Abstract
APETALA2/ethylene‐responsive factor (AP2/ERF) family transcription factors are well‐documented in plant responses to a wide range of biotic and abiotic stresses, but their roles in mediating elicitor‐induced disease resistance remains largely unexplored. PevD1 is a Verticillium dahliae secretory effector that can induce disease resistance in cotton and tobacco plants. In our previous work, Nicotiana benthamiana ERF114 (NbERF114) was identified in a screen of genes differentially expressed in response to PevD1 infiltration. Here, we found that the ortholog of NbERF114 in Arabidopsis thaliana (ERF114) also strongly responded to PevD1 treatment and transcripts were induced by Pseudomonas syringae pv. tomato (Pst) DC3000 infection. Loss of ERF114 function caused impaired disease resistance, while overexpressing ERF114 (OE‐ERF114) enhanced resistance to Pst DC3000. Moreover, ERF114 mediated PevD1‐induced disease resistance. RNA‐sequencing analysis revealed that the transcript level of phenylalanine ammonia‐lyase1 (PAL1) and its downstream genes were significantly suppressed in erf114 mutants compared with A. thaliana Col‐0. Reverse transcription‐quantitative PCR (RT‐qPCR) analysis further confirmed that the PAL1 mRNA level was significantly elevated in overexpressing OE‐ERF114 plants but reduced in erf114 mutants compared with Col‐0. Chromatin immunoprecipitation‐qPCR (ChIP‐qPCR) and electrophoretic mobility shift assay verified that ERF114 directly bound to the promoter of PAL1. The gene expression profiles of ERF114 and PAL1 in oestradiol‐inducible transgenic plants confirmed ERF114 could activate PAL1 transcriptional expression. Further investigation revealed that ERF114 positively modulated PevD1‐induced lignin and salicylic acid accumulation, probably by activating PAL1 transcription.
Keywords: disease resistance, ERF114, PevD1, Pst DC3000, Verticillium dahliae
Ethylene‐responsive factor ERF114 mediates Verticillium dahliae effector PevD1‐induced lignin and salicylic acid accumulation, probably by activating PAL1 transcription.
1. INTRODUCTION
Plants have developed sophisticated response mechanisms under pathogen infection. Generally, there are two layers of innate immunity against invading pathogens in plants: pathogen‐associated molecular pattern (PAMP)‐triggered immunity (PTI) and effector‐triggered immunity (ETI) (Dangl et al., 2013; Jones & Dangl, 2006). PAMPs are recognized by pattern recognition receptors (PRRs) located on the plant surface, resulting in the plant immune system PTI (Schwessinger & Ronald, 2012). ETI is elicited by recognition of pathogen‐derived effectors via intracellular nucleotide‐binding domain leucine‐rich repeat‐containing receptors (NLRs) (Dangl et al., 2013). PTI and ETI are associated with various plant immunity responses, including the hypersensitive response (HR), reactive oxygen species (ROS) production, and the expression of various defence‐related genes such as pathogenesis‐related (PR) genes and transcription factors (TFs) (Chandran et al., 2014; Greenberg & Yao, 2004). During the plant defence response, the phenylpropanoid pathway plays an important role in preventing pathogen infection (Li et al., 2020). The phenylpropanoid metabolic pathway positively regulates plant basal immunity to viruses, bacteria, and fungi (Konig et al., 2014; Zabala et al., 2006; Zhang et al., 2020). Phenylalanine ammonia‐lyase 1 (PAL1) is involved in the biosynthesis of multiple secondary metabolites, including lignin and salicylic acid (SA) in plants (Huang et al., 2010; Vanholme et al., 2019). Lignin as a defensive substance can limit the pathogen infection, therefore plant lignin accumulation can be used as a marker of the plant immune response (Sonbol et al., 2009).
Transcriptional regulation plays a key role in plant defence against pathogens (Gao et al., 2022; Zheng et al., 2019). During this process, TFs binding to DNA regulate normal growth and the defence response to pathogens in plants. The conserved TF superfamily APETALA2/ethylene‐responsive factor (AP2/ERF) has a special importance in the plant kingdom as its members are involved in growth, development, and responses to various biotic and abiotic stresses, including drought, high salinity, extreme temperature, and pathogen infection (Licausi et al., 2013; Yamaguchi‐Shinozaki & Shinozaki, 2006). ERFs regulate the expression of PR genes and confer tolerance to various biotic stresses. For example, several ERFs activate the transcription of basic type defence‐related genes, PR genes, osmotin, chitinase, and β‐1,3‐glucanase (Moffat et al., 2012). ERF11 activates BT4 transcription to regulate immunity to Pseudomonas syringae (Zheng et al., 2019).
PevD1 is an effector secreted by Verticillium dahliae. Our previous studies revealed that PevD1 can induce HR‐like necrosis, H2O2 production, NO generation, and systemic acquired resistance in cotton and tobacco plants (Bu et al., 2014; Liang et al., 2018; Wang et al., 2012). In addition, PevD1 also induces the expression of genes related to the phenylpropanoid metabolic pathway and enhanced lignin deposition, vessel reinforcement in cotton (Bu et al., 2014), and sesquiterpenoid phytoalexin capsidiol accumulation in tobacco (Liang et al., 2021). To reveal the mechanism of PevD1‐induced disease resistance, several differentially expressed genes (DEGs) induced by PevD1 have been identified in Nicotiana benthamiana. Among these DEGs, the most abundant genes are members of the AP2/ERF transcription factors family. N. benthamiana ERF114 strongly responded to PevD1 and its role in regulating disease resistance is unknown yet. In this study, we characterized the function of ERF114 in regulating disease resistance to P. syringae pv. tomato (Pst) DC3000 in Arabidopsis thaliana (Arabidopsis). ERF114 positively modulated disease resistance and mediated PevD1‐induced disease resistance. ERF114 targeted the promoter of PAL1 and activated PAL1 transcription, resulting in an increase in lignin and SA levels. Functional investigation of ERF114 helps us to understand the mechanism in PevD1‐induced disease resistance.
2. RESULTS
2.1. PevD1 triggers defence responses and enhances disease resistance to Pst DC3000 in A. thaliana
Previous studies showed PevD1 induces H2O2 accumulation, and enhances disease resistance in tobacco against tobacco mosaic virus (TMV) and in cotton against V. dahliae (Bu et al., 2014; Wang et al., 2012). Here, we investigated the PevD1‐triggered immune responses and disease resistance in Arabidopsis. We observed H2O2 and superoxide anion accumulation in Arabidopsis leaves by 3,3′‐diaminobenzidine (DAB) and nitroblue tetrazolium (NBT) staining, respectively. Compared to buffer‐treated leaves (Mock), PevD1 infiltration promoted the accumulation of H2O2 and superoxide anion (Figure 1a). Furthermore, the expression levels of defence genes PR1 and PR5 in Col‐0 leaves infiltrated with PevD1 was also significantly elevated compared to Mock (Figure 1b,c). To further examine the effect of PevD1 on plant resistance to bacteria, leaves were inoculated with Pst DC3000 24 h after infiltration with 30 µM PevD1. The results showed that PevD1‐pretreated leaves exhibited enhanced resistance to Pst DC3000 compared to mock‐treated leaves (Figure 1d,e). These results indicate that PevD1 activates the immune response and enhances disease resistance in Arabidopsis.
FIGURE 1.
