INTRODUCTION
If electronic health records (EHRs)1 are ubiquitous in health care, then why are EHR data so difficult to access and use? Since their inception in the 1970s, EHRs have been envisioned to transform how health systems generate and use knowledge,2-6 including in oncology.7,8 Federal investments from the 2010 Health Information Technology for Economic and Clinical Health (HITECH) Act9-11 led to near-universal EHR adoption by 2017 in nonfederal acute care settings (96%) and high adoption in clinics (80%).12-14 In parallel, the learning health system (LHS) model was born, which seeks to use EHR and other data on patients and patient care processes to increase quality at reduced cost.15,16
CONTEXT
Key Objective
To provide a narrative review of challenges and opportunities with electronic health record (EHR) data, including interoperability and integration with other data sources, impacts on cancer research and cancer care, and promising directions for next-generation EHR technologies.
Knowledge Generated
Although federal EHR technology investments had disappointing results, they were necessary to start digitizing the nation's health care data and set the stage for a promising future. An architecture focused on application programming interfaces and data standards provides a flexible approach that is highly likely to accommodate precision oncology and enable a rapid learning health system.
Relevance
New EHR technologies promise to support all levels of the data-information-knowledge-wisdom pyramid through efficient capture of clinical and molecular data on individuals, visualization of longitudinal context of care (information), use of real-world data to enable new and rapid modes of cancer research (knowledge), and clinical decision support driven by near real-time analyses of patient populations (wisdom).
The reality of EHRs has been mixed. Early studies showed potential to optimize work processes; provide information gathering, summarization, reminders, and clinical decision support (CDS)17; improve quality; and lower costs.18,19 However, recent studies have shown that EHRs require provider workarounds that negatively affect ability to access or write information reliably, leading to burnout20-23 and inefficient and lower quality care delivery.24,25
The LHS model has limited support from current EHRs. It needs readily available population data, but most EHRs only support querying a patient at a time. Pooling EHR data for regional and national LHS analyses is arduous. Oncology's precision medicine transformation from hundreds of diagnoses to thousands of distinct cancer subtypes driven by molecular testing26-29 places unique burdens on EHRs,8 which were not designed for molecular data.30 In addition, precision oncology requires synergy between the clinic and academic research that is ill supported by current EHRs.
This narrative review articulates how interoperability and other EHR technology issues have held back the oncology LHS, and it describes promising directions involving industry and academia for improving data and software integration and interoperability.
EHR LIMITATIONS
Incumbent EHR vendors in the United States used 1990s client-server software architecture31,32 for their products. Although innovative compared with previous terminal-based systems, today, these EHRs often lack functionality found in consumer software, were not designed for the open data exchange needed in modern clinical practice, and put hospitals at the center of care not patients. In addition, their centralized information technology (IT) management model restricts provider customization and experimentation, which was commonplace in paper charts, and makes customization slow and expensive.33-35 High purchase prices, disruption to health care operations from switching EHRs, vendor reluctance to implement interoperability with competing products, and complex regulations that new market entrants must implement have entrenched these technologies.
Furthermore, IT spending by US health care institutions has lagged other nations and other industries with complex needs,18,36-39 limiting institutions' ability to augment vendor EHRs or even fully implement what vendors provide. Rigorous evaluation through implementation science40 is needed to guide EHR implementations, but such evaluations can cost nearly as much as implementing the technology itself,41 which is likely part of why they are rarely done.42 Outsourcing EHR-related IT, as some health systems have attempted,43 is puzzling because digital transformation cannot happen if executives view informatics as outside of their organization's core competencies. As a result, most EHR implementations have numerous clinical and research limitations.
Clinical Limitations of Current EHRs
HITECH defined basic and comprehensive EHRs. Basic EHRs, shown in Figure 1, essentially duplicate the paper chart in electronic form. Comprehensive EHRs add simple data interpretations like threshold checks on numerical observations without regard for context. A total white count of 100,000 in a patient with no known blood cancer, a patient with acute leukemia, and a chronic lymphocytic leukemia patient with a steady white count for half a decade have drastically different implications, yet EHRs will detect this finding as critical in all three patients. Oncologists need dynamic longitudinal views that contextualize findings.
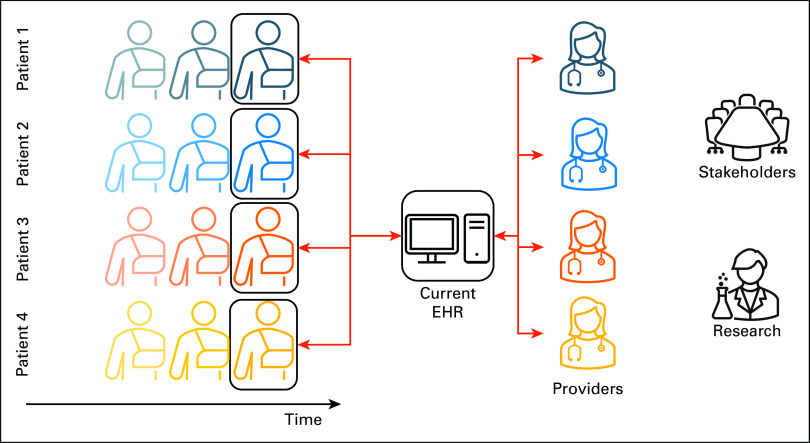
FIG 1.
Depiction of current EHRs in a LHS. Although current EHRs provide longitudinal patient data storage, they typically visualize only snapshots of the state of individual patients. In addition, they provide little support for research uses of the data or stakeholder decision making to improve care quality at lower cost. This figure depicts a snapshot view of four patients' medical records, accessed by their providers (red arrows), with no patient population view. EHR, electronic health record; LHS, learning health system.
Comprehensive EHRs also provide alerting44 that relies on clinical guideline development and manual curation of business rules that are triggered by patterns in laboratory test results, diagnosis codes, medications, and the like. This approach to authoring alerts cannot keep up with the pace of clinically actionable new knowledge.45 Furthermore, the accuracy of such rules is frequently low, leading many providers to override CDS alerts habitually.46
These visualizations and checks are limited partly because 80% of EHR data are in unstructured documents that EHRs cannot parse.47,48 A provider signing off on a note detailing a patient with shortness of breath because of end-of-life lung cancer will be asked by the EHR to duplicate the chief complaint in a structured field and order a prostate-specific antigen test. Cancer stage, histopathologic biomarkers, smoking status, clinical genetic testing, and other pertinent cancer data and provider interpretations of it are typically found in text notes that are unavailable to CDS.
Physician burnout has arisen from using these EHRs. Causes include information overload,49,50 increased complexity of cognitive work,51 excessive data entry driven by billing and administrative requirements, inefficient user interfaces, click fatigue,52 inadequate data exchange with other providers, and EHR-related workflows that interfere with the physician-patient relationship.22 Cancer is a complex set of diseases in which information overload in EHRs may be exacerbated.8 Patient safety measures like medication reconciliation, implemented through burdensome EHR workflows, have also contributed to burnout.19 The same problems result in information input and output errors and adversely affect care safety and efficiency.53,54 This has been noted in studies worldwide and for different provider roles and levels of training.55-57
Inadequate data exchange between providers is largely due to inadequate EHR data standardization. Most EHR implementations have site-specific codes that are ill documented. Where standards may be used, like Logical Observation Identifiers Names and Codes (LOINC) for clinical laboratory test results,58 codes may be applied inconsistently from one institution to another. Cancer-specific data are complex and would especially benefit from structured data representation and standardization. Disease classification standards used in EHRs, like the Systematic Nomenclature of Medicine—Clinical Terms (SNOMED-CT) used in EHR problem lists59 and structured pathology reporting,60 frequently classify cancer types by histology and location but not molecular testing although clinical genetic testing has become routine.61
Furthermore, data quality has plagued EHRs. Administrative coding systems like International Classification of Diseases version 10 have too many codes for providers and coders to document accurately without substantial computer support,62-64 and these codes document billing, not patient care. Problem lists, ostensibly a clinical representation of a patient's conditions, are frequently inaccurate, incomplete, and out-of-date because of burdensome data entry.65 EHRs were recognized early in their history to potentially cause the recording of a greater quantity of bad data compared with paper, leading to the first law of informatics in 1991 stating that data shall be used only for the purpose for which they were collected.66 In the decades since the first law, health care has found value in reusing EHR data, but the data's limitations are insufficiently understood.
Research Limitations of Current EHRs
EHR data are typically available for academic and operational research through a relational database called an Enterprise Data Warehouse (EDW),67-69 which may be refreshed daily to quarterly. Maintaining an EDW requires expertise that only larger health systems typically have, and even basic queries require substantial informatics support. EHR vendors increasingly provide self-service population query tools that are updated in near real time, but these tools often provide neither access to historical data before the EHR's implementation nor data stored in ancillary systems.
Data problems in EDWs affect the data's fitness for use in research.70,71 They include infrequent refreshes, left censoring (the first instance of disease may not be when the disease first manifested), right censoring (the data may not cover a long enough time interval), missing data from other clinical and nonclinical settings, institutional and personal variation in practice and documentation styles, and inconsistent use of medical codes.72 Misunderstanding these challenges has led to multiple retractions in major journals.73 Efforts to increase EDW data completeness and quality are frequently framed as a need for more structured data entry in EHRs, but research needs must be balanced with already-excessive provider documentation requirements.
