Abstract
Background: Bacteriophages may induce specific antibodies after natural exposure to phages or after phage therapy. As such, phage-specific antibodies might impact phage bioavailability in vivo, although limited non-neutralizing or insignificant effects have also been reported.
Materials and Methods: Here, we report antibody induction against PB1-related phages (Pseudomonas viruses LMA2, F8, DP1) in mice over an 80-day period, for a healthy population of humans, and in patients undergoing phage therapy (oral and/or topical treatment).
Results: All phages effectively induced specific immunoglobulin M and immunoglobulin G in mice. Phage-specific antibodies were observed in humans, whereas recombinant virion proteins (PB1 gp22, gp29) did not induce phage-neutralizing antibodies, either in mice or in humans. The healthy human population was differentiated for frequency of phage-neutralizing antibodies.
Conclusions: These data can hold key considerations for phage therapy cocktail design, as highly similar phages can still be highly complementary in cases where specific immune response hinders therapeutic use of phages.
Keywords: immune response, antibodies, PB1-related phages, phage therapy, virion proteins, Pseudomonas aeruginosa PAO1
Introduction
Bacteriophages (phages) are capable of inducing a specific antibody response, as demonstrated in animal models1–7 and as observed in humans.6,8–10 Animals and humans are naturally exposed to phages when phages ubiquitous in the environment and in natural microbiomes enter an organism. In phage therapy this exposition is artificial, as selected phages are given as therapeutic agents in high relative concentrations. Nevertheless, these phages from cocktails have also been selected from natural sources in the environment. The multiplicity of similar phages and phage groups that we are exposed to results in antibodies that may show cross-reactions. The specific response induced by an individual phage may affect another one, including both therapeutic and natural phage isolates.
Phage-specific antibodies are typically expected to affect pharmacokinetics and bioavailability of active phages in animals and humans. However, examples of both extensive11–13 and insignificant9,14–17 effects of specific antibodies on phages have been reported. This makes it difficult to predict phage bioavailability in the presence of specific antibodies. Since phage-specific antibodies are, in fact, antibodies specific to phage virions, that is, to many different structural proteins, phage immunogenicity may differ depending on the protein compositions of the phage capsids.6,7 Accordingly, the immune response to a phage is rather a sum of responses to different proteins of this phage. Further, as postulated by Jerne and Avegno,18 and Jerne,19 only a fraction of phage-specific antibodies can be neutralizing. Thus, individual identification of immunogenic proteins in phage virions, together with individual assessment of their ability to induce phage-neutralizing antibodies seems to be crucial for our understanding of how a particular phage (or a group of phages) interacts with the immune system, and how the response affects the phage.
Here, we present an analysis of antibody induction by three very closely related phages from PB1-like genus (Pbunavirus): LMA2, DP1 (full name: vB_PaeM_CEB_DP1), and F8 (or F-8). They are all active against Pseudomonas aeruginosa and belong to the therapeutic set of phages (in case of F8) used in the Phage Therapy Center, Hirszfeld Institute of Immunology and Experimental Therapy, Wrocław, Poland (PTU HIIET).20 As such, the analysis of antibodies specific to these phages is relevant to the phage therapy cocktail design for individual patient treatment.
Materials and Methods
Bacteriophages and bacterial host strain
Bacteriophage F8 (or F-8; NC_007810) was obtained from the Polish Collection of Microorganisms at the Hirszfeld Institute of Immunology and Experimental Therapy, Polish Academy of Science (HIIET PAS). Bacteriophages LMA2 (NC_011166) and DP1 (NC_041870; full name: vB_PaeM_CEB_DP1) were previously described.21,22 All three phages were propagated on Pseudomonas aeruginosa PAO1 (DSM 22644).
Preparation of phage lysates
Flasks containing enriched nutrient broth were inoculated with a 3-h culture of Pseudomonas aeruginosa PAO1. Phages were added to the flasks to the final concentration of 8 × 108 pfu/mL, and the cultures were incubated at 37°C with vigorous shaking for 5 h. After that time, the flasks were kept at 4°C for 1 day to clarify. Phage lysates were then centrifuged at 8000 rpm. The supernatants were filtered through 0.22 μm membrane filters (Merck Millipore) and purified by using size exclusion chromatography (gell filtration) on Sepharose 4B using fast protein liquid chromatography (FPLC) (Sigma-Aldrich); detailed information on timing and absorbance is presented in Supplementary Figure S1. This step was followed by dialysis through 1000 kDa membranes (Spectrum Laboratories, USA) against phosphate-buffered saline (PBS) (Na2HPO4 6.5 μM, KCl 3 mM, KH2PO4 1.5 mM, NaCl 137 mM, Na2HPO4 × 12H2O 8.1 × M). Phage titers in lysates and purified preparations were determined by using serial dilutions, and the spot plating technique. Lipopolysaccharide (LPS) content was determined by using EndoLISA (Hyglos GmbH, Germany) according to the manufacturer's instructions with minor modifications. Overnight incubation at the first step was applied. Plate reader EnSpire Multilabel Plate Reader Perkin Elmer was used. Detailed information on phage titer, endotoxin concentration, and relevant chromatograms are presented in Supplementary Figure S1 and Supplementary Table S1.
