Abstract
Understanding the plant immune system is crucial for using genetics to protect crops from diseases. Plants resist pathogens via a two-tiered innate immune detection-and-response system. The first plant Resistance (R) gene was cloned in 1992 . Since then, many cell-surface pattern recognition receptors (PRRs) have been identified, and R genes that encode intracellular nucleotide-binding leucine-rich repeat receptors (NLRs) have been cloned. Here, we provide a list of characterized PRRs and NLRs. In addition to immune receptors, many components of immune signaling networks were discovered over the last 30 years. We review the signaling pathways, physiological responses, and molecular regulation of both PRR- and NLR-mediated immunity. Recent studies have reinforced the importance of interactions between the two immune systems. We provide an overview of interactions between PRR- and NLR-mediated immunity, highlighting challenges and perspectives for future research.
A review of major research advances in plant immunity during the last three decades and individual characterized immune receptors, their immune signaling pathways, and interactions between immune systems
The plant immune system
Plants are constantly challenged by diverse organisms, including viruses, bacteria, fungi, oomycetes, herbivores, and parasitic plants. Disease ensues when a plant is susceptible to any of these organisms. Plants carry powerful defense mechanisms. To cause disease, pathogens usually need to evade detection by the host and/or to suppress these immune responses. Cell-surface pattern recognition receptors (PRRs) in plants recognize conserved pathogen-/damage-/microbe-/herbivore-associated molecular patterns (PAMPs/DAMPs/MAMPs/HAMPs) and activate pattern-triggered immunity (PTI), which restricts pathogenicity. PRRs are plasma membrane (PM)-associated and are usually either receptor-like kinases (RLKs) or receptor-like proteins (RLPs) that lack a protein kinase domain. Pathogens have evolved to evade or suppress PTI through secreted effector molecules, which results in effector-triggered susceptibility (ETS). Plants, in turn, have evolved intracellular nucleotide-binding leucine-rich repeat receptors (NLRs) to detect effectors, which are often encoded by Resistance (R) genes, and activate effector-triggered immunity (ETI) upon effector perception. Pathogens might then evolve or diversify or lose effectors to suppress or evade ETI. The interaction between PTI, ETS, and ETI was incorporated into the widely cited “zig-zag-zig” intellectual framework (Jones and Dangl, 2006).
The alphabet soup digested: nomenclatures applied to the plant immune system
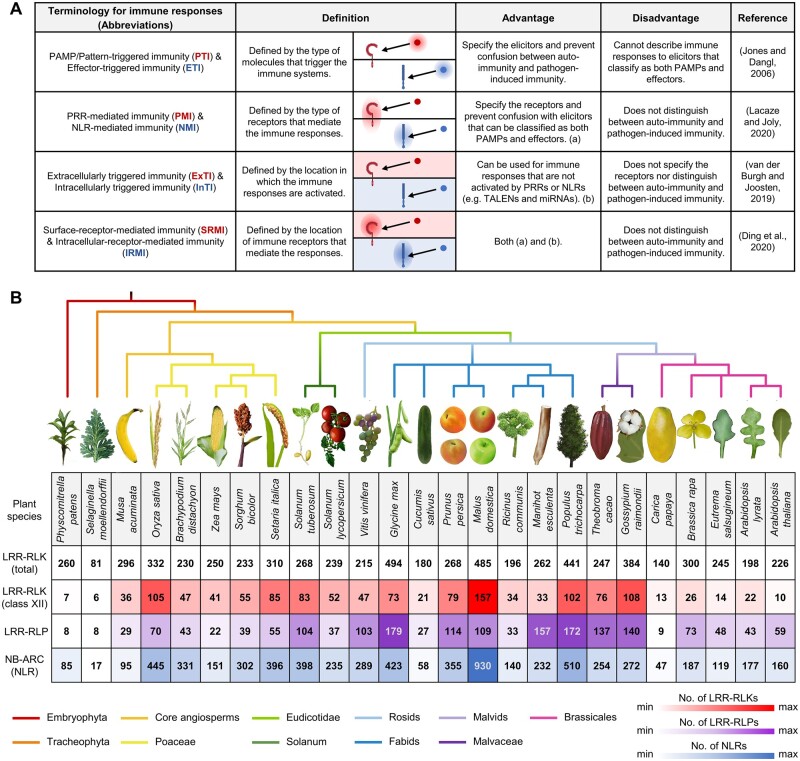
PTI was originally an abbreviation for “PAMP-triggered immunity”, mediated by PRRs such as Arabidopsis thaliana Flagellin-Sensing 2 (FLS2). ETI is an acronym for “effector-triggered immunity,” which is mostly mediated by NLRs (Jones and Dangl, 2006), but can also involve RLP-mediated detection of apoplastic effectors (Jones et al., 1994). While the terms PTI and ETI are frequently used in the literature, there are limitations to their use in describing specific immune responses (Thomma et al., 2011). For example, the apoplastic effector Avr4 from the tomato (Solanum lycopersicum) leaf mold pathogen Cladosporium fulvum binds to fungal chitin to retard cell wall degradation by host chitinases and thus the release of N-acetyl glucosamine oligomers that activate defense (Joosten et al., 1994; van den Burg et al., 2006). Avr4 is recognized by the tomato cell-surface RLP Cf-4 (Thomas et al., 1997). Thus, while immunity activated by some PRRs can be classified as PTI, others can be classified as ETI, since cell-surface receptors can recognize both PAMPs and apoplastic effectors (Thomma et al., 2011). Other terms have been introduced to classify immune responses based on receptors, such as PRR-mediated immunity and NLR-mediated immunity (Lacaze and Joly, 2020). Immune responses are best defined by the location of recognition by the initiating protein, such as extracellularly triggered immunity and intracellularly triggered immunity (van der Burgh and Joosten, 2019), or surface-receptor-mediated immunity and intracellular-receptor-mediated immunity (van der Burgh and Joosten, 2019; Ding et al., 2020). Each of these terms has its own advantages and should be used with caution (Figure 1A). In this review, we try to minimize the overuse of these acronyms and emphasize immune responses triggered by the corresponding receptors.
Figure 1.
Nomenclatures in plant immunity and the evolution of plant immune receptors. A, Terminology for plant immune responses. Tabular summary of the different terms used to describe plant immune responses. Definitions, advantages, and disadvantages for each of these are included. B, Number of LRR–RLKs, LRR–RLPs, and NLRs in different plant species. Phylogenetic tree illustrating different plant species with the corresponding numbers of LRR–RLKs, LRR–RLK XII (class or subgroup XII), LRR–RLPs, and NLRs. Red heatmap indicates the number of LRR–RLK XIIs, purple heatmap indicates the number of LRR–RLPs, and blue heatmap indicates the number of NLRs. The phylogenetic tree was generated using phyloT (https://phylot.biobyte.de/) based on the NCBI taxonomy database and visualized by iTOL (https://itol.embl.de/). LRR–RLK data were obtained from Dufayard et al. (2017), LRR–RLP data were obtained from Ngou et al. (2022), and NLR data were obtained from Baggs et al. (2020).
Structural and evolutionary overview of PRR proteins
Plant PRR proteins are either RLPs or RLKs. RLKs consist of an extracellular domain, a transmembrane domain, and cytoplasmic kinase domain. RLPs lack a cytoplasmic kinase domain, and both require co-receptors to transduce immune signals. PRRs are localized to the PM via a transmembrane α-helix or a glycophosphatidylinositol (GPI) anchor (Boutrot and Zipfel, 2017). Both RLPs and RLKs perceive ligands via a range of extracellular domains. These include leucine-rich repeat (LRR), lectin, malectin, lysin motif (LysM), and epidermal growth factor-like domains (Boutrot and Zipfel, 2017).
RLKs are found in Plasmodium, plants, and animals but not fungi (Shiu and Bleecker, 2003). Conceivably, RLKs were present in the common ancestors of these organisms but were later lost in the fungi. Plant RLKs underwent remarkable expansion and constitute 60% of the kinases in the Arabidopsis genome (Shiu and Bleecker, 2003). Arabidopsis RLKs can be classified into 44 subfamilies based on their kinase domains (Shiu and Bleecker, 2003). The LRR–RLKs represent the largest subfamily of RLKs and are the best characterized RLKs in plants. A phylogenetic study of 33 plant species concluded that the average number of LRR–RLKs in angiosperms is approximately 250 per species (Dufayard et al., 2017; Figure 1B). LRR–RLKs are further classified into 20 subgroups, with subgroup XII constituting genes involved in pathogen recognition, such as FLS2, EFR, and Xa21 (Dufayard et al., 2017). Interestingly, the gene number in the LRR–RLK subgroup XII is highly variable across plant species, indicating that these genes underwent either expansion or contraction in particular lineages (Dufayard et al., 2017; Ngou et al., 2022). Similarly, the LRR–RLPs represent the largest subfamily of RLPs in plants, and the size of this gene family is also highly variable across plant species (Ngou et al., 2022; Figure 1B).
Structural and evolutionary overview of NLR proteins
NLRs are grouped into three classes according to their N-terminal domains: coiled-coil (CC) NLRs (CNLs), Toll/Interleukin-1 receptor/Resistance (TIR) protein NLRs (TNLs), and RPW8-like CC domain (RPW8) NLRs (RNLs). Both CNLs and RNLs contain N-terminal CC-domains. Plant NLRs carry a nucleotide-binding (NB) domain shared by APAF-1, various plant R proteins and CED-4 (together, the NB-ARC domain), and LRR domains at their C-termini. These domains vary between NLRs, and additional noncanonical domains can be integrated into some NLRs (also known as NLR-integrated domains, or NLR-IDs; Sarris et al., 2016). The functions of these domains also vary among NLRs. The LRR domain is involved in direct or indirect recognition of effectors (Krasileva et al., 2010; Ma et al., 2020a; Martin et al., 2020). The NB-ARC domain exhibits ATP binding activity and acts as a switch for NLR activation (Wang et al., 2019b). The CC, TIR, and RPW8 domains function as signaling domains to downstream responses upon NLR activation (Adachi et al., 2019a; Bi et al., 2021; Duxbury et al., 2021; Jacob et al., 2021). Some CC-domains are involved in effector recognition and interact directly with effectors (Avr-Pik) as well as a “guardee” protein (such as RIN4), which is a target of pathogen effectors (Lukasik and Takken, 2009; Kanzaki et al., 2012). The α-helices in both the CC and RPW8 domains were recently shown to form cation channels required for defense signaling (Bi et al., 2021; Jacob et al., 2021). TIR domains can also self-associate or associate with the TIR domains from paired TNLs, which is crucial for their activation (Williams et al., 2014; Duxbury et al., 2020). TIR domains, upon oligomerization, exhibit NADase activity, which leads to the production of variant-cyclic-ADP-ribose (v-cADPR; Horsefield et al., 2019; Wan et al., 2019a). TIR domains also exhibit 2′,3′-cAMP/cGMP synthetase activity (Yu et al., 2021). These small molecules produced by TIR domains likely function in signaling. The ID domain in NLR-IDs functions as a decoy, which enables the NLR to detect effectors targeting proteins with homology to the ID (van der Hoorn and Kamoun, 2008; Sarris et al., 2016; Baggs et al., 2017).
NLR genes are present in the genomes of all land plants (Gao et al., 2018). CNLs, TNLs, and RNLs are present in basal angiosperm species such as Amborella and Nymphaea (Baggs et al., 2020; Liu et al., 2021). However, TNLs are absent from most monocot genomes, indicating that gene loss likely occurred before monocots diverged from dicots (Tarr and Alexander, 2009). The loss of TNLs was also accompanied by the loss of TNL-signaling components, such as ENHANCED DISEASE SUSCEPTIBILITY 1 (EDS1), PHYTOALEXIN DEFICIENT 4 (PAD4), and SENESCENCE-ASSOCIATED GENE 101 (SAG101; Baggs et al., 2020; Liu et al., 2021). The loss of these signaling components may have driven the contraction of TNLs in some angiosperm lineages, or vice versa (Liu et al., 2021). Similar to the LRR–RLK-XII and LRR–RLP, the number of NLRs (or NB-ARC containing proteins) is also highly variable across the angiosperms (Baggs et al., 2020; Liu et al., 2021). Furthermore, the LRR–RLK-XII, LRR–RLP, and NLR gene families have undergone lineage-specific co-expansion or co-contraction (Ngou et al., 2022; Figure 1B). The cause of these concerted expansions and/or contractions is currently unclear but has been proposed to be linked to pathogen pressure and ecological specialization (Plomion et al., 2018; Baggs et al., 2020; Liu et al., 2021; Ngou et al., 2022).
PRRs involved in pathogen recognition
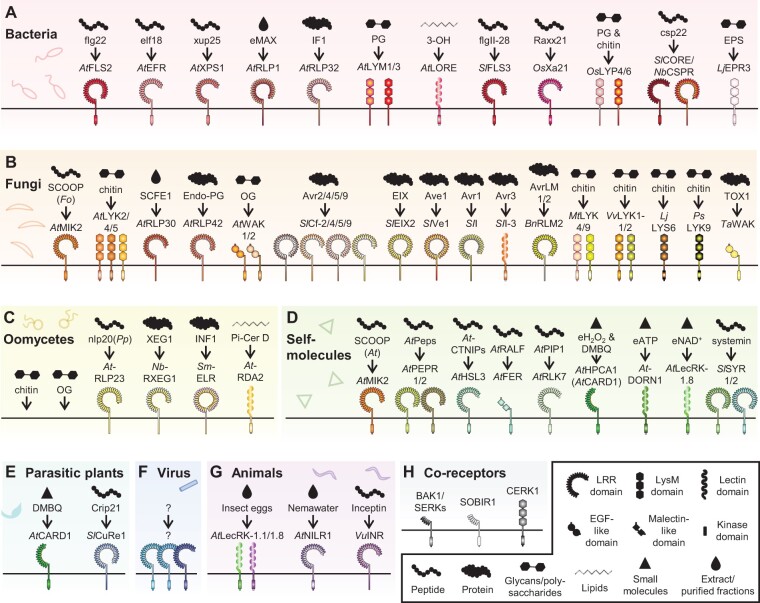
PRRs recognize PAMPs/MAMPs/HAMPs from bacteria, fungi, oomycetes, parasitic plants, and herbivores. Some PRRs also recognize self-molecules, such as DAMPs and other plant endogenous peptides (phytocytokines; Hou et al., 2021). Some PRRs are not involved in direct ligand recognition but function as PRR co-receptors and negative regulators of immune signaling. There are more than 60 characterized immunity-related PRRs with known elicitors, and we attempt here to list those PRRs with known elicitors that are involved in pathogen recognition (Figure 2). Due to space limitations, some PRR gene names are abbreviated: the full gene names can be found in Supplemental Data Set 1.
Figure 2.
PRRs involved in plant immunity. Characterized PRRs with known elicitors from (A) bacteria, (B) fungi, (C) oomycetes, (D) self-molecules, (E) parasitic plants, (F) viruses, and (G) herbivores. H, PRR co-receptors. Abbreviations for plant species: A. thaliana, At; S. lycopersicum, Sl; O. sativa, Os; N. benthamiana, Nb; L. japonicus, Lj; B. napus, Bn; M. truncatula, Mt; V. vinifera, Vv; L. japonicus, Lj; P. sativum, Ps; T. aestivum, Ta; S. microdontum, Sm; P. japonicum, Pj; V. unguiculata, Vu. Abbreviation for pathogens: F. oxysporum, Fo; P. parasitica, Pp. Number of LRR repeats in the LRR–RLKs and LRR–RLPs were predicted by phytoLRR (Chen, 2021). The full name of these PRR genes can be found in Supplemental Data Set 1.
PRRs involved in bacterial recognition
Plants perceive a range of PAMPs from bacteria, including peptides, lipids, peptidoglycans (PGs), and polysaccharides. Arabidopsis perceives the bacteria-derived peptides flg22, elf18, and xup25 via the LRR–RLKs AtFLS2, AtEFR, and AtXPS1 and the proteinaceous eMAX and translation initiation factor 1 via the LRR–RLPs AtRLP1 and AtRLP32, respectively (Chinchilla et al., 2006; Zipfel et al., 2006; Jehle et al., 2013; Mott et al., 2016; Fan et al., 2021). Other bacterial peptides such as RaxX21, flgII-28, and csp22 are perceived by rice (Oryza sativa) OsXa21, tomato SlFLS3, and SlCORE/NbCSPR (from tomato and Nicotiana benthamiana), respectively (Pruitt et al., 2015; Hind et al., 2016; Saur et al., 2016; Wang et al., 2016; Luu et al., 2019). The bacterial lipid 3-hydroxydecanoic acid is perceived through the lectin receptor kinase AtLORE (Kutschera et al., 2019). PGs from bacterial cell walls are perceived by the LysM-containing RLP AtLYM1/3 and rice OsLYP4/6 (Willmann et al., 2011; Liu et al., 2012). Bacterial exopolysaccharides are perceived by the LysM-containing RLK LjEPR3 from Lotus japonicus to control rhizobium infections (Kawaharada et al., 2015; Figure 2A).
PRRs involved in fungal recognition
The fungal cell wall comprises chitin and oligo-galacturonides (OGs), which are perceived by multiple PRRs. Chitin is perceived by LysM-containing RLKs such as AtLYM2/4/5, OsLYP4/6, Medicago truncatula MtLYK4/9, grapevine (Vitis vinifera) VvLYK1-1/2, L. japonicus LjLYS6, and pea (Pisum sativum) PsLYK9 (Wan et al., 2008, 2012; Liu et al., 2012; Faulkner et al., 2013; Cao et al., 2014; Bozsoki et al., 2017; Leppyanen et al., 2017; Brulé et al., 2019). OGs are perceived by the cell wall-associated kinases AtWAK1/2 (Brutus et al., 2010). AtWAK1/2 also perceive pectin from the plant cell wall (Kohorn and Kohorn, 2012). The common wheat (Triticum aestivum) wall-associated kinase TaWAK perceives the protein SnTox1 from the necrotrophic fungal pathogen Parastagonospora nodorum and induces cell death (Shi et al., 2016). In addition to the fungal cell wall, apoplastic effectors from fungal pathogens are recognized by multiple LRR–RLPs. These include SlCf-2, SlCf-4, SlCf-5, SlCf-9, SlEIX2, SlVe1, SlHrc9-4E, SlI, SlI-3, and Brassica napus BnRLM2 (Jones et al., 1994; Dixon et al., 1996, 1998; Thomas et al., 1997; Krüger et al., 2002; Westerink et al., 2004; Rep et al., 2004; Ron and Avni, 2004; Houterman et al., 2008; de Jonge et al., 2012; Larkan et al., 2013; Catanzariti et al., 2015). A proteinaceous elicitor from the fungal pathogen Sclerotinia sclerotiorum, sclerotinia culture filtrate elicitor 1, is perceived by AtRLP30, and fungal endopolygalacturonases (endo-PGs) are perceived by the LRR–RLP AtRLP42 (Zhang et al., 2013, 2014; Figure 2B).
PRRs involved in the recognition of oomycetes
The oomycete cell wall is also composed of chitin, endo-PGs, and OGs. Thus, plants also perceive oomycetes via PRRs described in the previous section. In addition, some PRRs recognize specific PAMPs from oomycetes. For example, the glycoside hydrolase XEG1 from Phytophthora sojae is recognized by the LRR–RLP NbRXEG1 (Wang et al., 2018d). INF1 elicitin from Phytophthora infestans is recognized by the LRR–RLP SmELR from Solanum microdontum (Kamoun et al., 1997; Du et al., 2015b; Domazakis et al., 2020). Arabidopsis AtRLP23 recognizes a conserved peptide (nlp20) in necrosis and ethylene (ET)-inducing peptide 1-like protein (NLP) from multiple pathogens, including Phytophthora parasitica (Böhm et al., 2014; Albert et al., 2015). The Arabidopsis lectin-receptor kinase AtRDA2 was recently shown to recognize 9-methyl sphingoid base, a PAMP derived from oomycete ceramide (Kato et al., 2021; Figure 2C).
PRRs involved in self-recognition
Plants perceive DAMPs and phytocytokines from damaged or infected tissues to amplify and modulate immune responses against pathogens. Damage-induced cytosolic calcium influx activates metacaspases, which cleave the DAMP precursor PROPEPs into PEPs (Hander et al., 2019). PEPs are then secreted and perceived by the LRR–RLKs AtPEPR1/2 (Yamaguchi et al., 2006, 2010). Multiple phytocytokines are upregulated during immunity (Hou et al., 2021). The stress-induced plant signaling peptides CTNIPs are upregulated during PTI and are perceived by the Arabidopsis LRR–RLK AtHSL3 (Rhodes et al., 2021a). Another defense-induced secreted peptide, PIP1, is recognized by AtRLK7 (Hou et al., 2014). The Arabidopsis LRR–RLK AtMIK2 perceives the phytocytokine SCOOP peptides and SCOOP-like peptides from Fusarium spp. (Coleman et al., 2021; Rhodes et al., 2021b). Thus, AtMIK2 is involved in both self and fungal recognition during immunity. Plant PRRs also perceive a range of extracellular (e) self-molecules, such as eH2O2, eATP, and eNAD. These molecules are perceived by AtHPCA1 (also known as AtCARD1), AtDORN1, and AtLecRK-1.8, respectively (Chen et al., 2017a; Wang et al., 2017; Wu et al., 2020a). In tomato, the hormone peptide systemin is perceived by SlSYR1/2 to enhance resistance against herbivores (Wang et al., 2018b; Figure 2D).
PRRs involved in the recognition of parasitic plants
In addition to eH2O2, AtCARD1 has also been shown to perceive the self-derived quinone compound 2,6-dimethoxy-1,4-benzoquinone (DMBQ; Laohavisit et al., 2020). Perception of DMBQ induces AtCARD1-dependent immune responses. On the other hand, the parasitic plant Phtheirospermum japonicum perceives DMBQ via AtCARD1 homologs PjCADL1/2/3, which leads to development of haustoria for parasitic infection (Laohavisit et al., 2020). Thus, CARD1 is involved in both immunity (for nonparasitic plants) and parasitic plant infection. Plants also perceive PAMPs from parasitic plants to restrict infection. The tomato LRR–RLP SlCuRe1 perceives the peptide Crip21 from the parasitic plant Cuscuta spp. (Hegenauer et al., 2020). Crip21 is derived from a Cuscuta glycine-rich cell wall protein. Activation of SlCuRe1 by Crip21 elicits cell death and defense responses in tomato (Hegenauer et al., 2020; Figure 2E).
PRRs involved in viral recognition
While some PRRs, such as AtNIK1, have been shown to be required for viral resistance, no PRR has been reported to directly perceive viral particles (Zorzatto et al., 2015). However, the Arabidopsis PRR co-receptor bak1 loss-of-function mutant exhibits enhanced susceptibility to multiple viruses (Kørner et al., 2013). In addition, exogenous application of double-stranded RNAs and viral coat protein (CP) elicits PTI responses in plants (Allan et al., 2001; Niehl et al., 2016). Conceivably, some uncharacterized PRR(s) are involved in the recognition of viral PAMPs (Figure 2F).
PRRs involved in the recognition of animals
In addition to eNAD+, AtLecRK-1.8 and AtLecRK-1.1 are involved in the perception of Pieris brassicae (cabbage moth) eggs (Gouhier-Darimont et al., 2019; Groux et al., 2021). The ligand from P. brassicae eggs that activates AtLecRK-1.8 remains to be identified and characterized. The Arabidopsis LRR–RLK AtNILR1 is involved in the perception of Heterodera schachtii (sugarbeet nematode) extracts, and nilr1 mutants are hypersusceptible to nematode infection (Mendy et al., 2017). The cowpea (Vigna unguiculata) LRR–RLP VuINR was shown to perceive inceptin, a proteolytic fragment of chloroplastic ATP synthase from the oral secretions of Lepidopteran herbivores (a HAMP; Steinbrenner et al., 2019). Whether PRRs can perceive ligands directly from herbivores remains to be determined (Figure 2G).
PRR co-receptors
Most, if not all, PRRs function with co-receptors to activate downstream immune responses. Multiple LRR–RLKs, such as FLS2, EFR, and PEPRs function with the co-receptors AtBAK1 and AtBKK1 (Chinchilla et al., 2007; Roux et al., 2011). LRR–RLPs function with the co-receptors SOBIR1 and BAK1, and the LysM-RLK LYKs and LysM-RLP LYMs function with the co-receptor CERK1 (Miya et al., 2007; Willmann et al., 2011; Liebrand et al., 2013; Cao et al., 2014). These co-receptors are highly conserved in land plants and are crucial for PRR-mediated immunity (Figure 2H).
NLRs involved in pathogen recognition
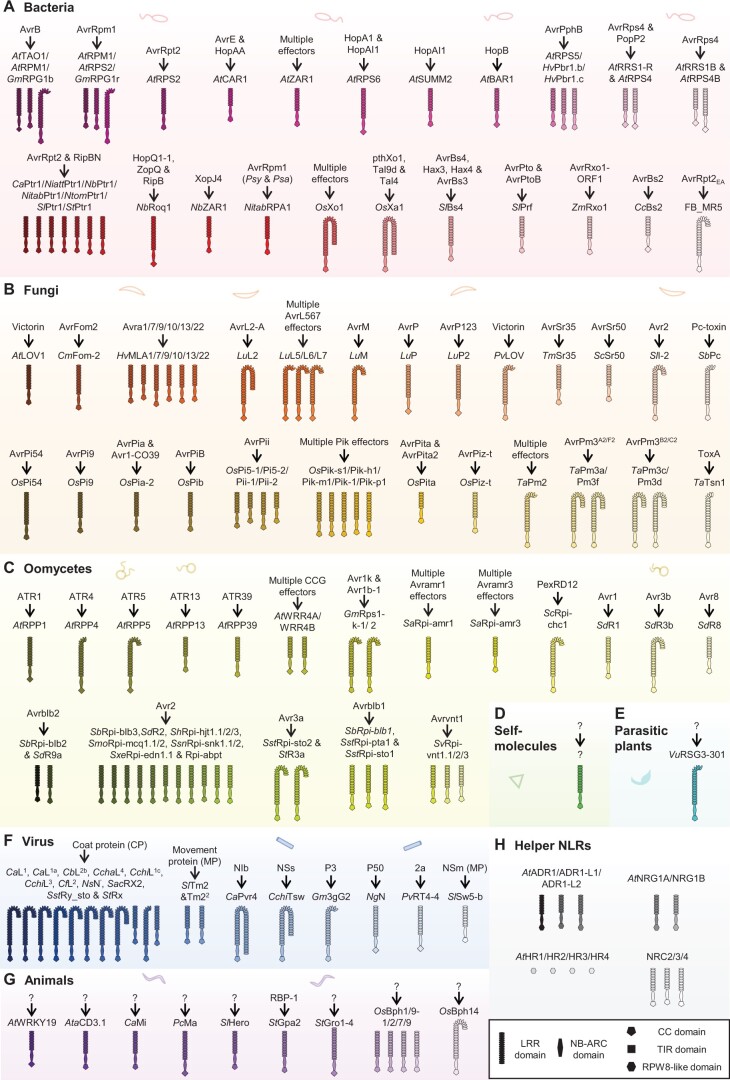
Sensor NLRs are involved in the recognition of effectors from viruses, bacteria, fungi, oomycetes, parasitic plants, and herbivores. Some NLRs act as helpers or co-receptors to transduce immune signals from sensor NLRs following effector recognition (Wu et al., 2018). Currently, there are more than 140 characterized NLRs with known recognized effectors (Kourelis and Kamoun, 2020). Here, we summarize a list of NLRs involved in effector recognition (Figure 3; Supplemental Data Set 2).
