Abstract
The decline of nicotinamide adenine dinucleotide (NAD) occurs in a variety of human pathologies including neurodegeneration. NAD-boosting agents can provide neuroprotective benefits. Here, we report the discovery and development of a class of potent activators (NATs) of nicotinamide phosphoribosyltransferase (NAMPT), the rate-limiting enzyme in the NAD salvage pathway. We obtained the crystal structure of NAMPT in complex with the NAT, which defined the allosteric action of NAT near the enzyme active site. The optimization of NAT further revealed the critical role of K189 residue in boosting NAMPT activity. NATs effectively increased intracellular levels of NAD and induced subsequent metabolic and transcriptional reprogramming. Importantly, NATs exhibited strong neuroprotective efficacy in a mouse model of chemotherapy-induced peripheral neuropathy (CIPN) without any overt toxicity. These findings demonstrate the potential of NATs in the treatment of neurodegenerative diseases or conditions associated with NAD level decline.
Subject terms: X-ray crystallography, Neural stem cells
Introduction
Nicotinamide adenine dinucleotide (NAD) is a widely distributed metabolite that has important cellular functions.1 NAD serves as a coenzyme for hundreds of dehydrogenases in redox reactions wherein the transfer of a hydride from a substrate to the oxidized form NAD (NAD+) to form the reduced form NADH, a universal electron carrier. This flow of electrons occurs in many metabolic pathways, in particular, driving energy production. For example, NAD accepts electrons to yield NADH during glycolysis or the TCA cycle. NADH formed from these catabolic pathways carries electrons to the first step of the electron transport chain in mitochondria, which initiates the proton pumping across membranes for maximal ATP generation via oxidative phosphorylation. NAD also serves as a non-redox cofactor or substrate for some NAD-dependent enzymes that modulate diverse cellular processes. Thus, tight maintenance of NAD homeostasis is pivotal for proper cellular functions. The decline in cellular or tissue NAD levels has been associated with aging and the pathophysiology of a variety of human diseases or conditions such as neurodegeneration, metabolic syndrome, and cancer.2
NAD can be synthesized de novo from tryptophan via the kynurenine pathway, as well as via the salvage pathway from nicotinamide (NAM) and the Preiss-Handler pathway from nicotinic acid (NA).3 Among these pathways, mammals rely primarily on NAD salvage from NAM, in which the rate-limiting enzyme nicotinamide phosphoribosyltransferase (NAMPT) catalyzes the condensation of NAM and phosphoribosyl pyrophosphate (PRPP) into nicotinamide mononucleotide (NMN), followed by the production of NAD via nicotinamide mononucleotide adenylyltransferase (NMNAT).4 NMNAT enzymes are involved in both de novo NAD synthesis and NAD salvage from nicotinamide. Thus, NAMPT and NMNAT are essential for maintaining adequate intracellular NAD levels.5,6
Drug discovery and development against neurodegenerative diseases are known to be challenging with an extremely low success rate. Recent studies have suggested NAD as a central modulator that controls the health of damaged or diseased neurons.2 Deletion of NAMPT or NMNAT in neurons of adult mice leads to neurodegeneration and death.7,8 Targeting NAD metabolism can otherwise improve neuronal cell health, memory, and cognitive function in disease animal models. To this end, supplementation of NAD precursors,9,10 enhancement of NAD biosynthesis,11,12 and inhibition of NAD consumption have been used.13,14 In this context, small-molecule NAMPT activators may hold promise in transforming the treatment of a broad range of neuronal disorders associated with impaired NAD production. Here, we report the discovery of a chemical class of NAMPT activator (NAT) that has the ability to promote the proliferation of neural stem cells in vitro and exhibit neuroprotective efficacy in a preclinical mouse chemotherapy-induced peripheral neuropathy (CIPN) model. We defined the structural basis for NAMPT activation and modulation by the NAT chemicals. This work provides a foundation for the development of new therapeutic agents against neurodegeneration.
Results
Identification of a NAT from a high-throughput screening
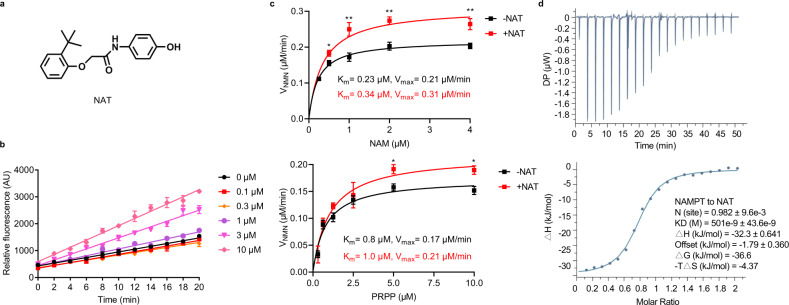
We embarked on target-based drug discovery using a high-throughput in vitro recombinant enzyme assay to identify compounds capable of activating NAMPT (Supplementary information, Fig. S1a). NAMPT enzyme reactions contained three enzymes, NAMPT, NMNAT1, and alcohol dehydrogenase (ADH). NAMPT catalyzes NMN that is subsequently converted into NAD by NMNAT1, and ADH converts NAD into the fluorescent end product, NADH. We screened roughly 50,000 synthetic chemicals, and several compounds were active in this triply-coupled NAMPT enzyme assay. Among them, we selected a compound as the potential NAT for further study because it robustly stimulated NAMPT activity, but was inactive in the NMNAT assay (Fig. 1a; Supplementary information, Fig. S1b, c). Furthermore, the NAT augmented NAMPT activity in a dose-dependent manner in both the triply-coupled NAMPT enzyme assay (Fig. 1b) and the direct NAMPT assay (Supplementary information, Fig. S1d). We measured the steady-state kinetics of NAMPT enzyme reactions. The addition of NAT markedly increased the catalytic activity (Vmax) of NAMPT with higher Michaelis constants (Km) for both substrates, NAM, and PRPP (Fig. 1c). We also performed an enzyme kinetic study on a recently reported NAMPT activator SBI-797812.15 As shown in Supplementary information, Fig. S1e, NAMPT enzyme reaction velocity peaked at around 5 μM NAM but decreased at a higher substrate concentration in the direct NAMPT assay. Interestingly, SBI-797812 reduced the enzyme reaction rate at low NAM concentrations. It only exhibited NAMPT activation at high NAM concentration, which appeared to be a relieving effect of substrate inhibition.
Fig. 1. NAT binds and directly activates NAMPT.
a Chemical structure of NAT. b Dose-dependent activation of NAMPT by NAT. A triply-coupled enzyme assay was performed to monitor the conversion of NAM to NADH. The first enzyme, NAMPT, converted NAM to NMN. The second enzyme, NMNAT1, converted NMN to NAD. The third enzyme, ADH, converted NAD to NADH. The reactions were carried out at room temperature for 20 min in the presence of increasing concentrations of NAT. Fluorescence of NADH was measured at Ex340/Em445 (y-axis). Progress curves of the enzyme reactions were plotted and data were analyzed with GraphPad Prism. Data are represented as means ± SEM from three replicates in a representative experiment. n = 3 independent experiments. c Effect of NAT on the NAMPT enzyme kinetics in the direct NAMPT assay. Michaelis–Menten curves for NAMPT reactions were plotted in the presence or absence of 10 μM NAT. The velocity of NMN production is shown on the y-axis as a function of substrate concentration (x-axis). Upper panel: Michaelis–Menten curves to determine the kinetic parameters for substrate NAM. Reactions contained the indicated concentrations of NAM and 10 μM PRPP. Lower panel: Michaelis–Menten curves to determine the kinetic parameters for substrate PRPP. Reactions contained the indicated concentrations of PRPP and 5 μM NAM. Vmax, maximum reaction rate; Km, substrate affinity. All error bars represent SEM from three replicates. Two-tailed t-test, *P < 0.05, **P < 0.01. d Binding of NAT to recombinant NAMPT as measured by isothermal titration calorimetry (ITC). A total of 200 μM recombinant NAMPT was titrated into the sample cell containing 20 μM NAT. The data shown here is a representative figure from three independent experiments. Top panel, the integrated heat signatures. Bottom panel, the fitted curves using one-site mode. KD, the dissociation constant; ΔG, change in Gibbs energy of binding; ΔH enthalpy; -TΔS, the entropy contribution to Gibbs energy; N, stoichiometry. Data were analyzed with MicroCal PEAQ-ITC Analysis software.
Next, we tested the direct binding of the NAT chemical to the purified NAMPT enzyme by isothermal titration calorimetry (ITC). The binding constant (KD) of the NAT chemical to NAMPT was determined to be about 500 nM. The stoichiometry analysis revealed that NAT bound to NAMPT at a 1:1 ratio (Fig. 1d). We also characterized the real-time binding kinetics of NAT to NAMPT using Surface Plasmon Resonance (SPR) assay. We failed to determine either Ka or KD, as both association of NAT to NAMPT and dissociation of NAT from NAMPT were too rapid, in a similar manner as in NMN binding to the enzyme (Supplementary information, Fig. S1f). This result suggests that NAT may function as a reversible activator during the kinetic assays.
The co-crystal structure of NAMPT with NAT
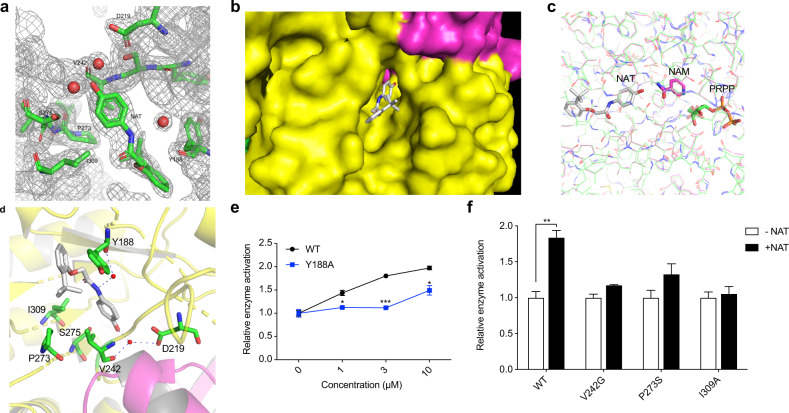
NAMPT is active in the dimeric form, and the active site is in a channel located at the dimer interface.16,17 We co-crystallized wild-type human NAMPT with NAT to determine the structural basis for NAT activation of NAMPT. The structure was solved and refined to resolutions of 2.2 Å (Supplementary information, Table S1; Fig. 2a). There were four copies of the NAMPT–NAT complex in the crystallographic unit (Supplementary information, Fig. S2a). NAT sits at one end of the channel (Fig. 2b). Our structural comparisons demonstrate that NAT is localized slightly away from the active site, overlapping with neither the substrates nor the product of the enzyme18 (Fig. 2c). The hydroxyl terminus of NAT is adjacent to the amide group of NAM and NMN, which slightly affects the enzyme binding to its substrate NAM, as shown in the above-mentioned kinetic results (Fig. 1c). This hydroxyl oxygen atom of NAT forms a hydrogen bond with a water molecule, which is also hydrogen-bonded with the side chains of residues V242 and D219. The interaction between the center amide nitrogen of NAT and Y188 is also via a water molecule (Fig. 2a, d). NAT binding does not cause the overall architectural change of the enzyme. However, some residues near NAT in the catalytic channel show conformational change upon activator binding (Supplementary information, Fig. S2b). Based on these structural observations, we hypothesized that the conformational changes seen in these residues might be necessary for NAT binding and NAMPT activation. Our mutagenesis and biochemical experiments confirmed the functional importance of some residues located in the NAT binding pocket. As shown in Fig. 2e, f, mutations in the binding site such as Y188A, V242G, P273S, and I309A largely impeded the enzyme activation by NAT, while these mutations did not affect the NAMPT enzymatic activity. Together, these data demonstrate that NAT may act as an allosteric activator of NAMPT, working adjacent to the active site.
Fig. 2. X-ray crystal structure of purified, recombinant NAMPT bound to the NAT chemical.
a The 2Fo-Fc electron density map for NAT and its surrounding molecules at 2.2 Å resolution. The contour level is at 1 σ. b A surface display for the NAT-binding at the dimer interface of NAMPT. NAT is shown as sticks in gray. Two monomers of NAMPT are colored in yellow and rose, respectively. c Comparison of the binding site of NAT on NAMPT with that of NAM and PRPP. NAT and the corresponding NAMPT protein structure are shown in gray. NAM, rose; PRPP, green. d Ribbon diagram showing the NAT-binding site of NAMPT. NAT is shown as sticks in gray with hydrogen bonds represented as dashed lines. The water molecules are shown as small red spheres. The nearby residues are shown as sticks in green. The two NAMPT monomers are colored yellow and pink, respectively. e, f Sensitivity of the wild-type NAMPT and mutants to NAT. Enzyme activity was measured by the direct NAMPT assay. 0.1 μM recombinant protein of wild type NAMPT or mutant Y188A was incubated with the indicated concentrations of NAT (e). The enzyme activities of wild-type or the indicated NAMPT mutants were assayed in the absence or presence of 3 μM NAT (f).
