Summary
Existing approaches to therapeutic gene transfer are marred by the transient nature of gene expression following non-integrative gene delivery and by safety concerns due to the random mechanism of viral-mediated genomic insertions. The disadvantages of these methods encourage future research in identifying human genomic sites that allow for durable and safe expression of genes of interest. We conducted a bioinformatic search followed by the experimental characterization of human genomic sites, identifying two that demonstrated the stable expression of integrated reporter and therapeutic genes without malignant changes to the cellular transcriptome. The cell-type agnostic criteria used in our bioinformatic search suggest widescale applicability of identified sites for engineering of a diverse range of tissues for clinical and research purposes, including modified T cells for cancer therapy and engineered skin to ameliorate inherited diseases and aging. In addition, the stable and robust levels of gene expression from identified sites allow for the industry-scale biomanufacturing of proteins in human cells.
Keywords: genomic safe harbor, criteria, expression, knock-in, transcriptomics, durability, safety
Graphical abstract
Highlights
-
•
Provides an integrated pipeline to identify human genomic safe harbor sites
-
•
Discovers two sites for long-term expression of genes of interest, Rogi1 and Rogi2
-
•
Verifies safety of gene expression from these sites by transcriptome profiling
-
•
Suggests use cases for identified sites and proposes further validation experiments
Motivation
The ability to express genes of interest in a reliable and safe manner in clinically relevant cells and tissues is critical for successful gene and cell therapies. Several tools allowing for targeted genomic insertions of DNA sequences have recently been developed; however, genomic regions suitable for safe, long-term hosting, and expression of these sequences have not been identified. We describe a pipeline to computationally predict and experimentally validate human genomic safe harbor sites and characterize two sites, Rogi1 and Rogi2, suitable for integrative gene delivery.
Aznauryan et al. establish a pipeline for computational prediction and experimental validation of human genomic safe harbor sites. Two genomic sites, Rogi1 and Rogi2, are characterized as capable of safe and durable expression of genes of interest following targeted insertions in a variety of cellular contexts.
Introduction
The development of technologies for predictable, durable, and safe expression of desired genetic constructs (i.e., transgenes) in human cells will contribute significantly to the improvement of gene and cell therapies (Bestor, 2000; Ellis, 2005), as well as to the advancement of protein manufacturing (Lee et al., 2019). One prominent beneficiary of such technologies is genetically engineered T cell therapies, which require the genomic integration of transgenes encoding novel immune receptors (Chen et al., 2020; Richardson et al., 2019); another example is gene therapies for highly proliferating tissues, such as inherited skin disorders, in which entire wild-type (WT) gene copies must be integrated into epidermal stem cells (Droz-Georget Lathion et al., 2015; Hirsch et al., 2017). Advances in genome editing using targeted integration tools (Maeder and Gersbach, 2016) already allow precise genomic delivery and sustained expression of transgenes in certain cellular contexts, such as chimeric antigen receptors (CARs) integrated into the T cell receptor alpha chain locus in T cells (Eyquem et al., 2017), and coagulation factors delivered to hepatocytes using recombinant adeno-associated viral (rAAV) vectors (Barzel et al., 2015). These applications, however, are limited to specific cell types and may cause disruption to the endogenous genes, confining the diversity of cellular engineering applications. Specific loci in the human genome that support stable and efficient transgene expression, without detrimentally altering cellular functions, are known as genomic safe harbor (GSH) sites. The precise integration of functional genetic constructs into GSH sites greatly enhances the safety and efficacy of genome engineering for clinical and biotechnology applications.
Empirical studies have identified three sites that support the long-term expression of transgenes—AAVS1, CCR5, and hRosa26—all of which were established without any a priori safety assessment of the genomic loci in which they reside (Papapetrou and Schambach, 2016). The AAVS1 site, located in an intron of the PPP1R12C gene region, has been observed to be a region for rare genomic integration events of the payload of the AAV (Oceguera-Yanez et al., 2016). Despite being successfully implemented for durable transgene expression in numerous cell types (Hong et al., 2017), the AAVS1 site location is in a gene-dense region, suggesting potential disruption of the expression profiles of genes located in the vicinity of this locus (Sadelain et al., 2012). In addition, studies have indicated frequent transgene silencing and decrease in growth rate following integration into AAVS1 (Ordovás et al., 2015; Shin et al., 2020), which represents a liability for clinical gene therapy. The second site lies within the CCR5 gene, which encodes a protein involved in chemotaxis and serves as a co-receptor for HIV cellular entry in T cells (Jiao et al., 2019). Serendipitously, researchers have identified that the naturally occurring CCR5-delta-32 mutation present in people of Scandinavian origin results in an HIV-resistant phenotype (Silva and Stumpf, 2004). This finding suggested disposability of this gene and applicability of the CCR5 locus for targeted genome engineering, especially for T cell therapies (Lombardo et al., 2011; Sather et al., 2015). However, similar to AAVS1, the CCR5 locus is located in a gene-rich region, surrounded by tumor-associated genes (Sadelain et al., 2012), thus severely limiting its safe use for therapeutic purposes. In addition, CCR5 expression has been associated with promoting functional recovery following stroke (Joy et al., 2019), thus disrupting CCR5, and may be undesirable in clinical practice. The third site, human Rosa26 (hRosa26) locus, was computationally predicted by mining the human genome for orthologous sequences of the mouse Rosa26 (mRosa26) locus (Irion et al., 2007). mRosa26 was originally identified in mouse embryonic stem cells by using random integration by lentiviral-mediated delivery of gene-trapping constructs consisting of promotorless transgenes (β-galactosidase and neomycin phosphotransferase), resulting in the sustainable expression of these transgenes throughout embryonic development (Friedrich and Soriano, 1991; Zambrowicz et al., 1997). Similar to the other two sites currently used, hRosa26 is located in an intron of a coding gene, THUMPD3 (Irion et al., 2007), the function of which is still not fully characterized. This site is also surrounded by proto-oncogenes in its immediate vicinity (Sadelain et al., 2012), which may be upregulated following transgene insertion, thus potentially limiting the use of hRosa26 in clinical settings.
Attempts have been made to identify human GSH sites that would satisfy various safety criteria, thus avoiding the disadvantages of existing sites. One approach developed by Sadelain and colleagues used lentiviral transduction of β-globin and green fluorescent protein (GFP) genes into induced pluripotent stem cells (iPSCs), followed by the assessment of the integration sites in terms of their linear distance from various coding and regulatory elements in the genome, such as cancer genes, micro RNAs (miRNAs), and ultraconserved regions (Papapetrou et al., 2011). They discovered one lentiviral integration site that satisfied all of the proposed criteria, demonstrating sustainable expression upon the erythroid differentiation of iPSCs. However, global transcriptome profile alterations of cells with transgenes integrated into this site were not assessed. A similar approach by Weiss and colleagues used lentiviral integrations in Chinese hamster ovary (CHO) cells to identify sites supporting long-term protein expression for biotechnological applications (e.g., recombinant monoclonal antibody production) (Gaidukov et al., 2018). Although this study led to the evaluation of multiple sites for durable, high-level transgene expression in CHO cells, no extrapolation to human genomic sites was carried out. Another study aimed at identifying GSHs through the bioinformatic search of mCreI sites, regions targeted by the monomerized version of I-CreI homing endonuclease found and characterized in green algae as capable of making targeted staggered double-strand DNA breaks (Ulge et al., 2011), residing in loci that satisfy GSH criteria (Pellenz et al., 2019). Like previous work, several stably expressing sites were identified and proposed for synthetic biology applications in humans. However, local and global gene expression profiling following integration events in these sites has not been conducted.
All of these potential GSH sites possess a shared limitation of being narrowed by lentiviral- or mCreI-based integration mechanisms. In addition, safety assessments of some of these identified sites, as well as previously established AAVS1, CCR5, and Rosa26, were carried out by evaluating the differential gene expression of genes located solely in the vicinity of these integration sites, without observing global transcriptomic changes following integration. A more comprehensive bioinformatic-guided and genome-wide search of GSH sites based on established criteria, followed by experimental assessment of transgene expression durability in various cell types and safety assessment using global transcriptome profiling, would thus lead to the identification of a more reliable and clinically useful genomic region.
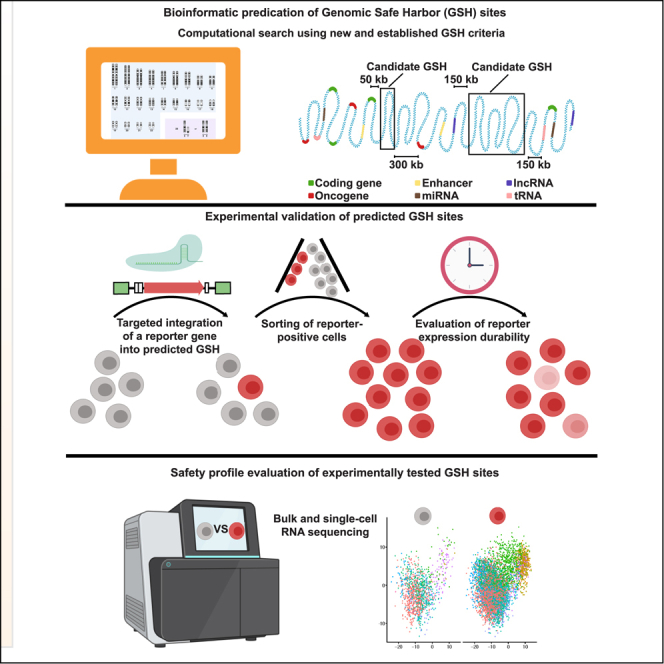
In this study, we used bioinformatic screening to rationally identify multiple sites that satisfy established as well as newly introduced GSH criteria. We then used CRISPR/Cas9 targeted genome editing to individually integrate a reporter gene into these predicted sites to monitor long-term expression of the transgene in HEK293T and Jurkat cells. This experimental evaluation in cell lines was followed by testing two promising candidate sites in primary human T cells and primary human dermal fibroblasts using reporter and therapeutic transgenes, respectively. Finally, bulk and single-cell RNA sequencing (RNA-seq) experiments were performed to analyze the transcriptomic effects of such integrations into these two established GSH sites.
