Abstract
Two new skipper butterfly (Hesperiidae) species are described from the United States: Staphylus floridus Grishin, sp. n. (type locality in Florida, Volusia Co.) and Staphylus ecos Grishin, sp. n. (type locality in Texas, Brewster Co.). They are cryptic and hence escaped recognition. They differ from their sister species by the relative size and morphology of genitalia and by genotype—including and beyond the COI barcode—thus suggesting genetic isolation that argues for their species-level status. A lectotype is designated for Helias ascalaphus Staudinger, 1876. Staphylus opites (Godman & Salvin, 1896), stat. rest. is a species-level taxon and not a synonym of Staphylus vincula (Plötz, 1886), while Pholisora iguala Williams & Bell, 1940, syn. n. is a junior subjective synonym of S. vincula.
Keywords: biodiversity, cryptic species, suture zones, genomics, phylogeny
INTRODUCTION
New methods bring new insights and reveal aspects not previously apparent. For centuries, zoology relied on phenotype-based research and classification. New species were traditionally discovered via phenotypic differences. Phenotypes are encoded by genotypes, and direct investigation of genotypes by the analysis of DNA sequences may be a more direct and powerful approach to classifying nature. Even short segments of DNA, such as COI barcodes, have been insightful in revealing species diversity not readily evident from phenotypes (Hebert et al. 2004).
Genome-scale phylogenetic approaches show promise in revolutionizing our understanding of butterfly evolution and refining their taxonomy (Toussaint et al. 2018; Allio et al. 2019; Li et al. 2019; Zhang et al. 2019a; Zhang et al. 2019b; Toussaint et al. 2021; Robbins et al. 2022; Toussaint et al. 2022). Accurate phylogenetic trees constructed from millions of base pairs give better confidence in the results and frequently reveal inconsistencies with the current classification (Cong et al. 2019b; Zhang et al. 2021; Toussaint et al. 2022; Zhang et al. 2022). While this approach is instrumental for higher classification and taxonomy above species level, its application to species discovery and description has been limited (Janzen et al. 2017; Cong et al. 2020). Even for COI barcodes, a major obstacle is the DNA analysis of extant primary type specimens which is required to properly place discovered new species in the context of previously named taxa (Pfeiler 2018).
Next-generation sequencing technology allows us to obtain whole genome shotgun sequences of some of the oldest type specimens, thus resolving this impediment (Cong et al. 2021). Sequencing of primary types is essential to confidently address nomenclatural questions. Imperative for cryptic species complexes, it could also be helpful in seemingly less difficult cases. Oftentimes type localities were incompletely documented or type specimens may have been mislabeled. Genomic comparison across a species’ range places type specimens among them and clarifies type localities (Cong et al. 2021). Sequencing of primary types on a large scale (Zhang et al. 2022) offers unprecedented opportunities for species-level taxonomy, both in correcting previous mistakes and new species discovery.
Our research group carries out large-scale butterfly genomic sequencing across taxonomic groups and continents. As a result, specimens currently assigned to one species sometimes partition into distinct and strongly supported clades in phylogenetic trees. We construct three maximum likelihood trees from all protein coding genes encoded by: nuclear genome partitioned into (1) autosomes and (2) the Z chromosome, and (2) mitochondrial genome. The Z chromosome holds a special place in the evolution of Lepidoptera (Sahara et al. 2012; Mongue and Walters 2018; Cong et al. 2019a; Mongue et al. 2022) and its phylogeny frequently agrees best with how species boundaries are defined based on previous work. Well-supported clades observed in all three trees within “species” prompts a more detailed investigation.
Here, we analyze two such cases dealing with North American species of Staphylus Godman & Salvin, 1896 (type species Helias ascalaphus Staudinger, 1876). Genomic trees that include primary type specimens of the relevant names reveal a split into two clades in each of the two current species of Staphylus. Following up on these results with morphological analyses, two new species are proposed.
MATERIALS AND METHODS
The specimens were inspected and photographed/sampled for DNA in the following collections: American Museum of Natural History, New York, NY, USA (AMNH), Natural History Museum, London, UK (BMNH), Carnegie Museum of Natural History, Pittsburgh, PA, USA (CMNH), Museum für Naturkunde, Berlin, Germany (MFNB), Texas A&M University Insect Collection, College Station, TX, USA (TAMU), National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), Zoologische Staatssammlung München, Germany (ZSMC), and the personal collection of John V. Calhoun, Palm Harbor, FL, USA. Historical documents, such as unpublished drawings, were inspected in the Natural History Museum, London, UK and National Museum of Natural History, Smithsonian Institution, Washington, DC, USA. Photographs of specimens and illustrations were taken with Nikon D800 camera through 105 mm macro lens; dissected genitalia were photographed in glycerol with the Nikon D200 camera without a lens and through a microscope at 4.5x magnification. Images were assembled and edited in Photoshop CS5. Genitalia photographs were taken in several focus slices and stacked in Photoshop to increase depth of field. Genomic DNA was extracted from a leg, libraries were prepared, and sequenced as previously reported (Li et al. 2019). The details of phylogenetic tree construction were given in Zhang et al. (2022). In brief, the maximum-likelihood tree was constructed from protein-coding regions in the Z chromosome using IQ-tree v1.6.12 under GTR+GAMMA model (Nguyen et al. 2015). To estimate the confidence of each node, we generated 100 replicates of 10,000 codons randomly sampled from the total set of codons in the Z chromosome and constructed maximum-likelihood trees for each replicate. The support values of each node were summarized from these replicate trees using sumtrees routine in dendropy package (Sukumaran and Holder 2010). The method to find diagnostic DNA characters is described in Cong et al. (Cong et al. 2019b). Sequence datasets obtained in this work are deposited in the NCBI database <https://www.ncbi.nlm.nih.gov/> as BioProject PRJNA831579 (BioSample entries of the project contain the locality and collection data of the sequenced specimens shown in the trees), and COI barcodes have GenBank accessions ON351018–ON351022. Exon sequences with diagnostic characters highlighted are also available from <https://osf.io/tygxc/>. DNA characters are given as abbreviations, e.g., aly728.44.1:G672C means position 672 in exon 1 of gene 44 from scaffold 728 of Cecropterus lyciades (formerly in Achalarus Scudder, 1872, thus aly) reference genome (Shen et al. 2017) is C, changed from G in the ancestor. Similar notations are used for the COI barcode characters, e.g., A78G means position 78 is G, changed from A in the ancestor, or T264T(not C) means position 264 is T, unchanged from the ancestor, and not C as in sister taxon.
