Abstract
As an oncogenic transcription factor, Yin Yang 1 (YY1) regulates enhancer and promoter connection. However, gaps still exist in understanding how YY1 coordinates coactivators and chromatin enhancer elements to assemble enhancers and super-enhancers. Here, we demonstrate that a histidine cluster in YY1’s transactivation domain is essential for its formation of phase separation condensates, which can be extended to additional proteins. The histidine cluster is also required for YY1-promoted cell proliferation, migration, clonogenicity and tumor growth. YY1-rich nuclear puncta contain coactivators EP300, BRD4, MED1 and active RNA polymerase II, and colocalize with histone markers of gene activation, but not that of repression. Furthermore, YY1 binds to the consensus motifs in the FOXM1 promoter to activate its expression. Wild-type YY1, but not its phase separation defective mutant, connects multiple enhancer elements and the FOXM1 promoter to form an enhancer cluster. Consistently, fluorescent in situ hybridization (FISH) assays reveal the colocalization of YY1 puncta with both the FOXM1 gene locus and its nascent RNA transcript. Overall, this study demonstrates that YY1 activates target gene expression through forming liquid-liquid phase separation condensates to compartmentalize both coactivators and enhancer elements, and the histidine cluster of YY1 plays a determinant role in this regulatory mechanism.
INTRODUCTION
Gene expression mediated by RNA polymerase II (Pol II) is a complex but well-regulated biological process. Transcription factors (TFs) and cofactors constitute the transcription machinery that recognizes specific elements on target promoters and coordinates concerted Pol II action (1,2). Among hundreds of TFs, many master TFs can establish enhancers or super-enhancers through associating with coactivators, such as mediator and BRD4, to activate key cell identity genes (3). During malignant transformation, deregulated TFs, especially when highly expressed, promote oncogenic signaling pathways and processes, and thus serve as both key regulators of cancer development and promising therapeutic targets (4,5). A TF protein generally consists of a DNA-binding domain(s) (DBD) and one or more activation domains (ADs). Many DBDs have been well-classified to have conserved sequences and structural motifs shared by different families of TFs, but structural features of ADs are much less well characterized (6,7). Recent studies revealed that multiple molecules of a master TF utilize its DBD to bind a target promoter and different enhancers, and simultaneously use the AD to recruit various coactivators, leading to the formation of an enhancer or super-enhancer complex in the spatial vicinity of transcription start sites (TSSs) of a target gene. A TF capable of using this mechanism typically contains intrinsically disordered regions (IDRs) in its AD(s), which allows it to form phase-separated droplets (8–13).
YY1 was first identified as a negative regulator of p53 by us and others in 2004 (14,15). Since then, the proliferative role of YY1 in oncogenesis has been frequently reported, and its increasing expression in tumor cells and tissues has been demonstrated in most cancer types (16). As a master TF, YY1 regulates many cancer-associated genes through various mechanisms that have not been completely delineated (17). YY1 promotes the expression of many key TFs with oncogenic activities, such as MYC and SNAIL1 (18–21). Stimulation of these TFs may trigger positive feedback loops to augment oncogenic signals in cancer cells.
A number of master TFs can form enhancer or super-enhancer complexes that comprise multiple TF molecules to compartmentalize several enhancer elements and many coactivator molecules to relatively separated condensates, leading to the utmost activation of target genes (8,9,22). This type of regulation requires DBDs of TFs to bind target promoters and different enhancer elements, and ADs to form phase-separated condensates with coactivators (8,10). The ADs of master TFs with phase separation capability typically possess intrinsically disordered regions (IDRs) characterized by either enrichment of acidic, proline, serine, threonine, or glutamine residues in their primary sequences, or special secondary structures (8,23–26).
YY1 promiscuously interacts with numerous transcriptional coactivators, such as EP300, CBP and PRMT1 (16), and plays a pivotal role in stabilizing enhancer-promoter loops (27). However, it still remains undetermined how YY1 coordinates the coactivators and chromatin elements to assemble enhancer or super-enhancer complexes. To answer this question, we interrogate whether YY1 exerts its transcriptional activity through a phase separation mechanism. Using a variety of biochemical and cell biology approaches, we discover that YY1 possesses bona fide ability to form phase separation condensates both in vitro and in cells. The histidine cluster of YY1 is indispensable for its ability to coordinate phase separation and maintain cell proliferation. YY1-rich nuclear puncta comprise major transcription coactivators and overlap with general histone markers of gene activation. With the forkhead box protein M1 (FOXM1) as an exemplary target gene, we demonstrate that YY1 compartmentalizes coactivators and enhancer elements in phase-separated condensates to activate gene expression.
MATERIALS AND METHODS
Reagents, antibodies and plasmids
Reagents and antibodies used in this study include: YY1 (H-10) (Santa Cruz, cat# sc-7341, 1:1000 for western blot (WB), 1:300 for immunofluorescent (IF) staining); YY1 (H414) (Santa Cruz, cat# sc-1703, 1:50 for chromatin immunoprecipitation (ChIP)); FOXM1 (Thermo Fisher Scientific, cat# 702664, 1:5000 for WB); Flag (Sigma, cat# F1804, 1:300 for IF); EP300 (Cell Signaling Technology, cat# 86377, 1:500 for IF, 1:50 for ChIP); BRD4 (Santa Cruz, cat# sc-518021, 1:200 for IF, 1:50 for ChIP); MED1 (Santa Cruz, cat# sc-74475, 1:200 for IF, 1:50 for ChIP); CDK9 (Cell Signaling Technology, cat# 2316, 1:100 for IF); RNA Pol II-S2P (Millipore, cat# 04-1571, 1:200 for IF); RNA Pol II-S5P (Millipore, cat# 04-1572, 1:200 for IF); H3K4me1 (Cell Signaling Technology, cat# 5326, 1:500 for IF); H3K4me3 (Cell Signaling Technology, cat# 9751, 1:200 for IF); H3K27ac (Abcam, cat# ab4729, 1:500 for IF); H3K9me3 (Cell Signaling Technology, cat# 13969, 1:500 for IF); pAKT-Thr308 (Cell Signaling Technology, cat# 13038S, 1:1000 for WB); pAKT-Ser473 (Cell Signaling Technology, cat# 4060S, 1:1000 for WB); AKT (Cell Signaling Technology, cat# 4685S, 1:1000 for WB); Ki-67 (Thermo Fisher Scientific, cat# 710229, 1:100 for IF); GAPDH (Acton, cat# 10R-G109A, 1:1000 for WB); Goat anti-Rabbit IgG (H + L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 (Thermo Fisher Scientific, cat# A32731, 1:500 for IF); Goat anti-Mouse IgG (H + L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 594 (Thermo Fisher Scientific, cat# A32742, 1:500 for IF); Goat anti-mouse IgG-HRP (Santa Cruz, cat# sc-2005,1:5000 for WB); Goat anti-rabbit IgG-HRP (Santa Cruz, cat# sc-2004, 1:5000 for WB); JQ-1 carboxylic acid (MedChemExpress, cat# 202592-23-2); 1,6-hexanediol (Aladdin, cat# H103708).
The full-length coding sequences of YY1, HOXA1, FOXG1B, ZIC3 and HNF6 (or their variants), as well as IDRs of EP300, BRD4, MED1 and RNA Pol II, were individually subcloned into a modified version of a pGEX vector with 6 × His and EGFP or mCherry at the N-terminus. A bacterial expression system was used to express these coding sequences and recombinant proteins were purified using Ni-NTA agarose. Meanwhile, the full lengths of YY1, its mutants and EP300 were individually subcloned into a eukaryotic EGFP or mCherry expression vector. YY1, its mutants and FOXM1 coding sequences were also individually subcloned into a lentiviral vector with a 3 × Flag-tag at the N-terminus. An shRNA, sh-YY1-3′-UTR, targeting the 3′-UTR of YY1 mRNA, and a control shRNA (sh-Cont) were designed as previously described (28). The promoter of FOXM1 was amplified by a nest PCR method and subcloned upstream of the Gaussia luciferase (Gluc) coding sequence to generate a pFOXM1-prmt-Gluc vector (WT). Also, reporter constructs S1M, S2M, S3M, S4M, S5M, S4/5M, S1/3M and S2/4/5M with correspondingly mutated YY1 binding sites were generated using the ClonExpress® II Recombination system (Vazyme Biotech Co., Ltd., Nanjing, China). The predicted enhancer regions were also individually amplified by nest PCR and subcloned upstream of the FOXM1 promoter or downstream of Gluc to generate reporter constructs for enhancer elements.
Protein expression and purification
All His × 6-tagged constructs were expressed in E. coli BL21 (DE3) cells. Bacteria were grown to an optical density at 600 nm (OD600 nm) of 0.6, and induced overnight with 0.15 mM IPTG at 18°C. Bacteria were pelleted and resuspended in a lysis buffer (20 mM HEPES, 0.2 mM EDTA, 100 mM KCl, 20% glycerol, 1% Triton, 2 mM PMSF, 1 mg/ml lysozyme). After sonication, the bacterial lysate was centrifuged at 12 000 g for 30 min. His × 6-tagged proteins in the supernatant were purified by Ni-NTA agarose beads (GE Healthcare). After extensive washing using a buffer containing 20 mM imidazole, the fractions eluted by 400 mM imidazole were collected and dialyzed. The size and purity of the purified proteins were monitored by SDS-PAGE.
Cell culture, transfection, lentiviral production, and infection
HeLa, HEK-293T, U2OS, MDA-MB-231, MCF-7 and MCF-10A cells were cultured according to the protocols of the ATCC. All culture media were purchased from Gibco, and fetal bovine serum (FBS) was from ExCell Bio. Transfection of cells was carried out using Lipofectamine 2000 (Thermo Fisher Scientific, Shanghai, China) according to the manufacturer's instructions. Lentiviral production and infection followed our published procedure (29).
In vitro phase separation assay
Recombinant EGFP- or mCherry-fusion proteins were diluted to appropriate concentrations using 50 mM Tris·HCl, pH 7.4. Recombinant proteins were added to solutions containing 125 mM NaCl and 10% PEG (polyethylene glycol) 8000 as a crowding agent unless otherwise specified. Protein solution (5 μl) was immediately loaded onto a glass slide, and covered with a coverslip. Slides were then imaged with a 60× objective using the GE Delta Vision Elite (GE, Boston, MA, USA).
Turbidity experiments were performed in Eppendorf tubes. Samples (60 μl) containing appropriate concentrations of proteins, NaCl and 10% PEGs with indicated molecular weights were left to stand for 10 s at room temperature. OD600 nm was measured using BioSpectrometer basic (Eppendorf, Hamburg, Germany).
Imaging of fluorescence recovery after photo-bleaching (FRAP)
FRAP was performed on a Zeiss LSM880 microscope using a 63× oil-immersion objective. Images were acquired using the ZEN software. For FRAP of the central region of protein droplets, three iterations of bleaching were performed with a 488 nm Argon laser at a 100% power with 3 frames being acquired prior to the bleach pulse. Fluorescence recovery was recorded every 2 s for 400 s after bleaching. U2OS cells cultured in glass-bottom dishes (NEST, China) were transfected for 24 h, and analyzed using FRAP studies. Three iterations of bleaching were performed with a 488 nm Argon laser at 30% power. Fluorescence recovery was recorded every 0.8 s for 20 s after bleaching. Analyses of the fluorescence intensity of the bleached region, reference region and background region were carried out using the FRAP module in the ZEN software.
Immunofluorescence staining and live-cell imaging
Cells were seeded on glass coverslips in 12-well plates and cultured overnight. Subsequently, cells were fixed with the Immunol Staining Fix Solution (Beyotime) for 30 min at room temperature. After blocking by 10% FBS for 30 min at room temperature, cells were incubated with a primary antibody for 30 min at room temperature. After washing thrice with PBS, cells were incubated with an Alex-Fluor-488 or 594-conjugated secondary antibody (Thermo Fisher Scientific, Shanghai, China) for 30 min at room temperature. Finally, after washing thrice with PBS, nuclei were counterstained with DAPI (Beyotime), and images were captured by the GE Delta Vision Elite (GE, Boston, MA, USA).
For immunofluorescent (IF) staining of xenograft tumor samples, frozen tumorous sections (10 μm) were sliced, and sections were fixed in 4% PFA for 20 min, followed by permeabilization with 1% Triton X-100 for 15 min. After being blocked with 10% goat serum at 37°C for 1 h, sections were incubated with primary antibodies overnight at 4°C. After extensive washes with PBS, sections were incubated with an Alex-Fluor-488 conjugated secondary antibody at 37°C for 1 h. After washes with PBS, sections were counterstained with DAPI (Beyotime), and images were captured by the GE Delta Vision Elite (GE, Boston, MA, USA).
For live-cell imaging, U2OS cells were seeded on glass-bottom dishes, and transfected by the plasmids expressing EGFP or mCherry fusion proteins. After 24 h of transfection, nuclei were counterstained with Hoechst (Beyotime), and cells were imaged using the GE Delta Vision Elite (GE, Boston, MA, USA) with a 60 × objective. During image acquisition, cells were incubated in a chamber at 37°C supplied with 5% CO2.
Western blot analysis
Cells were washed and lysed in a protein lysate buffer. Total protein concentrations were measured using the Bradford protein method. Protein samples were separated on the SDS-PAGE and transferred into poly-vinylidene difluoride transfer (PVDF) membranes. The membranes were blocked by 5% nonfat milk in TBST buffer at room temperature for 1 h, and incubated with primary antibodies diluted in TBST buffer containing 1% of BSA for 4°C overnight. After three washes with TBST buffer, the membranes were incubated with corresponding secondary antibodies at room temperature for 1 h. The membranes were washed, and then visualized using an ECL kit (Vazyme Biotech Co., Ltd., Nanjing, China).
