ABSTRACT
As a key bacterial second messenger, cyclic di-GMP (c-di-GMP) regulates various physiological processes, such as motility, biofilm formation, and virulence. Cellular c-di-GMP levels are regulated by the opposing activities of diguanylate cyclases (DGCs) and phosphodiesterases (PDEs). Beyond that, the enzymatic activities of c-di-GMP metabolizing proteins are controlled by a variety of extracellular signals and intracellular physiological conditions. Here, we report that pdcA (BTH_II2363), pdcB (BTH_II2364), and pdcC (BTH_II2365) are cotranscribed in the same operon and are involved in a regulatory cascade controlling the cellular level of c-di-GMP in Burkholderia thailandensis. The GGDEF domain-containing protein PdcA was found to be a DGC that modulates biofilm formation, motility, and virulence in B. thailandensis. Moreover, the DGC activity of PdcA was inhibited by phosphorylated PdcC, a single-domain response regulator composed of only the phosphoryl-accepting REC domain. The phosphatase PdcB affects the function of PdcA by dephosphorylating PdcC. The observation that homologous operons of pdcABC are widespread among betaproteobacteria and gammaproteobacteria suggests a general mechanism by which the intracellular concentration of c-di-GMP is modulated to coordinate bacterial behavior and virulence.
IMPORTANCE The transition from planktonic cells to biofilm cells is a successful strategy adopted by bacteria to survive in diverse environments, while the second messenger c-di-GMP plays an important role in this process. Cellular c-di-GMP levels are mainly controlled by modulating the activity of c-di-GMP-metabolizing proteins via the sensory domains adjacent to their enzymatic domains. However, in most cases how c-di-GMP-metabolizing enzymes are modulated by their sensory domains remains unclear. Here, we reveal a new c-di-GMP signaling cascade that regulates motility, biofilm formation, and virulence in B. thailandensis. While pdcA, pdcB, and pdcC constitute an operon, the phosphorylated PdcC binds the PAS sensory domain of PdcA to inhibit its DGC activity, with PdcB dephosphorylating PdcC to derepress the activity of PdcA. We also show this c-di-GMP regulatory model is widespread in the phylum Proteobacteria. Our study expands the current knowledge of how bacteria regulate intracellular c-di-GMP levels.
KEYWORDS: c-di-GMP, motility, biofilm, virulence, Burkholderia thailandensis
INTRODUCTION
Cyclic di-GMP (c-di-GMP), an important secondary messenger in bacteria, regulates many bacterial behaviors, including motility and biofilm formation (1–4). As a general rule, high levels of c-di-GMP stimulate biofilm formation, whereas low levels promote bacterial motility (1–5). c-di-GMP has also been shown to regulate virulence via the type III secretion system (T3SS) in diverse pathogenic bacteria, including Salmonella enterica serovar Typhimurium (6), Pseudomonas aeruginosa (7), and Burkholderia pseudomallei (8). To exert its diverse regulatory action, c-di-GMP interacts with various effectors, including transcriptional regulators (9), enzymes (10), adaptor proteins (11), and riboswitches (12).
The global concentration of c-di-GMP in bacterial cells is controlled by the rate of its synthesis and degradation (3, 13). c-di-GMP is synthesized from two GTP molecules by diguanylate cyclases (DGCs) containing a conserved GGDEF domain, and it is hydrolyzed into 5′-phosphoguanylyl-(3′–5′)-guanosine or GMP by c-di-GMP-specific phosphodiesterases (PDEs) with an EAL or HD-GYP domain (2–4). Bacterial genomes usually encode several c-di-GMP-metabolizing proteins (2). Although the catalytic domains are conserved, many c-di-GMP-metabolizing enzymes have an N-terminal sensory domain that is predicted to perceive specific signals (13–16). For example, in P. aeruginosa, WspR has an N-terminal receiver (REC) domain and a C-terminal GGDEF domain, and phosphorylation of the REC domain stimulates its DGC activity (17). It is worth noting that not all c-di-GMP-metabolizing proteins affect global cellular c-di-GMP levels; some only contribute to the local c-di-GMP pool (16, 18, 19).
The genus Burkholderia contains diverse species that occupy a wide range of ecological niches (20, 21). Burkholderia thailandensis is a nonfermenting motile Gram-negative bacterium found in tropical soils (22, 23). It is closely related to B. pseudomallei, but unlike B. pseudomallei, it only rarely causes disease in humans or animals (24). The genome of B. thailandensis E264 encodes 14 c-di-GMP-metabolizing proteins, B. cenocepacia H111 has 23, and B. pseudomallei K96243 has 16 (25–27). In B. cenocepacia, perception of the quorum-sensing signal cis-2-dodecenoic acid by RpfR stimulates its PDE activity, resulting in a reduction of cellular c-di-GMP (28–31). In B. pseudomallei, CdpA is a c-di-GMP-specific PDE affecting various phenotypes, such as bacterial motility, biofilm formation, and cytotoxicity (8), and the expression of cdpA is upregulated in the presence of nitrate (32). Moreover, CdpA regulates the intracellular level of c-di-GMP in response to amino acids in B. cenocepacia (27). However, the c-di-GMP regulatory network in B. thailandensis remains unknown.
In this study, we report a c-di-GMP signaling cascade in B. thailandensis. This c-di-GMP regulatory system contains a DGC (PdcA; BTH_II2363), a phosphatase (PdcB; BTH_II2364), and a phosphate-accepting response regulator (PdcC; BTH_II2365). We demonstrate that the DGC activity of PdcA is directly inhibited by phosphorylated PdcC and is indirectly regulated by PdcB, which can dephosphorylate PdcC. We reveal that PdcB and PdcC function together with PdcA to regulate motility, biofilm formation, and virulence by manipulating intracellular c-di-GMP levels. These findings may help further our understanding of the regulatory mechanism of c-di-GMP signaling in Burkholderia and other bacteria.
RESULTS
Genes BTH_II2365, BTH_II2364, and BTH_II2363 are cotranscribed as a homologous operon, which is widespread in the phylum Proteobacteria.
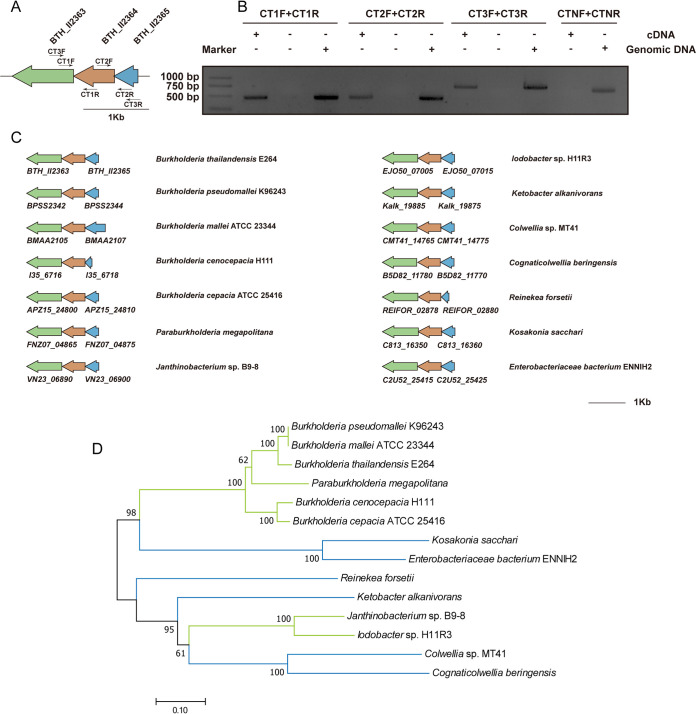
The genome of B. thailandensis E264 encodes six putative GGDEFF domain-containing DGC proteins, four proteins with GGDEF/EAL domains, and four putative PDEs with an EAL domain (see Fig. S1 in the supplemental material). Transmembrane helices of these proteins were predicted using the TMHMM server (33), which showed that eight are transmembrane proteins, whereas the others are located in the cytoplasm. The cytoplasmic putative c-di-GMP-metabolizing enzyme BTH_II2363 contains an N-terminal Per-ARNT-Sim (PAS) domain and a C-terminal GGDEF domain (Fig. S1). BTH_II2365 and BTH_II2364 are adjacent to BTH_II2363. (Fig. 1A). Using cDNA synthesized from B. thailandensis total RNA as a template, PCR analysis using specific primers revealed that BTH_II2365, BTH_II2364, and BTH_II2363 are cotranscribed as an operon (Fig. 1B). Based on the comparative analysis of homologous sequences and phylogenetic analysis, homologous operons of BTH_II2365–BTH_II2363 are widespread in betaproteobacteria and gammaproteobacteria (Fig. 1C and D).
FIG 1.
Cotranscription and wide distribution of homologous BTH_II2365–BTH_II2363 operon. (A) Gene organization of BTH_II2365–BTH_II2363 cluster in B. thailandensis E264. These genes are predicted to encode a single-domain response regulator (BTH_II2365), a phosphatase (BTH_II2364), and a diguanylate cyclase with PAS-GGDEF domain (BTH_II2363). (B) Agarose gel (1%, wt/vol) electrophoresis of the PCR-amplified products. cDNA synthesized from B. thailandensis E264 total RNA was used as a template to determine BTH_II2365–BTH_II2363 cotranscription with primers CTF1/CTR1, CTF2/CTR2, and CTF3/CTR3. CTNF and CTNR primers from an unexpressed region were used as a negative control to rule out possible genomic DNA contamination during RNA preparation. (C) Homologous operons of BTH_II2365–BTH_II2363 are widespread in betaproteobacteria and gammaproteobacteria. (D) The phylogenetic tree of BTH_II2363 homologs was conducted in MEGA 7 by using the neighbor-joining method (bootstrap, 1,000 replicates). Lines are colored based on the bacterial taxonomy of the corresponding organism. Green, betaproteobacteria; blue, gammaproteobacteria.
BTH_II2363 is a DGC that regulates motility and biofilm formation.
