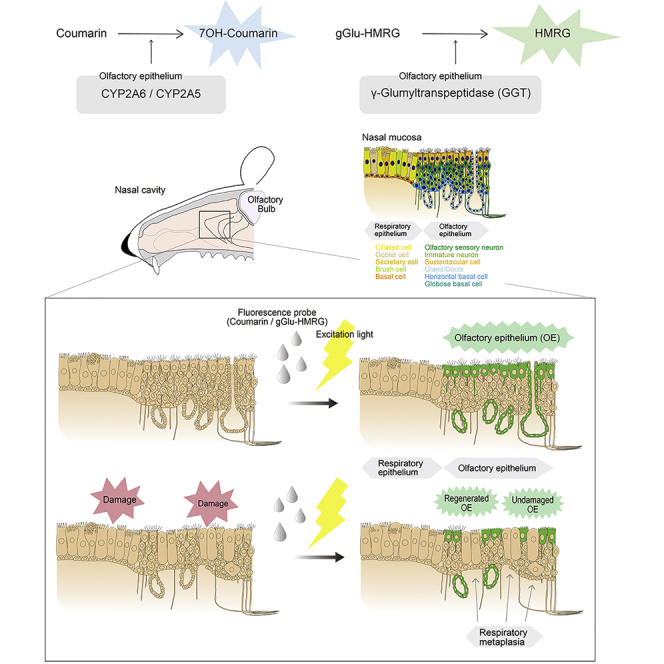
Summary
Olfactory epithelium (OE) undergoes degeneration in disorders such as age-related and post-viral olfactory dysfunction. However, methods for real-time in vivo detection of OE and assessment of total extent within the nasal cavity are currently unavailable. We identified two fluorescence probes for rapidly detecting and evaluating the entire extent of mice OE with topical application. Taking advantage of the differential expression of the enzymes cytochrome p450 (CYP) and γ-glutamyltranspeptidase (GGT) in OE relative to respiratory epithelium, we utilized the conversion of coumarin (a substrate of various CYP subtypes) and gGlu-HRMG (a substrate of GGT) by these enzymes to form metabolites with fluorescent emissions in the duct cells and sustentacular cells of neuron-containing OE. In depleted and regenerated OE model, the emission of these probes remained absent in respiratory metaplasia but appeared in regenerated OE. These substrates could be used to monitor OE degeneration and follow regenerative response to therapeutic interventions.
Subject areas: Medical imaging, Biological sciences, Biochemistry, Biological sciences research methodologies
Graphical abstract
Highlights
-
•
Enzymes Cyp2a5 and Ggt7 are expressed in olfactory epithelial cells
-
•
Substrates for Cyp2a5 and Ggt7 can label olfactory epithelium (OE) in situ
-
•
Lesion recovered, not damaged OE, is labeled with Cyp2a5 and Ggt7 substrates
Medical imaging; Biological sciences; Biochemistry; Biological sciences research methodologies
Introduction
The olfactory epithelium (OE) contains unique olfactory receptor neurons which connect the external environment through dendritic processes with the CNS through axonal processes of the same cells that synapse in the olfactory bulbs (Doty, 2015). The OE of vertebrates is a highly regenerative neuroepithelium that is maintained under normal conditions by a population of stem and progenitor cells throughout life (Graziadei and Graziadei, 1979; Schwob et al., 2017). However, pathologic conditions such as age-related or post-viral olfactory disorders lead to replacement of normal olfactory neuroepithelium with areas of aneuronal epithelium and respiratory metaplasia (Holbrook et al., 2011; Yamagishi et al., 1994).
Accurate in vivo identification of OE throughout the nasal cavity would facilitate quantification of degenerative epithelial changes occurring in olfactory disorders. This technique could improve disease diagnosis, prognosis for recovery, and monitor therapeutic efficacy. In particular, therapeutic interventions could be assessed objectively via either direct or indirect measures of neuronal regeneration in damaged areas. In addition, visualization of OE could provide a clear boundary for safe access during endoscopic sinus and skull base surgery by avoiding the OE and minimizing complications of temporary or permanent smell loss. However, techniques for real-time detection and discrimination of OE from respiratory epithelium (RE) or intact neuron-rich OE from neurogenically exhausted OE are not currently available. Surgical biopsy of the OE remains the only diagnostic method available for determining degenerative changes in OE (Holbrook et al., 2016), but this invasive procedure is limited in assessing only a small portion of the OE that may not reflect the entire neuroepithelium and also carries the risk, albeit minimal, of causing further olfactory dysfunction.
Fluorescent imaging is a powerful tool for in vivo visualization of regions of interest. Activatable fluorescent probes utilize various specific enzymatic reactions and can provide high-contrast images in a short time without any washout procedure (Urano et al., 2011). The OE, as well as RE, contains numerous tissue-specific enzymes that provide a high capacity for metabolizing endogenous and exogenous, or xenobiotic, substrates (Doty, 2015). Investigating enzymes that are differentially expressed in OE relative to RE for tissue-specific imaging is a suitable strategy to accomplish the goal of in vivo OE imaging.
Here, we describe a real-time vital imaging method for detecting and evaluating the neurogenic OE using two different fluorescence probes, coumarin and γ-glutamyl hydroxymethyl rhodamine green (gGlu-HMRG). These probes activate rapidly with enzymatic reaction in the presence of OE-specific enzymes cytochrome p450-2A6 (CYP2A6) and γ-glutamyltranspeptidase (GGT). The probes can be used topically by spraying onto nasal epithelium, and instant activation of the probe makes precise in situ detection of OE feasible.
Results
CYP2A6 is highly expressed in OE of mice and humans compared to RE
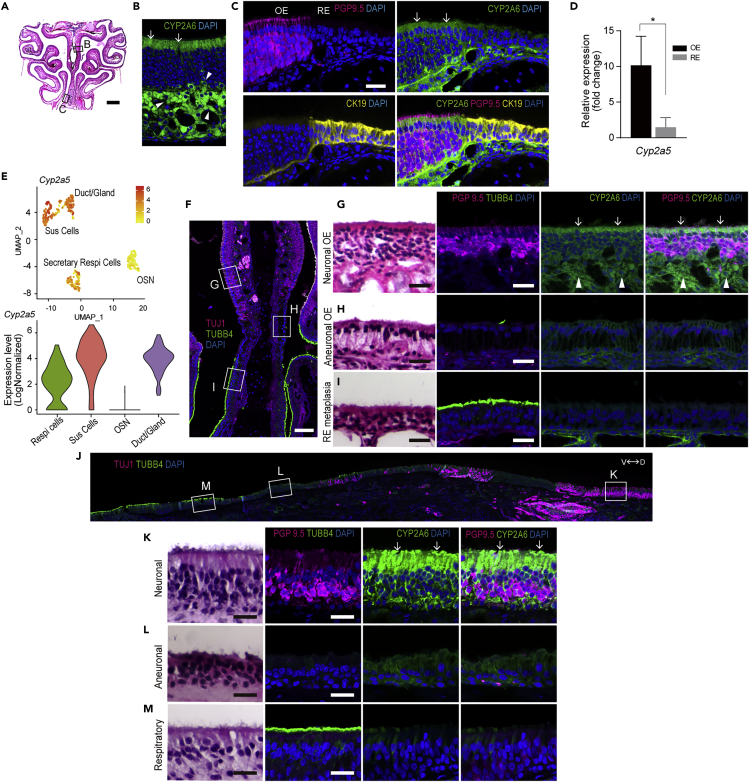
Cytochrome P450 (CYP) enzymes are ubiquitous catalysts of oxidative metabolism in biological systems (Lewis and Ito, 2008). Focusing on the 3.97-fold higher expression of CYP2A6 in OE compared to RE by whole genome analysis of human nasal mucosa (Olender et al., 2016), we investigated the expression of CYP2A6 in the nasal mucosa of human and the mouse ortholog Cyp2a5 in mouse nasal mucosa. Immunohistochemical staining of sections through nasal mucosa using anti-CYP2A6 antibodies demonstrated strong labeling at the apical surface of sustentacular cells and Bowman’s glands/ducts of mouse OE (Figure 1B). In contrast, the RE does not stain with anti-CYP2A6; the transition is sharp and is coincident with the boundary between the OE and the RE marked by the differential expression of PGP9.5 in OE and CK19 in RE (Figure 1C). Given the differential staining with anti-CYP2A6 in OE compared to RE, we confirmed increased gene expression of Cyp2a5, the mouse ortholog of human CYP2A6, in OE compared to RE using qPCR. The expression of Cyp2a5 was almost 10 times higher in OE than RE in mice (Figure 1D). In addition, single-cell RNA-seq of mouse nasal mucosa identified high expression of Cyp2a5 in sustentacular cells, Bowman’s glands, and ducts of OE but low expression in secretory RE cells (Figure 1E).
