Abstract
Mitochondrial dysfunction is one of the crucial factors involved in PD’s pathogenicity, which emerges from a combination of genetic and environmental factors. These factors cause differential molecular expression in neurons, such as varied transcriptional regulation of genes, elevated oxidative stress, α‐synuclein aggregation and endogenous neurotoxins release, which induces epigenetic modifications and triggers energy crisis by damaging mitochondria of the dopaminergic neurons (DN). So far, these events establish a complicated relationship with underlying mechanisms of mitochondrial anomalies in PD, which has remained unclear for years and made PD diagnosis and treatment extremely difficult. Therefore, in this review, we endeavored to discuss the complex association of epigenetic modifications and other associated vital factors in mitochondrial dysfunction. We propose a hypothesis that describes a vicious cycle in which mitochondrial dysfunction and oxidative stress act as a hub for regulating DA neuron's fate in PD. Oxidative stress triggers the release of endogenous neurotoxins (CTIQs) that lead to mitochondrial dysfunction along with abnormal α‐synuclein aggregation and epigenetic modifications. These disturbances further intensify oxidative stress and mitochondrial damage, amplifying the synthesis of CTIQs and works vice versa. This vicious cycle may result in the degeneration of DN to hallmark Parkinsonism. Furthermore, we have also highlighted various endogenous compounds and epigenetic marks (neurotoxic and neuroprotective), which may help for devising future diagnostic biomarkers and target specific drugs using novel PD management strategies.
Keywords: endogenous neurotoxins, epigenetics, mitochondrial dysfunction, neurodegenerative, oxidative stress, Parkinson's disease, α‐synuclein
Impact of the epigenetic regulator on mitochondrial dysfunction in Parkinson's disease.
Abbreviations
- 4‐HNE
4‐hydroxynonenal
- 5‐mC
methylated
- 5‐hmC
hydroxy methylated
- ALDH2
aldehyde dehydrogenase‐2
- AS Uchl1
antisense to the mouse Ubiquitin carboxy terminal hydrolase 1
- BBB
blood‐brain barrier
- CDKN2A
cyclin‐dependent kinase inhibitor 2A
- CDR1as
non‐coding antisense transcript to the protein coding gene CDR1
- Cerox1
cytoplasmic endogenous regulator of oxidative phosphorylation
- CpG‐2
CpG islands
- CTIQs
catechol TIQs
- DA
dopamine
- DJ‐1
Protein Deglycase
- DMDHIQ+
1,2‐dimethyl‐6,7‐dihydroxyisoquinolinium ions
- DN
dopaminergic neurons
- DNMT1
cytosine‐5‐methyltransferase 1
- DNMT3A
DNA methyltransferase 3 alpha
- DNMT3B
DNA methyltransferase 3 beta
- DOPAC
3,4‐dihydroxyphenylacetic acid
- DOPAL
3,4‐dihydroxyphenylacetaldehyde
- Drp1
dynamin‐related protein‐1
- Endo
endonuclease
- GPNMB
glycoprotein Nmb
- HAGLROS
HAGRL opposite strand LncRNA
- HOTAIR
HOX Transcript Antisense Intergenic RNA
- LINEs
long interspersed nuclear element
- LncRNA
long coding RNA
- LncRNA‐p21
long non‐coding RNA‐p21
- LRRK2
leucine‐rich repeat kinase 2
- MALAT1
metastasis‐associated lung adenocarcinoma transcript 1
- MDA
malondialdehyde
- MGO
methylglyoxal
- MPP+
1‐methyl‐4‐phenylpyridinium
- MPTP
1‐methyl‐4‐phenyl‐1,2,3,6‐tetrahydropyridine
- mtDNA
mitochondrial DNA
- MT‐ND6
NADH dehydrogenase gene 6
- ncRNAs
non‐coding RNAs
- Ndufs4
NADH: ubiquinone oxidoreductase iron‐sulfur protein 4
- NEAT1
nuclear enriched assembly transcript‐1
- NM‐Sal
N‐methyl‐salsolinol
- NORAD
non‐coding RNA activated by DNA damage
- Nrf2
NF‐E2‐related factor
- Opa1
optic atrophy 1
- OXPHOS
oxidative phosphorylation
- PARK2
Parkin
- PD
Parkinson's disease
- PEA
phenylethylamine
- PGC‐1α
peroxisome proliferator‐activated receptor gamma coactivator‐1 α
- PINK1
PTEN‐induced putative kinase 1
- piRNAs
piwi interacting RNAs
- POLG1
polymerase gamma 1
- PPARGC 1 A
PPARG Coactivator 1 alpha
- PRKAR2A
cAMP‐dependent protein kinase type II alpha regulatory subunit
- ROS
reactive oxygen specie
- Sal
salsolinol
- SEPW1
selenoprotein W
- SIAH1
Siah E3 Ubiquitin Protein Ligase 1
- SINEs
short interspersed nuclear element
- SNCA
α‐synuclein
- SNHG1
small nucleolar RNA host gene 1
- SNMT
salsolinol N‐methyltranseferase
- SNpc
substantia nigra pars compacta
- STX1B
syntaxin 1B
- TCE
Trichloroethylene
- TET
ten‐eleven translocation
- TFAM
mitochondrial transcription factor A
- TH
tyrosine hydroxylase
- TIQ
tetrahydroisoquinoline
- TNF
Tumor necrosis factor
- TRPC1
transient receptor potential protein 1
- UCA‐1
urothelial carcinoma‐associated‐1
- VPS35
vacuolar protein sorting 35
1. INTRODUCTION
Parkinson's disease (PD) is a complex, chronic and second‐most progressive neurodegenerative movement disorder after Alzheimer's disease. PD prevails in the old age population at a percentage of 1% (65 years old) to 4% (80 years old) (1, 2, 3) and has influenced more than ten million populations globally. Epidemiological studies have shown that PD affects ~3.9% of older people of the Chinese Han population aging ≥50 years old (3) and estimated to rise to 4.94 million by 2030, narrating half of the PD patients globally (4). This disease has an inevitable and progressive course, characterized by the loss of dopaminergic neurons (DA) in the substantia nigra pars compacta (SNpc) of the PD brain and the formation of Lewy bodies (α‐synuclein protein inclusions), leading to the dysregulation of various pathways, such as vesicle trafficking or activating neuroinflammation (5). However, on account of 50%–70% DA neurons loss, dopamine neurotransmitter production decreases, resulting in variable motor and non‐motor clinical symptoms (6). Significant motor symptoms include bradykinesia, resting tremor and rigidity, whereas non‐motor symptoms include insomnia, hallucinations and depression (7). Additionally, in PD other than SNpc, various neurodegenerative aberrations are also observed in other parts of the brain, making its diagnosis and treatment more complicated and challenging (8, 9). The etiopathogenesis of PD has emerged from the contribution of genetic factors (Familial PD) (10) and environmental factors (Sporadic PD) (10). Familiar PD occurs due to mutations in specific genes (SNCA, LRRK2, PARK2, PINK1, DJ‐1, VPS35) and accounts for only 10% PD (10). In contrast, sporadic PD occurs due to environmental factors such as poor lifestyle, exogenous neurotoxins, drug addiction, long‐term stress, diet and aging (10) (Figure 1).
FIGURE 1.
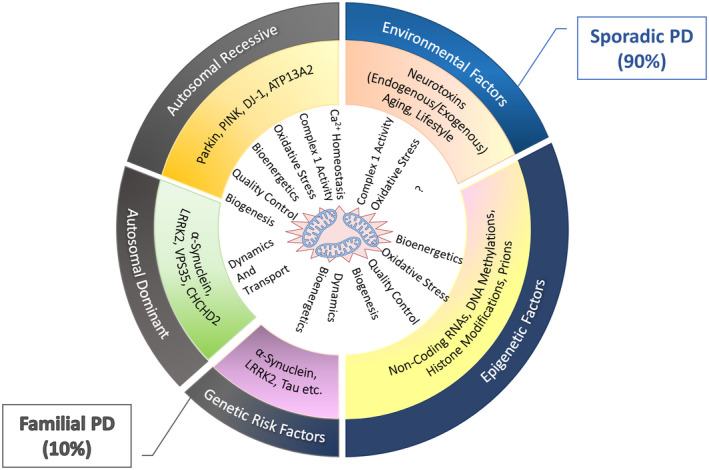
Aetiology of Parkinson's disease and possible risk factors with mitochondrial dysfunction. Familial PD accounts for 10% of PD and is caused by genetic variations inherited in an autosomal recessive or dominant manner. Sporadic PD accounts for 90% of PD, a complicated neurodegenerative disease caused by a combination of genetic and environmental factors. Genome‐wide association studies presented two susceptible loci of α‐synuclein and LRRK2 genes, which link classical PD genes and overlap the etiology of familial PD with sporadic PD. Both genetic and environmental factors induce epigenetic modifications that cause mitochondrial anomalies in various aspects such as oxidative stress, mitochondrial complex I activity, bioenergetics, dynamic, transport, biogenesis, and quality control.
Since the past few decades, mounting research has been carried out to understand the genetic architecture and cellular processes of PD. However, the exact mechanism involved in Parkinsonism remains unclear, which refers to complex etiology involving several molecular pathways such as apoptosis, endocytosis, immune response, lysosomal function, oxidative stress, and mitochondrial function (8, 10, 11). Increasing evidence has speculated an association of mitochondrial dysfunction with neuronal damage in PD although the exact relationship is unknown (8, 12). Moreover, uneven distribution of mitochondria throughout the living body fails to describe the specific neuronal damage that makes it more strenuous to explain why mitochondria are impaired explicitly in degenerative neurons and cause neuronal death in PD (13).
Various studies have shown that mitochondrial abnormalities may result from genetic, environmental, or a combination of both (epigenetic modifications). These modifications may either damage mitochondrial DNA (mtDNA) and impair mitochondrial functioning such as (i) abnormal electron transport chain complex I (ii) Ca2+ homeostasis aberrations, (iii) impaired biogenesis/bioenergetics, (iv) abnormal mitochondrial dynamics, (v) defective mitophagy, (vi) abnormal axonal transport and mitochondrial distribution in PD neurons, and (vii) increased oxidative stress (ROS production) (Figure 1) (14, 15). Further, it is observed that long‐lived individuals with prolonged stressful conditions are more prone to multiple molecular changes such as higher oxidative levels, the release of endogenous neurotoxins, and epigenetic modifications. These molecular changes cause an alteration in the transcriptional regulation of various genes, resulting in mitochondrial dysfunction and neuronal apoptosis to implicate Parkinsonism (13). However, the pathogenic role of these factors in mitochondrial dysfunction and their relationship with complex molecular mechanisms in PD progression still requires to be deciphered.
Moreover, partial research and the use of ineffective drugs have turned PD management and treatment more challenging. Unfortunately, most PD drugs work for a specific period or encounter various side effects on long‐term use, such as levodopa (16). That awakens the need for new PD‐specific drugs that efficiently treat PD at earlier stages in the aging population. Therefore, this review highlights various factors and their association in promoting oxidative stress and mitochondrial bioenergetic defects at the macro perspective level to mitigate PD research and clinical treatment impediments. Furthermore, we have briefly discussed how these epigenetic moderators and endogenous compounds based on their neurotoxic and neuroprotective properties could be used as future diagnostic biomarkers and may serve to design novel strategies for early PD diagnosis and therapeutics.
2. MITOCHONDRIAL DYSFUNCTION IN PARKINSON DISEASE
A mitochondrial dysfunction is an early event in PD progression, which remains a distinctive feature in all stages of PD. The first connection between mitochondrial dysfunction and PD was reported in the early 1980s, when a neurotoxin MPP+, the metabolite of MPTP (1‐methyl 4‐phenyl‐1,2,3,6‐tetrahydropyridine), caused Parkinsonism in patients by inhibiting NADH–ubiquinone oxidoreductase (complex I) of the mitochondrial respiratory chain (11, 17, 18). Later, Dr. Langston confirmed that MPTP‐induced mitochondrial dysfunction is associated with PD pathogenesis (19). Emerging evidence has reported that both forms of PD (sporadic and familial) share common pathways that link up with mitochondria (19). Therefore, speculating mitochondrial dysfunction as one of the cardinal characteristics of PD is well‐elaborated.
2.1. Mitochondrial dysfunction in familial Parkinson disease
To date, a handful of genes have been reported in familial PD, mainly including α‐synuclein (SNCA), leucine‐rich repeat kinase 2 (LRRK2), Parkin (PARK2), PTEN‐induced putative kinase 1 (PINK1), DJ‐1, and vacuolar protein sorting 35 (VPS35) (20). The proteins expressed by most of these genes connect with the mitochondrial membrane directly or indirectly. Variable expression of these genes causes various mitochondrial impairment; manifested by the altered morphology, imbalanced dynamics (fusion, fission, mitophagy, or transport), and mitochondrial dysfunction, including increased reactive oxygen species (ROS) production and decreased membrane potential, reduced ATP levels, and inhibition of respiratory complex activities. Thus, intensive study on these PD genes has contributed to our knowledge about the potential common pathway of mitochondrial function (21).
For instance, genetic mutations in the SNCA gene cause abnormal α‐synuclein expression (22), which leads to the development of Lewy bodies, a key pathological hallmark of PD. Emerging evidence has established the link between α‐synuclein (wild‐type and mutated forms) and mitochondrial function. It shows that the disruption of α‐synuclein induces the development of PD by impairing mitochondrial fusion, fission, transport, and mitophagy (23, 24). Moreover, the changes in α‐synuclein distribution between the cytoplasm and mitochondrial membrane cause mitochondrial function impairment: altered Ca2+ homeostasis, inhibition of ATP production, enhanced cytochrome c release, impaired complex I function, and increased oxidative stress (25, 26, 27). Higher oxidative stress was presumed to cause α‐synuclein abnormality. Later, this concept was modified by Kumar and his colleagues. They found that cytochrome c from leaky mitochondria as peroxidase triggered the formation of α‐synuclein radical and oligomerization, leading to the dopaminergic neuronal apoptosis (28). Therefore, through these pieces of evidence, it can be assumed that the consequences of mitochondrial damage could be one reason for the α‐synuclein abnormality.
Like α‐synuclein, LRRK2, a common genetic risk of an autosomal dominant form of PD is co‐localized with the dynamin‐superfamily GTPases (Drp1, Mfn1, Mfn2, and OPA1) at the mitochondrial membrane and mediates the mitochondrial dynamics (29, 30). A study on the PD mouse model showed that exposure to low oral rotenone resulted in mutant LRRK2 mice displaying significant locomotor defects. It was observed that locomotor defects have occurred due to low levels of striatal mitochondrial complex I (NDUFS4), striatal synaptosomes, and synaptic vesicular proton pump protein (V‐ATPase H) that lowered dopamine uptake in PD mouse than wild‐type mouse (31). Moreover, LRRK2 mutants reported significant mitochondrial abnormalities, including damaged mitochondrial DNA, abnormal morphology, movement, and homeostasis, low ATP production, decreased membrane potential, reduced respiratory chain complex IV activity, increased mitochondrial proton leakage, disturbed calcium handling, and impaired mitochondrial degradation (32, 33, 34, 35).
Interestingly, pathogenic modifications in the mitochondrial genome also play a significant role in PD development. These modifications can either be inherited or acquired during disease progression and divided into three major groups: (i) mtDNA point mutations (inherited or somatic), (ii) mtDNA deletions, and (iii) changes in mtDNA copy number (36). Growing evidence suggests that genetic mutations of mtDNA or nuclear genes (involved in mtDNA maintenance) increase with age and may participate in PD development. For instance, the POLG1 (polymerase gamma 1) gene, which regulates mtDNA synthesis, replication and repair, is observed dysregulated in PD patients. Several PD patients reported various mutations in this gene that lower mtDNA copy number and mitochondrial abnormalities (37).
Similarly, mutations in the TFAM (mitochondrial transcription factor A) resulted in abnormal mtDNA transcription and copy number (38, 39). A study performed on TFAM knockout DA neurons of MitoPark mice resulted in low mtDNA levels with progressive loss of neurons and PD pathogenesis (40). Moreover, in 2007, Baloh and his colleagues reported the first PD family with TWNK gene mutations. This gene is responsible for maintaining mtDNA replication and stability (41, 42). Research on mouse models exhibiting TWNK mutant DA neurons displayed more age‐related mtDNA deletions, DA neurodegeneration and Parkinson‐like phenotype (43). Taken together, the interaction between PD‐related genetic causes and mitochondrial dysfunction links several pathways that lead to PD‐associated neuronal damage. Therefore, it can be surmised that mitochondrial dysfunction is a common pathogenic pathway that should be under consideration to understand PD pathogenesis (44, 45, 46, 47).
2.2. Mitochondrial dysfunction in sporadic Parkinson disease
Sporadic PD occurs due to multiple undetermined genetic or environmental risk factors, including aging, drug usage, lifestyle, and exposure to harmful chemicals. These factors cause numerous molecular changes such as oxidative stress, the release of neurotoxins, and epigenetic modifications, affecting various cellular pathways linked with mitochondrial function. Notably, these factors induce different somatic mutations in mtDNA of PD models. For instance, several studies on PD patients reported higher somatic mt‐DNA deletions in SNpc than other brain parts (48, 49). Additional investigations revealed that PD patients exhibited increased 8‐oxoGuanine levels in the mt‐DNA of SNpc (DA neurons), imparting a role in PD progression (50). Moreover, various missense and heteroplasmic mutations in OXPHOS complex I of mt‐DNA in idiopathic PD patients resulted in OXPHOS complex I deficiency in PD (51, 52). Mutations in 7SDNA led to impaired mtDNA replication and transcription machinery, thus presenting a primary pathogenic mechanism in both forms of PD (53). Hence all these mt‐DNA lesions led to respiratory chain deficiency and exacerbated PD progression.
