Abstract
The proteasome has key roles in neuronal proteostasis, including the removal of misfolded and oxidized proteins, presynaptic protein turnover, and synaptic efficacy and plasticity. Proteasome dysfunction is a prominent feature of Alzheimer’s disease (AD). We show that prevention of proteasome dysfunction by genetic manipulation delays mortality, cell death, and cognitive deficits in fly and cell culture AD models. We developed a transgenic mouse with neuronal-specific proteasome overexpression that, when crossed with an AD mouse model, showed reduced mortality and cognitive deficits. To establish translational relevance, we developed a set of TAT-based proteasome-activating peptidomimetics that stably penetrated the blood-brain barrier and enhanced 20S/26S proteasome activity. These agonists protected against cell death, cognitive decline, and mortality in cell culture, fly, and mouse AD models. The protective effects of proteasome overexpression appear to be driven, at least in part, by the proteasome’s increased turnover of the amyloid precursor protein along with the prevention of overall proteostatic dysfunction.
Proteasome overexpression and proteasome-activating compounds alleviate AD-like pathology in cell culture, fly, and mouse models.
INTRODUCTION
Alzheimer’s disease (AD) affects millions worldwide, and its incidence increases exponentially after the age of 65. While the proximal cause of sporadic AD remains unknown, its histologic hallmarks are neurofibrillary tangles, β-amyloid (Aβ) plaques, synaptic loss, and neuronal death.
The proteasome represents the major, multifunctional, and multisubunit protease of the ubiquitin-proteasome pathway (1). Direct inhibition of the proteasome by protein aggregates is a feature of neurodegenerative diseases, including Parkinson’s and Huntington’s diseases in addition to AD (2). Many hallmarks of AD point to disrupted proteostasis, including reduced proteasome function in brain tissues from patients with AD (1, 3). Proteasome impairment is also observed in mouse and cell culture models of AD (4–6). Proteasome impairment is thought to be driven by direct inhibition of the proteasome complex by intracellular Aβ (4, 5, 7). Aβ plays a causative role in AD as a cleavage product of the Amyloid Precursor Protein (APP) (8), when the latter is processed by β-secretase 1 (BACE1), followed by γ-secretase (9). Aβ is neurotoxic and forms soluble oligomers and insoluble plaques that increase with AD progression (10, 11).
Many pathologic aspects of AD are thought to stem from the accumulation of intracellular Aβ (12), which may be internalized by membrane trafficking (13) or generated from APP on intracellular membranes (14). The proteasome is essential for presynaptic protein turnover, synaptic efficacy and plasticity, and many other functions that are impaired in AD (15). Thus, the toxicity of intracellular Aβ may come, in part, from impaired proteasome function. In support of this concept, mice with depleted neuronal proteasomes or injected with proteasome inhibitors show neurodegeneration, formation of neuronal aggregates, and cognitive deficits (16, 17). Furthermore, the microtubule-associated protein, tau, becomes hyperphosphorylated in AD and forms insoluble neurofibrillary tangles, which disrupt the cytoskeleton and cellular trafficking. In addition, hyperphosphorylated tau can form nonfibrillary aggregates in synapses, where its presence is associated with disrupted proteostasis (18).
In virtually all eukaryotic cells, multiple forms of the proteasome play a critical role in maintaining proteostasis. The core 20S proteasome degrades unorganized, misfolded, and oxidatively damaged proteins, while the 26S proteasome is capable of degrading proteins labeled with a polyubiquitin tag, which is a critical step in cell signaling and proteostasis. In neurons, the proteasome system has additional roles, especially in the synapse, where it is essential to establishing long-term potentiation, a key cellular mechanism underlying learning and memory (19, 20). These data underscore why the nervous system may be especially vulnerable to proteasome dysfunction and why pathologies of aging and neurodegenerative diseases, including AD, may be caused in part by impaired proteasome activity.
In this study, we demonstrate that proteasome dysfunction in multiple models is a prominent and early-stage marker of AD, preceding many other biological markers. We show that prevention of proteasome dysfunction through enhanced proteasome assembly can reduce deficits observed in cell culture, fly, and mouse models of AD. We also report that a set of novel TAT peptide–derived proteasome agonists reduced deficits observed in animal models of AD. Last, we present evidence that the protective effects observed were at least in part a product of increased turnover of the APP, leading to reduced levels of Aβ.
RESULTS
Proteasome dysfunction in patients with AD and AD animal models
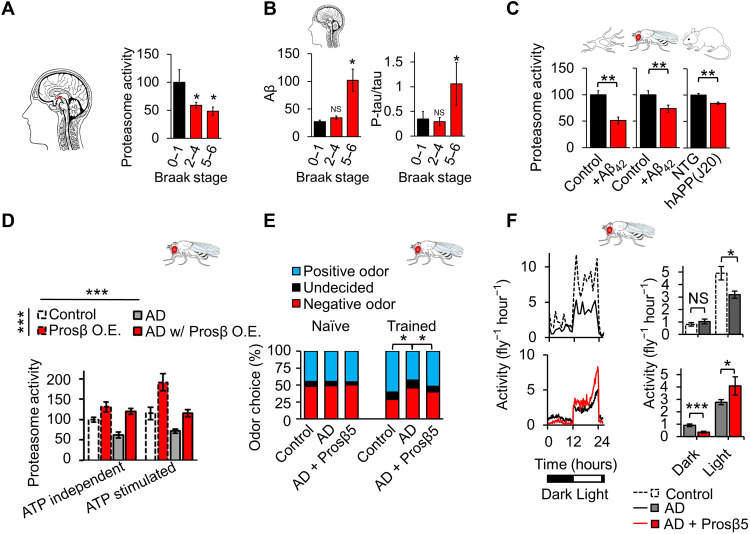
To investigate the impact of AD on proteasome function, we built on previous work showing proteasome impairment in patients with AD (1, 3) by examining when proteasome dysfunction occurs during the disease course of AD. To do this, we measured proteasome peptidolytic (chymotrypsin-like) activity in the hippocampus of patients with different stages of AD. We used hippocampal tissues from several repositories connected with the National Institutes of Health (NIH) NeuroBioBank (see Acknowledgments). Samples came from patients between 50 and 78 years old, with Braak stages ranging from 0 to 6. We divided samples into groups of controls (Braak 0 to 1, N = 11), early-stage AD (Braak 2 to 4, N = 12), and late-stage AD (Braak 5 to 6, N = 12) (fig. S1). We observed significant declines in proteasome activity in both early and late stages of AD (Fig. 1A), suggesting that impaired proteasome function could be an early-stage marker of AD. In contrast, other markers of AD pathologies, including increased soluble Aβ42 and elevated soluble tau phosphorylation relative to total tau, were seen only in late-stage patients (Fig. 1B and fig. S2). These results demonstrate that proteasome dysfunction may precede other pathologic markers of AD. We also observed proteasome dysfunction as a robust feature in animal models of AD. Proteasome function was impaired in cells treated with Aβ, in flies that overexpress preformed Aβ (UAS-Aβ3-42), and in brains of mice that overexpress a human familial variant of APP (hAPP-J20 mice with Swedish and Indiana mutations) (Fig. 1C).
Fig. 1. Proteasome dysfunction is a robust and early-stage marker of AD, and proteasome augmentation delays cognitive deficits in a fly model of AD.
(A) Proteasome peptidase (chymotrypsin-like) activity is impaired in the hippocampus of patients with AD in early and late stages of pathology. (B) Increases in tris-buffered saline (TBS)–soluble Aβ and tau phosphorylation were present only in late stages of pathology. Aβ42 enzyme-linked immunosorbent assay (ELISA), phospho-tau immunoblot normalized by total tau. Postmortem human hippocampal tissue (patients with AD versus age-matched unaffected controls; patient information in fig. S1). Patients divided into controls (Braak 0 to 1, N = 11), early-stage AD (Braak 2 to 4, N = 12), and late-stage AD (Braak 5 to 6, N = 12). (C) Proteasome (chymotrypsin-like) activity is impaired in AD models: B103 cells (±1 μM Aβ42, N = 3), 20-day-old Elav-GS-GAL4>UAS-Aβ3-42 flies (±200 μM RU-486, N = 12), and 7-month-old mice [hAPP(J20), N = 7 versus NTG (Non-Transgenic), N = 5). (D) Proteasome overexpression increased proteasome activity in control flies and prevented proteasome activity deficits in an AD fly model; chymotrypsin-like proteasome activity assay ± 0.5 μM ATP (basal/ATP stimulated). Flies maintained on SY10 medium with 400 μM RU-486 until day 30, N = 8 per group. Significance based on two-way analysis of variance (ANOVA). (E) Proteasome overexpression partially overcame deficits in olfactory aversion training in 30-day-old flies; control, N = 121; AD, N = 129; and AD with (w/) Prosβ5 O.E. (overexpression), N = 125; data combined from triplicate assays. Significance was based on chi-square test. Values shown as percentage of flies in each chamber at the end of the assay. (F) Proteasome overexpression partially restored deficits in circadian rhythmicity in 30-day-old flies, N = 150 per group. *P < 0.05, **P < 0.01, and ***P < 0.001. Student’s t test was used unless otherwise stated. N represents the number of animals or samples per group. NS, not significant.
UAS-APP;UAS-BACE1 Drosophila model of AD
To investigate the biological relevance of proteasome impairment in AD, we initially used a Drosophila model of the disease. Here, we used an already developed model with postdevelopmental pan-neuronal expression of hAPP and BACE1 (Elav-gs-GAL4>UAS-hAPP;UAS-hBACE1). The UAS-hAPP;UAS-hBACE1 transgene has been previously reported to produce a cleaved product of APP that is further cleaved by Drosophila γ-secretase to produce a peptide similar in size to Aβ40/42 (21). These flies have been shown to develop Aβ deposits (22), form plaques (21), and exhibit age-dependent deficits in courtship behavior and memory (23). To better model AD pathology, we limited transgene expression to adulthood (starting day 2 after eclosion). This is significant, since, although age is the single greatest risk factor for the development of sporadic AD, most models use lifelong overexpression and show pathology early in life. We demonstrated these flies to have Aβ accumulation in the mushroom body (learning and memory center) (fig. S3, A and B) (24). They also displayed cognitive deficits on olfaction aversion training. Flies were exposed to two neutral odors (3-octanol and 4-methylcyclohexanol) and trained to associate one with a negative sensation (mild electric shock). Odor choice was then evaluated by T maze (25). These flies also displayed a shorter life span (fig. S3C).
Proteasome augmentation prevents survival and cognitive deficits in a Drosophila model of AD
We enhanced proteasome function in the fly AD model through overexpression of Prosβ5 (Proteasome Subunit Beta 5), a major catalytic subunit bearing a catalytic center for chymotrypsin-like activity in the proteasome core (Fig. 1D) (26). We and others have previously reported that proteasome function can be enhanced by overexpression of Prosβ5, which increases the transcription of other core subunits and promotes whole-proteasome assembly. Although the mechanism whereby Prosβ5 up-regulates the other core subunits is unclear, it has been reported in mammalian cell cultures (27, 28), Caenorhabditis elegans (29), and Drosophila (30, 31).
Our fly AD model showed impaired adenosine 5′-triphosphate (ATP)–nonstimulated (“basal”) and ATP-stimulated proteasome chymotrypsin-like activities, which roughly approximate the activities of mostly 20S and 26S assemblies, respectively. The 20S and 26S proteasomes contribute to biologically distinct functions in the cell [reviewed in (32)]. Furthermore, overexpression of Prosβ5 in this AD model can stimulate basal and ATP-stimulated proteasome activities, overcoming AD-related impairments (Fig. 1D). Prevention of proteasome dysfunction abrogated AD-like deficits in associative learning and memory on olfaction aversion training. Control flies displayed a significant increase in avoidance of the “negative” odor after training, unlike AD model flies. However, when Prosβ5 was overexpressed in the AD model background, posttraining improvement returned, demonstrating the retention of associative learning and memory (Fig. 1E). Notably, Prosβ5 overexpression did not in itself alter odor preference or shock avoidance (fig. S4).
We have previously reported that neuronal proteasome overexpression extends life span in Drosophila (31), a finding that we recapitulated here. We showed the AD fly model to have reduced life span, but when proteasome overexpression was induced in the AD fly model, median life span was returned to a comparable life span of nondisease controls (Fig. 5). However, because neuronal proteasome overexpression itself increases life span, restoration of a wild-type life span may stem not from the prevention of AD-like deficits but from the benefits of proteasome overexpression independent of rescue from AD pathology. Prosβ5 overexpression did not appear to alter mRNA levels of hAPP. It is thus unlikely that the observed improvements were an artifact from GAL4 competition (fig. S3D). In addition, this AD model experiences deficits in both spontaneous activity and circadian rhythmicity, but enhanced proteasome function reduced these deficits in an AD background (Fig. 1F). We have previously reported that proteasome overexpression prevents circadian rhythmicity deficits in old wild-type animals but does not change circadian rhythmicity in young wild-type animals (31). Collectively, these data suggest that proteasome overexpression can reduce AD-like deficits in a fly model of AD.
Fig. 5. TAT1-8,9TOD and TAT1-DEN reduce cell death, cognitive deficits, and proteostatic dysfunction in a fly model of AD.