Figure 1.
Anti-CYP2A6 staining is identified in mouse and human OE
(A) Coronal section through the nasal cavity of a mouse stained with H&E. Scale bar = 1 mm.
(B) Anti-CYP2A6 staining of nasal septum OE; the area illustrated corresponds to the boxed area in Figure 1A. CYP2A6 staining is mainly located in sustentacular cells especially at the apical surface (arrows) and glands (arrowheads). Scale bar = 25 μm.
(C) IHC with anti-CYP2A6 staining recognizes the boundary between OE and RE. The area illustrated corresponds to the boxed area in Figure 1A. Antibodies against PGP 9.5 label olfactory sensory neurons (OSNs) of OE, while CK19 antibodies label columnar ciliated cells of the RE. CYP2A6 staining is found only in the sustentacular cells in OE (arrows) and is absent in RE. Scale bar = 25 μm.
(D) Relative mRNA expression of Cyp2a5 in mouse. The data are shown as the mean ± SEM (n = 4). ∗p < 0.05.
(E) Single-cell RNA-seq t-SNE and violin plots with expression levels of the Cyp2a5 gene in the mouse nasal epithelium.
(F) IHC of the nasal septum from an OMP-tTA; TetO-DTA transgenic mouse model. A mosaic image of a section labeled with anti-TUJ1 identifies OSNs (box G), and labeled with TUBB4 to identifies RE (box I). Areas that have become aneuronal are absent of labeling with both antibodies (box H). Scale bar = 100 μm.
(G) The neurogenic OE contains PGP9.5 (+) OSNs as well as CYP2A6 (+) sustentacular cells (arrow) and glands (arrowhead).
(H) The aneuronal OE lacks PGP9.5/TUBB4/CYP2A6 staining.
(I) Respiratory metaplastic epithelium contains TUBB4 (+) columnar respiratory cells but lacks PGP9.5/CYP2A6 staining. Scale bars = 25 μm.
(J) A mosaic image of a human section through the OE obtained at autopsy (74-year-old male) includes neuronal OE (TUJ1+/TUBB4-, box K), RE (TUJ1-/TUBB4+, box M), and patches of aneuronal OE (TUJ1-/TUBB4-, box L). D = dorsal, V = ventral. Scale bar = 250 μm.
(K) Neuronal OE contains PGP9.5(+) OSNs as well as CYP2A6(+) sustentacular cells with high labeling at the apical surface (arrows).
(L) Aneuronal OE lacks both PGP9.5 and TUBB4 staining.
(M) RE contains TUBB4(+) columnar epithelial cells but lacks both PGP9.5 and CYP2A6 labeling. Scale bars = 25 μm.
Our imunohistological analysis demonstrated differential staining with anti-CYP2A6 in OE vs. RE. However, in adult humans, the olfactory area is not a continuous sheet of neuron-rich epithelium as observed in young laboratory-housed rodents. Regions of respiratory metaplasia and aneuronal OE are patchily distributed within the boundaries of OE (Holbrook et al., 2011). We used our transgenic mouse model of OE aging to model the degeneration that is found in humans (OMP-tTA; TetO-DTA transgenic mouse). In this model, expression of diptheria toxin is under the indirect control of the olfactory marker protein gene, which abbreviates the lifespan and induces death of mature olfactory neurons (Figure S1). As a consequence, areas of the OE become aneuronal or undergo RE metaplasia (Child et al., 2018) (Figure 1F). Where aneuronal, the OE lacks globose basal cells and TUJ1/PGP9.5 (+) olfactory sensory neurons but retains horizontal basal cells and sustentacular cells establishing the epithelium as olfactory (cf. Figures 1G vs. 1H) (Holbrook et al., 2011). Where metaplastic, the epithelium becomes respiratory in composition, and the constituent ciliated columnar REcytes label with anti-beta-IV tubulin (TUBB4) (cf. Figures 1G vs. 1I). CYP2A6 antibody labeling of sections from the aging mouse model revealed expression in sustentacular cells of normal neurogenic OE, but not in the areas of aneuronal OE and RE metaplasia (Figures 1G–1I). Using the same battery of antibodies on sections of human nasal mucosa, we identified similar mixing of normal OE with aneuronal and respiratory epithelium (Figure 1J). On closer analysis, we found strong expression of CYP2A6 in sustentacular cells of OE, whereas aneuronal OE and metaplastic RE lacked staining for CYP2A6 (Figures 1J–1M). Thus, CYP2A6 marks neurogenic OE of both mice and humans and can be used to differentiate neurogenic OE from aneuronal OE or RE.
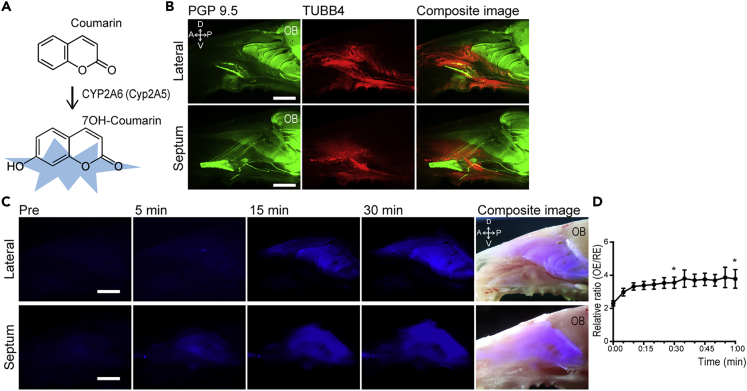
Vital imaging of OE with the CYP2A6 substrate, coumarin
Coumarin, a substrate of CYP2A6, is a naturally occurring odorous compound widely used in consumer products such as a fragrance ingredient, and also as a therapeutic agent such as herbal medical products (Lewis et al., 2006; Raunio et al., 2008). Coumarin is specifically catalyzed by CYP2A6 with subsequent formation of 7-hydroxycoumarin which fluoresces blue (440–460 nm) under UV excitation (Lewis et al., 2006; Pelkonen et al., 2000; Sherman and Robins, 1968) (Figure 2A).
Figure 2.
Vital imaging of nasal mucosa after application of coumarin demonstrates labeling of OE
(A) Coumarin is catalyzed to 7OH-coumarin by CYP2A6.
(B) Whole mount staining of a normal mouse nasal cavity using antibodies against PGP 9.5 to label OSNs, and TUBB4 to label respiratory cells reveal the demarcation between OE and RE on the lateral wall with turbinates and septum.
(C) Fluorescent imaging of whole mounts loaded with 10 mM coumarin captured before and 5, 15, and 30 min after probe application reveal signal in areas comparable to OE.
(D) Relative fluorescence intensity of OE vs. RE measured every 5 min after application of coumarin. The data are shown as the mean ± SEM (n = 5). Statistical differences were calculated in initial time vs 30 min and initial time vs 60 min ∗p < 0.05. D = dorsal, V = ventral, A = anterior, p = posterior, OB = olfactory bulb, scale bars = 2 mm.
