Summary
Admixture has been a pervasive phenomenon in human history, extensively shaping the patterns of population genetic diversity. There is increasing evidence to suggest that admixture can also facilitate genetic adaptation to local environments, i.e., admixed populations acquire beneficial mutations from source populations, a process that we refer to as “adaptive admixture.” However, the role of adaptive admixture in human evolution and the power to detect it remain poorly characterized. Here, we use extensive computer simulations to evaluate the power of several neutrality statistics to detect natural selection in the admixed population, assuming multiple admixture scenarios. We show that statistics based on admixture proportions, Fadm and LAD, show high power to detect mutations that are beneficial in the admixed population, whereas other statistics, including iHS and FST, falsely detect neutral mutations that have been selected in the source populations only. By combining Fadm and LAD into a single, powerful statistic, we scanned the genomes of 15 worldwide, admixed populations for signatures of adaptive admixture. We confirm that lactase persistence and resistance to malaria have been under adaptive admixture in West Africans and in Malagasy, North Africans, and South Asians, respectively. Our approach also uncovers other cases of adaptive admixture, including APOL1 in Fulani nomads and PKN2 in East Indonesians, involved in resistance to infection and metabolism, respectively. Collectively, our study provides evidence that adaptive admixture has occurred in human populations whose genetic history is characterized by periods of isolation and spatial expansions resulting in increased gene flow.
Keywords: admixture, positive selection, genetic adaptation, human population genetics, genome scan
Introduction
Over the last two decades, the search for molecular signatures of natural selection in the human genome has played an integral part in understanding human evolution and population differences in disease risk.1, 2, 3, 4, 5, 6 Genome scans for local adaptation have shed light on the environmental pressures that populations have faced for the last 100,000 years, including reduced exposure to sunlight, altitude-related hypoxia, new nutritional resources, or exposure to local pathogens. Candidate genes for local genetic adaptation have been identified on the basis of expected signatures of positive selection, such as extended haplotype homozygosity or strong differences in allele frequencies between geographically diverse populations. In doing so, selection studies have implicitly assumed that advantageous variation occurred in a single population that has remained isolated from other populations since their separation. Yet, ancient and modern genomics studies have clearly demonstrated that the last millennia of human history have been characterized by large-scale spatial expansions followed by extensive gene flow.1,7,8 These findings indicate that most human populations descend from admixture between formerly isolated groups, highlighting the need for detailed studies of the expected genomic signatures of natural selection in admixed populations.
Several studies have searched for evidence of genetic adaptation in admixed populations as a means to detect genes under positive selection in their ancestral sources prior to admixture.9, 10, 11, 12, 13, 14, 15 These studies showed that admixture can obscure signals of selective sweeps in the source populations and proposed approaches to alleviate this problem, such as local ancestry masking. Conversely, few studies have yet explored the patterns of diversity expected under admixture with selection as a means to detect genes under positive selection in the admixed population since admixture.16,17 Studying the genomic signatures of “adaptive admixture,” that is, positive selection in the admixed population of an allele that was beneficial in one of its ancestral sources, could shed light on the role of gene flow in spreading beneficial alleles among populations18 and the prevalence of recent, ongoing selection in humans.
While an increasing number of studies have revealed how introgression from ancient hominins, such as Neanderthals or Denisovans, facilitated genetic adaptation in modern humans,19 the occurrence of adaptive admixture among modern humans remains largely unexplored. Nonetheless, several empirical studies have reported candidate loci for positive selection in admixed populations.16,20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38 A striking example is the Duffy null FY∗BES allele, which confers protection against Plasmodium vivax malaria.39,40 Selection signals have been detected at the locus in diverse African-descent admixed populations from Madagascar, Cabo Verde, Sudan, and Pakistan,22,29,30,32,34 suggesting strong, ongoing selection owing to vivax malaria in these regions. A variety of methods has been used to detect the signatures of adaptive admixture, relying on classic neutrality statistics, such as iHS or FST, and deviations from allele frequencies22,41 or admixture proportions21,24,27,30,31,34, 35, 36, 37, 38 expected under admixture and neutrality.42 However, little is known about how these neutrality statistics behave under scenarios of admixture with selection and, therefore, about the power of these statistics to detect adaptive admixture. More worrying, it has been suggested that artifactual signals of adaptive admixture can be observed because of errors in local ancestry inference (LAI) in complex genomic regions43,44 and/or when the populations used as ancestral sources are poor proxies of the true source populations.16,31 Lastly, reported signals of adaptive admixture are still limited to few populations relative to the large number of admixture events reported in humans.1,7,8
In this study, we compared the power of various neutrality statistics to detect adaptive admixture through computer simulations under different admixture with selection scenarios. We then used a combination of the most powerful statistics to scan the genomes of 15 different admixed human populations from around the world and detect candidate loci for adaptive admixture. In doing so, we confirm several, iconic signals of ongoing positive selection since admixture and identify cases that highlight pathogens as key drivers of recent genetic adaptation in humans.
Material and methods
General simulation settings
All the simulations were computed with the SLiM 3.2 engine45 under the Wright-Fisher model. Each simulation consisted of a 2-Mb long locus characterized by varying recombination and mutation rates. For each simulation, we sampled the physical coordinates of a random 2-Mb genomic window in the human genome, excluding telomeric and centromeric regions, and assigned recombination rates based on the 1000 Genomes phase 3 genetic map46 and mutation rates based on Francioli et al. mutation map.47 To account for background selection, which is thought to be prevalent in the human genome and could affect the power of neutrality tests,48 we simulated exon-like genetic elements positioned according to the position of exons in the sampled 2-Mb genomic window. Each simulated exon is made of positions under negative selection or under neutrality, mimicking non-synonymous and synonymous positions, respectively. We set deleterious mutations to occur three times more frequently than neutral mutations to account for codon degeneracy. The fitness effects of deleterious mutations were sampled from the gamma distribution inferred in Europeans by Boyko and colleagues.49 For simulations that include positive selection, the beneficial mutation was set to appear in the middle of the 2-Mb simulated locus and assumed to be semi dominant. Because we used computationally intensive forward-in-time simulations, we rescaled population sizes and times according to and , with , and used rescaled mutation, recombination, and selection parameters, , , and .45 Of note, we found that simulating background selection has little impact on the power to detect alleles under strong positive selection in the admixed population (s ≥ 0.05; data not shown).
Admixture with selection models
We performed simulations of a population that originates from admixture between two source populations, referred to as P1 and P2 (Figure S1). We assumed that P1 and P2 contributed α1 and α2 admixture proportions to the admixed population, with α1 + α2 = 1. We also assumed that P1 and P2 diverged Tdiv generations ago and the single-pulse admixture event occurred Tadm generations ago. We simulated three scenarios of admixture with selection (Figures 1 and S2). For scenarios 1 and 2, a beneficial mutation was set to appear in the P1 source population and is transmitted to the admixed population with either the same selection coefficient (scenario 1) or a selection coefficient set to 0 (scenario 2). For scenario 3, we adapted a combination of recipes 9.6.2 and 14.7 from the SLiM manual,50 introducing a set of “ancestry marker” neutral mutations in the P1 source population, and randomly choosing one of them to become beneficial by setting its selection coefficient to s > 0 in the admixed population only. We computed 500 simulations for each admixture with selection scenario, as well as 500 simulations for the null scenario (i.e., no positive selection). Because the goal of these simulations was to compare the power of neutrality statistics to detect positive selection, only the selection coefficient of the beneficial mutation s was given different values, ranging from s = 0.01 to s = 0.05. All the other parameters were given fixed values: population sizes of source and admixed populations N = 10,000; divergence time between source populations Tdiv = 2,000 generations; admixture proportions α1 = 0.35 and α2 = 0.65; time of the single pulse admixture event Tadm = 70 generations; time when the beneficial mutation appears Tmut = 350 generations ago.
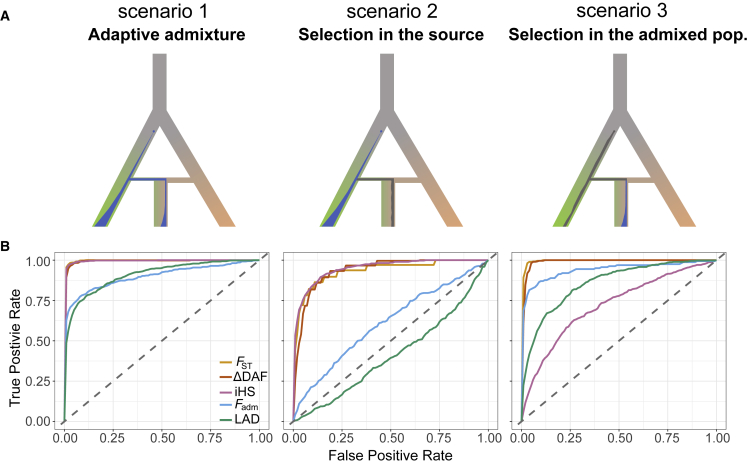
Figure 1.
Performance of neutrality statistics under different scenarios of admixture with selection
(A) Explored scenarios of admixture with selection (from left to right): adaptive admixture, positive selection in the source population only, and positive selection in the admixed population only. The blue and gray points indicate the appearance of a new beneficial or neutral mutation, respectively. The blue and gray areas indicate changes in frequency of the beneficial and neutral mutation, respectively.
(B) Receiver operating characteristic (ROC) curves comparing the performance of classic neutrality statistics FST, iHS, and ΔDAF and the admixture-specific statistics Fadm and LAD across the three explored scenarios. The selection coefficient was fixed to s = 0.05 to highlight the differences between statistics and between models (see Figure S2 for lower s values). False positive rate (FPR) is the fraction of simulated neutral sites that are incorrectly detected as under selection, and true positive rate (TPR) is the fraction of simulated adaptive mutations that are correctly detected as under selection.
Power of explored neutrality statistics
Neutrality statistics were computed for all genetic variants within the 2-Mb simulated loci under no positive selection (H0) and only for the selected mutation for simulated 2-Mb loci under positive selection (H1). We estimated detection power (i.e., the true positive rate [TPR]) for each statistic as the proportion of values under H1 that are above a varying threshold value under H0, corresponding to a given false positive rate (FPR). We computed FST, ΔDAF, and iHS by using selink.51 We computed FST and ΔDAF between the admixed population and the source population that does not experience positive selection. For iHS, we used a 200-kb window and normalized the values by bins of similar derived allele frequency (DAF).
For the admixture-specific statistics, we introduced an allele frequency-based statistic, Fadm, that measures the difference between , the observed frequency of allele i in the admixed population, and , the expected allele frequency under admixture and neutrality. It was shown that , which is the average of allele frequencies observed in the source populations weighted by estimated admixture proportions , where (Bernstein52). Under neutrality, the squared difference between and , , is the variance of allele frequencies in the admixed population due to genetic drift.42 Thus, can be interpreted as the genetic distance between the current admixed population and its ancestral population at the time of admixture. Analogously to FST, this genetic distance can be used to detect natural selection, as the change in frequency of a beneficial allele in time depends on its selection coefficient.53 Fadm is thus defined as follows:
where is the expected heterozygosity in the admixed population, used here to allow comparisons among SNPs.
