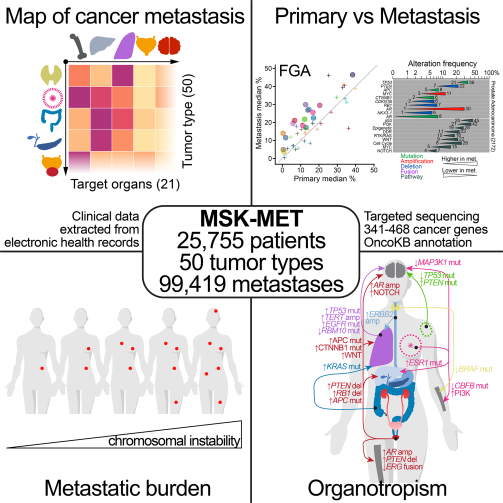
SUMMARY
Metastatic progression is the main cause of death in cancer patients, whereas the underlying genomic mechanisms driving metastasis remain largely unknown. Here, we assembled MSK-MET, a pan-cancer cohort of over 25,000 patients with metastatic diseases. By analyzing genomic and clinical data from this cohort, we identified associations between genomic alterations and patterns of metastatic dissemination across 50 tumor types. We found that chromosomal instability is strongly correlated with metastatic burden in some tumor types, including prostate adenocarcinoma, lung adenocarcinoma and HR+/HER2+ breast ductal carcinoma, but not in others, including colorectal cancer and high grade serous ovarian cancer, where copy-number alteration patterns may be established early in tumor development. We also identified somatic alterations associated with metastatic burden and specific target organs. Our data offer a valuable resource for the investigation of the biological basis for metastatic spread and highlight the complex role of chromosomal instability in cancer progression.
IN BRIEF
Clinico-genomic analysis of MSK-MET, a cohort of over 25,000 patients with metastasis across 50 cancer types, identifies somatic alterations associated with organ-specific metastasis and highlights that chromosomal instability correlates with metastatic burden in a cancer type-dependent manner.
Graphical Abstract
INTRODUCTION
Although most cancer deaths are due to metastatic spread, little is known about the genomic determinants of cancer metastasis. Once metastatic cancer cells have detached from the primary tumor site, they can invade all parts of the body (Lambert et al., 2017; Massagué and Obenauf, 2016). However, the distribution of metastatic sites for a given primary tumor is not random and is dictated by factors such as anatomical location, cell of origin and molecular subtype, among others (Gao et al., 2019; Nguyen et al., 2009). Furthermore, tumor cell-extrinsic factors such as treatment, target organ microenvironment and other systemic factors such as circulating chemokines and cytokines can also influence the pattern of metastatic progression (Massagué and Ganesh, 2021). The classical seed-and-soil hypothesis, according to which disseminated cancer cells preferentially colonize organs that enable and are compatible with their own growth, has been explored for more than a century (1889). Yet much remains unknown about the interplay between tumor genomic features and metastatic potential, as well as organ-specific patterns of metastasis.
Molecular profiling of tumors coupled with clinical annotation of metastatic events could help provide insight into this question. However, large-scale cancer sequencing efforts have so far focused on primary, untreated tumors (e.g., The Cancer Genome Atlas (Sanchez-Vega et al., 2018)), or they have characterized the overall genomic landscape of metastatic disease without explicitly interrogating specific metastatic organotropism (Priestley et al., 2019; Robinson et al., 2017; Zehir et al., 2017). Other studies have investigated the genomic complexity of cancer metastasis by reconstructing tumor evolution across different organs at varying levels of resolution, but they have been limited by small sample sizes (Brastianos et al., 2015; Brown et al., 2017; Eckert et al., 2016; Hu et al., 2020; Jiménez-Sánchez et al., 2017; Makohon-Moore et al., 2017; Naxerova et al., 2017; Noorani et al., 2020; Reiter et al., 2020; Shih et al., 2020). Identifying associations between genomic features and specific patterns of metastatic spread is an active area of research and several landmark studies on this topic have been published during the past few years (Birkbak and McGranahan, 2020). In particular, richly annotated datasets combining genomic features and detailed clinical history of metastases for individual patients have been recently made available through large collaborative efforts such as METABRIC in breast cancer (Rueda et al., 2019) and TRACERx in clear-cell renal cell carcinoma (Turajlic et al., 2018). However, a study involving thousands of participants across multiple tumor types in which clinical and genomic data has been homogeneously processed through a unified computational pipeline is still lacking.
We assembled a pan-cancer cohort of >25,000 patients with tumor genomic profiling and clinical information on metastatic events and outcomes, which we designate MSK-MET (Memorial Sloan Kettering - Metastatic Events and Tropisms). All samples were profiled using the MSK-IMPACT targeted sequencing platform (Cheng et al., 2015), which identifies somatic mutations, rearrangements and copy-number alterations in 341–468 cancer genes, as well as tumor mutational burden (TMB), chromosomal instability and microsatellite instability. Metastatic events were extracted from the electronic health records (EHR) and mapped to a reference set of 21 anatomic locations. We analyzed genomic differences between primary and metastatic samples and between primary tumors from metastatic and non-metastatic patients, stratified by tumor type and molecular subtypes. Our analysis identified associations between metastatic burden (defined as the number of distinct organs affected by metastases throughout a patient’s clinical course) and specific genomic features, including TMB, chromosomal instability, and somatic alterations in individual cancer genes. We also identified associations between genomic alterations and organ-specific patterns of metastatic dissemination and progression. The clinical and genomic data used in our study have been made publicly available and constitute a valuable resource that will help further our understanding of metastatic disease.
RESULTS
Overview of the MSK-MET cohort
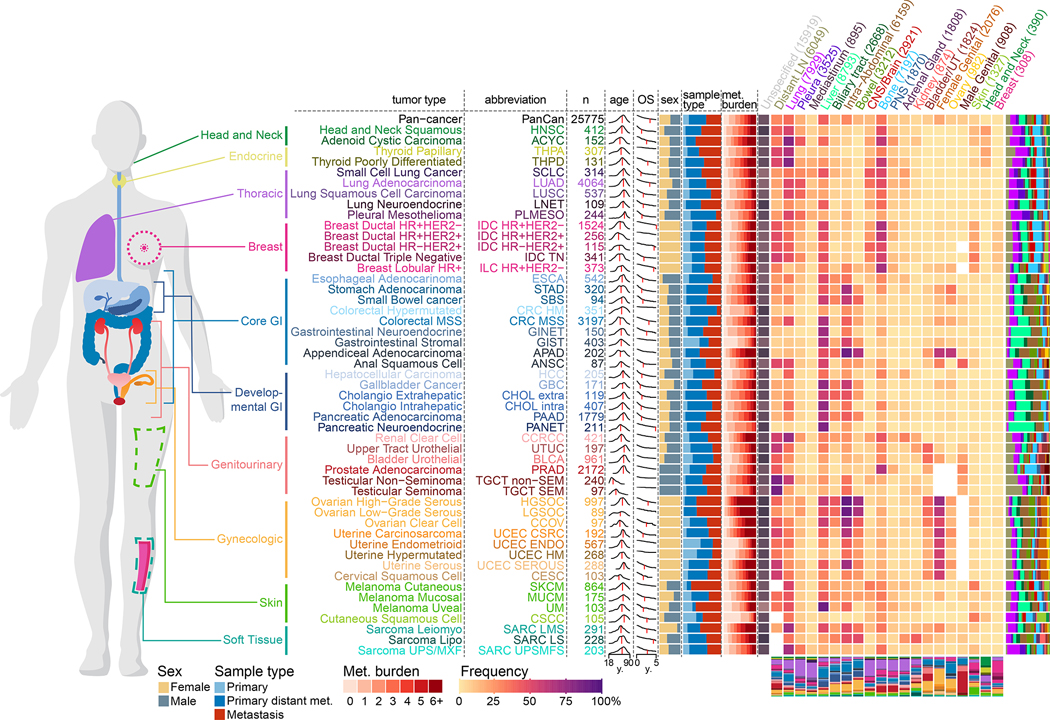
A total of 25,775 patients were included in the present study, consisting of 15,632 (61%) primary and 10,143 (39%) metastatic specimens spanning 50 different tumor types (Figure S1A–D; Table S1A). To ensure that samples are independent of each other, a unique representative sample was selected for the analysis of patients with multiple available sequenced samples (see Methods). The median interval between sample acquisition and sequencing was 62 days (interquartile range (IQR) = 0–287 days). The median sequencing coverage was 653x (IQR = 525–790x) and the median tumor purity assessed by pathologists was 40% (IQR = 20–50%) (Figure S1B). The majority of sequenced samples obtained from metastatic sites were from lymph nodes (n=2305, 23%), liver (n=2289, 23%), lung (n=982, 10%), or bone (n=726, 7%). Among primary tumors, 11,741 (75%) were from patients with metastatic disease at the time of sequencing or at a later time (Figure S1D). Overall, metastatic samples were more pre-treated than primary samples (39% vs 15%) with some cancer types showing a larger difference (prostate cancer, 72% vs 10%) than others (lung adenocarcinoma, 32% vs 24%) (Figure S1E, Table S1B). Over the entire course of the disease, a total of 99,419 metastatic events from 21,546 metastatic patients were retrieved from the EHR and mapped to 21 organ sites. The most common target organ sites were lung, liver and bone (Figure S1F). The frequencies of organ-specific metastasis of individual tumor types were similar to previous reports (Budczies et al., 2015; Gao et al., 2019) (Figure S1G). Internal validation using 4,859 (22.5%) patients included in previous studies with available metastatic events extracted through manual chart review (Abida et al., 2017; Jones et al., 2021; Razavi et al., 2018; Shoushtari et al., 2021; Yaeger et al., 2018) revealed a high concordance and sensitivity with metastatic events extracted from the EHR (Figure S1H–I). We used this data to map patterns of metastatic dissemination from 50 tumor types to 21 metastatic organ sites (Figure 1).
Figure 1. Overview of the MSK-MET cohort.
Metastatic patterns of 50 tumor types. For each tumor type, the following attributes are shown from left to right: tumor type abbreviation, number of patients, distribution of age at sequencing (red vertical line indicates the median), overall survival in years from time of sequencing (red vertical line indicates the median OS), sex ratio (female = gold, male = grey), distribution of metastatic burden across all patients (ranging from 0 to ≥6 distinct metastatic sites), and a heatmap with the percentage of metastatic patients with metastases at specific metastatic sites (the entire clinical course was taken into consideration). The number in each cell indicates the frequency of patients having at least one reported metastasis at that given site. For each tumor type, the distribution of all metastasis events by 21 organ sites is shown as a stacked bar chart to the right of the heatmap. For each metastatic site, the distribution of all 50 tumor types is shown as a stacked bar chart below the heatmap. For each metastatic site, the number of patients with at least one metastasis is indicated in parentheses. Frequencies for sex-specific target organs (female genital, ovary and male genital) were calculated using patients of the corresponding sex.
For the whole cohort, the median age at sequencing was 64y, ranging from a median of 33y for patients with testicular non-seminoma to a median of 70y for patients with cutaneous squamous cell carcinoma. Overall, the median follow-up time was 30 months and the five-year survival rate was 40%, ranging from 90% in testicular seminoma to 10% in pancreatic adenocarcinoma. There was a median of four metastatic events per patient, ranging from one in hypermutated colorectal cancer to eight in high-grade serous ovarian carcinoma. Metastatic patterns differed by tumor types and histological subtypes. For example, compared to lung adenocarcinoma, lung neuroendocrine cancer had a higher prevalence of liver metastasis (42% vs. 22%) but a lower prevalence of CNS/Brain metastasis (19% vs. 34%). Similarly, compared to ductal breast cancer, lobular breast cancer had a lower prevalence of lung metastasis (10% vs. 30%) but a higher prevalence of ovary(15% vs. 4%) and peritoneum metastasis (30% vs. 10%), as reported before (Borst and Ingold, 1993). Differences in metastatic patterns were also observed across molecular subtypes of the same tumor type. For example, and in line with a previous study (Kennecke et al., 2010), HR−/HER2+ ductal breast cancer had a higher prevalence of CNS/Brain metastasis than the HR+/HER2− subtype (38% vs. 20%), while the latter had a higher prevalence of bone metastasis (67% vs. 49%).