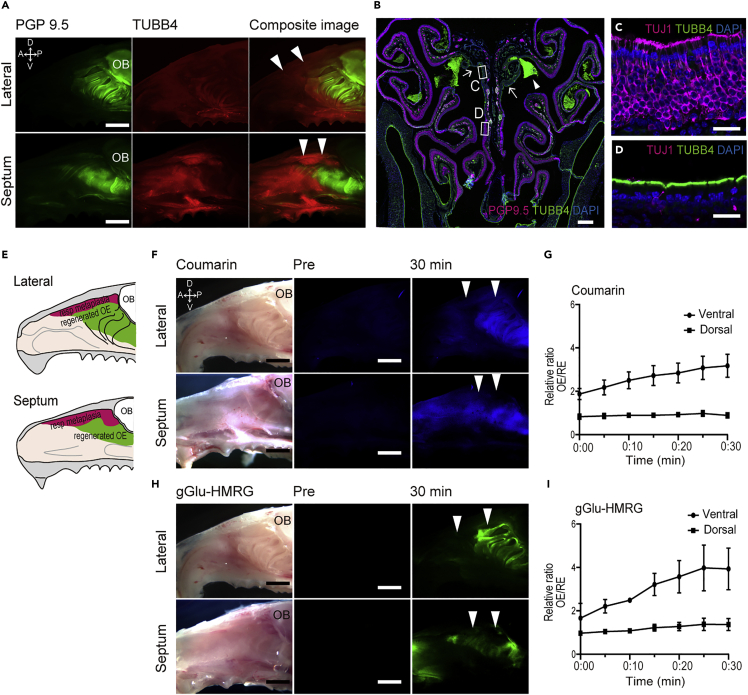
We stained whole mounts of the septum and lateral wall of the mouse nasal cavity with anti-PGP9.5 and anti-TUBB4 to identify the boundary of OE and RE against which the distribution of coumarin-derived fluorescence could be compared (Figure 2B).
A 10 mM solution of coumarin was applied topically to the mouse nasal mucosa immediately after dissection. After 5 min, blue fluorescence was observed in the area of OE under UV light (Figure 2C). The emission plateaued within 15 min (Figures 2C and 2D). Coumarin-generated fluorescence coincided with the OE as demonstrated by comparison to wholemount neuronal staining (Figure 2B). Likewise, the area marked by coumarin-derived fluorescence coincided with the distribution mature olfactory sensory neurons visualized in OMP-GFP mice (Figure S2).
In order to show substrate selectivity of CYP2A6 for coumarin, methoxypsoralen, an inhibitor of CYP2A6 (Mäenpää et al., 1994), was mixed with the coumarin solution before application to the nasal mucosa. The fluorescence signal was significantly reduced (Figure S3A). To rule out whether differences in coumarin-derived fluorescence between OE and RE were due to non-enzymatic reasons such as differential background coloration or epithelial thickness, we directly applied the ostensible metabolite of coumarin, 7OH-coumarin, to the nasal mucosa. The emission of 7OH-coumarin was observed equally in both OE and RE (Figure S3B) supporting coumarin as an OE-specific probe.
GGT7 and GGT1 are highly expressed in OE of mice and humans compared to RE
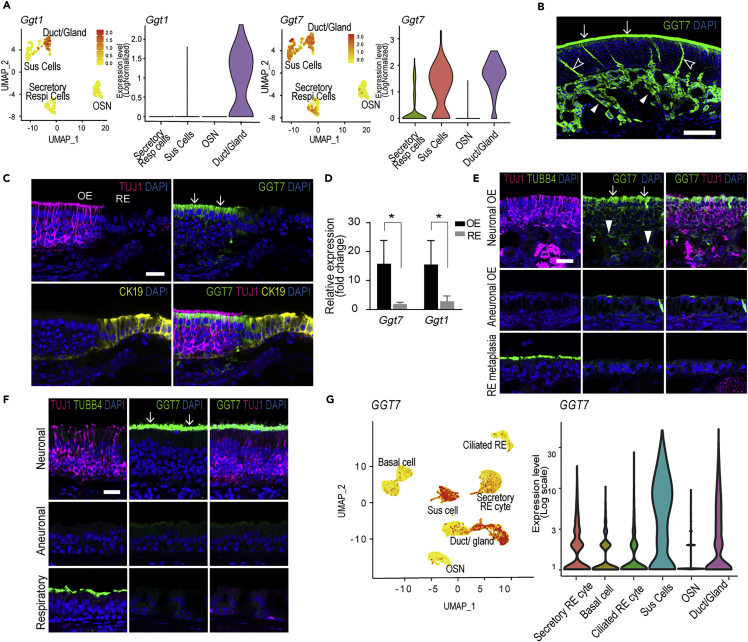
Whole transcriptome sequencing of human nasal mucosa demonstrates high expression of GGTs in OE compared to RE (Olender et al., 2016). GGTs are cell-surface-associated enzymes involved in cellular glutathione homeostasis in multiple tissues. However, the role of GGTs in OE is unclear. Of the several GGT types, single-cell RNA-seq of mouse nasal mucosa demonstrated that both GGT1 and GGT7 are expressed in cells of the nasal mucosa (Figure 3A). Specifically, GGT1 is highly expressed in duct/gland cells, and to a lesser extent in Sus cells, while GGT7 is highly expressed in both Sus cells and duct/gland cells (Figure 3A).
Figure 3.
Presence of GGT1 or GGT7 in mouse and human olfactory epithelium
(A) Single-cell RNA-seq t-SNE plots and violin plots with overlaid log normalized expression levels of the Ggt1 and Ggt7 genes in the mouse nasal epithelium.
(B) Anti-GGT7 staining of a coronal section through a mouse nasal septum corresponding to the boxed area labeled “B” in Figure 1A mainly labels the apical surface of sustentacular cells (arrows), glands (arrowheads), and duct cells (open arrowheads) in OE. Scale bar = 50 μm.
(C) Immunofluorescent staining of the boundary between OE and RE corresponding to the boxed area labeled “C” in Figure 1A with antibodies against TUJ1 to label OSNs and CK19 to label respiratory cells shows anti-GGT7 labeling confined to sustentacular cells (arrows) in OE. Scale bar = 25 μm.
(D) Relative mRNA expression of Ggt7 and Ggt1 in mouse OE and RE. The data are shown as the mean ± SEM (n = 4). ∗p < 0.05.
(E) Immunofluorescent staining of OE from an OMP-tTA; TetO-DTA transgenic mouse from the boxed areas labeled “G-I” in Figure 1F illustrates the selective staining of anti-GGT7 for neuronal OE. The neuronal OE contains TUJ1(+) OSNs as well as GGT7(+) sustentacular cells (arrows) and glands (arrowheads). The aneuronal OE lacks staining with TUJ1, TUBB4, and GGT7. Areas of respiratory metaplasia contain TUBB4(+) respiratory cells but lack TUJ1 and GGT7 staining. Scale bar = 25 μm.
(F) Sections of human autopsy samples corresponding with boxed areas labeled “K-M” in Figure 1J are labeled with anti-GGT7. The neuronal OE contains TUJ1(+) OSNs as well as GGT7(+) sustentacular cells (arrows). The aneuronal OE lacks TUJ1, TUBB4, and GGT7 staining. Respiratory metaplastic epitlelium consists of TUBB4 positive cells but lacks TUJ1 and GGT7 staining. Scale bar = 25 μm.
(G) Single-cell RNA-seq t-SNE and violin plots with expression levels of the GGT7 gene in the human nasal epithelium.
We then investigated the expression pattern of GGT1 and GGT7 through immunohistochemical staining of nasal mucosa. Anti-GGT7 labeling of the sustentacular cells, Bowman’s glands, and ducts in OE of mice matches the pattern of gene expression (cf. Figures 3B vs. 3C). Like CYP2A6, the expression of GGT7 coincides with the OE and falls sharply at the boundary between OE and RE in the posterior ventral part of the murine nasal cavity, as marked by the transition from neuronal staining with anti-TUJ1 to apical labeling of the respiratory epithelium for CK19 (Figure 3C). Staining with two different commercially available antibodies against GGT1 corresponded less well with the gene expression data. Both antibodies additionally label neurons resulting in high background (data not shown). Ggt1 was not identified in olfactory receptor neurons suggesting that the above antibody IHC labeling was a result of cross reactivity with a different antigen. In addition, qPCR comparison of mouse OE vs. RE demonstrated that expression of both Ggt7 and Ggt1 was approximately 15 times higher in OE (Figure 3D). We again utilized our aging mouse model (OMP-tTA; TetO-DTA) to assess the ability of anti-GGT7 antibodies to identify small areas of OE from non-neuronal epithelium. GGT7 was expressed in neurogenic OE, but not in areas of aneuronal OE or RE metaplasia (Figure 3E). Additionally, as observed with anti-CYP2A6, staining of human nasal mucosa with anti-GGT7 antibodies revealed staining in Sus cells of neurogenic OE but was absent in aneuronal and RE (Figure 3F). Furthermore, single-cell RNA-seq of human nasal mucosa identified high expression of GGT7 in sustentacular cells, Bowman’s glands, and ducts of OE but low expression in secretory RE cells (Figures 3G and S7).