Reverse transcription and quantitative PCR (RT-qPCR)
Total RNAs were extracted from cultured cells using the TRIzol reagent (Thermo Fisher Scientific Inc., Shanghai, China), and cDNA was synthesized using the M-MLV reverse-transcriptase (Vazyme Biotech Co., Ltd., Nanjing, China). In the reaction of reverse transcription, 1 μg of total RNA was mixed with 0.5 μg/μl of oligo(dT) primer, followed by incubation at 65°C for 5 min and 4°C for 2 min. The tubes were then immediately incubated at 42°C for 30 min, and then chilled at 4°C. For qPCR, cDNA was amplified using gene specific primers and the LightCycler 480 SYBR Green PCR Master Mix (Roche, Basel, Switzerland) on the Lightcycler 480 instrument (Roche). The conditions used for qPCR were as follows: 95°C for 3 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. All reactions were performed in triplicate. The results were analyzed using the 2–ΔΔCt method and normalized using β-actin. Primer sequences for qPCR were as follows: β-actin (5′-TTCCTTCCTGGGCATGGAGT and 5′-TCTTCATTGTGCTGGGTGCC), YY1 (5′-CCCACGGTCCCAGAGTCCA and 5′-GTGTGCGCAAATTGAAGTCC), and FOXM1 (5′-GCAGGCTGCACTATCAACAA and 5′-TCGAAGGCTCCTCAACCTTA).
Chromatin immunoprecipitation (ChIP)
ChIP analysis was performed as previously reported (30). In brief, cross-linking was completed after cell culture, followed by nuclei preparation and chromatin digestion. DNA gel electrophoresis was used to confirm adequate digestion. Samples immunoprecipitated by a normal IgG and a specific antibody were purified and subjected to both semiquantitative PCR and qPCR. For semiquantitative PCR, PCR products were analyzed using gel electrophoresis. The sequences of primers were listed in Supplementary Table S1.
Electrophoretic mobility shift assay (EMSA)
Oligonucleotides labeled by Cy5 at the 5′-end were synthesized and purified using HPLC by Genewiz (Suzhou, China), and sequences of oligonucleotide probes and competitors were listed in Supplementary Table S2. The EMSA was conducted as we previously described (31). Briefly, 1 μg of purified His × 6-tagged YY1 protein was incubated with 0.5 pmol of labeled double-stranded probe in a binding buffer (250 mM HEPES, 500 mM KCl, 20 mM MgSO4 and 10 mM DTT, pH 8.0) on ice for 30 min. In the competitive binding experiments, excessive unlabeled probes were added to the binding reaction of the labeled probe and His × 6-YY1. After the binding reaction, the samples were separated by 8% native PAGE at 100 V for 50 min at 4°C. The fluorescent intensity of the bands was immediately determined by Typhoon FLA7000 (GE, Boston, MA, USA).
Luciferase reporter assay
The reporter plasmids were constructed as described above. Cells in 24-well plates were cotransfected by 250 ng of a reporter construct, 500 ng of shRNA or expression construct, and 20 ng of pCMV-SEAP (secreted alkaline phosphatase) as a control. The Gluc activity was measured at 48 h after transfection and normalized to SEAP as described previously (31).
Chromosome conformation capture (3C) experiment
The 3C analysis was performed following a previously described procedure with minor modifications (32). Total of 1 × 106 MDA-MB-231 or MCF-7 cells were cross-linked by 2% formaldehyde in 10% (v/v) FBS/PBS for 10 min at room temperature, and the reaction was stopped by adding glycine to a final concentration of 0.125 M. After washing by PBS, the fixed cells were incubated for 15 min in cold lysis buffer (10 mM Tris·HCl, pH 8.0, 10 mM NaCl, 0.2% NP-40) with 1 × complete protease inhibitor cocktail (Roche). The nuclei were harvested and resuspended in a restriction buffer (50 μl 10 × CutSmart buffer and 0.3% SDS), and incubated for 1 h at 37°C. After incubation with 1.8% Triton X-100 for 1 h at 37°C to sequester the SDS, 600 units of restriction enzyme EcoRI or HindIII (NEB) was used to digest genomic DNAs by incubating at 37°C overnight. Then, 1.6% SDS was used to inactivate the restriction enzyme by incubation at 65°C for 20 min. The solution was diluted by adding 7 ml of ligation buffer (NEB) containing 1% Triton X-100 and 30 Units of T4 DNA ligase (NEB), followed by a ligation reaction at 16°C for 4.5 h, and then at room temperature for 30 min. Cross-linking was released by Proteinase K digestion at 65°C for 16 h. Finally, DNA fragments were purified by phenol-chloroform extraction and ethanol precipitation. The ligation products were analyzed by PCR using primers located adjacent to EcoRI- or HindIII-digested sites. PCR products were cloned into a pBluescript plasmid and sequenced to verify the ligated fragments. The sequences of primers used in the 3C assay are shown in Supplementary Tables S3 and S4.
DNA fluorescent in situ hybridization (FISH) assay combined with immunofluorescence
The assays were performed as previously reported (9,33). Cells were seeded on glass coverslips in 12-well plates and cultured overnight. An immunofluorescence assay using appropriate primary and secondary antibodies was carried out following a standard procedure described above. Prior to the FISH assays, cells on coverslips were washed thrice with PBS, and fixed in 4% paraformaldehyde (PFA) for 10 min at room temperature. After three washes with PBS, cells were permeabilized serially in 70%, 85% and 100% ethanol for 1 min of each, and then treated by 0.2 M HCl for 5 min. After RNase A treatment to remove all RNAs, cells were pre-hybridized with 80% formamide in 2 × SSC at 78°C for 5 min. Two steps of hybridization were carried out, and the probes, as well as genomic DNA of the cells, were denatured at 78°C for 5 min prior to use. The DNA FISH probes targeting the FOXM1 enhancers were designed by the genetics.med.harvard.edu/oligopaints website (34), and synthesized by Synbio Technologies. Probe sequences are shown in the Supplementary Table S5. The genomic region targeted by the probes covers Chr12: 2880430–3018116.
In the first hybridization, the cells with denatured genomic DNA were incubated with a hybridization solution containing 10% dextran sulfate, 30% formamide, 2 × SSC and a final concentration of 1 mM of the first probe targeting the genomic locus, and incubated at 37°C for 1 h. After washes by PBS, the second hybridization was carried out in a solution containing a final concentration of 1 mM of a Cy5-labeled readout probe specific to a sequence of the first probe, and incubated at 37°C overnight. After the hybridization, the coverslips were washed four times with 30% formamide in 2 × SSC at 42°C for 5 min, and once with PBS at room temperature. Then, the cells were stained with DAPI and imaged with the GE Delta Vision Elite (GE, Boston, MA, USA).
RNA-FISH assay combined with immunofluorescence
Cells were first treated by the procedure of immunofluorescence experiment as described above, and then subjected to the following RNA FISH assay. A set of Stellaris FISH probes targeting the introns of the FOXM1 gene was designed using the Stellaris™ Probe Designer software (Biosearch Technologies), and the Cy5-labeled probes were synthesized by Synbio Technologies (Supplementary Table S6). RNA FISH was carried out following previously reported procedures (35,36). Briefly, cells were fixed in 4% PFA for 10 min, washed thrice with PBS, and washed once with 70% ethanol. Prior to hybridization, cells were washed for 5 min in a washing buffer containing 10% formamide and 2 × SSC. A final concentration of 12.5 μM of the probe in a hybridization buffer containing 10% dextran sulfate, 10% formamide and 2 × SSC was added to cells, and incubated overnight at 37°C. Then, cells were washed twice in the washing buffer at 37°C for 30 min, and once with 2 × SSC for 5 min. Finally, the nuclei were stained with DAPI and imaged using the GE Delta Vision Elite (GE, Boston, MA, USA).
Cell viability, colony formation, and wound healing assays
These assays were performed as previously reported (37). In these experiments, cells were infected by lentivirus expressing either shRNAs or cDNAs. Each experiment was performed in triplicate.
Mouse xenograft study
Permission for animal experiments was obtained from the Animal Care and Ethics Committee of Northeast Forestry University. The experiments were carried out according to the guidelines of the Declaration of Helsinki, and following our previously reported procedure (38). MDA-MB-231 cells (4 × 106) were first stably infected by lentivirus bearing Doxycycline (DOX)-inducible sh-YY1-3′-UTR targeting the 3′-UTR of the YY1 mRNA (38), and the cells were then infected by lentivirus carrying an empty vector, or expressing wild-type (WT) YY1 or its mutant. After these manipulations, the cells were resuspended in 100 μl PBS, and mixed with 100 μl Matrigel (BD Biosciences). The cells in a final volume of 200 μl were subcutaneously inoculated into the right flank of 5-week-old female BALB/c nude mice purchased from Beijing Weitong Lihua Experimental Animal Technology Co. Ltd. (Beijing, China). Mice were continuously provided with drinking water containing 2.0 mg/ml DOX (n = 10 mice per group). The tumor sizes were measured twice a week using a Vernier caliper, and tumor volume (V) was calculated using the formula: V = length × (width2)/2. Five weeks after the inoculation, mice were humanely euthanized, and all tumor xenografts were collected, photographed, and analyzed by immunofluorescence staining and RT-qPCR.
Statistical analysis
All data were derived from at least three independent experiments. Results were presented as a mean with either standard deviation (SD) or standard error of mean (SEM), and sample sizes are indicated unless otherwise noted in the figure legends. Statistical significance calculations comparing two conditions were performed using a two-tailed unpaired Student's t-test. The criterion of statistical significance level was denoted as follows: *P< 0.05; **P< 0.01; ***P< 0.001.
RESULTS
YY1 undergoes liquid-liquid phase separation
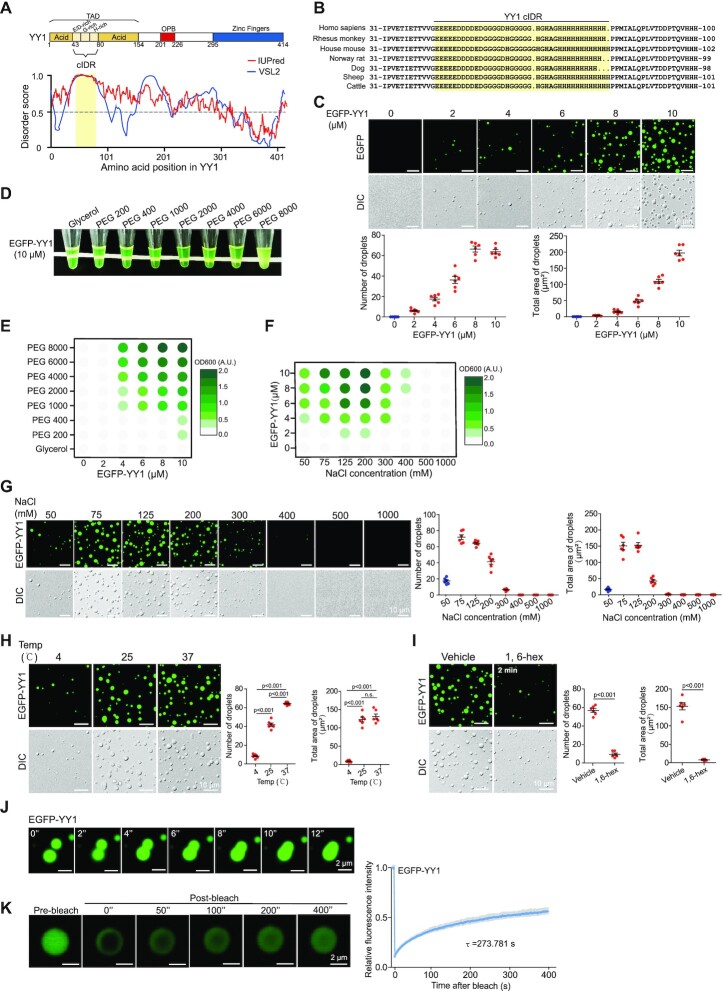
YY1 has a highly acidic transactivation domain (TAD) at its N-terminus. The 154 amino acids at the N-terminus contain 76 negatively charged glutamic/aspartic acid (E/D) residues, but only two positively charged residues (R109 and R122), as well as 18 histidines (Hs) and 20 glycines (Gs) (Supplementary Figure S1A). To date, the structure of the full length YY1 protein has not been reported, although the cocrystal structure of its C-terminus with a binding element was resolved (39). YY1 TAD embraces an eleven-E/D (11 × E/D) cluster and an 11 × H cluster flanked by two acidic regions (Figure 1A, top row). Consistently, inspection of the YY1 primary sequence by the IUPred and VSL2 algorithms (40,41) revealed a high propensity of structural disorder in its TAD (Figure 1A, bottom row). Amino acid composition examination revealed multiple potential IDRs in YY1, and the strongest region, amino acids 43–80, was predicted to have the highest propensity by both algorithms. We named this region as core the IDR (cIDR) that comprises an 11 × E/D and an 11 × H cluster separated by a G-rich stretch (Figure 1A and B). IDRs containing consecutive residues of the same or similar amino acids may have the strong ability to form phase separation condensates (26); therefore, we focused on the cIDR in following studies. Importantly, the E/D and H clusters, and their neighboring regions are highly conserved among different species of vertebrates (Figure 1B). Thus, bioinformatic analyses strongly support the presence of IDR in YY1’s TAD.
Figure 1.
YY1 undergoes liquid-liquid phase separation (LLPS) in vitro. (A) Domain structure and graphs of YY1 IDRs based on VSL2 and IUPred algorithms. Scores >0.5 indicate disorder. Yellow shade depicts the designated core IDR (cIDR). (B) The cIDRs and their flanking sequences of the YY1 proteins from different vertebrates. (C) Representative fluorescence and differential interference contrast (DIC) images of EGFP-YY1 droplets at different protein concentrations in a buffer containing 125 mM NaCl and 10% PEG-8000 (the same condition hereafter, if not specified). Quantification of droplets’ numbers and area is shown in the bottom row. (D) Turbidity visualization of EGFP-YY1 droplet formation. Tubes containing EGFP-YY1 (10 μM, the same concentration hereafter, if not specified) in the buffer containing PEGs with increasing molecular weights. (E and F) OD600nm phase diagram of turbidity changes caused by EGFP-YY1 droplet formation in different PEGs (E) and an increasing NaCl concentrations (F). (G) Representative fluorescence and DIC images of EGFP-YY1 droplets at increasing NaCl concentrations. Quantification of droplet's numbers and area is shown at right. (H and I) EGFP-YY1 droplet formation at different temperatures (H), and in the presence or absence of 5% of 1,6-hexanediol (1,6-hex) (I). Quantification is shown at right with P values indicated on top. n.s.: not significant. In (C), (G–I), quantification of droplets’ numbers and area was mean ± s.e.m. from six fields of view in each group. (J) Two EGFP-YY1 droplet coalescence over a time course. (K) Fluorescence recovery after photobleaching (FRAP) of EGFP-YY1 droplets. FRAP curve is shown at right. Data are presented as mean ± s.d. (n = 6 droplets). τ, half-time of recovery.
