Significance
Riboflavin (vitamin B2) is converted into flavin mononucleotide (FMN) and flavin adenine dinucleotide (FAD), which are essential cofactors for many redox reactions across all domains of life. Listeria monocytogenes is a facultative intracellular pathogen that cannot synthesize riboflavin and must therefore obtain flavins from the host. In this study, we show that a previously identified riboflavin transporter (RibU) is essential for virulence and intracellular growth, but rather than transporting riboflavin, RibU transports FMN and FAD directly from the host cell cytosol. Mutants unable to convert riboflavin to FMN and FAD retained their capacity to grow intracellularly and were virulent, but they were unable to grow extracellularly and were thus converted from facultative to obligate intracellular pathogens.
Keywords: riboflavin, bacteria, macrophage, inflammasome, intracellular pathogens
Abstract
Flavin mononucleotide (FMN) and flavin adenine dinucleotide (FAD) are essential riboflavin-derived cofactors involved in a myriad of redox reactions across all forms of life. Nevertheless, the basis of flavin acquisition strategies by riboflavin auxotrophic pathogens remains poorly defined. In this study, we examined how the facultative intracellular pathogen Listeria monocytogenes, a riboflavin auxotroph, acquires flavins during infection. A L. monocytogenes mutant lacking the putative riboflavin transporter (RibU) was completely avirulent in mice but had no detectable growth defect in nutrient-rich media. However, unlike wild type, the RibU mutant was unable to grow in defined media supplemented with FMN or FAD or to replicate in macrophages starved for riboflavin. Consistent with RibU functioning to scavenge FMN and FAD inside host cells, a mutant unable to convert riboflavin to FMN or FAD retained virulence and grew in cultured macrophages and in spleens and livers of infected mice. However, this FMN- and FAD-requiring strain was unable to grow in the gallbladder or intestines, where L. monocytogenes normally grows extracellularly, suggesting that these sites do not contain sufficient flavin cofactors to promote replication. Thus, by deleting genes required to synthesize FMN and FAD, we converted L. monocytogenes from a facultative to an obligate intracellular pathogen. Collectively, these data indicate that L. monocytogenes requires riboflavin to grow extracellularly in vivo but scavenges FMN and FAD to grow in host cells.
Riboflavin (vitamin B2) is a water-soluble vitamin essential to all organisms and the precursor of the biologically active flavin cofactors flavin mononucleotide (FMN) and flavin adenine dinucleotide (FAD) (Fig. 1A), which are necessary for a diverse array of oxidation-reduction reactions (1–3). While plants, fungi, and most bacteria and archaea synthesize riboflavin, some bacteria and all mammals lack the genes to make this vitamin de novo (4–6). Many organisms encode transporters that allow them to obtain riboflavin from the environment (5–8). Several bacterial and all eukaryotic intracellular pathogens require an exogenous source of riboflavin (9–11), and interestingly, some also lack the enzymes that catalyze the conversion of riboflavin to FMN and FAD (11, 12). How these riboflavin auxotrophic intracellular pathogens fulfill their flavin requirement in host cells is poorly understood. Most of these intracellular pathogens encode annotated riboflavin transporters. Interestingly, however, riboflavin is scarce in host cells while FMN and FAD are abundant (13–15). To better understand flavin acquisition and requirements for pathogenesis, here we focused on the riboflavin auxotrophic bacterium Listeria monocytogenes.
Fig. 1.
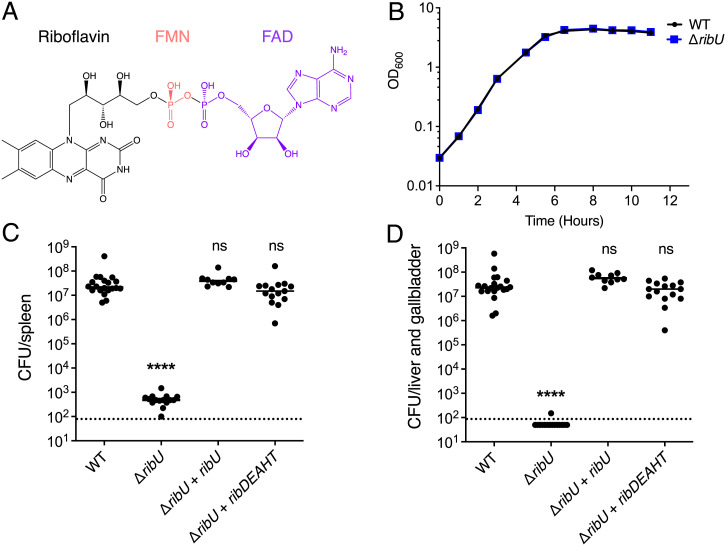
RibU is essential for virulence but dispensable for growth in nutrient-rich media. (A) Structures of riboflavin (black), FMN (red), and FAD (purple). Riboflavin is phosphorylated by riboflavin kinases to produce FMN. FAD synthetases adenylylate FMN to generate FAD. (B) Broth growth curves of L. monocytogenes strains grown in nutrient-rich media. OD600 was used to determine cell density. The means and SDs of three independent experiments are shown. Note: both the WT and ΔribU mutant growth curves are superimposable. (C and D) Bacterial burdens in CD-1 mice infected intravenously with 1 × 105 CFUs of indicated L. monocytogenes strains. At 48 h postinfection, the spleens (C) and livers (D) were harvested, homogenized, and plated to determine the CFUs per organ. The data show the combination of at least two independent experiments: WT and ΔribU (n = 20 mice), ΔribU + ribDEAHT (n = 15 mice), and ΔribU + ribU (n = 10 mice). The black lines represent the median CFUs for each strain. The dashed line represents the limit of detection. Statistical significance of logarithmically transformed CFU values was determined using one-way ANOVA and Dunnett’s posttest using WT as the control. ****P < 0.0001; ns, not significant, P > 0.05.
L. monocytogenes is a gram-positive bacterium that lives both as an environmental saprophyte and as a facultative intracellular pathogen of mammals including humans (16). Once L. monocytogenes enters a cell, it escapes from a phagosome and gains access to the cytosol, where it acquires nutrients and rapidly divides (17). Although L. monocytogenes has few growth requirements (18), it lacks the riboflavin biosynthetic genes. To obtain this vitamin, L. monocytogenes encodes an energy-coupling factor (ECF) transporter, RibU, that is annotated as a riboflavin transporter (19, 20). RibU is the substrate binding subunit of an ECF transporter, which is also composed of two ATPases and a transmembrane subunit that together form a complex for riboflavin import (21) (SI Appendix, Fig. S1). RibU from L. monocytogenes binds riboflavin (21) and rescues the growth of a riboflavin auxotrophic Bacillus subtilis strain (20). Additionally, L. monocytogenes encodes the enzymes RibC and RibF, which are involved in the biosynthesis of FMN and FAD (20, 22). RibC is a bifunctional enzyme that catalyzes the phosphorylation of riboflavin to FMN and the adenylylation of FMN to form FAD. RibF also converts FMN to FAD by adenylylation. RibU, RibC, and RibF are the only proteins known to control flavin metabolism in L.monocytogenes (SI Appendix, Fig. S1).
