Abstract
Fentanyl is an anesthetic/analgesic commonly used in surgical and recovery settings. CYP3A4 and CYP3A5 encode enzymes which metabolize fentanyl; genetic variants in these genes impact fentanyl pharmacokinetics in adults. Pharmacokinetic (PK) studies are difficult to replicate in children due to the burden of additional blood taken solely for research purposes. The aim of this study is to test the effect of CYP3A5 and CYP3A4 genetic variants on fentanyl PK in children using opportunistically collected samples. Fentanyl concentrations were measured from remnant blood specimens and dosing data were extracted from electronic health records. Variant data defining CYP3A4*1G and CYP3A5*3 and *6 alleles were available from prior genotyping; alleles with no variant were defined as *1. The study cohort included 434 individuals (median age 9 months, 52% male) and 1937 fentanyl concentrations were available. A two-compartment model was selected as the base model, and the final covariate model included age, weight, and surgical severity score. Clearance was significantly associated with either CYP3A5*3 or CYP3A5*6 alleles, but not the CYP3A4*1G allele. A genotype of CYP3A5*1/*3 or CYP3A5*1/*6 (i.e., intermediate metabolizer status) was associated with a 0.84-fold (95% confidence interval [CI]: 0.71 to 1.00) reduction in clearance vs. CYP3A5*1/*1 (i.e., normal metabolizer status). CYP3A5*3/*3, CYP3A5*3/*6, or CYP3A5*6/*6 (i.e., poor metabolizer status) was associated with a 0.76-fold (95% CI: 0.58 to 0.99) reduction in clearance. In the final model, expected clearance was 8.9 and 6.8 L/hr for a normal and poor metabolizer, respectively, with median population covariates (9 months old, 7.7 kg, low surgical severity).
Keywords: Pediatric, population pharmacokinetics, personalized medicine, pharmacogenetics
Introduction
Fentanyl is a potent opioid commonly used as an anesthetic and analgesic in children and infants.1 Previous pharmacokinetic (PK) studies of fentanyl in children have typically had small sample sizes.2 One of the challenges in conducting such a study is the need for serial blood collection solely for research purposes, which can raise practical and ethical concerns in such a vulnerable population. Residual blood plasma samples obtained from routine clinical care are a proposed source of opportunistic pediatric blood drug concentrations which avoid additional invasive sampling.2 This sampling scheme has already been used in published analyses of fentanyl pharmacokinetics in children.3–5 These irregular, sparse, opportunistic sampling strategies were the original motivation for application of nonlinear mixed effects models toward population pharmacokinetic (popPK) analyses6 and we have used such samples to perform a fentanyl popPK analysis in children.7 This prior study was the largest reported pediatric cohort (N = 130) for a popPK analysis of fentanyl to date but significant residual variability remained, unexplained by the model.
Fentanyl produces adverse effects including fatigue, nausea, vomiting, dizziness, respiratory depression, and bradycardia;8 chronic use can lead to withdrawal symptoms. It is therefore ideal to tailor individual dosage to maintain effective blood concentrations while minimizing risk of adverse effects. PopPK analyses have the potential to illuminate important individual factors to influence dosing decisions, part of which may include genomic data. Fentanyl is known to be metabolized by Cytochrome P450 (CYP) enzymes, CYP3A4 and CYP3A5. Specifically CYP3A4*1G, CYP3A5*3, and CYP3A5*6 alleles have been shown to affect fentanyl PK in adults,9–14 although not all studies have reported significant effects.15 Fentanyl clearance is reduced in neonates and rapidly matures in the first month of life,16 likely due in part to the maturation of CYP3A encoded enzymes. In children, these variants have been shown to be associated with tacrolimus blood concentrations17,18 but no studies to date have detected their effects on fentanyl disposition in pediatric populations using popPK analysis. The ABCB1 variant rs1045642 has also been shown to be associated with reduced fentanyl dosing in children.19 Further, children with congenital heart disease have varying degrees of cardiac insufficiency in the immediate postoperative period, leading to organ hypoperfusion and less effective drug distribution.20 This is partly offset by vasoactive drugs commonly used in the postoperative period which increase hepatic perfusion,21 but may have significant effects on fentanyl, as clearance is perfusion-limited.
The electronic health record (EHR) can be a critical resource in observational PK studies as it is a repository of information collected for clinical purposes. Further, non-clinical data recording can be burdensome to clinicians and unjustifiable during treatment of vulnerable populations such as pediatric intensive care unit (ICU) patients. We have been developing a system for popPK studies using EHRs called “EHR2PKPD”, which includes a set of tools for data extraction and processing from the EHR, implemented in the package EHR22 within the programming language R.23 The functions in this package can build PK datasets in a format common to PK modelling software such as NONMEM24 and Monolix25 using data extracted from EHRs (e.g., demographic, dosing, blood concentration data). This study is one of the first popPK studies performed using our system to generate PK datasets.
The goal of this study is to develop a pediatric fentanyl popPK model with this large cohort and assess the effect of genetic variants in genes CYP3A4, CYP3A5, and ABCB1 on fentanyl PK profile in a pediatric population.
Methods
Study Design
This study was approved by the Vanderbilt University Medical Center Institutional Review Board. Data collection is described in detail in a previous study.7 Briefly, any patient at this tertiary pediatric children’s hospital with congenital heart disease and scheduled for a corrective or palliative operative cardiac procedure was eligible for enrollment in this study. Parents or guardians provided written informed consent along with, when appropriate, informed assent from the patient. Participants were excluded if their surgery was cancelled, if they had known missing documentation for fentanyl dosing, if they required extracorporeal membrane oxygenation in the postoperative period, or if they did not survive to hospital discharge. If a patient had multiple procedures, data were used only from the first surgical procedure. All individuals included in the current study received post-operative care in the pediatric ICU from April 2013 to October 2017; when blood samples were obtained for clinical testing, any “leftover” blood was obtained from the clinical laboratory to be used for research.
Data Collection
Remnant blood plasma specimens of at least 100 μL were retrieved after clinical analyses were complete and processed for fentanyl concentration analysis using high-throughput tandem mass spectrometry as previously described.7 Briefly, after acetonitrile precipitation and addition of d5 fentanyl internal standard, drug concentrations were determined using the Agilent 6430 triple quadrupole fast-scanning electrospray ionization tandem mass spectrometry instrument (Santa Clara, CA). Fentanyl dosing (intravenous infusion and intermittent bolus) and comedication (e.g., CYP3A inducers/inhibitors) data were extracted from the EHR and the Enterprise Data Warehouse, a repository of medical records including drug dispensation and administration, which allows us to reconstruct the schedule of medication dosing during recovery in the ICU. Demographic data were documented by the study team at enrollment. Serum creatinine measurements were extracted from the EHR; given the remnant sample strategy, these laboratory data were generally available from the same time as fentanyl concentrations.
