Abstract
Carbapenem-resistant Acinetobacter baumannii are prevalent in low- and middle-income countries such as Egypt, but little is known about the molecular epidemiology and mechanisms of resistance in these settings. Here, we characterize carbapenem-resistant A. baumannii from Alexandria, Egypt, and place it in a regional context. Fifty-four carbapenem-resistant isolates from Alexandria Main University Hospital (AMUH), Alexandria, Egypt, collected between 2010 and 2015 were genome sequenced using Illumina technology. Genomes were de novo assembled and annotated. Genomes for 36 isolates from the Middle East region were downloaded from GenBank. The core-gene compliment was determined using Roary, and analyses of recombination were performed in Gubbins. Multilocus sequence typing (MLST) sequence type (ST) and antibiotic-resistance genes were identified. The majority of Egyptian isolates belonged to one of three major clades, corresponding to Pasteur MLST clonal complex (CCPAS) 1, CCPAS2 and STPAS158. Strains belonging to STPAS158 have been reported almost exclusively from North Africa, the Middle East and Pakistan, and may represent a region-specific lineage. All isolates carried an oxa23 gene, six carried bla NDM-1 and one carried bla NDM-2. The oxa23 gene was located on a variety of different mobile elements, with Tn2006 predominant in CCPAS2 strains, and Tn2008 predominant in other lineages. Of particular concern, in 8 of the 13 CCPAS1 strains, the oxa23 gene was located in a temperate bacteriophage phiOXA, previously identified only once before in a CCPAS1 clone from the USA military. The carbapenem-resistant A. baumannii population in AMUH is very diverse, and indicates an endemic circulating population, including a region-specific lineage. A major mechanism for oxa23 dissemination in CCPAS1 isolates appears to be a bacteriophage, presenting new concerns about the ability of these carbapenemases to spread throughout the bacterial population.
Keywords: antibiotic resistance, Egypt, MLST, phiOXA, transposon, whole-genome sequencing
Data Summary
The whole-genome shotgun sequences of the isolates from this study have been deposited at GenBank/ENA/DDBJ under the BioProject accession number PRJNA659545. The individual genome accession numbers for each isolate are as follows: A1a, JACSUC000000000; A2, JACSUB000000000; A4, JACSVQ000000000; A5, JACSUA000000000; A6, JACSTZ000000000; A7-T, JACSVP000000000; A8-T, JACSVO000000000; A8a, JACSTY000000000; A9, JACSTX000000000; A10, JACSTW000000000; A10a, JACSTV000000000; A11a, JACSTU000000000; A13a, JACSTT000000000; A14a, JACSTS000000000; A15, JACSTR000000000; A16, JACSTQ000000000; A18, JACSTP000000000; A21, JACSVN000000000; A22, JACSTO000000000; A27, JACSTN000000000; A30, JACSTM000000000; A31, JACSTL000000000; A34, JACSTK000000000; A35, JACSTJ000000000; A36, JACSTI000000000; A39, JACSTH000000000; A40, JACSTG000000000; A41, JACSTF000000000; A42, JACSTE000000000; A43, JACSTD000000000; A44, JACSTC000000000; A45, JACSTB000000000; A46, JACSTA000000000; A64, JACSSZ000000000; A68, JACSSY000000000; A69, JACSSX000000000; A70, JACSSW000000000; A71, JACSVM000000000; A72, JACSSV000000000; A73, JACSSU000000000; A74, JACSST000000000; A75, JACSSS000000000; A78, JACSSR000000000; A82, JACSSQ000000000; A83, JACSVL000000000; A84, JACSSP000000000; A85, JACSSO000000000; A86, JACSVK000000000; A87, JACSSN000000000; A88, JACSSM000000000; A89, JACSSL000000000; A92, JACSSK000000000; A5910, JACSSJ000000000; A6135, JACSVJ000000000.
Impact Statement.
In this study, we have analysed the whole genomes of a group of antibiotic-resistant bacteria – Acinetobacter baumannii – from Alexandria, Egypt, to identify why they are antibiotic resistant, and how resistance is being spread between bacteria. This is to help address the current knowledge gap regarding the mechanisms and spread of antibiotic resistance in low- and middle-income countries like Egypt. We found that for the vast majority of bacteria, resistance was due to a specific gene – oxa23. However, the bacteria carrying this gene were very varied, showing that they do not represent a specific outbreak, but rather the continuous circulation of multiple different antibiotic-resistant lineages. A significant number of bacteria belonged to a subgroup that has only been sporadically reported from North Africa, the Middle East and Pakistan, providing evidence that there may be a specific subgroup of A. baumannii from this geographical region. Of particular significance, in a number of bacteria the oxa23 gene was found to be carried by a bacteriophage – a virus that infects bacteria. We present evidence that it is likely that this bacteriophage is responsible for spreading the oxa23 gene between bacteria, which is not currently widely recognized as a major mechanism for antibiotic-resistance dissemination.
Introduction
The bacterium Acinetobacter baumannii is a major opportunistic hospital-acquired pathogen that is listed by the World Health Organization (WHO) as in critical need of new treatment options due to its multidrug-resistant nature [1]. In particular, the frequency of carbapenem-resistant A. baumannii has been steadily increasing over the last two decades, leaving very few treatment options available to combat this pathogen [2]. However, carbapenem-resistant A. baumannii are not uniformly distributed across the globe, with higher rates of resistance found in low- and middle-income countries [3–6], though rates in some southern and eastern European countries have now also reached very high levels [7]. In countries in the Middle East and North Africa, high levels of carbapenem-resistant A. baumannii are reported, with frequencies of 70% of isolates or greater being common [8]. Despite these very high rates of resistance, there are relatively few studies investigating the molecular epidemiology of the antibiotic-resistant strains.
Carbapenem resistance in A. baumannii is usually the result of the expression of an OXA-type β-lactamase, or occasionally metallo-β-lactamases such as the IMP, VIM and NDM groups [9]. The acquired OXA-type β-lactamases in A. baumannii are encoded by genes belonging to five main groups – oxa23 (or bla OXA-23-like), oxa40 (or bla OXA-40-like), oxa58 (or bla OXA-58-like), oxa134 (or bla OXA-134-like) and oxa143 (or bla OXA-143-like) [10, 11]. In addition, all A. baumannii carry an intrinsic OXA β-lactamase gene called oxaAb (or bla OXA-51-like), certain alleles of which, when highly expressed due to the presence of an ISAba1 insertion sequence upstream, can confer carbapenem resistance [12–14]. The most common of these resistance mechanisms globally is oxa23 [15]. In Egypt, and other countries in the region, oxa23 is so prevalent it can be found in up to 100% of carbapenem-resistant isolates, with frequencies greater than 70% being the norm [16–19]. In A. baumannii , oxa23 is usually located on a transposon mobilized by one or more insertion sequences, which has enabled the resistance gene to be spread to many different plasmids and many different lineages within the species [20]. Despite the particularly high prevalence of oxa23 in low- and middle-income countries such as Egypt, there are very few studies that have investigated the mobile genetic elements carrying the gene, which is crucial to gaining an understanding of the local population genetics of the species.
The majority of A. baumannii isolates belong to one of eight international clones (ICs), which correspond to specific multilocus sequence typing (MLST) sequence types (STs) and clonal complexes (CCs) [15, 21]. There are two MLST schemes for A. baumannii – the Pasteur scheme [22] and the Oxford scheme [23], with the Pasteur scheme containing genes that are less prone to recombination than those in the Oxford scheme [24]. Globally, isolates belonging to IC2, corresponding to CCPAS2, are the most common, though there are exceptions such as Latin American countries where isolates belonging to IC4 (CCPAS15), IC5 (CCPAS79) and IC7 (CCPAS25) are predominant [25, 26]. In many low- and middle-income countries, MLST is too costly to perform on large numbers of isolates, and so at present we often rely on a small number of studies to provide an indication of what the national epidemiology may be. In Egypt, studies have indicated that CCPAS2 is the most common CC, but that a large number of isolates from other CCs or that don’t belong to any of the defined CCs make up a substantial portion of the population [16, 18, 19, 27]. The aim of our study was to define the local population structure of A. baumannii in Alexandria Main University Hospital (AMUH), Alexandria, Egypt, and identify the mobile genetic elements responsible for resistance-gene dissemination.
Methods
Bacterial isolates and antimicrobial-susceptibility testing
A total of 54 carbapenem-resistant A. baumannii clinical isolates obtained from patients presenting at AMUH between 2010 and 2015 were included in the study. This is the largest hospital in the northern sector of Alexandria and a major referral hospital. The isolates were identified by conventional methods, including colony morphology, aerobic growth at 44 °C on MacConkey agar and species designations obtained using the Vitek system (bioMérieux). The identity of the isolates was further confirmed by PCR amplification of the intrinsic oxaAb (bla OXA-51-like) gene, as well as MALDI-TOF MS (Bruker Daltonik). The identified isolates were stored at −80 °C prior to subsequent characterization [28]. The susceptibility of the isolates to imipenem and meropenem was determined using agar dilution, and the results were interpreted according to Clinical and Laboratory Standards Institute guidelines (2018) [29].
