Abstract
Prostate cancer is one of the leading causes of mortality in men. The major cause of death in prostate cancer patients can be attributed to metastatic spread of disease or tumor recurrence after initial treatment. Prostate tumors are known to remain undetected or dormant for a long period of time before they progress locoregionally or at distant sites as overt tumors. However, the molecular mechanism of dormancy is yet poorly understood. In this study, we performed a differential gene expression analysis and identified a gene, Regucalcin (RGN), which promotes dormancy of prostate cancer. We found that cancer patients expressing higher level of RGN showed significantly longer recurrence-free and overall- survival. Using a doxycycline-inducible RGN expression system, we showed that ectopic expression of RGN in prostate tumor cells induced dormancy in vivo, while following suppression of RGN triggered recurrence of tumor growth. On the other hand, silencing RGN in LNCap cells promoted its outgrowth in the tibia of mice. Importantly, RGN promoted multiple known hallmarks of tumor dormancy including activation of p38 MAPK, decrease in Erk signaling and inhibition of FOXM1 expression. Furthermore, we found that RGN significantly suppressed angiogenesis by increasing secretory miR-23c level in the exosomes. Intriguingly, FOXM1 was found to negatively regulate miR-23c expression in prostate cancer. Additionally, we identified eleven RGN downstream target genes that independently predicted longer recurrence-free survival in patients. We found that expression of these genes was regulated by FOXM1 and/or p38 MAPK. These findings suggest a critical role of RGN in prostate cancer dormancy, and the utility of RGN signaling and exosomal miR-23c as biomarkers for predicting recurrence.
Keywords: Regucalcin, RGN, prostate cancer, Dormancy, Angiogenesis, miR-23c, cytoskeletal remodeling, FOXM1, p38 MAPK
INTRODUCTION:
Prostate cancer is deemed curable if diagnosed at an early stage of tumorigenesis. However, patients often experience disease recurrence, months or years after initial treatment, leading to reduced life expectancy6, 27. Despite significant attrition of tumor cells by treatment such as radical prostatectomy or hormone ablation therapy, a small fraction of cancer cells survives undetectable or dormant prior to their outgrowth as an overt tumor at local or distant sites35. One of the major sites of prostate cancer recurrence is bone7. Interestingly, prostate tumor cells are known to disseminate to bone even before diagnosis of primary disease35. A recent study has reported that more than 50% of patients with no evidence of disease after radical prostatectomy were found to be positive for disseminated tumor cells (DTC) in bone, indicating that cells disseminate and remain dormant in bone at a very early stage of tumorigenesis8 and these cells serve as the seed for recurrent tumor growth.
Tumor dormancy is thought to be the consequence of cancer-intrinsic or extrinsic factors. Two proposed mechanisms for cancer dormancy are cellular and environmental dormancy. Cellular dormancy results from inherent ability of cancer cells to remain in arrested state41. These cells intracellularly activate signaling such as p38 MAPK pathway that impairs growth ability of cells. On the other hand, environmental dormancy results from the crosstalk of tumor cells with microenvironment cells, such as stromal-, immune-, and endothelial-cells, which triggers partial cell death and molecular rewiring toward dormancy associated signaling1. A critical challenge in dormancy research is to identify key genetic mediators that regulate both cellular and environmental dormancy, and predict disease recurrence.
Regucalcin (RGN) is a highly conserved protein localized on X-chromosome and it is known to maintain intracellular calcium homeostasis55. RGN affects cellular growth and apoptosis by regulating protein kinases and repressing various signaling pathways at both transcription and translational level32, 56. RGN is also known to play a suppressive role on cancer initiation and progression33, 50. Ectopic RGN expression in cancer cell lines exerts growth suppressive effect by regulating various oncogenes and tumor suppressors50. Importantly, high RGN expression in cancer tissues is associated with prolonged survival of patients with liver, pancreatic, breast, colorectal and lung cancer58–61. Thus, RGN plays a prominent role in cellular processes associated with tumor progression.
In this report, we performed a screening of gene expression datasets of prostate cancer patients and identified a gene, RGN, as a strong prognostic indicator of longer disease recurrence-free survival. We demonstrated that RGN promoted dormancy by inducing both cellular dormancy via p38 MAPK activation and FOXM1 inhibition, and environmental dormancy by suppressing angiogenesis in the tumor microenvironment. The suppressive effect on angiogenesis was mediated by exosomal secretion of miR-23c. We also identified novel RGN target genes that independently predicted longer recurrence-free survival of patients.
RESULTS:
RGN expression correlates with late recurrence in patients
To identify genes that are associated with early- or late recurrence of prostate cancer, we utilized an existing dataset (GSE6919) and performed differential gene expression analysis between two groups of patients, (i) patients who had PSA recurrence before 3 years (designated as “early recurrence” group, n=16) and (ii) patients who showed disease recurrence only after 3 years (designated as “late recurrence” group, n=28) (Figure 1A and Supplementary Figure 1A). We identified 44 upregulated genes and 17 downregulated genes in late recurrence group when compared to early recurrence group. These 61 genes were further evaluated for their prognostic association with overall survival and recurrence-free survival using the TCGA dataset (n=566). We found that two genes, RGN and KIAA1462, were significantly associated with longer overall survival (Figure 1B and Supplementary Figure 1C–D); however, only RGN expression predicted longer recurrence-free survival (Figure 1C and Supplementary Figure 1E). Therefore, we selected RGN for further study. To substantiate the prognostic significance of RGN, we examined two additional datasets (GSE21034 and GSE6919) and found that prostate cancer patients expressing higher level of RGN showed longer recurrence-free survival (Figure 1D–E). Importantly, RGN expression did not correlate with disease severity as no progressive decrease in RGN was evident with increase in Gleason grade (Supplementary Figure 1F). We also examined RGN expression in patients who had incidence of either loco-regional recurrence or onset of new bone lesion after initial treatment of the disease (relapse group) and compared to patients who did not have such disease progression (no relapse group) using the TCGA dataset (Table 1). RGN expression was found to be significantly decreased in patients who experienced overt disease relapse (Figure 1F). Similar to RGN, BMP7, a protein that we have previously reported to promote dormancy26, was also significantly downregulated in the relapse group (Figure 1F). However, expression of established tumor or metastasis suppressor genes in prostate cancer such as, PTEN, PHLPP1 and CD8212, 16, 25, did not decrease in relapse group, suggesting a distinct role of RGN in tumor relapse (Figure 1F). Additionally, RGN level was significantly lower in patients with metastasis when compared to the patients with localized primary tumors (Figure 1G). This result was further verified by immunohistochemial analysis of patient’s tumor specimens. While strong RGN staining was observed in benign prostatic hyperplasia, RGN staining progressively decreased in primary and metastatic tumors (Figure 1H).
Figure 1: RGN expression correlates with late recurrence in patients.
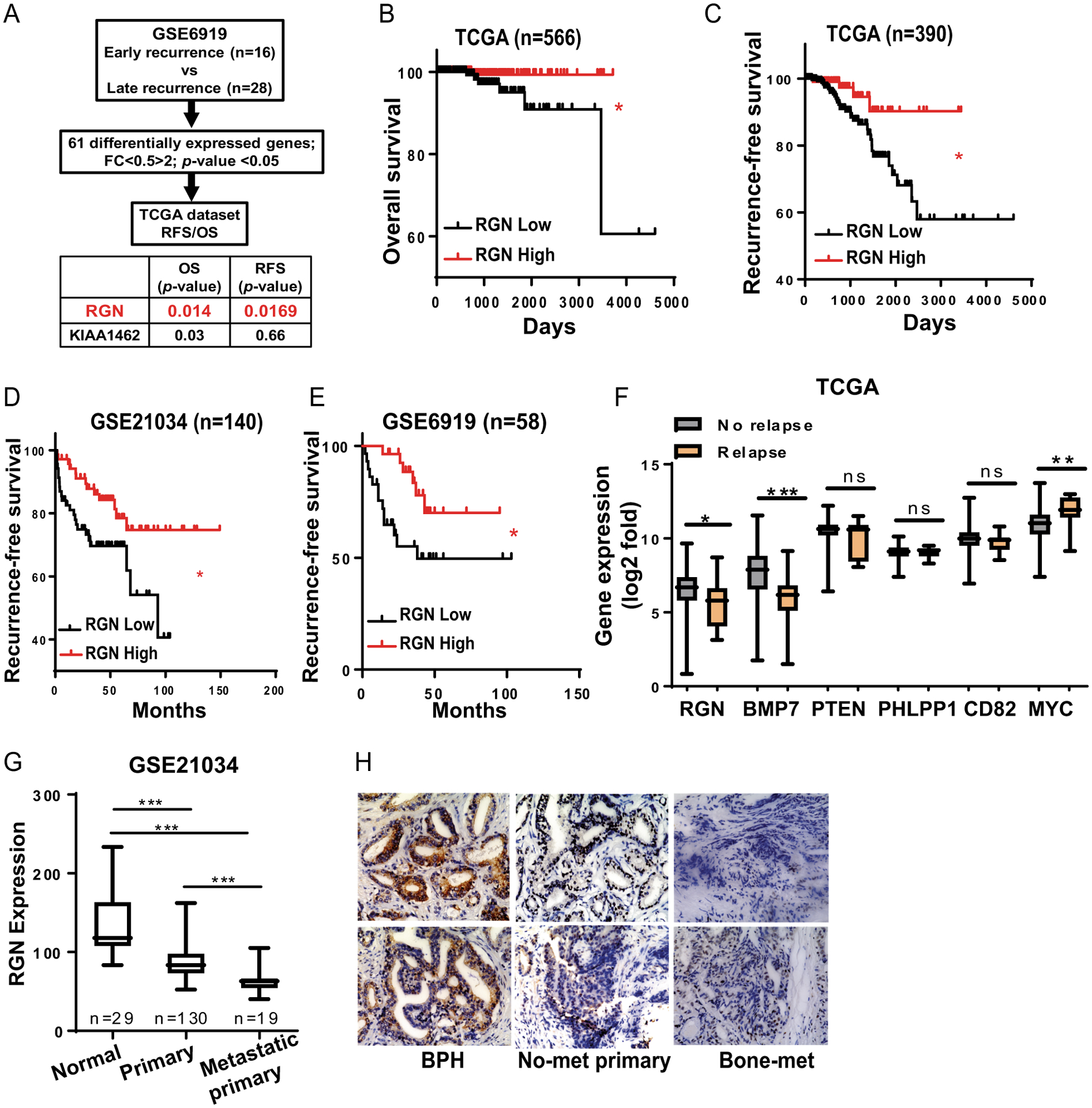
(A) Outline of differentially expressed genes analysis between Early recurrence and Late recurrence group. (B–C) Patients in TCGA were stratified based on their expression levels of RGN and overall- (B) and recurrence-free- survival (C) were examined. (D–E) Patients in GSE21034 (D) and GSE6919 (E) were stratified based on their RGN expression and its association with recurrence-free survival was examined. The overall- and recurrence-free survival plots in figure B–E were analyzed by log-rank (Mantel-cox) test. (F) Expression of indicated genes were examined in patients with No-relapse (n=541) and Relapse disease (n=11) using the TCGA dataset. Data were analyzed using unpaired t-test. (G) RGN expression was examined in patient normal, primary and metastatic primary tumors using the GSE21034 dataset. One way anova was performed to analyze the data (H) Immunohistochemistry staining was performed for patient tumor specimens to examine RGN expression. *, P-value<0.05, ** P-value<0.01 and ***, P-value<0.0001.
