Abstract
Integration of sensory and molecular inputs from the environment shapes animal behavior. A major site of exposure to environmental molecules is the gastrointestinal tract, where dietary components are chemically transformed by the microbiota1 and gut-derived metabolites are disseminated to all organs, including the brain2. In mice, the gut microbiota impacts behavior3, modulates neurotransmitter production in the gut and brain4,5, and influences brain development and myelination patterns6,7. Mechanisms mediating gut-brain interactions remain poorly defined, though broadly involve humoral or neuronal connections. We previously reported that levels of the microbial metabolite 4-ethylphenyl sulfate (4EPS) were elevated in a mouse model of atypical neurodevelopment8. Herein, we identified biosynthetic genes from the gut microbiome that mediate conversion of dietary tyrosine to 4-ethylphenol (4EP), and bioengineered gut bacteria to selectively produce 4EPS in mice. 4EPS entered the brain and was associated with changes in region-specific activity and functional connectivity. Gene expression signatures revealed altered oligodendrocyte function in the brain, and 4EPS impaired oligodendrocyte maturation in mice as well as decreased oligodendrocyte-neuron interactions in ex vivo brain cultures. Mice colonized with 4EP-producing bacteria exhibited reduced myelination of neuronal axons. Altered myelination dynamics in the brain have been associated with behavioral outcomes7,9–14,13,14. Accordingly, we observed that mice exposed to 4EPS displayed anxiety-like behaviors, and pharmacologic treatments that promote oligodendrocyte differentiation prevented the behavioral effects of 4EPS. These findings reveal that a gut-derived molecule influences complex behaviors in mice via effects on oligodendrocyte function and myelin patterning in the brain.
A microbial biosynthetic pathway for 4EP
Previously, the metabolite 4EPS was measured at higher relative abundance in a mouse model of atypical neurodevelopment, and systemic delivery of synthetic 4EPS to naïve mice altered behavior in the open-field test8. We recently reported that 4EPS is elevated in the plasma of individuals with autism spectrum disorder (ASD)15, and show here it is increased in the blood of the CNTNAP2 model of ASD (Extended Data Fig. 1f). The gut microbiome is predicted to harbor genes that convert tyrosine, the source of several mammalian neurotransmitters, to 4-ethylphenol (4EP), which could then be sulfated16 to 4EPS by the host (Fig. 1a). Consistent with this notion, germ-free (GF) mice devoid of a microbiota contain virtually no detectable levels of 4EPS (Extended Data Fig. 1f)8. Some rare bacterial species in the Firmicutes phylum produce 4EP using p-coumaric acid as a substrate17,18, and precursors of p-coumaric acid include tyrosine or plant-based molecules that can be metabolized by the gut microbiota to 4EP. Indeed, both a high-tyrosine fish-based diet and a soy-based diet resulted in measurable 4EPS levels in conventionally colonized mice (Extended Data Fig. 1g). By screening candidate gut bacterial isolates, we discovered that Bacteroides ovatus produces p-coumaric acid from tyrosine (Fig. 1b). Using the basic alignment search tool (BLAST), we identified a tyrosine ammonia lyase in B. ovatus (BACOVA_01194) that upon deletion, abrogated p-coumaric acid production (Fig. 1b).
Fig. 1 |. Discovery of a 4EP biosynthetic pathway, strain engineering of gut bacteria, and colonization of mice to produce 4EPS.
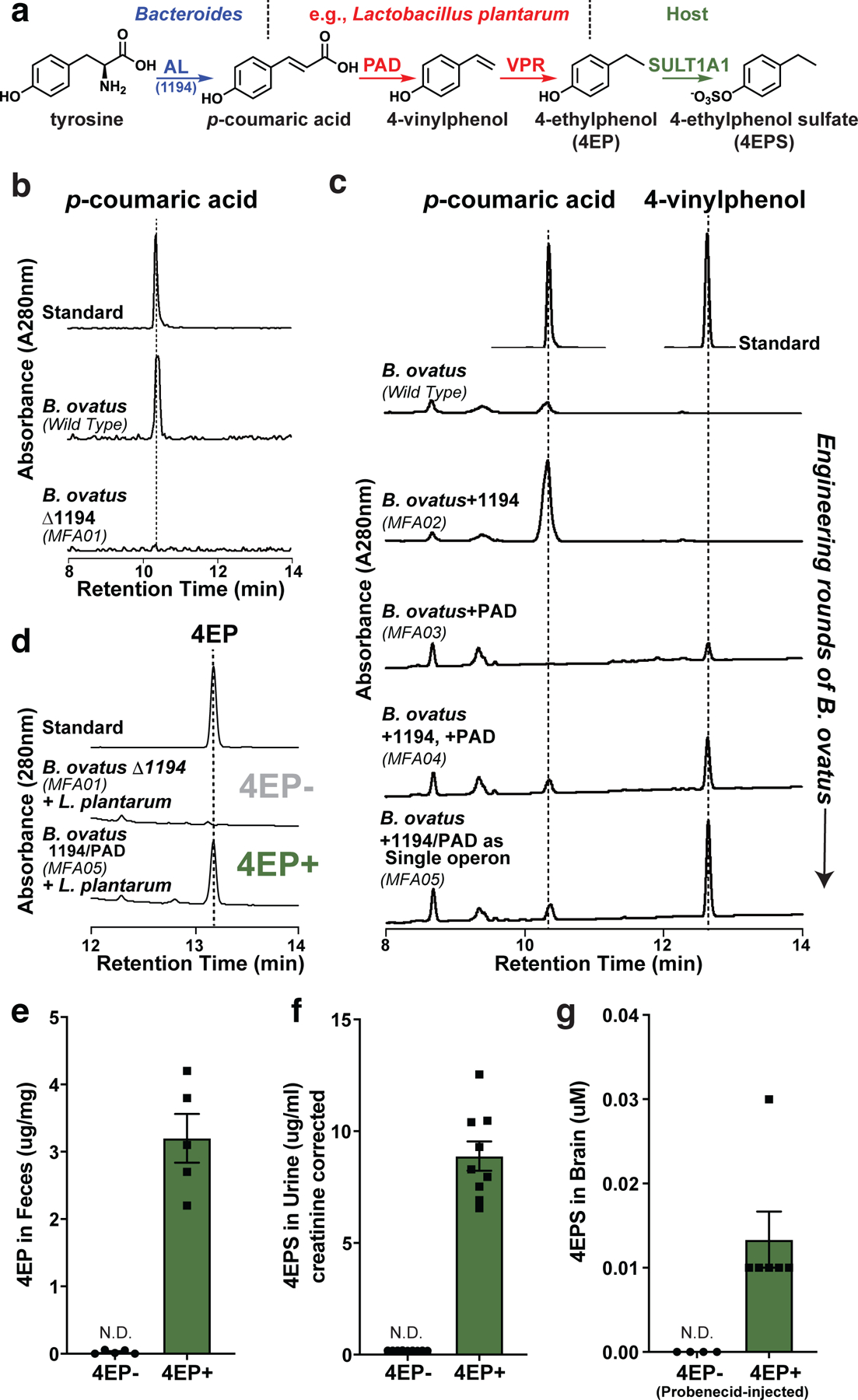
a, Proposed biosynthetic pathway of 4EP in gut bacteria via tyrosine ammonia lyase (AL), phenolic acid decarboxylase (PAD), and vinyl phenol reductase (VPR) enzymes, and subsequent sulfation to 4EPS by host sulfotransferase (SULT1A1). The genes identified and cloned for bacterial strain engineering can convert tyrosine to 4EP via two chemical intermediates. b and c, 4EP precursors, p-coumaric acid and 4-vinylphenol from in vitro cultures of Bacteroides ovatus strains provided tyrosine as substrate. b, Conversion, or lack thereof, of tyrosine to p-coumaric acid by BACOVA_01194 gene (AL/1194 in figure) and deletion mutant (MFA01), respectively. c, The BACOVA_01194 gene (AL/1194 in figure) was introduced separately (MFA02), in addition to the pad gene (MFA04), or both genes as a single, highly expressed operon (MFA05). Standards are shown at top. Further details in Extended Data Figure 1, Methods, and Supplementary Information Tables 1-3. d, 4EP levels from in vitro cultures (provided with tyrosine) of 4EP- strain pair (B. ovatus Δ1194 (MFA01) and Lactobacillus plantarum) or the highest 4-vinylphenol producing engineered strain, B. ovatus 1194/PAD (MFA05) with L. plantarum (4EP+ strain pair). A standard is shown at the top. e, Levels of 4EP in feces (μg/mg) of mice colonized with bacterial strains lacking (4EP-) and producing (4EP+) 4EP (n=5). f, Levels of 4EPS in urine of mice colonized with 4EP- or 4EP+ (n=9) bacterial strains. Additional urine data, Extended Data Fig. 1k. g, Brain levels of 4EPS (after injection with probenecid) in mice colonized with 4EP- (n=4) and 4EP+ (n=6) bacterial strains. Additional controls, Extended Data Fig. 1f. At least two biologically independent trials were performed for each experiment. Abbreviations: N.D., not detectable. Data in panels e-g represent mean ± SEM.
We co-colonized GF mice with B. ovatus and Lactobacillus plantarum, the latter of which can subsequently convert p-coumaric acid to 4EP18; however, the resulting 4EPS levels in urine were low (Extended Data Fig. 1f, h). To improve the efficiency of 4EP production, we carried out several rounds of strain engineering (see Methods). Briefly, an extra copy of the first two genes in the pathway, BACOVA_01194 and the phenolic acid decarboxylase (pad), were inserted into B. ovatus as a single, highly expressed operon (Fig. 1c). The engineered B. ovatus strain MFA05 robustly converts tyrosine to the intermediate, 4-vinylphenol (Fig. 1c, Extended Data Fig. 1a-e), and in co-culture with L. plantarum, 4-vinylphenol is quantitatively metabolized to 4EP (Fig. 1d). In contrast, when the B. ovatus ∆1194 mutant was co-cultured with L. plantarum, no 4EP was detected (Fig. 1d). We find homologs of each gene in ~25 genomes of sequenced human gut microbes, indicating common pathways may be intact in the human microbiome (Extended Data Fig. 1i).
Gut-derived 4EPS in the circulation and brain
We colonized separate groups of GF mice with either of the engineered strain pairs represented in Fig. 1d, generating 4EP+ or 4EP- animals. As expected, 4EP was detected in feces (Fig. 1e), and its host-sulfated derivative, 4EPS, was detected in the serum and urine of 4EP+ colonized mice (Fig. 1f, Extended Data Fig. 1j). 4EP was undetectable in the serum of 4EP+ mice, suggesting efficient sulfation to 4EPS (Extended Data Fig. 1j). Conversely, 4EP- mice do not have measurable 4EP or 4EPS levels (Fig. 1e-f, Extended Data Fig. 1j). 4EPS was detectable in brains of 4EP+ mice treated with probenecid, which inhibits organic anion transporters that mediate efflux of small molecules through the blood brain barrier, suggesting accumulation of 4EPS in the brain (Fig. 1g, Extended Data Fig. 1k-m). We observed sulfation of 4EP to 4EPS by the sulfotransferase SULT1A1 and others during in vitro biochemical reactions (Extended Data Fig. 2a-b). SULT1A1 is found in intestinal, liver, and brain tissues of mice (Extended Data Fig. 2c-d), though the site(s) of 4EP sulfation remains unknown.
4EP and 4EPS are phenolic molecules that may have toxic or inflammatory properties19. However, we observed no differences between 4EP+ and 4EP- groups in body weight or ambulatory activity (i.e., locomotion) (Extended Data Fig. 2f-g). No evidence of intestinal dysfunction was detected in 4EP+ mice when assessing epithelial permeability (Extended Data Fig. 2h), fecal output (Extended Data Fig. 2i), or gross histopathology (Extended Data Fig. 2j). Bacterial colonization levels and ultrastructural localization of bacteria were similar between groups of mice (Extended Data Fig. 2k-m). We did not observe pro-inflammatory cytokine responses in colonic tissue or serum (Extended Data Fig. 3a-b), and only modest changes in peripheral immune cell proportions (Extended Data Fig. 3c-d). Cytokine levels trended toward an anti-inflammatory profile in the brain with no signatures of microglial activation in 4EP+ mice (Extended Data Fig. 3e-g). Collectively, these studies establish a simplified animal model that reproduces the natural route of exposure to a gut microbial metabolite associated with altered behaviors (Extended Data Fig 2e).
4EPS-dependent brain activity patterns
While 4EP or 4EPS --designated here as 4EP(S)-- may have effects on various organs, we focused our study on the brain. Initially, to capture brain-wide differences between 4EP+ and 4EP- mice, we performed functional ultrasound imaging (fUSi), an in vivo method that measures resting state cerebral blood volume variation to assess functional connectivity (Extended Data Fig. 4a). We observed altered (mostly increased) correlation of signaling patterns within 4EP+ mice compared to 4EP- mice (Fig. 2a). These changes were primarily observed in subregions of the hippocampus, thalamus, amygdala, hypothalamus, piriform, and cortex (Fig. 2b, Extended Data Fig. 4b), indicating that elevated 4EPS is associated with aberrant functional connectivity between various brain regions in mice.
Fig. 2 |. Functional brain connectivity and regional activation is altered in response to colonization by 4EP-producing bacteria.

a, fUSi mean connectivity matrices correlating signal in brain regions43 for each group, 4EP+ and 4EP- (n=7), and each coronal plane, bregma −0.9mm, −1.6mm, and −2mm. Pearson correlation coefficient (r) is indicated by color according to the legend on right. Location of regions of interest (ROI) is categorized below, with further details found in Ext. Data Fig. 4a,b and Supplementary Information Table 4. b, Region pairs with significantly different connectivity in 4EP+ compared to 4EP- groups. Regions pairs are indicated by identical color of p-value (according to legend), and further indicated by dotted lines where necessary. Landmark brain regions are labeled in black, and specific ROIs are defined in Extended Data Fig. 4b and Supplementary Information Table 4. Analysis was performed using a paired t-test of each of the connectivity matrices with a Bonferroni correction to control for multiple comparisons. c-d, Color-coded overlays over representative coronal (c) and sagittal (d) sections of the mouse brain template showing significant differences in regional cerebral glucose uptake 4EP+ mice compared to 4EP- mice (n=11) (t-test, p ≤ 0.05, extent threshold > 200 contiguous voxels, with both conditions met to be deemed significant; red/blue: increase/decrease in glucose uptake in 4EP+ compared to 4EP- mice). Two independent cohorts of mice from multiple litters were used for each experiment in the figure. Abbreviations: AM, amygdala; TH, thalamus; HY, hypothalamus; CC, corpus callosum; HPC, hippocampus; CTX, cortex; GP, globus pallidus; LS, lateral septum; PVT, paraventricular nucleus of the thalamus; BNST, bed nucleus of the stria terminalis; RT/VPL: reticular/ventral posterolateral thalamic nuclei; Cb: cerebellar vermis; Pn: pontine reticular nucleus.
To compare neural activity across the brain, we mapped glucose uptake where a systemically injected radiolabeled tracer [(14C]-2-deoxyglucose, 2DG) is rapidly incorporated into active brain regions. Changes in brain activity were evaluated by autoradiography of brain sections comparing 4EP+ to 4EP- mice. 4EP(S) was associated with increased glucose uptake in subregions of the hypothalamus (anterior area; lateral; and paraventricular nucleus), amygdala (anterior, basolateral, central, cortical), and the bed nucleus of the stria terminalis (BNST), as well as in the paraventricular nucleus of the thalamus (PVT) (Fig. 2c,d). We also mapped uptake during a behavioral task (open-field exploration) where we observed overlap in increased activity in some regions (amygdala, hypothalamus, BNST, PVT), with differences in the spatial extent of these changes between stimuli conditions (Extended Data Fig. 4c-e). The regions highlighted by this analysis are important for a range of functions, including mediating appropriate responses to innate and learned fear stimuli20–23 and anxiety responses24,25. We conclude that gut exposure to 4EP in mice results in altered functional connectivity and activity in multiple brain regions, including several associated with the limbic system.
Altered oligodendrocyte maturation
To resolve molecular effects of 4EP(S) on the brain, we performed mRNA sequencing (QuantSeq) of six brain regions from 4EP+ and 4EP- mice, including the PVT, basolateral amygdala, hypothalamus, BNST, medial prefrontal cortex (mPFC), and ventral hippocampus, resulting in tight clustering of transcriptomic profiles by brain region (Extended Data Fig. 5a). 4EP(S) predominantly affected global gene expression in the PVT, and the BNST and basolateral amygdala to a lesser extent (Fig. 3a, Extended Data Fig. 5b, e). Differentially expressed genes were aggregated into functional categories using annotated Gene Ontology (GO) terms, disclosing that the Notch signaling pathway was elevated in the PVT of 4EP+ mice, while GO terms associated with dendrite and neuronal projection development were decreased (Extended Data Fig. 5c-d). Cell-specific enrichment analysis revealed decreased expression of genes specific to neurons, newly formed oligodendrocytes, and mature oligodendrocytes in the PVT of 4EP+ mice compared to 4EP- mice (Fig. 3b), suggesting that a potential decrease in development, abundance, and/or activity of these cell types is associated with exposure to 4EP(S). Increased proliferation of immature oligodendrocytes and decreased differentiation into mature oligodendrocytes has been associated with elevated Notch signaling26,27. Mature oligodendrocytes insulate neuronal projections with myelin, a fatty sheath that promotes conduction of action potentials along axons28. Accordingly, many genes that are hallmarks of mature oligodendrocytes such as the myelin oligodendrocyte glycoprotein (Mog) and Opalin genes were downregulated in the PVT of 4EP+ mice, while several genes associated with non-myelinating, oligodendrocyte progenitor cells (OPCs) were elevated (Fig. 3c, Ext. Data Fig. 5f, Supplementary Information). Using seed analysis of the 2DG-uptake data29, we observed fewer significant correlations between the PVT and the rest of the brain in the 4EP+ group (168,042 voxels) compared to the 4EP- group (271,392 voxels), an effect that is largely driven by the reduced number of positive correlations in the 4EP+ compared to the 4EP- group (61,572 vs 141,493 voxels) (Fig. 3d, Extended Data Fig. 5g).
