Abstract
Periodontal disease is an inflammatory reaction of the periodontal tissues to oral pathogens. In the present review we discuss the intricate effects of a regulatory network of gene expression modulators, microRNAs (miRs), as they affect periodontal morphology, function and gene expression during periodontal disease. These miRs are small RNAs involved in RNA silencing and post-transcriptional regulation and affect all stages of periodontal disease, from the earliest signs of gingivitis to the regulation of periodontal homeostasis and immunity and to the involvement in periodontal tissue destruction. MiRs coordinate periodontal disease progression not only directly but also through long non-coding RNAs (lncRNAs), which have been demonstrated to act as endogenous sponges or decoys that regulate the expression and function of miRs, and which in turn suppress the targeting of mRNAs involved in the inflammatory response, cell proliferation, migration and differentiation. While the integrity of miR function is essential for periodontal health and immunity, miR sequence variations (genetic polymorphisms) contribute toward an enhanced risk for periodontal disease progression and severity. Several polymorphisms in miR genes have been linked to an increased risk of periodontitis, and among those, miR-146a, miR-196, and miR-499 polymorphisms have been identified as risk factors for periodontal disease. The role of miRs in periodontal disease progression is not limited to the host tissues but also extends to the viruses that reside in periodontal lesions, such as herpesviruses (human herpesvirus, HHV). In advanced periodontal lesions, HHV infections result in the release of cytokines from periodontal tissues and impair antibacterial immune mechanisms that promote bacterial overgrowth. In turn, controlling the exacerbation of periodontal disease by minimizing the effect of periodontal HHV in periodontal lesions may provide novel avenues for therapeutic intervention. In summary, this review highlights multiple levels of miR-mediated control of periodontal disease progression, (i) through their role in periodontal inflammation and the dysregulation of homeostasis, (ii) as a regulatory target of lncRNAs, (iii) by contributing toward periodontal disease susceptibility through miR polymorphism, and (iv) as periodontal microflora modulators via viral miRs.
Keywords: microRNAs, long non-coding RNAs, viral microRNAs, polymorphism, inflammation, periodontal disease
Introduction
Inflammation is a broad term for the body’s immune response against an irritant, a stimulant, or a pathogen. While the attack of pathogens on a defenseless organism may have serious consequences, the defense itself, inflammation, may also be detrimental to the health of an organism, leading to disease, cancer, and death. The periodontal inflammatory process is the quintessential example of a prolonged immune response that often lasts for a lifetime [1,2], and in which connective tissues, barrier cells, and sophisticated epithelial boundaries mount a tenacious Sisyphus-like defense against continuously attacking pathogens from the oral cavity. Most often, the balance between attack and defense results in yet another treaty – homeostasis – and the remarkable stronghold against oral pathogens stands for another day. It may take weeks, months, or years until the prolonged attack of oral pathogens overwhelms the finely-knit response of leukocytes and macrophages, and damages the integrity of the remaining epithelial/connective tissue barrier, resulting in advanced periodontal disease. At that point, chronic inflammatory conditions result in a destruction of the periodontal ligament attachment apparatus and a loss of the alveolar apparatus [3].
The occlusal margin of the periodontium is formed by the junctional epithelium, a non-keratinized squamous epithelium that connects the sulcus epithelium with the underlying connective tissues of the periodontium. This junctional epithelium maintains a semipermeable attachment to the tooth surface and provides and initial barrier to the pathogens of the oral cavity. However, principal tissues responsible for the attachment of the tooth root to the jaw bone are three highly specialized connective tissues, root cementum, periodontal ligament, and alveolar bone. Root cementum is a mineralized tissue comprised of cellular and acellular cementum that forms a layer covering root dentin and provides anchorage for Sharpey’s fibers (Diekwisch 2001). The periodontal ligament contains collagen fibers, blood vessels and nerves and is responsible for the fibrous attachment between the cementum and the alveolar bone (Reed and Diekwisch 2015). Periodontal ligament fibers are anchored within sockets of alveolar bone which are responsible for stable attachment of the tooth to the jaw bone (Diekwisch 2016).
Experiments with gnotobiotic rats have established the essential role of bacteria residing within the dental plaque and in the peri-dental biofilm on periodontal disease including alveolar bone loss (Gibbons et al. 1966, Socransky et al. 1970). These bacteria then cause a cascade of tissue reactions summarized as gingivitis, including vasodilatation and new blood vessel formation, increased crevicular fluid secretion, as well as leucocyte and lymphocyte migration (Luan et al. 2016). Steps of progressing gingivitis are also characterized as initial, early, and established lesion and may remain stable as long as the host tissue maintains a successful defense against the attack of pathogens (Luan et al. 2017). Periodontal disease progresses toward the advanced periodontal lesion when the periodontal defense breaks down as evidenced by bone loss, loss of attachment, and increased pocket depth (Graves et al. 2011). This stage is characterized by an exacerbation of the periodontal immune response resulting in tissue destruction and loss Ivanyi and Lehner 1970, van Dyke et al. 2020).
The homeostasis between bacterial invasion and inflammatory response and between tissue loss and regeneration is regulated by a family of small RNAs, miRs, that not only control the physiological function of periodontal tissues in health, but also the pathobiology of invading bacteria and viruses and the tissue response to pathogens entering the sulcus from the oral cavity. Due to their complexity and nuanced response, the balance between periodontal disease and homeostasis is a quintessential example of a finely tuned miR regulatory response to external pathogens and the body’s prolonged defense against the microbes of the oral cavity [4–6]. MiRs ensure that gingiva and connective tissues orchestrate a proportionate immune response to microbial attacks to prevent wide-spread bacteremia and sepsis, while they also modulate the escalation of the mucosal immune response to counteract rapid tissue destruction and loss of teeth.
In this review we will provide an overview about the miRs in gingival and periodontal cells and tissues, about their function in periodontal infection and inflammation, and about how miRs collaborate to orchestrate the periodontal immune response: strong enough to fend off the viral and bacterial aggressors and gentle enough to avoid an escalation of inflammation and loss of tissue. Our review will discuss all aspects of the miR response to periodontal pathogens, including the control of oral microbial pathogens that initiate and sustain periodontal disease and the genetic and host microenvironmental factors that influence the degree of the disease. A summary of the major sites and pathways by which miRs affect periodontal immunity and tissue homeostasis during periodontal inflammation as well as the effects of miR polymorphism dependent variability and lncRNA sponging are presented in Figure 1.
Figure 1. MicroRNAs shape all aspects of periodontal inflammation.
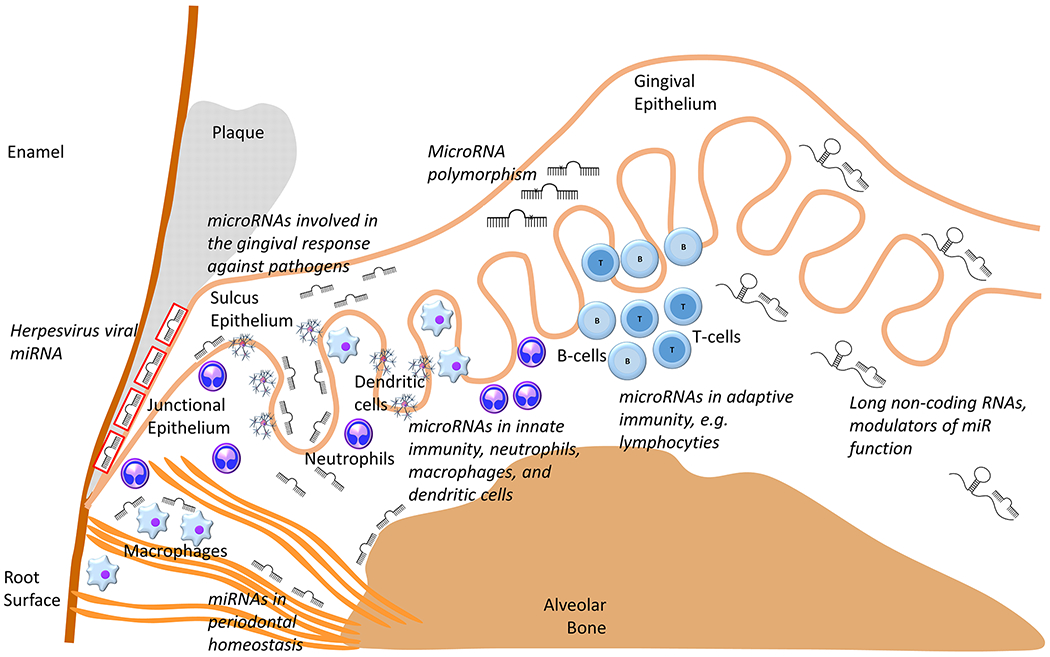
This sketch illustrates the multiple sites of action and diverse functions of miRs in the regulation of periodontal inflammation, some of which are reviewed in this article, including (i) miRs involved in the gingival response against pathogens, (ii) miRs involved in periodontal homeostasis, (iii) viral miRs. In turn miR function is altered by sponge-like long non-coding RNAs or through miR polymorphism. As part of the inflammatory response, miRs control the expression of sets of related genes and have an enormous impact on the periodontal host response against periodontal pathogens and even affect gene expression within some of the pathogens, such as herpesvirus populations residing within the periodontal lesion.
MicroRNA regulation of inflammation in gingival tissues
Anatomically, the gingiva forms a tight circumferential seal around the tooth cervix and covers the marginal connective tissues of the maxillary and mandibular alveolar processes. Gingival tissue consists of gingival epithelium and gingival connective tissue. Accumulation of a microbial biofilm on the tooth surface initiates an immune-inflammatory response in the gingival host tissue and results in gingival inflammation. Histopathologic changes in the inflamed gingiva include the elongation of rete ridges into the gingival connective tissue, a break-down of the collagen fiber network and infiltration with inflammatory and immune cells [7,8]. These changes in morphology and inflammatory state are accompanied by changes in gene expression related to host-microbial interaction, epithelial defense function, host cell chemotaxis, molecular signaling pathways as well as innate and adopted immune response after the transition from non-inflamed to inflamed gingiva [8–10].
MicroRNA regulation of proinflammatory cytokine and chemokine expression in gingival tissues
Cytokines and chemokines are the mediators of the host immune response to oral microbial infection and thus contribute to the initiation and progression of gingival inflammation [11,12]. At the onset of inflammation, cytokines are produced by resident cells including epithelial cells and fibroblasts, and by phagocytes such as neutrophils and macrophages. However, when inflammation becomes chronic, cytokines are also secreted by lymphocytes [13,14]. Prominent pro-inflammatory cytokines involved in the progression of periodontal disease include the Interleukin 1 (IL-1), Interleukin 6 (IL-6) and Tumor Necrosis Factor (TNF) families. Increased expression of IL-1, IL-6 and TNF-α results in enhanced periodontal disease pathogenesis and an upregulation of the inflammatory reaction [13,15]. Significant differences in miR expression profiles between inflamed and non-inflamed gingival tissues implicate miRs in the regulation of this cytokine network [16,17].
Individual miRs involved in gingival cytokine regulation include miR-21, miR-144, miR-146a, miR-128, miR-200b, miR-203 and let-7a [18–23], which are overexpressed as a result of inflammation and miR-205, miR-335, miR-100 and miR-125a [17,24,25], which are reduced in inflamed environments. Confirming the relationship between miRs and periodontal disease state, it has been demonstrated that down-regulation of miR-205 and miR-335 was accompanied by upregulation of IL-1, IL-6 and TNF-α expression in periodontitis patients and mice with experimental periodontitis [24,25]. MiR-205 targets the Interleukin 6 signal transducer (IL-6ST) which in combination with IL-6 activates the Janus kinase (JAK). The activation of JAK and its phosphorylation of signal transducers and activators of transcription 3 (STAT3) is an important event to trigger JAK/STAT3 signaling and resulting inflammatory cascades [26]. For example, in gingival epithelial cells, inhibition of miR-205 by P. gingivalis upregulates IL-6ST expression, activates JAK/STAT3 signaling and increases IL-1, IL-6 and TNF-α expression [24]. In contrast to miR-205, miR-203 is a miR upregulated in inflamed gingival tissues that functions through the suppressor of cytokine signaling (SOCS)/STAT3 pathway. In experimental studies, P. gingivalis induced an upregulation of miR-203, which resulted in a direct inhibition of SOCS3 and SOCS6 in gingival epithelial cells [23]. SOCS proteins function through the inhibition of JAKs and reduction of SOCS expression increases JAK activity on STAT3 phosphorylation and activation which in turn affects IL-1, IL-6 and TNF-α cytokine expression levels.
Several miRs with anti-inflammatory properties, including miR-21, miR-146a and miR-128 are also upregulated in gingival tissues of periodontitis patients and/or in animals with induced periodontal disease [18,19,21,27,28]. The increased expression level of miR-146 in gingival tissues has been associated with disease severity [27,28]. On a cellular level, overexpression of these miRs was reduced while inhibition increased IL-1, IL-6 and/or TNF-α expression in gingival fibroblasts and macrophages in response to lipopolysaccharide (LPS) challenge, indicating that these proinflammatory cytokines are downstream targets of miR-21, miR-146a and miR-128. Furthermore, in vivo administration of miR-146a suppressed the inflammatory reaction in periodontal tissues from periodontitis mice [21], while knockout of miR-21 exacerbated gingival inflammation and alveolar bone loss in a mouse periodontitis model [18]. Together, these studies underscore the anti-inflammatory properties of miR-146a and miR-21 in periodontal disease, a role that might also hold therapeutic potential.
MicroRNA regulation of autophagy in gingival tissues
Autophagy is a homeostatic mechanism involved in the degradation of damaged cellular organelles, denatured proteins and invaded pathogens in lysosomes [29]. Autophagy plays an important role in the pathogenesis of periodontal disease [30] and in the induction and suppression of inflammation by interacting with innate immune signaling pathways, influencing the survival of inflammatory cells, and regulating the production of proinflammatory cytokines [31]. Through a mutual regulatory loop, cytokines also affect autophagy activity. Cyclooxygenase 2 (COX2) and IL-17F are autophagy-related cytokines in the autophagy process and enhance the autophagy machinery. Confirming a potential regulatory function of miR-144 through autophagy on periodontal inflammation, increased miR-144 expression has been associated with decreased COX2 and IL-7F expression in the gingival tissues from periodontal patients [20]. Both COX-2 and IL-17F are target genes of miR-144, explaining the negative correlation between miR-144 and COX2/IL-7F [20,32].