Detailed time stamps for each metastatic event in 21,058 metastatic patients with available annotations were extracted from the EHR and are provided in Table S1B. While the order of metastatic colonization for individual organs varies widely across cancer types, we found that ovary and liver metastases were generally detected early, whereas CNS/Brain and peripheral nervous system metastasis tend to be detected later (Figure S1J). Similarly, when we used a Bradley-Terry model to derive a temporal ordering of target organs for each specific tumor type we observed that the order of metastatic colonization is not consistent across cancer types, likely influenced by a combination of anatomical, genomic and clinicopathological factors (Figure S1K).
Genomic differences between primary and metastatic tumors
To determine sample type specific genomic differences across 50 tumor types, we compared the genomic features of primary (n=15,632) and metastatic tumors (n=10,143) (independent of the metastatic status of the patients). The number of sequenced primaries was higher than the number of sequenced metastases for most tumor types, with some exceptions, such as cutaneous melanoma, high-grade serous ovarian cancer and adenoid cystic carcinoma. In 16 tumor types, metastases were significantly more chromosomally unstable, as inferred by a higher fraction of genome altered (FGA), compared to primary tumors, consistent with previous findings (Bakhoum et al., 2018; Ben-David and Amon, 2020; Hieronymus et al., 2018; Shukla et al., 2020; Stopsack et al., 2019; Watkins et al., 2020) (Figure 2A–B; Table S2A). The difference in tumor purity- and ploidy-adjusted FGA (adjusted FGA) was confirmed in 11 tumor types using a subset of samples with available FACETS data (n=17,224) (Table S2A). FACETS allowed us to estimate the frequency of whole-genome duplication (WGD) and assess the clonality of individual variants. As previously reported, WGD frequencies varied across tumor types (Bielski et al., 2018). In seven tumor types, we observed a significantly higher frequency of WGD in metastases compared to primary tumors (Figure 2A–B; Table S2A). The higher chromosomal instability (0% vs. 14%) and higher frequency of WGD (4% vs. 16%) were particularly marked in uterine endometrioid, which can be explained by differences in the distribution of genomic subtypes within these two groups (Cancer Genome Atlas Research Network et al., 2013). Tumor mutational burden (TMB) was significantly higher in metastases from 10 tumor types, while TMB was lower only in metastases from hypermutated uterine cancer (Figure 2A–B; Table S2A). Consistent with the evolutionary bottleneck hypothesis (Birkbak and McGranahan, 2020), metastases from 12 tumor types were significantly more homogeneous, with a higher fraction of clonal mutations compared to primary tumors.
Figure 2. Genomic differences between primary tumors and metastases.
Comparisons of the median fraction genome altered (FGA), median whole-genome duplication (WGD) frequency, median tumor mutation burden (TMB), and median clonal fraction for each tumor type in metastatic vs. primary tumors. Tumor types with statistically significant differences are labeled. For TMB both axes were limited to 10mut/Mb.
(A) The following clinical and genomic features are shown side-by-side for primary (top row within each cancer type) and metastatic (bottom row) sequenced samples using a combination of bar charts and violin plots; from left to right: sample counts, FGA, fraction of samples with WGD, TMB, clonality, fraction of samples with high TMB, and distribution of the highest actionable alteration levels. The black vertical line in each violin plots represents the median. The heatmap shows the frequency of individual arm level alterations in primary tumors and metastases (only the frequency of the more frequent event, gain or loss, is shown). Tumor types are ordered from top to bottom by decreasing FGA in metastasis and grouped by organ systems. * indicates q-value < 0.05. WGD and clonality were available for a subset of 17,224 samples with FACETS data.
(B) Statistically significant differences in the frequency of oncogenic alterations and pathways between primary tumors and metastases in individual tumor types. Triangles summarize oncogenic alteration frequencies in primary tumors vs. metastases and are colored according to alteration type. Gene names in italics refer to specific genes, those in regular font refer to pathways.
We further explored the clinical significance of TMB by comparing the percentage of patients with a high TMB (≥10 mut/Mb) and observed a higher percentage of TMB-high tumors in metastases from lung adenocarcinoma (19% vs. 27%, q-value < 0.001), HR+/HER2− ductal breast cancer (2% vs. 7%, q-value < 0.001) and lobular breast cancer patients (5% vs. 19%, q-value < 0.001). In six tumor types, we detected a significantly higher proportion of any actionable mutations (OncoKB levels 1 to 3, Methods) in metastases compared to primary tumors, but these differences were not significant after adjusting for differences in FGA and TMB (Figure 2B; Table S2A). Next, we investigated differences in the frequency of arm-level copy number alterations between primary tumors and metastases. Because FGA was generally higher in metastases, we used a multivariable model to adjust for FGA and found 26 statistically significant differences (Figure 2B; Table S2A). For example, in pancreatic adenocarcinoma, gain of chromosome 12p gain, where the oncogene KRAS is located, was more frequent in metastases than in primary tumors (17% vs. 4%, q-value = 0.002). In HR+/HER2− ductal and lobular breast cancer, loss of chromosome 16q, a feature of low-grade breast cancer (Natrajan et al., 2009), was more frequent in primary tumors than in metastases (41% vs. 30%, q-value < 0.001 and 68% vs. 56%, q-value = 0.002, respectively). Finally, we investigated the frequency of recurrent oncogenic alterations between primary tumors and metastases and identified a total of 67 statistically significant differences across 17 tumor types. We also investigated the frequency of oncogenic pathways and identified 47 statistically significant differences across 11 tumor types (Figure 2C; Table S2A). Amongst the statistically significant alterations, 53 were more frequent in metastases, while only 14 alterations were more frequent in primary tumors. The most commonly observed significant alteration was TP53 mutation, which was more frequent in metastases in 7 tumor types (lung adenocarcinoma, prostate adenocarcinoma, HR+/HER2− ductal breast, MSS colorectal, lobular breast cancer, pancreatic neuroendocrine and uterine endometrioid). A possible explanation is that TP53 mutation is a later event in some of these tumor types; in others, it may simply be a hallmark of more aggressive disease. The notable exception was head and neck cancer, where TP53 mutations were more frequent in primary tumors (62% vs. 45%, q-value = 0.01). Other genomic alterations that were most often enriched in metastases included CDKN2A deletion (significant in 5 tumor types), PTEN mutation and deletion (4 tumor types) and MYC amplification (4 tumor types). The most common significantly enriched oncogenic pathways in metastases were p53, Cell Cycle and DNA damage repair. The most significant differences were observed for alterations known to be associated with resistance to hormonal therapy in hormone-sensitive tumors. For example, AR amplification and AR mutations were significantly more frequent in prostate cancer metastases (1% vs. 30% and 0% vs. 6%, q-value < 0.001), and ESR1 mutations were more frequent in HR+/HER2− ductal breast cancer (2% vs. 19%, q-value < 0.001), lobular breast cancer (2% vs. 13%, q-value < 0.001), and endometrioid uterine cancer metastases (3% vs. 10%, q-value = 0.002). These differences can likely be attributed to positive selection due to therapy since most patients with prostate cancer and ER+ breast cancer receive hormone therapy. TERT mutations were more frequent in metastases from papillary thyroid cancer and cutaneous melanoma patients (46% vs. 69%, q-value = 0.001 and 70% vs. 81%, q-value = 0.02), but higher in primary tumors from head and neck squamous cell carcinoma patients (41% vs. 25%, q-value = 0.02). ALK fusions, a predictive biomarker for the use of ALK inhibitors, were slightly more frequent in lung adenocarcinoma metastases (3% vs. 6%, q-value < 0.001). KRAS mutations were more frequent in metastases from pancreatic neuroendocrine patients (1% vs. 10%, q-value = 0.03) as was the overall frequency of RTK/RAS pathway alteration in this tumor type (7% vs. 21%, q-value = 0.03). While KRAS mutation is a hallmark of pancreatic adenocarcinoma, this could suggest the existence of a transdifferentiation mechanism from neuroendocrine to an adenocarcinoma phenotype during metastatic progression. The results from our comparison of primaries vs. metastatic samples were largely replicated using an independent cohort of 9,215 patients sequenced with MSK-IMPACT since the data freeze date (Figure S2, Table S2B). Collectively, these data indicate that metastases have higher chromosomal instability across many tumor types and that mutations in a multitude of driver alterations occur at different frequencies in primary and metastatic tumors.
Genomic differences between primary samples from metastatic and non-metastatic patients
Many of the primary tumors included in the previous analysis were from patients with metastatic disease. To identify genomic determinants of metastatic disease present in primary tumors we compared the genomic features of primary tumors from metastatic patients (n=11,942) to primary tumors from non-metastatic patients (n=3,690). The median follow-up time for these two groups was 33 months and 27 months, respectively. In 10 tumor types, FGA was significantly higher in primary tumors from metastatic patients as compared to primary tumors from patients without metastases. Compared to non-metastatic patients, TMB was significantly higher in 7 tumor types but lower in head and neck squamous cell carcinoma (Figure S3A–B; Table S2C). When interrogating the frequencies of recurrent oncogenic alterations, we identified statistically significant frequency differences in 32 genes across 12 tumor types and 21 oncogenic pathways across 9 tumor types (Figure S3C; Table S2C), with the majority of events observed at higher frequencies in primary tumors from metastatic patients. Compared to non-metastatic patients, TP53 mutations were significantly more frequent in metastatic patients with lung adenocarcinoma (28% vs. 45%, q-value < 0.001), HR+/HER2− ductal breast cancer (17% vs. 29%, q-value < 0.001), urothelial bladder cancer (31% vs. 53%, q-value < 0.001), prostate adenocarcinoma (16% vs. 23%, q-value < 0.001), and endometrioid uterine cancer (9% vs. 20%, q-value = 0.01). TERT promoter mutations were more frequent in metastatic patients with papillary thyroid cancer (20% vs. 56%, q-value = 0.004). The frequency of MYC amplification was significantly higher in metastatic patients with prostate adenocarcinoma (1% vs. 4%, q-value = 0.03), microsatellite stable (MSS) colorectal cancer (1% vs. 4%, q-value = 0.03), and TN ductal breast cancer (3% vs. 17%, q-value = 0.03). On the other hand, SPOP mutations were less frequent in primary tumors from metastatic prostate adenocarcinoma patients (18% vs. 12%, q-value = 0.03), PIK3CA mutations were less frequent in the primary tumors of HR+/HER2− ductal breast cancer metastatic patients (49% vs. 38%, q-value = 0.003), and CDKN2A mutations were less frequent in the primary tumors of pancreatic cancer metastatic patients (17% vs. 7%, q-value = 0.02). These findings support the hypothesis that a higher chromosomal instability is associated with metastatic progression in multiple tumor types and that several individual driver mutations might inform metastatic risk. Only a few of these, such as SPOP mutations in prostate cancer, which has been previously reported to be more frequent in primary tumors (Armenia et al., 2018), are associated with decreased metastatic potential. The comparative analysis of genomic features differing between primary tumors vs metastases (“P vs M”) and genomic features differing between primary tumors from non-metastatic patients vs metastatic patients (“P-noM vs P-M”) can help to identify alterations that occur earlier or later in metastasis (Figure S3D–E). However, a detailed analysis of the timing of somatic events involved in metastatic progression will require additional data sources (such as comprehensive characterization of multiple lines of treatment, including surgical interventions) and is beyond the scope of our study.