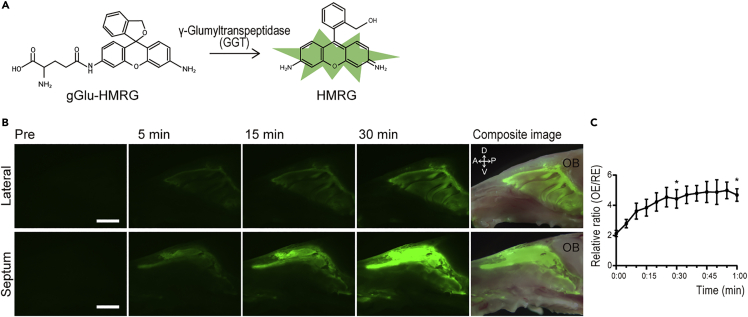
Vital imaging of OE with the GGT substrate, gGlu-HMRG
To take potential advantage of the differential expression of GGT1 and GGT7 in OE, we stained the nasal mucosa whole mounts with the GGT substrate gGlu-HMRG. gGlu-HMRG is non-fluorescent in its native state but is rapidly hydrolyzed by GGT to yield a highly fluorescent molecule, hydroxymethyl rhodamine green (HMRG) (Urano et al., 2011) (Figure 4A). Mouse nasal mucosa incubated with gGlu-HMRG soon after dissection displayed a strong green fluorescent signal corresponding to the area of OE within minutes (Figure 4B). The emission approached a plateau by 20 min (Figure 4C). The stained area clearly coincides with the region of OE that was indicated by whole mount antibody staining of the nasal cavity (Figure 2B) and by the distribution of mature olfactory sensory neurons in OMP-GFP mice (Figure S2).
Figure 4.
Vital imaging of nasal mucosa after application of gGlu-HMRG demonstrates labeling of OE
(A) gGlu-HMRG is hydrolyzed to HMRG by the catalytic activity of GGTs.
(B) Fluorescent imaging of nasal whole mounts loaded with 10 μM Glu-HMRG captured before and 5, 15, and 30 min after probe application reveal signal in areas comparable to OE.
(C) Relative fluorescence intensity of nasal mucosa of OE vs. RE measured every 5 min after application of gGlu-HMRG. The data are shown as the mean ± SEM (n = 5). Statistical differences were calculated in initial time vs 30 min and initial time vs 60 min ∗p < 0.05. D = dorsal, V = ventral, A = anterior, p = posterior, OB = olfactory bulb, scale bars = 2 mm.
Substrate selectivity of gGlu-HMRG for GGT was demonstrated using an inhibitor of GGT, GGsTop (Yamamoto et al., 2011). When the nasal mucosa was co-incubated with gGlu-HMRG and GgsTop, hydrolyzation of gGlu-HMRG was suppressed resulting in a significantly reduced fluorescence signal (Figure S4A). As with coumarin, possible signal artifact due to differences of background epithelial coloration and variation in epithelial thickness between OE and RE is possible. The metabolite of gGlu-HMRG, HMRG, was applied topically to the mucosal surface to assess for signal intensity that may be attributed to non-enzymatic differences. HMRG fluorescence was observed both in OE and RE with no significant variation at the OE-RE junction (Figure S4B).
Vital imaging of depleted and regenerated OE
Because small regions of respiratory and aneuronal epithelium in human olfactory mucosal are intermingled with neuron-containing OE in human biopsy or autopsy tissue, the spatial resolution with which a potential vital stain can generate a differential fluorescent signal becomes important. To confirm the ability of coumarin and gGlu-HMRG to distinguish between neuronal and non-neuronal regions of damaged nasal epithelium in whole mounts, individual mice were treated with a mixture of the olfactotoxic compounds dichlobenil (Brandt et al., 1990) and methimazole (Genter et al., 1995). The entire area of OE was depleted 1 week after administration (Figure S5A). After 5 weeks, OE regeneration was observed in much of the tissue with the exception of the lining of the dorsal recess of the nasal cavity (Figures 5A and 5B) In this region, whole mount staining of methimazole- and dichilobenil-treated mice at 5 weeks after administration demonstrated respiratory metaplasia, i.e., a lack of PGP9.5 labeling and abundant staining with anti-TUBB4 (Figure 5A). Respiratory metaplasia of the dorsal recess area was also confirmed by immunohistology (Figures 5B–5D).
Figure 5.
Vital imaging can define areas of neuronal OE regeneration after lesioning
(A) Whole mount staining of the nasal cavity from a methimazole/dichlobenil lesioned mouse using PGP 9.5/TUBB4 identifies areas on both the lateral wall and septum where respiratory metaplasia (arrowheads) in previous neuronal OE. D = dorsal, V = ventral, A = anterior, p = posterior, OB = olfactory bulb. Scale bars = 2 mm.
(B) A coronal section of the nasal cavity from a methimazole/dichlobenil lesioned mouse demonstrates respiratory metaplasia in the dorsal area of OE (arrows). The arrowhead indicates intracavitary excrescences from the injured OE. Scale bar = 1 mm.
(C) Regenerated ventral OE from the boxed area labeled “C” in Figure 5B contains a thick layer of TUJ1 labeled OSNs without TUBB4+ cells. Scale bar = 25 μm.
(D) A dorsal region of OE from the boxed area labeled “D” in Figure 5B is without neurons and is labeled for respiratory cells with TUBB4 antibodies. Scale bar = 25 μm.
(E) A cartoon illustrates wholemount staining in a methimazole/dichlobenil-injected mouse with respiratory metaplasia in the dorsal recess of the nasal cavity.
(F) Fluorescent imaging of whole mounts incubated with 10 mM coumarin captured before and 30 min after probe application reveals signal in the ventral area of OE but not in dorsal areas of what was formerly OE (arrowheads).
(G) Relative fluorescence intensity of dorsal vs. ventral mucosa measured every 5 min after application of coumarin. The data are shown as the mean ± SEM (n = 5).
(H) Fluorescent imaging of whole mounts incubated with 10 μM Glu-HMRG captured before and 30 min after probe application again reveals signal in the ventral area of OE but not the dorsal area (arrowheads).
(I) Relative fluorescence intensity of dorsal vs. ventral mucosa measured every 5 min after application of gGlu-HMRG. The data are shown as the mean ± SEM (n = 5). D = dorsal, V = ventral, A = anterior, p = posterior, OB = olfactory bulb, scale bars = 2 mm.
Coumarin and gGlu-HMRG was topically applied to the nasal mucosa of dichlobenil/methimazole-treated mice after 5 weeks of recovery. Fluorescence due to both coumarin and gGlu-HMRG was decreased in the dorsal area where neurons were depleted, whereas strong fluorescence was observed in more ventral areas where the OE underwent neuronal regeneration (Figures 5F and 5H). We quantified the signal intensity from each probe in the depleted dorsal OE and in the regenerated ventral region. The intensity of fluorescence was significantly decreased in the dorsal region compared to the regenerated ventral OE area (Figures 5G and 5I).
Discussion
The findings presented here establish two probes for rapid vital imaging that are specific for the neurogenic olfactory epithelium and, hence, may be useful for the assessment of epithelial status longitudinally in vivo. The two probes, coumarin and gGlu-HMRG, enable the visualization and evaluation of the OE in real time via the catalytic activity of CYP2A6 and GGT enzymes, respectively. Impairment of olfactory function correlates with the status of the olfactory periphery and is apparently due to the emergence of neurogenic exhaustion and of respiratory metaplasia (Holbrook et al., 2011; Yamagishi et al., 1994). We currently lack a means by which nasal endoscopic examination can be used to detect OE and to evaluate its changes directly. Biopsies of the olfactory area can give a detailed picture of epithelial composition, including neuronal maturity, but are unsuitable for a more global assessment, given geographic heterogeneity of epithelial status. Therefore, non-invasive OE-specific probes that allow real-time detection and diagnosis of the neurogenic status of OE would be beneficial. Although these probes indicate existence of OE via labeling of non-neuronal cells, they in fact do not label the olfactory receptor neurons and information regarding normal function cannot be inferred.
