Abstract
Emerging infectious diseases, especially if caused by bat-borne viruses, significantly affect public health and the global economy. There is an urgent need to understand the mechanism of interspecies transmission, particularly to humans. Viral genetics; host factors, including polymorphisms in the receptors; and ecological, environmental, and population dynamics are major parameters to consider. Here, we describe the taxonomy, geographic distribution, and unique traits of bats associated with their importance as virus reservoirs. Then, we summarize the origin, intermediate hosts, and the current understanding of interspecies transmission of Middle East respiratory syndrome coronavirus (MERS-CoV), severe acute respiratory syndrome coronavirus (SARS-CoV), SARS-CoV-2, Nipah, Hendra, Ebola, Marburg virus, and rotaviruses. Finally, the molecular interactions of viral surface proteins with host cell receptors are examined, and a comparison of these interactions in humans, intermediate hosts, and bats is conducted. This uncovers adaptive mutations in virus spike protein that facilitate cross-species transmission and risk factors associated with the emergence of novel viruses from bats.
Keywords: bat, RNA virus, interspecies transmission
Tian et al. review interspecies transmission of currently emerging viruses from bats. They focus on the molecular interactions of viral surface proteins with host cell receptors in humans, intermediate hosts, and bats, which provides an understanding of cross-species transmission of zoonotic viruses from bats.
Introduction: Systematics, distribution, and ecology of bats and their viruses
Bats (order Chiroptera) are a group of mammals with the second largest number of species. Although bats are believed to have emerged in the early Eocene period (∼56 million years ago), the scarcity and difficulty in identifying bat species from fossil records makes the evolutionary history of this order controversial (Simmons et al., 2008). Traditionally, based on morphological data, this order was divided into two suborders, Megachiroptera and Microchiroptera. According to the current view, which also includes molecular evidence, Chiroptera is divided into two suborders Yinpterochiroptera and Yangochiroptera (Teeling et al., 2002, 2005). Yinpterochiroptera consists of the family Pteropodidae and the superfamily Rhinolophoidea, which includes six families. Yangochiroptera is composed of Myzopodidae and three superfamilies: Vespertilionoidea, Noctilionoidea, and Emballonuroidea, which are further divided into 13 families. A total of 1,453 bat species belonging to 21 families have been recorded (retrieved on March 27, 2022, from https://batnames.org/). With the exception of some remote islands, bats populate every continent. However, their main habitat are the tropics and the subtropics, including tropical rain forests, grasslands, deserts, farmlands, and even cities. The widespread distribution of bat habitats indicates that they are a highly successful radiation in mammalian evolution. The Vespertilionidae contains the greatest number of species (n > 500). They are found in almost every geographical region where bats exist (Gunnell et al., 2017). Molossidae and Emballonuridae are present in all continents, except Antarctica and the Arctic (Figure 1 ). However, some bats inhabit only certain regions; e.g., some members of the Craseonycteridae are found only in Asia (Thailand), whereas Myzopodidae, Mystacinidae, and Noctilionoidea are found only in Africa (Madagascar and Egypt), Oceania, and the Americas respectively (Gunnell et al., 2014; Russell et al., 2008).
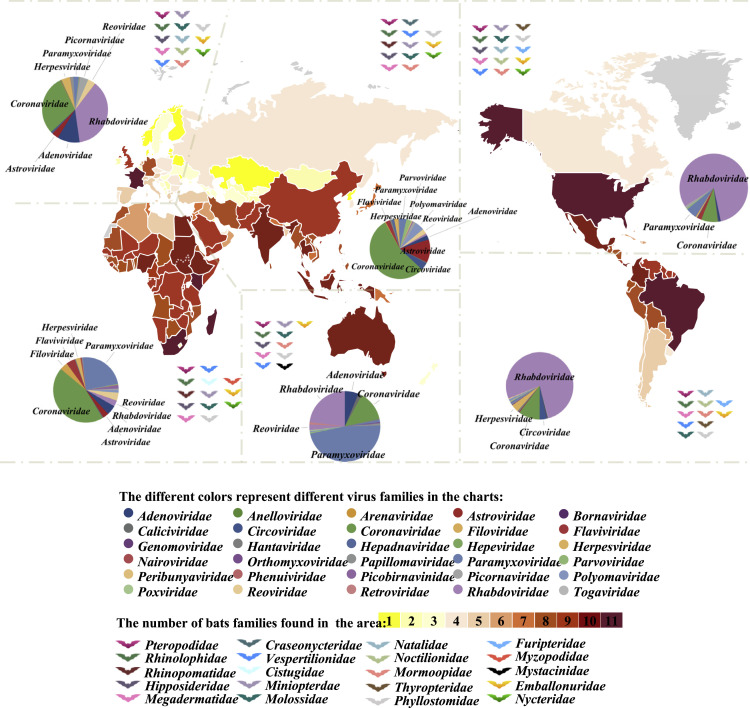
Figure 1.
Global distribution of bats and their viruses
The bat families present in each continent are symbolized by the color of the bat pictogram.
The number of different bat families identified in each country is represented by the color range (from yellow to red) as indicated. The pie charts show the percentage of bat-associated viruses from a certain family identified in the respective continents according to publicly available sequence data from a database of bat-associated viruses (http://www.mgc.ac.cn/DBatVir/) as of July 16, 2020, and GenBank data from July 16, 2020 to August 25, 2020.
Bats are herbivores, carnivores, or omnivores, with a variety of unique ecological characteristics (Teeling et al., 2018). As herbivores, they feed on fruits, flowers, leaves, nectar, and pollen. As carnivores, bats hunt insects, fishes, small amphibians, reptiles, and even small mammals. A large-bodied omnivorous bat (Noctilionoidea: Mystacinidae) has an omnivorous diet (Hand et al., 2018). A few species, e.g., vampire bats, feed on animal blood only, and some bats change their diet with the season (Gonsalves et al., 2013; Harper et al., 2013; Mello et al., 2004; Ripperger et al., 2015; Zepeda Mendoza et al., 2018). Bats vary in size, from only a few centimeters to up to 1 m of body length (Teeling et al., 2018). Most bats are nocturnal, being active during the night and sleeping during the day. They hang upside down by their feet in caves, attics, trees, or other refugees. Because of their nocturnal activity, many species of bats use echolocation—the emission of ultrasonic waves to produce echoes—in flight (Springer et al., 2001). Of note, this ultrasound generates aerosols, which may facilitate dispersion of viruses (Calisher et al., 2006; Constantine et al., 1972). Bats play an important role in maintaining the ecological balance by pest control, plant pollination, and seed dispersal (Baldwin et al., 2020; Fadini et al., 2018; López-Hoffman et al., 2014).
With these diverse ecology, biology, and unique traits, bats are perfectly equipped to be reservoirs for a large variety of viruses. Currently, more than 12,000 bat-associated viral sequences belonging to at least 30 (out of 168) viral families have been detected (see database of bat-associated viruses, http://www.mgc.ac.cn/DBatVir/) (Chen et al., 2014). Of note, viruses from 25 viral families were identified in bats in Asia and Africa. The high number of viral families identified in these regions may be the result of extensive epidemiological and virological investigations due to Ebola, severe acute respiratory syndrome (SARS), and Nipah virus (NiV) outbreaks.
DNA viruses appear to be the minority of bat-associated viruses. The most common of them are herpesviruses, circoviruses, and parvoviruses (Ge et al., 2011; James et al., 2020; Wu et al., 2016). Circoviruses are mainly found in Asia and Africa in Rhinolophidae and Vespertilionidae bats. They exhibit a relatively high mutation and recombination rate, resulting in high genetic diversity (Ge et al., 2011; Zhu et al., 2018). Recently, porcine circovirus type 3 has emerged in many pig herds around the world, and this virus is suspected to be of bat origin (Li et al., 2018). Interestingly, there is extensive diversity in bat polyomaviruses. With large numbers and multiple species of bats in the same roosting colonies in some habitats, the diversity of polyomaviruses is possibly due to intra- and interspecies transmission (Tan et al., 2020).
By contrast, RNA viruses account for the largest proportion of bat-associated viruses, and they cause more cross-species transmission events and with higher pathogenicity (Cui et al., 2019; Eaton et al., 2006). The most common RNA viruses found in bats are coronaviruses (CoVs), rhabdoviruses, paramyxoviruses, and astroviruses, followed by filoviruses, picobirnaviruses, caliciviruses, reoviruses, and flaviviruses (Figure 1). Furthermore, the distribution and composition of viruses vary between different bat populations and habitats. Among all bat-associated viral sequence data registered in the database (DBatVir), rhabdoviruses are the most frequently found virus in North and South America (81% and 77%, respectively), and in Europe (37%). Paramyxoviruses are the most common found bat-associated viruses in Oceania (47%), while CoVs account for the largest proportion in Asia (55%) and Africa (45%), as shown in Figure 1. However, these numbers are heavily influenced by sampling bias, and there are very limited studies for other viral types in those regions.
Around 80% of RNA viruses were identified in only three bat families: Vespertilionidae, Rhinolophidae and Pteropodidae. Most interesting is the members of the genus Rhinolophus within the Rhinolophidae, as they are the natural hosts of SARS-like CoVs (Ge et al., 2013; Lau et al., 2005; Li et al., 2005b; Rihtaric et al., 2010). Rhinolophus bats are small, insect-eating animals that are widely distributed in Southeast Asia, Africa, eastern Oceania, and Western Europe. The source of Middle East respiratory syndrome (MERS)-CoV are bats of the Vespertilionidae, the most diverse and widely distributed of all bat families, and Nycteridae (Cui et al., 2019; de Groot et al., 2013). Half of the bat-associated Paramyxoviridae sequences, including henipaviruses, were detected in pteropid bats (flying foxes). These are fruit-eating bats, are found in all six continents, and they fly long distances. Most filoviruses (90%), including Marburg and Ebola virus, were identified in members of Pteropodidae. These bats harbor also other zoonotic viruses, such as rotaviruses (RVs), members of Reoviridae, the cause of diarrheal disease in infants, other mammals, and birds. Bat rotavirus, which has a wide geographic dispersal and rich genetic diversity, may be the origin of several human and other animal RVs (Simsek et al., 2021). Other bat families, such as the Myzopodidae, have not been reported to carry viruses (as of August 25, 2020; http://www.mgc.ac.cn/DBatVir/). Whether this is due to insufficient epidemiological investigations, or these bats not being susceptible to viral infection, is unclear.
How bat-associated viruses evolve and spread is still largely unexplored. Similar to humans, most bats are social animals, and they have a relatively long lifespan (on average, 3.5 times that of non-flying placental mammals of similar size; Wilkinson and South, 2002). They live in colonies, some of them are composed of up to 1 million individuals, and might be intermixed with multiple bat species (Serra-Cobo and Lopez-Roig, 2017). These colonies allow viruses to be maintained, as susceptible hosts are abundant. Furthermore, viral infection is often asymptomatic in bats, which might be due to their strong innate immunity (Gorbunova et al., 2020; Jebb et al., 2020). Indeed, several immunity-related genes were identified to be under positive selection in bats, and may provide a mechanism for tolerance against viral pathogens (Jebb et al., 2020; Wang et al., 2021). Nevertheless, infected bats shed virus through blood, feces, nasal secretions, and saliva (Edson et al., 2015). Between bats, viruses are transmitted through the urine-oronasal mode, while other mammals may be infected by food or fomites contaminated by the above-mentioned bodily fluids, likely from infected bats roosting above or by feeding on the same foods (Arankalle et al., 2011; Edson et al., 2015; Islam et al., 2016; Nyakarahuka et al., 2019). The production of ultrasonic waves via the larynx and their emission through the mouth or nose might also spread viruses by aerosolizing the viruses (Calisher et al., 2006; Constantine et al., 1972). Many bats are territorial; i.e., they return day after day to the same refuge. However, the ever-increasing human expansion reduces the habitats available for bats (Frick et al., 2020). Livestock breeding and cultivation of arboreal landscapes near bat habitats not only affects the native wildlife and plant species but also increases the risk of contact to domesticated animals, or even to humans, thus increasing the risk of zoonotic spillover (Brearley et al., 2013; Field et al., 2016). Natural disasters may have the same effect. For example, the increasing incidence and severity of wildfires have decimated many bat colonies and their habitats (Blakey et al., 2019).
With their global distribution, unique biological characteristics, and strong immune system, bats emerge as important viral reservoirs (Gorbunova et al., 2020; Jebb et al., 2020; Wang et al., 2021). Among the wide range of viruses that bats carry, we focus the discussion here on viruses that have been of significant public health importance: CoVs, henipaviruses, filoviruses, and RVs. We review current knowledge on their evolution, epidemiology, and possible mechanisms of cross-species transmission. Using molecular data, we also examine the first step of interspecies transmission of virus from bats to intermediate hosts and to human; that is, the binding of viral surface protein to host cellular receptors. Importantly, we also discussed RVs, because zoonoses of bat RVs might occur more frequently than currently recognized, and likely will be of public health significance.
Emergence and cross-species transmission of pathogenic viruses from bats
Bat-associated viruses cause no or little disease in their primary animal reservoir. This is probably the result of the strong immune system of bats and long-term co-evolution (Gorbunova et al., 2020). However, successful transmission to humans often requires an intermediate or amplifying host. Ideal intermediate hosts are species that interact with the reservoir hosts, the bats, and with humans. Animals in close contact with humans, such as domestic animals, like pigs, horses, and dromedary camels, are important for zoonotic transmissions. Even wild animals, Himalayan palm civets, and raccoon dogs, which are raised as an exotic food sources by humans in Vietnam, Cambodia, Myanmar, other Southeast Asian countries, and in southern China, have been shown to serve as intermediate hosts for SARS-CoV (Guan et al., 2003). However, there is also evidence that bat-associated viruses might be able to jump directly to humans without prior adaptation to other hosts (Menachery et al., 2015; Wang et al., 2018b; Zheng et al., 2020).
One can assume that spillover events occur quite often, but they are recognized only if a virus acquires the ability for sustained transmission in its new host. We now describe the members of three virus families (CoVs, henipaviruses, and filoviruses) that have successfully crossed the species barrier causing substantial outbreaks and associated with significant pathogenicity in humans. We summarize current knowledge about the bat reservoir of these viruses, the putative intermediate hosts, and the effects of cross-species transmission.
CoVs
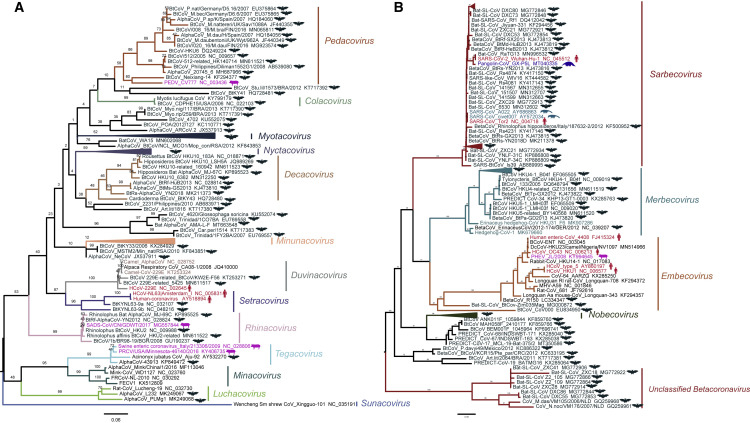
CoVs account for the largest proportion of known bat-associated viruses. They are enveloped, spherical viruses containing a positive stranded RNA genome of 26–32 kb (Brian and Baric, 2005). CoVs include several pathogens of clinical, veterinary, and agricultural importance. These viruses cause gastrointestinal or respiratory infections with a wide range of clinical manifestations (He et al., 2020; Weiss and Leibowitz, 2011). However, not all CoVs are pathogenic, and some CoVs cause only mild symptoms, such as four human “common cold” CoVs: HCoV-NL63, HCoV-229E, HCoV-OC43, and HCoV-HKU1. Orthocoronavirinae, a subfamily in Coronaviridae, are divided into four genera, Alphacoronavirus (alpha-CoV) and Betacoronavirus (beta-CoV), which mainly infect mammals, and Deltacoronavirus and Gammacoronavirus, which usually infect birds. There are 26 and 14 species in alpha-CoV and beta-CoV, respectively, 17 of which were found in bats (https://talk.ictvonline.org/taxonomy/). The alpha-CoVs have been identified in more than 130 bat species from the genera Miniopterus, Myotis, Rhinolophus, and Hipposideros, whereas beta-CoVs are found in approximately 100 bat species, the largest proportion in the genus Rhinolophus (http://www.mgc.ac.cn/DBatVir/). Importantly, these CoVs have a tendency to transmit among bat species: Alpha-CoVs are found more frequently and across more distantly related host taxa than beta-CoVs (Latinne et al., 2020). Phylogenetic analysis shows that bat CoVs display a high genetic diversity, which may play an important role in the evolution and emergence of these two genera (Figure 2 ). The subgenus Sarbecovirus of beta-CoVs, such as SARS-CoV and SARS-CoV-2, had spread to humans, most likely via intermediate hosts (Guan et al., 2003; Zhang and Holmes, 2020). The genomes of several human alpha-CoVs, including two of the common cold CoVs, namely HCoV-NL63 and HCoV-229E, and two porcine alpha-CoVs (described below) are closely related to CoVs identified in bats, suggesting that their common ancestors existed in bats (Corman et al., 2015; Tao et al., 2017) (Figure 2).