Phage proteins isolation and purification
Two sets of phage proteins were investigated in this study: gp22 (encoded by ORF22 for phages F8 and LMA22, and ORF23 for DP1) and gp29 (encoded by ORF29 for phages F8 and LMA22, and ORF30 for DP1); these proteins were putative structural proteins of the phages based on similar phage annotation. Due to inconsistencies in their names and numbers, proteins investigated herein were designed as gp22 and gp29. Their respective designations in the National Center for Biotechnology Information (NCBI) database are as follows: in F8 phage “hypothetical protein ORF022” and “hypothetical protein ORF029,” in LMA2 phage “ORF22,” “putative structural protein” and “ORF29,” “putative structural protein,” and in DP1 phage “FDG87_gp24,” “putative structural protein” and “FDG87_gp31,” and “putative structural protein.”
Protein-coding sequences were cloned by using Gateway technology (Thermo Fisher) to pDEST24 vector allowing for the expression of recombinant products with glutathione S-transferase (GST) affinity tags and expressed in the Escherichia coli expression system by using Escherichia coli B834(DE3) F− ompT hsdSB(rB− mB−) gal dcm met (DE3) (Novagen). Bacteria were grown in lysogeny broth (LB) high salt (10 g/L of NaCl) culture medium (Sigma-Aldrich) with ampicillin at 37°C overnight as a preculture; then, an expression culture was inoculated with 1 mL of preculture per 1 L of LB and it was cultured with ampicillin at 37°C to an optical density (OD600) of 0.8.
Next, isopropylthio-β-D-galactoside (Thermo Scientific) was added to a final concentration of 0.2 or 0.8 mM (for gp22 and gp29, respectively) to induce expression of phage proteins. Bacteria were further cultured overnight at 25°C. and centrifuged for 5 min at 8000 rpm. The collected bacteria were suspended in phosphate buffer (50 mM Na2HPO4, 300 mM NaCl, pH 8.0), treated with phenylmethylsulfonyl fluoride (1 mM), and incubated on ice for 15 min. Lysis was achieved by incubation with lysozyme (0.5 mg/mL) for 5 h on ice and by the freeze-thaw method (overnight at −80°C). Subsequently, the preparation was supplemented with Mg2+ (up to 0.5 mM), DNase I (10 μg/mL), and RNase A (20 μg/mL), and it was incubated on ice for 3–4 h. Fractions were separated by double centrifugations (12,000 rpm, 45 min, 15°C). The soluble fraction was filtered through 0.45 μm polyvinylidene difluoride (PVDF) filters (Millipore) and incubated with glutathione sorbent slurry (Glutathione Sepharose 4B; GE Healthcare Life Sciences, Poland).
The resin was subsequently washed with phosphate buffer, and proteins were released by proteolysis with rTev protease (5 U/mL) (Pure Biologics, Poland) at 10°C; GST tags remained bound in the resin. Protein preparations were purified: (1) initial LPS removal with EndoTrap HD (Hyglos GmbH), (2) gel filtration FPLC on a Superdex 75 10/300 GL column (GE Healthcare Life Sciences) for final separation of proteins, and (3) a final step of LPS removal with EndoTrap HD (Hyglos GmbH) followed by dialysis against PBS and filtration through 0.22-μm (PVDF) filters. Protein purity was inspected by sodium dodecyl sulfate–polyacrylamide gel electrophoresis, and concentrations were determined by using the Lowry chromogenic method (Thermo Scientific). The resulting proteins were positively verified by reaction with reference sera specific to whole phage virions. LPS content was determined by using EndoLISA (Hyglos GmbH) according to the manufacturer's instructions.
Murine protein-specific plasma
To obtain phage protein-specific plasma samples, C57BL/6J male mice (Center of Experimental Medicine, Medical University of Bialystok) were administered three doses of highly purified recombinant phage proteins intraperitoneally (IP) (200 μg/mouse each on days 0, 14, and 28). Blood was collected from the orbital vein to heparinized tubes, and plasma was separated by double centrifugation (2250 × g and 10,000 × g; 10 min each) and stored at −80°C for further use.
Animal immunization model
C57BL/6J male mice (N = 7) (Center of Experimental Medicine, Medical University of Bialystok) were administered IP three doses of PBS (control group) or highly purified phage preparations (F8, LMA2, DP1), at 1010 pfu per mouse each on days 0, 20, and 55. Blood samples were collected from the tail vein on days 1, 5, 10, 15, 20, 22, 25, 35, 40, 45, 50, 55, 60, 64, 70, 80; plasma was separated as described earlier and stored at −20°C for further use. Non-challenged mice from the same batch were included in each experiment to serve as the control for possible off-target antibodies in the murine plasma.
Human samples
Human sera samples representing a healthy population were collected from 55 healthy donors, both male and female (age range: 18–40 years), who had never been subjected to phage therapy or involved in phage work. Blood was collected into heparinized tubes, and serum was separated from the blood by double centrifugation at 2250 × g. The same sera (stored in HIIET's collection) were used as in previous studies for T4 phage6 and A3R and 676Z phage (Kaźmnierczak et al., manuscript under assessment).
Human sera of 11 patients undergoing phage therapy with bacteriophage F8 were collected at the Phage Therapy Unit of HIIET PAS. Blood samples were taken 1–5 days or just before starting phage therapy and during/after the treatment. The sera were separated from heparinized blood samples by centrifugation (10 min, 1500 × g) and stored at −70°C for further use.