Fig. 3 |. Reduced oligodendrocyte maturation in 4EP+ mice.

a, Number of differential genes in brain regions (n=5) of baseline mice; QuantSeq, one-way ANOVA Tukey test. b, Cell-specific enrichment analysis; p-values<0.05 in white. c, Representative subset of differential OPC and MO genes, one-way ANOVA Tukey with boxes showing 25–75th percentiles, median, and min/max points (n=5). d, Whole brain correlation to PVT seed using 2DG uptake data (n=11). PVT (red cross-section) is indicated. One-tailed multiple linear regression with minimum threshold of 200 contiguous voxels with p<0.05 was performed. e, Representative images of brain sections from 4EP- and 4EP+ mice; NG2 (red) and CC1 (green). Scale 100µm. f, Maturity quotient of oligodendrocytes in the PVT (n=8 cumulative totals used from biologically independent mice). p=0.004. g, Quantitative flow cytometry of PVT cells comparing the MOG+ quadrant 4 population over the NG2+ quadrant 1 population (n=4 pools of 5 PVTs). p=0.03. h, Representative flow plots. i-l, Organotypic brain slices treated with 10μM 4EPS. Each datapoint represents separate biological replicates. i, Maturity quotient of oligodendrocytes; using total cumulative counts from 3–5 images per replicate (vehicle, n=9; 4EPS, n=10). p=0.003. j, Percent colocalization of neurofilament (NF) and proteolipid protein (PLP) antibody stain (n=6). p=0.01. k, Representative images of panel j. Scale 20µm. l, Western blot for MBP of slices (n=6). p=0.04. Abbreviations: PVT, paraventricular nucleus of thalamus; BNST, bed nucleus of stria terminalis; BLA, basolateral amygdala; mPFC, medial prefrontal cortex; HY, hypothalamus; vHPC, ventral hippocampus; Astro, astrocyte; OPC, oligodendrocyte precursor cell; NFO, newly formed oligodendrocytes; MO, mature oligodendrocytes; Micro, microglia; Endo, endothelia; CC1, adenomatous polyposis coli; NG2, neural/glial antigen 2; MBP, myelin basic protein. Biologically independent mice from multiple randomized litters examined over one (a-c) or two respective experiments (d-l). Data represent mean ± SEM. Panels e-l analyzed by two-tailed Welch’s t-test. *p ≤ 0.05; **p ≤ 0.01.
We tested the hypothesis that 4EP(S) impacts oligodendrocyte maturation. Immunostaining in the PVT revealed increased expression of neural/glial antigen 2 (NG2) in 4EP+ mice, indicative of immature, oligodendrocyte precursor cells, along with decreased levels of a mature oligodendrocyte marker stained by a CC1 antibody (Fig. 3e-f, Extended Data Fig. 5h-o). Analysis by flow cytometry and Western blot corroborated a skewing toward immature oligodendrocytes in 4EP+ mice (Fig. 3g, h, Extended Data Fig. 6a-c). Levels of NeuN, a pan-neuronal marker, and OLIG2, a pan-oligodendrocyte marker, were unchanged (Extended Data Fig. 5k-n), indicating effects on oligodendrocyte maturation by 4EP(S) rather than a change in the total number of cells within this lineage. These data suggest that 4EP(S) exposure leads to reduced oligodendrocyte maturation.
Similar phenotypes were observed in organotypic brain slices cultured in the presence of 4EPS. 4EPS-treated ex vivo brain tissue showed increased levels of the early oligodendrocyte marker NG2 relative to the mature marker CC1 (Fig. 3i, Extended Data Fig. 6d), and reduced colocalization of myelin with neuronal axons (Fig. 3j-k, Extended Data Fig. 6e). Further, functional markers of mature oligodendrocytes (MOG and myelin basic protein, MBP), were lower in 4EPS-treated samples, while transcription of the gene for NG2 (Cspg4) was increased (Fig. 3l, Extended Data Fig. 6f-g). While 4EPS enters the brain and has direct effects on brain tissue, we cannot exclude peripheral influences.
Reduced neuronal myelination in 4EP+ mice
We employed electron microscope tomography (ET) to examine the ultrastructure of myelin in the dense and organized axonal tracts of the corpus callosum to facilitate myelin quantification. We observed a striking increase in the ratio of unmyelinated to myelinated axons in the brains of 4EP+ mice compared to 4EP- mice (Fig. 4a, d), and a decrease in normalized (indicated by an increased g-ratio) and actual myelin thickness in 4EP+ mice (Fig. 4b-d, Extended Data Fig. 7a-e). Thus, consistent with the decrease in mature oligodendrocytes, 4EP(S) exposure reduces myelination frequency and efficiency in the brain.
Fig. 4 |. 4EP(S) alters myelination and anxiety-like behavior.

a, Percent of unmyelinated axons (n=3; 4 images scored from each biological replicate). p=0.005. b, G-ratio (r/R) of all axons (4EP-, 56; 4EP+, 70 axons)(n=4; 4 images scored from each biological replicate). p=0.0001. c, Width of myelin; (4EP-, 56; 4EP+, 70 axons) (n=4; 4 images scored from each biological replicate). p=0.003. d, Representative electron tomography images. Several myelinated and unmyelinated axons marked by blue and orange arrows, respectively. e, Fractional anisotropy (FA) measured by diffusion tensor imaging (DTI) (n=4). p=0.04. f, Elevated plus maze: time spent in the terminus (outer one-third) of the open arms (4EP+ n=21; 4EP- n=24). p=0.003. g, Open field test (4EP+ n=21; 4EP- n=24). p=0.002. h, Number of marbles buried (4EP+ n=24; 4EP- n=23). p=0.02. i, Maturity quotient of oligodendrocytes in the PVT of 4EP- and 4EP+ mice ±clemastine fumarate (Cumulative totals used from individual mice: 4EP- control=7, treated=10; 4EP+ control=6, treated=6)(Cont 4EP-vs 4EP+ p=0.03; 4EP+Cont vs Treatment p=0.01). j, Representative images of brain sections from mice ±clemastine fumarate, stained for CC1 (green) and NG2 (red). k-m, Behavioral tests of mice (independent of cohorts used previously) ±clemastine fumarate. k, EPM (4EP- control n=13, treated n=13; 4EP+ control n=12, treated n=14)(4EP+Cont vs Treatment p=0.05). l, Open field test (4EP- control =15, treated n=17; 4EP+ control n=17, treated n=17) p=0.04. m, Number of marbles buried(4EP- control =15, treated n=17; 4EP+ control n=17, treated n=17)(Cont 4EP-vs 4EP+ p=0.009; 4EP+Cont vs Treatment p=0.03). Abbreviations: FA, fractional anisotropy; NG2; neural/glial antigen 2; CC1, adenomatous polyposis coli. Biologically independent mice from multiple randomized litters examined over two respective experiments used for all experiments. Data represent mean ± SEM analyzed by two-tailed Welch’s t-test (a-h) or two-way ANOVA with Dunnett multiple comparison to the 4EP+control group (i-m). *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001.
Diffusion tensor imaging (DTI), a magnetic resonance imaging modality that assesses diffusion along myelinated tracts in the brain, was used to investigate the structural connectivity of myelin between the PVT and the rest of the brain. We found lower fractional anisotropy (FA), an indication of more dispersive rather than linear/restricted diffusion, in 4EP+ compared to 4EP- mice (Fig. 4e, Extended Data Fig. 7f). A similar defect in myelination was observed in whole brains and trended in the corpus callosum (Extended Data Fig. 7g-i). The significance of altered myelin dynamics and brain connectivity is an emerging concept in behavioral neuroscience10–12,30,31.
4EP(S) increases anxiety-like behavior
Our previous study8 and the 4EP(S)-dependent changes in the limbic system observed here prompted us to investigate whether 4EP production in the gut can modulate complex behaviors in mice. 4EP(S) promoted robust anxiety-like behavior in several testing paradigms: 1) the elevated plus maze (EPM) where 4EP+ mice spend less time in the terminus of the open arms, 2) open-field exploration where mice ventured less into the more exposed zone of the arena, and 3) the light/dark box where 4EP+ mice spent more time in the dark (Fig. 4f, g, Extended Data Fig. 8a-c). 4EP+ mice also displayed increased marble burying, reflecting features of anxiety and/or stereotypic behaviors, but no increase in self-grooming (Fig. 4h, Extended Data Fig. 8d). Beyond anxiety-like behaviors, 4EP+ mice exhibited modestly altered social communication with increased anogenital sniffing in the direct social interaction assay (Extended Data Fig. 8e). In the adult ultrasonic vocalization (USV) test, male 4EP+ mice emit significantly fewer auditory communications to a novel age-matched female (Extended Data Fig. 8f). Interestingly, there were no significant differences in cognition or motor function between groups (via novel object recognition, Y-maze alternation, beam traversal, pole descent, or wire hang tests) (Extended Data Fig. 8g-k), further suggesting the effects of 4EP(S) are selective for emotional behaviors. Our reductionist model system employs an artificial microbiome to study the effects of a gut microbial metabolite (Extended Data Fig. 8l). Importantly however, oral administration of 4EP(S) to elevate levels in conventionally colonized mice also increased anxiety-like behaviors and reduced oligodendrocyte maturation (Extended Data Fig. 9a-j). 4EP- mice behaved similarly to conventionally colonized controls (Extended Data Fig. 9k,l), indicating the behavioral effects of 4EP(S) are not specific to gnotobiotic mice.
Finally, we sought to determine whether the 4EP(S)-mediated effects on oligodendrocytes contribute to altered behaviors. Administration of clemastine fumarate, a drug that promotes oligodendrocyte maturation32, increased mature oligodendrocyte ratios in 4EP+ mice as measured by CC1 and NG2 staining of brain sections (Fig. 4i,j, Extended Data Fig. 10a). Notably, enhancing maturation of oligodendrocytes prevented behavioral changes in 4EP+ mice, including alterations in EPM, open field, and the marble burying tests (Fig. 4k-m, Extended Data Fig. 10c,e,f). We observed similar improvements in anxiety-like behaviors with another myelination-inducing drug, miconazole (Extended Data Fig. 10b,d, g-i). We conclude that 4EP(S) impacts anxiety-like behaviors in mice in a manner that includes effects on oligodendrocyte maturation.
Discussion
Herein, we discovered a biosynthetic pathway for production of the gut microbial metabolite 4EP. While this pathway can utilize tyrosine as a precursor, we show that other dietary sources can also be metabolized by the gut microbiota into 4EP, and expect that in humans, diverse dietary and microbial community structures may impact circulating metabolite levels. Additionally, we show that 4EP is sulfated and 4EPS enters the brains of mice, is associated with altered activation and connectivity of specific brain regions, and disrupts the maturation of oligodendrocytes and myelination patterns in the brain. Environmental cues are known to have regional effects on oligodendrocyte-neural interactions, influencing brain circuits that govern particular behaviors6,7. While other brain regions are likely involved in the 4EP(S) response, as we detect broad changes to activation patterns, the PVT receives sensory and cognitive input, integrates these cues locally, and exports finely tuned signals to cortical and subcortical areas resulting in applied behavioral responses33. Indeed, we show gut exposure to 4EP alters several emotional, but not non-emotional, behaviors in mice. Future work will focus on uncovering how 4EPS leads to changes in oligodendrocyte maturation and myelination, defining brain regions that are causally affected, and exploring how myelination changes impact behavior. Our data do not resolve if 4EPS is the neuroactive metabolite, versus 4EP or unknown breakdown products. Identification of gut-derived microbial metabolites that enter the brain and affect brain activity defines a novel environmental influencer of anxiety-like behaviors.
Gut bacteria can synthesize classical neurotransmitters such as dopamine, norepinephrine, serotonin and gamma-aminobutyric acid, and the production of novel classes of neuroactive metabolites by the microbiome has been postulated34. Molecules with phenolic structures similar to 4EP(S) are dysregulated in several preclinical models of behavior8,35,36 as well as in certain neuropsychiatric disorders37–41. Intriguingly, a metabolite of tyrosine closely related to 4EP, p-cresol, has been suggested to influence oligodendrocyte function and affect social and depression-like behaviors in mice6,42. Increased 4EPS is associated with abnormal repetitive behavior in non-human primates36, and plasma levels of 4EPS are significantly increased in a subset of individuals with ASD15. Additionally, relative abundances of several related metabolites such as 2-ethylphenylsulfate, 4-allylphenyl sulfate, and 4-methylbenzenesulfonate are altered in this same cohort15. We propose the hypothesis that 4EP(S) represents the archetypical example of a neuroactive microbial molecule that impacts brain activity and complex behaviors in animals, conceptually akin to mammalian neurotransmitters that regulate nervous system function.
Materials and Methods
Bacterial strains and culture conditions
All bacterial strains and plasmids used in this study are shown in Supplementary Information Tables 1 and 2. B. ovatus was cultured in brain heart infusion (BHI) agar medium supplemented with 10% horse blood, TYG (tryptone-yeast extract-glucose) broth or minimal medium (MM) at 37 ˚C in an anaerobic chamber from Coy Laboratories44,45. Tyrosine was supplied as a substrate during investigation of 4EP production. L. plantarum was cultured anaerobically in MRS media (BD) at 37˚C. Escherichia coli strains were cultured aerobically in LB broth. When appropriate, the growth medium was supplemented with 100 µg/ml carbenicillin, 25 µg/ml erythromycin, 2 µg/ml tetracycline, 200 µg/ml gentamicin and 200µg/ml 5-fluoro-2’-deoxyuridine (FUdR).
Disruption of BACOVA_01194 in B. ovatus
Using NCBI BLAST alignment tools, we predicted that BACOVA_01194 was an ammonia lyase that could metabolize tyrosine. For all PCR amplification steps we used PrimeSTAR Max DNA polymerase (Takara Bio, Mountain View, CA) according to the manufacturer’s instructions. Primer sequences are shown in Extended Data Table 3. We used a previously reported double-crossover recombination method to construct B. ovatus Δ119446. Briefly, ~1 kb DNA fragments corresponding to the upstream and downstream regions of the target gene were PCR amplified and then digested with restriction endonucleases. The digested fragments were then ligated into the suicide plasmid pExchange-tdk using T4 DNA ligase (New England Biolabs, Ipswich, MA). The resulting plasmid was transformed into Escherichia coli S17–1 λ pir by electroporation and transformants were confirmed by PCR amplification of the junction regions. An E. coli clone harboring the plasmid was cultivated and plasmid DNA was isolated, purified, and verified by DNA sequencing.
For conjugation into B. ovatus, B. ovatus ∆tdk and E. coli S17–1 λ pir harboring the plasmid were cultivated and the cells were harvested by centrifugation. The cell pellets were washed with PBS to remove residual antibiotics and combined in TYG medium. The suspension was plated on BHI-blood agar medium without any antibiotics and cultivated aerobically at 37 ˚C for 1 day. The bacterial biomass was recovered by scraping and re-suspended in TYG medium. The suspension was then plated on BHI-blood agar medium supplemented with erythromycin and gentamicin and single-crossover integrants were selected. These strains cultured in TYG medium overnight and plated on BHI-blood agar medium supplemented with FUdR. The deletion mutant was screened by PCR amplification and verified by DNA sequencing.
B. ovatus strain engineering
When a culture of L. plantarum was supplemented with 4-vinylphenol, it was converted quantitatively to 4-ethylphenol; in contrast, when the culture was supplemented with p-coumarate, the conversion to 4-vinylphenol was slow. This suggested that the decarboxylation step might be rate-limiting for 4-ethylphenol biosynthesis in binary culture. To address this challenge, we introduced the pad gene from L. plantarum into B. ovatus in order to enable B. ovatus to produce 4-vinylphenol directly from tyrosine. However, the resulting strain did not produce 4-vinylphenol. We next introduced a second copy of the BACOVA_01194 gene into B. ovatus along with pad in case the intermediate p-coumarate was limiting, but this strain failed to produce 4-vinylphenol. To address the possibility that the pad gene from L. plantarum does not function robustly in B. ovatus, pad from Bacillus subtilis was introduced into B. ovatus; the engineered strain produced 4-vinylphenol robustly from tyrosine. To boost the level of 4-vinylphenol, a second copy of BACOVA_01194 was introduced into B. ovatus along with pad from B. subtilis using a second integration vector with a different antibiotic marker. This strain produced 4-vinylphenol more robustly. We then combined the second copy of BACOVA_01194 and pad into a single artificial operon driven by a strong phage promoter and introduced this construct into B. ovatus. Interestingly, this strain produced a much higher level of 4-vinylphenol and it was used throughout the manuscript for co-cultivation and colonization. Vector maps and sequences of primers used for B. ovatus engineering are shown in Extended Data Fig. 1 and Supplementary Information Table 3, respectively. The artificial operon was constructed in the following way: A phage promoter along with 5’ and 3’ flanking sequences from the target vector, pNBU2, and one of the target genes was amplified by PCR. The target gene with 5’ and 3’ flanking sequences containing sequences of the phage promoter and the pNBU2 vector were also amplified. These two PCR fragments were assembled by overlap PCR. The BO_01194 and pad genes were each assembled by one more round of overlap PCR. The fragment containing the target gene and the phage promoter was cloned into the fragment of pNBU2 vector using Gibson Assembly. The assembled plasmids were transformed into Escherichia coli S17–1 λ pir competent cells by electroporation and transformants were confirmed by PCR amplification of the junction regions. The positive clone harboring assembled plasmid was cultivated and the plasmid was isolated, purified, and confirmed by DNA sequencing. To conjugate the plasmid into B. ovatus, B. ovatus ∆tdk and E. coli S17–1 λ pir harboring the plasmid were cultivated and the cells were harvested. The cell pellets were washed with PBS to remove residual antibiotics and combined them in TYG medium. The suspension was plated on BHI-blood agar medium without any antibiotics and grown aerobically at 37 ˚C for 1 day. The bacterial biomass was recovered by scraping and re-suspended in TYG medium. The suspension was then plated on BHI-blood agar medium supplemented with erythromycin and gentamicin and antibiotic resistant strains were selected followed by PCR amplification of the junction regions.