Chart abstraction and harmonizing74 EDWs to standards are resource-intensive but increasingly necessary to participate in clinical research networks. Ill-documented local codes and mappings to standards require extensive informatics support to understand. Local codes tend to change without warning to the research community, resulting in corrupted data.15,75,76 Data located in text notes must be abstracted manually or parsed from the EHR's database using natural language processing (NLP).77
Other informatics challenges in EDWs include complicated data models that appear markedly different from how the same data appear in the EHR. Data visible in the EHR as scanned image files, such as clinical genetic test results, are usually unavailable in EDWs. Even when genetic testing is available, its use in research is often hampered by the absence of linkages to sequencing data. Extracting clinical concepts, exposures, and outcomes for studies and applying inclusion and exclusion criteria are challenging even for the most experienced data analysts.
Policies also hamper EDW use in research. Budget limitations and governance conflicts between the research and clinical sides of academic institutions tend to result in research needs getting low priority.78 When EHRs are outsourced as described above, the EHR vendor may charge for research access, thus increasing costs to researchers. In addition, although security and privacy concerns29,79 rightly necessitate carefully crafted access policies, many institutions simply restrict access. Safe harbor deidentification defined by the Health Insurance Portability and Accountability Act may alleviate privacy concerns but removes critical information such as seasonality.80 Furthermore, there is frequently an overlap between data capture forms used for patient care and research, but institutions may have separate clinical and research data capture policies, forcing investigators through a quagmire to get their work done and severely hampering integration between patient care and research needed to realize a LHS.81
THE LHS
The LHS encompasses infrastructure, governance, incentives, and shared values to harness data and analytics to learn from every patient, on the basis of real-world care process and outcomes data drawn primarily from EHRs.16,82 The LHS aims to feed this knowledge back to clinicians, informaticians, data scientists, public health professionals, patients, and other stakeholders to create cycles of continuous improvement.16,82,83 To realize this vision in oncology, EHRs must go beyond HITECH's comprehensive EHR requirements to improve patient care, better connect patients and their oncologists, and provide seamless data flows between clinical and research missions.7
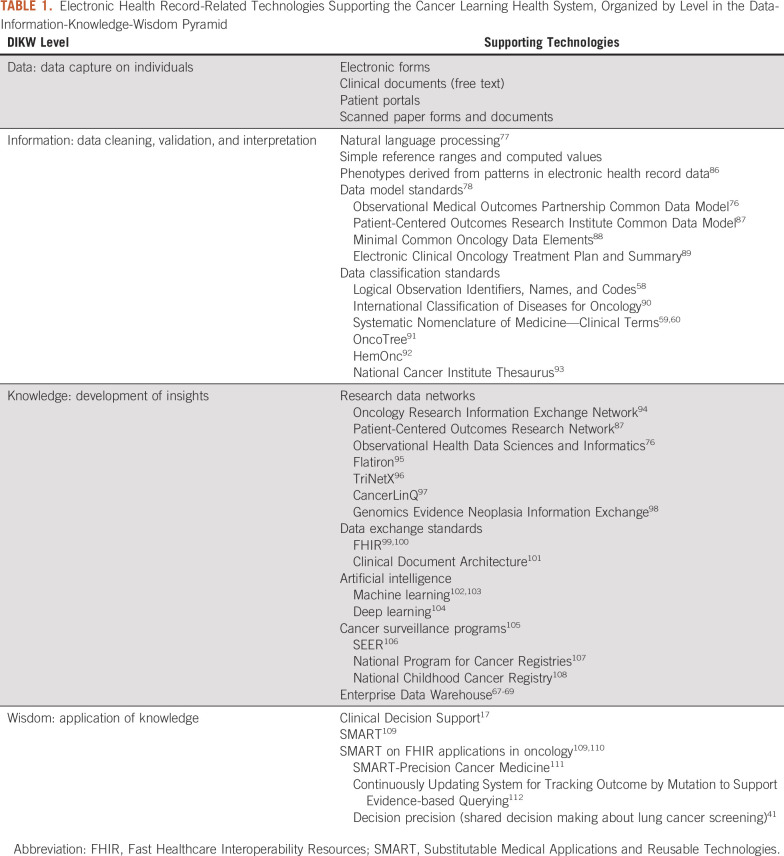
The data-information-knowledge-wisdom pyramid,84,85 shown in Figure 2, describes LHS data flows starting with capturing data on individuals (Fig 1); data cleaning, validation, and interpretation to form information, shown in Figure 3; developing insights that add to knowledge about similar patients, shown in Figure 4; and taking wise action, shown in Figure 5. EHR limitations create barriers at all levels of the pyramid, but with sufficient IT investments, opportunities exist to address them. Table 1 links topics in the research and clinical opportunities subsections below to each data-information-knowledge-wisdom level.
FIG 2.
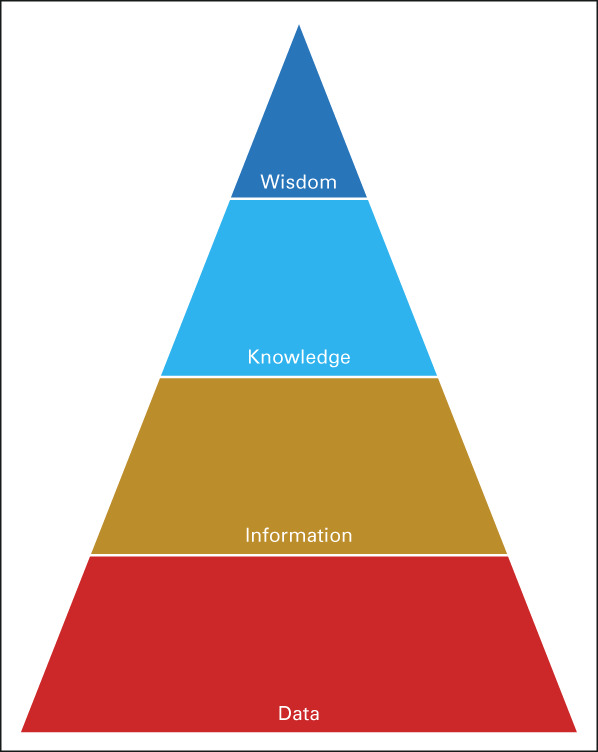
Illustration of the DIKW pyramid. It provides a framework for understanding how future EHRs need to better support abstraction of data into information, and inference of knowledge from information, to help providers and other stakeholders gain wisdom to realize the learning health system's overall goal of improving care quality at lower cost. DIKW, Data-Information-Knowledge-Wisdom; EHR, electronic health record; LHS, learning health system.
FIG 3.
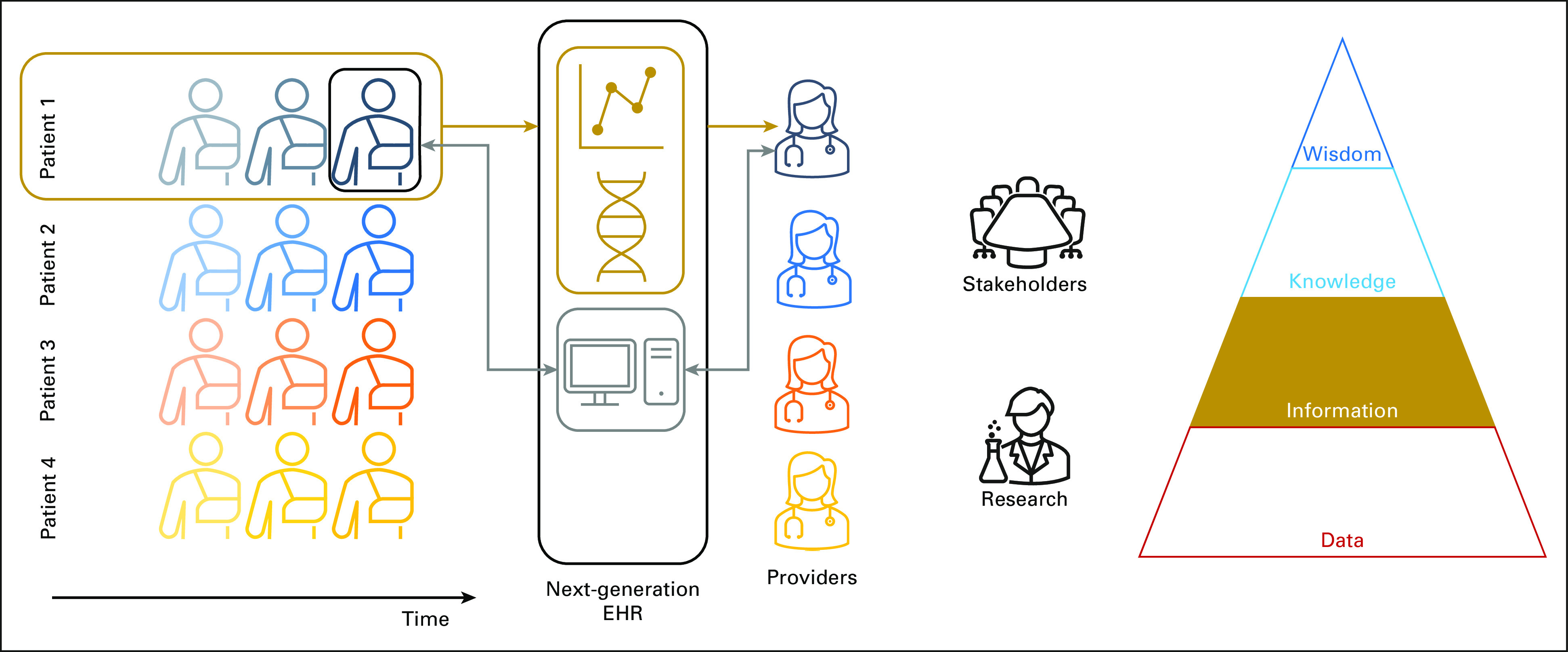
Longitudinal data abstraction on Patient 1's medical record (gold arrows) and interpretation of Patient 1's data to form information, the second level in the DIKW pyramid (Fig 2). Current EHRs typically display only snapshot views (thin gray arrows, Fig 1). With the ability to interpret longitudinal data, future EHRs could visualize a patient's cancer journey and provide basic support for correlating clinical features with genetic testing over time. DIKW, Data-Information-Knowledge-Wisdom; EHR, electronic health record.