(A) Proteasome peptidase (chymotrypsin-like) activity in W1118 flies fed with 1 μM TAT1-8,9TOD, TAT1-DEN, or vehicle for 24 hours, N = 12, ±proteasome inhibitor bortezomib. (B) Immunoblot against total polyubiquitinated proteins and K48-linked polyubiquitinated proteins of flies fed with TAT1-8,9TOD, TAT1-DEN, or vehicle for 24 hours, N = 7 to 8. (C) Schematic for (D) and (E). (D) Olfaction aversion training in W1118 or Elav-GS-GAL4>Elav-GS-GAL4>UAS-hAPP;UAS-hBACE1 flies treated for 10 days with 10 μM TAT1-8,9TOD, 10 μM TAT1-DEN, or vehicle mixed directly into food; control, N = 79; control with TAT1-8,9TOD, N = 80; control with TAT1-DEN, N = 69; AD, N = 60; AD with TAT1-8,9TOD, N = 68; AD model with TAT1-DEN, N = 69. Values shown as percentage of flies in each chamber at the end of the assay. (E) Life-span assay of Elav-GS-GAL4>UAS-hAPP;UAS-hBACE1 flies treated with 0.25 μM TAT1-8,9TOD, 0.25 μM TAT1-DEN, or vehicle mixed directly into food; AD, N = 189; AD with TAT1-8,9TOD, N = 195; AD model with TAT1-DEN, N = 195; Prosβ5 O.E., N = 194; AD with Prosβ5 O.E., N = 242. *P < 0.05, **P < 0.01, and ***P < 0.001. Significance was based on one- or two-way ANOVA in (A), (B), and (E). Chi-square test was used in (C). Log-rank test was used in (D).
Development of NSE-PSMB5, a mouse model of neuronal proteasome overexpression
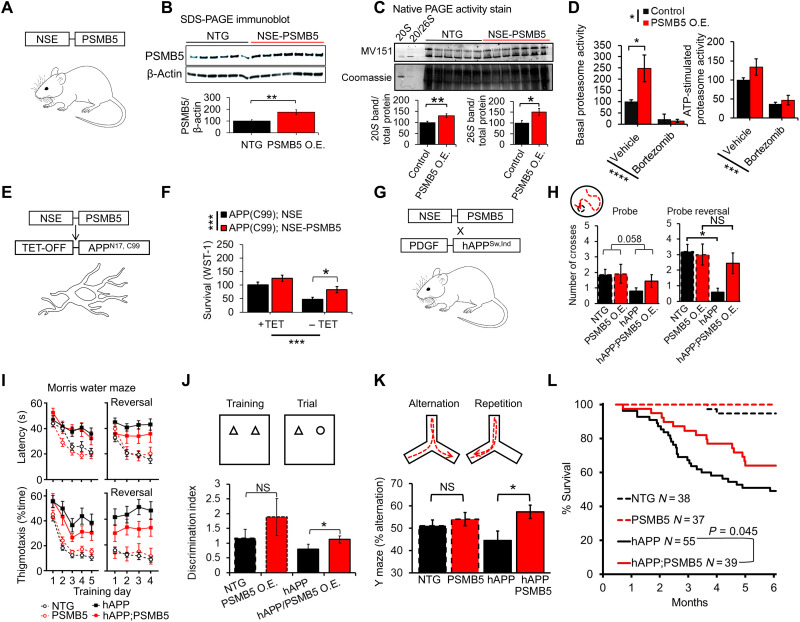
To investigate the translational potential of our findings, we generated a transgenic mouse model with neuronal-specific proteasome overexpression by inserting an additional copy of mouse PSMB5 (Prosβ5 ortholog) preceded by the neuron-specific enolase promoter (NSE-PSMB5) (Fig. 2A) (33). The resultant mice appear physiologically normal. Brains from 3-month-old mice showed a 70% increase in PSMB5 protein levels (Fig. 2B) and elevated expression of multiple proteasome subunits (fig. S5), consistent with findings from other model systems (27, 29–31). These changes resulted in increased content of active 20S and 26S proteasomes, demonstrated by native polyacrylamide gel electrophoresis (PAGE) analysis of mouse tissue incubated with the MV151 universal fluorescent proteasome inhibitor, which selectively binds and labels active proteasomes (Fig. 2C) (34). In addition, using plate-based activity measures, we show a twofold increase in basal peptidase proteasome activity, while ATP-stimulated proteasome activity (mostly the 26S proteasome) showed a nonsignificant increase (Fig. 2D). Furthermore, increased proteasome activity was blocked by adding the proteasome-specific inhibitor bortezomib, suggesting that these changes are not due to other proteases.
Fig. 2. PSMB5 overexpression increases proteasome function in mice and delays AD-like cognitive deficits and mortality.
(A) NSE-PSMB5 transgenic mouse model. (B) Immunoblot of PSMB5 normalized to β-actin, brains dissected from 3-month-old mice. NTG, N = 6; PSMB5, N = 7. (C) Native PAGE gel of brains dissected from 3-month-old mice; proteasome content and assembly assessed by incubation with the MV151 fluorescent proteasome probe. This is a broad proteasome inhibitor binding to active centers and producing fluorescence. Values adjusted to total protein based on Coomassie staining. Purified 20S and 26S proteasome used to identify bands. NTG, N = 6; PSMB5, N = 7. (D) Relative proteasome peptidase (chymotrypsin-like) activity, brains dissected from 3-month-old mice (N = 5 to 6). Replicate samples were incubated with the proteasome inhibitor bortezomib. (E and F) NSE-PSMB5 reduces AD-like cell death (WST-1 assay) in MC65 AD cell culture model transfected with NSE-PSMB5, 4 days after tetracycline (TET) withdrawal, N = 24 samples per group. (G) NSE-PSMB5 mice crossed with hAPP(J20) AD model. (H to K) NSE-PSMB5 reduces AD-like learning and memory deficits in Morris water maze, Y maze, and novel object recognition assay. All animals were 7- to 8-month-old littermates; NTG, N = 15; PSMB5, N = 11; hAPP, N = 14; hAPP;PSMB5, N = 9. (L) NSE-PSMB5 protects against early mortality in hAPP(J20) mice; NTG, N = 38; PSMB5, N = 37; hAPP, N = 55; hAPP;PSMB5, N = 39. Animals are littermates. Significance was based on Student’s t test in (B) and (C). Significance was based on two-way ANOVA, followed by Tukey or Bonferroni post hoc test in (D), (F), (H), (J), and (K). Significance was based on log-rank test; *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 were used unless otherwise stated. N represents the number of animals or samples per group.
These findings are consistent with our prior reports in flies that Prosβ5 overexpression leads to a significant elevation of basal proteasome activity (31). Thus, we conclude that overexpression of PSMB5/Prosβ5 leads to elevated 20S proteasome function, while the impact on 26S proteasome function remains unclear. Because of the close interplay between the proteasome and autophagy systems, we measured LC3 ratio as a marker of autophagic flux in brain tissues from our mouse model. We observed no significant changes, suggesting that autophagic flux is unaltered (fig. S6).
Proteasome augmentation prevents survival and cognitive deficits in a mouse model of AD
To test the AD-protective capacity of PSMB5 overexpression, we transiently transfected the NSE-PSMB5 construct into MC65 cells, which have a tetracycline (TET)–OFF APPN17-C99 overexpression construct. In this cell line, overexpression of a fragment of APP induced by TET withdrawal leads to cell death (35). PSMB5 overexpression significantly reduced cell death induced by APPN17-C99 fragment overexpression compared to empty vector transfection after 4 days, suggesting a protective effect against APP toxicity (Fig. 2, E and F).
To establish whether proteasome overexpression could protect against AD-like deficits in mice, we crossed our NSE-PSMB5 model with hAPP(J20) mice (36), which express the Swedish-Indiana familial-mutant hAPP and recapitulate key aspects of AD pathogenesis (Fig. 2G) (37–40). By 7 to 8 months of age, hAPP(J20) mice had profound spatial memory deficits in retention of a novel location for the escape platform during reversal testing on the Morris water maze. These deficits were significantly attenuated in hAPP(J20); NSE-PSMB5 mice (Fig. 2H). Latency to find the platform, percent time spent in thigmotactic swim (Fig. 2I), or floating time during reversal training did not differ between hAPP(J20) and hAPP(J20);NSE-PSMB5 mice. Thus, it is unlikely that changes in anxiety and/or motivation contributed to this improvement (Fig. 2H). We further evaluated spatial memory through novel object recognition and working memory (Y-maze test). In both, performance was significantly improved in hAPP(J20);NSE-PSMB5 mice relative to hAPP(J20) mice (Fig. 2, J and K). Although we detected a mild and not statistically significant improvement with PSMB5 overexpression in control mice, differences were significant between hAPP and hAPP;PSMB5 mice (Fig. 2, J and K). Still, we cannot completely differentiate between the general cognitive benefits of enhanced proteasome function and specific protection against AD-related deficits (Fig. 2, J and K). Total arm entries were unaltered between groups (fig. S7). In addition to improved learning and memory, hAPP(J20);NSE-PSMB5 mice had a 78% increase in survival time at the 75th percentile versus hAPP(J20) littermates (Fig. 2L).
Proteasome augmentation increases the turnover of APP in flies, cultured cells, and mice
Previous reports have shown that APP, BACE1, and γ-secretase–activating protein (GSAP) are all degraded by the proteasome and that treatment of cells with proteasome inhibitors increases levels of BACE1 and GSAP (41–43). Therefore, the protective effects of proteasome augmentation may stem from the degradation of the Aβ precursor machinery and substrate, leading to decreased Aβ production. In support of this concept, enhancing proteasome function in flies that express hAPP and hBACE1 resulted in significantly lower levels of hAPP protein (Fig. 3, A and B). Transient overexpression of PSMB5 in mammalian SK-N-SH neuroblastoma cells reduced APP levels and produced a lower–molecular weight band, which may represent an APP degradation product (Fig. 3, C and D). We observed less APPGFP fluorescence in cells when cotransfected with NSE-PSMB5 and APPGFP compared to cells transfected with APPGFP and the NSE empty vector, further supporting the enhanced turnover of the amyloid precursor machinery (Fig. 3E).
Fig. 3. PSMB5 overexpression increases degradation of APP machinery.
(A and B) Immunoblot of 70-day-old Elav-GS-GAL4>UAS-hAPP;UAS-hBACE1 and Elav-GS-GAL4>UAS-hAPP;UAS-hBACE1;UAS-Prosβ5 flies fed with 400 μM RU-486. AD model, N = 12; AD + Prosβ5, N = 12. Values normalized by β-actin. (C and D) Immunoblot of APP levels in SK-N-SH cells transfected with either NSE vector (control) or NSE-PSMB5, N = 3. (E) Images of SK-N-SH cells transfected with APPGFP and either the NSE vector or NSE-PSMB5, N = 6. (F) NSE-PSMB5 mice crossed with hAPP(J20) AD model. (G) Anti-APP immunoblots in 7- to 8-month-old hAPP(J20) ± NSE-PSMB5 mouse brains. APP, N = 9; APP;PSMB5, N = 9. Values normalized by β-actin with quantification (left) and Aβ42 ELISA (right); APP, N = 9; APP;PSMB5, N = 9. (H) X34 staining on brain coronal sections from hAPP(J20) ± NSE-PSMB5 mice, N = 3 per genotype. Number of plaques per hemibrain were quantified using ImageJ software. (I) Postmortem human hippocampal tissue (patients with AD versus age-matched unaffected controls). (J) Postmortem human hippocampal tissue sample lysate, proteasome activity plotted against soluble Aβ ELISA or anti-APP immunoblot (N = 35 in total, 11 controls and 24 patients with AD). (K to O) Postmortem human hippocampal tissue sample lysate, proteasome activity plotted against phosphorylated tau/total tau ratio (L), BACE1 (M), K48-polyubiquitinated (K48-UB) proteins (N), and total polyubiquitinated (poly-UB) proteins (O). N = 23. *P < 0.05 and ***P < 0.001. Significance is based on Student’s t test in (B), (E), (G), and (H). Significance was based on linear regression in (J) to (O). N represents the number of animals, samples, or patients per group.
In our mouse model, we detected significantly lower APP protein levels and an associated decline in Aβ in the brain (Fig. 3, F and G). Furthermore, hAPP;PSMB5 mice had significantly fewer brain plaques detected by X34 staining compared to hAPP mice (Fig. 3H). However, because hAPP(J20) mice artificially overexpress a mutated human variant of APP, we wanted to test the capacity for proteasome overexpression in a less artificial environment. To do this, we examined native APP abundance in brain tissue from our proteasome-overexpressing mice. This model also had depleted APP protein levels, supporting the idea that proteasome overexpression can increase APP turnover (fig. S8). We observed a nonsignificant trend toward reduced BACE1 levels with enhanced proteasome function (fig. S9). These findings suggest that proteasome-related protective effects stem at least in part from enhanced turnover/clearance of the Aβ precursor substrate.
Proteasome activity is correlated with APP and Aβ protein levels in human hippocampal tissues
To evaluate whether changes in proteasome function are biologically relevant to AD progression in humans, we evaluated hippocampal tissues from age-matched patients with AD and unaffected controls (patient details in fig. S1). Proteasome function and protein levels of APP were negatively correlated (Fig. 3, I and J), as were proteasome function and soluble Aβ42 levels (Fig. 3J). While we cannot establish causation in this cross-sectional study, these results agree with our animal model studies to suggest an interplay between proteasome function and levels of APP and Aβ.
