling in neurons. Nat Immunol 21, 1421–1429. 10.1038/s41590-020-0776-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asseman C, Read S, Powrie F, 2003. Colitogenic Th1 Cells Are Present in the Antigen-Experienced T Cell Pool in Normal Mice: Control by CD4+ Regulatory T Cells and IL-10. The Journal of Immunology 171, 971–978. 10.4049/jimmunol.171.2.971 [DOI] [PubMed] [Google Scholar]
- Atarashi K, Suda W, Luo C, Kawaguchi T, Motoo I, Narushima S, Kiguchi Y, Yasuma K, Watanabe E, Tanoue T, Thaiss CA, Sato M, Toyooka K, Said HS, Yamagami H, Rice SA, Gevers D, Johnson RC, Segre JA, Chen K, Kolls JK, Elinav E, Morita H, Xavier RJ, Hattori M, Honda K, 2017. Ectopic colonization of oral bacteria in the intestine drives TH1 cell induction and inflammation. Science 358, 359–365. 10.1126/science.aan4526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aujla SJ, Chan YR, Zheng M, Fei M, Askew DJ, Pociask DA, Reinhart TA, McAllister F, Edeal J, Gaus K, Husain S, Kreindler JL, Dubin PJ, Pilewski JM, Myerburg MM, Mason CA, Iwakura Y, Kolls JK, 2008. IL-22 mediates mucosal host defense against Gram-negative bacterial pneumonia. Nat Med 14, 275–281. 10.1038/nm1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacher P, Hohnstein T, Beerbaum E, Röcker M, Blango MG, Kaufmann S, Röhmel J, Eschenhagen P, Grehn C, Seidel K, Rickerts V, Lozza L, Stervbo U, Nienen M, Babel N, Milleck J, Assenmacher M, Cornely OA, Ziegler M, Wisplinghoff H, Heine G, Worm M, Siegmund B, Maul J, Creutz P, Tabeling C, Ruwwe-Glösenkamp C, Sander LE, Knosalla C, Brunke S, Hube B, Kniemeyer O, Brakhage AA, Schwarz C, Scheffold A, 2019. Human Anti-fungal Th17 Immunity and Pathology Rely on Cross-Reactivity against Candida albicans. Cell 176, 1340–1355.e15. 10.1016/j.cell.2019.01.041 [DOI] [PubMed] [Google Scholar]
- Backert I, Koralov SB, Wirtz S, Kitowski V, Billmeier U, Martini E, Hofmann K, Hildner K, Wittkopf N, Brecht K, Waldner M, Rajewsky K, Neurath MF, Becker C, Neufert C, 2014. STAT3 Activation in Th17 and Th22 Cells Controls IL-22–Mediated Epithelial Host Defense during Infectious Colitis. The Journal of Immunology 193, 3779–3791. 10.4049/jimmunol.1303076 [DOI] [PubMed] [Google Scholar]
- Bajaj JS, Hylemon PB, Ridlon JM, Heuman DM, Daita K, White MB, Monteith P, Noble NA, Sikaroodi M, Gillevet PM, 2012. Colonic mucosal microbiome differs from stool microbiome in cirrhosis and hepatic encephalopathy and is linked to cognition and inflammation. Am J Physiol Gastrointest Liver Physiol 303, G675–G685. 10.1152/ajpgi.00152.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bercik P, Denou E, Collins J, Jackson W, Lu J, Jury J, Deng Y, Blennerhassett P, Macri J, McCoy KD, Verdu EF, Collins SM, 2011. The Intestinal Microbiota Affect Central Levels of Brain-Derived Neurotropic Factor and Behavior in Mice. Gastroenterology 141, 599–609.e3. 10.1053/j.gastro.2011.04.052 [DOI] [PubMed] [Google Scholar]
- Buffington SA, Di Prisco GV, Auchtung TA, Ajami NJ, Petrosino JF, Costa-Mattioli M, 2016. Microbial Reconstitution Reverses Maternal Diet-Induced Social and Synaptic Deficits in Offspring. Cell 165, 1762–1775. 10.1016/j.cell.2016.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Itakura E, Nelson GM, Sheng M, Laurent P, Fenk LA, Butcher RA, Hegde RS, de Bono M, 2017. IL-17 is a neuromodulator of Caenorhabditis elegans sensory responses. Nature 542, 43–48. 10.1038/nature20818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Chou K, Fuchs E, Havran WL, Boismenu R, 2002. Protection of the intestinal mucosa by intraepithelial γδ T cells. PNAS 99, 14338–14343. 10.1073/pnas.212290499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiaro TR, Soto R, Stephens WZ, Kubinak JL, Petersen C, Gogokhia L, Bell R, Delgado JC, Cox J, Voth W, Brown J, Stillman DJ, O’Connell RM, Tebo AE, Round JL, 2017. A member of the gut mycobiota modulates host purine metabolism exacerbating colitis in mice. Science Translational Medicine 9. 10.1126/scitranslmed.aaf9044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikina AS, Nadalin F, Maurin M, San-Roman M, Thomas-Bonafos T, Li XV, Lameiras S, Baulande S, Henri S, Malissen B, Lacerda Mariano L, Barbazan J, Blander JM, Iliev ID, Matic Vignjevic D, Lennon-Duménil A-M, 2020. Macrophages Maintain Epithelium Integrity by Limiting Fungal Product Absorption. Cell 183, 411–428.e16. 10.1016/j.cell.2020.08.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi GB, Yim YS, Wong H, Kim S, Kim H, Kim SV, Hoeffer CA, Littman DR, Huh JR, 2016. The maternal interleukin-17a pathway in mice promotes autism-like phenotypes in offspring. Science. [DOI] [PMC free article] [PubMed]
