Summary:
Human pluripotent stem cell (hPSC) derived myogenic progenitor cell (MPC) transplantation is a promising therapeutic approach for a variety of degenerative muscle disorders. Here, using an MPC specific fluorescent reporter system (PAX7::GFP), we demonstrate that hPSC-derived MPCs can contribute to the regeneration of myofibers in mice following local injury and in mice deficient of dystrophin (mdx). We also demonstrate that a subset of PAX7::GFP MPCs engraft within the basal lamina of regenerated myofibers, adopt a quiescent state, and contribute to regeneration upon reinjury and in mdx mouse models. This subset of PAX7::GFP MPCs undergo a maturation process, and remodel their molecular characteristics to resemble late stage fetal MPCs/adult satellite cells following in vivo engraftment. These in vivo matured PAX7::GFP MPCs retain a cell autonomous ability to regenerate and can repopulate in niche of secondary recipient mice, providing a proof of principle for future hPSC-based cell therapy for muscle disorders.
Graphical Abstract
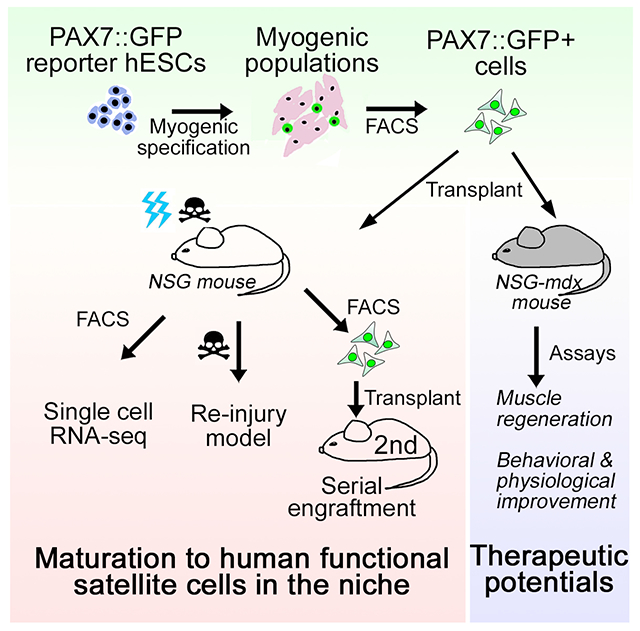
ETOC/In brief:
Sun et al., discover that myogenic progenitors derived from hPSCs could engraft and mature to become local satellite cells in vivo after transplantation. These engrafted MPCs not only function as satellite cells in muscle regeneration but also could ameliorate disease phenotype in DMD mouse model.
Introduction:
During vertebrate myogenesis, the transcription factors Pax3 and Pax7 mark myogenic progenitor cells (Relaix and Zammit, 2012).These cells give rise to Pax7 expressing muscle stem cells, termed satellite cells, which localize within the basal lamina of myofibers in the postnatal muscle (Gros et al., 2005). Satellite cells remain quiescent in healthy, postnatal muscle and can become activated upon injury giving rise to myoblasts that proliferate and repair damaged myofibers (Olguin et al., 2007; Zammit et al., 2006). At the same time, a fraction of satellite cell progeny maintain Pax7 expression and self-renew to reconstitute the satellite cell pool (Olguin and Olwin, 2004). Thus, transplanting satellite cells instead of transient amplifying myoblasts as a means of cell threapy would be advantageous due to their long-term self-renewal properties (Gussoni et al., 1999; Sacco et al., 2008).
Human induced pluripotent stem cells (hiPSCs) which can be derived from patients for autologous cell replacement therapy is a promising approach to avoid potential immune responses and cell rejection. Recently, methods of generating myogenic lineages from mouse and human pluripotent stem cells have been developed (Chal et al., 2015; Choi et al., 2016; Henrique et al., 2015; Loh et al., 2016; Sudheer et al., 2016; Xi et al., 2017). The cells that are generated from hiPSCs contain a mixed culture of myogenic progenitor cells identified by their expression of the transcription factors PAX3/PAX7, MYOD and Myogenin expressing myoblasts along with differentiated myotubes (Choi et al., 2016; Kim et al., 2017). Recent studies have demonstrated engraftment and differentiation of PAX3/PAX7 positive hiPSC-derived myogenic progenitor cells in immunocompromised mice (Hicks et al., 2018; Wu et al., 2018). However, whether hiPSC-derived MPCs can become quiescent and occupy the satellite cell niche, provide long-term regenerative ability and improve muscle function remains unclear.
Results:
PAX7::GFP MPCs derived from hESC engraft into mouse skeletal muscle
In order to study the efficiency of myogenic progenitor cell transplantation, we generated a PAX7::GFP hESC reporter line through a CRISPR-Cas9 mediated knock-in of GFP before 3’ UTR of PAX7 gene (Figure S1A), which was then subjected to our serum-free step-wise myogenic specification protocol to generate PAX7::GFP expressing MPCs (Choi et al., 2020; Choi et al., 2016). PAX7 expressing myogenic progenitor cells were detected on day 18, and the cell number gradually increased between day 18 and day 30 to reach 7.56 +/− 3.33% (Figure S1B–D) (Choi et al., 2020). We enriched PAX7::GFP expressing myogenic progenitor cells (PAX7::GFP+ MPCs) by fluorescence-activated cell sorting (FACS) and expanded them in vitro in the presence of FGF2. After expansion and passaging for 7 days, 30 +/− 10.0% of the MPCs in culture retained PAX7::GFP expression and a portion of cells entered the myogenic differentiation program and lost GFP expression (Figure S1E–F, S1H–I). We term these cells, in in vitro culture that include some non-GFP expressing cells, PAX7::GFP MPCs. We observed an increased and robust expression of PAX7 together with GFP and MYOD in freshly isolated PAX7::GFP+ cells, as well as in cells from a second passage, compared to non-myogenic cells (fibroblasts), representing their myogenic progenitor characteristics (Figure S1G). PAX7::GFP MPCs fused into myotubes in a fusion permissive condition (Figure S1J). Furthermore, compared to primary human myoblasts, the PAX7::GFP MPC culture exhibited three-fold more PAX7 expressing cells and had comparable fusion capability to human primary myoblasts (Figure S1K–M).
In order to establish a transplantation strategy, we compared two transplantation approaches: (1) a hESC-derived myogenic cell culture without FACS purification (a mixed population including myotube, myocytes, and myogenic progenitor cells) and (2) FACS-purified and twice passaged PAX7::GFP MPCs (Figure 1A). Prior to the transplantation, we irradiated the tibialis anterior muscles (TAs) of NOD Cg-Prkdcscid Il2rgtm1Wjl/SzJ immunodeficient mice (NSG) to deplete the host satellite cells. Host muscle fibers were damaged with 1.2% BaCl2 to allow de novo fiber formation from the transplanted cells. Four weeks after the transplantation, the TAs were cross sectioned, and the muscle fibers that were formed by the donor cells were labeled with human-specific dystrophin antibody and the secreted ECM was labeled with laminin antibody. Consistent with a previous study (Kim et al., 2017), in which cells were transplanted directly from a hESC myogenic derivation culture, the cells showed limited levels of differentiation into organized myofibers (Figure 1B). This could be due to the possibility that the unsorted cells contain fewer MPCs but more differentiated myogenic cells and partially non-myogenic cells like neurons (Kim et al., 2017). Meanwhile, we observed that PAX7::GFP MPCs extensively participate in regeneration of injured TAs reflected by muscle size compared with the control group, which was injected with only medium (Figure S2A). When the area of human-specific Dystrophin+ (huDystrophin+) fibers was quantified against the entire TA section, PAX7::GFP MPC transplanted TAs had significantly more fibers formed by the donor cells than by the unsorted cells (Figure 1B–C, Figure S2B). Moreover, the number of myofibers formed by PAX7::GFP MPCs were comparable to those of human primary myoblasts (Figure S2C–D). This supports the hypothesis that enriching PAX7::GFP MPCs in the myogenic derivation culture prior to transplantation enhances regeneration.
Figure 1. PAX7::GFP MPC derived from hPSCs can form new muscle fibers and localize to the muscle stem cell niche in vivo.