Although this study primarily focused on the impact of proteasome on APP and amyloidosis, we also evaluated levels of tau phosphorylation in patient tissues. Here, proteasome activity and tau phosphorylation were negatively correlated (Fig. 3, K and L). This work agrees with a prior pilot study of seven patients that also showed phosphorylated tau (tau phosphorylation) to be higher in patients with lower proteasome activity (3). However, further investigation of this finding is beyond the scope of the present study. We also noted a negative trend between BACE1 protein levels and proteasome activity, but, similar to rodent studies, this trend failed to reach statistical significance (Fig. 3, K and M).
To further investigate the role of the proteasome in AD, we examined the prevalence of K48-linked (proteasome-targeted) polyubiquitinated proteins, which we found to be negatively correlated with proteasome activity in patient samples (Fig. 3, K and N). In contrast, associations between proteasome activity and total polyubiquitinated proteins did not reach statistical significance (Fig. 3, K and O). Our findings suggest that impaired proteasome function leads to broad proteostatic dysfunction and accumulation of polyubiquitinated proteins.
Development of novel proteasome agonists, TAT1-8,9TOD and TAT1-Dendrite
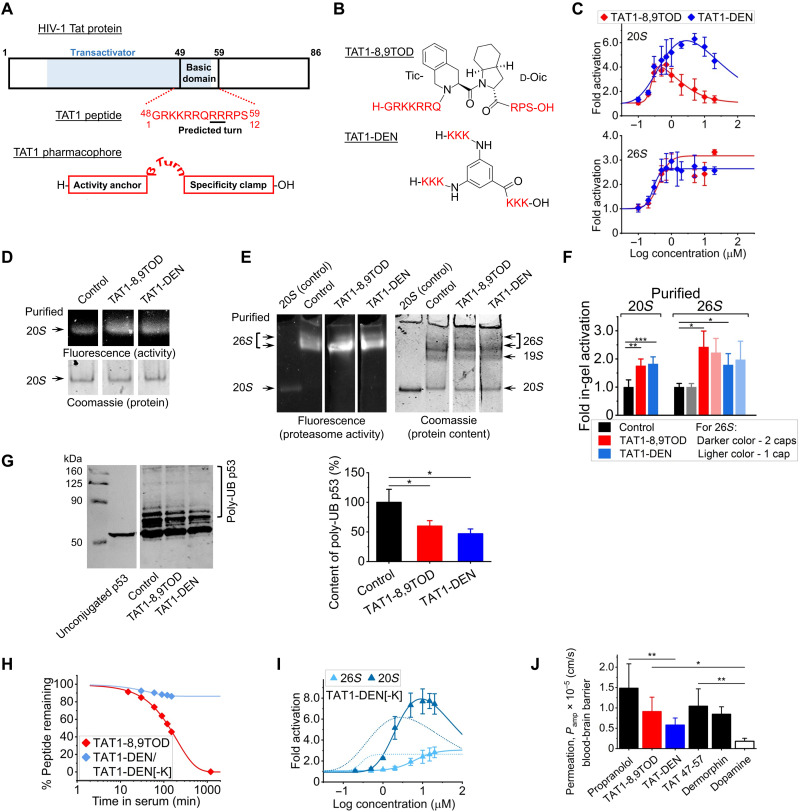
To determine whether pharmacologic manipulations that enhance proteasome function could also ameliorate AD-like progression, we developed a set of peptidomimetics that boost proteasome peptidolytic capacity. The rational design, optimization, and selected in vitro studies are described in detail in (44). Briefly, the design is based on the HIV-1 Transcriptional Activator TAR (Tat) protein (45), which contains a proteasome-binding RTP motif (REG/proteasome binding) shared with the 11S/REG/PA28 proteasome activator (Fig. 4A) (46). The full-length Tat protein prevents binding of the 11S regulator to the proteasome (46). Since proteasome activation by 11S plays an important role in the cellular immune response, the Tat protein competes with 11S as part of its anti-immune strategy (46). Manipulations of the Tat protein sequence led to certain products that could activate the proteasome core (44, 47). The goal of our design of Tat-derived peptides was to potentiate proteasome activation properties while obliterating function as a transcriptional transactivator. This was accomplished by generation of the TAT1 peptide, a fragment with the sequence 48GRKKRRQRRRPS59 (Fig. 4A) (44, 45, 47, 48). Very similar peptides derived from the HIV-1 Tat protein (Tat49–57, Tat47–57, or Tat48–60) have high capacity to cross the blood-brain barrier due to their structure and robust positive charge (49–52). These results suggested TAT1 as an attractive lead for the design of proteasome agonists with excellent absorption ability.
Fig. 4. TAT1-8,9TOD and TAT1-DEN enhance proteasome activity.
(A) HIV-1 Tat and TAT1 pharmacophore. (B) TAT1-8,9TOD and TAT1-DEN. (C) In vitro assays: chymotrypsin-like peptidase activity of purified 20S (top) and 26S (bottom) proteasomes treated with TAT1 compounds. (D and E) Representative native PAGE overlaid with Suc-LLVY-AMC: purified 20S (D) and 26S (E) substrate and vehicle (control) or 1 μM TAT1 compounds. Loading quantified by Coomassie. Arrows indicate proteasome assemblies. (E) From the top: 26S with two 19S caps, 26S single-capped, free 19S (no proteolytic activity), and free 20S. (F) Fluorescence signals corresponding to distinct proteasome assemblies quantified as in (D) and (E). (G) TAT1-8,9TOD and TAT1-DEN enhance degradation of polyubiquitinated p53 by purified 26S proteasome. p53 was in vitro polyubiquitinated with the MDM2 Ubiquitin Ligase Kit and incubated with 26S proteasome in the presence of vehicle or 1 μM TAT1-8,9TOD or TAT1-DEN. Free and polyubiquitinated p53 were separated by SDS-PAGE, detected by Western blotting with specific anti-p53 antibodies (left) (fragments of blots in a representative experiment), and their content was quantified from blot images (right); N = 5. (H) Serum stability of TAT1-8,9TOD and TAT-DEN. The latter compound was rapidly depleted in serum; however, a metabolite with two Lys residues removed from N termini, TAT1-DEN[-2K] was stable for >24 hours The blue trace represents a mixture of TAT1-DEN, [-K], and [-2K]. N = 5; SD covered by symbols. (I) The TAT1-DEN[-2K] metabolite of TAT1-DEN retained the power to activate both 20S and 26S proteasomes in a standard in vitro assay. (J) PAMPAs evaluating the blood-brain barrier penetrance capacity of TAT1-8,9TOD and TAT1-DEN (N = 5). Propranolol and dermorphine served as positive controls, in addition to Tat47–57 with proven blood-brain barrier permeability (49). Dopamine was included as a negative control (55). *P < 0.05, **P < 0.01, and ***P < 0.001. Student’s t test was used unless otherwise stated. N represents the number of samples per group or independent experiments.
Subsequent studies led us to propose a proteasome-targeting pharmacophore consisting of a strongly positively charged peptide moiety (“activity anchor”) connected by a β-type turn inducer to a short “specificity clamp” (44). We introduced the synthetic turn stabilizer Tic-d-Oic (TOD; Tic: l-1,2,3,4-tetrahydroisoquinoline-3-carboxylic acid; Oic: d-octahydroindole-2-carboxylic acid) (53) to the 8,9 position of the TAT1 structure to produce TAT1-8,9TOD (Fig. 4, A and B), which had superb proteasome activation potency (44). We then shortened the basic sequence to triple lysines, which we then multiplied into three fragments joined by 3,5-diaminobenzoic acid to produce TAT1-DEN (Fig. 4B) (44). The name Dendrite (abbreviated as DEN) refers to the branched structure of the compound and the turn-supporting diaminobenzoic acid. We demonstrated before that in vitro, both compounds strongly activated the chymotrypsin-like peptidase of the purified human core 20S. Under optimized assay conditions, TAT1-8,9TOD and TAT1-DEN maximally activated 20S almost 8- and 10-fold, respectively. The AC50 (concentration when activation reached 50% of the maximum) for these peptides ranged from 200 to 400 nM (44). We confirmed the 20S-activating power of the compounds in an independent set of experiments presented in Fig. 4C. TAT1-8,9TOD and TAT1-DEN also activated 26S proteasome assembly in vitro, with maximal fold of 3.2 and 2.6 and AC50 of 424 and 281 nM, respectively (Fig. 4C). Capacity to in vitro activate both proteasome forms was further confirmed using in-gel activity assays for proteasomes separated by native PAGE (Fig. 4, D to F, and fig. S10). Both single-capped and double-capped assemblies (20S core with one or two 19S regulatory particles) were activated (Fig. 4, E and F, and fig. S10). Incubation of a purified 26S proteasome with the peptidomimetics before electrophoretic separation did not appear to alter the stability of the 26S holoenzyme (fig. S11). We also demonstrated the enhanced ability of purified 26S proteasome to degrade p53, a model polyubiquitinated substrate, suggesting that TAT enhanced 26S functionality (Fig. 4G).
Before introducing the compounds to in vivo models, we performed two additional in vitro tests: stability in human serum and membrane permeability. The roughly 2-hour (136-min) half-life of TAT1-8,9TOD is relatively short (Fig. 4H); however, such half-lives are not uncommon among established drugs, i.e., acetaminophen (54). Moreover, stable metabolites retaining the basic activity anchor may still preserve biological activity, a notion open for further exploration. In turn, TAT1-DEN was found to rapidly lose a single Lys in serum then a second Lys from its N termini; however, the TAT1-DEN[-2K] metabolite was remarkably stable with a half-life of more than 24 hours (Fig. 4H). We found that TAT1-DEN[-2K] still activated both 20S and 26S proteasomes in vitro with comparable maximal activation folds but increased AC50 (Fig. 4I). To test the penetrance of these compounds across the blood-brain barrier, we performed parallel artificial membrane permeation assays (PAMPAs; Fig. 4J) with propranolol, Tat47–57, and dermorphin as positive controls, known for their ability to penetrate the blood-brain barrier (49). Dopamine was included as a negative control as it is considered a compound unable to efficiently cross the blood-brain barrier (55). Both TAT1-8,9TOD and TAT1-DEN had good ability to penetrate the artificial membrane similar to Tat47–57 and dermorphin, strongly suggesting an ability to cross the blood-brain barrier (Fig. 4J).
Proteasome agonists can reduce deficits in survival and cognitive functions in fly and cell culture AD models
After encouraging in vitro results indicating strong activation of the proteasome, we aimed to establish the efficacy of our compounds in AD models. We first demonstrated that flies fed with the compounds in their food showed increased proteasome (chymotrypsin-like) activity, which was prevented by the addition of the proteasome inhibitor bortezomib, indicating that changes were indeed proteasome driven (Fig. 5A). We also observed a significant decline in K48-linked polyubiquitinated proteins and a slight and nonsignificant decline in total polyubiquitinated proteins in flies fed with the compounds, suggesting improved proteostasis (Fig. 5B).
To test the capacity of the peptidomimetics to rescue AD-like deficits, we fed our fly AD model with the compounds for 7 days (Fig. 5C). Treated flies had reduced AD-like deficits in associative learning measured through olfaction aversion training (Fig. 5D). No significant changes in odor preference or learned response were observed in control flies fed with these compounds, and the drugs did not alter innate odor preference or shock reactivity (fig. S12). In addition, both compounds prevented AD-like mortality, with comparable survival curves to control (non-AD) flies (Fig. 5E).
Proteasome agonists can enter the mouse brain
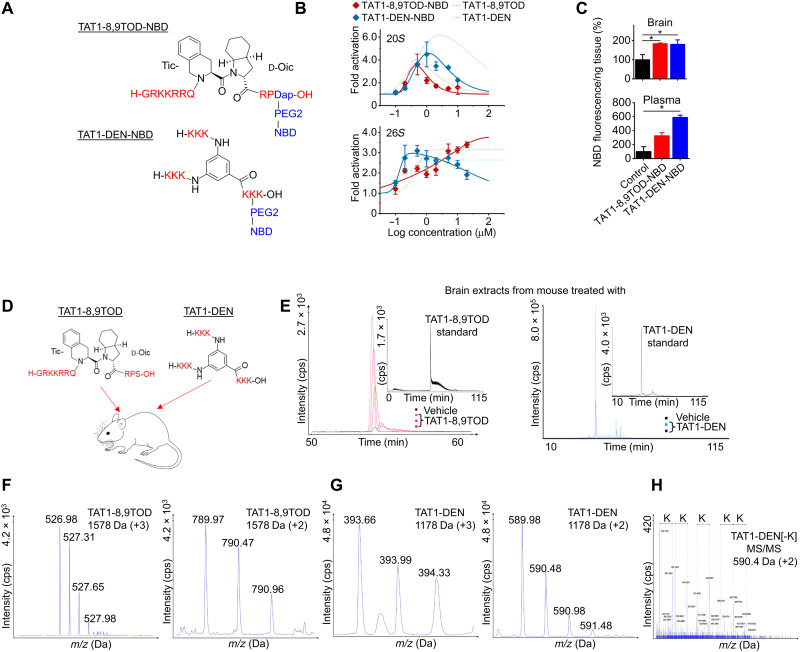
To follow on the encouraging PAMPA results (Fig. 4J) and to demonstrate the blood-brain barrier crossing ability in vivo, we attached a small (165-Da) nitrobenzoxadiazole (NBD) fluorescent label to the C terminus of TAT-8,9TOD (serine residue substituted with diaminopropionic acid) or the penultimate C-terminal Lys of TAT1-DEN (Fig. 6A). Addition of the label did not abolish the activation of 20S and 26S proteasomes in vitro (Fig. 6B), making the compounds suitable for relevant in vivo tests. Intraperitoneal injections of TAT1-8,9TOD-NBD or TAT1-DEN-NBD to mice produced detectable NBD fluorescence not only in the plasma (N = 3 mice) but also in the brain (N = 3) 1 hour later, as compared with control (N = 3) (Fig. 6C). This highly encouraging result was further validated by mass spectrometry (MS; Fig. 6, D to H). We performed a series of MS analyses of the peptidic fraction isolated from the brain tissue of six independent mice, euthanized 1 hour after intraperitoneal injection with either TAT1-8,9TOD or TAT1-DEN (Fig. 6, D to H). Our results revealed distinct and reproducible chromatographic profiles in brain extracts obtained from mice injected with TAT1-8,9TOD or TAT1-DEN when compared to vehicle-treated controls (Fig. 6E). The retention times of the signals in the extracted ion chromatograms of TAT1 compounds in brain samples were consistent with those observed for the reference samples (Fig. 6E, inserts). The molecular weight of putative TAT1-8,9TOD determined on the basis of +3 and +2 ions (Fig. 6F) was 1577.9427 and 1577.9356, respectively, which corresponds to the calculated monoisotopic mass of TAT1-8,9TOD standard (1577.9379).
