Abstract
Circular RNAs (circRNAs) are differentially expressed in various cardiovascular disease including myocardial ischemia-reperfusion (I/R) injury. However, their functional impact on cardiomyocyte cell death, in particular, in necrotic forms of death remains elusive. In this study, we found that the level of mmu_circ_000338, a cardiac- necroptosis-associated circRNA (CNEACR), was reduced in hypoxia-reoxygenation (H/R) exposed cardiomyocytes and I/R-injured mice hearts. The enforced expression of CNEACR attenuated the necrotic form of cardiomyocyte death caused by H/R and suppressed of myocardial necrosis in I/R injured mouse heart, which was accompanied by a marked reduction of myocardial infarction size and improved cardiac function. Mechanistically, CNEACR directly binds to histone deacetylase (HDAC7) in the cytoplasm and interferes its nuclear entry. This leads to attenuation of HDAC7-dependent suppression of forkhead box protein A2 (Foxa2) transcription, which can repress receptor-interacting protein kinase 3 (Ripk3) gene by binding to its promoter region. In addition, CNEACR-mediated upregulation of FOXA2 inhibited RIPK3-dependent necrotic/necroptotic death of cardiomyocytes. Our study reveals that circRNAs such as CNEACR can regulate the cardiomyocyte necroptosis associated activity of HDACs, promotes cell survival and improves cardiac function in I/R-injured heart. Hence, the CNEACR/HDAC7/Foxa2/ RIPK3 axis could be an efficient target for alleviating myocardial damage caused by necroptotic death in ischemia heart diseases.
Subject terms: Epigenetics, Cardiovascular diseases
Introduction
Cardiac diseases remain the leading cause of disability and mortality worldwide despite various strategies and advancement have been made in the treatment management of cardiovascular problems [1, 2]. In the heart, the restoration of blood flow (reperfusion) is the mainstay of treatment strategy in cardiac problems such as myocardial infarction (MI), atherosclerosis, coronary artery diseases, and cardiac surgery. However, this process inflicts ischemia/reperfusion (I/R) injury, which causes substantial damage to the myocardial tissue, cardiac dysfunction, and cardiac failure. The myocardial I/R imposes a gradual and/or acute loss of cardiomyocytes (CMs) by causing irreversible injury and cell death, which is primarily responsible for various cardiac pathology, ventricular remodeling and heart failure [3–5]. During cardiac I/R, a considerable amount of CMs undergoes cell death through the regulated non-apoptotic form of cell death including necroptosis, pyroptosis, and ferroptosis, in addition to the major forms of cell death such as apoptosis, necrosis and autophagy-dependent cell death [5, 6]. Given that the reduction of infarct size and the area at the risk of injury by blocking and/or attenuating cardiomyocyte cell death is a promising cardioprotective intervention in myocardial I/R, it is important to understand the molecules/signaling pathways associated with a specific mode of cell death and the regulatory mechanisms that promote cell survival and/or halt the cell death process in CMs.
Necrosis has traditionally been viewed as a passive and unregulated type of cell death, recent work clearly shows that necrosis is also highly regulated and orchestrated process, and it was involved in elaborate molecular circuitry [7, 8]. Necroptosis, as a form of programmed cell death, is highly regulated by receptor-interacting protein kinase 3 (RIPK3) and mixed lineage kinase domain-like (MLKL), and is generally manifested as a morphological feature of necrosis [9–12]. Luedde et al. discovered for the first time that the overexpression of RIPK3 alone was sufficient to induce necroptosis of CMs, and which was upregulated in cardiac tissue after MI [6]. Mice deficient in the RIPK3 gene exhibited significant reductions in infarct size than did wild-type mice in acute I/R injury models [13]. Phosphoglycerate mutase family member 5 (PGAM5), as a downstream signaling target of RIPK3, recruits the mitochondrial dynamin related protein-1 and provokes programmed necrotic death [14]. Furthermore, Ripk3-PGAM5-CypD-mPTP axis has been identified as one of the new pathway responsible for reperfusion-mediated microvascular damage [15]. Currently, targeting the necroptosis pathway is regarded as a promising new strategy for myocardial ischemia/reperfusion (MI/R) injury [16, 17].
NcRNAs such as microRNA (miRNA) and long noncoding RNA have been extensively studied for their functions in cardiomyocyte death under various cardiovascular diseases [1, 18]. Circular RNAs (circRNAs), a new type of single-stranded non-coding RNA that covalently forms a closed circular structure, was first discovered in viruses of plants in 1976 [19]. With the quick development and application of high-throughput sequencing technology and bioinformatics analyses, researchers have found that circRNAs are widely distributed, and its expression in a tissue- and developmental stage–specific manner [20–22]. Depending on the subcellular locality, circRNAs have diverse cellular functions including sponging of miRNAs, regulation of gene transcription or RNA splicing, regulating protein functions by acting as protein decoys or sponge, and regulation of protein [22–25]. Multiple studies have shown that circRNAs exhibit significant regulation functions in numerous physiologic and pathophysiologic processes of various diseases [26, 27]. However, no study to date has revealed the functions of circRNAs on cardiac myocyte necroptosis.
During myocardial I/R, the initiation and activation of the different modes of cell death in CMs are orchestrated by a complex network of molecules and signaling pathways that mainly associated with oxygen deprivation, inflammation and oxidative stress [8]. The I/R-injury causes epigenetic changes, including histone modification by histone acetyltransferase (HAT)/histone deacetylase (HDAC) [28–31] and dysregulation of noncoding RNA (ncRNA) [1, 18], which play a crucial role in altering the gene programs and expression/activities of transcription factors associated with cell survival and/or death, and that consequently results in myocardial tissue damage during I/R. However, the underlying mechanism of the dysregulated activities and/or expression of histone modifications including HDACs during I/R injury yet largely unknown.
In the present study, we show a cardiomyocyte-enriched CNEACR promotes cardiomyocyte cell survival by inhibiting HDAC7-dependent suppression of Foxa2 expression and necroptosis triggered by RIPK3. The reduction of CNEACR under I/R injury favours HDAC7 nuclear entry and derepression of Ripk3, a Foxa2-dependent gene. Together, our study demonstrates that CNEACR has a cardiomyocyte protective function by regulating the necroptotic pathway composed of HDAC7-FOXA2-RIPK3 and CNEACR could be an efficient therapeutic tool to protect the myocardial tissue and accelerating cardiac repair following ischemia and reperfusion injury.
Results
Identification and characterization of myocardial necrosis associated circRNAs
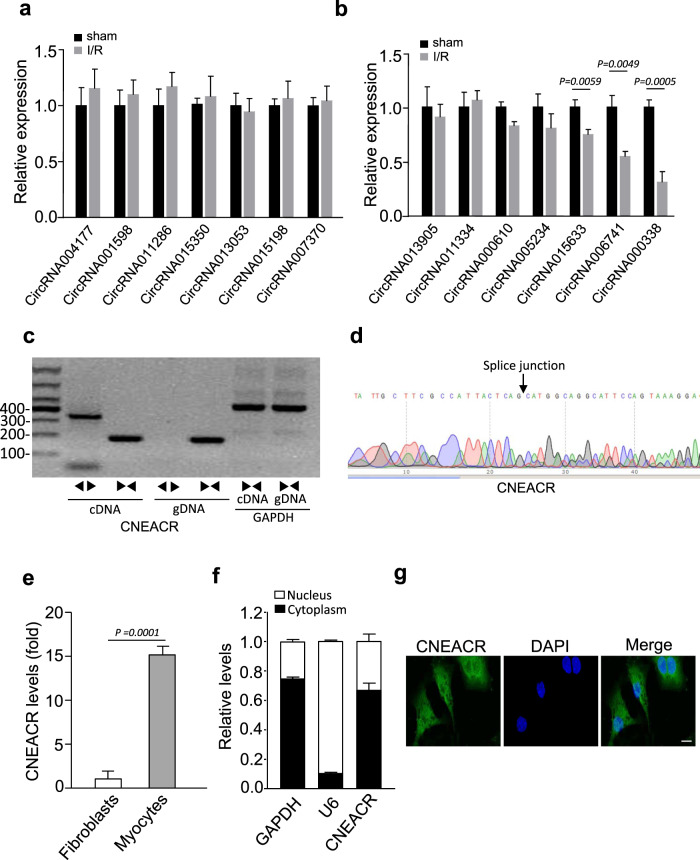
CircRNAs are involved in the regulation of genes associated with various cardiovascular diseases including ischemic heart failure and MI [24, 32, 33]. To identify and characterize the functions of circRNAs associated with necrotic cell death in CMs, we first performed circRNA microarray analysis in mice hearts with I/R injury and compared to sham treated heart (Supplementary Table 1). Among the identified and differentially expressed circRNAs, we selected 20 highly altered circRNAs (10 highly upregulated circRNAs and 10 highly downregulated circRNAs (at least 1.5-fold Up- and down-regulated; P < 0.05) and validated their expression levels using qRT-PCR. Among them, the level of circRNA000338 was remarkably reduced in I/R injured mouse hearts compared to sham hearts (Fig. 1a, b). Based on these results, we selected circRNA000338 for further investigation. We named this uncharacterized circRNA000338 as cardiac-necroptosis-associated circRNA (CNEACR) and verified its circular nature by RT-PCR using convergent and divergent primers. The divergent primers amplified CNEACR from cDNA but not from genomic DNA (Fig. 1c). Further, a specific back splice junction in CNEACR was verified using sanger sequencing after RNase R treatment (Fig. 1d). We then tested the expression levels of CNEACR in different tissues and found that it is also expressed in other organs (Supplementary Fig. 1a). In the mouse heart, the expression level of CNEACR was remarkably higher in CMs than in fibroblasts under physiological conditions (Fig. 1e) and CNEACR was predominantly found in the cytoplasm and a considerable level was found in the nucleus (Fig. 1f and 1g). Together, these results indicate that CNEACR is downregulated during myocardial I/R injury and its depletion may be associated with CMs injury and cell death.