The mammalian OE contains high levels and unique forms of microsomal CYP enzymes (Ding and Fang, 2015). One recent study identified 33 different CYPs in the OE of different mammals (Heydel et al., 2019). The purpose of these enzymes have been attributed to the maintenance of epithelial integrity, metabolic transformation of odorant molecules, and/or protection of the CNS from damage due to inhaled chemicals (Ling et al., 2004). Of these CYPs, the CYP2G1 gene is expressed in the olfactory mucosa at high level in rodents (Olender et al., 2016; Zhang and Ding, 2008); however, CYP2G1 is a pseudogene without function in human (Sheng et al., 2000). High levels of CYP2A13 expression are also found in the nasal mucosa (Olender et al., 2016; Raunio et al., 2008), but the main substrates of CYP2A13 are nicotine and steroid hormone (Raunio et al., 2008), which are disfavored for future human use due to their toxicity. In contrast, CYP2A6, which is also highly expressed in OE (Olender et al., 2016; Raunio et al., 2008), acts on coumarin, which is a naturally occurring compound contained in a large number of herbal medicinal products used for various disorders in some European countries (Raunio et al., 2008).
CYP2A6 catalyzes the conversion of coumarin to 7-hydroxy-coumarin which emits high-intensity fluorescence in the blue region (440–460 nm) on illumination with UV light (Sherman and Robins, 1968). Both in vitro and in vivo studies established that coumarin is specifically metabolized by CYP2A6, and consequently, it is the preferred probe for phenotyping CYP2A6 activity (Raunio et al., 2008). In this study, we confirmed CYP2A6 or the mouse ortholog Cyp2a5 is limited to neuron-containing OE, and is absent from aneuronal OE and RE. As a consequence, incubation with coumarin, the aforementioned substrate of CYP2A6, rapidly identifies the area of neurogenic OE by fluorescence.
GGT is a family of cell-surface-associated enzymes involved in cellular glutathione homeostasis. Although the role of the GGTs in OE is unclear, GGT7 is one of the most highly expressed biotransformation enzymes present in human OE (Olender et al., 2016). GGTs are also overexpressed in several human tumors, especially epithelial cancers (Fischer et al., 1990; Hanigan et al., 1999). gGlu-HMRG, which is catalyzed to a highly fluorescent product by GGTs, has been investigated as a rapid fluorescent probe for cancers, including lung and hepatic carcinomas (Hino et al., 2016; Miyata et al., 2017). The fluorescent probe is generated rapidly via a one-step enzymatic reaction catalyzed by GGT within 10 min of being applied topically to tissue. The ease of use and rapidity of fluorescent expression has great advantages compared to other imaging probes labeling by which can take hours to days to develop (Nagaya et al., 2017). In this study, we confirmed GGT7 is highly expressed in neuronal OE as compared to aneuronal OE or RE. As a consequence, GGT catalysis of gGlu-HMRG to its fluorescent product rapidly identifies areas of neuronal OE.
The utility of the two probes was established with reference to forms of experimental OE injury that approximate human peripheral olfactory pathology, in this case, systemic administration of olfactotoxins. Dichlobenil and methimazole are metabolized to toxicants that produce acute necrosis of the OE (Brittebo, 1997; Genter et al., 1995). Co-injection of dichlobenil and methimazole can be used to model the respiratory metaplasia characteristic of human olfactory pathology by inducing the dorsal area of the nasal lining to regenerate as respiratory-like epithelium (Brandt et al., 1990). In contrast, damage to the ventral epithelium is less severe, permitting a substantial degree of regeneration and a near-complete restoration of the numbers of olfactory sensory neurons within a few weeks (Suzukawa et al., 2011). Following recovery for 5 weeks after injecton of dichlobenil, the metaplastic dorsal epithelium lacked both CYP2A6/CYP2A5 and GGT7 protein and enzymatic activity. In contrast, ventral epithelium was robustly labeled by both probes. Thus, use of these probes in vivo offers the potential for assaying the neurogenic status of the olfactory area, globally, in patients with olfactory dysfunction before and after therapeutic intervention.
In conclusion, we identified two fluorescence probes for rapidly detecting and evaluating the entire extent of OE with topical application. Fluorescent imaging that selectively targets neurogenic OE has great potential for changing the clinical practice related to olfactory dysfunction because of its high sensitivity, low cost, and real-time capabilities.
Limitation of the study
This study has several limitations. First, small areas of changes between OE and non-OE may not be detectable with these substrates. We need to further investigate the spatial resolution of these probes in vitro. Second, the substrate probes were verified in mouse tissue only, and we can only infer based on location of antibody staining of the enzymes in human tissue sections that the substrates would work for human whole epithelium as well. However, as the substrate of these enzymes might differ among species, we need further investigation for applying the method to human tissue. In order to utilize these probes in clinical practice, there are things to overcome, such as the safety of the reagents and the use of fluorescent endoscopes. However, this rapid vital OE imaging for labeling for selective neurogenic epithelium support the potential for using these probes to assess the status of the epithelium across the whole of the olfactory area, eliminating some of the shortcomings in using the extremely limited sampling afforded by mucosal biopsies to characterize epithelial status. Moreover, the mapping of the olfactory area with these probes might even be used to direct the harvesting of biopsies for diagnostic and research purposes. The value of such an approach for clinical diagnosis, prognosis, evaluation of treatment efficacy, and further research into the pathophysiology of olfactory disorders is self-evident.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| PGP 9.5/UCHL1 | Proteintech | Cat# 14730-1-AP; RRID:AB_2210497 |
| TUJ1 | BioLegend | Cat# 801201; RRID: AB_2313773 |
| Tubulin beta Ⅳ (TUBB4) | Abcam | Cat# ab179509; RRID:AB_2716759 |
| Tubulin beta Ⅳ (TUBB4) | Sigma-Aldrich | Cat# T7941; RRID: AB_261775 |
| CK19 | DSHB | Cat# Troma-Ⅲ-c; RRID: AB_2133570 |
| CYP2A6 | Invitrogen | Cat# MA5-25758; RRID: AB_2723032 |
| GGT7 | Proteintech | Cat# 24674-1-AP; RRID: AB_2879667 |
| Donkey anti-Mouse IgG-Alexa Fluor 488 | Invitrogen | Cat# A-21202 RRID: AB_141607 |
| Donkey anti-Rabbit IgG-Alexa Cy3 | Jackson ImmunoResearch Laboratories | Cat# 711-165-152; RRID: AB_2307443 |
| Donkey anti-Rat IgG-Alexa Cy5 | Jackson ImmunoResearch Laboratories | Cat# 712-175-153: RRID: AB_2340672 |
| Chemicals, peptides, and recombinant proteins | ||
| Coumarin | Sigma-Aldrich | Cat# C4261 Cas 91-64-5 |
| ProteoGREEN™-gGlu | GORYO Chemical, Inc | Cat# GC801 |
| Hematoxylin Solution | Sigma-Aldrich | Cat# 105175 |
| Eosion Solution | Sigma-Aldrich | Cat# 2853 |
| 8-Methoxypsoralen | Sigma-Aldrich | Cat# M3501 Cas 298-81-7 |
| GGsTop | Tocris Bioscience | Cat# 4452 |
| Dichlobenil | Sigma-Aldrich | Cat# D57558 Cas 1194-65-6 |
| Methimazole | Sigma-Aldrich | Cat# M8506, Cas 60-56-0 |
| Critical commercial assays | ||
| Quick-RNA Miniprep | ZYMO RESEARCH | Cat# R1054 |
| GoScript Reverse Transcriptase | Promega | Cat# A5003 |
| 2X SYBR Green qPCR Master Mix | Genesee Scientific | Cat# 42-116PG |
| Deposited data | ||
| Single-cell RNA seq | Schwob Lab | GSE166612 |
| Single-cell RNA seq | Durante et al. | GSE139522 |
| Experimental models: Organisms/strains | ||
| Mouse: Crl:CD1(ICR) | Charles river | Strain #022 RRID:IMSR_CRL:022 |
| Mouse: B6;129P2-Omptm3Mom/MomJ (OMP-GFP) | Jackson Laboratory | Strain #006667 RRID:IMSR_JAX:006667 |
| OMP-tTA;TetO-DTA transgenic mouse | Child, et al. https://www.jneurosci.org/content/38/31/6806.long | N/A |
| Oligonucleotides | ||
| Primer for Cyp2a5 forward: 5′-TCG GAAGACGAACGGTGCTTTT-3′ |
This paper | N/A |
| Primer for Cyp2a5 reverse: 5′-GCTT CCCAGCATCATTCGAAGC-3′ |
This paper | N/A |
| Primer for Ggt1 forward: 5′-CTG GAAGAGCCATGAAGAATCG-3′ |
This paper | N/A |
| Primer for Ggt1 reverse: 5′-CACG GAACCACCTTCCTGTA-3′ |
This paper | N/A |
| Primer for Ggt7 forward: 5′-GTA GTCCGACCAGCAGAAGG-3′ |
This paper | N/A |
| Primer for Ggt7 reverse: 5′-GTG AACTCACTGTCAACCTGC-3′ |
This paper | N/A |
| Primer for Gapdh forward: 5′-TGT GTCCGTCGTGGATCTGA-3′ |
This paper | N/A |
| Primer for Gapdh reverse: 5′-TTGC TGTTGAAGTCGCAGGAG-3′ |
This paper | N/A |
| Software and algorithms | ||
| GraphPad Prism 8 | Graphpad | https://www.graphpad.com/scientificsoftware/prism/RRID:SCR_002798 |
| ImageJ | U.S. National Institutes of Health | https://imagej.nih.gov/ij/RRID:SCR_003070 |
| SPOT ver 5.2 | SPOT imaging | https://www.spotimaging.com/RRID:SCR_014313 |
| Adobe photoshop | Adobe | RRID:SCR_014199 |
| Adobe illustrator | Adobe | RRID:SCR_010279 |
| Zen 3.0 | Zeiss | RRID:SCR_013672 |
| Other | ||
| NanoDrop | Thermofisher Scientific | RRID:SCR_015804 |
| Confocal Microscope LSM800 | Zeiss | SCR_015963 |
| Nikon Microphot-SA | Nikon | https://www.microscope.healthcare.nikon.com/products/upright-microscopes/eclipse-ni-u |
| CFX 96 system | Bio-Rad Laboratories | Cat# 1855196J1 RRID:SCR_018064 |
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Eric H Holbrook (Eric_Holbrook@meei.harvard.edu).