Recently, there has been renewed interest in flavin metabolism in the context of pathogenesis stemming from the discovery that intermediates of riboflavin biosynthesis activate innate-like mucosal-associated invariant T (MAIT) cells (23). In addition, we recently discovered that flavins are implicated in distinct extracytosolic redox activities in thousands of bacterial species (24). These bacterial redox systems allow bacteria to transfer electrons from the cytosol to various electron acceptors. We previously described that L. monocytogenes possesses a conserved flavin-based extracellular electron transfer (EET) system that allows bacteria to respire anaerobically using various electron acceptors, such as iron and fumarate (25, 26). Among the L. monocytogenes transposon mutants that lacked EET were mutations in genes encoding the riboflavin transporter, an extracellular flavin transferase, and an FMNylated surface protein (25). Surprisingly, mutants in ribU lacked EET activity yet were able to grow in nutrient-rich media. In this study, we aimed to characterize how L. monocytogenes acquires flavins and their role during infection. We discovered that RibU was essential for intracellular growth and that its function is to transport FMN and FAD from the host cell cytosol.
Results
RibU Is Dispensable for Growth in Nutrient-Rich Media but Is Required for Virulence in Mice.
We previously isolated a mutant with a transposon insertion in the ribU gene, which encodes the sole annotated riboflavin transporter in L. monocytogenes (25). Since flavins are necessary for a myriad of essential processes, we were surprised that the only annotated riboflavin transporter was not essential. To confirm that RibU was not essential for growth, and possibly assess flavin acquisition and requirements during pathogenesis, we generated a L. monocytogenes strain with an in-frame deletion in ribU (ΔribU). Like the transposon mutant, the ΔribU strain had no detectable growth defect in nutrient-rich media compared to wild-type (WT) L. monocytogenes (Fig. 1B). In contrast, the ΔribU strain had a 5-log virulence defect in the spleens of mice compared to WT L. monocytogenes (Fig. 1C), and no colony-forming units (CFUs) could be recovered from the livers of infected mice (Fig. 1D). Complementation of the ΔribU mutant with a ribU gene with its endogenous promoter (ΔribU + ribU) fully restored virulence in vivo (Fig. 1 C and D).
To determine if the virulence defect of the ΔribU strain was caused by riboflavin starvation in vivo, we engineered the ΔribU mutant to synthesize riboflavin by inserting the riboflavin operon ribDEAHT from the closely related gram-positive bacterium B. subtilis (27) into the L. monocytogenes chromosome. This strain grew in colorless chemically defined synthetic media without riboflavin supplementation and turned the media yellow, the natural color of flavins (SI Appendix, Fig. S2). Expression of the ribDEAHT operon rescued the ΔribU strain’s virulence to WT L. monocytogenes levels in the spleens and livers of mice (Fig. 1 C and D). Based on these observations, we concluded that RibU is essential for L. monocytogenes pathogenesis and that its function relates to flavin acquisition, since de novo riboflavin production in the RibU-minus strain completely bypassed RibU’s essentiality in vivo.
L. monocytogenes Uses RibU to Grow in Macrophages.
To study why RibU was essential for growth of L. monocytogenes in mice, we performed infections in vitro using bone marrow-derived macrophages (BMMs). At 2 h postinfection, the ΔribU mutant had a small but significant growth advantage over WT L. monocytogenes (Fig. 2A), which was associated with an increase in phagosomal escape (SI Appendix, Fig. S3A). However, during exponential growth (2 to 5 h postinfection), the ΔribU mutant had an apparent defect in replication rate and showed a loss in CFUs during the late stages of infection (5 to 8 h postinfection) (Fig. 2A). Complementation of the ΔribU strain with the ribU gene or the ribDEAHT operon completely restored the growth defects in BMMs (SI Appendix, Fig. S3B).
Fig. 2.
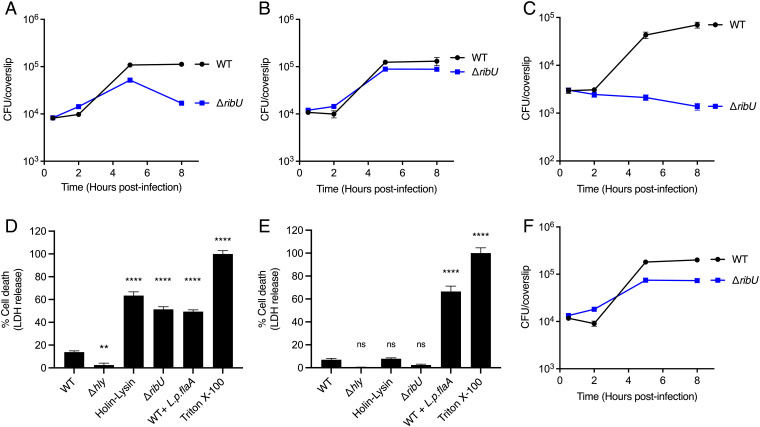
L. monocytogenes requires RibU to grow in riboflavin-starved macrophages. (A–C and F) Intracellular growth curves of L. monocytogenes strains in murine BMMs. BMMs were infected at a multiplicity of infection (MOI) of 0.1, and CFUs were enumerated at the indicated times. (A) Intracellular growth curves of indicated L. monocytogenes strains in WT BMMs. The data show the means and SEMs of four independent experiments. (B) Intracellular growth curves of indicated L. monocytogenes strains in WT BMMs incubated with cell culture media containing excess (10 µM) riboflavin during infection. The means and SEMs of three independent experiments are shown. (C) Intracellular growth curves of indicated flavin-starved L. monocytogenes strains in riboflavin-deficient WT BMMs. The data represent the means and SEMs of three independent experiments. (D and E) Cell death of WT (D) or AIM2 KO (E) BMMs infected with specified L. monocytogenes strains. LDH released to the cell culture media was used as an indicator of cell death. LDH release values were normalized to 1% Triton-X–treated cells which represent 100% lysis. BMMs were infected at an MOI of 4. The data show the means and SEMs of three technical replicates from at least two (D) and four (E) independent experiments. Statistical significance was determined using one-way ANOVA and Dunnett’s posttest using WT as the control. ****P < 0.0001; **P < 0.01; ns, not significant, P > 0.05. (F) Intracellular growth curves of indicated L. monocytogenes strains in AIM2 KO BMMs. The means and SEMs of five independent experiments are shown. WT + L.p.flaA, WT L. monocytogenes expressing Legionella pneumophila flagellin A under the control of the actA promoter (29). Holin-Lysin, WT L. monocytogenes expressing the bacteriophage proteins Holin and Lysin under the control of the actA promoter, leading to lysis of the bacteria in the cytosol of the host cell (28).