Figure 2.
Genetic evolution of coronaviruses in Alphacoronavirus and Betacoronavirus
(A and B) Maximum likelihood tree of Alphacoronavirus (A) and Betacoronavirus (B) reconstructed by RAxML based on the polymerase (RdRP) gene, using the PROTCATLG substitution model and 1,000 bootstrap value. Representative isolate sequences used in the present study were downloaded from GenBank and the RdRP gene was intercepted, and the corresponding GenBank accession numbers were provided in the tree. The hosts of the viruses are indicated by different colors (bat, black; human, dark red; pig, magenta; pangolin, blue; and civet, dark green) and by pictograms in the same color. Trees were rooted on midpoint, and branches of the tree labeled in different colors indicate the indicated subgenera.
The genomes of CoVs are highly variable. They often acquire deletions up to about 400 nucleotides, which do not compromise virus replication but might affect pathogenicity (Su et al., 2020). They also exhibit a high mutation rate (antigenic drift), which, however, is lower than many other RNA viruses, possibly due to the proof-reading activity of the viral polymerase complex (Posthuma et al., 2017). In addition, inter- and intra-typic recombination occurs frequently (Su et al., 2016). These genetic variations generate new virus populations with abundant genetic variations, similar to a viral gene pool. When these changes occur in or around the spike protein (S), they may critically affect the tissue or host tropism, hence a mechanism for interspecies transmission (Letko et al., 2020).
Evidence from veterinary medicine that CoVs can change tissue tropism
There is long-standing evidence from veterinary medicine that CoVs are highly flexible since they can change tissue tropism. One example is the transmissible gastroenteritis CoV (TGEV) of pigs. This virus was first isolated in 1946, and it infects mainly epithelial cells of the small intestine (Ristic et al., 1965). A related CoV (porcine respiratory CoV [PRCoV]) emerged in 1984, which replicates in the respiratory tract but causes only minor clinical signs. The major difference between the two porcine CoVs is the deletion of a sialic acid binding site in the S protein of PRCoV (Schwegmann-Wessels and Herrler, 2006). The sialic acid binding activity may allow TGEV to overcome the mucus barrier in the gut, hence enabling the virus to access the intestinal epithelium. Since the epitopes for neutralizing antibodies are still present in S of PRCoV, infections with this virus act like a natural vaccine against TGEV, resulting in a drastic reduction of TGE outbreaks in Europe (Laude et al., 1993). However, this unusual change in the tropism was not noticed by many virologists, since CoVs, prior to the SARS epidemics, were not considered significant human pathogens.
At least three pathogenic CoVs for humans have originated from bat-associated viruses. We summarize the essential features of the three viruses below, and they are also listed in Table 1 .
Table 1.
Essential features of SARS-CoV, MERS-CoV, and SARS-CoV-2
| First described human cases/country | Bat reservoir | Intermediate host or animal reservoir | Transmissibility between humans | Mode of transmission | Cellular receptor | Number of confirmed cases (5.3.21) | Case fatality rate (%) | |
|---|---|---|---|---|---|---|---|---|
| SARS-CoV | February 2003 Guangdong, China |
Rhinolophus species | masked palm civet (Paguma larvata) or raccoon dogs (Nyctereutes procyonoides) | moderate, R0: ∼1 | respiratory droplets, surfaces | ACE2 | 8,096 | 9.6 |
| MERS-CoV | April 2012 Saudi Arabia |
Neoromicia, Pipistrellus | dromedary (Camelus dromedaries) | low, R0: <1 | close contact | dipeptidylpeptidase 4 | 2,468 | 35 |
| SARS-CoV-2 | December 2019, Wuhan, China | Rhinolophus species? | Unknown | high, R0: ∼3 | aerosol, droplets | ACE2 | 115,655,792 | 2.3 |
Data from WHO:
MERS-CoV: https://www.who.int/health-topics/middle-east-respiratory-syndrome-coronavirus-mers#tab=tab_1
R0 is the basic reproduction number: expected number of cases generated by one case in a population where all individuals are susceptible to infection.
Case fatality rate is likely an overestimation since mild and asymptomatic cases are missed by surveillance systems.
SARS-CoV
CoVs came first into the spotlight in 2002–2003 during the SARS epidemics. The virus first appeared in Guangdong Province in southern China, in November of 2002, followed by spreading to Hong Kong in February, 2003 (Stadler et al., 2003). Epidemiological evidence indicated that early human cases were among restaurant workers involved in handling wild and exotic animals. The virus was introduced to Hong Kong by a physician who had treated patients with “atypical pneumonia.” From Hong Kong, by international air travel, the virus spread to 26 countries worldwide. A total of 8,096 confirmed cases with 774 deaths were recorded, which results in a case fatality rate of 9.6% (https://www.who.int/csr/sars/country/table2004_04_21/en/). Based on positive PCR tests and serological evidence, it was found that SARS-CoV was probably transmitted to humans via an intermediate host, either by Himalayan palm civets or raccoon dogs (Guan et al., 2003). However, these animals were not the natural host for this virus. These animals were mainly infected in the wet markets where they were sold, or during trade-related activities. Interestingly, this virus was not detected in the farms where they were raised or found in their natural habitats (Kan et al., 2005; Tu et al., 2004). A search for the origin of SARS-CoV led to the detection of CoVs in bats. Based on serological and molecular evidence, four species of horseshoe bats (genus Rhinolophus) were suspected as the natural reservoir for this virus. However, it took 10 more years before a bat-associated virus with a high sequence identity (95%) to SARS-CoV was isolated (Ge et al., 2013; Guan et al., 2003). This virus, designated as bat SL-CoV-WIV1, binds to angiotensin-converting enzyme 2 (ACE2) receptors from humans, civets, and Chinese horseshoe bats, the latter being its natural host. These findings are the strongest evidence that members of Rhinolophus like Rhinolophus sinicus are natural reservoirs for SARS-related CoV (Ge et al., 2013; He et al., 2014; Hu et al., 2017; Lau et al., 2005, 2010, 2015; Rihtaric et al., 2010; Yang et al., 2013). There are several genera of bats that are hosts of Sarbecovirus, not just Rhinolophus but other family members, including Hipposideros amiger, Hipposideros larvatus, Asseliscus stoliczkanus, and also others (less common) such as Chaerephon plicatus, Miniopterus schreibersii, and Nyctalus leisleri. (Drexler et al., 2010; Hu et al., 2017; Lin et al., 2017; Wacharapluesadee et al., 2015; Yang et al., 2013). Subsequent research has shown that bats in several caves, located in the southern Chinese province of Yunnan, harbor a large variety of SARS-related viruses. Some of these CoVs were shown also to bind to the human ACE2 receptor. Furthermore, limited serological data indicated that these CoVs occasionally infect humans, but without causing an epidemic (Hu et al., 2017), possibly lacking human-to-human transmissibility.
MERS-CoV
MERS-CoV, a beta-CoV, was first isolated in 2012 from human patients with acute pneumonia in Saudi Arabia (Zaki et al., 2012). There was strong evidence that these human cases were infected by close contact to dromedary camels, since both viral genomes and antibodies against MERS-CoV were detected in the camels associated with the human cases (Hemida et al., 2014; Reusken et al., 2013). Dromedary camels harbor several viral lineages, including one that is almost identical to viruses isolated from clinical samples (Haagmans et al., 2014; Sabir et al., 2016). Mounting evidence has indicated that MERS-CoV originated from Pipistrellus and Neoromicia bats belonging to the family Vespertilionidae and Nycteris bats belonging to the family Nycteridae (Annan et al., 2013; Corman et al., 2014; Cui et al., 2019; Ithete et al., 2013). A phylogenetic analysis of 65 MERS-CoV sequences revealed that they formed five clades (Cotten et al., 2014). However, only a few bat-related viruses recognize the human dipeptidylpeptidase 4 (DPP4) receptor, which is required for virus entry, and thus the bat-associated virus that contributed to MERS-CoV awaits its discovery (Cui et al., 2013; Lu et al., 2015). Phylodynamic analysis of viruses closely related to MERS-CoV revealed that substitutions of genetic elements had taken place either in bat or in camels before they were transmitted from camels to human. The introduction into camel population from bats might had occurred in sub-Saharan Africa (Corman et al., 2014). Furthermore, the MERS-CoV found in Arabian Peninsula camels seems to be imported from countries in the Greater Horn of Africa, as the majority of camels were imported from these regions (Corman et al., 2014; Muller et al., 2014). Camels serve as source of milk and meat, and are also used for transportation and for sport. Humans are infected with MERS-CoV by close contact with infected camels’ nasal secretions (Azhar et al., 2014). Multiple transmissions to humans have been described, mostly in the Arabian Peninsula, but occasionally the virus has spread to other regions through international travel; e.g., a large outbreak occurred in South Korea in 2015.
MERS-CoV infections in humans often results in severe pneumonia, with a case fatality rate of 35% (see update on cases and fatality rates on the World Health Organization [WHO] website: https://www.who.int/health-topics/middle-east-respiratory-syndrome-coronavirus-mers#tab=tab_1). By contrast, virus infections in camels results in mild upper respiratory tract infections (Adney et al., 2014). This might be explained by the inverted localization of the host receptor, DPP4, which the virus binds to penetrate host cells (Raj et al., 2013). This receptor is found at the nasal epithelium of camels. In contrast, DPP4 is mainly found in the epithelium of the lower respiratory tract in humans (Widagdo et al., 2019). The restriction of virus replication to the lung in human explains why transmission between humans is rare. Nonetheless, the frequency of sporadic outbreaks indicates that MERS remains a significant threat.
SARS-CoV-2
At the end of 2019, cases of unexplained pneumonia were reported from Wuhan, China. Thanks to the experience with SARS, the etiological agent was quickly identified as a novel beta-CoV, named SARS-CoV-2 by International Committee on Taxonomy of Viruses. Although most early-reported CoV disease 2019 (COVID-19) patients had contact history with wildlife animals at a wet seafood market (Sun et al., 2020), no virus was detected in the animals there. Therefore, there is debate over whether the seafood market is actually linked to zoonosis or simply a site for amplification. It is worth noting that there is some evidence that traces of SARS-CoV-2 appeared earlier in other regions, which makes the origin of this virus a mystery (Amendola et al., 2021; Basavaraju et al., 2021; Deslandes et al., 2020; Fongaro et al., 2021).
When comparing the full genome sequence of the human index virus, it exhibits 96.2% nucleotide identity to RaTG13, a bat CoV sampled previously from Rhinolophus affinis in the Yunnan Province (Zhou et al., 2020b). In addition, similar viruses were identified in Malaysian pangolins that were confiscated from illegal wildlife traders by Chinese customs (Lam et al., 2020; Xiao et al., 2020). However, pangolins are unlikely to be the intermediate host since the nucleotide identity with SARS-CoV-2 is only 85%–92%. Nonetheless, SARS-CoV-2 and pangolin viruses exhibit 97.4% amino acid identity in the viral spike protein, with just one amino acid difference in the receptor-binding motif (Xiao et al., 2020). By contrast, RatTG13 exhibits five substitutions (Zhou et al., 2020b). Another virus identified in bats, named RmYN02, has a nucleotide sequence identity of 93.3% to SARS-CoV-2. However, in some genomic regions, it is higher (96%) than in RaTG13 (Zhou et al., 2020a). The discovery of this virus may explain a unique feature of SARS-CoV-2: the insertion of four amino acids (PRRA) between the S1 and S2 subunit of the spike protein. This insertion creates a polybasic cleavage site, which can be recognized by the ubiquitous host protease furin. RmYN02 also contains an insertion of three amino acids (PAA) in this region. However, this insertion does not create a furin-cleavage site. Insertion of nucleotides in this genomic region of bat-associated viruses indicates the plasticity of the viral genome, and, in particular, at this receptor-binding viral protein (Zhou et al., 2020a). All of these viruses mentioned above, including SARS-CoV, form a distinctive Sarbecovirus clade in the phylogenetic tree for CoVs. In addition, sarbecoviruses undergo frequent recombination, and the substitutions of genetic material might explain the higher nucleotide homology between the S of the pangolin virus and SARS-CoV-2. However, there is no evidence yet to show that SARS-CoV-2 itself is a recent recombinant virus. Divergence dates between SARS-CoV-2 and other bat sarbecoviruses were estimated to be between 1948 and 1982, indicating that this lineage for SARS-CoV-2 had been circulating in bats for several decades (Boni et al., 2020). However, these estimates were calculated based on the genomic regions without recombination, and recombination events may have accelerated viral evolution to distort the divergence dating.
CoVs recently transmitted from bats to pigs
Bat-borne beta-CoVs are a significant public health threat to humans. Alpha-CoVs may also be a cause for concern, compared with just focusing on beta-CoVs. Furthermore, when the transmission affects livestock, it poses significant economic impact. Porcine epidemic diarrhea virus (PEDV) is an important example. This virus had been described in Europe in the 1970s. It reemerged in 2010–2011, affecting pig farms worldwide with substantial morbidity and mortality (He et al., 2022a). The receptor for PEDV is still under debate, but PEDV had been shown to infect cells from pigs, humans, monkeys, and bats, indicating its broad host range (Liu et al., 2015). PEDV is genetically more closely related to the bat-associated virus BtCoV/512/2005 than to TGEV, suggesting that PEDV may have originated by direct interspecies transmission from bats (Huang et al., 2013).
In 2017, China witnessed the emergence of a new virus in pig farms in the Guangdong Province, the birthplace of the SARS epidemic. This outbreaks of this novel virus resulted in death of 24,693 piglets (Pan et al., 2017; Zhou et al., 2018). The etiological agent was identified as a novel HKU2 (identified in Rhinolophus sinicus)-related bat alpha-CoV, named swine acute diarrhea syndrome (SADS) CoV (Lau et al., 2007; Zhou et al., 2018). The viral sequence is 98.48% identical to SADS-related CoV from Rhinolophus bats, a strong evidence that this new virus originated from bats (Zhou et al., 2018). Although the receptor of SADS-CoV has not been identified, SADS-CoV has a broad species tropism in vitro, including various rodent and human cell lines, indicating that alpha-CoVs might also be able to jump into humans (Edwards et al., 2020; Yang et al., 2019b). The outbreak was eventually controlled. However, this virus reappeared in February 2019, again in pig farms in southern China (Zhou et al., 2019). The frequent introduction of bat-associated viruses into pigs, plus the propensity of CoVs to undergo intra- and intertypic recombination, raises the concern that pigs may act as mixing vessels to generate new human pathogens (Wang et al., 2018a), a scenario similar to influenza virus.
Filoviruses: Ebola and Marburg virus
Filoviruses are enveloped, filamentous viruses with a negative-sense RNA genome (Kuhn et al., 2019). Two of their members, Marburg virus (MARV) and Ebola virus (EBOV), cause hemorrhagic fever with a very high case fatality rate from 25% to 90% (https://www.who.int/news-room/fact-sheets/detail/ebola-virus-disease). Filoviruses were first discovered in 1967 during a viral outbreak in the German town of Marburg, where laboratory workers were infected by monkeys that were imported from Africa by a pharmaceutical company. In 1976, the first Ebola outbreak occurred in proximate areas of Zaire (now the Democratic Republic of the Congo) and Sudan (Commission, 1978). Since then, 39 outbreaks of Ebola occurred across the entire equatorial belt of Africa, especially along riparian systems, most often in Central Africa (Baseler et al., 2017; Feldmann et al., 2020).
Based on viral RNA sequences similarity to EBOV, and coupling to serological studies, it has been shown that fruit bats, including Hypsignathus monstrosus, Epomops franqueti, Myonycteris torquata, and Rousettus aegyptiacus, are likely reservoirs of these viruses. However, a Zaire Ebola-like virus has never been isolated from bats, but similar Ebola-like viruses have been identified by nucleic acid identification (Leroy et al., 2005; Munster et al., 2018; Towner et al., 2009). By contrast, MARV was isolated from R. aegyptiacus (Towner et al., 2009), and RNA corresponding to another filovirus was detected in Rousettus bats in China (Yang et al., 2019a). The latter is phylogenetically distinct, and has low amino acid sequence identity (22%–39%) of the glycoprotein (GP) with other filoviruses, but the receptor-binding domain of the GP is relatively conserved, and hence presumably can use receptors from many species for viral infection (Yang et al., 2019a). Bats are generally asymptomatic carriers of filoviruses, but these viruses can occasionally spread to primates (gorilla, chimpanzee) and other wildlife animals such as duikers and antelope (Feldmann et al., 2020). The hunting of “bush meat” as a free protein source, or direct contact with fruit bats, have been proposed as risk factors for transmission to humans (Baudel et al., 2019; Mekibib and Arien, 2016). Secondary spread occurs by direct contact with infected patients’ blood and saliva, which triggers occasional outbreaks that might develop into epidemics (Subissi et al., 2018). However, wild animals usually represent dead-end hosts due to its severe pathology (Feldmann et al., 2020), and hence cannot sustain viral transmission.
Henipaviruses: Hendra and NiV
Henipaviruses are a distinct genus of the Paramyxoviridae. They are enveloped viruses containing a negative, single-stranded RNA genome (Murray et al., 1995). Hendra virus (HeV) was first identified in 1994 from a horse breeding farm in Australia, where it caused fatal encephalitis in several horses and humans, including a veterinarian tending the sick horses. Similar outbreaks have been reported thereafter (Murray et al., 1995). The natural hosts of HeV are pteropid bats (flying foxes), which are widely distributed in Australia as well as in tropical regions in Asia and Africa. Urine, blood, feces, nasal discharge, and saliva of flying foxes served as sources of oro-nasal infection of horses (Eaton et al., 2006). Transmission of HeV between horses occurs infrequently, and humans are infected by direct contact with horses (Khusro et al., 2020; Williamson et al., 1998). However, direct transmission from bats to humans has never been observed.