Compound optimization of NAT
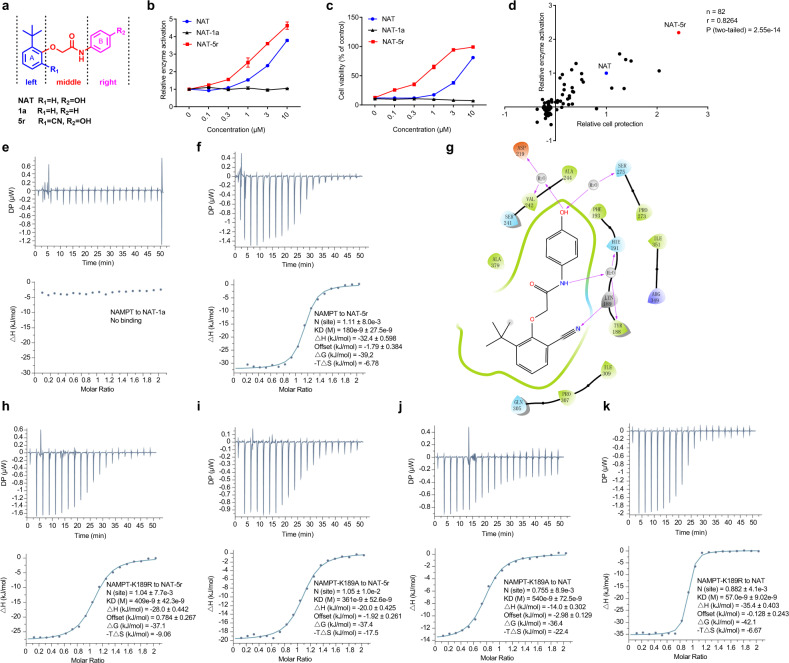
To improve the potency and pharmacologic properties of NAT, we conducted a systematic structural modification and optimization. Structurally, we divided NAT into three domains: the left A ring, the right B ring, and the middle linker (Fig. 3a). The structural variants on each domain were explored carefully, considering several factors such as hydrogen bonding effect, hydrophilic properties, and bioisosteric replacement. We synthesized 81 NAT derivatives in five rounds of structural optimization. These compounds were submitted to the structural-activity relationship study (SAR) to assess enzyme activation by these compounds as performed in the original screen (Supplementary information, Table S2). The majority were less active than NAT. Notably, elimination or replacement of the hydroxyl group (R2) from the right B ring generally abolished the activity. We used one such compound NAT-1a (Fig. 3a, b) as a negative control in later experiments. By contrast, several variants exhibited activity enhancement. The most active variant (NAT-5r) has an additional cyano group on the left A ring (Fig. 3a, b). Like NAT, NAT-5r had no effect on NMNAT1 enzyme activity (Supplementary information, Fig. S1c).
Fig. 3. Identification of NAT-5r led by a hit-to-lead optimization effort.
a Generalized structure of NATs with three regions denoted. b Dose-dependent activation of NAMPT by NAT and NAT-5r, but not NAT-1a. A triply-coupled NAMPT assay was performed in the presence of the indicated concentration of the compounds. Reaction rates were normalized to the vehicle control. Data were analyzed and dose-response curves were plotted with GraphPad Prism software. All error bars represent SEM from three replicates. c, d NATs relieve the cytotoxicity mediated by a NAMPT inhibitor FK866. c U2OS cells were treated with increasing concentrations of NAT, NAT-1a, or NAT-5r for 2 h prior to incubation with 10 nM FK866 for 72 h (n = 3 biological replicates). Cell viability is calculated by comparing the cell survival of each sample with that of DMSO-treated control. d Scatterplot reveals a significant correlation between the protective activity of 82 NAT compounds from FK866-mediated toxicity and their ability to activate NAMPT. The activity in both assays is represented by the relative Area Under Curve (AUC) of the dose-response curve of each test compound compared to that of NAT. Pearson correlation coefficient (r) and two-tailed P-value were determined by GraphPad Prism and Spearman Rank Correlation software. See also Supplementary information, Table S2. The binding affinity of NAT-1a (e) or NAT-5r (f) to recombinant NAMPT was measured using ITC as described in Fig. 1d. g Schematic drawing of the interactions between NAT-5r and NAMPT. Two-dimensional ligand-interaction diagrams of the induced fit docking model of NAMPT bound to NAT-5r were generated using Schrodinger Maestro software. Negatively charged residues are colored orange, positively charged residues are colored blue or gray, polar residues are colored light blue, hydrophobic residues are colored green, and water molecules are colored light gray. H-bond interactions to the amino acid side chain or main chain are shown as pink arrows, pointing towards the H-bond acceptor. ITC analysis of binding between NAT-5r and NAMPT-K189R (h) or NAMPT-K189A (i). ITC analysis of binding between NAT and NAMPT-K189A (j) or NAMPT-K189R (k). Top, raw data. Bottom, the integrated heat signatures and fitted curves. KD, the binding constant; ΔG, change in Gibbs energy of binding; ΔH enthalpy; −TΔS, the entropy contribution to Gibbs energy; N, stoichiometry. Data are analyzed with MicroCal PEAQ-ITC Analysis software.
We wondered whether active variants of NAT or NAT might counteract NAMPT inhibition. The compound FK866 is known to be a potent inhibitor of the NAMPT enzyme.19 Administration of FK866 in cultured cells causes the depletion of intracellular NAD levels and subsequent cell death. We compared the activity of NAT, NAT-1a, and NAT-5r in the FK866-mediated cytotoxicity assay. As shown in Fig. 3c, NAT blocked cell death induced by FK866 treatment in a dose-dependent manner, and NAT-5r was more active than NAT, whereas NAT-1a was essentially inactive. This same ranking of activity has been observed in both assays — stimulating NAMPT enzyme, and protecting cultured cells from FK866-mediated toxicity. Next, we assayed NAT and its 81 derivatives to measure the degree of protection from the NAMPT inhibitor FK866 and to test whether there might be a correlation between NAMPT activation and cell protection against FK866 (Supplementary information, Table S2). The scatter plot shown in Fig. 3d revealed a significant correlation between these two activities from the independent assays (Spearman’s rank correlation coefficient r = 0.83, P-value = 2.55e-14). Thus, we can predict with high confidence that active variants of NAT can block FK866-induced cell death but inactive variants cannot. Structural comparison of NAMPT-NAT and NAMPT-FK866 (PDB: 2GVJ) complexes predicts that NAT should be competitive against the binding of FK866 to NAMPT, as NAT and FK866 binding sites partially overlapped near the active site (Supplementary information, Fig. S2c). The protective activity of NAT variants might also reflect their abilities in binding to NAMPT.
NAT-1a showed no evidence of binding to NAMPT in the ITC assay, confirming the hydroxyl group as the key hydrogen-bonding contact in the X-ray crystal structure (Fig. 3e). By contrast, NAT-5r showed tighter binding to NAMPT than the original compound NAT, as its KD was reduced nearly threefold to about 180 nM (Fig. 3f). Molecular docking was used based on the crystal structure of the NAMPT-NAT complex to examine the binding mode of NAT-5r. In the docking model of the NAMPT-NAT-5r complex, besides the existing interactions between NAMPT and NAT, the cyano group of NAT-5r is directly hydrogen-bonded to residue K189, causing a different orientation of the A ring of the compound (Fig. 3g; Supplementary information, Fig. S3). Notably, K189 exhibited a conformational change upon binding with NAT in comparison to its structure in the free enzyme (Supplementary information, Fig. S2b). Furthermore, this residue was critical for NAMPT enzyme activity, as both mutants of K189R and K189A were inactive. Using ITC as a binding assay, we observed that both mutations reduced the binding affinity of NAT-5r (Fig. 3h, i). These data confirm that the additional hydrogen bond formed between the cyano group on NAT-5r and the K189 on NAMPT contributes to the enhanced binding. Strikingly, K189A mutation did not affect the binding affinity for NAT compared with wild-type NAMPT (Fig. 3j), whereas introducing a longer side chain as K189R led to about eight-fold higher affinity to NAT (KD = 60 nM) (Fig. 3k). It appears that the extra side chain on the arginine may bring K189R closer to NAT, like what the cyano group of NAT-5r does to wild-type NAMPT, hence strengthening the interactions. Taken together, our SAR results, together with the structural and mutagenesis studies, demonstrate that phenolic hydroxyl in the right fragment plays a vital role in interacting with NAMPT and the middle linker is incompatible with structural modification. Whereas the addition of an optimal group on the left A ring of NAT can lead to much tighter binding and more potent activators, providing evidence that modifications on the left domain are promising.
Elevation of cellular NAD salvage and dynamic metabolic reprogramming in naïve cells
To assess the cellular effects of NATs, we first tested whether active NATs have overt cytotoxicity under normal conditions. As shown in Supplementary information, Fig. S4a–d, when the compounds were administered up to a concentration of 10 μM, for the concentration range we had observed robust NAMPT activation, NAT and NAT-5r exerted no effect on the viability of multiple cell lines such as liver carcinoma (HepG2), osteosarcoma (U2OS), glioblastoma (T98G), and neuroblastoma (SH-SY5Y). Neither NAT nor NAT-5r affected cell proliferation (Supplementary information, Fig. S4e).
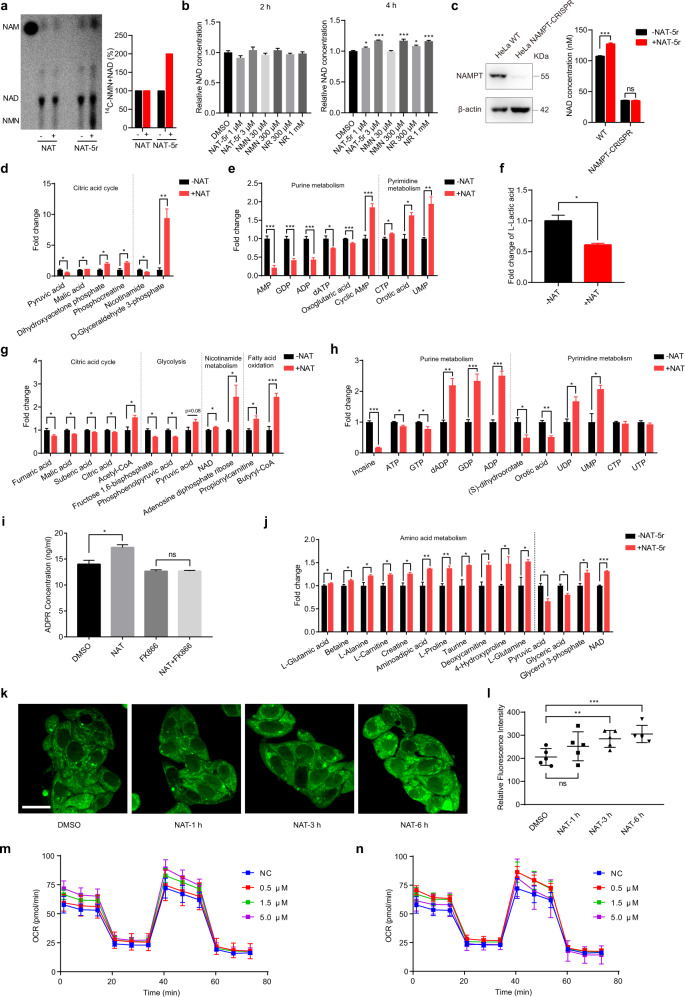
NAM can be sequentially converted into NMN and NAD in cells via the NAMPT-controlled salvage pathway.3 Having observed that active NAT variants bind to NAMPT to stimulate its enzymatic activity, we then tested whether these compounds could enhance the flux from NAM to NAD through the salvage pathway in naïve cells without any stress stimulus. 14C-labeled NAM was administered to cultured HepG2 cells and assayed by thin-layer chromatography (TLC) followed by autoradiography. Generated NMN might be rapidly converted to NAD, and thus was not detected on the TLC plate. We observed little if any change in NAD production in NAT-treated cells compared to control cells. However, upon treatment of a more active variant NAT-5r, a significant increase in the conversion of radiolabeled NAM into NMN and NAD was observed (Fig. 4a) while cellular NAMPT protein levels were unaffected (Supplementary information, Fig. S5a). We next compared NAT-5r with NAD precursors such as NMN and NR in boosting NAD in HepG2 cells. Treatment of NAT-5r at 3 μM concentrations for 4 h induced significant elevation of cellular NAD, to the extent comparable to NMN and NR administration at 300 μM and 1 mM, respectively (Fig. 4b). To investigate whether the NAD-boosting effects induced by NAT-5r are indeed dependent on NAMPT, we knocked out the NAMPT gene in HeLa cells using CRISPR-Cas9. We used lentiviral delivery of Cas9 and sgRNA targeting NAMPT in HeLa cells, followed by selection with puromycin for 14 days. CRISPRs depleted NAMPT in the majority of cells (Fig. 4c). We found that NAD levels in NAMPT-CRISPR cells were much lower than that of wild-type cells (NAMPT-WT) (Fig. 4c). NAT-5r treatment significantly enhanced NAD content in NAMPT-WT cells, but not in NAMPT-CRISPR cells (Fig. 4c), confirming that NATs elevate cellular NAD through activating NAMPT. However, we later failed to establish a pure NAMPT-KO cell line from a single clone. The western blot in Fig. 4c suggested that the pool of NAMPT-CRISPR cells appeared to contain a very small fraction of NAMPT-expressing cells. These cells might secret extracellular NAMPT (eNAMPT),20 which could convert the NAM in the culture medium to NMN, in turn supporting the cell survival of neighboring NAMPT-KO cells.
