Abstract
Background
CIRCADIAN CLOCK ASSOCIATED 1 like (CCA1L) proteins are important components that participate in plant growth and development, and now have been characterized in multiple plant species. However, information on mungbean CCA1L genes is limited.
Results
In this study, we identified 27 VrCCA1L genes from the mungbean genome. VrCCA1L genes were unevenly distributed on 10 of the 11 chromosomes and showed one tandem and two interchromosomal duplication events. Two distinct kinds of conserved MYB domains, MYB 1 and MYB 2, were found, and the conserved SHAQK(Y/F) F sequence was found at the C terminus of each MYB 2 domain. The VrCCA1Ls displayed a variety of exon-intron organizations, and 24 distinct motifs were found among these genes. Based on phylogenetic analysis, VrCCA1L proteins were classified into five groups; group I contained the most members, with 11 VrCCA1Ls. VrCCA1L promoters contained different types and numbers of cis-acting elements, and VrCCA1Ls showed different expression levels in different tissues. The VrCCA1Ls also displayed distinct expression patterns under different photoperiod conditions throughout the day in leaves. VrCCA1L26 shared greatest homology to Arabidopsis CCA1 and LATE ELONGATED HYPOCOTYL (LHY). It delayed the flowering time in Arabidopsis by affecting the expression levels of CONSTANS (CO), FLOWERING LOCUS T (FT), and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1).
Conclusion
We identified and characterized 27 VrCCA1L genes from mungbean genome, and investigated their spatio-temporal expression patterns. Further analysis revealed that VrCCA1L26 delayed flowering time in transgenic Arabidopsis plants. Our results provide useful information for further functional characterization of the VrCCA1L genes.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12864-022-08620-7.
Keywords: Mungbean, Flowering time, CCA1, VrCCA1L26, Photoperiod
Background
Mungbean [Vigna radiata (L.) Wilczek] is an important legume crop grown mainly in Asian countries and consumed as a common food worldwide. Mungbean seeds contain many kinds of proteins and nutrients and are used to alleviate heat shock and reduce swelling in summer [1–3]. Because of the breakdown of proteins, vitamins, and minerals, the nutritive value of mungbean seeds increases after germination [4]. Mungbean sprouts have high nutritive value and are a common vegetable food in many countries [5]. However, the production of mungbean seeds and sprouts is affected by many endogenous and environmental factors, and the investigation of functional genes based on genomic information will provide essential genetic resources for modifying mungbean plants to obtain high yield [6].
The characterization of functional genes in mungbean is limited. However, many functional genes have been identified in the past decades in multiple plant species, which provides important information for mungbean gene function analysis. Among these functional genes, transcription factors such as B-box, MADS-box, heat shock transcription factor (Hsf), and MYELOBLASTOSIS ONCOGENE (MYB) family members are important components that regulate plant growth [7–15]. Transcription factors bind to the cis-acting elements of their target gene promoters to regulate their expression. The MYB transcription factor family has the greatest number of members among the Arabidopsis gene families [16]. MYBs contain a DNA binding domain and an activation domain, and are characterized by MYB DNA binding domains at the N terminus [11]. The MYB proteins can be further grouped into five groups based on their gene structures: CIRCADIAN CLOCK ASSOCIATED 1 (CCA1)-like, CAPRICE-like, telomeric DNA binding proteins-like, I-box binding factor-like, and R-R-type proteins [7, 17]. The CCA1-like (CCA1L) proteins, which are identified based on the SHAQK(Y/F) F consensus sequence in the MYB domain, constitute the major subfamily of MYB proteins. Among these MYB proteins, CCA1L proteins have been identified to exert important functions in the control of circadian clock and flowering time, and have been identified in multiple plant species, such as Arabidopsis, soybean, and peach [17–20].
Among these CCA1L proteins, CCA1 and its close homologous gene LATE ELONGATED HYPOCOTYL (LHY) have been well studied [21–24]. In Arabidopsis, the expression of CCA1 or LHY shows a diurnal rhythm under light/dark cycle conditions and constant light or dark conditions. Overexpression of CCA1 or LHY delays flowering time by regulating FLOWERING LOCUS T (FT) [25, 26]. CCA1 and LHY suppresses the expression of a central circadian clock gene, TIMING OF CAB EXPRESSION 1 (TOC1), by binding to the evening element (AAATATCT) in the promoter region [27]. Mutation of CCA1 or LHY, disrupts circadian rhythms such as leaf movement and hypocotyl elongation [28]. Moreover, TOC1 in turn represses the expression of CCA1 and LHY from its induction at dusk until slightly before dawn [29]. Loss of function of the soybean CCA1 and LHY homologs GmLCLa1, GmLCLa2, GmLCLb1, and GmLCLb2 results in a short-period circadian rhythm and a late flowering phenotype [30]. In addition, some other CCA1L genes have been found to be involved in isoflavonoid biosynthesis, leaf senescence, seed germination, and stress response [31–35].
Although CCA1L genes have important roles in plant growth and development, and have been studied in many plant species [17–19], the characterization of mungbean VrCCA1L genes is limited. Genome-wide identification of VrCCA1L genes based on mungbean genomic information will provide essential information for understanding the circadian clock and flowering time regulation in mungbean [6]. In this study, we identified and characterized 27 mungbean VrCCA1L family members based on the conserved MYB and SHAQK(Y/F) F domains. We characterized many aspects and expression profiles of the VrCCA1Ls. In addition, we also investigated the function of VrCCA1L26 in flowering time regulation. Our study provides essential information for further functional characterization of mungbean VrCCA1L genes.
Methods
Plant growth conditions
The draft genome of mungbean variety VC1973A provided by Suk-Ha Lee at Seoul National University, Seoul, South Korea, was used in this study [6]. To collect different tissues, VC1973A plants were grown in the field in Qingdao, China. Mungbean seeds were planted in the field at the end of May, and different tissues were collected when mungbean plants had produced full-length pods. Eight tissues (roots, nodule roots, shoot apices, stems, leaves, flowers, pods, and seeds) were sampled and immediately frozen in liquid nitrogen in the late afternoon (approximately 10–12 hours after dawn, approximate photoperiod conditions: 15 h light/9 h dark) in early July 2018 [36, 37]. For different photoperiod treatments, mungbean seeds were germinated in water for 1 day and then grown in soil in the growth chamber for 5 weeks. The growth conditions were set as follows: 16 h 24 °C light/8 h 24 °C dark cycles for long-day (LD) conditions, and 10 h 24 °C light/14 h 24 °C dark cycles for short-day (SD) conditions [36]. The humidity of the growth chamber was set at approximately 30%. Leaves of the mungbean plants were sampled every 4 h from lights-on at six time points under both LD and SD conditions. Arabidopsis plants were grown in the growth chamber under 16 h 23 °C light/8 h 21 °C dark cycle conditions. The shoots of 2-week-old Arabidopsis plants grown on MS agar medium were sampled every 4 h from lights-on at six time points throughout the day for gene expression analysis.
Identification of mungbean VrCCA1L genes
The amino acid sequences of 20 Arabidopsis and 54 soybean CCA1L proteins were used as blast queries against the mungbean genome databases in Seoul National University (http://plantgenomics.snu.ac.kr/mediawiki-1.21.3/index.php/Main_Page), and National Center for Biotechnology Information (NCBI) to search for candidate genes. The presence of conserved MYB domains was verified using the Pfam database and the InterPro program [38, 39]. The presence of the SHAQK(Y/F) F motif within the MYB domain was also confirmed using blast. Predicted proteins that contained both conserved MYB and SHAQK(Y/F) F domains were designated as VrCCA1L proteins. The ProtParam program (https://web.expasy.org/protparam/) was used to predict the molecular weight (Mol. Wt) and theoretical iso-electric point (pI) of each VrCCA1L protein.
Chromosomal locations and gene structure analyses
The physical positions of the VrCCA1L genes on each chromosome were obtained from NCBI and used to construct the chromosomal location map using MapInspect software (Mike Lischke, Berlin, Germany). The genomic and coding sequences (CDS) of the VrCCA1L genes were obtained from NCBI, and the Gene Structure Display Server program was used to analyze their gene structures [40].
Analyses of conserved domains, conserved motifs, and sequence logos
The physical positions of the MYB domains in the VrCCA1L amino acid sequences were identified using the Pfam database and the InterPro program [38, 39]. The sequences of the conserved MYB domains were isolated from each VrCCA1L protein and used to create a sequence logo with the WebLogo platform [41]. The conserved motifs in each VrCCA1L protein were analyzed using MEME tools with default parameters [42].
Gene duplication analysis
The OrthoMCL software was used to identify the duplicated VrCCA1L gene pairs as described [43, 44]. Specifically, the amino acid sequences of the VrCCA1L proteins were aligned with one another, and VrCCA1L proteins with sequence similarities greater than 70% were considered to be encoded by duplicated gene pairs.
Analysis of cis-acting elements in the VrCCA1L promoter regions
VrCCA1L promoter regions 2 kb upstream of the initiation codon were obtained from NCBI and their cis-acting elements were analyzed using PlantCARE [45]. Cis-acting elements were clustered into six different types based on their potential functions as described by Hou et al. [46].