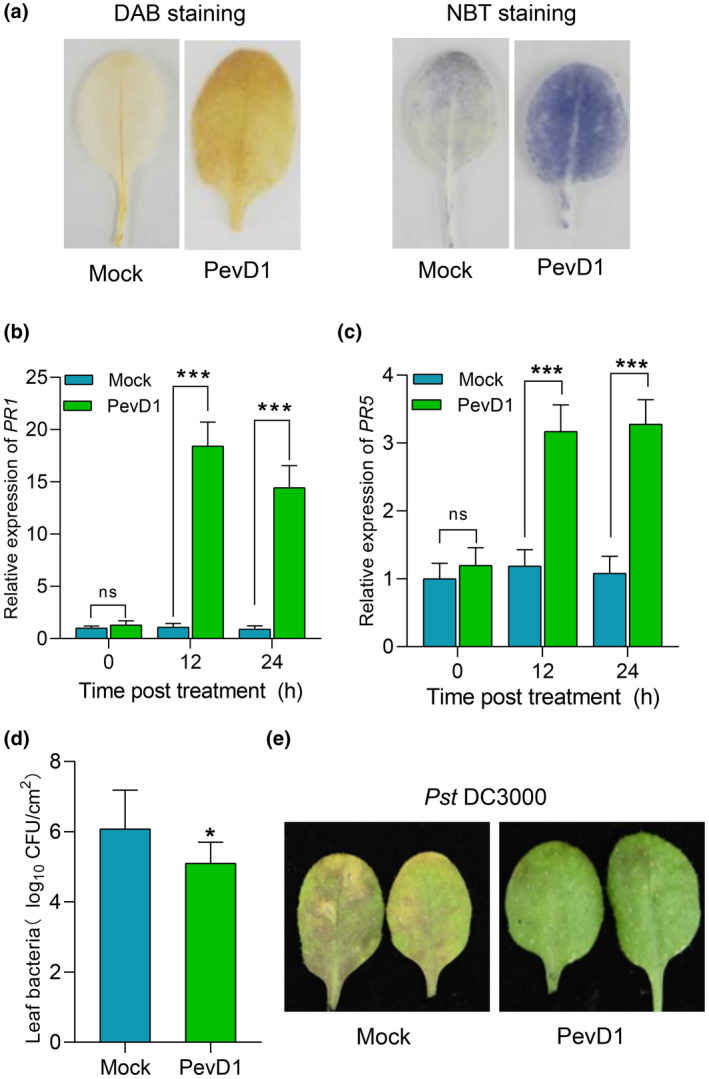
PevD1 triggers a defence response and enhances disease resistance to Pseudomonas syringae pv. tomato (Pst) DC3000 in Arabidopsis thaliana (Col‐0). (a) Photographs of 3,3′‐diaminobenzidine (DAB)‐ and nitroblue tetrazolium (NBT)‐stained leaves infiltrated with buffer (Mock) or PevD1. Four‐week‐old wild‐type Col‐0 leaves were infiltrated with PevD1 or Tris buffer (Mock) and stained with DAB and NBT at 24 h postinfiltration (hpi). (b,c) Relative expression levels of PR1 and PR5 in 4‐week‐old wild‐type Col‐0 leaves at 0, 12, and 24 hpi. UBC21 was used as the internal control, the PR1 and PR5 expression is represented relative to the UBC21 transcription level. The bars were calculated based on three independent experiments. The values are mean ± SD (n = 3). ***p < 0.001, one‐way analysis of variance (ANOVA). (d) Leaf bacteria was measured by colony counting. Col‐0 leaves were infiltrated with PevD1 or Tris buffer (Mock) and inoculated with Pst DC3000 at 24 hpi. Bacterial colonies were counted at 48 hpi. Data from three separate experiments are shown (mean ± SD, n = 6). *p < 0.05, one‐way ANOVA. (e) Photographs of Pst DC3000‐infected disease symptoms in leaves. Col‐0 leaves were infiltrated with PevD1 or Tris buffer (Mock) then inoculated with Pst DC3000 at 24 hpi. Photograph was taken at 48 hpi
2.2. ERF114 transcript is induced by PevD1 and Pst DC3000 infection
NbERF114 is strongly induced by PevD1 infiltration (Figure S1) (Liang et al., 2021), suggesting that it might be involved in PevD1‐induced defence. To study the function of ERF114 in plant defence, we identified the ortholog of NbERF114 in Arabidopsis by BLAST analysis. We then checked the induction of ERF114 on PevD1 treatment in Arabidopsis using reverse transcription‐quantitative PCR (RT‐qPCR). The transcript level of ERF114 was significantly elevated at the indicated time points postinfiltration with PevD1 (Figure 2a). Notably, ERF114 was also induced on Pst DC3000 infection, further supporting that hypothesis that ERF114 could be associated with plant defence (Figure 2c). To further test if the promoter activity of ERF114 responds to PevD1 and Pst DC3000 infection, a chimeric promoter fusion with the β‐glucuronidase gene (GUS) was constructed and introduced into Arabidopsis Col‐0 plants (PromERF114:GUS). GUS staining of 4‐week‐old PromERF114:GUS plants was performed under PevD1 infiltration or Pst DC3000 inoculation. The treated PromERF114:GUS transgenic leaves stained dark blue compared to mock‐treated leaves, indicating that the ERF114 promoter activity was significantly induced (Figure 2b,d). These data suggest that ERF114 transcription is induced by PevD1 and Pst DC3000 infection.
FIGURE 2.
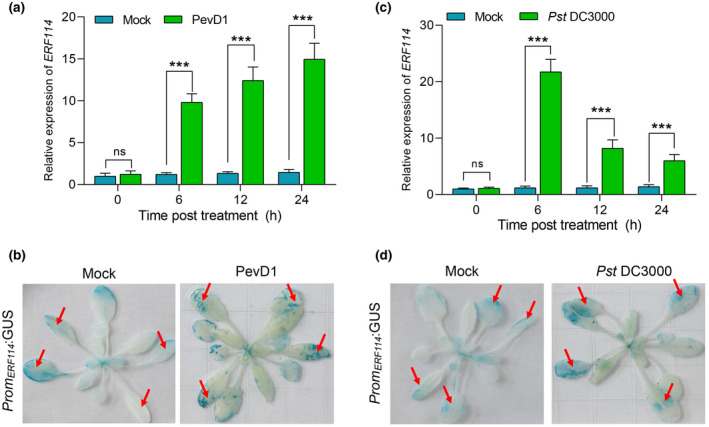
ERF114 expression is induced by PevD1 infiltration and Pseudomonas syringae pv. tomato (Pst) DC3000 infection. (a,c) Relative expression level of ERF114 in 4‐week‐old wild‐type Col‐0 leaves at 0, 6, 12 and 24 h after PevD1 infiltration or Pst DC3000 inoculation. UBC21 was used as the internal control. The bars were calculated based on three independent experiments. The values are mean ± SD (n = 3). ***p < 0.001, one‐way analysis of variance. (b,d) ERF114 expression pattern was analysed by β‐glucuronidase (GUS) enzymatic activity in 4‐week‐old PromERF114:GUS leaves infiltrated with PevD1 or inoculated with Pst DC3000
2.3. ERF114 positively modulates Arabidopsis defence responses and disease resistance to Pst DC3000
To investigate the function of ERF114 in Pst DC3000 infection in Arabidopsis, we generated ERF114 gene deletion mutants by CRISPR/Cas9‐mediated genome editing. Two target sites in the exon of ERF114 were chosen, and the corresponding sgRNA/Cas9 vectors were transformed into the wild‐type Col‐0 via Agrobacterium tumefaciens‐mediated transformation. The mutations at the target sites in the CRISPR/Cas9 transformants were examined by sequencing (Figure 3a). Two individual homozygous knockout lines (erf114#1 and erf114#2) were chosen for further analysis, containing a 1‐bp insertion in the coding region of ERF114 resulting in a frameshift mutation (Figure 3a). Furthermore, the ERF114 coding sequence was overexpressed under the driver of the cauliflower mosaic virus 35S promoter in Arabidopsis (OE‐ERF114). Two OE‐ERF114 lines (OE‐ERF114#2 and OE‐ERF114#4) were selected for further analysis (Figure 3b).
FIGURE 3.