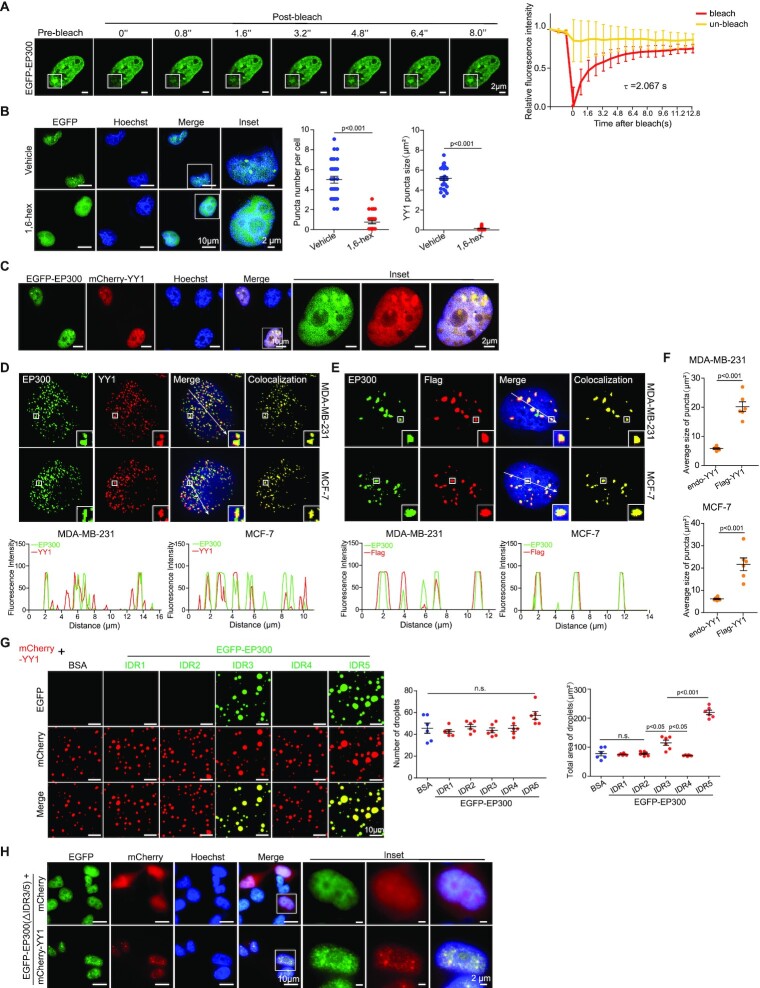
Many IDR-containing TFs form dynamic liquid-like droplets, or gel-like phase-separated condensates, due to multivalent and weak interactions among IDRs (9,25,26). Fluorescent droplets and turbidity created by gel-like condensates are used to evaluate phase separation, while polyethylene glycol (PEG) mimicking crowded cellular conditions promotes this in vitro process (9). When purified recombinant EGFP-YY1 (Supplementary Figure S1B) was incubated with PEG of a molecular weight (MW) of 8000 Daltons (PEG 8000) in the presence of 125 mM NaCl, we observed droplet formation, of which the density and size scaled up with increased protein concentrations (Figure 1C). Meanwhile, as measured by optical density at 600 nm (OD600 nm), the turbidity of solutions monotonically intensified, which was proportional to MWs of PEG in the buffer (Figure 1D and E). To evaluate the biological relevance of EGFP-YY1 droplet formation, we carried out the assays in solutions with increasing salt concentrations. The highest turbidity and most droplet formation by EGFP-YY1 were observed at 125–200 mM NaCl, in the range of physiological saline levels, but vanished as the salt concentration reached 500 mM (Figure 1F, G and Supplementary Figure S1C). In addition, the ability of EGFP-YY1 to form droplets was higher at 37°C than that at 25°C, and markedly descended at 4°C (Figure 1H). Importantly, EGFP-YY1 droplets were sensitive to 1,6-hexanediol, a chemical disrupting liquid-liquid phase separated condensates (Figure 1I). We also observed fusion events between two adjacent droplets (Figure 1J and Supplementary Video 1) and quick green fluorescence recovery of droplets to about 57% within 400 s after targeted photobleaching treatment (Figure 1K), indicating a dynamic feature of the EGFP-YY1 condensates.
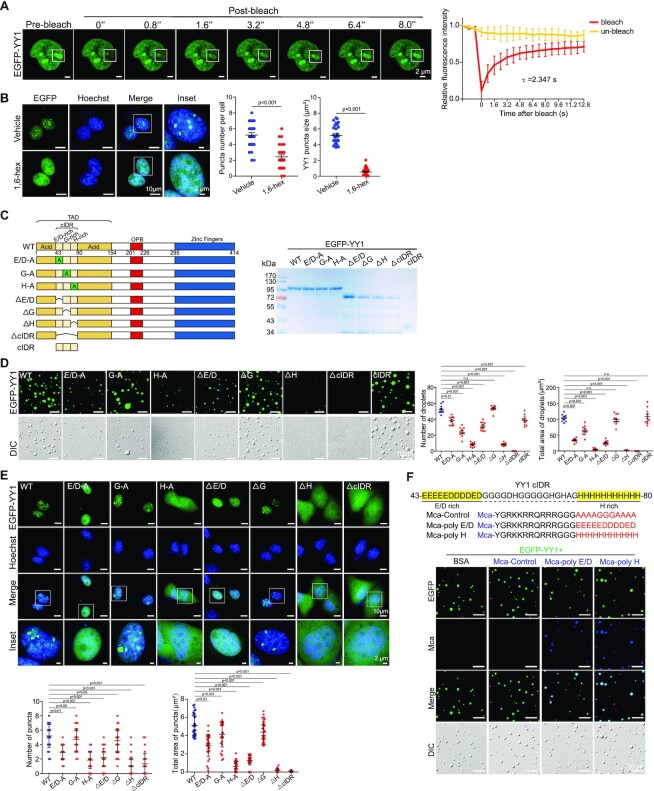
When EGFP-YY1 was expressed in U2OS cells, we detected green fluorescent puncta that also exhibited the ability of prompt fluorescence recovery to about 71% within 8 s after photobleaching (Figure 2A), consistent with its droplet formation in vitro. Furthermore, treatment of EGFP-YY1 transfected cells by 1,6-hexanediol greatly diffused green fluorescent puncta in nuclei, suggesting their liquid condensate properties (Figure 2B). These results strongly suggest that YY1 is capable of forming phase-separated condensates. To evaluate how special amino acid clusters in the predicted cIDR of YY1 (Figure 2C, left panel, and Supplementary Figure S1D) contributed to phase separation, we generated EGFP-YY1 mutants E/D-A, G-A and H-A (with E/D-, G- and H-rich regions replaced by corresponding numbers of alanines, respectively), and their deletion mutants (denoted as Δ), as well as ΔcIDR and cIDR of EGFP-YY1 fusion mutants (Figure 2C, left panel), and purified recombinant proteins from a bacterial expression system (Figure 2C, right panel). We tested their droplet formation capability in a droplet formation buffer containing PEG 8000 and 125 mM NaCl, and also transfected their eukaryotic expression vectors into U2OS cells. EGFP-cIDR could steadily form droplets with comparable numbers and sizes to those of the WT, while EGFP-YY1-ΔcIDR completely lost this ability (Figure 2D). Consistently, EGFP-YY1-ΔcIDR did not generate nuclear puncta, but instead showed diffusive distribution in both nuclei and cytoplasm (Figure 2E). Strikingly, both H-A and ΔH mutants of EGFP-YY1 exhibited very similar phenomena to the cIDR-deleted mutant, indicating a critical role of the 11 × H tract in YY1 phase separation. Meanwhile, E/D-A and ΔE/D mutants formed droplets with markedly reduced numbers and sizes, and displayed diffused green fluorescence in nuclei; however, G-cluster mutation or deletion had much less impact on the droplet forming ability of EGFP-YY1 than that of H and D/E cluster changes, and consistently G-A and ΔG mutants could still generate puncta in cells with numbers and sizes comparable to WT (Figure 2E). The turbidity of EGFP-YY1 WT and its mutants in PEGs reflected their droplet forming capability, with the least condensates formed by H-A, ΔH and ΔcIDR mutants (Supplementary Figure S1E). In addition, in transfection of U2OS cells by Flag-tagged YY1 vectors, the effects of cIDR alterations on punctum formation were virtually identical to that of the EGFP-YY1 proteins (Supplementary Figure S1F). These data strongly suggested that both H and E/D clusters, especially the former one, are important elements of YY1’s cIDR in phase separation. We also designed (7-Methoxycoumarin-4-yl) acetic acid (Mca)-labeled peptides, with a transmembrane sequence (TAT) fused to an A/G-rich control sequence, E/D and H cluster sequences of YY1 (Figure 2F). Importantly, both TAT-E/D and TAT-H formed fusion droplets with EGFP-YY1, suggesting their ability to promote phase separation. Contribution of E/D clusters to liquid condensate formation has been frequently observed (9,24), but the role of H clusters in phase separation was only reported in P-TEFb (42). In addition to YY1, we also verified an indispensable role of H clusters to the phase separation ability of HOXA1, FOXG1B, ZIC3 and HNF6 proteins (Supplementary Figure S2A–D).
Figure 2.
Characterization of YY1 LLPS properties. (A) FRAP of EGFP-YY1 puncta in U2OS cells. White squares depict photobleached puncta undergoing fluorescence recovery. FRAP quantification is shown at right; bleaching event occurred at t = 0 s. Data represent mean ± s.d. (n = 6). τ, half-time of recovery. (B) EGFP-YY1 puncta in live U2OS cells treated in buffers with or without 5% 1,6-hexanediol. Nuclei were visualized by Hoechst staining. Quantification of numbers and sizes of puncta is shown at right with P values indicated on top. (C) Domain structure of the YY1 protein. E/D, G and H represent E/D-, G- and H-rich regions, respectively. SDS-PAGE of recombinant YY1 wild-type (WT) and mutant proteins purified from E. coli is shown at right. (D) Droplet formation of EGFP-YY1 WT and mutants. Quantification of droplets’ numbers and area is shown at right as mean ± s.e.m. (n = 6). (E) Live U2OS cells transfected with EGFP-YY1 WT and mutants. In the quantifications of (D and E), P value of each YY1 mutant versus WT is labeled; n.s.: not significant. In (B) and (E), 30 cells were quantified for each sample. Data represent mean ± s.e.m. (F) Representative droplet formation by EGFP-YY1 and peptides. EGFP-YY1 and (7-methoxycoumarin-4-yl) acetic acid (Mca)-labeled poly E/D, poly H and control (A/G repeat) peptides or BSA were incubated.
YY1 residues required for phase separation are essential to its cell proliferative activity and tumor growth
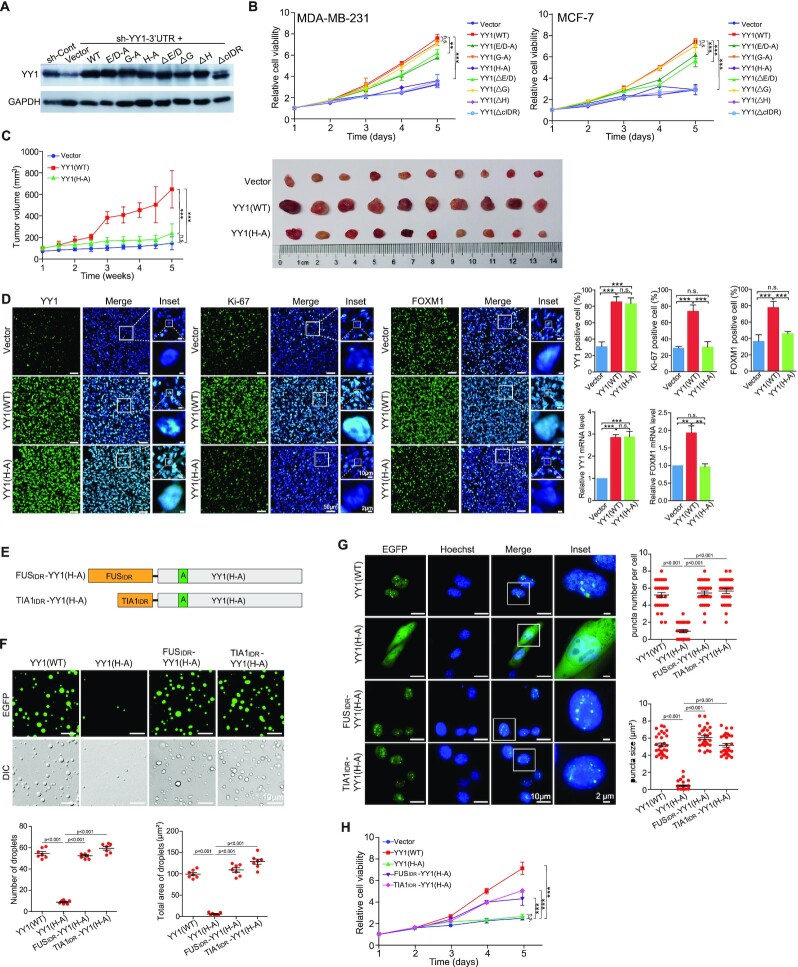
To test whether the mutations or deletions in its cIDR could adversely affect YY1’s function, we expressed the mutants in MDA-MB-231 and MCF-7 cells, while simultaneously silencing endogenous YY1 using a DOX-inducible shRNA targeting the 3′-UTR of the endogenous YY1 mRNA (sh-YY1-3′-UTR, Figure 3A). YY1 depletion reduced breast cancer cell viability, consistent with our previous report (38), while ectopically expressed WT YY1 could largely restore it (Figure 3B). In accordance with droplet and punctum formation studies, cIDR deletion and H-cluster mutation or deletion completely abolished YY1’s ability to restore cell viability (Figure 3B), suggesting indispensability of the cIDR and H-cluster to YY1’s function. Similarly, both E/D-A and ΔE/D mutants only partially rescued the reduced viability caused by YY1 depletion, while G-A and ΔG mutants virtually retained YY1’s function in reinstating cell viability (Figure 3B). In addition, results of the YY1 mutants in promoting cell migration and clonogenicity were consistent with the cell viability data (Supplementary Figure S2E and F). To evaluate the biological function of the H-cluster in vivo, we conducted tumor xenograft studies in nude mice using MDA-MB-231 cells with endogenous YY1 silenced by the sh-YY1-3′-UTR and infected by lentiviruses carrying an empty vector, YY1 WT and its H-A mutant. YY1 WT, but not the H-A mutant, could rescue the retarded tumor growth, reduced tumor cell proliferation and attenuated FOXM1 expression caused by endogenous YY1 knockdown (Figure 3C and D). Overall, the stretches required for YY1 phase separation, including the cIDR and the H-cluster inside the cIDR, are essential to YY1’s activity in maintaining basic cellular activities and xenograft tumor growth of breast cancer cells.
