SUMMARY
Adaptive CD4+ T helper cells and their innate counterparts, innate lymphoid cells, utilize an identical set of transcription factors (TFs) for their differentiation and functions. However, similarities and differences in the induction of these TFs in related lymphocytes are still elusive. Here we show that T helper-1 (Th1) cells and natural killer (NK) cells displayed distinct epigenomes at the Tbx21 locus, which encodes T-bet, a critical TF for regulating type 1 immune responses. The initial induction of T-bet in NK precursors was dependent on the NK-specific DNase I hypersensitive site Tbx21-CNS-3, and the expression of the interleukin-18 (IL-18) receptor; IL-18 induced T-bet expression through the transcription factor RUNX3, which bound to Tbx21-CNS-3. By contrast, STAT-binding motifs within Tbx21-CNS-12 were critical for IL-12-induced T-bet expression during Th1 cell differentiation both in vitro and in vivo. Thus, type 1 innate and adaptive lymphocytes utilize distinct enhancer elements for their development and differentiation.
Graphical Abstract
eTOC blurb:
Transcription factor T-bet is critically involved in type 1 immune responses. Fang et al. report that type 1 innate and adaptive lymphocytes utilize distinct cis-regulatory elements at the Tbx21 locus for T-bet induction during their development, differentiation, and activation.
INTRODUCTION
Adaptive CD4+ T helper (Th) cells and innate lymphoid cells (ILCs) play critical roles in host defense against invading pathogens and are involved in different forms of inflammatory diseases including allergy and autoimmunity. While Th cell differentiation occurs upon the engagement of T cell receptor (TCR) in a specific cytokine environment, ILCs are pre-developed and can promptly respond to inflammatory signals in an antigen non-specific manner. Th cell subsets and their ILC counterparts with a similar cytokine-producing capability utilize an identical set of lineage-determining transcription factors (LDTFs): T-bet for group 1 innate lymphoid cells (ILC1s) and T helper-1 (Th1) cells; GATA3 for ILC2s and Th2 cells; RORγt for ILC3s and Th17 cells (Fang and Zhu, 2017; Vivier et al., 2018). T-bet is also critical for the maturation of conventional natural killer (cNK) cells (Townsend et al., 2004). However, whether these cells adopt similar or different mechanisms in inducing the expression of these LDTFs during their development remains elusive. Furthermore, while cytokine-mediated activation of signal transducer and activator of transcription (STAT) proteins plays a critical role during T cell differentiation, there is less known about the signals involved in the induction of these LDTFs during ILC development.
T-bet (encoded by the Tbx21 gene) is critical for Th1 cell differentiation and optimal expression of Th1 effector cytokine interferon-γ (IFN-γ) (Finotto et al., 2002; Szabo et al., 2002; Zhu et al., 2012). Interleukin (IL)-12 directly acts on CD4+ T cells and activates the transcription factor STAT4 to induce T-bet expression; IFN-γ may also be involved in T-bet induction that is independent of IL-12-STAT4 signaling (Christie and Zhu, 2014; Lighvani et al., 2001; Mullen et al., 2001). It has been shown that activated STAT4 can bind to the Tbx21 locus (Madera et al., 2018; Rapp et al., 2017; Vahedi et al., 2012; Wei et al., 2010; Yang et al., 2007), however, the importance of such binding in T-bet induction in vivo is unknown.
T-bet is also critical for ILC1 development and cNK cell maturation (Mujal et al., 2021). In the absence of T-bet, ILC1s and cNK cells fail to either develop or undergo further maturation (Gordon et al., 2012; Sojka et al., 2014). Some tissue-resident ILC1s have been originally regarded as a subset of NK cells until recent studies which separate them into different lineages based on their distinct expression of cell surface markers and transcription factors (Daussy et al., 2014; Klose et al., 2014; Peng et al., 2013; Pikovskaya et al., 2016; Sojka et al., 2014; Vivier et al., 2018). Both T-bet and Eomes (another T-box transcription factor) are expressed by cNK cells and are required for their development and/or maturation, whereas ILC1s only express T-bet which is critical for their development.
In the bone marrow (BM), common lymphoid progenitors (CLPs) develop into pre-NK precursors (Pre-NKPs), then transit to refined-NKPs (rNKPs) with the expression of IL-2 receptor β chain (CD122) allowing their responsiveness to IL-15. Subsequent expression of the receptors NKG2D followed by NK1.1 occurs when the progenitors reach the immature NK (iNK) stage (Abel et al., 2018; Carotta et al., 2011; Fathman et al., 2011; Sharrock, 2019; Stokic-Trtica et al., 2020). Transcription factors RUNX3, CBF-β, STAT5, NFIL3, PU.1, Notch and TCF-1 are critical for the development of Pre-NKPs and/or rNKPs, whereas T-bet has been widely reported to play an important role during NK cell maturation (Collins et al., 2017; Gordon et al., 2012; Townsend et al., 2004).
Chromatin accessibility, which can be assessed by either assay for transposase-accessible chromatin using sequencing (ATAC-Seq) or DNase I hypersensitive site (DHS) sequencing (DNase-Seq), is highly associated with the binding of specific transcription factors required for gene regulation. In addition, the epigenetic “imprints” as a result of chromatin remolding at a particular region often reflect the history of cell’s response to previous signaling events (Fang et al., 2018; Gury-BenAri et al., 2016; Lau et al., 2018; Sciume et al., 2020; Shih et al., 2016; Shih et al., 2014). During lymphoid cell lineage determination and commitment, LDTF binds to many cis-regulatory elements in the lineage-specific gene loci, regulates chromatin accessibility at these sites and induces their gene expression at the genome-wide level (Heinz and Glass, 2012; Kang and Malhotra, 2015; Rogers and Bulyk, 2018; Shih et al., 2014; Zaret and Carroll, 2011). While it is critical to understand how LDTFs dictate lineage commitment, it is equally important to understand how these LDTFs are being regulated during cell development and differentiation.
To investigate the regulation of LDTF induction during innate and adaptive lymphocyte development, we performed DNase-Seq and chromatin immunoprecipitation sequencing (ChIP-Seq) analyses to compare the epigenetic landscapes in different Th cell and ILC subsets. We then focused on the Tbx21 locus as an example for in depth study. Our data revealed that a DHS located at 3 kb upstream of the Tbx21 transcriptional start site (Tbx21-CNS-3) was preferentially accessible in cNK cells, bound by the RUNX-CBF-β complex, and important for optimal T-bet induction during cNK cell development, while this DHS was not required for Th1 cell differentiation. IL-18-mediated RUNX3 induction up-regulated T-bet expression in a Tbx21-CNS-3-dependent manner in cNK cells. The initial expression of T-bet was detected as early as the rNKP stage correlated with high amounts of IL-18Rα expression in these cells. T-bet also bound to the Tbx21-CNS-3 element and regulated its own expression in NK cells. On the other hand, Tbx21-CNS-12 was the major accessible region in Th1 cells; this site was bound by STAT4 and was responsible for IL-12-STAT4-mediated T-bet induction during Th1 cell differentiation both in vitro and in vivo. However, STAT-binding sites in Tbx21-CNS-12 were dispensable for T-bet expression during the development of cNK cells and ILC1s in steady state, although they responded to IL-12 stimulation to promote T-bet expression during cNK cell activation. Therefore, innate and adaptive lymphocytes may utilize distinct regulatory elements in response to environmental stimulus for the induction of key transcription factors required for their differentiation and functions.
RESULTS
Differential epigenomes are found at the Tbx21 locus in CD4+ T cell and ILC subsets
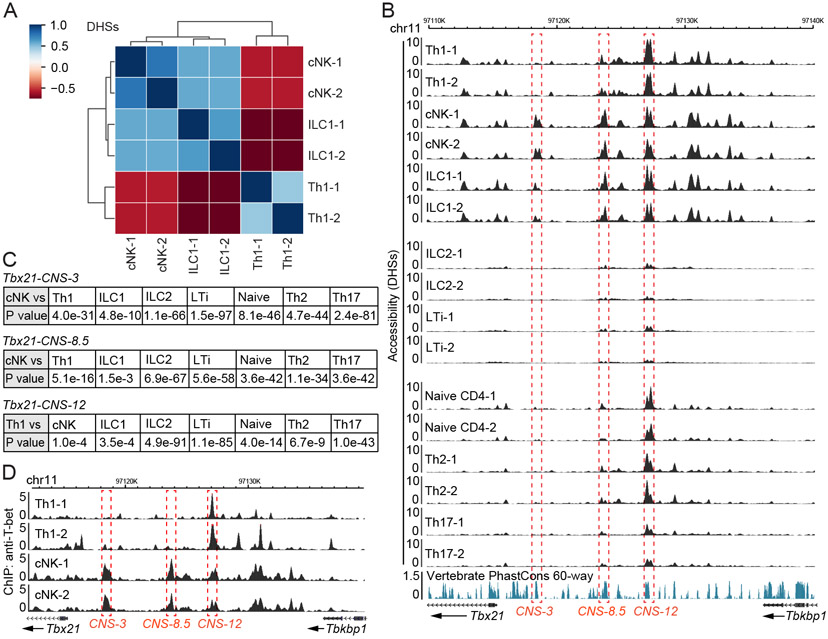
We first performed DNase-Seq to assess the similarities and differences in genome-wide chromatin accessibility among distinct innate and adaptive lymphocytes. A series of cNK-, ILC1-, ILC1+cNK-, ILC2- and LTi-specific DHSs as well as the shared ones were identified in the ILC component (Figure S1A-S1C and Table S1). Similarly, naïve CD4+ T cell-, Th1-, Th2- and Th17 cell-specific DHSs as well as the shared one in CD4+ T cell component were also detected (Figure S1D and Table S1). After excluding all the shared DHSs, we focused on Th1 cells, cNK cells and ILC1s for more detailed data analysis. Clustering analysis indicated that the DHSs in cNK cells and ILC1s were closer to each other, while Th1 cells displayed a more distinct pattern (Figure 1A).
Figure 1. Epigenomes at the Tbx21 locus in Th1 cells, cNK cells and ILC1s.
(A-C) DHS analysis of chromatin accessibility was performed through DNase-Seq. Th1, Th2 and Th17 cells were prepared in vitro; naïve CD4+ T cells were harvested from LNs; cNK cells and ILC1s were harvested from livers; ILC2s and LTi cells were harvested from small intestine. Heatmap showed the correlation of DHSs between Th1 cell, cNK cell and ILC1 samples after excluding the ILC-shared and CD4+ T cell-shared DHSs (A). Chromatin accessibility at the Tbx21 locus was viewed by Wash U genome browser (B). The accessibility of Tbx21-CNS-3 in cNK cells, Tbx21-CNS-8.5 in cNK cells and Tbx21-CNS-12 in Th1 cells was compared with that in other cell subsets (C). Samples are in biological duplicates.
(D) Anti-T-bet ChIP-Seq was performed using Th1 cells and splenic cNK cells. The binding of T-bet at the Tbx21 locus was viewed by Wash U genome browser. The dataset of Th1-2 was derived from the GEO: GSE38808. The cNK samples are in biological duplicates. See also Figure S1, Table S1 and Table S2.
In the Tbx21 locus, multiple DHSs with differential accessibilities were identified upstream of the transcription starting site Tbx21-TSS (Figure 1B, top). Tbx21-CNS-3 (conserved non-coding sequence 3 kb upstream of TSS) was found preferentially accessible in cNK cells, partially accessible in ILC1s but not accessible in Th1 cells. Tbx21-CNS-8.5 was preferentially accessible in both cNK cells and ILC1s, whereas Tbx21-CNS-12 was more accessible in Th1 cells and it represented the major DHS at the Tbx21 locus in Th1 cells. In ILC2s and LTi cells, which do not express T-bet, all of these three sites, Tbx21-CNS-3, Tbx21-CNS-8.5 and Tbx21-CNS-12, were either not accessible or displayed much reduced accessibility compared to those in type 1 lymphocytes (Figure 1B middle). We also compared DHSs at the Tbx21 locus in naïve CD4+ T cells and other Th cell subsets (Figure 1B, bottom). Although naïve CD4+ T cells do not express T-bet (Fang et al., 2018), Tbx21-CNS-12 was already open in naïve CD4+ T cells. The accessibility of this site was maintained in Th2 cells, increased in Th1 cells but reduced in Th17 cells. A Poisson test of the differential accessibilities at these CNS regions between different cell types (considered as statistically significant when P value < 1e-5) confirmed that the accessibility at the Tbx21-CNS-3 was found preferentially in cNK cells whereas the Tbx21-CNS-12 was more accessible in all type 1 lymphocytes compared to other cell types (Figure 1C).
Differential accessibilities at the Gata3 locus were also observed in Th2 cells and ILC2s (Figure S1E). Close to the T cell enhancer region at +280 kb (280 kb downstream of the Gata3 structural gene) (Hosoya-Ohmura et al., 2011), which was accessible in both Th2 cells and ILC2s, there was a cluster of Th2-specific accessible regions (at +290-314 kb). By contrast, a cluster of accessible regions (at +684-731 kb) was found to be ILC2-specific. The differential functions of those two regions for GATA3 expression during Th2 cell differentiation and ILC development have been recently reported (Kasal et al., 2021). Similarly, the accessibility at the Rorc locus also showed some differences between Th17 cells and LTi cells (Figure S1F). While the Rorc locus was generally more accessible in LTi cells compared to that in Th17 cells and this correlated with the higher RORγt expression found in the former subset, a recent report has demonstrated that deletion of CNS6 or CNS9 results in a defect in Th17 cell differentiation without affecting the development of RORγt-expressing innate lymphocytes (Chang et al., 2020).
To investigate the mechanisms involved in potentially differential regulation of LDTF expression via distinct cis-regulatory elements in adaptive and innate lymphocytes, we further analyzed T-bet regulation in type 1 lymphocytes by the three CNS regions mentioned above. Since T-bet is able to bind to the Tbx21 locus in Th1 cells (Zhu et al., 2012), we further performed ChIP-Seq analysis of T-bet binding in cNK cells and Th1 cells (Figure S1G and Table S2). While T-bet strongly bound to Tbx21-CNS-12 in Th1 cells, it strongly bound to Tbx21-CNS-3 and Tbx21-CNS-8.5 in cNK cells (Figure 1D). The fact that the epigenomes at the Tbx21 locus are distinct in innate and adaptive lymphoid cell subsets suggests that Tbx21-CNS-3 and/or Tbx21-CNS-8.5 could be involved in T-bet expression during the development of cNK cells and ILC1s, whereas Tbx21-CNS-12 may function as a major cis-regulatory element for T-bet induction in Th1 cells.
Tbx21-CNS-3 but not Tbx21-CNS-8.5 is critical for T-bet induction during cNK cell development
To investigate the importance of Tbx21-CNS-3 and Tbx21-CNS-8.5 in T-bet induction during cNK cell and ILC1 development, we obtained the Tbx21ΔCNS-3 and Tbx21ΔCNS-8.5 strains by deleting ~400 bp sequence at these two regions (Table S3), respectively, using CRISPR-Cas9 technology. Since Tbx21-CNS-3 is not accessible in Th1 cells, the protein amounts of T-bet in Tbx21ΔCNS-3 Th1 cells was similar to those in wildtype (WT) Th1 controls as expected (Figure 2A). However, deleting Tbx21-CNS-3 significantly reduced T-bet protein amounts in cNK cells from the liver, spleen and lung (Figure 2B, 2C and S2A). Nevertheless, the reduction of T-bet protein amounts in Tbx21ΔCNS-3 ILC1s was only moderate, and the total cell number of ILC1s in the Tbx21ΔCNS-3 mice was comparable to that in WT controls (Figure 2C and 2D). Consistent with previous studies suggesting a critical role of T-bet in NK cell maturation, Tbx21−/− cNK cells were blocked at the CD11b+CD27+ and CD11b−CD27+ immature stages (Figure S2B and S2C). In addition, Tbx21+/− cNK cells also exhibited a maturation defect (Figure S2D), indicating that optimal expression of T-bet protein regulates cNK cell maturation. Similarly, a reduction of T-bet expression in Tbx21ΔCNS-3 cNK cells resulted in an accumulation of immature CD11b+CD27+ and CD11b−CD27+ population (Figure 2E), and a decrease in total cell number of mature CD11b+CD27− NK cells in the liver, spleen and lung compared to WT controls (Figure 2D, 2F, S2E and S2F). In the Tbx21ΔCNS-3 BM, the CD27− mature NK cells were also reduced in number compared to those in the WT bone marrow (Figure S2G). Ectopic expression of T-bet in sorted lymphoid-primed multipotent progenitors (LMPPs) promoted NK cell maturation in vitro in both WT and Tbx21ΔCNS-3 group (Figure S2H). To confirm the cell intrinsic effect of Tbx21-CNS-3 on T-bet expression and cNK cell maturation, we generated chimeric mice by co-transferring CD45.1 Tbx21+/+ BM cells together with CD45.2 Tbx21+/+ BM cells or CD45.2 Tbx21ΔCNS-3 BM cells into sub-lethally irradiated Rag2−/−γc−/− recipient mice. T-bet expression was reduced and cNK cell maturation was impaired within the CD45.2 Tbx21ΔCNS-3 cNK cells compared to the CD45.1 WT controls in the same animal (Figure S2I-S2K).
Figure 2. Tbx21-CNS-3 is critical for T-bet induction during cNK cell development.
