Abstract
Grass pea (Lathyrus sativus L.) is a grain legume commonly grown in Asia and Africa for food and forage. It is a highly nutritious and robust crop, capable of surviving both droughts and floods. However, it produces a neurotoxic compound, β-N-oxalyl-L-α,β-diaminopropionic acid (β-ODAP), which can cause a severe neurological disorder when consumed as a primary diet component. While the catalytic activity associated with β-ODAP formation was demonstrated more than 50 years ago, the enzyme responsible for this activity has not been identified. Here, we report on the identity, activity, 3D structure, and phylogenesis of this enzyme—β-ODAP synthase (BOS). We show that BOS belongs to the benzylalcohol O-acetyltransferase, anthocyanin O-hydroxycinnamoyltransferase, anthranilate N-hydroxycinnamoyl/benzoyltransferase, deacetylvindoline 4-O-acetyltransferase superfamily of acyltransferases and is structurally similar to hydroxycinnamoyl transferase. Using molecular docking, we propose a mechanism for its catalytic activity, and using heterologous expression in tobacco leaves (Nicotiana benthamiana), we demonstrate that expression of BOS in the presence of its substrates is sufficient for β-ODAP production in vivo. The identification of BOS may pave the way toward engineering β-ODAP–free grass pea cultivars, which are safe for human and animal consumption.
Keywords: food security, Lathyrus sativus, grass pea, neurolathyrism, oxalyl-CoA, diaminopropionic acid, beta-ODAP, beta-ODAP synthetase, BAHD acyltransferase, hydroxycinnamoyl transferase
Abbreviations: β-ODAP, L-β-N-oxalyl-α-β-diaminopropionic acid; BAHD, Benzylalcohol O-acetyltransferase, Anthocyanin O-hydroxycinnamoyltransferase, anthranilate N-Hydroxycinnamoyl/benzoyltransferase, Deacetylvindoline 4-O-acetyltransferase; BIA, β-(isoxazolin-5-on-2-yl)alanine; BOS, β-ODAP synthetase; cDNA, complementary DNA; DDW, dextrose in distilled water; DTNB, 5,5′-dithio-bis-(2-nitrobenzoic acid); GO, gene ontology; HCT, hydroxycinnamoyl transferases; L-DAPA, L-α,β-diaminopropionic acid; LsOCS, Lathyrus sativus oxalyl CoA-synthetase; MWCO, molecular weight cutoff; O/N, overnight; OPT, o-phthalaldehyde; PDB, Protein Data Bank; RT, room temperature; UPLC/MS, ultra-performance liquid chromatography–mass spectrometry
Grass pea (Lathyrus sativus L.[Ls]) is an annual legume crop grown for food and forage, mainly in South Asia and Sub-Saharan Africa (1, 2). In addition to its high grain yield (3) and the high nutritional value of its seeds (4, 5), its attractiveness as a farming crop stems from its remarkable tolerance to harsh environmental conditions such as drought, high salinity, and flooding (6, 7, 8), as well as its resistance to insects and fungal diseases (9, 10). Unfortunately, it produces a neurotoxic glutamate analog, named β-N-oxalyl-L-α,β-diaminopropionic acid (β-ODAP) (11, 12). This neurotoxin may cause neurolathyrism, a nutritional neurodegenerative disorder characterized by lower limb paralysis (13, 14). Neurolathyrism results from a chronic intoxication caused by the long-term ingestion of seeds or flour made from grass pea as a primary diet component (15). While grass pea cultivars with reduced concentrations of β-ODAP have been bred (16), none is devoid of β-ODAP. Furthermore, β-ODAP levels may increase under stress conditions such as drought (17). As an alternative approach to conventional breeding, a constitutively expressed fungal oxalate decarboxylase gene was introduced into grass pea to degrade oxalate and, subsequently, reduce the concentration of oxalyl-CoA, a precursor of β-ODAP (18). This attempt reduced β-ODAP concentrations up to 73% in the seeds of the transgenic plant but was unable to prevent β-ODAP production completely. Thus, despite its high nutritional value and enormous agricultural potential, grass pea remains an underutilized crop of limited economic importance in global markets. Yet, it constitutes an essential source of food and income security for resource-poor farmers in developing countries (2, 19).
To ensure safe consumption of grass pea seeds and increase its use as a food crop, a cultivar that does not contain β-ODAP is required. For such a cultivar to be developed by genetic engineering using, for example, CRISPR/CRISPR-associated protein 9(20), the biosynthetic pathway leading to β-ODAP production needs to be elucidated.
Studies aiming to identify the enzymes responsible for the biosynthesis of β-ODAP in grass pea were initiated more than 50 years ago (21, 22, 23). They indicated that β-ODAP is synthesized by the ligation of oxalyl-CoA and L-α,β-diaminopropionic acid (L-DAPA), catalyzed by a dedicated synthase (23). This synthase, however, has hitherto not been identified, precluding the use of genome editing tools to generate β-ODAP–free cultivars.
Here, we report on the isolation, identification, and characterization of a β-ODAP synthase (BOS) from grass pea. The identification of BOS paves the path toward the application of genome editing techniques to generate grass pea cultivars devoid of β-ODAP.
Results and discussion
Isolation and identification of BOS synthase from grass pea
Grass pea seeds, seedlings, and developing leaves accumulate the highest levels of β-ODAP in the plant (24). We, therefore, used seeds and seedlings to identify and isolate BOS by means of fractionation and protein purification. To trace the enzyme during purification, we assayed protein fractions for their ability to ligate oxalyl-CoA to L-DAPA (Fig. 1), using a colorimetric assay that detects the presence of free L-DAPA following its derivatization by o-phthalaldehyde (OPT) (25). While L-DAPA is commercially available, the availability of oxalyl-CoA is quite limited (26). We, therefore, synthesized it in vitro from oxalic acid and Co-A using a recombinant oxalyl-CoA synthetase (OCS) (27). OCS is an ATP-dependent enzyme that catalyzes the ligation of oxalate to CoA, forming oxalyl-CoA (Scheme S1). Its activity and involvement in the production of β-ODAP in grass pea were discovered more than 50 years ago (21, 22). We cloned the OCS gene from grass pea, expressed and purified it from Escherichia coli (27), and used it to produce oxalyl-CoA as a substrate for β-ODAP activity assays.
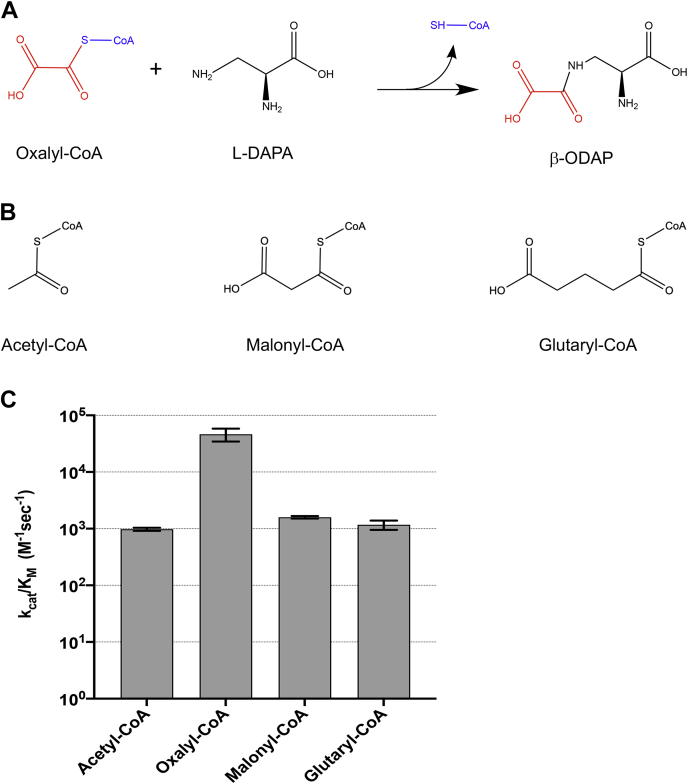
Figure 1.
Reaction scheme and catalytic efficiencies of BOS. A, a scheme of the reaction catalyzed by BOS. B, additional CoA substrates analyzed in this work. C, apparent catalytic efficiencies of BOS with different acyl-CoA substrates. The data for oxalyl-CoA were derived from a fit to the Michaelis–Menten equation; error bars denote SD of three independent repeats. Data for all other substrates were fitted to the linear regime of the Michaelis–Menten model, and kcat/Km was deduced from the slope; error bars denote SE of the fit. BOS, β-ODAP synthetase.
Following sequential chromatographic separations of soluble grass pea extracts on multiple columns (Experimental procedures section), we obtained a protein fraction enriched in BOS activity. The process was repeated four times, and the proteins present in the final fractions were then subjected to proteolysis and peptide sequencing using liquid chromatography-mass spectrometry (LC–MS/MS) analysis (Experimental procedures section).
To correlate between the peptide sequences and their encoding genes, we sequenced the grass pea transcriptome using long-read PacBio sequencing. The constructed set of annotated full-length mRNA transcripts was translated and used as a database for the identification of the proteins in the enzymatically active fractions (Table S1). Between 244 and 1960 different proteins, including BOS, were identified, of which only 219 were found in all active fractions (Table S2). Of these, 45 were predicted to be ligases (gene ontology [GO]:0016874), transferases (GO:0016740), synthases (e.g., GO:0004019), or CoA-binding enzymes (GO:0120225). From this latter set, we selected 18 sequences for functional analysis, based on the fact that they were annotated as CoA-binding enzymes or shikimate pathway ligases and transferases and seemed likely to bind the relevant substrates and catalyze the ligation reaction. The corresponding genes were cloned, expressed, and purified from E. coli cells, and the level of BOS-like activity was assayed in vitro. Of the 18 clones tested, only one exhibited a significant level of such activity (Fig. S1).
The catalytic activity of BOS
To verify that the observed activity was that of a bona fide BOS, BOS was purified from E. coli and the catalytic activity of the recombinant enzyme, an untagged monomer of 439 aa (49.3 kDa, Fig. S2), was assayed using several CoA substrates. The catalytic efficiencies of BOS toward L-DAPA were determined using fixed concentrations of the CoA substrates in excess over varying concentrations of L-DAPA (Figs. 1 and S3). With oxalyl-CoA as substrate, BOS exhibited moderate efficiency, with an apparent turnover rate (kcat) and Michaelis (KmL-DAPA) constants of 118 ± 15 (sec˗1) and 2.5 ± 0.6 (mM), respectively, giving rise to an apparent catalytic efficiency (kcat/KmL-DAPA) of 4.7 ± 1.3 × 104 (sec−1M−1) (Fig. S3) and specific activity of 13.2 ± 1.6 (mmol⋅sec1⋅mg˗1). However, when oxalyl-CoA was substituted by other CoA substrates, such as acetyl-CoA, malonyl-CoA, or glutaryl-CoA, the catalytic efficiency dropped by 30− to 48-fold (Figs. 1 and S3), indicating a catalytic specificity of BOS toward oxalyl-CoA. The enzyme used for synthesizing the substrate oxalyl-CoA, OCS (LsOCS), did not produce any β-ODAP in the presence of L-DAPA and oxalyl-CoA (data not shown).