Fig. 6. TAT1-8,9TOD and TAT1-DEN enter the mouse brain.
(A) The TAT compounds tagged with fluorescent label NBD (excitation/emission, 460/590) via a PEG2 linker. (B) NBD label did not abolish the potency of TAT1 compounds to activate 20S and 26S proteasomes in vitro. (C) NBD fluorescence detectable in plasma and brain lysates from mice injected intraperitoneally with labeled TAT1 compounds euthanized 1 hour after TAT1-8,9TOD (12.6 mg/kg) or TAT1-DEN (9.4 mg/kg; or vehicle) intraperitoneal injection. N = 3. (D) Mice received an intraperitoneal injection of TAT1-8,9TOD (12.6 mg/kg) or TAT1-DEN (9.4 mg/kg; or vehicle) and were euthanized after 1 hour. (E) Extracted ion chromatograms for the brain supernatants of TAT1-8,9TOD (left; m/z 526.90 to 526.99) and TAT1-DEN (right; m/z 590.40 to 590.50). Inserts represent standards for extracted ion chromatograms for TAT1-8,9TOD standard (1 pmol; diluted with 9 μl of the brain supernatant of vehicle-treated mouse) and TAT1-DEN standard. The retention time for TAT1-8,9TOD samples was 54.0 ± 0.5 min. The m/z signal corresponds to +3 ion, which deconvolutes to 1577.9356 Da. The retention time for TAT1-DEN–digested samples was 32.0 ± 0.5 min. (F) Time-of-flight (TOF) MS spectrum of +3 (left) and +2 (right) charged ions of TAT1-8,9TOD before deconvolution corresponding to experimental molecular mass of 1578 Da. (G) TOF MS spectrum of +3 (left) and +2 (right) charged ions of TAT1-DEN[-K] deconvolute to 1177.9474 and 1177.9492 Da, respectively (calculated mass of TAT1-DEN[-K], 1177.8261 Da). The corresponding masses are the same for the control sample (trypsin-digested TAT1-DEN standard) and digested brain extracts. (H) MS/MS spectra of m/z 590.40 ± 0.6 Da recorded for 32 min at room temperature (RT), which corresponds to TAT1-DEN[-K] double-charged ion. Dotted lines indicate signals differing by 129 Da (lysine residue). About 244 peaks of >24 cps (counts per second) were detected, among which 126 were identified (52.5%).
Because of extreme polarity and poor detectability of TAT1-DEN by liquid chromatography (LC)–MS, we performed digestion with trypsin to be able to confirm the presence of TAT1-DEN or its metabolites in brain tissue. The deconvolution of mass/charge ratio (m/z) signals of double- and triple-charged ions (Fig. 6G) enabled the detection of peaks representing molecular weights of 1177.9474 and 1177.9492 Da, corresponding to the calculated monoisotopic mass of TAT1-DEN devoid of one lysine residue (calculated molecular weight is 1177.8261 Da). Tandem MS (MS/MS) spectrum of +2 ions revealed a clear fragmentation pattern, confirming the presence of TAT1-DEN metabolite in the brain samples and thus its ability to cross the blood-brain barrier (Fig. 6H).
Proteasome agonists prevent cell death in APP fragment overexpression cell line
All subsequent investigations were focused on TAT1-8,9TOD because of its deeper characterization. We have reported previously that treatment with TAT1-8,9TOD enhances proteasome activity in human neuroblastoma SK-N-SH cells (50). Building on that work, we now treated TET-OFF APPN17,C99 overexpression (MC65) cells with 1 μM TAT1-8,9TOD; treatment reduced APPN17,C99-induced cell death versus cells treated with vehicle alone (Fig. 7A).
Fig. 7. TAT1-8,9TOD reduces cognitive deficits, proteostatic dysfunction, and abundance of Aβ machinery in hAPP(J20) mice.
(A) Treatment of MC65 cells with TAT1-8,9TOD reduces AD-related cell death, based on WST-1 viability assay, N = 24. (B) Proteasome activity in the brain of nontransgenic mice 24 hours after intraperitoneal injection with TAT1-8,9TOD, N = 5. (C) Treatment schematic; 6-month-old hAPP(J20) mice were treated every 2 days for 14 days with intraperitoneal injections of TAT1-8,9TOD (1.26 mg/kg) or vehicle (N = 5). (D) Novel object recognition assay in TAT1-8,9TOD–treated hAPP(J20) mice. (E) Representative immunoblot. (F and G) Immunoblots and ELISA of brain tissue from TAT1-8,9TOD–treated hAPP(J20) against (F) anti-APP, (G) anti-BACE1, (H) Aβ42 ELISA, (I) anti–K48-polyubiquitinated proteins, and (J) anti–K63-polyubiquitinated proteins. Values normalized by β-actin. *P < 0.05 and **P < 0.01. Significance was based on one- or two-way ANOVA in (A) and (B). Student’s t test was used in (D) to (J). N represents the number of animals or samples per group.
On the basis of our in vitro data and in vivo MS data that TAT1-8,9TOD is entering the brain, as well as our fly and cell culture data that the agonist can reduce AD-like deficits, we tested whether TAT1-8,9TOD could reduce AD-like deficits in a mouse model of the disease. First, to test functionality, we demonstrated that TAT1-8,9TOD enhanced proteasome activity in mouse brain 24 hours after treatment (Fig. 7B). We verified in a separate experiment that increased proteasome activity could be blocked by the proteasome inhibitor N-carbobenzyloxy-l-leucyl-l-leucyl-l-leucinal (MG132) (fig. S13A) and that activation occurred in other non-neuronal organs (fig. S13B).
Proteasome agonists can reduce AD-like deficits in hAPP(J20) mice
To test whether our agonists protect mice from AD-like deficits, 6-month-old hAPP(J20) mice received intraperitoneal injections of TAT1-8,9TOD on alternate days for 2 weeks (Fig. 7C). Treated mice showed reduced deficits in associative learning and memory (novel object recognition assay) versus vehicle-treated hAPP littermate controls (Fig. 7D). Furthermore, TAT1-8,9TOD produced a decline in APP and BACE1 protein levels (Fig. 7, E to G), likely from increased turnover by the proteasome, with associated decline in Aβ (Fig. 7H). We also observed lower levels of K48-linked polyubiquitinated proteins in treated mice with a negative trend in K63-linked polyubiquitinated proteins (Fig. 7, I and J), suggesting lessened proteostatic deficits. These results suggest that our compounds can attenuate AD-like deficits and could potentially be tested in new therapeutic treatments.
We additionally evaluated the impacts of peptidomimetics on autophagic flux. This was accomplished through investigation of LC3 ratio. We observed a slight increase in LC3-I and a slight decline in LC3-II. This perhaps suggests an increased autophagial flux in hAPP(J20) mice treated with TAT1-8,9TOD (fig. S14). Because of the interplay between proteasome and autophagy, this is not completely unexpected. This result indicates that some of the effects that we observe could occur through the interplay between the proteasome and autophagy pathways.
Our results demonstrate that proteasome dysfunction is a robust and early feature of AD pathology and that its prevention can alleviate AD-like symptoms in fly and mouse models of AD. Protective effects may be produced at least in part through increased turnover of Aβ precursor proteins in animal models, and proteasome function and abundance of these factors were negatively correlated in patient tissues. Last, our novel proteasome agonists can stably penetrate the blood-brain barrier and protect against AD-like deficits in fly and mouse models.
DISCUSSION
Impaired proteasome function profoundly affects neuronal proteostasis and is a robust feature of AD, as reported in cell culture and mouse models of AD (3–7, 56). Both Aβ, either internal or internalized, and aggregated tau may inhibit the proteasome’s activity (3, 12, 56). Impaired proteasome function has also been reported in the brains of patients with AD, in whom the most significant declines occurred in the hippocampus, superior and middle temporal gyri, and inferior parietal lobule (1, 3). Our results corroborate these findings and expand them. We demonstrate that stage-dependent impaired proteasome function in the hippocampus precedes increases in Aβ levels and tau phosphorylation, with a significant decline in proteasome activity arising early in the disease course. We and others have previously shown that impaired proteasome function in the brain is a robust feature of aging in various model organisms (31, 57). Accordingly, we conclude that age-related proteasome functional decline may add to AD risk factors. During AD development, proteasome function and proteostasis are further compromised in a positive feedback loop fashion with deleterious effects.
Numerous studies have demonstrated that proteasome inhibition replicates many aspects of AD-like neuropathologies. However, prior studies have not investigated whether prevention of AD-linked proteasome dysfunction may protect against AD-like deficits [reviewed in (58)]. Because of the size and complexity of the proteasome assemblies, manipulating their expression and activity in experimental models has posed a challenge. This study used genetic approaches and pharmacological intervention to augment proteasome functions, which we show to prevent cell death in cell culture models of AD, as well as reduce cognitive and survival deficits in Drosophila and mouse models of AD.
We achieved augmentation of proteasome functions by genetically increasing proteasome levels or enhancing catalytic activity with novel pharmacologic agents. For the former, we used overexpression of PSMB5/Prosβ5, the 20S proteasome core subunit bearing the catalytic center for peptidase activity. This approach was initially used by Chondrogianni et al. (27) and recapitulated by several groups, including our own (28–31). Numerous studies have suggested that levels of PSMB5/Prosβ5 are rate limiting and that increased proteasome function is driven by the concomitant up-regulation of other proteasome subunits and increased content of proteasome assemblies (27–29). Here, we show increased content of other proteasome subunits with PSMB5 overexpression. Proteasome activation may contribute to these observed effects, as suggested by a study that used PSMB5 overexpression (30, 31). Our quantification of the proteasome assemblies labeled with the specific fluorescent competitive inhibitor MV151 and separated by native PAGE reflects primarily the content of proteasomes. However, it may also be affected by the accessibility of active sites to the substrate-mimicking label inhibitor, thus, to some extent, reflecting assembly activity.
Proteasome inhibition by small molecules is an anticancer strategy used for almost 20 years (59). To the contrary, specific proteasome activation is still at a preclinical stage, with promising but nevertheless limited reports concerning mostly the 20S core (44, 60–64). Recent notable examples include fluspirilene analogs that activate the 20S proteasome in vitro and prevent its inhibition by Aβ oligomers (64). We developed a set of novel peptidomimetics that allosterically activate the proteasome in vitro (44). These compounds activate the core proteasome (44) and the 26S holoenzyme. Previously reported enhancement of 26S proteasome activity occurred via regulation of proteasome phosphorylation by small molecules (65), not directly via an activator, as we present here. Our TAT1-8,9TOD and TAT1-DEN compounds showed potent effects, were nontoxic, and could penetrate the blood-brain barrier. Furthermore, they ameliorated AD-like pathologies in model systems, with effects comparable to those achieved by genetic augmentation of the proteasome.
The proteasome has critical roles in many cellular functions, including the prevention of apoptosis, presynaptic vesicle transport, and synaptic plasticity (4–6, 66). Proteasome impairment results in altered cleavage and abundance of different parts of the amyloid precursor pathway (41–43). Thus, we focused here on proteasome function in relation to the turnover of APPs and downstream Aβ. Prevention of proteasome dysfunction in our models reduced the abundance of APP and, in turn, prevented the accumulation of Aβ. These findings appear robust and reproducible across varied APP overexpression models of the AD.
A critical question in future research will be whether augmentation of proteasome function can also improve outcomes in tau models of AD. As with Aβ, tau oligomers directly interact with and inhibit proteasome function (3, 56), and Aβ-induced inhibition of the proteasome appears to drive tau accumulation (67). In this study, higher proteasome activity was correlated with lower tau phosphorylation in patient tissues (fig. S5). Last, studies wherein tau was specifically targeted for increased degradation by the 26S proteasome have shown promising results (68, 69), further suggesting that up-regulating proteasome activity could be an effective treatment for AD. It is also both possible and plausible that some protective effects of proteasome overexpression stem from mechanisms independent of APP/tau turnover.
We have previously reported that preventing age-related proteasome dysfunction in the brains of flies blocked age-related declines in cognitive function (31). Thus, certain improvements that we observed in the present study may represent prevention of age-related declines. However, we observed prevention of cognitive deficits (evaluated by Morris water maze tests) in AD mouse models to a level comparable to that of nondisease control with proteasome overexpression, but we did not observe further improvements in non-AD mice. Likewise, although we did observe increases in novel object recognition with proteasome overexpression in mice from both AD and non-AD backgrounds, the effects only reached significance in AD mouse models. Thus, we conclude that AD-relevant improvements in cognitive functions occurred independently of aging-related benefits, although we cannot rule out some AD-independent functional improvements.
