SUMMARY
In recent years, the nuclear pore complex (NPC) has emerged as a key player in genome regulation and cellular homeostasis. New discoveries have revealed that the NPC has multiple cellular functions besides mediating the molecular exchange between the nucleus and the cytoplasm. In this review, we discuss non-transport aspects of the NPC focusing on the NPC-genome interaction, the extreme longevity of the NPC proteins, and NPC dysfunction in age-related diseases. The examples summarized herein demonstrate that the NPC, which first evolved to enable the biochemical communication between the nucleus and the cytoplasm, now doubles as the gatekeeper of cellular identity and aging.
Introduction
The nuclear pore complex (NPC) is central to all forms of eukaryotic life as it mediates biomolecular communication between the nucleoplasm and the cytoplasm. The NPC consists of ~ 30 subunits called nucleoporins (Nups). Some of them are very static and spend most of their time stably incorporated in the NPC, while others are soluble and shuttle between the NPC and the nucleoplasm. For its foundational role, the NPC is directly or indirectly involved in essentially all cellular activities. Surprisingly, starting in the late 2000s, two major biological processes, genome regulation and aging - long thought to be indirectly affected by NPCs - have been shown to be under the direct control of these nuclear transport channels. (Ahmed et al., 2010; Brown et al., 2008; Capelson et al., 2010; Casolari et al., 2005; D’Angelo et al., 2009; Kalverda et al., 2010; Taddei et al., 2006). Recent studies have revealed that the NPC is in fact a key cellular component for the transcriptional control of cell identity genes and cellular health.
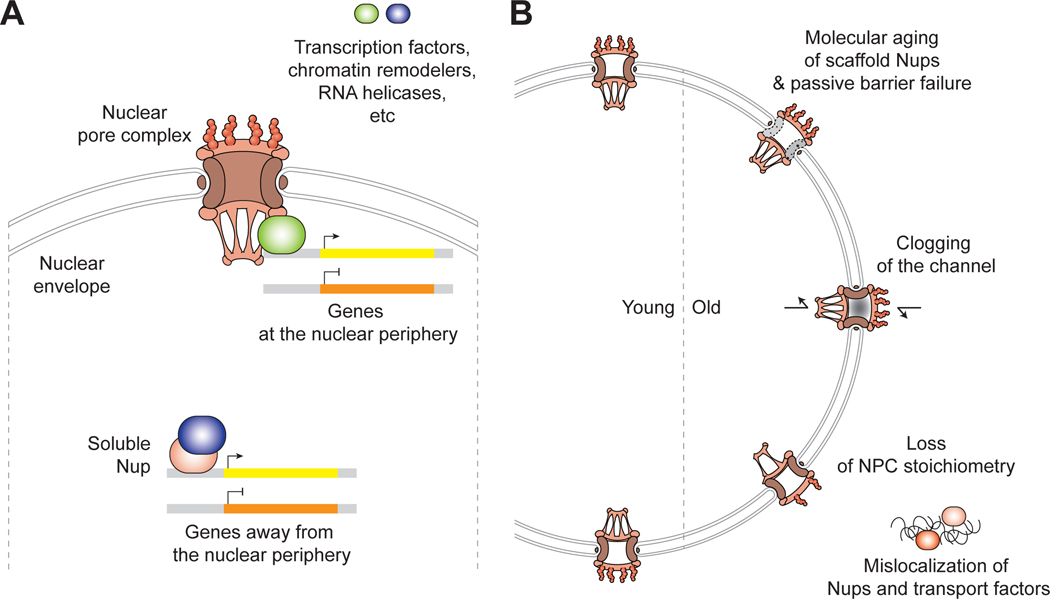
Evidence that the NPC might directly regulate gene activity dates back to the 1950s. Pioneering electron microscopy studies uncovered that the genome at the nuclear envelope is condensed into heterochromatin with the exception of the regions below NPCs. The discovery of these so-called ‘heterochromatin exclusion zones’ was the first evidence that hinted at the role of the NPC in genome organization, and by extension gene regulation (Swift, 1959; Watson, 1959). In 1985, Günter Blobel proposed the ‘gene gating’ hypothesis which stated that these patches of open euchromatin are generated by the interactions between NPCs and transcriptionally active portion of the genome (Blobel, 1985). Subsequent studies in Saccharomyces cerevisiae have demonstrated that NPCs not only determine the three-dimensional locations of the genes within the nucleus but also affect their activities (Randise-Hinchliff and Brickner, 2018). In Drosophila and mammalian cells, the NPC has evolved to enable more intricate genome regulation. Soluble Nups that have significant residence time in the nucleoplasm can regulate gene activity even when they are away from the NPC. In other words, Nups function both at and away from the NPC as genome regulating proteins (Figure 1A) (Raices and D’Angelo, 2017). Latest studies on the genome regulatory roles of Nups illustrate two convergent themes: (1) Nups are mostly involved in the activation or silencing of cell type-specific genes, and in line with this function, (2) Nup expression levels vary greatly in different cell types. We discuss how these features enable cell differentiation and safeguard the identity of a cell.
Figure 1. NPC in gene regulation and aging.
(A) Nups can regulate genes both at and away from the NPC, typically by recruiting transcription factors, chromatin remodelers, RNA helicases, or other auxiliary proteins. They can either activate or suppress genes. (B) The NPC can be compromised by various mechanisms during aging.
In addition to their dual functionality in nucleocytoplasmic transport and transcriptional control, another major focus of the studies on the NPC in the last two decades has been the dynamics of Nups. Soon after the gene-activating capability of Nup98 was reported in the context of acute myeloid leukemia (AML) in 1999, it was revealed that Nup98 is very mobile (Griffis et al., 2002; Kasper et al., 1999). This observation raised a question whether the NPC comprises subunits of widely varying residence times. It is now well established that the dwell time of the ~30 Nups that make up the NPC ranges from tens of seconds to several years, spanning seven orders of magnitude in time (Rabut et al., 2004; Toyama et al., 2013). The discovery of long-lived Nups, together with the finding that NPC biogenesis is minimal in postmitotic cells (D’Angelo et al., 2009), suggested that NPCs may accrue significant amount of biochemical damage over time and be a major player in aging (Figure 1B). Unlike other long-lived proteins that have been shown to deteriorate with age, such as eye lens crystallin, myelin, and collagen, Nups are strictly intracellular and thus can have greater impact on cellular health. The NPCs in physiological aging section details the results that have confirmed the age-dependent decline in the NPC’s transport and barrier functions.
Physiological aging is not the only biological context in which disruption of the NPC is observed. During neurodegeneration, progeria, oncogene-induced senescence, viral infection, and apoptosis, the NPC can be compromised via proteolysis, phosphorylation, displacement, and/or mislocalization of Nups. Although the mechanisms vary, the consequences are invariably catastrophic, reflecting the paramount importance of this protein complex in eukaryotic cells. NPC dysfunction is also seen during replicative aging of yeast, suggesting that the NPC has long been a weak spot in eukaryotic cellular homeostasis. Recently, a few NPC surveillance mechanisms have been discovered in yeast, raising an exciting possibility that metazoans may have developed similar measures to cope with the same problem. We will mainly focus on the NPC failure in age-related conditions and diseases (Figure 1B), but also briefly describe the NPC modification and disintegration by viral proteins and caspases for comparison. As our understanding of the molecular details of NPC disruption deepens, we will be able to better prevent, slow down, or cure the fatal human pathologies involving the NPC like amyotrophic lateral sclerosis (ALS) and Huntington’s disease. Altogether, we are now beginning to understand the fundamental importance of the NPC in both development and aging.
NPCs in maintaining and changing cell identity
While many aspects of NPC-mediated transcriptional control remain to be determined, we have come a long way in our understanding of gene regulation at the nuclear envelope. Like many other research topics in the field of biology, NPC-mediated genome regulation was first investigated in yeast and flies. For example, the very first genome-wide screening for NPC-interacting genes (Casolari et al., 2004), the identification of genes that are recruited to the NPC upon induction and their DNA sequence requirements (Ahmed et al., 2010; Brickner and Walter, 2004; Cabal et al., 2006; Casolari et al., 2005; Taddei et al., 2006), and the discovery of the NPC-mediated transcriptional memory and subtelomeric gene silencing (Galy et al., 2000; Light et al., 2010) were carried out in Saccharomyces cerevisiae and Schizosaccharomyces pombe. Drosophila melanogaster was use to demonstrate that gene gating occurs via SAGA (Spt-Ada-Gcn5-Acetyltransferase) complex (Kurshakova et al., 2007), to study nuclear pore basket Nup-interacting genes (Mendjan et al., 2006; Vaquerizas et al., 2010), and to show that Nups interact with the genome not only at the nuclear periphery but also in the nuclear interior (Capelson et al., 2010; Kalverda et al., 2010). One common theme that emerged from these studies in the 2000s was that Nups, in general, target cell identity genes. Here, we discuss how Nups activate or silence these genes in mammalian systems. Genome regulation by Nups in yeast, flies, and Caenorhabditis elegans (C. elegans) has been reviewed elsewhere (Kuhn and Capelson, 2019).
Genome regulation by Nup93 at the NPC
Nup93 was among the first mammalian Nups whose genome regulatory function was examined (Brown et al., 2008). Nup93 is a scaffold Nup that forms the core module (Nup93–205 complex) of the NPC. Unlike mobile Nups such as Nup98, Nup153, and Nup50 that shuttle between the NPC and the nucleoplasm, Nup93 resides almost exclusively at the nuclear envelope and is suitable for studying NPC-genome interactions confined to the nuclear periphery. Soon after the interaction between Nup93 and CREB-binding protein was confirmed (Ryan et al., 2006), the Silver group, who used ChIP-chip (chromatin immunoprecipitation with DNA microarray) to find genes that come in contact with nucleoporins in yeast (Casolari et al., 2004), employed the same technique to map the interactions between Nup93 and chromosome 5, 7, and 16 in HeLa cells (Brown et al., 2008). Among 86 genes adjacent to Nup93-binding sites were HOXA1, 3, and 5. The role of Nup93 in HOXA1 gene regulation was investigated in more depth several years later (Labade et al., 2016; Labade et al., 2019). In mice, Hoxa1 is transiently expressed in the presumptive hindbrain, and is necessary for proper development of the brainstem, inner ear, and heart (Makki and Capecchi, 2011). In DLD1 (diploid colorectal cancer) cells, when Nup93 was depleted by siRNA, HOXA1 transcript level increased ~4 fold and histones became modified with activation marks. At the same time, the gene loci moved away from the nuclear edge. Taken together, these results suggest that Nup93 on the NPC is involved in gene silencing. A recent study in fact showed that Nup93 preferentially associates with polycomb-silenced regions of the genome in flies, whereas Nup107 largely targets active genes (Gozalo et al., 2019). Interestingly, repression and de-repression of the HOXA1 gene though Nup93 binding is observed during neuronal differentiation. When NTERA2/D1 (pluripotent embryonal carcinoma) cells were treated with retinoic acid, the HOXA1 loci moved away from the nuclear edge early during differentiation, but returned back to their initial positions as cells matured into neurons. Accordingly, HOXA1 transcript level surged ~15 fold upon retinoic acid addition and decreased afterward. Current understanding is that the majority of cell identity-related genes are controlled by Nup93 and other NPC components. Nup93 DNA adenine methyltransferase identification - a ChIP-chip equivalent that does not require ChlP-grade antibody - revealed that 30.1% of the super-enhancers (SEs) in U2OS cells are associated with Nup93 at the nuclear periphery (Ibarra et al., 2016). Given that SEs regulate genes that determine cell fate (Hnisz et al., 2013), Nup93 can be considered a structural platform for cell identity maintenance and change.
