Abstract
Epidemiological studies have found an increased incidence of colorectal cancer (CRC) in people who undergo cholecystectomy compared to healthy individuals. After cholecystectomy, bile enters the duodenum directly, unregulated by the timing of meals. Disruption of the balance of bile acid metabolism and increased production of primary bile acids, which in turn affects the composition and abundance of intestinal microorganisms. The link among cholecystectomy, the gut microbiota, and the occurrence and development of CRC is becoming clearer. However, due to the complexity of the microbial community, the mechanistic connections are less well understood. In this review, we summarize the changes of gut microbiota after cholecystectomy and illuminate the potential mechanisms on CRC, such as inflammation and immune regulation, production of genotoxins, metabolism of dietary ingredients, activation of signaling pathways, and so on. By reviewing these, we aimed to unravel the interactions between the gut microbiota and its host and be better positioned to develop treatments for CRC after cholecystectomy.
Keywords: colorectal cancer, gut microbiota, bile acid, genotoxin, diet, epidemiology, cholecystectomy
Introduction
As one of the most common malignant tumors worldwide, colorectal cancer (CRC) ranks second and third in morbidity and mortality rates, respectively, and its incidence is gradually increasing in developing countries (1). It has been estimated that by 2030, the global disease burden of CRC will increase by 60%, and there will be more than 2.2 million new cases and 1.1 million deaths worldwide (2). Moreover, the morbidity and mortality rates of CRC are increasing yearly, particularly, in individuals under the age of 50 (3). Furthermore, there is continuous growth in the overall number of diagnosed cases, which contributes to the increasing disease burden of CRC (2). Therefore, research on the risk factors and pathogenesis of CRC is becoming increasingly essential.
CRC is associated with multiple factors, including genetic susceptibility and environmental factors, which play a greater role in its occurrence and development (4). The gut microbiota is among the various environmental factors recognized in cancer biology. There are more than 3×1013 bacterial cells in the human colorectum, which interact with host cells to regulate many physiological processes. Disruption of the gut microbiota affects the balance of physiological processes, contributing to the development and progression of many diseases, such as inflammatory bowel disease (IBD) and CRC (5–8).
Changes in lifestyle and eating habits are directly linked to an increase in gallbladder diseases (9). Although cholecystectomy is an acceptable standard treatment for gallbladder diseases (2016), there is evidence that it is likely to increase the incidence of CRC (10–13). However, little is known about the mechanisms responsible for this process. There are two major theories on the effects of cholecystectomy on CRC development (1): cholecystectomy may alter the concentration, composition, and excretion rhythm of bile acids, leading to an increase in the content of secondary bile acids, which can continuously stimulate intestinal cells. For example, deoxycholic acid and lithocholic acid directly induce DNA damage and activate signaling pathways, including epidermal growth factor receptor-, Wnt-β-catenin-, and protein kinase C pathways, thereby promoting the occurrence and development of CRC (14) (2). Cholecystectomy causes changes in the composition and abundance of the gut microbiota in both stool and tumor tissues. The main manifestations are a decline in the diversity of the microbiota, particularly, a decrease in beneficial bacteria and an increase in pathogenic bacteria (15–19), including carcinogenic bacteria, such as Fusobacterium nucleatum, enterotoxigenic Bacteroides fragilis (ETBF), Clostridium difficile, and Escherichia coli, which promote CRC development (20–23). A recent study showed that the gut microbiota of patients who underwent cholecystectomy was significantly different from that of healthy people, but similar to that of patients with CRC, which suggests that changes in the gut microbiota in patients undergoing cholecystectomy may activate CRC occurrence and progression (24).
In this article, we described the current knowledge on changes in the gut microbiota after cholecystectomy, the interaction between cholecystectomy and CRC, and potential mechanisms, aiming to clarify the role of gut microbiota alteration on the occurrence and development of CRC after a cholecystectomy.
Epidemiological Relationship Between Cholecystectomy and CRC
In 1978, Capron was the first to report that cholecystectomy could increase the incidence of CRC (25). Subsequently, scholars worldwide conducted a series of studies on the relationship between cholecystectomy and the incidence of CRC, and two studies published in 1981 involving patients in Finland and the United States provided evidence of a significant link between cholecystectomy and CRC (10, 11).
A retrospective study in 2005 analyzed more than 8 million people on a general medicine research database in the United Kingdom and found that cholecystectomy was associated with increased risk of colon cancer, but not rectal cancer (12). Some meta-analyses have also indicated a correlation between cholecystectomy and the increasing risk of CRC. Among these, one analysis included ten cohort studies which indicated a strong correlation between the proximal colon with a history of cholecystectomy and carcinogenesis (13, 26, 27). These results indicate that a history of cholecystectomy is closely associated with the occurrence and progression of CRC.
Effect of Cholecystectomy on Gut Microbiota
Trillions of microorganisms, such as bacteria, viruses, fungi, and other life forms, live inside every person. Various organs show distinct microbial inhabitants, but the inhabitants that have drawn the most attention are those in the colorectum (28). The gut microbiota is a key player in physiological activities, including metabolism of food residues, synthesis of micronutrients (such as vitamins), metabolism of primary bile acids, synthesis of secondary bile acids, regulation of immune responses, and metabolism and production of butyric acid and other substances which provide substances for epithelial cell renewal and mucosal integrity maintenance (29). Of note, the gut microbiota is related to the development of a wide range of digestive diseases, such as IBD, irritable bowel syndrome, and CRC (30–32).
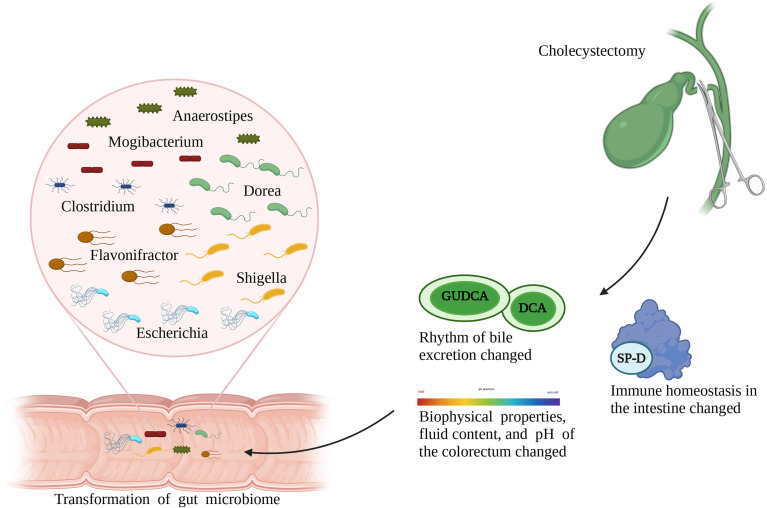
Cholecystectomy induces dramatic changes in intestinal microecology, including the composition and function of the gut microbiota. The changes in the gut microbiota after cholecystectomy are shown in Table 1 (24, 33–37). At the phylum level, the abundance of Fusobacteria increased, whereas that of Proteobacteria decreased. Other phyla, including Bacteroidetes, Firmicutes, and Actinobacteria, showed distinct variations in different studies. Interestingly, the changes in bacterial abundance of Firmicutes and Actinobacteria were similar in all studies, in contrast to the alteration in the bacterial abundance of Bacteroidetes. Different Bacteroides species affect the health of the host in different ways (38–40). For example, ETBF can induce colitis and promote the occurrence of intestinal tumors, while other species, such as Bacteroides vulgatus and Bacteroides fragilis, are associated with the protection of the intestinal barrier. At the genus level, existing research has not reached a consensus. Genera reported with increasing abundance mainly include Anaerostipes, Dorea, Clostridium, Mogibacterium, Flavonifractor, Shigella, and Escherichia, whilst those with reduced abundance include Paraprevotella, Prevotella, Barnesiella, Alistipes, Faecalibacterium, Haemophilus, and Desulfovibrio. Few studies have focused on the species level; Blautia obeum, Veillonella parvula, Bacteroides ovatus, Parabacteroides distasonis, and Fusobacterium varium were found to increase, and Eubacterium rectale, Roseburia faecis, and Bifidobacterium adolescentis were reported to decrease.
Table 1.