To test whether BTH_II2363 is a functional DGC, in vitro enzymatic activity assays were performed with GTP as the substrate, and the synthesized products were separated by high-performance liquid chromatography (HPLC). It was observed that purified BTH_II2363 produces c-di-GMP in the presence of GTP, while c-di-GMP was not detected in the presence of ATP (see Fig. 3C and Fig. S3). These results indicate that BTH_II2363 is a functional DGC protein, and we named it PdcA (PAS domain-containing diguanylate cyclase A). We then sought to determine whether pdcA affects the intracellular levels of c-di-GMP in B. thailandensis. We measured the cellular concentration of c-di-GMP via liquid chromatography-tandem mass spectrometry (LC-MS/MS), which showed that the ΔpdcA mutant produced significantly less c-di-GMP than the wild-type and complemented strains (P < 0.001) (Fig. 2G).
FIG 2.
BTH_II2363, BTH_II2364, and BTH_II2365 regulate biofilm formation and motility by indirectly modulating intracellular c-di-GMP levels. (A and B) Deletion of BTH_II2363, BTH_II2364, or BTH_II2365 regulates swimming motility (A) and biofilm formation (B) of B. thailandensis. (C and D) Overexpression of BTH_II2363, BTH_II2364, or BTH_II2365 regulates swimming motility (C) and biofilm formation (D) of B. thailandensis. Swimming motility of the indicated strains was tested in the semisolid agar medium (up), and the sizes of swimming zones were measured (down). Biofilm formation of the indicated strains was displayed with crystal violet staining (up) and quantified with optical density measurement (down). Data shown are the averages and standard deviations (SD) from three independent experiments. (E and F) Growth curves of the indicated strains under normal conditions. Growth of the indicated strains in LB (E) or M63 (F) medium was monitored by measuring OD600 at indicated time points at 37°C. (G) The intracellular concentrations of c-di-GMP in WT, Δ2363, Δ2364, Δ2365, and the corresponding complemented strains were detected by LC-MS/MS. Data shown are the averages and SD from six independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
c-di-GMP is an important signaling molecule that regulates motility and biofilm formation in diverse bacterial species (1, 3). To determine the effect of pdcA on motility and biofilm formation in B. thailandensis, the ΔpdcA deletion mutant was constructed by a homologous recombination method and confirmed by PCR analysis as previously described (34). To complement the pdcA mutant, the pBBR1MCS1 derivative carrying full-length pdcA under a constitutive lac promoter was transformed into the pdcA mutant by electroporation, and the empty plasmid pBBR1MCS1 was used as a vector control in all experiments. Quantitative real-time PCR (qRT-PCR) data showed that the expression level of pdcA in the complemented strain was 7.8-fold higher than that of the wild-type strain (Fig. S2). Swimming motility assays were tested by seeding stationary-phase cells (2.5 μL; 2.0 × 108 CFU) onto the center of semisolid agar media at 30°C, and swim diameters were measured after 24 h. As shown in Fig. 2A, the ΔpdcA mutant strain showed a significant increase in motility compared with the wild-type and complemented strains (P < 0.001). To assay biofilm formation, cultures were grown for 48 h with shaking (140 rpm) at 30°C in glass tubes, and biofilms attached to the surfaces of the glass tubes were washed with sterile water and stained with 0.1% (wt/vol) crystal violet. Crystal violet-stained biofilm was dissolved by 95% ethanol, and the solution was measured at an absorbance of 595 nm. The ΔpdcA mutant strain produced significantly less biofilm than the wild-type and complemented strains (P < 0.001) (Fig. 2B). The phenotypic changes in motility and biofilm formation were not caused by differences in growth rates (Fig. 2E and F). Taken together, these results demonstrated that PdcA possesses DGC activity and plays a role in the regulation of motility and biofilm formation in B. thailandensis.
BTH_II2364 and BTH_II2365 regulate motility and biofilm formation by indirectly modulating intracellular c-di-GMP levels.
Cotranscribed genes usually exhibit related functions and roles in the same signaling pathways or macromolecular protein apparatus (35). Cotranscription of BTH_II2364, BTH_II2365, and pdcA led us to speculate that BTH_II2364 and BTH_II2365 also play a role in the regulation of motility and biofilm formation. To this end, the Δ2364 and Δ2365 deletion mutants and complemented strains were constructed in a way similar to that for the ΔpdcA strain and its complemented strain. qRT-PCR data showed that deletion of BTH_II2364 or BTH_II2365 did not cause any polar effect in the corresponding mutant (Fig. S2), and the expression levels of BTH_II2364 and BTH_II2365 in the corresponding complemented strain were 11.6-fold and 9.7-fold higher than that of the wild-type strain, respectively (Fig. S2). Indeed, deletion of BTH_II2364 resulted in significantly enhanced swimming motility and reduced biofilm formation (P < 0.001) (Fig. 2A and B). In contrast, the Δ2365 mutant exhibited significantly reduced swimming motility and increased biofilm formation compared with the wild-type strain (P < 0.001) (Fig. 2A and B). Expression of BTH_II2364 and BTH_II2365 restored these phenotypes in the corresponding mutant (Fig. 2A and B). Moreover, overexpression of BTH_II2363 or BTH_II2364 in the wild-type strain significantly decreased swimming motility and promoted biofilm formation, while overexpression of BTH_II2365 in the wild-type strain led to significantly increased swimming motility and reduced biofilm formation (P < 0.05) (Fig. 2C and D). The phenotypic changes observed were not caused by differences in growth rates (Fig. 2E and F). These results suggest that BTH_II2364 and BTH_II2365 are involved in c-di-GMP modulation. Thus, we measured the intracellular concentration of c-di-GMP in the Δ2364 and Δ2365 mutants using LC-MS/MS as previously described (36). As expected, the Δ2364 mutant had significantly less (P < 0.01), while the Δ2365 mutant had significantly more (P < 0.001), c-di-GMP than the wild-type strain; levels were restored upon complementation (Fig. 2G). Altogether, these results demonstrated that BTH_II2364 and BTH_II2365 regulate motility and biofilm formation by modulating intracellular c-di-GMP levels, and we named it PdcB and PdcC, respectively.
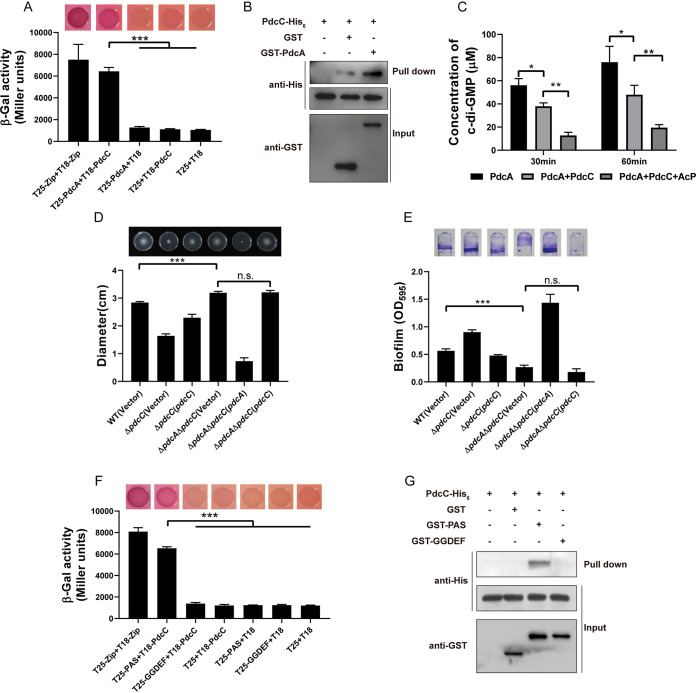
PdcC interacts with PdcA to inhibit c-di-GMP synthesis.
The gene pdcC encodes a single-domain phosphate-accepting response regulator. Response regulators are commonly composed of an N-terminal phosphoacceptor REC domain and a variable C-terminal functional domain, such as enzymatic domains (37). Single-domain response regulators composed of only a REC domain can directly interact with downstream effector proteins and allosterically regulate their enzymatic activity (38, 39). PdcC decreases the intracellular c-di-GMP concentration, leading us to question whether PdcC affects the intracellular c-di-GMP level by influencing the DGC activity of PdcA. A bacterial two-hybrid assay showed that PdcA interacts with PdcC in vivo (Fig. 3A). Glutathione-S-transferase (GST) pulldown assay, which is commonly used to determine the direct protein-protein interaction in vitro, also revealed that PdcC-His6 binds GST-PdcA rather than GST (Fig. 3B). Next, we determined the impact of PdcC on the DGC activity of PdcA. Indeed, production of c-di-GMP was significantly decreased when PdcA was incubated with PdcC, suggesting that the DGC activity of PdcA was inhibited by PdcC (P < 0.05) (Fig. 3C and Fig. S3). To assess the impact of phosphorylated PdcC on PdcA activity, PdcC was pretreated with acetyl phosphate, which is commonly used to phosphorylate response regulators in vitro (17, 40). We found that the production of c-di-GMP by PdcA was further significantly reduced in acetyl phosphate-pretreated PdcC compared with that without treatment (P < 0.01) (Fig. 3C and Fig. S3). These results suggest that phosphorylated PdcC inhibits the DGC activity of PdcA. The ΔpdcA ΔpdcC double mutant exhibited significantly increased swimming motility and reduced biofilm formation compared with the wild-type strain (P < 0.001), while expression of pdcC restored swimming motility and biofilm formation in the ΔpdcC mutant but had no effect in the ΔpdcA ΔpdcC double mutant (Fig. 3D and E). Taken together, these results suggest that PdcC-mediated phenotypic changes are dependent on PdcA.
FIG 3.