Table 1:
Clinical details of patients with new tumor event after initial treatment in TCGA dataset.
| Patient ID | Days to new tumor event after initial treatment | New neoplasm event occurrence anatomic site |
New tumor event after initial treatment | New neoplasm event type |
|---|---|---|---|---|
| TCGA-VP-A878-01 | 2354 | Bone | YES | Distant metastasis |
| TCGA-YL-A9WY-01 | 975 | Bone | YES | Distant metastasis |
| TCGA-EJ-5518-01 | 2104 | Bone | YES | Distant metastasis |
| TCGA-YL-A9WJ-01 | 1476 | Other, Specify | YES | Distant metastasis |
| TCGA-EJ-5525-01 | 546 | Bone | YES | Distant metastasis |
| TCGA-G9-A9S0-01 | 650 | Other, Specify | YES | Distant metastasis |
| TCGA-CH-5751-01 | 365 | Bone | YES | Distant metastasis |
| TCGA-KK-A59X-01 | 2233 | Other, Specify | YES | Locoregional Recurrence |
| TCGA-KK-A7B4-01 | 963 | Bone | YES | Locoregional Recurrence |
| TCGA-EJ-8469-01 | 1925 | YES | Locoregional Recurrence | |
| TCGA-YL-A8S9-01 | 1006 | YES | Locoregional Recurrence |
Our group and others have previously demonstrated that tumor recurrence is controlled by epigenetic mechanisms, such as promoter-methylation dependent silencing of genes that inhibits recurrent outgrowth of tumors29, 42, 51. Therefore, we investigated whether RGN can be regulated by promoter DNA methylation using the TCGA methylation dataset. The RGN promoter was found to be significantly methylated in prostate tumors compared to paired normal tissues (Supplementary Figure 1G). We then treated PC3mm prostate cancer cells with Decitabine, a drug that inhibits methylation of cytosine residues in DNA, and found that decitabine increased RGN level (Supplementary Figure 1H), indicating that RGN is epigenetically silenced but the expression of this gene is reversible. These findings strongly indicate that RGN plays a significant role in regulation of disease relapse of prostate cancer.
RGN induces dormancy in vivo
Based on our findings, we hypothesized that RGN may induce dormancy of prostate cancer. To study the role of RGN in dormancy in vivo, we first generated bone metastatic PC3mm and C42B cell lines that expressed RGN in a doxycycline-inducible manner (Supplementary Figure 2A). We then transplanted these cells by intracardiac injection into nude mice and controlled RGN expression by adding or withdrawing doxycycline in the drinking water. We found that continuous RGN expression led to a significant decrease in the onset of bone metastasis of both C42B and PC3mm cells (Figure 2A–B, −/− vs +/+ group). Importantly, withdrawal of doxycycline and concomitant reduction of RGN expression after 14 days rescued the suppressive effect of RGN and increased the onset of bone metastasis (Figure 2A–B, +/+ vs +/− group). In our histological analysis, a significant decrease in bone metastasis area was evident in tumors that continuously expressed RGN (Figure 2C). These results indicate the reversible effect of RGN in inducing dormancy of prostate cancer in bone. We further studied the effect of RGN on the growth of metastatic lesion by treating mice with doxycycline after formation of metastatic lesions in bone. RGN significantly reduced the growth of bone lesions demonstrating its suppressive effect in progression of metastasis (Supplementary Figure 2B). We also performed a loss of function study by silencing RGN expression in LNCap cells (Supplementary Figure 2C). We implanted 150,000 LNCap cells, which is less than the number of cells required to establish tumor growth in the bone14, 20, to examine the effect of RGN knockdown in tibial growth ability. While scrambled shRNA infected LNCap cells showed baseline bioluminescence signal, knockdown of RGN significantly accelerated tumor growth (Figure 2D). We then sought of a possibility that LNCap-shScrambled cells injected mice harbor dormant cells that lacked in vivo growth potential. We therefore flushed the tibial bones of these mice and examined the presence of LNCap cells by quantifying human-specific Prostate Specific Membrane Antigen (PSMA) by flow cytometry. As shown in Figure 2E LNCap-shScrambled cells injected tibia harbored PSMA positive cells, while age-matched tumor-free tibia were negative for PSMA expressing cells. Importantly, tibia from RGN silenced LNCap cells injected mice retained significantly higher number of PSMA positive cells indicating that these cells acquired ability to grow in the bone. Together, our results strongly demonstrate that RGN promotes dormancy prostate cancer cells in bone.
Figure 2: RGN promotes dormancy in vivo.
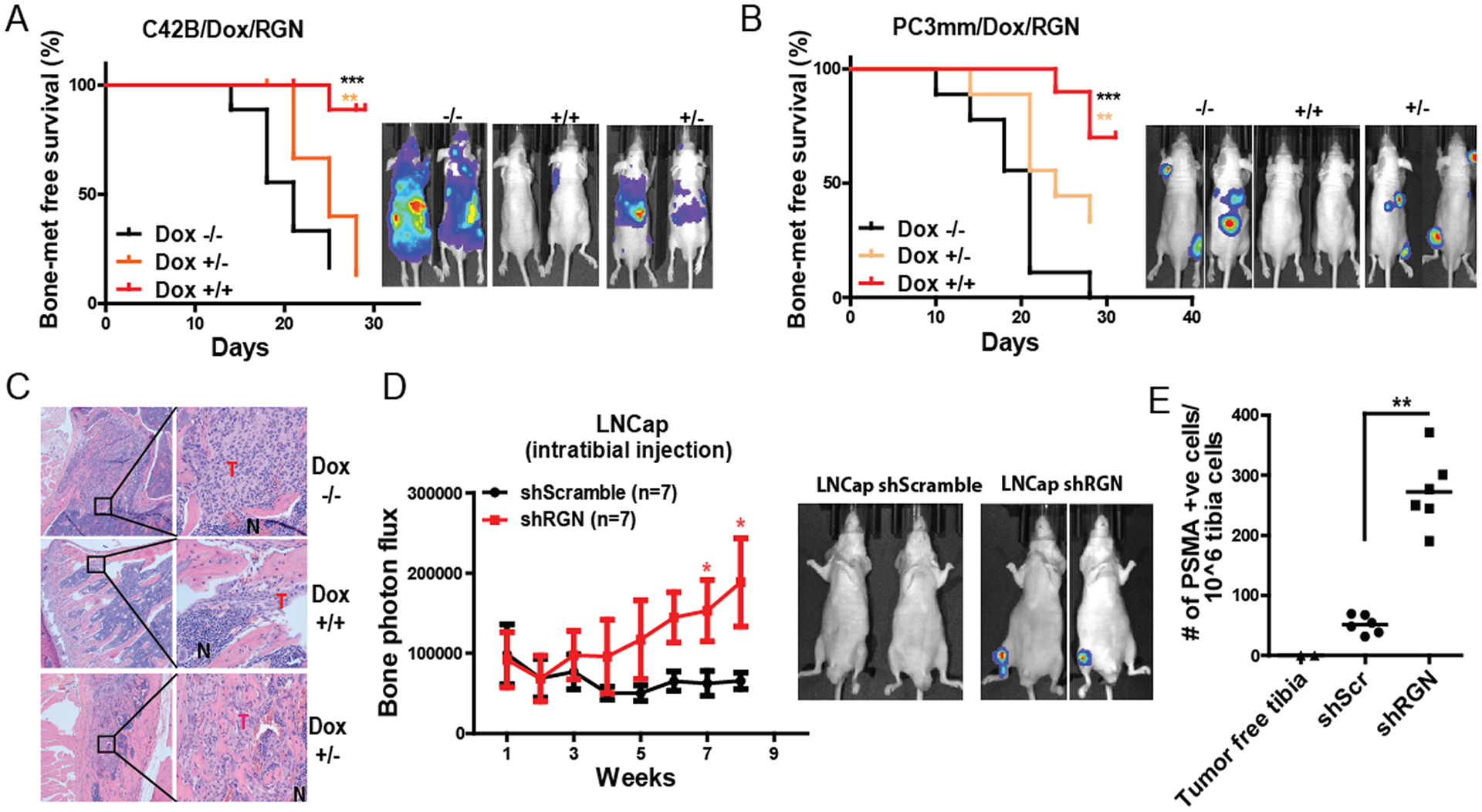
(A) C42B cells were modified to express RGN in doxycycline dependent manner. These cells were systemically implanted into nude mice by intracardiac injection, and kaplan-meier bone metastasis-free survival curve was plot (5 × 105 cells/mouse, n=9/group). −/− group was fed with 5% sucrose water, +/+ group received doxycycline water (2mg/ml in 5% sucrose) and +/− group was fed with doxycycline water for 14 days followed by feeding mice with 5% sucrose water. Panel on the right shows representative bioluminescence image of mice at endpoint. (B) PC3mm cells were modified to express RGN in a doxycycline dependent manner and transplanted to nude mice via intracardiac injection (5 × 105 cells/mouse, n=9/group). −/− group was fed with 5% sucrose water, +/+ group received doxycycline water (2mg/ml in 5% sucrose) and +/− group was fed with doxycycline water for 12 days followed by feeding mice with 5% sucrose water. Panel on the right shows representative bioluminescence image of mice at endpoint. The bone-metastasis free survival plots in Figure A–B were analyzed by log-rank (Mantel-cox) test. (C) Representative H&E staining of bone sections from mice in panel A. (D) LNCap shScramble and LNCap shRGN cells were injected in nude mice and tumor growth in tibia was examined by IVIS imaging (n=7/group, 150,000 cells/tibia). Representative images of IVIS imaging are shown in the right panel. The data collected at each time point was analyzed using unpaired t-test. (E) Human-specific PSMA antibody staining was performed in the tibia flushed cells from mice in panel E by flow cytometry. Unpaired t-test was used to analyze the data. *, P-value<0.05, ** P-value<0.01 and ***, P-value<0.0001.