Comparison of gut microbiota between patients who have undergone cholecystectomy and healthy individuals.
| Country (Author & Year) | Sample size | Sequencing method | Changes of gut microbiota after cholecystectomy | |
|---|---|---|---|---|
| 1 | Israel (Keren et al., 2015) (33) | 20 | 16S rRNA | The diversity of microbiome is basically stable. Phylum Bacteroides↑ Family Bacteroides, Parabacteraceae↑ |
| 2 | China (Wang et al., 2018) (34) | 135 | 16S rRNA | The diversity of microbiome declined. Phylum Actinomycetes, Firmicutes↑ Bacteroides, Proteobacteria↓ Genus Bifidobacterium, Dallella, Anaerobic↑ Palapuella, Prevotella, Barnesella, Alternaria, Desulfovibrio↓ |
| 3 | Korea (Yoon et al., 2019) (35) | 108 | 16S rRNA | The diversity of microbiome declined. Phylum Firmicutes↑ Bacteroides↓ Species Broutella ovale, Veillonella parvula↑ |
| 4 | China (Ren et al., 2020) (24) | 104 | 16S rRNA | The diversity of microbiome increased. Phylum Bacteroides, Fusobacteria↑ Firmicutes, Actinomycetes↓ Genus Prevotella↑ Faecalibacterium↓ Species Bacteroides ovatus, Parabacteroides diundi, Fusobacterium proteus↑ Eubacterium rectale, Roseburia faecis, Bifidobacterium adolescentis↓ |
| 5 | Germany (Frost et al., 2021) (36) | 1968 | 16S rRNA | The diversity of microbiome declined. Genus Clostridium XIVa, Flavonoids, Clostridium difficile, Escherichia, Shigella↑ Faecalibacterium, Haemophilus↓ |
It was widely reported that Escherichia and ETBF had increased in abundance of CRC patients, promoting CRC development through damaged DNA, and produced toxins. While beneficial bacteria, including Alistipes and Faecalibacterium, which can produce active metabolites, such as butyrate and folic acid and inhibit the occurrence and development of CRC, were significantly reduced in patients with cholecystectomy history (24, 33–37). Due to differences in race, diet, and experimental conditions, changes in the gut microbiota after cholecystectomy are inconsistent. However, all the relevant studies confirm that alterations in the gut microbiota promote CRC occurrence and progression.
Transformation of the gut microbiota after cholecystectomy can be attributed to the following reasons ( Figure 1 ): first, bile excretion regulation weakens or disappears after cholecystectomy; as a result, the bile flows into the intestine continuously (41). This changed pattern is conducive to the growth of bacteria that metabolize bile acid or live through bile-dependent fat decomposition but has adverse effects on the growth of other bacteria, thereby reshaping the gut microbiota. For example, experiments have shown that deoxycholic acid inhibits the growth of Lactobacillus, Bifidobacterium, and other bile-sensitive bacteria (42). Second, cholecystectomy alters bowel movements by changing the biophysical properties, fluid content, and pH of the colorectum, thereby providing favorable or harmful growth conditions for certain bacteria. For example, persistent secretion of bile, which is alkaline, after cholecystectomy, increases the pH value in the intestines, thereby inhibiting the proliferation of acidic-adapted bacteria, including Lactobacillus and Bifidobacterium. Third, changes in immune homeostasis in the intestines after cholecystectomy should be considered. For example, surfactant protein D, an important substance secreted by the gallbladder, can be transported to the intestinal lumen with the entered bile and inhibits the growth of symbiotic bacteria through direct binding. Cholecystectomy unavoidably decreases the level of surfactant protein D in the human intestines, which leads to disorders of bacterial and host-bacterial interactions and affects the natural environment of the gut microbiota (43).
Figure 1.
Role of cholecystectomy in the alternation of gut microbiota.
Role of Gut Microbiota in CRC
CRC development and progression is a multi-factor interaction, in which the role of the gut microbiota is now attracting increasing attention. A study that analyzed the Health Care Claims Database from the United States confirmed the relationship between the recurrence of CRC and disorders of the gut microbiota (44). Widespread use of antibiotics, alterations in diet, obesity, stress, and other risk factors are attributed to disorders of the gut microbiota in young people, which may partly explain their increased risk of CRC (45). A gradual increase in some bacteria and a constant decrease in some bacteria in normal, para-adenoma, adenoma, pericarcinomatous, and cancerous tissues have been found, suggesting that bacterial distribution may act as an essential factor in CRC development (46). Yu et al. conducted metagenomic sequencing of the stool samples of patients with CRC and healthy individuals and found significant differences in the composition of the gut microbiota between the two groups (47). Fecal bacterial transplantation studies were conducted, and the incidence of CRC in mice inoculated with stool samples from patients with CRC increased. The gut microbiota in the stool of patients with CRC can activate the intestinal mucosal immunity of mice and induce inflammation, so as to promote the proliferation of epithelial cells and induce the development of CRC (48). A number of clinical and animal studies have clarified the relationship between the gut microbiota and CRC and identified specific bacteria as key factors that affect the occurrence and development of CRC (19, 46, 47, 49–58), as summarized in Table 2 , including relevant studies in the past decade, reflecting the differences in the gut microbiota between patients with CRC and healthy individuals. The diversity of the gut microbiota in patients with CRC was lower than in healthy individuals, with a decrease in beneficial bacteria and an increase in pathogenic bacteria. For example, Fusobacterium nucleatum, Campylobacter, ETBF, and Escherichia coli that express the polyketide synthase gene (pks+ Escherichia coli) were enriched in the intestines of patients with CRC, induced inflammation, damaged DNA, and produced toxins, thereby promoting CRC development. Beneficial bacteria, including Bifidobacterium, Lachnospira, Alistipes, and Faecalibacterium, which can produce active metabolites, such as butyrate and folic acid and inhibit the occurrence and development of CRC, were significantly reduced.
Table 2.
Comparison of gut microbiota between patients with colorectal cancer and healthy individuals.
| Country (Author & Year) | Sample size | Sample type | Sequencing method | Changes of gut microbiota after cholecystectomy | |
|---|---|---|---|---|---|
| 1 | China (Wang et al., 2012) (19) | 102 | Stool | 16S rRNA | The diversity of microbiome is basically stable. Phylum Firmicutes, Proteobacteria, Actinomycetes↑ Bacteroides, Fusobacteria↓ Genus Porphyromonas, Escherichia, Shigella, Enterococcus, Streptococcus, Peptostreptococcus↑ Bacteroides, Rossella, Alternaria, Eubacteria, Trichospirillum↓ |
| 2 | China (Wu et al., 2013) (50) | 39 | Stool | 16S rRNA | The diversity of microbiome is basically stable. Phylum Fusobacterium↑ Family Eubacteriaceae, Clostridiaceae, Staphylococcus, Enterococcus, Fusobacteria, Campylobacter, Porphyridaceae↑ Genus Bacteroides, Alternaria, Blautella, Dallella, Fusobacterium, Campylobacter, Escherichia, Shigella, Odorbacterium, Oscillatoria, Testa Lactobacillus, Rumenococcus↑ Rossella, Faecalibacterium↓ |