Figure 3.
NLRs involved in plant immunity. Characterized NLRs with known effectors from (A) bacteria, (B) fungi, (C) oomycetes, (D) self-molecules, (E) parasitic plants, (F) viruses, (G) herbivores, and (H) Helper NLRs. Abbreviations for plant species: G. max, Gm; H. vulgare, Hv; C. annuum, Ca; Nicotiana attenuate, Niatt; N. tabacum, Nitab; Nicotiana tomentosiformis, Ntom; S. tuberosum, St; Z. mays, Zm; C. chacoense, Cch; C. melo, Cm; L. usitatissimum, Lu; P. vulgaris, Pv; Triticum monococcum, Tm; S. cereale, Sc; S. bicolor, Sb; S. americanum, Sa; S. bulbocastanum, Sbu; S. chacoense, Sch; S. demissum, Sd; Solanum hjertingii, Sh; Solanum mochicense, Smo; Solanum nigrescens, Ssn; Solanum × edinense, Sxe; S. stoloniferum, Sst; S. venturi, Sv; C. baccatum, Cb; C. chinense, Cchi; C. frutescens, Cf; N. sylvestris, Ns; S. acaule, Sac; N. glutinosa, Ng; A. tauschii, Ata; P. cerasifera, Pc. Number of LRR repeats in the NLRs were predicted by LRRpredictor (Martin et al., 2020a). The full list of NLRs can be found in Supplemental Data Set 2.
NLRs involved in bacterial recognition usually act by guarding host components
Bacterial effectors have been selected that target PRR signaling components and suppress host immunity. Plants have evolved multiple NLRs to guard host immune components, which indirectly detect bacteria and induce ETI. For example, the Pseudomonas syringae effector AvrPto suppresses PTI by inhibiting host kinase activity (Li et al., 2005; He et al., 2006; Xing et al., 2007; Xiang et al., 2008; Wu et al., 2017b). The tomato decoy kinase Pto is guarded by the CNL Prf, which detects the perturbation of Pto kinase activity by AvrPto and activates ETI (Wu et al., 2004; Mucyn et al., 2006; Ntoukakis et al., 2013). Since plants have evolved multiple NLRs to guard central immune signaling pathways, some effectors from P. syringae are recognized by multiple NLRs from different plant species (Jones and Dangl, 2006). Examples include the following: AvrB is recognized by AtTAO1, AtRPM1, and Glycine max GmRPG1b (Grant et al., 1995; Ashfield et al., 2004; Eitas et al., 2008). AvrRpm1 from P. syringae pv. maculicola (Pma) is recognized by AtRPM1, AtRPS2, and GmRPG1r (Ashfield et al., 1995; Grant et al., 1995; Kim et al., 2009a). AvrPphB is recognized by AtRPS5, Hordeum vulgare HvPbr1.b and HvPbr1.c (DeYoung et al., 2012; Carter et al., 2019; Laflamme et al., 2020). AvrRpt2 from P. syringae and RipBN from Ralstonia pseudosolanacearum are recognized by the CNL Ptr1 from multiple Solanaceous species (Mazo-Molina et al., 2020). In addition, AvrRpt2 is recognized by the CNL AtRPS2, and AvrRpt2_EA from Erwinia amylovora is recognized by FB_MR5 from Malus × robusta 5 (Axtell and Staskawicz, 2003; Mackey et al., 2003; Peil et al., 2019). HopA1 is recognized by AtRPS6, and HopAI1 is recognized by both AtSUMM2 and AtRPS6 (Kim et al., 2009b; Zhang et al., 2012; Takagi et al., 2019).
On the other hand, central hubs of the immune system are targeted by multiple effectors. Correspondingly, NLRs, which guard central immune signaling components, can recognize multiple effectors (Khan et al., 2016). For example, the CNL AtZAR1 functions with the pseudokinase RKS1 to guard the receptor-like cytoplasmic kinase (RLCK) PBL2 (Wang et al., 2015). By guarding RLCKs or decoy pseudokinases, AtZAR1 indirectly recognizes HopZ1a, HopF2, HopBA1, HopO1, HopX1, and AvrAC from P. syringae or Xanthomonas campestris, and potentially more effectors that target RLCKs (Wang et al., 2015; Laflamme et al., 2020). NbZAR1 is also required to recognize XopJ4 from Xanthomonas perforans via the pseudokinase JIM2 (Schultink et al., 2019). Other examples include the following: AvrRpm1Psa and AvrRpm1Psy from P. syringae pv. actinidiae biovar 3 (Psa) and P. syringae pv. syringae strain B728a (Psy) are recognized by Nicotiana tabacum NitabRPA1 (Yoon and Rikkerink, 2020). AvrE and HopAA are both recognized by Arabidopsis CAR1 (Laflamme et al., 2020). AvrRps4 from P. syringae pv. pisi and PopP2 from Ralstonia solanacearum are recognized by the paired-TNLs AtRRS1-R and AtRPS4 (Narusaka et al., 2009; Sarris et al., 2015). In addition, AvrRps4 can also be recognized by the paired-TNLs AtRRS1B and AtRPS4B (Saucet et al., 2015). The TNL NbRoq1 recognizes HopQ1-1, XopQ, and RipB from P. syringae, Xanthomonas, and R. solanacearum, respectively (Schultink et al., 2017; Thomas et al., 2020). Multiple TRANSCRIPTION ACTIVATOR-LIKE (TAL) effectors from Xanthomonas oryzae are recognized by the CNLs OsXo1 and OsXa1 (Yoshimura et al., 1998; Triplett et al., 2016; Read et al., 2020a, 2020b).The tomato TNL SlBs4 also recognizes multiple Xanthomonas effectors (Schornack et al., 2004, 2005). AvrRxo1-ORF1 from X. oryzae and Burkholderia andropogonis are recognized by the CNL ZmRxo1 from maize (Zea mays; Zhao et al., 2004; Figure 3A).
NLRs involved in fungal recognition
Plant NLRs recognize multiple effectors and molecules from fungal pathogens. Victorin, a secondary metabolite from Cochliobolus victoriae, is recognized by LOV1 from Arabidopsis and Phaseolus vulgaris (Sweat et al., 2008; Lorang et al., 2018). AvrFom2 from Fusarium oxysporum is recognized by the CNL CmFom-2 from Cucumis melo (Schmidt et al., 2016). Hordeum vulgare RESISTANCE LOCUS A NLRs recognize a range of effectors from Blumeria graminis (Ridout et al., 2006; Lu et al., 2016; Saur et al., 2019) and can even recognize races of wheat stripe rust (Bettgenhaeuser et al., 2021). Multiple TNLs from Linum usitatissimum recognize effectors from Melampsora lini (Dodds et al., 2004; Dodds and Thrall, 2009; Catanzariti et al., 2010; Anderson et al., 2016). Effectors from the rice blast fungus Magnaporthe oryzae are recognized by multiple CNLs from O. sativa (Jia et al., 2000; Ashikawa et al., 2008, 2012; Li et al., 2009, 2019; Zeng et al., 2011; Rai et al., 2011; Sone et al., 2013; Zhai et al., 2014; Devanna et al., 2014; Zhang et al., 2015; Wu et al., 2015; Vo et al., 2019). Effectors from B. graminis, P. nodorum, Pyrenophora tritici-repentis, and Puccinia graminis are recognized by multiple CNLs from Triticum species (Srichumpa et al., 2005; Liu et al., 2006; Salcedo et al., 2017; Bourras et al., 2019; Navathe et al., 2020; Manser et al., 2021). AvrSr50 from P. graminis is recognized by ScSr50 from Secale cereale (Chen et al., 2017b). Avr2 from F. oxysporum is recognized by the CNL SlI2, and Pc-toxin from Periconia circinata is recognized by the CNL SbPc from Sorghum bicolor (Nagy et al., 2007; Nagy and Bennetzen, 2008; Houterman et al., 2009; Figure 3B).
NLRs involved in the recognition of oomycetes
Multiple effectors from Hyaloperonospora arabidopsidis (Hpa) are recognized by Arabidopsis NLRs. ATR1, ATR4, ATR5, ATR13, and ATR39 are recognized by AtRPP1, AtRPP4, AtRPP5, AtRPP13, and AtRPP39, respectively (Rentel et al., 2008; Krasileva et al., 2010; Bailey et al., 2011; Goritschnig et al., 2012; Asai et al., 2018). CX2CX5G effector-like proteins (CCG effectors) from Albugo candida are recognized by AtWRR4A and AtWRR4B (Redkar et al., 2021).
The oomycete genus Phytophthora carries multiple phytopathogenic species that cause enormous crop losses worldwide. Identification of NLRs that recognize Phytophthora effectors provides resources for crop resistance. The P. sojae effectors Avr1k and Avr1b-1 are recognized by GmRps1-k (Song et al., 2013). Effectors from P. infestans are also recognized by NLRs from multiple Solanaceae species. For example, the effectors Avramr1 and Avramr3, with homologs in many Phytophthora species, are recognized by Rpi-amr1 (from Solanum americanum) and Rpi-amr3, respectively (Lin et al., 2020, 2021; Witek et al., 2021). Avrblb1 is recognized by Rpi-blb1 (from Solanum bulbocastanum), Rpi-pta1, and Rpi-sto1 (from Solanum stoloniferum; Vleeshouwers et al., 2008; Oh et al., 2009). Avrblb2 is recognized by Rpi-blb2 and R9a (from S. bulbocastanum and Solanum demissum, respectively; Oh et al., 2009; Jo, 2013). PexRD12 is recognized by Rpi-chc1 (from Solanum chacoense; Monino-Lopez et al., 2021; Petre et al., 2021). Avr1, Avr3b, and Avr8 are recognized by R1, R3b, and R8, respectively (Ballvora et al., 2002; Li et al., 2011; Jo, 2013; Du et al., 2015a; Vossen et al., 2016). PiAvr2 is recognized by multiple NLRs from Solanaceae (Park et al., 2005; Lokossou et al., 2009; Champouret, 2010; Aguilera-Galvez et al., 2018). Avr3a is recognized by Rpi-sto2 and R3a (from Solanum tuberosum; Bos et al., 2010; Champouret, 2010; Vleeshouwers et al., 2011; Chapman et al., 2014). Avrvnt1 is recognized by Rpi-vnt1 from Solanum venturi (Foster et al., 2009; Pel, 2010; Figure 3C).
Apparent absence of NLRs involved in self-recognition in plants
In mammals, DAMPs can be indirectly recognized the intracellular NOD-, LRR-, and pyrin domain-containing protein 3-inflammasome in macrophages (Swanson et al., 2019). However, no plant NLRs have been reported to detect self-molecules so far (Figure 3D).
NLRs involved in the recognition of parasitic plants
Virus-induced silencing of the CNL VuRSG3-301 from V. unguiculata leads to enhanced susceptibility to the parasitic plant Striga gesnerioides race 3 (Li and Timko, 2009). The effector recognized by VuRSG3-301 has not yet been identified (Figure 3E).
NLRs involved in viral recognition
The CPs from different viruses are recognized by pepper (Capsicum annuum) CaL1, CaL1a, Capsicum baccatum CbL2b, Capsicum chacoense CchaL4, Capsicum chinense CchiL1c, CchiL3, Capsicum frutescens CfL2, Nicotiana sylvestris NsN′, Solanum acaule Rx2, S. stoloniferum Rysto, and potato (S. tuberosum) Rx (Saito et al., 1987; Bendahmane et al., 1995; Berzal-Herranz et al., 1995; Gilardi et al., 2004; Tameling and Baulcombe, 2007; Matsumoto et al., 2008; Tomita et al., 2011; Mizumoto et al., 2012; Grech-Baran et al., 2021). Viral movement proteins are recognized by Tm2, SlTm22, and SlSw5-b (Pelham, 1966; Hall, 1980; Weber and Pfitzner, 1998; Peiró et al., 2014). The RNA-dependent RNA Polymerase (NIb) of potyviruses is recognized by the Ca Pvr4 (Kim et al., 2015). The RNA silencing suppressor protein NSs from tomato spotted wilt virus is recognized by CchiTsw (de Ronde et al., 2013). P3 cistrons from soybean mosaic virus are recognized by Gm3gG2 (Wen et al., 2013). The helicase domain of the tobacco mosaic virus replicase (p50) is recognized by Nicotiana glutinosa N (Whitham et al., 1994; Erickson et al., 1999). Cucumber mosaic virus 2a protein is recognized by P. vulgaris PvRT4-4 (Seo et al., 2006). To summarize, multiple components involved in the process of viral infection are recognized by NLRs (Figure 3F).
NLRs involved in the recognition of animals
Multiple NLRs were shown to be involved in resistance against herbivores. NLRs involved in nematode resistance include the TIR–NB–LRR pair AtDSC1 and AtWRKY19, Aegilops tauschii AtaCD3.1, CaMi, Prunus cerasifera PcMa, SlHero, StGpa-2, and StGro1-4 (Lagudah et al., 1997; van der Voort et al., 1997; Milligan et al., 1998; Paal et al., 2004; Sobczak et al., 2005; Chen et al., 2007; Claverie et al., 2011; Warmerdam et al., 2020). In addition, the tomato Mi gene confers resistance to multiple herbivores, such as nematodes, aphids, and whiteflies (Kaloshian et al., 1995; Milligan et al., 1998; Rossi et al., 1998; Neiva et al., 2019). Other NLRs have been shown to confer resistance against the arthropod Nilaparvata lugens (brown planthopper). These include the rice OsBph1/9 and OsBph14 (Du et al., 2009; Zhao et al., 2016). While multiple NLRs are involved in herbivore resistance, more work is needed to identify the recognized effectors (Figure 3G).
Helper NLRs
While some sensor NLRs do not require helper NLRs, many NLRs function with helper NLRs to transduce immune signals. In Arabidopsis, some CNLs and/or most TNLs require the RNLs ACTIVATED DISEASE RESISTANCE 1 (collectively known as ADR1s, which includes AtADR1, AtADR1-L1, and AtADR1-L2) and/or N REQUIREMENT GENE 1 (collectively known as NRG1s, which includes AtNRG1A and AtNRG1B; Bonardi et al., 2011; Castel et al., 2019a; Wu et al., 2019; Saile et al., 2020). In Arabidopsis accession Col-0, the four RPW8 homologs, AtHR1, AtHR2, AtHR3, and AtHR4, also contribute to resistance against bacterial and fungal pathogens (Barragan et al., 2019; Castel et al., 2019b). In Solanaceous plants, the CNLs NB-LRR REQUIRED FOR HR-ASSOCIATED CELL DEATH-2 (NRC2), NRC3, and NRC4 function as helper NLRs for multiple sensor NLRs (Wu et al., 2017a; Figure 3H). The contribution of the NRC network to the functions of sensor NLRs has been extensively discussed (Wu et al., 2018; Ngou et al., 2021c).
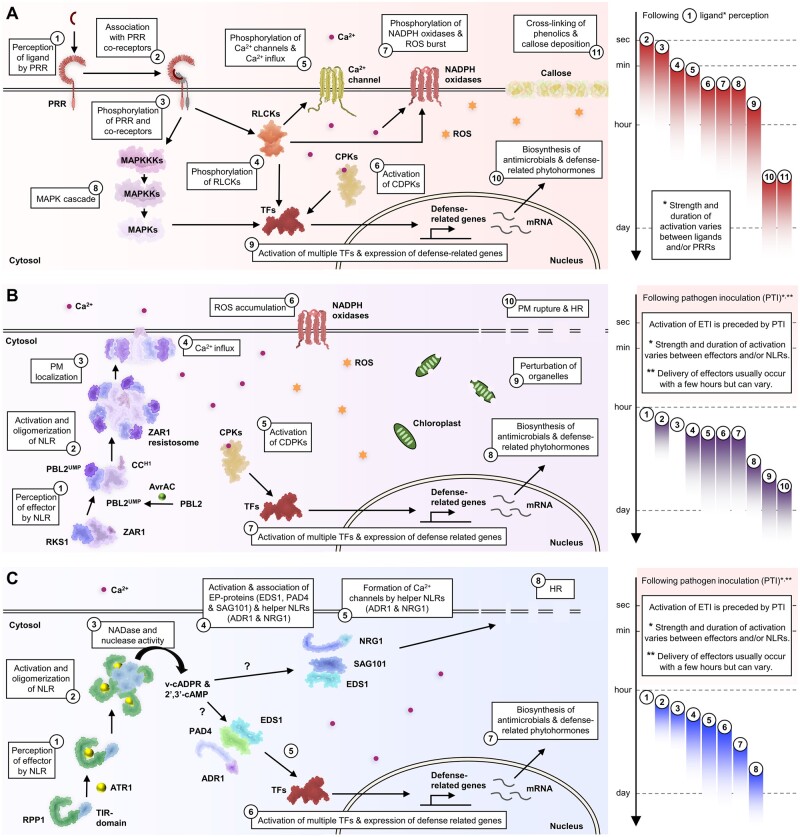
The PRR signaling pathway
The extracellular domains of plant PRRs perceive diverse ligands (Boutrot and Zipfel, 2017). Binding of ligands leads to heterodimeric receptor complex formation between PRRs and their co-receptors, such as BAK1 and CERK1 (Miya et al., 2007; Ma et al., 2016; Hohmann et al., 2017). On the other hand, RLPs constitutively interact with SOBIR1 and recruit BAK1 upon ligand recognition (Liebrand et al., 2013; Albert et al., 2015). In Arabidopsis, the bacterial flagellin peptide flg22 is perceived by the LRR–RLK FLS2 (Felix et al., 1999; Chinchilla et al., 2006). Flg22 acts as a “molecular glue” and interacts with and brings together the extracellular LRR domains of FLS2 and BAK1 (Sun et al., 2013; Hohmann et al., 2017). Heterodimeric complex formation between the LRR domains of FLS2 and BAK1 brings their cytoplasmic kinase domains into close proximity, which leads to a series of auto- and trans-phosphorylation events (Schwessinger et al., 2011; Cao et al., 2013; Sun et al., 2013). This activated receptor complex then phosphorylates RLCKs (Lin et al., 2013; Liang and Zhou, 2018). RLCK subfamily VII members (collectively known as RLCK-VIIs) were first shown to be important for surface receptor-mediated immunity in tomato and tobacco and to be required for Cf-4 and Cf-9 to confer fungal resistance (Rowland et al., 2005). In Arabidopsis, RLCKs play particularly important roles during PRR-mediated immunity (Lu et al., 2010; Lin et al., 2014; Liang and Zhou, 2018; Rao et al., 2018). BAK1 associates with and phosphorylates the RLCK-VII BIK1 at the Try243 and Try250 residues (Lu et al., 2010; Lin et al., 2014).
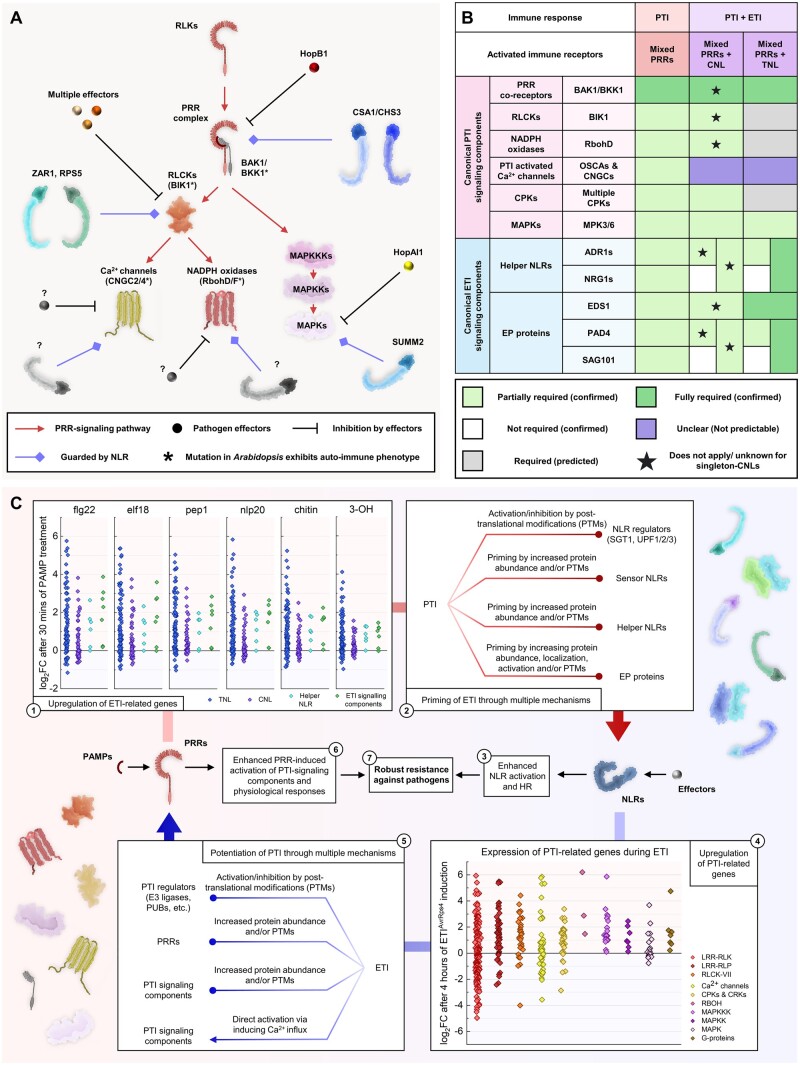
The activation of RLCK-VIIs promotes the phosphorylation of multiple signaling components, including the calcium channels CNGC2/4 and OSCA1.3, the NADPH oxidase respiratory burst oxidase protein D (RbohD), and the mitogen-activated protein kinase kinase kinase (MAPKKK5) (Kadota et al., 2014; Li et al., 2014; Bi et al., 2018; Tian et al., 2019; Thor et al., 2020). The activation of multiple calcium channels by BIK1 leads to cytosolic calcium influx, which activates calcium-dependent protein kinases (CPKs). In Arabidopsis, CPK4/5/6/11, together with BIK1, phosphorylate and activate RbohD, which leads to reactive oxygen species (ROS) production (Kadota et al., 2014, 2015; Li et al., 2014). The phosphorylation of multiple ion channels by RLCKs also leads to stomatal closure in response to PAMPs (Liu et al., 2019; Thor et al., 2020). In parallel, MAPKKK3 and MAPKKK5 phosphorylate the MAPKKs MKK4 and MKK5, which then phosphorylate the MAPKs MPK3 and MPK6 in Arabidopsis. In parallel, MKK1/MKK2 also phosphorylate MPK4 (Asai et al., 2002; Rasmussen et al., 2012). RLCK-VIIs, CPKs, and MPKs phosphorylate and activate multiple defense-related transcription factors, such as WRKY transcription factors, resulting in the upregulation of defense-related genes (Boudsocq et al., 2010; Gao et al., 2013; Lal et al., 2018). PTI-induced transcriptional reprogramming leads to the biosynthesis of antimicrobial compounds and defense-related hormones, such as ET and salicylic acid (SA; Macho et al., 2014; Bigeard et al., 2015; Guan et al., 2015; Bjornson et al., 2021). Hydrogen peroxide (a type of ROS) promotes protein and phenolic cross-linking, which result in callose deposition and restricts fungal and oomycete infection (Luna et al., 2011; Voigt, 2014; Figure 4A).
Figure 4.
Plant immune signaling pathways. A, PRR signaling pathway. Ligand perception by PRRs activates multiple kinases, which leads to calcium influx to the cytosol, ROS production, transcriptional reprogramming, and callose deposition. B, Singleton NLR signaling pathway. The ZAR1/RKS1 heterodimer detects the effector AvrAC via association with uridylylated PBL2 by AvrAC. This leads to the activation and oligomerization of ZAR1. The ZAR1 resistosome localizes to the PM and triggers calcium influx, which leads to the HR and cell rupture. C, Helper-NLR-dependent sensor NLR signaling pathway. Recognition of ATR1 by the TNL RPP1 leads to oligomerization and the induced proximity of TIR domains. The TIR domain exhibits NADase activity and produces v-cADPR, which might activate EP-proteins and the helper NLRs (RNLs). Following TNL activation, EP-proteins and RNLs associate with each other and activate downstream immune responses, likely via cation channel activity from the helper NLRs. Timeline on the right indicates the order and duration of each signaling event following ligand/effector perception. Numbers indicate the corresponding signaling events in the figure on the left. Note that the activation of ETI is usually preceded by PTI activation, and the strength and duration of each event vary and are dependent on the PRRs/NLRs that are activated.
Signaling pathway of singleton NLRs
NLR-mediated immunity is triggered by the detection of effectors through intracellular NLRs. NLRs detect effectors either via direct interactions with effectors, guarding effector targets, or guarding decoy proteins (Van der Biezen and Jones, 1998; Dangl and Jones, 2001; van der Hoorn and Kamoun, 2008). In Arabidopsis, CNLs and TNLs act as sensor NLRs that recognize effectors, while RNLs act as helper NLRs to transduce immune signals (Feehan et al., 2020). While the majority of sensor NLRs in Arabidopsis require helper NLRs to mediate immunity, some CNLs mediate immune responses alone. These are known as singleton NLRs, such as ZAR1 and RPM1 (Adachi et al., 2019b). ZAR1 recognizes a range of effectors by monitoring pseudokinases such as RKS1 and PBL2, which mimic authentic RLCK targets of effectors (Wang et al., 2019a). The bacterial effector AvrAC from X. campestris uridylylates the RLCK PBL2. The ZAR1/RKS1 heterodimer associates with uridylylated PBL2 (PBL2UMP), which leads to conformational changes in the heterodimer. ADP in the NB-ARC domain in ZAR1 is ejected and replaced by ATP (Wang et al., 2019b). This results in the oligomerization of ZAR1/RKS1/PBL2UMP oligomers into pentameric resistosomes (Wang et al., 2019a) that localize to the PM to trigger downstream immune responses (Wang et al., 2019a; Bi et al., 2021).