2.2.1. Exogenous neurotoxin
Exogenous neurotoxins possess a slightly lower risk in PD development due to a lower population exposure rate. The exogenous neurotoxins include MPTP, pesticides, heavy metals, and solvents, wildly employed to develop animal models of PD to explore the pathological mechanisms of PD (54, 55). Environmental neurotoxins cause similar PD clinical manifestations that share common pathways of mitochondrial dysfunction (47). MPTP and pesticides showed the attenuation of the mitochondrial function by imbalancing the mitochondrial fission and fusion, lowering mitochondrial transport, and inducing autophagy (56, 57). These neurotoxins also adversely impact the respiratory chain function by inhibiting mitochondrial complex I (58, 59, 60). However, a previous study also showed that the deletion of the Ndufs4 (complex I subunit, NADH: ubiquinone oxidoreductase iron‐sulfur protein 4) did not inhibit dopaminergic neuronal death induced by rotenone, MPP+, or paraquat, indicating that toxicity of these neurotoxins may not be solely caused by the inhibition of complex I (61). Instead, other factors are also involved; for instance, dysfunction of mitochondrial Lon protease after MPTP treatment in mice has resulted from the elevated oxidative stress, known as the pathological driver in PD development (62).
In addition, exposure to heavy metals such as iron (Fe), manganese (Mn), and lead (Pb) is also associated with mitochondrial dysfunction in PD, suggesting that mitochondria display a significant role in cellular iron metabolism. The disequilibrium of mitochondrial iron homeostasis leads to the accumulation of cellular Fe, which is the crucial element in degenerated DA neurons in PD (63). Due to similar structural and chemical properties with Fe, Mn also preferentially accumulates in the mitochondria and disrupts cellular energy metabolism to induce dopaminergic neurotoxicity (64). Previous research expounded the association between Mn and mitochondrial dysfunction, such that Mn accumulation hampered the efflux of calcium, inhibited MAO activity, as well as caused imbalances in the mitochondrial fusion/fission equilibrium (65, 66). However, several studies involving lead‐induced neurotoxicity also showed similar mitochondrial damage (67). Of the many different complex I inhibitors, some solvents have been used to establish the animal models of Parkinson's disease (68). For example, Trichloroethylene (TCE) is a typical solvent widely used industrially for metal degreasing and dry cleaning. The exposure to TCE significantly decreased Complex I‐driven state III and state V activity in the mitochondria of SNpc that altered mitochondrial bioenergetics and induced nigral dopaminergic neuronal loss (69, 70).
2.2.2. Endogenous neurotoxins
For decades, endogenous neurotoxins have gained momentum owing to their crucial role in PD. Recently, dopamine‐derived alkaloids, structurally or functionally like MPTP, and tetrahydroisoquinoline compounds (TIQ), have been proposed as the potential pathogenic factors of PD (71, 72). TIQ reported significant inhibitory activity on the mitochondrial respiration, complex I, and ATP synthesis, and the methylation at the 3‐position of TIQ derivatives potentiate complex I inhibitory activities (73, 74). TIQs are categorized into two types: catechol and non‐catechol structure. Non‐catechol TIQs:1‐BnTIQ and 1MeTIQ can easily cross the blood‐brain barrier (BBB), migrate into the brain, and induce some positive or negative effects on the nervous system. Although 1‐BnTIQ and 1MeTIQ are both TIQ derivatives and endogenously detected in the human brain, they possess opposite pharmacological properties (1‐MeTIQ is neuroprotective, and 1‐BnTIQ is neurotoxic). Behavioral analyses and neurochemical experiments demonstrated that 1MeTIQ completely antagonized 1BnTIQ‐induced changes in rat locomotor activity and reduced the dopamine (DA) concentration in the rat brain (75). Wasik et al. pointed out that the neurotoxic effect of 1BnTIQ is dose‐dependent. For instance, in lower concentrations (50 μM), it displayed neuroprotective properties, and in higher concentrations (500 μM), it showed neurotoxic properties (76). Further, it is observed that the neurotoxic effect of 1‐BnTIQ in monkeys and mice occurs due to inhibition of mitochondrial NADH‐ubiquinone oxidoreductase (complex I) (77). Contrarily, 1‐MeTIQ reported various neuroprotective actions, such as reducing oxidative stress, inhibiting MAO activity, and reducing DA release reduction (78, 79, 80). Interestingly, besides endogenous synthesis of 1‐MeTIQ in the human brain, it can be synthesized enzymatically with phenylethylamine (PEA) and pyruvate and catalyzed by 1MeTIQase in the mitochondrial–synaptosomal fraction.
Differing from non‐catechol TIQs, there is still controversy about whether Catechol TIQs (CTIQs) such as salsolinol (Sal) can cross BBB or not (81). Therefore, CTIQs have been extensively investigated for their neurotoxic effects on the DA neurons, especially in the mitochondria (82). Sal was reported as the first CTIQs in the urine and brain of PD patients after L‐DOPA treatment, which is synthesized actively with DA and acetaldehyde under the catalysis of Sal synthase (83, 84). The confirmed peculiarity of Sal synthesis in the DA neurons proposes its prominent role in the occurrence of PD. In some circumstances, the formed Sal compounds are catalyzed into N‐methyl‐salsolinol (NM‐Sal) by N‐methyltransferase and then further oxidized into 1,2‐dimethyl‐6,7‐dihydroxyisoquinolinium ions (DMDHIQ+) (84). Similar to the exogenous neurotoxins, Sal and its derivatives (e.g., NM‐Sal, Norsal) also inhibited mitochondrial electron transport chain (complex I), mitochondrial membrane potential, ATP levels, and conversely increased the cytochrome c release (84, 85, 86). Further, the (R)‐enantiomer of NM‐Sal (NM‐(R)‐Sal) caused severe neurotoxicity and is regarded as the strongest toxic CTIQ so far (87, 88). Based on the properties of Sal derivatives, our team deeply investigated their pathogenic roles in PD in the recent years. We evaluated the concentration of Sal and NM‐Sal under oxidative stress such as exposure to iron, manganese, ethanol, and H2O2 or alterative expression of α‐synuclein and parkin, which could be the hub between mitochondrial dysfunction and development of PD (88, 89, 90, 91). One well‐established example of mitochondrial dysfunction induced by Sal and NM‐Sal is that the cytotoxicity was attenuated by the transient receptor potential protein 1 (TRPC1). Its overexpression prevented neurons from apoptosis by inhibiting the release of cytochrome c from the mitochondria (92). It was assumed that Sal could be synthesized in situ of the substantia nigra and striatum of the brain. This was further confirmed by our research group that purified sal synthase from rat brain successfully, demonstrated it as a novel protein with 77 amino acids (83), and verified that salsolinol N‐methyltransferase (SNMT) activity in the substantia nigra of rat brain was significantly increased after treatment with 6‐OHDA (93).
According to these findings, Sal and its derivatives synthesis were proposed to depend on the initial concentration of their substrates (e.g., DA and acetaldehyde). Under physiological conditions, acetaldehyde may convert into acetate through aldehyde dehydrogenase‐2 (ALDH2) metabolism. ALDH2 is widely present in the brain and performs its functions in the mitochondria matrix; therefore, its inhibition can exacerbate the PD risk (94). However, decreased ALDH2 activity resulted in acetaldehyde accumulation in the blood or other tissues such as the brain, forming CTIQs like Sal (83, 95, 96). A study by Kinoshita's group has speculated that the levels of Sal increased in the dorsal striatum after administration of acetaldehyde in the Aldh2‐knockout mice (96). Interestingly, acetaldehyde is not only one of the toxic aldehydes that result from lipid peroxidation of membrane‐rich mitochondria. In fact, multiple lipid peroxidation‐derived α,β‐unsaturated aldehydes can be generated in DA neurons, such as 4‐hydroxynonenal (4‐HNE), malondialdehyde (MDA), and 3,4‐dihydroxyphenylacetaldehyde (DOPAL, MAO product of dopamine). 4‐HNE and MDA are highly expressed in the substantia nigra of PD patients and are the substrates of ALDH2 and can inhibit the ALDH2 activity (94, 95). As discussed above, these processes could lead to the accumulation of acetaldehyde in the DA neurons, which is probably the reason for Sal's production.
On the other hand, we have noticed that Sal formation is also dependent on the presence of DA, which is an essential neuromodulator in several brain regions and present in lower levels in the substantia nigra and striatum of PD patients (75). Moreover, the generation of CTIQs is dependent on the related enzyme's distribution and activity, such as (R)‐salsolinol synthase and N‐methyltransferase. The most toxic NM‐(R)‐Sal is present in the nigro‐striatum, which might be the reason for Sal toxicity. Therefore, the production of CTIQs may primarily rely on the aldehydes levels induced by oxidative stress and enzyme activity (13). However, MAO performs synergistic action for damaging DA neurons. In contrast, DOPAL is reported as highly toxic and induced the loss of DA neurons and the accumulation of oligomeric forms of α‐synuclein after injection into the substantia nigra of rats (97).
Moreover, some neurotoxins such as benomyl, rotenone, and MPTP inhibit the activity of ALDH, especially ALDH2, which leads to the DOPAL accumulation, degeneration of DA neurons, and development of PD. Thus, speculating that mitochondrial ALDH2 could serve as a neuroprotective therapy for PD (98, 99). Contrarily, chronic administration of Sal decreases the production of dopamine and DOPAC concentrations in the substantia nigra and the striatum, suggesting that Sal might disturb DA storage and release (80). Further, ADTIQ is another Sal‐like, highly expressed neurotoxin in the putamen of PD patients, which might be a novel compound related to PD (100). Analogously, ADTIQ is also endogenously synthesized from the condensation of DA and methylglyoxal (MGO). MGO is a glycolytic by‐product and is present in higher concentrations in diabetic patients. Indeed, elevated concentrations of glucose might lead to increased production of MGO and trigger the occurrence of PD in patients with diabetes. Therefore, it can be inferred that mitochondrial dysfunctions are involved in both diabetes and PD induced by MGO and ADTIQ. Besides, MGO also increased the concentration of DA and Sal in SH‐SY5Y cells. So, ADTIQ should be the central hub between diabetes and PD (101, 102, 103).
3. MITOCHONDRIAL‐RELATED EPIGENETICS IN PD
Epigenetic modifications have been reported to bring various changes in gene expression and protein levels in PD. Various environmental factors induce epigenetic changes that trigger significant molecular variations and alter the normal function of the locus or chromosome despite altering the DNA sequence (14, 15). Converging evidence has supported that these epigenetic modifications account for significant expression differences at mitochondrial levels in Parkinson's disease (Figure 2). Thus, owing to their essential role in PD, it is speculated that these epigenetic modifications may fill the gap for our further understanding between mitochondrial impairment and the onset of PD progression. And might serve as a potential alternative therapeutic strategy for mitochondrial‐dependent PD.
FIGURE 2.
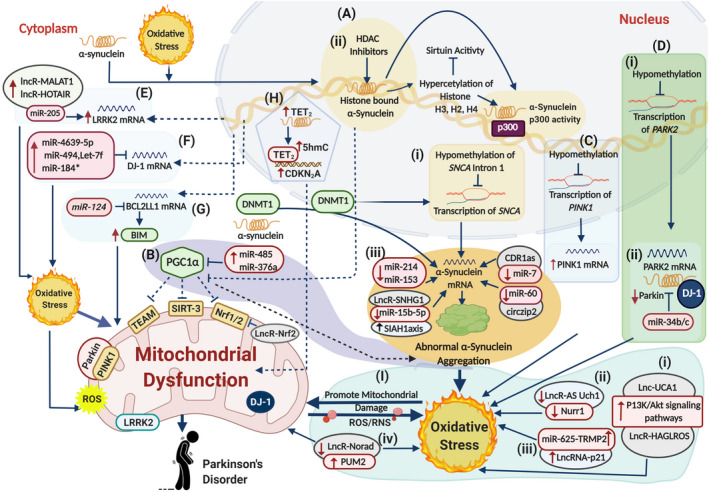
The landscape of neurotoxic epigenetic modifications inducing mitochondrial dysfunction in PD. (A) Epigenetic modifications of SNCA gene include multiple events that cause abnormal α‐synuclein expression. (i) Hypomethylation of the intron 1 of SNCA locus dysregulates SNCA gene transcription, leading to abnormal α‐synuclein aggregation in the cytoplasm. This allows abnormal α‐synuclein to enter the nucleus following oxidative stress and sequester DNMT1 into the cytoplasm, enhancing the hypomethylation of SNCA gene and causing dysregulated transcription. (ii) Meanwhile, HDAC inhibitors cause hyperacetylation of α‐synuclein‐linked histones H2, H3, H4, and p300 promoter region of SNCA. This results in reduced Sirtuin 1/2 activity, which leads to higher α‐synuclein aggregation accompanied by impaired mitochondrial biogenesis. (iii) Various noncoding RNAs dysregulate SNCA gene expression, such that, CDR1as sponge with miR‐7, circzip2 with miR‐60, lowered levels of miR‐124 and miR‐153 expression and higher expression of LncR‐SNHG lowers miR‐15b‐5p levels, which upregulates SIAH1 axis, thus contributing to the over‐expression of mRNA (SNCA) to trigger α‐synuclein aggregation and oxidative stress. (B) Reduced Sirtuin activity results in increased histone acetylation and reduced expression of nuclear‐encoded mitochondrial gene PGC1‐α that further downregulate TEAM, SIRT‐3, and Nrf1/2, impairing the mitochondrial biogenesis and promoting α‐synuclein accumulation. Upregulated miR‐485 and miR‐366a also downregulate the expression of PGC1 α to fuel the process. Further, LncR‐Nrf2 also inhibits Nrf1/2 expression to promote mitochondrial dysfunction. (C) Hypomethylation of the PINK gene stimulates elevated ROS production and disturbed mitochondrial metabolism. (D) Epigenetic modifications in PARK2 gene. (i) Hypomethylation of PARK2 gene results in dysregulated parkin expression. (ii) Upregulated miR‐34b/c causes downregulation of parkin protein with DJ‐1, leading to higher oxidative levels, abridged mitochondrial membrane potential, and reduced ATP levels along with higher mitochondrial fragmentation. (E) Upregulated lncRNA‐MALAT1 and lncRNA‐HOTAIR sponge with miR‐205‐5p to increase the LRRK2 expression that promotes neuronal apoptosis through oxidative stress and mitochondrial dysfunction. (F) Upregulated miR‐4639‐5p, miR‐494, miR‐184*, and let‐7f downregulate DJ‐1 expression that triggers oxidative stress‐induced neuronal lesions. (G) Downregulated miR‐124 upregulates BCL2L11 (BIM protein) that induces mitochondrial apoptotic signaling pathways. (H) Overexpression of TET2 promotes hydroxy‐methylation (5hmC) in the promoter region of CDKN2A gene that causes α‐syn aggregation and mitochondrial dysfunction. (I) Various noncoding RNAs target multiple genes and pathways to enhance oxidative stress‐simulated mitochondrial damage. (i) lncRNA HAGLROS and lncRNA UCA‐1 activate P13K/Akt pathway that induces oxidative stress and damages mitochondria, resulting in neuronal apoptosis and autophagy. (ii) Downregulated LncRNA AS Uchl1 lowers Nurr1 activity that links with mitochondrial dysfunction due to higher oxidative stress. (iii) LncRNA‐p2 sponge with miR‐625 exaggerating oxidative levels and inducing cytotoxicity and neuronal apoptosis. (iv) lncRNA NORAD target PUM2 gene to induce genomic instability and mitochondrial dysfunction. These events cause higher oxidative stress levels and mitochondrial anomalies, leading to DA neurons’ degenerative death, hallmarks of PD. The solid lines illustrate confirmed pathways, whereas the dotted lines illustrate pathways that need to be confirmed further. ↑ illustrate upregulated and ↓ illustrate downregulated
3.1. Mitochondrial related non‐coding RNA in PD
Regulation of non‐coding RNAs (ncRNAs) is one aspect of epigenetics. It mainly includes lncRNAs of longer than 200 bp, circular ncRNAs, mid‐size ncRNAs (snoRNAs, PASRs, etc.), and short ncRNAs (microRNAs, piwi‐interacting RNAs, etc.), all of which are distinguished based on the transcript length (104). Although ncRNAs do not translate into proteins, they can regulate many genes at the transcriptional level regulating various cellular processes. However, there is increasing evidence that ncRNAs exert multiple roles in many underlying mechanisms of PD pathogenesis. Their functions have been detected in the mitochondria, particularly susceptible to environmental factors, which are vital elements in PD (105).
3.1.1. The crosstalk of microRNA and mitochondria
MicroRNAs (miRNAs) are the most abundant, endogenous, small non‐coding RNAs (20, 21), involved in cascades of biological processes to preserve normal cellular functions (106). Usually, they act as negative regulators of gene expression at post‐transcriptional levels (107), and their dysregulated expression leads to the development of various pathologies ranging from brain disease to cancer. From the past few years, miRNAs have reported inducing mitochondrial impairment in PD by targeting genes involved in regulating mitochondrial and cellular functioning.