Phylogenetic relationship analysis
The amino acid sequences of VrCAA1L, GmCAA1L, and AtCAA1L proteins were aligned using ClustalW2 [17, 19, 47]. The alignment result was used to construct a phylogenetic tree using MEGA 7.0, and the neighbor-joining method and default parameters were used for analysis [48]. Another phylogenetic tree was constructed with MEGA 7.0 using only VrCCA1L proteins to analyze their evolutionary relationships.
Gene expression analysis
Total RNA was isolated from Arabidopsis shoots or mungbean leaves using the RNeasy mini kit (Qiagen) according to the manufacturer’s instructions. For each sample, 1 μg RNA was used for cDNA synthesis with SuperScript II reverse transcriptase (Promega). Quantitative real-time PCR (qRT–PCR) was performed as described by Li et al. [49]. Gene expression was normalized to a mungbean Actin gene (Vradi03g00210) or an Arabidopsis Actin (At3g18780) gene [50]. Three technical replicates and three biological replicates were used for each treatment to analyze gene expression. All primers used in this study are listed in Additional file 1.
Plasmid construction and Arabidopsis transformation
To make a 35S:VrCCA1L26 construct, the CDS of VrCCA1L26 was amplified from the cDNA of VC1973A and then ligated to a modified pCAMBIA1300 vector using T4 DNA ligase (Promega) as described by Li et al. [51]. The constructed plasmid was transformed into Arabidopsis using the floral dip method [52]. The VrCCA1L26 transgenic Arabidopsis plants were checked using PCR with specific primers, and the PCR products were sent for sequencing to confirm the sequence of the VrCCA1L26 fragment. All primers used in this study are listed in Additional file 1.
Results
Identification of VrCCA1L genes in the mungbean genome
To identify mungbean VrCCA1L genes, we used the amino acid sequences of CCA1L proteins from Arabidopsis and soybean as blast queries against mungbean genome. Candidate genes that lacked conserved MYB and SHAQK(Y/F) F domains were discarded, and 27 VrCCA1L genes were confirmed in mungbean (Table 1). The VrCCA1L genes differed in genomic length, CDS length, and amino acid number (Table 1). The genomic length ranged from 906 (XP_014517291 and XP_014517292) to 15,635 bp (XP_014499065), and the CDS length ranged from 594 (XP_014499363) to 2253 bp (XP_014521593). As a result, the amino acid number of the VrCCA1L proteins varied from 197 to 750. The molecular weight of VrCCA1Ls ranged from 21,774.66 (XP_014499363) to 82,272.45 Da (XP_014521593).
Table 1.
VrCCA1L genes identified in mungbean genome
| Gene ID | Genomic Length (bp) | CDS length (bp) | No. of AA | Mol. Wt (Da) | pI | Chr | Gene names |
|---|---|---|---|---|---|---|---|
| XP_014511774 | 4574 | 1434 | 477 | 52,344.16 | 5.32 | 1 | VrCCA1L1 |
| XP_014503116 | 7975 | 924 | 307 | 34,010.7 | 9.33 | 1 | VrCCA1L2 |
| XP_014495340 | 3123 | 843 | 280 | 31,439.83 | 9.28 | 3 | VrCCA1L3 |
| XP_014497263 | 6384 | 900 | 299 | 34,056.66 | 9.66 | 4 | VrCCA1L4 |
| XP_014498253 | 2975 | 909 | 302 | 32,547.43 | 8.94 | 4 | VrCCA1L5 |
| XP_014499065 | 15,635 | 2064 | 687 | 75,656.9 | 6.06 | 5 | VrCCA1L6 |
| XP_014499363 | 2012 | 594 | 197 | 21,774.66 | 9.7 | 5 | VrCCA1L7 |
| XP_014502167 | 4280 | 1062 | 353 | 38,457.15 | 8.95 | 5 | VrCCA1L8 |
| XP_014502591 | 3476 | 1263 | 420 | 46,786.58 | 8.37 | 6 | VrCCA1L9 |
| XP_014505359 | 2349 | 915 | 304 | 34,265.21 | 8.73 | 6 | VrCCA1L10 |
| XP_014508244 | 1148 | 642 | 213 | 23,941.21 | 10 | 7 | VrCCA1L11 |
| XP_014510415 | 1999 | 888 | 295 | 32,333.38 | 8.75 | 7 | VrCCA1L12 |
| XP_014507110 | 1599 | 888 | 295 | 32,234.85 | 6.98 | 7 | VrCCA1L13 |
| XP_014507579 | 1297 | 708 | 235 | 26,211.71 | 9.51 | 7 | VrCCA1L14 |
| XP_014512388 | 3584 | 918 | 305 | 33,977.52 | 9.04 | 8 | VrCCA1L15 |
| XP_014512566 | 2030 | 726 | 241 | 27,351.48 | 6.14 | 8 | VrCCA1L16 |
| XP_014516756 | 6136 | 1005 | 334 | 37,152.27 | 9.04 | 9 | VrCCA1L17 |
| XP_014517291 | 906 | 768 | 255 | 30,185.95 | 6.3 | 10 | VrCCA1L18 |
| XP_014517292 | 906 | 768 | 255 | 30,167.15 | 6.22 | 10 | VrCCA1L19 |
| XP_014518898 | 2789 | 963 | 320 | 36,181.58 | 9.07 | 10 | VrCCA1L20 |
| XP_014517128 | 3104 | 1038 | 345 | 38,087.74 | 8.06 | 10 | VrCCA1L21 |
| XP_014520829 | 9559 | 882 | 293 | 31,226.46 | 6.37 | 11 | VrCCA1L22 |
| XP_014490537 | 2021 | 912 | 303 | 32,472.26 | 9.67 | N/A | VrCCA1L23 |
| XP_014491847 | 1416 | 600 | 199 | 22,257.63 | 6.09 | N/A | VrCCA1L24 |
| XP_014491921 | 5543 | 891 | 296 | 32,540.82 | 9.01 | N/A | VrCCA1L25 |
| XP_014521593 | 9651 | 2253 | 750 | 82,272.45 | 6.04 | N/A | VrCCA1L26 |
| XP_014523608 | 9308 | 885 | 294 | 31,231.63 | 6.8 | N/A | VrCCA1L27 |
Chr Chromosome number, AA Amino acid, Mol. Wt Molecular weight, pI Isoelectric point
Chromosomal location and duplication analyses of VrCCA1L genes
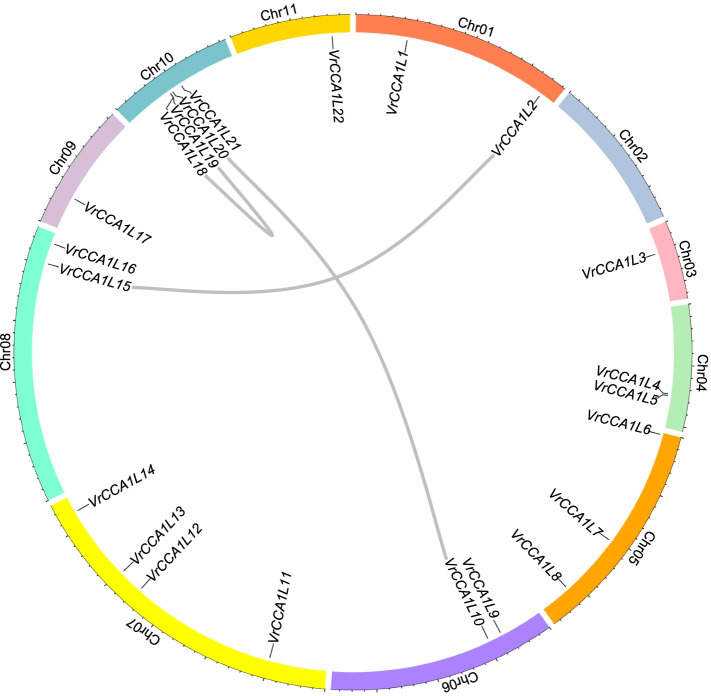
Mungbean genome experienced one round of whole genome duplication during evolution and produced multiple duplicated gene pairs [6, 12]. The investigation of VrCCA1L chromosomal locations can provide insight into gene distributions after duplication, and a chromosomal distribution map of VrCCA1L genes was therefore constructed based on their physical positions (Additional file 2, Table 1). XP_014490537, XP_014491847, XP_014491921, XP_014521593, and XP_014523608 were not included in Additional file 2 due to a lack of positional information. The VrCCA1L genes were designated VrCCA1L1 to VrCCA1L22 based on their chromosome positions (Table 1). The five VrCCA1L genes that lacked chromosomal position information were randomly named VrCCA1L23 to VrCCA1L27 (Table 1). Ten of the eleven mungbean chromosomes contained VrCCA1L genes, with the exception of chromosome 2. Chromosomes 7 and 10 contained the most VrCCA1L members, with 4 VrCCA1L genes on each, followed by chromosome 5, with 3 VrCCA1L genes (Additional file 2). VrCCA1L18 and VrCCA1L19 were located close together on chromosome 10.
