Abstract
With recent advances in high-throughput next-generation sequencing, it is possible to describe the regulation and expression of genes at multiple levels. An assay for transposase-accessible chromatin using sequencing (ATAC-seq), which uses Tn5 transposase to sequence protein-free binding regions of the genome, can be combined with chromatin immunoprecipitation coupled with deep sequencing (ChIP-seq) and ribonucleic acid sequencing (RNA-seq) to provide a detailed description of gene expression. Here, we reviewed the literature on ATAC-seq and described the characteristics of ATAC-seq publications. We then briefly introduced the principles of RNA-seq, ChIP-seq and ATAC-seq, focusing on the main features of the techniques. We built a phylogenetic tree from species that had been previously studied by using ATAC-seq. Studies of Mus musculus and Homo sapiens account for approximately 90% of the total ATAC-seq data, while other species are still in the process of accumulating data. We summarized the findings from human diseases and other species, illustrating the cutting-edge discoveries and the role of multi-omics data analysis in current research. Moreover, we collected and compared ATAC-seq analysis pipelines, which allowed biological researchers who lack programming skills to better analyze and explore ATAC-seq data. Through this review, it is clear that multi-omics analysis and single-cell sequencing technology will become the mainstream approach in future research.
Keywords: ATAC-seq, ChIP-seq, RNA-seq, gene expression, multi-omics analysis
Introduction
The study of genes is a perennial topic in biology. Our understanding of genes—the nucleotide sequence, the gene structure, the gene function and expression—has reached an unprecedented level [1]. In eukaryotes, the genetic material DNA binds to histones to form nucleosomes, which fold and condense to form chromatin [2, 3]. In the processes of DNA replication and transcription, some regions of chromatin are opened [4], i.e. depleted of nucleosomes. Regulatory elements [such as transcription factors (TFs)] can bind to the exposed DNA sites in open regions and regulate DNA replication or transcription [5]. In addition, the chromatin structure can undergo dynamic epigenetic modifications, such as DNA methylation [6], histone modification [7] and chromatin remodelling [8], which are difficult to fully detect by traditional molecular techniques.
Because the interaction between protein and DNA influences gene expression, it has been the focus of intense interest. Immunoprecipitation is a technique for protein enrichment using the principle of antibody binding, which is the foundation of chromatin immunoprecipitation coupled with deep sequencing (ChIP-seq) [9]. The DNA bound to the target is then sequenced to reveal its binding sites in the genome [10, 11]. More recently, ATAC-seq, which uses Tn5 transposase to cleave only those regions of DNA that are not protected by bound proteins [12, 13], has been used to assay dynamic changes of chromatin structure. ATAC-seq and ChIP-seq are therefore highly consistent and complementary. ATAC-seq and ChIP-seq can be integrated to explore the mechanism of protein regulation of gene expression, making it possible to identify transcriptional differences caused by transcription initiation regulatory factors. Together with RNA-seq, which determines the expression level of specific RNA, ATAC-seq and ChIP-seq can be integrated to provide a unified picture of gene expression regulation [14]. Integrated analysis of multiple sequencing methods provides a powerful means for annotating the functional features of the genome, revealing the mechanisms of gene regulation at the frontiers of biology [15].
Here, we mainly discuss ATAC-seq, ChIP-seq and RNA-seq, which focus on the epigenome and transcriptome. We begin with a literature review of ATAC-seq and analyze the main topics of ATAC-seq through keyword extraction. Then, we describe the basic principles of three sequencing methods, focusing on their integrated usage and present a series of examples from diseases and species. Finally, we compare the analytical pipelines used in ATAC-seq data analysis in order to provide references for biological scientists who lack programming skills to better process their data and try to use established computational pipelines.
Development and growth of ATAC-seq
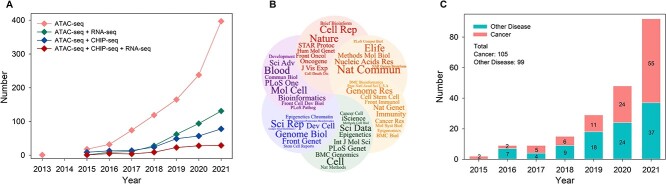
An exponentially increasing number of articles using ATAC-seq technology have been published since its first description in 2013 (Figure 1A). A total of 1016 papers were published from 2013 to 2021, indicating that ATAC-seq technology is a widely and increasingly used method. Studies that use ChIP-seq and RNA-seq together with ATAC-seq have also steadily increased, indicating a growing trend in comprehensive multi-omics data analysis. Specifically, we identified 336 articles in which ATAC-seq was used in combination with RNA-seq from 2015 to 2021 and 244 articles where it was used in combination with ChIP-seq from 2015 to 2021 (Figure 1A). The number of articles per year combining the three technologies has also grown rapidly (Figure 1A).
Figure 1.
Bibliometrics and datasets statistics of ATAC-seq. (A) The number of articles using combinations of ATAC-seq, ChIP-seq and RNA-seq published from 2013 to 2021 from PubMed database. Keywords of ATAC-seq, ChIP-seq and RNA-seq are searched, respectively, under the Title/Abstract term in the query box. (B) The names of top 50 journals published ATAC-seq articles. (C) The stacked bar plot shows the number of cancer studies versus other diseases using ATAC-seq and/or other sequencing techniques from 2015 to 2021.
ATAC-seq studies have appeared in more than 200 journals in diverse fields. The top 50 journals are shown in Figure 1B. The two most frequent journals are Nature Communications and Scientific Reports, each with more than 30 articles. In addition to these two journals, Nucleic Acids Research, Genome Biology, eLife, Cell Reports, Genome Research, Cell, Methods in Molecular Biology and Bioinformatics were the top 10 published journals, each with more than 18 articles.
One of the main topics in ATAC-seq research is the identification of chromatin structure and accessibility. Epigenetic changes are closely related to the occurrence and development of cancer and other diseases. Therefore, we counted publications that studied cancers and other diseases using ATAC-seq and/or other sequencing techniques, resulting in 204 studies, among which there were 36 types of cancer and 57 diseases. We counted the number of cancer studies and other diseases in each year, which shows an exponential increase in recent years (Figure 1C). The 204 publications used for Figure 1C are detailed in Supplementary Table 1 (see Supplementary Data available online at https://academic.oup.com/bib).
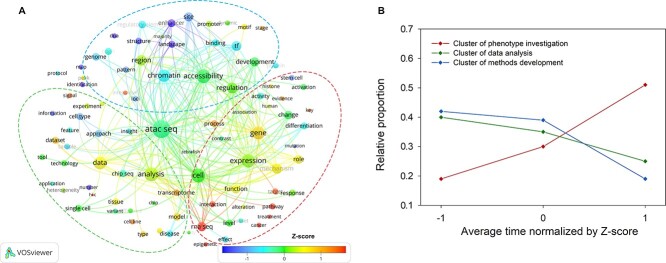
To obtain a general impression of the main topics of ATAC-seq research, keywords were extracted from the abstracts of the ATAC-seq articles (Figure 2A), and their co-occurrence with the term ‘ATAC-seq’ was analyzed. The most frequently co-occurring words (larger circles), such as genes, chromatin and analysis, are the main targets of ATAC-seq, while the words in the smaller circles indicate more general topics of ATAC-seq. The lines between different circles connect two words, indicating that they are from the same article. As a result, words that occur more often have more connections to other words.