Extraction of bacterial culture metabolites
For metabolite analysis, B. ovatus was cultured in TYG medium overnight and the cells were harvested and washed with MM. The cell pellet was re-suspended in MM containing 0.5 mg/ml tyrosine to a density of OD600 = 1.0 and incubated anaerobically for 1 day. The culture was extracted with acetone (20% v/v) and centrifuged. The supernatant was analyzed by LC/MS as described below. For LC-MS analysis of co-culture experiments, B. ovatus and L. plantarum were cultivated anaerobically in TYG and MRS, respectively overnight and the cells were harvested and washed with MM. The cell pellet was re-suspended in MM containing 0.5 mg/ml tyrosine to a density of OD600 = 1.0 and combined in the same culture tube. After 1-day incubation, samples for HPLC analysis were prepared as described above.
LC/MS analysis of bacterial culture metabolites
Metabolite extracts were analyzed using an Agilent 1260 LC system coupled to an Agilent 6120 quadrupole mass spectrometer with a 3 µm, 4.6 × 75 mm Unison UK-C18 column (Imtakt, Portland, OR). Water with 10mM ammonium acetate and 0.1% formic acid (A) and acetonitrile with 0.1% formic acid (B) was used as the mobile phase at a flow rate of 1.0 ml/min with the following 20 min gradient: 0–5 min, 0% B; 5–17 min, 0–90% B; 17–20 min, 95% B. P-coumaric acid and 4-VP were detected at 280nm and these retention times were ~10.3 min and ~12.6 min, respectively. Retention time of 4-EP was ~7.6 min. Standards of p-coumaric acid and 4-ethyl phenol were purchased (Sigma, St. Louis, MO).
Gene sequence alignment
Sequences of the genes used for strain engineering were aligned against the reference genomes in the WoL database, which were pre-annotated using UniRef release 2019_07. The annotation files are publicly available at: https://biocore.github.io/wol/download. The alignment used DIAMOND v0.9.25 with all default parameters (`diamond blastx-d/path/to/db-qinput.fa-o output.txt`). The output files were then processed using Woltka (https://github.com/qiyunzhu/woltka), which generated the taxonomic and functional (UniRef) profile of the three genes against the entire WoL genome catalog.
Mouse Husbandry
All animal husbandry and experiments were approved by the Caltech Institutional Animal Care and Use Committee. Throughout the study, animals were maintained in autoclaved microisolator cages with autoclaved bedding (Aspen Chip Bedding, Northeastern Products Corp, Warrensburg, NY), water, and chow (Laboratory Autoclavable Rodent Diet - 5010, LabDiet; St. Louis, MO, USA). Diets used in Extended Data Fig. 1g were purchased from Envigo Teklad (Madison, WI), carbohydrate, and fat content. The soy protein has ~300ppm total isoflavones. The tyrosine levels are 4.6g/kg in soy diet and 17.1g/kg in fish diet. 8-wk, SPF C57BL/6J male mice (Jackson Laboratory, Bar Harbor, ME), were provided these special diets for 2 weeks ad libitum prior to urine collection for metabolite analysis as described below.
Mice were maintained at an ambient temperature of 71–75F, 30% - 70% humidity, at a cycle of 13 hours light & 11 hours dark.
Experimental Design of Mouse Experiments
Germ-free (GF) C57BL/6J male weanlings (3 weeks of age) from the Mazmanian laboratory colony were colonized by gavage of 100ul of 1:1 mixture of 109 CFU/ml B. ovatus (+/− 4EP pathway genes) and wild type L. plantarum. This process was continually performed, yielding a steady schedule of cohorts for continued study throughout this work. Size of animal groups was determined by the largest number of pups that could be born within the space of the germ-free isolators. Weekly tests to confirm gnotobiotic status were performed and 4EPS levels were confirmed in urine regularly. All experiments in the study were repeated on at least two independent cohorts. Preliminary behavioral analysis identified a stronger phenotype in males (Extended Data Fig. 10j-l), so they were used for the remainder of the study in effort to limit animal use.
16S sequencing was performed by Laragen (Culver City, CA) and analysis was done with a 16S V6 library. Paired-end fastq files were processing using Qiime247. Briefly, sequences were quality control processed with dada248, truncating reads to 150 bp. Taxonomic classification was then conducted using the greengenes database. Analysis was conducted on samples rarefied to 24,000 reads.
In the conventionally colonized (SPF) experiment, male weanlings (3 weeks of age) were provided 250mM 4EP and 4EPS in drinking water (or vehicle). In these experiments, oral administration continued until endpoint. 4EPS was synthesized as previously described8, and 4EP was purchased from Sigma, (St. Louis, MO). When appropriate, beginning at 4–5 weeks of age, sterile clemastine fumarate (0.03mg/ml) or DMSO vehicle (Sigma, St. Louis, MO) was added to drinking water and water was changed every other day, or miconazole solution (40mg/kg) or 3% DMSO vehicle was gavaged once daily. In all cases, behavior testing started at 6 weeks of age.
Extraction of urine, feces, serum and brain metabolites
Urine was collected by placing autoclaved aluminum foil under the mouse while briefly scruffing it, and then pipetting the urine from the foil. The urine was diluted 5-fold with aqueous acetonitrile (50% v/v) and centrifuged. The supernatant was analyzed by LC/MS to detect 4EPS. 4EPS for a standard was prepared as previously described8. Ethyl acetate was added to the supernatant to create a 1:1:1 mixture (v/v) of water, acetonitrile, and ethyl acetate. After mixing and centrifugation, the organic layer was analyzed by GC/MS to detect 4EP.
Fecal pellets were collected by placing the mouse briefly into a sterile plastic beaker. A 5x volume of 50% acetonitrile/water was added and the pellets were homogenized by bead beating, followed by centrifugation. The supernatant was analyzed by LC/MS to detect 4EPS. To detect 4EP, samples were extracted with ethyl acetate as described above.
Blood was collected by cardiac puncture followed by separation using Sarstedt Serum-Gel microtubes (Thermo Fisher Scientific, Waltham, MA) according to the manufacturer’s specifications.
Brain tissue was collected by cardiac perfusion as described for immunohistochemistry but with PBS instead of PFA, then dissected. A 2x volume of 50% acetonitrile/water was added and the tissue was homogenized by bead beating, followed by centrifugation. The supernatant was directly injected for analysis by LC/MS to detect 4EPS.
LC/MS analysis of metabolites from urine, serum, and brain
Samples were analyzed using an Agilent 1290 LC system coupled to an Agilent 6530 QTOF with a 3 µm, 4.6 × 75 mm Unison UK-C18 column using the same method described above. All data were collected in negative ion mode. 4EPS was detected as [M-H]- (calculated m/z is 201.0227) and retention time was ~7.9 min. 4EPS prepared previously was used as a standard8.
GC/MS analysis of metabolites from feces
Samples were analyzed with a split ratio of 10:1 using an Agilent 7890 GC coupled to an Agilent 5977 MSD with a HP-5MS fused silica capillary column (30m x 250 µm x 0.25 µm). The injector temperature was set at 250˚C and high purity helium gas was used as carrier at a constant flow rate of 1.0 ml/min. The column temperature was initially kept on 40˚C for 2 min, then increased to 100˚C at a rate of 40˚C/min, then went up to 105˚C at a rate of 2˚C/min and then raised to 320˚C at a rate of 30˚C/min, held for 3 min, giving 16.367 min in total. Retention time of 4-EP (calculated m/z is 122.07) was ~7.6 min.
Creatinine measurement
Concentration of creatinine was measured using Colorimetric Creatinine Assay Kit (Abcam, Cambridge, UK) according to the manufacturer’s instructions.
Brain Levels of 4EPS
Probenecid (Invitrogen, Carlsbad, CA), an organic anion transporter (OAT) inhibitor that works on OAT1 and OAT3 was injected intraperitoneally (i.p.) into mice (120mg/kg). After 1-hour, whole brains were removed, homogenized, and analyzed as described above by LCMS. For SPF animals, 30 minutes after the probenecid injection, either an i.p. injection of 100ul of 8mM 4EPS or an oral gavage of 160mg/kg 4EP (Sigma, St. Louis, MO) was administered, then brains were harvested after 30 additional minutes or along 30-minute time points. For mice colonized with 4EP- and 4EP+ strains, mice were perfused with PBS before tissue collection to ensure any metabolite detected was not simply due to levels in the blood.
qPCR
RNA was extracted using the RNeasy Mini Kit (Qiagen, Hilden, Germany) and cDNA was transcribed using the iScript cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, MA). qPCR was performed with the SYBR master mix (Thermo Fisher Scientific, Waltham, MA) using the primers found in Supplementary Information Table 5.
In vitro assays of 4EP sulfation
Recombinant sulfotransferases (sult1b1, 1c2, 1e1, 2a1, and 1a1) and the Universal Sulfotransferase Activity Kit (R&D Systems, Minneapolis, MN) were used according to manufacturer’s recommendations. Following analysis by plate reader, samples were analyzed by LCMS as described above to confirm that the sulfated product was indeed 4EPS. Cytosolic fractions from 50–200ug tissue containing endogenous sulfotransferases were extracted and tested for SULT activity on 4EP as previously described49,50 and analyzed by LCMS as described above.
Intestinal Permeability
The FITC-dextran intestinal permeability assay was performed as described previously51 on 4EP- and 4EP+ mice at nine weeks.
Hematoxylin and eosin (H&E) staining
Gut tissue was dissected immediately after sacrifice at nine weeks of age and fixed in neutral 10% formalin, paraffin embedded, sectioned, and stained with hematoxylin and eosin (H&E) by Pacific Pathology, Inc, San Diego, CA.
Cytokine Analysis
Tissue samples collected at nine weeks of age were homogenized by bead beating in lysing matrix D tubes (MP Biomedicals, Irvine, CA) in RIPA buffer (Millipore, Burlington, MA) containing protease inhibitor tablets (Roche, Basel, Switzerland) followed by protein quantification and normalization using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific, Waltham, MA). Blood samples were collected by cardiac puncture followed by serum isolation in clotting tubes (Sarstedt, Newton, NC). Using the Bio-Plex Pro Mouse Cytokine 23-plex assay (Bio-Rad, Hercules, CA) according to manufacturer’s recommendations, cytokine and chemokine levels were determined using a Bio-Plex 200 Systems instrument. Undetected values were rare, and in those cases were imputed with the lowest detected value from that cytokine. One-way ANOVA with multiple comparisons was performed and significance is indicated with asterisks (*<0.05, **<0.01, ***<0.001).
Microglial enrichment for qPCR
Brain samples were collected and single cell suspensions were generated as described in flow cytometry section below. Microglial-enriched populations were obtained using CD11b microbeads (Miltenyi Biotec, San Diego, CA).
Animal protocol for functional ultrasound imaging (fUSi) session
14 mice were imaged in total: 7 mice for the 4EP- group and 7 for the 4EP+ group. Images were acquired through intact skull and skin after hair removal using a commercial depilatory cream (Nair™ , Church&Dwigth, USA) without any contrast agent injection. During handling (shaving, positionning) mice were anaesthetized with isoflurane (2%) administered in a mixture of 30% O2 and 70% N2. During the resting state experiment, animals were sedated using dexmedetomidine (Tocris Bioscience, Minneapolis, MN). A bolus of 0.10 mg/kg was injected subcutaneously, and isoflurane was discontinued after 5 minutes. Mice were head fixed on a stereotaxic frame to minimize brain motion during imaging. After a 90 min. imaging session, animals were euthanized by cervical dislocation.
Functional ultrasound imaging
Functional ultrasound imaging (fUSi) visualizes neural activity by mapping local changes in cerebral blood volume (CBV). CBV variations are tightly linked to neuronal activity through the neurovascular coupling52 and are evaluated by calculated power doppler variations in the brain53. fUSi was performed transcranially as described in54 using a 15 MHz ultrasonic ultralight probe prototype (15 MHz, 64 elements, 0.110 mm pitch, Vermon, Tours, France) connected to a Verasonics Vantage ultrasound system (Verasonics Inc., Redmond, WA, USA) driven by custom Matlab® (MathWorks, USA) transmission scripts (https://github.com/brittanyneedham/Needham_Nature2022)55. Each Power Doppler image was obtained from the temporal integration of 220 compounded frames acquired at 500 Hz frame rate, using 5 tilted plane waves separated by 3° (−6°, −3°, 0°, 3°, 6°) acquired at a 2500 Hz pulse repetition frequency (PRF). Power Doppler images were then repeated every second (1Hz image framerate). Each block of 220 images was processed using a SVD clutter filter56 to separate tissue signal from blood signal to obtain a final Power Doppler image exhibiting cerebral blood volume (CBV) in the whole imaging plane. Three coronal planes per mice were scanned at a rate of 15min imaging time per plane, respectively Bregma −0.9mm, Bregma −1.6mm, and Bregma −2mm.
Functional ultrasound data processing and statistics
Power Doppler data were collected continuously during the imaging session and connectivity process was applied afterwards. We followed the functional connectivity process on fUSi data described in Osmanski, et. al57. For each coronal plane, for each acquisition, and each mouse: first, a low-pass filter (cutting-frequency: 0.2 Hz) was performed on the Power Doppler temporal signals for each individual pixel of the image to remove high frequency signals while preserving the resting-state frequency band. The signal was then detrended with a polynomial fit of order 4 to remove low frequencies which could bias the correlation value. Finally, the normal score of the temporal filtered signals were calculated to make possible correlation calculation. In order to build functional connectivity matrices, we determined within each coronal plane regions of interest (ROIs) defined from the Paxinos Atlas43 (See Extended Data Fig. 2a for ROIs mapping in each plane). The Pearson correlation of the filtered signals of each pair of ROIs’ within a same plane were then calculated and the corresponding correlation values were stored in the cells where regions intersect in the connectivity matrix. Fig.2 shows the mean connectivity matrices from each coronal plane (Bregma-0.9mm, Bregma-1.6mm and Bregma-2mm) of each studied group (4EP- and 4EP+). Within the same coronal plane, cells of the connectivity matrices from the 4EP- and 4EP+ groups were statistically analyzed individually using a paired t-test. Multiple comparison correction was ensured with a Bonferroni correction58. Region pairs that showed significant differences between groups are shown in Fig. 2c.
ROIs distribution graphs of each coronal plane are provided. Coronal plane B-0.9mm: ROIs #1 to #8 are located in the left cortex, ROI#9 is the left hippocampus, ROIs#11 to #21 are located in the thalamus, ROI#22 is the right hippocampus, ROIs#23 to #30 are located in the right cortex and finally ROIs#31 to #48 are subthalamic regions. Coronal plane B-1.6mm: ROIs #1 to #20 are located in the cortex, ROIs#21 #22 are the left and right hippocampi, ROIs#23 to #38 are located in the thalamus and ROIs#39 to #42 are subthalamic regions. Coronal plane B-2mm: ROIs #1 to #9 are located in the left cortex, ROI#10 is the left hippocampus, ROIs#11 to #22 are located in the thalamus, ROI#23 is the right hippocampus, ROIs#24 to #32 are located in the right cortex and finally ROIs#33 to #50 are subthalamic regions.
Autoradiography Brain Mapping
The autoradiographic 2DG uptake method is a well-established, time-tested approach to functional brain mapping based on a tight coupling between neural activity and metabolism. It is particularly suitable in awake, free-moving animals, and complements the fUSi approach. Male mice colonized as described above were housed in pairs from weaning. Mapping of cerebral glucose metabolism was performed as described previously59,60 in four groups: 4EP+/Home cage (n = 11), 4EP+/Open field (n = 11), 4EP-/Home cage (n = 10), 4EP-/Open field (n = 11). The experiment was performed in two cohorts with balanced group assignment in each cohort. At 7 weeks mice were habituated to handling for 5 minutes each day for 3 days prior to 2DG mapping. They were brought in their home cages to the experimental suite 16 hours prior to mapping and were fasted of food overnight with water ad libitum. A pair of mice from the same home cage were administered i.p. [14C]-2-deoxy-D-glucose (Cat # MC355, radiochemical purity > 97%, specific activity 45 – 60 mCi/mmol, Moravek Inc., Brea, CA, USA) at 0.3 µCi/g bodyweight in 0.5 ml normal saline. Animals were placed back in their home cage for five minutes. One mouse was then placed into the Open Field arena and allowed to explore the arena for 45 minutes to allow uptake of the tracer, while the other remained in the home cage. At the end of exposure, following cervical dislocation, brains were extracted and flash frozen in methylbutane over dry ice (~ −55oC) and later serially sectioned into 20-μm slices in a cryostat at −20oC (Mikron HM550 OMP, Thermofisher Scientific, Waltham, MA, USA). Slices were heat dried on glass slides and exposed to Kodak Biomax MR diagnostic film (Eastman Kodak, Rochester, NY, USA) for 3 days at room temperature. Autoradiographs were then digitized on an 8-bit gray scale using a voltage stabilized light box (Northern Lights Illuminator, InterFocus Ltd., England) and a Retiga 4000R charge-coupled device monochrome camera (Qimaging, Canada).