RESULTS
All three phylogenetic trees—based on nuclear genome: (1) autosomes and (2) Z chromosome, and (3) mitochondrial genome—consistently reveal a well-supported split into two clades in each of the two North American species of Staphylus: S. hayhurstii (W. H. Edwards, 1870) and S. ceos (W. H. Edwards, 1882). These two names have no known synonyms (Pelham 2008; Pelham 2022), and therefore one of the clades in each species is likely to represent a new taxon. A combination of genomic and morphological analyses suggests that these unnamed taxa are new species, which are herein described. In the first case, nuclear genome diverged stronger than the mitogenome. In the second case, the opposite occurred. Furthermore, genomic comparison of old primary type specimens placed among more recently collected specimens suggests a number of taxonomic changes that are detailed below.
Staphylus floridus Grishin, new species.
http://zoobank.org/B7CE2D54-7E0F-4FBD-96B6-4A96BCE02C6D (Figs. 1 part, 2, 3a, b, e)
Fig. 1. Phylogenetic analysis of Staphylus floridus sp. n. and S. hayhurstii.

The trees are constructed from protein-coding regions in: a. autosomes, b. Z chromosome, c. mitochondrial genome. Different species are shown in different colors. Names of primary type specimens are highlighted in yellow.
Definition and diagnosis.
Genomic analysis of specimens identified as Staphylus hayhurstii (W. H. Edwards, 1870) (type locality USA: Missouri, possibly in Pettis Co.) from across its range, including the lectotype (NVG-15096H05), reveals their split into two prominent clades with 100% statistical support in all three trees (Fig. 1 red and blue clades). The red clade consists of specimens from Florida, and the blue clade includes the rest, where the lectotype of S. hayhurstii belongs. Genetic differentiation between the two clades is more pronounced in the nuclear genome than in the mitogenome: Fst/Gmin computed on the Z chromosome are 0.49/0.02, suggesting that the two clades are two distinct species (Cong et al. 2019a); but mitogenomes of the two species diverged less, e.g., their COI barcodes differ by 1.1% (7 bp), a value by itself insufficient to substantiate species-level distinction. Comparison of male and female genitalia offers additional support for the species-level status of the new taxon, and we conclude that the red clade (Fig. 1) represents a new species. This species (Fig. 2) is superficially similar to its sister S. hayhurstii, and we were not able to find wing pattern characters that distinguish them. This is hardly surprising for Staphylus, a genus that includes dozens of species nearly impossible to identify by facies even for more distant relatives, such as Staphylus mazans (Reakirt, [1867]) (type locality in Mexico: Veracruz) (Fig. 1 green), which is sympatric with S. hayhurstii in central Texas (e.g., in Comal Co., Fig. 1).
Fig. 2.

Staphylus floridus sp. n. holotype, male, NVG-18058E05 (a, b), USA: Florida, Volusia Co. and a female paratype NVG-20049A08 (c, d), USA: Florida, Sumter Co., in dorsal (a, c) and ventral (b, d) views. Other data are in text.
The new species differs from S. hayhurstii by less robust and relatively smaller genitalia, in both males (Fig. 3a–d) and females (Fig. 3e, f): illustrated genitalia are of specimens with similar forewing length. In male genitalia, tegumen longer and narrower, vs. more compact but broader tegumen in S. hayhurstii; harpe narrower (in lateral view), straighter, and bent less from the body of valva at its junction (best seen in dorsal view). In female genitalia, lamella postvaginalis less sculptured, typically with fewer and shorter ridges; lamella antevaginalis with comparatively smaller tooth on each side and more expanded middle section protruding caudad, with a central notch, vs. nearly crescent-shaped (tooth to tooth) lamella with irregular middle margin in S. hayhurstii. These genitalic characters are rather subtle, and due to individual variation the most confident identification can be achieved using DNA sequences. Combinations of the following DNA characters are diagnostic: in COI barcode: C271T, T322A, C401T, and T613C, and in the nuclear genome: aly1849.21.2:G3621A, aly1849.21.2:G3078A, aly1113.5.4: T1282A, aly1849.21.2:A702G, aly1656.10.4:T414G. Each of the barcode characters taken individually distinguishes the new species from S. hayhurstii in a sample of specimens we sequenced.
Fig. 3.