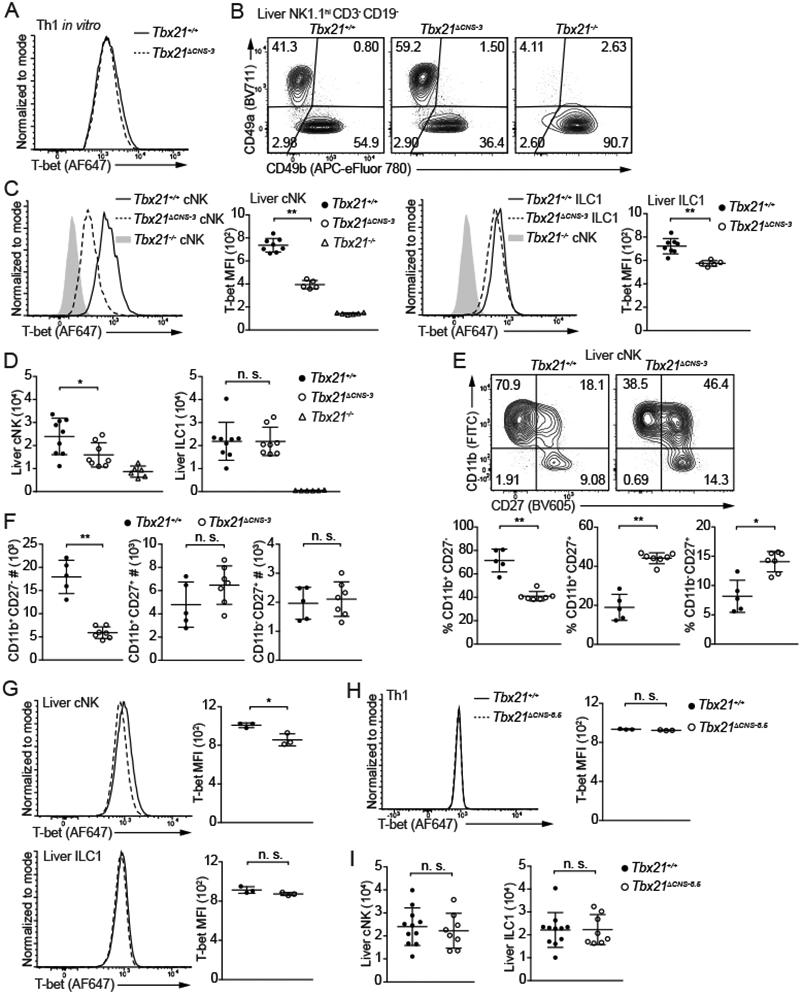
(A) Naïve CD4+ T cells were cultured in Th1-skewing conditions for 3 days and then in resting conditions (IL-2-containing medium) for 1 day. T-bet protein amounts in Tbx21+/+ and Tbx21ΔCNS-3 Th1 cells were measured.
(B and C) T-bet protein amounts in hepatic cNK cells and ILC1s from Tbx21+/+ (n=8), Tbx21ΔCNS-3 (n=5) and Tbx21−/− (n=6) mice were measured, and T-bet MFI was calculated. Mean ± SD.
(D) cNK and ILC1 cell number in the livers from Tbx21+/+ (n=9), Tbx21ΔCNS-3 (n=8), and Tbx21−/− (n=6) mice was calculated. Mean ± SD.
(E and F) Maturation of hepatic cNK cells from the Tbx21+/+ (n=5) and Tbx21ΔCNS-3 (n=7) mice was assessed by the expression of cell surface proteins CD11b and CD27. The percentage and the cell number of CD11b+CD27−, CD11b+CD27+ and CD11b−CD27+ population were calculated. Mean ± SD.
(G) T-bet protein amounts in hepatic cNK cells and ILC1s from Tbx21+/+ (n=3) and Tbx21ΔCNS-8.5 (n=3) mice were measured. Mean ± SD.
(H) T-bet protein amounts in Tbx21+/+ (n=3) and Tbx21ΔCNS-8.5 (n=3) Th1 cells differentiated in vitro were measured.
(I) cNK and ILC1 cell number in livers from Tbx21+/+ (n=11) and Tbx21ΔCNS-8.5 (n=8) mice were calculated. Mean ± SD.
Data are representative of two (A, G-I) or three (B-F) independent experiments. See also Figure S2, Figure S3 and Table S3.
By contrast, the Tbx21-CNS-8.5 was dispensable for T-bet expression in ILC1s and Th1 cells, and there was only a subtle reduction of T-bet in Tbx21ΔCNS-8.5 cNK cells compared to WT controls (Figure 2G and 2H). Consequently, total cell number of both ILC1 and cNK cells was normal in the Tbx21ΔCNS-8.5 mice (Figure 2I). In addition, T-bet expression in the Tbx21-CNS-3 and Tbx21-CNS-8.5 doubly deficient cNK cells was similar to that in Tbx21-CNS-3 deficient cNK cells (Figure S2L). Hence, cis-regulatory element Tbx21-CNS-3 but not Tbx21-CNS-8.5 is important for the optimal induction of T-bet during cNK cell development and thus required for cNK cell maturation.
NK1.1+ cells play critical roles in host defense against T. gondii infection (Denkers et al., 1993). The parasite load in the liver and peritoneal cavity (Figure S3A), as well as the IFN-γ in the serum (Figure S3B), were comparable between Tbx21+/+ and Tbx21ΔCNS-3 mice five days after infection, and the Tbx21ΔCNS-3 mice survived infection as the WT mice (Figure S3C), indicating that the defect in NK cell maturation in steady state in the Tbx21ΔCNS-3 mice has no obvious physiological consequence during T. gondii infection. However, while cNK cells from the infected Tbx21ΔCNS-3 mice expressed lower amounts of T-bet than cNK cells from the infected WT mice, they expressed higher amounts of T-bet than WT cNK cells from naïve mice (Figure S3D). Accordingly, the maturation defect of cNK cells in the Tbx21ΔCNS-3 mice was restored after infection (Figure S3E). This is consistent with previous reports that some infection conditions may promote cNK cell maturation (Kamimura and Lanier, 2015; Muller et al., 2016).
We then tested the responses of Tbx21ΔCNS-3 NK cells to acute poly (I:C) stimulation in vivo (Longhi et al., 2009). The cNK cells from the Tbx21ΔCNS-3 mice after poly (I:C) treatment produced lower amounts of IFN-γ compared to their WT counterparts (Figure S3F). Furthermore, lower amounts of IFN-γ in the sera from the Tbx21ΔCNS-3 mice were detected six hours after poly (I:C) injection compared with the sera from similarly treated WT mice (Figure S3G).
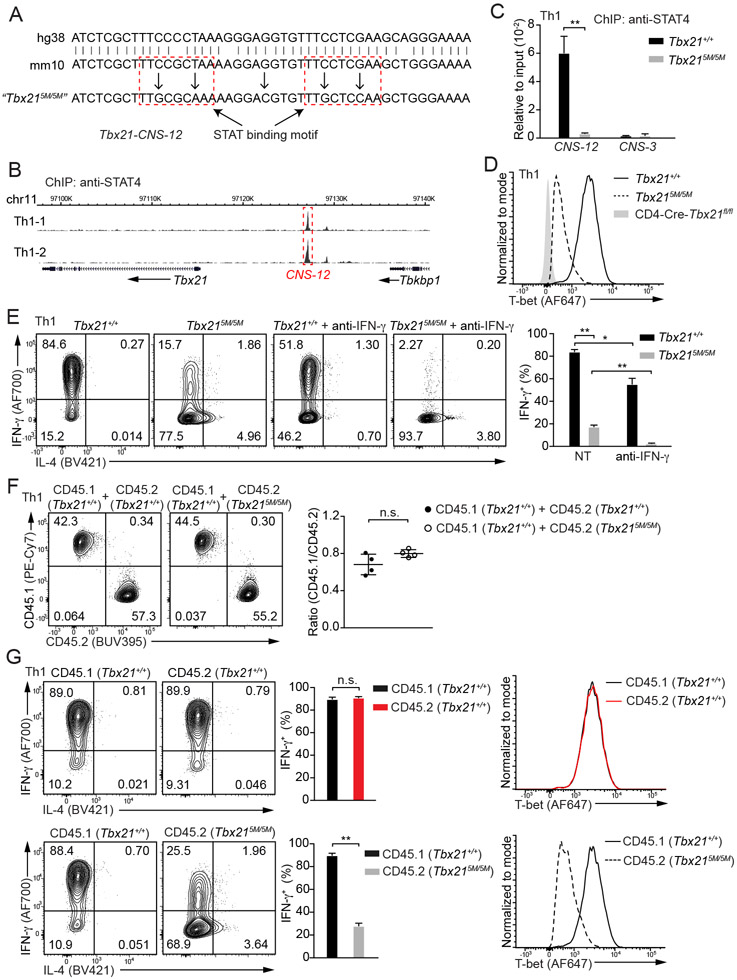
STAT binding motifs at Tbx21-CNS-12 are critical for T-bet induction during Th1 cell differentiation
ChIP-Seq analyses of STAT4 and T-bet binding to the genome in Th1 cells showed that approximate half of STAT4 binding sites were also bound by T-bet, although T-bet bound to many more sites than STAT4 did (Figure S4A and Table S2). Two typical STAT-binding motifs were identified at the Tbx21-CNS-12 element (Figure 3A). STAT4 primarily bound to Tbx21-CNS-12 at the Tbx21 locus in Th1 cells and no STAT4 binding was detected at Tbx21-CNS-3 or Tbx21-CNS-8.5 (Figure 3B). To test whether Tbx21-CNS-12 is the key element for T-bet induction mediated by IL-12-STAT4 signaling during Th1 cell differentiation, we mutated these two motifs, which are conserved between human and mouse, and generated a Tbx215M/5M mouse strain by using CRISPR-Cas9 strategy (Figure 3A and Table S3). ChIP-PCR analysis confirmed that STAT4 was no longer able to bind to Tbx21-CNS-12 in Tbx215M/5M “Th1” cells (Figure 3C). Consequently, the protein amounts of T-bet and the capability of IFN-γ production in Tbx215M/5M cells cultured under Th1 cell conditions in vitro were reduced compared to WT cells cultured under the same conditions (Figure 3D and 3E). Furthermore, Tbx215M/5M “Th1” cells acquired IL-4-producing capability, a feature of T-bet-deficient “Th1” cells. While neutralizing endogenous IFN-γ with antibodies diminished IFN-γ production by WT Th1 cells, it completely abolished the IFN-γ production by Tbx215M/5M “Th1” cells (Figure 3E). We further co-cultured congenic CD45.1 Tbx21+/+ naïve CD4+ T cells with CD45.2 Tbx21+/+ or Tbx215M/5M naïve CD4+ T cells in Th1 cell-skewing conditions to determine the cell intrinsic deficiency of Tbx215M/5M. The expansion of Tbx215M/5M Th1 cells was comparable to that of WT Th1 cells (Figure 3F). CD45.2 Tbx215M/5M Th1 cells exhibited significant reduction in IFN-γ production and T-bet expression compared to either CD45.2 Tbx21+/+ controls or CD45.1 Tbx21+/+ cells in the same culture (Figure 3G). Overall, our results indicate that the binding of STAT4 at Tbx21-CNS-12 is essential for T-bet induction during Th1 cell differentiation.
Figure 3. STAT4 binding at Tbx21-CNS-12 is essential for T-bet induction during Th1 cell differentiation in vitro.
(A) Two highlighted STAT binding motifs at Tbx21-CNS-12 were mutated through CRISPR-Cas9 technology, and the homozygous mutant strain was named as Tbx215M/5M. Both human and mouse sequences around the conserved STAT binding sites were shown.
(B) Anti-STAT4 ChIP-Seq was performed using Th1 cells. The binding of STAT4 at the Tbx21 locus was viewed by Wash U genome browser. Cell samples are in biological duplicates.
(C) Anti-STAT4 ChIP-PCR was performed using Tbx21+/+ (n=3) and Tbx215M/5M (n=3) Th1 cells. The primer pairs targeting Tbx21-CNS-12 and Tbx21-CNS-3 were used to measure STAT4 binding. Mean ± SD.
(D) T-bet protein amounts in Tbx21+/+, Tbx215M/5M and CD4-Cre-Tbx21fl/fl Th1 cells were measured.
(E) Naïve CD4+ T cells from Tbx21+/+ (n=3) and Tbx215M/5M (n=4) mice were used to prepare Th1 cells with or without the addition of anti-IFN-γ antibodies in the culture. In the presence of monensin, cells were stimulated with phorbol 12-myristate 13-acetate (PMA) and ionomycin for 4 hours. The percentage of IFN-γ-producing cells was calculated. Mean ± SD.
(F and G) CD45.1 Tbx21+/+ (n=4) naïve CD4+ T cells were co-cultured with equal numbers of CD45.2 Tbx21+/+ (n=4) or CD45.2 Tbx215M/5M (n=4) naïve CD4+ T cells under Th1-skewing conditions. The ratio of CD45.1 and CD45.2 cells in the co-cultured conditions was calculated (F); The percentage of IFN-γ-producing cells was calculated after PMA and ionomycin treatment in the presence of monensin (G, left); T-bet protein amounts were measured (G, right). Mean ± SD.
Data are representative of two (C-G) independent experiments. See also Figure S4, Table S2 and Table S3.
STAT binding to Tbx21-CNS-12 is critical for Th1 cell differentiation during T. gondii infection
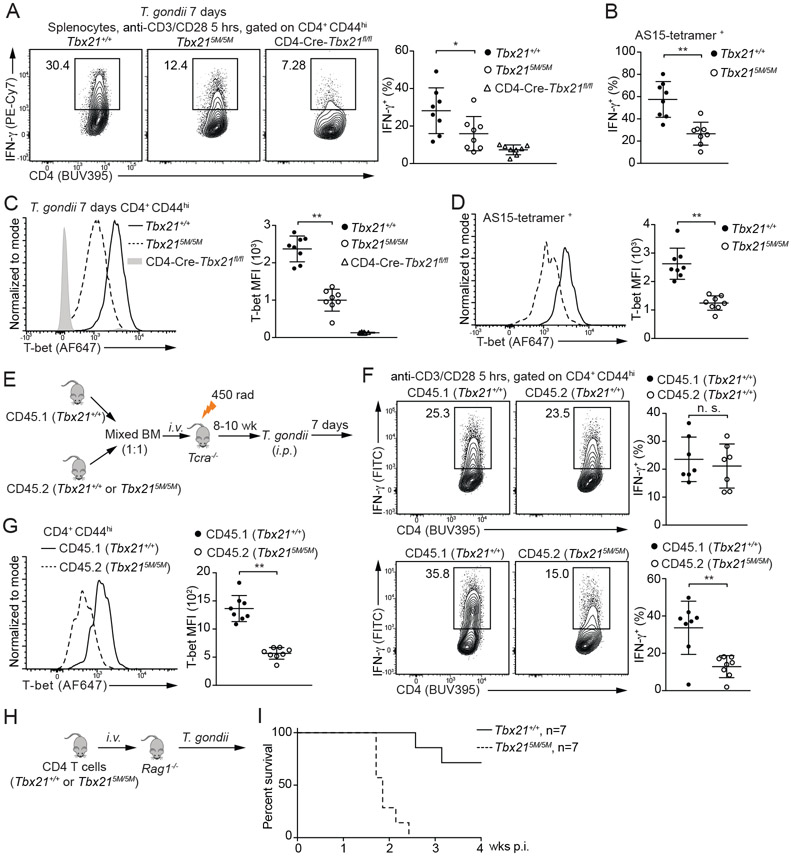
IL-12 secreted by dendritic cells drives Th1 cell-mediated immunity against T. gondii infection. To assess the importance of STAT4 binding to Tbx21-CNS-12 during Th1 responses in vivo, we infected the Tbx21+/+, Tbx215M/5M and CD4-Cre-Tbx21fl/fl mice with T. gondii. After seven days, splenocytes were re-stimulated with anti-CD3 and anti-CD28 ex vivo. Consistent with the results from in vitro culture, lower IFN-γ-producing capacity was found in the Tbx215M/5M CD4+CD44hi cells compared to the WT controls (Figure 4A). Such reduction was also observed in T. gondii antigen AS15-specific CD4+ T cells (Figure 4B). Moreover, T-bet protein amounts were found to be substantially lower in the Tbx215M/5M CD4+CD44hi cells compared to the WT controls (Figure 4C and 4D). Therefore, IL-12-mediated STAT4 (and/or IFN-γ-mediated STAT1) binding to Tbx21-CNS-12 is critical for optimal T-bet induction and thus Th1 cell differentiation in response to T. gondii infection.
Figure 4. Defective Th1 cell differentiation in the Tbx215M/5M mice in response to T. gondii infection.
(A-D) Tbx21+/+ (n=8), Tbx215M/5M (n=8) and CD4-Cre-Tbx21fl/fl (n=8) mice were infected with T. gondii for 7 days. Splenocytes were stimulated with anti-CD3 and anti-CD28 antibodies for 5 hours in the presence of monensin. The percentage of IFN-γ-producing cells in the CD4+CD44hi population and in the T. gondii antigen AS15-specific CD4+ T cells was calculated (A and B). T-bet protein amounts were measured, and T-bet MFI was calculated (C and D). Mean ± SD.
(E-G) Experimental procedure of BM chimeras infected with T. gondii for 7 days (E). The percentage of IFN-γ-producing cells was calculated after anti-CD3 and anti-CD28 antibodies stimulation in the presence of monensin (F). T-bet protein amounts in the CD4+CD44hi T cells from CD45.1 Tbx21+/+ and CD45.2 Tbx215M/5M chimeras (n=8) were measured, and T-bet MFI was calculated (G). Mean ± SD.
(H and I) Negatively selected Tbx21+/+ or Tbx215M/5M CD4+ T cells were transferred into Rag1−/− recipients. The reconstituted mice (n=7 for each group) were infected with T. gondii, and then monitored for their survival.
Data are representative of two (A-I) independent experiments. See also Figure S3.
IFN-γ production associated with Th1 cell differentiation is critical for the elimination of intracellular parasite T. gondii (Blanchard et al., 2008; Brown and McLeod, 1990; Denkers el al., 1993; Grover et al., 2012; Parker et al., 1991; Scharton-Kersten et al., 1998; Yarovinsky, 2014). Consequently, differential survival rate was noted between WT and Tbx215M/5M mice after T. gondii infection (Figure S3C). We also created chimeric mice by co-transferring CD45.1 Tbx21+/+ BM cells together with CD45.2 Tbx21+/+ BM cells or CD45.2 Tbx215M/5M BM cells into sub-lethally irradiated Tcra−/− recipient mice (Figure 4E). Reconstituted chimeras were challenged with T. gondii. Seven days after infection, the capacity of IFN-γ production by CD45.2 Tbx215M/5M Th1 cells was significantly lower than CD45.1 Tbx21+/+ Th1 cells in the same animal (Figure 4F) and CD45.2 Tbx215M/5M Th1 cells expressed lower T-bet protein amounts compared with the WT controls (Figure 4G).