Genome regulation by Nup98 away from the NPC
The gene regulatory functions of some Nups were discovered through the studies of human diseases. For example, in hematopoietic malignancies, Nup98, Nup214, and Nup358 genes are often found to be fused with genes that encode DNA-binding domains (Fahrenkrog, 2014; Franks and Hetzer, 2013; Gough et al., 2011; Mendes and Fahrenkrog, 2019). The fusion proteins aberrantly activate genes to promote abnormal proliferation of blood cells. More specifically, in AML, a Nup98-Hoxa9 fusion protein is generated by chromosomal translocation, and the mutation and deletion studies of the fusion protein confirmed that the NUP98 FG-repeat domain recruits CREB-binding protein and p300 to function as a transactivator (Kasper et al., 1999). The gene activation by Nup98 gained more interest when it was shown to be a soluble Nup that spends significant amount of time in the nucleoplasm (Griffis et al., 2002). Since then, four other co-activators were identified: Exportin-1 (Oka et al., 2016), Histone-lysine N-methyltransferase 2A (Shima et al., 2017), DExH-Box Helicase 9 (Capitanio et al., 2017), and Wdr82 (Franks et al., 2017). However, histone deacetylase (HDAC) 1 was also found to interact with Nup98 (Bai et al., 2006), suggesting that Nup98 can mediate both gene activation and silencing. Nup98 ChlP-seq (chromatin immunoprecipitation sequencing) and DNA FISH (fluorescence in situ hybridization) data from human embryonic stem cells (ESCs) and neural progenitor cells (NeuPCs) explains how both gene up- and down-regulation can be achieved by the same protein (Liang et al., 2013). During ESC-to-NeuPC differentiation, the expression of Nup98-target genes remains stable in one subset (‘Group I’), and increases >5 fold in the other subset (‘Group II’). Interestingly, DNA FISH revealed that Group I gene loci were mostly near the nuclear edge in NeuPCs, while those of Group II were predominantly in the interior of the nucleus. This indicates that the NPC provides a biochemical environment that promotes gene silencing by Nup98, and the nucleoplasm supports the gene activation capability of Nup98. Accordingly, it was later discovered that Nup98 binds to intranuclear promoters, recruits Wdr82-Set1A/COMPASS complex, and deposits H3K4 trimethylation activating marks (Franks et al., 2017). Of note, out of 1962 and 2081 Nup98-binding regions in ESCs and NeuPCs, only 321 overlapped, validating that Nup98 dynamically associates with developmental genes as Nup93. Nup98 targets in NeuPCs were enriched for neurogenesis genes, such as NRG1, SOX5, ROBO, and ERBB4. In line with the findings in mammalian cells, in Drosophila, Nup98 regulates genes under the control of ecdysone, an insect hormone that is critical for molting and metamorphosis (Pascual-Garcia et al., 2017).
Nups in differentiation
Generally, Nups modulate gene activity by recruiting transcription factors, chromatin remodelers, RNA helicases, or other types of auxiliary proteins (Figure 1A, Table 1). The latest example is the recruitment of Olig2 and Brd7 by the Y-complex Nup Seh1 during oligodendrocyte differentiation (Liu et al., 2019). Olig2 is a transcription factor and Brd7 is a bromodomain protein that binds to acetylated histones. Upon induction of differentiation, Seh1 in oligodendrocyte progenitor cells, interacts with the two proteins at the NPC, and enables the expression of myelination-related genes such as Mbp, Cnp, Plp1, and Mog. Accordingly, mice lacking Seh1 in the progenitor cells show impaired adult myelinogenesis and remyelination after injury. In C2C12 cells undergoing myoblast-to-myotube transition, a similar but slightly more complex mechanism was observed (D’Angelo et al., 2012; Raices et al., 2017). Nup210, also known as gp210 and one of the three transmembrane Nups, is absent in C2C12 myoblasts, but is rapidly upregulated during differentiation into myotubes. Subsequently, Nup210 recruits the transcription factor Mef2C and the transcriptional co-regulator Trip6 to the nuclear periphery, and induces myogenic genes such as Tnnt1, Myl2, Tmod1, miR133a1, and miR206. Nup210-depleted myoblasts fail to become myotubes. While Seh1 in oligodendrocytes and Nup210 in myotubes upregulate new genes, in other systems, Nups repress yet-to-be expressed developmental genes in progenitor cells. For example, in mouse NeuPCs, Nup153 and Sox2 co-occupy genes necessary for the stemness of the cells at both nuclear periphery and interior. The shRNA-mediated knockdown of Nup153 induced neurite extension and neuronal genes such as Syn1, Tubb3, and Hes5, suggesting that Nup153 is critical for the cell identity homeostasis of NeuPCs (Toda et al., 2017). In mouse ESCs, Nup153 represses developmental genes by recruiting the Polycomb-repressive complex 1 (PRC1) (Jacinto et al., 2015). As in the case of NeuPCs, Nup153 knockdown induced differentiation. In fact, both transcript and protein levels of Nup153 are significantly reduced during ESC-to-NeuPC differentiation. Intriguingly, Polycomb proteins Pc and E(z) interact with Nup93 for gene silencing in Drosophila, hinting that the partnership between Nups and Polycomb proteins arose early during metazoan evolution (Gozalo et al., 2019). Lastly, in resting cardiomyocytes, sarcomeric and Ca2+-handling genes are repressed by Nup155 and HDAC4; however, when hypertrophic growth starts, HDAC4 is exported from the nucleus and Nup155/HDAC4-target genes are de-repressed (Kehat et al., 2011). Unlike the aforementioned Nup98-HDAC1 interaction, Nup155 and HDAC4 work together predominantly at the nuclear periphery. In addition to the Nup155 and HDAC4 pair, in cardiomyocytes, Nup153 and HDAC5 downregulate Cacnalc (Cav1.2 calcium channel) and other genes that can cause cardiac dysfunction when de-repressed (Nanni et al., 2016). To summarize, Nups interact with various transcription factors and coregulators to achieve cell type-specific control of the genome.
Table 1.
Genome-regulating Nups and their binding partners.
| Nucleoporin | Binding partner | Cell type | References |
|---|---|---|---|
| Nup98 | CREB-binding protein p300 | NIH 3T3 and HeLa | Kasper et al., 1999 |
| Exportin-1 | Embryonic stem cell | Oka et al., 2016 | |
| Histone-lysine N-methyltransferase 2A | HEK293T | Shima et al., 2017 | |
| DExH-Box Helicase 9 | HEK293T | Capitanio et al., 2017 | |
| WDR82 | Hematopoietic progenitor cell | Franks et al., 2017 | |
| CREB-binding protein Histone deacetylase 1 | HEK293T | Bai et al., 2006 | |
| sPom121 | HeLa, U2OS, and IMR90 | Franks et al., 2016 | |
| Seh1 | Olig2 and Brd7 | Oligodendrocyte | Liu et al., 2019 |
| Nup210 | Mef2C and Trip6 | Myotube | Raices et al., 2017 |
| Nup153 | Sox2 | Neural progenitor cell | Toda et al., 2017 |
| Nup153 | Polycomb repressive complex 1 | Embryonic stem cell | Jacinto et al., 2015 |
| Nup153 | Histone deacetylase 5 | Cardiomyocyte | Nanni et al., 2016 |
| Nup155 | Histone deacetylase 4 | Cardiomyocyte | Kehat et al., 2011 |
Compositional variation of the NPC in various cell types
The role of Nups in modulating the activities of developmental genes is to some extent reflected in the compositional variation of the NPC in different cell types. As mentioned above, Nup210, which enables the expression of myogenic proteins, is absent in myoblasts, but highly induced during their differentiation into myotubes. In this section, we summarize the recent efforts to quantitatively determine NPC stoichiometry in various cells and tissues.
It was known as early as the late 1990s that the expression level of Nups can be cell and tissue type-dependent. For example, during the characterization of Nup45, which turned out to be an isoform of Nup58 generated by alternative mRNA splicing, Nup45 and Nup58 protein levels were found to be dissimilar in murine melanoma, teratocarcinoma, hepatoma, and liver cells (Hu and Gerace, 1998). Similarly, Nup50 expression was found to be significantly higher in testis than in kidney, liver, and spleen, and was undetectable in heart (Guan et al., 2000). In situ hybridization for Nup210 mRNA in a whole mouse embryo section further validated the tissue-specific expression of NPC subunits (Olsson et al., 1999). Before the 2010s, only low throughput methods were available for examining NPC assembly variation; however, with the advent of advanced mass spectrometry and transcriptomic technologies, it became possible to study the cell type-specificity of the NPC not only with greater throughput but also with better precision.