Fig. 1. Identification of CNEACR in mice hearts with ischemia/reperfusion (I/R) injury.
a and b The expression levels of circRNAs (selected from circRNA microarray data) in I/R injured mice hearts determined by qPCR (n = 5 independent experiments). c The representative agarose gel image showing the linear or back-splicing products amplified using convergent and divergent primers of CNEACR in mice hearts. d Sanger sequencing was performed to confirm head-to-tail back-spliced site in CNEACR (black arrow). e QPCR analysis showing the expression level of CNEACR in cardiomyocytes and fibroblasts isolated from mice hearts (n = 3 independent experiments). f The level of CNEACR in the cytoplasmic or nuclear fractions of isolated cardiomyocytes as determined by qPCR. U6 and GAPDH used as internal controls (n = 3 independent experiments). g Representative images of fluorescence in situ hybridization with junction-specific probes of CNEACR indicates its subcellular localization. Green represents CNEACR and blue labels nuclei. Scale bar, 20 μm. Data in graphs are presented as Mean ± s.d. P values were calculated using two-sided Student’s t test.
CNEACR protects cardiomyocytes from cardiac I/R induced necrotic cell death
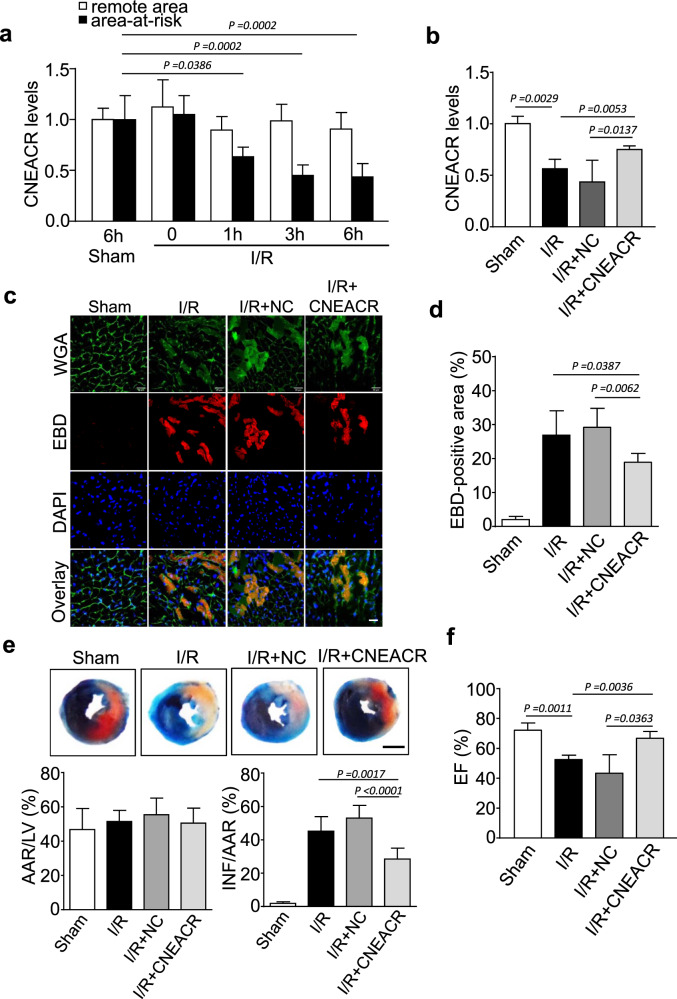
Cardiomyocyte cell death, due to necrosis, is one of the primary contributors to the pathogenesis and progression of various cardiac problems, including I/R injury [8, 34]. The expression of CNEACR was decreased in mice model of I/R injury (Fig. 2a). To investigate whether the depletion of CNEACR is associated with CMs injury and its functional role in necrotic death, the cardiac-specific overexpression of CNEACR was carried out by injecting adenovirus expressing CNEACR into WT mice (Supplementary Fig. 1b) and subjected to I/R. The overexpression of CNEACR markedly increased its level in mouse hearts with I/R injury compared to WT hearts (Fig. 2b). Cardiomyocyte necrosis was obvious in hearts section with I/R injury by the Evans blue dye (EBD) penetration and the overexpression of CNEACR suppressed the necrotic cell death as indicated by a significant reduction of EBD-positive cells in hearts with I/R injury (Fig. 2c and 2d). Evans blue-TTC staining showed that I/R-induced increase of infarct size reduced in CNEACR overexpressing hearts (Fig. 2e). The left ventricular ejection fraction, indicating cardiac function, also improved in I/R-injured hearts overexpressing CNEACR compared to WT mice hearts with I/R injury (Fig. 2f). Together, these results indicate that CNEACR blocks the necrotic death of CMs and attenuates myocardial dysfunction caused by I/R injury.
Fig. 2. CNEACR inhibits necrotic cell death in the heart.
a Mice were subjected to ischemia/reperfusion (I/R) at indicated time. Area-at-risk and the remote zone were prepared for qRT-PCR analysis of CNEACR levels. (n = 6 mice per group). b–f Adenoviral vector carrying mouse CNEACR or negative control (NC) were used for the cardiac-specific overexpression of CNEACR and then subjected to 60 min ischemia and 3 h reperfusion (I/R) or Sham in mice. b QPCR analysis showing the level of CNEACR in mice hearts. c Representative fluorescence images showing the Evans blue dye (EBD) (red) incorporation in LV sections counterstained with wheat germ agglutinin (WGA) (green) and DAPI (blue); scales bar, 20 μm. d Statistical analysis of the proportion of the EBD-positive cells in each group (n = 5 mice per group). e The representative images showing Evans blue-TTC stained midventricular heart sections (upper panel, blue-healthy and viable area, white-infarcted area, red-area at risk, Scale Bar, 2 mm). Quantification of the left ventricle infarct size measured as the percentage of left ventricle area (LV) and area at risk (AAR) after I/R or Sham (lower panel, n = 6 mice per group). INF, Infarct area. f Cardiac function (Ejection fraction, EF%) measured using echocardiography analysis after I/R (n = 6 mice per group).
CNEACR blocks cardiomyocytes necrosis in vitro
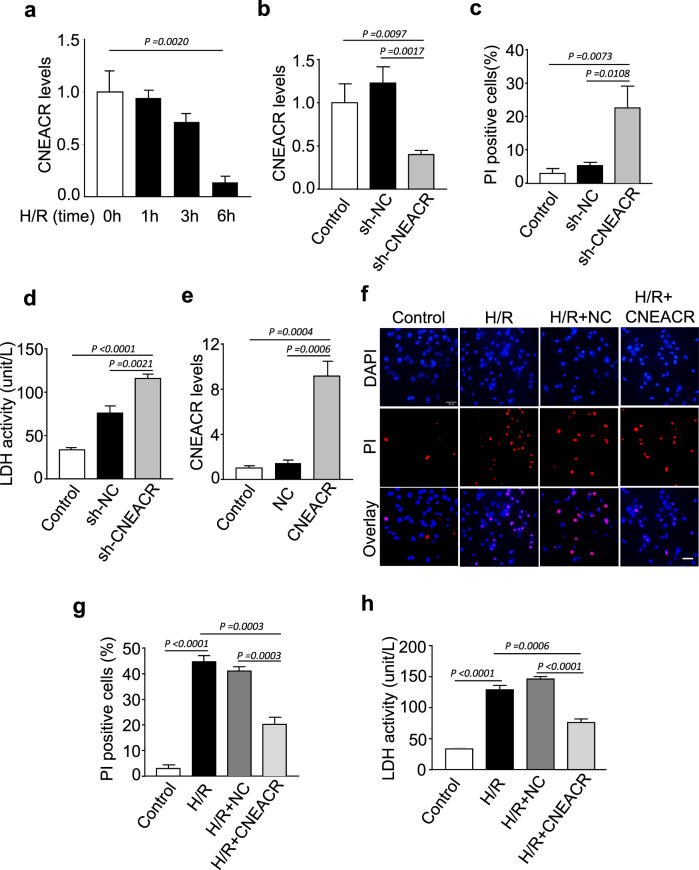
To confirm the role of CNEACR in the protection of CMs from necrotic cell death, we used the hypoxia-reoxygenation (H/R) model, an in vitro model that mimics I/R injury. In isolated neonatal mouse CMs, the expression level of CNEACR time-dependently decreased following H/R exposure (Fig. 3a). Under the normoxic condition, the depletion of CNEACR caused necrosis in CMs as indicated by a marked increase of PI-positive cells and LDH activity (Fig. 3b-d). In contrast, H/R-induced increase of PI-positive cells and LDH activity were suppressed upon overexpression of CNEACR in CMs (Fig. 3e-h). These results confirm that CNEACR can block necrotic death of CMs and it is involved in the protection of CMs from myocardial I/R injury.
