Significance
Photoinhibitory high light stress in plants leads to increases in markers of protein degradation and transcriptional up-regulation of proteases and proteolytic machinery, but protein homeostasis (proteostasis) of most enzymes is largely maintained under high light, so we know little about the metabolic consequences of it beyond photosystem damage. We developed a technique to look for rapid protein turnover events in response to high light through 13C partial labeling and detailed peptide mass spectrometry. This analysis reveals a light-induced transcriptional program for nuclear-encoded genes, beyond the regulation of photosystem II, to replace key protein degradation targets in plants and ensure proteostasis under high light stress.
Keywords: protein turnover, high light, proteostasis, translation
Abstract
Photoinhibitory high light stress in Arabidopsis leads to increases in markers of protein degradation and transcriptional up-regulation of proteases and proteolytic machinery, but proteostasis is largely maintained. We find significant increases in the in vivo degradation rate for specific molecular chaperones, nitrate reductase, glyceraldehyde-3 phosphate dehydrogenase, and phosphoglycerate kinase and other plastid, mitochondrial, peroxisomal, and cytosolic enzymes involved in redox shuttles. Coupled analysis of protein degradation rates, mRNA levels, and protein abundance reveal that 57% of the nuclear-encoded enzymes with higher degradation rates also had high light–induced transcriptional responses to maintain proteostasis. In contrast, plastid-encoded proteins with enhanced degradation rates showed decreased transcript abundances and must maintain protein abundance by other processes. This analysis reveals a light-induced transcriptional program for nuclear-encoded genes, beyond the regulation of the photosystem II (PSII) D1 subunit and the function of PSII, to replace key protein degradation targets in plants and ensure proteostasis under high light stress.
Protein homeostasis (proteostasis) requires strictly controlled protein synthesis and degradation through coordinated gene expression, translational controls, and protein degradation (1, 2). Protein turnover rates have been typically measured through a pulse-chase strategy by feeding plants or isolated organelles radioactive precursors and monitoring the rates of appearance and disappearance of labeling (3, 4). The identification of the photosystem II (PSII) D1 subunit as the protein undergoing rapid turnover in chloroplasts and the basis of photoinhibition under high (H) light stress was originally found using radioactive labeling of proteins in isolated chloroplasts (3, 5–7). Using more recently developed discovery tools based on stable isotope labeling to measure turnover rates of many proteins, D1 was also noted as undergoing very rapid turnover in both Arabidopsis and barley; however, these studies also identified other chloroplastic proteins being rapidly degraded under standard (Std) light conditions (2, 8). These studies have raised the prospect that a combination of direct or indirect photodegradation targets may underlie photoinhibition and its consequences in plants (9).
The degradation of soluble cytosolic proteins in plants typically occurs through the ubiquitin–proteasome pathway guided by selective ubiquitination of targeted proteins (10). Plastids were commonly considered to be separated from this system by their membranes, thus relying on independent mechanisms of protein degradation (11). However, recent research has uncovered an interconnection of these systems with specific plastid-localized proteins being tagged by ubiquitination to be degraded by the proteasome, a pathway termed chloroplast-associated degradation (CHLORAD) (12). Chloroplasts damaged by ultraviolet exposure or overaccumulation of oxygen radicals are also degraded whole by globular vacuoles or by central vacuoles via selective autophagy (13, 14). Specific protein degradation by selective autophagy has also been studied, but mainly for plastid stromal proteins such as RuBisCo (15). The proteolysis network inside chloroplasts works to differentially break down specific damaged proteins. CtpA and CtpA1 peptidase, CLP, DEG, and FTSH family proteases have all been found or proposed to play specialized roles in maturation, processing, and cleavage of plastid-localized proteins (16, 17). As a consequence, there is ample opportunity for different rates of protein degradation to be initiated for specific plastid-localized proteins, which raises the question of how proteostasis is controlled when specific proteolytic processes are initiated.
High throughput studies have revealed rapid and robust changes in the metabolome, transcriptome, and proteome in plants during light and dark transitions or high light stresses (18–22). Such studies typically confirm the lack of positive correlations between changes in steady-state mRNA and protein abundance; i.e., compared with the rapid and robust changes in mRNA, protein abundances are often very stable, and statistically significant changes in abundance are rare.
Here, we use high light–induced photoinhibition to trigger protein degradation and explore the relationship between protein degradation rate, transcriptional responses, and protein abundance for enzymes that participate in the metabolic response to high light. In so doing, we have found direct or indirect targets of photodamage in plants and shed light on how transcriptional processes counteract protein degradation to mask light-response changes in the proteome and enable proteostasis.
Results
High Light Leads to PSII Photodamage and Metabolic Changes Indicative of Protein Degradation.
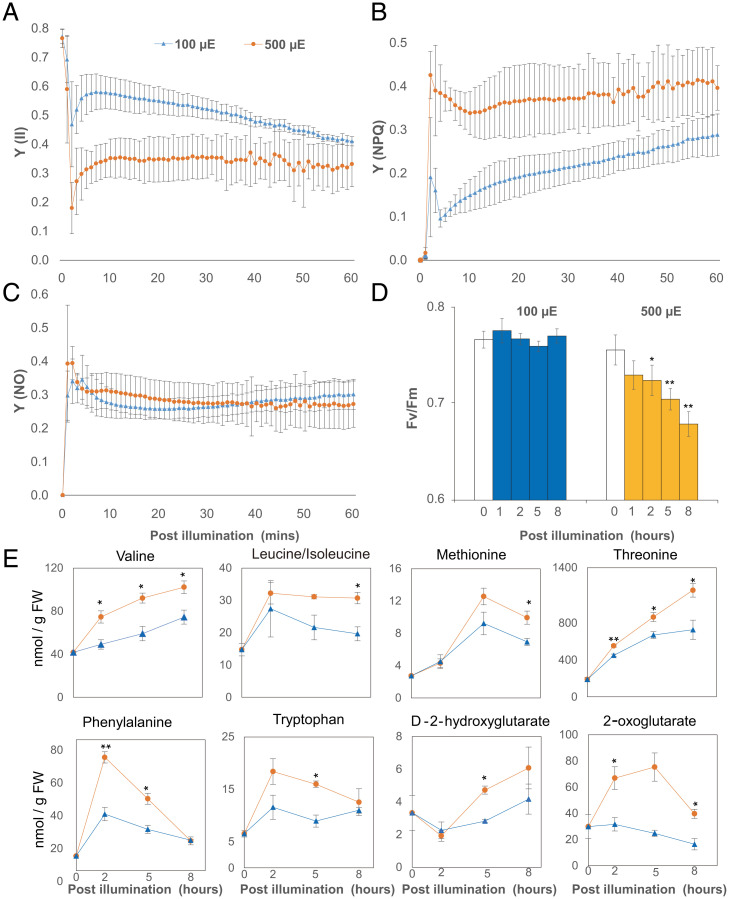
To analyze protein homeostasis under light stress, we performed a high light treatment of Arabidopsis plants aimed at inducing photoinhibition in conditions we could subsequently use to rapidly label proteins for analysis. We used a modified whole-plant growth chamber system (23) and replaced the Plexiglas lid with glass to increase light transmittance from an external light-emitting diode light source to an Arabidopsis rosette inside (see Fig. 3A). Light intensity was held at 100 µmol protons m−2·S−1 (standard light) or escalated to 500 µmol protons m−2·S−1 (high light), and fluorescence pulse-amplitude modulation (PAM) was utilized to evaluate PSII-associated photochemical parameters inside leaves (Fig. 1). After 1 h of high light exposure, PSII parameters, including Y(II)-quantum yields of photochemical energy conversion in PSII and Y(NPQ)-quantum yields of regulated nonphotochemical energy loss in PSII, showed significant changes under high light compared to standard light conditions (Fig. 1 A and B) while Y(NO)-quantum yields of nonregulated nonphotochemical energy loss in PSII remained steady under both light conditions (Fig. 1C), consistent with previous reports of high light treatment in Arabidopsis (20). This indicated 500 µmol protons m−2·S−1 exceeded the maximum capacity of PSII, thus requiring energy dissipation through nonphotochemical quenching. Dark adaptation could rescue the maximum quantum yield of PSII (Fv/Fm) after 1 h of high light exposure but failed to restore Fv/Fm after 2, 5, or 8 h of high light exposure (Fig. 1D). Heat can contribute to nonphotochemical quenching, so we measured the leaf surface temperature using an infrared thermometer. We could not detect statistically significant changes in leaf surface temperature on either the adaxial or abaxial leaf surface (SI Appendix, Fig. S1). The continuous room temperature air that was vented into the growth chamber during our measurements likely cooled the plant surface as shown in another recent high light stress study (21).