Genomic features associated with metastatic burden
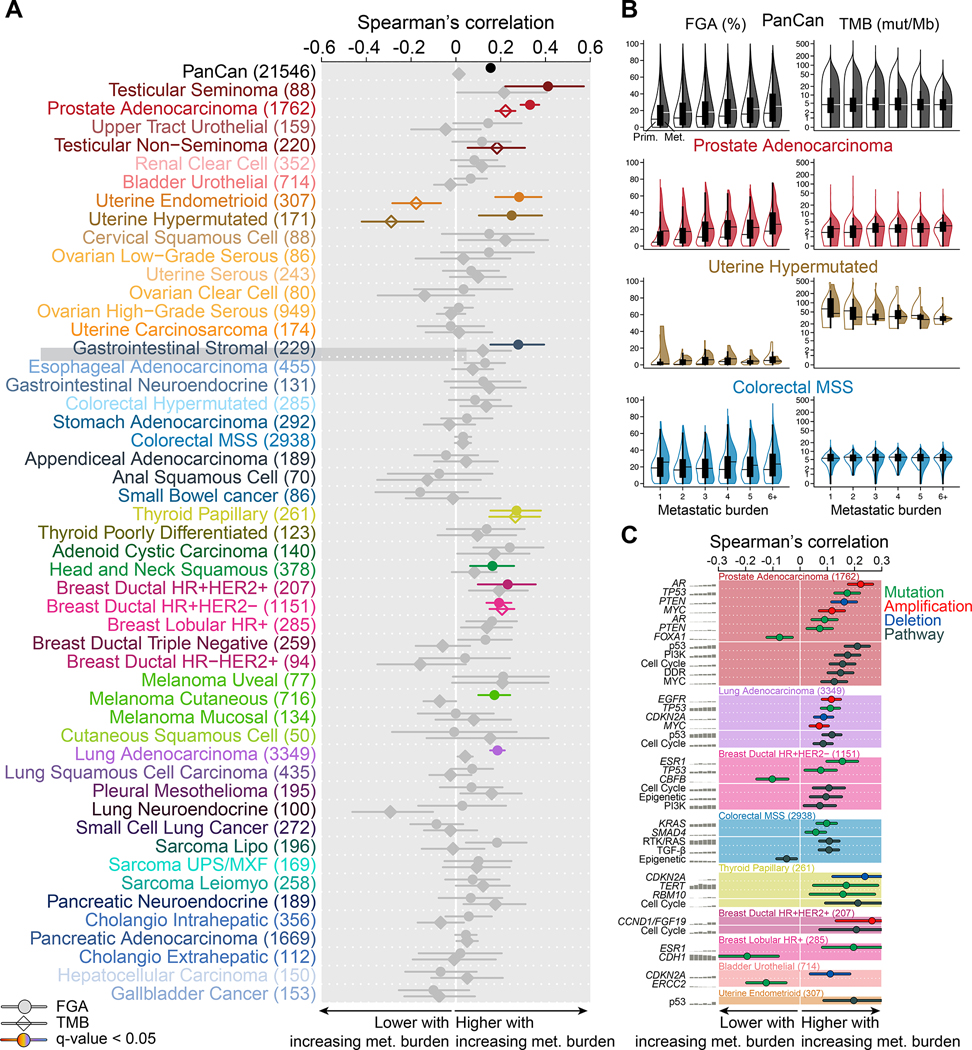
To explore the genomic determinants of metastatic burden, we analyzed the relationship between genomic alterations and the number of metastatic sites per patient (n=21,546). Not surprisingly, a higher metastatic burden was significantly associated with shorter overall survival in most (39/50, 78%) tumor types (Table S3A). We observed that chromosomal instability, as inferred by FGA, was positively correlated with metastatic burden on a pan-cancer level and in 11 individual tumor types. TMB, on the other hand, was not associated with metastatic burden on a pan-cancer level; it was positively correlated with metastatic burden in 3 tumor types, and negatively associated with metastatic burden in endometrioid and hypermutated uterine cancer (Figure 3A–B; Table S3B). One of the strongest correlations between FGA and metastatic burden was observed in prostate cancer (rho = 0.33, q-value = 7.0E-45), which is in line with previous studies (Hieronymus et al., 2018; Taylor et al., 2010). Conversely, we did not observe such association in many tumor types, including MSS colorectal cancer, where chromosomal instability is already high in patients with low metastatic burden.
Figure 3. Genomic features associated with metastatic burden.
(A) Spearman’s correlation coefficient between FGA (circle) and TMB (diamond) with metastatic burden. Associations without a significant trend are shown in grey, and the lines indicate 95% CI.
(B) Correlation between FGA and TMB with metastatic burden in the entire data set, prostate adenocarcinoma, hypermutated uterine cancer, and MSS colorectal cancer. Boxplots display median point, IQR boxes and 1.5 × IQR whiskers for all samples. Split violin plots show the distribution of FGA and TMB in primary tumors (left, not filled) and metastases (right, filled).
(C) Statistically significant oncogenic alterations and pathways associated with metastatic burden in individual tumor types. Spearman’s correlation coefficient is shown for each event, and the lines indicate 95% CI. Gene names in italics refer to specific genes, those in regular font refer to pathways.
Next, we investigated the association between recurrent oncogenic alterations and metastatic burden and identified a total of 24 statistically significant associations across 8 tumor types. We also investigated the association with oncogenic pathways and identified 16 statistically significant differences across 7 tumor types (Figure 3C; Table S3B). Consistent with its role as a gatekeeper against chromosomal instability (Bieging et al., 2014), we observed a significant positive correlation between TP53 mutations and metastatic burden in prostate adenocarcinoma, lung adenocarcinoma and HR+/HER2− ductal breast cancer. There was also a significant positive correlation between p53 pathway alterations and metastatic burden in endometrioid uterine cancer. In metastatic prostate adenocarcinoma, AR amplifications were positively associated with metastatic burden. The frequency of ESR1 mutation increased with metastatic burden in HR+/HER2− ductal and lobular breast cancer. CDKN2A deletion frequency was positively correlated with metastatic burden in bladder urothelial cancer, lung adenocarcinoma and papillary thyroid cancer, while MYC amplification frequency was associated with increasing metastatic burden in lung adenocarcinoma and prostate adenocarcinoma. Of note, the frequency of four oncogenic alterations and one oncogenic pathway were negatively correlated with metastatic burden; FOXA1 in prostate adenocarcinoma, CBFB in HR+/HER2− ductal breast cancer, CDH1 in lobular breast cancer, ERCC2 in urothelial bladder cancer and the epigenetic pathway in MSS colorectal cancer (Figure 3C; Table S3B). These results demonstrate that the relationship between higher chromosomal instability and increasing metastatic burden is tumor lineage dependent and that several driver mutations are associated with metastatic burden in both directions.
Genomic differences of metastases according to their organ location
Next, we investigated the genomic characteristics of metastases (n=10,143) according to their organ location. As expected, the location of the sequenced metastases differed by tumor type (Figure S4A). We found 17 significant associations between FGA and the metastatic site in six tumor types, 10 of which were also significant when using adjusted FGA (Figure S4B; Table S4A). CNS/Brain metastases from patients with lung adenocarcinoma, MSS colorectal cancer and cutaneous melanoma had a significantly higher FGA, while lymph node metastases from patients with lung adenocarcinoma, pancreatic adenocarcinoma, bladder urothelial and cutaneous melanoma had a significantly lower FGA. There were seven significant associations between TMB and the metastatic site. A total of 31 genomic alterations in nine tumor types were significantly associated with specific metastatic sites and 25 oncogenic pathways across six tumor types (Figure S3C; Table S4A). TP53 mutations were significantly more frequent in CNS/Brain metastasis from lung adenocarcinoma (55% vs. 66%, q-value = 0.002) and liver metastasis from pancreatic adenocarcinoma (63% vs. 84%, q-value < 0.001), but less frequent in lung metastasis from urothelial bladder cancer (57% vs. 29%, q-value = 0.04) and liver metastasis from neuroendocrine lung cancer (83% vs. 25%, q-value = 0.005), as well as in intra-abdominal metastasis from pancreatic adenocarcinoma (X% vs. X%, q-value = ). In HR+/HER2− ductal breast cancer, ESR1 mutations were significantly more frequent in liver metastasis (79% vs. 58%, q-value = 0.003). In lobular breast cancer, RHOA mutations were significantly more frequent in ovarian metastasis (3% vs. 36%, q-value = 0.02) and FOXA1 mutations were enriched in liver metastasis (3% vs. 33%, q-value = 0.02). In lung adenocarcinoma, CDKN2A deletion was more frequent in skin (16% vs. 60%, q-value = 0.03) and liver metastases (16% vs. 27%, q-value = 0.03) but less frequent in lymph nodes (19% vs. 12%, q-value = 0.002). PTEN mutation, as well as PI3K pathway alterations, were higher in brain metastases from melanoma (7% vs. 23%, q-value = 0.01 and 19% vs. 39%, q-value = 0.02, respectively), which is in line with a previous melanoma-specific study (Bucheit et al., 2014). Among others, we found that ERG fusions were less frequent in bone metastasis of prostate cancer patients (30% vs. 15%, q-value = 0.002), NF1 mutations were more frequent in lung metastasis of melanoma patients (20% vs. 38%, q-value = 0.003) and that FGFR3 mutations were more frequent in lung metastasis of bladder urothelial patients (11% vs. 39%, q-value = 0.005). Taken together, our results show that metastases from different organs can have different genomic makeup.
Genomic features associated with metastasis to specific target organs
We analyzed the relationship between genomic features of metastatic patients and their organ-specific patterns of metastasis (n=21,546). We found 13 significant associations between FGA and organotropisms in 11 tumor types, seven of which were also significant when using adjusted FGA (Table S4B). We observed a significant positive association between FGA and patients with liver metastasis in four tumor types (HR+/HER2− ductal breast, prostate adenocarcinoma, pancreatic adenocarcinoma and head and neck squamous), patients with lung metastasis in two tumor types (endometrioid uterine and cutaneous melanoma), and bone metastasis in two tumor types (HR+/HER2− ductal breast and prostate adenocarcinoma). For TMB, we found eight significant associations between TMB and organ-specific patterns of metastasis in six tumor types, including four positive associations (lung adenocarcinoma to brain and adrenal gland, pancreatic adenocarcinoma to liver, head and neck squamous to head and neck) and four negative associations (prostate adenocarcinoma to bone, cutaneous melanoma to intra-abdominal, lung adenocarcinoma to pleura and lung neuroendocrine to liver).