Artificial overexpression of APP, used to model AD-like deficits in both our fly and mouse systems, causes axonal transport defects, leads to accumulations of C-terminal fragments of APP, and induces neurotoxicity. To date, it is not clear whether these events are related to the pathogenesis of AD.
In conclusion, we show that proteasome dysfunction is a robust and early feature of AD in human tissues and animal models. Prevention of proteasome dysfunction in cell culture, fly, and mouse models of AD ameliorated cognitive deficits and survival. Protective effects that we observed stem, at least in part, from increased degradation of APP, resulting in reduced Aβ. Last, we developed a set of novel proteasome agonists that hold therapeutic potential for AD.
MATERIALS AND METHODS
Fly lines and strain maintenance
UAS-Prosβ5 (21794) (70), UAS-hAPP.Abeta3-42 (64217), and Elav-GS-GAL4;UAS-hAPP;UAS-hBACE1 (56756) stocks were obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537). All lines were maintained on agar-cornmeal-dextrose-yeast growth medium (71) in a humidified 24°C incubator with 12-hour light/dark cycles. All crosses were set up with female virgins of the respective GAL4 driver line and male UAS-Prosβ5 or W1118 flies. Progeny were collected within 48 hours of eclosion and allowed to mate on 10% sugar/yeast (SY10) medium (72) for another 48 hours. Females were then separated and sorted into sets of 25 flies per vial containing SY10 medium supplemented with either 400 μM mifepristone (RU-486) or ethanol vehicle, mixed directly into the food. Blue dye #1 (8 μM) was added to food containing RU-486 for the purpose of identification. Carbon dioxide was used to briefly anesthetize flies for sorting. Flies were moved to vials of fresh medium every 2 to 3 days.
Mice
hAPP(J20) (36, 73, 74) and NSE-PSMB5 mice were maintained by heterozygous crosses with C57BL/6J mice (the Jackson Laboratories, Bar Harbor, ME). Nontransgenic littermates were used as controls. Mice were housed in ventilated cage racks with up to five animals per cage under 12-hour light/dark cycles at 24°C. Mice were monitored daily by UT Health San Antonio Laboratory Animal Resources staff and were transferred to new cages weekly.
Mouse dissection
Mice were anesthetized through inhalation of isoflurane until breathing could no longer be observed. After animals were decapitated, the brain was then isolated, divided into hemispheres, and flash-frozen. Tissues were then powdered using a liquid nitrogen–chilled pestle and mortar. Following powdering, tissue powder was weighed then resuspended and lysed in an assay-dependent buffer.
Fly brain dissection and immunostaining
Flies were anesthetized through inhalation of CO2. Flies were then decapitated, and brains were isolated and fixed in 4% paraformaldehyde. Samples were washed twice with phosphate-buffered saline (PBS) and then incubated overnight at 4°C with an antibody to Aβ (15126, Cell Signaling Technology), washed thoroughly, and incubated with secondary anti-mouse antibodies. Brains were mounted using 4′,6-diamidino-2-phenylindole–containing mounting medium (VECTASHIELD).
Cell culture
Cells were cultured in EMEM (Eagle’s Minimum Essential Media) (SK-N-SH) or Dulbecco’s modified Eagle’s medium (MC65) supplemented with 10% heat-inactivated fetal bovine serum and antimicrobials [penicillin (100 U/ml), streptomycin (100 μg/ml), and amphotericin B (0.25 μg/ml); Gibco-Invitrogen). Incubators were maintained at 5% CO2 and 37°C. Medium was replaced every 3 to 4 days. For most experiments, cells were seeded at 100,000 cells/ml in either 6-well or 96-well plates 24 hours before assay. In most cases, medium was replaced with serum-free Opti-MEM medium 24 hours before assay.
Transfections and imaging
Cells were seeded at 75,000 cells per well in six-well plates. They were transfected the next day with 1.4 μg of NSE-PSMB5 vector or NSE empty vector control plus 1 μg of APPGFP vector per well using Lipofectamine LTX and Plus reagent (#15338030, Thermo Fisher Scientific) following the manufacturer’s instructions. Cells were passaged to 96-well plates the day after transfection for some experiments. Cells were imaged with the Incucyte system (Sartorius), and images were analyzed with the manufacturer’s software for green fluorescent protein (GFP) fluorescence intensity normalized by cell confluence.
Neuron-specific enolase PSMB5
A full-length mouse PSMB5 plasmid was used (MR203485, Origene). PSMB5 was excised to remove the Myc-DDK tag then cloned into the ShuttleNSE empty vector (50958, Addgene) adjacent to the NSE promoter. The NSE-PSMB5 region was excised and microinjected into (C57BL/6 X SJL) F2 mouse eggs at the University of Michigan Transgenic Animal Model Core. Mice were then bred into a C57BL/6J background for three generations. To control for the mixed backgrounds of mice, all experimental comparisons were made between littermates.
X34 staining
Mouse hemibrains were cryopreserved by covering them slowly with powdered dry ice and then stored at −80°C. Serial coronal brain sections (10 μm; approximate bregma, −1.82 mm) were cut using a cryostat, mounted on positively charged (Superfrost Plus) slides, and stained using X34, a fluorescent marker of β-sheet structures that intensely labels amyloid plaques and vascular amyloid. Briefly, slides were fixed using 4% paraformaldehyde for 30 min, washed with PBS, and stained with 10 μM X34 (pH 10) in 40% ethanol for 10 min, and then washed and differentiated with 0.2% NaOH in 80% ethanol for 2 min. Last, slides were washed again and mounted using VECTASHIELD mounting medium (Vector Laboratories) with propidium iodide. Images were taken using a Keyence BZX fluorescence microscope (20× Nikon objective). Amyloid plaques per hemibrain were quantified using ImageJ software and plotted using the GraphPad Prism program.
Quantitative polymerase chain reaction
mRNA was isolated using standard TRIzol methods, and cDNA was prepared using a high-capacity cDNA reverse transcriptase kit (Applied Biosystems). Quantitative polymerase chain reaction was carried out using SYBR Green and normalized to β-actin.
Peptide synthesis
Synthesis and purification were performed as described previously for TAT1-8,9TOD (53) and TAT1-DEN (44). Briefly, each synthesis was carried out on a solid support in a microwave-assisted mode using the Fmoc/tBu methodology. Incorporation of NBD fluorescent tag was performed by attaching it to the C-terminal (TAT1-8,9TOD) or penultimate (TAT1-TOD) residue of the agonist’s sequence. To provide orthogonal protection during the sequence assembling, Lys(ivDde) and Dap(ivDde) derivatives [diaminopropionic acid (Dap)] were used in TAT1-DEN and TAT1-8,9TOD, respectively, and the N-terminal amino acid residues were incorporated as the N-α-Boc derivatives. The ivDde moiety was selectively removed using a 5% solution of hydrazine in N,N′-dimethylformamide and microwave heating at 60°C for 10 min. The PEG2 spacer was attached to the deprotected side-chain amine group of Lys or Dap, followed by coupling of NBD tag. The compounds were cleaved from the solid support, purified by reversed-phase high-performance LC, and characterized with MS. Each compound was of >99% purity. For mice experiments, the peptides were solubilized in dimethyl sulfoxide (DMSO) and then diluted in filtered PBS. For NBD experiments, mice were injected with compounds by intraperitoneal injections then euthanized 1 hour later. Tissue was dissected as described in the “Mouse dissection” section then lysed in ice-cold PBS.
Patient samples
Flash-frozen hippocampal tissue was provided by the NIH NeuroBioank. Tissues were provided by multiple repositories; thus, some sites provided brain slices, while others provided powderized tissues. Samples were requested to have a postmortem interval of less than 24 hours, from patients between 50 and 80 years old. Samples were powderized if needed and the materials weighed. For proteasome activity assays, samples were resuspended and homogenized in proteasome activity buffer (see the Supplementary Materials for assay details). For immunoblot and enzyme-linked immunosorbent assays (ELISAs), samples were homogenized in tris-buffered saline (TBS) supplemented with protease inhibitors. The soluble fraction was then removed for assays. We observed no significant links between proteasome activity and postmortem interval (fig. S15).
In vitro proteasome activity assay
Purified human 20S (housekeeping) and 26S proteasomes were purchased from Boston Biochem Inc. (Cambridge, MA; 20S) and Enzo Life Sciences (Farmingdale, NY; 20S and 26S), with at least three distinct batches used for separate experiments. For purified protein studies, the chymotrypsin-like activity was tested with 50 μM Suc-LLVY-AMC model substrate, in a 96-well plate format, as previously described (47). The reaction buffer consisted of 50 mM tris-HCl (pH 8.0). For testing the activity of the 26S holoenzyme, the buffer was supplemented with “MAD”: 1 mM MgCl2, 2 mM ATP, and 1 mM DTT (dithiothreitol).
For studies of lysates, the reaction buffer used to test ATP-stimulated activity was 50 mM tris-HCl (pH 7.8) supplemented with 1 mM MgCl2, 0.5 mM ATP, and 1 mM DTT. The reaction buffer used to measure ATP-independent proteasome activity consisted of 50 mM tris-HCl (pH 8.0) supplemented with 1 mM MgCl2 and 1 mM DTT.
Purified proteasomes were preincubated with the substrate in reaction buffers for 10 min at room temperature (RT) before the TAT compounds or vehicle (DMSO; 1%, v/v) was added and the reaction carried for 1 hour at 37°C, with fluorescence readings at 1-min intervals (Fluoroskan Ascent plate reader). The reaction rates were calculated from a linear segment of kinetic curves. The rates and titration curve fittings were computed using Slope Analyzer and Enzyme Kinetics applications within Origin Pro 2019. The data presented are averages ± SD from three to five independent experiments.
For testing of activity in tissue lysates, animals were intraperitoneally injected (mice) or fed with TAT1 peptidomimetics mixed into their food stock (flies). Animals were euthanized, and tissues were flash-frozen and then powdered with liquid nitrogen–chilled pestle and mortar. Tissues were then lysed by mechanical lysis in reaction buffer. The resulting supernatant was used for further experiments. The volume of buffer used was based on tissue weight. Assays were initiated by the addition of 50 μM Suc-LLVY-AMC. For some assays, 1 μM proteasome inhibitor, MG132 or bortezomib, was added 5 min before incubation with Suc-LLVY-AMC. The reaction was carried out with readings at 5-min intervals for a period of 4 hours at 37°C.
Separation of proteasomes by nondenaturing PAGE for activity and stability assessment
Purified human 20S and 26S proteasomes (as above) were separated by tris-glycine PAGE, in 4% acrylamide gels (casted in-house) or 6% gels (Novex, Invitrogen). Two micrograms of 20S and 6 μg of 26S per line were loaded. For stability experiments, proteasomes before separation were preincubated with TAT1-8,9TOD, TAT1-DEN, or DMSO at RT for 10 min. Electrophoresis was run at 4°C for 3 hours at 70 V. The gels were then stained with colloidal Coomassie Brilliant Blue (stability experiments, after in-gel activity experiments) or subjected to in-gel activity assay. For the latter, the gels were overlayed with 50 mM tris-HCl (pH 8.0) containing 100 μM Suc-LLVY-AMC, MAD, and 1 μM TAT1-8,9TOD, Tat1-DEN, or vehicle. After 30 min of incubation at RT, the gels were photographed in ultraviolet (UV) light, and the intensity of fluorescence of the degradation product was quantitated in ImageJ. For proteasome stability assessment, the intensity of Coomassie-stained proteasome bands in photographed gels was quantitated in ImageJ. Quantitative results obtained with 4% (images in Fig. 4) and 6% gels (images in fig. S10) were undistinguishable and were processed together.
For tissue experiments, samples were individually homogenized by pestle in 100 μl of chilled proteasome buffer [50 mM tris, 5 mM MgCl2, 1 mM DTT, and 0.5 mM ATP (pH 7.4)]. Samples were vortexed and then centrifuged at 21,000g at 4°C for 15 min. Supernatant was incubated with 1 μM proteasome probe MV151 for 1 hour at 4°C and then run on 10% tris-glycine nondenaturing polyacrylamide gels (Bio-Rad) in Native Gel Buffer (diluted from 20×; Life Technologies) supplemented with 5 mM MgCl2, 1 mM DTT, and 0.5 mM ATP to maintain proteasome assembly. Gels boxes were surrounded by ice and run in a 4°C refrigerator at 100 V for 1 hour, followed by an additional 3 hours at 250 V. MV151 labeling was then measured by fluorescence under UV and ImageQuant 4000 (GE Healthcare), using 312-nm excitation and measuring emissions between 585 and 625 nm. Coomassie stain was used as a total protein loading control.
Drosophila life span assays
Flies were transferred to fresh medium, and survival was scored every 2 to 3 days. dLife software (75) was used to record survival and to compare median and maximum life span via log-rank analysis. Vials were randomized in terms of tray position and semiblinded to reduce the impacts of environment or investigator bias.
Olfactory aversion training
Experiments were performed as described in (25). Animals were exposed (via an air pump) in alternation to two neutral odors (3-octanol and 4-methylcyclohexanol, prepared as a 1:10 dilution in mineral oil) for 5 min under low red light, and a 100-V 60-Hz shock was applied during exposure to one of the two odors. The odor associated with the electric shock was alternated between vials. After three training rounds per odor, animals were given 1 hour to recover then placed in a T maze (CelExplorer Labs) with opposing odors from either side. Flies were allowed 2 min to explore the maze, after which the maze sections were sealed, and the number of flies in each chamber was scored.