ZAR1 resistosomes were recently shown to exhibit cation channel activity (Bi et al., 2021). The N-terminal α-helices in ZAR1 form a funnel-shaped structure with a negatively charged carboxylate ring, which allows cations to pass through into the cytosol. Co-expression of ZAR1 with RKS1, PBL2, and AvrAC in plant protoplasts results in cytosolic calcium influx, ROS accumulation, and the perturbation of chloroplasts and vacuoles (Bi et al., 2021). Robust ROS accumulation during ZAR1 activation is likely caused by the activation of multiple downstream signaling components, such as the NADPH oxidases, since the CPKs are activated by cytosolic calcium influx (Gao et al., 2013). In addition, multiple CPKs and RbohD have been shown to be phosphorylated during RPS2 activation (Gao et al., 2013; Kadota et al., 2019). Defense-related transcription factors are also likely activated by cytosolic calcium influx (Boudsocq et al., 2010; Gao et al., 2013). The perturbation of chloroplasts and vacuoles is quickly followed by the loss of PM integrity and cellular rupture (Bi et al., 2021; Figure 4B). How these processes are regulated by immune signaling components and their relationships to transcriptional reprogramming are currently unclear.
The signaling pathway of helper-NLR-dependent sensor NLRs
The majority of sensor NLRs requires helper NLRs to mediate immunity. In solanaceous plants, the NB-LRR REQUIRED FOR HR-ASSOCIATED CELL DEATH proteins (collectively known as NRCs) are required for immunity and hypersensitive cell death response (HR) mediated by multiple sensor NLRs (Wu et al., 2017a). Interestingly, the N-terminal CC domain in ZAR1 contains a “MADA motif” that is also present in NRCs (Adachi et al., 2019a). This suggests that perhaps NRCs also form cation channels with α-helices following activation. In Arabidopsis, ADR1s and NRG1s are required for resistance and HR mediated by some CNLs and many TNLs (Bonardi et al., 2011; Castel et al., 2019a; Wu et al., 2019; Saile et al., 2020). Following effector recognition, TNLs also oligomerize into resistosomes to mediate resistance (Ma et al., 2020a; Martin et al., 2020). The Arabidopsis RPP1 recognizes the Hpa effector ATR1, and N. benthamiana ROQ1 recognizes the Xanthomonas effector XopQ. These effectors are recognized by the LRR and post-LRR domain, which likely leads to conformational changes and oligomerization of these TNLs into tetrameric resistosomes (Ma et al., 2020a; Martin et al., 2020).
The TIR domains of TNLs are brought into close proximity following oligomerization, activating NADase activity and producing v-cADPR (Horsefield et al., 2019; Wan et al., 2019a; Duxbury et al., 2020; Ma et al., 2020a; Martin et al., 2020). TIR domains also exhibit 2′,3′-cAMP/cGMP synthetase activity by hydrolyzing RNA or DNA (Yu et al., 2021). v-cADPR and 2′,3′-cAMP/cGMP are proposed to be signaling molecules that activate downstream signaling components (Horsefield et al., 2019; Wan et al., 2019a; Yu et al., 2021). Following the activation of TNLs, the EP-domain containing proteins (EP-proteins) SAG101 and EDS1 associate with NRG1 (Sun et al., 2021). Similarly, the activation of TNLs also leads to the association of the EP-proteins PAD4 and EDS1 with ADR1 (Wu et al., 2021b). These associations lead to the activation of these signaling components, which in turn activate downstream immune responses, such as defense-related gene expression and HR (Lapin et al., 2019; Sun et al., 2021). The RNLs ADR1 and NRG1 were also recently shown to function as calcium channels to activate immunity (Jacob et al., 2021). It is conceivable that the association and activation of helper RNLs and EP-proteins induces calcium influx and triggers downstream immune responses (Figure 4C).
Physiological responses induced by RLKs
Following ligand perception, the PRR co-receptor BAK1 and the RLCK BIK1 are phosphorylated (Lin et al., 2014; Perraki et al., 2018). This leads to the phosphorylation and activation of multiple signaling components (Macho and Zipfel, 2014). The activation of multiple calcium channels and NADPH oxidases leads to calcium influx, stomatal closure, ROS production, and callose deposition (Luna et al., 2011; Kadota et al., 2014; Li et al., 2014; Thor et al., 2020). The activation of CPKs and MAPKs leads to transcriptional reprograming and the biosynthesis of defense-related hormones (Boudsocq et al., 2010). In Arabidopsis, MPK3/MPK6 activate 1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID SYNTHASE (ACS) isoforms ACS2 and ACS6, which are involved in ET biosynthesis (Liu and Zhang, 2004; Han et al., 2010). The transcription factors SYSTEMIC-ACQUIRED RESISTANCE DEFICIENT 1 (SARD1) and CALMODULIN-BINDING PROTEIN 60 G (CBP60g) are required for PTI-induced upregulation of SA biosynthesis genes, such as ISOCHORISMATE SYNTHASE 1 (ICS1), EDS5, and AVRPPHB SUSCEPTIBLE 3 (PBS3; Zhang et al., 2010b; Sun et al., 2015). SARD1 and CBP60g are also required for the upregulation of pipecolic acid (N-hydroxyl-pipecolic acid [NHP])-biosynthesis genes, such as FLAVIN-CONTAINING MONOOXYGENASE 1 (Sun et al., 2015; Liu et al., 2020; Figure 5).
Figure 5.
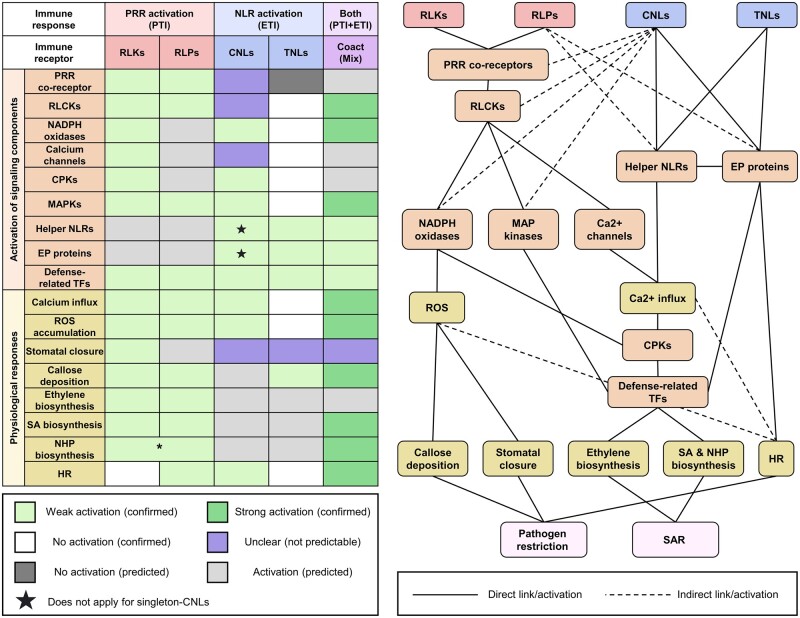
Signaling components and physiological responses activated by different modes of action of immune receptors. (Left) Tabular summary of signaling components and physiological responses activated by RLKs, RLPs, CNLs, TNLs, and coactivation of PRRs and NLRs. Green (weak or strong activation) and white (no activation) shading represent confirmed responses from publications. Gray shading indicates predicted responses. Purple shading represents unclear responses that cannot be predicted. Asterisks indicate inoculation with the bacterial pathogen P. syringae pv. maculicola (Psm) leads to NHP accumulation (Wang et al., 2018c; Liu et al., 2020). (Right) PRR and NLR signaling network. Activation of PRRs (red) and NLRs (blue) lead to the activation of downstream signaling components (orange) and physiological responses (yellow), which result in resistance against pathogens (pink). Note that the activation of physiological responses can vary between immune receptors and are dependent on specific PRRs/NLRs.
Physiological responses induced by RLPs
Similar to RLKs, RLPs also require PRR co-receptors, RLCKs, CPKs, and MAPKs to transduce immune signals (Piedras et al., 1998; Romeis et al., 1999, 2000; Rowland et al., 2005; González-LamotHe et al., 2006; Yang et al., 2006; van den Burg et al., 2008). In Arabidopsis, nlp20-induced immune responses mediated by RLP23 require the co-receptors BAK1, SOBIR1, and multiple RLCKs such as PBL19/20/30/31/32 (Albert et al., 2015; Pruitt et al., 2021; Tian et al., 2020). The activation of RLP23 leads to changes in PM potential, an ROS burst, the phosphorylation of BIK1 and MAPKs, callose deposition, and SA and ET production, similar to the activation of FLS2 (Wan et al., 2019b). In addition, flg22 and nlp20 induce highly overlapping transcriptional reprogramming in Arabidopsis (Wan et al., 2019b; Bjornson et al., 2021). Thus, RLKs and RLPs induce overlapping responses due to the activation of similar downstream signaling components. However, the individual activation of multiple RLPs, such as SlCf-4, SlCf-9, and AtRLP23, leads to the HR, perhaps due to the prolonged activation of downstream signaling components (Jones et al., 1994; Thomas et al., 1997; Rowland et al., 2005; Albert et al., 2015). PAD4, EDS1, and ADR1 are required for both RLK- and RLP-mediated immunity (Pruitt et al., 2021; Tian et al., 2021). Thus, EP-proteins and helper NLRs might also be activated during some PTI signaling, although it remains to be established whether EP proteins play a primary or secondary role in defense signaling (Figure 5).
Physiological responses induced by CNLs alone
Activation of the Arabidopsis CNL RPS2 in the absence of PTI leads to the phosphorylation of RbohD (in Ser343/347), CPKs, and MAPKs (Gao et al., 2013; Tsuda et al., 2013; Kadota et al., 2019; Ngou et al., 2021a; Yuan et al., 2021). RPS2-induced RbohD phosphorylation and ROS production are dependent on BAK1/BKK1 and BIK1 (Yuan et al., 2021). However, it is currently unclear whether BAK1/BKK1 and BIK1 are directly or indirectly activated by CNLs. While the ZAR1 resistosome directly triggers calcium influx, other calcium channels may also be activated by CNLs (Bi et al., 2021). The activation of RPM1, RPS2, and RPS5 leads to MAPK activation and the HR (Ngou et al., 2021a). In addition, the activation of many CNLs leads to the upregulation of SA- and NHP-biosynthesis genes (Jacob et al., 2018; Ngou et al., 2021a). Thus, ET, SA, and NHP are likely to be produced during CNL activation (Figure 5).
Physiological responses induced by TNLs alone
Activation of the Arabidopsis TNL RRS1/RPS4 does not lead to the phosphorylation of BIK1, RbohD (in Ser39/343/347), MAPKs, calcium influx, ROS accumulation, or the HR (Ngou et al., 2020, 2021a). Thus, RLCKs, NADPH oxidases, calcium channels, or CPKs are unlikely to be activated by RRS1/RPS4 alone. Activation of RRS1/RPS4 induces weak callose deposition, perhaps via SA accumulation (Tateda et al., 2014; Ngou et al., 2021a). Activation of TNLs leads to the association of EP-proteins with helper NLRs, which induces transcriptional reprogramming (Saile et al., 2020; Sun et al., 2021; Wu et al., 2021b). Similar to CNLs, the activation of TNLs leads to the upregulation of SA- and NHP-biosynthesis genes (Ding et al., 2020; Ngou et al., 2021a). Thus, SA and NHP are likely to be produced during TNL activation (Figure 5).
Physiological responses induced by the co-activation of PRRs and NLRs
Co-activation of PRRs and NLRs (“PTI + ETI”) leads to the robust activation of BIK1, RbohD, and MPK3 (Tsuda et al., 2013; Su et al., 2018; Ngou et al., 2021a; Yuan et al., 2021). This results in stronger calcium influx, ROS accumulation, and callose deposition compared to PTI or ETI alone (Ngou et al., 2021a; Yuan et al., 2021). In addition, “PTI + ETI” leads to stronger accumulation of SA and NHP compared to PTI alone, which is likely due to the stronger expression of SA- and NHP-biosynthesis genes during ETI (Wang et al., 2018c; Castel et al., 2019a; Ding et al., 2020; Liu et al., 2020; Figure 5).
Regulation of PRR-mediated immunity
The PRR-signaling pathway is tightly regulated as the excessive activation of PRRs leads to autoimmunity and growth inhibition (Navarro et al., 2006; Albrecht et al., 2012; Huot et al., 2014).
Regulation of PRRs
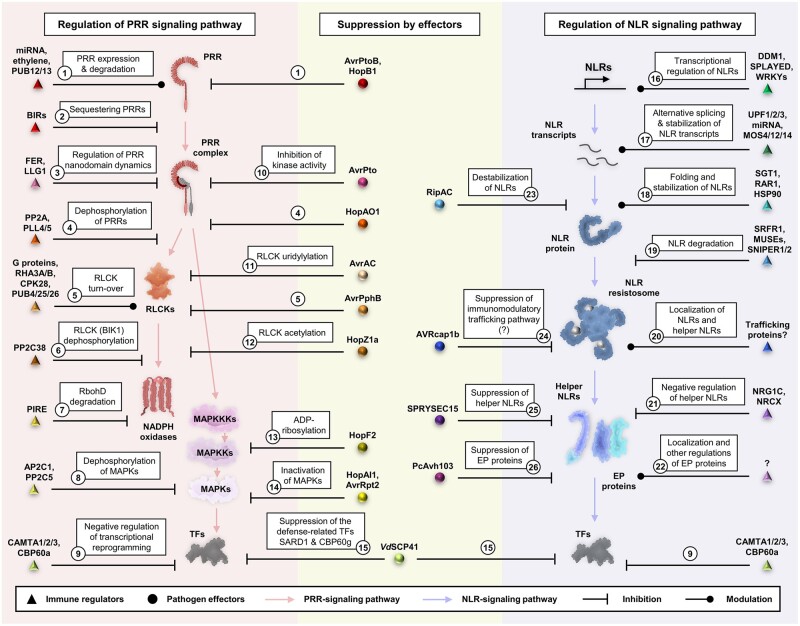
Both the transcript and protein levels of PRRs are regulated by multiple mechanisms. For example, the expression of FLS2 is regulated by the microRNA miR172b (Zou et al., 2018). The expression of FLS2 is also upregulated by ET (Boutrot et al., 2010). U-BOX DOMAIN-CONTAINING PROTEIN 12 (PUB12) and PUB13 mediate the polyubiquitination of FLS2, which leads to the endocytosis and degradation of this protein (Lu et al., 2011). Cf-4 also undergoes endocytosis upon Avr4 recognition (PostMa et al., 2016). The activation of PRRs and their co-receptors must also be regulated. BAK1-INTERACTING RECEPTOR (BIR)-LIKE KINASE 1 is an RLK that associates with and sequesters BAK1 to prevent the auto-activation of BAK1-associated PRRs (Gao et al., 2009; Ma et al., 2017; Hohmann et al., 2018). Following PAMP perception, the peptide RAPID ALKALINIZATION FACTOR 23 (RALF23) is perceived by a PRR complex composed of the CrRLK1L FERONIA (FER) and the LORELEI-LIKE-GPI ANCHORED PROTEIN 1. The perception of RALF23 by FER negatively regulates the formation of the FLS2–BAK1 complex (Stegmann et al., 2017; Xiao et al., 2019). FER regulates PM nanodomain organization to modulate PRR signaling (Gronnier et al., 2020). In addition, the phosphorylation status of PRRs is regulated by multiple protein phosphatases. In Arabidopsis, POLTERGEIST-LIKE 4 (PLL4) and PLL5 associate with EFR and negatively regulate elf18-induced responses (Holton et al., 2015). PROTEIN PHOSPHATASE 2A negatively regulates the phosphorylation status of BAK1 (Segonzac et al., 2014; Figure 6).
Figure 6.
Regulation and suppression of immunity by plant proteins and pathogen-derived effectors. (Left; red shading) regulation of the PRR signaling pathway by host proteins. Protein abundance and PTMs of PRRs and PRR signaling components are tightly regulated. (Middle; yellow shading) suppression of immunity by pathogen effectors. Many identified effectors suppress PTI via multiple mechanisms. Very few effectors that target the NLR signaling pathway have been identified so far. (Right; blue shading) regulation of the NLR signaling pathway by host proteins. Both the transcript and protein level of NLRs are tightly regulated by multiple processes. The regulation of signaling events post-NLR activation has not been well characterized. Numbers indicate the corresponding mechanisms of immune regulation.
Regulation of PRR-signaling components
In addition to PRRs, downstream signaling components are also regulated to prevent prolonged activation. As a central signaling component in the PRR-signaling pathway, the Arabidopsis RLCK BIK1 is regulated by multiple mechanisms. EXTRA-LARGE G PROTEIN 2 (XLG2) functions with other heterotrimeric G proteins to attenuate proteasome-mediated degradation of BIK1 (Liang et al., 2016). The turnover of BIK1 is regulated by CPK28, PUB4/25/26, and the E3 ubiquitin ligases RING-H2 FINGER A3A/B (Monaghan et al., 2014; Wang et al., 2018a; Derkacheva et al., 2020; Ma et al., 2020b). The phosphorylation status of BIK1 is also negatively regulated by the protein phosphatase PP2C38 (Couto et al., 2016). In addition to RLCKs, other PRR-signaling components must also be regulated. RbohD is ubiquitinated by the E3 ubiquitin ligase PIRE (PBL13 interacting RING domain E3 ligase), which leads to proteasome-mediated degradation (Lee et al., 2020). PHAGOCYTOSIS OXIDASE/ BEM1P (PB1) DOMAIN-CONTAINING PROTEIN negatively regulates ROS production by controlling the localization of RbohD (Goto et al., 2020). The PP2C phosphatases PP2C5 and AP2C1 negatively regulate the phosphorylation of MPK3 and MPK6 (Brock et al., 2010; Figure 6).
Regulation of NLR-mediated immunity
Similar to PRRs, the prolonged activation of NLRs also leads to autoimmunity. Thus, the regulation of both NLRs and downstream signaling components is important to prevent autoimmunity.
Regulation of NLRs
The expression of NLRs is regulated at multiple levels (van Wersch et al., 2020). The transcription of NLRs is regulated by chromatin-remodeling proteins such as DECREASE IN DNA METHYLATION 1, SWI/SNF CHROMATIN REMODELER SYD, and multiple WRKY transcription factors (Li et al., 2010b; Johnson et al., 2015; Lai and Eulgem, 2018). NLR transcript stability is also regulated by microRNAs and NONSENSE-MEDIATED mRNA DECAY factors, such as UP-FRAMESHIFT1/2/3 (Shivaprasad et al., 2012; Jung et al., 2020). NLR transcripts also undergo alternative splicing, which is regulated by some MODIFIER OF SUPPRESSOR OF NPR1-1 (SNC1; MOS) proteins such as MOS4/12/14 (Zhang and Gassmann, 2007; Xu et al., 2011, 2012).
REQUIRED FOR MLA12 RESISTANCE 1 (RAR1), SUPPRESSOR OF THE G2 ALLELE OF SKP1 (SGT1), and HEAT SHOCK PROTEIN 90 (HSP90) function together as protein chaperones to regulate the folding, localization, and turnover of NLRs (Azevedo et al., 2002; Peart et al., 2002; Takahashi et al., 2003; Shirasu, 2009). In addition, NLR protein turnover is regulated by the SGT1-interacting protein SUPPRESSORS OF RPS4-RLD, multiple MUTANT SNC1-ENHANCING proteins, and the E3 ligases SNIPER1 and SNIPER2 (Li et al., 2010a; Huang et al., 2016; Dong et al., 2018; Wu et al., 2020c).
The localization of the ZAR1 resistosome to the PM is required for ZAR1-mediated resistance (Wang et al., 2019a; Bi et al., 2021). In addition, the Arabidopsis importin-α nuclear transport receptor protein IMP-α3/MOS6 is required for SUPPRESSOR OF SNC1-mediated immunity (Lüdke et al., 2021). Thus, the localization of NLRs is important and is likely regulated by proteins involved in trafficking (Figure 6).
Regulation of NLR-signaling components
The correct localization of helper NLRs is likely important for signaling. For example, the helper NLR NRC4 accumulates at the extra-haustorial membrane following P. infestans infection (Duggan et al., 2021). In addition, the balanced activity of both cytosolic- and nuclear-EDS1 is required for full immunity (García et al., 2010). Thus, the localization of helper NLRs and NLR-signaling components is important for defense. The activity of NLR signaling components is also negatively regulated. The Arabidopsis RNL NRG1C functions as a negative regulator in NLR-mediated immunity; overexpressing NRG1C compromised TNL-mediated HR and resistance (Wu et al., 2021a). In addition, an atypical member of the NRC family, NRCX, negatively regulates other NRC members to modulate immunity (Adachi et al., 2021). Posttranslational modifications (PTMs) are important for the functions of both PRRs and NLRs. For example, the phosphorylation of the C-terminus of the TNL RRS1-R is crucial for its recognition of the effector PopP2 (Guo et al., 2020). It is currently unclear whether PTMs are important for the activation and/or stability of NLR-signaling components. Perhaps, EP-proteins and helper NLRs must also undergo PTMs in order to function properly. The additional regulation of NLR-signaling components pre-NLR activation and postNLR activation remains to be investigated (Figure 6).
Suppression of immunity by effectors
Multiple effectors have been shown to target both the PRR- and NLR-signaling pathways. Here, we summarize our knowledge of effectors reported to target PTI or ETI. Unless specified, the effectors mentioned in this section are from various P. syringae strains. AvrPtoB is an E3 ubiquitin ligase that induces the degradation of FLS2 (Göhre et al., 2008; Lu et al., 2011). HopB1 specifically degrades activated BAK1 (Li et al., 2016). AvrPto targets SOBIR1 and the FLS2–BAK1 complex by inhibiting their kinase activities (Xing et al., 2007; Shan et al., 2008; Xiang et al., 2008; Meng and Zhang, 2013; Wu et al., 2017b). Similarly, the conserved Colletotrichum effector NIS1 also targets receptor kinase complexes (Irieda et al., 2019). The tyrosine phosphatase HopAO1 directly dephosphorylates EFR (Macho et al., 2014). As RLCKs are central immune regulators, they are targeted by multiple effectors. AvrAC from X. campestris uridylylates BIK1 and PBL2 (Feng et al., 2012; Wang et al., 2015). HopZ1a acetylates RLCKs, and AvrPphB is a cysteine protease that degrades RLCKs such as BIK1, PBS1, and PBL1 (Zhang et al., 2010a; Bastedo et al., 2019). Other downstream PRR signaling components are also targeted by effectors. The ADP-ribosyltransferase HopF2 targets both BAK1 and MKK5 to suppress PTI signaling (Wang et al., 2010; ZHou et al., 2014). HopAI1 inactivates MPK3, MPK4, and MPK6 via its phosphothreonine lyase activity (Zhang et al., 2007). AvrRpt2 suppresses MPK4/11 activation (Eschen-Lippold et al., 2016). Interestingly, many parallel mechanisms are employed to suppress the same PRR-signaling node in different hosts by different pathogens (Figure 6).
Phosphorylation of SGT1 by MAPKs is required for NLR activation, implying that NLRs are regulated by SGT1 following PTI-induced MAPK activation (Hoser et al., 2013; Yu et al., 2020). The R. solanacearum effector RipAC prevents MAPK-mediated phosphorylation of SGT1, which suppresses NLR-mediated immunity (Yu et al., 2020). Two effectors were recently shown to suppress NRC-mediated HR. The P. infestans effector AVRcap1b and the cyst nematode effector SPRYSEC15 can suppress autoimmunity induced by autoactive alleles of NRC2 and NRC3 (Derevnina et al., 2021). Suppression of NRC2 and NRC3 by AVRcap1b is dependent on the membrane trafficking-associated protein TARGET OF MYB 1-LIKE PROTEIN 9A (NbTOL9a; Derevnina et al., 2021). AVRcap1b suppresses NRC2 and NRC3 by directly interacting with their NB-ARC domains (Derevnina et al., 2021). Another Phytophthora effector (from Phytophthora capsici), PcAvh103, suppresses immunity by promoting the disassociation of the EDS1–PAD4 complex (Li et al., 2020). More studies are needed to identify pathogen effectors that target the NLR signaling pathway.
In Arabidopsis, the transcription factors CALMODULIN-BINDING TRANSCRIPTION ACTIVATOR 1/2/3 (CAMTA1/2/3) and CBP60a negatively regulate defense-induced transcriptional reprogramming (Truman et al., 2013; Kim et al., 2020; Sun et al., 2020). Pathogens also target defense-related transcription factors to suppress immunity. For example, the R. solanacearum effector PopP2 acetylates and inhibits WRKY transcription factors to suppress immunity (Le Roux et al., 2015; Sarris et al., 2015; Zhang et al., 2017b). In addition, the Verticillium dahliae effector VdSCP41 inhibits SARD1 and CBP60g to facilitate its proliferation (Qin et al., 2018; Figure 6).
The interactions between PTI and ETI
While PRR- and NLR-mediated immunity has been extensively studied for the last 20 years, it has not been clear how or if these defense mechanisms interact. NLR-mediated immunity is mostly activated in the presence of microbes or PAMPs. Most studies on NLR-mediated immunity have involved transient expression-based comparisons between PTI and “PTI + ETI.” The activation of NLRs in the absence of PTI has not been extensively studied until recently. There have been multiple reports on the different interactions between these two immune systems. Here, we describe three situations in which PTI and ETI interact with each other.
NLRs guard the PRR-signaling pathway
Many effectors target the PRR-signaling pathway. Plants have evolved multiple NLRs to detect these effectors via the guarding of PRR-signaling components or decoys. As a result, many PRRs and PRR-signaling loss-of-function mutants, such as the Arabidopsis mutants bak1-4 bkk1-1, bik1, cngc2/4, rbohd/f, mekk1, mkk1/2, mpk4, and camta3, exhibit autoimmune phenotypes (Torres et al., 2002; Roux et al., 2011; Zhang et al., 2012; Chen et al., 2016; Liu et al., 2017; Lolle et al., 2017; Kadota et al., 2019; Tian et al., 2019). The autoimmunity observed in some of these mutants is caused by the activation of multiple NLRs. The TNL CONSTITUTIVE SHADE-AVOIDANCE 1 guards both BIR3 and BAK1 (Schulze et al., 2021). In addition, bak1-3 bkk-1-autoimmunity and HopB1-triggered immunity are dependent on ADR1s (Wu et al., 2020b). RLCKs are targeted by multiple effectors. The CNL ZAR1 together with the RLCK RKS1 monitor PBL2, and the CNL RPS5 monitors PBS1, to reverse ETS (Shao et al., 2003; Zhang et al., 2010a; Wang et al., 2015). The CNL SUMM2 guards and senses the disruption of the MEKK1–MKK1/2–MPK4 kinase cascade via CALMODULIN-BINDING RECEPTOR-LIKE CYTOPLASMIC KINASE 3, a substrate protein of MPK4 (Zhang et al., 2012, 2017a). SUMM2 also detects the P. syringae effector HopAI1, which inhibits MPK4 kinase activity (Zhang et al., 2012). The TNL RPS6 also contributes to HopAI1-triggered immunity (Takagi et al., 2019). Whether the autoimmunity in bik1, cgnc2/4, and rbohd/f is dependent on NLRs remains unclear. Other NLRs that guard the PRR-signaling pathway remain to be identified (Figure 7A).
Figure 7.