RGN promotes dormancy in vitro
As the in vivo experiments suggested that RGN retains cells in dormant state, we studied the effect of ectopic RGN expression on cell proliferation and cell cycle progression. RGN significantly decreased cell proliferation and induced cell cycle arrest, which was reversed when its expression was downregulated by doxycycline withdrawal (Figure 3A–B, Supplementary Figure 3A–B). To further validate the effect of RGN in cell proliferation, we labeled cells with fluorescent staining Celltrace dye. The signal of this dye dilutes to half with each cell division and non-proliferating cells retain the fluorescent signal. We found that RGN expression significantly increased the number of label-retaining cells while withdrawal of doxycycline decreased label retention ability of these cells (Figure 3C). The RGN dependent decrease in cell growth was also validated by examining Ki67 positive population by flow cytometry (Figure 3D). Additionally, the expression of cell cycle inhibitor proteins, p21 and p18, were found to be augmented in cells overexpressed with RGN (Figure 3E). Conversely, silencing RGN in LNCap cells significantly increased cell proliferation and decreased p21 and p27 expression (Figure 3F–H). We further investigated the effect of RGN in p21 expression in vivo, by performing immunohistochemical staining of the tumors. As shown in Figure 3I, cells with reduced RGN expression showed decreased p21 staining. These results suggest that RGN promotes tumor cell dormancy by blocking cell cycle in vitro.
Figure 3: RGN promotes dormancy in vitro.
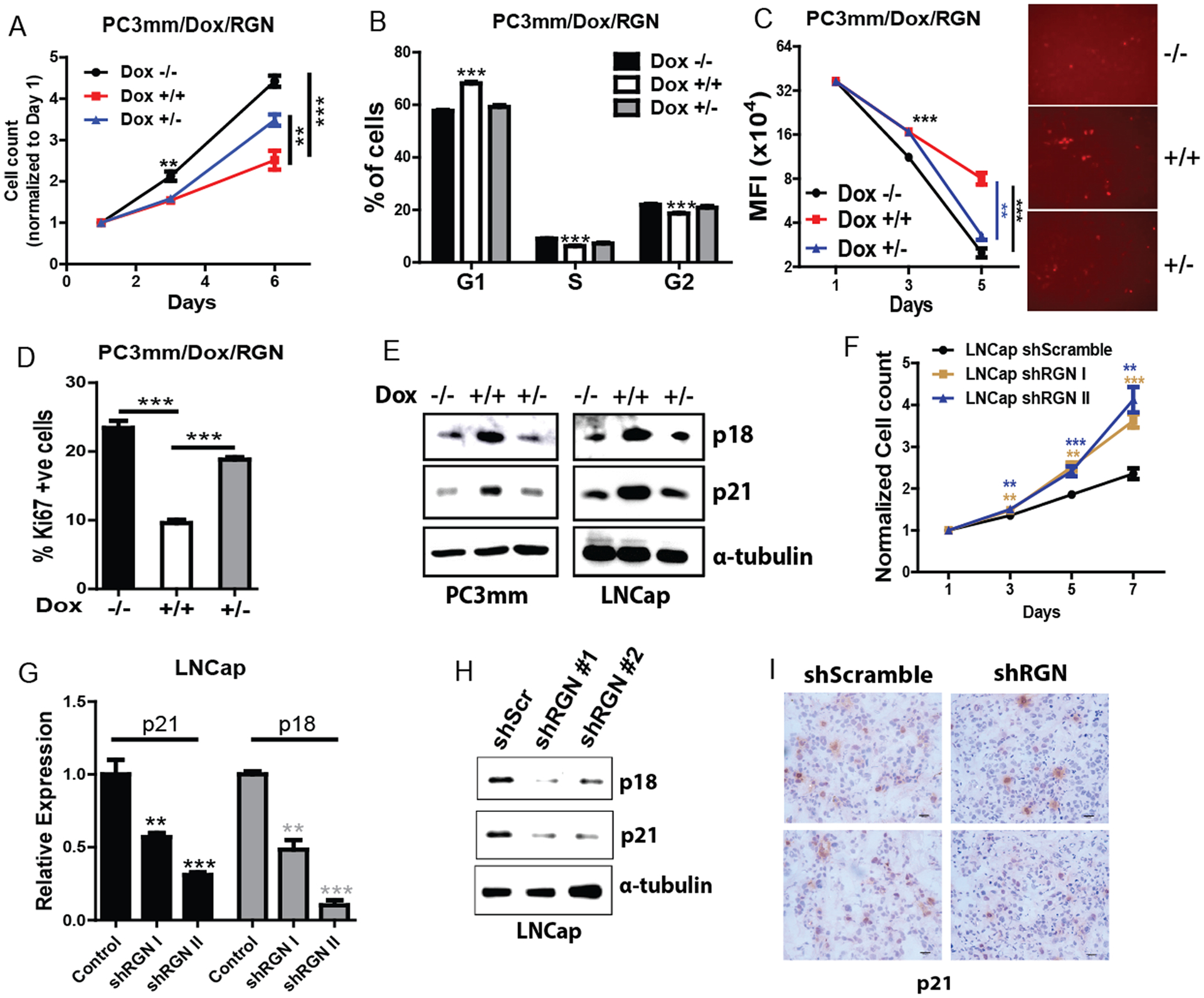
(A) PC3mm cells were seeded in 96-well plates and treated with or without doxycycline as indicated, and cell growth was examined by MTS assay (500 cells/well and n=8). −/−, no doxycycline treatment till day 6, +/+, continuous treatment with doxycycline for six days, +/–, 3 days of doxycycline treatment followed by culturing cells in no doxycycline conditions. (B) PC3mm cells treated with or without doxycline were subjected to cell cycle analysis using flow cytometry. % of cells in G1, S and G2 phase is shown (n=4/group). −/−, no doxycycline treatment, +/+, doxycycline treatment for 72 hrs, +/−, 24 hrs of doxycycline treatment followed by culturing cells in no doxycycline conditions. Data was analyzed using one-way anova and tukey’s multiple comparison test. (C) PC3mm/Dox/RGN cells were stained with CellTrace Far Red dye and treated with or without doxycycline as indicated for 5 days followed by quantifying mean fluorescence of dye staining positive cells by flow cytometry (n=4/group). The right panel shows the representative image of CellTrace dye staining cells. Data was analyzed using one-way anova and tukey’s multiple comparison test. (D) PC3mm cells harboring the Ki67 reporter plasmid were treated with or without doxycycline, and population of Ki67 positive cells was quantified by flow cytometry (n=4/group). Data was analyzed using one-way anova and tukey’s multiple comparison test. (E) PC3mm and LNCap cells were treated with doxycycline to induce RGN expression, and expression of p18, p21 was examined by western blot. α-tubulin was used as a loading control. (F) LNCap shScramble, LNCap shRGN I and LNCap shRGN II cells were seeded in 96 well plate and subjected to cell proliferation analysis at day 1, 3, 5 and 7 by MTS assay (n=8/group). Data was analyzed using one-way anova and tukey’s multiple comparison test. (G–H) Cells in panel F were used to examine the expression of p18 and p21 by qRT-PCR (G) and western blot (H). α-tubulin was used as a loading control. (I) IHC was performed for p21 in tumors from mice transplanted with LNCap shScrambled or LNCap shRGN. Representative images are shown. Bar: 20uM. ** P-value<0.01 and ***, P-value<0.0001.
RGN promotes dormancy signaling
Multiple cellular pathways and paracrine signaling in the metastatic microenvironment regulate tumor dormancy1. In prostate cancer, p38/Erk signaling and FOXM1 expression are known to induce dormancy or indolent growth1, 5, 44. We therefore examined these signaling in prostate cancer cells expressing RGN in a doxycycline-dependent manner. We found that RGN expression significantly increased activation of p38 signaling. However, this activation was reversed when RGN expression was reduced in the cells by doxycycline withdrawal from the culture media (Figure 4A). p38 activation was also observed in the cells stably expressing RGN (Supplementary Figure 4A). In support of this finding, gene set enrichment analysis (GSEA) analysis also revealed that the p38MAPK signature is significantly enriched in patients expressing high levels of RGN (Figure 4B). Conversely, RGN expression reduced activation of Erk, indicating an increase in p38 to Erk ratio (Figure 4C). On the other hand, RGN silencing decreased p38 activation while increasing Erk phosphorylation thereby decreasing p38 to Erk ratio (Figure 4D). We then examined the effect of RGN on FOXM1 expression. As shown in Figure 4E, RGN expression decreased FOXM1 level and activation of its upstream AKT pathway (Figure 4E). The negative correlation between RGN and FOXM1 was even evident in the clinical samples (Figure 4F). Importantly, patients expressing higher levels of FOXM1 had significantly shorter recurrence-free survival time (Figure 4G). We have previously shown that the self-renewal property of prostate cancer cells are compromised during dormancy26. We examined the effect of RGN on self-renewal by sphere formation assay. Indeed, RGN significantly decreased the sphere forming ability of PC3mm and LNCap cells (Figure 4H–I). Taken together, these results strongly suggest that RGN activates dormancy signaling and inhibits stem cell property of prostate cancer.
Figure 4: RGN promotes dormancy signaling.