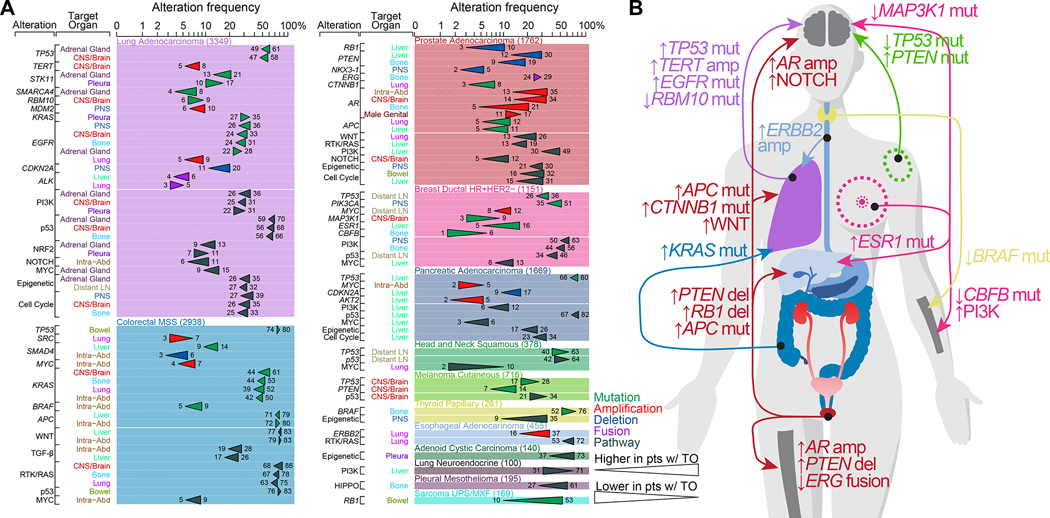
We found 57 significant recurrent oncogenic alterations associated with specific patterns of metastasis in 10 tumor types. When interrogating oncogenic pathway alterations, we found 48 significant associations in 12 tumor types (Figure 4A; Table S4B). Lung adenocarcinoma, MSS colorectal cancer, and prostate cancer were associated with the highest number of significant associations. These results are summarized in Figure 4B. For example, lung adenocarcinoma patients with CNS/Brain metastasis had a higher frequency of TP53 mutations, TERT amplification, and EGFR mutations, but a lower frequency of RBM10 mutations. MSS colorectal cancer patients with lung metastasis had a higher frequency of KRAS mutations (39% vs. 52%, q-value < 0.001), as previously reported (Cejas et al., 2009; Pereira et al., 2015; Tie et al., 2011) but a lower frequency of SRC amplification (7% vs. 3%, q-value < 0.001). Prostate cancer patients with bone metastasis had a higher frequency of AR amplification (5% vs. 21%, q-value < 0.001) and PTEN deletion (9% vs. 19%, q-value < 0.001) but a lower frequency of ERG fusions (29% vs. 24%, q-value = 0.04); those with liver metastasis had a higher frequency of PTEN loss (11% vs. 30%, q-value < 0.001), RB1 loss (3% vs. 10%, q-value < 0.001) and APC mutations (5% vs. 11%, q-value = 0.001); those with brain metastasis had a higher frequency of AR amplification (14% vs. 34%, q-value < 0.001) and NOTCH pathway alterations (5% vs. 12%, q-value = 0.02); and those with lung metastasis had a higher frequency of APC mutations (5% vs. 12%, q-value < 0.001) and CTNNB1 mutations (3% vs. 8%, q-value = 0.007). Experimental work has revealed the role of WNT pathway activation in driving prostate cancer metastasis (Leibold et al., 2020) and discovered a vulnerability to tankyrase inhibition in WNT altered prostate cancer. When interrogating the association between oncogenic pathways and organotropisms, we found that 26% of prostate cancer patients with lung metastasis had WNT pathway alterations, compared to 13% of patients without lung metastasis (q-value < 0.001, Figure 4A). As previously reported (Gerratana et al., 2020), ESR1 mutations were more frequent in HR+/HER2− ductal breast cancer patients with liver metastasis (5% vs. 16%, q-value < 0.001). CBFB mutations were less frequent in HR+/HER2− ductal breast cancer patients with bone metastasis (5% vs. 1%, q-value = 0.009), which was demonstrated in a mouse model (Ran et al., 2020) while alterations in the PI3K pathway were more frequent in patients with bone metastasis (44% vs. 56%, q-value = 0.003). HR+/HER2− ductal breast cancer patients with brain metastasis had a lower frequency of MAP3K1 mutation (9% vs. 3%, q-value = 0.02), which was recently shown to be a surrogate for the less aggressive luminal A breast cancer subtype (Nixon et al., 2019). In line with a previous study (Bucheit et al., 2014), PTEN mutations were more frequent in cutaneous melanoma patients with brain metastases (7% vs. 14%, q-value = 0.04), while TP53 mutations were less frequent in those patients (28% vs. 17%, q-value = 0.04). Thyroid papillary cancer patients with bone metastasis had a lower frequency of BRAF mutations (73% vs. 53%, q-value = 0.02), and esophageal cancer patients with lung metastasis had a higher frequency of ERBB2 amplification (16% vs. 37%, q-value < 0.001). Our analysis of arm-level events also revealed that that chromosome 22q loss was more frequent in thyroid papillary patients with bone metastasis (13% vs 40%, q-value = 0.0002), even though broader chromosomal instability was not significantly different in those patients (median FGA; 32% vs. 45%, q-value = 0.07, Table S4B). In sum, while we did not observe gene or pathway alterations associated with specific target organs that were shared consistently across different tumor types at a pan-cancer level (Figure S4), our analysis revealed specific genomic alterations linked to specific organotropisms in individual tumor types.
Figure 4. Genomic features associated with metastasis to specific target organs.
(A) Statistically significant oncogenic alterations and pathways associated with organ-specific patterns of metastatic spread. Gene names in italics refer to specific genes, those in regular font refer to pathways.
(B) Schematic drawing summarizing the main findings from (A).
DISCUSSION
We present MSK-MET, a unique, curated cohort of cancer patients with available genomic sequencing data and clinical information on metastatic disease and cancer outcome. Our study expands a previous pan-cancer dataset (Zehir et al., 2017) by including a larger number of patients with longer follow-up and by including a comprehensive description of metastatic events at the patient level. We demonstrate that mining of electronic health records can be used to extract relevant clinical information, and we present a pan-cancer map of metastasis in a contemporary cohort of patients treated at a single tertiary referral center.
Our analysis of genomic alterations from unpaired primary and metastatic samples revealed that metastases generally had a higher level of chromosomal instability, along with a higher frequency of WGD and TP53 mutations. These results are consistent with previous studies that have shown an association between chromosomal instability and cancer progression (Bakhoum et al., 2018; Ben-David and Amon, 2020; Hieronymus et al., 2018; Shukla et al., 2020; Stopsack et al., 2019; Watkins et al., 2020). Our results also suggest that metastases generally have a higher fraction of clonal mutations. This lower intra-tumor heterogeneity could be attributed to clonal selection and selective pressure from cancer therapy (Birkbak and McGranahan, 2020). We also identified several genomic alterations and signaling pathways enriched in metastatic samples. As described before (Hu et al., 2020; Pareja et al., 2020; Razavi et al., 2018), the most significant enrichments were associated with known drug resistance mechanisms (e.g., AR alterations in prostate cancer, and ESR1 mutations in breast cancer). We also compared primary tumor samples from metastatic and non-metastatic patients. In several tumor types, we observed a higher chromosomal instability and a higher frequency of TP53 mutations amongst other drivers in primary samples from metastatic patients whereas the clonal fraction was generally similar.
In an analysis aimed at identifying genomic alterations associated with metastatic burden, we found that higher chromosomal instability was correlated with metastatic burden in several tumor types. This association, however, was absent in many other tumor types, including colorectal cancer, where copy-number alteration patterns may be established early in tumor development. Several mechanisms can explain the pro-metastatic effects of chromosomal instability and have been reviewed before (Ben-David and Amon, 2020). It is believed that chromosomal instability can promote tumor progression by increasing subclonal diversity and tumor evolution (Watkins et al., 2020), but aneuploidy itself is not a universal promoter of transformation and recent studies suggest that aneuploidy is cancer-type-specific (Ben-David and Amon, 2020), which is in line with our observations. Beyond global chromosomal instability, we also identified several specific genomic alterations and signaling pathways associated with metastatic burden. The majority, including alterations associated with drug resistance, were enriched in samples from patients with higher metastatic burden. Few were associated with lower metastatic burden, including FOXA1 mutations in prostate cancer and CBFB mutations in breast cancer.
Lastly, we investigated associations between genomic alterations and metastatic colonization of specific target organs. We compared independent metastatic samples according to their organ sites and we observed that the genomic landscape of metastasis differed according to their target organs. Previous studies have also interrogated the differences between primary tumors and metastatic sites using either independent samples (Armenia et al., 2018; Priestley et al., 2019; Robinson et al., 2017; Shih et al., 2020) or paired samples (Brastianos et al., 2015; Brown et al., 2017; Eckert et al., 2016; Hu et al., 2020; Jiménez-Sánchez et al., 2017; Makohon-Moore et al., 2017; Naxerova et al., 2017; Noorani et al., 2020; Reiter et al., 2020). Clinical data extraction from the EHR allowed us to explore the genomic alterations of metastatic patients by taking into consideration a greater part of the metastatic events occurring in a patient’s clinical course. We have generated a variety of hypotheses linking specific genomic alterations to specific organotropisms occurring in a cancer-specific manner. Future functional characterization of these alterations could result in the identification of novel biomarkers and therapeutic approaches that will have the potential to influence the clinical management of patients. Our results highlight the importance of chromosomal instability in progression and metastasis, and drugs targeting this hallmark could represent an attractive strategy in several tumor types. MSK-MET is publicly available via the cBioPortal for Cancer Genomics (https://www.cbioportal.org/study?id=msk_met_2021) (Cerami et al., 2012; Gao et al., 2013). We hope that it will be a valuable resource for the community and will stimulate further research and applications in cancer care.
Limitations of the study
Our study has several limitations. Firstly, while the overall cohort is large, sample size varied significantly between tumor types, which prevented us from drawing robust conclusions in less common tumor types. Therefore, the lack of significant differences in those tumor types might be due to a lack of statistical power and should be interpreted with caution. Also, our definition of tumor types could be further refined in some cases, to account, for example, for different predominant histologic subtypes in lung adenocarcinomas (Caso et al., 2020). This might provide additional valuable insights, but would also result in decreased sample sizes and lower statistical power for those refined groups. Secondly, the ICD billing codes used in our study likely do not fully capture all metastatic events and may be affected by inter-physician variability. Future improvements to the clinical data extraction process could come from the use of natural-language processing and machine learning approaches, which will be required to mine the wealth of data contained in EHR systems at scale. Thirdly, because of our use of a targeted sequencing panel, we may be missing biologically or clinically relevant signals that could be discovered using alternative approaches such as whole-exome or whole-genome sequencing. Finally, all analyses presented here have been performed using a single representative sample for each patient. In the future, longitudinal sampling of multiple anatomical locations at different time points from the same patient will allow us to investigate additional questions about the timing of genomic events and the genomic heterogeneity across different organs throughout metastatic progression. Although this study represents a first step towards understanding how genomic alterations shape tumor progression, metastatic burden and organotropisms, more integrated studies are needed to fully investigate the impact of tumor cell-extrinsic effects, such as cancer therapy, target organ microenvironment and systemic factors. These studies will require comprehensive clinical timelines with accurate information about all lines of therapy and metastatic events. Additionally, single-cell profiling methods may be required to fully understand the cross-talk between tumor cells and the metastatic niche.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Nikolaus Schultz (schultzn@mskcc.org).
Materials availability
This study did not generate new unique reagents.
Data and code availability
The raw sequencing data for the MSK-IMPACT cohort are protected and are not broadly available due to privacy laws. Raw data may be requested from schultzn@mskcc.org with appropriate institutional approvals.
Our full dataset including clinical and genomic data is publicly available at Zenodo https://doi.org/10.5281/zenodo.5801902 and through the cBioPortal for Cancer Genomics https://www.cbioportal.org/study?id=msk_met_2021
Original code to do the organ site mapping for metastatic cancer is available at https://github.com/clinical-data-mining/organ-site-mapping and has been deposited at figshare. DOIs is listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human subjects
This study was approved by the Memorial Sloan Kettering Cancer Center Institutional review board and all patients provided written informed consent for tumor sequencing and review of patient medical records for detailed demographic, pathologic, and treatment information (NCT01775072). Characteristics of each subject, including age and sex, are available in Table S1B.