Despite the fact that L-DAPA is, by itself, a stable compound in vitro, attempts to isolate it from grass pea have failed to do so, suggesting that it is a short-lived metabolic intermediate (28). Its precursor β-(isoxazolin-5-on-2-yl)alanine (BIA) and its product (β-ODAP) (Scheme S1), on the other hand, accumulate to high concentrations in the seeds and seedlings of grass pea: up to 2% of the dry weight for BIA (29) and 0.5 to 2.5% for β-ODAP (11). Notwithstanding the large variabilities in seed weight (34–350 mg) (30, 31) and water content (7.5–30.7%) (12, 32) of grass pea seeds, their effective concentrations of BIA and β-ODAP may reach 0.25 to 1.7 M, suggesting that L-DAPA is produced in grass pea at concentrations well above the observed KmL-DAPA of BOS. The magnitude of the latter is similar to that of other enzymes on the biosynthetic pathway of β-ODAP in grass pea, such as cysteine synthase A and B (Km = 6.1 mM and 7.2 mM, respectively) (33), enzymes involved in secondary metabolism (34) and many other enzymes (35).
The formation of β-ODAP and its nontoxic isomer α-ODAP in the reaction, along with the consumption of L-DAPA, were monitored by LC–MS. The results showed that >99% of the L-DAPA in the BOS-catalyzed reaction had been converted to β-ODAP, while the concentration of α-ODAP was in order of magnitude less (<1%) and similar to those obtained in the uncatalyzed control reaction (Fig. S4). BOS, thus, displays a high degree of regioselectivity toward acylating the β-amino group of L-DAPA in vitro.
Production of β-ODAP by BOS in tobacco leaves
To determine if BOS activity is sufficient to produce β-ODAP in vivo, we transiently expressed BOS in Nicotiana benthamiana leaves using Agrobacterium tumefaciens infiltration and injected them with L-DAPA (Experimental procedures section). The presence of the other precursor, oxalyl-CoA, in N. benthamiana leaves was expected due to the identification of putative OCSs in database searches of several Nicotiana species, including benthamiana (data not shown). The concentrations of α- and β-ODAP, as well as of L-DAPA, were then determined in leaf samples using LC–MS (Fig. S5). The results revealed that only leaves that expressed BOS and were injected with L-DAPA produced α- and β-ODAP (Fig. S5). The relative concentrations of the two isomers in the leaves could not be determined since the extraction process converts part of β-ODAP to α-ODAP (36). Wild-type N. benthamiana leaves or leaves infiltrated with A. tumefaciens carrying an empty vector and injected with L-DAPA did not produce α- or β-ODAP. Thus, BOS expression in the presence of its substrates is both necessary and sufficient to produce β-ODAP in planta.
Phylogeny
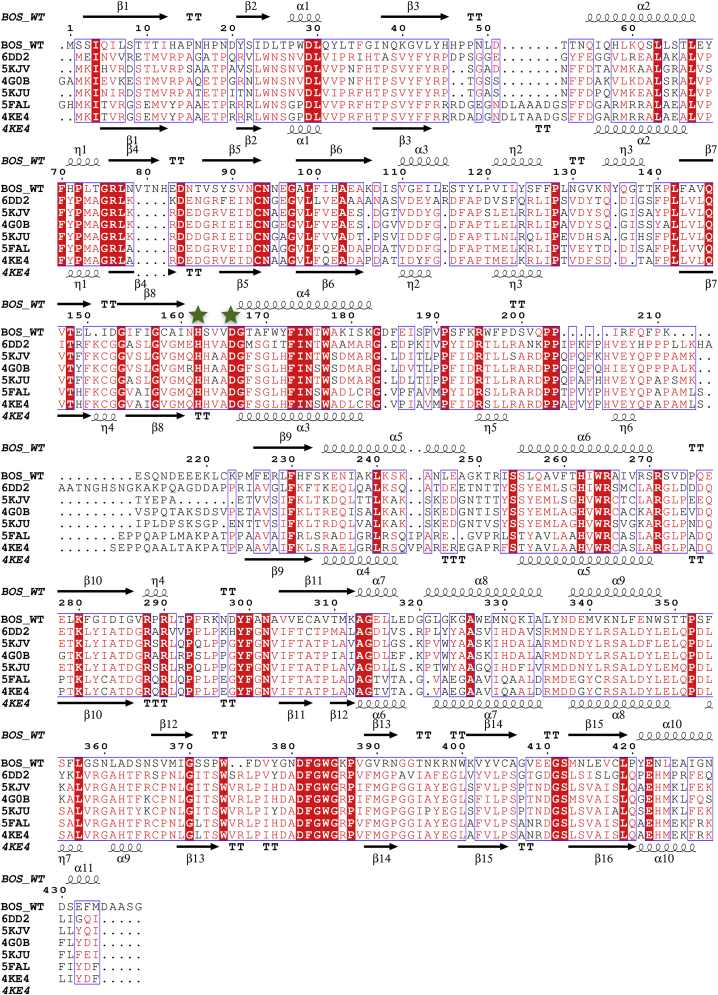
Sequence analysis of BOS revealed that almost its entire sequence (residues 3–434 of 439) corresponds to a transferase domain (PF02458) of a family found predominantly in plants and fungi. The closest relative identified was enhanced Pseudomonas susceptibility 1—an Arabidopsis thaliana protein that belongs to the benzylalcohol O-acetyltransferase, anthocyanin O-hydroxycinnamoyltransferase, anthranilate N-Hydroxycinnamoyl/benzoyltransferase, deacetylvindoline 4-O-acetyltransferase (BAHD) acyltransferase superfamily (37). Correspondingly, BOS was found to possess the two hallmarks of this superfamily: a conserved active site HXXXD motif (residues 162–166, HSVVD, Fig. 2) and a structural DFGWG motif (residues 381–385, Fig. 2) (37, 38, 39). We note that the sequence we determined is identical to that of a transcript that was identified as a BOS candidate and published in a PhD thesis (40).
Figure 2.
Secondary structure–based sequence alignment of BOS and HCT homologs. The sequences of hydroxycinnamoyl:CoA-shikimate hydroxycinnamoyl transferases (HCTs) from different species were aligned to that of BOS. Sequences are labeled by their PDB codes: Selaginella moellendorffii (6DD2); Plectranthus scutellarioides (5KJV); Coffea canephora (4G0B); Arabidopsis thaliana (5KJU); Panicum virgatum (5FAL); and Sorghum bicolor (4KE4). BOS secondary structure elements are noted above the sequences and those of Sorghum bicolor (4KE4) - at the bottom. α-Helices and η-helices are shown as spirals, and β-strands are indicated by arrows. Conserved residues are shown in red blocks. Green stars denote BOS His162 and Asp166, which are part of the conserved BAHD acyltransferase HXXXD motif. BAHD, Benzylalcohol O-acetyltransferase, Anthocyanin O-hydroxycinnamoyltransferase, anthranilate N-Hydroxycinnamoyl/benzoyltransferase, Deacetylvindoline 4-O-acetyltransferase; BOS, β-ODAP synthetase.
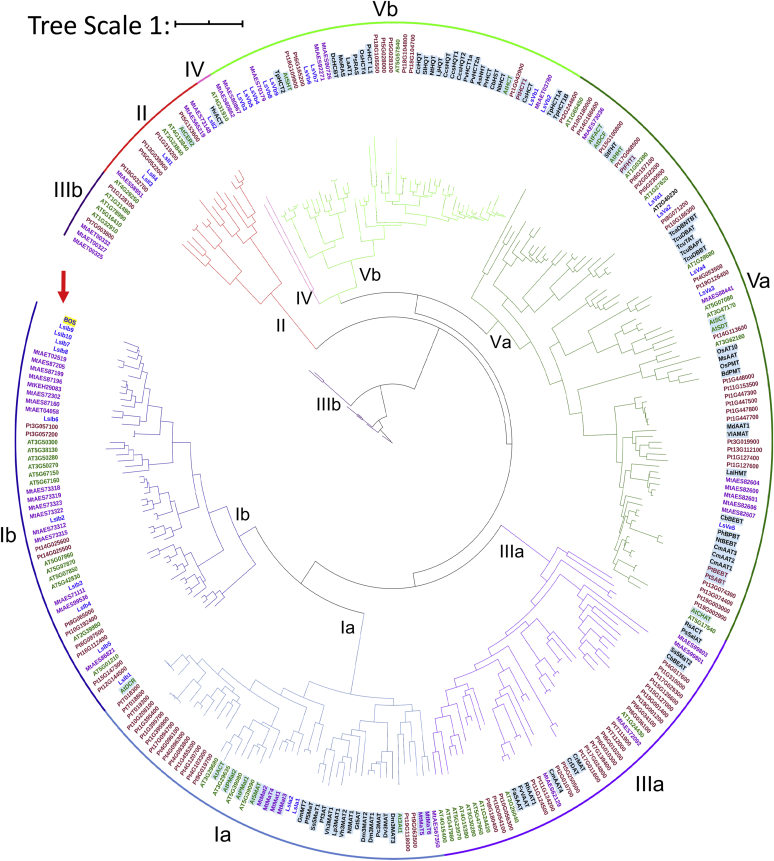
A search for other BAHD acyltransferases in the grass pea transcriptome that we generated identified 30 additional members of this superfamily (SI Appendix A). A phylogenetic tree of BAHD proteins was constructed to determine the clade to which BOS belongs (Fig. 3). In total, 299 sequences were aligned, 93 of which had known biological functions (SI Appendix A and B). The tree (Fig. 3) shows the typical clades of the superfamily, namely, Ia, Ib, II, IIIa IIIb, IV, Va, and Vb (37, 41, 42) with many branches exhibiting species-specific expansions (Fig. S6). BOS, along with 10 other grass pea proteins, forms part of clade Ib (Figs. 3 and S6) and is the second protein in that clade, apart from A. thaliana defective in cuticular ridges (43), to be characterized. Of these 10 proteins (LsIb1–10), three (LsIb3, LsIb4, and LsIb9) were not found in any of the four enzymatically active fractions of BOS. Four sequences (LsIb7–10) constitute the closest homologs of BOS (Fig. 3). Nonetheless, all clade Ib members were cloned, purified, and assayed for BOS activity. Besides BOS, none of them had BOS activity (Fig. S1). Additional BAHD proteins we identified in grass pea belong to other, more distant clades (Fig. 3), have much lower sequence similarity to BOS, and are highly unlikely to have BOS activity. BOS is, thus, likely to be the sole enzyme capable of producing β-ODAP in grass pea.