NiV, a closely related paramyxovirus, was first identified in 1998 in Malaysia in pig farm workers that had developed severe encephalitis (Chua et al., 1999, 2000; Goh et al., 2000). Serological data plus the isolation of NiV from three Pteropus bat species provided evidence that these bats are the natural virus hosts (Chua et al., 2002; Epstein et al., 2020; Reynes et al., 2005; Sendow et al., 2006; Yob et al., 2001). Outbreaks of NiV disease occur almost every year in Bangladesh, and sporadically in other regions of South Asia, besides Malaysia, in Singapore and India (Hsu et al., 2004; Yadav et al., 2019). However, NiV has a more widespread distribution, essentially in the regions where pteropid bats exist, including China and Southeast Asia (Breed et al., 2010, 2013; Enchéry and Horvat, 2017; Hasebe et al., 2012; Iehlé et al., 2007; Li et al., 2008; Reynes et al., 2005; Sendow et al., 2010; Wacharapluesadee et al., 2005). The predominant mode of human infection is by direct contact with infected pigs, where it causes a respiratory syndrome. However, recent outbreaks of NiV in Bangladesh and India were caused by direct transmission from bats to humans (i.e., by drinking contaminated palm sap), and limited human-to-human transmission has been reported (de Wit et al., 2014; Kulkarni et al., 2013; Luby et al., 2006; Weatherman et al., 2018). Phylogenetic and genetic analysis showed that NiV can be divided into two lineages: the Malaysia lineage and the Bangladesh lineage (Li et al., 2020). The initial outbreak of Malaysia lineage resulted in a mortality rate of approximate 40%, with very low incidence of human-to-human transmission. However, the mortality rate in outbreaks in Bangladesh lineages was as high as 70%, and person-to-person transmission has been observed (Chua et al., 2000; Gurley et al., 2007; Lo and Rota, 2008).
In summary, zoonotic viruses have been identified in various bat species living in the subtropical and tropical regions of the world. Modes of transmission to humans, either directly or via intermediate hosts, include direct contact with infected animals, through contaminated food, or through respiratory droplets or aerosol. However, most of the above-described viruses have not established themselves permanently in human populations. Initial outbreaks were controlled by public health measures, and because viruses did not transmit easily between humans. An exception is SARS-CoV-2, the cause of COVID-19 pandemic, which was already easily transmissible between humans soon after the initial outbreak. We will discuss this unique property below.
Receptor recognition as an important limiting step for cross-species transmission
An essential precondition for virus infection is binding to a specific receptor on the cell surface. Understanding the molecular characteristics of this interaction is helpful to infer the possible host range of a bat-associated virus and to understand the mechanism of interspecies transmission. We examine and describe the essential features of the spike proteins (in the broader sense of long surface projections) of CoVs, henipaviruses, and filoviruses, plus their respective cellular receptors using available data on their 3D structures. We compare virus binding sites in the receptors from different species, and describe possible adaptive mutations in the spike protein that facilitate cross-species transmission.
Essential features of the spike protein of CoVs
The S protein is cleaved by host proteases into the receptor-binding S1 subunit, and S2, which mediates membrane fusion. Proteolytic cleavage of S usually occurs at two sites (S1/S2 and S2′), and it is required to prime the protein to execute its fusion activity. Cleavage can be performed by members of the host serine transmembrane protease family (mainly TMPRSS2), or an endosomal cathepsin. S proteins having a polybasic cleavage site are processed by the ubiquitous host enzyme furin. The presence of these enzymes also determines the cell tropism of CoVs. The S1 subunit is divided into the N-terminal domain, which is usually responsible for binding carbohydrate residues, and a C-terminal domain, which binds to receptor. The receptor-binding domain (RBD) contains two subdomains: a conserved core structure and a variable receptor-binding motif (RBM). The RBM determines the receptor-binding specificity: ACE2 in the case of SARS-CoV, and DPP4 for MERS-CoV. The diversity of receptor binding is an outstanding feature of CoVs, and it might have been instrumental to increase their host range during evolution (Li, 2016; Millet and Whittaker, 2015).
SARS-CoV interaction with the ACE2 receptor
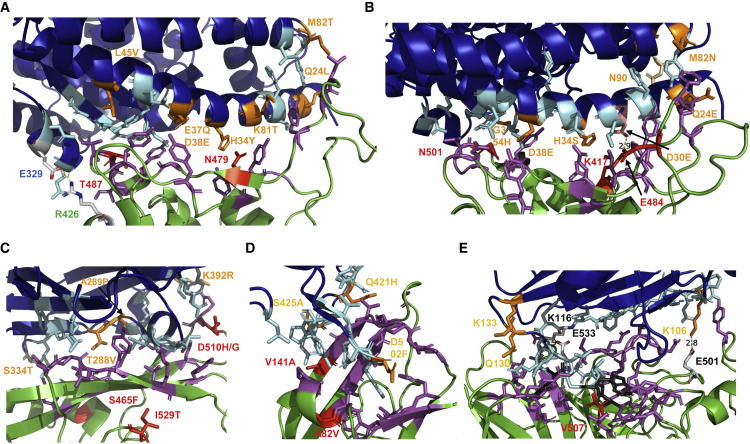
The RBD of S contains two subdomains, a core and an extended loop, which presents a concave surface (parts of its main chain are shown as green cartoon in the lower part of Figure 3A) that binds to an N-terminal helix of ACE2 (shown as blue cartoon in the upper part of Figure 3A). The RBM contains 16 amino acids (their side chains are highlighted as sticks, either magenta or red) that have direct contact with 20 amino acids in ACE2 (side chains are shown as light blue or orange sticks) (Lan et al., 2020; Li et al., 2005a).
Figure 3.
Interaction of SARS-CoV, SARS-CoV-2, MERS-CoV, EBOV, and NiV with their receptors
(A) Interaction surface between ACE2 and S of SARS-CoV: ACE2 is colored blue, S in green. The contacting amino acids are shown as cyan (ACE2) or magenta (S) sticks. The seven amino acids substituted in civet ACE2 are labeled in orange. In ACE2 of mice, M82 is substituted to N, thereby creating an N-glycosylation site. The two amino acid substitutions during adaptation of SARS-CoV from civets to humans (N479) and from humans to humans (T487) are shown in red. Glu329 in S and Arg426 in ACE2 form a salt bridge at the periphery of the interaction surface. This figure was generated with PyMol 2.1.1. using the pdb file 2AJF. The contacting amino acids shown here are from Lan et al. (2020), which differ in four peripheral residues from the first publication Li et al. (2005a).
(B) Interaction surface between ACE2 and S of SARS-CoV-2: ACE2 is colored blue, S in green. The contacting amino acids are shown as cyan (ACE2) or magenta (S) sticks. The seven amino acids substituted in pangolin ACE2 are labeled in orange. Lys417 in S and Glu30 in ACE2 form a salt bridge in the middle of the interaction surface. Lys417 (labeled red) is substituted by a Thr in variant P1 and by an Asn in variant B 1.351. Highlighted as a red stick is also N501, which is substituted by Tyr in variants B1.1.1, B1.351. and P1. The figure was generated with PyMol 2.1.1. using the pdb file 6M0J.
(C) Interaction surface between human DPP4 and S of MERS-CoV: DPP4 is colored blue, S in green. The contacting amino acids are shown as cyan (DPP4) or magenta (S) sticks. The four amino acids substituted in camel DPP4 are labeled in orange. Residue 334 is not directly contacting the spike but is an N-glycosylation site in mice DPP4, which needs to be removed to make mice susceptible to MERS-CoV infection. The two amino acid substitutions (S465F, D510H) during adaptation of S to a suboptimal bat-associated virus receptor and during MERS outbreak in South Korea (I529T, D510G) are shown in red. The figure was generated with PyMol 2.1.1. using the pdb file 4KRO.
(D) Interaction surface between human NPC1 and GP of EBOV. NPC1 is colored blue, GP in green. The contacting amino acids are shown as cyan (NPC1) or magenta (GP) sticks. The three amino acids substituted in pig NPC1 are labeled in orange. Amino acid substitutions restoring EBOV binding to refractory receptors (V141A) and during the large Ebola epidemic (A82V) are labeled red. The figure was generated with PyMol 2.1.1. using the pdb file 5F1B.
(E) Interaction surface between human ephrin-b2 and G of Nipah virus. Ephrin-B2 is colored blue, G in green. Most of the contacting amino acids are shown as sticks. Three hydrophobic amino acids (F120, L124, W125) in the loop of ephrin-B2 essential for binding are colored in gray. The two salt bridges (K106 with E501, K116 with E533) are indicated. The only amino acid substitution between G of Nipah and Hendra virus is shown in red. Receptor amino acids near the binding site variable between species (K106, Q130, K133) are labeled in orange. The figure was generated with PyMol 2.1.1. using the pdb file 2VSM.
Although SARS-CoV can infect civets, the interaction surface of its ACE2 receptor contains seven amino acid substitutions (shown as orange sticks, the position is indicated) relative to human ACE2 demonstrating the plasticity of the spike protein in receptor usage (Table S1). ACE2 of rodents, which cannot (rat) or can only inefficiently (mice) be infected by SARS-CoV, contains nine and seven substitutions, respectively. The substitution that prevents infection is an N-glycosylation site at position 82 in rat ACE2 (Lu et al., 2015). SARS-CoV experimentally adapted to replicate in mice had a Tyr to His substitution at position 436 (Roberts et al., 2007). The N-terminal helix of ACE2 is also an attachment site for bat-associated CoVs, but it exhibits amino acid variation among different bat species. Even bats from the species R. sinicus, sampled from three Chinese provinces, exhibit up to eight amino acid variations in the S protein contact region. Their overall amino acid identity, however, is very high (99%) (Ge et al., 2013; Hou et al., 2010). The ACE2 variants support SARS-CoV infection and infection with bat SARS-like viruses, but with different binding affinities to different S proteins. Molecular evolution analysis indicated that the key residues at the binding site are under positive selection, suggesting that the S protein of bat-associated viruses and ACE2 may have co-evolved over a long time and adapted to selection pressure from each other (Guo et al., 2020).
Despite this receptor usage being flexible, the S protein of the ancestral lineages of SARS-CoV had also acquired mutations in the RBM for cross-species transmission (Cui et al., 2019). The mutation Lys479Asn in the RBM has played an important role in the civet-to-human jump, whereas the mutation Ser487Thr facilitated human-to-human transmission, and both are indicated as red sticks in Figure 3A (Cui et al., 2019; Li, 2013). However, some bat-associated viruses recognize human ACE2, although they exhibit several changes in the amino acids contacting the receptor compared with SARS-CoV. This suggests that they can infect humans directly without using an intermediate host (Ge et al., 2013; Menachery et al., 2015).
SARS-CoV-2 interaction with the ACE2 receptor
SARS-CoV-2 and SARS-CoV share the same receptor, and most of the residues in ACE2 involved in binding to S are identical (Figure 3B). Some contact points are unique, especially the location of a salt bridge, which is located in a central position in the SARS-CoV-2 ACE2 complex, suggesting that CoVs can adapt by multiple pathways to bind human ACE2 (Lan et al., 2020; Shang et al., 2020). From the approximately 60 mammalian ACE2 proteins tested, only five did not support virus entry if transfected into cells, namely ACE2 from mice, koala, and three New World monkeys (Liu et al., 2021). This broad host tropism, at least in cell culture, is not due to conservation of the amino acids in ACE2 contacting the viral spike. Up to seven (out of 20) amino acids can be substituted in ACE2 from civets and pangolin, which can still serve as virus receptor. ACE2 proteins that do not function as receptors contain more substitutions: eight in mice, nine in koala, or ten in chicken (Liu et al., 2021). An exception is ACE2 from New World monkeys, which contain only four substitutions, but at residues that are rarely substituted in other ACE2 proteins (Table S2). In accordance with the plasticity in receptor binding of SARS-CoV-2, many mammals can be infected, either by human COVID-19 patients (cats, tigers, minks, dogs) and/or experimentally (rhesus and cynomolgus macaques, African green monkeys, Syrian and dwarf hamsters, ferrets, tree shrews, raccoon dogs, and cattle), whereas chickens and ducks are apparently resistant (Freuling et al., 2020; Munoz-Fontela et al., 2020; Shi et al., 2020; Trimpert et al., 2020; Ulrich et al., 2020; Zhao et al., 2020). Even the fruit bat R. aegyptiacus is susceptible to intranasal inoculation, and it transmits the virus to contact animals, despite R. aegyptiacus not being the original reservoir species for SARS-CoV-2 (Schlottau et al., 2020). Pigs are unique in this regard, since they are not susceptible to infection, although their ACE2 can serve as SARS-CoV-2 receptor (Munoz-Fontela et al., 2020; Shi et al., 2020; Zhai et al., 2020).
Interestingly, the S proteins of SARS-CoV-2 and human ACE2 are not a perfect match. Deep mutational scanning of human ACE2 revealed 122 substitutions at 35 positions that enhance binding to the viral spike, including substitutions of Thr27 and a substitution of Lys31 by aromatic residues, and these substitutions do not occur in mammalian ACE2 protein. All substitutions that cause a deletion of the N-glycosylation site at position 90 are favorable for RBM binding (Chan et al., 2020). However, the N-glycosylation motif Asn90XSer/Thr is present in ACE2 proteins of many susceptible species, including humans, but is absent in the ACE2 proteins of the species that do not serve as SARS-CoV-2 receptors (see Table S2).
A similar deep mutational scanning approach with the RBD of the SARS-CoV-2 S protein (3,800 mutations; i.e., almost every residue was substituted by any of the other amino acids) revealed that many substitutions are tolerated, and that some mutations even enhance binding (Starr et al., 2020). However, initial isolates of SARS-CoV-2 do not contain mutations in the RBM, suggesting that SARS-CoV-2 already possessed adequate affinity for human ACE2 at the beginning of the pandemic. Nevertheless, SARS-CoV-2 can adapt to a new receptor. Mice became susceptible to SARS-CoV-2 after serial virus passage by an Asn501Tyr mutation, or by mutations Gln498Tyr and Pro499Thr in the RBD of its S protein (Dinnon et al., 2020; Gu et al., 2020).
However, it should be noted that a more efficient binding to the receptor does not necessarily result in improved virus replication. After completing the replication cycle, virus particles must be released from infected cells to initiate infection of new cells. However, it is detrimental if the receptor-binding activity is too strong, as it prevents release of viruses from infected cells. An example is influenza virus, which possesses both a receptor-binding viral protein (hemagglutinin [HA]) and a receptor-destroying protein (neuraminidase [NA]). An optimal balance of both activities is required for efficient virus replication (Wagner et al., 2002). In fact, oseltamivir, an antiviral drug that blocks the enzymatic activity of NA, inhibits virus release, and hence reduces disease severity.
A mutation occurring in the S protein early during the pandemic was Asp614Gly, which emerged in early March 2020. Viruses harboring this mutation had rapidly become the dominant form (Korber et al., 2020). The substituted residue is located in the S1 subunit at the interface between two monomers of the trimeric S protein. The carboxyl group of Asp614 forms a hydrogen bond with the OH group of Thr859 present in the other monomer, which is no longer available if substituted by a glycine residue (Figure S1). Functional studies had shown that this mutation enhances virus replication in cell culture and in the upper, but not in the lower, respiratory tract in animal models and in COVID-19 patients. This preference for the upper respiratory epithelium probably results in higher transmissibility. However, the mutation does not affect disease severity and does not compromise the neutralizing activity of antibodies elicited by D614 spike-based vaccines or convalescent plasma (Hou et al., 2020; Plante et al., 2020; Weissman et al., 2021; Yurkovetskiy et al., 2020).
In December 2020, new SARS-CoV-2 variants were independently detected in the United Kingdom. These variants have several mutations in the S protein and in other viral genes (Faria et al., 2021; Tegally et al., 2021; Volz et al., 2021). Most interesting are the three mutations in the RBD (red sticks in Figure 3B); one of them, Asn501Tyr, is present in all three variants, arguing in favor of convergent evolution. Variants B1.351 (now termed Beta) and P.1 (now termed Gamma) contain two additional substitutions in the RBD, residue Lys417 is a Thr in P.1 and an Asn in B1.351, and the negatively charged Glu484 is substituted by a positively charged Lys in both variants. However, residue 484 is not directly involved in the binding to ACE2. Besides the mutations in the RBD, five to eight additional substitutions are present in the S protein of these variants, such as short deletions in the N-terminal domain (Figure S1) and substitution of Pro681His in B.1.1.7 (now termed Alpha), which alters the furin-cleavage site from PRRAR to HRRAR. Epidemiological, immunological, and functional studies with these variants are ongoing. B.1.1.7. might be more transmissible by its rapid increase in frequency in every country where it was introduced (Volz et al., 2021). B1.351 and P1 exhibit high apparent prevalence rate of 40%–50% in South Africa and in Brazil, respectively, but these variants are much less abundant in other countries (https://outbreak.info/situation-reports). Why this unusually large number of genetic changes, particularly in the S protein, occur at the same time but apparently at different locations remains a mystery. Rapid virus evolution within a chronically infected individual, particularly immunocompromised, might contribute to this phenomenon (Clark et al., 2021).