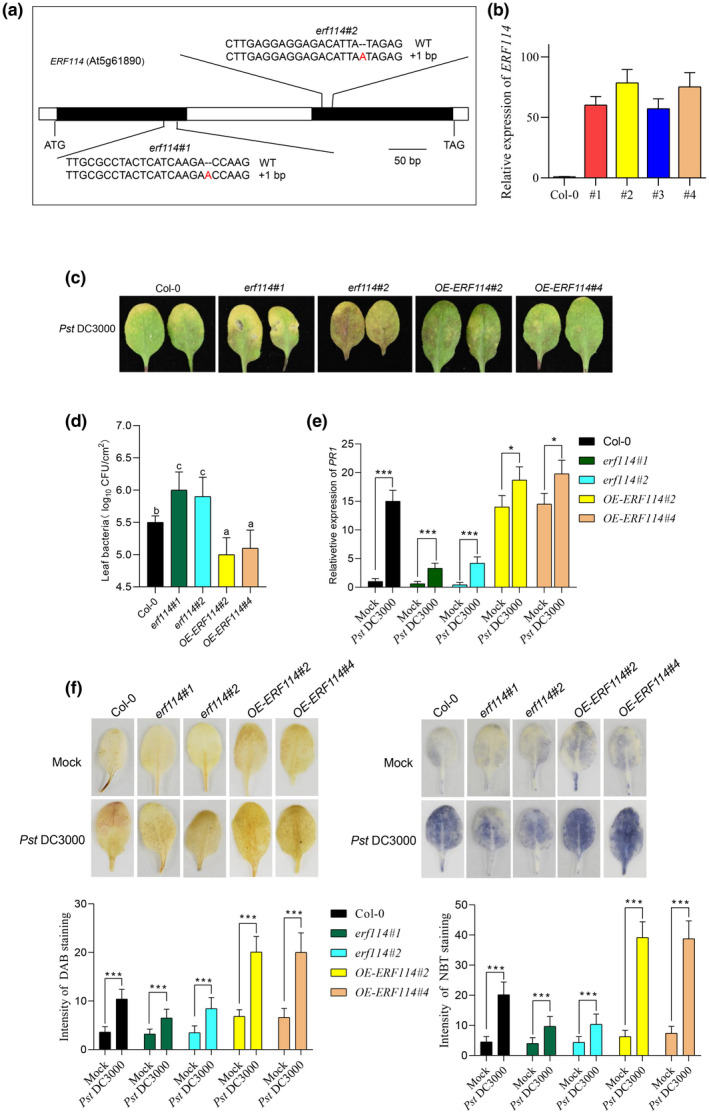
ERF114 contributes to Arabidopsis disease resistance to Pseudomonas syringae pv. tomato (Pst) DC3000. (a) Construction of CRISPR/Cas9‐based erf114 knockout transgenic lines. Two sgRNA sequences that specifically target ERF114 were used, generating two mutants, erf114#1 and erf114#2, both with an “A” insertion (indicated in red font). (b) Relative expression level of ERF114 in four‐week‐old transgenic lines overexpressing ERF114 (OE‐ERF114) and wild‐type Col‐0 leaves. UBC21 was used as the internal control. Data represent the ratio of ERF114 expression between OE‐ERF114 plants and wild‐type Col‐0. The bars were calculated based on three independent experiments. The values are mean ± SD (n = 3). (c) Typical Pst DC3000 disease symptoms in Col‐0, erf114#1, erf114#2, OE‐ERF114#2, and OE‐ERF114#4 leaves. Four‐week‐old leaves were inoculated with Pst DC3000 bacterial suspension or 10 mM MgCl2. Photographs were taken at 48 h postinoculation (hpi). (d) Bacterial growth in Col‐0, erf114, and OE‐ERF114 leaves. Bacteria were isolated from leaves at 48 hpi and counted with gradient dilution assays. Data from three separate experiments are shown (mean ± SD, n = 6). Different letters above the bars indicate statistically significant differences at p < 0.05 (one‐way analysis of variance, ANOVA). (e) Relative expression levels of PR1 in the leaves of 4‐week‐old wild‐type (Col‐0), erf114, OE‐ERF114 plants at 24 hpi. UBC21 was used as the internal control. Relative expression is indicated compared to the transcript level of UBC21. The bars were calculated based on three independent experiments. The values are mean ± SD (n = 3). *p < 0.05, ***p < 0.001, one‐way ANOVA. (f) Leaves were inoculated with Pst DC3000 or 10 mM MgCl2 and stained with 3,3′‐diaminobenzidine (DAB) and nitroblue tetrazolium (NBT) at 24 hpi. Photographs were taken (top) and H2O2 and superoxide anion accumulation was quantified in nine leaves by measuring the intensity of staining with ImageJ (bottom). ***p < 0.001, one‐way ANOVA
We then determined the bacterial growth in Col‐0, erf114 mutants, and OE‐ERF114 plants. The results showed that erf114 mutants were more susceptible, while OE‐ERF114 lines displayed enhanced resistance to Pst DC3000 compared to Col‐0 (Figure 3c,d). This observation was further supported by the expression of PR1, which is a pathogen response reporter gene, observed in the erf114 mutants and OE‐ERF114 lines (Figure 3e). ROS accumulation in the indicated genotypes upon Pst DC3000 infection was detected by DAB and NBT staining. We found higher ROS levels in OE‐ERF114 plants on pathogen attack compared to Col‐0, whereas erf114 mutants showed the opposite (Figure 3f). These data suggest that ERF114 promotes plant defence responses and enhances disease resistance to Pst DC3000.
2.4. ERF114 mediates PevD1‐induced disease resistance
To explore whether ERF114 mediates PevD1‐induced disease resistance to Pst DC3000, we observed the typical chlorotic disease symptoms that appeared at 48 h postinoculation (hpi) and showed more resistance in PevD1‐infiltreated OE‐ERF114#2 and OE‐ERF114#4 than in erf114#1 and erf114#2 mutant plants (Figure 4a). We also examined the growth of Pst DC3000 in PevD1‐pretreated Col‐0, erf114#1, erf114#2, OE‐ERF114#2, and OE‐ERF114#4 leaves inoculated with the bacteria. Higher bacterial counts were found in the two erf114 mutants while Pst DC3000 was inhibited in the two OE‐ERF114 lines (Figure 4b). We then compared the expression level of the PR1 gene in PevD1‐infiltrated leaves of Col‐0, erf114#1, erf114#2, OE‐ERF114#2, and OE‐ERF114#4 plants using RT‐qPCR. The results showed that the PR1 gene expression level was increased in OE‐ERF114#2 and OE‐ERF114#4, while it was decreased in erf114#1 and erf114#2 compared with Col‐0 (Figure 4c). These results indicate that ERF114 mediates PevD1‐induced disease resistance against Pst DC3000.
FIGURE 4.
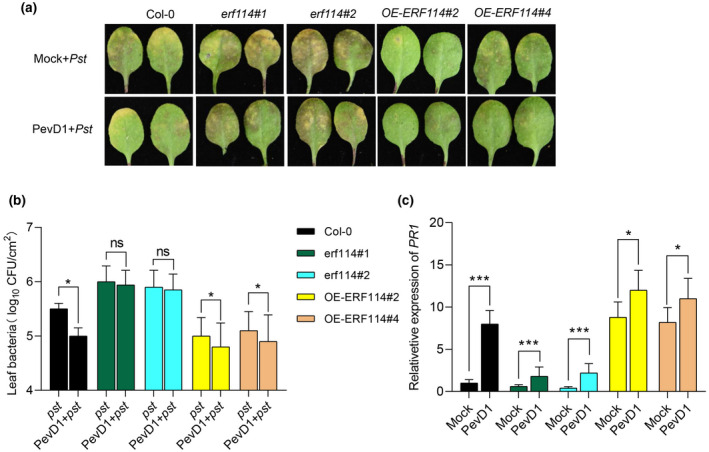
ERF114‐mediated PevD1‐induced disease resistance. (a) The disease symptoms of Pseudomonas syringae pv. tomato (Pst) DC3000 in Col‐0, erf114#1, erf114#2, OE‐ERF114#2, and OE‐ERF114#4 leaves. The photograph was taken at 48 h after Pst DC3000 inoculation. (b) Bacteria in PevD1‐infiltrated leaves were measured by colony counting. Col‐0, erf114 and OE‐ERF114 leaves were infiltrated with Tris buffer (Mock) or PevD1 then inoculated with Pst DC3000 at 24 h postinfiltration (hpi). Bacterial titres were counted at 48 hpi. Data from three separate experiments are shown (mean ± SD, n = 6). *p < 0.05, ns, not significant, one‐way analysis of variance (ANOVA). (c) Relative expression level of PR1 in 4‐week‐old wild‐type (Col‐0), erf114, and OE‐ERF114 leaves at 12 hpi. UBC21 was used as the internal control. The bars were calculated based on three independent experiments. The values are mean ± SD (n = 3). *p < 0.05, ***p < 0.001, one‐way ANOVA
2.5. ERF114 targets PAL1 and activates its transcription
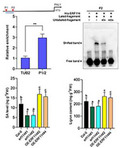
To explore the underlying mechanism of ERF114 in regulating defence responses, RNA‐sequencing (RNA‐Seq) was performed using Col‐0 and the erf114#1 mutant. In total, six samples of the erf114 mutant and Col‐0 with three biological replicates were used for RNA‐Seq analysis. On average, this generated about 48.77 million reads per sample. After read filtering, the average mapping ratio with the reference genome was 97.97% and the average mapping ratio with the gene was 98.24%. A total of 27,444 genes were detected. All of the DEGs were aligned to public databases. Comparative analysis revealed that 661 genes were affected by the ERF114 gene (fold‐change [FC] > 1.5, p < 0.1). Of these, 199 genes were down‐regulated and 462 genes were up‐regulated (Figure S2). Given that ERF114 positively regulates plant disease against Pst DC3000, our data analysis focused on the down‐regulated genes. To determine the cellular processes and function distribution associated with the down‐regulated DEGs, we carried out enrichment analysis of Gene Ontology (GO) biological processes and the Kyoto Encyclopedia of Genes and Genomes (KEGG) functional pathway. GO category analysis revealed that secondary metabolite biosynthetic process and cellulose synthesis activity were dominant in the top 30 GO terms (Figure S3a). KEGG analysis showed a large number of DEGs significantly enriched in the biosynthesis of secondary metabolite pathways, including anthocyanin, flavone and flavonol biosynthesis, and phenylpropanoid biosynthesis (Figure S3). Anthocyanins and flavonoids are major secondary metabolites derived from the phenylpropanoid pathway (Buer et al., 2010; Liu et al., 2021; Wang et al., 2020). PAL1 is the key enzyme of phenylpropanoid biosynthesis; it also affects multiple secondary metabolites, including lignin and SA biosynthesis (Huang et al., 2010). We speculated that ERF114 may affect the synthesis of secondary metabolites through modulating PAL1 transcript expression. Our analysis revealed that the transcript level of PAL1 is down‐regulated in the erf114 mutant and up‐regulated in OE‐ERF114 lines (Figure 5a), and that the PAL downstream genes were down‐regulated in the erf114 mutant compared with Col‐0 (Figure S4). Based on this data, we speculate that PAL1 is a potential target candidate for ERF114. Given that ERF proteins specifically bind to dehydration responsive/C‐repeat (DRE/CRT) elements and a GCCGCC motif (GCC‐box) in the promoter of downstream target genes (Eklund et al., 2011; Ohme‐Takagi & Shinshi, 1995), we analysed the sequence of the promoter of PAL1. We found that the promoter of PAL1 contains two GCC‐boxes (P1 and P2) upstream of the ATG codon (Figure 5b). We speculate that ERF114 might target the PAL1 gene to regulate the phenylpropanoid biosynthesis pathway. To investigate whether ERF114 physically interacts with the PAL1 promoter, we then employed chromatin immunprecipitation‐qunatitative PCR (ChIP‐qPCR) analysis with 35S:ERF114‐GFP transgenic plants, along with Col‐0 as a control. Primer pair P1/2 in the promoter of the PAL1 gene was designed for ChIP‐qPCR (Table S1). As shown in Figure 5b, significant enrichment was found in the P1/2 region. Region P2 was used for an electrophoretic mobility shift assay (EMSA). The results demonstrated the binding of ERF114 to the PAL1 promoter in vitro (Figure 5c).