When calculating Fadm in the simulated and observed data, the allele frequencies at the time of admixture were estimated by the allele frequencies in the current generation, which is accurate when genetic drift in source populations is weak or when admixture is recent. We used as admixture proportions the simulated proportions αsim, for the simulated data, and the estimated proportions , for the observed data, obtained by running ADMIXTURE v.1.23 (Alexander et al.;54 see empirical detection of adaptive admixture). We verified with simulations that errors in the estimation of admixture proportions do not affect Fadm detection power (Figure S3A) by computing Fadm with α sampled from a normal distribution (μ = αsim, σ2 = 0.0262); 0.026 is the highest root-mean-square deviation of the ADMIXTURE estimation.54 Additionally, we excluded sites where the observed allele frequency in the admixed population is higher (or lower) than the maximum (or minimum) of the frequencies in the source populations. Although this can reduce the detection power in scenario 3, this filter increases power for the adaptive admixture scenario (Figure S3B), which is the focus of this study.
We also computed an LAI-based neutrality statistic, LAD, which measures the local ancestry deviation from the average genome-wide ancestry, defined as follows:
where is the admixture proportion from source population p for a given window and is the estimated genome-wide admixture proportion. Natural selection has been proposed to bias the estimation of admixture proportions since the first estimates of this parameter were obtained.55, 56, 57 The rationale is that, when a beneficial allele is transmitted from a source population to the admixed population, estimated admixture proportions from this source population are expected to increase at the locus relative to neutral loci. As single-marker estimates of admixture proportions are sensitive to errors in the estimation of allele frequencies, more powerful haplotype-based methods have preferentially been used to detect natural selection since admixture.38
We used RFMix v1.5.4 to estimate local ancestry,58 with default parameter values (except for –G, which was replaced with the simulated Tadm value) and using the forward-backward option with three expectation maximization steps. Because LAD is sensitive to phasing errors,58 we incorporated potential phasing errors in our simulations by phasing, with SHAPEIT v.4.2.1 (Delaneau et al.59), unphased diploid individuals obtained from the combination of two simulated haploid individuals. Admixture proportions were estimated as the local ancestry inferred by RFMix averaged across loci for both the simulated and observed data.
Sample size and source population choice scenarios
We explored five different values of sample sizes for the two source populations and the admixed population: n = 20, 50, 100, 200, and 500 individuals (Figures 2A and S4). When exploring the values for a given population, sample size for the other two was fixed to n = 50 individuals. For the use of a proxy source population (Figure 2B), we simulated two additional populations that diverge 400 generations ago from each of the two source populations. We then used these proxy populations for Fadm and LAD calculations. To explore the effect of the genetic distance (estimated by FST) between the proxy population and the true source population on detection power, we set the population size of the proxies to 10,000, 4,000, 1,000, and 500, resulting in FST values of 0.005, 0.01, 0.02, and 0.03, respectively.
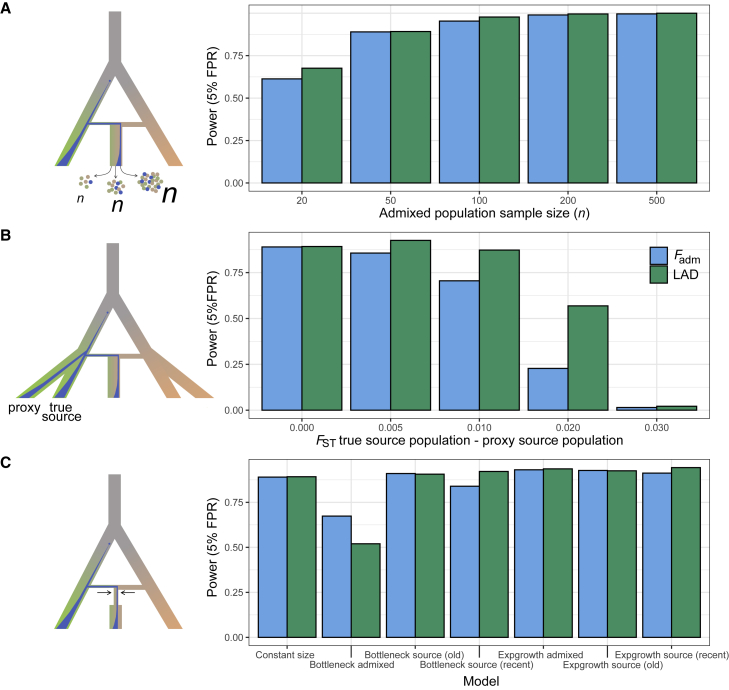
Figure 2.
Effects of study design and non-stationary demography on the power to detect adaptive admixture
(A) Effects of the sample size of the admixed population (n) on the detection power of Fadm and LAD at a fixed FPR = 5%. The simulated model is shown on the left, illustrating different values of n.
(B) Effects of the use of proxy source populations on the detection power of Fadm and LAD at a fixed FPR = 5%. The genetic distance between the true source and its proxy was measured by FST. The simulated model is shown on the left, including the true source and its proxy.
(C) Effects of non-stationary demography on the detection power of Fadm and LAD at a fixed FPR = 5%. The simulated model is shown on the left, including a bottleneck in the admixed population.
For the scenario of selection in the proxy source population only (Figure S5), we simulated two additional populations that diverge 600 generations ago from each of the two source populations. We randomly selected a mutation that occurred in the ancestral population of the P1 source population and its related proxy population and assigned it a selection coefficient of s = 0.02 in the proxy population only, 599 generations ago. Under the latter scenario, Fadm and LAD detect mutations that are not beneficial in the admixed population and wrongly support positive selection in the P2 source population (Figures S5A and S5B). For comparison purposes, we thus compared Fadm and LAD distributions under this scenario to those obtained under a simple scenario of adaptive admixture (scenario 1, Figure 1) where the beneficial mutation is transmitted to the admixed population from the P2 source population (Figure S5D). In all these scenarios, the following parameters were given a fixed value: N = 10,000; Tdiv = 2,000 generations; α1 = 0.35; α2 = 0.65; Tadm = 70 generations; s = 0.02; and Tmut = 1,400 generations ago.
Complex admixture scenarios
We estimated detection power under two additional admixture scenarios: a double pulse model and a constant continuous model (Figures S6A and S6B). For these scenarios to be comparable to the single pulse admixture scenario, we set the sum of the admixture proportions contributed by each pulse to be equal to α1 = 35% and the average of the admixture dates to be equal to 70 generations. Namely, under the double pulse model, the admixed population originates from an admixture event that occurs 130 generations ago between two source populations, with α1 = 17.5%, and receives a second admixture pulse from P1 10 generations ago with α1 = 17.5%. Under the constant continuous model, the admixed population also originates from an admixture event occurring 130 generations ago between two source populations with α1 = 35%/130 = 0.27%, when Tadm is not rescaled, and a1 = 2.7%, when Tadm is rescaled, but receives an additional pulse from P1 of α1 = 2.7% at each generation until present. In all scenarios, the following parameters were given a fixed value: N = 10,000; Tdiv = 2,000 generations; s = 0.02; and Tmut = 1,400 generations ago.
Admixture parameters
Under the single pulse admixture model (Figure S1), we explored detection power as a function of different model parameters (Figures 3 and S7–S11; Table S1). In total, 32,956 compatible parameter combinations were explored. We thus reduced the number of simulations per combination from 500 to 100 to limit computational burden. For the frequency of the beneficial mutation in the source population at the time of admixture, instead of conditioning on the frequency within simulations (which would have drastically increased computations), we introduced the beneficial mutation Tmut generations ago in the source population on the basis of previous results.60 For each statistic and parameter combination, we calculated the proportion of simulated sites under selection that were recovered by using a threshold of FPR = 5%. We then averaged the power across demographic parameter values to obtain a single value for each combination of Tadm, α1, and s. We performed a similar procedure to obtain a single value for each combination of Tadm, α1, and one of the other parameters (e.g., Tdiv and N; Figures S7–S11).
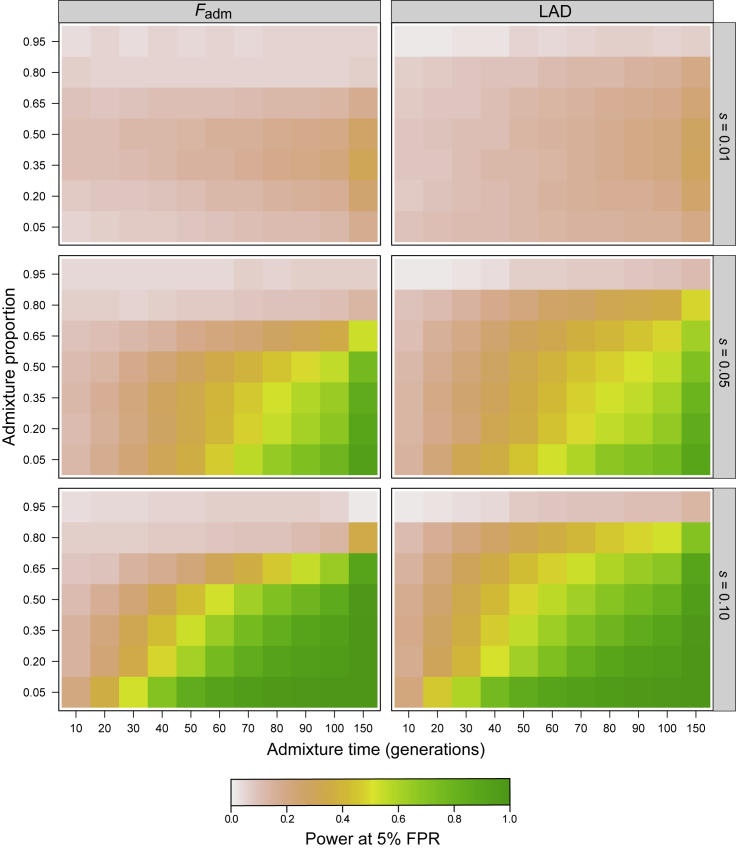
Figure 3.
Effects of model parameters on the power to detect adaptive admixture
Color represents average detection power for a fixed FPR = 5% across parameter combinations. The effects of other parameters, such as population sizes and divergence time (Figure S1 and Table S1), are shown in Figures S7–S11.
