HISTORY OF METRONIDAZOLE USE: PARASITES CAME FIRST
Metronidazole is one of the rare examples of a drug developed against a parasite which has since gained broad use as an antibacterial agent (24). Briefly, at Rhone-Poulenc labs in France, extracts of Streptomyces spp. were screened for activity against Trichomonas vaginalis, a cause of vaginal itching. Azomycin, a nitroimidazole, was identified, and metronidazole, a synthetic derivative, was used to treat chronic trichomonad infections, beginning in 1959 (66). Metronidazole was shown to be efficacious against Entamoeba histolytica, the cause of amebic dysentery and liver abscess, in 1966 (67). Giardia lamblia (also known as G. duodenalis) was treated with metronidazole after this luminal parasite was recognized as a cause of malabsorption and epigastric pain in the 1970s (102).
The antibacterial activity of metronidazole was discovered by accident in 1962 when metronidazole cured a patient of both trichomonad vaginitis and bacterial gingivitis (78). However, it was not until the 1970s that metronidazole was popularized for treatment of infections caused by gram-negative anaerobes such as bacteroides or gram-positive anaerobes such as clostridia (24, 48). Presently, metronidazole, which is inexpensive, has good tissue penetration, and produces relatively mild side effects, is on the formulary at most hospitals for prophylaxis against anaerobic infection after bowel surgery, for treatment of wound abscess, and for treatment of antibiotic-associated colitis caused by Clostridium difficile (85, 94). Metronidazole is an important part of combination therapy against Helicobacter pylori, a major cause of gastritis and a risk factor for stomach cancer (21, 57).
NEW IDEAS ABOUT METRONIDAZOLE-SENSITIVE PARASITES
Luminal parasites have two characteristics which distinguish them from other eukaryotes: (i) they live under anaerobic conditions, and (ii) they lack mitochondria and enzymes of oxidative phosphorylation (61). Indeed, a widely held view is that luminal parasites are “living fossils,” which reflect eukaryotic lifestyles prior to oxygenation of the planet and acquisition of the mitochondrial endosymbiont (2, 9, 68). This conclusion is supported by the presence of amebae, giardia, and trichomonads at or near the bases of the eukaryotic phylogenetic trees constructed from small subunit rRNA sequences (82). Further, these luminal parasites lack centrioles, Golgi with tight lamellae (giardia and amebae), and introns (giardia and trichomonads) (1, 26, 51, 55, 81).
Recent studies of metronidazole-sensitive parasites suggest that these organisms are not living fossils but instead are diverse eukaryotes with novel adaptations to their anaerobic niche. First, amebae and giardia lack fermentation enzymes (lactate dehydrogenase and pyruvate decarboxylase), which are present in yeast and other eukaryotes (60, 90). Second, luminal parasites appear to have acquired by horizontal transfer bacterial genes which encode fermentation enzymes (72, 80). These include an iron-sulfur protein called pyruvate:ferredoxin oxidoreductase (POR), which is involved in metronidazole activation (36, 64, 88). Third, all of these “amitochondriate” parasites have a gene encoding a homologue of the mitochondrial 60-kDa heat shock protein (Hsp60) (11, 15, 32, 34, 70, 71). The trichomonad mitochondrion has been converted into a fermentation factory called “hydrogenosome” (5, 8, 11–13, 36, 40, 62). The mitochondrion-derived organelle of amebae is atrophic, while that of giardia may have been lost (26, 52, 71, 84).
The goals of this review are to (i) demonstrate how luminal parasites are similar to and different from each other, (ii) discuss the biochemistry and phylogeny of bacterium-like fermentation enzymes involved in metronidazole activation, (iii) explore the peculiar compartmentalization of these fermentation enzymes, and (iv) discuss mechanisms of metronidazole resistance in these parasites. Readers are referred to recent reviews of the clinical uses of metronidazole and of metronidazole resistance in H. pylori and other anaerobic bacteria (24, 30, 48, 57, 64, 85, 89, 94, 102).
DIVERSE MORPHOLOGY OF METRONIDAZOLE-SENSITIVE ORGANISMS
The bacteria and parasites treated with metronidazole, which share an anaerobic niche in the lumen of the bowel or vagina and in tissue abscesses, show little resemblance to each other (61). For example, amebae have a large cytosol that is filled with vesicles and vacuoles that resemble those of macrophages (55). Like macrophages, amebae phagocytose bacteria, including anaerobic bacteria such as Clostridia spp. or facultative anaerobes such as Escherichia coli (59). Giardias have two identical nuclei, multiple flagellae, and a sucker disc, which is composed of a set of unique cytoskeletal proteins called “giardins” (1, 26). Trichomonads alternate between an ameboid form, which phagocytoses bacteria like entamoebae, and a flagellated form, which moves somewhat as do giardias (66). In the pouch or rumen of bicloved mammals (e.g., sheep or cows), cellulose is degraded by anaerobic bacteria, as well as anaerobic fungi and ciliates (14). The rumen fungi (e.g., Neocallimastix, Piromyces, and Orpinomyces) are filamentous and resemble the more familiar hyphal fungi, which are facultative anaerobes, such as Aspergillus and Candida (92). The rumen ciliates (e.g., Polyplastron and Dasytricha), which are covered with cilia, resemble their aerobic counterparts, such as Paramecium and Tetrahymena (20, 33, 65).
DIVERSE PHYLOGENY OF METRONIDAZOLE-SENSITIVE EUKARYOTES
In the absence of fossil evidence, which might be used to dissect the history of metronidazole-sensitive eukaryotes, comparisons of small subunit rRNA (ssRNA) sequences may be used to reconstruct the phylogeny of these organisms and their aerobic counterparts (53, 82). Remarkably, the amitochondriate eukaryotes are distributed almost as broadly throughout the ssRNA tree as are the aerobic eukaryotes (Fig. 1). Giardia and trichomonads, which are distinct from each other, as evidenced by their long branch lengths, are at the root of the ssRNA tree, along with microsporidia. Microsporidia are opportunistic, intracellular parasites, which lack a mitochondrion (35). Amebae and rumen ciliates are in the middle of the tree, along with apicomplexa (cause of malaria and toxoplasmosis), kinetoplastids (cause of sleeping sickness and visceral leishmaniasis), and slime molds. Anaerobic and aerobic ciliates are not separated into separate groups or clades (20). Rumen fungi are at the crown of the ssRNA tree, along with other fungi, plants, and animals. It appears then that the luminal protozoan parasites, rumen fungi, and rumen ciliates do not have an immediate common ancestor. Similar adaptations made by these organisms to their anaerobic environment, therefore, are most likely examples of convergent evolution (5, 72).
FIG. 1.
Diverse locations of metronidazole-sensitive eukaryotic organisms in a phylogenetic tree constructed by using small ssRNA sequences. An aligned set of ssRNA sequences was downloaded from the web site of the Ribosomal DNA Project, and a parsimony tree was drawn by using the program PAUP (22, 53, 82). Branch lengths have no information in parsimony trees. Organisms containing hydrogenosomes and bacterium-like fermentation enzymes in the cytosol, which are metronidazole sensitive, are indicated to distinguish them from organisms with mitochondria (unmarked) or chloroplasts. The ssRNA tree includes the luminal diplomonad G. lamblia and the free-living diplomonad Hexamita inflata, microsporidia Vairimorpha necatrix and Encephalitozoon hellem, the vaginal trichomonad T. vaginalis and the intestinal trichomonad Dientamoeba fragilis, the microaerophilic ameba E. histolytica and the aerobic amebae Naegleria gruberi and Acanthamoeba castellanii, the slime mold Dictyostelium discoideum, kinetoplastids Leishmania donovani and Euglena gracilis, apicomplexa Plasmodium falciparum and Toxoplasma gondii, anaerobic ciliates Metopus contortus and Dasytricha ruminantium and the aerobic ciliate Tetrahymena pyriformis, plants Glycine max and Arabidopsis thaliana, animals Homo sapiens and Caenorhabditis elegans, and anaerobic fungi Neocallimastix frontalis and Piromonas communis and aerobic fungi Saccharomyces cerevisiae and Candida albicans. A similar tree was obtained by neighbor-joining methods (data not shown).