To test if the ΔribU strain has an inherent intracellular virulence defect not related to riboflavin, BMMs were incubated with excess riboflavin (10 µM) prior to infection to increase the concentration of intracellular riboflavin. In this condition of riboflavin excess, the ΔribU mutant replicated to WT levels (Fig. 2B). To assess if the growth of the ΔribU strain observed during the exponential growth phase in BMMs (Fig. 2A) is due to residual flavins from the medium, we infected riboflavin-deficient BMMs with riboflavin-starved bacteria. In this experiment, the ΔribU mutant and WT L. monocytogenes strains were incubated in chemically defined synthetic media lacking flavins for 16 to 18 h prior to infection. The BMM cell culture media was replaced with media lacking riboflavin, and the macrophages were incubated for 3 h prior to infection with the riboflavin-starved bacteria. We observed that riboflavin-starved WT L. monocytogenes were able to grow in riboflavin-deficient BMMs. However, the riboflavin-starved ΔribU mutant was unable to replicate in riboflavin-depleted BMMs (Fig. 2C). The riboflavin-starved ΔribU mutant was able to grow in riboflavin-deficient BMMs supplemented with 1 µM riboflavin just prior to infection (SI Appendix, Fig. S3C).
To further characterize the growth dynamics of the ΔribU mutant, we focused on the late stage of infection and the loss of CFUs observed between 5 and 8 h postinfection (Fig. 2A). We hypothesized that the loss of CFUs was due to the inability of the ΔribU mutant to obtain riboflavin from the cytosol of host cells and that riboflavin starvation led to bacterial and/or host cell death. To test if infection with the ΔribU strain led to host cell death, we performed a lactate dehydrogenase (LDH) release assay. The results demonstrated that ΔribU caused a significant increase in host cell death (Fig. 2D). In dying host cells, gentamicin from the media can enter the cytoplasm and kill intracellular bacteria. Based on previous studies (28), we hypothesized that the ΔribU strain was lysing in the macrophage cytosol and releasing DNA that activated the DNA-dependent AIM2 inflammasome, resulting in pyroptotic cell death. To test if ΔribU was triggering AIM2-dependent pyroptosis, we performed LDH release assays using AIM2 knockout (KO) BMMs and observed that the ΔribU mutant did not lead to LDH release (Fig. 2E). As a control, L. monocytogenes secreting flagellin (WT + L.p.flaA), which activates the NLRC4 inflammasome (29), still mediated cell death in infected AIM2 KO BMMs (Fig. 2E). Since the ΔribU mutant did not lead to cell death in AIM2 KO BMMs, we hypothesized that the loss of CFUs in the ΔribU mutant during the later stages of infection in WT BMMs should be rescued in AIM2 KO BMMs. Indeed, there was no loss of CFUs of the ΔribU mutant at 8 h postinfection in AIM2 KO BMMs (Fig. 2F). These data suggest that the RibU-minus strain lysed to some extent in vivo and activated AIM2-dependent pyroptosis, which negatively impacted the virulence of the strain.
L. monocytogenes Uses RibU to Scavenge FMN and FAD from the Cytosol of Host Cells.
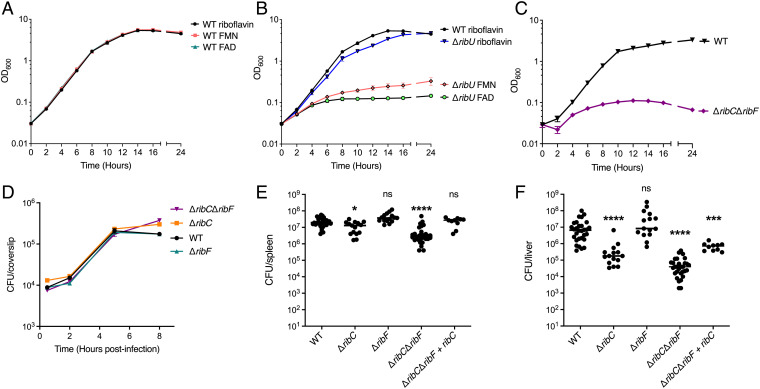
Our observation that riboflavin-starved WT L. monocytogenes grew in riboflavin-deprived macrophages (Fig. 2C) led us to question how L. monocytogenes fulfills its flavin requirements under these conditions. Interestingly, we did not observe any difference in the ability of WT L. monocytogenes to replicate intracellularly in the absence of riboflavin. The doubling time of riboflavin-starved WT L. monocytogenes in riboflavin-deficient BMMs was very similar to the doubling time of WT L. monocytogenes growing in BMMs with riboflavin: 48.6 and 51.5 min, respectively (SI Appendix, Fig. S3D). Based on these results, and the fact that mammalian cells rapidly convert riboflavin to FMN and FAD upon import (13, 30), we hypothesized that WT L. monocytogenes imports FMN and/or FAD intracellularly to grow, using RibU for their transport. To test if L. monocytogenes can import FMN and FAD to support growth, we used chemically defined synthetic media supplemented with riboflavin, FMN, or FAD as the sole flavin source and found that WT L. monocytogenes grew in media containing each of the three flavins (Fig. 3A). By contrast, the ΔribU strain did not replicate in chemically defined media with FMN or FAD (Fig. 3B) and had only a slight defect in growth in media containing riboflavin. These results suggested that RibU is responsible for growth on FMN and FAD and that riboflavin can enter cells using RibU and/or another, yet to be identified, riboflavin transporter.
Fig. 3.