Whole-genome sequencing and analyses
Genomic DNA was extracted using the Wizard genomic DNA purification kit (Promega), according to the manufacturer’s instructions. A Qubit fluorometer (Life Technologies) was used to quantify the extracted DNA. Dual indexing library preparation was carried out using the Nextera XT DNA preparation kit (Illumina). Whole-genome sequencing of the library was performed on an Illumina MiSeq using the 2×250 bp paired-end protocol. Following quality filtering of the reads using Trimmomatic v0.36 [30] and FastQC v0.11.5 [31], genomes were de novo assembled with Spades v3.11.1 [32], and annotated using Prokka v1.11 [33]. The assemblies were quality checked using quast [34]. The Sequence Read Archive (SRA) was searched using keywords of Middle Eastern countries, and the genomes of an additional 36 strains were downloaded and included in subsequent analyses. The genomes of a further 17 geographically and genomically diverse strains [35] were also downloaded and included in subsequent analyses. The core-genome content of the strain collection was determined using Roary v3.12.0 [36], and the core-gene phylogeny estimated using FastTree v2.1.10 [37]. Isolates A8-T and A74 were chosen to be the reference genomes for CCPAS1 and CCPAS2, respectively, for subsequent variant calling. Sequences belonging to CCPAS1 and CCPAS2 were mapped to the reference genomes and variant called using PHEnix v1.3 [38]. A SnapperDB v1.0.6 for each CC was created, allowing inclusion of SNPs with a minimum mean read depth of 10 [39]. Whole-genome alignments were generated including isolates 10 000 SNPs from the CC1 reference and 20 000 SNPs from the CC2 reference, which were used as input for Gubbins. Estimates of recombination within clades identified in the phylogeny were conducted with Gubbins v2.3.1 using default settings [40]. The Pasteur MLST ST of each isolate was determined from the whole-genome sequence using the online Center for Genomic Epidemiology’s MLST software [41], and antibiotic-resistance genes were determined using ariba [42] with the card [43] and srst2 [44] databases. All oxaAb alleles were confirmed using the blast function on the Beta-Lactamase DataBase [11] (http://www.bldb.eu/). Analyses of the accessory genome were conducted using panini [45].
Annotation of phiOXA-A35 from the A35 genome assembly
The phiOXA-A35 sequence was constructed by alignment of three contigs and manual resolution of overlaps from assembly data obtained for A. baumannii A35. phiOXA ORFs were initially annotated using prokka v1.12 and then refined using blastp, InterProScan [46] and HHpred [47]. Prediction of tRNAs was performed using tRNAscan-SE 2.0 [48]. Alignments of the portal vertex and major capsid proteins were performed using Clustal Omega [49] and phylogenetic trees reconstructed using iq-tree v1.6.12 with ModelFinder, SH-aLRT test and ultrafast bootstrap with 1000 replicates [50–52]. Read coverage of phiOXA-A35 was calculated using QualiMap v.2.2.2 [53].
Genetic environment of oxa23
In the program Geneious R10 (https://www.geneious.com/), for each oxa23-positive strain, the contig containing oxa23 was identified, then all these contigs were aligned. The alignment was used to group strains by similarity of the sequence surrounding oxa23. Where appropriate, the presence of insertion sequences ISAba1 and ISAba125 surrounding the oxa23 gene were confirmed by PCR using combinations of primers ISAba1-B [54], OXA-23-F and OXA-23-R [55], and ISAba125-F (5′-TAAAACTATTCATGAGCGCC-3′). To obtain the complete sequences of the prophages containing oxa23, contigs were aligned against the phiOXA sequence from strain AB5075-UW (GenBank accession no. CP008706.1). PCRs with primers Phi-F (5′-CGT TGT TGG GCT TCT AGT GC-3′) and OXA-23-R [55] were used to confirm the contig joins either side of the ISAba1 insertion sequence.
Bacteriophage induction
Bacterial cultures were grown overnight in LB at 37 °C and shaking at 180 r.p.m. Overnight cultures were diluted to an OD600 of approximately 0.05 using pre-warmed LB, then incubated at 37 °C and 180 r.p.m. until the OD600 reached 0.2. Cultures were then divided to generate two treatment cultures and two control cultures per strain. Mitomycin C was added to a final concentration of 2 µg ml−1 to the treatment cultures before they were wrapped in foil to block out light, then both treatment and control cultures were incubated at 37 °C. The OD600 of cultures was recorded every 30 min to identify a marked drop in the optical density in the mitomycin C-treated cultures, representing bacteriophage induction. Once this was observed, all cultures were centrifuged at 10 000 g for 5 min, filter sterilized through a 0.22 µm filter, and treated with DNase (TURBO DNase; Invitrogen) and RNase A (Thermo Scientific) to remove all bacteria and free nucleic acid from the cell lysate. The presence of intact bacteriophage carrying oxa23 in the bacterial cell lysate was determined by PCR using primers OXAphi-F (5′-GGAAATGCGGTCAGAAATGC-3′) situated within oxa23 and OXAphi-R (5′-TGGACCCTGTAGATTTTGCC-3′) situated within a phage tail protein gene, giving a 1032 bp product size. PCR conditions were 95 °C for 10 min; followed by 40 cycles of 95 °C for 30 s, 55 °C for 30 s and 72 °C for 1 min; with a final extension of 72 °C for 5 min. A 1 µl culture volume from a phiOXA-positive strain was used as a positive control.
Bacteriophage purification and sequencing
Clarified lysates were prepared from batch cultures of A35 treated with mitomycin C by centrifugation at 10 000 g for 10 min and filter sterilization (0.22 µm). Bacteriophages were precipitated by addition of NaCl and PEG 8000 to final concentrations of 1 M and 10 % (w/v), respectively. After storage overnight at 4 °C, precipitate was recovered by centrifugation at 11 000 g for 15 min at 4 °C and pellets resuspended in SM buffer (50 mM Tris-HCl, 8 mM MgSO4, 100 mM NaCl, pH 7.5). Residual PEG was removed by the addition of an equal volume of chloroform and the aqueous phase recovered after centrifugation at 3000 g for 10 min at 4 °C. For the extraction of bacteriophage genomic DNA, samples were treated with DNase I and RNase A (Sigma Aldrich) for 1 h at 37 °C, before the addition of EDTA, SDS and proteinase K to final concentrations of 20 mM, 0.5% (w/v) and 50 µg ml−1, respectively [56]. DNA was then purified using phenol:chloroform:isoamyl alcohol extraction. Preparation of libraries and sequencing of DNA was performed at the Genomic Services and Development Unit (now called the Central Sequencing Laboratory) (Public Health England) using an Illumina HiSeq1000 and 100 bp paired-end reads. Assembly was performed using SPAdes version 3.11.1 [32]. Sequence reads were mapped to the de novo assembled contigs and the A35 genome assembly using bwa-mem [57], and assembly statistics obtained using quast [34] and Qualimap [53]. Annotation was performed as previously described.