Relative regional cerebral glucose uptake (rCGU) was measured and analyzed on a whole-brain basis using Statistical Parametric Mapping (SPM, version 5, Wellcome Centre for Neuroimaging, University College London, London, UK) as previously described59,61. Here briefly, each three-dimensional (3D) brain was reconstructed from 68 digitized autoradiographs (voxel size: 40 × 140 × 40 μm) using TurboReg, an automated pixel-based registration algorithm implemented in ImageJ (version 1.35, http://rsbweb.nih.gov/ij/). This algorithm registered each section sequentially to the previous section using a nonwarping geometric model that included rotations, rigid-body transformation, and nearest-neighbor interpolation. One “artifact free” mouse brain was selected as reference, and all brains were spatially normalized to the reference in SPM. Spatial normalization consisted of applying a 12-parameter affine transformation followed by a nonlinear spatial normalization using 3D discrete cosine transforms. All normalized brains were then averaged to create a final mouse brain template. Each original 3D-reconstructed brain was then spatially normalized to the template. Normalized brains were smoothed with a Gaussian kernel (full width at half maximum = 3x voxel dimension in the coronal plane). Voxels for each brain failing to reach a specified threshold in optical density (70% of the mean voxel value) were masked out to eliminate the background and ventricular spaces without masking gray or white matter. Differences in the absolute amount of radiotracer uptake in the brain were normalized in SPM for each animal by scaling the voxel optical densities such that the whole-brain mean for each brain was the same (proportional scaling). For each condition (open field and home cage exposure), one-tailed t-tests were performed voxel-by-voxel comparing 4EP+ and 4EP- animals. Threshold for significance was set at P < 0.05 at the voxel level and an extent threshold of 200 contiguous voxels to eliminate false positive statistically significant results. Color-coded functional overlays showing statistically significant changes in rCGU were displayed over coronal sections of the template brain in MRIcro (version 1.40, https://people.cas.sc.edu/rorden/mricro/mricro.html). This combination reflected a balanced approach to control both type I and type II errors. The minimum cluster criterion was applied to avoid basing our results on significance at a single or small number of suprathreshold voxels. Brain regions were identified according to a mouse brain atlas(43 and atlas.brain-map.org).
A seed correlation approach was applied to assess 4EP-related differences in the functional connectivity of the PVT. A structural region of interest (ROI) was hand drawn in MRIcro over the template brain according to the mouse brain atlas for the PVT between bregma −1.0 and −1.6mm. Mean optical density of the seed ROI was extracted for each animal using the MarsBaR toolbox for SPM (version 0.42, http://marsbar.sourceforge.net). Correlation analysis was performed in SPM for each home cage group. T statistics were calculated using a linear regression model with the seed value as the only covariate (regressor). Threshold for significance of directional correlation was set at p < 0.05 (one-tailed t-test) at the voxel level and an extent threshold of 200 contiguous voxels to serve as a proxy for multiple comparison correction, which is standard for the field. Regions showing statistically significant correlations (positive or negative) in rCGU with the seed are considered functionally connected with the seed. Color-coded functional overlays were displayed in the template brain after 3D rendering in MRIcro to allow visual comparison of overall level of functional connectivity.
Brain sample collection for immunohistochemistry
Mice were perfused via the cardiovascular system with PBS followed by 4% paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA). Brains were removed and post-fixed in 4% paraformaldehyde 1 day at 4°C. The brains were kept in PBS with 0.02% sodium azide at 4°C until sectioning. For sectioning, the brains were embedded in 4% UltraPure low melting point agarose (Thermo Fisher Scientific, Waltham, MA) and were coronally sectioned by vibratome (VT1000S; Leica Microsystems, Wetzlar, Germany) at a thickness of 50 µm. Brain sections of 50 µm were collected and stained every 0.15 mm. The brain sections were stored as free-floating in PBS with 0.02% sodium azide at 4°C until staining.
The free-floating sections were incubated with primary antibody in blocking solution (10% horse serum, 0.3% triton X-100, and 0.02% sodium azide in PBS) overnight at room temperature. The next day, sections were incubated with fluorescence-conjugated secondary antibody for 1.5–2 hours at room temperature. Between each step and after secondary antibody staining, sections were thoroughly washed with PBS or PBS with 0.1% triton-X-100 at least three times for 10 minutes each. The stained free-floating sections were then mounted onto the Superfrost Plus microscope slides (Fisher Scientific, Hampton, NH) in PBS. Excess PBS from adhered sections were carefully removed. Slides were dried at room temperature for 2–5 minutes. 150–200 µl of ProLong Diamond, anti-fade mountant with DAPI (Thermo Fisher Scientific, Waltham, MA) was applied to the slides before placing the coverslip. The slides were left to set overnight before imaging.
Primary antibodies used for imaging throughout and their dilutions were: mouse anti-NeuN (1:1000; MAB377; Millipore Sigma, Burlington, MA), goat anti-Olig2 (1:500; AF2418; R&D Systems, Minneapolis, MN); mouse anti-CC1 (1:250; NB600–1021; Novus Biologicals, Littleton, CO); rabbit anti-NG2 (1:300; AB5320; Millipore Sigma, Burlington, MA); chicken anti-MBP (1:250; CH22112; Neuromics, Edina, MN); mouse anti-neurofilament (1:250; 837802; Biolegend, San Diego, CA), rabbit anti-PLP (ab183493, Abcam, Cambridge, UK). The fluorescent-conjugated secondary antibodies were donkey anti-goat (1:1000; A-32814, A-21082, A11057; ThermoFisher Scientific, Waltham, MA), donkey anti-rabbit (1:1000; A-21206, A-10042, A-31573; ThermoFisher Scientific, Waltham, MA), and donkey anti-mouse (1:1000; A-21202, A-10037, A-31571; ThermoFisher Scientific, Waltham, MA), and donkey anti-chicken (1:1000; A-11041; A-11039; A-21449, Thermo Fisher Scientific, Waltham, MA).
Microscopic imaging and image analysis
Imaging was performed using the Zeiss LSM 800 inverted confocal laser scanning microscope (Carl Zeiss, Oberkochen, Germany) with Zen software (Carl Zeiss, Oberkochen, Germany). Confocal images were obtained by Z-stacks covering the entire Z-axis range of the sections. The interval for each focal plane was 2 µm intervals. The images were then projected in the visualization plane with maximum intensity voxels (3D pixel) by maximum intensity projection using Zen software. Positively stained cells were quantified using a manual cell counter in ImageJ software (NIH). All images were minimally processed with brightness and contrast adjustment. The adjustment was applied equally across the entire image and consistent in the corresponding controls. Regions of interest were selected by a segmented line based on the anatomical features of each region. The final number of positive cells reported is averaged from 4 images.
Coordinates for imaging relative to Bregma (bilateral) were: BLA −1.06 to −2.06mm; BNST: +0.38 to +0.26mm; PVN and AH: −0.70 to −0.94mm; mPFC: +1.10mm; PVT: −0.70 to −1.58mm; LHB and MBH: −1.06 to −1.34mm; SM: −0.46 to −1.34mm; ACA: 0.26mm; CC: 0.26mm; LS: ; MS: 0.26mm; ME: −0.82mm.
QuantSeq
Following cervical dislocation, brains were extracted and placed in iced PBS for five minutes. The brains were placed upside down into a brain matrix (SA-2175; Roboz, Gaithersburg, MD) and sliced by single edge blades (Personna, Verona, VA). Brain slices were left on the blades and placed on ice to maintain RNA integrity. Specific brain regions were isolated by gross dissection or brain punches by using 1.0 mM Biopsy Punches (Miltex, VWR, Radnor, PA) and were immediately frozen in RNAlater (Qiagen, Hilden, Germany) until RNA collection following manufacturers recommendations using the Split RNA extraction kit (Lexogen, Greenland, NH).
Coordinates for mPFC brain slices spanned from anterior to posterior (AP) +1.94 mm to +1.10 mm relative to bregma. The gross anatomy of the mPFC was based on the morphology of corpus callosum and the appearance of lateral septum. Coordinates for BNST brain slices spanned from anterior to posterior (AP) +0.62 mm to +0.14 mm relative to bregma (bilateral). The gross anatomy of the BNST was based on the features of caudate putamen, lateral ventricle, and anterior commissure. Coordinates for PVT brain slices spanned from anterior to posterior (AP) −0.94 mm to −1.58 mm relative to bregma. The gross anatomy of the PVT was based on the appearance of dorsal hippocampus. Coordinates for the hypothalamus spanned from anterior to posterior (AP) −0.58 mm to −2.92 mm relative to bregma. The gross anatomy of the hypothalamus was based on the medioventral part of the brain and covered numerous hypothalamic subregions. Coordinates for BLA brain slices spanned from anterior to posterior (AP) −1.06 mm to −2.06 mm relative to bregma (bilateral). The gross anatomy of the BLA was based on the terminal of external capsule branches nearby the piriform cortex. Coordinates for vHPC brain slices spanned from anterior to posterior (AP) −2.06 mm to −4.04 mm relative to bregma (bilateral). All coordinates and diagrams were based on the Paxinos and Franklin atlas43.
Isolated brain tissue was immediately frozen in RNAlater (Qiagen, Hilden, Germany) until RNA collection following manufacturers recommendations using the Split RNA extraction kit (Lexogen, Greenland, NH). Quality control, library prep, and sequencing was performed by the Penn State College of Medicine’s genome sciences facility as follows. The cDNA libraries were prepared using the QuantSeq 3’mRNA-Seq Library Prep Kit FWD for Illumina (Lexogen, Greenland, NH) as per the manufacturer’s instructions. Briefly, total RNA was reverse transcribed using oligo (dT) primers. The second cDNA strand was synthesized by random priming, in a manner that DNA polymerase is efficiently stopped when reaching the next hybridized random primer, so only the fragment closed to the 3’ end gets captured for later indexed adapter ligation and PCR amplification. The processed libraries were assessed for its size distribution and concentration using BioAnalyzer High Sensitivity DNA Kit (Agilent Technologies, Santa Clara, CA- Cat. No. 5067–4626 and −4627). Pooled libraries were diluted to 2 nM in EB buffer (Qiagen, Hilden, Germany, Cat. No. 19086) and then denatured using the Illumina protocol. The libraries were pooled and diluted to 2 nM using 10 mM Tris-HCl, pH 8.5 and then denatured using the Illumina protocol. The denatured libraries were diluted to 10 pM by pre-chilled hybridization buffer and loaded a TruSeq SR v3 flow cell on an Illumina HiSeq 2500 and run for 50 cycles using a single-read recipe according to the manufacturer’s instructions. Single-end 50 bp reads were obtained. De-multiplexed sequencing reads were generated using Illumina bcl2fastq (version 2.18.0.12).
After the quality and polyA trimming by BBDuk (Version 37.90; http://jgi.doe.gov/data-and-tools/bb-tools/) and alignment by STAR (Version 2.5.2b)62, read counts were calculated from BAM files using Salmon (Version 0.7.2)63. The Ensembl genome was used with GENCODE annotation (GRCm38, M11). Quality statistics were also gathered from genome aligned BAM files using Picard (Version 2.5.0) to use in downstream differential gene expression analysis. Genes were filtered such that only genes with a CPM (counts per million) > 0.1 in at least 10% of samples were retained. Normalized expression data was obtained through taking log2(CPM + 0.001). Then, six outliers were removed such that 114 samples were carried forward for differential gene expression analysis. Two of these outliers contained less than one million reads, and four samples were removed based on the principal components plot of the normalized expression data (these samples did not cluster with other samples from the same brain region).
A linear regression was then conducted to test for differential gene expression in four groups (4EP+PostEPM, 4EP+ Baseline, 4EP- PostEPM, 4EP- Baseline) separately within each brain region. The first five sequencing principal components (derived from the Picard sequencing statistics) were used as covariates in the linear regression to account for technical effects in the gene expression data. A one-way ANOVA test was then used to identify genes with any difference across our four test groups. This was followed by a post-hoc Tukey HSD test, which was performed on the genes with a significant p-adjusted (FDR) value less than 0.05 for the group ANOVA. Three specific comparisons were then made with the groups using the significantly differentially expressed genes from the post-hoc Tukey test (p<0.05): 1) 4EP differences in the PostEPM, but not in homecage, baseline condition. 2) 4EP differences at baseline, but not in the PostEPM. 3) Baseline and PostEPM differences with 4EP. To gain a functional understanding of genes in these comparisons, cell type enrichment was done with the pSI package64 in R using cell type markers from Zhang et al.65, and gene ontology enrichment was done with Go-Elite66 and Metascape (metascape.org).
For visualization of the normalized gene expression data without the influence of technical factors, the effects of the five sequencing principal components were removed with a linear regression, such that only the model intercept, residual, behavior, metabolite, and regional gene expression effects remained. This ‘regressed’ dataset was used to generate principal components for sample visualization. Metascape (metascape.org) was used to determine enriched gene ontology terms in brain regions with sufficient differentially regulated genes, with p-value < 0.01 and biological processes as output.
Flow Cytometry
Spleens or brain tissue dissections of the PVT, hypothalamus, cerebellum or cortex were collected. Single cell suspensions were generated by passing the tissue through a 100um cell strainer (Corning, Corning, NY) with syringe plunger and rinsed with HBSS (Corning, Corning, NY) containing 10% FBS (Gibco, Life Technologies, Thermo Fisher Scientific, Waltham, MA ). Red blood cell lysis was performed (Sigma, St. Louis, MO), diluted with HBSS 10% FBS, and tissue suspension was spun at 350xg for 5 minutes at 4°C. For brain regions, papain digestion in 1ml (200U papain, 0.1mg/ml DNase I) was performed for 20 minutes at 37°C, samples were triturated gently through a 1000ul pipette tip, and then passed through a 40uM filter. Samples were then mixed with 5–10 mls of 25% percoll in HBSS and spun at 1000xg for 25 minutes in a swinging rotor. Myelin debris layer was aspirated from the top, cell pellet was washed in HBSS 10% FBS and then resuspended in HBSS buffer containing Fc block (Anti-Mo CD16/CD32, Ebioscience, San Diego, CA; Invitrogen, Carlsbad, CA) for 20 minutes at 4°C. Further staining was done as previously described for flow cytometry. Primary stains and antibodies included: Aqua Dead cell stain (Thermo Fisher Scientific, Waltham, MA), anti-NG2 (1:400; Alexa488; Ebioscience, San Diego, CA) and anti-MOG (1:100; biotinylated, Novus Biologicals, Littleton, CO), anti-CX3Cr1 (1:500, PE-Cy7, Biolegend, San Diego, CA), anti-CD45.2 (1:500, PerCP-Cy5.5, Tonbo, San Diego, CA), anti-CD11b (1:500, PE, Biolegend San Diego, CA), anti-CD19 (1:500, FITC, Biolegend, San Diego, CA), anti-CD3e (1:500, PE, Thermofisher, Waltham, MA), anti-CD4 (1:500, APC, Biolegend, San Diego, CA), anti-TCRb (1:500, PerCP-Cy5.5, Tonbo, San Diego, CA), anti-CD8a (1:500, APC-Cy7, Thermofisher, Waltham, MA) and were incubated with cells for 30 minutes at 4°C. After washing, secondary streptavidin antibody (1:1000, SAV Alexa 647, 405237, Biolegend, San Diego, CA), was incubated with cells for 30 minutes at 4°C. After washing, cells were fixed with 2% PFA for 20 minutes, washed, and analyzed on a 4 laser (Violet, Blue, Yellow, and Red) CytoFLEX S Flow Cytometer in the Caltech Flow Cytometry Core Facility and analyzed in Flowjo v10.6.2.
MRI mouse brain scanning and Data analysis
Brain samples were collected by cardiac perfusion as described above for immunohistochemistry. Intact, defleshed skulls were collected and fixed in 4% PFA overnight, followed by incubation for 14 days at 4C in gadoteridol (Prohance®, Bracco Diagnostics Inc., Princeton NJ) and 0.01% sodium azide in PBS as previously described67. Samples were scanned immersed in Krytox perfluoropolymer vacuum oil (Sigma, St. Louis, MO), supported by cotton wool, in 15 ml conical tubes.
All MRI scans were performed using a Bruker Avance Neo 7T/30 scanner and a Bruker gradient and shim unit (B-GA12SHP insert for BGA20) supported by a Bruker PV360 software package. A Bruker quadrature volume coil (B-GA12s HP) and a mouse brain 2×2 receive-only four element array coil were used for 1H RF pulse transmission and signal receiving, respectively, at its operating frequency of 300.33 MHz. A Bruker standard DWSE (diffusion weighted spin echo) sequence was used to collect images with b value of 4000 s/mm2, gradient duration=5.6 ms, gradient separation=12.54 ms, repetition time TR=200 ms, echo time TE=23.34 ms, and using 60 gradient different directions. T1 and T2 weighted images were also collected using standard Bruker sequences (T1_FLASH_3D, T2_TurboRARE). Image resolution was fixed for all scanning at a field of view (FoV)= 14 mm and a resolution of 175 μm at each dimension. All GQI based tractographic images were acquired after data analysis that was performed using DSI Studio software package (68). The total surface area (TSA), dfi_FA, MD, AD, and RD of tracts were calculated using built-in functions of the DSI studio after individual ROI (i.e, PVT) was first calculated using the implemented routine under CIVM mouse atlas (Duke radiology, https://www.civm.duhs.duke.edu/). The GQI based diffusion data reconstruction was performed with a diffusion sampling length ratio of 1.2. A deterministic fiber tracking algorithm96 was used. In track calculations, a seeding region was placed at whole brain and the change threshold was set to 20% uniformly and the angular threshold was randomly selected. The tracking anisotropy threshold was varied from 0.06–0.1, depending on ROIs employed (Extended Data Figure 7). The step size was randomly selected from 0.5 voxel to 1.5 voxels. Various Min and Max track lengths were tested, but we have not seen any significant difference among track parameter values, and data reported here are limited for tracks within 5 mm < L<10 mm range. A total of 20000 tracts were calculated.
Electron Microscopy
Mice were anesthetized using 150 ul Euthasol and perfused with 30 ml of 37°C PBS followed by 40–50 ml 37°C 4% PFA at 6ml/minute flow rate. Brains and intestinal tissue were harvested and continuously hydrated with cold (4°C) fixative solution of 3% glutaraldehyde, 1% paraformaldehyde, 5% sucrose in 0.1M sodium cacodylate trihydrate throughout the dissection process. Fixed brains were rapidly dissected out and fitted into. 1.0mm brain matrix (Roboz SA-2175, Gaithersburg, MD) where two gross cuts were made at +0.75mm and −0.25mm relative to bregma. Brain tissue anterior to +0.75mm and posterior to −0.25mm was discarded. The remaining tissue block was cut at positions ±0.5mm lateral to the midline and the flanking tissues discarded. Remaining tissue dorsal and ventral to the visible corpus callosum was excised and discarded. Fixed intestines were rapidly dissected out and cuts were made to harvest the proximal colon. The 2 mm3 of intestinal tissue or 1 mm3 brain tissue containing the corpus callosum were immediately stored in the cold fixative solution until proceeding to the next step.