Numerous studies presented various dysregulated miRNA expression profiles, which display their peculiar behavior toward PD development and progression. In 2017, Martinez et al. reported miRNAs expression profiling in postmortem brain samples (cingulate gyri, cerebellum, putamen, midbrain, SN, prefrontal and frontal cortex, amygdala, and striatum) of IPD‐affected individuals. The results of this study have revealed considerable levels of dysregulated miRNAs targeting PD‐associated genes (PARK2, SNCA, LRRK2, TNFSF13B, LTA, SLC5A3, PSMB2, SR, GBA, LAMP‐2A, HAS). However, most miRNAs were reported downregulated and confirmed in PD models through agomirs (108). Several studies showed that miRNAs exert neurotoxic roles in PD development by targeting genes associated with mitochondrial functioning and oxidative stress. For instance, miR‐214 has upregulated the SNCA gene in PD models, which resulted in the α‐synuclein aggregation, oxidative stress, and mitochondrial impairment (Figure 2A‐iii) (109).
Strikingly, a subset of miRNAs mobilizing from the nucleus to mitochondria, termed mitochondrial miRNAs (mitomiRNAs), has gained researcher's attention. It is believed that mitomiRNAs play a significant role in PD pathogenesis (110), though research in this domain is nascent. Several PD studies have reported various miRNAs that exclusively target genes involved in maintaining mitochondrial integrity. Alterations in mitomiRNAs expression result in abnormal mitochondrial function and biogenesis. For example, upregulated miR‐34b and miR‐34c have reported downregulating DJ‐1 and Parkin genes. The deregulated miR‐34b and miR‐34c resulted in elevated oxidative levels, abridged mitochondrial membrane potential, reduced ATP levels, and higher mitochondrial fragmentation (Figure 2D‐ii) (111, 112). Other studies showed that miR‐4639‐5p (113) and miR‐494 (114) directly target DJ‐1 to trigger oxidative stress‐induced neuronal lesions (Figure 2F) (113, 114). Similarly, overexpressed LRRK2 inhibited the expression of let‐7 and miR‐184, resulting in the upregulation of E2F1 and DP1 genes (Figure 2F), respectively. Thus, it mediated the toxicity like dysregulated LRRK2 and stimulated the dopaminergic neuronal degeneration (115).
miR‐124 plays a crucial role in neuronal differentiation, neurogenesis, mitochondria apoptotic‐signaling pathways, and autophagy (116, 117). Downregulated miR‐124 resulted in higher Bim protein expression that enhanced translocation of BAX protein to mitochondria and lysosome, resulting in the activation of mitochondria apoptotic signaling pathways and impaired autophagy process (Figure 2G) (116). Another investigation on the PD model showed that miR‐485 targets the PGC‐1α gene (peroxisome proliferator‐activated receptor‐gamma coactivator‐1 α), a potent stimulator of mitochondrial biogenesis and respiration. Upregulated miR‐485 resulted in the suppression of PGC‐1α, which mediated neurotoxicity through α‐synuclein accumulation and mitochondrial dysfunction (Figure 2B) (118, 119). Recently, upregulated miR‐376a in PD patients inversely downregulated the PGC1α and TFAM gene expressions, impacting PD pathogenesis through mitochondrial dysfunction (Figure 2B) (120).
Further, in addition to the pathogenic behavior of miRNAs, various studies reported several miRNAs with neuroprotective properties exclusively target PD‐associated genes and impact the mitochondrial function in PD, summarized in Table 1. It is worth mentioning that these miRNAs displayed therapeutic potential in PD models by protecting against intercellular ROS generation, mitochondrial dysfunction, and neuronal injury. Though the research in this domain is still in infancy, as most of the casual agents inducing mitochondrial dysfunction are interlinked, at this point, it is difficult to predict the exact function of related miRNAs and requires extensive research. However, neurotoxicity and neuroprotection properties of miRNAs may provide valuable insights to understand PD progression and open new lines of investigations to determine their role as a biomarker for diagnostics and therapeutics avenues against PD to mitigate brain damage.
TABLE 1.
MicroRNAs involved in mitochondrial dysfunction in Parkinson's disease
| microRNAs | Model | Target | miRNA expression | Function | References |
|---|---|---|---|---|---|
| miRNA with neuroprotective effect | |||||
| miR‐7 | PD patients, MPTP mice model, MPP+‐SH‐SY5Y cell model | SNCA | ↑ | Downregulates α‐synuclein and act as a neuroprotective agent against oxidative stress and mitochondrial impairment | (121) |
| miR‐7 and miR‐153 | MPP+ HEK293 | SNCA | ↑ | Downregulates α‐synuclein and act as a neuroprotective agent against mitochondrial reactive oxygen species and inhibit neuronal death | (122) |
| miR‐7 | MPP+‐SH‐SY5Y cell model | VDAC1 | ↑ | Downregulates VDAC1 expression and stimulate neuroprotection by inhibiting ROS and mitochondrial dysfunction | (123) |
| miR‐27a/b and miR 26 | Human cervical HeLa and dopaminergic‐like M17 cells | PINK1 | ↑ | Suppress PINK1 gene expression by preventing the mitophagic influx | (124) |
| miR‐30a* and let‐7f | LRRK2‐knockout mice model | LRRK2 | ↑ | Acts as a neuroprotective agent against PD progression | (125) |
| miR‐205 | PD patients (frontal cortex + corpus striatum) | LRRK2 | ↑ | Acts as a therapeutic agent by upregulating LRRK2 gene expression to rescue neurite outgrowth phenotype | (126) |
| miR‐137 | PD patients’ exosomes (serum) | OXR1 | ↑ | Downregulate OXR1and protect by reducing oxidative stress injury and reduce neuronal apoptosis in PD patients | (127) |
| miRNA with neurotoxic effect | |||||
| miR‐124 | PD patients, MPTP mice model, MPP+‐SH‐SY5Y cell model | BCL2L11 | ↓ | Upregulate BIM protein expression to trigger mitochondrial apoptotic signaling pathways and Autophagy | (116, 117) |
| miR‐485 | PGC‐1α null mice | PGC‐1α | ↑ | Downregulate PGC‐1α and results in the accumulation of α‐synuclein, leading the neurotoxicity through mitochondrial dysfunction | (119) |
| miR‐376a | PD patients (blood samples), MPP+‐SH‐SY5Y cells | PGC‐1α | ↑ | Downpregulate PGC1α and TFAM gene expressions impacting on PD pathogenesis through mitochondrial dysfunction | (120) |
| miR‐214 | MPTP‐PD mouse model and MPP+‐SH‐SY5Y cell model | SNCA | ↓ | Upregulates α‐synuclein accumulation and triggers mitochondrial impairment | (109) |
| miR‐34b/c | PD patients, MPP+‐SH‐SY5Y cells | DJ‐1 and Parkin | ↓ | Downregulate DJ‐1 and Parkin, which result in oxidative stress, reduced ATP concentration, and elevated mitochondrial fragmentation | (111, 112) |
| miR‐4639‐5p and miR‐494 | PD patients | DJ‐1 | ↑ | Downregulates DJ‐1 expression to stimulate oxidative stress‐induced neuronal lesions | (113, 114) |
| let‐7 and miR‐184* | LRRK2 overexpressed Drosophila model | E2F1 and DP | ↑ | Mediate toxic effects similar to the dysregulated LRRK2 expression and stimulate dopaminergic neuronal degeneration | (115) |
↑ shows upregulation and ↓ shows downregulation.
3.1.2. The crosstalk of long non‐coding RNA and mitochondria
Long non‐coding RNA (lncRNAs) are RNA molecules that consist of more than 200 nucleotides derived from various genomic resources such as intergenic regions, gene regulatory regions, chromosomal regions, and mitochondrial genomes (128, 129, 130). About 40% lncRNAs are expressed in specific neuronal tissues, regulating various neurobiological processes such as neural plasticity, synaptic transmission, neurogenesis, and brain development. Any dysregulation in lncRNAs results in various neurodegenerative diseases (131). Accumulating evidence has pointed out that multiple changes in lncRNA may play a crucial role in oxidative stress and mitochondrial dysfunction in PD progression, summarized in Table 2.
TABLE 2.
Long non‐coding RNAs involved in mitochondrial dysfunction in Parkinson's disease
| LncRNAs | Study model | LncRNA expression | Target | Function | References |
|---|---|---|---|---|---|
| LncRNA‐Norad | Norad knockout‐mice model | ↓ | PUM2 | Upregulates PUM2 gene expression that results in genomic instability and mitochondrial dysfunction | (132) |
| LncRNA‐NEAT1 | PD patients (SNc), cell line model (SH‐SY5Y), HEK‐293T cells | ↑ | PINK | Positively upregulates PINK gene expression by preventing PINK1 protein deterioration and enhanced autophagy along with neuroprotection against oxidative stress | (133, 134) |
| LncRNA‐UCA1 | Substantia Nigra striatum 6‐OHDA PD model, MPP+ mouse model, MPP+ induced SH‐SY5Y cell model | ↑ | P13K/Akt signaling pathways | Activated P13K/Akt signaling pathways and induced mitochondrial dysfunction followed with apoptosis, whereas downregulation of lncRNA‐UCA1 acts in the opposite way | (135, 136, 137) |
| LncRNA‐p21 | MPP+ induced SH‐SY5Y cell model, Knockdown‐lncR‐p21 MPP+ induced SH‐SY5Y cell model | ↑ | miR‐625‐TRMP2 | Sponge with miR‐625 and upregulates TRMP2 to induce cytotoxicity and promotes cell apoptosis (neuronal injury), whereas p21 knockdown acts oppositely | (138) |
| LncRNA‐Cerox1 | Human and mouse model | ↑ | miR‐488‐3‐OXPHOS | Upregulates mitochondrial oxidative phosphorylation (OXPHOS) and protect against oxidative stress | (139) |
| Nrf2 related LncRNAs | PD patients (SNc), MPTP induced mouse model PD | ‐ | Nrf2 | Promotes neurodegeneration through oxidative stress | (140) |
| LncRNA‐HAGLROS | MPP+ induced SH‐SY5Y cell model, MPTP‐mouse model | ↑ | 100/ATG10 axis and P13K/Akt/mTOR | Upregulates 100/ATG10 and P13K/Akt/mTOR and contribute to Parkinsonism via apoptosis and autophagy | (141) |
| LncRNA‐MALAT1 | MPP+ induced SH‐SY5Y cell model, MPTP‐mice model | ↑ | miR‐205‐5p‐LRRK2 | Upregulates LRRK2 expression by miR‐205 inhibition, thus induces mitochondrial dysfunction and apoptosis (neuronal damage) | (142) |
| LncRNA‐HOTAIR | MPP+ induced SH‐SY5Y cell model, MPTP‐mice model | ↑ | miR‐205‐5p‐LRRK2 | Upregulates LRRK2 expression by miR‐205 inhibition, thus induces mitochondrial dysfunction and apoptosis (neuronal damage) | (143) |
| LncRNA‐HOTAIRM1 | PD patients (circulating leukocytes), 6‐OHDA PD model, SH‐SY5Y cell model | ↑ | ‐ | Overexpressed HOTAIRM1 plays a part in Parkinsonism by promoting dopaminergic neurons apoptosis | (144) |
| LncRNA‐SNHG1 | MPP+ induced SH‐SY5Y cell model, MPTP‐mice model | ↑ | miR‐15b‐5p/SIAH1 axis | Downregulates miR‐15b‐5p that overexpress the SIAH1 axis, leads to α‐synuclein accumulation and neuronal toxicity | (145) |
| LncRNA‐AS Uch1 | iMN9D cells, MPTP‐mice model | ↓ | Nurr1 | Downregulated AS Uch1 expression is regulated by downregulated Nurr1 activity, results in dopaminergic dysfunction | (146) |
↑ shows upregulation and ↓ shows downregulation.
Recently, lncRNA NORAD (long non‐coding RNA activated by DNA damage) targeting the PUM2 gene is reported to induce genomic instability and mitochondrial dysfunction. That resulted in premature aging and other deformities in the PD model, proposing that mitochondrial‐related lncRNAs play a crucial role in PD progression (Figure 2I‐iv) (132). In another study, lncRNA UCA‐1 (urothelial carcinoma‐associated‐1) induced apoptosis with mitochondrial impairment in the PD model through targeting P13K/Akt‐signaling pathways (Figure 2I‐i). Its silencing resulted in lower destruction of dopaminergic neurons and oxidative stress along with inflammatory response linked with PD (135, 136, 137). LncRNA‐p21 (long non‐coding RNA‐p21) sponge with miR‐625 and induce cytotoxicity and neuronal apoptosis (Figure 2I‐iii). In contrast, its knockdown weakened cytotoxicity, apoptosis, ROS production, oxidative stress, and neuroinflammation (138). LncRNA MALAT1 (metastasis‐associated lung adenocarcinoma transcript 1) sponge miR‐205‐5p and upregulated LRRK2 expression. That resulted in mitochondrial dysfunction and apoptosis (Figure 2E) (142). Conversely, overexpression of lncRNA HOTAIR (HOX Transcript Antisense Intergenic RNA) promoted PD progression by upregulating LRRK2 gene expression (Figure 2E) (143). Another LncRNA SNHG1 (small nucleolar RNA host gene 1) triggered α‐synuclein accumulation and toxicity by targeting the miR‐15b‐5p/SIAH1 axis (Siah E3 Ubiquitin Protein Ligase 1), leading to dopaminergic neurons injury (Figure 2A‐iii) (145).
LncRNA AS Uchl1 (antisense to the mouse Ubiquitin carboxy‐terminal hydrolase 1) an antisense transcript of Uchl1 gene (synthetic locus of UCHL1/PARK5) work in association with Nurr1 gene to regulate dopaminergic cell differentiation and maintenance. During Parkinsonism, AS Uchl1 is found downregulated due to downregulated Nurr1 (Nuclear receptor related‐1) activity, resulted in dopaminergic dysfunction and neuronal injury. It is proposed that downregulated Nurr1 activity is linked with mitochondrial dysfunction that mediates neuronal injury in PD (Figure 2I‐ii) (146). Another study on PD models reported that lncRNA HAGLROS (HAGRL opposite strand LncRNA) induced neuronal apoptosis and autophagy through activation of P13K/Akt/mTOR pathway along with miR‐100/ATG10 (Figure 2I‐i) (141). Although a plethora of dysregulated lncRNA was observed in PD patients, still, this data is not enough to predict the underlying mechanism of LncRNAs role in PD progression. Therefore, detailed investigations are needed to elucidate their mechanism and in vivo function in PD progression.
Apart from the neurotoxic role of LncRNAs in PD, some LncRNAs presented therapeutic significance by protecting neurons against oxidative stress and mitochondrial damage in PD models. For instance, lncRNA NEAT (nuclear‐enriched assembly transcript‐1), which works in association with nucleus and mitochondria, upregulates PINK1 gene expression to inhibit mitochondrial induced autophagy along with PINK1 protein deterioration in PD models and reduced neuronal injury (133). In another study on PD patients (SNc), upregulated lncRNA NEAT1 provided similar protection against oxidative stress like neuroprotective agents fenofibrate and simvastatin (134). Similarly, upregulated LncRNA Cerox1 (cytoplasmic endogenous regulator of oxidative phosphorylation) in association with miR‐488‐3p has upregulated complex I subunit protein and reduced ROS levels, which ultimately protected complex I inhibitor rotenone (139). Nrf2 (NF‐E2‐related factor), known as essential signaling molecules, protects against oxidative stress. On accountancy to oxidative stress, Nrf2 detaches itself and translocates from cytoplasm to the nucleus to enhance the expression of antioxidant protein genes to enable cell survival (140). Owing to the Nrf2 defensive role, Wang et al. reported various Nrf2‐linked lncRNAs in the substantia nigra of paraquat and MPTP PD models with their defensive role against oxidative stress‐associated mitochondrial anomalies (140). Although only a few LncRNAs reported neuroprotective properties, they still have a new avenue toward the practical application of LncRNAs in mitochondrial‐associated deformities in PD.
3.1.3. The crosstalk of circRNAs and mitochondria
Circular RNA belongs to a class of endogenous long non‐coding RNAs that stabilize themselves in a circle due to covalent linkage of 3′–5′ heads (147). They act as miRNA regulators that make sponges with miRNAs, suppress their activity (148), impact gene transcription, mRNA splicing, RNA decay, and translation (147). It is reported that 20% of brain coding genes produce circular RNAs compared to other tissues. Therefore, due to high expression in mammalian brains, especially in neuropils and dendrites, they regulate synaptic functions and neural plasticity (149, 150). Up till now, very few studies have reported the involvement of circRNAs in the pathophysiology of various neuropsychiatric and neurological diseases, including PD (Table 3). Therefore, it emphasizes more research to determine underlying biological mechanisms in connection with circular RNA.
TABLE 3.
circRNAs involved in mitochondrial dysfunction in Parkinson's disease
| Circular RNA | miRNA sponge | Expression | Target | Study model | Function | Reference |
|---|---|---|---|---|---|---|
| CDR1as | miR‐7 | ↑ | SNCA | Zebrafish (midbrain) | Downregulates miR‐7 and upregulate SNCA leads to neuronal damage | (151) |
| circSNCA | miR‐7 | ↓ | SNCA | MPTP‐mouse model MPP+‐SH‐SY5Y | Upregulate miR‐7 that downregulate SNCA and reduce cell apoptosis and improves autophagy | (152) |
| circzip2 | miR‐60 | ↓ | SNCA | Transgenic C‐elegans model of PD | Enhances α‐synuclein accumulation by upregulating SNCA, results in neuronal damage in PD | (153) |
| circDLGAP4 | miR‐134‐5p | ↓ | CREB | MPP+ SH‐SY5Y, MN9D | Reduce neurotoxic effect by promoting cell viability, lowers apoptosis along with mitochondrial damage, and improved autophagy | (154) |
↑ shows upregulation and ↓ shows downregulation.