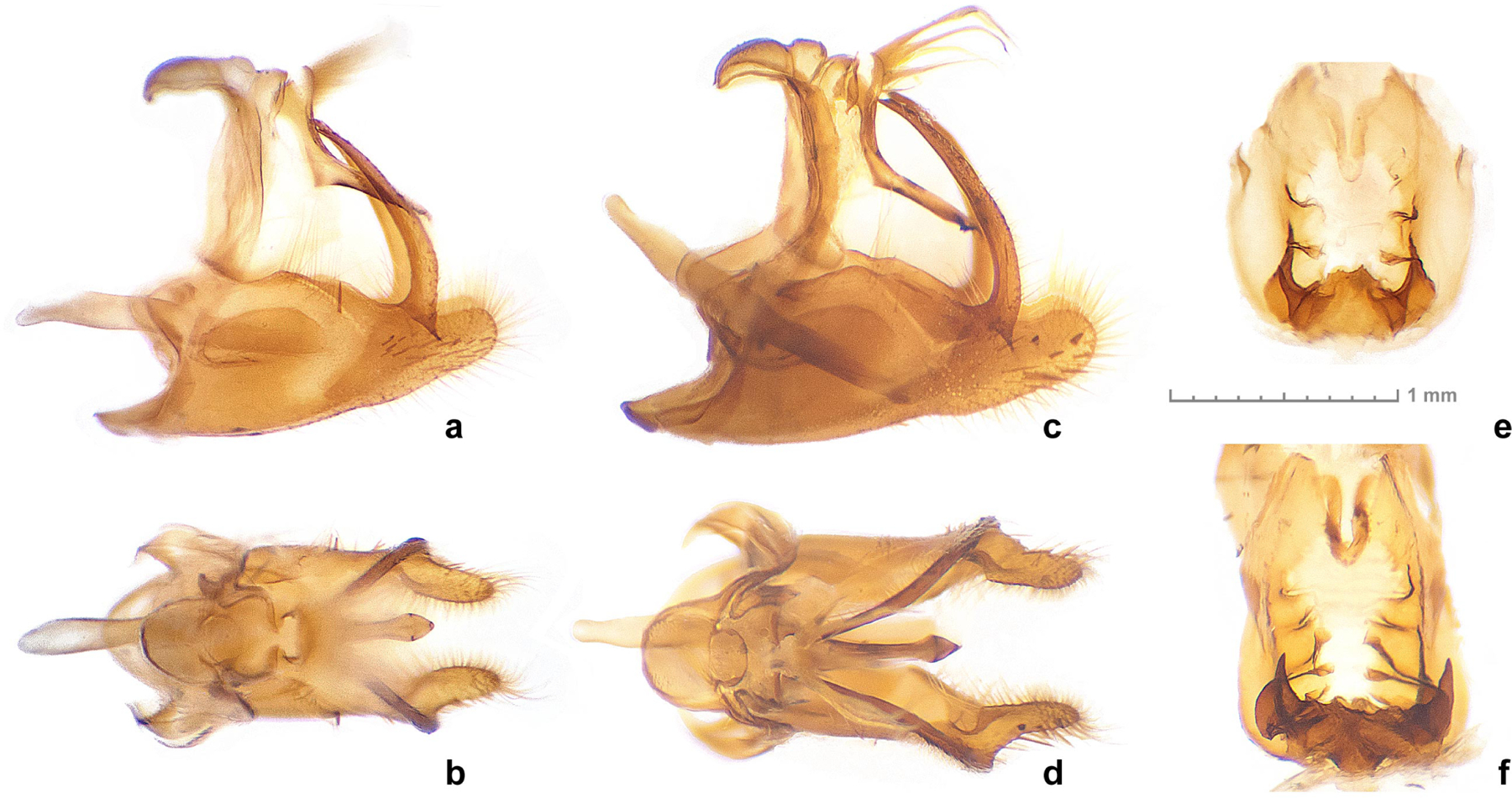
Genitalia of Staphylus floridus sp. n. paratypes (a, b. NVG-20049A03 and e. NVG-20049A07, data in text) and S. hayhurstii from USA: Texas (c, d. NVG-12597, Dallas Co. and f. NVG-7042, Delta Co.), males (a–d) and females (e, f), in left lateral (a, c), dorsal (b, d), and ventral (e, f) views. Whole genitalic capsule is shown for males and sterigma for females.
Barcode sequence of the holotype:
Sample NVG-18058E05, GenBank ON351018, 658 base pairs:
AACTTTATATTTTATTTTTGGTATTTGATCAGGAATAGTAGGAACTTCTTTAAGTATTCTTATTCGTTCTGAATTAGGAACACCTGGATCTTTAATTGGAGATGATCAAATTTATAATACTATTGTAACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGATTCGGAAATTGACTTGTTCCTCTTATATTAGGAGCCCCTGATATAGCTTTTCCTCGAATAAATAATATAAGATTTTGATTATTACCTCCATCTTTAATACTTTTAATTTCAAGAAGAATTGTAGAAAATGGAGCAGGAACTGGATGAACAGTTTACCCCCCCCTTTCAGCCAATATTGCTCATCAAGGTTCTTCTGTAGATTTAGCTATTTTTTCCTTACATTTAGCAGGTATTTCTTCTATTTTAGGAGCAATTAATTTTATTACAACTATTATTAATATACGAATTAATAATTTATCATTTGATCAAATACCTTTATTTGTTTGAGCTGTTGGAATTACAGCATTACTTTTACTTTTATCTTTACCAGTATTAGCAGGTGCTATTACTATACTTTTAACAGATCGAAATCTTAATACATCATTCTTCGATCCTGCTGGTGGAGGAGATCCTATTTTATATCAACATCTATTC
Type material.
Holotype: ♂ (Fig. 2a, b), deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), bears the following three rectangular white printed (date partly handprinted) labels: [ FLORIDA | Volusia Co, | New Smyrna Beach | 13 June 1969 | G. W. Rawson leg. ], [ DNA sample ID: | NVG-18058E05 | c/o Nick V. Grishin ], [ USNMENT | {QR Code} | 01466715 ] and one red printed [ HOLOTYPE ♂ | Staphylus | floridus Grishin ]. Paratypes: 5♂♂ and 2♀♀, all from USA: Florida: 1♂ NVG-18058E06, Alachua Co., Alachua, leg. Scott W. Gross, 29-Aug-1987; others leg. John V. Calhoun: 3♂♂ Volusia Co.: NVG-20049A04, New Smyrna Beach, 2495 N Dixie Fwy, 5-Mar-1987; NVG-20049A03, W side of US1, 1 mi N of Brevard Co. line, 7-Mar-1993; and NVG-20049A06, Reed Grove Rd. N of Stacey Grove Rd., SW of Oak Hill, 20-Mar-1994; 1♂ NVG-20049A05 & 1♀ NVG-20049A07, Lee Co., W of IH75, off Carrell Rd. nr. Bonita Springs, 25-Mar-1983; 1♀ NVG-20049A08, Sumter Co., E side SR301, N side of Shady Brook, 14-May-1993 (coll. J. V. Calhoun, Palm Harbor, FL, USA).
Type locality.
USA: Florida, Volusia County, New Smyrna Beach.
Distribution.
Currently known only from USA: Florida with specimens sequenced from Alachua, Sumter, Volusia, and Lee Counties. This species may be allopatric with S. hayhurstii, and geographically separated from it by the lack of records in northern Florida and nearby parts of other states. This apparent gap in the distribution of Staphylus would be interesting to investigate further.
Etymology.
Being a reference to Florida, the state from which the type series is known, a Latin word translated as flowery or blooming was chosen as the name for this not so garish species from Florida that nevertheless flaunts rather elaborate pattern of browns, nearly as flamboyant as gothic styles. The name is a masculine adjective.
Suggested English name.
Florida Sootywing.