Results
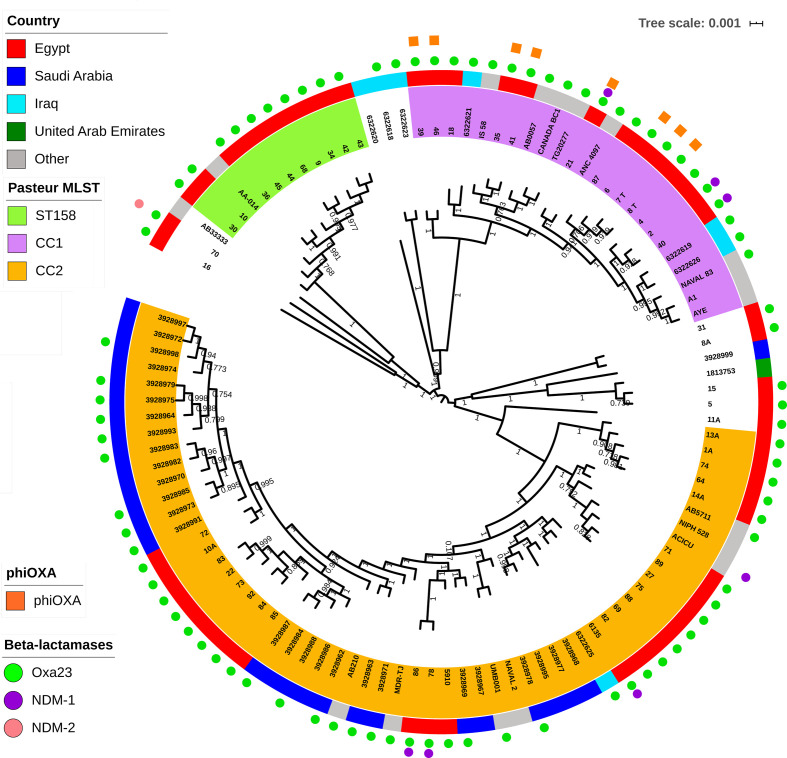
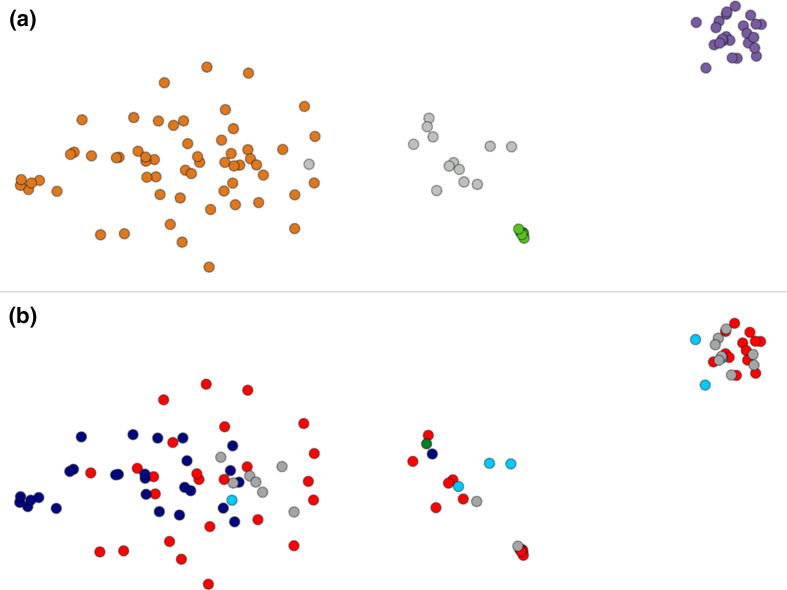
Analysis of the antibiotic susceptibilities of the Egyptian isolates showed that, as expected due to the isolates being selected for their carbapenem resistance, they were all resistant to imipenem and meropenem (Table 1). The majority of Egyptian isolates belonged to one of three major well-supported clades based upon core-gene sequences, corresponding to Pasteur MLST CC (CCPAS) 1 (13 isolates), CCPAS2 (24 isolates) and STPAS158 (10 isolates) (Table 1, Fig. 1). CCPAS1 isolates belong to IC1, while CCPAS2 isolates belong to IC2 [21, 58]. In addition, two isolates belonged to STPAS15, which are members of IC4, and two isolates belonged to STPAS25, which are members of IC7 (Table 1). All isolates belonging to the ICs carried the oxaAb allele previously shown to be associated with their respective IC, with isolates in CCPAS1 (IC1) carrying oxaAb(69), isolates in CCPAS2 (IC2) carrying oxaAb(66), isolates in STPAS15 (IC4) carrying oxaAb(51), and isolates in STPAS25 (IC7) carrying the oxaAb(64) allele [59]. Isolates in STPAS158 carried oxaAb(65) alleles, which are usually associated with CCPAS79 and IC5. However, it should be noted that the oxaAb(65) allele in the STPAS158 isolates differed from the original oxaAb(65) allele (GenBank accession no. AY750908) by three silent substitutions (T90C, C636T and A663G). We compared our STPAS158 isolates with other published or publicly available data, which demonstrated that this particular oxaAb(65) variant is a feature of STPAS158 isolates in general (Table 2), and is distinct from CCPAS79 (IC5) isolates. Isolates from CCPAS1 and CCPAS2 were more diverse than STPAS158 isolates. This was evident in gene conservation analysis with 3069 genes shared by 90% of STPAS158 isolates, whereas only 2394 genes were shared by 90% of CCPAS2 isolates, and 2600 genes shared by 90% of CCPAS1 isolates. For all three of the major clades identified, the isolates from AMUH did not form their own specific sub-clades, but were interspersed with the strains both from other Middle Eastern countries, as well as with the globally distributed strains (Fig. 1). Interestingly a similar pattern was observed with respect to the accessory genome (Fig. 2). Based upon their accessory genomes, CCPAS1, CCPAS2 and STPAS158 isolates clustered together. The only exception was isolate 11a, which clustered with the CCPAS2 isolates (the grey dot found on the right-hand edge of the orange CCPAS2 cluster in Fig. 2a). However, this is not too surprising given that of all the non-CCPAS2 isolates, 11a is most closely related to CCPAS2 at the core-genome level (Fig. 1). The accessory genome clusters did not show any geographical signal (Fig. 2b), in agreement with the lack of geographical signal in the core-gene tree (Fig. 1). Together these data demonstrate two points: firstly, that there are both multiple circulating clonal lineages, and multiple circulating sub-lineages within each clonal lineage, that are responsible for infecting patients in AMUH; and, secondly, that while the accessory genome is shared across isolates from several different countries within a clonal lineage, there is little sharing of the accessory genome between clonal lineages.
Table 1.
Carbapenem-susceptibility data, MLST assignments, carbapenem-resistance genes and associated mobile genetic elements of Egyptian isolates
|
Strain |
MICs (mg l−1) |
MLST (Pasteur) |
Resistance gene |
oxa23 mobile element* |
|||
|---|---|---|---|---|---|---|---|
|
IMI |
MER |
ST |
CC |
oxaAb |
oxa23/bla NDM-1/2 |
|
|
|
1a |
4 |
32 |
664 |
2 |
66 |
oxa23 |
A |
|
2 |
8 |
64 |
1 |
1 |
69 |
oxa23, bla NDM-1 |
nd |
|
4 |
16 |
256 |
1 |
1 |
69 |
oxa23 |
nd |
|
5 |
32 |
64 |
25 |
25 |
64 |
oxa23 |
A |
|
6 |
8 |
32 |
1 |
1 |
69 |
oxa23 |
phiOXA |
|
7-T |
8 |
64 |
1 |
1 |
69 |
oxa23 |
phiOXA |
|
8-T |
8 |
64 |
1 |
1 |
69 |
oxa23 |
phiOXA |
|
8a |
16 |
32 |
15 |
15 |
51 |
oxa23 |
A |
|
9 |
8 |
64 |
158 |
158 |
65 |
oxa23 |
H |
|
10 |
8 |
32 |
158 |
158 |
65† |
oxa23 |
C |
|
10a |
16 |
32 |
2 |
2 |
66 |
oxa23 |
A |
|
11a |
16 |
32 |
1535 |
− |
65‡ |
oxa23 |
E |
|
13a |
4 |
32 |
664 |
2 |
66 |
oxa23 |
A |
|
14a |
16 |
16 |
664 |
2 |
66 |
oxa23 |
A |
|
15 |
8 |
64 |
25 |
25 |
64 |
oxa23 |
A |
|
16 |
8 |
64 |
85 |
− |
94 |
oxa23 |
nd |
|
18 |
8 |
64 |
19 |
1 |
69 |
oxa23 |
J |
|
21 |
32 |
64 |
1 |
1 |
69 |
oxa23, bla NDM-1 |
phiOXA |
|
22 |
8 |
32 |
2 |
2 |
66 |
oxa23 |
A |
|
27 |
8 |
32 |
2 |
2 |
66 |
oxa23 |
F |
|
30 |
8 |
32 |
158 |
158 |
65 |
oxa23 |
nd |
|
31 |
16 |
64 |
15 |
15 |
51 |
oxa23 |
G |
|
34 |
8 |
64 |
158 |
158 |
65 |
oxa23 |
nd |
|
35 |
4 |
64 |
1 |
1 |
69 |
oxa23 |
phiOXA |
|
36 |
8 |
64 |
158 |
158 |
65 |
oxa23 |
nd |
|
39 |
4 |
64 |
1 |
1 |
69 |
oxa23 |
phiOXA |
|
40 |
32 |
64 |
1 |
1 |
69 |
oxa23, bla NDM-1 |
nd |
|
41 |
16 |
32 |
– | – |
69 |
oxa23 |
phiOXA |
|
42 |
32 |
64 |
158 |
158 |
65 |
oxa23 |
B |
|
43 |
16 |
64 |
158 |
158 |
65 |
oxa23 |
B |
|
44 |
8 |
64 |
158 |
158 |
65 |
oxa23 |
B |
|
45 |
32 |
32 |
158 |
158 |
65 |
oxa23 |
nd |
|
46 |
8 |
16 |
1 |
1 |
69§ |
oxa23 |
phiOXA |
|
64 |
8 |
64 |
664 |
2 |
66 |
oxa23 |
A |
|
68 |
8 |
32 |
158 |
158 |
65 |
oxa23 |
D |
|
69 |
8 |
32 |
2 |
2 |
66 |
oxa23 |
C |
|
70 |
16 |
32 |
103 |
− |
70 |
oxa23, bla NDM-2 |
nd |
|
71 |
8 |
32 |
2 |
2 |
66 |
oxa23 |
F |
|
72 |
16 |
64 |
2 |
2 |
66 |
oxa23 |
A |
|
73 |
8 |
64 |
2 |
2 |
66 |
oxa23 |
A |
|
74 |
8 |
32 |
664 |
2 |
66 |
oxa23 |
A |
|
75 |
8 |
16 |
2 |
2 |
66 |
oxa23 |
F |
|
78 |
8 |
16 |
570 |
2 |
+|| |
oxa23, bla NDM-1 |
A |
|
82 |
64 |
64 |
2 |
2 |
66 |
oxa23 |
C |
|
83 |
64 |
64 |
2 |
2 |
66 |
oxa23 |
A |
|
84 |
64 |
64 |
2 |
2 |
66 |
oxa23 |
A |
|
85 |
64 |
32 |
2 |
2 |
66 |
oxa23 |
A |
|
86 |
64 |
>256 |
570 |
2 |
66 |
oxa23, bla NDM-1 |
A |
|
87 |
64 |
64 |
1 |
1 |
69 |
oxa23 |
I |
|
88 |
64 |
64 |
2 |
2 |
66 |
oxa23 |
A |
|
89 |
64 |
64 |
2 |
2 |
66 |
oxa23 |
F |
|
92 |
32 |
64 |
2 |
2 |
66¶ |
oxa23 |
A |
|
5910 |
128 |
>256 |
2 |
2 |
66 |
oxa23 |
A |
|
6135 |
8 |
32 |
600 |
2 |
66 |
oxa23, bla NDM-1 |
A |
IMI, Imipenem; MER, meropenem.