| 3 | America (Ahn et al., 2013) (49) | 151 | Stool | 16S rRNA | The diversity of microbiome declined. Phylum Bacteroides↑ Firmicutes↓ Genus Porphyromonas, Fusobacterium, Mirabilis↑ Faecococcus, Trichospirillum↓ |
| 4 | America (Zackular et al., 2014) (51) | 60 | Stool | 16S rRNA |
Family
Porphyridaceae, Enterobacteriaceae↑ Lacetospiraceae↓ Genus Porphyromonas, Fusobacterium↑ Bacteroides↓ |
| 5 | France (Zeller et al., 2014) (52) | 114 | Stool | Metagenomic sequencing | The diversity of microbiome is basically stable. Phylum Bacteroides, Fusobacteria, Proteobacteria↑ Actinomycetes, Firmicutes↓ Species Saccharolytic Porphyromonas, Oral Peptostreptococcus, Fusobacterium nucleatum↑ |
| 6 | Australia (Feng et al., 2015) (53) | 109 | Stool | Metagenomic sequencing | The diversity of microbiome declined. Genus Bacteroides, Alternaria, Bileophilus, Trichospira, Escherichia, Micromonas, Fusobacterium↑ Bifidobacterium, Streptococcus, Rumenococcus↓ |
| 7 | China (Nakatsu et al., 2015) (46) | 113 | Tissue | 16S rRNA |
Genus
Fusobacterium, Gemini, Peptostreptococcus, Micromonas, Streptococcus granulosus↑ Species Bacteroides fragilis↑ |
| 8 | America (Baxter et al., 2016) (54) | 292 | Stool | 16S rRNA |
Genus
Porphyria, Peptostreptococcus, Fusobacterium, Micromonas, Prevotella, Gemini↑ Species Saccharolytic Porphyromonas, Fusobacterium nucleatum, Micromonas parvum, Oral Peptostreptococcus↑ |
| 9 | Ireland (Flemer et al., 2017) (55) | 115 | Stool/ Tissue |
16S rRNA | Genus Bacteroides, Rossella, Rumenococcus, Oscillatoria, Porphyromonas, Peptostreptococcus, Micromonas, Fusobacterium↑ |
| 10 | China (Yu et al., 2017) (47) | 128 | Stool | Metagenomic sequencing | The diversity of microbiome declined. Phylum Fusobacteria, Basidiomycota↑ Species Micromonas parvum, Oral Peptostreptococcus, Fusobacterium nucleatum, Bacteroides fragilis, Solobacterium moorei↑ |
| 11 | Saudi Arabia (Alomair et al., 2018) (56) | 58 | Tissue | Metagenomic sequencing | The diversity of microbiome is basically stable. Genus Peptostreptococcus, Porphyromonas, Listeria, Atopobium, Burkholderia, Collins, Comamonas, Fusobacterium↑ |
| 12 | Japan (Yachida et al., 2019) (58) | 616 | Stool | Metagenomic sequencing | The diversity of microbiome increased. Phylum Firmicutes, Fusobacteria, Bacteroides↑ Genus Bacteroides, Peptostreptococcus↑ Prevotella, Bifidobacterium↓ Species Fusobacterium nucleatum, Micromonas parvum, Streptococcus oralis, Solobacterium moorei↑ Phascolarctobacterium succinatutens, Desulfovibrio longreachensis, Atopobium parvulum↓ |
| 13 | Italy (Thomas et al., 2019) (57) | 826 | Stool | Metagenomic sequencing | The diversity of microbiome increased. Genus Peptostreptococcus, Clostridium, Porphyromonas, Escherichia coli↑ Bifidobacterium, Trichospirillum, Alternative Mycobacterium↓ Species Fusobacterium nucleatum, Porphyromonas saccharolyticus, Micromonas parvum, Oral Peptostreptococcus, Escherichia coli↑ |
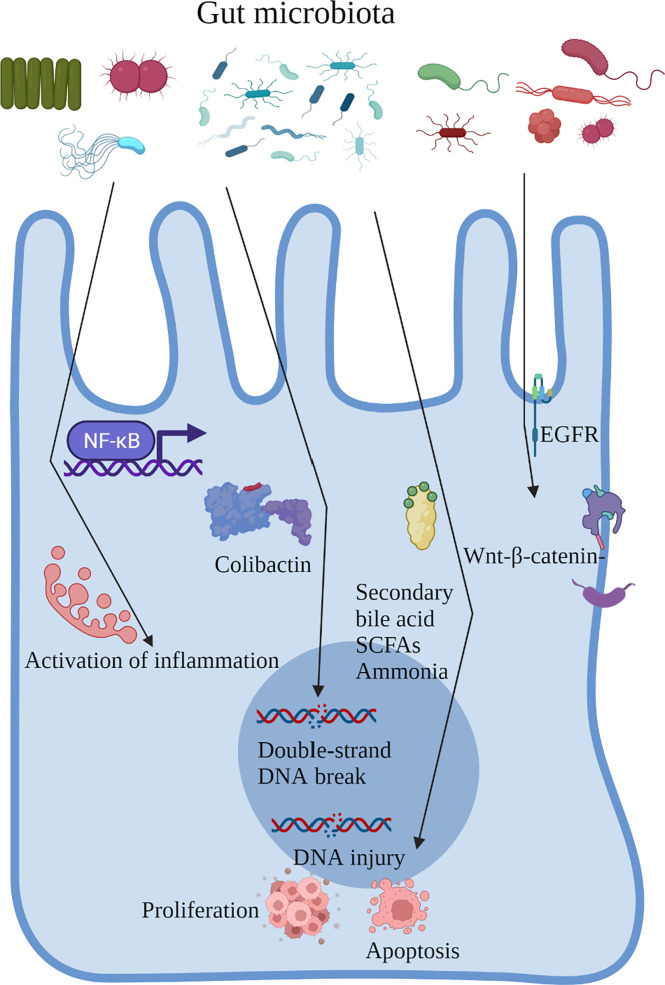
Patients who underwent cholecystectomy and those with CRC had similar gut microbial changes, an increased abundance of pathogenic bacteria, including Escherichia, Clostridium, and Dorea, and a decrease in beneficial bacteria, including Prevotella, Alistipes, and Faecalibacterium (24, 33–37). The gut microbiota can regulate the biological behavior of the host through direct cell interactions and in a metabolite-dependent manner. In addition, intestinal inflammation caused by the gut microbiota, secretion of flora-derived factors, such as genotoxins to induce DNA damage, production of metabolites, and direct activation of carcinogenic signaling pathways are major factors in CRC development. Next, we proposed to focus on the role of the gut microbiota in the process of CRC and the carcinogenic mechanism of gut microbiota alterations after cholecystectomy ( Figure 2 ).
Figure 2.
Role of the gut microbiota in the process of CRC and the carcinogenic mechanism of gut microbiota alterations after cholecystectomy.
Inflammation and Immune Regulation
Inflammation is an established risk factor of CRC carcinogenesis. Patients with IBD are more susceptible to CRC than the general population (59, 60). Inflammation plays a key role in the development of colitis-associated cancer, even in CRC unrelated to IBD, and the levels of pro-inflammatory cytokines are increased (61). The gut microbiota has the potential to form an inflammatory microenvironment and, vice versa, inflammation may affect gut microbiota composition. Colon polyposis in Apcmin/+ mice is accompanied by the accumulation of microorganisms in the polyps, triggering a local inflammatory response (Dennis etal. (62). Besides, defective expression of alarmin/IL-33 renders mice highly susceptible to probiotic microbiota-promoted IL-1α-dependent colitis and colitis-associated cancer (63). Gavage with stool samples from patients with CRC caused enhanced inflammation and intestinal adenoma development in a sterile mouse model (48), indicating that specific components of the gut microbiota promote the occurrence and development of CRC through the activation of inflammation. For example, enriched Fusobacterium nucleatum and Escherichia coli in the intestines of patients with CRC can activate the nuclear factor kappa B (NF-κB) signaling pathway and drive the infiltration of myeloid cells in the tumor, producing a pro-inflammatory environment that is conducive to the progression of colorectal tumors in Apcmin/+ mice (64, 65). ETBF can trigger an inflammatory cascade involving interleukin 17, signal transducer and activator of transcription 3, and NF-κB conduction in colonic epithelial cells via the production of a metalloproteinase toxin, promoting the local inflammatory environment in the intestines and inducing carcinogenesis (66). Cholecystectomy increases the abundance of Escherichia coli and decreases the abundance of Faecalibacterium, which can secrete small-molecule anti-inflammatory substances to inhibit intestinal inflammation (36, 50).
Intestinal homeostasis is achieved by the continuous interaction between the intestinal microbiome and the host immune system. Once this balance is disrupted, a variety of diseases, such as IBD, appear due to immune system dysfunction (67). In mice, BFT+ B. fragilis colonization was able to induce Th-17-mediated colitis and distal CRC in an IL17-mediated NF-κB upregulation-dependent manner in the APCmin/+ mouse model (40), as demonstrated by Chung et al. (68) who observed repressed BFT-induced tumor formation in APCmin IL17/IL17 mice. Furthermore, it is reported that an accumulation of regulatory T-expressing cells (Treg) cells in APCmin/+ mice after BFT colonization, which could be a trigger for IL17-mediated pro-oncogenic inflammatory responses. Certain probiotics, such as Bifidobacterium infantis (69) and Bifidobacterium breve (70), are able to activate intestinal dendritic cells (DCs) by interacting with Toll-like receptors (TLRs) and inducing retinoid metabolism, leading to the release of Foxp3+ Treg and type 1 regulatory T cells (Tr1) and IL-10 (71).