We analyzed the amino acid sequence similarities of the VrCCA1L proteins and found five duplicated gene pairs: VrCCA1L2/VrCCA1L15, VrCCA1L3/VrCCA1L25, VrCCA1L10/VrCCA1L20, VrCCA1L18/VrCCA1L19, and VrCCA1L22/VrCCA1L27. Three duplication events are shown in Fig. 1; VrCCA1L3/VrCCA1L25 and VrCCA1L22/VrCCA1L27 were not included due to a lack of positional information. The duplicated genes were located on chromosomes 1, 6, 8, and 10, and chromosome 10 contained the largest number of duplicated genes (VrCCA1L18, VrCCA1L19, and VrCCA1L20). VrCCA1L2/VrCCA1L15 formed an interchromosomal duplicated gene pair, as did VrCCA1L10/VrCCA1L20, whereas VrCCA1L18/VrCCA1L19 appeared to have arisen from a tandem duplication event (Fig. 1).
Fig. 1.
Duplication analysis of VrCCA1L genes. Different chromosomes are represented using different colored lines, and duplicated gene pairs are connected by grey lines. Chr indicate chromosomes
Phylogenetic relationships among the VrCCA1L genes
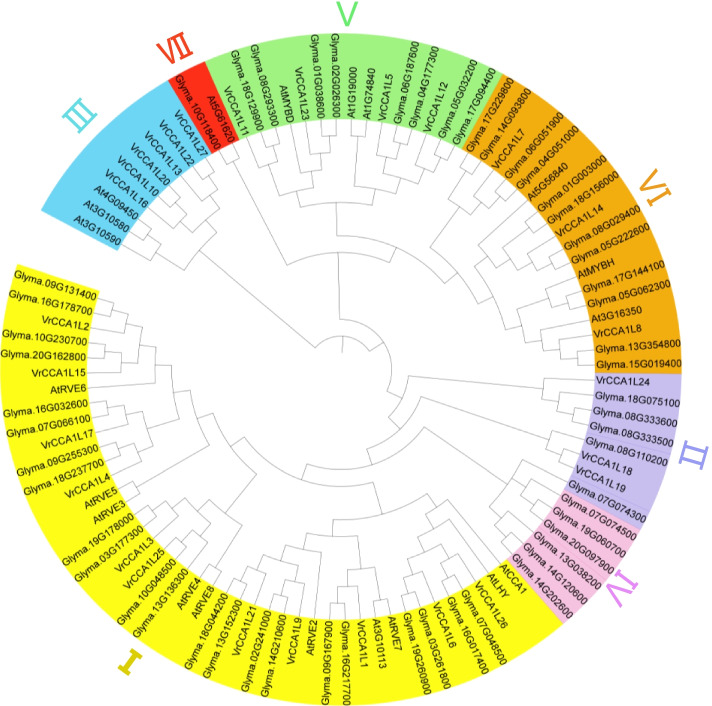
To investigate the evolutionary relationships among the VrCCA1L genes and their homology to well-studied CCA1L genes from other plant species, we constructed a phylogenetic tree using 101 CCA1L proteins from Arabidopsis, soybean, and mungbean (Fig. 2). The CCA1L genes were classified into seven groups as described in soybean [19]. Group I was the largest subfamily and contained 11 VrCCA1L members, whereas groups IV and VII had no VrCCA1L members (Fig. 2). Groups II, III, V and VI contained 3, 6, 4 and 3 VrCCA1L members, respectively (Fig. 2). Among the VrCCA1L members, VrCCA1L6 and VrCCA1L26 showed close relationships with the well-studied Arabidopsis circadian clock and flowering time genes CCA1 and LHY, as well as the soybean genes GmLCLa1 (Glyma16g01980), GmLCLa2 (Glyma07g05410), GmLCLb1 (Glyma03g42260), and GmLCLb2 (Glyma19g45030) [30]. These results suggest that VrCCA1L6 and VrCCA1L26 may be key factors involved in circadian clock and flowering time regulation in mungbean. We also constructed a phylogenetic tree using only VrCCA1L genes and found that all genes from the same group were clustered together (Fig. 3a). Moreover, the genes from each pair of duplicated genes were placed into the same clades in the phylogenetic tree (Fig. 3a).
Fig. 2.
Evolutionary relationship analysis of the VrCCA1L proteins. The amino acid sequences of CCA1L proteins from Arabidopsis, soybean, and mungbean were used to construct a phylogenetic tree with the neighbor-joining method. Groups I to VII are presented in different colored bars. Glyma16g01980, Glyma07g05410, Glyma03g42260, and Glyma19g45030 indicate GmLCLa1, GmLCLa2, GmLCLb1, and GmLCLb2, respectively
Fig. 3.
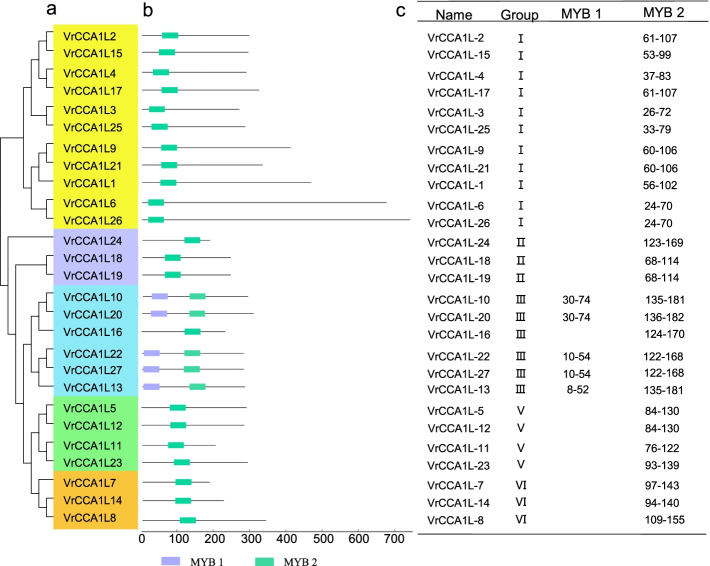
Phylogenetic relationships and conserved domains of the VrCCA1L proteins. a Phylogenetic analysis of the VrCCA1L proteins. Groups I to VII of VrCCA1L proteins are presented in different colored bars as described in (b) The positions of conserved MYB 1 and MYB 2 domains in each VrCCA1L protein. The purple and green boxes indicate MYB 1 and MYB 2 domains, respectively. c Classification of the VrCCA1L proteins and the positions of each MYB domain in the VrCCA1L proteins. Groups I to VII of VrCCA1L proteins, and conserved MYB 1 and MYB 2 domains are presented
Analysis of VrCCA1L conserved domains
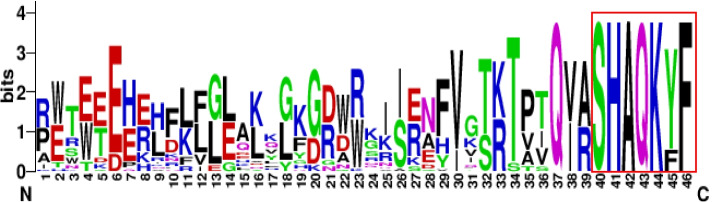
The CCA1L protein is a member of the MYB gene family and contains MYB domains [7, 17]. Different CCA1L proteins contain different numbers and types of conserved MYB domains [17–20], and we therefore analyzed the MYB domain numbers and types in the VrCCA1L proteins. Two distinct kinds of conserved MYB domain, MYB 1 and MYB 2, were found (Fig. 3b, Additional file 3). All members of groups I, II, V, and VI contained only one MYB 2 domain. By contrast, most group III members contained one MYB 1 and one MYB 2 domain, with the exception of VrCCA1L16, which had only one MYB 2 domain (Fig. 3b, c). All the MYB 1 domains were found close to the N termini of the VrCCA1L proteins (Fig. 3b, c). In addition, the conserved SHAQK(Y/F) F sequence of the VrCCA1L proteins was found at the C termini of the MYB 2 domains (Fig. 4, Additional file 3).
Fig. 4.