All experiments were conducted in accordance with the Declaration of Helsinki and approved by the local Commission of Bioethics, Wroclaw Medical University.
Specific antibody level measurement by enzyme-linked immunosorbent assay
MaxiSorp flat-bottom 96-well plates (Nunc; Thermo Scientific) were coated with 100 μL of highly purified proteins (1 μg/well) or phage preparations (5 × 108 pfu/well), or with albumin as the negative control, overnight at 4°C. Irrelevant serum was used as the specificity control. Since no universal (for the three phages) standard serum was available, only relative comparisons were applied. Plates were then washed five times with PBS and blocked with 1% (w/v) albumin (Sigma) in PBS at room temperature for 1 h. Blocking solution was then removed, and plates were washed five times with PBS with 0.05% (v/v) Tween 20 (BD Biosciences). Murine or human plasma samples diluted in PBS were then added to wells in duplicate at 100 μL per well and incubated at 37°C for 2 h. Subsequently, plates were washed five times with PBS with 0.05% (v/v) Tween 20, and 100 μL per well of diluted horseradish peroxidase-conjugated detection antibody was added: goat anti-mouse immunoglobulin M (IgM) (Jackson ImmunoResearch Laboratories), immunoglobulin G (IgG) (Jackson ImmunoResearch Laboratories), or goat anti-human IgG (Abcam). Plates were incubated for 1 h at room temperature in the dark and then again washed five times with PBS with 0.05% (v/v) Tween 20. 3,3′,5,5′-Tetramethylbenzidine (TMB) (50 μL/well) was used as a substrate reagent for horseradish peroxidase (HRP) according to the manufacturer's instructions (R&D Systems) and incubated for 30 min. Finally, 25 μL of 2N H2SO4 was added to stop the reaction, and the absorbance was measured at 450 nm (main reading) and normalized by subtracting the background absorbance at 570 nm.
Reverse cumulative distribution plots for structural proteins gp22 and gp29 were assigned from direct OD570nm values, according to Reed et al.23
Protein localization in phage capsids in transmission electron microscopy
Fifty microliters of phage preparations was deposited on nickel formvar/carbon coated transmission electron microscopy (TEM) grids (400-mesh) and incubated for 1 min. Then, the excess liquid was removed by using the side of a Whatman blotting paper, and the grids were blocked with 1% (w/v) bovine serum albumin. Ammonium sulfate-precipitated antibodies from phage-protein specific murine plasma samples were then added, and the grids were incubated for 1 h. Subsequently, the grids were extensively washed in PBS, and 1:50 diluted immunoconjugate gold-labeled secondary antibody (10 nm; Sigma) was deposited on the grids. The grids were incubated for 15 min and then again extensively washed in PBS, followed by distilled water. Finally, the samples were stained with 2% uranyl acetate and were observed with a Zeiss EM900 transmission electron microscope.
Ethics approval statement
All animal experiments were performed according to EU Directive 2010/63/EU for animal experimentations and were approved by the first Local Committee for Experiments with the Use of Laboratory Animals, Wroclaw, Poland (No. 64/2009). The authors followed the Animal Research: Reporting of In Vivo Experiments guidelines.24
Results
Specific immune response to phages F8, LMA2, and P1 in mice
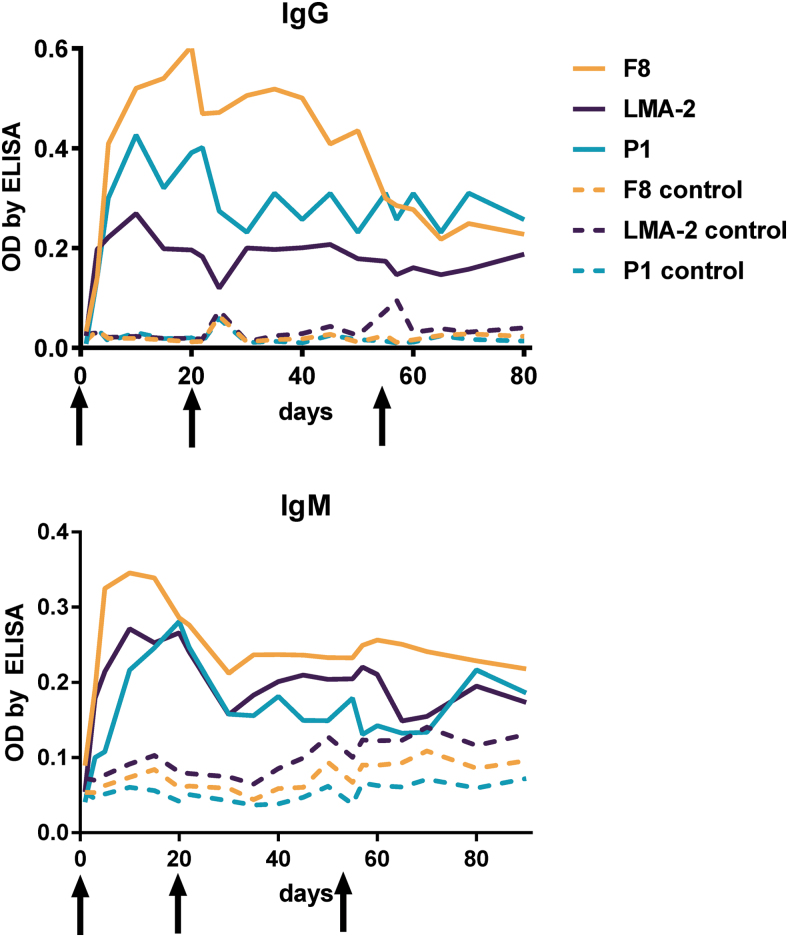
Kinetics of specific immune response induction by phages F8, LMA2, and DP1 (morphology by TEM: Supplementary Fig. S2) was assessed in mice by identification of phage-specific IgM and IgG in murine sera for an 80-day period. Phages were injected IP as purified preparations 1010 pfu per mouse on days 0, 20, and 55. All phages effectively induced specific IgM and specific IgG; antibodies increased within the first week after injection, reaching their maximum peaks around days 5–20 (IgM) and 10–20 (IgG). Later, serum levels of phage-specific antibodies slightly decreased, but they remained markedly higher than those observed in control (PBS-injected) mice.