POR IS INVOLVED IN ACTIVATING METRONIDAZOLE IN ANAEROBIC BACTERIA AND LUMINAL PARASITES
Like many drugs, metronidazole is relatively inactive until it is metabolized within host or microbial cells (24, 48). Metronidazole is activated when reduced, and reduction occurs only under strongly reducing conditions (28, 43). In anaerobic or microaerophilic bacteria or luminal parasites, metronidazole is activated when it receives an electron from ferredoxin or flavodoxin that was reduced by POR (9, 36, 37, 40, 47, 61, 68, 72, 87, 88). POR, which may be detected by measuring the coenzyme A (CoA)-dependent reduction of methyl viologen, is inactivated by oxygen. Among microorganisms, there is nearly a one-to-one correspondence between the presence of POR activity and metronidazole sensitivity (64). An exception may be Mycobacterium tuberculosis under anaerobic conditions, where mycobacteria stop dividing and become susceptible to metronidazole (93). Most eukaryotes and eubacteria fail to activate metronidazole, because POR is absent (58). In humans, pyruvate dehydrogenase substitutes for POR, and only NADH is produced rather than reduced ferredoxin. Metronidazole is also reduced and activated in poorly oxygenated tissues, such as those in abscesses or necrotic centers of tumors (43). Activated metronidazole damages cells by forming protein and DNA adducts (28). DNA damage may be the basis for the carcinogenicity of metronidazole in lab animals, although the carcinogenicity of metronidazole in humans has not been demonstrated (21).
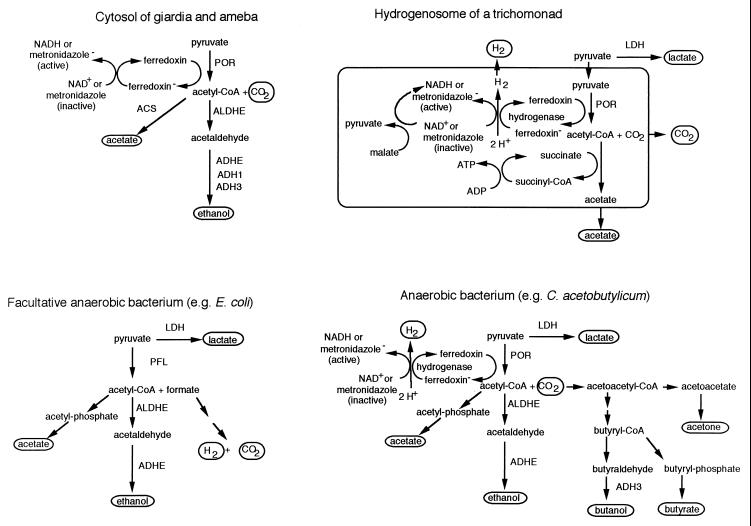
FERMENTATION PATHWAYS OF LUMINAL PARASITES ARE SIMPLER THAN THOSE OF BACTERIA
The fermentation enzymes of parasites, bacteria, cilia, and fungi living under anaerobic conditions are necessary to dispose of reducing equivalents (NADH or NADPH) which are generated during glycolysis (9, 16, 27, 61, 68). Amebic and giardia fermentation enzymes are present in the cytosol, while most fermentation enzymes of trichomonads are in hydrogenosomes (Fig. 2) (4, 5, 8, 9, 12, 36, 37, 40, 46, 61, 62, 68, 69, 77, 87, 88, 97). The fermentation of pyruvate to ethanol, CO2, and acetate (amebae and giardia) or to CO2, acetate, and H2 (trichomonads) begins in each parasite with POR (Fig. 2). POR catalyzes the oxidative decarboxylation of pyruvate to acetyl-CoA, producing CO2 and a reduced ferredoxin radical. Ferredoxins of giardia and amebae are 6 kDa (like those of anaerobic bacteria), while the ferredoxin of trichomonads is 12 kDa (like those of mitochondria) (37, 40, 87). In the absence of metronidazole or oxygen as an electron acceptor, reduced ferredoxin, which was produced in the reaction catalyzed by POR, donates its electron to NAD (amebae and giardia) and/or to protons to produce hydrogen gas (trichomonads) (12).
FIG. 2.
Metabolic pathways of E. histolytica and T. vaginalis contrasted with those of E. coli and C. acetobutylicum (redrawn from references 16, 27, 62, and 68). Fermentation products are indicated by ellipses. PORs, ferredoxins, ADHEs, and hydrogenases are labeled, while other enzymes are not. LDH, lactic dehydrogenase; ACS, acetyl-CoA synthase; PFL, pyruvate-formate lyase.
Some luminal parasites and anaerobic bacteria convert acetyl-CoA to acetaldehyde via a CoA-dependent acetaldehyde dehydrogenase (ALDHE), which is absent in humans and most other eukaryotes (Fig. 2) (16, 27, 29, 63, 68, 72, 77, 83, 97). A CoA-independent ALDH (ALDH2), which is located in human mitochondria, is inhibited by disulfuram (antabuse) (56). Amebae are able to convert acetaldehyde to ethanol via one of three alcohol dehydrogenases (ADHs), which include an NAD-dependent ADHE and NADP-dependent ADH1 and ADH3 (46, 68, 97). Amebic ALDHE and ADHE are potential targets for new drugs. A novel assay for antiamebic compounds uses an E. coli adhe mutant, which is complemented with the amebic adhe gene (99). Giardias have ALDHE and ADHE, which may be targets for new drugs, while trichomonads have a bacterium-like hydrogenase, which may be a target for new drugs (12, 77).
The fermentation pathways of facultative anaerobes, such as E. coli, and obligate anaerobes, such as Clostridium acetobutylicum, are much more complex than those of the luminal protozoa (Fig. 2) (16, 27). Although E. coli has a por gene, pyruvate is, for the most part, converted to acetyl-CoA and formate via a pyruvate-formate lyase (6). Anaerobic bacteria have the most complex sets of fermentation products (carbon dioxide, hydrogen, ethanol, butanol, acetate, butyrate, lactate, and acetone). An ongoing sequencing project of the G. lamblia genome and a proposed sequencing project of the E. histolytica genome are likely to reveal many more bacterium-like fermentation enzymes in these parasites (81).