Fig. 3. CNEACR attenuates hypoxia-reoxygenation (H/R)-induced necrosis in cardiomyocytes.
a–e Isolated neonatal mouse cardiomyocytes were infected with adenoviral vector carrying shRNA targeting CNEACR (sh-CNEACR) or negative control (sh-NC) for silencing CNEACR as described in the methods section. a QPCR analysis showing expression level of CNEACR at indicated time points (n = 3 independent experiments). b The level of CNEACR as determined by qPCR in cardiomyocytes infected with sh-CNEACR or sh-NC (n = 3 independent experiments). c The percentage of PI positive cells calculated from cardiomyocytes infected with sh-CNEACR or sh-NC. (n = 3 independent experiments). d The lactate dehydrogenase (LDH) activity in sh-CNEACR or sh-NC transfected cardiomyocytes (n = 3 independent experiments). e–h Adenoviral vector harboring mouse CNEACR or negative control (NC) were used overexpression of CNEACR in cardiomyocytes and subjected to H/R as described in the methods section. e QPCR analysis showing the level of CNEACR in cardiomyocytes infected with CNEACR or NC (n = 3 independent experiments). f Representative fluorescence images showing PI staining (red) and DAPI (blue, labels nucleus) in cardiomyocytes overexpressing CNEACR or NC and subjected to H/R. scales bar, 20 μm. g The percentage of PI positive cells calculated from experiment shown in (f) (n = 3 independent experiments). h The lactate dehydrogenase (LDH) activity in cardiomyocytes overexpressing CNEACR or NC and subjected to H/R (n = 4 independent experiments).
CNEACR binds to histone deacetylase 7 (HDAC7) and restricts its nuclear translocation
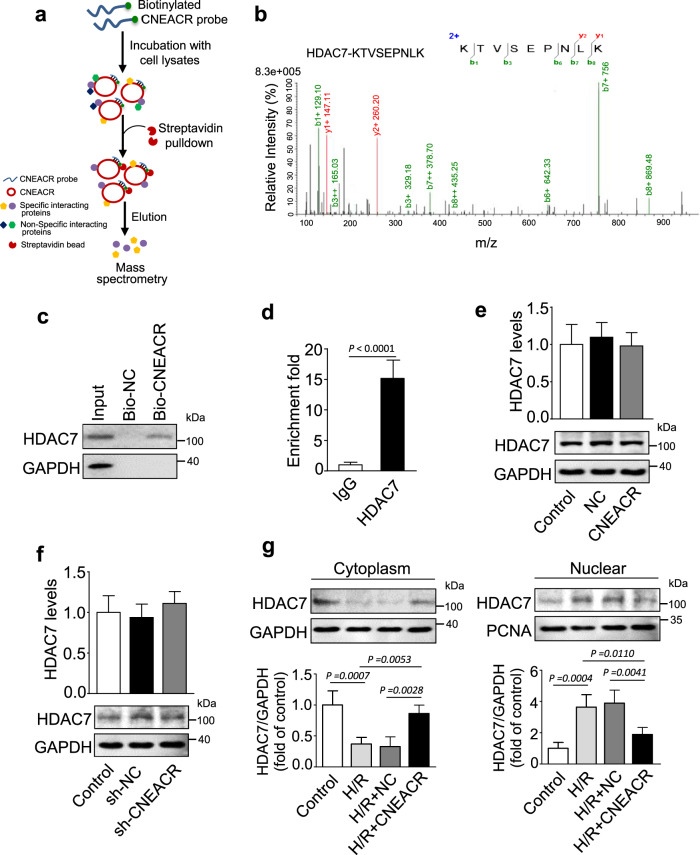
To explicate the molecular mechanism of CNEACR mediated protection of CMs necrotic death, we first examined the molecules interacting with CNEACR in CMs using biotinylated CNEACR and performed RNA pull-down assay as shown in Fig. 4a. The proteins bound with CNEACR were identified using liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis. Among those proteins, we chose histone deacetylases 7 (HDAC7), whose function was unknown in the regulation of necroptosis (Fig. 4b). In vitro RNA pull-down using biotinylated CNEACR followed by immunoblotting confirmed that CNEACR binds with HDAC7 (Fig. 4c). RIP (using HDAC7 antibody) followed by qPCR validated the interaction of CNEACR with HDAC7 in CMs (Fig. 4d). Also, the overexpression or silencing of CNEACR did not affect the expression levels of mRNA and protein of HDAC7 in CMs (Fig. 4e and 4f). Next, we examined whether CNEACR binding affects the function of HDAC7 protein. Given that a class II HDAC, HDAC7, generally functions as a suppressor or de-repressor of gene transcription and that shuttles between the cytoplasm and nucleus [35], we assessed the subcellular distribution of HDAC7. HDAC7 was located in both cytoplasm and nucleus under normal conditions and the nuclear accumulation of HDAC7 was markedly increased after H/R exposure. The overexpression of CNEACR reduced its nuclear import and retained in the cytoplasm in H/R exposed CMs (Fig. 4g). Together, these results indicate that the activity of HDAC7 is regulated by CNEACR and the depletion of CNEACR might be associated with the nuclear import of HDAC7 and its pathological function during I/R injury.
Fig. 4. CNEACR binds to HDAC7 and affects its nuclear import.
a A scheme showing the detection of CNEACR binding proteins using biotinylated CNEACR pull-down assay in isolated neonatal mouse cardiomyocytes. b Identification of CNEACR binding protein (detected HDAC7) using LC-MS/MS. c In cardiomyocytes, RNA pull-down assay was carried out using biotinylated CNEACR (Bio-CNEACR) or negative control (Bio-NC) and western blot analysis showing CNEACR binds with HDAC7 protein (n = 3 independent experiments). d RNA immunoprecipitation using HDAC7 specific antibody followed by qPCR analysis showing CNEACR enriched in HDAC7 fraction (n = 4 independent experiments). e The levels of Hdac7 mRNA (upper) and protein (lower) in cardiomyocytes overexpressing CNEACR or negative control (NC) (n = 3 independent experiments). f Expression levels of Hdac7 mRNA (upper) and protein (lower) in cardiomyocytes infected with adenovirus harboring CNEACR shRNA (sh-CNEACR) or negative control (sh-NC) (n = 3 independent experiments). g The subcellular distribution of HDAC7 in H/R injured cardiomyocytes infected with or without adenoviral vector carrying CNEACR or negative control (NC). Representative western blots (upper) and statistical data (lower) (n = 4 independent experiments) showing the expression of HDAC7 in the cytoplasmic or nuclear fractions. PCNA and GAPDH were used as an internal control for nuclear and cytoplasmic fractions, respectively.