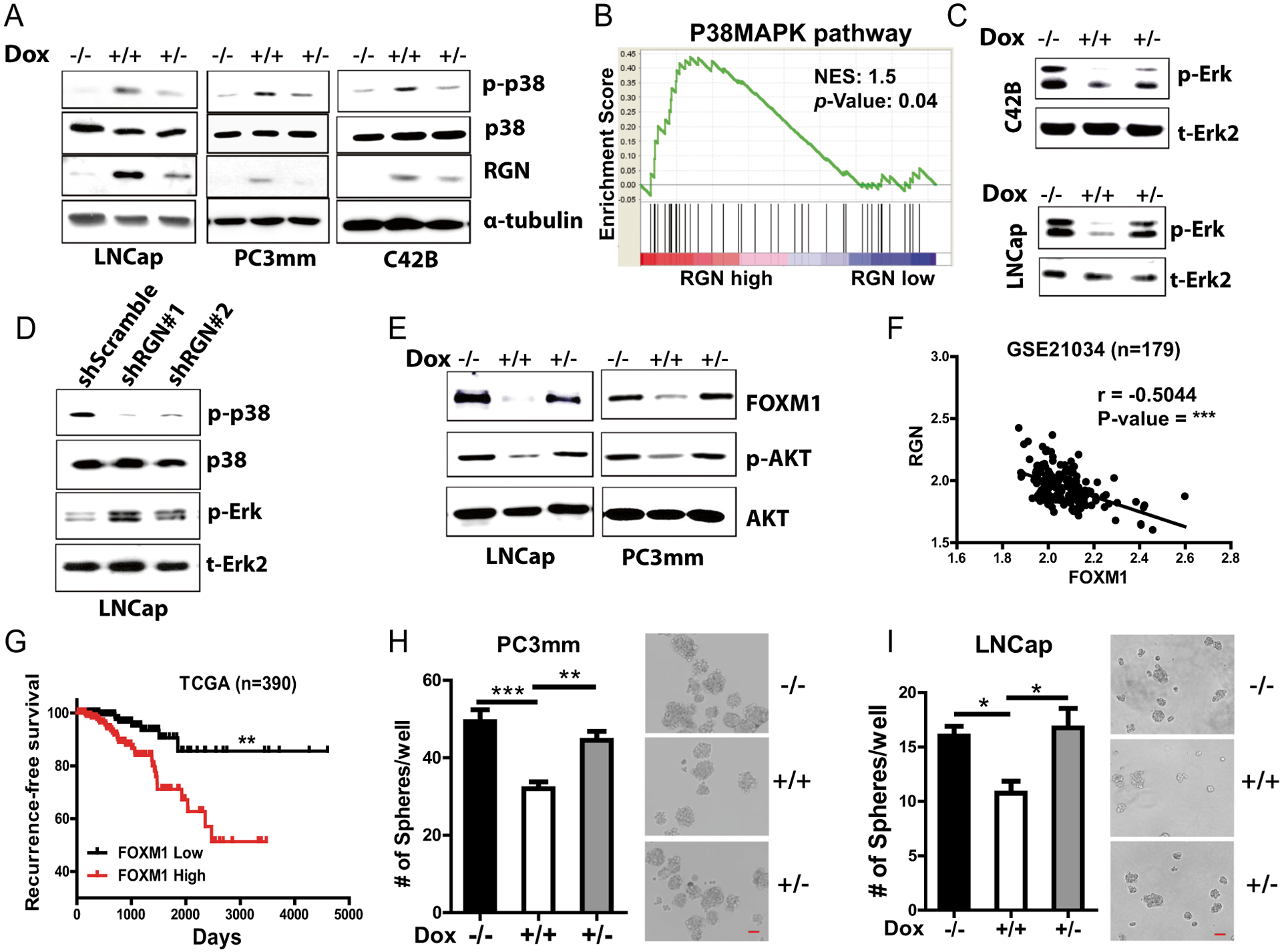
(A) RGN expression was induced in LNCap, PC3mm and C42B cells by treating them with or without doxycycline (0.2ug/ml), and p-p38, p38 and RGN expression was examined by western blot. −/−, no doxycycline treatment, +/+, continuous treatment with doxycycline for 48 hours, +/−, 24 hours of doxycycline followed by withdrawal of doxycycline. α-tubulin was used as a loading control. (B) Enrichment of Biocarta p38 MAPK pathway signature was examined in patients expressing low or high level of RGN. The RGN-low group includes patients with RGN expression in the lower quartile while the RGN-high group includes patients expressing RGN in the upper quartile in the TCGA dataset. (C) C42B/Dox/RGN and LNCap/Dox/RGN cells were treated with doxycycline and expression of p-Erk and total Erk2 was examined by western blot. −/−, no doxycycline treatment, +/+, continuous treatment with doxycycline for 48 hours, +/−, 24 hours of doxycycline followed by withdrawal of doxycycline. (D) p-p38, t-p38, p-Erk and t-Erk2 expression was examined in LNCap shScramble, LNCap shRGN I and LNCap shRGN II cells by western blot. (E) LNCap and PC3mm cells were induced for RGN expression, and the expressions of FOXM1, p-AKT and AKT expression were examined by western blot. (F) Correlation of RGN with FOXM1 was examined using the GSE21034 cohort dataset. (G) Patients in the TCGA cohort were stratified based on their FOXM1 expression levels, and recurrence-free- survival was examined. The recurrence-free survival plot was analyzed by log-rank (Mantel-cox) test. (H–I) RGN expression in LNCap and PC3mm cells by doxycycline treatment as in panels A and B, and cells were seeded in low binding plate (200/well for PC3mm and 400/well for LNCap; n=8/group) followed by counting the number of spheres at day 5. Representative images are shown in right panels. Data were analyzed using one way anova and tukey test. Bar: 50uM. *, P-value<0.05, ** P-value<0.01 and ***, P-value<0.0001.
Identification of RGN downstream target genes
To explore the clinically relevant downstream target genes of RGN, we separated patients in three different datasets (TCGA (n=498), GSE6919 (n=90) and GSE21034 (n=140)) based on their RGN expression (lower quartile- vs upper quartile- RGN expression groups) irrespective of their clinical parameters and performed differential gene expression analysis between these two groups. This analysis revealed that 93 genes were commonly and significantly upregulated in RGN upper quartile cohorts while no gene was found to be commonly downregulated (Figure 5A). We further screened these 93 genes by two criteria – (i) their association with relapse status of patient and (ii) their prognostic ability to predict recurrence-free survival. This screening resulted in 11 genes that were significantly downregulated in relapsed tumors and strongly correlated to longer recurrence-free survival (Figure 5B and Table 2). We then investigated whether these genes are directly regulated by RGN in prostate cancer cell lines. We found that, ectopic RGN expression significantly elevated expression of majority of these genes in PC3mm and LNCap cells whereas, silencing RGN in LNCap cells decreased their expression (Figure 5C–E). This result indicates that these genes are downstream targets of RGN. As our previous findings suggested that RGN stimulated p38 activation and inhibited FOXM1 expression, we explored whether signaling from p38 and FOXM1 mediate the upregulation of RGN target genes. We therefore expressed activated p38 and FOXM1 in PC3mm cells (Supplementary Figures 5A–B). As shown in Figure 5F, expression of activated p38 in PC3mm cells significantly increased expression of all target genes. On the other hand, ectopic expression of FOXM1 downregulated five genes (Figure 5G). These results imply that RGN stimulates expression of these genes via either p38 or FOXM1 or both, and these genes are positive downstream regulators of dormancy signaling.
Figure 5: RGN expression enhances expression of cytoskeletal genes via p38 and FOXM1 signaling.
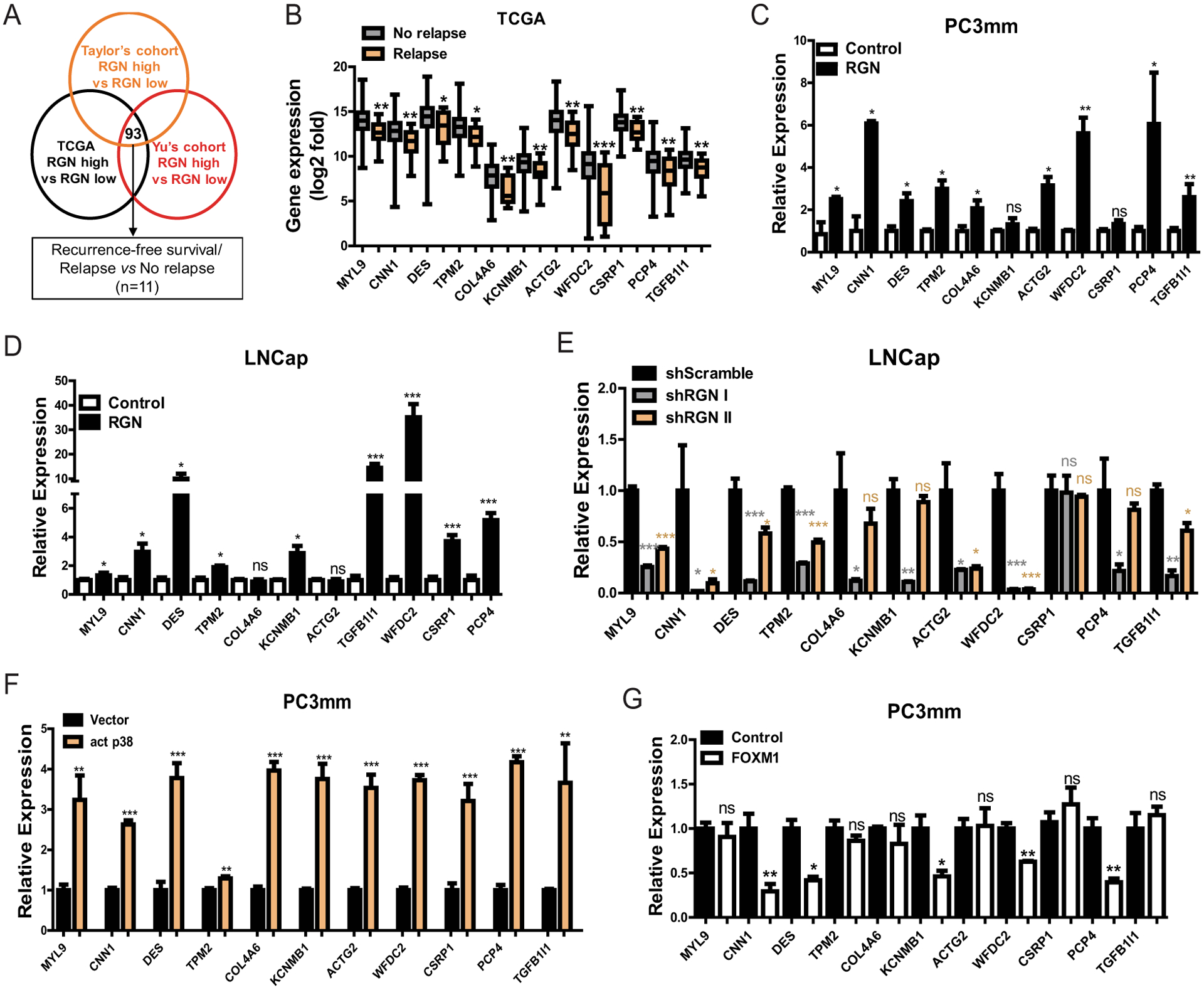
(A) Scheme outlining the approach to identify differentially expressed genes between lower- and upper-quartile RGN- expressing patients. 93 genes were found to be commonly upregulated in all 3 datasets (n=125/group for TCGA, n=23/group for GSE6919 and n=35/group for GSE21034). These genes were then screened by the TCGA dataset for their prognostic significance of recurrence-free survival and differential expression between patients with or without incidence of new tumor onset, (disease relapse vs no-relapse). 11 genes were selected based on these criteria. (B) Expression of 11 genes identified by screening shown in (A) was examined between No-relapse and Relapse groups. Unpaired t-test was used for Statistical analysis of each gene. (C–D) RGN was ectopically expressed in PC3mm (C) and LNCap (D) cells in a doxycycline dependent manner and the expression of 11 genes were examined by qRT-PCR. Unpaired t-test was used for statistical analysis of each gene. (E) RGN expression was suppressed in LNCap cells by infecting with two different shRNAs (#1 and #2), followed by performing qRT-PCR for indicated gene expression. One-way anova and tukey’s multiple comparison test was used for statistical analysis of each gene. (F) PC3mm cells were transfected with either vector only or activated p38 expressing vector followed by examining the levels of indicated gene by qRT-PCR. Unpaired t-test was used for statistical analysis of each gene. (G) Expression levels of indicated genes were examined in PC3mm cells expressing vector control or FOXM1. Unpaired t-test was used for statistical analysis of each gene. *, P-value<0.05, ** P-value<0.01 and ***, P-value<0.0001.