Differences between the investigated phages in IgG levels (Fig. 1, IgG) may suggest differences in their overall immunogenicity. Specifically, the highest enzyme-linked immunosorbent assay (ELISA) signal was detected for phage F8, followed by phage DP1, whereas phage LMA2 had the lowest signal. The signal observed for LMA2 was also weak when compared with other phages investigated to date: T4 phage6 and staphylococcal phages A3R and 676Z (Kaźmierczak et al., under assessment). This suggests that LMA2 is a phage characterized by relatively lower immunogenicity under the conditions studied.
FIG. 1.
Specific immune response to phages F8, LMA2, DP1 in mice. Purified phages were injected IP, 1010 pfu per mouse (N = 7), on days 0, 20, and 55 (arrows). Figure legend is given as follows: F8-serum level of F8 phage-specific antibodies in phage-challenged mice, F8 control-serum level of F8 phage-specific antibodies in control mice injected with PBS, LMA2-serum level of LMA2 phage-specific antibodies in phage-challenged mice, LMA2 control-serum level of LMA2 phage-specific antibodies in control mice injected with PBS, P1-serum level of P1 phage-specific antibodies in phage-challenged mice, and P1 control-serum level of P1 phage-specific antibodies in control mice injected with PBS. This experiment was repeated three times, and the exemplary result is presented. IP, intraperitoneally; PBS, phosphate-buffered saline.
Induction of specific antibodies in mice by structural proteins gp22 and gp29
To assess the contribution of individual structural proteins to the immunogenicity, we selected two important structural proteins for recombinant expression and analysis, gp22 and gp29. These proteins are highly conserved in F8, LMA2, and DP1. Homological proteins in the related phage SN were previously identified as head decorating (gp22) and tail sheath forming (gp29) proteins.21,25 We confirmed this structural function and localization, by performing immunomicroscopy on DP1, using gp22- and gp29-specific sera in mice, developed by using recombinant highly purified proteins gp22 and gp29 (Supplementary Fig. S3).
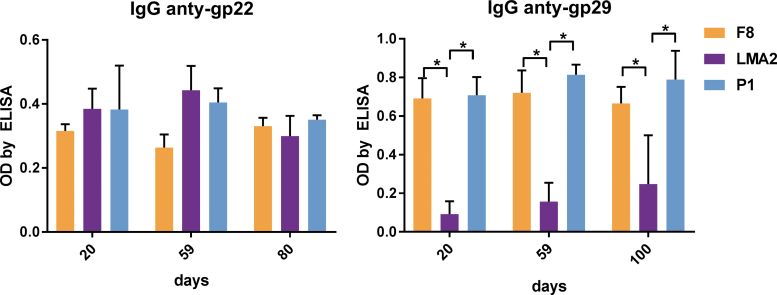
The immunogenicity of recombinant gp22 and gp29 as structural elements of the phage virions was investigated in murine sera from animals challenged with bacteriophages F8, LMA2, or DP1 (Fig. 1). Relative IgG levels were compared in ELISA with isolated recombinant proteins of all investigated phages as bottom antigens (Fig. 2). Gp22 induced marked levels of specific IgG in all phages, but gp29 induced a markedly stronger response in animals treated with F8 or DP1, but a weaker response in animals treated with the LMA2 phage. This is consistent with the lower antibody level observed for LMA2 in the analysis of the entire phage particles (Fig. 1). It suggests that weaker response to gp29 may be the reason for the lower reactivity of the sera with the entire virion particle. Nevertheless, both proteins (gp22 and gp29) contribute significantly to the development of phage-specific sera in mice. We also compared the neutralizing potential of specific anti-gp22 and anti-gp29 sera, but no significant neutralizing activity could be observed in any phage (data not shown).
FIG. 2.
Individual immunogenicity of two capsid proteins: gp22 and gp29 in phages F8, LMA2, and DP1. Purified phages were injected IP 1010 pfu per mouse (N = 7) on days 0, 20, and 55. Blood sera from these mice were tested for IgG antibodies specific to structural proteins gp22 and gp29. Results are presented as OD by ELISA. Statistically significant differences (p < 0.001 Kruskal–Wallis analysis of variance) between groups are indicated with asterisks. ELISA, enzyme-linked immunosorbent assay; IgG, immunoglobulin G; IgM, immunoglobulin M; OD, optical density.