Non-stationary demography
We estimated detection power under five alternative demographic scenarios (Figures 2C and S6C), each with 500 simulations under adaptive admixture and 500 simulations with no positive selection. Demographic scenarios include: (1) a recent expansion of the source population, where the source population undergoes an expansion with a 5% growth rate since Tadm, from an initial N = 10,000; (2) a recent expansion of the admixed population, where the admixed population undergoes an expansion with a 5% growth rate since Tadm, from an initial N = 10,000; (3) an old expansion of the source population, where the source population undergoes an expansion with a 5% growth rate since Tadm + 500 generations, from an initial N = 10,000; (4) an old bottleneck in the source population, where the source population undergoes a 10-fold size reduction from Tdiv – 50 to Tdiv, from an initial N = 10,000; and (5) a recent bottleneck in the admixed population, where the admixed population undergoes a 10-fold size reduction from Tadm – 50 to Tadm, from an initial N = 10,000. We compared these scenarios to a constant population size scenario and the size of all populations fixed to N = 10,000. In all scenarios, the following parameters were given a fixed value: Tdiv = 2,000 generations; α1 = 0.35; α2 = 0.65; Tadm = 70 generations; s = 0.02; and Tmut = 1,400 generations ago.
Empirical detection of adaptive admixture
We analyzed the genomes of 15 admixed populations to search for signals of adaptive admixture. The datasets and references for all admixed and source populations can be found in Table S2, as well as the final number of SNPs used after merging the datasets for admixed and source populations. For each merged dataset, we (1) excluded sites with a proportion of missing genotypes > 5% via PLINK v.2.0 (Chang et al.61), (2) excluded A/T and C/G variant sites, (3) excluded first- and second-degree-related individuals (kinship coefficient > 0.08 computed with KING v2.2.2; Manichaikul et al.62), and (4) performed phasing by using SHAPEIT v.4.2.1 with default parameter values. Additionally, we verified the validity of the admixture model for each set of source/admixed populations (Table S2) by computing admixture f3 statistics with admixr package v.0.7.1 (Petr et al.63).
We obtained admixture proportions by running ADMIXTURE v.1.23, considering the K value producing the lowest cross-validation error and a set of “independent” SNPs obtained by running the “--indep-pairwise” command with PLINK v.2.0 with the following parameters: 50-SNP window, 5-SNP step, and r2 threshold of 0.5. We verified for each studied admixed population that the K value with the fewest cross-validation errors matches the number of source populations. Local ancestry was inferred with RFMix v.1.5.4, after excluding 2 Mb at telomeres and centromeres of each chromosome, as well as invariant sites and singletons, and with default parameter values except for the generation time “-G,” which was given a value based on literature (Table S2).
We combined the SNP ranks for Fadm and LAD statistics by using Fisher’s method,64 defined as follows:
where ri is defined as the rank of a given SNP for the statistic i, divided by the total number of analyzed SNPs (i.e., the empirical p value), and k = 2 is the number of statistics.
Using simulations, we verified that this statistic followed a chi-squared distribution with 2k = 4 degrees of freedom under no positive selection (Figure 4A), including when the admixed population experienced a 10-fold bottleneck. In these simulations, we used the same parameter values as those in Figure 2C for the “constant size” and “bottleneck in the admixed population” scenarios. Statistical significance was defined based on Bonferroni correction: we considered a p value threshold of 0.05 divided by the number of 0.2-cM RFMix windows analyzed (all SNPs within the same window had the same local ancestry value), which yielded, on average, a p value threshold of 3.5 × 10−6 (Table S3). To reduce the number of false positives due to positive selection in a proxy source population only (Figure S5), we computed iHS in the two source populations and excluded from the list of candidate genes any locus that includes SNPs with both an |iHS| > 2 in one source population and an excess of local ancestry from the other source population (Figure S5). To annotate the different signals that passed this threshold, we chose the protein-coding gene within 250 kb of the variant with the highest V2G score.65
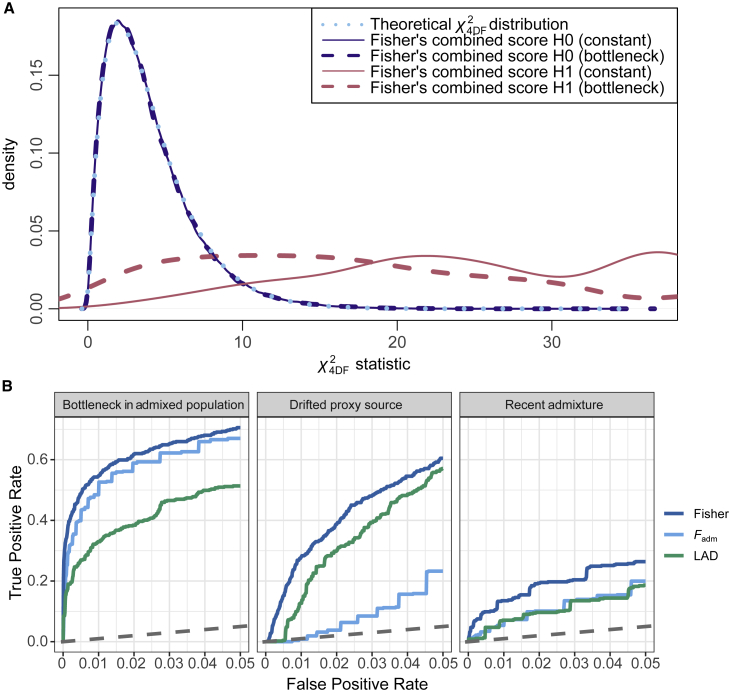
Figure 4.
Performance of the Fisher’s method to detect adaptive admixture
(A) Distributions of the combined Fisher’s score under the null hypothesis of no positive selection (H0, blue lines) and under adaptive admixture (H1, pink lines), compared to the theoretical chi-squared distribution with 4 degrees of freedom (dotted light blue line). Solid and dashed lines indicate distributions under a constant population size and a 10-fold bottleneck in the admixed population, respectively.
(B) ROC curves for Fadm, LAD, and the combined Fisher’s score under unfavorable scenarios for detecting adaptive admixture: a 10-fold bottleneck introduced in the admixed population, the use of a proxy source population having experienced strong drift (FST between the true source and proxy populations of 0.02), and recent admixture (Tadm = 10 generations). Only FPR < 5% are shown.
Results
Power estimation under different models of admixture with selection
To estimate the power to detect positive selection in admixed populations, we performed extensive forward-in-time simulations of a population that originates from admixture between two source populations (Figure S1). We introduced a beneficial mutation in one of the source populations, with a varying selection coefficient (material and methods). We considered three different scenarios of admixture with selection (Figures 1A and S2). Scenario 1 corresponds to adaptive admixture, where the admixed population inherits an allele that is beneficial in one of its source populations: the mutation is under positive selection in the source population, is transmitted to the admixed population, and remains beneficial—with the same selection coefficient—in the admixed population. In scenario 2, the beneficial allele is under positive selection in the source population, is transmitted to the admixed population, and becomes neutral in the admixed population only. We simulated this scenario to verify whether some neutrality statistics wrongly support positive selection in the admixed population because of a residual signal inherited from the source population. At the same time, this scenario is also useful for evaluating the power to detect residual signals of positive selection in the admixed population, as a means to detect genes under positive selection in source populations that no longer exist in an unadmixed form.9, 10, 11, 12, 13, 14, 15 Finally, in scenario 3, a neutral mutation in the source population becomes beneficial in the admixed population only, at the time of admixture. This case is used for determining how neutrality statistics behave when natural selection operates since admixture on standing neutral variation.
We evaluated the performance, under each scenario, of three classic neutrality statistics, FST, ΔDAF, and iHS, as well as two statistics that are specifically designed to detect selection in the admixed population: Fadm, which is proportional to the squared difference between the observed and the expected allele frequency in the admixed population,22,42,57 and LAD, the difference between the admixture proportion at the locus and its genome-wide average,38 estimated on the basis of local ancestry inference (LAI) by RFMix (material and methods).58 Receiver operating characteristic (ROC) curves indicate that both the classic neutrality statistics and Fadm and LAD are powerful to detect adaptive admixture (scenario 1) when the selection coefficient s = 0.05 (>70% detection power for a false positive rate [FPR] of 5%; Figures 1B and S2), in agreement with a previous study.16 Nevertheless, the power of FST, ΔDAF, and iHS is also high when the mutation is beneficial in the source population and is no longer selected in the admixed population (scenario 2), indicating that these statistics wrongly detect selection in the source population as selection in the admixed population. In contrast, Fadm and LAD detect adaptive admixture specifically, as their power under scenario 2 is low or nil (Figure 1B). Of note, our simulations also imply that the power of classic statistics is substantial when using the admixed population as a means to detect selection in the source populations (>65% detection power when s = 0.05 and FPR = 5%). Finally, LAD and iHS showed a reduced power to detect selection in the admixed population when the mutation is neutral in the source populations (scenario 3), relative to the adaptive admixture case (scenario 1). This may stem from the fact that, under scenario 3, the beneficial mutation has been selected for fewer generations than in scenario 1, resulting in a weaker signal. Furthermore, this scenario is similar to selection on standing variation, where the adaptive mutation may be present on several haplotypes, making it harder to detect.66
Collectively, our simulations indicate that Fadm and LAD are the only studied statistics that have substantial power to specifically detect strong, ongoing selection in the admixed population and have more power to detect adaptive admixture than post-admixture selection on standing neutral variation. Because our objective is to detect the signatures of positive selection in the admixed population, and not in the source populations, we based all subsequent analyses on the Fadm and LAD statistics.
Effects of the study design
We investigated how sample size and the choice of source populations affect the power of Fadm and LAD to detect adaptive admixture signals (material and methods). We explored sample sizes ranging from n = 20 to n = 500 for both the admixed and the source populations. We found that n = 100 already provides optimal power because the variance of neutrality statistics is virtually unchanged when n ≥ 100 (Figures 2A and S4A). Conversely, we found that when n < 50, sampling error increases the variance of Fadm and LAD null distributions by as much as 5 times and ultimately decreases detection power by up to 40% (FPR = 5%). Interestingly, LAD detection power is not affected when the sample size of the source populations is low, even when n = 20 (Figure S4B). Consistently, RFMix accuracy was shown to be only minimally reduced when the sample size of reference panels is as small as n = 3, as it uses both source and admixed individuals for LAI.58
Because obtaining genotype data for the true source populations of an admixed population is difficult, if not impossible, population geneticists often use related, present-day populations as proxies, which may lead to false adaptive admixture signals.16,31 We explored how detection power is affected by the genetic distance between the true source population and a related population used as a proxy for Fadm and LAD computations (material and methods). We observed a difference in performance between Fadm and LAD, the latter’s being more robust to the use of a proxy (Figure 2B). LAD maintains similar detection power even if the divergence between the true and proxy populations is FST = 0.01, whereas power decreases by 25% for Fadm. Such a difference in power may result from the nature of the two statistics. In the case of Fadm, the expected allele frequency is directly estimated from the allele frequencies observed in the proxy, and these frequencies are decreasingly correlated with those in the true source population as their divergence increases. On the other hand, LAD is derived from LAI by RFMix, which has been shown to be robust to the use of proxy reference populations.58
Nonetheless, we identified a potentially problematic scenario for both Fadm and LAD involving population proxies: when the selection event occurs specifically in the proxy source population (i.e., the mutation is not selected in both the true source and the admixed populations; Figure S5), spurious deviations in local ancestry and in allele frequencies were observed in the admixed population. Specifically, this generates an excess of local ancestry from the other source population and expected allele frequencies higher than those observed in the admixed population (Figures S5A and S5B). We found that this scenario produces weaker LAD values (i.e., lower detection power) but larger Fadm values (i.e., higher detection power) relative to an adaptive admixture event (Figures S5C–S5F). To remediate this, we performed a selection scan in the proxy population by using a single-population statistic, iHS, and excluded the top 1% values. In doing so, we managed to exclude approximately 90% of the outlier values of Fadm and LAD generated by this scenario. More importantly, because there is no correlation between iHS in the proxy population and Fadm or LAD in the case of adaptive admixture, none of the outlier values generated by a true adaptive admixture event were excluded by this analysis step (Figures S5G and S5H).