To assess the physiological importance of the reduced IFN-γ production by Tbx215M5M CD4+ T cells, we transferred Tbx21+/+ CD4+ T cells or Tbx215M/5M CD4+ T cells into Rag1−/− recipients followed by T. gondii infection (Figure 4H). Most of the Rag1−/− mice receiving WT CD4+ T cells survived at four weeks post infection, consistent with previous reports (Jankovic et al., 2007; Kugler et al., 2013), all Rag1−/− mice that received Tbx215M/5M CD4+ T cells were found dead around two to three weeks after infection (Figure 4I). Taken together, these findings indicate that a cell intrinsic deficiency of T-bet induction in Tbx215M/5M CD4+ T cells renders the host vulnerable to T. gondii infection.
cNK cell and ILC1 development is independent of STAT binding to Tbx21-CNS-12
In constrast to the importance of STAT binding at Tbx21-CNS-12 in Th1 cell differentiation, the protein amounts of T-bet in both cNK cells and ILC1s from Tbx215M/5M mice were not different from those from WT mice (Figure 5A). Hepatic cNK cell numbers were also comparable between Tbx21+/+ and Tbx215M/5M mice, while there was a slight increase of ILC1s in the Tbx215M/5M mice compared with the WT controls (Figure 5B). Furthermore, IFN-γ-producing capability of the Tbx215M/5M cNK cells or ILC1s upon IL-12 and IL-18 stimulation ex vivo was intact (Figure 5C). Since NK cells from the Tbx21ΔCNS-3 mice still expressed low amounts of T-bet, we further assessed whether IL-12-mediated STAT4 activation contributes to T-bet induction during NK cell development in the absence of Tbx21-CNS-3. To this end, we prepared mixed BM chimeric mice that were treated with either anti-IL-12 or control IgG. Anti-IL-12p40 antibody treatment was able to block the IL-12-STAT4 signaling and decrease the T-bet protein amounts in “Th1” cells during T. gondii infection (Figure S4B), suggesting this reagent functions properly in vivo. However, in these BM chimeras, both CD45.1 or CD45.2 Tbx21+/+ cNK cells expressed same amounts of T-bet with or without blocking of IL-12 signaling (Figure S4C and S4D). More importantly, blocking of IL-12 signaling did not further reduce T-bet expression in CD45.2 Tbx21ΔCNS-3 cNK cells. These results indicate that IL-12-STAT4 signaling does not contribute to NK cell development in the steady state. Nevertheless, T-bet expression in cNK cells could still be regulated by IL-12 during in vitro culture through the STAT binding sites within Tbx21-CNS-12 (Figure S5A).
Figure 5. STAT binding motifs at Tbx21-CNS-12 are dispensable for T-bet induction during cNK cell and ILC1 development.
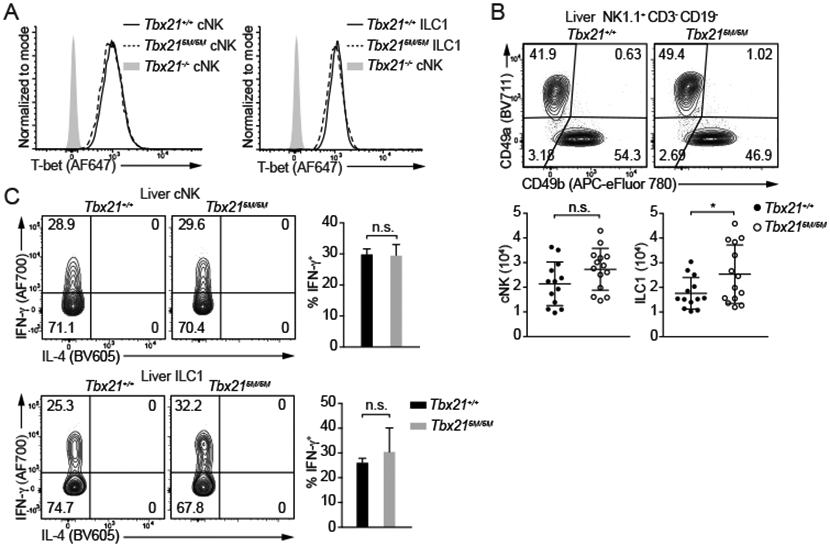
(A and B) T-bet protein amounts in cNK cells and ILC1s from Tbx21+/+, Tbx215M/5M and Tbx21−/− mice were measured (A). Total cNK and ILC1 cell number in livers from Tbx21+/+ (n=13) and Tbx215M/5M (n=14) was calculated (B). Mean ± SD.
(C) Purified cNK cells and ILC1s from Tbx21+/+ (n=4) and Tbx215M/5M (n=3) mice were stimulated with IL-12 and IL-18 for 5 hours in the presence of monensin. The percentage of IFN-γ-producing cells was calculated. Mean ± SD.
Data are representative of two (A-C) independent experiments. See also Figure S4 and S5.
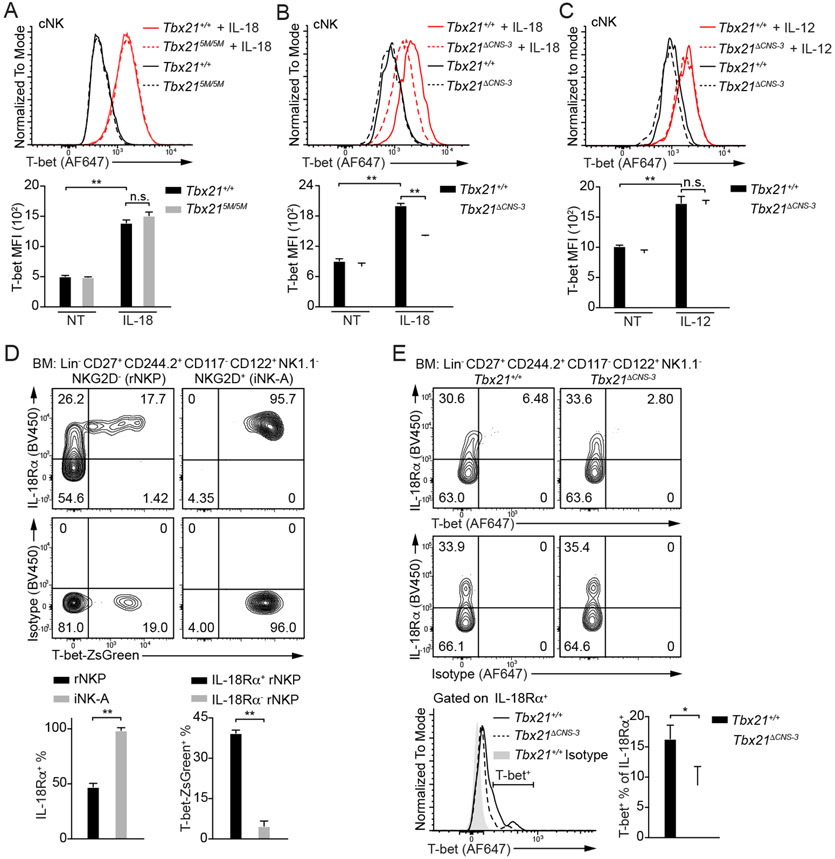
IL-18 up-regulates T-bet protein amounts through Tbx21-CNS-3 in cNK cells
Since Tbx21-CNS-3 but not Tbx21-CNS-12 is involved in cNK cell development, we further investigated which signaling(s) is responsible for T-bet induction via Tbx21-CNS-3 during cNK cell development. We first tested the effect of several possible regulators (Abel et al., 2018; Simonetta et al., 2016; Wu et al., 2017) on T-bet expression in cNK cells in vitro. IL-2 was supplemented for maintaining NK cell survival, and it had very little effect on sustaining T-bet expression (Figure S5B). Given that Tbx21-CNS-12 was also accessible in cNK cells (Figure 1B) and STAT4 may bind to this element after IL-12 stimulation (Madera et al., 2018), to minimize the effect of STAT signaling on T-bet expression through Tbx21-CNS-12 (Figure S5A), we used cNK cells from the Tbx215M/5M mice and incubated them with IL-1β, IL-6, IL-7, IL-15, IL-18, IL-21, CXCL12, Flt3L, SCF, IFN-γ or TGF-β1 for three days (Figure 6A and S5C-S5G). Among these cytokines and growth factors tested, IL-18 up-regulated T-bet protein amounts, and IL-21 also had some effects on T-bet expression (Figure 6A and S5E). T-bet induction by IL-18 was equally efficient in WT and Tbx215M/5M cNK cells (Figure 6A). Although IL-15 is considered as a key cytokine for NK cell development and homeostasis (Abel el al., 2018; Lieberman et al., 2004; Waldmann and Tagaya, 1999; Wu et al., 2017), it did not strongly induce T-bet expression (Figure S5D). Notch signaling induces T-bet expression in CD4+ T cells, and Tbx21 has been reported as a potential target of Notch pathway in cNK cells (Chaves et al., 2018; Maekawa et al., 2003; Minter et al., 2005; Perchet et al., 2018). However, while co-culturing of Tbx215M/5M cNK cells with stromal OP9 cell line helped T-bet expression, additional Notch signaling supplied by OP9-DLL1 cell line failed to further up-regulate T-bet protein amounts (Figure S5H). By contrast, Ki-67 staining indicated that DLL1-Notch signaling promoted cNK cell proliferation (Figure S5I). While the vast majority of cNK cells expressed IL-18Rα, ILC1s did not express this receptor (Figure S5J).
Figure 6. IL-18 up-regulates T-bet expression in cNK cells.
(A) Tbx21+/+ (n=3) and Tbx215M/5M (n=3) cNK cells were incubated with or without IL-18 for 3 days in the presence of IL-2. T-bet protein amounts were measured, and T-bet MFI was calculated. Mean ± SD.
(B and C) Tbx21+/+ (n=3) and Tbx21ΔCNS-3 (n=3) cNK cells were incubated with or without IL-18 (B) or with or without IL-12 (C) for 3 days in the presence of IL-2. T-bet protein amounts were measured, and T-bet MFI was calculated. Mean ± SD.
(D) The expression of IL-18Rα and T-bet reporter ZsGreen in the rNKPs and iNK-A cells from T-bet-ZsGreen reporter mouse BM was measured. The percentage of IL-18Rα+ cells in the rNKPs and iNK-A cells was calculated. The percentage of T-bet-ZsGreen+ cells in the IL-18Rα+ and IL-18Rα− rNKPs was calculated. Mean ± SD, n=3.
(E) The expression of T-bet in the bulk of rNKPs and iNK-A cells (Lin−CD27+CD244.2+CD117− CD122+NK1.1−) from Tbx21+/+ (n=3) and Tbx21ΔCNS-3 (n=3) mice was measured through intracellular staining. The percentage of T-bet+ cells in the IL-18Rα+ population was calculated. Mean ± SD.
Data are representative of two (A, C-E) or three (B) independent experiments. See also Figure S5 and S6.
Importantly, the protein amounts of T-bet in Tbx21ΔCNS-3 cNK cells treated with IL-18 were diminished compared to similarly treated WT controls (Figure 6B), however, both WT and Tbx21ΔCNS-3 cNK cells responded equally well to IL-12 stimulation in upregulating T-bet expression (Figure 6C). Low amounts of Tbx21 mRNA were also detected in Tbx21ΔCNS-3 cNK cells ex vivo, as well as in the cells stimulated with IL-18 in vitro, compared to the WT counterparts (Figure S5K). Therefore, the element Tbx21-CNS-3 plays a critical role in IL-18-mediated T-bet induction in cNK cells.
IL-18Rα expression is correlated with T-bet induction during NK cell development
To investigate whether IL-18 signaling is related to T-bet induction during early NK cell development (Figure S6A), we assessed the expression of IL-18Rα and T-bet in rNKPs and iNK-A cells in the T-bet-ZsGreen reporter mice (Zhu et al., 2012). While IL-18Rα expression was found in approximate half of rNKPs, all iNK-A and Post-iNK-A cells, but much less in Pre-NKPs (Figure 6D and S6B), T-bet-ZsGreen positive rNKPs were only found within the IL-18Rαhi population but not the IL-18Rα-negative population. All iNK-A cells expressed both IL-18Rα and T-bet-ZsGreen.
RUNXs and CBF-β transcription factors are indispensable for cNK cell and ILC1 development (Ebihara et al., 2015; Rapp et al., 2017); RUNX3, rather than RUNX1, is highly expressed in NKPs and critical for CD122 expression (Ohno et al., 2008); RUNX3 is induced before T-bet expression during NK cell development (Sharrock, 2019; Wang and Malarkannan, 2020). By using the RUNX3-YFP mouse strain (Egawa and Littman, 2008), we found that IL-18Rαhi NKPs, in which T-bet expression was detected, expressed much higher amounts of RUNX3-YFP than the IL-18Rαlo NKPs (Figure S6C). The Tbx21 mRNA was also significantly induced during the conversion from NK1.1−IL-18Rα− to NK1.1−IL-18Rα+ stage (Figure S6D). Therefore, the early induction of T-bet seems to be at the transition from rNKP to iNK-A stage correlated with the induction of IL-18Rα and RUNX3 expression. Although IL-21 was also capable of up-regulating T-bet protein amounts in developed cNK cells in vitro, neither rNKPs nor iNK-A cells expressed IL-21R (Figure S6E).
Anti-T-bet staining was not as sensitive as T-bet-ZsGreen reporter, nevertheless, the expression of T-bet was preferentially detected in the IL-18Rαhi population (Figure 6E). Importantly, lower protein amounts of T-bet and reduced percentage of T-bet positive cells within the IL-18Rα+ population were observed in the Tbx21ΔCNS-3 NK progenitors compared to the WT counterparts. Tbx21 mRNA was also significantly reduced in the Tbx21ΔCNS-3 NKPs compared with WT controls (Figure S6F). To rule out the possibility that the reduction of Tbx21 transcripts in the Tbx21ΔCNS-3 NKPs was due to a reduction in T-bet-expressing cells rather than a reduction of Tbx21 transcripts in “T-bet”-expressing cells, we generated the T-bet-ZsGreen-Tbx21−/+ and T-bet-ZsGreen-Tbx21−/ΔCNS-3 mice. We were able to find and sort T-bet-ZsGreenhiNK1.1− NKPs from both mouse strains. The fact that the Tbx21 mRNA amounts were lower in the ZsGreen-expressing Tbx21−/ΔCNS-3 NKPs compared to the ZsGreen-expressing Tbx21−/+ controls (Figure S6G), confirmed that the regulation of T-bet expression by Tbx21-CNS-3 was indeed at the transcription level. T-bet expression in Post-iNK-A, CD11b−CD27+, CD11b+CD27+ and CD11b+CD27− NK subsets from the Tbx21ΔCNS-3 mice was also lower than that in their WT counterparts (Figure S6H and S6I).
Although NK cell number was relatively normal in the spleen of Il18r1−/− and Il18−/− mice in steady state, defective NK cell maturation and functions have been found in these mice (Chaix et al., 2008; Hoshino et al., 1999; Muller et al., 2016; Takeda et al., 1998). To assess the importance of IL-18 signaling on T-bet induction during early NK cell development, we further analyzed Il18r1−/− mice. Indeed, the percentage of T-bet-expressing cells within the rNKPs and iNK-A cells was diminished in the Il18r1−/− mice compared to the WT controls (Figure S6J). Thus, our results indicate that early T-bet induction is highly correlated with IL-18 signaling, and that the cis-regulatory element Tbx21-CNS-3 is important for optimal induction of T-bet expression starting from an early stage of NK cell development.
IL-18-induced RUNX3 expression directly regulates T-bet in cNK
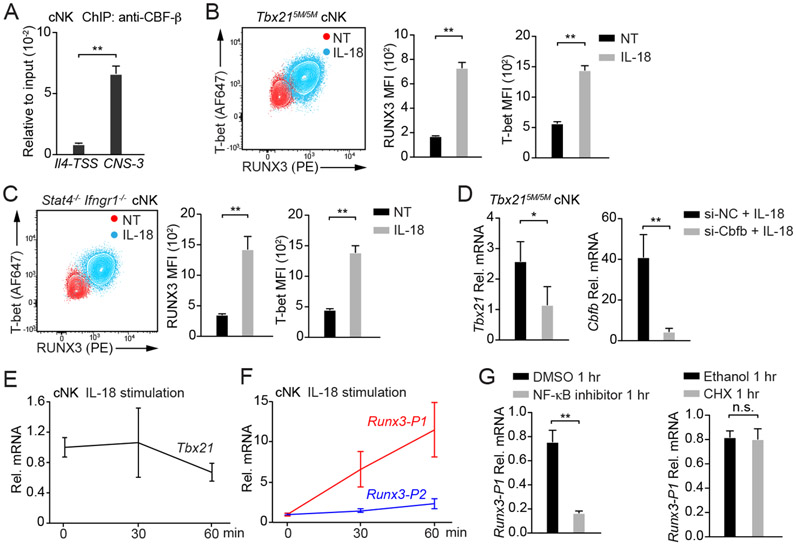
It has been reported that RUNX proteins and T-bet may form a complex to regulate the expression of several genes including Rorc, Ifng or Il4 (Djuretic et al., 2007; Lazarevic et al., 2011; Wang et al., 2014). In addition, T-box motifs are enriched in RUNX3 binding peaks (Levanon et al., 2014). Indeed, both T-bet binding motifs and RUNX binding motifs were identified within Tbx21-CNS-3 (Figure S7A), and two conserved core-sequence of RUNX binding motif CCAC (Seo et al., 2017) were also found near the T-bet binding motifs. Through ChIP-PCR analysis, we further confirmed that CBF-β bound to Tbx21-CNS-3 (Figure 7A). Therefore, we hypothesized that RUNX3 and T-bet may be involved in IL-18-mediated T-bet induction through Tbx21-CNS-3 in cNK cells. Indeed, the protein amounts of both RUNX3 and T-bet were up-regulated in Tbx215M/5M cNK cells incubated with IL-18 (Figure 7B). The STAT-independent effect of IL-18 in T-bet induction was further confirmed by using Stat4−/− Ifngr1−/− cNK cells in which both IL-12-STAT4 and IFN-γ-STAT1 signaling pathways are defective (Figure 7C). Furthermore, by silencing CBF-β expression through siRNA delivered by Nucleofector, we found that in the presence of IL-18, lower amounts of Tbx21 mRNA and T-bet protein were observed in Cbfo siRNA-treated cells in comparison with the cells transfected with control siRNA (Figure 7D, S7B and S7C), suggesting that IL-18-mediated T-bet up-regulation in cNK cells depends on the RUNX3/CBF-β complex. Consistent with the induction of RUNX3 being an upstream event of T-bet induction, when cNK cells were acutely stimulated with IL-18, the Tbx21 transcripts were not induced (Figure 7E). However, the Runx3 transcripts using the distal promoter (Runx3-P1) but not the transcripts using proximal promoter (Runx3-P2) were significantly induced by acute IL-18 stimulation (Figure 7F and Table S3). Blocking the NF-κB activity abolished the IL-18-mediated induction of Runx3 transcripts (Figure 7G). However, the protein synthesis inhibitor cycloheximide had no effect on Runx3 induction (Figure 7G). Taken together, IL-18-mediated upregulation of T-bet expression is likely through RUNX3 induction and its binding to Tbx21-CNS-3 in cNK cells.