Results
Bioinformatic search of GSH sites
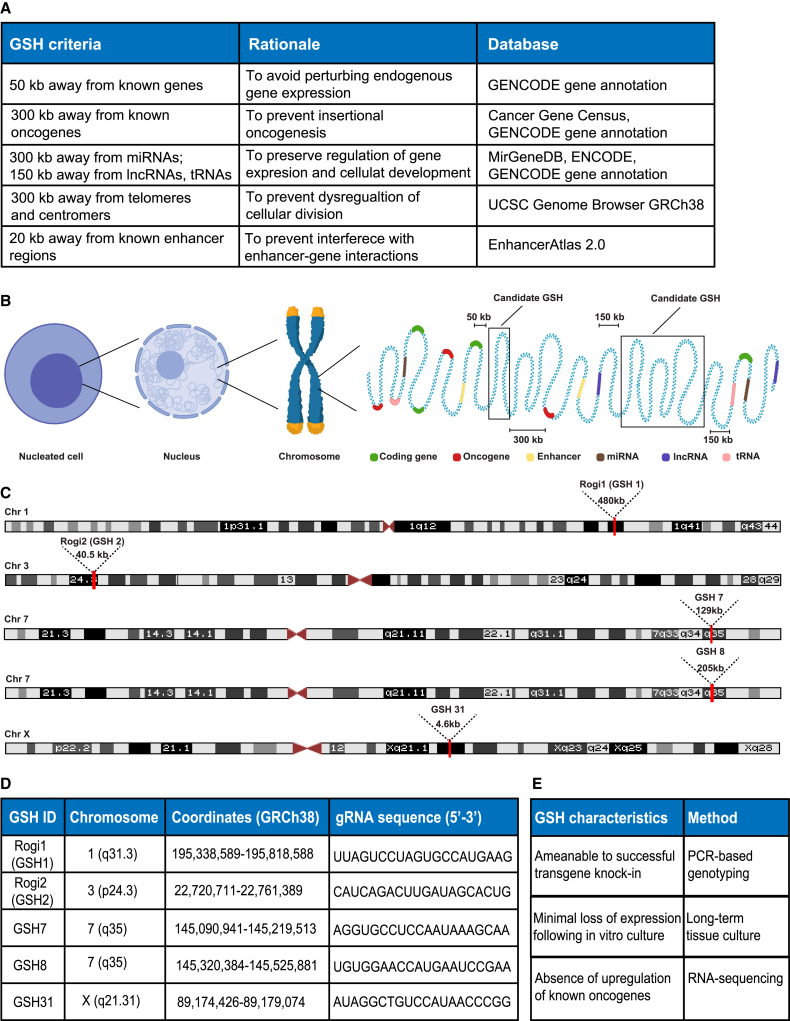
To identify sites that could serve as potential GSHs, we conducted a genome-wide bioinformatic search based on previously established, widely accepted (Sadelain et al., 2012), and newly introduced criteria that would satisfy safe and stable gene expression (Figures 1A and 1B). We started by eliminating gene-encoding sequences and their flanking regions of 50 kb, a distance suggested by previous studies in the field (Sadelain et al., 2012), to avoid disruption of the functional regions of gene expression and contrast GSHs to typical genomic insertion patterns of integrative viral deliveries. We then identified oncogenes and eliminated regions of 300 kb upstream and downstream to prevent insertional oncogenesis, a complication of γ-retroviral and lentiviral integrations that may arise through unintended upregulation of an oncogene in the vicinity of the integration site, previously reported to reach such long distances (Hacein-Bey-Abina et al., 2008; Bushman, 2020). We used oncogenes from both tier 1 (extensive evidence of association with cancer available) and tier 2 (strong indications of the association exist) to decrease the likelihood of oncogene activation upon integration. In addition, genes can be substantially regulated by microRNAs (miRNAs), which cleave and decay mature transcripts as well as inhibit translation machinery, thus modulating protein abundance (Filipowicz et al., 2008). We, therefore, excluded miRNA-encoding regions and 300-kb-long regions around them, using the information from retroviral tagged cancer gene databases as well as guidelines developed in prior studies (Akagi, 2004; Sadelain et al., 2012). Apart from promoters and miRNAs, gene expression may depend on the presence of enhancers that could be located kilobases away (Schoenfelder and Fraser, 2019; Vangala et al., 2020). We therefore excluded enhancers as well as 20-kb regions around them, which excludes proximal enhancer-gene interactions (Mora et al., 2015) and provides an overall distance of up to 320 kb from regions involved in oncogenesis, decreasing the likelihood of malignant gene activation. In addition to the previously established criteria described above, we excluded regions surrounding long non-coding RNAs and tRNAs as well as 150 kb around them because they are involved in nuclear architecture, differentiation, and developmental programs determining cell fate and are essential for normal protein translation, respectively; thus, avoiding changes to their physiological expression is desired (Guttman et al., 2009; Chen et al., 2016; Schimmel, 2018). Finally, we excluded centromeric and telomeric regions to prevent alterations in DNA replication, cellular division, and normal aging (Villasante et al., 2007).
Figure 1.
Bioinformatic identification of genomic safe harbor sites
(A) Table shows GSH criteria, rationale, and databases used to computationally predict GSH sites in the human genome.
(B) Schematic representation of candidate GSH sites, showing linear distances from different coding and regulatory elements in the genome according to the established and newly introduced criteria.
(C) Chromosomal locations and lengths of 5 candidate GSH sites, which were subsequently experimentally tested.
(D) Chromosomal coordinates of 5 candidate GSH sites and the gRNA sequences used for subsequent CRISPR/Cas9 genome editing. See also Table S1 for the list of all of the computationally predicted sites.
(E) Characterization of experimentally tested GSH sites includes genotyping of the targeted locus to verify transgene insertion, long-term culture of GSH integrated cells to observe the durability of transgene expression, and bulk and single-cell RNA sequencing (RNA-seq) to validate safety of integration by absence of upregulation of genes associated with oncogenic cellular proliferation.
Based on our bioinformatic screening, we identified close to 2000 sites that satisfied all of our criteria (Table S1). We chose five sites that varied significantly in size (GSH1, -2, -7, -8, and -31) and designed guide RNAs (gRNA) targeting these sites as well as possessing high on- and off-target scores (high on-target and low off-target activities) (Figures 1C and 1D). Characterization of the evaluated GSHs involved experimental assessment of the durability and safety of transgene expression from these sites (Figure 1E). First, successful GSH integrations were confirmed by PCR-based genotyping of the targeted locus to confirm the presence of the gene of interest. Second, the durability of expression was verified by long-term in vitro culture of studied primary and immortalized cells bearing transgene integration in predicted GSHs. Finally, the safety of the predicted sites was assessed in the context of insertional oncogenesis. For that, we used bulk and single-cell RNA-seqg to study changes in gene expression following the long-term culture of GSH-integrated cells. The absence of the upregulation of genes driving oncogenic cellular proliferation, an occasional devastating side effect of discussed integrative viral gene delivery methods, was used as a criterion for GSH safety.
Experimental validation of bioinformatically identified GSH sites by targeted transgene integration in human cell lines
To experimentally assess transgene expression from the five predicted GSH sites, we performed targeted integration of a gene construct encoding a red fluorescence reporter protein (mRuby) into two common human cell lines: HEK293T and Jurkat cells. HEK293 are used frequently for medium- to large-scale production of recombinant proteins (Chin et al., 2019). Thus, identifying GSH in HEK293 would be relevant for protein manufacturing. The Jurkat cell line was derived from the T cells of a pediatric patient with acute lymphoblastic leukemia (Abraham and Weiss, 2004) and has been used extensively for assessing the functionality of engineered immune receptors; therefore, the discovery of GSH in this cell line supports applications in T cell therapies (Roybal et al., 2016; Vazquez-Lombardi et al., 2020). For the integration of mRuby, we used a CRISPR/Cas9-based genome editing strategy that uses the precise integration into target chromosome (PITCh) method (Nakade et al., 2014; Sakuma et al., 2016), assisted by microhomology-mediated end-joining (MMEJ) (Sfeir and Symington, 2015). This approach uses a reporter-bearing plasmid possessing short microhomology sequences flanked by gRNA-binding sites. Once inside the cell, the reporter gene, together with microhomologies directed against the candidate GSH site, are liberated from the plasmid by Cas9-generated double-stranded breaks (DSBs) at gRNA-binding sites on the PITCh donor plasmid. A different gRNA-Cas9 pair generates DSBs at the candidate GSH locus, and the freed reporter gene with flanking microhomologies is integrated by exploiting the MMEJ repair pathway (Figures 2A and 2B). This PITCh MMEJ approach allowed us to rapidly generate donor plasmids targeted against different predicted safe harbor sites, in contrast to the more elaborate process of cloning long homology arms (i.e., >300 bp) required for homology-directed repair (HDR). The error-prone mechanism of MMEJ-mediated integration did not represent a substantial concern since the targeted sites are distanced from any identified coding or regulatory element, and thus mutations arising following the integration are unlikely to invoke any detrimental changes.
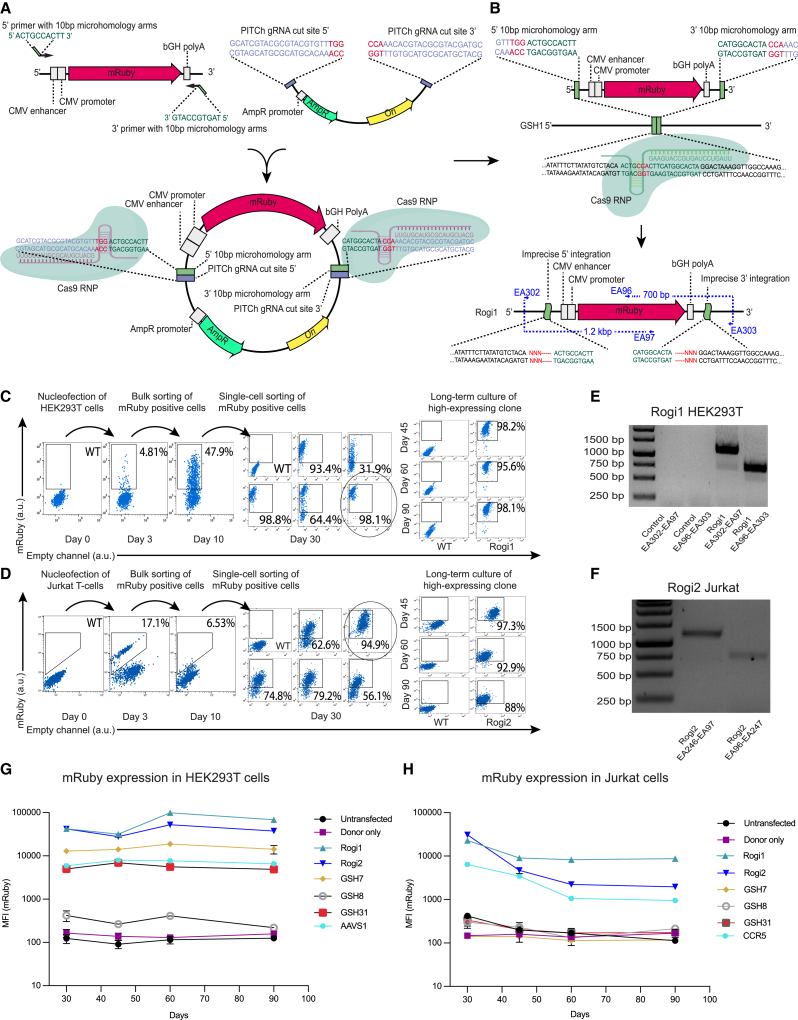
Figure 2.
Experimental validation of candidate GSH sites by targeted genome insertions in HEK293T and Jurkat cells
(A) Generation of PITCh plasmid by cloning an mRuby-bearing insert with microhomologies against specific GSH into a backbone possessing PITCh gRNA target sites, required for the Cas9-based liberation of the insert.
(B) mRuby insert is integrated into a desired site by the MMEJ pathway following a Cas9-induced double-stranded break.
(C and D) Flow cytometry demonstrating the isolation of clonal populations expressing the mRuby transgene from Rogi1 locus in HEK293T cells and Rogi2 locus in Jurkat cells using pooled and single-cell flow cytometry-mediated sorting. The highest expressing Rogi1-HEK293T clone and the Rogi2-Jurkat clone were expanded in cell culture, and flow cytometry measurements at days 45, 60, and 90 demonstrated stable levels of transgene expression.