METHOD DETAILS
Samples and patients
A total of 43,400 solid tumor samples from 38,933 patients sequenced at Memorial Sloan Kettering Cancer Center from 2013–11-18 to 2020–01-06 (6.1y) and included in the AACR Project Genomics Evidence Neoplasia Information Exchange (GENIE) (AACR Project GENIE Consortium 2017) 9.0-public database were considered for this study. A total of 9,215 samples sequenced at Memorial Sloan Kettering Cancer Center from 2020–01-07 to 2021–08-18 (1.6y) were used for validation. All tumors were profiled using the Memorial Sloan Kettering Integrated Molecular Profiling of Actionable Cancer Targets (MSK-IMPACT) clinical sequencing assay, a hybridization capture-based, next-generation sequencing platform (Cheng et al. 2015). Tumor types were defined using a unique cancer type and one or more cancer type detailed (Table S1A). For endometrial and colorectal cancers, we defined a subset of hypermutated (HM) tumors as those having an oncogenic POLE mutation or exhibiting more than 25 mutations/Mb or having MSIsensor score (Niu et al. 2014) > 10. Exclusion criteria were as follows: unavailable matched normal; low sequencing coverage (<100x); low tumor purity as defined by the absence of somatic alterations (including silent); pediatric patients (<18y at time of sequencing); patients with more than one unique sequenced tumor type; cancer of unknown primary; tumor type in which metastasis are rare (e.g. Gliomas); breast cancer with unavailable molecular subtype information; tumor types with small sample size (i.e. n <80 and either primary n <30 or metastasis n <30). Finally, one sample per patient was selected using a set of priority rules as follows: the presence of a FACETS fit that passed qc > highest purity > highest sample coverage > most recent gene panel. A total of 25,775 samples spanning 50 tumor types were used for analysis (Figure S1A–D, Table S1A–B). This set included samples that were sequenced with three generations of the MSK-IMPACT panel, containing 341 genes (n = 1,801 samples), 410 genes (n = 6,372 samples), and 468 genes (n = 17,602 samples).
Clinical data extraction procedures for the identification and mapping of metastatic events
Clinical data were retrieved from the institutional electronic health records (EHR) database on 2020–11-05. Metastatic events were extracted from the pathology report of the sequenced samples and patients’ electronic health records. The anatomic location of the sequenced samples is described in the sample pathology reports as a free-text description by pathologists. The EHR includes International Classification of Diseases (ICD) billing codes which classify a comprehensive list of diseases, disorders, injuries and other health conditions including metastatic events. Metastatic events from the sample pathology report and the ICD billing codes from the EHR were systematically mapped to a curated list of 21 organs (Table S1C). Lymph nodes were also classified as distant or regional given the anatomic location of the primary tumor (link). Of note, the classification of distant vs. regional was not possible for tumor types in which the anatomic location of the primary tumor is not well defined (e.i. melanoma cutaneous, cutaneous squamous cell, sarcoma lipo and sarcoma UPS/MXF). The organ site mapping for metastatic cancer is available at https://github.com/clinical-data-mining/organ-site-mapping. For a user providing a table of organ site descriptions or ICD Billing codes, annotations of the 21 organ sites will be generated. Furthermore, additional annotations recognizing local extension and distant lymph node spread can be created. Metastatic burden was defined as the number of distinct organs (excluding regional lymph nodes) affected by metastases throughout a patient’s clinical course (ranging from 1 to 15 in the present study). Patients with more than six affected organ sites were grouped for analyses of metastatic burden.
Comparison of metastatic sites automatically extracted from electronic health records vs. manual chart review
A total of 4,859 patients (22.5%) with metastatic sites extracted through manual chart review and previously published were available (Abida et al., 2017; Jones et al., 2021; Razavi et al., 2018; Shoushtari et al., 2021; Yaeger et al., 2018). Ten tumor types were represented including the most frequent (prostate, lung, breast, colorectal, and melanoma). There was a strong correlation between the number of metastatic sites retrieved from manual chart review and the number of metastatic sites automatically extracted from electronic health records (Figure S1H). For colorectal hyper mutant and MSS only the first metastatic events were reported so we restricted the comparison to the first metastatic event extracted from EHR. It is also important to note that the manual chart review was done before this study. Therefore, the present study has a longer follow-up which resulted in a higher number of metastatic sites. We also calculated the sensitivity for each metastatic site and each tumor type (Figure S1I). The median sensitivity was 77% across tumor types and metastatic sites.
Genomic analysis
Tumor mutational burden (TMB) was calculated for each sample as the total number of nonsynonymous mutations, divided by the number of bases sequenced. Fraction of genome altered (FGA) was calculated for each sample as the percentage of the genome with absolute log2 copy ratios >0.2. Log2 copy-number ratios were derived as previously described (Cheng et al., 2015). Chromosome arm-level copy number alterations were computed using the ASCETS tool (Spurr et al., 2020) using default parameters. Allele-specific analyses of copy number alterations were performed using the FACETS tool (Shen and Seshan, 2016), which infers purity- and ploidy-corrected integer DNA copy number calls from sequencing data. The quality of FACETS fits was determined using a set of criteria as described in facets-preview (https://github.com/taylor-lab/facets-preview). To estimate a tumor purity- and ploidy-adjusted version of the FGA, we defined “adjusted FGA” as the fraction of the genome different from the major integer copy number (Mcn), where Mcn is defined as the integer total copy number spanning the largest portion of the genome. Tumor samples were considered to have undergone whole-genome doubling (WGD) if more than 50% of their autosomal genome had Mcn >2. The clonality of each mutation (clonal or subclonal or indeterminate) was determined as described in facets-suite (https://github.com/mskcc/facets-suite). For each tumor sample, the fraction of clonal mutations (clonal fraction) was determined by dividing the total number of clonal mutations by the sum of clonal and subclonal mutations. MSI-H status was defined by an MSIsensor score >10 (Niu et al., 2014). Somatic alterations were annotated using OncoKB for oncogenicity and clinical actionability (Chakravarty et al., 2017) (Data version: v2.8, released on 2020–09-17). For hypermutated colorectal and hypermutated uterine cancer, only genes that were recurrently mutated based on MutSig-CV (q-value<0.1) were considered for association analyses. For each tumor type, recurrent oncogenic alterations were defined as those considered oncogenic or likely oncogenic by OncoKB and present in at least 5% of either primary or metastatic samples (median of 15 per tumor type, Table S1A). Canonical oncogenic pathway-level alterations were computed using curated pathway templates as previously reported (Ding et al., 2018; Sanchez-Vega et al., 2018). Segmented copy-number data were processed using the CNtools package v1.4.
Quantification and statistical analysis
The relative temporal order of target organs for each tumor type was assessed using a Bradley-Terry model. For each tumor type, we included patients with at least two metastases in different organs. For each patient, metastases can be timed relative to one another and these pairwise comparisons were aggregated (as implemented by the R package BradleyTerryScalable) to give an overall ordering of metastasis per tumor type. Comparisons between groups (primary vs. metastatic tumors, primary samples from metastatic vs. non-metastatic patients, and metastases according to their organ location) were performed using the non-parametric Mann-Whitney U test for continuous variables or the Fisher’s exact test for categorical variables. Differences in the frequency of actionable mutations (Levels 1 to 3, as defined by OncoKB) between groups (primary tumors vs. metastases and primary tumors from metastatic vs. non-metastatic patients) were further tested using a multivariable logistic regression model adjusted for TMB and FGA. Differences in the frequency of arm-level copy number alterations between groups (primary vs. metastatic tumors and primary samples from metastatic vs. non-metastatic patients) were tested using a multivariable logistic regression model adjusted for FGA. A genomic feature was considered to be significantly correlated with metastatic burden if (a) the Spearman’s correlation between the two variables was statistically significant (q-value < 0.05) and (b) the coefficient associated with the genomic feature as a predictive variable in a multivariable linear regression model adjusted for sample type (metastatic vs. primary tumor) was statistically significant (p-value < 0.05). The second condition was required because the ratio of metastatic samples to primary samples was associated with metastatic burden and could otherwise act as a confounding factor. We assessed genomic features associated with the presence or absence of metastasis in a target organ using only target organs present in at least 5% of the patients. A genomic feature was considered to be significantly associated with metastasis to specific target organs if (a) the Mann-Whitney U test for continuous variables or the Fisher’s exact test for categorical variables was statistically significant (q-value < 0.05) and (b) the coefficient associated with the genomic feature as a predictive variable in a multivariable logistic regression model adjusted for sample type (metastatic vs. primary tumor, categorical) and metastatic burden (1 to ≥6, numerical) was statistically significant (p-value < 0.05). The second condition was required because the ratio of metastatic samples to primary samples and metastatic burden were associated with metastasis to specific target organs and could otherwise act as a confounding factor. When TMB and FGA were used in a generalized linear model (linear and logistic model), their distributions were harmonized using a normal transformation as described before (Vokes et al., 2019) then scaled from 0 to 1 by subtracting the minimum and dividing by the maximum. Logistic regression was performed using Firth’s bias-reduction method as implemented in the R package brglm (Kosmidis and Firth, 2020). Overall survival (OS) was measured from the time of sequencing to death and was censored at the last time the patient was known to be alive. If a patient had more than one sequenced sample, the first time of sequencing was used. Median follow-up time was calculated using the reverse Kaplan-Meier method. Median overall survival and five-year survival rate were calculated by the Kaplan-Meier method. The association between metastatic burden and overall survival was assessed using univariable Cox proportional hazards regression models. All reported p-values are two-tailed. Multiple testing correction was applied within each tumor type using the false discovery rate (q-value) method and q-value < 0.05 was considered significant. All analyses were performed using R v3.5.2 (www.R-project.org) and Bioconductor v3.4.
Supplementary Material
Figure S1. Study design and characteristics of the patients and samples included in MSK-MET, related to Figure 1.
(A) CONSORT flow diagram of the study.
(B) Distribution of age at time of sequencing, time interval between surgical procedure and sequencing, tumor sample coverage and tumor purity assessed by the pathologist.
(C) Distribution of 25,775 tumors across 50 tumor types grouped by ten organ systems.
(D) Distribution of the 25,775 tumors according to the sample type (primary vs. metastasis), site of metastatic sample and whether the primary sample was from a patient with evidence of distant metastasis at the time of the study or not.
(E) Frequency of primary and metastatic samples previously exposed to any treatment for each tumor type.
(F) Distribution of the 99,220 metastatic events mapped to 21 organ sites.
(G) Comparison of the frequency of metastasis in several target organs from different tumor types reported in (Gao et al., 2019) and in (Budczies et al., 2015) vs. the present study.
(H) Comparison of the number of metastasis using data from manual chart reviews and clinical data automatically extracted from the EHR (This study).
(I) Heatmap showing the recall rate (sensitivity) across several target organs from different tumor types using patients retrieved from manual chart reviews.
(J) Median time to metastasis (months) relative to the first metastatic event. The number in each cell indicates the median time to metastasis at that given site in months. Only metastases with at least 3 representative patients are shown here. Note that for patients with multiple metastases to the same organ only the first one was taken into consideration. For each metastatic site, the distribution of all 50 tumor types is shown as a stacked bar chart below the heatmap.
(K) Results from the Bradley-Terry model showing the order of metastatic events observed in the top 10 most common tumor types. For each tumor type, the order of metastasis from first to last is represented from top to bottom.
Figure S2. Genomic differences between primary tumors and metastases using an independent cohort of 9,215 patients sequenced with MSK-IMPACT since the data freeze date related to Figure 2.
(A) Comparisons of the median fraction genome altered (FGA), median tumor mutation burden (TMB), and median frequency of tumor mutation burden high (TMB-hi) for each tumor type in metastatic vs. primary tumors. Tumor types with statistically significant differences are labeled.