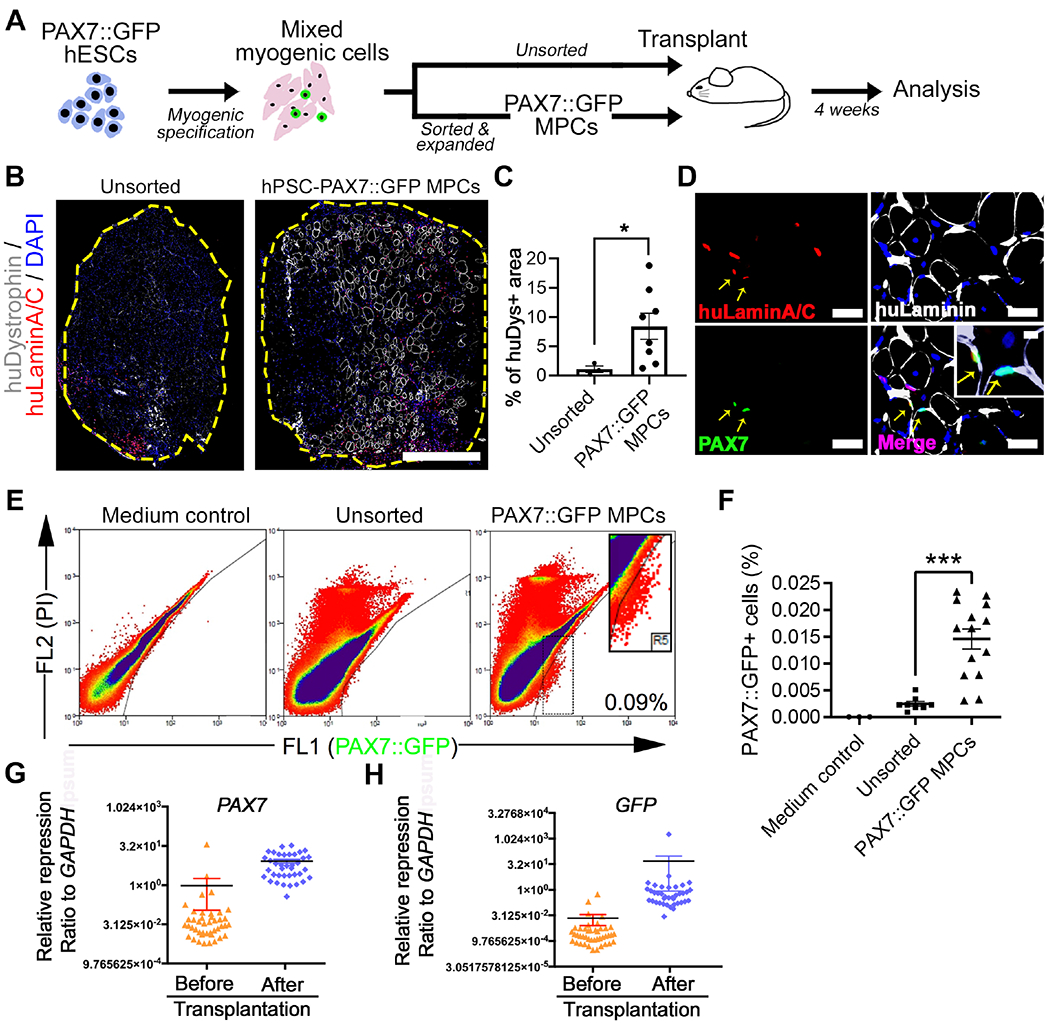
(A) Schematic of PAX7::GFP MPC derivation, cell transplantation, and downstream analysis. (B) Representative images of cross-section of tibialis anterior (TA) muscle of NSG mouse 4 weeks after transplantation of either sorted and expanded (PAX7::GFP MPC) or unsorted myogenic cells derived from PAX7::GFP hESCs. These cells participated in regeneration and formed new muscle fibers (labeled by huDystrophin) that have donor cell origin. All donor originated cell nuclei were labeled with huLaminA/C+ antibody (red). Edge of TA was highlighted by yellow. Scale bar=500μm (C) Quantification of percentage of huDytrophin+ fibers area against the entire TA area (n(Unsorted)=3, n(Expanded)=8; Student’s t-test, *p<0.05). (D) Images of sectioned TAs 4 weeks after the PAX7::GFP MPC transplantation.PAX7::GFP MPC progenies could be identified by double labeling of huLaminA/C (red) and PAX7 (green) proteins. They locate under basal Lamina (yellow arrow). Scale bar=100μm (E) FACS plot of ‘in vivo PAX7::GFP+ cells’ isolated 4 weeks after either being sorted and expanded or unsorted. TAs injected with vector medium was used as control. (F) Quantification of PAX7::GFP+ cells as a proportion of all isolated mononucleated cells from a single TA (n(Control)=4, n(Sorted & expanded)=14, n(Unsorted)=8; One-way ANOVA, *p<0.001). (G-H) Single cell Q-PCR showing quiescent muscle stem cell markers (PAX7) and confirmative marker GFP were expressed at higher level in in vivo PAX7::GFP+ cells compared with in vitro PAX7::GFP+ cells (n=48; Student’s t-test, *p<0.05).Result=Mean±SEM.
We next tested whether hESC-PAX7::GFP MPCs could populate the satellite cell niche, in addition to giving rise to the myofibers. We examined the localization of PAX7::GFP+ cells in vivo four weeks post transplantation with double labelling of PAX7 and human-specific Lamin A/C (huLaminA/C) antibodies to trace the progenies of PAX7::GFP MPCs. Among the human cells recognized by huLaminA/C, some of the cells were found to reside within the myofiber basal lamina where adult muscle stem cells are typically located (Figure 1D, Movie S1). Meanwhile a small portion of the transplanted PAX7::GFP MPCs become interstitial myoblasts and fibroblasts (Figure S2E–F). Next, we isolated the progenies of the transplanted PAX7::GFP MPCs that maintained PAX7::GFP expression four weeks post transplantation by FACS. The percentage of GFP+ cells recovered from all the mononucleated cells was as high as 0.0146 +/− 0.0006% when isolated from a single TA or 0.066 +/− 0.022% when four TAs were pooled together (Figure 1E–F). These isolated GFP+ cells retained their high level expression of PAX7 and GFP as well as other satellite cell specific gene markers (CALCR and PMP22) when compared with PAX7::GFP MPCs before transplantation (Figure 1G, Figure S2G–H) (Fukada et al., 2007). Finally, we confirmed that the PAX7::GFP+ cells isolated from transplanted mice were progenies of hESC-PAX7::GFP MPCs, and not of mouse origin, by performing human specific primer PCR and also comparing their RNA expression profile to the human genome (Figure S2L–M). Collectively, these studies demonstrate the successful engraftment of hPSC-MPCs into the endogenous satellite cell niche.
PAX7::GFP+ cells adopt a satellite cell molecular profile upon engraftment
To further understand the transcriptomic changes of engrafted PAX7::GFP+ cells isolated from the in vivo environment, we performed single cell RNA-sequencing (scRNA-seq) on 176 PAX7::GFP+ cells four weeks subsequent to the in vivo transplantation. For comparison, we sequenced three additional cell populations: in vitro PAX7::GFP+ cells (117 cells), 1 month in vitro PAX7::GFP+ cells (PAX7::GFP+ cells after one month of in vitro cell culture, 83 cells), and undifferentiated human embryonic stem cells (76 cells) (Figure 2A). It is noteworthy that during the reverse transcription step we added a known amount of exogenous RNA spike-ins (from the External RNA Controls Consortium (ERCC)). This enabled us to investigate if the overall RNA content had changed per cell over the course of the in vivo transplantation and in vitro maintenance(Lun et al., 2017). We approximated the RNA content by normalizing the number of sequenced RNA molecules per cell to the detected spike-ins. Intriguingly, we found that in vivo PAX7::GFP+ cells consistently displayed lower RNA content than their in vitro-cultured counterparts, in vitro PAX7::GFP+ and 1 month in vitro PAX7::GFP+ cells (Figure 2B), which indicated a shift to a more quiescent state(Fukada et al., 2007). This observation is corroborated with previous transcriptomic studies of satellite cells(Charville et al., 2015; Dell’Orso et al., 2019; Fukada et al., 2007). Previous studies normalized the transcriptomic data with the assumption of constant RNA content across populations, which could, in turn, lead to the identification of different sets of differentially regulated genes. Therefore, we performed the normalization relative to spike-ins (using the scran package) to account for the differences in the RNA content between the samples(Lun et al., 2017).
Figure 2. Single-cell RNA sequencing of PAX7::GFP+ cells isolated from TA of NSG mice four weeks after PAX7::GFP MPC transplantation.
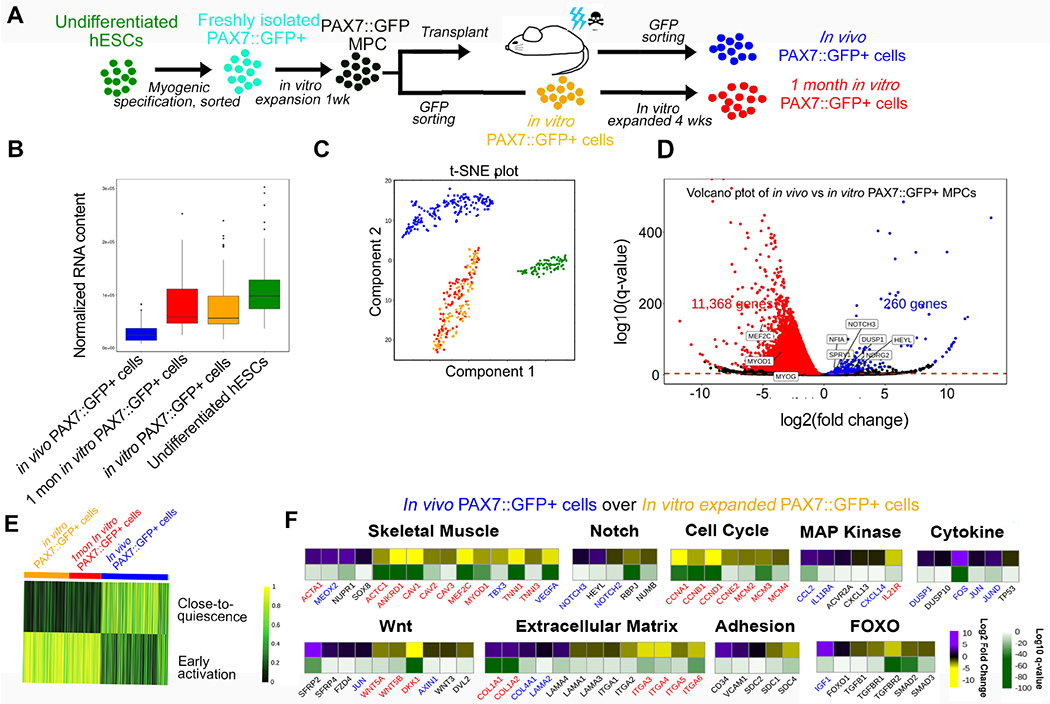
(A) Schematic of the origin of cells undergoing single cell RNA sequencing. (B) Normalized RNA content of in vivo PAX7::GFP cells, in vitro PAX7::GFP cells, 1 month in vitro PAX7::GFP+, and hESCs showing in vivo PAX7::GFP+ cells have less RNA content compared with the other three groups. (C) t-SNE plot showing the distinctive RNA expression profiles among undifferentiated hESCs (green), in vitro PAX7::GFP+ cells (yellow), 1 month in vitro PAX7::GFP+ cells (red), and in vivo PAX7::GFP+ cells (blue). (D) Volcano plot of differentially expressed genes in PAX7::GFP+ in vivo cells vs. in vitro PAX7::GFP+ cells. (E) Heat map showing in vivo PAX7::GFP+ cells share molecular signatures of quiescent mouse satellite cells (Dell’Orso et al., 2019). (F) Heat map of differentially expressed genes of in vivo PAX7::GFP+ cells over in vitro PAX7::GFP+ cells. Log2 and log10 values were both presented. Blue highlighted genes were upregulated and red highlighted genes downregulated in human satellite cell studies when comparing quiescent to activated satellite cells (Charville et al., 2015). See also Figure S3 and Table S1.