F8, LMA2, and DP1 phage-specific sera in a healthy human population without phage therapy
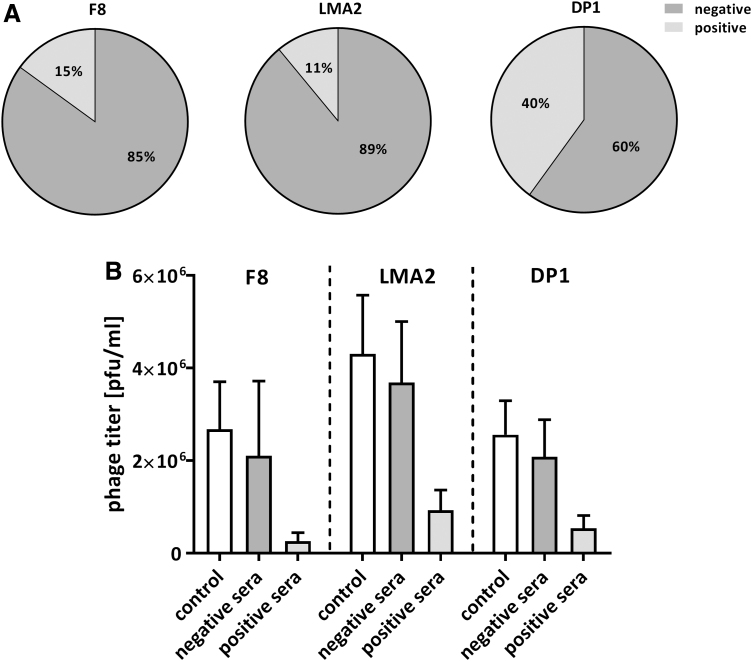
Humans are naturally exposed to phages that are commonly present in food and water, as these viruses constitute a major part of the natural human virome.26,27 Antibodies that result from continuous contact with phages are often called natural antibodies. Here, we determine the frequency of natural antibodies that are specific to F8, LMA2, and DP1 in healthy human volunteers who have not been subjected to phage therapy (N = 55, the same collection of sera as previously used to assess immune response to T4 phage).6 Sera were incubated with bacteriophages F8, LMA2, or DP1 (2 × 106 – 4 × 106 pfu/mL) to identify the neutralizing activity of phage-specific antibodies. Neutralizing activity was found as follows: 15% (8 out of 55) for phage F8, 11% (6 out of 55) for phage LMA2, and 40% (22 out of 55) for phage DP1. The differences between DP1 phage and F8 phage or LMA2 phage were statistically significant (McNemar's test, p = 0.0005 and 0.0008, respectively); no significant difference between F8 and LMA2 phage was observed (p = 0.7266). Inhibition of phage activity by positive sera, although evident, was not strong: phage titer on average remained in the same order of magnitude as controls (12% of initial phage titer in F8 and 25% in LMA2 and DP1) (Fig. 3). Interestingly, despite the fact that F8, LMA2, and DP1 phages are closely related and highly similar, the reactivity of specific individuals' sera was different between phages in 42% (23 out of 55) of studied individuals (Supplementary Table S2). This demonstrates antigenic differences between these three closely related phages, and it suggests their complementary potential if used in human phage therapy.
FIG. 3.
F8, LMA2, and DP1 phage-inhibitory effect of healthy humans' sera; sera were collected from 55 healthy donors never subjected to phage therapy, (A) frequency of phage-inhibitory effect in the investigated donors, (B) the strength of phage inhibition by the investigated sera: average phage titer after incubation with positive and negative sera. Negative sera—not inactivating phage (phage titer after incubation was within the range: mean value of the control ±2 × standard deviation); 8, 6, and 22 samples could be classified into this group in the case of F8, LMA2, and DP1, respectively. Positive sera—inactivating the phage (phage titer after incubation was lower than the range of the negative samples); 47, 49, and 33 samples could be classified into this group in the case of F8, LMA2, and DP1, respectively. The control was calculated from titers of the same phage preparation incubated with fetal bovine serum (N = 6).
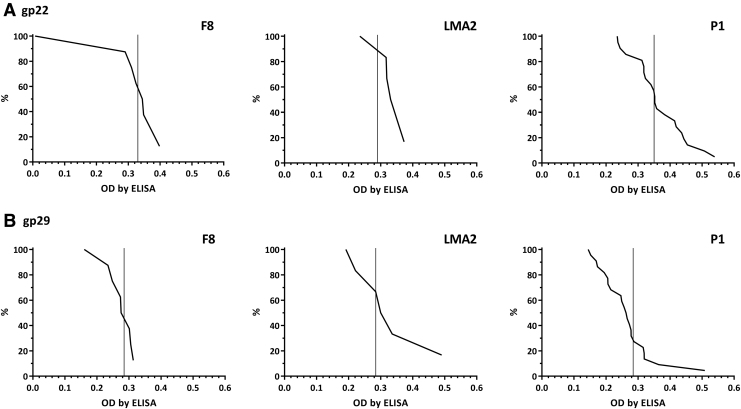
Also, IgG fractions specific to our selected structural phage proteins (gp22 and gp29) were assessed in the same collection of healthy humans' sera. The levels of antibodies specific to both investigated proteins were tested in sera previously identified as “positive” (Fig. 3). They were compared by ELISA with reverse cumulative plotting, but no significant differences in population immunity to proteins gp22 and gp29 could be detected (Fig. 4). Thus, no differences between these proteins' contribution to PB1-like phages' immunogenicity at the population level (that is: in healthy human volunteers as representatives for the human population) were detected, although specific antibodies for all proteins were observed in the investigated population. The relatively high cutoff values observed in the plots argue that proteins gp22 and gp29 are not involved in the induction of phage-neutralizing antibodies. Neutralization can probably be elicited by other virion proteins, most likely those engaged in the process of phage-infecting bacteria; this issue could be investigated further in the future, but remains dependent on the availability of purified recombinant structural proteins and corresponding sera for all virion-associated proteins.