Production of Genotoxins
Another carcinogenic mechanism of the gut microbiota is the production of genotoxins, which may interact with intracellular signal cascades or result in mutations by binding to particular cell surface receptors and it could also damage DNA. Colibactin is a characteristic toxin produced by Escherichia coli, which induces double-strand DNA breaks in intestinal cells, causing cancer through its deoxyribonuclease activity (72–74). In addition, the enriched ETBF in the intestine of patients with CRC can produce a metalloproteinase toxin, which initiates cell proliferation, activates c-Myc expression, increases polyamine metabolism, and induces DNA damage, thereby promoting the occurrence and development of CRC (75). Furthermore, Salmonella typhi secretes virulence protein A, which enhances the development and proliferation of colon tumors (76). Interestingly, although many genotoxins can cause tumors, recent research has indicated their potential use in cancer therapy (77). For example, Clostridium perfringens enterotoxin is a pore-forming toxin with selective cytotoxicity, which rapidly and effectively kills tumor cells (78). Several genotoxins have been studied as therapeutic tools for cancer, including CRC (79); however, their role as a cancer promoter is beyond doubt.
Metabolism of Dietary Ingredients
Metabolism is an essential process in the interaction between the host and the microbiome. Genes encoded by the microorganisms can metabolize several dietary nutrients, including host-indigestible carbohydrates, such as dietary fiber, and host endogenous compounds, such as bile acids. Bacteria in the intestines produce a series of metabolites, including secondary bile acids, sulfides, ammonia, nitrosamines, and short-chain fatty acids (SCFAs), which are involved in the occurrence and development of CRC.
A substantial accumulation of primary bile acids in the intestines was discovered after cholecystectomy, and the enriched Bacteroides ovatus and Parabacteroides diundi due to cholecystectomy metabolized primary bile acids into secondary ones, which participated in cell proliferation, apoptosis, DNA injury, and other processes, promoting CRC carcinogenesis (24, 80, 81). Dietary fiber is metabolized and decomposed into SCFAs, including acetate, propionate, and butyrate, in the colon. Among them, butyrate (the most widely studied SCFA) regulates cell proliferation, apoptosis, and differentiation to inhibit CRC (82, 83). Cholecystectomy drastically reduced the abundance of intestinal bacteria responsible for metabolizing butyrate, including Faecalibacterium and Roseburia faecis, thereby decreasing the expression level of butyrate and promoting the occurrence of CRC (24). Broutella ovale and Veillonella parvula, which can activate azo reductase and produce toxic ammonia substances which promote the occurrence of CRC, were observed in patients who underwent cholecystectomy (35). Additionally, the gut microbiota can also destroy the mucus barrier function by producing sulfide, thereby intensifying the stimulation of intestinal cells (84). For instance, cholecystectomy significantly decreased the abundance of Desulfovibrio, producing more sulfides and leading to metabolic disorders, thereby stimulating intestinal epithelial cells and promoting carcinogenesis (34).
Activation of Signaling Pathways
Multiple signaling pathways, such as the epithelial growth factor receptor (EGFR), Wnt/β-catenin, NF-κB, and transforming growth factor-beta pathways, are involved in CRC development. Notably, the gut microbiota activates host carcinogenic signaling pathways.
The EGFR signaling pathway is closely related to the proliferation, apoptosis, and survival of colonic epithelial cells. Activation of the EGFR signaling pathway by secondary bile acids is achieved mainly by disturbing the structure of the cell membrane (reduced membrane fluidity, altered membrane cholesterol distribution), binding to natural ligands (e.g., epidermal growth factor), or inducing calcium signaling-mediated non-dependent activation of ligands (85). Activation of EGFR activates downstream MAPK/RAS/RAF/MEX/extracellular signal-regulated kinase/proto-oncogene activator protein-1, which in turn mediates cell proliferation and activates RAS/RAF1/extracellular signal-regulated kinase signaling pathway leading to upregulation of mucin 2, also activates the phosphatidylinositol 3 kinase/Akt signaling pathway, which regulates downstream target molecules such as Caspase-8, leading to apoptosis (86, 87). When it comes to practical clinical applications, using biomarkers to target anti-EGFR treatments for metastatic CRC is well established, while the anti-EGFR antibody cetuximab is only effective against a subgroup of CRC (88, 89).
Wnt/β-linked protein signaling plays a key role not only in maintaining intestinal homeostasis but also in regulating the proliferation of CRC cells. The Wnt/β-linked protein classical signaling pathway regulates the expression of Wnt/β-linked proteins through the binding of Wnt ligands to Frizzleds receptors. The Wnt/β-linked protein can transfer to the nucleus and interact with T-cell factor and lymphatic enhancer transcription factors to modulate the transcription of downstream gene targets (survivin, Cyclin D1, and c-Myc) and affect the cell cycle pathway (90). A marked accumulation of Fusobacterium nucleatum, which expresses FadA adhesin on its surface, was found in patients with CRC. FadA adhesin stimulates CRC cell growth by increasing the expression of inflammatory genes, oncogenes, and transcription factors by binding to E-cadherin, activating the Wnt/β-catenin signaling pathway, and promoting the transcription of oncogenes (91). Fusobacterium nucleatum can also directly activate toll-like receptor signaling to promote tumor development (92).
NF-κB is a key regulator associated with inflammation and cancer on multiple levels (93). The NF-κB signaling pathway regulates many genes involved in different cellular processes, such as cell differentiation, proliferation, genomic stability, and immune responses (94), and its activation is involved in the occurrence and development of CRC. Escherichia coli, Fusobacterium nucleatum, and ETBF, which are enriched in patients with CRC, are involved in the modulation of this pathway (95). Mechanistically, Escherichia coli activates NF-κB through increased phosphorylation of transcription factor 65 and inhibitor of NF-κB kinase alpha, inactivation of inhibitor of NF-κB alpha, and induction of the Wnt/β-catenin pathway by upregulation of β-catenin and its downstream genes (96). The hyperactivation of NF-κB was also found in CRC tissues with abundant Fusobacterium nucleatum. Furthermore, ETBF activates the NF-κB pathway by stimulating intracellular interleukin 17 secretion in Apc min/+ mice (68).
However, most of the existing research on cholecystectomy is focused on clinical studies, and there is a lack of in-depth mechanistic investigation after cholecystectomy. More studies are needed in the future to elucidate the changes in signaling pathways after cholecystectomy.
Conclusion and Perspective
Epidemiological studies on CRC and cholecystectomy have proved the correlation between these two parameters and indicated that changes in the gut microbiota may be a vital intermediate link. In this review, we summarized the changes in the gut microbiota after cholecystectomy. With the variability of sequencing technologies and the complexity of bacterial populations, the conclusions were not unanimous among the studies. Despite this, the differences in the gut microbiota between patients who underwent cholecystectomy and healthy individuals have been proven, and alterations of the gut microbiota affect the development of CRC. Based on previous studies, alterations in the gut microbiota after cholecystectomy may lead to intestinal inflammation, increased metabolism of harmful substances (such as secondary bile acids), and reduction of secretion of beneficial substances (such as butyrate), resulting in the progression of CRC.
There are some potential problems with the current research on the relationship between altered gut microbiota and CRC after cholecystectomy, the results of studies on gut microbiota may vary by race, and different sequencing methods could also affect the results. In addition, alterations of gut microbiota in stool samples cannot fully reflect the tumor microenvironment. And few studies have focused on the role of beneficial bacteria in CRC. In the future, it is necessary to pay attention to the gut microbiota in cancerous and pericarcinomatous tissues and detect the changes in the local microbiome on the occurrence and development of CRC. And a decrease in abundance of beneficial bacteria should also be watched, which may be a potential therapeutic target in CRC. With the rapid development of high-throughput sequencing technology, in-depth information can be provided to understand the links between CRC, cholecystectomy, and the gut microbiota. Notably, different bacterial species of the same genus had mixed efficacy. Therefore, further studies should focus on the changes and function of the gut microbiota in patients who have undergone cholecystectomy at the species level. Hopefully, this approach will elucidate how CRC, cholecystectomy, and gut microbiota interact, allowing therapies to be targeted to individual microbiological, cancer, and lifestyle factors.