Lectotype designation for Helias ascalaphus Staudinger, 1876
Helias ascalaphus Staudinger, 1876 (type locality in Panama), the type species of Staphylus, was described from a series of specimens (Staudinger 1876), and several syntypes are in the MFNB collection.
We sequenced a male syntype already bearing a red “Lectotypus” label (designation unpublished), and the results agreed with the current application of this name (Fig. 1). To stabilize nomenclature, N.V.G. hereby designates this sequenced syntype, in the Museum für Naturkunde, Berlin, Germany bearing the following 8 rectangular labels [ Lectotypus ], [ Origin. ], [ Panama | Ribbe ], [ ascalaphus ], [ Ascalaphus | Stdgr. ], [ GEN.PREP., | MIELKE | 1996 ], [ {QR Code} http://coll.mfn-berlin.de/u/ | 908619 ], and [ DNA sample ID: | NVG-15033F12 | c/o Nick V. Grishin ] as the lectotype of Helias ascalaphus Staudinger, 1876. The COI barcode sequence of the lectotype (GenBank ON351019) is:
AACTTTATATTTTATCTTTGGTATTTGATCTGGAATAGTAGGAACTTCTTTAAGTATTCTTATTCGTTCTGAATTAGGAACTCCTGGATCTTTAATTGGAGATGATCAAATTTATAATACTATTGTAACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGTTTTGGAAATTGACTTGTTCCTCTTATATTAGGAGCCCCTGATATAGCTTTTCCCCGAATAAATAATATAAGATTTTGATTATTACCCCCATCTTTAATACTTTTAATTTCAAGTAGAATCGTAGAAAATGGAGCAGGTACTGGATGAACAGTTTATCCCCCCCTTTCAGCTAACATTGCCCATCAAGGTTCTTCTGTAGATTTAGCTATTTTTTCTTTACATTTAGCAGGTATTTCTTCTATTTTAGGAGCAATTAATTTTATTACAACTATTATCAATATACGAATTAATAATTTATCCTTTGATCAAATACCCTTATTTGTTTGAGCAGTTGGAATTACAGCATTATTATTACTTTTATCTTTACCAGTATTAGCAGGTGCTATTACTATACTTTTAACAGATCGAAATCTTAATACATCATTTTTTGATCCTGCTGGTGGAGGAGATCCTATTTTATATCAACATTTATTC
Staphylus ecos Grishin, new species.
http://zoobank.org/33FF40E5-1E83-4E3E-8A0F-C85A81A0521A (Figs. 4 part, 5, 6a, b)
Fig. 4. Phylogenetic analysis of Staphylus ecos sp. n. and S. ceos.

The trees are constructed from protein-coding regions in: a. autosomes, b. Z chromosome, c. mitochondrial genome. Different species are shown in different colors. Names of primary type specimens are highlighted in yellow.
Definition and diagnosis.
Genomic analysis of specimens identified as Staphylus ceos (W. H. Edwards, 1882) (type locality in USA: Arizona, Graham Co.) from across its range, including the lectotype (NVG-15096H06), reveals their split into two prominent clades with 100% statistical support in the mitochondrial genome tree (Fig. 4c red and blue clades), substantiated by a shallow but consistent split in both nuclear genome trees (Fig, 4a, b). The red clade consists of specimens from the eastern part of the range and the blue clade includes the rest, where the lectotype of S. ceos belongs. Genetic differentiation between the two clades is limited in the nuclear genome, but is substantial in the mitogenome. Fst/Gmin computed on the Z chromosome are 0.08/0.15, well within the range of one species (Cong et al. 2019a). Mitogenomes of the two groups of specimens (red and blue) diverged strongly, e.g., their COI barcodes differ by 2.7% (18 bp), a value typically sufficient to support species-level distinction (Hebert et al. 2003), but only in the presence of phenotypic differences. Comparison of male genitalia reveals the differences that are typical of closely related but distinct species, and therefore we conclude that the red clade (Fig. 4) represents a new species. This species (Fig. 5) is superficially similar to its sister S. ceos, and we were not able to find meaningful wing pattern characters to differentiate them. This is typical for Staphylus, a genus that includes a number of species with similar appearance, even for more distant relatives.
Fig. 5.

Staphylus ecos sp. n. holotype, male, NVG-21113E02 (a, b) and a female paratype NVG-21113E03 (c, d), from USA: Texas, Brewster Co., Big Bend National Park, Chisos Mountains, in dorsal (a, c) and ventral (b, d) views. Other data are in text.
The new species differs from S. ceos by more robust and relatively larger male genitalia (Fig. 6): illustrated genitalia are of specimens with similar forewing length. Ampulla of valva with nearly straight distal margin (in lateral view), not concave as in typical S.ceos; harpe longer relatively to its height (in lateral view), less curved dorsad, with longer dorsal ridge that projects farther anteriad (towards vinculum) connecting to valva at several points that look like “teeth” in lateral view, and with thicker short spines at the distal end, vs. S. ceos, which has a shorter, more square-shaped harpe that is terminally more curved dorsad, the dorsal ridge that connects to valva closer to the ampulla and well before the anterior portion of valva along ventral margin, the “teeth” formed by connecting points of the ridge with valva that appear smaller, and distal spines that are more minute. These genitalic characters are rather subtle, and due to individual variation, the most confident identification can be achieved using DNA sequences. Combinations of the following DNA characters are diagnostic: in COI barcode: T67C, T205C, T337A, T478C, and T514C, and in the nuclear genome: aly1113.5.4:A1684G, aly1849.21.2:A8067G, aly1849.21.2:A8055G, aly1113.5.4:A1338C. Each of the barcode characters taken individually distinguishes the new species from S. ceos in a sample of specimens we sequenced.
Fig. 6.

Male genitalia of Staphylus ecos sp. n. (a, b), paratype NVG-11137, USA: Texas, Jeff Davis Co. and S. ceos (c, d), NVG-10941, USA: Texas, El Paso Co. in left lateral (a, c) and dorsal (b, d) views, whole genitalic capsule.