Sequence logo of the conserved MYB 2 domain. The red box at the C terminus indicates the conserved SHAQK(Y/F) F domain
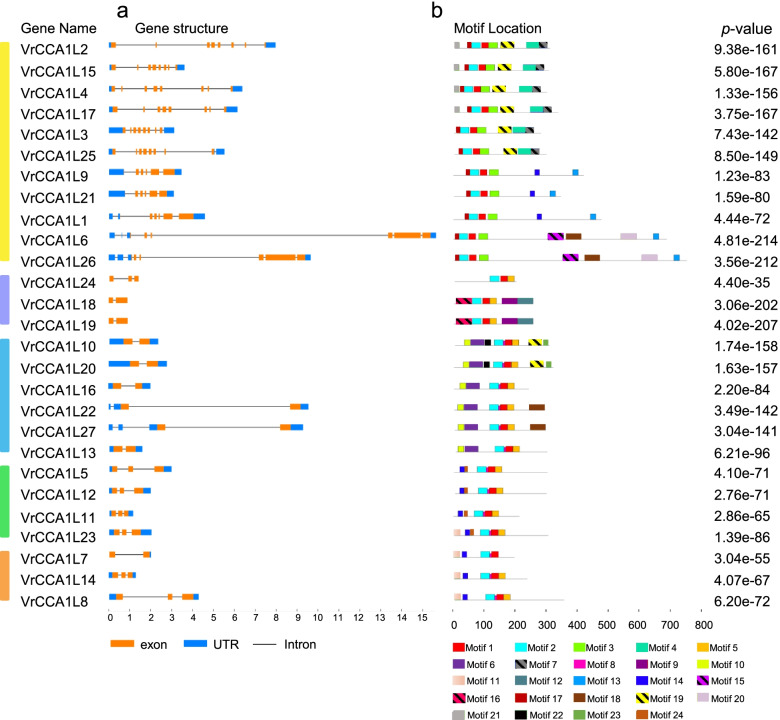
Exon-intron organization and conserved motif analyses of the VrCCA1L genes
To investigate the similarity and diversity in gene structures of the VrCCA1L genes, we analyzed their full-length genomic and CDS using the Gene Structure Display Server program [40]. Twenty-four of the 27 VrCCA1L genes contained UTRs, with the exception of all group II members (Fig. 5a). All group I members contained a relatively large number of exons, ranging from 5 to 8 (Fig. 5a). Most group II and III genes had 2 exons, with the exception of VrCCA1L24, which had 3 exons. All group V VrCCA1Ls had 3 exons, and 1 and 2 members of group VI contained 2 and 3 exons, respectively. To further analyze the conservation and diversity of the VrCCA1L gene structures, we analyzed conserved motifs in their encoded proteins using MEME tools [42]. Twenty-four distinct motifs were identified (Fig. 5b, Additional file 4). All VrCCA1L proteins contained motif 2, which appeared to represent the conserved MYB 2 domain. Most members within a group shared some motifs; for example, group V members contained motifs 1, 2, 5, 10, and 11 (Fig. 5b), indicating that these genes may share some common gene structures. Other motifs were found only in specific groups; for example, motif 4 was found only in group I, and motif 10 was found only in group III (Fig. 5b), highlighting the structure diversity of VrCCA1L proteins from different groups.
Fig. 5.
Gene structures and conserved motifs of the VrCCA1L genes. a Exon-intron organization of the VrCCA1L genes. The orange, blue, and black boxes indicate exons, UTRs, and introns, respectively. (b) Conserved motifs in the VrCCA1L proteins. Motifs 1–24 are presented in different colored boxes
Cis-acting elements in the VrCCA1L promoter regions
To predict the cis-acting elements in the VrCCA1L promoter regions, 2 kb of sequence upstream of the ATG initiation codon for each gene was downloaded from NCBI and analyzed using PlantCare [45]. Eighty-seven cis-acting elements were obtained across all the VrCCA1L promoters, and 56 were predicted to have potential functions (Additional file 5). The VrCCA1L genes contained various numbers and types of cis-acting elements, again highlighting their functional diversity (Additional file 5). Based on their predicted functions, the cis-acting elements were classified into six groups as described by Jin et al. [44]: development-related elements, environmental stress-related elements, hormone-responsive elements, light-responsive elements, promoter-related elements, and site binding-related elements (Table 2). All VrCCA1L genes contained light-responsive elements and promoter-related elements. The light-responsive element was the most abundant element in each VrCCA1L promoter, and the number of light-responsive elements ranged from 3 (VrCCA1L24) to 11 (VrCCA1L13) (Table 2). The promoter-related elements CAAT-box and TATA-box, which are basic promoter elements, and the light-responsive element Box 4 were found in all VrCCA1L promoters (Additional file 5). The number of cis-acting elements in each duplicated gene pair varied, indicating that these duplicated genes may exhibit distinct expression responses under specific conditions (Table 2).
Table 2.
Numbers and types of cis-acting elements in each VrCCA1L promoter region
| Gene name | Development related elements | Environmental stress related elements | Hormone-responsive elements | Light-responsive elements | Promoter related elements | Site-binding related elements | Others |
|---|---|---|---|---|---|---|---|
| VrCCA1L1 | 0 | 1 | 2 | 9 | 2 | 0 | 9 |
| VrCCA1L2 | 1 | 1 | 3 | 7 | 2 | 0 | 10 |
| VrCCA1L3 | 3 | 4 | 1 | 5 | 2 | 0 | 8 |
| VrCCA1L4 | 0 | 2 | 4 | 10 | 2 | 0 | 9 |
| VrCCA1L5 | 2 | 1 | 2 | 5 | 2 | 0 | 12 |
| VrCCA1L6 | 3 | 3 | 5 | 9 | 2 | 1 | 13 |
| VrCCA1L7 | 3 | 2 | 4 | 5 | 2 | 0 | 9 |
| VrCCA1L8 | 2 | 0 | 1 | 9 | 2 | 0 | 14 |
| VrCCA1L9 | 2 | 3 | 4 | 7 | 2 | 0 | 14 |
| VrCCA1L10 | 1 | 2 | 2 | 6 | 2 | 0 | 11 |
| VrCCA1L11 | 0 | 3 | 4 | 9 | 3 | 1 | 11 |
| VrCCA1L12 | 1 | 2 | 2 | 8 | 3 | 0 | 12 |
| VrCCA1L13 | 0 | 1 | 1 | 11 | 2 | 1 | 14 |
| VrCCA1L14 | 1 | 2 | 4 | 8 | 2 | 0 | 12 |
| VrCCA1L15 | 2 | 1 | 3 | 8 | 2 | 0 | 17 |
| VrCCA1L16 | 1 | 3 | 4 | 8 | 2 | 0 | 14 |
| VrCCA1L17 | 3 | 0 | 3 | 6 | 2 | 2 | 12 |
| VrCCA1L18 | 1 | 0 | 3 | 6 | 2 | 0 | 8 |
| VrCCA1L19 | 1 | 0 | 3 | 8 | 2 | 0 | 7 |
| VrCCA1L20 | 0 | 1 | 5 | 6 | 2 | 0 | 14 |
| VrCCA1L21 | 2 | 4 | 4 | 7 | 2 | 2 | 15 |
| VrCCA1L22 | 1 | 3 | 3 | 8 | 2 | 2 | 10 |
| VrCCA1L23 | 1 | 1 | 4 | 6 | 2 | 0 | 13 |
| VrCCA1L24 | 0 | 0 | 0 | 3 | 2 | 0 | 5 |
| VrCCA1L25 | 3 | 1 | 4 | 6 | 2 | 2 | 14 |
| VrCCA1L26 | 3 | 3 | 4 | 5 | 2 | 1 | 10 |
| VrCCA1L27 | 0 | 2 | 5 | 9 | 2 | 1 | 9 |
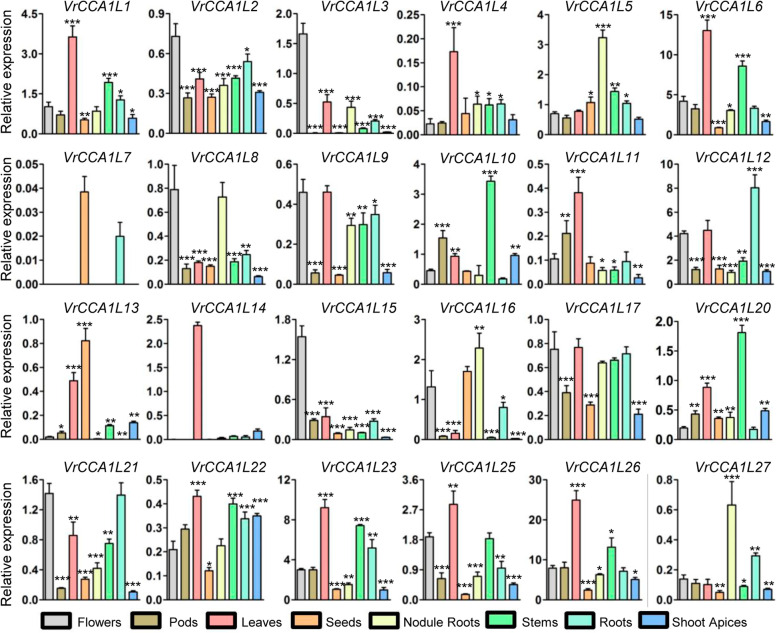
Expression of VrCCA1L genes in different tissues
To address the potential functions of VrCCA1L genes, eight tissues were sampled from the reference genome variety VC1973A at a single time point and used for gene expression analysis: roots, nodule roots, shoot apices, stems, leaves, flowers, pods, and seeds [36, 37]. VrCCA1L genes showed distinct expression levels in different tissues at the tested time point. For example, VrCCA1L12 showed relatively high expression levels in most tissues examined (expression level > 1) (Fig. 6). By contrast, VrCCA1L18, VrCCA1L19, and VrCCA1L24 could not be detected in any tissues, indicating that their transcript abundances were extremely low (data not shown). Some VrCCA1L genes were expressed at high abundance only in specific tissues at the tested time point. For example, VrCCA1L14 showed high expression levels in leaves but low levels elsewhere, indicating that it may have a critical function in leaves. In addition, the expression of orthologs of the well-studied circadian clock genes CCA1 and LHY (VrCCA1L6 and VrCCA1L26) was also analyzed at the tested time point. VrCCA1L26 was expressed at relatively high levels in all tissues; VrCCA1L6 showed relatively high abundance in most tissues, with the exception of seeds (Fig. 6).