MiRs are also involved in the cellular decision making between cell survival and cell death and between apoptosis and necroptosis. One miR at the heart of this decision making process is miR-214 [33] as it targets the Activation transcription factor 4 (ATF4), which is at the center of controlling life-death decisions [34] as well as the switching between apoptosis and necroptosis [35]. This key role of miR-214 in cell death fate decisions was elucidated by an elevation of miR-214, receptor-interacting serine-threonine protein (RIP)1, RIP3 and phosphor-mixed lineage kinase domain-like (p-MLKL) proteins and a reduction in ATF4 expression in gingival tissues of diabetic periodontitis patients associated with necroptosis [33]. The RIP1/RIP3/MLKL signaling pathway is required for necroptosis and activated to generate the necrosome for the initiation of necroptosis [36,37]. Confirming the relationship between miR-214 and ATF4, experimental studies demonstrated that inflammatory conditions upregulated miR-214 expression and subsequently downregulated ATF4 expression [33].
microRNA regulation of angiogenesis in gingival tissues
Angiogenesis is one of the primary co-factors of chronic inflammation. Chronic inflammation is almost always accompanied by angiogenesis, and angiogenesis can facilitate inflammation as well [38]. Inflammation-induced aberrant angiogenesis often prolongs or intensifies the inflammatory response [39]. During gingival inflammation, angiogenesis is initiated by pro-angiogenic growth factors and cytokines including Vascular endothelial growth factor (VEGF), IL-1, IL-6 and TNF-α released from gingival resident cells. MiRs exert pro-angiogenic or anti-angiogenic functions through regulating the expression of these pro-inflammatory and pro-angiogenic cytokines and signaling molecules. One of the miRs upregulated in inflamed gingival tissues is miR-200b [22]. MiR-200b regulates a set of gene targets and biological pathways associated with angiogenesis such as VEGF receptor 2 (VEGFR2, also called KDR), GATA-binding protein 2 (GATA2) and Zinc finger E-box-binding homeobox 1 (ZEB1) [22]. Both GATA2 and ZEB1 are transcription factors and control the VEGF/VEGFR signaling axis. GATA2 regulates the expression of VEGF receptors [40], while ZEB1 regulates VEGF expression [41]. As a result, miR-200 inhibits angiogenesis by inducing endothelial cell apoptosis via ZEB1 inhibition [42] and/or by silencing GATA2 and VEGFR2 [43]. We have summarized the changes in the transcription of well-known miRs in inflamed gingival tissues in Table 1 and presented original data related to miR-100, miR-125a, miR-146a, and miR-155 in inflamed gingival tissues in Figure 2.
Table 1.
miRNA regulation of Inflammation in gingival tissues
| miRNAs | Expression | Targets | Function | References |
|---|---|---|---|---|
| miR-205 | Downregulated | IL-6st | Increase IL-1, IL-6 and TNF-a expression, Activate JAK/STAT3 signaling | Li et al. 2020 |
| miR-335 | Downregulated | TPX2 | Increase IL-1, IL-6 and TNF-a expression, Activate JAK/STAT3 signaling | Lian et al. 2019 Gu et al. 2020 |
| miR-21 | Upregulated | PDCD4 | Increase IL-1, IL-6 and TNF-a expression | Zhou et al. 2018 |
| miR-144 | Upregulated | COX2, IL-7F | Inhibit autophagy | Li et al. 2019, Yao et al. 2018 |
| miR-146a | Upregulated | IRAK1 | Increase IL-1, IL-6 and TNF-a expression |
Sanada et al. 2020
Jiang et al. 2018 |
| miR-214 | Upregulated | ATF-4 | Cell life-death decision, and apoptosis-necroptosis switch | Ou et al. 2019 |
| miR-200b | Upregulated | VEGF, EZB1, GATA2 | Inhibit angiogenesis | Magental et al. 2011, Chan et al. 2012 |
| miR-203 | Upregulated | SOCS3/6 | Increase IL-1, IL-6 and TNF-a expression, Activate SOCS/STAT3 signaling | Moffatt and Lamont, 2011 |
Figure 2. Inflammation-related microRNA expression in healthy and diseased gingiva and periodontal ligament, human gingival fibroblasts and PDL fibroblasts, human monocytes, and mouse RAW cells.
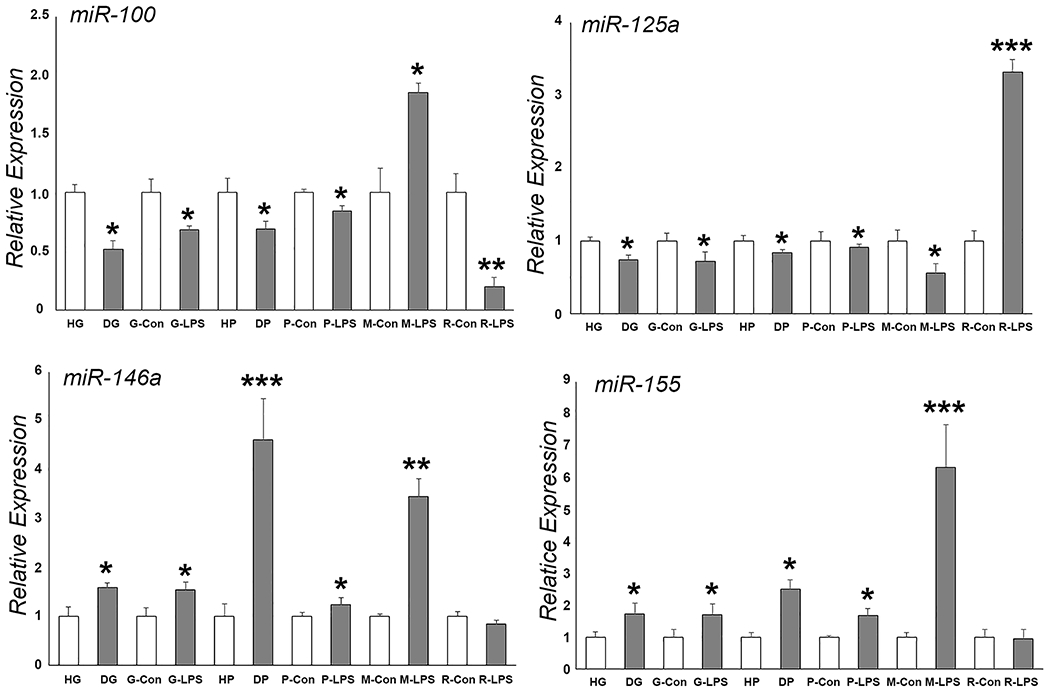
There was a significant upregulation or downregulation of select miRs (miR-100, miR-125a, miR-146a, and miR-155) in response to inflammatory conditions. * p <0.05, ** p < 0.01, *** p < 0.001. HG = healthy gingiva, DG = diseased gingiva, G-Con = human gingival fibroblast control, G-LPS = LPS treated gingival fibroblasts, HP = healthy periodontal ligament, DP = diseased periodontal ligament, P-Con = human PDL cells, P-LPS = LPS treated PDL cells, M-Con = human monocyte control, M-LPS = LPS treated human monocytes, R-Con = RAW264.7 murine monocyte/macrophage like cells, R-LPS = LPS treated RAW264.7 cells.
MicroRNA regulation of periodontal cell functions and osteogenic differentiation under inflammatory conditions
The periodontal ligament (PDL) is a unique connective tissue at the interface between the tooth root cementum and the alveolar bone. In addition to its primary function as a mediator of masticatory force transmission, the PDL plays important roles in periodontal homeostasis and regeneration. Histologically, the PDL consists of various cell types including fibroblasts, cementoblasts and osteoblasts, and hosts periodontal ligament stem cells (PDLSCs) [44]. Cell lineage tracing experiments in rodents have revealed that PDLSCs differentiate into cementoblasts, osteoblasts and periodontal ligament fibroblasts during cell turn-over as well as following injury [45–48]. Within the periodontium, PDLSCs respond to surrounding microenvironmental stimuli, such as inflammation, mechanical force, and chemical challenges, which influence their cell fate decisions and affect periodontal homeostasis and regeneration. Numerous studies have established the essential role of miRs in regulating PDLSC function in vivo and in vitro.
PDLSC culture systems are commonly employed to study the mechanisms by which miRs regulate PDLSC function under inflammatory conditions. In these in vitro environments, total heat-inactivated or sonicated bacteria, bacterial virulent factor LPS, inflammatory cytokines or chemicals are often used to mimic the in vivo microenvironment resulting from bacterial infection or chronic inflammation. These in vitro systems have been successfully applied to study the involvement of miRs in the cellular inflammatory response as it relates to PDLSC proliferation/apoptosis and osteogenic differentiation. Several studies have implicated miR-132, miR-143 and miR-302b in the inhibition of proliferation and the promotion of apoptosis through the ATK signaling pathway [49–51]. LPS challenge has resulted in an upregulation of all three miRs, miR-132, miR-143 and miR-302b in PDLSCs. MiR-132 and miR-143 have been demonstrated to interact with the long noncoding RNAs taurine-upregulated gene 1 (TUG1) and maternally expressed gene 3 (MEG3) respectively, two regulators of the ATK signaling pathway [52]. Other studies have revealed that TUG1/miR-132 and MEG3/miR-143 interactions suppress AKT signaling and contribute to LPS-induced proliferation and apoptosis of PDLSCs [49,50]. In contrast, miR-302b has been shown to directly target insulin-like growth factor type 1 receptor (IGF-1R). Explaining the effect of miR-302b, overexpression of miR-302b and silencing of IGF-1R decreases Protein kinase B (PKB, also known as AKT) phosphorylation, which then modulates AKT related cell cycle regulators (cyclin A2, cyclin D1, Cyclin dependent kinase 2 and 6) and apoptotic proteins B-cell lymphoma 2 (BCL2)/Bcl-2 Associated X-protein (BAX) resulting in apoptosis [51,53].
Microenvironmental changes in chemical or physical conditions may also affect PDLSC cell fate. For example, hydrogen peroxide (H2O2), a wide-used dental clinical chemical for tooth bleaching, induces severe PDLC apoptosis via miR-24 upregulation. In experimental studies, H2O2 treatment has been demonstrated to significantly increase miR-24 expression [54]. In these studies, bioinformatics analysis and dual luciferase reporter gene assay revealed that miR-24 interacted with X chromosome-linked inhibitor of apoptosis (XIAP) and inhibited XIAP gene expression [54]. As a result, the gene silencing of XIAP reduced AKT activity, suggesting that the miR-24/XIAP/AKT pathway is involved in H2O2-induced cell apoptosis [54]. Hyperglycemic conditions as they occur in diabetic patients are another microenvironmental challenge for PDLC survival that profoundly affect periodontal health. Explaining the miR contributions to PDLC cell fate in diabetic patients, high glucose (30mM) conditions in PDLC culture medium have been demonstrated to increase apoptosis and caspase-3 expression while decreasing miR-222 and miR-223 expression. Caspase-3 is an established target for miR-222 and miR-223 and facilitates apoptosis [55]. A third microenvironmental condition destined to affect periodontal cell fate is hypoxia, which is also associated with apoptosis. It has been demonstrated that low O2 concentrations downregulate miR-646 in vitro while they upregulate IGF-1 expression in PDLSCs. The hypoxia mediated regulation of PDLSC apoptosis occurs through the miR-646/IGF-1 signaling pathway [56].
Inflammatory conditions that occur during periodontal disease not only impair cell proliferation but also suppress cell differentiation. When cultured PDLSCs were exposed to the proinflammatory cytokine Tumor necrosis factor α (TNF-α), Sprouty 1 (Spry1) gene expression was upregulated and miR-21 expression was downregulated. Spry1 is a miR-21 target gene, and increasing Spry1 gene expression inhibits the osteogenic differentiation of PDLSCs [57], explaining the detrimental effect of inflammatory conditions on periodontal homeostasis [4,6]. Recently, our group reported that miR-138 functions as an inflammatory miR and a potential regulator of periodontal stem cells as they affect periodontal homeostasis under inflammatory conditions [58]. Treatment with inflammatory modulators LPS and IL-6 resulted in a significant increase in miR-138 expression, while Osteocalcin (OCN) and Runt-related transcription factor 2 (Runx2) expression was significantly decreased. Our studies demonstrated via a dual luciferase reporter gene assay that miR-138 directly targets the OCN gene. Application of a miR-138 inhibitor or of OCN protein in PDLSC culture partially rescued inflammatory modulator-induced suppression of PDLSC differentiation, establishing miR-138 as a key miR modulator of periodontal homeostasis under inflammatory conditions [58]. Another miR, miR-148, might also act as a modulator of periodontal mineralization under inflammatory conditions, as it has been demonstrated that LPS exposure inhibited the mineralization capability, reduced Neuropilin 1 (NRP1) expression and increased the expression of miR-148 in PDLSCs. In this study, the expression level of miR-148 in PDLSCs was negatively correlated with the expression of NRP1 and the osteogenic ability of PDLSCs [59].
Next to PDLSCs and alveolar bone progenitors, cementoblasts are the third progenitor cell population subtype in the periodontal ligament. Cementoblasts participate in cementum homeostasis and regeneration. Recent studies have implicated several miRs in the regulation of cementoblast function under inflammatory conditions, resulting in changes in cementoblast viability and differentiation through targeting downstream genes. As an example, miR-181b promoted cementoblast apoptosis under inflammatory conditions through the miR-181b/IL-6/ Nuclear factor kappa B (NF-κB) signaling pathway [60]; miR-325 inhibited cementoblast differentiation by targeting the RunX2 gene [61]; and miR-155 suppressed cementoblast differentiation and mineralization through the miR-155/KCTD1/Wnt pathway [62]. KCTD1 (potassium channel tetramerization domain containing 1) is a negative regulator of the canonical Wnt signaling pathway [63], explaining the effect of inflammation-induced miR-155 on periodontal mineralized tissue homeostasis. A summary of changes in key miR changes upon introduction of inflammatory or osteogenic conditions is presented in Table 2. Corresponding original data related to miR-100, miR-125a, miR-146a, and miR-155 under inflammatory conditions are displayed in Figure 2.
Table 2.
miRNA regulation of PDL cell function
| miRNAs | Treatments | Targets | Functions | References |
|---|---|---|---|---|
| miR-132 | LPS | AKT | AKT signaling PDL Proliferation/apoptosis |
Han et al. 2019
Dong et al. 2020 |
| miR-143 | LPS | AKT | AKT signaling PDL Proliferation/apoptosis |
Duan et al. 2020 |
| miR-302b | LPS | AKT | AKT signaling PDL Proliferation/apoptosis |
Duan et al. 2020
Guo et al. 2017 |
| miR-24 | H2O2 | XIAP | AKT signaling PDL apoptosis |
Liu et al. 2016 |
| miR-222 | High glucose | Caspase 3 | PDL apoptosis | Monteiro et al. 2020 |
| miR-223 | High glucose | Caspase 3 | PDL apoptosis | Monteiro et al. 2020 |
| miR-646 | Low O2 | IGF-1 | PDL apoptosis | Yang et al. 2018 |
| miR-21 | TNF-a | SPRY-1 | PDL differentiation | Yang et al. 2017 |
| miR-138 | LPS, IL-6 | OCN | PDL differentiation | Zhou et al. 2016 |
| miR-148 | LPS | NRP1 | PDL differentiation | Bao et al. 2019 |
| miR-181b | TNF-a | IL-6 | Cementoblast apoptosis | Wang et al. 2019 |
| miR-155 | TNF-a | KCTD1 | Cementoblast differentiation | Wang et al. 2017 |
| miR-325 | Osteogenic induction | RUNX2 | Cementoblast differentiation | Wang et al. 2020 |
MicroRNA regulation of the immune inflammatory response in periodontal diseases.