FIGURE 5.
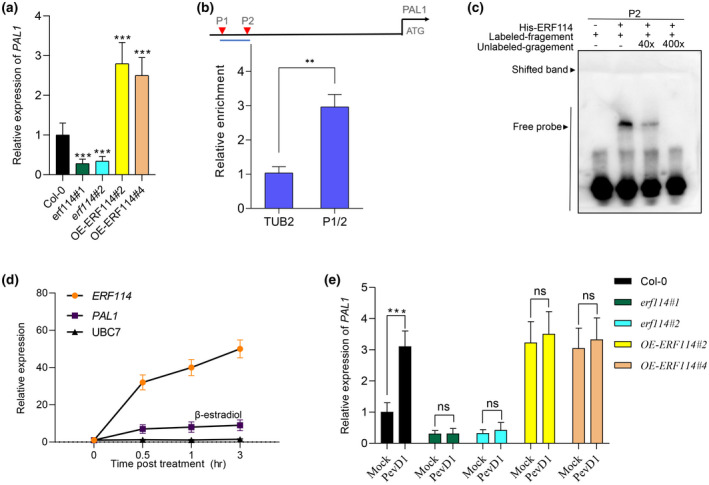
ERF114 binds to the promoter of PAL1 and activates PAL1 expression. (a) Relative expression level of PAL1 in erf114 and OE‐ERF114 leaves compared to wild type Col‐0. UBC21 was used as the internal control. The bars were calculated based on three independent experiments. The values are mean ± SD (n = 3). ***p < 0.001, one‐way analysis of variance (ANOVA). (b) Schematic diagrams of ERF114‐binding cis‐elements in the promoter region of PAL1 (P1 and P2). ATG represents the translational start codon. The lines below the binding sites indicate the sequences detected in the chromatin immunoprecipitation‐quantitative PCR (ChIP‐qPCR) assay. An anti‐GFP monoclonal antibody was used for DNA immunoprecipitation from 4‐week‐old 35S:GFP‐ERF114#2 transgenic plants. The relative enrichment of ERF114 binding to the PAL1 promoter was normalized to TUBULIN2 (TUB2). Each experiment was repeated at least three times with similar results. The values are mean ± SD (n = 3). **p < 0.01, one‐way ANOVA. (c) Electrophoretic mobility shift assays (EMSA) for the binding of ERF114 to the PAL1 promoter in vitro. A biotin‐labelled probe was used for the EMSA experiments and unlabelled fragments were used as competitors. The “+” and “−” symbols represent the presence and absence of components, respectively. Band shift is indicated by an arrow. (d) ERF114 induces the transcript of PAL1. Reverse transcription‐quantitative PCR analysis of ERF114, PAL1, and UBC7 expression in the iERF114 plants treated with 20 μM β‐oestradiol for the indicated time points. The bars were calculated based on three independent experiments. The values are mean ± SD (n = 3). (e) Relative expression level of PAL1 in PevD1‐induced wild‐type Col‐0, erf114, and OE‐ERF114 leaves at 24 h postinoculation (hpi). UBC21 was used as the internal control. Relative expression is indicated compared to the transcript level of UBC21. The bars were calculated based on three independent experiments. The values are mean ± SD (n = 3). ns, not significant. ***p < 0.001, one‐way ANOVA
To further analyse whether ERF114 could activate PAL1 expression, pER8‐ERF114 oestradiol‐inducible transgenic plants were generated and briefly treated with oestradiol in a time course (0.5, 1, or 3 h) to clarify the regulatory relationship between PAL1 and ERF114. The RT‐qPCR results showed that PAL1 was rapidly expressed along with ERF114 expression (Figure 5d), further supporting the result that ERF114 directly regulates PAL1 expression. These findings suggest that ERF114 can bind to the promoter of PAL1 and activate its expression.
To investigate whether ERF114 mediates PevD1‐induced PAL1 expression, we compared the expression level of the PAL1 gene in Col‐0, erf114, and OE‐ERF114 plants with or without PevD1 induction using RT‐qPCR. The results showed that the expression of PAL1 exhibited no difference in PevD1‐induced erf114 mutant and OE‐ERF114 plants compared with mock treatment. Conversely, the PAL1 expression level in PevD1‐induced plants was significantly higher than in buffer‐treated Col‐0 plants (Figure 5e). The results reveal that ERF114 mediates PevD1‐induced PAL1 expression.
2.6. ERF114 positively modulates lignin and SA accumulation
The phenylpropanoid pathway in plants is responsible for the synthesis of a variety of structural and defence compounds. PAL catalyses the first step in phenylpropanoid biosynthesis, which produces precursors to a variety of important secondary metabolites. Lignin, an important phenylalanine‐derived metabolite, has functions in both structural support and plant defence (Huang et al., 2010). SA is a crucial plant hormone for mediating Arabidopsis defence to Pst DC3000 (Chen et al., 2019), and the synthesis of SA is partially derived from the phenylpropanoid pathway (Lefevere et al., 2020). To investigate whether ERF114 regulates SA and lignin accumulation, we compared total lignin deposition and free SA concentration in Col‐0, erf114, and OE‐ERF114. The results showed that erf114 was deficient in SA and lignin accumulation compared with Col‐0; in contrast, SA and lignin accumulation increased in OE‐ERF114 (Figure 6a,b), indicating a role of ERF114 in regulating the SA and lignin accumulation. To further investigate whether ERF114 mediated PevD1‐induced lignin and SA accumulation, Col‐0, erf114, and OE‐ERF114 leaves were treated with PevD1 or buffer before measuring SA and lignin content. The results showed that SA and lignin notably accumulated in PevD1‐induced Col‐0 plants. Meanwhile, SA and lignin accumulation decreased and showed no difference in PevD1‐induced erf114 compared to mock treatment, indicating PevD1 could not induce SA and lignin accumulation in erf114 mutants (Figure 6c,d). These results suggest that ERF114 mediated PevD1‐induced disease resistance against Pst DC3000 mainly through regulating lignin and SA accumulation.
FIGURE 6.