Studies on gut microbiota and CRC aim to clarify the mechanisms employed by gut microbiota in the development of CRC and further apply them to the screening, diagnosis, treatment, and prevention of CRC. For instance, Yu et al. discovered a new fecal bacterial marker (‘m3’ from a Lachnoclostridium) that can be used for the diagnosis of colorectal adenoma and CRC. This is superior to other stool-based tests such as fecal microbiota transplantation (FMT) and may be used for the early screening of CRC in the future (97). In addition, there are more clinical studies on techniques such as oral probiotics and FMT. Recent studies have found that the probiotic bacterium Lactobacillus reuteri and its produced antimicrobial compound, reuterin, can inhibit the development of CRC by depleting glutathione and inducing oxidative stress in CRC cells, resulting in protein oxidation and impaired ribosome activity. When CRC mice were orally administered Lactobacillus reuteri, remission, tumor shrinkage, and prolonged survival were observed in the mice (98). Recently, experimental studies on the efficacy of FMT have focused on animal models. A recent study reported that FMT from wild to laboratory mice improved host adaptation and resistance to dextran sodium sulfate/azoxymethane-induced colorectal tumorigenesis, and thus a normal gut microbiome plays a protective role in the development of CRC (99). Furthermore, the effectiveness of immunotherapy seems to be strongly influenced by the composition of the gut microbiota. Oral administration of probiotics, such as Bifidobacterium (100) and Akkermansia muciniphila (101), or FMT (102) from treatment-responsive patients greatly enhanced PD1-based immunotherapy and eliminated tumor growth through enhancing dendritic cell and T-cell responses. At present, there are several ongoing international clinical trials to validate the effect of gut microbiota on CRC chemotherapy (NCT04021589, NCT04131803, NCT01579591).
Currently, cholecystectomy is still the preferred treatment option for gallbladder stones, gallbladder polyps, and cholecystitis. Cholecystectomy is a very routine procedure and more and more patients are undergoing cholecystectomy. However, the possible induction of colorectal after cholecystectomy is getting attention, and its specific mechanism has not been elucidated yet. To clarify the specific mechanism of CRC induced after cholecystectomy can interrupt the development of CRC in a targeted way. The change of gut microbiota after cholecystectomy is an important cause of CRC, and further research on specific species of bacteria and their mechanisms will provide important methods to prevent CRC in the future. As research progresses, dietary intervention with probiotics or prebiotics, or changes in diet could potentially be an effective way to prevent CRC in patients with cholecystectomy history in the future.
Author Contributions
All authors listed have contributed to the article and approved its publication. YM, RQ, and YZ: designing, writing, and figure plotting. CJ, ZZ, and WF: designing, funding acquisition, and review and editing. All authors contributed to the article and approved the submitted version.
Funding
This research was supported in part by the National Key Research and Development Program of China (2018YFA0800700 and 2018YFC1003200), the National Natural Science Foundation of the P. R. of China (No. 91857115, 31925021, 81921001, 81972702 and 91959110), National multidisciplinary cooperative diagnosis and treatment capacity building project for major diseases: comprehensive diagnosis and treatment of gastrointestinal tumors, National Health and Family Planning Commission Foundation of China (Grant No. 2020YB57), and “Clinical Medicine + X” Foundation of Peking University (Grant No. PKU2021LCXQ001).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
CRC, colorectal cancer; ETBF, enterotoxigenic Bacteroides fragilis; IBD, inflammatory bowel disease; NF-κB, nuclear factor kappa B; SCFAs, short-chain fatty acids.
References
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA: Cancer J Clin (2018) 68(6):394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2. Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Patterns and Trends in Colorectal Cancer Incidence and Mortality. Gut (2017) 66(4):683–91. doi: 10.1136/gutjnl-2015-310912 [DOI] [PubMed] [Google Scholar]
- 3. Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. Cancer Statistics in Chin. CA: Cancer J Clin (2016) 66(2):115–32. doi: 10.3322/caac.21338 [DOI] [PubMed] [Google Scholar]
- 4. Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Koskenvuo M, et al. Environmental and Heritable Factors in the Causation of Cancer–Analyses of Cohorts of Twins From Sweden, Denmark, and Finland. New Engl J Med (2000) 343(2):78–85. doi: 10.1056/NEJM200007133430201 [DOI] [PubMed] [Google Scholar]
- 5. Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An Obesity-Associated Gut Microbiome With Increased Capacity for Energy Harvest. Nature (2006) 444(7122):1027–31. doi: 10.1038/nature05414 [DOI] [PubMed] [Google Scholar]
- 6. Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A Human Gut Microbial Gene Catalogue Established by Metagenomic Sequencing. Nature (2010) 464(7285):59–65. doi: 10.1038/nature08821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chung H, Pamp SJ, Hill JA, Surana NK, Edelman SM, Troy EB, et al. Gut Immune Maturation Depends on Colonization With a Host-Specific Microbiota. Cell (2012) 149(7):1578–93. doi: 10.1016/j.cell.2012.04.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wong SH, Yu J. Gut Microbiota in Colorectal Cancer: Mechanisms of Action and Clinical Applications. Nat Rev Gastroenterol Hepatol (2019) 16(11):690–704. doi: 10.1038/s41575-019-0209-8 [DOI] [PubMed] [Google Scholar]
- 9. Lammert F, Gurusamy K, Ko CW, Miquel J-F, Méndez-Sánchez N, Portincasa P, et al. Gallstones. Nat Rev Dis Primers (2016) 2:16024. doi: 10.1038/nrdp.2016.24 [DOI] [PubMed] [Google Scholar]
- 10. Linos D, Beard CM, O'Fallon WM, Dockerty MB, Beart RW, Kurland LT. Cholecystectomy and Carcinoma of the Colon. Lancet (London England) (1981) 2(8243):379–81. doi: 10.1016/S0140-6736(81)90829-1 [DOI] [PubMed] [Google Scholar]
- 11. Turunen MJ, Kivilaakso EO. Increased Risk of Colorectal Cancer After Cholecystectomy. Ann Surg (1981) 194(5):639–41. doi: 10.1097/00000658-198111000-00014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shao T, Yang Y-X. Cholecystectomy and the Risk of Colorectal Cancer. Am J Gastroenterol (2005) 100(8):1813–20. doi: 10.1111/j.1572-0241.2005.41610.x [DOI] [PubMed] [Google Scholar]
- 13. Zhang Y, Liu H, Li L, Ai M, Gong Z, He Y, et al. Cholecystectomy can Increase the Risk of Colorectal Cancer: A Meta-Analysis of 10 Cohort Studies. PloS One (2017) 12(8):e0181852. doi: 10.1371/journal.pone.0181852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Farhana L, Nangia-Makker P, Arbit E, Shango K, Sarkar S, Mahmud H, et al. Bile Acid: A Potential Inducer of Colon Cancer Stem Cells. Stem Cell Res Ther (2016) 7(1):181. doi: 10.1186/s13287-016-0439-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Scanlan PD, Shanahan F, Clune Y, Collins JK, O'Sullivan GC, O'Riordan M, et al. Culture-Independent Analysis of the Gut Microbiota in Colorectal Cancer and Polyposis. Environ Microbiol (2008) 10(3):789–98. doi: 10.1111/j.1462-2920.2007.01503.x [DOI] [PubMed] [Google Scholar]