Genotype Data Extraction
Study participants provided anticoagulated blood, saliva, or a buccal swab for genetic analysis and genomic DNA was extracted through the Vanderbilt Technologies for Advanced Genomics (VANTAGE) Core laboratory. Study participants had been previously genotyped using either the Affymetrix Axiom™ Precision Medicine Research Array or the Precision Medicine Diversity Array (Thermo Fisher Scientific, Waltham, MA). As part of genotype data quality control, variants were removed if genotype call rate was <98%, if minor allele frequency was >20% different from 1000 Genomes phase 3 European reference populations, or deviation from Hardy-Weinberg Equilibrium (p-value < 1×10−10). Data for individuals were removed if their genotype call rate was <98%, if the genetically estimated sex differed from parental-reported sex, or if they are related (2nd degree or closer). Genotype data were imputed to the 1000 Genomes phase 3 reference panel. For this study, we extracted data for specific variants in CYP3A5 (*3, 6981A>G, rs776746, NC_000007.14:g.99672916T>C; and *6, 14685G>A, rs10264272, NC_000007.14:g.99665212C>T), CYP3A4 (*1G, 20239G>A, rs2242480, NC_000007.14:g.99763843C>T), and ABCB1 (rs1045642) from the study database.
Assessment of Population Substructure
Principal components (PCs) of ancestry were calculated from the genetic relationship matrix using directly genotyped data with the software package EIGENSOFT,26 incorporating allele frequencies from 1000 Genomes phase 3 reference panel. The first five PCs were included in the PK analysis as covariates to adjust for population substructure.
Data Processing
Data processing was performed using functions from the EHR R package22 within EHR2PKPD. This system collects fentanyl dosing data, demographics, laboratory data, comedication data, and drug concentration data into a form appropriate for popPK analysis. Due to the remnant sample collection strategy, many individuals had serial fentanyl measurements below the fentanyl assay lower limit of quantification (BLOQ) of 0.05 ng/mL as samples taken after fentanyl treatment was completed. We included only the first of these BLOQ measurements for each individual, excluding the rest. We excluded individuals with only BLOQ drug concentration measurements. We also excluded drug concentration measurements above the upper limit of quantification of 50 ng/mL.
Population PK Analysis
We performed a popPK analysis in Monolix® 2021R with the stochastic approximation of expectation and maximization (SAEM) method. To handle the BLOQ observations, we used a likelihood method that treats the BLOQ observations as censored,27 equivalent to the NONMEM® M3 method. We first selected the base model, assuming a lognormal distribution for the random effects PK parameters. After selecting the base model, we prespecified a fixed allometric scaling factor centered at 70 kg on each of the main PK parameters as recommended by Holford et al.28 With this model, we further performed covariate modeling with a priori selected candidate covariates based on previous research and biological plausibility: age (months, recorded at study entry), sex, race (per parent report at study enrollment), serum creatinine, Society of Thoracic Surgery–European Association for Cardio-Thoracic Surgery (STAT) Congenital Heart Surgery Mortality Score,29 cardiopulmonary bypass time, and co-medications for CYP3A inducer (coded 2 if any drug classified as an inducer is present, 1 otherwise) and CYP3A inhibitor (coded 2 if any drug classified as an inhibitor is present, 1 otherwise). In addition, because CYP3A allele frequencies covary with both ancestral origins and PK outcomes, the first five ancestry PCs were included as covariates to account for confounding by population substructure. Continuous covariates were standardized by dividing by their median while ordered covariates such as genotypes were coded additively as 1, 2, …, m where m is the number of unique levels in the variable. Once covariates were selected, effects of genetic variants on clearance were tested. CYP3A5*3, CYP3A5*6, CYP3A4*1G, and rs1045642 were assessed separately. For CYP3A5, we also assigned those individuals who had no variant alleles as normal metabolizers (i.e., CYP3A5*1/*1), one variant allele as intermediate metabolizers (i.e., CYP3A5*1/*3 or *1/*6), and 2 variant alleles as poor metabolizers (i.e., CYP3A5*3/*3, *3/*6, or *6/*6). Age was also tested against a maturation model using gestational age, a coefficient for half-adult-clearance-age, and a Hill coefficient as described in Holford et al.28
Model covariate selection was performed based on the difference in the objective function (−2 × log-likelihood) and the number of parameters, which would approximately follow a χ2 distribution. An objective function value decrease of 3.84 (with 1 degree of freedom) corresponds to a p-value of 0.05 in a likelihood ratio test, which is the cutoff for covariate inclusion as a significant model improvement. Race and ancestry PC covariates were tested with cutoffs reflecting the degrees of freedom in each. Genetic effects were added separately to the covariate model and the Wald test determined inclusion based on a p-value of 0.05, which was used to estimate the 95% confidence interval (CI). As the SAEM is a stochastic method, it could yield varying results especially when data are sparse.30 For this reason, all models were repeatedly fit with 5 distinct seeds and the estimates corresponding to the minimum objective function value were used for model selection as well as final reported results. For model assessment, we examined goodness-of-fit plots using population and individual predicted fentanyl blood concentrations, and residual plots using individual weighted residuals (IWRES). In addition, we used a visual predictive check (VPC) to compare model predicted concentrations across time. Model assessment plots were generated using R version 4.0.2.23
Simulation
Using the final model with covariates including genetic variants (CYP3A5*3 or *6 variants), we checked for clinical significance using simulation. We established a set of profiles typical for younger (2 months, 5 kg), average age (9 months, 8 kg), and older (120 months, 32 kg) individuals. For each of these profiles, we simulated 1000 random effects for Cl, V1, Q, and V2 according to our model estimates, making 3000 simulated PK profiles. We further replicate each profile into 3 individuals with normal, intermediate, or poor metabolizer status but who are otherwise identical. Using these PK profiles, we calculated the expected response to a 1 mcg/kg intravenous (IV) bolus followed by a 1, 2, 3, and 4 mcg/kg/hour IV infusion over 12 hours. We reported the proportion of individuals which is expected to reach or exceed the therapeutic drug range (TDR, 1 to 3 ng/mL)31 as well as the mean times to reach those cutoffs among the subset of patients who reach them within the 12 hour window.
Results
Study Population and Sample Characteristics
Characteristics of our study cohort are summarized in Table 1 and further details are in Table S1 of the Supplemental Materials. We collected demographics, blood concentrations, and dosing data from 519 individuals. Twenty individuals were excluded, who did not survive to discharge (n=9) or required extracorporeal membrane oxygenation postoperatively (n=11). This left 2928 fentanyl blood concentrations from 499 individuals. Of those concentrations, 704 were censored as BLOQ measurements (after the first BLOQ measurement). Twenty-two individuals were excluded as they had only BLOQ concentrations. Forty-three additional individuals were excluded for having no genetic data available. This yielded 434 individuals with 1937 fentanyl blood concentrations, of which 21% were BLOQ. The number of blood concentrations per individual was a range of 1 to 30 with a median of 4 (interquartile range [IQR] 3 – 5). There were 4131 fentanyl dosing events for the final analysis. The population medians for weight and age are 7.7 kg (IQR 5.3 – 16.9) and 9.0 months (IQR 3.8 – 58.5). The median time from first ICU-administered fentanyl dose to first blood concentration sample was 6.9 hours (IQR 5.3 – 16.0).