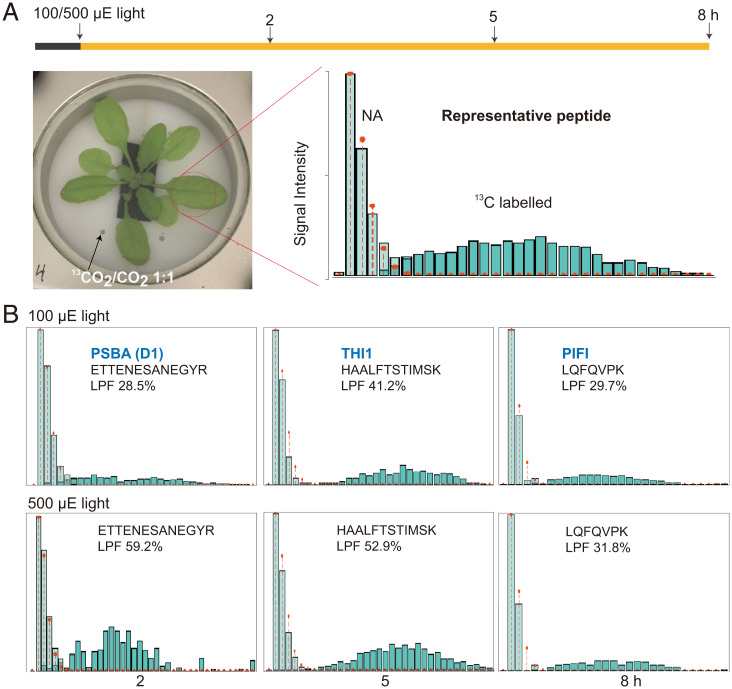
Fig. 3.
Measurement protein turnover rates in Arabidopsis shoots by partial 13CO2 labeling of the proteome. (A) Air containing 13CO2 was supplied at the end of the night to a sealed growth chamber with a transparent glass lid allowing efficient light entry. Total proteins extracted from labeled shoots were analyzed by peptide mass spectrometry. A representative mass spectrum of one peptide from labeled shoot shows the natural abundance (NA) population and the new peptide synthesized using 13C-labeled amino acids. (B) The mass spectra and calculated percentage LPF for peptides derived from PSBA (D1; ATCG00020), THI1 (AT5G54770), and PIFI (AT3G15840) after 2, 5, and 8 h of 13C labeling. The NA population is colored light green and the newly synthesized peptide population is colored dark green in each case. Red dots and lines show the distribution of the natural abundance mass spectrum.
Fig. 1.
High light–induced changes in photochemical responses and metabolite abundances in Arabidopsis. Arabidopsis in a growth chamber (Fig. 3A) were dark adapted for at least 20 min before being exposed to 100 and 500 μmol protons m−2·S−1 LED light. Chlorophyll fluorescence measurements were transformed to three parameters that describe the fate of excitation energy in PSII, including Y(II)-quantum yields of photochemical energy conversion in PSII (A), Y(NPQ)-quantum yields of regulated nonphotochemical energy loss in PSII (B), and Y(NO)-quantum yields of nonregulated nonphotochemical energy loss in PSII (C). Arabidopsis plants were exposed to 100 and 500 μmol protons m−2·S−1 LED light for 1, 2, 5, and 8 h before their Fv/Fm values were determined by maxi-PAM (D). Specific amino acids (triple quadrupole mass spectrometry, QQQ) and organic acids (quadrupole time-of-flight mass spectrometry, Q-TOF) that increased in abundance in response to high light treatment (E). Error bars show SDs for photochemical parameter measurements (biological replicates n = 4) and SEs for metabolite measurements (biological replicates n = 3). Statistical significance tests were performed with a Student’s t test (**P < 0.01, *P < 0.05).
To measure the impact of photoinhibition of PSII on cellular metabolism, we measured amino acids, organic acids, and sugar concentration in plants grown under standard and high light conditions (Fig. 1E and SI Appendix, Fig. S2). Six of 14 amino acids increased significantly (P < 0.05) in abundance after high light treatment. Stress-induced protein degradation products, including branched chain amino acids (Val, Leu, and Ile) and aromatic amino acids (Phe, Trp, and Tyr), were more abundant under high light. During plant stress, these protein degradation products serve as alternative respiratory substrates by being metabolized to D-2-hydroxyglutarate and 2-oxoglutarate (24). Accordingly, both D-2-hydroxyglutarate and 2-oxoglutarate increased in abundance with high light treatment. These two metabolites are also known markers of high light–dependent photorespiration (25). Aspartate abundance decreased significantly (P < 0.05) after high light treatment (SI Appendix, Fig. S2). However, threonine and methionine, which are biosynthesized from aspartate and contribute to isoleucine biosynthesis (26), showed higher abundance after high light treatment. Sugars (sucrose, glucose, and fructose) and tricarboxylic acid cycle metabolites, other than 2-oxoglutarate, had comparable abundances over time between standard and high light conditions, with only citrate showing decreased abundance under high light.
High Light Responses in the Transcriptome Correlate Poorly with Proteomic Changes.
To investigate the wider cellular response, we assessed changes in the transcriptome and the proteome under high light. Arabidopsis plants were transferred into the aforementioned growth chamber and left overnight to acclimate before plants were treated for 2, 5, and 8 h in standard or high light conditions, then harvested for protein or RNA. Total RNA sequencing (RNA-seq) detected 18,575 transcripts (SI Appendix, Data S1) and quantitative proteomic experiments measured 1,548 protein abundances (SI Appendix, Data S2).
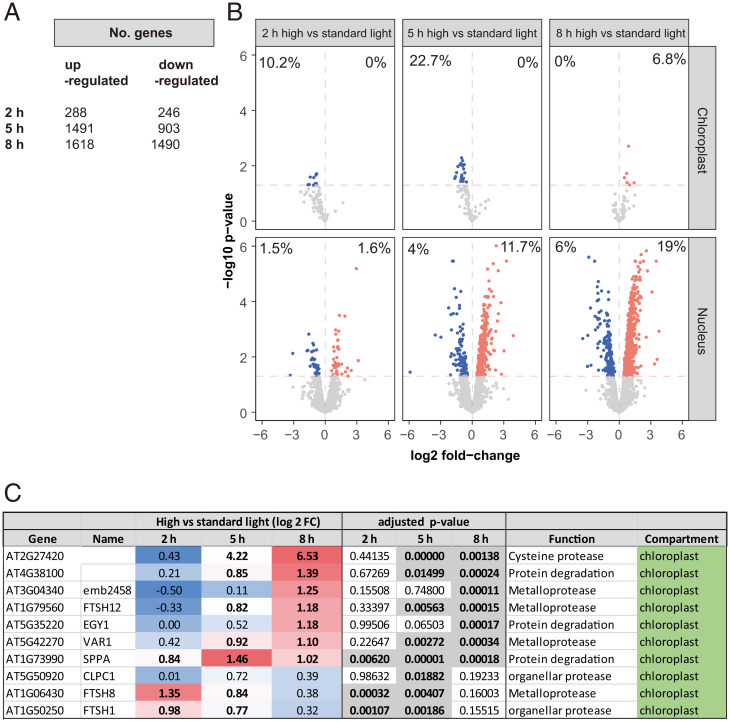
Two hours of high light treatment led to the up- or down-regulation of 288 and 246 genes, respectively, in Arabidopsis shoots (Fig. 2A). Gene Ontology (GO) overrepresentation tests reveal enrichment of ontologies related to stress and unfolded protein responses (SI Appendix, Data S1). By 5 h and 8 h of high light, there were 1,491 and 1,618 up-regulated genes, and 903 and 1,490 down-regulated genes, respectively (Fig. 2A). We compared these results to similar high light treatments, which recorded a total of 1,358 (21) and 3,151 differentially expressed genes (20). In both cases, we observed significant overlaps (as determined by Fisher's exact test) of 24% [odds ratio = 1.67, P value <0.001 (20)] and 16% [odds ratio = 2.92, P value <0.001 (21)]. GO overrepresentation tests showed up-regulation of genes encoding proteins involved in RNA metabolism, translation, and nucleotide synthesis. At 8 h, up-regulated enrichment was also evident for proteolysis, proteasome, and cellular catabolic processes (SI Appendix, Data S1). While high light appeared to directly affect chlorophyll fluorescence (Fig. 1), transcriptional responses, especially up-regulations, were most pronounced for nuclear-encoded genes despite a comparable proportion of down-regulated chloroplast-encoded genes (Fig. 2B). Given the increase in amino acid abundances indicative of protein degradation (Fig. 1E), and the GO enrichment analysis (SI Appendix, Data S1), we further investigated the expression of nuclear genes encoding proteases and proteolytic machinery. We identified 264 nuclear-encoded genes in this ontological group that were differentially expressed in response to high light; most only reached significance after 8 h of treatment (SI Appendix, Data S1). Among these, key genes encoding for chloroplast-localized proteases were up-regulated (Fig. 2C).