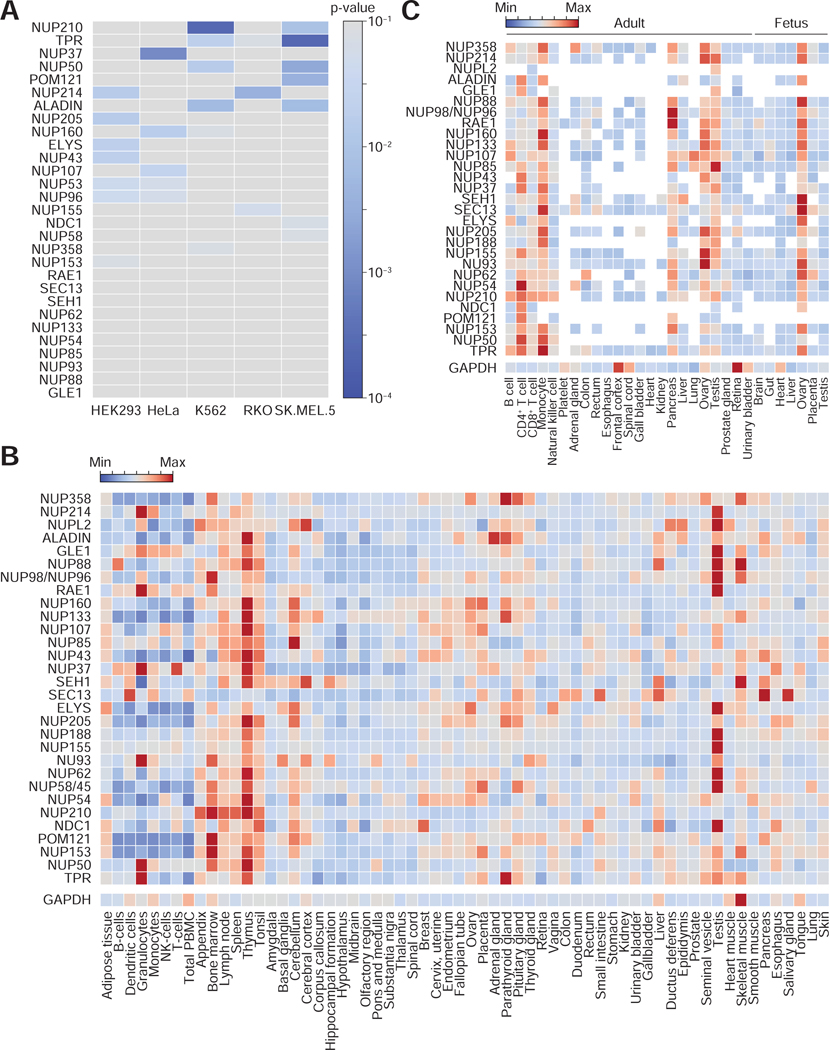
In 2013, the relative copy numbers of 29 Nup proteins in human cells were analyzed using mass spectrometry (Ori et al., 2013). Seven Nups (Nup214, Aladin, Nup210, Pom121, Tpr, Nup50, and Nup37) showed varying protein abundance in HEK293 and four cancer cell lines (Figure 2A). The mRNA levels of these Nups were analyzed in 44 tissue types and 30 disease states, and also exhibited significant variance. A similar survey was carried out for tens of other protein complexes, including the anaphase promoting complex, RNA polymerase I and III core complexes, cohesion complex, linker of nucleoskeleton and cytoskeleton complex, and spliceosomes. About two thirds showed stable stoichiometry across the aforementioned five human cell lines, and among those whose composition was cell type-dependent like the NPC were transcription-export complex, nucleosome remodeling deacetylase complex, and polycomb repressive complex 1. (A more comprehensive study on the compositional variation of 182 mammalian protein complexes was published three years later (Ori et al., 2016)). Taken together, these results establish that the NPC is a cellular machinery tailored to meet the particularities of a cell.
Figure 2. NPC stoichiometry and Nup expression vary in different cell types and tissues.
(A) A p-value heatmap that visualizes the differential expression of Nups in HEK293 and four different human cancer cell lines. A low p-value means that Nup protein level in a given cell line significantly differs from that in others. Image reproduced from (Ori et al., 2013) under a Creative Commons license (https://creativecommons.org/licenses/by/4.0/). (B) Nup mRNA (Uhlen et al., 2015) and (C) protein expression (Kim et al., 2014) in various cells and tissues.
While the study described above clearly demonstrates the differential expression of Nups across cell lines, a similar survey for human tissues, which can be more meaningful from a developmental biology point of view, is still lacking. Therefore, we turned to publicly available transcriptomics and proteomics data to compile this information. Using the RNA expression consensus dataset from the Tissue Atlas curated by the Human Protein Atlas consortium (www.proteinatlas.org) (Uhlen et al., 2015), we generated a heatmap visualizing the mRNA abundance of 30 Nups in various cells and tissues (Figure 2B). As a control, we included GAPDH. Similarly, using the proteomics data that is available through the European Bioinformatics Institute Expression Atlas (https://www.ebi.ac.uk/gxa/) (Kim et al., 2014), the expression patterns of 29 Nups were plotted onto a heatmap (Figure 2C). The data is less comprehensive compared to its RNA counterpart, presumably due to the low abundance and solubility of Nup proteins. Nevertheless, the two heatmaps reveals a few intriguing points: (1) In adults, testis, ovary, pancreas, monocytes, and T cells have high levels of Nup proteins overall, suggesting that these tissues and cells require a greater number of NPCs. (It should be noted that not all cell types in testis, ovary, and pancreas will be abundant in NPCs.) (2) In blood cells, although the transcript levels are low in general, the protein levels are relatively high, hinting that Nup transcripts can be efficiently translated or that Nup proteins have long half-lives in these cells. (3) The expression profile of Nups drastically diverges during the hematopoietic cell differentiation (B cell, T cell, monocyte, versus NK cell). Analogous analyses can be performed for many different biological contexts - such as human brain development, aging, and dementia with the data generated from the Allen Brain Atlas project (www://portal.brain-map.org). While one needs to use caution in interpreting these large-scale data, they can be utilized as an initial guide for exploring the cell- and tissue-specificity of the NPC.
NPCs in physiological aging
During the last decade, the NPC was revealed to be a major weak spot of aging eukaryotic cells. It has been shown that NPC integrity and functions deteriorate during physiological, pathological, and premature aging. This is reminiscent of the NPC disintegration by caspases and viral proteins, and presumably signifies that undermining NPC integrity is among the most unfailing ways to make cells dysfunctional. In cells undergoing apoptosis, the core functions of the NPC, nucleocytoplasmic transport and passive permeability barrier, are abolished by caspases which can cleave at least seven Nups (Tpr, Nup358, Nup214, Nup153, Nup96, Nup93, and Nup50) (Faleiro and Lazebnik, 2000; Ferrando-May et al., 2001; Patre et al., 2006). The NPC is similarly compromised by viruses (Gomez et al., 2019; Le Sage and Mouland, 2013; Yarbrough et al., 2014). They quickly disrupt NPCs to inhibit host mRNA export and protein import using proteases, kinases, and other mechanisms. For example, poliovirus 2A protease cleaves Nup98, Nup153, and Nup62, and encephalomyocarditis virus L protein hyperphosphorylates Nup214, Nup153, and Nup62. Severe acute respiratory syndrome coronavirus nonstructural protein 1 disrupts Nup93 localization. In short, NPC integrity needs to be well maintained for a cell to function normally. In this section, we describe how NPCs becomes defective during normal aging, and briefly summarize the NPC quality control mechanisms identified in yeast, which can guide the discovery of similar protective and repair measures in metazoans.
Extreme longevity of scaffold Nups
Scaffold Nups (Nup93, Nup155, and Nup205) have extremely long residence times and low degradation rates. In fact, they are the most long-lived proteins alongside histone H3.1 (Toyama et al., 2013). These Nups have to persist for the entire lifespan of an animal in neurons, cardiomyocytes, myotubes, etc. since the biogenesis of new NPCs is negligible in post-mitotic cells (D’Angelo et al., 2009). Although not as long-lived as these, a few Y-complex Nups (Nup96, Nup160, and Nup107) also have long half-lives. The minimal turnover of long-lived Nups in the rat brain and heart was shown by labeling newborns with 15N until 6 weeks after birth and switching to 14N diet afterward. In neurons of 1-year-old rats, one fourth of scaffold Nups still contained 15N, proving their extreme longevity. Considering these samples also contained nonpost-mitotic cells from brain and heart that continue to divide in adulthood, the actual replacement rate in post-mitotic cells could be even lower. In cultured cells, the lifetimes of scaffold Nups were determined to be about 100 hours, much shorter than in a whole animal (Mathieson et al., 2018; Rabut et al., 2004). However, they are still longer than the average lifespan of the whole proteome, which is 1–2 days depending on the cell lines (Cambridge et al., 2011). Consistently, using the recombination-induced tag exchange technology (Verzijlbergen et al., 2010), we were able to confirm that the majority of long-lived Nups are not replaced over the course of 2 weeks in C2C12 myotubes (Toyama et al., 2019). During the same time period, short-lived Nups, such as Nup133 and Pom121, were mostly replenished with newly synthesized copies, suggesting that the NPC in old cells are composed of Nups with varying lifespans.
Like other long-lived proteins such as eye lens crystallin, myelin, and collagen, long-lived Nups can accrue damage over time and can be difficult to replace once lost from the complex (Figure 1B) (Toyama and Hetzer, 2013). We indeed discovered an aging phenotype in C. elegans and rat brains that can be attributed to Nup degeneration (D’Angelo et al., 2009). Nup93 and Nup205 are essential for NPC passive permeability barrier (Galy et al., 2003), and their breakdown is expected to make nuclei permeable to macromolecules that are normally excluded. Using a fluorescently labeled 70-kDa dextran, we confirmed that the nuclei isolated from post-mitotic adult worms and old rat brains are significantly ‘leakier’ than those obtained from younger counterparts. Impaired permeability barrier can lead to unregulated mixing of nuclear and cytoplasmic contents, jeopardizing cellular homeostasis. This aging phenotype was more pronounced in C. elegans grown in the presence of paraquat, a chemical that generates reactive oxygen species, and suppressed in worms with increased lifespan (daf-2 RNAi), indicating that NPC integrity is strongly linked to organismal longevity. In nuclei isolated from rat brains, Nup93 immunofluorescence signal was two-fold higher in old-but-intact than in old-and- leaky nuclei, further corroborating that the nuclear barrier decline is due to irreversible Nup damage or loss. In a separate study, the same dextran exclusion assay revealed that cardiomyocyte nuclei from old rats are more permeable than those from young rats (Kajstura et al., 2010). More recently, the age-dependent reduction of Nup93 immunofluorescence intensity and deterioration of permeability barrier were also confirmed in human pancreases (Arrojo e Drigo et al., 2019a). Interestingly, the basal body of the primary cilium, which contains Nup93 (del Viso et al., 2016; Kee et al., 2012; McClure-Begley and Klymkowsky, 2017), was found to be long-lived by 15N pulse-chase labeling in mice (Arrojo e Drigo et al., 2019b). This raises an interesting possibility that primary cilia, analogous to NPCs, may experience an age-related decline in function.