Barcode sequence of a paratype:
Sample NVG-19126A06, GenBank ON351020, 658 base pairs:
AACTTTATATTTTATTTTTGGTATTTGATCTGGAATAGTAGGAACTTCTTTAAGTATTCTTATTCGCTCAGAATTAGGAACCCCAGGATCTTTAATTGGAGATGATCAAATTTATAATACTATTGTAACAGCTCATGCTTTTATTATAATTTTTTTTATAGTAATACCTATTATAATCGGAGGTTTTGGAAATTGATTAGTACCCCTAATATTAGGAGCCCCAGATATAGCTTTCCCTCGAATAAATAATATAAGTTTCTGATTATTACCCCCCTCTCTTATACTTTTAATTTCAAGTAGTATTGTAGAAAATGGAGCAGGTACAGGATGAACTGTATACCCCCCTCTTTCAGCTAATATTGCTCATCAAGGATCATCTGTAGATTTAGCTATTTTTTCCCTTCATTTAGCCGGAATTTCTTCAATTTTAGGGGCAATTAATTTTATTACAACTATTATTAATATACGAATTAATAACTTATCATTTGATCAAATACCTTTATTTGTTTGAGCCGTAGGTATTACAGCATTACTTTTACTTTTATCTCTCCCAGTATTAGCTGGTGCTATTACTATACTTTTAACAGATCGAAATCTTAATACATCATTTTTTGACCCTGCAGGAGGTGGAGATCCTATTTTATACCAACATTTATTT
Type material.
Holotype: ♂ (Fig. 5a, b), deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), bears the following two rectangular white printed labels: [ Green Gulch | Big Bend Nat. Park | Brewster Co. TX | 12-Jun-2004 USA | leg. Grishin N. V. ], [ DNA sample ID: | NVG-21113E02 | c/o Nick V. Grishin ], and one red printed [ HOLOTYPE ♂ | Staphylus | ecos Grishin ]. Paratypes: 8♂♂ and 5♀♀, most collected by Roy O. Kendall & C. A. Kendall (ROK) and Nick V. Grishin (NVG), from USA: Texas: 1♂ NVG-19125H10, Comal Co., New Braunfels, Panther Canyon, leg. W. W. McGuire, 6-May-1972; Bexar Co.: 1♂ NVG-19125H08, USH281 north at Salado Creek, ROK, 4-Jun-1966; 1♀ NVG-18058B05, O. C. Poling, no date, estimated around 1900; 1♀ NVG-19125H12, Val Verde Co., Del Rio, leg. Hugh Avery Freeman, 12-Jul-1949; Brewster Co., Big Bend National Park: 1♂ NVG-19125H09, 22-Sep-1971 & 1♀ NVG-19126A06, 14-Sep-1971, K-Bar Research Station, ROK; 1♀ NVG-21113E03, Chisos Basin Campground, NVG, 29-May-2004; Jeff Davis Co.: 1♂ NVG-19125H11, 10-Jul-1969 & 1♀ NVG-19126A07, 7-Jul-1969, Musquiz Canyon, SH118, ROK; 1♂ NVG-11137, Jeff Davis Co., Fort Davis city limits, NVG, 18-May-2018; 2♂♂ NVG-21113E04 & 5, Jeff Davis Co., Davis Mts. State Park, nr. Indian Lodge, NVG, 25-Mar-2005; and 1♂ NVG-19126A01, Mexico: Coahuila, Hwy 57 ca. 18 mi S of Monclova, ROK, 14-Sep-1977.
Type locality.
USA: Texas, Brewster County, Big Bend National Park, Green Gulch, elevation 1960 m, GPS 29.2769, −103.2836.
Distribution.
Confirmed from USA: Texas, in the following counties: Jeff Davis, Brewster, Val Verde, Bexar, and Comal; and from Mexico: Coahuila, ca. 18 mi S of Monclova. Specimens we sequenced from El Paso County Texas were S. ceos (Fig. 4). The two sister species are currently allopatric, and the boundary between them in the USA lies somewhere between the Davis Mountains and the Franklin Mountains in Texas with no known intervening populations. Further studies of these species may be productive in Chihuahua, Mexico, where they may closely approach one another.
Etymology.
The species that ec[h]o[e]s ceos with distortion and the name is its anagram. For a simple mnemonic, think of “e” for “east” for this more eastern species. The name is a noun in apposition.
Suggested English name.
Texas Sootywing.
Staphylus opites (Godman & Salvin, 1896), reinstated status
Scantilla opites Godman & Salvin, 1896 (type locality in Guatemala) was placed as a junior subjective synonym of Staphylus vincula (Plötz, 1886) (type locality in Panama) by Evans (1953). Genomic sequencing of Tagiades vincula Plötz, 1886 syntype in ZSMC (NVG-18056G01, Fig. 7a–c) reveals that it is in a different clade from the two specimens of S. opites from Mexico: Oaxaca (Fig. 4). This syntype, a female (Fig. 7a–c), agrees with the original description of T. vincula. It is close in appearance to the specimen from Costa Rica in BMNH that Godman considered to be similar to the original Plötz illustration (Godman 1907), and is curated as a type in ZSMC. Therefore, we accept it as a true syntype, and due to significant genetic differentiation between the two taxa, reinstate species-level status for Staphylus opites (Godman & Salvin, 1896), stat. rest. The COI barcode sequence of the T. vincula syntype (GenBank ON351021) is:
Fig. 7.

Staphylus vincula syntype, female, NVG-18056G01 in ZSMC: a. dorsal and b. ventral views, c. labels (lectotype designation was not published and the specimen remains a syntype); d. an illustration pinned in a Staphylus drawer in USNM, possibly of a specimen from Costa Rica in BMNH, which Godman (1907) considered to be similar to the original illustration of Tagiades vincula by Plötz. Larger scale bar refers to the specimen and illustration and smaller scale bar refers to labels (reduced compared to the specimen).