- 16. O'Keefe SJD, Ou J, Aufreiter S, O'Connor D, Sharma S, Sepulveda J, et al. Products of the Colonic Microbiota Mediate the Effects of Diet on Colon Cancer Risk. J Nutr (2009) 139(11):2044–8. doi: 10.3945/jn.109.104380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Azcárate-Peril MA, Sikes M, Bruno-Bárcena JM. The Intestinal Microbiota, Gastrointestinal Environment and Colorectal Cancer: A Putative Role for Probiotics in Prevention of Colorectal Cancer? Am J Physiol Gastrointestinal liver Physiol (2011) 301(3):G401–24. doi: 10.1152/ajpgi.00110.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen W, Liu F, Ling Z, Tong X, Xiang C. Human Intestinal Lumen and Mucosa-Associated Microbiota in Patients With Colorectal Cancer. PloS One (2012) 7(6):e39743. doi: 10.1371/journal.pone.0039743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang T, Cai G, Qiu Y, Fei N, Zhang M, Pang X, et al. Structural Segregation of Gut Microbiota Between Colorectal Cancer Patients and Healthy Volunteers. ISME J (2012) 6(2):320–9. doi: 10.1038/ismej.2011.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Toprak NU, Yagci A, Gulluoglu BM, Akin ML, Demirkalem P, Celenk T, et al. A Possible Role of Bacteroides Fragilis Enterotoxin in the Aetiology of Colorectal Cancer. Clin Microbiol infect Off Publ Eur Soc Clin Microbiol Infect Dis (2006) 12(8):782–6. doi: 10.1111/j.1469-0691.2006.01494.x [DOI] [PubMed] [Google Scholar]
- 21. Bonnet M, Buc E, Sauvanet P, Darcha C, Dubois D, Pereira B, et al. Colonization of the Human Gut by E. Coli and Colorectal Cancer Risk. Clin Cancer Res an Off J Am Assoc Cancer Res (2014) 20(4):859–67. doi: 10.1158/1078-0432.CCR-13-1343 [DOI] [PubMed] [Google Scholar]
- 22. Abed J, Emgård JEM, Zamir G, Faroja M, Almogy G, Grenov A, et al. Fap2 Mediates Fusobacterium Nucleatum Colorectal Adenocarcinoma Enrichment by Binding to Tumor-Expressed Gal-GalNAc. Cell Host Microbe (2016) 20(2):215–25. doi: 10.1016/j.chom.2016.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jahani-Sherafat S, Azimirad M, Alebouyeh M, Ahmadi Amoli H, Hosseini P, Ghasemian-Safaei H, et al. The Rate and Importance of in Colorectal Cancer Patients. Gastroenterol Hepatol bed to bench (2019) 12(4):358–63. [PMC free article] [PubMed] [Google Scholar]
- 24. Ren X, Xu J, Zhang Y, Chen G, Zhang Y, Huang Q, et al. Bacterial Alterations in Post-Cholecystectomy Patients Are Associated With Colorectal Cancer. Front Oncol (2020) 10:1418. doi: 10.3389/fonc.2020.01418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Capron JP, Delamarre J, Canarelli JP, Brousse N, Dupas JL. [Does Cholecystectomy Predispose to Colo-Rectal Cancer?]. Gastroenterologie clin biol (1978) 2(4):383–9. [PubMed] [Google Scholar]
- 26. Giovannucci E, Colditz GA, Stampfer MJ. A Meta-Analysis of Cholecystectomy and Risk of Colorectal Cancer. Gastroenterology (1993) 105(1):130–41. doi: 10.1016/0016-5085(93)90018-8 [DOI] [PubMed] [Google Scholar]
- 27. Schernhammer ES, Leitzmann MF, Michaud DS, Speizer FE, Giovannucci E, Colditz GA, et al. Cholecystectomy and the Risk for Developing Colorectal Cancer and Distal Colorectal Adenomas. Br J Cancer (2003) 88(1):79–83. doi: 10.1038/sj.bjc.6600661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sender R, Fuchs S, Milo R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PloS Biol (2016) 14(8):e1002533. doi: 10.1371/journal.pbio.1002533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Almeida A, Mitchell AL, Boland M, Forster SC, Gloor GB, Tarkowska A, et al. A New Genomic Blueprint of the Human Gut Microbiota. Nature (2019) 568(7753):499–504. doi: 10.1038/s41586-019-0965-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shanahan F, van Sinderen D, O'Toole PW, Stanton C. Feeding the Microbiota: Transducer of Nutrient Signals for the Host. Gut (2017) 66(9):1709–17. doi: 10.1136/gutjnl-2017-313872 [DOI] [PubMed] [Google Scholar]
- 31. Vich Vila A, Imhann F, Collij V, Jankipersadsing SA, Gurry T, Mujagic Z, et al. Gut Microbiota Composition and Functional Changes in Inflammatory Bowel Disease and Irritable Bowel Syndrome. Sci Trans Med (2018) 10(472):eaap8914. doi: 10.1126/scitranslmed.aap8914 [DOI] [PubMed] [Google Scholar]
- 32. Saus E, Iraola-Guzmán S, Willis JR, Brunet-Vega A, Gabaldón T. Microbiome and Colorectal Cancer: Roles in Carcinogenesis and Clinical Potential. Mol Aspects Med (2019) 69:93–106. doi: 10.1016/j.mam.2019.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Keren N, Konikoff FM, Paitan Y, Gabay G, Reshef L, Naftali T, et al. Interactions Between the Intestinal Microbiota and Bile Acids in Gallstones Patients. Environ Microbiol Rep (2015) 7(6):874–80. doi: 10.1111/1758-2229.12319 [DOI] [PubMed] [Google Scholar]
- 34. Wang W, Wang J, Li J, Yan P, Jin Y, Zhang R, et al. Cholecystectomy Damages Aging-Associated Intestinal Microbiota Construction. Front Microbiol (2018) 9:1402. doi: 10.3389/fmicb.2018.01402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yoon WJ, Kim H-N, Park E, Ryu S, Chang Y, Shin H, et al. The Impact of Cholecystectomy on the Gut Microbiota: A Case-Control Study. J Clin Med (2019) 8(1):79. doi: 10.3390/jcm8010079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Frost F, Kacprowski T, Rühlemann M, Weiss S, Bang C, Franke A, et al. Carrying Asymptomatic Gallstones is Not Associated With Changes in Intestinal Microbiota Composition and Diversity But Cholecystectomy With Significant Dysbiosis. Sci Rep (2021) 11(1):6677. doi: 10.1038/s41598-021-86247-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hepner GW, Hofmann AF, Malagelada JR, Szczepanik PA, Klein PD. Increased Bacterial Degradation of Bile Acids in Cholecystectomized Patients. Gastroenterology (1974) 66(4):556–64. doi: 10.1016/S0016-5085(74)80044-2 [DOI] [PubMed] [Google Scholar]
- 38. Waidmann M, Bechtold O, Frick J-S, Lehr H-A, Schubert S, Dobrindt U, et al. Bacteroides Vulgatus Protects Against Escherichia Coli-Induced Colitis in Gnotobiotic Interleukin-2-Deficient Mice. Gastroenterology (2003) 125(1):162–77. doi: 10.1016/S0016-5085(03)00672-3 [DOI] [PubMed] [Google Scholar]
- 39. Mazmanian SK, Round JL, Kasper DL. A Microbial Symbiosis Factor Prevents Intestinal Inflammatory Disease. Nature (2008) 453(7195):620–5. doi: 10.1038/nature07008 [DOI] [PubMed] [Google Scholar]
- 40. Wu S, Rhee K-J, Albesiano E, Rabizadeh S, Wu X, Yen H-R, et al. A Human Colonic Commensal Promotes Colon Tumorigenesis via Activation of T Helper Type 17 T Cell Responses. Nat Med (2009) 15(9):1016–22. doi: 10.1038/nm.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sauter GH, Moussavian AC, Meyer G, Steitz HO, Parhofer KG, Jüngst D. Bowel Habits and Bile Acid Malabsorption in the Months After Cholecystectomy. Am J Gastroenterol (2002) 97(7):1732–5. doi: 10.1111/j.1572-0241.2002.05779.x [DOI] [PubMed] [Google Scholar]
- 42. Floch MH, Binder HJ, Filburn B, Gershengoren W. The Effect of Bile Acids on Intestinal Microflora. Am J Clin Nutr (1972) 25(12):1418–26. doi: 10.1093/ajcn/25.12.1418 [DOI] [PubMed] [Google Scholar]