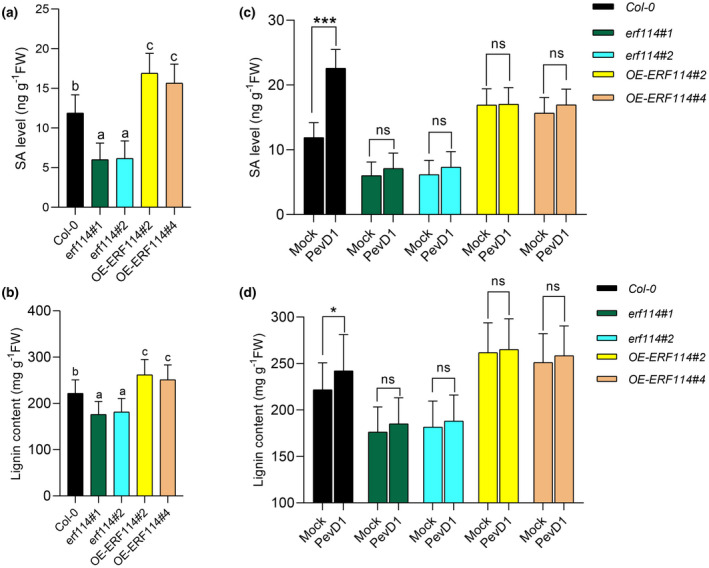
ERF114 contributes to salicylic acid (SA) and lignin accumulation and mediates PevD1‐induced lignin and SA production in Arabidopsis thaliana. (a,b) Levels of SA and lignin in Col‐0, erf114, and OE‐ERF114. Data from three separate experiments are shown. The bars were calculated based on three independent experiments. The values are mean ± SD (n = 4). Different letters above the bars indicate statistically significant differences at p < 0.05 (one‐way analysis of variance, ANOVA). (c,d) Levels of SA and lignin in PevD1‐induced Col‐0, erf114 and OE‐ERF114 at 12 h postinoculation. Each experiment was repeated at least three times. The bars were calculated based on three independent experiments. The values are mean ± SD (n = 4). ns, not significant. *p < 0.05, ***p < 0.001, one‐way ANOVA
3. DISCUSSION
AP2/ERF TFs constitute one of the largest plant‐specific TF families, which contains 147 members in Arabidopsis (Nakano et al., 2006). ERF TFs have a variety of roles in plant processes including development and responses to biotic and abiotic stresses (Licausi et al., 2013). Previous studies demonstrated that one ERF TF, ERF114, plays an important role in callus production and tissue regeneration (Mehrnia et al., 2013), and acts as a ROS‐responsive factor that maintains root stem cell niche identity (Kong et al., 2018). However, the function of ERF114 in the regulation of plant defence to pathogen infection is mostly unknown. Our previous research revealed that the transcript level of NbERF114 is significantly elevated in response to the V. dahliae effector PevD1, suggesting a possible role of ERF114 in pathogen–plant interactions. In this study, the function of Arabidopsis ERF114 was further characterized. We found that both PevD1 and the bacterial pathogen Pst DC3000 could strongly induce the expression of ERF114 (Figure 2). Knockout lines of ERF114 exhibited enhanced susceptibility, while the ERF114‐overexpressing plants were more resistant to Pst DC3000 (Figure 3). Based on RNA‐Seq and biochemical assays, we further revealed that ERF114 positively activated PAL1 by directly binding to its promoter (Figures 5 and S4), resulting in the accumulation of lignin and SA. Thus, our study provides definitive evidence to reveal a positive role of ERF114 in the regulation of plant responses to pathogen infection. In addition, we have demonstrated that ERF114 mediates PevD1‐induced disease resistance. Our findings and previous studies showed that PevD1 could induce ROS, systemic acquired resistance, and lignin accumulation in plants (Bu et al., 2014; Liang et al., 2018; Wang et al., 2012). Pretreatment of PevD1 conferred enhanced resistance to Pst DC3000 in Arabidopsis (Figure 1d,e). Notably, loss‐of‐function of ERF114 compromised the PevD1‐induced accumulation of lignin and SA, as well as attenuated resistance phenotypes (Figure 4), indicating that ERF114 mediates PevD1‐triggered defence responses. Our data give an explanation for PevD1‐induced plant disease resistance.
Phenylpropanoid compounds are precursors to a wide range of compounds with many functions in plant defence against pathogens, such as lignin, phenolic compounds, flavonoids, isoflavonoids, coumarins, and stilbenes (Ferrer et al., 2008). Lignin determines plant cell wall mechanical strength and rigidity. When plants are infected with pathogens, lignin is accumulated in the cell wall and provides a basic barrier against pathogen spread (Liu et al., 2018; Yadav et al., 2020). PAL catalyses the conversion of phenylalanine to trans‐cinnamic acid, which is the first step in the phenylpropanoid pathway and an important regulation point between primary and secondary metabolism (Huang et al., 2010; Malamy et al., 1990). Many studies have shown that PAL expression is responsive to a variety of environmental stimuli, including pathogen infection (Dixon & Paiva, 1995; Lawton et al., 1983; Liang et al., 1989). PAL has also been shown to mediate plant resistance to the brown planthopper by regulating the biosynthesis and accumulation of SA and lignin in rice. OsMYB30 directly up‐regulates the expression of OsPAL8 in response to brown planthopper attack (He et al., 2020). Although PAL1 plays important roles in plant defence components, its transcriptional regulation has rarely been reported. In this research using ChIP‐qPCR and EMSA assays, we found ERF114 could directly bind to the PAL1 promoter and regulate PAL1 expression (Figure 5b,c). This is the first report that an ERF family member positively modulates phenylpropanoid biosynthesis and enhances disease resistance. Further studies on the transcriptional regulation on PAL1 are needed to uncover its significance in plant–pathogen interactions.
SA is a critical component for plant–pathogen interactions (Cui et al., 2017; Vlot et al., 2009) and acts a signal that increases in response to pathogen infection (Malamy et al., 1990). SA accumulation is often accompanied by elevated expression of PR genes and enhanced disease resistance (Yalpani et al., 1991). It is widely believed that the isochorismate synthase pathway is important for SA synthesis in Arabidopsis. The PAL inhibitor 2‐aminoindan‐2‐phosphonic acid significantly reduces pathogen‐induced SA accumulation in Arabidopsis (Chen et al., 2009). Therefore, the PAL pathway is also important for SA accumulation in Arabidopsis. In the present research, the level of SA significantly reduced in the PevD1‐induced erf114 mutant and increased in PevD1‐induced ERF114 overexpressing plants (Figure 6c), indicating that ERF114 mediates PevD1‐induced SA accumulation.
Many studies have shown that ERF transcription factors play a role in regulating phytoalexin production. An ERF2‐like transcription factor positively regulates production of the sesquiterpene phytoalexin capsidiol and plant resistance through the direct transactivation of a capsidiol biosynthetic gene, EAS12 (Song et al., 2019). Our RNA‐Seq data show that ERF114 regulates the expression of various genes in the flavonoid biosynthesis pathway derived from the phenylpropanoid pathway. Flavonoids can act as chemical messengers, physiological regulators, and inhibitors against organisms, including phytopathogens, Fusarium oxysporum and Gymnosporangium yamadai (Lu et al., 2017; Zhou et al., 2017; Zhu et al., 2013). However, whether ERF114 is involved in the synthesis of antibacterial substances that suppress pathogen infection remains to be researched.
In conclusion, we reveal here that ERF114 mediates PevD1‐induced accumulation of lignin and SA, probably by activating the expression of PAL1. Our results provide evidence that ERF114 positively modulates plant defence responses and mediates PevD1‐induced disease resistance to Pst DC3000 in Arabidopsis.
4. EXPERIMENTAL PROCEDURES
4.1. Plant growth conditions
All seeds from wild‐type A. thaliana (Col‐0) and transgenic lines of plants were surface sterilized using ethanol for 10 min, washed with sterile water five times, and placed on Murashige and Skoog medium (4.3 g/L MS salts, 1% sucrose [pH 5.7–5.8], and 6 g/L agar). After incubation at 4℃ in darkness for 3–5 days, the plates were transferred to a growth chamber and cultivated at 22℃ under a 16 h light/8 h dark cycle.
4.2. Generation of transgenic plants
For construction of CRISPR/cas9‐mediated ERF114 knockout plants, two target sites of ERF114 designed by CRISPR‐P (http://crispr.hzau.edu.cn/CRISPR2/) were used. Homozygous plants were identified by sequencing. All primers used are listed in Table S1. To construct constitutive overexpressing transgenic plants, the corresponding coding sequence was amplified and introduced into the pEGAD vector (Cutler et al., 2000). To generate the inducible overexpressing transgenic plant iERF114, ERF114 was amplified and inserted into the pER8 vector (Zuo et al., 2000). To generate PromERF114:GUS/Col‐0, a 1.4‐kb genomic promoter sequence was amplified and introduced into the pBI101 vector. The constructs were transformed into Agrobacterium tumefaciens GV3101, which was used to transform Col‐0 plants with the floral dip method (Clough & Bent, 1998).
4.3. Pst DC3000 culture and inoculation
Pst DC3000 was cultured overnight at 28℃ in Luria‐Bertani (LB) medium. When the bacterial cell concentration of OD600 = 0.8–1.0, the cells were centrifuged and resuspended in 10 mM MgCl2. Final bacterial suspensions in 10 mM MgCl2 with OD600 = 0.0002 were used in the infection assays. Four‐week‐old Col‐0 and transgenic Arabidopsis leaves were used, and the bacterial suspension was infiltrated using 1‐ml syringes without a needle; 10 mM MgCl2 was used as mock treatment. For each independent experiment, at least six plants were assayed for each data point. The experiments were repeated three times (Zipfel et al., 2004).