First, we observed that the in vivo environment exposure modulated the transcriptome of PAX7::GFP MPCs dramatically while the one month in vitro culture did not (Fig 2C). When comparing the in vivo PAX7::GFP+ cells with the in vitro PAX7::GFP+ cells prior to transplantation, we found that most of the differentially regulated genes showed decreased expression (Fig 2D and Table S1). In a previous study, we observed at baseline a concordance between in vivo PAX7::GFP+ cells and human satellite cells when we normalized our data using the constant RNA assumption (Figure S2N)(Charville et al., 2015). However, the concordance was not maintained when we did not assume constant RNA (Figure S2O). These results suggest that further consideration should be given to the normalization approaches in scRNA-seq of satellite cells where changing RNA content is an inherent aspect of the biology and may affect identification of differentially expressed genes. To further confirm the quiescent nature of our transplanted cells, we used the SingleCellNet (Tan and Cahan, 2019) algorithm. After training a classifier on endogenous activated and quiescent mouse stem cell scRNA-seq data from Dell’Orso et al. and subsequently classifying the cells from our dataset (Dell’Orso et al., 2019), we found that in vivo PAX7::GFP+ cells were classified as having a ‘quiescent signature’ while in vitro cells, in vitro PAX7::GFP+ and 1 month in vitro PAX7::GFP+ cells, were classified as having an ‘activated signature’ (Figure 2E).
We looked closer into the genes that are important for myogenic differentiation. DUSP1, NF1A, NDRG2, and SPRY1, which are all associated with satellite cell quiescence and were all upregulated in the in vivo PAX7::GFP+ cells (Figure 2D)(Charville et al., 2015; Fukada et al., 2007). PLAGL1 which is a PAX7 target genes, is shown to be significantly upregulated after being exposed to the in vivo environment (Figure S3A) (Cornelison and Wold, 1997; McKinnell et al., 2008). MYF5 which is expressed in some but not all satellite cells was upregulated in some of the in vivo PAX7::GFP+ cells (Figure S3B) (Beauchamp et al., 2000). Furthermore, MYOD1, whose expression was reported to be undetectable when satellite cells return to a quiescent status (Zammit et al., 2004) was found to be downregulated in the in vivo PAX7::GFP+ cells, in line with the considerable decrease of differentiation marker transcription factors MEF2C and MYOG (Ridgeway et al., 2000) (Figure S3C–E). We also found that in vivo PAX7::GFP+ cells showed the molecular signatures of certain pathways that are functionally similar to those found in quiescent human satellite cells(Bjornson et al., 2012; Otto et al., 2008; Segales et al., 2016) (Figure 2F and Figure S3F). In the in vivo cells, genes that control cell cycle progression were downregulated, indicating that these cells had exited the cell cycle, as reported in mouse quiescent satellite cells in vivo (Fukada et al., 2007). Meanwhile, a number of transcripts critical to stem cell homing to the niche were distinctively upregulated, including extracellular matrix related genes (e.g. COL1A1 and LAMA4) and notch signaling related genes (e.g. NOTCH3 and HEYL) (Figure 2F)(Brohl et al., 2012). Moreover, genes associated with adhesion proteins, such as CD34 and VCAM1, well recognized cell surface markers for quiescent satellite cells (Beauchamp et al., 2000; Choo et al., 2017), were significantly upregulated (Figure 2F). Overall, we developed a single cell RNA-sequencing strategy to analyze the molecular change of the PAX7::GFP+ cells during the engraftment process and observed they gained the molecular signature of satellite cells.
PAX7::GFP MPCs become quiescent and developmentally mature after being exposed to the in vivo environment.
We further studied the pathways affected by the transplantation process through overrepresentation of gene ontology terms. We found that genes upregulated in the in vivo PAX7::GFP+ cells were mainly genes related to muscle development, skeletal muscle tissue/organ development and ECM structure and maintenance (Figure 3A). The downregulated genes included genes related to striated muscle cell differentiation, mitotic cell cycle process, and protein metabolism (Figure 3B), indicating the prohibition of differentiation of the in vivo PAX7::GFP+ cells as well as entry into quiescence. Moreover, we confirmed that most of the in vivo PAX7::GFP+ cells were in the G0 phase in FACS-based cell cycle analysis, while control groups mostly contained proliferating cells (Figure 3C). Taken as a whole, these results support a transition of proliferating in vitro PAX7::GFP+ cells to quiescent in vivo PAX7::GFP+ cells with similar molecular signatures of muscle stem cells.
Figure 3. PAX7::GFP+ cells become quiescent and developmentally mature after being exposed to the in vivo environment.
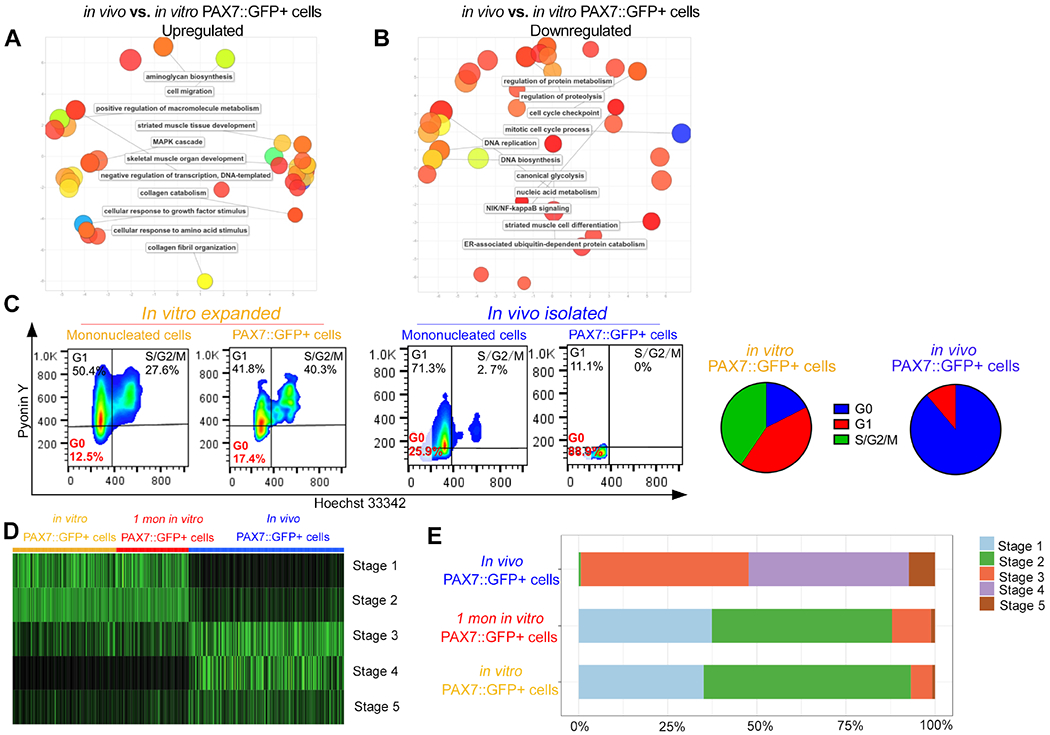
(A-B) Clustering analysis of the Gene Ontology (GO) Biological Process of up- and down-regulated genes in in vivo PAX7::GFP+ cells compared with in vitro PAX7::GFP+ cells. (C) Pyronin Y and Hoechst cell cycle analysis of in vivo PAX7::GFP+ cells and in vitro PAX7::GFP+ cells. (D) Heat map distribution of developmental score across in vitro PAX7::GFP+ cells, 1 month in vitro PAX7::GFP+ cells, and in vivo PAX7::GFP+ cells. (E) Proportions of in vitro PAX7::GFP+ cells, 1 month in vitro PAX7::GFP+ cells, and in vivo PAX7::GFP+ cells assigned to each computational developmental stage. See also Figure S4 and Table S2.
Next, we examined the developmental stage of the various cell populations. In a recent study, Xi et al. generated a developmental score system, which clustered single cell RNA-seq results of individual human skeletal muscle samples in a series of developmental stages (Xi et al., 2020). We applied this SingleCellNet algorithm to align our data to the developmental stages. While the in vitro PAX7::GFP+ and 1 month in vitro PAX7::GFP+ cells were aligned to developmental stages 1-2, corresponding to the embryonic stage (up to 6-7 weeks gestation), in vivo PAX7::GFP+ cells were aligned to developmental stages 3-4, corresponding to the fetal stage (12-18 weeks gestation), and developmental stage 5, corresponding to the postnatal stage (Figure 3D–E). This evidence supports that the in vivo environment modulates the transplanted human PAX7::GFP+ cells to undergo a transition from an embryonic developmental stage to a fetal as well as postnatal satellite cell stage.
To further understand the timeline of when PAX7::GFP+ cells enter into a quiescent muscle stem cell status, we performed a single cell RNA-seq analysis on PAX7::GFP+ cells isolated from TAs at 1- (135 cells), 2- (192 cells), 3- (137 cells), and 4- (20 cells) week time points following transplantation. We pooled together cells from both scRNA-seq studies, correcting for batch effects using mnnCorrect through scran (Figure S3G). Interestingly, the samples harvested from 1-, 2-, 3-, and 4-week time points had comparable transcriptomic profiles as well as developmental status (Figure S3G–H). This data indicate that the transplanted human PAX7::GFP+ cells immediately start the transition toward quiescent satellite cell status (approximately less than a week after injection), as described previously in mouse satellite cell experiments (De Micheli et al., 2020). These results suggest the PAX7+ cells mature and become quiescent within one week of engraftment in the in vivo environment.