Effects of the admixture model and non-stationary demography
Several studies have shown that admixture in humans has often involved multiple admixture pulses from two or more source populations.8,51,67, 68, 69, 70, 71 We thus estimated the detection performance of Fadm and LAD under admixture models that are more complex than the single admixture pulse. We found that the power to detect adaptive admixture is only moderately reduced under a two-pulse admixture model or a constant, continuous admixture model: the true positive rate (TPR) decreases by <11% at a FPR = 5%, relative to the single pulse model (Figures S6A and S6B). This suggests that our power estimations are valid for a variety of admixture models.
Assuming a single-pulse admixture model, we then explored how detection power is impacted by key parameters of the adaptive admixture model, including the strength of selection s, the admixture time Tadm, the admixture proportion α, and the divergence time between source populations Tdiv (Figures 3 and S7–S11; Table S1). As expected, we found that detection power is high only when the selection coefficient s is strong; the TPR is up to 94% and 27% when s = 0.05 and 0.01, respectively (FPR = 5%; Figure 3). Power is also determined by the admixture time Tadm, as it affects the duration of selection; the TPR is up to 94% and 21% when Tadm ≥ 70 and ≤20 generations, respectively. Interestingly, we observed that the higher the admixture proportion α (from the source population where the selected mutation appeared), the lower the detection power. Power decreases particularly when α > 0.65, probably because of a threshold effect: if the beneficial allele is at high frequency and, e.g., α = 0.9, there is little room for the observed allele frequency or local ancestry to deviate from its expectation, making it hard to detect. Finally, as the divergence time between source populations decreases, the detection power of LAD is reduced by ∼15% (51% versus 67% when Tdiv = 500 or 2,000 generations, respectively; Figure S7), whereas that of Fadm is not affected (59% versus 55%, when Tdiv = 500 or 2,000 generations, respectively). The reduced detection power of LAD is probably due to the decreased accuracy of RFMix when Tdiv decreases.72
We also estimated power under scenarios where demography deviates from a constant population size model. Indeed, demographic events, such as bottlenecks, have been shown to alter the performance of several neutrality statistics.73, 74, 75, 76, 77, 78, 79 We simulated five demographic scenarios, including 10-fold bottlenecks and 5% growth rate expansions in either the admixed or the source populations (material and methods). We found that detection power is minimally affected under all expansion models (TPR decrease of 5% at a FPR = 5%; Figure 2C). In contrast, detection power is reduced by as much as 50% under the scenario where a 10-fold bottleneck is introduced in the admixed population, relative to the stationary model. This is mirrored by the increased variance of Fadm and LAD null distributions under this scenario (Figure S6C). Finally, detection power of both Fadm and LAD is minimally affected when the 10-fold bottleneck is introduced in the source populations, either few generations after their divergence or before the admixture pulse (TPR decrease of 5% at a FPR = 5%; Figure 2C), suggesting that both statistics are relatively robust to increased genetic drift occurring in the source populations.
Empirical detection of adaptive admixture in humans
We next sought to detect candidate genes for adaptive admixture in humans by scanning, with both Fadm and LAD statistics, the genomes of 15 worldwide populations (Table S2) that have experienced at least one admixture event in the last 5,000 years (i.e., the upper detection limit set for accurate local ancestry inference80). To improve detection power and facilitate candidate prioritization, we combined the empirical p values of both statistics with Fisher’s method,64 used here as a combined test for positive selection since admixture. We confirmed with simulations that the Fisher’s score follows a chi-squared distribution with 4 degrees of freedom under the null hypothesis of absence of positive selection and when assuming different demographic scenarios (Figure 4A). Consistently, we found that Fadm and LAD statistics are not correlated under the null hypothesis (Spearman’s coefficient = 0.03), whereas they are correlated under adaptive admixture (Spearman’s coefficient = 0.96). Importantly, we found that Fisher’s method increases detection power under unfavorable scenarios, relative to each individual statistic (Figure 4B). In particular, Fisher’s method improves power when the admixed population experienced a 10-fold bottleneck, when admixture is recent (Tadm = 10 generations), or when using a proxy population that experienced strong drift (FST with the true source population = 0.02). Given the limited knowledge on the past population sizes of the studied populations, which could increase FPR (Figure S6C), we applied a conservative Bonferroni correction on Fisher’s p values, considering the number of RFMix genomic windows as the number of tests (all SNPs within a given window have the same value for LAD). This yielded a p value threshold of approximately p = 3.5 × 10−6 (Table S3). Finally, we verified that the empirical distribution of Fisher’s p values is uniform in all studied populations and found an excess of low p values for several populations (Figure S12), suggesting that adaptive admixture has occurred in these groups.
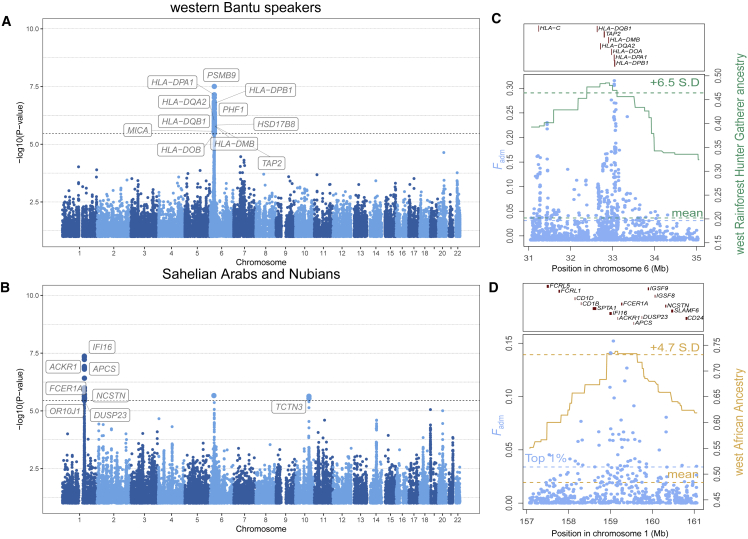
Our genome scans identified a number of previously reported signals of adaptive admixture. Among these, we found the HLA class II locus in Bantu-speaking populations from Gabon31 (Figures 5A and 5C; top ranking SNP identified in HLA-DPA1 [MIM: 142880]; p = 7.9 × 10−8; expected frequency of 0.33 versus observed frequency of 0.70), the HLA class I locus in Mexicans27,35,37,81 (Figure S13; top ranked SNP identified in ABCF1; p = 2.2 × 10−6; expected frequency of 0.013 versus observed frequency of 0.039), the lactase persistence-associated LCT/MCM6 locus (MIM: 223100) in the Fulani nomads of Burkina Faso82 (Figure 6A; top ranked SNP identified in CCNT2 [MIM: 603862]; p = 1.1 × 10−6; expected frequency of 0.12 versus observed frequency of 0.47), and ACKR1 (previously referred as DARC [MIM: 613665]) in African-descent populations from Madagascar, the Sahel, and Pakistan29,30,34 (Figures 5B and 5D and S13). For the latter locus, the top-ranking variant is rs12075 in the Malagasy (p = 3.4 × 10−9; expected frequency 0.45 versus observed frequency of 0.93), as previously found.30 This variant, also known as the Duffy null FY∗BES allele (MIM: 110700), confers resistance against Plasmodium vivax infection in sub-Saharan Africans.39,40 Together, these results confirm that our conservative approach can recover strong, well-documented signals of adaptive admixture.
Figure 5.
Iconic genomic signals of adaptive admixture
(A) Genome-wide signals of adaptive admixture in Bantu-speaking populations from Gabon.
(B) Genome-wide signals of adaptive admixture in Sahelian Arabs and Nubians.
(A and B) Highlighted blue points indicate variants that passed the Bonferroni significance threshold (shown by a horizontal dotted line). Gene labels were attributed on the basis of the gene with the highest V2G score within 250 kb of the candidate variant. p values were obtained assuming that combined Fisher's scores follow a chi-squared distribution under no selection.
(C) Local signatures of adaptive admixture for the HLA region in Bantu-speaking populations from Gabon.
(D) Local signatures of adaptive admixture for the ACKR1 region in Sahelian Arabs and Nubians.
(C and D) Light blue points indicate Fadm values for individual variants. The green and gold solid lines indicate average local ancestry from African rainforest hunter-gatherers and West Africans respectively.
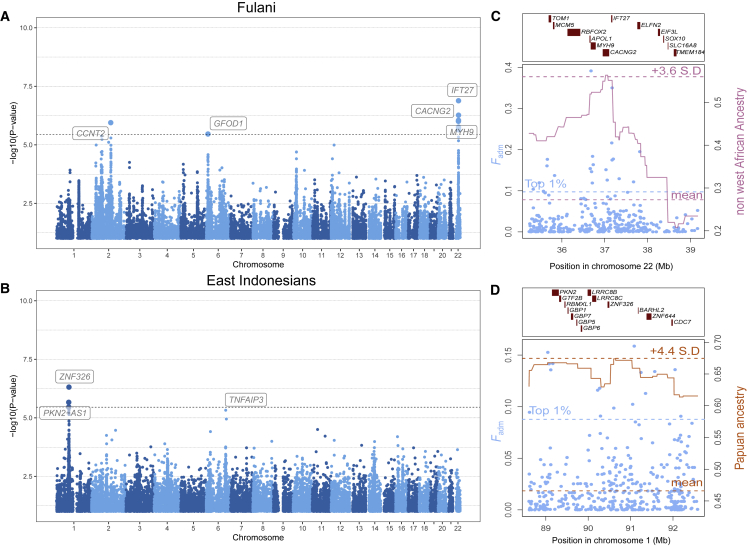
Figure 6.
Genomic signals of adaptive admixture discovered in this study
(A) Genome-wide signals of adaptive admixture in the Fulani nomads of West Africa.
(B) Genome-wide signals of adaptive admixture in East Indonesians.