POR AND OTHER REDOX PROTEINS OF LUMINAL PROTOZOA ARE FUSION PEPTIDES
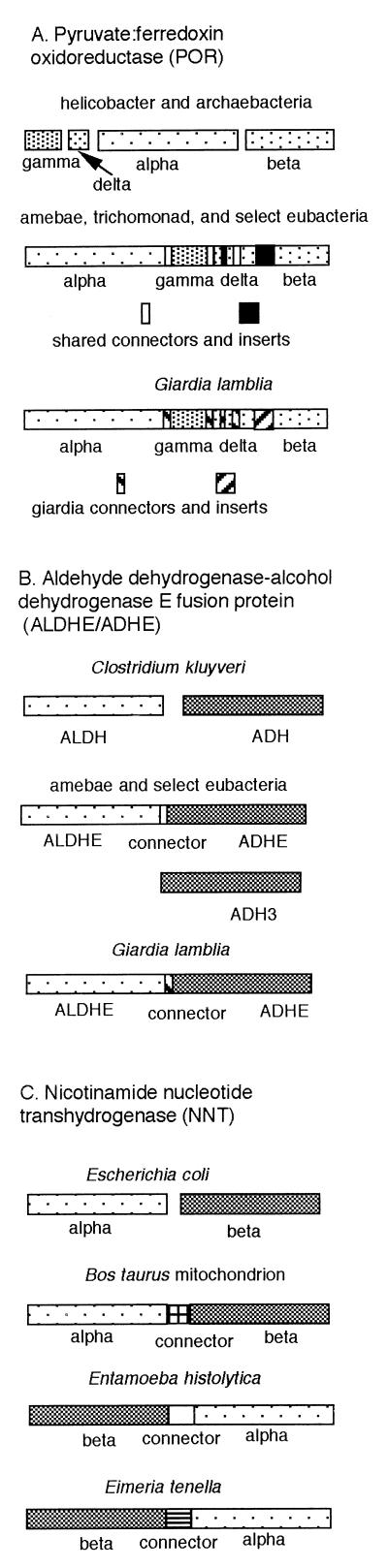
Genes encoding longer peptides of higher eukaryotes are often composed of multiple fused exons (exon-shuffling hypothesis) (19a). Luminal parasites have rare introns (amebae) or no introns (giardia and trichomonads) (51, 81). This is consistent with secondary loss of introns and the splicing apparatus from giardia and trichomonads (introns-early hypothesis) or with development of introns and the splicing apparatus after giardia and trichomonads diverged from the main trunk of the eukaryotic tree (introns-late hypothesis) (73). Genes encoding longer peptides of fermentation enzymes of the luminal protozoa appear to be constructed by fusion of bacterial genes within an operon (42, 72). For example, the PORs of the luminal parasites and of some eubacteria are homodimers, composed of a 110-kDa peptide, which appears to be the result of the fusion of four POR peptides present in archaebacteria and H. pylori (Fig. 3). The group III microbial ADHs of some bacteria, amebae, and giardia are in the C half of a fusion enzyme, which contains the CoA-dependent ALDHE in the N half (29, 63, 72, 97). In Clostridium kluyveri, the ALDHE and ADHE are encoded by separate genes on the same operon (83). Other microbial group III ADHs are encoded by genes independent of ALDHE in amebae (ADH3), E. coli, and C. acetobutylicum (16, 27, 69). The amebic NNT, which is homologous to mitochondrial and bacterial enzymes, is a fusion peptide composed of two peptides that are homologous to α- and β-subunits of the bacterial nicotinamide nucleotide transhydrogenase (NNT) (100). However, homologues of α- and β-subunits of bacterial NNT are reversed in NNTs of amebae and apicomplexa versus those of higher eukaryotes (Fig. 4) (95). This surprising result suggests different origins of the amebic and mitochondrial NNTs (see below).
FIG. 3.
POR, ADHE, and NNT are fusion proteins (29, 36, 42, 63, 69, 72). Connector regions of POR, which correspond to untranslated regions between nonfused bacterial genes, are similar for amebic, trichomonad, and bacterial enzymes but unalignable for the G. lamblia POR. Similarly, insert regions of POR, which are absent from the beta subunit of the helicobacter and archaebacterium genes, are similar for amebic and bacterial enzymes but unalignable for the G. lamblia POR. The connector regions of ALDHE and ADHE, which correspond to untranslated regions between nonfused C. kluyveri genes, are also similar for amebic and bacterial enzymes and unalignable for the G. lamblia enzyme. The connector regions of the NNT, which correspond to untranslated regions between nonfused E. coli genes, are dissimilar for amebic, eimeria, and bovine enzymes. Further, the order of the alpha and beta subunits of the amebic and eimeria NNTs is opposite to that of the bovine enzyme.
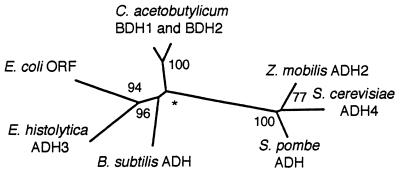
FIG. 4.
Neighbor-joining tree of group 3 microbial ADHs of C. acetobutylicum (GenBank accession numbers M96945 and M96946), Bacillus subtilis (bsz939347), E. coli (U28377), E. histolytica (D49910), Zymomonas mobilis (M15394), Schizosaccaromyces pombe (Q09669), and Saccharomyces cerevisiae (Z2778). Branch lengths are proportionate to differences between adjacent sequences, while bootstrap values at each node derive from 100 replicates (28). Asterisk, node where the analysis failed to identify the same bifurcating branch in over 50% of the bootstrap replicates. A similar tree was obtained by parsimony methods (data not shown).
GENES ENCODING FERMENTATION ENZYMES OF LUMINAL PARASITES MAY HAVE ORIGINATED FROM MULTIPLE BACTERIA
Genes encoding the bacterium-like fermentation enzymes of the luminal protozoa appear to have been horizontally transferred from bacteria, a phenomenon highly unusual for nonmitochondrial proteins (see below) (12, 31, 72, 80). Evidence for this horizontal transfer includes the following. First, parasite enzymes show remarkable amino acid sequence identities to bacterial fermentation enzymes. For example, amebic ADH1 showed >60% amino acid sequence identities with secondary ADHs of Clostridium beijerinckii and Thermoanaerobacter brockii (39, 45, 46). The amebic POR and ADHE showed 49% amino acid sequence identities to those of Klebsiella pneumoniae and E. coli, respectively (29, 72, 97). Second, in phylogenetic trees parasite enzymes are present in groups or clades which contain bacterial enzymes and no other eukaryotic enzymes (72). For example, the amebic ADH3 was paired with an ADH predicted by an E. coli open reading frame (Fig. 4) (6, 22, 69). The very high bootstrap value (94 at node A) indicates that the genes encoding the amebic ADH3 and the E. coli ADH share a recent ancestor. Third, genes encoding fermentation proteins of the same luminal parasite appear to derive from different bacteria. Amebic ADH3 is most similar to a gram-negative rod, amebic ADH1 is most similar to gram-positive rods, while amebic ADHE is equally similar to gram-negative and gram-positive rods (46, 72). Fourth, genes encoding homologous peptides in different parasites appear to derive from different bacteria. Amebic ferredoxin is most similar to that of the archaebacterium Methanosarcina barkeri, giardia ferredoxin is most similar to that of the δ-proteobacterium Desulfovibrio desulfuricans, while trichomonad ferredoxin is most similar to that of the γ-proteobacterium Pseudomonas putida (37, 40, 72, 87). Fifth, connector regions of POR, which correspond to untranslated regions in nonfused bacterial genes, are completely unalignable for amebae and giardia (Fig. 3) (42, 72). This result is consistent with differences in appearance and phylogeny between these luminal parasites.
It seems most likely then that genes encoding fermentation enzymes of luminal parasites were horizontally transferred from multiple bacteria a long time ago (72). Identification of these bacterial donors may be aided by an explosion of new microbial enzyme sequences predicted from numerous ongoing whole-genome sequencing projects (6). It is possible but less likely that multiple genes encoding homologous fermentation enzymes were present in a common eukaryotic ancestor and then were lost from all organisms except luminal parasites (61, 68). Whether these gene transfers occurred with a bacterial endosymbiont or by means of plasmids, phages, or virus cannot be determined (13, 31, 54). The luminal parasites are surrounded by bacteria, which they phagocytose (59). Further, some protozoa have unusual endosymbionts, while bacterial plasmids replicate under antibiotic selection in transfected amebae, giardia, and trichomonads (18, 25, 33, 44, 54, 79). Genes that encode pyrophosphate-dependent glycolytic enzymes of amebae and giardia and cellulose-degrading proteins of rumen ciliates also appear to have been horizontally transferred from bacteria (10, 14). The mammalian triose-phosphate isomerase gene may also have been horizontally transferred from bacteria (41). Conversely, eukaryotic gene segments encoding fibronectin 3 domains appear to have been horizontally transferred into eubacteria (50).