PdcC interacts with PdcA to inhibit c-di-GMP synthesis. (A) Interaction between PdcC and PdcA was assessed using MacConkey plates (up), and the strength of interaction was quantified by measurement of β-galactosidase activity (down). (B) GST pulldown assay confirmed the interaction between PdcC and PdcA. Recombinant protein PdcC-His6 was incubated with GST or GST-PdcA individually, and protein complexes were captured by glutathione beads and detected by Western blotting. (C) PdcC inhibits the DGC activity of PdcA. PdcA was incubated with GTP in the presence or absence of PdcC at 30°C for 0, 30, and 60 min, and the products were analyzed by HPLC (Fig. S2). The levels of synthesized c-di-GMP in the samples were determined from a standard curve established with a serially diluted c-di-GMP solution. To evaluate the effect of the phosphorylation of PdcC on PdcA DGC activity, PdcC was pretreated by 25 mM acetyl phosphate (AcP) for 30 min at 30°C. (D) Swimming motility of the indicated strains was tested in the semisolid agar medium (up), and the diameters of swimming zones were measured (down). (E) Biofilm formation of the indicated strains was displayed with crystal violet staining (up) and quantified with optical density measurement (down). (F) Interactions between PdcC and the PAS domain of PdcA were assessed using MacConkey plates (up), and the strength of interaction was quantified by measurement of β-galactosidase activity (down). (G) GST pulldown assay confirmed the interaction between PdcC and the PAS domain of PdcA. Recombinant protein PdcC-His6 was incubated with GST, GST-PAS, or GST-GGDEF individually, and protein complexes were captured by glutathione beads and detected by Western blotting. Data shown are the averages and SD from three independent experiments. *, P < 0.05; ***, P < 0.001; n.s., not significant.
In addition to its enzymatic domain, PdcA contains a PAS domain. PAS domains act as sensors to detect signals in proteins and regulate their activity (15, 41, 42). The bacterial two-hybrid assay suggested that PdcC interacts with the PAS domain but not the GGDEF domain (Fig. 3F). Consistent with this, the GST pulldown assay confirmed that GST-PAS, but not GST-GGDEF or GST, interacts with PdcC-His6 (Fig. 3G). These results suggest that PdcC regulates the DGC activity of PdcA via direct interaction with its PAS domain.
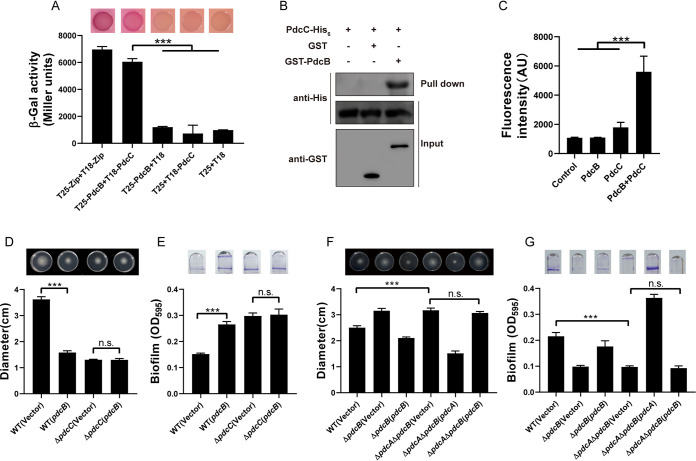
PdcB dephosphorylates PdcC.
The gene pdcB encodes a phosphatase with a CheC/CheX domain, while BTH_II2365 encodes a phosphoryl-accepting response regulator, which led us to speculate that PdcB dephosphorylates PdcC. The bacterial two-hybrid assay showed that PdcB interacted with PdcC but not with PdcA (Fig. 4A and Fig. S4). The interaction between PdcB and PdcC was further confirmed by a GST pulldown assay using purified GST-PdcB and PdcC-His6 proteins in vitro (Fig. 4B). Furthermore, samples containing purified PdcB were incubated with PdcC at 30°C for 20 min, and released inorganic phosphate was detected using a phosphate sensor assay kit that uses a fluorescent phosphate sensor to detect the inorganic phosphate. The reaction buffer and samples containing only PdcC were used as controls to exclude inorganic phosphate in the reaction buffer and that released by autodephosphorylated PdcC. As shown in Fig. 4C, the fluorescence intensity was significantly increased after incubating purified PdcB with PdcC compared to controls (P < 0.001), suggesting that PdcB is a phosphatase that dephosphorylates PdcC. Overexpression of pdcB in the wild-type strain caused significantly decreased swimming motility and enhanced biofilm formation (P < 0.001); these effects were not observed with pdcB overexpression in the ΔpdcC mutant, indicating that PdcB-mediated phenotypic changes are dependent on PdcC (Fig. 4D and E). The ΔpdcA ΔpdcB double mutant showed significantly enhanced swimming motility and decreased biofilm formation compared with the wild-type strain (P < 0.001) (Fig. 4F and G). Expression of pdcA, but not pdcB, restored these phenotypes in the ΔpdcA ΔpdcB double mutant (Fig. 4F and G). Collectively, these results suggest that PdcB dephosphorylates PdcC to modulate the DGC activity of PdcA.
FIG 4.
PdcB dephosphorylates PdcC. (A) Interaction between PdcB and PdcC was assessed using MacConkey plates (up), and the strength of interaction was quantified by measurement of β-galactosidase activity (down). (B) GST pulldown assay confirmed the interaction between PdcB and PdcC. Recombinant protein PdcC-His6 was incubated with GST or GST-PdcB individually, and protein complexes were captured by glutathione beads and detected by Western blotting. (C) PdcB can dephosphorylate PdcC in vitro. The inorganic phosphate released from PdcC was detected by using a phosphate sensor assay kit. The reaction buffer was used as a negative control to exclude the inorganic phosphate in the reaction buffer. Fluorescence signals were measured using a SpectraMax M2 plate reader (Molecular Devices) with excitation/emission wavelengths of 420/450 nm. (D) Swimming motility of the indicated strains was tested in the semisolid agar medium (up) and the diameters of swimming zones were measured (down). (E) Biofilm formation of the indicated strains was displayed with the crystal violet staining (up) and quantified with optical density measurement (down). (F) The swimming motility of WT, mutant, and complemented strains was tested (up) and measured (down). (G) Biofilm formation of the indicated strains was displayed (up) and quantified (down). Data shown are the averages and SD from three independent experiments. **, P < 0.01; ***, P < 0.001; n.s., not significant.
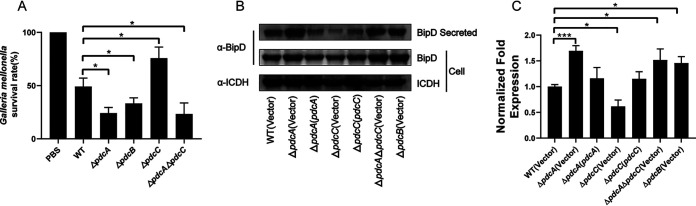
The PdcABC signaling cascade contributes to the virulence of B. thailandensis.
To investigate the role of the PdcABC signaling cascade in pathogenesis, Galleria mellonella (wax moth) larvae were used to determine the virulence of different B. thailandensis strains. Larvae were injected with 50 μL of 105 CFU in the secondary right proleg using a Hamilton H syringe as previously described (34), and 50 μL phosphate-buffered saline (PBS) was injected as a control. Twenty larvae were injected for each B. thailandensis strain. Infection with the wild-type strain caused a modest survival rate (average, 48%), whereas challenge with ΔpdcA, ΔpdcB, ΔpdcC, and ΔpdcA ΔpdcC strains resulted in mean survival rates of 24%, 33%, 75%, and 23% in larvae, respectively (Fig. 5A). These results suggest that a mutation in pdcA or pdcB results in significant elevation of virulence of B. thailandensis (P < 0.05). However, the loss of pdcC causes significant attenuation of virulence in B. thailandensis (P < 0.05), which may result from the elevation of cellular c-di-GMP levels. High levels of c-di-GMP have been shown to reduce virulence via T3SS in a variety of bacterial pathogens, including B. pseudomallei (18, 43–46). We measured the expression and secretion of a T3SS effector, BipD (BTH_II0844), using qRT-PCR and Western blot analysis. BipD in the culture supernatants of relevant B. thailandensis strains was probed for using specific anti-BipD antibody as previously reported (47). For the pellet fraction, isocitrate dehydrogenase (ICDH) was used as a loading control. Deletion of pdcA or pdcB resulted in significantly increased secretion of BipD and deletion of pdcC leads to significantly reduced secretion of BipD compared with the wild-type strain, whereas the expression levels of bipD were slightly affected in the ΔpdcA, ΔpdcB, and ΔpdcC mutants (Fig. 5B and C). The expression of pdcA or pdcC in the corresponding mutant restored the secretion of BipD to wild-type levels. Moreover, the ΔpdcA ΔpdcC double mutant showed similar effects compared with the ΔpdcA and ΔpdcB mutants with respect to the secretion of BipD (Fig. 5B). Taken together, these results suggest that this c-di-GMP signaling cascade modulates the virulence of B. thailandensis by affecting the secretion of T3SS effectors.
FIG 5.
PdcABC signaling cascade contributes to the virulence of B. thailandensis. (A) Virulence survival of relative B. thailandensis strains in G. mellonella larvae. Ordinate represents the mean percent survival rate of G. mellonella infected with different strains after 16 h. (B) Western blot detection of BipD secretion in relative B. thailandensis strains. Proteins in the culture supernatant of relevant B. thailandensis strains were probed for specific anti-BipD rabbit polyclonal antibody. For the pellet fraction, isocitrate dehydrogenase (ICDH) was used as a loading control. (C) The expression of bipD in relative B. thailandensis strains. Data shown are the averages and SD from three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
DISCUSSION
Previous studies have found that interruption of pdcA homologs results in increased motility in B. pseudomallei and B. cenocepacia (25, 26). However, the DGC activity of PdcA and the regulatory mechanism of its catalytic activity remain obscure. Here, we report that PdcA, PdcB, and PdcC constitute a c-di-GMP regulatory cascade that modulates intracellular c-di-GMP levels and regulates motility, biofilm formation, and virulence in B. thailandensis. In this c-di-GMP regulatory model, phosphorylated PdcC inhibits the DGC activity of PdcA by directly binding to the PAS domain, while PdcB dephosphorylates the phosphoryl group of PdcC, relieving the inhibition of PdcA (Fig. 6). This c-di-GMP signaling cascade is widely distributed in betaproteobacteria and gammaproteobacteria, such as Ketobacter alkanivorans, Reinekea forsetii, and Kosakonia sacchari, suggesting a general mechanism underlying the modulation of c-di-GMP levels (Fig. 1C and D).