Fig. 4. NATs enhance NAD salvage and promote dynamic metabolic reprogramming.
a The effect of NATs on the flux of nicotinamide through the salvage pathway. HepG2 cells were pretreated with 5 μM NAT or NAT-5r for 24 h, followed by 4 h treatment with 14C-NAM. Metabolites were extracted and analyzed by thin-layer chromatography. A representative result from two independent experiments is shown. The first lane is the free 14C-NAM as a standard. Quantification of the relative intensities of NAD and NMN from the thin-layer chromatogram is shown in the right panel. b NAD-boosting effects of NAT-5r, NMN, and NR. HepG2 cells were treated with the indicated concentrations of NAT-5r, NMN, or NR for 2 h and 4 h. Cellular NAD was extracted with HCIO4 and measured with fluorometric NAD assay. All error bars represent SEM from three replicates. Two-tailed t-test, *P < 0.05, **P < 0.01, ***P < 0.005. c NAD enhancement induced by NAT-5r is dependent on NAMPT. Left panel, total cell lysates of WT HeLa and NAMPT-CRISPR HeLa were subjected to western blotting with an anti-NAMPT antibody. Actin served as a loading control. Right panel, WT HeLa and NAMPT-CRISPR HeLa cells were treated with DMSO or 10 μM NAT-5r for 6 h before cellular NAD was measured with fluorometric NAD assay. The change of cellular metabolic profile in HepG2 cells treated with NAT for 1.5 h (d–f), NAT for 6 h (g–i), or NAT-5r for 6 h (j). HepG2 cells in a 10-cm cell culture dish were treated with DMSO, 5 μM NAT or NAT-5r. In i, HepG2 cells were treated with either 5 μM NAT or 1 μM FK866 alone or both. Metabolites were extracted by frozen-thaw protocols and measured by mass spectrometry. Data were normalized to DMSO control and analyzed with GraphPad Prism. All error bars represent SEM from three replicates. Two-tailed t-test, *P < 0.05, **P < 0.01, ***P < 0.005. k Fluorescence imaging of fatty acid β-oxidation (FAO) activity in live HepG2 cells. The cells were treated with DMSO, 5 μM NAT for the indicated time before incubation with FAO probe for 30 min in HBS+ buffer at 37 °C. Scale bar, 20 μm. l Statistical analysis of FAO activity in HepG2 cells shown in k. The bar represents means ± SEM, n = 5 fields, **P < 0.01, ***P < 0.005. The OCR in NAT-5r-treated HepG2 cells. The real-time OCR of HepG2 cells treated with the indicated concentration of NAT-5r for 1 h (m) or 3 h (n). Each time point represents 8 replicates with means ± SEM.
Given that NAD plays a pivotal role in coordinating metabolic reprogramming,1 activation of NAMPT by NAT chemicals might lead to dynamic and sensitive changes of metabolic profile in cells. We tested this idea using an unbiased comparative metabolomics analysis by LC-MS. We quantitatively measured roughly two hundred representative metabolites extracted from HepG2 cells. The cells such as hepatocytes maintain total NAD and NADH levels fairly constant. We did not observe a significant change in cellular NAD levels upon NAT treatment for 1.5 h. However, we observed a significant increase of some metabolites in glycolysis and TCA, and a decrease of lactic acid (Fig. 4d–f), consistent with the role of NAD in energy metabolism. Interestingly, we also observed a significant increase of some pyrimidine nucleotides but a decrease of some purine nucleotides such as AMP, ADP, and GDP at this time point. Among them, ADP might be used for ATP production during glycolysis and oxidative phosphorylation. Lower concentrations of ADP and GDP otherwise favor the synthesis of PRPP, one of the substrates for NAMPT enzyme reaction. Treatment of NAT for 6 h induced a slight but significant elevation of intracellular NAD levels. As expected, we observed a statistically significant increase of pyruvic acid, acetyl-CoA, and butyryl-CoA, the key intermediates in the bioenergetic pathways (Fig. 4g). Notably, an accumulation of purine nucleotides such as ADP, GMP, GDP concurred with the decline of ATP, GTP and inosine levels, suggesting that the breakdown of ATP and GTP and the decrease of purine degradation might contribute to the increase of their corresponding mono- and diphosphate nucleotides (Fig. 4h). Interestingly, NAT treatment led to an increase of NAD catabolic product adenosine diphosphate ribose (ADPR), and this increase was abolished by co-treatment with an excess of FK866 (Fig. 4i), indicating that NAT induced increased NAD turnover. Consistent with the enhanced flux of nicotinamide through the salvage pathway, NAT-5r boosted NAD levels higher than NAT after 6 h treatment. Notably, NAT-5r induced significant elevation of various amino acids such as glutamine, proline, alanine, and creatine (Fig. 4j). We also observed the concomitant decrease of pyruvate, which might be consumed for amino acid synthesis, as amino acids such as alanine and valine are derived from pyruvate (Fig. 4j). Levels of autophagosome marker LC3-I and LC3-II remained unaffected by treatment with NAT-5r, excluding the involvement of autophagy in the increased levels of the amino acids (Supplementary information, Fig. S5b). However, increasing intracellular amino acids in NAT-5r-treated cells did not lead to enhanced protein synthesis, as the phosphorylation of ribosomal protein S6 kinase (S6K) remained unchanged (Supplementary information, Fig. S5b). Furthermore, neither NAT nor NAT-5r had any impact on total protein levels (Supplementary information, Fig. S5c). Thus, NAD augmentation appears to promote energy and amino acid metabolism, but not protein synthesis or cell proliferation in naïve cells.
We further quantify the kinetics of the metabolic reprogramming by NAT. Metabolites related to fatty acid beta-oxidation (FAO) are robustly increased under NAT treatment (Fig. 4g). We wonder whether this reflects an increase in FAO metabolism. We synthesized a fluorescent probe to detect the metabolic activity of the FAO pathway in living cells.21 Consistent with our metabolomic data, NAT treatment induced a time-dependent increase in FAO metabolism in live HepG2 cells, indicated by significantly enhanced fluorescent probe signal in mitochondria (Fig. 4k, l). We next investigate the effect of NATs on the mitochondrial function of oxidative phosphorylation. Measurement of the oxygen consumption rate (OCR) using Seahorse revealed a small but dose-dependent increase of both basal and maximal mitochondrial respiration in NAT-5r-treated cells compared to control cells in 1 h treatment. But we observed no such change in 3 h treatment (Fig. 4m, n). Taken together, these results demonstrate the time course of metabolic remodeling, which is very dynamic and dependent on the potency and concentration of the activators.
Stimulation of proliferation and self-renewal of neural stem cells (NSCs)
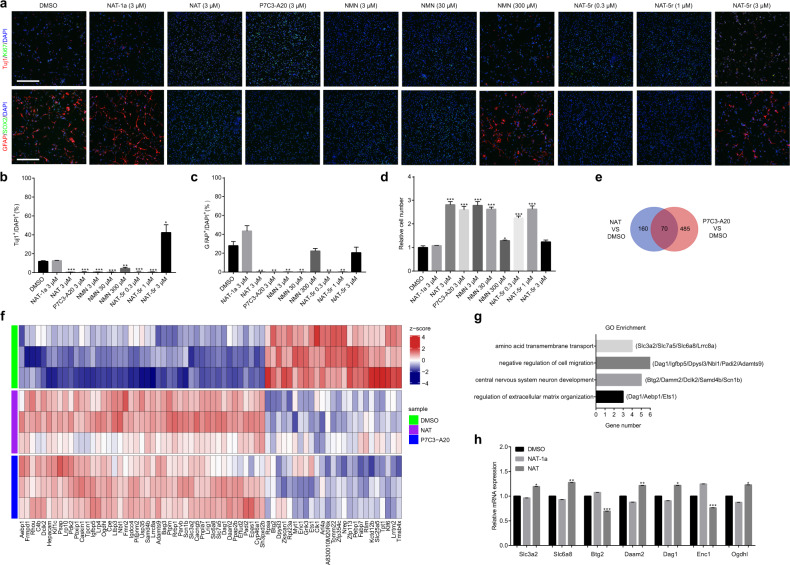
NSCs serve as a lifelong source of neurons and glia in various mammals.22,23 NAMPT helps maintain NAD levels in NSCs, which is critical for cell cycle progression.24 The NAMPT activator P7C3 chemical was initially discovered to be capable of enhancing hippocampal neurogenesis in adult mice,25 but the downstream effectors of NAMPT activation by P7C3 are not known. We sought to evaluate the pro-neurogenic activity of NAT and investigate the drug action in a facile system, in which we isolated and propagated NSCs/progenitors, and later induced them to differentiate in culture conditions.26,27 Briefly, we harvested cells from dissected hippocampus of mouse pup, seeded the cells to form what was known as neurospheres in vitro, mechanically dissociated after three divisions (DIV 3), and then maintained the cells as monolayers (Supplementary information, Fig. S6a). When grown in complete medium, all cells express proliferation marker, Ki67, and markers for the neuroepithelial stem cells, Nestin and SOX2. Upon withdrawal of basic fibroblast growth factor (bFGF) and epidermal growth factor (EGF) from the culture medium, cell proliferation was dramatically decreased (Supplementary information, Fig. S6b). We observed that many remaining NSCs differentiated to β3-Tubulin (Tuj1)-positive neurons, glial fibrillary acidic protein (GFAP)-positive astrocytes, or O4-positive oligodendrocytes (Supplementary information, Fig. S6c). Under such conditions, both NAT and P7C3-A20 largely inhibited the differentiation of NSCs to neurons (Tuj1+) or astrocytes (GFAP+), whereas inactive variant NAT-1a-treated NSCs proceeded with differentiation (Fig. 5a–c). NSCs maintained the self-renewal when exposed to low concentrations of NAT-5r but tilted to differentiation at higher concentrations, a perplexed hook-effect shared by NMN (Fig. 5a–c). Likewise, NAT, NAT-5r, NMN, and P7C3-A20 dramatically fostered NSCs proliferation compared with vehicle control and NAT-1a (Fig. 5d). Both NAT-5r and NMN elicited a downturn of cell proliferation at the concentration of 3 μM and 300 μM, respectively, when cell differentiation occurred (Fig. 5a–d), suggesting that this hook-effect of NAT-5r is likely due to the overactivation of NAMPT.