Figure 3.
Phylogenetic tree of BAHD superfamily proteins. Sequence names are prefixed by a two-letter species code and colored by species: L. sativus (Ls), blue; A. thaliana (At), green; M. truncutula (Mt), purple; P. trichocarpa (Pt), brown; other species are colored black. The outer ring and clades are colored as follows (clockwise from the open end on the left): putative clade IIIb (dark purple), clade II (red), clade IV (pink), clade Vb (light green), clade Va (dark green), clade IIIa (light purple), clade Ia (light blue), and clade Ib (dark blue). Sequences of proteins with known functions are shaded in light blue; BOS is marked by a red arrow and shaded in yellow. BAHD, Benzylalcohol O-acetyltransferase, Anthocyanin O-hydroxycinnamoyltransferase, anthranilate N-Hydroxycinnamoyl/benzoyltransferase, Deacetylvindoline 4-O-acetyltransferase.
Crystal structure
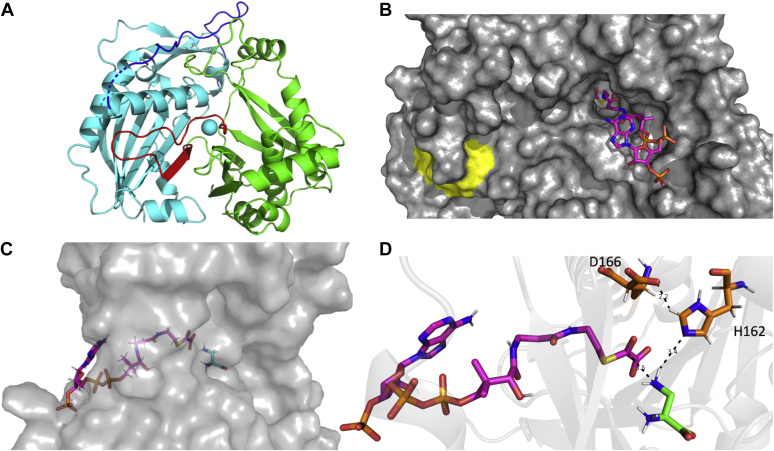
BOS was crystallized in its apo state, and its structure was solved to 2.35-Å resolution (Table S3). It consists of 15 β-strands and 11 α-helices and is organized into two equally sized domains of ∼200 aa. Each domain centers around a six-stranded β-sheet flanked by α-helices (Fig. 4A). The two domains are connected by two loops, one (residues 183–209) linking the N- and C-terminal domains (Fig. 4A) and the other (residues 371–388) joining β-strand 13 from the C-terminal domain (residues 389–393) to the β-sheet of the N-terminal domain (Fig. 4A). The latter loop contains the aforementioned DFGWG motif (Fig. 4B).
Figure 4.
Structure of BOS and docked substrates. A, ribbon diagram of BOS (PDB ID: 6ZBS). The N- (cyan) and C-terminal (green) domains are connected by loop (residues 183–209; blue) and segment comprised of a β-strand and a short loop (residues 371–393, red). B, surface representation with oxalyl-CoA (magenta) docked into the active site. The conserved DFGWG motif is colored yellow. C, side view showing both oxalyl-CoA (magenta) and L-DAPA (cyan) docked into the active site. D, a close-up view showing oxalyl-CoA (magenta) and L-DAPA (green) docked into the active site along with the conserved catalytic residues His162 and Asp166 (orange). A rotamer of Asp166 that places its residue in hydrogen-bonding distance to His166 was chosen manually to demonstrate a possible interaction. H-bonds are denoted by dashed lines. BOS, β-ODAP synthetase; L-DAPA, L-α,β-diaminopropionic acid.
A search for structural homologs of BOS yielded six enzymes (Fig. 2), all of which are plant hydroxycinnamoyl transferases (HCTs; EC:2.3.1.133; Fig. S7). The average RMSD between the structure of BOS and HCTs from Selaginella moellendorffii, A. thaliana, Coffea canephora, Panicum virgatum, Sorghum bicolor, and Plectranthus scutellarioides is 2.7 ± 0.1 Å. The average sequence identity between BOS and these HCTs is only 22 ± 2%, while the sequence identity among the HCTs themselves ranges from 56% to 93%. The main differences between BOS and these enzymes are the lengths and conformations of some of its loops (Fig. S8, A and B) and the geometry of the active site tunnel, which is approximately straight in HCTs (Fig. S8, C and D), but bent to ∼15° in its middle in BOS (Fig. S8, E and F). Thus, despite its overall structural similarity, BOS is different from other plant HCTs, both in sequence and in key structural features.
Catalytic mechanism
Our attempts to cocrystallize BOS with either of its substrates or products were unsuccessful. To gain insight into the catalytic mechanism of BOS, we docked its two substrates to the active site, using the structure of P. virgatum HCT (44) (Protein Data Bank [PDB] ID: 5FAL) as a guide. The docking result showed that oxalyl-CoA resides within the active-site tunnel with the CoA nucleotide moiety situated at the entrance to the tunnel and its oxalyl moiety buried deep inside the tunnel (Fig. 4, B–D). As predicted, the other substrate, L-DAPA, is positioned within binding distance from the thioester carbonyl carbon, with which it interacts (Fig. 4D). Histidine 162 of the conserved catalytic HXXXD motif is located 3.5 Å away from the β amine group of L-DAPA (Fig. 4D). Thus, similar to its role in other BAHD acetyltransferases (38), His162 likely acts to deprotonate the terminal amino nitrogen of L-DAPA, enabling a nucleophilic attack on the carbonyl carbon. This, in turn, results in the formation of a tetrahedral intermediate that subsequently collapses to release CoA and the product β-ODAP (Figs. 4D and S9). The role of the conserved aspartic acid residue of the HXXXD motif, Asp166, may be structural (38) or may serve to activate His162 through hydrogen bonding (Fig. 4D). Our analysis also suggests that other residues, including Lys40, Asp284, Ser371, Ile369, and Lys401, may participate in substrate binding in the active site (Fig. S10). To verify the requirement of His162 and Asp166 to catalysis, we mutated them, individually, to residues with similar side-chain volumes and polarities (His162Gln and Asp166Ser). Both single mutants expressed and purified as soluble proteins but exhibited little to no activity (Fig. S1), supporting their essential roles in the catalytic mechanism of BOS.
Molecular docking simulations
We used molecular docking simulations to predict the likelihood that different CoA substrates will occupy the active site of BOS in a catalytically productive manner. The simulations generated different ligand orientations, and for each orientation, a docking score was calculated. The ligand orientations were sorted by their docking score, and the top-scoring (most negative, thus favorable to binding) orientations were analyzed to estimate their catalytic efficiency. We ran 385 docking simulations using CoA, acetyl-CoA, oxalyl-CoA, malonyl-CoA, glutaryl-CoA, and 4-coumaroyl-CoA as acyl substrates. The simulations were carried out using the structure of BOS predocked with L-DAPA. The rotamer of Asp166 that places it in close, hydrogen bonding distance with His162 (Fig. 4D) was manually chosen in accordance with our proposed catalytic mechanism (Fig. S9). We surmised that optimal substrate binding would place the CoA substrate at a distance and an angle that would favor a productive nucleophilic attack by the β-amine group of L-DAPA on the thioester carbonyl carbon. The optimal angle of attack for a nucleophile on an unsaturated carbonyl, that is, the Bürgi–Dunitz angle (45), is generally taken to be 105 ± 5°, and the initial distance between the nucleophile and the electrophile is expected to range between 2.5 and 3.4 Å (46, 47). Accordingly, we analyzed the top 10% docking results (data not shown) by the proximity and angle of their carbonyl group to the nucleophilic amine of L-DAPA. Of the seven best-ranking binding simulations (Table S4), five were of oxalyl-CoA. Malonyl-CoA received the highest docking score, yet its carbonyl was found a long distance (4.58 Å) away from its nucleophile and at an attack angle of only 59.5 degrees. Binding in this mode is less likely to be productive than that of oxalyl-CoA. Acetyl-CoA, on the other hand, received the lowest docking score of the top seven results, yet its carbonyl was positioned at the shortest distance from the nucleophile (3.2 Å) and at a favorable angle of 102 degrees. Two substrates were not included in the top seven ranking solutions: glutaryl-CoA and 4-coumaryl-CoA. The former was chosen since it is a three-carbon backbone analog of oxalyl-CoA, and the latter is the substrate of the HCT structural analogs of BOS. In both cases, the simulations resulted in their thioester carbonyl groups positioned at long distances (10 ± 5 Å) from the β-amine group of L-DAPA. Such orientations are less likely to lead to a productive ligation reaction. As a reference, we docked CoA itself and obtained docking scores similar to that of other substrates (Table S4), indicating that much of the binding interaction between BOS and the CoA substrates is mediated by their large CoA moiety. In our simulations, the average distance of the oxalyl-CoA carbonyl from its attacking nucleophile was 3.49 ± 0.1 Å (Table S4). It is longer than the expected distance limit for bond formation yet was derived from rigid-body simulations, which do not reflect distance changes in the enzyme following substrate binding or enzyme dynamics. Despite their limitations, our docking simulations may help explain the observed substrate specificity profile of BOS and predict it will be inefficient in catalyzing a reaction with 4-coumaryl-CoA as a substrate.
Conclusions
This work presents the isolation, identification, and characterization of a BOS from grass pea, more than 50 years after its activity was first demonstrated and its existence proposed (21, 22, 23). BOS is part of a superfamily of BAHD acyltransferases that has at least 30 more members in grass pea. However, none of the closest sequence homologs of BOS we identified in grass pea have similar enzymatic activity. The crystal structure of BOS and the results of our molecular docking analysis suggest that both of its substrates interact within a long V-shaped active-site tunnel. They further suggest that upon binding, the thioester carbonyl of oxalyl-CoA is positioned within hydrogen-bonding distance to the terminal β-amine group of L-DAPA and to the conserved His162 residue of BOS. This likely facilitates catalysis through a proton shuttle between the highly conserved Asp166 and His162 residues and is supported by the fact that mutating either of them results in a soluble, folded yet inactive enzyme. Aside from grass pea, β-ODAP has been found in the seeds of 13 species of Crotalaria, 17 species of Acacia, and 20 additional species of Lathyrus (48, 49), all from the legume family (Fabaceae), as well as in several species of Panax (29, 50). We searched for BOS orthologs in the available transcriptomes of Panax ginseng, Panax notoginseng, Crotalaria juncea, and Acacia koa in the NCBI transcriptome shotgun assembly database. However, while we were able to find BAHD family Ib members in all of these species, none branched closely enough to BOS to consider them as orthologs (data not shown). Thus, it is unknown if these and other species that produce β-ODAP utilize a different synthetic pathway that evolved independently for this purpose or use the same pathway and express a BOS-like gene that evolved in parallel and has little sequence similarity to LsBOS.