PAX7::GFP in vivo cells participate in muscle regeneration similar to satellite cells
Given that transplanted PAX7::GFP MPCs were capable of engrafting into the stem cell niche and turning on muscle stem cell signature gene expression, we were interested to know if they could be reactivated following subsequent muscle reinjury or by transplantation into a secondary recipient mouse. To perform the reinjury experiments, we first injured TAs and transplanted PAX7::GFP MPCs as previously done. We then re-injured the muscle four weeks after the first injury. (Figure 4A). Four weeks after the reinjury, we observed robust de novo muscle fiber regeneration from progenies of PAX7::GFP MPCs labeled by huDystrophin antibody (Figure 4B, right panel and Figure 4C–D), despite the fact that the initial huDystrophin+ muscle fibers that formed after the first injury were depleted during the reinjury process (Figure 4B, left panel). Next, we tested if the in vivo PAX7::GFP+ cells could function like muscle stem cells upon transplantation and whether they had better transplantation efficacy than PAX7::GFP MPCs. Therefore, we isolated in vivo PAX7::GFP+ cells and then re- transplanted them into a secondary recipient mouse (Figure 4E). After four weeks, we found that the serially transplanted PAX7::GFP MPCs could form new myofibers as well as become PAX7 expressing cells residing in the satellite cell niche (Figure 4F and Figure S4A–B). While the muscle fiber formation measured by huDystrophin+ fibers was more robust after the first transplantation (probably due to the number of injected cells, 1 million MPCs in the first transplantation versus a few hundred MPCs in the second), the transplantation efficiency, calculated by the ratio of the number of PAX7::GFP+ cells per transplanted cells (as measured by FACS), was significantly higher in the secondary mice compared to the primary recipient mice (Figure 4G–H, S4C). Moreover, when the rate of PAX7::GFP+ cells residing in the satellite cell niche was calculated according to the injected cell number, the serially transplanted PAX7::GFP+ in vivo cells had a higher rate of homing to the niche than that of primarily transplanted PAX7::GFP MPCs and human primary myoblasts (Figure 4I, Figure S4D). Since a small number of cells in the serial transplantation experiment resulted in a higher contribution to muscle regeneration and in a better engraftment rate, we separately measured the transplantation efficiency of 400 PAX7::GFP MPCs. In this scenario, a very limited amount of huDystrophin+ fibers were observed, and the ratio of huDystrophin+ fibers to the cell number transplanted were significantly lower compared to the serial transplantation group (Figure S4E–F). In order to further understand the mechanism of why in vivo PAX7::GFP+ cells have better engraftment capability, an additional bulk RNA sequencing experiment was performed with the in vivo PAX7::GFP+ cells that were isolated from primary recipient mice and cultured in vitro for 4 days (right before 2nd transplantation) in comparison to PAX7::GFP MPC maintained in vitro (before 1st transplantation). Our RNA-seq analysis data shows that the cultured in vivo PAX7::GFP+ cells have upregulated gene expression profiles of extracellular matrix reorganization as well as muscle structure development (Figure S4G–I). Collectively, the upregulated genes (e.g. MMP3, COMP and IL13RA2) as well as the GO terms indicate an enhanced stem cell niche homing process (Table S2). The above evidence shows that the in vivo PAX7::GFP+ cells function similar to muscle stem cells and the exposure to the in vivo environment increased transplantation efficiency by enabling the cellular function of stem cell homing.
Figure 4. Functionality of PAX7::GFP MPCs in muscle regeneration of NSG and NSG-mdx mice.
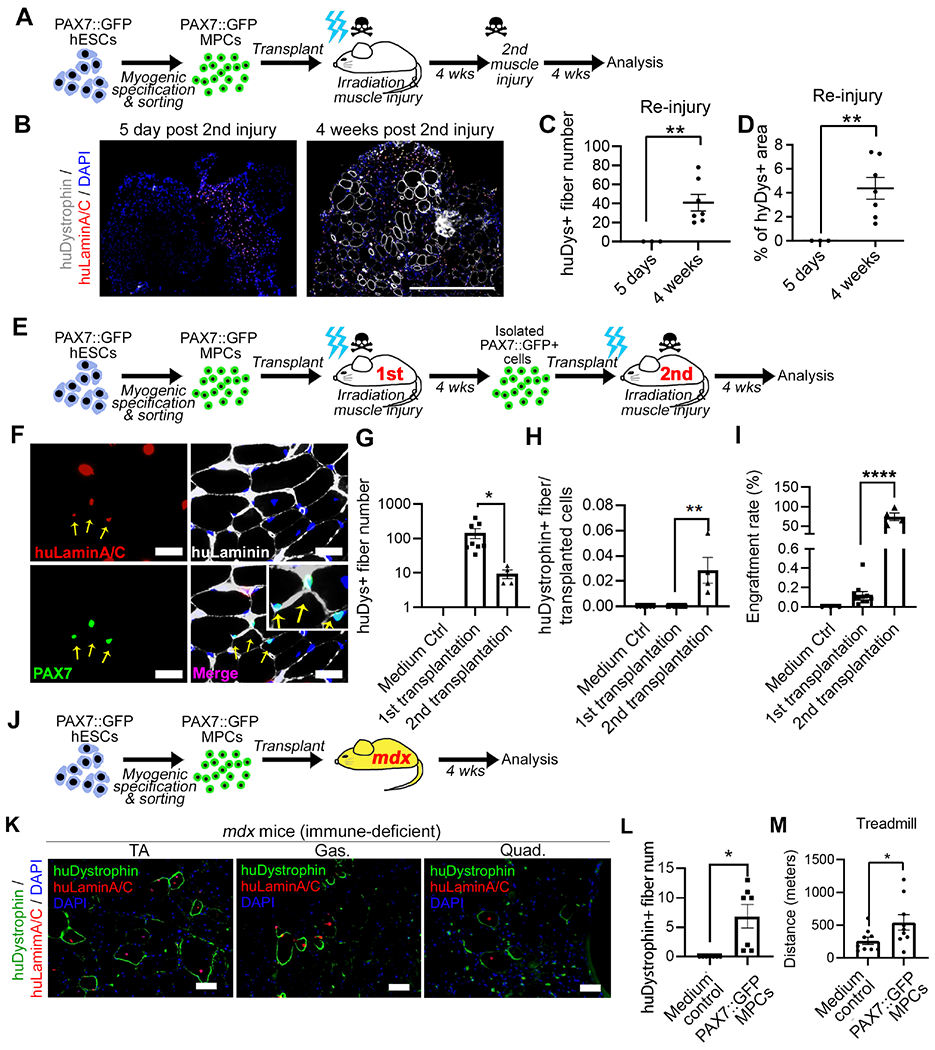
(A) Schematic of reinjury experiment of NSG mice. (B) huDystrophin and huLaminA/C expression in TAs engrafted with PAX7::GFP+ cells and repetitively injured. Scale bar=500μm (C-D) Quantification of huDystrophin+ fiber number and the percentage of area in TA in reinjury experiment (n(PAX7::GFP MPC)=7, n(Control)=3; Student’s t-test, *p<0.05). (E) Schematic of serial transplantation using NSG mice. (F) PAX7::GFP+ cells were isolated from the first recipient mouse and transplanted into the second recipient mouse. TAs of the second recipient mice were sectioned and Pax7 and huLaminA/C were used to trace transplanted cells. The co-labeled cells were found to reside within the basal lamina (yellow arrow). Scale bar=100μm (G) Quantification of huDystrophin+ fibers from the entire TA (n(Control)=3, n(1st transplantation)=6, n(2nd transplantation)=4; One-way ANOVA, *p<0.05). (H) Quantification of ratio of huDystrophin+ fibers to cell number being injected (n(Control)=3, n(1st transplantation)=6, n(2nd transplantation)=4; One-way ANOVA, **p<0.01). (I) Quantification of cells located between muscle fiber and basal lamina (huLaminin labeled) that originated from PAX7::GFP MPCs (Pax7+, huLaminA/C+) after 1st or 2nd transplantation, as a proportion of the total number of PAX7::GFP MPCs transplanted. (n=3; Student’s t-test, **p<0.001). (J) Schematic of PAX7::GFP MPCs transplantation into NSG-mdx mice. (K) Images of PAX7::GFP MPCs formed huDystrophin+ muscle fibers in TAs, gastrocnemius (Gas), and quadriceps (Quad). Scale bar=50μm (L) Quantification of huDystrophin+ fibers in TA, Gas, and Quad in NSG-mdx mice. (n(Control)=4, n(PAX7::GFP MPC)=5; Student’s t-test, *p<0.05). (M) Treadmill running exhaustion test showing PAX7::GFP MPC transplanted NSG-mdx mice ran longer distance than medium-injected control mice (n(PAX7::GFP MPC)=9, n(Control)=8; Student’s t-test, *p<0.05). Result=Mean±SEM. See also Figure S4.
Finally, we transplanted PAX7::GFP+ MPCs to the TA, gastrocnemius, and quadriceps of both hindlimbs in the DMD mouse model (NSG-mdx4cv) without injury or irradiation, to more closely resemble a potential clinical scenario (Figure 4J). Even without muscle injury, PAX7::GFP+ MPCs formed huDystrophin muscle fibers (Figure 4K–L). We examined if there was functional improvement of NSG-mdx mice following PAX7::GFP+ MPC transplantation. In a treadmill assay, mice which received transplantation ran longer distances compared to sham injected controls (Figure 4M). To more accurately model DMD, which has a more severe phenotype than the mdx mouse, we added an exercise protocol to induce muscle degeneration before cell transplantation in a second cohort (Cohort-2) of the NSG-mdx transplantation experiments (Bouchentouf et al., 2006; De Luca et al., 2003). In the Cohort-2, we detected significant improvement of the treadmill test result (data not shown). Furthermore, an improved resistance to fatigue of TA muscle was demonstrated by decreased percentage of force loss after repeated contractions (Figure S4J–L). Importantly, we found that some of the transplanted human PAX7::GFP MPCs homed to niche area in cohort-2 (Figure S4M–N). These results show the functional similarity of PAX7::GFP+ in vivo cells to satellite cells that can result in long-term regeneration and ameliorate some aspects of the muscular dystrophy disease phenotype.