Figure 7. IL-18 induces RUNX3 expression in cNK cells.
(A) Anti-CBF-β ChIP-PCR was performed using cNK cells sorted ex vivo. Primer pairs targeting to Tbx21-CNS-3 and the Il4 transcription start site (Il4-TSS, negative control) were used. Mean ± SD, n=3.
(B and C) Tbx215M/5M cNK cells (n=3) and Stat4−/−Ifngr1−/− cNK cells (n=3) were incubated with or without IL-18 for 3 days in the presence of IL-2. T-bet and RUNX3 protein amounts were measured, and MFI was calculated. Mean ± SD.
(D) Tbx215M/5M cNK cells were co-transfected with siRNA targeting to Cbfb (n=3) or negative control (NC, n=3) and Green indicator by using Amaxa™ 4D-Nucleofector™. After 24 hours, the Greenhi population was sorted and cultured for additional 12 hours. Cells were then incubated with IL-18 and IL-2 for 2 days. The transcripts of Tbx21 and Cbfb were assessed by qRT-PCR. Mean ± SD.
(E and F) cNK cells were stimulated with IL-18 for various time points as indicated. The relative Tbx21 (E), Runx3-P1 (distal) and Runx3-P2 (proximal) mRNA (F) were measured by using qRT-PCR and normalized to Hprt mRNA. Mean ± SD, n=3.
(G) cNK cells were pre-treated with NF-κB inhibitor or cycloheximide (CHX) for 10 minutes and stimulated with IL-18 for 1 hour. The relative Runx3-P1 mRNA was measured. Mean ± SD, n=3.
Data are representative of two (A-G) independent experiments. See also Figure S7 and Table S3.
IL-18 and RUNX3 fail to directly induce T-bet expression in Th1 cells
IL-18 together with IL-12 has been shown to promote the expression of Th1-related genes including Ifng (Tominaga et al., 2000), however, whether IL-18 can directly regulate T-bet expression in CD4+ T cells is not clear. Our results showed that IL-12 induced T-bet in WT cells activated with anti-CD3 and anti-CD28, but IL-18 failed to do so in either WT or in Tbx215M/5M cells (Figure S7D). Although the combination of IL-12 and IL-18 induced higher amounts of T-bet in WT cells, such induction was greatly diminished in the Tbx215M/5M cells. Furthermore, neutralization of IFN-γ in the culture completely abolished the induction of T-bet expression by IL-12 and IL-18 in the Tbx215M/5M cells, indicating that the synergistic effect of IL-18 and IL-12 on T-bet expression in T cells is through inducing IFN-γ expression.
Ectopic expression of RUNXs may up-regulate Tbx21 mRNA in Th cells (Wang et al., 2014), however, enforced RUNX3 expression has a limited effect on T-bet induction when IFN-γ is neutralized, i.e. under Th2 skewing conditions (Yagi et al., 2010). To exclude the effect of IL-12-STAT4 and IFN-γ-STAT1 signaling through Tbx21-CNS-12 in T-bet induction, we enforced the expression of RUNX3 in the Tbx215M/5M “Th1” cells. Indeed, while ectopic expression of RUNX3 promoted IFN-γ production in the Thx215M/5M “Th1” cells (Figure S7E), it failed to up-regulate T-bet expression in these cells (Figure S7F). WT Th1 cells already expressed high amounts of T-bet and retroviral over-expression of RUNX3 had no additional effect (Figure S7F). Furthermore, RUNX3 did not promote T-bet expression in Stat4−/−Ifngr1−/− “Th1” cells (Figure S7G). Therefore, although CBF-β-RUNX3 can bind to Tbx21-CNS-3 and regulate T-bet expression in cNK cells, RUNX3 does not directly induce T-bet expression in Th1 cells consistent with the fact that Tbx21-CNS-3 is not accessible in Th1 cells.
T-bet regulates its own expression in NK cells
By using a BAC transgenic reporter mouse strain T-bet-ZsGreen on the Tbx21−/− background, we have previously reported that T-bet does not regulate its own expression in “Th1” cells (Zhu et al., 2012). However, lower amounts of T-bet-ZsGreen were observed in the T-bet-ZsGreen-Tbx21−/− Post-iNK-A cells compared to the T-bet-ZsGreen-WT controls (Figure S7H). Furthermore, hepatic cNK cells from the T-bet-ZsGreen-Tbx21−/− mice also expressed lower amounts of T-bet-ZsGreen compared with their WT counterparts (Figure S7I). Thus, T-bet regulates its own expression during NK cell development.
DISCUSSION
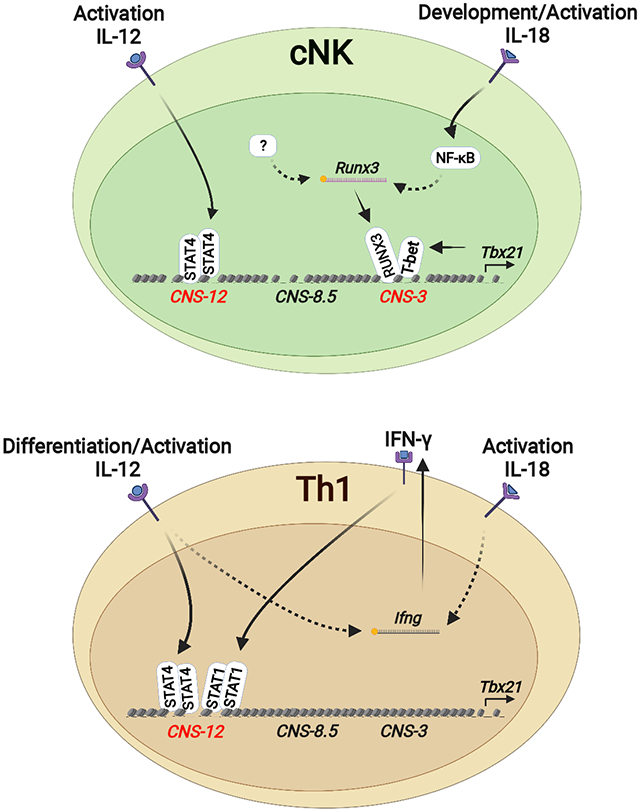
While T-bet is critical for the development and functions of type 1 innate and adaptive lymphocytes, little is known about its regulation at the genomic level. Through DHS analysis of the Tbx21 locus in Th and ILC subsets, we identified three cis-regulatory elements upstream of the Tbx21 TSS: Tbx21-CNS-3, Tbx21-CNS-8.5 and Tbx21-CNS-12 that were preferentially accessible in innate or adaptive type 1 lymphocytes and clearly demonstrated that distinct cis-regulatory elements at the Tbx21 locus were utilized for the induction of T-bet expression during the development and differentiation of type 1 innate and adaptive lymphocytes.
The differential induction of T-bet between Th1 cells and group 1 innate lymphoid cells we have reported in this study may also apply to GATA3 induction in Th2 cells and ILC2s, and RORγt induction in Th17 cells and ILC3s since these gene loci also displayed differential accessibilities in innate and adaptive lymphocytes of the same class. Therefore, while innate lymphocytes and their adaptive counterparts share the same set of LDTFs in determining their similar functionality after development or differentiation, there appear to be different routes to reach the same destination, providing important insights into understanding the evolutionary relationship between innate and adaptive lymphocytes.
IL-12 signaling-mediated STAT4 binding to the element Tbx21-CNS-12 was critical for the T-bet induction during Th1 cell differentiation, but this mechanism is not utilized for the induction of T-bet during cNK cell and ILC1 development in steady state. Nevertheless, the element Tbx21-CNS-12 is also accessible in NK cells. Indeed, the IL-12-STAT4 signaling through acting on the element Tbx21-CNS-12 can induce T-bet expression in cNK cells in vitro, suggesting that Tbx21-CNS-12 may also play an important role in T-bet regulation during NK cell activation in response to infection. The fact that Tbx21-CNS-12 is partially accessible even in naïve CD4+ T cells suggests that this is the critical element for lymphocytes responding to cytokine environment during immune responses. In addition, this site is also accessible in Th2 cells and Th17 cells, which may explain CD4+ T cell plasticity (Lee et al., 2009; Wei et al., 2009).
IL-18 signaling is a potent T-bet inducer in cNK cells. As a member of IL-1 family cytokines, IL-18 can be processed through inflammasome cleavage both in hematopoietic cells and non-hematopoietic cells in BM without involving an inflammatory response (Mantovani et al., 2019; Ratajczak et al., 2020; Silberstein et al., 2016). Although we cannot exclude a potential role of IL-18 in the posttranscriptional regulation of Tbx21 mRNA, IL-18 does induce T-bet at the transcriptional level. It is unlikely that IL-18 directly induces T-bet via the NF-κB pathway since the Tbx21 mRNA is not induced immediately after IL-18 stimulation. Instead, IL-18 through activating NF-κB significantly induces the expression of RUNX3, which in turn up-regulates T-bet expression in cNK cells. In the NK progenitors, RUNX3 expression precedes T-bet induction. However, T-bet expression is detected only in IL-18Rαhi NK progenitors which also express high amounts of RUNX3. Not only the initial expression of T-bet is highly correlated with the expression of IL-18Rα in NK progenitors in BM, such early T-bet induction in NK progenitors is diminished in the Il18r1-deficient mice. However, the lack of IL-18 or IL-18R has little effect on NK cell development although their functionalities may have been altered (Chaix et al., 2008; Hoshino et al., 1999; Takeda et al., 1998).
Tbx21-CNS-3 is preferentially accessible in cNK cells and this element plays an important role in optimal T-bet induction in cNK cells and NK progenitors, and thus promotes NK cell maturation. While RUNX3 alone may directly or indirectly regulate T-bet expression in NK cells, RUNX3 and T-bet could form a complex to bind to the Tbx21-CNS-3 element in promoting T-bet expression. However, Tbx21-CNS-3 is not accessible in Th1 cells and its deletion has no effect on T-bet expression during Th1 cell differentiation. Furthermore, IL-18 and RUNX3 are unable to directly induce T-bet expression during CD4+ T cell differentiation. Therefore, in T cells, the main mechanism of IL-18- and RUNX3-mediated T-bet induction is through regulating IFN-γ production.
The binding of T-bet at the Tbx21 locus may constitute a positive feedback loop for T-bet induction. T-bet self-regulates in NK cells, however, T-bet does not seem to be needed for regulating its own expression during Th1 cell differentiation (Zhu et al., 2012). ChIP-Seq analysis of T-bet binding showed that T-bet binds to its own gene but in a cell type-specific manner, that is, it bound to Tbx21-CNS-3 in cNK cells but bound to Tbx21-CNS-12 in Th1 cells. Since STAT4 also bound to Tbx21-CNS-12 in Th1 cells, which may compensate the function of T-bet binding to the same region, it could explain why T-bet is not needed for self-regulation in Th1 cells.
The cNK cell lineage development branches off the Id2-negative early innate lymphoid progenitor whereas the ILC1 lineage is derived from Id2+ ILC progenitors (ILCPs) (Colonna, 2018; Constantinides et al., 2014; Diefenbach et al., 2014; Klose et al., 2014), although the ILCPs still have the potential to become cNK cells (Xu et al., 2019; Zhong et al., 2020). ILC1s, highly enriched in the adult liver, start to arise during fetal liver hematopoiesis prior to the development of cNK cells during ontogeny (Chiossone et al., 2009; Daussy et al., 2014; Gordon et al., 2012). There are also reports showing a conversion of cNK cells into ILC1-like cells in the tumor microenvironment or during T. gondii infection (Gao et al., 2017; Park et al., 2019). Our study indicates that in the absence of Tbx21-CNS-3, T-bet induction in cNK cells but not in ILC1s is severely impaired. The differential regulation of T-bet expression in ILC1s and cNK cells supports the concept that they are distinct lineages both in mice and in humans (McFarland et al., 2021). Our results also strongly indicate that even these two closely related type 1 innate lymphocytes may utilize different mechanisms in T-bet induction during their development.
Although Th cell subsets and their ILC counterparts have similar characteristics, distinct gene regulatory pathways have been identified (Colonna, 2018; Koues et al., 2016; Shih et al., 2016). Th cells and ILCs experience different environments during their development and differentiation. ILCs largely develop in the BM and/or fetal liver without an inflammatory response. It has been proposed that while ILCs require STAT signaling for their functionality, STAT signaling is dispensable for their development. However, the latter concept has not been explicitly proven due to the possible redundancy of different STAT proteins. Here in this study, by abolishing the critical STAT-binding sites at the Tbx21 locus, we clearly demonstrated that STAT signaling through the Tbx21-CNS-12, which is essential for Th1 cell differentiation, is not required for the induction of T-bet in type 1 innate lymphocytes.
In summary, we have reported that type 1 innate and adaptive lymphocytes utilize distinct cis-regulatory elements for the induction of T-bet. Particularly, we have shown that cytokine-mediated STAT signaling is critical for Th1 cell differentiation but not for NK cell development, although STAT signaling is also crucical for NK cell functional activation through regulating the expression of other genes such as Ifng.
Limitations of Study
None of the three elements we investigated in this study seems to be required for T-bet induction during ILC1 development. Thus, whether there is an ILC1-specific element for T-bet induction requires further investigation. Further investigation is also needed to identify other signaling(s) which may regulate RUNX3 expression before and/or after IL-18Rα is induced during NK cell development. In addition, while T-bet can be further induced through STAT-binding sites at the Tbx21-CNS-12 during NK cell activation, the physiological importance of such T-bet regulation is unexplored. Finally, T-bet has been recently shown to play an important role in the development and functions of type 1 lymphocytes in humans (Yang et al., 2020; Yang et al., 2021). Whether there are loss- or gain-of-function mutations within the cis-regulatory elements at the TBX21 locus in humans, which may differentially affect the development and functions of innate and adaptive cells in patients, requires further investigation.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Jinfang Zhu (jfzhu@niaid.nih.gov).
Materials availability
Tbx215M/5M, Tbx21ΔCNS-3, Tbx21ΔCNS-8.5 and Tbx21ΔCNS-3ΔCNS-8.5 mouse strains are generated in this study.