- Chu C, Murdock MH, Jing D, Won TH, Chung H, Kressel AM, Tsaava T, Addorisio ME, Putzel GG, Zhou L, Bessman NJ, Yang R, Moriyama S, Parkhurst CN, Li A, Meyer HC, Teng F, Chavan SS, Tracey KJ, Regev A, Schroeder FC, Lee FS, Liston C, Artis D, 2019. The microbiota regulate neuronal function and fear extinction learning. Nature 574, 543–548. 10.1038/s41586-019-1644-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke G, Grenham S, Scully P, Fitzgerald P, Moloney RD, Shanahan F, Dinan TG, Cryan JF, 2013. The microbiome-gut-brain axis during early life regulates the hippocampal serotonergic system in a sex-dependent manner. Mol Psychiatry 18, 666–673. 10.1038/mp.2012.77 [DOI] [PubMed] [Google Scholar]
- Collins JW, Keeney KM, Crepin VF, Rathinam VAK, Fitzgerald KA, Finlay BB, Frankel G, 2014. Citrobacter rodentium : infection, inflammation and the microbiota. Nat Rev Microbiol 12, 612–623. 10.1038/nrmicro3315 [DOI] [PubMed] [Google Scholar]
- Coombes JL, Powrie F, 2008. Dendritic cells in intestinal immune regulation. Nat Rev Immunol 8, 435–446. 10.1038/nri2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello SP, Hughes PA, Waters O, Bryant RV, Vincent AD, Blatchford P, Katsikeros R, Makanyanga J, Campaniello MA, Mavrangelos C, Rosewarne CP, Bickley C, Peters C, Schoeman MN, Conlon MA, Roberts-Thomson IC, Andrews JM, 2019. Effect of Fecal Microbiota Transplantation on 8-Week Remission in Patients With Ulcerative Colitis: A Randomized Clinical Trial. JAMA 321, 156–164. 10.1001/jama.2018.20046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning TL, Wang Y, Patel SR, Williams IR, Pulendran B, 2007. Lamina propria macrophages and dendritic cells differentially induce regulatory and interleukin 17–producing T cell responses. Nat Immunol 8, 1086–1094. 10.1038/ni1511 [DOI] [PubMed] [Google Scholar]
- Desbonnet L, Clarke G, Shanahan F, Dinan TG, Cryan JF, 2014. Microbiota is essential for social development in the mouse. Mol Psychiatry 19, 146–148. 10.1038/mp.2013.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desbonnet L, Clarke G, Traplin A, O’Sullivan O, Crispie F, Moloney RD, Cotter PD, Dinan TG, Cryan JF, 2015. Gut microbiota depletion from early adolescence in mice: Implications for brain and behaviour. Brain, Behavior, and Immunity 48, 165–173. 10.1016/j.bbi.2015.04.004 [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR, 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson GP, Lee SM, Mazmanian SK, 2016. Gut biogeography of the bacterial microbiota. Nat Rev Microbiol 14, 20–32. 10.1038/nrmicro3552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doron I, Mesko M, Li XV, Kusakabe T, Leonardi I, Shaw DG, Fiers WD, Lin W-Y, Bialt-DeCelie M, Románm E, Longman RS, Pla J, Wilson PC,Iliev ID, 2021. Mycobiota-induced IgA antibodies regulate fungal commensalism in the gut and are dysregulated in Crohn’s disease. Nature Microbiol. 6, 1493–1504, 10.1038/s41564-021-00983-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doron I, Leonardi I, Li XV, Fiers WD, Semon A, Bialt-DeCelie M, Migaud M, Gao IH, Lin W-Y, Kusakabe T, Puel A, Iliev ID, 2021. Human gut mycobiota tune immunity via CARD9-dependent induction of anti-fungal IgG antibodies. Cell 184, 1017–1031.e14. 10.1016/j.cell.2021.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC, 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, Guiot Y, Derrien M, Muccioli GG, Delzenne NM, de Vos WM, Cani PD, 2013. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. PNAS 110, 9066–9071. 10.1073/pnas.1219451110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontenot JD, Gavin MA, Rudensky AY, 2003. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol 4, 330–336. 10.1038/ni904 [DOI] [PubMed] [Google Scholar]
- Fung TC, Artis D, Sonnenberg GF, 2014. Anatomical localization of commensal bacteria in immune cell homeostasis and disease. Immunological Reviews 260, 35–49. 10.1111/imr.12186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung TC, Bessman NJ, Hepworth MR, Kumar N, Shibata N, Kobuley D, Wang K, Ziegler CGK, Goc J, Shima T, Umesaki Y, Sartor RB, Sullivan KV, Lawley TD, Kunisawa J, Kiyono H, Sonnenberg GF, 2016. Lymphoid-Tissue-Resident Commensal Bacteria Promote Members of the IL-10 Cytokine Family to Establish Mutualism. Immunity 44, 634–646. 10.1016/j.immuni.2016.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffen SL, Jain R, Garg AV, Cua DJ, 2014. The IL-23–IL-17 immune axis: from mechanisms to therapeutic testing. Nat Rev Immunol 14, 585–600. 