FIG. 4.
Frequency of antibodies specific to F8, LMA2, and DP1 phage structural proteins gp22 and gp29 in the human population. Serum samples (N = 55) were tested at 1:100 dilution in ELISA. Reverse cumulative distribution plots for positive sera (Fig. 3) are presented, and cutoff values for negative sera (Fig. 3) are represented by vertical lines in each plot.
Anti-F8 phage antibodies and anti-phage protein (gp22 and gp29) antibodies in patients subjected to phage therapy
Among the investigated phages, F8 belongs to the group of therapeutic phages used in the Phage Therapy Unit of HIIET.20 Antibodies specific to structural proteins gp22 and gp29 were further investigated in patients treated for Pseudomonas infections at the PTU HIIET (for description of the patients see: Supplementary Table S3). In general, we did not observe increased specific immune responses to the F8 phage. First, the efficiency of plating phages before and after phage therapy revealed no significant increase in phage-neutralizing activity of patients' sera (data not shown), though only 11 patients were available for this study. These patients were not injected with phage preparations, but they were treated either orally or topically, which makes an important limitation for comparisons to the animal model. However, assessment of IgG fractions specific to proteins gp22 and gp29 (in the same sera from patients undergoing phage therapy) revealed no significant differences when compared before and after phage therapy (Supplementary Fig. S3).
Discussion
Consistent with other phages, bacteriophages F8, LMA2, and DP1 can induce phage-specific antibodies.1–6,10,28 In our animal model, these phages also display similar kinetics of IgM and IgG induction as those of other bacteriophage isolates (A3R, 676Z, T4)7,29 (Kaźmierczak et al., under assessment). Since these observations are very similar for taxonomically different phages, with different host bacteria, the observed kinetics of immune responses to phages seems to be universal and might also apply to other phages. At least phages characterized with similar structure and chemical composition (dsDNA, tailed) may share similar immunogenicity and resulting kinetics of immune response to phages. To date, variations in administration schedules and doses levels have not yet been studied, but they could result in differences in kinetics.
Among the investigated phages, LMA2 appears to be the least immunogenic in our analyses (Fig. 1). Further, structural protein gp29 was less immunogenic in the LMA2 than in the F8 and DP1 phage (Fig. 2), which offers an interesting consistency. Indeed, these data suggest that differences in “total immunogenicity” of PB1-like phages (response induced by whole virions) may result from specific differences in the ability of gp29 to induce specific IgG. Protein gp29 in LMA2 differs from its homologs in F8 and DP1 in only two positions: Thr37→Pro and Ile67→Thr (F8 or DP1→LMA2, respectively) (Supplementary Fig. S4). Reports on immunogenicity of similar peptides are extremely scarce, but one report from in silico models developed for eukaryotic viruses suggested that threonine deletion might result in decreased immunogenicity of viral peptides.30 Thus, it is possible that substitution in the position 37 mediated observed lower immunogenicity of the LMA2 phage in this model. However, it should be noted that there is a limitation for comparisons of phage immunogenicity (between phage isolates) due to differences in phage adherence to detection plates in ELISA31 Since specific testing by quantitative PCR (qPCR) for very small quantities of PB1-like phages (like those immobilized on plastic surfaces) has not been validated yet, detailed comparison between F8, LMA2, and DP1 phage is not available. As long as reference genes for qPCR in this group of phages have not been revealed, unverified adherence of the investigated phages to plates should be considered as the study limitation. An overall (with whole phage virions) assessment of how F8, LMA2, and DP1 can be detected by ELISA was conducted by cross-reaction testing (Supplementary Fig. S5) and it revealed no differences between these three phages.
In the healthy human population, the lowest frequency of phage-neutralizing antibodies (11%) was also observed for LMA2. This frequency was slightly higher for F8 (15%) and markedly higher for DP1 (40%) (Fig. 3). Despite the fact that the investigated phages are highly similar, reactivity of a particular human's sera differed between phages in 42% of individuals (23 people out of 55) (Supplementary Table S2). This demonstrates antigenic differences between closely related phage strains, suggesting that in cases where specific immune response hinders therapeutic use of phage, similar phages have a complementary potential. Further, sera used in this study were the same as those used in the identification of Escherichia phage T4 neutralizing sera6 and staphylococcal phages A3R and 676Z neutralizing sera (Kaźmierczak et al., under assessment), for which neutralization frequencies of 82% and about 35% were observed, respectively. This demonstrates that pre-immunization of the human population to phages is highly differentiated between phage isolates. Probably, antibodies specific to phages that are common in the environment and human gastrointestinal tracts (like T4) are more frequent than those neutralizing other phages.