The inflammatory response that occurs during the response of the periodontal apparatus to pathogens is an essential function of the immune system. This inflammatory response involves both innate and acquired immunity, and includes all major immune effector cells such as neutrophils, macrophages, T lymphocytes and B lymphocytes [13]. Tissue-specific types of immune cells during the progression of periodontal inflammation correspond to the severity of inflammation [64–67]. Recent studies have identified the infiltrating immune cell subtypes from healthy controls and periodontitis derived tissues from the GENE Expression Omnibus (GEO) database using CIBERSORT gene signature files [68,69]. Based on these studies, resting dendritic cells, M1 and M2 macrophages, T cells and B cells were mainly present in healthy periodontal tissues, whereas neutrophils increased in gingival tissues in response to the level of inflammation, and plasma and naïve B cells were elevated in diseased tissues from periodontitis patients [68,69]. This unique signature of immune cell distribution in periodontal tissues indicates that distinct immune cells play discrete roles in periodontal inflammation and disease. The regulation of the immune inflammatory response by miRs is executed through integrated miR-mRNA regulatory networks in innate and acquired immune cells.
MicroRNA regulation of innate immunity
The initial period of defense against oral bacteria is called the innate immune response as it contributes and functionalizes cells such as neutrophils, macrophages, and dendritic cells in the primary host response against infection. Among these three cell types, neutrophils are the most important phagocytic cells and the primary effectors of the host defense against acute bacterial infection. The miRs active in periodontal neutrophils fine-tune gene expression within individual gene networks and buffer transcriptional variation [70]. Functionally relevant neutrophil miRs include miR-155, and miR-223, which are upregulated in periodontal disease, and miR-17 and miR-31, which are downregulated in periodontal disease [5].
In addition to neutrophils, macrophages constitute a second layer of the innate immune response against bacteria as they digest and present antigens to other immune cells and facilitate interactions with the adaptive immune system [71]. In inflamed periodontal tissues, macrophages respond to LPS and activate multiple host defense functions through the production of inflammatory mediators [5]. The regulation of macrophage differentiation and activation and their response to LPS is finely regulated by miR networks [72,73] and consists of individual miRs such as miR-24, miR-30b and miR-142-3p which have antagonistic effects against periodontal inflammation and inflammation-related tissue destruction [5]. Other periodontal miRs involved in the macrophage response include miR-29b and let-7f and the inflammatory miRs miR-146a and miR-155 involved in the macrophage activation in inflamed gingival tissues [74–78]. The expression of these miRs is NF-κB dependent. MiR-146 is considered a “fine-tune” negative feedback regulator in innate immunity, while miR-155 affects both macrophage activation and function. Moreover, miR-155 as well as two other miRs, miR-125b and miR-21 affect M1/M2 macrophage ratios and macrophage polarization [79].
Dendritic cells are professional antigen-presenting cells involved in the presentation of antigens. They also contribute to the activation of T cells, killer T cells, and B cells in periodontal and other tissues, and comprise the third major cell type involved in the periodontal innate immune response. During the progression of periodontal disease, dendritic cells serve as a link between innate and adaptive immunity [80,81]. In dendritic cells, miRs affect primary and secondary immune responses by regulating the gene expression of transcription factor networks [5]. Earlier studies reported a number of dendritic cell-related miRs including miR-24, miR-30, miR-126, miR-142, miR-146, miR-155, and let-7i that were upregulated in a mouse periodontitis model, and the role of these miRs in periodontal disease was confirmed in published human studies [5].
MiR regulation of adaptive immunity
Adaptive immunity is an acquired immune response to invading pathogens. Adaptive immunity begins with an initial trigger event that leads to an immunological recognition of a pathogen which is then followed by an enhanced immunological response upon repeat encounters with the same pathogen [5]. In general, the adaptive immune system is based on the ability of lymphocytes to eliminate pathogens or interfere with their growth. There are two types of adaptive immunity, antibody-mediated immune responses against freely circulating pathogens facilitated by B cells and cell-mediated immune responses against intracellular pathogens that are carried out by T cells [5].
T cells are core member cells of the cellular (macrophage/lymphocyte) immune response and are essential for both specific antibody production and polyclonal B-cell activation. T-cells carry T-cell receptors on their cell surface. MiRs also affect T cell function both directly and indirectly as it has been demonstrated that upregulation of the miR-214 and miR-17-92 cluster promotes T cell activation and proliferation while miR-146 functions as a feedback regulator of NF-κB signaling and modulates the productive immune response, division and growth of T cells [82]. So far, three miRs, miR-155, let-7 family members and miR-126 have been shown to affect Th2 differentiation by regulating cytokine production [83].
The functional antagonism between regulatory T cells (Treg cells) and the IL-17 producing T helper 17 cells (Th17) has been proposed to be a major factor in the pathogenesis of periodontitis [84]. Th17 cells secrete IL-17 and recruit neutrophils and macrophages to participate in and amplify the inflammatory reaction during periodontal disease [85–87]. Treg cells are characterized by high levels of miR-155 and miR-146 and reduced levels of miR-24, miR-31 and miR-125 expression [88,89]. In contrast, Th17 cell differentiation is promoted by the miR-17-92 cluster member miR-19b [90], as well as miR-301 and miR-155. Supportive of a participatory role of miRs in the Treg/Th17 functional antagonism during periodontal disease progression, previous studies have identified an upregulation of miR-17, miR-19, miR-155 and miR-301 in the gingival tissues of periodontitis patients and animals when compared to healthy controls [74,75,91].
In addition to T cells, B cells are the second major cellular component of the adaptive immune system. B cells contribute to humoral immunity by secreting antibodies. They also contribute a substantial proportion of the inflammatory infiltrate in diseased periodontal tissues. [92,93]. Remarkably, the expression of the five periodontal inflammation-related miRs associated with periodontal disease, miR-125, miR-148, miR-155, miR-181 and miR-217 [74,75,91] are elevated in advanced periodontal lesions. These five miRs regulate the progression of B cell terminal differentiation by targeting a coordinated network of transcription factors, suggestive of a correlation between periodontal disease progression and B cell differentiation, [5].
Together, these studies highlight some of the miRs involved in the innate and adaptive immune response against periodontal pathogens and the initial periodontal disease progression as it involves neutrophils, macrophages, and dendritic cells as well as T and B lymphocytes.
LncRNA-miR interactions in the periodontal inflammatory response
Long non-coding RNAs (lncRNAs) are transcribed RNA molecules with a length of more than 200 nucleotides. LncRNAs mediate their functions through interactions with proteins, RNA, DNA, or a combination of these [94]. LncRNAs can bind to complementary RNA and affect RNA processing, turnover or localization. During the interaction between a lncRNA and a miR molecule, the lncRNA functions as a miR sponge which indirectly de-represses the expression of a mRNA targeted by the miR [94]. Numerous studies have identified lncRNAs acting as endogenous sponges or decoys that regulate the expression and function of miRs in periodontal cells and tissues, which in turn suppress the targeting of mRNAs and affect diverse cellular processes such as inflammatory response, cell proliferation, migration and differentiation [49,50,95–100].
Lnc RNA expression is often significantly altered in response to periodontal inflammation. For example, the expression of lncRNA 01126 (LINC01126) and metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) was highly increased in inflamed gingival tissues or in human PDLSCs subjected to inflammatory conditions. In vitro experiments have demonstrated that LINC01126 inhibits proliferation and promotes apoptosis and inflammatory pathways of PDLSCs via sponging of miR-518a, which directly targets Hypoxia-Inducible Factor (HIF)-1a [95]. Similarly, MALAT1 acts as a sponge for miR-20a. In turn, miR-20a inhibits TLR4 via binding to the TLR4 3’-UTR in an RNA-Protein complex together with Ago2 in a cell-type specific manner [96], establishing the MALAT1 lncRNA/miR-20a axis as a regulatory mechanism for periodontal innate immunity. Together, these studies indicate that lncRNAs promote periodontitis pathogenesis through LINC01126/miR-518a/HIF-1a and MALAT1/miR-20a/TLR4 pathways. In contrast, other lncRNAs such as maternally-expressed gene 3 (MEG3), taurine-upregulated gene 1 (TUG1) and FYVE, RhoGEF and PH domain containing 5 antisense RNA 1 (FGD5-AS1) were downregulated in gingival tissues of periodontitis patients and in LPS-treated human PDLSCs. Further studies have confirmed the sponging effects of lncRNAs on miRs, resulting in a downregulation of miR-143 through the lncRNA MEG3, a downregulation of miR-132 through TUG1 and a reduction in miR-142 via FGD5-AS1 [49,50,97]. Downregulation of MEG3, TUG1 and FGD5-AS1 through their downstream miRs leads to an activation of AKT/IKK and NF-kB pathways, which then contribute to the pathogenesis of periodontitis [49,50,97].
lncRNAs also participate in the regulation of PDLSC proliferation and differentiation. A number of studies have elucidated the effects of lncRNA-miR interactions on PDLSC differentiation using PDLSCs cultured under osteogenic induction conditions. Osteogenic differentiation conditions resulted in an increase in the expression of lncRNAs Fer-1-like family member 4 (FER1L4), prostate cancer-associated ncRNA transcript-1 (lncPCAT1), antisense to the cerebellar degeneration-protein 1 transcript (CDR1as), TUG1, X-inactive specific transcript (XIST), human periodontal ligament stem cell (hPDLSC) osteogenesis impairment-related lncRNA (lncRNA-POIR) in cultured PDLSCs [98–103] and a decrease in Antidifferentiation noncoding RNA (ANCR) expression [104]. In these studies, changes in lncRNA expression resulted in the changes of their miR sponging activities, resulting in altered expression of downstream miR-targeted osteogenic growth factors, transcription factors, and signaling molecules. For example, in one study overexpression of FER1L4 promoted osteogenic differentiation of PDLSCs. FER1L4 has been shown to directly interact with miR-874 targeting VEGF, a crucial gene for angiogenesis-mediated osteogenesis [98]. By targeting another osteogenic growth factor BMP2, PCAT1 sponges miR-106a and regulates the miR-106a-target gene BMP2, resulting in changes in the expression of the major bone growth factor BMP2 and directly impacting osteogenesis. Furthermore, miR-106a regulates the expression of E2F5, which binds to the promoter of the lncRNA PCAT1 and forms a feed-forward regulatory network targeting BMP2 [99]. Thus, multiple lncRNA pathways involving miR-106a or miR-874 affect upregulation of osteogenesis, either through VEGF or through BMP2.
Similar to BMPs, GDF5 is another member of the TGF-β growth factor superfamily. Osteogenic differentiation of PDLSCs is associated with an upregulation of CDR1as and GDF5 lncRNAs expression and a downregulation of miR-7 expression [100]. Mechanistic studies have revealed that CDR1as is an upstream sponge whereas GDF5 is a downstream target of miR-7. These studies have demonstrated that the CDR1as/miR-7/GDF5 pathway regulates the phosphorylation of Smad 1/5/8, the major signaling molecules of the TFG-β pathway involved in osteogenesis [100]. LncRNA-miR interactions not only regulate Smad 1/5/8, but also the activity of Smad 2/7. This includes the TUG1/miR-222 and Smad 2/7 pathways as they enhance the expression of RunX2, ALP and OCN, and facilitate osteogenic differentiation of PDLSCs [101]. These studies have demonstrated that lncRNAs positively regulate osteogenesis through lncRNA-miR interactions that affect Smad signaling and target osteogenic downstream gene expression.
Interestingly, a novel lncRNA named periodontal ligament stem cell osteogenesis impairment-related lncRNA (lncRNA-POIR) was downregulated in PDLSCs from periodontitis patients while upregulated upon osteogenic induction. LncRNA-POIR acted as a competing endogenous RNA for miR-182 and led to derepression of the miR-182 target gene FoxO1. FoxO1 promotes PDLSC osteogenic differentiation by competing with Transcription factor 4 (TCF-4) for β-Catenin and inhibiting the canonical Wnt pathway [103]. In contrast to these osteogenesis-enhancing lncRNAs, lncRNA-ANCR suppressed osteogenic differentiation of PDLSCs. LncRNA-ANCR targeted miR-758 while miR-758 regulated Notch2 expression by targeting Notch2 3’-UTR, resulting in a reduced expression of ALP, RunX2 and Osterix (Osx) [104]. These studies demonstrate that lncRNA affect osteogenic differentiation by regulating miR targets related to osteogenesis. Some of the known interactions between miRs and lncRNAs in gingival and periodontal cells and tissues are summarized in Table 3.
Table 3.
miRNA-lncRNA interaction
| miRNAs | Tissues/Cells | Sponges | Targets | Functions | References |
|---|---|---|---|---|---|
| miR-20a | Gingival | MALAT1 | TLR4 | Innate immunity | Li et al. 2019 |
| miR-132 | Gingival | TUG1 | PTEN FOXO3a | AKT/IKK and NF-kB pathway |
Han et al. 2019
Wong et al. 2013 |
| miR-142 | Gingival | FGD5as | COX2 MLE-12 | AKT/IKK and NF-kB pathway |
Chen et al. 2019
Guo et al. 2017 |
| miR-143 | Gingival | MEG3 | ITGA6 MSI2 | AKT/IKK and NF-kB pathway |
Dong et al. 2020
Jin et al. 2018 Wang et al. 2019 |
| miR-518a | Gingival | LINC01126 | HIF-1a | PDL Proliferation/apoptosis | Zhou et al. 2020 |
| miR-7 | PDL cells | CDR1as | GDF5 | PDL Differentiation | Li et al. 2018 |
| miR-106a | PDL cells | PCAT1 | BMP2 | PDL Differentiation | Jia et al. 2019 |
| miR-182 | PDL cells | POIR | FOXO1 | PDL Differentiation | Wang et al. 2016 |
| miR-222 | PDL cells | TUG1 | Smad2/7 | PDL Differentiation | Wu et al. 2020 |
| miR-758 | PDL cells | ANCR | Notch2 | PDL Differentiation | Peng et al. 2018 |
| miR-874 | PDL cells | FERIL4 | VEGF | PDL Differentiation | Huang et al. 2020 |
MiR gene polymorphism associated with chronic inflammatory periodontitis susceptibility
The host inflammatory response plays a significant role in the progression of periodontal disease. The intensity, tissue-specific pathogenesis, and the duration of the host response are determined by the innate and acquired immunity of the host and affected by genetic factors. As a complex disease, periodontitis is associated with genetic variations in multiple genes, each of which has a defined role and a relative risk during disease progression [105,106]. The outcome of periodontal chronic inflammation is also affected by the interactions among genetic, behavioral and environmental factors [107]. To identify the genetic risk factors involved in periodontitis, several techniques have been used including linkage analysis, haplotype analysis, candidate gene association studies and genome-wide association studies. Candidate gene association studies are frequently used to identify genes that may present specific risk factors for periodontitis [105]. There are three types of candidate genes: functional candidate genes, positional candidate genes and expressional candidate genes [106,108]. Genetic research in periodontitis has mostly focused on functional candidate genes and their polymorphisms related to the immune system, tissue destruction process and metabolic mechanisms [105]. In addition to genetic factors, geographical and ethnic factors also play a significant role as contributing factors toward periodontal disease susceptibility, as the prevalence of specific polymorphisms varies greatly depending on the population studied [109]. Racial differences in genes encoding IL-1, IL-6, TNF-a, Vitamin D receptor (VDR), CD14, Fcγ receptors Fcγ RIIa, FcγRIIIa, FcγRIIIb, and matrix metalloproteinase-1 (MMP-1) have been discovered and the single nucleotide polymorphisms (SNPs) of these genes have been associated with periodontitis in African-American, Caucasian and Asian populations [106].