Fig. 2.
High light–induced changes to the transcriptome in Arabidopsis shoots. Gene expression differences were induced by high light treatment of Arabidopsis shoot tissue. (A) Numbers of differentially expressed genes (adjusted P value <0.05) at 2, 5, and 8 h of high light treatment. (B) Volcano plots of differential gene expression (DGE) (horizontal red line denotes adjusted P value <0.05) of nuclear vs. chloroplast-encoded genes encoding chloroplast-localized proteins at 2, 5, and 8 h of high light treatment. Red and blue dots/percentages denote up- and down-regulated genes or proportion of DGEs in chloroplast or nuclear genomes, respectively. (C) Differential expression of selected genes encoding proteases and components of the proteolytic machinery, in chloroplasts. Full list can be found in SI Appendix, Data S1.
To compare transcriptomic and proteomic responses, the abundance of proteins measured across all samples and their corresponding transcripts were extracted for principal component analysis (PCA) (SI Appendix, Fig. S3 A and B). While samples showed clustering by both time point and light treatment based on transcript abundance, there was far less separation based on protein abundances. We also performed correlation analysis between the fold changes in protein and transcript abundance between high and standard light conditions (SI Appendix, Fig. S3 C–E), which were found to be negligible (Pearson’s r: T2 = 0.08, T5 = 0.04, and T8 = 0.03). The same pattern was also found for 66 proteins selected as components in protein homeostasis machinery, except that their transcript abundances were only separated at 5 and 8 h (SI Appendix, Fig. S4).
Direct Measurement of Protein Turnover Rates by Partial 13CO2 Labeling in Arabidopsis.
To determine whether proteins were being degraded in response to high light but then replaced, we sought to isotopically label new proteins and thus allow degradation of preexisting proteins to be tracked by mass spectrometry. While we have previously used 15N labeling to assess protein degradation rates, the 4- to 6-h lag in this technique due to uptake by roots and translocation to leaves (2, 8) limited its utility to assess the impact of high light within 8 h. 13CO2 fixation via photosynthesis is reported to allow rapid stable isotope incorporation in leaf amino acids and proteins (27, 28). However, as the number of C atoms greatly exceeds N atoms in a tryptic peptide, the large mass shifts from 13C labeling greatly increase the complexity of the resulting peptide mass spectra (29). This was also confirmed with a preparatory experiment by supplementing air with 98% 13CO2 and considering modeling of enrichment levels (SI Appendix, Fig. S5 A and B). To minimize this effect, we calculated that lowering the 13C incorporation rate into Arabidopsis plants by supplementing air with 50% 13CO2 at 400 ppm could allow peptide mass spectra to be more readily interpreted (SI Appendix, Fig. S5). To conduct 13CO2 experiments, Arabidopsis plants were transferred into the growth chamber and left overnight to acclimate before labeling began in the morning. Arabidopsis plants were labeled for 2, 5, and 8 h in standard or high light conditions, then harvested, and protein samples were isolated and digested to measure isotopic incorporation and protein turnover rates. A representative mass spectrum of a tryptic peptide (ANLGMEVMHER) from a 2-h sample shows the clear separation of the preexisting peptide population (a typical natural abundance 13C-labeling pattern with 1, 2, or 3 13C atoms in the peptide) and a newly synthesized peptide population derived from the newly fixed 13CO2 (13C-labeled pattern containing a median of 16 13C atoms in the peptide) (Fig. 3A). The technique itself is robust against the influence of differences in enrichment level because it measures labeled proteins of different enrichments as a group (2, 8). However, to determine whether such differences exist between the two light regimes, we determined the 13C carbon content of the amino acids in the labeled peptide populations as described previously (2, 8). There were no differences in 13C enrichment between standard and high light conditions over the time course, i.e., 2, 5, and 8 h (SI Appendix, Fig. S6, median enrichment: Std light 29%, 25%, and 30%; H light 29%, 27%, and 34%). Mass spectra derived from peptides from three well-known rapidly turned over proteins (D1, THI1, and PIFI) showed that the 13C-labeled peptide fraction (LPF) was one-third to one-half of the total peptide population under standard light conditions, indicating the rapid half-lives of these proteins (Fig. 3B). A twofold higher LPF was detected for D1 peptides under high light compared with standard light. Calculations of protein turnover rates over the time course measured 13C-derived protein turnover rates for 202 proteins in standard light and 269 proteins in high light (SI Appendix, Data S3). The proteins measured had degradation rates (KD) of 0.15 to 10 per day, representing a half-life range from 1.6 h to 5 d.
High Light Leads to Faster Turnover of Photosynthetic Proteins and Associated Enzymes in Metabolic Cascade Reactions.
Light stress can cause direct photodamage to D1 and change the turnover of proteins including D1 as well as other subunits of PSII and adenosine triphosphate (ATP) synthase (4, 9). Using our 13C-derived protein turnover rates, we compared the difference in protein turnover rates between standard and high light conditions. This allowed us to confirm expected, and discover new, direct targets of photodamage or proteins that are indirectly degraded under high light. Protein turnover rates of 140 proteins could be compared between two light conditions (SI Appendix, Data S4). Compared to measurement under standard light conditions, 74 of 140 proteins showed statistically significant changes in degradation rate under high light, 73 showed faster turnover, while 1 protein (protochlorophyllide oxidoreducase B-PORB, At4g27440) turned over more slowly. Placing the proteins with measured degradation rates in their functional, metabolic, and subcellular contexts shows the depth of impact that high light has on protein degradation rates in Arabidopsis rosettes (Fig. 4). D1 (PSBA) showed the fastest rate of degradation overall and a threefold increase in degradation rate under high light, while other PSII subunits, PSB28 and PSBP, showed lower median degradation rates but still a two- to threefold increase in degradation rate under high light. ATP synthase subunits (α, ε, and b/b′) also showed significantly faster turnover under high light. A similar degree of degradation rate induction was seen for a series of molecular chaperones in the chloroplast and mitochondria, and also specific enzymes involved in the Calvin–Benson cycle (CBC) and glycolysis. All the major members of the malate dehydrogenase (MDH) family, which catalyze the malate shuttle between organelles, as well as thioredoxin- and glutaredoxin-linked enzymes in the chloroplast and mitochondria, also degraded more rapidly under high light conditions.
Fig. 4.
Changes in protein turnover rates in response to high light treatment. Changes in protein degradation rate are shown as log2 fold changes between high and standard light. For visualization, 61 proteins with annotated functions and localized in four major cellular compartments, i.e., chloroplast, cytosol, mitochondrion, and peroxisome, were extracted from the set of 74 proteins with significant changes in rate. Protein subunits in photosynthetic complexes, import apparatus, chaperonin, and protease were colored according to the values of log2 fold changes (SI Appendix, Data S4). Protein subunits with nonsignificant changes or nonavailable (NA) data were colored gray.
Notably, there was no change in degradation rate of LHC-II, PSI, NDH, or Cyt b6f subunits (Fig. 4 and SI Appendix, Data S4). However, NIR1 and FTR, which take electrons from PSI Fd to reduce oxidized thioredoxin and nitrite, clearly degrade faster in high light. It was reported that reduced thioredoxin from FTR can serve as a reductant for activation of MDH and CBC, which links PSI Fd with metabolic enzymes in the chloroplast (30–35). Here we show that Trxm1 and Trxm4 turned over faster, as did chloroplast MDH and CBC enzymes, in response to high light. Furthermore, faster turnover of metabolic enzymes involved in malate shuttles between cellular compartments was observed. There was a faster turnover of MTHFR1, ATMS1, and BCAT4 in the cytosol that catalyze the reductive conversion of 5,10-methylenetetrahydrofolate (CH2-THF) to 5-methyltetrahydrofolate (CH3-THF), which then serves as a methyl donor for methionine biosynthesis and the following chain elongation pathway. In mitochondria, SHMT1, which catalyses the production of serine from glycine, degraded faster under high light. A number of enzymes involved in the consumption of nicotinamide adenine dinucleotide phosphate (NADPH), nicotinamide adenine dinucleotide (NADH), ATP, and glutathione also show faster degradation rates (Fig. 4 and SI Appendix, Data S4). Examples of this group include PORC, which catalyses the conversion of protochlorophyllide to chlorophyllide, and geranylgeranyl (GG) chlorophyll a reductase-GGR, which catalyzes the formation of chlorophyll a in the thylakoid membrane; cytosolic NDPK1 and mitochondrial NDPK3 that catalyse the production of nucleotides by consuming ATP; cytosolic NADPH:quinone reductase (NQR) that converts quinone to semiquinone; and Gpx2, GsTU19, GrxS15, and lactoyl-GS lyase that reduce oxidative metabolites by consuming glutathione. Taken together, we can see a clear pattern of faster turnover of enzymes involved in metabolic reactions responding to high light.