Table 1:
Summary of Study Cohort.
| Whole cohort |
CYP3A5 normal metabolizers |
CYP3A5 intermediate metabolizers | CYP3A5 poor metabolizers |
CYP3A4
*1/*1 |
CYP3A4
*1/*1G |
CYP3A4
*1G/*1G |
|
|---|---|---|---|---|---|---|---|
|
|
|||||||
| N | 434 | 10 | 76 | 348 | 311 | 90 | 33 |
| Male sex | 226 (52%) | 6 (60%) | 32 (42%) | 188 (54%) | 166 (53%) | 44 (49%) | 16 (48%) |
| Weight at enrollment (kg) | 7.7 [5.3, 16.9] {1.9, 138.0} |
14.2 [6.7, 17.5] {3.4, 37.8} |
7.6 [5.1, 15.8] {2.1, 82.3} |
7.7 [5.3, 17.0] {1.9, 138.0} |
7.8 [5.3, 17.8] {1.9, 138.0} |
7.6 [5.5, 13.6] {2.1, 102.7} |
9.2 [4.6, 17.1] {2.8, 82.3} |
| Age at enrollment (months) | 9.0 [3.8, 58.5] {0.1, 270.9} |
41.1 [5.5, 43.9] {0.4, 126.9} |
8.2 [3.8, 50.6] {0.1, 232.3} |
9.0 [3.8, 59.4] {0.1, 270.9} |
8.9 [3.8, 61.7] {0.1, 270.9} |
8.2 [4.2, 37.2] {0.1, 246.2} |
18.3 [4.0, 62.8] {0.2, 214.1} |
| Race | |||||||
| White | 357 (82.3%) | 0 (0.0%) | 46 (60.5%) | 311 (89.4%) | 292(94.9%) | 59 (65.6%) | 6 (18.2%) |
| Black | 52 (12.0%) | 10 (100%) | 24 (31.6%) | 18 (5.2%) | 7 (2.3%) | 20 (22.2%) | 25 (75.8%) |
| Asian | 6 (1.4%) | 0 (0.0%) | 0 (0.0%) | 6 (1.7%) | 3 (1.0%) | 3 (3.3%) | 0 (0.0%) |
| American Indian or Alaska Native | 2 (0.5%) | 0 (0.0%) | 0 (0.0%) | 2 (0.5%) | 2 (0.6%) | 0 (0.0%) | 0 (0.0%) |
| Other | 6 (1.4%) | 0 (0.0%) | 2 (2.6%) | 4 (1.1%) | 2 (0.6%) | 3 (3.3%) | 1 (3.0%) |
| Unknown | 11 (2.5%) | 0 (0.0%) | 4 (5.2%) | 7 (2.0%) | 5 (1.6%) | 5 (5.6%) | 1 (3.0%) |
| Cardiac bypass time (hours) | 1.7 [1.1, 2.4] {0.0, 7.1} |
1.4 [1.0, 1.8] {0.0, 3.2} |
1.6 [1.0, 2.2] {0.0, 5.8} |
1.7 [1.2, 2.5] {0.0, 7.1} |
1.7 [1.1, 2.4] {0.0, 6.2} |
1.8 [1.2, 2.7] {0.0, 7.1} |
1.3 [0.7, 1.6] {0.0, 4.1} |
| Length of ICU stay (days) | 3 [2, 6] {0, 119} |
3 [1, 4] {1, 119} |
3 [2, 4] {1, 54} |
3 [2, 6] {0, 87} |
3 [2, 6] {0, 79} |
3 [2, 5] {1, 87} |
3 [2, 5] {1, 119} |
| STAT category | |||||||
| 1 | 164 (37.8%) | 4 (40.0%) | 28 (36.8%) | 132 (37.9%) | 120(38.6%) | 30 (33.3%) | 14 (42.4%) |
| 2 | 124 (28.6%) | 3 (30.0%) | 24 (31.6%) | 97 (27.9%) | 78 (25.1%) | 33 (36.7%) | 13 (39.4%) |
| 3 | 56 (12.9%) | 2 (20.0%) | 10 (13.2%) | 44 (12.6%) | 42 (13.5%) | 12 (13.3%) | 2 (6.1%) |
| 4 | 74 (17.1%) | 1 (10.0%) | 10 (13.2%) | 63 (18.1%) | 59 (19.0%) | 12 (13.3%) | 3 (9.1%) |
| 5 | 16 (3.7%) | 0 (0.0%) | 4 (5.3%) | 12 (3.4%) | 12 (3.9%) | 3 (3.3%) | 1 (3.0%) |
| CYP3A4*1G | |||||||
| 0 | 311 (71.7%) | ||||||
| 1 | 90 (20.7%) | ||||||
| 2 | 33 (7.6%) | ||||||
| CYP3A5*6 | |||||||
| 0 | 411 (94.7%) | ||||||
| 1 | 21 (4.8%) | ||||||
| 2 | 2 (0.5%) | ||||||
| CYP3A5*3 | |||||||
| 0 | 22 (5.1%) | ||||||
| 1 | 77 (17.7%) | ||||||
| 2 | 335 (77.2%) | ||||||
| CYP3A5 metabolizer status | |||||||
| Normal | 10 (2.3%) | ||||||
| Intermediate | 76 (17.5%) | ||||||
| Poor | 348 (80.2%) | ||||||
| ABCB1 rs1045642 | |||||||
| GG | 121 (27.8%) | ||||||
| AG | 213 (49.1%) | ||||||
| AA | 100 (23.0%) | ||||||
| Fentanyl blood concentrations per individual | 4 [3, 5] {1, 30} |
||||||
| Fentanyl blood concentration (ng/mL) | 0.25 [0.06, 1.31] {0.00, 34.25} |
||||||
| Fentanyl dosing events per individual | 7 [5, 11] {1, 56} |
||||||
Summary statistics are presented as count (percent) for categorical variables, and median [interquartile range] {range} for continuous variables. Selected covariates are also tabulated according to CYP3A5 metabolizer status and CYP3A4*1G variants. ICU, intensive care unit; STAT, Society of Thoracic Surgery–European Association for Cardio-Thoracic Surgery Congenital Heart Surgery Mortality Score.
There were 311 individuals (71.1%) with no CYP3A4*1G alleles, 90 (20.7%) with 1 allele and 33 (7.6%) with 2 alleles. For CYP3A5*6, 411 individuals (94.7%) had no alleles, 21 (4.8%) had 1 allele, and 2 (0.5%) had 2 alleles. For CYP3A5*3, 22 individuals (5.1%) had no alleles, and 77 (17.7%) and 335 (77.2%) had 1 and 2 alleles, respectively. Based on both CYP3A5*3 and *6, 10 (2.3%) of the cohort were normal metabolizers, 76 (17.5%) were intermediate metabolizers, and 348 (80.2%) were poor metabolizers. For the rs1045642 variant in ABCB1, 213 (49.1%) were heterozygous, and 100 (23.0%) were homozygous for the A allele.
Population PK Analysis
A two-compartment model was selected as the base model (objective function = 2850 vs. 3545 for the two- vs. one-compartment model), which has also been used in previous work for fentanyl PK analysis in children. The two-compartment model was parameterized in terms of total clearance (CL, L/hr), volume of distribution for the central compartment (V1, L), inter-compartmental clearance (Q, L/hr), and volume of distribution for the peripheral compartment (V2, L). We allowed for random effects on these four major PK parameters and selected a combined proportional and additive residual error model. The results of the population PK analysis are presented in Table 2. PK parameter estimates from the base model along with coefficient of variation (%CV) were CL = 6.2 L/hr (63.7%), V1 = 12.9 L (87.5%), Q = 3.2 L/hr (49.7%), and V2 = 43.5 L (67.6%).