Corpus collosum tissue was placed into petri dishes with 0.1M cacodylate buffer and cut into ~1mm2 × 0.5 mm thick slices using a microsurgical scalpel. These were placed individually into brass high-pressure freezing planchettes (Ted Pella, Inc., Redding, CA) with cacodylate buffer containing 10% Ficoll (an extracellular cryoprotectant). Samples were ultra-rapidly frozen with an HPM-010 high pressure freezing machine and transferred to liquid nitrogen. Planchettes containing vitrified tissue were transferred under liquid nitrogen to cryotubes (Nunc, Roskilde, Denmark) prefilled with 2% OsO4, 0.05% uranyl acetate in acetone. Samples were placed in a AFS-2 freeze-substitution machine (Leica Microsystems, Wetzlar, Germany), processed at −90°C for 72 h, warmed to −20°C over 12 h, further processed at that temperature for 24 h, then rinsed with acetone and flat-embedded in Epon-Araldite resin (Electron Microscopy Sciences, Port Washington, PA) between Teflon-coated glass slides. Resin was polymerized at 60° for 48 h.
Embedded tissue blocks were observed by phase-contrast microscopy to select well-preserved and optimally oriented regions, then extracted with a scalpel and glued to plastic sectioning stubs. Semi-thick (300–400 nm) serial sections were cut with an EM UC-6 ultramicrotome (Leica Microsystems, Wetzlar, Germany) using a diamond knife (Diatome U.S., Hatfield, PA). Sections were collected onto Formvar-coated copper/rhodium slot grids and stained with 3% uranyl acetate and lead citrate. Colloidal gold particles (10 nm) were placed on both surfaces of the grids to serve as fiducial markers for tomographic image alignment.
Tomography
Grids were placed in a dual-axis tomography holder (Model 2040, E.A. Fischione Instruments, Inc., Export, PA) and imaged with a TF-30ST transmission electron microscope (Thermo Fisher Scientific, Waltham, MA) at 300KeV. For tomography, grids were tilted +/−64° and images acquired at 1° increments. The grid was then rotated 90° and a similar tilt-series was taken about the orthogonal axis. Tilt-series were acquired automatically using the SerialEM software package68 and recorded with a CCD camera (US1000, Gatan, Inc., Pleasanton, CA). Tomographic datasets were processed and analyzed with the IMOD software package69,70 on MacPro and iMacPro computers (Apple, Inc.).
Western Blots
Western blots were performed on the PVT using brain punches collected using 1.0mM Biopsy Punches (Miltex, VWR, Radnor, PA), or on ex vivo organotypic brain slices. Tissue samples were homogenized by bead beating in lysing matrix D tubes (MP Biomedicals, Irvine, CA) in RIPA buffer (Millipore Sigma, Burlington, MA) containing Protease Inhibitor tablets (Roche, Basel, Switzerland) followed by protein quantification and normalization using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific, Waltham, MA). Proteins were separated by SDS page using 4–20% tris-glycine Wedgewell gels (Invitrogen, Carlsbad, CA) and blotted onto 0.45um PVDF membrane (Millipore Sigma, Burlington, MA). B-actin loading controls were used, and images were taken of the same blot under specific excitation wavelengths for the various protein visualization. Antibody titrations were 1:500 for primary antibodies and 1:5,000 for fluorescent conjugated secondary antibodies. Antibodies included: mouse anti-MOG (CL2858, Thermo Fisher Scientific, Waltham, MA), chicken anti-MBP (CH22112, Neuromics, Edina, MN), rabbit anti-ß-actin (13E5, Cell Signaling, Danvers, MA), 2° anti-chicken-488 (ab150173, abcam, Cambridge, UK); 2° anti-mouse-488 (ab150113, abcam, Cambridge, UK); 2° anti-rabbit-647 (A31573, Thermo Fisher Scientific, Waltham, MA).
Organotypic brain slice culturing
Brains from P8 mice were sectioned by Vibratome into 300μm sections in HBSS with 3mM HEPES while bubbling in carbogen gas. Sections were cultured at 37°C with 5% CO2 on 12-well transwell membrane plates in 50% DMEM/HEPES (Gibco, Gaithersburg MD) containing 25% heat inactivated horse serum (Gibco), 25% Hank’s solution (Gibco), 2mM sodium bicarbonate (Merck, Rahway, NJ), 6.5 mg/ml glucose, 2mM glutamine, 1x pen/strep (Gibco), pH 7.2. Samples were visually observed for general signs of health (adherence, spread on membrane) and viability tested by standard lactate dehydrogenase and alamarblue assays as previously described71. After 10 days culture in the presence of 10uM 4EPS or vehicle (PBS), samples were taken for downstream western blot and qPCR as described above. For immunohistochemistry, samples were fixed in 4% paraformaldehyde for 3 hours at 4°C and stained for neurofilament and PLP as described above. Colocalization was quantified using IMARIS software (South Windsor, CT).
Analysis of myelination and calculation of g-ratio
Tomograms containing cross-sectional images of myelinated and unmyelinated axons were visualized in three dimensions using IMOD and analyzed using the 3dmod image processing program. Myelinated and unmyelinated axons were differentiated by eye and counted accordingly. To calculate g-ratio (ratio of axon radius to its outer, myelinated radius), the edge of the axon and the edge of the outer myelin sheath were traced using the Object tool. Traces were analyzed for area, yielding one inner and one outer area per myelinated axon. Radii were determined using the formula raxon=square root of (Aaxon/π) for the bare axon, and raxon+myelin=square root of (Aaxon+myelin/π) for the myelinated axon. Final g-ratios were calculated as raxon/raxon+myelin. Every axon apparent in the tomogram was included in all analyses of myelination.
Behavior Testing
Behavior testing was performed as previously described8,51,72–76. All mice were tested by a blinded researcher starting at six weeks of age, in the following order: EPM, light/dark box, open-field testing, marble burying, grooming, social behavior, and USV (male-female context). Separate cohorts of mice were used to test cognitive and motor function. These cohorts were tested in elevated plus maze to confirm phenotype followed by novel object recognition and Y-maze or beam traversal, pole descent, and wire hang. Mice were allowed to settle for at least two days after cage changing before they were tested, and tests were performed 2–3 days apart to allow mice to rest between tests. Mice were acclimated to the behavior testing room for one hour prior to testing. Mice were tested during the light phase of the light cycle.
Elevated Plus Maze (EPM)
EPM was performed in a maze with 25cm by 5cm arms and a 5cm by 5cm center, recorded using an overhead camera, and tracked and analyzed using the EthoVision XT 4 software package (Noldus Information Technology; Leesburg, VA, USA). Prior to testing, the maze was disinfected using Rescue disinfectant (Virox technologies, Oakville, ON, Canada) then allowed to evaporate. Mice were then introduced to the arena and allowed to explore for 5 min while being tracked. The number of entries into and the time spent in open and closed arms as well as the outer third of the open arms (the terminus) were analyzed. If a mouse fell or jumped from the apparatus during the test it was removed from the dataset.
Light/Dark Box
The light/dark box test was performed in a 50×30cm2 arena with a 30×30 cm2 open section and a black, lidded section sized 20×30 cm2 with an open doorway for free passage from light to dark areas. Prior to testing, the arena was disinfected using Rescue disinfectant (Virox technologies, Oakville, ON, Canada) then allowed to evaporate. Mice were placed into the dark box and lid replaced, and then recorded using an overhead camera, and tracked and analyzed using the EthoVision XT 10 software package (Noldus Information Technology; Leesburg, VA, USA). Entry and duration in the light was analyzed over a 10-minute period.
Open-field Test
The open-field test was performed in 50 × 50 cm2 white Plexiglas arenas, recorded using an overhead camera, and tracked and analyzed using the EthoVision XT 4 software package (Noldus Information Technology; Leesburg, VA, USA). Prior to testing, the arena was disinfected using Rescue disinfectant (Virox technologies, Oakville, ON, Canada) then allowed to evaporate. Mice were then introduced to the arena and allowed to explore for 10 min while being tracked. The total distance traveled, and the number of entries and time spent in a 17 × 17 cm2 center square were analyzed. Fecal pellets left during the assay were quantified.
Marble Burying
Marble burying was performed in a normal cage bottom (Lab Products; Seaford, DE) filled with 3–4 cm of fresh, autoclaved wood chip bedding (Aspen chip bedding, Northeastern Products Corp; Warrensburg, NY). Mice were first habituated to the cage for 10 min, and subsequently transferred to a holding cage while the bedding was leveled and 20 glass marbles (4 × 5) were placed on top. Mice were then returned to their own cage and removed after 10 min. The number of buried marbles (50% or more covered) was then recorded and photographed for reference and scored by an additional blinded researcher. A fresh cage was used for each mouse, and marbles were soaked in Rescue disinfectant (Virox technologies, Oakville, ON, Canada) and dried in bedding in between tests.
Grooming
Mice were placed in autoclaved, empty standard cages (Lab Products; Seaford, DE) and video recorded from the side for 15 minutes. The final 10 minutes were scored manually by two independent blinded, trained researchers for grooming behavior.
Social Interaction
Each mouse was introduced to a fresh, empty, standard, autoclaved cage (Lab Products; Seaford, DE) and allowed to habituate for 10 min before a novel mouse matched in age and sex was introduced to the cage for 5 additional minutes for scoring of social activity. Two blinded, trained researchers scored videos for any social behavior using the ETHOM software77.
Ultrasonic Vocalization
Mice were single-housed and exposed to a new SPF C57BL/6J female for 10 min every day in the five days prior to the test. On the fourth day, mice were habituated to an empty cage (no bedding) with a filter soaked with 10ul of fresh pooled female urine for 10 min. Subsequently, a novel female was introduced to the cage and ultrasonic vocalizations were recorded using Avisoft UltraSoundGate 116Hme microphone (Avisoft Bioacoustics, Nordbahn, Germany) and the Avisoft Sas-lab Recorder software (Avisoft Bioacoustics, Nordbahn, Germany). Total vocalization and vocalization counts were recorded during 3-minute sessions of male-female interaction.
Novel Object Recognition
Novel object recognition (NOR) was performed as described previously with sand filled flasks and Duplo towers72. Mice were habituated to the open field arenas (50 × 50cm2 white Plexiglass) for 10 minutes on day 1. On day 2, mice were reintroduced to the arena with duplicate copies of one of two objects. On day 3, mice were reintroduced to the arena with one copy of the familiar object and one copy of a novel object. Within each group of mice, mice were split so that both objects were used as the familiar object to control for inherent interest levels in the different objects. Time spent with the nose cone directed toward the objects and within 2cm from the objects was scored the EthoVision XT 10 software package (Noldus Information Technology; Leesburg, VA). NOR index was calculated as time at novel object / total time at objects * 100.
Y-maze
Y-maze alternations test was performed in a white Y-maze (Maze Engineers, Boston, MA) video-taped from above. Entries into each arm were scored manually by a blinded researcher and the number of repetitive, spontaneous alternations were calculated.
Beam Traversal
Beam traversal was performed as previously described73 on a 1 meter plexiglass beam (Stark’s Plastics, Forest Park, OH) constructed of four segments of 0.25m in length, with each segment decreasing in widths from 3.5cm, 2.5cm, 1.5cm, and 0.5cm, with 1cm overhangs placed 1cm below the surface of the beam. The widest segment acted as a loading platform for the animals and the narrowest end placed into home cage. Animals had two days of training to traverse the length of the beam before testing. On the first day of training, animals received 1 trial with the home cage positioned close to the loading platform and guided the animals forward along the narrowing beam. Animals received two more trials with limited or no assistance to encourage forward movement and stability on the beam. On the second day of training, animals had three trials to traverse the beam and generally did not require assistance in forward movement. On the third day, animals were timed using a stopwatch to traverse from the loading platform and to the home cage. Timing began when the animals placed their forelimbs onto the 2.5cm segment and ended when one forelimb reached the home cage.
Pole Descent
Pole descent was performed as previously described73 with a 0.5m long pole, 1cm in diameter, wrapped with non-adhesive shelf liner to facilitate the animals grip, which was placed into the home cage. Animals received two days of training to descend from the top of the pole and into the home cage. On day one of training, animals received 3 trials. The first trial, animals were placed head-down 1/3 the distance above the floor, the second trial from 2/3 the distance, and the third trial animals were placed at the top. The second day of training, animals were given 3 trials to descend, head-down, from the top of the pole. On the test day, animals were placed head-down on the top of the pole and timed to descend back into the home cage. Timing began when the experimenter released the animal and ended when one hind-limb reached the home cage base.
Wire Hang
Wire hang was performed as previously described73. Animals were placed in the center of a 30 × 30 cm2 screen with 1cm wide mesh. The screen was inverted head-over-tail and placed on supports ∼40cm above an open, clean cage with bedding. Animals were timed until they released their grip or remained for 60s.
Statistical Information
All data are represented as mean ± standard error mean (SEM). Two-tailed unpaired Welch’s t-tests were used to compare data between two independent groups (ie 4EP- vs 4EP+). Data with more than two independent groups were analyzed by one-way ANOVA with Dunnett’s multiple comparison correction or Sidak multiple comparison correction, according to specific experimental design. All data was analyzed using Graphpad Prism, with a p-value (or adjusted p-value in the case of ANOVA with multiple comparison correction) of less than 0.05 was deemed statistically significant. The number of biological replicates is indicated in all figures and the statistical methods are noted in the figure legends and Methods. The number of asterisks indicates the difference in the figures, with exact p-values in the legends. For comprehensive statistical information regarding the autoradiography, fUSi, and QuantSeq experiments, see the corresponding detailed paragraphs above in the Methods section.
Data Availability
All data analyzed for this study is included in this published article with source data for all graphs in the supplementary information files, with additional 2DG data available at https://gin.g-node.org/bneedham/Needham_Nature_2022. The WoL database annotation files are publicly available at: https://biocore.github.io/wol/download. Greengenes database is publicly available at: https://greengenes.secondgenome.com/.
Code Availability
Custom scripts used in fUSi analysis can be accessed at https://github.com/brittanyneedham/Needham_Nature2022.
Extended Data
Extended Data Fig. 1. Strain engineering and 4EPS quantification.
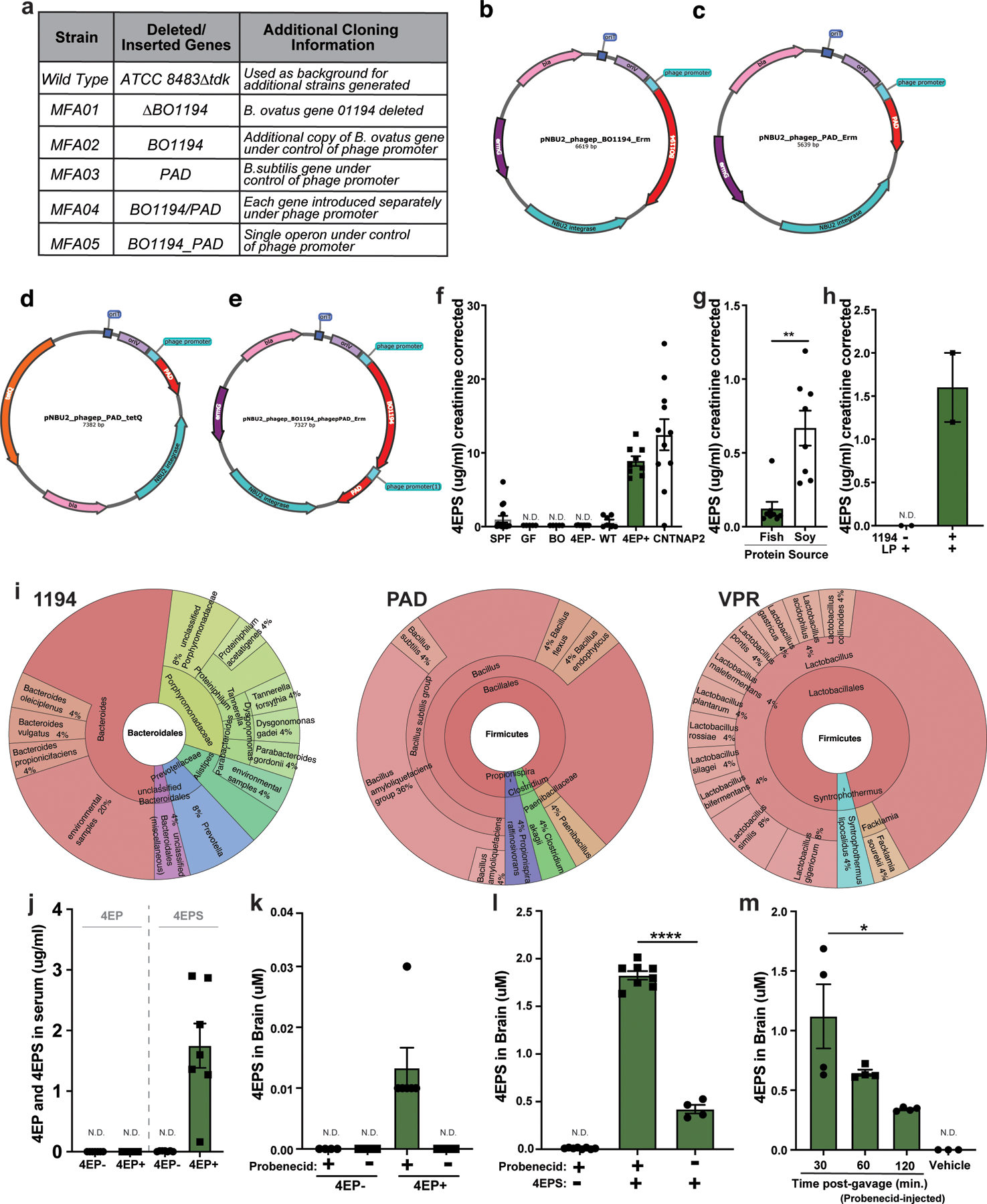
a, Strain legend corresponding to Figure 1b-d, containing further details for strain names and cloning strategy. Additional information can be found in Methods and Supplementary Information Tables 1-3. b-e, Maps for integration vectors. b, The BO1194 gene was cloned into the pNBU2 vector with an erythromycin resistant marker under the phage promoter. c, The PAD gene was cloned into pNBU2 vector with an erythromycin resistant marker under the phage promoter. d, The PAD gene was cloned into the pNBU2 vector with a tetracycline resistant marker under the phage promoter. e, BO1194 and PAD were tandemly connected, putting the phage promoter in front of both genes, then icloned into the pNBU2 vector with the erythromycin resistant marker. f, Creatinine corrected urinary levels of 4EPS from SPF, GF, gnotobiotic mice colonized with strains as labeled, and a mouse model of atypical behavior including anxiety-like phenotypes. This data corresponds to abbreviated data in Fig. 1f. SPF, specific pathogen free mice; GF, germ-free mice; BO, Bacteroides ovatus mono-colonized mice; 4EP-, GF mice colonized with non-producing strain pair (B. ovatus Δ1194 and WT L. plantarum with endogenous VPR) used throughout; WT, GF mice colonized with wild type strain pair (WT B. ovatus and WT L. plantarum with endogenous VPR) before optimal engineering; 4EP+, colonized with strain pair engineered for higher production of 4EP+ (B. ovatus+1194/PAD and WT L. plantarum with endogenous VPR), used throughout; CNTNAP2, conventionally colonized mouse model of atypical behavior (left to right columns: n=15, 5, 5, 9, 7, 9, 11). g, 4EPS levels (creatinine corrected) in urine of wild type, specific pathogen free (SPF) mice fed one of two isocaloric diets matched for protein, mineral, carbohydrate, and fat levels but differing in either high tyrosine fish meal (fish diet), or a high plant protein (soy diet) (n=8 each group)p=0.002. h, 4EPS levels (creatinine corrected) in urine of gnotobiotic mice colonized with isogenic strain pairs of B. ovatus and wild type L. plantarum, which differ only in the presence or absence of gene 1194, and thus the tyrosine lyase activity requires to convert tyrosine to p-coumaric acid (n=2 each group). i, Alignments of genes used to engineer the 4EP synthesis pathway to the reference genomes in the WoL database, showing that these genes are found in ~25 genomes each, most of which are common human gut lineages. j, 4EP and 4EPS levels (ug/ml) in serum of colonized mice (n=7 each group). k, 4EP+ and 4EP- colonized mice were injected with the organic anion transporter to inhibit potential 4EPS transport out of the brain, then analyzed by LCMS (n=6 each group). Extended data from Figure 1G. l, Conventionally colonized (SPF) mice were injected with probenecid and then injected with high dose 4EPS (n=8) or saline vehicle (n=8), and whole brain lysate was analyzed by LCMS compared to 4EPS injection alone (n=4)p<0.0001. m, Conventionally colonized (SPF) mice were injected with probenecid and then gavaged with 4EP, then whole brains were harvested at 30-minute intervals and 4EPS levels were quantified by LCMS (n=4; vehicle group n=3)p=0.01. Two independent trials on biological replicates were used for the experiments in the figure. Abbreviations: SPF, specific pathogen-free; GF, germ-free; BO, Bacteroides ovatus; WT, wild type; LP, Lactobacillus plantarum. Data represent mean ± SEM analyzed by a two-tailed Welch’s t-test or one-way ANOVA with Dunnett multiple comparisons test as appropriate. * p ≤ 0.05; ** p ≤ 0.01; *** p ≤ 0.001; **** p ≤0.0001.