CDR1as (non‐coding antisense transcript to the protein‐coding gene CDR1) is reported as a negative regulator of miR‐7. Concerning the PD studies, CDR1as downregulated the miR‐7 expression, which resulted in α‐synuclein aggregation, oxidative stress, and mitochondrial dysfunction (Figure 2A‐iii) (151). In the C‐elegans model of PD, circzip2 was found downregulated, which lead to the upregulation of SNCA and α‐synuclein accumulation with associated mitochondrial anomalies (Figure 2A‐iii) (153). Few circular RNAs presented a defensive role against oxidative stress and mitochondrial dysfunction. CircSNCA was observed downregulated upon PPX (pramipexole) treatment in the PD model, which reduced cell apoptosis and improved autophagy by targeting miR‐7, illustrating circSNCA as a ceRNA of miR‐7 in PD (152). Furthermore, recently, circDLGAP4 was investigated for its neuroprotective effect by regulating the miR‐134‐5P/CREB pathway in PD models. The results of this study presented reduced circDLGAP4 in MPTP‐induced PD mouse model and MPP+‐induced PD cell models. Further in vitro studies affirm that circDLGAP4 boosts cell viability, lowers apoptosis, reduces mitochondrial damage, elevated autophagy, and debilitates the neurotoxic effects of MPP+ in SH‐SY5Y and MN9D cells (154). Hence due to their stability and precise involvement in PD pathogenesis, it is predicted that circular RNAs can serve as a potent tool for devising early diagnostic and targeted therapeutics.
3.1.4. Other non‐coding RNA and mitochondria
Apart from the above‐discussed non‐coding RNAs, piwi interacting RNAs (piRNAs) have emerged as another class of non‐coding RNAs, entitled genomic guardians. Although poorly understood, they protect the genome by facilitating transcriptional and post‐transcriptional silencing of transposable elements through DNA methylation and RNA interference. piRNAs are linked with PIWI proteins (a subfamily of Argonaute proteins), including Ago3, piwi, and Aub (155, 156) and involved in regulating the genomic modifications such as histone modification (H3k9ME3) or DNA modification (157). Initially, it was proposed that this mechanism is only confined to germ cells due to transgenerational inheritance. However, later, it was disproved when similar genomic instability due to transposable elements was observed in somatic cells, including post‐mitotic neurons, which insists that piRNAs are involved in various disorders’ pathogenesis (155, 158). Recently, piRNAs play a significant role in regulating brain function and being involved in multiple neurological diseases, including PD (157).
Up till now, there is only a single study reporting piRNAs expressions in neuronal cells from sporadic PD patients (159). In this study, piRNA expression was found dysregulated from SINEs (short interspersed nuclear element) and LINEs (long interspersed nuclear element) of PD patients. Due to which neuronal cells of PD patients presented prominent variations in various pathways such as PGC‐1α and CREB‐pathways, involved in mitochondrial dysfunction in a disease‐specific manner (159). It is further reported that piRNAs may have a methylated CREB2 gene's promoter region in Aplysia neurons, which regulates synaptic plasticity during memory (160). Thus, it infers a possible connection between piRNA and CREB pathways inactivation (157). Though this domain is barely investigated for Parkinsonism, further research is suggested that may help to understand the mitochondrial‐associated mechanisms in PD.
3.2. Mitochondrial‐related DNA methylation in PD
Mitochondrial DNA (mtDNA) constitutes 1% of total cellular DNA and synthesizes mitochondrial gene products essential for normal cellular function. Due to their prominent role in cellular functioning and disease pathogenesis, mtDNA has gained significant attention for various epigenetic changes that limelight research from nuclear DNA to mtDNA. In addition to nDNA methylation, mtDNA methylation has always been a remaining question (161), with arguments like mtDNA is more prone to oxidative stress and has a higher mutation rate due to lack of mtDNA histones and methylases accession to mitochondria (162). However, later, this theory was disapproved when both methylated (5‐mC) and hydroxymethylated (5‐hmC) cytosine residues in mtDNA along with DNA methyltransferase DNMT1 (cytosine‐5‐methyltransferase 1) were observed in mitochondria (163, 164). Mitochondria in the central nervous system contain de novo methyltransferase DNMT3A (DNA methyltransferase 3 alpha) and DNMT3B (DNA methyltransferase 3 beta) proteins along with DNMTI, 5‐mC, and 5‐hmC (165, 166, 167). Increasing evidence suggests that various aberrations in mtDNA, such as methylation and hydroxymethylation, are associated with various environmental toxins, oxidative stress, treatment strategies, drug, ailment, and aging that induce Parkinsonism (168).
3.2.1. DNA methylation
In general, little information is available on the association of mitochondrial methylation with Parkinsonism. Several research groups investigated the 5‐mC and 5‐hmC levels in the D loop region of mitochondria (involved in mitochondrial transcription and replication) (169) and NADH dehydrogenase gene 6 (MT‐ND6) gene in the SNpc of PD patients and healthy individuals (168, 170). Surprisingly, they found that the D loop region of mtDNA of PD patients presented a loss of methylation in almost all CpG and non‐CpG sites compared to healthy individuals. In contrast, 5‐mC levels in MT‐ND6 gene and 5‐hmC levels in the D‐loop region remain unchanged in PD patients than healthy individuals. Thus, postulating that the lower methylation levels in the D‐loop were not due to the shrinking of neuronal content in SNpc of PD patients; instead, these resulted from epigenetic dysregulation of essential mitochondrial functional sites (168, 170, 171). Therefore, despite substantial evidence regarding impaired mtDNA D‐loop region methylation in both humans and animal models of neurodegeneration, further studies are required to clarify the biological significance of these epigenetic changes and their cause/consequence relationship with the neurodegenerative process.
Several studies reported that methylation in various genes (nDNA) might directly contribute to mitochondrial dysfunction as a hallmark of PD. Thirty percent of global hypomethylation is correlated with the upregulation of the SNCA gene and sequestering of DNMT1 outside the nucleus to feed‐forward Parkinsonism (172). Due to the prominent role of α‐Syn levels in PD, various animal models and cell lines have been studied for SNCA methylation. Matsumoto et al. analyzed both PD cell lines and postmortem brain samples for methylation in the SNCA gene. Cell lines reported methylation in the SNCA CpG islands (CpG‐2), which result in SNCA overexpression. Post‐mortem brain samples revealed SNCA hypomethylation in the SNc, along with elevated levels of α‐Syn (173). Contrarily, methylation of human SNCA intron 1 abridges SNCA gene expression, whereas inhibition of DNA methylation has upregulated SNCA expression, thus proposing SNCA intron1, an attractive target to fine‐tuned SNCA downregulation (174). In another study, intron one region of SNCA was found hypermethylated in hiPSC‐derived dopaminergic neurons from PD patients, which resulted in the downregulation of SNCA expression level, suggesting a potential target for PD therapeutics (175).
Another research on PD brain samples revealed that lower levels of nuclear DNMT1 (DNA methylation stabilizer enzyme) are involved in the accumulation of α‐Syn in the cells that prevent the penetration of DNMT1 enzyme into the nucleus. It may result in the abnormal distribution of DNMT1 in the cells that lead to a reduction in global methylation in both human and mouse brains, including CpG islands upstream of SNCA, SEPW1 (selenoprotein W), and PRKAR2A genes (cAMP‐dependent protein kinase type II alpha regulatory subunit) (172). Additionally, other than the SNCA gene, genome‐wide association studies on PD patients revealed various abnormalities in DNA methylation on three different genes, including PARK16/1q32, GPNMB (glycoprotein Nmb), and STX1B (syntaxin 1B) (176). Similarly, methylation of the TNF (tumor necrosis factor) promoter region in the SNpc region caused overexpression of TNF levels. That elevated inflammatory reactions in dopaminergic neurons along with oxidative stress (172). Furthermore, various studies reported DNA methylation in PARK2 and PINK1 genes due to age‐related changes in mitochondria that resulted in higher ROS production levels and disturbed mitochondrial metabolism (177, 178).
Till yet, PD research is restricted due to limited access to brain samples, which raises concerns for identifying a new source of biomarkers. Recently, a study on serum samples of female PD patients reported higher mtDNA concentrations along with discernible mtDNA methylation (CpGs) in platelet‐derived mtDNA (179). Likewise, longitudinal genome‐wide methylation of blood samples of PD patients revealed significant changes in DNA methylation, proposing how abnormalities in DNA methylation can be helpful to track PD progression and drug's response in blood methylome in the future [100]. In another study, the PPARGC1A (PPARG Coactivator 1 alpha) gene was found hypermethylated at CpG‐1, CpG‐13.14, CpG‐17.18, and CpG‐20 sites in peripheral blood leukocytes of PD patients with reduced PGC‐ α expression, thus proposing that these gene dysregulations may be instilled in disturbed cellular energy metabolism, microglial plasticity, and mitochondrial dysfunction implicating PD pathogenesis (180). Interestingly, DNA hypomethylation, which is the unmethylation of DNA that is normally methylated, compromises the regular expression of various genes resulting in multiple pathologies. In this context, DNA hypomethylation of the TNF‐α gene (tumor necrosis factor) was reported as a cause of vulnerability in the substantia nigra of the PD brain (181). Overexpression of the TNF‐α gene resulted in reduced mitochondrial complex I and oxidative stress that triggers a caspase cascade leading to mitochondrial impairment and apoptosis (182). In a recent study, Ndayisaba et al. proposed that TNF‐α inhibitors act as neuroprotective agents against α‐synuclein accumulation and can be used as targets against oxidative stress and mitochondrial dysfunction, suggesting a possible future therapy (183). Overall, a comprehensive study about DNA methylation of various genes and their transcription factors strongly suggests its potential role in inducing impaired mitochondrial biogenesis leading to PD pathogenesis and is predicted to assist researchers in designing early PD diagnostic strategies for better disease management and treatment.
3.2.2. DNA hydroxymethylation
In 2009, hydroxymethylation was brought to light after the identification of ten‐eleven translocation (TET) (184). It is a family of enzymes (TET1, TET2, and TET3) which reverse DNA methylation 5mC (5‐methyl cytosine) to 5hmC (5‐hydroxymethyl cytosine) through an oxidative process (185, 186). It is reported that oxidative stress is the key factor that disrupts the performance of TET proteins and leads to hydroxymethylation, thus describing TET as a crucial and stable epigenetic mark (187, 188). 5‐hmC is highly enriched in the brain, particularly in neuronal cells, and is involved in regulating various biological functions such as aging, neurodevelopment, neurological disorders, and cancer (189, 190). The role of 5hmC in regulating gene expression and as a new diagnostic marker in Parkinsonism has gained attention. Initially, it was investigated in the 6‐OHDA rat model (191, 192); later, research on RNA for 5hmC modifications in different brain regions (brain stem, hippocampus, and cerebellum) presented a higher level of 5hmC modifications in RNA. Further investigation on the MPTP‐induced PD model revealed reduced 5hmC modifications (193). Therefore, demonstrating that 5hmC modifications in RNA might play a significant role in protein or miRNA expression. However, further investigations are still needed to elucidate the exact role of 5hmC variations in RNA in neurological pathologies.
Research on the human brain reported higher levels of 5hmC in the cerebellum in both male and female PD individuals than healthy individuals, thus raising the question; whether a higher level of 5‐hmC is a driver or consequence of Parkinsonism? (194, 195). Recently, various environmental factors, including exogenous chemicals, stress, and imbalanced lifestyle result in changes in 5‐hmC in the brain, making it more susceptible to neurogenerative disorders (196). Of significance to PD, TET3 is reported as a regulator of autophagy and lysosomal genes in neurons. After nDNA, there is an emerging trend to explore the methylation effects on the mtDNA regulation through mtDNMT and TET proteins, which is proposed to be higher in aging neurons due to higher mtDNA hydroxymethylation. Recently, researchers reported an impact of TET2‐mediated CDKN2A (cyclin‐dependent kinase inhibitor 2A) DNA hydroxymethylation in Parkinsonism. In this study, TET2 was found upregulated in dopaminergic neurons of MPTP‐induced mice that promote 5‐hmC in the promotor region of the CDKN2A gene, resulting in dysregulated cell‐cycle apoptosis. Interestingly, upregulated 5‐hmC in another PD model (A53T cell line) presented higher levels of α‐syn levels. Conversely, downregulated TET2 expression in the MPP+‐stimulated SH‐SY5Y model has rescued neuronal cell damage and cell‐cycle arrest. Further, knockdown of TET2 in SNpc of MPTP‐induced mice PD model resulted in lesser motor loss and dopaminergic neuronal injury through CDKN2A‐p16 suppression, suggesting a novel strategy for PD epigenetic‐based therapy (197). Hence, with the link to this research, it is hypothesized that neuronal deformities induced by elevated TET2 levels may disturb regular CDKN2A activity that further links with cascades of reactions involved in dysregulated mitophagy and mitochondrial dysfunction, implicating Parkinsonism (Figure 2) (197, 198). However, due to scarce research, it is quite challenging to predict the exact role of 5‐hmC in PD neurodegeneration, thus opening the gate for more research on the impact of DNA methylation, especially 5‐hmC, a nascent factor in controlling neuronal homeostasis.
3.3. Mitochondrial‐related histone modifications in PD
The association of histone modifications in mitochondrial impairment and PD has been a subject of dynamic discussion for the past few years. Aberrations in histones may disturb the access of the transcription machinery to the promoter of various genes, resulting in dysregulated gene expressions. Histone modifications consist of acetylation, methylation, phosphorylation, ubiquitination, sumoylation, and ADP‐ribosylation, along with other post‐translational modifications (199, 200). Apart from the strong association between abnormal mitochondrial modification and defective histone modifications, very few investigations have been reported on this epigenetic mechanism modulating PD‐associated risk.
Aforementioned, the PD brain is characterized by multiple mitochondrial defects that impart aberrant mitochondrial DNA maintenance (48, 201), deviant mitochondrial quality control, and abnormal mitochondrial respiration in the form of complex I deficiency (14, 15, 202). It is reported that mitochondrial complex I deficiency may lead to histone hyperacetylation (203) due to decreased NAD+/NADH ratio and disturbed activity of NAD+‐dependent class III histone deacetylase termed as sirtuins (204, 205, 206). Several other studies demonstrated that hyperacetylation of various histone sites such as H3K9, H3K27, and H2BK15 followed by treating neuronal cells with PD‐linked mitochondrial complex I inhibitors had reduced sirtuin activity (203, 207). It was found that lower sirtuin activity, especially SIRT1/SIRT2/SIRT3 activity, led to higher p300‐mediated histone acetylation at the promoter region of the SCNA gene, which resulted in the higher α‐synuclein aggregation accompanied by impaired mitochondrial biogenesis (Figure 2A‐ii). To further investigate, recently, Toker et al. reported a genome‐wide association of histone acetylation at various PD‐associated genes (promoter or enhancer region) in PD patients. This study revealed elevated histone acetylation of H2B, H3, and H4 (multiple histone sites) with a noticeable change in H3K27 and increased sirtuin proteins (SIRT‐1/SIRT‐3) in the PD brain. These hyperacetylations in genomic regions may result in dysregulated deacetylation machinery that predisposes various somatic variations. It is proposed that these genomic rearrangements may lead to mitochondrial complex I deficiency followed by reduced NAD+/NADH ratio, lowered sirtuin activity, along increased SIRT‐1 and SIRT‐3 protein levels. That may trigger abnormal cellular processes synergistically with neuronal injury and oxidative stress, a prerequisite to PD‐specific pathology (208).
However, concerning PD, various studies directly correlate histone modification of various genes with mitochondrial dysfunction. It was observed that aggregation of α‐synuclein triggers histone H3 hypoacetylation, chromatin compression, and gene expression in PD, resulting in neurotoxicity and apoptosis (209, 210). Furthermore, in a study on the C‐Elegans PD model, overexpression of wild type and A53T human SNCA resulted in the downregulation of nine histone genes forming linker H1, H2B, and H4. Thus, it supports the significant role of histone modification in mediating neurotoxicity by disturbing mitochondrial complexes (211).
In another study, sirtuin 2 (histone deacetylase) was investigated for its neuroprotective effect against α‐synuclein toxicity in nerve cells of the animal model (212). It was found that upregulation of the SNCA gene leads to the binding of α‐synuclein with the promoter region of PGC‐1α gene that causes PGC‐1α hypoacetylation followed by transcriptional inhibition of PGC‐1α and mitochondrial impairment (213). Histone deacetylase 6 was reported as a neuroprotective enzyme and stimulated the synthesis of α‐synuclein inclusions in the TH‐GAL4 cell line of the Drosophila PD model, thus presenting its defensive role against α‐synuclein accumulation. Therefore, it is proposed that mutations in the histone deacetylase 6 gene may induce toxic α‐synuclein accumulation and neurotoxicity (214).
3.4. Crosstalk of Prion and Mitochondrial dysfunction
Prions are fatal, infectious protein agents of prion diseases that share critical biophysical and biochemical characteristics with various neurodegenerative disorders, including PD (215, 216). At present, several researchers have speculated “prions” as another epigenetic agent that transmit genetic information to other cells leading to various genetic changes in phenotype without underlying alterations in genomic sequence (217). Parkinsonism is characterized by aberrant aggregation of α‐synuclein accompanied by mitochondrial dysfunction. It is proposed that abnormal α‐synuclein can spread to acquainting brain parts and lead to the accumulation of endogenous α‐synuclein in these regions as prions (PrPsc) to impact pathogenicity (218).