CNEACR dependent inhibition of HDAC7 upregulates Forkhead box protein A2 (Foxa2) expression
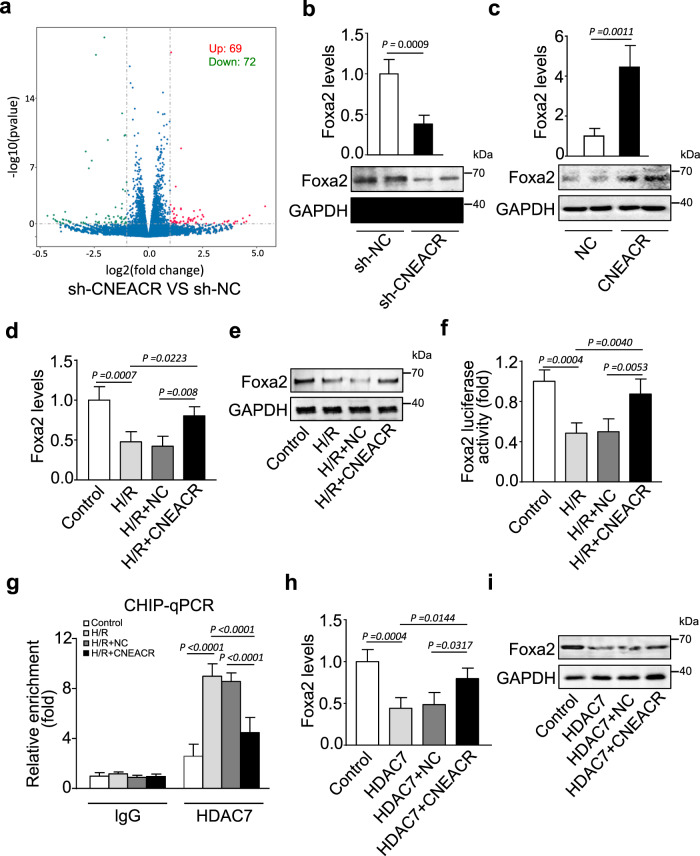
To identify the genes altered by the depletion of CNEACR and its associated increase of nuclear HDAC7 in conditions such as H/R, we performed transcriptome profiling using microarray in neonatal mouse CMs transfected with shRNA specifically silencing CNEACR (sh-CNEACR). We observed that 141 genes (69 upregulated and 72 downregulated) were differentially expressed upon knockdown of CNEACR (Fig. 5a and Supplementary Table 2). We selected highly altered genes (at least fivefold up- and down-regulated; P < 0.05) for further validation using qRT-PCR. Among those genes, the mRNA levels of Gm10309, Rdh1, Gm15839, A830005F24Rik, Hspa1a and Foxa2 were markedly altered in CNEACR silenced CMs (Supplementary Figs. 1c-g, Fig. 5b). Considering the functions of the FOX family of transcription factors, including Foxa2, in heart development and cardiac diseases [36, 37], we selected Foxa2 (also known as HNF-3β) and further assessment was carried out to delineate whether Foxa2 functions as a downstream target of CNEACR and HDAC7. The knockdown of CNEACR in CMs reduced the expression levels of Foxa2 mRNA and protein (Fig. 5b), whereas overexpression of CNEACR enforced the Foxa2 mRNA and protein levels (Fig. 5c).
Fig. 5. CNEACR promotes Foxa2 expression through HDAC7 inhibition in cardiomyocytes.
Isolated cardiomyocytes from neonatal mouse were infected with adenovirus harboring sh-CNEACR or negative control (sh-NC). a Volcanic plot from transcriptome microarray analysis showing the significant changes in the expression levels of genes (sh-CNEACR vs sh-NC). Red dots represent upregulated and green dots represent downregulated genes. b The expression levels of Foxa2 mRNA determined by qPCR (n = 3 independent experiments) and protein determined by western blot in cardiomyocytes infected with adenovirus harboring sh-CNEACR or sh-NC. c The expression levels of Foxa2 mRNA determined by qPCR (n = 3 independent experiments) and protein determined by western blot in cardiomyocytes infected with adenovirus harboring CNEACR or NC. d–f Cardiomyocytes were infected with adenoviral vector harboring mouse CNEACR or negative control (NC) and subjected to H/R. d QPCR analysis showing the expression level of Foxa2 mRNA in cardiomyocytes (n = 3 independent experiments). e Immunoblot showing the level of Foxa2 protein in cardiomyocytes. f Luciferase reporter assay results showing the transcription level of Foxa2 in H/R-exposed cardiomyocytes infected with or without overexpressing CNEACR (n = 3 independent experiments). g ChIP-qPCR analysis showing the enrichment level (the level of binding) of HDAC7 on Foxa2 promoter (n = 3 independent experiments). h Cardiomyocytes co-infected with adenoviral vector harboring Hdac7 and CNEACR or its negative control (NC) and expression levels of Foxa2 mRNA (h) and Foxa2 protein (i) were determined (n = 3 independent experiment).
Further, H/R exposure markedly reduced mRNA and protein levels of Foxa2, and overexpression of CNEACR suppressed H/R-induced reduction of Foxa2 mRNA and protein in CMs (Fig. 5d and 5e). HDACs regulate gene transcription through the modification of histones and chromatin remodeling [38, 39]. To examine whether CNEACR and HDAC7 influence expression of Foxa2 at the transcriptional level, we used luciferase reporter assay and found that H/R exposure reduced transcription of Foxa2 gene and overexpression of CNEACR reversed Foxa2 transcription in H/R exposed CMs (Fig. 5f). To confirm that suppression of Foxa2 transcription is mediated by HDAC7, we performed ChIP-qPCR analysis using HDAC7 specific antibody and found that the enrichment of HDAC7 on Foxa2 promoter was drastically increased in H/R exposed CMs and overexpression of CNEACR reduced its enrichment (Fig. 5g). In addition, the overexpression of HDAC7 reduced Foxa2 mRNA level while the silencing of HDAC7 increased the expression level of Foxa2 mRNA in CMs (Supplementary Fig. 2a and 2b). These results confirm that HDAC7 binds to the promoter region of Foxa2 and interferes with its transcription. To validate whether Foxa2 is the direct downstream target of the CNEACR-HDAC7 axis, we overexpressed both CNEACR and HDAC7 in CMs. The co-expression of CNEACR along with HDAC7 suppressed HDAC7-induced reduction in the levels of Foxa2 mRNA and protein in CMs (Fig. 5h and 5i). These results indicate that HDAC7 suppresses Foxa2 expression under H/R condition and CNEACR blocks the activity of HDAC7. Together, our data suggest that CNEACR can promote Foxa2 expression by blocking the nuclear entry of HDAC7, however, the depletion of CNEACR during H/R injury leads to the nuclear accumulation of HDAC7, which negatively regulates Foxa2 transcription in CMs.
Foxa2 protects cardiomyocytes from myocardial I/R induced necrotic form of cell death
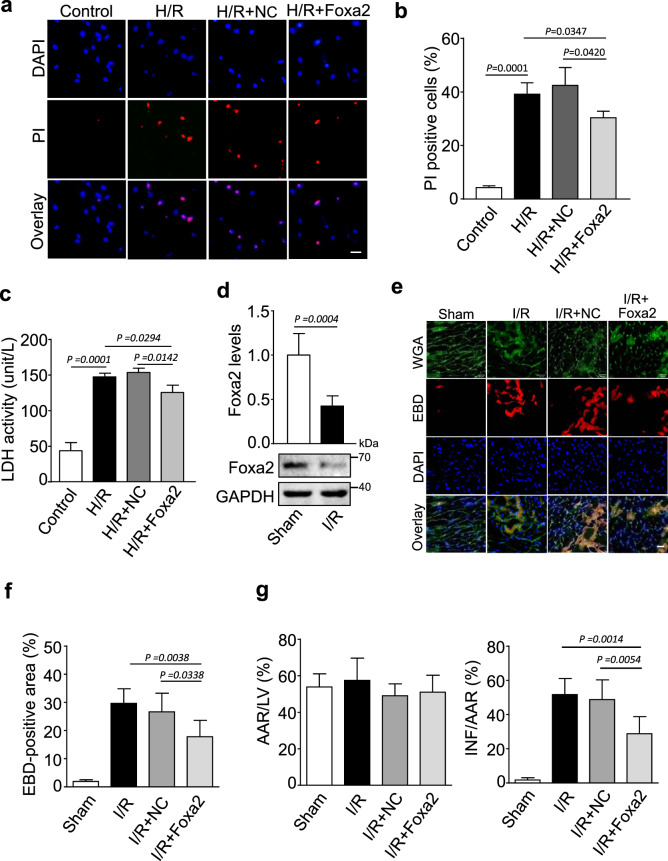
To substantiate that Foxa2 is involved in CNEACR mediated cardiomyocyte protection, we overexpressed Foxa2 in neonatal mouse CMs and subjected them H/R injury (Supplementary Fig. 3a and 3b). As shown in Fig. 6a-c, H/R-induced increase of PI-positive cells and LDH activity were reduced in CMs overexpressing Foxa2. Further, the levels of Foxa2 mRNA and protein were markedly reduced following myocardial I/R injury (Fig. 6d) and cardiac-specific overexpression of Foxa2 using adenoviral vector system (Supplementary Fig. 3c and 3d) reduced the level of EBD-positive cells (Fig. 6e and 6f), reduced infarct size (Fig. 6g) and improved the ventricular function as indicated by increased ejection fraction (Supplementary Fig. 3e). These data reveal that Foxa2 is involved in cardiomyocyte protection and its expression is suppressed during the progression of myocardial I/R injury.