4.4. Protein preparation and treatments
PevD1 was expressed and purified according to a previously described protocol (Liang et al., 2021). Protein concentration was quantified using the Easy II Protein Quantitative Kit (BCA; TransGen Biotech). Four‐week‐old Arabidopsis leaves were infiltrated with 30 μM PevD1 using 1‐ml syringes without a needle; 30 mM Tris‐HCl (pH 8.0) buffer was used as mock treatment. Plants were then inoculated with Pst DC3000 at 24 h after PevD1 treatment. The experiments were repeated three times.
4.5. RNA extraction and gene expression analysis
Total RNA was extracted from 4‐week‐old plants treated with protein or the pathogen using plant RNA kits (ER301; TransGen Biotech). The total RNA was reverse transcribed to complementary DNA (cDNA) using M‐MLV reverse transcriptase according to the manufacturer's instructions. qPCR analyses were performed as previously described (Zhang et al., 2019). The relative mRNA quantity was calculated from the average values using the ΔΔC t method (Schmittgen & Livak, 2008). UBC21 was used as the internal reference to normalize the expression value in each sample. The primers used for RT‐qPCR are shown in Table S1.
4.6. Measurement of ROS accumulation
To detect H2O2 accumulation in situ, NBT staining and DAB staining were used as described by Jambunathan (2010). Four‐week‐old leaves were treated with PevD1 or Pst DC3000 and then transferred to 1 mg/ml DAB or 0.5 mg/ml NBT solution and vacuum‐infiltrated at 37℃ for 30 min. Subsequently, the leaves were decolourized with 95% ethanol. H2O2 and superoxide anion accumulation was quantified by measuring the intensity with ImageJ.
4.7. Histochemical analysis of GUS activity
GUS staining was performed as described previously (Jefferson et al., 1987). Leaves inoculated with Pst DC3000 were incubated with GUS staining solution (100 mM Na3PO4 [pH 7.0], 0.5 mg/ml 5‐bromo‐4‐chloro‐3‐indolyl‐β‐d‐glucuronide, 1198 mM EDTA, 1 mM potassium ferrocyanide, 1 mM potassium ferricyanide, 1% Triton X‐100) overnight at 37℃ and then incubated in ethanol to eliminate chlorophyll before photographing.
4.8. RNA‐Seq and DEGS analysis
The third/fourth rosette leaves of 24‐day‐old Col‐0 and erf114#1 mutant plants that were cultivated at 22℃ under a 16 h light/8 h dark cycle were used for RNA‐Seq. Total RNA was extracted using the TRIzol reagent according to the manufacturer's protocol. In total, six samples, including erf114#1 mutant and Col‐0 with three biological replicates per genotype combination, were used for RNA‐Seq analysis. The transcriptome sequencing and analysis were conducted by OE Biotech Co., Ltd (Shanghai, China).
About 49.23 million raw reads for each sample were generated. Raw data (raw reads) of fastq format were first processed using Trimmomatic (Bolger et al., 2014) and the low‐quality reads were removed to obtain the clean reads. About 48.42 million clean reads for each sample were retained for subsequent analyses. Differential expression analysis was performed using the DESeq (2012) R package. p < 0.1 and fold change >1.5 were set as the threshold for significant differential expression. Hierarchical cluster analysis of DEGs was performed to demonstrate the expression pattern of genes in different groups and samples. GO enrichment and KEGG (Kanehisa et al., 2008) pathway enrichment analysis of DEGs were performed using R based on the hypergeometric distribution.
4.9. Protein expression and EMSA
The coding region of the ERF114 gene was cloned into vector pET‐32. Recombinant plasmid with glutathione S‐transferases (His) tag was transformed into Escherichia coli BL21 (DE3) and then induced with 0.2 mM isopropyl‐β‐d‐thiogalactoside (IPTG) at 20°C for 12 h. Cell pellets were collected and lysed by sonication in Tris‐HCl. His‐tagged protein was purified with His‐bind resin according to the manufacturer's instructions. Protein purification and quantification were performed using Ni‐NTA Resin (DP101; TransGen Biotech) and a BCA Protein Assay Kit (DQ111; TransGen Biotech), respectively. Next, 40‐bp probes within the indicated DNA fragment in the ERF114 promoter were labelled with biotin (Table S1). EMSA was conducted using a Light Shift Chemiluminescent Kit (Thermo Scientific).
4.10. ChIP‐qPCR
Two grams of 4‐week‐old 35S:GFP‐ERF114 leaves was used for ChIP experiments as previously described (Saleh et al., 2008). Anti‐GFP antibody was used to immunoprecipitate the protein–DNA complex. The enrichment of DNA fragments was determined by qPCR. All oligonucleotide sequences used are listed in Table S1.
4.11. Lignin analysis
Lignin assay was carried out according to the protocols described in the Solarbio kit. To measure the lignin content, the samples were dried at 80℃ to constant weight, crushed, and passed through a 40‐mesh sieve, weighing about 5 mg. Reagent Ⅰ 500 μl and perchloric acid 500 μl were successively added to the 1.5‐ml centrifuge tube, the sealing film sealed, and the solution thoroughly mixed. Acetylation was carried out in a water bath at 80℃ for 40 min. It was shaken every 10 min and then cooled naturally. Then 500 μl of reagent Ⅱ was added, and the solution thoroughly mixed and centrifuged for 10 min at 8000 × g at room temperature, and finally 980 μl of glacial acetic acid was added to the supernatant to determine the absorbance at 280 nm. Each data point had three replicates.
4.12. SA detection
Twenty‐eight‐day‐old leaves were harvested for SA extraction. Samples of 200 mg were weighed, then 1 ml methanol was added, followed by eddy mixing and ultrasonic extraction for 2 h. The mixture was centrifuged for 10 min at 8000 × g at room temperature, then the supernatant was taken for analysis. A Waters Acquity UPLC I‐CLASS instrument and Xevo TQ‐S Micro with an HSS T3 C18 column (1.7 μm, 100 × 2.1 mm) were used to separate and detect SA. The mobile phase included a gradient of water (0.1% formic acid) and hexane nitrile. The standard curve was made from SA at concentrations of 50, 100, 250, 500, and 1000 ng/ml, and was used to calculate the final concentration in each sample with Excel software. Each data point had three replicates.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ACCESSION NUMBERS
The genes mentioned in the study are as follows: ERF114 (AT5G61890), NbERF114 (Niben101Scf12210g08011), PAL1 (AT2G37040), PAL2 (AT3G53260), C4H (AT2G30490), FAR5 (AT3G44550), 4CL3 (AT1G65060), TT4 (AT5G13930), DFR (AT5G42800), TT7 (AT5G07990), PRR2 (AT4G13660), F3H (AT3G51240).
Supporting information
FIGURE S1 The log2 ratio (Treatment/Control) of NbERF114 expression at 0 h, 6 h, 12 h, 24 h and 36 h. Four‐week‐old Nicotiana benthamiana leaves of wild‐type were treated with 10 μM PevD1 as treatment, and the buffer (20 mM Tris‐HCl, pH 8.0) was used as mock treatment
FIGURE S2 Genome‐wide transcriptome analysis of erf114 mutant plants. (a) Volcano plots indicate the number of differentially expressed genes (DEGs): 199 down‐regulated genes or 462 up‐regulated genes in three biological replicates of erf114 mutant plant rosette leaves vs. Col‐0 with RNA‐seq. DEGs were identified with fold change ≥ 1.5, p ≤ 0.1. (b) Heat map showing DEGs in erf114 mutant plant rosette leaves compared to Arabidopsis thaliana Col‐0. Means of three experiments are shown. The log2 fold change scale is indicated on the right side of the heat map
FIGURE S3 GO and KEGG analysis of differentially expressed genes (DEGs) between wild type (WT) and erf114. (a) GO classification of differentially expressed genes (DEGs). The x axis indicates the GO groups, and the y axis indicates the p value of DEGs. (b) Top 20 of pathway enrichment (down). The x axis indicates the enrichment score corresponding to each pathway, and the y axis indicates name of the KEGG pathway. The colour of the point represents the p values of the enrichment analysis. The size and colour of bubbles represent the number and degree of enrichment of DEGs, respectively
FIGURE S4 RNA‐seq analysis and reverse transcription‐quantitative PCR (RT‐qPCR) analysis of transcripts of genes in phenylpropanoid and downstream pathways in 4‐week‐old wild‐type (Arabidopsis thaliana Col‐0) and erf114#1 leaves. The relative repression in RT‐qPCR analysis is represented in comparison to the UBC21 transcription level. The bars were calculated based on three independent experiments. The values are means ± SD (n = 3)
TABLE S1 Primers used in this study
ACKNOWLEDGEMENTS
This study was supported by the by the National Natural Science Foundation of China (grant no. 31772151). The funding body did not play any role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript. We would like to thank OE Biotech Co., Ltd (Shanghai, China, http://www.oebiotech.com/) for providing RNA‐Seq.