Materials availability
This study did not generate any unique reagents.
Experimental model and subject details
Animals
Adult male CD-1 (Charles river, Wilmington, MA) and OMP-GFP mice (Jackson Laboratory, Inc., Bar Harbor, ME) aged 7 weeks (30-35 g) were kept in a heat and humidity controlled, AALAC-accredited vivarium operating under a standard light/dark cycle. Male mice were chosen to reduce any confounding effects related to hormonal influences on the olfactory epithelium. All protocols were approved by the Committee for the Humane Use of Animals at Tufts University School of Medicine, where the mice were housed and the experiments were conducted.
The OMP-tTA;TetO-DTA transgenic mouse was developed as an model for accelerated aging of the OE as previously reported (Child et al., 2018). Briefly, OMP-tTA mice were crossed with the TetO-DTA mice (Figure S1). Mice of the desired genotype (OMP-tTA;TetO-DTA) were either maintained ad libitum on standard rodent chow and water or on chow containing 200 mg of doxycycline and sacrificed at 4 months of age.
Chemicals and animal treatments
As a depleted and regenerated OE model, mice were treated with a single intraperitoneal co-injection of dichlobenil (2, 6-dichlorobenzonitrile), and methimazole (Sigma-Aldrich, ST. Louis, MO). Dichlobenil was dissolved in a mixture (1:4; v/v) of dimethyl sulfoxide (DMSO; Sigma-Aldrich) and corn oil, and administered at 50mg/kg. Methimazole was dissolved in phosphate-buffered saline (PBS), pH 7.4, and administered at 50mg/kg. Five weeks after intraperitoneal injection, nasal tissues were harvested for vital staining (n = 4) and histological analysis (n = 4).
Method details
Tissue processing of mice for histology
At time points indicated for the experiments, mice were anesthetized by intraperitoneal injection of a lethal dose of a mixture of ketamine (37.5 mg/kg), xylazine (7.5 mg/kg), and acepromazine (1.25 mg/kg). Deeply anesthetized mice were then transcardially perfused with PBS-10% neutral buffered formalin solution, and decapitated. Nasal cavities were locally irrigated with the same fixative. After dissection, the tissue was postfixed under vacuum for 1 h in formalin fixative, washed in PBS, and placed in saturated EDTA overnight. The tissue was then cryoprotected in 30% sucrose in PBS, embedded in OTC medium (Tissue-Tek OTC Compound; Sakura Finetek, Torrance, CA), and frozen in liquid nitrogen. Coronal sections were cut on a Leica cryostat at 10um, mounted on “Plus” slides (Thermo Fisher Scientific), and stored at −20°C.
Human tissue for histology
Nasal specimens were obtained at autopsy through the National Disease Research Interchange (NDRI, Philadelphia, PA) and fixed in 10% formalin in saline. The nasal mucosa was stripped off the underlying bone and cartilage. A rectangular dorsal-ventral piece of the olfactory area, intended to bisect the OE was harvested and prepared for cryosectioning and immunohistochemistry as with the mouse tissue.
Immunostaining
The following primary antibodies were used: Anti-PGP 9.5/UCHL (rabbit, 1:500, Proteintech, Cat# 14730-1-AP), Anti-TUJ1 (mouse, 1:500, BioLegend, Cat# 801201), Anti-Tubulin beta Ⅳ (TUBB4) (rabbit, 1:400, Abcam, Cat# ab179509), Anti-Tubulin beta Ⅳ (TUBB4) (mouse, 1:300, Sigma-Aldrich, Cat# T7941), Anti-CK19 (rat, 1:300, DSHB, Troma-Ⅲ-c), Anti-CYP2A6 (mouse, 1:400, Invitrogen, Cat# PIMA525758), Anti-GGT7 (rabbit, 1:500, Proteintech, Cat# 24674-1-AP). Before immunostaining, tissue sections were rinsed in PBS for 5 min to remove OCT and then placed in 3% hydrogen peroxide-MeOH solution for 10 min to quench intrinsic peroxidase activity. Sections were then puddled with 0.01 M citrate buffer, pH 6.0, and steam heated for 10 min in a commercial food steamer for antigen retrieval. Sections were rinsed in PBS and then blocked with 10% donkey serum 5% nonfat dry milk/4% bovine serum albumin/0.1% Triton X-100 in PBS for 10 min and incubated overnight with primary antibody at 4°C in a humid chamber. The following day, the staining was visualized by incubating with Alexa 488, Cy3, or Cy5-conjugated secondary antibodies (Invitrogen, Carlsbad, CA; Jackson ImmunoResearch Laboratories, West Grove, PA) at 1:200 dilution. Immunofluorescent-stained sections were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) to label nuclei and coverslipped with glycerol/n-propyl gallate mounting medium.
Whole-mount staining of mouse nasal cavity
To visualize and assess the condition of the nasal epithelium globally, the divided nasal lateral and septal cavity were immune-stained using a modification of an existing protocol (Masselink et al., 2019)31. Blocks were fixed in formalin at 4°C overnight, washed in PBS, and placed in PBS-TxDBN solution (2% TX100, 20%DMSO, 5% BSA, 0.05% NaN in PBS) at room temperature (RT) for 2 days on a shaker. The tissue was then incubated with rabbit PGP9.5 (anti-PGP 9.5; Proteintech, 1:100) and mouse anti tubulin betaIV (Sigma-Aldrich, 1:100) on a tube rotator at RT for 5 days. After washing with PBS-TxDBN at RT for 2 days, tissues were then incubated with Alexa 488 (1:50), 594 (1:50)-conjugated secondary antibody (Invitrogen, Carlsbad, CA) on a tube rotator at RT for 5 days. After washing, the tissue was imaged on Nikon Microphot-SA.