Figure 3.
Effects of YY1 cIDR and H-cluster mutagenesis on its biological functions. (A) Ectopic YY1 expression with simultaneous endogenous YY1 knockdown. MDA-MB-231 cells with DOX-induced sh-YY1 targeting the 3′-UTR of the YY1 mRNA were infected by lentivirus expressing 3 × Flag-YY1 WT and mutants, or a vector. Cell lysates were analyzed by Western blot using indicated antibodies. (B) Effects of YY1 mutations on cell viability. MDA-MB-231 and MCF-7 cells expressing 3 × Flag-YY1 WT and mutants with endogenous YY1 knockdown were evaluated by WST-1 assays to determine cell viability. **P < 0.01; ***P < 0.001. (C and D) Mouse xenograft tumor formation to evaluate the function of YY1(H-A) mutant. MDA-MB-231 cells (4 × 106) carrying DOX-inducible sh-YY1-3′-UTR and infected by lentivirus carrying an empty vector, or expressing WT YY1 or YY1(H-A). In (C), the growth curves of mouse xenograft tumors were generated by measuring tumor sizes twice a week and then calculating tumor volumes. In (D), IF staining and RT-qPCR analyses of frozen xenograft tumor samples. The IF images of YY1, Ki-67 and FOXM1 antibodies and their merged images with DAPI are presented, with the quantitation shown at the upper right panel. The RT-qPCR data of YY1 and FOXM1 mRNA expression normalized to GAPDH levels at the lower right panel. (E–H) IDR fusion studies to evaluate the function of the H-cluster of YY1. In (E), the diagrams of fusion proteins by conjugating the FUS IDR (FUSIDR) and TIA1 IDR (TIA1IDR) to the N-terminus of YY1(H-A) mutant are displayed. In (F), representative fluorescence and DIC images of EGFP-conjugated proteins as labeled on top are presented. Quantification of droplet's numbers and area is shown at the low panel. In (G), live U2OS cells transfected with EGFP-conjugated proteins as labeled at left are shown. In the quantifications of (F) and (G), P values for the comparison of different groups are labeled. In (H), cell viability was determined by WST-1 assays for the MDA-MB-231 cells with endogenous YY1 knocked down by sh-YY1-3′-UTR and infected by lentivirus carrying an empty vector or expressing Flag-YY1 WT, H-A mutant, or its fusion proteins. **P < 0.01; ***P < 0.001; n.s.: not significant.
To further test whether the H-A mutant's inability to form phase separation was the cause for its deficiency in rescuing cell proliferation, we designed two fusion proteins with the N-terminus of the YY1(H-A) mutant linked to the IDRs of the FUS and TIA1 proteins that were known to drive phase separation (43–45), to generate FUSIDR-YY1(H-A) and TIA1IDR-YY1(H-A), respectively (Figure 3E). When attached to EGFP, both fusion proteins, but not YY1(H-A) alone, could form droplets in vitro, and create puncta in cells with numbers and sizes comparable to those of WT YY1 (Figure 3F and G). In addition, when compared to WT YY1, both FUSIDR-YY1(H-A) and TIA1IDR-YY1(H-A) could partially, but still highly significantly, restore the loss of YY1(H-A) in rescuing the reduced cell viability caused by sh-YY1-3′-UTR (Figure 3H). Therefore, our data support that notion that the phase separation characteristic of YY1 plays a critical role in its proliferative activity.
YY1 compartmentalizes coactivators to nuclear puncta
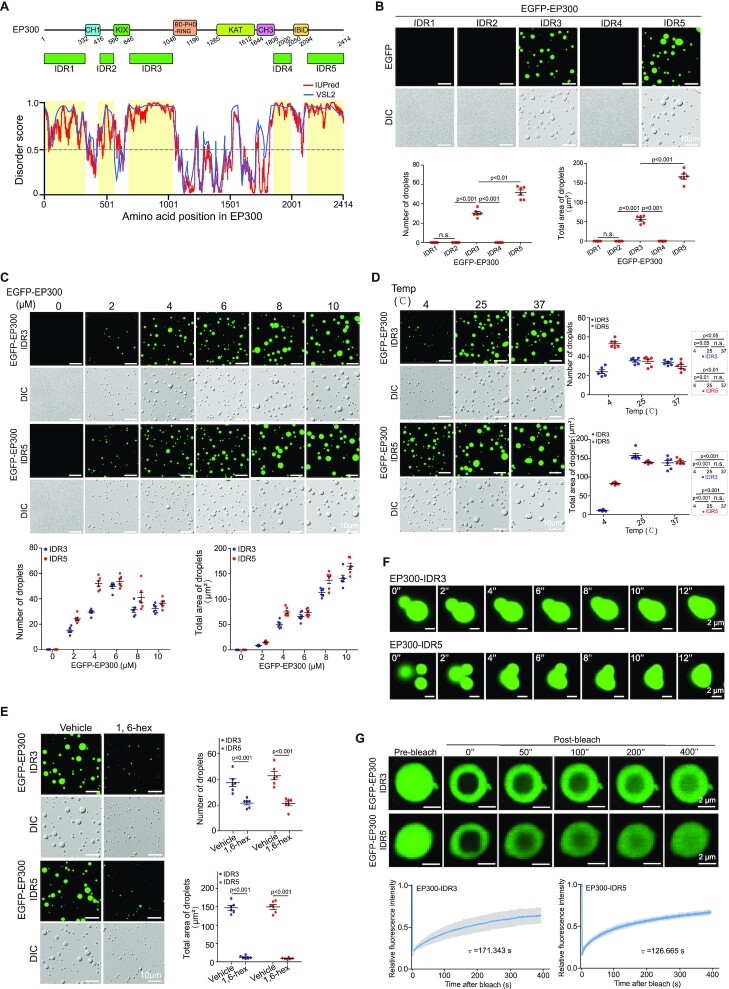
The name Yin Yang 1 represents YY1’s ability to mediate both the repression and activation of target genes, depending on the recruited cofactor (46). In the past decade, the role of YY1 in promoting gene expression has been frequently reported. Consistently, YY1 interacts with several histone acetyltransferases, including EP300, CBP and PCAF (47,48). Among them, EP300 is a well-recognized coactivator. We analyzed the primary sequence of human EP300 using the IUPred and VSL2 algorithms, and discovered five potential IDRs, of which the IDR3 and IDR5 are larger than the others, and have relatively high scores (Figure 4A). Using a bacterial expression system, we purified recombinant EGFP fusion proteins with these IDRs (Supplementary Figure S3A). EGFP-EP300-IDR3 and -IDR5, but not the other three proteins, could generate droplets (Figure 4B), with dependencies on PEG MWs and protein concentrations (Figure 4C, Supplementary Figure S3B and S3C), and formed relatively large droplets at 25°C and 37°C versus 4°C (Figure 4D). The two EP300-IDRs formed the most droplets at NaCl concentrations between 50 and 125 mM (Supplementary Figure S3D and E). Droplets formed by EGFP-EP300-IDR3 and -IDR5 also showed properties of 1,6-hexanediol sensitivity (Figure 4E), adjacent droplet fusion (Figure 4F, Supplementary Videos 2 and 3), and recovery to 63–68% within 400 s from photobleaching treatment (Figure 4G), characteristics of phase separation condensates.
Figure 4.
YY1 coactivator EP300 undergoes LLPS in vitro. (A) EP300 domain structure and predicted IDRs (yellow shades). (B) Droplets of EGFP-EP300-IDRs (top row) and their quantification of numbers and area (bottom row). (C) Droplets of EGFP-EP300-IDR3 and -IDR5 at different protein concentrations. Quantification is at the bottom row. (D and E) EGFP-EP300-IDR3 and -IDR5 droplet formation at different temperatures (D), and in the presence or absence of 5% 1,6-hexanediol (E). In (B–E), representative images are presented, and quantification is shown as mean ± s.e.m. from six droplets’ fields in each group. (F) EGFP-EP300-IDR3 and -IDR5 droplets coalescence. (G) FRAP recovery of EGFP-EP300-IDR3 and -IDR5 droplets. FRAP recovery curves at right are the data of mean ± s.d. (n = 6 droplets). τ, half-time of recovery.
Consistent with in vitro data, immunostaining analyses also presented endogenous EP300 puncta with fluorescence recovery to ∼75% within 8 s from photobleaching and sensitivity to 1,6-hexanediol (Figure 5A and B), suggesting EP300 phase separation in a cellular environment. With EGFP-EP300 and mCherry-YY1 cotransfected into U2OS cells, we observed colocalization of the green and red fluorescent signal (Figure 5C). When examining the endogenous proteins in MDA-MB-231 and MCF-7 cells, we observed that most EP300 puncta stained by its antibody overlapped with YY1 signal (Figure 5D). Additionally, with endogenous YY1 knockdown, when Flag-YY1 was transfected into the cells, endogenous EP300 showed intensely stained puncta well overlapping with the Flag tag signal (Figure 5E). The puncta formed by Flag-YY1 were markedly larger in size than those by endogenous YY1 (Figure 5F). Consistently, mCherry-YY1 could form droplets with EGFP-EP300-IDR3 and -IDR5 with overlapped color and increased sizes, especially the latter, compared to other predicted EP300-IDRs (Figure 5G). The data suggest that EP300 is a coactivator present in YY1 nuclear puncta. Noteworthily, an EGFP-EP300 mutant lacking both IDR3 and IDR5 did not form any nuclear punctum by itself, but could still be detected in the mCherry-YY1 puncta (Figure 5H). The phenomenon could be attributed to the presence of multiple previously identified YY1 binding sites in this EP300 mutant (49,50). Nevertheless, our data suggest that YY1 is the primary driving force for the initiation of the YY1-associated nuclear condensates.
Figure 5.
EP300 and YY1 condensates have LLPS properties. (A) FRAP recovery of EGFP-EP300 puncta in U2OS cells. White squares highlight photobleached puncta, and quantification is at right. Bleaching occurs at t = 0 s. Data represent mean ± s.d. (n = 6). τ, half-time of recovery. (B) EGFP-EP300 puncta in live U2OS cells in the presence or absence of 5% 1,6-hexanediol. Nuclei were detected by Hoechst staining. For each sample, 30 cells were quantified. Data are presented as mean ± s.e.m. (C) EGFP-EP300 and mCherry-YY1 puncta in live U2OS cells. Expression vectors were transfected into U2OS cells followed by fluorescent microscopy. Nuclei were detected by Hoechst staining. (D and E) Colocalization of endogenous EP300 with YY1 (D) or ectopic Flag-YY1 (E) in nuclear puncta in breast cancer cells. In (E), endogenous YY1 was knocked down by sh-YY1-3′-UTR. Immunofluorescence staining was used with nuclei detected by DAPI. Line scans of colocalization images are depicted by white arrows (bottom row). (F) Quantified average sizes of merged puncta in MDA-MB-231 (D) and MCF-7 (E) cells. Data are presented as mean ± s.e.m. with puncta in six fields of each group. (G) Representative images of droplets of mCherry-YY1 incubated with EGFP-EP300-IDR mutants. Quantified droplets' numbers and sizes in merged images is shown at right. Data are presented as mean ± s.e.m. with droplets in six fields for each group. (H) EGFP-EP300(ΔIDR3/5) mutant and mCherry-YY1 puncta in live U2OS cells. Expression vectors were transfected into U2OS cells followed by fluorescent microscopy. Nuclei were detected by Hoechst staining.
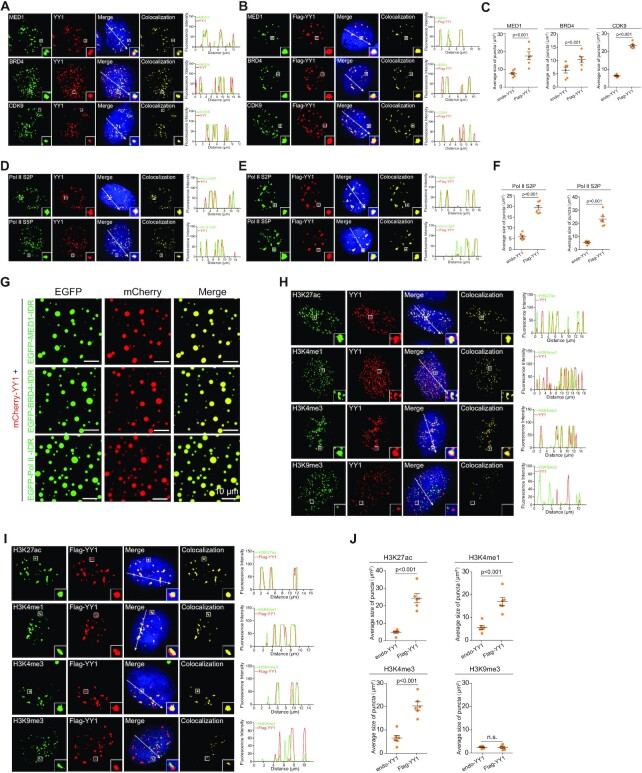
Furthermore, we tested for the presence of additional coactivators in YY1 puncta. In immunostained MDA-MB-231 cells, endogenous YY1 colocalized with MED1, BRD4 and CDK9 (Figure 6A). Consistently, with endogenous YY1 knockdown, ectopic Flag-YY1 also presented puncta overlapping with endogenous proteins of these three coactivators (Figure 6B), but with significantly increased sizes versus those of endogenous YY1 (Figure 6C). Ser2 and Ser5 phosphorylation of the C-terminal heptapeptide repeats of RNA polymerase II (Pol II S2P and S5P) are general markers of gene transcription (51). Endogenous YY1 colocalized with Pol II S2P and S5P signals (Figure 6D), suggesting that YY1 puncta also contained active RNA polymerase II. Similar to coactivators, with endogenous YY1 knockdown, Flag-YY1 showed relatively intensified puncta overlapping with Pol II S2P and S5P signals (Figure 6E) with increased sizes compared to those by endogenous YY1 (Figure 6F). In line with these data, mCherry-YY1 could form droplets that overlapped with signals from EGFP-fused IDRs of MED1, BRD4 and Pol II in vitro (Figure 6G). Based on these results, we propose that YY1 plays a key role in promoting gene transcription through recruiting major coactivators to form phase-separated condensates. Consistent with this hypothesis, both endogenous YY1 and exogenous Flag-YY1 showed puncta that overlapped with gene activation histone markers H3K27ac, H3K4me1 and H3K4me3, but not the repressive marker H3K9me3 (Figure 6H and I). In addition, with endogenous YY1 knockdown, Flag YY1 markedly increased the sizes of the puncta overlapping with the three activation markers but not H3K9me3 (Figure 6J). Immunostaining assays were also conducted in MCF-7 cells with virtually the same results (Supplementary Figure S4A–I). Overall, our data suggested that YY1 compartmentalized transcriptional coactivators to phase-separated nuclear puncta, which is potentially relevant to YY1-mediated gene transcription.