Recently, new SARS-CoV-2 variants have emerged and became predominant, especially Delta (B.1.617.2) and subsequently Omicron (B.1.1.529). Their properties and substitutions in the S proteins have been reviewed recently (Ghosh et al., 2022; Jung et al., 2022; McLean et al., 2022). See also https://outbreak.info/situation-reports for the always up-to-date tracking of SARS-CoV-2 variant prevalence and https://jbloomlab.github.io/SARS-CoV-2-RBD_DMS_variants/RBD-heatmaps/ for the mutational effects in variant RBDs on ACE2-binding affinity.
MERS-CoV interaction with the DPP4 receptor
The RBD of MERS-CoV, similar to SARS-CoV and SARS-CoV-2, is divided into a core domain with identical folding in all CoVs, and an external subdomain containing the RBM. The latter is markedly different, leading to recognition of the DPP4 receptor instead of ACE2. The contact to DPP4 is mainly mediated by hydrophilic side chain interactions (Figure 3C) (Lu et al., 2013). DPP4 from mammalian cell lines susceptible to MERS-CoV infection, e.g., from camel and pig, contain four amino acid substitutions relative to human DPP4, whereas DPP4 from non-permissive cell lines (hamster and mice) contain five and six substitutions, respectively (Table S3) (Lu et al., 2015). Genomic substitutions of two amino acids in the mouse DPP4 receptor (Asn294Leu, Thr336Arg; numbering according to the human DPP4, the substitution Thr336Arg removes the N-glycosylation site at position 334 from mouse DPP4) makes mice susceptible to MERS-CoV infection (Cockrell et al., 2014, 2016; Peck et al., 2015).
DPP4 receptors from bats differ substantially in the amino acids that have direct contacting interaction with the S protein, and evolutionary analysis shows a pattern of “long-term arms race” between bats and MERS-related CoVs (Cui et al., 2013). A detailed analysis of bat receptors from different species revealed that four of them conferred better virus replication—higher titer—relative to using human DPP4, suggesting that MERS-CoV is not optimally adapted to human DPP4 (Letko et al., 2018). DPP4 proteins from seven bat species are equally or slightly less effective, and four barely support virus replication. Almost every animal DPP4 protein contains substitutions at residues 288 and 392 (which are also substituted in camel and in pig DPP4). DPP4 proteins that do not confer MERS-CoV susceptibility contain one or more substitutions at various positions. The least effective receptor was from Desmodus rotundi, which contains two substitutions, Ile295Thr (which is also a substitution in DPP4 form hamster and mice) and a unique substitution Arg317Gln. Interestingly, MERS-CoV can be adapted to this suboptimal DPP4 receptor by acquiring two mutations in the RBD, Ser465Phe and Asp510His (Letko et al., 2018).
A MERS-like CoV isolated from Pipistrellus bats in Uganda exhibits 86% amino acid identity to MERS-CoV across the full genome but has only 46% identity in the S protein. This difference is probably due to a recombination and this inhibits attachment to the DPP4 receptor (Anthony et al., 2017). Recombination involving the S protein also occurred among co-circulating MERS-CoV lineages in camels, and this might have led to the MERS outbreak in South Korea in 2015 (Sabir et al., 2016). During that outbreak, most sequences obtained from patients in South Korea contained mutations in the RBD, mostly Ile529Thr, but also in Asp510Gly. Contrary to what was expected, the mutations decreased binding to the receptor and reduced viral entry, suggesting that these substitutions were driven by neutralizing antibodies (Kim et al., 2016).
EBOV and MARV interaction with the NPC1 receptor
The GP mediates all stages of virus entry, attachment, entry, and fusion. It is synthesized as a precursor that is cleaved by furin-like proteases into subunits GP1 and GP2. Priming occurs through cathepsin-mediated cleavage upon uptake of the virus into endosomes, which removes the largest part of GP1, including a mucin-like domain that mediates the initial virus attachment to the cell surface receptors (Davey et al., 2017). Only the cleaved GP (termed GPcl) can bind to the Niemann-Pick C1 (NPC1) receptor, which is a ubiquitous polytypic membrane protein that regulates cholesterol trafficking (Davey et al., 2017; Kirchdoerfer et al., 2017).
The second loop of NPC1, which is composed of seven α helices and seven β strands, binds to a hydrophobic groove at the head of GPcl of EBOV via two protruding loops containing mainly aromatic residues (Figure 3D). Mutagenesis and affinity measurements revealed that five residues (Tyr423, Pro424, Phe503, Phe504, and Tyr506) contributed to most of the interactions (Wang et al., 2016). However, the affinity of GPcI for its receptor is exceptionally low (150 μM), about four orders of magnitude lower than determined for other spike-receptor complexes (Wang et al., 2016). All contacting residues are conserved in NPC1 of primates, including various monkeys, in ferrets, and in dogs. There are three substitutions in porcine NPC1 (Gln421His, Ser425Ala, and D502Phe), but pigs are still susceptible to EBOV infection (Table S4) (Weingartl et al., 2013).
There is a large diversity in the amino acids in NPC1 in contact with the viral spike protein among bat species. R. aegyptiacus, which is susceptible to EBOV infection, contains the same substitutions (Ser425Ala) as the pig receptor. NPC1 from Pteropus dasymallus serves as receptor for EBOV, although it contains three amino acid substitutions (Ser425Thr, Gly426Glu, Phe504Tyr). Cells from Eidolon helvum are non-permissive to EBOV infection and they contain three different amino acid substitutions, Ser425Ala, Val505Thr, and Asp502Phe, of which residue 502 is critical (Ng et al., 2015). Since MARV infects cells derived from E. helvum, but not from P. dasymallus (which is opposite to the infectivity of EBOV), it was suggested that the heterogeneity of NPC1 affects filovirus tropism (Takadate et al., 2020). Indeed, bioinformatics evidence had shown that residue 502 was a positive selected site in bats (Ng et al., 2015). Likewise, in many mammals, virus-interacting residues in NPC1 are under positive selection (Kondoh et al., 2018; Pontremoli et al., 2016). However, deficiencies in virus binding are counteracted by mutations in the viral GP. Binding to a receptor having a Phe at residue 502 can be restored by a mutation at Val141Ala (labeled red) in GP, a substitution that occurs in certain filoviruses (Ng et al., 2015).
During the large Ebola epidemic in West Africa, a mutation in GP (Ala82Val near the RBD, labeled red) arose early and dominated the viral population. Three studies had reported a fitness advantage of 82Val over 82Ala in cell culture (Diehl et al., 2016; Dietzel et al., 2017; Urbanowicz et al., 2016), and that this site was positively selected (Ladner et al., 2015). However, it is difficult to distinguish between virus selections within or among infected patients. In the first case, a faster-replicating virus would out-compete the wild type and is therefore preferentially transmitted. In the second case, mutant virus is more transmissible and thus causes more secondary infections. In addition, stochastic processes, for example, founder effects, might also contribute to the enrichment of certain virus variants (Bedford and Malik, 2016).
HeV and NiV interaction with ephrin receptors
Cell entry of henipaviruses involves the concerted action of its two GPs, the receptor-binding G protein and the fusion protein F. Upon receptor binding, G triggers F to induce a conformational change to catalyze membrane fusion, either by releasing previously bound or by newly associating with the F protein (Azarm and Lee, 2020). Another prerequisite for fusion is the cleavage of F into two subunits by cathepsins, which occurs after endocytosis of F from the plasma membrane in the virus-producing cell (Diederich and Maisner, 2007).
The functional receptors of both viruses are ephrin-B2 and ephrin-B3, members of a large family of membrane-bound tyrosine kinases (Pernet et al., 2012; Xu et al., 2012). Both proteins insert a protruding loop into the central cavity of the head domain of the viral G protein, which is folded into a β propeller with six blades (Figure 3E). G protein from other paramyxoviruses exhibits a similar folding of their head domain, despite them using carbohydrates as receptor (Cox and Plemper, 2017). NiV-G and HeV-G are extremely similar in structure and hence use almost the same amino acid residues for attachment; only valine 507 is substituted to threonine in HeV-G (Bowden et al., 2008; Pernet et al., 2012; Xu et al., 2012). The receptor interaction is of very high affinity (1 nm) due to the large interface containing 24 hydrogen bonds, plus two salt bridges and several key hydrophobic interactions (Phe 120, Leu124, Trp125) in the protruding loop. The amino acids in this loop from both ephrin-B2 and ephrin-B3 are identical in bats, humans, intermediate hosts, and in other mammals and even in chicken. Therefore, many mammalian receptors can mediate cell entry of both NiV and HeV with similar efficiency, and many small animals can be experimentally infected (Bossart et al., 2008). An exception is mice, which are resistant, although their receptor confers susceptibility in cell culture, indicating that virus replication is blocked at a later step (Thibault et al., 2017). For ephrin-B2, mostly conservative substitutions occur outside the protruding loop (106, 130, 133), especially between some bat species (Table S5). Therefore, since there is no variation between animals in the amino acids that contact the viral spike proteins, strong adaptation to a different receptor is not required for the host jump. However, by analyzing all available NiV sequences (including clinical isolates), we found seven adaptive sites in the G protein, three of which are located close to the receptor-binding region, and they may modulate the binding (Li et al., 2020).
In summary, bat-derived viruses face different conditions in the host receptors to enter cells. The amino acids in ephrin-B2 and ephrin-B3 interacting with the G protein of henipaviruses exhibit no sequence variation among humans, bats, and intermediate hosts. Thus, bat-associated viruses can easily cross the species barrier as far as receptor attachment is concerned. By contrast, the receptors used by filoviruses and CoVs show variability at the interacting surface among different species. EBOV and MERS-CoV can tolerate up to three substitutions, whereas the S protein of SARS-CoV and of SARS-CoV-2 is even more flexible, as it can bind to receptors having up to seven substitutions. The contacting amino acid residues in the receptors used by filoviruses and CoVs show large variation among (and even within) bat species, and there is evidence for an evolutionary arms race between bats and their viruses (Guo et al., 2020; Hu et al., 2017; Ng et al., 2015; Takadate et al., 2020).
Potential of interspecies transmissions of bat RVs to mammalian hosts
RV infections are a major cause of life-threatening gastroenteritis in infants and children, and the young of many mammalian and avian species worldwide (Crawford et al., 2017). RVs are non-enveloped double-stranded RNA viruses with a genome of 11 segments. According to the difference of VP6 amino acid sequence, RVs are currently grouped into nine species (RVs A–D and RVs F–J), among which RVA is the most common and pathogenic (https://talk.ictvonline.org/taxonomy/). Based on nucleotide sequence identities in VP7 and VP4 genes, RVA is further classified into G and P genotypes, respectively. The P[8], P[4], and P[6] genotypes in the P[II] genogroup are responsible for over 90% of human infections worldwide, and are considered to have originated from P [I] RVs with an animal host origin and evolved the ability to infect humans (Xu et al., 2021). Currently, RVs have been reported from a variety of bat species belonging to Rhinolophidae, Vespertilionidae, Hipposideridae, Pteropodidae, Emballonuridae, and Rhinopomatidae (Esona et al., 2010; He et al., 2013, 2017; Mishra et al., 2019; Waruhiu et al., 2017; Xia et al., 2014).
Some RVA isolates may be interspecies transmission of bat RVA strains. A child with acute gastrointestinal in Thailand was infected with RVA of genotype G3P[10], bearing bat-like VP7 and VP4 proteins (Jampanil et al., 2021). Okitsu et al. (2018) reported that infection with a reassortment belonging to G3P[13] involving a bat RVA strain leads to severe gastrointestinal symptoms in a child. Recent studies have shown that human RVA strain G20 also exists in bats, which suggests a potential bat reservoir (Esona et al., 2018; Solberg et al., 2009). In addition, the rotaviruses of some livestock may also be related to bats. An equine G3P[3] RVA strain has high nucleotide similarities with a bat-derived rotavirus, indicating that it originated from bats (He et al., 2017; Mino et al., 2013; Xia et al., 2014). Recently, some purple SA11 (simian RVA)-like RVs were found in multiple bat individuals in Gabon, suggesting that bats are the prime suspect for being the major hosts of these viruses (Simsek et al., 2021). In fact, the genome structure of rotavirus determines its characteristics for easy reassortment between human, animal, and bat viruses (Esona et al., 2010). Our previous studies revealed that RV shows high prevalence and signatures of cross-species transmission in some game animals, indicating that its host range is much wider than we know (He et al., 2022b). The close evolutionary links between human and animal RVs, especially porcine RVs, suggest that they may have a common ancestor, which allows interspecies transmission and establishment in human populations and animals, emphasizing the need for simultaneous monitoring of rotaviruses in animals.
Host factors or cellular receptors that mediate RVs’ successful infection are also diverse. Some host cell surface molecules, such as heat shock cognate protein 70 and integrins, have been identified as potential RV receptors (Lopez and Arias, 2004). Previous studies have shown that some RVs recognize sialic acid-containing glycoconjugates and are NA sensitive, but the majority of human and animal RVs are sialidase insensitive (Banda et al., 2009; Ciarlet et al., 2002). Li et al. (2021) reported that the single amino acid mutation in the VP8∗ protein of bat P[3] genotype rotavirus obtained the ability to hemagglutinate the red blood cells, and this residue played an important role in the ligand recognition and may contribute to the cross-species transmission. However, other specific glycans, such as mucin and histo-blood group antigens (HBGAs), can interact with VP8∗ and affect RVs tropism in different hosts (Xu et al., 2021). Thus, the role of species-specific glycans in the cross-species transmission need to be further determined, which is of great significance for determining host range and epidemiology, species barrier mechanism, and cross-species transmission.
Outlook
Given the large variety of viruses in bats and their propensity to cross the species barrier, the question regarding which measures should be taken to prevent the emergence of novel virus to humans needs to be addressed with urgency, as it can become a major public health crisis, as exemplified by the COVID-19 pandemic. Since bats and other wildlife animal species serve as food sources, banning their hunting, trading, and consumption should be considered. Another measure is to close wet markets, where numerous living wildlife and farm animals are clustered and traded. In addition, based on the concept of “One Health,” wildlife protection and epidemiological research on bat-associated viruses should be conducted. Having identified viruses in a rural region with the potential to spill over, surveillance of the local population, including their livestock, would reveal whether these viruses have the potential to emerge as human pathogens, hence stopping an emerging pandemic at the source before it spreads to larger cities with larger population. Furthermore, only a small fraction of the 72,000 viruses estimated to occur in bats has been identified or studied. A clever, but costly, strategy of how to identify a majority of them has been outlined with the aim to predict viruses that represent a public health threat (Anthony et al., 2013). However, based on the inherent flexibility of the viral S protein, it is difficult to predict whether a new CoV will bind to the human receptor. In addition, the identified virus population represents only a transient snapshot, as viruses evolve rapidly by mutations and by recombination. Furthermore, even if potentially harmful viruses have been identified, it is hard to predict when a virus outbreak will occur. There are so many parameters to consider.
Another pandemic preparedness strategy is to develop drugs against conserved viral targets. One candidate is the RNA-dependent RNA polymerase, which shares structural similarity among all RNA viruses. Indeed, remdesivir was originally developed against EBOV. It had been shown to have positive effect for COVID-19 treatment (Beigel et al., 2020). Since there is little commercial incentive to develop drugs against viruses that might never emerge, public and private partnership is required to generate prototype drugs. They can be tested for pharmacological properties and for safety in animal models, and perhaps in small groups of humans. Once a new virus has emerged, these drugs could be rapidly tested in phase III clinical trials, and, if required, the compound can be modified once the 3D-structure of its target—viral receptor protein—becomes available. Other potential targets are the cellular enzymes essential for virus replication; i.e., protein kinases and palmitoyl transferases (Gadalla and Veit, 2020; Lan et al., 2020; Totura and Bavari, 2019). If the same enzyme is used by several or groups of viruses, such drugs might even be effective against outbreaks caused by members of different virus families. In summary, a combination of the above suggested measures might be helpful in preventing future virus outbreaks, or at least mitigate their effect on human health with positive impact to global economy. It will be a small investment in comparison.
Acknowledgments
This work was financially supported by the Bioinformatics Center of Nanjing Agricultural University, the Fundamental Research Funds for the Central Universities Y0201900459, the National Key Research and Development Program of China (grant no. 2021YFD1801105), and the National Natural Science Foundation of China (grant no. 31770172). Experimental work in the lab of M.V. is funded by the German Research Foundation (DFG).
Author contributions
S.S., M.V., J.T., and J.S. are co-senior authors of this paper. Conceptualization, S.S., M.V., and J.T.; methodology, M.V., J.S., and L.W.; validation, J.T. and J.S.; formal analysis, J.T., J.S., and M.V.; investigation, J.T., J.S., and M.V.; writing – original draft, J.S. and M.V.; writing – review & editing, J.T., J.S., S.S., M.V., D.L., N.W., L.W., C.Z., X.M., X.J., M.A.S., X.Z., and A.L.; supervision, S.S., M.V., and J.T.; funding acquisition, S.S., J.T., and M.V.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.110969.