While it is still unknown whether metazoan cells have a dedicated protein quality control mechanism for the NPC, several such processes have been identified in yeast, and it is tempting to assume that similar pathways exist in multicellular organisms. For example, during budding in yeast, dysfunctional NPCs are preferentially retained in the mother cells to maximize daughter lifespan (Webster et al., 2014). Defective NPCs in mothers become buried in the nuclear envelope, which presumably is a measure to prevent faulty nucleocytoplasmic transport activities (Rempel et al., 2019). During meiosis of budding yeast, gametes inherit short-lived nuclear basket Nups but not long-lived core Nups (King et al., 2019). This ensures that progeny start their lives only with fully functional, recently synthesized proteins. It will be interesting to investigate whether the second polar body generated in the course of meiosis II in mammals also sequester away the damaged Nups or NPCs from the mature ovum. In fact, during C. elegans gametogenesis, aged proteins are cleared by lysosomes (Bohnert and Kenyon, 2017), and mammals are likely to have a similar renewal checkpoint.
NPC stoichiometry and density change during aging
Another form of molecular aging for large protein assemblies like the NPC is the loss of optimal stoichiometry. For example, the transcript and protein levels of NUP88, NUP107, NUP155, and NUP50 decrease with passage number in human fibroblasts (Kim et al., 2010). The decrease in several Nups explains the reduction in nuclear translocation of phosphorylated ERK1/2 and NF-kB p50 and related aging phenotypes such as the insensitivity to growth factors and apoptotic stress. More recently, in a study where age-dependent changes in the transcriptomes of human fibroblasts, fibroblast-derived induced neurons, and prefrontal cortex cells were analyzed, RANBP17, a transport receptor that belongs to the importin b family (Koch et al., 2000; Kutay et al., 2000), was discovered to decline chronologically at both RNA and protein levels (Mertens et al., 2015). Accordingly, when RFP-NLS and GFP-NES (NLS: nuclear localization signal; NES: nuclear export signal) were co-expressed in fibroblasts, GFP-to-RFP ratio in the nucleus well correlated with the donor age, demonstrating that nucleocytoplasmic transport becomes defective in old cells. Yeast NPCs also suffer from stoichiometry loss during replicative aging (Janssens et al., 2015; Rempel et al., 2019), further testifying to the strong link between the NPC and cellular health. Yet another cellular aging phenotype is an increase in NPC density at the nuclear envelope (Maeshima et al., 2006). It is unclear what induces NPC density to surge and what the long-term consequences are, but in oncogene-induced senescence, NPC density increase is necessary for the formation of senescence-associated heterochromatin foci (SAHFs), which are required for the silencing of several proliferation-related genes (Boumendil et al., 2019). In short, aging brings about changes to NPCs at multiple levels, from individual Nups to their assembly to density.
NPCs in premature and pathological aging
NPC in premature aging
In the past decade, the NPC has been studied widely not only in the context of physiological aging but also in premature and pathological forms of aging. It is now well accepted that the NPC is directly involved in the latter as well. Hutchinson-Gilford progeria syndrome (HGPS) is a premature aging genetic disorder caused by a mutant form of LMNA which generates a protein called Progerin (De Sandre-Giovannoli et al., 2003). This protein weakens the nuclear lamina and promotes nuclear blebbing. Thus, early studies on HGPS thus had focused on nuclear lamina defects and the concomitant reduction of perinuclear heterochromatin. However, in the 2010s, several studies have shown that nucleocytoplasmic transport is also affected. For example, the ratio of Ran protein in the nucleus versus cytoplasm is normally 3:1, but it is reduced to 1:1 in fibroblasts from HGPS patients (Kelley et al., 2011). As a consequence, the translocation of large cargos like Tpr and ATM is markedly diminished although importin b-dependent nuclear import is still functional. (Dworak et al., 2019; Snow et al., 2013). Nup153 import is also decreased as its binding partner Transportin-1 is sequestered by an abnormally stable microtubule network in HGPS cells. Exportin-1-dependent nuclear export, on the other hand, is enhanced (Garcia-Aguirre et al., 2019). Interestingly, small molecule studies indicate that several cellular defects in HGPS can be rescued simultaneously by correcting one phenotype - whether it be NPC transport defect, permanent farnesylation of Progerin, or anomalously sturdy microtubule network. For example, Leptomycin B, a covalent inhibitor of Exportin-1, not only attenuates nuclear blebbing but also prevents fusiform morphology and nucleolar expansion, both of which are characteristic HGPS fibroblast phenotypes (Buchwalter and Hetzer, 2017; Garcia-Aguirre et al., 2019). Farnesyl transferase inhibitors restore nuclear architecture, the Ran gradient, Tpr import, and heterochromatin level (Capell et al., 2005; Kelley et al., 2011). Remodelin, a small molecule that inhibits N-acetyltransferase 10 and subsequently destabilizes microtubules, reverts Transportin-1 and Nup153 sequestration in the cytoplasm and normalizes nuclear shape, Ran localization, and gene regulation (Larrieu et al., 2014; Larrieu et al., 2018). Of note, normally aged cells share several phenotypes with HGPS cells, and these small molecules may mitigate physiological aging as well. Taken together, the studies on HGPS demonstrate that maintaining functional NPCs is essential for the longevity and robustness of eukaryotic cells.
NPC in neurodegenerative diseases
The vulnerability of the NPC is also seen in neurodegenerative diseases, such as ALS, frontotemporal dementia, Huntington’s disease, and Alzheimer’s diseases (Table 2) (Kim and Taylor, 2017; Li and Lagier-Tourenne, 2018). The convergent theme is that mutated or repeat-expanded proteins or RNAs (hnRNP A1, FUS, TDP-43, huntingtin, C9orf72, etc) either induce the mislocalization of Nups and transport factors or localize to the NPC and clog the channel (Figure 1B). This has been ascribed to low-complexity domains (LCDs) in NPC-related proteins, which enable passive permeability barrier and active transport through phase separation (Schmidt and Gorlich, 2016). In fact, other membrane-less organelles that contain several LCD proteins and make use of phase separation like nucleolus, nuclear speckle, Cajal body, and stress granule also become defective in the presence of these pathogenic macromolecules (Lee et al., 2016). In other words, LCDs are a double-edged sword in disease context.
Table 2.
Nups and other nucleocytoplasmic transport proteins in premature aging and neurodegeneration.
| Disease | Pathogenic macromolecules of interest | Experimental systems (excluding non-mammalian models) | Mislocalized or coaggregated proteins | Reference |
|---|---|---|---|---|
| HGPS | Progerin | Patient fibroblasts | Tpr and Ran | Kelley et al., 2011 |
| Nup153 and Transportin-1 | Larrieu et al., 2018 | |||
| ALS | N/A | iPSC-derived neurons and patient brains | Nup107, Nup205, Ran, and RanGAP1 | Zhang et al., 2015 |
| N/A | iPSC-derived neurons | RCC1 | Jovicic et al., 2015 | |
| (GR)n and (PR)n | HEK293T | Nup155, Nup205, Ran, RanGAP1, and several karyopherins | Lee et al., 2016 | |
| (GA)n | Mouse models | Pom121 and RanGAP1 | Zhang et al., 2016 | |
| (PR)n | Recombinant proteins | Nup98 and Nup54 | Shi et al., 2017 | |
| Arginine-containing DPRs | HEK293T lysate | >20 Nups and transport factors | Hayes et al., 2020 | |
| (G4C2)n and (G2G4)n RNAs | iPSC-derived neurons and patient brains | Pom121, Ndc1, Nup210, Tpr, Nup50, Nup98, Nup107,and Nup133 | Coyne et al., 2020 | |
| TDP-43 | Neuro-2a cell line and patient brains | >20 Nups and transport factors | Chou et al., 2018 | |
| Huntington’s | Polyglutamine- expanded |
Mouse models, iPSC-derived neurons, and patient brains | Gle1 and RanGAP1 | Gasset-Rosa et al., 2017 |
| HTT | Mouse models, iPSC-derived neurons, and patient brains | Nup62, Nup88, Ran, and RanGAP1 | Grima et al., 2017 | |
| Alzheimer’s | Tau | Mouse models and patient brains | Nup98 and Ran | Eftekharzadeh et al., 2018 |
NPC transport failure has been most extensively studied in C9orf72 ALS, which is caused by the GGGGCC-repeat expansion in the C9orf72 gene. The hexanucleotide repeats give rise to dipeptide-repeat proteins (DPRs; (GA)n, (GP)n, (GR)n, (PA)n, or (PR)n) through repeat-associated, non-AUG translation and (G4C2)n and (G2G4)n RNAs from bidirectional transcription. The link between nucleocytoplasmic transport and these gene products was first discovered through unbiased genetic screens in yeast and flies where several Nups and transport-related proteins were shown to enhance or suppress disease phenotypes (Freibaum et al., 2015; Jovicic et al., 2015; Zhang et al., 2015). In addition, reduced mRNA export, protein import and disrupted Ran gradient was observed in C9orf72 patient induced pluripotent stem cell (iPSC)-derived neurons, further supporting NPC dysfunction. Since this pioneering work, there have been significant efforts to determine the relative contribution of DPRs versus (G4C2)n and (G2G4)n RNAs to the pathogenesis of ALS. For example, arginine-containing DPRs have been shown to interact with several LCD proteins including Nups and to be cytotoxic in nonneuronal cells and in vitro systems (Hayes et al., 2020; Lee et al., 2016; Lin et al., 2016; Shi et al., 2017). Similar results were obtained in primary neurons and mouse brains overexpressing (GA)n (Khosravi et al., 2017; Zhang et al., 2016). Furthermore, antibodies against DPRs decrease neurodegeneration and increase survival in ALS mouse models (Nguyen et al., 2019; Zhou et al., 2017). Most recent report, however, provides an additional layer suggesting that repeat RNAs might be the root cause and DPRs are likely to be a secondary factor (Coyne et al., 2020). In C9orf72 patient iPSC-derived spinal neurons and postmortem motor cortex and thoracic spinal cord, the levels of Pom121, NDC1, Tpr, Nup50, Nup98, Nup107, and Nup133 at NPCs were found to be significantly and specifically reduced. This phenotype can be recapitulated in control iPSC-derived neurons by knocking down Pom121 alone, or, more importantly, by transfecting a stop codon-optimized plasmid that produces (G4C2)n RNA but not DPRs. In line with these results, treating patient-derived neurons with antisense oligonucleotides against (G4C2)n RNA for five days, which is long enough to induce the degradation of the RNA but not DPRs, rescues the disease phenotype. Overexpression of Pom121 alone also has the same effect. While this study does not rule out the contribution of DPRs in the later stages of ALS, using a model system that closely mimics neurons in patients and actual postmortem tissues, it demonstrates that the hexanucleotide-repeat RNAs initiate the disease process.