FIG 6.

Proposed model of the PdcABC signaling cascade in B. thailandensis. PdcA, PdcB, and PdcC cooperatively regulate biofilm formation, motility, and virulence in B. thailandensis. Phosphorylated PdcC acts as an inhibitor controlling the DGC activity of PdcA. The levels of phosphorylation of PdcC were affected by the phosphatase PdcB, while the phosphoryl group donors of PdcC remain unknown.
Response regulators encoded by bacteria play an essential role in signal transduction, as they can fuse various output domains, such as DNA-binding, RNA-binding, and enzymatic domains, and can function as a stand-alone module (48). Single-domain response regulators can transfer the phosphoryl group to other proteins (49) or modulate the activity of its protein partner by direct interaction (39). Here, we show that the phosphorylated single-domain response regulator PdcC regulates the DGC activity of PdcA by direct interaction (Fig. 3C). No phosphorylation site has been predicted in PdcA, and the DGC activity of PdcA is not affected by treatment with acetyl phosphate (see Fig. S3 in the supplemental material), indicating that PdcC does not transfer the phosphoryl group to PdcA. Nevertheless, the phosphoryl group donors of PdcC need to be further ascertained.
It is well known that c-di-GMP is produced by DGCs and hydrolyzed or minimized by PDEs (3, 50). Moreover, many c-di-GMP-metabolizing proteins have an N-terminal sensory domain, such as the PAS domain (15). Generally, PAS domains serve as sensors to detect a wide range of signals or function via interaction with other proteins (41). In Escherichia coli, the PAS domain of DgcE contributes to its dimerization and DGC activity because GGDEF functions as a dimer (51). In Campylobacter jejuni, the single PAS domain protein CetB controls the kinase activity of CetA by direct binding to its HAMP domain (52). In B. cenocepacia, cis-2-dodecenoic acid binds to the PAS domain of RpfR and allosterically stimulates its PDE activity (30). In this study, we revealed that the PAS domain of PdcA is bound by phosphorylated PdcC, leading to allosteric inhibition of the DGC activity of PdcA (Fig. 3C). Thus, it is possible that bacteria use this type of c-di-GMP signaling cascade to rapidly modulate intracellular c-di-GMP levels and adapt to environmental changes.
As a signaling molecule in bacteria, c-di-GMP is crucial for regulating the transition from the planktonic state to the biofilm state (1, 3, 4). Consistent with previous reports, our results indicate that the PdcABC signaling cascade controls motility and biofilm formation via c-di-GMP turnover in B. thailandensis (Fig. 2A and B). c-di-GMP also regulates virulence in many pathogenic bacteria (8, 26, 44). In B. pseudomallei, high cellular levels of c-di-GMP reduce mammalian cell invasion and expression of the T3SS (8). In B. cenocepacia, high levels of c-di-GMP attenuate virulence to Caenorhabditis elegans and G. mellonella (26, 44). Consistent with this, our data show that a high intracellular c-di-GMP level attenuates the virulence of B. thailandensis to G. mellonella by inhibiting the secretion of T3SS (Fig. 5).
In summary, we identified a novel c-di-GMP signaling cascade that regulates motility, biofilm formation, and virulence by manipulating the intracellular concentration of c-di-GMP (Fig. 6). Widespread distribution of this c-di-GMP signaling cascade in the phylum Proteobacteria reveals a general strategy used by bacteria to modify their behaviors.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. B. thailandensis E264 and derivatives were grown in Luria-Bertani (LB; 1% tryptone, 0.5% yeast extract, 0.5% NaCl) or M63 minimal medium [100 mM KH2PO4, 75 mM KOH, 15 mM (NH4)2SO4, 1 mM MgSO4, 3.9 μM FeSO4, 22 mM glucose] with shaking at 220 rpm at 37°C or 30°C. E. coli strains were cultured at 37°C in LB medium. Antibiotics were added at the following concentrations: ampicillin, 100 μg/mL; kanamycin, 50 μg/mL; and chloramphenicol, 20 μg/mL, for E. coli; and streptomycin, 100 μg/mL, and chloramphenicol, 50 μg/mL, for B. thailandensis.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype descriptiona | Reference or source |
|---|---|---|
| E. coli | ||
| BL21(DE3) | Host for expression vector pET21a and pET28a | Novagen |
| XL1-Blue | Host for expression vector pGEX6p-1 | Novagen |
| TG1 | Host for cloning | Novagen |
| SM10 λ pir | λ-pir lysogen of SM10, thi pro hsdR hsdM+ recA RP4 2-Tc::Mu-Km::Tn7 | 34 |
| BTH101 | Host for the bacterial two-hybrid system | 62 |
| B. thailandensis | ||
| E264 | Wild-type strain (ATCC 700388) environmental isolate from Thailand, Strr | 34 |
| ΔpdcA | pdcA gene deleted in B. thailandensis E264, Strr | This study |
| ΔpdcB | pdcB gene deleted in B. thailandensis E264, Strr | This study |
| ΔpdcC | pdcC gene deleted in B. thailandensis E264, Strr | This study |
| ΔpdcA ΔpdcC | pdcA and pdcC genes deleted in B. thailandensis E264, Strr | This study |
| ΔpdcA ΔpdcB | pdcA and pdcB genes deleted in B. thailandensis E264, Strr | This study |
| Plasmids | ||
| pBBR1MCS1 | Expression vector contains a lac promoter, Cmr | Novagen |
| pBBR1MCS1-pdcA | pBBR1MCS1 carries pdcA coding region at down-stream of the lac promoter, Cmr | This study |
| pBBR1MCS1-pdcB | pBBR1MCS1 carries pdcB coding region at down-stream of the lac promoter, Cmr | This study |
| pBBR1MCS1-pdcC | pBBR1MCS1 carries pdcC coding region at down-stream of the lac promoter, Cmr | This study |
| pET28a | Expression vector with N-terminal hexahistidine affinity tag, Kmr | Novagen |
| pET28a-pdcA | pET28a carrying pdcA coding region, Kmr | This study |
| pET28a-pdcB | pET28a carrying pdcB coding region, Kmr | This study |
| pET21a | Expression vector with C-terminal hexahistidine affinity tag, Ampr | Novagen |
| pET21a-pdcC | pET21a carrying pdcC coding region, Ampr | This study |
| pGEX-6P-1 | Expression vector with N-terminal GST tag, Ampr | Novagen |
| pGEX-6P-1-pdcA | pGEX-6P-1 carrying pdcA coding region, Ampr | This study |
| pGEX-6P-1-pas | pGEX-6P-1 carrying pas of pdcA coding region, Ampr | This study |
| pGEX-6P-1-ggdef | pGEX-6P-1 carrying ggdef of pdcA coding region, Ampr | This study |
| pGEX-6P-1-pdcB | pGEX-6P-1 carrying pdcB coding region, Ampr | This study |
| pDM4-phes | Suicide vector, mobRK2, oriR6K, pir, phes, Cmr | 34 |
| pDM4-phes-ΔpdcA | Construct used for in-frame deletion of pdcA, Cmr | This study |
| pDM4-phes-ΔpdcB | Construct used for in-frame deletion of pdcB, Cmr | This study |
| pDM4-phes-ΔpdcC | Construct used for in-frame deletion of pdcC, Cmr | This study |
| pDM4-phes-ΔpdcAΔpdcB | Construct used for in-frame deletion of pdcA and pdcB, Cmr | This study |
| pKT25 | p15A origin of replication encoding CyaA1224, Kmr | 62 |
| pUT18C | ColE1 origin of replication encoding CyaA225399, Ampr | 62 |
| pKT25M | Modified pKT25 | 56 |
| pUT18CM | Modified pUT18C | 56 |
| pKT25-zip | Leucine zipper of GCN1 (BTH positive control), Kmr | 62 |
| pUT18C-zip | Leucine zipper of GCN1 (BTH positive control), Ampr | 62 |
| pKT25M-pdcA | pdcA in pKT25M, Kmr | This study |
| pKT25M-pas | pas of BTH_II2363 in pKT25M, Kmr | This study |
| pKT25M-ggdef | ggdef of BTH_II2363 in pKT25M, Kmr | This study |
| pKT25M-pdcB | pdcB in pKT25M, Kmr | This study |
| pUT18CM-pdcB | pdcB in pUT18CM, Ampr | This study |
| pUT18CM-pdcC | pdcC in pUT18CM, Ampr | This study |
Strr, Cmr, Kmr, and Ampr represent resistance to streptomycin, chloramphenicol, kanamycin, and ampicillin at 100, 20, 50, and 100 μg/mL, respectively.
Plasmid construction.