FIG 4.
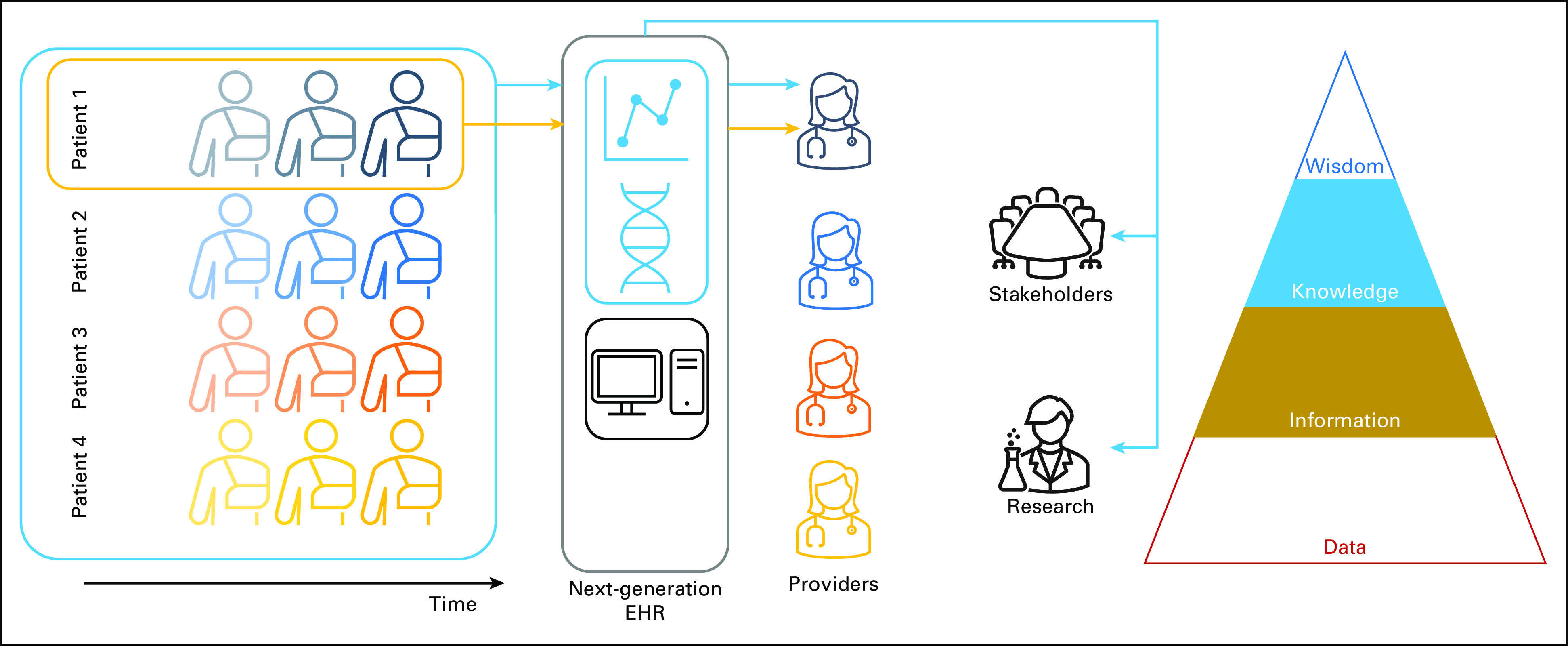
Future EHRs providing longitudinal individual information (Fig 3, gold arrows) and population information (light blue arrows) to better guide patient care and inform continuous quality improvement efforts. With such EHRs, providers, stakeholders, and researchers may more easily generate knowledge through insights gained from advanced visualizations and clinical decision support. Knowledge is the third level in the DIKW pyramid. DIKW, Data-Information-Knowledge-Wisdom; EHR, electronic health record.
FIG 5.
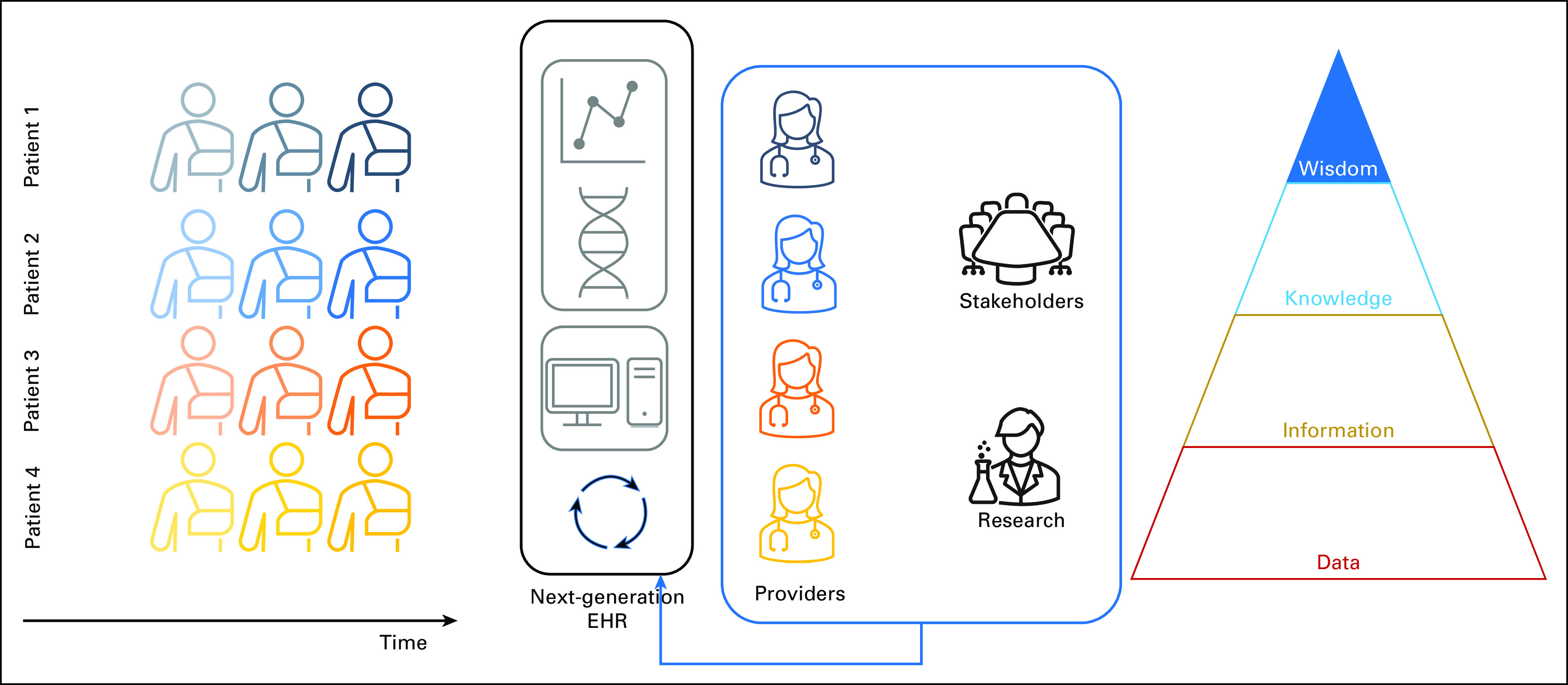
Implementation of new care processes in a LHS (blue arrow), in part through evolving the EHR (cyclic arrows) in response to knowledge about a patient population gained from advanced visualizations and clinical decision support (Fig 4). Transforming knowledge into action is the top level of the DIKW pyramid (Wisdom). DIKW, Data-Information-Knowledge-Wisdom; EHR, electronic health record; LHS, learning health system.