Fig. 5. NATs promote proliferation and maintain the self-renewal of NSCs.
a–d The effects of NATs, P7C3-A20, and NMN on the differentiation and proliferation of NSCs. NSCs were treated with the indicated concentrations of NAT-1a, NAT, P7C3-A20, NMN, or NAT-5r, and cultured in the growth factors drop-out medium for 3 days. a Representative images of NSCs in the differentiation assays. Newly formed neurons are identified as Tuj1+ cells and glial cells as GFAP+ cells. Scale bar, 200 μm. b Quantification of the proportion of Tuj1+ cells. c Quantification of the proportion of GFAP+ cells. d The relative cell number in each sample compared to that of DMSO-treated control. All data are presented as means ± SEM (n = 3 biological replicates, two-tailed t-test). *P < 0.05; **P < 0.01; ***P < 0.005. e NSCs treated with NAT or P7C3-A20 were subjected to RNA-seq analysis for differential expression of genes compared with vehicle control. The Venn chart summarizes the findings as common genes whose expression was significantly changed by both NAT and P7C3-A20 (P < 0.01). f Heatmap of the common genes in e. (n = 3 biological replicates). g GO Enrichment analysis of the common genes in e. h qRT-PCR validation of several genes (Slc3a2, Slc6a8, Btg2, Daam2, Dag1, Enc1, and Ogdhl) from e–g. NSCs were treated with DMSO, 3 μM NAT-1a, or NAT (n = 3 biological replicates, two-tailed t-test). Error bars represent SEM. *P < 0.05; **P < 0.01; ***P < 0.005.
To understand how NAMPT activation affects cell proliferation and cell fate decision of the NSCs in response to the cues of growth factors deficiency, we compared expression profiles of the NSCs treated with NAT and P7C3-A20 vs vehicle control by RNA sequencing (RNA-seq) analysis. There was only a slight overall change of transcriptome. As shown in Fig. 5e, f, seventy genes exhibit similar expression changes (P value < 0.01) caused by both NAT- and P7C3-A20-treatment. Consistent with the role of NAD in central energy metabolism, gene expression of oxoglutarate dehydrogenase-like (Ogdhl, α-ketoglutarate dehydrogenase-like) and pyruvate dehydrogenase kinase 2 (PDK2), which are important in the TCA cycle, were upregulated in response to NAMPT activation. Next, we performed Gene Ontology (GO) analysis to find gene categories most affected by these two compounds’ treatment. Gene Ontology categories ranked in the top 3 were amino acid transmembrane transport, negative regulation of cell migration, and central nervous system neuron development (Fig. 5g). The upregulation of a set of amino acids transmembrane transporters such as Slc3a2, Slc7a5, and Slc6a8, along with the above evidence of NAT-5r-enhanced production of selective amino acids in HepG2 cells, underscored the role of NAMPT activation in promoting amino acid availability for biosynthesis and energy. Notably, the expression of Btg2, an NGF-inducible anti-proliferative protein,28 was significantly decreased in both NAT- and P7C3-A20-treated NSCs. Besides, some regulators in nervous system development, such as Enc1,29 Dag1,30 and Daam1a,31 experienced gene expression changes, consistent with the inhibition of differentiation of NSCs by either NAT or P7C3-A20 (Fig. 5 f, g). These alterations were further confirmed by quantitative real-time PCR analysis using NAT-1a as the negative control (Fig. 5h). These results may also reflect the other critical signaling role of NAD via acting as a cofactor for various NAD-dependent enzymes such as sirtuins. Deacetylase activity of sirtuins changes the histone modifications and thus epigenetically regulates gene expression.32 Sirtuins also regulate the downstream transcriptional regulators including Forkhead box O (FOXOs) that play important roles in metabolism, stress resistance, and cell death.33,34 We treated NSCs with NATs, P7C3-A20, or NMN for 3 days. The acetylated/total FOXO1 ratio was reduced in NSCs treated with a high concentration of NAT-5r, P7C3-A20, and NMN (Supplementary information, Fig. S7), indicating activation of sirtuins by these NAD boosters.
Neuroprotective efficacy in a mouse model of CIPN
The improved derivatives of P7C3, including the P7C3-A20 and P7C3-S321 variants, have shown efficacy in various animal models of neurodegeneration.2,35 These studies suggest that small molecules activating NAMPT to maintain or elevate cellular NAD levels may be expected to display protective effects towards neurons. We, therefore, set out to assess the neuroprotective efficacy of NAT chemicals in an animal model of CIPN, a debilitating side effect of cancer treatment. CIPN-associated cancer drugs such as paclitaxel (PTX) and vinblastine can cause damage to the peripheral nerve. CIPN features a set of progressive, enduring, and disabling symptoms including pain, numbness, tingling, and sensitivity to cold in the hands and feet. CIPN afflicts between 30% and 40% of patients undergoing chemotherapy. But there is no effective treatment for CIPN, and current treatment can only help manage some of the symptoms.36
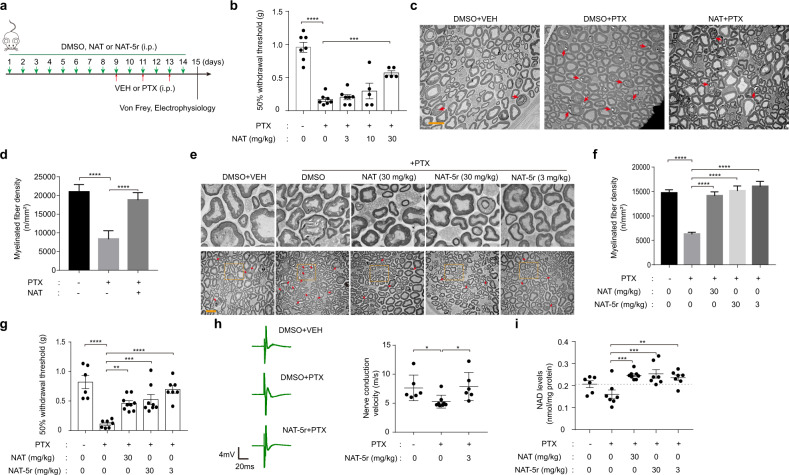
Pharmacokinetic analysis of NAT in plasma was undertaken when administered to mice via intraperitoneal, intravenous, or oral routes, for determination of its half-life, rate of clearance, and bioavailability in mice (Supplementary information, Fig. S8, a–d). NAT was 39.2% intraperitoneally bioavailable and 33.2% orally bioavailable. The terminal plasma half-life was 3.00, and 2.97 h after IP and oral delivery, respectively. These pharmacokinetic properties allowed us to conduct a dose-response experiment for the assessment of the neuroprotective efficacy of NAT in the mouse CIPN model. NAT was intraperitoneally injected daily at doses of 3, 10, or 30 mg/kg in C57BL/6 J mice for one week prior to the initiation of PTX treatment. Mice then received IP injection with PTX (18.3 mg/kg) on three alternate days. Two days after the last dose of PTX, we conducted a behavioral test to evaluate mechanical allodynia, a hallmark of CIPN, in mice (Fig. 6a). PTX treatments led to a marked reduction of the mechanical withdrawal threshold of the paw measured with the von Frey hair test. We observed an increased withdrawal threshold in a dose-dependent manner in mice pretreated with compound NAT (Fig. 6b). After behavioral analysis, sciatic nerves were recovered, stained, and sectioned for electron microscopy to capture images of the nerve sections in order to assess the morphology changes in the axon. As shown in Fig. 6c, d, mice treated with PTX manifested evident damage in axonal myelin sheaths, with significantly reduced density of myelinated fibers compared with vehicle control. However, pretreatment with 30 mg/kg NAT largely restored the density of myelinated fibers relative to the vehicle control group. These protective effects were not due to any drug interaction between NAT and PTX, as NAT did not affect PTX’s potency in killing cancer cells (Supplementary information, Fig. S8e). We next conducted a double-blinded test of NAT and a more active variant NAT-5r. Remarkably, the histology analysis of sciatic nerves demonstrated that treatment with both 3 mg/kg and 30 mg/kg NAT-5r fully protected myelin sheaths from damage, as effective as the treatment with 30 mg/kg NAT (Fig. 6e, f). The drug-mediated protection manifested in histology analysis was consistent with the behavioral assessment (Fig. 6g). Myelin sheaths facilitate the transmission of electrical impulses quickly and efficiently along the axon. Figure 6h showed that the nerve conduction velocities (NCVs) of these impulses in sciatic nerves were significantly slowed down in the PTX treatment group, which is likely due to myelin damage. By contrast, NCVs of the NAT-5r-treatment group were as quick as that of the vehicle-treated group. This functional result further supports the neuroprotective benefit of NATs.
Fig. 6. NAT protects peripheral sensory neurons from paclitaxel-(PTX) induced damage.
a Illustration of the experimental design of the CIPN model. b Mechanical withdrawal threshold of the hind paw of adult male C57BL/6 J mice determined from their response to von Frey filaments. Mice were treated with vehicle (DMSO) or 3 mg/kg, 10 mg/kg, or 30 mg/kg NAT (every day, intraperitoneal injection) for two weeks combined with vehicle or 18.3 mg/kg PTX (intraperitoneal injection) on day 9, 11, and 13. Data represent the mean sensitivity threshold. Bars represent mean ± SEM. Each symbol represents data from an individual mouse (n = 5–7). c Transmission electron microscopy (TEM) images of nerve fibers in the cross-sections of mouse sciatic nerves. Mice were treated with vehicle or 30 mg/kg NAT combined with vehicle or PTX. Red arrows indicate the injured nerve fiber. Scale bar, 10 μm. d Quantification of the density of the intact nerve fibers in the TEM images in c. Data represent mean fiber density. Bars represent means ± SEM. e TEM images of the cross-sections of mouse sciatic nerves. Mice were treated with vehicle or 30 mg/kg NAT, 3 mg/kg NAT-5r, or 30 mg/kg NAT-5r in combination with vehicle or PTX. Arrows indicate the injured nerve fiber. Scale bar, 10 μm. The upper images are the magnified TEM image from the indicated regions within the boxes on the corresponding lower images. f Quantification of the myelinated fiber density in the TEM images in e. Data represent mean fiber density. Bars represent means ± SEM. g Paw withdrawal threshold of adult male C57BL/6J mice treated as in e. Data represent the mean sensitivity threshold. Bars represent means ± SEM. Each symbol represents data from an individual mouse (n = 6–9). h Sciatic nerve conduction velocity (NCV) of adult male C57BL/6J mice treated with vehicle or 3 mg/kg NAT-5r in combination with vehicle or PTX. Left panel, representative recording of nerve action potentials. Right panel, the mean NCV. Each symbol represents data from an individual mouse (n = 6–8). Bars represent means ± SEM. i Tissue NAD levels of the sciatic nerves of adult male C57BL/6 J mice treated as in e. Data represent the mean NAD levels. Bars represent the means ± SEM. Each symbol represents data from an individual mouse (n = 6–9). *P < 0.05; **P < 0.01; ***P < 0.005; ****P < 0.001 vs PTX-only treatment group, determined by one-way ANOVA and Dunnett’s multiple comparisons test.
Importantly, mice injected with 30 mg/kg NAT or NAT-5r daily for two weeks showed little if any change of body weight (Supplementary information, Fig. S8f), and no signs of acute or chronic liver toxicity were observed (Supplementary information, Fig. S8g). We extracted NAD from a homogenized sciatic nerve to assess the in vivo NAMPT-stimulating effect by NAT chemicals. PTX treatment caused slightly reduced NAD levels compared to vehicle control. As expected, pretreatment with NAT or NAT-5r significantly elevated NAD in the sciatic nerve to the levels even slightly higher than the control group (Fig. 6i). These observations suggest that NAT chemicals protect peripheral sensory neurons against the neuropathic effects of PTX through enhanced NAD production.
Discussion
Here we report the discovery of a new chemical class (NATs) that binds and directly activates the nicotinamide phosphoribosyltransferase (NAMPT), the rate-limiting enzyme in the NAD salvage pathway. NAT was discovered from a target-based drug screen using the triply-coupled NAMPT enzyme assay in a high-throughput format. We show that NATs promoted the flux rate of NAM to NAD in the triply-coupled enzyme assay. Likewise, NAT activated NAMPT in a direct assay for the conversion of nicotinamide into NMN. NAT directly bound to NAMPT as revealed by ITC, and bound near the active site within the core channel of the enzyme as defined by X-ray crystallography of the NAMPT-NAT complex. This is, to our knowledge, the first report of the cocrystal structure of a small-molecule activator bound to NAMPT. This binding causes a slight conformational change in the active site, which may favor the condensation of nicotinamide and PRPP to form NMN. How this cooperative process happens awaits a further study of the enzyme reaction, such as pre-steady-state kinetics, in the presence of the activator. SAR studies of 81 derivatives of the NAT chemical yielded the NAT-5r variant, which has improved binding affinity to NAMPT, improved activation properties, and improved capacity to protect cultured U2OS cells from toxicity elicited by the FK866 inhibitor of NAMPT. The model of the NAMPT-NAT-5r complex from molecular docking indicates that NAT-5r binds to the enzyme analogously to NAT while featuring additional hydrogen-bonded interaction with K189, which may explain its enhanced activity and point to a critical direction for future structure-guided compound optimization. The binding mode between P7C3 and NAMPT is not known yet. Another NAMPT activator SBI-797812 structurally evolved from a potent competitive NAMPT inhibitor to a NAMPT activator, such that the compound is expected to bind to the active site of NAMPT. We showed that SBI-797812 has a dual action on NAMPT, inhibitory and stimulatory, dependent on the concentration of enzyme-substrate. As the chemical structure of NAT is distinct from either P7C3 or SBI-797812, we deduce that the mode of these compounds binding to NAMPT might be different. These independent studies led to the revelation of multiple ways to activate the NAMPT enzyme.