10.1038/nri3707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevers D, Kugathasan S, Denson LA, Vázquez-Baeza Y, Van Treuren W, Ren B, Schwager E, Knights D, Song SJ, Yassour M, Morgan XC, Kostic AD, Luo C, González A, McDonald D, Haberman Y, Walters T, Baker S, Rosh J, Stephens M, Heyman M, Markowitz J, Baldassano R, Griffiths A, Sylvester F, Mack D, Kim S, Crandall W, Hyams J, Huttenhower C, Knight R, Xavier RJ, 2014. The treatment-naïve microbiome in new-onset Crohn’s disease. Cell Host Microbe 15, 382–392. 10.1016/j.chom.2014.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronke K, Hernández PP, Zimmermann J, Klose CSN, Kofoed-Branzk M, Guendel F, Witkowski M, Tizian C, Amann L, Schumacher F, Glatt H, Triantafyllopoulou A, Diefenbach A, 2019. Interleukin-22 protects intestinal stem cells against genotoxic stress. Nature 566, 249–253. 10.1038/s41586-019-0899-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall AB, Tolonen AC, Xavier RJ, 2017. Human genetic variation and the gut microbiome in disease. Nat Rev Genet 18, 690–699. 10.1038/nrg.2017.63 [DOI] [PubMed] [Google Scholar]
- Hallen-Adams HE, Suhr MJ, 2016. Fungi in the healthy human gastrointestinal tract. Virulence 8, 352–358. 10.1080/21505594.2016.1247140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoarau G, Mukherjee PK, Gower-Rousseau C, Hager C, Chandra J, Retuerto MA, Neut C, Vermeire S, Clemente J, Colombel JF, Fujioka H, Poulain D, Sendid B, Ghannoum MA, 2016. Bacteriome and Mycobiome Interactions Underscore Microbial Dysbiosis in Familial Crohn’s Disease. mBio 7, e01250–16. 10.1128/mBio.01250-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeffer CA, Tang W, Wong H, Santillan A, Patterson RJ, Martinez LA, Tejada-Simon MV, Paylor R, Hamilton SL, Klann E, 2008. Removal of FKBP12 Enhances mTOR-Raptor Interactions, LTP, Memory, and Perseverative/Repetitive Behavior. Neuron 60, 832–845. 10.1016/j.neuron.2008.09.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper LV, Littman DR, Macpherson AJ, 2012. Interactions Between the Microbiota and the Immune System. Science 336, 1268–1273. 10.1126/science.1223490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T, Codelli JA, Chow J, Reisman SE, Petrosino JF, Patterson PH, Mazmanian SK, 2013. Microbiota Modulate Behavioral and Physiological Abnormalities Associated with Neurodevelopmental Disorders. Cell 155, 1451–1463. 10.1016/j.cell.2013.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliev ID, Funari VA, Taylor KD, Nguyen Q, Reyes CN, Strom SP, Brown J, Becker CA, Fleshner PR, Dubinsky M, Rotter JI, Wang HL, McGovern DPB, Brown GD, Underhill DM, 2012. Interactions Between Commensal Fungi and the C-Type Lectin Receptor Dectin-1 Influence Colitis. Science 336, 1314–1317. 10.1126/science.1221789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliev ID, Leonardi I, 2017. Fungal dysbiosis: immunity and interactions at mucosal barriers. Nature Reviews Immunology 17, 635–646. 10.1038/nri.2017.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, Tanoue T, Imaoka A, Itoh K, Takeda K, Umesaki Y, Honda K, Littman DR, 2009. Induction of Intestinal Th17 Cells by Segmented Filamentous Bacteria. Cell 139, 485–498. 10.1016/j.cell.2009.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain U, Heul AMV, Xiong S, Gregory MH, Demers EG, Kern JT, Lai C-W, Muegge BD, Barisas DAG, Leal-Ekman JS, Deepak P, Ciorba MA, Liu T-C, Hogan DA, Debbas P, Braun J, McGovern DPB, Underhill DM, Stappenbeck TS, 2021. Debaryomyces is enriched in Crohn’s disease intestinal tissue and impairs healing in mice. Science. 10.1126/science.abd0919 [DOI] [PMC free article] [PubMed]
- Jiang TT, Shao T-Y, Ang WXG, Kinder JM, Turner LH, Pham G, Whitt J, Alenghat T, Way SS, 2017. Commensal Fungi Recapitulate the Protective Benefits of Intestinal Bacteria. Cell Host & Microbe 22, 809–816.e4. 10.1016/j.chom.2017.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson MEV, Phillipson M, Petersson J, Velcich A, Holm L, Hansson GC, 2008. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. PNAS 105, 15064–15069. 10.1073/pnas.0803124105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayyal M, Javkar T, Firoz Mian M, Binyamin D, Koren O, McVey Neufeld K-A, Forsythe P, 2020. Sex dependent effects of post-natal penicillin on brain, behavior and immune regulation are prevented by concurrent probiotic treatment. Sci Rep 10, 10318. 10.1038/s41598-020-67271-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kernbauer E, Ding Y, Cadwell K, 2014. An enteric virus can replace the beneficial function of commensal bacteria. Nature 516, 94–98. 10.1038/nature13960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopec AM, Smith CJ, Ayre NR, Sweat SC, Bilbo SD, 2018. Microglial dopamine receptor elimination defines sex-specific nucleus accumbens development and social behavior in adolescent rats. Nat Commun 9, 3769. 10.1038/s41467-01806118-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korotkevich G, Sukhov V, Budin N, Shpak B, Artyomov MN, Sergushichev A, 2021. Fast gene set enrichment analysis. bioRxiv 060012. 10.1101/060012 [DOI]