Supplemental information
References
- Adney D.R., van Doremalen N., Brown V.R., Bushmaker T., Scott D., de Wit E., Bowen R.A., Munster V.J. Replication and shedding of MERS-CoV in upper respiratory tract of inoculated dromedary camels. Emerg. Infect. Dis. 2014;20:1999–2005. doi: 10.3201/eid2012.141280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amendola A., Bianchi S., Gori M., Colzani D., Canuti M., Borghi E., Raviglione M.C., Zuccotti G.V., Tanzi E. Evidence of SARS-CoV-2 RNA in an oropharyngeal swab specimen, milan, Italy, early December 2019. Emerg. Infect. Dis. 2021;27:648–650. doi: 10.3201/eid2702.204632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annan A., Baldwin H.J., Corman V.M., Klose S.M., Owusu M., Nkrumah E.E., Badu E.K., Anti P., Agbenyega O., Meyer B., et al. Human betacoronavirus 2c EMC/2012-related viruses in bats, Ghana and Europe. Emerg. Infect. Dis. 2013;19:456–459. doi: 10.3201/eid1903.121503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony S.J., Epstein J.H., Murray K.A., Navarrete-Macias I., Zambrana-Torrelio C.M., Solovyov A., Ojeda-Flores R., Arrigo N.C., Islam A., Ali Khan S., et al. A strategy to estimate unknown viral diversity in mammals. mBio. 2013;4:e00598-13. doi: 10.1128/mbio.00598-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony S.J., Gilardi K., Menachery V.D., Goldstein T., Ssebide B., Mbabazi R., Navarrete-Macias I., Liang E., Wells H., Hicks A., et al. Further evidence for bats as the evolutionary source of middle east respiratory syndrome coronavirus. mBio. 2017;8:e00373-17. doi: 10.1128/mBio.00373-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arankalle V.A., Bandyopadhyay B.T., Ramdasi A.Y., Jadi R., Patil D.R., Rahman M., Majumdar M., Banerjee P.S., Hati A.K., Goswami R.P., et al. Genomic characterization of Nipah virus, West Bengal, India. Emerg. Infect. Dis. 2011;17:907–909. doi: 10.3201/eid1705.100968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azarm K.D., Lee B. Differential features of fusion activation within the Paramyxoviridae. Viruses. 2020;12:161. doi: 10.3390/v12020161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azhar E.I., El-Kafrawy S.A., Farraj S.A., Hassan A.M., Al-Saeed M.S., Hashem A.M., Madani T.A. Evidence for camel-to-human transmission of MERS coronavirus. N. Engl. J. Med. 2014;370:2499–2505. doi: 10.1056/nejmoa1401505. [DOI] [PubMed] [Google Scholar]
- Baldwin J.W., Dechmann D.K.N., Thies W., Whitehead S.R. Defensive fruit metabolites obstruct seed dispersal by altering bat behavior and physiology at multiple temporal scales. Ecology. 2020;101:e02937. doi: 10.1002/ecy.2937. [DOI] [PubMed] [Google Scholar]
- Banda K., Kang G., Varki A. 'Sialidase sensitivity' of rotaviruses revisited. Nat. Chem. Biol. 2009;5:71–72. doi: 10.1038/nchembio0209-71. [DOI] [PubMed] [Google Scholar]
- Basavaraju S.V., Patton M.E., Grimm K., Rasheed M.A.U., Lester S., Mills L., Stumpf M., Freeman B., Tamin A., Harcourt J., et al. Serologic testing of US blood donations to identify severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)-reactive antibodies: December 2019-January 2020. Clin. Infect. Dis. 2021;72:e1004–e1009. doi: 10.1093/cid/ciaa1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baseler L., Chertow D.S., Johnson K.M., Feldmann H., Morens D.M. The pathogenesis of Ebola virus disease. Annu. Rev. Pathol. 2017;12:387–418. doi: 10.1146/annurev-pathol-052016-100506. [DOI] [PubMed] [Google Scholar]
- Baudel H., De Nys H., Mpoudi Ngole E., Peeters M., Desclaux A. Understanding Ebola virus and other zoonotic transmission risks through human-bat contacts: exploratory study on knowledge, attitudes and practices in Southern Cameroon. Zoonoses Public Health. 2019;66:288–295. doi: 10.1111/zph.12563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford T., Malik H.S. Did a single amino acid change make Ebola virus more virulent? Cell. 2016;167:892–894. doi: 10.1016/j.cell.2016.10.032. [DOI] [PubMed] [Google Scholar]
- McCaw Z.R., Kim D.H., Wei L.J., Mehta A.K., Zingman B.S., Kalil A.C., Hohmann E., Chu H.Y., Luetkemeyer A., Kline S., et al. Remdesivir for the treatment of Covid-19 - preliminary report. N. Engl. J. Med. 2020;383:993. doi: 10.1056/NEJMc2022236. [DOI] [PubMed] [Google Scholar]
- Blakey R.V., Webb E.B., Kesler D.C., Siegel R.B., Corcoran D., Johnson M. Bats in a changing landscape: linking occupancy and traits of a diverse montane bat community to fire regime. Ecol. Evol. 2019;9:5324–5337. doi: 10.1002/ece3.5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boni M.F., Lemey P., Jiang X., Lam T.T.Y., Perry B.W., Castoe T.A., Rambaut A., Robertson D.L. Evolutionary origins of the SARS-CoV-2 sarbecovirus lineage responsible for the COVID-19 pandemic. Nat Microbiol. 2020;5:1408–1417. doi: 10.1038/s41564-020-0771-4. [DOI] [PubMed] [Google Scholar]
- Bossart K.N., Tachedjian M., McEachern J.A., Crameri G., Zhu Z., Dimitrov D.S., Broder C.C., Wang L.F. Functional studies of host-specific ephrin-B ligands as Henipavirus receptors. Virology. 2008;372:357–371. doi: 10.1016/j.virol.2007.11.011. [DOI] [PubMed] [Google Scholar]
- Bowden T.A., Aricescu A.R., Gilbert R.J.C., Grimes J.M., Jones E.Y., Stuart D.I. Structural basis of Nipah and Hendra virus attachment to their cell-surface receptor ephrin-B2. Nat. Struct. Mol. Biol. 2008;15:567–572. doi: 10.1038/nsmb.1435. [DOI] [PubMed] [Google Scholar]
- Brearley G., Rhodes J., Bradley A., Baxter G., Seabrook L., Lunney D., Liu Y., McAlpine C. Wildlife disease prevalence in human-modified landscapes. Biol Rev Camb Philos Soc. 2013;88:427–442. doi: 10.1111/brv.12009. [DOI] [PubMed] [Google Scholar]
- Breed A.C., Meers J., Sendow I., Bossart K.N., Barr J.A., Smith I., Wacharapluesadee S., Wang L., Field H.E. The distribution of henipaviruses in Southeast Asia and Australasia: is Wallace's line a barrier to Nipah virus? PLoS One. 2013;8:e61316. doi: 10.1371/journal.pone.0061316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breed A.C., Yu M., Barr J.A., Crameri G., Thalmann C.M., Wang L.F. Prevalence of henipavirus and rubulavirus antibodies in pteropid bats, Papua New Guinea. Emerg. Infect. Dis. 2010;16:1997–1999. doi: 10.3201/eid1612.100879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brian D.A., Baric R.S. Coronavirus genome structure and replication. Curr. Top. Microbiol. Immunol. 2005;287:1–30. doi: 10.1007/3-540-26765-4_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calisher C.H., Childs J.E., Field H.E., Holmes K.V., Schountz T. Bats: important reservoir hosts of emerging viruses. Clin. Microbiol. Rev. 2006;19:531–545. doi: 10.1128/cmr.00017-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K.K., Dorosky D., Sharma P., Abbasi S.A., Dye J.M., Kranz D.M., Herbert A.S., Procko E. Engineering human ACE2 to optimize binding to the spike protein of SARS coronavirus 2. Science. 2020;369:1261–1265. doi: 10.1126/science.abc0870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Liu B., Yang J., Jin Q. DBatVir: the database of bat-associated viruses. Database. 2014;2014:bau021. doi: 10.1093/database/bau021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua K.B., Bellini W.J., Rota P.A., Harcourt B.H., Tamin A., Lam S.K., Ksiazek T.G., Rollin P.E., Zaki S.R., Shieh W.J., et al. Nipah virus: a recently emergent deadly paramyxovirus. Science. 2000;288:1432–1435. doi: 10.1126/science.288.5470.1432. [DOI] [PubMed] [Google Scholar]
- Chua K.B., Goh K.J., Wong K.T., Kamarulzaman A., Tan P.S., Ksiazek T.G., Zaki S.R., Paul G., Lam S.K., Tan C.T. Fatal encephalitis due to Nipah virus among pig-farmers in Malaysia. Lancet. 1999;354:1257–1259. doi: 10.1016/s0140-6736(99)04299-3. [DOI] [PubMed] [Google Scholar]
- Chua K.B., Lek Koh C., Hooi P.S., Wee K.F., Khong J.H., Chua B.H., Chan Y.P., Lim M.E., Lam S.K. Isolation of Nipah virus from Malaysian island flying-foxes. Microbes Infect. 2002;4:145–151. doi: 10.1016/s1286-4579(01)01522-2. [DOI] [PubMed] [Google Scholar]
- Ciarlet M., Ludert J.E., Iturriza-Gomara M., Liprandi F., Gray J.J., Desselberger U., Estes M.K. Initial interaction of rotavirus strains with N-acetylneuraminic (sialic) acid residues on the cell surface correlates with VP4 genotype, not species of origin. J. Virol. 2002;76:4087–4095. doi: 10.1128/jvi.76.8.4087-4095.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark S.A., Clark L.E., Pan J., Coscia A., McKay L.G.A., Shankar S., Johnson R.I., Brusic V., Choudhary M.C., Regan J., et al. SARS-CoV-2 evolution in an immunocompromised host reveals shared neutralization escape mechanisms. Cell. 2021;184:2605–2617.e18. doi: 10.1016/j.cell.2021.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockrell A.S., Peck K.M., Yount B.L., Agnihothram S.S., Scobey T., Curnes N.R., Baric R.S., Heise M.T. Mouse dipeptidyl peptidase 4 is not a functional receptor for Middle East respiratory syndrome coronavirus infection. J. Virol. 2014;88:5195–5199. doi: 10.1128/jvi.03764-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockrell A.S., Yount B.L., Scobey T., Jensen K., Douglas M., Beall A., Tang X.C., Marasco W.A., Heise M.T., Baric R.S. A mouse model for MERS coronavirus-induced acute respiratory distress syndrome. Nat Microbiol. 2016;2:16226. doi: 10.1038/nmicrobiol.2016.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Commission R.o.a.I. Ebola haemorrhagic fever in Zaire, 1976. Bull. World Health Organ. 1978;56:271–293. [PMC free article] [PubMed] [Google Scholar]
- Constantine D.G., Emmons R.W., Woodie J.D. Rabies virus in nasal mucosa of naturally infected bats. Science. 1972;175:1255–1256. doi: 10.1126/science.175.4027.1255. [DOI] [PubMed] [Google Scholar]
- Corman V.M., Baldwin H.J., Tateno A.F., Zerbinati R.M., Annan A., Owusu M., Nkrumah E.E., Maganga G.D., Oppong S., Adu-Sarkodie Y., et al. Evidence for an ancestral association of human coronavirus 229E with bats. J. Virol. 2015;89:11858–11870. doi: 10.1128/jvi.01755-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Ithete N.L., Richards L.R., Schoeman M.C., Preiser W., Drosten C., Drexler J.F. Rooting the phylogenetic tree of Middle East respiratory syndrome coronavirus by characterization of a conspecific virus from an African bat. J. Virol. 2014;88:11297–11303. doi: 10.1128/jvi.01498-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotten M., Watson S.J., Zumla A.I., Makhdoom H.Q., Palser A.L., Ong S.H., Al Rabeeah A.A., Alhakeem R.F., Assiri A., Al-Tawfiq J.A., et al. Spread, circulation, and evolution of the Middle East respiratory syndrome coronavirus. mBio. 2014;5:e01062-13. doi: 10.1128/mbio.01062-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox R.M., Plemper R.K. Structure and organization of paramyxovirus particles. Curr Opin Virol. 2017;24:105–114. doi: 10.1016/j.coviro.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford S.E., Ramani S., Tate J.E., Parashar U.D., Svensson L., Hagbom M., Franco M.A., Greenberg H.B., O'Ryan M., Kang G., et al. Rotavirus infection. Nat Rev Dis Primers. 2017;3:17083. doi: 10.1038/nrdp.2017.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Eden J.S., Holmes E.C., Wang L.F. Adaptive evolution of bat dipeptidyl peptidase 4 (dpp4): implications for the origin and emergence of Middle East respiratory syndrome coronavirus. Virol. J. 2013;10:304. doi: 10.1186/1743-422x-10-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey R.A., Shtanko O., Anantpadma M., Sakurai Y., Chandran K., Maury W. Mechanisms of filovirus entry. Curr. Top. Microbiol. Immunol. 2017;411:323–352. doi: 10.1007/82_2017_14. [DOI] [PubMed] [Google Scholar]
- de Groot R.J., Baker S.C., Baric R.S., Brown C.S., Drosten C., Enjuanes L., Fouchier R.A.M., Galiano M., Gorbalenya A.E., Memish Z.A., et al. Commentary: Middle East respiratory syndrome coronavirus (MERS-CoV): announcement of the coronavirus study group. J. Virol. 2013;87:7790–7792. doi: 10.1128/jvi.01244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E., Prescott J., Falzarano D., Bushmaker T., Scott D., Feldmann H., Munster V.J. Foodborne transmission of Nipah virus in Syrian hamsters. PLoS Pathog. 2014;10:e1004001. doi: 10.1371/journal.ppat.1004001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslandes A., Berti V., Tandjaoui-Lambotte Y., Alloui C., Carbonnelle E., Zahar J.R., Brichler S., Cohen Y. SARS-CoV-2 was already spreading in France in late December 2019. Int. J. Antimicrob. Agents. 2020;55:106006. doi: 10.1016/j.ijantimicag.2020.106006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diederich S., Maisner A. Molecular characteristics of the Nipah virus glycoproteins. Ann. N. Y. Acad. Sci. 2007;1102:39–50. doi: 10.1196/annals.1408.003. [DOI] [PubMed] [Google Scholar]
- Diehl W.E., Lin A.E., Grubaugh N.D., Carvalho L.M., Kim K., Kyawe P.P., McCauley S.M., Donnard E., Kucukural A., McDonel P., et al. Ebola virus glycoprotein with increased infectivity dominated the 2013-2016 epidemic. Cell. 2016;167:1088–1098.e6. doi: 10.1016/j.cell.2016.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzel E., Schudt G., Krähling V., Matrosovich M., Becker S. Functional characterization of adaptive mutations during the west African Ebola virus outbreak. J. Virol. 2017;91:e01913-16. doi: 10.1128/jvi.01913-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinnon K.H., Leist S.R., Schäfer A., Edwards C.E., Martinez D.R., Montgomery S.A., West A., Yount B.L., Hou Y.J., Adams L.E., et al. A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature. 2020;586:560–566. doi: 10.1038/s41586-020-2708-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler J.F., Gloza-Rausch F., Glende J., Corman V.M., Muth D., Goettsche M., Seebens A., Niedrig M., Pfefferle S., Yordanov S., et al. Genomic characterization of severe acute respiratory syndrome-related coronavirus in European bats and classification of coronaviruses based on partial RNA-dependent RNA polymerase gene sequences. J. Virol. 2010;84:11336–11349. doi: 10.1128/jvi.00650-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton B.T., Broder C.C., Middleton D., Wang L.F. Hendra and Nipah viruses: different and dangerous. Nat. Rev. Microbiol. 2006;4:23–35. doi: 10.1038/nrmicro1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edson D., Field H., McMichael L., Vidgen M., Goldspink L., Broos A., Melville D., Kristoffersen J., de Jong C., McLaughlin A., et al. Routes of Hendra virus excretion in naturally-infected flying-foxes: implications for viral transmission and spillover risk. PLoS One. 2015;10:e0140670. doi: 10.1371/journal.pone.0140670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards C.E., Yount B.L., Graham R.L., Leist S.R., Hou Y.J., Dinnon K.H., Sims A.C., Swanstrom J., Gully K., Scobey T.D., et al. Swine acute diarrhea syndrome coronavirus replication in primary human cells reveals potential susceptibility to infection. Proc. Natl. Acad. Sci. USA. 2020;117:26915–26925. doi: 10.1073/pnas.2001046117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enchéry F., Horvat B. Understanding the interaction between henipaviruses and their natural host, fruit bats: paving the way toward control of highly lethal infection in humans. Int. Rev. Immunol. 2017;36:108–121. doi: 10.1080/08830185.2016.1255883. [DOI] [PubMed] [Google Scholar]
- Epstein J.H., Anthony S.J., Islam A., Kilpatrick A.M., Ali Khan S., Balkey M.D., Ross N., Smith I., Zambrana-Torrelio C., Tao Y., et al. Nipah virus dynamics in bats and implications for spillover to humans. Proc. Natl. Acad. Sci. USA. 2020;117:29190–29201. doi: 10.1073/pnas.2000429117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esona M.D., Mijatovic-Rustempasic S., Conrardy C., Tong S., Kuzmin I.V., Agwanda B., Breiman R.F., Banyai K., Niezgoda M., Rupprecht C.E., et al. Reassortant group A rotavirus from straw-colored fruit bat (Eidolon helvum) Emerg. Infect. Dis. 2010;16:1844–1852. doi: 10.3201/eid1612.101089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esona M.D., Roy S., Rungsrisuriyachai K., Gautam R., Hermelijn S., Rey-Benito G., Bowen M.D. Molecular characterization of a human G20P[28] rotavirus a strain with multiple genes related to bat rotaviruses. Infect. Genet. Evol. 2018;57:166–170. doi: 10.1016/j.meegid.2017.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadini R.F., Fischer E., Castro S.J., Araujo A.C., Ornelas J.F., de Souza P.R. Bat and bee pollination in Psittacanthus mistletoes, a genus regarded as exclusively hummingbird-pollinated. Ecology. 2018;99:1239–1241. doi: 10.1002/ecy.2140. [DOI] [PubMed] [Google Scholar]