Figure 2.
Keywords correlation analysis and the changes of clusters in ATAC-seq articles. (A) Keyword correlation analysis. The colour of the words represents the average time of occurrence of each keyword in the literature corpus. The colour bar represents the normalized Z-score of average time. The size of the circle behind each word represents its frequency of occurrence. The three ellipses indicate three clusters of highly correlated keywords (abbreviation of TF). (B) The change of clusters over time. Keywords are divided into three clusters according to their relevance. The horizontal axis represents the average time normalized by Z-score. The vertical axis shows its proportion to all the keywords in each cluster.
While extracting these keywords, the publication time of the articles containing the words was also analyzed; the average time of occurrence of each term is indicated by its colour in Figure 2A. The earliest articles (dark blue) focused on functional exploration (landscape, site, pattern), while later articles (green) tended to focus more on data analysis (tools, data, analysis), and the most recent articles (orange) expanded to include various biological topics (process, pathway, treatment). In addition, these are often integrated with ATAC-seq to provide a more comprehensive and persuasive edition, and the presence of the two keywords ChIP-seq and RNA-seq revealed -these technique descriptions of transcriptional regulation.
Words that often co-occur were divided into three clusters (dotted ellipses). The blue cluster (Cluster of methods development) focuses on the methods and uses of ATAC-seq at the molecular level, such as DNA accessibility, enhancers and TFs. The green cluster (Cluster of data analysis) focuses on the protocol and process of ATAC-seq, from upstream sequencing to downstream data analysis, such as sample preparation, experimental protocols and data analysis. The red cluster (Cluster of phenotype investigation) focuses more on practical applications, such as genes, regulation, pathways and diseases. The changes of these three clusters over time were counted (Figure 2B). The cluster of phenotype investigation is growing, while the other two are declining, suggesting that ATAC-seq is developing its value in more practical applications.
Introduction to RNA-seq, ChIP-seq and ATAC-seq
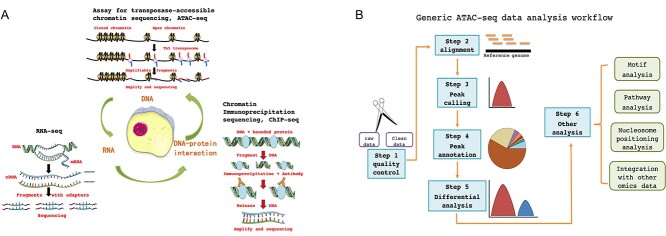
The genetic material of life is DNA, which is made up of four nucleotides with four different bases: adenine, thymine, cytosine and guanine. DNA is transcribed to form RNA, which is then translated into polypeptide chains [16]. The process of DNA replication is the basis of next-generation sequencing (NGS) technology (i.e. sequencing by synthesis). In NGS sequencing approaches, adapters are added to both ends of the DNA template, serving to locate and bind DNA polymerase, allowing them to hybridize to complementary oligonucleotides attached to a solid substrate, on which each fragment is amplified using PCR to form a cluster of identical DNA molecules derived from the original DNA fragment. The addition of each fluorescently labelled deoxyribonucleotide base to the template chain is detected to determine the sequence of bases in the DNA strand. The detailed principle of NGS has been described previously in a simple and clear way [17], and there is a comprehensive reference standard for how to interpret NGS data [18]. The development and progress of NGS technology have laid a foundation for the emergence of other technologies [19], including RNA-seq, ChIP-seq and ATAC-seq. We briefly describe the principles of these three techniques and their abilities to advance our understanding of epigenomics and transcriptomics (Figure 3A). These technologies are maturing, and the protocols are being refined, so we will not go into detail but instead focus on the features that make them potentially integrable with other technologies.
Figure 3.
Principles and workflow of ATAC-seq. (A) Principles of ATAC-seq, ChIP-seq and RNA-seq. (B) The generic ATAC-seq data analysis workflow.
RNA-seq
Messenger RNA provides a blueprint for building proteins, but the transcription of genes varies from cell to cell and over time. Selective expression of genes allows a single genome to produce different proteins, which leads to differences in cell morphology and function. The description of the transcriptome is therefore an important step in the exploration of gene function. RNA-seq provides a far more precise and comprehensive measurement of transcript and isoform levels than earlier methods such as microarray and low-throughput sequencing technology [20].
Compared with DNA sequencing, RNA-seq differs only in the additional sample preparation steps [21] since the RNA needs to be converted to DNA and then treated in the same way as a DNA sample. Sample quality control and sequencing are performed for the DNA samples, but the downstream analysis is significantly different. Briefly, the sequence reads are aligned with a reference genome or transcriptome, and the expression level is quantified in terms of the number of reads per gene or isoform. At present, pipelines for determining differential gene/isoform expression are well developed [20, 22]. RNA-seq has become one of the most commonly used tools in biology, and it has altered our view of the complexity of transcriptomes, which provides novel information about transcriptional and posttranscriptional gene regulation [23]. In addition, RNA-seq has advanced our understanding of RNA biology, i.e. it has allowed the accurate description of transcription and the intermolecular interactions that control RNA function [24].
Traditional bulk RNA-seq collects the transcripts of all cells, ignoring the characteristics of individual cells. Through techniques such as single-cell isolation [25, 26], transcriptome changes in small samples of defined cell types can be examined. Single-cell RNA sequencing (scRNA-seq) contributes to an in-depth understanding of the regulation of gene expression in cells and provides a way to address cellular heterogeneity [27]. Haque et al. [28] provide a guide to the use of scRNA-seq for biomedical and clinical purposes that lists the problems that may be encountered when using scRNA-seq, including protocol selection and biological interpretation. In particular, it is essential for researchers to understand the steps and principles of data processing in order to obtain correct and reproducible results [29]. ScRNA-seq has been applied in a wide range of studies, such as cancer treatment [30], infectious diseases [31] and the regulation of stem cell differentiation [32].
ChIP-seq
Proteins play an important role in gene expression. Protein–DNA interactions affect transcription levels at the source, and ChIP-seq technology allows genome-wide detection of DNA fragments that interact with histones and TFs [33]. In ChIP-seq [9], chromatin is first immunoprecipitated to specifically enrich the sample with DNA fragments bound to the target protein. The DNA fragments are then purified, a sequencing library is constructed (Figure 3A), and the DNA library is sequenced by NGS. By accurately locating the sequence fragments on the genome, researchers can obtain genome-wide information about DNA segments that interact with histones, TFs and other proteins. A set of working standards and guidelines have been developed for ChIP-seq, and they are regularly updated [34]. ChIP-seq used to require a large amount of input material, but now it has been optimized to be possible with as few as hundreds of cells [35]. Because of its ability to detect protein–DNA interactions, it has several applications: (i) localization of TF-binding sites and potential differential regulation of genes mediated by TFs [36]; (ii) study of protein modifications to dissect epigenetic characteristics and biological functions [36] and (iii) obtaining a map of nucleosome positioning [37]. Through the interpretation of the sequencing data, ChIP-seq can link epigenetic changes to the occurrence and progression of the disease [38].