Genetic susceptibility affects periodontal pathogenesis not only due to genetic variations among extracellular matrix genes or genes of the immune system but also affects regulatory genes such as miRs. MiRs regulate gene expression through post-transcriptional modulation and play multiple roles in all aspects of periodontal homeostasis and are involved in periodontal inflammation and disease [6,105,110]. Genetic variants in miR genes affect many aspects of miR biology, including transcription and maturation, binding affinity, the specificity of target gene interactions and the degree of disease risk [111–114]. Over the recent decade, several studies have confirmed and defined the contribution of individual miRs toward the initiation and progression of periodontal disease [111,112,115].
MiR-125 and the risk of periodontitis
The human miR-125 family consists of three homologs, hsa-miR-125a, hsa-miR-125b-1 and hsa-miR-125b-2. The miR-125a gene is located on chromosome 19q13, hsa-miR-125b-1 on chromosomes 11q23 and hsa-miR-125b-2 on chromosome 21q21 [116]. All three homologs share the same seed sequence. The miR-125 family contributes to inflammation and both miR-125a and miR-125b constitutively activate NF-kB signaling [117], which is consistent with the finding that miR-125a expression is upregulated in the gingival tissues of periodontitis patients [6,17].
In an effort to elucidate the role of miR-125 polymorphism in periodontitis, five miR-125a SMPs, rs12976445 (T>C), rs41275794, rs10404453, rs12975333, rs78758318, were genotyped by direct dye-terminator sequencing in a sample population from South India [112]. These polymorphisms are located about 200 bp upstream and downstream of the pri-miR-125a sequence, suggesting that they may affect the maturation of the pre-miR-125 sequence [112,118]. The recessive model for miR-125a polymorphism rs12976445 was found to be statistically significant in periodontitis subjects when compared to healthy control individuals. Haplotype analysis revealed that the haplotype “GCGGCA” was higher in the periodontitis group than control group. In addition, pairwise linkage disequilibrium analysis demonstrated that the polymorphisms rs41275794 and rs12976445 in miR-125a were in a strong linkage equilibrium. However, statistical evaluation did not reveal a significant difference between clinical parameters and the genotypes of the miR-125a polymorphisms [112]. Together, these data indicate that even though several SNPs were detected in the miR-125a gene, their effect on periodontal disease parameters remains to be established.
MiR-146a and the risk of periodontitis
The human miR-146 family is composed of two homologs, miR-146a and miR-146b. The miR-146a gene is located on chromosome 5 while the miR-146b gene is located on chromosome 10. Even though the mature sequences of these two miRs only differ by two nucleotides at the 3’ end, their biological functions do not seem to be redundant [119]. MiR-146a is primarily involved in the regulation of inflammation and functions in the innate immune system [120]. The expression of miR-146a is upregulated in periodontitis and the level of miR-146a expression is positively correlated with the severity of periodontal diseases [6,28,121].
The association between miR-146a polymorphism and periodontitis has been studied using polymorphism evaluation, haplotype and linkage analyses as well as logistic regression analysis in an Iranian population and an Indian population [111,115]. The miR-146a polymorphisms are located at the 60th nucleotide position in the seed region of pre-miR-146a with a genotype C>G (rs2910146, C>G), the −386 nucleotide position in the promoter region (rs57095323, A>G), and the −690 nucleotide position in the promoter region (rs73318382, A>C) [122]. Polymorphism studies comparing healthy, periodontitis and peri-implantitis patients from Iran revealed a significant association between miR-146a (rs2910146) and susceptibility to periodontitis and peri-implantitis [115]. However, no statistically significant association between miR-146a and periodontitis susceptibility was detected in a similar comparison in an Indian population, likely due to environmental factors and lifestyle [111]. However, haplotype analysis of the three SNPs in the miR-146a gene revealed that specific haplotypes were detected at significantly higher levels in control groups compared to periodontitis groups, exhibiting an inverse association with periodontitis [111].
miR-196 and the risk of periodontitis
MiR-196 family has three members, miR-196a-1, miR-196a-2 and miR-196b, and their genes are located on chromosome 17, chromosome 12, and chromosome 7, respectively. The genes encoding miR-196a-1 and miR-196a-2 share the same functional mature sequence, while there was only a single nucleotide difference between miR196a and miR-196b. MiR-196a-2 contains two types of mature miRs, miR-196a2-5p and miR-196a2-3p, processed from the same stem loop. Functional studies demonstrated that miRs from the miR-196 family play important roles in development and immunity by targeting specific genes rather than affecting a broad spectrum of genes [111,123]. Supportive of its role in periodontal disease, miR-196a expression was downregulated in periodontitis patients with obesity [22].
Association analysis of miR-196a2 variants with the risk of periodontitis was carried out in the same Indian population that was subjected to miR-146 polymorphism studies as mentioned above [111]. The miR-196a2 polymorphisms were located in the mature sequence of miR-196a2-3p (passenger strand, rs11614913, C>T). The variant genotype (TT) in the miR-196a2 polymorphism (rs11614913) was statistically higher in controls compared to periodontitis patients, suggesting a significant inverse association with chronic periodontal disease. Haplotype analysis revealed 13 different combinations, five of which demonstrated an inverse association with periodontitis [111].
miR-499 and the risk of periodontitis
In general, miR-499 acts as an inflammation suppressor as it ameliorates inflammatory damage by targeting inflammatory cytokines [124]. Possible targets of miR-499 include IL-17Rβ, IL-23α, IL-2R, IL-6, IL-2, and IL-18R implicating the miR-499 gene family in inflammation-related signaling pathways. In support of an immunomodulatory role of miR-499 in periodontal disease, miR-499a expression was downregulated in gingival tissues of periodontitis patients [6,91].
Human miR-499 rs3746444 (T > C) polymorphism is located in the seed region of the mature miR-499 sequence. This T > C polymorphism-resulted mismatch may affect target mRNA expression [125] and has been associated with asthma susceptibility and airway inflammation [126]. Studies on the association of miR-499 variants in an Indian and an Iranian population [115] demonstrated a significant association of the miR-499a polymorphism (rs3746444) variant allele with periodontitis. These results suggest that miR-499 rs3746444 contributes to the risk of periodontitis.
miR-17 and the risk of persistent apical periodontitis
MiR-17 is a member of the miR-17-92 cluster, which is composed of miR-17, miR-18a, miR-10a, miR-19b, miR-20a and miR-92-1. The miR-17-92 host gene was originally identified as chromosome 13 open reading frame 25 (C13orf25) [127,128]. Recent studies have identified miR-17 as an important player in the development and homeostasis of the immune system. MiR-17 regulates T cell activation by targeting Ikaros, Phosphatase and tensin homolog (PTEN) as well as Forkhead box protein P3 (FOXP3) co-regulators [90,129]. In addition, miR-17 also participates in macrophage activation by targeting signal-regulatory protein a (SIRPa) [130]. Underscoring the role of miR-17 as a regulator of immunity, miR-17 expression was increased in patients with chronic periodontal disease [131]. However, a clinical association between miR-17 gene polymorphism and periapical lesions has not yet been established. A recent Brazilian study trying to link miR-17 rs4284505 allelic and genotypic polymorphism with persistent apical periodontitis models based on salivary samples failed to detect a statistically significant association in a relatively small patient cohort [132].
MiRs at the microorganism/host interface and cross-species gene regulation in periodontal inflammation and disease
Periodontal inflammation and periodontal disease are the consequences of an invasion of periodontopathic microorganisms into the periodontal region and the reaction of periodontal tissues against this invasive threat [8,133,134]. At the onset of periodontal disease, bacterial plaque or microbial biofilm accumulate at and below the gingival margin and cause a limited and inflammatory infiltrate as a physiological immune surveillance [135]. As the microbial biofilms persist, the inflammatory reaction expands continuously and affects surrounding gingival and periodontal tissues. The effect of continuous inflammation on the integrity of the tooth root support tissues results in the formation of a clinically detectable periodontal lesion. In addition to periodontal bacteria, periodontal lesions contain multiple viruses (primarily herpesvirus, HHV), which have been associated with the risk of periodontal disease [136,137]. It has been proposed that periodontal HHV infections result in the release of pro-inflammatory cytokines and subsequently impair antibacterial immune mechanisms, causing an overgrowth of periodontopathic bacteria [133,138]. Viral miRs from periodontal lesions may not be restricted to the HHV host but may also be transmitted from one species to another, opening the door to a novel route-of-entry for viral miRs into periodontal tissues [139,140]. In turn, controlling the exacerbation of periodontal disease by minimizing the effect of periodontal HHV in periodontal lesions may provide novel avenues for therapeutic intervention.
MiRs play crucial roles in the interplay between pathogens and host as part of the host defense against invading pathogens as well as microorganism-derived modulators interfering with host cell function during pathogenesis [141]. Based on this relationship, miRs from both pathogens and host may function through four different mechanisms: (i) pathogen-based miRs regulate pathogen gene expression; (ii) pathogen-based miRs regulate host gene expression, (iii) host miRs regulate pathogen gene expression and (iv) pathogen infection alters host miR and mRNA expression profiles [141]. Several studies have unveiled that regulatory sRNA are transferred to host cells via extracellular vesicles (EVs), including sRNAs from gram-positive and gram-negative bacteria [142], viral miRs [143,144], sRNAs from parasites [145,146], and plant- and food-derived RNAs [147–149]. These bacterial, viral, parasitic and plant-derived RNAs have been identified in the human circulatory system, often encapsulated in EVs [150].
Bacterial small RNAs (sRNAs) with miR characteristics from periodontopathic bacteria have not been reported. However, secretable small RNAs released via outer membrane vesicles have been detected in periodontal pathogens [151]. Similar to their cells of origin, P. gingivalis vesicles containing RNAs and DNAs are able to invade oral epithelial cells and gingival fibroblasts to promote aggregation of specific oral bacteria and to induce host immune responses [152], representing a novel gene transfer mechanism between P. gingivalis strains and periodontal tissues. In addition, secreted extracellular RNAs (exRNAs) encapsulated in outer membrane vesicles (OMVs) from Aggregatibacter actino-mycetemcomitants (Aa) were infected into cytoplasm of human macrophage-like cells (U937) and regulate hostTNF-α gene expression via the TLR-8 and NF-kB signaling pathways [142].
Herpesviruses have been associated with severe types of periodontal disease including aggressive periodontitis, periodontitis and periodontal abscess [133,138,153–155]. Herpesvirus infections upregulate IL-1β and TNF-α gene expression in monocytes and macrophages and impair the host antibacterial immune response [156]. Herpesviridae comprise a family of enveloped double-stranded DNA viruses, among them three subgroups infectious to humans, including α-herpesviruses encompassing herpes simplex virus (HSV) type 1 and 2, β-herpesviruses including cytomegalovirus (CMV) and human herpesviruses 6 and 7, as well as γ-herpesviruses such as the Epstein-Barr virus and human herpesvirus 8 [157–159]. HSV, CMV and Epstein-Barr virus are the most commonly studied viruses infecting periodontal tissues [153,154,160,161]. It has been demonstrated that herpesviruses encode miRs, and the miRs of α-, β-, and γ-herpesviruses are implicated in targeting host cellular genes and regulating the transition from latent to lytic gene expression throughout the herpesvirus life cycle [159,162].
HSV-1 and HSV-2 encode 18 precursor miRs which ultimately produce 27 and 24 mature miRs, respectively [162]. These viral miRs regulate the life cycle of the virus itself, including the replication, latency and reactivation, and host immune responses [163,164]. HSV-1 miR-H1 is abundantly expressed during lytic infection, while miR-H2, -H3, and -H4 are most predominantly expressed during latency [163,165–167]. MiR-H2 is completely complementary to the infected cell polypeptide 0 (ICP0) mRNA, and miR-H3 and -H4 target ICP 34.5, the proteins that promote viral replication and reactivation [168–170]. In contrast, miR-H1 does not affect viral replication [163] but targets ubiquitin protein ligase E3 component n-recognin 1 (Ubr1), a RING-type E3 ubiquitin ligase of the Arg/N-end rule pathway, which causes the degradation of proteins bearing destabilizing N-terminal residues [171]. Additionally, miR-H28 induces Interferon-γ expression to limit the viral spread to uninfected neighboring cells [164].
Recent data have demonstrated that Cytomegalovirus encodes 26 mature miRs originating from 15 precursor transcripts [162]. These miRs function during immune system evasion, cell cycle regulation and vascular transport [172]. An in vitro study has demonstrated that HCMV miR-US4 regulates Glutaminyl-tRNA Synthetase (QARS), which modulates signal transduction pathways for cellular apoptosis [173]. Furthermore, miR-UL70-3p and UL148D directly target the proapoptotic genes Modulator of Apoptosis 1 (MOAP1), PHA granule-associated protein PHAP, and Endoplasmic Reticulum To Nucleus Signaling 1 (ERN1), respectively [174]. In regulating host innate immune response, miR-UL112 decreases the natural killer (NK) cells recognizing virus infected cells by targeting the major histocompatibility complex class-I related chain B (MICB) [174,175]. MiR-UL112 also reduces NF-кB signaling during late infection by targeting IkB kinase (IKK) complex members IKKa and IKKb [176]. These studies indicate that HCMV employs its miR repertoire to counter cellular apoptosis and autophagy, and to benefit the discharge of infectious virus particles [173,177].