Table 2:
Pharmacokinetic Model Parameter Estimates.
| Base Model | Base+Weight Model | CYP3A4*1G Model | Final Model | ||||
|---|---|---|---|---|---|---|---|
| Obj = 2850 | Obj = 2384 | Obj = 2371 | Obj = 2369 | ||||
|
| |||||||
| Parameter | Est (SE) [CI] | Parameter | Est (SE) [CI] | Parameter | Est (SE) [CI] | Parameter | Est (SE) [CI] |
| CL= θ1* × (wt/70)0.75 | CL = θ1 × (wt/70)0.75 × (age/9)θ2 × (STAT)θ3 × (CYP3A4*1G)θ4 | CL = θ1 × (wt/70)0.75 × (age/9)θ2 × (STAT)θ3 × (CYP3A5)θ4 | |||||
| CL | 6.2 (0.2) [5.8,6.6] |
θ1* | 34.7 (1.0) [32.7,36.7] |
θ1 | 35.9 (1.7) [32.6,39.2] |
θ1 | 46.6 (6.2) [34.4, 58.8] |
| θ2 | 0.03 (0.01) [0.01,0.05] |
θ2 | 0.04 (0.01) [0.02, 0.06] |
||||
| θ3 | −0.08 (0.05) [−0.18,0.02] |
θ3 | −0.06 (0.05) [−0.16, 0.04] |
||||
| θ4 | 0.03 (0.07) [−0.11,0.17] |
θ4 | −0.25 (0.12) [−0.49, -0.01] |
||||
| V1 = θ2* × (wt/70)1 | V1 = θ5 × (wt/70)1 | V1 = θ5 × (wt/70)1 | |||||
| V1 | 12.9 (1.3) [10.4,15.4] |
θ2* | 269.3 (20.2) [229.7,308.9] |
θ5 | 261.9 (20.0) [222.7,301.1] |
θ5 | 277.5 (19.4) [239.5,315.5] |
| Q = θ3* × (wt/70)0.75 | Q = θ6 × (wt/70)0.75 | Q = θ6 × (wt/70)0.75 | |||||
| Q | 3.2 (0.2) [2.8,3.6] |
θ3* | 19.9 (1.4) [17.2,22.6] |
θ6 | 20.5 (1.5) [17.6,23.4] |
θ6 | 20.7 (1.5) [17.8, 23.6] |
| V2 = θ4* × (wt/70)1 | V2 = θ7 × (wt/70)1 | V2 = θ7 × (wt/70)1 | |||||
| V2 | 43.5 (2.8) [38.0,49.0] |
θ4* | 517.9 (37.9) [443.6,592.2] |
θ7 | 532.4 (33.0) [467.7,597.1] |
θ7 | 528.8 (37.3) [455.7,601.9] |
| ωCL (%CV) | 63.7 (2.7) [58.3,68.9] |
ωCL (%CV) | 49.3 (2.2) [45.0,53.6] |
ωCL (%CV) | 47.9 (2.1) [43.8,52.0] |
ωCL (%CV) | 47.9 (2.1) [43.8,52.0] |
| ωV1 (%CV) | 87.5 (8.2) [71.4,103.6] |
ωV1 (%CV) | 65.1 (5.5) [54.3,75.9] |
ωV1 (%CV) | 57.4 (5.3) [47.0,67.8] |
ωV1 (%CV) | 59.3 (5.5) [48.5,70.1] |
| ωQ (%CV) | 49.7 (8.5) [33.0,66.4] |
ωQ (%CV) | 50.7 (12.9) [25.4,76.0] |
ωQ (%CV) | 73.4 (7.5) [58.7,88.1] |
ωQ (%CV) | 70.0 (8.8) [52.8,87.2] |
| ωV2 (%CV) | 67.6 (5.0) [57.9,77.5] |
ωV2 (%CV) | 76.9 (8.0) [61.2,92.6] |
ωV2 (%CV) | 68.9 (5.8) [57.5,80.3] |
ωV2 (%CV) | 70.0 (5.8) [58.6,81.4] |
| σprop (%CV) | 47.0 (1.8) [43.5,50.5] |
σprop (%CV) | 45.4 (1.6) [42.3,48.5] |
σprop (%CV) | 45.5 (1.6) [42.4,48.6] |
σprop (%CV) | 44.9 (1.6) [41.8,48.0] |
| σadd (ng/mL) |
0.02 (0.01) [0.00,0.04] |
σadd (ng/mL) | 0.01 (<0.01) [0.00,0.02] |
σadd (ng/mL) | 0.01 (<0.01) [0.00,0.02] |
σadd (ng/mL) | 0.02 (<0.01) [0.01,0.03] |
Pharmacokinetic parameter estimates are presented as estimate (standard error) [Wald 95% confidence interval] and Obj denotes model objective function value. The wt, age, and STAT represent covariates values. CYP3A4*1G is coded continuously as 1, 2, 3, for 0, 1, and 2 CYP3A4*1G variants. CYP3A5 is coded continuously as 1, 2, 3 for 0, 1, and 2 no function (CYP3A5*3 or CYP3A5*6) variants, respectively. CL, V1, Q, and V2 are total clearance (L/hr), central compartment volume of distribution (L), intercompartmental clearance (L/hr), and peripheral compartment volume of distribution (L). The ω parameters are between-individual variance components for their respective pharmacokinetic parameters, presented as %CV. The σprop and σadd represent the proportional and additive residual errors in the combined residual error model, presented as %CV and the standard deviation, respectively. The θs denote model parameters as typically used in statistical models.
Inclusion of weight on all the main PK parameters with fixed allometric scaling parameters markedly improved the model fit (objective function reduction of 466 from the base model). Additional inclusion of age and STAT significantly improved the model fit (objective function reduction of 9.8 and 6.9, respectively), while other covariates including inducer- and inhibitor-comedication, race, ancestry PCs, and cardiopulmonary bypass time were not selected as they did not improve the model fit. A maturation factor for gestational age did not yield any improvement in objective function over age, so was not included in the final model. Independent inclusion of CYP3A5*3 and CYP3A5 normal/intermediate/poor metabolizer status were statistically significant (p=0.043 and p=0.046, respectively). CYP3A5 metabolizer status, denoted by CYP3A5 in the model, was included in our final model.
The structure of the final model is as follows:
and
where CLi, V1i, Qi, and V2i are the individual-specific CL, V1, Q, and V2 for individual i, wt is body weight in kilogram (kg), age is individual age in months, STAT is individual surgical severity score ranging from 1 to 5, and CYP3A5 is metabolizer status, coded by 1, 2, and 3 for normal, intermediate, and poor metabolizer, respectively. The ηiCL, ηiV1, ηiQ, and ηiV2 are random effects explaining between-individual variability for CL, V1, Q, and V2, which follow a lognormal distribution with means zero and covariance matrix with ω2CL, ω2V1, ω2Q, and ω2V2 in the diagonal. The θs in the equations denote model parameters as typically used in statistical models. Estimates (%CV) for typical parameter values for an individual of weight 7.7 kg, age 9 month, with STAT score 1 and normal metabolizer function are CL = 8.9 L/hr (47.9%), V1 = 30.5 L (59.3%), Q = 4.0 L/hr (70.0%), and V2 = 58.2 L (70.0%). Compared to the %CV of 63.7% for CL in the base model, the %CV in the final model, 47.9%, is substantially reduced due to the inclusion of covariates which explain some of the interindividual variability in CL.