Table 2:
Hazard ratio, correlation with RGN expression and p-value of recurrence-free survival analysis of indicated genes in three datasets.
| Taylor’s Cohort | Yu’s Cohort | TCGA | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene Name | Survival | Hazard ratio | Correlation | Survival | Hazard ratio | Correlation | Survival | Hazard ratio | Correlation | No-relapse vs Relapse (Fold Change) | No-relapse vs Relapse (P-value) |
| (P-value) | (HR) | (r) | (P-value) | (HR) | (r) | (P-value) | (HR) | (r) | |||
| MYL9 | *** | 0.31 | 0.78 | ns | 0.48 | 0.59 | * | 0.45 | 0.79 | 6.74 | *** |
| CNN1 | *** | 0.3 | 0.84 | 0.08 | 0.45 | 0.57 | *** | 0.42 | 0.8 | 6.4 | ** |
| DES | * | 0.5 | 0.74 | * | 0.4 | 0.6 | *** | 0.41 | 0.78 | 8.23 | *** |
| TPM2 | ** | 0.39 | 0.78 | * | 0.36 | 0.62 | *** | 0.4 | 0.75 | 6.36 | ** |
| COL4A6 | 0.08 | 0.56 | 0.67 | * | 0.37 | 0.55 | * | 0.5 | 0.64 | 4.47 | ** |
| KCNMB1 | ns | 0.52 | 0.78 | ns | 0.59 | 0.65 | ** | 0.5 | 0.82 | 4.41 | ** |
| ACTG2 | * | 0.42 | 0.77 | * | 0.37 | 0.55 | * | 0.41 | 0.78 | 8.08 | *** |
| WFDC2 | * | 0.52 | 0.36 | 0.07 | 0.45 | 0.4 | * | 0.47 | 0.45 | 10.66 | **** |
| CSRP1 | ** | 0.36 | 0.79 | 0.09 | 0.46 | 0.46 | * | 0.47 | 0.78 | 5.79 | *** |
| PCP4 | * | 0.42 | 0.78 | * | 0.3 | 0.46 | 0.05 | 0.52 | 0.69 | 5.1 | ** |
| TGFB1I1 | 0.06 | 0.53 | 0.7 | 0.17 | 0.54 | 0.57 | * | 0.47 | 0.7 | 4.21 | ** |
MYL9: Myosin light chain 9; CNN1: Calponin 1; DES: Desmin; TPM2: Tropomyosin 2; COL4A6: Collagen type IV alpha 6 chain; KCNMB1: Potassium calcium-activated channel subfamily M regulatory beta subunit 1; ACTG2: Actin gamma 2, smooth muscle; WFDC2: WAP four disulfide core domain 2; CSRP1: Cysteine and glycine rich protein 1; PCP4: Purkinje cell protein 4; TGFB1I1: Transforming growth factor beta 1 induced transcript 1.
Several RGN target genes are implicated with the cellular cytoskeleton and extracellular matrix4, 45, 62 (Table 2), which is responsible for regulating the cell morphology thereby affecting cell migration and invasion. Therefore, we examined the effect of RGN in migration and invasion ability of cancer cells. RGN significantly reduced invasion and migration ability of cancer cells (Supplementary Figure 5C–D). These results suggest that RGN downstream genes potentially inhibit invasion and migration thereby limiting aggressiveness of prostate cancer cells.
RGN suppresses angiogenesis in tumor microenvironment by exosomal miRNA-23c
Angiogenic dormancy is one prominent mechanism of environmental dormancy40. Therefore, we sought of a possibility that RGN affects angiogenesis. We examined the bone metastatic lesions from mice that were systemically implanted with C42B cells and performed immunohistochemistry for the expression of CD31. As shown in Figure 6A, tumors expressing RGN (Dox +/+ group) showed significantly reduced vessel formation compared to the lesions in which cancer cells did not express RGN (Dox −/− and Dox +/−). This finding prompted us to examine the effect of RGN in angiogenesis in vitro. We treated human umbilical vein endothelial (HUVEC) cells with conditioned medium (CM) derived from prostate cancer cell lines with or without RGN expression. The tube forming ability of HUVEC cells significantly decreased when incubated with CM prepared from RGN expressing cells (Figure 6B–C). It has been reported that tumor cells regulate angiogenesis by communicating with endothelial cells through exosomes secretion3, 24, 43. Therefore, we investigated if exosomes in CM prepared from RGN expressing cells were involved in the suppression of angiogenesis. We depleted exosomes from CM and treated HUVEC cells with CM deprived of exosomes. Interestingly, the exosome-depleted CM collected from RGN expressing cells did not decrease the tube formation ability of HUVEC cells (Figure 6D). We then isolated exosomes from PC3mm cells, characterized its size by Nanoparticle tracking analysis (NTA) and electron microscopy. We also validated exosome uptake by HUVEC cells (Supplementary Figure 6A–C). When HUVEC cells were treated with exosomes isolated from RGN expressing cells, their tube-forming ability significantly decreased, indicating that factors in exosomes mediates suppression of angiogenesis.
Figure 6: RGN suppresses angiogenesis in the tumor microenvironment by exosomal miR23c.
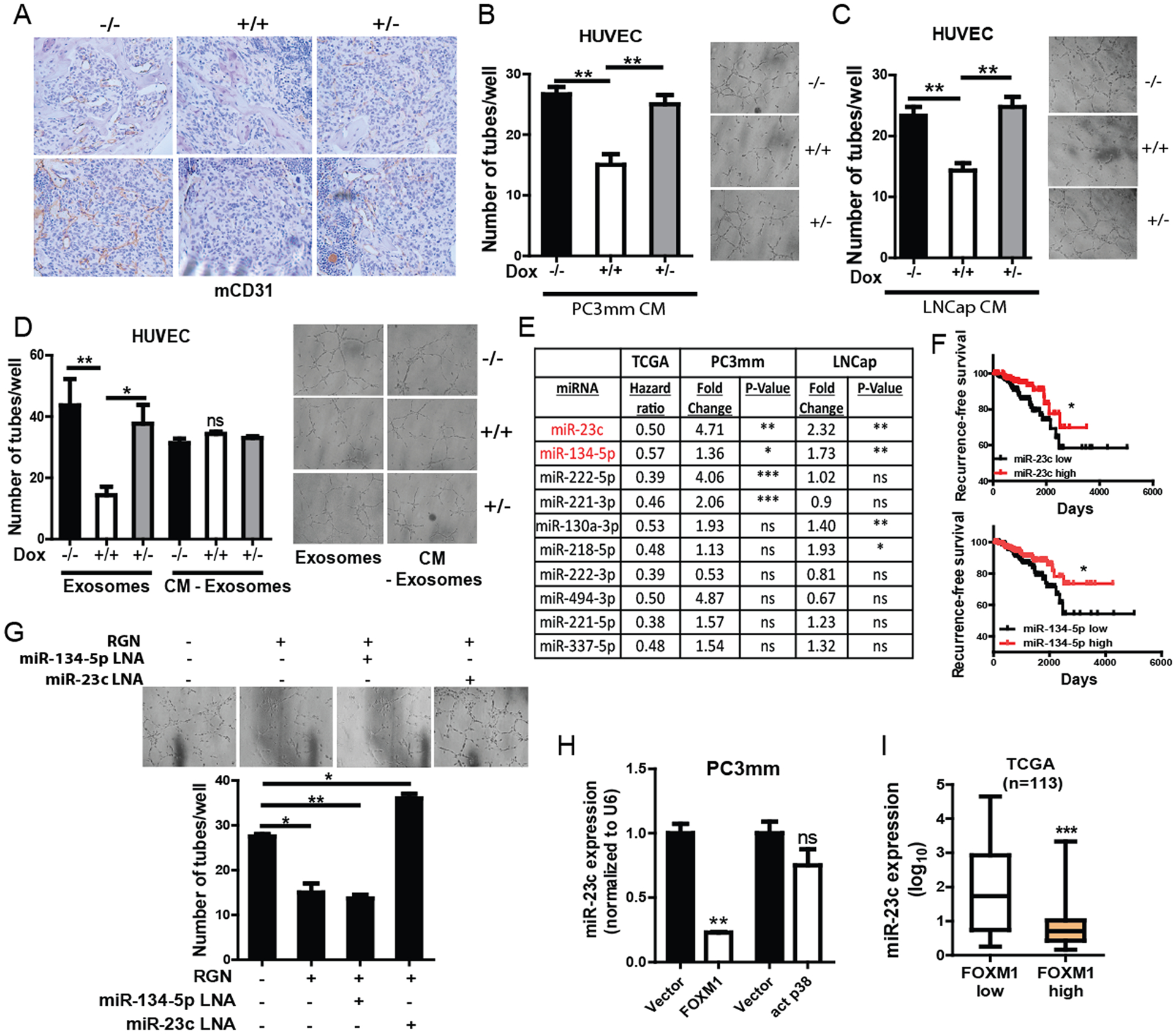
(A) IHC analysis for CD31 was performed for bone metastasis samples from mice that were intracardially injected with C42B cells. Representative images are shown. (B–C) HUVEC cells were treated with CM collected from PC3mm (A) and LNCap (B) cells treated with or without doxycycline to induce RGN expression (−/−, no doxycycline treatment, +/+, continuous treatment with doxycycline for 48 hrs, +/−, doxycycline treatment for 18 hrs followed by culturing cells in no doxycycline conditions). After 24 hrs of treatment with CMs, cells were seeded in matrigel coated 96-well plates followed by the counting number of tubes formed after 6 hrs (n=3/group). Representative images are shown in the right panels. Data was analyzed using one-way anova and tukey’s multiple comparison test. (D) Exosomes were extracted from conditioned medium (CM) of PC3mm/Dox/RGN cells treated with or without doxycycline as in (A), and HUVEC cells were treated with either exosomes or exosomes-depleted CM for 24 hrs followed by seeding cells in matrigel coated 96-well plate. Tube formation ability of these cells was examined 6 hours after seeding (n=3/group). Representative images are shown. Data was analyzed using one-way anova and tukey’s multiple comparison test. (E) Table showing the hazard ratio of indicated miRNAs in TCGA database recurrence free survival, and fold change of indicated miRNAs in PC3mm and LNCap cells overexpressing RGN. (F) Patients in the TCGA dataset were stratified based on their level of miR-23c and miR-134-5p expression, and Kaplan Meier curve was plotted for recurrence-free survival (n=398). The recurrence-free survival plots were analyzed by log-rank (Mantel-cox) test. (G) Exosomes were prepared from PC3mm cells with or without RGN expression. Exosomes were also isolated from RGN-expressing cells transfected with miR-23c and miR-134-5p LNAs. HUVEC cells were treated with these exosomes for 24 hrs followed by performing tube formation assay (n=3/group). Number of tubes was counted 6 hours after cell seeding. Representative images are shown in right panels. One-way anova and tukey’s multiple comparison test was used for statistical analysis of each gene. (H) The FOXM1 gene was ectopically expressed in PC3mm cells by lentiviral expression system or transiently transfected with vector control or activated p38 containing vector, and miR-23c expression was examined by qRT-PCR. Unpaired t-test was used for statistical analysis. (I) miR-23c expression was examined in patients with low or high levels of FOXM1 using TCGA. Unpaired t-test was used for statistical analysis. *, P-value<0.05, ** P-value<0.01 and ***, P-value<0.0001.