FERMENTATION ENZYMES OF LUMINAL PARASITES RESEMBLE THOSE OF BACTERIA IN SPECIFICITY AND TERTIARY STRUCTURE
The substrate and cofactor specificities of bacterium-like fermentation enzymes have been remarkably well conserved. For example, amebic, giardia, and bacterial ADHEs are iron and NAD dependent, prefer short-chain primary alcohols, and form paracrystalline arrays (29, 63, 77, 97). Amebic ADH1, which is a 40-kDa, zinc-dependent enzyme distantly related to liver ADHs, has the same specificity for NADP and secondary alcohols as the C. beijerinckii and T. brockii enzymes (39, 46, 98). These specificities are distinct from those of liver enzymes, which prefer NAD and primary alcohols. The tertiary structure of amebic ADH1, determined by X-ray diffraction of crystals of the recombinant enzyme, closely resembles those of the bacterial enzymes (45). For example, 16 of 17 residues present in the active site of amebic ADH1 are identical to those of the T. brockii enzyme (76). In contrast, the substrate specificities and tertiary structures of human ADHs differ dramatically from each other (38).
HYDROGENOSOMES ARE MODIFIED MITOCHONDRIA IN WHICH ANAEROBIC FERMENTATION ENZYMES HAVE REPLACED ENZYMES OF OXIDATIVE PHOSPHORYLATION
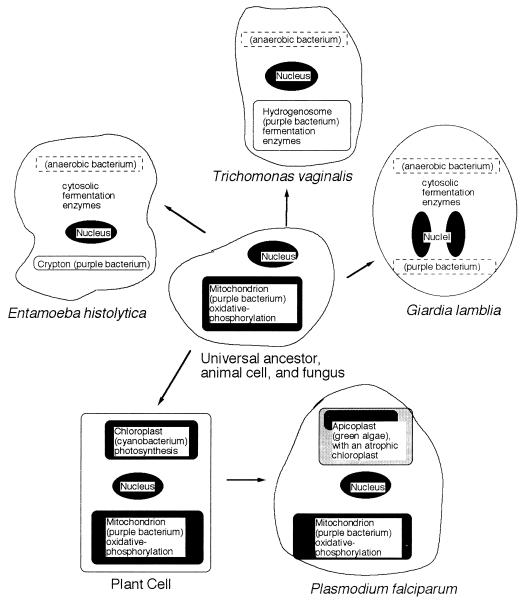
The endosymbiont hypothesis, one of the great ideas in cell biology, suggests that mitochondria and chloroplasts derive from a phagocytosed α-proteobacterium and a phagocytosed cyanobacterium, respectively (Fig. 5) (13, 31, 54, 96). Evidence for this idea includes sequences of rRNA genes, which remain in the matrices of mitochondria, and sequences of heat shock proteins such as Hsp60, which are targeted to mitochondria (32, 34, 53). In T. vaginalis and in rumen fungi, bacterium-like fermentation enzymes have replaced enzymes of oxidative phosphorylation in a modified mitochondrion, the hydrogenosome (5, 11, 13, 33, 62). Like mitochondria, most hydrogenosomes have double membranes and divide by segmentation, and one hydrogenosomal genome has been identified (4). Further, trichomonad hydrogenosomal heat shock proteins (Hsp10, Hsp60, and Hsp70) align with mitochondrial heat shock proteins in phylogenetic trees (11). Hydrogenosomal proteins are synthesized and directed to hydrogenosomes via presequences which are similar to those of nucleus-encoded mitochondrial proteins (8, 12, 17, 36, 40). Although hydrogenosomal function is not essential for viability, metronidazole-resistant trichomonads, which lack POR and hydrogenase activities, grow very slowly (47). Hydrogenosomes of rumen fungi have similar fermentation enzymes and presequences to those of trichomonads (5, 33, 65, 92).
FIG. 5.
Revised endosymbiont hypothesis for animal cells (same as common ancestor), E. histolytica, T. vaginalis (same as rumen fungi or rumen ciliates), G. lamblia, plant cells, and P. falciparum (same as T. gondii). Structures in black (nuclei, mitochondria, and chloroplasts) contain DNA, while no DNA has yet been isolated from unshaded structures (hydrogenosomes and crypton). Structures indicated by dashed rectangles (anaerobic bacteria and the purple bacterium in giardia) are absent now, but their past presence is suggested by bacterium-like nuclear genes.
The presence of a mitochondrion-derived organelle in trichomonads, mitochondrial Hsp60 in giardia and amebae, and mitochondrial Hsp70 in microsporidia, all of which are at the root of the eukaryotic phylogenetic tree, suggests that all extant eukaryotes have a common ancestor that had the same mitochondrion-derived endosymbiont (Fig. 5) (5, 11, 15, 35, 70, 71). Although the structural proteins of the mitochondrion appear to have been conserved in hydrogenosomes, some of the enzymes inside are likely not of mitochondrial origin (e.g., POR and hydrogenase) (12, 36). As discussed above, it is likely that these fermentation enzymes derive from anaerobic bacteria (72).
CRYPTIC MITOCHONDRION-DERIVED ORGANELLE OF AMEBAE
E. histolytica has an Hsp60-associated, mitochondrion-derived organelle, called “crypton” because it was previously hidden and its function remains unclear (Fig. 5) (52). Evidence for the amebic crypton includes the following. The amebic Hsp60 is grouped with those of mitochondria and hydrogenosomes in phylogenetic trees (11, 15, 70, 71). Amebic Hsp60 is functional, as suggested by its induction by heat shock, its ability to complement an E. coli groEL mutant, and its organellar localization (34, 52). The necessity of the presequence for targeting the Hsp60 to the organelle suggests conserved mitochondrion-like transporters in the crypton (8, 17, 52). Conversely, cleavage of the Hsp60 presequence suggests a conserved mitochondrion-like endopeptidase in the crypton. As the amebic cryptons are small and few, the organelles most closely resemble the atrophic mitochondria of bloodstream trypanosomes or anaerobically grown yeast (91). The difference between the crypton and these atrophic mitochondria is that the latter become competent organelles, which are capable of oxidative phosphorylation, when trypanosomes move to the insect vector or yeasts are exposed to oxygen.
The apicoplast of toxoplasma and P. falciparum is another recently discovered parasite organelle (Fig. 5) (44). The apicoplast, which is surrounded by four membranes, derives from an endosymbiont alga that contained a chloroplast. Antibiotics such as clindamycin target protein-synthetic machinery within the apicoplast, which is absent in host cells (23). To date, giardia and microsporidia are the only eukaryotes which do not have an endosymbiont-derived organelle (Fig. 5) (1, 26). The giardia Hsp60 lacks the usual organelle-targeting presequences at its N terminus and has been localized to the cytosol (71, 84).
MECHANISMS OF METRONIDAZOLE RESISTANCE AMONG LUMINAL PROTOZOA
Resistance of bacteria to antibiotics is quick to develop, rapid in its geographic spread, and oftentimes complete, rendering the particular antibiotic and related compounds no longer useful (49). For example, the widespread use of metronidazole, which is cheap and available without prescription in the developing world, has led to metronidazole-resistant H. pylori (57). The mechanism of metronidazole resistance appears to be a null mutation in the H. pylori rdxA gene, which encodes an oxygen-insensitive NADPH nitroreductase (30). In contrast, nitroimidazole resistance in bacteroides is plasmid mediated (89).