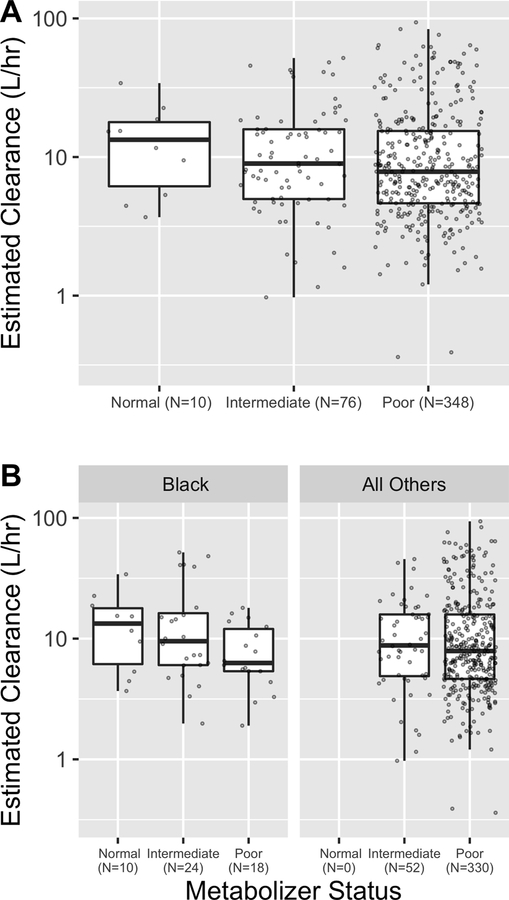
The estimates for the effect on CL (with 95% Wald CI) for CYP3A4*1G, CYP3A5*3, CYP3A5*6, and rs1045642 variants are 0.03 [−0.11, 0.17], −0.20 [−0.39, −0.01], 0.13 [−0.19, 0.45], and 0.01 [−0.12, 0.14], respectively. These correspond to 1.03-fold [0.89, 1.21], 0.80-fold [0.65, 0.99], 1.15-fold [0.81, 1.64], and 1.01-fold [0.88, 1.17] changes to CL in the presence of two CYP3A4*1G, CYP3A5*3, CYP3A5*6, or rs1045642 AA variants. The estimate and 95% CI for a CYP3A5 metabolizer status is −0.25 [−0.49, −0.01], corresponding to a 0.76-fold [0.58, 0.99] change to CL in the presence of two CYP3A5 variants. We proceed to report results in terms of this composite metabolizer status because it is clinically meaningful and has the greater estimated effect size (−0.25 vs. −0.20). Figure 1 (A) shows that estimated CL for normal, intermediate, and poor metabolizers have medians (horizontal lines) of 13.5 L/hr, 9.0 L/hr, and 7.8 L/hr, respectively. Median estimated CL for the entire population is 8.2 L/hr.
Figure 1: Individual estimated clearance by CYP3A5 metabolizer status.
Clearance is summarized for normal, intermediate, and poor metabolizers. The hinge is set at the median, the boxes span the 25th to 75th percentile, and the whiskers extend to the most extreme datum which is at most 1.5 times the interquartile range. Clearances are shown for (A) the entire cohort and (B) separately for individuals whose self-reported race was Black vs. the rest of the cohort.
Inclusion of the first five ancestry PCs did not significantly reduce the objective value function and did not impact the major parameters of interest such as the coefficient for CYP3A5 metabolizer status, and so the PCs did not remain in the final model. Figure 1 (B) shows estimated clearance from the final model by CYP3A5 metabolizer status, separately for the individuals with self-reported race of Black. Addition of either race or ancestry PCs to the final model did not impact estimation of the size of the CYP3A5 metabolizer effect (−0.24 with race and −0.26 with ancestry PCs).
The estimated effect for STAT is −0.06 [−0.16, 0.04] corresponding to a 0.91-fold [0.77, 1.07] change in CL when STAT is 5 compared to 1 (i.e., two extreme categories, comparing highest mortality risk to lowest mortality risk for the congenital heart surgery procedure). The estimated effect for age is 0.04 [0.02, 0.06] corresponding to a 1.08-fold [1.04, 1.12] change in CL when age is 5 years compared to 9 months.
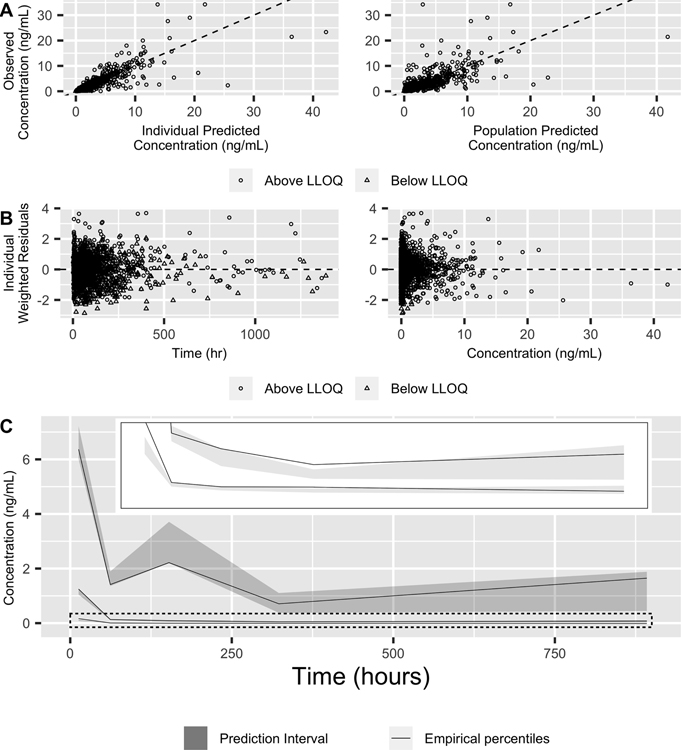
Goodness-of-fit plots for the final model support that overall the model fit is appropriate; Observed vs. predicted values (Figure 2A) are not systematically above or below the diagonal line of equality, although there were few outlying predictions. The individual weighted residuals (Figure 2B) show symmetric distribution around the zero line, also indicating good model fit. In general, the visual predictive check (Figure 2C) reveals that simulated percentiles from the model are reasonably well aligned with observed data, especially at higher concentrations.
Figure 2: Model diagnostics for the final population pharmacokinetic model.
(A) Observed vs. predicted concentrations for both individual (left) and population (right) predicted concentrations with the identity line (dashed). Open circles and triangles represent above- and below-lower limit of quantification observations, respectively. (B) Individual weighted residuals as a function of time (left) and individual predicted concentrations (right) with a dashed line for zero residuals. (C) The visual predictive check. Solid lines represent empirical quartiles of concentration and shaded regions represent the 10th and 90th quantiles of each quartile as determined by simulations. The dotted rectangle is enlarged in the inset to better visualize the median and lower quartile.