- Ladinsky MS, Araujo LP, Zhang X, Veltri J, Galan-Diez M, Soualhi S, Lee C, Irie K, Pinker EY, Narushima S, Bandyopadhyay S, Nagayama M, Elhenawy W, Coombes BK, Ferraris RP, Honda K, Iliev ID, Gao N, Bjorkman PJ, Ivanov II, 2019. Endocytosis of commensal antigens by intestinal epithelial cells regulates mucosal T cell homeostasis. Science 363. 10.1126/science.aat4042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Tato CM, Joyce-Shaikh B, Gulen MF, Cayatte C, Chen Y, Blumenschein WM, Judo M, Ayanoglu G, McClanahan TK, Li X, Cua DJ, 2015. Interleukin-23-Independent IL-17 Production Regulates Intestinal Epithelial Permeability. Immunity 43, 727–738. 10.1016/j.immuni.2015.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardi I, Li X, Semon A, Li D, Doron I, Putzel G, Bar A, Prieto D, Rescigno M, McGovern DPB, Pla J, Iliev ID, 2018. CX3CR1+ mononuclear phagocytes control immunity to intestinal fungi. Science 359, 232–236. 10.1126/science.aao1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardi I, Paramsothy S, Doron I, Semon A, Kaakoush NO, Clemente JC, Faith JJ, Borody TJ, Mitchell HM, Colombel J-F, Kamm MA, Iliev ID, 2020. Fungal Trans-kingdom Dynamics Linked to Responsiveness to Fecal Microbiota Transplantation (FMT) Therapy in Ulcerative Colitis. Cell Host & Microbe 27, 823–829.e3. 10.1016/j.chom.2020.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MO, Wan YY, Flavell RA, 2007. T Cell-Produced Transforming Growth Factor-β1 Controls T Cell Tolerance and Regulates Th1- and Th17-Cell Differentiation. Immunity 26, 579–591. 10.1016/j.immuni.2007.03.014 [DOI] [PubMed] [Google Scholar]
- Li X, Leonardi I, Semon A, Doron I, Gao IH, Putzel GG, Kim Y, Kabata H, Artis D, Fiers WD, Ramer-Tait AE, Iliev ID, 2018. Response to Fungal Dysbiosis by Gut-Resident CX3CR1+ Mononuclear Phagocytes Aggravates Allergic Airway Disease. Cell Host & Microbe 24, 847–856.e4. 10.1016/j.chom.2018.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XV, Leonardi I, Putzel GG, Semon A, Fiers WD, Kusakabe T, Doron I, Gao IH, Lin W-Y, Gutierrez-Guerrero A, Bialt-DeCelie M, Yang C, Naglik JR, Hube B, Scherl E, Iliev ID, 2022. Strain-level diversity defines host-mycobiota interactions and inflammation in inflammatory bowel disease. Nature in press.
- Liao Y, Smyth GK, Shi W, 2019. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Research 47, e47. 10.1093/nar/gkz114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limon JJ, Tang J, Li D, Wolf AJ, Michelsen KS, Funari V, Gargus M, Nguyen C, Sharma P, Maymi VI, Iliev ID, Skalski JH, Brown J, Landers C, Borneman J, Braun J, Targan SR, McGovern DPB, Underhill DM, 2019. Malassezia Is Associated with Crohn’s Disease and Exacerbates Colitis in Mouse Models. Cell Host & Microbe 25, 377–388.e6. 10.1016/j.chom.2019.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S, 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology 15, 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupien SJ, McEwen BS, Gunnar MR, Heim C, 2009. Effects of stress throughout the lifespan on the brain, behaviour and cognition. Nat Rev Neurosci 10, 434–445. 10.1038/nrn2639 [DOI] [PubMed] [Google Scholar]
- Maloy KJ, Salaun L, Cahill R, Dougan G, Saunders NJ, Powrie F, 2003. CD4+CD25+ TR Cells Suppress Innate Immune Pathology Through Cytokine-dependent Mechanisms. Journal of Experimental Medicine 197, 111–119. 10.1084/jem.20021345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Guryn K, Leone V, Chang EB, 2019. Regional Diversity of the Gastrointestinal Microbiome. Cell Host & Microbe 26, 314–324. 10.1016/j.chom.2019.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morarach K, Mikhailova A, Knoflach V, Memic F, Kumar R, Li W, Ernfors P, Marklund U, 2021. Diversification of molecularly defined myenteric neuron classes revealed by single-cell RNA sequencing. Nat Neurosci 24, 34–46. 10.1038/s41593-020-00736-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moy SS, Nadler JJ, Perez A, Barbaro RP, Johns JM, Magnuson TR, Piven J, Crawley JN, 2004. Sociability and preference for social novelty in five inbred strains: an approach to assess autistic-like behavior in mice. Genes, Brain and Behavior 3, 287–302. 10.1111/j.1601-1848.2004.00076.x [DOI] [PubMed] [Google Scholar]