Interactions between PRR- and NLR-mediated immunity. A, NLRs guarding the PRR-signaling pathway. Multiple PRR-signaling components are suppressed by effectors. NLRs guard these signaling components and reverse susceptibility triggered by these effectors. Question marks indicate unidentified effectors or NLRs. B, Tabular summary of signaling components required for PRR- and NLR-mediated immunity. Green shading represents confirmed requirement from publications. Gray shading indicates predicted requirement. Purple shading represents unclear requirement that cannot be predicted. C, Mechanisms involved in the mutual potentiation between PRR- and NLR-mediated immunity. Transcriptomic data were obtained from previously published data (Bjornson et al., 2021; Ngou et al., 2021a). Numbers indicate the corresponding mechanisms to potentiate PRR- or NLR-mediated immunity to achieve robust resistance against pathogens.
Interdependency of signaling components between PRRs and NLRs
PRR co-receptors, RLCKs, NADPH oxidases, calcium channels, CPKs, and MAPKs are considered to be canonical PRR-signaling components, while EP proteins and helper NLRs are considered to be canonical NLR-signaling components. However, recent studies indicated that PRR-mediated resistance is dependent on canonical NLR-signaling components and vice versa (Ngou et al., 2021a; Pruitt et al., 2021; Tian et al., 2021; Yuan et al., 2021; Figure 7B). As mentioned, flg22- and nlp20-induced resistance is partially dependent on EDS1, PAD4, SAG101, ADR1s, and NRG1s (Pruitt et al., 2021; Tian et al., 2021). Pruitt et al. (2021) proposed that EP-proteins and helper NLRs are activated by RLPs through interactions between RLP co-receptors (SOBIR1), EP-proteins, and helper NLRs, although it remains to be determined whether EP-proteins play a primary or secondary role in RLP defense signaling. Another report, however, suggested that the activation of PRRs leads to increased expression of multiple NLRs and other TIR-domain-containing proteins, promoting downstream signaling (Tian et al., 2021). These two hypotheses are not mutually exclusive, and the exact mechanisms by which PRR-mediated immunity involves NLR-signaling components remain to be determined.
NLR-mediated immunity is also dependent on PRRs and multiple PRR-signaling components. In Arabidopsis, RPS2-, RPS5-, and RRS1/RPS4-mediated resistance is dependent on BAK1 and BKK1 (Ngou et al., 2021a; Yuan et al., 2021). RPS2-mediated resistance is also dependent on BIK1 and RbohD (Kadota et al., 2019; Yuan et al., 2021). Both RPM1- and RPS2-mediated resistance and the HR are dependent on CPK1/2/5/6 (Gao et al., 2013). The activation of MPK3 and MPK6 is also required for the HR and resistance mediated by multiple NLRs including RPM1, RPS2, RPS5, and RRS1/RPS4 (Su et al., 2018). One of the proposed key mechanisms by which ETI halts pathogen infection is to potentiate and restore PTI from turnover and the action of pathogen effectors (Ngou et al., 2021a; Yuan et al., 2021). As a result, PRRs and PRR-signaling components are required for NLR-mediated resistance. The molecular mechanisms by which ETI potentiates PTI will be discussed in the next section.
Mutual potentiation between PRR- and NLR-mediated immunity
Activation of the TNLs RRS1/RPS4 and RPP4 using an estradiol-inducible recognized effector (ETI without PTI) did not trigger the HR. The presence of PAMPs/MAMPs restored the HR induced by these TNLs (Ngou et al., 2020, 2021a). Similarly, the HR induced by the CNLs RPM1, RPS2, and RPS5 was also potentiated by the activation of PRRs (Ngou et al., 2021a). In addition, the HR and resistance induced by RPS2 are compromised in PRR mutants (Ma et al., 2012; Yuan et al., 2021). There are a few possible mechanisms by which PRRs potentiate NLR-induced immunity. First, the activation of PRRs could induce the expression of NLRs and NLR-signaling components (Navarro et al., 2004; Bonardi et al., 2011; Brendolise et al., 2018; Jung et al., 2020). A recent transcriptomics study suggested that the activation of different PRRs induces highly overlapping transcriptional changes (Bjornson et al., 2021). Indeed, the activation of six distinct PRRs led to the upregulation of genes encoding most TNLs, CNLs, EP-proteins, and helper NLRs in Arabidopsis (Bjornson et al., 2021; Figure 7C; Supplemental Data Set 3). The increased abundance of these proteins might therefore “prime” the activation of NLRs upon effector recognition. Second, the activation of PRRs might prime NLR-mediated immunity via PTMs. Upon PAMP perception, SGT1 is phosphorylated by MAPKs, which is important for the stability of NLRs (Yu et al., 2020). In addition, nonsense-mediated decay of NLR transcripts is inhibited upon PAMP recognition (Jung et al., 2020). Thus, the stability of NLRs can be affected by both transcriptional and posttranscriptional modifications activated by PTI. Conceivably, EP proteins and helper NLRs might also be primed via PTMs induced by PTI. Flg22 treatment led to reduced polyubiquitination levels of EDS1 (Grubb et al., 2021; Ma et al., 2021). Whether and how PTI primes NLR-signaling components remain to be investigated.
The activation of NLRs potentiates PAMP-induced cellular responses, such as ROS production, callose deposition, and defense-related gene expression (Ngou et al., 2021a). The activation of multiple PRR signaling components, such as BIK1, RbohD, and MPK3, is also potentiated by ETI (Ngou et al., 2021a; Yuan et al., 2021). ETI induces the transcript and protein accumulation of SOBIR1, BAK1, BIK1, RbohD, and MPK3 (Ngou et al., 2021a). Transcriptomic analysis confirmed that multiple PRR signaling components are also upregulated upon the activation of RRS1/RPS4. These include CPK1/2/5/6, XLG2, and the calcium channels OSCA1.3, CNGC19/20, GLR2.7/2.8/2.9 (Ngou et al., 2021a; Figure 7C; Supplemental Data Set 4). Interestingly, the transcript levels of BIK1, MPK3, and RbohD are only transiently upregulated during ETI. However, the protein levels of these genes remain upregulated for an extensive period of time (Ngou et al., 2021a). This implies that PTMs or other posttranscriptional mechanisms might also influence the stability of PRR-signaling components during ETI. The protein abundance of PRR signaling components, such as BAK1, BIK1, and RbohD, is tightly regulated by multiple processes (Figure 6). How ETI regulates or affects these processes remains unclear. In addition, calcium influx induced by NLRs might contribute to the potentiation of PTI through CPKs (Bi et al., 2021; Jacob et al., 2021; Ngou et al., 2021b). To summarize, PTI and ETI mutually potentiate each other through multiple mechanisms to induce robust immunity against pathogens (Figure 7C).
Historic overview of research in PTI and future challenges
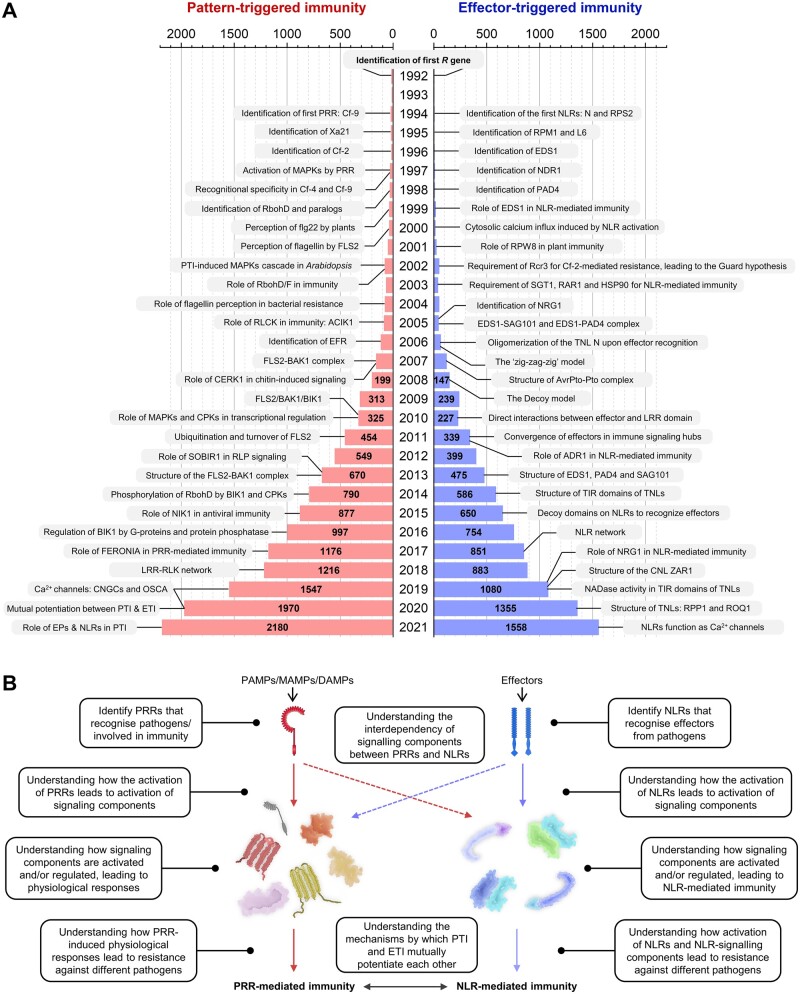
Researchers identified the first PRR-encoding gene, Cf-9, back in 1994 (Jones et al., 1994). Multiple PRR genes, such as Xa21, Cf-2, Cf-4, FLS2, EFR, and RLP23, were subsequently identified and used as models to study PTI (Song et al., 1995; Dixon et al., 1996; Thomas et al., 1997; Gómez-Gómez and Boller, 2000; Zipfel et al., 2006). Researchers then explored PRR-induced physiological responses and identified multiple signaling components. The activation of MAPKs by cell-surface receptors were reported back in 1997 (Ligterink et al., 1997) and was verified for Cf- genes 2 years later (Romeis et al., 1999). In tobacco (N. tabacum), the perception of PAMPs leads to the activation of wounding-induced protein kinase (WIPK) and SA-induced protein kinase (SIPK; Zhang and Klessig, 1998; Yang et al., 2001). WIPKs and SIPKs are orthologs of the subsequently identified Arabidopsis MPK3 and MPK6, respectively (Asai et al., 2002). Accumulation of ROS and callose deposition during infection were also reported in 1997 (Thordal-Christensen et al., 1997), and for Cf-initiated responses (Piedras et al., 1998). Researchers identified the human Rbohs in Arabidopsis and showed that two of these (RbohD and RbohF) are required for ROS production during infection (Torres et al., 1998, 2002). It was unclear how these signaling components were activated by PRRs until the identification of the PRR co-receptors and RLCKs. BAK1 was identified as a co-receptor essential for FLS2-mediated resistance in 2007 (Chinchilla et al., 2007). In the same year, CERK1 was also shown to be essential for chitin-mediated immunity (Miya et al., 2007). In 2013, SOBIR1 was identified as a co-receptor of RLPs, and the structure of the FLS2/BAK1 receptor complex was also defined (Liebrand et al., 2013; Sun et al., 2013). In 2018, a genome-wide analysis of Arabidopsis LRR–RLKs interactions was reported, further supporting the theory that PRRs interact with each other to modulate and transduce signals (Smakowska-Luzan et al., 2018). Tomato ACIK1 was the first RLCK shown to be an essential signaling component in PRR-mediated immunity (Rowland et al., 2005). The Arabidopsis ortholog BIK1 was subsequently shown to be a central PRR-signaling component (Lu et al., 2010; Zhang et al., 2010a). RbohD, MAPKKKs, and multiple calcium channels were shown to be phosphorylated by RLCKs, which leads to downstream immune responses (Boudsocq et al., 2010; Kadota et al., 2014; Li et al., 2014; Yamada et al., 2016; Bi et al., 2018; Tian et al., 2019; Thor et al., 2020; Figure 8A).
Figure 8.
Historic overview of PTI and ETI and future challenges. A, Discoveries in PTI (left) and ETI (right) in the past 30 years. Bar charts represent the number of “plant biology” publications that mentioned “pattern-trigger immunity” (red) and “effector-triggered immunity” (blue). Data obtained from Dimensions (https://www.dimensions.ai/). B, Future challenges and outlook in plant immunity research.
More than 60 immunity-related PRRs with known ligands have now been identified. Arabidopsis EFR has been introduced into multiple plant species, such as tomato, rice, orange, and apple, providing broadspectrum resistance to many bacteria (Lacombe et al., 2010; Schwessinger et al., 2015; Mitre et al., 2021; Piazza et al., 2021). Therefore, the identification of novel PRRs that recognize PAMPs or other elicitors would provide resources to engineer disease-resistant crops. Other challenges in PRR biology include trying to understand how PRRs activate downstream signaling components and physiological responses, how these processes are regulated and suppressed by effectors, and how resistance against pathogens is achieved (Figure 8B).
Historic overview of research in ETI and future challenges
Arabidopsis RPS2 and the tobacco N gene were the first reported NLR genes (Bent et al., 1994; Mindrinos et al., 1994; Whitham et al., 1994). Multiple NLRs, including RPM1 and L6, were subsequently identified (Grant et al., 1995; Lawrence et al., 1995). Understanding how NLRs detect effectors has led to multiple models. The guard hypothesis was proposed to explain how the protein kinase Pto confers Prf-dependent recognition of AvrPto (Van der Biezen and Jones, 1998). Many other examples have emerged that are consistent with this hypothesis, such as the requirement of the protease Rcr3 for Cf-2-mediated resistance (Van der Biezen and Jones, 1998; Dangl and Jones, 2001; Krüger et al., 2002). The decoy model was then proposed, which is further supported by the discovery of integrated decoy domains in NLRs (van der Hoorn and Kamoun, 2008; Cesari et al., 2014; Le Roux et al., 2015; Sarris et al., 2015, 2016). The discovery of NRCs led to the concept of NLR networks (Gabriëls et al., 2007; Wu et al., 2017a, 2018). Following the identification of multiple NLRs, researchers identified multiple genetic components required for NLR-mediated immunity. These include EDS1, NDR1, PAD4, RPW8, SGT1, RAR1, HSP90, SAG101, NRG1s, and ADR1s (Parker et al., 1996; Century et al., 1997; Zhou et al., 1998; Falk et al., 1999; Xiao et al., 2001; Azevedo et al., 2002; Takahashi et al., 2003; Feys et al., 2005; Peart et al., 2005; Bonardi et al., 2011). EDS1 was later shown to co-function with SAG101 and PAD4 to mediate HR and resistance during ETI (Feys et al., 2001, 2005; Wagner et al., 2013; Sun et al., 2021; Wu et al., 2021b). Similarly, ADR1 and NRG1 have been shown to function downstream of multiple sensor NLRs to mediate the HR and resistance (Castel et al., 2019a; Wu et al., 2019; Saile et al., 2020). How sensor NLRs activate these signaling components is currently under investigation. v-cADPR produced by TIR domains might contribute to the activation of EP-proteins and helper NLRs (Horsefield et al., 2019; Wan et al., 2019a, 2019b). NLRs were shown to oligomerize and trigger cytosolic calcium influx following effector recognition (Grant et al., 2000; Mestre and Baulcombe, 2006). The discovery of the structures of multiple NLR resistosomes proved that the oligomerization of NLRs is required for resistance, likely through the formation of cation channels (Wang et al., 2019a; Ma et al., 2020a; Martin et al., 2020; Bi et al., 2021; Jacob et al., 2021). However, oligomerization of TIR domains imposed by an NLRC4 scaffold is sufficient to activate defense (Duxbury et al., 2020; Figure 8A).
More than 140 NLRs with known recognized effectors have been identified (Kourelis and Kamoun, 2020). Cross-species transfer of NLR “stacks” provides durable resistance against pathogens (Jones et al., 2003; Mukhtar, 2013; Ghislain et al., 2019; Luo et al., 2021; Witek et al., 2021). Identification of novel NLRs will provide resources to engineer crop resistance against multiple pathogens. Current challenges in NLR biology include understanding how NLRs activate downstream signaling components, how these signaling components then trigger immune responses, how these processes are regulated and suppressed by effectors, and how NLRs and PRRs co-function to achieve resistance against pathogens (Figure 8B).
Conclusion and perspectives
Plants respond to pathogens using a two-tier innate immune system activated by both cell-surface and intracellular immune receptors. The perception of PAMPs/MAMPs/DAMPs/HAMPs on the cell surface leads to PRR-mediated immunity, and the recognition of effectors leads to intracellular NLR-mediated immunity. The first plant Resistance (R) gene, Hm1, was cloned back in 1992 (Johal and Briggs, 1992). Many immune receptors have been identified since 1994, when the first PRR and NLRs were identified. Tremendous efforts have been made to understand the PRR- and NLR-signaling pathways. PRRs and NLRs utilize some overlapping but also unique signaling components to activate each of their downstream physiological responses, which thwart pathogen proliferation. Both signaling pathways are tightly regulated to prevent autoimmunity, while being suppressed by pathogen effectors. Recent studies have shown that PRR- and NLR-mediated immunity can be mutually potentiated and are dependent on each other. Great opportunities for novel discoveries remain in addressing the following challenges in the research of plant immunity: (1) identifying novel immune receptors; (2) understanding the signaling pathways and physiological responses triggered by both cell-surface and intracellular immune receptors; (3) understanding how immunity is intrinsically regulated and manipulated by external biotic and/or abiotic factors; (4) understanding the vastly diverse mechanisms by which plants resist pathogen infections; and (5) understanding how different immune systems function synergistically during infections. These challenges overlap with some of the “top 10 unanswered questions in molecular plant-microbe interactions” (Harris et al., 2020) and will shape our understanding of plant immunity in the coming decades (Figure 8B).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Data Set 1. PRRs involved in plant immunity.
Supplemental Data Set 2. NLRs involved in plant immunity.
Supplemental Data Set 3. Expression of ETI-related genes during PTI.
Supplemental Data Set 4. Expression of PTI-related genes during ETI.
Supplementary Material
Acknowledgments
We thank Samuel Warner, Shanshan Wang, and Jack Rhodes for discussions and suggestions. We thank the Gatsby Foundation for funding to the J.D.G.J. laboratory. B.P.M.N was supported by the Norwich Research Park Biosciences Doctoral Training Partnership from the Biotechnology and Biological Sciences Research Council (BBSRC) (grant agreement BB/M011216/1); P.D. acknowledges support from the the Future Leader Fellowship from BBSRC (grant agreement BB/R012172/1).
Conflict of interest statement. None declared.
Contributor Information
Bruno Pok Man Ngou, The Sainsbury Laboratory, University of East Anglia, Norwich NR4 7UH, UK; RIKEN Center for Sustainable Resource Science, Yokohama, Japan.
Pingtao Ding, The Sainsbury Laboratory, University of East Anglia, Norwich NR4 7UH, UK; Institute of Biology Leiden, Leiden University, Leiden 2333 BE, The Netherlands.
Jonathan D G Jones, The Sainsbury Laboratory, University of East Anglia, Norwich NR4 7UH, UK.
B.P.M.N., P.D., and J.D.G.J. conceptualized the review outline. B.P.M.N. curated information, prepared the figures, and wrote the original draft, and B.P.M.N., P.D., and J.D.G.J. reviewed and edited the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: Bruno Pok Man Ngou (pokman.ngou@riken.jp).