Small RNAs such as miRNAs are known to be abundantly present in exosomes52, and our group and others have recently demonstrated that exosomes-entrapped miRNAs regulate angiogenesis in the tumor microenvironment43, 52. To identify exosomal miRNAs that regulate angiogenesis, we first focused on 86 miRNAs that were previously reported to regulate angiogenesis. These miRNAs were then screened for their prognostic significance in predicting prostate cancer recurrence-free survival using the TCGA PRAD miRNA dataset. Based on this screening, 10 miRNAs that significantly correlated with longer relapse-free survival in prostate cancer patients were selected for further studies (Figure 6E–F and Supplementary Figure 7A). We then examined whether RGN directly regulated expression of these miRNAs. As shown in Figure 6F, we found that two miRNAs, miR-23c and miR-134-5p, were commonly and significantly upregulated in exosomes of PC3mm and LNCap cells ectopically expressing RGN. To investigate whether these two miRNAs are involved in anti-angiogenic effect of RGN, we transfected PC3mm cells with locked nucleic acid (LNA) against these miRNAs and isolated exosomes from transfected cells. When HUVEC cells were treated with exosomes isolated from cells transfected with miR-23c LNA, but not with miR-134-5p, the angiogenesis inhibitory effect of RGN was significantly rescued (Figure 6G and Supplementary Figure 7B). Collectively, these results indicate that RGN suppresses angiogenesis in the tumor microenvironment by exosomal miR-23c.
To investigate how RGN expression upregulate miR-23c gene, we examined whether miR-23c was regulated by signaling from p38 or FOXM1. We found that ectopic expression of FOXM1 significantly decreased miR-23c expression while expression of activated p38 did not alter the miR-23c level (Figure 6H). In support of this finding, a significant negative correlation between miR-23c and FOXM1 was evident in clinical specimens (Figure 6I). These results strongly indicate that FOXM1 negatively regulates miR-23c expression and that RGN decreases FOXM1 to stimulate miR-23c expression.
DISCUSSION:
RGN is a multifunctional protein that regulates calcium homeostasis, intracellular cell signaling, proliferation and apoptosis57. RGN is also known as Senescence Marker Protein 30 (SMP30) as its level decreases with aging and the transgenic rats reverses age-associated cellular changes22, 48. Extracellular and intracellular calcium levels and protein kinase C (PKC) pathway regulate RGN expression36, 49, 54. The tumor suppressive role of RGN in multiple tumor types, including prostate cancer has been previously reported31, 50. RGN regulates expression of oncogenes and tumor suppressors to inhibit rat hepatoma cells50. Although loss of RGN was previously reported to promote prostate cancer, this is the first report demonstrating the pathological mechanism involved in metastatic dormancy of prostate cancer at preclinical level.
Our study revealed that RGN promotes dormancy-associated signaling. It has been well documented that higher p38/Erk ratio dictates the dormant phenotype of cancer cells1. In fact, disseminated tumor cells (DTC) isolated from bone of prostate cancer patients with no evidence of disease were found to be enriched for p38 MAPK associated genes13. Similarly, we also found that p38 MAPK signature was significantly enriched in patients with high RGN expression. In addition, ectopic RGN expression by doxycycline treatment activated p38 MAPK in a reversible manner in both androgen-sensitive and -resistant cell lines, suggesting that the activation of p38 MAPK pathway by RGN is independent of androgen signaling. It should be noted that RGN expression using non-inducible system significantly suppressed cell growth in vitro; therefore, we used doxycycline inducible system, in which cells were selected in the absence of doxycycline, to study the mechanism of RGN. However, RGN was reported to suppress p38 MAPK pathway in HepG2 liver cancer cell line60, which indicates that the role of RGN is tumor-type dependent. By comparing the indolent/non-recurrent tumors with aggressive/recurrent ones, Abate Shen’s group have previously identified that transcription factor FOXM1 is a master regulator that drives aggressive/recurrent tumor growth5. Our findings demonstrate that RGN reversibly suppresses FOXM1 expression possibly by inhibiting upstream AKT activation. It is noteworthy that FOXM1 expression was reported to be lower in quiescent HNSCC cells17. Activation of p38 signaling, and inhibition of AKT/FOXM1 signaling are known to induce cell cycle arrest, which was manifested in RGN expressed cells. Our finding of this novel role of RGN in activating p38 pathway and inhibiting FOXM1 expression strongly indicates that RGN activates multiple hallmarks of cellular dormancy.
Androgen receptor (AR) signaling and PI3K/AKT pathway are two major drivers of prostate cancer15. Androgen deprivation therapy (ADT) remains the mainstay for hormone sensitive prostate cancer23. Cancer cells frequently overcome ADT and lead to castration resistant prostate cancer (CRPC). AKT pathway activation is known to stimulate CRPC growth and survival. On the other hand, inhibition of AKT pathway promotes AR signaling dependent disease progression15. This inverse correlation between AR and AKT pathway suggested that targeting both PI3K/AKT and AR signaling is ideal for treating prostate cancer9. Interestingly, RGN expression is known to be negatively regulated by androgen signaling as dihydrotestosterone (DHT) treatment of LNCap cells significantly decreased RGN level30. However, how RGN regulates AR signaling has not been studied yet. Our findings suggest that RGN suppresses AKT signaling in both hormone-sensitive and –resistant cell lines. Therefore, further studies examining the effect of RGN to AR signaling in castration-sensitive and CRPC disease context will shed further mechanistic insight into the role of RGN in tumor dormancy.
Angiogenic switch plays a prominent role in expansion of tumor mass at metastatic/recurrent site40. Additionally, stability of target microvasculature regulates metastatic outgrowth of disseminated dormant cells. Ghajar et al found that sprouting endothelial vessels secrete factors to accelerate cancer cell growth in the microvasculature while stable microvasculature promoted dormancy via thrombospondin-1 secretion18. Exosomes are one of the major communicator between tumor cells and vasculature39. Cargoes carried by exosomes modulate various cellular functions of environmental cells52. We found that RGN suppresses angiogenesis by augmenting miRNA-23c level in the exosomes. Interestingly, we found that miR-23c level was negatively regulated by FOXM1. Interestingly, miR-23c target, SDF1α, is known to promote neoangiogenesis in bone which is the primary site for prostate cancer dormancy2, 38. Therefore, it is plausible that RGN expressing dormant cells inhibit overt recurrence by secreting miR-23c in exosomes to suppress neoangiogenesis in bone microenvironment.
Our results indicate that multiple RGN target genes were functionally linked to the cellular cytoskeleton and extracellular matrix (ECM)4, 45, 62. It is noteworthy that MYL9, COL4A6 and TPM2 expression was reported to negatively correlate with survival, recurrence or metastasis status of prostate cancer patients21, 47. CNN1, DES, ACTG2, KCNMB1, CSRP1 are known to be expressed by smooth muscle cells19, 28, 34, 37, 62. Indeed, the increased presence of smooth muscle within prostate carcinoma is known to hamper micro-invasion thereby suppressing tumor progression62. However, our study indicated that RGN increased expression of cytoskeleton and ECM genes in prostate cancer cell lines and concomitantly reduced the invasion and migration ability of these cells. Furthermore, p38 signaling promoted expression of these genes whereas FOXM1 reduced their expression, which implies that these genes are downstream regulators of dormancy. It is possible that FOXM1 reduces expression of these genes by directly binding to their promoter region as reported previously10 although this notion needs to be directly tested. Collectively, our study has indicated that RGN serves as a central node to regulate prominent genes and signaling associated with dormancy of prostate cancer. We also showed the potential utility of the signaling axis of RGN and exosomal miR-23c as biomarkers of predicting prostate cancer recurrence (Figure 7).
Figure 7:
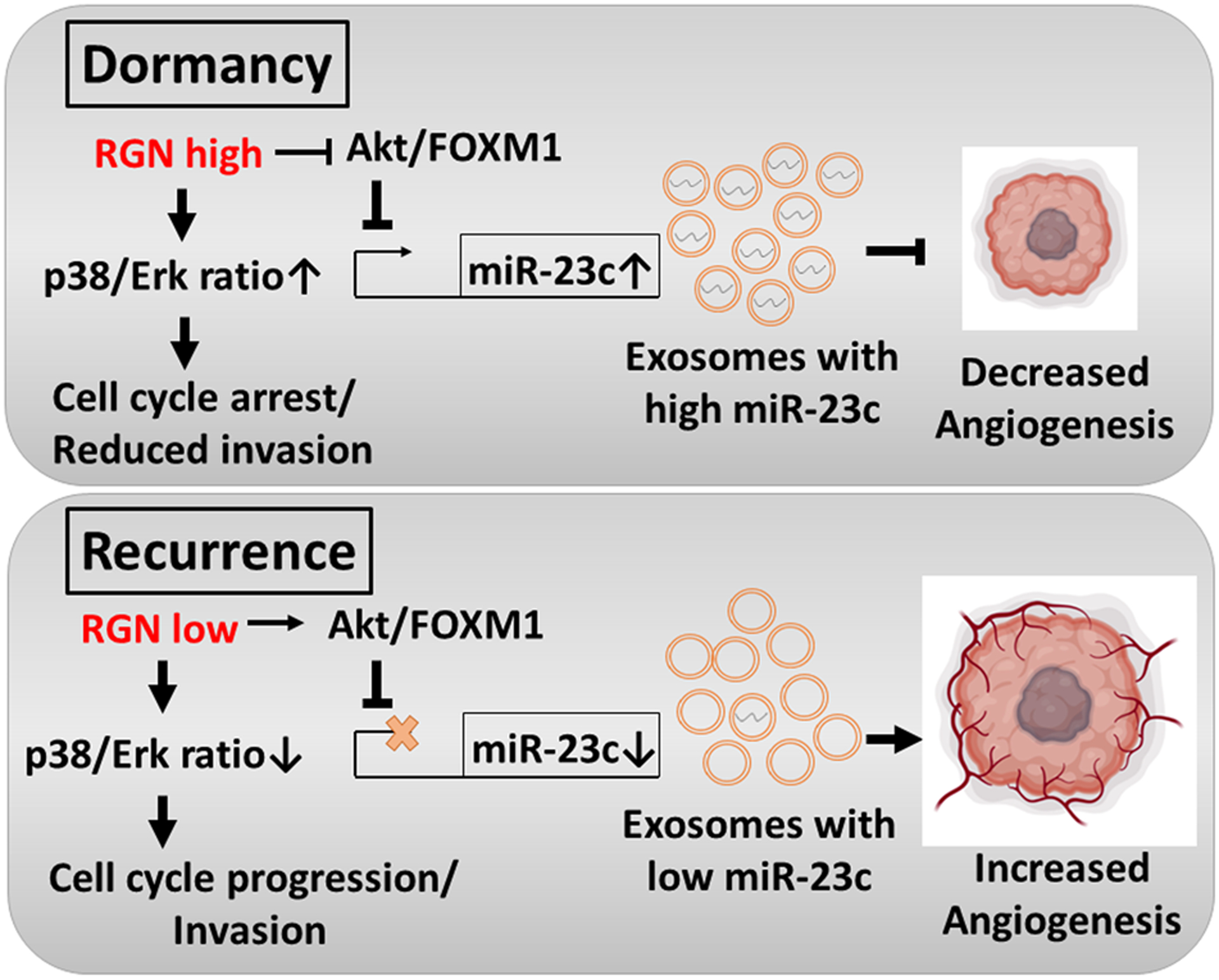
The proposed mechanism of action of RGN in inducing prostate cancer dormancy.