Beyond metabolic enzymes, faster degradation was also observed for a number of elongation factors, chaperonins, and proteases in response to high light (Fig. 4 and SI Appendix, Data S4). Proteins in this group are essential for protein synthesis, folding, assembly, and degradation to maintain proteostasis. This group includes RNA binding proteins (CSP41B and RBPs) and elongation factors (EF-P/G, RPS1, SCO1, and RABE1b) in the chloroplast; cpHSP70 and HSP93-V (ClpC) that are involved in chloroplast protein import (36); ClpC that forms a protein complex with CLP protease to unfold selected proteins for degradation (17, 36); chloroplast CPN20 and CPN60 that form a protein complex for the assembly of RuBisCo (37, 38); mitochondrial chaperonin CPN10 and mtHsc70-1 involved in electron transport chain protein complex assembly (39); and FtsH2 that forms a protein complex with FtsH1/5/8 and is involved in D1 degradation (40, 41).
Transcriptional Responses Counteract Increased Protein Turnover to Maintain Proteostasis of Many Major Cellular Enzymes.
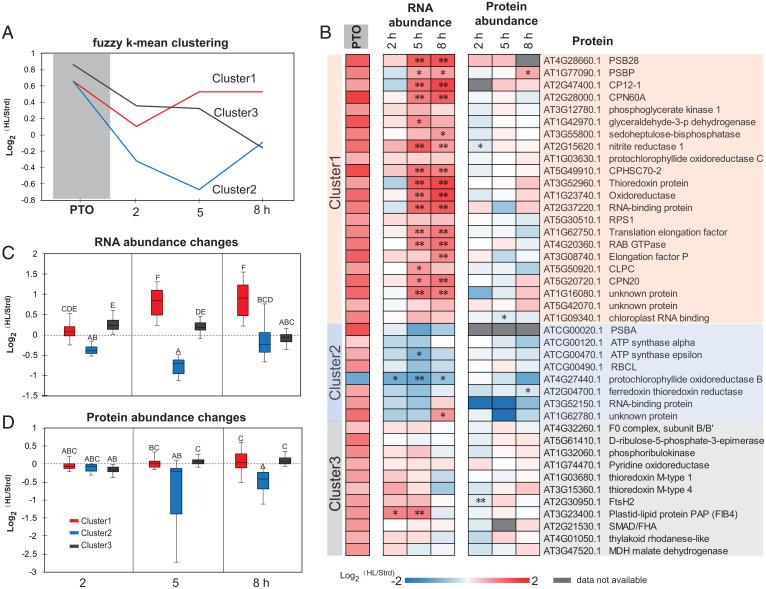
To further examine the 74 proteins exhibiting light-induced changes in degradation rate, we performed fuzzy k-means clustering based on their changes in degradation rate and transcript abundance (Fig. 5A and SI Appendix, Data S5). This approach grouped the 74 proteins into three clusters. Proteins in clusters 1 and 2 had the same change in degradation rate; however, cluster 1 genes were up-regulated by high light, whereas those in cluster 2 were down-regulated. Cluster 3 contained proteins with greater changes in degradation rate whose encoding genes were first up-regulated and then down-regulated by high light.
Fig. 5.
Changes in transcript and protein abundance for proteins with significant changes in protein turnover rate during high light treatment. Based on patterns of protein degradation and transcript changes, a fuzzy k-means clustering method was utilized to cluster the 74 proteins with significant changes in protein turnover rate. Representative curves of the three clusters were plotted (A) and values of distance to centroid for specific proteins are provided in SI Appendix, Data S5. Forty-one plastid proteins were extracted from the whole set to show their protein turnover rate alongside fold changes in transcript and protein abundance (B). Boxplots of changes in transcript (C) and protein abundance (D) of each cluster over the time course are shown. PTO, change in protein turnover rate. * shows P for proteins or adjusted P value for transcripts <0.05, ** shows P for proteins or adjusted P value for transcripts <0.01.
A total of 41 of the 74 proteins in the light-induced degradation set were chloroplast-localized proteins (SI Appendix, Data S5). To further investigate the coordination between protein turnover and RNA and protein abundance in the chloroplast, we aligned the changes (log2 fold change) in these traits between standard and high light conditions, then ranked them by functional categories in the three clusters (Fig. 5B). In contrast to their faster turnover in response to high light treatment, chloroplast protein abundance for these proteins remained unchanged. We only found FtsH2 protease, RNA binding protein At1g09340, and NIR1 that showed statistically significant abundance decreases after 2 or 5 h of high light treatment (Fig. 5B). PSII subunit PSBP even showed a statistically significant increase in abundance after 8 h. This indicated proteostasis of fast turnover chloroplast proteins is maintained even after high light treatment for 8 h. Transcriptional up-regulation for the genes encoding proteins in clusters 1 and 3 masked their faster turnover, thus maintaining protein levels. Cluster 2 consisted of eight proteins, with half of them being encoded by the chloroplast genome. All chloroplast-encoded proteins with measured turnover in this study, namely PSII D1, ATP synthase subunits (α and ε), and RuBisCo large subunit, follow the cluster 2 pattern (Fig. 5B). Expression of these chloroplast-encoded genes is not induced by high light stress treatment, so their proteostasis in the face of increased protein degradation rates must be governed by the posttranscriptional process.
To investigate the timing of coordination between transcription and proteostasis for chloroplast proteins, we plotted RNA and protein abundance changes by members in each cluster (Fig. 5 C and D). Consistently, smaller net changes were observed in protein abundance (statistical grouping A–C) than RNA abundance (statistical grouping A–E). Net protein abundances started to decrease after 2 h of high light exposure. However, protein abundance for clusters 1 and 3 started to recover while the abundance of members of cluster 2 continued decreasing over the time course of high light treatment. It is evident that clusters 1 and 3 complemented their faster protein turnover through enhanced transcription. Increased transcript abundance would facilitate continued translation to counteract light-induced protein degradation. In contrast, cluster 2 members were more inclined to drop in both transcription and protein abundance after high light treatment. Their proteostasis is likely to be recovered more slowly through posttranscriptional responses involving translational control.
Discussion
A Multiomics Analysis Reveals Targets of Light-Dependent Protein Degradation.
It is well known that high light stress causes damage to the PSII reaction center protein, D1, and leads to impaired PSII efficiency. In this study, we found high light led to a PSII efficiency [Y(II)] decrease and tripled the degradation rate of D1 (Fig. 1 and SI Appendix, Data S4). We observed that although Y(II) dropped at the beginning of the high light treatment it gradually recovered to the level observed under standard light over the first hour of high light exposure (Fig. 1A). This suggests Arabidopsis plants can cope with the increased turnover rate of D1 under high light by maintaining proteostasis and PSII function after a short time course of high light exposure. Consistent with this, a recent study utilizing ribosomal profiling and pulse labeling found that D1 photodamage can trigger recruitment of its mRNA to the ribosome to enhance D1 synthesis (4). This demonstrates that D1 degradation and synthesis are matched to maintain proteostasis for short-term high light acclimation. However, we found that Fv/Fm, an indicator of PSII maximum efficiency after dark adaption, declined after longer high light exposure. This suggests that longer periods of high light caused irreversible damage, from which Arabidopsis PSII efficiency cannot recover even after dark adaptation, likely due to the uncoupling of D1 degradation from its synthesis rate (Fig. 5).
Beyond the D1 protein, we found high light significantly increased the degradation of another 72 proteins (SI Appendix, Data S4). Protein degradation in our high light experiments is supported by our measures of the accumulation of amino acids through protein degradation (Fig. 1 and SI Appendix, Fig. S2) and up-regulation of protease gene expression (Fig. 2). To investigate how Arabidopsis coped with this enhanced degradation to maintain proteostasis, we investigated changes in transcript abundances between standard and high light conditions (Figs. 2 and 5). Nuclear-encoded genes encoding proteins with high turnover rates (clusters 1 and 3) demonstrated transcriptional responses that masked protein turnover changes, resulting in proteostasis under high light (Fig. 5 A–C). These strong correlations between faster protein turnover and higher transcript abundances help explain the purpose of high light–triggered transcript induction without apparent protein abundance changes.