Metronidazole resistance among luminal parasites has been slow to develop and has not yet become of great clinical importance. The reasons for this may include the following. First, luminal parasites are most likely diploid, so that changes in a single gene are not sufficient to confer drug resistance (101). Bacteria are haploid, as is the bloodstream stage of Plasmodium falciparum, which is notorious for its resistance to chloroquine and other antimalarials (3). Second, luminal parasites have few metabolic alternatives to POR, which activates metronidazole. Amebae and giardia lack lactate dehydrogenase and pyruvate decarboxylase, which are present in other eukaryotes (9, 60, 68, 90). Trichomonads have cytosolic lactate dehydrogenase, which may substitute for POR in parasites selected for high levels of metronidazole resistance in the lab (47). However, these metronidazole-resistant trichomonads grow extremely slowly, making their competitiveness in nature uncertain. Decreased expression of ferredoxin confers low-level resistance to metronidazole on clinical isolates of trichomonads, which is easiest to detect under microaerophilic conditions (47). Decreases in POR activity and changes in permeability are associated with metronidazole resistance in giardia (86). Increased expression of superoxide dismutase by amebae is associated with low-level metronidazole resistance and with stress (74). Third, overexpression of ATP-binding cassette (ABC) family transporters, which are also known as “multidrug resistance” (mdr) gene products, confers resistance to some hydrophobic drugs but does not confer resistance to metronidazole (7, 25). For example, amebae step selected for resistance to emetine overexpress at least two ABC family transporters (19). However, these emetine-resistant parasites are not more resistant to metronidazole, which is hydrophilic and therefore not transported by P-glycoprotein pumps (75).
CONCLUSIONS
Luminal protozoa and bacteria are killed by metronidazole because they share the same fermentation enzyme, POR. An implication of this observation is that other bacterium-like fermentation enzymes (e.g., ADHE and hydrogenase) may be targets for antiparasitic drugs.
Luminal protozoan parasites targeted by metronidazole are not living fossils of eukaryotic life under anaerobic conditions but, instead, are diverse eukaryotes which have adapted bacterial solutions to life in an anaerobic niche.
Genes encoding fermentation enzymes of luminal eukaryotes were most likely horizontally transferred from bacteria, even if the identity of the bacterial donor(s) and the timing of the transfer(s) are not clear.
All extant eukaryotes have a common ancestor, which had a mitochondrion. In luminal protozoa, this endosymbiont-derived organelle may be remodeled into a hydrogenosome (e.g., trichomonads and rumen fungi and ciliates), may be atrophic (e.g., amebic crypton), or may be lost (e.g., giardia and microsporidia).
Metronidazole resistance among clinical isolates of luminal protozoa is rare. The slow development of metronidazole resistance among luminal protozoa is likely secondary to their polyploidy and the absence of alternative fermentation pathways.
ACKNOWLEDGMENT
This work was supported in part by National Institutes of Health grant AI-33492.
REFERENCES
- 1.Adam R D. The biology of Giardia spp. Microbiol Rev. 1991;55:706–732. doi: 10.1128/mr.55.4.706-732.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bakker-Grunwald T, Wostmann C. Entamoeba histolytica as a model for the primitive eukaryotic cell. Parasitol Today. 1993;9:27–31. doi: 10.1016/0169-4758(93)90161-8. [DOI] [PubMed] [Google Scholar]
- 3.Barat L M, Bloland P B. Drug resistance among malaria and other parasites. Infect Dis Clin N Am. 1997;11:969–987. doi: 10.1016/s0891-5520(05)70400-1. [DOI] [PubMed] [Google Scholar]
- 4.Benchimol M, Johnson P J, de Souza W. Morphogenesis of the hydrogenosome: an ultrastructural study. Biol Cell. 1996;87:197–205. [PubMed] [Google Scholar]
- 5.Biagini G A, Findlay B J, Lloyd D J. Evolution of the hydrogenosome. FEMS Microbiol Lett. 1997;155:133–140. doi: 10.1016/s0378-1097(97)00333-9. [DOI] [PubMed] [Google Scholar]
- 6.Blattner F R, Plunkett G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 7.Borst P, Ouellette M. New mechanisms of drug resistance in parasitic protozoa. Annu Rev Microbiol. 1995;49:427–460. doi: 10.1146/annurev.mi.49.100195.002235. [DOI] [PubMed] [Google Scholar]
- 8.Bradley P J, Lahti C J, Plumper E, Johnson P J. Targeting and translocation of proteins into the hydrogenosome of the protist Trichomonas: similarities with mitochondrial protein import. EMBO J. 1997;16:3484–3493. doi: 10.1093/emboj/16.12.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown D M, Upcroft J A, Edwards M R, Upcroft P. Anaerobic bacterial metabolism in the ancient eukaryote Giardia duodenalis. Int J Parasitol. 1998;28:149–164. doi: 10.1016/s0020-7519(97)00172-0. [DOI] [PubMed] [Google Scholar]
- 10.Bruchhaus I, Tannich E. Primary structure of the pyruvate phosphate dikinase in Entamoeba histolytica. Mol Biochem Parasitol. 1993;62:153–156. doi: 10.1016/0166-6851(93)90193-2. [DOI] [PubMed] [Google Scholar]
- 11.Bui E T N, Bradley P J, Johnson P J. A common evolutionary origin for mitochondria and hydrogenosomes. Proc Natl Acad Sci USA. 1996;93:9651–9656. doi: 10.1073/pnas.93.18.9651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bui E T, Johnson P J. Identification and characterization of [Fe]-hydrogenases in the hydrogenosome of Trichomonas vaginalis. Mol Biochem Parasitol. 1996;76:305–310. doi: 10.1016/0166-6851(96)02567-4. [DOI] [PubMed] [Google Scholar]
- 13.Cavalier-Smith T. The simultaneous symbiotic origin of mitochondria, chloroplasts, and microbodies. Ann N Y Acad Sci. 1987;503:55–72. doi: 10.1111/j.1749-6632.1987.tb40597.x. [DOI] [PubMed] [Google Scholar]
- 14.Chen H, Li X L, Ljungdahl L G. Sequencing of a 1,3-1,4-beta-d-glucanase (lichenase) from the anaerobic fungus Orpinomyces strain PC-2: properties of the enzyme expressed in Escherichia coli and evidence that the gene has a bacterial origin. J Bacteriol. 1997;179:6028–6034. doi: 10.1128/jb.179.19.6028-6034.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clark C G, Roger A J. Direct evidence for secondary loss of mitochondria in Entamoeba histolytica. Proc Natl Acad Sci USA. 1995;92:6518–6521. doi: 10.1073/pnas.92.14.6518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clark D P. The fermentation pathways of Escherichia coli. FEMS Microbiol Rev. 1989;5:223–234. doi: 10.1016/0168-6445(89)90033-8. [DOI] [PubMed] [Google Scholar]