TABLE 1.
Electronic Health Record-Related Technologies Supporting the Cancer Learning Health System, Organized by Level in the Data-Information-Knowledge-Wisdom Pyramid
Clinical Opportunities
The Quality Oncology Practice Initiative113 took initial steps toward creating an oncology LHS by combining patient-reported surveys and manually abstracted EHR data from seven practice groups to study clinical practice variation. Improvements in compliance with practice benchmarks were highest when relevant information was in the EHR,114 but the heterogeneity of cancer cases limited the effectiveness of chart abstraction-based data capture.
A rapid LHS aims to replace manual chart abstraction with near-real-time data flow from EHRs. In this future vision, EHRs will automatically mine and render genotypic and phenotypic data86 together with the latest evidence for clinical decision making.27 Clinical outcomes will lead to iterative knowledge refinement and new research studies that may ultimately result in further practice changes.8,29,115 This approach may be of particular value for patients with late-stage cancer for whom treatment benefits are less clear.
Early rapid LHS adopters developed a proof-of-concept iPad-based CDS system called Substitutable Medical Applications and Reusable Technologies (SMART) Precision Cancer Medicine (PCM).111 SMART PCM supports providers and patients in collaboratively reviewing the patient's mutation status in the context of a continuously updated database of patients with similar mutations and linkages to other data and knowledge. CUSTOM-SEQ (Continuously Updating System for Tracking Outcome by Mutation to Support Evidence-based Querying) calculates and displays mutation-specific survival statistics.112 Decision precision is a lung cancer screening application for shared decision making between patients and providers about screening.41 These applications are tightly integrated into EHRs, and they rely on availability of structured clinical genetic test results, which have begun to be supported by major EHR vendors, for example, Epic's genomics module.116 Similar technologies may allow automated capture of patient-reported data in EHRs, for example, Apple HealthKit, thus reducing provider data entry and potentially increasing patient engagement.19,117,118
A prerequisite of this tight integration is shared data models between EHRs and apps.78 Early oncology data models include electronic Clinical Oncology Treatment Plan and Summary, which builds upon a health care data exchange standard from Health Level Seven International119 called Clinical Document Architecture.101 Its successor, Minimal Common Oncology Data Elements (mCODE), is built upon a Clinical Document Architecture replacement called FHIR (Fast Healthcare Interoperability Resources) that is new but is already used by SMART PCM, HealthKit, and other apps.88 As important as shared data models are, structured data are unlikely to replace all text notes as information sources for CDS. Thus, EHR vendors have begun introducing NLP-based document indexing that extracts clinical events and observations from text notes automatically.
Machine learning may further accelerate creating apps that reason with continuously updated knowledge inferred from EHRs.102,103 Potential applications include cohort identification120 and prediction of future events, prognosis, and outcomes.102 In addition, machine learning may simplify provider interactions with EHRs through speech recognition102,121 and reducing data entry through improved NLP.122 Unsolved challenges include burdensome provider time to label cases, trained algorithms' lack of generalizability from one disease condition to another, no way to run clinical trials of trained models in the clinical environment, and no way to incorporate validated models into EHRs. Newer machine learning methods like deep learning require less data preprocessing and training than prior techniques,104 but more work is needed. Integrated academic and clinical missions are required to support algorithm development, evaluation, and implementation.
Research Opportunities
Clinical practice knowledge inferred from EHRs may complement traditional scientific discovery, embodied by the clinical trial.123 Two to three percent of patients participate in trials.29 Although the participation rate in cancer trials may be somewhat higher,124 such low participation may result in under-representing older patients, racial and ethnic minorities, comorbidities, and low socioeconomic status, which are present in real-world EHR data.125,126 In addition, data collection in clinical trials is expensive, and trials are slow to yield results and affect patient care.97 Real-world evidence-based studies will not fully replace clinical trials, and careful attention to established guidelines in using such data is needed,73 but appropriate use of EHR data may yield results faster and at lower cost.
Potential studies include comparing anticancer drug combinations and their timing and identifying unexpected associations between clinical events, prognoses, and outcomes, for example, comorbidities associated with poorer chemotherapeutic response.115 Population-based disparities such as geographic differences in cancer outcomes could point toward epidemiologic and performance hypotheses. Such associations can support predictive modeling of individual prognosis and outcomes.
Furthermore, integrating EHR data with health department, social media, and other data could increase statistical power and increase the likelihood of new genetic variant associations and other discoveries.127,128 EHR data could boost cancer registries105 such as the SEER program106 and the National Program for Cancer Registries107 through enhanced data capture and linkage. Early examples include CancerLinQ, which harmonizes EHR and SEER data for quality improvement and research,97 and the National Childhood Cancer Registry,108 which links tumor registry data with EHR data from National Cancer Institute (NCI)–designated cancer centers.
Integrating regional and national data from many EHRs also shows promise. Examples that support oncology research include the Oncology Research Information Exchange Network,94 the Patient-Centered Outcomes Research Network,87 the Observational Health Data Sciences and Informatics (OHDSI) initiative,76 the All of Us Research Program,129 and the Genomics Evidence Neoplasia Information Exchange (GENIE) network.98 Oncology Research Information Exchange Network, All of Us, and GENIE capture genetic data and link them with clinical data. OHDSI provides an open-source repository of statistical analysis scripts that make correct use of multisite EHR data.130,131
Research networks use many of the same vocabularies as CDS, but more work is needed to expand and augment them. For example, like SNOMED-CT, the International Classification of Diseases for Oncology used by tumor registries90 classifies cancer types by histology and location but not molecular testing. OncoTree, used by GENIE, shows promise for additionally classifying cancers by genotype.91 The NCI Thesaurus aims to provide broad coverage of clinical and molecular concepts in oncology and relationships between them93 and is used by the National Childhood Cancer Registry for annotating data contributions. HemOnc aims to represent chemotherapeutic regimens at OHDSI sites,92 expanding upon SNOMED-CT and the NCI Thesaurus. Continuous development of vocabularies for both patient care and research is needed to move data through capture, interpretation, knowledge generation, and CDS (wisdom) steps of the LHS.
REIMAGINING THE EHR
The above EHR opportunities represent substantial changes and are unlikely to be realized through purely incremental improvement of existing EHRs. The American Medical Informatics Association EHR-2020 Task Force19 and the JASON advisory group report127 proposed similar visions for next-generation EHRs that advance the state-of-the-art by enabling new market entrants to invent advanced functionality that integrates seamlessly with hospital and clinic databases.
Their recommendations focus on reducing manual data entry through multiple approaches, including NLP47; alternative data entry modes like voice and handwriting recognition; patient portal and mobile device integration with EHRs; interoperability between EHRs and ancillary systems132; and broadening the settings in which EHRs operate to home health, pharmacy, population health, long-term care, and physical and behavioral therapy centers. With these changes, providers could refocus manual documentation on data elements that contribute to patient outcomes rather than billing and administration.
EHR data are among the most granular available, and sequencing data are even more so.128 Patient privacy concerns with health care big data29 legitimately increase as the volume of data held in EHRs increases, as do security concerns when a breach could affect so many patients. Such detailed data could be nearly impossible to deidentify because of unique data patterns that only one individual has.127 Next-generation EHRs will require advanced encryption to ensure security, privacy, and patients' trust in the clinical and research communities.
Additional recommendations include restoring the academic informatics community's role in EHR technology advancement, which was common in early EHR implementations but lost when academic centers adopted vendor systems. Many promising CDS and other methods from the informatics community remain unimplemented by vendors, and site-specific customization requests to vendors tend to receive low priority, creating suboptimal fit with local operational processes.
Both reports propose an open EHR architecture composed of third-party best-of-breed applications, available in an app store for health systems to purchase in different combinations. Health systems would also purchase a software foundation that provides apps with authentication, authorization, encryption, databases, protocols for reading and writing data, terminologies, and patient and encounter data models. The foundation would have application programming interfaces (APIs) for apps to access its capabilities, connect to other apps and comprise a cohesive system. Like how the Apple app store catalyzed the popularity of the iPhone on the basis of its iOS foundation, EHR APIs would allow a software marketplace to form. If all software foundation vendors shared the same APIs, new market entrants could write apps once for use anywhere with a standards-compliant foundation.
Early versions of this architecture extended current EHRs with sidecar133 software that connects to any EHR's database and presents standardized data views on the basis of a common data model (CDM),96 primarily in support of research. Sidecars include i2b2, using a flexible star schema134; OHDSI, using the Observational Medical Outcomes Partnership (OMOP) CDM135; Patient-Centered Outcomes Research Network, using the Patient-Centered Outcomes Research Institute CDM87; and TriNetX, using a proprietary CDM.96 Sidecars access data the same way as EDWs and require similar levels of provider resourcing. Data movement is typically one way from EHRs into the sidecar, limiting support for patient care. Services like CancerLinQ,97 Flatiron,95 and Health Catalyst Data Operating System136 moved the sidecar into the cloud but similarly rely on one-way EDW-style data movement from EHRs into the services.