References
- Adachi H, Contreras MP, Harant A, Wu CH, Derevnina L, Sakai T, Duggan C, Moratto E, Bozkurt TO, Maqbool A, et al. (2019a) An N-terminal motif in NLR immune receptors is functionally conserved across distantly related plant species. eLife 8: e49956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adachi H, Derevnina L, Kamoun S (2019b) NLR singletons, pairs, and networks: evolution, assembly, and regulation of the intracellular immunoreceptor circuitry of plants. Curr Opin Plant Biol 50: 121–131 [DOI] [PubMed] [Google Scholar]
- Adachi H, Sakai T, Harant A, Duggan C, Bozkurt TO, Wu C, Kamoun S (2021) An atypical NLR protein modulates the NRC immune receptor network. BioRxiv doi: 10.1101/2021.11.15.468391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera-Galvez C, Champouret N, Rietman H, Lin X, Wouters D, Chu Z, Jones JDG., Vossen JH, Visser RGF, Wolters PJ, et al. (2018) Two different R gene loci co-evolved with Avr2 of Phytophthora infestans and confer distinct resistance specificities in potato. Stud Mycol 89: 105–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert I, Albert M, Feiler C, Imkampe J, Brancato C, Raaymakers TM, Oome S, Wallmeroth N, Zhang H, Hedrich R, et al. (2015) An RLP23-SOBIR1-BAK1 complex mediates NLP-triggered immunity. Nat Plants 1: 15140. [DOI] [PubMed] [Google Scholar]
- Albrecht C, Boutrot F, Segonzac C, Schwessinger B, Gimenez-Ibanez S, Chinchilla D, Rathjen JP, de Vries SC, Zipfel C (2012) Brassinosteroids inhibit pathogen-associated molecular pattern-triggered immune signaling independent of the receptor kinase BAK1. Proc Natl Acad Sci USA 109: 303–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan AC, Lapidot M, Culver JN, Fluhr R (2001) An early tobacco mosaic virus-induced oxidative burst in tobacco indicates extracellular perception of the virus coat protein. Plant Physiol 126: 97–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson C, Khan MA, Catanzariti AM, Jack CA, Nemri A, Lawrence GJ, Upadhyaya NM, Hardham AR, Ellis JG, et al. (2016) Genome analysis and avirulence gene cloning using a high-density RADseq linkage map of the flax rust fungus, Melampsora lini. BMC Genomics 17: 667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai S, Furzer OJ, Cevik V, Kim DS, Ishaque N, Goritschnig S, Staskawicz BJ, Shirasu K, Jones JDG (2018) A downy mildew effector evades recognition by polymorphism of expression and subcellular localization. Nat Commun 9: 5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983 [DOI] [PubMed] [Google Scholar]
- Ashfield T, Keen NT, Buzzell RI, Innes RW (1995) Soybean resistance genes specific for different Pseudomonas syringae avirulence genes are allelic, or closely linked, at the RPG1 locus. Genetics 141: 1597–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashfield T, Ong LE, Nobuta K, Schneider CM, Innes RW (2004) Convergent evolution of disease resistance gene specificity in two flowering plant families. Plant Cell 16: 309–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashikawa I, Hayashi N, Abe F, Wu J, Matsumoto T (2012) Characterization of the rice blast resistance gene Pik cloned from Kanto51. Mol Breeding 30: 485–494 [Google Scholar]
- Ashikawa I, Hayashi N, Yamane H, Kanamori H, Wu J, Matsumoto T, Ono K, Yano M (2008) Two adjacent nucleotide-binding site-leucine-rich repeat class genes are required to confer Pikm-specific rice blast resistance. Genetics 180: 2267–2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ, Staskawicz BJ (2003) Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 112: 369–377 [DOI] [PubMed] [Google Scholar]
- Azevedo C, Sadanandom A, Kitagawa K, Freialdenhoven A, Shirasu K, Schulze-Lefert P (2002) The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science 295: 2073–2076 [DOI] [PubMed] [Google Scholar]
- Baggs E, Dagdas G, Krasileva KV (2017) NLR diversity, helpers and integrated domains: making sense of the NLR IDentity. Curr Opin Plant Biol 38: 59–67 [DOI] [PubMed] [Google Scholar]
- Baggs EL, Monroe JG, Thanki AS, O’Grady R, Schudoma C, Haerty W, Krasileva KV (2020) Convergent loss of an EDS1/PAD4 signaling pathway in several plant lineages reveals coevolved components of plant immunity and drought response. Plant Cell 32: 2158–2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey K, Cevik V, Holton N, Byrne-Richardson J, Sohn KH, Coates M, Woods-Tör A, Aksoy HM, Hughes L, Baxter L, et al. (2011) Molecular cloning of ATR5(Emoy2) from Hyaloperonospora arabidopsidis, an avirulence determinant that triggers RPP5-mediated defense in Arabidopsis. Mol Plant Microbe Interact 24: 827–838 [DOI] [PubMed] [Google Scholar]
- Ballvora A, Ercolano MR, Weiss J, Meksem K, Bormann CA, Oberhagemann P, Salamini F, Gebhardt C (2002) The R1 gene for potato resistance to late blight (Phytophthora infestans) belongs to the leucine zipper/NBS/LRR class of plant resistance genes. Plant J 30: 361–371 [DOI] [PubMed] [Google Scholar]
- Barragan CA, Wu R, Kim ST, Xi W, Habring A, Hagmann J, Van de Weyer AL, Zaidem M, Ho WH, Wang G et al. (2019) RPW8/HR repeats control NLR activation in Arabidopsis thaliana. PLoS Genet 15: e1008313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastedo DP, Khan M, Martel A, Seto D, Kireeva I, Zhang J, Masud W, Millar D, Lee JY, Lee AHY, et al. (2019) Perturbations of the ZED1 pseudokinase activate plant immunity. PLoS Pathog 15: e1007900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendahmane A, Köhn BA, Dedi C, Baulcombe DC (1995) The coat protein of potato virus X is a strain-specific elicitor of Rx1-mediated virus resistance in potato. Plant J 8: 933–941 [DOI] [PubMed] [Google Scholar]
- Bent AF, Kunkel BN, Dahlbeck D, Brown KL, Schmidt R, Giraudat J, Leung J, Staskawicz BJ (1994) RPS2 of Arabidopsis thaliana: a leucine-rich repeat class of plant disease resistance genes. Science 265: 1856–1860 [DOI] [PubMed] [Google Scholar]
- Berzal-Herranz A, de la Cruz A, Tenllado F, Díaz-Ruíz JR, López L, Sanz AI, Vaquero C, Serra MT, García-Luque I (1995) The Capsicum L3 gene-mediated resistance against the tobamoviruses is elicited by the coat protein. Virology 209: 498–505 [DOI] [PubMed] [Google Scholar]
- Bettgenhaeuser J, Hernández-Pinzón I, Dawson AM, Gardiner M, Green P, Taylor J, Smoker M, Ferguson JN, Emmrich P, Hubbard A, et al. (2021) The barley immune receptor Mla recognizes multiple pathogens and contributes to host range dynamics. Nat Commun 12: 6915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi G, Su M, Li N, Liang Y, Dang S, Xu J, Hu M, Wang J, Zou M, Deng Y et al. (2021) The ZAR1 resistosome is a calcium-permeable channel triggering plant immune signaling. Cell 184 [DOI] [PubMed] [Google Scholar]
- Bi G, Zhou Z, Wang W, Li L, Rao S, Wu Y, Zhang X, Menke FLH, Chen S, Zhou JM (2018) Receptor-like cytoplasmic kinases directly link diverse pattern recognition receptors to the activation of mitogen-activated protein kinase cascades in Arabidopsis. Plant Cell 30: 1543–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigeard J, Colcombet J, Hirt H (2015) Signaling mechanisms in pattern-triggered immunity (PTI). Mol Plant 8: 521–539. [DOI] [PubMed] [Google Scholar]
- Bjornson M, Pimprikar P, Nürnberger T, Zipfel C (2021) The transcriptional landscape of Arabidopsis thaliana pattern-triggered immunity. Nat Plants 7: 579–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhm H, Albert I, Oome S, Raaymakers TM, Van den Ackerveken G, Nürnberger T (2014) A conserved peptide pattern from a widespread microbial virulence factor triggers pattern-induced immunity in Arabidopsis. PLoS Pathog 10: e1004491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonardi V, Tang S, Stallmann A, Roberts M, Cherkis K, Dangl JL (2011) Expanded functions for a family of plant intracellular immune receptors beyond specific recognition of pathogen effectors. Proc Natl Acad Sci USA 108: 16463–16468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos JIB, Armstrong MR, Gilroy EM, Boevink PC, Hein I, Taylor RM, Zhendong T, Engelhardt S, Vetukuri RR, Harrower B, et al. (2010) Phytophthora infestans effector AVR3a is essential for virulence and manipulates plant immunity by stabilizing host E3 ligase CMPG1. Proc Natl Acad Sci USA 107: 9909–9914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudsocq M, Willmann MR, McCormack M, Lee H, Shan L, He P, Bush J, Cheng SH, Sheen J (2010) Differential innate immune signalling via Ca(2+) sensor protein kinases. Nature 464: 418–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourras S, Kunz L, Xue M, Praz C, Müller M, Kälin C, Schläfli M, Ackermann PM, Flückiger S, Parlange F, et al. (2019) The AvrPm3-Pm3 effector-NLR interactions control both race-specific resistance and host-specificity of cereal mildews on wheat. Nat Commun 10: 2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutrot F, Zipfel C (2017) Function, discovery, and exploitation of plant pattern recognition receptors for broad-spectrum disease resistance. Annu Rev Phytopathol 55: 257–286 [DOI] [PubMed] [Google Scholar]
- Boutrot F, Segonzac C, Chang KN, Qiao H, Ecker JR, Zipfel C, Rathjen JP (2010) Direct transcriptional control of the Arabidopsis immune receptor FLS2 by the ethylene-dependent transcription factors EIN3 and EIL1. Proc Natl Acad Sci USA 107: 14502–14507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozsoki Z, Cheng J, Feng F, Gysel K, Vinther M, Andersen KR, Oldroyd G, Blaise M, Radutoiu S, Stougaard J (2017) Receptor-mediated chitin perception in legume roots is functionally separable from Nod factor perception. Proc Natl Acad Sci USA 114: E8118–E8127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brendolise C, Martinez-Sanchez M, Morel A, Chen R, Dinis R, Deroles S, Peeters N, Rikkerink EHA, Montefiori M (2018) NRG1-mediated recognition of HopQ1 reveals a link between PAMP and Effector-triggered Immunity. BioRxiv doi: 10.1101/293050 [Google Scholar]
- Brock AK, Willmann R, Kolb D, Grefen L, Lajunen HM, Bethke G, Lee J, Nürnberger T, Gust AA (2010) The Arabidopsis mitogen-activated protein kinase phosphatase PP2C5 affects seed germination, stomatal aperture, and abscisic acid-inducible gene expression. Plant Physiol 153: 1098–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brulé D, Villano C, Davies LJ, Trdá L, Claverie J, Héloir MC, Chiltz A, Adrian M, Darblade B, Tornero P, et al. (2019) The grapevine (Vitis vinifera) LysM receptor kinases VvLYK1-1 and VvLYK1-2 mediate chitooligosaccharide-triggered immunity. Plant Biotechnol J 17: 812–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutus A, Sicilia F, Macone A, Cervone F, De Lorenzo G (2010) A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc Natl Acad Sci USA 107: 9452–9457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Aceti DJ, Sabat G, Song J, Makino SI, Fox BG, Bent AF (2013) Mutations in FLS2 Ser-938 dissect signaling activation in FLS2-mediated Arabidopsis immunity. PLoS Pathog 9: e1003313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Liang Y, Tanaka K, Nguyen CT, Jedrzejczak RP, Joachimiak A, Stacey G (2014) The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin-induced complex with related kinase CERK1. Elife 3: e03766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter ME, Helm M, Chapman AVE, Wan E, Restrepo Sierra AM, Innes RW, Bogdanove AJ, Wise RP (2019) Convergent evolution of effector protease recognition by arabidopsis and barley. Mol Plant Microbe Interact 32: 550–565 [DOI] [PubMed] [Google Scholar]
- Castel B, Ngou PM, Cevik V, Redkar A, Kim DS, Yang Y, Ding P, Jones JDG (2019a) Diverse NLR immune receptors activate defence via the RPW8-NLR NRG1. New Phytol 222: 966–980 [DOI] [PubMed] [Google Scholar]
- Castel B, Wu Y, Xiao S, Jones JD (2019b) An rpw8 quadruple mutant of Arabidopsis Col-0 is partially compromised in bacterial and fungal resistance. BioRxiv doi: 10.1101/839308 [Google Scholar]
- Catanzariti AM, Dodds PN, Ve T, Kobe B, Ellis JG, Staskawicz BJ (2010) The AvrM effector from flax rust has a structured C-terminal domain and interacts directly with the M resistance protein. Mol Plant Microbe Interact 23: 49–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catanzariti AM, Lim GTT, Jones DA (2015) The tomato I-3 gene: a novel gene for resistance to Fusarium wilt disease. New Phytol 207: 106–118 [DOI] [PubMed] [Google Scholar]
- Century KS, Shapiro AD, Repetti PP, Dahlbeck D, Holub E, Staskawicz BJ (1997) NDR1, a pathogen-induced component required for Arabidopsis disease resistance. Science 278: 1963–1965 [DOI] [PubMed] [Google Scholar]
- Cesari S, Bernoux M, Moncuquet P, Kroj T, Dodds PN (2014) A novel conserved mechanism for plant NLR protein pairs: the “integrated decoy” hypothesis. Front Plant Sci 5: 606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champouret N (2010) Functional genomics of Phytophthora infestans effectors and Solanum resistance genes. https://research.wur.nl/en/publications/functional-genomics-of-phytophthora-infestans-effectors-and-solan
- Chapman S, Stevens LJ, Boevink PC, Engelhardt S, Alexander CJ, Harrower B, Champouret N, McGeachy K, Van Weymers PSM, Chen X, et al. (2014) Detection of the virulent form of AVR3a from Phytophthora infestans following artificial evolution of potato resistance gene R3a. PLoS One 9: e110158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Cao Y, Li H, Kim D, Ahsan N, Thelen J, Stacey G (2017a) Extracellular ATP elicits DORN1-mediated RBOHD phosphorylation to regulate stomatal aperture. Nat Commun 8: 2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Upadhyaya NM, Ortiz D, Sperschneider J, Li F, Bouton C, Breen S, Dong C, Xu B, Zhang X, et al. (2017b) Loss of AvrSr50 by somatic exchange in stem rust leads to virulence for Sr50 resistance in wheat. Science 358: 1607–1610 [DOI] [PubMed] [Google Scholar]
- Chen R, Li H, Zhang L, Zhang J, Xiao J, Ye Z (2007) CaMi, a root-knot nematode resistance gene from hot pepper (Capsium annuum L.) confers nematode resistance in tomato. Plant Cell Rep 26: 895–905 [DOI] [PubMed] [Google Scholar]
- Chen T, Bi K, He Z, Gao Z, Zhao Y, Fu Y, Cheng J, Xie J, Jiang D (2016) Arabidopsis mutant bik1 exhibits strong resistance to Plasmodiophora brassicae. Front Physiol 7: 402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T (2021) Identification and characterization of the LRR repeats in plant LRR-RLKs. BMC Mol and Cell Biol 22: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D, Bauer Z, Regenass M, Boller T, Felix G (2006) The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell 18: 465–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D, Zipfel C, Robatzek S, Kemmerling B, Nürnberger T, Jones JDG, Felix G, Boller T (2007) A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448: 497–500 [DOI] [PubMed] [Google Scholar]
- Claverie M, Dirlewanger E, Bosselut N, Van Ghelder C, Voisin R, Kleinhentz M, Lafargue B, Abad P, Rosso MN, Chalhoub B, et al. (2011) The Ma gene for complete-spectrum resistance to Meloidogyne species in Prunus is a TNL with a huge repeated C-terminal post-LRR region. Plant Physiol 156: 779–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman AD, Maroschek J, Raasch L, Takken FLW, Ranf S, Hückelhoven R (2021) The Arabidopsis leucine-rich repeat receptor-like kinase MIK2 is a crucial component of early immune responses to a fungal-derived elicitor. New Phytol 229: 3453–3466 [DOI] [PubMed] [Google Scholar]
- Couto D, Niebergall R, Liang X, Bücherl CA, Sklenar J, Macho AP, Ntoukakis V, Derbyshire P, Altenbach D, Maclean D. et al. (2016) The arabidopsis protein phosphatase PP2C38 negatively regulates the central immune kinase BIK1. PLoS Pathog 12: e1005811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl JL, Jones JD (2001) Plant pathogens and integrated defence responses to infection. Nature 411: 826–833 [DOI] [PubMed] [Google Scholar]
- de Jonge R, van Esse HP, Maruthachalam K, Bolton MD, Santhanam P, Saber MK, Zhang Z, Usami T, Lievens B, Subbarao KV, et al. (2012) Tomato immune receptor Ve1 recognizes effector of multiple fungal pathogens uncovered by genome and RNA sequencing. Proc Natl Acad Sci USA 109: 5110–5115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Ronde D, Butterbach P, Lohuis D, Hedil M, van Lent JWM, Kormelink R (2013) Tsw gene-based resistance is triggered by a functional RNA silencing suppressor protein of the Tomato spotted wilt virus. Mol. Plant Pathol 14: 405–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derevnina L, Contreras MP, Adachi H, Upson J, Cruces AV, Xie A, Skłenar J, Menke FLH, Mugford ST, MacLean D, et al. (2021) Plant pathogens convergently evolved to counteract redundant nodes of an NLR immune receptor network. PLoS Biol 19: e3001136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkacheva M, Yu G, Rufian JS, Jiang S, Derbyshire P, Morcillo RJL, Stransfeld L, Wei Y, Menke FLH, Zipfel C, et al. (2020) The Arabidopsis E3 ubiquitin ligase PUB4 regulates BIK1 homeostasis and is targeted by a bacterial type-III effector. BioRxiv doi: 10.1101/2020.10.25.354514 [Google Scholar]
- Devanna NB, Vijayan J, Sharma TR (2014) The blast resistance gene Pi54of cloned from Oryza officinalis interacts with Avr-Pi54 through its novel non-LRR domains. PLoS One 9: e104840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeYoung BJ, Qi D, Kim SH, Burke TP, Innes RW (2012) Activation of a plant nucleotide binding-leucine rich repeat disease resistance protein by a modified self protein. Cell Microbiol 14: 1071–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding P, Sakai T, Shrestha RK, Perez NM, Guo W, Ngou BPM, He S, Liu C, Feng X, Zhang R, et al. (2020) Chromatin accessibility landscapes activated by cell surface and intracellular immune receptors. J Exp Bot 72: 7927–7941 [DOI] [PubMed] [Google Scholar]
- Dixon MS, Hatzixanthis K, Jones DA, Harrison K, Jones JD (1998) The tomato Cf-5 disease resistance gene and six homologs show pronounced allelic variation in leucine-rich repeat copy number. Plant Cell 10: 1915–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon MS, Jones DA, Keddie JS, Thomas CM, Harrison K, Jones JD (1996) The tomato Cf-2 disease resistance locus comprises two functional genes encoding leucine-rich repeat proteins. Cell 84: 451–459 [DOI] [PubMed] [Google Scholar]
- Dodds P, Thrall P (2009) Recognition events and host-pathogen co-evolution in gene-for-gene resistance to flax rust. Funct Plant Biol 36: 395–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds PN, Lawrence GJ, Catanzariti AM, Ayliffe MA, Ellis JG (2004) The Melampsora lini AvrL567 avirulence genes are expressed in haustoria and their products are recognized inside plant cells. Plant Cell 16: 755–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domazakis E, Wouters D, Lochman J, Visser R, Joosten MHAJ, Vleeshouwers VGAA (2020) ELR is a true pattern recognition receptor that associates with elicitins from diverse Phytophthora species. BioRxiv doi: 10.1101/2020.09.21.305813 [Google Scholar]
- Dong OX, Ao K, Xu F, Johnson KCM, Wu Y, Li L, Xia S, Liu Y, Huang Y, Rodriguez E, et al. (2018) Individual components of paired typical NLR immune receptors are regulated by distinct E3 ligases. Nat Plants 4: 699–710 [DOI] [PubMed] [Google Scholar]
- Du B, Zhang W, Liu B, Hu J, Wei Z, Shi Z, He R, Zhu L, Chen R, Han B, et al. (2009) Identification and characterization of Bph14, a gene conferring resistance to brown planthopper in rice. Proc Natl Acad Sci USA 106: 22163–22168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Berg J, Govers F, Bouwmeester K (2015a) Immune activation mediated by the late blight resistance protein R1 requires nuclear localization of R1 and the effector AVR1. New Phytol 207: 735–747 [DOI] [PubMed] [Google Scholar]
- Du J, Verzaux EChaparro-Garcia A, Bijsterbosch G, Keizer LCP, Zhou J, Liebrand TWH, Xie C, Govers F, Robatzek S, et al. (2015b) Elicitin recognition confers enhanced resistance to Phytophthora infestans in potato. Nat Plants 1: 15034. [DOI] [PubMed] [Google Scholar]
- Dufayard JF, Bettembourg M, Fischer I, Droc G, Guiderdoni E, Périn C, Chantret N, Diévart A (2017) New insights on leucine-rich repeats receptor-like kinase orthologous relationships in angiosperms. Front Plant Sci 8: 381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggan C, Moratto E, Savage Z, Hamilton E, Adachi H, Wu CH, Leary AY, Tumtas Y, Rothery S, Maqbool A, et al. (2021) Dynamic accumulation of a helper NLR at the plant-pathogen interface underpins pathogen recognition. BioRxiv doi: 10.1101/2021.03.15.435521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duxbury Z, Wang S, MacKenzie CI, Tenthorey JL, Zhang X, Huh SU, Hu L, Hill L, Ngou PM, Ding P, et al. (2020) Induced proximity of a TIR signaling domain on a plant-mammalian NLR chimera activates defense in plants. Proc Natl Acad Sci USA 117: 18832–18839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duxbury Z, Wu CH, Ding P (2021) A comparative overview of the intracellular guardians of plants and animals: nlrs in innate immunity and beyond. Annu Rev Plant Biol 72: 155–184 [DOI] [PubMed] [Google Scholar]
- Eitas TK, Nimchuk ZL, Dangl JL (2008) Arabidopsis TAO1 is a TIR-NB-LRR protein that contributes to disease resistance induced by the Pseudomonas syringae effector AvrB. Proc Natl Acad Sci USA 105: 6475–6480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson FL, Holzberg S, Calderon-Urrea A, Handley V, Axtell M, Corr C, Baker B (1999) The helicase domain of the TMV replicase proteins induces the N-mediated defence response in tobacco. Plant J 18: 67–75 [DOI] [PubMed] [Google Scholar]
- Eschen-Lippold L, Jiang X, Elmore JM, Mackey D, Shan L, Coaker G, Scheel D, Lee J (2016) Bacterial AvrRpt2-like cysteine proteases block activation of the Arabidopsis mitogen-activated protein kinases, MPK4 and MPK11. Plant Physiol 171: 2223–2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk A, Feys BJ, Frost LN, Jones JD, Daniels MJ, Parker JE (1999) EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proc Natl Acad Sci USA 96: 3292–3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L, Fröhlich K, Melzer E, Albert I, Pruitt RN, Zhang L, Albert M, Kim ST, Chae E, Weigel D, et al. (2021) Genotyping-by-sequencing-based identification of Arabidopsis pattern recognition receptor RLP32 recognizing proteobacterial translation initiation factor IF1. BioRxiv doi: 10.1101/2021.03.04.433884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner C, Petutschnig E, Benitez-Alfonso Y, Beck M, Robatzek S, Lipka V, Maule AJ (2013) LYM2-dependent chitin perception limits molecular flux via plasmodesmata. Proc Natl Acad Sci USA 110: 9166–9170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feehan JM, Castel B, Bentham AR, Jones JD (2020) Plant NLRs get by with a little help from their friends. Curr Opin Plant Biol 56: 99–108 [DOI] [PubMed] [Google Scholar]
- Felix G, Duran JD, Volko S, Boller T (1999) Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J 18: 265–276 [DOI] [PubMed] [Google Scholar]
- Feng F, Yang F, Rong W, Wu X, Zhang J, Chen S, He C, Zhou JM (2012) A Xanthomonas uridine 5’-monophosphate transferase inhibits plant immune kinases. Nature 485: 114–118 [DOI] [PubMed] [Google Scholar]
- Feys BJ, Moisan LJ, Newman MA, Parker JE (2001) Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J 20: 5400–5411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys BJ, Wiermer M, Bhat RA, Moisan LJ, Medina-Escobar N, Neu C, Cabral A, Parker JE (2005) Arabidopsis SENESCENCE-ASSOCIATED GENE101 stabilizes and signals within an ENHANCED DISEASE SUSCEPTIBILITY1 complex in plant innate immunity. Plant Cell 17: 2601–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster SJ, Park TH, Pel M, Brigneti G, Sliwka J, Jagger L, van der Vossen E, Jones JDG (2009) Rpi-vnt1.1, a Tm-2(2) homolog from Solanum venturii, confers resistance to potato late blight. Mol Plant Microbe Interact 22: 589–600 [DOI] [PubMed] [Google Scholar]
- Gabriëls SHEJ, Vossen JH, Ekengren SK, van Ooijen G, Abd-El-Haliem AM, van den Berg GCM, Rainey DY, Martin GB, Takken FLW, de Wit PJGM, et al. (2007) An NB-LRR protein required for HR signalling mediated by both extra- and intracellular resistance proteins. Plant J 50: 14–28 [DOI] [PubMed] [Google Scholar]
- Gao M, Wang X, Wang D, Xu F, Ding X, Zhang Z, Bi D, Cheng YT, Chen S, Li X, et al. (2009) Regulation of cell death and innate immunity by two receptor-like kinases in Arabidopsis. Cell Host Microbe 6: 34–44 [DOI] [PubMed] [Google Scholar]
- Gao X, Chen X, Lin W, Chen S, Lu D, Niu Y, Li L, Cheng C, McCormack M, Sheen J, et al. (2013) Bifurcation of Arabidopsis NLR immune signaling via Ca2+-dependent protein kinases. PLoS Pathog 9: e1003127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Wang W, Zhang T, Gong Z, Zhao H, Han GZ (2018) Out of water: the origin and early diversification of plant R-genes. Plant Physiol 177: 82–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García AV, Blanvillain-Baufumé S, Huibers RP, Wiermer M, Li G, Gobbato E, Rietz S, Parker JE (2010) Balanced nuclear and cytoplasmic activities of EDS1 are required for a complete plant innate immune response. PLoS Pathog 6: e1000970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghislain M, Byarugaba AA, Magembe E, Njoroge A, Rivera C, Román ML, Tovar JC, Gamboa S, Forbes GA, Kreuze JF, et al. (2019) Stacking three late blight resistance genes from wild species directly into African highland potato varieties confers complete field resistance to local blight races. Plant Biotechnol J 17: 1119–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilardi P, García-Luque I, Serra MT (2004) The coat protein of tobamovirus acts as elicitor of both L2 and L4 gene-mediated resistance in Capsicum. J Gen Virol 85: 2077–2085 [DOI] [PubMed] [Google Scholar]
- Göhre V, Spallek T, Häweker H, Mersmann S, Mentzel T, Boller T, de Torres M, Mansfield JW, Robatzek S (2008) Plant pattern-recognition receptor FLS2 is directed for degradation by the bacterial ubiquitin ligase AvrPtoB. Curr Biol 18: 1824–1832 [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L, Boller T (2000) FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell 5: 1003–1011 [DOI] [PubMed] [Google Scholar]
- González-Lamothe R, Tsitsigiannis DI, Ludwig AA, Panicot M, Shirasu K, Jones JDG (2006) The U-box protein CMPG1 is required for efficient activation of defense mechanisms triggered by multiple resistance genes in tobacco and tomato. Plant Cell 18: 1067–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goritschnig S, Krasileva KV, Dahlbeck D, Staskawicz BJ (2012) Computational prediction and molecular characterization of an oomycete effector and the cognate Arabidopsis resistance gene. PLoS Genet 8: e1002502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto Y, Maki N, Sklenar J, Derbyshire P, Menke FLH, Zipfel C, Kadota Y, Shirasu K (2020) The phagocytosis oxidase/Bem1p (PB1) domain-containing protein PB1CP negatively regulates the NADPH oxidase RBOHD in plant immunity. BioRxiv doi: 10.1101/2020.12.28.423414 [DOI] [PubMed] [Google Scholar]
- Gouhier-Darimont C, Stahl E, Glauser G, Reymond P (2019) The Arabidopsis lectin receptor kinase LecRK-I.8 is involved in insect egg perception. Front Plant Sci 10: 623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant M, Brown I, Adams S, Knight M, Ainslie A, Mansfield J (2000) The RPM1 plant disease resistance gene facilitates a rapid and sustained increase in cytosolic calcium that is necessary for the oxidative burst and hypersensitive cell death. Plant J 23: 441–450 [DOI] [PubMed] [Google Scholar]
- Grant M.R., Godiard L., Straube E., Ashfield T., Lewald J., Sattler A., Innes R.W., Dangl J.L. (1995) Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science 269: 843–846. [DOI] [PubMed] [Google Scholar]
- Grech-Baran M, Witek K, Poznański J, Grupa-Urbańska A, Malinowski T, Lichocka M, Jones JD, Hennig J (2021) The Rysto immune receptor recognizes a broadly conserved feature of potyviral coat proteins. BioRxiv [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronnier J, Franck CM, Stegmann M, DeFalco TA, Cifuentes AA, Dünser K, Lin W, Yang Z, Kleine-Vehn J, Ringli C, Zipfel C (2020) FERONIA regulates FLS2 plasma membrane nanoscale dynamics to modulate plant immune signaling. BioRxiv [Google Scholar]
- Groux R, Stahl E, Gouhier-Darimont C, Kerdaffrec E, Jimenez-Sandoval P, Santiago J, Reymond P (2021) Arabidopsis natural variation in insect egg-induced cell death reveals a role for LECTIN RECEPTOR KINASE-I.1. Plant Physiol 185: 240–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubb LE, Derbyshire P, Dunning KE, Zipfel C, Menke FLH, Monaghan J (2021) Large-scale identification of ubiquitination sites on membrane-associated proteins in Arabidopsis thaliana seedlings. Plant Physiol 185: 1483–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan R, Su J, Meng X, Li S, Liu Y, Xu J, Zhang S (2015) Multilayered regulation of ethylene induction plays a positive role in Arabidopsis resistance against Pseudomonas syringae. Plant Physiol 169: 299–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ahn HK, Sklenar J, Huang J, Ma Y, Ding P, Menke FLH, Jones JDG (2020) Phosphorylation-regulated activation of the Arabidopsis RRS1-R/RPS4 immune receptor complex reveals two distinct effector recognition mechanisms. Cell Host Microbe 27: 769–781.e6 [DOI] [PubMed] [Google Scholar]
- Hall TJ (1980) Resistance at the TM-2 locus in the tomato to tomato mosaic virus. Euphytica 29: 189–197 [Google Scholar]
- Han L, Li GJ, Yang KY, Mao G, Wang R, Liu Y, Zhang S (2010) Mitogen-activated protein kinase 3 and 6 regulate Botrytis cinerea-induced ethylene production in Arabidopsis. Plant J 64: 114–127 [DOI] [PubMed] [Google Scholar]
- Hander T, Fernández-Fernández AD, Kumpf RP, Willems P, Schatowitz H, Rombaut D, Staes A, Nolf J, Pottie R, Yao P, et al. (2019) Damage on plants activates Ca2+-dependent metacaspases for release of immunomodulatory peptides. Science 363: eaar7486 [DOI] [PubMed] [Google Scholar]
- Harris JM, Balint-Kurti P, Bede JC, Day B, Gold S, Goss EM, Grenville-Briggs LJ, Jones KM, Wang A, et al. (2020) What are the top 10 unanswered questions in molecular plant-microbe interactions? Mol Plant Microbe Interact 33: 1354–1365 [DOI] [PubMed] [Google Scholar]
- He P, Shan L, Lin NC, Martin GB, Kemmerling B, Nürnberger T, Sheen J (2006) Specific bacterial suppressors of MAMP signaling upstream of MAPKKK in Arabidopsis innate immunity. Cell 125: 563–575 [DOI] [PubMed] [Google Scholar]
- Hegenauer V, Slaby P, Körner M, Bruckmüller JA, Albert I, Burggraf R, Kaiser B, Droste-Borel I, Sklenar J, Menke FLH, et al. (2020) The tomato receptor CuRe1 senses a cell wall protein to identify Cuscuta as a pathogen. Nat Commun 11: 5299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hind SR, Strickler SR, Boyle PC, Dunham DM, Bao Z, O'Doherty IM, Baccile JA, Hoki JS, Viox EG, Clarke CR, et al. (2016) Tomato receptor FLAGELLIN-SENSING 3 binds flgII-28 and activates the plant immune system. Nat Plants 2: 16128. [DOI] [PubMed] [Google Scholar]
- Hohmann U, Lau K, Hothorn M (2017) The structural basis of ligand perception and signal activation by receptor kinases. Annu Rev Plant Biol 68: 109–137 [DOI] [PubMed] [Google Scholar]
- Hohmann U, Nicolet J, Moretti A, Hothorn LA, Hothorn M (2018) Mechanistic analysis of the SERK3 elongated allele defines a role for BIR ectodomains in brassinosteroid signaling. BioRxiv doi: 10.1101/257543 [DOI] [PubMed] [Google Scholar]
- Holton N, Nekrasov V, Ronald PC, Zipfel C (2015) The phylogenetically-related pattern recognition receptors EFR and XA21 recruit similar immune signaling components in monocots and dicots. PLoS Pathog 11: e1004602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsefield S, Burdett H, Zhang X, Manik MK, Shi Y, Chen J, Qi T, Gilley J, Lai JS, Rank MX et al. (2019) NAD+ cleavage activity by animal and plant TIR domains in cell death pathways. Science 365: 793–799 [DOI] [PubMed] [Google Scholar]
- Hoser R, Zurczak M, Lichocka M, Zuzga S, Dadlez M, Samuel MA, Ellis BE, Stuttmann J, Parker JE, Hennig J, et al. (2013) Nucleocytoplasmic partitioning of tobacco N receptor is modulated by SGT1. New Phytol 200: 158–171 [DOI] [PubMed] [Google Scholar]
- Hou S, Liu D, He P (2021) Phytocytokines function as immunological modulators of plant immunity. Stress Biol 1: 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou S, Wang X, Chen D, Yang X, Wang M, Turrà D, Di Pietro A, Zhang W (2014) The secreted peptide PIP1 amplifies immunity through receptor-like kinase 7. PLoS Pathog 10: e1004331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houterman PM, Cornelissen BJC, Rep M (2008) Suppression of plant resistance gene-based immunity by a fungal effector. PLoS Pathog 4: e1000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houterman PM, Ma L, van Ooijen G, de Vroomen MJ, Cornelissen BJC, Takken FLW, Rep M (2009) The effector protein Avr2 of the xylem-colonizing fungus Fusarium oxysporum activates the tomato resistance protein I-2 intracellularly. Plant J 58: 970–978 [DOI] [PubMed] [Google Scholar]
- Huang S, Chen X, Zhong X, Li M, Ao K, Huang J, Li X (2016) Plant TRAF proteins regulate NLR immune receptor turnover. Cell Host Microbe 19: 204–215 [DOI] [PubMed] [Google Scholar]
- Huot B, Yao J, Montgomery BL, He SY (2014) Growth-defense tradeoffs in plants: a balancing act to optimize fitness. Mol Plant 7: 1267–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irieda H, Inoue Y, Mori M, Yamada K, Oshikawa Y, Saitoh H, Uemura A, Terauchi R, Kitakura S, Kosaka A, et al. (2019) Conserved fungal effector suppresses PAMP-triggered immunity by targeting plant immune kinases. Proc Natl Acad Sci USA 116: 496–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob F, Kracher B, Mine A, Seyfferth C, Blanvillain-Baufumé S, Parker JE, Tsuda K, Schulze-Lefert P, Maekawa T (2018) A dominant-interfering camta3 mutation compromises primary transcriptional outputs mediated by both cell surface and intracellular immune receptors in Arabidopsis thaliana. New Phytol 217: 1667–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob P, Kim NH, Wu F, El-Kasmi F, Chi Y, Walton WG, Furzer OJ, Lietzan AD, Sunil S, Kempthorn K, et al. (2021) Plant “helper” immune receptors are Ca2+-permeable nonselective cation channels. Science 373: 420–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jehle AK, Lipschis M, Albert M, Fallahzadeh-Mamaghani V, Fürst U, Mueller K, Felix G (2013) The receptor-like protein ReMAX of Arabidopsis detects the microbe-associated molecular pattern eMax from Xanthomonas. Plant Cell 25: 2330–2340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y, McAdams SA, Bryan GT, Hershey HP, Valent B (2000) Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J 19: 4004–4014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo KR (2013) Unveiling and deploying durability of late blight resistance in potato : from natural stacking to cisgenic stacking.