(A and B) Highlighted blue points indicate variants that pass the Bonferroni significance threshold (shown by a horizontal dotted line). Gene labels were attributed on the basis of the gene with the highest V2G score within 250 kb of the candidate variant. p values were obtained assuming that combined Fisher's scores follow a chi-squared distribution under no selection.
(C) Local signatures of adaptive admixture for the IFT27/MYH9/APOL1 region in the Fulani nomads.
(D) Local signatures of adaptive admixture for the PKN2/LRR8CB region in East Indonesians.
(C and D) Light blue points indicate Fadm values for individual variants. The pink solid line indicates the local ancestry from Europeans and North Africans, and the orange solid line indicates the local ancestry from Papuans.
Candidate genes for adaptive admixture
We found several candidate loci for adaptive admixture (Figures 6 and S14), among which was the MYH9/APOL1 (MIM: 603743) locus in the Fulani (Figures 6A and 6C; p = 1.3 × 10−7; top ranked SNP in IFT27 [MIM: 615870]; expected frequency of 0.15 versus observed frequency of 0.45). Common APOL1 variants confer both protection against human African trypanosomiasis (HAT, or sleep sickness) and susceptibility to common kidney diseases (MIM: 612551) in African-descent individuals.83 Another candidate is the PKN2 (MIM: 602549) locus in East Indonesians (p = 1.1 × 10−6; top ranked SNP in ZNF326 [MIM: 614601]; expected frequency of 0.27 versus observed frequency of 0.46), which shows a large excess of Papuan ancestry (Figures 6B and 6D). PKN2 plays a role in cellular signal transduction responses and has been reported as involved in the regulation of glucose metabolism in skeletal muscle.84 A nearby locus, LRRC8B (MIM: 612888), has been reported as a candidate for positive selection in Solomon Islanders,51 although it did not show signals for adaptive admixture in this population. A unique, strong signal was detected at the ARRDC4/IGF1R (MIM: 147370) locus in Solomon Islanders (p = 7.4 × 10−9; top ranked SNP close to ARRDC4; expected frequency of 0.09 versus observed frequency of 0.58), where an excess of East Asian-related ancestry was observed (Figures S14B and S14F). This locus was previously identified as a candidate for positive selection in near and western remote Oceanians.51 ARRDC4 is an arrestin that plays important roles in glucose metabolism and immune response to enterovirus infection,85 whereas IGF1R, the receptor for the insulin-like growth factor, is a key determinant of body size and growth.86,87 A last example is CXCL13 (MIM: 605149) in the Nama pastoralists from South Africa (Figures S14A and S14E; p = 2.3 × 10−6; top ranked SNP identified in CXCL13; expected frequency of 0.51 versus observed frequency of 0.80). The CNOT6L/CXCL13 locus has previously been reported as suggestively associated with tuberculosis (TB) risk in South African populations with San ancestry.88 However, we found that the top-ranking variants show outlier extended haplotype homozygosity in the Ju/’hoansi San, used as source population (iHS = −3.12), while European ancestry is in excess at the locus in the Nama, suggesting a spurious signal due to positive selection in the proxy source population (Figure S5).
Lastly, we detected suggestive signals of adaptive admixture at genes shown to be strong candidates for positive selection, including the MCM6/LCT locus in the Bantu-speaking Bakiga of Uganda (Figure S15; p = 4.3 × 10−6; top ranked SNP in CCNT2; expected frequency of 0.15 versus observed frequency of 0.31) and TNFAIP3 (MIM: 191163) in East Indonesians, who show an excess of Papuan-related ancestry at the locus (Figure 6B; p = 5.0 × 10−6; top ranked SNP in TNFAIP3; expected frequency of 0.27 versus observed frequency of 0.43). The TNFAIP3 locus has not only been reported as evolving under positive selection in Papuans51 but also as adaptively introgressed from Denisovans.51,89, 90, 91 TNFAIP3 plays an important role in human immune tolerance to pathogen infections.92 Collectively, these results indicate that adaptive admixture has occurred in various admixed populations around the world and highlight the immune system and nutrient metabolism as important targets of recent genetic adaptation.
Discussion
In this study, we evaluated the power of several neutrality statistics to detect loci under positive selection in admixed populations and used these statistics to explore cases of adaptive admixture in the genomes of 15 worldwide human populations. Although Fadm and LAD, or closely related statistics based on the difference between observed and expected allele frequencies and admixture proportions, have been used in several empirical studies, their power has not been thoroughly evaluated. Here, we showed that these statistics are powerful to detect adaptive admixture and have little power to detect residual signals of positive selection in the source populations. Thus, Fadm and LAD are suited to search for loci under positive selection in admixed populations since admixture, particularly when selection is strong (i.e., s ≥ 0.05), admixture is relatively old (i.e., Tadm > 2,000 years) and the admixture proportion is moderate-to-low (i.e., α < 0.6). Notably, we found that power is marginally affected when admixture has been recurrent, a feature that is convenient given the difficulty to distinguish between single-pulse, double-pulse, or more complex admixture models from the genetic data.8,51,67, 68, 69, 70, 71 Furthermore, Fadm is more powerful than LAD when selection occurs in the admixed population only and when the divergence time between source populations is low (Tdiv = 500 generations), whereas LAD is more powerful than Fadm when source sample sizes are low (i.e., n = 20) and when the true and proxy source populations are distantly related (i.e., FST ≥ 0.01; Table S4). The latter result is consistent with the known robustness of LAI to cases where the populations used as reference sources are poor proxies of the true source populations.58 Nonetheless, caution must be taken when handling population proxies, as selection occurring only in the proxy population can produce artifactual genomic signals, for both LAD and Fadm, that might be misinterpreted as adaptive admixture.16,31,51 We suggest that performing selection scans on the proxy source populations can help distinguish false from true adaptive admixture signals. We also caution that Fadm calculation relies on the accurate estimation of admixture proportions, which can be biased under certain scenarios.93 Finally, we found that combining Fadm and LAD statistics into a unique statistic, based on the Fisher’s method, provides well-calibrated p values under different models and substantially increases power under several realistic admixture with selection scenarios, relative to individual statistics.
When applying this combined method on the empirical data, we identified several previously reported candidate variants for adaptive admixture. These include the ACKR1 Duffy null allele detected in admixed populations from Madagascar,30 the Sahel,34 and Pakistan,29 the lactase persistence c.−13910C>T LCT allele in the Fulani from West Africa,82 and HLA alleles in Bantu-speaking populations from western Central Africa31 and Mexicans.27,35,37,81 These candidate loci were detected previously on the basis of LAD only or in combination with classic neutrality statistics. However, the detection of natural selection with the LAD statistic has previously been questioned because deviations in local ancestry can be explained as artifacts of long-range linkage disequilibrium (LD), which was not properly modeled by the first-generation LAI methods.43 Our analyses reveal that these genomic regions not only show outlier LAD values but also outlier Fadm values. Because Fadm only depends on allele frequencies at the SNP of interest, these results support the view that the observed signals of adaptive admixture are true and unlikely to be explained by incorrectly modeled LD.
Our results also highlight novel signals of adaptive admixture, such as the APOL1/MYH9 locus in the Fulani nomads of West Africa. Interestingly, an APOL1 haplotype of non-African origin, named G3, was shown to be under positive selection in the Fulani of Cameroon,94 in line with the excess of non-African ancestry that we detected at the locus in the Fulani from Burkina Faso. Nevertheless, the physiological effect of the G3 variants is still debated: experimental work suggests that the G3 haplotype has no lytic activity against Trypanosoma parasites and is not associated with increased susceptibility to common kidney diseases in African Americans.95 Alternatively, the significant excess of non-African ancestry observed at the locus may be due to strong negative selection against HAT-resistance APOL1 alleles (i.e., G1 and G2 haplotypes) in regions where the incidence of sleeping sickness is low, such as Burkina Faso.96 As they do not confer a selective advantage in Trypanosoma brucei-free regions, the G1 and G2 haplotypes only strongly increase the risk for chronic kidney diseases83 and thus become disadvantageous. Further epidemiological and experimental work will be needed to confirm this hypothesis.
In accordance with our simulation study, several of the putatively selected alleles detected here are known to be under strong positive selection in humans, including alleles in ACKR1,97, 98, 99 LCT,100,101 or HLA.81 Given that we focused on admixture events occurring during the five last millennia, only alleles that confer a very strong selective advantage can leave detectable signatures in the genomes of the studied admixed individuals. In addition to their confirmatory nature, these results improve our understanding of the selective advantage conferred by these well-known beneficial alleles. First, because Fadm and LAD detect natural selection since admixture only, selection studies in recently admixed populations represent a valuable tool to detect recent ongoing selection. Second, admixed and source populations have often lived in different environments, so evolutionary studies of adaptive admixture can help refine correlations between signatures of natural selection and environmental pressures. An illustrative example is the Duffy null FY∗BES allele, which is fixed or nearly fixed in most sub-Saharan African populations.99 It has long been proposed that natural selection has favored this allele because it protects against malaria due to Plasmodium vivax.102 Indeed, cellular experiments have shown that the parasite depends on the ACKR1 protein for erythrocytic infection.39,40 However, recent studies have casted doubt on this result because P. vivax has been detected in FY∗BES homozygous carriers,103,104 suggesting that parasite invasion is possible when its human receptor ACKR1 is absent. We and others have found signatures of adaptive admixture for the FY∗BES allele in African-descent admixed populations from Madagascar,22,30 Cabo Verde,32 the Sahel,34 and Pakistan29 but not in North Americans or South Africans.16,31 Evidence of ongoing positive selection for Duffy negativity is thus confined to regions where the current incidence of P. vivax malaria is estimated to be high.105 These findings thus support the view that resistance to vivax malaria is the main evolutionary force driving the frequency of the FY∗BES allele in humans.