Fig. 6. Foxa2 inhibits myocardial necrosis.
a–c Isolated cardiomyocytes were infected with adenoviral vector harboring mouse Foxa2 (Foxa2) or negative control (NC) and subjected to H/R. a Representative fluorescence images showing the propidium iodide (PI) staining (red) and DAPI (blue, labels nucleus). Scales bar, 20 μm. b The percentage of PI positive cells calculated from the experiment (a) (n = 4 independent experiments). c The lactate dehydrogenase (LDH) activity in Foxa2 or NC infected cardiomyocytes after H/R exposure (n = 4 independent experiments). d–g The adenoviral vector harboring mouse Foxa2 (Foxa2) or negative control (NC) were injected into mouse and subjected to I/R or sham as described in the methods section. d The expression levels of Foxa2 mRNA (upper) and protein (lower) were determined in hearts with Sham or I/R injury. GAPDH were used as an internal control (n = 5 biological replicates). e Representative image of Evans blue dye (EBD, red) incorporation in LV sections counterstained with wheat germ agglutinin (WGA, green and DAPI, blue); scale bar, 20 μm. f Statistical analysis of the proportion of the EBD-positive cells in each group (n = 5 independent experiments). g The ventricular heart sections were stained with Evans blue-TTC and the left ventricle infarct size measured as the percentage of left ventricle area (LV), Infarct area (INF) and area at risk (AAR) after I/R or sham (n = 5 mice each group).
CNEACR dependent upregulation of Foxa2 suppresses receptor-interacting protein kinase 3 (RIPK3) and necroptosis
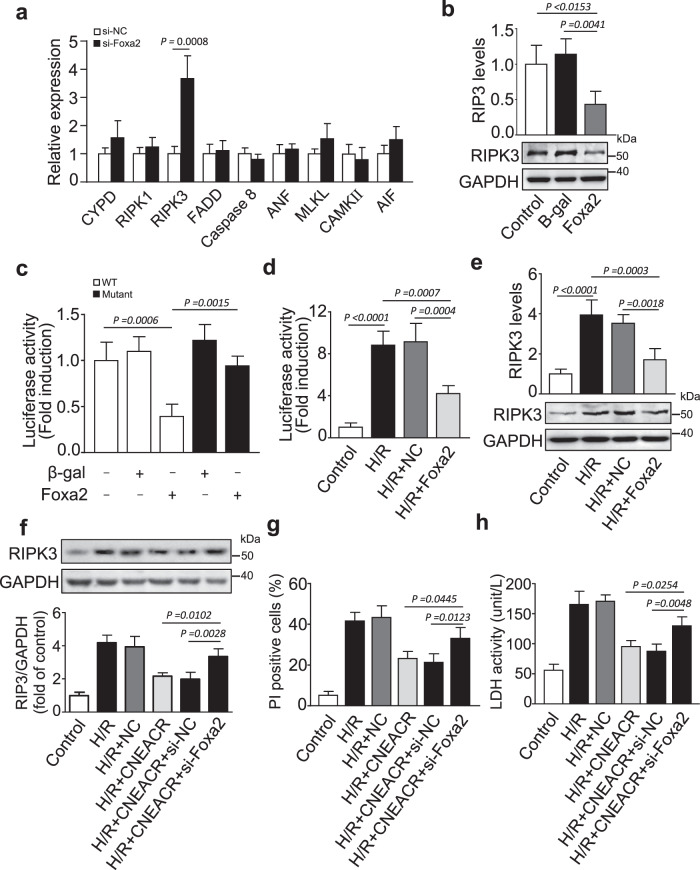
We next investigated the potential mechanism of Foxa2-dependent protection of necrotic death of CMs. We assessed the expression levels of key players associated with the necrotic/necroptotic mode of cell death. Among them, the expression level of RIPK3, the key necroptosis molecule, was increased upon silencing of Foxa2 in CMs (Supplementary Fig. 4a, Fig. 7a and Supplementary Fig. 4b). Conversely, the overexpression of Foxa2 remarkably reduced the levels of RIPK3 mRNA and protein (Fig. 7b), which indicates the inverse relationship between Foxa2 and RIPK3. Given that Foxa2 can control gene expression through its DNA binding activity [40, 41]. We next assessed whether Foxa2 directly targets Ripk3 gene. We observed that RIPK3 promoter region has a potential Foxa2 binding site (Supplementary Fig. 4c). We constructed Ripk3 promoter containing a Foxa2 binding site and analyzed the effect of Foxa2 on its activity using the luciferase reporter assay. The overexpression of Foxa2 reduced the luciferase activity of the wild-type Ripk3 construct, but not by the mutant Ripk3 construct (Fig. 7c and Supplementary Fig.4d). H/R-induced increase of Ripk3 gene transcription, indicated by increased luciferase activity of wild-type Ripk3 construct, was attenuated upon overexpression of Foxa2 in CMs (Fig. 7d). Also, Foxa2 overexpression inhibited H/R-induced increase of RIPK3 mRNA and protein in CMs (Fig. 7e). These results indicate that Foxa2 suppresses RIPK3 expression through its DNA binding activity and direct binding to the promoter region of RIPK3.
Fig. 7. Foxa2 regulates receptor-interacting protein kinase 3 (RIPK3) expression in mouse hearts with I/R injury.
a Neonatal mouse cardiomyocytes were transfected with siRNA of Foxa2 (siFoxa2) or its negative control (NC) and expression levels of key genes involved in myocyte necrosis was determined by qPCR (n = 3 independent experiments). b Cardiomyocytes were infected with adenoviral vector harboring Foxa2 (Foxa2) or its negative control (NC) and expression levels of RIPK3 mRNA assessed by qPCR (upper) and RIPK3 protein assessed by western blot (lower) (n = 3 independent experiments). c HEK293 cells were treated with the adenoviral NC or Foxa2, the constructs of vector carring the wild-type promoter (RIPK3-wt) or the promoter with mutations in the binding site (RIPK3-mut) respectively. Luciferase activity was carried out (n = 3 independent experiments). d Isolated cardiomyocytes were cotransfected with pGL-4.17 vector carrying Foxa2 binding sites of Ripk3 promoter region and adenovirus harboring Foxa2 and subjected to H/R. Luciferase activity was carried out (n = 3 independent experiments). e The expression levels of Ripk3 mRNA (upper) and protein (lower) in cardiomyocytes infected with adenovirus harboring Foxa2 (Foxa2) or its negative control (NC) and subjected to H/R (n = 3 independent experiments). f–h Cardiomyocytes co-infected with adenovirus carrying CNEACR, its negative control (NC), Foxa2 siRNA (siFoxa2), scrambled RNA (siNC) or its combination and exposed to H/R. f Representative western blots (upper) and statistical data (lower) showing the level of RIPK3 in cardiomyocytes. GAPDH was used as an internal control (n = 3 independent experiments with similar results). g The percentage of propidium iodide (PI) positive cells were calculated (n = 3 independent experiments). h The lactate dehydrogenase (LDH) activity was assessed in cardiomyocytes (n = 3 independent experiments).
We next validated the association between CNEACR and Foxa2 in the regulation of RIPK3-mediated necrotic cell death of CMs. The overexpression of CNEACR decreased the level of RIPK3 protein (Supplementary Fig. 4e) while knockdown of CNEACR increased the level of RIPK3 protein in CMs (Supplementary Fig. 4f). H/R-induced increase of RIPK3 protein was suppressed upon enforced expression of CNEACR and this effect was counteracted by silencing of Foxa2 in CMs (Fig. 7f). In addition, the overexpression of CNEACR markedly reduced the level of PI-positive cells and LDH activity and silencing of Foxa2 reversed these effects in CMs exposed to H/R injury (Fig. 7g and 7h). To further test if RIPK3 is indeed the target of CNEACR, we examined the effect of RIPK3 knockdown on I/R-induced myocardial injury in CNEACR knockdown mouse hearts using AAV9 carrying RIPK3-shRNA (Supplementary Fig. 5a). The results showed that the inhibition of CNEACR enhanced I/R-induced necrotic death of CMs and infarct size, and knockdown of RIPK3 counteracted the effects of CNEACR silencing in I/R-treated mice hearts (Supplementary Fig. 5b and 5c). Similarly, echocardiography analysis revealed that the left ventricular function was further decreased in CNEACR knockdown mice with I/R, whereas administration of RIPK3-shRNA attenuated the effect of CNEACR knockdown as indicated by increased ejection fraction after I/R (Supplementary Fig. 5d). In addition, we examined the effect of CNEACR knockdown on I/R-induced myocardial injury in MLKL KO mice, and also MLKL knockout reversed the effect of CNEACR knockdown on myocardial injury (Supplementary Fig. 6a-c). These results indicated that RIPK3 and MLKL function as the downstream molecules of CNEACR during I/R injury.