(B) The following clinical and genomic features are shown side-by-side for primary (top row within each cancer type) and metastatic (bottom row) sequenced samples using a combination of bar charts and violin plots; from left to right: sample counts, FGA, TMB, fraction of samples with high TMB, and distribution of the highest actionable alteration levels. The black vertical line in each violin plot represents the median. The heatmap shows the frequency of individual arm level alterations in primary tumors and metastases (only the frequency of the more frequent event, gain or loss, is shown). Tumor types are ordered from top to bottom by decreasing FGA in metastasis and grouped by organ systems. * indicates q-value < 0.05.
(C) Statistically significant differences in the frequency of oncogenic alterations and pathways between primary tumors and metastases in individual tumor types. Triangles summarize oncogenic alteration frequencies in primary tumors vs. metastases and are colored according to alteration type. Gene names in italics refer to specific genes, those in regular font refer to pathways.
(D) Comparison of the effect size of FGA and TMB between the original and the new dataset. Each point is colored according to significance (green; significant in the new dataset only, blue; significant in the original dataset only; pink significant in both the original and the new dataset).
(E) Comparison of the effect size of the difference in oncogenic alteration between the original and the new dataset. Each point is colored according to tumor type (left) or significance (right). The comparison was tested using a Spearman correlation.
Figure S3. Genomic differences between primary samples from metastatic and non-metastatic patients, related to Figure 2.
(A) Scatterplot showing the comparison of the median FGA, median WGD, median TMB and median clonality for each tumor type in primary samples from metastatic and non-metastatic patients.
(B) The following clinical and genomic features are shown side-by-side for primary samples from metastatic and non-metastatic patients using a combination of bar charts and violin plots; from left to right: sample counts, FGA, fraction of samples with WGD, TMB, clonality, fraction of samples with TMB-high status and distribution of highest actionable alteration. The black vertical line in each violin plots represents the median. Heatmap shows the frequency of arm level alterations in primary tumors and metastases. Tumor types are ordered from top to bottom by decreasing FGA in metastasis and grouped by organ systems. * indicates q-value < 0.05. WGD and clonality were available for a subset of 10,106 samples with FACETS data.
(C) Statistically significant differences in the frequency of oncogenic alterations between primary tumors and metastases across all tumor types. Triangles summarize oncogenic alterations frequencies in primary samples from metastatic vs. non-metastatic patients and are colored according to alteration type.
(D) Comparison of genomic features differing between primary tumors vs metastases (“P vs M”) and genomic features differing between primary tumors from non-metastatic vs metastatic patients (“P-noM vs P-M”). Comparison of the effect size for FGA and TMB (D) (log2 fold change) between the two analyses. Top; each point represents a tumor type and is color-coded according to the three statistical significance scenarios (pink; significant in both analyses, green; significant in P vs M only, blue; significant in P-noM vs P-M only). Bottom; boxplots showing FGA and TMB according to the three sample types (primary tumors from non-metastatic vs primary tumors from metastatic patients vs metastatic samples) for two representative tumor types and every three scenarios.
(E) Comparison of the effect size for oncogenic alterations (log odds ratio) between the two analyses. Top; each point represents an oncogenic alteration that was significantly different in either analysis and is color-coded according to the three statistical significance scenarios (left) (pink; significant in both analyses, green; significant in P vs M only, blue; significant in P-noM vs P-M only,) and the tumor type in which it belongs (left). Bottom; bar graphs showing the alterations frequency according to the three sample types (primary tumors from non-metastatic vs primary tumors from metastatic patients vs metastatic samples) for two representative tumor types and every three scenarios. For each comparison, a Venn diagram shows the number of common and exclusive significant associations between the two analyses (top-left corner). The comparisons were tested using a Spearman correlation.
Figure S4. Genomic differences of metastases according to organ location, related to Figure 4.
(A) Distribution of sequenced metastasis according to organ location with the heatmap showing the percentage of metastatic samples where each row represents a tumor type and each column represents the organ location. For each tumor type, the distribution of all 21 organ locations is shown as a stacked bar chart to the right of the heatmap. For each organ, the distribution of all 50 tumor types is shown as a stacked bar chart below the heatmap. For each tumor type, the number of metastasis samples is indicated in parentheses. For each organ, the number of metastasis samples is indicated in parentheses.
(B) Statistically significant association between FGA (black circle) and TMB (white diamond) and specific metastatic sites.
(C) Statistically significant oncogenic alterations associated with specific metastatic sites.
(D) Heatmap showing 69 genomic features associated with metastasis to specific target organs and significant in at least two cancer types. The shade of red represents the three significance thresholds (dark red, q-value < 0.05 & logit < 0.05; red, q-value < 0.05; light red, p-value < 0.05).
Table S1. Summary of the 50 tumor types included in the MSK-MET cohort (A), Individual patient data included in the MSK-MET cohort (B), Mapping between free-text description from pathology reports and ICD billing codes from EHR to a curated list of 21 organs (C), Related to Figure 1
Table S2. Genomic differences between primary tumors and metastases (A), Genomic differences between primary tumors and metastases using an independent cohort (B), Genomic differences between primary samples from metastatic and non-metastatic patients (C), Related to Figure 2, Figure S2 and Figure S3
Table S3. Association between metastatic burden and overall survival assessed using Cox proportional hazards model (A), Genomic associations with metastatic burden (B), Related to Figure 3
Table S4. Genomic differences of metastases according to their organ location, Related to Figure S4 (A), Genomic features associated with metastasis to specific target organs (B), Related to Figure S4 and Figure 4
HIGHLIGHTS.
A large clinico-genomic database to study metastatic patterns across 50 tumor types
Oncogenic alteration frequency and chromosomal instability are increased in metastases
Correlations between chromosomal instability and metastatic burden depend on cancer type
Genomic features associated with metastasis are identified for specific target organs
ACKNOWLEDGMENTS
This study was supported by the MSK Cancer Center Support Grant (P30 CA008748), the MSK MIND initiative, Cycle for Survival, the Alan and Sandra Gerry Metastasis and Tumor Ecosystems Center, the Fund for Innovation in Cancer Informatics (ICI), the Robertson Foundation, a Prostate Cancer Foundation Challenge Award, and the Marie-Josée and Henry R. Kravis Center for Molecular Oncology.
Footnotes
DECLARATION OF INTERESTS
S.C. receives consulting fees from Novartis, Lilly, Sanofi and research funding from Daiichi-Sankyo and Paige.ai. J.J.H. receives consulting fees from Bristol Myers Squibb, Merck, Exelexis, Eisai, QED, Cytomx, Zymeworks, Adaptiimmune, ImVax and research funding from Bristol Myers Squibb. C.I.D. receives research funding from Bristol Myers Squibb. P.R. received consultation/Ad board/Honoraria from Novartis, Foundation Medicine, AstraZeneca, Epic Sciences, Inivata, Natera, and Tempus and institutional grant/funding from Grail, Illumina, Novartis, Epic Sciences, ArcherDx. C.M.R. has consulted regarding oncology drug development with AbbVie, Amgen, Astra Zeneca, Epizyme, Genentech/Roche, Ipsen, Jazz, Lilly, and Syros, and serves on the scientific advisory boards of Bridge Medicines, Earli, and Harpoon Therapeutics. D.Z. receives research funding from Astra Zeneca, Plexxikon, and Genentech and consulting fees from Merck, Synlogic Therapeutics, GSK, Bristol Myers Squibb, Genentech, Xencor, Memgen, Immunos, Agenus, Hookipa, Calidi, Synthekine. B.W. has a ad hoc membership advisory board Repare Therapeutics. W.A. receives speaking honoraria from Roche, Medscape, Aptitude Health, Clinical Education Alliance and consulting fees from Clovis Oncology, Janssen, ORIC pharmaceuticals, Daiichi Sankyo and reserach funding from AstraZeneca, Zenith Epigenetics, Clovis Oncology, ORIC pharmaceuticals, Epizyme. G.K.A. receives research funding from Arcus, Agios, Astra Zeneca, Bayer, BioNtech, BMS, Celgene, Flatiron, Genentech/Roche, Genoscience, Incyte, Polaris, Puma, QED, Sillajen, Yiviva, and consulting fees from Adicet, Agios, Astra Zeneca, Alnylam, Autem, Bayer, Beigene, Berry Genomics, Cend, Celgene, CytomX, Eisai, Eli Lilly, Exelixis, Flatiron, Genentech/Roche, Genoscience, Helio, Incyte, Ipsen, Legend Biotech, Loxo, Merck, MINA, Nerviano,QED, Redhill, Rafael, Silenseed, Sillajen, Sobi, Surface Oncology, Therabionics, Twoxar, Vector, Yiviva. E.M.O. receives research Funding from Genentech/Roche, Celgene/BMS, BioNTech, BioAtla, AstraZeneca, Arcus, Elicio, Parker Institute, AstraZeneca and consulting fees from Cytomx Therapeutics (DSMB), Rafael Therapeutics (DSMB), Sobi, Silenseed, Tyme, Seagen, Molecular Templates, Boehringer Ingelheim, BioNTech, Ipsen, Polaris, Merck, IDEAYA, Cend, AstraZeneca, Noxxon, BioSapien, Bayer (spouse), Genentech-Roche (spouse), Celgene-BMS (spouse), Eisai (spouse). M.A.P. receive consulting fees from BMS, Merck, Array BioPharma, Novartis, Incyte, NewLink Genetics, Aduro, Eisai, Pfizer and honoraria from BMS and Merck and research support from RGenix, Infinity, BMS, Merck, Array BioPharma, Novartis, AstraZeneca. G.J.R. has insitutional research funding from Mirait, Takeda, Merck, Roche, Novartis, and Pfizer. SFB holds a patent related to some of the work described targeting CIN in advanced cancer. He owns equity in, receives compensation from, and serves as a consultant and the Scientific Advisory Board and Board of Directors of Volastra Therapeutics Inc. A.N.S. has advisory board / personal fees from Bristol-Myers Squibb, Immunocore, Castle Biosciences and research support from Bristol-Myers Squibb, Immunocore, Xcovery, Polaris, Novartis, Pfizer, Checkmate Pharmaceuticals and research funding from OBI-Pharma, GSK, Silenseed, BMS, Lilly. R.Y. receives consulting fees from Array BioPharma/Pfizer, Mirati Therapeutics, Natera and research funding from Pfizer, Boehringer Ingelheim. D.R.J. is member of the Advisory Council for Astra Zeneca and member of the Clinical Trial Steering Committee for Merck. D.M. reports disclosures from AstraZeneca, Johnson & Johnson, Boston Scientific, Bristol-Myers Squibb, Merck. S.P.D. is shareholder and consultant for Canexia Health Inc. M.F.B receives consulting fees from Roche, Eli Lilly, PetDx and research funding from Grail. D.B.S. has received consulted for and received honoraria from Pfizer, Lilly/Loxo Oncology, Vividion Therapeutics, Scorpion Therapetuics and BridgeBio. B.N. is an employee of Loxo Oncology at Lilly.