Primers used in this study are listed in Table 2. To express His6-tagged PdcA, PdcB, and PdcC, primers pdcAHF-BamHI/pdcAHR-HindIII, pdcBHF-BamHI/pdcBHR-HindIII, and pdcCHF-BamHI/pdcCHR-HindIII were used to amplify pdcA, pdcB, and pdcC fragments from genomic DNA of B. thailandensis. The PCR products of pdcA, pdcB, and pdcC were inserted into BamHI and HindIII restriction sites of pET28a or pET21a by Seamless Cloning, resulting in plasmids pET28a-pdcA, pET28a-pdcB, and pET21a-pdcC. To express GST-tagged PdcA, primers pdcAGF-BamHI and pdcAGR-EcoRI were used to amplify the pdcA gene from the genomic DNA of B. thailandensis. The PCR product of pdcA was inserted into the BamHI/EcoRI sites of pGEX6P-1 by Seamless Cloning, resulting in plasmid pGEX6P-1-pdcA. The expression plasmids pGEX6P-1-pas, pGEX6P-1-ggdef, and pGEX6P-1-pdcB were constructed in similar manners by using the primers listed in Table 2. The suicide plasmid pDM4-phes-ΔpdcA (BTH_II2363) was constructed to prepare the ΔpdcA in-frame deletion mutant. Briefly, the 831-bp upstream fragment and 814-bp downstream fragment of pdcA were amplified with primer pairs pdcAUF-XbaI/pdcAUR and pdcADF/pdcADR-BglII. The upstream and downstream fragments then were fused by overlap PCR with pdcAUF-XbaI/pdcADR-BglII. The fused PCR products were inserted between XbaI and BglII restriction sites of pDM4-phes by Seamless Cloning, resulting in pDM4-phes-ΔpdcA. The knockout plasmids pDM4-phes-ΔpdcB (BTH_II2364), pDM4-phes-ΔpdcC (BTH_II2365), and pDM4-phes-ΔpdcAΔpdcB were constructed in similar manners by using the primers listed in Table 2. To complement the pdcA mutant, primers pdcABF-KpnI/pdcABR-HindIII were used to amplify the pdcA gene fragment from B. thailandensis genomic DNA. The PCR product of pdcA was inserted into the KpnI/HindIII sites of pBBR1MCS1 by Seamless Cloning, resulting in plasmid pBBR1MCS1-pdcA. The complementary plasmids pBBR1MCS1-pdcB and pBBR1MCS1-pdcC were constructed in similar manners by using the primers listed in Table 2. To construct the bacterial two-hybrid assay plasmids, pdcA-25-F-BamHI and pdcA-25-R-SalI primers were used to amplify pdcA from the genomic DNA of B. thailandensis. The pdcA gene fragment was digested with BamHI/SalI and then inserted into similarly digested pKT25M, yielding pKT25M-pdcA. The bacterial hybrid plasmids pKT25M-pas, pKT25M-ggdef, pKT25M-pdcB, pUT18CM-pdcB, and pUT18CM-pdcC were constructed in similar manners by using the primers listed in Table 2. The validity of all the plasmids constructed above was confirmed by DNA sequencing.
TABLE 2.
Primers used in this studya
| Primer | Sequence (5′ to 3′) | Function |
|---|---|---|
| pdcABF-KpnI | CCTCACTAAAGGGAACAAAAGCTGGGTACCATGAACCTGCTTTCCTCGCT | To generate pBBR1MCS1-pdcA, pBBR1MCS1-pdcB, pBBR1MCS1-pdcC |
| pdcABR-HindIII | CCCGGGCTGCAGGAATTCGATATCAAGCTTTCAGCCGAACGCGACCGAG | |
| pdcBBF-KpnI | CCTCACTAAAGGGAACAAAAGCTGGGTACCATGTCTGAGACGGTGTTGACGG | |
| pdcBBR-HindIII | CCCGGGCTGCAGGAATTCGATATCAAGCTTTCATAAGCTGGACAGCAACAAA | |
| pdcCBF-KpnI | CCTCACTAAAGGGAACAAAAGCTGGGTACCATGCCCCTGCCGATTGTGATC | |
| pdcCBR-HindIII | CCCGGGCTGCAGGAATTCGATATCAAGCTTTCAGACATACAACCCATACTC | |
| pdcAHF-BamHI | ACTGGTGGACAGCAAATGGGTCGCGGATCCATGAACCTGCTTTCCTCGCT | To generate pET-28a-pdcA |
| pdcAHR-HindIII | GTGGTGGTGCTCGAGTGCGGCCGCAAGCTTTCAGCCGAACGCGACCGAG | |
| pdcBHF-BamHI | ACTGGTGGACAGCAAATGGGTCGCGGATCCATGTCTGAGACGGTGTTGACGG | To generate pET-28a-pdcB |
| pdcBHR-HindIII | GTGGTGGTGCTCGAGTGCGGCCGCAAGCTTTCATAAGCTGGACAGCAACAAA | |
| pdcCHF-BamHI | ACTGGTGGACAGCAAATGGGTCGCGGATCCATGCCCCTGCCGATTGTGATC | To generate pET-21a-pdcC |
| pdcCHR-HindIII | GTGGTGGTGCTCGAGTGCGGCCGCAAGCTTGACATACAACCCATACTC | |
| pdcAGF-BamHI | GAAGTTCTGTTCCAGGGGCCCCTGGGATCCATGAACCTGCTTTCCTCGCT | To generate pGEX6p-1-pdcA |
| pdcAGR-EcoRI | ATGCGGCCGCTCGAGTCGACCCGGGAATTCTCAGCCGAACGCGACCGAG | |
| pasGF-BamHI | GAAGTTCTGTTCCAGGGGCCCCTGGGATCCATGAACCTGCTTTCCTCGCT | To generate pGEX6p-1-pas |
| pasGR-EcoRI | ATGCGGCCGCTCGAGTCGACCCGGGAATTCTCACAGTTTCGCCACGGC | |
| ggdefGF-BamHI | GAAGTTCTGTTCCAGGGGCCCCTGGGATCCATGCAGGAATACGCGAACC | To generate pGEX6p-1-ggdef |
| ggdefGR-EcoRI | ATGCGGCCGCTCGAGTCGACCCGGGAATTCTCAGCCGAACGCGACC | |
| pdcBGF-BamHI | GAAGTTCTGTTCCAGGGGCCCCTGGGATCCATGTCTGAGACGGTGTTGACG | To generate pGEX6p-1-pdcB |
| pdcBGR-EcoRI | ATGCGGCCGCTCGAGTCGACCCGGGAATTCTCATAAGCTGGACAGCAACAAA | |
| pdcAUF-XbaI | AGTACGCGTCACTAGTGGGGCCCTTCTAGAGTCGCCAAGCCCGTCACCTC | To generate pDM4-phes-ΔpdcA |
| pdcAUR | TCGCCTCGTCCCGAGCTGGAACACGCTCTC | |
| pdcADF | TCCAGCTCGGGACGAGGCGATGCACGTCGC | |
| pdcADR-BglII | GAGAGCTCAGGTTACCCGCATGCAAGATCTGCTGCACACCATCGATCAGG | |
| pdcBUF-XbaI | AGTACGCGTCACTAGTGGGGCCCTTCTAGAGAACGAATGCGGAAGGGATT | To generate pDM4-phes-ΔpdcB |
| pdcBUR | CGAGCGCGTCTCTCCTGCAGGGCGTCACGT | |
| pdcBDF | CTGCAGGAGAGACGCGCTCGATTTGTTGCT | |
| pdcBDR-BglII | GAGAGCTCAGGTTACCCGCATGCAAGATCTATCGTCTGCCTGATCTTCTC | |
| pdcCUF-XbaI | AGTACGCGTCACTAGTGGGGCCCTTCTAGACGCACCGCGGCGTCGTCGAG | To generate pDM4-phes-ΔpdcC |
| pdcCUR | TCTCAGACATGGCTGTCGTAGCGGTAAACG | |
| pdcCDF | TACGACAGCCATGTCTGAGACGGTGTTGAC | |
| pdcCDR-BglII | GAGAGCTCAGGTTACCCGCATGCAAGATCTGATCGAAGCGGAACAGATAC | |
| pdcABUF-XbaI | AGTACGCGTCACTAGTGGGGCCCTTCTAGAGATTCGATTTCGCGCTAATC | To generate pDM4-phes-ΔpdcAΔpdcB |
| pdcABUR | CGACCGAGCATTGCTCCGCCGTCAACACCG | |
| pdcABDF | GGCGGAGCAATGCTCGGTCGCGTTCGGCTG | |
| pdcABDR-BglII | GAGAGCTCAGGTTACCCGCATGCAAGATCTACGGCATCAACGCCCGCTTC | |
| CT1F | CGTTCGTGATCGTCGTGTCAGC | For the cotranscription assay |
| CT1R | GCCGAATTCGTCATCCGCGTAC | |
| CT2F | CGATTTGTTGCTGTCCAGCT | |
| CT2R | TATTCCTGCAGTTTCGCCAC | |
| CT3F | AAGCCCGTCACCTCGGAAGCAT | |
| CT3R | GCACGAAAATGCCGAAGCTGAC | |
| CTNF | AACGCCACCGGAAAGCCTTG | |
| CTNR | CACTCACGGCCCAGCATCCA | |
| pdcA-25-F-BamHI | CGCGGATCCATGAACCTGCTTTCCTCGCT | To generate pKT25M-pdcA |
| pdcA-25-R-SalI | ACGCGTCGACTCAGCCGAACGCGACCGAG | |
| pas-25-F-BamHI | CGCGGATCCATGAACCTGCTTTCCTCGCT | To generate pKT25M-pas |
| pas-25-R-SalI | ACGCGTCGACTCACAGTTTCGCCACGGC | |
| ggdef-25-F-BamHI | CGCGGATCCATGCAGGAATACGCGAACC | To generate pKT25M-ggdef |
| ggdef-25-R-SalI | ACGCGTCGACTCAGCCGAACGCGACC | |
| pdcB-25-F-BamHI | CGCGGATCCATGTCTGAGACGGTGTTGACGG | To generate pKT25M-pdcB |
| pdcB -25-R-XhoI | CCGCTCGAGTCATAAGCTGGACAGCAACAAA | |
| pdcB -18-F-BamHI | CGCGGATCCATGTCTGAGACGGTGTTGACGG | To generate pUT18CM-pdcB |
| pdcB -18-R-XhoI | CCGCTCGAGTCATAAGCTGGACAGCAACAAA | |
| pdcC-18-F-BamHI | CGCGGATCCATGCCCCTGCCGATTGTGATC | To generate pUT18CM-pdcC |
| pdcC -18-R-SalI | CCGCTCGAGTCAGACATACAACCCATACTC | |
| bipD-QF | CGATGGGAAACCGAGCACC | qRT-PCR |
| bipD-QR | TCTTTCGCCGAGATGTAGTCC | |
| 16S RNA-F | AACCTTACCTACCCTTGA | |
| 16S RNA-R | GCTCGTTGCGGGACTTA | |
| pdcA-QF | TCTGTCGGTGCTGCTGTTCG | |
| pdcA-QR | GATCGTCTGCCTGATCTTCTCG | |
| pdcB-QF | ATGGCGAACGTGCTGATGG | |
| pdcB-QR | ATGTGGCGAATCGAATCTTCC | |
| pdcC-QF | GCAAGCTGCTGACGAAGGC | |
| pdcC-QR | GCTGACACGACGATCACGAAC |
Underlined sites indicate overlap fragments added for seamless cloning. Letters in boldface denote the restriction enzyme cutting sites.