Next, we further explored the downstream mechanism of CNEACR-mediated cardiomyocyte necroptosis. Phosphorylation of RIPK1 at S166 and phosphorylation of MLKL at S345 are considered the hallmark of their activation and have been widely used as biomarkers for necroptotic death [42–45], and thus we detected the changes in the biochemical hallmarks of necroptosis, including the phosphorylation of RIPK1 and MLKL. The results showed that RIPK3, RIPK3 phosphorylation and MLKL phosphorylation were elevated in CNEACR-knockdown cells, but no increase in RIPK1, RIPK1 phosphorylation and MLKL were observed (Supplementary Fig. 7a). Furthermore, overexpression of CNEACR reduced the phosphorylation of MLKL and RIPK3 under H/R treatment in cardiomyocyte (Supplementary Fig. 7b). These data indicate that CNEACR negatively regulates RIPK3/ MLKL phosphorylation and thus H/R-induced necroptosis. Furthermore, recent reports have highlighted the role of ESCRT-III (Endosomal Sorting Complex Required for Transport-III) in repairing damaged membranes in necroptosis [40, 46, 47], and ESCRT-III supports cell survival and functions downstream of active MLKL to regulate necrotic cell death [40]. To determine whether ESCRT-III can act downstream of CNEACR to mediate necroptosis induced by H/R in cardiomyocyte, we silenced the ESCRT-III components CHMP2A and CHMP4B. The results showed that silencing of CHMP2A or CHMP4B attenuated the inhibitory effect of CNEACR on cardiomyocyte necroptosis upon H/R treatment (Supplementary Fig. 7c and 7d), indicating that ESCRT-III-dependent plasma membrane repair mechanism might be involved in the pathway of H/R-CNEACR-mediated cardiomyocyte necroptosis. Together these results indicate that CNEACR/HDAC7/Foxa2 axis targets RIPK3 and MLKL and protects CMs from necrotic/necroptotic cell death.
Discussion
In this study, we identified that CNEACR, a circRNA enriched in CMs, is reduced after myocardial I/R and its depletion leads to necrotic/necroptotic death of CMs. However, the enforced expression of CNEACR in I/R injured heart or H/R exposed CMs promotes cardiomyocyte survival, reduces cardiac injury and improved cardiac function. CNEACR can directly interact with HDAC7 and interfering its nuclear entry and that results in increased expression of Foxa2 and suppression of RIPK3 transcription, which is a key player involved in the necrosis/necroptosis. Our findings provide new insights into how the activities of epigenetic machinery such as HDAC7 are dysregulated during the progression of myocardial injury. Thus, CNEACR/HDAC7/FOXA2/ RIPK3 pathway adding a new dimension to learn the programmed necrosis of CMs, an important contributor of cardiac myocyte loss, and CNEACR could be used as a tool to halt the programmed necrotic cell death in severe pathological conditions such as cardiac I/R injury and MI.
The dysregulation of circRNAs is closely associated with CMs death during ischemic injury of the heart [48–51]. However, their functional role in myocardial cell necrosis and/or necroptosis is still largely unknown. Here, we show that the levels of CNEACR is reduced during the progression of myocardial injury, which is confirmed in H/R exposed CMs and I/R injured myocardial tissue. However, the restoration of CNEACR level using adenoviral vector carrying CNEACR alleviates CMs necroptotic cell death and improves cardiac function. Currently, it is unclear the mechanism by which CNEACR level is reduced during I/R injury. Given that inflammation is the primary contributor in triggering necrotic/necroptotic mode cell death [52, 53] and that is critically involved in the extensive cardiomyocyte damage and myocardial injury following ischemia insults [54], it could be possible that pro-inflammatory cytokines and chemokine dependent signaling might be associated with the suppression/reduction of CNEACR in I/R injured CMs and myocardial tissue. Further studies are warranted to address this notion.
CircRNAs exert diverse biological functions depending on their tissue specificity and subcellular distribution. The majority of circRNAs are localized in the cytoplasm in which they confiscate miRNAs and/or proteins and interfering their functions [22, 23, 25]. In our study, we found that CNEACR is mainly distributed in the cytoplasm of CMs where it directly interacts with HDAC7 and blocks its nuclear translocation. HDAC7 is abundantly localized to the nucleus following I/R injury and its nuclear translocation is associated with the lack of CNEACR. However, the enforced expression of CNEACR promotes cytoplasmic retention of HDAC7 in H/R exposed CMs, which clearly show that the circRNAs such as CNEACR has the ability to control the activity and nuclear translocation of HDAC7, and thereby CNEACR might upregulate the expression of cell survival genes and suppression of cell death-associated genes in CMs. It is well defined that the activities of multiple forms of HDACs, including class II HDACs, are highly increased in myocardial I/R and their persistent activities in the nucleus favour the gene program associated with cell death and progression of myocardial injury. The chemical or pharmacological inhibition of HDACs promote cardiomyocyte survival, reduce I/R stress and myocardial infarct size, and improve cardiac function [55–57]. However, it is yet largely unknown, how the activities and subcellular distribution of HDACs, including HDAC7 are dysregulated during the progression of I/R-induced cellular and myocardial damage. Our study provides novel evidence that the cytoplasmic circRNA, CNEACR, functions as a regulator of necrosis/necroptosis associated HDACs such as HDAC7.
HDAC7 is a member of class IIa HDAC subfamily, which exhibit tissue- and cell-specific expression pattern. HDAC 7 can shuttle between the cytoplasm and nucleus, which is a tightly regulated event. The nuclear HDAC7 is in the non-phosphorylated form that represses gene transcription by histone modifications, while the phosphorylated HDAC7 is translocated to the cytosol, which results in the derepression of genes targeted by HDAC7 [35, 58]. Since the nuclear HDAC7 functions as a gene suppressor, we found that CNEACR-dependent inhibition of HDAC7 nuclear entry upregulates Foxa2 transcription, which suppresses the expression of RIPK3, a key mediator of necroptosis, and promotes the survival of CMs. However, the myocardial I/R-induced depletion of CNEACR leads to nuclear accumulation of HDAC7 where it binds to the Foxa2 promoter region and represses Foxa2 transcription. Despite we found that CNEACR detained the nuclear import of HDAC7, currently it is unknown whether CNEACR interaction affects the dephosphorylation/phosphorylation status of HDAC7. A further detailed study will be carried out to address this notion.
Foxa2 is a member of the forkhead transcription factor family that has a conserved DNA binding domain. It can bind to both promoter and enhancer regions of target genes and regulates their transcription. In general, the binding of Foxa2 to the promoter region inhibits gene expression [59, 60]. At the developmental stage, Foxa2 is required for the normal development of ventricular cells, heart fields and cardiac morphogenesis [37]. Foxa2 being a hepatocyte-enriched transcription factor, is essential for the hepatocyte cell survival and protection against various toxic and stress insults [61, 62]. In the myoblast and skeletal muscle, the increased expression and activity of Foxa2 attenuates inflammatory response and inflammation-associated tissue damage [63]. However, the functional relevance of Foxa2 in cell survival/death, in particular, its influence on the necrotic/necroptotic death of CMs remains unknown. In this study, we found that CNEACR mediated upregulation of Foxa2 reduces I/R-induced necroptosis of CMs and myocardial tissue through suppressing the transcription of Ripk3, a central player involved in necroptotic signalling. Notably, Ripk3 promoter region contains Foxa2 binding site, which facilitate Foxa2 binding and repression of Ripk3 transcription. In CMs, the co-ordinated function of CNEACR and Foxa2 negatively regulates RIPK3 expression, which is confirmed by the diminution of RIPK3 level in H/R-exposed CMs overexpressing CNEACR or Foxa2 as well as the augmentation of CNEACR-mediated reduction of RIPK3 by silencing of Foxa2 in CMs with H/R injury. In severe cardiac pathological conditions such as I/R and MI, the increased expression of RIPK3 triggers the necroptosis by forming a complex with RIPK1, which is known as necrosome. Notably, RIPK3-dependent necroptosis hastens inflammatory response as well as oxidative stress-mediated myocardial remodeling and heart failure [6, 13]. Thus, our findings divulge the mechanism responsible for the relentless expression of RIPK3 and necroptotic damage of CMs during the ischemic heart diseases.
In summary, our study reveals that CNEACR functions as a positive regulator of CMs survival by controlling the activity of HDAC7 and promoting Foxa2-dependent suppression of a key molecule (RIPK3) associated with necroptotic cell death. The myocardial ischemic injury leads to the reduction of CNEACR and increase of nuclear activity of HDAC7, which consequently elicits necroptotic mode of cardiac myocytes death by suppressing Foxa2 and derepression of Ripk3 during the progression of I/R injury. Our study not only found that CNEACR is a master regulator of a non-apoptotic form of programmed cell death but also provides a new mechanism that connects the dysregulated activities of HDACs such as HDAC7 and upregulation of death-associated molecules such as Ripk3 in I/R-injured CMs. Further, the silencing of HDAC7 or overexpression of CNEACR/Foxa2 can slow and/or halt the progression of myocardial injury and improved the cardiac function, which is accompanied by a significant reduction of necroptotic death of CMs. This suggests that the therapeutic manipulation of the pathway composed of CNEACR /HDAC7/Foxa2/ RIPK3 could be a valuable strategy for the attenuation of the loss of CMs, due to necroptosis, in ischemia associated heart diseases and cardiac failure.