Emerging evidence suggests that misfolded α‐synuclein share common characteristic with prions in their mode of action such as (i) both (α‐syn and prions) exist in α‐helix or undefined structure and can misfold under certain circumstances, especially when confirmation is enriched with β‐sheets; (ii) misfolded proteins can aggregate normal proteins and disturb their conformation by acting as a template; (iii) these misfolded proteins can be engulfed by recipient cells and transferred from one cell to another (216, 218). However, the process of how α‐syn is taken in by and induces misfolding and endogenous assembly remains unclear. Therefore, this phenomenon raises several unanswered questions regarding the pathogenesis of abnormal α‐syn as a prion such that (i) What is the specific mechanism involved in the misfolding, discharge, uptake, and spread of α‐syn; (ii) Is there a direct relationship in inducing misfolding between abnormal protein and healthy neighbor cell protein leading mitochondrial damage; (iii) How physicochemical and environmental factors affect proteins biogenesis and propagate the release of endogenous neurotoxins to aggravate the pathological process of PD; (iv) Whether Parkinson disease has a similar infectious mode as prions. Thus, addressing all the above questions may stratify a new field of research in neurodegenerative disorders.
4. THE VICIOUS CYCLE OF MITOCHONDRIAL DYSFUNCTION IN PD
Up till now, we have observed five common factors involved in oxidative stress simulated mitochondrial dysfunction in PD: endogenous neurotoxins, epigenetic factors, abnormal α‐synuclein accumulation, oxidative stress, and mitochondrial dysfunction. All these factors are found inter‐related through three cycles and seem to work alone to induce Parkinsonism, as illustrated in Figure 3.
The first cycle consists of endogenous neurotoxin (CTIQs), oxidative stress, and mitochondrial dysfunction. Prolonged oxidative stress conditions stimulate the aldehyde synthesis from lipid peroxidation, which interacts with dopamine to produce series of CTIQs such as Sal under the catalysis of a series of enzymes in DA neurons and inhibit oxide‐scavenging mechanisms by restricting mitochondrial complex I activity (219). This impairs mitochondria and increases oxidative stress levels, which, in turn, promotes the release of more CTIQs.
After the first cycle, the second cycle between epigenetic factors, oxidative stress, and mitochondrial function may establish. Under higher oxidative stress levels, various epigenetic modifications such as ncRNAs (miRNAs, LncRNAs, CircRNAs, and snRNAs), DNA modifications such as histone modification, and DNA methylation (hypermethylation and hypomethylation) are triggered that target Parkinson‐associated genes and pathways (177). These epigenetic modifications dysregulate normal proteome expression, which damages mitochondrial activities (complex I and III) and promotes excessive oxidative stress levels in DNs, leading to neuronal damage and LB development. Altogether, these epigenetic factors and endogenous neurotoxins cause energy crises by damaging mitochondria of DNs (177, 219) and induce the production of excessive oxidative stresses in DA neurons, perpetuating epigenetic modifications and production of CTIQs.
The biological effect of these two cycles results in the formation of the third cycle of abnormal α‐synuclein accumulation. Oxidative stress, endogenous neurotoxins, and epigenetic modifications may induce an abnormal modification in α‐synuclein such as nitrated α‐synuclein and ATP depletion malfunction protein degradation pathways, and phosphorylated or carbonylated α‐synuclein (220, 221, 222). These modifications impair α‐synuclein normal function and impede subsequent ubiquitination. It also causes cellular degradation and promotes α‐synuclein protein aggregation, which intensifies oxidative stress levels (223) and again enhances epigenetic modifications, synthesizing endogenous neurotoxins, and abnormal α‐synuclein aggregation.
FIGURE 3.
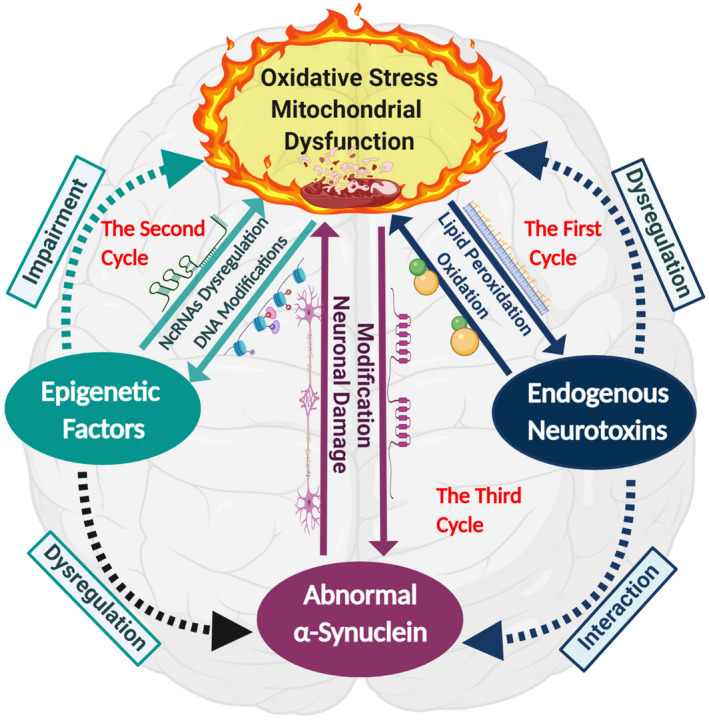
The vicious cycle combines three inter‐related cycles that consist of oxidative stress, mitochondrial dysfunction, endogenous neurotoxins, epigenetic factors, and abnormal α‐synuclein aggregation in the etiopathogenesis of PD. The first cycle begins with prolonged oxidative stress in the body and triggers aldehydes’ synthesis through lipid peroxidation, which interacts with dopamine to produce endogenous neurotoxins. Endogenous neurotoxins cause mitochondrial dysfunction by inhibiting oxide‐scavenging mechanisms by restricting mitochondrial complex I activity and increasing oxidative stress. After the first cycle, the second cycle is formed. Oxidative stress and endogenous neurotoxins trigger epigenetic modifications that dysregulate the expression of various genes and induces damage to the mitochondrial activities that again raise oxidative stress levels. These two cycles lead to the third cycle of abnormal α‐synuclein aggregation; oxidative stress, endogenous neurotoxins, and epigenetic modifications may induce abnormal α‐synuclein modifications. These impaired proteins may not be degraded; instead, they interact to promote α‐synuclein protein aggregation. Abnormal proteins act as antigens and induce intracellular damage through mitochondrial anomalies and intensify oxidative stress levels, which further promotes the release of endogenous neurotoxins, epigenetic modifications, and abnormal α‐synuclein aggregation. The three cycles in the vicious cycle work alone or together and induce DA neuronal degeneration and PD development. The solid lines present confirmed pathways, whereas dotted lines present assumed pathways that need to be confirmed
Owing to the amplifying effects of these three cycles, we propose a vicious cycle feedback loop that interlinks these three cycles in PD’s associated mitochondrial dysfunction. However, the discussed above three cycles require further verifications. Usually, endogenous neurotoxin production and epigenetic modifications may not occur instantly, especially in familial PD. The abnormal protein expression in familial PD is associated with genetic mutations in SNCA and UCHL1, which causes abnormal α‐synuclein aggregation, triggering mitochondrial dysfunction with higher oxidative stress, leading to endogenous neurotoxins synthesis and epigenetic modifications. In a nutshell, each of the three cycles can be a starting point; however, they work together in a vicious cycle to induce mitochondrial dysfunction that leads to neuronal death, development of Lewy bodies, and Parkinsonism.
5. CONCLUSION AND FURTHER CONSIDERATIONS
Over the past decades, besides extensive research, the underlying mechanisms involved in the pathophysiology of PD remain unclear. It is narrated that both forms of PD are strongly associated with mitochondrial dysfunction and oxidative stress, triggered by various factors, including genetics and epigenetics (8). Therefore, within the framework of this article, we have briefly reviewed the impact of genetic and epigenetic regulators primarily involved in the mitochondrial dysfunction associated with PD progression. Mitochondrial dysfunction is a complicated and multifaceted process involving various key processes such as mitochondrial respiratory chain, mitochondrial genome, interaction with α‐syn, and mitophagy/mitochondrial (219). Malfunctioning these processes may cause an energy crisis in the cells, leading to a terrible fate of neuronal apoptosis. Therefore, keeping in view of the finding of previous studies, we have inferred that, in PD, oxidative stress, endogenous neurotoxins (CTIQs), α‐synuclein proteins, and epigenetic regulators work in a vicious cycle to impair mitochondrial biogenesis, which, in turn, exaggerates oxidative levels to induce the production of CTIQs and α‐synuclein aggregation and work vice versa as described in Figure 3. Most of these processes are interlinked at various stages, making PD a very complicated neurodegenerative disorder and have gained the researcher's attention to find key elements involved in mitochondrial dysfunction. Nevertheless, many mitochondrial‐based therapies have been identified in the laboratories, but, unfortunately, none of the therapy proved effective against PD (224). Therefore, our proposed vicious cycle components may serve as a new hope to identify key disease mechanisms and help to design target‐based therapeutics under these scenarios. Although various studies have contributed to this aspect, still, this field is in its infancy phase and requires more research to uncover PD pathogenesis.
Unfortunately, due to disease complexity, there is no treatment for Parkinson's disease, and present medical interventions are not enough to control PD progression. However, clinicians have no choice other than to rely on symptomatic treatments that alleviate symptoms for the time being (225). One of the significant hindrances in the PD treatment is that most patients display clinical symptoms upon 60% loss of dopaminergic neurons. Hence, early‐stage diagnosis and treatment have become a dire need of time for effective disease management. In previous studies, our research group reported various novel components of the damaging cycle, such as salsolinol synthase (83), that can be utilized as targets for PD treatment. On the other hand, various antioxidants have also been reported as another PD treatment but restricted by lower efficiency against PD. Therefore, it suggests more verifications to draw conclusive results concerning PD treatment (226).
Strenuous limitations of PD drugs have awakened the need for target‐specific therapeutics with minimal side effects. The epigenetic marks discussed in this review article are proposed as potential candidates for designing future strategies for early PD diagnosis and therapeutics. These epigenetic regulators act as a double‐edged sword, displaying neurotoxic and neuroprotective characteristics, as shown in Figure 4. Several neurotoxic epigenetic regulators (discussed above) have confirmed their specific role in mitochondrial dysfunction in PD development and are hypothesized as future diagnostic markers for early PD diagnosis. For example, upregulated miR‐376a targeting the PGC‐1α gene in peripheral blood samples of PD patients can serve as PD diagnostic marker. Similarly, the panel of neurotoxic non‐coding RNAs or/with a combination of other neurotoxic epigenetic marks may also aid clinicians in identifying differential PD stages. Additionally, various neuroprotective epigenetic modifications in PD may actualize personalized medicines. Especially RNA‐based therapeutics has revolutionized the medical industry due to low cost, simple manufacturing, and targeting undruggable pathways. Several neuroprotective epigenetic regulators in the PD model have shown a defensive role against PD progression. For instance, upregulated miR‐7 has reduced α‐synuclein in DA neurons and protected against oxidative stress and neuronal injury (121). Furthermore, various modifications at DNA methylation levels, such as hypermethylated SNCA intron 1 (175), may also serve as a neuroprotective agent in devising future PD therapeutics. Altogether, it is hypothesized that these neuroprotective epigenetic marks can be utilized to make epidrugs that explicitly target specific sites without posing side effects. Though, for clinical implication, these epigenetic regulators require further clinical trials to determine their efficacy.
FIGURE 4.
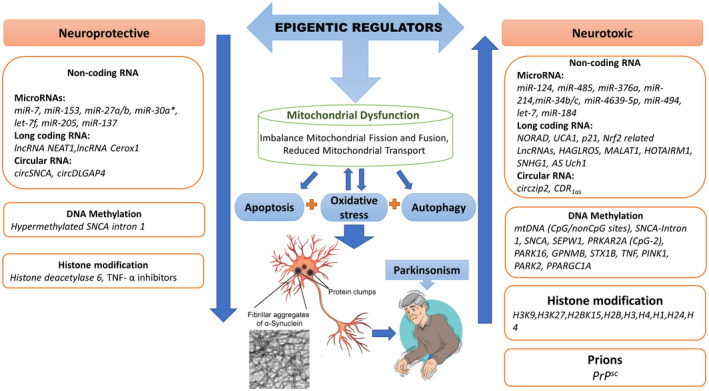
Impact of the epigenetic regulator on mitochondrial dysfunction in Parkinson's disease. Epigenetic regulators are categorized as neuroprotective and neurotoxic based on their reported characteristics. Epigenetic regulators with neurotoxic characteristics upregulate mitochondrial dysfunction (imbalance mitochondrial fission and fusion reaction and reduced mitochondrial function) and oxidative stress to trigger neuronal injury, apoptosis, and disrupt autophagy due to α‐synuclein protein inclusions, the hallmark of Parkinsonism. Whereas, epigenetic regulators owing to their neuroprotective characteristics downregulate mitochondrial dysfunction to alleviate Parkinsonism
Practical clinical implementation of these epigenetic agents encounters various limitations: (i) determination of key elements in PD pathological pathways that could serve as a target, (ii) designing a stable delivery vehicle that could cross the BBB and target specific site in neuronal cells, (iii) identification of precise targets, as these epigenetic regulators, such as miRNAs, may target more than one gene and alter various signaling pathways that can switch cellular physiology from cell death to cell survival state and vice versa. Thus, have clear chances to target unspecific sites that could be disastrous. Therefore, it dramatically emphasizes the on‐target specificity and efficient delivery systems. Another major obstacle is that most researchers rely on animal or cell line models, which has become a great challenge due to the scarcity of the human model to draw consistent results. Therefore, these inconsistencies make neurological studies a little dubious and need verifications by successive trials. Consequently, it is suggested to design more specific and low‐cost cell‐base chips for target‐based PD therapies to identify specific roles of vital elements in PD prognosis. Moreover, the reliance on brain samples in PD research has narrow down this field. Therefore, it is suggested to identify more peripheral blood‐based epigenetic marks that could serve as better diagnostic markers and help in future therapies. Additionally, in view of the above study, it is suggested that a combination of epigenetic regulators with other symptomatic treatments may provide an efficient way for PD treatment. Though further study is needed to verify the efficiency and accuracy of these feedback mechanisms associated with epigenetic regulators, it still promises a new hope for PD as a curable disorder.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
ACKNOWLEDGMENTS
The authors thank the management of the Beijing Institute of Technology and Professor Hong Ma for her assistance during manuscript preparation. This work was supported by the National Natural Science Foundation of China [Grant No. 81601114] and the Excellent Young Scholars Research Fund of Beijing Institute of Technology.
Chen Z, Rasheed M, Deng Y. The epigenetic mechanisms involved in mitochondrial dysfunction: Implication for Parkinson’s disease. Brain Pathology. 2022;32:e13012. 10.1111/bpa.13012
Zixuan Chen and Madiha Rasheed are the first co‐authors of this manuscript and contributed equally.
Funding information
This work was supported by the National Natural Science Foundation of China [Grant No. 81601114] and the Excellent Young Scholars Research Fund of Beijing Institute of Technology
Contributor Information
Zixuan Chen, Email: zx-chen@bit.edu.cn.
Madiha Rasheed, Email: madiharasheed@bit.edu.cn.
Yulin Deng, Email: deng@bit.edu.cn.
DATA AVAILABILITY STATEMENT
Not applicable.