Li, Z. , Zhang, Y. , Ren, J. , Jia, F. , Zeng, H. , Li, G. & et al (2022) Ethylene‐responsive factor ERF114 mediates fungal pathogen effector PevD1‐induced disease resistance in Arabidopsis thaliana . Molecular Plant Pathology, 23, 819–831. 10.1111/mpp.13208
DATA AVAILABILITY STATEMENT
The raw data have been deposited in the Sequence Read Archive (SRA) ( https://www.ncbi.nlm.nih.gov/sra/) with the accession number PRJNA807385.
REFERENCES
- Bolger, A.M. , Lohse, M. & Usadel, B. (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics, 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu, B. , Qiu, D. , Zeng, H. , Guo, L. , Yuan, J. & Yang, X. (2014) A fungal protein elicitor PevD1 induces Verticillium wilt resistance in cotton. Plant Cell Reports, 33, 461–470. [DOI] [PubMed] [Google Scholar]
- Buer, C.S. , Imin, N. & Djordjevic, M.A. (2010) Flavonoids: new roles forold molecules. Journal of Integrative Plant Biology, 52, 98–111. [DOI] [PubMed] [Google Scholar]
- Chandran, D. , Rickert, J. , Huang, Y. , Steinwand, M.A. , Marr, S.K. & Wildermuth, M.C. (2014) A typical E2F transcriptional repressor DEL1 acts at the intersection of plant growth and immunity by controlling the hormone salicylic acid. Cell Host & Microbe, 15, 506–513. [DOI] [PubMed] [Google Scholar]
- Chen, L.U. , Wang, W.‐S. , Wang, T. , Meng, X.‐F. , Chen, T.‐T. , Huang, X.‐X. et al. (2019) Methyl salicylate glucosylation regulates plant defense signaling and systemic acquired resistance. Plant Physiology, 180, 2167–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Z. , Zheng, Z. , Huang, J. , Lai, Z. & Fan, B. (2009) Biosynthesis of salicylic acid in plants. Plant Signaling & Behavior, 4, 493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J. & Bent, A.F. (1998) Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana . The Plant Journal, 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Cui, H. , Gobbato, E. , Kracher, B. , Qiu, J. , Bautor, J. & Parker, J.E. (2017) A core function of EDS1 with PAD4 is to protect the salicylic acid defense sector in Arabidopsis immunity. New Phytologist, 213, 1802–1817. [DOI] [PubMed] [Google Scholar]
- Cutler, S.R. , Ehrhardt, D.W. , Griffitts, J.S. & Somerville, C.R. (2000) Random GFP∷cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proceedings of the National Academy of Sciences of the United States of America, 97, 3718–3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl, J.L. , Horvath, D.M. & Staskawicz, B.J. (2013) Pivoting the plant immune system from dissection to deployment. Science, 341, 746–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon, R.A. & Paiva, N.L. (1995) Stress‐induced phenylpropanoid metabolism. The Plant Cell, 7, 1085–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eklund, D.M. , Cierlik, I. , Staldal, V. , Claes, A.R. , Vestman, D. , Chandler, J. et al. (2011) Expression of Arabidopsis SHORT INTERNODES/STYLISH family genes in auxin biosynthesis zones of aerial organs is dependent on a GCC box‐like regulatory element. Plant Physiology, 157, 2069–2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer, J.L. , Austin, M.B. , Stewart, C. Jr. & Noel, J.P. (2008) Structure and function of enzymes involved in the biosynthesis of phenylpropanoids. Plant Physiology and Biochemistry, 46, 356–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, Y. , Li, Z. , Yang, C. , Li, G. , Zeng, H. , Li, Z. et al. (2022) Pseudomonas syringae activates ZAT18 to inhibit salicylic acid accumulation by repressing EDS1 transcription for bacterial infection. New Phytologist, 233, 1274–1288. [DOI] [PubMed] [Google Scholar]
- Greenberg, J.T. & Yao, N. (2004) The role and regulation of programmed cell death in plant–pathogen interactions. Cellular Microbiology, 6, 201–211. [DOI] [PubMed] [Google Scholar]
- He, J. , Liu, Y. , Yuan, D. , Duan, M. , Liu, Y. , Shen, Z. et al. (2020) An R2R3 MYB transcription factor confers brown plant hopper resistance by regulating the phenylalanine ammonia‐lyase pathway in rice. Proceedings of the National Academy of Sciences of the United States of America, 117, 271–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, J. , Gu, M. , Lai, Z. , Fan, B. , Shi, K. , Zhou, Y.H. et al. (2010) Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiology, 153, 1526–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambunathan, N. (2010) Determination and detection of reactive oxygen species (ROS), lipid peroxidation, and electrolyte leakage in plants. Methods in Molecular Biology, 639, 292–298. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A. , Kavanagh, T.A. & Bevan, M.W. (1987) GUS fusions: β‐glucuronidase as a sensitive and versatile gene fusion marker in higher plants. The EMBO Journal, 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.D.G. & Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kanehisa, M. , Araki, M. , Goto, S. , Hattori, M. , Hirakawa, M. , Itoh, M. et al. (2008) KEGG for linking genomes to life and the environment. Nucleic Acids Research, 36, D480–D484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, X. , Tian, H. , Yu, Q. , Zhang, F. , Wang, R. , Gao, S. et al. (2018) PHB3 maintains root stem cell niche identity through ROS‐responsive AP2/ERF transcription factors in Arabidopsis. Cell Reports, 22, 1350–1363. [DOI] [PubMed] [Google Scholar]
- Konig, S. , Feussner, K. , Kaever, A. , Landesfeind, M. , Thurow, C. , Karlovsky, P. et al. (2014) Soluble phenylpropanoids are involved in the defense response of Arabidopsis against Verticillium longisporum . New Phytologist, 202, 823–837. [DOI] [PubMed] [Google Scholar]
- Lawton, M.A. , Dixon, R.A. , Hahlbrock, K. & Lamb, C. (1983) Rapid induction of the synthesis of phenylalanine ammonia‐lyase and of chalcone synthase in elicitor‐treated plant cells. European Journal of Biochemistry, 129, 593–601. [DOI] [PubMed] [Google Scholar]
- Lefevere, H. , Bauters, L. & Gheysen, G. (2020) Salicylic acid biosynthesis in plants. Frontiers in Plant Science, 11, 338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Yu, T. , Wu, T. , Wang, R. , Wang, H. , Du, H.U. et al. (2020) The dynamic transcriptome of pepper (Capsicum annuum) whole roots reveals an important role for the phenylpropanoid biosynthesis pathway in root resistance to Phytophthora capsici . Gene, 728, 144288. [DOI] [PubMed] [Google Scholar]
- Liang, X. , Dron, M. , Cramer, C.L. , Dixon, R.A. & Lamb, C.J. (1989) Differential regulation of phenylalanine ammonia‐lyase genes during plant development and by environmental cues. Journal of Biological Chemistry, 264, 14486–14492. [PubMed] [Google Scholar]
- Liang, Y. , Cui, S. , Tang, X. , Zhang, Y.I. , Qiu, D. , Zeng, H. et al. (2018) An asparagine‐rich protein Nbnrp1 modulate Verticillium dahliae protein PevD1‐induced cell death and disease resistance in Nicotiana benthamiana . Frontiers in Plant Science, 9, 303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, Y. , Li, Z.E. , Zhang, Y.I. , Meng, F. , Qiu, D. , Zeng, H. et al. (2021) Nbnrp1 mediates Verticillium dahliae effector PevD1‐triggered defense responses by regulating sesquiterpenoid phytoalexins biosynthesis pathway in Nicotiana benthamiana . Gene, 768, 145280. [DOI] [PubMed] [Google Scholar]
- Licausi, F. , Ohme‐Takagi, M. & Perata, P. (2013) APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: mediators of stress responses and developmental programs. New Phytologist, 199, 639–649. [DOI] [PubMed] [Google Scholar]
- Liu, H. , Liu, Z. , Wu, Y. , Zheng, L. & Zhang, G. (2021) Regulatory mechanisms of anthocyanin biosynthesis in apple and pear. International Journal of Molecular Sciences, 22, 8441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Q. , Luo, L. & Zheng, L. (2018) Lignins: biosynthesis and biological functions in plants. International Journal of Molecular Sciences, 19(2), 335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, Y. , Chen, Q. , Bu, Y. , Luo, R. , Hao, S. , Zhang, J. et al. (2017) Flavonoid accumulation plays an important role in the rust resistance of Malus plant leaves. Frontiers in Plant Science, 8, 1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy, J. , Carr, J.P. , Klessig, D.F. & Raskin, I. (1990) Salicylic acid: a likely endogenous signal in the resistance response of tobacco to viral infection. Science, 250, 1002–1004. [DOI] [PubMed] [Google Scholar]