- Mueller KD, Zhang H, Serrano CR, Billmyre RB, Huh EY, Wiemann P, Keller NP, Wang Y, Heitman J, Lee SC, 2019. Gastrointestinal microbiota alteration induced by Mucor circinelloides in a murine model. J Microbiol. 57, 509–520. 10.1007/s12275-019-8682-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller PA, Schneeberger M, Matheis F, Wang P, Kerner Z, Ilanges A, Pellegrino K, del Mármol J, Castro TBR, Furuichi M, Perkins M, Han W, Rao A, Pickard AJ, Cross JR, Honda K, de Araujo I, Mucida D, 2020. Microbiota modulate sympathetic neurons via a gut–brain circuit. Nature 583, 441–446. 10.1038/s41586-020-2474-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundy R, MacDonald TT, Dougan G, Frankel G, Wiles S, 2005. Citrobacter rodentium of mice and man. Cellular Microbiology 7, 1697–1706. 10.1111/j.1462-5822.2005.00625.x [DOI] [PubMed] [Google Scholar]
- Nagalakshmi ML, Rascle A, Zurawski S, Menon S, de Waal Malefyt R, 2004. Interleukin-22 activates STAT3 and induces IL-10 by colon epithelial cells. International Immunopharmacology, IL-Teenagers; the family of the IL-10 homologue comes of age 4, 679–691. 10.1016/j.intimp.2004.01.008 [DOI] [PubMed] [Google Scholar]
- Neil JA, Matsuzawa-Ishimoto Y, Kernbauer-Hölzl E, Schuster SL, Sota S, Venzon M, Dallari S, Galvao Neto A, Hine A, Hudesman D, Loke P, Nice TJ, Cadwell K, 2019. IFN-I and IL-22 mediate protective effects of intestinal viral infection. Nat Microbiol 4, 1737–1749. 10.1038/s41564-019-0470-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ost KS, O’Meara TR, Stephens WZ, Chiaro T, Zhou H, Penman J, Bell R, Catanzaro JR, Song D, Singh S, Call DH, Hwang-Wong E, Hanson KE, Valentine JF, Christensen KA, O’Connell RM, Cormack B, Ibrahim AS, Palm NW, Noble SM, Round JL, 2021. Adaptive immunity induces mutualism between commensal eukaryotes. Nature 596, 114–118. 10.1038/s41586-021-03722-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelaseyed T, Bergström JH, Gustafsson JK, Ermund A, Birchenough GMH, Schütte A, van der Post S, Svensson F, Rodríguez-Piñeiro AM, Nyström EEL, Wising C, Johansson MEV, Hansson GC, 2014. The mucus and mucins of the goblet cells and enterocytes provide the first defense line of the gastrointestinal tract and interact with the immune system. Immunological Reviews 260, 8–20. 10.1111/imr.12182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham TAN, Clare S, Goulding D, Arasteh JM, Stares MD, Browne HP, Keane JA, Page AJ, Kumasaka N, Kane L, Mottram L, Harcourt K, Hale C, Arends MJ, Gaffney DJ, Dougan G, Lawley TD, 2014. Epithelial IL-22RA1-Mediated Fucosylation Promotes Intestinal Colonization Resistance to an Opportunistic Pathogen. Cell Host & Microbe 16, 504–516. 10.1016/j.chom.2014.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pull SL, Doherty JM, Mills JC, Gordon JI, Stappenbeck TS, 2005. Activated macrophages are an adaptive element of the colonic epithelial progenitor niche necessary for regenerative responses to injury. PNAS 102, 99–104. 10.1073/pnas.0405979102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed MD, Yim YS, Wimmer RD, Kim H, Ryu C, Welch GM, Andina M, King HO, Waisman A, Halassa MM, Huh JR, Choi GB, 2020. IL-17a promotes sociability in mouse models of neurodevelopmental disorders. Nature 577, 249–253. 10.1038/s41586-019-1843-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowan F, Docherty NG, Murphy M, Murphy TB, Coffey JC, O’Connell PR, 2010. Bacterial Colonization of Colonic Crypt Mucous Gel and Disease Activity in Ulcerative Colitis. Annals of Surgery 252, 869–875. 10.1097/SLA.0b013e3181fdc54c [DOI] [PubMed] [Google Scholar]
- Salvador AF, de Lima KA, Kipnis J, 2021. Neuromodulation by the immune system: a focus on cytokines. Nat Rev Immunol 21, 526–541. 10.1038/s41577-02100508-z [DOI] [PubMed] [Google Scholar]