ChIP-seq and RNA-seq were quickly integrated due to the clear impact of proteins on transcriptional levels. Details were discussed when analysing these two sequencing datasets including location clustering of TF-binding sites, transcript discovery, quantification of expression, etc. [39]. A recent study provided a standard integration process for ChIP-seq and RNA-seq, and it also supported the application of deep learning methods in the prediction of gene regulation [40].
ATAC-seq
With a deepening understanding of gene regulation mechanisms, changes in epigenetics have been found to play an important role [41]. These changes are seen as DNA base modifications [42] and chromatin structure [43]. We can use a wide array of sequencing techniques to measure changes in epigenetics from low to high levels of chromatin structure. For example, bisulphite sequencing for DNA methylation [44], high-throughput chromosome conformation capture (Hi-C) for the 3D structure of genome [45], DNase I coupled with deep sequencing (DNase-seq) [46], micrococcal nuclease digestion coupled with deep sequencing (MNase-seq) [47], formaldehyde-assisted isolation of regulatory elements (FAIRE-seq) [48] and ATAC-seq [12, 13]. In DNase-seq, DNase I has endonuclease activity used to cut DNA in enzyme sensitive parts. In MNase-seq, enzymes digest the open region directly, and any DNA that binds to the protein is retained. The former can identify open chromatin based on the presence of nuclease sensitive sites, while the latter directly identifies the protein–DNA-binding regions. FAIRE-seq uses formaldehyde to fix the exposed DNA in the chromatin, ultrasound to disrupt the chromatin and phenolic chloroform extraction to separate the fragmented DNA. FAIRE-seq uses only physics and no enzymes, which makes the experiment relatively simple but brings more noise.
In ATAC-seq, the nuclei are isolated from cells or tissue samples after nucleoplasm separation, and the nuclear chromatin is cut by a transposase [49]. Closely wrapped chromatin DNA is not cut by the transposase, whereas open regions of chromatin DNA are randomly fragmented. The particular advantage of using a transposase is that it can cut DNA and then attach it directly to sequencing adaptors, simplifying the process and reducing artefacts and noise during the experiment. Then, the fragmented DNA is purified, a sequencing library is constructed, and the sample is sequenced by NGS. After mapping the sequenced fragments to a reference genome, regions of open chromatin can be identified. The detailed process of ATAC-seq data analysis has been described in a recent paper [50]. Briefly, a typical ATAC-seq data analysis includes six steps: (i) quality control of raw sequencing reads, (ii) alignment of the reads to a reference genome, (iii) peak calling, (iv) peak annotation, (v) differential analysis and (vi) other downstream analysis such as motif enrichment analysis (Figure 3B). As no antibodies are required for ATAC-seq, it provides an effective method to characterize epigenetic landscapes and identify potential cis-regulatory modules [51].
Compared with other technologies, ATAC-seq has the advantages of requiring fewer samples, a shorter sample preparation time and higher reliability [52]. With its incomparable advantages, ATAC-seq quickly became a useful approach for studying open regulatory regions. However, some cautions are required during this process. (i) Extra steps are often needed to remove the contamination of mitochondrial DNA, which can be done both experimentally [53, 54] and analytically [55]. (ii) Some high-throughput sequencing technologies inevitably produce errors or biases, including ATAC-seq [56]. It has been found that Tn5 transposase preferentially targets the entry-exit sites of nucleosome DNA [57], which may lead to a biased sequencing result. However, this transposase bias can be corrected by developing computational tools or improved statistical models, such as using position dependency models to correct for bias [58].
Similar to RNA-seq and ChIP-seq, ATAC-seq is also moving to the single-cell level. ATAC-seq has been integrated into a programmable microfluidics platform for measuring mammalian DNA regulatory region variation [59]. However, single-cell ATAC-seq data require more careful data analysis [60]. By integrating a single-cell assay for transposase-accessible chromatin sequencing (scATAC-seq) and scRNA-seq, differential accessibility and differential gene expression can be linked together [61]. The use of scATAC-seq and single-cell DNA methylation profiling has been discussed as an approach to exploring gene regulatory programmes [62].
In a word, the combination of epigenome and transcriptome is necessary. Fundamentally, the transcriptome is the result of transcription, posttranscriptional regulation and RNA degradation, while the epigenome provides a perspective on transcriptional initiation. Moreover, gene regulatory networks characterized by the combination of active TFs and their target genes can result in cell type-specific transcriptional states that determine cell heterogeneity. Thus, single-cell multi-omics will assist us in better understanding cellular identity and developmental pathways.
Application of multi-omics analysis in different species
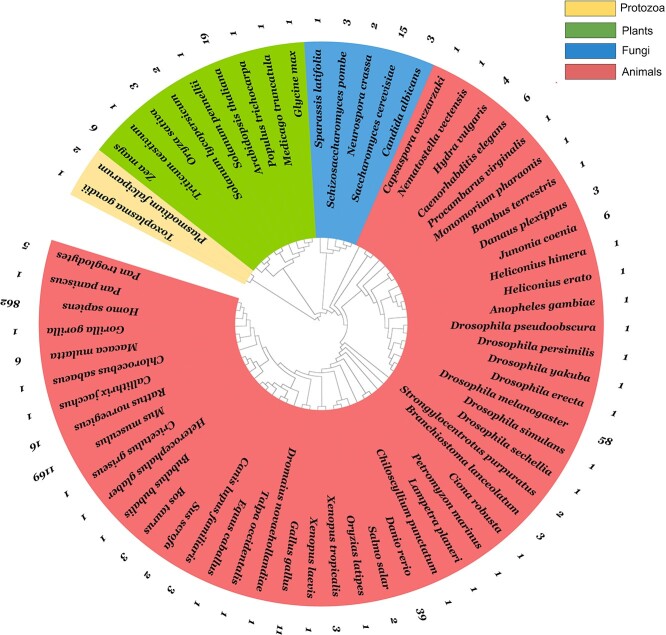
A large amount of ATAC-seq data have been produced from a variety of species since the first use of ATAC-seq in 2013. We collected species information from the Gene Expression Omnibus (GEO) of the National Center for Biotechnology Information from experiments that used ATAC-seq and this resulted in more than 2000 datasets from 65 species. Then, we built a phylogenetic tree based on evolutionary relationships among these species. There were four clades from the tree, which represented four kingdoms: animals, plants, fungi and protozoa (Figure 4). The tree included 49 animals, Mus musculus, Homo sapiens, Drosophila melanogaster, Danio rerio, Rattus norvegicus, Macaca mulatta, etc.; 9 plant species, Zea mays, Triticum aestivum, Oryza sativa, Arabidopsis thaliana, Populus trichocarpa, etc.; 5 fungal species Saccharomyces cerevisiae, Neurospora crassa, etc.; and only 2 (Toxoplasma gondii and Plasmodium falciparum) protozoa.