- 17.Claros M G, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- 18.Delgadillo M G, Liston D R, Niazi K, Johnson P J. Transient and selectable transformation of the parasitic protist Trichomonas vaginalis. Proc Natl Acad Sci USA. 1997;94:4716–4720. doi: 10.1073/pnas.94.9.4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Descoteaux S, Ayala P, Samuelson J, Orozco E. Increase in mRNA of multiple Eh pgp genes encoding P-glycoprotein homologues in emetine-resistant Entamoeba histolytica parasites. Gene. 1995;164:179–184. doi: 10.1016/0378-1119(95)00533-c. [DOI] [PubMed] [Google Scholar]
- 19a.de Souza S J, Long M, Klein R J, Roy S, Lin S, Gilbert W. Toward a resolution of the introns early/late debate: only phase zero introns are correlated with the structure of ancient proteins. Proc Natl Acad Sci USA. 1998;95:5094–5099. doi: 10.1073/pnas.95.9.5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Embley T M, Finlay B J, Dyal P L, Hirt R P, Wilkinson M, Williams A G. Multiple origins of anaerobic ciliates with hydrogenosomes within the radiation of aerobic ciliates. Proc R Soc Lond Ser B. 1995;262:87–93. doi: 10.1098/rspb.1995.0180. [DOI] [PubMed] [Google Scholar]
- 21.Falagas M E, Walker A M, Jick H, Ruthazer R, Griffith J, Snydman D R. Late incidence of cancer after metronidazole use: a matched metronidazole user/nonuser study. Clin Infect Dis. 1998;26:384–388. doi: 10.1086/516306. [DOI] [PubMed] [Google Scholar]
- 22.Felsenstein J. PHYLIP—phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 23.Fichera M E, Roos D S. A plastid organelle as a drug target in apicomplexan parasites. Nature. 1997;390:407–409. doi: 10.1038/37132. [DOI] [PubMed] [Google Scholar]
- 24.Freeman C D, Klutman N E, Lamp K C. Metronidazole. A therapeutic review and update. Drugs. 1997;54:679–708. doi: 10.2165/00003495-199754050-00003. [DOI] [PubMed] [Google Scholar]
- 25.Ghosh S K, Lohia A, Kumar A, Samuelson J. Overexpression of P-glycoprotein gene 1 by transfected Entamoeba histolytica confers emetine-resistance. Mol Biochem Parasitol. 1996;82:257–260. doi: 10.1016/0166-6851(96)02733-8. [DOI] [PubMed] [Google Scholar]
- 26.Gillin F D, Reiner D S, McCaffery J M. Cell biology of the primitive eukaryote Giardia lamblia. Annu Rev Microbiol. 1996;50:679–705. doi: 10.1146/annurev.micro.50.1.679. [DOI] [PubMed] [Google Scholar]
- 27.Girbal L, Croux C, Vasconcelos I, Soucaille P. Regulation of metabolic shifts in Clostridium acetobutylicum ATCC 824. FEMS Microbiol Rev. 1995;17:287–297. [Google Scholar]
- 28.Goldman P, Koch R L, Yeung T C, Chrystal E J, Beaulieu B B, Jr, McLafferty M A, Sudlow G. Comparing the reduction of nitroimidazoles in bacteria and mammalian tissues and relating it to biological activity. Biochem Pharmacol. 1986;35:43–51. doi: 10.1016/0006-2952(86)90553-8. [DOI] [PubMed] [Google Scholar]
- 29.Goodlove P E, Cunningham P R, Parker J, Clark D P. Cloning and sequence analysis of the fermentative alcohol-dehydrogenase-encoding gene of Escherichia coli. Gene. 1989;85:209–214. doi: 10.1016/0378-1119(89)90483-6. [DOI] [PubMed] [Google Scholar]
- 30.Goodwin A, Kersulyte D, Sisson G, Veldhuyzen van Zanten S J, Berg D E, Hoffman P S. Metronidazole resistance in Helicobacter pylori is due to null mutations in a gene (rdxA) that encodes an oxygen-insensitive NADPH nitroreductase. Mol Microbiol. 1998;28:383–393. doi: 10.1046/j.1365-2958.1998.00806.x. [DOI] [PubMed] [Google Scholar]
- 31.Gray M W, Doolittle W F. Has the endosymbiont hypothesis been proven? Microbiol Rev. 1982;46:1–42. doi: 10.1128/mr.46.1.1-42.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gupta R S. Evolution of the chaperonin families (Hsp60, Hsp10 and Tcp-1) of proteins and the origin of eukaryotic cells. Mol Microbiol. 1995;15:1–11. doi: 10.1111/j.1365-2958.1995.tb02216.x. [DOI] [PubMed] [Google Scholar]
- 33.Hackstein J H, Vogels G D. Endosymbiotic interactions in anaerobic protozoa. Antonie Leeuwenhoek. 1997;71:151–158. doi: 10.1023/a:1000154526395. [DOI] [PubMed] [Google Scholar]
- 34.Hartl F U. Molecular chaperones in cellular protein folding. Nature. 1996;381:571–580. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- 35.Hirt R P, Healy B, Vossbrinck C R, Canning C R, Embley T M. A mitochondrial Hsp70 orthologue in Vairimorpha necatrix: evidence that microsporidia once contained mitochondria. Curr Biol. 1997;7:995–998. doi: 10.1016/s0960-9822(06)00420-9. [DOI] [PubMed] [Google Scholar]
- 36.Hrdy I, Muller M. Primary structure and eubacterial relationship of the pyruvate:ferredoxin oxidoreductase of the amitochondriate eukaryote Trichomonas vaginalis. J Mol Evol. 1995;41:388–396. [PubMed] [Google Scholar]
- 37.Huber M, Garfinkel L, Gitler C, Mirelman D, Revel M, Rozenblatt S. Nucleotide sequence analysis of an Entamoeba histolytica ferredoxin gene. Mol Biochem Parasitol. 1988;31:27–34. doi: 10.1016/0166-6851(88)90142-9. [DOI] [PubMed] [Google Scholar]
- 38.Hurley T D, Steinmetz C G, Xie P, Yang Y N. Three-dimensional structures of human alcohol dehydrogenase isoenzymes reveal the molecular basis for their functional diversity. Adv Exp Med Biol. 1997;414:291–302. doi: 10.1007/978-1-4615-5871-2_34. [DOI] [PubMed] [Google Scholar]
- 39.Ismaiel A A, Zhu C X, Colby G D, Chen J S. Purification and characterization of a primary-secondary alcohol dehydrogenase from two strains of Clostridium beijerinckii. J Bacteriol. 1993;175:5097–5105. doi: 10.1128/jb.175.16.5097-5105.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson P J, D’Oliveira C E, Gorrell T E, Muller M. Molecular analysis of the hydrogenosomal ferredoxin of the anaerobic protist Trichomonas vaginalis. Proc Natl Acad Sci USA. 1990;87:6097–6101. doi: 10.1073/pnas.87.16.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keeling P J, Doolittle W F. Evidence that eukaryotic triosephosphate isomerase is of alpha-proteobacterial origin. Proc Natl Acad Sci USA. 1997;94:1270–1275. doi: 10.1073/pnas.94.4.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kletzin A, Adams M W W. Molecular and phylogenetic characterization of pyruvate and 2-ketoisovalerate ferredoxin oxidoreductases from Pyrococcus furiosus and pyruvate ferredoxin oxidoreductase from Thermotoga maritima. J Bacteriol. 1996;178:248–257. doi: 10.1128/jb.178.1.248-257.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koch C J, Lord E M, Shapiro I M, Clyman R I, Evans S M. Imaging hypoxia and blood flow in normal tissues. Adv Exp Med Biol. 1997;428:585–593. doi: 10.1007/978-1-4615-5399-1_83. [DOI] [PubMed] [Google Scholar]
- 44.Kohler S, Delwiche C F, Denny P W, Tilney L G, Webster P, Wilson R J, Palmer J D, Roos D S. A plastid of probable green algal origin in apicomplexan parasites. Science. 1997;275:1485–1489. doi: 10.1126/science.275.5305.1485. [DOI] [PubMed] [Google Scholar]
- 45.Korkhin Y, Frolow F, Bogin O, Peretz M, Kalb A J, Burstein Y. Crystalline alcohol dehydrogenases from the mesophilic bacterium Clostridium beijerinckii and the thermophilic bacterium Thermoanaerobium brockii—preparation, characterization and molecular symmetry. Acta Crystallogr Sect D. 1996;52:882–886. doi: 10.1107/S0907444996001461. [DOI] [PubMed] [Google Scholar]