(E) Genotyping of the Rogi1 site in HEK293T cells using primers spanning the junction between integration site and the transgene. Lane names refer to the primers used from (B). Control and Rogi1 EA302-EA97 lanes correspond to the 5′ integration junction in untransfected HEK293T cells and cells that were transfected using the PITCh CRISPR/Cas9 method, respectively. Control and Rogi1 EA97-EA303 lanes correspond to the 3′ integration junction in untransfected HEK293T cells and cells that were transfected using the PITCh CRISPR/Cas9 method, respectively.
(F) Genotyping of the Rogi2 site in Jurkat cells using primers spanning the junction between integration site and the transgene. The Rogi2 EA246-EA97 lane corresponds to the 5′ integration junction, while EA96-EA247 corresponds to the 3′ integration junction in HEK293T cells that were transfected using the PITCh CRISPR/Cas9 method.
(G) Continuous assessment of mRuby expression levels following the integration into each of the tested GSH sites as well as into AAVS1 control in HEK293T cells. Data are represented as means ± SEMs of the highest expressing clonal population of each tested site; N = 2.
(H) Continuous assessment of mRuby expression levels following the integration into each of the tested GSH sites as well as into CCR5 control in Jurkat cells. Data are represented as means ± SEMs of the highest expressing clonal population of each tested site; N = 2.
Using the PITCh approach, we transfected the mRuby transgene into five candidate GSH sites using the best predicted gRNA sequence for each site (see STAR Methods). We then conducted a pooled selection of mRuby-expressing HEK293T and Jurkat cells by fluorescence-activated cell sorting (FACS), followed by expansion for 1 week and single-cell sorting to produce monoclonal populations of mRuby-expressing cells. To determine sites that support long-term stable transgene expression, we monitored clones with homogeneous and high mRuby expression levels by performing flow cytometry at days 30, 45, 60, and 90 after integration.
Of 5 candidate GSH sites, 4 sites in HEK293T cells—GSH1, -2, -7 and -31 (Figures 2C and 2G)—and two sites in Jurkat cells—GSH1 and GSH2 (Figures 2D and 2H)—demonstrated stable mRuby expression levels 90 days after integration. Interestingly, two sites in HEK293T cells, GSH1 and GSH2, allowed for over an order of magnitude higher transgene expression levels as compared to the widely used AAVS1 site throughout the 90-day duration of cell culture (Figure 2G). Similarly, in Jurkat cells, expression levels from GSH1 exceeded those from CCR5, another commonly used gene integration site (Figure 2H). Transgene integration into GSH1 and GSH2 sites was confirmed by genotyping using primer pairs amplifying the junction between tested GSH and the transgene (Figures 2E and 2F). We also observed a high degree of transgene expression heterogeneity between different clonal populations on day 30. This can be attributed to substantial genetic variability and the instability of the studied immortalized cell lines, which can lead to non-uniform gene expression across different clones. Two sites, GSH1 and GSH2, allowed for stable transgene expression in both HEK293T and Jurkat cells. We called these two sites Region Optimal for Gene Insertions 1 and 2 (Rogi1 and Rogi2), respectively, and continued to evaluate their safety and applicability for primary cell engineering.
Transcriptome profiling of cell lines following targeted integration in GSH sites
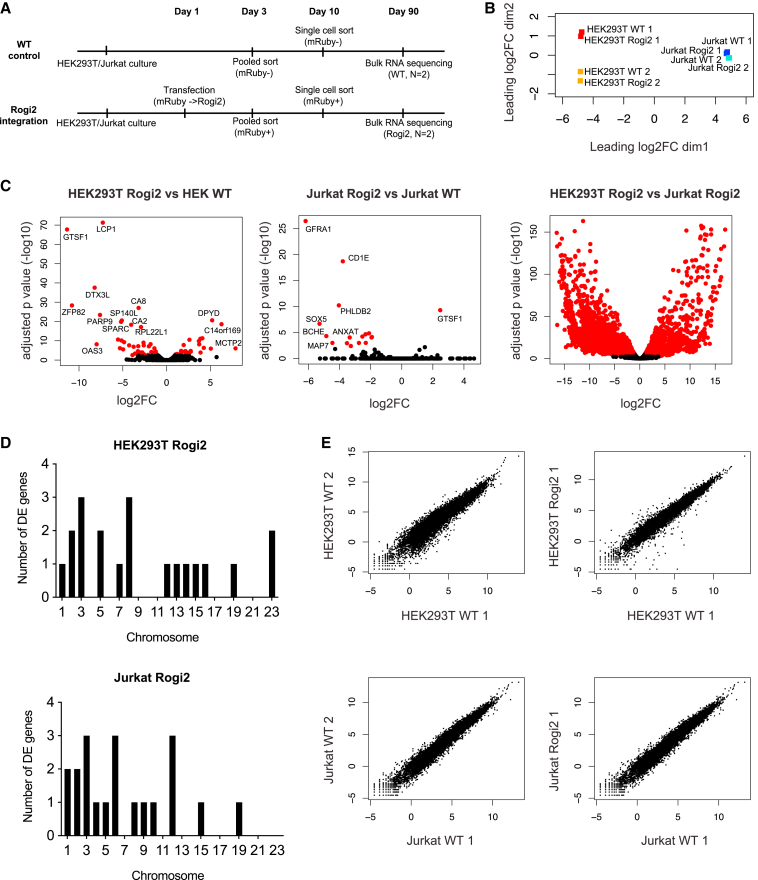
To assess whether targeted integration into the candidate GSH sites resulted in the aberration of the global transcriptome profiles, we performed a bulk RNA-seq and analysis. Following 90 days in culture, the clone showing the highest Rogi2-integrated mRuby levels was compared with untreated cells from the same culture for both HEK293T and Jurkat cells (Figure 3A). Paired-end sequencing on Ilumina NextSeq500 with an average read length of 100 bp and 30 million reads per sample was used on 2 biological replicates of untreated and Rogi2-mRuby cultures of HEK293T and Jurkat cells. We performed a principal-component analysis (PCA) and visualized each sample in two dimensions using the first two PCs. This immediately revealed transcriptional similarity within the integrated and WT samples of the same biological replicate for both cell lines (Figure 3B). While biological variation was observed between the HEK293T samples, the Jurkat samples, both treated and untreated, maintained conserved transcriptional profiles. Performing differential gene expression analysis revealed significant differences between several genes in integrated and unintegrated samples for both cell lines relative to the differences between the two cell types (Figure 3C). It was promising that the most differentially expressed (DE) genes were not shared between Jurkat and HEK293T cell lines, further suggesting that integration in Rogi2 does not systematically alter gene expression. Interestingly, DE genes were scattered across different chromosomes, as opposed to being concentrated within the integrated chromosome where more local contacts exist, again pointing at biological variation (Figure 3D). Furthermore, performing Gene Ontology analysis revealed no significant enrichment of cancer-associated genes or pathways in both HEK and Jurkat cells (Figures S1 and S2), again supporting the potential safety of the Rogi2 site. Lastly, we quantified the differences in gene expression for both cell lines either across biological replicates without Rogi2 integration versus within a biological replicate with or without Rogi2 integration (Figure 3E). Mirroring our PCA (Figure 3B), this analysis again supports that the differences in gene expression we observe arise from biological variation between clones and are not due to integration at Rogi2.
Figure 3.
RNA-seq and transcriptome analysis of HEK293T and Jurkat cells following mRuby integration into Rogi2
(A) Pipeline for bulk RNA-seq experiment on Rogi2-integrated and -non-integrated HEK293T and Jurkat cells.
(B) PCA of 2 biological replicates of HEK293T and Jurkat cells with and without mRuby integration into Rogi2.
(C) Differential expression of genes following Rogi2 integration in HEK293T and Jurkat and comparison of HEK293T and Jurkat non-integrated cells.
(D) Chromosomal distribution of differentially expressed (DE) genes in HEK293T and Jurkat cells. Genes with an adjusted p < 0.05 were considered DE.
(E) Correlation of gene expression between cells with and without mRuby integration into Rogi2 as well as unintegrated biological replicates. See also Figures S1 and S2 for the functional classification of DE genes in HEK and Jurkat, respectively. DE, differentially expressed; FC, fold change.
Targeted integration into GSH sites in primary human T-cells and primary human dermal fibroblasts
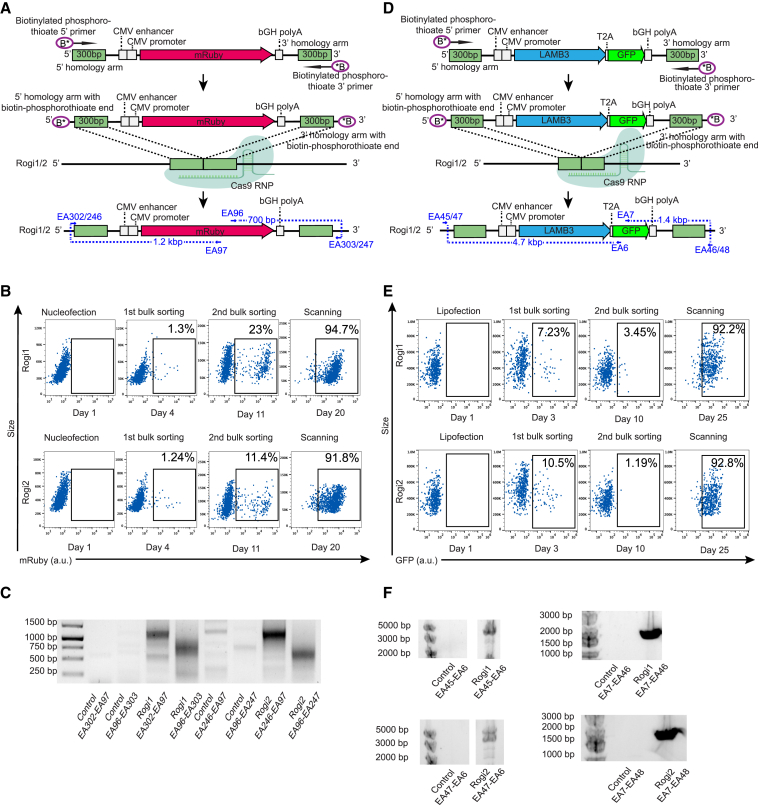
We next sought to characterize targeted integration into Rogi1 and Rogi2 sites in primary human cells. One of the potential applications of targeted integration into GSH sites is for the ex vivo engineering of human T cells, which are being extensively explored for adoptive cell therapies in cancer and autoimmune disease. Thus, we tested Rogi1 and Rogi2 in primary human T cells isolated from the peripheral blood of a healthy donor. This time, we targeted these sites with an HDR-based integration approach using a linear double-stranded DNA donor template, which contained the mRuby transgene driven by a cytomegalovirus (CMV) promoter and with 300-bp homology arms (Figure 4A; Table S2). Phosphorothioate bonds and biotin groups were also added to the 5′ and 3′ ends of the HDR template to increase its stability and prevent concatemerization, respectively (Gutierrez-Triana et al., 2018). Nucleofection of Cas9-gRNA ribonucleoprotein (RNP) complexes and HDR templates into primary T cells resulted in mRuby+ expression in 1.3% of cells for Rogi1 and 1.24% of cells for Rogi2. These mRuby-expressing cells were isolated by FACS on day 4, cultured for another 7 days, when the second round of sorting was performed on the mRuby+ populations. The first sorting step allowed us to isolate all of the cells that possessed double-stranded mRuby donor (either stably integrated or as an extrachromosomal DNA), while the subsequent sort enriched the rare CRISPR knockin events that occurred in the tested GSHs, and extrachromosomal donors were diluted during cellular division. Following these two rounds of pooled sorting, a highly enriched population of T cells stably expressing the mRuby transgene was isolated and cultured for the duration of T cell ex vivo culture (up to day 20), with mRuby expression from Rogi1 and Rogi2 in 94.7% and 91.8% of cells, respectively (Figure 4B). Correct integration into Rogi1 and Rogi2 was confirmed by genotyping and Sanger sequencing using primers amplifying the junction between Rogi1/Rogi2 loci and the mRuby donor (Figure 4C).