Figure 4.
Phylogenetic tree of 65 species to which ATAC-seq data are applied. The four colours correspond to four kingdoms: protozoa, animals, plants and fungi. The outer circle indicates the number of GEO experiments for each species.
Given that M. musculus and H. sapiens mainly account for approximately 90% of the total data, while most of the other species are still in the process of accumulating data (mainly mapping the chromatin accessibility landscape because the functional control elements in their genomes are less well annotated), we summarized and listed the discoveries as shown below.
Examples in human diseases
Epigenetic alterations have been considered to be one of the causes of cancer [63]. Cancer studies including pancreatic cancer, liver cancer and bladder cancer, which used ATAC-seq and other sequencing techniques such as RNA-seq and ChIP-seq, have discovered novel regulatory mechanisms and pointed out that the regulation of epigenetics might be a promising anticancer target. In leukaemia, one recent study reviewed the role of the Ikaros protein [64]. The use of ChIP-seq, RNA-seq, ATAC-seq and other functional experiments revealed that Ikaros regulates both the global epigenomic landscape and the epigenetic signature in the promoter regions of its target genes. In addition, public datasets have provided a growing number of practical data for multi-omics analysis. One study obtained public data from the Cancer Genome Atlas and integrated different sequencing data across 23 cancer types [65]. They attempted to use ATAC-seq to assist in the identification of distal regulatory elements and promoted the classification of cancer types. One integration of RNA-seq and ATAC-seq revealed factors related to motif protection and nucleosome repositioning. Another integration of RNA-seq and ATAC-seq with paired datasets provided a quantitative model linking the accessibility of a regulatory element to the expression of the predicted target genes. Finally, whole-genome sequencing (WGS-seq) and ATAC-seq were used to identify somatic mutations that occur in regulatory regions, leading to significant increases in chromatin accessibility. In addition to cancer, changes in the chromatin landscape have been studied in several types of diseases, such as type 2 diabetes, Alzheimer’s disease, osteoarthritis, coronary artery disease, rheumatoid arthritis, Parkinson’s disease and scleroderma (Supplementary Table 1, see Supplementary Data available online at https://academic.oup.com/bib). These findings provide key insights into the pathogenic mechanisms and therapeutic targets. Meanwhile, epiCOLOC, a web portal that collected tens of thousands of bulk or single-cell epigenome analysis covering 53 human tissue/cell types, may be able to help explain biological problems on chromatin accessibility and transcriptional event [66].
Examples in other animal species
A large amount of epigenetic data have been produced in a variety of animals in addition to humans. In mice, a comprehensive view of chromatin dynamics during mouse foetal development has been provided by performing 1128 ChIP-seq analyses for histone modifications and 132 ATAC-seq analyses for chromatin accessibility in the Encyclopedia of DNA Elements project [67]. A large amount of information can be obtained by fully interpreting these data, e.g. the chromatin state can be modelled based on combinatorial patterns of histone modifications, characterization of the spatial and temporal dynamics of chromatin states, polycomb-mediated repression and changes in chromatin accessibility are all consistent with changes in enhancer chromatin status and often precede changes in nearby H3K27Ac levels. This offers us a standard use of multi-omics data in genome annotation. In zebrafish, to generate a comprehensive map of transcriptomes, cis-regulatory elements (CREs), heterochromatin, methylomes, 3D genome organization, RNA-seq, ATAC-seq, ChIP-seq, whole-genome bisulphite sequencing and Hi-C have been performed [68]. In mosquitoes, techniques such as ATAC-seq have provided important insights into chromatin structure and regulation, and the association of chromatin-associated proteins and 3D genome structure has been reviewed [69]. For example, by integrating ATAC-seq and RNA-seq in Anopheles gambiae tissues, an accurate genome-wide map of CREs was mapped, and the in vivo binding sites of related TFs were predicted. Additional works, such as gene editing techniques and ChIP-seq experiments, are still needed to confirm these CREs. Moreover, they also mapped the transcription regulation mechanism of mosquitoes, and the corresponding chromatin structure with different sequencing results, such as the transcription level measured by RNA-seq, chromatin accessibility level measured by ATAC-seq and histone modification level measured by ChIP-seq.
Chromatin accessibility data can also be compared between species. Researchers performed genome-wide assessments of accessible chromatin regions during embryogenesis in three vertebrates (mouse, chicken and medaka) and estimated the evolutionary ages of these regions to determine their evolutionary origins in the phylogenetic tree [70]. The use of whole-embryo ATAC-seq data provided a reliable landscape of chromatin accessibility, by which they found that genomic regions tend to become accessed in a similar order to the phylogenetic history, with new gene regulation activated at later developmental stages.
Examples in plants
Chromatin accessibility has been extensively explored in plants. The integration of single-nucleus RNA sequencing and single-nucleus assays for transposase accessible chromatin sequencing data demonstrated that cell-type-specific marker genes display cell-type-specific patterns of chromatin accessibility in Arabidopsis roots. The data also suggested that differential chromatin accessibility is a critical mechanism regulating gene activity at the cell-type level [71]. In addition to model plants, there have been studies of the chromatin structure of crops. Using Hi-C, RNA-seq and ATAC-seq, it has been shown that chromatin loops act as components of the gene regulation machinery in maize, with a restricted number of chromatin loop anchors as the core structural unit [72]. The colocalization analysis of ATAC-seq nucleosome-free peaks with chromatin loops showed that among 19 532 peaks, up to 13 026 overlapped primarily with anchors. Another study examined the nuclear architecture of hexaploid wheat by using RNA-seq, ChIP-seq, ATAC-seq, Hi-C and in situ Hi-C followed by chromatin immunoprecipitation (Hi-ChIP) data [73]. They provided some new insights into the physical chromosome organization of a polyploid genome and the key factors governing gene transcription in polyploids. They analyzed the relationship between histone modifications, chromatin accessibility and chromatin interaction. The results showed that while the interaction of genes with neighbouring sequences was generally weak, genes with neutral histone marks had particularly little physical contact with surrounding regions.
Examples in fungi and protozoa
In fungi and protozoa, the study of environmental stimuli is particularly important. The first integrative analysis of ATAC-seq and RNA-seq was performed in macrofungi [74]. These results provide a new perspective on the regulation of key pathways and hub genes in light-induced primordia formation (LIPF) of Sparassis latifolia. In particular, the integration of ATAC-seq and RNA-seq identified 30 genes in LIPF. An ATAC-seq protocol adapted for the opportunistic pathogen Candida albicans has been presented and used to gain further insight into the interplay of chromatin accessibility and gene expression during fungal adaptation to oxidative stress [75]. Chromatin accessibility is increased promoters associated with oxidative stress genes, including CAT1, ICL1, OYE32 and TSA1. ATAC-seq read signals were found to be increased around the Cap1 motif, which is a key driver of transcriptional induction during the oxidative stress response. This indicates that ATAC-seq data are suitable for motif discovery. In summary, the use of ATAC-seq enabled the identification of chromatin signatures and the dynamics of regulatory mechanisms mediating the environmental adaptation of C. albicans. In addition, ATAC-seq and RNA-seq were used to study the cnidarian-dinoflagellate model Exaiptasia pallida (Aiptasia). This study revealed the role of chromatin dynamics in response to thermal stress [76]. They identified the characteristic patterns of accessibility, which contained 1309 genomic sites. Through the analysis of TF motifs, they also found the expressed genes related to immunological pathways and oxidative stress pathways.