Data and code availability
All sequencing data are available in the GEO database listed in the key resources table. This paper analyzed publicly available data and the accession code is listed in the key resources table.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-B220-Biotin (Clone RA3-6B2) | BD | Cat#553086; RRID: AB_ 394615 |
| Anti-CD3e-FITC (Clone 145-2C11) | BD | Cat#553062; RRID: AB_394595 |
| Anti-CD3e-Biotin (Clone 145-2C11) | eBioscience | Cat#13-0031-85; RRID: AB_466319 |
| Anti-CD4-BUV395 (Clone GK1.5) | BD | Cat#563790; RRID: AB_2738426 |
| Anti-CD4-Biotin (Clone GK1.5) | eBioscience | Cat#13-0041-85; RRID: AB_466326 |
| Anti-CD4-BUV737 (Clone GK1.5) | BD | Cat#564298 |
| Anti-CD5-Biotin (Clone 53-7.3) | BD | Cat#553019; RRID: AB_394557 |
| Anti-CD8-Biotin (Clone 53-6.7) | eBioscience | Cat#13-0081-85; RRID: AB_466347 |
| Anti-CD11b-FITC (Clone M1/70) | BD | Cat#553310; RRID: AB_396679 |
| Anti-CD11b-PerCP-Cy™5.5 (Clone M1/70) | BD | Cat#550993; RRID: AB_394002 |
| Anti-CD11b-BUV737 (Clone M1/70) | BD | Cat#612800; RRID: AB_2738811 |
| Anti-CD11b-Biotin (Clone M1/70) | BD | Cat#553309; RRID: AB_394773 |
| Anti-CD14-Biotin (Clone Sa2-8) | eBioscience | Cat#13-0141-85; RRID: AB_466371 |
| Anti-CD19-PE (Clone 1D3) | BD | Cat#553786; RRID: AB_395050 |
| Anti-CD19-Biotin (Clone 1D3) | eBioscience | Cat#13-0193-85; RRID: AB_657658 |
| Anti-CD25-FITC (Clone 7D4) | BD | Cat#553072; RRID: AB_394603 |
| Anti-CD27-BV605 (Clone LG.3A10) | BD | Cat#563365; RRID: AB_2738160 |
| Anti-CD44-eFluor 450 (Clone IM7) | eBioscience | Cat#48-0441-82; RRID: AB_1272246 |
| Anti-CD44-APC-eFluor 780 (Clone IM7) | eBioscience | Cat#47-0441-82; RRID: AB_1272244 |
| Anti-CD45.1-PE-Cy™7 (Clone A20) | BD | Cat#560578; RRID: AB_1727488 |
| Anti-CD45.2-BUV395 (Clone 104) | BD | Cat#564616; RRID: AB_2738867 |
| Anti-CD45.2-BUV737 (Clone 104) | BD | Cat#612778; RRID: AB_2870107 |
| Anti-CD49a-BV421 (Clone Ha31/8) | BD | Cat#740046; RRID: AB_2739815 |
| Anti-CD49a-BV711 (Clone Ha31/8) | BD | Cat#564863; RRID: AB_2738987 |
| Anti-CD49b-APC (Clone DX5) | BD | Cat#560628; RRID: AB_1727502 |
| Anti-CD49b-eFluor 780 (Clone DX5) | eBioscience | Cat#47-5971-82; RRID: AB_11218895 |
| Anti-CD62L-PE (Clone MEL-14) | eBioscience | Cat#12-0621-82; RRID: AB_465721 |
| Anti-CD62L-APC (Clone MEL-14) | eBioscience | Cat#17-0621-82; RRID: AB_469410 |
| Anti-CD117-APC-H7 (Clone 2B8) | BD | Cat#560185; RRID: AB_1645231 |
| Anti-CD117-PE (Clone 2B8) | BD | Cat#553355; RRID: AB_394806 |
| Anti-CD122-BV786 (Clone TM-β1) | BD | Cat#740869; RRID: AB_2740521 |
| Anti-CD122-FITC (Clone 5H4) | BD | Cat#554452; RRID: AB_395401 |
| Anti-CD127-PE-Cyanine7 (Clone A7R34) | eBioscience | Cat#25-1271-82; RRID: AB_469649 |
| Anti-CD244.2-BUV395 (Clone 2B4) | BD | Cat#740226; RRID: AB_2739974 |
| Anti-CD244.2-PE-Cyanine7 (eBio244F4) | eBioscience | Cat#25-2441-82; RRID: AB_2573432 |
| Anti-CCR6-BV421 (Clone 140706) | BD | Cat#564736; RRID: AB_2738926 |
| Anti-F4/80-Biotin (Clone BM8) | eBioscience | Cat#13-4801-85; RRID: AB_466658 |
| Anti-FceR1 alpha-Biotin (Clone MAR-1) | eBioscience | Cat#13-5898-82; RRID: AB_466783 |
| Anti-IFN-γ-Alexa Fluor® 700 (Clone XMG1.2) | BD | Cat#557998; RRID: AB_396979 |
| Anti-IFN-γ-PE-Cy™7 (Clone XMG1.2) | BD | Cat#557649; RRID: AB_396766 |
| Anti-IFN-γ-FITC (Clone XMG1.2) | BD | Cat#554411; RRID: AB_395375 |
| Anti-IL-4-Brilliant Violet 421™ (Clone 11B11) | BioLegend | Cat#504120; RRID: AB_2562102 |
| Anti-IL-4-Brilliant Violet 605™ (Clone 11B11) | BioLegend | Cat#504126; RRID: AB_2686971 |
| Anti-IL-18Rα-eFluor 450 (Clone P3TUNYA) | eBioscience | Cat#48-5183-82; RRID: AB_2574069 |
| Anti-IL-21R-PE (Clone 4A9) | BioLegend | Cat#131906; RRID: AB_1279430 |
| Anti-KLRG1-APC-eFluor 780 (Clone 2F1) | eBioscience | Cat#47-5893-82; RRID: AB_2573988 |
| Anti-Ly-6G/Ly-6C-Biotin (Clone RB6-8C5) | eBioscience | Cat#13-5931-85; RRID: AB_466801 |
| Anti-NK1.1-PE-Cy™7 (Clone PK136) | BD | Cat#552878; RRID: AB_394507 |
| Anti-NK1.1-BV650 (Clone PK136) | BD | Cat#564143; RRID: AB_2738617 |
| Anti-NKG2D-PE-CF594 (Clone CX5) | BD | Cat#562614; RRID: AB_2737677 |
| Anti-NKp46-PE (Clone 29A1.4) | BD | Cat#560757; RRID: AB_1727466 |
| Anti-TER-119-Biotin (Clone TER-119) | eBioscience | Cat#13-5921-85; RRID: AB_466798 |
| Anti-CBFβ Rabbit mAb (Clone D4N2N) | CST | Cat#62184 |
| Anti-Stat4 Rabbit mAb (Clone C46B10) | CST | Cat#2653 |
| Anti-T-bet-Alexa Fluor® 647 (Clone O4-46) | BD | Cat#561267; RRID: AB_10564093 |
| Anti-EOMES-eFluor 450 (Clone Dan11mag) | eBioscience | Cat#48-4875-82; RRID: AB_2574062 |
| Anti-Ki-67-BV786 (Clone B56) | BD | Cat#563756; RRID: AB_2732007 |
| Anti-RUNX3-PE (Clone R3-5G4) | BD | Cat#564814; RRID: AB_2738969 |
| Anti-NGFR-PE (Clone C40-1457) | BD | Cat#557196; RRID: AB_396599 |
| Anti-CD3e (Clone 2C11) | Harlan | Lot#A2022931 |
| Anti-CD28 (Clone 37.51) | Harlan | Lot#A2022929 |
| Anti-IL-4 (Clone 11B.11) | NCI-Frederick | Lot#L0412006 |
| Anti-IFN-γ (Clone XMG1.2) | Harlan | Lot#A2022930 |
| Anti-CD16/CD32 (Clone 2.4G2) | Harlan | Lot#A2083015 |
| Anti-Biotin MicroBeads | Miltenyi Biotec | Cat#130-090-485 |
| Anti-CD3/CD28 Dynabeads | Gibco | Cat#11453D |
| Alexa Fluor® 647 Mouse IgG1 κ | BD | Cat#557732; RRID: AB_396840 |
| Rat IgG2a Isotype-eFluor 450 (eBR2a) | eBioscience | Cat#48-4321-82; RRID: AB_1271999 |
| Anti-mouse IgG coated magnetic beads | Diagenode | Cat#C03010022 |
| InVivoMAb anti-mouse IL-12 p40 | BioXCell | Cat#BE0051; RRID: AB_1107698 |
| InVivoMAb rat IgG2a isotype control, anti-trinitrophenol | BioXCell | Cat#BE0089; RRID: AB_1107769 |
| Bacterial and virus strains | ||
| Toxoplasma gondii ME49 | Kawabe et al., 2017 | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| IL-1β | R&D systems | Cat#401-ML |
| IL-2 | R&D systems | Cat#402-ML |
| IL-6 | PeproTech | Cat#216-16 |
| IL-7 | R&D systems | Cat#407-ML |
| IL-12 | PeproTech | Cat#210-12 |
| IL-15 | R&D systems | Cat#447-ML |
| IL-18 | R&D systems | Cat#9139-IL |
| IL-21 | R&D systems | Cat#594-ML |
| CXCL12 | R&D systems | Cat#460-SD |
| Flt-3 Ligand | R&D systems | Cat#427-FL |
| IFN-γ | PeproTech | Cat#315-05 |
| SCF | R&D systems | Cat#455-MC |
| TGF-beta 1 | R&D systems | Cat#7666-MB |
| Fixable Viability Dye eFluor™ 506 | eBioscience | Cat#65-0866-14 |
| Fixable Viability Dye eFluor™ 780 | eBioscience | Cat#65-0865-14 |
| Streptavidin-BV605 | BD | Cat#563260; RRID: AB_2869476 |
| Streptavidin-APC-R700 | BD | Cat#565144; RRID: AB_2869657 |
| Protein A-coated magnetic beads | Diagenode | Cat#C03010020 |
| Percoll Plus | GE Healthcare | Cat#17544501 |
| Polybrene | Sigma | Cat#TR-1003-G |
| ACK Lysing Buffer | Gibco | Cat#A1049201 |
| AS15-Tetramer-BV421 | NIH TCF | Order#42412 |
| AS15-Tetramer-APC | NIH TCF | Order#42413 |
| Monensin Solution | eBioscience | Cat#00-4505-51 |
| L-Glutamine | Gibco | Cat#25030 |
| Sodium pyruvate | Gibco | Cat#11360 |
| β-mercaptoethanol | Gibco | Cat#21985 |
| NEAA | Gibco | Cat#11140 |
| Penicillin and streptomycin | Gibco | Cat#15140 |
| RPMI medium 1640 | Gibco | Cat#21870-076 |
| MEM | Gibco | Cat#11095080 |
| FspI | NEB | Cat#R0135L |
| PMA (Phorbol 12-myristate 13-acetate) | Sigma | Cat#P8139 |
| Ionomycin | Sigma | Cat#407952 |
| InSolution NF-κB Activation Inhibitor | Sigma | Cat#481407-1MG |
| Polyinosinic–polycytidylic acid sodium salt | Sigma | Cat#P0913-50MG |
| EasySep™ Buffer | Stem cell | Cat#20144 |
| Cycloheximide | Sigma | Cat#C1988-1G |
| DMSO | Sigma | Cat#D2650 |
| Critical commercial assays | ||
| Foxp3 Staining Buffer Set | eBioscience | Cat#00-5523-00 |
| QuantiTect Rev. Transcription Kit | Qiagen | Cat#205313 |
| MinElute Reaction Cleanup Kit | Qiagen | Cat#28206 |
| QIAquick Gel Extraction Kit | Qiagen | Cat#28704 |
| RNeasy Plus Mini Kit | Qiagen | Cat#74136 |
| PrimeScript™ RT Master Mix | TAKARA | Cat#RR036A |
| FastStart Universal SYBR Green Master (Rox) | Roche | Cat#04913850001 |
| P4 Primary Cell 4D-Nucleofector | LONZA | Cat#V4XP-4024 |
| CD4+ T Cell Isolation Kit | Miltenyi Biotec | Cat#130-104-454 |
| EasySep™ Mouse Streptavidin RapidSpheres™ Isolation Kit | Stem cell | Cat#19860 |
| RNase-Free DNase Set | Qiagen | Cat#79256 |
| DNeasy Blood & Tissue Kit | Qiagen | Cat#69506 |
| IFN gamma Mouse Uncoated ELISA Kit | Invitrogen | Cat#88-7314-86 |
| Deposited data | ||
| ChIP-Seq and DNase-Seq data | This manuscript | GEO: GSE172358 |
| ChIP-Seq (T-bet in Th1-2) | Zhu et al., 2012 | GEO: GSE38808 |
| Experimental models: Cell lines | ||
| OP9 | Schmitt and Zúñiga-Pflücker, 2002 | N/A |
| OP9-DLL1 | Schmitt and Zúñiga-Pflücker, 2002 | N/A |
| Experimental models: Organisms/strains | ||
| Mouse: Tbx215M/5M | This manuscript | N/A |
| Mouse: Tbx21ΔC-3 | This manuscript | N/A |
| Mouse: Tbx21ΔC-8.5 | This manuscript | N/A |
| Mouse: Tbx21ΔC-3ΔC-8.5 | This manuscript | N/A |
| Mouse: Tcra−/− | NIAID-Taconic | Taconic line 98 |
| Mouse: CD45.1 congenic | NIAID-Taconic | Taconic line 7 |
| Mouse: Rag1−/− | NIAID-Taconic | Taconic line 146 |
| Mouse: Rag2−/−γc−/− | NIAID-Taconic | Taconic line 111 |
| Mouse: T-bet-ZsGreen | NIAID-Taconic | Taconic line 8419 |
| Mouse: T-bet-ZsGreen-Tbx21−/− | NIAID-Taconic | Taconic line 8451 |
| Mouse: T-bet-ZsGreen-Stat4−/−Ifngr1−/− | NIAID-Taconic | Taconic line 8450 |
| Mouse: Il18r1−/− | The Jackson Laboratory | Stock#004131 |
| Mouse: C57BL/6J | The Jackson Laboratory | Stock#000664 |
| Mouse: RUNX3-YFP | Egawa and Littman, 2008 | Now also available at Jackson Laboratory, Stock#008774 |
| Oligonucleotides | ||
| See Excel file Table S3 | ||
| Recombinant DNA | ||
| NGFR-RUNX3 retrovirus | Yagi et al., 2010 | N/A |
| GFP-T-bet retrovirus | Yagi et al., 2010 | N/A |
| Software and algorithms | ||
| FlowJo V10 | FlowJo LLC | https://www.flowjo.com |
| Prism 7 | Graphpad | https://www.graphpad.com |
| MEME Suite | N/A | https://meme-suite.org |
| WashU Epigenome Browser | Washington University in St. Louis | http://epigenomegateway.wustl.edu |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
All the mouse strains are on C57BL/6 background. The single-stranded Tbx215M/5M oligonucleotides, serving as the repair template, and the Tbx215M/5M sgRNA, serving as the single guide RNA for targeting the Tbx21 locus, were used to generate the Tbx215M/5M mouse strain through the CRISPR-Cas9 technology. The restriction endonuclease FspI site “TGCGCA” was induced in the mutant to facilitate genotyping. The PCR products using the primer pair Tbx215M were purified and digested with FspI to screen the knock-in mutants, and the desired mutation was then confirmed by sequencing. To generate the Tbx21ΔCNS-3 mouse strain, two sgRNAs, Tbx21ΔC-3-5’ and Tbx21ΔC-3-3’, were used, and the primer pair Tbx21C-3 was used for genotyping. To generate the Tbx21ΔCNS-8.5 mouse strain, two sgRNAs, Tbx21ΔC-8.5-5’ and Tbx21ΔC-8.5-3’, were used, and the primer pair Tbx21C-8.5 was used for genotyping. To generate the Tbx21ΔCNS-3ΔCNS-8.5 mouse strain, two sgRNAs, Tbx21ΔC-8.5-5’ and Tbx21ΔC-8.5-3’, were used on the Tbx21ΔCNS-3 mouse strain. All these strains were back crossed to wildtype C57BL/6 mice more than 2 generations before they were used in the study. CD4-Cre-Tbx21fl/fl mice were previously described (Fang et al., 2018). Tcra−/−, Rag1−/−, Rag2−/−γc−\−, T-bet-ZsGreen, T-bet-ZsGreen-Tbx21−/−, Stat4−/−Ifngr1−/−, and CD45.1 congenic mice were obtained from the NIAID-Taconic repository. T-bet-ZsGreen-Tbx21−/− mice and T-bet-ZsGreen mice were crossed to wildtype C57BL/6 mice to generate Tbx21+/− mice and controls respectively. T-bet-ZsGreen-Tbx21−/− mice were crossed to wildtype C57BL/6 mice and Tbx21ΔCNS-3 mice to generate T-bet-ZsGreen-Tbx21−/+ and T-bet-ZsGreen-Tbx21−/ΔCNS-3 mice, respectively. The Il18r1−− mice and the control C57BL/6J mice were purchased from the Jackson Laboratory. RUNX3-YFP mice (Egawa and Littman, 2008) were kindly provided by Drs. Takeshi Egawa and Dan Littman. Mice were bred and/or maintained in the NIAID specific pathogen-free animal facilities, and they were used at 8-12 weeks of age. All the animal experiments were performed under a protocol approved by the NIAID Animal Care and Use Committee.
Parasites
Parasite T. gondii ME49 was described in previous studies (Kawabe et al., 2017; Yu et al., 2018).
METHOD DETAILS
T. gondii infection
T. gondii infection was described previously (Kawabe et al., 2017). Briefly, 8-12 weeks old mice of the indicated strain were inoculated (i.p.) with 15 ME49 cysts harvested from the brains of chronically infected C57BL/6 mice. To assess the relative pathogen load, 0.2 ml suspension from 4 ml PC and 40 ml liver homogenate was used to prepared total DNA followed by real-time PCR. The mouse gene Gapdh was used to normalize the T. gondii DNA (Kawabe et al., 2017). The supernatants from serum were used for IFN-γ ELISA assay. In addition to measuring immune responses, mortality of the infected animals was monitored in some experiments.
Poly (I:C) treatment
8-10 weeks old mice of the indicated strain were inoculated (i.p.) with 100 μg poly (I:C) or control saline for 2.5 hours and the hepatic lymphocytes were harvested and incubated with monensin for 3.5 hours followed by IFN-γ intracellular staining. Mice were inoculated (i.p.) with 100 μg poly (I:C) or control saline for 6 hours and the serum was collected for IFN-γ ELISA assay.
Bone marrow chimeras and cell transfer
The Tcra−/− and Rag2−/−γc−/− mice were sub-lethally irradiated (450 Rad) followed by bone marrow reconstitution (i.v.) with 5 million cells from femurs and then kept for 8-10 weeks before use. In some experiments, total CD4+ T cells purified from the spleens and lymph nodes via negative selection were transferred (i.v.) into Rag1−/− recipients (7.5 million/mouse).
In vivo IL-12 neutralization
WT mice were pre-treated (i.p.) with control IgG or anti-IL-12 p40 antibodies (200 μg/mouse) 2 days before T. gondii infection (i.p.). The infected mice were treated with control IgG or anti-IL-12 p40 antibodies (200 μg/mouse) at day 1 and day 4 after infection and the mice were analyzed at day 7. Rag2−/−γc−/− mice were pre-treated (i.p.) with control IgG or anti-IL-12 p40 antibodies (200 μg/mouse) 2 days before BM transfer. The chimeras were treated with control IgG or anti-IL-12 p40 antibodies (200 μg/mouse) at day 1 and day 6, and further treated (100 μg/mouse) every 5 days for additional 6-7 weeks.
Cell isolation
Spleen, LN and BM were dissociated and put through the 40 μm cell strainer to make single cell suspensions in ice-cold HBSS with 3% FBS. To isolate lymphocytes from the liver, the perfused liver was mechanically dissociated by gentleMACS™ Dissociator and put through the 70 μm cell strainer. ILC2s and LTi cells from small intestine and cNK cells from lung were harvested following a protocol described previously (Moro et al., 2015). The liver and small intestine suspensions were spun down, re-suspended in 40% Percoll Plus, and centrifuged at 1800 rpm for 20 minutes at room temperature (RT). The ACK treatment was applied to get rid of red blood cells when necessary.
Measurement of cytokine production
To measure cytokine production by Th1 cells generated in vitro, cells were stimulated with PMA (10 ng/ml) and ionomycin (1 μM) for 4 hours. To measure cytokine production by T cells from T. gondii-infected mice ex vivo, cells were stimulated with anti-CD3 and anti-CD28 antibody-coated beads for 5 hours. To measure cytokine production by cNK cells and ILC1s ex vivo, cells were stimulated with IL-12 (10 ng/ml) and IL-18 (20 ng/ml) for 5 hours. Monensin (2 μM) was added during stimulation to block cytokine secretion. To measure cytokine secreted in the serum, commercial ELISA kit was used.