- 43. Sarashina-Kida H, Negishi H, Nishio J, Suda W, Nakajima Y, Yasui-Kato M, et al. Gallbladder-Derived Surfactant Protein D Regulates Gut Commensal Bacteria for Maintaining Intestinal Homeostasis. Proc Natl Acad Sci USA (2017) 114(38):10178–83. doi: 10.1073/pnas.1712837114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chen H, Zheng X, Zong X, Li Z, Li N, Hur J, et al. Metabolic Syndrome, Metabolic Comorbid Conditions and Risk of Early-Onset Colorectal Cancer. Gut (2021) 70(6):1147–54. doi: 10.1136/gutjnl-2020-321661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hofseth LJ, Hebert JR, Chanda A, Chen H, Love BL, Pena MM, et al. Early-Onset Colorectal Cancer: Initial Clues and Current Views. Nat Rev Gastroenterol Hepatol (2020) 17(6):352–64. doi: 10.1038/s41575-019-0253-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nakatsu G, Li X, Zhou H, Sheng J, Wong SH, Wu WK, et al. Gut Mucosal Microbiome Across Stages of Colorectal Carcinogenesis. Nat Commun (2015) 6(8727):8727. doi: 10.1038/ncomms9727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yu J, Feng Q, Wong SH, Zhang D, Liang QY, Qin Y, et al. Metagenomic Analysis of Faecal Microbiome as a Tool Towards Targeted non-Invasive Biomarkers for Colorectal Cancer. Gut (2017) 66(1):70–8. doi: 10.1136/gutjnl-2015-309800 [DOI] [PubMed] [Google Scholar]
- 48. Wong SH, Zhao L, Zhang X, Nakatsu G, Han J, Xu W, et al. Gavage of Fecal Samples From Patients With Colorectal Cancer Promotes Intestinal Carcinogenesis in Germ-Free and Conventional Mice. Gastroenterology (2017) 153(6):1621–33. doi: 10.1053/j.gastro.2017.08.022 [DOI] [PubMed] [Google Scholar]
- 49. Ahn J, Sinha R, Pei Z, Dominianni C, Wu J, Shi J, et al. Human Gut Microbiome and Risk for Colorectal Cancer. J Natl Cancer Inst (2013) 105(24):1907–11. doi: 10.1093/jnci/djt300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wu N, Yang X, Zhang R, Li J, Xiao X, Hu Y, et al. Dysbiosis Signature of Fecal Microbiota in Colorectal Cancer Patients. Microb Ecol (2013) 66(2):462–70. doi: 10.1007/s00248-013-0245-9 [DOI] [PubMed] [Google Scholar]
- 51. Zackular JP, Rogers MAM, Ruffin MT, Schloss PD. The Human Gut Microbiome as a Screening Tool for Colorectal Cancer. Cancer Prev Res (Philadelphia Pa.) (2014) 7(11):1112–21. doi: 10.1158/1940-6207.CAPR-14-0129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zeller G, Tap J, Voigt AY, Sunagawa S, Kultima JR, Costea PI, et al. Potential of Fecal Microbiota for Early-Stage Detection of Colorectal Cancer. Mol Syst Biol (2014) 10:766. doi: 10.15252/msb.20145645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Feng Q, Liang S, Jia H, Stadlmayr A, Tang L, Lan Z, et al. Gut Microbiome Development Along the Colorectal Adenoma-Carcinoma Sequence. Nat Commun (2015) 6:6528. doi: 10.1038/ncomms7528 [DOI] [PubMed] [Google Scholar]
- 54. Baxter NT, Ruffin MT, Rogers MAM, Schloss PD. Microbiota-Based Model Improves the Sensitivity of Fecal Immunochemical Test for Detecting Colonic Lesions. Genome Med (2016) 8(1):37. doi: 10.1186/s13073-016-0290-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Flemer B, Lynch DB, Brown JMR, Jeffery IB, Ryan FJ, Claesson MJ, et al. Tumour-Associated and non-Tumour-Associated Microbiota in Colorectal Cancer. Gut (2017) 66(4):633–43. doi: 10.1136/gutjnl-2015-309595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Alomair AO, Masoodi I, Alyamani EJ, Allehibi AA, Qutub AN, Alsayari KN, et al. Colonic Mucosal Microbiota in Colorectal Cancer: A Single-Center Metagenomic Study in Saudi Arabia. Gastroenterol Res Pract (2018) 2018:5284754. doi: 10.1155/2018/5284754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Thomas AM, Manghi P, Asnicar F, Pasolli E, Armanini F, Zolfo M, et al. Metagenomic Analysis of Colorectal Cancer Datasets Identifies Cross-Cohort Microbial Diagnostic Signatures and a Link With Choline Degradation. Nat Med (2019) 25(4):667–78. doi: 10.1038/s41591-019-0405-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yachida S, Mizutani S, Shiroma H, Shiba S, Nakajima T, Sakamoto T, et al. Metagenomic and Metabolomic Analyses Reveal Distinct Stage-Specific Phenotypes of the Gut Microbiota in Colorectal Cancer. Nat Med (2019) 25(6):968–76. doi: 10.1038/s41591-019-0458-7 [DOI] [PubMed] [Google Scholar]
- 59. Beaugerie L, Itzkowitz SH. Cancers Complicating Inflammatory Bowel Disease. N Engl J Med (2015) 372(15):1441–52. doi: 10.1056/NEJMra1403718 [DOI] [PubMed] [Google Scholar]
- 60. Lasry A, Zinger A, Ben-Neriah Y. Inflammatory Networks Underlying Colorectal Cancer. Nat Immunol (2016) 17(3):230–40. doi: 10.1038/ni.3384 [DOI] [PubMed] [Google Scholar]
- 61. Terzić J, Grivennikov S, Karin E, Karin M. Inflammation and colon cancer. Gastroenterology (2010) 138(6):2101–14. doi: 10.1053/j.gastro.2010.01.058 [DOI] [PubMed] [Google Scholar]
- 62. Dennis KL, Wang Y, Blatner NR, Wang S, Saadalla A, Trudeau E, et al. Adenomatous Polyps are Driven by Microbe-Instigated Focal Inflammation and are Controlled by IL-10-Producing T Cells. Cancer Res (2013) 73(19):5905–13. doi: 10.1158/0008-5472.CAN-13-1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Malik A, Sharma D, Zhu Q, Karki R, Guy CS, Vogel P, et al. IL-33 Regulates the IgA-Microbiota Axis to Restrain IL-1α-Dependent Colitis and Tumorigenesis. J Clin Invest (2016) 126(12):4469–81. doi: 10.1172/JCI88625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Savkovic SD, Koutsouris A, Hecht G. Activation of NF-kappaB in Intestinal Epithelial Cells by Enteropathogenic Escherichia Coli. Am J Physiol (1997) 273(4):C1160–7. doi: 10.1152/ajpcell.1997.273.4.C1160 [DOI] [PubMed] [Google Scholar]
- 65. Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, et al. Fusobacterium Nucleatum Potentiates Intestinal Tumorigenesis and Modulates the Tumor-Immune Microenvironment. Cell Host Microbe (2013) 14(2):207–15. doi: 10.1016/j.chom.2013.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sears CL, Geis AL, Housseau F. Bacteroides Fragilis Subverts Mucosal Biology: From Symbiont to Colon Carcinogenesis. J Clin Invest (2014) 124(10):4166–72. doi: 10.1172/JCI72334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Gophna U, Sommerfeld K, Gophna S, Doolittle WF, Veldhuyzen van Zanten SJO. Differences Between Tissue-Associated Intestinal Microfloras of Patients With Crohn's Disease and Ulcerative Colitis. J Clin Microbiol (2006) 44(11):4136–41. doi: 10.1128/JCM.01004-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Chung L, Thiele Orberg E, Geis AL, Chan JL, Fu K, DeStefano Shields CE, et al. Bacteroides Fragilis Toxin Coordinates a Pro-Carcinogenic Inflammatory Cascade via Targeting of Colonic Epithelial Cells. Cell Host Microbe (2018) 23(2):203–14. doi: 10.1016/j.chom.2018.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Konieczna P, Groeger D, Ziegler M, Frei R, Ferstl R, Shanahan F, et al. Bifidobacterium Infantis 35624 Administration Induces Foxp3 T Regulatory Cells in Human Peripheral Blood: Potential Role for Myeloid and Plasmacytoid Dendritic Cells. Gut (2012) 61(3):354–66. doi: 10.1136/gutjnl-2011-300936 [DOI] [PubMed] [Google Scholar]
- 70. Jeon SG, Kayama H, Ueda Y, Takahashi T, Asahara T, Tsuji H, et al. Probiotic Bifidobacterium Breve Induces IL-10-Producing Tr1 Cells in the Colon. PloS Pathog (2012) 8(5):e1002714. doi: 10.1371/journal.ppat.1002714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Geis AL, Fan H, Wu X, Wu S, Huso DL, Wolfe JL, et al. Regulatory T-Cell Response to Enterotoxigenic Bacteroides Fragilis Colonization Triggers IL17-Dependent Colon Carcinogenesis. Cancer Discov (2015) 5(10):1098–109. doi: 10.1158/2159-8290.CD-15-0447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Nougayrède J-P, Homburg S, Taieb F, Boury M, Brzuszkiewicz E, Gottschalk G, et al. Escherichia Coli Induces DNA Double-Strand Breaks in Eukaryotic Cells. Sci (New York NY) (2006) 313(5788):848–51. doi: 10.1126/science.1127059 [DOI] [PubMed] [Google Scholar]