Taken together, these studies have indicated that ATAC-seq can play a powerful and effective role in the study of disease mechanisms, molecular targeting, chromatin accessibility mapping, chromatin structure detection and environmental pressure on organisms.
ATAC-seq data analysis and pipelines
As we have shown above, ATAC-seq provides an efficient tool to obtain epigenetic information and explore the transcriptional regulation mechanism, and it has demonstrated broad application prospects. Analysing ATAC-seq data would require a certain degree of computational and programming skills, knowledge of command line operations under the Unix-based or Linux-based operating system, and statistical knowledge. However, biologists often lack these skills, which makes it difficult to fully understand and interpret ATAC-seq results.
Recently, several pipelines, programmes and websites have been developed to help researchers analyze and visualize ATAC-seq data. Single-cell ATAC-seq pipelines have already been discussed in another recent review paper entitled ‘Profiling Chromatin Accessibility at Single-cell Resolution’ [77], which has collected 19 bioinformatics tools for scATAC-seq analysis, discussing the algorithms, key features, limitations and platforms for each tool. Therefore, we focused more on the bulk ATAC-seq data analysis pipelines and summarized 16 existing analysis tools (Table 1). We collected information for each pipeline, including its data type, function, advantages, programming languages, the website for download and platform. Among these 16 pipelines, 7 pipelines were used for ATAC-seq data analysis only and 9 for multi-omics data analysis, such as ATAC-seq, DNase-seq and ChIP-seq. Most of the pipelines start from quality control and end with differential analysis, but some focus only on one or two steps, such as Ataqv [78] for quality control or MMARGE [79] for TF and motif analysis. To facilitate downloading by researchers, the websites for each software package are shown in Table 1.
Table 1.
Pipelines for bulk ATAC-seq data analysis
| Name | Data type | Input format | Function | Advantages | Download website | Language | Platform |
|---|---|---|---|---|---|---|---|
| ALTRE [80] | ATAC-seq | CSV BAM BED |
(i) Peak merging and annotation, (ii) differential analysis, (iii) pathway enrichment analysis | (i) Easy-to-use (ii) Allows for parameters change (iii) A shiny website platform |
https://github.com/Mathelab/ALTRE | R | Windows, Linux, MacOS |
| ATAC-pipe [84] | ATAC-seq | FASTQ | (i) QC, (ii) alignment, (iii) peak calling, (iv) differential analysis, (v) search for motifs, (vi) TF footprinting, (vii) Regulatory network reconstruction | (i) Integrated pipeline with multiple toolkits (ii) High-quality figures |
https://github.com/QuKunLab/ATAC-pipe | Python | Linux, MacOS |
| atacR [81] | ATAC-seq | BAM CSV |
(i) Normalization, (ii) differential analysis | (i) Allows for normalization (ii) Diagnostic plots |
https://github.com/TeamMacLean/atacr | R | Platform independent |
| Ataqv [78] | ATAC-seq | BAM | (i) QC with visualization | (i) Diverse QC metrics (ii) Quick and convenient visualization |
https://github.com/ParkerLab/ataqv/ | C++ | Linux, MacOS |
| CIPHER [88] | ATAC-seq ChIP-seq RNA-seq MNase-seq DNase-seq GRO-seq |
FASTQ | (i) QC, (ii) alignment, (iii) peak calling, (iv) peak annotation and visualization, (v) differential analysis, (vi) motif identification | (i) Stand-alone workflow platform (ii) Integrates multiple NGS data (iii) Extensive and detailed QC reports |
https://github.com/c-guzman/cipher-workflow-platform | Nextflow, R | Linux, MacOS |
| COCOA [95] | ATAC-seq DNA methylation data |
Counts matrix | (i) quantify inter-sample variation (ii) annotating variations |
(i) Supports supervised and unsupervised analysis (ii) Integrated with multi-omics analyses |
http://bioconductor.org/packages/COCOA | R | Windows, MacOS |
| DEBrowser [86] | ATAC-seq RNA-seq |
Counts matrix | (i) QC, (ii) differential analysis, (iii) pathway analysis | (i) A shiny website platform (ii) Interactive analysis (iii) Diverse plots |
https://bioconductor.org/packages/release/bioc/html/debrowser.html | R | Platform independent |
| diffTF [96] | ATAC-seq ChIP-seq RNA-seq |
BAM BED |
(i) calculate differential TF activity, (ii) classify TF with RNA-seq data | Classify TF into activators or repressors | https://git.embl.de/grp-zaugg/diffTF | Snakemake | Cluster system |
| esATAC [82] | ATAC-seq | FASTQ | (i) QC, (ii) alignment, (iii) peak calling, (iv) peak annotation, (v) motif analysis | (i) Perform ‘one command line for results’ analysis (ii) Maximize memory control and parallel computing |
https://www.bioconductor.org/packages/release/bioc/html/esATAC.html | R and C++ | Windows, Linux, MacOS |
| GUAVA [89] | ATAC-seq | FASTQ | (i) QC, (ii) alignment, (iii) peak calling, (iv) peak annotation, (v) differential analysis, (vi) functional annotation | (i) Standalone software (ii) Diverse plots |
https://github.com/MayurDivate/GUAVA | JAVA | Linux, MacOS |
| I-ATAC [83] | ATAC-seq ChIP-seq WGS-seq |
FASTQ | (i) QC, (ii) alignment, (iii) peak calling | (i) Standalone software (ii) Easy-to-use (iii) One-click operation |
https://github.com/UcarLab/I-ATAC | JAVA | UNIX, Linux, Windows, MacOS |
| MMARGE [79] | ATAC-seq ChIP-seq DNase-seq |
FASTQ VCF |
(i) alignment, (ii) TF and motif analysis | Identify combinations of TFs | https://github.com/vlink/marge | Perl and R | UNIX |
| Octopus-toolkit [90] | ATAC-seq ChlP-seq DNase-seq MeDIP-seq MNase-seq RNA-seq |
SRA FASTQ |
(i) QC, (ii) alignment, (iii) peak calling, (iv) peak annotation, (v) motif analysis | (i) Standalone software (ii) Retrieving SRA files from the GEO database (iii) Provides several model genomes (iv) Allows meta-analysis |
https://github.com/kangk1204/Octopustoolkit2 | JAVA | Linux, MacOS |
| Recoup [85] | ATAC-seq ChIP-seq RNA-seq |
BAM BED BIGWIG |
(i) Signal normalization , (ii) coverage profile analysis | (i) High-quality figures (ii) Fast and quick output |
https://bioconductor.org/packages/recoup | R | Linux, Windows, MacOS |
| snakePipes [87] | ATAC-seq ChIP-seq RNA-seq Bisulfite-seq Hi-C RNA-seq |
FASTQ BAM |
(i) QC, (ii) alignment, (iii) peak calling, (iv) differential analysis | (i) Integrates multiple NGS data, (ii) Extensive and detailed QC reports (iii) Allows for parameters change |
https://github.com/maxplanck-ie/snakepipes | Snakemake | Cluster system |
| TOBIAS [97] | ATAC-seq | BAM FASTA PWM |
(i) Bias correction, (ii) footprint analysis, (iii) differential analysis | (i) Focus on footprint analysis (ii) Time-series data analysis (iii) Data visualization within one framework |
https://github.com/loosolab/TOBIAS | Python | Cluster system |
The advantages of the pipelines are summarized in Table 1. For example, some pipelines are easy to use (ALTRE, atacR, esATAC, I-ATAC) [80–83]; some produce high-quality plots (ATAC-pipe, Recoup) [84, 85]; some allow for interactive analysis, which allows users to iteratively modify parameters (DEBrowser, snakePipes) [86, 87] and some are stand-alone software (CIPHER, GUAVA, Octopus-toolkit) [88–90] that integrate several tools into one package. However, there are also disadvantages to these pipelines. Most of the tools require the input files as the raw reads file, of which the file size can reach up to hundreds of gigabytes for a single experiment. Therefore, users may not be able to analyze the data on a laptop or workstation. In addition, some pipelines are designed for specific purposes. To analyze all of the steps of ATAC-seq, users may need to choose other tools, and how to merge the results from different platforms represents another problem. Some programs integrate several tools into one package, which would require large computational resources such as multiple CPUs and large memories.