Fig. 6.
Expression analyses of the VrCCA1L genes in different tissues. The expression patterns of VrCCA1L genes were analyzed using qRT–PCR. All the VrCCA1L genes were firstly normalized to a mungbean Actin gene (Vradi03g00210). Then the expression level of VrCCA1L1 in flowers was set to 1, and the values of all the VrCCA1L genes in different tissues were adjusted accordingly. Significant differences relative to the expression in flowers are indicated by asterisks (***P < 0.001; **P < 0.01; and *P < 0.05)
Duplicated genes may retain some common functions from the original gene and may also acquire new functions during evolution [53]. We therefore analyzed the expression of duplicated VrCCA1L genes in multiple tissues at the tested time point. Some duplicated pairs showed similar expression patterns in some tissues but distinct expression levels in others (Fig. 6). For example, the tandem duplicates VrCCA1L18 and VrCCA1L19 showed extremely low expression levels in all the tissues examined at the tested time point, suggesting that these two genes may have similar transcriptional regulatory mechanisms. By contrast, VrCCA1L10 and VrCCA1L20 exhibited similar expression levels in leaves and roots but different expression patterns in flowers, stems, pods, seeds, nodule roots, and shoot apices (Fig. 6), indicating that they acquired different transcriptional regulatory mechanisms in these tissues after genome duplication.
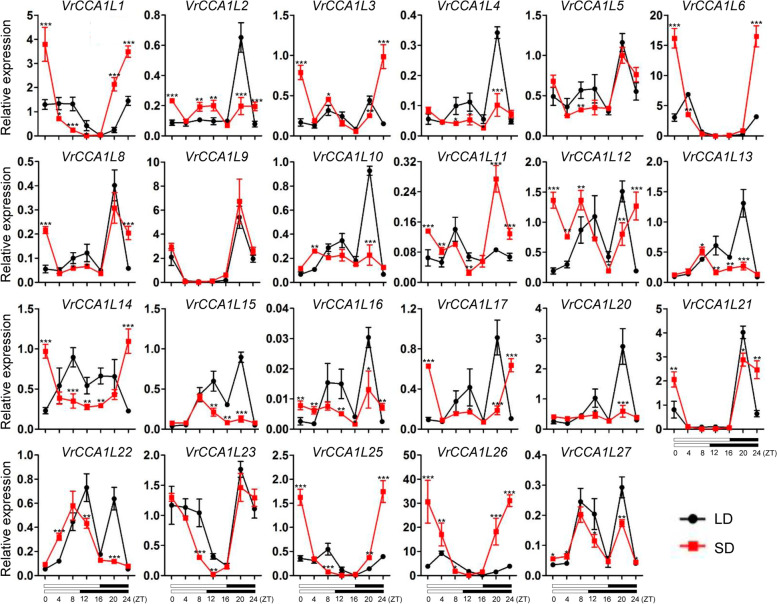
Expression patterns of VrCCA1Ls under different photoperiod conditions in leaves
Because the expression of Arabidopsis CCA1 and LHY shows different expression levels under different photoperiod conditions, we investigated the expression patterns of VrCCA1L genes in mungbean leaves during the day and night under LD and SD conditions (Fig. 7). VrCCA1L7, VrCCA1L18, VrCCA1L19, and VrCCA1L24 were not detected in leaves during the day under either LD or SD conditions. Most VrCCA1L genes showed different expression levels in leaves under LD and SD conditions, with the exception of VrCCA1L9, whose expression was not regulated by day length (Fig. 7), indicating that VrCCA1L genes are important factors in response to different photoperiod conditions. Several VrCCA1L genes might be diurnal rhythm genes in leaves under both LD and SD conditions. For example, the expression of VrCCA1L23 decreased during the day and increased during the night under both LD and SD conditions (Fig. 7). By contrast, some VrCCA1L genes might be diurnal rhythm genes in leaves only under specific day length conditions. For example, VrCCA1L2 exhibited increased expression during the night, and decreased during the day in leaves under LD conditions but not SD conditions, indicating that these genes may be involved in photoperiod-dependent regulatory pathways. Some duplicated genes showed similar expression patterns in leaves under specific photoperiod conditions. For example, VrCCA1L10 and VrCCA1L20 showed similar expression patterns in leaves under LD conditions. By contrast, some duplicated gene pairs, such as VrCCA1L2 and VrCCA1L15, exhibited distinct expression patterns in leaves under LD and SD conditions, indicating functional differentiation of these duplicated genes (Fig. 7).
Fig. 7.
Expression analysis of the VrCCA1L genes throughout the day under LD and SD conditions. The photoperiod treatments were 16 h light/8 h dark and 10 h light/14 h dark cycles for LD and SD conditions, respectively. ZT, Zeitgeber Time. The x-axis indicates the light and dark cycle throughout the day. Open boxes indicate the time of lights-on, and black boxes indicate the time of darkness. Red lines indicate SD and black lines mean LD. Expression levels of the VrCCA1Ls were normalized to that of an Actin gene from mungbean. Significant differences relative to the expression under LD conditions are indicated with asterisks (***P < 0.001; **P < 0.01; and *P < 0.05)
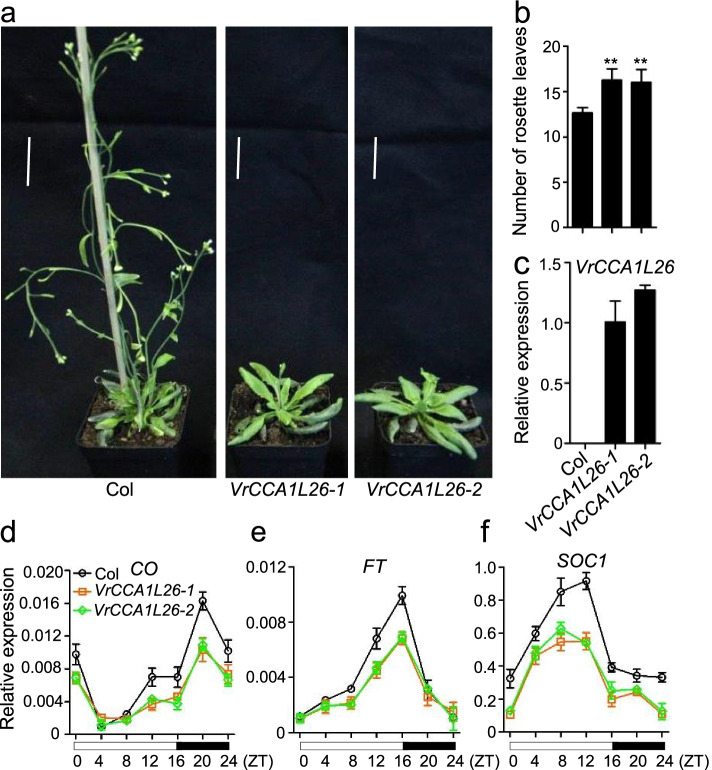
VrCCA1L26 influenced flowering time in Arabidopsis
VrCCA1L6 and VrCCA1L26 showed a close phylogenetic relationship with Arabidopsis CCA1 and LHY, and the expression levels of VrCCA1L26 were higher than those of VrCCA1L6 in most tested tissues. We therefore selected VrCCA1L26 for functional characterization in this study, and we transformed VrCCA1L26 into Arabidopsis for further analysis (Additional file 6). VrCCA1L26 exhibited high expression levels in the transgenic lines, whereas VrCCA1L26 was not detected in wild-type Arabidopsis plants (Fig. 8). VrCCA1L26 transgenic lines showed greater numbers of rosette leaves than wild-type Arabidopsis plants (Fig. 8), indicating that VrCCA1L26 delayed flowering time in Arabidopsis. To search for the factors responsible for the delayed flowering time in VrCCA1L26 transgenic plants, we investigated the expression of several flowering time–related genes, including CO, FT, and SOC1, all of which accelerate flowering in Arabidopsis [54, 55]. The expression levels of CO, FT, and SOC1 were reduced in VrCCA1L26 transgenic Arabidopsis lines at several time point throughout the day, indicating that VrCCA1L26 may regulate flowering time by affecting expression of these flowering time genes (Fig. 8).
Fig. 8.
Flowering time phenotypes of the VrCCA1L26 transgenic Arabidopsis plants. a Flowering time of VrCCA1L26 transgenic and wild-type Arabidopsis plants. Arabidopsis plants were grown in the growth chamber under 16 h 23 °C light/8 h 21 °C dark cycle conditions. Bars = 4 cm. b The number of rosette leaves in VrCCA1L26 transgenic and wild-type plants. The rosette leaf numbers of VrCCA1L26 transgenic and wild-type plants were recorded after the bolting of Arabidopsis plants. c The expression levels of VrCCA1L26 in VrCCA1L26 transgenic and wild-type plants. The expression level of VrCCA1L26 in transgenic line 1 was set to 1, and the values for other transgenic and wild-type plants were adjusted accordingly. Significant differences relative to the corresponding wild-type plants are indicated with asterisks (**P < 0.01). d-f The expression levels of CO (d), FT (e), and SOC1 (f) in VrCCA1L26 transgenic and wild-type plants. The x-axis indicates the light and dark cycle throughout the day. The expression levels of CO, FT, and SOC1 were normalized to AtActin. Open boxes indicate the time of lights-on, and black boxes indicate the time of darkness. ZT, Zeitgeber Time (ZT)
Discussion
CCA1L transcription factors, especially the circadian clock and flowering time regulation genes CCA1 and LHY, play critical roles in plant growth and development and have been identified in several plant species [17–19]. Investigation of VrCCA1L genes in mungbean will increase our understanding of its growth and development. In this study, we identified 27 mungbean VrCCA1L genes and investigated many of their characteristics.