Overall, our study reports evidence that recent admixture has facilitated human genetic adaptation to varying environmental conditions. It has been proposed that gene flow can promote rapid evolution when the demographic structure of a species is unstable.18 Our findings support this view, as Homo sapiens is a structured species that has settled a large variety of ecological niches and has undergone large-scale, massive dispersals followed by extensive gene flow.1,7,8 We thus anticipate that more cases of adaptive admixture in humans will soon be uncovered, thanks to methodological and technological advances. Importantly, given the highly conservative nature of our approach, it is very likely that we do not recover variants that have probably been weakly to mildly selected since admixture, such as TNFAIP3 in Indonesian populations of Papuan-related ancestry51,89, 90, 91 or the MCM6/LCT locus in the Bantu-speaking Bakiga from Uganda.31 The use of new, accurate LAI methods80,106 and the development of novel powerful neutrality statistics, such as the integrated decay in ancestry tracts (iDAT),32 and model-based probabilistic frameworks107 are promising paths to improve the power to detect adaptive admixture while better accounting for the demography of admixed populations. Furthermore, many human traits are known to be highly polygenic, suggesting that polygenic adaptation is a key driver of phenotypic evolution,108 highlighting the need for new methods to detect polygenic selection since admixture.109 Finally, genomic studies of adaptive admixture are expected to be more powerful when admixture is ancient, but statistical tests for admixture in modern genomes have low power when admixture time is older than 5,000 years.8 Ancient genomics studies offer a great opportunity to circumvent this limitation by revealing how human populations interacted in the past and how beneficial alleles have spread in time and space.41,110
Acknowledgments
We thank all volunteers participating in this research; Sophie Créno and the HPC Core Facility of Institut Pasteur (Paris) for the management of computational resources; the two anonymous reviewers for their useful comments; and Omar Alva Sanchez, Denis Pierron, Thierry Letellier, Mario Vicente, Carina Schlebusch, Andres Moreno-Estrada, Andres Ruiz-Linares, and the Health Aging and Body Composition (Health ABC) Study for kindly providing access to their data. We also thank Javier Bougeard, Lara Rubio Arauna, Jérémy Choin, Maxime Rotival, Paul Verdu, and Olivier Tenaillon for helpful discussions. S.C.-E. is supported by Sorbonne Université Doctoral College, the Inception program (Investissement d’Avenir grant ANR-16-CONV-0005), and the Institut Pasteur. The laboratory of human evolutionary genetics is supported by the Institut Pasteur, the Collège de France, the CNRS, the Fondation Allianz-Institut de France, the French Government’s Investissement d’Avenir program, Laboratoires d’Excellence “Integrative Biology of Emerging Infectious Diseases” (ANR-10-LABX-62-IBEID) and “Milieu Intérieur” (ANR-10-LABX-69-01), the Fondation de France (No. 00106080), the Fondation pour la Recherche Médicale (équipe FRM DEQ20180339214), and the French National Research Agency (ANR-19-CE35-0005).
Declaration of interests
The authors declare no competing interests.
Published: March 7, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2022.02.011.
Contributor Information
Lluis Quintana-Murci, Email: quintana@pasteur.fr.
Etienne Patin, Email: epatin@pasteur.fr.
Data and code availability
Accession numbers for the genotype data used in this study are listed in Table S2. All SLiM parameter files can be found at the following: https://github.com/h-e-g/ADAD.
Web resources
1000 Genomes Phase 3 and HGDP genomic data, https://www.internationalgenome.org/data
admixr R package, https://cran.r-project.org/web/packages/admixr/index.html
ADMIXTURE software, https://dalexander.github.io/admixture/index.html
dbGAP database, https://dbgap.ncbi.nlm.nih.gov/
Estonian Biocenter public genomic data, https://evolbio.ut.ee
European Genome-Phenome archive, https://ega-archive.org/
Jakobsson Lab genomic data, http://jakobssonlab.iob.uu.se/data/
OMIM, http://www.omim.org/
PLINK software, https://www.cog-genomics.org/plink/2.0/
RFMix software, https://www.dropbox.com/s/cmq4saduh9gozi9/RFMix_v1.5.4.zip
selink software, https://github.com/h-e-g/selink
SHAPEIT software, https://odelaneau.github.io/shapeit4/
SLiM software, https://messerlab.org/slim/
Supplemental information
References
- 1.Nielsen R., Akey J.M., Jakobsson M., Pritchard J.K., Tishkoff S., Willerslev E. Tracing the peopling of the world through genomics. Nature. 2017;541:302–310. doi: 10.1038/nature21347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Novembre J., Di Rienzo A. Spatial patterns of variation due to natural selection in humans. Nat. Rev. Genet. 2009;10:745–755. doi: 10.1038/nrg2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Quintana-Murci L. Human Immunology through the Lens of Evolutionary Genetics. Cell. 2019;177:184–199. doi: 10.1016/j.cell.2019.02.033. [DOI] [PubMed] [Google Scholar]
- 4.Rees J.S., Castellano S., Andrés A.M. The Genomics of Human Local Adaptation. Trends Genet. 2020;36:415–428. doi: 10.1016/j.tig.2020.03.006. [DOI] [PubMed] [Google Scholar]
- 5.Fan S., Hansen M.E.B., Lo Y., Tishkoff S.A. Going global by adapting local: A review of recent human adaptation. Science. 2016;354:54–59. doi: 10.1126/science.aaf5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mathieson I. Human adaptation over the past 40,000 years. Curr. Opin. Genet. Dev. 2020;62:97–104. doi: 10.1016/j.gde.2020.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pickrell J.K., Reich D. Toward a new history and geography of human genes informed by ancient DNA. Trends Genet. 2014;30:377–389. doi: 10.1016/j.tig.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hellenthal G., Busby G.B.J., Band G., Wilson J.F., Capelli C., Falush D., Myers S. A genetic atlas of human admixture history. Science. 2014;343:747–751. doi: 10.1126/science.1243518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson N.A., Coram M.A., Shriver M.D., Romieu I., Barsh G.S., London S.J., Tang H. Ancestral components of admixed genomes in a Mexican cohort. PLoS Genet. 2011;7:e1002410. doi: 10.1371/journal.pgen.1002410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vicuña L., Fernandez M.I., Vial C., Valdebenito P., Chaparro E., Espinoza K., Ziegler A., Bustamante A., Eyheramendy S. Adaptation to Extreme Environments in an Admixed Human Population from the Atacama Desert. Genome Biol. Evol. 2019;11:2468–2479. doi: 10.1093/gbe/evz172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yelmen B., Mondal M., Marnetto D., Pathak A.K., Montinaro F., Gallego Romero I., Kivisild T., Metspalu M., Pagani L. Ancestry-Specific Analyses Reveal Differential Demographic Histories and Opposite Selective Pressures in Modern South Asian Populations. Mol. Biol. Evol. 2019;36:1628–1642. doi: 10.1093/molbev/msz037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lohmueller K.E., Bustamante C.D., Clark A.G. Detecting directional selection in the presence of recent admixture in African-Americans. Genetics. 2011;187:823–835. doi: 10.1534/genetics.110.122739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reynolds A.W., Mata-Míguez J., Miró-Herrans A., Briggs-Cloud M., Sylestine A., Barajas-Olmos F., Garcia-Ortiz H., Rzhetskaya M., Orozco L., Raff J.A., et al. Comparing signals of natural selection between three Indigenous North American populations. Proc. Natl. Acad. Sci. USA. 2019;116:9312–9317. doi: 10.1073/pnas.1819467116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ávila-Arcos M.C., McManus K.F., Sandoval K., Rodríguez-Rodríguez J.E., Villa-Islas V., Martin A.R., Luisi P., Peñaloza-Espinosa R.I., Eng C., Huntsman S., et al. Population History and Gene Divergence in Native Mexicans Inferred from 76 Human Exomes. Mol. Biol. Evol. 2020;37:994–1006. doi: 10.1093/molbev/msz282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huerta-Sánchez E., Degiorgio M., Pagani L., Tarekegn A., Ekong R., Antao T., Cardona A., Montgomery H.E., Cavalleri G.L., Robbins P.A., et al. Genetic signatures reveal high-altitude adaptation in a set of ethiopian populations. Mol. Biol. Evol. 2013;30:1877–1888. doi: 10.1093/molbev/mst089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhatia G., Tandon A., Patterson N., Aldrich M.C., Ambrosone C.B., Amos C., Bandera E.V., Berndt S.I., Bernstein L., Blot W.J., et al. Genome-wide scan of 29,141 African Americans finds no evidence of directional selection since admixture. Am. J. Hum. Genet. 2014;95:437–444. doi: 10.1016/j.ajhg.2014.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Refoyo-Martínez A., da Fonseca R.R., Halldórsdóttir K., Árnason E., Mailund T., Racimo F. Identifying loci under positive selection in complex population histories. Genome Res. 2019;29:1506–1520. doi: 10.1101/gr.246777.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Slatkin M. Gene flow and the geographic structure of natural populations. Science. 1987;236:787–792. doi: 10.1126/science.3576198. [DOI] [PubMed] [Google Scholar]
- 19.Racimo F., Sankararaman S., Nielsen R., Huerta-Sánchez E. Evidence for archaic adaptive introgression in humans. Nat. Rev. Genet. 2015;16:359–371. doi: 10.1038/nrg3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pagani L., Kivisild T., Tarekegn A., Ekong R., Plaster C., Gallego Romero I., Ayub Q., Mehdi S.Q., Thomas M.G., Luiselli D., et al. Ethiopian genetic diversity reveals linguistic stratification and complex influences on the Ethiopian gene pool. Am. J. Hum. Genet. 2012;91:83–96. doi: 10.1016/j.ajhg.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bryc K., Velez C., Karafet T., Moreno-Estrada A., Reynolds A., Auton A., Hammer M., Bustamante C.D., Ostrer H. Colloquium paper: genome-wide patterns of population structure and admixture among Hispanic/Latino populations. Proc. Natl. Acad. Sci. USA. 2010;107(Suppl 2):8954–8961. doi: 10.1073/pnas.0914618107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hodgson J.A., Pickrell J.K., Pearson L.N., Quillen E.E., Prista A., Rocha J., Soodyall H., Shriver M.D., Perry G.H. Natural selection for the Duffy-null allele in the recently admixed people of Madagascar. Proc. Biol. Sci. 2014;281:20140930. doi: 10.1098/rspb.2014.0930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Breton G., Schlebusch C.M., Lombard M., Sjödin P., Soodyall H., Jakobsson M. Lactase persistence alleles reveal partial East African ancestry of southern African Khoe pastoralists. Curr. Biol. 2014;24:852–858. doi: 10.1016/j.cub.2014.02.041. [DOI] [PubMed] [Google Scholar]