- Johal GS, Briggs SP (1992) Reductase activity encoded by the HM1 disease resistance gene in maize. Science 258: 985–987 [DOI] [PubMed] [Google Scholar]
- Johnson KCM, Xia S, Feng X, Li X (2015) The chromatin remodeler SPLAYED negatively regulates SNC1-mediated immunity. Plant Cell Physiol 56: 1616–1623 [DOI] [PubMed] [Google Scholar]
- Jones DA, Thomas CM, Hammond-Kosack KE, Balint-Kurti PJ, Jones JDG (1994) Isolation of the tomato Cf-9 gene for resistance to Cladosporium fulvum by transposon tagging. Science 266: 789–793 [DOI] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Jones JDG, Brigneti G, Smilde D (2003) Putting plant disease resistance genes to work. In Vasil IK, ed, Plant Biotechnology 2002 and Beyond, Springer Netherlands, Dordrecht, the Netherlands, pp 10–17 [Google Scholar]
- Joosten MH, Cozijnsen TJ, De Wit PJ (1994) Host resistance to a fungal tomato pathogen lost by a single base-pair change in an avirulence gene. Nature 367: 384–386 [DOI] [PubMed] [Google Scholar]
- Jung HW, Panigrahi GK, Jung GY, Lee YJ, Shin KH, Sahoo A, Choi ES, Lee E, Kim KM, Yang SH, et al. (2020) Pathogen-associated molecular pattern-triggered immunity involves proteolytic degradation of core nonsense-mediated mRNA decay factors during the early defense response. Plant Cell 32: 1081–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadota Y, Liebrand TWH, Goto Y, Sklenar J, Derbyshire P, Menke FLH, Torres MA, Molina A, Zipfel C, Coaker G, et al. (2019) Quantitative phosphoproteomic analysis reveals common regulatory mechanisms between effector- and PAMP-triggered immunity in plants. New Phytol 221: 2160–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadota Y, Shirasu K, Zipfel C (2015) Regulation of the NADPH oxidase RBOHD during plant immunity. Plant Cell Physiol 56: 1472–1480 [DOI] [PubMed] [Google Scholar]
- Kadota Y, Sklenar J, Derbyshire P, Stransfeld L, Asai S, Ntoukakis V, Jones JDG, Shirasu K, Menke F, Jones A, et al. (2014) Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol Cell 54: 43–55 [DOI] [PubMed] [Google Scholar]
- Kaloshian I, Lange WH, Williamson VM (1995) An aphid-resistance locus is tightly linked to the nematode-resistance gene, Mi, in tomato. Proc Natl Acad Sci USA 92: 622–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamoun S, van West P, de Jong AJ, de Groot KE, Vleeshouwers V, Govers F (1997) A gene encoding a protein elicitor of Phytophthora infestans is down-regulated during infection of potato. Mol. Plant Microbe Interact 10: 13–20 [DOI] [PubMed] [Google Scholar]
- Kanzaki H, Yoshida K, Saitoh H, Fujisaki K, Hirabuchi A, Alaux L, Fournier E, Tharreau D, Terauchi R (2012) Arms race co-evolution of Magnaporthe oryzae AVR-Pik and rice Pik genes driven by their physical interactions. Plant J 72: 894–907 [DOI] [PubMed] [Google Scholar]
- Kato H, Nemoto K, Shimizu M, Abe A, Asai S, Ishihama N, Daimon T, Ojika M, Kawakita K, Onai K, et al. (2021) Pathogen-derived 9-methyl sphingoid base is perceived by a lectin receptor kinase in Arabidopsis. BioRxiv doi: 10.1101/2021.10.18.464766 [Google Scholar]
- Kawaharada Y, Kelly S, Nielsen MW, Hjuler CT, Gysel K, Muszyński A, Carlson RW, Thygesen MB, Sandal N, Asmussen MH, et al. (2015) Receptor-mediated exopolysaccharide perception controls bacterial infection. Nature 523: 308–312 [DOI] [PubMed] [Google Scholar]
- Khan M, Subramaniam R, Desveaux D (2016) Of guards, decoys, baits and traps: pathogen perception in plants by type III effector sensors. Curr Opin Microbiol 29: 49–55 [DOI] [PubMed] [Google Scholar]
- Kim MG, Geng X, Lee SY, Mackey D (2009a) The Pseudomonas syringae type III effector AvrRpm1 induces significant defenses by activating the Arabidopsis nucleotide-binding leucine-rich repeat protein RPS2. Plant J 57: 645–653 [DOI] [PubMed] [Google Scholar]
- Kim SB, Lee HY, Seo S, Lee JH, Choi D (2015) RNA-dependent RNA polymerase (NIb) of the potyviruses is an avirulence factor for the broad-spectrum resistance gene Pvr4 in Capsicum annuum cv. CM334. PLoS One 10: e0119639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Kwon SI, Saha D, Anyanwu NC, Gassmann W (2009b) Resistance to the Pseudomonas syringae effector HopA1 is governed by the TIR-NBS-LRR protein RPS6 and is enhanced by mutations in SRFR1. Plant Physiol 150: 1723–1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Gilmour SJ, Chao L, Park S, Thomashow MF (2020) Arabidopsis CAMTA transcription factors regulate pipecolic acid biosynthesis and priming of immunity genes. Mol Plant 13: 157–168 [DOI] [PubMed] [Google Scholar]
- Kohorn BD, Kohorn SL (2012) The cell wall-associated kinases, WAKs, as pectin receptors. Front Plant Sci 3:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kørner CJ, Klauser D, Niehl A, Domínguez-Ferreras A, Chinchilla D, Boller T, Heinlein M, Hann DR (2013) The immunity regulator BAK1 contributes to resistance against diverse RNA viruses. Mol Plant Microbe Interact 26: 1271–1280 [DOI] [PubMed] [Google Scholar]
- Kourelis J, Kamoun S (2020) RefPlantNLR: a comprehensive collection of experimentally validated plant NLRs. BioRxiv doi: 10.1101/2020.07.08.193961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasileva KV, Dahlbeck D, Staskawicz BJ (2010) Activation of an Arabidopsis resistance protein is specified by the in planta association of its leucine-rich repeat domain with the cognate oomycete effector. Plant Cell 22: 2444–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger J, Thomas CM, Golstein C, Dixon MS, Smoker M, Tang S, Mulder L, Jones JDG (2002) A tomato cysteine protease required for Cf-2-dependent disease resistance and suppression of autonecrosis. Science 296: 744–747 [DOI] [PubMed] [Google Scholar]
- Kutschera A, Dawid C, Gisch N, Schmid C, Raasch L, Gerster T, Schäffer M, Smakowska-Luzan E, Belkhadir Y, Vlot AC, et al. (2019) Bacterial medium-chain 3-hydroxy fatty acid metabolites trigger immunity in Arabidopsis plants. Science 364: 178–181 [DOI] [PubMed] [Google Scholar]
- Lacaze A, Joly DL (2020) Structural specificity in plant-filamentous pathogen interactions. Mol Plant Pathol 21: 1513–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacombe S, Rougon-Cardoso A, Sherwood E, Peeters N, Dahlbeck D, van Esse HP, Smoker M, Rallapalli G, Thomma BPHJ, Staskawicz B, et al. (2010) Interfamily transfer of a plant pattern-recognition receptor confers broad-spectrum bacterial resistance. Nat Biotechnol 28: 365–369 [DOI] [PubMed] [Google Scholar]
- Laflamme B, Dillon MM, Martel A, Almeida RND, Desveaux D, Guttman DS (2020) The pan-genome effector-triggered immunity landscape of a host-pathogen interaction. Science 367: 763–768 [DOI] [PubMed] [Google Scholar]
- Lagudah ES, Moullet O, Appels R (1997) Map-based cloning of a gene sequence encoding a nucleotide-binding domain and a leucine-rich region at the Cre3 nematode resistance locus of wheat. Genome 40: 659–665 [DOI] [PubMed] [Google Scholar]
- Lai Y, Eulgem T (2018) Transcript-level expression control of plant NLR genes. Mol Plant Pathol 19: 1267–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal NK, Nagalakshmi U, Hurlburt NK, Flores R, Bak A, Sone P, Ma X, Song G, Walley J, Shan L, et al. (2018) The receptor-like cytoplasmic kinase BIK1 localizes to the nucleus and regulates defense hormone expression during plant innate immunity. Cell Host Microbe 23: 485–497.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laohavisit A, Wakatake T, Ishihama N, Mulvey H, Takizawa K, Suzuki T, Shirasu K (2020) Quinone perception in plants via leucine-rich-repeat receptor-like kinases. Nature 587: 92–97 [DOI] [PubMed] [Google Scholar]
- Lapin D, Kovacova V, Sun X, Dongus JA, Bhandari D, von Born P, Bautor J, Guarneri N, Rzemieniewski J, Stuttmann J, et al. (2019) A coevolved EDS1-SAG101-NRG1 module mediates cell death signaling by TIR-domain immune receptors. Plant Cell 31: 2430–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkan NJ, Lydiate DJ, Parkin IAP, Nelson MN, Epp DJ, Cowling WA, Rimmer SR, Borhan MH (2013) The Brassica napus blackleg resistance gene LepR3 encodes a receptor-like protein triggered by the Leptosphaeria maculans effector AVRLM1. New Phytol 197: 595–605 [DOI] [PubMed] [Google Scholar]
- Lawrence GJ, Finnegan EJ, Ayliffe MA, Ellis JG (1995) The L6 gene for flax rust resistance is related to the Arabidopsis bacterial resistance gene RPS2 and the tobacco viral resistance gene N. Plant Cell 7: 1195–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Roux C, Huet G, Jauneau A, Camborde L, Trémousaygue D, Kraut A, Zhou B, Levaillant M, Adachi H, Yoshioka H, et al. (2015) A receptor pair with an integrated decoy converts pathogen disabling of transcription factors to immunity. Cell 161: 1074–1088 [DOI] [PubMed] [Google Scholar]
- Lee D, Lal NK, Lin ZD, Ma S, Liu J, Castro B, Toruño T, Dinesh-Kumar SP, Coaker G (2020) Regulation of reactive oxygen species during plant immunity through phosphorylation and ubiquitination of RBOHD. Nat Commun 11: 1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leppyanen IV, Shakhnazarova VY, Shtark OY, Vishnevskaya NA, Tikhonovich IA, Dolgikh EA (2017) Receptor-like kinase LYK9 in Pisum sativum L. Is the CERK1-like receptor that controls both plant immunity and AM symbiosis development. Int J Mol Sci 19: 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Huang S, Guo X, Li Y, Yang Y, Guo Z, Kuang H, Rietman H, Bergervoet M, Vleeshouwers VGGA, et al. (2011) Cloning and characterization of r3b; members of the r3 superfamily of late blight resistance genes show sequence and functional divergence. Mol Plant Microbe Interact 24: 1132–1142 [DOI] [PubMed] [Google Scholar]
- Li J, Timko MP (2009) Gene-for-gene resistance in Striga-cowpea associations. Science 325: 1094. [DOI] [PubMed] [Google Scholar]
- Li J, Wang Q, Li C, Bi Y, Fu X, Wang R (2019) Novel haplotypes and networks of AVR-Pik alleles in Magnaporthe oryzae. BMC Plant Biol 19: 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Kim P, Yu L, Cai G, Chen S, Alfano JR, Zhou JM (2016) Activation-dependent destruction of a co-receptor by a Pseudomonas syringae effector dampens plant immunity. Cell Host Microbe 20: 504–514 [DOI] [PubMed] [Google Scholar]
- Li L, Li M, Yu L, Zhou Z, Liang X, Liu Z, Cai G, Gao L, Zhang X, Wang Y, et al. (2014) The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe 15: 329–338 [DOI] [PubMed] [Google Scholar]
- Li Q, Wang J, Bai T, Zhang M, Jia Y, Shen D, Zhang M, Dou D (2020) A Phytophthora capsici effector suppresses plant immunity via interaction with EDS1. Mol Plant Pathol 21: 502–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Wang B, Wu J, Lu G, Hu Y, Zhang X, Zhang Z, Zhao Q, Feng Q, Zhang H, et al. (2009) The Magnaporthe oryzae avirulence gene AvrPiz-t encodes a predicted secreted protein that triggers the immunity in rice mediated by the blast resistance gene Piz-t. Mol Plant Microbe Interact 22: 411–420 [DOI] [PubMed] [Google Scholar]
- Li X, Lin H, Zhang W, Zou Y, Zhang J, Tang X, Zhou JM (2005) Flagellin induces innate immunity in nonhost interactions that is suppressed by Pseudomonas syringae effectors. Proc Natl Acad Sci USA 102: 12990–12995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Li S, Bi D, Cheng YT, Li X, Zhang Y (2010a) SRFR1 negatively regulates plant NB-LRR resistance protein accumulation to prevent autoimmunity. PLoS Pathog 6: e1001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Tessaro MJ, Li X, Zhang Y (2010b) Regulation of the expression of plant resistance gene SNC1 by a protein with a conserved BAT2 domain. Plant Physiol 153: 1425–1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Zhou JM (2018) Receptor-like cytoplasmic kinases: central players in plant receptor kinase-mediated signaling. Annu Rev Plant Biol 69: 267–299 [DOI] [PubMed] [Google Scholar]
- Liang X, Ding P, Lian K, Wang J, Ma M, Li L, Li L, Li M, Zhang X, Chen S, et al. (2016) Arabidopsis heterotrimeric G proteins regulate immunity by directly coupling to the FLS2 receptor. eLife 5: e13568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebrand TWH, van den Berg GCM, Zhang Z, Smit P, Cordewener JHG, America AHP, Sklenar J, Jones AME, Tameling WIL, Robatzek S, et al. (2013) Receptor-like kinase SOBIR1/EVR interacts with receptor-like proteins in plant immunity against fungal infection. Proc Natl Acad Sci USA 110: 10010–10015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligterink W, Kroj T, zur Nieden U, Hirt H, Scheel D (1997) Receptor-mediated activation of a MAP kinase in pathogen defense of plants. Science 276: 2054–2057 [DOI] [PubMed] [Google Scholar]
- Lin W, Li B, Lu D, Chen S, Zhu N, He P, Shan L (2014) Tyrosine phosphorylation of protein kinase complex BAK1/BIK1 mediates Arabidopsis innate immunity. Proc Natl Acad Sci USA 111: 3632–3637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Lu D, Gao X, Jiang S, Ma X, Wang Z, Mengiste T, He P, Shan L (2013) Inverse modulation of plant immune and brassinosteroid signaling pathways by the receptor-like cytoplasmic kinase BIK1. Proc Natl Acad Sci USA 110: 12114–12119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Olave-Achury A, Heal R, Witek K, Karki HS, Song T, Wu C, Adachi H, Kamoun S, Vleeshouwers VGAA, et al. (2021) Rpi-amr3 confers resistance to multiple Phytophthora species by recognizing a conserved RXLR effector. BioRxiv doi: 10.1101/2021.06.10.447899 [DOI] [PubMed] [Google Scholar]
- Lin X, Song T, Fairhead S, Witek K, Jouet A, Jupe F, Witek AI, Karki HS, Vleeshouwers VGAA, Hein I, et al. (2020). Identification of Avramr1 from Phytophthora infestans using long read and cDNA pathogen-enrichment sequencing (PenSeq). Mol Plant Pathol 21: 1502–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Li JF, Ao Y, Qu J, Li Z, Su J, Zhang Y, Liu J, Feng D, Qi K, et al. (2012) Lysin motif-containing proteins LYP4 and LYP6 play dual roles in peptidoglycan and chitin perception in rice innate immunity. Plant Cell 24: 3406–3419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Chen S, Chen L, Zhou Q, Wang M, Feng D, Li JF, Wang J, Wang HB, Liu B (2017) BIK1 cooperates with BAK1 to regulate constitutive immunity and cell death in Arabidopsis. J Integr Plant Biol 59: 234–239 [DOI] [PubMed] [Google Scholar]
- Liu Y, Zhang S (2004) Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell 16: 3386–3399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Maierhofer T, Rybak K, Sklenar J, Breakspear A, Johnston MG, Fliegmann J, Huang S, Roelfsema MRG, Felix G, et al. (2019) Anion channel SLAH3 is a regulatory target of chitin receptor-associated kinase PBL27 in microbial stomatal closure. eLife 8: e44474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Sun T, Sun Y, Zhang Y, Radojičić A, Ding Y, Tian H, Huang X, Lan J, Chen S, et al. (2020) Diverse roles of the salicylic acid receptors NPR1 and NPR3/NPR4 in plant immunity. Plant Cell 32: 4002–4016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zeng Z, Zhang YM, Li Q, Jiang XM, Jiang Z, Tang JH, Chen D, Wang Q, Chen JQ, et al. (2021) An angiosperm NLR Atlas reveals that NLR gene reduction is associated with ecological specialization and signal transduction component deletion. Mol. Plant 14: 2015–2031 [DOI] [PubMed] [Google Scholar]
- Liu Z, Friesen TL, Ling H, Meinhardt SW, Oliver RP, Rasmussen JB, Faris JD (2006) The Tsn1-ToxA interaction in the wheat-Stagonospora nodorum pathosystem parallels that of the wheat-tan spot system. Genome 49: 1265–1273 [DOI] [PubMed] [Google Scholar]
- Lokossou AA, Park T, van Arkel G, Arens M, Ruyter-Spira C, Morales J, Whisson SC, Birch PRJ, Visser RGF, Jacobsen E, et al. (2009) Exploiting knowledge of R/Avr genes to rapidly clone a new LZ-NBS-LRR family of late blight resistance genes from potato linkage group IV. Mol Plant Microbe Interact 22: 630–641 [DOI] [PubMed] [Google Scholar]
- Lolle S, Greeff C, Petersen K, Roux M, Jensen MK, Bressendorff S, Rodriguez E, Sømark K, Mundy J, Petersen M (2017) Matching NLR immune receptors to autoimmunity in camta3 mutants using antimorphic NLR alleles. Cell Host Microbe 21: 518–529.e4 [DOI] [PubMed] [Google Scholar]
- Lorang JM, Hagerty CH, Lee R, McClean PE, Wolpert TJ (2018) Genetic analysis of victorin sensitivity and identification of a causal nucleotide-binding site leucine-rich repeat gene in Phaseolus vulgaris. Mol Plant Microbe Interact 31: 1069–1074 [DOI] [PubMed] [Google Scholar]
- Lu D, Lin W, Gao X, Wu S, Cheng C, Avila J, Heese A, Devarenne TP, He P, Shan L (2011) Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science 332: 1439–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D, Wu S, Gao X, Zhang Y, Shan L, He P (2010) A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc Natl Acad Sci USA 107: 496–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Kracher B, Saur IML, Bauer S, Ellwood SR, Wise R, Yaeno T, Maekawa T, Schulze-Lefert P (2016) Allelic barley MLA immune receptors recognize sequence-unrelated avirulence effectors of the powdery mildew pathogen. Proc Natl Acad Sci USA 113: E6486–E6495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüdke D, Roth C, Kamrad SA, Messerschmidt J, Hartken D, Appel J, Hörnich BF, Yan Q, Kusch S, Klenke M, et al. (2021) Functional requirement of the Arabidopsis importin-α nuclear transport receptor family in autoimmunity mediated by the NLR protein SNC1. Plant J 105: 994–1009 [DOI] [PubMed] [Google Scholar]
- Lukasik E, Takken FLW (2009) STANDing strong, resistance proteins instigators of plant defence. Curr Opin Plant Biol 12: 427–436 [DOI] [PubMed] [Google Scholar]
- Luna E, Pastor V, Robert J, Flors V, Mauch-Mani B, Ton J (2011) Callose deposition: a multifaceted plant defense response. Mol Plant Microbe Interact 24: 183–193 [DOI] [PubMed] [Google Scholar]
- Luo M, Xie L, Chakraborty S, Wang A, Matny O, Jugovich M, Kolmer JA, Richardson T, Bhatt D, Hoque M, et al. (2021) A five-transgene cassette confers broad-spectrum resistance to a fungal rust pathogen in wheat. Nat. Biotechnol 39: 1–6 [DOI] [PubMed] [Google Scholar]
- Luu DD, Joe A, Chen Y, Parys K, Bahar O, Pruitt R, Chan LJG, Petzold CJ, Long K, Adamchak C, et al. (2019) Biosynthesis and secretion of the microbial sulfated peptide RaxX and binding to the rice XA21 immune receptor. Proc Natl Acad Sci USA 116: 8525–8534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C, Liu Y, Bai B, Han Z, Tang J, Zhang H, Yaghmaiean H, Zhang Y, Chai J (2017) Structural basis for BIR1-mediated negative regulation of plant immunity. Cell Res 27: 1521–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Lapin D, Liu L, Sun Y, Song W, Zhang X, Logemann E, Yu D, Wang J, Jirschitzka J, et al. (2020a) Direct pathogen-induced assembly of an NLR immune receptor complex to form a holoenzyme. Science 370: eabe3069. [DOI] [PubMed] [Google Scholar]
- Ma X, Claus LAN, Leslie ME, Tao K, Wu Z, Liu J, Yu X, Li B, Zhou J, Savatin DV, et al. (2020b) Ligand-induced monoubiquitination of BIK1 regulates plant immunity. Nature 581: 199–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Xu G, He P, Shan L (2016) SERKing coreceptors for receptors. Trends Plant Sci 21: 1017–1033 [DOI] [PubMed] [Google Scholar]
- Ma X, Zhang C, Kim DY, Huang Y, Chatt E, He P, Vierstra RD, Shan L (2021) Ubiquitylome analysis reveals a central role for the ubiquitin-proteasome system in plant innate immunity. Plant Physiol 185: 1943–1965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Walker RK, Zhao Y, Berkowitz GA (2012) Linking ligand perception by PEPR pattern recognition receptors to cytosolic Ca2+ elevation and downstream immune signaling in plants. Proc Natl Acad Sci USA 109: 19852–19857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macho AP, Zipfel C (2014) Plant PRRs and the activation of innate immune signaling. Mol Cell 54: 263–272 [DOI] [PubMed] [Google Scholar]
- Macho AP, Schwessinger B, Ntoukakis V, Brutus A, Segonzac C, Roy S, Kadota Y, Oh M-H, Sklenar J, Derbyshire P, et al. (2014) A bacterial tyrosine phosphatase inhibits plant pattern recognition receptor activation. Science 343: 1509–1512 [DOI] [PubMed] [Google Scholar]
- Mackey D, Belkhadir Y, Alonso JM, Ecker JR, Dangl JL (2003) Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 112: 379–389 [DOI] [PubMed] [Google Scholar]
- Manser B, Koller T, Praz CR, Roulin AC, Zbinden H, Arora S, Steuernagel B, Wulff BBH, Keller B, Sánchez-Martín J (2021) Identification of specificity-defining amino acids of the wheat immune receptor Pm2 and powdery mildew effector AvrPm2. Plant J 106: 993–1007 [DOI] [PubMed] [Google Scholar]
- Martin R, Qi T, Zhang H, Liu F, King M, Toth C, Nogales E, Staskawicz BJ (2020) Structure of the activated ROQ1 resistosome directly recognizing the pathogen effector XopQ. Science 370: eabd9993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto K, Sawada H, Matsumoto K, Hamada H, Yoshimoto E, Ito T, Takeuchi S, Tsuda S, Suzuki K, Kobayashi K, et al. (2008) The coat protein gene of tobamovirus P 0 pathotype is a determinant for activation of temperature-insensitive L 1a-gene-mediated resistance in Capsicum plants. Arch Virol 153: 645–650 [DOI] [PubMed] [Google Scholar]
- Mazo-Molina C, Mainiero S, Haefner BJ, Bednarek R, Zhang J, Feder A, Shi K, Strickler SR, Martin GB (2020) Ptr1 evolved convergently with RPS2 and Mr5 to mediate recognition of AvrRpt2 in diverse solanaceous species. Plant J 103: 1433–1445 [DOI] [PubMed] [Google Scholar]
- Mendy B, Wang’ombe MW, Radakovic ZS, Holbein J, Ilyas M, Chopra D, Holton N, Zipfel C, Grundler FMW, Siddique S (2017) Arabidopsis leucine-rich repeat receptor-like kinase NILR1 is required for induction of innate immunity to parasitic nematodes. PLoS Pathog 13: e1006284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Zhang S (2013) MAPK cascades in plant disease resistance signaling. Annu Rev Phytopathol 51: 245–266 [DOI] [PubMed] [Google Scholar]
- Mestre P, Baulcombe DC (2006) Elicitor-mediated oligomerization of the tobacco N disease resistance protein. Plant Cell 18: 491–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan SB, Bodeau J, Yaghoobi J, Kaloshian I, Zabel P, Williamson VM (1998) The root knot nematode resistance gene Mi from tomato is a member of the leucine zipper, nucleotide binding, leucine-rich repeat family of plant genes. Plant Cell 10: 1307–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindrinos M, Katagiri F, Yu GL, Ausubel FM (1994) The A. thaliana disease resistance gene RPS2 encodes a protein containing a nucleotide-binding site and leucine-rich repeats. Cell 78: 1089–1099 [DOI] [PubMed] [Google Scholar]
- Mitre LK, Teixeira-Silva NS, Rybak K, Magalhães DM, de Souza-Neto RR, Robatzek S, Zipfel C, de Souza AA (2021) The Arabidopsis immune receptor EFR increases resistance to the bacterial pathogens Xanthomonas and Xylella in transgenic sweet orange. Plant Biotechnol J 19: 1294–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miya A, Albert P, Shinya T, Desaki Y, Ichimura K, Shirasu K, Narusaka Y, Kawakami N, Kaku H, Shibuya N (2007) CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc Natl Acad Sci USA 104: 19613–19618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizumoto H, Nakamura I, Shimomoto Y, Sawada H, Tomita R, Sekine KT, Kiba A, Nishiguchi M, Kobayashi K, Hikichi Y (2012) Amino acids in Tobamovirus coat protein controlling pepper L(1a)' ' gene-mediated resistance. Mol Plant Pathol 13: 915–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan J, Matschi S, Shorinola O, Rovenich H, Matei A, Segonzac C, Malinovsky FG, Rathjen JP, MacLean D, Romeis T, et al. (2014) The calcium-dependent protein kinase CPK28 buffers plant immunity and regulates BIK1 turnover. Cell Host Microbe 16: 605–615 [DOI] [PubMed] [Google Scholar]