Grass pea is a nutritious and robust crop whose seeds have a high-protein and low-fat content (4, 5). It is inexpensive, easy to grow, and tolerant to drought and high salinity as well as flooding (2) and possesses a hardy and penetrating root system that enables it to grow on a wide range of soils, including poor soils and heavy clays (51). Sometimes, it is the only surviving crop in drought-prone (and often famine-ravaged) areas in Africa and Asia (52). It has, therefore, a potential to significantly improve food security, particularly in arid and semiarid regions, which are likely to expand due to a looming climate change (53).
These attributes, however, are overshadowed by the hazards associated with the production of β-ODAP, which greatly limit its use. Indeed, the sale and storage of grass pea, in any form, have been banned in various jurisdictions in India since 1961 (2). Since the biological role of β-ODAP in grass pea is still unknown, the consequences of eliminating its production in the plant are not clear. β-ODAP has been proposed to transport zinc in soils depleted of zinc or rich in iron (54), to act as a radical scavenger, or to enhance symbiosis with Rhizobium bacteria (17, 55, 56), and while it was shown to accumulate under water stress (57, 58), its role, if any, in grass pea drought tolerance is unknown. The latter has, in fact, been attributed to upregulation of antioxidant enzymes and increased concentrations of osmoprotectants, such as proline and soluble sugars (59), as well as to an extensive root system and leaf rolling during water shortage (2). It is hoped that the identification of BOS, along with its encoding gene, will pave the way toward the generation of β-ODAP–free grass pea cultivars. These would, hopefully, preserve the various beneficial traits of grass pea while being safe for consumption, adding a valuable resource to the world’s battery of food crops.
Experimental procedures
Materials
Grass pea (L. sativus L) seeds were obtained from Fratelli Ingegnoli Milano© (https://www.ingegnoli.it/) and from Plant World Seeds© (www.plant-world-seeds.com). Protease inhibitor cocktail for plant cells and for bacterial cells, Miracloth, Hepes, EDTA, ammonium sulfate, sodium phosphate monobasic and dibasic, sodium oxalate, magnesium chloride, CoA sodium salt, 5,5′-dithio-bis-(2-nitrobenzoic acid) (DTNB), acetyl-CoA, malonyl-CoA, glutaryl-CoA, β-ODAP (SML1836), sodium tetraborate, 13C6-arginine hydrochloride, 2,3-diaminopropionic acid, benzonase, lysozyme, Amicon Ultra-2, and 15 centrifugal filters were obtained from Sigma–Aldrich© and MERCK©. 6-Aminoquinolyl-N-hydroxy-succinimyl carbamate was synthesized following Cohen and Michaud (60). α-ODAP was obtained by partial thermal isomerization of β-ODAP (61). L-DAPA was obtained from AEchem Scientific©. GELRITE and Murashige and Skoog medium, including vitamins, were obtained from Duchefa©. RNeasy plant mini kit and DNeasy plant mini kit were obtained from QIAGEN©. RNase-free DNase I was purchased from New England BioLabs©. Snakeskin dialysis tubing was obtained from Thermo Fisher Scientific©. HiTrap Q FF 10/16, HiLoad 26/600 Superdex 200 pg, HiTrap Blue HP, and Superdex 200 increase 10/300 Gl columns were obtained from GE Healthcare©. Zenix SEC-300 column was purchased from Sepax Technologies©. Synthetic DNA constructs were obtained from Twist Bioscience©. 96-deep-well plates were obtained from Axygen. Ninety-six-well ELISA plates were obtained from Greiner. Ultra-performance liquid chromatography–mass spectrometry (UPLC/MS)–grade solvents were used for all chromatographic steps. The pDGB3Ω1 cloning vector was a generous gift from Dr Diego Orzáez (Instituto de Biología Molecular y Celular de Plantas, Consejo Superior de Investigaciones Científicas, and Universidad Politécnica de Valencia).
Plant seed germination
Grass pea (L. sativus) seeds were surface sterilized by a brief wash with 70% ethanol and soaking in 3% sodium hypochlorite for 20 min at constant, gentle (50 rpm) shaking. Then, they were rinsed four times with autoclaved dextrose in distilled water (DDW) and kept in DDW overnight (O/N) at 4 °C to synchronize their germination. For RNA extraction, the seeds were inoculated in a container supplemented with half-strength Murashige and Skoog salts and 30 g/l sucrose and gelled with 0.3% GELRITE at pH 5.8. For protein extraction, the seeds were inoculated in plastic pots containing 250 cm3 of commercial potting mixture. Seeds were germinated under 12-h light (150 μmol m−2 s−1)/dark cycle at 25 °C for 1 week.
Isolation of BOS from grass pea
Prior to processing, seeds (30 g) were soaked in sterile water for 3 to 4 days, while sprouts (25 g) were used following a brief wash with DDW. Both seeds and sprouts were frozen in liquid nitrogen. Seeds and sprouts (thawed) were ground using an electric hand blender and ceramic mortar and pestle, respectively. The ground tissue was resuspended in plant extraction buffer (50 mM Hepes, pH 7.5; 1 mM EDTA, pH 8.3; 250 mM sucrose; protease inhibitor cocktail 1:250), homogenized using glass homogenizer, and pelleted (20,000g, 1 h, 4 °C). The (viscous) supernatant was filtered through Miracloth and pelleted by ultracentrifugation (190,000g, 1 h, 4 °C). It was then precipitated by ammonium sulfate at 50% saturation (4 °C, O/N) and repelleted (20,000g, 1 h, 4 °C). Pellets were resuspended in buffer A (50 mM Hepes, pH 7.5; 1 mM EDTA, pH 8.3; 50 mM NaCl), dialyzed extensively (10K molecular weight cutoff [MWCO], 22 mm) against buffer A, and the solution was pelleted (20,000g, 1 h, 4 °C). The supernatant was loaded onto a HiTrap-Q-FF-10/16 column connected to an AKTA pure 25M HPLC (GE Healthcare©) and eluted using a gradient from buffer A to buffer B (50 mM Hepes, pH 7.5; 1 mM EDTA, pH 8.3; 1 M NaCl). Fractions exhibiting BOS activity were pooled, concentrated, and washed with buffer A using Amicon Ultra-15 centrifugal filters (10 kDa MWCO). The concentrates were loaded onto a HiLoad 26/600 Superdex 200 pg column and eluted using buffer C (50 mM Hepes, pH 7.5; 1 mM EDTA, pH 8.3; 150 mM NaCl). Eluted fractions exhibiting BOS activity were pooled again, concentrated and washed with buffer D (20 mM Na-phosphate, pH 7.5; 1 mM EDTA, pH 8.3), and loaded onto a HiTrap Blue HP (5 ml) column. BOS-containing fractions were eluted using a gradient from buffer D to buffer E (20 mM Na-phosphate, pH 7.5; 1 mM EDTA, pH 8.3; 2 M NaCl). Fractions exhibiting BOS activity were pooled, washed with buffer A, and reconcentrated. The concentrates were loaded on a Superdex 200 increase 10/300 Gl and eluted using buffer C or loaded onto a Zenix SEC-300 and eluted with PBS buffer. Fractions exhibiting BOS activity were pooled, rinsed with 50 mM NaCl, concentrated using Amicon Ultra 2 ml centrifugal filters (10 kDa MWCO), and sent for peptide sequencing.
In vitro synthesis of oxalyl-CoA
The gene encoding OCS was amplified from extracted genomic DNA of L. sativus, cloned into a bacterial expression vector, expressed in E. coli, and purified (27). Purified OCS (0.25 μM) was added to a reaction solution (125 mM Hepes, pH 7.5; 2 mM MgCl2; 10 mM ATP) containing oxalic acid (5 mM) and CoA (1.5 mM) and incubated at 37 °C for 1 h. The reaction mixture was sampled before the addition of OCS and at the end of the incubation. The samples, diluted twofold, were mixed with an equal volume of buffered DTNB solution (50 mM Hepes, pH 7.5; 1 mM DTNB) and incubated for 15 min at room temperature (RT). The amount of unreacted CoA was determined by comparing the absorbance of the sample solutions at 412 nm, before and after incubation with OCS. Typically, under these conditions, more than 95% of CoA reacted to form oxalyl-CoA. Reaction mixtures containing in vitro synthesized oxalyl-CoA were used directly as substrates for in vitro β-ODAP synthesis, as described in the following.
L-α,β-Diaminopropionic acid elimination assay
The activity of BOS was determined in purified protein fractions as described (25). Oxalyl-CoA was freshly prepared in vitro as described previously. Samples containing oxalyl-CoA (1.5 mM, 40 μl) were mixed with buffered DAPA solution (50 mM Hepes, pH 7.5; 0.7 mM, 30 μl DAPA) and purified grass pea protein fractions (30 μl). The mixtures were incubated at 37 °C for 15 to 30 min in 96-deep-well plates and then mixed with OPT reagent solution (50 mM boric acid, pH 9.9; 60 mM β-mercaptoethanol; 7.5 mM, 400 μl OPT). DAPA concentrations were determined by measuring solution absorbance at 422 nm. The reaction produces a yellow color with free DAPA and other vicinal diamine compounds, but not with primary amines or with β-ODAP. The formation of β-ODAP in the reactions, performed in plant or E. coli extracts, or in vitro, resulted in the reduction in free DAPA concentrations and was verified independently using LC–MS (detailed in “Determination of α/β-ODAP and L-DAPA concentrations by LC–MS”).
Grass pea complementary DNA library preparation and sequencing
Total RNA from five-week-old plants was isolated using RNeasy plant mini kit, and residual DNA was removed with RNase-free DNase I. The RNA was quantified using a Qubit 3 fluorometer and an RNA broad-range kit (Invitrogen©), and its integrity was assessed using an Agilent 2100 Bioanalyzer. Only RNA with an integrity number greater than 8 was used for (SMRTbell) library preparation. An Iso-Seq library was prepared according to the isoform sequencing protocol, using Clontech SMARTer PCR complementary DNA (cDNA) synthesis kit as described by Pacific Biosciences with the following modifications: 1 mg of RNA, combined from two different tissues, was used as an input. PCR cycle optimization was performed by amplification of first-strand cDNA by 10 to 18 cycles, using PrimeSTAR GXL polymerase (Takara). Large-scale PCR was performed by 11 amplification cycles to generate double-stranded cDNA for SMRTbell library construction. One single-molecule real-time cell was sequenced using the PacBio Sequel platform.