- 46.Kumar A, Shen P-S, Descoteaux S, Pohl J, Bailey G, Samuelson J. Cloning and expression of an NADP+-dependent alcohol dehydrogenase gene of Entamoeba histolytica. Proc Natl Acad Sci USA. 1992;85:1782–1786. doi: 10.1073/pnas.89.21.10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Land K M, Johnson P J. Molecular mechanisms underlying metronidazole resistance in trichomonads. Exp Parasitol. 1997;87:305–308. doi: 10.1006/expr.1997.4223. [DOI] [PubMed] [Google Scholar]
- 48.Lau A H, Lam N P, Piscitelli S C, Wilkes L, Danziger L H. Clinical pharmokinetics of metronidazole and other nitroimidazole anti-infectives. Clin Pharmacokinet. 1992;23:328–364. doi: 10.2165/00003088-199223050-00002. [DOI] [PubMed] [Google Scholar]
- 49.Levy S B. The challenge of antibiotic resistance. Sci Am. 1998;278:46–53. doi: 10.1038/scientificamerican0398-46. [DOI] [PubMed] [Google Scholar]
- 50.Little E, Bork P, Doolittle R F. Tracing the spread of fibronectin type III domains in bacterial glycohydrolases. J Mol Evol. 1994;39:631–643. doi: 10.1007/BF00160409. [DOI] [PubMed] [Google Scholar]
- 51.Lohia A, Samuelson J. Cloning of the Eh cdc2 gene from Entamoeba histolytica encoding a protein kinase p34cdc2 homologue. Gene. 1993;127:203–207. doi: 10.1016/0378-1119(93)90720-n. [DOI] [PubMed] [Google Scholar]
- 52.Mai Z, Ghosh S, Frisardi M, Rosenthal B, Rogers R, Samuelson J. Hsp60 is targeted to a cryptic mitochondrion-derived organelle (“crypton”) in the microaerophilic protozoan parasite Entamoeba histolytica. Mol Cell Biol. 1999;19:2198–2205. doi: 10.1128/mcb.19.3.2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The RDP (Ribosomal Database Project) Nucleic Acids Res. 1997;25:109–110. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Margulis L. Symbiosis in cell evolution. W. H. New York, N.Y: Freeman & Co.; 1981. [Google Scholar]
- 55.Martinez-Palomo A. Cell biology. In: Brown K N, editor. The biology of Entamoeba histolytica. New York, N.Y: John Wiley Research Studies Press; 1982. pp. 5–59. [Google Scholar]
- 56.Mays D C, Ortiz-Bermudez P, Lam J P, Tong I H, Fauq A H, Lipsky J J. Inhibition of recombinant human mitochondrial aldehyde dehydrogenase by two intermediate metabolites of disulfiram. Biochem Pharmacol. 1998;55:1099–1103. doi: 10.1016/s0006-2952(97)00686-2. [DOI] [PubMed] [Google Scholar]
- 57.Megraud F, Doermann H P. Clinical relevance of resistant strains of Helicobacter pylori: a review of current data. Gut. 1998;43:S61–S65. doi: 10.1136/gut.43.2008.s61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mewes H W, Albermann K, Bahr M, Frishman D, Gleissner A, Hani J, Heumann K, Kleine K, Maierl A, Oliver S G, Pfeiffer F, Zollner A. Overview of the yeast genome. Nature. 1997;387:7–65. doi: 10.1038/42755. [DOI] [PubMed] [Google Scholar]
- 59.Mirelman D. Ameba-bacterium relationship in amebiasis. Microbiol Rev. 1987;51:272–284. doi: 10.1128/mr.51.2.272-284.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mucke U, Wohlfarth T, Fiedler U, Baumlein H, Rucknagel K P, Konig S. Pyruvate decarboxylase from Pisum sativum. Eur J Biochem. 1996;237:373–382. doi: 10.1111/j.1432-1033.1996.0373k.x. [DOI] [PubMed] [Google Scholar]
- 61.Muller M. Energy metabolism of ancestral eukaryotes: a hypothesis based on the biochemistry of amitochondriate parasitic proteins. BioSystems. 1992;28:33–40. doi: 10.1016/0303-2647(92)90005-j. [DOI] [PubMed] [Google Scholar]
- 62.Muller M. The hydrogenosome. J Gen Microbiol. 1993;139:2879–2889. doi: 10.1099/00221287-139-12-2879. [DOI] [PubMed] [Google Scholar]
- 63.Nair R V, Bennett G N, Papoutsakis E. Molecular characterization of an aldehyde/alcohol dehydrogenase gene from Clostridium acetobutylicum ATCC 824. J Bacteriol. 1994;176:871–885. doi: 10.1128/jb.176.3.871-885.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Narikawa S. Distribution of metronidazole susceptibility factors in obligate anaerobes. J Antimicrob Chemother. 1986;18:565–574. doi: 10.1093/jac/18.5.565. [DOI] [PubMed] [Google Scholar]
- 65.Paul R G, Williams A G, Butler R D. Hydrogenosomes in the rumen entodiniomorphid ciliate Polyplastron multivesiculatum. J Gen Microbiol. 1990;136:1981–1999. doi: 10.1099/00221287-136-10-1981. [DOI] [PubMed] [Google Scholar]
- 66.Petrin D, Delgaty K, Bhatt R, Garber G. Clinical and microbiological aspects of Trichomonas vaginalis. Clin Microbiol Rev. 1998;11:300–317. doi: 10.1128/cmr.11.2.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ravdin J I. Amebiasis. State-of-the-art clinical article. Clin Infect Dis. 1995;20:1453–1464. doi: 10.1093/clinids/20.6.1453. [DOI] [PubMed] [Google Scholar]
- 68.Reeves R E. Metabolism of Entamoeba histolytica Schaudinn, 1903. Adv Parasitol. 1984;23:105–142. doi: 10.1016/s0065-308x(08)60286-9. [DOI] [PubMed] [Google Scholar]
- 69.Rodriguez M A, Baez-Camargo M, Delgadillo D M, Orozco E. Cloning and expression of an Entamoeba histolytica NADP+-dependent alcohol dehydrogenase gene. Biochim Biophys Acta. 1996;1306:23–26. doi: 10.1016/0167-4781(96)00014-0. [DOI] [PubMed] [Google Scholar]
- 70.Roger A J, Clark C G, Doolittle W F. A possible mitochondrial gene in the early-branching amitochondriate protist Trichomonas vaginalis. Proc Natl Acad Sci USA. 1996;93:14618–14622. doi: 10.1073/pnas.93.25.14618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Roger A J, Svard S G, Tovar J, Clark C G, Smith M W, Gillen F D, Sogin M L. A mitochondrial-like chaperonin 60 gene in Giardia lamblia: evidence that diplomonads once harbored an endosymbiont related to the progenitor of mitochondria. Proc Natl Acad Sci USA. 1998;95:229–234. doi: 10.1073/pnas.95.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rosenthal B, Mai Z, Caplivski D, Ghosh S, de la Vega H, Graf T, Samuelson J. Evidence for the bacterial origin of genes encoding fermentation enzymes of the amitochondriate protozoan parasite Entamoeba histolytica. J Bacteriol. 1997;179:3736–3745. doi: 10.1128/jb.179.11.3736-3745.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rzhetsky A, Ayala F J, Hsu L C, Chang C, Yoshida A. Exon/intron structure of aldehyde dehydrogenase genes supports the “introns-late” theory. Proc Natl Acad Sci USA. 1997;94:6820–6825. doi: 10.1073/pnas.94.13.6820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Samarawickrema N A, Brown D M, Upcroft J A, Thammapalerd N, Upcroft P. Involvement of superoxide dismutase and pyruvate:ferredoxin oxidoreductase in mechanisms of metronidazole resistance in Entamoeba histolytica. J Antimicrob Chemother. 1997;40:833–840. doi: 10.1093/jac/40.6.833. [DOI] [PubMed] [Google Scholar]
- 75.Samuelson J, Burke A, Courval J. Susceptibility of an emetine-resistant mutant of Entamoeba histolytica to multiple drugs and to channel blockers. Antimicrob Agents Chemother. 1992;36:2392–2397. doi: 10.1128/aac.36.11.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Samuelson, J., and J. F. Head. Unpublished data.