In contrast, chloroplast-encoded genes (D1, Rubisco large subunit, and ATP synthase subunits) do not respond to high light at the transcriptional level, and their RNA levels even dropped to some extent. This limited transcription response in the chloroplast under light stress was also reported in tobacco (22). It appears that chloroplast-encoded genes largely rely on posttranscriptional controls to counteract rapid protein turnover under high light. Previous studies focusing on in vitro or in vivo chloroplast translation observed translation elongation rate stimulated by light (4, 42–44). The activation of protein synthesis by elongation is also supported by faster turnover of different RNA binding proteins and elongation factors in this study (Fig. 4). For short-term high light exposure, rapid protein synthesis from translation elongation can complement rapid protein degradation due to photodamage to maintain proteostasis. However, we found chloroplast translation failed to keep pace with protein degradation after longer periods of high light exposure. This is supported by the failure of dark adaptation to recover PSII (Fig. 1D) and the tendency toward protein abundance decreases after a longer high light exposure (Fig. 5D). Recently, a salvaging strategy to circumvent inefficient chloroplast translation by expressing D1 protein from the nuclear genome was found to enhance Arabidopsis, tobacco, and rice performance under stress conditions (45). It would be attractive to perform a wider salvaging operation involving other photodamage targets discovered in this study to maintain their proteostasis under high light or other stresses.
Many Fast Turnover Proteins Are Unaffected by High Light.
We found that high light does not affect turnover rates for nearly half of the 140 proteins that we could assess between standard and high light conditions (SI Appendix, Data S4). Some of these are rapid turnover proteins such as PIFI (Post-Illumination Chlorophyll Fluorescence Increase) and CCD4 (Carotenoid Cleavage Dioxygenase 4). PIFI is an ancillary subunit of the chloroplast NDH complex, and we previously proposed PIFI’s rapid turnover could relate to the putative role of the NDH complex in photoprotection (2, 9). But our high light data suggest the control of PIFI turnover is independent of light stress. CCD4 is a plastoglobuli-localized enzyme that cleaves carotenoids, such as β-carotene (11, 46). Its degradation was proposed to associate with a plastoglobuli M48 peptidase PGM48 (47). In silico modeling of CCD4 suggests it has lower stability compared with other members of the CCD gene family (48). Its rapid turnover may reflect its suborganelle location, which is distinct from the other CCDs or this modeled intrinsic lower stability rather than light stress. Rapid degradation of other proteins, such as CML10, THI1, GRP2, and BAM3, show only small rate changes in high light. It is probable, at least for these proteins, that their rapid turnover rates are due to their function, sequence, protein domains, or cellular location rather than light stress (2).
We also observed that the rapidly turning over enzyme, protochlorophyllide oxidoreductase (PORB) exhibited a significant slowing of its degradation rate under high light (Fig. 4). Protochlorophyllide oxidoreductase is a light-activated enzyme, which catalyzes the transformation of protochlorophyllide to chlorophyllide. In barley and rice, there are two isoforms of protochlorophyllide oxidoreductases whose expression are regulated differently by normal and high light (49–51). In Arabidopsis, there are three protochlorophyllide oxidoreductases namely PORA, PORB, and PORC (52). Repression of Arabidopsis PORB gene expression by light has been reported (53, 54). In this study, we also found PORB gene expression is repressed after high light treatment. In contrast, PORC showed faster protein turnover in high light and slightly induced gene expression. It is conceivable that PORC plays a specific role in chlorophyll biogenesis under high light conditions. For PORB and PORC, transcription plays a key role in maintaining proteostasis, and their protein turnover rates appear to be responsive to changes in transcript abundance.
Metabolic Explanation of Increased Protein Turnover Rates.
The turnover of D1 is typically explained as a response to photoinactivation of the protein. Research suggests that photodamage to PSII may involve the disintegration of the Mn2+ center in PSII that leads to an energy imbalance and so-far ill-defined oxidative damage of residues in D1. Photodamage to D1 impairs PSII function and leads to cleavage of the damaged D1 subunit by proteases in a two-step model (55–57). Turnover of another rapidly degrading protein, thiamin synthase (THI1), is explained by its suicide mechanism that means the enzyme has a single catalytic cycle before it is inactivated and needs to be replaced (58, 59). Recently we showed across a wide range of enzymes in Arabidopsis, yeast, and bacteria, that the number of catalytic cycles until replacement varied according to the chemical risk of the reaction they undertook, including enzymes with photoactivatable substrates or with reactive oxygen producing roles in metabolism (60). It is evident from our protein turnover measurements that high light leads to faster degradation of PSII D1, PSB28, PSBP, PORC, and GGR, which all catalyze light-activated reactions (Fig. 4 and SI Appendix, Data S4). PSI was also activated by high light, yet seemingly its rate of protein degradation was unaffected. FTR and NIR1, which take electrons from PSI Fd to reduce thioredoxin and nitrite, turned over faster. Thioredoxins can serve as reductants to activate CBC and MDH catalyst activities (61). Moreover, transient overproduction of NADPH and ATP as substrates may further accelerate the usage of CBC and MDH enzymes and also accelerate their turnover. MDH activation in the chloroplast acts as a stimulus to malate circulation to the cytosol, mitochondrion, and peroxisome in so-called malate shuttles of reductant from sites of synthesis to cellular sinks (62). In terms of risk, high light and photoinhibition is likely to lead to increased reactive oxygen species production and an elevated need for shuttling of reductant out of the plastid to other cellular compartments (63). The increased turnover of redox shuttling systems, namely glutathione- and thioredoxin-linked systems and the MDH enzymes involved in malate shuttles throughout the cell (Fig. 4), may be due to increased flux through these pathways and thus a consequence of an increased rate of wear-out damage of these enzymes (60, 64).
Conclusion.
We have discovered a range of proteins with enhanced rates of degradation in response to high light. Light-activated electron transport pathways and metabolic fluxes likely stimulate the usage of metabolic enzymes and accelerate their degradation. Potential protein targets of photodamage, many of which are chloroplast localized, have been revealed, and a differential role of nuclear and plastid transcriptional control to maintain proteostasis has been highlighted.
Materials and Methods
Arabidopsis Plant Preparation and 13CO2 Labeling.
Arabidopsis thaliana accession Columbia-0 plants were grown under 16-h/8-h light/dark conditions with cool white T8 tubular fluorescent lamps 4000K 3350 lm (Osram) with the intensity of 100 to 125 μmol m−2⋅s−1 at 22 °C. Arabidopsis plants were grown in soil pots for 21 d until they reached leaf production stage 1.10 (65). Shoots of Arabidopsis at the leaf production stage 1.10 were positioned into the sealed growth chamber with the soil pots kept underneath (Fig. 3). Six tandem growth chambers were supplied with air at a continuous flow rate 6 L/min and kept overnight before the labeling experiment (T0). A homemade water column was connected to the air hose to keep the air humidity inside the growth chamber. A commercial LED (Heliospectra) was used as the light source for the labeling experiment, and the light spectra were set as (420 nm-25%, 450 nm-63.8%, 530 nm-75%, 630 nm-100%, 660 nm-25%, and 735 nm-2.5% of maximum light intensity). Normal and high light intensity at 100 and 500 μmol protons m−2·S−1 was achieved by adjusting the distance between the growth chamber and the light source. 13CO2 labeling was started at dawn by supplying the growth chamber with a mixture of CO2 air and 13CO2 air at equal volume for a continuous flow rate of 6 L/min. The Arabidopsis plants were labeled for 2, 5, and 8 h (T2, T5, and T8) before their shoots were cut and snap frozen in liquid nitrogen to stop all biological activities immediately. Three biological replicates were collected at each time point.
Protein Extraction, In-Gel/Solution Digestion, High pH High-Performance Liquid Chromatography (HPLC) Separation and Liquid Chromatography-Mass Spectrometry (LC-MS) Analysis of Tryptic Peptides.