Supplementary information
Acknowledgements
We thank Prof Ling Li for assistance with immunohistochemistry work. We also thank Dr Murugavel Ponnusamy for polishing the paper.
Author contributions
KW, ZC, and YW designed research. XG, CL, YZ, YW, LZ, XL, KW, XC, TW, JJ, FW, SW performed experiments. XG, KW, ZC and YW analyzed the data. XG, KW and ZC wrote the paper.
Funding
This work was supported by National Natural Science Foundation of China (81770275, 82070313, 81800272, 81870236, 82070314, 81770406, 81873472, 31671447); Key Projects in the National Science and Technology 6 Pillar Program of the 13th Five-Year Plan Period (2017YFC1308300, 2017YFC1308305); Taishan Scholar Program of Shandong Province and Qingdao public domain science and technology support plan project (19-6-1-6-nsh).
Data availability
All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
Competing interests
The authors declare no competing interests.
Ethical approval
Ethical approval was obtained from Ethics Committee of Qingdao University.
Footnotes
Edited by: A Oberst
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Xiang-Qian Gao, Cui-Yun Liu, Yu-Hui Zhang, Yun-Hong Wang
Contributor Information
Yin Wang, Email: wangyin200139@yeah.net.
Zhao-Yang Chen, Email: chenzhaoy2006809@yeah.net.
Kun Wang, Email: wangk696@qdu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41418-021-00872-2.
References
- 1.Gomes CPC, Schroen B, Kuster GM, Robinson EL, Ford K, Squire IB, et al. Regulatory RNAs in Heart Failure. Circulation. 2020;141:313–28. doi: 10.1161/CIRCULATIONAHA.119.042474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roth GA, Mensah GA, Johnson CO, Addolorato G, Ammirati E, Baddour LM, et al. Global Burden of Cardiovascular Diseases and Risk Factors, 1990-2019: Update From the GBD 2019 Study. J Am Coll Cardiol. 2020;76:2982–3021. doi: 10.1016/j.jacc.2020.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Derek M. Yellon, Derek J. Hausenloy. Myocardial Reperfusion Injury. N Engl J Med. 2007;357:1121–35. doi: 10.1056/NEJMra071667. [DOI] [PubMed] [Google Scholar]
- 4.Hausenloy DJ, Yellon DM. Myocardial ischemia-reperfusion injury: a neglected therapeutic target. J Clin Investig. 2013;123:92–100. doi: 10.1172/JCI62874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heusch G. Myocardial ischaemia-reperfusion injury and cardioprotection in perspective. Nat Rev Cardiol. 2020;17:773–89. doi: 10.1038/s41569-020-0403-y. [DOI] [PubMed] [Google Scholar]
- 6.Luedde M, Lutz M, Carter N, Sosna J, Jacoby C, Vucur M, et al. RIP3, a kinase promoting necroptotic cell death, mediates adverse remodelling after myocardial infarction. Cardiovasc Res. 2014;103:206–16. doi: 10.1093/cvr/cvu146. [DOI] [PubMed] [Google Scholar]
- 7.Vandenabeele P, Galluzzi L, Vanden Berghe T, Kroemer G. Molecular mechanisms of necroptosis: an ordered cellular explosion. Nat Rev Mol cell Biol. 2010;11:700–14. doi: 10.1038/nrm2970. [DOI] [PubMed] [Google Scholar]
- 8.Del ReDP, Amgalan D, Linkermann A, Liu Q, Kitsis RN. Fundamental Mechanisms of Regulated Cell Death and Implications for Heart Disease. Physiol Rev. 2019;99:1765–817. doi: 10.1152/physrev.00022.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang DW, Shao J, Lin J, Zhang N, Lu BJ, Lin SC, et al. RIP3, an energy metabolism regulator that switches TNF-induced cell death from apoptosis to necrosis. Science. 2009;325:332–6. doi: 10.1126/science.1172308. [DOI] [PubMed] [Google Scholar]
- 10.He S, Wang L, Miao L, Wang T, Du F, Zhao L, et al. Receptor interacting protein kinase-3 determines cellular necrotic response to TNF-α. Cell. 2009;137:1100–11. doi: 10.1016/j.cell.2009.05.021. [DOI] [PubMed] [Google Scholar]
- 11.Wang H, Sun L, Su L, Rizo J, Liu L, Wang LF, et al. Mixed Lineage Kinase Domain-like Protein MLKL Causes Necrotic Membrane Disruption upon Phosphorylation by RIP3. Mol Cell. 2014;54:133–46. doi: 10.1016/j.molcel.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 12.Chen X, Li W, Ren J, Huang D, He WT, Song Y, et al. Translocation of mixed lineage kinase domain-like protein to plasma membrane leads to necrotic cell death. Cell Res. 2014;24:105–21. doi: 10.1038/cr.2013.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang T, Zhang Y, Cui M, Jin L, Wang Y, Lv F, et al. CaMKII is a RIP3 substrate mediating ischemia- and oxidative stress–induced myocardial necroptosis. Nat Med. 2016;22:175–82. doi: 10.1038/nm.4017. [DOI] [PubMed] [Google Scholar]
- 14.Wang Z, Jiang H, Chen S, Du F, Wang X. The mitochondrial phosphatase PGAM5 functions at the convergence point of multiple necrotic death pathways. Cell. 2012;148:228–43. doi: 10.1016/j.cell.2011.11.030. [DOI] [PubMed] [Google Scholar]
- 15.Zhou H, Li D, Zhu P, Ma Q, Toan S, Wang J, et al. Inhibitory effect of melatonin on necroptosis via repressing the Ripk3-PGAM5-CypD-mPTP pathway attenuates cardiac microvascular ischemia-reperfusion injury. J Pineal Res. 2018;65:e12503. doi: 10.1111/jpi.12503. [DOI] [PubMed] [Google Scholar]
- 16.Declercq W, Vanden Berghe T, Vandenabeele P. RIP kinases at the crossroads of cell death and survival. Cell. 2009;138:229–32. doi: 10.1016/j.cell.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 17.Christofferson DE, Yuan J. Necroptosis as an alternative form of programmed cell death. Curr Opin Cell Biol. 2010;22:263–8. doi: 10.1016/j.ceb.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dong Y, Liu C, Zhao Y, Ponnusamy M, Li P, Wang K. Role of noncoding RNAs in regulation of cardiac cell death and cardiovascular diseases. Cell Mol Life Sci. 2018;75:291–300. doi: 10.1007/s00018-017-2640-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–8. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 20.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PloS ONE. 2012;7:e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. Rna. 2013;19:141–57. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qu S, Zhong Y, Shang R, Zhang X, Song W, Kjems J, et al. The emerging landscape of circular RNA in life processes. RNA Biol. 2017;14:992–9. doi: 10.1080/15476286.2016.1220473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846–58. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lim TB, Lavenniah A, Foo RS. Circles in the heart and cardiovascular system. Cardiovasc Res. 2020;116:269–78. doi: 10.1093/cvr/cvz227. [DOI] [PubMed] [Google Scholar]
- 25.Zhou MY, Yang JM, Xiong XD. The emerging landscape of circular RNA in cardiovascular diseases. J Mol Cell Cardiol. 2018;122:134–9. doi: 10.1016/j.yjmcc.2018.08.012. [DOI] [PubMed] [Google Scholar]
- 26.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–8. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 27.Holdt LM, Stahringer A, Sass K, Pichler G, Kulak NA, Wilfert W, et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun. 2016;7:12429. doi: 10.1038/ncomms12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xie M, Hill JA. HDAC-dependent ventricular remodeling. Trends Cardiovasc Med. 2013;23:229–35. doi: 10.1016/j.tcm.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gang H, Shaw J, Dhingra R, Davie JR, Kirshenbaum LA. Epigenetic regulation of canonical TNFalpha pathway by HDAC1 determines survival of cardiac myocytes. Am J Physiol Heart Circ Physiol. 2013;304:H1662–1669. doi: 10.1152/ajpheart.00093.2013. [DOI] [PubMed] [Google Scholar]
- 30.Zhang L, Wang H, Zhao Y, Wang J, Dubielecka PM, Zhuang S, et al. Myocyte-specific overexpressing HDAC4 promotes myocardial ischemia/reperfusion injury. Mol Med. 2018;24:37. doi: 10.1186/s10020-018-0037-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shaw J, Zhang T, Rzeszutek M, Yurkova N, Baetz D, Davie JR, et al. Transcriptional silencing of the death gene BNIP3 by cooperative action of NF-kappaB and histone deacetylase 1 in ventricular myocytes. Circ Res. 2006;99:1347–54. doi: 10.1161/01.RES.0000251744.06138.50. [DOI] [PubMed] [Google Scholar]