Genetic Effects from the Simulation Study
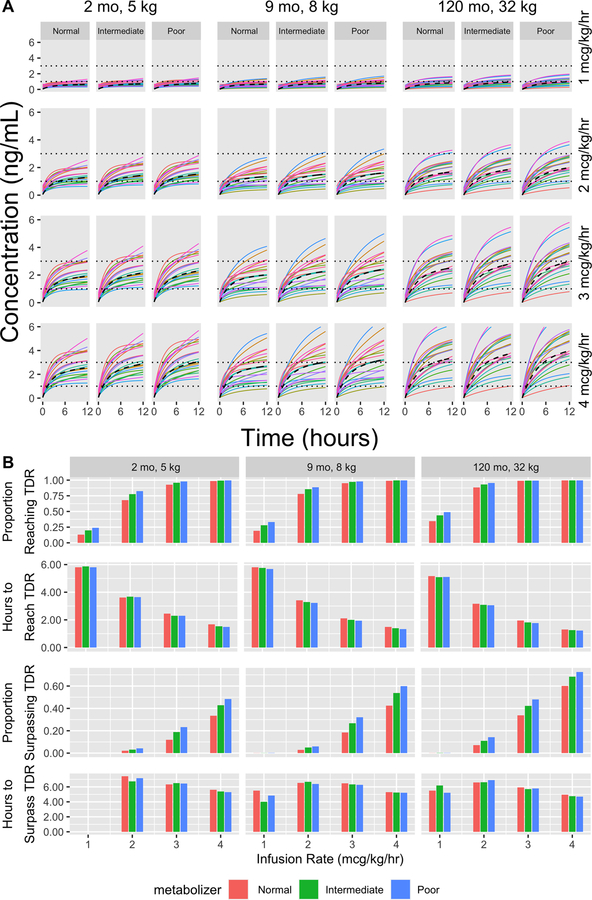
Figure 3A shows the dose response for a random sample of 20 simulated individuals, grouped by age/weight and infusion rate. Each individual is simulated to receive a 1 mcg/kg IV bolus at time 0 and then receives the specified IV infusion for 12 hours. The TDR cutoffs are shown as horizontal dotted lines. The population average response is shown as a dashed line. It is clinically desirable to reach the TDR quickly and avoid surpassing it. Variability shown in the response curves is indicative of estimated inter-individual variability in PK parameters as represented by the ω parameters in Table 2. Figure 3B shows the summary of the full simulation with 1000 individuals in each scenario receiving a 1 mcg/kg loading bolus followed by 1, 2, 3, or 4 mcg/kg/hr infusions over 12 hours, representing a reasonable but longer continuous infusion time in our dataset. The estimated effect of metabolizer status is apparent in how the normal metabolizer group is typically the last to enter/leave the TDR and the least likely to pass those thresholds. For example, in the 9 month/8 kg population receiving 4 mcg/kg/hr infusions, the percentage of normal metabolizers surpassing the TDR upper limit of 3ng/mL is 42% compared to 60% for poor metabolizers. For the same population receiving 1 mcg/kg/hr, 19% of normal metabolizers compared to 33% of poor metabolizers reach the lower TDR cutoff of 1 ng/mL during the 12 hour infusion. Generally, per kg dose response is more modulated by age/weight than by metabolizer status, with older and heavier individuals requiring lower doses to achieve similar responses. Note that in some instances, none of the simulated individuals surpassed the TDR and no bar is displayed for “Hours to surpass TDR”. In other instances a small proportion of individuals in a group surpass the TDR, and hence “Hours to Surpass TDR” is subject to chance variation due to the small sample.
Figure 3: Simulation results.
(A) Expected response to varying intravenous (IV) infusion rates, 1 – 4 mcg/kg/hr, following a 1mcg/kg IV bolus for populations from three hypothetical groups with varying age and weight (from left to right, younger: 2 months, 5 kg; average: 9 months, 8 kg); older: 120 months, 32 kg). Within each scenario, we simulate 20 normal, intermediate, and poor metabolizers (left to right) and calculate their expected dose response over 12 hours, which is presented in different color for each individual. Horizontal dotted lines represent the therapeutic drug range (TDR) for fentanyl blood concentration and a dashed line represents the population average response. Each of the panels under their respective age/weight categories represent the same 20 simulated random effects but applied to the indicated metabolizer status/infusion rate to allow for easy comparison of the genetic and dosage effects. (B) Summary of complete simulation results for the three hypothetical groups with normal/intermediate/poor metabolizers receiving 1/2/3/4 mcg/kg/hr infusions. For each scenario, 1000 individuals were simulated. We report the proportion of individuals who reach the TDR, the average time to reach the TDR (among those who do reach it), the proportion of individuals who surpass the TDR, and the average time to surpass the TDR (among those who do so). When no bar is present in “Hours to Surpass TDR” none of the 1000 simulated patents reached the cutoff within 12 hours.
Discussion
We successfully developed a fentanyl popPK model for children using only opportunistically collected blood samples which minimized the burden of study participation on the enrolled individuals. Our PK parameter estimates are in line with published results in adults and children, and this study is the first to demonstrate the effect of CYP3A5 no function variants on clearance in a popPK analysis of children. Through the development of our EHR2PKPD system and the increasing role of EHRs in automating data collection as part of clinical care, these kinds of opportunistic studies should become more feasible over time to collect data with the large sample sizes necessary for popPK analysis while minimizing the burden of data collection in vulnerable populations. As reported by Choi et al.32 estimates for a complete analysis of fentanyl population pharmacokinetics are comparable between manually built data and our automized system built data. Given that all datasets including blood concentration measurements and dosages were collected outside of our EHR2PKPD system and we only used our system implemented in the EHR R package to process and build the dataset, there is little possibility for our automated data building algorithms to introduce data errors. Regarding the data building algorithms in our system, they were validated during their development in such a way that we iteratively revised them by comparing the algorithm generated data with gold standard data until both datasets were identical. Model fit was overall good (Figure 2) but there was considerable interindividual variability in the four PK parameters of interest. Future studies done on populations with longer exposure to fentanyl may be suitable for population PK profiling to gain a better sense of the true interindividual variability in PK parameters in the population. For now, it is difficult to make clinical recommendations on the basis of the genetic effect in the presence of so much unexplained interindividual variability, although one study has reported that CYP3A5 genotypes are associated with fentanyl dosing, but not drug levels, in a multiple regression analysis.33
There are a few papers published using opportunistic samples from children (e.g., studies with 98 preterm neonates,3 14 pediatric burn patients,34 and 66 critically ill children).4 The study performed by Hagos et al.4 with 278 samples from 66 critically ill children is most relevant to our study. They presented typical CL for a 70 kg individual with no CYP3A4/5 inducers as 34.6 L/hr. We estimated the population-level clearance for a 70kg individual as 34.7 L/hr from our Base+Weight Model (without inducer, which was not included in our final model). Notably, the authors did not find a significant CYP3A5 genetic effect on clearance and speculated that their smaller sample size was likely the reason. Our sample size (1937 observations in 434 individuals) was larger than theirs and our confidence interval for the genetic effect is close to including 0, suggesting that we would likely not have detected the effect either if our sample size were smaller.