The shoot samples (∼0.1 g) were snap frozen in liquid nitrogen and homogenized using Qiagen tissue lysis beads (2 mm). A total plant protein extraction kit (PE0230-1KT, Sigma Chemicals) was used to extract total proteins. The final pellet of total protein was dissolved in solution 4 and then reduced and alkylated by tributylphosphine (TBP) and iodoacetamide (IAA) as described in the Sigma manual. The suspension was centrifuged at 16,000 × g for 30 min, and the supernatant was assayed for protein concentration by amido black quantification as described previously (66). Protein (100 µg) in solution from each sample was then mixed with an equal volume of 2× sample buffer (4% sodium dodecyl sulfate (SDS), 125 mM Tris, 20% glycerol, 0.005% bromophenol blue, and 10% mercaptoethanol, pH 6.8) before being separated on a Bio-Rad protean II electrophoresis system with a 4% (vol/vol) polyacrylamide stacking gel and 12% (vol/vol) polyacrylamide separation gel. Proteins were visualized by colloidal Coomassie Brilliant Blue G250 staining. One gel lane from one single biological replicate was excised into 11 fractions. Gel lanes from 10 samples (T0 as a control, three biological replicates for a 13C-labeled sample at each time point) were fractioned into 110 and in-gel digested as described previously (2). For protein abundance measurements, total proteins (50 µg) from 14N grown plants were combined with the fully 15N-labeled protein reference (50 µg) and then in-solution trypsin digested. Each sample was separated into 96 fractions by high pH HPLC separation and further pooled into 6 fractions. Twenty-one total protein samples (three biological samples from T0; 2, 5, and 8 under both standard and high light conditions kept in the same growth chamber as 13C labeling) were in-solution digested and separated into 126 fractions. Tryptic peptides from in-gel/in-solution digested were lyophilized in a Labconco centrifugal vacuum concentrator. Lyophilized samples were first resuspended in loading buffer (5% acetonitrile, 0.% formic acid) and filtered through a 0.22-µm Millipore column before being run in an Orbitrap Fusion (Thermo Fisher Scientific) mass spectrometer over the course of 95 min over 2 to 30% (vol/vol) acetonitrile in 0.1% (vol/vol) formic acid (Dionex UltiMate 3000) on a 250 × 0.075 mm column (Dr. Maisch ReproSil-Pur 1.9 mm).
Mass Spectrometry Data Analysis.
Orbitrap fusion raw (.raw) files were first converted to mzML using the Msconvert package from the Proteowizard project, and mzML files were subsequently converted to Mascot generic files (.mgf) using the mzxml2 search tool from the Trans Proteomic Pipeline (TPP). Mascot generic file peak lists were searched against an in-house Arabidopsis database comprising ATH1.pep (release 10) from The Arabidopsis Information Resource (TAIR) and the Arabidopsis mitochondrial and plastid protein sets (33,621 sequences and 13,487,170 residues) (67), using the Mascot search engine version 2.3 and utilizing error tolerances of 10 ppm for MS and 0.5 Da for MS/MS, “Max Missed Cleavages” set to 1, and variable modifications of oxidation (Met) and carbamidomethyl (Cys). All mzML files and dat files are provided in ProteomeXchange. We used iProphet and ProteinProphet from the TPP to analyze peptide and protein probability and global false discovery rate (FDR) (68–70). The reported peptide lists with P = 0.8 have FDRs of <3%, and protein lists with P = 0.95 have FDRs of <0.5%. Quantification of LPFs (13C-labeled protein fraction) and protein abundance (14N/15N ratios) were accomplished by an in-house script written in R as described previously (2, 8, 71). An averaged turnover rate over the 2-, 5-, and 8-h time course was calculated following methods described previously (2). Mass spectrometry data can be accessed through ProteomeXchange through two entries: protein abundance changes in response to high light treatment (PXD010888) (72) and protein turnover rates under high light treatment (PXD010889) (73).
Total RNA Sequencing.
Arabidopsis plants were grown under identical conditions as per the 13C-labeling experiment, except that normal air was supplied to the growth chamber. Shoot tissues were harvested (as above) in biological triplicate after differing light transitions from dark: T0D, end of night (dark control); T2H, 2 h high light; T2L, 2 h standard light; T5H, 5 h high light; T5L, 5 h standard light; T8H, 8 h high light; and T8L, 8 h standard light. Total RNA was isolated using TRI Reagent based on an adapted protocol (20). Full details are available at protocols.io: dx.doi.org/10.17504/protocols.io.bt8wnrxe. Total RNA-sequencing libraries were prepared using the TruSeq Stranded Total RNA with Ribo-Zero Plant kit (RS-122-2402, Illumina) as per manufacturer’s instructions but with input RNA and reaction volumes adjusted by one-third. PCR amplified libraries were pooled equal molar and sequenced (75 bp, single end) on one lane of the NextSeq500.
Raw read quality was first diagnosed using FastQC (v0.11.7). Trim Galore! (v0.4.4) was used for adapter and low-quality read trimming with PHRED score <20 (-q 20). Trimmed reads were input for single-end splice-aware alignments using Subjunc from the Subread package v1.5.0 (74), retaining only reads that uniquely aligned to the Arabidopsis TAIR10 reference genome. Uniquely aligned reads were sorted and indexed using Samtools v1.3.1 (75). Aligned reads were summarized to gene-level loci using the Araport11 annotation (76) using featureCounts (-s 2 for reverse stranded libraries) (77). Differential gene expression was tested using the edgeR quasi-likelihood pipeline (78, 79). Reads mapping to ribosomal RNA were removed; only loci containing counts per million >1 in at least three samples were examined. The trimmed mean of M values method was used for library normalization to account for sequencing depth and composition (78, 79). Generalized linear models were fitted using normalized counts to estimate dispersion (glmQLFit) followed by employing quasi-likelihood F tests (glmQLFTest) to test for differential expression while controlling for false discovery rates due to multiple hypothesis testing (FDR adjusted P value <0.05). Protein subcellular localization data were acquired from SUBA (80).
RNA-seq data are summarized in SI Appendix, Data S1 and can be accessed at the Gene Expression Omnibus (GEO) repository GSE131545 (81). Code used for analyses are available on GitHub: https://github.com/dtrain16/NGS-scripts.
PSII Fluorescence Parameters Measured by Mini-PAM and IMAGING-PAM.
The Arabidopsis plants grown under the same condition for the labeling experiment were used for PSII fluorescence parameter measurements except that normal air was supplied to the growth chamber. After darkness adaption for at least 20 min, PSII parameters of T0 plants were measured using LED as the light source at 100 and 500 μmol protons m−2·S−1 light intensity with a mini-PAM (Heinz Walz GmbH). Basal (F0,measuring light only) fluorescence, instantaneous (F, actinic light), and maximum (Fm, after saturating pulse) fluorescence was obtained at 1 minute intervals over 1 hour. These measures allowed determination of Y(II), Y(NPQ), and Y(NO) values with the WinControl-3.25 data acquisition software. For Fv/Fm measurements, T0 and T1, T2, T5, and T8 plants light exposed at 100 and 500 μmol protons m−2·S−1 were darkness adapted at least 20 min before being measured by a MAXI version of the IMAGING-PAM (Heinz Walz GmbH). A color gradient was used to demonstrate the Fv/Fm (maximum quantum yield of PSII) values measured by IMAGING-PAM in leaves of the whole rosette. One biological replicate was a combination of measured Fv/Fm values in three leaves in Arabidopsis plants.
Metabolite Extraction.
The Arabidopsis plants grown under the same condition for the labeling experiment were used for metabolite extraction except that normal air was supplied to the growth chamber. Plant tissues (15 to 50 mg) were collected at specified time points and immediately snap frozen in liquid nitrogen. Samples were ground to fine powder and 500 μL of cold metabolite extraction solution (90% [vol/vol] methanol, spiked with 2 mg/mL ribitol, 6 mg/mL adipic acid, and 2 mg/mL and 13C-leucine as internal standards). Samples were immediately vortexed and shaken at 1,400 rpm for 20 min at 75 °C. Cell debris was removed by centrifugation at 20,000 × g for 5 min. For each sample, 100 or 400 μL of supernatant was transferred to a new tube and either proceeded to derivatization for LC-MS analysis or dried using a SpeedVac.
Analyses of Organic Acids and Amino Acids by Selective Reaction Monitoring Using Triple Quadrupole (QQQ) Mass Spectrometry.
For LC-MS analysis of organic acids, sample derivatization was carried out based on previously published methods with modifications (82). Briefly, for each 100 µL of sample, 50 µL of 250 mM 3-nitrophenylhydrazine in 50% methanol, 50 µL of 150 mM 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide in methanol, and 50 µL of 7.5% pyridine in 75% methanol were mixed and allowed to react on ice for 60 min. To terminate the reaction, 50 µL of 2 mg/mL butylated-hydroxytoluene in methanol was added, followed by the addition of 700 µL of water. Derivatized organic acids were separated on a Phenomenex Kinetex XB-C18 column (50 × 2.1 mm, 5-µm particle size) using 0.1% formic acid in water (solvent A) and methanol with 0.1% formic acid (solvent B) as the mobile phase. The elution gradient was 18% B at 1 min, 90% B at 10 min, 100% B at 11 min, 100% B at 12 min, 18% B at 13 min, and 18% B at 20 min. The column flow rate was 0.3 mL/min and the column temperature was maintained at 40 °C. The QQQ-MS was operated in the negative ion mode with multiple reaction monitoring mode.