REFERENCES
- 1. Han S, Kim S, Kim H, Shin HW, Na KS, Suh HS. Prevalence and incidence of Parkinson’s disease and drug‐induced Parkinsonism in Korea. BMC Public Health. 2019;22(1):19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tysnes OB, Storstein A. Epidemiology of Parkinson’s disease. J Neural Transm. 2017;124(8):901–5. [DOI] [PubMed] [Google Scholar]
- 3. Li G, Ma J, Cui S, He Y, Xiao Q, Liu J, et al. Parkinson’s disease in China: a forty‐year growing track of bedside work. Transl Neurodegener. 2019;8(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dorsey ER, Constantinescu R, Thompson JP, Biglan KM, Holloway RG, Kieburtz K, et al. Projected number of people with Parkinson disease in the most populous nations, 2005 through 2030. Neurology. 2007;68(5):384–6. [DOI] [PubMed] [Google Scholar]
- 5. Wirdefeldt K, Adami HO, Cole P, Trichopoulos D, Mandel J. Epidemiology and etiology of Parkinson’s disease: a review of the evidence. Eur J Epidemiol. 2011;26(S1):1–58. [DOI] [PubMed] [Google Scholar]
- 6. Renani PG, Taheri F, Rostami D, Farahani N, Abdolkarimi H, Abdollahi E, et al. Involvement of aberrant regulation of epigenetic mechanisms in the pathogenesis of Parkinson’s disease and epigenetic‐based therapies. J Cell Physiol. 2019;234(11):19307–19. [DOI] [PubMed] [Google Scholar]
- 7. Mantri S, Fullard ME, Duda JE, Morley JF. Physical activity in early Parkinson disease. J Parkinsons Dis. 2018;8(1):107–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jamebozorgi K, Taghizadeh E, Rostami D, Pormasoumi H, Barreto GE, Hayat SMG, et al. Cellular and molecular aspects of Parkinson treatment: future therapeutic perspectives. Mol Neurobiol. 2019;56:4799–811. [DOI] [PubMed] [Google Scholar]
- 9. Toulorge D, Schapira AHV, Hajj R. Molecular changes in the postmortem parkinsonian brain. J Neurochem. 2016;139:27–58. [DOI] [PubMed] [Google Scholar]
- 10. Koros C, Simitsi A, Stefanis L. Genetics of Parkinson’s disease: genotype‐phenotype correlations. Int Rev Neurobiol. 2017;132:197–231. [DOI] [PubMed] [Google Scholar]
- 11. Davie CA. A review of Parkinson’s disease. Br Med Bull. 2008;86(1):109–27. [DOI] [PubMed] [Google Scholar]
- 12. Li Z, Chalazonitis A, Huang YY, Mann JJ, Margolis KG, Yang QM, et al. Essential roles of enteric neuronal serotonin in gastrointestinal motility and the development/survival of enteric dopaminergic neurons. J Neurosci. 2011;31(24):8998–9009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sun F, Deng Y, Han X, Liu Q, Zhang P, Manzoor R, et al. A secret that underlies Parkinson’s disease: the damaging cycle. Neurochem Int. 2019;129:104484. [DOI] [PubMed] [Google Scholar]
- 14. Billingsley KJ, Barbosa IA, Bandrés‐Ciga S, Quinn JP, Bubb VJ, Deshpande C, et al. Mitochondria function associated genes contribute to Parkinson’s disease risk and later age at onset. NPJ Parkinsons Dis. 2019;5(1):8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Georgiou A, Demetriou CA, Heraclides A, Christou YP, Leonidou E, Loukaides P, et al. Mitochondrial superclusters influence age of onset of Parkinson’s disease in a gender specific manner in the Cypriot population: a case‐control study. PLoS One. 2017;12(9):e0183444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Walton‐Hadlock JL. Levodopa and the progression of Parkinson’s disease. N Engl J Med. 2005;352(13):1386. [DOI] [PubMed] [Google Scholar]
- 17. Langston JW, Irwin I, Langston EB, Forno LS. 1‐Methyl‐4‐phenylpyridinium ion (MPP+): identification of a metabolite of MPTP, a toxin selective to the substantia nigra. Neurosci Lett. 1984;48(1):87–92. [DOI] [PubMed] [Google Scholar]
- 18. Ramsay RR, Salach JI, Singer TP. Uptake of the neurotoxin 1‐methyl‐4‐phenylpyridine (MPP+) by mitochondria and its relation to the inhibition of the mitochondrial oxidation of NAD+‐linked substrates by MPP+. Biochem Biophys Res Commun. 1986;134(2):743–8. [DOI] [PubMed] [Google Scholar]
- 19. Langston JW. The MPTP story. J Parkinsons Dis. 2017;7:S11–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lill CM. Genetics of Parkinson's disease. Mol Cell Probes. 2016;30(6):386–96. [DOI] [PubMed] [Google Scholar]
- 21. Scott L, Dawson VL, Dawson TM. Trumping neurodegeneration: targeting common pathways regulated by autosomal recessive Parkinson's disease genes. Exp Neurol. 2017;298:191–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, Dutra A, et al. Mutation in the α‐synuclein gene identified in families with Parkinson's disease. Science. 1997;276(5321):2045–7. [DOI] [PubMed] [Google Scholar]
- 23. Pozo Devoto VM, Falzone TL. Mitochondrial dynamics in Parkinson's disease: a role for α‐synuclein? Dis Model Mech. 2017;10(9):1075–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Guardia‐Laguarta C, Area‐Gomez E, Schon EA, Przedborski S. A new role for α‐synuclein in Parkinson’s disease: alteration of ER–mitochondrial communication. Mov Disord. 2015;30(8):1026–33. [DOI] [PubMed] [Google Scholar]
- 25. Chinta SJ, Mallajosyula JK, Rane A, Andersen JK. Mitochondrial alpha‐synuclein accumulation impairs complex I function in dopaminergic neurons and results in increased mitophagy in vivo. Neurosci Lett. 2010;486(3):235–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Paillusson S, Gomez‐Suaga P, Stoica R, Little D, Gissen P, Devine MJ, et al. α‐Synuclein binds to the ER–mitochondria tethering protein VAPB to disrupt Ca2+ homeostasis and mitochondrial ATP production. Acta Neuropathol. 2017;134(1):129–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Parihar MS, Parihar A, Fujita M, Hashimoto M, Ghafourifar P. Mitochondrial association of alpha‐synuclein causes oxidative stress. Cell Mol Life Sci. 2008;65(7–8):1272–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kumar A, Ganini D, Mason RP. Role of cytochrome c in α‐synuclein radical formation: implications of α‐synuclein in neuronal death in Maneb‐ and paraquat‐induced model of Parkinson’s disease. Mol Neurodegener. 2016;11(1):70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang X, Yan MH, Fujioka H, Liu J, Wilson‐delfosse A, Chen SG, et al. LRRK2 regulates mitochondrial dynamics and function through direct interaction with DLP1. Hum Mol Genet. 2012;21(9):1931–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stafa K, Tsika E, Moser R, Musso A, Glauser L, Jones A, et al. Functional interaction of Parkinson’s disease‐associated LRRK2 with members of the dynamin GTPase superfamily. Hum Mol Genet. 2014;23(8):2055–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu HF, Ho PWL, Leung GCT, Lam CSC, Pang SYY, Li L, et al. Combined LRRK2 mutation, aging and chronic low dose oral rotenone as a model of Parkinson’s disease. Sci Rep. 2017;7:40887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mortiboys H, Furmston R, Bronstad G, Aasly J, Elliott C, Bandmann O. UDCA exerts beneficial effect on mitochondrial dysfunction in LRRK2 G2019S carriers and in vivo. Neurology. 2015;85(10):846–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Esteves AR, Cardoso SM. LRRK2 at the crossroad between autophagy and microtubule trafficking: insights into Parkinsons disease. Neuroscientist. 2017;23:16–26. [DOI] [PubMed] [Google Scholar]
- 34. Esteves AR, Swerdlow RH, Cardoso SM. LRRK2, a puzzling protein: insights into Parkinson's disease pathogenesis. Exp Neurol. 2014;261:206–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cherra SJ, Steer E, Gusdon AM, Kiselyov K, Chu CT. Mutant LRRK2 elicits calcium imbalance and depletion of dendritic mitochondria in neurons. Am J Pathol. 2013;182(2):474–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Martín‐Jiménez R, Lurette O, Hebert‐Chatelain E. Damage in mitochondrial DNA associated with Parkinson’s disease. DNA Cell Biol. 2020;39(8):1421–30. [DOI] [PubMed] [Google Scholar]
- 37. Luoma PT, Eerola J, Ahola S, Hakonen AH, Hellström O, Kivistö KT, et al. Mitochondrial DNA polymerase gamma variants in idiopathic sporadic Parkinson disease. Neurology. 2007;69(11):1152–9. [DOI] [PubMed] [Google Scholar]
- 38. Gaweda‐Walerych K, Safranow K, Maruszak A, Bialecka M, Klodowska‐Duda G, Czyzewski K, et al. Mitochondrial transcription factor A variants and the risk of Parkinson’s disease. Neurosci Lett. 2010;469(1):24–9. [DOI] [PubMed] [Google Scholar]
- 39. Gatt AP, Jones EL, Francis PT, Ballard C, Bateman JM. Association of a polymorphism in mitochondrial transcription factor A (TFAM) with Parkinson’s disease dementia but not dementia with Lewy bodies. Neurosci Lett. 2013;557:177–80. [DOI] [PubMed] [Google Scholar]
- 40. Ekstrand MI, Terzioglu M, Galter D, Zhu S, Hofstetter C, Lindqvist E, et al. Progressive Parkinsonism in mice with respiratory‐chain‐deficient dopamine neurons. Proc Natl Acad Sci U S A. 2007;104(4):1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kiferle L, Orsucci D, Mancuso M, Lo Gerfo A, Petrozzi L, Siciliano G, et al. Twinkle mutation in an Italian family with external progressive ophthalmoplegia and Parkinsonism: a case report and an update on the state of art. Neurosci Lett. 2013;556:1–4. [DOI] [PubMed] [Google Scholar]
- 42. Baloh RH, Salavaggione E, Milbrandt J, Pestronk A. Familial Parkinsonism and ophthalmoplegia from a mutation in the mitochondrial DNA helicase twinkle. Arch Neurol. 2007;64(7):998–1000. [DOI] [PubMed] [Google Scholar]
- 43. Song L, Shan Y, Kent Lloyd KC, Cortopassi GA. Mutant twinkle increases dopaminergic neurodegeneration, mtDNA deletions and modulates Parkin expression. Hum Mol Genet. 2012;21(23):5147–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pickrell AM, Youle RJ. The roles of PINK1, Parkin, and mitochondrial fidelity in Parkinson’s disease. Neuron. 2015;85(2):257–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Trancikova A, Tsika E, Moore DJ. Mitochondrial dysfunction in genetic animal models of Parkinson’s disease. Antioxid Redox Signal. 2012;16:896–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Williams ET, Chen X, Moore DJ. VPS35, the retromer complex and Parkinson’s disease. J Parkinsons Dis. 2017;7:219–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ruan Y, Tecott L, Jiang MM, Jan LY, Jan YN. Ethanol hypersensitivity and olfactory discrimination defect in mice lacking a homolog of Drosophila neuralized. Proc Natl Acad Sci U S A. 2001;98(17):9907–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bender A, Krishnan KJ, Morris CM, Taylor GA, Reeve AK, Perry RH, et al. High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nat Genet. 2006;38(5):515–7. [DOI] [PubMed] [Google Scholar]
- 49. Kraytsberg Y, Kudryavtseva E, McKee AC, Geula C, Kowall NW, Khrapko K. Mitochondrial DNA deletions are abundant and cause functional impairment in aged human substantia nigra neurons. Nat Genet. 2006;38(5):518–20. [DOI] [PubMed] [Google Scholar]
- 50. Nakabeppu Y, Tsuchimoto D, Yamaguchi H, Sakumi K. Oxidative damage in nucleic acids and Parkinson’s disease. J Neurosci Res. 2007;85(5):919–34. [DOI] [PubMed] [Google Scholar]
- 51. Simon DK, Pulst SM, Sutton JP, Browne SE, Beal MF, Johns DR. Familial multisystem degeneration with Parkinsonism associated with the 11778 mitochondrial DNA mutation. Neurology. 1999;53(8):1787–93. [DOI] [PubMed] [Google Scholar]
- 52. Parker WD, Parks JK. Mitochondrial ND5 mutations in idiopathic Parkinson’s disease. Biochem Biophys Res Commun. 2005;326(3):667–9. [DOI] [PubMed] [Google Scholar]
- 53. Podlesniy P, Puigròs M, Serra N, Fernández‐Santiago R, Ezquerra M, Tolosa E, et al. Accumulation of mitochondrial 7S DNA in idiopathic and LRRK2 associated Parkinson’s disease. EBioMedicine. 2019;48:554–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chin‐Chan M, Navarro‐Yepes J. Environmental pollutants as risk factors for neurodegenerative disorders: Alzheimer and Parkinson diseases. Front Cell Neurosci. 2015;9:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Deng Y, Zhang Y, Duan J, Xiong Y, Qing H. An overview of endogenous catechol‐isoquinolines and their related enzymes: possible biomarkers for Parkinson’s disease. Curr Transl Geriatr Exp Gerontol Rep. 2012;1(2):59–67. [Google Scholar]
- 56. Rappold PM, Cui M, Grima JC, Fan RZ, De Mesy‐Bentley KL, Chen L, et al. Drp1 inhibition attenuates neurotoxicity and dopamine release deficits in vivo. Nat Commun. 2014;5:5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Dukes AA, Bai Q, Van Laar VS, Zhou Y, Ilin V, David CN, et al. Live imaging of mitochondrial dynamics in CNS dopaminergic neurons in vivo demonstrates early reversal of mitochondrial transport following MPP+ exposure. Neurobiol Dis. 2016;95:238–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gatt AP, Duncan OF, Attems J, Francis PT, Ballard CG, Bateman JM. Dementia in Parkinson’s disease is associated with enhanced mitochondrial complex I deficiency. Mov Disord. 2016;31(3):352–9. [DOI] [PubMed] [Google Scholar]
- 59. Schapira AHV, Cooper JM, Dexter D, Clark JB, Jenner P, Marsden CD. Mitochondrial complex I deficiency in Parkinson’s disease. J Neurochem. 1990;54(3):823–7. [DOI] [PubMed] [Google Scholar]
- 60. Sherer TB, Betarbet R, Greenamyre JT. Environment, mitochondria, and Parkinson’s disease. Neuroscientist. 2002;8:192–7. [DOI] [PubMed] [Google Scholar]
- 61. Choi WS, Kruse SE, Palmiter RD, Xia Z. Mitochondrial complex I inhibition is not required for dopaminergic neuron death induced by rotenone, MPP+, or paraquat. Proc Natl Acad Sci U S A. 2008;105(39):15136–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bulteau AL, Mena NP, Auchère F, Lee I, Prigent A, Lobsiger CS, et al. Dysfunction of mitochondrial Lon protease and identification of oxidized protein in mouse brain following exposure to MPTP: implications for Parkinson disease. Free Radic Biol Med. 2017;108:236–46. [DOI] [PubMed] [Google Scholar]
- 63. Jiang H, Wang J, Rogers J, Xie J. Brain iron metabolism dysfunction in Parkinson’s disease. Mol Neurobiol. 2017;54:3078–101. [DOI] [PubMed] [Google Scholar]
- 64. Smith MR, Fernandes J, Go YM, Jones DP. Redox dynamics of manganese as a mitochondrial life‐death switch. Biochem Biophys Res Commun. 2017;482:388–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Alaimo A, Gorojod RM, Beauquis J, Muñoz MJ, Saravia F, Kotler ML. Deregulation of mitochondria‐shaping proteins Opa‐1 and Drp‐1 in manganese‐induced apoptosis. PLoS One. 2014;9(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhang S, Zhou Z, Fu J. Effect of manganese chloride exposure on liver and brain mitochondria function in rats. Environ Res. 2003;93(2):149–57. [DOI] [PubMed] [Google Scholar]
- 67. Dabrowska A, Venero JL, Iwasawa R, Hankir MK, Rahman S, Boobis A, et al. PGC‐1a controls mitochondrial biogenesis and dynamics in leadinduced neurotoxicity. Aging. 2015;7(9):629–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lock EA, Zhang J, Checkoway H. Solvents and Parkinson disease: a systematic review of toxicological and epidemiological evidence. Toxicol Appl Pharmacol. 2013;266:345–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Gash DM, Rutland K, Hudson NL, Sullivan PG, Bing G, Cass WA, et al. Trichloroethylene: Parkinsonism and complex 1 mitochondrial neurotoxicity. Ann Neurol. 2008;63(2):184–92. [DOI] [PubMed] [Google Scholar]
- 70. Zaheer F, Slevin JT. Trichloroethylene and Parkinson disease. Neurol Clin. 2011;29:657–65. [DOI] [PubMed] [Google Scholar]
- 71. DeCuypere M, Lu Y, Miller DD, LeDoux MS. Regional distribution of tetrahydroisoquinoline derivatives in rodent, human, and parkinson’s disease brain. J Neurochem. 2008;107(5):1398–413. [DOI] [PubMed] [Google Scholar]
- 72. Herraiz T. N‐methyltetrahydropyridines and pyridinium cations as toxins and comparison with naturally‐occurring alkaloids. Food Chem Toxicol. 2016;97:23–39. [DOI] [PubMed] [Google Scholar]
- 73. Kotake Y, Sekiya Y, Okuda K, Ohta S. Cytotoxicity of 17 tetrahydroisoquinoline derivatives in SH‐SY5Y human neuroblastoma cells is related to mitochondrial NADH‐ubiquinone oxidoreductase inhibition. Neurotoxicology. 2007;28(1):27–32. [DOI] [PubMed] [Google Scholar]
- 74. Suzuki K, Mizuno Y, Yoshida M. Inhibition of mitochondrial NADH‐ubiquinone oxidoreductase activity and ATP synthesis by tetrahydroisoquinoline. Neurosci Lett. 1988;86(1):105–8. [DOI] [PubMed] [Google Scholar]
- 75. Wąsik A, Romańska I, Michaluk J, Zelek‐Molik A, Nalepa I, Antkiewicz‐Michaluk L. Neuroprotective effect of the endogenous amine 1MeTIQ in an animal model of Parkinson’s disease. Neurotox Res. 2016;29(3):351–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wąsik A, Kajta M, Lenda T, Antkiewicz‐Michaluk L. Concentration‐dependent opposite effects of 1‐Benzyl‐1,2,3,4‐ tetrahydroisoquinoline on markers of apoptosis: in vitro and ex vivo studies. Neurotox Res. 2014;25(1):90–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kotake Y, Sekiya Y, Okuda K, Ohta S. Detection of a novel neurotoxic metabolite of parkinson’s disease‐related neurotoxin, 1‐benzyl‐1,2,3,4‐tetrahydroisoquinoline. J Toxicol Sci. 2014;38:749–54. [DOI] [PubMed] [Google Scholar]