*nd, Not determined; the Illumina sequence data was not able to resolve contigs showing the genetic environment of oxa23 in these isolates.
†Contig break giving incomplete gene, with 243/243 amino match to oxaAb(65).
‡Contig break giving incomplete gene, with 265/266 amino acid match to oxaAb(65).
§Contig break giving incomplete gene, with 266/266 amino acid match to oxaAb(69).
||oxaAb gene was not identified in genome sequence, but was positive by PCR.
¶Contig break giving incomplete gene, with 266/266 amino acid match to oxaAb(66).
Fig. 1.
Core-gene tree of all isolates. In the centre is the core-gene tree generated in FastTree [37] using a core-gene alignment output from Roary [36]. The tree is scaled by genetic distance, and branch labels indicate level of support based upon the Shimodaira–Hasegawa test using 1000 resamples. Leaves are labelled with isolate names or SRA (Sequence Read Archive) accession numbers, and are colour coded to highlight the three major Pasteur MLST scheme CCs or STs identified in this study. The ST/CC of isolates that are not coloured can be seen in Table 1. The outer solid coloured ring indicates the geographical source of the isolates. The outer rings of shapes indicate β-lactamases and phiOXA encoded by the isolates.
Table 2.
CCPAS158 and CCOX499 isolates reported in the literature or in public databases
|
Isolate† |
STPAS |
STOX |
Country |
Year |
oxaAb‡ |
Accession no. |
Reference |
|---|---|---|---|---|---|---|---|
|
10 isolates |
158 |
499§ |
Egypt |
2010–15 |
65* |
– |
This study |
|
1309; 2226C |
158 |
– |
Turkey |
2009 |
– |
– |
PubMLST |
|
2313; AA-014 |
158 |
960 |
Iraq |
2008 |
65* |
GCA_000335595 |
[89] |
|
3826; 778944; ABC002 |
158 |
1717 |
Egypt |
2012 |
– |
– |
PubMLST |
|
K50 |
158 |
– |
Kuwait |
2008 |
65* |
[90] |
|
|
Unnamed |
158 |
– |
Lebanon |
2013 |
65* |
– |
[91] |
|
Ab-Pak-Pesh-01 |
158 |
– |
Pakistan |
2015 |
65* |
[68] |
|
|
Ab-Pak-Pesh-07 |
158 |
– |
Pakistan |
2015 |
65* |
[68] |
|
|
Ab-Pak-Pesh-28 |
158 |
– |
Pakistan |
2015 |
65* |
[68] |
|
|
AMA 341 |
158 |
499 |
Denmark|| |
2012 |
65¶ |
SAMN03160609 |
[92] |
|
2 isolates |
158 |
– |
Kuwait |
2011–12 |
– |
– |
[93] |
|
ACB69C |
158 |
– |
Turkey |
2009–11 |
– |
– |
[94] |
|
30 isolates |
158 |
– |
Kuwait |
2007–08 |
66 |
– |
[95] |
|
7 isolates |
158 |
499 |
Tunisia |
2008–09 |
– |
– |
[96] |
|
1830; J17 |
342 |
– |
China |
2011 |
– |
– |
[97] |
|
3840; ACIN00151 |
342 |
1776 |
USA |
2016 |
694 |
PubMLST# |
PubMLST |
|
2178; A.baumannii64 |
615 |
– |
Egypt |
2012 |
– |
– |
[16] |
|
2180; A.baumannii85 |
615 |
– |
Egypt |
2013 |
– |
– |
[16] |
|
2182; A.baumannii108 |
618 |
– |
Egypt |
2013 |
– |
– |
[16] |
|
3950; TR112 |
1241 |
– |
Turkey |
2016 |
– |
– |
PubMLST |
|
8 isolates |
– |
499 |
Egypt |
2015 |
– |
– |
[27] |
|
2 isolates |
– |
499 |
Saudi Arabia |
2011–13 |
– |
– |
[19] |
|
1 isolate |
– |
499 |
Kuwait |
2011–13 |
– |
– |
[19] |
†If an isolate is known by more than one name, all names are provided separated by semicolons.
‡The oxaAb(65*) alleles differ from the original oxaAb(65) sequence by three silent substitutions.
§One isolate did not have its STOX determined.
||This isolate was likely imported from Egypt.
¶The authors did not state whether the nucleotide sequence differed from the original oxaAb(65) sequence.
#This genome is available through the PubMLST website.
Fig. 2.
Clustering of isolates by similarity of their non-core genomes using panini. Each dot represents an isolate, and the distance between isolates indicates the similarity of their accessory genomes. (a) The network is coloured according to the MLST data as in Fig. 1 (CC1 is purple, CC2 is orange, ST158 is green and other STs are grey). (b) The network is coloured according to the country of origin of the isolates as in Fig. 1 (Egypt is red, Saudi Arabia is dark blue, Iraq is light blue, the United Arab Emirates is green and other countries outside the Middle East are grey).
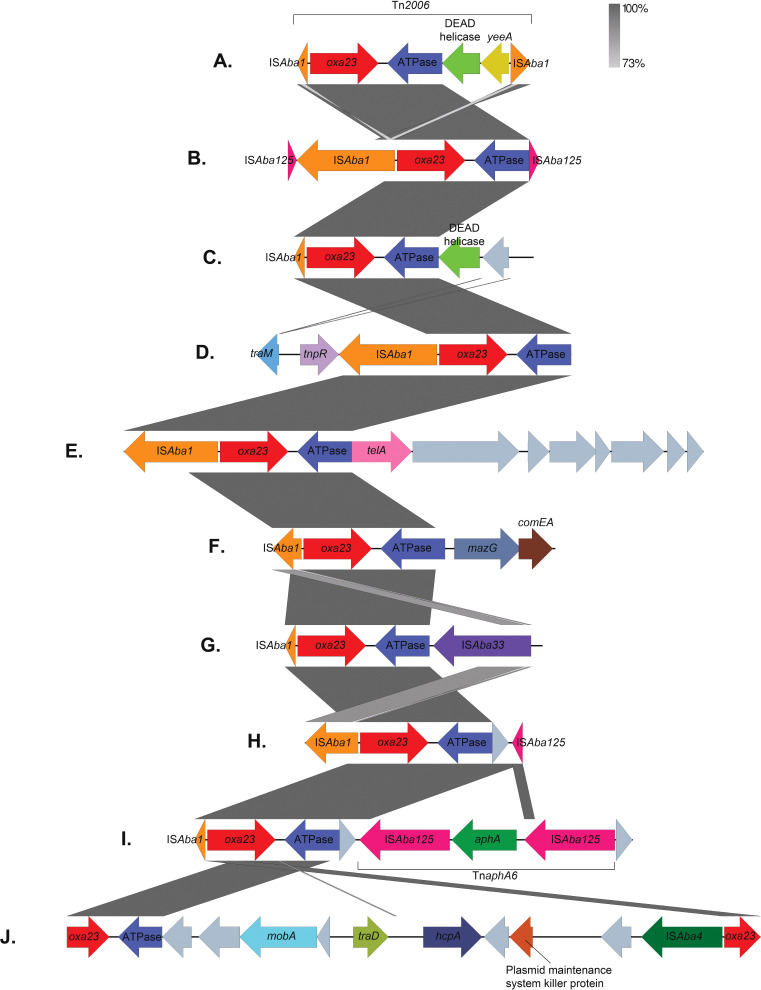
All 54 isolates carried an oxa23 gene (Table 1). In addition, six isolates also carried a bla NDM-1 gene, and one isolate carried bla NDM-2. The bla NDM genes were not clustered in one particular bacterial ST, with three of the bla NDM-1 genes located in CCPAS1 isolates (in one ST), while the other three were located in CCPAS2 isolates (across two STs) (Table 1, Fig. 1). While complete transposons could not be identified due to the limitations of short-read sequence data, all bla NDM-1 genes had an ISAba125 insertion sequence upstream and a ble bleomycin-resistance gene followed by a trpF phosphoribosylanthranilate isomerase gene downstream, as has typically been found in A. baumannii in other studies [60]. In two isolates (A86 and A6135) where longer contigs containing the bla NDM-1 gene were assembled, it appears likely that the bla NDM-1 gene is carried on a transposon similar to ΔTn125 as described by Bonnin et al. [61] as an ISAba14 fragment was detected following the dct gene. However, further investigation using technology such as long-read sequencing is required to completely resolve these mobile elements. The bla NDM-2 gene in isolate A70 was located within the previously described transposon Tn125 [60]. The oxa23 gene was located on a variety of different mobile genetic elements, with 11 different structures identified (Table 1, Figs 3 and 4). Several of these structures were found in multiple isolates: structure A, representing Tn2006 [62], was the most common and was found in 21 isolates, 18 of which belonged to CCPAS2, 2 belonged to CCPAS25 and 1 to CCPAS15; structure B was found exclusively in 3 STPAS158 isolates; structure C in 1 STPAS158 and 2 CCPAS2 isolates; and structure F in 4 CCPAS2 isolates and appeared to be borne on the chromosome (Table 1, Fig. 3). Of particular concern, in 8 of the 13 CCPAS1 strains carrying oxa23, the carbapenemase gene was located in prophage called phiOXA. This prophage has been identified only once before in the CCPAS1 isolate AB5075-UW, derived from a strain isolated in 2008 from a USA soldier at the Walter Reed Army Medical Centre, USA, but to our knowledge has not been shown to be viable [63]. In order to determine whether phiOXA can form viable viral particles that contain the oxaAb gene, four isolates encoding phiOXA (A8-T, A21, A35 and A39) and one isolate that did not (A18) were treated with mitomycin C to induce bacteriophage, followed by DNase and RNase treatment to remove any DNA that is not contained within a virus particle. Then, a PCR for oxa23, with an extended initial denaturation phase to lyse bacteriophage particles, was used to identify the carriage of the antibiotic-resistance gene by the bacteriophage. Cultures of three of the four isolates tested (A8-T, A35 and A39) that had been treated with mitomycin C were found to have produced intact bacteriophage carrying oxa23. No PCR products for oxa23 were detected for these strains when they were not induced, nor for isolate A18 (phiOXA negative) with either the presence or absence of mitomycin C treatment. To confirm these data, virions from cultures of A35 exposed to mitomycin C were purified and sequenced. Two contigs corresponding to phiOXA and a second predicted prophage were identified. Due to extremely high coverage of the second prophage, an assembly was performed using a random subset of 10% of the paired-end reads. Alignment of the complete dataset showed that the majority of sequence reads mapped to this prophage (89%) yielding a coverage of 5651×. For phiOXA-A35, a contig was identified in all assemblies, regardless of the proportion of reads employed, representing 6% of the total reads and a coverage of 477×. Comparison of this contig to the AB5075-UW genome using blastn showed 100% coverage and identity to the phiOXA prophage in this strain, and annotation confirmed the presence of the oxa23 carbapenemase. Collectively, these data demonstrate that the phiOXA prophage in these isolates can be induced and form intact bacteriophage particles, and that these bacteriophages carry the oxa23 gene. Further work is required to identify a susceptible host for phiOXA-A35 in order to demonstrate lysogenic conversion to a carbapenem-resistance phenotype.