Discussion:
Previous studies of hPSC-derived myogenic cell transplantation have demonstrated the formation of myofibers originated from donor cells as well as the localization of donor cells within the basal lamina where they maintain Pax7 expression (Chal et al., 2015; Hicks et al., 2018; Incitti et al., 2019; Wu et al., 2018). However, the fate of these donor Pax7 expressing cells in the satellite cell niche has remained unclear. In 2019, Incitti et al. demonstrated that the engraftment process remodels Pax3/7-induced myogenic progenitor cells derived from mouse iPSCs, and induces in the transplanted cells a molecular signature of the mouse satellite cells (Incitti et al., 2019). Our study used a single cell strategy to explore the fate of the human iPSC-derived myogenic progenitor cells setting a step forward towards utilizing hPSC derived MPCs for clinic. A significant advantage of our approach is not relying on ectopic gene overexpression to generate the ‘engraftable’ lineage-committed cells as in previous studies (Al Tanoury et al., 2020; Xi et al., 2020), like HoxB4 in hematopoietic cells (Kyba et al., 2002; Rideout et al., 2002) and PAX3/7 in human myogenic cells (Darabi et al., 2011). Of significance in our study was the difference in the repopulation rate between primary and secondary populating MPCs (Figure 3F), implying two possible hypotheses: (1) an ‘educational communication’ process between the satellite cell niche and the quiescent MPCs and (2) a selective advantage of ‘pre-determined’ or ‘pre-matured’ cells with greater repopulation capability, which warrants further studies.
Limitations of study
Due to the paucity of human engrafted MPCs harvested from the mouse host, we could not perform a large-scale single cell transcriptome analysis. In the future, we would like to perform a larger scale single cell transcriptome analysis of engrafted human PAX7::GFP+ cells with multiple time points. Following the transplantation, most of MPCs did not engraft to become local stem cells as observed in our study (0.066 +/− 0.022% engraftment rate), which is comparable to hematopoietic stem cell transplantation (Bhattacharya et al., 2006). While most of the human PSCs become myofibers to regenerate the damaged muscle tissue, some cells could differentiate into myoblasts without becoming muscle stem-like celles, while other cells could transdifferentiate into additional cell types (e.g. fibrogenic and/or adipogenic cells) (Aguiari et al., 2008; Asakura et al., 2001; Hashimoto et al., 2008; Sacco et al., 2008). The fate of transplanted MPCs will require further study. In addition, an antibody-based purification approach for MPC is needed for future cell therapy application.
STAR Methods
RESOURCE AVAILABILITY
Lead contact:
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Gabsang Lee (glee48@jhmi.edu).
Materials availability:
The genetic reporter lines used in his study will be available upon request with a proper MTA.
Data and code availability:
RNA-sequencing datasets generated during this study are available at Gene Expression Omnibus (GEO), accession number GEO: GSE165075(single cell) and GEO: GSE190233(bulk). Code generated for single cell (SC) RNA-seq data analysis is deposited at Github, accession: https://github.com/skannan4/satellite-cell-transplantation (DOI:10.5281/zenodo.6330148). Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animal and cell transplantation strategy:
All animal experiments were approved by the Institutional Animal Care and Use Committee (IACUC) of The Johns Hopkins University, School of Medicine (Baltimore, Maryland). 8-week old NSG (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ, Jackson Lab) and 8- to 12-week old NSG-mdx4CV (generously provided by Dr. Michael Kyba, University of Minnesota) mice were maintained in a 12-h light cycle (7 am −7 pm) with ad libitum access to food and water. In the experiments of cell transplantation to NSG mice, left TA of mice were irradiated (18g) 72 hours prior to 50 μl 1.2% BaCl2 (Sigma) injury. 1 million PAX7::GFP MPCs were resuspended in 50 μl culture medium supplemented with rock inhibitor Y-27632 (Tocris) and injected into TA. For reinjury experiment, at 4-week post first transplantation, 50 μl 1.2% BaCl2 was used to reinjure the same TA. For serial transplantation experiment, PAX7::GFP+ cells were isolated at 4-week post transplantation using FACS and donor cells were transplanted into the secondary recipient mouse TA which were irradiated and injured as previously described. In the experiments of cell transplantation to NSG-mdx4CV mice, PAX7::GFP MPCs were resuspend with 50 μl culture medium with rock inhibitor and injected into TA, Gastrocnemius and Quadricep (1 million cells each muscle tissue). In the 2nd cohort of NSG-mdx4CV experiment, mice were exercised on the treadmill (10 m/min for 10 mins, 3 times a week) for 2 weeks to induce injury prior to the cell transplantation.
hPSC maintenance:
hESCs (H9, WiCell) and hiPSCs were cultured using standard protocols. The hESC lines and hiPSC lines were cultured with mouse embryonic fibroblasts (MEFs) (Gibco) pre-plated at 12,000–15,000 cells/ cm2. hPSC culture medium contained DMEM/F12, 20% knockout serum replacement, 1 mM L-glutamine, 100 mM MEM non-essential amino acids, and 0.1 mM b-mercaptoethanol. 10 ng/mL of FGF-2 was added after sterile filtration, and cells were fed daily, and passaged weekly, using 6 U/mL dispase or mechanically.
PAX7::GFP hESC cell line was generated using CRISPR-Cas9 (Choi et al., 2020).
Myogenic cell derivation and cell culture:
For myogenic linage derivation, hPSCs were plated as single cells on Geltrex (Gibco) treated dishes, at a density of 1.5 x 105 cells per well in a 24-well plate, in the presence of MEF-conditioned N2 media containing 10 ng/ml of FGF-2 (PeproTech) and 10 μM of Y-27632 (Tocris). The cells were induced to differentiate into myoblasts by adding CHIR99021 (3 μM, STEMCELL technologies) in N2 medium for 4 days and by DAPT (10 μM, PeproTech) for the following 8 days. Cells continued to differentiate and mature in N2 medium for the next 13 days. PAX7::GFP+ cells were isolated by FACS (FACS core at Johns Hopkins School of Public health) with GFP signal. Human primary myoblasts were isolated from healthy individual’s muscle biopsy with FACS enrichment of CD56+ cells. Human tissue collection was approved by Johns Hopkins University IRB.
The PAX7::GFP MPCs were maintained in a humidified incubator containing 5% CO2 at 37°C and grew in N2 medium supplemented with 20% FBS, FGF-2. To induce myotube formation, expanded myogenic cells were plated to confluency, and switched to N2 media without serum. Human primary muscle cells were grown in a growth medium composed of Ham’s F-10 (Mediatech10-070-CV) supplemented with 20% fetal bovine serum (FBS) (Hyclone, #SH30071.03), 0.5% chicken embryo extract, 1% Penicillin/Streptomycin (Invitrogen, #10569010), 0.5% amphotericin-B (Sigma, #A2942), and 1.2 mM CaCl2.
METHOD DETAILS
Isolation of mononuclear cells from TA for FACS:
TAs were harvested and tissues were minced until a slurry formed with no more large muscle pieces. Minced tissue was further digested with PBS containing 2 mg/ml Collagenase A (Roche), 2.4 U/ml Dispase II (Roche), 10 μg/ml DNase I (Roche) and 0.38mM CaCl2 for 90 minutes at 37°C. Digestion was terminated by adding 3 times ice cold PBS containing 0.2%BSA and DNasel. The suspension was filtered through 70μm filter and 40μm filter. Finally, cells were centrifuged and resuspended in PBS containing 2%FBS and DNasel for downstream FACS or other analysis.
Pyronin Hoechst cell cycle analysis:
For cell cycle analysis, cells were fixed in 1% PFA for 15 minutes at 37°C and permeabilized in 100% methanol over night at −20°C.The next day, cells were washed and blocked in blocking buffer (0.75% Saponin, Sigma, 1%FBS, 1%BSA in PBS). Primary GFP antibody (Invitrogen) and secondary antibody (goat anti chicken 488) was stained for 30 mins at room temperature. Then cells were suspended in blocking buffer containing Hoechst 33342 (2 μg/mL) and Pyronin-Y (4 μg/mL) for 30mins for DNA and RNA binding. Finally, cells were suspended in FACs buffer containing Pyronin-Y (2 μg/mL) for flow cytometry analysis (BD, LSDII).
Immunocytochemistry:
Muscle pieces were collected and frozen using −60°C isopentane. Frozen muscles were cryosectioned into 8μm sections. Cryosections were fixed with 4% PFA and washed with PBS. To detect Pax7, Primary antibodies against human lamin A+C (Mouse IgG2b, 1:200, Leica), pan-species Pax7 (IgG1, 1:200, DSHB), and human laminin (IgG2a, 1:200, DSHB) were incubated on the slides at 4°C overnight. Antigen retrieval was done by incubating slides in sodium citrate buffer at 85°C for 20 mins. HRP-conjugated secondary antibodies were applied for 60 minutes at RT and Tyramide signal amplification was performed using the Tyramide Signal Amplification with SuperBoost Kits (Thermo). To detect dystrophin, cryosections were fixed with cold methanol, washed with PBS, and blocked in 20% normal goat serum (Vector Laboratories) and 2% bovineserum albumin (Sigma Aldrich). a combination of primary antibodies against human dystrophin (Millipore, 1:200) and human Lamin A+C were incubated on the slides at 4°C overnight, followed by incubation with mouse anti-mouse secondary antibodies at room temperature for 1 hour. Other antibody used was mouse Laminin (1:300, Sigma). Sections were mounted using Vector Shield mounting medium with DAPI (Vector Laboratories), and imaged using fluorescent microscopy (Zeiss). Engraftment rate was calculated by cells located between muscle fiber and basal lamina (huLaminin labeled) that originated from PAX7::GFP MPCs (PAX7+, huLaminA/C+) after transplantation, as a proportion of the total number of PAX7::GFP MPCs being transplanted.