- Schaedler RW, Dubos R, Costello R, 1965. ASSOCIATION OF GERMFREE MICE WITH BACTERIA ISOLATED FROM NORMAL MICE. Journal of Experimental Medicine 122, 77–82. 10.1084/jem.122.1.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schieber AMP, Lee YM, Chang MW, Leblanc M, Collins B, Downes M, Evans RM, Ayres JS, 2015. Disease tolerance mediated by microbiome E. coli involves inflammasome and IGF-1 signaling. Science 350, 558–563. 10.1126/science.aac6468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao T-Y, Ang WXG, Jiang TT, Huang FS, Andersen H, Kinder JM, Pham G, Burg AR, Ruff B, Gonzalez T, Hershey GKK, Haslam DB, Way SS, 2019. Commensal Candida albicans Positively Calibrates Systemic Th17 Immunological Responses. Cell Host & Microbe 25, 404–417.e6. 10.1016/j.chom.2019.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol H, Leducq V, Aschard H, Pham H-P, Jegou S, Landman C, Cohen D, Liguori G, Bourrier A, Nion-Larmurier I, Cosnes J, Seksik P, Langella P, Skurnik D, Richard ML, Beaugerie L, 2017. Fungal microbiota dysbiosis in IBD. Gut 66, 1039–1048. 10.1136/gutjnl-2015-310746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenberg GF, Monticelli LA, Alenghat T, Fung TC, Hutnick NA, Kunisawa J, Shibata N, Grunberg S, Sinha R, Zahm AM, Tardif MR, Sathaliyawala T, Kubota M, Farber DL, Collman RG, Shaked A, Fouser LA, Weiner DB, Tessier PA, Friedman JR, Kiyono H, Bushman FD, Chang K-M, Artis D, 2012. Innate Lymphoid Cells Promote Anatomical Containment of Lymphoid-Resident Commensal Bacteria. Science 336, 1321–1325. 10.1126/science.1222551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenberg GF, Monticelli LA, Elloso MM, Fouser LA, Artis D, 2011. CD4+ Lymphoid Tissue-Inducer Cells Promote Innate Immunity in the Gut. Immunity 34, 122–134. 10.1016/j.immuni.2010.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spadoni I, Zagato E, Bertocchi A, Paolinelli R, Hot E, Sabatino A, Caprioli F, Bottiglieri L, Oldani A, Viale G, Penna G, Dejana E, Rescigno M, 2015. A gut-vascular barrier controls the systemic dissemination of bacteria. Science 350, 830–834. 10.1126/science.aad0135 [DOI] [PubMed] [Google Scholar]
- Speakman EA, Dambuza IM, Salazar F, Brown GD, 2020. T Cell Antifungal Immunity and the Role of C-Type Lectin Receptors. Trends in Immunology 41, 61–76. 10.1016/j.it.2019.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strati F, Cavalieri D, Albanese D, De Felice C, Donati C, Hayek J, Jousson O, Leoncini S, Renzi D, Calabrò A, De Filippo C, 2017. New evidences on the altered gut microbiota in autism spectrum disorders. Microbiome 5, 24. 10.1186/s40168-017-0242-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudo N, Chida Y, Aiba Y, Sonoda J, Oyama N, Yu X-N, Kubo C, Koga Y, 2004. Postnatal microbial colonization programs the hypothalamic–pituitary–adrenal system for stress response in mice. The Journal of Physiology 558, 263. 10.1113/jphysiol.2004.063388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C-M, Hall JA, Blank RB, Bouladoux N, Oukka M, Mora JR, Belkaid Y, 2007. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. Journal of Experimental Medicine 204, 1775–1785. 10.1084/jem.20070602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Iliev ID, Brown J, Underhill DM, Funari VA, 2015. Mycobiome: Approaches to analysis of intestinal fungi. Journal of Immunological Methods, Gut immune system in health and disease 421, 112–121. 10.1016/j.jim.2015.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng F, Goc J, Zhou L, Chu C, Shah MA, Eberl G, Sonnenberg GF, 2019. A circadian clock is essential for homeostasis of group 3 innate lymphoid cells in the gut. Science Immunology 4. 10.1126/sciimmunol.aax1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tropini C, Earle KA, Huang KC, Sonnenburg JL, 2017. The Gut Microbiome: Connecting Spatial Organization to Function. Cell Host & Microbe 21, 433–442. 10.1016/j.chom.2017.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veiga-Fernandes H, Mucida D, 2016. Neuro-Immune Interactions at Barrier Surfaces. Cell 165, 801–811. 10.1016/j.cell.2016.04.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler ML, Limon JJ, Bar AS, Leal CA, Gargus M, Tang J, Brown J, Funari VA, Wang HL, Crother TR, Arditi M, Underhill DM, Iliev ID, 2016. Immunological Consequences of Intestinal Fungal Dysbiosis. Cell Host & Microbe 19, 865–873. 10.1016/j.chom.2016.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler ML, Limon JJ, Underhill DM, 2017. Immunity to Commensal Fungi: Detente and Disease. Annual Review of Pathology: Mechanisms of Disease 12, 359–385. 10.1146/annurev-pathol-052016-100342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong ARC, Pearson JS, Bright MD, Munera D, Robinson KS, Lee SF, Frankel G, Hartland EL, 2011. Enteropathogenic and enterohaemorrhagic Escherichia coli: even more subversive elements. Mol Microbiol 80, 1420–1438. 10.1111/j.13652958.2011.07661.x [DOI] [PubMed] [Google Scholar]