L. monocytogenes scavenges FMN and FAD from the host cytosol using RibU. (A) Broth growth curves of WT L. monocytogenes grown in chemically defined synthetic media with riboflavin, FMN, or FAD as the only flavin source. OD600 was used to determine cell density. The data show the means and SDs of four independent experiments. (B) Broth growth curves of the ΔribU mutant L. monocytogenes strain grown in chemically defined synthetic media with riboflavin, FMN, or FAD as the sole flavin source. WT L. monocytogenes grown in chemically defined media with riboflavin is used as a reference. OD600 was used to determine cell density. The data show the means and SDs of four independent experiments. (C) Broth growth curves of indicated L. monocytogenes strains grown in chemically defined media with riboflavin. OD600 was used to determine cell density. The data show the means and SDs of two independent experiments. (D) Intracellular growth curves of L. monocytogenes strains in murine BMMs. BMMs were infected at an MOI of 0.1, and CFUs were enumerated at the indicated times. The means and SEMs of two independent experiments are shown. (E and F) Bacterial burdens in CD-1 mice infected intravenously with 1 × 105 CFUs of indicated L. monocytogenes strains. At 48 h postinfection, the spleens (E) and livers (F) were harvested, homogenized, and plated to determine the CFUs per organ. The data show the combination of at least two independent experiments: WT and ΔribCΔribF (n = 30 mice), ΔribC and ΔribF (n = 15 mice), and ΔribCΔribF + ribC (n = 10 mice). The black lines represent the median CFUs for each strain. Statistical significance of logarithmically transformed CFU values was determined using one-way ANOVA and Dunnett’s posttest using WT as the control. ****P < 0.0001; ***P < 0.001; *P < 0.01; ns, not significant, P > 0.05.
To test the hypothesis that L. monocytogenes utilizes RibU to scavenge FMN and FAD from the cytosol of host cells, we sought to generate a L. monocytogenes FMN and FAD auxotroph by constructing strains lacking ribC, ribF, or both (ΔribCΔribF), enzymes responsible for converting riboflavin to FMN and FAD. Since FMN and FAD are essential cofactors, construction of this strain was performed in nutrient-rich media containing excess FMN and FAD to circumvent synthetic lethality. As predicted, the ΔribCΔribF mutant was unable to replicate in chemically defined media with riboflavin as the sole flavin source (Fig. 3C).
We reasoned that if L. monocytogenes imports FMN and FAD from the host cytosol, the ΔribCΔribF mutant should not be impaired for intracellular growth. Indeed, these strains replicated intracellularly in BMMs to WT L. monocytogenes levels (Fig. 3D). To test if the ΔribC, ΔribF, and the ΔribCΔribF mutant L. monocytogenes strains grew in vivo, we performed mouse virulence assays. The ΔribC, ΔribF, and ΔribCΔribF mutants maintained their virulence and grew to high levels in both the spleens and livers of mice, although the ΔribC and ΔribCΔribF strains had statistically significant 2-log defects in the liver (Fig. 3 E and F). Complementation of the ΔribCΔribF strain with the ribC gene with its endogenous promoter was able to restore most of the growth in the spleens and livers of infected mice (Fig. 3 E and F). Thus, these results support a model in which L. monocytogenes uses RibU to import FMN and FAD from the cytosol of host cells.
The ΔribCΔribF Mutant Cannot Grow in Blood, Gallbladders, or the Gastrointestinal Tract.
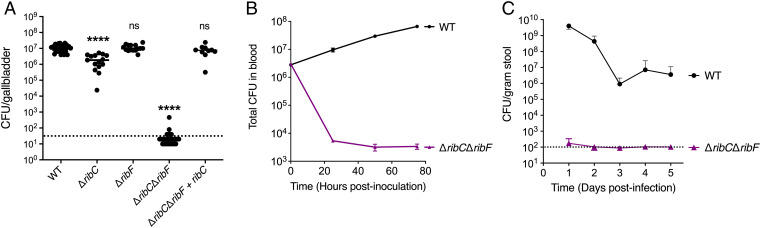
The observation that the ΔribCΔribF strain replicated similarly to WT L. monocytogenes in the spleens but not in the livers of mice (Fig. 3 E and F) prompted us to examine if this strain was able to colonize and grow in other sites of infection in mice and use growth as an indicator of flavin availability. WT L. monocytogenes can grow extracellularly in the gallbladder, blood, and the gastrointestinal (GI) tract of mice (31–34). During infection, L. monocytogenes colonizes the lumen of the gallbladder, which is connected to the liver through biliary ducts, and rapidly replicates extracellularly in the bile, establishing this organ as a bacterial reservoir (31, 32). The ΔribCΔribF mutant was unable to colonize the gallbladder, while the ΔribC and ΔribF strains grew to WT L. monocytogenes levels (Fig. 4A). Complementation of the ΔribCΔribF mutant with a ribC gene with its endogenous promoter completely rescued the growth of the ΔribCΔribF strain in the gallbladder.
Fig. 4.
An FMN and FAD auxotrophic L. monocytogenes strain is restricted to intracellular growth in vivo. (A) Bacterial burdens in the gallbladder of CD-1 mice infected intravenously with 1 × 105 CFUs of indicated L. monocytogenes strains. At 48 h postinfection, the gallbladders were harvested, homogenized, and plated to determine the CFUs per organ. The data show the combination of at least two independent experiments: WT and ΔribCΔribF (n = 30 mice), ΔribC and ΔribF (n = 15 mice), and ΔribCΔribF + ribC (n = 10 mice). The black lines represent the median CFUs for each strain. The dashed line represents the limit of detection. Statistical significance of logarithmically transformed CFU values was determined using one-way ANOVA and Dunnett’s posttest using WT as the control. ****P < 0.0001; ns, not significant, P > 0.05. (B) In vitro growth of L. monocytogenes strains in defibrinated sheep’s blood. Bacterial growth was determined by plating at the indicated times to determine the total CFUs in blood. The means and SEMs of three independent experiments are shown. (C) Bacterial burdens in the GI tract of CD-1 mice infected orally with 1 × 108 CFUs of indicated L. monocytogenes strains. Mice were pretreated orally with streptomycin prior to the infection. Stool samples were collected from day 1 to day 5 and plated to determine the CFUs per gram of stool. The data show the means and SDs of the mean from a combination of three independent experiments, n = 15 mice per L. monocytogenes strain. The dashed line represents the limit of detection.
To assess if the ΔribCΔribF mutant strain can grow extracellularly in blood, we performed a growth curve in defibrinated sheep’s blood and found that the ΔribCΔribF mutant did not replicate and had a 2- to 3-log CFU loss by 24 h postinoculation (Fig. 4B). Finally, to test if the ΔribCΔribF mutant can grow extracellularly in the lumen of the GI tract, mice were pretreated for 2 d prior to the infection with streptomycin and were infected with 1 × 108 CFU/mouse. Stool pellets were collected daily for 5 d and plated to assess bacterial burden. The ΔribCΔribF mutant had a 7-log defect in fecal pellet CFUs compared to WT L. monocytogenes at 24 h postinfection (Fig. 4C). No CFUs were recovered from the ΔribCΔribF mutant–infected mice following day 1 postinfection (Fig. 4C). Collectively, these observations suggest that the ΔribCΔribF strain cannot grow extracellularly in the gallbladder, blood, or the GI tract and that this mutant is restricted to intracellular growth in vivo.