Fig. 3.
Genetic environments surrounding oxa23 genes. Arrows represent genes, which are colour-coded by their type. Unlabelled grey genes represent hypothetical proteins. The size of the genes and the distances between them are drawn to scale. Vertical grey boxes indicate homology between sequences ranging between 73 and 100% identity (blastn). The diagram was created using Easyfig [98] and annotated in Adobe Photoshop.
Fig. 4.
Schematic genome map of phiOXA-A35 and related prophages. Prophages are orientated as they appear in their host genome. Arrows depict ORFs and are coloured according to function. Homologues to gene products in Escherichia phage P2 are indicated in parentheses. ORFs encoding hypothetical proteins are shown as black outlines. The tRNA-Leu, representing the attL site, is shown as a dark red rectangle. Shading between entries represents the per cent identity (blastn) from 90% (light grey) to 100% (dark grey). The map was constructed using Easyfig [98] and annotated in Adobe Illustrator.
Analysis of the sequence of phiOXA-A35 showed it is identical to the bacteriophage/prophage reported in strain AB5075-UW (Fig. 4), with mean read coverage of 22 (sd=7). The phiOXA-A35 prophage consists of a contiguous 32 kb region comprising 48 ORFs with the attL site residing within a tRNA-Leu, as was seen previously in strain AB5075-UW. The genomic architecture of phiOXA-A35 is similar to that of members of the Peduovirinae, a widespread subfamily of temperate bacteriophages that infect ɣ- and β-proteobacteria and includes Escherichia phage P2 and Pseudomonas phage phiCTX. The genome can be divided into four modules, representing genes involved in virion morphogenesis and assembly that contains the diagnostic Q-P-O-N-M-L capsid gene cluster [64], lysis, replication, and control of lysogeny. This relationship is further supported by phylogenetic analysis of the portal vertex and major capsid protein (Figs S1 and S2, available with the online version of this article). Apart from a single syntenic break with ISAba1, oxa23 and a gene encoding a DUF815 domain protein, phiOXA is nearly identical to predicted prophage regions found in A. baumannii strains A85, AYE, DA33382, USA15 and WCHAB005078. Comparison of these regions using viridic [65] suggests that they represent a single species of temperate bacteriophage, as each exhibit >95% sequence similarity [66]. We propose that phiOXA-A35 represents a new genus within the subfamily Peduovirinae. A total of eight phiOXA ORFs are annotated as hypothetical proteins and whether these represent additional proteins that influence the pathobiology or environmental fitness of their host lysogen remains to be elucidated.
The two most likely scenarios that could explain the presence of phiOXA in multiple CCPAS1 isolates are (i) that the bacteriophage inserted once into a CCPAS1 isolate and has then spread via vertical transmission, or (ii) that phiOXA has independently infected multiple isolates. In order to investigate this, we examined the apparent insertion site (tRNA-Leu) for all isolates included in the CCPAS1 clade in Fig. 1. Our analyses showed that within the CCPAS1 clade, isolates carrying phiOXA are not monophyletic and are found in four separate sub-clades, indicating acquisition of phiOXA is likely to have occurred on at least four independent occasions (Fig. S3). These data, combined with the demonstration that complete bacteriophage particles carrying oxa23 are released by the bacteria, suggest that phiOXA has been spreading through the CCPAS1 population via horizontal transmission of the bacteriophage.
Discussion
In this study, we aimed to use genomics to characterize the molecular epidemiology and carbapenem resistance of A. baumannii isolates from Alexandria, Egypt. Genome-level studies of this nature from low- and middle-income countries are not common, despite the fact that these countries bear the highest burden of antibiotic resistance. By using genomics, we can simultaneously characterize antibiotic-resistance genes and the genetic environment supporting them, and the fine-scale epidemiological relationships between isolates. It also has the added benefit of being backward-compatible with previous typing methods such as MLST. In the context of Egypt, there are a few studies that have used one of the MLST schemes for A. baumannii – either the Pasteur scheme [22] (as used in this study) or the Oxford scheme [23] – to investigate the relatedness of isolates. Where studies have used MLST, the most commonly identified CC is CCPAS2 (CCOX208). However, a considerable proportion of isolates are often found to belong to less common CCs or are singletons [16, 19, 27, 67]. This is entirely consistent with the results from our study, where 44% of isolates belonged to CCPAS2, 24% of isolates belonged to CCPAS1 and 19% belonged to STPAS158. The core-genome analysis we conducted demonstrated that even within MLST STs there was a lot of diversity. This shows that multiple carbapenem-resistant strains are present within AMUH, suggesting that rather than facing an outbreak, the bacterium is endemic. Whether patients are acquiring these strains once admitted to the hospital or whether there is widespread circulation of carbapenem-resistant A. baumannii in the community is an open question that we hope to address in the future.
While CCPAS1 and CCPAS2 strains are globally distributed and frequently encountered, strains belonging to STPAS158 have been reported far less frequently and from a more focused geographical area. STPAS158 belongs to CCPAS158 (CCOX499) [68], and is usually found in isolates from North Africa, the Middle East and Pakistan (Table 2). Most previous studies that have identified CCPAS158 isolates have found them to carry the OxaAb variant OxaAb(65). However, in CCPAS158 strains, the oxaAb(65) allele differs from the original allele (GenBank accession no. AY750908) by three synonymous substitutions. As the oxaAb genes are intrinsic to A. baumannii and specific alleles are associated with certain ICs, the gene can be used as a useful epidemiological marker to identify the IC an isolate belongs to [59, 69, 70]. However, under this scheme, OxaAb(65) is associated with IC5. Isolates belonging to IC5 are members of CCPAS79 and are found at particularly high frequency in Latin America [15, 59, 71, 72]. The allele profiles of the founder STs of CCPAS158 and CCPAS79 (STPAS158 and STPAS79, respectively) are quite different, sharing only one of the seven alleles (rplB allele 4), which at the nucleotide level translates to 13 SNPs. It is clear, therefore, that in this instance numbering the oxaAb alleles based upon their amino acid sequence can mask important epidemiological information and that, as suggested by Karah et al. [68], these genes should be numbered according to their nucleotide sequences as has been done for the Acinetobacter ampC genes [73].