- 24.Jeong C., Alkorta-Aranburu G., Basnyat B., Neupane M., Witonsky D.B., Pritchard J.K., Beall C.M., Di Rienzo A. Admixture facilitates genetic adaptations to high altitude in Tibet. Nat. Commun. 2014;5:3281. doi: 10.1038/ncomms4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Macholdt E., Lede V., Barbieri C., Mpoloka S.W., Chen H., Slatkin M., Pakendorf B., Stoneking M. Tracing pastoralist migrations to southern Africa with lactase persistence alleles. Curr. Biol. 2014;24:875–879. doi: 10.1016/j.cub.2014.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rishishwar L., Conley A.B., Wigington C.H., Wang L., Valderrama-Aguirre A., Jordan I.K. Ancestry, admixture and fitness in Colombian genomes. Sci. Rep. 2015;5:12376. doi: 10.1038/srep12376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou Q., Zhao L., Guan Y. Strong Selection at MHC in Mexicans since Admixture. PLoS Genet. 2016;12:e1005847. doi: 10.1371/journal.pgen.1005847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Busby G., Christ R., Band G., Leffler E., Le Q.S., Rockett K., Kwiatkowski D., Spencer C. Inferring adaptive gene-flow in recent African history. Preprint at bioRxiv. 2017 doi: 10.1101/205252. [DOI] [Google Scholar]
- 29.Laso-Jadart R., Harmant C., Quach H., Zidane N., Tyler-Smith C., Mehdi Q., Ayub Q., Quintana-Murci L., Patin E. The Genetic Legacy of the Indian Ocean Slave Trade: Recent Admixture and Post-admixture Selection in the Makranis of Pakistan. Am. J. Hum. Genet. 2017;101:977–984. doi: 10.1016/j.ajhg.2017.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pierron D., Heiske M., Razafindrazaka H., Pereda-Loth V., Sanchez J., Alva O., Arachiche A., Boland A., Olaso R., Deleuze J.-F., et al. Strong selection during the last millennium for African ancestry in the admixed population of Madagascar. Nat. Commun. 2018;9:932. doi: 10.1038/s41467-018-03342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patin E., Lopez M., Grollemund R., Verdu P., Harmant C., Quach H., Laval G., Perry G.H., Barreiro L.B., Froment A., et al. Dispersals and genetic adaptation of Bantu-speaking populations in Africa and North America. Science. 2017;356:543–546. doi: 10.1126/science.aal1988. [DOI] [PubMed] [Google Scholar]
- 32.Hamid I., Korunes K.L., Beleza S., Goldberg A. Rapid adaptation to malaria facilitated by admixture in the human population of Cabo Verde. eLife. 2021;10:e63177. doi: 10.7554/eLife.63177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schlebusch C.M., Skoglund P., Sjödin P., Gattepaille L.M., Hernandez D., Jay F., Li S., De Jongh M., Singleton A., Blum M.G.B., et al. Genomic variation in seven Khoe-San groups reveals adaptation and complex African history. Science. 2012;338:374–379. doi: 10.1126/science.1227721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Triska P., Soares P., Patin E., Fernandes V., Cerny V., Pereira L. Extensive Admixture and Selective Pressure Across the Sahel Belt. Genome Biol. Evol. 2015;7:3484–3495. doi: 10.1093/gbe/evv236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deng L., Ruiz-Linares A., Xu S., Wang S. Ancestry variation and footprints of natural selection along the genome in Latin American populations. Sci. Rep. 2016;6:21766. doi: 10.1038/srep21766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin W., Xu S., Wang H., Yu Y., Shen Y., Wu B., Jin L. Genome-wide detection of natural selection in African Americans pre- and post-admixture. Genome Res. 2012;22:519–527. doi: 10.1101/gr.124784.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Norris E.T., Rishishwar L., Chande A.T., Conley A.B., Ye K., Valderrama-Aguirre A., Jordan I.K. Admixture-enabled selection for rapid adaptive evolution in the Americas. Genome Biol. 2020;21:29. doi: 10.1186/s13059-020-1946-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang H., Choudhry S., Mei R., Morgan M., Rodriguez-Cintron W., Burchard E.G., Risch N.J. Recent genetic selection in the ancestral admixture of Puerto Ricans. Am. J. Hum. Genet. 2007;81:626–633. doi: 10.1086/520769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller L.H., Mason S.J., Clyde D.F., McGinniss M.H. The resistance factor to Plasmodium vivax in blacks. The Duffy-blood-group genotype, FyFy. N. Engl. J. Med. 1976;295:302–304. doi: 10.1056/NEJM197608052950602. [DOI] [PubMed] [Google Scholar]
- 40.Tournamille C., Colin Y., Cartron J.P., Le Van Kim C. Disruption of a GATA motif in the Duffy gene promoter abolishes erythroid gene expression in Duffy-negative individuals. Nat. Genet. 1995;10:224–228. doi: 10.1038/ng0695-224. [DOI] [PubMed] [Google Scholar]
- 41.Mathieson I., Lazaridis I., Rohland N., Mallick S., Patterson N., Roodenberg S.A., Harney E., Stewardson K., Fernandes D., Novak M., et al. Genome-wide patterns of selection in 230 ancient Eurasians. Nature. 2015;528:499–503. doi: 10.1038/nature16152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Long J.C. The genetic structure of admixed populations. Genetics. 1991;127:417–428. doi: 10.1093/genetics/127.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Price A.L., Weale M.E., Patterson N., Myers S.R., Need A.C., Shianna K.V., Ge D., Rotter J.I., Torres E., Taylor K.D., et al. Long-range LD can confound genome scans in admixed populations. Am. J. Hum. Genet. 2008;83:132–135. doi: 10.1016/j.ajhg.2008.06.005. author reply 135–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pasaniuc B., Sankararaman S., Torgerson D.G., Gignoux C., Zaitlen N., Eng C., Rodriguez-Cintron W., Chapela R., Ford J.G., Avila P.C., et al. Analysis of Latino populations from GALA and MEC studies reveals genomic loci with biased local ancestry estimation. Bioinformatics. 2013;29:1407–1415. doi: 10.1093/bioinformatics/btt166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haller B.C., Messer P.W. SLiM 3: Forward Genetic Simulations Beyond the Wright-Fisher Model. Mol. Biol. Evol. 2019;36:632–637. doi: 10.1093/molbev/msy228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R., 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Francioli L.C., Polak P.P., Koren A., Menelaou A., Chun S., Renkens I., van Duijn C.M., Swertz M., Wijmenga C., van Ommen G., et al. Genome-wide patterns and properties of de novo mutations in humans. Nat. Genet. 2015;47:822–826. doi: 10.1038/ng.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lohmueller K.E., Albrechtsen A., Li Y., Kim S.Y., Korneliussen T., Vinckenbosch N., Tian G., Huerta-Sanchez E., Feder A.F., Grarup N., et al. Natural selection affects multiple aspects of genetic variation at putatively neutral sites across the human genome. PLoS Genet. 2011;7:e1002326. doi: 10.1371/journal.pgen.1002326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boyko A.R., Williamson S.H., Indap A.R., Degenhardt J.D., Hernandez R.D., Lohmueller K.E., Adams M.D., Schmidt S., Sninsky J.J., Sunyaev S.R., et al. Assessing the evolutionary impact of amino acid mutations in the human genome. PLoS Genet. 2008;4:e1000083. doi: 10.1371/journal.pgen.1000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Haller B.C., Messer P.W. 2016. SLiM: An Evolutionary Simulation Framework.http://benhaller.com/slim/SLiM_Manual.pdf [Google Scholar]
- 51.Choin J., Mendoza-Revilla J., Arauna L.R., Cuadros-Espinoza S., Cassar O., Larena M., Ko A.M.-S., Harmant C., Laurent R., Verdu P., et al. Genomic insights into population history and biological adaptation in Oceania. Nature. 2021;592:583–589. doi: 10.1038/s41586-021-03236-5. [DOI] [PubMed] [Google Scholar]
- 52.Bernstein F. Comitato Weak Representations of the Data. Italiano per Lo Studio Dei Problemi Della Popolazione. Istituto Poligrafico dello Stato; Roma: 1931. Die geographische Verteilung der Blutgruppen und ihre anthropologische Bedeutung; pp. 227–243. [Google Scholar]
- 53.Charlesworth B., Charlesworth D. W. H. Freeman; 2010. Elements of Evolutionary Genetics. [Google Scholar]
- 54.Alexander D.H., Novembre J., Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Workman P.L., Blumberg B.S., Cooper A.J. Selection, Gene Migration and Polymorphic Stability in a U. S. White and Negro Population. Am. J. Hum. Genet. 1963;15:429–437. [PMC free article] [PubMed] [Google Scholar]
- 56.Reed T.E. Caucasian genes in American Negroes. Science. 1969;165:762–768. doi: 10.1126/science.165.3895.762. [DOI] [PubMed] [Google Scholar]
- 57.Cavalli-Sforza L.L., Bodmer W.F. Freeman & Co; 1971. The Genetics of Human Populations. [Google Scholar]
- 58.Maples B.K., Gravel S., Kenny E.E., Bustamante C.D. RFMix: a discriminative modeling approach for rapid and robust local-ancestry inference. Am. J. Hum. Genet. 2013;93:278–288. doi: 10.1016/j.ajhg.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Delaneau O., Zagury J.-F., Robinson M.R., Marchini J.L., Dermitzakis E.T. Accurate, scalable and integrative haplotype estimation. Nat. Commun. 2019;10:5436. doi: 10.1038/s41467-019-13225-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stephan W., Wiehe T.H.E., Lenz M.W. The effect of strongly selected substitutions on neutral polymorphism: Analytical results based on diffusion theory. Theor. Popul. Biol. 1992;41:237–254. [Google Scholar]
- 61.Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Manichaikul A., Mychaleckyj J.C., Rich S.S., Daly K., Sale M., Chen W.-M. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26:2867–2873. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Petr M., Vernot B., Kelso J. admixr-R package for reproducible analyses using ADMIXTOOLS. Bioinformatics. 2019;35:3194–3195. doi: 10.1093/bioinformatics/btz030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fisher R.A. Oliver and Boyd; Edinburgh: 1925. Statistical Methods for Research Workers. [Google Scholar]