- Wu LW, Agirman G, Hsiao EY, 2022. The Gut Microbiome as a Regulator of the Neuroimmune Landscape. Ann Rev Immunol 40. 10.1146/annurevimmunol-101320-014237 [DOI] [PubMed] [Google Scholar]
- Yang A-M, Inamine T, Hochrath K, Chen P, Wang L, Llorente C, Bluemel S, Hartmann P, Xu J, Koyama Y, Kisseleva T, Torralba MG, Moncera K, Beeri K, Chen C-S, Freese K, Hellerbrand C, Lee SML, Hoffman HM, Mehal WZ, Garcia-Tsao G, Mutlu EA, Keshavarzian A, Brown GD, Ho SB, Bataller R, Stärkel P, Fouts DE, Schnabl B, 2017. Intestinal fungi contribute to development of alcoholic liver disease. J Clin Invest 127, 2829–2841. 10.1172/JCI90562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XO, Chang SH, Park H, Nurieva R, Shah B, Acero L, Wang Y-H, Schluns KS, Broaddus RR, Zhu Z, Dong C, 2008. Regulation of inflammatory responses by IL-17F. Journal of Experimental Medicine 205, 1063–1075. 10.1084/jem.20071978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap CX, Henders AK, Alvares GA, Wood DLA, Krause L, Tyson GW, Restuadi R, Wallace L, McLaren T, Hansell NK, Cleary D, Grove R, Hafekost C, Harun A, Holdsworth H, Jellett R, Khan F, Lawson LP, Leslie J, Frenk ML, Masi A, Mathew NE, Muniandy M, Nothard M, Miller JL, Nunn L, Holtmann G, Strike LT, de Zubicaray GI, Thompson PM, McMahon KL, Wright MJ, Visscher PM, Dawson PA, Dissanayake C, Eapen V, Heussler HS, McRae AF, Whitehouse AJO, Wray NR, Gratten J, 2021. Autism-related dietary preferences mediate autism-gut microbiome associations. Cell 0. 10.1016/j.cell.2021.10.015 [DOI] [PubMed]
- Zeisel A, Hochgerner H, Lönnerberg P, Johnsson A, Memic F, van der Zwan J, Häring M, Braun E, Borm LE, La Manno G, Codeluppi S, Furlan A, Lee K, Skene N, Harris KD, Hjerling-Leffler J, Arenas E, Ernfors P, Marklund U, Linnarsson S, 2018. Molecular Architecture of the Mouse Nervous System. Cell 174, 999–1014.e22. 10.1016/j.cell.2018.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan X, Cao M, Yoo AS, Zhang Z, Chen L, Crabtree GR, Wu JI, 2015. Generation of BAF53b-Cre transgenic mice with pan-neuronal Cre activities. genesis 53, 440–448. 10.1002/dvg.22866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Zhang H, Liu S, Luo W, Jiang Y, Gao J, 2021. Association of Peripheral Blood Levels of Cytokines With Autism Spectrum Disorder: A Meta-Analysis. Frontiers in Psychiatry 12, 1006. 10.3389/fpsyt.2021.670200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Valdez PA, Danilenko DM, Hu Y, Sa SM, Gong Q, Abbas AR, Modrusan Z, Ghilardi N, de Sauvage FJ, Ouyang W, 2008. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med 14, 282–289. 10.1038/nm1720 [DOI] [PubMed] [Google Scholar]
- Zhou C, Wu D, Jawale C, Li Y, Biswas PS, McGeachy MJ, Gaffen SL, 2021. Divergent functions of IL-17-family cytokines in DSS colitis: Insights from a naturallyoccurring human mutation in IL-17F. Cytokine 148, 155715. 10.1016/j.cyto.2021.155715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou R, Wang Y, Duan M, Guo M, Zhang Q, Zheng H, 2021. Dysbiosis of Gut Fungal Microbiota in Children with Autism Spectrum Disorders. J Autism Dev Disord 51, 267–275. 10.1007/s10803-020-04543-y [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A. rt-qPCR of mucosal scraping of mice colonized with the mucosal fungal consortium 10 days post-colonization. Data shown are mean values, dots represent individual mice. B. Gating strategy of myeloid cells in the colonic lamina propria. Lineage: CD3, NK1.1, Gr1, TCRβ, CD5, CD19). B. Gating strategy of lymphocytes in the colonic lamina propria. CD4+ T cells are gated as Live (Fixable Viability Dye negative), CD45+ CD3+ CD90.2+ CD4+. Type 3 innate lymphoid cells are gated as Live, CD45+ Lin- CD3- CD90.2+ CD127+ CD127+ RORγt+. Lin: CD11c, CD11b, NK1.1, Gr1, TCRβ, CD5, CD19).
SPF mice were treated with antibiotics were colonized with consortia of mucosal (MUC) or luminal (LUM) fungi. A. Representative flow cytometry plot and quantification of the numbers of FoxP3+ RORγt-, FoxP3+ RORγt+, and FoxP3- RORγt+ CD4+ T cells among CD45+ hematopoietic cells in the mLN. B. Representative flow cytometry plot and quantification of the total numbers of IL-17A and/or IL-22 producing CD4+ Th cells. B. Representative flow cytometry plot and quantification of the total numbers of IL-17A and IL-17F producing CD4+ Th cells. C. Representative flow cytometry plot and quantification of the total numbers of IFNγ producing CD4+ Th cells. Experiment was repeated three times with n>4 per group. *P<0.05, **P<0.01, ***P<0.001. Data are shown as Tukey Box Whiskers plots. Mann-Whitney T Test (F, G).