No significant differences were found in population immunity (specific IgG in healthy humans) to proteins gp22 and gp29 (Fig. 4). These proteins were selected for the study due to their structural function and good exposition on phage capsids, as previously demonstrated21,25 and confirmed herein for the DP1 phage (Supplementary Fig. S3). Their contribution to the immunogenicity of PB1-like phages at the population level seems to be of limited importance, although specific antibodies (to these proteins) were observed in the investigated population. Importantly, “positive” and “negative” human sera (Fig. 3) were determined by their ability to neutralize the phages, whereas gp22- and gp-29 specific sera developed in mice did not affect the activity of the investigated phages (data not shown). Further, relatively high cutoff values were observed in the anti-gp22 and anti-gp29 plots from healthy human populations (Fig. 4), which means that a correlation between their occurrence and phage-blocking activity of the sera was weak. This further indicates that proteins gp22 and gp29 are not involved in the induction of phage-neutralizing antibodies. Neutralization might probably be elicited by other virion proteins, or by their combination. Such proteins must probably be directly involved in the process of phage infection of bacteria, since antibodies specific to gp29 do not affect phage infectivity. Also, in patients treated for Pseudomonas infections with F8 phage (Phage Therapy Unit of HIIET), no significant increase in the levels of gp22- and gp29-specific IgG was observed (Supplementary Table S3). So far, no immunological data on other (than F8) PB1-like phage used in phage therapy in humans are available. For this reason, possible cross-reactions and cross-neutralization cannot be fully excluded. An important caveat here, which limits the comparison, is also the difference in administration route, between the therapeutic application (to humans) and model application (to animals), as patients were treated orally. For this reason, further investigation of other administration routes and schedules should be considered one of the major directions in immunological studies of these therapeutic phages.
Conclusion
Important structural phage proteins, present in virions in many copies (e.g., tail sheath assembly), are potentially capable of inducing specific antibodies (as demonstrated in mice), but they proved much less immunogenic in humans when applied in the therapeutic schedule. In addition, induced anti-gp22 and anti-gp29 antibodies had no direct neutralizing effect on phage activity. In the future, other structural proteins of PB1-like phages should to be investigated, together with other schedules of immunization and exposition to phages.
Overall, these observations contribute to our limited understanding of how specific immune response determines phage bioavailability in animals and in humans. However, the impact toward phage cocktail design and the individual success rate of phage therapy applications may be directly linked to these properties and should be incorporated as a key parameter in clinical trial studies on phage therapy.
Supplementary Material
Authors' Contributions
K.H.S. performed major part of the experimental work, including gene cloning, protein expression and purification, animal experiments, immunological assays, and participated in data analysis. Z.K., J.M., D.L., M.H., P.W., and B.O. participated in experimental work including gene cloning, protein expression and purification, animal experiments, and immunological assays. S.S. and J.A. delivered DP1 phage, participated in and guided DP1 phage gene cloning, and participated in concluding results and writing the article. R.L. delivered LMA2 phage, participated in and guided LMA2 gene cloning, concluding results and in writing the article. S.N. performed TEM immunolocalization. A.G. and R.M. covered all aspects of phage therapy, including patients' interaction and supervision, treatment, and related information analysis. A.G. reviewed the article; K.G. participated in the preparation of the article; K.D. conceived and planned the study, participated in gene cloning, protein expression and purification, animal experiments, immunological assays, analyzed and concluded data, and wrote the article. All authors have reviewed and approved of the article before submission.
Author Disclosure Statement
No other competing financial interests exist.
Funding Information
This work was supported by the National Science Centre in Poland grant no. UMO-2012/05/E/NZ6/03314 (granted to K.D.). S.M.S. acknowledges the European Union's Horizon 2020 research and innovation programme under grant agreement No. 713640.