Iso-Seq data analysis
Raw data were processed using SMRTlink 4.0 software (PacBio). Circular consistency sequences were generated into full-length reads, which were clustered into full-length nonchimeric transcripts by IsoSeq3. From these transcripts, high-quality isoforms (Qscore > 30) were selected for further analysis. Since a reference genome for grass pea was not available at the time, the Coding Genome Reconstruction Tool (Cogent) was used to generate a fake coding genome on which Cupcake ToFU was used to remove redundant sequences. This pipeline yielded a nonredundant high-likelihood gene assembly. The ‘Dumb’ algorithm was used to find the longest open reading frames (ORFs). We then used the ‘ANGEL’ (62) algorithm to predict the most likely ORFs based on the length and coding potential of each given sequence. GO annotation was performed using Blast2GO (http://www.blast2go.org/), version 2.5.0. After gene-ID mapping, GO term assignment, annotation augmentation, and generic GO-slim process, the final annotation file was generated. Data were submitted as BioProject ID PRJNA662941 (http://www.ncbi.nlm.nih.gov/bioproject/662941).
Proteomic analysis
Protein fractions were subjected to in-solution tryptic digestion, using suspension trapping as described (63). Briefly, protein concentration was measured using the bicinchoninic acid assay (Thermo Scientific). Protein fractions were supplemented with SDS (5%, final concentration) in 50 mM Tris–HCl, reduced with 5 mM dithiothreitol, and alkylated with 10 mM iodoacetamide in the dark. Samples were loaded onto suspension trapping microcolumns (ProtiFi) according to the manufacturer’s instructions, and the columns were washed with 90:10% methanol/50 mM ammonium bicarbonate. Samples were digested with trypsin (1:50 trypsin/protein) for 1.5 h at 47 °C. The digested peptides were eluted using 50 mM ammonium bicarbonate. Trypsin was added to this fraction, which was further incubated O/N at 37 °C. Two more elutions were made using 0.2% formic acid and 0.2% formic acid in 50% acetonitrile. The three eluates were pooled together and vacuum-centrifuged to dryness. Samples were kept at −80 °C until further analysis. Dry digested samples were dissolved in 97:3% H2O/acetonitrile + 0.1% formic acid. Peptides were separated by chromatography using nano-UPLC (10 kpsi nanoAcquity; Waters). The mobile phase was H2O + 0.1% formic acid and acetonitrile + 0.1% formic acid. Desalting of the samples was performed online using a reversed-phase Symmetry C18 trapping column (180 μm internal diameter, 20 mm length, 5 μm particle size; Waters). Peptides were separated using a T3 high-strength silica nanocolumn (75 μm internal diameter, 250 mm length, 1.8 μm particle size; Waters) at 0.35 μl/min and were eluted into the mass spectrometer using the following gradient: 4% to 30% B in 50 min, 30% to 90% B in 5 min, maintained at 90% for 5 min, and then back to initial conditions. The nano-UPLC was coupled online through a nanoelectrospray ionization emitter (10 μm tip; New Objective) to a quadrupole Orbitrap mass spectrometer (sample BOS1: Q Exactive Plus, samples BOS2-3: Fusion Lumos, BOS4: Q Exactive HFX, Thermo Scientific) using a FlexIon nanospray apparatus (Proxeon). Data were acquired in data-dependent acquisition mode, using the Top10 method. MS1 resolution was set to 120,000 (at 200 m/z), mass range of 375 to 1650 m/z, automatic gain control of 3e6, and the maximum injection time was set to 60 msec. MS2 resolution was set to 15,000, quadrupole isolation 1.7 m/z, automatic gain control of 1e5, dynamic exclusion of 20 s, and maximum injection time of 60 msec. Raw data were processed with Proteome Discoverer V2.2 (Thermo Fisher Scientific) and searched using SequestHT (64) and Mascot (v. 2.5.1) (65) engines against the translated transcriptome as the protein database, appended with common lab protein contaminants. The number of database sequence entries used for the search was 18,649 for the Dumb database and 49,324 for the Angel database. Enzyme specificity was set to trypsin, and up to two missed cleavages were allowed. Fixed modification was set to carbamidomethylation of cysteines, and variable modifications were set to oxidation of methionines, protein N-terminal acetylation, and deamidation of glutamines and asparagines. Peptide precursor ions were searched using a maximum mass deviation of 10 ppm and fragment ions using maximum mass deviation of 0.02 Da. Peptide and protein identifications were filtered at a false discovery rate of 0.01 using the decoy database strategy.
BOS activity assays of recombinant proteins
Selected grass pea genes and mutated BOS variants were codon-optimized for expression in E. coli synthesized and cloned into a pET–His–SUMO expression vector. The vector was constructed by transferring the His14-bdSUMO cassette from expression vector K151, generously provided by Prof. Dirk Görlich (Max-Planck Institute) (66), into the expression vector pET28-TevH (67). Plasmids were transformed into electrocompetent BL21/DE3 cells harboring a chaperone-expressing plasmid pGJKE8 (Takara©) and plated on LB agar plates with 35 μg/ml chloramphenicol, 50 μg/ml kanamycin, and 1% w/v glucose. Following O/N incubation at 37 °C, individual colonies from each transformation were randomly picked into 5-ml culture tubes (Falcon) containing growth media (2YT + Cap 35 μg/ml + Kanamycin 50 μg/ml; 1 ml) and grown O/N (at 37 °C; shaking, 250 rpm). The resulting cultures were used to inoculate 10-ml growth media cultures (1:100 dilution) and grown at 37 °C to A600 ≈ 0.6. Chaperone expression was induced by addition of arabinose (0.5 mg/ml) for 2 h at 37 °C. Protein expression was induced by IPTG (1 mM), and cultures were grown O/N (37 °C; shaking 250 rpm). Subsequently, the cells were pelleted (4000 rpm, 4 °C, 20 min), frozen at −80 °C, thawed at RT for 10 min, and resuspended in 500 μl of cell lysis buffer (20 mM PBS, pH 7.4; 300 mM NaCl; 0.6 mg/ml lysozyme; 10 U/ml benzonase nuclease; bacterial protease inhibitor cocktail without EDTA 1:50). Cells were lysed by sonication, and the lysate was clarified by centrifugation (30 min, 4 °C, 15,000g). The clarified lysates were loaded on Ni–nitriloacetic acid spin columns (NEBExpress), and the proteins were purified according to the manufacturer’s protocol. Purified protein solutions (20 μl) were mixed with oxalyl-CoA (4 mM; 20 μl) and buffered L-DAPA solution (50 mM Hepes, pH 7.5; 1 mM, 20 μl, DAPA) and incubated for 1 h at 37 °C. The mixtures were transferred to individual wells in a 96-well ELISA plate, combined with 200 μl of OPT reagent solution (50 mM boric acid, pH 9.9; 60 mM β-mercaptoethanol; 7.5 mM OPT), and incubated for 10 min at RT, and the solution absorbance was read at 422 nm using a plate reader (BioTek©). Negative control samples contained elution buffer only (20 μl).
Large-scale cloning, expression, and purification of BOS
BOS was expressed in BL21 (DE3), using the pET28-bdSUMO expression vector. A 5-L culture was induced with 200 μM IPTG and grown O/N at 15 °C. The cells were harvested and lysed by a cell disrupter (Constant Systems©) in lysis buffer (50 mM Tris, pH 7.5, 0.1 M NaCl, 1 mM DTT, 2 mM MgCl2, 8% glycerol) containing 200 KU/100 ml lysozyme, 20 μg/ml DNase, 1 mM phenylmethylsulfonyl fluoride (PMSF), and protease inhibitor cocktail. Following centrifugation, 1% Triton was added to the supernatant and the lysate was incubated with Ni-beads (Adar Biotech©) for 1 h at 4 °C. The beads were washed four times with 50 ml of lysis buffer after which the enzyme was eluted by incubation with 5 ml of lysis buffer containing 0.4 mg of bdSUMO protease for 2 h at RT. The eluate, containing untagged BOS, was concentrated and applied to a size exclusion column (HiLoad 16/60 Superdex75 prep-grade, GE Healthcare©) equilibrated with 50 mM Tris pH 7.5, 0.1 M NaCl, 2 mM MgCl2, and 8% glycerol. Purified BOS was eluted as a single peak at 58 ml and pooled, concentrated to 28 mg/ml, and frozen in aliquots at −80 °C.
Kinetic analysis of BOS
Oxalyl-CoA (6 mM; synthesized in situ), acetyl-CoA (2 mM), malonyl-CoA (2 mM), or glutaryl-CoA (2 mM) were mixed with purified BOS (0.1 μM) and varying concentrations of L-DAPA (0.09–4.8 mM) in buffer A (50 mM Hepes, pH 7.5; 1 mM EDTA, pH 8.3; 50 mM NaCl) at RT. The reaction mixtures were sampled at different time intervals during the first 5 min, and samples were quenched by mixing with an OPT solution (50 mM boric acid, pH 9.9; 60 mM β-mercaptoethanol; 7.5 mM OPT) at a 1:9 (v/v) ratio. The absorbance of sample solutions was measured at 422 nm in 96-well plates using an ELISA plate reader (BioTek©). Initial velocities obtained for reactions with oxalyl-CoA were fit using nonlinear regression to the Michaelis–Menten equation to derive kcat and Km of BOS. For other CoA substrates, fits were made to the linear part of the equation. Errors in kcat and Km of BOS with oxalyl-CoA or with other Co-A substrates were derived from the fit to the Michaelis–Menten curve or the slope of its linear part, respectively. Values were determined at saturating concentrations of the acyl-CoA substrates and should, therefore, be considered apparent.
Determination of α/β-ODAP and L-DAP concentrations by liquid chromatography–mass spectrometry
Leaf samples were ground using a bead beater, mixed with 1 ml of 0.1% formic acid, shaken (Eppendorf shaker 2000 rpm, 10 °C, 10 min), and then pelleted (14000g, 10 °C, 10 min). The supernatant was collected, and the extraction was repeated once more. The combined extracts were dried using a lyophilizer, resuspended in 50 ul of 0.1% formic acid, and then derivatized using 6-aminoquinolyl-N-hydroxy-succinimyl carbamate (AQC) as described (60, 68). Briefly, a 10-μl sample aliquot or amino acid standard solution was mixed with 1 μl of 13C6-Arg solution (60 μM) as internal standard and with 69 μl of 0.15 M sodium borate solution, pH 8.8. Samples were derivatized by adding 20 μl of AQC in acetonitrile (2.7 mg/ml), followed by incubation at 55 °C for 10 min. The reaction mixtures were cooled and subjected to LC–MS/MS analysis, using Acquity I-class UPLC system (Waters©) and Xevo TQ-S triple quadrupole mass spectrometer (Waters©) equipped with an electrospray ion source. Liquid chromatography was performed using a 100 × 2.1 mm internal diameter, 1.8 μm UPLC high-strength silica T3 column (Waters©) with mobile phases A (0.1% aqueous formic acid) and B (0.1% formic acid in acetonitrile) at a flow rate of 0.4 ml/min and column temperature of 35 °C. The gradient was as follows: 2% B for 1 min, then an increase to 30% B (7 min), an increase to 100% B (0.5 min), 100% B for 1.5 min, then reduced to 2% B (0.5 min), and kept at 2% B for additional 1.5 min. Samples were kept at 4 °C and automatically injected. Sample volume was 1 μl. MS was performed in positive-ion mode, with capillary voltage of 3 kV, cone voltage of 23 V, desolvation temperature of 350 °C, source temperature of 120 °C, desolvation gas flow of 500 L/h, and collision gas flow of 0.10 ml/min. The following MS/MS transitions were monitored: m/z 347.0 → 171.0 for α- and β-ODAP (retention time: 4.02 and 3.81 min, respectively), 275.1 → 171.0 and 223.1 → 171.0 for DAPA (as 1× AQC and 2× AQC adducts, respectively), and 351.0 → 171.0 for 13C6-Arg as internal standard. In vitro reaction samples of 10 μl were similarly derivatized and analyzed.