Flow cytometry and cell sorting
Cells were blocked with anti-mouse CD16/CD32 antibodies before incubated with fluorochrome-conjugated antibodies and Live/Dead dye. Cell surface staining was usually performed in HBSS with 3% FBS at 4°C for 15 minutes if not specified otherwise. For intracellular cytokine staining, cells were fixed in 4% paraformaldehyde for 15 minutes at RT, and then permeabilized and stained with antibodies at 4°C in 0.5% Triton X-100/PBS for 15 minutes. For intracellular transcription factor staining, the Foxp3 Staining Buffer Set was used following the manufacturer’s instruction. For T. gondii peptide AS15-tetramer staining, cells were incubated with the tetramer at 37°C in RPMI 1640 medium for 30 minutes. To analyze NKPs in BM, the Lin antibodies (CD3, CD4, CD5, CD8, CD11b, B220, TER119, Ly6G/6C) were used. Flow cytometry data were collected from FORTESSA or LSR II (BD Biosciences), and the results were analyzed with the FlowJo software 10 (Tree Star). Cells were sorted on an FACS Aria cell sorter (BD Biosciences). Naïve CD4+ T cells were sorted as CD4+CD44loCD62L+CD25− population from the LNs. To sort cNK cells from the spleen, cells were depleted with Lin antibodies (CD3, CD4, CD8, CD14, CD19, Ly-6G/Ly-6C and TER-119) as described (Pak-Wittel et al., 2014), then sorted as NK1.1+Lin− population. To sort NK progenitors from BM, cells were depleted with Lin antibodies (CD3, CD4, CD5, CD8, CD11b, B220, TER119, Ly6G/6C, c-Kit) before sorting. To sort lymphoid-primed multipotent progenitors (LMPPs) from BM, cells were depleted with Lin antibodies (CD3, CD5, CD11b, CD19, B220, TER119, Ly6G/6C, NK1.1) before sorting. To sort cNK cells and ILC1s from the liver, cNK cells were sorted as CD3−CD19−NK1.1+CD49a−CD49b+ whereas ILC1s were sorted as CD3−CD19−NK1.1+CD49a+CD49b−. ILC2s and LTi cells from the small intestine were sorted as Lin (CD3, CD4, CD5, CD8, CD11c, CD19, MAR-1, TER119, F4/80, Ly6G/6C)− CD127+KLRG1+ and Lin−CD127+KLRG1−CCR6+NKp46−, respectively.
Cell culture
CD4+ T cells and cNK cells were cultured in the complete RPMI 1640 medium (10% FBS, 200 mM glutamine, 100 mM sodium pyruvate, 50 μM β-mercaptoethanol, 100 U/ml penicillin and 100 μg/ml streptomycin). Th1-skewing conditions: α-CD3 (1 μg/ml), α-CD28 (3 μg/ml), α-IL-4 (10 μg/ml), IL-12 (10 ng/ml) and IL-2 (100 U/ml); Th2-skewing conditions: μ-CD3 (1 μg/ml), α-CD28 (3 μg/ml), α-IFN-γ (10 μg/ml), α-IL-12 (10 μg/ml) and IL-4 (5000 U/ml); Th17-skewing conditions: α-CD3 (1 μg/ml), α-CD28 (3 μg/ml), α-IL-4 (10 μg/ml), α-IFN-γ (10 μg/ml), α-IL-12 (10 μg/ml), TGFβ1 (1 ng/ml), IL-6 (10 ng/ml) and IL-1β (10 ng/ml). The irradiated T cell-depleted splenocytes were used as antigen-presenting cells in vitro. In some experiments, α-IFN-γ (10 μg/ml) was added to neutralize the supernatant IFN-γ. cNK cells were incubated with IL-1 (10 ng/ml), IL-6 (10 ng/ml), IL-7 (10 ng/ml), IL-12 (2 ng/ml), IL-15 (10 ng/ml), IL-18 (10 ng/ml), IL-21 (20 ng/ml), CXCL12 (10 ng/ml), Flt3L (10 ng/ml), SCF (20 ng/ml), IFN-γ (10 ng/ml) or TGF-β1 (1 ng/ml) in the presence of IL-2 (100 U/ml). OP9 and OP9-DL1 cell line (Schmitt and Zuniga-Pflucker, 2002) were cultured in the MEM medium (5% FBS, 100 U/ml penicillin and 100 μg/ml streptomycin).
Retroviral infection and nucleofection
Ectopic gene expression in CD4+ T cells was achieved through retroviral infection. Naïve CD4+ T cells were cultured under Th1-skewing conditions for 1 day, and then the virus-containing supernatants and polybrene were added into the cell culture. The mixture was spun down at 3000 rpm for 45 minutes at RT and then cultured for another day. Cells were then cultured with fresh Th1 skewing medium for additional 3 days. The negatively selected cNK cells (Pak-Wittel el al., 2014) from the spleen were transfected with siRNA and indicator (5:1) using Amaxa™ P4 Primary Cell 4D-Nucleofector™ X Kit L under the program CZ-167 and cultured in the complete RPMI 1640 medium without antibiotics. After 1 day, cells were sorted according to the intensity of the indicator and then cultured in the complete medium for additional 12 hours. Cells were further incubated with IL-18 for 2 days (for mRNA) or 3 days (for protein) with IL-2 supplemented in the medium. Ectopic gene expression in LMPPs was achieved through retroviral infection. Sorted LMPPs (Lin−CD117+Sca1+Flt3+CD127−) were cultured in DMEM medium supplemented with SCF (20 ng/ml), Flt3L (10 ng/ml), IL-7 (5 ng/ml) for 2.5 days before retroviral infection with GFP-T-bet or GFP-Vector control. On day 6, the infected cells were further co-cultured with OP9 cell line in DMEM medium supplemented with IL-15 (10 ng/ml) for additional 8 days.
qRT–PCR
The total RNAs were extracted using RNeasy Plus Mini Kit, and reverse transcribed with PrimeScript RT Master Mix (TAKARA) or QuantiTect Rev Transcription Kit (QIAGEN). The cDNAs were analyzed with QuantStudio 7 Flex Real-Time PCR System (Applied Biosystems) using FastStart Universal SYBR Green Master (Roche). The following primer pairs were used to quantify specific gene transcripts: Tbx21-CDS for Tbx21, Runx3-P1 for Runx3 from distal promoter, Runx3-P2 for Runx3 from proximal promoter, Cbfb-CDS for Cbfb. Hprt was used for normalization.
ChIP-PCR
Two million Th1 cells or one million cNK cells were used in the ChIP assay. Cells were cross-linked with 1% formaldehyde for 10 minutes at RT and sonicated in the shearing buffer (0.4% SDS in TE buffer) to generate 100 to 400 bp DNA fragments. 10% of total DNA was used as input, and the rest was used in the subsequent immunoprecipitation. DNA-protein complexes were pulled down with specific antibodies in the RIPA buffer overnight. DiaMag anti-mouse IgG coated magnetic beads or DiaMag protein A coated magnetic beads were added into the buffer for additional 4 hours. After washing, reverse-crosslinking and DNA purification (100 to 400 bp) were performed. The DNA fragments were analyzed through real-time PCR. Primer pair Tbx21-CNS-12-CP was used for targeting Tbx21-CNS-12, Tbx21-CNS-3-CP for targeting Tbx21-CNS-3, and Il4-TSS-CP for targeting Il4-TSS.
ChIP-Seq, DNase-Seq and data analysis
Single-cell DNase sequencing protocol (Jin et al., 2015) was used to perform low-cell number DHS assay. ChIP-Seq analysis of T-bet and STAT4 binding (using 1 million cells) was performed as described previously (Hu et al., 2018). DNA samples were blunt ended, ligated to the Solexa adaptors, amplified, and sequenced with Illumina HiSeq system through the NHLBI DNA Sequencing and Computational Biology Core. ChIP-Seq and DNase-Seq reads (50 bp) were mapped to mm10 genome with Bowtie2 (Langmead and Salzberg, 2012). Only non-redundant reads with MAPQ >=10 were used for follow-up analysis. DNase-Seq peaks were called by MACS2 (Zhang et al., 2008) with settings of --nomodel--extsize 75 from down-sampling reads of 10 million for each replicate. Then only overlapped DHSs from two replicates were kept for following analysis. Poisson test implemented as scipy.stats.poisson.sf function was used to evaluate the statistical significances of enriched reads located in CNS regions among different cell types, and P value < 1e-5 was used as the significant cutoff. T-bet and STAT4 ChIP-seq peaks were called by callPeaks module in cLoops2 package (https://github.com/YaqiangCao/cLoops2, an extension from cLoops (Cao et al., 2020)), with key parameters of -eps 150 -minPts 10, 20, and only overlapped peaks from two replicates were kept for following analysis. Peaks correlation analysis were carried out by Intervene (Khan and Mathelier, 2017) with key parameters of intervene pairwise--compute frac--corr--htype dendrogram. Peak annotations were obtained by annotatePeak.pl in HOMER package (Heinz et al., 2010). Washington University Genome Browser with mm10 was used to visualize peaks, identify conserved regions and compare the sequence between human and mouse. Cell samples are in biological duplicates.
QUANTIFICATION AND STATISTICAL ANALYSIS
Differences between two groups were determined by two-tailed unpaired or paired Student’s t-test using Prism 7 software (GraphPad). P<0.05 was considered significant. Mean ± SD; n.s., none-significant, *P < 0.05, **P < 0.01.
Supplementary Material
Table S1. Cell type-specific DHSs in CD4+ T cell and ILC subsets, related to Figure 1
Table S3. Oligonucleotides, related to key resources table, Figure 2, Figure 3 and Figure 7
Highlights:
Differential chromatin accessibility identified in innate and adaptive lymphocytes.
Tbx21-CNS-3 is important for optional T-bet expression and NK cell maturation.
IL-18-RUNX3 and T-bet itself activate Tbx21-CNS-3 element in NK cells.
STAT binding at Tbx21-CNS-12 is critical for T-bet induction in Th1 cells.
ACKNOWLEDGEMENTS
We thank Drs. John J. O’Shea and Ronald N. Germain for their critical reading of the manuscript. We thank Steven L. Reiner for providing the Tbx21fl/fl mice, Takeshi Egawa and Dan R. Littman for providing the RUNX3-YFP mice, Juan Carlos Zúñiga-Pflücker for providing OP9 and OP9-DL1 cell lines, Ke Weng and the NIAID Flow Cytometry Section for cell sorting, the NIH Tetramer Core Facility for the tetramer reagents, and the NHLBI DNA Sequencing Core facility for sequencing the ChIP-Seq and DNase-Seq libraries. This work is supported by the Division of Intramural Research of the NIAID (grant 1ZIA-AI-001169) and the NHLBI (grant 1ZIA-HL-006030).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information includes 7 figures and 3 Excel files.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Abel AM, Yang C, Thakar MS, and Malarkannan S (2018). Natural Killer Cells: Development, Maturation, and Clinical Utilization. Front Immunol 9, 1869. 10.3389/fimmu.2018.01869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard N, Gonzalez F, Schaeffer M, Joncker NT, Cheng T, Shastri AJ, Robey EA, and Shastri N (2008). Immunodominant, protective response to the parasite Toxoplasma gondii requires antigen processing in the endoplasmic reticulum. Nat Immunol 9, 937–944. 10.1038/ni.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CR, and McLeod R (1990). Class I MHC genes and CD8+ T cells determine cyst number in Toxoplasma gondii infection. J Immunol 145, 3438–3441. [PubMed] [Google Scholar]
- Cao YQ, Chen ZX, Chen XW, Ai DS, Chen GY, McDermott J, Huang Y, Guo XX, and Han JDJ (2020). Accurate loop calling for 3D genomic data with cLoops. Bioinformatics 36, 666–675. 10.1093/bioinformatics/btz651. [DOI] [PubMed] [Google Scholar]
- Carotta S, Pang SH, Nutt SL, and Belz GT (2011). Identification of the earliest NK-cell precursor in the mouse BM. Blood 117, 5449–5452. 10.1182/blood-2010-11-318956. [DOI] [PubMed] [Google Scholar]
- Chaix J, Tessmer MS, Hoebe K, Fuseri N, Ryffel B, Dalod M, Alexopoulou L, Beutler B, Brossay L, Vivier E, and Walzer T (2008). Cutting edge: Priming of NK cells by IL-18. J Immunol 181, 1627–1631. 10.4049/jimmunol.181.3.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D, Xing Q, Su Y, Zhao X, Xu W, Wang X, and Dong C (2020). The Conserved Non-coding Sequences CNS6 and CNS9 Control Cytokine-Induced Rorc Transcription during T Helper 17 Cell Differentiation. Immunity 53, 614–626 e614. 10.1016/j.immuni.2020.07.012. [DOI] [PubMed] [Google Scholar]
- Chaves P, Zriwil A, Wittmann L, Boukarabila H, Peitzsch C, Jacobsen SEW, and Sitnicka E (2018). Loss of Canonical Notch Signaling Affects Multiple Steps in NK Cell Development in Mice. J Immunol 201, 3307–3319. 10.4049/jimmunol.1701675. [DOI] [PubMed] [Google Scholar]
- Chiossone L, Chaix J, Fuseri N, Roth C, Vivier E, and Walzer T (2009). Maturation of mouse NK cells is a 4-stage developmental program. Blood 113, 5488–5496. 10.1182/blood-2008-10-187179. [DOI] [PubMed] [Google Scholar]
- Christie D, and Zhu J (2014). Transcriptional regulatory networks for CD4 T cell differentiation. Curr Top Microbiol Immunol 381, 125–172. 10.1007/82_2014_372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins A, Rothman N, Liu K, and Reiner SL (2017). Eomesodermin and T-bet mark developmentally distinct human natural killer cells. JCI Insight 2, e90063. 10.1172/jci.insight.90063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colonna M (2018). Innate Lymphoid Cells: Diversity, Plasticity, and Unique Functions in Immunity. Immunity 48, 1104–1117. 10.1016/j.immuni.2018.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinides MG, McDonald BD, Verhoef PA, and Bendelac A (2014). A committed precursor to innate lymphoid cells. Nature 508, 397–401. 10.1038/nature13047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daussy C, Faure F, Mayol K, Viel S, Gasteiger G, Charrier E, Bienvenu J, Henry T, Debien E, Hasan UA, et al. (2014). T-bet and Eomes instruct the development of two distinct natural killer cell lineages in the liver and in the bone marrow. J Exp Med 211, 563–577. 10.1084/jem.20131560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denkers EY, Gazzinelli RT, Martin D, and Sher A (1993). Emergence of NK1.1+ cells as effectors of IFN-gamma dependent immunity to Toxoplasma gondii in MHC class I-deficient mice. J Exp Med 178, 1465–1472. 10.1084/jem.178.5.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diefenbach A, Colonna M, and Koyasu S (2014). Development, differentiation, and diversity of innate lymphoid cells. Immunity 41, 354–365. 10.1016/j.immuni.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djuretic IM, Levanon D, Negreanu V, Groner Y, Rao A, and Ansel KM (2007). Transcription factors T-bet and Runx3 cooperate to activate Ifng and silence Il4 in T helper type 1 cells. Nature immunology 8, 145–153. [DOI] [PubMed] [Google Scholar]
- Ebihara T, Song C, Ryu SH, Plougastel-Douglas B, Yang L, Levanon D, Groner Y, Bern MD, Stappenbeck TS, Colonna M, et al. (2015). Runx3 specifies lineage commitment of innate lymphoid cells. Nature immunology 16, 1124–1133. 10.1038/ni.3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egawa T, and Littman DR (2008). ThPOK acts late in specification of the helper T cell lineage and suppresses Runx-mediated commitment to the cytotoxic T cell lineage. Nature immunology 9, 1131–1139. 10.1038/ni.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang D, Cui K, Mao K, Hu G, Li R, Zheng M, Riteau N, Reiner SL, Sher A, Zhao K, and Zhu J (2018). Transient T-bet expression functionally specifies a distinct T follicular helper subset. The Journal of experimental medicine 215, 2705–2714. 10.1084/jem.20180927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang D, and Zhu J (2017). Dynamic balance between master transcription factors determines the fates and functions of CD4 T cell and innate lymphoid cell subsets. The Journal of experimental medicine 214, 1861–1876. 10.1084/jem.20170494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fathman JW, Bhattacharya D, Inlay MA, Seita J, Karsunky H, and Weissman IL (2011). Identification of the earliest natural killer cell-committed progenitor in murine bone marrow. Blood 118, 5439–5447. 10.1182/blood-2011-04-348912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finotto S, Neurath MF, Glickman JN, Qin S, Lehr HA, Green FH, Ackerman K, Haley K, Galle PR, Szabo SJ, et al. (2002). Development of spontaneous airway changes consistent with human asthma in mice lacking T-bet. Science 295, 336–338. 10.1126/science.1065544. [DOI] [PubMed] [Google Scholar]
- Gao Y, Souza-Fonseca-Guimaraes F, Bald T, Ng SS, Young A, Ngiow SF, Rautela J, Straube J, Waddell N, Blake SJ, et al. (2017). Tumor immunoevasion by the conversion of effector NK cells into type 1 innate lymphoid cells. Nat Immunol 18, 1004–1015. 10.1038/ni.3800. [DOI] [PubMed] [Google Scholar]