- Faria N.R., Mellan T.A., Whittaker C., Claro I.M., Candido D.D.S., Mishra S., Crispim M.A.E., Sales F.C.S., Hawryluk I., McCrone J.T., et al. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science. 2021;372:815–821. doi: 10.1126/science.abh2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann H., Sprecher A., Geisbert T.W. Ebola. New Engl J Med. 2020;382:1832–1842. doi: 10.1056/nejmra1901594. [DOI] [PubMed] [Google Scholar]
- Field H.E., Smith C.S., de Jong C.E., Melville D., Broos A., Kung N., Thompson J., Dechmann D.K.N. Landscape utilisation, animal behaviour and Hendra virus risk. Ecohealth. 2016;13:26–38. doi: 10.1007/s10393-015-1066-8. [DOI] [PubMed] [Google Scholar]
- Fongaro G., Stoco P.H., Souza D.S.M., Grisard E.C., Magri M.E., Rogovski P., Schörner M.A., Barazzetti F.H., Christoff A.P., de Oliveira L.F.V., et al. The presence of SARS-CoV-2 RNA in human sewage in Santa Catarina, Brazil, November 2019. Sci. Total Environ. 2021;778:146198. doi: 10.1016/j.scitotenv.2021.146198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freuling C.M., Breithaupt A., Müller T., Sehl J., Balkema-Buschmann A., Rissmann M., Klein A., Wylezich C., Höper D., Wernike K., et al. Susceptibility of raccoon dogs for experimental SARS-CoV-2 infection. Emerg. Infect. Dis. 2020;26:2982–2985. doi: 10.3201/eid2612.203733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick W.F., Kingston T., Flanders J. A review of the major threats and challenges to global bat conservation. Ann Ny Acad Sci. 2020;1469:5–25. doi: 10.1111/nyas.14045. [DOI] [PubMed] [Google Scholar]
- Gadalla M.R., Veit M. Toward the identification of ZDHHC enzymes required for palmitoylation of viral protein as potential drug targets. Expert Opin Drug Discov. 2020;15:159–177. doi: 10.1080/17460441.2020.1696306. [DOI] [PubMed] [Google Scholar]
- Ge X., Li J., Peng C., Wu L., Yang X., Wu Y., Zhang Y., Shi Z. Genetic diversity of novel circular ssDNA viruses in bats in China. J. Gen. Virol. 2011;92:2646–2653. doi: 10.1099/vir.0.034108-0. [DOI] [PubMed] [Google Scholar]
- Ge X.Y., Li J.L., Yang X.L., Chmura A.A., Zhu G., Epstein J.H., Mazet J.K., Hu B., Zhang W., Peng C., et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503:535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh N., Nandi S., Saha I. A review on evolution of emerging SARS-CoV-2 variants based on spike glycoprotein. Int Immunopharmacol. 2022;105:108565. doi: 10.1016/j.intimp.2022.108565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh K.J., Tan C.T., Chew N.K., Tan P.S., Kamarulzaman A., Sarji S.A., Wong K.T., Abdullah B.J.J., Abdullah B.J., Chua K.B., Lam S.K. Clinical features of Nipah virus encephalitis among pig farmers in Malaysia. N. Engl. J. Med. 2000;342:1229–1235. doi: 10.1056/nejm200004273421701. [DOI] [PubMed] [Google Scholar]
- Gonsalves L., Law B., Webb C., Monamy V. Foraging ranges of insectivorous bats shift relative to changes in mosquito abundance. PLoS One. 2013;8:e64081. doi: 10.1371/journal.pone.0064081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbunova V., Seluanov A., Kennedy B.K. The world goes bats: living longer and tolerating viruses. Cell Metab. 2020;32:31–43. doi: 10.1016/j.cmet.2020.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H., Chen Q., Yang G., He L., Fan H., Deng Y.Q., Wang Y., Teng Y., Zhao Z., Cui Y., et al. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science. 2020;369:1603–1607. doi: 10.1126/science.abc4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y., Zheng B.J., He Y.Q., Liu X.L., Zhuang Z.X., Cheung C.L., Luo S.W., Li P.H., Zhang L.J., Guan Y.J., et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- Gunnell G.F., Simmons N.B., Seiffert E.R. New Myzopodidae (Chiroptera) from the late Paleogene of Egypt: emended family diagnosis and biogeographic origins of Noctilionoidea. PLoS One. 2014;9:e86712. doi: 10.1371/journal.pone.0086712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunnell G.F., Smith R., Smith T. 33 million year old Myotis (Chiroptera, Vespertilionidae) and the rapid global radiation of modern bats. PLoS One. 2017;12:e0172621. doi: 10.1371/journal.pone.0172621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Hu B.J., Yang X.L., Zeng L.P., Li B., Ouyang S.Y., Shi Z.L. Evolutionary arms race between virus and host drives genetic diversity in bat severe acute respiratory syndrome-related coronavirus spike genes. J. Virol. 2020;94:e00902-20. doi: 10.1128/jvi.00902-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurley E.S., Montgomery J.M., Hossain M.J., Bell M., Azad A.K., Islam M.R., Molla M.A.R., Carroll D.S., Ksiazek T.G., Rota P.A., et al. Person-to-person transmission of Nipah virus in a Bangladeshi community. Emerg. Infect. Dis. 2007;13:1031–1037. doi: 10.3201/eid1307.061128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haagmans B.L., Al Dhahiry S.H.S., Reusken C.B.E.M., Raj V.S., Galiano M., Myers R., Godeke G.J., Jonges M., Farag E., Diab A., et al. Middle East respiratory syndrome coronavirus in dromedary camels: an outbreak investigation. Lancet Infect. Dis. 2014;14:140–145. doi: 10.1016/s1473-3099(13)70690-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hand S.J., Beck R.M.D., Archer M., Simmons N.B., Gunnell G.F., Scofield R.P., Tennyson A.J.D., De Pietri V.L., Salisbury S.W., Worthy T.H. A new, large-bodied omnivorous bat (Noctilionoidea: Mystacinidae) reveals lost morphological and ecological diversity since the Miocene in New Zealand. Sci. Rep. 2018;8:235. doi: 10.1038/s41598-017-18403-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper C.J., Swartz S.M., Brainerd E.L. Specialized bat tongue is a hemodynamic nectar mop. Proc. Natl. Acad. Sci. USA. 2013;110:8852–8857. doi: 10.1073/pnas.1222726110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasebe F., Thuy N.T.T., Inoue S., Yu F., Kaku Y., Watanabe S., Akashi H., Dat D.T., Mai L.T.Q., Morita K. Serologic evidence of Nipah virus infection in bats, Vietnam. Emerg. Infect. Dis. 2012;18:536–537. doi: 10.3201/eid1803.111121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B., Huang X., Zhang F., Tan W., Matthijnssens J., Qin S., Xu L., Zhao Z., Yang L., Wang Q., et al. Group A rotaviruses in Chinese bats: genetic composition, serology, and evidence for bat-to-human transmission and reassortment. J. Virol. 2017;91:e02493-16. doi: 10.1128/jvi.02493-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B., Yang F., Yang W., Zhang Y., Feng Y., Zhou J., Xie J., Feng Y., Bao X., Guo H., et al. Characterization of a novel G3P[3] rotavirus isolated from a lesser horseshoe bat: a distant relative of feline/canine rotaviruses. J. Virol. 2013;87:12357–12366. doi: 10.1128/jvi.02013-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B., Zhang Y., Xu L., Yang W., Yang F., Feng Y., Xia L., Zhou J., Zhen W., Feng Y., et al. Identification of diverse alphacoronaviruses and genomic characterization of a novel severe acute respiratory syndrome-like coronavirus from bats in China. J. Virol. 2014;88:7070–7082. doi: 10.1128/jvi.00631-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W.T., Bollen N., Xu Y., Zhao J., Dellicour S., Yan Z., Gong W., Zhang C., Zhang L., Lu M., et al. Phylogeography reveals association between swine trade and the spread of porcine epidemic diarrhea virus in China and across the world. Mol. Biol. Evol. 2022;39:msab364. doi: 10.1093/molbev/msab364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W.T., Hou X., Zhao J., Sun J., He H., Si W., Wang J., Jiang Z., Yan Z., Xing G., et al. Virome characterization of game animals in China reveals a spectrum of emerging pathogens. Cell. 2022;185:1117–1129.e8. doi: 10.1016/j.cell.2022.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W.T., Ji X., He W., Dellicour S., Wang S., Li G., Zhang L., Gilbert M., Zhu H., Xing G., et al. Genomic epidemiology, evolution, and transmission dynamics of porcine deltacoronavirus. Mol. Biol. Evol. 2020;37:2641–2654. doi: 10.1093/molbev/msaa117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemida M.G., Chu D.K.W., Poon L.L.M., Perera R.A.P.M., Alhammadi M.A., Ng H.Y., Siu L.Y., Guan Y., Alnaeem A., Peiris M. MERS coronavirus in dromedary camel herd, Saudi Arabia. Emerg. Infect. Dis. 2014;20:1231–1234. doi: 10.3201/eid2007.140571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y., Peng C., Yu M., Li Y., Han Z., Li F., Wang L.F., Shi Z. Angiotensin-converting enzyme 2 (ACE2) proteins of different bat species confer variable susceptibility to SARS-CoV entry. Arch. Virol. 2010;155:1563–1569. doi: 10.1007/s00705-010-0729-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y.J., Chiba S., Halfmann P., Ehre C., Kuroda M., Dinnon K.H., Leist S.R., Schäfer A., Nakajima N., Takahashi K., et al. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science. 2020;370:1464–1468. doi: 10.1126/science.abe8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu V.P., Hossain M.J., Parashar U.D., Ali M.M., Ksiazek T.G., Kuzmin I., Niezgoda M., Rupprecht C., Bresee J., Breiman R.F. Nipah virus encephalitis reemergence, Bangladesh. Emerg. Infect. Dis. 2004;10:2082–2087. doi: 10.3201/eid1012.040701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B., Zeng L.P., Yang X.L., Ge X.Y., Zhang W., Li B., Xie J.Z., Shen X.R., Zhang Y.Z., Wang N., et al. Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. PLoS Pathog. 2017;13:e1006698. doi: 10.1371/journal.ppat.1006698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.W., Dickerman A.W., Piñeyro P., Li L., Fang L., Kiehne R., Opriessnig T., Meng X.J. Origin, evolution, and genotyping of emergent porcine epidemic diarrhea virus strains in the United States. mBio. 2013;4 doi: 10.1128/mbio.00737-13. e00737-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iehlé C., Razafitrimo G., Razainirina J., Andriaholinirina N., Goodman S.M., Faure C., Georges-Courbot M.C., Rousset D., Reynes J.M. Henipavirus and Tioman virus antibodies in pteropodid bats, Madagascar. Emerg. Infect. Dis. 2007;13:159–161. doi: 10.3201/eid1301.060791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam M.S., Sazzad H.M., Satter S.M., Sultana S., Hossain M.J., Hasan M., Rahman M., Campbell S., Cannon D.L., Ströher U., et al. Nipah virus transmission from bats to humans associated with drinking traditional liquor made from date palm sap, Bangladesh, 2011-2014. Emerg. Infect. Dis. 2016;22:664–670. doi: 10.3201/eid2204.151747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ithete N.L., Stoffberg S., Corman V.M., Cottontail V.M., Richards L.R., Schoeman M.C., Drosten C., Drexler J.F., Preiser W. Close relative of human Middle East respiratory syndrome coronavirus in bat, South Africa. Emerg. Infect. Dis. 2013;19:1697–1699. doi: 10.3201/eid1910.130946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James S., Donato D., de Thoisy B., Lavergne A., Lacoste V. Novel herpesviruses in neotropical bats and their relationship with other members of the Herpesviridae family. Infect. Genet. Evol. 2020;84:104367. doi: 10.1016/j.meegid.2020.104367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jampanil N., Kumthip K., Yodmeeklin A., Kanai Y., Okitsu S., Kobayashi T., Ukarapol N., Ushijima H., Maneekarn N., Khamrin P. Epidemiology and genetic diversity of group A rotavirus in pediatric patients with acute gastroenteritis in Thailand, 2018-2019. Infect. Genet. Evol. 2021;95:104898. doi: 10.1016/j.meegid.2021.104898. [DOI] [PubMed] [Google Scholar]
- Jebb D., Huang Z., Pippel M., Hughes G.M., Lavrichenko K., Devanna P., Winkler S., Jermiin L.S., Skirmuntt E.C., Katzourakis A., et al. Six reference-quality genomes reveal evolution of bat adaptations. Nature. 2020;583:578–584. doi: 10.1038/s41586-020-2486-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung C., Kmiec D., Koepke L., Zech F., Jacob T., Sparrer K.M.J., Kirchhoff F. Omicron: what makes the latest SARS-CoV-2 variant of concern so concerning? J. Virol. 2022;96:e0207721. doi: 10.1128/jvi.02077-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan B., Wang M., Jing H., Xu H., Jiang X., Yan M., Liang W., Zheng H., Wan K., Liu Q., et al. Molecular evolution analysis and geographic investigation of severe acute respiratory syndrome coronavirus-like virus in palm civets at an animal market and on farms. J. Virol. 2005;79:11892–11900. doi: 10.1128/jvi.79.18.11892-11900.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khusro A., Aarti C., Pliego A.B., Cipriano-Salazar M. Hendra virus infection in horses: a review on emerging mystery paramyxovirus. J. Equine Vet. Sci. 2020;91:103149. doi: 10.1016/j.jevs.2020.103149. [DOI] [PubMed] [Google Scholar]
- Kim Y., Cheon S., Min C.K., Sohn K.M., Kang Y.J., Cha Y.J., Kang J.I., Han S.K., Ha N.Y., Kim G., et al. Spread of mutant Middle East respiratory syndrome coronavirus with reduced affinity to human CD26 during the South Korean outbreak. mBio. 2016;7:e00019. doi: 10.1128/mbio.00019-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchdoerfer R.N., Wasserman H., Amarasinghe G.K., Saphire E.O. Filovirus structural biology: the molecules in the machine. Curr Top Microbiol. 2017;411:381–417. doi: 10.1007/82_2017_16. [DOI] [PubMed] [Google Scholar]
- Kondoh T., Letko M., Munster V.J., Manzoor R., Maruyama J., Furuyama W., Miyamoto H., Shigeno A., Fujikura D., Takadate Y., et al. Single-nucleotide polymorphisms in human NPC1 influence filovirus entry into cells. J. Infect. Dis. 2018;218:S397–S402. doi: 10.1093/infdis/jiy248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn J.H., Amarasinghe G.K., Basler C.F., Bavari S., Bukreyev A., Chandran K., Crozier I., Dolnik O., Dye J.M., Formenty P.B.H., et al. ICTV virus taxonomy profile: filoviridae. J. Gen. Virol. 2019;100:911–912. doi: 10.1099/jgv.0.001252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni D.D., Tosh C., Venkatesh G., Senthil Kumar D. Nipah virus infection: current scenario. Indian J. Virol. 2013;24:398–408. doi: 10.1007/s13337-013-0171-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladner J.T., Wiley M.R., Mate S., Dudas G., Prieto K., Lovett S., Nagle E.R., Beitzel B., Gilbert M.L., Fakoli L., et al. Evolution and spread of Ebola virus in Liberia, 2014-2015. Cell Host Microbe. 2015;18:659–669. doi: 10.1016/j.chom.2015.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam T.T.Y., Jia N., Zhang Y.W., Shum M.H.H., Jiang J.F., Zhu H.C., Tong Y.G., Shi Y.X., Ni X.B., Liao Y.S., et al. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature. 2020;583:282–285. doi: 10.1038/s41586-020-2169-0. [DOI] [PubMed] [Google Scholar]