AACTTTATATTTTATTTTTGGTATTTGATCCGGTATAGTAGGAACTTCTTTAAGTATTCTTATTCGTTCAGAACTAGGAACTCCTGGATCTTTAATTGGAGATGATCAAATTTACAATACTATTGTAACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGATTTGGAAATTGATTAGTACCTTTAATATTAGGAGCTCCTGATATAGCTTTCCCTCGAATAAATAATATAAGTTTTTGATTATTACCCCCTTCTCTTATACTTTTAATTTCAAGTAGTATTGTAGAAAATGGAGCAGGTACAGGATGAACTGTTTACCCACCTCTTTCAGCCAATATTGCTCATCAAGGAGCATCTGTAGATTTAGCTATTTTTTCCCTTCATTTAGCAGGAATTTCTTCAATTTTAGGAGCAATTAATTTTATTACAACTATTATTAATATACGAATTAATAATTTATCATTTGATCAAATACCTTTATTCGTTTGAGCTGTAGGAATTACAGCTTTACTTTTACTTTTATCTTTACCAGTATTAGCAGGTGCTATTACTATACTTTTAACTGATCGAAATCTTAATACATCATTTTTTGATCCAGCAGGAGGAGGAGATCCAATTTTATATCAACATTTATTT
Additionally, we found an illustration of T. vincula (Fig. 7d) pinned in a Staphylus drawer in USNM. This color illustration depicts the left dorsal surface and is rather similar to a Costa Rican specimen from the Godman & Salvin collection in BMNH, which lacks the abdomen and is spread in a similar manner. Furthermore, the illustration shows three subapical hyaline spots, as the BMNH specimen, but the syntype has two, as stated in the original description. Hence, this illustration is possibly of the BMNH specimen, rather than a copy of the original Plötz’s drawing.
Pholisora iguala Williams & Bell, 1940 is a junior subjective synonym of Staphylus vincula (Plötz, 1886)
As we have shown above, Staphylus opites (Godman & Salvin, 1896), stat. rest. (type locality in Guatemala) and Staphylus vincula (Plötz, 1886) (type locality in Panama) are distinct, non-sister species (Fig. 4). While S. vincula is sister to Staphylus cartagoa (Williams & Bell, 1940) (type locality in Costa Rica), S. opites is sister to the clade consisting of Staphylus ceos (W. H. Edwards, 1882) (type locality in USA: Arizona, Graham Co.) and Staphylus ecos sp. n. (type locality in USA: Texas, Brewster Co.). Genomic sequencing of the holotype of Pholisora iguala Williams & Bell, 1940 (type locality in Mexico: Guerrero) in the AMNH (NVG-18024G11), together with a more recently collected specimen identified as S. iguala (NVG-18059A11), reveals that they are genetically close to the syntype of S. vincula, in both nuclear and mitochondrial genomes. Moreover, their COI barcodes differ by only 0.3% (2 bp). Therefore, we propose that Pholisora iguala Williams & Bell, 1940 syn. n. is a junior subjective synonym of Staphylus vincula (Plötz, 1886). As a result of this change, current synonyms of P. iguala become junior subjective synonyms of S. vincula. This genetic similarity with Mexican specimens casts certain doubt on the Panamanian origin of the only known S. vincula syntype, and this question should be investigated further by sequencing of additional specimens. Furthermore, the syntype bears two conflicting locality labels: “Panama” and “S[outh] America” (Fig. 7d), but the original description gives Panama as the locality, where it was supposedly collected by Ribbe. Ribbe is not mentioned on the labels of the syntype. Genomic sequencing suggests that its South American (and maybe even Panamanian) origin is unlikely, and therefore the specimen was possibly mislabeled. For these reasons, we refrain from designating it a lectotype (it already bears a lectotype label, but this designation remains unpublished) waiting for further research. However, this is the only credible syntype we located thus far, and we are using it to define the identity of S. vincula. The COI barcode sequence of the P. iguala holotype (GenBank ON351022) is:
AACTTTATATTTTATTTTTGGTATTTGATCCGGTATAGTAGGAACTTCTTTAAGTATTCTTATTCGTTCAGAACTAGGAACTCCTGGATCTTTAATTGGAGATGATCAAATTTACAATACTATTGTAACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGATTTGGAAATTGATTAGTACCTTTAATATTAGGAGCTCCTGATATAGCTTTCCCTCGAATAAATAATATAAGTTTTTGATTATTACCTCCTTCTCTTATACTTTTAATTTCAAGTAGTATTGTAGAAAATGGAGCAGGTACAGGATGAACTGTTTACCCACCCCTTTCAGCCAATATTGCTCATCAAGGAGCATCTGTAGATTTAGCTATTTTTTCCCTTCATTTAGCAGGAATTTCTTCAATTTTAGGAGCAATTAATTTTATTACAACTATTATTAATATACGAATTAATAATTTATCATTTGATCAAATACCTTTATTCGTTTGAGCTGTAGGAATTACAGCTTTACTTTTACTTTTATCTTTACCAGTATTAGCAGGTGCTATTACTATACTTTTAACTGATCGAAATCTTAATACATCATTTTTTGATCCAGCAGGAGGAGGAGATCCAATTTTATATCAACATTTATTT
DISCUSSION
Screening of butterfly populations across their distribution ranges by genomic sequencing to probe genetic diversification is productive in revealing new taxa. Addition of primary type specimens to these datasets puts this genetic approach on a solid nomenclatural footing. Traditionally done using only COI barcodes, such screens have been insightful (Hebert et al. 2004; Burns and Janzen 2005; Burns et al. 2007; Burns et al. 2008), but total genomic coverage alleviates the problems with introgression and other irregularities of COI barcodes. Here, we applied the genomic approach that led to the discovery of two new, partly cryptic butterfly species in the United States. Genetic differences are reflected in genitalic differences, although these are rather subtle and therefore difficult to evaluate without genetic differentiation.
Geographically, the two allopatric species in each Staphylus pair are separated by well-documented suture zones (Remington 1968; Rising 1983), which form known boundaries for other butterfly taxa. For instance, the North Florida suture zone separates Erynnis somnus (Lintner, 1881) from Erynnis brizo (Boisduval & Le Conte, [1837]) (Burns 2020), as well as Staphylus floridus sp. n. from S. hayhurstii, among many subspecies-level taxa (Warren et al. 2016). The Central New Mexico suture zone separates a number of species pairs, such as Megathymus violae D. Stallings & Turner, 1956 from Megathymus ursus Poling, 1902 (Zhang et al. 2020), Poladryas minuta (W. H. Edwards, 1861) from Poladryas arachne (W. H. Edwards, 1869), Microtia elada (Hewitson, 1868) from Microtia perse (W. H. Edwards, 1882), and Junonia coenia Hübner, [1822] from Junonia grisea Austin & J. Emmel, 1998 (Lalonde and Marcus 2019), among others, together with Staphylus ecos sp. n. from S. ceos. Notably, in this case, the exact geographic position of the boundary between the two species in every pair varies somewhat: it could be to the west (as for Megathymus) or to the east (as for Staphylus) of El Paso.