Spontaneous activity and circadian rhythm
Spontaneous activity was monitored using a TriKinetic activity monitor, in which vials containing 20 to 25 flies were secured, and activity was recorded in a humidified 24°C incubator with 12-hour light/dark cycles as described above. Flies were allowed to acclimate for 8 hours before data collection. Activity was averaged for each 12-hour cycle and normalized per fly.
Cell viability
Cells were maintained in a clear 96-well plate. On the day of assay, 10 μl of WST-1 reagent (11644807001, Sigma-Aldrich) was added to each well, and cells were incubated in a 37°C, 5% CO2 incubator for 2 to 4 hours. Absorbance was measured at 450 nm using a Gemini series spectrophotometer.
Morris water maze
This test provides measures of hippocampal-dependent spatial learning and memory (76–79). A 121-cm water maze was used. Animals were given a series of four trials, ~30 min apart, per day for 5 days to find a submerged platform (~1 cm below water level) in a large tank filled with water made opaque through the addition of white tempera-based nontoxic paint at 23.0° ± 1.0°C, in a room that the operators entered only to introduce and remove the animals from the pool. The pool was surrounded by large panels with geometric black and white designs that serve as distal cues. Maximum trial time was limited to 60 s, whereupon mice were guided to the platform. Mice were allowed to remain on the platform for 5 s and then were gently towel-dried and moved to their home cage under a heating lamp until dry. At the end of training, a probe trial was conducted where the platform was unavailable to measure the retention of the former platform location. The time each animal spent in the quadrant formerly containing the platform and the number of passes over that location provided a measure of memory. At the end of the probe trial, the platform was raised to its previous location to maintain response reinforcement contingency. On week 2 of training, reversal learning was tested by changing the location of the platform to the quadrant opposite to the one used during initial learning, followed by a second probe trial. Data were collected using TopScan (CleverSys) by operators blinded to genotype.
Novel object recognition test
To measure recognition of a previously encountered object, animals are placed in an opacified rat cage with bedding for 10 min. The following (training) day, mice are returned to the chamber, which now contains two identical objects (3 cm by 3 cm by 8 cm), and allowed to explore the arena for 5 min. The percent of time exploring each object is recorded using TopScan. During testing (4 hours after training), one of the objects in the box is replaced with a new object, and the side of the replaced object is randomized among animals. Mice are given 5 min to explore the two objects, and the percent time exploring each object is recorded using TopScan. A discrimination ratio calculated as (tnovel − tfamiliar)/(tnovel + tfamiliar) is used as a measure of retention of the previously encountered object (positive and negative discrimination ratio values indicate a preference for exploration of the novel and familiar objects, respectively).
Y maze
Working memory is assessed by placing animals in a Y-shaped maze made of white Plexiglas with three arms, with equal angles between all arms. Each animal is placed in an arm of the maze and allowed to move freely around the apparatus, while the sequence and number of arm entries for each animal during a 5-min period are recorded manually by an experimenter blinded to the genotype. The number of spontaneous alternations, which occur when a mouse enters a different arm of the maze in each of three consecutive arm entries (e.g., ABC, CAB, or BCA but not BAB), was counted, and percent alternation was calculated as: (# of spontaneous alternations)/(total arm entries − 2) × 100.
Stability of TAT peptides in plasma
Stock solutions (10 mM) of the peptides were mixed at a ratio of 1:1 with human plasma (Merck) to a final concentration of 500 μM. The samples were incubated in a ThermoMixer (Eppendorf AG, Hamburg, Germany) at 37°C with continuous shaking at 300 rpm. At each time point, in the range of 0 to 48 hours for TAT1-DEN and 0 to 150 min for TAT1-8-9TOD, 100 μl of aliquots were taken and mixed with 15% trichloroacetic acid [3% (w/v) final concentration]. Samples were centrifuged at 18,000 rpm for 15 min, after which precipitates were removed, and 5 μl of the resulting supernatant was subjected to ultrafast high-performance LC (Nexera-I UHPLC, Shimadzu, Kyoto, Japan). The samples were analyzed using a Luna Omega (polar) column (Phenomenex) with an internal diameter of 2.1 mm, a length of 100 mm, and a particle size of 1.6 μm. The mobile phase A was Milli-Q water/0.1% trifluoroacetic acid (TFA), and the mobile phase B was 80% aqueous solution of acetonitrile (ACN) containing 0.1% TFA. The flow was 0.3 ml/min, and the column temperature was 40°C. A linear, 15-min gradient of 2 to 50% B was applied for an analysis of TAT1-DEN, and 2 to 100% B was chosen for TAT1-8,9TOD. The peptides were detected at 223 nm and quantified with the LC Solution software (Shimadzu, Kyoto, Japan). The stability tests were performed in triplicate. The half-life time (T1/2) of the peptides was calculated using the SigmaPlot 12.3 software. The exponential equation estimates based on fitting of two or three logistic parameters were used. The original TAT1-DEN sequence was stable for less than 4 min and subsequently transformed into metabolites with one or two Lys residues clipped from N termini; the latter metabolite was stable for more than 24 hours.
Parallel Artificial Membrane Permeability Assay (PAMPA)
PAMPA was performed in 96-well MultiScreen-IP filter plates (Millipore Corp., Bedford, MA), consisting of a donor and an acceptor plate, assembled in a sandwich format. Propranolol hydrochloride (Sigma-Aldrich), dermorphin (Bachem), and Tat47–57 (synthesized in-house) were used as positive controls. Dopamine hydrobromide (Sigma-Aldrich) was included as a negative control. The 10 mM stock solutions of the tested and control compounds were prepared in water. The 500 μM working solutions of each compound were obtained by dissolving the stocks in 0.1 M PBS (pH 7.4), prepared from PBS tablets (Sigma-Aldrich), and supplemented with DMSO. The final DMSO concentration in the samples was 5%. In separate tubes, 200 μM equilibrium standards in 5% DMSO in PBS were prepared for each test and control compound (if the compound is able to permeabilize the membrane and fully reach equilibrium, then 200 μM will be the final concentration of solution in the donor and acceptor wells).
The porcine brain lipid membrane (PBL; Avanti Polar Lipids Inc.) was prepared as a 2% (20 mg/ml) solution in anhydrous dodecane (Sigma-Aldrich). The polyvinylidene difluoride filter surface in the donor wells was covered with 5 μl of the PBL solution, and the microtiter plate was shaken for 30 s at 400 rpm in a plate shaker to ensure even distribution of the PBL membrane. The working solution (200 μl) of each sample was added to the PBL-coated donor wells in four to five repetitions. The blank control was 5% DMSO in PBS. The acceptor wells were filled with 300 μl of 5% DMSO/PBS. The donor plate was placed on the top of the acceptor plate to create a sandwich. The assembled plate was disposed in a zip-lock bag, together with a water-wetted filter paper, and tightly closed to limit the evaporation of the solution. The assembly was incubated for 20 hours at 37°C, shaking at 250 rpm to guarantee good solvent mixing. Six replicates of the experiment were conducted.
After the incubation, the sandwich was disassembled, and 100 μl of the acceptor solutions was transferred to a 96-well UV transparent plate (Greiner flat-bottom UV-star microplate). Because peptides display maximum absorption in the range completely obscured by DMSO absorption, the absorbance was measured for propranolol only at 290 nm (Infinite M200 Pro plate reader, Tecan). For the remaining samples, fluorescence measurements were performed after the reaction of the compounds with 0.02% fluorescamine in anhydrous acetone. Briefly, 60 μl of PBS, 30 μl of fluorescamine solution, and 10 μl of the acceptor solution were mixed in wells of a Costar flat-bottom black plate. The plate was incubated for 5 min at RT while shaken at 400 rpm in a plate shaker. Then, the fluorescence intensity was measured (excitation at 388 nm and emission at 462 nm; Infinite M200 Pro plate reader, Tecan). In a similar way, the absorbance and fluorescence intensity were measured for equilibrium standards of propranolol and the remaining compounds, respectively.
The rate of passive diffusion was calculated as linear velocity of permeation (Pe), using the following equation
where ODA is the absorbance/fluorescence of the acceptor solution, ODE is the absorbance/fluorescence of the equilibrium standard, and C was calculated as follows
where VD is the volume (in cubic centimeters) of the donor well, VA is the volume (in cubic centimeters) of the acceptor well, area is the effective area of the membrane (0.24 cm2), and time is the incubation time (in seconds). The data represent an average of six independent biological repeats.
In vitro degradation of polyubiquitinated substrate
To prepare a substrate for purified human 26S proteasome, p53 was polyubiquitinated with a MDM2 Ubiquitin Ligase Kit–p53 Substrate from Boston Biochem Inc. (Cambridge, MA), under conditions recommended by the manufacturer. After 15 min of polyubiquitination, 26S proteasome in buffer containing 1 μM TAT1-8,9TOD, TAT1-DEN, or vehicle was added to the reaction mixtures, and the reaction was carried out for 1 hour at 37°C. The reaction was stopped by the addition of SDS-PAGE buffer, as recommended. The samples were separated by SDS-PAGE (10% acrylamide tris-glycine precast Novex gels from Invitrogen, Carlsbad, CA), transferred to nitrocellulose membrane in a semidry system, and probed with anti-p53–specific monoclonal antibodies provided in the kit. Molecular weight standards Chameleon Duo (LI-COR Biosciences, Lincoln, NE) were run alongside reaction samples in each gel. Visualization of p53 was performed with an Odyssey Infrared Imaging System (LI-COR, Lincoln, NE) with secondary antibodies from Cell Signaling Technology (Danvers, MA).
Immunoblotting
Tissues were chilled in liquid nitrogen and then homogenized using a pestle and mortar, which had also been chilled with liquid nitrogen. The homogenate was incubated in radioimmunoprecipitation assay buffer supplemented with protease inhibitors at 4°C for 15 min and was subjected to vortexing every 5 min. The homogenate was then centrifuged at 15,000g for 5 min, and the supernatant was removed. Lysate protein content was determined using a BCA assay. Samples were mixed with Laemmli sample buffer supplemented with 5% β-mercaptoethanol and boiled at 95°C for 5 min before immunoblotting. Blots were evaluated against anti-PSMB5 (ab140450, Abcam), anti-APP (ab126732, Abcam), anti-BACE1 (ab183612, Abcam), anti-tau (MAB361, EMD Millipore), anti–phospho-tau (NM1060, Thermo Fisher Scientific), and anti–β-actin (ab8227, Abcam).
Mass spectrometry
Intact protein sample preparation
Brain samples were homogenized in a cold methanol:water 1:1 solution and centrifuged. Ten microliters of the brain supernatant was diluted with 20 μl of a denaturing solution (7 M urea, 2 M thiourea, and 20 mM DTT) and added dropwise to 900 μl of ice-cold acetone. The resulting suspension was shaken at 4°C for 1 hour and centrifuged at 19,000g. The supernatant was discarded, and the air-dried pellet was taken up in 70% ACN containing 0.1% TFA and 12 mM HCl. This mixture was shaken at 4°C for 1 hour and then centrifuged again. The supernatant was lyophilized, and the powder was resuspended in 0.1% formic acid (FA) and analyzed by LC-MS.
Digested protein sample preparation
The brain supernatant samples were added dropwise at 1:4 ratio to ice-cold acetone, incubated for 1 hour at −20°C, and then centrifuged at 15,000g. The supernatant was discarded, and the air-dried pellet was dissolved in a mixture of 6 M urea and 10 mM thiourea. The samples were incubated for 1 hour at 60°C, with continuous shaking, then diluted with ammonium bicarbonate (pH 7.6) (final buffer concentration, 50 mM), and centrifuged at 12,000g. One microgram of either trypsin or chymotrypsin prepared according to the manufacturer’s instructions (Promega Co., USA) was added to the supernatant, and the resulting solution was incubated for 20 hours at 37°C in a thermoshaker at 300 rpm. The samples were then centrifuged, and the supernatant was analyzed by LC-MS.
LC-MS analysis
LC-MS was performed using a Thermo Fisher Scientific UltiMate 3000 (Thermo Fisher Scientific, USA) nano-LC system connected in-line to a TripleTOF apparatus (Sciex, Framingham, MA). The samples were concentrated and desalted using trap cartridges and a 3-min flow of a loading buffer (10 μl/min; 2% ACN in 0.1% TFA). The nondigested and the trypsin-digested samples were concentrated on a C18 5-μm PepMap100 sorbent (300 μm × 5 mm; Thermo Fisher Scientific, USA) and then applied to a fused-silica analytical column packed with PepMap 2-μm sorbent (75 μm × 250 mm; Thermo Fisher Scientific, USA). The trap cartridges with C18–RP1 sorbent (75 μm × 10 mm; Phenomenex, USA) were used to concentrate and desalt the chymotrypsin-digested samples, which were then separated on a core-shell column with BioZen Polar 3-μm sorbent (75 μm × 150 mm; Phenomenex, USA).