CYP3A5*6 has been shown in previous studies to reduce fentanyl clearance but our estimate for the effect is positive because the effect is being compared to all variants in the population, not just to the CYP3A5*1 allele. Because the CYP3A5*3 no function variant dominates in this dataset (77% of individuals have the CYP3A5*3/*3 genotype) the positive effect of the CYP3A5*6 allele indicates mostly that the variant does not reduce clearance as much as the CYP3A5*3 allele. Regardless, we suggest that the two variants should be considered jointly, as the physiological loss-of-function effect of both variants is well-established and testing for both variants is routinely performed on panel-based pharmacogenetic tests. CYP3A4*1G and rs1045642 have been associated with fentanyl clearance and dosing13,19 but neither showed evidence of impacting clearance in our analysis.
The difference in estimated clearance based on CYP3A5 metabolizer status is obscured in Figure 1 (A) by the differential distribution of weight in the subgroups, with normal metabolizers being older and heavier (median weight 14.2 kg vs. 7.6 kg and 7.7 kg for intermediate and poor metabolizers, respectively). Figure S1 provides clearance standardized by a fixed weight of 7.7 kg for each individual. The weight-standardized clearances show a similar trend by metabolizer status, supporting CYP3A5 metabolizer effects. Inter-individual variation in clearance is estimated at 47.9 %CV so the level of unexplained variability is still substantial. To further illustrate this, Figure 3A compares expected blood concentrations during typical infusions given in a clinical setting by simulating individual PK parameters from the final model for various age/weight populations. The expected concentration profile shows the effect of CYP3A5 function, but it is not uncommon for some individuals with normal metabolizer status to reach or exceed the TDR more quickly than individuals with intermediate/poor metabolizer status. If residual variability were also considered, the impact of CYP3A5 metabolizer status would be obscured further. Even though this is the largest cohort used in the study of pediatric fentanyl PK, a clinically meaningful characterization of fentanyl popPK may require an even larger cohort. This may only be feasible through the opportunistic sampling strategy presented in this work. This work serves as an example of how such data can be used to perform clinically relevant pharmacogenomic studies when the burden of structured PK data collection is too great.
The fact that each of the 10 normal metabolizers and a disproportionate number of the intermediate metabolizers in the cohort had a self-reported race of “Black or African American” reflects the potential confounding between race and CYP3A5 metabolizer status.35 As a covariate, race was not significant but the possibility remains that the apparent effect of CYP3A5 metabolizer status on clearance reflects factors mediated through race. Further, it is possible that the effect of race was reduced due to the categorical classification of self-reported race. Ancestry PCs are continuous covariates that account for genetic ancestry and are correlated with self-reported race (Figure S2). In our data, neither categorical race nor continuous ancestry PCs improved model fit or impacted the effect of CYP3A5 metabolizer status on clearance.
While sample size does not permit analyses stratified by race, Figure 1 (B) allows examination of model estimated clearance by race, which confirms that the apparent effect of CYP3A5 metabolizer status is consistent. Further, we observed that addition of either race or ancestry PCs in the final model did not impact the scale of the observed genetic effect. All of this supports that the CYP3A5 metabolizer effect observed in our analysis would not be spurious, however the analysis would be considerably strengthened by increased sample size for underrepresented groups.
There are some notable limitations in this study. The major limitation is the use of an EHR data source with opportunistic samples, which may not provide optimal data. The number of samples per patient is limited, and the sampling times are not optimized to estimate PK parameters. In addition, rare instances of EHR downtime or emergent bedside procedures can lead to dosing documentation on paper forms which are scanned into the EHR and not extractable by EHR data pulls or natural language processing. In our manual data review of over 650 fentanyl concentrations and associated fentanyl dosing, we identified 3 instances of inaccurate dosing, all due to emergent beside procedures. The data for these 3 individuals were truncated to include only observations with accurate dosing data. Although this study design allowed for a large study cohort, a combination of sparse sampling and data errors would increase interindividual and residual variability, making it difficult to make clinical recommendations regarding individualized dosing even when the estimated effect of age, weight, STAT score, and genetics are significant. There may be other covariates not available for our dataset (e.g., markers of liver function) that can improve the model. Our goal was to assess a variety of genetic effects on clearance, but our populations were not optimized for detection of such effects. Some allele frequencies were small (18% CYP3A4*1G, 3% CYP3A5*6) and hence power to detect their effects on clearance was reduced.
This study is evidence that CYP3A5 no function variants impact fentanyl clearance in pediatric populations, but the scale of this effect is small compared to the level of unexplained variability in the model. Some possibilities for this are that the genetic effect is smaller in children than in adults, variability in dose-response is greater in children than in adults, or that genetic effects appear relatively smaller in children due to the higher impact of weight and age on PK profile. Beyond those we tested, there are further genetic variants such as more in the ABCB1 transporter gene which have been shown to influence fentanyl dosing and pharmacokinetics.9,19 Opportunistic sampling schemes may empower genome wide association studies which can explore these effects exhaustively and suggest further areas of study. Future researchers should be aware of these possibilities when considering the benefits and drawbacks of opportunistic sampling schemes to produce well-powered studies which yield results that are relevant to clinical care.
Supplementary Material
Study Highlights.
What is the current knowledge on the topic?
CYP3A5*3/*6 and CYP3A4*1G variants impair clearance of a wide range of drugs in adults, including fentanyl. These findings have not been replicated in a pediatric population where small sample sizes typically limit the power to detect such an effect.
What question did this study address?
Do these variants impact fentanyl clearance in a population of children recovering from cardiac procedures? Is the estimated effect large enough to impact clinical dosing decisions given unexplained interindividual and residual variability?
What does this study add to our knowledge?
This study is the largest reported cohort of children for fentanyl pharmacokinetics and the resulting model can be a benchmark for future studies. There is little evidence for a CYP3A4*1G effect, but CYP3A5*3/*6 variants significantly reduce fentanyl clearance. Considering the unexplained variability in our model, the reduction in clearance may not be large enough to change fentanyl dosing decisions.
How might this change clinical pharmacology or translational science?
Clinicians may consult our simulation study and decide whether they want to consider CYP3A5*3/*6 variants when making dosing decisions.
Acknowledgements
The authors have no acknowledgments to make.
Funding Information
This work is supported by NIH GM124109. The work is supported in part by R01HD084461 from the Eunice Kennedy Shriver National Institute of Child Health & Human Development and Clinical Scientist Development Award 2017075 from the Doris Duke Foundation. Dr. Leena Choi is the principal investigator for grant R01-GM124109. This study used REDCap, supported by UL1 TR000445 from NCATS/NIH.
Footnotes
Conflict of Interest
The authors declared no competing interests for this work. As an Associate Editor for Clinical Pharmacology and Therapeutics, Sara L. Van Driest was not involved in the review or decision process for this paper.