Textbooks can be written about species delimitation and its practice. For practical applications, we reason that genetic differentiation is by itself sufficient to substantiate species-level status of a taxon under certain conditions. This differentiation should be: 1) strongly supported by statistics, e.g., bootstrap-like values near 100%; 2) consistent throughout the genomes, both nuclear and mitochondrial, e.g., Fig. 1 red and blue clades; and 3) of a magnitude documented for well-studied species (Cong et al. 2019a). This is even more meaningful when coupled with differences in genitalia, although these differences would necessarily be subtle for closely related species. The second case (Fig. 4 red and blue clades) provides more food for thought. Although mitochondrial genome differentiation is prominent (Fig. 4c), and agrees with that of closely related but distinct species (2.7% COI barcode difference), nuclear genomes did not diverge much, and this divergence does not pass our conservative criteria for speciation. However, nuclear genomic differences are consistent between autosomes and the Z chromosome (Fig. 4a, b) and therefore are a reflection of distinct evolutionary paths of these clades. Due to the small divergence, we considered proposing S. ecos sp. n. as a subspecies of S. ceos. However, this opinion changed after genitalic comparison. The differences in male genitalia are consistent in magnitude with those used to support new species even in the absence of DNA evidence. Further studies of S. ceos species complex in Mexico are likely to enrich our understanding of speciation.
ACKNOWLEDGMENTS
We acknowledge Ping Chen and Ming Tang for excellent technical assistance. We are grateful to David Grimaldi and Courtney Richenbacher (AMNH: American Museum of Natural History, New York, NY, USA), Blanca Huertas, David Lees, and Geoff Martin (BMNH: Natural History Museum, London, UK), Jim Fetzner, Bob Androw, Vanessa Verdecia, Cat Giles, and the late John Rawlins (CMNH: Carnegie Museum of Natural History, Pittsburgh, PA, USA), Wolfram Mey, Viola Richter, and Theo Leger (MFNB: Museum für Naturkunde, Berlin, Germany), Edward G. Riley, Karen Wright, and John Oswald (TAMU: Texas A&M University Insect Collection, College Station, TX, USA), Robert K. Robbins, John M. Burns, and Brian Harris (USNM: National Museum of Natural History, Smithsonian Institution, Washington, DC, USA), and Axel Hausmann and Ulf Buchsbaum (ZSMC: Zoologische Staatssammlung München, Germany) for granting access to the collections under their care and for stimulating discussions; to Bernard Hermier for exchange of opinions, to John V. Calhoun for the loan of specimens, suggestions, and critical review of the manuscript. We are indebted to Texas Parks and Wildlife Department (Natural Resources Program Director David H. Riskind) for the research permit 08-02Rev, to U. S. National Park Service for the research permits: Big Bend (Raymond Skiles) for BIBE-2004-SCI-0011. We acknowledge the Texas Advanced Computing Center (TACC) at The University of Texas at Austin for providing HPC resources. The study has been supported in part by grants from the National Institutes of Health GM127390 and the Welch Foundation I-1505.
Footnotes
ZooBank registration: http://zoobank.org/CCF018F4-0188-451A-BE75-006F64938595
LITERATURE CITED
- Allio R, Scornavacca C, Benoit N, Clamens AL, Sperling FAH, and Condamine FL. 2019. Whole genome shotgun phylogenomics resolves the pattern and timing of swallowtail butterfly evolution. Systematic Biology 69(1): 38–60. [DOI] [PubMed] [Google Scholar]
- Burns JM 2020. Taxonomic status of a Florida differentiate in the Erynnis brizo species group: classical evidence (Lepidoptera: Hesperiidae: Pyrginae). Proceedings of the Entomological Society of Washington 122(1): 25–41. [Google Scholar]
- Burns JM and Janzen DH. 2005. Pan-neotropical genus Venada (Hesperiidae: Pyrginae) is not monotypic: Four new species occur on one volcano in the Area de Conservación Guanacaste, Costa Rica. Journal of the Lepidopterists’ Society 59(1): 19–34. [Google Scholar]
- Burns JM, Janzen DH, Hajibabaei M, Hallwachs W, and Hebert PD. 2008. DNA barcodes and cryptic species of skipper butterflies in the genus Perichares in Area de Conservacion Guanacaste, Costa Rica. Proceedings of the National Academy of Sciences of the United States of America 105(17): 6350–6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns JM, Janzen DH, Hajibabaei M, Hallwachs W, and Hebert PDN. 2007. DNA barcodes of closely related (but morphologically and ecologically distinct) species of skipper butterflies (Hesperiidae) can differ by only one to three nucleotides. Journal of the Lepidopterists’ Society 61(3): 138–153. [Google Scholar]
- Cong Q, Shen J, Zhang J, Li W, Kinch LN, Calhoun JV, Warren AD, and Grishin NV. 2021. Genomics reveals the origins of historical specimens. Molecular Biology and Evolution 38(5): 2166–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Q, Zhang J, and Grishin NV. 2019a. Genomic determinants of speciation. bioRxiv BIORXIV/2019/837666. [Google Scholar]
- Cong Q, Zhang J, Shen J, Cao X, Brevignon C, and Grishin NV. 2020. Speciation in North American Junonia from a genomic perspective. Systematic Entomology 45(4): 803–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Q, Zhang J, Shen J, and Grishin NV. 2019b. Fifty new genera of Hesperiidae (Lepidoptera). Insecta Mundi 0731: 1–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans WH 1953. A catalogue of the American Hesperiidae indicating the classification and nomenclature adopted in the British Museum (Natural History). Part III. Pyrginae. Section 2 The Trustees of the British Museum (Natural History); London. v + 246 pp., pls. 26–53. [Google Scholar]
- Godman FD 1907. Notes on the American species of Hesperiidae described by Plötz. Annals and Magazine of natural History (7) 20(16): 132–155. [Google Scholar]