- 78. Antkiewicz‐Michaluk L, Wąsik A, Michaluk J. 1‐Methyl‐1,2,3,4‐tetrahydroisoquinoline, an endogenous amine with unexpected mechanism of action: new vistas of therapeutic application. Neurotox Res. 2014;25:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wąsik A, Antkiewicz‐Michaluk L. The mechanism of neuroprotective action of natural compounds. Pharmacol Rep. 2017;69:851–60. [DOI] [PubMed] [Google Scholar]
- 80. Wąsik A, Romańska I, Zelek‐Molik A, Antkiewicz‐Michaluk L. Multiple administration of endogenous amines TIQ and 1MeTIQ protects against a 6‐OHDA‐induced essential fall of dopamine release in the rat striatum. In vivo microdialysis study. Neurotox Res. 2018;33(3):523–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Peana AT, Bassareo V, Acquas E. Not just from ethanol. tetrahydroisoquinolinic (TIQ) derivatives: from neurotoxicity to neuroprotection. Neurotox Res. 2019;36:653–68. [DOI] [PubMed] [Google Scholar]
- 82. Kurnik‐Łucka M, Panula P, Bugajski A, Gil K. Salsolinol: an unintelligible and double‐faced molecule—lessons learned from in vivo and in vitro experiments. Neurotox Res. 2018;33:485–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Chen X, Zheng X, Ali S, Guo M, Zhong R, Chen Z, et al. Isolation and sequencing of salsolinol synthase, an enzyme catalyzing salsolinol biosynthesis. ACS Chem Neurosci. 2018;9(6):1388–98. [DOI] [PubMed] [Google Scholar]
- 84. Sandler M, Carter SB, Hunter KR, Stern GM. Tetrahydroisoquinoline alkaloids: in vivo metabolites of L‐dopa in man. Nature. 1973;241(5390):439–43. [DOI] [PubMed] [Google Scholar]
- 85. Kobayashi H, Fukuhara K, Tada‐Oikawa S, Yada Y, Hiraku Y, Murata M, et al. The mechanisms of oxidative DNA damage and apoptosis induced by norsalsolinol, an endogenous tetrahydroisoquinoline derivative associated with Parkinson’s disease. J Neurochem. 2009;108(2):397–407. [DOI] [PubMed] [Google Scholar]
- 86. Możdżeń E, Kajta M, Wąsik A, Lenda T, Antkiewicz‐Michaluk L. Salsolinol, an endogenous compound triggers a two‐phase opposing action in the central nervous system. Neurotox Res. 2015;27(3):300–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Maruyama W, Naoi M, Kasamatsu T, Hashizume Y, Takahashi T, Kohda K, et al. An endogenous dopaminergic neurotoxin, N‐methyl‐(R)‐salsolinol, induces DNA damage in human dopaminergic neuroblastoma SH‐SY5Y cells. J Neurochem. 1997;69(1):322–9. [DOI] [PubMed] [Google Scholar]
- 88. Su Y, Duan J, Ying Z, Hou Y, Zhang Y, Wang R, et al. Increased vulnerability of parkin knock down PC12 cells to hydrogen peroxide toxicity: the role of salsolinol and NM‐salsolinol. Neuroscience. 2013;233:72–85. [DOI] [PubMed] [Google Scholar]
- 89. Mao J, Ma H, Xu Y, Su Y, Zhu H, Wang R, et al. Increased levels of monoamine‐derived potential neurotoxins in fetal rat brain exposed to ethanol. Neurochem Res. 2013;38(2):356–63. [DOI] [PubMed] [Google Scholar]
- 90. Wang R, Qing H, Liu XQ, Zheng XL, Deng YL. Iron contributes to the formation of catechol isoquinolines and oxidative toxicity induced by overdose dopamine in dopaminergic SH‐SY5Y cells. Neurosci Bull. 2008;24(3):125–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Zhang Y, Ma H, Xie B, Han C, Wang C, Qing H, et al. Alpha‐synuclein overexpression induced mitochondrial damage by the generation of endogenous neurotoxins in PC12 cells. Neurosci Lett. 2013;547:65–9. [DOI] [PubMed] [Google Scholar]
- 92. Arshad A, Chen X, Cong Z, Qing H, Deng Y. TRPC1 protects dopaminergic SH‐SY5Y cells from MPP+, salsolinol, and N‐methyl‐(R)‐salsolinol‐induced cytotoxicity. Acta Biochim Biophys Sin. 2014;46(1):22–30. [DOI] [PubMed] [Google Scholar]
- 93. Zhang Y, Wang L, Mu X, Duan J, Zhu Y, Qing H, et al. Assessment of salsolinol N‐methyltransferase activity in rat peripheral lymphocytes by liquid chromatography‐electrospray time‐of‐flight mass spectrometry. Anal Bioanal Chem. 2011;399(10):3541–5. [DOI] [PubMed] [Google Scholar]
- 94. Deza‐Ponzio R, Herrera ML, Bellini MJ, Virgolini MB, Hereñú CB. Aldehyde dehydrogenase 2 in the spotlight: the link between mitochondria and neurodegeneration. Neurotoxicology. 2018;68:19–24. [DOI] [PubMed] [Google Scholar]
- 95. Chen CH, Ferreira JCB, Gross ER, Mochly‐Rosen D. Targeting aldehyde dehydrogenase 2: new therapeutic opportunities. Physiol Rev. 2014;94(1):1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Ito A, Jamal M, Ameno K, Tanaka N, Takakura A, Kawamoto T, et al. Acetaldehyde administrationinduces salsolinol formation in vivo in the dorsal striatum of Aldh2‐knockout and C57BL/6N mice. Neurosci Lett. 2018;685:50–4. [DOI] [PubMed] [Google Scholar]
- 97. Emamzadeh FN, Surguchov A. Parkinson’s disease: biomarkers, treatment, and risk factors. Front Neurosci. 2018;12:612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Fitzmaurice AG, Rhodes SL, Lulla A, Murphy NP, Lam HA, O’Donnell KC, et al. Aldehyde dehydrogenase inhibition as a pathogenic mechanism in Parkinson disease. Proc Natl Acad Sci U S A. 2013;110(2):636–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Chiu CC, Yeh TH, Lai SC, Wu‐Chou YH, Chen CH, Mochly‐Rosen D, et al. Neuroprotective effects of aldehyde dehydrogenase 2 activation in rotenone‐induced cellular and animal models of Parkinsonism. Exp Neurol. 2015;263:244–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Deng Y, Zhang Y, Li Y, Xiao S, Song D, Qing H, et al. Occurrence and distribution of salsolinol‐like compound, 1‐acetyl‐6,7‐dihydroxy‐1,2,3,4‐tetrahydroisoquinoline (ADTIQ) in Parkinsonian brains. J Neural Transm. 2012;119(4):435–41. [DOI] [PubMed] [Google Scholar]
- 101. Xie B, Lin F, Ullah K, Peng L, Ding W, Dai R, et al. A newly discovered neurotoxin ADTIQ associated with hyperglycemia and Parkinson’s disease. Biochem Biophys Res Commun. 2015;459(3):361–6. [DOI] [PubMed] [Google Scholar]
- 102. Xie B, Lin F, Peng L, Ullah K, Wu H, Qing H, et al. Methylglyoxal increases dopamine level and leads to oxidative stress in SH‐SY5Y cells. Acta Biochim Biophys Sin. 2014;46(11):950–6. [DOI] [PubMed] [Google Scholar]
- 103. Biosa A, Outeiro TF, Bubacco L, Bisaglia M. Diabetes mellitus as a risk factor for parkinson’s disease: a molecular point of view. Mol Neurobiol. 2018;55:8754–63. [DOI] [PubMed] [Google Scholar]
- 104. Esteller M. Non‐coding RNAs in human disease. Nature Reviews Genetics. Nat Rev Genet. 2011;12:861–74. [DOI] [PubMed] [Google Scholar]
- 105. Labbé C, Lorenzo‐Betancor O, Ross OA. Epigenetic regulation in Parkinson’s disease. Acta Neuropath. 2016;132:515–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Leggio L, Vivarelli S, L’Episcopo F, Tirolo C, Caniglia S, Testa N, et al. MicroRNAs in parkinson’s disease: from pathogenesis to novel diagnostic and therapeutic approaches. Int J Mol Sci. 2017;18(12):2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Li M, Marin‐Muller C, Bharadwaj U, Chow K‐H, Yao Q, Chen C. MicroRNAs: control and loss of control in human physiology and disease. Word J Surg. 2009;33(4):667–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Martinez B, Peplow PV. MicroRNAs in Parkinson’s disease and emerging therapeutic targets. Neural Regen Res. 2017;12:1945–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Wang ZH, Zhang JL, Duan YL, Zhang QS, Li GF, Zheng DL. MicroRNA‐214 participates in the neuroprotective effect of Resveratrol via inhibiting α‐synuclein expression in MPTP‐induced Parkinson’s disease mouse. Biomed Pharmacother. 2015;74:252–6. [DOI] [PubMed] [Google Scholar]
- 110. John A, Kubosumi A, Reddy PH. Mitochondrial microRNAs in aging and neurodegenerative diseases. Cells. 2020;9(6):1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Miñones‐Moyano E, Porta S, Escaramís G, Rabionet R, Iraola S, Kagerbauer B, et al. MicroRNA profiling of Parkinson’s disease brains identifies early downregulation of miR‐34b/c which modulate mitochondrial function. Hum Mol Genet. 2011;20(15):3067–78. [DOI] [PubMed] [Google Scholar]
- 112. Villar‐Menéndez I, Porta S, Buira SP, Pereira‐Veiga T, Díaz‐Sánchez S, Albasanz JL, et al. Increased striatal adenosine A2A receptor levels is an early event in Parkinson’s disease‐related pathology and it is potentially regulated by miR‐34b. Neurobiol Dis. 2014;69:206–14. [DOI] [PubMed] [Google Scholar]
- 113. Chen Y, Gao C, Sun Q, Pan H, Huang P, Ding J, et al. MicroRNA‐4639 is a regulator of DJ‐1 expression and a potential early diagnostic marker for Parkinson’s disease. Front Aging Neurosci. 2017;21:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Xiong R, Wang Z, Zhao Z, Li H, Chen W, Zhang B, et al. MicroRNA‐494 reduces DJ‐1 expression and exacerbates neurodegeneration. Neurobiol Aging. 2014;35(3):705–14. [DOI] [PubMed] [Google Scholar]
- 115. Gehrke S, Imai Y, Sokol N, Lu B. Pathogenic LRRK2 negatively regulates microRNA‐mediated translational repression. Nature. 2010;466(7306):637–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Wang H, Ye Y, Zhu Z, Mo L, Lin C, Wang Q, et al. MiR‐124 regulates apoptosis and autophagy process in MPTP model of Parkinson’s disease by targeting to bim. Brain Pathol. 2016;26(2):167–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Kanagaraj N, Beiping H, Dheen ST, Tay SSW. Downregulation of miR‐124 in MPTP‐treated mouse model of Parkinson’s disease and MPP iodide‐treated MN9D cells modulates the expression of the calpain/cdk5 pathway proteins. Neuroscience. 2014;272:167–79. [DOI] [PubMed] [Google Scholar]
- 118. St‐Pierre J, Drori S, Uldry M, Silvaggi JM, Rhee J, Jäger S, et al. Suppression of reactive oxygen species and neurodegeneration by the PGC‐1 transcriptional coactivators. Cell. 2006;127(2):397–408. [DOI] [PubMed] [Google Scholar]
- 119. Lou C, Xiao M, Cheng S, Lu X, Jia S, Ren Y, et al. MiR‐485‐3p and mir‐485‐5p suppress breast cancer cell metastasis by inhibiting PGC‐1α expression. Cell Death Dis. 2016;7(3):e2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Baghi M, Rostamian Delavar M, Yadegari E, Peymani M, Pozo D, Hossein Nasr‐Esfahani M, et al. Modified level of miR‐376a is associated with Parkinson’s disease. J Cell Mol Med. 2020;24(4):2622–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Junn E, Lee KW, Byeong SJ, Chan TW, Im JY, Mouradian MM. Repression of α‐synuclein expression and toxicity by microRNA‐7. Proc Natl Acad Sci U S A. 2009;106(31):13052–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Fragkouli A, Doxakis E. miR‐7 and miR‐153 protect neurons against MPP+‐induced cell death via upregulation of mTOR pathway. Front Cell Neurosci. 2014;8:132. https://www.frontiersin.org/articles/10.3389/fncel.2014.00182/full. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Chaudhuri AD, Choi DC, Kabaria S, Tran A, Junn XE. MicroRNA‐7 regulates the function of mitochondrial permeability transition pore by targeting vdac1 expression. J Biol Chem. 2016;291(12):6483–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Kim J, Fiesel FC, Belmonte KC, Hudec R, Wang WX, Kim C, et al. MiR‐27a and miR‐27b regulate autophagic clearance of damaged mitochondria by targeting PTEN‐induced putative kinase 1 (PINK1). Mol Neurodegener. 2016;11(1):55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Dorval V, Mandemakers W, Jolivette F, Coudert L, Mazroui R, De Strooper B, et al. Gene and microRNA transcriptome analysis of Parkinson’s related LRRK2 mouse models. PLoS One. 2014;9(1):e85510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Cho HJ, Liu G, Jin SM, Parisiadou L, Xie C, Yu J, et al. Microrna‐205 regulates the expression of Parkinson’s disease‐related leucine‐rich repeat kinase 2 protein. Hum Mol Genet. 2013;22(3):608–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Jiang Y, Liu J, Chen L, Jin Y, Zhang G, Lin Z, et al. Serum secreted miR‐137‐containing exosomes affects oxidative stress of neurons by regulating OXR1 in Parkinson’s disease. Brain Res. 2019;1722:146331. [DOI] [PubMed] [Google Scholar]
- 128. Ma L, Bajic VB, Zhang Z. On the classification of long non‐coding RNAs. RNA Biol. 2013;10:924–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Qureshi IA, Mehler MF. Emerging roles of non‐coding RNAs in brain evolution, development, plasticity and disease. Nat Rev Neurosci. 2012;13:528–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Wu H, Yang L, Chen LL. The diversity of long noncoding RNAs and their generation. Trend Genet. 2017;33:540–52. [DOI] [PubMed] [Google Scholar]
- 131. Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22(9):1775–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Kopp F, Elguindy MM, Yalvac ME, Zhang H, Chen B, Gillett FA, et al. PUMILIO hyperactivity drives premature aging of norad‐deficient mice. Elife. 2019;8:e42650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Yan W, Chen ZY, Chen JQ, Chen HM. LncRNA NEAT1 promotes autophagy in MPTP‐induced Parkinson’s disease through stabilizing PINK1 protein. Biochem Biophys Res Commun. 2018;496(4):1019–24. [DOI] [PubMed] [Google Scholar]
- 134. Simchovitz A, Hanan M, Niederhoffer N, Madrer N, Yayon N, Bennett ER, et al. NEAT1 is overexpressed in Parkinson’s disease substantia nigra and confers drug‐inducible neuroprotection from oxidative stress. FASEB J. 2019;33(10):11223–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Cai L, Tu L, Li T, Yang X, Ren Y, Gu R, et al. Downregulation of lncRNA UCA1 ameliorates the damage of dopaminergic neurons, reduces oxidative stress and inflammation in Parkinson’s disease through the inhibition of the PI3K/Akt signaling pathway. Int Immunopharmacol. 2019;75:105734. [DOI] [PubMed] [Google Scholar]
- 136. Goan YG, Wu WT, Liu CI, Neoh CA, Wu YJ. Involvement of mitochondrial dysfunction, endoplasmic reticulum stress, and the PI3K/AKT/mTOR pathway in nobiletin‐induced apoptosis of human bladder cancer cells. Molecules. 2019;24(16):2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Lu M, Sun WL, Shen J, Wei M, Chen B, Qi YJ, et al. LncRNA‐UCA1 promotes PD development by upregulating SNCA. Eur Rev Med Pharmacol Sci. 2018;22(22):7908–15. [DOI] [PubMed] [Google Scholar]
- 138. Ding XM, Zhao LJ, Qiao HY, Wu SL, Wang XH. Long non‐coding RNA‐p21 regulates MPP+‐induced neuronal injury by targeting miR‐625 and derepressing TRPM2 in SH‐SY5Y cells. Chem Biol Interact. 2019;307:73–81. [DOI] [PubMed] [Google Scholar]
- 139. Sirey TM, Roberts K, Haerty W, Bedoya‐Reina O, Granados SR, Tan JY, et al. The long non‐coding RNA cerox1 is a post transcriptional regulator of mitochondrial complex I catalytic activity. Elife. 2019;8:e45051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Wang L, Yang H, Wang Q, Zhang Q, Wang Z, Zhang Q, et al. Paraquat and MPTP induce alteration in the expression profile of long noncoding RNAs in the substantia nigra of mice: role of the transcription factor Nrf2. Toxicol Lett. 2018;291:11–28. [DOI] [PubMed] [Google Scholar]
- 141. Peng T, Liu X, Wang J, Liu Y, Fu Z, Ma X, et al. Long noncoding RNA HAGLROS regulates apoptosis and autophagy in Parkinson’s disease via regulating miR‐100/ATG10 axis and PI3K/Akt/mTOR pathway activation. Artif Cells Nanomedicine Biotechnol. 2019;47(1):2764–74. [DOI] [PubMed] [Google Scholar]
- 142. Liu W, Zhang Q, Zhang J, Pan W, Zhao J, Xu Y. Long non‐coding RNA MALAT1 contributes to cell apoptosis by sponging miR‐124 in Parkinson disease. Cell Biosci. 2017;7(1):19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Wang S, Zhang X, Guo Y, Rong H, Liu T. The long noncoding RNA HOTAIR promotes Parkinson’s disease by upregulating LRRK2 expression. Oncotarget. 2017;8(15):24449–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Fan Y, Li J, Yang Q, Gong C, Gao H, Mao Z, et al. Dysregulated long non‐coding RNAs in Parkinson’s disease contribute to the apoptosis of human neuroblastoma cells. Front Neurosci. 2019;13:1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Chen Y, Lian Y, Ma Y, Wu C, Zheng Y, Xie N. LncRNA SNHG1 promotes α‐synuclein aggregation and toxicity by targeting miR‐15b‐5p to activate SIAH1 in human neuroblastoma SH‐SY5Y cells. Neurotoxicol. 2018;68:212–21. [DOI] [PubMed] [Google Scholar]
- 146. Carrieri C, Forrest ARR, Santoro C, Persichetti F, Carninci P, Zucchelli S, et al. Expression analysis of the long non‐coding RNA antisense to Uchl1 (AS Uchl1) during dopaminergic cells’ differentiation in vitro and in neurochemical models of Parkinson’s disease. Front Cell Neurosci. 2015;9:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Li TR, Jia YJ, Wang Q, Shao XQ, Lv RJ. Circular RNA: a new star in neurological diseases. Int J Neurosci. 2017;127:726–34. [DOI] [PubMed] [Google Scholar]