Single cell RNA sequencing of mouse and human olfactory epithelium
Single cell transcriptomic data of the murine olfactory epithelium was obtained from the Gene Expression Omnibus under accession number GSE166612. Briefly, the olfactory mucosa was isolated from wild-type mice and dissociated to single cells. Single cell gene expression was measured using the Single Cell Gene Expression Solution and the Cell Ranger pipeline by 10X Genomics. OMP(+)/Gng13(+)/Gnal(+) mature olfactory sensory neurons, Sox9(+)/Aqp5(+)/Msln(+) duct/gland cells, Sox2(+)/Cyp2g1(+)/Muc2(+) sustentacular cells, and Muc5b(+)/Reg3g(+)/Krt19(+) cells from the adjacent respiratory epithelium were identified using Loupe Cell Browser v4.2.0. Due to the great excess of olfactory cell types relative to respiratory, the aforementioned cells were down-sampled to generate a dataset with 100 cells per type using R v4.0.2 and RStudio v1.3.1073. Seurat v3.2.1 was used to normalize single cell data (using the LogNormalize method) and identify the top 2000 genes with highest variability. Principal component analysis was performed on these genes and the first 10 components were used for dimensionality reduction and clustering (Figure S6).
Single cell transcriptomic data of the human olfactory epithelium was obtained from GSE139522 and analyzed according to the methods in the paper (Durante et al., 2020).
Quantitative real-time PCR
Total RNA was isolated separately from mouse nasal septal OE and RE using the Quick-RNA kit (ZYMO RESEARCH, Irvine, CA). First-strand cDNA was reverse transcribed using GoScript Reverse Transcriptase (Promega, Madison, WI) based on the manufacturer’s instructions. Quantitative polymerase chain reaction (qPCR) was performed using Apex qPCR 2X GREEN Master Mix (Genesee Scientific, San Diego, CA) and an CFX 96 Real-time system (BIO-RAD, Hercules, CA). The primer sequences for qPCR are listed in key resources table. The gene expression levels were calculated as the difference between the cycle threshold (Ct) value of the target gene and the Ct value of GAPDH (ΔCt) using the comparative threshold cycle (2−ΔΔCT) method (n = 4 per group).
Activatable fluorescence probe: Coumarin
Coumarin (C4261, Sigma-Aldrich) was used as an activatable fluorescence probe targeting CYP2A6. 1 mM DMSO stock solution of coumarin was prepared and diluted in PBS to the final concentration of 10 mM (PH8.5).
Activatable fluorescence probe: gGlu-HMRG
gGlu-HMRG (ProteoGREEN-gGlu, Gc801, GORYO CHEMICAL, Sapporo, Japan) was used to detect GGT catalysis via the generation of a fluorescenct product (Urano et al., 2011). A 10 mM stock solution of gGlu-HMRG was prepared in DMSO and diluted to a final concentration of 20 μM in PBS.
Inhibitor analysis for CYP2A6 and GGT
To demonstrate substrate selectivity of coumarin for CYP2A6, and gGlu-HMRG for GGT, 10mM of 8-Methoxypsoralen (M3501, Sigma-Aldrich), a known inhibitor of CYP2A6 (Pelkonen et al., 2000), or 50μM of GGsTop (4452, Tocris Bioscience, Bristol, UK), a known inhibitor of GGT (Han et al., 2007) were applied to the mouse nasal mucosa 5 min before administrating activatable fluorescence probes, respectively.
Analysis using metabolites of coumarin and gGlu-HMRG
To control for the contribution of background autofluorescence by the targeted epithelium, 1 mM 7-OH coumarin (generous gift from Dr. Yasuteru Urano, The University of Tokyo), the CYP2A6-catalyzed metabolite of coumarin (Pelkonen et al., 2000), or 1 μM HMRG (generous gift from Dr. Urano), the GGT-catalyzed metabolite of gGlu-HMRG (Urano et al., 2011) were applied to nasal mucosa 5 min before administrating activatable fluorescence probes.
Vital fluorescence imaging of mice nasal mucosa
Mice were anesthetized, decapitated, then bisected to the left of the midline, thereby exposing the left lateral and septal aspects of the nasal mucosa. After washing with PBS, the tissues were immobilized on the microscope stage and visualized.
50 μL of a mixture of the two probes, with final concentrations of coumarin (10 mM) and gGlu-HMRG (20 μM), was topically applied to both surfaces of the nasal cavity, and observed by fluorescent microscopy at designated intervals for 30-60 min at RT. Fluorescent images were captured before and every 5 min after probe administration. An additional 30 μL of probe were administered in every 10 min. Excitation and emission filters wavelength for coumarin were 375 and 460 nm, and for gGlu-HMRG were 470 and 535 nm, respectively
All fluorescence imaging studies started within 20 min after decapitation. All experiments were carried out under the same conditions using a Nikon Microphot-SA upright microscope, equipped with a PLAN 1X/0.04 objective lens and SPOT Advanced Imaging software ver5.2 (SPOT imaging, Sterling Heights, MI).
Image analysis
Immuonstained sections were imaged on a Zeiss LSM800 confocal microscope and Nikon Microphot-SA. These images were assembled using Adobe Photoshop, Illustrator and ImageJ. In all photos, only balance and contrast were adjusted.
Image analysis and quantification of vital staining were performed using ImageJ. Regions of interest were drawn two times for OE and RE, and the mean fluorescence intensity of both regions were measured in every 5 min, then ratio of the intensity, then OE/RE, were calculated.
Quantification and statistical analysis
Statistical differences were calculated by two-tailed unpaired t-test for two datasets using GraphPad Prism8 (GraphPad Inc., La Jolla, CA). p < 0.05 was considered statistically significant. ∗p < 0.05; ns; not statistically significant.
Acknowledgments
The authors thank Po Kwok-Tse for her outstanding technical assistance. We are thankful to Y.Urano and M.Kamiya for the HRMG, 7OH coumarin, and critical contribution of the discussion. We also thank D. Xingxing and B. Shimizu for suggestions and contributions to the discussion. This work was supported by Fund for the Promotion of Joint International Research (grant number 18KK0448) to H.N. from the Japan Society for the Promotion of Science, Grant from Society for Promotion of International Oto-Rhino-Laryngology (SPIO) to H.N., Grant for Young researcher’s international deployment from the University of Tokyo to H.N., Grant R01 DC014217 from National Institutes of Health to J.E.S.
Author contributions
H.N., J.E.S., and E.H.H., designed all experiments and wrote the manuscript. H.N. performed all experiments and analyses except single-cell RNA-seq analysis. M.Z. performed single-cell RNA-seq analysis. M.Y. and K.K. designed gGlu-HMRG vital staining experiments. T.Y. commented on the manuscript.
Declaration of interests
The authors have declared that no conflict of interest exists.
Published: May 20, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104222.