In-frame deletion and complementation in B. thailandensis.
To construct in-frame deletion mutants, the suicide plasmid pDM4-phes derivatives were transformed into relevant B. thailandensis strains through E. coli SM10(λpir)-mediated conjugation, and the transconjugants were selected by plating on LB agar plates supplemented with chloramphenicol (50 μg/mL) and streptomycin (100 μg/mL). The in-frame deletion mutants were subsequently screened on M9 minimal medium (6 g/liter Na2HPO4, 3 g/liter KH2PO4, 0.5 g/liter NaCl, 1 g/liter NH4Cl, 1 mM MgSO4, 0.1 mM CaCl2, 0.2% glucose) agar plates with 0.1% (wt/vol) p-chlorophenylalanine. All the mutants were confirmed by PCR and DNA sequencing. For complementation, the pBBR1MCS1 derivatives were transformed into relevant B. thailandensis strains by electroporation as previously described (53).
Protein expression and purification.
GST- and His6-tagged recombinant proteins were purified as previously described (54). The expression plasmid pGEX6p-1 and pET28a or pET21a derivatives were transformed into XL1-Blue and E. coli BL21(DE3) competent cells, respectively. For protein production, a single colony was cultured in 5 mL LB at 37°C overnight and diluted 100-fold into 500 mL LB. The culture was shifted to 22°C when the optical density at 600 nm (OD600) was 0.5 and induced with 0.3 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) and then further cultivated for 12 h to produce the recombinant proteins. Harvested cells were washed and resuspended in His or GST binding buffer and lysed by sonication. The GST- and His6-tagged proteins were purified with the GST·Bind resin and His·Bind nickel-nitrilotriacetic acid resin (Novagen), respectively, according to the manufacturer’s instructions. Eluted recombinant proteins were dialyzed against the appropriate buffer at 4°C and stored at −80°C until used.
Motility assay and biofilm formation assays.
Swimming motility assay was carried out on semisolid agar medium (1% tryptone, 0.5% NaCl, 0.3% Difco Bacto agar) (55). Briefly, B. thailandensis strains were grown overnight at 37°C in LB medium and then diluted (1:10) into fresh LB medium. A volume of 2.5 μL of bacterial suspension was injected into the center of the semisolid agar medium and incubated at 30°C for 24 h before observation. Biofilm formation assay was measured in glass tubes as previously described (54). B. thailandensis strains were grown in LB broth and then diluted (1:100) into 3 mL of M63 medium containing 0.4% glucose on a rotary shaker (140 rpm) at 30°C. After 48 h, the glass tube was washed gently two times with sterile water, stained with 0.1% (wt/vol) crystal violet for 15 min, and then washed two times again with sterile water to remove redundant crystal violet. The remaining crystal violet was solubilized in ethanol and measured at an absorbance of 590 nm by a microplate reader (BioTek Instruments, Inc.). Assays were repeated at least three times independently.
RNA extraction and reverse transcription-PCR.
cDNA synthesized from B. thailandensis total RNA was used as a template to perform the cotranscription assay. B. thailandensis was grown to midexponential phase, and total RNA was purified by the total RNA isolation kit (Tiangen) according to the manufacturer’s protocol. The purity and concentration of sample RNA were determined using gel electrophoresis and a spectrophotometer (NanoDrop, Thermo Scientific). The cDNA was synthesized from the RNA template by using cDNA Synthesis SuperMix (TransGen Biotech). For the cotranscription assay, the PCR fragment was further amplified with specific primers listed in Table 2.
qRT-PCR.
Quantitative real-time PCR (qRT-PCR) was used to quantify gene expression of bipD. B. thailandensis strains were harvested and washed with PBS during the mid-exponential phase, and total RNA was extracted using the RNAprep pure cell/bacterial kit and treated with RNase-free DNase (TIANGEN, Beijing, China). The purity and concentration of sample RNA were determined using gel electrophoresis and a spectrophotometer (NanoDrop; Thermo Scientific). A volume of 2 μg of total RNA was reverse transcribed to synthesize the first-strand cDNA using the TransScript first-strand cDNA Synthesis SuperMix (TransGen Biotech). qRT-PCR was performed in a CFX96 real-time PCR detection system (Bio-Rad, USA) with TransStart green qPCR SuperMix (TransGen Biotech, Beijing, China). The qRT-PCR primers are listed in Table 2, and the qRT-PCR parameters were 95°C for 30 s followed by 40 cycles of 94°C for 15 s and 54°C for 30 s. The relative abundance of 16S rRNA was used as the internal standard for standardization of results.
Inorganic phosphate detection assays.
The inorganic phosphate released from PdcC was detected by using a Phosphate Sensor assay kit (Beyotime Biotech) according to the manufacturer’s instructions; 5 μM PdcB was added to the reaction buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 5 mM MgCl2) with or without 25 μM PdcC. Reaction solutions were incubated for 20 min at 30°C, and 20-μL samples were used to detect inorganic phosphate. Fluorescence signals were measured using a SpectraMax M2 plate reader (Molecular Devices) with excitation/emission wavelengths of 430/450 nm. The results shown represent the means from one representative assay performed in triplicate, and error bars represent the standard deviations (SD). Statistical analysis was carried out with one-way analysis of variance (ANOVA), followed by Tukey multiple-comparison test (GraphPad Software).
Bacterial two-hybrid assays.
Bacterial two-hybrid assays were carried out as previously described (56). Briefly, the bait and prey plasmids were cotransformed into E. coli BTH101 competent cells and selected by LB plates containing ampicillin and kanamycin. The colonies were spotted on MacConkey plates (IPTG plus maltose) and cultured at 30°C. The strength of interaction was quantified by measurement of β-galactosidase activity as described by Miller (57). All experiments were performed at least three times independently.
GST pulldown assay.
The GST pulldown assay was carried out as previously described (58). To test the protein interactions with purified proteins, purified GST-PdcA, GST-PAS, GST-GGDEF, and GST-PdcB were mixed with PdcC-His6 in PBS buffer on a rotator for 1 h at 4°C, and GST was adopted as a negative control. After 1 h of incubation, 50 μL prewashed glutathione beads was added to the reaction mix and incubated for another 1 h. The beads then were washed five times with PBS buffer, and the associated proteins were resolved with SDS loading buffer, separated by 15% SDS-PAGE, and detected by Western blotting.
Protein secretion assay.
Secretion of BipD was detected according to a previous report (47). Briefly, B. thailandensis strains were grown in 200 mL LB and incubated with continuous shaking until the OD600 reached 1.0 at 30°C. A 2-mL culture was centrifuged and the cell pellet was resuspended in 100 μL SDS-loading buffer to serve as the whole-cell lysate sample. A 180-mL bacterial culture was centrifuged at 8,000 rpm to separate the bacterial cells and culture supernatants. The culture supernatant was further filtered through a 0.22-μm filter (Millipore) to remove residual bacterial cells and then filtered three times through a nitrocellulose filter (BA85) (Whatman) to collect secreted proteins. The filter was cut into pieces in 1.5-mL tubes and resuspended in 100 μL SDS loading buffer for 15 min at 65°C to recover the proteins. All samples were normalized to the OD600 of the culture and volume used in the preparation, separated by 15% SDS-PAGE, and detected by Western blotting using specific antibodies. Isocitrate dehydrogenase (ICDH) was used as a loading control.
Western blot analysis.
Western blot analysis was performed as previously reported (59). Samples were resuspended by SDS loading buffer, separated by 15% SDS-PAGE, and then transferred onto polyvinylidene difluoride membranes (Millipore). The membrane was blocked by 5% bovine serum albumin at room temperature for 4 h and then incubated with primary antibodies overnight at 4°C. The primary antibodies were anti-His, 1:1,000; anti-GST, 1:1,000; anti-BipD rabbit polyclonal antibody, 1:1,000; anti-ICDH, 1:6,000. The ICDH antisera were made in our previous study (60). The membrane was washed five times with TBST buffer (50 mM Tris, 150 mM NaCl, 0.05% Tween 20, pH 7.4) and incubated with a 1:5,000 dilution of horseradish peroxidase-conjugated secondary antibody (Shanghai Genomics) for 4 h at 4°C. Signals were detected using the ECL plus kit (GE Healthcare, Piscataway, NJ, United States) by following the manufacturer’s specified protocol.
In vitro DGC activity assays.
The DGC activity of PdcA was measured as previously described (61). Briefly, 5 μM PdcA was added into reaction buffer (50 mM Tris–HCl, pH 7.5, 150 mM NaCl, and 5 mM MgCl2) with or without 25 μM PdcC at 30°C for 30 min. After incubating, 200 μM GTP was added into the buffer to initiate the reaction. At 0, 30, and 60 min, 50-μL samples were heated at 100°C for 5 min and centrifuged for 10 min at 12,000 rpm to remove protein pellets. To evaluate the effect of phosphorylation of PdcC on PdcA DGC activity, PdcC was pretreated by 25 mM acetyl phosphate for 30 min at 30°C. Samples were separated by an HPLC (Agilent 1260 Infinity II) system equipped with a C18 reverse-phase column and a UV detector. The c-di-GMP of samples was eluted with 98% phase A (150 mM Na2HPO4, pH 5.2) and 2% phase B (acetonitrile) in 15 min, and the flow rate was 1 mL/min. The wavelength of the UV detector was 254 nm. GTP (number G8877; Sigma) and c-di-GMP (number SML1228; Sigma) were purchased from Sigma and used as standards. The concentration of produced c-di-GMP was determined from a standard curve established with a serially diluted c-di-GMP solution.
Quantification of intracellular c-di-GMP levels.