Additionally, this study offers progress toward an understanding of the functionality of activating NAD salvage and altering NAD metabolism under different cell states. While only slightly elevating intracellular NAD levels, NAMPT activation by NATs in naïve cells appears to significantly promote multiple catabolic pathways, including glycolysis, TCA cycle, and fatty acid oxidation, consistent with the pivotal role of NAD by Krebs et al.37 Oxidative phosphorylation is consequently increased, but only at the early stage of NATs treatment. The distinct metabolic profiles induced by the treatment of different NAT variants at different time points reflect the complex and dynamic metabolic reprogramming in response to NAMPT activation. For example, after NAT treatment the concentrations of purine nucleotides (GDP, ADP) were decreased in 1.5 h but increased in 6 h. Notably, we observed the significant accumulation of some nucleoside mono- and diphosphates in 6 h, without the concordant increase of the nucleoside triphosphates such as ATP, GTP, UTP, TTP, or CTP or cell proliferation. It is possible that the triphosphate nucleotides may be synthesized more but consumed more, resulting in little net change in the pool. Alternatively, the cells may lower those nucleotides in order to maintain normal REDOX. We also observed enhancement in amino acid metabolism upon NAMPT activation by NAT-5r. Yet neither NAT nor NAT-5r promotes cell proliferation in naïve or healthy cells, suggesting that the metabolic reprogramming in these cells may represent a preparatory state for energy production and biosynthesis when there is no need for energy replenishment or cell proliferation even upon NAMPT activation. On the other hand, we found that boosting NAD by NATs promotes proliferation of the neural stem cells under stress conditions, i.e., growth factors deprivation. The observed hook-effect of highly active NAT variants and the excess of NMN might be due to the feedback inhibition mechanisms for keeping NAD homeostasis. Thus, it is imperative to carefully determine the safe and efficacious dose of NAT-5r or NMN for the treatment of neurodegenerative disease. Besides, we observed the expression change of a handful of genes involved in amino acid transport, energy metabolism, cell cycle, stress response, cell differentiation, and cell death. Further investigation will be needed to fully understand the molecular basis underlying the intricate but concerted metabolic and transcriptional reprogramming in response to altered NAD metabolism.
NATs showed protective efficacy in an aggressive mouse model of CIPN, in agreement with a previous report of the resistance to PTX-induced neuropathy by the Wallerian degeneration slow (WldS) mice.38 Wallerian degeneration, first characterized in 1850,39 can be described as the degeneration process of the distal part of the axon when a nerve fiber is cut. The mutation in the WldS mice allows the ectopic expression of a chimeric fusion protein of NMNAT1 in the cytosol instead of the nucleus, leading to the enhanced NAD production in the axonal region of the nerve cell. The neuroprotective characteristics of the WldS mice are likely due to this mutation.40 Indeed, the overexpression of NAMPT and NMNAT1 can prevent axon degeneration.41,42 It has also been reported that sterile alpha and TIR motif-containing protein 1 (SARM1) mediates vincristine- and PTX-induced CIPN, as genetic deletion of SARM1 provides axon protection.43,44 SARM1 was discovered from a forward genetic screen in Drosophila by showing a similar phenotype of strong axon protection as WldS mutants. SARM1 acts as the executor of Wallerian degeneration,45 as it harbors the intrinsic NADase activity in its C-terminal Toll/interleukin-1 receptor (TIR) domain, which is activated to rapidly destruct NAD upon axonal damage.41 Our results suggest that NAD augmentation via NAMPT activation can oppose SARM1 activation, supporting the “induced NADase activation and NAD loss” model in SARM1-mediated axon degeneration. Additionally, compared to other cell types, neurons have high-energy demands and unique polarized structures. They face tremendous challenges for maintaining energy homeostasis, particularly in some peripheral nerves whose axons extend from several centimeters long to even up to a meter. Mitochondria function as powerhouses to generate ATP. Efficient long-distance mitochondrial mobilization and transport, which are driven by microtubule (MT)-based motors, is critical for neurons to meet altered energy requirements.46 Defects in mitochondrial dynamics and transport have been implicated in several major neurodegenerative diseases,47 possibly also in neuropathy induced by the MT-polymerizing drug PTX, which disrupts MT dynamics. NATs-mediated NAD replenishment may drive mitochondrial bioenergy production to sustain the energy needs of the injured neurons, via not only its redox carrier function for ATP production, but also the substrate-based activation of sirtuins, which acts on PGC-1α, the main co-activator of mitochondrial biogenesis.48 As important as understanding these mechanisms in vivo, exploring the possibility of NATs administration to mitigate other neurological lesions, including lesions in the CNS, will offer new therapeutic opportunities. Given the neuroprotective activity of P7C3 chemicals in several animal models of neurodegeneration in CNS, NATs are expected to have similar activities.
The replenishment of NAD is a plausible strategy in correcting bioenergetic or metabolic defects, which has prompted the direct supplementation of NAD precursors (e.g. niacin, NMN, nicotinamide riboside), and the modulation of NAD metabolism to boost NAD levels. Niacin and NMN have been used in clinical trials49,50 and NR has been successfully used in mouse models of ETC impairment.51 Administration of NMN, NR, and NAMPT activators replenishes NAD through the same factor, NMN. What is the advantage to use NAMPT activators instead of nutriceuticals such as nicotinamide riboside and NMN? First, though NR and NMN are readily incorporated into NAD in cell culture,52 NAT-5r appears to be much more potent than these nutriceuticals in boosting NAD in HepG2 cells. Second, bioavailability studies suggest that these nutriceuticals are metabolized quickly in mammalian blood and tissues.53,54 Oral administration fails to deliver intact NR or NMN molecules to tissues without breaking the nicotinamide-ribose bond, as NR is slowly converted into NAM, and NMN is first rapidly hydrolyzed to NR and then into NAM by brush border cells in the small intestine.55 Small-molecule NAMPT activators like NATs already have better bioavailability and have expanded space for further chemical modification to achieve more favorable ADME properties. Third, NMN and NR are relatively expensive even on the market as nutrient supplements due to limited resources. NATs can be easily synthesized at a low cost to be more affordable. Last but not least, NAMPT activity appears to be tightly controlled and subjected to negative feedback regulation by NMN and NAD18,56. Likewise, the action of NAMPT activators may be subjected to regulation at the rate-limiting step to meet cells’ needs under different physiological conditions, thus may prevent the side effects caused by the excess of NMN or NAD. NAMPT activators NATs have shown good efficacy in the animal model of CIPN, reinforcing that NAMPT can be targeted to intervene in NAD metabolism for therapeutic use. Given the guidance of the structure-based rational drug design, the NAT class of NAMPT-activating chemicals may be optimized to have improved potency and pharmacologic properties, thus holding promise in the treatment of diseases and conditions associated with impaired NAD homeostasis.
Materials and methods
Reagents
Chemical reagents HEPES (Cat. # H4034), Trizma base (Cat. # T1503), MgCl2 (Cat. # M8266), K2CO3 (Cat. # V900112), ATP (Cat. # A7699), PRPP (Cat. # P8296), nicotinamide (Cat. # 72340), NAD+ (Cat. # S8045), KOH (Cat. # 221473), acetophenone (Cat. # A10701), alcohol dehydrogenase (Cat. # A3263), resazurin (Cat. # R7017), semicarbazide (Cat. # S2201) were purchased from Sigma Aldrich. Formic acid (Cat. # F0513) was purchased from TCI chemicals. HClO4 (Cat. # 423096) was purchased from J&K Scientific. NMN (Cat. # BD116593) was purchased from Bidepharm. TRIzol (Cat. # 15596026) was purchased from Thermo Fisher Scientific. PrimeScript RT reagent Kit (Cat. # RR037A) was purchased from Takara Biomedical Technology. 2x SYBR Green qPCR Master Mix (Cat. # B21202) was purchased from Bimake. Mut Express II Fast Mutagenesis Kit (Cat. # C214) was purchased from Nanjing Vazyme biotechnology. M5 HiPer ECL Western HRP Substrate (Cat. # MF074) was purchased from Beijing Mei5bio. FK866 (Cat. # S2799) was purchased from Selleck. SBI-797812(Cat. # HY-126255) was purchased from MedChem Express.
Animals
Mice were 8–9-week-old males of the strain C57BL/6J from Beijing Vital River Laboratory Animal Technology Co., Ltd. All mice were housed in isolated ventilated cages (maximal six mice per cage) in a barrier facility at Tsinghua University. The mice were maintained on a 12/12-h light/dark cycle, 22–26 °C with sterile pellet food and water ad libitum. The laboratory animal facility has been accredited by Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC). The Institutional Animal Care and Use Committee (IACUC) of Tsinghua University approved all animal protocols used in this study.
Cell lines
All the cell lines (HepG2, U2OS, T98G, SH-SY5Y, and HeLa) used in this study were from ATCC and tested negative for mycoplasma contamination. The HepG2, U2OS, T98G, and HeLa cells were grown in DMEM medium (Sigma) supplemented with 10% fetal bovine serum (FBS, Gibco), 100 U/mL penicillin, and 100 μg/mL penicillin-streptomycin (Thermo Fisher). The SH-SY5Y cells were grown in DMEM-F12 medium (Sigma) supplemented with 15% FBS, 100 U/mL penicillin, and 100 μg/mL penicillin-streptomycin. The Cells were maintained in a humidified incubator at 37 °C and 5% CO2.
Brain-derived NSCs
We used newborn (2-day-old) C57BL/6J mice from the THU-LARC. The hippocampus from each hemisphere was micro-dissected and dissociated with enzymatic digestion with 0.25% trypsin-EDTA (Life Technologies) for 10 min at 37 °C. Cells were grown in Neurobasal™-A Medium (Thermo Fisher) supplemented with B27 (Thermo Fisher, 1:50), N2 (Thermo Fisher, 1:100), 0.6% glucose, penicillin-streptomycin–glutamine (Life Technologies, 1:100), GlutaMAX™ (Thermo Fisher, 1:100), insulin–transferrin–selenium (Life Technologies, 1:1,000), recombinant Human Epidermal Growth Factor (EGF, Gibco, 20 ng/mL), and recombinant Human Fibroblast Growth Factor Basic (bFGF, Gibco, 20 ng/mL). The cells were seeded on Poly-l-ornithine (Sigma, 50 μg/mL) and laminin (Sigma, 5 μg/mL) coated plate. For promoting differentiation, the culture medium was switched to a drop-out medium without bFGF or EGF.
Screening for small-molecule NAMPT activators
A high-throughput screen was performed in search of small-molecule activators of NAMPT enzyme from a chemical library composed of over 50,000 diverse compounds. Each compound was tested in a triply-coupled NAMPT assay that contains three enzymes, NAMPT, NMNAT1, and ADH. The first enzyme, NAMPT, converted NAM to NMN. The second enzyme, NMNAT1, converted NMN to NAD. The third enzyme, ADH, converted NAD to NADH. The reaction was prepared to contain 50 mM Tris (pH 8.0), 12 mM MgCl2, 15 μM phosphoribosyl pyrophosphate (PRPP), 1.5% ethanol, 2.5 mM ATP, 10 mM semicarbazide, 0.2% bovine serum albumin (BSA), 2.4 μg/mL NMNAT1, and 60 units/mL ADH. The mixture was dispensed into 384-well plates. Individual compounds were added to each well at a final concentration of 10 μM using an Echo 550 robotic liquid handler. The reaction was initiated by the addition of nicotinamide at 200 μM followed by gentle mixing in the plate. Fluorescence of NADH at Ex340nm/Em445nm was measured continuously for 30 min to establish the reaction velocity.