Figure 6.
YY1 compartmentalizes additional coactivators to nuclear puncta. (A and B) Colocalization of BRD4, MED1 and CDK9 with endogenous YY1 (A) or with Flag-YY1 (B) in nuclear puncta in MDA-MB-231 cells. In (B), endogenous YY1 was knocked down by sh-YY1-3′-UTR. Endogenous proteins were detected by their corresponding antibodies. Flag-YY1 was detected by a Flag epitope antibody. Nuclei were visualized by DAPI staining. Line scans of the colocalization images are depicted by white arrows with quantification shown at right. (C) Quantified average sizes of merged puncta in MDA-MB-231 cells with endogenous YY1 (A) and Flag-YY1 (B). Data are mean ± s.e.m. of puncta in 6 fields in each group. (D and E) Colocalization of active RNA Pol II with YY1 (D) or Flag-YY1 (E) in nuclear puncta of MDA-MB-231 cells. In (E), endogenous YY1 was knocked down by sh-YY1-3′-UTR. Active RNA Pol II was detected by antibodies for phosphorylation of Ser 5 (S5P) or Ser 2 (S2P), with nuclei detected by DAPI. Line scans of colocalization images are depicted by white arrows with quantification shown at right. (F) Quantification of average sizes of merged puncta in MDA-MB-231 cells with endogenous YY1 (D) and Flag-YY1 (E). Data are presented as mean ± s.e.m. from puncta of six fields in each group. (G) Representative images of droplet formation of mCherry-YY1 with EGFP-MED1-IDR, EGFP-BRD4-IDR, or EGFP-Pol II-IDR. (H and I) Localization of active (H3K27ac, H3K4me1 and H3K4me3) and repressive (H3K9me3) histone markers with endogenous YY1 (H) or Flag-YY1 (I) in MDA-MB-231 cells. In (I), endogenous YY1 was knocked down by sh-YY1-3′-UTR. Histone markers were determined using corresponding antibodies. Nuclei were detected by DAPI. Line scans of colocalization images are depicted by white arrows with quantification shown at right. (J) Quantified average sizes of merged puncta in MDA-MB-231 cells with endogenous YY1 (H) and Flag-YY1 (I). Data are presented as mean ± s.e.m. of puncta in six fields in each group. All experiments in this figure were independently repeated at least 6 times with similar results. In (C), (F) and (J), P values are indicated on top of the quantification analyses. n.s.: not significant.
YY1 activates FOXM1 gene expression through recruiting general coactivators
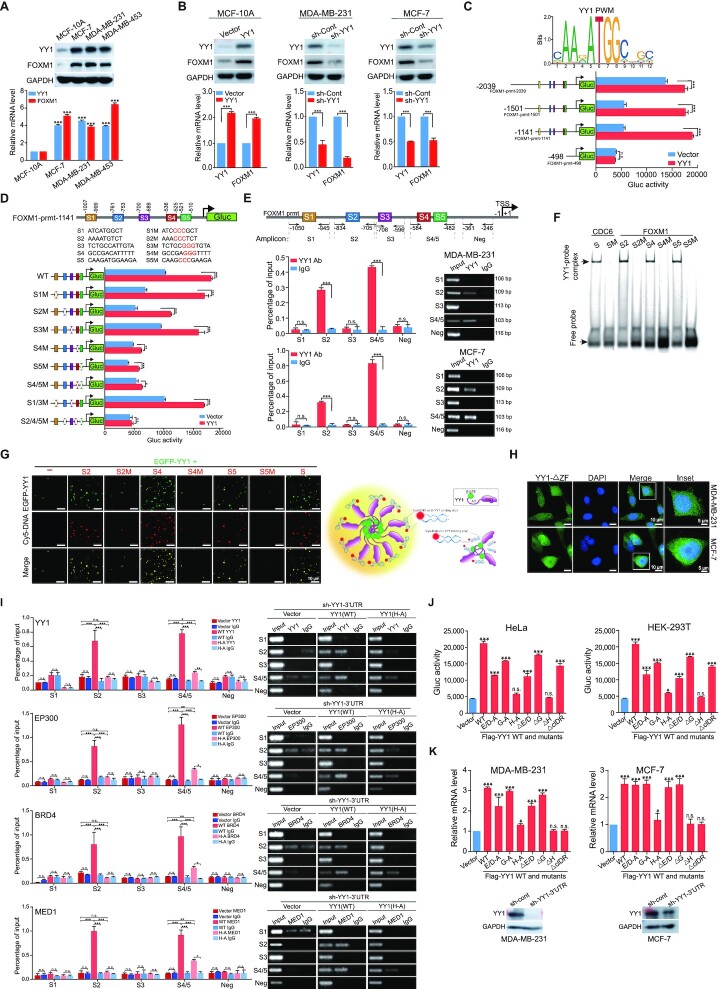
YY1 conditional knockout in mouse embryonic fibroblast cells revealed many genes as potential targets of YY1 (52). Among them, FOXM1 exhibited over 2-fold reduction in response to YY1 depletion. Both YY1 and FOXM1 play stimulative or proliferative roles in oncogenesis (16,53). Consistently, they were overexpressed in breast cancer and associated with patients’ poor prognosis (38,54). Analyses of a TCGA dataset of mammary samples indicated positive YY1 and FOXM1 correlations (Supplementary Figure S5A), especially in normal tissues, suggesting physiological significance of their functional interplay. In mammary cells, we detected concurrent increase of YY1 and FOXM1 expression in breast cancer cells versus nontumorigenic MCF-10A cells (Figure 7A). At both mRNA and protein levels, ectopically expressed YY1 in MCF-10A cells increased endogenous FOXM1 expression, while shRNA-mediated YY1 knockdown reduced it in breast cancer cells (Figure 7B). All these data strongly suggest a positive regulation of the FOXM1 gene by YY1.
Figure 7.
YY1 binds to the FOXM1 promoter and activates its expression. (A) YY1 and FOXM1 expression in mammary cell lines. Nontumorigenic MCF-10A cells, and three indicated breast cancer cell lines were analyzed by Western blot using indicated antibodies (top row). YY1 and FOXM1 mRNA levels were quantified by RT-qPCR (bottom row). (B) Effects of YY1 on FOXM1 in mammary cells. MCF-10A cells were infected by lentivirus expressing YY1, while MDA-MB-231 and MCF-7 cells were infected by lentivirus carrying sh-YY1. YY1 and FOXM1 protein and mRNA levels were analyzed by Western blot (top row) and RT-qPCR (bottom row). (C) Mapping the YY1-regulated region in the FOXM1 promoter. Conserved YY1 binding elements with a core sequence of ATGG (Jaspar Matrix ID: MA0095.2, top row). Reporters with different FOXM1 promoter lengths driving Gluc were generated. Data of reporter assays in response to ectopic YY1 in HeLa cells were examined (bottom row). Data represent mean ± s.d. (n = 3). (D) Evaluation of putative YY1 binding sites in the FOXM1 promoter. Putative YY1 binding sites S1–S5 were identified in the 1141-bps region upstream of the transcription start site (TSS) of the FOXM1 gene, with WT and mutants presented (top panel). Data of reporter assays in response to YY1 in HeLa cells are presented in the bottom row. (E) ChIP-qPCR assays to examine YY1 binding to the FOXM1 promoter. The FOXM1 promoter is presented in the top row with five qPCR primer pairs. YY1 antibody and a control IgG were used in ChIP assays. Data quantification and agarose gels of ChIP-qPCR in MDA-MB-231 and MCF-7 cells were presented in the middle and bottom rows, respectively. (F) EMSA to test YY1 binding to the FOXM1 promoter. His × 6-YY1 was incubated with Cy5-labeled S2, S4, S5 probes, and mutants (S2M, S4M and S5M) with a CDC6 promoter probe and its mutant as positive and negative controls, respectively. YY1-probe complex and free probe positions are labeled on the left. (G) Left: representative images of droplet formation by EGFP-YY1 with Cy5-labeled probes in Figure 7F and Supplementary Table S2. Right: schematic model of droplet formation promoted by DNA probes. (H) Image of live MDA-MB-231 and MCF-7 cells transfected with EGFP-YY1(ΔZF) plasmid. (I) ChIP-qPCR assays to test the effects of YY1(H-A) mutant expression on coactivators’ binding to the FOXM1 promoter. MDA-MB-231 cells with endogenous YY1 knocked down by sh-YY1-3′-UTR and infected by lentivirus carrying an empty vector, or expressing Flag-YY1 WT or its (H-A) mutant were collected. EP300, BRD4 and MED1 antibodies, and a control IgG were used in ChIP assays, and qPCR primers are shown in Supplementary Table S1. Both quantification and agarose gels of ChIP-qPCR were presented. (J and K) Evaluation of YY1 mutations’ effects on the FOXM1 promoter. In (J), FOXM1 promoter reporter (pFOXM1-prmt-Gluc) and pCMV-SEAP vector were cotransfected with YY1 WT and mutant vectors into HeLa (left) and HEK-293T (right) cells in 24-well plates. Gluc activity in each well was measured and normalized against its SEAP activity. In (K), YY1 vectors were transfected into MDA-MB-231 (left) and MCF-7 (right) cells with endogenous YY1 knockdown, followed by RT-qPCR to quantify FOXM1 mRNA levels. Western blot analyses showing shRNA-mediated endogenous YY1 knockdown are presented at bottom panels.
To examine the mechanism regulating FOXM1 gene expression, we first mapped the essential region of the FOXM1 promoter. Four reporter constructs were generated with Gaussia luciferase (Gluc) driven by different lengths of the upstream sequence from the TSS of the FOXM1 gene (Figure 7C). Based on the response of the reporters to cotransfected YY1, YY1-regulated essential elements reside within the 1141-bp region upstream of the TSS (Figure 7C). To explore potential regulation of FOXM1 expression by YY1, we examined the human FOXM1 promoter for YY1 consensus binding elements using the JASPAR (55) and Tfsitescan (http://www.ifti.org/cgi-bin/ifti/Tfsitescan.pl) databases. Within the 1,141-bps FOXM1 promoter, we identified five potential YY1 binding sites (Supplementary Figure S5B), and mutagenesis of the #2, #4 and #5 sites, especially the latter two, caused remarkable reduction of FOXM1 promoter activity in reporter assays (Figure 7D). YY1 binding to the FOXM1 promoter through these sites was verified by a chromatin immunoprecipitation (ChIP) assay (Figure 7E). To test YY1 binding in vitro, we synthesized double-stranded (ds) oligonucleotides S1 to S5 based on the corresponding YY1 binding sites in the FOXM1 promoter. In EMSA studies, a Cy5-labeled probe based on a YY1 consensus motif in the CDC6 promoter (56) could be out-competed in His × 6-YY1 binding by S2, S4 and S5, but not S1 and S3, or mutants of S2, S4 and S5 (Supplementary Figure S5C). In addition, Cy5-labeled S2, S4 and S5, but not their mutants, could bind YY1 to form slowly migrating bands in EMSA (Figure 7F). To evaluate how DNA fragments harboring YY1 consensus motifs could affect the formation of YY1 phase-separated condensates in vitro, we incubated Cy5-labeled oligonucleotides with a relatively low concentration (2 μM) of EGFP-YY1. Strikingly, the presence of Cy5-labeled S2, S4 and S5, but not their mutants, could associate with EGFP-YY1 to form droplets with relatively large sizes (Figure 7G, left panel). The data suggested that YY1 phase-separated condensate formation in vitro can be promoted by binding to DNA containing its consensus motifs (Figure 7G, right panel). Furthermore, when cotransfected into cells, Cy5-labeled S2/S4/S5, but not their mutants, could mostly colocalize with YY1, MED1 and BRD4 nuclear puncta; however, no significant change was observed in punctum sizes and numbers between the two transfected groups (Supplementary Figure S6A and S6B). These results indicated that the oligonucleotides containing YY1 binding motifs could not significantly improve YY1 condensate formation in the nuclear environment. We next created EGFP-YY1(ΔZF) lacking the zinc finger (ZF) domain to test whether DNA binding ability was needed for YY1 punctum formation. When transfected in breast cancer cells, this mutant did not form any punctum, but instead distributed in both the nucleus and cytoplasm (Figure 7H). The results indicated that the DNA-binding domain of YY1 plays a determinant role in its phase-separated condensate formation in cells.
To evaluate the effects of YY1 on the binding of coactivators to the FOXM1 promoter, we carried out ChIP assays for several coactivators. Both semi-quantitative and quantitative PCR analyses revealed that EP300, BRD4 and MED1 could bind to the regions containing YY1’s S2 and S4/5 consensus sites, but not S1 and S3 sites, in the FOXM1 promoter of breast cancer cells (Supplementary Figure S7A and S7B). Importantly, in MDA-MB-231 cells with sh-YY1-3′-UTR-mediated endogenous YY1 knockdown, exogenous WT YY1, but not the YY1(H-A) mutant defective in phase separation, could restore the binding of these coactivators to these YY1 consensus sites in the FOXM1 promoter (Figure 7I), indicating that YY1-mediated punctum formation is essential for coactivator recruitment to the FOXM1 promoter.