Previous studies of A. baumannii in Egypt have found that rates of carbapenem resistance are high, typically >70% [17, 74], and that this is usually associated with isolates carrying the oxa23 gene with carriage frequencies reaching as high as 100% in carbapenem-resistant isolates [16–18]. This was reflected in our study, where oxa23 was carried by 100% of carbapenem-resistant isolates. Reports of the metallo-β-lactamases NDM-1 and NDM-2 being encoded by isolates from Egypt indicate frequencies of bla NDM-1 can typically reach up to 30% [18, 75, 76], though reports from specific hospitals can occasionally report higher frequencies [16, 77]. This is in line with our study where six isolates (11%) carried a bla NDM-1 gene and only one isolate (2%) carried a bla NDM-2 gene. It is possible that the almost ubiquitous nature of the oxa23 gene has reduced the selective advantage for subsequent acquisition and retention of bla NDM genes, limiting their spread within A. baumannii . The oxa23 gene in A. baumannii is typically carried on a transposon mobilized by insertion sequences, usually ISAba1 [10]. The insertion sequence elements are located immediately upstream of the oxa23 gene, where they provide a promotor sequence that drives high-level expression of oxa23 [13, 62]. The most commonly reported transposons carrying oxa23 are Tn2006, which is a composite transposon where oxa23 and three other genes are bracketed by two ISAba1 elements [62], and Tn2008, which is a one-ended transposon with a single ISAba1 element upstream of the oxa23 gene [78]. While the limitations of short-read sequencing in enabling the assembly of transposons is well known, in our study we were nevertheless able to identify a large number of different genetic arrangements surrounding the oxa23 gene. In line with what is reported in the literature, a structure likely to be Tn2006 was the most common arrangement in our isolates. However, the large number of different structures we have identified involving ISAba125, ISAba33 and ISAba4 in addition to ISAba1 demonstrate that the carbapenem-resistant A. baumannii population in AMUH is not dominated by a single mobile element that is disseminating oxa23. Rather, a multitude of different mobile elements are hosting the gene, consistent with the apparent endemic nature of oxa23 in the bacterial population, where multiple A. baumannii lineages co-circulate and there is the opportunity for persistent transfer, re-arrangement and selection to occur over an extended period of time.
The carriage of antibiotic-resistance genes on transposons is common, and is the typical genetic context for OXA-type carbapenemases in A. baumannii . However, in isolates belonging to CCPAS1 in our study, the most commonly identified mobile element carrying oxa23 was a bacteriophage phiOXA. Reports of the carriage of antibiotic-resistance genes in prophages have become more common in recent years [79–81], but it is thought that this is generally a rare occurrence [82]. However, recent evidence from studies focusing on A. baumannii have suggested that carriage of both virulence and antibiotic-resistance genes by prophages is relatively common in this species and may be a major mechanism of horizontal transfer of these genes [83–87]. It was recently noted that prophages appeared to be more common in IC5 isolates than in those belonging to IC1 or IC2 [85], and it is an intriguing possibility that prophages may be a major factor in the evolution of different ICs. The carriage of OXA-type carbapenemases in prophages has been observed previously, with oxa58 identified on a prophage in a Proteus mirabilis strain [88], and oxa23 identified on a prophage in A. baumannii strain ANC 4097 [83, 84] and on the phage phiOXA in isolate AB5075-UW [63]. However, López-Leal et al. [85] recently indicated that OXA carbapenemases in prophages may be more widespread, with evidence for potential OXA prophage carriage in approximately 25% of isolates studied. Similarly, we found oxa23 carried on phiOXA in 15% of our isolates. Moreover, these isolates were not clonally related within CCPAS1 but were spread throughout the CCPAS1 clade, indicating that phiOXA is widely disseminated amongst CCPAS1 isolates in AMUH. Furthermore, we demonstrated that phiOXA can be induced and that the induced phage particles are carrying the oxa23 gene. It is clear, therefore, that in A. baumannii , bacteriophages could be a major mechanism for the mobilization of antibiotic-resistance genes, including those of greatest clinical concern such as the carbapenemases. As more genomic studies using long-read sequencing are conducted that can properly resolve complex mobile element structures, the true magnitude of bacteriophage-mediated antibiotic-resistance gene carriage will be revealed.
Supplementary Data
Funding information
A.A. was funded by a Newton-Mosharafa Program Researcher Links Travel Grant (Egyptian STDF project ID 26235). J.M. was funded by a PhD studentship from the University of East Anglia. D.T. was funded by the University of the West of England. E.L. was funded by a University of East Anglia Norwich Medical School/Faculty of Medicine and Health Sciences PhD studentship. B.A.E. was funded by the University of East Anglia.
Author contributions
B.A.E. and A.A. were involved in the conceptualization, methodology, investigation, writing (original draft preparation, review and editing) and funding of the study. J.M. and D.T. were involved in the methodology, investigation and writing (original draft preparation, review and editing) of the study. E.L. was involved in the investigation and writing (review and editing) of the study.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: AMUH, Alexandria Main University Hospital; CC, clonal complex; IC, international clone; MLST, multilocus sequence typing; ST, sequence type.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Three supplementary figures are available with the online version of this article.
References
- 1.WHO Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. Geneva: World Health Organization; 2017. [Google Scholar]
- 2.Antunes LCS, Visca P, Towner KJ. Acinetobacter baumannii: evolution of a global pathogen. Pathog Dis. 2014;71:292–301. doi: 10.1111/2049-632X.12125. [DOI] [PubMed] [Google Scholar]
- 3.Rodríguez CH, Nastro M, Famiglietti A. Carbapenemases in Acinetobacter baumannii: review of their dissemination in Latin America. Rev Argent Microbiol. 2018;50:327–333. doi: 10.1016/j.ram.2017.10.006. [DOI] [PubMed] [Google Scholar]
- 4.Kempf M, Rolain JM. Emergence of resistance to carbapenems in Acinetobacter baumannii in Europe: clinical impact and therapeutic options. Int J Antimicrob Agents. 2012;39:105–114. doi: 10.1016/j.ijantimicag.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 5.Hu F, Zhu D, Wang F, Wang M. Current status and trends of antibacterial resistance in China. Clin Infect Dis. 2018;67:S128–S134. doi: 10.1093/cid/ciy657. [DOI] [PubMed] [Google Scholar]
- 6.Hsu LY, Apisarnthanarak A, Khan E, Suwantarat N, Ghafur A, et al. Carbapenem-resistant Acinetobacter baumannii and Enterobacteriaceae in South and Southeast Asia. Clin Microbiol Rev. 2017;30:1–22. doi: 10.1128/CMR.masthead.30-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kostyanev T, Vilken T, Lammens C, Timbermont L, Van’t Veen A, et al. Detection and prevalence of carbapenem-resistant Gram-negative bacteria among European laboratories in the COMBACTE network: a COMBACTE LAB-Net survey. Int J Antimicrob Agents. 2019;53:268–274. doi: 10.1016/j.ijantimicag.2018.10.013. [DOI] [PubMed] [Google Scholar]
- 8.Moghnieh RA, Kanafani ZA, Tabaja HZ, Sharara SL, Awad LS, et al. Epidemiology of common resistant bacterial pathogens in the countries of the Arab League. Lancet Infect Dis. 2018;18:e379–e394. doi: 10.1016/S1473-3099(18)30414-6. [DOI] [PubMed] [Google Scholar]
- 9.Potron A, Poirel L, Nordmann P. Emerging broad-spectrum resistance in Pseudomonas aeruginosa and Acinetobacter baumannii: mechanisms and epidemiology. Int J Antimicrob Agents. 2015;45:568–585. doi: 10.1016/j.ijantimicag.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 10.Evans BA, Amyes SGB. OXA β-lactamases. Clin Microbiol Rev. 2014;27:241–263. doi: 10.1128/CMR.00117-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Naas T, Oueslati S, Bonnin RA, Dabos ML, Zavala A, et al. Beta-lactamase database (BLDB) – structure and function. J Enzyme Inhib Med Chem. 2017;32:917–919. doi: 10.1080/14756366.2017.1344235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takebayashi Y, Findlay J, Heesom KJ, Warburton PJ, Avison MB, et al. Variability in carbapenemase activity of intrinsic OxaAb (OXA-51-like) β-lactamase enzymes in Acinetobacter baumannii . J Antimicrob Chemother. 2021;76:587–595. doi: 10.1093/jac/dkaa502. [DOI] [PubMed] [Google Scholar]
- 13.Turton JF, Ward ME, Woodford N, Kaufmann ME, Pike R, et al. The role of ISAba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii . FEMS Microbiol Lett. 2006;258:72–77. doi: 10.1111/j.1574-6968.2006.00195.x. [DOI] [PubMed] [Google Scholar]
- 14.Nigro SJ, Hall RM. Does the intrinsic oxaAb (blaOXA-51-like) gene of Acinetobacter baumannii confer resistance to carbapenems when activated by ISAba1? J Antimicrob Chemother. 2018;73:3518–3520. doi: 10.1093/jac/dky334. [DOI] [PubMed] [Google Scholar]
- 15.Higgins PG, Dammhayn C, Hackel M, Seifert H. Global spread of carbapenem-resistant Acinetobacter baumannii . J Antimicrob Chemother. 2010;65:233–238. doi: 10.1093/jac/dkp428. [DOI] [PubMed] [Google Scholar]