The number of CCA1L genes varies among different plant species. Of the seven CCA1L groups, group I has the largest number of CCA1L genes in soybean, mungbean, and Arabidopsis (Fig. 2), indicating the evolutionary conservation of CCA1L genes in different plant species. Most of the seven groups are shared among multiple plant species, but some groups have been lost during evolution. For example, mungbean has no group IV or VII members, and soybean has no group III GmCCA1L members, indicating functional divergence of CCA1L genes in different plant species (Fig. 2). Moreover, the number of CCA1Ls in mungbean is half that in soybean. This reflects the fact that soybean has experienced two rounds of whole genome duplication [56], whereas mungbean has experienced only one [6]. However, the numbers of VrCCA1L genes in most of the seven groups in mungbean are not half of those in soybean, with the exception of group I (11 members in mungbean, 22 in soybean), indicating that the gene structures of many CCA1L proteins in legumes have changed during evolution.
Cis-acting elements are essential factors for gene expression, and several circadian clock related cis-acting elements have been found in VrCCA1L promoters (Additional file 5). For example, the G-box cis-acting element is necessary for the transcriptional regulation of CCA1 in Arabidopsis [57], and have been found in many VrCCA1L promoters, including VrCCA1L6 and VrCCA1L26, indicating that these VrCCA1L genes play important roles in circadian clock regulation. The expression of VrCCA1L genes in different tissues provides insight into their potential functions. The numbers and types of cis-acting elements varied among the promoter regions of different VrCCA1L genes (Table 2), and this may be responsible for their different expression patterns in various tissues at the tested time point and under contrasting photoperiod conditions in leaves (Figs. 6 and 7). Although all the VrCCA1L promoters contained cis-acting elements, the expression of several VrCCA1L members could not be detected in any tissues examined at the tested time point; these included VrCCA1L18, VrCCA1L19, and VrCCA1L24. The expression of many CCA1L genes is controlled by the circadian clock and changes during the day and night in leaves. For example, the expression of CCA1 and LHY shows a diurnal rhythm [21, 22, 28]. However, the expression of VrCCA1L18, VrCCA1L19, and VrCCA1L24 was not detected throughout the day under either LD or SD conditions in leaves. Many factors affect gene expression in addition to cis-acting elements. For example, epigenetic modifications such as DNA methylation have a substantial effect on gene expression [58, 59]. The extremely low expression of VrCCA1L18, VrCCA1L19, and VrCCA1L24 in all tissues at the tested time point and under all photoperiod conditions in leaves may therefore reflect epigenetic modification. These genes may be expressed at high levels under other growth conditions.
Mungbean genome has experienced one round of whole genome duplication, which may have produced many novel genes [6, 12, 53]. Five duplicated gene pairs were found among the 27 VrCCA1L genes (Fig. 1). Duplicated genes have evolved from a single ancestor and may retain some common functions. For example, the duplicated genes VrCCA1L18/VrCCA1L19 have similar exon-intron structures and conserved motifs (Fig. 5). Moreover, VrCCA1L18 and VrCCA1L19 both showed extremely low expression in different tissues at the tested time point and under different photoperiod conditions in leaves, suggesting that they may share some common functions. By contrast, some duplicated genes appeared to have obtained new functions and showed functional diversity. For example, VrCCA1L10 and VrCCA1L20 had different expression levels in several tissues at the tested time point, indicating different potential functions (Fig. 6). In addition, the different motifs in these VrCCA1L proteins indicate structure diversity of VrCCA1L genes. Some VrCCA1L genes shared some common motifs, such as motif 2, indicating that these motifs might be historical structures among these genes (Fig. 5). Some motifs were found in some specific groups, suggesting that theVrCCA1L genes in these groups obtained new gene structures during evolution.
In Arabidopsis, CCA1 and LHY are critical components involved in circadian clock and flowering time regulation [21, 22]. Loss of function of the soybean homologs GmLCLa1, GmLCLa2, GmLCLb1, and GmLCLb2 results in a short-period circadian rhythm and a late flowering phenotype [30]. The expression of many VrCCA1L genes was regulated by photoperiod and showed different expression levels during the day and night under either LD or SD conditions in leaves (Fig. 7). Moreover, most of the VrCCA1L genes showed different expression patterns under LD and SD conditions throughout the day in leaves (Fig. 7), indicating that they may have distinct roles under different photoperiod conditions. Whether VrCCA1L genes are diurnal rhythm factors still need further investigation under a full forty-eight-hour time course and constant (free-running) conditions. VrCCA1L26 showed a close relationship with Arabidopsis CCA1 and LHY, as well as soybean GmLCLa1, GmLCLa2, GmLCLb1, and GmLCLb2 (Fig. 2). It also showed relatively high expression levels in most tissues at the tested time point (Fig. 6). When expressed in transgenic Arabidopsis, it delayed flowering time by suppressing CO, FT, and SOC1 expression (Fig. 8). VrCCA1L26 exhibited distinct expression patterns under different photoperiod conditions in leaves, but whether it has different functions in flowering time regulation under different photoperiod conditions requires further investigation. Arabidopsis CCA1 and LHY delay flowering time, and VrCCA1L26 also delayed flowering time in transgenic Arabidopsis plants. By contrast, soybean GmLCLa1, GmLCLa2, GmLCLb1, and GmLCLb2 are essential for accelerating flowering in soybean [21, 22, 30]. CCA1 and LHY homologs therefore have distinct functions in flowering time regulation in different plant species. Our study provides evidence that VrCCA1L26 suppresses flowering in Arabidopsis, but we do not yet know its function in mungbean. Whether VrCCA1L26 has similar functions in regulation of flowering time in mungbean and Arabidopsis will require further investigation. Arabidopsis CCA1 and LHY are also involved in the regulation of hypocotyl elongation [21, 22, 28], and VrCCA1L26 overexpression plants showed elongated hypocotyl in Arabidopsis (Additional file 7), thus VrCCA1L26 may be a key factor that influences mungbean sprout production. In addition, VrCCA1L26 may interact with LHY or CCA1 to regulate the expression of circadian clock genes in Arabidopsis, thereby influencing circadian clock regulation. The mechanisms by which VrCCA1L26 affects circadian clock regulation require further investigation.
Conclusion
In summary, we identified 27 VrCCA1L genes from mungbean and investigated many of their characteristics, such as chromosomal locations, exon-intron organization, conserved domains, conserved motifs, cis-acting elements, duplicated gene pairs, and expression patterns in different tissues at a single time and under different photoperiod conditions in leaves. In addition, our analysis revealed that VrCCA1L26 delayed flowering time in transgenic Arabidopsis plants. Our results provide essential information for further functional characterization of VrCCA1L genes and circadian clock regulation in mungbean.
Supplementary Information
Acknowledgments
We thank Suk-Ha Lee at Seoul National University, Seoul, South Korea, for supplying mungbean VC1973A seeds.
Abbreviations
- Vr
Vigna radiate
- LD
long-day
- SD
Short-day
- LHY
LATE ELONGATED HYPOCOTYL
- CO
CONSTANS
- FT
FLOWERING LOCUS T
- SOC1
SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1
- CCA1
CIRCADIAN CLOCK ASSOCIATED 1
- Hsf
Heat Shock transcription Factor
- MYB
MYELOBLASTOSIS ONCOGENE
- TOC1
TIMING OF CAB EXPRESSION 1
- ZT
Zeitgeber Time
- CDS
Coding sequences
Authors’ contributions
SL and HZ conceived and designed the research. CL, QZ, JD and CC conducted the experiments and analyzed the data. SL and HZ wrote the manuscript. All authors read and approved the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (grant 31971898, 32171979), the National Science Foundation of Shandong Province (grant ZR2020QC120), the Opening Foundation of Shandong Dry-land Farming Technology Key Laboratory (grant 2219006), and the Qingdao Agricultural University Scientific Research Foundation (grant 6631119010). The funders had no role in the design of the study, collection, analysis, and interpretation of data, or in writing the manuscript.
Availability of data and materials
All data used in this study are included in this article and additional files. The genome sequence and annotation datasets that supported our findings are available in: Arabidopsis (http://www.arabidopsis.org), soybean (https://soybase.org/) and mungbean (http://plantgenomics.snu.ac.kr/mediawiki-1.21.3/index.php/Main_Page) (https://www.ncbi.nlm.nih.gov/). All the mungbean genes used in this study for phylogeny and subsequent analysis are mentioned in Table 1 and can be downloaded from NCBI.