NPC defects in ALS have also been linked to cytoplasmic mislocalization of an RNA-binding protein TDP-43, which is predominantly nuclear in healthy cells. Although mutations in the gene of TDP-43 protein, TARDBP, accounts for less than 5% of ALS, cytoplasmic aggregation of wildtype/mutated TDP-43 is observed in 97% of ALS (Ling et al., 2013). Proximity-dependent biotin identification in Neuro-2a cells using a C-terminal fragment of wildtype TDP-43 has revealed that cytoplasmic TDP-43 aggregates are enriched for Nups and nucleocytoplasmic transport factors (Chou et al., 2018). An ALS-associated missense TDP-43 mutant also bring about a similar problem in nucleocytoplasmic transport in the same cell line. These findings require further confirmation since TDP-43 variants were expressed at abnormally high levels in a neuroblastoma cell line rather than at endogenous levels in patient-derived neurons. Nevertheless, in line with the results, the motor cortex from TARDBP mutation-associated and sporadic ALS patients shows reduced Nup205 nuclear immunoreactivity and large Nup205-positive cytoplasmic inclusions. The same tissue from C9orf72 mutation-associated ALS exhibits abnormal perinuclear punctate Nup205 staining. On the other hand, in the TARDBP ALS cerebellum and SOD1 mutation-associated ALS motor cortex, where TDP-43 does not aggregate, Nup205 localization is not perturbed. It remains to be determined whether TDP-43 aggregation represents the earliest event that disrupts NPC or a secondary symptom (for example, in C9orf72 ALS) that further aggravates NPC failure.
The results from ALS studies prompted researchers to examine NPC integrity in other neurodegenerative diseases as well. Huntington’s disease, for instance, has a very similar pathogenic mechanism to C9orf72 ALS. It is caused by a CAG-repeat expansion in the HTT gene and concomitant elongation of the polyglutamine tract. Polyglutamine-expanded HTT, which typically forms a huge insoluble aggregate inside the nucleus, co-localize with Nup62, Nup88, Gle1, and RanGAP1, impairing both active transport and passive permeability barrier (Gasset-Rosa et al., 2017; Grima et al., 2017). Consistently, pharmacological agents like Thiamet-G and KPT-350 that mend NPC transport dysfunction, as well as the overexpression of RanGAP1 and Ran and immunotherapy, were shown to reduce neurotoxicity in Huntington’s disease (Denis et al., 2019; Grima et al., 2017). While more studies in animal models and human patients are required, this finding suggests that the NPC can be an effective therapeutic target not only for HGPS but also for neurodegenerative diseases. Tau protein in Alzheimer’s disease was confirmed to compromise nucleocytoplasmic transport by interacting with Nup98 (Eftekharzadeh et al., 2018), again illustrating that NPC impairment is central to and shared by most pathological neurodegeneration. One remaining question in the field is whether non-transport functions of the NPC, e.g., gene activation/silencing and genome folding, are comprised as well. This could be another major factor undermining cellular homeostasis in diseased neurons.
Conclusions and perspectives
In this review, we have summarized non-transport functions of the NPC, which we are just beginning to understand. The NPC is an essential structure for the non-random three-dimensional organization of the nuclear genome, and it is particularly important for the regulation of cell type-specific genes. In line with this cellular function, the NPC stoichiometry and Nup expression levels vary greatly depending on the cell and tissue types. Nups can control gene activity both at the nuclear periphery and inside the nucleus, and they have been shown to double as transcription factors in yeast, worms, flies, and vertebrates, suggesting that they have long been multifunctional proteins during eukaryotic evolution. One exciting hypothesis arose soon after the dual role of Nups was revealed: Can there be a subpopulation of NPCs in a cell that is specialized for genome regulation? In other words, is there a subset of NPCs that have forsaken their primary function of the NPC as transport channel to better interact with the genome? In fact, it has been shown that NPCs in the same nucleus exhibit varying levels of transport activity (Grunwald and Singer, 2010). Another circumstantial evidence is that gene-regulating Nups such as Nup93 and Nup210 appear virtually inaccessible in the NPC structures obtained by cryo-electron microscopy (Beck and Hurt, 2017; Bui et al., 2013; Kosinski et al., 2016; von Appen et al., 2015). These data are acquired from purified, DNase-treated nuclear envelope, and presumably is a representation of NPCs that are not interacting with chromatins. We surmise that NPCs participating in gene regulation will adopt a different conformation or stoichiometry to expose Nup93 and Nup210 to their binding partners, and may have reduced transport capability. Single-molecule fluorescence resonance energy transfer imaging in live cells can be employed to examine whether NPCs alternate between quaternary structures attuned for nucleocytoplasmic transport versus genome regulation.
Another intriguing hypothesis that has not been addressed is whether the NPC- and/or soluble Nup-genome interaction is mediated by phase separation. SEs, the major target sites for Nups, are often found in phase-separated liquid droplets with high concentrations of transcription machineries (Sabari et al., 2018). Given that many Nups have LCDs, it is tempting to assume that phase-separating capabilities of Nups enable genome binding. The NPC, with a high local concentration of FG-Nups, indeed seems to be an ideal venue for phase separation. Soluble Nups like Nup98 can also form liquid-like droplets on their own (Celetti et al., 2020), suggesting phase separation may be utilized inside the nucleus as well.
We have also discussed how the molecular integrity and function of the NPC decline during normal, premature, and pathological aging. The NPC is a huge protein complex with dozens of dynamic and static components, and many of them can molecularly senesce or mislocalize during aging. In addition, Nup expression levels can deviate from the normal range due to genome dysregulation. Since the NPC is a fundamental apparatus for eukaryotic forms of life, it is not too surprising that cells with NPC dysfunction quickly become debilitated. Yet, there remains one poorly understood aspect regarding the NPC-aging link. Multiple studies have shown that NPC passive barrier and transport is compromised by aging conditions, but it has not been examined whether NPC- and/or soluble Nup-genome interactions are disrupted at the same time. Their loss may result in unwanted (de-)differentiation - one of the major aging phenotypes - as they are essential for the regulation of cell identity genes. If this turns out to be the case, it would mean that the NPC can contribute to aging in two different ways. Similarly, in many other biological contexts where the NPC is disturbed, such as viral infection and cellular stress, most research has focused only on transport failure. Given what we have learned from a decade of studies on the genome regulatory roles of the NPC and Nups, alterations in transcriptional programs may be another major mechanism by which cells lose identity and homeostasis upon NPC disruption.
Box 1. Visualizing NPC stoichiometry with super-resolution microscopy.
With the maturation of super-resolution microscopy techniques, in the 2010s, it became feasible to directly probe the quaternary structure of the NPC. In other words, the NPC composition can now be examined by counting the copy numbers of Nups in a single pore. Individual Nups within the NPC were successfully resolved for the first time in 2012 by dSTORM (direct stochastic optical reconstruction microscopy) imaging of Xenopus oocytes (Loschberger et al., 2012). Super-resolution micrographs of Nup210 showed clearly the eightfold rotational symmetry of the NPC ring. Its luminal diameter was determined to be 146 nm, and the innermost diameter, 40 nm. In addition, Nup210 dSTORM imaging demonstrated that both luminal and central channel ring diameters decrease by 3–7 nm as Xenopus oocytes develop (stage II vs VI) (Selles et al., 2017). The NPC density concomitantly reduces from 54 to 37 NPC per mm2, suggesting that not only the quaternary structure but also the density of the NPC can change to meet the need of a cell.
In 2013, the Ellenberg group conducted a more exhaustive study (Szymborska et al., 2013). They too adopted dSTORM, but imaged Nup133, Nup107, Nup96, Nup160, Seh1 and Nup62; the first five are components of the Y-complex (the Nup107-Nup160 complex) and the last is a central channel Nup. The radii of the rings formed by eight copies of each Nup was calculated, and this information was utilized to determine their locations on the three-dimensional model of the NPC generated by cryogenic electron microscopy. Furthermore, it enabled them to map the stacking orientation of the Y-complexes on the NPC. Of note, central channel Nup62 appeared as a dot, rather than a ring, in the super-resolution micrographs, validating their method. The latest technology to image the NPC at a subdiffraction limit involves the use of SNAP or Halo-tagged Nups in conjunction with an O6-benzylguanine- or O2-benzylcytosine-functionalized fluorophore or GFP-tagged Nups in conjunction with GFP nanobody (Schlichthaerle et al., 2019; Thevathasan et al., 2019). Compared to antibodies, these labeling modules enable a smaller offset between the protein of interest and the imaging probe, enabling a greater localization precision. In short, latest imaging approaches allow us to spatially probe the NPC at a single protein level.
Acknowledgments
We thank Jongmin Kim (Massachusetts General Hospital) and the members of the Hetzer lab for critical reading of the manuscript. Alessandro Ori (Fritz Lipmann Institute) kindly provided the raw data for Figure 2A. This work was supported by a Glenn Foundation for Medical Research Postdoctoral Fellowship in Aging Research (U.H.C.), Nomis Foundation, W. M. Keck Foundation, and the National Institutes of Health (R01 NS096786, M.W.H.).
Footnotes
Cho et al. discuss recent studies that have established the nuclear pore complex as a key regulator of transcription control of cell identity genes and have linked its functional decline to premature, physiological, and pathological aging.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahmed S, Brickner DG, Light WH, Cajigas I, McDonough M, Froyshteter AB, Volpe T, and Brickner JH (2010). DNA zip codes control an ancient mechanism for gene targeting to the nuclear periphery. Nat. Cell Biol 12, 111–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrojo e Drigo R, Erikson G, Tyagi S, Capitanio J, Lyon J, Spigelman AF, Bautista A, Manning Fox JE, Shokhirev M, MacDonald PE, et al. (2019a). Aging of human endocrine pancreatic cell types is heterogeneous and sex-specific. bioRxiv, 729541.