Overall, we listed the function of each pipeline, which data type the pipeline can analyze, and the advantages for each pipeline. Therefore, we suggest that biologists choose the most efficient pipeline according to their experimental purposes.
Conclusion
The emergence of ATAC-seq provides a new perspective for understanding biological mechanisms in terms of chromatin accessibility. It can construct different models associated with various omics approaches according to the actual problems and be combined with different technologies. For the applications, we mentioned above, ATAC-seq provides chromatin accessibility maps when used alone, insight into gene regulation mechanisms when integrated with ChIP-seq and RNA-seq, and a description of chromatin macrostructure when integrated with Hi-C. Different schemes have been applied to different organisms, aiming to focus on disease treatment and species research.
However, the integration of different sequencing methods and results is still challenging. There are several problems: (i) high cost for multi-omics sequencing; (ii) difficulty in unifying or standardizing the results of different techniques and (iii) multiple sequencing cannot be applied simultaneously to the same cells, but the batch effect and interference from different experiments may influence the sequencing results.
New sequencing technologies are emerging, and current research using these new sequencing methods aims to explain the process of gene expression at different levels and expand our view of biological problems. Examples of new methods include RNA immunoprecipitation sequencing (RIP-seq) is similar to ChIP-seq but detects interactions between proteins and RNA [91], and RNA hybrid and individual-nucleotide resolution UV cross-linking and immunoprecipitation which detects protein–RNA interactions [92]. In particular, ribosome profiling (Ribo-seq) provides a complete description of the translation process by directly studying the ribosome-protected mRNA fragments that are being translated [93]. It can capture the speed of RNA translation and provide insights into protein expression. Another emerging technology, cleavage under targets and tagging (CUT and Tag), is targeted at studying the interaction between DNA and protein using transposase [94]. Compared with traditional ChIP-seq, it eliminates the need for cross-linking, improves the signal-to-noise ratio and reduces the number of cells needed.
Overall, the current sequencing technology is still in its early days and will be further refined and integrated to provide a detailed temporal and spatial perspective on gene regulation. We are also expecting the development of new analytics software and pipelines with better compatibility, flexible parameter settings and higher levels of visualization. As we continue to explore the mechanism of gene regulation, we will be able to describe life processes in full detail throughout the cell cycle, and our understanding of life will reach levels that have never been achieved before.
Key Points
We reviewed the literature on ATAC-seq and described the characteristics of ATAC-seq publications, then introduced the principles of the RNA-seq, ChIP-seq and ATAC-seq, focusing on the main features of techniques.
We built a phylogenetic tree from species that had been studied using ATAC-seq. It has been found that M. musculus and H. sapiens mainly account for about 90% of the total ATAC-seq data, while most other species are still in the process of accumulating data stage. We summarized the findings from human diseases and other species, illustrating the cutting-edge discoveries and the role of multi-omics data analysis in current research.
We collected and compared ATAC-seq analysis pipelines, which allowed biological researchers who lack programming skills to better analyze and explore ATAC-seq data.
Through this review, it is clear that multi-omics analysis and single-cell sequencing technology will become mainstream techniques in future research.
Supplementary Material
Author Biographies
Liheng Luo is a fourth-year undergraduate student, major in Biotechnology in School of Life Sciences at Northwestern Polytechnical University.
Michael Gribskov is a professor in Department of Biological Sciences at Purdue University. He received his PhD from University of Wisconsin-Madison. His research interests focus on computational genomics, systems biology and bioinformatics.
Sufang Wang is a associate professor in School of Life Sciences at Northwestern Polytechnical University. She received her PhD from Purdue University. Her research interests are primarily in genomics, transcriptomics and genome-wide association study.
Authors’ contributions
L.L. and S.W. wrote the manuscript and M.G. revised manuscript.
Funding
National Natural Science Foundation of China (31800781).