Analytical gradient was performed by a linear increase in mobile phase B (0.1% FA in 80% ACN) in mobile phase A (0.1% FA in water), from 2 to 40% B in 20 min, and then to 99% B for 40 min, with a flow rate of 300 nl/min. The column oven temperature was set to 35°C. Peptides eluting from the column were ionized in Optiflow nano-ion source (electrospray ionization) and introduced to Sciex TT6600+ mass spectrometer at positive ionization mode. MS operation parameters were as follows: spray voltage, +5.5 kV; nebulizer gas (N2) pressure, 96.5 kPa; collision energy, +10 V; declustering potential, +90 V; source temperature, 210°C. Full-scan range was set at m/z 350 to 1500 and 100 to 1500 for the intact and digested samples, respectively. Each full scan was followed by fragmentation of the top 10 most intense precursor ions with charge from +2 to +5. The collision energy was from 25 to 50 eV, with 10-eV spread. The Analyst 1.8 (Sciex, Canada) software was used for data collection, and PeakView 2.2 (Sciex, Canada) was used for their visualization.
Statistics
Statistics were performed with OriginPro 2020 (OriginLab), Prism 9.0 (GraphPad Software), and Microsoft Excel. We compared groups of data using two-tailed Student’s t tests or one-way analysis of variance (ANOVA) after we confirmed the normal data distribution with Kolmogorov-Smirnov tests. Post hoc Tukey test was applied for comparisons of multiple groups. If the normality test failed, the nonparametric Kruskal-Wallis test was applied, and the Dunn test was used for between-group comparison. Differences in animal life spans were analyzed using log-rank tests. Results of olfaction aversion training in flies were analyzed with chi-square tests. Linear regression analysis was used to establish significance of interaction between proteasome function and levels of APP, BACE1, and Aβ in hippocampal tissue samples from patients. Response of proteasomes to increasing compound concentrations was approximated with nonlinear fit functions (OriginPro 2020). If the fit converged at the established chi-square tolerance value, then we calculated the corresponding AC50 and maximum fold of activation. The same approach was also used for fitting TAT stability data. Statistically significant differences between populations were assumed if P < 0.05.
Study approval
All mouse studies performed were approved by the Institutional Animal Care and Use Committee at the University of Texas Health Science Center at San Antonio (protocol 20170040AR; Pickering, PI).
Acknowledgments
We thank J. Cropper and L. Quintanilla, who performed animal genotyping and husbandry and ran many of the molecular biology measures. We thank B. Sopher and G. Martin for the donation of MC65 cells. We thank H. Overkleeft for the donation of MV151. We thank X. Han for the use of the Keyence microscope for X34 imaging. Morris water maze studies were performed and data analyzed in the Healthspan and Functional Assessment Core of the San Antonio Nathan Shock Center (P30AG013319-21). Transgenic NSE-PSMB5 mice were created at the University of Michigan Transgenic Animal Core. The ShuttleNSE vector was donated to Addgene by J. Bamburg. Drosophila artwork is used with the permission of I. Salecker (80). Mouse artwork is used under the Creative Commons license: commons.wikimedia.org/wiki/File:Vector_diagram_of_laboratory_mouse_(black_and_white).svg. Human tissue was received from the NIH NeuroBioBank.
Funding: This work was supported by the National Institute of Aging R56 AG061051 (to A.M.P.), National Institute of Aging RF1 AG065301 (to A.M.P.), 2018 Glenn Foundation for Medical Research and AFAR Grants for Junior Faculty (to A.M.P.), Voelcker Young Investigator Award (to A.M.P.), Perry & Ruby Stevens Parkinson’s Disease Center of Excellence (to A.M.P.), San Antonio Nathan Shock Center Pilot Grant P30AG013319-21 (to A.M.P.), San Antonio Pepper Center P30-AG044271-03 (to A.M.P.), National Institute on Aging T32 AG021890 (to E.M. and H.S.K.) UTHSCSA M.D. Ph.D. Program T32, NIH GM113896 (to H.S.K.), National Institute of General Medical Science R01 GM069819 (to M.G.), William and Ella Owens Medical Research Foundation (to M.G. and P.A.O.), National Science Center 2019/33/B/NZ7/00112 and 2019/35/O/NZ7/00227 (to E.J.), and Polish Ministry of Education and Science grant no. SPUB 22-0006/22/531 (to P.W. and L.K.).
Author contributions: Fly experiments: A.M.P., E.M., H.S.K., and N.J. Cell culture: E.S.C. and A.M.P. Histology: E.S.C. and J.P.P. Compound development: P.A.O., M.G., E.J., and P.K. Mouse experiments: A.M.P., E.S.C., C.E.V.S., A.B., and N.D. Analysis: A.M.P., E.S.C., V.G., M.G., P.A.O., E.J., and P.K. MS: P.K., P.W., L.K., and E.J. Supervision: A.M.P. Writing: A.M.P., E.S.C., M.G., P.A.O., and E.M. WB: A.M.P., and E.S.C.
Competing interests: A.M.P., M.G., P.A.O., E.J., and P.K. are inventors on a patent application related to this work filed by The University of Texas Health Science Center at San Antonio (HSC1567, filed 13 September 2019). The authors declare that they have no other competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S15
REFERENCES AND NOTES
- 1.Keller J. N., Hanni K. B., Markesbery W. R., Impaired proteasome function in Alzheimer’s disease. J. Neurochem. 75, 436–439 (2000). [DOI] [PubMed] [Google Scholar]
- 2.Thibaudeau T. A., Anderson R. T., Smith D. M., A common mechanism of proteasome impairment by neurodegenerative disease-associated oligomers. Nat. Commun. 9, 1097 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keck S., Nitsch R., Grune T., Ullrich O., Proteasome inhibition by paired helical filament-tau in brains of patients with Alzheimer’s disease. J. Neurochem. 85, 115–122 (2003). [DOI] [PubMed] [Google Scholar]
- 4.Almeida C. G., Takahashi R. H., Gouras G. K., β-Amyloid accumulation impairs multivesicular body sorting by inhibiting the ubiquitin-proteasome system. J. Neurosci. 26, 4277–4288 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shringarpure R., Grune T., Sitte N., Davies K. J., 4-Hydroxynonenal-modified amyloid-β peptide inhibits the proteasome: Possible importance in Alzheimer’s disease. Cell. Mol. Life Sci. 57, 1802–1809 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosen K. M., Moussa C. E.-H., Lee H.-K., Kumar P., Kitada T., Qin G., Fu Q., Querfurth H. W., Parkin reverses intracellular β-amyloid accumulation and its negative effects on proteasome function. J. Neurosci. Res. 88, 167–178 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gregori L., Fuchs C., Figueiredo-Pereira M. E., Van Nostrand W. E., Goldgaber D., Amyloid β-protein inhibits ubiquitin-dependent protein degradation in vitro. J. Biol. Chem. 270, 19702–19708 (1995). [DOI] [PubMed] [Google Scholar]
- 8.Kang J., Lemaire H. G., Unterbeck A., Salbaum J. M., Masters C. L., Grzeschik K. H., Multhaup G., Beyreuther K., Müller-Hill B., The precursor of Alzheimer’s disease amyloid A4 protein resembles a cell-surface receptor. Nature 325, 733–736 (1987). [DOI] [PubMed] [Google Scholar]
- 9.Rossner S., Sastre M., Bourne K., Lichtenthaler S. F., Transcriptional and translational regulation of BACE1 expression—Implications for Alzheimer’s disease. Prog. Neurobiol. 79, 95–111 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Pike C. J., Burdick D., Walencewicz A. J., Glabe C. G., Cotman C. W., Neurodegeneration induced by beta-amyloid peptides in vitro: The role of peptide assembly state. J. Neurosci. 13, 1676–1687 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lambert M. P., Barlow A. K., Chromy B. A., Edwards C., Freed R., Liosatos M., Morgan T. E., Rozovsky I., Trommer B., Viola K. L., Wals P., Zhang C., Finch C. E., Krafft G. A., Klein W. L., Diffusible, nonfibrillar ligands derived from Aβ1–42 are potent central nervous system neurotoxins. Proc. Natl. Acad. Sci. U.S.A. 95, 6448–6453 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.LaFerla F. M., Green K. N., Oddo S., Intracellular amyloid-β in Alzheimer’s disease. Nat. Rev. Neurosci. 8, 499–509 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Bahr B. A., Hoffman K. B., Yang A. J., Hess U. S., Glabe C. G., Lynch G., Amyloid β protein is internalized selectively by hippocampal field CA1 and causes neurons to accumulate amyloidogenic carboxyterminal fragments of the amyloid precursor protein. J. Comp. Neurol. 397, 139–147 (1998). [PubMed] [Google Scholar]
- 14.Greenfield J. P., Tsai J., Gouras G. K., Hai B., Thinakaran G., Checler F., Sisodia S. S., Greengard P., Xu H., Endoplasmic reticulum and trans-Golgi network generate distinct populations of Alzheimer β-amyloid peptides. Proc. Natl. Acad. Sci. U.S.A. 96, 742–747 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Upadhya S. C., Hegde A. N., Role of the ubiquitin proteasome system in Alzheimer’s disease. BMC biochemistry 8( suppl. 1), S12 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bedford L., Hay D., Devoy A., Paine S., Powe D. G., Seth R., Gray T., Topham I., Fone K., Rezvani N., Mee M., Soane T., Layfield R., Sheppard P. W., Ebendal T., Usoskin D., Lowe J., Mayer R. J., Depletion of 26S proteasomes in mouse brain neurons causes neurodegeneration and Lewy-like inclusions resembling human pale bodies. J. Neurosci. 28, 8189–8198 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Romero-Granados R., Fontan-Lozano A., Aguilar-Montilla F. J., Carrion A. M., Postnatal proteasome inhibition induces neurodegeneration and cognitive deficiencies in adult mice: A new model of neurodevelopment syndrome. PLOS ONE 6, e28927 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tai H.-C., Serrano-Pozo A., Hashimoto T., Frosch M. P., Spires-Jones T. L., Hyman B. T., The synaptic accumulation of hyperphosphorylated tau oligomers in Alzheimer disease is associated with dysfunction of the ubiquitin-proteasome system. Am. J. Pathol. 181, 1426–1435 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cai F., Frey J. U., Sanna P. P., Behnisch T., Protein degradation by the proteasome is required for synaptic tagging and the heterosynaptic stabilization of hippocampal late-phase long-term potentiation. Neuroscience 169, 1520–1526 (2010). [DOI] [PubMed] [Google Scholar]
- 20.Hegde A. N., Haynes K. A., Bach S. V., Beckelman B. C., Local ubiquitin-proteasome-mediated proteolysis and long-term synaptic plasticity. Front Mol. Neurosci. 7, 96 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greeve I., Kretzschmar D., Tschäpe J.-A., Beyn A., Brellinger C., Schweizer M., Nitsch R. M., Reifegerste R., Age-dependent neurodegeneration and Alzheimer-amyloid plaque formation in transgenic Drosophila. J. Neurosci. 24, 3899–3906 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chakraborty R., Vepuri V., Mhatre S. D., Paddock B. E., Miller S., Michelson S. J., Delvadia R., Desai A., Vinokur M., Melicharek D. J., Utreja S., Khandelwal P., Ansaloni S., Goldstein L. E., Moir R. D., Lee J. C., Tabb L. P., Saunders A. J., Marenda D. R., Characterization of a Drosophila Alzheimer’s disease model: Pharmacological rescue of cognitive defects. PLOS ONE 6, e20799 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mhatre S. D., Michelson S. J., Gomes J., Tabb L. P., Saunders A. J., Marenda D. R., Development and characterization of an aged onset model of Alzheimer’s disease in Drosophila melanogaster. Exp. Neurol. 261, 772–781 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Gronenberg W., Lopez-Riquelme G. O., Multisensory convergence in the mushroom bodies of ants and bees. Acta Biol. Hung. 55, 31–37 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Malik B. R., Hodge J. J., Drosophila adult olfactory shock learning. J. Vis. Exp., e50107 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heinemeyer W., Fischer M., Krimmer T., Stachon U., Wolf D. H., The active sites of the eukaryotic 20S proteasome and their involvement in subunit precursor processing. J. Biol. Chem. 272, 25200–25209 (1997). [DOI] [PubMed] [Google Scholar]
- 27.Chondrogianni N., Tzavelas C., Pemberton A. J., Nezis I. P., Rivett A. J., Gonos E. S., Overexpression of proteasome β5 assembled subunit increases the amount of proteasome and confers ameliorated response to oxidative stress and higher survival rates. J. Biol. Chem. 280, 11840–11850 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Liu Y., Liu X., Zhang T., Luna C., Liton P. B., Gonzalez P., Cytoprotective effects of proteasome β5 subunit overexpression in lens epithelial cells. Mol. Vis. 13, 31–38 (2007). [PMC free article] [PubMed] [Google Scholar]