Extended Data Fig. 2. 4EP sulfation, experimental timeline, normal weight gain, colonization and intestinal barrier of 4EP+ mice.
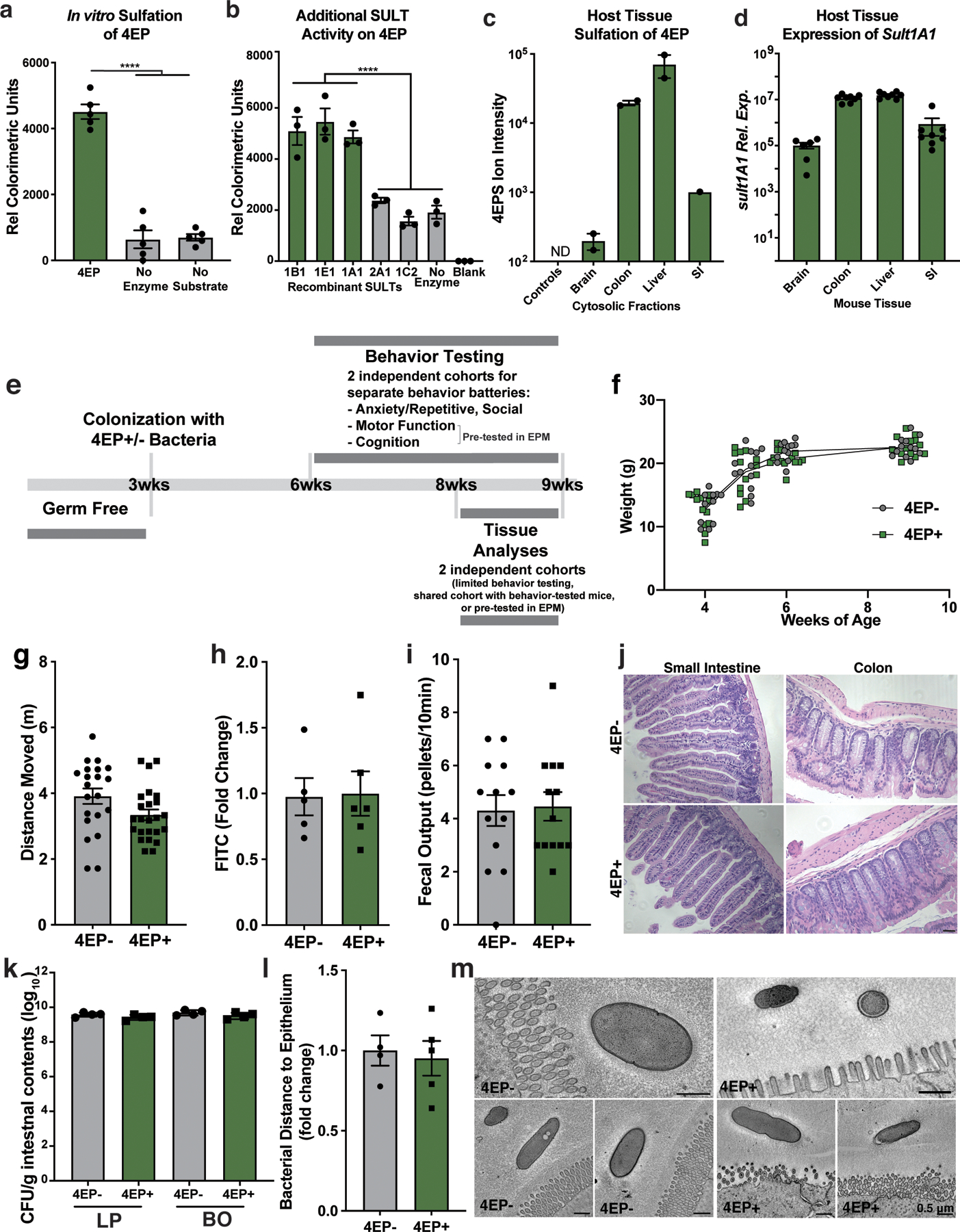
a, In vitro recombinant SULT1A1 sulfotransferase assay with 4EP as potential sulfate acceptor (n=5)p<0.0001. b, Results of the sulfotransferase assay using various recombinant SULTs (n=3)p<0.0001. c, 4EP sulfation capacity of cytosolic fractions of brain, colon, liver, and small intestinal tissue, each containing endogenous sulfotransferases (n=2 datapoints of samples pooled from triplicate biological replicates, with only 1 pool for SI). Ion intensity of 4EPS measured by LCMS is plotted along the y-axis. d, Confirmation of expression of the Sult1a1 gene measured by qPCR in tissue from the brain, colon, liver, and small intestine of colonized mice (n=8; brain n=6)p<0.0001. e, Schematic of mouse experimental timeline, showing ages of mice at colonization of GF mice with 4EP+/− strains, behavior testing and tissue collection. f, Weights of mice (grams) after colonization of GF mice with 4EP- or 4EP+ bacteria (n=13). g, Ambulatory activity of 4EP- and 4EP+ mice over ten minutes, measured by distance moved when mice were placed in an open arena and allowed to explore (4EP- n=21; 4EP+ n=24). h, FITC-dextran levels in serum as a measure of intestinal permeability (4EP- n=5, 4EP+ n=6). i, Fecal output of colonized mice over 10 minutes (n=13). j, Images of hematoxylin and eosin (H&E) stained small intestine and colon of 4EP+/− mice (representative images of n=4). Scale bar 50μm. k, Colonization of ex-GF mice with engineered Bacteroides ovatus (BO) and Lactobacillus plantarum (LP), plotted as colony forming units (CFU) per gram of intestinal contents (n=4). l, Quantification of bacterial distance from the intestinal epithelium imaged by electron tomography, where each data point represents a separate animal comprised of an average of 5–10 bacterial cells per image per mouse (4EP- n=4; 4EP+ n=5). m, Example images of bacterial cells near the intestinal epithelial layer of 4EP+/− mice. Two independent trials using multiple randomized litters were used for the experiments in the figure. Abbreviations: SULT, sulfotransferase; SI, small intestine. Data represent mean ± SEM. Panel f was analyzed by a 2-way ANOVA with a Bonferroni multiple comparison correction. Panels g-i and l were analyzed using two-tailed Welch’s t-tests and panels a-d, k by a one-way ANOVA with Dunnett multiple comparisons test.
Extended Data Fig. 3. Lack of inflammatory signals in peripheral and brain cytokine and immune profiles of 4EP+/− mice.

a-b, Cytokine and chemokine levels presented in bar graph where grey (−) is 4EP- and green (+) is 4EP+, measured by bioplex in colon (n=9) a, and serum (n=9) b. No significant differences were observed. c-d, Flow cytometry of spleens of 4EP+ and 4EP- mice. For gating strategy see Supplementary Information Figure 2. c, Percentages of CD4+ or CD8+ T cells (4EP- n=11, 4EP+ n=10). d, Percentage of B cells (4EP- n=11, 4EP+ n=10)p<0.0001. e, Cytokine and chemokine levels presented in bar graph where grey (−) is 4EP- and green (+) is 4EP+, measured by bioplex (n=9) in brain tissue (Il-3 p=0.046; Il-5 p=0.01; KC p=0.02; TNF-a p=0.01). f, Flow cytometry percentages of microglia in the brain of 4EP- and 4EP+ mice. For gating strategy see Supplementary Information Figure 2. g, Relative expression of microglial genes in microglial-enriched samples (n=8). Multiple randomized litters were used for the experiments in the figure. Each data point represents biologically independent mice from multiple randomized litters examined over one (a-b) or two (c-h) respective experiments. Data represent mean ± SEM. Panels a, b, d, f, and h were analyzed by one-way ANOVA with Dunnett multiple comparisons test comparing each 4EP+/4EP- pair, and panels g and e by a two-tailed Welch’s t-test. * p ≤ 0.05; ** p ≤ 0.01; *** p ≤ 0.001.
Extended Data Fig. 4. Extended information on functional connectivity experiments fUSi and autoradiography.
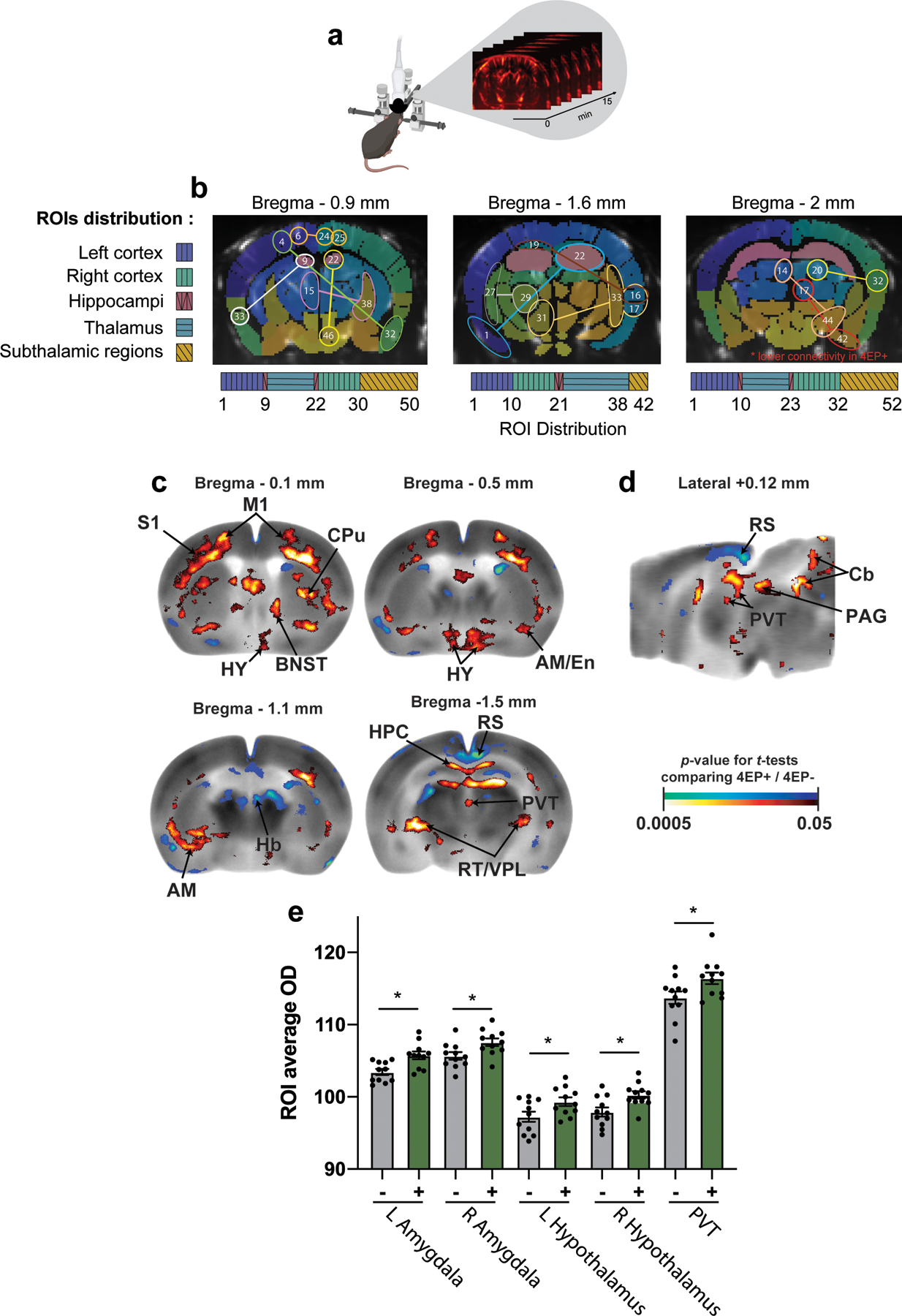
a, Schematic representation of the functional ultrasound (fUSi) set-up. Functional acquisitions are acquired non-invasively through intact skull and scalp in anesthetized mice during 15 minutes per coronal plane. Created with BioRender.com. b, Maps overlaid with regions of interest colored in a gradient for easier visualization. Significant pairs are indicated, corresponding to the data in main Fig. 2b. Three coronal planes per mouse were studied: Bregma −0.9 mm, Bregma −1.6 mm, Bregma −2mm. 50, 52 and 52 ROIS are respectively delineated for each plane according the Paxinos Atlas. Coronal plane B-0.9mm: ROIs #1 to #8 are located in the left cortex, ROI#9 is the left hippocampus, ROIs#11 to #21 are located in the thalamus, ROI#22 is the right hippocampus, ROIs#23 to #30 are located in the right cortex and finally ROIs#31 to #48 are subthalamic regions. Coronal plane B-1.6mm: ROIs #1 to #20 are located in the cortex, ROIs#21 #22 are the left and right hippocampi, ROIs#23 to #38 are located in the thalamus and ROIs#39 to #42 are subthalamic regions. Coronal plane B-2mm: ROIs #1 to #9 are located in the left cortex, ROI#10 is the left hippocampus, ROIs#11 to #22 are located in the thalamus, ROI#23 is the right hippocampus, ROIs#24 to #32 are located in the right cortex and finally ROIs#33 to #50 are subthalamic regions. c-d, Color-coded overlays over representative coronal (c) and sagittal (d) sections of the mouse brain template showing significant differences in regional cerebral glucose uptake following open field exposure in 4EP+ mice compared to 4EP- mice (n=11) (t-test, p ≤ 0.05, extent threshold > 200 contiguous voxels, with both conditions met to be deemed significant; red/blue: increase/decrease in glucose uptake in 4EP+ compared to 4EP- mice). e, Quantitated 2DG-uptake by region of interest, including the amygdala, hypothalamus and PVT, confirming changes in relative 2DG uptake in the open field groups (n=11 each group) From left to right, p=0.01; 0.03; 0.02; 0.009; 0.003). Average optical density of each ROI in each animal was normalized to whole-brain average of that animal. Abbreviations: AM, amygdala; HY, hypothalamus; PVT, paraventricular nucleus of the thalamus; BNST, bed nucleus of the stria terminalis; S1, primary somatosensory cortex; CPu, caudate putamen; Hb, habenular nucleus; RT, reticular nucleus of the thalamus; VPL/VPM, Posterior Lateral/Ventral Posterior Medial Thalamus; R, right; L, left. Two cohorts of mice from multiple litters were used for each experiment in this figure. Panel e was analyzed by one-way ANOVA with multiple comparison correction and two-stage linear step-up procedure of Benjamini, Krieger and Yekutieli. * p ≤ 0.05.
Extended Data Fig. 5. Extended results of mRNA high throughput sequencing (QuantSeq), and oligodendrocyte analysis by immunofluorescence implicating oligodendrocyte differences in 4EP+ mice.