References
- 1. Green ED, Gunter C, Biesecker LG, et al. . Strategic vision for improving human health at the forefront of genomics. Nature 2020;586:683–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Richmond TJ, Davey CA. The structure of DNA in the nucleosome core. Nature 2003;423:145–50. [DOI] [PubMed] [Google Scholar]
- 3. Zhou BR, Bai Y. Chromatin structures condensed by linker histones. Essays Biochem 2019;63:75–87. [DOI] [PubMed] [Google Scholar]
- 4. Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell 2007;128:707–19. [DOI] [PubMed] [Google Scholar]
- 5. Maston GA, Evans SK, Green MR. Transcriptional regulatory elements in the human genome. Annu Rev Genom Hum Genet 2006;7:29–59. [DOI] [PubMed] [Google Scholar]
- 6. Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology 2013;38:23–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Goll MG, Bestor TH. Histone modification and replacement in chromatin activation. Genes Dev 2002;16:1739–42. [DOI] [PubMed] [Google Scholar]
- 8. Tsukiyama T, Wu C. Chromatin remodeling and transcription. Curr Opin Genet Dev 1997;7:182–91. [DOI] [PubMed] [Google Scholar]
- 9. Schmidt D, Wilson MD, Spyrou C, et al. . ChIP-seq: using high-throughput sequencing to discover protein-DNA interactions. Methods 2009;48:240–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Robertson G, Hirst M, Bainbridge M, et al. . Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Method 2007;4:651–7. [DOI] [PubMed] [Google Scholar]
- 11. Park PJ. ChIP-seq: advantages and challenges of a maturing technology. Nat Rev Genet 2009;10:669–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Buenrostro JD, Giresi PG, Zaba LC, et al. . Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Method 2013;10:1213–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Buenrostro JD, Wu B, Chang HY, et al. . ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr Protoc Mol Biol 2015;109:29.21–1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhou W, Ji Z, Fang W, et al. . Global prediction of chromatin accessibility using small-cell-number and single-cell RNA-seq. Nucleic Acid Res 2019;47:e121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hawkins RD, Hon GC, Ren B. Next-generation genomics: an integrative approach. Nat Rev Genet 2010;11:476–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Thieffry D, Sarkar S. Forty years under the central dogma. Trends Biochem Sci 1998;23:312–6. [DOI] [PubMed] [Google Scholar]
- 17. Yohe S, Thyagarajan B. Review of clinical next-generation sequencing. Arch Pathol Lab Med 2017;141:1544–57. [DOI] [PubMed] [Google Scholar]
- 18. Hardwick SA, Deveson IW, Mercer TR. Reference standards for next-generation sequencing. Nat Rev Genet 2017;18:473–84. [DOI] [PubMed] [Google Scholar]
- 19. Dijk EL, Auger H, Jaszczyszyn Y, et al. . Ten years of next-generation sequencing technology. Trends Genet 2014;30:418–26. [DOI] [PubMed] [Google Scholar]
- 20. Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 2009;10:57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chatterjee A, Ahn A, Rodger EJ, et al. . A guide for designing and analyzing RNA-Seq data. Methods Mol Biol 2018;1783:35–80. [DOI] [PubMed] [Google Scholar]
- 22. Conesa A, Madrigal P, Tarazona S, et al. . A survey of best practices for RNA-seq data analysis. Genome Biol 2016;17:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Marguerat S, Bahler J. RNA-seq: from technology to biology. Cell Mol Life Sci 2010;67:569–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stark R, Grzelak M, Hadfield J. RNA sequencing: the teenage years. Nat Rev Genet 2019;20:631–56. [DOI] [PubMed] [Google Scholar]
- 25. Choi JR, Yong KW, Choi JY, et al. . Single-cell RNA sequencing and its combination with protein and DNA analyses. Cell 2020;9:1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hwang B, Lee JH, Bang D. Single-cell RNA sequencing technologies and bioinformatics pipelines. Exp Mol Med 2018;50:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Birnbaum KD. Power in numbers: single-cell RNA-Seq strategies to dissect complex tissues. Annu Rev Genet 2018;52:203–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Haque A, Engel J, Teichmann SA, et al. . A practical guide to single-cell RNA-sequencing for biomedical research and clinical applications. Genome Med 2017;9:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wu Y, Zhang K. Tools for the analysis of high-dimensional single-cell RNA sequencing data. Nat Rev Nephrol 2020;16:408–21. [DOI] [PubMed] [Google Scholar]
- 30. Gonzalez-Silva L, Quevedo L, Varela I. Tumor functional heterogeneity unraveled by scRNA-seq technologies. Trend Cancer 2020;6:13–9. [DOI] [PubMed] [Google Scholar]
- 31. Luo G, Gao Q, Zhang S, et al. . Probing infectious disease by single-cell RNA sequencing: progresses and perspectives. Comput Struct Biotechnol J 2020;18:2962–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Picelli S. Single-cell RNA-sequencing: the future of genome biology is now. RNA Biol 2017;14:637–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mahony S, Pugh BF. Protein-DNA binding in high-resolution. Crit Rev Biochem Mol Biol 2015;50:269–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Landt SG, Marinov GK, Kundaje A, et al. . ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res 2012;22:1813–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fosslie M, Manaf A, Lerdrup M, et al. . Going low to reach high: small-scale ChIP-seq maps new terrain. Wiley Interdiscip Rev Syst Biol Med 2020;12:e1465. [DOI] [PubMed] [Google Scholar]
- 36. Mundade R, Ozer HG, Wei H, et al. . Role of ChIP-seq in the discovery of transcription factor binding sites, differential gene regulation mechanism, epigenetic marks and beyond. Cell Cycle 2014;13:2847–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jiang C, Pugh BF. Nucleosome positioning and gene regulation: advances through genomics. Nat Rev Genet 2009;10:161–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yan H, Tian S, Slager SL, et al. . ChIP-seq in studying epigenetic mechanisms of disease and promoting precision medicine: progresses and future directions. Epigenomics 2016;8:1239–58. [DOI] [PubMed] [Google Scholar]
- 39. Pepke S, Wold B, Mortazavi A. Computation for ChIP-seq and RNA-seq studies. Nat Method 2009;6:S22–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hollbacher B, Balazs K, Heinig M, et al. . Seq-ing answers: current data integration approaches to uncover mechanisms of transcriptional regulation. Comput Struct Biotechnol J 2020;18:1330–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cavalli G, Heard E. Advances in epigenetics link genetics to the environment and disease. Nature 2019;571:489–99. [DOI] [PubMed] [Google Scholar]
- 42. Zhao LY, Song J, Liu Y, et al. . Mapping the epigenetic modifications of DNA and RNA. Protein Cell 2020;11:792–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chang P, Gohain M, Yen MR, et al. . Computational methods for assessing chromatin hierarchy. Comput Struct Biotechnol J 2018;16:43–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gouil Q, Keniry A. Latest techniques to study DNA methylation. Essays Biochem 2019;63:639–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Berkum NL, Lieberman-Aiden E, Williams L, et al. . Hi-C: a method to study the three-dimensional architecture of genomes. J Vis Exp 2010;6:1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Crawford GE, Holt IE, Whittle J, et al. . Genome-wide mapping of DNase hypersensitive sites using massively parallel signature sequencing (MPSS). Genome Res 2006;16:123–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cui K, Zhao K. Genome-wide approaches to determining nucleosome occupancy in metazoans using MNase-Seq. Method Mol Biol 2012;833:413–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Giresi PG, Kim J, McDaniell RM, et al. . FAIRE (formaldehyde-assisted isolation of regulatory elements) isolates active regulatory elements from human chromatin. Genome Res 2007;17:877–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li N, Jin K, Bai Y, et al. . Tn5 transposase applied in genomics research. Int J Mol Sci 2020;21:8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yan F, Powell DR, Curtis DJ, et al. . From reads to insight: a hitchhiker’s guide to ATAC-seq data analysis. Genome Biol 2020;21:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Suryamohan K, Halfon MS. Identifying transcriptional cis-regulatory modules in animal genomes. Wiley Interdiscip Rev Dev Biol 2015;4:59–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sun Y, Miao N, Sun T. Detect accessible chromatin using ATAC-sequencing, from principle to applications. Hereditas 2019;156:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Corces MR, Trevino AE, Hamilton EG, et al. . An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat Methods 2017;14:959–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rickner HD, Niu SY, Cheng CS. ATAC-seq assay with low mitochondrial DNA contamination from primary human CD4+ T lymphocytes. J Vis Exp 2019;145:e59120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ou J, Liu H, Yu J, et al. . ATACseqQC: a Bioconductor package for post-alignment quality assessment of ATAC-seq data. BMC Genomics 2018;19:169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wang JR, Quach B, Furey TS. Correcting nucleotide-specific biases in high-throughput sequencing data. BMC Bioinformat 2017;18:357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sato S, Arimura Y, Kujirai T, et al. . Biochemical analysis of nucleosome targeting by Tn5 transposase. Open Biol 2019;9:190116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Li Z, Schulz MH, Look T, et al. . Identification of transcription factor binding sites using ATAC-seq. Genome Biol 2019;20:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Buenrostro JD, Wu B, Litzenburger UM, et al. . Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 2015;523:486–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Baek S, Lee I. Single-cell ATAC sequencing analysis: from data preprocessing to hypothesis generation. Comput Struct Biotechnol J 2020;18:1429–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kashima Y, Sakamoto Y, Kaneko K, et al. . Single-cell sequencing techniques from individual to multiomics analyses. Exp Mol Med 2020;52:1419–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Fiers M, Minnoye L, Aibar S, et al. . Mapping gene regulatory networks from single-cell omics data. Brief Funct Genom 2018;17:246–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Jones PA, Issa J-PJ, Baylin S. Targeting the cancer epigenome for therapy, nature reviews. Genetics 2016;17:630–41. [DOI] [PubMed] [Google Scholar]
- 64. Gowda C, Song C, Ding Y, et al. . Cellular signaling and epigenetic regulation of gene expression in leukemia. Adv Biol Regul 2020;75:100665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Corces MR, Granja JM, Shams S, et al. . The chromatin accessibility landscape of primary human cancers. Science 2018;362:eaav1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhou Y, Sun Y, Huang D, et al. . epiCOLOC: integrating large-scale and context-dependent epigenomics features for comprehensive colocalization analysis. Front Genet 2020;11:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Gorkin DU, Barozzi I, Zhao Y, et al. . An atlas of dynamic chromatin landscapes in mouse fetal development. Nature 2020;583:744–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Yang H, Luan Y, Liu T, et al. . A map of cis-regulatory elements and 3D genome structures in zebrafish. Nature 2020;588:337–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Lezcano OM, Sanchez-Polo M, Ruiz JL, et al. . Chromatin structure and function in mosquitoes. Front Genet 2020;11:602949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Uesaka M, Kuratani S, Takeda H, et al. . Recapitulation-like developmental transitions of chromatin accessibility in vertebrates. Zoological Lett 2019;5:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Farmer A, Thibivilliers S, Ryu KH, et al. . Single-nucleus RNA and ATAC sequencing reveals the impact of chromatin accessibility on gene expression in Arabidopsis roots at the single-cell level. Mol Plant 2021;14:372–83. [DOI] [PubMed] [Google Scholar]
- 72. Deschamps S, Crow JA, Chaidir N, et al. . Chromatin loop anchors contain core structural components of the gene expression machinery in maize. BMC Genom 2021;22:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Concia L, Veluchamy A, Ramirez-Prado JS, et al. . Wheat chromatin architecture is organized in genome territories and transcription factories. Genome Biol 2020;21:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Yang C, Ma L, Xiao D, et al. . Integration of ATAC-Seq and RNA-Seq identifies key genes in light-induced primordia formation of Sparassis latifolia. Int J Mol Sci 2019;21:185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Jenull S, Tscherner M, Mair T, et al. . ATAC-Seq identifies chromatin landscapes linked to the regulation of oxidative stress in the human fungal pathogen Candida albicans. J Fungi 2020;6:182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Weizman E, Levy O. The role of chromatin dynamics under global warming response in the symbiotic coral model Aiptasia. Commun Biol 2019;2:282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Sinha S, Satpathy AT, Zhou W, et al. . Profiling chromatin accessibility at single-cell resolution. Genom Proteom Bioinformat 2021;S1672-0229:00011–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Orchard P, Kyono Y, Hensley J, et al. . Quantification, dynamic visualization, and validation of bias in ATAC-seq data with ataqv. Cell Syst 2020;10:298–306 e294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Link VM, Romanoski CE, Metzler D, et al. . MMARGE: motif mutation analysis for regulatory genomic elements. Nucleic Acid Res 2018;46:7006–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Baskin E, Farouni R, Mathe EA. ALTRE: workflow for defining altered regulatory elements using chromatin accessibility data. Bioinformatics 2017;33:740–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Shrestha RK, Ding P, Jones JDG, et al. . A workflow for simplified analysis of ATAC-cap-seq data in R. Gigascience 2018;7:giy080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Wei Z, Zhang W, Fang H, et al. . esATAC: an easy-to-use systematic pipeline for ATAC-seq data analysis. Bioinformatics 2018;34:2664–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Ahmed Z, Ucar D. I-ATAC: interactive pipeline for the management and pre-processing of ATAC-seq samples. PeerJ 2017;5:e4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Zuo Z, Jin Y, Zhang W, et al. . ATAC-pipe: general analysis of genome-wide chromatin accessibility. Brief Bioinform 2019;20:1934–43. [DOI] [PubMed] [Google Scholar]
- 85. Moulos P. Recoup: flexible and versatile signal visualization from next generation sequencing. BMC Bioinformatics 2021;22:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Kucukural A, Yukselen O, Ozata DM, et al. . DEBrowser: interactive differential expression analysis and visualization tool for count data. BMC Genom 2019;20:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Bhardwaj V, Heyne S, Sikora K, et al. . snakePipes: facilitating flexible, scalable and integrative epigenomic analysis. Bioinformatics 2019;35:4757–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Guzman C, D'Orso I. CIPHER: a flexible and extensive workflow platform for integrative next-generation sequencing data analysis and genomic regulatory element prediction. BMC Bioinformat 2017;18:363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Divate M, Cheung E. GUAVA: a graphical user interface for the analysis and visualization of ATAC-seq data. Front Genet 2018;9:250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Kim T, Seo HD, Hennighausen L, et al. . Octopus-toolkit: a workflow to automate mining of public epigenomic and transcriptomic next-generation sequencing data. Nucleic Acid Res 2018;46:e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Zhao J, Ohsumi TK, Kung JT, et al. . Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell 2010;40:939–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Sugimoto Y, Chakrabarti AM, Luscombe NM, et al. . Using hiCLIP to identify RNA duplexes that interact with a specific RNA-binding protein. Nat Protoc 2017;12:611–37. [DOI] [PubMed] [Google Scholar]
- 93. Ingolia NT, Ghaemmaghami S, Newman JR, et al. . Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 2009;324:218–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Kaya-Okur HS, Wu SJ, Codomo CA, et al. . CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat Commun 2019;10:1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Lawson JT, Smith JP, Bekiranov S, et al. . COCOA: coordinate covariation analysis of epigenetic heterogeneity. Genome Biol 2020;21:240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Berest I, Arnold C, Reyes-Palomares A, et al. . Quantification of differential transcription factor activity and multiomics-based classification into activators and repressors: diffTF. Cell Rep 2019;29:3147–3159 e3112. [DOI] [PubMed] [Google Scholar]
- 97. Bentsen M, Goymann P, Schultheis H, et al. . ATAC-seq footprinting unravels kinetics of transcription factor binding during zygotic genome activation. Nat Commun 2020;11:4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.