- Monino-Lopez D, Nijenhuis M, Kodde L, Kamoun S, Salehian H, Schentsnyi K, Stam RLokossou A, Abd-El-Haliem A, Visser RGF, et al. (2021) Allelic variants of the NLR protein Rpi-chc1 differentially recognize members of the Phytophthora infestans PexRD12/31 effector superfamily through the leucine-rich repeat domain. Plant J 107: 182–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mott GA, Thakur S, Smakowska E, Wang PW, Belkhadir Y, Desveaux D, Guttman DS (2016) Genomic screens identify a new phytobacterial microbe-associated molecular pattern and the cognate Arabidopsis receptor-like kinase that mediates its immune elicitation. Genome Biol 17: 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucyn TS, Clemente A, Andriotis VME, Balmuth AL, Oldroyd GED, Staskawicz BJ, Rathjen JP (2006) The tomato NBARC-LRR protein Prf interacts with Pto kinase in vivo to regulate specific plant immunity. Plant Cell 18: 2792–2806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhtar MS (2013) Engineering NLR immune receptors for broad-spectrum disease resistance. Trends Plant Sci 18: 469–472 [DOI] [PubMed] [Google Scholar]
- Nagy ED, Bennetzen JL (2008) Pathogen corruption and site-directed recombination at a plant disease resistance gene cluster. Genome Res 18: 1918–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy ED, Lee TC, Ramakrishna W, Xu Z, Klein PE, SanMiguel P, Cheng CP, Li J, Devos KM, Schertz K, et al. (2007) Fine mapping of the Pc locus of Sorghum bicolor, a gene controlling the reaction to a fungal pathogen and its host-selective toxin. Theor Appl Genet 114: 961–970 [DOI] [PubMed] [Google Scholar]
- Narusaka M, Shirasu K, Noutoshi Y, Kubo Y, Shiraishi T, Iwabuchi M, Narusaka Y (2009) RRS1 and RPS4 provide a dual Resistance-gene system against fungal and bacterial pathogens. Plant J 60: 218–226 [DOI] [PubMed] [Google Scholar]
- Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, Voinnet O, Jones JDG (2006) A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312: 436–439 [DOI] [PubMed] [Google Scholar]
- Navarro L, Zipfel C, Rowland O, Keller I, Robatzek S, Boller T, Jones JDG (2004) The transcriptional innate immune response to flg22. Interplay and overlap with Avr gene-dependent defense responses and bacterial pathogenesis. Plant Physiol 135: 1113–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navathe S, Yadav PS, Chand R, Mishra VK, Vasistha NK, Meher PK, Joshi AK, Gupta PK (2020) ToxA-Tsn1 interaction for spot blotch susceptibility in Indian wheat: an example of inverse gene-for-gene relationship. Plant Dis 104: 71–81 [DOI] [PubMed] [Google Scholar]
- Neiva IP, Silva AA da, Resende JF, Carvalho R, de C, Oliveira AMS, de, Maluf WR (2019) Tomato genotype resistance to whitefly mediated by allelochemicals and Mi gene. Chilean J Agric Res 79: 124–130 [Google Scholar]
- Ngou BPM, Ahn HK, Ding P, Jones JDG (2021a) Mutual potentiation of plant immunity by cell-surface and intracellular receptors. Nature 592: 110–115 [DOI] [PubMed] [Google Scholar]
- Ngou BPM, Ahn HK, Ding P, Redkar A, Brown H, Ma Y, Youles M, Tomlinson L, Jones JDG (2020) Estradiol-inducible AvrRps4 expression reveals distinct properties of TIR-NLR-mediated effector-triggered immunity. J Exp Bot 71: 2186–2197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngou BPM, Ding P, Jones JDG (2021b) Channeling plant immunity. Cell 184: 3358–3360 [DOI] [PubMed] [Google Scholar]
- Ngou BPM, Heal R, Wyler M, Schmid MW, Jones JD (2022) Concerted expansion and contraction of immune receptor gene repertoires in plant genomes. BioRxiv doi: 10.1101/2022.01.01.474684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngou BPM, Jones JDG, Ding P (2021c) Plant immune networks. Trends Plant Sci 27: 255–273 [DOI] [PubMed] [Google Scholar]
- Niehl A, Wyrsch I, Boller T, Heinlein M (2016) Double-stranded RNAs induce a pattern-triggered immune signaling pathway in plants. New Phytol 211: 1008–1019 [DOI] [PubMed] [Google Scholar]
- Ntoukakis V, Balmuth AL, Mucyn TS, Gutierrez JR, Jones AME, Rathjen JP (2013) The tomato Prf complex is a molecular trap for bacterial effectors based on Pto transphosphorylation. PLoS Pathog 9: e1003123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SK, Young C, Lee M, Oliva R, Bozkurt TO, Cano LM, Win J, Bos JIB, Liu H-Y, van Damme M, et al. (2009) In planta expression screens of Phytophthora infestans RXLR effectors reveal diverse phenotypes, including activation of the Solanum bulbocastanum disease resistance protein Rpi-blb2. Plant Cell 21: 2928–2947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paal J, Henselewski H, Muth J, Meksem K, Menéndez CM, Salamini F, Ballvora A, Gebhardt C (2004) Molecular cloning of the potato Gro1-4 gene conferring resistance to pathotype Ro1 of the root cyst nematode Globodera rostochiensis, based on a candidate gene approach. Plant J 38: 285–297 [DOI] [PubMed] [Google Scholar]
- Park TH, Vleeshouwers VGAA, Huigen DJ, van der Vossen EAG, van Eck HJ, Visser RGF (2005) Characterization and high-resolution mapping of a late blight resistance locus similar to R2 in potato. Theor Appl Genet 111: 591–597 [DOI] [PubMed] [Google Scholar]
- Parker JE, Holub EB, Frost LN, Falk A, Gunn ND, Daniels MJ (1996) Characterization of eds1, a mutation in Arabidopsis suppressing resistance to Peronospora parasitica specified by several different RPP genes. Plant Cell 8: 2033–2046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peart JR, Lu R, Sadanandom A, Malcuit I, Moffett P, Brice DC, Schauser L, Jaggard DAW, Xiao S, Coleman MJ, et al. (2002) Ubiquitin ligase-associated protein SGT1 is required for host and nonhost disease resistance in plants. Proc Natl Acad Sci USA 99: 10865–10869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peart JR, Mestre P, Lu R, Malcuit I, Baulcombe DC (2005) NRG1, a CC-NB-LRR protein, together with N, a TIR-NB-LRR protein, mediates resistance against tobacco mosaic virus. Curr Biol 15: 968–973 [DOI] [PubMed] [Google Scholar]
- Peil A, Hübert C, Wensing A, Horner M, Emeriewen OF, Richter K, Wöhner T, Chagné D, Orellana-Torrejon C, Saeed M, et al. (2019) Mapping of fire blight resistance in Malus ×robusta 5 flowers following artificial inoculation. BMC Plant Biol 19: 532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiró A, Cañizares MC, Rubio L, López C, Moriones E, Aramburu J, Sánchez-Navarro J (2014) The movement protein (NSm) of Tomato spotted wilt virus is the avirulence determinant in the tomato Sw-5 gene-based resistance. Mol Plant Pathol 15: 802–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pel M (2010) Mapping, isolation and characterization of genes responsible for late blight resistance in potato.
- Pelham J (1966) Resistance in tomato to tobacco mosaic virus. Euphytica 15: 258–267 [Google Scholar]
- Perraki A, DeFalco TA, Derbyshire P, Avila J, Séré D, Sklenar J, Qi X, Stransfeld L, Schwessinger B, Kadota Y, et al. (2018) Phosphocode-dependent functional dichotomy of a common co-receptor in plant signalling. Nature 561: 248–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petre B, Contreras MP, Bozkurt TO, Schattat MH, Sklenar J, Schornack S, Abd-El-Haliem A, Castells-Graells R, Lozano-Duran R, Dagdas YF, et al. (2021) Host-interactor screens of Phytophthora infestans RXLR proteins reveal vesicle trafficking as a major effector-targeted process. Plant Cell 33: 1447–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piazza S, Campa M, Pompili V, Costa LD, Salvagnin U, Nekrasov V, Zipfel C, Malnoy M (2021) The Arabidopsis pattern recognition receptor EFR enhances fire blight resistance in apple. BioRxiv doi: 10.1101/2021.01.22.427734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piedras P, Hammond-Kosack KE, Harrison K, Jones JDG (1998) Rapid, Cf-9 - and Avr9-dependent production of active oxygen species in tobacco suspension cultures. Mol Plant Microbe Interact 11: 1155–1166 [Google Scholar]
- Plomion C, Aury J-M, Amselem J, Leroy T, Murat F, Duplessis S, Faye S, Francillonne N, Labadie K, Le Provost G, et al. (2018) Oak genome reveals facets of long lifespan. Nat Plants 4: 440–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postma J, Liebrand TWH, Bi G, Evrard A, Bye RR, Mbengue M, Kuhn H, Joosten MHAJ, Robatzek S (2016) Avr4 promotes Cf-4 receptor-like protein association with the BAK1/SERK3 receptor-like kinase to initiate receptor endocytosis and plant immunity. New Phytol 210: 627–642 [DOI] [PubMed] [Google Scholar]
- Pruitt RN, Schwessinger B, Joe A, Thomas N, Liu F, Albert M, Robinson MR, Chan LJG, Luu DD, Chen H, et al. (2015) The rice immune receptor XA21 recognizes a tyrosine-sulfated protein from a Gram-negative bacterium. Sci Adv 1: e1500245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt RN, Zhang L, Saile SC, Karelina D, Fröhlich K, Wan W-L, Rao S, Gust AA, Locci F, Joosten MHAJ, et al. (2020) Arabidopsis cell surface LRR immune receptor signaling through the EDS1-PAD4-ADR1 node. BioRxiv doi: 10.1101/2020.11.23.391516 [Google Scholar]
- Pruitt RN, Locci F, Wanke F, Zhang L, Saile SC, Joe A, Karelina D, Hua C, Fröhlich K, Wan W-L, et al. (2021) The EDS1-PAD4-ADR1 node mediates Arabidopsis pattern-triggered immunity. Nature 598: 495–499 [DOI] [PubMed] [Google Scholar]
- Qin J, Wang K, Sun L, Xing H, Wang S, Li L, Chen S, Guo HS, Zhang J (2018) The plant-specific transcription factors CBP60g and SARD1 are targeted by a Verticillium secretory protein VdSCP41 to modulate immunity. eLife 7: e34902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai AK, Kumar SP, Gupta SK, Gautam N, Singh NK, Sharma TR (2011) Functional complementation of rice blast resistance gene Pi-k h (Pi54) conferring resistance to diverse strains of Magnaporthe oryzae. J Plant Biochem Biotechnol 20: 55–65 [Google Scholar]
- Rao S, Zhou Z, Miao P, Bi G, Hu M, Wu Y, Feng F, Zhang X, Zhou JM (2018) Roles of receptor-like cytoplasmic kinase VII members in pattern-triggered immune signaling. Plant Physiol 177: 1679–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen MW, Roux M, Petersen M, Mundy J (2012) MAP kinase cascades in arabidopsis innate immunity. Front Plant Sci 3: 169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read AC, Hutin M, Moscou MJ, Rinaldi FC, Bogdanove AJ (2020a) Cloning of the rice Xo1 resistance gene and interaction of the Xo1 protein with the defense-suppressing xanthomonas effector Tal2h. Mol Plant Microbe Interact 33: 1189–1195 [DOI] [PubMed] [Google Scholar]
- Read AC, Moscou MJ, Zimin AV, Pertea G, Meyer RS, Purugganan MD, Leach JE, Triplett LR, Salzberg SL, Bogdanove AJ (2020b) Genome assembly and characterization of a complex zfBED-NLR gene-containing disease resistance locus in Carolina Gold Select rice with Nanopore sequencing. PLoS Genet 16: e1008571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redkar A, Cevik V, Bailey K, Furzer OJ, Fairhead S, Borhan MH, Holub EB, Jones JDG (2021) The Arabidopsis WRR4A and WRR4B paralogous NLR proteins both confer recognition of multiple Albugo candida effectors. BioRxiv doi: 10.1101/2021.03.29.436918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentel MC, Leonelli L, Dahlbeck D, Zhao B, Staskawicz BJ (2008) Recognition of the Hyaloperonospora parasitica effector ATR13 triggers resistance against oomycete, bacterial, and viral pathogens. Proc Natl Acad Sci USA 105: 1091–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rep M, van der Does HC, Meijer M, van Wijk R, Houterman PM, Dekker HL, de Koster CG, Cornelissen BJC (2004) A small, cysteine-rich protein secreted by Fusarium oxysporum during colonization of xylem vessels is required for I-3-mediated resistance in tomato. Mol Microbiol 53: 1373–1383 [DOI] [PubMed] [Google Scholar]
- Rhodes J, Roman AO, Bjornson M, Brandt B, Derbyshire P, Wyler M, Schmid M, Menke FLH, Santiago J, Zipfel C (2021a) Perception of a conserved family of plant signalling peptides by the receptor kinase HSL3. BioRxiv doi: 10.1101/2021.10.25.465685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes J, Yang H, Moussu S, Boutrot F, Santiago J, Zipfel C (2021b) Perception of a divergent family of phytocytokines by the Arabidopsis receptor kinase MIK2. Nat Commun 12: 705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridout CJ, Skamnioti P, Porritt O, Sacristan S, Jones JDG, Brown JKM (2006) Multiple avirulence paralogues in cereal powdery mildew fungi may contribute to parasite fitness and defeat of plant resistance. Plant Cell 18: 2402–2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeis T, Piedras P, Jones JD (2000) Resistance gene-dependent activation of a calcium-dependent protein kinase in the plant defense response. Plant Cell 12: 803–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeis T, Piedras P, Zhang S, Klessig DF, Hirt H, Jones JD (1999) Rapid Avr9- and Cf-9 -dependent activation of MAP kinases in tobacco cell cultures and leaves: convergence of resistance gene, elicitor, wound, and salicylate responses. Plant Cell 11: 273–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron M, Avni A (2004) The receptor for the fungal elicitor ethylene-inducing xylanase is a member of a resistance-like gene family in tomato. Plant Cell 16: 1604–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi M, Goggin FL, Milligan SB, Kaloshian I, Ullman DE, Williamson VM (1998) The nematode resistance gene Mi of tomato confers resistance against the potato aphid. Proc Natl Acad Sci USA 95: 9750–9754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux M, Schwessinger B, Albrecht C, Chinchilla D, Jones A, Holton N, Malinovsky FG, Tör M, de Vries S, Zipfel C (2011) The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell 23: 2440–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland O, Ludwig AA, Merrick CJ, Baillieul F, Tracy FE, Durrant WE, Fritz-Laylin L, Nekrasov V, Sjölander K, Yoshioka H, et al. (2005) Functional analysis of Avr9/Cf-9 rapidly elicited genes identifies a protein kinase, ACIK1, that is essential for full Cf-9-dependent disease resistance in tomato. Plant Cell 17: 295–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saile SC, Jacob P, Castel B, Jubic LM, Salas-Gonzalez I, Bäcker M, Jones JDG, Dangl JL, El Kasmi F (2020) Two unequally redundant “helper” immune receptor families mediate Arabidopsis thaliana intracellular “sensor” immune receptor functions. PLoS Biol 18: e3000783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T, Meshi T, Takamatsu N, Okada Y (1987) Coat protein gene sequence of tobacco mosaic virus encodes a host response determinant. Proc Natl Acad Sci USA 84: 6074–6077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salcedo A, Rutter W, Wang S, Akhunova A, Bolus S, Chao S, Anderson N, De Soto MF, Rouse M, Szabo L, et al. (2017) Variation in the AvrSr35 gene determines Sr35 resistance against wheat stem rust race Ug99. Science 358: 1604–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarris PF, Duxbury Z, Huh SU, Ma Y, Segonzac C, Sklenar J, Derbyshire P, Cevik V, Rallapalli G, Saucet SB, et al. (2015) A plant immune receptor detects pathogen effectors that target WRKY transcription factors. Cell 161: 1089–1100 [DOI] [PubMed] [Google Scholar]
- Sarris PF, Cevik V, Dagdas G, Jones JDG, Krasileva KV (2016) Comparative analysis of plant immune receptor architectures uncovers host proteins likely targeted by pathogens. BMC Biol 14: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saucet SB, Ma Y, Sarris PF, Furzer OJ, Sohn KH, Jones JDG (2015) Two linked pairs of Arabidopsis TNL resistance genes independently confer recognition of bacterial effector AvrRps4. Nat Commun 6: 6338. [DOI] [PubMed] [Google Scholar]
- Saur IM, Bauer S, Kracher B, Lu X, Franzeskakis L, Müller MC, Sabelleck B, Kümmel F, Panstruga R, Maekawa T, et al. (2019) Multiple pairs of allelic MLA immune receptor-powdery mildew AVRA effectors argue for a direct recognition mechanism. eLife 8: e44471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saur IML, Kadota Y, Sklenar J, Holton NJ, Smakowska E, Belkhadir Y, Zipfel C, Rathjen JP (2016) NbCSPR underlies age-dependent immune responses to bacterial cold shock protein in Nicotiana benthamiana. Proc Natl Acad Sci USA 113: 3389–3394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt SM, Lukasiewicz J, Farrer R, van Dam P, Bertoldo C, Rep M (2016) Comparative genomics of Fusarium oxysporum f. sp. melonis reveals the secreted protein recognized by the Fom-2 resistance gene in melon. New Phytol 209: 307–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schornack S, Ballvora A, Gürlebeck D, Peart J, Baulcombe D, Ganal M, Baker B, Bonas U, Lahaye T (2004) The tomato resistance protein Bs4 is a predicted non-nuclear TIR-NB-LRR protein that mediates defense responses to severely truncated derivatives of AvrBs4 and overexpressed AvrBs3. Plant J 37: 46–60 [DOI] [PubMed] [Google Scholar]
- Schornack S, Peter K, Bonas U, Lahaye T (2005) Expression levels of avrBs3-like genes affect recognition specificity in tomato Bs4- but not in pepper Bs3-mediated perception. Mol Plant Microbe Interact 18: 1215–1225 [DOI] [PubMed] [Google Scholar]
- Schultink A, Qi T, Bally J, Staskawicz B (2019) Using forward genetics in Nicotiana benthamiana to uncover the immune signaling pathway mediating recognition of the Xanthomonas perforans effector XopJ4. New Phytol 221: 1001–1009 [DOI] [PubMed] [Google Scholar]
- Schultink A, Qi T, Lee A, Steinbrenner AD, Staskawicz B (2017) Roq1 mediates recognition of the Xanthomonas and Pseudomonas effector proteins XopQ and HopQ1. Plant J 92: 787–795 [DOI] [PubMed] [Google Scholar]
- Schulze S, Yu L, Ehinger A, Kolb D, Saile S, Stahl M, Franz-Wachtel M, Li L, El Kasmi F, Cevik V, et al. (2021) The TIR-NBS-LRR protein CSA1 is required for autoimmune cell death in Arabidopsis pattern recognition co-receptor bak1 and bir3 mutants. BioRxiv doi: 10.1101/2021.04.11.438637 [Google Scholar]
- Schwessinger B, Bahar O, Thomas N, Holton N, Nekrasov V, Ruan D, Canlas PE, Daudi A, Petzold CJ, Singan VR, et al. (2015) Transgenic expression of the dicotyledonous pattern recognition receptor EFR in rice leads to ligand-dependent activation of defense responses. PLoS Pathog 11: e1004809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwessinger B, Roux M, Kadota Y, Ntoukakis V, Sklenar J, Jones A, Zipfel C (2011) Phosphorylation-dependent differential regulation of plant growth, cell death, and innate immunity by the regulatory receptor-like kinase BAK1. PLoS Genet 7: e1002046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segonzac C, Macho AP, Sanmartín M, Ntoukakis V, Sánchez-Serrano JJ, Zipfel C (2014) Negative control of BAK1 by protein phosphatase 2A during plant innate immunity. EMBO J 33: 2069–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo YS, Rojas MR, Lee JY, Lee SW, Jeon JS, Ronald P, Lucas WJ, Gilbertson RL (2006) A viral resistance gene from common bean functions across plant families and is up-regulated in a non-virus-specific manner. Proc Natl Acad Sci USA 103: 11856–11861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan L, He P, Li J, Heese A, Peck SC, Nürnberger T, Martin GB, Sheen J (2008) Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host Microbe 4: 17–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao F, Golstein C, Ade J, Stoutemyer M, Dixon JE, Innes RW (2003) Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science 301: 1230–1233 [DOI] [PubMed] [Google Scholar]
- Shi G, Zhang Z, Friesen TL, Raats D, Fahima T, Brueggeman RS, Lu S, Trick HN, Liu Z, Chao W, et al. (2016) The hijacking of a receptor kinase-driven pathway by a wheat fungal pathogen leads to disease. Sci Adv 2: e1600822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasu K (2009) The HSP90-SGT1 chaperone complex for NLR immune sensors. Annu Rev Plant Biol 60: 139–164 [DOI] [PubMed] [Google Scholar]
- Shiu SH, Bleecker AB (2003) Expansion of the receptor-like kinase/Pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol 132: 530–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaprasad PV, Chen HM, Patel K, Bond DM, Santos BACM, Baulcombe DC (2012) A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. Plant Cell 24: 859–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smakowska-Luzan E, Mott GA, Parys K, Stegmann M, Howton TC, Layeghifard M, Neuhold J, Lehner A, Kong J, Grünwald K, et al. (2018) An extracellular network of Arabidopsis leucine-rich repeat receptor kinases. Nature 553: 342–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobczak M, Avrova A, Jupowicz J, Phillips MS, Ernst K, Kumar A (2005) Characterization of susceptibility and resistance responses to potato cyst nematode (Globodera spp.) infection of tomato lines in the absence and presence of the broad-spectrum nematode resistance Hero gene. Mol Plant Microbe Interact 18: 158–168 [DOI] [PubMed] [Google Scholar]
- Sone T, Takeuchi S, Miki S, Satoh Y, Ohtsuka K, Abe A, Asano K (2013) Homologous recombination causes the spontaneous deletion of AVR-Pia in Magnaporthe oryzae. FEMS Microbiol Lett 339: 102–109 [DOI] [PubMed] [Google Scholar]
- Song T, Kale SD, Arredondo FD, Shen D, Su L, Liu L, Wu Y, Wang Y, Dou D, Tyler BM (2013) Two RxLR avirulence genes in Phytophthora sojae determine soybean Rps1k-mediated disease resistance. Mol Plant Microbe Interact 26: 711–720 [DOI] [PubMed] [Google Scholar]
- Song WY, Wang GL, Chen LL, Kim HS, Pi LY, Holsten T, Gardner J, Wang B, Zhai WX, Zhu LH, et al. (1995) A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270: 1804–1806 [DOI] [PubMed] [Google Scholar]
- Srichumpa P, Brunner S, Keller B, Yahiaoui N (2005) Allelic series of four powdery mildew resistance genes at the Pm3 locus in hexaploid bread wheat. Plant Physiol 139: 885–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmann M, Monaghan J, Smakowska-Luzan E, Rovenich H, Lehner A, Holton N, Belkhadir Y, Zipfel C (2017) The receptor kinase FER is a RALF-regulated scaffold controlling plant immune signaling. Science 355: 287–289 [DOI] [PubMed] [Google Scholar]
- Steinbrenner AD, Munoz-Amatriain M, Venegas JMA, Lo S, Shi D, Holton N, Zipfel C, Abagyan R, Huffaker A, Close TJ, et al. (2019) A receptor for herbivore-associated molecular patterns mediates plant immunity. BioRxiv doi: 10.1101/679803 [Google Scholar]
- Su J, Yang L, Zhu Q, Wu H, He Y, Liu Y, Xu J, Jiang D, Zhang S (2018) Active photosynthetic inhibition mediated by MPK3/MPK6 is critical to effector-triggered immunity. PLoS Biol 16: e2004122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T, Huang J, Xu Y, Verma V, Jing B, Sun Y, Ruiz Orduna A, Tian H, Huang X, Xia S, et al. (2020) Redundant CAMTA transcription factors negatively regulate the biosynthesis of salicylic acid and N-hydroxypipecolic acid by modulating the expression of SARD1 and CBP60g. Mol Plant 13: 144–156 [DOI] [PubMed] [Google Scholar]
- Sun T, Zhang Y, Li Y, Zhang Q, Ding Y, Zhang Y (2015) ChIP-seq reveals broad roles of SARD1 and CBP60g in regulating plant immunity. Nat Commun 6: 10159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Lapin D, Feehan JM, Stolze SC, Kramer K, Dongus JA, Rzemieniewski J, Blanvillain-Baufumé S, Harzen A, Bautor J, et al. (2021) Pathogen effector recognition-dependent association of NRG1 with EDS1 and SAG101 in TNL receptor immunity. Nat Commun 12: 3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Li L, Macho AP, Han Z, Hu Z, Zipfel C, Zhou JM, Chai J (2013) Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex. Science 342: 624–628 [DOI] [PubMed] [Google Scholar]
- Swanson KV, Deng M, Ting JPY (2019) The NLRP3 inflammasome: molecular activation and regulation to therapeutics. Nat Rev Immunol 19: 477–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweat TA, Lorang JM, Bakker EG, Wolpert TJ (2008) Characterization of natural and induced variation in the LOV1 gene, a CC-NB-LRR gene conferring victorin sensitivity and disease susceptibility in Arabidopsis. Mol Plant Microbe Interact 21: 7–19 [DOI] [PubMed] [Google Scholar]
- Takagi M, Hamano K, Takagi H, Morimoto T, Akimitsu K, Terauchi R, Shirasu K, Ichimura K (2019) Disruption of the MAMP-induced MEKK1-MKK1/MKK2-MPK4 pathway activates the TNL immune receptor SMN1/RPS6. Plant Cell Physiol 60: 778–787 [DOI] [PubMed] [Google Scholar]