Protein expression and purification
cDNA corresponding to full-length human NAMPT was cloned into pET21a to produce fusion proteins with a 6× His tag at C-terminus. Mutagenic DNA primers to mutate individual base pairs were designed to obtain NAMPT mutants. Mutagenesis was carried out by using Quikchange XL Kit (Agilent). The recombinant proteins were expressed in E. coli BL21 (DE3) strain. Cells were induced with 0.3 mM IPTG at 18 °C overnight for protein expression and lysed in lysis buffer containing 50 mM Tris, pH 7.5, 500 mM NaCl, 10 mM imidazole, 5 mM β-ME. The procedure for protein purification included Ni-NTA agarose beads (Qiagen), Hi-trap Q column, and gel filtration column.
Direct NAMPT enzyme assay
NAMPT activity was determined by a fluorometric assay as described previously.57 The reaction buffer contained 50 mM Tris (pH 8.0), 12 mM MgCl2, 15 μM PRPP, 2.5 mM ATP, and 0.2% bovine serum albumin (BSA). 0.1 μM purified recombinant NAMPT enzyme was added to the reaction, which was initiated by adding 5 μM nicotinamide before running for 2, 4, 6, and 8 min intervals at 24 °C. The NMN product was measured by adding 25 μL of the reaction mixture to 10 μL of 20% acetophenone in dimethyl sulfoxide (DMSO), and 10 μL of 2 M potassium hydroxide. The mixture was incubated on ice for 2 min before the addition of 45 μL of 88% formic acid. After incubation at 37 °C for 20 min, the solution was transferred into a black, flat-bottom 96-well microtiter plate. Fluorescence was measured using a BioTek Cytation 3 plate reader with excitation and emission wavelengths set to 382 nm and 445 nm, respectively.
ITC
Thermodynamic parameters of NATs binding to NAMPT were measured using a MicroCal PEAQ-ITC calorimeter. Purified NAMPT protein was concentrated in an Amicon Ultra-4 Centrifugal Filter (Merck Millipore) against binding buffer (50 mM Tris pH 8.0, 200 mM NaCl, 1 mM TCEP, 1 mM MgCl2). The ultrafiltrate was subsequently used to prepare a matched compound solution. The titrations were carried out in the reverse mode by injecting 200 μM NAMPT into 20 μM NATs at 30 °C with the spacing of 150 sec between injections. The reference power of the instrument was set at 10 μCal/sec. The data were analyzed using MicroCal PEAQ-ITC Analysis Software.
SPR
SPR assays were performed on the Biacore 8 K + system. The CM5 sensor chip was purchased from GE Cytiva. NAMPT was immobilized at 50 μg/mL in sodium acetate buffer (pH 4.5) to form 12,000 immobilized resonance units in the CM5 chip. SPR experiments were carried out in TBS-T buffer (20 mM Tris, 200 mM NaCl, 1 mM TCEP, 0.05% Tween-20, 5% DMSO, pH 8.0) with a contact time of 60 s and a dissociation time of 120 s at a flow rate of 30 μL/min. The binding of NAT to NAMPT was monitored by applying NAT at varying concentrations (0.3125-10 μM) to the NAMPT-immobilized chip. The data were analyzed using Biacore Insight Evaluation Software.
Protein crystallization and data collection
Purified wild-type human NAMPT and NAT were mixed at 1:1 stoichiometry and concentrated to 10 mg/mL. Crystals of the complex were obtained using a precipitant solution containing 50 mM phosphate buffer (pH 9.0), 27% (w/v) PEG 3,350, 200 mM NaCl, 5 mM DTT, and 5 mM BaCl2. Crystals were grown at 20 °C using the hanging drop vapor diffusion method. Diffraction data were collected under cryo-conditions at 0.9792 Å at the Shanghai Synchrotron Radiation Facility (SSRF) beamline BL17U1 of the National Center for Protein Science, Shanghai.
Structure determination and refinement
The diffraction images were processed and scaled with the HKL 2000. The structure of NAT-bound NAMPT was determined by molecular replacement in CCP4 using the published structure of human NAMPT (PDB ID: 2e5b)18 as the search model. The models were then refined using the Refmac package in CCP4 and Phenix.refine. The data processing and structural refinement statistics are summarized in Supplementary information, Table S1. Structural figures were created using PyMOL (https://pymol.org).
Molecular docking
The protein structure of NAMPT with NAT was built from crystal X-ray diffraction. The Schrodinger software was employed for optimization and docking. Protein Preparation Wizard was used to assign bond orders, delete excess water, and correct overlaps. The structure of NAT-5r was built in ChemDraw and then prepared with the LigPred module. Induced fit docking was employed to dock NAT-5r into the NAMPT structure.
Immunostaining of cells
Cells were rinsed with PBS before fixation with 4% PFA for 15 min at room temperature. The cells were then stained with primary antibodies that were diluted in PBS with 5% BSA and 0.01% Triton X-100 overnight at 4 °C in a humidified chamber. The cells were washed with wash buffer (PBS with 0.01% Triton X-100) and stained with fluorescent secondary antibodies (Jackson) for 1 h. Nuclei DNA was stained by DAPI (Sigma). Slides were mounted with VectaMount medium (Vector). Images were taken with a laser scanning confocal microscope (Dragonfly, Andor). Further image processing and analysis were performed using the Imaris 9.3.1 software package. The information for commercially available antibodies was provided in the following table.
| Antibody | Source | Identifier |
|---|---|---|
| Mouse anti-Nestin | Millipore | Cat. # MAB353 |
| Rabbit anti-Ki67 | Abcam | Cat. # ab16667 |
| Rabbit anti-PAX6 | Millipore | Cat. # AB2237 |
| Mouse anti-Tuj1 | R&D | Cat. # MAB1195 |
| Mouse anti-GFAP | Millipore | Cat. # MAB360 |
| Mouse anti-O4 | R&D | Cat. # MAB1326 |
| Rabbit anti-SOX2 | Thermo Fisher | Cat. # 481400 |
| Goat anti-rabbit IgG (H + L), Alexa Fluor 488 | Thermo Fisher | Cat. # A-11008 |
| Donkey anti-mouse IgG (H + L), Alexa Fluor 594 | Thermo Fisher | Cat. # A-21203 |
Western blotting
The cells were lysed with lysis buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA, 2.5 mM sodium pyrophosphate, 1% Triton X-100, 1 mM PMSF, and protease inhibitor cocktail), and the samples were separated by SDS-PAGE, followed by transfer to polyvinylidene difluoride (PVDF) membranes (Millipore). The membrane was probed with primary antibodies and then incubated with the secondary antibodies. Antibodies were listed in the following table. The immune-reactive products were detected by enhanced chemiluminescence.
| Antibody | Source | Identifier |
|---|---|---|
| Mouse anti-NAMPT | Adipogen clone OMNI379 | Cat. # AG-20A-0034-C100 |
| Rabbit anti-LC3B | Cell Signaling Technology | Cat. # 3868 |
| Rabbit anti-p-S6K | Cell Signaling Technology | Cat. # 9234 |
| Rabbit anti-FOXO1 | Cell Signaling Technology | Cat. # 2880 |
| Rabbit anti-Ac FOXO1 | ABcolonal | Cat. # A17406 |
| Mouse anti-β-actin | Abcepta | Cat. # MAB1326 |
| goat anti-mouse HRP-conjugated secondary antibody | Easybio | Cat. # BE0102 |
| goat anti-rabbit HRP-conjugated secondary antibody | Easybio | Cat. # BE0101 |
RNA-Seq and analysis
Transcriptomic analysis of NSCs was carried out using a protocol for RNA-Seq. In brief, three sets of samples were collected for each group in Trizol. Total RNA was extracted from NSCs using RNeasy Mini Kit and reversed to cDNA using PrimeScript RT Master Mix. oligo(dT) was used to prime cDNA synthesis. cDNA was then amplified by PCR to enrich nucleic acid. The library was constructed after purification of amplified products, including DNA fragmentation, end-repair, adding “A” and joint, PCR amplification, and library quality control. The constructed library was sequenced with the Illumina platform. The original sequence (raw reads) obtained from HiSeq sequencing was subjected to the process of removing low-quality sequences and connector pollution to get the processed high-quality sequences (clean reads). All subsequent analysis was based on the clean reads. Quantitative real-time PCR was conducted using SYBR Green PCR Master Mix with Bio-Rad Real-Time PCR System. Actin served as the control for data normalization, and fold changes were calculated by the comparative CT method. Experiments were repeated at least three times. Primer sequences were listed in the following table.
| Gene | Primer sequence | NCBI reference sequence |
|---|---|---|
| Actin |
F: CATTGCTGACAGGATGCAGAAGG R: TGCTGGAAGGTGGACAGTGAGG |
NM_007393 |
| Btg2 |
F: GAGCGAGCAGAGACTCAAGGTT R: CGATAGCCAGAACCTTTGGATGG |
NM_007570 |
| Dag1 |
F: GTGGTTGGCATTCCAGACGGTA R: CAGTGTAGCCAAGACGGTAAGG |
NM_010017 |
| Enc1 |
F: CGAAGGTGTCAGCAATGCTGCT R: GCACTGACCATCTGTTCTCGCA |
NM_007930 |
| Ogdhl |
F: ATCCGCAAGGACCTGGAACAGA R: GTTCCTGTTGGTGACACGGTTG |
NM_001081130 |
| Daam2 |
F: GAGAATGAGGTGAAGCAGTGGAG R: CTTGTTGAGCGTCCGCATCATC |
NM_001008231 |
| SLC3a2 |
F: GAGCGTACTGAATCCCTAGTCAC R: GCTGGTAGAGTCGGAGAAGATG |
NM_008577 |
| SLC6a8 |
F: ATCCTGGCACTCATCAACAGCG R: GGTAGGCAATGAAGGCTAGACC |
NM_133987 |
Cell viability assay
Cell viability assay was performed in 96-well plates using CellTiter-Glo Luminescent Cell Viability Assay kit (Promega) that measures cellular ATP content. 50 μL of CellTiter-Glo reagent was directly added to 100 μL of cell culture per well. The plates were incubated at room temperature for 10 min. Luminescence was recorded using a multi-mode microplate reader (BioTek, Cytation 3). Cell viability was presented as the percentage of viable cells compared to the untreated control, and the means ± SEM was calculated from experimental triplicates.
TLC analysis of NAD salvage from 14C-labeled NAM
HepG2 cells cultured in 6-well plates were treated with compounds for 24 h. Afterward, 14C-labeled NAM (0.1 mCi/mL, specific activity: 50 mCi/mmol; American Radiolabeled Chemicals, Inc.) were added at 0.5 μCi/mL and incubated for 6 h. The cells were then washed once with PBS and collected by trypsin digestion. Metabolites were extracted from cells with 50 μL of perchloric acid (0.5 M) by incubating on ice for 20 min. The extracts were then neutralized with 13.75 μL of KCl/KOH (0.5 M/2.0 M) and centrifuged at 2500 × g for 10 min at 4 °C. 10 µL of extracted metabolites were dotted and run on silicon thin-layer chromatography plates with 1 M ammonium acetate: ethanol (3:7) as the solvent. For autoradiography, the chromatograms were exposed to a storage Phospho screen for 7 days. Afterward, the screen was scanned using the Typhoon scanner.
Generation of NAMPT CRISPR-Cas9 knockout cell line
sgRNA (5′-CCGGCCCGAGATGAATCCTG-3′) targeting the human NAMPT gene was designed by using the online tool (GPP sgRNA Designer, BROAD Institute). The sgRNA was inserted into the lentiCRISPRv2 vector. The lentiviral plasmids (10 μg/10-cm dishes) were co-transfected into 293 T cells with the packing plasmids pSPAX2(7.5 μg/10-cm dishes) and pMD2G (5 μg/10-cm dishes). Lentivirus was collected from culture medium after 48 h transfection and was used to infect HeLa cells. 48 h after infection, the cells were grown in the puromycin-containing medium for the selection of NAMPT knockout cells.
Measurement of cellular NAD abundance
Cellular NAD was measured using a coupled assay as described by Graeff et al.58 with some modifications. Briefly, cells were washed with PBS twice and 100 μL 0.5 M HClO4 was then added to each well. The plates were incubated at 4 °C for 15 min and then neutralized with 100 μL 0.55 M K2CO3. The plates were shaken for 2 min and the mixture was transferred to 1.5 mL tubes, followed by centrifugation for 10 min at 20,000 g at 4°C. The reaction buffer contained 100 mM Tris (pH 8.0), 20 μM resazurin, 10 μM FMN, 10 mM nicotinamide, 0.001% BSA, 0.2 μM rat NQO1. The reaction was initiated by the addition of a 5 μL sample to a 45 μL reaction buffer. Fluorescence at Ex544nm/Em590nm was measured continuously for 30 min to establish the reaction velocity.