The SMART project109 went a step further by creating a standardized API on top of these sidecars for building apps.137 SMART apps use FHIR for data exchange,99,100 and existing EHRs can eliminate the sidecar and implement the API directly. Cerner, Epic, Athenahealth, and others support SMART on FHIR to some degree, and more than 500 hospitals have begun using SMART on FHIR apps,110 including ours.41
The above SMART PCM application is an example of SMART on FHIR in oncology. Compass linked mCODE data elements in an EHR with data from similar patients extracted from CancerLinQ.138 The Integrating Clinical Trials and Real-World Endpoints app supported an Alliance for Clinical Trials in Oncology study using real-world data.138 Compass and Integrating Clinical Trials and Real-World Endpoints were pilot projects leading to the development of mCODE, which future SMART apps in oncology will likely rely upon. Epic's App Orchard effectively provides an app store in the health care space.
Over time, a new software foundation would replace current EHRs and EDWs. Functionality would include a fully encrypted database permitting selective data decryption according to fine-grained access granted by patients together with their providers. Flexible data storage would permit high-performance query, extraction, and visualization of patient and population data in a variety of clinical and research contexts. Existing apps would continue to function as before because the new foundation would implement extensions of existing APIs that add functionality without breaking current capabilities. Cost-effective means of formally evaluating these technologies are needed to ensure that they have the desired impact on patient care and research.
In summary, EHRs began as highly customized tools developed by academic medical centers.139 Initial systems were replaced by one-size-fits-all vendor EHRs that are deployed widely but have limited support for oncology. The informatics community has envisioned a new architecture that returns to permitting the innovation and customization required to realize the LHS. The adoption of SMART on FHIR by vendor EHRs will allow this new architecture to be studied and fine-tuned in the real world before ultimately replacing current EHRs with new technologies.
Challenges include scaling SMART to a vision of EHRs as a collection of applications that providers compose according to need. Apps are envisioned as independent and connected to a foundation. However, it is easy to imagine a CDS app needing to connect directly to a data visualization app because the foundation does not yet support the necessary integrations. Unless informatics architectures and governance support interoperability between apps in an organized fashion, chaos could ensue as many-to-many relationships form between apps.
Before one-size-fits-all EHRs, many institutions mixed paper charts with best-of-breed applications, typically for billing and pathology and radiology reports.4 Institutions migrated to legacy EHRs in part to simplify purchasing and obtain vendor support for integrations between EHR components.140 Perhaps vendors will emerge who package and support sets of interoperable SMART on FHIR apps for oncology and other subspecialties.
The SMART and FHIR standards need performant population query capability to realize the LHS. FHIR was recently extended with EDW-style bulk query,141 but EHR vendors have yet to implement it. EHRs have data gaps and performance bottlenecks when querying large data volumes using FHIR, and thus, the sidecar remains the only working solution for bulk queries. Interoperability between EHR data and high-value oncology data like SEER and National Program for Cancer Registries is also needed.
Despite these challenges, this new API-centric and interoperability-focused architecture is a flexible approach that is highly likely to accommodate the needs of precision oncology. Results from HITECH have been disappointing, but HITECH was a necessary step toward digitizing the nation's health care data and setting the stage for a promising future.
ACKNOWLEDGMENT
The research reported in this publication used the Research Informatics Shared Resource at Huntsman Cancer Institute at the University of Utah.
Zachary Burningham
Research Funding: AbbVie, Genentech/Roche, Pharmacyclics
Ahmad S. Halwani
Research Funding: Bristol Myers Squibb (Inst), Kyowa Hakko Kirin (Inst), Roche/Genentech (Inst), AbbVie/Genentech (Inst), AbbVie (Inst), Immune Design (Inst), miRagen (Inst), Amgen (Inst), Seattle Genetics (Inst), Genentech (Inst), Takeda (Inst), Pharmacyclics (Inst), Bayer (Inst)
Travel, Accommodations, Expenses: Pharmacyclics, AbbVie, Seattle Genetics, Immune Design
No other potential conflicts of interest were reported.
DISCLAIMER
The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
SUPPORT
Supported by the National Cancer Institute of the National Institutes of Health under Award No. P30CA042014.
AUTHOR CONTRIBUTIONS
Conception and design: All authors
Financial support: Andrew R. Post
Administrative support: Andrew R. Post
Provision of study material or patients: Andrew R. Post
Collection and assembly of data: All authors
Data analysis and interpretation: All authors
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/cci/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Zachary Burningham
Research Funding: AbbVie, Genentech/Roche, Pharmacyclics
Ahmad S. Halwani
Research Funding: Bristol Myers Squibb (Inst), Kyowa Hakko Kirin (Inst), Roche/Genentech (Inst), AbbVie/Genentech (Inst), AbbVie (Inst), Immune Design (Inst), miRagen (Inst), Amgen (Inst), Seattle Genetics (Inst), Genentech (Inst), Takeda (Inst), Pharmacyclics (Inst), Bayer (Inst)
Travel, Accommodations, Expenses: Pharmacyclics, AbbVie, Seattle Genetics, Immune Design
No other potential conflicts of interest were reported.
REFERENCES
- 1.Institute of Medicine (US) Committee on Data Standards for Patient Safety . Key Capabilities of an Electronic Health Record System: Letter Report. Washington, DC: National Academies Press (US); 2003. [PubMed] [Google Scholar]
- 2. Middleton B. Achieving the vision of EHR--take the long view. Front Health Serv Manage. 2005;22:37–42. discussion 43-45. [PubMed] [Google Scholar]
- 3. Wyatt J. Computer-based knowledge systems. Lancet. 1991;338:1431–1436. doi: 10.1016/0140-6736(91)92731-g. [DOI] [PubMed] [Google Scholar]
- 4.Institute of Medicine . Computer-Based Patient Record: An Essential Technology for Health Care. Washington, DC: The National Academies Press; 1991. [PubMed] [Google Scholar]
- 5. Powsner SM, Wyatt JC, Wright P. Opportunities for and challenges of computerisation. Lancet. 1998;352:1617–1622. doi: 10.1016/S0140-6736(98)08309-3. [DOI] [PubMed] [Google Scholar]
- 6. Evans RS. Electronic health records: Then, now, and in the future. Yearb Med Inform. 2016:S48–S61. doi: 10.15265/IYS-2016-s006. suppl 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Warner J, Hochberg E. Where is the EHR in oncology? J Natl Compr Canc Netw. 2012;10:584–588. doi: 10.6004/jnccn.2012.0060. [DOI] [PubMed] [Google Scholar]
- 8. Yu P, Artz D, Warner J. Electronic health records (EHRs): Supporting ASCO's vision of cancer care. Am Soc Clin Oncol Educ Book. 2014;34:225–231. doi: 10.14694/EdBook_AM.2014.34.225. [DOI] [PubMed] [Google Scholar]
- 9. Jha AK, DesRoches CM, Campbell EG, et al. Use of electronic health records in U.S. hospitals. N Engl J Med. 2009;360:1628–1638. doi: 10.1056/NEJMsa0900592. [DOI] [PubMed] [Google Scholar]
- 10. DesRoches CM, Campbell EG, Rao SR, et al. Electronic health records in ambulatory care—A national survey of physicians. N Engl J Med. 2008;359:50–60. doi: 10.1056/NEJMsa0802005. [DOI] [PubMed] [Google Scholar]
- 11. Blumenthal D. Launching HITECH. N Engl J Med. 2010;362:382–385. doi: 10.1056/NEJMp0912825. [DOI] [PubMed] [Google Scholar]
- 12.The Office of the National Coordinator for Health Information Technology (ONC) Office-Based Physician Health IT Adoption and Use. https://www.healthit.gov/data/datasets/office-based-physician-health-it-adoption-and-use [Google Scholar]
- 13.The Office of the National Coordinator for Health Information Technology (ONC) Non-federal Acute Care Hospital Electronic Health Record Adoption. https://www.healthit.gov/data/quickstats/non-federal-acute-care-hospital-electronic-health-record-adoption [Google Scholar]
- 14. Adler-Milstein J, Jha AK. HITECH act drove large gains in hospital electronic health record adoption. Health Aff (Millwood) 2017;36:1416–1422. doi: 10.1377/hlthaff.2016.1651. [DOI] [PubMed] [Google Scholar]
- 15. Friedman CP, Wong AK, Blumenthal D. Achieving a nationwide learning health system. Sci Transl Med. 2010;2:1–3. doi: 10.1126/scitranslmed.3001456. [DOI] [PubMed] [Google Scholar]
- 16. Greene SM, Reid RJ, Larson EB. Implementing the learning health system: From concept to action. Ann Intern Med. 2012;157:207–210. doi: 10.7326/0003-4819-157-3-201208070-00012. [DOI] [PubMed] [Google Scholar]