- Arrojo e Drigo R, Lev-Ram V, Tyagi S, Ramachandra R, Deerinck T, Bushong E, Phan S,Orphan V, Lechene C, Ellisman MH, et al. (2019b). Age mosaicism across multiple scales in adult tissues. Cell Metab. 30, 343–351 e343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai XT, Gu BW, Yin T, Niu C, Xi XD, Zhang J, Chen Z, and Chen SJ (2006). Trans-repressive effect of NUP98-PMX1 on PMX1-regulated c-FOS gene through recruitment of histone deacetylase 1 by FG repeats. Cancer Res. 66, 4584–4590. [DOI] [PubMed] [Google Scholar]
- Beck M, and Hurt E. (2017). The nuclear pore complex: understanding its function through structural insight. Nat. Rev. Mol. Cell Biol 18, 73–89. [DOI] [PubMed] [Google Scholar]
- Blobel G. (1985). Gene gating: a hypothesis. Proc. Natl. Acad. Sci. U.S.A 82, 8527–8529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnert KA, and Kenyon C. (2017). A lysosomal switch triggers proteostasis renewal in the immortal C. elegans germ lineage. Nature 551, 629–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boumendil C, Hari P, Olsen KCF, Acosta JC, and Bickmore WA (2019). Nuclear pore density controls heterochromatin reorganization during senescence. Genes Dev. 33, 144–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner JH, and Walter P. (2004). Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS Biol. 2, e342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CR, Kennedy CJ, Delmar VA, Forbes DJ, and Silver PA (2008). Global histone acetylation induces functional genomic reorganization at mammalian nuclear pore complexes. Genes Dev. 22, 627–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchwalter A, and Hetzer MW (2017). Nucleolar expansion and elevated protein translation in premature aging. Nat. Commun 8, 328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui KH, von Appen A, DiGuilio AL, Ori A, Sparks L, Mackmull MT, Bock T, Hagen W, Andres-Pons A, Glavy JS, et al. (2013). Integrated structural analysis of the human nuclear pore complex scaffold. Cell 155, 1233–1243. [DOI] [PubMed] [Google Scholar]
- Cabal GG, Genovesio A, Rodriguez-Navarro S, Zimmer C, Gadal O, Lesne A, Buc H, Feuerbach-Fournier F, Olivo-Marin JC, Hurt EC, et al. (2006). SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature 441, 770–773. [DOI] [PubMed] [Google Scholar]
- Cambridge SB, Gnad F, Nguyen C, Bermejo JL, Kruger M, and Mann M. (2011). Systems-wide proteomic analysis in mammalian cells reveals conserved, functional protein turnover. J. Proteome Res 10, 5275–5284. [DOI] [PubMed] [Google Scholar]
- Capell BC, Erdos MR, Madigan JP, Fiordalisi JJ, Varga R, Conneely KN, Gordon LB, Der CJ, Cox AD, and Collins FS (2005). Inhibiting farnesylation of progerin prevents the characteristic nuclear blebbing of Hutchinson-Gilford progeria syndrome. Proc. Natl. Acad. Sci. U.S.A 102, 12879–12884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capelson M, Liang Y, Schulte R, Mair W, Wagner U, and Hetzer MW (2010). Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell 140, 372–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capitanio JS, Montpetit B, and Wozniak RW (2017). Human Nup98 regulates the localization and activity of DExH/D-box helicase DHX9. eLife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casolari JM, Brown CR, Drubin DA, Rando OJ, and Silver PA (2005). Developmentally induced changes in transcriptional program alter spatial organization across chromosomes. Genes Dev. 19, 1188–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casolari JM, Brown CR, Komili S, West J, Hieronymus H, and Silver PA (2004). Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell 117, 427–439. [DOI] [PubMed] [Google Scholar]
- Celetti G, Paci G, Caria J, VanDelinder V, Bachand G, and Lemke EA (2020). The liquid state of FG-nucleoporins mimics permeability barrier properties of nuclear pore complexes. J. Cell Biol 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou CC, Zhang Y, Umoh ME, Vaughan SW, Lorenzini I, Liu F, Sayegh M, Donlin- Asp PG, Chen YH, Duong DM, et al. (2018). TDP-43 pathology disrupts nuclear pore complexes and nucleocytoplasmic transport in ALS/FTD. Nat. Neurosci 21, 228–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne AN, Zaepfel BL, Hayes L, Fitchman B, Salzberg Y, Bowen K, Trost H, Rigo F, Harel A, Svendsen CN, et al. (2020). G4C2 repeat RNA mediates the disassembly of the nuclear pore complex in C9orf72 ALS/FTD. bioRxiv, 947721.
- D’Angelo MA, Gomez-Cavazos JS, Mei A, Lackner DH, and Hetzer MW (2012). A change in nuclear pore complex composition regulates cell differentiation. Dev. Cell 22, 446–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angelo MA, Raices M, Panowski SH, and Hetzer MW (2009). Age-dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell 136, 284–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Sandre-Giovannoli A, Bernard R, Cau P, Navarro C, Amiel J, Boccaccio I, Lyonnet S,Stewart CL, Munnich A, Le Merrer M, et al. (2003). Lamin a truncation in Hutchinson-Gilford progeria. Science 300, 2055. [DOI] [PubMed] [Google Scholar]
- del Viso F, Huang F, Myers J, Chalfant M, Zhang Y, Reza N, Bewersdorf J, Lusk CP, and Khokha MK (2016). Congenital heart disease genetics uncovers context-dependent organization and function of nucleoporins at cilia. Dev. Cell 38, 478–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis HL, David LS, and Cicchetti F. (2019). Antibody-based therapies for Huntington’s disease: current status and future directions. Neurobiol. Dis 132, 104569. [DOI] [PubMed] [Google Scholar]
- Dworak N, Makosa D, Chatterjee M, Jividen K, Yang CS, Snow C, Simke WC, Johnson IG, Kelley JB, and Paschal BM (2019). A nuclear lamina-chromatin-Ran GTPase axis modulates nuclear import and DNA damage signaling. Aging Cell 18, e12851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eftekharzadeh B, Daigle JG, Kapinos LE, Coyne A, Schiantarelli J, Carlomagno Y, Cook C, Miller SJ, Dujardin S, Amaral AS, et al. (2018). Tau protein disrupts nucleocytoplasmic transport in Alzheimer’s disease. Neuron 99, 925–940 e927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrenkrog B. (2014). Nucleoporin gene fusions and hematopoietic malignancies. New J. Sci 2014.
- Faleiro L, and Lazebnik Y. (2000). Caspases disrupt the nuclear-cytoplasmic barrier. J. Cell Biol 151, 951–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrando-May E, Cordes V, Biller-Ckovric I, Mirkovic J, Gorlich D, and Nicotera P. (2001). Caspases mediate nucleoporin cleavage, but not early redistribution of nuclear transport factors and modulation of nuclear permeability in apoptosis. Cell Death Differ. 8, 495–505. [DOI] [PubMed] [Google Scholar]
- Franks TM, Benner C, Narvaiza I, Marchetto MC, Young JM, Malik HS, Gage FH, and Hetzer MW (2016). Evolution of a transcriptional regulator from a transmembrane nucleoporin. Genes Dev. 30, 1155–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks TM, and Hetzer MW (2013). The role of Nup98 in transcription regulation in healthy and diseased cells. Trends Cell Biol. 23, 112–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks TM, McCloskey A, Shokirev MN, Benner C, Rathore A, and Hetzer MW (2017). Nup98 recruits the Wdr82-Set1A/COMPASS complex to promoters to regulate H3K4 trimethylation in hematopoietic progenitor cells. Genes Dev. 31, 2222–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freibaum BD, Lu Y, Lopez-Gonzalez R, Kim NC, Almeida S, Lee KH, Badders N, Valentine M, Miller BL, Wong PC, et al. (2015). GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature 525, 129–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy V, Mattaj IW, and Askjaer P. (2003). Caenorhabditis elegans nucleoporins Nup93 and Nup205 determine the limit of nuclear pore complex size exclusion in vivo. Mol. Biol. Cell 14, 5104–5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy V, Olivo-Marin JC, Scherthan H, Doye V, Rascalou N, and Nehrbass U. (2000). Nuclear pore complexes in the organization of silent telomeric chromatin. Nature 403, 108–112. [DOI] [PubMed] [Google Scholar]
- Garcia-Aguirre I, Alamillo-Iniesta A, Rodriguez-Perez R, Velez-Aguilera G, Amaro-Encarnacion E, Jimenez-Gutierrez E, Vasquez-Limeta A, Samuel Laredo-Cisneros M, Morales-Lazaro SL, Tiburcio-Felix R, et al. (2019). Enhanced nuclear protein export in premature aging and rescue of the progeria phenotype by modulation of CRM1 activity. Aging Cell 18, e13002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasset-Rosa F, Chillon-Marinas C, Goginashvili A, Atwal RS, Artates JW, Tabet R, Wheeler VC, Bang AG, Cleveland DW, and Lagier-Tourenne C. (2017). Polyglutamine-expanded huntingtin exacerbates age-related disruption of nuclear integrity and nucleocytoplasmic transport. Neuron 94, 48–57 e44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez GN, Abrar F, Dodhia MP, Gonzalez FG, and Nag A. (2019). SARS coronavirus protein nsp1 disrupts localization of Nup93 from the nuclear pore complex. Biochem Cell Biol 97, 758–766. [DOI] [PubMed] [Google Scholar]
- Gough SM, Slape CI, and Aplan PD (2011). NUP98 gene fusions and hematopoietic malignancies: common themes and new biologic insights. Blood 118, 6247–6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozalo A, Duke A, Lan Y, Pascual-Garcia P, Talamas JA, Nguyen SC, Shah PP, Jain R, Joyce EF, and Capelson M. (2019). Core components of the nuclear pore bind distinct states of chromatin and contribute to polycomb repression. Mol. Cell [DOI] [PMC free article] [PubMed]