Metabolomic profiling analysis
HepG2 cells were plated in 10-cm cell culture dishes in triplicates. The fresh medium supplemented with 5 μM compounds was added on the next day. After incubation at 37 °C for 1.5 h or 6 h, each dish was washed twice with cold PBS, and metabolites were extracted using cold 80% methanol by multiple freeze-thaw cycles. Metabolite extracts were then centrifuged and the supernatant was dried in a speed-Vac. The dried materials were dissolved in 0.1% formic acid and subjected to mass spectrometric analysis. For ADPR validation, HepG2 cells were treated with 5 μM NAT or 1 μM FK866 or co-treated for 6 h, others are as same as above.
Fluorescence detection of FAO activity
A fluorescent probe was synthesized, and the FAO in living cells was determined as described previously.21 Briefly, HepG2 cells (2 × 105 cells) were seeded on 35-mm glass-base-dish. The cells were treated with 5 μM NAT for 1, 3, or 6 h the next day. After incubation, the cells were washed with HBS+ (20 mM HEPES, 107 mM NaCl, 6 mM KCl, 1.2 mM MgSO4, 2 mM CaCl2, 11.5 mM glucose, adjusted to pH 7.4 with NaOH) twice and incubated with 10 μM FAO-probe for 30 min in HBS+ buffer at 37 °C. The cells were then subjected to fluorescence imaging with Zeiss LSM980 Airyscan2 confocal microscopy. Fluorescence images were acquired at 405 nm excitation. Data was collected using Zen Blue software and analyzed by Imaris 9.7 software.
OCR measurement
OCR was measured at 37 °C in an XF96 extracellular analyzer (Seahorse Bioscience). Briefly, HepG2 cells (1 × 104 cells/well) were seeded into a 96 well Agilent Seahorse XF Cell Culture Microplate and incubated in DMEM with 10% FBS overnight. Next day, cells were pre-treated with NAT-5r (0, 0.5, 1.5, or 5 μM) for 1 h or 3 h and changed to OCR conditional medium (XF base medium + 10 mM Glucose + 2 mM Glutamine + 1 mM Sodium pyruvate) before measurement. HepG2 cells were then incubated in a 37 °C CO2-free pre-station for 1 h and sequentially exposed to 2 mM oligomycin, 2 mM FCCP, and 0.5 mM rotenone and antimycin A at the indicated time point for OCR determination.
Drug preparation and the mouse model of CIPN
PTX (Selleck) was dissolved in ethanol, and mixed with an equal volume of Kolliphor to make a 12 mg/mL stock solution. The resulting solution was diluted with 3 volumes of sterile PBS before use. The final ratio of ethanol, Kolliphor, and PBS is 1:1:6. The test compounds were dissolved in DMSO, mixed with 4 volumes of Kolliphor, and diluted with 35 volumes of sterile 5% dextrose as the working solutions.
The mice were intraperitoneally injected with PTX or vehicle at a dose of 18.3 mg/kg on 3 alternated days (days 9, 11, and 13). The accumulative dose of PTX was 55 mg/kg. To evaluate the neuroprotective effect of the candidate compounds, the mice were daily administrated with compounds or vehicle control one week prior to the PTX treatment.
For the data in Fig. 6b–d, mice were randomly allocated into five groups. For the data in Fig. 6e–i, mice were prescreened by Von Fray tests before the experiment. Mice with the paw withdrawal threshold (PWT) greater than 0.88 or less than 0.40 were excluded. The remaining mice were randomly allocated into experimental groups.
Measurement of tissue NAD abundance
The sciatic nerve was dissected as quickly as possible following decapitation and frozen on dry ice. Samples were kept at −80 °C until ready for assay. To determine tissue NAD levels, samples were transferred from −80 °C to a container with dry ice. Frozen tissue samples were weighed and transferred immediately into ice-cold homogenization buffer (approximately 5 mg tissue in 1 mL buffer), homogenized with a Tissue Tearor homogenizer for 15–20 s, then placed on ice. Samples were vortexed every 5 min for 15 min then frozen on dry ice until all samples have been completed. 50 μL aliquots of each sample were rapidly frozen in liquid nitrogen and preserved under −80 °C. The aliquots were rapidly thawed, 25 μL of each sample was added into 384-well plates, and 25 μL NAD/NADH-Glo reagent (Promega) was added to each well. A serial dilution of NAD, as the standard, was assayed using the same procedure. Relative luminescence units for each sample were interpolated to determine the NAD concentration (nmol/L). Protein content (μg/mL) was determined for each sample using the BCA protein assay kit (Beyotime) to normalize tissue NAD levels (nmol/mg protein).
Von Frey tests
To assess the sensitivity of an animal’s hind paw to noxious mechanical stimulation, the paw withdrawal threshold (PWT) was evaluated using von Frey filaments (Aesthesio). The mice were transferred to the test room one day earlier for environmental adaption. Mice were placed in acrylic chambers (17 × 16 × 13 cm) suspended above a wire mesh grid and allowed to acclimatize to the testing apparatus for half an hour before the experiments. When the mouse was not moving, the von Frey filaments were pressed against the plantar surface of the paw until the filament buckled and held for a maximum of 3 s. A positive response was noted if the paw was sharply withdrawn upon the application of the filament. Flinching immediately upon the removal of the filament was also considered a positive response. The paw withdrawal threshold was determined using a simplified up and down method as described by Bonin et al.59
Measurement of nerve conduction velocity
The nerve conduction velocity was measured as described by Chiorazzi et al.60 with some modification. Mice were anesthetized with 2% isoflurane and placed on a warm pad (37 °C). Subdermal needle electrodes were used for stimulation and recording. The stimulation electrodes were placed in the base of the second toe of the hind paw, and the recording electrodes were placed on the same side sciatic nerve. The distance between stimulation and recording electrodes was measured by caliper. Stimuli were imposed with an electrical pulse of 10 mA intensity (AMPI Master-8/CP/VP pulse stimulator). Each mouse was subjected to pulse stimulation up to 6 times. The latency of nerve potential was measured from stimulus onset with Spike2 (version 10).
Tissue staining and axon quantification
One day after von Frey tests, mice were sacrificed, and the sciatic nerves were immediately dissected, and fixed by immersion in freshly prepared 3% glutaraldehyde in 0.1 M PBS overnight at 4 °C. The nerves were fixed by 1% osmium tetroxide and stained with uranyl acetate. Selected nerve blocks were cut into ultrathin cross-sections (60 nm thick) using a Leica EM UC6 ultramicrotome. Electron microscopy images were obtained with a HITACHI-7650 TEM. The density of the axon was measured using Image J 1.51j8.
Toxicity panels and pharmacokinetic analyses
For toxicity panels, 8–9-week-old males of the strain C57BL/6J obtained from Beijing Vital River Laboratory Animal Technology Co., Ltd. were used. Mice were daily injected intraperitoneally with either vehicle or 30 mg/kg test compounds for two weeks. Body weight was measured once daily. Blood samples were taken for analysis on day 15.
For the pharmacokinetic profiles, male CD-1 mice were used. The test compounds were administrated at a single dose of 1 mg/kg intravenously, 30 mg/kg orally, or 30 mg/kg intraperitoneally. Plasma samples were obtained over a 24 h interval, and test compound concentrations were analyzed by LC-MS/MS (API 5500 QTRAP, SCIEX). Briefly, test compounds were extracted from plasma by protein precipitation using ACN and separated on a C18 column with an ACN and water mixture containing 0.1% formic acid as solvent. Chromatographic conditions and mass spectrometer parameters were optimized for each test compound before the analysis of plasma samples. The PK parameters in plasma, according to the concentration-time data and dose level, were estimated using WinNonlin® Professional 8.1 with a non-compartmental analysis model. The oral bioavailability was calculated as: F (%) = (Dose IV × AUC PO) / (Dose PO × AUC IV) X 100.
Statistical analysis
Data are presented as means ± SEM, as indicated in the figure legends. Details of the number of biological replicates and independent experiments are provided in the figure legends. All the graphs presented in this study were plotted, and statistical tests were performed using GraphPad Prism 7 software. A two-sided unpaired Student’s t-test was used for comparisons between the means of two groups. ANOVA test was used for the analysis of means of three or more groups. P < 0.05 was considered to be statistically significant. The correlation data analysis was performed in GraphPad Prism 7, and the exact p-value was obtained using Spearman Rank Correlation (v1.0.1) in Free Statistics Software (v1.1.23-r7), Office for Research Development and Education (http://www.wessa.net/rwasp_spearman.wasp).
Data and code availability
X-ray crystal structure data for the NAMPT-NAT complex have been deposited in the PDB with the accession code 7ENQ. All RNA sequencing data can be found at the NCBI Sequence Read Archive (GEO accession number: GSE174114, BioProject: PRJNA728447, SRA: SRP319055).
Supplementary information
Acknowledgements
We thank Xiaodong Wang for critical reading of the manuscript; Kosaku Uyeda and Bailong Xiao for helpful discussion; Yi Ding and Ting Wang for assistance in the compound screen; Yanli Zhang and Bingyu Liu for help with fluorescence microscopy; Haiteng Deng, Xiaohui Liu, Yu Tang, and Weihua Wang for help with metabolite analysis; Shilong Fan for assistance in X-ray crystallography; and Tao Cai (National Institute of Biological Sciences, NIBS) for assistance in RNA-seq. This work was supported by grants to G.W. and Y.T. from the National Natural Science Foundation of China (81872874, 91949101, 21772109, 21971140), Tsinghua-Peking Center for Life Sciences, Beijing Advanced Innovation Center for Structural Biology, and Tsinghua-Toyota Joint Research Fund.
Author contributions
G.W. and Y.T. designed and supervised the study. H.Y., M.L., L.W., Y.Z., and C.W. performed most of the experiments and analyzed the results. H.L. and S.X. assisted in the compound synthesis. R.Z. and S.C. conducted the compound screen. C.L., M.Y. and T.L. assisted in solving the crystal structure of the NAMPT-NAT complex. F.L., J.C., S.W., W.X. and G.X. assisted in mouse experiments. X.G. assisted in the preparation of recombinant proteins. L.H. assisted in molecular docking. G.W. wrote the manuscript with input from H.Y., M.L., Y.Z., C.W., Y.T., and L.W.
Competing interests
G.W., Y.T., H.Y., M.L., L.W., C.L., R.Z., Y.Z., C.W., F.L., and S.C. filed a patent application for NAMPT activator NATs. The other authors have no conflicts of interest to declare.
Footnotes
These authors contributing equally: Hong Yao, Minghui Liu, Leibo Wang, Yumeng Zu, Chou Wu.
Contributor Information
Yefeng Tang, Email: yefengtang@tsinghua.edu.cn.
Gelin Wang, Email: gelinwang@tsinghua.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41422-022-00651-9.