- 148. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–8. [DOI] [PubMed] [Google Scholar]
- 149. Rybak‐Wolf A, Stottmeister C, Glažar P, Jens M, Pino N, Giusti S, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2014;58(5):870–85. [DOI] [PubMed] [Google Scholar]
- 150. You X, Vlatkovic I, Babic A, Will T, Epstein I, Tushev G, et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci. 2015;18(4):603–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Shao Y, Chen Y. Roles of circular RNAs in neurologic disease. Front Mol Neurosci. 2016;9:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Sang Q, Liu X, Wang L, Qi L, Sun W, Wang W, et al. CircSNCA downregulation by pramipexole treatment mediates cell apoptosis and autophagy in Parkinson’s disease by targeting miR‐7. Aging. 2018;10(6):1281–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Kumar L, Shamsuzzama, Jadiya P, Haque R, Shukla S, Nazir A. Functional characterization of novel circular RNA molecule, circzip‐2 and Its synthesizing gene zip‐2 in C. elegans model of Parkinson’s disease. Mol Neurobiol. 2018;55(8):6914–26. [DOI] [PubMed] [Google Scholar]
- 154. Feng Z, Zhang L, Wang S, Hong Q. Circular RNA circDLGAP4 exerts neuroprotective effects via modulating miR‐134‐5p/CREB pathway in Parkinson’s disease. Biochem Biophys Res Commun. 2020;522(2):388–94. [DOI] [PubMed] [Google Scholar]
- 155. Mani SR, Juliano CE. Untangling the web: the diverse functions of the PIWI/piRNA pathway. Mol Reprod Dev. 2013;80(8):632–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Ozata DM, Gainetdinov I, Zoch A, O’Carroll D, Zamore PD. PIWI‐interacting RNAs: small RNAs with big functions. Nat Rev Genet. 2019;20:89–108. [DOI] [PubMed] [Google Scholar]
- 157. Wakisaka KT, Imai Y. The dawn of pirna research in various neuronal disorders ‐ PubMed. Front Biosci. 2019;24(1):1440–51. [DOI] [PubMed] [Google Scholar]
- 158. Cavalcante GC, Magalhães L, Ribeiro‐Dos‐santos Â, Vidal AF. Mitochondrial epigenetics: non‐coding RNAs as a novel layer of complexity. Int J Mol Sci. 2020;21(5):1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Schulze M, Sommer A, Plötz S, Farrell M, Winner B, Grosch J, et al. Sporadic Parkinson’s disease derived neuronal cells show disease‐specific mRNA and small RNA signatures with abundant deregulation of piRNAs. Acta Neuropathol Commun. 2018;6(1):58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Rajasethupathy P, Antonov I, Sheridan R, Frey S, Sander C, Tuschl T, et al. A role for neuronal piRNAs in the epigenetic control of memory‐related synaptic plasticity. Cell. 2012;149(3):693–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Groot GSP, Kroon AM. Mitochondrial DNA from various organisms does not contain internally methylated cytosine in ‐CCGG‐ sequences. Biochim Biophys Acta. 1979;564(2):355–7. [DOI] [PubMed] [Google Scholar]
- 162. Satoh M, Kuroiwa T. Organization of multiple nucleoids and DNA molecules in mitochondria of a human cell. Exp Cell Res. 1991;196(1):137–40. [DOI] [PubMed] [Google Scholar]
- 163. Liu B, Du Q, Chen L, Fu G, Li S, Fu L, et al. CpG methylation patterns of human mitochondrial DNA. Sci Rep. 2016;6:23421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Shock LS, Thakkar PV, Peterson EJ, Moran RG, Taylor SM. DNA methyltransferase 1, cytosine methylation, and cytosine hydroxymethylation in mammalian mitochondria. Proc Natl Acad Sci U S A. 2011;108(9):3630–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Wong M, Gertz B, Chestnut BA, Martin LJ. Mitochondrial DNMT3A and DNA methylation in skeletal muscle and CNS of transgenic mouse models of ALS. Front Cell Neurosci. 2013;7:279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Chestnut BA, Chang Q, Price A, Lesuisse C, Wong M, Martin LJ. Epigenetic regulation of motor neuron cell death through DNA methylation. J Neurosci. 2011;31(46):16619–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Bellizzi D, D'Aquila P, Scafone T, Giordano M, Riso V, Riccio A, et al. The control region of mitochondrial DNA shows an unusual CpG and non‐CpG methylation pattern. DNA Res. 2013;20(6):537–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Byun HM, Baccarelli AA. Environmental exposure and mitochondrial epigenetics: study design and analytical challenges. Hum Genet. 2014;133(3):247–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Iacobazzi V, Castegna A, Infantino V, Andria G. Mitochondrial DNA methylation as a next‐generation biomarker and diagnostic tool. Mol Genet Metab. 2013;110:25–34. [DOI] [PubMed] [Google Scholar]
- 170. Jakubowski JL, Labrie V. Epigenetic biomarkers for Parkinson’s disease: from diagnostics to therapeutics. J Parkinson Dis. 2017;7:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Coppedè F, Stoccoro A. Mitoepigenetics and neurodegenerative diseases. Front Endocrinol. 2019;10:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Desplats P, Spencer B, Coffee E, Patel P, Michael S, Patrick C, et al. α‐synuclein sequesters Dnmt1 from the nucleus: a novel mechanism for epigenetic alterations in Lewy body diseases. J Biol Chem. 2011;286(11):9031–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Matsumoto L, Takuma H, Tamaoka A, Kurisaki H, Date H, Tsuji S, et al. CpG demethylation enhances alpha‐synuclein expression and affects the pathogenesis of Parkinson’s disease. PLoS One. 2010;5(11):e15522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Jowaed A, Schmitt I, Kaut O, Wüllner U. Methylation regulates alpha‐synuclein expression and is decreased in Parkinson’s disease patients’ brains. J Neurosci. 2010;30(18):6355–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Kantor B, Tagliafierro L, Gu J, Zamora ME, Ilich E, Grenier C, et al. Downregulation of SNCA expression by targeted editing of DNA methylation: a potential strategy for precision therapy in PD. Mol Ther. 2018;26(11):2638–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Masliah E, Dumaop W, Galasko D, Desplats P. Distinctive patterns of DNA methylation associated with Parkinson disease: identification of concordant epigenetic changes in brain and peripheral blood leukocytes. Epigenetics. 2013;8(10):1030–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Pavlou MAS, Outeiro TF. Epigenetics in parkinson’s disease. Adv Exp Med Biol. 2017;978:363–90. [DOI] [PubMed] [Google Scholar]
- 178. Zinovkina LA, Zinovkin RA. DNA methylation, mitochondria, and programmed aging. Biochem. 2015;80:1571–7. [DOI] [PubMed] [Google Scholar]
- 179. Sharma A, Schaefer ST, Sae‐Lee C, Byun HM, Wüllner U. Elevated serum mitochondrial DNA in females and lack of altered platelet mitochondrial methylation in patients with Parkinson’s disease. Int J Neurosci. 2020;3:279–82. [DOI] [PubMed] [Google Scholar]
- 180. Yang X, Xu S, Qian Y, He X, Chen S, Xiao Q. Hypermethylation of the gene coding for PGC‐1α in peripheral blood leukocytes of patients with Parkinson’s disease. Front Neurosci. 2020;14:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Henderson‐Smith A, Fisch KM, Hua J, Liu G, Ricciardelli E, Jepsen K, et al. DNA methylation changes associated with Parkinson’s disease progression: outcomes from the first longitudinal genome‐wide methylation analysis in blood. Epigenetics. 2019;14(4):365–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Prajapati P, Sripada L, Singh K, Bhatelia K, Singh R, Singh R. TNF‐α regulates miRNA targeting mitochondrial complex‐I and induces cell death in dopaminergic cells. Biochim Biophys Acta‐Mol Basis Dis. 2015;1852(3):451–61. [DOI] [PubMed] [Google Scholar]
- 183. Ndayisaba A, Jellinger K, Berger T, Wenning GK. TNFα inhibitors as targets for protective therapies in MSA: a viewpoint. J Neuroinflammation. 2019;16:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. Conversion of 5‐methylcytosine to 5‐hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(5929):930–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Pastor WA, Aravind L, Rao A. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat Rev Mol Cell Biol. 2013;14:341–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Wu X, Zhang Y. TET‐mediated active DNA demethylation: mechanism, function and beyond. Nat Rev Genet. 2017;18:517–34. [DOI] [PubMed] [Google Scholar]
- 187. Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, et al. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341(6146):1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Szulwach KE, Li X, Li Y, Song CX, Wu H, Dai Q, et al. 5‐hmC‐mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat Neurosci. 2011;14(12):1607–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Liang J, Yang F, Zhao L, Bi C, Cai B. Physiological and pathological implications of 5‐hydroxymethylcytosine in diseases. Oncotarget. 2016;7:48813–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Wang T, Pan Q, Lin L, Szulwach KE, Song CX, He C, et al. Genome‐wide DNA hydroxymethylation changes are associated with neurodevelopmental genes in the developing human cerebellum. Hum Mol Genet. 2012;21(26):5500–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Wüllner U, Kaut O, deBoni L, Piston D, Schmitt I. DNA methylation in Parkinson’s disease. J Neurochem. 2016;139:108–20. [DOI] [PubMed] [Google Scholar]
- 192. Al‐Mahdawi S, Virmouni SA, Pook MA. The emerging role of 5‐hydroxymethylcytosine in neurodegenerative diseases. Front Neurosci. 2014;8:397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Miao Z, Xin N, Wei B, Hua X, Zhang G, Leng C, et al. 5‐hydroxymethylcytosine is detected in RNA from mouse brain tissues. Brain Res. 2016;1642:546–52. [DOI] [PubMed] [Google Scholar]
- 194. Kaut O, Kuchelmeister K, Moehl C, Wüllner U. 5‐methylcytosine and 5‐hydroxymethylcytosine in brains of patients with multiple system atrophy and patients with Parkinson’s disease. J Chem Neuroanat. 2019;96:41–8. [DOI] [PubMed] [Google Scholar]
- 195. Stöger R, Scaife PJ, Shephard F, Chakrabarti L. Elevated 5hmC levels characterize DNA of the cerebellum in Parkinson’s disease. NPJ Parkinsons Dis. 2017;3(1):6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Kochmanski J, Bernstein AI. The impact of environmental factors on 5‐hydroxymethylcytosine in the brain. Cur Environ Health Rep. 2020;7:109–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Wu TT, Liu T, Li X, Chen YJ, Chen TJ, Zhu XY, et al. TET2‐mediated Cdkn2A DNA hydroxymethylation in midbrain dopaminergic neuron injury of Parkinson’s disease. Hum Mol Genet. 2020;29(8):1239–52. [DOI] [PubMed] [Google Scholar]
- 198. Korolchuk VI, Miwa S, Carroll B, von Zglinicki T. Mitochondria in cell senescence: is mitophagy the weakest link? EBioMedicine. 2017;21:7–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447(7143):407–12. [DOI] [PubMed] [Google Scholar]
- 200. Maze I, Noh KM, Allis CD. Histone regulation in the CNS: basic principles of epigenetic plasticity. Neuropsychopharmacol. 2013;38:3–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Dölle C, Flønes I, Nido GS, Miletic H, Osuagwu N, Kristoffersen S, et al. Defective mitochondrial DNA homeostasis in the substantia nigra in Parkinson disease. Nat Commun. 2016;7:13548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Ryan BJ, Hoek S, Fon EA, Wade‐Martins R. Mitochondrial dysfunction and mitophagy in Parkinson’s: from familial to sporadic disease. Trends Biochem Sci. 2015;40:200–10. [DOI] [PubMed] [Google Scholar]
- 203. Karamanlidis G, Lee CF, Garcia‐Menendez L, Kolwicz SC, Suthammarak W, Gong G, et al. Mitochondrial complex i deficiency increases protein acetylation and accelerates heart failure. Cell Metab. 2013;18(2):239–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Anderson KA, Madsen AS, Olsen CA, Hirschey MD. Metabolic control by sirtuins and other enzymes that sense NAD+, NADH, or their ratio. Biochim Biophys Acta Bioenerg. 2017;1858:991–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Imai SI, Guarente L. It takes two to tango: Nad+ and sirtuins in aging/longevity control. NPJ Aging Mech Dis. 2016;2(1):16017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Verdin E. NAD+ in aging, metabolism, and neurodegeneration. Science. 2015;350:1208–13. [DOI] [PubMed] [Google Scholar]
- 207. Park G, Tan J, Garcia G, Kang Y, Salvesen G, Zhang Z. Regulation of histone acetylation by autophagy in Parkinson disease. J Biol Chem. 2016;291(7):3531–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Toker L, Tran GT, Sundaresan J, Tysnes OB, Alves G, Haugarvoll K, et al. Genome‐wide dysregulation of histone acetylation in the Parkinson’s disease brain. bioRxiv. 2019;785550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Dietz KC, Casaccia P. HDAC inhibitors and neurodegeneration: at the edge between protection and damage. Pharmacol Res. 2010;62:11–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Kontopoulos E, Parvin JD, Feany MB. α‐synuclein acts in the nucleus to inhibit histone acetylation and promote neurotoxicity. Hum Mol Genet. 2006;15(20):3012–23. [DOI] [PubMed] [Google Scholar]
- 211. Vartiainen S, Pehkonen P, Lakso M, Nass R, Wong G. Identification of gene expression changes in transgenic C. elegans overexpressing human α‐synuclein. Neurobiol Dis. 2006;22(3):477–86. [DOI] [PubMed] [Google Scholar]
- 212. Outeiro TF, Kontopoulos E, Altmann SM, Kufareva I, Strathearn KE, Amore AM, et al. Sirtuin 2 inhibitors rescue α‐synuclein‐mediated toxicity in models of Parkinson’s disease. Science. 2007;317(5837):516–9. [DOI] [PubMed] [Google Scholar]
- 213. Siddiqui A, Chinta SJ, Mallajosyula JK, Rajagopolan S, Hanson I, Rane A, et al. Selective binding of nuclear alpha‐synuclein to the PGC1alpha promoter under conditions of oxidative stress may contribute to losses in mitochondrial function: Implications for Parkinson’s disease. Free Radic Biol Med. 2012;53(4):993–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Du G, Liu X, Chen X, Song M, Yan Y, Jiao R, et al. Drosophila histone deacetylase 6 protects dopaminergic neurons against α‐synuclein toxicity by promoting inclusion formation. Mol Biol Cell. 2010;21(13):2128–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Acquatella‐Tran Van Ba I, Imberdis T, Perrier V. From prion diseases to prion‐like propagation mechanisms of neurodegenerative diseases. Int J Cell Biol. 2013;2013:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Ma J, Gao J, Wang J, Xie A. Prion‐like mechanisms in Parkinson’s disease. Front Neurosci. 2019;13:552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Lema Tomé CM, Tyson T, Rey NL, Grathwohl S, Britschgi M, Brundin P. Inflammation and α‐synuclein’s prion‐like behavior in Parkinson’s disease–is there a link? Mol Neurobiol. 2013;47:561–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Karpowicz RJ, Trojanowski JQ, Lee VMY. Transmission of α‐synuclein seeds in neurodegenerative disease: recent developments. Nature. 2019;99:971–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Bhat AH, Dar KB, Anees S, Zargar MA, Masood A, Sofi MA, et al. Oxidative stress, mitochondrial dysfunction and neurodegenerative diseases; a mechanistic insight. Biomed Pharmacother. 2015;74:101–10. [DOI] [PubMed] [Google Scholar]
- 220. Bae ON, Kim YD, Lim KM, Noh JY, Chung SM, Kim K, et al. Salsolinol, an endogenous neurotoxin, enhances platelet aggregation and thrombus formation. Thromb Haemost. 2008;100(1):52–9. [DOI] [PubMed] [Google Scholar]
- 221. Nam YJ, Lee DH, Lee MS, Lee CS. KATP channel block prevents proteasome inhibitor‐induced apoptosis in differentiated PC12 cells. Eur J Pharmacol. 2015;764:582–91. [DOI] [PubMed] [Google Scholar]
- 222. Miyake H, Kadoya A, Ohyashiki T. Increase in molecular rigidity of the protein conformation of brain Na+‐K+‐ATPase by modification with 4‐hydroxy‐2‐nonenal. Biol Pharm Bull. 2003;26(12):1652–6. [DOI] [PubMed] [Google Scholar]
- 223. Roberts HL, Brown DR. Seeking a mechanism for the toxicity of oligomeric α‐synuclein. Biomolecules. 2015;5(2):282–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Choong CJ, Mochizuki H. Gene therapy targeting mitochondrial pathway in Parkinson’s disease. J Neural Transm. 2017;124:193–207. [DOI] [PubMed] [Google Scholar]
- 225. Carrera I, Cacabelos R. Current drugs and potential future neuroprotective compounds for Parkinson’s disease. Curr Neuropharmacol. 2018;17(3):295–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Hughes KC, Gao X, Kim IY, Rimm EB, Wang M, Weisskopf MG, et al. Intake of antioxidant vitamins and risk of Parkinson’s disease. Mov Disord. 2016;31(12):1909–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.