- 32.Altesha MA, Ni T, Khan A, Liu K, Zheng X. Circular RNA in cardiovascular disease. J Cell Physiol. 2019;234:5588–600. doi: 10.1002/jcp.27384. [DOI] [PubMed] [Google Scholar]
- 33.Mester-Tonczar J, Hasimbegovic E, Spannbauer A, Traxler D, Kastner N, Zlabinger K, et al. Circular RNAs in Cardiac Regeneration: Cardiac Cell Proliferation, Differentiation, Survival, and Reprogramming. Front Physiol. 2020;11:580465. doi: 10.3389/fphys.2020.580465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu H, Sun A. Programmed necrosis in heart disease: molecular mechanisms and clinical implications. J Mol Cell Cardiol. 2018;116:125–34. doi: 10.1016/j.yjmcc.2018.01.018. [DOI] [PubMed] [Google Scholar]
- 35.Parra M. Class IIa HDACs - new insights into their functions in physiology and pathology. FEBS J. 2015;282:1736–44. doi: 10.1111/febs.13061. [DOI] [PubMed] [Google Scholar]
- 36.Zhu H. Forkhead box transcription factors in embryonic heart development and congenital heart disease. Life Sci. 2016;144:194–201. doi: 10.1016/j.lfs.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 37.Bardot E, Calderon D, Santoriello F, Han S, Cheung K, Jadhav B, et al. Foxa2 identifies a cardiac progenitor population with ventricular differentiation potential. Nat Commun. 2017;8:14428. doi: 10.1038/ncomms14428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Delcuve GP, Khan DH, Davie JR. Roles of histone deacetylases in epigenetic regulation: emerging paradigms from studies with inhibitors. Clin Epigenet. 2012;4:5. doi: 10.1186/1868-7083-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu C, Chen Q, Xie Z, Ai J, Tong L, Ding J, et al. The role of histone deacetylase 7 (HDAC7) in cancer cell proliferation: regulation on c-Myc. J Mol Med. 2010;89:279–89. doi: 10.1007/s00109-010-0701-7. [DOI] [PubMed] [Google Scholar]
- 40.Gong YN, Guy C, Olauson H, Becker JU, Yang M, Fitzgerald P, et al. ESCRT-III Acts Downstream of MLKL to Regulate Necroptotic Cell Death and Its Consequences. Cell. 2017;169:286–300.e216. doi: 10.1016/j.cell.2017.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bochkis IM, Schug J, Ye DZ, Kurinna S, Stratton SA, Barton MC, et al. Genome-wide location analysis reveals distinct transcriptional circuitry by paralogous regulators Foxa1 and Foxa2. PLoS Genet. 2012;8:e1002770. doi: 10.1371/journal.pgen.1002770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Degterev A, Hitomi J, Germscheid M, Ch’en IL, Korkina O, Teng X, et al. Identification of RIP1 kinase as a specific cellular target of necrostatins. Nat Chem Biol. 2008;4:313–21. doi: 10.1038/nchembio.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laurien L, Nagata M, Schünke H, Delanghe T, Wiederstein JL.Kumari S,Autophosphorylation at serine 166 regulates RIP kinase 1-mediated cell death and inflammation.2020;11:1747. [DOI] [PMC free article] [PubMed]
- 44.Sun L, Wang H, Wang Z, He S, Chen S, Liao D, et al. Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell. 2012;148:213–27. doi: 10.1016/j.cell.2011.11.031. [DOI] [PubMed] [Google Scholar]
- 45.Rodriguez DA, Weinlich R.Characterization of RIPK3-mediated phosphorylation of the activation loop of MLKL during necroptosis.2016;23:76–88. [DOI] [PMC free article] [PubMed]
- 46.Gong YN, Guy C, Crawford JC, Green DR. Biological events and molecular signaling following MLKL activation during necroptosis. Cell Cycle (Georget, Tex) 2017;16:1748–60. doi: 10.1080/15384101.2017.1371889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoon S, Kovalenko A, Bogdanov K, Wallach D. MLKL, the Protein that Mediates Necroptosis, Also Regulates Endosomal Trafficking and Extracellular Vesicle Generation. Immunity. 2017;47:51–65.e57. doi: 10.1016/j.immuni.2017.06.001. [DOI] [PubMed] [Google Scholar]
- 48.Geng HH, Li R, Su YM, Xiao J, Pan M, Cai XX, et al. The Circular RNA Cdr1as Promotes Myocardial Infarction by Mediating the Regulation of miR-7a on Its Target Genes Expression. PloS ONE. 2016;11:e0151753. doi: 10.1371/journal.pone.0151753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang S, Chen J, Yu W, Deng F. Circular RNA DLGAP4 ameliorates cardiomyocyte apoptosis through regulating BCL2 via targeting miR-143 in myocardial ischemia-reperfusion injury. Int J Cardiol. 2019;279:147. doi: 10.1016/j.ijcard.2018.09.023. [DOI] [PubMed] [Google Scholar]
- 50.Zhang CL, Long TY, Bi SS, Sheikh SA, Li F. CircPAN3 ameliorates myocardial ischaemia/reperfusion injury by targeting miR-421/Pink1 axis-mediated autophagy suppression. Lab Investig. 2021;101:89–103. doi: 10.1038/s41374-020-00483-4. [DOI] [PubMed] [Google Scholar]
- 51.Zhou LY, Zhai M, Huang Y, Xu S, An T, Wang YH, et al. The circular RNA ACR attenuates myocardial ischemia/reperfusion injury by suppressing autophagy via modulation of the Pink1/ FAM65B pathway. Cell Death Differ. 2019;26:1299–315. doi: 10.1038/s41418-018-0206-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kearney CJ, Martin SJ. An Inflammatory Perspective on Necroptosis. Mol Cell. 2017;65:965–73. doi: 10.1016/j.molcel.2017.02.024. [DOI] [PubMed] [Google Scholar]
- 53.Dhuriya YK, Sharma D. Necroptosis: a regulated inflammatory mode of cell death. J Neuroinflammation. 2018;15:199. doi: 10.1186/s12974-018-1235-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang Z, Zingarelli B, Szabo C. Effect of genetic disruption of poly (ADP-ribose) synthetase on delayed production of inflammatory mediators and delayed necrosis during myocardial ischemia-reperfusion injury. Shock. 2000;13:60–66. doi: 10.1097/00024382-200013010-00011. [DOI] [PubMed] [Google Scholar]
- 55.Granger A, Abdullah I, Huebner F, Stout A, Wang T, Huebner T, et al. Histone deacetylase inhibition reduces myocardial ischemia-reperfusion injury in mice. FASEB J. 2008;22:3549–60. doi: 10.1096/fj.08-108548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao TC, Cheng G, Zhang LX, Tseng YT, Padbury JF. Inhibition of histone deacetylases triggers pharmacologic preconditioning effects against myocardial ischemic injury. Cardiovasc Res. 2007;76:473–81. doi: 10.1016/j.cardiores.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 57.Xie M, Kong Y, Tan W, May H, Battiprolu PK, Pedrozo Z, et al. Histone deacetylase inhibition blunts ischemia/reperfusion injury by inducing cardiomyocyte autophagy. Circulation. 2014;129:1139–51. doi: 10.1161/CIRCULATIONAHA.113.002416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kao H-Y, Verdel A, Tsai C-C, Simon C, Juguilon H, Khochbin S. Mechanism for Nucleocytoplasmic Shuttling of Histone Deacetylase 7. J Biol Chem. 2001;276:47496–507. doi: 10.1074/jbc.M107631200. [DOI] [PubMed] [Google Scholar]
- 59.Friedman JR, Kaestner KH. The Foxa family of transcription factors in development and metabolism. Cell Mol Life Sci. 2006;63:2317–28. doi: 10.1007/s00018-006-6095-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sekiya T, Muthurajan UM, Luger K, Tulin AV, Zaret KS. Nucleosome-binding affinity as a primary determinant of the nuclear mobility of the pioneer transcription factor FoxA. Genes Dev. 2009;23:804–9. doi: 10.1101/gad.1775509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang W, Yao LJ, Shen W, Ding K, Shi PM, Chen F, et al. FOXA2 alleviates CCl4-induced liver fibrosis by protecting hepatocytes in mice. Sci Rep. 2017;7:15532. doi: 10.1038/s41598-017-15831-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang K, Brems JJ, Gamelli RL, Holterman AX. Foxa2 may modulate hepatic apoptosis through the cIAP1 pathway. Cell Signal. 2013;25:867–74. doi: 10.1016/j.cellsig.2012.12.012. [DOI] [PubMed] [Google Scholar]
- 63.Phua WWT, Tan WR, Yip YS, Hew ID, Wee JWK, Cheng HS. PPARbeta/delta Agonism Upregulates Forkhead Box A2 to Reduce Inflammation in C2C12 Myoblasts and in Skeletal Muscle. Int J Mol Sci. 2020;21:1747–62. doi: 10.3390/ijms21051747. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.