References
- 1.Yang Y, Sauve AA. NAD(+) metabolism: bioenergetics, signaling and manipulation for therapy. Biochim. Biophys. Acta. 2016;1864:1787–1800. doi: 10.1016/j.bbapap.2016.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Verdin E. NAD(+) in aging, metabolism, and neurodegeneration. Science. 2015;350:1208–1213. doi: 10.1126/science.aac4854. [DOI] [PubMed] [Google Scholar]
- 3.Preiss J, Handler P. Biosynthesis of diphosphopyridine nucleotide. I. Identification of intermediates. J. Biol. Chem. 1958;233:488–492. doi: 10.1016/S0021-9258(18)64789-1. [DOI] [PubMed] [Google Scholar]
- 4.Bogan KL, Brenner C. Nicotinic acid, nicotinamide, and nicotinamide riboside: a molecular evaluation of NAD+ precursor vitamins in human nutrition. Annu. Rev. Nutr. 2008;28:115–130. doi: 10.1146/annurev.nutr.28.061807.155443. [DOI] [PubMed] [Google Scholar]
- 5.Revollo JR, Grimm AA, Imai S. The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates Sir2 activity in mammalian cells. J. Biol. Chem. 2004;279:50754–50763. doi: 10.1074/jbc.M408388200. [DOI] [PubMed] [Google Scholar]
- 6.Stromland O, et al. Keeping the balance in NAD metabolism. Biochem. Soc. Trans. 2019;47:119–130. doi: 10.1042/BST20180417. [DOI] [PubMed] [Google Scholar]
- 7.Wang X, et al. Deletion of Nampt in projection neurons of adult mice leads to motor dysfunction, neurodegeneration, and death. Cell Rep. 2017;20:2184–2200. doi: 10.1016/j.celrep.2017.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gilley J, Orsomando G, Nascimento-Ferreira I, Coleman MP. Absence of SARM1 rescues development and survival of NMNAT2-deficient axons. Cell Rep. 2015;10:1974–1981. doi: 10.1016/j.celrep.2015.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Long AN, et al. Effect of nicotinamide mononucleotide on brain mitochondrial respiratory deficits in an Alzheimer’s disease-relevant murine model. BMC Neurol. 2015;15:19. doi: 10.1186/s12883-015-0272-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hou Y, et al. NAD(+) supplementation normalizes key Alzheimer’s features and DNA damage responses in a new AD mouse model with introduced DNA repair deficiency. Proc. Natl. Acad. Sci. USA. 2018;115:E1876–E1885. doi: 10.1073/pnas.1718819115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang G, et al. P7C3 neuroprotective chemicals function by activating the rate-limiting enzyme in NAD salvage. Cell. 2014;158:1324–1334. doi: 10.1016/j.cell.2014.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katsyuba E, et al. De novo NAD(+) synthesis enhances mitochondrial function and improves health. Nature. 2018;563:354–359. doi: 10.1038/s41586-018-0645-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Escande C, et al. Flavonoid apigenin is an inhibitor of the NAD+ ase CD38: implications for cellular NAD+ metabolism, protein acetylation, and treatment of metabolic syndrome. Diabetes. 2013;62:1084–1093. doi: 10.2337/db12-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hughes RO, et al. Small molecule SARM1 inhibitors recapitulate the SARM1(-/-) phenotype and allow recovery of a metastable pool of axons fated to degenerate. Cell Rep. 2021;34:108588. doi: 10.1016/j.celrep.2020.108588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gardell SJ, et al. Boosting NAD(+) with a small molecule that activates NAMPT. Nat. Commun. 2019;10:3241. doi: 10.1038/s41467-019-11078-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khan JA, Tao X, Tong L. Molecular basis for the inhibition of human NMPRTase, a novel target for anticancer agents. Nat. Struct. Mol. Biol. 2006;13:582–588. doi: 10.1038/nsmb1105. [DOI] [PubMed] [Google Scholar]
- 17.Burgos ES, Ho MC, Almo SC, Schramm VL. A phosphoenzyme mimic, overlapping catalytic sites and reaction coordinate motion for human NAMPT. Proc. Natl. Acad. Sci. USA. 2009;106:13748–13753. doi: 10.1073/pnas.0903898106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takahashi R, et al. Structure and reaction mechanism of human nicotinamide phosphoribosyltransferase. J. Biochem. 2010;147:95–107. doi: 10.1093/jb/mvp152. [DOI] [PubMed] [Google Scholar]
- 19.Hasmann M, Schemainda I. FK866, a highly specific noncompetitive inhibitor of nicotinamide phosphoribosyltransferase, represents a novel mechanism for induction of tumor cell apoptosis. Cancer Res. 2003;63:7436–7442. [PubMed] [Google Scholar]
- 20.Revollo JR, et al. Nampt/PBEF/Visfatin regulates insulin secretion in beta cells as a systemic NAD biosynthetic enzyme. Cell Metab. 2007;6:363–375. doi: 10.1016/j.cmet.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uchinomiya S, et al. Fluorescence detection of metabolic activity of the fatty acid beta oxidation pathway in living cells. Chem. Commun. 2020;56:3023–3026. doi: 10.1039/C9CC09993J. [DOI] [PubMed] [Google Scholar]
- 22.Gross CG. Neurogenesis in the adult brain: death of a dogma. Nat. Rev. Neurosci. 2000;1:67–73. doi: 10.1038/35036235. [DOI] [PubMed] [Google Scholar]
- 23.Gage FH, Temple S. Neural stem cells: generating and regenerating the brain. Neuron. 2013;80:588–601. doi: 10.1016/j.neuron.2013.10.037. [DOI] [PubMed] [Google Scholar]
- 24.Stein LR, Imai SI. Specific ablation of Nampt in adult neural stem cells recapitulates their functional defects during aging. EMBO J. 2014;33:937–1085. doi: 10.1002/embj.201488303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pieper AA, McKnight SL, Ready JM. P7C3 and an unbiased approach to drug discovery for neurodegenerative diseases. Chem. Soc. Rev. 2014;43:6716–6726. doi: 10.1039/C3CS60448A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kilpatrick TJ, Bartlett PF. Cloning and growth of multipotential neural precursors: requirements for proliferation and differentiation. Neuron. 1993;10:255–265. doi: 10.1016/0896-6273(93)90316-J. [DOI] [PubMed] [Google Scholar]
- 27.Reynolds BA, Weiss S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science. 1992;255:1707–1710. doi: 10.1126/science.1553558. [DOI] [PubMed] [Google Scholar]
- 28.Tirone F, Farioli-Vecchioli S, Micheli L, Ceccarelli M, Leonardi L. Genetic control of adult neurogenesis: interplay of differentiation, proliferation and survival modulates new neurons function, and memory circuits. Front. Cell Neurosci. 2013;7:59. doi: 10.3389/fncel.2013.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim TA, et al. NRP/B, a novel nuclear matrix protein, associates with p110(RB) and is involved in neuronal differentiation. J. Cell Biol. 1998;141:553–566. doi: 10.1083/jcb.141.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sugita S, et al. A stoichiometric complex of neurexins and dystroglycan in brain. J. Cell Biol. 2001;154:435–445. doi: 10.1083/jcb.200105003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matusek T, et al. Formin proteins of the DAAM subfamily play a role during axon growth. J. Neurosci. 2008;28:13310–13319. doi: 10.1523/JNEUROSCI.2727-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang T, Kraus WL. SIRT1-dependent regulation of chromatin and transcription: linking NAD(+) metabolism and signaling to the control of cellular functions. Biochim. Biophys. Acta. 2010;1804:1666–1675. doi: 10.1016/j.bbapap.2009.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Canto C, Auwerx J. Targeting sirtuin 1 to improve metabolism: all you need is NAD(+)? Pharmacol. Rev. 2012;64:166–187. doi: 10.1124/pr.110.003905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vilchez D, et al. FOXO4 is necessary for neural differentiation of human embryonic stem cells. Aging Cell. 2013;12:518–522. doi: 10.1111/acel.12067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.LoCoco, P. M. et al. Pharmacological augmentation of nicotinamide phosphoribosyltransferase (NAMPT) protects against paclitaxel-induced peripheral neuropathy. Elife6.10.7554/eLife.29626 (2017). [DOI] [PMC free article] [PubMed]
- 36.Fukuda Y, Li Y, Segal RA. A mechanistic understanding of axon degeneration in chemotherapy-induced peripheral neuropathy. Front. Neurosci. 2017;11:481. doi: 10.3389/fnins.2017.00481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krebs HA, Veech RL. Equilibrium relations between pyridine nucleotides and adenine nucleotides and their roles in the regulation of metabolic processes. Adv. Enzyme Regul. 1969;7:397–413. doi: 10.1016/0065-2571(69)90030-2. [DOI] [PubMed] [Google Scholar]
- 38.Wang MS, Davis AA, Culver DG, Glass JD. WldS mice are resistant to paclitaxel (taxol) neuropathy. Ann. Neurol. 2002;52:442–447. doi: 10.1002/ana.10300. [DOI] [PubMed] [Google Scholar]
- 39.Waller A. Experiments on the section of the glossopharyngeal and hypoglossal nerves of the frog, and observations of the alterations produced thereby in the structure of their primitive fibres. Philos. Trans. R. Soc. Lond. 1850;140:423–429. [Google Scholar]
- 40.Araki T, Sasaki Y, Milbrandt J. Increased nuclear NAD biosynthesis and SIRT1 activation prevent axonal degeneration. Science. 2004;305:1010–1013. doi: 10.1126/science.1098014. [DOI] [PubMed] [Google Scholar]
- 41.Gerdts J, Brace EJ, Sasaki Y, DiAntonio A, Milbrandt J. SARM1 activation triggers axon degeneration locally via NAD(+) destruction. Science. 2015;348:453–457. doi: 10.1126/science.1258366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sasaki, Y., Nakagawa, T., Mao, X., DiAntonio, A. & Milbrandt, J. NMNAT1 inhibits axon degeneration via blockade of SARM1-mediated NAD(+) depletion. Elife5.10.7554/eLife.19749 (2016). [DOI] [PMC free article] [PubMed]
- 43.Geisler S, et al. Prevention of vincristine-induced peripheral neuropathy by genetic deletion of SARM1 in mice. Brain. 2016;139:3092–3108. doi: 10.1093/brain/aww251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Turkiew E, Falconer D, Reed N, Hoke A. Deletion of Sarm1 gene is neuroprotective in two models of peripheral neuropathy. J. Peripher. Nerv. Syst. 2017;22:162–171. doi: 10.1111/jns.12219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Osterloh JM, et al. dSarm/Sarm1 is required for activation of an injury-induced axon death pathway. Science. 2012;337:481–484. doi: 10.1126/science.1223899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sheng ZH. The interplay of axonal energy homeostasis and mitochondrial trafficking and anchoring. Trends Cell Biol. 2017;27:403–416. doi: 10.1016/j.tcb.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen H, Chan DC. Mitochondrial dynamics-fusion, fission, movement, and mitophagy-in neurodegenerative diseases. Hum. Mol. Genet. 2009;18:R169–R176. doi: 10.1093/hmg/ddp326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bai P, et al. PARP-1 inhibition increases mitochondrial metabolism through SIRT1 activation. Cell Metab. 2011;13:461–468. doi: 10.1016/j.cmet.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pirinen E, et al. Niacin cures systemic NAD(+) deficiency and improves muscle performance in adult-onset mitochondrial myopathy. Cell Metab. 2020;32:144. doi: 10.1016/j.cmet.2020.05.020. [DOI] [PubMed] [Google Scholar]
- 50.Yoshino M, et al. Nicotinamide mononucleotide increases muscle insulin sensitivity in prediabetic women. Science. 2021;372:1224–1229. doi: 10.1126/science.abe9985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cerutti R, et al. NAD(+)-dependent activation of Sirt1 corrects the phenotype in a mouse model of mitochondrial disease. Cell Metab. 2014;19:1042–1049. doi: 10.1016/j.cmet.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ratajczak J, et al. NRK1 controls nicotinamide mononucleotide and nicotinamide riboside metabolism in mammalian cells. Nat. Commun. 2016;7:13103. doi: 10.1038/ncomms13103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Canto C, Menzies KJ, Auwerx J. NAD(+) metabolism and the control of energy homeostasis: a balancing act between mitochondria and the nucleus. Cell Metab. 2015;22:31–53. doi: 10.1016/j.cmet.2015.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu L, et al. Quantitative analysis of NAD synthesis-breakdown fluxes. Cell Metab. 2018;27:1067–1080. doi: 10.1016/j.cmet.2018.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gross CJ, Henderson LM. Digestion and absorption of NAD by the small intestine of the rat. J. Nutr. 1983;113:412–420. doi: 10.1093/jn/113.2.412. [DOI] [PubMed] [Google Scholar]
- 56.Burgos ES, Schramm VL. Weak coupling of ATP hydrolysis to the chemical equilibrium of human nicotinamide phosphoribosyltransferase. Biochemistry. 2008;47:11086–11096. doi: 10.1021/bi801198m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang RY, et al. A fluorometric assay for high-throughput screening targeting nicotinamide phosphoribosyltransferase. Anal. Biochem. 2011;412:18–25. doi: 10.1016/j.ab.2010.12.035. [DOI] [PubMed] [Google Scholar]
- 58.Graeff R, Lee HC. A novel cycling assay for cellular cADP-ribose with nanomolar sensitivity. Biochem. J. 2002;361:379–384. doi: 10.1042/bj3610379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bonin RP, Bories C, De Koninck Y. A simplified up-down method (SUDO) for measuring mechanical nociception in rodents using von Frey filaments. Mol. Pain. 2014;10:26. doi: 10.1186/1744-8069-10-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chiorazzi A, et al. Ghrelin agonist HM01 attenuates chemotherapy-induced neurotoxicity in rodent models. Eur. J. Pharmacol. 2018;840:89–103. doi: 10.1016/j.ejphar.2018.09.029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
X-ray crystal structure data for the NAMPT-NAT complex have been deposited in the PDB with the accession code 7ENQ. All RNA sequencing data can be found at the NCBI Sequence Read Archive (GEO accession number: GSE174114, BioProject: PRJNA728447, SRA: SRP319055).