The Epstein-Barr virus encodes more than 40 mature miRs originating from 25 precursor miRs [178], including BamHI A rightward transcripts (BARTs) and Bam HI fragment H rightward open reading frame 1 (BHRF1) regions that encode clusters of EBV miRs [179,180]. These viral miRs regulate genes involved in cell apoptosis, antigen presentation and recognition, as well as B cell transformation. The majority of these miRs have been studied in the context of EBV associated cancers. However, recent studies have correlated Epstein–Barr virus (EBV) in periodontal lesions with periodontitis and identified EBV as a promising pathogenic candidate for periodontal disease, suggesting that EBV antivirals may be necessary for the successful treatment of periodontal disease [181].
Other studies related to the role of herpesvirus miRs in periodontal disease have focused on the expression and function of herpesvirus miRs by comparing healthy human subjects and periodontal patients as well as using oral and periodontal cell lines [136,137,140]. These studies demonstrated that three viral miRs from the human herpesvirus family, HSV miR-H1, HCMA miR-US4 and Kaposi Sarcoma-Associated Virus (KSHV) miR-K12-3 were increased in diseased periodontal tissues when compared with healthy tissues [140]. Overexpression of miR-H1 and miR-K12-3 in human oral keratinocytes (HOK), a common target of various HHV, altered the expression of more than 1300 genes. Bioinformatic analysis revealed hundreds of potential v-miR binding sites on genes downregulated by miR-H1 and miR-K12-3. Three of the novel target sites involved in endocytic and intracellular trafficking pathways have been validated [137,140]. Moreover, high levels of human cytomegalic virus (HCMV) miR-H1, miR-K12-3 and miR-UL70 were detected in exosomes from v-miR-transfected HOK and KSHV-infected cell lines. These studies have demonstrated that the HOK-derived exosomes release their contents into macrophages and change the expression of endogenous miRs [137,140]. Illustrating the effects of trans-species viral transfer, phagocytic uptake of labeled bacteria revealed significant attenuation of bacterial phagocytosis in miR-H1 and miR-K12-3 transfected primary human macrophages. Phagocytic uptake of viral miRs in this study also caused a remarkable reduction in cytokine secretion from bacterial-challenged macrophages [137,140]. These data confirm that v-miRs play a significant role in the immune subversion that contributes to the pathogenesis of inflammatory periodontal diseases.
References
- [1].Hasturk H, Kantarci A, Activation and resolution of periodontal inflammation and its systemic impact, Periodontology 2000. 69 (2015) 255–273. 10.1111/prd.12105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Ebersole JL, Graves CL, Gonzalez OA, Dawson D, Morford LA, Huja PE, Hartsfield JK, Huja SS, Pandruvada S, Wallet SM, Aging, inflammation, immunity and periodontal disease, Periodontology 2000. 72 (2016) 54–75. 10.1111/prd.12135. [DOI] [PubMed] [Google Scholar]
- [3].Könönen E, Gursoy M, Gursoy UK, Periodontitis: A Multifaceted Disease of Tooth-Supporting Tissues. Journal of Clinical Medicine. 8 (2019) 1135. 10.3390/jcm8081135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Diekwisch TGH, Novel approaches toward managing the micromanagers: “Non-toxic” but effective, Gene Therapy. 23 (2016) 697–698. 10.1038/gt.2016.49. [DOI] [PubMed] [Google Scholar]
- [5].Luan X, Zhou X, Naqvi A, Francis M, Foyle D, Nares S, Diekwisch TGH, MicroRNAs and immunity in periodontal health and disease, International Journal of Oral Science. 10 (2018). 10.1038/s41368-018-0025-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Luan X, Zhou X, Trombetta-Esilva J, Francis M, Gaharwar AK, Atsawasuwan P, Diekwisch TGH, MicroRNAs and Periodontal Homeostasis, Journal of Dental Research. 96 (2017) 491–500. 10.1177/0022034516685711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Page RC, Schroeder HE, Pathogenesis of inflammatory periodontal disease: a summary of current work, Laboratory Investigation. 34 (1976) 235–249. [PubMed] [Google Scholar]
- [8].Murakami S, Mealey BL, Mariotti A, Chapple ILC, Dental plaque-induced gingival conditions, Journal of Periodontology. 89 (2018) S17–S27. 10.1002/JPER.17-0095. [DOI] [PubMed] [Google Scholar]
- [9].Offenbacher S, Beck JD, Jared HL, Mauriello SM, Mendoza LC, Couper DJ, Stewart DD, Murtha AP, Cochran DL, Dudley DJ, Reddy MS, Geurs NC, Hauth JC, Effects of periodontal therapy on rate of preterm delivery: A randomized controlled trial, Obstetrics and Gynecology. 114 (2009) 551–559. 10.1097/AOG.0b013e3181b1341f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Jönsson D, Ramberg P, Demmer RT, Kebschull M, Dahlén G, Papapanou PN, Gingival tissue transcriptomes in experimental gingivitis, Journal of Clinical Periodontology. 38 (2011) 599–611. 10.1111/j.1600-051X.2011.01719.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Graves DT, Fine D, Teng YTA, van Dyke TE, Hajishengallis G, The use of rodent models to investigate host-bacteria interactions related to periodontal diseases, Journal of Clinical Periodontology. 35 (2008) 89–105. 10.1111/j.1600-051X.2007.01172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Graves DT, Li J, Cochran DL, Critical review in oral biology & medicine: Inflammation and uncoupling as mechanisms of periodontal bone loss, Journal of Dental Research. 90 (2011) 143–153. 10.1177/0022034510385236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Cekici A, Kantarci A, Hasturk H, van Dyke TE, Inflammatory and immune pathways in the pathogenesis of periodontal disease, Periodontology 2000. 64 (2014) 57–80. 10.1111/prd.12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Ara T, Kurata K, Hirai K, Uchihashi T, Uematsu T, Imamura Y, Furusawa K, Kurihara S, Wang PL, Human gingival fibroblasts are critical in sustaining inflammation in periodontal disease, Journal of Periodontal Research. 44 (2009) 21–27. 10.1111/j.1600-0765.2007.01041.x. [DOI] [PubMed] [Google Scholar]
- [15].Pan W, Wang Q, Chen Q, The cytokine network involved in the host immune response to periodontitis, International Journal of Oral Science. 11 (2019). 10.1038/s41368-019-0064-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ogata Y, Matsui S, Kato A, Zhou L, Nakayama Y, Taka H, MicroRNA expression in inflamed and noninflamed gingival tissues from Japanese patients, Journal of Oral Science. 56 (2014) 253–260. 10.2334/josnusd.56.253. [DOI] [PubMed] [Google Scholar]
- [17].Venugopal P, Koshy T, Lavu V, Ranga Rao S, Ramasamy S, Hariharan S, Venkatesan V, Differential expression of microRNAs let-7a, miR-125b, miR-100, and miR-21 and interaction with NF-kB pathway genes in periodontitis pathogenesis, Journal of Cellular Physiology. 233 (2018) 5877–5884. 10.1002/jcp.26391. [DOI] [PubMed] [Google Scholar]
- [18].Zhou W, Su L, Duan X, Chen X, Hays A, Upadhyayula S, Shivde J, Wang H, Li Y, Huang D, Liang S, MicroRNA-21 down-regulates inflammation and inhibits periodontitis, Molecular Immunology. 101 (2018) 608–614. 10.1016/j.molimm.2018.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Na HS, Park MH, Song YR, Kim S, Kim H-J, Lee JY, Choi J-I, Chung J, Elevated MicroRNA-128 in Periodontitis Mitigates Tumor Necrosis Factor-α Response via p38 Signaling Pathway in Macrophages, Journal of Periodontology. 87 (2016) e173–e182. 10.1902/jop.2016.160033. [DOI] [PubMed] [Google Scholar]
- [20].Li J, Wang R, Ge Y, Chen D, Wu B, Fang F, Assessment of microRNA-144–5p and its putative targets in inflamed gingiva from periodontitis patients, Journal of Periodontal Research. 54 (2019) 266–277. 10.1111/jre.12627. [DOI] [PubMed] [Google Scholar]
- [21].Sanada T, Sano T, Sotomaru Y, Alshargabi R, Yamawaki Y, Yamashita A, Matsunaga H, Iwashita M, Shinjo T, Kanematsu T, Asano T, Nishimura F, Anti-inflammatory effects of miR-146a induced in adipose and periodontal tissues, Biochemistry and Biophysics Reports. 22 (2020). 10.1016/j.bbrep.2020.100757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kalea AZ, Hoteit R, Suvan J, Lovering RC, Palmen J, Cooper JA, Khodiyar VK, Harrington Z, Humphries SE, D’Aiuto F, Upregulation of gingival tissue miR-200b in obese periodontitis subjects, Journal of Dental Research. 94 (2015) 59S–69S. 10.1177/0022034514568197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Moffatt CE, Lamont RJ, Porphyromonas gingivalis induction of microRNA-203 expression controls suppressor of cytokine signaling 3 in gingival epithelial cells, Infection and Immunity. 79 (2011) 2632–2637. 10.1128/IAI.00082-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Li J, Li L, Wang X, Xiao L, Porphyromonas gingivalis Inhibition of MicroRNA-205-5p Expression Modulates Proinflammatory Cytokines in Gingival Epithelial Cells, Biochemical Genetics. 58 (2020) 566–579. 10.1007/s10528-020-09957-y. [DOI] [PubMed] [Google Scholar]
- [25].Lian J, Wu X, Liu Y, Qiu W, Zhu X, Wang X, Meng S, Valverde P, Steffensen B, Tu Q, Pan J, Chen J, Potential roles of miR-335-5p on pathogenesis of experimental periodontitis, Journal of Periodontal Research. 55 (2020) 191–198. 10.1111/jre.12701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Song L, Rawal B, Nemeth JA, Haura EB, JAK1 activates STAT3 activity in non-small-cell lung cancer cells and IL-6 neutralizing antibodies can suppress JAK1-STAT3 signaling, Molecular Cancer Therapeutics. 10 (2011) 481–494. 10.1158/1535-7163.MCT-10-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Ghotloo S, Motedayyen H, Amani D, Saffari M, Sattari M, Assessment of microRNA-146a in generalized aggressive periodontitis and its association with disease severity, Journal of Periodontal Research. 54 (2019) 27–32. 10.1111/jre.12538. [DOI] [PubMed] [Google Scholar]
- [28].Motedayyen H, Ghotloo S, Saffari M, Sattari M, Amid R, Evaluation of MicroRNA-146a and Its Targets in Gingival Tissues of Patients With Periodontitis, Journal of Periodontology. 86 (2015) 1380–1385. 10.1902/jop.2015.150319. [DOI] [PubMed] [Google Scholar]
- [29].Qian G, Liu D, Hu J, Gan F, Hou L, Chen X, Huang K, Ochratoxin A-induced autophagy in vitro and in vivo promotes porcine circovirus type 2 replication, Cell Death & Disease. 8 (2017) e2909. 10.1038/cddis.2017.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Greabu M, Giampieri F, Melescanu Imre M, Mohora M, Totan A, Pituru SM, Ionescu E, Autophagy, one of the main steps in periodontitis pathogenesis and evolution, Molecules. 25 (2020). 10.3390/molecules25184338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Deretic V, Saitoh T, Akira S, Autophagy in infection, inflammation and immunity, Nature Reviews Immunology. 13 (2013) 722–737. 10.1038/nri3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Yao Q, Gu A, Wang Z, Xue Y, MicroRNA-144 functions as a tumor suppressor in gastric cancer by targeting cyclooxygenase-2, Experimental and Therapeutic Medicine. 15 (2018) 3088–3095. 10.3892/etm.2018.5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Ou L, Sun T, Cheng Y, Huang L, Zhan X, Zhang P, Yang J, Zhang Y, Zhou Z, MicroRNA-214 contributes to regulation of necroptosis via targeting ATF4 in diabetes-associated periodontitis, Journal of Cellular Biochemistry. 120 (2019) 14791–14803. 10.1002/jcb.28740. [DOI] [PubMed] [Google Scholar]
- [34].Iurlaro R, Muñoz-Pinedo C, Cell death induced by endoplasmic reticulum stress, FEBS Journal. (2016) 2640–2652. 10.1111/febs.13598. [DOI] [PubMed] [Google Scholar]
- [35].Goodall EF, Leach V, Wang C, Cooper-Knock J, Heath PR, Baker D, Drew DR, Jill Saffrey M, Simpson JE, Romero IA, Wharton SB, Age-associated mRNA and miR expression changes in the blood-brain barrier, International Journal of Molecular Sciences. 20 (2019). 10.3390/ijms20123097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Chen X, Li W, Ren J, Huang D, He WT, Song Y, Yang C, Li W, Zheng X, Chen P, Han J, Translocation of mixed lineage kinase domain-like protein to plasma membrane leads to necrotic cell death, Cell Research. 24 (2014) 105–121. 10.1038/cr.2013.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Pasparakis M, Vandenabeele P, Necroptosis and its role in inflammation, Nature. 517 (2015) 311–320. 10.1038/nature14191. [DOI] [PubMed] [Google Scholar]
- [38].Bonnet CS, Walsh DA, Osteoarthritis, angiogenesis and inflammation, Rheumatology. 44 (2005) 7–16. 10.1093/rheumatology/keh344. [DOI] [PubMed] [Google Scholar]
- [39].Frantz S, Vincent KA, Feron O, Kelly RA, Innate immunity and angiogenesis, Circulation Research. 96 (2005) 15–26. 10.1161/01.RES.0000153188.68898.ac. [DOI] [PubMed] [Google Scholar]