Consistent with the observation above, both YY1(H-A) and (ΔH) mutants, defective in phase separation, exhibited the least ability to promote both promoter reporter and endogenous transcripts of the FOXM1 gene (Figure 7J and K). The observation that YY1 mutants lacking the H-cluster did not activate the FOXM1 promoter revealed a potential dependence of FOXM1 gene activation on YY1 phase separation. However, we could not completely exclude the possibility that the H-cluster's interaction with coactivators or components of the transcription machinery would also play an important role in driving the FOXM1 promoter, especially in the scenario of transfected Gluc reporters.
Interestingly, in reporter assays of Figure 7J, transfection of YY1(ΔcIDR) still repeatedly retained significant Gluc activity, but its expression in two breast cancer cell lines could not activate the FOXM1 gene (Figure 7K). The discrepancy between the two experiments was likely attributed to the different actions of YY1(ΔcIDR) with, an undisrupted DNA binding domain, in the two experimental settings of Gluc assays involving transfected reporters versus endogenous FOXM1 expression on its chromatin locus.
YY1 promotes enhancer cluster formation to drive FOXM1 gene expression
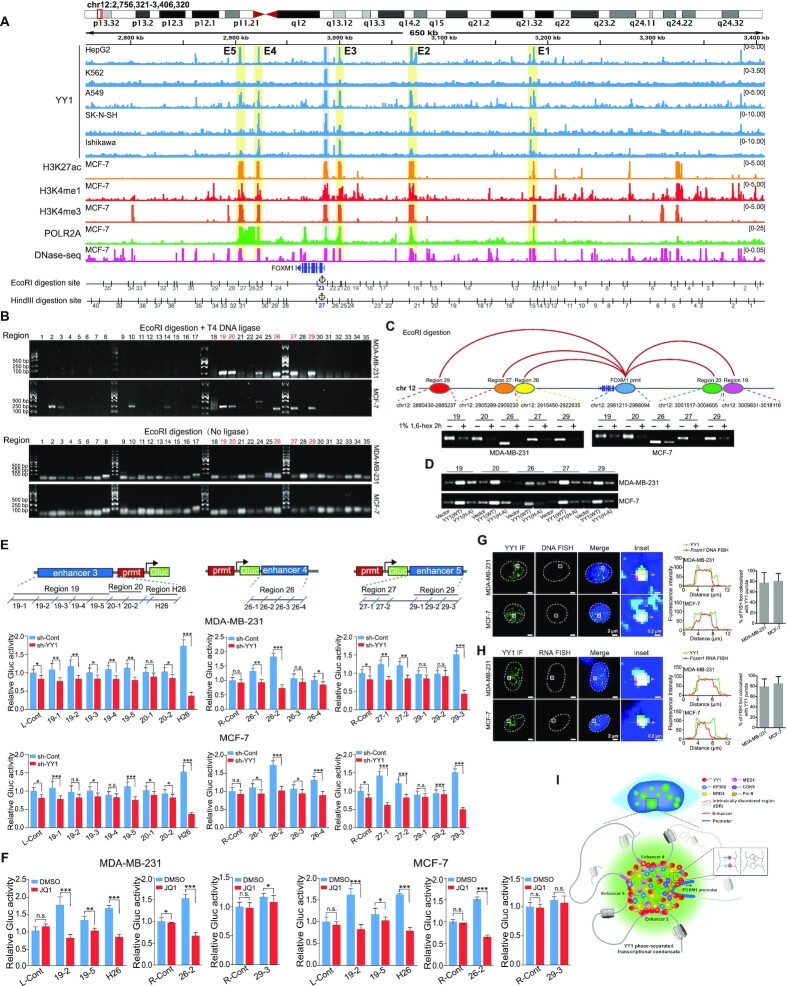
YY1 has been reported to regulate gene expression through promoting enhancer activity (27,57,58), and phase separation is a characteristic feature of the enhancer mechanism (59). To interrogate whether enhancers were involved in YY1-regulated FOXM1 expression, we surveyed the regions of overlapping YY1 binding enrichment and gene activation markers in the vicinity of the FOXM1 gene in the human genome. Within 650 kb of the FOXM1 promoter, we identified five candidate enhancer regions E1 to E5, based on the enrichments of multiple enhancer markers, especially H3K27ac (Figure 8A). The chromosome conformation capture (3C) approach (32) with restriction enzyme digestion, digested genomic DNA ligation, and PCR amplification of ligated DNA, was used to examine neighbouring regions in forming enhancer complexes through interacting with the FOXM1 promoter. Most ligation-dependent PCR amplifications were manifested by the EcoRI- and HindIII-digested segments around enhancers E3, E4 and E5 (Figure 8B and Supplementary Figure S8A). However, we observed inconsistency of PCR amplified regions between EcoRI- and HindIII-digested samples. Samples of the two cell lines with the same digestion also showed discrepancies, but their overall patterns or trends were similar. For example, EcoRI-digested samples showed PCR bands for its fragments 19/20, but not the E3-overlapped fragments 21/22 (Figure 8B). Actually, fragments 19/20 are adjacent to E3, and enriched with decent signal of H3K4me1, another enhancer marker (Figure 8A). Importantly, this region showed positive PCR amplification in HindIII-digested samples (Supplementary Figure S8A), suggesting the connection of both E3 and fragments 19/20 to the FOXM1 promoter. Additionally, different digestion efficiencies of the two enzymes could contribute to their inconsistency of PCR amplified regions. Overall, most PCR amplified regions in EcoRI- and HindIII-digested samples were overlapping with E3, E4 and E5 (Figure 8B and Supplementary Figure S8A), which offered us clues in designing subsequent experiments to validate their enhancer identify.
Figure 8.
YY1 phase separation is required for the connection between distal enhancer elements and the FOXM1 promoter. (A) Schematic view across the FOXM1 gene locus (chr12:2 768 321–3 408 320 [hg19]) with genomic and epigenetic information. Graphic active regulatory regions were generated using the ENCODE database, and potential enhancers (E1 to E5) are shaded in yellow. Fragments digested by EcoRI or HindIII are numbered and shown below the graph. (B) Chromatin conformation capture (3C) analysis to examine direct physical interactions between the FOXM1 promoter and enhancer elements. The 3C analyses followed the protocol in Materials and Methods, using EcoRI in genomic DNA digestion. Ligation step was performed with or without T4 DNA ligase as indicated. Numbers on the top of the gel correspond to ‘EcoRI digestion site’ in the bottom row of (A). PCR to examine direct physical interactions between the FOXM1 promoter and each of EcoRI fragments used the primer sets in Supplementary Table S3. Numbers of fragments overlapping with enhancers and interacting with the FOXM1 promoter are in red text. (C) Schematic interactions between enhancers and the FOXM1 promoter on chromosome 12 (top row), and PCR product images of ligated DNA in 3C assays conducted in the absence or presence of 1% of 1,6-hexanediol (bottom row). (D) PCR product images of ligated DNA in 3C assays in MDA-MB-231 and MCF-7 cells infected by lentivirus carrying an empty vector, or expressing Flag-YY1 WT or its (H-A) mutant. (E) Reporter assays to examine the effects of enhancers on the FOXM1 promoter. Sectionalized enhancers 3, 4 or 5 were individually placed adjacent to the FOXM1 promoter to create reporter vectors, as schematically shown in the top row. R-Cont and L-Cont indicate control fragments that are located to right and left of the FOXM1 TSS, respectively. Reporter vectors were transfected into MDA-MB-231 and MCF-7 cells with or without YY1 knockdown (bottom row). Data are shown as mean ± s.d. (n = 3). (F) Reporter assays to assess the effects of JQ1. Reporter vectors with enhancer fragments were transfected into MDA-MB-231 and MCF-7 cells with or without JQ1, followed by examining Gluc activity. Data are shown as mean ± s.d. (n = 3). (G and H) DNA FISH (G) and RNA FISH (H) assays to determine the colocalization of YY1 puncta with the FOXM1 genomic locus and FOXM1 nascent hnRNA, respectively, in MDA-MB-231 and MCF-7 cells. In G and H, each inset shows a view zoomed-in from the white box in a merge image. Line graph showed the fluorescence intensity, and histogram shows the quantification of DNA-FISH (G) or RNA-FISH (H) foci colocalized with YY1 puncta in the nucleus (n = 18 cells). (I) Schematic model of an enhancer cluster formed by YY1-mediated phase separation with incorporation of coactivators and stabilization by three distal enhancers to activate FOXM1 gene transcription.
Furthermore, shRNA-mediated YY1 knockdown eliminated most of these PCR bands (Supplementary Figure S8B), indicating that physical closeness of the enhancer elements to the FOXM1 promoter was contingent on the presence of YY1. Importantly, we confirmed the precise ligation between digested segments of the FOXM1 promoter and the enhancers by DNA sequencing analysis (Supplementary Figure S8C). In addition, 1,6-hexanediol treatment greatly reduced PCR products (Figure 8C), suggesting that phase separation is a prerequisite for FOXM1 promoter's proximity to enhancers. Consistently, with endogenous YY1 knocked down, ectopically expressed WT YY1, but not the YY1(H-A) mutant defective in phase separation, could markedly increase the connection between the enhancer segments and the FOXM1 promoter (Figure 8D). The results indicated that the ability to undergo phase separation is required for YY1-mediated contacts between the enhancers and the FOXM1 promoter. To assess enhancer potential of EcoRI-digested segments 19, 20, 26, 27 and 29, and HindIII-digested segment 26 (H26) (Figure 8A), we generated FOXM1 promoter reporters with sub-segmented sequences of these regions located either upstream or downstream according to their natural positions relative to the FOXM1 TSS in the genome (Figure 8E, top row). In reporter assays, several fragments from these segments showed a response to YY1’s ectopic expression or knockdown (Figure 8E, bottom row, Supplementary Figure S8D), consistent with their enhancer identity. In line with these data, treatment of transfected cells by JQ1, a BRD4-targeting inhibitor, dampened the reporter activities (Figure 8F and Supplementary Figure S8E).
To interrogate whether the candidate enhancer segments at the FOXM1 locus were present in YY1 puncta, we designed DNA probes specific to these enhancers. In DNA fluorescent in situ hybridization (FISH) assays, the signal detected by these FOXM1 probes mostly resided in the YY1 puncta in both MDA-MB-231 and MCF-7 cells (Figure 8G). Furthermore, we created probes specific to the FOXM1 pre-mRNA. In nascent RNA FISH assays, the nascent heterogeneous nuclear RNA (hnRNA) of the FOXM1 gene preferentially colocalized with YY1 nuclear puncta in breast cancer cells (Figure 8H).
Therefore, our data strongly supported the idea that YY1 forms nuclear puncta through phase-separated transcriptional condensates with compartmentalized coactivators, and subsequently promotes enhancer cluster formation at the FOXM1 gene locus to activate its expression.
We further evaluated the importance of YY1-regulated FOXM1 expression in mammary cells. As we previously reported, ectopic YY1 promoted proliferation of primary mammary epithelial cells, while its depletion reduced it (38). In the current study, FOXM1 knockdown or its ectopic expression could significantly counteract cell viability changes in MCF-10A cells (by exogenous YY1 expression) or breast cancer cells (by YY1 silencing), respectively (Supplementary Figure S9A). We reported YY1-promoted AKT activation (60); consistently, FOXM1 could reinstate AKT-T308 and -S473 phosphorylation reduced by YY1 knockdown (Supplementary Figure S9B). FOXM1 depletion attenuated YY1-promoted migration and clonogenicity of MCF-10A cells in wound healing and colony formation assays, while, in these assays, ectopic FOXM1 could significantly rescue the deficiencies of breast cancer cells caused by YY1 silencing (Supplementary Figure S9C and S9D).
Based on our data, we propose a model of YY1-regulated gene activation (Figure 8I). In this model, an enhancer cluster is formed by YY1-mediated phase separation condensates that compartmentalize major coactivators and is stabilized by three distal enhancers to activate the FOXM1 promoter and stimulate its gene transcription.
DISCUSSION
Phase separation is a general phenomenon in polymer chemistry, but has recently been developed into a concept or mechanism of biological regulation (61). Liquid-liquid phase separation in different subcellular sections creates membrane-less condensates that compartmentalize biomolecules, such as proteins, RNAs and DNAs, with pertinent biological activities, and allows a specific biological event to be processed in a relatively undisturbed manner (13). Applications of phase separation in transcriptional regulation are the seminal discovery of Young's group through demonstrating the formation of phase-separated condensates that confine various coactivators, and function as super-enhancers (8,9). These discoveries largely extended our view of transcriptional regulation and revolutionized the mechanism or concept of sustained gene expression through super-enhancers.
As a key transcription factor, YY1 interacts with numerous epigenetic writers and erasers (16). In addition to recognizing its consensus sites, YY1 binds G-quadruplex structures (62). Importantly, YY1’s homodimerization (27,62) allows it to bridge promoters and enhancer elements. Weintraub et al. reported that YY1 generally occupies active enhancers and promoters in different cell types, and perturbation of YY1 binding disrupts enhancer-promoter looping (27). The promiscuous interaction of YY1 with coactivators can promote their recruitment into the enhancer complexes. Consistently, a number of studies demonstrated YY1’s participation in forming enhancers or super-enhancers to regulate genes involved in various biological processes (27,58,63–65). These studies strongly support a key role of YY1 in promoting enhancer cluster or super-enhancer assembly to activate gene expression. However, despite these exciting indications, molecular evidence is still lacking for detailed mechanisms of YY1-regulated enhancer formation.
The YY1 protein primary structure has several unique features scarcely observed in other proteins, including a highly acidic N-terminus, consecutive E/D-, G- and H-rich regions in the TAD, and four tandem zinc fingers at a basic C-terminus, as well as a high lysine content (32 versus total 414 amino acids) but none of them found in the first 157 residues. When scanning the YY1 sequence, we discovered a high propensity toward structural disorder in its TAD. Subsequently, our experimental data unequivocally demonstrate YY1’s competence in undertaking liquid-liquid phase separation both in vitro and in cell. Interestingly, we identify the 11 × H cluster as a critical motif of YY1 to promote its phase separation, which can be extended to four additional histidine cluster-containing proteins.