Figure 4.
Targeted transgene integration into Rogi1 and Rogi2 in primary human cells
(A) Targeted integration of mRuby into Rogi1 and Rogi2 in primary human T cells by Cas9 HDR.
(B) Flow cytometry plots showing 2 rounds of pooled sorting following mRuby integration into Rogi1 and Rogi2 in primary human T cells.
(C) PCR-based genotyping of Rogi1 and Rogi2 sites using primers spanning the junctions of targeted site and the inserted mRuby transgene in primary human T cells. Lane names correspond to primers shown in (A). Control refers to untransfected T cells, and Rogi1 and Rogi2 refer to T cells transfected with mRuby donors and CRISPR/Cas9 targeting these 2 loci. EA302-EA97 refers to the 5′ integration junction in the Rogi1 locus, EA96-EA303 refers to the 3′ integration junction in the Rogi1 locus, EA246-EA97 refers to the 5′ integration junction in the Rogi2 locus, EA96-EA247 refers to the 3′ integration junction in the Rogi2 locus.
(D) Targeted integration of LAMB3-T2A-GFP into Rogi1 and Rogi2 in primary human dermal fibroblasts by Cas9 HDR.
(E) Flow cytometry plots showing 2 rounds of pooled sorting following LAMB3-T2A-GFP integration into Rogi1 and Rogi2 in primary human dermal fibroblasts.
(F) PCR-based genotyping of Rogi1 and Rogi2 sites using primers spanning the junction of the targeted site and the inserted LAMB3-T2A-GFP transgene in primary human dermal fibroblasts. Lane names correspond to the primers used from (D). Control refers to untransfected dermal fibroblasts, and Rogi1 and Rogi2 refer to dermal fibroblasts transfected with LAMB3-T2A-GFP donors and CRISPR/Cas9 targeting these 2 loci. EA45-EA6 refers to the 5′ integration junction in the Rogi1 locus, EA7-EA46 refers to the 3′ integration junction in the Rogi1 locus, the EA47-EA6 refers to the 5′ integration junction in the Rogi2 locus, and EA8-EA48 refers to the 3′ integration junction in the Rogi2 locus.
Another possible ex vivo application of identified GSH sites includes engineering dermal fibroblasts and keratinocytes for autologous skin grafting in people with burns or inherited skin disorders. A group of genetic skin disorders called junctional epidermolysis bullosa (JEB) is associated primarily with mutations in a family of multi-subunit laminin proteins, which are involved in anchoring the epidermis layer of the skin to derma (Bardhan et al., 2020). Certain variants of JEB are specifically related to mutations in a β subunit of laminin-5 protein, encoded by the LAMB3 gene (Robbins et al., 2001). Using a similar dsDNA HDR donor with 300-bp homology arms possessing phosphorothioate bonds and biotin, we used Cas9 HDR to integrate the LAMB3 gene with GFP (total insert size 5,409 bp) into Rogi1 and Rogi2 sites in primary human dermal fibroblasts isolated from neonatal skin (Figure 4D, Table S2). After the lipofection of fibroblasts with Cas9 and HDR templates, the expression of GFP, which is indicative of LAMB3 expression, was observed in 7.23% (Rogi1) and 10.5% (Rogi2) of cells. These cells were sorted at day 3, cultured for 7 days, and the GFP+ population (3.45% for Rogi1 and 1.19% for Rogi2) was sorted again. Similar to T cells, 2 rounds of pooled sorting led to >92% enrichment of GFP+ cells, with the expression of LAMB3-GFP transgene maintained for the duration of cell culture (>25 days) (Figure 4E). Genotyping and Sanger sequencing confirmed successful integration into both loci by using primers amplifying the junction between Rogi1/Rogi2 and the LAMB3-GFP donor (Figure 4F).
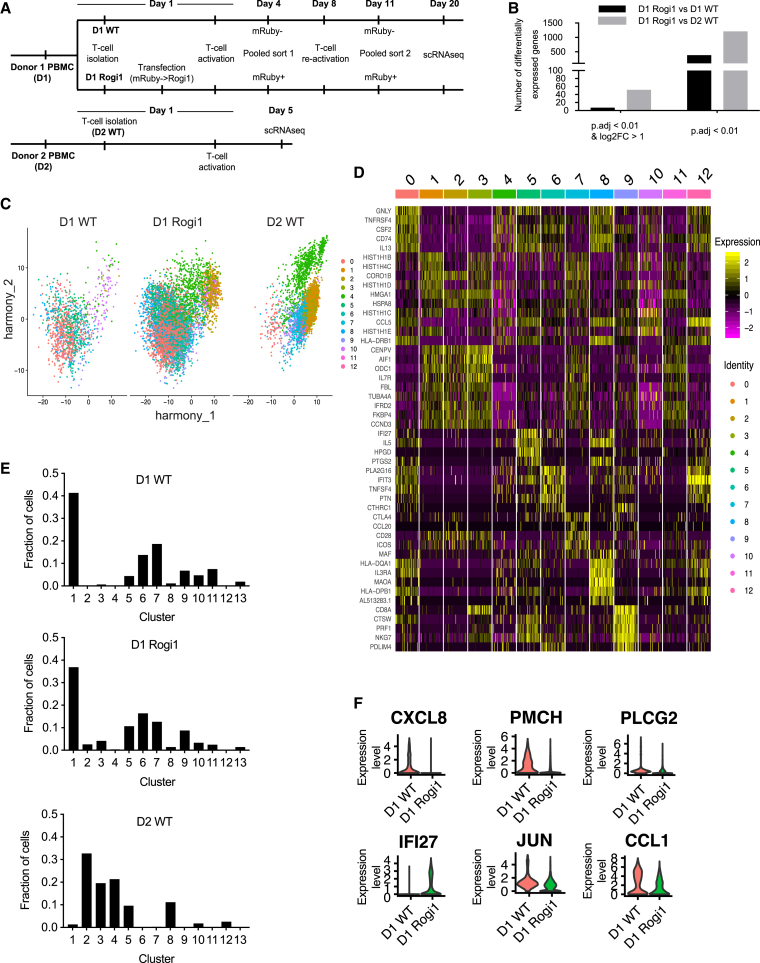
Single-cell RNA-seq and analysis of primary human T cells following transgene integration into a GSH site
Lastly, we assessed transcriptome-wide effects on a single-cell level following transgene integration into Rogi1 in primary T cells. We performed single-cell RNA-seq using the 10X Genomics protocol, which consists of encapsulating cells in gel beads bearing a reverse transcription (RT) reaction mix with unique cell primers. Following the RT reaction, the cDNA was pooled, and the library was amplified for subsequent next-generation sequencing.
This single-cell sequencing workflow was applied to human T cells expressing mRuby in Rogi1 after 25 days in culture; wild-type (non-transfected) cells from the same donor were used as a control. We also compared these cells with WT controls from a different donor to again compare whether GSH integration resulted in more variability in gene expression relative to a biological replicate (Figure 5A). Performing differential gene expression analysis across three samples revealed a trend toward fewer up- or downregulated genes between Rogi1-integrated and non-integrated cells from the same donor relative to the non-integrated, second donor sample (Figure 5B). We performed uniform manifold approximation projection (UMAP) paired with an unbiased clustering based on global gene expression, which resulted in 13 distinct clusters (Figure 5C). Many genes defining these clusters corresponded to typical T cell markers such as IL7R, ICOS, CD28, CCL5, CD74, and NKG7 (Figure 5D). We subsequently quantified the proportion of cells per cluster for each sample, again demonstrating congruent gene expression signatures from cells arising from a single patient, regardless of whether integration into Rogi1 occurred (Figure 5E). Furthermore, similar to bulk RNA-seq results on cell lines, none of the most DE genes that were upregulated in cells with Rogi1 transgene integration were associated with any cancer-related pathways (Figure 5F). Interestingly, the expression of the Jun gene encoding the oncogenic c-Jun transcription factor is decreased in cells bearing transgene integration into Rogi1. Despite the association of this gene with several cancer types, the pathogenesis of these diseases involves excess c-Jun protein presence in the cells (Blau et al., 2012; Brennan et al., 2020). Taken together, both single-cell and bulk RNA-seq data suggest that transcriptomic changes that occur following integration into our computationally determined and experimentally validated GSHs have minimal potential of insertional oncogenesis.
Figure 5.
Single-cell RNA-seq of primary human T cells following targeted transgene integration into Rogi1 site
(A) Pipeline of the RNA-seq experiment following Cas9 HDR targeted integration of mRuby into Rogi1 (Rogi1-mRuby cells) and T cell activation.
(B) Number of DE genes between Rogi1-mRuby T cells and WT T cells (non-integrated) from donor 1, and Rogi1-mRuby T cells from donor 1 and WT T cells from donor 2.
(C) UMAP analysis comparing transcriptional clusters of Rogi1-mRuby and WT T cells from donor 1 and WT T cells from donor 2. Each point represents a unique cell barcode, and each color corresponds to cluster identity.
(D) Expression of genes determining the 7 largest clusters. Intensity corresponds to normalized gene expression.
(E) Distribution of Rogi1-mRuby and WT T cells from donor 1 and WT T cells from donor 2 across different clusters.
(F) Normalized expression for selected differentially expressed genes between Rogi1-mRuby and WT T cells from donor 1.
Discussion
In this study, we used bioinformatic screening to identify GSH sites and performed phenotypic validation by targeted transgene integration in human cell lines and primary cells followed by global transcriptomic analysis. The durability and safety of the identified GSH sites, Rogi1 and Rogi2, was confirmed by long-term transgene expression and the absence of the upregulation of cancer pathway-associated genes following transgene insertion. These findings make the identified sites preferable to currently used AAVS1, CCR5, and hRosa26, which have the drawbacks of being located within functional genes, in gene-dense regions, and surrounded by oncogenes (Sadelain et al., 2012). Although previous studies have also resulted in the discovery of sites capable of long-term expression of transgenes, they were limited by the integration mechanism researchers used. Changes to the entire transcriptome following integration events were not evaluated, as they were focused on differential expression of a handful of genes in the vicinity of the discovered site (Papapetrou et al., 2011). Finally, generalizability of the criteria used to establish our GSH sites suggests their possible applicability to different cell types, expanding the genome engineering toolkit for diverse cell therapy and synthetic biology applications (Nielsen and Voigt, 2014). This cell-type agnostic nature of the predicted sites could be evaluated subsequently by GSH integration of reporter genes in iPSCs followed by differentiation into various cell lineages while observing the stability of the reporter expression.