- 17.Greenes RA. Clinical Decision Support: The Road Ahead. New York: Elsevier; 2007. [Google Scholar]
- 18. Payne TH, Bates DW, Berner ES, et al. Healthcare information technology and economics. J Am Med Inform Assoc. 2013;20:212–217. doi: 10.1136/amiajnl-2012-000821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Payne TH, Corley S, Cullen TA, et al. Report of the AMIA EHR-2020 Task Force on the status and future direction of EHRs. J Am Med Inform Assoc. 2015;22:1102–1110. doi: 10.1093/jamia/ocv066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gardner RL, Cooper E, Haskell J, et al. Physician stress and burnout: The impact of health information technology. J Am Med Inform Assoc. 2019;26:106–114. doi: 10.1093/jamia/ocy145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shanafelt TD, Dyrbye LN, West CP. Addressing physician burnout: The way forward. JAMA. 2017;317:901–902. doi: 10.1001/jama.2017.0076. [DOI] [PubMed] [Google Scholar]
- 22. Kroth PJ, Morioka-Douglas N, Veres S, et al. The electronic elephant in the room: Physicians and the electronic health record. JAMIA Open. 2018;1:49–56. doi: 10.1093/jamiaopen/ooy016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shanafelt T, Dyrbye L. Oncologist burnout: Causes, consequences, and responses. J Clin Oncol. 2012;30:1235–1241. doi: 10.1200/JCO.2011.39.7380. [DOI] [PubMed] [Google Scholar]
- 24. Bodenheimer T, Sinsky C. From triple to quadruple aim: Care of the patient requires care of the provider. Ann Fam Med. 2014;12:573–576. doi: 10.1370/afm.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wallace JE, Lemaire JB, Ghali WA. Physician wellness: A missing quality indicator. Lancet. 2009;374:1714–1721. doi: 10.1016/S0140-6736(09)61424-0. [DOI] [PubMed] [Google Scholar]
- 26.National Research Council . Toward Precision Medicine: Building a Knowledge Network for Biomedical Research and a New Taxonomy of Disease. Washington, DC: The National Academies Press; 2011. [PubMed] [Google Scholar]
- 27. Van Allen EM, Wagle N, Levy MA. Clinical analysis and interpretation of cancer genome data. J Clin Oncol. 2013;31:1825–1833. doi: 10.1200/JCO.2013.48.7215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Garraway LA, Lander ES. Lessons from the cancer genome. Cell. 2013;153:17–37. doi: 10.1016/j.cell.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 29. Schmidt C. Cancer: Reshaping the cancer clinic. Nature. 2015;527:S10–S11. doi: 10.1038/527S10a. [DOI] [PubMed] [Google Scholar]
- 30. Conway JR, Warner JL, Rubinstein WS, et al. Next-generation sequencing and the clinical oncology workflow: Data challenges, proposed solutions, and a call to action. JCO Precis Oncol. 2019;3 doi: 10.1200/PO.19.00232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hripcsak G. IAIMS architecture. J Am Med Inform Assoc. 1997;4:S20–S30. [PMC free article] [PubMed] [Google Scholar]
- 32. Huff SM, Haug PJ, Stevens LE, et al. HELP the next generation: A new client-server architecture. Proc Annu Symp Comput Appl Med Care. 1994:271–275. [PMC free article] [PubMed] [Google Scholar]
- 33.Allen A. A 40-Year 'Conspiracy' at the VA. https://www.politico.com/agenda/story/2017/03/vista-computer-history-va-conspiracy-000367/ [Google Scholar]
- 34. Mandl KD, Kohane IS. Escaping the EHR trap—The future of health IT. N Engl J Med. 2012;366:2240–2242. doi: 10.1056/NEJMp1203102. [DOI] [PubMed] [Google Scholar]
- 35. Weaver CA, Teenier P. Rapid EHR development and implementation using web and cloud-based architecture in a large home health and hospice organization. Stud Health Technol Inform. 2014;201:380–387. [PubMed] [Google Scholar]
- 36. Moses H, 3rd, Matheson DH, Dorsey ER, et al. The anatomy of health care in the United States. JAMA. 2013;310:1947–1963. doi: 10.1001/jama.2013.281425. [DOI] [PubMed] [Google Scholar]
- 37. Anderson GF, Frogner BK, Johns RA, et al. Health care spending and use of information technology in OECD countries. Health Aff (Millwood) 2006;25:819–831. doi: 10.1377/hlthaff.25.3.819. [DOI] [PubMed] [Google Scholar]
- 38.Weins K. IT Spending by Industry. https://www.flexera.com/blog/technology-value-optimization/it-spending-by-industry/ [Google Scholar]
- 39.JMark.com . Analysis of I.T. in the Healthcare Industry. https://www.jmark.com/wp-content/uploads/2019/09/JMARK-HEALTHCARE-ANALYSIS-lo.pdf [Google Scholar]
- 40. Eccles MP, Mittman BS. Welcome to implementation science. Implement Sci. 2006;1:1. [Google Scholar]
- 41. Kawamoto K, Kukhareva PV, Weir C, et al. Establishing a multidisciplinary initiative for interoperable electronic health record innovations at an academic medical center. JAMIA Open. 2021;4:ooab041. doi: 10.1093/jamiaopen/ooab041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Richardson JE, Abramson EL, Pfoh ER, et al. Bridging informatics and implementation science: Evaluating a framework to assess electronic health record implementations in community settings. AMIA Annu Symp Proc. 20122012:770–778. [PMC free article] [PubMed] [Google Scholar]
- 43.Snell E. EHR Implementation Support Top Health IT Outsourcing Request. https://ehrintelligence.com/news/ehr-implementation-support-top-health-it-outsourcing-request [Google Scholar]
- 44. Elias P, Peterson E, Wachter B, et al. Evaluating the impact of interruptive alerts within a health system: Use, response time, and cumulative time burden. Appl Clin Inform. 2019;10:909–917. doi: 10.1055/s-0039-1700869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Middleton B, Sittig DF, Wright A. Clinical decision support: A 25 year retrospective and a 25 year vision. Yearb Med Inform Suppl. 2016;1:S103–S116. doi: 10.15265/IYS-2016-s034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Baysari MT, Tariq A, Day RO, et al. Alert override as a habitual behavior—A new perspective on a persistent problem. J Am Med Inform Assoc. 2017;24:409–412. doi: 10.1093/jamia/ocw072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Koleck TA, Dreisbach C, Bourne PE, et al. Natural language processing of symptoms documented in free-text narratives of electronic health records: A systematic review. J Am Med Inform Assoc. 2019;26:364–379. doi: 10.1093/jamia/ocy173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kong HJ. Managing unstructured big data in healthcare system. Healthc Inform Res. 2019;25:1–2. doi: 10.4258/hir.2019.25.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pivovarov R, Elhadad N. Automated methods for the summarization of electronic health records. J Am Med Inform Assoc. 2015;22:938–947. doi: 10.1093/jamia/ocv032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Semanik MG, Kleinschmidt PC, Wright A, et al. Impact of a problem-oriented view on clinical data retrieval. J Am Med Inform Assoc. 2021;28:899–906. doi: 10.1093/jamia/ocaa332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dunn Lopez K, Chin CL, Leitao Azevedo RF, et al. Electronic health record usability and workload changes over time for provider and nursing staff following transition to new EHR. Appl Ergon. 2021;93:103359. doi: 10.1016/j.apergo.2021.103359. [DOI] [PubMed] [Google Scholar]
- 52. Collier R. Rethinking EHR interfaces to reduce click fatigue and physician burnout. CMAJ. 2018;190:E994–E995. doi: 10.1503/cmaj.109-5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Magrabi F, Baker M, Sinha I, et al. Clinical safety of England's national programme for IT: A retrospective analysis of all reported safety events 2005 to 2011. Int J Med Inform. 2015;84:198–206. doi: 10.1016/j.ijmedinf.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 54. Magrabi F, Ong MS, Runciman W, et al. Using FDA reports to inform a classification for health information technology safety problems. J Am Med Inform Assoc. 2012;19:45–53. doi: 10.1136/amiajnl-2011-000369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dabliz R, Poon SK, Ritchie A, et al. Usability evaluation of an integrated electronic medication management system implemented in an oncology setting using the unified theory of the acceptance and use of technology. BMC Med Inform Decis Mak. 2021;21 doi: 10.1186/s12911-020-01348-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gephart S, Carrington JM, Finley B. A systematic review of nurses' experiences with unintended consequences when using the electronic health record. Nurs Adm Q. 2015;39:345–356. doi: 10.1097/NAQ.0000000000000119. [DOI] [PubMed] [Google Scholar]
- 57. Carayon P, Hundt AS, Hoonakker P. Technology barriers and strategies in coordinating care for chronically ill patients. Appl Ergon. 2019;78:240–247. doi: 10.1016/j.apergo.2019.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. McDonald CJ, Huff SM, Suico JG, et al. LOINC, a universal standard for identifying laboratory observations: A 5-year update. Clin Chem. 2003;49:624–633. doi: 10.1373/49.4.624. [DOI] [PubMed] [Google Scholar]
- 59. Agrawal A, He Z, Perl Y, et al. The readiness of SNOMED problem list concepts for meaningful use of electronic health records. Artif Intell Med. 2013;58:73–80. doi: 10.1016/j.artmed.2013.03.008. [DOI] [PubMed] [Google Scholar]