- Hebert PD, Cywinska A, Ball SL, and deWaard JR. 2003. Biological identifications through DNA barcodes. Proceedings of the Royal Society B: Biological Sciences 270(1512): 313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert PD, Penton EH, Burns JM, Janzen DH, and Hallwachs W. 2004. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proceedings of the National Academy of Sciences of the United States of America 101(41): 14812–14817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janzen DH, Burns JM, Cong Q, Hallwachs W, Dapkey T, Manjunath R, Hajibabaei M, Hebert PDN, and Grishin NV. 2017. Nuclear genomes distinguish cryptic species suggested by their DNA barcodes and ecology. Proceedings of the National Academy of Sciences of the United States of America 114(31): 8313–8318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalonde MML and Marcus JM. 2019. Getting western: biogeographical analysis of morphological variation, mitochondrial haplotypes and nuclear markers reveals cryptic species and hybrid zones in the Junonia butterflies of the American southwest and Mexico. Systematic Entomology 44(3): 465–489. [Google Scholar]
- Li W, Cong Q, Shen J, Zhang J, Hallwachs W, Janzen DH, and Grishin NV. 2019. Genomes of skipper butterflies reveal extensive convergence of wing patterns. Proceedings of the National Academy of Sciences of the United States of America 116(13): 6232–6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongue AJ, Hansen ME, and Walters JR. 2022. Support for faster and more adaptive Z chromosome evolution in two divergent lepidopteran lineages. Evolution 76(2): 332–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongue AJ and Walters JR. 2018. The Z chromosome is enriched for sperm proteins in two divergent species of Lepidoptera. Genome 61(4): 248–253. [DOI] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, and Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution 32(1): 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham JP 2008. Catalogue of the Butterflies of the United States and Canada. Journal of Research on the Lepidoptera 40: 1–658. [Google Scholar]
- Pelham JP 2022. Catalogue of the Butterflies of the United States and Canada. Revised 2 February 2022. <http://www.butterfliesofamerica.com/US-Can-Cat.htm> Accessed 24 April 2022. [Google Scholar]
- Pfeiler E 2018. DNA barcoding and taxonomic challenges in describing new putative species: examples from sootywing and cloudywing butterflies (Lepidoptera: Hesperiidae). Diversity 10(4): 111. [Google Scholar]
- Remington CL 1968. Suture-zones of hybrid interaction between recently joined biotas. In: Dobzhansky T, Hecht MK, and Steere WC (Eds.). Evolutionary Biology. Springer; Boston, pp. 321–428. [Google Scholar]
- Rising JD 1983. The great plains hybrid zones. In: Johnston R (Ed.). Current Ornithology. pp. 131–158. [Google Scholar]
- Robbins RK, Cong Q, Zhang J, Shen J, Busby RC, Faynel C, Duarte M, Martins ARP, Prieto C, Lamas G, and Grishin NV. 2022. Genomics-based higher classification of the species-rich hairstreaks (Lepidoptera: Lycaenidae: Eumaeini). Systematic Entomology syen.12541, Early View: 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahara K, Yoshido A, and Traut W. 2012. Sex chromosome evolution in moths and butterflies. Chromosome Research 20(1): 83–94. [DOI] [PubMed] [Google Scholar]
- Shen J, Cong Q, Borek D, Otwinowski Z, and Grishin NV. 2017. Complete genome of Achalarus lyciades, the first representative of the Eudaminae subfamily of skippers. Current Genomics 18(4): 366–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudinger O 1876. Neue Lepidopteren des südamerikanischen Faunengebiets. Verhandlungen der kaiserlich-königlichen zoologisch-botanischen Gesellschaft in Wien 25(1): 89–124. [Google Scholar]
- Sukumaran J and Holder MT. 2010. DendroPy: a Python library for phylogenetic computing. Bioinformatics 26(12): 1569–1571. [DOI] [PubMed] [Google Scholar]
- Toussaint EFA, Braby MF, Müller CJ, Petrie EA, and Kawahara AY. 2022. Molecular phylogeny, systematics and generic classification of the butterfly subfamily Trapezitinae (Lepidoptera: Papilionoidea: Hesperiidae). Zoological Journal of the Linnean Society zlab086, ahead of print: 1–15. [Google Scholar]
- Toussaint EFA, Breinholt JW, Earl C, Warren AD, Brower AVZ, Yago M, Dexter KM, Espeland M, Pierce NE, Lohman DJ, and Kawahara AY. 2018. Anchored phylogenomics illuminates the skipper butterfly tree of life. BMC Evolutionary Biology 18(1): 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toussaint EFA, Chiba H, Yago M, Dexter KM, Warren AD, Storer C, Lohman DJ, and Kawahara AY. 2021. Afrotropics on the wing: phylogenomics and historical biogeography of awl and policeman skippers. Systematic Entomology 46(1): 172–185. [Google Scholar]
- Warren AD, Davis KJ, Stangeland EM, Pelham JP, Willmott KR, and Grishin NV. 2016. Illustrated Lists of American Butterflies. [21-XI-2017]. [Google Scholar]
- Zhang J, Cong Q, Shen J, Brockmann E, and Grishin NV. 2019a. Genomes reveal drastic and recurrent phenotypic divergence in firetip skipper butterflies (Hesperiidae: Pyrrhopyginae). Proceedings of the Royal Society B: Biological Sciences 286(1903): 20190609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Cong Q, Shen J, Brockmann E, and Grishin NV. 2019b. Three new subfamilies of skipper butterflies (Lepidoptera, Hesperiidae). Zookeys 861: 91–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Cong Q, Shen J, and Grishin NV. 2022. Taxonomic changes suggested by the genomic analysis of Hesperiidae (Lepidoptera). Insecta Mundi 0921: 1–135. [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Cong Q, Shen J, Opler PA, and Grishin NV. 2020. Genomic evidence suggests further changes of butterfly names. The Taxonomic Report of the International Lepidoptera Survey 8(7): 1–40. [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Cong Q, Shen J, Opler PA, and Grishin NV. 2021. Genomics-guided refinement of butterfly taxonomy. The Taxonomic Report of the International Lepidoptera Survey 9(3): 1–54. [DOI] [PMC free article] [PubMed] [Google Scholar]