In addition to compiling new and existing sets of criteria desirable for safe harbor sites, we also proposed a sequence of validation experiments that should be used to confirm the durability and safety of transgene expression from identified sites. These characterization steps involve long-term culture of GSH-integrated cells to prove the stability of gene expression from studied locus over time, as well as transcriptome evaluation of GSH-integrated cells using RNA-seq to confirm the absence of the upregulation of genes involved in the oncogenic proliferation of cells. These approaches allow for qualitative and quantitative validation of computationally predicted sites and can be used for the evaluation of other sites listed in this study. Additional assessments of safe harbor properties of identified sites following GSH-based transgene integration may involve epigenomic and metabolomic studies, comparing changes in chromatin architecture as well as cellular metabolic state using Hi-C and high-throughput mass spectrometry, respectively.
The most immediate use of identified GSH sites may involve the safe and predictable engineering of human T cells for adoptive cell therapy applications (Schwarz and Leonard, 2016). Copious endeavors to design, modify, and augment functions of T cells ex vivo have been successfully initiated in research labs (Baeuerle et al., 2019; Eyquem et al., 2017). However, most strategies have relied on viral-mediated delivery, which results in random transgene integration and is thus associated with the risk of insertional oncogenesis, potentially leading to cancerous transformations of engineered cells and the unpredictability of transgene expression levels associated with the nature of the integration locus and frequent silencing of the integrated construct. Performing targeted integration into GSH sites would enable long-term transgene expression in a safe manner and would support advanced efforts in engineered T cell therapies such as armored chimeric antigen receptor (CAR)-T cells, capable of overcoming hostile tumor microenviroments (Yeku et al., 2017), as well as T cells bearing synthetic receptors that introduce logic gates into the behavior of a cell, allowing for safer and more effective treatments (Morsut et al., 2016; Roybal and Lim, 2017). In addition, given the demonstrated efficiency in dermal fibroblasts, we envision a rapid application of the discovered sites to skin engineering, particularly in the context of treatment of the inherited skin disorders, wound healing, and skin rejuvenation. Finally, the enhancement of the transgene integration efficiency into sites we identified can be achieved by engineering different naturally occurring site-specific recombinases, known for the high on-target knockin activity, and directing them against our GSHs.
Another exciting aspect of the identified GSH sites is the level of transgene expression observed, especially in HEK293T cells, which are known to be suitable for large-scale production of therapeutic proteins. High levels of reporter gene expression from Rogi1 and Rogi2 in HEK293T that was sustained for >3 months and exceeded expression levels from the AAVS1 site was observed in several clonal populations of integrated cells. This high expression level can be enhanced further by multiple biallelic integration events into identified loci and can thus be exploited for the durable large-scale production of commercially valuable proteins.
In summary, two human genomic safe harbor sites, Rogi1 and Rogi2, identified and validated in this study may serve as a robust and safe platform for a variety of clinically and industrially relevant cell engineering efforts, culminating in safer and more reliable gene and cell therapies.
Limitations of the study
In this work, we presented two genomic regions capable of the safe and durable expression of genes of interest in a variety of cellular contexts. We used CRISPR-based knockin for targeted gene insertions and genotyped on-target integration events; however, we did not assess the potential occurrence of non-specific integrations. Given the low efficiency of CRIPSR knockins for large DNA donors, observed in this and previous studies, we predict the likelihood of such off-target events to be extremely low. Conducting a genome-wide search of non-specific insertions would further alleviate this concern. We have also observed a clonal heterogeneity of the reporter expression from the identified GSHs in investigated HEK293 and Jurkat T cell lines. This can be attributed to substantial genetic variability between different clonal populations of these immortalized, heavily mutated cell lines. We observe a lower degree of gene expression heterogeneity in cells with more homogeneous genetic backgrounds, such as primary donor-derived cells. Finally, given the complexity as well as resource-intensive nature of the single-cell RNA-seq experiments, we were only able to process single replicates of each of the primary T cell samples. Nevertheless, we believe that the single-cell RNA-seq data we generated in this study is representative of a trend for fewer gene expression changes between integrated and non-integrated cells from the same donor as compared to non-integrated cells from a different donor. The inclusion of additional replicates would augment the safety claim of the discovered sites.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| NEB 5-alpha | New England Biolabs | C2987 |
| pcDNA3-mRuby2 | Addgene | 40260 |
| pEF-GFP | Addgene | 11154 |
| pENTR CMV-mRuby-bGH | Twist Biosciences | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Q5 High-Fidelity 2X Master Mix | New England Biolabs | M0492S |
| Gibson Assembly Master Mix | New England Biolabs | E2611L |
| Human IL-2 | Peprotech | 200-02 |
| Alt-R crRNA | IDT | N/A |
| Alt-R tracrRNA | IDT | 1072534 |
| Alt-R SpCas9 Nuclease V3 | IDT | 1081059 |
| CRISPRevolution sgRNA EZ Kit | Synthego | N/A |
| SpCas9 2NLS Nuclease | Synthego | N/A |
| Critical commercial assays | ||
| SPRIselect | Beckman Coulter | B23318 |
| SF Cell line kit | Lonza | V4XC-2012 |
| SE Cell line kit | Lonza | V4XC-1012 |
| P3 Primary Cell kit | Lonza | V4XP-3032 |
| Dynabeads Human T-Activator CD3/CD28 | Thermo Fischer Scientific | 11161D |
| EasySep Human T Cell Isolation kit | Stemcell Technologies | 17951 |
| Lipofectamine CRISPRMAX Cas9 Transfection Reagent | Thermo Fischer Scientific | CMAX00001 |
| PureLink Genomic DNA extraction kit | Thermo Fischer Scientific | K1820-01 |
| Zymoclean Gel DNA Recovery Kit | Zymo Research | D4001 |
| TOPO-vector using Zero-blunt TOPO PCR Cloning Kit | Thermo Fischer Scientific | 450245 |
| PureLink RNA Mini Kit | Thermo Fischer Scientific | 12183018A |
| RiboCop rRNA Depletion Kit | Lexogen | 144 |
| SuperScript Double-Stranded cDNA Synthesis Kit | Thermo Fischer Scientific | 11917010 |
| ZR Plasmid Miniprep - Classic kit | Zymo Research | D4015 |
| Chromium Single Cell 3ʹ GEM, Library & Gel Bead Kit v3 | 10-X Genomics | PN-1000075 |
| Chromium Single Cell B Chip | 10-X Genomics | 1000074 |
| Deposited data | ||
| Predicted GSH sites | This paper-Table S1 | N/A |
| Bulk RNA-sequencing data | This paper | ENA: E-MTAB-11289 |
| Single-cell RNA-sequencing data | This paper | ENA: E-MTAB-11289 |
| Protein coding gene coordinates | GENCODE gene annotation (Release 24) | https://www.gencodegenes.org/human/release_24.html |
| Oncogenes coordinates | Cancer Gene Census | https://cancer.sanger.ac.uk/census |
| miRNA coordinates | MirGeneDB | Fromm et al. (2020) |
| Enhancer coordinates | EnhancerAtlas 2.0 | Gao and Qian (2019) |
| tRNA coordinates | GENCODE gene annotation (Release 24) | https://www.gencodegenes.org/human/release_24.html |
| lncRNA coordinates | GENCODE gene annotation (Release 24) | https://www.gencodegenes.org/human/release_24.html |
| Telomere coordinates | UCSC genome browser GRCh38/hg38 | https://genome.ucsc.edu |
| Centromere coordinates | UCSC genome browser GRCh38/hg38 | https://genome.ucsc.edu |
| Experimental models: Cell lines | ||
| HEK293T cell line | ATCC | CRL-3216 |
| Jurkat E6-1 T cell line | ATCC | TIB152 |
| Human PBMC | Stemcell Technologies | 70025 |
| Neonatal human dermal fibroblasts | Coriell Institute | GM03377 |
| Oligonucleotides | ||
| Rogi1/2 genotyping primers | This paper-Table S3 | N/A |
| Recombinant DNA | ||
| LAMB3 cDNA | Genscript | NM_000228.3 |
| Rogi1 HDR mRuby donor | This paper | Addgene #179860 |
| Rogi2 HDR mRuby donor | This paper | Addgene #179861 |
| Rogi1 HDR LAMB3-T2A-GFP donor | This paper | Addgene #179864 |
| Rogi2 HDR LAMB3-T2A-GFP donor | This paper | Addgene #179865 |
| AAVS1/CCR5 MMEJ PITCh mRuby donors | This paper | N/A |
| GSH1, 2, 7, 8, 31 MMEJ PITCh mRuby donors | This paper | N/A |
| Software and algorithms | ||
| GSH prediction algorithm | This paper | https://doi.org/10.5281/zenodo.5786825 |
| BEDtools | Quinlan and Hall (2010) | http://github.org/pezmaster31/bamtools |
| Platypus | Yermanos et al. (2021) | https://github.com/alexyermanos/Platypus |
| GraphPad Prism v8.3.1 for MacOS | GraphPad software | N/A |
| Subread v1.6.2 | Liao et al. (2013) | http://subread.sourceforge.net |
| Rsubread | Liao et al. (2014) | https://bioconductor.org/packages/release/bioc/html/Rsubread.html |
| edgeR | Robinson et al. (2010) | https://bioconductor.org/packages/release/bioc/html/edgeR.html |
| CellRanger | 10XGenomics | https://support.10xgenomics.com/single-cell-gene-expression/software/downloads/latest |
| Seurat | Stuart et al. (2019) | https://satijalab.org/seurat/ |
| Harmony | Korsunsky et al. (2019) | https://portals.broadinstitute.org/harmony/ |
| Genenious Prime 2019.2.3 | Biomatters Ltd. | N/A |
| FlowJo 10.8 | Becton Dickinson & Company | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr Sai Reddy, sai.reddy@bsse.ethz.ch.
Materials availability
Plasmid used to produce HDR donors targeting Rogi1 and Rogi2 sites have been deposited to Addgene. ID numbers are provided in the Key Resources Table. PITCh plasmids used for the initial GSH screen will be provided upon request.
Experimental model and subject details
HEK293T cell line
-
•
HEK293T cells were obtained from the American Type Culture Collection (ATCC) (#CRL-3216).
-
•
Cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) (ATCC 30-2002) supplemented with 2 mM L-glutamine (ATCC 30-2214), 10% FBS (Thermo Fischer Scientific, #A4766801), 50 U ml−1 penicillin and 50 μg mL−1 streptomycin (Thermo Fischer Scientific, #15070063).
-
•
Detachment of HEK cells for passaging was performed using the TrypLE reagent (Thermo Fisher Scientific, #12605010).
-
•
Cells were cultured at 37°C, 5% CO2 in a humidified atmosphere.
Jurkat leukemia E6-1 T cell line
-
•
Jurkat cell line was obtained from ATCC (#TIB152).
-
•
Jurkat cells were cultured in ATCC-modified RPMI-1640 (Thermo Fisher Scientific, #A1049101), 10% FBS (Thermo Fischer Scientific, #A4766801), 50 U ml−1 penicillin and 50 μg mL−1 streptomycin (Thermo Fischer 15070063).