Crystallization, data collection, and refinement
Purified BOS was crystallized using the hanging-drop vapor diffusion method and a mosquito robot (TTP LabTech©) at 19 °C, utilizing the precipitants 0.1 M MgCl2 and 10% PEG 6000 in 50 mM Hepes (pH 7.0). BOS crystals formed in the tetragonal space group P43212, with one monomer per asymmetric unit and diffracted to 2.35-Å resolution. Diffraction data were collected under cryogenic conditions (100 K), in-house, using a Rigaku RU-H3R X-ray instrument. All diffraction images were indexed and integrated using the Mosflm program (69), and the integrated reflections were scaled using the SCALA program (70). Structure factor amplitudes were calculated using TRUNCATE (71) from the CCP4 program suite. The structure of BOS was solved by molecular replacement with the program PHASER (University of Cambridge) (72), using the distantly related (23% sequence identity) HCT structure from S. bicolor (PDB ID: 4KE4). All steps of refinement were performed using PHENIX.refine and Parallel PHENIX.phaser programs (NIH) (73). The model was built into 2mFobs-DFcalc and mFobs-DFcalc maps using COOT (74), optimized using PDB_REDO (75) and evaluated using MolProbity (76). The crystal structure was deposited in the PDB with the accession number 6ZBS.
Phylogenetic analysis
Database searches (BlastP) were performed at the NCBI website against the nonredundant database with default parameters. The full complement of Medicago truncatula and A. thaliana (41) BAHD protein sequences were compared to the PacBio transcripts using TBlastN in a local installation (version 2.6.0+ (77)). Hits with an E-score <10˗30 were taken for further analysis. DNA sequence redundancy was examined using the Sequencher software (version 5.4.6, Gene Codes Corporation©). Domain analysis was performed at the NCBI, using the Batch-CDD search tool (78). Multiple alignment was performed with ClustalW (version 2.1 (79)). The full complement of sequences from Populus trichocarpa was added (41). The phylogenetic tree of BAHD proteins was constructed using the 31 BAHD proteins from grass pea, 47 BAHD proteins from Medicago truncutula, 58 BAHD proteins from A. thaliana, 96 from P. trichocarpa (41), and 67 sequences with known functions from other species (37, 42). Sequences that, for various reasons, were omitted from the alignments are listed with reasons at the bottom of SI Appendix A. Alignments were performed with ClustalW. Phylogenetic analysis was performed with neighbor-joining in ClustalW, PHYLIP (version 3.697 (80)) and PhyML online (version 3 (81)), using default parameters. The PhyML tree was visualized with iTOL (82). The multiple sequence alignment in Figure 2 was produced using Multalin (83), and the figure was created using ESPript (84).
Molecular docking
The 3D crystal structure of BOS (PDB ID: 6ZBS) was optimized prior to docking using the Protein Preparation Wizard in Schrödinger Maestro Suite 2019 (Schrödinger Suite 2019-2 Protein Preparation Wizard; Epik, Schrödinger, LLC, 2019; Impact, Schrödinger, LLC, 2018; Prime, Schrödinger, LLC, 2018). Inconsistencies in the structure, such as missing side chains or hydrogens, incorrect bond orders, or side-chain orientation, were rectified during optimization (85) and the resulting structure was used for Glide docking (86, 87, 88). To support the proposed catalytic mechanism of BOS, a rotamer of Asp166 facing His162 was manually selected. The LigPrep module in Schrödinger Maestro Suite 2019 (LigPrep, Schrödinger, 2019-2) was used to convert the structures of the oxalyl-CoA, L-DAPA, β-ODAP, 4-coumaroyl-CoA, malonyl-CoA, and glutaryl-CoA from 2D to 3D, to correct improper bond distances and bond orders, to generate the ionization states of the ligands, and to minimize their energy. The optimized structures were then used for rigid ligand docking using the standard precision mode of Schrödinger’s Glide docking tool. Of BOS′ two substrates, oxalyl-CoA was docked first, using the crystal structure of PvHCT in complex with CoA and p-coumaroyl-shikimate (PDB ID: 5FAL) to generate the receptor grid, which represents the active site of the receptor for Glide ligand-docking jobs. For the docking of L-DAPA and β-ODAP, the best oxalyl-CoA docking pose was selected and truncated to a four-carbon chain, which was used as a dummy ligand for grid generation. The poses were sorted based on the Glide docking score. For docking the CoA-derivative ligands library, L-DAPA is the nucleophile that attacks the carbonyl carbon of the CoA-derivatives ligand thioester moiety; hence, L-DAPA is included as part of the BOS protein. For three-body molecular docking, L-DAPA was docked and merged as BOS–L-DAPA, with a negative charge to the nucleophilic β-amine group (NH-). For the second-grid generation, the docking result of oxalyl-CoA is used. The second grid was then used for docking the ligands library of 4-coumaroyl-CoA, malonyl-CoA, glutaryl-CoA, oxalyl-CoA, CoA, and acetyl-CoA using Extra Precision, XP-Glide docking algorithm.
Transient expression of BOS in N. benthamiana leaves
The BOS-encoding gene was cloned into the binary vector pART27 (89) and transformed into A. tumefaciens (GV3101) electrocompetent cells. Transformed cells were selected on LB agar plates containing 200 μg/ml spectinomycin and 50 μg/ml gentamicin at 28 °C. Single colonies were then used to inoculate 10 ml of LB media containing the respective antibiotics and grown at 28 °C O/N. Cells were harvested (3500 rpm, 10 min), washed with 10 ml of infiltration solution (100 mM Mes buffer, 2 mM Na3PO4, 100 μM acetosyringone), and repelleted. The cell pellet was resuspended in infiltration solution to a final A600 ≈ 0.3 and incubated at RT for 2 h. The resulting Agrobacterium suspension (2 ml) was infiltrated into 4- to 5-week-old N. benthamiana leaves. A. tumefaciens cells transformed with an empty cloning vector pDGB3Ω1 were used as a control. Plants were grown under 16-h light/8-h dark conditions. After 3 days of agroinfiltration, the same leaves were infiltrates with a 2-ml substrate solution (1 mM, L-DAPA, pH 7.5). Leaves were collected 2 days after substrate infiltration and frozen in liquid nitrogen. Ground tissue was further used for metabolic extraction. Six individual plants were used for the BOS expression group and four for each of the other controls (vector only and wt).
Data and materials availability
Refined coordinates were deposited in the Protein Data Bank Database under accession code 6ZBS (BOS). Transcriptomic data are available on GenBank, BioProject ID PRJNA662941. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (90) partner repository (http://www.ebi.ac.uk/pride/) with the data set identifier PXD030847. Details of all proteins identified using the “Dumb” and “Angel” databases are specified in Appendices C and D, respectively. Materials are available from the authors upon request.
Supporting information
This article contains supporting information (64, 65).
Conflict of interests
The authors declare no competing interests.
Acknowledgments
We thank Drs Hadas Keren-Shaul and Inbal Neta-Sharir and Mr Jossef Jacobovitch for their kind assistance with mRNA sequencing, plant growth and protein purification, respectively.
Author contributions
M. G. investigation, formal analysis, methodology, writing–original draft, review & editing, visualization, project administration; S. B. investigation, writing–original draft. data curation; M. K. formal analysis. writing–original draft. data curation; A. S. investigation, formal analysis, writing–original draft; S. B.-D. formal analysis, data curation, writing–original draft, review & editing, visualization; A. B. investigation, writing–original draft; T. M., investigation; Y. P., resources, writing – original draft; S. A. resources, writing–original draft, visualization; O. D. investigation, writing–original draft, visualization, data curation; E. B.-Z. investigation, writing–original draft, visualization; R. S. B. investigation, writing–original draft; A. A. supervision, writing- review & editing; Z. R. conceptualization, writing–review & editing, supervision, funding acquisition.
Funding and additional information
This work was supported by a grant from the Israeli Ministry of Agriculture and Rural Development (#15–37–0003). S. A. is the incumbent of Prof. David Casson Research Fellowship. R. S. B was supported by a Planning and Budgeting Committee (VATAT) fellowship to the Weizmann Institute of Science.
Edited by Joseph Jez
Contributor Information
Moshe Goldsmith, Email: moshe.goldsmith@weizmann.ac.il.
Ziv Reich, Email: ziv.reich@weizmann.ac.il.