The intracellular concentration of c-di-GMP was measured as previously reported (36). B. thailandensis strain cultures were prepared in M63 medium at 30°C in a shaker (220 rpm) until the late exponential growth phase for extracting c-di-GMP. A volume of 20 mL of bacterial culture was collected, washed, and resuspended in 100 μL prechilled double-distilled hydrogen peroxide and incubated at 100°C for 10 min. After incubation, we added 186 μL prechilled 100% ethanol to a final concentration of 65% and vortexed it for 20 s. The sample was centrifuged for 10 min at 14,000 rpm to remove the cell pellet. The cell pellet was extracted twice without heat. The supernatants extracted were collected and dried by speed-vacuum centrifugation, and the protein concentration of cell pellets was measured through bicinchoninic acid protein assay for sample normalization. The dry residue was dissolved in 100 μL HPLC-grade water and analyzed by LC-MS/MS on an AB SCIEX Triple Quad 6500+ LC-MS/MS system (AB SCIEX, USA). A Synergi Hydro-RP 80A LC column (4 μm, 150 by 2 mm; Phenomenex, Torrance, CA, USA) was used to separate 10-μL samples with phase A (0.1% acetic acid in 10 mM ammonium acetate) and phase B (0.1% formic acid in methanol), and the flow rate was 0.3 mL/min. The gradient was 0 to 4 min, 98% phase A, 2% phase B; 10 to 15 min, 5% phase A, 95% phase B. c-di-GMP (SML1228; Sigma) was used as a standard.
G. mellonella infection model.
The G. mellonella infection model was used to assess the virulence of B. thailandensis mutants as previously described (34). G. mellonella larvae were purchased from Livefood JiaYing Ltd. (TianJin). B. thailandensis strains were cultured in LB at 37°C. Until an OD600 of 1.2, bacteria were centrifuged at 4,500 rpm for 10 min, washed, and resuspended in PBS buffer to a final concentration of 105 bacteria for G. mellonella infection model; 20 larvae were injected with 50 μL of 105 CFU by using a Hamilton H syringe, and 50 μL PBS was injected as a control. After injection, G. mellonella larvae were incubated statically at 37°C for 18 h before determining survival rates. Each experiment was performed in triplicate.
Statistical analysis.
Statistical analyses of biofilm formation assay, swimming motility assay, c-di-GMP content determination, β-galactosidase activity, and gene expression data were performed using one-way ANOVA, followed by Tukey multiple-comparison test (GraphPad Software). Statistical analyses of the virulence assay were analyzed using two sided Mann–Whitney test (GraphPad Software).
ACKNOWLEDGMENTS
This work was supported by grants from the National Key R&D Program of China (2018YFA0901200 to X.S. and L.Z.) and the National Natural Science Foundation of China (32170048 to L.Z. and 31725003 to X.S.).
We thank Feng Shao (National Institute of Biological Sciences, Beijing) for kindly providing strains and plasmids and Ying Fu (Public Technology Service Center, Institute of Microbiology, Chinese Academy of Sciences) for c-di-GMP detection by LC-MS/MS. We also thank the Teaching and Research Core Facility at College of Life Sciences, NWAFU, for their technical support.
We declare no conflict of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
Xihui Shen, Email: xihuishen@nwsuaf.edu.cn.
Lei Zhang, Email: zhanglei0075@nwsuaf.edu.cn.
Gladys Alexandre, University of Tennessee at Knoxville.
REFERENCES
- 1.Hengge R. 2009. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol 7:263–273. 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- 2.D'Argenio DA, Miller SI. 2004. Cyclic di-GMP as a bacterial second messenger. Microbiology (Reading) 150:2497–2502. 10.1099/mic.0.27099-0. [DOI] [PubMed] [Google Scholar]
- 3.Romling U, Galperin MY, Gomelsky M. 2013. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev 77:1–52. 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jenal U, Reinders A, Lori C. 2017. Cyclic di-GMP: second messenger extraordinaire. Nat Rev Microbiol 15:271–284. 10.1038/nrmicro.2016.190. [DOI] [PubMed] [Google Scholar]
- 5.Floyd KA, Lee CK, Xian W, Nametalla M, Valentine A, Crair B, Zhu S, Hughes HQ, Chlebek JL, Wu DC, Hwan Park J, Farhat AM, Lomba CJ, Ellison CK, Brun YV, Campos-Gomez J, Dalia AB, Liu J, Biais N, Wong GCL, Yildiz FH. 2020. c-di-GMP modulates type IV MSHA pilus retraction and surface attachment in Vibrio cholerae. Nat Commun 11:1549. 10.1038/s41467-020-15331-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng Y, Sambou T, Bogomolnaya LM, Cirillo JD, McClelland M, Andrews-Polymenis H. 2013. The EAL domain containing protein STM2215 (rtn) is needed during Salmonella infection and has cyclic di-GMP phosphodiesterase activity. Mol Microbiol 89:403–419. 10.1111/mmi.12284. [DOI] [PubMed] [Google Scholar]
- 7.Moscoso JA, Mikkelsen H, Heeb S, Williams P, Filloux A. 2011. The Pseudomonas aeruginosa sensor RetS switches type III and type VI secretion via c-di-GMP signalling. Environ Microbiol 13:3128–3138. 10.1111/j.1462-2920.2011.02595.x. [DOI] [PubMed] [Google Scholar]
- 8.Lee HS, Gu F, Ching SM, Lam Y, Chua KL. 2010. CdpA is a Burkholderia pseudomallei cyclic di-GMP phosphodiesterase involved in autoaggregation, flagellum synthesis, motility, biofilm formation, cell invasion, and cytotoxicity. Infect Immun 78:1832–1840. 10.1128/IAI.00446-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hickman JW, Harwood CS. 2008. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol Microbiol 69:376–389. 10.1111/j.1365-2958.2008.06281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morgan JL, McNamara JT, Zimmer J. 2014. Mechanism of activation of bacterial cellulose synthase by cyclic di-GMP. Nat Struct Mol Biol 21:489–496. 10.1038/nsmb.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan XF, Xin L, Yen JT, Zeng Y, Jin S, Cheang QW, Fong R, Chiam KH, Liang ZX, Gao YG. 2018. Structural analyses unravel the molecular mechanism of cyclic di-GMP regulation of bacterial chemotaxis via a PilZ adaptor protein. J Biol Chem 293:100–111. 10.1074/jbc.M117.815704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bordeleau E, Purcell EB, Lafontaine DA, Fortier LC, Tamayo R, Burrus V. 2015. Cyclic di-GMP riboswitch-regulated type IV pili contribute to aggregation of Clostridium difficile. J Bacteriol 197:819–832. 10.1128/JB.02340-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ryan RP, Fouhy Y, Lucey JF, Dow JM. 2006. Cyclic di-GMP signaling in bacteria: recent advances and new puzzles. J Bacteriol 188:8327–8334. 10.1128/JB.01079-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Valentini M, Filloux A. 2016. Biofilms and cyclic di-GMP (c-di-GMP) signaling: lessons from Pseudomonas aeruginosa and other bacteria. J Biol Chem 291:12547–12555. 10.1074/jbc.R115.711507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moglich A, Ayers RA, Moffat K. 2009. Structure and signaling mechanism of Per-ARNT-Sim domains. Structure 17:1282–1294. 10.1016/j.str.2009.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Srivastava D, Waters CM. 2012. A tangled web: regulatory connections between quorum sensing and cyclic Di-GMP. J Bacteriol 194:4485–4493. 10.1128/JB.00379-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hickman JW, Tifrea DF, Harwood CS. 2005. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc Natl Acad Sci USA 102:14422–14427. 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valentini M, Filloux A. 2019. Multiple roles of c-di-GMP signaling in bacterial pathogenesis. Annu Rev Microbiol 73:387–406. 10.1146/annurev-micro-020518-115555. [DOI] [PubMed] [Google Scholar]
- 19.Hengge R. 2021. High-specificity local and global c-di-GMP signaling. Trends Microbiol 29:993–1003. 10.1016/j.tim.2021.02.003. [DOI] [PubMed] [Google Scholar]
- 20.Eberl L, Vandamme P. 2016. Members of the genus Burkholderia: good and bad guys. F1000Res 5:1007. 10.12688/f1000research.8221.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coenye T, Vandamme P. 2003. Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ Microbiol 5:719–729. 10.1046/j.1462-2920.2003.00471.x. [DOI] [PubMed] [Google Scholar]
- 22.Stone JK, DeShazer D, Brett PJ, Burtnick MN. 2014. Melioidosis: molecular aspects of pathogenesis. Expert Rev Anti Infect Ther 12:1487–1499. 10.1586/14787210.2014.970634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wiersinga WJ, Virk HS, Torres AG, Currie BJ, Peacock SJ, Dance DAB, Limmathurotsakul D. 2018. Melioidosis. Nat Rev Dis Primers 4:17107. 10.1038/nrdp.2017.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia EC. 2017. Burkholderia thailandensis: genetic manipulation. Curr Protoc Microbiol 45:4C.2.1–4C.2.15. 10.1002/cpmc.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Plumley BA, Martin KH, Borlee GI, Marlenee NL, Burtnick MN, Brett PJ, AuCoin DP, Bowen RA, Schweizer HP, Borlee BR. 2017. Thermoregulation of biofilm formation in Burkholderia pseudomallei is disrupted by mutation ofa putative diguanylate cyclase. J Bacteriol 199:e00780-16. 10.1128/JB.00780-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richter AM, Fazli M, Schmid N, Shilling R, Suppiger A, Givskov M, Eberl L, Tolker-Nielsen T. 2018. Key players and individualists of cyclic-di-GMP signaling in Burkholderia cenocepacia. Front Microbiol 9:3286. 10.3389/fmicb.2018.03286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar B, Sorensen JL, Cardona ST. 2018. A c-di-GMP-modulating protein regulates swimming motility of Burkholderia cenocepacia in response to arginine and glutamate. Front Cell Infect Microbiol 8:56. 10.3389/fcimb.2018.