- 16.El-Sayed-Ahmed MAE-G, Amin MA, Tawakol WM, Loucif L, Bakour S, et al. High prevalence of bla(NDM-1) carbapenemase-encoding gene and 16S rRNA armA methyltransferase gene among Acinetobacter baumannii clinical Isolates in Egypt. Antimicrob Agents Chemother. 2015;59:3602–3605. doi: 10.1128/AAC.04412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fouad M, Attia AS, Tawakkol WM, Hashem AM. Emergence of carbapenem-resistant Acinetobacter baumannii harboring the OXA-23 carbapenemase in intensive care units of Egyptian hospitals. Int J Infect Dis. 2013;17:e1252–4. doi: 10.1016/j.ijid.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 18.Al-Hassan L, Zafer MM, El-Mahallawy H. Multiple sequence types responsible for healthcare-associated Acinetobacter baumannii dissemination in a single centre in Egypt. BMC Infect Dis. 2019;19:829. doi: 10.1186/s12879-019-4433-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zowawi HM, Sartor AL, Sidjabat HE, Balkhy HH, Walsh TR, et al. Molecular epidemiology of carbapenem-resistant Acinetobacter baumannii isolates in the Gulf Cooperation Council States: dominance of OXA-23-type producers. J Clin Microbiol. 2015;53:896–903. doi: 10.1128/JCM.02784-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nigro SJ, Hall RM. Structure and context of Acinetobacter transposons carrying the oxa23 carbapenemase gene. J Antimicrob Chemother. 2016;71:1135–1147. doi: 10.1093/jac/dkv440. [DOI] [PubMed] [Google Scholar]
- 21.Zarrilli R, Pournaras S, Giannouli M, Tsakris A. Global evolution of multidrug-resistant Acinetobacter baumannii clonal lineages. Int J Antimicrob Agents. 2013;41:11–19. doi: 10.1016/j.ijantimicag.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 22.Diancourt L, Passet V, Nemec A, Dijkshoorn L, Brisse S. The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS One. 2010;5:e10034. doi: 10.1371/journal.pone.0010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bartual SG, Seifert H, Hippler C, Luzon MAD, Wisplinghoff H, et al. Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii . J Clin Microbiol. 2005;43:4382–4390. doi: 10.1128/JCM.43.9.4382-4390.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaiarsa S, Batisti Biffignandi G, Esposito EP, Castelli M, Jolley KA, et al. Comparative analysis of the two Acinetobacter baumannii multilocus sequence typing (MLST) schemes. Front Microbiol. 2019;10:930. doi: 10.3389/fmicb.2019.00930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamidian M, Nigro SJ. Emergence, molecular mechanisms and global spread of carbapenem-resistant Acinetobacter baumannii . Microb Genom. 2019;5:e000306. doi: 10.1099/mgen.0.000306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodríguez CH, Balderrama Yarhui N, Nastro M, Nuñez Quezada T, Castro Cañarte G, et al. Molecular epidemiology of carbapenem-resistant Acinetobacter baumannii in South America. J Med Microbiol. 2016;65:1088–1091. doi: 10.1099/jmm.0.000328. [DOI] [PubMed] [Google Scholar]
- 27.Ghaith DM, Zafer MM, Al-Agamy MH, Alyamani EJ, Booq RY, et al. The emergence of a novel sequence type of MDR Acinetobacter baumannii from the intensive care unit of an Egyptian tertiary care hospital. Ann Clin Microbiol Antimicrob. 2017;16:34. doi: 10.1186/s12941-017-0208-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abouelfetouh A, Torky AS, Aboulmagd E. Phenotypic and genotypic characterization of carbapenem-resistant Acinetobacter baumannii isolates from Egypt. Antimicrob Resist Infect Control. 2019;8:185. doi: 10.1186/s13756-019-0611-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abouelfetouh A, Torky AS, Aboulmagd E. Role of plasmid carrying blaNDM in mediating antibiotic resistance among Acinetobacter baumannii clinical isolates from Egypt. 3 Biotech. 2020;10:170. doi: 10.1007/s13205-020-2157-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wingett SW, Andrews S. FastQ Screen: a tool for multi-genome mapping and quality control. F1000Res. 2018;7:1338. doi: 10.12688/f1000research.15931.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 34.Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Viale AM, Evans BA. Microevolution in the major outer membrane protein OmpA of Acinetobacter baumannii . Microb Genom. 2020;6:e000381. doi: 10.1099/mgen.0.000381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Price MN, Dehal PS, Arkin AP. FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jironkin A, Al-Shahib A, Painset A, Underwood A, Kapatai G, et al. PHEnix 1.3 ed; 2017
- 39.Dallman T, Ashton P, Schafer U, Jironkin A, Painset A, et al. SnapperDB: a database solution for routine sequencing analysis of bacterial isolates. Bioinformatics. 2018;34:3028–3029. doi: 10.1093/bioinformatics/bty212. [DOI] [PubMed] [Google Scholar]
- 40.Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43:e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, et al. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol. 2012;50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hunt M, Mather AE, Sánchez-Busó L, Page AJ, Parkhill J, et al. ARIBA: rapid antimicrobial resistance genotyping directly from sequencing reads. Microb Genom. 2017;3:e000131. doi: 10.1099/mgen.0.000131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McArthur AG, Waglechner N, Nizam F, Yan A, Azad MA, et al. The Comprehensive Antibiotic Resistance Database. Antimicrob Agents Chemother. 2013;57:3348–3357. doi: 10.1128/AAC.00419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Inouye M, Dashnow H, Raven L-A, Schultz MB, Pope BJ, et al. SRST2: rapid genomic surveillance for public health and hospital microbiology labs. Genome Med. 2014;6:11. doi: 10.1186/s13073-014-0090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abudahab K, Prada JM, Yang Z, Bentley SD, Croucher NJ, et al. PANINI: pangenome neighbour identification for bacterial populations. Microb Genom. 2019;5:e000220. doi: 10.1099/mgen.0.000220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jones P, Binns D, Chang H-Y, Fraser M, Li W, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Söding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lowe TM, Chan PP. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016;44:W54–W57. doi: 10.1093/nar/gkw413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 2018;35:518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Okonechnikov K, Conesa A, García-Alcalde F. Qualimap 2: advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics. 2016;32:292–294. doi: 10.1093/bioinformatics/btv566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Héritier C, Poirel L, Nordmann P. Cephalosporinase over-expression resulting from insertion of ISAba1 in Acinetobacter baumannii . Clin Microbiol Infect. 2006;12:123–130. doi: 10.1111/j.1469-0691.2005.01320.x. [DOI] [PubMed] [Google Scholar]
- 55.Woodford N, Ellington MJ, Coelho JM, Turton JF, Ward ME, et al. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int J Antimicrob Agents. 2006;27:351–353. doi: 10.1016/j.ijantimicag.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 56.Sambrook J, Russell DW. Extraction of bacteriophage λ DNA from large-scale cultures using proteinase K and SDS. CSH Protoc. 2006;2006:pdb.prot3972. doi: 10.1101/pdb.prot3972. [DOI] [PubMed] [Google Scholar]
- 57.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv. 2013:1303.3997 [Google Scholar]
- 58.Tomaschek F, Higgins PG, Stefanik D, Wisplinghoff H, Seifert H. Head-to-head comparison of two multi-locus sequence typing (MLST) schemes for characterization of Acinetobacter baumannii outbreak and sporadic isolates. PLoS One. 2016;11:e0153014. doi: 10.1371/journal.pone.0153014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zander E, Nemec A, Seifert H, Higgins PG. Association between β-lactamase-encoding bla(OXA-51) variants and DiversiLab rep-PCR-based typing of Acinetobacter baumannii isolates. J Clin Microbiol. 2012;50:1900–1904. doi: 10.1128/JCM.06462-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Poirel L, Bonnin RA, Boulanger A, Schrenzel J, Kaase M, et al. Tn125-related acquisition of blaNDM-like genes in Acinetobacter baumannii . Antimicrob Agents Chemother. 2012;56:1087–1089. doi: 10.1128/AAC.05620-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bonnin RA, Poirel L, Naas T, Pirs M, Seme K, et al. Dissemination of New Delhi metallo-β-lactamase-1-producing Acinetobacter baumannii in Europe. Clin Microbiol Infect. 2012;18:E362–E365. doi: 10.1111/j.1469-0691.2012.03928.x. [DOI] [PubMed] [Google Scholar]
- 62.Corvec S, Poirel L, Naas T, Drugeon H, Nordmann P. Genetics and expression of the carbapenem-hydrolyzing oxacillinase gene blaOXA-23 in Acinetobacter baumannii . Antimicrob Agents Chemother. 2007;51:1530–1533. doi: 10.1128/AAC.01132-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gallagher LA, Ramage E, Weiss EJ, Radey M, Hayden HS, et al. Resources for genetic and genomic analysis of emerging pathogen Acinetobacter baumannii . J Bacteriol. 2015;197:2027–2035. doi: 10.1128/JB.00131-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Casjens SR, Grose JH. Contributions of P2- and P22-like prophages to understanding the enormous diversity and abundance of tailed bacteriophages. Virology. 2016;496:255–276. doi: 10.1016/j.virol.2016.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moraru C, Varsani A, Kropinski AM. VIRIDIC – a novel tool to calculate the intergenomic similarities of prokaryote-infecting viruses. Viruses. 2020;12:1268. doi: 10.3390/v12111268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Adriaenssens E. How to name and classify your phage: an informal guide. Viruses. 2017;9:E70. doi: 10.3390/v9040070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Al-Hassan L, El Mehallawy H, Amyes SGB. Diversity in Acinetobacter baumannii isolates from paediatric cancer patients in Egypt. Clin Microbiol Infect. 2013;19:1082–1088. doi: 10.1111/1469-0691.12143. [DOI] [PubMed] [Google Scholar]