Discussion
The riboflavin derivatives FMN and FAD are redox active cofactors essential to all organisms (1–3), including pathogens which either synthesize riboflavin or import it from their host (35–37). Little is known about how pathogens that cannot synthesize riboflavin acquire this vitamin during infection. Here we show that the riboflavin auxotrophic, facultative intracellular bacterium L. monocytogenes uses the riboflavin transporter RibU to acquire FMN and FAD, but not riboflavin, from the cytoplasm of host cells. The finding that L. monocytogenes imports FMN and FAD to grow in host cells using RibU is supported by our data showing that a mutant lacking RibU is avirulent in vivo (Fig. 1 C and D) but grew in nutrient-rich media and chemically defined media supplemented with riboflavin and not with FMN and FAD (Fig. 3B). Furthermore, a mutant lacking the enzymes that convert riboflavin to FMN and FAD (ribC/ribF double mutant) grew like WT L. monocytogenes in BMMs (Fig. 3D) and in vivo in the spleen (Fig. 3E). Collectively, these observations led us to conclude that (1) RibU is a flavin transporter that, in addition to riboflavin, can import FMN and FAD; (2) L. monocytogenes might encode another, unknown riboflavin transporter that is unable to import FMN and FAD; and (3) L. monocytogenes primarily acquires FMN and FAD in vivo to support its intracellular growth.
The reliance of L. monocytogenes on host cell FMN and FAD is not surprising given the fact that the concentration of these flavins inside cells far surpasses that of their precursor riboflavin (13–15). We report that L. monocytogenes can obtain FMN and FAD from the host cell cytoplasm and we infer that this may be the case for other intracellular pathogens. For example, obligate intracellular pathogens in the Rickettsia and Cryptosporidium genera do not synthesize riboflavin and lack FMN and FAD synthetases (11, 12). Thus, we speculate that these pathogens must acquire FMN and FAD from the host to satisfy their flavin requirements. Similarly, we hypothesize that riboflavin auxotrophic vacuolar parasites, like members of the Apicomplexa phylum and Leishmania species, might rely on FMN and FAD import to grow intracellularly as well. These vacuolar parasites likely employ different mechanisms to scavenge these flavins from the cytosol, as has been shown for other micronutrients (38–40).
The evolutionary path to becoming an obligate intracellular pathogen often results in genome reduction (41–43) involving loss of a pathogen’s ability to synthesize its own metabolites and rely on import of nutrients from their hosts. The active cofactors FMN and FAD are highly prevalent in host cells (13–15), which might explain why some intracellular pathogens do not synthesize riboflavin or encode enzymes that convert it to FMN and FAD. In this study, we demonstrate that a L. monocytogenes mutant lacking the RibC and RibF enzymes required supplementation of FMN and FAD to grow and was unable to replicate in the blood (Fig. 4B), gallbladder (Fig. 4A) and GI tract (Fig. 4C) of mice, which represent environments where WT L. monocytogenes grows extracellularly (31–34). In contrast, the ribC/ribF double mutant grew intracellularly in macrophages and the spleen and liver of mice. Thus, we show the apparent conversion of a facultative intracellular pathogen into an obligate intracellular pathogen by eliminating two genes involved in the flavin biosynthesis pathway. This study provides an example of how pathogens, as an evolutionary adaptation, might lose their biosynthetic capabilities, by discarding genes involved in the production of metabolites prevalent in their hosts, and instead rely on acquisition of nutrients directly from the host, ultimately becoming obligate intracellular pathogens. Our findings may also have practical applications by providing an additional safety measure to L. monocytogenes strains used as therapeutic cancer vaccines by preventing extracellular growth and limiting dissemination (44).
Considering that L. monocytogenes thrives as a ubiquitous environmental saprophyte and has few nutritional requirements, it is curious that these bacteria lack the capacity to synthesize riboflavin. We hypothesize that the lack of riboflavin biosynthesis may provide L. monocytogenes an evolutionary advantage during infection of mammalian hosts, which almost universally have a prevalent population of MAIT cells (45) that are activated by an intermediate of bacterial riboflavin biosynthesis (23) and kill host cells harboring riboflavin-synthesizing bacteria such as francisellae, salmonellae, and legionellae (46–50). We are currently testing our hypothesis by examining the pathogenesis of L. monocytogenes strains engineered to synthesize riboflavin and the activation status of MAIT cells in infected mice.
Materials and Methods
Bacterial Culture and Strains.
All strains of L. monocytogenes used in this study (SI Appendix, Table S1) were derived from the WT 10403S strain and were cultured in filter-sterilized nutrient-rich brain heart infusion (BHI) media (BD) containing 200 µg/mL streptomycin (Sigma-Aldrich). Construction of the ΔribU (lmo1945), ΔribC (lmo1329), ΔribF (lmo0728), and ΔribCΔribF strains was done using allelic exchange with the temperature-sensitive plasmid pKSV7, as previously described (51). During the process of generating the ΔribC, ΔribF, and ribC/ribF double-mutant strains, the bacteria were always cultured in BHI media with 2.5 µM FMN (Sigma-Aldrich) and 2.5 µM FAD (Sigma-Aldrich) to circumvent synthetic lethality. For all procedures in which the ΔribC, ΔribF, and ΔribCΔribF strains were used, the bacteria were always grown in BHI media with 2.5 µM FMN and 2.5 µM FAD.
Generation of the ΔribU strain expressing the ribDEAHT operon or complementation of the strain expressing ribU was done by amplifying the ribDEAHT operon with its native promoter from B. subtilis and the ribU gene with its native promoter from WT L. monocytogenes, respectively, and cloning into the site-specific pPL2 integrating vector. Similarly, complementation of the ΔribCΔribF mutant was done by amplifying the ribC gene with its native promoter, cloning into the pPL2 vector, and introduction into L. monocytogenes by conjugation, as previously described (52).
Broth growth curves were performed with L. monocytogenes strains from overnight cultures grown at 37 °C with shaking (200 rpm). Nutrient-rich (BHI) and chemically defined synthetic media growth curves were started at an optical density (OD600) of 0.03. Chemically defined synthetic media was prepared as previously reported (53). Growth curves were spectrophotometrically measured by optical density at a wavelength of 600 nm (OD600).