Publisher's Disclaimer: This is a PDF file of an article that has undergone enhancements after acceptance, such as the addition of a cover page and metadata, and formatting for readability, but it is not yet the definitive version of record. This version will undergo additional copyediting, typesetting and review before it is published in its final form, but we are providing this version to give early visibility of the article. Please note that, during the production process, errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- AACR Project GENIE Consortium (2017). AACR Project GENIE: Powering Precision Medicine through an International Consortium. Cancer Discov. 7, 818–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abida W, Armenia J, Gopalan A, Brennan R, Walsh M, Barron D, Danila D, Rathkopf D, Morris M, Slovin S, et al. (2017). Prospective Genomic Profiling of Prostate Cancer Across Disease States Reveals Germline and Somatic Alterations That May Affect Clinical Decision Making. JCO Precis Oncol 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armenia J, Wankowicz SAM, Liu D, Gao J, Kundra R, Reznik E, Chatila WK, Chakravarty D, Han GC, Coleman I, et al. (2018). The long tail of oncogenic drivers in prostate cancer. Nat. Genet. 50, 645–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhoum SF, Ngo B, Laughney AM, Cavallo J-A, Murphy CJ, Ly P, Shah P, Sriram RK, Watkins TBK, Taunk NK, et al. (2018). Chromosomal instability drives metastasis through a cytosolic DNA response. Nature 553, 467–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-David U, and Amon A (2020). Context is everything: aneuploidy in cancer. Nat. Rev. Genet. 21, 44–62. [DOI] [PubMed] [Google Scholar]
- Bieging KT, Mello SS, and Attardi LD (2014). Unravelling mechanisms of p53-mediated tumour suppression. Nat. Rev. Cancer 14, 359–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielski CM, Zehir A, Penson AV, Donoghue MTA, Chatila W, Armenia J, Chang MT, Schram AM, Jonsson P, Bandlamudi C, et al. (2018). Genome doubling shapes the evolution and prognosis of advanced cancers. Nat. Genet. 50, 1189–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkbak NJ, and McGranahan N (2020). Cancer Genome Evolutionary Trajectories in Metastasis. Cancer Cell 37, 8–19. [DOI] [PubMed] [Google Scholar]
- Borst MJ, and Ingold JA (1993). Metastatic patterns of invasive lobular versus invasive ductal carcinoma of the breast. Surgery 114, 637–641; discussion 641–642. [PubMed] [Google Scholar]
- Brastianos PK, Carter SL, Santagata S, Cahill DP, Taylor-Weiner A, Jones RT, Van Allen EM, Lawrence MS, Horowitz PM, Cibulskis K, et al. (2015). Genomic Characterization of Brain Metastases Reveals Branched Evolution and Potential Therapeutic Targets. Cancer Discov. 5, 1164–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D, Smeets D, Székely B, Larsimont D, Szász AM, Adnet P-Y, Rothé F, Rouas G, Nagy ZI, Faragó Z, et al. (2017). Phylogenetic analysis of metastatic progression in breast cancer using somatic mutations and copy number aberrations. Nat. Commun. 8, 14944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucheit AD, Chen G, Siroy A, Tetzlaff M, Broaddus R, Milton D, Fox P, Bassett R, Hwu P, Gershenwald JE, et al. (2014). Complete loss of PTEN protein expression correlates with shorter time to brain metastasis and survival in stage IIIB/C melanoma patients with BRAFV600 mutations. Clin. Cancer Res. 20, 5527–5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budczies J, von Winterfeld M, Klauschen F, Bockmayr M, Lennerz JK, Denkert C, Wolf T, Warth A, Dietel M, Anagnostopoulos I, et al. (2015). The landscape of metastatic progression patterns across major human cancers. Oncotarget 6, 570–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network, Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, Shen H, Robertson AG, Pashtan I, Shen R, et al. (2013). Integrated genomic characterization of endometrial carcinoma. Nature 497, 67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caso R, Sanchez-Vega F, Tan KS, Mastrogiacomo B, Zhou J, Jones GD, Nguyen B, Schultz N, Connolly JG, Brandt WS, et al. (2020). The Underlying Tumor Genomics of Predominant Histologic Subtypes in Lung Adenocarcinoma. J. Thorac. Oncol. 15, 1844–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejas P, López-Gómez M, Aguayo C, Madero R, de Castro Carpeño J, Belda-Iniesta C, Barriuso J, Moreno García V, Larrauri J, López R, et al. (2009). KRAS mutations in primary colorectal cancer tumors and related metastases: a potential role in prediction of lung metastasis. PLoS One 4, e8199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. (2012). The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, Wang J, Rudolph JE, Yaeger R, Soumerai T, Nissan MH, et al. (2017). OncoKB: A Precision Oncology Knowledge Base. JCO Precis Oncol 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng DT, Mitchell TN, Zehir A, Shah RH, Benayed R, Syed A, Chandramohan R, Liu ZY, Won HH, Scott SN, et al. (2015). Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J. Mol. Diagn. 17, 251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Bailey MH, Porta-Pardo E, Thorsson V, Colaprico A, Bertrand D, Gibbs DL, Weerasinghe A, Huang K-L, Tokheim C, et al. (2018). Perspective on Oncogenic Processes at the End of the Beginning of Cancer Genomics. Cell 173, 305–320.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert MA, Pan S, Hernandez KM, Loth RM, Andrade J, Volchenboum SL, Faber P, Montag A, Lastra R, Peter ME, et al. (2016). Genomics of Ovarian Cancer Progression Reveals Diverse Metastatic Trajectories Including Intraepithelial Metastasis to the Fallopian Tube. Cancer Discov. 6, 1342–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. (2013). Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, l1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Bado I, Wang H, Zhang W, Rosen JM, and Zhang XH-F (2019). Metastasis Organotropism: Redefining the Congenial Soil. Dev. Cell 49, 375–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerratana L, Davis AA, Polano M, Zhang Q, Shah AN, Lin C, Basile D, Toffoli G, Wehbe F, Puglisi F, et al. (2020). Understanding the organ tropism of metastatic breast cancer through the combination of liquid biopsy tools. Eur. J. Cancer 143, 147–157. [DOI] [PubMed] [Google Scholar]
- Hieronymus H, Murali R, Tin A, Yadav K, Abida W, Moller H, Berney D, Scher H, Carver B, Scardino P, et al. (2018). Tumor copy number alteration burden is a pan-cancer prognostic factor associated with recurrence and death. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Li Z, Ma Z, and Curtis C (2020). Multi-cancer analysis of clonality and the timing of systemic spread in paired primary tumors and metastases. Nat. Genet. 52, 701–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez-Sánchez A, Memon D, Pourpe S, Veeraraghavan H, Li Y, Vargas HA, Gill MB, Park KJ, Zivanovic O, Konner J, et al. (2017). Heterogeneous Tumor-Immune Microenvironments among Differentially Growing Metastases in an Ovarian Cancer Patient. Cell 170, 927–938.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones GD, Brandt WS, Shen R, Sanchez-Vega F, Tan KS, Martin A, Zhou J, Berger M, Solit DB, Schultz N, et al. (2021). A Genomic-Pathologic Annotated Risk Model to Predict Recurrence in Early-Stage Lung Adenocarcinoma. JAMA Surg. 156, e205601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennecke H, Yerushalmi R, Woods R, Cheang MCU, Voduc D, Speers CH, Nielsen TO, and Gelmon K (2010). Metastatic behavior of breast cancer subtypes. J. Clin. Oncol. 28, 3271–3277. [DOI] [PubMed] [Google Scholar]
- Kosmidis I, and Firth D (2020). Jeffreys-prior penalty, finiteness and shrinkage in binomial-response generalized linear models. Biometrika 108, 71–82. [Google Scholar]
- Lambert AW, Pattabiraman DR, and Weinberg RA (2017). Emerging Biological Principles of Metastasis. Cell 168, 670–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibold J, Ruscetti M, Cao Z, Ho Y-J, Baslan T, Zou M, Abida W, Feucht J, Han T, Barriga FM, et al. (2020). Somatic Tissue Engineering in Mouse Models Reveals an Actionable Role for WNT Pathway Alterations in Prostate Cancer Metastasis. Cancer Discov. 10, 1038–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makohon-Moore AP, Zhang M, Reiter JG, Bozic I, Allen B, Kundu D, Chatterjee K, Wong F, Jiao Y, Kohutek ZA, et al. (2017). Limited heterogeneity of known driver gene mutations among the metastases of individual patients with pancreatic cancer. Nat. Genet. 49, 358–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massagué J, and Ganesh K (2021). Metastasis-Initiating Cells and Ecosystems. Cancer Discov. 11, 971–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massagué J, and Obenauf AC (2016). Metastatic colonization by circulating tumour cells. Nature 529, 298–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natrajan R, Lambros MBK, Geyer FC, Marchio C, Tan DSP, Vatcheva R, Shiu K-K, Hungermann D, Rodriguez-Pinilla SM, Palacios J, et al. (2009). Loss of 16q in high grade breast cancer is associated with estrogen receptor status: Evidence for progression in tumors with a luminal phenotype? Genes Chromosomes Cancer 48, 351–365. [DOI] [PubMed] [Google Scholar]
- Naxerova K, Reiter JG, Brachtel E, Lennerz JK, van de Wetering M, Rowan A, Cai T, Clevers H, Swanton C, Nowak MA, et al. (2017). Origins of lymphatic and distant metastases in human colorectal cancer. Science 357, 55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen DX, Bos PD, and Massagué J (2009). Metastasis: from dissemination to organ-specific colonization. Nat. Rev. Cancer 9, 274–284. [DOI] [PubMed] [Google Scholar]
- Niu B, Ye K, Zhang Q, Lu C, Xie M, McLellan MD, Wendl MC, and Ding L (2014). MSIsensor: microsatellite instability detection using paired tumor-normal sequence data. Bioinformatics 30, 1015–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon MJ, Formisano L, Mayer IA, Estrada MV, González-Ericsson PI, Isakoff SJ, Forero-Torres A, Won H, Sanders ME, Solit DB, et al. (2019). PIK3CA and MAP3K1 alterations imply luminal A status and are associated with clinical benefit from pan-PI3K inhibitor buparlisib and letrozole in ER+ metastatic breast cancer. NPJ Breast Cancer 5, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noorani A, Li X, Goddard M, Crawte J, Alexandrov LB, Secrier M, Eldridge MD, Bower L, Weaver J, Lao-Sirieix P, et al. (2020). Genomic evidence supports a clonal diaspora model for metastases of esophageal adenocarcinoma. Nat. Genet. 52, 74–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pareja F, Ferrando L, Lee SSK, Beca F, Selenica P, Brown DN, Farmanbar A, Da Cruz Paula A, Vahdatinia M, Zhang H, et al. (2020). The genomic landscape of metastatic histologic special types of invasive breast cancer. NPJ Breast Cancer 6, 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira AAL, Rego JFM, Morris V, Overman MJ, Eng C, Garrett CR, Boutin AT, Ferrarotto R, Lee M, Jiang Z-Q, et al. (2015). Association between KRAS mutation and lung metastasis in advanced colorectal cancer. Br. J. Cancer 112, 424–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priestley P, Baber J, Lolkema MP, Steeghs N, de Bruijn E, Shale C, Duyvesteyn K, Haidari S, van Hoeck A, Onstenk W, et al. (2019). Pan-cancer whole-genome analyses of metastatic solid tumours. Nature 575, 210–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran R, Harrison H, Syamimi Ariffin N, Ayub R, Pegg HJ, Deng W, Mastro A, Ottewell PD, Mason SM, Blyth K, et al. (2020). A role for CBFβ in maintaining the metastatic phenotype of breast cancer cells. Oncogene 39, 2624–2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razavi P, Chang MT, Xu G, Bandlamudi C, Ross DS, Vasan N, Cai Y, Bielski CM, Donoghue MTA, Jonsson P, et al. (2018). The Genomic Landscape of Endocrine-Resistant Advanced Breast Cancers. Cancer Cell 34, 427–438.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter JG, Hung W-T, Lee I-H, Nagpal S, Giunta P, Degner S, Liu G, Wassenaar ECE, Jeck WR, Taylor MS, et al. (2020). Lymph node metastases develop through a wider evolutionary bottleneck than distant metastases. Nat. Genet. 52, 692–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson DR, Wu Y-M, Lonigro RJ, Vats P, Cobain E, Everett J, Cao X, Rabban E, Kumar-Sinha C, Raymond V, et al. (2017). Integrative clinical genomics of metastatic cancer. Nature 548, 297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueda OM, Sammut S-J, Seoane JA, Chin S-F, Caswell-Jin JL, Callari M, Batra R, Pereira B, Bruna A, Ali HR, et al. (2019). Dynamics of breast-cancer relapse reveal late-recurring ER-positive genomic subgroups. Nature 567, 399–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Vega F, Mina M, Armenia J, Chatila WK, Luna A, La KC, Dimitriadoy S, Liu DL, Kantheti HS, Saghafinia S, et al. (2018). Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell 173, 321–337.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen R, and Seshan VE (2016). FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res. 44, e131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih DJH, Nayyar N, Bihun I, Dagogo-Jack I, Gill CM, Aquilanti E, Bertalan M, Kaplan A, D’Andrea MR, Chukwueke U, et al. (2020). Genomic characterization of human brain metastases identifies drivers of metastatic lung adenocarcinoma. Nat. Genet. 52, 371–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoushtari AN, Chatila WK, Arora A, Sanchez-Vega F, Kantheti HS, Rojas Zamalloa JA, Krieger P, Callahan MK, Betof Warner A, Postow MA, et al. (2021). Therapeutic Implications of Detecting MAPK-Activating Alterations in Cutaneous and Unknown Primary Melanomas. Clin. Cancer Res. 27, 2226–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla A, Nguyen THM, Moka SB, Ellis JJ, Grady JP, Oey H, Cristino AS, Khanna KK, Kroese DP, Krause L, et al. (2020). Chromosome arm aneuploidies shape tumour evolution and drug response. Nat. Commun. 11, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurr LF, Touat M, Taylor AM, Dubuc AM, Shih J, Meredith DM, Pisano WV, Meyerson ML, Ligon KL, Cherniack AD, et al. (2020). Quantification of aneuploidy in targeted sequencing data using ASCETS. Bioinformatics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stopsack KH, Whittaker CA, Gerke TA, Loda M, Kantoff PW, Mucci LA, and Amon A (2019). Aneuploidy drives lethal progression in prostate cancer. Proc. Natl. Acad. Sci. U. S. A. 116, 11390–11395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B, et al. (2010). Integrative genomic profiling of human prostate cancer. Cancer Cell 18, 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tie J, Lipton L, Desai J, Gibbs P, Jorissen RN, Christie M, Drummond KJ, Thomson BNJ, Usatoff V, Evans PM, et al. (2011). KRAS mutation is associated with lung metastasis in patients with curatively resected colorectal cancer. Clin. Cancer Res. 17, 1122–1130. [DOI] [PubMed] [Google Scholar]