- 68.Karah N, Khalid F, Wai SN, Uhlin BE, Ahmad I. Molecular epidemiology and antimicrobial resistance features of Acinetobacter baumannii clinical isolates from Pakistan. Ann Clin Microbiol Antimicrob. 2020;19:2. doi: 10.1186/s12941-019-0344-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Evans BA, Hamouda A, Towner KJ, Amyes SGB. OXA-51-like beta-lactamases and their association with particular epidemic lineages of Acinetobacter baumannii . Clin Microbiol Infect. 2008;14:268–275. doi: 10.1111/j.1469-0691.2007.01919.x. [DOI] [PubMed] [Google Scholar]
- 70.Hamouda A, Evans BA, Towner KJ, Amyes SGB. Characterization of epidemiologically unrelated Acinetobacter baumannii isolates from four continents by use of multilocus sequence typing, pulsed-field gel electrophoresis, and sequence-based typing of bla(OXA-51-like) genes. J Clin Microbiol. 2010;48:2476–2483. doi: 10.1128/JCM.02431-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stietz MS, Ramírez MS, Vilacoba E, Merkier AK, Limansky AS, et al. Acinetobacter baumannii extensively drug resistant lineages in Buenos Aires hospitals differ from the international clones I-III. Infect Genet Evol. 2013;14:294–301. doi: 10.1016/j.meegid.2012.12.020. [DOI] [PubMed] [Google Scholar]
- 72.Graña-Miraglia L, Evans BA, López-Jácome LE, Hernández-Durán M, Colín-Castro CA, et al. Origin of OXA-23 variant OXA-239 from a recently emerged lineage of Acinetobacter baumannii international clone V. mSphere. 2020;5:e00801-19. doi: 10.1128/mSphere.00801-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Karah N, Jolley KA, Hall RM, Uhlin BE. Database for the ampC alleles in Acinetobacter baumannii . PLoS One. 2017;12:e0176695. doi: 10.1371/journal.pone.0176695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Al-Agamy MH, Khalaf NG, Tawfick MM, Shibl AM, El Kholy A. Molecular characterization of carbapenem-insensitive Acinetobacter baumannii in Egypt. Int J Infect Dis. 2014;22:49–54. doi: 10.1016/j.ijid.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 75.Kaase M, Nordmann P, Wichelhaus TA, Gatermann SG, Bonnin RA, et al. NDM-2 carbapenemase in Acinetobacter baumannii from Egypt. J Antimicrob Chemother. 2011;66:1260–1262. doi: 10.1093/jac/dkr135. [DOI] [PubMed] [Google Scholar]
- 76.Benmahmod AB, Said HS, Ibrahim RH. Prevalence and mechanisms of carbapenem resistance among. Microb Drug Resist. 2019;25:480–488. doi: 10.1089/mdr.2018.0141. [DOI] [PubMed] [Google Scholar]
- 77.Gomaa FAM, Helal ZH, Khan MI. High Prevalence of blaNDM-1, blaVIM, qacE, and qacEΔ1 genes and their association with decreased susceptibility to antibiotics and common hospital biocides in clinical isolates of Acinetobacter baumannii . Microorganisms. 2017;5:E18. doi: 10.3390/microorganisms5020018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Adams-Haduch JM, Paterson DL, Sidjabat HE, Pasculle AW, Potoski BA, et al. Genetic basis of multidrug resistance in Acinetobacter baumannii clinical isolates at a tertiary medical center in Pennsylvania. Antimicrob Agents Chemother. 2008;52:3837–3843. doi: 10.1128/AAC.00570-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Colomer-Lluch M, Imamovic L, Jofre J, Muniesa M. Bacteriophages carrying antibiotic resistance genes in fecal waste from cattle, pigs, and poultry. Antimicrob Agents Chemother. 2011;55:4908–4911. doi: 10.1128/AAC.00535-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Quirós P, Colomer-Lluch M, Martínez-Castillo A, Miró E, Argente M, et al. Antibiotic resistance genes in the bacteriophage DNA fraction of human fecal samples. Antimicrob Agents Chemother. 2014;58:606–609. doi: 10.1128/AAC.01684-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gómez-Gómez C, Blanco-Picazo P, Brown-Jaque M, Quirós P, Rodríguez-Rubio L, et al. Infectious phage particles packaging antibiotic resistance genes found in meat products and chicken feces. Sci Rep. 2019;9:13281. doi: 10.1038/s41598-019-49898-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Enault F, Briet A, Bouteille L, Roux S, Sullivan MB, et al. Phages rarely encode antibiotic resistance genes: a cautionary tale for virome analyses. ISME J. 2017;11:237–247. doi: 10.1038/ismej.2016.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Touchon M, Cury J, Yoon E-J, Krizova L, Cerqueira GC, et al. The genomic diversification of the whole Acinetobacter genus: origins, mechanisms, and consequences. Genome Biol Evol. 2014;6:2866–2882. doi: 10.1093/gbe/evu225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Costa AR, Monteiro R, Azeredo J. Genomic analysis of Acinetobacter baumannii prophages reveals remarkable diversity and suggests profound impact on bacterial virulence and fitness. Sci Rep. 2018;8:15346. doi: 10.1038/s41598-018-33800-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.López-Leal G, Santamaria RI, Cevallos MÁ, Gonzalez V, Castillo-Ramírez S. Prophages encode antibiotic resistance genes in Acinetobacter baumannii . Microb Drug Resist. 2020;26:1275–1277. doi: 10.1089/mdr.2019.0362. [DOI] [PubMed] [Google Scholar]
- 86.Wachino JI, Jin W, Kimura K, Arakawa Y. Intercellular transfer of chromosomal antimicrobial resistance genes between Acinetobacter baumannii strains mediated by prophages. Antimicrob Agents Chemother. 2019;63:e00334-19. doi: 10.1128/AAC.00334-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kenyon JJ, Arbatsky NP, Shneider MM, Popova AV, Dmitrenok AS, et al. The K46 and K5 capsular polysaccharides produced by Acinetobacter baumannii NIPH 329 and SDF have related structures and the side-chain non-ulosonic acids are 4-O-acetylated by phage-encoded O-acetyltransferases. PLoS One. 2019;14:e0218461. doi: 10.1371/journal.pone.0218461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Girlich D, Bonnin RA, Bogaerts P, De Laveleye M, Huang DT, et al. Chromosomal amplification of the blaOXA-58 carbapenemase gene in a Proteus mirabilis clinical isolate. Antimicrob Agents Chemother. 2017;61:e01697-16. doi: 10.1128/AAC.01697-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chan AP, Sutton G, DePew J, Krishnakumar R, Choi Y, et al. A novel method of consensus pan-chromosome assembly and large-scale comparative analysis reveal the highly flexible pan-genome of Acinetobacter baumannii . Genome Biol. 2015;16:143. doi: 10.1186/s13059-015-0701-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wibberg D, Salto IP, Eikmeyer FG, Maus I, Winkler A, et al. Complete genome sequencing of Acinetobacter baumannii strain K50 discloses the large conjugative plasmid pK50a encoding carbapenemase OXA-23 and extended-spectrum β-lactamase GES-11. Antimicrob Agents Chemother. 2018;62:e00212-18. doi: 10.1128/AAC.00212-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rafei R, Pailhoriès H, Hamze M, Eveillard M, Mallat H, et al. Molecular epidemiology of Acinetobacter baumannii in different hospitals in Tripoli, Lebanon using bla(OXA-51-like) sequence based typing. BMC Microbiol. 2015;15:103. doi: 10.1186/s12866-015-0441-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hammerum AM, Hansen F, Skov MN, Stegger M, Andersen PS, et al. Investigation of a possible outbreak of carbapenem-resistant Acinetobacter baumannii in Odense, Denmark using PFGE, MLST and whole-genome-based SNPs. J Antimicrob Chemother. 2015;70:1965–1968. doi: 10.1093/jac/dkv072. [DOI] [PubMed] [Google Scholar]
- 93.Vali L, Dashti K, Opazo-Capurro AF, Dashti AA, Al Obaid K, et al. Diversity of multi-drug resistant Acinetobacter baumannii population in a major hospital in Kuwait. Front Microbiol. 2015;6:743. doi: 10.3389/fmicb.2015.00743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Castanheira M, Costello SE, Woosley LN, Deshpande LM, Davies TA, et al. Evaluation of clonality and carbapenem resistance mechanisms among Acinetobacter baumannii-Acinetobacter calcoaceticus complex and Enterobacteriaceae isolates collected in European and Mediterranean countries and detection of two novel β-lactamases, GES-22 and VIM-35. Antimicrob Agents Chemother. 2014;58:7358–7366. doi: 10.1128/AAC.03930-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bonnin RA, Rotimi VO, Al Hubail M, Gasiorowski E, Al Sweih N, et al. Wide dissemination of GES-type carbapenemases in Acinetobacter baumannii isolates in Kuwait. Antimicrob Agents Chemother. 2013;57:183–188. doi: 10.1128/AAC.01384-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mabrouk A, Grosso F, Botelho J, Achour W, Ben Hassen A, et al. GES-14-producing Acinetobacter baumannii isolates in a neonatal intensive care unit in Tunisia are associated with a typical Middle East clone and a transferable plasmid. Antimicrob Agents Chemother. 2017;61:e00142-17. doi: 10.1128/AAC.00142-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang X, Qiao F, Yu R, Gao Y, Zong Z. Clonal diversity of Acinetobacter baumannii clinical isolates revealed by a snapshot study. BMC Microbiol. 2013;13:234. doi: 10.1186/1471-2180-13-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sullivan MJ, Petty NK, Beatson SA. Easyfig: a genome comparison visualizer. Bioinformatics. 2011;27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.