Tissue Culture and Growth Media.
BMMs were prepared by collecting bone marrow from 8-wk-old female WT (Jackson Laboratory) and AIM2 KO (a gift from Kate Fitzgerald, University of Massachusetts Chan Medical School, Worcester, MA) C57BL/6J mice and differentiated as previously described (54). All BMMs used in the experiments were cultured in high-glucose Gibco Dulbecco’s modified Eagle’s medium (DMEM) (Thermo Fisher Scientific) with 20% fetal bovine serum (FBS) (Avantor-Seradigm), 10% macrophage colony-stimulating factor (M-CSF)–producing 3T3 cell supernatant, 1% L-glutamine (Corning), 1% sodium pyruvate (Corning), 14 mM 2-mercaptoethanol (Gibco Thermo Fisher Scientific).
Intracellular Growth Curves.
From 16 to 18 h prior to infection, 3 × 106 BMMs were seeded in 60-mm non-tissue culture–treated dishes (MIDSCI) containing 14 12-mm glass coverslips (Thermo Fischer Scientific) in each dish. L. monocytogenes strains were grown at 30 °C overnight in 14-mL round polypropylene tubes (Thermo Fisher Scientific) at a slanted position. The bacteria were washed and diluted in sterile 1X phosphate-buffered saline (PBS), and BMMs were infected at an MOI of 0.25. At 3 min postinfection, the cells were washed twice with 1X PBS. At 1 h postinfection, 50 µg/mL gentamicin sulfate (Sigma-Aldrich) was added to the cell culture media to kill/prevent bacteria from growing extracellularly. The growth curves then proceeded as previously described (55).
Intracellular Growth Curves in Media Lacking Riboflavin.
To deplete the intracellular flavins in L. monocytogenes, bacterial cultures were started 2 d prior to BMM infection in chemically defined media containing 1 µM riboflavin and grown at 37 °C with shaking. The bacteria were washed twice 16 to 18 h prior to infection with 1X PBS and then diluted into chemically defined media lacking flavins and grown at 37 °C with shaking.
Macrophages were washed twice, 3 h prior to BMM infection, with 1X PBS, and the cell culture media was replaced with high-glucose DMEM lacking riboflavin (Millipore Sigma), a gift from Erin Benanti at Aduro Biotech, with 20% dialyzed FBS using SnakeSkin dialysis tubing, 3.5K molecular weight cutoff (MWCO) (Thermo Fisher Scientific), and other components as described in Tissue Culture and Growth Media. The riboflavin-starved L. monocytogenes were washed and diluted in sterile 1X PBS, and the BMMs were infected at an MOI of 0.25. These growth curves were performed without the addition of riboflavin unless otherwise stated in the legend. The growth curve experiments then proceeded as previously described (55).
Cell Death (LDH Release) Assay.
From 16 to 18 h before infection, 5 × 105 BMMs per well were seeded in 24-well plates with 100 ng/mL Pam3CSK4 (InvivoGen) in DMEM. Before infecting the BMMs, the cell culture media was replaced with DMEM with 5% FBS. L. monocytogenes strains were grown overnight, slanted, at 30 °C. For the infection, bacteria were diluted in 1X PBS, and BMMs were infected at an MOI of 4. At 30 min postinfection, the BMMS were washed twice with 1X PBS and DMEM with 5% FBS and 50 µg/mL gentamicin was added to wells. The experiment was conducted as previously described (28).
Mouse Intravenous Infections.
Each 8-wk-old female CD-1 mouse (Charles River Laboratories) was infected via the tail vain with 200 µL of PBS containing 1 × 105 logarithmically growing bacteria. The mice were euthanized 48 h postinfection, and the spleen, liver, and gallbladder were collected, homogenized, and plated to determine the number of CFU per organ.
Blood Growth Curve.
The growth of L. monocytogenes strains in blood was determined using defibrinated sheep’s blood (HemoStat Laboratories). The bacteria were grown logarithmically for 2.5 h, washed, and resuspended in 3 mL of defibrinated sheep’s blood at a concentration of 1 × 106 per milliliter. Blood cultures were incubated at 37 °C with shaking. The growth of L. monocytogenes in blood was monitored for 3 d by diluting the blood in 1X PBS and plating to determine the number of CFU in total blood.
Mouse Oral Infections.
Mice were given 5 mg/mL streptomycin sulfate salt (Sigma-Aldrich) in drinking water 48 h prior to infection, as previously described (32). Mice were transferred to clean cages 18 to 24 h prior to infection, and the food source (mouse colony chow) was removed to start the overnight fast. The day of the infection, a 3-mm piece of bread was inoculated with 1 × 108 logarithmically growing bacteria in 1X PBS and covered with 3 µL of butter. Each 8-wk-old female CD-1 mouse (Charles River Laboratories) was then fed a single piece of infected bread. The streptomycin sulfate water was replaced with standard drinking water and the chow was restored. Fecal samples were collected everyday postinfection for 5 d, weighted, vortexed at 4 °C for 10 min, and plated to determine the number of CFU per gram of stool.
Phagosomal Escape Assay.
BMMs were seeded in 24-well plates containing 12-mm glass coverslips (Thermo Fischer Scientific) and cultured overnight. BMMs were treated with 250 ng/mL cytochalasin D (Sigma-Aldrich) 30 min prior to infection (MOI of 15) with L. monocytogenes strains grown at 30 °C overnight in a slanted position. At 1 h 15 min postinfection, the BMMs were washed twice with 1X PBS and fixed with 4% paraformaldehyde (Electron Microscopy Sciences) for 15 min. The immunofluorescence staining, microscopy, and image analysis then proceeded as previously described (56, 57). The primary antibodies used were rabbit anti-Listeria (catalog no. 223021, BD Difco) at a 1:1,000 dilution and guinea pig anti-p62 (catalog no. 20R-PP001, Fitzgerald) at a 1:200 dilution. The secondary antibodies used were rhodamine red-X goat anti-rabbit immunoglobulin G (IgG) (catalog no. R6394, Invitrogen-Thermo Fisher Scientific) at 1:2,000 dilution and Alexa Fluor-647 goat anti-guinea pig IgG (catalog no. A21450, Invitrogen-Thermo Fisher Scientific) at a 1:2,000 dilution. At least 100 bacteria per condition were quantified for analysis.
Animal Use Ethics Statement.
The mice were maintained by University of California, Berkeley Office of Laboratory Animal Care, personnel according to institutional guidelines. Animal studies were performed in accordance with the guidance and recommendations of the University of California, Berkeley Office of Laboratory Animal Care, and the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocols used in this study were reviewed and approved by the Animal Care and Use Committee at the University of California, Berkeley (AUP-2016-05-8811).