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang C, Cui C, Ye Q, Kan J, Fu S, Song S, Huang Y, He F, Zhang LH, Jia Y, Gao YG, Harwood CS, Deng Y. 2017. Burkholderia cenocepacia integrates cis-2-dodecenoic acid and cyclic dimeric guanosine monophosphate signals to control virulence. Proc Natl Acad Sci USA 114:13006–13011. 10.1073/pnas.1709048114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deng Y, Lim A, Wang J, Zhou T, Chen S, Lee J, Dong YH, Zhang LH. 2013. Cis-2-dodecenoic acid quorum sensing system modulates N-acyl homoserine lactone production through RpfR and cyclic di-GMP turnover in Burkholderia cenocepacia. BMC Microbiol 13:148. 10.1186/1471-2180-13-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deng Y, Schmid N, Wang C, Wang J, Pessi G, Wu D, Lee J, Aguilar C, Ahrens CH, Chang C, Song H, Eberl L, Zhang LH. 2012. Cis-2-dodecenoic acid receptor RpfR links quorum-sensing signal perception with regulation of virulence through cyclic dimeric guanosine monophosphate turnover. Proc Natl Acad Sci USA 109:15479–15484. 10.1073/pnas.1205037109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mhatre E, Snyder DJ, Sileo E, Turner CB, Buskirk SW, Fernandez NL, Neiditch MB, Waters CM, Cooper VS. 2020. One gene, multiple ecological strategies: a biofilm regulator is a capacitor for sustainable diversity. Proc Natl Acad Sci USA 117:21647–21657. 10.1073/pnas.2008540117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mangalea MR, Plumley BA, Borlee BR. 2017. Nitrate sensing and metabolism inhibit biofilm formation in the opportunistic pathogen Burkholderia pseudomallei by reducing the intracellular concentration of c-di-GMP. Front Microbiol 8:1353. 10.3389/fmicb.2017.01353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580. 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 34.Si M, Zhao C, Burkinshaw B, Zhang B, Wei D, Wang Y, Dong TG, Shen X. 2017. Manganese scavenging and oxidative stress response mediated by type VI secretion system in Burkholderia thailandensis. Proc Natl Acad Sci USA 114:E2233–E2242. 10.1073/pnas.1614902114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu L, Li QZ, Jin W, Lv H, Lin H. 2019. Revealing gene function and transcription relationship by reconstructing gene-level chromatin interaction. Comput Struct Biotechnol J 17:195–205. 10.1016/j.csbj.2019.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen Y, Yuan M, Mohanty A, Yam JK, Liu Y, Chua SL, Nielsen TE, Tolker-Nielsen T, Givskov M, Cao B, Yang L. 2015. Multiple diguanylate cyclase-coordinated regulation of pyoverdine synthesis in Pseudomonas aeruginosa. Environ Microbiol Rep 7:498–507. 10.1111/1758-2229.12278. [DOI] [PubMed] [Google Scholar]
- 37.Gao R, Bouillet S, Stock AM. 2019. Structural basis of response regulator function. Annu Rev Microbiol 73:175–197. 10.1146/annurev-micro-020518-115931. [DOI] [PubMed] [Google Scholar]
- 38.Welch M, Oosawa K, Aizawa S, Eisenbach M. 1993. Phosphorylation-dependent binding of a signal molecule to the flagellar switch of bacteria. Proc Natl Acad Sci USA 90:8787–8791. 10.1073/pnas.90.19.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paul R, Jaeger T, Abel S, Wiederkehr I, Folcher M, Biondi EG, Laub MT, Jenal U. 2008. Allosteric regulation of histidine kinases by their cognate response regulator determines cell fate. Cell 133:452–461. 10.1016/j.cell.2008.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCleary WR, Stock JB. 1994. Acetyl phosphate and the activation of two-component response regulators. J Biol Chem 269:31567–31572. 10.1016/S0021-9258(18)31731-9. [DOI] [PubMed] [Google Scholar]
- 41.Stuffle EC, Johnson MS, Watts KJ. 2021. PAS domains in bacterial signal transduction. Curr Opin Microbiol 61:8–15. 10.1016/j.mib.2021.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Upadhyay AA, Fleetwood AD, Adebali O, Finn RD, Zhulin IB. 2016. Cache domains that are homologous to, but different from PAS domains comprise the largest superfamily of extracellular sensors in prokaryotes. PLoS Comput Biol 12:e1004862. 10.1371/journal.pcbi.1004862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hall CL, Lee VT. 2018. Cyclic-di-GMP regulation of virulence in bacterial pathogens. Wiley Interdiscip Rev RNA 9:10.1002/wrna.1454. 10.1002/wrna.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmid N, Suppiger A, Steiner E, Pessi G, Kaever V, Fazli M, Tolker-Nielsen T, Jenal U, Eberl L. 2017. High intracellular c-di-GMP levels antagonize quorum sensing and virulence gene expression in Burkholderia cenocepacia H111. Microbiology (Reading) 163:754–764. 10.1099/mic.0.000452. [DOI] [PubMed] [Google Scholar]
- 45.Tamayo R, Pratt JT, Camilli A. 2007. Roles of cyclic diguanylate in the regulation of bacterial pathogenesis. Annu Rev Microbiol 61:131–148. 10.1146/annurev.micro.61.080706.093426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jung HI, Kim YJ, Lee YJ, Lee HS, Lee JK, Kim SK. 2017. Mutation of the cyclic di-GMP phosphodiesterase gene in Burkholderia lata SK875 attenuates virulence and enhances biofilm formation. J Microbiol 55:800–808. 10.1007/s12275-017-7374-7. [DOI] [PubMed] [Google Scholar]
- 47.Jitprasutwit S, Thaewpia W, Muangsombut V, Lulitanond A, Leelayuwat C, Lertmemongkolchai G, Korbsrisate S. 2010. Effect of acidic pH on the invasion efficiency and the type III secretion system of Burkholderia thailandensis. J Microbiol 48:526–532. 10.1007/s12275-010-0078-x. [DOI] [PubMed] [Google Scholar]
- 48.Galperin MY. 2006. Structural classification of bacterial response regulators: diversity of output domains and domain combinations. J Bacteriol 188:4169–4182. 10.1128/JB.01887-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lori C, Kaczmarczyk A, de Jong I, Jenal U. 2018. A single-domain response regulator functions as an integrating hub to coordinate general stress response and development in Alphaproteobacteria. mBio 9:e01534-18. 10.1128/mBio.01534-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sultan SZ, Pitzer JE, Miller MR, Motaleb MA. 2010. Analysis of a Borrelia burgdorferi phosphodiesterase demonstrates a role for cyclic-di-guanosine monophosphate in motility and virulence. Mol Microbiol 77:128–142. 10.1111/j.1365-2958.2010.07191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pfiffer V, Sarenko O, Possling A, Hengge R. 2019. Genetic dissection of Escherichia coli's master diguanylate cyclase DgcE: role of the N-terminal MASE1 domain and direct signal input from a GTPase partner system. PLoS Genet 15:e1008059. 10.1371/journal.pgen.1008059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Elliott KT, Dirita VJ. 2008. Characterization of CetA and CetB, a bipartite energy taxis system in Campylobacter jejuni. Mol Microbiol 69:1091–1103. 10.1111/j.1365-2958.2008.06357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Panta PR, Kumar S, Stafford CF, Billiot CE, Douglass MV, Herrera CM, Trent MS, Doerrler WT. 2019. A DedA family membrane protein is required for Burkholderia thailandensis colistin resistance. Front Microbiol 10:2532. 10.3389/fmicb.2019.02532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Z, Wang T, Cui R, Zhang Z, Chen K, Li M, Hua Y, Gu H, Xu L, Wang Y, Yang Y, Shen X. 2020. HpaR, the repressor of aromatic compound metabolism, positively regulates the expression of T6SS4 to resist oxidative stress in Yersinia pseudotuberculosis. Front Microbiol 11:705. 10.3389/fmicb.2020.00705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guan J, Xiao X, Xu S, Gao F, Wang J, Wang T, Song Y, Pan J, Shen X, Wang Y. 2015. Roles of RpoS in Yersinia pseudotuberculosis stress survival, motility, biofilm formation and type VI secretion system expression. J Microbiol 53:633–642. 10.1007/s12275-015-0099-6. [DOI] [PubMed] [Google Scholar]
- 56.Xu S, Peng Z, Cui B, Wang T, Song Y, Zhang L, Wei G, Wang Y, Shen X. 2014. FliS modulates FlgM activity by acting as a non-canonical chaperone to control late flagellar gene expression, motility and biofilm formation in Yersinia pseudotuberculosis. Environ Microbiol 16:1090–1104. 10.1111/1462-2920.12222. [DOI] [PubMed] [Google Scholar]
- 57.Miller JH. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press. [Google Scholar]
- 58.Si M, Wang Y, Zhang B, Zhao C, Kang Y, Bai H, Wei D, Zhu L, Zhang L, Dong TG, Shen X. 2017. The type VI secretion system engages a redox-regulated dual-functional heme transporter for zinc acquisition. Cell Rep 20:949–959. 10.1016/j.celrep.2017.06.081. [DOI] [PubMed] [Google Scholar]
- 59.Wang T, Si M, Song Y, Zhu W, Gao F, Wang Y, Zhang L, Zhang W, Wei G, Luo ZQ, Shen X. 2015. Type VI secretion system transports Zn2+ to combat multiple stresses and host immunity. PLoS Pathog 11:e1005020. 10.1371/journal.ppat.1005020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu L, Shen X, Bryan A, Banga S, Swanson MS, Luo ZQ. 2010. Inhibition of host vacuolar H+-ATPase activity by a Legionella pneumophila effector. PLoS Pathog 6:e1000822. 10.1371/journal.ppat.1000822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang L, Li S, Liu X, Wang Z, Jiang M, Wang R, Xie L, Liu Q, Xie X, Shang D, Li M, Wei Z, Wang Y, Fan C, Luo ZQ, Shen X. 2020. Sensing of autoinducer-2 by functionally distinct receptors in prokaryotes. Nat Commun 11:5371. 10.1038/s41467-020-19243-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Karimova G, Pidoux J, Ullmann A, Ladant D. 1998. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci USA 95:5752–5756. 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S4. Download aem.02529-21-s0001.pdf, PDF file, 0.5 MB (494.7KB, pdf)