The YY1-rich nuclear phase-separated condensates are likely to contain enhancer clusters or super-enhancers based on their inclusion of major coactivators, including EP300, MED1, BRD4 and CDK9, as well as active Pol II. With FOXM1 expression as a regulatory model, we also demonstrate the role of YY1 in convening three distal enhancer elements and the FOXM1 promoter to assemble an enhancer cluster for gene activation. Strikingly, oligonucleotides containing YY1 consensus sites in the FOXM1 promoter can participate in and steadily facilitate YY1 droplet formation in vitro (Figure 7G), suggesting that YY1-mediated phase-separated nuclear condensates are likely involved and promoted by genomic DNA. However, after being transfected into cells, the oligonucleotides only colocalized with YY1, MED1 and BRD4 nuclear puncta, but did not affect their sizes and numbers (Supplementary Figure S6A and S6B). Therefore, DNA length is possibly a determinant factor to promote punctum formation in the nuclear environment, and the detected S2/S4/S5 signal was likely enriched by YY1 proteins that were unbound to chromatin in the puncta. Interestingly, Sigova, et al. demonstrated that YY1 could bind both gene regulatory elements and their associated RNA species transcribed from enhancers and promoters in a genome-wide manner (17). Thus, it is logical to predict that cognate RNAs may participate in YY1-coordinated phase separation condensates, which deserves future investigation.
Despite mounting studies showing that YY1 either activates or represses gene expression, we observe YY1 colocalization with coactivators and histone markers for gene activation, but not for repression. Our data are consistent with previous studies showing YY1’s general association with active promoters and enhancers (17,27). Thus, although YY1 was reported to suppress gene expression through recruiting corepressors, such as HDACs, EZH2 and DNMTs (66–68), our data support its primary role as a transcriptional activator.
Ectopically expressed Flag-YY1 formed relatively large puncta compared to those of endogenous YY1 (Figures 5F, 6C, F and J, Supplementary Figure S4C, F and I). Similar observations were also reported in the studies of other nuclear proteins, including TAZ, YAP, SRC3 and BRD4S (12,33,69,70). This phenomenon is reminiscent of the positive correlation between YY1 concentration and droplets’ sizes (or area) in the in vitro studies of Figure 1C. The relatively large punctum sizes were likely due to overexpressed Flag-YY1, but whether increased amounts of coactivators were incorporated deserves future investigation.
To date, no definitive nuclear localization signal (NLS) has been identified in the YY1 protein. Austen et al. generated a series of YY1 mutants to determine its NLS, but found that deletions of many regions at either its N- or C-terminus could abolish the dominant nuclear localization of YY1 (71). The YY1(H-A) mutant was detected in both nucleus and cytoplasm of cultured cells (Figure 2E and Supplementary Figure S1F), but its localization was mostly nuclear in the staining of xenograft tumors (Figure 3D); additionally, two fusion proteins of FUS/TIA1-IDRs with YY1(H-A) in cultured cells were also contained in the nucleus (Figure 3D), while no NLS was identified in either FUS- or TIA1-IDR (data not shown). Therefore, YY1 nuclear localization is likely determined by different mechanisms, including its interactions with DNA or other proteins, and physiological conditions. Moreover, the YY1(ΔZF) mutant defective in DNA binding was dispersed all over the cells without forming any detectable nuclear punctum (Figure 7H), although the YY1 cIDR alone could still generate droplets in vitro (Figure 2D). Therefore, YY1-mediated condensates in a nuclear environment probably need chromatin anchoring. It is possible that multiple YY1 proteins act as seeding molecules to initially bind a genomic locus through the consensus binding sites, and subsequently form nuclear puncta with the involvement of various coactivators.
EGFP-YY1 proteins with E/D-cluster mutation or deletion still formed relatively small droplets in vitro, but showed no punctum formation in cells (Figure 2D and E), which was likely due to the high green fluorescence background in transfected cells. Consistently, Flag-YY1(E/D-A) and (ΔE/D) mutants still formed puncta, despite their significantly reduced numbers and sizes compared to those of Flag-YY1 WT (Supplementary Figure S1F). Additionally, YY1(E/D-A) and (ΔE/D) mutants were still located in the nucleus, but YY1(H-A) and (ΔH) were both nuclear and cytoplasmic, suggesting more hostile effects on YY1 activities caused by H-cluster changes. This may explain the phenomenon that both Flag-YY1(E/D-A) and (ΔE/D), but not the two H-cluster mutants, retained a significant ability to restore cell viability reduced by endogenous YY1 knockdown (Figure 3B).
FOXM1 is recognized as a critical proliferation-associated transcription factor, regulating cell proliferation, self-renewal and oncogenesis. As a proliferative gene, FOXM1 overexpression was reported in almost all cancer types, and correlated with patients’ poor prognoses (53). Consistently, the FOXM1 gene is regulated by a variety of oncogenic TFs, such as E2Fs, MYC, ERα, STAT3 and CREB, as well as FOXM1 itself (53). In the current study, we provide the first evidence to demonstrate that FOXM1 expression is promoted by an enhancer cluster formed by YY1-containing phase-separated condensates, which extends molecular mechanisms driving FOXM1 overexpression in cancers. Interestingly, FOXM1 has also been reported to participate in enhancer regulation of other genes (72,73). Based on its self-regulated feature, FOXM1 itself may potentially get involved in the formation of the enhancers regulating its expression.
We provided both in vitro and cell-based data to demonstrate the ability of EP300 to form phase separation condensates. Based on algorithm prediction, we identify five potential IDRs in the EP300 primary sequence, and verify that IDR3 (in the middle region) and IDR5 (at the C-terminus) form droplets in vitro. In two recent studies, the EP300 C-terminus was reported to undertake phase separation (33,74), consistent with our results, but Ma et al. showed the ability of its N-terminus (1–566 amino acids) to form droplets in vitro, which was not the case in our study. Nevertheless, we provided ample evidence to demonstrate the competence of EP300 in forming phase-separated condensates, including the optimal parameters, and proved its role as a primary coactivator of the YY1-mediated enhancer complex.
Reporter assays have been used in recent studies to evaluate enhancer activities of a subcloned DNA fragment (75–77); however, their non-natural characteristics and limitations are very obvious. A DNA vector containing a relatively short enhancer fragment cannot fully recapitulate the activation of a promoter by enhancers in genomic DNA, which may need cooperation or assembly of multiple enhancers. It is even unclear whether any enhancer can be efficiently assembled on a promoter of transfected plasmids, which are mostly dissociative from the genomic DNA. These factors could account for relatively weak effects in our reporter assays (Figure 8E). Nevertheless, enhancer reporters can provide reference data for the results obtained from other approaches.
In this study, we proposed a novel model that YY1 promotes enhancer cluster formation through a phase separation mechanism to compartmentalize various coactivators and activate FOXM1 gene expression. Noteworthily, Weintraub et al. proposed that homodimerized YY1 could act as a structural regulator of enhancer-promoter loops (27). In this model, direct interaction of two YY1 protein molecules can bridge a distal enhancer element to a target promoter. The interacting sites between two YY1 molecules were first predicted to be its zinc fingers (78), and then mapped to a stretch of its spacer region (62). However, neither of these two regions overlaps with the IDRs of YY1. Whether YY1 homodimerization may directly contribute to its phase separation identified in our study is unclear. Importantly, the model of homodimerized YY1-mediated enhancer formation was reported in 2017 (27), which did not incorporate any concept of phase separation-mediated enhancer formation that was tested in 2018 (8,9). Therefore, it is plausible to predict that YY1-mediated connection between enhancer elements and a target promoter, especially in the case of forming an enhancer cluster or super-enhancer, is mostly through its phase-separated condensates.
Supplementary Material
ACKNOWLEDGEMENTS
We sincerely thank Dr Yang Shi for critical reading the manuscript. We are grateful for the generous technical supports from Dr Yujie Sun and Dr Mengling Zhang in designing the FISH probes. We sincerely thank Ms. Haijiao Huang for providing the technical assistance with the confocal laser scanning microscopy.
Contributor Information
Wenmeng Wang, College of Life Science, Northeast Forestry University, Harbin 150040, China.
Shiyao Qiao, College of Life Science, Northeast Forestry University, Harbin 150040, China.
Guangyue Li, College of Life Science, Northeast Forestry University, Harbin 150040, China.
Jiahui Cheng, College of Life Science, Northeast Forestry University, Harbin 150040, China.
Cuicui Yang, College of Life Science, Northeast Forestry University, Harbin 150040, China.
Chen Zhong, College of Life Science, Northeast Forestry University, Harbin 150040, China.
Daniel B Stovall, College of Arts and Sciences, Winthrop University, Rock Hill, SC 29733, USA.
Jinming Shi, College of Life Science, Northeast Forestry University, Harbin 150040, China.
Chunbo Teng, College of Life Science, Northeast Forestry University, Harbin 150040, China.
Dangdang Li, College of Life Science, Northeast Forestry University, Harbin 150040, China.
Guangchao Sui, College of Life Science, Northeast Forestry University, Harbin 150040, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [81802798 to D.L., 81872293 to G.S.]; startup fund from Northeast Forestry University (to G.S.); Fundamental Research Funds for the Central Universities [2572020AW34 to W.W.]; National Natural Science Foundation of Heilongjiang, China [LH2020H001 to D.L.]. Funding for open access charge: National Natural Science Foundation of China [81802798 to D.L., 81872293 to G.S.]; startup fund from Northeast Forestry University (to G.S.); Fundamental Research Funds for the Central Universities [2572020AW34 to W.W.]; National Natural Science Foundation of Heilongjiang, China [LH2020H001 to D.L.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Levine M., Cattoglio C., Tjian R.. Looping back to leap forward: transcription enters a new era. Cell. 2014; 157:13–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hnisz D., Day D.S., Young R.A.. Insulated neighborhoods: structural and functional units of mammalian gene control. Cell. 2016; 167:1188–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Whyte W.A., Orlando D.A., Hnisz D., Abraham B.J., Lin C.Y., Kagey M.H., Rahl P.B., Lee T.I., Young R.A.. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013; 153:307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hanahan D., Weinberg R.A.. Hallmarks of cancer: the next generation. Cell. 2011; 144:646–674. [DOI] [PubMed] [Google Scholar]
- 5. Hahn W.C., Bader J.S., Braun T.P., Califano A., Clemons P.A., Druker B.J., Ewald A.J., Fu H., Jagu S., Kemp C.J.et al.. An expanded universe of cancer targets. Cell. 2021; 184:1142–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Keegan L., Gill G., Ptashne M.. Separation of DNA binding from the transcription-activating function of a eukaryotic regulatory protein. Science. 1986; 231:699–704. [DOI] [PubMed] [Google Scholar]
- 7. Vaquerizas J.M., Kummerfeld S.K., Teichmann S.A., Luscombe N.M.. A census of human transcription factors: function, expression and evolution. Nat. Rev. Genet. 2009; 10:252–263. [DOI] [PubMed] [Google Scholar]
- 8. Sabari B.R., Dall’Agnese A., Boija A., Klein I.A., Coffey E.L., Shrinivas K., Abraham B.J., Hannett N.M., Zamudio A.V., Manteiga J.C.et al.. Coactivator condensation at super-enhancers links phase separation and gene control. Science. 2018; 361:eaar3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Boija A., Klein I.A., Sabari B.R., Dall’Agnese A., Coffey E.L., Zamudio A.V., Li C.H., Shrinivas K., Manteiga J.C., Hannett N.M.et al.. Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell. 2018; 175:1842–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hnisz D., Shrinivas K., Young R.A., Chakraborty A.K., Sharp P.A.. A phase separation model for transcriptional control. Cell. 2017; 169:13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shrinivas K., Sabari B.R., Coffey E.L., Klein I.A., Boija A., Zamudio A.V., Schuijers J., Hannett N.M., Sharp P.A., Young R.A.et al.. Enhancer features that drive formation of transcriptional condensates. Mol. Cell. 2019; 75:549–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lu Y., Wu T., Gutman O., Lu H., Zhou Q., Henis Y.I., Luo K.. Phase separation of TAZ compartmentalizes the transcription machinery to promote gene expression. Nat. Cell Biol. 2020; 22:453–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Boija A., Klein I.A., Young R.A.. Biomolecular condensates and cancer. Cancer Cell. 2021; 39:174–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sui G., Affar el B., Shi Y., Brignone C., Wall N.R., Yin P., Donohoe M., Luke M.P., Calvo D., Grossman S.R.et al.. Yin yang 1 is a negative regulator of p53. Cell. 2004; 117:859–872. [DOI] [PubMed] [Google Scholar]
- 15. Gronroos E., Terentiev A.A., Punga T., Ericsson J.. YY1 inhibits the activation of the p53 tumor suppressor in response to genotoxic stress. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:12165–12170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang Q., Stovall D.B., Inoue K., Sui G.. The oncogenic role of yin yang 1. Crit. Rev. Oncog. 2011; 16:163–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sigova A.A., Abraham B.J., Ji X., Molinie B., Hannett N.M., Guo Y.E., Jangi M., Giallourakis C.C., Sharp P.A., Young R.A.. Transcription factor trapping by RNA in gene regulatory elements. Science. 2015; 350:978–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Riggs K.J., Saleque S., Wong K.K., Merrell K.T., Lee J.S., Shi Y., Calame K.. Yin-yang 1 activates the c-myc promoter. Mol. Cell. Biol. 1993; 13:7487–7495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Palmer M.B., Majumder P., Cooper J.C., Yoon H., Wade P.A., Boss J.M.. Yin yang 1 regulates the expression of snail through a distal enhancer. Mol. Cancer Res. 2009; 7:221–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shi J., Hao A., Zhang Q., Sui G.. The role of YY1 in oncogenesis and its potential as a drug target in cancer therapies. Curr. Cancer Drug Targets. 2015; 15:145–157. [DOI] [PubMed] [Google Scholar]