EXPERIMENTAL PROCEDURES:
Cell culture and reagents:
LNCap and C42B cells were obtained from MD Anderson Cancer Center. PC3mm cell line was provided by I.J. Fidler (The University of Texas MD Anderson Cancer Center). HUVEC cells were obtained from American Type Culture Collection (ATCC). The RGN was expressed in PC3mm, LNCap and C42B cells by sub-cloning RGN gene in pCMV6-XL4 plasmid (Catalog no. SC101207) to tet-on 3G lentiviral system obtained from Clonetech (Catalog no. 631187). PC3mm, LNCap and C42B cells were maintained in RPMI 1640 mediium with 10% fetal bovine serum, 100μg/ml streptomycin, and 100 U/ml penicillin at 37°C in a 5% CO2 atmosphere. HUVEC cells were maintained in Ham’s F12K medium supplemented with 0.1mg/ml Heparin, 1.5g/L NaHCo3, 10% endothelial cell growth supplement) and 10% fetal bovine serum at 37°C and 5% CO2 atmosphere. For bioluminescence tracking, PC3mm, LNCap, and C42B cells were transduced with firefly luciferase carrying lentivirus vector. The RGN expression was knocked down in LNCap cells using lentiviral shRNAs obtained from Dharmacon.
qRT-PCR:
Total RNAs were extracted from indicated cells using TRIzol reagent (Qiagen). The RNA was reverse transcribed into complementary DNA (cDNA) by using a Reverse Transcription Supermix kit (Bio-Rad, USA). The cDNA was then amplified with a pair of forward and reverse primers for the indicated genes using Biorad CFX-Connect device. For miRNAs, primers were designed as previously reported11. Total RNAs were reverse transcribed using reverse transcription supermix kit (Bio-Rad, USA) in the presence of 50nM stem-loop RT primer that contained 4–6 bases of sequence complementary to 3’ end of the mature miRNA sequence. The forward primer for PCR included 12–17 nucleotide of mature miRNA sequence. The RT primer excluding the complementary sequences to mature miRNA was used as a common reverse primer for all miRNAs.
Animal experiments:
All animal experiments were performed in compliance with protocol approved by laboratory animal care and use committee of Wake Forest University. For bone metastasis experiment, we implanted 5–6 weeks old athymic nude male mice with 5 × 105 C42B/Dox/RGN/luc or PC3mm/Dox/RGN/luc cells into the left cardiac ventricle in a total volume of 100μl. For inducing RGN gene expression, mice were fed doxycycline containing water (2mg/ml doxycycline and 5% sucrose). The control group (−/−) was treated with water without doxycycline (5% sucrose only). The incidence of bone metastasis was monitored by IVIS bioluminescence imaging (BLI). For intratibial injection, 150,000 LNCap/shScrambled or LNCap/shRGN cells labeled with luciferase in 10ul PBS were injected into the left tibia of anesthetized nude mice, and the growth of tumor in tibia was monitored by IVIS bioluminescence imaging.
Exosome isolation and treatment:
Exosomes were isolated using differential centrifugation method as previously described53. These exosomes were characterized by electron microscopy and nanoparticle tracking analysis. For treating endothelial cells with exosomes, the protein content of exosomes was quantified by the Bradford assay (Biorad) and 20μg of each sample was used to treat endothelial cells. For exosome uptake assay, exosomes were labeled with Vybrant™ DiD cell-labeling solution (InVitrogen) for 20 mins and washed twice in PBS. HUVEC cells seeded in 8-well glass chamber slides were then incubated with the labeled exosomes overnight and imaged using Keyence fluorescence microscope (BZ-X700).
Bioinformatics:
For differential gene expression analysis between Early- and Late- recurrence patients (Figure 1a), we downloaded the expression file from Gene Expression Omnibus (GEO) database. The PSA based recurrence time and incidence data was downloaded from a previous report46. Patients were divided based on the 3 years cutoff for recurrence and genes that were differentially expressed were identified. For identification of RGN target genes, we used the TCGA, GSE6919 and GSE21034 dataset and compared gene expression between patients in the lower and upper quartile of RGN expression for each dataset. For TCGA dataset 1266 differentially expressed genes with >2 fold increase or decrease and p-value <1e–5 were identified by comparing 125 patients in each quartile. For GSE6919, 225 genes with >2 fold increase or decrease and p-value <0.01 were identified by comparing 23 patients each group. For Taylor’s cohort, differentially expressed genes with p-value <1e–5 identified 454 genes. To maximize the number of common genes, we did not set the fold change cutoff for Taylor’s cohort. Patients with all clinical annotation were included in the differential gene analysis.
Supplementary Material
Acknowledgment—
This work was supported by NIH grant RO1CA173499, RO1CA185650 and RO1CA205067 to K.W. X.P. was supported by State Scholarship fund (2014084100701) by China Scholarship Council. F.L was supported by State Scholarship fund 2018093 by China Scholarship Council. The tumor tissue and pathology shared resources, biostatistics/bioinformatics shared resources, flow cytometry, and cell engineering shared resources are supported by comprehensive cancer center of Wake Forest University and, National Institutes of Health Grant (P30CA012197). The funding agency had no role in study design, data collection, data analysis, interpretation and writing of the report. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute.
Footnotes
Conflict of interest: The authors declare that they have no conflicts of interest with the contents of this article.
REFERENCES:
- 1.Aguirre-Ghiso JA. Models, mechanisms and clinical evidence for cancer dormancy. Nature reviews Cancer 2007; 7: 834–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amin KN, Umapathy D, Anandharaj A, Ravichandran J, Sasikumar CS, Chandra SKR et al. miR-23c regulates wound healing by targeting stromal cell-derived factor-1alpha (SDF-1alpha/CXCL12) among patients with diabetic foot ulcer. Microvasc Res 2020; 127: 103924. [DOI] [PubMed] [Google Scholar]
- 3.Aslan C, Maralbashi S, Salari F, Kahroba H, Sigaroodi F, Kazemi T et al. Tumor-derived exosomes: Implication in angiogenesis and antiangiogenesis cancer therapy. J Cell Physiol 2019; 234: 16885–16903. [DOI] [PubMed] [Google Scholar]
- 4.Ayala G, Tuxhorn JA, Wheeler TM, Frolov A, Scardino PT, Ohori M et al. Reactive stroma as a predictor of biochemical-free recurrence in prostate cancer. Clinical cancer research : an official journal of the American Association for Cancer Research 2003; 9: 4792–4801. [PubMed] [Google Scholar]
- 5.Aytes A, Mitrofanova A, Lefebvre C, Alvarez MJ, Castillo-Martin M, Zheng T et al. Cross-species regulatory network analysis identifies a synergistic interaction between FOXM1 and CENPF that drives prostate cancer malignancy. Cancer Cell 2014; 25: 638–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boorjian SA, Thompson RH, Tollefson MK, Rangel LJ, Bergstralh EJ, Blute ML et al. Long-term risk of clinical progression after biochemical recurrence following radical prostatectomy: the impact of time from surgery to recurrence. Eur Urol 2011; 59: 893–899. [DOI] [PubMed] [Google Scholar]
- 7.Bubendorf L, Schopfer A, Wagner U, Sauter G, Moch H, Willi N et al. Metastatic patterns of prostate cancer: an autopsy study of 1,589 patients. Hum Pathol 2000; 31: 578–583. [DOI] [PubMed] [Google Scholar]
- 8.Cackowski FC, Wang Y, Decker JT, Sifuentes C, Weindorf S, Jung Y et al. Detection and isolation of disseminated tumor cells in bone marrow of patients with clinically localized prostate cancer. Prostate 2019; 79: 1715–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carver BS, Chapinski C, Wongvipat J, Hieronymus H, Chen Y, Chandarlapaty S et al. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell 2011; 19: 575–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chand V, Pandey A, Kopanja D, Guzman G, Raychaudhuri P. Opposing Roles of the Forkhead Box Factors FoxM1 and FoxA2 in Liver Cancer. Molecular cancer research : MCR 2019; 17: 1063–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic acids research 2005; 33: e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen M, Pratt CP, Zeeman ME, Schultz N, Taylor BS, O’Neill A et al. Identification of PHLPP1 as a tumor suppressor reveals the role of feedback activation in PTEN-mutant prostate cancer progression. Cancer Cell 2011; 20: 173–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chery L, Lam HM, Coleman I, Lakely B, Coleman R, Larson S et al. Characterization of single disseminated prostate cancer cells reveals tumor cell heterogeneity and identifies dormancy associated pathways. Oncotarget 2014; 5: 9939–9951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corey E, Quinn JE, Bladou F, Brown LG, Roudier MP, Brown JM et al. Establishment and characterization of osseous prostate cancer models: intra-tibial injection of human prostate cancer cells. Prostate 2002; 52: 20–33. [DOI] [PubMed] [Google Scholar]
- 15.Crumbaker M, Khoja L, Joshua AM. AR Signaling and the PI3K Pathway in Prostate Cancer. Cancers (Basel) 2017; 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong JT, Lamb PW, Rinker-Schaeffer CW, Vukanovic J, Ichikawa T, Isaacs JT et al. KAI1, a metastasis suppressor gene for prostate cancer on human chromosome 11p11.2. Science 1995; 268: 884–886. [DOI] [PubMed] [Google Scholar]
- 17.Eckers JC, Kalen AL, Sarsour EH, Tompkins VS, Janz S, Son JM et al. Forkhead box M1 regulates quiescence-associated radioresistance of human head and neck squamous carcinoma cells. Radiat Res 2014; 182: 420–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghajar CM, Peinado H, Mori H, Matei IR, Evason KJ, Brazier H et al. The perivascular niche regulates breast tumour dormancy. Nat Cell Biol 2013; 15: 807–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henderson JR, Macalma T, Brown D, Richardson JA, Olson EN, Beckerle MC. The LIM protein, CRP1, is a smooth muscle marker. Dev Dyn 1999; 214: 229–238. [DOI] [PubMed] [Google Scholar]