Single cell QPCR:
The detailed protocol was described previously(Lim et al., 2014). Briefly, single cell was isolated by FACS for cells expressing GFP and lysed directly by using cell lysis buffer. The mRNA was selected from total RNA in Reverse Transcription using SMA-T15, with anchoring nucleotide at the 5′ end SMA-A. The product from the RT reaction was PCR amplified using universal sequence as SMA-p2 primer. RT-PCR mixtures were prepared with SYBR Green PCR Master Mix. Primer sets used were: huPAX7 forward-CTGGCTACCAGTACGACCAGT, huPAX7 reverse-GCACGCCGGTTACTGAAC, huCalC forward-ACACCGGCTGGAGTATTGTC, huCalC reverse-AAAATCCCCAGGGAAATCAC, huPmp22 forward- ACATAGGGAAGGGAGGAAGG, huPmp22 reverse- TTGGTTTGGTTTGAGTTTGG, eGFP forward- ACGTAAACGGCCACAAGT, eGFP reverse- AAGTCGTGCTGCTTCATGTG.
Library construction and sequencing for Single cell as well as bulk RNA sequencing
PAX7::GFP+ cells were sorted by FACS as single cells in 96-well or 384-well capture plates using MoFlo Legacy sorter. Capture plate wells contained 5 ul of capture solution (1:500 Phusion High-Fidelity Reaction Buffer, New England Biolabs; 1:250 RnaseOUT Ribonuclease Inhibitor, Invitrogen). Single cell libraries were then prepared using the previously described SCRB-seq protocol (Soumillon et al., 2014; Ziegenhain et al., 2017). Briefly, cells were subjected to proteinase K treatment followed by RNA desiccation to reduce the reaction volume. RNA was subsequently reverse transcribed using a custom template-switching primer as well as a barcoded adapter primer. The customized SCRB-seq barcode primers contain a unique 6 base pair cell-specific barcode as well as a 10 base pair unique molecular identifier (UMI). Transcribed products were pooled and concentrated, with unincorporated barcode primers subsequently digested using Exonuclease I treatment. cDNA was PCR-amplified using Terra PCR Direct Polymerase (Takara Bio). Final libraries were prepared using 1ng of cDNA per library with the Nextera XT kit (Ilumina) using a custom P5 primer as previously described. Pooled libraries were sequenced on two high-output lanes of the Illumina NextSeq500 with a 16 base pair barcode read, 8 base pair i7 index read, and a 66 base pair cDNA read design.
For bulk RNA-seq, FACS isolated 1400 in vivo PAX7::GFP+ cells were seeded into 2 wells of 96-well plate coated with Geltrex and cultured in the PAX7::GFP MPC maintenance medium described above. The steps were repeated for in vitro PAX7::GFP+ cells. Cells were harvested 2 days later and RNA was extracted for sequencing.
Genomic diagnostic PCR:
Cells (50-100 cells) were sorted by FACs into Phusion direct PCR master mix (Thermo) and PCR was performed with human specific primer pair (17-specific-satellite locus of genome): Fw-ACACTCTTTTTGCAGGATCTA, Rv- AGCAATGTGAAACTCTGGGA. Thermo cycle applied was: 98°C 5mins, 40 cycles of 98°C 5s, 65°C 5s, 72°C 25s and final extension at 72°C for1min.
Fluorescence-activated cell sorting (FACS):
Suspension was then acquired and sorted using a 100 micron nozzle on a MoFlo Legacy sorter (Beckman Coulter, Miami, USA). The gating was performed to include most of the intact and viable cells, was set based on events measured in the Forward versus Side scatter detectors. Measurement of GFP and auto fluorescence from cells were done using detectors of GFP and FL2. The filter settings were 530/30nm for GFP and 580/30nm for FL2 (C). Cells were defined as GFP positive based on negative control. For in vivo experiments, propidium iodide (PI) was used sometimes to facilitate the determination of the GFP positive cell population. GFP positive cells were either sorted as bulk for cell culture or single cell per well into 96 well or 384 well PCR plates for RNAseq.
Treadmill exhaustion test
Treadmill testing was performed using a motor-driven treadmill (Columbus Instruments). Prior to the test day, acclimatization was performed 5 times over a period of 2 weeks at 10 m/min. On the test day, mice ran at 5 m/min for 5 min (warm up) and the speed was increased 1 m/min every min up to 10 m/min. Mice were considered exhausted when they sat for more than 10 sec on a shock pad for the third time.
Physiology:
Tibialis anterior (TA) strength and susceptibility to fatigue (loss in force after repeated contractions) were assessed in vivo. Susceptibility to fatigue was measured by loss in force with repeated contraction, every other second for 4 minutes in TA muscle. The detailed experimental procedure was as described previously (Sun et al., 2020).
Tissue 3D imaging:
Two samples of freshly fixed mouse TAs were sectioned into 200 um thick slices using a KD-400 Vibration Microtome. The slices subsequently underwent a dehydration and rehydration process in gradient methanol and permeabilized with 0.2% Triton as well as Visikol® HISTO™ Penetration Buffer. Sections were then blocked in 5% goat serum and incubated in primary antibodies for Lamin A/C (Mouse anti-Lamin A/C , Invitrogen MA3-1000, 1:100) and Laminin (Rabbit anti-Laminin, Sigma L9393, 1:100) for 24 hours at 37°C. The next day, sections were washed in PBS and incubated in Goat anti Rabbit Alexa Fluor 647 (Thermofisher, A32733) and Goat anti Mouse IgG2b AF 555 (Life technologies, A21147) at 1:400 dilution for overnight at 37°C. Sections were then incubated in mouse IgG blocking buffer (Vector, MKB-2213) and incubated in a third primary antibody (Mouse anti-Pax7, DSHB, 1:100) for 24 hours at 37°C. Secondary antibody (Alexa Fluor® 488 AffiniPure Goat Anti-Mouse IgG, Fcγ subclass 1 specific, Jackson ImmunoResearch, 115-545-205) and DAPI was added the following day for overnight incubation at 37°C. Sections were then washed in PBS and cleared with large volumes of Visikol® HISTO-1™ and Visikol® HISTO-2™ for 12 hours each.
For imaging, each section was mounted in Visikol ClearWell™ Silicon Imaging Chambers using HISTO-2 as imaging media, and imaged using ImageXpress Micro confocal imager (Molecular Devices) at 10X magnification (1 pixel = 0.65 um) across 10 um confocal z steps in 4 channels (DAPI, PAX7, Lamin A/C and Laminin). Images were stitched together using a custom written imageJ macro. Images were processed into animation with Imaris software.
QUANTIFICATION AND STATISTICAL ANALYSIS
RNA-seq analysis
To analyze the single cell sequencing data, reads were mapped and counted using zUMIs 2.2.3(Parekh et al., 2018) with default settings and barcodes provided as a list. zUMIs utilizes STAR (2.5.4b)(Dobin et al., 2013) to map reads to an input reference genome and feature Counts through Rsubread (1.28.1) to tabulate counts and UMI tables. We used GRCh38 from Ensembl concatenated with ERCC spike-in references for the reference genome and gene annotations. Dimensionality reduction and cluster analysis were performed with Seurat (2.3.4)(Butler et al., 2018), while differential gene expression analysis was done in Monocle (2.4.0)(Qiu et al., 2017). We perform GO Enrichment Analysis on differentially expressed genes through the Gene Ontology Consortium tool(Ashburner et al., 2000; Mi et al., 2017; The Gene Ontology, 2019), and visualized enriched terms and pathways with REVIGO(Supek et al., 2011). To correct for batch effects, 2000 genes with highest expression were selected and used to extract expression matrices for the mnnCorrect function in the batchelor package (v1.6.3).
To analyze the bulk RNA sequencing data, raw sequencing data (FASTQ files) were imported into the AltAnalyze software (v2.1.4), which uses the embedded program Kallisto and Ensembl 72 annotations for pseudoalignment and isoform quantification. Processed expression files, including transcript-level expression (transcripts per million) values summed at the gene-level and read counts, generated by the AltAnalyze were imported into R (v4.0.3) to perform differential expression (DE) analysis and gene ontology (GO) enrichment analysis. The Ensembl annotations having the maximum mean expression levels across the samples were collapsed to the gene symbols using the mapIds function in the AnnotationDbi package (v1.52.0). The edgeR (v3.32.1) and limma (v3.46.0) packages were used to identify DE genes with log2 fold change > 2 and adjusted p-value < 0.05. Gene set enrichment analysis (GSEA) was performed using the fgsea package (v1.16.0) to assess the enrichment of gene sets of interest in PAX7::GFP+ cells isolated from transplanted mice. Gene ontology (GO) analysis was performed using the enrichGO function in the clusterProfiler package (v3.18.1) and the enrichr function in the enrichR package (v3.0).
Statistics
Statistical testing was performed using the unpaired two-tailed Student’s t-test or ANOVA, as stated in the figure legends. The method used, P-values and N numbers are indicated in the figure legends. Sample size of NSG-mdx mice was determined by using sample size tool on www.biomath.info website.
Supplementary Material
Movie S1, related with Figure 1. Structure and location of in vivo PAX7::GFP+ cell (co-stained with PAX7 (green) and huLaminA/C (red)) and Laminin (white).
Table S1, related with Figure 2. The list of differentially expressed genes in the scRNA-seq analyses with in vivo PAX7::GFP+ cells and in vitro PAX7::GFP+ samples.