Supplementary Material
Acknowledgments
We thank Jacob Petereit and Sandra Kebler for advice on plant growth conditions and lighting the chamber with LEDs. We acknowledge the Biomolecular Resource Facility at The Australian National University for assistance with Illumina sequencing and computational resources provided by the National Computational Infrastructure, which is supported by the Australian Government. Mass spectrometry was performed on instruments managed by the Western Australian Proteomics Facility as a node of Proteomics Australia and was supported by infrastructure funding from the Western Australian State Government in partnership with Bioplatforms Australia under the Commonwealth Government National Collaborative Research Infrastructure Strategy. This work was supported through funding by the Australian Research Council (CE140100008 and FL200100057) (to A.H.M. and B.J.P.), the National Natural Science Foundation of China (31970294), Tianjin Natural Science Foundation (19JCYBJC24100) (to L.L.), and the CSIRO Synthetic Biology Future Science Platform (to D.R.G.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2121362119/-/DCSupplemental.
Data Availability
Proteomics and transcriptomics data have been deposited in the ProteomeXchange Consortium (PXD010888 and PXD010889) (72, 73) and GEO (GSE131545) (81).
References
- 1.Millar A. H., et al. , The scope, functions, and dynamics of posttranslational protein modifications. Annu. Rev. Plant Biol. 70, 119–151 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Li L., et al. , Protein degradation rate in Arabidopsis thaliana leaf growth and development. Plant Cell 29, 207–228 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sundby C., McCaffery S., Anderson J. M., Turnover of the photosystem II D1 protein in higher plants under photoinhibitory and nonphotoinhibitory irradiance. J. Biol. Chem. 268, 25476–25482 (1993). [PubMed] [Google Scholar]
- 4.Chotewutmontri P., Barkan A., Light-induced psbA translation in plants is triggered by photosystem II damage via an assembly-linked autoregulatory circuit. Proc. Natl. Acad. Sci. U.S.A. 117, 21775–21784 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ohad I., Kyle D. J., Arntzen C. J., Membrane protein damage and repair: Removal and replacement of inactivated 32-kilodalton polypeptides in chloroplast membranes. J. Cell Biol. 99, 481–485 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohad I., Kyle D. J., Hirschberg J., Light-dependent degradation of the Q(B)-protein in isolated pea thylakoids. EMBO J. 4, 1655–1659 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Long S. P., Humphries S., Falkowski P. G., Photoinhibition of photosynthesis in nature. Annu. Rev. Plant Physiol. 45, 633–662 (1994). [Google Scholar]
- 8.Nelson C. J., Alexova R., Jacoby R. P., Millar A. H., Proteins with high turnover rate in barley leaves estimated by proteome analysis combined with in planta isotope labeling. Plant Physiol. 166, 91–108 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li L., Aro E. M., Millar A. H., Mechanisms of photodamage and protein turnover in photoinhibition. Trends Plant Sci. 23, 667–676 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Guo H., Ecker J. R., Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell 115, 667–677 (2003). [DOI] [PubMed] [Google Scholar]
- 11.van Wijk K. J., Kessler F., Plastoglobuli: Plastid microcompartments with integrated functions in metabolism, plastid developmental transitions, and environmental adaptation. Annu. Rev. Plant Biol. 68, 253–289 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Ling Q., et al. , Ubiquitin-dependent chloroplast-associated protein degradation in plants. Science 363, eaav4467 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Izumi M., Ishida H., Nakamura S., Hidema J., Entire photodamaged chloroplasts are transported to the central vacuole by autophagy. Plant Cell 29, 377–394 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woodson J. D., et al. , Ubiquitin facilitates a quality-control pathway that removes damaged chloroplasts. Science 350, 450–454 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Michaeli S., Galili G., Genschik P., Fernie A. R., Avin-Wittenberg T., Autophagy in plants—What’s new on the menu? Trends Plant Sci. 21, 134–144 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Gururani M. A., Venkatesh J., Tran L. S., Regulation of photosynthesis during abiotic stress-induced photoinhibition. Mol. Plant 8, 1304–1320 (2015). [DOI] [PubMed] [Google Scholar]
- 17.van Wijk K. J., Protein maturation and proteolysis in plant plastids, mitochondria, and peroxisomes. Annu. Rev. Plant Biol. 66, 75–111 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Vogel M. O., et al. , Fast retrograde signaling in response to high light involves metabolite export, MITOGEN-ACTIVATED PROTEIN KINASE6, and AP2/ERF transcription factors in Arabidopsis. Plant Cell 26, 1151–1165 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liang C., et al. , Transcriptomic, proteomic and metabolic changes in Arabidopsis thaliana leaves after the onset of illumination. BMC Plant Biol. 16, 43 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crisp P. A., et al. , Rapid recovery gene downregulation during excess-light stress and recovery in Arabidopsis. Plant Cell 29, 1836–1863 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang J., Zhao X., Chory J., The Arabidopsis transcriptome responds specifically and dynamically to high light stress. Cell Rep. 29, 4186–4199.e3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schuster M., Gao Y., Schöttler M. A., Bock R., Zoschke R., Limited responsiveness of chloroplast gene expression during acclimation to high light in tobacco. Plant Physiol. 182, 424–435 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kölling K., George G. M., Künzli R., Flütsch P., Zeeman S. C., A whole-plant chamber system for parallel gas exchange measurements of Arabidopsis and other herbaceous species. Plant Methods 11, 48 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Araújo W. L., Tohge T., Ishizaki K., Leaver C. J., Fernie A. R., Protein degradation—An alternative respiratory substrate for stressed plants. Trends Plant Sci. 16, 489–498 (2011). [DOI] [PubMed] [Google Scholar]
- 25.Kuhn A., et al. , D-2-hydroxyglutarate metabolism is linked to photorespiration in the shm1-1 mutant. Plant Biol. 15, 776–784 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Hildebrandt T. M., Nunes Nesi A., Araújo W. L., Braun H. P., Amino acid catabolism in plants. Mol. Plant 8, 1563–1579 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Ishihara H., Obata T., Sulpice R., Fernie A. R., Stitt M., Quantifying protein synthesis and degradation in Arabidopsis by dynamic 13CO2 labeling and analysis of enrichment in individual amino acids in their free pools and in protein. Plant Physiol. 168, 74–93 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishihara H., et al. , Growth rate correlates negatively with protein turnover in Arabidopsis accessions. Plant J. 91, 416–429 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Nelson C. J., Li L., Millar A. H., Quantitative analysis of protein turnover in plants. Proteomics 14, 579–592 (2014). [DOI] [PubMed] [Google Scholar]
- 30.Dai S., Schwendtmayer C., Schürmann P., Ramaswamy S., Eklund H., Redox signaling in chloroplasts: Cleavage of disulfides by an iron-sulfur cluster. Science 287, 655–658 (2000). [DOI] [PubMed] [Google Scholar]
- 31.Collin V., et al. , The Arabidopsis plastidial thioredoxins: New functions and new insights into specificity. J. Biol. Chem. 278, 23747–23752 (2003). [DOI] [PubMed] [Google Scholar]
- 32.Marri L., et al. , Prompt and easy activation by specific thioredoxins of Calvin cycle enzymes of Arabidopsis thaliana associated in the GAPDH/CP12/PRK supramolecular complex. Mol. Plant 2, 259–269 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Michelet L., et al. , Redox regulation of the Calvin-Benson cycle: Something old, something new. Front Plant Sci 4, 470 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guinea Diaz M., Nikkanen L., Himanen K., Toivola J., Rintamäki E., Two chloroplast thioredoxin systems differentially modulate photosynthesis in Arabidopsis depending on light intensity and leaf age. Plant J. 104, 718–734 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu J., et al. , Plant chloroplast stress response: Insights from thiol redox proteomics. Antioxid. Redox Signal. 33, 35–57 (2020). [DOI] [PubMed] [Google Scholar]
- 36.Nakai M., New perspectives on chloroplast protein import. Plant Cell Physiol. 59, 1111–1119 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Aigner H., et al. , Plant RuBisCo assembly in E. coli with five chloroplast chaperones including BSD2. Science 358, 1272–1278 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Vitlin Gruber A., Feiz L., Rubisco assembly in the chloroplast. Front. Mol. Biosci. 5, 24 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wei S. S., et al. , Arabidopsis mtHSC70-1 plays important roles in the establishment of COX-dependent respiration and redox homeostasis. J. Exp. Bot. 70, 5575–5590 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Nishimura K., Kato Y., Sakamoto W., Chloroplast proteases: Updates on proteolysis within and across suborganellar compartments. Plant Physiol. 171, 2280–2293 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zaltsman A., Ori N., Adam Z., Two types of FtsH protease subunits are required for chloroplast biogenesis and Photosystem II repair in Arabidopsis. Plant Cell 17, 2782–2790 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mühlbauer S. K., Eichacker L. A., Light-dependent formation of the photosynthetic proton gradient regulates translation elongation in chloroplasts. J. Biol. Chem. 273, 20935–20940 (1998). [DOI] [PubMed] [Google Scholar]