-
•
Cells were cultured at 37°C, 5% CO2 in a humidified atmosphere.
Primary human T cells
-
•
Human peripheral blood mononuclear cells were purchased from Stemcell Technologies (#70025).
-
•
T cells isolated using the EasySep Human T Cell Isolation kit (Stemcell Technologies, #17951).
-
•
Primary human T cells were cultured for up to 20 days in X-VIVO 15 Serum-free Hematopoietic Cell Medium (Lonza, #BE02-060Q), 50 U ml−1 penicillin and 50 μg ml−1 streptomycin (Thermo Fischer Scientific, #15070063), as well as freshly added 20 ng ml−1 recombinant human IL-2, (Peprotech, #200-02).
-
•
Cells were cultured at 37°C, 5% CO2 in a humidified atmosphere.
Neonatal human dermal fibroblasts
-
•
Neonatal human dermal fibroblasts were purchased from Coriell Institute (Catalog ID GM03377).
-
•
Primary fibroblasts were cultured for up to 25 days in Prime Fibroblast media (CELLNTEC, CnT-PR-F).
-
•
Cells were passaged at 70% confluency using Accutase (CELLNTEC, CnT-Accutase-100). Detached cells were centrifuged for 5 min, 200 × g at room temperature and seeded at 2,000 cells per cm2.
-
•
Fibroblasts were cultured at 37°C, 5% CO2 in a humidified atmosphere.
Method details
Computational search for GSH sites
Previously established criteria (Sadelain et al., 2012) as well as newly introduced ones were used to predict genomic locations of GSHs. Specifically, coordinates of all known genes were extracted from GENCODE gene annotation (Release 24). A set of tier 1 and tier 2 oncogenes was obtained from Cancer Gene Census. The miRNA coordinates were obtained from MirGeneDB (Fromm et al., 2020). Enhancer regions were obtained from the EnhancerAtlas 2.0 database (Gao and Qian, 2019), coordinates were transposed into GRCh38/hg38 genome and union of enhancer sites was used. Genomic locations of tRNA and lncRNA were extracted from GENCODE gene annotation (Release 24). UCSC genome browser GRCh38/hg38 was used to obtain the coordinates of telomeres and centromeres as well as unannotated regions. BEDTools (Quinlan and Hall, 2010) were used to determine flanking regions of each element of the criteria as well as to obtain union or difference between sets of coordinates. The custom source code developed for the computational identification of human genomic safe harbor sites is available at https://github.com/erikaznauryan/GSH-1.
Plasmids, guide RNA design and HDR donor generation
PITCh plasmids were generated through standard cloning methods. CMV-mRuby-bGH insert was amplified using Q5 High-Fidelity 2X Master Mix (New England Biolabs, #M0492S) from pcDNA3-mRuby2 plasmid (Addgene, Plasmid #40260) with primers containing mircohomology sequences against specific GSHs, AAVS1 and CCR5 sites with 10bp of overlapping ends for the pcDNA3 backbone. The pcDNA3 backbone was amplified with primers containing sequences of PITCh gRNA cut site (GCATCGTACGCGTACGTGTTTGG) on both 5′ and -3′ ends of the backbone. The insert and the backbone were assembled using Gibson Assembly Master Mix (New England Biolabs, #E2611L) and NEB 5-alpha (New England Biolabs, #C2987) cells were transformed.
Guide RNA sequences for five tested GSH sites were predicted using Geneious gRNA design tool. Briefly, coordinates of the predicted GSH sites were pasted into UCSC Genome Browser GRCh38/hg38 and DNA sequences were extracted and transferred into Geneious. An internal gRNA design tool was used to identify gRNA sequences located in the predicted GSHs against the entire human genome. The evaluation of the efficacy of double-stranded break generation (on-target activity) was based on Doench et al. (2016), while the specificity of the gRNA-induced break (off-target activity) was assessed based on Hsu et al. (2013). Guide RNAs with high on-target and off-target scores were used to target predicted GSHs.
Plasmids encoding CMV-mRuby-bGH flanked by Rogi1/Rogi2 300bp homology arms were ordered from Twist Biosciences in pENTR vector. HDR donors were amplified from these plasmids using Q5 High-Fidelity 2X Master Mix (New England Biolabs, #M0492S) and biotinylated primers with phosphorothioate bonds between the first 5 nucleotides on both 5′ and -3′ ends. Plasmid encoding CMV-LAMB3-T2A-GFP-bGH was generated by overlap extension PCR of LAMB3 cDNA, purchased from Genscript (NM_000228.3) and GFP-bGH sequence from Addgene (Plasmid #11154). T2A sequence was added to 5’primer amplifying GFP-bGH. Produced insert was cloned into the abovementioned pENTR vector from Twist Biosciences bearing Rogi1 and Rogi2 300bp homology arms as well as CMV promoter sequence using Gibson Assembly Master Mix (NEB, #E2611L). HDR donors were amplified from these plasmids using biotinylated primers with phosphorothioate bonds between the first 5 nucleotides on both 5′ and -3′ ends. HDR donors were then purified from PCR mix using SPRIselect beads (Beckman Coulter, #B23318) at 0.4X beads to PCR mix ratio.
HEK293T and Jurkat cell transfection and sorting
Prior to transfection of HEK293T and Jurkat, gRNA molecules were assembled by mixing 4 μl of custom Alt-R crRNA (200 μM, IDT) with 4 μL of Alt-R tracrRNA (200 μM, IDT, #1072534), incubating the mix at 95°C for 5 min and cooling it to room temperature. 2 μL of assembled gRNA molecules were mixed with 2 μL of recombinant Alt-R SpCas9 (61 μM, IDT, #1081059) and incubated for 10 min at room temperature to generate Cas9 RNP complexes.
For transfection of HEK cells 100 μL format SF Cell line kit (Lonza, V4XC-2012) and electroporation program CM-130 was used on the 4D-Nucleofector. 1 × 106 HEK cells were transfected with 2 μg of PITCh donor, 2 μl of Cas9 RNP complex against specific GSH and 2 μl of Cas9 RNP complex against PITCh plasmid to liberate MMEJ insert.
For transfection of Jurkat cells 100 μL format SE Cell line kit (Lonza, V4XC-1012) and electroporation program CL-120 was used on the 4D-Nucleofector. 1 × 106 Jurkat cells were transfected with 2 μg of PITCh donor, 2 μl of Cas9 RNP complex against specific GSH and 2 μl of Cas9 RNP complex against PITCh plasmid to liberate MMEJ insert.
Transfected HEK and Jurkat cells were bulk sorted on day 3 and single-cell sorted on day 10 following transfection using Sony SH800S sorter. Best expressing clone was selected on day 30, split into two wells and cultured for another 2 months. mRuby expression of the best expressing clone was analyzed on BD LSRFortessa Flow Cytometer on day 45, 60 and 90 following transfection.
Human T-cells transfection and sorting
On day 1 of culture, transfection of primary T cells with Cas9 RNP complexes and Rogi1/Rogi2-mRuby HDR templates was performed using the 4D-Nucleofector and a 20 uL format P3 Primary Cell kit (Lonza, V4XP-3032). Briefly, gRNA molecules were assembled by mixing 4 μl of custom Alt-R crRNA (200 μM, IDT) with 4 μL of Alt-R tracrRNA (200 μM, IDT, #1072534), incubating the mix at 95°C for 5 min and cooling it to room temperature. 2 μL of assembled gRNA molecules were mixed with 2 μL of recombinant Alt-R SpCas9 (61 μM, IDT, #1081059) and incubated for 10 min at room temperature to generate Cas9 RNP complexes. 1 × 106 primary T cells were transfected with 1 μg of HDR template, 1 μl of Rogi1/Rogi2 Cas9 RNP complex using the EO115 electroporation program. T cells were activated with Dynabeads Human T-Activator CD3/CD28 (Thermo Fischer Scientific, #11161D) 3–4 hours following transfection. mRuby-positive T-cells were bulk sorted on day 4 using Sony SH800S sorter, re-activated with the new beads on day 8, sorted again on day 11 and analyzed on BD LSRFortessa Flow Cytometer on day 20.
Human dermal fibroblasts transfection and sorting
Fibroblasts were transfected using Lipofectamine CRISPRMAX Cas9 Transfection Reagent (ThermoFisher Scientific, CMAX00001). Briefly, cells were transfected at 50% confluency with 1:1 ratio of custom sgRNA (40 pmoles, Synthego) and SpCas9 (40pmoles, Synthego) and 2.5 μg of Rogi1/Rogi2 LAMB3-T2A-GFP HDR template. GFP-positive fibroblasts were bulk sorted on day 3 and 10 using Sony SH800S sorter and analyzed on BD LSRFortessa Flow Cytometer on day 25.
Genotypic analysis of GSH integration
Genomic DNA was extracted from 1 × 106 cells using PureLink Genomic DNA extraction kit (ThermoFischer Scientific, #K1820-01). 5 μL of genomic DNA extract were then used as templates for 25 μL Q5 High-Fidelity 2X Master Mix (New England Biolabs, #M0492S) PCR reactions using a primer with one primer residing outside of the homology arm of the integrated sequence and the other primer inside the integrated sequence. Obtained bands were gel extracted using Zymoclean Gel DNA Recovery Kit (Zymo Research, #D4001), 4ul of eluted DNA was cloned into a TOPO-vector using Zero-blunt TOPO PCR Cloning Kit (Thermo Fischer Scientific, #450245), incubated for 1 hour, transformed into NEB 5-alpha Competent E. coli cells (New England Biolabs, C2987H) and plated on agar plates containing kanamycin at 50 μg/ml. Produced clones were picked and inoculated for overnight culture in 5ml of liquid broth supplemented with kanamycin at 50 μg/ml. Liquid cultures were mini-prepped the following morning using ZR Plasmid Miniprep - Classic kit (Zymo Research, #D4015) and Sanger sequenced by Microsynth using M13-forward and M13-reverse standard primers.
Bulk RNA-sequencing of HEK293T and Jurkat cells Rogi2 and WT
Following single-cell sort, the best expressing clone (Rogi2) and wild-type (WT) of HEK293T and Jurkat cells were split into 2 wells (1 and 2) and cultured for 80 days, after which total RNA was extracted using PureLink RNA Mini Kit (Thermo Fischer Scientific, #12183018A). Extracted total RNA was depleted of rRNA using RiboCop rRNA Depletion Kit (Lexogen, #144), first and second strands of cDNA were generated with SuperScript Double-Stranded cDNA Synthesis Kit (Thermo Fischer Scientific, #11917010) using random hexamers and flow cell adapters were ligated to the produced double-stranded cDNA. DNA fragments were enriched by PCR using Q5 High-Fidelity 2X Master Mix (New England Biolabs, #M0492S) and sequenced by the Illumina NextSeq 500 system in the Genomics Facility Basel. Sequencing reads were aligned to the human reference genome (GRCh38) using Subread (v1.6.2) using unique mapping (Liao et al., 2013). Expression levels were quantified using the featureCounts function in the Rpackage Rsubread at gene-level (Liao et al., 2014). Normalization across the samples was performed using default parameters in the Rpackage edgeR (Robinson et al., 2010). Differential expression analysis was performed using the exactTest function in the edgeR package. Gene ontology was performed by supplying those differentially expressed genes (adjusted p value < 0.05) to the goana function (Young et al., 2010).