Table S2, related with Figure 4 The list of differentially expressed genes in the bulk RNA-seq analyses with Cultured in vivo PAX7::GFP+ cells and cultured in vitro PAX7::GFP+ samples.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| PAX7 | DSHB | RRID:AB_528428 |
| MyoD | Santa Cruz | Cat# sc-760, RRID:AB_2148870 |
| Myogenin | DSHB | Cat# F5D, RRID:AB_2146602 |
| Anti-human dystrophin | Millipore | MABT827 |
| GFP Polyclonal Antibody | Invitrogen | Cat# A10262, RRID:AB_2534023 |
| Dystrophin | Abcam | Cat# 12577 |
| Lamin A/C Monoclonal Antibody | Thermo Fisher | Cat# MA3-1000, RRID:AB_325377 |
| Anti-Laminin antibody | Sigma | Cat# L9393, RRID:AB_477163 |
| Anti-human Laminin antibody | DSHB | Cat# 2E8, RRID:AB_528343 |
| Desmin Antibody | DAKO | Cat# M0760, RRID:AB_2335684 |
| ERTR-7 Antibody | Santa Cruz | Cat# sc-73355 RRID:AB 1122890 |
| Bacterial and virus strains | ||
| Biological samples | ||
| Chemicals, peptides, and recombinant proteins | ||
| CHIR99021 | Cayman | Cat# 13122 |
| DAPT | Cayman | Cat# 13197 |
| FGF-2 | Pepro Tech | Cat# 100-18B |
| FGF8 | Peprotech | Cat# 100-25 |
| BaCl2 | Sigma | Cat# 342920 |
| Y-27632 | Tocris | Cat# 1524 |
| Hoechst 33342 | Invitrogen | Cat# H3570 |
| Pyronin-Y | Thermo Scientific | Cat# J61068.09 |
| Critical commercial assays | ||
| SuperScript™ III One-Step RT-PCR System with Platinum™ Taq DNA Polymerase | Invitorgen | Cat# 12574018 |
| KAPA SYBR® FAST qPCR Master Mix (2X) Kit |
Kapabiosystems | Cat# KR0389 |
| Phire Tissue Direct PCR Master Mix | Thermo Scientific | Cat# F170S |
| Deposited data | ||
| SC RNA-seq data | GEO | GSE165075 |
| Code for SC RNA-seq data analysis | Github | https://github.com/skannan4/satellite-cell-transplantation |
| Bulk RNA-seq data | GEO | GSE190233 |
| Experimental models: cell lines | ||
| Human H9 ESC | WiCell | Cat# WA09, RRID:CVCL_9773 |
| Human primary myoblasts | N/A | |
| Gibco CF1 Mouse Embryonic Fibroblasts, Irradiated cell line | Gibco | Cat# A34181 RRID:CVCL_RB05 |
| Experimental models: organisms/strains | ||
| NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ | JAX | Cat# JAX:005557, RRID:IMSR_JAX:005557 |
| NSG-mdx4Cv mice | (Arpke et al., 2013) | N/A |
| Oligonucleotides | ||
| Primer: eGFP Forward- ACGTAAACGGCCACAAGT, Reverse- AAGTCGTGCTGCTTCATGTG | This paper | |
| Primer: huPmp22 forward- ACATAGGGAAGGGAGGAAGG, reverse- TTGGTTTGGTTTGAGTTTGG | This paper | |
| Primer: huCalC forward- ACACCGGCTGGAGTATTGTC, reverse- AAAATCCCCAGGGAAATCAC | This paper | |
| Primer: huPAX7 forward- CTGGCTACCAGTACGACCAGT, reverse- GCACGCCGGTTACTGAAC | This paper | |
| Recombinant DNA | ||
| Software and algorithms | ||
| Imaris | Oxford Instrument | |
| FlowJo | FlowJo Software | RRID:SCR_008520; https://www.flowjo.com |
| GraphPad Prism | GraphPad Software Inc. | RRID:SCR_002798; https://www.graphpad.com/scientific-software/prism/ |
| Fiji | https://doi.org/10.1038/nmeth.2019 | RRID:SCR_002285; https://imagej.net/Fiji |
| R | https://www.r-project.org/ | |
| Other | ||
| Quick-RNA Microprep Kit | Zymo Research | Cat# R1050 |
Highlights:
hPSC-derived MPCs engraft into muscle stem cell niche area after transplantation
scRNA-seq depicted satellite cell-like molecular profile of transplanted MPCs
Quiescent MPCs mature developmentally with significant regeneration capability
Transplanted MPCs behave as local satellite cells in vivo in NSG-mdx mice
Acknowledgements:
We thank members of the Wagner lab and the Lee lab for providing valuable discussions. We also thank Dr. Michael Kyba who generously shared the NSG-mdx mice line with us. This work was supported by NIH grants R01NS093213 (GL), UG3TR003271 (GL), R01-AR076390 (TEL), K01-AR074048 (SRI) and R01AR070751 (GL, KRW), the Maryland Stem Cell Research Fund (MSCRF) (GL, KRW), the Maryland Stem Cell Fellowship (GL, CS), Vita Therapeutics (GL), the Muscular Dystrophy Association (MDA) (GL, KRW, TEL, SRI), the Peter and Carmen Lucia Buck Foundation (TEL, GL), the (AHA) Predoctoral Fellowship (SK), Career Development Award 19CDA34760161 (PA), Established Investigator (CK) Awards and National Research Foundation of Korea grants 2020R1A6A1A03043539 (SBL) and 2020M3A9D8037604 (SBL).
Declaration of Interests
CS, PA, KRW and GL are scientific founders and shareholders, and PA and GL serve as scientific advisors of Vita Therapeutics. KRW is a current employee of F. Hoffmann La-Roche. CS is a current employee of Vita Therapeutics. CS, IYC, HL, PA, KRW, and GL are inventors of a patent related with this study. Lee lab receives a funding from Vita Therapeutics.
RML is a current employee at National Institutes of Health but this work was prepared while RML was employed at University of Maryland School of Medicine. The opinions expressed in this article are the author’s own and do not reflect the view of the National Institutes of Health, the Department of Health and Human Services, or the United States government.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- Aguiari P, Leo S, Zavan B, Vindigni V, Rimessi A, Bianchi K, Franzin C, Cortivo R, Rossato M, Vettor R, et al. (2008). High glucose induces adipogenic differentiation of muscle-derived stem cells. Proc Natl Acad Sci U S A 105, 1226–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Tanoury Z, Rao J, Tassy O, Gobert B, Gapon S, Garnier JM, Wagner E, Hick A, Hall A, Gussoni E, et al. (2020). Differentiation of the human PAX7-positive myogenic precursors/satellite cell lineage in vitro. Development. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakura A, Komaki M, and Rudnicki M (2001). Muscle satellite cells are multipotential stem cells that exhibit myogenic, osteogenic, and adipogenic differentiation. Differentiation 68, 245–253. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. (2000). Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauchamp JR, Heslop L, Yu DS, Tajbakhsh S, Kelly RG, Wernig A, Buckingham ME, Partridge TA, and Zammit PS (2000). Expression of CD34 and Myf5 defines the majority of quiescent adult skeletal muscle satellite cells. J Cell Biol 151, 1221–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya D, Rossi DJ, Bryder D, and Weissman IL (2006). Purified hematopoietic stem cell engraftment of rare niches corrects severe lymphoid deficiencies without host conditioning. J Exp Med 203, 73–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjornson CR, Cheung TH, Liu L, Tripathi PV, Steeper KM, and Rando TA (2012). Notch signaling is necessary to maintain quiescence in adult muscle stem cells. Stem Cells 30, 232–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchentouf M, Benabdallah BF, Mills P, and Tremblay JP (2006). Exercise improves the success of myoblast transplantation in mdx mice. Neuromuscul Disord 16, 518–529. [DOI] [PubMed] [Google Scholar]
- Brohl D, Vasyutina E, Czajkowski MT, Griger J, Rassek C, Rahn HP, Purfurst B, Wende H, and Birchmeier C (2012). Colonization of the satellite cell niche by skeletal muscle progenitor cells depends on Notch signals. Dev Cell 23, 469–481. [DOI] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chal J, Oginuma M, Al Tanoury Z, Gobert B, Sumara O, Hick A, Bousson F, Zidouni Y, Mursch C, Moncuquet P, et al. (2015). Differentiation of pluripotent stem cells to muscle fiber to model Duchenne muscular dystrophy. Nat Biotechnol 33, 962–969. [DOI] [PubMed] [Google Scholar]
- Charville GW, Cheung TH, Yoo B, Santos PJ, Lee GK, Shrager JB, and Rando TA (2015). Ex Vivo Expansion and In Vivo Self-Renewal of Human Muscle Stem Cells. Stem Cell Reports 5, 621–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi IY, Lim H, Cho HJ, Oh Y, Chou BK, Bai H, Cheng L, Kim YJ, Hyun S, Kim H, et al. (2020). Transcriptional landscape of myogenesis from human pluripotent stem cells reveals a key role of TWIST1 in maintenance of skeletal muscle progenitors. Elife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi IY, Lim H, Estrellas K, Mula J, Cohen TV, Zhang Y, Donnelly CJ, Richard JP, Kim YJ, Kim H, et al. (2016). Concordant but Varied Phenotypes among Duchenne Muscular Dystrophy Patient-Specific Myoblasts Derived using a Human iPSC-Based Model. Cell Rep 15, 2301–2312. [DOI] [PubMed] [Google Scholar]
- Choo HJ, Canner JP, Vest KE, Thompson Z, and Pavlath GK (2017). A tale of two niches: differential functions for VCAM-1 in satellite cells under basal and injured conditions. Am J Physiol Cell Physiol 313, C392–C404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelison DD, and Wold BJ (1997). Single-cell analysis of regulatory gene expression in quiescent and activated mouse skeletal muscle satellite cells. Dev Biol 191, 270–283. [DOI] [PubMed] [Google Scholar]
- Darabi R, Santos FN, Filareto A, Pan W, Koene R, Rudnicki MA, Kyba M, and Perlingeiro RC (2011). Assessment of the myogenic stem cell compartment following transplantation of Pax3/Pax7-induced embryonic stem cell-derived progenitors. Stem Cells 29, 777–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Luca A, Pierno S, Liantonio A, Cetrone M, Camerino C, Fraysse B, Mirabella M, Servidei S, Ruegg UT, and Conte Camerino D (2003). Enhanced dystrophic progression in mdx mice by exercise and beneficial effects of taurine and insulin-like growth factor-1. J Pharmacol Exp Ther 304, 453–463. [DOI] [PubMed] [Google Scholar]