- [40].Coma S, Allard-Ratick M, Akino T, van Meeteren LA, Mammoto A, Klagsbrun M, GATA2 and Lmo2 control angiogenesis and lymphangiogenesis via direct transcriptional regulation of neuropilin-2, Angiogenesis. 16 (2013) 939–952. 10.1007/s10456-013-9370-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Liu L, Tong Q, Liu S, Cui J, Zhang Q, Sun W, Yang S, ZEB1 upregulates VEGF expression and stimulates angiogenesis in breast cancer, PLoS ONE. 11 (2016). 10.1371/journal.pone.0148774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Magenta A, Cencioni C, Fasanaro P, Zaccagnini G, Greco S, Sarra-Ferraris G, Antonini A, Martelli F, Capogrossi MC, MiR-200c is upregulated by oxidative stress and induces endothelial cell apoptosis and senescence via ZEB1 inhibition, Cell Death and Differentiation. 18 (2011) 1628–1639. 10.1038/cdd.2011.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Chan YC, Roy S, Khanna S, Sen CK, Downregulation of endothelial MicroRNA-200b supports cutaneous wound angiogenesis by desilencing GATA binding protein 2 and vascular endothelial growth factor receptor 2, Arteriosclerosis, Thrombosis, and Vascular Biology. 32 (2012) 1372–1382. 10.1161/ATVBAHA.112.248583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Seo BM, Miura M, Gronthos S, Bartold PM, Batouli S, Brahim J, Young M, Robey PG, Wang CY, Shi S, Investigation of multipotent postnatal stem cells from human periodontal ligament, Lancet. 364 (2004) 149–155. 10.1016/S0140-6736(04)16627-0. [DOI] [PubMed] [Google Scholar]
- [45].Diekwisch TGH, Pathways and fate of migratory cells during late tooth organogenesis, Connective Tissue Research. 43 (2002) 246–256. 10.1080/03008200290001221. [DOI] [PubMed] [Google Scholar]
- [46].Roguljic H, Matthews BG, Yang W, Cvija H, Mina M, Kalajzic I, In vivo identification of periodontal progenitor cells, Journal of Dental Research. 92 (2013) 709–715. 10.1177/0022034513493434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Zhao H, Feng J, Seidel K, Shi S, Klein O, Sharpe P, Chai Y, Secretion of shh by a neurovascular bundle niche supports mesenchymal stem cell homeostasis in the adult mouse incisor, Cell Stem Cell. 14 (2014) 160–173. 10.1016/j.stem.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Hosoya A, Shalehin N, Takebe H, Fujii S, Seki Y, Mizoguchi T, Shimo T, Iijima M, Irie K, Stem cell properties of Gli1-positive cells in the periodontal ligament, Journal of Oral Biosciences. 62 (2020) 299–305. 10.1016/j.job.2020.08.002. [DOI] [PubMed] [Google Scholar]
- [49].Han Y, Wang F, Shao L, Huang P, Xu Y, LncRNA TUG1 mediates lipopolysaccharide-induced proliferative inhibition and apoptosis of human periodontal ligament cells by sponging miR-132, Acta Biochimica et Biophysica Sinica. 51 (2019) 1208–1215. 10.1093/abbs/gmz125. [DOI] [PubMed] [Google Scholar]
- [50].Dong Y, Feng S, Dong F, Maternally-expressed gene 3 (MEG3)/miR-143-3p regulates injury to periodontal ligament cells by mediating the AKT/inhibitory κkB Kinase (IKK) pathway, Medical Science Monitor. 26 (2020). 10.12659/MSM.922486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Duan Y, An W, Wu Y, Wang J, Tetramethylpyrazine reduces inflammation levels and the apoptosis of LPS-stimulated human periodontal ligament cells via the downregulation of miR-302b, International Journal of Molecular Medicine. 45 (2020) 1918–1926. 10.3892/ijmm.2020.4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Zhou Q, Hu T, Xu Y, Anticancer potential of TUG1 knockdown in cisplatin-resistant osteosarcoma through inhibition of MET/Akt signalling, Journal of Drug Targeting. 28 (2020) 204–211. 10.1080/1061186X.2019.1644651. [DOI] [PubMed] [Google Scholar]
- [53].Guo B, Zhao Z, Wang Z, Li Q, Wang X, Wang W, Song T, Huang C, MicroRNA-302b-3p Suppresses Cell Proliferation Through AKT Pathway by Targeting IGF-1R in Human Gastric Cancer, Cellular Physiology and Biochemistry. 42 (2017) 1701–1711. 10.1159/000479419. [DOI] [PubMed] [Google Scholar]
- [54].Liu C, Chen Z, Wang J, Hu H, Overexpression of X chromosome-linked inhibitor of apoptosis by inhibiting microRNA-24 protects periodontal ligament cells against hydrogen peroxide-induced cell apoptosis, Cellular and Molecular Biology. 62 (2016) 6–13. 10.14715/cmb/2016.62.4.2. [DOI] [PubMed] [Google Scholar]
- [55].Monteiro MM, Lima CR, Gomes CC, Cruz MC, Horliana ACRT, Santos MF, Lowered Expression of MicroRNAs 221 and 222 Mediate Apoptosis Induced by High Glucose in Human Periodontal Ligament Cells, Cell Biochemistry and Biophysics. 78 (2020) 391–398. 10.1007/s12013-020-00932-3. [DOI] [PubMed] [Google Scholar]
- [56].Yang J, Zhou J, Cui B, Yu T, Evaluation of hypoxia on the expression of miR-646/IGF-1 signaling in human periodontal ligament cells (hPDLCs), Medical Science Monitor. 24 (2018) 5282–5291. 10.12659/MSM.910163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Yang N, Li Y, Wang G, Ding Y, Jin Y, Xu Y, Tumor necrosis factor-α suppresses adipogenic and osteogenic differentiation of human periodontal ligament stem cell by inhibiting miR-21/Spry1 functional axis, Differentiation. 97 (2017) 33–43. 10.1016/j.diff.2017.08.004. [DOI] [PubMed] [Google Scholar]
- [58].Zhou X, Luan X, Chen Z, Francis M, Gopinathan G, Li W, Lu X, Li S, Wu C, Diekwisch TGH, MicroRNA-138 Inhibits Periodontal Progenitor Differentiation under Inflammatory Conditions, Journal of Dental Research. 95 (2016) 230–237. 10.1177/0022034515613043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Bao L, Zhang X, Xu Y, Wang M, Song Y, Gu Y, Zheng Y, Xiao J, Wang Y, Zhou Q, Qian J, Liang Y, Ji L, Feng X, Dysfunction of MiR-148a-NRP1 Functional Axis Suppresses Osteogenic Differentiation of Periodontal Ligament Stem Cells under Inflammatory Microenvironment, Cellular Reprogramming. 21 (2019) 314–322. 10.1089/cell.2019.0026. [DOI] [PubMed] [Google Scholar]
- [60].Wang X, Sun H, Liu H, Ma L, Jiang C, Liao H, Xu S, Xiang J, Cao Z, MicroRNA-181b-5p modulates tumor necrosis factor-α-induced inflammatory responses by targeting interleukin-6 in cementoblasts, Journal of Cellular Physiology. 234 (2019) 22719–22730. 10.1002/jcp.28837. [DOI] [PubMed] [Google Scholar]
- [61].Wang Y, Li Y, Shao P, Wang L, Bao X, Hu M, IL1β inhibits differentiation of cementoblasts via microRNA-325-3p, Journal of Cellular Biochemistry. 121 (2020) 2606–2617. 10.1002/jcb.29482. [DOI] [PubMed] [Google Scholar]
- [62].Wang X, Sun H, Liao H, Wang C, Jiang C, Zhang Y, Cao Z, MicroRNA-155-3p Mediates TNF-α-Inhibited Cementoblast Differentiation, Journal of Dental Research. 96 (2017) 1430–1437. 10.1177/0022034517718790. [DOI] [PubMed] [Google Scholar]
- [63].Li X, Chen C, Wang F, Huang W, Liang Z, Xiao Y, Wei K, Wan Z, Hu X, Xiang S, Ding X, Zhang J, KCTD1 suppresses canonical wnt signaling pathway by enhancing b-catenin degradation, PLoS ONE. 9 (2014). 10.1371/journal.pone.0094343. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [64].Mahanonda R, Champaiboon C, Subbalekha K, Sa-Ard-Iam N, Yongyuth A, Isaraphithakkul B, Rerkyen P, Charatkulangkun O, Pichyangkul S, Memory T cell subsets in healthy gingiva and periodontitis tissues, Journal of Periodontology. 89 (2018) 1121–1130. 10.1002/JPER.17-0674. [DOI] [PubMed] [Google Scholar]
- [65].Mahanonda R, Champaiboon C, Subbalekha K, Sa-Ard-Iam N, Rattanathammatada W, Thawanaphong S, Rerkyen P, Yoshimura F, Nagano K, Lang NP, Pichyangkul S, Human Memory B Cells in Healthy Gingiva, Gingivitis, and Periodontitis, The Journal of Immunology. 197 (2016) 715–725. 10.4049/jimmunol.1600540. [DOI] [PubMed] [Google Scholar]
- [66].Dutzan N, Konkel JE, Greenwell-Wild T, Moutsopoulos NM, Characterization of the human immune cell network at the gingival barrier, Mucosal Immunology. 9 (2016) 1163–1172. 10.1038/mi.2015.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Thorbert-Mros S, Larsson L, Berglundh T, Cellular composition of long-standing gingivitis and periodontitis lesions, Journal of Periodontal Research. 50 (2015) 535–543. 10.1111/jre.12236. [DOI] [PubMed] [Google Scholar]
- [68].Li W, Zhang Z, min Wang Z, Differential immune cell infiltrations between healthy periodontal and periodontitis tissues, BMC Oral Health. 20 (2020). 10.1186/s12903-020-01287-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Zhang X, Wang Q, Yan X, Shan Y, Xing L, Li M, Long H, Lai W, Immune landscape of periodontitis unveils alterations of infiltrating immunocytes and molecular networks-aggregating into an interactive web-tool for periodontitis related immune analysis and visualization, Journal of Translational Medicine. 18 (2020). 10.1186/s12967-020-02616-1. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [70].Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP, The impact of microRNAs on protein output, Nature. 455 (2008) 64–71. 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Silva MT, Correia-Neves M, Neutrophils and macrophages: The main partners of phagocyte cell systems, Frontiers in Immunology. 3 (2012). 10.3389/fimmu.2012.00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Roy S, MiR in macrophage development and function, Antioxidants and Redox Signaling. 25 (2016) 795–804. 10.1089/ars.2016.6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Zhou H, Zhang J, Eyers F, Xiang Y, Herbert C, Tay HL, Foster PS, Yang M, Identification of the microRNA networks contributing to macrophage differentiation and function, Oncotarget. 7 (2016) 28806–28820. 10.18632/oncotarget.8933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Stoecklin-Wasmer C, Guarnieri P, Celenti R, Demmer RT, Kebschull M, Papapanou PN, MicroRNAs and their target genes in gingival tissues, Journal of Dental Research. 91 (2012) 934–940. 10.1177/0022034512456551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Xie YF, Shu R, Jiang SY, Liu DL, Zhang XL, Comparison of microRNA profiles of human periodontal diseased and healthy gingival tissues, International Journal of Oral Science. 3 (2011) 125–134. 10.4248/IJOS11046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Jiang SY, Xue D, Xie YF, Zhu DW, Dong YY, Wei CC, Deng JY, The negative feedback regulation of microRNA-146a in human periodontal ligament cells after Porphyromonas gingivalis lipopolysaccharide stimulation, Inflammation Research. 64 (2015) 441–451. 10.1007/s00011-015-0824-y. [DOI] [PubMed] [Google Scholar]
- [77].O’Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D, MicroRNA-155 is induced during the macrophage inflammatory response, Proceedings of the National Academy of Sciences of the United States of America. 104 (2007) 1604–1609. 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Taganov KD, Boldin MP, Chang KJ, Baltimore D, NF-κB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses, Proceedings of the National Academy of Sciences of the United States of America. 103 (2006) 12481–12486. 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Wang Z, Brandt S, Medeiros A, Wang S, Wu H, Dent A, Serezani CH, MicroRNA 21 Is a homeostatic regulator of macrophage polarization and prevents prostaglandin e2 -mediated M2 generation, PLoS ONE. 10 (2015). 10.1371/journal.pone.0115855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Venkatesan G, Uppoor A, Naik DG, Redefining the role of dendritic cells in periodontics, Journal of Indian Society of Periodontology. 17 (2013) 700–705. 10.4103/0972-124X.124467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Wilensky A, Segev H, Mizraji G, Shaul Y, Capucha T, Shacham M, Hovav AH, Dendritic cells and their role in periodontal disease, Oral Diseases. 20 (2014) 119–126. 10.1111/odi.12122. [DOI] [PubMed] [Google Scholar]
- [82].Yang L, Boldin MP, Yu Y, Liu CS, Ea CK, Ramakrishnan P, Taganov KD, Zhao JL, Baltimore D, miR-146a controls the resolution of T cell responses in mice, Journal of Experimental Medicine. 209 (2012) 1655–1670. 10.1084/jem.20112218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Podshivalova K, Salomon DR, MicroRNA regulation of T-lymphocyte immunity: Modulation of molecular networks responsible for T-cell activation, differentiation, and development, Critical Reviews in Immunology. 33 (2013) 435–476. 10.1615/CritRevImmunol.2013006858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Dezerega A, Maggiolo S, Garrido M, Dutzan N, The TH17 vs. TREG Imbalance in the Pathogenesis of Periodontitis: New Approach for Dichotomy TH1 vs. TH2, Revista Clínica de Periodoncia, Implantología y Rehabilitación Oral. 1 (2008) 70–72. 10.1016/s0718-5391(08)70012-0. [DOI] [Google Scholar]
- [85].Allam JP, Duan Y, Heinemann F, Winter J, Götz W, Deschner J, Wenghoefer M, Bieber T, Jepsen S, Novak N, IL-23-producing CD68 + macrophage-like cells predominate within an IL-17-polarized infiltrate in periodontitis lesions, Journal of Clinical Periodontology. 38 (2011) 879–886. 10.1111/j.1600-051X.2011.01752.x. [DOI] [PubMed] [Google Scholar]
- [86].Zhao L, Zhou Y, Xu Y, Sun Y, Li L, Chen W, Effect of non-surgical periodontal therapy on the levels of Th17/Th1/Th2 cytokines and their transcription factors in Chinese periodontitis patients, Journal of Clinical Periodontology. 38 (2011) 509–516. 10.1111/j.1600-051X.2011.01712.x. [DOI] [PubMed] [Google Scholar]
- [87].Laurence A, O’Shea JJ, TH-17 differentiation: Of mice and men, Nature Immunology. 8 (2007) 903–905. 10.1038/ni0907-903. [DOI] [PubMed] [Google Scholar]
- [88].Rouas R, Fayyad-Kazan H, el Zien N, Lewalle P, Rothé F, Simion A, Akl H, Mourtada M, el Rifai M, Burny A, Romero P, Martiat P, Badran B, Human natural Treg microRNA signature: Role of microRNA-31 and microRNA-21 in FOXP3 expression, European Journal of Immunology. 39 (2009) 1608–1618. 10.1002/eji.200838509. [DOI] [PubMed] [Google Scholar]
- [89].Sethi A, Kulkarni N, Sonar S, Lal G, Role of miRs in CD4 T cell plasticity during inflammation and tolerance, Frontiers in Genetics. 4 (2013). 10.3389/fgene.2013.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Liu SQ, Jiang S, Li C, Zhang B, Li QJ, Mir-17–92 cluster targets phosphatase and tensin homology and ikaros family zinc finger 4 to promote th17-mediated inflammation, Journal of Biological Chemistry. 289 (2014) 12446–12456. 10.1074/jbc.M114.550723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Lee YH, Na HS, Jeong SY, Jeong SH, Park HR, Chung J, Comparison of inflammatory microRNA expression in healthy and periodontitis tissues, Biocell. 35 (2011) 43–49. [PubMed] [Google Scholar]