- Lan J., Ge J.W., Yu J.F., Shan S.S., Zhou H., Fan S.L., Zhang Q., Shi X.L., Wang Q.S., Zhang L.Q., Wang X.Q. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Latinne A., Hu B., Olival K.J., Zhu G., Zhang L., Li H., Chmura A.A., Field H.E., Zambrana-Torrelio C., Epstein J.H., et al. Origin and cross-species transmission of bat coronaviruses in China. Nat. Commun. 2020;11:4235. doi: 10.1038/s41467-020-17687-3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Lau S.K.P., Feng Y., Chen H., Luk H.K.H., Yang W.H., Li K.S.M., Zhang Y.Z., Huang Y., Song Z.Z., Chow W.N., et al. Severe acute respiratory syndrome (SARS) coronavirus ORF8 protein is acquired from SARS-related coronavirus from greater horseshoe bats through recombination. J. Virol. 2015;89:10532–10547. doi: 10.1128/jvi.01048-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K.P., Li K.S.M., Huang Y., Shek C.T., Tse H., Wang M., Choi G.K.Y., Xu H., Lam C.S.F., Guo R., et al. Ecoepidemiology and complete genome comparison of different strains of severe acute respiratory syndrome-related Rhinolophus bat coronavirus in China reveal bats as a reservoir for acute, self-limiting infection that allows recombination events. J. Virol. 2010;84:2808–2819. doi: 10.1128/jvi.02219-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K.P., Woo P.C.Y., Li K.S.M., Huang Y., Tsoi H.W., Wong B.H.L., Wong S.S.Y., Leung S.Y., Chan K.H., Yuen K.Y. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. USA. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K., Woo P.C., Li K.S., Huang Y., Wang M., Lam C.S., Xu H., Guo R., Chan K.H., Zheng B.J., Yuen K.Y. Complete genome sequence of bat coronavirus HKU2 from Chinese horseshoe bats revealed a much smaller spike gene with a different evolutionary lineage from the rest of the genome. Virology. 2007;367:428–439. doi: 10.1016/j.virol.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laude H., Van Reeth K., Pensaert M. Porcine respiratory coronavirus: molecular features and virus-host interactions. Veterinary Res. 1993;24:125–150. [PubMed] [Google Scholar]
- Leroy E.M., Kumulungui B., Pourrut X., Rouquet P., Hassanin A., Yaba P., Délicat A., Paweska J.T., Gonzalez J.-P., Swanepoel R. Fruit bats as reservoirs of Ebola virus. Nature. 2005;438:575–576. doi: 10.1038/438575a. [DOI] [PubMed] [Google Scholar]
- Letko M., Miazgowicz K., McMinn R., Seifert S.N., Sola I., Enjuanes L., Carmody A., van Doremalen N., Munster V. Adaptive evolution of MERS-CoV to species variation in DPP4. Cell Rep. 2018;24:1730–1737. doi: 10.1016/j.celrep.2018.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letko M., Seifert S.N., Olival K.J., Plowright R.K., Munster V.J. Bat-borne virus diversity, spillover and emergence. Nat. Rev. Microbiol. 2020;18:461–471. doi: 10.1038/s41579-020-0394-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Wang M., Mao T., Wang M., Zhang Q., Wang H., Pang L., Sun X., Duan Z. The functional characterization of bat and human P[3] rotavirus VP8∗s. Virol. Sin. 2021;36:1187–1196. doi: 10.1007/s12250-021-00400-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Receptor recognition and cross-species infections of SARS coronavirus. Antiviral Res. 2013;100:246–254. doi: 10.1016/j.antiviral.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- Li G., He W., Zhu H., Bi Y., Wang R., Xing G., Zhang C., Zhou J., Yuen K.Y., Gao G.F., Su S. Origin, genetic diversity, and evolutionary dynamics of novel porcine circovirus 3. Adv. Sci. 2018;5:1800275. doi: 10.1002/advs.201800275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K., Yan S., Wang N., He W., Guan H., He C., Wang Z., Lu M., He W., Ye R., et al. Emergence and adaptive evolution of Nipah virus. Transbound Emerg Dis. 2020;67:121–132. doi: 10.1111/tbed.13330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H., Wang H., Crameri G., Hu Z., Zhang H., et al. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- Li Y., Wang J., Hickey A.C., Zhang Y., Li Y., Wu Y., Zhang H., Yuan J., Han Z., McEachern J., et al. Antibodies to Nipah or Nipah-like viruses in bats, China. Emerg. Infect. Dis. 2008;14:1974–1976. doi: 10.3201/eid1412.080359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X.D., Wang W., Hao Z.Y., Wang Z.X., Guo W.P., Guan X.Q., Wang M.R., Wang H.W., Zhou R.H., Li M.H., et al. Extensive diversity of coronaviruses in bats from China. Virology. 2017;507:1–10. doi: 10.1016/j.virol.2017.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Tang J., Ma Y., Liang X., Yang Y., Peng G., Qi Q., Jiang S., Li J., Du L., Li F. Receptor usage and cell entry of porcine epidemic diarrhea coronavirus. J. Virol. 2015;89:6121–6125. doi: 10.1128/jvi.00430-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Hu G., Wang Y., Ren W., Zhao X., Ji F., Zhu Y., Feng F., Gong M., Ju X., et al. Functional and genetic analysis of viral receptor ACE2 orthologs reveals a broad potential host range of SARS-CoV-2. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2025373118. e2025373118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo M.K., Rota P.A. The emergence of Nipah virus, a highly pathogenic paramyxovirus. J. Clin. Virol. 2008;43:396–400. doi: 10.1016/j.jcv.2008.08.007. [DOI] [PubMed] [Google Scholar]
- López-Hoffman L., Wiederholt R., Sansone C., Bagstad K.J., Cryan P., Diffendorfer J.E., Goldstein J., Lasharr K., Loomis J., McCracken G., et al. Market forces and technological substitutes cause fluctuations in the value of bat pest-control services for cotton. PLoS One. 2014;9:e87912. doi: 10.1371/journal.pone.0087912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López S., Arias C.F. Multistep entry of rotavirus into cells: a Versaillesque dance. Trends Microbiol. 2004;12:271–278. doi: 10.1016/j.tim.2004.04.003. [DOI] [PubMed] [Google Scholar]
- Lu G., Hu Y., Wang Q., Qi J., Gao F., Li Y., Zhang Y., Zhang W., Yuan Y., Bao J., et al. Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature. 2013;500:227–231. doi: 10.1038/nature12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G., Wang Q., Gao G.F. Bat-to-human: spike features determining 'host jump' of coronaviruses SARS-CoV, MERS-CoV, and beyond. Trends Microbiol. 2015;23:468–478. doi: 10.1016/j.tim.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luby S.P., Rahman M., Hossain M.J., Blum L.S., Husain M.M., Gurley E., Khan R., Ahmed B.N., Rahman S., Nahar N., et al. Foodborne transmission of Nipah virus, Bangladesh. Emerg. Infect. Dis. 2006;12:1888–1894. doi: 10.3201/eid1212.060732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean G., Kamil J., Lee B., Moore P., Schulz T.F., Muik A., Sahin U., Türeci Ö., Pather S. The impact of evolving SARS-CoV-2 mutations and variants on COVID-19 vaccines. mBio. 2022;13:e0297921. doi: 10.1128/mbio.02979-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekibib B., Ariën K. Aerosol transmission of filoviruses. Viruses. 2016;8:148. doi: 10.3390/v8050148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello M.A.R., Schittini G.M., Selig P., Bergallo H.G. Seasonal variation in the diet of the bat Carollia perspicillata (Chiroptera : phyllostomidae) in an Atlantic forest area in southeastern Brazil. Mammalia. 2004;68:49–55. doi: 10.1515/mamm.2004.006. [DOI] [Google Scholar]
- Menachery V.D., Yount B.L., Debbink K., Agnihothram S., Gralinski L.E., Plante J.A., Graham R.L., Scobey T., Ge X.Y., Donaldson E.F., et al. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat. Med. 2015;21:1508–1513. doi: 10.1038/nm.3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet J.K., Whittaker G.R. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120–134. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miño S., Matthijnssens J., Badaracco A., Garaicoechea L., Zeller M., Heylen E., Van Ranst M., Barrandeguy M., Parreño V. Equine G3P[3] rotavirus strain E3198 related to simian RRV and feline/canine-like rotaviruses based on complete genome analyses. Vet. Microbiol. 2013;161:239–246. doi: 10.1016/j.vetmic.2012.07.033. [DOI] [PubMed] [Google Scholar]
- Mishra N., Fagbo S.F., Alagaili A.N., Nitido A., Williams S.H., Ng J., Lee B., Durosinlorun A., Garcia J.A., Jain K., et al. A viral metagenomic survey identifies known and novel mammalian viruses in bats from Saudi Arabia. PLoS One. 2019;14:e0214227. doi: 10.1371/journal.pone.0214227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller M.A., Corman V.M., Jores J., Meyer B., Younan M., Liljander A., Bosch B.J., Lattwein E., Hilali M., Musa B.E., et al. MERS coronavirus neutralizing antibodies in camels, Eastern Africa, 1983-1997. Emerg. Infect. Dis. 2014;20:2093–2095. doi: 10.3201/eid2012.141026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Fontela C., Dowling W.E., Funnell S.G.P., Gsell P.S., Riveros-Balta A.X., Albrecht R.A., Andersen H., Baric R.S., Carroll M.W., Cavaleri M., et al. Animal models for COVID-19. Nature. 2020;586:509–515. doi: 10.1038/s41586-020-2787-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munster V.J., Bausch D.G., de Wit E., Fischer R., Kobinger G., Muñoz-Fontela C., Olson S.H., Seifert S.N., Sprecher A., Ntoumi F., et al. Outbreaks in a rapidly changing central Africa - lessons from Ebola. N. Engl. J. Med. 2018;379:1198–1201. doi: 10.1056/nejmp1807691. [DOI] [PubMed] [Google Scholar]
- Murray K., Selleck P., Hooper P., Hyatt A., Gould A., Gleeson L., Westbury H., Hiley L., Selvey L., Rodwell B., Ketterer P. A morbillivirus that caused fatal fisease in horses and humans. Science. 1995;268:94–97. doi: 10.1126/science.7701348. [DOI] [PubMed] [Google Scholar]
- Ng M., Ndungo E., Kaczmarek M.E., Herbert A.S., Binger T., Kuehne A.I., Jangra R.K., Hawkins J.A., Gifford R.J., Biswas R., et al. Filovirus receptor NPC1 contributes to species-specific patterns of ebolavirus susceptibility in bats. Elife. 2015;4:e11785. doi: 10.7554/elife.11785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyakarahuka L., Shoemaker T.R., Balinandi S., Chemos G., Kwesiga B., Mulei S., Kyondo J., Tumusiime A., Kofman A., Masiira B., et al. Marburg virus disease outbreak in Kween District Uganda, 2017: epidemiological and laboratory findings. PLoS Negl Trop Dis. 2019;13:e0007257. doi: 10.1371/journal.pntd.0007257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okitsu S., Hikita T., Thongprachum A., Khamrin P., Takanashi S., Hayakawa S., Maneekarn N., Ushijima H. Detection and molecular characterization of two rare G8P[14] and G3P[3] rotavirus strains collected from children with acute gastroenteritis in Japan. Infect. Genet. Evol. 2018;62:95–108. doi: 10.1016/j.meegid.2018.04.011. [DOI] [PubMed] [Google Scholar]
- Pan Y., Tian X., Qin P., Wang B., Zhao P., Yang Y.L., Wang L., Wang D., Song Y., Zhang X., Huang Y.W. Discovery of a novel swine enteric alphacoronavirus (SeACoV) in southern China. Vet. Microbiol. 2017;211:15–21. doi: 10.1016/j.vetmic.2017.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck K.M., Cockrell A.S., Yount B.L., Scobey T., Baric R.S., Heise M.T. Glycosylation of mouse DPP4 plays a role in inhibiting Middle East respiratory syndrome coronavirus infection. J. Virol. 2015;89:4696–4699. doi: 10.1128/jvi.03445-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernet O., Wang Y.E., Lee B. Henipavirus receptor usage and tropism. Curr. Top. Microbiol. Immunol. 2012;359:59–78. doi: 10.1007/82_2012_222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plante J.A., Liu Y., Liu J.Y., Xia H.J., Johnson B.A., Lokugamage K.G., Zhang X.W., Muruato A.E., Zou J., Fontes-Garfias C.R., et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2020;592:116–121. doi: 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontremoli C., Forni D., Cagliani R., Filippi G., De Gioia L., Pozzoli U., Clerici M., Sironi M. Positive selection drives evolution at the host-filovirus interaction surface. Mol. Biol. Evol. 2016;33:2836–2847. doi: 10.1093/molbev/msw158. [DOI] [PubMed] [Google Scholar]
- Posthuma C.C., Te Velthuis A.J.W., Snijder E.J. Nidovirus RNA polymerases: complex enzymes handling exceptional RNA genomes. Virus Res. 2017;234:58–73. doi: 10.1016/j.virusres.2017.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj V.S., Mou H.H., Smits S.L., Dekkers D.H.W., Müller M.A., Muller M.A., Dijkman R., Muth D., Demmers J.A.A., Zaki A., et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reusken C.B.E.M., Haagmans B.L., Müller M.A., Muller M.A., Gutierrez C., Godeke G.J., Meyer B., Muth D., Raj V.S., Vries L.S.D., et al. Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: a comparative serological study. Lancet Infect. Dis. 2013;13:859–866. doi: 10.1016/s1473-3099(13)70164-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynes J.M., Counor D., Ong S., Faure C., Seng V., Molia S., Walston J., Georges-Courbot M.C., Deubel V., Sarthou J.L. Nipah virus in Lyle's flying foxes, Cambodia. Emerg. Infect. Dis. 2005;11:1042–1047. doi: 10.3201/eid1107.041350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rihtarič D., Hostnik P., Steyer A., Grom J., Toplak I. Identification of SARS-like coronaviruses in horseshoe bats (Rhinolophus hipposideros) in Slovenia. Arch. Virol. 2010;155:507–514. doi: 10.1007/s00705-010-0612-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripperger S.P., Kalko E.K.V., Rodríguez-Herrera B., Mayer F., Tschapka M. Frugivorous bats maintain functional habitat connectivity in agricultural landscapes but rely strongly on natural forest fragments. PLoS One. 2015;10:e0120535. doi: 10.1371/journal.pone.0120535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristic M., Sibinovic S., Alberts J.O. Electron microscopy and ether sensitivity of transmissible gastroenteritis virus of swine. Am. J. Vet. Res. 1965;26:609–616. [PubMed] [Google Scholar]
- Roberts A., Deming D., Paddock C.D., Cheng A., Yount B., Vogel L., Herman B.D., Sheahan T., Heise M., Genrich G.L., et al. A mouse-adapted SARS-coronavirus causes disease and mortality in BALB/c mice. PLoS Pathog. 2007;3:e5. doi: 10.1371/journal.ppat.0030005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell A.L., Goodman S.M., Fiorentino I., Yoder A.D. Population genetic analysis of myzopoda (Chiroptera : Myzopodidae) in Madagascar. J. Mammal. 2008;89:209–221. doi: 10.1644/07-mamm-a-044.1. [DOI] [Google Scholar]
- Sabir J.S.M., Lam T.T.Y., Ahmed M.M.M., Li L., Shen Y., E M Abo-Aba S., Qureshi M.I., Abu-Zeid M., Zhang Y., Khiyami M.A., et al. Co-circulation of three camel coronavirus species and recombination of MERS-CoVs in Saudi Arabia. Science. 2016;351:81–84. doi: 10.1126/science.aac8608. [DOI] [PubMed] [Google Scholar]
- Schlottau K., Rissmann M., Graaf A., Schön J., Sehl J., Wylezich C., Höper D., Mettenleiter T.C., Balkema-Buschmann A., Harder T., et al. SARS-CoV-2 in fruit bats, ferrets, pigs, and chickens: an experimental transmission study. Lancet Microbe. 2020;1:e218–e225. doi: 10.1016/s2666-5247(20)30089-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwegmann-Wessels C., Herrler G. Transmissible gastroenteritis virus infection: a vanishing specter. Dtsch Tierarztl Wochenschr. 2006;113:157–159. [PubMed] [Google Scholar]
- Sendow I., Field H.E., Adjid A., Ratnawati A., Breed A.C., Darminto, Morrissy C., Daniels P., Daniels P. Screening for Nipah virus infection in West kalimantan province, Indonesia. Zoonoses Public Health. 2010;57:499–503. doi: 10.1111/j.1863-2378.2009.01252.x. [DOI] [PubMed] [Google Scholar]
- Sendow I., Field H.E., Curran J., Darminto, Morrissy C., Meehan G., Buick T., Daniels P. Henipavirus in Pteropus vampyrus bats, Indonesia. Emerg. Infect. Dis. 2006;12:711–712. doi: 10.3201/eid1204.051181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra-Cobo J., Lopez-Roig M. Bats and emerging infections: an ecological and virological puzzle. Adv. Exp. Med. Biol. 2017;972:35–48. doi: 10.1007/5584_2016_131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J.Z., Wen Z.Y., Zhong G.X., Yang H.L., Wang C., Huang B.Y., Liu R.Q., He X.J., Shuai L., Sun Z.R., et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science. 2020;368:1016–1020. doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons N.B., Seymour K.L., Habersetzer J., Gunnell G.F. Primitive Early Eocene bat from Wyoming and the evolution of flight and echolocation. Nature. 2008;451:818–821. doi: 10.1038/nature06549. [DOI] [PubMed] [Google Scholar]