- De Micheli AJ, Laurilliard EJ, Heinke CL, Ravichandran H, Fraczek P, Soueid-Baumgarten S, De Vlaminck I, Elemento O, and Cosgrove BD (2020). Single-Cell Analysis of the Muscle Stem Cell Hierarchy Identifies Heterotypic Communication Signals Involved in Skeletal Muscle Regeneration. Cell Rep 30, 3583–3595 e3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell’Orso S, Juan AH, Ko KD, Naz F, Perovanovic J, Gutierrez-Cruz G, Feng X, and Sartorelli V (2019). Single cell analysis of adult mouse skeletal muscle stem cells in homeostatic and regenerative conditions. Development 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukada S, Uezumi A, Ikemoto M, Masuda S, Segawa M, Tanimura N, Yamamoto H, Miyagoe-Suzuki Y, and Takeda S (2007). Molecular signature of quiescent satellite cells in adult skeletal muscle. Stem Cells 25, 2448–2459. [DOI] [PubMed] [Google Scholar]
- Gros J, Manceau M, Thome V, and Marcelle C (2005). A common somitic origin for embryonic muscle progenitors and satellite cells. Nature 435, 954–958. [DOI] [PubMed] [Google Scholar]
- Gussoni E, Soneoka Y, Strickland CD, Buzney EA, Khan MK, Flint AF, Kunkel LM, and Mulligan RC (1999). Dystrophin expression in the mdx mouse restored by stem cell transplantation. Nature 401, 390–394. [DOI] [PubMed] [Google Scholar]
- Hashimoto N, Kiyono T, Wada MR, Umeda R, Goto Y, Nonaka I, Shimizu S, Yasumoto S, and Inagawa-Ogashiwa M (2008). Osteogenic properties of human myogenic progenitor cells. Mech Dev 125, 257–269. [DOI] [PubMed] [Google Scholar]
- Henrique D, Abranches E, Verrier L, and Storey KG (2015). Neuromesodermal progenitors and the making of the spinal cord. Development 142, 2864–2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks MR, Hiserodt J, Paras K, Fujiwara W, Eskin A, Jan M, Xi H, Young CS, Evseenko D, Nelson SF, et al. (2018). ERBB3 and NGFR mark a distinct skeletal muscle progenitor cell in human development and hPSCs. Nat Cell Biol 20, 46–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Magli A, Chan SSK, Oliveira VKP, Wu J, Darabi R, Kyba M, and Perlingeiro RCR (2017). Expansion and Purification Are Critical for the Therapeutic Application of Pluripotent Stem Cell-Derived Myogenic Progenitors. Stem Cell Reports 9, 12–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyba M, Perlingeiro RC, and Daley GQ (2002). HoxB4 confers definitive lymphoid-myeloid engraftment potential on embryonic stem cell and yolk sac hematopoietic progenitors. Cell 109, 29–37. [DOI] [PubMed] [Google Scholar]
- Lim H, Choi IY, and Lee G (2014). Profiling individual human embryonic stem cells by quantitative rt-PCR. J Vis Exp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh KM, Chen A, Koh PW, Deng TZ, Sinha R, Tsai JM, Barkal AA, Shen KY, Jain R, Morganti RM, et al. (2016). Mapping the Pairwise Choices Leading from Pluripotency to Human Bone, Heart, and Other Mesoderm Cell Types. Cell 166, 451–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun ATL, Calero-Nieto FJ, Haim-Vilmovsky L, Gottgens B, and Marioni JC (2017). Assessing the reliability of spike-in normalization for analyses of single-cell RNA sequencing data. Genome Res 27, 1795–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinnell IW, Ishibashi J, Le Grand F, Punch VG, Addicks GC, Greenblatt JF, Dilworth FJ, and Rudnicki MA (2008). Pax7 activates myogenic genes by recruitment of a histone methyltransferase complex. Nat Cell Biol 10, 77–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Huang X, Muruganujan A, Tang H, Mills C, Kang D, and Thomas PD (2017). PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res 45, D183–D189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olguin HC, and Olwin BB (2004). Pax-7 up-regulation inhibits myogenesis and cell cycle progression in satellite cells: a potential mechanism for self-renewal. Dev Biol 275, 375–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olguin HC, Yang Z, Tapscott SJ, and Olwin BB (2007). Reciprocal inhibition between Pax7 and muscle regulatory factors modulates myogenic cell fate determination. J Cell Biol 177, 769–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto A, Schmidt C, Luke G, Allen S, Valasek P, Muntoni F, Lawrence-Watt D, and Patel K (2008). Canonical Wnt signalling induces satellite-cell proliferation during adult skeletal muscle regeneration. J Cell Sci 121, 2939–2950. [DOI] [PubMed] [Google Scholar]
- Parekh S, Ziegenhain C, Vieth B, Enard W, and Hellmann I (2018). zUMIs - A fast and flexible pipeline to process RNA sequencing data with UMIs. Gigascience 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA, and Trapnell C (2017). Reversed graph embedding resolves complex single-cell trajectories. Nat Methods 14, 979–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relaix F, and Zammit PS (2012). Satellite cells are essential for skeletal muscle regeneration: the cell on the edge returns centre stage. Development 139, 2845–2856. [DOI] [PubMed] [Google Scholar]
- Rideout WM 3rd, Hochedlinger K, Kyba M, Daley GQ, and Jaenisch R (2002). Correction of a genetic defect by nuclear transplantation and combined cell and gene therapy. Cell 109, 17–27. [DOI] [PubMed] [Google Scholar]
- Ridgeway AG, Wilton S, and Skerjanc IS (2000). Myocyte enhancer factor 2C and myogenin up-regulate each other’s expression and induce the development of skeletal muscle in P19 cells. J Biol Chem 275, 41–46. [DOI] [PubMed] [Google Scholar]
- Sacco A, Doyonnas R, Kraft P, Vitorovic S, and Blau HM (2008). Self-renewal and expansion of single transplanted muscle stem cells. Nature 456, 502–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segales J, Perdiguero E, and Munoz-Canoves P (2016). Regulation of Muscle Stem Cell Functions: A Focus on the p38 MAPK Signaling Pathway. Front Cell Dev Biol 4, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soumillon M, Cacchiarelli D, Semrau S, van Oudenaarden A, and Mikkelsen TS (2014). Characterization of directed differentiation by high-throughput single-cell RNA-Seq. bioRxiv, 003236. [Google Scholar]
- Sudheer S, Liu J, Marks M, Koch F, Anurin A, Scholze M, Senft AD, Wittler L, Macura K, Grote P, et al. (2016). Different Concentrations of FGF Ligands, FGF2 or FGF8 Determine Distinct States of WNT-Induced Presomitic Mesoderm. Stem Cells 34, 1790–1800. [DOI] [PubMed] [Google Scholar]
- Sun C, Choi IY, Rovira Gonzalez YI, Andersen P, Talbot CC Jr., Iyer SR, Lovering RM, Wagner KR, and Lee G (2020). Duchenne muscular dystrophy hiPSC-derived myoblast drug screen identifies compounds that ameliorate disease in mdx mice. JCI Insight 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F, Bosnjak M, Skunca N, and Smuc T (2011). REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 6, e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y, and Cahan P (2019). SingleCellNet: A Computational Tool to Classify Single Cell RNA-Seq Data Across Platforms and Across Species. Cell Syst 9, 207–213 e202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology C (2019). The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res 47, D330–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Matthias N, Lo J, Ortiz-Vitali JL, Shieh AW, Wang SH, and Darabi R (2018). A Myogenic Double-Reporter Human Pluripotent Stem Cell Line Allows Prospective Isolation of Skeletal Muscle Progenitors. Cell Rep 25, 1966–1981 e1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi H, Fujiwara W, Gonzalez K, Jan M, Liebscher S, Van Handel B, Schenke-Layland K, and Pyle AD (2017). In Vivo Human Somitogenesis Guides Somite Development from hPSCs. Cell Rep 18, 1573–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi H, Langerman J, Sabri S, Chien P, Young CS, Younesi S, Hicks M, Gonzalez K, Fujiwara W, Marzi J, et al. (2020). A Human Skeletal Muscle Atlas Identifies the Trajectories of Stem and Progenitor Cells across Development and from Human Pluripotent Stem Cells. Cell Stem Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zammit PS, Golding JP, Nagata Y, Hudon V, Partridge TA, and Beauchamp JR (2004). Muscle satellite cells adopt divergent fates: a mechanism for self-renewal? J Cell Biol 166, 347–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zammit PS, Relaix F, Nagata Y, Ruiz AP, Collins CA, Partridge TA, and Beauchamp JR (2006). Pax7 and myogenic progression in skeletal muscle satellite cells. J Cell Sci 119, 1824–1832. [DOI] [PubMed] [Google Scholar]
- Ziegenhain C, Vieth B, Parekh S, Reinius B, Guillaumet-Adkins A, Smets M, Leonhardt H, Heyn H, Hellmann I, and Enard W (2017). Comparative Analysis of Single-Cell RNA Sequencing Methods. Mol Cell 65, 631–643 e634. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1, related with Figure 1. Structure and location of in vivo PAX7::GFP+ cell (co-stained with PAX7 (green) and huLaminA/C (red)) and Laminin (white).
Table S1, related with Figure 2. The list of differentially expressed genes in the scRNA-seq analyses with in vivo PAX7::GFP+ cells and in vitro PAX7::GFP+ samples.
Table S2, related with Figure 4 The list of differentially expressed genes in the bulk RNA-seq analyses with Cultured in vivo PAX7::GFP+ cells and cultured in vitro PAX7::GFP+ samples.
Data Availability Statement
RNA-sequencing datasets generated during this study are available at Gene Expression Omnibus (GEO), accession number GEO: GSE165075(single cell) and GEO: GSE190233(bulk). Code generated for single cell (SC) RNA-seq data analysis is deposited at Github, accession: https://github.com/skannan4/satellite-cell-transplantation (DOI:10.5281/zenodo.6330148). Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