- 65.Ghoussaini M., Mountjoy E., Carmona M., Peat G., Schmidt E.M., Hercules A., Fumis L., Miranda A., Carvalho-Silva D., Buniello A., et al. Open Targets Genetics: systematic identification of trait-associated genes using large-scale genetics and functional genomics. Nucleic Acids Res. 2021;49(D1):D1311–D1320. doi: 10.1093/nar/gkaa840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Peter B.M., Huerta-Sanchez E., Nielsen R. Distinguishing between selective sweeps from standing variation and from a de novo mutation. PLoS Genet. 2012;8:e1003011. doi: 10.1371/journal.pgen.1003011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Baharian S., Barakatt M., Gignoux C.R., Shringarpure S., Errington J., Blot W.J., Bustamante C.D., Kenny E.E., Williams S.M., Aldrich M.C., Gravel S. The Great Migration and African-American Genomic Diversity. PLoS Genet. 2016;12:e1006059. doi: 10.1371/journal.pgen.1006059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fortes-Lima C.A., Laurent R., Thouzeau V., Toupance B., Verdu P. Complex genetic admixture histories reconstructed with Approximate Bayesian Computation. Mol. Ecol. Resour. 2021;21:1098–1117. doi: 10.1111/1755-0998.13325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Malaspinas A.-S., Westaway M.C., Muller C., Sousa V.C., Lao O., Alves I., Bergström A., Athanasiadis G., Cheng J.Y., Crawford J.E., et al. A genomic history of Aboriginal Australia. Nature. 2016;538:207–214. doi: 10.1038/nature18299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Medina P., Thornlow B., Nielsen R., Corbett-Detig R. Estimating the Timing of Multiple Admixture Pulses During Local Ancestry Inference. Genetics. 2018;210:1089–1107. doi: 10.1534/genetics.118.301411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pickrell J.K., Patterson N., Loh P.-R., Lipson M., Berger B., Stoneking M., Pakendorf B., Reich D. Ancient west Eurasian ancestry in southern and eastern Africa. Proc. Natl. Acad. Sci. USA. 2014;111:2632–2637. doi: 10.1073/pnas.1313787111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Molinaro L., Marnetto D., Mondal M., Ongaro L., Yelmen B., Lawson D.J., Montinaro F., Pagani L. A Chromosome-Painting-Based Pipeline to Infer Local Ancestry under Limited Source Availability. Genome Biol. Evol. 2021;13:evab025. doi: 10.1093/gbe/evab025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tajima F. The effect of change in population size on DNA polymorphism. Genetics. 1989;123:597–601. doi: 10.1093/genetics/123.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wakeley J., Aliacar N. Gene genealogies in a metapopulation. Genetics. 2001;159:893–905. doi: 10.1093/genetics/159.2.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fu Y.X., Li W.H. Statistical tests of neutrality of mutations. Genetics. 1993;133:693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Przeworski M. The signature of positive selection at randomly chosen loci. Genetics. 2002;160:1179–1189. doi: 10.1093/genetics/160.3.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Coop G., Pickrell J.K., Novembre J., Kudaravalli S., Li J., Absher D., Myers R.M., Cavalli-Sforza L.L., Feldman M.W., Pritchard J.K. The role of geography in human adaptation. PLoS Genet. 2009;5:e1000500. doi: 10.1371/journal.pgen.1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ferrer-Admetlla A., Liang M., Korneliussen T., Nielsen R. On detecting incomplete soft or hard selective sweeps using haplotype structure. Mol. Biol. Evol. 2014;31:1275–1291. doi: 10.1093/molbev/msu077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dias-Alves T., Mairal J., Blum M.G.B. Loter: A Software Package to Infer Local Ancestry for a Wide Range of Species. Mol. Biol. Evol. 2018;35:2318–2326. doi: 10.1093/molbev/msy126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Meyer D., C Aguiar V.R., Bitarello B.D.C., C Brandt D.Y., Nunes K. A genomic perspective on HLA evolution. Immunogenetics. 2018;70:5–27. doi: 10.1007/s00251-017-1017-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vicente M., Priehodová E., Diallo I., Podgorná E., Poloni E.S., Černý V., Schlebusch C.M. Population history and genetic adaptation of the Fulani nomads: inferences from genome-wide data and the lactase persistence trait. BMC Genomics. 2019;20:915. doi: 10.1186/s12864-019-6296-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Genovese G., Friedman D.J., Ross M.D., Lecordier L., Uzureau P., Freedman B.I., Bowden D.W., Langefeld C.D., Oleksyk T.K., Uscinski Knob A.L., et al. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science. 2010;329:841–845. doi: 10.1126/science.1193032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ruby M.A., Riedl I., Massart J., Åhlin M., Zierath J.R. Protein kinase N2 regulates AMP kinase signaling and insulin responsiveness of glucose metabolism in skeletal muscle. Am. J. Physiol. Endocrinol. Metab. 2017;313:E483–E491. doi: 10.1152/ajpendo.00147.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Meng J., Yao Z., He Y., Zhang R., Zhang Y., Yao X., Yang H., Chen L., Zhang Z., Zhang H., et al. ARRDC4 regulates enterovirus 71-induced innate immune response by promoting K63 polyubiquitination of MDA5 through TRIM65. Cell Death Dis. 2017;8:e2866. doi: 10.1038/cddis.2017.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wit J.M., Walenkamp M.J. Role of insulin-like growth factors in growth, development and feeding. World Rev. Nutr. Diet. 2013;106:60–65. doi: 10.1159/000342546. [DOI] [PubMed] [Google Scholar]
- 87.Warrington N.M., Beaumont R.N., Horikoshi M., Day F.R., Helgeland Ø., Laurin C., Bacelis J., Peng S., Hao K., Feenstra B., et al. Maternal and fetal genetic effects on birth weight and their relevance to cardio-metabolic risk factors. Nat. Genet. 2019;51:804–814. doi: 10.1038/s41588-019-0403-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chimusa E.R., Zaitlen N., Daya M., Möller M., van Helden P.D., Mulder N.J., Price A.L., Hoal E.G. Genome-wide association study of ancestry-specific TB risk in the South African Coloured population. Hum. Mol. Genet. 2014;23:796–809. doi: 10.1093/hmg/ddt462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vernot B., Tucci S., Kelso J., Schraiber J.G., Wolf A.B., Gittelman R.M., Dannemann M., Grote S., McCoy R.C., Norton H., et al. Excavating Neandertal and Denisovan DNA from the genomes of Melanesian individuals. Science. 2016;352:235–239. doi: 10.1126/science.aad9416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gittelman R.M., Schraiber J.G., Vernot B., Mikacenic C., Wurfel M.M., Akey J.M. Archaic Hominin Admixture Facilitated Adaptation to Out-of-Africa Environments. Curr. Biol. 2016;26:3375–3382. doi: 10.1016/j.cub.2016.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jacobs G.S., Hudjashov G., Saag L., Kusuma P., Darusallam C.C., Lawson D.J., Mondal M., Pagani L., Ricaut F.-X., Stoneking M., et al. Multiple Deeply Divergent Denisovan Ancestries in Papuans. Cell. 2019;177:1010–1021.e32. doi: 10.1016/j.cell.2019.02.035. [DOI] [PubMed] [Google Scholar]
- 92.Zammit N.W., Siggs O.M., Gray P.E., Horikawa K., Langley D.B., Walters S.N., Daley S.R., Loetsch C., Warren J., Yap J.Y., et al. Denisovan, modern human and mouse TNFAIP3 alleles tune A20 phosphorylation and immunity. Nat. Immunol. 2019;20:1299–1310. doi: 10.1038/s41590-019-0492-0. [DOI] [PubMed] [Google Scholar]
- 93.Toyama K.S., Crochet P.-A., Leblois R. Sampling schemes and drift can bias admixture proportions inferred by structure. Mol. Ecol. Resour. 2020;20:1769–1785. doi: 10.1111/1755-0998.13234. [DOI] [PubMed] [Google Scholar]
- 94.Ko W.-Y., Rajan P., Gomez F., Scheinfeldt L., An P., Winkler C.A., Froment A., Nyambo T.B., Omar S.A., Wambebe C., et al. Identifying Darwinian selection acting on different human APOL1 variants among diverse African populations. Am. J. Hum. Genet. 2013;93:54–66. doi: 10.1016/j.ajhg.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Limou S., Nelson G.W., Lecordier L., An P., O’hUigin C.S., David V.A., Binns-Roemer E.A., Guiblet W.M., Oleksyk T.K., Pays E., et al. Sequencing rare and common APOL1 coding variants to determine kidney disease risk. Kidney Int. 2015;88:754–763. doi: 10.1038/ki.2015.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Franco J.R., Simarro P.P., Diarra A., Jannin J.G. Epidemiology of human African trypanosomiasis. Clin. Epidemiol. 2014;6:257–275. doi: 10.2147/CLEP.S39728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hamblin M.T., Di Rienzo A. Detection of the signature of natural selection in humans: evidence from the Duffy blood group locus. Am. J. Hum. Genet. 2000;66:1669–1679. doi: 10.1086/302879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hamblin M.T., Thompson E.E., Di Rienzo A. Complex signatures of natural selection at the Duffy blood group locus. Am. J. Hum. Genet. 2002;70:369–383. doi: 10.1086/338628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.McManus K.F., Taravella A.M., Henn B.M., Bustamante C.D., Sikora M., Cornejo O.E. Population genetic analysis of the DARC locus (Duffy) reveals adaptation from standing variation associated with malaria resistance in humans. PLoS Genet. 2017;13:e1006560. doi: 10.1371/journal.pgen.1006560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tishkoff S.A., Reed F.A., Ranciaro A., Voight B.F., Babbitt C.C., Silverman J.S., Powell K., Mortensen H.M., Hirbo J.B., Osman M., et al. Convergent adaptation of human lactase persistence in Africa and Europe. Nat. Genet. 2007;39:31–40. doi: 10.1038/ng1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bersaglieri T., Sabeti P.C., Patterson N., Vanderploeg T., Schaffner S.F., Drake J.A., Rhodes M., Reich D.E., Hirschhorn J.N. Genetic signatures of strong recent positive selection at the lactase gene. Am. J. Hum. Genet. 2004;74:1111–1120. doi: 10.1086/421051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Livingstone F.B. The Duffy blood groups, vivax malaria, and malaria selection in human populations: a review. Hum. Biol. 1984;56:413–425. [PubMed] [Google Scholar]
- 103.Ménard D., Barnadas C., Bouchier C., Henry-Halldin C., Gray L.R., Ratsimbasoa A., Thonier V., Carod J.-F., Domarle O., Colin Y., et al. Plasmodium vivax clinical malaria is commonly observed in Duffy-negative Malagasy people. Proc. Natl. Acad. Sci. USA. 2010;107:5967–5971. doi: 10.1073/pnas.0912496107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Popovici J., Roesch C., Rougeron V. The enigmatic mechanisms by which Plasmodium vivax infects Duffy-negative individuals. PLoS Pathog. 2020;16:e1008258. doi: 10.1371/journal.ppat.1008258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Battle K.E., Lucas T.C.D., Nguyen M., Howes R.E., Nandi A.K., Twohig K.A., Pfeffer D.A., Cameron E., Rao P.C., Casey D., et al. Mapping the global endemicity and clinical burden of Plasmodium vivax, 2000-17: a spatial and temporal modelling study. Lancet. 2019;394:332–343. doi: 10.1016/S0140-6736(19)31096-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Guan Y. Detecting structure of haplotypes and local ancestry. Genetics. 2014;196:625–642. doi: 10.1534/genetics.113.160697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sugden L.A., Atkinson E.G., Fischer A.P., Rong S., Henn B.M., Ramachandran S. Localization of adaptive variants in human genomes using averaged one-dependence estimation. Nat. Commun. 2018;9:703. doi: 10.1038/s41467-018-03100-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sella G., Barton N.H. Thinking About the Evolution of Complex Traits in the Era of Genome-Wide Association Studies. Annu. Rev. Genomics Hum. Genet. 2019;20:461–493. doi: 10.1146/annurev-genom-083115-022316. [DOI] [PubMed] [Google Scholar]
- 109.Racimo F., Berg J.J., Pickrell J.K. Detecting Polygenic Adaptation in Admixture Graphs. Genetics. 2018;208:1565–1584. doi: 10.1534/genetics.117.300489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dehasque M., Ávila-Arcos M.C., Díez-Del-Molino D., Fumagalli M., Guschanski K., Lorenzen E.D., Malaspinas A.-S., Marques-Bonet T., Martin M.D., Murray G.G.R., et al. Inference of natural selection from ancient DNA. Evol. Lett. 2020;4:94–108. doi: 10.1002/evl3.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Accession numbers for the genotype data used in this study are listed in Table S2. All SLiM parameter files can be found at the following: https://github.com/h-e-g/ADAD.