Declarations
Ethics approval and consent to participate
All methods in this research were carried out in accordance with relevant guidelines and regulations of Qingdao Agricultural University, Qingdao, China. No specific permits are required for sample collection in this study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Chenyang Liu, Qianqian Zhang and Jing Dong contributed equally to this work.
Contributor Information
Chenyang Liu, Email: lcy931115@163.com.
Qianqian Zhang, Email: zqqsxn123@163.com.
Jing Dong, Email: d13399507598@163.com.
Chunmei Cai, Email: caichunmei0902@163.com.
Hong Zhu, Email: zhuhong198812@126.com.
Shuai Li, Email: li2014shuai@qau.edu.cn.
References
- 1.Lambrides C, Godwin I. Mungbean. In: Kole C, editor. Genome mapping and molecular breeding in plants. Berlin: Springer; 2017. pp. 69–90. [Google Scholar]
- 2.Keatinge JDH, Easdown WJ, Yang RY, Chadha ML, Shanmugasundaram S. Overcoming chronic malnutrition in a future warming world: the key importance of mungbean and vegetable soybean. Euphytica. 2011;180(1):129–141. doi: 10.1007/s10681-011-0401-6. [DOI] [Google Scholar]
- 3.Ganesan K, Xu BJ. A critical review on phytochemical profile and health promoting effects of mung bean (Vigna radiata) Food Sci Hum Well. 2018;7(1):11–33. doi: 10.1016/j.fshw.2017.11.002. [DOI] [Google Scholar]
- 4.El-Adawy TA, Rahma EH, El-Bedawey AA, El-Beltagy AE. Nutritional potential and functional properties of germinated mung bean, pea and lentil seeds. Plant Food Hum Nutr. 2003;58(3):1–13. doi: 10.1023/B:QUAL.0000040339.48521.75. [DOI] [Google Scholar]
- 5.Tang D, Dong Y, Ren H, Li L, He C. A review of phytochemistry, metabolite changes, and medicinal uses of the common food mung bean and its sprouts (Vigna radiata) Chem Cent J. 2014;8(1):1–9. doi: 10.1186/1752-153X-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kang YJ, Kim SK, Kim MY, Lestari P, Kim KH, Ha BK, Jun TH, Hwang WJ, Lee T, Lee J, Shim S, Yoon MY, Jang YE, Han KS, Taeprayoon P, Yoon N, Somta P, Tanya P, Kim KS, Gwag JG, Moon JK, Lee YH, Park BS, Bombarely A, Doyle JJ, Jackson SA, Schafleitner R, Srinives P, Varshney RK, Lee SH. Genome sequence of mungbean and insights into evolution within Vigna species. Nat Commun. 2014;5:5443. doi: 10.1038/ncomms6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Riechmann JL, Ratcliffe OJ. A genomic perspective on plant transcription factors. Curr Opin Plant Biol. 2000;3(5):423–434. doi: 10.1016/S1369-5266(00)00107-2. [DOI] [PubMed] [Google Scholar]
- 8.Nam JM, dePamphilis CW, Ma H, Nei M. Antiquity and evolution of the MADS-box gene family controlling flower development in plants. Mol Biol Evol. 2003;20(9):1435–1447. doi: 10.1093/molbev/msg152. [DOI] [PubMed] [Google Scholar]
- 9.Parenicová L, de Folter S, Kieffer M, Horner DS, Favalli C, Busscher J, Cook HE, Ingram RM, Kater MM, Davies B, Angenent GC, Colombo L. Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: new openings to the MADS world. Plant Cell. 2003;15(7):1538–1551. doi: 10.1105/tpc.011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gangappa SN, Botto JF. The BBX family of plant transcription factors. Trends Plant Sci. 2014;19(7):460–470. doi: 10.1016/j.tplants.2014.01.010. [DOI] [PubMed] [Google Scholar]
- 11.Liu JY, Osbourn A, Ma PD. MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Mol Plant. 2015;8(5):689–708. doi: 10.1016/j.molp.2015.03.012. [DOI] [PubMed] [Google Scholar]
- 12.Li S, Wang R, Jin H, Ding Y, Cai C. Molecular characterization and expression profile analysis of heat shock transcription factors in mungbean. Front Genet. 2019;9:736. doi: 10.3389/fgene.2018.00736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou Y, Zhu H, He S, Zhai H, Zhao N, Xing S, Wei Z, Liu Q. A novel sweetpotato transcription factor gene IbMYB116 enhances drought tolerance in transgenic Arabidopsis. Front Plant Sci. 2019;10:1025. doi: 10.3389/fpls.2019.01025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin H, Xing M, Cai C, Li S. B-box proteins in Arachis duranensis: genome-wide characterization and expression profiles analysis. Agronomy. 2020;10(1):23. doi: 10.3390/agronomy10010023. [DOI] [Google Scholar]
- 15.Zhu H, Zhou Y, Zhai H, He S, Zhao N, Liu Q. A novel sweetpotato WRKY transcription factor, IbWRKY2, positively regulates drought and salt tolerance in transgenic Arabidopsis. Biomolecules. 2020;10(4):506. doi: 10.3390/biom10040506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, Creelman R, Pilgrim M, Broun P, Zhang JZ, Ghandehari D, Sherman BK, Yu G. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science. 2000;290(5499):2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- 17.Chen YH, Yang XY, He K, Liu MH, Li JG, Gao ZF, Lin ZQ, Zhang YF, Wang XX, Qiu XM, Shen YP, Zhang L, Deng XH, Luo JC, Deng XW, Chen ZL, Gu HY, Qu LJ. The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol Biol. 2006;60(1):107–124. doi: 10.1007/s11103-005-2910-y. [DOI] [PubMed] [Google Scholar]
- 18.Du H, Wang YB, Xie Y, Liang Z, Jiang SJ, Zhang SS, Huang YB, Tang YX. Genome-wide identification and evolutionary and expression analyses of MYB-related genes in land plants. DNA Res. 2013;20(5):437–448. doi: 10.1093/dnares/dst021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bian S, Jin DH, Li RH, Xie X, Gao GL, Sun WK, Li YJ, Zhai LL, Li XY. Genome-wide analysis of CCA1-like proteins in soybean and functional characterization of GmMYB138a. Int J Mol Sci. 2017;18(10):2040. doi: 10.3390/ijms18102040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang C, Ma R, Xu J, Yan J, Guo L, Song J, Feng R, Yu M. Genome-wide identification and classification of MYB superfamily genes in peach. PLoS One. 2018;13(6):e0199192. doi: 10.1371/journal.pone.0199192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang W, Perez-Garcia P, Pokhilko A, Millar AJ, Antoshechkin I, Riechmann JL, Mas P. Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science. 2012;336(6077):75–79. doi: 10.1126/science.1219075. [DOI] [PubMed] [Google Scholar]
- 22.Wang L, Kim J, Somers DE. Transcriptional corepressor TOPLESS complexes with pseudoresponse regulator proteins and histone deacetylases to regulate circadian transcription. Proc Natl Acad Sci. 2013;110(2):761–766. doi: 10.1073/pnas.1215010110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anwer MU, Davis A, Davis SJ, Quint M. Photoperiod sensing of the circadian clock is controlled by EARLY FLOWERING 3 and GIGANTEA. Plant J. 2020;101(6):1397–1410. doi: 10.1111/tpj.14604. [DOI] [PubMed] [Google Scholar]
- 24.Romanowski A, Schlaen RG, Perez-Santangelo S, Mancini E, Yanovsky MJ. Global transcriptome analysis reveals circadian control of splicing events in Arabidopsis thaliana. Plant J. 2020;103(2):889–902. doi: 10.1111/tpj.14776. [DOI] [PubMed] [Google Scholar]
- 25.Wang Z, Tobin E. Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell. 1998;93(7):1207–1217. doi: 10.1016/S0092-8674(00)81464-6. [DOI] [PubMed] [Google Scholar]
- 26.Lu SX, Webb CJ, Knowles SM, Kim SH, Wang ZY, Tobin EM. CCA1 and ELF3 interact in the control of hypocotyl length and flowering time in Arabidopsis. Plant Physiol. 2012;158(2):1079–1088. doi: 10.1104/pp.111.189670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alabadí D, Oyama T, Yanovsky MJ, Harmon FG, Más P, Kay SA. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science. 2001;293(5531):880–883. doi: 10.1126/science.1061320. [DOI] [PubMed] [Google Scholar]
- 28.Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carré IA, Coupland G. The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell. 1998;93(7):1219–1229. doi: 10.1016/S0092-8674(00)81465-8. [DOI] [PubMed] [Google Scholar]
- 29.Nakamichi N, Kiba T, Henriques R, Mizuno T, Chua NH, Sakakibara H. PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell. 2010;22(3):594–605. doi: 10.1105/tpc.109.072892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y, Yuan L, Su T, Wang Q, Gao Y, Zhang S, Jia Q, Yu G, Fu Y, Cheng Q, Liu B, Kong F, Zhang X, Song C, Xu X, Xie Q. Light- and temperature-entrainable circadian clock in soybean development. Plant Cell Environ. 2020;43(3):637–648. doi: 10.1111/pce.13678. [DOI] [PubMed] [Google Scholar]