Supplemental information
Data and code availability
-
•
The RNAseq dataset has been deposited at GenBank and is publicly available at https://www.ncbi.nlm.nih.gov/bioproject/. The accession link is listed in the key resources table. All other data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Brandt I., Brittebo E., Feil V., Bakke J. Irreversible binding and toxicity of the herbicide dichlobenil (2, 6-dichlorobenzonitrile) in the olfactory mucosa of mice. Toxicol. Appl. Pharmacol. 1990;103:491–501. doi: 10.1016/0041-008X(90)90322-L. [DOI] [PubMed] [Google Scholar]
- Brittebo E.B. Metabolism-dependent activation and toxicity of chemicals in nasal glands. Mutat. Research/Fundamental Mol. Mech. Mutagenesis. 1997;380:61–75. doi: 10.1016/S0027-5107(97)00127-9. [DOI] [PubMed] [Google Scholar]
- Child K.M., Herrick D.B., Schwob J.E., Holbrook E.H., Jang W. The neuroregenerative capacity of olfactory stem cells is not limitless: implications for aging. J. Neurosci. J. Neurosci. 2018;38:6806–6824. doi: 10.1523/JNEUROSCI.3261-17.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X., Fang X. In: Handbook of Olfaction and Gustation. Doty R.L., editor. John Wiley & Sons; 2015. Olfactory mucosa: composition, enzymatic localization, and metabolism; pp. 63–92. [Google Scholar]
- Doty R.L. John Wiley & Sons; 2015. Handbook of Olfaction and Gustation. [Google Scholar]
- Durante M.A., Kurtenbach S., Sargi Z.B., Harbour J.W., Choi R., Kurtenbach S., Goss G.M., Matsunami H., Goldstein B.J. Single-cell analysis of olfactory neurogenesis and differentiation in adult humans. Nat. Neurosci. 2020;23:323–326. doi: 10.1038/s41593-020-0587-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer P., Scherberich J.E., Schoeppe W. Comparative biochemical and immunological studies on gamma-glutamyltransferases from human kidney and renal cell carcinoma applying monoclonal antibodies. Clinica Chim. Acta. 1990;191:185–200. doi: 10.1016/0009-8981(90)90020-S. [DOI] [PubMed] [Google Scholar]
- Genter M.B., Deamer N.J., Blake B.L., Wesley D.S., Levi P.E. Olfactory toxicity of methimazole: dose-response and structure-activity studies and characterization of flavin-containing monooxygenase activity in the Long-Evans rat olfactory mucosa. Toxicologic Pathol. 1995;23:477–486. doi: 10.1177/019262339502300404. [DOI] [PubMed] [Google Scholar]
- Graziadei P., Graziadei G.M. Neurogenesis and neuron regeneration in the olfactory system of mammals. I. Morphological aspects of differentiation and structural organization of the olfactory sensory neurons. J. Neurocytol. 1979;8:1–18. doi: 10.1007/BF01206454. [DOI] [PubMed] [Google Scholar]
- Han L., Hiratake J., Kamiyama A., Sakata K. Design, synthesis, and evaluation of γ-phosphono diester analogues of glutamate as highly potent inhibitors and active site probes of γ-glutamyl transpeptidase. Biochemistry. 2007;46:1432–1447. doi: 10.1021/bi061890j. [DOI] [PubMed] [Google Scholar]
- Hanigan M.H., Frierson H.F., Jr., Swanson P.E., De Young B.R. Altered expression of gamma-glutamyl transpeptidase in human tumors. Hum. Pathol. 1999;30:300–305. doi: 10.1016/S0046-8177(99)90009-6. [DOI] [PubMed] [Google Scholar]
- Heydel J.-M., Faure P., Neiers F. Nasal odorant metabolism: enzymes, activity and function in olfaction. Drug Metab. Rev. 2019;51:224–245. doi: 10.1080/03602532.2019.1632890. [DOI] [PubMed] [Google Scholar]
- Hino H., Kamiya M., Kitano K., Mizuno K., Tanaka S., Nishiyama N., Kataoka K., Urano Y., Nakajima J. Rapid cancer fluorescence imaging using a γ-glutamyltranspeptidase-specific probe for primary lung cancer. Transl. Oncol. 2016;9:203–210. doi: 10.1016/j.tranon.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbrook E.H., Rebeiz L., Schwob J.E. Office-based olfactory mucosa biopsies. Int. Forum Allergy Rhinol. 2016;6:646–653. doi: 10.1002/alr.21711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbrook E.H., Wu E., Curry W.T., Lin D.T., Schwob J.E. Immunohistochemical characterization of human olfactory tissue. Laryngoscope. 2011;121:1687–1701. doi: 10.1002/lary.21856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis D.F., Ito Y. In: Cytochrome P450: Role in the Metabolism and Toxicity of Drugs and Other Xenobiotics. Ioannides C., editor. Advancing the Chemical Sciences Publishing; 2008. Cytochrome P450 structure and function: An evolutionary perspective; pp. 4–39. [Google Scholar]
- Lewis D.F., Ito Y., Lake B.G. Metabolism of coumarin by human P450s: a molecular modelling study. Toxicol. Vitro. 2006;20:256–264. doi: 10.1016/j.tiv.2005.08.001. [DOI] [PubMed] [Google Scholar]
- Ling G., Gu J., Genter M.B., Zhuo X., Ding X. Regulation of cytochrome P450 gene expression in the olfactory mucosa. Chem. Biol. Interact. 2004;147:247–258. doi: 10.1016/j.cbi.2004.02.003. [DOI] [PubMed] [Google Scholar]
- Mäenpää J., Juvonen R., Raunio H., Rautio A., Pelkonen O. Metabolic interactions of methoxsalen and coumarin in humans and mice. Biochem. Pharmacol. 1994;48:1363–1369. doi: 10.1016/0006-2952(94)90558-4. [DOI] [PubMed] [Google Scholar]
- Masselink W., Reumann D., Murawala P., Pasierbek P., Taniguchi Y., Bonnay F., Meixner K., Knoblich J.A., Tanaka E.M. Broad applicability of a streamlined ethyl cinnamate-based clearing procedure. Development. 2019;146:dev166884. doi: 10.1242/dev.166884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata Y., Ishizawa T., Kamiya M., Yamashita S., Hasegawa K., Ushiku A., Shibahara J., Fukayama M., Urano Y., Kokudo N. Intraoperative imaging of hepatic cancers using γ-glutamyltranspeptidase-specific fluorophore enabling real-time identification and estimation of recurrence. Sci. Rep. 2017;7:3542. doi: 10.1038/s41598-017-03760-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaya T., Nakamura Y.A., Choyke P.L., Kobayashi H. Fluorescence-guided surgery. Front. Oncol. 2017;7:314. doi: 10.3389/fonc.2017.00314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olender T., Keydar I., Pinto J.M., Tatarskyy P., Alkelai A., Chien M.-S., Fishilevich S., Restrepo D., Matsunami H., Gilad Y. The human olfactory transcriptome. BMC Genomics. 2016;17:619. doi: 10.1186/s12864-016-2960-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelkonen O., Rautio A., Raunio H., Pasanen M. CYP2A6: a human coumarin 7-hydroxylase. Toxicology. 2000;144:139–147. doi: 10.1016/S0300-483X(99)00200-0. [DOI] [PubMed] [Google Scholar]
- Raunio H., Hakkola J., Pelkonen O. In: The CYP2A Subfamily. Cytochrome P450: Role in the Metabolism and Toxicity of Drugs and Other Xenobiotics. Ioannides C., editor. Advancing the Chemical Sciences Publishing; 2008. pp. 150–171. [Google Scholar]
- Schwob J.E., Jang W., Holbrook E.H., Lin B., Herrick D.B., Peterson J.N., Hewitt Coleman J. Stem and progenitor cells of the mammalian olfactory epithelium: taking poietic license. J. Comp. Neurol. 2017;525:1034–1054. doi: 10.1002/cne.24105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng J., Guo J., Hua Z., Caggana M., Ding X. Characterization of human CYP2G genes: widespread loss-of-function mutations and genetic polymorphism. Pharmacogenetics. 2000;10:667–678. doi: 10.1097/00008571-200011000-00001. [DOI] [PubMed] [Google Scholar]
- Sherman W.R., Robins E. Fluorescence of substituted 7-hydroxycoumarins. Anal. Chem. 1968;40:803–805. doi: 10.1021/ac60260a045. [DOI] [PubMed] [Google Scholar]
- Suzukawa K., Kondo K., Kanaya K., Sakamoto T., Watanabe K., Ushio M., Kaga K., Yamasoba T. Age-related changes of the regeneration mode in the mouse peripheral olfactory system following olfactotoxic drug methimazole-induced damage. J. Comp. Neurol. 2011;519:2154–2174. doi: 10.1002/cne.22611. [DOI] [PubMed] [Google Scholar]
- Urano Y., Sakabe M., Kosaka N., Ogawa M., Mitsunaga M., Asanuma D., Kamiya M., Young M.R., Nagano T., Choyke P.L. Rapid cancer detection by topically spraying a γ-glutamyltranspeptidase–activated fluorescent probe. Sci. Transl. Med. 2011;3:110ra119. doi: 10.1126/scitranslmed.3002823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi M., Fujiwara M., Nakamura H. Olfactory mucosal findings and clinical course in patients with olfactory disorders following upper respiratory viral infection. Rhinology. 1994;32:113–118. [PubMed] [Google Scholar]
- Yamamoto S., Watanabe B., Hiratake J., Tanaka R., Ohkita M., Matsumura Y. Preventive effect of GGsTop, a novel and selective γ-glutamyl transpeptidase inhibitor, on ischemia/reperfusion-induced renal injury in rats. J. Pharmacol. Exp. Ther. 2011;339:945–951. doi: 10.1124/jpet.111.183004. [DOI] [PubMed] [Google Scholar]
- Zhang Q.-Y., Ding X. In: Cytochrome P450: Role in the Metabolism and Toxicity of Drugs and Other Xenobiotics. Ioannides C., editor. Advancing the Chemical Sciences Publishing; 2008. The CYP2F, CYP2G and CYP2J Subfamilies; pp. 309–353. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The RNAseq dataset has been deposited at GenBank and is publicly available at https://www.ncbi.nlm.nih.gov/bioproject/. The accession link is listed in the key resources table. All other data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.