A-B: C57Bl/6J mice were treated with antibiotics and inoculated with the Mucosal consortium (MUC), the individual members of the consortium (Sc: S. cerevisiae, Ca: C. albicans, Sf: S. fibuligera), or left untreated. A. Frequency of IL-17A+ CD3+ CD4+ T cells among the CD45+ cLP population. B. Frequency of IL-22A+ CD3+ CD4+ T cells among the CD45+ cLP population. C-D: IL-22 reporter mice (expressing eGFP under the control of the IL-22 promoter) were treated with antibiotics and inoculated with the Mucosal consortium (MUC) or left untreated. cLP were harvested at day 3 and 7. C. Frequency of eGFP+ CD3+ CD4+ T cells among the CD45+ cLP population. D. Frequency of eGFP+ Lin-CD90.2+ CD127+ ILC3 among the CD45+ cLP population. E-F. Fate mapper mice were colonized with the MUC consortium. Expression of IL-22 by CD45+ cells in the colonic lamina propria (cLP, E) and mesenteric lymph nodes (F). G-H. Fate mapper mice were colonized with a MUC consortium (MUC) composed of mucosa-associated C. albicans and S. cerevisiae, isolated from healthy individuals. Control mice were administered PBS (PBS). G. Expression of IL-22 by CD4+ T cells in the cLP. H. Expression of IL-22 by CD4+ T cells in the mLN. Data shown are mean values +/− SEM, dots represent individual mice. *P<0.05, **P<0.01, ***P<0.001; Mann-Whitney T est.
A-B. Representative flow cytometry plots and quantification of IL-22, IL17A, IL-17F, and IFNγ production by CD3+ CD4+ T cells in the cLP. C. Quantification of IL-22, IL-17A, IL-17F, and IFNγ production by CD3+ CD4+ T cells in the mesenteric lymph nodes (mLN). Data shown are mean values, dots represent individual mice. *P<0.05, **P<0.01, ***P<0.001; Mann Whitney Test. E. WT and Il17a−/− mice were treated with broad spectrum antibiotics prior to fungal colonization (MUC) or not (NT) and administration of Dextran Sodium Sulphate as described in Figure 2 G. F. Percent survival. Log-rank (Mantel-Cox) test.
A-E. 3 weeks old male C57Bl/6J mice (n=10 per group) were administered with an antibiotic cocktail (ABX: ampicillin 0.4g/l, vancomycin 0.4g/l, metronidazole 0.3g/l, cefoperazone 0.4g/l, 10% sucrose) or not treated (NT). ABX mice were colonized with a fungal consortium (MUC) or administered PBS (PBS) every 4 days until testing at 8 weeks of age. A. Marble burying behavior in the marble burying test. Graph shows the total number of buried marbles (out of 20) during a 10-minute trial. B. Experimental layout of the elevated plus-maze test and total distance travelled (cm) by individual mice. C. Frequency of visits to the closed arms of the elevated plus-maze. D. Percent time spent in the closed arms. E. Percent time spent in the open arms of the elevated plus maze. Data shown are mean values, dots represent individual mice. *P<0.05, **P<0.01, ***P<0.001; Mann-Whitney T Test.
A. Representative flow cytometry plot and quantification of the production of cytokines by CD4+ T helper cells in the mouse spleen following colonization with a luminal consortium (LUM) or mucosal consortium (MUC). B. Levels of Cytokines in the serum of mice following colonization with the MUC consortium. Mice treated with a cocktail of antibiotics (ABX, 0.3g/l Metronidazole, 0.4 g/l Cefoperazone, 0.4g/l Ampicillin, 0.4 g/l Vancomycin) or germ-free mice (GF) were colonized with the MUC consortium (MUC) or not treated (NT). Circulating levels of cytokines in the serum were measured using a bead-based immune assay. Data shown are mean values, dots represent individual mice. *P<0.05, **P<0.01, ***P<0.001; Mann Whitney Test. C-F. Baf53bCre/+ mice were crossed with Il17rafl/fl mice to selectively abrogate IL-17ra signaling on neurons (Baf53bΔIl17ra) littermates were used as controls (Litt). Mice were treated with antibiotics and colonized with a fungal consortium (MUC) or not (NT). C. Total fungal loads expressed as colony forming unit (cfu) per gram of feces in Litt and Baf53bΔIl17ra mice. D. Direct interaction index during the social phase. E. Total distance travelled and moving time in the habituation phase. F. Percent time spent in the chamber center or border of the open field test chamber. Dots represent individual mice (triangles: females, diamonds males). Mann-Whitney T test.
Supplemental Table 1. Related to Figure 1. Metadata for human subjects’ samples.
Supplemental Table 2. Related to Figure 2. RNA quality
Supplemental Table 3. Related to Figure 6. Publicly available RNA-Seq data obtained from Zeisel, 2018 (Zeisel et al., 2018)
Data Availability Statement
The ITS sequencing data are deposited in NCBI Sequence Read Archive (SRA, http://www.ncbi.nlm.nih.gov/Traces/sra, PRJNA594055). The RNA sequencing data are deposited in NCBI Gene expression omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/ GSE141622). This paper does not report original code. All other data needed to evaluate the conclusions in the manuscript are available within the main text or supplementary materials.