- Gordon SM, Chaix J, Rupp LJ, Wu J, Madera S, Sun JC, Lindsten T, and Reiner SL (2012). The transcription factors T-bet and Eomes control key checkpoints of natural killer cell maturation. Immunity 36, 55–67. 10.1016/j.immuni.2011.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grover HS, Blanchard N, Gonzalez F, Chan S, Robey EA, and Shastri N (2012). The Toxoplasma gondii peptide AS15 elicits CD4 T cells that can control parasite burden. Infect Immun 80, 3279–3288. 10.1128/IAI.00425-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gury-BenAri M, Thaiss CA, Serafini N, Winter DR, Giladi A, Lara-Astiaso D, Levy M, Salame TM, Weiner A, David E, et al. (2016). The Spectrum and Regulatory Landscape of Intestinal Innate Lymphoid Cells Are Shaped by the Microbiome. Cell 166, 1231–1246 e1213. 10.1016/j.cell.2016.07.043. [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, and Glass CK (2010). Simple Combinations of Lineage-Determining Transcription Factors Prime cis-Regulatory Elements Required for Macrophage and B Cell Identities. Molecular Cell 38, 576–589. 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, and Glass CK (2012). Roles of lineage-determining transcription factors in establishing open chromatin: lessons from high-throughput studies. Curr Top Microbiol Immunol 356, 1–15. 10.1007/82_2011_142. [DOI] [PubMed] [Google Scholar]
- Hoshino K, Tsutsui H, Kawai T, Takeda K, Nakanishi K, Takeda Y, and Akira S (1999). Cutting edge: generation of IL-18 receptor-deficient mice: evidence for IL-1 receptor-related protein as an essential IL-18 binding receptor. J Immunol 162, 5041–5044. [PubMed] [Google Scholar]
- Hosoya-Ohmura S, Lin YH, Herrmann M, Kuroha T, Rao A, Moriguchi T, Lim KC, Hosoya T, and Engel JD (2011). An NK and T cell enhancer lies 280 kilobase pairs 3' to the gata3 structural gene. Mol Cell Biol 31, 1894–1904. 10.1128/MCB.05065-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Cui K, Fang D, Hirose S, Wang X, Wangsa D, Jin W, Ried T, Liu P, Zhu J, et al. (2018). Transformation of Accessible Chromatin and 3D Nucleome Underlies Lineage Commitment of Early T Cells. Immunity 48, 227–242 e228. 10.1016/j.immuni.2018.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankovic D, Kullberg MC, Feng CG, Goldszmid RS, Collazo CM, Wilson M, Wynn TA, Kamanaka M, Flavell RA, and Sher A (2007). Conventional T-bet(+)Foxp3(−) Th1 cells are the major source of host-protective regulatory IL-10 during intracellular protozoan infection. The Journal of experimental medicine 204, 273–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W, Tang Q, Wan M, Cui K, Zhang Y, Ren G, Ni B, Sklar J, Przytycka TM, Childs R, et al. (2015). Genome-wide detection of DNase I hypersensitive sites in single cells and FFPE tissue samples. Nature 528, 142–146. 10.1038/nature15740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamimura Y, and Lanier LL (2015). Homeostatic control of memory cell progenitors in the natural killer cell lineage. Cell Rep 10, 280–291. 10.1016/j.celrep.2014.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, and Malhotra N (2015). Transcription factor networks directing the development, function, and evolution of innate lymphoid effectors. Annu Rev Immunol 33, 505–538. 10.1146/annurev-immunol-032414-112025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasal DN, Liang Z, Hollinger MK, O'Leary CY, Lisicka W, Sperling AI, and Bendelac A (2021). A Gata3 enhancer necessary for ILC2 development and function. Proc Natl Acad Sci U S A 118. 10.1073/pnas.2106311118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabe T, Jankovic D, Kawabe S, Huang Y, Lee PH, Yamane H, Zhu J, Sher A, Germain RN, and Paul WE (2017). Memory-phenotype CD4(+) T cells spontaneously generated under steady-state conditions exert innate TH1-like effector function. Sci Immunol 2, eaam9304. 10.1126/sciimmunol.aam9304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A, and Mathelier A (2017). Intervene: a tool for intersection and visualization of multiple gene or genomic region sets. BMC Bioinformatics 18, 287. 10.1186/s12859-017-1708-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose CSN, Flach M, Mohle L, Rogell L, Hoyler T, Ebert K, Fabiunke C, Pfeifer D, Sexl V, Fonseca-Pereira D, et al. (2014). Differentiation of type 1 ILCs from a common progenitor to all helper-like innate lymphoid cell lineages. Cell 157, 340–356. 10.1016/j.cell.2014.03.030. [DOI] [PubMed] [Google Scholar]
- Koues OI, Collins PL, Cella M, Robinette ML, Porter SI, Pyfrom SC, Payton JE, Colonna M, and Oltz EM (2016). Distinct Gene Regulatory Pathways for Human Innate versus Adaptive Lymphoid Cells. Cell 165, 1134–1146. 10.1016/j.cell.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugler DG, Mittelstadt PR, Ashwell JD, Sher A, and Jankovic D (2013). CD4+ T cells are trigger and target of the glucocorticoid response that prevents lethal immunopathology in toxoplasma infection. J Exp Med 210, 1919–1927. 10.1084/jem.20122300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–359. 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau CM, Adams NM, Geary CD, Weizman OE, Rapp M, Pritykin Y, Leslie CS, and Sun JC (2018). Epigenetic control of innate and adaptive immune memory. Nat Immunol 19, 963–972. 10.1038/s41590-018-0176-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarevic V, Chen X, Shim JH, Hwang ES, Jang E, Bolm AN, Oukka M, Kuchroo VK, and Glimcher LH (2011). T-bet represses T(H)17 differentiation by preventing Runx1-mediated activation of the gene encoding RORgammat. Nature immunology 12, 96–104. 10.1038/ni.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YK, Turner H, Maynard CL, Oliver JR, Chen D, Elson CO, and Weaver CT (2009). Late developmental plasticity in the T helper 17 lineage. Immunity 30, 92–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levanon D, Negreanu V, Lotem J, Bone KR, Brenner O, Leshkowitz D, and Groner Y (2014). Transcription factor Runx3 regulates interleukin-15-dependent natural killer cell activation. Mol Cell Biol 34, 1158–1169. 10.1128/MCB.01202-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman LA, Villegas EN, and Hunter CA (2004). Interleukin-15-deficient mice develop protective immunity to Toxoplasma gondii. Infect Immun 72, 6729–6732. 10.1128/IAI.72.11.6729-6732.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lighvani AA, Frucht DM, Jankovic D, Yamane H, Aliberti J, Hissong BD, Nguyen BV, Gadina M, Sher A, Paul WE, and O'Shea JJ (2001). T-bet is rapidly induced by interferon-gamma in lymphoid and myeloid cells. Proceedings of the National Academy of Sciences of the United States of America 98, 15137–15142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longhi MP, Trumpfheller C, Idoyaga J, Caskey M, Matos I, Kluger C, Salazar AM, Colonna M, and Steinman RM (2009). Dendritic cells require a systemic type I interferon response to mature and induce CD4+ Th1 immunity with poly IC as adjuvant. J Exp Med 206, 1589–1602. 10.1084/jem.20090247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madera S, Geary CD, Lau CM, Pikovskaya O, Reiner SL, and Sun JC (2018). Cutting Edge: Divergent Requirement of T-Box Transcription Factors in Effector and Memory NK Cells. J Immunol 200, 1977–1981. 10.4049/jimmunol.1700416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa Y, Tsukumo S, Chiba S, Hirai H, Hayashi Y, Okada H, Kishihara K, and Yasutomo K (2003). Delta1-Notch3 interactions bias the functional differentiation of activated CD4+ T cells. Immunity 19, 549–559. [DOI] [PubMed] [Google Scholar]
- Mantovani A, Dinarello CA, Molgora M, and Garlanda C (2019). Interleukin-1 and Related Cytokines in the Regulation of Inflammation and Immunity. Immunity 50, 778–795. 10.1016/j.immuni.2019.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarland AP, Yalin A, Wang SY, Cortez VS, Landsberger T, Sudan R, Peng V, Miller HL, Ricci B, David E, et al. (2021). Multi-tissue single-cell analysis deconstructs the complex programs of mouse natural killer and type 1 innate lymphoid cells in tissues and circulation. Immunity. 10.1016/j.immuni.2021.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minter LM, Turley DM, Das P, Shin HM, Joshi I, Lawlor RG, Cho OH, Palaga T, Gottipati S, Telfer JC, et al. (2005). Inhibitors of γ-secretase block in vivo and in vitro T helper type 1 polarization by preventing Notch upregulation of Tbx21. Nature Immunology 6, 680–688. 10.1038/ni1209x. [DOI] [PubMed] [Google Scholar]
- Moro K, Ealey KN, Kabata H, and Koyasu S (2015). Isolation and analysis of group 2 innate lymphoid cells in mice. Nat Protoc 10, 792–806. 10.1038/nprot.2015.047. [DOI] [PubMed] [Google Scholar]
- Mujal AM, Delconte RB, and Sun JC (2021). Natural Killer Cells: From Innate to Adaptive Features. Annual review of immunology 39, 417–447. 10.1146/annurev-immunol-101819-074948. [DOI] [PubMed] [Google Scholar]
- Mullen AC, High FA, Hutchins AS, Lee HW, Villarino AV, Livingston DM, Kung AL, Cereb N, Yao TP, Yang SY, and Reiner SL (2001). Role of T-bet in commitment of TH1 cells before IL-12-dependent selection. Science 292, 1907–1910. 10.1126/science.1059835. [DOI] [PubMed] [Google Scholar]
- Muller AA, Dolowschiak T, Sellin ME, Felmy B, Verbree C, Gadient S, Westermann AJ, Vogel J, LeibundGut-Landmann S, and Hardt WD (2016). An NK Cell Perforin Response Elicited via IL-18 Controls Mucosal Inflammation Kinetics during Salmonella Gut Infection. PLoS pathogens 12, e1005723. 10.1371/journal.ppat.1005723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S, Sato T, Kohu K, Takeda K, Okumura K, Satake M, and Habu S (2008). Runx proteins are involved in regulation of CD122, Ly49 family and IFN-gamma expression during NK cell differentiation. Int Immunol 20, 71–79. 10.1093/intimm/dxm120. [DOI] [PubMed] [Google Scholar]
- Pak-Wittel MA, Piersma SJ, Plougastel BF, Poursine-Laurent J, and Yokoyama WM (2014). Isolation of murine natural killer cells. Curr Protoc Immunol 105, 3 22 21–23 22 29. 10.1002/0471142735.im0322s105. [DOI] [PubMed] [Google Scholar]
- Park E, Patel S, Wang Q, Andhey P, Zaitsev K, Porter S, Hershey M, Bern M, Plougastel-Douglas B, Collins P, et al. (2019). Toxoplasma gondii infection drives conversion of NK cells into ILC1-like cells. Elife 8. 10.7554/eLife.47605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker SJ, Roberts CW, and Alexander J (1991). CD8+ T cells are the major lymphocyte subpopulation involved in the protective immune response to Toxoplasma gondii in mice. Clin Exp Immunol 84, 207–212. 10.1111/j.1365-2249.1991.tb08150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H, Jiang X, Chen Y, Sojka DK, Wei H, Gao X, Sun R, Yokoyama WM, and Tian Z (2013). Liver-resident NK cells confer adaptive immunity in skin-contact inflammation. J Clin Invest 123, 1444–1456. 10.1172/JCI66381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perchet T, Petit M, Banchi EG, Meunier S, Cumano A, and Golub R (2018). The Notch Signaling Pathway Is Balancing Type 1 Innate Lymphoid Cell Immune Functions. Front Immunol 9, 1252. 10.3389/fimmu.2018.01252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikovskaya O, Chaix J, Rothman NJ, Collins A, Chen YH, Scipioni AM, Vivier E, and Reiner SL (2016). Cutting Edge: Eomesodermin Is Sufficient To Direct Type 1 Innate Lymphocyte Development into the Conventional NK Lineage. J Immunol 196, 1449–1454. 10.4049/jimmunol.1502396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapp M, Lau CM, Adams NM, Weizman OE, O'Sullivan TE, Geary CD, and Sun JC (2017). Core-binding factor beta and Runx transcription factors promote adaptive natural killer cell responses. Sci Immunol 2. 10.1126/sciimmunol.aan3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratajczak MZ, Bujko K, Cymer M, Thapa A, Adamiak M, Ratajczak J, Abdel-Latif AK, and Kucia M (2020). The Nlrp3 inflammasome as a "rising star" in studies of normal and malignant hematopoiesis. Leukemia 34, 1512–1523. 10.1038/s41375-020-0827-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers JM, and Bulyk ML (2018). Diversification of transcription factor-DNA interactions and the evolution of gene regulatory networks. Wiley Interdiscip Rev Syst Biol Med, e1423. 10.1002/wsbm.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharton-Kersten T, Nakajima H, Yap G, Sher A, and Leonard WJ (1998). Infection of mice lacking the common cytokine receptor gamma-chain (gamma(c)) reveals an unexpected role for CD4+ T lymphocytes in early IFN-gamma-dependent resistance to Toxoplasma gondii. J Immunol 160, 2565–2569. [PubMed] [Google Scholar]
- Schmitt TM, and Zuniga-Pflucker JC (2002). Induction of T cell development from hematopoietic progenitor cells by delta-like-1 in vitro. Immunity 17, 749–756. 10.1016/s1074-7613(02)00474-0. [DOI] [PubMed] [Google Scholar]
- Sciume G, Mikami Y, Jankovic D, Nagashima H, Villarino AV, Morrison T, Yao C, Signorella S, Sun HW, Brooks SR, et al. (2020). Rapid Enhancer Remodeling and Transcription Factor Repurposing Enable High Magnitude Gene Induction upon Acute Activation of NK Cells. Immunity 53, 745–758 e744. 10.1016/j.immuni.2020.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo W, Muroi S, Akiyama K, and Taniuchi I (2017). Distinct requirement of Runx complexes for TCRbeta enhancer activation at distinct developmental stages. Sci Rep 7, 41351. 10.1038/srep41351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrock J (2019). Natural Killer Cells and Their Role in Immunity. EMJ Allergy & Immunology 4, 108–116. [Google Scholar]
- Shih HY, Sciume G, Mikami Y, Guo L, Sun HW, Brooks SR, Urban JF Jr., Davis FP, Kanno Y, and O'Shea JJ (2016). Developmental Acquisition of Regulomes Underlies Innate Lymphoid Cell Functionality. Cell 165, 1120–1133. 10.1016/j.cell.2016.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih HY, Sciume G, Poholek AC, Vahedi G, Hirahara K, Villarino AV, Bonelli M, Bosselut R, Kanno Y, Muljo SA, and O'Shea JJ (2014). Transcriptional and epigenetic networks of helper T and innate lymphoid cells. Immunol Rev 261, 23–49. 10.1111/imr.12208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberstein L, Goncalves Kevin A., Kharchenko Peter V., Turcotte R, Kfoury Y, Mercier F, Baryawno N, Severe N, Bachand J, Spencer Joel A., et al. (2016). Proximity-Based Differential Single-Cell Analysis of the Niche to Identify Stem/Progenitor Cell Regulators. Cell Stem Cell 19, 530–543. 10.1016/j.stem.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonetta F, Pradier A, and Roosnek E (2016). T-bet and Eomesodermin in NK Cell Development, Maturation, and Function. Front Immunol 7, 241. 10.3389/fimmu.2016.00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sojka DK, Plougastel-Douglas B, Yang L, Pak-Wittel MA, Artyomov MN, Ivanova Y, Zhong C, Chase JM, Rothman PB, Yu J, et al. (2014). Tissue-resident natural killer (NK) cells are cell lineages distinct from thymic and conventional splenic NK cells. eLife 3, e01659. 10.7554/eLife.01659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokic-Trtica V, Diefenbach A, and Klose CSN (2020). NK Cell Development in Times of Innate Lymphoid Cell Diversity. Front Immunol 11, 813. 10.3389/fimmu.2020.00813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo SJ, Sullivan BM, Stemmann C, Satoskar AR, Sleckman BP, and Glimcher LH (2002). Distinct effects of T-bet in TH1 lineage commitment and IFN-gamma production in CD4 and CD8 T cells. Science 295, 338–342. 10.1126/science.1065543. [DOI] [PubMed] [Google Scholar]