Supporting information
References
- 1.Yan Z.Y., Spencer P.S., Li Z.X., Liang Y.M., Wang Y.F., Wang C.Y., Li F.M. Lathyrus sativus (grass pea) and its neurotoxin ODAP. Phytochemistry. 2006;67:107–121. doi: 10.1016/j.phytochem.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 2.Lambein F., Travella S., Kuo Y.H., Van Montagu M., Heijde M. Grass pea (Lathyrus sativus L.): Orphan crop, nutraceutical or just plain food? Planta. 2019;250:821–838. doi: 10.1007/s00425-018-03084-0. [DOI] [PubMed] [Google Scholar]
- 3.Mikic A., Cupina B., Rubiales D., Mihailovic V., Sarunaitek L., Fustec J., Antanasovic S., Krstic D., Bedoussac L., Zoric L., Dordevic V., Peric V., Srebric M. Models, developments, and perspectives of mutual legume intercropping. Adv. Agron. 2015;130:337–419. [Google Scholar]
- 4.Mondal M.M.A., Puteh A.B. Spectrum of variability in seed size and content of protein and odap in grass pea (Lathyrus sativus L.) germplasm. Legume Res. 2014;37:479–482. [Google Scholar]
- 5.Arslan M. Diversity for vitamin and amino acid content in grass pea (Lathyrus sativus L.) Legume Res. 2017;40:803–810. [Google Scholar]
- 6.Mahdavi B., Sanavy S.A. Germination and seedling growth in grasspea (Lathyrus sativus) cultivars under salinity conditions. Pak J. Biol. Sci. 2007;10:273–279. doi: 10.3923/pjbs.2007.273.279. [DOI] [PubMed] [Google Scholar]
- 7.Zhou L.N., Cheng W., Hou H.Z., Peng R.Y., Hai N., Bian Z.Y., Jiao C.J., Wang C.Y. Antioxidative responses and morpho-anatomical alterations for coping with flood-induced hypoxic stress in grass pea (Lathyrus sativus L.) in comparison with pea (Pisum Sativum) J. Plant Growth Regul. 2016;35:690–700. [Google Scholar]
- 8.Boukecha D., Laouar M., Mekliche-Hanifi L., Harek D. Drought tolerance in some populations of grass pea (Lathyrus sativus L.) Legume Res. 2018;41:12–19. [Google Scholar]
- 9.Tiwari K.R., Campbell C.G. Inheritance of neurotoxin (ODAP) content, flower and seed coat colour in grass pea (Lathyrus sativus L) Euphytica. 1996;91:195–203. [Google Scholar]
- 10.Almeida N.F., Krezdorn N., Rotter B., Winter P., Rubiales D., Vaz Patto M.C. Lathyrus sativus transcriptome resistance response to Ascochyta lathyri investigated by deepSuperSAGE analysis. Front. Plant Sci. 2015;6:178. doi: 10.3389/fpls.2015.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiong J.L., Xiong Y.C., Bai X., Kong H.Y., Tan R.Y., Zhu H., Siddique K.H., Wang J.Y., Turner N.C. Genotypic variation in the concentration of beta-N-Oxalyl-L-alpha,beta-diaminopropionic acid (beta-ODAP) in grass pea (Lathyrus sativus L.) seeds is associated with an accumulation of leaf and pod beta-ODAP during vegetative and reproductive stages at three levels of water stress. J. Agric. Food Chem. 2015;63:6133–6141. doi: 10.1021/acs.jafc.5b01729. [DOI] [PubMed] [Google Scholar]
- 12.Xu Q., Liu F., Chen P., Jez J.M., Krishnan H.B. beta-N-Oxalyl-l-alpha,beta-diaminopropionic acid (beta-ODAP) content in Lathyrus sativus: The integration of nitrogen and sulfur metabolism through beta-cyanoalanine synthase. Int. J. Mol. Sci. 2017;18:526. doi: 10.3390/ijms18030526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ngudi D.D., Kuo Y.H., Van Montagu M., Lambein F. Research on motor neuron diseases konzo and neurolathyrism: Trends from 1990 to 2010. PLoS Negl. Trop. Dis. 2012;6 doi: 10.1371/journal.pntd.0001759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tan R.-Y., Xing G.-Y., Zhou G.-M., Li F.-M., Hu W.-T., Lambein F., Xiong J.-L., Zhang S.-X., Kong H.-Y., Zhu H., Li Z.-X., Xiong Y.-C. Plant toxin β-ODAP activates integrin β1 and focal adhesion: A critical pathway to cause neurolathyrism. Scientific Rep. 2017;7:40677. doi: 10.1038/srep40677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fearon C., Murray B., Mitsumoto H. 7th edn. Elsevier Inc.; Philadelphia, PA: 2016. Disorders of Upper and Lower Motor Neurons. Bradley’s Neurology in Clinical Practice; pp. 1484–1518. [Google Scholar]
- 16.Kumar S., Bejiga G., Ahmed S., Nakkoul H., Sarker A. Genetic improvement of grass pea for low neurotoxin (beta-ODAP) content. Food Chem. Toxicol. 2011;49:589–600. doi: 10.1016/j.fct.2010.06.051. [DOI] [PubMed] [Google Scholar]
- 17.Jiao C.J., Jiang J.L., Ke L.M., Cheng W., Li F.M., Li Z.X., Wang C.Y. Factors affecting beta-ODAP content in Lathyrus sativus and their possible physiological mechanisms. Food Chem. Toxicol. 2011;49:543–549. doi: 10.1016/j.fct.2010.04.050. [DOI] [PubMed] [Google Scholar]
- 18.Kumar V., Chattopadhyay A., Ghosh S., Irfan M., Chakraborty N., Chakraborty S., Datta A. Improving nutritional quality and fungal tolerance in soya bean and grass pea by expressing an oxalate decarboxylase. Plant Biotechnol. J. 2016;14:1394–1405. doi: 10.1111/pbi.12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cullis C., Kunert K.J. Unlocking the potential of orphan legumes. J. Exp. Bot. 2017;68:1895–1903. doi: 10.1093/jxb/erw437. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y., Ma X., Xie X., Liu Y.G. CRISPR/Cas9-Based genome editing in plants. Prog. Mol. Biol. Transl. Sci. 2017;149:133–150. doi: 10.1016/bs.pmbts.2017.03.008. [DOI] [PubMed] [Google Scholar]
- 21.Malathi K., Padmanaban G., Rao S.L., Sarma P.S. Studies on the biosynthesis of beta-N-oxalyl-L-alpha, beta-diaminopropionic acid, the Lathyrus sativus neurotoxin. Biochim. Biophys. Acta. 1967;141:71–78. doi: 10.1016/0304-4165(67)90246-2. [DOI] [PubMed] [Google Scholar]
- 22.Johnston G.A., Lloyd H.J. Oxalyl-coenzyme a synthetase and the neurotoxin beta-n-oxalyl-l-alpha, beta-diaminopropionate. Aust. J. Biol. Sci. 1967;20:1241–1244. doi: 10.1071/bi9671241. [DOI] [PubMed] [Google Scholar]
- 23.Malathi K., Padmanaban G., Sarma P.S. Biosynthesis of beta-N-Oxalyl-L-Alpha,Beta-Diaminopropionic acid, Lathyrus-sativus neurotoxin. Phytochemistry. 1970;9:1603–+. doi: 10.1016/0304-4165(67)90246-2. [DOI] [PubMed] [Google Scholar]
- 24.Jiao C.J., Xu Q.L., Wang C.Y., Li F.M., Li Z.X., Wang Y.F. Accumulation pattern of toxin beta-ODAP during lifespan and effect of nutrient elements on beta-ODAP content in Lathyrus sativus seedlings. J. Agr. Sci-cambridge. 2006;144:369–375. [Google Scholar]
- 25.Rao S.L. A sensitive and specific colorimetric method for the determination of alpha, beta-diaminopropionic acid and the Lathyrus sativus neurotoxin. Anal. Biochem. 1978;86:386–396. doi: 10.1016/0003-2697(78)90762-5. [DOI] [PubMed] [Google Scholar]
- 26.Peter D.M., Vogeli B., Cortina N.S., Erb T.J. A chemo-enzymatic road map to the synthesis of CoA esters. Molecules. 2016;21:517. doi: 10.3390/molecules21040517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goldsmith M., Barad S., Peleg Y., Albeck S., Dym O., Brandis A., Mehlman T., Reich Z. The identification and characterization of an oxalyl-CoA synthetase from grass pea (Lathyrus sativus L.) RSC Chem. Biol. 2022;3:320–333. doi: 10.1039/d1cb00202c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuo Y.H., Lambein F. Biosynthesis of the neurotoxin beta-N-Oxalyl-Alpha,Beta-Diaminopropionic acid in callus-tissue of Lathyrus-sativus. Phytochemistry. 1991;30:3241–3244. [Google Scholar]
- 29.Lambein F., Kuo Y.H., Kusama-Eguchi K., Ikegami F. 3-N-oxalyl-L-2,3-diaminopropanoic acid, a multifunctional plant metabolite of toxic reputation. Arkivoc. 2007;2007:45–52. [Google Scholar]
- 30.Ulloa P., Mera M. Inheritance of seed weight in large-seed grass pea Lathyrus sativus L. Chil J. Agr Res. 2010;70:357–364. [Google Scholar]
- 31.Kumar S., Gupta P., Barpete S., Sarker A., Amri A., Mathur P.N., Baum M. In: Genetic and genomic resources of grain legume improvement. Singh M., Upadhyaya H.D., Bisht I.S., editors. Elsevier; Amsterdam: 2013. Grass Pea; pp. 269–292. [Google Scholar]
- 32.Zewdu A.D., Solomon W.K. Moisture-dependent physical properties of grass pea (Lathyrus sativus L.) seeds. Agric. Eng. Int. 2008;X:1–14. [Google Scholar]
- 33.Ikegami F., Ongena G., Sakai R., Itagaki S., Kobori M., Ishikawa T., Kuo Y.H., Lambein F., Murakoshi I. Biosynthesis of beta-(isoxazolin-5-on-2-Yl)-L-alanine by cysteine synthase in Lathyrus-sativus. Phytochemistry. 1993;33:93–98. [Google Scholar]
- 34.Amiola R.O., Ademakinwa A.N., Ayinla Z.A., Ezima E.N., Agboola F.K. Purification and biochemical characterization of a β-cyanoalanine synthase expressed in germinating seeds of Sorghum bicolor (L.) moench. Turkish J. Biochem. 2018;43:638–650. [Google Scholar]
- 35.Bar-Even A., Noor E., Savir Y., Liebermeister W., Davidi D., Tawfik D.S., Milo R. The moderately efficient enzyme: Evolutionary and physicochemical trends shaping enzyme parameters. Biochemistry. 2011;50:4402–4410. doi: 10.1021/bi2002289. [DOI] [PubMed] [Google Scholar]
- 36.Wu G., Bowlus S.B., Kim K.S., Haskell B.E. L-2-Oxalylamino-3-Aminopropionic acid, an isomer of Lathyrus-sativus neurotoxin. Phytochemistry. 1976;15:1257–1259. [Google Scholar]
- 37.D'Auria J.C. Acyltransferases in plants: A good time to be BAHD. Curr. Opin. Plant Biol. 2006;9:331–340. doi: 10.1016/j.pbi.2006.03.016. [DOI] [PubMed] [Google Scholar]
- 38.Molina I., Kosma D. Role of HXXXD-motif/BAHD acyltransferases in the biosynthesis of extracellular lipids. Plant Cell Rep. 2015;34:587–601. doi: 10.1007/s00299-014-1721-5. [DOI] [PubMed] [Google Scholar]
- 39.Ma X., Koepke J., Panjikar S., Fritzsch G., Stockigt J. Crystal structure of vinorine synthase, the first representative of the BAHD superfamily. J. Biol. Chem. 2005;280:13576–13583. doi: 10.1074/jbc.M414508200. [DOI] [PubMed] [Google Scholar]
- 40.Emmrich P.M.F. 2017. Genetic Improvement of Grass Pea (Lathyrus Sativus) for Low β-L-ODAP Content. Ph.D. Ph.D, The university of east anglia. [Google Scholar]
- 41.Tuominen L.K., Johnson V.E., Tsai C.J. Differential phylogenetic expansions in BAHD acyltransferases across five angiosperm taxa and evidence of divergent expression among Populus paralogues. BMC Genomics. 2011;12:236. doi: 10.1186/1471-2164-12-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bontpart T., Cheynier V., Ageorges A., Terrier N. BAHD or SCPL acyltransferase? What a dilemma for acylation in the world of plant phenolic compounds. New Phytol. 2015;208:695–707. doi: 10.1111/nph.13498. [DOI] [PubMed] [Google Scholar]