a, Principal component analysis of all samples analyzed by QuantSeq, with tight clustering by brain region. Each brain region is colored according to the legend. b, Differential gene analysis summary showing number of significantly different genes in 6 tested brain regions. Top pie chart refers to total number of differential genes by one-way group ANOVA with all the 4EP- and 4EP+, at baseline and post-EPM. The bottom pie chart was generated according to the specific significant (p<0.05) contrasts between 4EP+ and 4EP- conditions of post-EPM conditions as calculated with the post-hoc Tukey HSD test. c-d, Gene ontology terms enriched in genes that are upregulated (c) or downregulated (d) in the PVT in 4EP+ relative to 4EP- mice in the baseline, home cage condition. X-axis represents -log10(P) of enrichment analysis. e, p-value histogram of PVT sequencing at baseline, graphed as p-values binned by 0.05 along the x-axis and number of genes along the y-axis. f, Adjusted p-values for 4EP+/4EP- comparison of expression of oligodendrocyte specific genes in all 6 brain regions analyzed by QuantSeq, corresponding to data presented in Main Figure 3c for the PVT. p<0.05 in bold. g, PVT seed analysis correlating 2DG uptake in the PVT to the rest of the brain. Images show 3D rendered, average whole brain correlation data (n=11). The center of the PVT is indicated at the cross section of the red lines, and the number of negatively correlated voxels is shown below in each. Significance is illustrated according to the legend. A minimum threshold of 200 contiguous voxels with p<0.05 was used. h, Schematic of immature OPC and mature, myelinating oligodendrocyte markers used for immunostaining, flow cytometry, or western blots in this study. i, Raw counts of mature oligodendrocytes, measured by CC1+ and OLIG2+ staining in 4EP+ and 4EP- mice in the PVT, used to calculate maturity quotient in Figure 3f (n=8). p=0.04. j, Raw counts of immature oligodendrocytes, measured by NG2+ and OLIG2+ staining in 4EP+ and 4EP- mice in the PVT, used to calculate maturity quotient in Figure 3f (n=8 each group). p=0.055. k, Representative images (corresponding to main Fig. 3e-f) of the PVT in brain sections from 4EP- and 4EP+ mice, stained for OLIG2, using total cumulative counts from 3–5 images per replicate (n=8). Scale bar 100µm. l, Raw counts of OLIG2+ cells in the PVT of 4EP+/− mice. Each data point represents an individual mouse, with total cumulative counts from the PVT (n=8) p=0.8. m, Raw counts of NeuN+ staining in the PVT (4EP- n=10, 4EP+ n=9), using average counts from 3–5 images per replicate. n, Representative images of pan-neuronal NeuN staining in the PVT, with 3–5 images per replicate (4EP- n=10, 4EP+ n=9). Scale bar 100µm. o, NG2+ staining in extended regions of the brain (n=3 each group) p=0.009. Abbreviations: PVT, the paraventricular nucleus of the thalamus; BNST, bed nucleus of the stria terminalis; BLA, basolateral amygdala; mPFC, medial prefrontal cortex; HY, hypothalamus; vHPC, ventral hippocampus; OLIG2, oligodendrocyte transcription factor 2; NG2, neural/glial antigen 2; CC1, antibody (anti-adenomatous polyposis coli (APC) clone) that binds mature oligodendrocyte marker; MOG, myelin oligodendrocyte glycoprotein; MBP, myelin basic protein; ACA, anterior commissure; CC, corpus callosum; LHB, lateral habenula; LS, lateral septum; MHB, medial habenula; MS, medial septum; ME, median eminence; PFC, prefrontal cortex; PVN, paraventricular nucleus of the hypothalamus; SM, stria medullaris of thalamus. Two cohorts of mice from multiple litters were used for experiments in panels g-o in this figure. Data represent mean ± SEM. Analysis performed using two-tailed Welch’s t-tests (i-m) or one-way ANOVA with Dunnett multiple comparison between 4EP+/− pairs (o). * p ≤ 0.05.
Extended Data Fig. 6. Extended results of oligodendrocyte analysis implicating oligodendrocyte differences in 4EP+ mice in vivo and in vitro.

a, Extended quantitation of flow cytometry with MOG+/NG2+ ratio by quadrant in the cerebellum, cortex, and hypothalamus (n=4). b-c, Western blot analysis of MOG (p=0.03) and MBP (p=0.002) markers (respectively) of mature oligodendrocytes from the PVT brain region. PVT punches from two mice were pooled per data point in quantitative data (n=6 pooled samples each group). For gel source data, see Supplementary Information Figure 1. d-g, Organotypic brain slices were cultured in the presence of 10uM 4EPS. d, Representative images of CC1, NG2 and Olig2 staining. Two cohorts of mice were used, with each data point in Fig 3i representing quantified images from samples from individual mice. Scale bar 20µm. e, Example high-magnification image of axon in organotypic brain slices, stained with antibodies specific to NF (red), and PLP (green), with DAPI (blue), taken from image set used for quantification in 3i and Extended Data Fig 6d. Scale bar 8µm. f, qPCR results of oligodendrocyte genes, Cspg4 (p=0.04), Mog (p=0.03), and Mpb from organotypic brain slices (n=7). g, Western blot image of mature oligodendrocyte marker MBP (quantified in main Fig. 3l). For gel source data, see Supplementary Figure 1. Abbreviations: NG2, neural/glial antigen 2; CC1, antibody (anti-adenomatous polyposis coli (APC) clone) that binds mature oligodendrocyte marker; MOG, myelin oligodendrocyte glycoprotein; MBP, myelin basic protein. Two cohorts of mice from multiple litters were used for experiments in this figure. Data represent mean ± SEM. Statistics were performed using two-tailed Welch’s t-tests. * p ≤ 0.05; ** p ≤ 0.01.
Extended Data Fig. 7. Extended results of myelin analysis by electron microscopy and DTI.
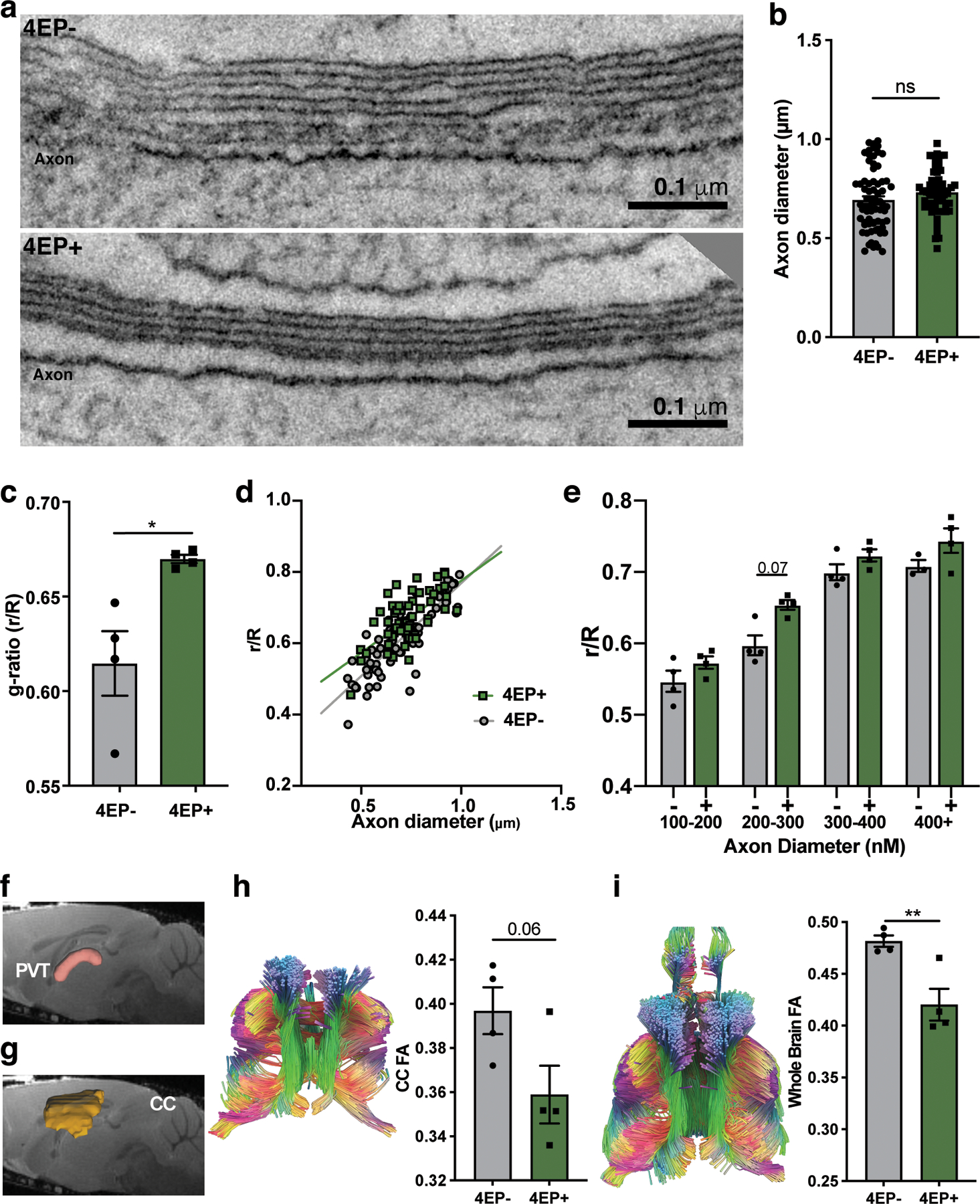
a, Additional ET of myelinated axons in 4EP- (top) and 4EP+ (bottom) mice visualized longitudinally along an axon (n=3; 4 images each). These longitudinal images were used only for qualitative visual assessment of myelin to accompany the other quantitative measures. b, Axon diameters (measured from cell membrane, not including myelin layer) from all axons used to calculate g-ratio (4EP-, 56; 4EP+, 70 axons) (n=4; 4 images each). p=0.1. c, g-ratio (r/R), the inner axon diameter/outer diameter of the myelin sheath, of 4EP+ and 4EP- mice. Each data point represents a mouse (n=4), which is an average of the g-ratio of all axons quantified from 4 images per mouse, where a larger g-ratio indicates a thinner layer of myelin. p=0.046. d, Plot of g-ratio (r/R) on the y-axis and axon diameter on the x-axis, with linear regression noted by lines (4EP-, 56; 4EP+, 70 axons) (n=4; 4 images each). e, Average g-ratio of each animal, binned by axon size, indicating that mid-sized range of axons are driving the change in overall g-ratio phenotype observed in the mice (n=4). f-g, Defined regions of interest for the PVT and corpus callosum (CC) overlaid on representative sagittal MRI image. h-i, representative tracts observed from a bilateral coronal plane view (left) and fractional anisotropy (FA) analysis (right) for the corpus callosum (CC), and whole brain (p=0.009), respectively (n=4). Two independent trials using multiple litters were used for experiments in this figure. Data represent mean ± SEM. Statistics were performed using two-tailed Welch’s t-tests (b,c,h,i), simple linear regression test (d), or one-way ANOVA with Dunnett multiple comparison between 4EP+/− groups at each size group. * p ≤ 0.05; ** p ≤ 0.01.
Extended Data Fig. 8. Additional behavior tests in 4EP- and 4EP+ mice.

a, Distance traveled (p=0.06), time in center (p=0.01), and thigmotaxis time (p=0.007), during open field test over a period of 10 minutes (distance traveled data was also shown in Extended Data Fig. 2g) (4EP- n=21, 4EP+ n=24). b, Time spent in the open arms of the elevated plus maze (EPM) (left)(p=0.02) and ratio of time spent in the open/time spent in the closed arms of the EPM (right) (4EP- n=21, 4EP+ n=24). c, Light/dark box: time mice spent in the open, lit portion of the arena. Test time, 10 minutes (4EP- n=25, 4EP+ n=23) p=0.02. d, Grooming: total time mice spent self-grooming over a period of 10 minutes (4EP- n=27, 4EP+ n=24). e, Social interaction: with an unfamiliar, age-matched male intruder. Total time socializing (left) and percent of total socializing that is anogenital sniffing (right) (4EP- n=25, 4EP+ n=22) p=0.0009. f, Ultrasonic vocalization: time spent vocalizing to an unfamiliar, age-matched female for 3 minutes (4EP- n=23, 4EP+ n=21) p=0.01. g, Novel object recognition (NOR): time spent investigating a novel object when presented with a novel and a familiar object (n=22). h, Y-maze alternations: percent of times mice repeated entry into an arm it had just visited rather than alternate to all arms in succession (4EP- n=15, 4EP+ n=16). i, Beam traversal: time required for the mouse to cross the narrowing beam (4EP- n=19, 4EP+ n=22). j, Pole descent: time required for the mouse to descend from the pole to the home cage (4EP- n=19, 4EP+ n=22). k, Wire hang: time the mouse hung on and explored the underside of the wire grid before releasing into cage below (4EP- n=19, 4EP+ n=22). l, Fecal 16s profiles of 4EP+/− mice after behavior tests, where tan is the colonization group and contamination from exposure to behavior tests is colored according to the legend (left panel) and a magnified view of contaminants only (right panel) (4EP- n=8, 4EP+ n=10). Two independent cohorts of mice from multiple litters were used for each experiment in this figure. Data a-k represent mean ± SEM analyzed by two-tailed Welch’s t-test. * p ≤ 0.05; ** p ≤ 0.01.
Extended Data Fig. 9. Behavior results from 4EP or 4EPS administration by drinking water and anxiety behavior tests of 4EP+/− mice relative to GF and SPF mice.

a-f, Behavior tests of conventionally colonized mice administered 4EP or 4EPS by drinking water. a, Left panel: EPM, time spent in open arms over time spent in closed arms (seconds). Right panel: time spent at terminus (outer 1/3 of open arms)(Veh. n=15; 4EPS n=17; 4EP n=16) (Open/closed: Veh. vs 4EPS p<0.0001; Veh vs 4EP p=0.007; 4EPS vs 4EP p=0.03; Terminus: Veh vs 4EPS p=0.004). b, Open field test, from left to right: time spent in center area over thigmotaxis, time spent in center (seconds), distance moved in open field test, showing an increase in activity in 4EP-treated animals (Veh. n=16; 4EPS n=16; 4EP n=18) p=02. c, Grooming test, total percent of time spent grooming, showing an increase in grooming in the 4EPS-treated animals (Veh. n=16; 4EPS n=18; 4EP n=17). d, Number of marbles buried (Veh. n=16; 4EPS n=17; 4EP n=18). e, Social interaction. Left panel: total socialization time (seconds). Right panel: % of total socialization time spent as anogenital sniffing (Veh. n=8; 4EPS n=7; 4EP n=7). f, USV test. Left panel: Vocalization events. Right panel: Total duration spent vocalizing (seconds) (Veh. n=16; 4EPS n=15; 4EP n=18). g, 4EPS levels in urine of mice administered 4EP or 4EPS in the drinking water (creatinine corrected)(Veh. n=12; 4EPS n=9; 4EP n=11)(Veh. vs 4EPS p=0.0005; Veh vs 4EP p=0.008; 4EPS vs 4EP p=0.03). h, 4EPS levels in serum of mice administered 4EP or 4EPS in the drinking water (Veh. n=4; 4EPS n=4; 4EP n=3). i, Quantified flow cytometry data of PVT of mice administered 4EP or 4EPS in the drinking water, with ratio of MOG+ quartile percentages/NG2+ quartile percentages presented (Veh. n=5; 4EPS n=5; 4EP n=4) p=0.03. For gating strategy see Supplementary Information Figure 2 and main Fig. 3h. j, Quantified western blot data for MOG and MBP in the PVT of mice administered 4EP or 4EPS in the drinking water, with blots shown below (n=4 each group). (MBP: H2O vs 4EPS p=0.04; vs 4EP p=0.02). For gel source data, see Supplementary Figure 1. k-m, Anxiety behavior tests contextualizing 4EP+/− mice to GF and SPF mice. k, Open field test, from left to right: time spent in center area over time spent along walls (4EP- vs 4EP+ p=0.04; 4EP+ vs GF p=0.02; 4EP+ vs SPF p=0.04), time spent in center (seconds), distance moved in open field test(4EP+ vs SPF p=0.047; GF vs SPF p-0.01) (4EP- n=8; 4EP+ n=9; GF n=9; SPF n=11). l, Elevated plus maze, time spent in the open arms/time spent in the closed arms ration (left)(4EP- vs 4EP+ p=0.03; 4EP+ vs SPF p=0.03) and time spent at the terminus (right) (4EP- n=9; 4EP+ n=9; GF n=11; SPF n=13). m, Marble burying (4EP- n=9; 4EP+ n=9; GF n=9; SPF n=112)(4EP- vs 4EP+ p=0.02; 4EP- vs SPF p=0.03; 4EP+ vs GF p=0.02; GF vs SPF p=0.03). Abbreviations: Veh., vehicle control; MOG, myelin oligodendrocyte glycoprotein; MBP, myelin basic protein. Two independent cohorts of mice from multiple litters were used for the experiments in this figure. Data represent mean ± SEM analyzed by one-way ANOVA with Dunnett multiple comparison between all groups. * p ≤ 0.05; ** p ≤ 0.01; *** p ≤ 0.001; **** p ≤0.0001.
Extended Data Fig. 10. Fluorescence imaging and extended behavior test results of clemastine fumarate and miconazole-treated mice, and preliminary behavior tests performed on both sexes, used to determine continued studies on males for the study.