Statistical Analysis.
All statistical analyses were performed using GraphPad Prism version 9.2 for MacOS, GraphPad Software.
Supplementary Material
Acknowledgments
We thank Valerie Vargas-Zapata (University of California, Berkeley) for helpful discussion and editing of this manuscript. We also thank Brittney Nguyen (University of California, Berkeley), Frank Lee (University of California, Berkeley), and Han Yin (University of California, Berkeley) for experimental assistance and Kate Fitzgerald (University of Massachusetts Chan Medical School) and Erin Benanti (Aduro Biotech) for providing AIM2 KO mouse femurs and riboflavin-free DMEM, respectively. The research reported in this publication was supported by funding from the National Academies of Sciences, Engineering, and Medicine, Ford Foundation Fellowship, to R.R.-L.; the University of California Dissertation-Year Fellowship to R.R.-L.; and the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (1P01AI063302 and 1R01AI27655 to D.A.P., and F32AI136389 and K22AI144031 to S.H.L.). The model figure was created with https://BioRender.com.
Footnotes
Reviewers: J.M., Harvard University; and S.M., University of Washington.
Competing interest statement: R.R.-L. and D.A.P. have filed a provisional patent application with the US Patent and Trademark Office based on two L. monocytogenes strains (DP-L7381 and DP-L7378) constructed for this study.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2122173119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Mansoorabadi S. O., Thibodeaux C. J., Liu H. W., The diverse roles of flavin coenzymes – Nature’s most versatile thespians. J. Org. Chem. 72, 6329–6342 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Massey V., The chemical and biological versatility of riboflavin. Biochem. Soc. Trans. 28, 283–296 (2000). [PubMed] [Google Scholar]
- 3.Ghisla S., Edmondson D. E., Flavin coenzymes. eLS, 10.1002/9780470015902.A0000654.PUB3 (2014). [DOI] [Google Scholar]
- 4.Abbas C. A., Sibirny A. A., Genetic control of biosynthesis and transport of riboflavin and flavin nucleotides and construction of robust biotechnological producers. Microbiol. Mol. Biol. Rev. 75, 321–360 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rodionova I. A., et al. , A novel bifunctional transcriptional regulator of riboflavin metabolism in Archaea. Nucleic Acids Res. 45, 3785–3799 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yonezawa A., Inui K., Novel riboflavin transporter family RFVT/SLC52: Identification, nomenclature, functional characterization and genetic diseases of RFVT/SLC52. Mol. Aspects Med. 34, 693–701 (2013). [DOI] [PubMed] [Google Scholar]
- 7.García-Angulo V. A., Overlapping riboflavin supply pathways in bacteria. Crit. Rev. Microbiol. 43, 196–209 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Gutiérrez-Preciado A., et al. , Extensive identification of bacterial riboflavin transporters and their distribution across bacterial species. PLoS One 10, e0126124 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balcazar D. E., et al. , The superfamily keeps growing: Identification in trypanosomatids of RibJ, the first riboflavin transporter family in protists. PLoS Negl. Trop. Dis. 11, e0005513 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deka R. K., Brautigam C. A., Biddy B. A., Liu W. Z., Norgard M. V, Evidence for an ABC-type riboflavin transporter system in pathogenic spirochetes. mBio 4, e00615-12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Driscoll T. P., et al. , Wholly rickettsia! Reconstructed metabolic profile of the quintessential bacterial parasite of eukaryotic cells. mBio 8, e00859-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu Z., Guo Y., Roellig D. M., Feng Y., Xiao L., Comparative analysis reveals conservation in genome organization among intestinal Cryptosporidium species and sequence divergence in potential secreted pathogenesis determinants among major human-infecting species. BMC Genomics 20, 406 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aw T. Y., Jones D. P., McCormick D. B., Uptake of riboflavin by isolated rat liver cells. J. Nutr. 113, 1249–1254 (1983). [DOI] [PubMed] [Google Scholar]
- 14.Hustad S., et al. , Riboflavin, flavin mononucleotide, and flavin adenine dinucleotide in human plasma and erythrocytes at baseline and after low-dose riboflavin supplementation. Clin. Chem. 48, 1571–1577 (2002). [PubMed] [Google Scholar]
- 15.Vasilaki A. T., et al. , Relation between riboflavin, flavin mononucleotide and flavin adenine dinucleotide concentrations in plasma and red cells in patients with critical illness. Clin. Chim. Acta 411, 1750–1755 (2010). [DOI] [PubMed] [Google Scholar]
- 16.Freitag N. E., Port G. C., Miner M. D., Listeria monocytogenes – From saprophyte to intracellular pathogen. Nat. Rev. Microbiol. 7, 623–628 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Radoshevich L., Cossart P., Listeria monocytogenes: Towards a complete picture of its physiology and pathogenesis. Nat. Rev. Microbiol. 16, 32–46 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Whiteley A. T., Pollock A. J., Portnoy D. A., The PAMP c-di-AMP is essential for Listeria monocytogenes growth in rich but not minimal media due to a toxic increase in (p)ppGpp. [corrected]. Cell Host Microbe 17, 788–798 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mansjö M., Johansson J., The riboflavin analog roseoflavin targets an FMN-riboswitch and blocks Listeria monocytogenes growth, but also stimulates virulence gene-expression and infection. RNA Biol. 8, 674–680 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matern A., Pedrolli D., Großhennig S., Johansson J., Mack M., Uptake and metabolism of antibiotics roseoflavin and 8-demethyl-8-aminoriboflavin in riboflavin-auxotrophic Listeria monocytogenes. J. Bacteriol. 198, 3233–3243 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karpowich N. K., Song J. M., Cocco N., Wang D.-N., ATP binding drives substrate capture in an ECF transporter by a release-and-catch mechanism. Nat. Struct. Mol. Biol. 22, 565–571 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sebastián M., Arilla-Luna S., Bellalou J., Yruela I., Medina M., The biosynthesis of flavin cofactors in Listeria monocytogenes. J. Mol. Biol. 431, 2762–2776 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Kjer-Nielsen L., et al. , MR1 presents microbial vitamin B metabolites to MAIT cells. Nature 491, 717–723 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Méheust R., Huang S., Rivera-Lugo R., Banfield J. F., Light S. H., Post-translational flavinylation is associated with diverse extracytosolic redox functionalities throughout bacterial life. eLife 10, e66878 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Light S. H., et al. , A flavin-based extracellular electron transfer mechanism in diverse Gram-positive bacteria. Nature 562, 140–144 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Light S. H., et al. , Extracellular electron transfer powers flavinylated extracellular reductases in Gram-positive bacteria. Proc. Natl. Acad. Sci. U.S.A. 116, 26892–26899 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vitreschak A. G., Rodionov D. A., Mironov A. A., Gelfand M. S., Regulation of riboflavin biosynthesis and transport genes in bacteria by transcriptional and translational attenuation. Nucleic Acids Res. 30, 3141–3151 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sauer J. D., et al. , Listeria monocytogenes triggers AIM2-mediated pyroptosis upon infrequent bacteriolysis in the macrophage cytosol. Cell Host Microbe 7, 412–419 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sauer J.-D., et al. , Listeria monocytogenes engineered to activate the Nlrc4 inflammasome are severely attenuated and are poor inducers of protective immunity. Proc. Natl. Acad. Sci. U.S.A. 108, 12419–12424 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gastaldi G., et al. , Riboflavin phosphorylation is the crucial event in riboflavin transport by isolated rat enterocytes. J. Nutr. 130, 2556–2561 (2000). [DOI] [PubMed] [Google Scholar]