- 43.Panikashvili D., Shi J.X., Schreiber L., Aharoni A. The Arabidopsis DCR encoding a soluble BAHD acyltransferase is required for cutin polyester formation and seed hydration properties. Plant Physiol. 2009;151:1773–1789. doi: 10.1104/pp.109.143388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eudes A., Pereira J.H., Yogiswara S., Wang G., Teixeira Benites V., Baidoo E.E., Lee T.S., Adams P.D., Keasling J.D., Loque D. Exploiting the substrate promiscuity of hydroxycinnamoyl-CoA:shikimate hydroxycinnamoyl transferase to reduce lignin. Plant Cell Physiol. 2016;57:568–579. doi: 10.1093/pcp/pcw016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Burgi H.B., D J.D., Lehn J.M., Wipff G. Stereochemistry of reaction paths at carbonyl centers. Tetrahedron. 1974;30:1563–1572. [Google Scholar]
- 46.Lubkowski J., Wlodawer A. Geometric considerations support the double-displacement catalytic mechanism of l-asparaginase. Protein Sci. 2019;28:1850–1864. doi: 10.1002/pro.3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chakrabarti P., Pal D. An electrophile-nucleophile interaction in metalloprotein structures. Protein Sci. 1997;6:851–859. doi: 10.1002/pro.5560060412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yasin Quereshi M., Pilbeam David J., Evans Christine S., Arthur Bell E. The neurolathyrogen, α-amino-β-oxalylaminopropionic acid in legume seeds. Phytochemistry. 1977;16:477–479. [Google Scholar]
- 49.Bell E.A. Relevance of biochemical taxonomy to the problem of lathyrism. Nature. 1964;203:378–380. doi: 10.1038/203378a0. [DOI] [PubMed] [Google Scholar]
- 50.Long Y.C., Ye Y.H., Xing Q.Y. Studies on the neuroexcitotoxin beta-N-oxalo-L-alpha,beta-diaminopropionic acid and its isomer alpha-N-oxalo-L-alpha,beta-diaminopropionic acid from the root of Panax species. Int. J. Pept. Protein Res. 1996;47:42–46. doi: 10.1111/j.1399-3011.1996.tb00808.x. [DOI] [PubMed] [Google Scholar]
- 51.Campbell C.G., Mehra R.B., Agrawal S.K., Chen Y.Z., Elmoneim A.M.A., Khawaja H.I.T., Yadov C.R., Tay J.U., Araya W.A. Current status and future strategy in breeding grasspea (Lathyrus-Sativus) Euphytica. 1994;73:167–175. [Google Scholar]
- 52.Patto M.C.V., Skiba B., Pang E.C.K., Ochatt S.J., Lambein F., Rubiales D. Lathyrus improvement for resistance against biotic and abiotic stresses: From classical breeding to marker assisted selection. Euphytica. 2006;147:133. [Google Scholar]
- 53.Asadieh B., Krakauer N.Y. Global change in streamflow extremes under climate change over the 21st century. Hydrol. Earth Syst. Sc. 2017;21:5863–5874. [Google Scholar]
- 54.Lambein F., Haque R., Khan J.K., Kebede N., Kuo Y.H. From soil to brain: Zinc deficiency increases the neurotoxicity of Lathyrus sativus and may affect the susceptibility for the motorneurone disease neurolathyrism. Toxicon. 1994;32:461–466. doi: 10.1016/0041-0101(94)90298-4. [DOI] [PubMed] [Google Scholar]
- 55.Zhang J., Xing G.M., Yan Z.Y., Li Z.X. beta-N-oxalyl-L-alpha,beta-diaminopropionic acid protects the activity of glycolate oxidase in Lathyrus sativus seedlings under high light. Russ. J. Plant Physl. 2003;50:618–622. [Google Scholar]
- 56.Jiao C.J., Jiang J.L., Li C., Ke L.M., Cheng W., Li F.M., Li Z.X., Wang C.Y. beta-ODAP accumulation could be related to low levels of superoxide anion and hydrogen peroxide in Lathyrus sativus L. Food Chem. Toxicol. 2011;49:556–562. doi: 10.1016/j.fct.2010.05.054. [DOI] [PubMed] [Google Scholar]
- 57.Xing G., Cui K., Ji L., Wang Y., Li Z. Water stress and accumulation of beta-N-oxalyl-L-alpha,beta-diaminopropionic acid in grass pea (Lathyrus sativus) J. Agric. Food Chem. 2001;49:216–220. doi: 10.1021/jf000912k. [DOI] [PubMed] [Google Scholar]
- 58.Xiong Y.C., Xing G.M., Li F.M., Wang S.M., Fan X.W., Li Z.X., Wang Y.F. Abscisic acid promotes accumulation of toxin ODAP in relation to free spermine level in grass pea seedlings (Lathyrus sativus L.) Plant Physiol. Biochem. 2006;44:161–169. doi: 10.1016/j.plaphy.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 59.Jiang J.L., Su M., Chen Y.R., Gao N., Jiao C.J., Sun Z.X., Li F.N., Wang C.Y. Correlation of drought resistance in grass pea (Lathyrus sativus) with reactive oxygen species scavenging and osmotic adjustment. Biologia. 2013;68:231–240. [Google Scholar]
- 60.Cohen S.A., Michaud D.P. Synthesis of a fluorescent derivatizing reagent, 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate, and its application for the analysis of hydrolysate amino acids via high-performance liquid chromatography. Anal. Biochem. 1993;211:279–287. doi: 10.1006/abio.1993.1270. [DOI] [PubMed] [Google Scholar]
- 61.Belay A., Ruzgas T., Csoregi E., Moges G., Tessema M., Solomon T., Gorton L. LC-biosensor system for the determination of the neurotoxin beta-N-Oxalyl-l-alpha,beta-diaminopropionic acid. Anal. Chem. 1997;69:3471–3475. doi: 10.1021/ac961098w. [DOI] [PubMed] [Google Scholar]
- 62.Shimizu K., Adachi J., Muraoka Y. Angle: A sequencing errors resistant program for predicting protein coding regions in unfinished cDNA. J. Bioinform. Comput. Biol. 2006;4:649–664. doi: 10.1142/s0219720006002260. [DOI] [PubMed] [Google Scholar]
- 63.Elinger D., Gabashvili A., Levin Y. Suspension trapping (S-trap) is compatible with typical protein extraction buffers and detergents for bottom-up proteomics. J. Proteome Res. 2019;18:1441–1445. doi: 10.1021/acs.jproteome.8b00891. [DOI] [PubMed] [Google Scholar]
- 64.Eng J.K., McCormack A.L., Yates J.R. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 65.Perkins D.N., Pappin D.J., Creasy D.M., Cottrell J.S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 66.Frey S., Gorlich D. A new set of highly efficient, tag-cleaving proteases for purifying recombinant proteins. J. Chromatogr. A. 2014;1337:95–105. doi: 10.1016/j.chroma.2014.02.029. [DOI] [PubMed] [Google Scholar]
- 67.Peleg Y., Unger T. Application of high-throughput methodologies to the expression of recombinant proteins in E. coli. Methods Mol. Biol. 2008;426:197–208. doi: 10.1007/978-1-60327-058-8_12. [DOI] [PubMed] [Google Scholar]
- 68.Chen X., Wang F., Chen Q., Qin X.C., Li Z. Analysis of neurotoxin 3-N-oxalyl-L-2,3-diaminopropionic acid and its alpha-isomer in Lathyrus sativus by high-performance liquid chromatography with 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate (AQC) derivatization. J. Agric. Food Chem. 2000;48:3383–3386. doi: 10.1021/jf000033y. [DOI] [PubMed] [Google Scholar]
- 69.Leslie A.G.W., Powell H.R. In: Evolving Methods for Macromolecular Crystallography. Read R.J., Sussman J.L., editors. Springer Netherlands; Dordrecht: 2007. Processing diffraction data with mosflm. [Google Scholar]
- 70.Evans P. Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 71.French S., Wilson K. Treatment of negative intensity observations. Acta Crystallogr. A. 1978;34:517–525. [Google Scholar]
- 72.McCoy A.J. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Afonine P.V., Grosse-Kunstleve R.W., Echols N., Headd J.J., Moriarty N.W., Mustyakimov M., Terwilliger T.C., Urzhumtsev A., Zwart P.H., Adams P.D. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. Section D-Structural Biol. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Emsley P., Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 75.Joosten R.P., Joosten K., Cohen S.X., Vriend G., Perrakis A. Automatic rebuilding and optimization of crystallographic structures in the Protein Data Bank. Bioinformatics. 2011;27:3392–3398. doi: 10.1093/bioinformatics/btr590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen V.B., Arendall W.B., Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. Section D-Structural Biol. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L. BLAST+: Architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Marchler-Bauer A., Lu S., Anderson J.B., Chitsaz F., Derbyshire M.K., DeWeese-Scott C., Fong J.H., Geer L.Y., Geer R.C., Gonzales N.R., Gwadz M., Hurwitz D.I., Jackson J.D., Ke Z., Lanczycki C.J., et al. Cdd: A conserved domain database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., Thompson J.D., Gibson T.J., Higgins D.G. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 80.Felsenstein J. Taylor & Francis; Seattle: 2005. PHYLIP (Phylogeny Inference Package) Version 3.6. Distributed by Author. Department of Genome Sciences, University of Washington, Seattle. [Google Scholar]
- 81.Guindon S., Dufayard J.F., Lefort V., Anisimova M., Hordijk W., Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 82.Letunic I., Bork P. Interactive tree of life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019;47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Robert X., Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sastry G.M., Adzhigirey M., Day T., Annabhimoju R., Sherman W. Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 2013;27:221–234. doi: 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- 86.Friesner R.A., Banks J.L., Murphy R.B., Halgren T.A., Klicic J.J., Mainz D.T., Repasky M.P., Knoll E.H., Shelley M., Perry J.K., Shaw D.E., Francis P., Shenkin P.S. Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 87.Halgren T.A., Murphy R.B., Friesner R.A., Beard H.S., Frye L.L., Pollard W.T., Banks J.L. Glide: A new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J. Med. Chem. 2004;47:1750–1759. doi: 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- 88.Friesner R.A., Murphy R.B., Repasky M.P., Frye L.L., Greenwood J.R., Halgren T.A., Sanschagrin P.C., Mainz D.T. Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem. 2006;49:6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- 89.Gleave A.P. A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol. Biol. 1992;20:1203–1207. doi: 10.1007/BF00028910. [DOI] [PubMed] [Google Scholar]
- 90.Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D.J., Inuganti A., Griss J., Mayer G., Eisenacher M., Perez E., Uszkoreit J., Pfeuffer J., Sachsenberg T., Yilmaz S., et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019;47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.