- Turajlic S, Xu H, Litchfield K, Rowan A, Chambers T, Lopez JI, Nicol D, O’Brien T, Larkin J, Horswell S, et al. (2018). Tracking Cancer Evolution Reveals Constrained Routes to Metastases: TRACERx Renal. Cell 173, 581–594.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vokes NI, Liu D, Ricciuti B, Jimenez-Aguilar E, Rizvi H, Dietlein F, He MX, Margolis CA, Elmarakeby HA, Girshman J, et al. (2019). Harmonization of Tumor Mutational Burden Quantification and Association With Response to Immune Checkpoint Blockade in Non-Small-Cell Lung Cancer. JCO Precis Oncol 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins TBK, Lim EL, Petkovic M, Elizalde S, Birkbak NJ, Wilson GA, Moore DA, Grönroos E, Rowan A, Dewhurst SM, et al. (2020). Pervasive chromosomal instability and karyotype order in tumour evolution. Nature 587, 126–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaeger R, Chatila WK, Lipsyc MD, Hechtman JF, Cercek A, Sanchez-Vega F, Jayakumaran G, Middha S, Zehir A, Donoghue MTA, et al. (2018). Clinical Sequencing Defines the Genomic Landscape of Metastatic Colorectal Cancer. Cancer Cell 33, 125–136.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, Srinivasan P, Gao J, Chakravarty D, Devlin SM, et al. (2017). Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med. 23, 703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1889). THE DISTRIBUTION OF SECONDARY GROWTHS IN CANCER OF THE BREAST. Lancet 133, 571–573. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Study design and characteristics of the patients and samples included in MSK-MET, related to Figure 1.
(A) CONSORT flow diagram of the study.
(B) Distribution of age at time of sequencing, time interval between surgical procedure and sequencing, tumor sample coverage and tumor purity assessed by the pathologist.
(C) Distribution of 25,775 tumors across 50 tumor types grouped by ten organ systems.
(D) Distribution of the 25,775 tumors according to the sample type (primary vs. metastasis), site of metastatic sample and whether the primary sample was from a patient with evidence of distant metastasis at the time of the study or not.
(E) Frequency of primary and metastatic samples previously exposed to any treatment for each tumor type.
(F) Distribution of the 99,220 metastatic events mapped to 21 organ sites.
(G) Comparison of the frequency of metastasis in several target organs from different tumor types reported in (Gao et al., 2019) and in (Budczies et al., 2015) vs. the present study.
(H) Comparison of the number of metastasis using data from manual chart reviews and clinical data automatically extracted from the EHR (This study).
(I) Heatmap showing the recall rate (sensitivity) across several target organs from different tumor types using patients retrieved from manual chart reviews.
(J) Median time to metastasis (months) relative to the first metastatic event. The number in each cell indicates the median time to metastasis at that given site in months. Only metastases with at least 3 representative patients are shown here. Note that for patients with multiple metastases to the same organ only the first one was taken into consideration. For each metastatic site, the distribution of all 50 tumor types is shown as a stacked bar chart below the heatmap.
(K) Results from the Bradley-Terry model showing the order of metastatic events observed in the top 10 most common tumor types. For each tumor type, the order of metastasis from first to last is represented from top to bottom.
Figure S2. Genomic differences between primary tumors and metastases using an independent cohort of 9,215 patients sequenced with MSK-IMPACT since the data freeze date related to Figure 2.
(A) Comparisons of the median fraction genome altered (FGA), median tumor mutation burden (TMB), and median frequency of tumor mutation burden high (TMB-hi) for each tumor type in metastatic vs. primary tumors. Tumor types with statistically significant differences are labeled.
(B) The following clinical and genomic features are shown side-by-side for primary (top row within each cancer type) and metastatic (bottom row) sequenced samples using a combination of bar charts and violin plots; from left to right: sample counts, FGA, TMB, fraction of samples with high TMB, and distribution of the highest actionable alteration levels. The black vertical line in each violin plot represents the median. The heatmap shows the frequency of individual arm level alterations in primary tumors and metastases (only the frequency of the more frequent event, gain or loss, is shown). Tumor types are ordered from top to bottom by decreasing FGA in metastasis and grouped by organ systems. * indicates q-value < 0.05.
(C) Statistically significant differences in the frequency of oncogenic alterations and pathways between primary tumors and metastases in individual tumor types. Triangles summarize oncogenic alteration frequencies in primary tumors vs. metastases and are colored according to alteration type. Gene names in italics refer to specific genes, those in regular font refer to pathways.
(D) Comparison of the effect size of FGA and TMB between the original and the new dataset. Each point is colored according to significance (green; significant in the new dataset only, blue; significant in the original dataset only; pink significant in both the original and the new dataset).
(E) Comparison of the effect size of the difference in oncogenic alteration between the original and the new dataset. Each point is colored according to tumor type (left) or significance (right). The comparison was tested using a Spearman correlation.
Figure S3. Genomic differences between primary samples from metastatic and non-metastatic patients, related to Figure 2.
(A) Scatterplot showing the comparison of the median FGA, median WGD, median TMB and median clonality for each tumor type in primary samples from metastatic and non-metastatic patients.
(B) The following clinical and genomic features are shown side-by-side for primary samples from metastatic and non-metastatic patients using a combination of bar charts and violin plots; from left to right: sample counts, FGA, fraction of samples with WGD, TMB, clonality, fraction of samples with TMB-high status and distribution of highest actionable alteration. The black vertical line in each violin plots represents the median. Heatmap shows the frequency of arm level alterations in primary tumors and metastases. Tumor types are ordered from top to bottom by decreasing FGA in metastasis and grouped by organ systems. * indicates q-value < 0.05. WGD and clonality were available for a subset of 10,106 samples with FACETS data.
(C) Statistically significant differences in the frequency of oncogenic alterations between primary tumors and metastases across all tumor types. Triangles summarize oncogenic alterations frequencies in primary samples from metastatic vs. non-metastatic patients and are colored according to alteration type.
(D) Comparison of genomic features differing between primary tumors vs metastases (“P vs M”) and genomic features differing between primary tumors from non-metastatic vs metastatic patients (“P-noM vs P-M”). Comparison of the effect size for FGA and TMB (D) (log2 fold change) between the two analyses. Top; each point represents a tumor type and is color-coded according to the three statistical significance scenarios (pink; significant in both analyses, green; significant in P vs M only, blue; significant in P-noM vs P-M only). Bottom; boxplots showing FGA and TMB according to the three sample types (primary tumors from non-metastatic vs primary tumors from metastatic patients vs metastatic samples) for two representative tumor types and every three scenarios.
(E) Comparison of the effect size for oncogenic alterations (log odds ratio) between the two analyses. Top; each point represents an oncogenic alteration that was significantly different in either analysis and is color-coded according to the three statistical significance scenarios (left) (pink; significant in both analyses, green; significant in P vs M only, blue; significant in P-noM vs P-M only,) and the tumor type in which it belongs (left). Bottom; bar graphs showing the alterations frequency according to the three sample types (primary tumors from non-metastatic vs primary tumors from metastatic patients vs metastatic samples) for two representative tumor types and every three scenarios. For each comparison, a Venn diagram shows the number of common and exclusive significant associations between the two analyses (top-left corner). The comparisons were tested using a Spearman correlation.
Figure S4. Genomic differences of metastases according to organ location, related to Figure 4.
(A) Distribution of sequenced metastasis according to organ location with the heatmap showing the percentage of metastatic samples where each row represents a tumor type and each column represents the organ location. For each tumor type, the distribution of all 21 organ locations is shown as a stacked bar chart to the right of the heatmap. For each organ, the distribution of all 50 tumor types is shown as a stacked bar chart below the heatmap. For each tumor type, the number of metastasis samples is indicated in parentheses. For each organ, the number of metastasis samples is indicated in parentheses.
(B) Statistically significant association between FGA (black circle) and TMB (white diamond) and specific metastatic sites.
(C) Statistically significant oncogenic alterations associated with specific metastatic sites.
(D) Heatmap showing 69 genomic features associated with metastasis to specific target organs and significant in at least two cancer types. The shade of red represents the three significance thresholds (dark red, q-value < 0.05 & logit < 0.05; red, q-value < 0.05; light red, p-value < 0.05).
Table S1. Summary of the 50 tumor types included in the MSK-MET cohort (A), Individual patient data included in the MSK-MET cohort (B), Mapping between free-text description from pathology reports and ICD billing codes from EHR to a curated list of 21 organs (C), Related to Figure 1
Table S2. Genomic differences between primary tumors and metastases (A), Genomic differences between primary tumors and metastases using an independent cohort (B), Genomic differences between primary samples from metastatic and non-metastatic patients (C), Related to Figure 2, Figure S2 and Figure S3
Table S3. Association between metastatic burden and overall survival assessed using Cox proportional hazards model (A), Genomic associations with metastatic burden (B), Related to Figure 3
Table S4. Genomic differences of metastases according to their organ location, Related to Figure S4 (A), Genomic features associated with metastasis to specific target organs (B), Related to Figure S4 and Figure 4
Data Availability Statement
The raw sequencing data for the MSK-IMPACT cohort are protected and are not broadly available due to privacy laws. Raw data may be requested from schultzn@mskcc.org with appropriate institutional approvals.
Our full dataset including clinical and genomic data is publicly available at Zenodo https://doi.org/10.5281/zenodo.5801902 and through the cBioPortal for Cancer Genomics https://www.cbioportal.org/study?id=msk_met_2021
Original code to do the organ site mapping for metastatic cancer is available at https://github.com/clinical-data-mining/organ-site-mapping and has been deposited at figshare. DOIs is listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.