- [92].Seymour GJ, Powell RN, Cole KL, Aitken JF, Brooks D, Beckman I, Zola H, Bradley J, Burns GF, Experimental gingivitis in humans: A histochemical and immunological characterization of the lymphoid cell subpopulations, Journal of Periodontal Research. 18 (1983) 375–385. 10.1111/j.1600-0765.1983.tb00373.x. [DOI] [PubMed] [Google Scholar]
- [93].Stoufi ED, Taubman MA, Ebersole JL, Smith DJ, Preparation and characterization of human gingival cells, Journal of Periodontal Research. 22 (1987) 144–149. 10.1111/j.1600-0765.1987.tb01554.x. [DOI] [PubMed] [Google Scholar]
- [94].Robinson EK, Covarrubias S, Carpenter S, The how and why of lncRNA function: An innate immune perspective, Biochimica et Biophysica Acta - Gene Regulatory Mechanisms. 1863 (2020). 10.1016/j.bbagrm.2019.194419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Zhou M, Hu H, Han Y, Li J, Zhang Y, Tang S, Yuan Y, Zhang X, Long non-coding RNA 01126 promotes periodontitis pathogenesis of human periodontal ligament cells via miR-518a-5p/HIF-1α/MAPK pathway, Cell Proliferation. 54 (2020). 10.1111/cpr.12957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Li J, Wang M, Song L, Wang X, Lai W, Jiang S, LncRNA MALAT1 regulates inflammatory cytokine production in lipopolysaccharide-stimulated human gingival fibroblasts through sponging miR-20a and activating TLR4 pathway, Journal of Periodontal Research. 55 (2020) 182–190. 10.1111/jre.12700. [DOI] [PubMed] [Google Scholar]
- [97].Chen H, Lan Z, Li Q, Li Y, Abnormal expression of long noncoding RNA FGD5-AS1 affects the development of periodontitis through regulating miR-142-3p/SOCS6/NF-κB pathway, Artificial Cells, Nanomedicine and Biotechnology. 47 (2019) 2098–2106. 10.1080/21691401.2019.1620256. [DOI] [PubMed] [Google Scholar]
- [98].Huang Y, Han Y, Guo R, Liu H, Li X, Jia L, Zheng Y, Li W, Long non-coding RNA FER1L4 promotes osteogenic differentiation of human periodontal ligament stromal cells via miR-874-3p and vascular endothelial growth factor A, Stem Cell Research and Therapy. 11 (2020). 10.1186/s13287-019-1519-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Jia B, Qiu X, Chen J, Sun X, Zheng X, Zhao J, Li Q, Wang Z, A feed-forward regulatory network lncPCAT1/miR-106a-5p/E2F5 regulates the osteogenic differentiation of periodontal ligament stem cells, Journal of Cellular Physiology. 234 (2019) 19523–19538. 10.1002/jcp.28550. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [100].Li X, Zheng Y, Zheng Y, Huang Y, Zhang Y, Jia L, Li W, Circular RNA CDR1as regulates osteoblastic differentiation of periodontal ligament stem cells via the miR-7/GDF5/SMAD and p38 MAPK signaling pathway, Stem Cell Research and Therapy. 9 (2018). 10.1186/s13287-018-0976-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Wu D, Yin L, Sun D, Wang F, Wu Q, Xu Q, Xin B, Long noncoding RNA TUG1 promotes osteogenic differentiation of human periodontal ligament stem cell through sponging microRNA-222-3p to negatively regulate Smad2/7, Archives of Oral Biology. 117 (2020). 10.1016/j.archoralbio.2020.104814. [DOI] [PubMed] [Google Scholar]
- [102].Feng Y, Wan P, Yin L, Long noncoding RNA X-inactive specific transcript (XIST) promotes osteogenic differentiation of periodontal ligament stem cells by sponging microRNA-214-3p, Medical Science Monitor. 26 (2020). 10.12659/MSM.918932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Wang L, Wu F, Song Y, Li X, Wu Q, Duan Y, Jin Z, Long noncoding RNA related to periodontitis interacts with miR-182 to upregulate osteogenic differentiation in periodontal mesenchymal stem cells of periodontitis patients, Cell Death and Disease. 7 (2016). 10.1038/cddis.2016.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Peng W, Deng W, Zhang J, Pei G, Rong Q, Zhu S, Long noncoding RNA ANCR suppresses bone formation of periodontal ligament stem cells via sponging miR-758, Biochemical and Biophysical Research Communications. 503 (2018) 815–821. 10.1016/j.bbrc.2018.06.081. [DOI] [PubMed] [Google Scholar]
- [105].Laine ML, Moustakis V, Koumakis L, Potamias G, Loos BG, Modeling susceptibility to periodontitis, Journal of Dental Research. 92 (2013) 45–50. 10.1177/0022034512465435. [DOI] [PubMed] [Google Scholar]
- [106].Zhang J, Sun X, Xiao L, Xie C, Xuan D, Luo G, Gene polymorphisms and periodontitis, Periodontology 2000. 56 (2011) 102–124. 10.1111/j.1600-0757.2010.00371.x. [DOI] [PubMed] [Google Scholar]
- [107].Slayton RL, Genetics and environmental factors play important roles in the risk for periodontal disease and edentulism, Journal of Evidence-Based Dental Practice. 6 (2006) 238–239. 10.1016/j.jebdp.2006.06.011. [DOI] [PubMed] [Google Scholar]
- [108].Huang QY, Recker RR, Deng HW, Searching for osteoporosis genes in the post-genome era: Progress and challenges, Osteoporosis International. 14 (2003) 701–715. 10.1007/s00198-003-1445-9. [DOI] [PubMed] [Google Scholar]
- [109].Brodzikowska A, Górska R, Kowalski J, Interleukin-1 Genotype in Periodontitis, Archivum Immunologiae et Therapiae Experimentalis. 67 (2019) 367–373. 10.1007/s00005-019-00555-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Irwandi RA, Vacharaksa A, The role of microRNA in periodontal tissue: A review of the literature, Archives of Oral Biology. 72 (2016) 66–74. 10.1016/j.archoralbio.2016.08.014. [DOI] [PubMed] [Google Scholar]
- [111].Venugopal P, Lavu V, Rangarao S, Venkatesan V, Evaluation of a panel of single-nucleotide polymorphisms in miR-146a and miR-196a2 genomic regions in patients with periodontitis, Genetic Testing and Molecular Biomarkers. 21 (2017) 228–235. 10.1089/gtmb.2016.0358. [DOI] [PubMed] [Google Scholar]
- [112].Venugopal P, Lavu V, Rao SR, Venkatesan V, Association of microRNA-125a and microRNA-499a polymorphisms in periodontitis in a sample south Indian population: A hospital-based genetic association study, Gene. 631 (2017) 10–15. 10.1016/j.gene.2017.07.053. [DOI] [PubMed] [Google Scholar]
- [113].Muiños-Gimeno M, Guidi M, Kagerbauer B, Martín-Santos R, Navinés R, Alonso P, Menchón JM, Gratacòs M, Estivill X, Espinosa-Parrilla Y, Allele variants in functional microRNA target sites of the neurotrophin-3 receptor gene (NTRK3) as susceptibility factors for anxiety disorders, Human Mutation. 30 (2009) 1062–1071. 10.1002/humu.21005. [DOI] [PubMed] [Google Scholar]
- [114].Muĩos-Gimeno M, Montfort M, Bayés M, Estivill X, Espinosa-Parrilla Y, Design and evaluation of a panel of single-nucleotide polymorphisms in microRNA genomic regions for association studies in human disease, European Journal of Human Genetics. 18 (2010) 218–226. 10.1038/ejhg.2009.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Kadkhodazadeh M, Jafari AR, Amid R, Ebadian AR, Alipour MM, Mollaverdi F, Lafzi A, MiR146a and MiR499 gene polymorphisms in Iranian periodontitis and peri-implantitis patients, Journal of Long-Term Effects of Medical Implants. 23 (2013) 9–16. 10.1615/JLongTermEffMedImplants.2013007073. [DOI] [PubMed] [Google Scholar]
- [116].Sun YM, Lin KY, Chen YQ, Diverse functions of miR-125 family in different cell contexts, Journal of Hematology and Oncology. 6 (2013). 10.1186/1756-8722-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Kim SW, Ramasamy K, Bouamar H, Lin AP, Jiang D, Aguiar RCT, MicroRNAs miR-125a and miR-125b constitutively activate the NF-κB pathway by targeting the tumor necrosis factor alpha-induced protein 3 (TNFAIP3, A20), Proceedings of the National Academy of Sciences of the United States of America. 109 (2012) 7865–7870. 10.1073/pnas.1200081109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Hu Y, Liu CM, Qi L, He TZ, Liu SG, Hao CJ, Cui Y, Zhang N, Xia HF, Ma X, Two common SNPs in pri-miR-125a alter the mature miR expression and associate with recurrent pregnancy loss in a Han-Chinese population, RNA Biology. 8 (2011). 10.4161/rna.8.5.16034. [DOI] [PubMed] [Google Scholar]
- [119].Labbaye C, Testa U, The emerging role of MIR-146A in the control of hematopoiesis, immune function and cancer, Journal of Hematology and Oncology. 5 (2012). 10.1186/1756-8722-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Saba R, Sorensen DL, Booth SA, MicroRNA-146a: A dominant, negative regulator of the innate immune response, Frontiers in Immunology. 5 (2014). 10.3389/fimmu.2014.00578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Bagavad Gita J, George A. v., Pavithra N, Chandrasekaran SC, Latchumanadhas K, Gnanamani A, Dysregulation of miR-146a by periodontal pathogens: A risk for acute coronary syndrome, Journal of Periodontology. 90 (2019) 756–765. 10.1002/JPER.18-0466. [DOI] [PubMed] [Google Scholar]
- [122].Luo X, Yang W, Ye DQ, Cui H, Zhang Y, Hirankarn N, Qian X, Tang Y, Lau YL, de Vries N, Tak PP, Tsao BP, Shen N, A functional variant in microRNA-146a promoter modulates its expression and confers disease risk for systemic lupus erythematosus, PLoS Genetics. 7 (2011). 10.1371/journal.pgen.1002128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Chen CJ, Zhang Y, Zhang L, Weakley SM, Yao Q, MicroRNA-196: Critical roles and clinical applications in development and cancer, Journal of Cellular and Molecular Medicine. 15 (2011) 14–23. 10.1111/j.1582-4934.2010.01219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Zhang YH, He K, Shi G, Effects of MicroRNA-499 On the Inflammatory Damage of Endothelial Cells during Coronary Artery Disease Via the Targeting of PDCD4 Through the NF-Kβ/TNF-α Signaling Pathway, Cellular Physiology and Biochemistry. 44 (2017) 110–124. 10.1159/000484588. [DOI] [PubMed] [Google Scholar]
- [125].Chen C, Yang S, Chaugai S, Wang Y, Wang DW, Meta-analysis of Hsa-mir-499 polymorphism (rs3746444) for cancer risk: Evidence from 31 case-control studies, BMC Medical Genetics. 15 (2014). 10.1186/s12881-014-0126-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Toraih EA, Hussein MH, al Ageeli E, Riad E, AbdAllah NB, Helal GM, Fawzy MS, Structure and functional impact of seed region variant in MIR-499 gene family in bronchial asthma, Respiratory Research. 18 (2017). 10.1186/s12931-017-0648-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Ota A, Tagawa H, Karnan S, Tsuzuki S, Karpas A, Kira S, Yoshida Y, Seto M, Identification and Characterization of a Novel Gene, C13orf25, as a Target for 13q31-q32 Amplification in Malignant Lymphoma, Cancer Research. 64 (2004) 3087–3095. 10.1158/0008-5472.CAN-03-3773. [DOI] [PubMed] [Google Scholar]
- [128].Kuo G, Wu CY, Yang HY, MiR-17-92 cluster and immunity, Journal of the Formosan Medical Association. 118 (2019) 2–6. 10.1016/j.jfma.2018.04.013. [DOI] [PubMed] [Google Scholar]
- [129].Yang HY, Barbi J, Wu CY, Zheng Y, Vignali PDA, Wu X, Tao JH, Park B. v., Bandara S, Novack L, Ni X, Yang X, Chang KY, Wu RC, Zhang J, Yang CW, Pardoll DM, Li H, Pan F, MicroRNA-17 Modulates Regulatory T Cell Function by Targeting Co-regulators of the Foxp3 Transcription Factor, Immunity. 45 (2016) 83–93. 10.1016/j.immuni.2016.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Zhu D, Pan C, Li L, Bian Z, Lv Z, Shi L, Zhang J, Li D, Gu H, Zhang CY, Liu Y, Zen K, MicroRNA-17/20a/106a modulate macrophage inflammatory responses through targeting signal-regulatory protein α, Journal of Allergy and Clinical Immunology. 132 (2013). 10.1016/j.jaci.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Liu Y, Liu W, Hu C, Xue Z, Wang G, Ding B, Luo H, Tang L, Kong X, Chen X, Liu N, Ding Y, Jin Y, MiR-17 modulates osteogenic differentiation through a coherent feed-forward loop in mesenchymal stem cells isolated from periodontal ligaments of patients with periodontitis, Stem Cells. 29 (2011) 1804–1816. 10.1002/stem.728. [DOI] [PubMed] [Google Scholar]
- [132].Silva-Sousa AC, Mazzi-Chaves JF, Freitas JV, Salles AG, da RAB, Segato S, da Silva LAB, Antunes LAA, Antunes LS, Baratto-Filho F, Sousa-Neto MD, Küchler EC, Association between estrogen, vitamin D and microrna17 gene polymorphisms and periapical lesions, Brazilian Dental Journal. 31 (2020) 19–24. 10.1590/0103-644020200. [DOI] [PubMed] [Google Scholar]
- [133].Slots J, Human viruses in periodontitis, Periodontology 2000. 53 (2010) 89–110. 10.1111/j.1600-0757.2009.00325.x. [DOI] [PubMed] [Google Scholar]
- [134].Lamont RJ, Koo H, Hajishengallis G, The oral microbiota: dynamic communities and host interactions, Nature Reviews Microbiology. 16 (2018) 745–759. 10.1038/s41579-018-0089-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Lang NP, Bartold PM, Periodontal health, Journal of Periodontology. 89 (2018) S9–S16. 10.1002/JPER.16-0517. [DOI] [PubMed] [Google Scholar]
- [136].Naqvi AR, Shango J, Seal A, Shukla D, Nares S, Herpesviruses and MicroRNAs: New Pathogenesis Factors in Oral Infection and Disease?, Frontiers in Immunology. 9 (2018) 2099. 10.3389/fimmu.2018.02099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Naqvi AR, Shango J, Seal A, Shukla D, Nares S, Viral miRs alter host cell miR profiles and modulate innate immune responses, Frontiers in Immunology. 9 (2018). 10.3389/fimmu.2018.00433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Slots J, Herpesviruses in periodontal diseases, Periodontology 2000. 38 (2005) 33–62. 10.1111/j.1600-0757.2005.00109.x. [DOI] [PubMed] [Google Scholar]