a-i, 4EP+/− mice were administered vehicle (control), clemastine fumarate (clem.), or miconazole and then behavior tested and imaged.. a, Representative images of CC1 (green), NG2 (red), and OLIG2 (blue) staining in 4EP+/− mice with or without treatment. Representative of group of individual mice used for quantification: 4EP- control=7, treated=10; 4EP+ control=6, treated=6, which came from two cohorts (Scale bar 100µm). b, Quantification of fluorescent imaging, presented as a ratio of CC1+/NG2+ cells (from left to right n=7, 10, 6, 6, 4, 4). The first four columns are also shown in Main Fig. 4i (Cont 4EP-vs 4EP+ p=0.04; 4EP+Cont vs 4EP+Clem p=0.01). c-d, Confirmation that treatment does not reduce 4EPS levels with clemastine (c) (Control. 4EP- n=4; Control. 4EP+ n=6, Clem 4EP- n=3; Clem 4EP+ n=4) or miconazole (d) (Control. 4EP- n=4; Control. 4EP+ n=4, Mic 4EP- n=4; Mic 4EP+ n=5) treatment. e, Extended EPM results (Control 4EP- n=13; Control 4EP+ n=13; Clem 4EP- n=12; Clem 4EP+ n=14). f, Extended open field results (Control 4EP- n=15; Control 4EP+ n=17; Clem 4EP- n=17; Clem 4EP+ n=17). g-i, Behavioral results for mice treated with miconazole, including g, Open field (Control 4EP- n=24; Control 4EP+ n=25; Mic. 4EP- n=18; Mic. 4EP+ n=20)(left graph: 4EP-Cont vs 4EP+ Cont p=0.02; 4EP+cont vs 4EP+Mic p=0.008)(middle graph: 4EP-Cont vs 4EP+ Cont p=0.02; 4EP+cont vs 4EP+Mic p=0.005). h, EPM (Control 4EP- n=24; Control 4EP+ n=25; Mic. 4EP- n=15; Mic. 4EP+ n=17)(left graph: 4EP-Cont vs 4EP+ Cont p=0.002; 4EP+cont vs 4EP+Mic p=0.03)(right graph: 4EP-Cont vs 4EP+Cont p=0.0009). i, Marble Burying (Control 4EP- n=24; Control 4EP+ n=26; Mic. 4EP- n=21; Mic. 4EP+ n=21)(p=0.001) j, Left panel: EPM, time spent in open arms over time spent in closed arms. Right panel: time spent at terminus (outer 1/3 of open arms) (Males 4EP- n=17; Males 4EP+ n=17; Females 4EP- n=20; Females 4EP+ n=21)p=0.02. k, Open field test, time spent in center area over time spent in thigmotaxis left), time spent in center (right) (Males 4EP- n=16; Males 4EP+ n=14; Females 4EP- n=18; Females 4EP+ n=13)p-values left to right: 0.005, 0.003. l, Number of marbles buried in marble burying test (Males 4EP- n=24; Males 4EP+ n=23; Females 4EP- n=17; Females 4EP+ n=26)p=0.03. Abbreviations: NG2, neural/glial antigen 2; CC1, antibody (anti-adenomatous polyposis coli (APC) clone) that binds mature oligodendrocyte marker; OLIG2, oligodendrocyte transcription factor 2; clem, clemastine fumarate; mic, miconazole. Two independent cohorts of mice from multiple litters were used for each experiment in this figure. Data represent mean ± SEM analyzed by two-way ANOVA with Dunnett multiple comparison to 4EP+ group (panels b-i) or Sidak multiple comparison between 4EP+/− groups within each sex (panels j-l). * p ≤ 0.05, ** p ≤ 0.01.
Supplementary Material
Acknowledgements:
We thank members of the Mazmanian laboratory for critical evaluation of the manuscript, Sarah Reisman and Lauren Chapman (Caltech) for synthesizing 4EPS, the Caltech Kavli Nanoscience Institute for aid in maintaining the TF30 electron microscope, and the Gordon and Betty Moore and Beckman Foundations for gifts to Caltech to support electron microscopy.
Funding:
This work was supported by funds from the Center for Environmental Microbial Interactions to B.D.N., National Science Foundation MRI grant 1920364 for Bruker console upgrading, the Human Frontier Science Program (Grant No. LT000217/2020-C) to C.R., the National Institutes of Health (NIH) (2 P50 GM082545–08) to P.J.B., the Ministry of Science and Technology in Taiwan (MOST107– 2320-B-006–072-MY3; MOST108–2321-B-006–025-MY2) to W-L.W, and the Heritage Medical Research Institute, Lynda and Blaine Fetter, and the NIH (MH100556 and AG063744) to S.K.M.
Footnotes
Competing interests: S.K.M. has financial interests in Axial Biotherapeutics. All other authors declare no competing interests related to this work.
References
- 1.Sharon G et al. Specialized metabolites from the microbiome in health and disease. Cell Metab 20, 719–730 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Swann JR et al. Application of 1H NMR spectroscopy to the metabolic phenotyping of rodent brain extracts: A metabonomic study of gut microbial influence on host brain metabolism. J Pharm Biomed Anal 143, 141–146 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Vuong HE, Yano JM, Fung TC & Hsiao EY The Microbiome and Host Behavior. Annu. Rev. Neurosci 40, 21–49 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Needham BD, Kaddurah-Daouk R & Mazmanian SK Gut microbial molecules in behavioural and neurodegenerative conditions. Nature Reviews Neuroscience 1–15 (2020) doi: 10.1038/s41583-020-00381-0. [DOI] [PubMed] [Google Scholar]
- 5.Huang F & Wu X Brain Neurotransmitter Modulation by Gut Microbiota in Anxiety and Depression. Front Cell Dev Biol 9, 649103 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gacias M et al. Microbiota-driven transcriptional changes in prefrontal cortex override genetic differences in social behavior. eLife 5, e13442 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoban AE et al. Regulation of prefrontal cortex myelination by the microbiota. Transl Psychiatry 6, e774 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsiao EY et al. Microbiota Modulate Behavioral and Physiological Abnormalities Associated with Neurodevelopmental Disorders. Cell 155, 1451–1463 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berer K et al. Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature 479, 538–541 (2011). [DOI] [PubMed] [Google Scholar]
- 10.Pan S, Mayoral SR, Choi HS, Chan JR & Kheirbek MA Preservation of a remote fear memory requires new myelin formation. Nature Neuroscience 23, 487–499 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maheras KJ et al. Absence of Claudin 11 in CNS Myelin Perturbs Behavior and Neurotransmitter Levels in Mice. Sci Rep 8, 3798 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bonnefil V et al. Region-specific myelin differences define behavioral consequences of chronic social defeat stress in mice. Elife 8, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galvez-Contreras AY, Zarate-Lopez D, Torres-Chavez AL & Gonzalez-Perez O Role of Oligodendrocytes and Myelin in the Pathophysiology of Autism Spectrum Disorder. Brain Sci 10, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cassoli JS et al. Disturbed macro-connectivity in schizophrenia linked to oligodendrocyte dysfunction: from structural findings to molecules. npj Schizophrenia 1, 1–10 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Needham BD et al. Plasma and Fecal Metabolite Profiles in Autism Spectrum Disorder. Biol Psychiatry (2020) doi: 10.1016/j.biopsych.2020.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gamage N et al. Human Sulfotransferases and Their Role in Chemical Metabolism. Toxicol Sci 90, 5–22 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Chamkha M, Garcia JL & Labat M Metabolism of cinnamic acids by some Clostridiales and emendation of the descriptions of Clostridium aerotolerans, Clostridium celerecrescens and Clostridium xylanolyticum. Int J Syst Evol Microbiol 51, 2105–2111 (2001). [DOI] [PubMed] [Google Scholar]
- 18.Santamaría L, Reverón I, Felipe F. L. de, Rivas B. de las & Muñoz R Ethylphenol Formation by Lactobacillus plantarum: Identification of the Enzyme Involved in the Reduction of Vinylphenols. Appl. Environ. Microbiol 84, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Agency for Toxic Substances and Disease Registry (ATSDR). Toxicological profile for Cresols https://wwwn.cdc.gov/TSP/ToxProfiles/ToxProfiles.aspx?id=946&tid=196 (2008). [PubMed]
- 20.Penzo MA et al. The paraventricular thalamus controls a central amygdala fear circuit. Nature 519, 455–459 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahrens S et al. A Central Extended Amygdala Circuit That Modulates Anxiety. J. Neurosci 38, 5567–5583 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duval ER, Javanbakht A & Liberzon I Neural circuits in anxiety and stress disorders: a focused review. Ther Clin Risk Manag 11, 115–126 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robinson OJ, Pike AC, Cornwell B & Grillon C The translational neural circuitry of anxiety. J Neurol Neurosurg Psychiatry 90, 1353–1360 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Barson JR, Mack NR & Gao W-J The Paraventricular Nucleus of the Thalamus Is an Important Node in the Emotional Processing Network. Front. Behav. Neurosci 14, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirouac GJ The Paraventricular Nucleus of the Thalamus as an Integrating and Relay Node in the Brain Anxiety Network. Front Behav Neurosci 15, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park H-C & Appel B Delta-Notch signaling regulates oligodendrocyte specification. Development 130, 3747–3755 (2003). [DOI] [PubMed] [Google Scholar]
- 27.Wang S et al. Notch Receptor Activation Inhibits Oligodendrocyte Differentiation. Neuron 21, 63–75 (1998). [DOI] [PubMed] [Google Scholar]
- 28.Williamson JM & Lyons DA Myelin Dynamics Throughout Life: An Ever-Changing Landscape? Front. Cell. Neurosci 12, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holschneider DP, Wang Z & Pang RD Functional connectivity-based parcellation and connectome of cortical midline structures in the mouse: a perfusion autoradiography study. Front Neuroinform 8, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaller MS, Lazari A, Blanco-Duque C, Sampaio-Baptista C & Johansen-Berg H Myelin plasticity and behaviour — connecting the dots. Curr Opin Neurobiol 47, 86–92 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barak B et al. Neuronal deletion of Gtf2i , associated with Williams syndrome, causes behavioral and myelin alterations rescuable by a remyelinating drug. Nature Neuroscience 22, 700–708 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Xie D et al. Clemastine improves hypomyelination in rats with hypoxic–ischemic brain injury by reducing microglia-derived IL-1β via P38 signaling pathway. Journal of Neuroinflammation 17, 57 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kirouac GJ Placing the paraventricular nucleus of the thalamus within the brain circuits that control behavior. Neurosci Biobehav Rev 56, 315–329 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Sharon G, Sampson TR, Geschwind DH & Mazmanian SK The Central Nervous System and the Gut Microbiome. Cell 167, 915–932 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tian P, Wang G, Zhao J, Zhang H & Chen W Bifidobacterium with the role of 5-hydroxytryptophan synthesis regulation alleviates the symptom of depression and related microbiota dysbiosis. The Journal of Nutritional Biochemistry 66, 43–51 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Koyama H et al. Effects of housing conditions on behaviors and biochemical parameters in juvenile cynomolgus monkeys (Macaca fascicularis). Exp. Anim 68, 195–211 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tian J-S et al. A GC-MS urinary quantitative metabolomics analysis in depressed patients treated with TCM formula of Xiaoyaosan. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci 1026, 227–235 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Sankowski B et al. Higher cerebrospinal fluid to plasma ratio of p-cresol sulfate and indoxyl sulfate in patients with Parkinson’s disease. Clin. Chim. Acta 501, 165–173 (2020). [DOI] [PubMed] [Google Scholar]
- 39.Gabriele S et al. Urinary p-cresol is elevated in young French children with autism spectrum disorder: a replication study. Biomarkers 19, 463–470 (2014). [DOI] [PubMed] [Google Scholar]
- 40.Neul JL et al. Metabolic Signatures Differentiate Rett Syndrome From Unaffected Siblings. Front Integr Neurosci 14, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takesada M, Miyamoto E, Kakimoto Y, Sano I & Kaneko Z Phenolic and Indole Amines in the Urine of Schizophrenics. Nature 207, 1199–1200 (1965). [DOI] [PubMed] [Google Scholar]
- 42.Sun C-Y et al. p-Cresol Sulfate Caused Behavior Disorders and Neurodegeneration in Mice with Unilateral Nephrectomy Involving Oxidative Stress and Neuroinflammation. Int J Mol Sci 21, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paxinos G & PhD, K. B. J. F., MA. The Mouse Brain in Stereotaxic Coordinates (Elsevier Science, 2007). [Google Scholar]
Method References:
- 44.Holdeman Lillian V. & Moore WEC Anaerobe laboratory manual (4th ed). (Virginia Polytechnic Institute and State University, 1977). [Google Scholar]
- 45.Martens EC, Chiang HC & Gordon JI Mucosal Glycan Foraging Enhances Fitness and Transmission of a Saccharolytic Human Gut Bacterial Symbiont. Cell Host Microbe 4, 447–457 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koropatkin NM, Martens EC, Gordon JI & Smith TJ Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices. Structure 16, 1105–1115 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bolyen E et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nature Biotechnology 37, 852–857 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Callahan BJ et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nature Methods 13, 581–583 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.BANOGLU E & KING RS Sulfation of indoxyl by human and rat aryl (phenol) sulfotransferases to form indoxyl sulfate. Eur J Drug Metab Pharmacokinet 27, 135–140 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Teubner W, Meinl W, Florian S, Kretzschmar M & Glatt H Identification and localization of soluble sulfotransferases in the human gastrointestinal tract. Biochem J 404, 207–215 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sharon G et al. Human Gut Microbiota from Autism Spectrum Disorder Promote Behavioral Symptoms in Mice. Cell 177, 1600–1618.e17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Iadecola C The Neurovascular Unit Coming of Age: A Journey through Neurovascular Coupling in Health and Disease. Neuron 96, 17–42 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mace E et al. Functional ultrasound imaging of the brain: theory and basic principles. IEEE Transactions on Ultrasonics, Ferroelectrics, and Frequency Control 60, 492–506 (2013). [DOI] [PubMed] [Google Scholar]
- 54.Tiran E et al. Transcranial Functional Ultrasound Imaging in Freely Moving Awake Mice and Anesthetized Young Rats without Contrast Agent. Ultrasound in Medicine & Biology 43, 1679–1689 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. github.com/brittanyneedham/Needham_Nature2022. [Google Scholar]
- 56.Demené C et al. Spatiotemporal Clutter Filtering of Ultrafast Ultrasound Data Highly Increases Doppler and fUltrasound Sensitivity. IEEE Transactions on Medical Imaging 34, 2271–2285 (2015). [DOI] [PubMed] [Google Scholar]
- 57.Osmanski B-F, Pezet S, Ricobaraza A, Lenkei Z & Tanter M Functional ultrasound imaging of intrinsic connectivity in the living rat brain with high spatiotemporal resolution. Nat Commun 5, 1–14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Armstrong RA When to use the Bonferroni correction. Ophthalmic Physiol Opt 34, 502–508 (2014). [DOI] [PubMed] [Google Scholar]
- 59.Holschneider DP, Guo Y, Wang Z, Vidal M & Scremin OU Positive Allosteric Modulation of Cholinergic Receptors Improves Spatial Learning after Cortical Contusion Injury in Mice. J. Neurotrauma 36, 2233–2245 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sokoloff L Localization of functional activity in the central nervous system by measurement of glucose utilization with radioactive deoxyglucose. J. Cereb. Blood Flow Metab 1, 7–36 (1981). [DOI] [PubMed] [Google Scholar]
- 61.Wang Z, Pang RD, Hernandez M, Ocampo MA & Holschneider DP Anxiolytic-like effect of pregabalin on unconditioned fear in the rat: an autoradiographic brain perfusion mapping and functional connectivity study. Neuroimage 59, 4168–4188 (2012). [DOI] [PubMed] [Google Scholar]
- 62.STAR: ultrafast universal RNA-seq aligner | Bioinformatics | Oxford Academic. https://academic.oup.com/bioinformatics/article/29/1/15/272537. [DOI] [PMC free article] [PubMed]
- 63.Salmon provides fast and bias-aware quantification of transcript expression | Nature Methods. https://www.nature.com/articles/nmeth.4197. [DOI] [PMC free article] [PubMed]
- 64.Xu X, Wells AB, O’Brien DR, Nehorai A & Dougherty JD Cell Type-Specific Expression Analysis to Identify Putative Cellular Mechanisms for Neurogenetic Disorders. J. Neurosci 34, 1420–1431 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Y et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci 34, 11929–11947 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zambon AC et al. GO-Elite: a flexible solution for pathway and ontology over-representation. Bioinformatics 28, 2209–2210 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tyszka JM, Readhead C, Bearer EL, Pautler RG & Jacobs RE Statistical diffusion tensor histology reveals regional dysmyelination effects in the shiverer mouse mutant. Neuroimage 29, 1058–1065 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mastronarde DN Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol 152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 69.Kremer JR, Mastronarde DN & McIntosh JR Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol 116, 71–76 (1996). [DOI] [PubMed] [Google Scholar]
- 70.Mastronarde DN Correction for non-perpendicularity of beam and tilt axis in tomographic reconstructions with the IMOD package. J Microsc 230, 212–217 (2008). [DOI] [PubMed] [Google Scholar]
- 71.Carranza-Torres IE et al. Organotypic Culture of Breast Tumor Explants as a Multicellular System for the Screening of Natural Compounds with Antineoplastic Potential. Biomed Res Int 2015, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Leger M et al. Object recognition test in mice. Nature Protocols 8, 2531–2537 (2013). [DOI] [PubMed] [Google Scholar]
- 73.Sampson TR et al. Gut Microbiota Regulate Motor Deficits and Neuroinflammation in a Model of Parkinson’s Disease. Cell 167, 1469–1480.e12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Komada M, Takao K & Miyakawa T Elevated Plus Maze for Mice. J Vis Exp (2008) doi: 10.3791/1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Takao K & Miyakawa T Light/dark Transition Test for Mice. J Vis Exp (2006) doi: 10.3791/104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Miedel CJ, Patton JM, Miedel AN, Miedel ES & Levenson JM Assessment of Spontaneous Alternation, Novel Object Recognition and Limb Clasping in Transgenic Mouse Models of Amyloid-β and Tau Neuropathology. J Vis Exp (2017) doi: 10.3791/55523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shih H-T & Mok H-K ETHOM: Event-Recording Computer Software for the Study of Animal Behavior in (2000). doi: 10.6576/AZT.2000.11.(1).4. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data analyzed for this study is included in this published article with source data for all graphs in the supplementary information files, with additional 2DG data available at https://gin.g-node.org/bneedham/Needham_Nature_2022. The WoL database annotation files are publicly available at: https://biocore.github.io/wol/download. Greengenes database is publicly available at: https://greengenes.secondgenome.com/.