- 43.Trebitsh T., Danon A., Translation of chloroplast psbA mRNA is regulated by signals initiated by both photosystems II and I. Proc. Natl. Acad. Sci. U.S.A. 98, 12289–12294 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chotewutmontri P., Barkan A., Dynamics of chloroplast translation during chloroplast differentiation in maize. PLoS Genet. 12, e1006106 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen J.-H., et al. , Nuclear-encoded synthesis of the D1 subunit of photosystem II increases photosynthetic efficiency and crop yield. Nat. Plants 6, 570–580 (2020). [DOI] [PubMed] [Google Scholar]
- 46.Gonzalez-Jorge S., et al. , Carotenoid cleavage dioxygenase4 is a negative regulator of β-carotene content in Arabidopsis seeds. Plant Cell 25, 4812–4826 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bhuiyan N. H., Friso G., Rowland E., Majsec K., van Wijk K. J., The plastoglobule-localized metallopeptidase PGM48 is a positive regulator of senescence in Arabidopsis thaliana. Plant Cell 28, 3020–3037 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Priya R., et al. , Molecular modeling and dynamic simulation of Arabidopsis thaliana carotenoid cleavage dioxygenase gene: A comparison with Bixa orellana and Crocus sativus. J. Cell. Biochem. 118, 2712–2721 (2017). [DOI] [PubMed] [Google Scholar]
- 49.Sakuraba Y., et al. , The rice faded green leaf locus encodes protochlorophyllide oxidoreductase B and is essential for chlorophyll synthesis under high light conditions. Plant J. 74, 122–133 (2013). [DOI] [PubMed] [Google Scholar]
- 50.Lebedev N., Timko M. P., Protochlorophyllide oxidoreductase B-catalyzed protochlorophyllide photoreduction in vitro: Insight into the mechanism of chlorophyll formation in light-adapted plants. Proc. Natl. Acad. Sci. U.S.A. 96, 9954–9959 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Holtorf H., Reinbothe S., Reinbothe C., Bereza B., Apel K., Two routes of chlorophyllide synthesis that are differentially regulated by light in barley (Hordeum vulgare L.). Proc. Natl. Acad. Sci. U.S.A. 92, 3254–3258 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gabruk M., Mysliwa-Kurdziel B., Light-dependent protochlorophyllide oxidoreductase: Phylogeny, regulation, and catalytic properties. Biochemistry 54, 5255–5262 (2015). [DOI] [PubMed] [Google Scholar]
- 53.Hoecker U., Quail P. H., The phytochrome A-specific signaling intermediate SPA1 interacts directly with COP1, a constitutive repressor of light signaling in Arabidopsis. J. Biol. Chem. 276, 38173–38178 (2001). [DOI] [PubMed] [Google Scholar]
- 54.Matsumoto F., et al. , Gene expression profiling of the tetrapyrrole metabolic pathway in Arabidopsis with a mini-array system. Plant Physiol. 135, 2379–2391 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kato Y., Ozawa S., Takahashi Y., Sakamoto W., D1 fragmentation in photosystem II repair caused by photo-damage of a two-step model. Photosynth. Res. 126, 409–416 (2015). [DOI] [PubMed] [Google Scholar]
- 56.Lindahl M., et al. , The thylakoid FtsH protease plays a role in the light-induced turnover of the photosystem II D1 protein. Plant Cell 12, 419–431 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kley J., et al. , Structural adaptation of the plant protease Deg1 to repair photosystem II during light exposure. Nat. Struct. Mol. Biol. 18, 728–731 (2011). [DOI] [PubMed] [Google Scholar]
- 58.Joshi J., et al. , Bioinformatic and experimental evidence for suicidal and catalytic plant THI4s. Biochem. J. 477, 2055–2069 (2020). [DOI] [PubMed] [Google Scholar]
- 59.Chatterjee A., et al. , Saccharomyces cerevisiae THI4p is a suicide thiamine thiazole synthase. Nature 478, 542–546 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hanson A. D., et al. , The number of catalytic cycles in an enzyme’s lifetime and why it matters to metabolic engineering. Proc. Natl. Acad. Sci. U.S.A. 118, e2023348118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nikkanen L., Rintamäki E., Chloroplast thioredoxin systems dynamically regulate photosynthesis in plants. Biochem. J. 476, 1159–1172 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Selinski J., Scheibe R., Malate valves: Old shuttles with new perspectives. Plant Biol. 21 (suppl. 1), 21–30 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jung H. S., et al. , Subset of heat-shock transcription factors required for the early response of Arabidopsis to excess light. Proc. Natl. Acad. Sci. U.S.A. 110, 14474–14479 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tivendale N. D., Fenske R., Duncan O., Millar A. H., In vivo homopropargylglycine incorporation enables sampling, isolation and characterization of nascent proteins from Arabidopsis thaliana. Plant J. 107, 1260–1276 (2021). [DOI] [PubMed] [Google Scholar]
- 65.Boyes D. C., et al. , Growth stage-based phenotypic analysis of Arabidopsis: A model for high throughput functional genomics in plants. Plant Cell 13, 1499–1510 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li L., Nelson C. J., Solheim C., Whelan J., Millar A. H., Determining degradation and synthesis rates of Arabidopsis proteins using the kinetics of progressive 15N labeling of two-dimensional gel-separated protein spots. Mol. Cell Prot. 11, M111.010025 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lamesch P., et al. , The Arabidopsis Information Resource (TAIR): Improved gene annotation and new tools. Nucleic Acids Res. 40, D1202–D1210 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Deutsch E. W., et al. , A guided tour of the Trans-Proteomic Pipeline. Proteomics 10, 1150–1159 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nesvizhskii A. I., Keller A., Kolker E., Aebersold R., A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 (2003). [DOI] [PubMed] [Google Scholar]
- 70.Shteynberg D., et al. , iProphet: Multi-level integrative analysis of shotgun proteomic data improves peptide and protein identification rates and error estimates. Mol. Cell Prot. 10, M111.007690 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Salih K. J., et al. , Impact of oxidative stress on the function, abundance, and turnover of the Arabidopsis 80S cytosolic ribosome. Plant J. 103, 128–139 (2020). [DOI] [PubMed] [Google Scholar]
- 72.Li L., Millar A. H., Protein abundance changes in response to high light treatment. ProteomeXchange Consortium via the PRIDE database. https://www.ebi.ac.uk/pride/archive/projects/PXD010888/. Deposited 28 August 2018.
- 73.Li L., Millar A. H., Protein turnover rates under high light treatment. ProteomeXchange Consortium via the PRIDE database. https://www.ebi.ac.uk/pride/archive/projects/PXD010889/. Deposited 28 August 2018.
- 74.Liao Y., Smyth G. K., Shi W., The Subread aligner: Fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 41, e108 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li H., et al. ; 1000 Genome Project Data Processing Subgroup, The sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cheng C. Y., et al. , Araport11: A complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 89, 789–804 (2017). [DOI] [PubMed] [Google Scholar]
- 77.Liao Y., Smyth G. K., Shi W., featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 78.Robinson M. D., Oshlack A., A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11, R25 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen Y., Lun A. T., Smyth G. K., From reads to genes to pathways: Differential expression analysis of RNA-Seq experiments using Rsubread and the edgeR quasi-likelihood pipeline. F1000 Res. 5, 1438 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hooper C. M., Castleden I. R., Tanz S. K., Aryamanesh N., Millar A. H., SUBA4: The interactive data analysis centre for Arabidopsis subcellular protein locations. Nucleic Acids Res. 45 (D1), D1064–D1074 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li L., Ganguly D. R., Pogson B. J., Millar A. H., Data from "Enzymes degraded under high light maintain proteostasis by transcriptional regulation in Arabidopsis." NCBI Gene Expression Omnibus (GEO) database. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE131545. Deposited 21 May 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Han J., Gagnon S., Eckle T., Borchers C. H., Metabolomic analysis of key central carbon metabolism carboxylic acids as their 3-nitrophenylhydrazones by UPLC/ESI-MS. Electrophoresis 34, 2891–2900 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Proteomics and transcriptomics data have been deposited in the ProteomeXchange Consortium (PXD010888 and PXD010889) (72, 73) and GEO (GSE131545) (81).