- 60. Renshaw AA, Mena-Allauca M, Gould EW, et al. Synoptic reporting: Evidence-based review and future directions. JCO Clin Cancer Inform. 2018;2:1–9. doi: 10.1200/CCI.17.00088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hicks JK, Howard R, Reisman P, et al. Integrating somatic and germline next-generation sequencing into routine clinical oncology practice. JCO Precis Oncol. 2021;5:884–895. doi: 10.1200/PO.20.00513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Farzandipour M, Sheikhtaheri A, Sadoughi F. Effective factors on accuracy of principal diagnosis coding based on International Classification of Diseases, the 10th revision (ICD-10) Int J Inf Manage. 2010;30:78–84. [Google Scholar]
- 63. O'Malley KJ, Cook KF, Price MD, et al. Measuring diagnoses: ICD code accuracy. Health Serv Res. 2005;40:1620–1639. doi: 10.1111/j.1475-6773.2005.00444.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhou L, Cheng C, Ou D, et al. Construction of a semi-automatic ICD-10 coding system. BMC Med Inform Decis Mak. 2020;20:67. doi: 10.1186/s12911-020-1085-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wright A, McCoy AB, Hickman TT, et al. Problem list completeness in electronic health records: A multi-site study and assessment of success factors. Int J Med Inform. 2015;84:784–790. doi: 10.1016/j.ijmedinf.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. van der Lei J. Use and abuse of computer-stored medical records. Methods Inf Med. 1991;30:79–80. [PubMed] [Google Scholar]
- 67.Kimball R, Ross M. The Data Warehouse Toolkit: The Complete Guide to Dimensional Modeling. 2. New York: Wiley Computer Publishing; 2002. [Google Scholar]
- 68. Einbinder JS, Scully KW, Pates RD, et al. Case study: A data warehouse for an academic medical center. J Healthc Inf Manag. 2001;15:165–175. [PubMed] [Google Scholar]
- 69. Gagalova KK, Leon Elizalde MA, Portales-Casamar E, et al. What you need to know before implementing a clinical research data warehouse: Comparative review of integrated data repositories in health care institutions. JMIR Form Res. 2020;4:e17687. doi: 10.2196/17687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Juran JM, Godfrey AB, Hoogstoel RE, et al. Juran's Quality Handbook. 5. New York: McGraw-Hill; 1999. p. 1808. [Google Scholar]
- 71. Kahn MG, Callahan TJ, Barnard J, et al. A harmonized data quality assessment terminology and framework for the secondary use of electronic health record data. EGEMS (Wash DC) 2016;4:1244. doi: 10.13063/2327-9214.1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Yadav P, Steinbach M, Kumar V, et al. Mining electronic health records (EHRs): A survey. ACM Comput Surv. 2018;50:1–40. [Google Scholar]
- 73. Kohane IS, Aronow BJ, Avillach P, et al. What every reader should know about studies using electronic health record data but may be afraid to ask. J Med Internet Res. 2021;23:e22219. doi: 10.2196/22219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Schmidt BM, Colvin CJ, Hohlfeld A, et al. Definitions, components and processes of data harmonisation in healthcare: A scoping review. BMC Med Inform Decis Mak. 2020;20:222. doi: 10.1186/s12911-020-01218-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. McMurry AJ, Murphy SN, MacFadden D, et al. SHRINE: Enabling nationally scalable multi-site disease studies. PLoS One. 2013;8:e55811. doi: 10.1371/journal.pone.0055811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Hripcsak G, Duke JD, Shah NH, et al. Observational Health Data Sciences and Informatics (OHDSI): Opportunities for observational researchers. Stud Health Technol Inform. 2015;216:574–578. [PMC free article] [PubMed] [Google Scholar]
- 77. Friedman C, Rindflesch TC, Corn M. Natural language processing: State of the art and prospects for significant progress, a workshop sponsored by the National Library of Medicine. J Biomed Inform. 2013;46:765–773. doi: 10.1016/j.jbi.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 78. Gamal A, Barakat S, Rezk A. Standardized electronic health record data modeling and persistence: A comparative review. J Biomed Inform. 114:103670. 2021. doi: 10.1016/j.jbi.2020.103670. [DOI] [PubMed] [Google Scholar]
- 79. Sandhu E, Weinstein S, McKethan A, et al. Secondary uses of electronic health record data: Benefits and barriers. Jt Comm J Qual Patient Saf. 2012;38:34–40. doi: 10.1016/s1553-7250(12)38005-7. [DOI] [PubMed] [Google Scholar]
- 80. Meystre SM, Friedlin FJ, South BR, et al. Automatic de-identification of textual documents in the electronic health record: A review of recent research. BMC Med Res Methodol. 2010;10:70. doi: 10.1186/1471-2288-10-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Weng C, Appelbaum P, Hripcsak G, et al. Using EHRs to integrate research with patient care: Promises and challenges. J Am Med Inform Assoc. 2012;19:684–687. doi: 10.1136/amiajnl-2012-000878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Etheredge LM. A rapid-learning health system. Health Aff (Millwood) 2007;26:w107–w118. doi: 10.1377/hlthaff.26.2.w107. [DOI] [PubMed] [Google Scholar]
- 83.Institute of Medicine (US) Roundtable on Evidence-Based Medicine . : The Learning Healthcare System. Washington, DC: National Academies Press (US); 2007. [PubMed] [Google Scholar]
- 84. Rowley J. The wisdom hierarchy: representations of the DIKW hierarchy. J Inf Sci. 2007;33:163–180. [Google Scholar]
- 85. Zins C. Conceptual approaches for defining data, information, and knowledge. J Am Soc Inf Sci Technol. 2007;58:479–493. [Google Scholar]
- 86. Pathak J, Kho AN, Denny JC. Electronic health records-driven phenotyping: Challenges, recent advances, and perspectives. J Am Med Inform Assoc. 2013;20:e206–11. doi: 10.1136/amiajnl-2013-002428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Forrest CB, McTigue KM, Hernandez AF, et al. PCORnet® 2020: Current state, accomplishments, and future directions. J Clin Epidemiol. 2021;129:60–67. doi: 10.1016/j.jclinepi.2020.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Osterman TJ, Terry M, Miller RS. Improving cancer data interoperability: The promise of the minimal common oncology data elements (mCODE) initiative. JCO Clin Cancer Inform. 2020;4:993–1001. doi: 10.1200/CCI.20.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Warner JL, Maddux SE, Hughes KS, et al. Development, implementation, and initial evaluation of a foundational open interoperability standard for oncology treatment planning and summarization. J Am Med Inform Assoc. 2015;22:577–586. doi: 10.1093/jamia/ocu015. [DOI] [PubMed] [Google Scholar]
- 90.World Health Organization . International Classification of Diseases for Oncology. https://www.who.int/standards/classifications/other-classifications/international-classification-of-diseases-for-oncology [Google Scholar]
- 91. Kundra R, Zhang H, Sheridan R, et al. OncoTree: A cancer classification system for precision oncology. JCO Clin Cancer Inform. 2021;5:221–230. doi: 10.1200/CCI.20.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Warner JL, Dymshyts D, Reich CG, et al. HemOnc: A new standard vocabulary for chemotherapy regimen representation in the OMOP common data model. J Biomed Inform. 2019;96:103239. doi: 10.1016/j.jbi.2019.103239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Sioutos N, de Coronado S, Haber MW, et al. NCI Thesaurus: A semantic model integrating cancer-related clinical and molecular information. J Biomed Inform. 2007;40:30–43. doi: 10.1016/j.jbi.2006.02.013. [DOI] [PubMed] [Google Scholar]
- 94.M2Gen . ORIEN. https://www.oriencancer.org/ [Google Scholar]
- 95.Flatiron Health. https://flatiron.com/ [Google Scholar]
- 96. Weeks J, Pardee R. Learning to share health care data: A brief timeline of influential common data models and distributed health data networks in U.S. Health Care Research. EGEMS (Wash DC) 2019;7:4. doi: 10.5334/egems.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Rubinstein SM, Warner JL. CancerLinQ: Origins, implementation, and future directions. JCO Clin Cancer Inform. 2018;2:1–7. doi: 10.1200/CCI.17.00060. [DOI] [PubMed] [Google Scholar]
- 98. Micheel CM, Sweeney SM, LeNoue-Newton ML, et al. American Association for Cancer Research project Genomics Evidence Neoplasia Information Exchange: From inception to first data release and beyond-lessons learned and member institutions' perspectives. JCO Clin Cancer Inform. 2018;2:1–14. doi: 10.1200/CCI.17.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Braunstein ML. Health care in the age of interoperability Part 6: The future of FHIR. IEEE Pulse. 2019;10:25–27. doi: 10.1109/MPULS.2019.2922575. [DOI] [PubMed] [Google Scholar]
- 100.Choi M, Starr R, Braunstein M, et al. OHDSI on FHIR Platform Development with OMOP CDM Mapping to FHIR Resources, OHDSI Symposium. Washington, DC: Observational Health Data Sciences and Informatics; 2016. [Google Scholar]