- [139].Plaza JJG, Small RNAs in cell-to-cell communications during bacterial infection, FEMS Microbiology Letters. 365 (2018). 10.1093/femsle/fny024. [DOI] [PubMed] [Google Scholar]
- [140].Naqvi AR, Seal A, Shango J, Brambila MF, Martinez G, Chapa G, Hasan S, Yadavalli T, Jaishankar D, Shukla D, Nares S, Herpesvirus-encoded microRNAs detected in human gingiva alter host cell transcriptome and regulate viral infection, Biochimica et Biophysica Acta - Gene Regulatory Mechanisms. 1861 (2018) 497–508. 10.1016/j.bbagrm.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Aguilar C, Mano M, Eulalio A, MicroRNAs at the Host–Bacteria Interface: Host Defense or Bacterial Offense, Trends in Microbiology. 27 (2019) 206–218. 10.1016/j.tim.2018.10.011. [DOI] [PubMed] [Google Scholar]
- [142].Han EC, Choi SY, Lee Y, Park JW, Hong SH, Lee HJ, Extracellular RNAs in periodontopathogenic outer membrane vesicles promote TNF-α production in human macrophages and cross the blood-brain barrier in mice, FASEB Journal : Official Publication of the Federation of American Societies for Experimental Biology. 33 (2019) 13412–13422. 10.1096/fj.201901575R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Yoshikawa FSY, Teixeira FME, Sato MN, da S LM. Oliveira, Delivery of microRNAs by Extracellular Vesicles in Viral Infections: Could the News be Packaged?, Cells. 8 (2019) 611. 10.3390/cells8060611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Liang H, Zen K, Zhang J, Zhang CY, Chen X, New roles for microRNAs in cross-species communication, RNA Biology. 10 (2013) 367–370. 10.4161/rna.23663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Quintana JF, Kumar S, Ivens A, Chow FWN, Hoy AM, Fulton A, Dickinson P, Martin C, Taylor M, Babayan SA, Buck AH, Comparative analysis of small RNAs released by the filarial nematode Litomosoides sigmodontis in vitro and in vivo, PLoS Neglected Tropical Diseases. 13 (2019). 10.1371/journal.pntd.0007811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Hoy AM, Lundie RJ, Ivens A, Quintana JF, Nausch N, Forster T, Jones F, Kabatereine NB, Dunne DW, Mutapi F, MacDonald AS, Buck AH, Parasite-Derived MicroRNAs in Host Serum As Novel Biomarkers of Helminth Infection, PLoS Neglected Tropical Diseases. 8 (2014). 10.1371/journal.pntd.0002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Beatty M, Guduric-Fuchs J, Brown E, Bridgett S, Chakravarthy U, Hogg RE, Simpson DA, Small RNAs from plants, bacteria and fungi within the order Hypocreales are ubiquitous in human plasma, BMC Genomics. 15 (2014). 10.1186/1471-2164-15-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Zhang L, Hou D, Chen X, Li D, Zhu L, Zhang Y, Li J, Bian Z, Liang X, Cai X, Yin Y, Wang C, Zhang T, Zhu D, Zhang D, Xu J, Chen Q, Ba Y, Liu J, Wang Q, Chen J, Wang J, Wang M, Zhang Q, Zhang J, Zen K, Zhang CY, Exogenous plant MIR168a specifically targets mammalian LDLRAP1: Evidence of cross-kingdom regulation by microRNA, Cell Research. 22 (2012) 107–126. 10.1038/cr.2011.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Zempleni J, Baier SR, Howard KM, Cui J, Gene regulation by dietary microRNAs, Canadian Journal of Physiology and Pharmacology. 93 (2015) 1097–1102. 10.1139/cjpp-2014-0392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Cai Q, He B, Weiberg A, Buck AH, Jin H, Small RNAs and extracellular vesicles: New mechanisms of cross-species communication and innovative tools for disease control, PLoS Pathogens. 15 (2019). 10.1371/journal.ppat.1008090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Choi JW, Kim SC, Hong SH, Lee HJ, Secretable Small RNAs via Outer Membrane Vesicles in Periodontal Pathogens, Journal of Dental Research. 96 (2017) 458–466. 10.1177/0022034516685071. [DOI] [PubMed] [Google Scholar]
- [152].Ho MH, Chen CH, Goodwin JS, Wang BY, Xie H, Functional advantages of Porphyromonas gingivalis vesicles, PLoS ONE. 10 (2015). 10.1371/journal.pone.0123448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Kamma JJ, Contreras A, Slots J, Herpes viruses and periodontopathic bacteria in early-onset periodontitis, Journal of Clinical Periodontology. 28 (2001) 879–885. 10.1034/j.1600-051x.2001.028009879.x. [DOI] [PubMed] [Google Scholar]
- [154].Kubar A, Saygun I, Özdemir A, Yapar M, Slots J, Real-time polymerase chain reaction quantification of human cytomegalovirus and Epstein-Barr virus in periodontal pockets and the adjacent gingiva of periodontitis lesions, Journal of Periodontal Research. 40 (2005) 97–104. 10.1111/j.1600-0765.2005.00770.x. [DOI] [PubMed] [Google Scholar]
- [155].Ting M, Contreras A, Slots J, Herpesviruses in localized juvenile periodontitis, Journal of Periodontal Research. 35 (2000) 17–25. 10.1034/j.1600-0765.2000.035001017.x. [DOI] [PubMed] [Google Scholar]
- [156].Contreras A, Slots J, Herpesviruses in human periodontal disease, Journal of Periodontal Research. 35 (2000) 3–16. 10.1034/j.1600-0765.2000.035001003.x. [DOI] [PubMed] [Google Scholar]
- [157].Wagner EK, Bloom DC, Experimental investigation of herpes simplex virus latency., Clinical Microbiology Reviews. 10 (1997) 419–443. 10.1128/cmr.10.3.419-443.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Carter C, Savic S, Cole J, Wood P, Natural killer cell receptor expression in patients with severe and recurrent Herpes simplex virus-1 (HSV-1) infections, Cellular Immunology. 246 (2007) 65–74. 10.1016/j.cellimm.2007.06.002. [DOI] [PubMed] [Google Scholar]
- [159].Barbu MG, Condrat CE, Thompson DC, Bugnar OL, Cretoiu D, Toader OD, Suciu N, Voinea SC, MicroRNA Involvement in Signaling Pathways During Viral Infection., Frontiers in Cell and Developmental Biology. 8 (2020) 143. 10.3389/fcell.2020.00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Konstantinidis A, Sakellari D, Papa A, Antoniadis A, Real-time polymerase chain reaction quantification of Epstein-Barr virus in periodontitis patients, Journal of Periodontal Research. 40 (2005) 294–298. 10.1111/j.1600-0765.2005.00796.x. [DOI] [PubMed] [Google Scholar]
- [161].Ling L-J, Ho C-C, Wu C-Y, Chen Y-T, Hung S-L, Association Between Human Herpesviruses and the Severity of Periodontitis, Journal of Periodontology. 75 (2004) 1479–1485. 10.1902/jop.2004.75.11.1479. [DOI] [PubMed] [Google Scholar]
- [162].Kozomara A, Birgaoanu M, Griffiths-Jones S, MiRBase: From microRNA sequences to function, Nucleic Acids Research. 47 (2019) D155–D162. 10.1093/nar/gky1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Jurak I, Kramer MF, Mellor JC, van Lint AL, Roth FP, Knipe DM, Coen DM, Numerous Conserved and Divergent MicroRNAs Expressed by Herpes Simplex Viruses 1 and 2, Journal of Virology. 84 (2010) 4659–4672. 10.1128/jvi.02725-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Huang R, Zhou X, Ren S, Liu X, Han Z, Zhou GG, Effect of Loss-of-function of the Herpes Simplex Virus-1 microRNA H6-5p on Virus Replication, Virologica Sinica. 34 (2019) 386–396. 10.1007/s12250-019-00111-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Kramer MF, Jurak I, Pesola JM, Boissel S, Knipe DM, Coen DM, Herpes simplex virus 1 microRNAs expressed abundantly during latent infection are not essential for latency in mouse trigeminal ganglia, Virology. 417 (2011) 239–247. 10.1016/j.virol.2011.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Umbach JL, Nagel MA, Cohrs RJ, Gilden DH, Cullen BR, Analysis of Human Alphaherpesvirus MicroRNA Expression in Latently Infected Human Trigeminal Ganglia, Journal of Virology. 83 (2009) 10677–10683. 10.1128/jvi.01185-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Cui C, Griffiths A, Li G, Silva LM, Kramer MF, Gaasterland T, Wang X-J, Coen DM, Prediction and Identification of Herpes Simplex Virus 1-Encoded MicroRNAs, Journal of Virology. 80 (2006) 5499–5508. 10.1128/jvi.00200-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Pan D, Flores O, Umbach JL, Pesola JM, Bentley P, Rosato PC, Leib DA, Cullen BR, Coen DM, A neuron-specific host MicroRNA targets herpes simplex virus-1 ICP0 expression and promotes latency, Cell Host and Microbe. 15 (2014) 446–456. 10.1016/j.chom.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Tang S, Patel A, Krause PR, Novel Less-Abundant Viral MicroRNAs Encoded by Herpes Simplex Virus 2 Latency-Associated Transcript and Their Roles in Regulating ICP34.5 and ICP0 mRNAs, Journal of Virology. 83 (2009) 1433–1442. 10.1128/jvi.01723-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Tang S, Bertke AS, Patel A, Wang K, Cohen JI, Krause PR, An acutely and latently expressed herpes simplex virus 2 viral microRNA inhibits expression of ICP34.5, a viral neurovirulence factor, Proceedings of the National Academy of Sciences of the United States of America. 105 (2008) 10931–10936. 10.1073/pnas.0801845105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Zheng K, Liu Q, Wang S, Ren Z, Kitazato K, Yang D, Wang Y, HSV-1-encoded microRNA miR-H1 targets Ubr1 to promote accumulation of neurodegeneration-associated protein, Virus Genes. 54 (2018) 343–350. 10.1007/s11262-018-1551-6. [DOI] [PubMed] [Google Scholar]
- [172].Hook LM, Grey F, Grabski R, Tirabassi R, Doyle T, Hancock M, Landais I, Jeng S, McWeeney S, Britt W, Nelson JA, Cytomegalovirus miRs target secretory pathway genes to facilitate formation of the virion assembly compartment and reduce cytokine secretion, Cell Host and Microbe. 15 (2014) 363–373. 10.1016/j.chom.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Shao Y, Qi Y, Huang Y, Liu Z, Ma Y, Guo X, Jiang S, Sun Z, Ruan Q, Human cytomegalovirus-encoded miR-US4-1 promotes cell apoptosis and benefits discharge of infectious virus particles by targeting QARS, Journal of Biosciences. 41 (2016) 183–192. 10.1007/s12038-016-9605-1. [DOI] [PubMed] [Google Scholar]
- [174].Babu SG, Pandeya A, Verma N, Shukla N, Kumar RV, Saxena S, Role of HCMV miR-UL70-3p and miR-UL148D in overcoming the cellular apoptosis, Molecular and Cellular Biochemistry. 393 (2014) 89–98. 10.1007/s11010-014-2049-8. [DOI] [PubMed] [Google Scholar]
- [175].Stern-Ginossar N, Elefant N, Zimmermann A, Wolf DG, Saleh N, Biton M, Horwitz E, Prokocimer Z, Prichard M, Hahn G, Goldman-Wohl D, Greenfield C, Yagel S, Hengel H, Altuvia Y, Margalit H, Mandelboim O, Host immune system gene targeting by a viral miR, Science. 317 (2007) 376–381. 10.1126/science.1140956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Hancock MH, Hook LM, Mitchell J, Nelson JA, Human cytomegalovirus microRNAs miR-US5-1 and miR-UL112-3p block proinflammatory cytokine production in response to NF-κB-activating factors through direct downregulation of IKKα and IKKβ, MBio. 8 (2017). 10.1128/mBio.00109-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Fruci D, Rota R, Gallo A, The role of HCMV and HIV-1 MicroRNAs: Processing, and mechanisms of action during viral infection, Frontiers in Microbiology. 8 (2017). 10.3389/fmicb.2017.00689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Hassani A, Khan G, Epstein-Barr virus and miRs: Partners in crime in the pathogenesis of multiple sclerosis?, Frontiers in Immunology. 10 (2019). 10.3389/fimmu.2019.00695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Fan C, Tang Y, Wang J, Xiong F, Guo C, Wang Y, Xiang B, Zhou M, Li X, Wu X, Li Y, Li X, Li G, Xiong W, Zeng Z, The emerging role of Epstein-Barr virus encoded microRNAs in nasopharyngeal carcinoma., Journal of Cancer. 9 (2018) 2852–2864. 10.7150/jca.25460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Sunagawa K, Hishima T, Fukumoto H, Hasegawa H, Katano H, Conserved sequences of BART and BHRF regions encoding viral microRNAs in Epstein-Barr virus-associated lymphoma, BMC Research Notes. 10 (2017). 10.1186/s13104-017-2603-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Imai K, Ogata Y, How does Epstein–Barr virus contribute to periodontitis?, International Journal of Molecular Sciences. 21 (2020). 10.3390/ijms21061940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Gu X, Yao X, Liu D, Up-regulation of microRNA-335-5p reduces inflammation via negative regulation of the TPX2-mediated AKT/GSK3β signaling pathway in a chronic rhinosinusitis mouse model, Cellular Signalling. 70 (2020). 10.1016/j.cellsig.2020.109596. [DOI] [PubMed] [Google Scholar]
- [183].Jiang S, Hu Y, Deng S, Deng J, Yu X, Huang G, Kawai T, Han X, miR-146a regulates inflammatory cytokine production in Porphyromonas gingivalis lipopolysaccharide-stimulated B cells by targeting IRAK1 but not TRAF6, Biochimica et Biophysica Acta - Molecular Basis of Disease. 1864 (2018) 925–933. 10.1016/j.bbadis.2017.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Wong HKA, Veremeyko T, Patel N, Lemere CA, Walsh DM, Esau C, Vanderburg C, Krichevsky AM, De-repression of FOXO3a death axis by microRNA-132 and −212 causes neuronal apoptosis in Alzheimer’s disease, Human Molecular Genetics. 22 (2013) 3077–3092. 10.1093/hmg/ddt164. [DOI] [PubMed] [Google Scholar]
- [185].Jin YP, Hu YP, Wu XS, Wu YS, Ye YY, Li HF, Liu YC, Jiang L, Liu FT, Zhang YJ, Hao YJ, Liu XY, bin Liu Y, MiR-143-3p targeting of ITGA6 suppresses tumour growth and angiogenesis by downregulating PLGF expression via the PI3K/AKT pathway in gallbladder carcinoma, Cell Death and Disease. 9 (2018). 10.1038/s41419-017-0258-2. [DOI] [PMC free article] [PubMed] [Google Scholar]