- Mehrnia, M. , Balazadeh, S. , Zanor, M. & Mueller‐Roeber, B. (2013) EBE, an AP2/ERF transcription factor highly expressed in proliferating cells, affects shoot architecture in Arabidopsis. Plant Physiology, 162, 842–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat, C.S. , Ingle, R.A. , Wathugala, D.L. , Saunders, N.J. , Knight, H. & Knight, M.R. (2012) ERF5 and ERF6 play redundant roles as positive regulators of JA/Et‐mediated defense against Botrytis cinerea in Arabidopsis. PLoS One, 7, e35995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano, T. , Suzuki, K. , Fujimura, T. & Shinshi, H. (2006) Genome‐wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiology, 140, 411–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohme‐Takagi, M. & Shinshi, H. (1995) Ethylene‐inducible DNA binding proteins that interact with an ethylene‐responsive element. The Plant Cell, 7, 173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh, A. , Alvarez‐Venegas, R. & Avramova, Z. (2008) An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nature Protocols, 3, 1018–1025. [DOI] [PubMed] [Google Scholar]
- Schmittgen, T.D. & Livak, K.J. (2008) Analyzing real‐time PCR data by the comparative C T method. Nature Protocols, 3, 1101–1108. [DOI] [PubMed] [Google Scholar]
- Schwessinger, B. & Ronald, P.C. (2012) Plant innate immunity: perception of conserved microbial signatures. Annual Review of Plant Biology, 63, 451–482. [DOI] [PubMed] [Google Scholar]
- Sonbol, F.‐M. , Fornalé, S. , Capellades, M. , Encina, A. , Touriño, S. , Torres, J.‐L. et al. (2009) The maize ZmMYB42 represses the phenylpropanoid pathway and affects the cell wall structure, composition and degradability in Arabidopsis thaliana . Plant Molecular Biology, 70, 283–296. [DOI] [PubMed] [Google Scholar]
- Song, N.A. , Ma, L. , Wang, W. , Sun, H. , Wang, L. , Baldwin, I.T. et al. (2019) An ERF2‐like transcription factor regulates production of the defense sesquiterpene capsidiol upon Alternaria alternata infection. Journal of Experimental Botany, 70, 5895–5908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme, R. , De Meester, B. , Ralph, J. & Boerjan, W. (2019) Lignin biosynthesis and its integration into metabolism. Current Opinion in Biotechnology, 56, 230–239. [DOI] [PubMed] [Google Scholar]
- Vlot, A.C. , Dempsey, D.A. & Klessig, D.F. (2009) Salicylic acid, a multifaceted hormone to combat disease. Annual Review of Phytopathology, 47, 177–206. [DOI] [PubMed] [Google Scholar]
- Wang, B. , Yang, X. , Zeng, H. , Liu, H. , Zhou, T. , Tan, B. et al. (2012) The purification and characterization of a novel hypersensitive‐like response‐inducing elicitor from Verticillium dahliae that induces resistance responses in tobacco. Applied Microbiology and Biotechnology, 93, 191–201. [DOI] [PubMed] [Google Scholar]
- Wang, X. , Wu, J. , Guan, M. , Zhao, C. , Geng, P. & Qx, Z. (2020) Arabidopsis MYB4 plays dual roles in flavonoid biosynthesis. The Plant Journal, 101, 637–652. [DOI] [PubMed] [Google Scholar]
- Yadav, V. , Wang, Z. , Wei, C. , Amo, A. , Ahmed, B. , Yang, X. et al. (2020) Phenylpropanoid pathway engineering: an emerging approach towards plant defense. Pathogens, 9, 312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalpani, N. , Silverman, P. , Wilson, T.M. , Kleier, D.A. & Raskin, I. (1991) Salicylic acid is a systemic signal and an inducer of pathogenesis‐related proteins in virus‐infected tobacco. The Plant Cell, 3, 809–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi‐Shinozaki, K. & Shinozaki, K. (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annual Review of Plant Biology, 57, 781–803. [DOI] [PubMed] [Google Scholar]
- Zabala, G. , Zou, J. , Tuteja, J. , Gonzalez, D.O. , Clough, S.J. & Vodkin, L.O. (2006) Transcriptome changes in the phenylpropanoid pathway of Glycine max in response to Pseudomonas syringae infection. BMC Plant Biology, 6, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, P. , Du, H. , Wang, J. , Pu, Y. , Yang, C. , Yan, R. et al. (2020) Multiplex CRISPR/Cas9‐mediated metabolic engineering increases soya bean isoflavone content and resistance to soya bean mosaic virus. Plant Biotechnology Journal, 18, 1384–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Gao, Y. , Liang, Y. , Dong, Y. , Yang, X. & Qiu, D. (2019) Verticillium dahliae PevD1, an Alt a 1‐like protein, targets cotton PR5‐like protein and promotes fungal infection. Journal of Experimental Botany, 70, 613–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, X. , Xing, J. , Zhang, K. , Pang, X. , Zhao, Y. , Wang, G. et al. (2019) Ethylene Response Factor ERF11 activates BT4 transcription to regulate immunity to Pseudomonas syringae . Plant Physiology, 180, 1132–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Z. , Schenke, D. , Miao, Y. & Cai, D. (2017) Investigation of the crosstalk between the flg22 and the UV‐B‐induced flavonol pathway in Arabidopsis thaliana seedlings. Plant, Cell and Environment, 40, 453–458. [DOI] [PubMed] [Google Scholar]
- Zhu, J. , Li, Y. & Liao, J. (2013) Involvement of anthocyanins in the resistance to chilling‐induced oxidative stress in Saccharum officinarum leaves. Plant Physiology and Biochemistry, 73, 427–433. [DOI] [PubMed] [Google Scholar]
- Zipfel, C. , Robatzek, S. , Navarro, L. , Oakeley, E.J. , Jones, J.D.G. , Felix, G. et al. (2004) Bacterial disease resistance in Arabidopsis through flagellin perception. Nature, 428, 764–767. [DOI] [PubMed] [Google Scholar]
- Zuo, J. , Niu, Q. & Chua, N. (2000) Technical advance: an estrogen receptor‐based transactivator XVE mediates highly inducible gene expression in transgenic plants. The Plant Journal, 24, 265–273. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1 The log2 ratio (Treatment/Control) of NbERF114 expression at 0 h, 6 h, 12 h, 24 h and 36 h. Four‐week‐old Nicotiana benthamiana leaves of wild‐type were treated with 10 μM PevD1 as treatment, and the buffer (20 mM Tris‐HCl, pH 8.0) was used as mock treatment
FIGURE S2 Genome‐wide transcriptome analysis of erf114 mutant plants. (a) Volcano plots indicate the number of differentially expressed genes (DEGs): 199 down‐regulated genes or 462 up‐regulated genes in three biological replicates of erf114 mutant plant rosette leaves vs. Col‐0 with RNA‐seq. DEGs were identified with fold change ≥ 1.5, p ≤ 0.1. (b) Heat map showing DEGs in erf114 mutant plant rosette leaves compared to Arabidopsis thaliana Col‐0. Means of three experiments are shown. The log2 fold change scale is indicated on the right side of the heat map
FIGURE S3 GO and KEGG analysis of differentially expressed genes (DEGs) between wild type (WT) and erf114. (a) GO classification of differentially expressed genes (DEGs). The x axis indicates the GO groups, and the y axis indicates the p value of DEGs. (b) Top 20 of pathway enrichment (down). The x axis indicates the enrichment score corresponding to each pathway, and the y axis indicates name of the KEGG pathway. The colour of the point represents the p values of the enrichment analysis. The size and colour of bubbles represent the number and degree of enrichment of DEGs, respectively
FIGURE S4 RNA‐seq analysis and reverse transcription‐quantitative PCR (RT‐qPCR) analysis of transcripts of genes in phenylpropanoid and downstream pathways in 4‐week‐old wild‐type (Arabidopsis thaliana Col‐0) and erf114#1 leaves. The relative repression in RT‐qPCR analysis is represented in comparison to the UBC21 transcription level. The bars were calculated based on three independent experiments. The values are means ± SD (n = 3)
TABLE S1 Primers used in this study
Data Availability Statement
The raw data have been deposited in the Sequence Read Archive (SRA) ( https://www.ncbi.nlm.nih.gov/sra/) with the accession number PRJNA807385.