- 101. Dolin RH, Alschuler L, Boyer S, et al. HL7 clinical document architecture, release 2. J Am Med Inform Assoc. 2006;13:30–39. doi: 10.1197/jamia.M1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Negro-Calduch E, Azzopardi-Muscat N, Krishnamurthy RS, et al. Technological progress in electronic health record system optimization: Systematic review of systematic literature reviews. Int J Med Inform. 152:104507. 2021. doi: 10.1016/j.ijmedinf.2021.104507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Ngiam KY, Khor IW. Big data and machine learning algorithms for health-care delivery. Lancet Oncol. 2019;20:e262–e273. doi: 10.1016/S1470-2045(19)30149-4. [DOI] [PubMed] [Google Scholar]
- 104. Xiao C, Choi E, Sun J. Opportunities and challenges in developing deep learning models using electronic health records data: A systematic review. J Am Med Inform Assoc. 2018;25:1419–1428. doi: 10.1093/jamia/ocy068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Penberthy L, Rivera DR, Ward K. The contribution of cancer surveillance toward real world evidence in oncology. Semin Radiat Oncol. 2019;29:318–322. doi: 10.1016/j.semradonc.2019.05.004. [DOI] [PubMed] [Google Scholar]
- 106.Cronin KA, Ries LA, Edwards BK.The Surveillance, Epidemiology, and end results (SEER) program of the National Cancer Institute Cancer 3755–3757.2014 [DOI] [PubMed]
- 107. Wingo PA, Jamison PM, Hiatt RA, et al. Building the infrastructure for nationwide cancer surveillance and control—a comparison between the National Program of Cancer Registries (NPCR) and the Surveillance, Epidemiology, and end results (SEER) program (United States) Cancer Causes Control. 2003;14:175–193. doi: 10.1023/a:1023002322935. [DOI] [PubMed] [Google Scholar]
- 108.National Cancer Institute . NCCR*Explorer: An Interactive Website for NCCR Cancer Statistics. https://NCCRExplorer.ccdi.cancer.gov/ [Google Scholar]
- 109. Mandel JC, Kreda DA, Mandl KD, et al. SMART on FHIR: A standards-based, interoperable apps platform for electronic health records. J Am Med Inform Assoc. 2016;23:899–908. doi: 10.1093/jamia/ocv189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mandl K. SMART on FHIR: Big Gains, Big Challenges | Executive Education at HMS. https://executiveeducation.hms.harvard.edu/industry-insights/smart-fhir-big-gains-big-challenges [Google Scholar]
- 111. Warner JL, Rioth MJ, Mandl KD, et al. SMART Precision Cancer Medicine: A FHIR-based app to provide genomic information at the point of care. J Am Med Inform Assoc. 2016 doi: 10.1093/jamia/ocw015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Warner JL, Wang L, Pao W, et al. CUSTOM-SEQ: A prototype for oncology rapid learning in a comprehensive EHR environment. J Am Med Inform Assoc. 2016;23:692–700. doi: 10.1093/jamia/ocw008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Neuss MN, Desch CE, McNiff KK, et al. A process for measuring the quality of cancer care: The quality oncology practice initiative. J Clin Oncol. 2005;23:6233–6239. doi: 10.1200/JCO.2005.05.948. [DOI] [PubMed] [Google Scholar]
- 114. Blayney DW, McNiff K, Hanauer D, et al. Implementation of the quality oncology practice initiative at a university comprehensive cancer center. J Clin Oncol. 2009;27:3802–3807. doi: 10.1200/JCO.2008.21.6770. [DOI] [PubMed] [Google Scholar]
- 115. Abernethy AP, Etheredge LM, Ganz PA, et al. Rapid-learning system for cancer care. J Clin Oncol. 2010;28:4268–4274. doi: 10.1200/JCO.2010.28.5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Lau-Min KS, Asher SB, Chen J, et al. Real-world integration of genomic data into the electronic health record: The PennChart Genomics Initiative. Genet Med. 2021;23:603–605. doi: 10.1038/s41436-020-01056-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. North F, Chaudhry R. Apple HealthKit and health app: Patient uptake and barriers in primary care. Telemed J E Health. 2016;22:608–613. doi: 10.1089/tmj.2015.0106. [DOI] [PubMed] [Google Scholar]
- 118. Chiauzzi E, Rodarte C, DasMahapatra P. Patient-centered activity monitoring in the self-management of chronic health conditions. BMC Med. 2015;13:77. doi: 10.1186/s12916-015-0319-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Health Level Seven International http://www.hl7.org
- 120. Bates DW, Saria S, Ohno-Machado L, et al. Big data in health care: Using analytics to identify and manage high-risk and high-cost patients. Health Aff (Millwood) 2014;33:1123–1131. doi: 10.1377/hlthaff.2014.0041. [DOI] [PubMed] [Google Scholar]
- 121. Hodgson T, Magrabi F, Coiera E. Efficiency and safety of speech recognition for documentation in the electronic health record. J Am Med Inform Assoc. 2017;24:1127–1133. doi: 10.1093/jamia/ocx073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Kreimeyer K, Foster M, Pandey A, et al. Natural language processing systems for capturing and standardizing unstructured clinical information: A systematic review. J Biomed Inform. 2017;73:14–29. doi: 10.1016/j.jbi.2017.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Umscheid CA, Margolis DJ, Grossman CE. Key concepts of clinical trials: A narrative review. Postgrad Med. 2011;123:194–204. doi: 10.3810/pgm.2011.09.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Unger JM, Vaidya R, Hershman DL, et al. Systematic review and meta-analysis of the magnitude of structural, clinical, and physician and patient barriers to cancer clinical trial participation. J Natl Cancer Inst. 2019;111:245–255. doi: 10.1093/jnci/djy221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Meropol NJ, Wong YN, Albrecht T, et al. Randomized trial of a web-based intervention to address barriers to clinical trials. J Clin Oncol. 2016;34:469–478. doi: 10.1200/JCO.2015.63.2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Sledge GW, Hudis CA, Swain SM, et al. ASCO's approach to a learning health care system in oncology. J Oncol Pract. 2013;9:145–148. doi: 10.1200/JOP.2013.000957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.JASON . A Robust Health Data Infrastructure (Prepared by JASON at the MITRE Corporation under Contract No. 13-717F-13) Rockville, MD: Agency for Healthcare Research and Quality; 2013. [Google Scholar]
- 128. Weber GM, Mandl KD, Kohane IS. Finding the missing link for big biomedical data. JAMA. 2014;311:2479. doi: 10.1001/jama.2014.4228. [DOI] [PubMed] [Google Scholar]
- 129. Klann JG, Joss MAH, Embree K, et al. Data model harmonization for the All of Us Research Program: Transforming i2b2 data into the OMOP common data model. PLoS One. 2019;14:e0212463. doi: 10.1371/journal.pone.0212463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Hripcsak G, Schuemie MJ, Madigan D, et al. Drawing reproducible conclusions from observational clinical data with OHDSI. Yearb Med Inform. 2021;30:283–289. doi: 10.1055/s-0041-1726481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.OHDSI . HADES Health Analytics Data-To-Evidence Suite. https://ohdsi.github.io/Hades/index.html [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Blobel B. Interoperable EHR systems—challenges, standards and solutions. Eur J Biomed Inform. 2018;14:10–19. [Google Scholar]
- 133. Mandl KD, Kohane IS, McFadden D, et al. Scalable Collaborative Infrastructure for a Learning Healthcare System (SCILHS): Architecture. J Am Med Inform Assoc. 2014;21:615–620. doi: 10.1136/amiajnl-2014-002727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Murphy SN, Weber G, Mendis M, et al. Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2) J Am Med Inform Assoc. 2010;17:124–130. doi: 10.1136/jamia.2009.000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Observational Health Data Sciences and Informatics (OHDSI) OMOP Common Data Model. http://www.ohdsi.org/data-standardization/the-common-data-model/ [Google Scholar]
- 136.Qureshi I. Healthcare Analytics Platform: DOS Delivers the 7 Essential Components. https://www.healthcatalyst.com/insights/healthcare-analytics-platform-seven-core-elements-dos/ [Google Scholar]
- 137. Wagholikar KB, Mandel JC, Klann JG, et al. SMART-on-FHIR implemented over i2b2. J Am Med Inform Assoc. 2017;24:398–402. doi: 10.1093/jamia/ocw079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Bertagnolli MM, Anderson B, Norsworthy K, et al. Status update on data required to build a learning health system. J Clin Oncol. 2020;38:1602–1607. doi: 10.1200/JCO.19.03094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Colicchio TK, Cimino JJ. Twilighted homegrown systems: The experience of six traditional electronic health record developers in the post-meaningful use era. Appl Clin Inform. 2020;11:356–365. doi: 10.1055/s-0040-1710310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Gaynor M, Yu F, Andrus CH, et al. A general framework for interoperability with applications to healthcare. Health Pol Technol. 2014;3:3–12. [Google Scholar]
- 141. Mandl KD, Gottlieb D, Mandel JC, et al. Push button population health: The SMART/HL7 FHIR bulk data access application programming interface. NPJ Digit Med. 2020;3:151. doi: 10.1038/s41746-020-00358-4. [DOI] [PMC free article] [PubMed] [Google Scholar]