Single-cell RNA sequencing of human T-cells
Single-cell RNA sequencing was conducted on day 20 of culture for Donor 1 WT (D1 WT) and Donor 1 Rogi1 (D1 Rogi1) and on day 5 for Donor 2 WT (D2 WT). Single cell 10X libraries were constructed from the isolated single cells following the Chromium Single Cell 3ʹ GEM, Library & Gel Bead Kit v3 (10X Genomics, PN-1000075). Briefly, single cells were co- encapsulated with gel beads (10X Genomics, 2000059) in droplets using Chromium Single Cell B Chip (10X Genomics, 1000074). Final D1 WT, D1 Rogi1 and D2 WT libraries were pooled and sequenced on the Illumina NovaSeq platform (26/8/0/93 cycles). Raw sequencing files were supplied to cellranger (v3.1.0) using the count argument under default parameters and the human reference genome (GRCh38-3.0.0). Filtering, normalization and transcriptome analysis was performed using a previously described pipeline in the R package Platypus (Yermanos et al., 2021). Briefly, filtered gene expression matrices from cellranger were supplied as input into the Read10x function in the R package Seurat (Stuart et al., 2019). Cells containing more than 5% mitochondrial genes, or less than 150 unique genes detected were filtered out before using the RunPCA function and subsequent normalization using the function RunHarmony from the Harmony package under default parameters (Korsunsky et al., 2019). Uniform manifold approximation projection was performed with Seurat’s RunUMAP function using the first 20 dimensions and the previously computed Harmony reduction. Clustering was performed by the Seurat functions FindNeighbors and FindClusters using the Harmony reduction and first 20 principal components and the default cluster resolution of 0.5, respectively (Satija et al., 2015). Cluster-specific genes were determined by Seurat’s FindMarkers function for those genes expressed in at least 25% of cells in one of the two groups. Differential genes between samples were calculated using the FindMarkers function from Seurat using the default Wilcoxon Rank Sum Test with Bonferroni multiple hypothesis correction. The source code for the analysis of scRNA-seq data is available at https://github.com/alexyermanos/Platypus.
Quantification and statistical analysis
Specific quantification and statistical analysis details for each experiment can be found in the method details section and in the figure legends. Statistical analysis was performed using the software GraphPad. For each of the timepoints in Figures 2G and 2H, two biological replicates were used to produce mean and SEM. For Figure 3, two biological replicates of WT and Rogi2 integrated HEK293T and Jurkat cells were used at day 90 post-transfection to generate bulk RNA-sequencing data. For Figure 5, one replicate of each T cell sample – D1 WT, D1 Rogi1 and D2 – were used to generate single-cell RNA-sequencing data. Biological replicates are cells that were transfected together and sorted together, and subsequently cultured individually.
Acknowledgments
The authors would like to thank Genomics Facility Basel, particularly Dr Christian Beisel, Mrs Katja Eschbach, and Mrs Elodie Burcklen, and the Single Cell Facility of the BSSE department of ETH Zürich, particularly Dr Thomas Horn and Dr Mariangela Di Tacchio. The authors also thank Mr Eric Zigon from the Imaging Core at the Wyss Institute. This study is supported by ETH Research Grants (to S.T.R.), Helmut Horten Stiftung (to S.T.R.) and Aging and Longevity-Related Research Fund at HMS (to G.M.C.). E.A. is the recipient of the Genome Engineer Innovation Grant 2019 from Synthego.
Author contributions
E.A. and S.T.R. designed the study; E.A., A.Y., A.D., E. Kapetanovic, D.M., G.M.C., and S.T.R. contributed to the experimental design; E.A. performed the experiments; E.A. and E. Kinzina developed the bioinformatic pipeline for GSH identification; E.A. and A.Y. analyzed the data; G.M.C. and S.T.R. supervised the work; E.A., A.Y., and S.T.R. wrote the manuscript, with input from all of the authors.
Declaration of interests
ETH Zürich and Harvard University have filed for patent protection on the technology described herein, and E.A., D.M., S.T.R., and G.M.C are named as co-inventors on the patent. The full disclosure for G.M.C. is available at http://arep.med.harvard.edu/gmc/tech.html.
Published: January 14, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.crmeth.2021.100154.
Supplemental information
Data and code availability
-
•
All original code has been deposited at GitHub and the release pertaining this publication is publicly available through Zenodo. DOIs are listed in the key resources table.
-
•
Bulk and single-cell RNA-sequencing data reported in this paper are available at the European Nucleotide Archive. Accession numbers are listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Abraham R.T., Weiss A. Jurkat T cells and development of the T-cell receptor signalling paradigm. Nat. Rev. Immunol. 2004;4:301–308. doi: 10.1038/nri1330. [DOI] [PubMed] [Google Scholar]
- Akagi K. RTCGD: retroviral tagged cancer gene database. Nucleic Acids Res. 2004;32:523D–527. doi: 10.1093/nar/gkh013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeuerle P.A., Ding J., Patel E., Thorausch N., Horton H., Gierut J., Scarfo I., Choudhary R., Kiner O., Krishnamurthy J., et al. Synthetic TRuC receptors engaging the complete T cell receptor for potent anti-tumor response. Nat. Commun. 2019;10:2087. doi: 10.1038/s41467-019-10097-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardhan A., Bruckner-Tuderman L., Chapple I.L.C., Fine J.-D., Harper N., Has C., Magin T.M., Marinkovich M.P., Marshall J.F., McGrath J.A., et al. Epidermolysis bullosa. Nat. Rev. Dis. Primer. 2020;6:78. doi: 10.1038/s41572-020-0210-0. [DOI] [PubMed] [Google Scholar]
- Barzel A., Paulk N.K., Shi Y., Huang Y., Chu K., Zhang F., Valdmanis P.N., Spector L.P., Porteus M.H., Gaensler K.M., et al. Promoterless gene targeting without nucleases ameliorates haemophilia B in mice. Nature. 2015;517:360–364. doi: 10.1038/nature13864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestor T.H. Gene silencing as a threat to the success of gene therapy. J. Clin. Invest. 2000;105:409–411. doi: 10.1172/JCI9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blau L., Knirsh R., Ben-Dror I., Oren S., Kuphal S., Hau P., Proescholdt M., Bosserhoff A.-K., Vardimon L. Aberrant expression of c-Jun in glioblastoma by internal ribosome entry site (IRES)-mediated translational activation. Proc. Natl. Acad. Sci. U S A. 2012;109:E2875–E2884. doi: 10.1073/pnas.1203659109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan A., Leech J.T., Kad N.M., Mason J.M. Selective antagonism of cJun for cancer therapy. J. Exp. Clin. Cancer Res. 2020;39:184. doi: 10.1186/s13046-020-01686-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman F.D. Retroviral insertional mutagenesis in humans: evidence for four genetic mechanisms promoting expansion of cell clones. Mol. Ther. 2020;28:352–356. doi: 10.1016/j.ymthe.2019.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.-K., Blanco M., Jackson C., Aznauryan E., Ollikainen N., Surka C., Chow A., Cerase A., McDonel P., Guttman M. Xist recruits the X chromosome to the nuclear lamina to enable chromosome-wide silencing. Science. 2016;354:468–472. doi: 10.1126/science.aae0047. [DOI] [PubMed] [Google Scholar]
- Chen W., Tan L., Zhou Q., Li W., Li T., Zhang C., Wu J. AAVS1 site-specific integration of the CAR gene into human primary T cells using a linear closed-ended AAV-based DNA vector. J. Gene Med. 2020;22:e3157. doi: 10.1002/jgm.3157. [DOI] [PubMed] [Google Scholar]
- Chin C.L., Goh J.B., Srinivasan H., Liu K.I., Gowher A., Shanmugam R., Lim H.L., Choo M., Tang W.Q., Tan A.H.-M., et al. A human expression system based on HEK293 for the stable production of recombinant erythropoietin. Sci. Rep. 2019;9:16768. doi: 10.1038/s41598-019-53391-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench J.G., Fusi N., Sullender M., Hegde M., Vaimberg E.W., Donovan K.F., Smith I., Tothova Z., Wilen C., Orchard R., et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 2016;34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droz-Georget Lathion S., Rochat A., Knott G., Recchia A., Martinet D., Benmohammed S., Grasset N., Zaffalon A., Besuchet Schmutz N., Savioz-Dayer E., et al. A single epidermal stem cell strategy for safe ex vivo gene therapy. EMBO Mol. Med. 2015;7:380–393. doi: 10.15252/emmm.201404353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis D.J. Silencing and variegation of gammaretrovirus and lentivirus vectors. Hum. Gen. Ther. 2005;16:1241–1246. doi: 10.1089/hum.2005.16.1241. [DOI] [PubMed] [Google Scholar]
- Eyquem J., Mansilla-Soto J., Giavridis T., van der Stegen S.J.C., Hamieh M., Cunanan K.M., Odak A., Gönen M., Sadelain M. Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature. 2017;543:113–117. doi: 10.1038/nature21405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W., Bhattacharyya S.N., Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Friedrich G., Soriano P. Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev. 1991;5:1513–1523. doi: 10.1101/gad.5.9.1513. [DOI] [PubMed] [Google Scholar]
- Fromm B., Domanska D., Høye E., Ovchinnikov V., Kang W., Aparicio-Puerta E., Johansen M., Flatmark K., Mathelier A., Hovig E., et al. MirGeneDB 2.0: the metazoan microRNA complement. Nucleic Acids Res. 2020;48:D132–D141. doi: 10.1093/nar/gkz885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaidukov L., Wroblewska L., Teague B., Nelson T., Zhang X., Liu Y., Jagtap K., Mamo S., Tseng W.A., Lowe A., et al. A multi-landing pad DNA integration platform for mammalian cell engineering. Nucleic Acids Res. 2018;46:4072–4086. doi: 10.1093/nar/gky216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao T., Qian J. EnhancerAtlas 2.0: an updated resource with enhancer annotation in 586 tissue/cell types across nine species. Nucleic Acids Res. 2019;48:D58–D64. doi: 10.1093/nar/gkz980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Triana J.A., Tavhelidse T., Thumberger T., Thomas I., Wittbrodt B., Kellner T., Anlas K., Tsingos E., Wittbrodt J. Efficient single-copy HDR by 5’ modified long dsDNA donors. ELife. 2018;7:e39468. doi: 10.7554/eLife.39468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M., Amit I., Garber M., French C., Lin M.F., Feldser D., Huarte M., Zuk O., Carey B.W., Cassady J.P., et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S., Garrigue A., Wang G.P., Soulier J., Lim A., Morillon E., Clappier E., Caccavelli L., Delabesse E., Beldjord K., et al. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J. Clin. Invest. 2008;118:3132–3142. doi: 10.1172/JCI35700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch T., Rothoeft T., Teig N., Bauer J.W., Pellegrini G., De Rosa L., Scaglione D., Reichelt J., Klausegger A., Kneisz D., et al. Regeneration of the entire human epidermis using transgenic stem cells. Nature. 2017;551:327–332. doi: 10.1038/nature24487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S.G., Yada R.C., Choi K., Carpentier A., Liang T.J., Merling R.K., Sweeney C.L., Malech H.L., Jung M., Corat M.A.F., et al. Rhesus iPSC safe harbor gene-editing platform for stable expression of transgenes in differentiated cells of all germ layers. Mol. Ther. 2017;25:44–53. doi: 10.1016/j.ymthe.2016.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O., et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irion S., Luche H., Gadue P., Fehling H.J., Kennedy M., Keller G. Identification and targeting of the ROSA26 locus in human embryonic stem cells. Nat. Biotechnol. 2007;25:1477–1482. doi: 10.1038/nbt1362. [DOI] [PubMed] [Google Scholar]