- 21. Meliala I.T.S., Hosea R., Kasim V., Wu S.. The biological implications of yin yang 1 in the hallmarks of cancer. Theranostics. 2020; 10:4183–4200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Soutourina J. Transcription regulation by the mediator complex. Nat. Rev. Mol. Cell Biol. 2018; 19:262–274. [DOI] [PubMed] [Google Scholar]
- 23. Wang Y., Zolotarev N., Yang C.Y., Rambold A., Mittler G., Grosschedl R.. A Prion-like domain in transcription factor EBF1 promotes phase separation and enables b cell programming of progenitor chromatin. Immunity. 2020; 53:1151–1167. [DOI] [PubMed] [Google Scholar]
- 24. Shin Y., Brangwynne C.P.. Liquid phase condensation in cell physiology and disease. Science. 2017; 357:eaaf4382. [DOI] [PubMed] [Google Scholar]
- 25. Chong S., Dugast-Darzacq C., Liu Z., Dong P., Dailey G.M., Cattoglio C., Heckert A., Banala S., Lavis L., Darzacq X.et al.. Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science. 2018; 361:eaar2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Basu S., Mackowiak S.D., Niskanen H., Knezevic D., Asimi V., Grosswendt S., Geertsema H., Ali S., Jerkovic I., Ewers H.et al.. Unblending of transcriptional condensates in human repeat expansion disease. Cell. 2020; 181:1062–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Weintraub A.S., Li C.H., Zamudio A.V., Sigova A.A., Hannett N.M., Day D.S., Abraham B.J., Cohen M.A., Nabet B., Buckley D.L.et al.. YY1 is a structural regulator of enhancer-promoter loops. Cell. 2017; 171:1573–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stovall D.B., Wan M., Zhang Q., Dubey P., Sui G.. DNA vector-based RNA interference to study gene function in cancer. J. Vis. Exp. 2012; e4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Deng Z., Wan M., Sui G.. PIASy-mediated sumoylation of yin yang 1 depends on their interaction but not the RING finger. Mol. Cell. Biol. 2007; 27:3780–3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Huang W., Smaldino P.J., Zhang Q., Miller L.D., Cao P., Stadelman K., Wan M., Giri B., Lei M., Nagamine Y.et al.. Yin yang 1 contains G-quadruplex structures in its promoter and 5′-UTR and its expression is modulated by G4 resolvase 1. Nucleic Acids Res. 2012; 40:1033–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Deng Z., Wan M., Cao P., Rao A., Cramer S.D., Sui G.. Yin yang 1 regulates the transcriptional activity of androgen receptor. Oncogene. 2009; 28:3746–3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hagege H., Klous P., Braem C., Splinter E., Dekker J., Cathala G., de Laat W., Forne T.. Quantitative analysis of chromosome conformation capture assays (3C-qPCR). Nat. Protoc. 2007; 2:1722–1733. [DOI] [PubMed] [Google Scholar]
- 33. Yu M., Peng Z., Qin M., Liu Y., Wang J., Zhang C., Lin J., Dong T., Wang L., Li S.et al.. Interferon-gamma induces tumor resistance to anti-PD-1 immunotherapy by promoting YAP phase separation. Mol. Cell. 2021; 81:1216–1230. [DOI] [PubMed] [Google Scholar]
- 34. Beliveau B.J., Kishi J.Y., Nir G., Sasaki H.M., Saka S.K., Nguyen S.C., Wu C.T., Yin P.. OligoMiner provides a rapid, flexible environment for the design of genome-scale oligonucleotide in situ hybridization probes. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:E2183–E2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Raj A., van den Bogaard P., Rifkin S.A., van Oudenaarden A., Tyagi S.. Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods. 2008; 5:877–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bousard A., Raposo A.C., Żylicz J.J., Picard C., Pires V.B., Qi Y., Gil C., Syx L., Chang H.Y., Heard E.et al.. The role of Xist-mediated polycomb recruitment in the initiation of X-chromosome inactivation. EMBO Rep. 2019; 20:e48019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cao P., Deng Z., Wan M., Huang W., Cramer S.D., Xu J., Lei M., Sui G.. MicroRNA-101 negatively regulates ezh2 and its expression is modulated by androgen receptor and HIF-1alpha/HIF-1beta. Mol. Cancer. 2010; 9:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wan M., Huang W., Kute T.E., Miller L.D., Zhang Q., Hatcher H., Wang J., Stovall D.B., Russell G.B., Cao P.D.et al.. Yin yang 1 plays an essential role in breast cancer and negatively regulates p27. Am. J. Pathol. 2012; 180:2120–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Houbaviy H.B., Usheva A., Shenk T., Burley S.K.. Cocrystal structure of YY1 bound to the adeno-associated virus P5 initiator. Proc. Natl. Acad. Sci. U.S.A. 1996; 93:13577–13582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dosztanyi Z., Csizmok V., Tompa P., Simon I.. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005; 21:3433–3434. [DOI] [PubMed] [Google Scholar]
- 41. Uversky V.N. Analyzing IDPs in interactomes. Methods Mol. Biol. 2020; 2141:895–945. [DOI] [PubMed] [Google Scholar]
- 42. Lu H., Yu D., Hansen A.S., Ganguly S., Liu R., Heckert A., Darzacq X., Zhou Q.. Phase-separation mechanism for C-terminal hyperphosphorylation of RNA polymerase iI. Nature. 2018; 558:318–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Qamar S., Wang G., Randle S.J., Ruggeri F.S., Varela J.A., Lin J.Q., Phillips E.C., Miyashita A., Williams D., Ströhl F.et al.. FUS phase separation is modulated by a molecular chaperone and methylation of arginine cation-π interactions. Cell. 2018; 173:720–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lin Y., Protter D.S., Rosen M.K., Parker R.. Formation and maturation of phase-separated liquid droplets by RNA-Binding proteins. Mol. Cell. 2015; 60:208–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li R.H., Tian T., Ge Q.W., He X.Y., Shi C.Y., Li J.H., Zhang Z., Liu F.Z., Sang L.J., Yang Z.Z.et al.. A phosphatidic acid-binding lncRNA SNHG9 facilitates LATS1 liquid-liquid phase separation to promote oncogenic YAP signaling. Cell Res. 2021; 31:1088–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shi Y., Seto E., Chang L.S., Shenk T.. Transcriptional repression by YY1, a human GLI-Kruppel-related protein, and relief of repression by adenovirus E1A protein. Cell. 1991; 67:377–388. [DOI] [PubMed] [Google Scholar]
- 47. Lee J.S., Galvin K.M., See R.H., Eckner R., Livingston D., Moran E., Shi Y.. Relief of YY1 transcriptional repression by adenovirus E1A is mediated by E1A-associated protein p300. Genes Dev. 1995; 9:1188–1198. [DOI] [PubMed] [Google Scholar]
- 48. Yao Y.L., Yang W.M., Seto E.. Regulation of transcription factor YY1 by acetylation and deacetylation. Mol. Cell. Biol. 2001; 21:5979–5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Giles R.H., Peters D.J., Breuning M.H.. Conjunction dysfunction: CBP/p300 in human disease. Trends Genet. 1998; 14:178–183. [DOI] [PubMed] [Google Scholar]
- 50. Vo N., Goodman R.H.. CREB-binding protein and p300 in transcriptional regulation. J. Biol. Chem. 2001; 276:13505–13508. [DOI] [PubMed] [Google Scholar]
- 51. Jeronimo C., Bataille A.R., Robert F.. The writers, readers, and functions of the RNA polymerase II C-terminal domain code. Chem. Rev. 2013; 113:8491–8522. [DOI] [PubMed] [Google Scholar]
- 52. Affar el B., Gay F., Shi Y., Liu H., Huarte M., Wu S., Collins T., Li E., Shi Y.. Essential dosage-dependent functions of the transcription factor yin yang 1 in late embryonic development and cell cycle progression. Mol. Cell. Biol. 2006; 26:3565–3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Liao G.B., Li X.Z., Zeng S., Liu C., Yang S.M., Yang L., Hu C.J., Bai J.Y.. Regulation of the master regulator FOXM1 in cancer. Cell Commun. Signal. 2018; 16:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Myatt S.S., Lam E.W.. The emerging roles of forkhead box (Fox) proteins in cancer. Nat. Rev. Cancer. 2007; 7:847–859. [DOI] [PubMed] [Google Scholar]
- 55. Wasserman W.W., Sandelin A.. Applied bioinformatics for the identification of regulatory elements. Nat. Rev. Genet. 2004; 5:276–287. [DOI] [PubMed] [Google Scholar]
- 56. Chen K., Lu Y., Shi K., Stovall D.B., Li D., Sui G.. Functional analysis of YY1 zinc fingers through cysteine mutagenesis. FEBS Lett. 2019; 593:1392–1402. [DOI] [PubMed] [Google Scholar]
- 57. Patten D.K., Corleone G., Gyorffy B., Perone Y., Slaven N., Barozzi I., Erdos E., Saiakhova A., Goddard K., Vingiani A.et al.. Enhancer mapping uncovers phenotypic heterogeneity and evolution in patients with luminal breast cancer. Nat. Med. 2018; 24:1469–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Han J., Meng J., Chen S., Wang X., Yin S., Zhang Q., Liu H., Qin R., Li Z., Zhong W.et al.. YY1 complex promotes quaking expression via super-enhancer binding during EMT of hepatocellular carcinoma. Cancer Res. 2019; 79:1451–1464. [DOI] [PubMed] [Google Scholar]
- 59. Hahn S. Phase separation, protein disorder, and enhancer function. Cell. 2018; 175:1723–1725. [DOI] [PubMed] [Google Scholar]
- 60. Zhang Q., Wan M., Shi J., Horita D.A., Miller L.D., Kute T.E., Kridel S.J., Kulik G., Sui G.. Yin yang 1 promotes mTORC2-mediated AKT phosphorylation. J. Mol. Cell Biol. 2016; 8:232–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Boeynaems S., Alberti S., Fawzi N.L., Mittag T., Polymenidou M., Rousseau F., Schymkowitz J., Shorter J., Wolozin B., Van Den Bosch Let al.. Protein phase separation: a new phase in cell biology. Trends Cell Biol. 2018; 28:420–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Li L., Williams P., Ren W., Wang M.Y., Gao Z., Miao W., Huang M., Song J., Wang Y.. YY1 interacts with guanine quadruplexes to regulate DNA looping and gene expression. Nat. Chem. Biol. 2021; 17:161–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Qiu X., Kumari G., Gerasimova T., Du H., Labaran L., Singh A., De S., Wood W.H., Becker K.G., Zhou W.et al.. Sequential enhancer sequestration dysregulates recombination center formation at the IgH locus. Mol. Cell. 2018; 70:21–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Beagan J.A., Duong M.T., Titus K.R., Zhou L., Cao Z., Ma J., Lachanski C.V., Gillis D.R., Phillips-Cremins J.E.. YY1 and CTCF orchestrate a 3D chromatin looping switch during early neural lineage commitment. Genome Res. 2017; 27:1139–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hua J.T., Ahmed M., Guo H., Zhang Y., Chen S., Soares F., Lu J., Zhou S., Wang M., Li H.et al.. Risk SNP-Mediated promoter-enhancer switching drives prostate cancer through lncRNA PCAT19. Cell. 2018; 174:564–575. [DOI] [PubMed] [Google Scholar]
- 66. Yang W.M., Yao Y.L., Seto E.. The FK506-binding protein 25 functionally associates with histone deacetylases and with transcription factor YY1. EMBO J. 2001; 20:4814–4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Caretti G., Di Padova M., Micales B., Lyons G.E., Sartorelli V.. The polycomb ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 2004; 18:2627–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ko C.Y., Hsu H.C., Shen M.R., Chang W.C., Wang J.M.. Epigenetic silencing of CCAAT/enhancer-binding protein delta activity by YY1/polycomb group/DNA methyltransferase complex. J. Biol. Chem. 2008; 283:30919–30932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Liu J., Xie Y., Guo J., Li X., Wang J., Jiang H., Peng Z., Wang J., Wang S., Li Q.et al.. Targeting NSD2-mediated SRC-3 liquid-liquid phase separation sensitizes bortezomib treatment in multiple myeloma. Nat. Commun. 2021; 12:1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Han X., Yu D., Gu R., Jia Y., Wang Q., Jaganathan A., Yang X., Yu M., Babault N., Zhao C.et al.. Roles of the BRD4 short isoform in phase separation and active gene transcription. Nat. Struct. Mol. Biol. 2020; 27:333–341. [DOI] [PubMed] [Google Scholar]
- 71. Austen M., Lüscher B., Lüscher-Firzlaff J.M.. Characterization of the transcriptional regulator YY1. The bipartite transactivation domain is independent of interaction with the TATA box-binding protein, transcription factor IIB, TAFII55, or cAMP-responsive element-binding protein (CPB)-binding protein. J. Biol. Chem. 1997; 272:1709–1717. [DOI] [PubMed] [Google Scholar]
- 72. Kang K., Choi Y., Kim H.H., Yoo K.H., Yu S.. Predicting FOXM1-Mediated gene regulation through the analysis of genome-wide FOXM1 binding sites in MCF-7, K562, SK-N-SH, GM12878 and ECC-1 cell lines. Int. J. Mol. Sci. 2020; 21:6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Sanders D.A., Ross-Innes C.S., Beraldi D., Carroll J.S., Balasubramanian S.. Genome-wide mapping of FOXM1 binding reveals co-binding with estrogen receptor alpha in breast cancer cells. Genome Biol. 2013; 14:R6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ma L., Gao Z., Wu J., Zhong B., Xie Y., Huang W., Lin Y.. Co-condensation between transcription factor and coactivator p300 modulates transcriptional bursting kinetics. Mol. Cell. 2021; 81:1682–1697. [DOI] [PubMed] [Google Scholar]
- 75. Liu Q., Thoms J.A.I., Nunez A.C., Huang Y., Knezevic K., Packham D., Poulos R.C., Williams R., Beck D., Hawkins N.J.et al.. Disruption of a -35 kb enhancer impairs CTCF binding and MLH1 expression in colorectal cells. Clin. Cancer Res. 2018; 24:4602–4611. [DOI] [PubMed] [Google Scholar]
- 76. Liu Q., Kulak M.V., Borcherding N., Maina P.K., Zhang W., Weigel R.J., Qi H.H.. A novel HER2 gene body enhancer contributes to HER2 expression. Oncogene. 2018; 37:687–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Inoue F., Kircher M., Martin B., Cooper G.M., Witten D.M., McManus M.T., Ahituv N., Shendure J.. A systematic comparison reveals substantial differences in chromosomal versus episomal encoding of enhancer activity. Genome Res. 2017; 27:38–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. López-Perrote A., Alatwi H.E., Torreira E., Ismail A., Ayora S., Downs J.A., Llorca O.. Structure of yin yang 1 oligomers that cooperate with ruvbl1-ruvbl2 ATPases. J. Biol. Chem. 2014; 289:22614–22629. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.