- Takahashi A, Casais C, Ichimura K, Shirasu K (2003) HSP90 interacts with RAR1 and SGT1 and is essential for RPS2-mediated disease resistance in Arabidopsis. Proc Natl Acad Sci USA 100: 11777–11782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tameling WIL, Baulcombe DC (2007) Physical association of the NB-LRR resistance protein Rx with a Ran GTPase-activating protein is required for extreme resistance to Potato virus X. Plant Cell 19: 1682–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarr DEK, Alexander HM (2009) TIR-NBS-LRR genes are rare in monocots: evidence from diverse monocot orders. BMC Res Notes 2: 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateda C, Zhang Z, Shrestha J, Jelenska J, Chinchilla D, Greenberg JT (2014) Salicylic acid regulates Arabidopsis microbial pattern receptor kinase levels and signaling. Plant Cell 26: 4171–4187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CM, Jones DA, Parniske M, Harrison K, Balint-Kurti PJ, Hatzixanthis K, Jones JD (1997) Characterization of the tomato Cf-4 gene for resistance to Cladosporium fulvum identifies sequences that determine recognitional specificity in Cf-4 and Cf-9. Plant Cell 9: 2209–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas NC, Hendrich CG, Gill US, Allen C, Hutton SF, Schultink A (2020) The immune receptor Roq1 confers resistance to the bacterial pathogens xanthomonas, Pseudomonas syringae, and ralstonia in tomato. Front Plant Sci 11: 463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma BPHJ, Nürnberger T, Joosten MHAJ (2011) Of PAMPs and effectors: the blurred PTI-ETI dichotomy. Plant Cell 23: 4–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thor K, et al. (2020) The calcium-permeable channel OSCA1.3 regulates plant stomatal immunity. Nature 585: 569–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thordal-Christensen H, Zhang Z, Wei Y, Collinge DB (1997) Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J 11: 1187–1194 [Google Scholar]
- Tian H, Chen S, Wu Z, Ao K, Yaghmaiean H, Sun T, Huang W, Xu F, Zhang Y, Wang S, et al. (2020) Activation of TIR signaling is required for pattern-triggered immunity. BioRxiv doi: 10.1101/2020.12.27.424494 [Google Scholar]
- Tian H, Wu Z, Chen S, Ao K, Huang W, Yaghmaiean H, Sun T, Xu F, Zhang Y, Wang S, et al. (2021) Activation of TIR signalling boosts pattern-triggered immunity. Nature 598: 500–503 [DOI] [PubMed] [Google Scholar]
- Tian W, Hou C, Ren Z, Wang C, Zhao F, Dahlbeck D, Hu S, Zhang L, Niu Q, Li L, et al. (2019) A calmodulin-gated calcium channel links pathogen patterns to plant immunity. Nature 572: 131–135 [DOI] [PubMed] [Google Scholar]
- Tomita R, Sekine KT, Mizumoto H, Sakamoto M, Murai J, Kiba A, Hikichi Y, Suzuki K, Kobayashi K (2011) Genetic basis for the hierarchical interaction between Tobamovirus spp. and L resistance gene alleles from different pepper species. Mol Plant Microbe Interact 24: 108–117 [DOI] [PubMed] [Google Scholar]
- Torres MA, Dangl JL, Jones JDG (2002) Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc Natl Acad Sci USA 99: 517–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres MA, Onouchi H, Hamada S, Machida C, Hammond-Kosack KE, Jones JD (1998) Six Arabidopsis thaliana homologues of the human respiratory burst oxidase (gp91phox). Plant J 14: 365–370 [DOI] [PubMed] [Google Scholar]
- Triplett LR, Cohen SP, Heffelfinger C, Schmidt CL, Huerta AI, Tekete C, Verdier V, Bogdanove AJ, Leach JE (2016) A resistance locus in the American heirloom rice variety Carolina Gold Select is triggered by TAL effectors with diverse predicted targets and is effective against African strains of Xanthomonas oryzae pv. oryzicola. Plant J 87: 472–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman W, Sreekanta S, Lu Y, Bethke G, Tsuda K, Katagiri F, Glazebrook J (2013) The CALMODULIN-BINDING PROTEIN60 family includes both negative and positive regulators of plant immunity. Plant Physiol 163: 1741–1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K, Mine A, Bethke G, Igarashi D, Botanga CJ, Tsuda Y, Glazebrook J, Sato M, Katagiri F (2013) Dual regulation of gene expression mediated by extended MAPK activation and salicylic acid contributes to robust innate immunity in Arabidopsis thaliana. PLoS Genet 9: e1004015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Burg HA, Harrison SJ, Joosten MHAJ, Vervoort J, de Wit PJGM (2006) Cladosporium fulvum Avr4 protects fungal cell walls against hydrolysis by plant chitinases accumulating during infection. Mol Plant Microbe Interact 19: 1420–1430 [DOI] [PubMed] [Google Scholar]
- van den Burg HA, Tsitsigiannis DI, Rowland O, Lo J, Rallapalli G, Maclean D, Takken FLW, Jones JDG (2008) The F-box protein ACRE189/ACIF1 regulates cell death and defense responses activated during pathogen recognition in tobacco and tomato. Plant Cell 20: 697–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Biezen EA, Jones JD (1998) Plant disease-resistance proteins and the gene-for-gene concept. Trends Biochem Sci 23: 454–456 [DOI] [PubMed] [Google Scholar]
- van der Burgh AM, Joosten MHAJ (2019) Plant immunity: thinking outside and inside the box. Trends Plant Sci 24: 587–601 [DOI] [PubMed] [Google Scholar]
- van der Hoorn RAL, Kamoun S (2008) From Guard to Decoy: a new model for perception of plant pathogen effectors. Plant Cell 20: 2009–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Voort JR, Wolters P, Folkertsma R., Hutten R., van Zandvoort P, Vinke H, Kanyuka K, Bendahmane A, Jacobsen E, Janssen R, et al. (1997) Mapping of the cyst nematode resistance locus Gpa2 in potato using a strategy based on comigrating AFLP markers. Theor Appl Genet 95: 874–880 [Google Scholar]
- van Wersch S, Tian L, Hoy R, Li X (2020) Plant nlrs: the whistleblowers of plant immunity. Plant Commun 1: 100016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vleeshouwers VGAA, Rietman H, Krenek P, Champouret N, Young C, Oh S-K, Wang M, Bouwmeester K, Vosman B, Visser RGF, et al. (2008) Effector genomics accelerates discovery and functional profiling of potato disease resistance and Phytophthora infestans avirulence genes. PLoS One 3: e2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vleeshouwers VGAA, Raffaele S, Vossen JH, Champouret N, Oliva R, Segretin ME, Rietman H, Cano LM, Lokossou A, Kessel G, et al. (2011) Understanding and exploiting late blight resistance in the age of effectors. Annu Rev Phytopathol 49: 507–531 [DOI] [PubMed] [Google Scholar]
- Vo KTX, Lee SK, Halane MK, Song MY, Hoang TV, Kim CY, Park SY, Jeon J, Kim ST, Sohn KH, et al. (2019) Pi5 and pii paired nlrs are functionally exchangeable and confer similar disease resistance specificity. Mol Cells 42: 637–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt CA (2014) Callose-mediated resistance to pathogenic intruders in plant defense-related papillae. Front Plant Sci 5: 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vossen JH, van Arkel G, Bergervoet M, Jo KR, Jacobsen E, Visser RGF (2016) The Solanum demissum R8 late blight resistance gene is an Sw-5 homologue that has been deployed worldwide in late blight resistant varieties. Theor Appl Genet 129: 1785–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner S, Stuttmann J, Rietz S, Guerois R, Brunstein E, Bautor J, Niefind K, Parker JE (2013) Structural basis for signaling by exclusive EDS1 heteromeric complexes with SAG101 or PAD4 in plant innate immunity. Cell Host Microbe 14: 619–630 [DOI] [PubMed] [Google Scholar]
- Wan J, Tanaka K, Zhang XC, Son GH, Brechenmacher L, Nguyen THN, Stacey G (2012) LYK4, a lysin motif receptor-like kinase, is important for chitin signaling and plant innate immunity in Arabidopsis. Plant Physiol 160: 396–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J, Zhang XC, Neece D, Ramonell KM, Clough S, Kim SY, Stacey MG, Stacey G (2008) A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell 20: 471–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L, Essuman K, Anderson RG, Sasaki Y, Monteiro F, Chung EH, Osborne Nishimura E, DiAntonio A, Milbrandt J, Dangl JL, et al. (2019a) TIR domains of plant immune receptors are NAD+-cleaving enzymes that promote cell death. Science 365: 799–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan WL, Zhang L, Pruitt R, Zaidem M, Brugman R, Ma X, Krol E, Perraki A, Kilian J, Grossmann G, et al. (2019b) Comparing Arabidopsis receptor kinase and receptor protein-mediated immune signaling reveals BIK1-dependent differences. New Phytol 221: 2080–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Zhou M, Zhang X, Yao J, Zhang Y, Mou Z (2017) A lectin receptor kinase as a potential sensor for extracellular nicotinamide adenine dinucleotide in Arabidopsis thaliana. eLife 6: e25474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Roux B, Feng F, Guy E, Li L, Li N, Zhang X, Lautier M, Jardinaud M-F, Chabannes M, et al. (2015) The decoy substrate of a pathogen effector and a pseudokinase specify pathogen-induced modified-self recognition and immunity in plants. Cell Host Microbe 18: 285–295 [DOI] [PubMed] [Google Scholar]
- Wang J, Grubb LE, Wang J, Liang X, Li L, Gao C, Ma M, Feng F, Li M, Li L, et al. (2018a) A regulatory module controlling homeostasis of a plant immune kinase. Mol Cell 69: 493–504.e6 [DOI] [PubMed] [Google Scholar]
- Wang J, Hu M, Wang J, Qi J, Han Z, Wang G, Qi Y, Wang HW, Zhou JM, Chai J (2019a) Reconstitution and structure of a plant NLR resistosome conferring immunity. Science 364: eaav5870. [DOI] [PubMed] [Google Scholar]
- Wang J, Wang J, Hu M, Wu S, Qi J, Wang G, Han Z, Qi Y, Gao N, Wang HW, et al. (2019b) Ligand-triggered allosteric ADP release primes a plant NLR complex. Science 364: eaav5868. [DOI] [PubMed] [Google Scholar]
- Wang L, Albert M, Einig E, Fürst U, Krust D, Felix G (2016) The pattern-recognition receptor CORE of Solanaceae detects bacterial cold-shock protein. Nat Plants 2: 16185. [DOI] [PubMed] [Google Scholar]
- Wang L, Einig E, Almeida-Trapp M, Albert M, Fliegmann J, Mithöfer A, Kalbacher H, Felix G (2018b) The systemin receptor SYR1 enhances resistance of tomato against herbivorous insects. Nat Plants 4: 152–156 [DOI] [PubMed] [Google Scholar]
- Wang Y, Li J, Hou S, Wang X, Li Y, Ren D, Chen S, Tang X, Zhou JM (2010) A Pseudomonas syringae ADP-ribosyltransferase inhibits Arabidopsis mitogen-activated protein kinase kinases. Plant Cell 22: 2033–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Schuck S, Wu J, Yang P, Döring AC, Zeier J, Tsuda K (2018c) A MPK3/6-WRKY33-ALD1-pipecolic acid regulatory loop contributes to systemic acquired resistance. Plant Cell 30: 2480–2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Xu Y, Sun Y, Wang H, Qi J, Wan B, Ye W, Lin Y, Shao Y, Dong S, Tyler BM, Wang Y (2018d) Leucine-rich repeat receptor-like gene screen reveals that Nicotiana RXEG1 regulates glycoside hydrolase 12 MAMP detection. Nat Commun 9: 594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warmerdam S, Sterken MG, Sukarta OCA, van Schaik CC, Oortwijn MEP, Lozano-Torres JL, Bakker J, Smant G, Goverse A (2020) The TIR-NB-LRR pair DSC1 and WRKY19 contributes to basal immunity of Arabidopsis to the root-knot nematode Meloidogyne incognita. BMC Plant Biol 20: 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber H, Pfitzner AJ (1998) Tm-2(2) resistance in tomato requires recognition of the carboxy terminus of the movement protein of tomato mosaic virus. Mol Plant Microbe Interact 11: 498–503 [DOI] [PubMed] [Google Scholar]
- Wen RH, Khatabi B, Ashfield T, Saghai Maroof MA, Hajimorad MR (2013) The HC-Pro and P3 cistrons of an avirulent Soybean mosaic virus are recognized by different resistance genes at the complex Rsv1 locus. Mol Plant Microbe Interact 26: 203–215 [DOI] [PubMed] [Google Scholar]
- Westerink N, Brandwagt BF, de Wit PJGM, Joosten MHAJ (2004) Cladosporium fulvum circumvents the second functional resistance gene homologue at the Cf-4 locus (Hcr9-4E) by secretion of a stable avr4E isoform. Mol Microbiol 54: 533–545 [DOI] [PubMed] [Google Scholar]
- Whitham S, Dinesh-Kumar SP, Choi D, Hehl R, Corr C, Baker B (1994) The product of the tobacco mosaic virus resistance gene N: similarity to toll and the interleukin-1 receptor. Cell 78: 1101–1115 [DOI] [PubMed] [Google Scholar]
- Williams SJ, Sohn KH, Wan L, Bernoux M, Sarris PF, Segonzac C, Ve T, Ma Y, Saucet SB, Ericsson DJ, et al. (2014) Structural basis for assembly and function of a heterodimeric plant immune receptor. Science 344: 299–303 [DOI] [PubMed] [Google Scholar]
- Willmann R, Lajunen HM, Erbs G, Newman M-A, Kolb D, Tsuda K, Katagiri F, Fliegmann J, Bono J-J, Cullimore JV, et al. (2011) Arabidopsis lysin-motif proteins LYM1 LYM3 CERK1 mediate bacterial peptidoglycan sensing and immunity to bacterial infection. Proc Natl Acad Sci USA 108: 19824–19829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witek K, Lin X, Karki HS, Jupe F, Witek AI, Steuernagel B, Stam R, van Oosterhout C, Fairhead S, Heal R, et al. (2021) A complex resistance locus in Solanum americanum recognizes a conserved Phytophthora effector. Nat Plants 7: 198–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu AJ, Andriotis VME, Durrant MC, Rathjen JP (2004) A patch of surface-exposed residues mediates negative regulation of immune signaling by tomato Pto kinase. Plant Cell 16: 2809–2821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CH, Abd-El-Haliem A, Bozkurt TO, Belhaj K, Terauchi R, Vossen JH, Kamoun S (2017a) NLR network mediates immunity to diverse plant pathogens. Proc Natl Acad Sci USA 114: 8113–8118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CH, Derevnina L, Kamoun S (2018) Receptor networks underpin plant immunity. Science 360: 1300–1301 [DOI] [PubMed] [Google Scholar]
- Wu F, Chi Y, Jiang Z, Xu Y, Xie L, Huang F, Wan D, Ni J, Yuan F, Wu X, et al. (2020a) Hydrogen peroxide sensor HPCA1 is an LRR receptor kinase in Arabidopsis. Nature 578: 577–581 [DOI] [PubMed] [Google Scholar]
- Wu J, Kou Y, Bao J, Li Y, Tang M, Zhu X, Ponaya A, Xiao G, Li J, Li C, et al. (2015) Comparative genomics identifies the Magnaporthe oryzae avirulence effector AvrPi9 that triggers Pi9-mediated blast resistance in rice. New Phytol 206: 1463–1475 [DOI] [PubMed] [Google Scholar]
- Wu J, van der Burgh A, Bi G, Zhang L, Alfano JR, Martin GB, Joosten MHAJ (2017b) The bacterial effector AvrPto targets the regulatory co-receptor SOBIR1 and suppresses defence signalling mediated by the receptor-like protein Cf-4 . Mol Plant Microbe Interact 31: 75–85 [DOI] [PubMed] [Google Scholar]
- Wu Y, Gao Y, Zhan Y, Kui H, Liu H, Yan L, Kemmerling B, Zhou JM, He K, Li J (2020b) Loss of the common immune coreceptor BAK1 leads to NLR-dependent cell death. Proc Natl Acad Sci USA 117: 27044–27053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Li M, Dong OX, Xia S, Liang W, Bao Y, Wasteneys G, Li X (2019) Differential regulation of TNL-mediated immune signaling by redundant helper CNLs. New Phytol 222: 938–953 [DOI] [PubMed] [Google Scholar]
- Wu Z, Tian L, Li X (2021a) N-terminally truncated helper NLR NRG1C antagonizes immunity mediated by its full-length neighbors NRG1A and NRG1B. BioRxiv doi: 10.1101/2021.01.27.428547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Tian L, Liu X, Zhang Y, Li X (2021b) TIR signaling promotes the interactions between EDS1/PAD4 and ADR1-L1 and oligomerization of ADR1-L1. BioRxiv doi: 10.1101/2021.05.23.445317 [Google Scholar]
- Wu Z, Tong M, Tian L, Zhu C, Liu X, Zhang Y, Li X (2020c) Plant E3 ligases SNIPER1 and SNIPER2 broadly regulate the homeostasis of sensor NLR immune receptors. EMBO J 39: e104915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang T, Zong N, Zou Y, Wu Y, Zhang J, Xing W, Li Y, Tang X, Zhu L, Chai J, et al. (2008) Pseudomonas syringae effector AvrPto blocks innate immunity by targeting receptor kinases. Curr Biol 18: 74–80 [DOI] [PubMed] [Google Scholar]
- Xiao S, Ellwood S, Calis O, Patrick E, Li T, Coleman M, Turner JG (2001) Broad-spectrum mildew resistance in Arabidopsis thaliana mediated by RPW8. Science 291: 118–120 [DOI] [PubMed] [Google Scholar]
- Xiao Y, Stegmann M, Han Z, DeFalco TA, Parys K, Xu L, Belkhadir Y, Zipfel C, Chai J (2019) Mechanisms of RALF peptide perception by a heterotypic receptor complex. Nature 572: 270–274 [DOI] [PubMed] [Google Scholar]
- Xing W, Zou Y, Liu Q, Liu J, Luo X, Huang Q, Chen S, Zhu L, Bi R, Hao Q, et al. (2007) The structural basis for activation of plant immunity by bacterial effector protein AvrPto. Nature 449: 243–247 [DOI] [PubMed] [Google Scholar]
- Xu F, Xu S, Wiermer M, Zhang Y, Li X (2012) The cyclin L homolog MOS12 and the MOS4-associated complex are required for the proper splicing of plant resistance genes. Plant J 70: 916–928 [DOI] [PubMed] [Google Scholar]
- Xu S, Zhang Z, Jing B, Gannon P, Ding J, Xu F, Li X, Zhang Y (2011) Transportin-SR is required for proper splicing of resistance genes and plant immunity. PLoS Genet 7: e1002159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Yamaguchi K, Shirakawa T, Nakagami H, Mine A, Ishikawa K, Fujiwara M, Narusaka M, Narusaka Y, Ichimura K, et al. (2016) The Arabidopsis CERK1-associated kinase PBL27 connects chitin perception to MAPK activation. EMBO J 35: 2468–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Huffaker A, Bryan AC, Tax FE, Ryan CA (2010) PEPR2 is a second receptor for the Pep1 and Pep2 peptides and contributes to defense responses in Arabidopsis. Plant Cell 22: 508–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Pearce G, Ryan CA (2006) The cell surface leucine-rich repeat receptor for AtPep1, an endogenous peptide elicitor in Arabidopsis, is functional in transgenic tobacco cells. Proc Natl Acad Sci USA 103: 10104–10109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CW, González-Lamothe R, Ewan RA, Rowland O, Yoshioka H, Shenton M, Ye H, O’Donnell E, Jones JDG, Sadanandom A (2006) The E3 ubiquitin ligase activity of arabidopsis PLANT U-BOX17 and its functional tobacco homolog ACRE276 are required for cell death and defense. Plant Cell 18: 1084–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang KY, Liu Y, Zhang S (2001) Activation of a mitogen-activated protein kinase pathway is involved in disease resistance in tobacco. Proc Natl Acad Sci USA 98: 741–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon M, Rikkerink EHA (2020) Rpa1 mediates an immune response to avrRpm1Psa and confers resistance against Pseudomonas syringae pv. actinidiae. Plant J 102: 688–702 [DOI] [PubMed] [Google Scholar]
- Yoshimura S, Yamanouchi U, Katayose Y, Toki S, Wang ZX, Kono I, Kurata N, Yano M, Iwata N, Sasaki T (1998) Expression of Xa1, a bacterial blight-resistance gene in rice, is induced by bacterial inoculation. Proc Natl Acad Sci USA 95: 1663–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Song W, Tan EYJ, Liu L, Cao Y, Jirschitzka J, Li E, Logemann E, Xu C, Huang S, et al. (2021) TIR domains of plant immune receptors are 2’,3'-cAMP/cGMP synthetases mediating cell death. BioRxiv doi: 10.1101/2021.11.09.467869 [DOI] [PubMed] [Google Scholar]
- Yu G, Xian L, Xue H, Yu W, Rufian JS, Sang Y, Morcillo RJL, Wang Y, Macho AP (2020) A bacterial effector protein prevents MAPK-mediated phosphorylation of SGT1 to suppress plant immunity. PLoS Pathog 16: e1008933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M, Jiang Z, Bi G, Nomura K, Liu M, Wang Y, Cai B, Zhou JM, He SY, Xin XF (2021) Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature 592: 1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Yang X, Zhao Z, Lin F, Wang L, Pan Q (2011) Characterization and fine mapping of the rice blast resistance gene Pia. Sci China Life Sci 54: 372–378 [DOI] [PubMed] [Google Scholar]
- Zhai C, Zhang Y, Yao N, Lin F, Liu Z, Dong Z, Wang L, Pan Q (2014) Function and interaction of the coupled genes responsible for Pik-h encoded rice blast resistance. PLoS One 9: e98067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Shao F, Li Y, Cui H, Chen L, Li H, Zou Y, Long C, Lan L, Chai J, et al. (2007) A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP-induced immunity in plants. Cell Host Microbe 1: 175–185 [DOI] [PubMed] [Google Scholar]
- Zhang J, Li W, Xiang T, Liu Z, Laluk K, Ding X, Zou Y, Gao M, Zhang X, Chen S, . et al. (2010a) Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe 7: 290–301 [DOI] [PubMed] [Google Scholar]
- Zhang L, Kars I, Essenstam B, Liebrand TWH, Wagemakers L, Elberse J, Tagkalaki P, Tjoitang D, van den Ackerveken G, et al. (2014) Fungal endopolygalacturonases are recognized as microbe-associated molecular patterns by the Arabidopsis receptor-like protein RESPONSIVENESS TO BOTRYTIS POLYGALACTURONASES1. Plant Physiol 164: 352–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Klessig DF (1998) Resistance gene N-mediated de novo synthesis and activation of a tobacco mitogen-activated protein kinase by tobacco mosaic virus infection. Proc Natl Acad Sci USA 95: 7433–7438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Wang L, Wu W, He L, Yang X, Pan Q (2015) Function and evolution of Magnaporthe oryzae avirulence gene AvrPib responding to the rice blast resistance gene Pib. Sci Rep 5: 11642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Fraiture M, Kolb D, Löffelhardt B, Desaki Y, Boutrot FFG, Tör M, Zipfel C, Gust AA, Brunner F (2013) Arabidopsis receptor-like protein30 and receptor-like kinase suppressor of BIR1-1/EVERSHED mediate innate immunity to necrotrophic fungi. Plant Cell 25: 4227–4241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XC, Gassmann W (2007) Alternative splicing and mRNA levels of the disease resistance gene RPS4 are induced during defense responses. Plant Physiol 145: 1577–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Xu S, Ding P, Wang D, Cheng YT, He J, Gao M, Xu F, Li Y, Zhu Z, et al. (2010b) Control of salicylic acid synthesis and systemic acquired resistance by two members of a plant-specific family of transcription factors. Proc Natl Acad Sci USA 107: 18220–18225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Liu Y, Huang H, Gao M, Wu D, Kong Q, Zhang Y (2017a) The NLR protein SUMM2 senses the disruption of an immune signaling MAP kinase cascade via CRCK3. EMBO Rep 18: 292–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Wu Y, Gao M, Zhang J, Kong Q, Liu Y, Ba H, Zhou J, Zhang Y (2012) Disruption of PAMP-induced MAP kinase cascade by a Pseudomonas syringae effector activates plant immunity mediated by the NB-LRR protein SUMM2. Cell Host Microbe 11: 253–263 [DOI] [PubMed] [Google Scholar]
- Zhang ZM, Ma KW, Gao L, Hu Z, Schwizer S, Ma W, Song J (2017b) Mechanism of host substrate acetylation by a YopJ family effector. Nat Plants 3: 17115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Ardales EY, Raymundo A, Bai J, Trick HN, Leach JE, Hulbert SH (2004) The avrRxo1 gene from the rice pathogen Xanthomonas oryzae pv. oryzicola confers a nonhost defense reaction on maize with resistance gene Rxo1. Mol Plant Microbe Interact 17: 771–779 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Huang J, Wang Z, Jing S, Wang Y, Ouyang Y, Cai B, Xin X-F, Liu X, Zhang C, et al. (2016) Allelic diversity in an NLR gene BPH9 enables rice to combat planthopper variation. Proc Natl Acad Sci USA 113: 12850–12855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wu S, Chen X, Liu C, Sheen J, Shan L, He P (2014) The Pseudomonas syringae effector HopF2 suppresses Arabidopsis immunity by targeting BAK1. Plant J 77: 235–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou N, Tootle TL, Tsui F, Klessig DF, Glazebrook J (1998) PAD4 functions upstream from salicylic acid to control defense responses in Arabidopsis. Plant Cell 10: 1021–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JDG, Boller T, Felix G (2006) Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125: 749–760 [DOI] [PubMed] [Google Scholar]
- Zorzatto C, Machado JPB, Lopes KVG, Nascimento KJT, Pereira WA, Brustolini OJB, Reis PAB, Calil IP, Deguchi M, Sachetto-Martins G, et al. (2015) NIK1-mediated translation suppression functions as a plant antiviral immunity mechanism. Nature 520: 679–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y, Wang S, Zhou Y, Bai J, Huang G, Liu X, Zhang Y, Tang D, Lu D (2018) Transcriptional regulation of the immune receptor FLS2 controls the ontogeny of plant innate immunity. Plant Cell 30: 2779–2794 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.