- 29.Chondrogianni N., Georgila K., Kourtis N., Tavernarakis N., Gonos E. S., 20S proteasome activation promotes life span extension and resistance to proteotoxicity in Caenorhabditis elegans. FASEB J. 29, 611–622 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nguyen N. N., Rana A., Goldman C., Moore R., Tai J., Hong Y., Shen J., Walker D. W., Hur J. H., Proteasome β5 subunit overexpression improves proteostasis during aging and extends lifespan in Drosophila melanogaster. Sci. Rep. 9, 3170 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Munkácsy E., Chocron E. S., Quintanilla L., Gendron C. M., Pletcher S. D., Pickering A. M., Neuronal-specific proteasome augmentation via Prosβ5 overexpression extends lifespan and reduces age-related cognitive decline. Aging Cell 18, e13005 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tanaka K., The proteasome: Overview of structure and functions. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 85, 12–36 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sakimura K., Kushiya E., Takahashi Y., Suzuki Y., The structure and expression of neuron-specific enolase gene. Gene 60, 103–113 (1987). [DOI] [PubMed] [Google Scholar]
- 34.Verdoes M., Florea B. I., Menendez-Benito V., Maynard C. J., Witte M. D., van der Linden W. A., van den Nieuwendijk A. M. C. H., Hofmann T., Berkers C. R., van Leeuwen F. W. B., Groothuis T. A., Leeuwenburgh M. A., Ovaa H., Neefjes J. J., Filippov D. V., van der Marel G. A., Dantuma N. P., Overkleeft H. S., A fluorescent broad-spectrum proteasome inhibitor for labeling proteasomes in vitro and in vivo. Chem. Biol. 13, 1217–1226 (2006). [DOI] [PubMed] [Google Scholar]
- 35.Sopher B. L., Fukuchi K., Kavanagh T. J., Furlong C. E., Martin G. M., Neurodegenerative mechanisms in Alzheimer disease. Mol. Chem. Neuropathol. 29, 153–168 (1996). [DOI] [PubMed] [Google Scholar]
- 36.Mucke L., Masliah E., Yu G. Q., Mallory M., Rockenstein E. M., Tatsuno G., Hu K., Kholodenko D., Wood K. J., McConlogue L., High-level neuronal expression of Aβ1–42 in wild-type human amyloid protein precursor transgenic mice: Synaptotoxicity without plaque formation. J. Neurosci. 20, 4050–4058 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wright A. L., Zinn R., Hohensinn B., Konen L. M., Beynon S. B., Tan R. P., Clark I. A., Abdipranoto A., Vissel B., Neuroinflammation and neuronal loss precede Aβ plaque deposition in the hAPP-J20 mouse model of Alzheimer’s disease. PLOS ONE 8, e59586 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hong S., Beja-Glasser V. F., Nfonoyim B. M., Frouin A., Li S., Ramakrishnan S., Merry K. M., Shi Q., Rosenthal A., Barres B. A., Lemere C. A., Selkoe D. J., Stevens B., Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science 352, 712–716 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheng I. H., Scearce-Levie K., Legleiter J., Palop J. J., Gerstein H., Bien-Ly N., Puoliväli J., Lesné S., Ashe K. H., Muchowski P. J., Mucke L., Accelerating amyloid-β fibrillization reduces oligomer levels and functional deficits in Alzheimer disease mouse models. J. Biol. Chem. 282, 23818–23828 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Saganich M. J., Schroeder B. E., Galvan V., Bredesen D. E., Koo E. H., Heinemann S. F., Deficits in synaptic transmission and learning in amyloid precursor protein (APP) transgenic mice require C-terminal cleavage of APP. J. Neurosci. 26, 13428–13436 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qing H., Zhou W., Christensen M. A., Sun X., Tong Y., Song W., Degradation of BACE by the ubiquitin-proteasome pathway. FASEB J. 18, 1571–1573 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Nunan J., Williamson N. A., Hill A. F., Sernee M. F., Masters C. L., Small D. H., Proteasome-mediated degradation of the C-terminus of the Alzheimer’s disease β-amyloid protein precursor: Effect of C-terminal truncation on production of β-amyloid protein. J. Neurosci. Res. 74, 378–385 (2003). [DOI] [PubMed] [Google Scholar]
- 43.Chu J., Li J. G., Hoffman N. E., Madesh M., Pratico D., Degradation of gamma secretase activating protein by the ubiquitin-proteasome pathway. J. Neurochem. 133, 432–439 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Osmulski P. A., Karpowicz P., Jankowska E., Bohmann J., Pickering A. M., Gaczyńska M., New peptide-based pharmacophore activates 20S proteasome. Molecules 25, 1439 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jeang K. T., Xiao H., Rich E. A., Multifaceted activities of the HIV-1 transactivator of transcription, Tat. J. Biol. Chem. 274, 28837–28840 (1999). [DOI] [PubMed] [Google Scholar]
- 46.Huang X., Seifert U., Salzmann U., Henklein P., Preissner R., Henke W., Sijts A. J., Kloetzel P. M., Dubiel W., The RTP site shared by the HIV-1 Tat protein and the 11S regulator subunit α is crucial for their effects on proteasome function including antigen processing. J. Mol. Biol. 323, 771–782 (2002). [DOI] [PubMed] [Google Scholar]
- 47.Jankowska E., Gaczynska M., Osmulski P., Sikorska E., Rostankowski R., Madabhushi S., Tokmina-Lukaszewska M., Kasprzykowski F., Potential allosteric modulators of the proteasome activity. Biopolymers 93, 481–495 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rana T. M., Jeang K. T., Biochemical and functional interactions between HIV-1 Tat protein and TAR RNA. Arch. Biochem. Biophys. 365, 175–185 (1999). [DOI] [PubMed] [Google Scholar]
- 49.Stalmans S., Bracke N., Wynendaele E., Gevaert B., Peremans K., Burvenich C., Polis I., De Spiegeleer B., Cell-penetrating peptides selectively cross the blood-brain barrier in vivo. PLOS ONE 10, e0139652 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Copolovici D. M., Langel K., Eriste E., Langel U., Cell-penetrating peptides: Design, synthesis, and applications. ACS Nano 8, 1972–1994 (2014). [DOI] [PubMed] [Google Scholar]
- 51.Rizzuti M., Nizzardo M., Zanetta C., Ramirez A., Corti S., Therapeutic applications of the cell-penetrating HIV-1 Tat peptide. Drug Discov. Today 20, 76–85 (2015). [DOI] [PubMed] [Google Scholar]
- 52.Bravo J. A., Forsythe P., Chew M. V., Escaravage E., Savignac H. M., Dinan T. G., Bienenstock J., Cryan J. F., Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc. Natl. Acad. Sci. U.S.A. 108, 16050–16055 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Karpowicz P., Osmulski P. A., Witkowska J., Sikorska E., Giżyńska M., Belczyk-Ciesielska A., Gaczynska M. E., Jankowska E., Interplay between structure and charge as a key to allosteric modulation of human 20S proteasome by the basic fragment of HIV-1 tat protein. PLOS ONE 10, e0143038 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Forrest J. A., Clements J. A., Prescott L. F., Clinical pharmacokinetics of paracetamol. Clin. Pharmacokinet. 7, 93–107 (1982). [DOI] [PubMed] [Google Scholar]
- 55.Singh J. K., Chettiar S. S., Jhala D., Optimization and validation of an in vitro blood brain barrier permeability assay using artificial lipid membrane. J. Bioequivalence Bioavailab. 14, 1–6 (2011). [Google Scholar]
- 56.Myeku N., Clelland C. L., Emrani S., Kukushkin N. V., Yu W. H., Goldberg A. L., Duff K. E., Tau-driven 26S proteasome impairment and cognitive dysfunction can be prevented early in disease by activating cAMP-PKA signaling. Nat. Med. 22, 46–53 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Keller J. N., Hanni K. B., Markesbery W. R., Possible involvement of proteasome inhibition in aging: Implications for oxidative stress. Mech. Ageing Dev. 113, 61–70 (2000). [DOI] [PubMed] [Google Scholar]
- 58.Ding Q., Keller J. N., Does proteasome inhibition play a role in mediating neuropathology and neuron death in Alzheimer’s disease? J. Alzheimers Dis. 5, 241–245 (2003). [DOI] [PubMed] [Google Scholar]
- 59.Crawford L. J., Irvine A. E., Targeting the ubiquitin proteasome system in haematological malignancies. Blood Rev. 27, 297–304 (2013). [DOI] [PubMed] [Google Scholar]
- 60.Njomen E., Osmulski P. A., Jones C. L., Gaczynska M., Tepe J. J., Small molecule modulation of proteasome assembly. Biochemistry 57, 4214–4224 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Giżyńska M., Witkowska J., Karpowicz P., Rostankowski R., Chocron E. S., Pickering A. M., Osmulski P., Gaczynska M., Jankowska E., Proline- and arginine-rich peptides as flexible allosteric modulators of human proteasome activity. J. Med. Chem. 62, 359–370 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jones C. L., Njomen E., Sjogren B., Dexheimer T. S., Tepe J. J., Small molecule enhancement of 20S proteasome activity targets intrinsically disordered proteins. ACS Chem. Biol. 12, 2240–2247 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fiolek T. J., Magyar C. L., Wall T. J., Davies S. B., Campbell M. V., Savich C. J., Tepe J. J., Mosey R. A., Dihydroquinazolines enhance 20S proteasome activity and induce degradation of α-synuclein, an intrinsically disordered protein associated with neurodegeneration. Bioorg. Med. Chem. Lett. 36, 127821 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fiolek T. J., Keel K. L., Tepe J. J., Fluspirilene analogs activate the 20S proteasome and overcome proteasome impairment by intrinsically disordered protein oligomers. ACS Chem. Nerosci. 12, 1438–1448 (2021). [DOI] [PubMed] [Google Scholar]
- 65.Leestemaker Y., de Jong A., Witting K. F., Penning R., Schuurman K., Rodenko B., Zaal E. A., van de Kooij B., Laufer S., Heck A. J. R., Borst J., Scheper W., Berkers C. R., Ovaa H., Proteasome activation by small molecules. Cell Chem. Biol. 24, 725–736.e7 (2017). [DOI] [PubMed] [Google Scholar]
- 66.Bernassola F., Ciechanover A., Melino G., The ubiquitin proteasome system and its involvement in cell death pathways. Cell Death Differ. 17, 1–3 (2010). [DOI] [PubMed] [Google Scholar]
- 67.Tseng B. P., Green K. N., Chan J. L., Blurton-Jones M., LaFerla F. M., Aβ inhibits the proteasome and enhances amyloid and tau accumulation. Neurobiol. Aging 29, 1607–1618 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cai N., Chen J., Bi D., Gu L., Yao L., Li X., Li H., Xu H., Hu Z., Liu Q., Xu X., Specific degradation of endogenous tau protein and inhibition of tau fibrillation by tanshinone IIA through the ubiquitin-proteasome pathway. J. Agric. Food Chem. 68, 2054–2062 (2020). [DOI] [PubMed] [Google Scholar]
- 69.Chu T.-T., Gao N., Li Q.-Q., Chen P.-G., Yang X.-F., Chen Y.-X., Zhao Y.-F., Li Y.-M., Specific knockdown of endogenous tau protein by peptide-directed ubiquitin-proteasome degradation. Cell Chem. Biol. 23, 453–461 (2016). [DOI] [PubMed] [Google Scholar]
- 70.Staudt N., Molitor A., Somogyi K., Mata J., Curado S., Eulenberg K., Meise M., Siegmund T., Häder T., Hilfiker A., Brönner G., Ephrussi A., Rørth P., Cohen S. M., Fellert S., Chung H.-R., Piepenburg O., Schäfer U., Jäckle H., Vorbrüggen G., Gain-of-function screen for genes that affect Drosophila muscle pattern formation. PLOS Genet. 1, e55 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ren C., Finkel S. E., Tower J., Conditional inhibition of autophagy genes in adult Drosophila impairs immunity without compromising longevity. Exp. Gerontol. 44, 228–235 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Skorupa D. A., Dervisefendic A., Zwiener J., Pletcher S. D., Dietary composition specifies consumption, obesity, and lifespan in Drosophila melanogaster. Aging Cell 7, 478–490 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hsia A. Y., Masliah E., McConlogue L., Yu G. Q., Tatsuno G., Hu K., Kholodenko D., Malenka R. C., Nicoll R. A., Mucke L., Plaque-independent disruption of neural circuits in Alzheimer’s disease mouse models. Proc. Natl. Acad. Sci. U.S.A. 96, 3228–3233 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Roberson E. D., Scearce-Levie K., Palop J. J., Yan F., Cheng I. H., Wu T., Gerstein H., Yu G. Q., Mucke L., Reducing endogenous tau ameliorates amyloid β-induced deficits in an Alzheimer’s disease mouse model. Science 316, 750–754 (2007). [DOI] [PubMed] [Google Scholar]
- 75.Linford N. J., Bilgir C., Ro J., Pletcher S. D., Measurement of lifespan in Drosophila melanogaster. J. Vis. Exp. 71, 50068 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Morris R., Developments of a water-maze procedure for studying spatial learning in the rat. J. Neurosci. Methods 11, 47–60 (1984). [DOI] [PubMed] [Google Scholar]
- 77.Galvan V., Gorostiza O. F., Banwait S., Ataie M., Logvinova A. V., Sitaraman S., Carlson E., Sagi S. A., Chevallier N., Jin K., Greenberg D. A., Bredesen D. E., Reversal of Alzheimer’s-like pathology and behavior in human APP transgenic mice by mutation of Asp664. Proc. Natl. Acad. Sci. U.S.A. 103, 7130–7135 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Butterfield D. A., Galvan V., Lange M. B., Tang H., Sowell R. A., Spilman P., Fombonne J., Gorostiza O., Zhang J., Sultana R., Bredesen D. E., In vivo oxidative stress in brain of Alzheimer disease transgenic mice: Requirement for methionine 35 in amyloid β-peptide of APP. Free Radic. Biol. Med. 48, 136–144 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pierce A., Podlutskaya N., Halloran J. J., Hussong S. A., Lin P. Y., Burbank R., Hart M. J., Galvan V., Over-expression of heat shock factor 1 phenocopies the effect of chronic inhibition of TOR by rapamycin and is sufficient to ameliorate Alzheimer’s-like deficits in mice modeling the disease. J. Neurochem. 124, 880–893 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shimosako N., Hadjieconomou D., Salecker I., Flybow to dissect circuit assembly in the Drosophila brain. Methods Mol. Biol. 1082, 57–69 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S15