- Jiao X., Nawab O., Patel T., Kossenkov A.V., Halama N., Jaeger D., Pestell R.G. Recent advances targeting CCR5 for cancer and its role in immuno-oncology. Cancer Res. 2019;79:4801–4807. doi: 10.1158/0008-5472.CAN-19-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joy M.T., Ben Assayag E., Shabashov-Stone D., Liraz-Zaltsman S., Mazzitelli J., Arenas M., Abduljawad N., Kliper E., Korczyn A.D., Thareja N.S., et al. CCR5 is a therapeutic target for recovery after stroke and traumatic brain injury. Cell. 2019;176:1143–1157.e13. doi: 10.1016/j.cell.2019.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korsunsky I., Millard N., Fan J., Slowikowski K., Zhang F., Wei K., Baglaenko Y., Brenner M., Loh P., Raychaudhuri S. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods. 2019;16:1289–1296. doi: 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.S., Kildegaard H.F., Lewis N.E., Lee G.M. Mitigating clonal variation in recombinant mammalian cell lines. Trends Biotechnol. 2019;37:931–942. doi: 10.1016/j.tibtech.2019.02.007. [DOI] [PubMed] [Google Scholar]
- Liao Y., Smyth G.K., Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013;41:e108. doi: 10.1093/nar/gkt214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Lombardo A., Cesana D., Genovese P., Di Stefano B., Provasi E., Colombo D.F., Neri M., Magnani Z., Cantore A., Lo Riso P., et al. Site-specific integration and tailoring of cassette design for sustainable gene transfer. Nat. Methods. 2011;8:861–869. doi: 10.1038/nmeth.1674. [DOI] [PubMed] [Google Scholar]
- Maeder M.L., Gersbach C.A. Genome-editing technologies for gene and cell therapy. Mol. Ther. 2016;24:430–446. doi: 10.1038/mt.2016.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora A., Sandve G.K., Gabrielsen O.S., Eskeland R. In the loop: promoter–enhancer interactions and bioinformatics. Brief. Bioinform. 2015;17:980–995. doi: 10.1093/bib/bbv097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morsut L., Roybal K.T., Xiong X., Gordley R.M., Coyle S.M., Thomson M., Lim W.A. Engineering customized cell sensing and response behaviors using synthetic notch receptors. Cell. 2016;164:780–791. doi: 10.1016/j.cell.2016.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakade S., Tsubota T., Sakane Y., Kume S., Sakamoto N., Obara M., Daimon T., Sezutsu H., Yamamoto T., Sakuma T., et al. Microhomology-mediated end-joining-dependent integration of donor DNA in cells and animals using TALENs and CRISPR/Cas9. Nat. Commun. 2014;5:5560. doi: 10.1038/ncomms6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen A.A., Voigt C.A. Multi-input CRISPR/Cas genetic circuits that interface host regulatory networks. Mol. Syst. Biol. 2014;10:763. doi: 10.15252/msb.20145735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oceguera-Yanez F., Kim S.-I., Matsumoto T., Tan G.W., Xiang L., Hatani T., Kondo T., Ikeya M., Yoshida Y., Inoue H., et al. Engineering the AAVS1 locus for consistent and scalable transgene expression in human iPSCs and their differentiated derivatives. Methods. 2016;101:43–55. doi: 10.1016/j.ymeth.2015.12.012. [DOI] [PubMed] [Google Scholar]
- Ordovás L., Boon R., Pistoni M., Chen Y., Wolfs E., Guo W., Sambathkumar R., Bobis-Wozowicz S., Helsen N., Vanhove J., et al. Efficient recombinase-mediated cassette exchange in hPSCs to study the hepatocyte lineage reveals AAVS1 locus-mediated transgene inhibition. Stem Cell Rep. 2015;5:918–931. doi: 10.1016/j.stemcr.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papapetrou E.P., Schambach A. Gene insertion into genomic safe harbors for human gene therapy. Mol. Ther. 2016;24:678–684. doi: 10.1038/mt.2016.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papapetrou E.P., Lee G., Malani N., Setty M., Riviere I., Tirunagari L.M.S., Kadota K., Roth S.L., Giardina P., Viale A., et al. Genomic safe harbors permit high β-globin transgene expression in thalassemia induced pluripotent stem cells. Nat. Biotechnol. 2011;29:73–78. doi: 10.1038/nbt.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellenz S., Phelps M., Tang W., Hovde B.T., Sinit R.B., Fu W., Li H., Chen E., Monnat R.J. New human chromosomal sites with “safe harbor” potential for targeted transgene insertion. Hum. Gene Ther. 2019;30:814–828. doi: 10.1089/hum.2018.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson N.H., Luttrell J.B., Bryant J.S., Chamberlain D., Khawaja S., Neeli I., Radic M. Tuning the performance of CAR T cell immunotherapies. BMC Biotechnol. 2019;19:84. doi: 10.1186/s12896-019-0576-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins P.B., Lin Q., Goodnough J.B., Tian H., Chen X., Khavari P.A. In vivo restoration of laminin 5 3 expression and function in junctional epidermolysis bullosa. Proc. Natl. Acad. Sci. U S A. 2001;98:5193–5198. doi: 10.1073/pnas.091484998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roybal K.T., Lim W.A. Synthetic immunology: hacking immune cells to expand their therapeutic capabilities. Ann. Rev. Immunol. 2017;35:229–253. doi: 10.1146/annurev-immunol-051116-052302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roybal K.T., Williams J.Z., Morsut L., Rupp L.J., Kolinko I., Choe J.H., Walker W.J., McNally K.A., Lim W.A. Engineering T cells with customized therapeutic response programs using synthetic notch receptors. Cell. 2016;167:419–432.e16. doi: 10.1016/j.cell.2016.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadelain M., Papapetrou E.P., Bushman F.D. Safe harbours for the integration of new DNA in the human genome. Nat. Rev. Cancer. 2012;12:51–58. doi: 10.1038/nrc3179. [DOI] [PubMed] [Google Scholar]
- Sakuma T., Nakade S., Sakane Y., Suzuki K.-I.T., Yamamoto T. MMEJ-assisted gene knock-in using TALENs and CRISPR-Cas9 with the PITCh systems. Nat. Protoc. 2016;11:118–133. doi: 10.1038/nprot.2015.140. [DOI] [PubMed] [Google Scholar]
- Sather B.D., Ibarra G.S.R., Sommer K., Curinga G., Hale M., Khan I.F., Singh S., Song Y., Gwiazda K., Sahni J., et al. Efficient modification of CCR5 in primary human hematopoietic cells using a megaTAL nuclease and AAV donor template. Sci. Transl. Med. 2015;7:307ra156. doi: 10.1126/scitranslmed.aac5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satija R., Farrell J.A., Gennert D., Schier A.F., Regev A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmel P. The emerging complexity of the tRNA world: mammalian tRNAs beyond protein synthesis. Nat. Rev. Mol. Cell Biol. 2018;19:45–58. doi: 10.1038/nrm.2017.77. [DOI] [PubMed] [Google Scholar]
- Schoenfelder S., Fraser P. Long-range enhancer–promoter contacts in gene expression control. Nat. Rev. Genet. 2019;20:437–455. doi: 10.1038/s41576-019-0128-0. [DOI] [PubMed] [Google Scholar]
- Schwarz K.A., Leonard J.N. Engineering cell-based therapies to interface robustly with host physiology. Adv. Drug Deliv. Rev. 2016;105:55–65. doi: 10.1016/j.addr.2016.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfeir A., Symington L.S. Microhomology-mediated end joining: a back-up survival mechanism or dedicated pathway? Trends Biochem. Sci. 2015;40:701–714. doi: 10.1016/j.tibs.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S., Kim S.H., Shin S.W., Grav L.M., Pedersen L.E., Lee J.S., Lee G.M. Comprehensive analysis of genomic safe harbors as target sites for stable expression of the heterologous gene in HEK293 cells. ACS Synth. Biol. 2020;9:1263–1269. doi: 10.1021/acssynbio.0c00097. [DOI] [PubMed] [Google Scholar]
- Silva E., Stumpf M.P.H. HIV and the CCR5-Δ32 resistance allele. FEMS Microbiol. Lett. 2004;241:1–12. doi: 10.1016/j.femsle.2004.09.040. [DOI] [PubMed] [Google Scholar]
- Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulge U.Y., Baker D.A., Monnat R.J. Comprehensive computational design of mCreI homing endonuclease cleavage specificity for genome engineering. Nucleic Acids Res. 2011;39:4330–4339. doi: 10.1093/nar/gkr022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vangala P., Murphy R., Quinodoz S.A., Gellatly K., McDonel P., Guttman M., Garber M. High-resolution mapping of multiway enhancer-promoter interactions regulating pathogen detection. Mol. Cell. 2020;80:359–373.e8. doi: 10.1016/j.molcel.2020.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Lombardi R., Jung J.S., Bieberich F., Kapetanovic E., Aznauryan E., Weber C.R., Reddy S.T. CRISPR-targeted display of functional T cell receptors enables engineering of enhanced specificity and prediction of cross-reactivity. 2020. [DOI]
- Villasante A., Abad J.P., Mendez-Lago M. Centromeres were derived from telomeres during the evolution of the eukaryotic chromosome. Proc. Natl. Acad. Sci. U S A. 2007;104:10542–10547. doi: 10.1073/pnas.0703808104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeku O.O., Purdon T.J., Koneru M., Spriggs D., Brentjens R.J. Armored CAR T cells enhance antitumor efficacy and overcome the tumor microenvironment. Sci. Rep. 2017;7:10541. doi: 10.1038/s41598-017-10940-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yermanos A., Agrafiotis A., Kuhn R., Robbiani D., Yates J., Papadopoulou C., Han J., Sandu I., Weber C., Bieberich F., et al. Platypus: an open-access software for integrating lymphocyte single-cell immune repertoires with transcriptomes. NAR Genom. Bioinform. 2021;3:lqab023. doi: 10.1093/nargab/lqab023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M.D., Wakefield M.J., Smyth G.K., Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11:R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambrowicz B.P., Imamoto A., Fiering S., Herzenberg L.A., Kerr W.G., Soriano P. Disruption of overlapping transcripts in the ROSA geo 26 gene trap strain leads to widespread expression of -galactosidase in mouse embryos and hematopoietic cells. Proc. Natl. Acad. Sci. U S A. 1997;94:3789–3794. doi: 10.1073/pnas.94.8.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All original code has been deposited at GitHub and the release pertaining this publication is publicly available through Zenodo. DOIs are listed in the key resources table.
-
•
Bulk and single-cell RNA-sequencing data reported in this paper are available at the European Nucleotide Archive. Accession numbers are listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.