- Simsek C., Corman V.M., Everling H.U., Lukashev A.N., Rasche A., Maganga G.D., Binger T., Jansen D., Beller L., Deboutte W., et al. At least seven distinct rotavirus genotype Constellations in bats with evidence of reassortment and zoonotic transmissions. mBio. 2021;12:e02755-20. doi: 10.1128/mbio.02755-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solberg O.D., Hasing M.E., Trueba G., Eisenberg J.N. Characterization of novel VP7, VP4, and VP6 genotypes of a previously untypeable group A rotavirus. Virology. 2009;385:58–67. doi: 10.1016/j.virol.2008.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer M.S., Teeling E.C., Madsen O., Stanhope M.J., de Jong W.W. Integrated fossil and molecular data reconstruct bat echolocation. Proc. Natl. Acad. Sci. USA. 2001;98:6241–6246. doi: 10.1073/pnas.111551998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler K., Masignani V., Eickmann M., Becker S., Abrignani S., Klenk H.D., Rappuoli R. SARS - beginning to understand a new virus. Nat. Rev. Microbiol. 2003;1:209–218. doi: 10.1038/nrmicro775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T.N., Greaney A.J., Hilton S.K., Ellis D., Crawford K.H.D., Dingens A.S., Navarro M.J., Bowen J.E., Tortorici M.A., Walls A.C., et al. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals Constraints on folding and ACE2 binding. Cell. 2020;182:1295–1310.e20. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S., Wong G., Shi W.F., Liu J., Lai A.C.K., Zhou J.Y., Liu W.J., Bi Y.H., Gao G.F. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y.C.F., Anderson D.E., Young B.E., Linster M., Zhu F., Jayakumar J., Zhuang Y., Kalimuddin S., Low J.G.H., Tan C.W., et al. Discovery and genomic characterization of a 382-nucleotide deletion in ORF7b and ORF8 during the early evolution of SARS-CoV-2. mBio. 2020;11:e01610-20. doi: 10.1128/mbio.01610-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subissi L., Keita M., Mesfin S., Rezza G., Diallo B., Van Gucht S., Musa E.O., Yoti Z., Keita S., Djingarey M.H., et al. Ebola virus transmission caused by persistently infected survivors of the 2014-2016 outbreak in West Africa. J. Infect. Dis. 2018;218:S287–S291. doi: 10.1093/infdis/jiy280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., He W.T., Wang L., Lai A., Ji X., Zhai X., Li G., Suchard M.A., Tian J., Zhou J., et al. COVID-19: epidemiology, evolution, and cross-disciplinary perspectives. Trends Mol. Med. 2020;26:483–495. doi: 10.1016/j.molmed.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takadate Y., Kondoh T., Igarashi M., Maruyama J., Manzoor R., Ogawa H., Kajihara M., Furuyama W., Sato M., Miyamoto H., et al. Niemann-pick C1 heterogeneity of bat cells controls filovirus tropism. Cell Rep. 2020;30:308–319.e5. doi: 10.1016/j.celrep.2019.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Z., Gonzalez G., Sheng J., Wu J., Zhang F., Xu L., Zhang P., Zhu A., Qu Y., Tu C., et al. Extensive genetic diversity of polyomaviruses in sympatric bat Communities: host switching versus coevolution. J. Virol. 2020;94:e02101-19. doi: 10.1128/jvi.02101-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y., Shi M., Chommanard C., Queen K., Zhang J., Markotter W., Kuzmin I.V., Holmes E.C., Tong S. Surveillance of bat coronaviruses in Kenya identifies relatives of human coronaviruses NL63 and 229E and their recombination history. J. Virol. 2017;91:e01953-16. doi: 10.1128/jvi.01953-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeling E.C., Madsen O., Van den Bussche R.A., de Jong W.W., Stanhope M.J., Springer M.S. Microbat paraphyly and the convergent evolution of a key innovation in Old World rhinolophoid microbats. Proc. Natl. Acad. Sci. USA. 2002;99:1431–1436. doi: 10.1073/pnas.022477199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeling E.C., Springer M.S., Madsen O., Bates P., O'Brien S.J., Murphy W.J. A molecular phylogeny for bats illuminates biogeography and the fossil record. Science. 2005;307:580–584. doi: 10.1126/science.1105113. [DOI] [PubMed] [Google Scholar]
- Teeling E.C., Vernes S.C., Dávalos L.M., Ray D.A., Gilbert M.T.P., Myers E., Consortium B.K. Bat biology, genomes, and the Bat1K project: to generate Chromosome-level genomes for all living bat species. Annu Rev Anim Biosci. 2018;6:23–46. doi: 10.1146/annurev-animal-022516-022811. [DOI] [PubMed] [Google Scholar]
- Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., Doolabh D., Pillay S., San E.J., Msomi N., et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature. 2021;592:438–443. doi: 10.1038/s41586-021-03402-9. [DOI] [PubMed] [Google Scholar]
- Thibault P.A., Watkinson R.E., Moreira-Soto A., Drexler J.F., Lee B. Zoonotic potential of emerging paramyxoviruses: knowns and unknowns. Adv. Virus Res. 2017;98:1–55. doi: 10.1016/bs.aivir.2016.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totura A.L., Bavari S. Broad-spectrum coronavirus antiviral drug discovery. Expert Opin Drug Discov. 2019;14:397–412. doi: 10.1080/17460441.2019.1581171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towner J.S., Amman B.R., Sealy T.K., Carroll S.A.R., Comer J.A., Kemp A., Swanepoel R., Paddock C.D., Balinandi S., Khristova M.L., et al. Isolation of genetically diverse Marburg viruses from Egyptian fruit bats. PLoS Pathog. 2009;5:e1000536. doi: 10.1371/journal.ppat.1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimpert J., Vladimirova D., Dietert K., Abdelgawad A., Kunec D., Dökel S., Voss A., Gruber A.D., Bertzbach L.D., Osterrieder N. The roborovski dwarf hamster is A highly susceptible model for a rapid and fatal course of SARS-CoV-2 infection. Cell Rep. 2020;33:108488. doi: 10.1016/j.celrep.2020.108488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu C., Crameri G., Kong X., Chen J., Sun Y., Yu M., Xiang H., Xia X., Liu S., Ren T., et al. Antibodies to SARS coronavirus in civets. Emerg. Infect. Dis. 2004;10:2244–2248. doi: 10.3201/eid1012.040520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich L., Wernike K., Hoffmann D., Mettenleiter T.C., Beer M. Experimental infection of cattle with SARS-CoV-2. Emerg. Infect. Dis. 2020;26:2979–2981. doi: 10.3201/eid2612.203799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanowicz R.A., McClure C.P., Sakuntabhai A., Sall A.A., Kobinger G., Müller M.A., Holmes E.C., Rey F.A., Simon-Loriere E., Ball J.K. Human adaptation of Ebola virus during the West African outbreak. Cell. 2016;167:1079–1087.e5. doi: 10.1016/j.cell.2016.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz E., Mishra S., Chand M., Barrett J.C., Johnson R., Geidelberg L., Hinsley W.R., Laydon D.J., Dabrera G., O’Toole Á., et al. Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature. 2021;593:266–269. doi: 10.1038/s41586-021-03470-x. [DOI] [PubMed] [Google Scholar]
- Wacharapluesadee S., Duengkae P., Rodpan A., Kaewpom T., Maneeorn P., Kanchanasaka B., Yingsakmongkon S., Sittidetboripat N., Chareesaen C., Khlangsap N., et al. Diversity of coronavirus in bats from Eastern Thailand. Virol. J. 2015;12:57. doi: 10.1186/s12985-015-0289-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacharapluesadee S., Lumlertdacha B., Boongird K., Wanghongsa S., Chanhome L., Rollin P., Stockton P., Rupprecht C.E., Ksiazek T.G., Hemachudha T. Bat Nipah virus, Thailand. Emerg. Infect. Dis. 2005;11:1949–1951. doi: 10.3201/eid1112.050613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Shi Y., Song J., Qi J., Lu G., Yan J., Gao G.F. Ebola viral glycoprotein bound to its endosomal receptor Niemann-pick C1. Cell. 2016;164:258–268. doi: 10.1016/j.cell.2015.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Su S., Bi Y., Wong G., Gao G.F. Bat-origin coronaviruses expand their host range to pigs. Trends Microbiol. 2018;26:466–470. doi: 10.1016/j.tim.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner R., Matrosovich M., Klenk H.D. Functional balance between haemagglutinin and neuraminidase in influenza virus infections. Rev. Med. Virol. 2002;12:159–166. doi: 10.1002/rmv.352. [DOI] [PubMed] [Google Scholar]
- Wang L.F., Gamage A.M., Chan W.O.Y., Hiller M., Teeling E.C. Decoding bat immunity: the need for a coordinated research approach. Nat. Rev. Immunol. 2021;21:269–271. doi: 10.1038/s41577-021-00523-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Li S.Y., Yang X.L., Huang H.M., Zhang Y.J., Guo H., Luo C.M., Miller M., Zhu G., Chmura A.A., et al. Serological evidence of bat SARS-related coronavirus infection in humans, China. Virol. Sin. 2018;33:104–107. doi: 10.1007/s12250-018-0012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waruhiu C., Ommeh S., Obanda V., Agwanda B., Gakuya F., Ge X.Y., Yang X.L., Wu L.J., Zohaib A., Hu B., Shi Z.L. Molecular detection of viruses in Kenyan bats and discovery of novel astroviruses, caliciviruses and rotaviruses. Virol. Sin. 2017;32:101–114. doi: 10.1007/s12250-016-3930-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weatherman S., Feldmann H., de Wit E. Transmission of henipaviruses. Curr Opin Virol. 2018;28:7–11. doi: 10.1016/j.coviro.2017.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingartl H.M., Nfon C., Kobinger G. Review of Ebola virus infections in domestic animals. Dev. Biol. 2013;135:211–218. doi: 10.1159/000178495. [DOI] [PubMed] [Google Scholar]
- Weiss S.R., Leibowitz J.L. Coronavirus pathogenesis. Adv. Virus Res. 2011;81:85–164. doi: 10.1016/B978-0-12-385885-6.00009-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman D., Alameh M.G., de Silva T., Collini P., Hornsby H., Brown R., LaBranche C.C., Edwards R.J., Sutherland L., Santra S., et al. D614G spike mutation increases SARS CoV-2 susceptibility to neutralization. Cell Host Microbe. 2021;29:23–31.e4. doi: 10.1016/j.chom.2020.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widagdo W., Sooksawasdi Na Ayudhya S., Hundie G.B., Haagmans B.L. Host determinants of MERS-CoV transmission and pathogenesis. Viruses. 2019;11:280. doi: 10.3390/v11030280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson G.S., South J.M. Life history, ecology and longevity in bats. Aging Cell. 2002;1:124–131. doi: 10.1046/j.1474-9728.2002.00020.x. [DOI] [PubMed] [Google Scholar]
- Williamson M.M., Hooper P.T., Selleck P.W., Gleeson L.J., Daniels P.W., Westbury H.A., Murray P.K. Transmission studies of Hendra virus (equine morbillivirus) in fruit bats, horses and cats. Aust. Vet. J. 1998;76:813–818. doi: 10.1111/j.1751-0813.1998.tb12335.x. [DOI] [PubMed] [Google Scholar]
- Wu Z., Yang L., Ren X., He G., Zhang J., Yang J., Qian Z., Dong J., Sun L., Zhu Y., et al. Deciphering the bat virome catalog to better understand the ecological diversity of bat viruses and the bat origin of emerging infectious diseases. ISME J. 2016;10:609–620. doi: 10.1038/ismej.2015.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia L., Fan Q., He B., Xu L., Zhang F., Hu T., Wang Y., Li N., Qiu W., Zheng Y., et al. The complete genome sequence of a G3P[10] Chinese bat rotavirus suggests multiple bat rotavirus inter-host species transmission events. Infect. Genet. Evol. 2014;28:1–4. doi: 10.1016/j.meegid.2014.09.005. [DOI] [PubMed] [Google Scholar]
- Xiao K., Zhai J., Feng Y., Zhou N., Zhang X., Zou J.J., Li N., Guo Y., Li X., Shen X., et al. Isolation of SARS-CoV-2-related coronavirus from Malayan pangolins. Nature. 2020;583:286–289. doi: 10.1038/s41586-020-2313-x. [DOI] [PubMed] [Google Scholar]
- Xu K., Broder C.C., Nikolov D.B. Ephrin-B2 and ephrin-B3 as functional henipavirus receptors. Semin. Cell Dev. Biol. 2012;23:116–123. doi: 10.1016/j.semcdb.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S., McGinnis K.R., Liu Y., Huang P., Tan M., Stuckert M.R., Burnside R.E., Jacob E.G., Ni S., Jiang X., Kennedy M.A. Structural basis of P[II] rotavirus evolution and host ranges under selection of histo-blood group antigens. Proc. Natl. Acad. Sci. U.S.A. 2021;118 doi: 10.1073/pnas.2107963118. e2107963118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav P.D., Shete A.M., Kumar G.A., Sarkale P., Sahay R.R., Radhakrishnan C., Lakra R., Pardeshi P., Gupta N., Gangakhedkar R.R., et al. Nipah virus sequences from humans and bats during Nipah outbreak, Kerala, India, 2018. Emerg. Infect. Dis. 2019;25:1003–1006. doi: 10.3201/eid2505.181076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Wu Z., Ren X., Yang F., He G., Zhang J., Dong J., Sun L., Zhu Y., Du J., et al. Novel SARS-like betacoronaviruses in bats, China, 2011. Emerg. Infect. Dis. 2013;19:989–991. doi: 10.3201/eid1906.121648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.L., Tan C.W., Anderson D.E., Jiang R.D., Li B., Zhang W., Zhu Y., Lim X.F., Zhou P., Liu X.L., et al. Characterization of a filovirus (Měnglà virus) from Rousettus bats in China. Nat Microbiol. 2019;4:390–395. doi: 10.1038/s41564-018-0328-y. [DOI] [PubMed] [Google Scholar]
- Yang Y.L., Qin P., Wang B., Liu Y., Xu G.H., Peng L., Zhou J., Zhu S.J., Huang Y.W. Broad cross-species infection of cultured cells by bat HKU2-related swine acute diarrhea syndrome coronavirus and identification of its replication in murine dendritic cells in vivo highlight its potential for diverse interspecies transmission. J. Virol. 2019;93:e01448-19. doi: 10.1128/jvi.01448-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yob J.M., Field H., Rashdi A.M., Morrissy C., van der Heide B., Rota P., bin Adzhar A., White J., Daniels P., Jamaluddin A., Ksiazek T. Nipah virus infection in bats (order Chiroptera) in peninsular Malaysia. Emerg. Infect. Dis. 2001;7:439–441. doi: 10.3201/eid0703.017312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurkovetskiy L., Wang X., Pascal K.E., Tomkins-Tinch C., Nyalile T.P., Wang Y.T., Baum A., Diehl W.E., Dauphin A., Carbone C., et al. Structural and functional analysis of the D614G SARS-CoV-2 spike protein variant. Cell. 2020;183:739–751.e8. doi: 10.1016/j.cell.2020.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D.M.E., Fouchier R.A.M. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. New Engl J Med. 2012;367:1814–1820. doi: 10.1056/nejmoa1211721. [DOI] [PubMed] [Google Scholar]
- Zepeda Mendoza M.L., Xiong Z., Escalera-Zamudio M., Runge A.K., Thézé J., Streicker D., Frank H.K., Loza-Rubio E., Liu S., Ryder O.A., et al. Hologenomic adaptations underlying the evolution of sanguivory in the common vampire bat. Nat Ecol Evol. 2018;2:659–668. doi: 10.1038/s41559-018-0476-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai X., Sun J., Yan Z., Zhang J., Zhao J., Zhao Z., Gao Q., He W.T., Veit M., Su S. Comparison of severe acute respiratory syndrome coronavirus 2 spike protein binding to ACE2 receptors from human, pets, farm animals, and putative intermediate hosts. J. Virol. 2020;94:e00831-20. doi: 10.1128/jvi.00831-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.Z., Holmes E.C. A genomic perspective on the origin and emergence of SARS-CoV-2. Cell. 2020;181:223–227. doi: 10.1016/j.cell.2020.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Wang J.B., Kuang D.X., Xu J.W., Yang M.L., Ma C.X., Zhao S.W., Li J.M., Long H.T., Ding K.Y., et al. Susceptibility of tree shrew to SARS-CoV-2 infection. Sci. Rep. 2020;10:16007. doi: 10.1038/s41598-020-72563-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M., Zhao X., Zheng S., Chen D., Du P., Li X., Jiang D., Guo J.T., Zeng H., Lin H. Bat SARS-Like WIV1 coronavirus uses the ACE2 of multiple animal species as receptor and evades IFITM3 restriction via TMPRSS2 activation of membrane fusion. Emerg Microbes Infect. 2020;9:1567–1579. doi: 10.1080/22221751.2020.1787797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Chen X., Hu T., Li J., Song H., Liu Y.R., Wang P.H., Liu D., Yang J., Holmes E.C., et al. A novel bat coronavirus closely related to SARS-CoV-2 contains natural insertions at the S1/S2 cleavage site of the spike protein. Curr. Biol. 2020;30:3896. doi: 10.1016/j.cub.2020.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Li Q.N., Su J.N., Chen G.H., Wu Z.X., Luo Y., Wu R.T., Sun Y., Lan T., Ma J.Y. The re-emerging of SADS-CoV infection in pig herds in Southern China. Transbound Emerg Dis. 2019;66:2180–2183. doi: 10.1111/tbed.13270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Fan H., Lan T., Yang X.L., Shi W.F., Zhang W., Zhu Y., Zhang Y.W., Xie Q.M., Mani S., et al. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature. 2018;556:255–258. doi: 10.1038/s41586-018-0010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu A., Jiang T., Hu T., Mi S., Zhao Z., Zhang F., Feng J., Fan Q., He B., Tu C. Molecular characterization of a novel bat-associated circovirus with a poly-T tract in the 3′ intergenic region. Virus Res. 2018;250:95–103. doi: 10.1016/j.virusres.2018.04.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.