References
- 1.Ziesenitz VC, Vaughns JD, Koch G, Mikus G & Anker JN van den Pharmacokinetics of fentanyl and its derivatives in children: a comprehensive review. Clin. Pharmacokinet 57, 125–149 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leroux S et al. Pharmacokinetic Studies in Neonates: The Utility of an Opportunistic Sampling Design. Clin. Pharmacokinet 54, 1273–1285 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Völler S et al. Rapidly maturing fentanyl clearance in preterm neonates. Arch. Dis. Child Fetal Neonatal Ed 104, F598–F603 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Hagos FT et al. Factors Contributing to Fentanyl Pharmacokinetic Variability Among Diagnostically Diverse Critically Ill Children. Clin. Pharmacokinet 58, 1567–1576 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maharaj AR et al. Dosing of Continuous Fentanyl Infusions in Obese Children: A Population Pharmacokinetic Analysis. J. Clin. Pharmacol 60, 636–647 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sheiner LB, Rosenberg B & Marathe VV Estimation of population characteristics of pharmacokinetic parameters from routine clinical data. J. Pharmacokinet. Biopharm 5, 445–479 (1977). [DOI] [PubMed] [Google Scholar]
- 7.Van Driest SL et al. Pragmatic pharmacology: population pharmacokinetic analysis of fentanyl using remnant samples from children after cardiac surgery. Br. J. Clin. Pharmacol 81, 1165–1174 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stanley TH The Fentanyl Story. J. Pain 15, 1215–1226 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Saiz-Rodríguez M et al. Polymorphisms associated with fentanyl pharmacokinetics, pharmacodynamics and adverse effects. Basic Clin. Pharmacol. Toxicol 124, 321–329 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Zhang W et al. Influence of CYP3A5*3 polymorphism and interaction between CYP3A5*3 and CYP3A4*1G polymorphisms on post-operative fentanyl analgesia in Chinese patients undergoing gynaecological surgery: Eur. J. Anaesthesiol 28, 245–250 (2011). [PubMed] [Google Scholar]
- 11.Ishida T, Naito T, Sato H & Kawakami J Relationship between the plasma fentanyl and serum 4β-hydroxycholesterol based on CYP3A5 genotype and gender in patients with cancer pain. Drug Metab. Pharmacokinet 31, 242–248 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Takashina Y et al. Impact of CYP3A5 and ABCB1 Gene Polymorphisms on Fentanyl Pharmacokinetics and Clinical Responses in Cancer Patients Undergoing Conversion to a Transdermal System. Drug Metab. Pharmacokinet 27, 414–421 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Yuan J-J et al. CYP3A4*1G Genetic Polymorphism Influences Metabolism of Fentanyl in Human Liver Microsomes in Chinese Patients. Pharmacology 96, 55–60 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Yan Q et al. Impact of CYP3A4*1G Polymorphism on Fentanyl Analgesia Assessed by Analgesia Nociception Index in Chinese Patients Undergoing Hysteroscopy. Chin. Med. J 131, 2693–2698 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saiz-Rodríguez M et al. Effect of the Most Relevant CYP3A4 and CYP3A5 Polymorphisms on the Pharmacokinetic Parameters of 10 CYP3A Substrates. Biomedicines 8, 94 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gauntlett IS et al. Pharmacokinetics of Fentanyl in Neonatal Humans and Lambs: Effects of Age. Anesthesiology 69, 683–687 (1988). [DOI] [PubMed] [Google Scholar]
- 17.Huang L et al. Impact of CYP3A4/5 and ABCB1 polymorphisms on tacrolimus exposure and response in pediatric primary nephrotic syndrome. Pharmacogenomics 20, 1071–1083 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Liu F, Jagannatha A & Yu H Towards Drug Safety Surveillance and Pharmacovigilance: Current Progress in Detecting Medication and Adverse Drug Events from Electronic Health Records. Drug Saf. 42, 95–97 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horvat CM et al. ABCB1 genotype is associated with fentanyl requirements in critically ill children. Pediatr. Res 82, 29–35 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koren G, Goresky G, Crean P, Klein J & MacLeod SM Unexpected alterations in fentanyl pharmacokinetics in children undergoing cardiac surgery: age related or disease related? Dev. Pharmacol. Ther 9, 183–191 (1986). [DOI] [PubMed] [Google Scholar]
- 21.Mitchell IM, Pollock JC & Jamieson MP Effects of dopamine on liver blood flow in children with congenital heart disease. Ann. Thorac. Surg 60, 1741–1744 (1995). [DOI] [PubMed] [Google Scholar]
- 22.Choi L, Beck C, Weeks H, McNeer E & James N EHR: Electronic Health Record (EHR) Data Processing and Analysis Tool. (2020).at <https://cran.r-project.org/web/packages/EHR/EHR.pdf>
- 23.R Core Team R: A Language and Environment for Statistical Computing. (R Foundation for Statistical Computing, Vienna, Austria, 2020).at <https://www.R-project.org/> [Google Scholar]
- 24.Sheiner LB & Beal SL Evaluation of methods for estimating population pharmacokinetic parameters. I. Michaelis-menten model: Routine clinical pharmacokinetic data. J. Pharmacokinet. Biopharm 8, 553–571 (1980). [DOI] [PubMed] [Google Scholar]
- 25.Monolix. (Lixoft SAS, Antony, France, 2020).at <https://monolix.lixoft.com> [Google Scholar]
- 26.Price AL et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet 38, 904–909 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Beal SL Ways to Fit a PK Model with Some Data Below the Quantification Limit. J. Pharmacokinet. Pharmacodyn 28, 481–504 (2001). [DOI] [PubMed] [Google Scholar]
- 28.Holford N, Heo Y-A & Anderson B A Pharmacokinetic Standard for Babies and Adults. J. Pharm. Sci 102, 2941–2952 (2013). [DOI] [PubMed] [Google Scholar]
- 29.O’Brien SM et al. An empirically based tool for analyzing mortality associated with congenital heart surgery. J. Thorac. Cardiovasc. Surg 138, 1139–1153 (2009). [DOI] [PubMed] [Google Scholar]
- 30.Sukarnjanaset W, Wattanavijitkul T & Jarurattanasirikul S Evaluation of FOCEI and SAEM Estimation Methods in Population Pharmacokinetic Analysis Using NONMEM® Across Rich, Medium, and Sparse Sampling Data. Eur. J. Drug Metab. Pharmacokinet 43, 729–736 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Katz R & H William K Pharmacokinetics of continuous infusions of fentanyl in critically ill children. Crit. Care Med 21, 995–1000 (1993). [DOI] [PubMed] [Google Scholar]
- 32.Choi L et al. Development of a System for Postmarketing Population Pharmacokinetic and Pharmacodynamic Studies Using Real-World Data From Electronic Health Records. Clin. Pharmacol. Ther 107, 934–943 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horvat C et al. ARDA2A and CYP3A5 genotypes are associated with fentanyl requirements in critically ill children. Crit. Care Med 46, 424 (2018). [Google Scholar]
- 34.Grimsrud KN, Lima KM, Tran NK & Palmieri TL Characterizing Fentanyl Variability Using Population Pharmacokinetics in Pediatric Burn Patients. J. Burn Care Res 41, 8–14 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roy J-N et al. CYP3A5 Genetic Polymorphisms in Different Ethnic Populations. Drug Metab. Dispos 33, 884–887 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.