- 20.Huang W, Fridman Y, Bonfil RD, Ustach CV, Conley-LaComb MK, Wiesner C et al. A novel function for platelet-derived growth factor D: induction of osteoclastic differentiation for intraosseous tumor growth. Oncogene 2012; 31: 4527–4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang YQ, Han ZD, Liang YX, Lin ZY, Ling XH, Fu X et al. Decreased expression of myosin light chain MYL9 in stroma predicts malignant progression and poor biochemical recurrence-free survival in prostate cancer. Med Oncol 2014; 31: 820. [DOI] [PubMed] [Google Scholar]
- 22.Ishigami A, Masutomi H, Handa S, Maruyama N. Age-associated decrease of senescence marker protein-30/gluconolactonase in individual mouse liver cells: Immunohistochemistry and immunofluorescence. Geriatr Gerontol Int 2015; 15: 804–810. [DOI] [PubMed] [Google Scholar]
- 23.Jones CU, Hunt D, McGowan DG, Amin MB, Chetner MP, Bruner DW et al. Radiotherapy and short-term androgen deprivation for localized prostate cancer. N Engl J Med 2011; 365: 107–118. [DOI] [PubMed] [Google Scholar]
- 24.Kalluri R The biology and function of exosomes in cancer. J Clin Invest 2016; 126: 1208–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim MJ, Cardiff RD, Desai N, Banach-Petrosky WA, Parsons R, Shen MM et al. Cooperativity of Nkx3.1 and Pten loss of function in a mouse model of prostate carcinogenesis. Proceedings of the National Academy of Sciences of the United States of America 2002; 99: 2884–2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kobayashi A, Okuda H, Xing F, Pandey PR, Watabe M, Hirota S et al. Bone morphogenetic protein 7 in dormancy and metastasis of prostate cancer stem-like cells in bone. The Journal of experimental medicine 2011; 208: 2641–2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kupelian P, Katcher J, Levin H, Zippe C, Klein E. Correlation of clinical and pathologic factors with rising prostate-specific antigen profiles after radical prostatectomy alone for clinically localized prostate cancer. Urology 1996; 48: 249–260. [DOI] [PubMed] [Google Scholar]
- 28.Long X, Tharp DL, Georger MA, Slivano OJ, Lee MY, Wamhoff BR et al. The smooth muscle cell-restricted KCNMB1 ion channel subunit is a direct transcriptional target of serum response factor and myocardin. The Journal of biological chemistry 2009; 284: 33671–33682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lyu T, Jia N, Wang J, Yan X, Yu Y, Lu Z et al. Expression and epigenetic regulation of angiogenesis-related factors during dormancy and recurrent growth of ovarian carcinoma. Epigenetics 2013; 8: 1330–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maia C, Santos C, Schmitt F, Socorro S. Regucalcin is under-expressed in human breast and prostate cancers: Effect of sex steroid hormones. J Cell Biochem 2009; 107: 667–676. [DOI] [PubMed] [Google Scholar]
- 31.Maia CJ, Santos CR, Schmitt F, Socorro S. Regucalcin is expressed in rat mammary gland and prostate and down-regulated by 17beta-estradiol. Mol Cell Biochem 2008; 311: 81–86. [DOI] [PubMed] [Google Scholar]
- 32.Marques R, Maia CJ, Vaz C, Correia S, Socorro S. The diverse roles of calcium-binding protein regucalcin in cell biology: from tissue expression and signalling to disease. Cell Mol Life Sci 2014; 71: 93–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marques R, Vaz CV, Maia CJ, Gomes M, Gama A, Alves G et al. Histopathological and in vivo evidence of regucalcin as a protective molecule in mammary gland carcinogenesis. Exp Cell Res 2015; 330: 325–335. [DOI] [PubMed] [Google Scholar]
- 34.Miwa T, Manabe Y, Kurokawa K, Kamada S, Kanda N, Bruns G et al. Structure, chromosome location, and expression of the human smooth muscle (enteric type) gamma-actin gene: evolution of six human actin genes. Mol Cell Biol 1991; 11: 3296–3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morgan TM, Lange PH, Porter MP, Lin DW, Ellis WJ, Gallaher IS et al. Disseminated tumor cells in prostate cancer patients after radical prostatectomy and without evidence of disease predicts biochemical recurrence. Clinical cancer research : an official journal of the American Association for Cancer Research 2009; 15: 677–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakagawa T, Yamaguchi M. Nuclear localization of regucalcin is enhanced in culture with protein kinase C activation in cloned normal rat kidney proximal tubular epithelial NRK52E cells. Int J Mol Med 2008; 21: 605–610. [PubMed] [Google Scholar]
- 37.Paulin D, Li Z. Desmin: a major intermediate filament protein essential for the structural integrity and function of muscle. Exp Cell Res 2004; 301: 1–7. [DOI] [PubMed] [Google Scholar]
- 38.Petit I, Jin D, Rafii S. The SDF-1-CXCR4 signaling pathway: a molecular hub modulating neoangiogenesis. Trends Immunol 2007; 28: 299–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ribeiro MF, Zhu H, Millard RW, Fan GC. Exosomes Function in Pro- and Anti-Angiogenesis. Curr Angiogenes 2013; 2: 54–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shaked Y, McAllister S, Fainaru O, Almog N. Tumor dormancy and the angiogenic switch: possible implications of bone marrow- derived cells. Curr Pharm Des 2014; 20: 4920–4933. [DOI] [PubMed] [Google Scholar]
- 41.Sharma S, Watabe K. Biomarkers and mechanisms associated with recurrent prostate cancer. Frontiers in bioscience (Landmark edition) 2014; 19: 339–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sharma S, Xing F, Liu Y, Wu K, Said N, Pochampally R et al. Secreted Protein Acidic and Rich in Cysteine (SPARC) Mediates Metastatic Dormancy of Prostate Cancer in Bone. The Journal of biological chemistry 2016; 291: 19351–19363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sharma S, Wu SY, Jimenez H, Xing F, Zhu D, Liu Y et al. Ca(2+) and CACNA1H mediate targeted suppression of breast cancer brain metastasis by AM RF EMF. EBioMedicine 2019; 44: 194–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sosa MS, Avivar-Valderas A, Bragado P, Wen HC, Aguirre-Ghiso JA. ERK1/2 and p38alpha/beta signaling in tumor cell quiescence: opportunities to control dormant residual disease. Clinical cancer research : an official journal of the American Association for Cancer Research 2011; 17: 5850–5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tanjore H, Kalluri R. The role of type IV collagen and basement membranes in cancer progression and metastasis. Am J Pathol 2006; 168: 715–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Varambally S, Yu J, Laxman B, Rhodes DR, Mehra R, Tomlins SA et al. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell 2005; 8: 393–406. [DOI] [PubMed] [Google Scholar]
- 47.Varisli L Identification of new genes downregulated in prostate cancer and investigation of their effects on prognosis. Genet Test Mol Biomarkers 2013; 17: 562–566. [DOI] [PubMed] [Google Scholar]
- 48.Vaz CV, Marques R, Maia CJ, Socorro S. Aging-associated changes in oxidative stress, cell proliferation, and apoptosis are prevented in the prostate of transgenic rats overexpressing regucalcin. Transl Res 2015; 166: 693–705. [DOI] [PubMed] [Google Scholar]
- 49.Vaz CV, Rodrigues DB, Socorro S, Maia CJ. Effect of extracellular calcium on regucalcin expression and cell viability in neoplastic and non-neoplastic human prostate cells. Biochimica et biophysica acta 2015; 1853: 2621–2628. [DOI] [PubMed] [Google Scholar]
- 50.Vaz CV, Correia S, Cardoso HJ, Figueira MI, Marques R, Maia CJ et al. The Emerging Role of Regucalcin as a Tumor Suppressor: Facts and Views. Curr Mol Med 2016; 16: 607–619. [DOI] [PubMed] [Google Scholar]
- 51.Wang J, Hua L, Guo M, Yang L, Liu X, Li Y et al. Notable roles of EZH2 and DNMT1 in epigenetic dormancy of the SHP1 gene during the progression of chronic myeloid leukaemia. Oncology letters 2017; 13: 4979–4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu K, Xing F, Wu SY, Watabe K. Extracellular vesicles as emerging targets in cancer: Recent development from bench to bedside. Biochim Biophys Acta Rev Cancer 2017; 1868: 538–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xing F, Liu Y, Wu SY, Wu K, Sharma S, Mo YY et al. Loss of XIST in Breast Cancer Activates MSN-c-Met and Reprograms Microglia via Exosomal miRNA to Promote Brain Metastasis. Cancer research 2018; 78: 4316–4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yamaguchi M, Mori S. Inhibitory effect of calcium-binding protein regucalcin on protein kinase C activity in rat liver cytosol. Biochem Med Metab Biol 1990; 43: 140–146. [DOI] [PubMed] [Google Scholar]
- 55.Yamaguchi M Role of regucalcin in maintaining cell homeostasis and function (review). Int J Mol Med 2005; 15: 371–389. [PubMed] [Google Scholar]
- 56.Yamaguchi M The anti-apoptotic effect of regucalcin is mediated through multisignaling pathways. Apoptosis 2013; 18: 1145–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yamaguchi M Involvement of regucalcin as a suppressor protein in human carcinogenesis: insight into the gene therapy. J Cancer Res Clin Oncol 2015; 141: 1333–1341. [DOI] [PubMed] [Google Scholar]
- 58.Yamaguchi M, Osuka S, Weitzmann MN, El-Rayes BF, Shoji M, Murata T. Prolonged survival in pancreatic cancer patients with increased regucalcin gene expression: Overexpression of regucalcin suppresses the proliferation in human pancreatic cancer MIA PaCa-2 cells in vitro. Int J Oncol 2016; 48: 1955–1964. [DOI] [PubMed] [Google Scholar]
- 59.Yamaguchi M, Osuka S, Weitzmann MN, Shoji M, Murata T. Increased regucalcin gene expression extends survival in breast cancer patients: Overexpression of regucalcin suppresses the proliferation and metastatic bone activity in MDA-MB-231 human breast cancer cells in vitro. Int J Oncol 2016; 49: 812–822. [DOI] [PubMed] [Google Scholar]
- 60.Yamaguchi M, Osuka S, Shoji M, Weitzmann MN, Murata T. Survival of lung cancer patients is prolonged with higher regucalcin gene expression: suppressed proliferation of lung adenocarcinoma A549 cells in vitro. Mol Cell Biochem 2017; 430: 37–46. [DOI] [PubMed] [Google Scholar]
- 61.Yamaguchi M, Osuka S, Murata T. Prolonged survival of patients with colorectal cancer is associated with a higher regucalcin gene expression: Overexpression of regucalcin suppresses the growth of human colorectal carcinoma cells in vitro. Int J Oncol 2018; 53: 1313–1322. [DOI] [PubMed] [Google Scholar]
- 62.Yang Z, Peng YC, Gopalan A, Gao D, Chen Y, Joyner AL. Stromal hedgehog signaling maintains smooth muscle and hampers micro-invasive prostate cancer. Dis Model Mech 2017; 10: 39–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