- Takeda K, Tsutsui H, Yoshimoto T, Adachi O, Yoshida N, Kishimoto T, Okamura H, Nakanishi K, and Akira S (1998). Defective NK cell activity and Th1 response in IL-18-deficient mice. Immunity 8, 383–390. [DOI] [PubMed] [Google Scholar]
- Tominaga K, Yoshimoto T, Torigoe K, Kurimoto M, Matsui K, Hada T, Okamura H, and Nakanishi K (2000). IL-12 synergizes with IL-18 or IL-1 beta for IFN-gamma production from human T cells. Int. Immunol 12, 151–160. [DOI] [PubMed] [Google Scholar]
- Townsend MJ, Weinmann AS, Matsuda JL, Salomon R, Farnham PJ, Biron CA, Gapin L, and Glimcher LH (2004). T-bet regulates the terminal maturation and homeostasis of NK and Valpha14i NKT cells. Immunity 20, 477–494. [DOI] [PubMed] [Google Scholar]
- Vahedi G, Takahashi H, Nakayamada S, Sun HW, Sartorelli V, Kanno Y, and O'Shea JJ (2012). STATs shape the active enhancer landscape of T cell populations. Cell 151, 981–993. 10.1016/j.cell.2012.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivier E, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, Koyasu S, Locksley RM, McKenzie ANJ, Mebius RE, et al. (2018). Innate Lymphoid Cells: 10 Years On. Cell 174, 1054–1066. 10.1016/j.cell.2018.07.017. [DOI] [PubMed] [Google Scholar]
- Waldmann TA, and Tagaya Y (1999). The multifaceted regulation of interleukin-15 expression and the role of this cytokine in NK cell differentiation and host response to intracellular pathogens. Annu Rev Immunol 17, 19–49. 10.1146/annurev.immunol.17.1.19. [DOI] [PubMed] [Google Scholar]
- Wang D, and Malarkannan S (2020). Transcriptional Regulation of Natural Killer Cell Development and Functions. Cancers (Basel) 12. 10.3390/cancers12061591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Godec J, Ben-Aissa K, Cui K, Zhao K, Pucsek AB, Lee YK, Weaver CT, Yagi R, and Lazarevic V (2014). The transcription factors T-bet and Runx are required for the ontogeny of pathogenic interferon-gamma-producing T helper 17 cells. Immunity 40, 355–366. 10.1016/j.immuni.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei G, Wei L, Zhu J, Zang C, Hu-Li J, Yao Z, Cui K, Kanno Y, Roh TY, Watford WT, et al. (2009). Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity 30, 155–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L, Vahedi G, Sun HW, Watford WT, Takatori H, Ramos HL, Takahashi H, Liang J, Gutierrez-Cruz G, Zang C, et al. (2010). Discrete roles of STAT4 and STAT6 transcription factors in tuning epigenetic modifications and transcription during T helper cell differentiation. Immunity 32, 840–851. 10.1016/j.immuni.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Tian Z, and Wei H (2017). Developmental and Functional Control of Natural Killer Cells by Cytokines. Front Immunol 8, 930. 10.3389/fimmu.2017.00930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Cherrier DE, Chea S, Vosshenrich C, Serafini N, Petit M, Liu P, Golub R, and Di Santo JP (2019). An Id2RFP-Reporter Mouse Redefines Innate Lymphoid Cell Precursor Potentials. Immunity 50, 1054–1068.e1053. 10.1016/j.immuni.2019.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi R, Junttila IS, Wei G, Urban JF Jr., Zhao K, Paul WE, and Zhu J (2010). The transcription factor GATA3 actively represses RUNX3 protein-regulated production of interferon-gamma. Immunity 32, 507–517. 10.1016/j.immuni.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R, Mele F, Worley L, Langlais D, Rosain J, Benhsaien I, Elarabi H, Croft CA, Doisne JM, Zhang P, et al. (2020). Human T-bet Governs Innate and Innate-like Adaptive IFN-gamma Immunity against Mycobacteria. Cell 183, 1826–1847 e1831. 10.1016/j.cell.2020.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R, Weisshaar M, Mele F, Benhsaien I, Dorgham K, Han J, Croft CA, Notarbartolo S, Rosain J, Bastard P, et al. (2021). High Th2 cytokine levels and upper airway inflammation in human inherited T-bet deficiency. The Journal of experimental medicine 218. 10.1084/jem.20202726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Ochando JC, Bromberg JS, and Ding Y (2007). Identification of a distant T-bet enhancer responsive to IL-12/Stat4 and IFNgamma/Stat1 signals. Blood 110, 2494–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarovinsky F (2014). Innate immunity to Toxoplasma gondii infection. Nat Rev Immunol 14, 109–121. 10.1038/nri3598. [DOI] [PubMed] [Google Scholar]
- Yu F, Sharma S, Jankovic D, Gurram RK, Su P, Hu G, Li R, Rieder S, Zhao K, Sun B, and Zhu J (2018). The transcription factor Bhlhe40 is a switch of inflammatory versus antiinflammatory Th1 cell fate determination. The Journal of experimental medicine 215, 1813–1821. 10.1084/jem.20170155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS, and Carroll JS (2011). Pioneer transcription factors: establishing competence for gene expression. Genes Dev 25, 2227–2241. 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, and Liu XS (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol 9, R137. 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong C, Zheng M, Cui K, Martins AJ, Hu G, Li D, Tessarollo L, Kozlov S, Keller JR, Tsang JS, et al. (2020). Differential Expression of the Transcription Factor GATA3 Specifies Lineage and Functions of Innate Lymphoid Cells. Immunity 52, 83–95 e84. 10.1016/j.immuni.2019.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Jankovic D, Oler AJ, Wei G, Sharma S, Hu G, Guo L, Yagi R, Yamane H, Punkosdy G, et al. (2012). The transcription factor T-bet is induced by multiple pathways and prevents an endogenous Th2 cell program during Th1 cell responses. Immunity 37, 660–673. 10.1016/j.immuni.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Cell type-specific DHSs in CD4+ T cell and ILC subsets, related to Figure 1
Table S3. Oligonucleotides, related to key resources table, Figure 2, Figure 3 and Figure 7
Data Availability Statement
All sequencing data are available in the GEO database listed in the key resources table. This paper analyzed publicly available data and the accession code is listed in the key resources table.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-B220-Biotin (Clone RA3-6B2) | BD | Cat#553086; RRID: AB_ 394615 |
| Anti-CD3e-FITC (Clone 145-2C11) | BD | Cat#553062; RRID: AB_394595 |
| Anti-CD3e-Biotin (Clone 145-2C11) | eBioscience | Cat#13-0031-85; RRID: AB_466319 |
| Anti-CD4-BUV395 (Clone GK1.5) | BD | Cat#563790; RRID: AB_2738426 |
| Anti-CD4-Biotin (Clone GK1.5) | eBioscience | Cat#13-0041-85; RRID: AB_466326 |
| Anti-CD4-BUV737 (Clone GK1.5) | BD | Cat#564298 |
| Anti-CD5-Biotin (Clone 53-7.3) | BD | Cat#553019; RRID: AB_394557 |
| Anti-CD8-Biotin (Clone 53-6.7) | eBioscience | Cat#13-0081-85; RRID: AB_466347 |
| Anti-CD11b-FITC (Clone M1/70) | BD | Cat#553310; RRID: AB_396679 |
| Anti-CD11b-PerCP-Cy™5.5 (Clone M1/70) | BD | Cat#550993; RRID: AB_394002 |
| Anti-CD11b-BUV737 (Clone M1/70) | BD | Cat#612800; RRID: AB_2738811 |
| Anti-CD11b-Biotin (Clone M1/70) | BD | Cat#553309; RRID: AB_394773 |
| Anti-CD14-Biotin (Clone Sa2-8) | eBioscience | Cat#13-0141-85; RRID: AB_466371 |
| Anti-CD19-PE (Clone 1D3) | BD | Cat#553786; RRID: AB_395050 |
| Anti-CD19-Biotin (Clone 1D3) | eBioscience | Cat#13-0193-85; RRID: AB_657658 |
| Anti-CD25-FITC (Clone 7D4) | BD | Cat#553072; RRID: AB_394603 |
| Anti-CD27-BV605 (Clone LG.3A10) | BD | Cat#563365; RRID: AB_2738160 |
| Anti-CD44-eFluor 450 (Clone IM7) | eBioscience | Cat#48-0441-82; RRID: AB_1272246 |
| Anti-CD44-APC-eFluor 780 (Clone IM7) | eBioscience | Cat#47-0441-82; RRID: AB_1272244 |
| Anti-CD45.1-PE-Cy™7 (Clone A20) | BD | Cat#560578; RRID: AB_1727488 |
| Anti-CD45.2-BUV395 (Clone 104) | BD | Cat#564616; RRID: AB_2738867 |
| Anti-CD45.2-BUV737 (Clone 104) | BD | Cat#612778; RRID: AB_2870107 |
| Anti-CD49a-BV421 (Clone Ha31/8) | BD | Cat#740046; RRID: AB_2739815 |
| Anti-CD49a-BV711 (Clone Ha31/8) | BD | Cat#564863; RRID: AB_2738987 |
| Anti-CD49b-APC (Clone DX5) | BD | Cat#560628; RRID: AB_1727502 |
| Anti-CD49b-eFluor 780 (Clone DX5) | eBioscience | Cat#47-5971-82; RRID: AB_11218895 |
| Anti-CD62L-PE (Clone MEL-14) | eBioscience | Cat#12-0621-82; RRID: AB_465721 |
| Anti-CD62L-APC (Clone MEL-14) | eBioscience | Cat#17-0621-82; RRID: AB_469410 |
| Anti-CD117-APC-H7 (Clone 2B8) | BD | Cat#560185; RRID: AB_1645231 |
| Anti-CD117-PE (Clone 2B8) | BD | Cat#553355; RRID: AB_394806 |
| Anti-CD122-BV786 (Clone TM-β1) | BD | Cat#740869; RRID: AB_2740521 |
| Anti-CD122-FITC (Clone 5H4) | BD | Cat#554452; RRID: AB_395401 |
| Anti-CD127-PE-Cyanine7 (Clone A7R34) | eBioscience | Cat#25-1271-82; RRID: AB_469649 |
| Anti-CD244.2-BUV395 (Clone 2B4) | BD | Cat#740226; RRID: AB_2739974 |
| Anti-CD244.2-PE-Cyanine7 (eBio244F4) | eBioscience | Cat#25-2441-82; RRID: AB_2573432 |
| Anti-CCR6-BV421 (Clone 140706) | BD | Cat#564736; RRID: AB_2738926 |
| Anti-F4/80-Biotin (Clone BM8) | eBioscience | Cat#13-4801-85; RRID: AB_466658 |
| Anti-FceR1 alpha-Biotin (Clone MAR-1) | eBioscience | Cat#13-5898-82; RRID: AB_466783 |
| Anti-IFN-γ-Alexa Fluor® 700 (Clone XMG1.2) | BD | Cat#557998; RRID: AB_396979 |
| Anti-IFN-γ-PE-Cy™7 (Clone XMG1.2) | BD | Cat#557649; RRID: AB_396766 |
| Anti-IFN-γ-FITC (Clone XMG1.2) | BD | Cat#554411; RRID: AB_395375 |
| Anti-IL-4-Brilliant Violet 421™ (Clone 11B11) | BioLegend | Cat#504120; RRID: AB_2562102 |
| Anti-IL-4-Brilliant Violet 605™ (Clone 11B11) | BioLegend | Cat#504126; RRID: AB_2686971 |
| Anti-IL-18Rα-eFluor 450 (Clone P3TUNYA) | eBioscience | Cat#48-5183-82; RRID: AB_2574069 |
| Anti-IL-21R-PE (Clone 4A9) | BioLegend | Cat#131906; RRID: AB_1279430 |
| Anti-KLRG1-APC-eFluor 780 (Clone 2F1) | eBioscience | Cat#47-5893-82; RRID: AB_2573988 |
| Anti-Ly-6G/Ly-6C-Biotin (Clone RB6-8C5) | eBioscience | Cat#13-5931-85; RRID: AB_466801 |
| Anti-NK1.1-PE-Cy™7 (Clone PK136) | BD | Cat#552878; RRID: AB_394507 |
| Anti-NK1.1-BV650 (Clone PK136) | BD | Cat#564143; RRID: AB_2738617 |
| Anti-NKG2D-PE-CF594 (Clone CX5) | BD | Cat#562614; RRID: AB_2737677 |
| Anti-NKp46-PE (Clone 29A1.4) | BD | Cat#560757; RRID: AB_1727466 |
| Anti-TER-119-Biotin (Clone TER-119) | eBioscience | Cat#13-5921-85; RRID: AB_466798 |
| Anti-CBFβ Rabbit mAb (Clone D4N2N) | CST | Cat#62184 |
| Anti-Stat4 Rabbit mAb (Clone C46B10) | CST | Cat#2653 |
| Anti-T-bet-Alexa Fluor® 647 (Clone O4-46) | BD | Cat#561267; RRID: AB_10564093 |
| Anti-EOMES-eFluor 450 (Clone Dan11mag) | eBioscience | Cat#48-4875-82; RRID: AB_2574062 |
| Anti-Ki-67-BV786 (Clone B56) | BD | Cat#563756; RRID: AB_2732007 |
| Anti-RUNX3-PE (Clone R3-5G4) | BD | Cat#564814; RRID: AB_2738969 |
| Anti-NGFR-PE (Clone C40-1457) | BD | Cat#557196; RRID: AB_396599 |
| Anti-CD3e (Clone 2C11) | Harlan | Lot#A2022931 |
| Anti-CD28 (Clone 37.51) | Harlan | Lot#A2022929 |
| Anti-IL-4 (Clone 11B.11) | NCI-Frederick | Lot#L0412006 |
| Anti-IFN-γ (Clone XMG1.2) | Harlan | Lot#A2022930 |
| Anti-CD16/CD32 (Clone 2.4G2) | Harlan | Lot#A2083015 |
| Anti-Biotin MicroBeads | Miltenyi Biotec | Cat#130-090-485 |
| Anti-CD3/CD28 Dynabeads | Gibco | Cat#11453D |
| Alexa Fluor® 647 Mouse IgG1 κ | BD | Cat#557732; RRID: AB_396840 |
| Rat IgG2a Isotype-eFluor 450 (eBR2a) | eBioscience | Cat#48-4321-82; RRID: AB_1271999 |
| Anti-mouse IgG coated magnetic beads | Diagenode | Cat#C03010022 |
| InVivoMAb anti-mouse IL-12 p40 | BioXCell | Cat#BE0051; RRID: AB_1107698 |
| InVivoMAb rat IgG2a isotype control, anti-trinitrophenol | BioXCell | Cat#BE0089; RRID: AB_1107769 |
| Bacterial and virus strains | ||
| Toxoplasma gondii ME49 | Kawabe et al., 2017 | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| IL-1β | R&D systems | Cat#401-ML |
| IL-2 | R&D systems | Cat#402-ML |
| IL-6 | PeproTech | Cat#216-16 |
| IL-7 | R&D systems | Cat#407-ML |
| IL-12 | PeproTech | Cat#210-12 |
| IL-15 | R&D systems | Cat#447-ML |
| IL-18 | R&D systems | Cat#9139-IL |
| IL-21 | R&D systems | Cat#594-ML |
| CXCL12 | R&D systems | Cat#460-SD |
| Flt-3 Ligand | R&D systems | Cat#427-FL |
| IFN-γ | PeproTech | Cat#315-05 |
| SCF | R&D systems | Cat#455-MC |
| TGF-beta 1 | R&D systems | Cat#7666-MB |
| Fixable Viability Dye eFluor™ 506 | eBioscience | Cat#65-0866-14 |
| Fixable Viability Dye eFluor™ 780 | eBioscience | Cat#65-0865-14 |
| Streptavidin-BV605 | BD | Cat#563260; RRID: AB_2869476 |
| Streptavidin-APC-R700 | BD | Cat#565144; RRID: AB_2869657 |
| Protein A-coated magnetic beads | Diagenode | Cat#C03010020 |
| Percoll Plus | GE Healthcare | Cat#17544501 |
| Polybrene | Sigma | Cat#TR-1003-G |
| ACK Lysing Buffer | Gibco | Cat#A1049201 |
| AS15-Tetramer-BV421 | NIH TCF | Order#42412 |
| AS15-Tetramer-APC | NIH TCF | Order#42413 |
| Monensin Solution | eBioscience | Cat#00-4505-51 |
| L-Glutamine | Gibco | Cat#25030 |
| Sodium pyruvate | Gibco | Cat#11360 |
| β-mercaptoethanol | Gibco | Cat#21985 |
| NEAA | Gibco | Cat#11140 |
| Penicillin and streptomycin | Gibco | Cat#15140 |
| RPMI medium 1640 | Gibco | Cat#21870-076 |
| MEM | Gibco | Cat#11095080 |
| FspI | NEB | Cat#R0135L |
| PMA (Phorbol 12-myristate 13-acetate) | Sigma | Cat#P8139 |
| Ionomycin | Sigma | Cat#407952 |
| InSolution NF-κB Activation Inhibitor | Sigma | Cat#481407-1MG |
| Polyinosinic–polycytidylic acid sodium salt | Sigma | Cat#P0913-50MG |
| EasySep™ Buffer | Stem cell | Cat#20144 |
| Cycloheximide | Sigma | Cat#C1988-1G |
| DMSO | Sigma | Cat#D2650 |
| Critical commercial assays | ||
| Foxp3 Staining Buffer Set | eBioscience | Cat#00-5523-00 |
| QuantiTect Rev. Transcription Kit | Qiagen | Cat#205313 |
| MinElute Reaction Cleanup Kit | Qiagen | Cat#28206 |
| QIAquick Gel Extraction Kit | Qiagen | Cat#28704 |
| RNeasy Plus Mini Kit | Qiagen | Cat#74136 |
| PrimeScript™ RT Master Mix | TAKARA | Cat#RR036A |
| FastStart Universal SYBR Green Master (Rox) | Roche | Cat#04913850001 |
| P4 Primary Cell 4D-Nucleofector | LONZA | Cat#V4XP-4024 |
| CD4+ T Cell Isolation Kit | Miltenyi Biotec | Cat#130-104-454 |
| EasySep™ Mouse Streptavidin RapidSpheres™ Isolation Kit | Stem cell | Cat#19860 |
| RNase-Free DNase Set | Qiagen | Cat#79256 |
| DNeasy Blood & Tissue Kit | Qiagen | Cat#69506 |
| IFN gamma Mouse Uncoated ELISA Kit | Invitrogen | Cat#88-7314-86 |
| Deposited data | ||
| ChIP-Seq and DNase-Seq data | This manuscript | GEO: GSE172358 |
| ChIP-Seq (T-bet in Th1-2) | Zhu et al., 2012 | GEO: GSE38808 |
| Experimental models: Cell lines | ||
| OP9 | Schmitt and Zúñiga-Pflücker, 2002 | N/A |
| OP9-DLL1 | Schmitt and Zúñiga-Pflücker, 2002 | N/A |
| Experimental models: Organisms/strains | ||
| Mouse: Tbx215M/5M | This manuscript | N/A |
| Mouse: Tbx21ΔC-3 | This manuscript | N/A |
| Mouse: Tbx21ΔC-8.5 | This manuscript | N/A |
| Mouse: Tbx21ΔC-3ΔC-8.5 | This manuscript | N/A |
| Mouse: Tcra−/− | NIAID-Taconic | Taconic line 98 |
| Mouse: CD45.1 congenic | NIAID-Taconic | Taconic line 7 |
| Mouse: Rag1−/− | NIAID-Taconic | Taconic line 146 |
| Mouse: Rag2−/−γc−/− | NIAID-Taconic | Taconic line 111 |
| Mouse: T-bet-ZsGreen | NIAID-Taconic | Taconic line 8419 |
| Mouse: T-bet-ZsGreen-Tbx21−/− | NIAID-Taconic | Taconic line 8451 |
| Mouse: T-bet-ZsGreen-Stat4−/−Ifngr1−/− | NIAID-Taconic | Taconic line 8450 |
| Mouse: Il18r1−/− | The Jackson Laboratory | Stock#004131 |
| Mouse: C57BL/6J | The Jackson Laboratory | Stock#000664 |
| Mouse: RUNX3-YFP | Egawa and Littman, 2008 | Now also available at Jackson Laboratory, Stock#008774 |
| Oligonucleotides | ||
| See Excel file Table S3 | ||
| Recombinant DNA | ||
| NGFR-RUNX3 retrovirus | Yagi et al., 2010 | N/A |
| GFP-T-bet retrovirus | Yagi et al., 2010 | N/A |
| Software and algorithms | ||
| FlowJo V10 | FlowJo LLC | https://www.flowjo.com |
| Prism 7 | Graphpad | https://www.graphpad.com |
| MEME Suite | N/A | https://meme-suite.org |
| WashU Epigenome Browser | Washington University in St. Louis | http://epigenomegateway.wustl.edu |