- Griffis ER, Altan N, Lippincott-Schwartz J, and Powers MA (2002). Nup98 is a mobile nucleoporin with transcription-dependent dynamics. Mol. Biol. Cell 13, 1282–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grima JC, Daigle JG, Arbez N, Cunningham KC, Zhang K, Ochaba J, Geater C, Morozko E, Stocksdale J, Glatzer JC, et al. (2017). Mutant huntingtin disrupts the nuclear pore complex. Neuron 94, 93–107 e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunwald D, and Singer RH (2010). In vivo imaging of labelled endogenous beta-actin mRNA during nucleocytoplasmic transport. Nature 467, 604–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan T, Kehlenbach RH, Schirmer EC, Kehlenbach A, Fan F, Clurman BE, Arnheim N, and Gerace L. (2000). Nup50, a nucleoplasmically oriented nucleoporin with a role in nuclear protein export. Mol. Cell Biol 20, 5619–5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes LR, Duan L, Bowen K, Kalab P, and Rothstein JD (2020). C9orf72 arginine-rich dipeptide repeat proteins disrupt karyopherin-mediated nuclear import. eLife 9. [DOI] [PMC free article] [PubMed]
- Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, Hoke HA, and Young RA (2013). Super-enhancers in the control of cell identity and disease. Cell 155, 934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu T, and Gerace L. (1998). cDNA cloning and analysis of the expression of nucleoporin p45. Gene 221, 245–253. [DOI] [PubMed] [Google Scholar]
- Ibarra A, Benner C, Tyagi S, Cool J, and Hetzer MW (2016). Nucleoporin-mediated regulation of cell identity genes. Genes Dev. 30, 2253–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacinto FV, Benner C, and Hetzer MW (2015). The nucleoporin Nup153 regulates embryonic stem cell pluripotency through gene silencing. Genes Dev. 29, 1224–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens GE, Meinema AC, Gonzalez J, Wolters JC, Schmidt A, Guryev V, Bischoff R, Wit EC, Veenhoff LM, and Heinemann M. (2015). Protein biogenesis machinery is a driver of replicative aging in yeast. eLife 4, e08527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovicic A, Mertens J, Boeynaems S, Bogaert E, Chai N, Yamada SB, Paul JW 3rd, Sun S, Herdy JR, Bieri G, et al. (2015). Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nat. Neurosci 18, 1226–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajstura J, Urbanek K, Perl S, Hosoda T, Zheng H, Ogorek B, Ferreira-Martins J, Goichberg P, Rondon-Clavo C, Sanada F, et al. (2010). Cardiomyogenesis in the adult human heart. Circ. Res 107, 305–315. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kalverda B, Pickersgill H, Shloma VV, and Fornerod M. (2010). Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell 140, 360–371. [DOI] [PubMed] [Google Scholar]
- Kasper LH, Brindle PK, Schnabel CA, Pritchard CE, Cleary ML, and van Deursen JM (1999). CREB binding protein interacts with nucleoporin-specific FG repeats that activate transcription and mediate NUP98-HOXA9 oncogenicity. Mol. Cell Biol 19, 764–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee HL, Dishinger JF, Blasius TL, Liu CJ, Margolis B, and Verhey KJ (2012). A size-exclusion permeability barrier and nucleoporins characterize a ciliary pore complex that regulates transport into cilia. Nat. Cell Biol 14, 431–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehat I, Accornero F, Aronow BJ, and Molkentin JD (2011). Modulation of chromatin position and gene expression by HDAC4 interaction with nucleoporins. J. Cell Biol 193, 21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley JB, Datta S, Snow CJ, Chatterjee M, Ni L, Spencer A, Yang CS, Cubenas-Potts C, Matunis MJ, and Paschal BM (2011). The defective nuclear lamina in Hutchinson-gilford progeria syndrome disrupts the nucleocytoplasmic Ran gradient and inhibits nuclear localization of Ubc9. Mol. Cell Biol 31, 3378–3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosravi B, Hartmann H, May S, Mohl C, Ederle H, Michaelsen M, Schludi MH, Dormann D, and Edbauer D. (2017). Cytoplasmic poly-GA aggregates impair nuclear import of TDP-43 in C9orf72 ALS/FTLD. Hum. Mol. Genet 26, 790–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, and Taylor JP (2017). Lost in transportation: nucleocytoplasmic transport defects in ALS and other neurodegenerative diseases. Neuron 96, 285–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MS, Pinto SM, Getnet D, Nirujogi RS, Manda SS, Chaerkady R, Madugundu AK, Kelkar DS, Isserlin R, Jain S, et al. (2014). A draft map of the human proteome. Nature 509, 575–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Ryu SJ, Ahn HJ, Choi HR, Kang HT, and Park SC (2010). Senescence-related functional nuclear barrier by down-regulation of nucleo-cytoplasmic trafficking gene expression. Biochem. Biophys. Res. Commun 391, 28–32. [DOI] [PubMed] [Google Scholar]
- King GA, Goodman JS, Schick JG, Chetlapalli K, Jorgens DM, McDonald KL, and Unal E. (2019). Meiotic cellular rejuvenation is coupled to nuclear remodeling in budding yeast. eLife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch P, Bohlmann I, Schafer M, Hansen-Hagge TE, Kiyoi H, Wilda M, Hameister H, Bartram CR, and Janssen JW (2000). Identification of a novel putative Ran-binding protein and its close homologue. Biochem. Biophys. Res. Commun 278, 241–249. [DOI] [PubMed] [Google Scholar]
- Kosinski J, Mosalaganti S, von Appen A, Teimer R, DiGuilio AL, Wan W, Bui KH, Hagen WJ, Briggs JA, Glavy JS, et al. (2016). Molecular architecture of the inner ring scaffold of the human nuclear pore complex. Science 352, 363–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn TM, and Capelson M. (2019). Nuclear pore proteins in regulation of chromatin state. Cells 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurshakova MM, Krasnov AN, Kopytova DV, Shidlovskii YV, Nikolenko JV, Nabirochkina EN, Spehner D, Schultz P, Tora L, and Georgieva SG (2007). SAGA and a novel Drosophila export complex anchor efficient transcription and mRNA export to NPC. EMBO J. 26, 4956–4965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay U, Hartmann E, Treichel N, Calado A, Carmo-Fonseca M, Prehn S, Kraft R, Gorlich D, and Bischoff FR (2000). Identification of two novel RanGTP-binding proteins belonging to the importin beta superfamily. J. Biol. Chem 275, 40163–40168. [DOI] [PubMed] [Google Scholar]
- Labade AS, Karmodiya K, and Sengupta K. (2016). HOXA repression is mediated by nucleoporin Nup93 assisted by its interactors Nup188 and Nup205. Epigenetics Chromatin 9, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labade AS, Salvi A, Karmodiya K, and Sengupta K. (2019). Nup93 and CTCF co-modulate spatiotemporal dynamics and function of the HOXA gene cluster during differentiation. bioRxiv, 646224. [DOI] [PubMed]
- Larrieu D, Britton S, Demir M, Rodriguez R, and Jackson SP (2014). Chemical inhibition of NAT10 corrects defects of laminopathic cells. Science 344, 527–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrieu D, Vire E, Robson S, Breusegem SY, Kouzarides T, and Jackson SP (2018). Inhibition of the acetyltransferase NAT10 normalizes progeric and aging cells by rebalancing the Transportin-1 nuclear import pathway. Sci. Signal 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Sage V, and Mouland AJ (2013). Viral subversion of the nuclear pore complex. Viruses 5, 2019–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KH, Zhang P, Kim HJ, Mitrea DM, Sarkar M, Freibaum BD, Cika J, Coughlin M, Messing J, Molliex A, et al. (2016). C9orf72 dipeptide repeats impair the assembly, dynamics, and function of membrane-less organelles. Cell 167, 774–788 e717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, and Lagier-Tourenne C. (2018). Nuclear pores: the gate to neurodegeneration. Nat. Neurosci 21, 156–158. [DOI] [PubMed] [Google Scholar]
- Liang Y, Franks TM, Marchetto MC, Gage FH, and Hetzer MW (2013). Dynamic association of NUP98 with the human genome. PLoS Genet. 9, e1003308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Light WH, Brickner DG, Brand VR, and Brickner JH (2010). Interaction of a DNA zip code with the nuclear pore complex promotes H2A.Z incorporation and INO1 transcriptional memory. Mol. Cell 40, 112–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Mori E, Kato M, Xiang S, Wu L, Kwon I, and McKnight SL (2016). Toxic PR poly-dipeptides encoded by the C9orf72 repeat expansion target LC domain polymers. Cell 167, 789–802 e712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling SC, Polymenidou M, and Cleveland DW (2013). Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron 79, 416–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Yan M, Liang Y, Liu M, Zhang K, Shao D, Jiang R, Li L, Wang C, Nussenzveig DR, et al. (2019). Nucleoporin Seh1 interacts with Olig2/Brd7 to promote oligodendrocyte differentiation and myelination. Neuron 102, 587–601 e587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loschberger A, van de Linde S, Dabauvalle MC, Rieger B, Heilemann M, Krohne G, and Sauer M. (2012). Super-resolution imaging visualizes the eightfold symmetry of gp210 proteins around the nuclear pore complex and resolves the central channel with nanometer resolution. J. Cell Sci 125, 570–575. [DOI] [PubMed] [Google Scholar]
- Maeshima K, Yahata K, Sasaki Y, Nakatomi R, Tachibana T, Hashikawa T, Imamoto F, and Imamoto N. (2006). Cell-cycle-dependent dynamics of nuclear pores: pore-free islands and lamins. J. Cell Sci 119, 4442–4451. [DOI] [PubMed] [Google Scholar]
- Makki N, and Capecchi MR (2011). Identification of novel Hoxa1 downstream targets regulating hindbrain, neural crest and inner ear development. Dev. Biol 357, 295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieson T, Franken H, Kosinski J, Kurzawa N, Zinn N, Sweetman G, Poeckel D, Ratnu VS, Schramm M, Becher I, et al. (2018). Systematic analysis of protein turnover in primary cells. Nat. Commun 9, 689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure-Begley TD, and Klymkowsky MW (2017). Nuclear roles for cilia-associated proteins. Cilia 6, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes A, and Fahrenkrog B. (2019). NUP214 in leukemia: it’s more than transport. Cells 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendjan S, Taipale M, Kind J, Holz H, Gebhardt P, Schelder M, Vermeulen M, Buscaino A, Duncan K, Mueller J, et al. (2006). Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol. Cell 21, 811–823. [DOI] [PubMed] [Google Scholar]