- 31.Zhang T., et al. , Deciphering the landscape of host barriers to Listeria monocytogenes infection. Proc. Natl. Acad. Sci. U.S.A. 114, 6334–6339 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Louie A., Zhang T., Becattini S., Waldor M. K., Portnoy D. A., A multiorgan trafficking circuit provides purifying selection of Listeria monocytogenes virulence genes. mBio 10, e02948-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mehmood H., Marwat A. D. J. K., Khan N. A. J., Invasive Listeria monocytogenes gastroenteritis leading to stupor, bacteremia, fever, and diarrhea: A rare life-threatening condition. J. Investig. Med. High Impact Case Rep. 5, 2324709617707978 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burke T. P., et al. , Listeria monocytogenes is resistant to lysozyme through the regulation, not the acquisition, of cell wall-modifying enzymes. J. Bacteriol. 196, 3756–3767 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang H., et al. , Dual-targeting small-molecule inhibitors of the Staphylococcus aureus FMN riboswitch disrupt riboflavin homeostasis in an infectious setting. Cell Chem. Biol. 24, 576–588.e6 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Fuentes Flores A., Sepúlveda Cisternas I., Vásquez Solis de Ovando J. I., Torres A., García-Angulo V. A., Contribution of riboflavin supply pathways to Vibrio cholerae in different environments. Gut Pathog. 9, 64 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bonomi H. R., et al. , An atypical riboflavin pathway is essential for Brucella abortus virulence. PLoS One 5, e9435 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krishnan A., Kloehn J., Lunghi M., Soldati-Favre D., Vitamin and cofactor acquisition in apicomplexans: Synthesis versus salvage. J. Biol. Chem. 295, 701–714 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Valigurová A., Florent I., Nutrient acquisition and attachment strategies in basal lineages: A tough nut to crack in the evolutionary puzzle of Apicomplexa. Microorganisms 9, 1430 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gold D. A., et al. , The toxoplasma dense granule proteins GRA17 and GRA23 mediate the movement of small molecules between the host and the parasitophorous vacuole. Cell Host Microbe 17, 642–652 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sakharkar K. R., Dhar P. K., Chow V. T. K., Genome reduction in prokaryotic obligatory intracellular parasites of humans: A comparative analysis. Int. J. Syst. Evol. Microbiol. 54, 1937–1941 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Khachane A. N., Timmis K. N., Martins dos Santos V. A. P., Dynamics of reductive genome evolution in mitochondria and obligate intracellular microbes. Mol. Biol. Evol. 24, 449–456 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Merhej V., Royer-Carenzi M., Pontarotti P., Raoult D., Massive comparative genomic analysis reveals convergent evolution of specialized bacteria. Biol. Direct 4, 13 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Flickinger J. C., Rodeck U., Snook A. E., Listeria monocytogenes as a vector for cancer immunotherapy: Current understanding and progress. Vaccines 6, 48 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kurioka A., Walker L. J., Klenerman P., Willberg C. B., MAIT cells: New guardians of the liver. Clin. Transl. Immunology 5, e98 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meierovics A., Yankelevich W.-J. C., Cowley S. C., MAIT cells are critical for optimal mucosal immune responses during in vivo pulmonary bacterial infection. Proc. Natl. Acad. Sci. U.S.A. 110, E3119–E3128 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang H., et al. , MAIT cells protect against pulmonary Legionella longbeachae infection. Nat. Commun. 9, 3350 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hinks T., et al. , MAIT cells protect against fatal intracellular pulmonary legionella infection via IFN-γ. Eur. Respir. J. 50, PA4122 (2017). [Google Scholar]
- 49.Howson L. J., et al. , MAIT cell clonal expansion and TCR repertoire shaping in human volunteers challenged with Salmonella Paratyphi A. Nat. Commun. 9, 253 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ussher J. E., Klenerman P., Willberg C. B., Mucosal-associated invariant T-cells: New players in anti-bacterial immunity. Front. Immunol. 5, 450 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Camilli A., Tilney L. G., Portnoy D. A., Dual roles of plcA in Listeria monocytogenes pathogenesis. Mol. Microbiol. 8, 143–157 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lauer P., Chow M. Y. N., Loessner M. J., Portnoy D. A., Calendar R., Construction, characterization, and use of two Listeria monocytogenes site-specific phage integration vectors. J. Bacteriol. 184, 4177–4186 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Whiteley A. T., et al. , c-di-AMP modulates Listeria monocytogenes central metabolism to regulate growth, antibiotic resistance and osmoregulation. Mol. Microbiol. 104, 212–233 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nguyen B. N., et al. , TLR2 and endosomal TLR-mediated secretion of IL-10 and immune suppression in response to phagosome-confined Listeria monocytogenes. PLoS Pathog. 16, e1008622 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Portnoy D. A., Jacks P. S., Hinrichs D. J., Role of hemolysin for the intracellular growth of Listeria monocytogenes. J. Exp. Med. 167, 1459–1471 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cheng M. I., Chen C., Engström P., Portnoy D. A., Mitchell G., Actin-based motility allows Listeria monocytogenes to avoid autophagy in the macrophage cytosol. Cell. Microbiol. 20, e12854 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Peterson B. N., Portman J. L., Feng Y., Wang J., Portnoy D. A., Secondary structure of the mRNA encoding listeriolysin O is essential to establish the replicative niche of L. monocytogenes. Proc. Natl. Acad. Sci. USA. 117, 23774–23781 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.