- 73. Cuevas-Ramos G, Petit CR, Marcq I, Boury M, Oswald E, Nougayrède J-P. Escherichia Coli Induces DNA Damage In Vivo and Triggers Genomic Instability in Mammalian Cells. Proc Natl Acad Sci USA (2010) 107(25):11537–42. doi: 10.1073/pnas.1001261107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. He Z, Gharaibeh RZ, Newsome RC, Pope JL, Dougherty MW, Tomkovich S, et al. Promotes Colorectal Tumorigenesis Through the Action of Cytolethal Distending Toxin. Gut (2019) 68(2):289–300. doi: 10.1136/gutjnl-2018-317200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Valguarnera E, Wardenburg JB. Good Gone Bad: One Toxin Away From Disease for Bacteroides Fragilis. J Mol Biol (2020) 432(4):765–85. doi: 10.1016/j.jmb.2019.12.003 [DOI] [PubMed] [Google Scholar]
- 76. Lu R, Wu S, Zhang YG, Xia Y, Liu X, Zheng Y, et al. Enteric Bacterial Protein AvrA Promotes Colonic Tumorigenesis and Activates Colonic Beta-Catenin Signaling Pathway. Oncogenesis (2014) 3:e105. doi: 10.1038/oncsis.2014.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Zahaf N-I, Schmidt G. Bacterial Toxins for Cancer Therapy. Toxins (2017) 9(8):236. doi: 10.3390/toxins9080236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Pahle J, Menzel L, Niesler N, Kobelt D, Aumann J, Rivera M, et al. Rapid Eradication of Colon Carcinoma by Clostridium Perfringens Enterotoxin Suicidal Gene Therapy. BMC Cancer (2017) 17(1):129. doi: 10.1186/s12885-017-3123-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Karpiński TM, Adamczak A. Anticancer Activity of Bacterial Proteins and Peptides. Pharmaceutics (2018) 10(2):54. doi: 10.3390/pharmaceutics10020054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Bernstein C, Holubec H, Bhattacharyya AK, Nguyen H, Payne CM, Zaitlin B, et al. Carcinogenicity of Deoxycholate, a Secondary Bile Acid. Arch Toxicol (2011) 85(8):863–71. doi: 10.1007/s00204-011-0648-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Gérard P. Metabolism of Cholesterol and Bile Acids by the Gut Microbiota. Pathog (Basel Switzerland) (2013) 3(1):14–24. doi: 10.3390/pathogens3010014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Chang PV, Hao L, Offermanns S, Medzhitov R. The Microbial Metabolite Butyrate Regulates Intestinal Macrophage Function via Histone Deacetylase Inhibition. Proc Natl Acad Sci USA (2014) 111(6):2247–52. doi: 10.1073/pnas.1322269111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Wang G, Yu Y, Wang Y-Z, Wang J-J, Guan R, Sun Y, et al. Role of SCFAs in Gut Microbiome and Glycolysis for Colorectal Cancer Therapy. J Cell Physiol (2019) 234(10):17023–49. doi: 10.1002/jcp.28436 [DOI] [PubMed] [Google Scholar]
- 84. Ijssennagger N, Belzer C, Hooiveld GJ, Dekker J, van Mil SWC, Müller M, et al. Gut Microbiota Facilitates Dietary Heme-Induced Epithelial Hyperproliferation by Opening the Mucus Barrier in Colon. Proc Natl Acad Sci USA (2015) 112(32):10038–43. doi: 10.1073/pnas.1507645112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Centuori SM, Gomes CJ, Trujillo J, Borg J, Brownlee J, Putnam CW, et al. Deoxycholic Acid Mediates non-Canonical EGFR-MAPK Activation Through the Induction of Calcium Signaling in Colon Cancer Cells. Biochim Biophys Acta (2016) 1861(7):663–70. doi: 10.1016/j.bbalip.2016.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Dong W, Liu L, Dou Y, Xu M, Liu T, Wang S, et al. Deoxycholic Acid Activates Epidermal Growth Factor Receptor and Promotes Intestinal Carcinogenesis by ADAM17-Dependent Ligand Release. J Cell Mol Med (2018) 22(9):4263–73. doi: 10.1111/jcmm.13709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Napolitano S, Matrone N, Muddassir AL, Martini G, Sorokin A, De Falco V, et al. Triple Blockade of EGFR, MEK and PD-L1 has Antitumor Activity in Colorectal Cancer Models With Constitutive Activation of MAPK Signaling and PD-L1 Overexpression. J Exp Clin Cancer Res CR (2019) 38(1):492. doi: 10.1186/s13046-019-1497-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Bhullar DS, Barriuso J, Mullamitha S, Saunders MP, O'Dwyer ST, Aziz O. Biomarker Concordance Between Primary Colorectal Cancer and its Metastases. EBioMedicine (2019) 40:363–74. doi: 10.1016/j.ebiom.2019.01.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Woolston A, Khan K, Spain G, Barber LJ, Griffiths B, Gonzalez-Exposito R, et al. Genomic and Transcriptomic Determinants of Therapy Resistance and Immune Landscape Evolution During Anti-EGFR Treatment in Colorectal Cancer. Cancer Cell (2019) 36(1):35–50. doi: 10.1016/j.ccell.2019.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Nusse R, Clevers H. Wnt/β-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell (2017) 169(6):985–99. doi: 10.1016/j.cell.2017.05.016 [DOI] [PubMed] [Google Scholar]
- 91. Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium Nucleatum Promotes Colorectal Carcinogenesis by Modulating E-Cadherin/β-Catenin Signaling via its FadA Adhesin. Cell Host Microbe (2013) 14(2):195–206. doi: 10.1016/j.chom.2013.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Yang Y, Weng W, Peng J, Hong L, Yang L, Toiyama Y, et al. Fusobacterium Nucleatum Increases Proliferation of Colorectal Cancer Cells and Tumor Development in Mice by Activating Toll-Like Receptor 4 Signaling to Nuclear Factor-κb, and Up-Regulating Expression of MicroRNA-21. Gastroenterology (2017) 152(4):851–66. doi: 10.1053/j.gastro.2016.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. DiDonato JA, Mercurio F, Karin M. NF-κb and the Link Between Inflammation and Cancer. Immunol Rev (2012) 246(1):379–400. doi: 10.1111/j.1600-065X.2012.01099.x [DOI] [PubMed] [Google Scholar]
- 94. Hoesel B, Schmid JA. The Complexity of NF-κb Signaling in Inflammation and Cancer. Mol Cancer (2013) 12:86. doi: 10.1186/1476-4598-12-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Peng C, Ouyang Y, Lu N, Li N. The NF-κb Signaling Pathway, the Microbiota, and Gastrointestinal Tumorigenesis: Recent Advances. Front Immunol (2020) 11:1387. doi: 10.3389/fimmu.2020.01387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Sahu U, Choudhury A, Parvez S, Biswas S, Kar S. Induction of Intestinal Stemness and Tumorigenicity by Aberrant Internalization of Commensal non-Pathogenic E. Coli. Cell Death Dis (2017) 8(3):e2667. doi: 10.1038/cddis.2017.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Liang JQ, Li T, Nakatsu G, Chen Y-X, Yau TO, Chu E, et al. A Novel Faecal Marker for the non-Invasive Diagnosis of Colorectal Adenoma and Cancer. Gut (2020) 69(7):1248–57. doi: 10.1136/gutjnl-2019-318532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Bell HN, Rebernick RJ, Goyert J, Singhal R, Kuljanin M, Kerk SA, et al. Reuterin in the Healthy Gut Microbiome Suppresses Colorectal Cancer Growth Through Altering Redox Balance. Cancer Cell (2021). doi: 10.1016/j.ccell.2021.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Rosshart SP, Vassallo BG, Angeletti D, Hutchinson DS, Morgan AP, Takeda K, et al. Wild Mouse Gut Microbiota Promotes Host Fitness and Improves Disease Resistance. Cell (2017) 171(5):1015–28. doi: 10.1016/j.cell.2017.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, et al. Commensal Bifidobacterium Promotes Antitumor Immunity and Facilitates Anti-PD-L1 Efficacy. Sci (New York NY) (2015) 350(6264):1084–9. doi: 10.1126/science.aac4255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillère R, et al. Gut Microbiome Influences Efficacy of PD-1-Based Immunotherapy Against Epithelial Tumors. Sci (New York NY) (2018) 359(6371):91–7. doi: 10.1126/science.aan3706 [DOI] [PubMed] [Google Scholar]
- 102. Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, et al. Gut Microbiome Modulates Response to Anti-PD-1 Immunotherapy in Melanoma Patients. Sci (New York NY) (2018) 359(6371):97–103. doi: 10.1126/science.aan4236 [DOI] [PMC free article] [PubMed] [Google Scholar]