- Mertens J, Paquola ACM, Ku M, Hatch E, Bohnke L, Ladjevardi S, McGrath S, Campbell B, Lee H, Herdy JR, et al. (2015). Directly reprogrammed human neurons retain aging-associated transcriptomic signatures and reveal age-related nucleocytoplasmic defects. Cell Stem Cell 17, 705–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanni S, Re A, Ripoli C, Gowran A, Nigro P, D’Amario D, Amodeo A, Crea F, Grassi, Pontecorvi A, et al. (2016). The nuclear pore protein Nup153 associates with chromatin and regulates cardiac gene expression in dystrophic mdx hearts. Cardiovasc Res 112, 555–567. [DOI] [PubMed] [Google Scholar]
- Nguyen L, Montrasio F, Pattamatta A, Tusi SK, Bardhi O, Meyer KD, Hayes L, Nakamura K, Banez-Coronel M, Coyne A, et al. (2019). Antibody therapy targeting RAN proteins rescues C9 ALS/FTD phenotypes in C9orf72 mouse model. Neuron. [DOI] [PMC free article] [PubMed]
- Oka M, Mura S, Yamada K, Sangel P, Hirata S, Maehara K, Kawakami K, Tachibana T, Ohkawa Y, Kimura H, et al. (2016). Chromatin-prebound Crm1 recruits Nup98-HoxA9 fusion to induce aberrant expression of Hox cluster genes. eLife 5, e09540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson M, Ekblom M, Fecker L, Kurkinen M, and Ekblom P. (1999). cDNA cloning and embryonic expression of mouse nuclear pore membrane glycoprotein 210 mRNA. Kidney Int. 56, 827–838. [DOI] [PubMed] [Google Scholar]
- Ori A, Banterle N, Iskar M, Andres-Pons A, Escher C, Khanh Bui H, Sparks L, Solis-Mezarino V, Rinner O, Bork P, et al. (2013). Cell type-specific nuclear pores: a case in point for context-dependent stoichiometry of molecular machines. Mol. Syst. Biol 9, 648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori A, Iskar M, Buczak K, Kastritis P, Parca L, Andres-Pons A, Singer S, Bork P, and Beck M. (2016). Spatiotemporal variation of mammalian protein complex stoichiometries. Genome Biol. 17, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual-Garcia P, Debo B, Aleman JR, Talamas JA, Lan Y, Nguyen NH, Won KJ, and Capelson M. (2017). Metazoan nuclear pores provide a scaffold for poised genes and mediate induced enhancer-promoter contacts. Mol. Cell 66, 63–76 e66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patre M, Tabbert A, Hermann D, Walczak H, Rackwitz HR, Cordes VC, and Ferrando-May E. (2006). Caspases target only two architectural components within the core structure of the nuclear pore complex. J. Biol. Chem 281, 1296–1304. [DOI] [PubMed] [Google Scholar]
- Rabut G, Doye V, and Ellenberg J. (2004). Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat. Cell Biol 6, 1114–1121. [DOI] [PubMed] [Google Scholar]
- Raices M, Bukata L, Sakuma S, Borlido J, Hernandez LS, Hart DO, and D’Angelo MA (2017). Nuclear pores regulate muscle development and maintenance by assembling a localized Mef2C complex. Dev. Cell 41, 540–554 e547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raices M, and D’Angelo MA (2017). Nuclear pore complexes and regulation of gene expression. Curr. Opin. Cell Biol 46, 26–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randise-Hinchliff C, and Brickner JH (2018). Nuclear pore complex in genome organization and gene expression in yeast. In Nuclear Pore Complexes in Genome Organization, Function and Maintenance, D’Angelo M, ed. (Cham: Springer International Publishing; ), pp. 87–109. [Google Scholar]
- Rempel IL, Crane MM, Thaller DJ, Mishra A, Jansen DP, Janssens G, Popken P, Aksit A, Kaeberlein M, van der Giessen E, et al. (2019). Age-dependent deterioration of nuclear pore assembly in mitotic cells decreases transport dynamics. eLife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan CM, Harries JC, Kindle KB, Collins HM, and Heery DM (2006). Functional interaction of CREB binding protein (CBP) with nuclear transport proteins and modulation by HDAC inhibitors. Cell Cycle 5, 2146–2152. [DOI] [PubMed] [Google Scholar]
- Sabari BR, Dall’Agnese A, Boija A, Klein IA, Coffey EL, Shrinivas K, Abraham BJ, Hannett NM, Zamudio AV, Manteiga JC, et al. (2018). Coactivator condensation at super-enhancers links phase separation and gene control. Science 361, eaar3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlichthaerle T, Strauss MT, Schueder F, Auer A, Nijmeijer B, Kueblbeck M, Jimenez Sabinina V, Thevathasan JV, Ries J, Ellenberg J, et al. (2019). Direct visualization of single nuclear pore complex proteins using genetically-encoded probes for DNA-PAINT. Angew. Chem. Int. Ed. Engl 58, 13004–13008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt HB, and Gorlich D. (2016). Transport selectivity of nuclear pores, phase separation, and membraneless organelles. Trends Biochem. Sci 41, 46–61. [DOI] [PubMed] [Google Scholar]
- Selles J, Penrad-Mobayed M, Guillaume C, Fuger A, Auvray L, Faklaris O, and Montel F. (2017). Nuclear pore complex plasticity during developmental process as revealed by super-resolution microscopy. Sci. Rep 7, 14732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi KY, Mori E, Nizami ZF, Lin Y, Kato M, Xiang S, Wu LC, Ding M, Yu Y, Gall JG, et al. (2017). Toxic PRn poly-dipeptides encoded by the C9orf72 repeat expansion block nuclear import and export. Proc. Natl. Acad. Sci. U.S.A 114, E1111–E1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima Y, Yumoto M, Katsumoto T, and Kitabayashi I. (2017). MLL is essential for NUP98-HOXA9-induced leukemia. Leukemia 31, 2200–2210. [DOI] [PubMed] [Google Scholar]
- Snow CJ, Dar A, Dutta A, Kehlenbach RH, and Paschal BM (2013). Defective nuclear import of Tpr in Progeria reflects the Ran sensitivity of large cargo transport. J. Cell Biol 201, 541–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift H. (1959). Studies on nuclear fine structure. Brookhaven Symp. Biol 12, 134–152. [PubMed] [Google Scholar]
- Szymborska A, de Marco A, Daigle N, Cordes VC, Briggs JA, and Ellenberg J. (2013). Nuclear pore scaffold structure analyzed by super-resolution microscopy and particle averaging. Science 341, 655–658. [DOI] [PubMed] [Google Scholar]
- Taddei A, Van Houwe G, Hediger F, Kalck V, Cubizolles F, Schober H, and Gasser SM (2006). Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature 441, 774–778. [DOI] [PubMed] [Google Scholar]
- Thevathasan JV, Kahnwald M, Cieslinski K, Hoess P, Peneti SK, Reitberger M, Heid, Kasuba KC, Hoerner SJ, Li Y, et al. (2019). Nuclear pores as versatile reference standards for quantitative super-resolution microscopy. Nat. Methods 16, 1045–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T, Hsu JY, Linker SB, Hu L, Schafer ST, Mertens J, Jacinto FV, Hetzer MW, and Gage FH (2017). Nup153 interacts with Sox2 to enable bimodal gene regulation and maintenance of neural progenitor cells. Cell Stem Cell 21, 618–634 e617. [DOI] [PubMed] [Google Scholar]
- Toyama BH, Arrojo EDR, Lev-Ram V, Ramachandra R, Deerinck TJ, Lechene C, Ellisman MH, and Hetzer MW (2019). Visualization of long-lived proteins reveals age mosaicism within nuclei of postmitotic cells. J. Cell Biol 218, 433–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyama BH, and Hetzer MW (2013). Protein homeostasis: live long, won’t prosper. Nat. Rev. Mol. Cell Biol 14, 55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyama BH, Savas JN, Park SK, Harris MS, Ingolia NT, Yates JR 3rd, and Hetzer MW (2013). Identification of long-lived proteins reveals exceptional stability of essential cellular structures. Cell 154, 971–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, et al. (2015). Tissue-based map of the human proteome. Science 347, 1260419. [DOI] [PubMed] [Google Scholar]
- Vaquerizas JM, Suyama R, Kind J, Miura K, Luscombe NM, and Akhtar A. (2010). Nuclear pore proteins nup153 and megator define transcriptionally active regions in the Drosophila genome. PLoS Genet. 6, e1000846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verzijlbergen KF, Menendez-Benito V, van Welsem T, van Deventer SJ, Lindstrom DL, Ovaa H, Neefjes J, Gottschling DE, and van Leeuwen F. (2010). Recombination-induced tag exchange to track old and new proteins. Proc. Natl. Acad. Sci. U.S.A 107, 64–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Appen A, Kosinski J, Sparks L, Ori A, DiGuilio AL, Vollmer B, Mackmull MT, Banterle N, Parca L, Kastritis P, et al. (2015). In situ structural analysis of the human nuclear pore complex. Nature 526, 140–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson ML (1959). Further observations on the nuclear envelope of the animal cell. J. Biophys. Biochem. Cytol 6, 147–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster BM, Colombi P, Jager J, and Lusk CP (2014). Surveillance of nuclear pore complex assembly by ESCRT-III/Vps4. Cell 159, 388–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarbrough ML, Mata MA, Sakthivel R, and Fontoura BM (2014). Viral subversion of nucleocytoplasmic trafficking. Traffic 15, 127–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Donnelly CJ, Haeusler AR, Grima JC, Machamer JB, Steinwald P, Daley L, Miller SJ, Cunningham KM, Vidensky S, et al. (2015). The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature 525, 56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YJ, Gendron TF, Grima JC, Sasaguri H, Jansen-West K, Xu YF, Katzman RB, Gass J, Murray ME, Shinohara M, et al. (2016). C9ORF72 poly(GA) aggregates sequester and impair HR23 and nucleocytoplasmic transport proteins. Nat. Neurosci 19, 668–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Lehmer C, Michaelsen M, Mori K, Alterauge D, Baumjohann D, Schludi MH, Greiling J, Farny D, Flatley A, et al. (2017). Antibodies inhibit transmission and aggregation of C9orf72 poly-GA dipeptide repeat proteins. EMBO Mol. Med 9, 687–702. [DOI] [PMC free article] [PubMed] [Google Scholar]