Supplementary Material
References
- 1. Uhr JW, Finkelstein MS, Baumann JB. Antibody formation. III. The primary and secondary antibody response to bacteriophage phi × 174 in guinea pigs. J Exp Med. 1962;115:655–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Uhr JW, Finkelstein S, Franklin EC. Antibody response to bacteriophage phi-X-174 in non-mammalian vertebrates. Proc Soc Exp Biol Med. 1962;111:13–15. [DOI] [PubMed] [Google Scholar]
- 3. Huff WE, Huff GR, Rath NC, et al. . Evaluation of aerosol spray and intramuscular injection of bacteriophage to treat an Escherichia coli respiratory infection. Poult Sci. 2003;82:1108–1112. [DOI] [PubMed] [Google Scholar]
- 4. Huff WE, Huff GR, Rath NC, et al. . Prevention of Escherichia coli infection in broiler chickens with a bacteriophage aerosol spray. Poult Sci. 2002;81:1486–1491. [DOI] [PubMed] [Google Scholar]
- 5. Huff WE, Huff GR, Rath NC, et al. . Immune interference of bacteriophage efficacy when treating colibacillosis in poultry. Poult Sci. 2010;89:895–900. [DOI] [PubMed] [Google Scholar]
- 6. Dąbrowska K, Miernikiewicz P, Piotrowicz A, et al. . Immunogenicity studies of proteins forming the T4 phage head surface. J Virol. 2014;88:12551–12557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Majewska J, Beta W, Lecion D, et al. . Oral application of T4 phage induces weak antibody production in the gut and in the blood. Viruses. 2015;7:4783–4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kucharewicz-Krukowska A, Slopek S. Immunogenic effect of bacteriophage in patients subjected to phage therapy. Arch Immunol Ther Exp (Warsz) 1987;35:553–561. [PubMed] [Google Scholar]
- 9. Lusiak-Szelachowska M, Zaczek M, Weber-Dabrowska B, et al. . Antiphage activity of sera during phage therapy in relation to its outcome. Future Microbiol. 2017;12:109–117. [DOI] [PubMed] [Google Scholar]
- 10. Fogelman I, Davey V, Ochs HD, et al. . Evaluation of CD4+ T cell function In vivo in HIV-infected patients as measured by bacteriophage phiX174 immunization. J Infect Dis. 2000;182:435–441. [DOI] [PubMed] [Google Scholar]
- 11. Bradley SG, Watson DW. Production of neutralizing antibody by mice injected with actinophage. J Immunol. 1963;90:782–787. [PubMed] [Google Scholar]
- 12. Nelstrop AE, Taylor G, Collard P. Studies on phagocytosis. II. In vitro phagocytosis by macrophages. Immunology. 1968;14:339–346. [PMC free article] [PubMed] [Google Scholar]
- 13. Hodyra-Stefaniak K, Miernikiewicz P, Drapala J, et al. . Mammalian host-versus-phage immune response determines phage fate in vivo. Sci Rep. 2015;5:14802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zaczek M, Lusiak-Szelachowska M, Jonczyk-Matysiak E, et al. . Antibody production in response to staphylococcal MS-1 phage cocktail in patients undergoing phage therapy. Front Microbiol. 2016;7:1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Capparelli R, Nocerino N, Iannaccone M, et al. . Bacteriophage therapy of Salmonella enterica: A fresh appraisal of bacteriophage therapy. J Infect Dis. 2010;201:52–61. [DOI] [PubMed] [Google Scholar]
- 16. Capparelli R, Parlato M, Borriello G, et al. . Experimental phage therapy against Staphylococcus aureus in mice. Antimicrob Agents Chemother. 2007;51:2765–2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang J, Hu B, Xu M, et al. . Use of bacteriophage in the treatment of experimental animal bacteremia from imipenem-resistant Pseudomonas aeruginosa. Int J Mol Med. 2006;17:309–317. [PubMed] [Google Scholar]
- 18. Jerne NK, Avegno P. The development of the phage-inactivating properties of serum during the course of specific immunization of an animal: Reversible and irreversible inactivation. J Immunol. 1956;76:200–208. [PubMed] [Google Scholar]
- 19. Jerne NK. The presence in normal serum of specific antibody against bacteriophage T4 and its increase during the earliest stages of immunization. J Immunol. 1956;76:209–216. [PubMed] [Google Scholar]
- 20. Miedzybrodzki R, Borysowski J, Weber-Dabrowska B, et al. . Clinical aspects of phage therapy. Adv Virus Res. 2012;83:73–121. [DOI] [PubMed] [Google Scholar]
- 21. Ceyssens PJ, Miroshnikov K, Mattheus W, et al. . Comparative analysis of the widespread and conserved PB1-like viruses infecting Pseudomonas aeruginosa. Environ Microbiol. 2009;11:2874–2883. [DOI] [PubMed] [Google Scholar]
- 22. Pires DP, Sillankorva S, Kropinski AM, et al. . Complete genome sequence of Pseudomonas aeruginosa phage vB_PaeM_CEB_DP1. Genome Announc. 2015;3:e00918-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Reed GF, Meade BD, Steinhoff MC. The reverse cumulative distribution plot: A graphic method for exploratory analysis of antibody data. Pediatrics. 1995;96:600–603. [PubMed] [Google Scholar]
- 24. Kilkenny C, Browne WJ, Cuthill IC, et al. . Improving bioscience research reporting: The ARRIVE guidelines for reporting animal research. PLoS Biology. 2010;8:e1000412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fil'chikov MV, Osmakov DI, Logovskaia LV, et al. . [Pseudomonas aeruginosa bacteriophage SN: 3D-reconstruction of the capsid and identification of surface proteins by electron microscopy]. Bioorg Khim. 2009;35:808–815. [PubMed] [Google Scholar]
- 26. Reyes A, Semenkovich NP, Whiteson K, et al. . Going viral: Next-generation sequencing applied to phage populations in the human gut. Nat Rev Microbiol. 2012;10:607–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Manrique P, Bolduc B, Walk ST, et al. . Healthy human gut phageome. Proc Natl Acad Sci U S A. 2016;113:10400–10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ochs HD, Davis SD, Wedgwood RJ. Immunologic responses to bacteriophage phi-X 174 in immunodeficiency diseases. J Clin Invest. 1971;50:2559–2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Majewska JKZ, Lahutta KLD, Szymczak A, et al. . Induction of phage-specific antibodies by two therapeutic staphylococcal bacteriophages administered per os. Front Immunol. 2019;10:2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hecht L, Dormer A. The evolutionary significance of certain amino acid substitutions and their consequences for HIV-1 immunogenicity toward HLA's A*0201 and B*27. Bioinformation. 2013;9:315–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Klopot A, Zakrzewska A, Lecion D, et al. . Real-time qPCR as a method for detection of antibody-neutralized phage particles. Front Microbiol. 2017;8:2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.