- 77.Sanchez L B. Aldehyde dehydrogenase (CoA-acetylating) and the mechanisms of ethanol formation in the amitochondriate protist, Giardia lamblia. Arch Biochem Biophys. 1998;354:57–64. doi: 10.1006/abbi.1998.0664. [DOI] [PubMed] [Google Scholar]
- 78.Shinn D L S. Metronidazole in acute ulcerative gingivitis. Lancet. 1962;i:1191. [Google Scholar]
- 79.Singer S M, Yee J, Nash T E. Episomal and integrated maintenance of foreign DNA in Giardia lamblia. Mol Biochem Parasitol. 1998;92:59–69. doi: 10.1016/s0166-6851(97)00225-9. [DOI] [PubMed] [Google Scholar]
- 80.Smith M W, Feng D-F, Doolittle R F. Evolution by acquisition: the case for horizontal gene transfers. Trends Biochem Sci. 1992;17:489–493. doi: 10.1016/0968-0004(92)90335-7. [DOI] [PubMed] [Google Scholar]
- 81.Smith M W, Aley S B, Sogin M, Gillin F D, Evans G A. Sequence survey of the Giardia lamblia genome. Mol Biochem Parasitol. 1998;95:267–280. doi: 10.1016/s0166-6851(98)00113-3. [DOI] [PubMed] [Google Scholar]
- 82.Sogin M L, Silberman J D. Evolution of the protists and protistan parasites from the perspective of molecular systematics. Int J Parasitol. 1998;28:11–20. doi: 10.1016/s0020-7519(97)00181-1. [DOI] [PubMed] [Google Scholar]
- 83.Sohling B, Gottschalk G. Molecular analysis of the anaerobic succinate degradation pathway in Clostridium kluyveri. J Bacteriol. 1996;178:871–880. doi: 10.1128/jb.178.3.871-880.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Soltys B J, Gupta R S. Presence and cellular distribution of a 60-kDa protein related to mitochondrial HSP60 in Giardia lamblia. J Parasitol. 1994;80:580–590. [PubMed] [Google Scholar]
- 85.Song F, Glenny A M. Antimicrobial prophylaxis in colorectal surgery: a systematic review of randomized controlled trials. Br J Surg. 1998;85:1232–1241. doi: 10.1046/j.1365-2168.1998.00883.x. [DOI] [PubMed] [Google Scholar]
- 86.Townson S M, Borehman P F L, Upcroft P, Upcroft J A. Resistance to nitroheterocyclic drugs. Acta Trop. 1994;56:173–194. doi: 10.1016/0001-706x(94)90062-0. [DOI] [PubMed] [Google Scholar]
- 87.Townson S M, Hanson G R, Upcroft J A, Upcroft P. A purified ferredoxin from Giardia duodenalis. Eur J Biochem. 1994;220:439–446. doi: 10.1111/j.1432-1033.1994.tb18641.x. [DOI] [PubMed] [Google Scholar]
- 88.Townson S M, Upcroft J A, Upcroft P. Characterization and purification of pyruvate:ferredoxin oxidoreductase from Giardia duodenalis. Mol Biochem Parasitol. 1996;79:183–193. doi: 10.1016/0166-6851(96)02661-8. [DOI] [PubMed] [Google Scholar]
- 89.Trinh S, Reysset G. Detection by PCR of the nim genes encoding 5-nitroimidazole resistance in Bacteroides spp. J Clin Microbiol. 1996;34:2078–2084. doi: 10.1128/jcm.34.9.2078-2084.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tsuji S, Qureshi M A, Hou E W, Fitch W M, Li S S-L. Evolutionary relationships of lactate dehydrogenases (LDHs) from mammals, birds, an amphibian, fish, barley, and bacteria: LDH cDNA sequences from Xenopus, pig, and rat. Proc Natl Acad Sci USA. 1994;91:9392–9396. doi: 10.1073/pnas.91.20.9392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tyler K M, Matthews K R, Gull K. The bloodstream differentiation-division of Trypanosoma brucei studied using mitochondrial markers. Proc R Soc Lond Ser B. 1997;264:1481–1490. doi: 10.1098/rspb.1997.0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.van der Giezen M, Rechinger K B, Svendsen I, Durand R, Hirt R P, Fevre M, Embley T M, Prins R A. A mitochondrial-like targeting signal on the hydrogenosomal malic enzyme from the anaerobic fungus Neocallimastix frontalis: support for hypothesis that hydrogenosomes are modified mitochondria. Mol Microbiol. 1997;23:11–21. doi: 10.1046/j.1365-2958.1997.1891553.x. [DOI] [PubMed] [Google Scholar]
- 93.Wayne L G, Sramek H A. Metronidazole is bactericidal to dormant cells of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1994;38:2054–2058. doi: 10.1128/aac.38.9.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wilcox M H. Treatment of Clostridium difficile infection. J Antimicrob Chemother. 1998;41C:41–46. doi: 10.1093/jac/41.suppl_3.41. [DOI] [PubMed] [Google Scholar]
- 95.Yamaguchi M, Hatefi Y, Trach K, Hoch J A. The primary structure of the mitochondrial energy-linked nicotinamide nucleotide transhydrogenase deduced from the sequence of cDNA clones. J Biol Chem. 1988;263:2761–2767. [PubMed] [Google Scholar]
- 96.Yang D, Oyaizu Y, Oyaizu H, Olsen G J, Woese C R. Mitochondrial origins. Proc Natl Acad Sci USA. 1985;82:4443–4447. doi: 10.1073/pnas.82.13.4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yang W, Li E, Kairong T, Stanley S L., Jr Entamoeba histolytica has an alcohol dehydrogenase homologous to the multifunctional adhE gene product of Escherichia coli. Mol Biochem Parasitol. 1994;64:253–260. doi: 10.1016/0166-6851(93)00020-a. [DOI] [PubMed] [Google Scholar]
- 98.Yokoyama S, Yokoyama R, Kinlaw C S, Harry D E. Molecular evolution of zinc-containing long-chain alcohol genes. Mol Biol Evol. 1990;7:143–154. doi: 10.1093/oxfordjournals.molbev.a040593. [DOI] [PubMed] [Google Scholar]
- 99.Yong T S, Li E, Clark D, Stanley S L., Jr Complementation of an Escherichia coli adhE mutant by the Entamoeba histolytica EhADH2 gene provides a method for the identification of new anti-amebic drugs. Proc Natl Acad Sci USA. 1996;93:6464–6469. doi: 10.1073/pnas.93.13.6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yu Y, Samuelson J. Primary structure of an Entamoeba histolytica nicotinamide nucleotide transhydrogenase. Mol Biochem Parasitol. 1994;68:323–328. doi: 10.1016/0166-6851(94)90178-3. [DOI] [PubMed] [Google Scholar]
- 101.Yuh Y S, Liu J Y, Shaio M F. Chromosome number of Trichomonas vaginalis. J Parasitol. 1997;83:551–533. [PubMed] [Google Scholar]
- 102.Zaat J O M, Mank T G, Assendelft W J J. A systematic review on the treatment of giardiasis. Trop Med Int Health. 1997;2:63–82. doi: 10.1046/j.1365-3156.1997.d01-132.x. [DOI] [PubMed] [Google Scholar]