- 31.Zhang X, Chen Y, Wang Z, Chen Z, Gu H, Qu L. Constitutive expression of CIR1 (RVE2) affects several circadian-regulated processes and seed germination in Arabidopsis. Plant J. 2007;51(3):512–552. doi: 10.1111/j.1365-313X.2007.03156.x. [DOI] [PubMed] [Google Scholar]
- 32.Penfield S, Hall A. A role for multiple circadian clock genes in the response to signals that break seed dormancy in Arabidopsis. Plant Cell. 2009;21(6):1722–1732. doi: 10.1105/tpc.108.064022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yi J, Derynck M, Li X, Telmer P, Marsolais F, Dhaubhadel S. A single-repeat MYB transcription factor, GmMYB176, regulates CHS8 gene expression and affects isoflavonoid biosynthesis in soybean. Plant J. 2010;62(2):1019–1034. doi: 10.1111/j.1365-313X.2010.04214.x. [DOI] [PubMed] [Google Scholar]
- 34.Lai AG, Doherty CJ, Mueller-Roeber B, Kay SA, Schippers JH, Dijkwel PP. CIRCADIAN CLOCK-ASSOCIATED 1 regulates ROS homeostasis and oxidative stress responses. Proc Natl Acad Sci. 2012;109(42):17129–17134. doi: 10.1073/pnas.1209148109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nagel DH, Doherty CJ, Pruneda-Paz JL, Schmitz RJ, Ecker JR, Kay SA. Genome-wide identification of CCA1 targets uncovers an expanded clock network in Arabidopsis. Proc Natl Acad Sci. 2015;112(34):E4802–E4810. doi: 10.1073/pnas.1513609112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu C, Zhang Q, Zhu H, Cai C, Li S. Characterization of mungbean CONSTANS-LIKE genes and functional analysis of CONSTANS-LIKE 2 in the regulation of flowering time in Arabidopsis. Front Plant Sci. 2021;12:608603. doi: 10.3389/fpls.2021.608603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi R, Xu W, Liu T, Cai C, Li S. VrLELP controls flowering time under short-day conditions in Arabidopsis. J Plant Res. 2021;134:141–149. doi: 10.1007/s10265-020-01235-7. [DOI] [PubMed] [Google Scholar]
- 38.Finn RD, Attwood TK, Babbitt PC, Bateman A, Bork P, Bridge AJ, Chang HY, Dosztányi Z, El-Gebali S, Fraser M, Gough J, Haft D, Holliday GL, Huang HZ, Huang XS, Letunic I, Lopez R, Lu S, Marchler-Bauer A, Mi HY, Mistry J, Natale DA, Necci M, Nuka G, Orengo CA, Park Y, Pesseat S, Piovesan D, Potter SC, Rawlings ND, Redaschi N, Richardson L, Rivoire C, Sangrador-Vegas A, Sigrist C, Sillitoe I, Smithers B, Squizzato S, Sutton G, Thanki N, Thomas PD, cTosatto SCE, Wu CH, Xenarios I, Yeh LS, Young SY, Mitchell AL. InterPro in 2017–beyond protein family and domain annotations. Nucleic Acids Res. 2017;45(D1):D190–D199. doi: 10.1093/nar/gkw1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, Qureshi M, Richardson LJ, Salazar GA, Smart A, Sonnhammer ELL, Hirsh L, Paladin L, Piovesan D, Tosatto SCE, Finn RD. The Pfam protein families database in 2019. Nucleic Acids Res. 2019;47(D1):D427–D432. doi: 10.1093/nar/gky995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu B, Jin JP, Guo AY, Zhang H, Luo JC, Gao G. GSDS 2.0. An upgraded gene feature visualization server. Bioinformatics. 2015;31(8):1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14(6):1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME suite: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fischer S, Brunk BP, Chen F, Gao X, Harb OS, odice JB, Shanmugam D, Roos DS, Stoeckert CJ. Using OrthoMCL to assign proteins to OrthoMCL-DB groups or to cluster proteomes into new ortholog groups. Curr Protoc Bioinformatics. 2011;35:1–19. doi: 10.1002/0471250953.bi0612s35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jin H, Tang X, Xing M, Zhu H, Sui J, Cai C, Li S. Molecular and transcriptional characterization of phosphatidyl ethanolamine-binding proteins in wild peanuts Arachis duranensis and Arachis ipaensis. BMC Plant Biol. 2019;19(1):484. doi: 10.1186/s12870-019-2113-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30(1):325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hou L, Zhang Z, Dou S, Zhang Y, Pang X, Li Y. Genome-wide identification, characterization, and expression analysis of the expansin gene family in Chinese jujube (Ziziphus jujuba mill.) Planta. 2019;49(3):815–829. doi: 10.1007/s00425-018-3020-9. [DOI] [PubMed] [Google Scholar]
- 47.Oliver T, Schmidt B, Nathan D, Clemens R, Maskell D. Using reconfigurable hardware to accelerate multiple sequence alignment with clustalw. Bioinformatics. 2005;21(16):3431–3432. doi: 10.1093/bioinformatics/bti508. [DOI] [PubMed] [Google Scholar]
- 48.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li S, Wang X, Xu W, Liu T, Cai C, Chen L, Clark CB, Ma J. Unidirectional movement of small RNAs from shoots to roots in interspecific heterografts. Nat Plants. 2021;7:50–59. doi: 10.1038/s41477-020-00829-2. [DOI] [PubMed] [Google Scholar]
- 50.Li S, Ding Y, Zhang D, Wang X, Tang X, Dai D, Jin H, Lee S, Cai C, Ma J. Parallel domestication with broad mutational spectrum of determinate stem growth habit in leguminous crops. Plant J. 2018;96:761–771. doi: 10.1111/tpj.14066. [DOI] [PubMed] [Google Scholar]
- 51.Li S, Ying Y, Secco D, Wang C, Narsai R, Whelan J, Shou H. Molecular interaction between PHO2 and GIGANTEA reveals a new crosstalk between flowering time and phosphate homeostasis in Oryza sativa. Plant Cell Environ. 2017;40(8):1487–1499. doi: 10.1111/pce.12945. [DOI] [PubMed] [Google Scholar]
- 52.Bent A. Arabidopsis thaliana floral dip transformation method. Methods Mol Biol. 2006;343:87–103. doi: 10.1385/1-59745-130-4:87. [DOI] [PubMed] [Google Scholar]
- 53.Kondrashov FA, Rogozin IB, Wolf YI, Koonin EV. Selection in the evolution of gene duplications. Genome Biol. 2002;3(2):RESEARCH0008. doi: 10.1186/gb-2002-3-2-research0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jack T. Molecular and genetic mechanisms of floral control. Plant Cell. 2004;16(Suppl):S1–17. doi: 10.1105/tpc.017038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baurle I, Dean C. The timing of developmental transitions in plants. Cell. 2006;125(4):655–664. doi: 10.1016/j.cell.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 56.Schmutz J, Cannon SB, Schlueter J, Ma JX, Mitros T, Nelson W, Hyten DL, Song QJ, Thelen JJ, Cheng JL, Xu D, Hellsten U, May GD, Yu Y, Sakurai T, Umezawa T, Bhattacharyya MK, Sandhu D, Valliyodan B, Lindquist E, Peto M, Grant D, Shu SQ, Goodstein D, Barry K, Futrell-Griggs M, Abernathy B, Du JC, Tian ZX, Zhu LC, Gill N, Joshi T, Libault M, Sethuraman A, Zhang XC, Shinozaki K, Nguyen HT, Wing RA, Cregan P, Specht J, Grimwood J, Rokhsar D, Stacey G, Shoemaker RC, Jackson SA. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463(7278):178–183. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- 57.Liu TL, Newton L, Liu MJ, Shiu SH, Farré EM. A G-box-like motif is necessary for transcriptional regulation by circadian pseudo-pesponse pegulators in Arabidopsis. Plant Physiol. 2016;170(1):528–539. doi: 10.1104/pp.15.01562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li X, Xing T, Du D. Identification of top-ranked proteins within a directional protein interaction network using the pagerank algorithm: applications in humans and plants. Curr Issues Mol Biol. 2016;20:13–28. [PubMed] [Google Scholar]
- 59.Zhang Q, Zhe L, Cui X, Ji C, Li Y, Zhang P, Liu J, Riaz A, Yao P, Liu M, Wang Y, Lu T, Yu H, Yang D, Zheng H, Gu X. N6-methyladenine DNA methylation in japonica and indica rice genomes and its association with gene expression, plant development, and stress responses. Mol Plant. 2018;11(12):1492–1508. doi: 10.1016/j.molp.2018.11.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data used in this study are included in this article and additional files. The genome sequence and annotation datasets that supported our findings are available in: Arabidopsis (http://www.arabidopsis.org), soybean (https://soybase.org/) and mungbean (http://plantgenomics.snu.ac.kr/mediawiki-1.21.3/index.php/Main_Page) (https://www.ncbi.nlm.nih.gov/). All the mungbean genes used in this study for phylogeny and subsequent analysis are mentioned in Table 1 and can be downloaded from NCBI.