SUMMARY
Argonaute (AGO) proteins are central players in RNA interference in eukaryotes. They associate with small RNAs (sRNA) and lead to transcriptional or posttranscriptional silencing of targets, thereby regulating diverse biological processes. The molecular and biological functions of AGO proteins have been extensively characterized, particularly in a few angiosperm species, leading to the recognition that the AGO family has expanded to accommodate diverse sRNAs thereby performing diverse biological functions. However, understanding of the expansion of AGO proteins in plants is still limited, due to a dearth of knowledge of AGO proteins in green algal groups. Here, we identified more than 2900 AGO proteins from 244 plant species, including green algae, and performed a large-scale phylogenetic analysis. The phylogeny shows that the plant AGO family gave rise to four clades after the emergence of hydrobiontic algae and prior to the emergence of land plants. Subsequent parallel expansion in ferns and angiosperms resulted in eight main clades in angiosperms: AGO2, AGO7, AGO6, AGO4, AGO1, AGO10a, AGO10b and AGO5. On the basis of this phylogeny, we identified two novel AGO4 orthologs that Arabidopsis does not have, and redefined AGO10, which is composed of AGO10a and AGO10b. Finally, we propose a hypothetical evolutionary model of AGO proteins in plants. Our studies provide a deeper understanding of the phylogenetic relationships of AGO family members in the green lineage, which would help to further reveal their roles as RNAi effectors.
Keywords: Argonaute protein, molecular evolution, origin, diversification, hydrobiontic algae, land plants, angiosperm
INTRODUCTION
In eukaryotes, RNA interference (RNAi) is an important genetic regulatory mechanism mediated by small RNAs (sRNAs; Hutvagner and Simard, 2008). As the major effector in RNAi, ARGONAUTE proteins (AGOs) associate with sRNAs to form RNA-induced silencing complexes, which in turn repress the targets of sRNAs at the transcriptional or posttranscriptional levels (Hutvagner and Simard, 2008). The AGO family in eukaryotes can be phylogenetically divided into four groups: Trypanosoma Ago family, WAGO family, Ago-like family, and PIWI family, with all plant AGOs belonging to the Ago-like family (Swarts et al., 2014). The eukaryotic AGOs are highly conserved in structure and generally contain four domains: N-terminal domain, PAZ, MID and PIWI, which together contribute to sRNA loading, target recognition and target regulation (Gu et al., 2012; Rodriguez-Leal et al., 2016) The PAZ domain binds the 3′-end of the sRNA (Lingel et al., 2004; Song et al., 2003; Yan et al., 2003); the MID domain binds the 5′ phosphate (Frank et al., 2012) and together with the PIWI domain forms a pocket for the first base of the sRNA (Parker et al., 2005); the PIWI domain contains D-E-D-H/D sites, which are critical for the RNase H-like endonuclease activity (Fang and Qi, 2016; Liu et al., 2004; Song et al., 2004).
In plants, sRNAs and AGOs impact multiple biological processes by serving as sequence-specific regulators of genes and genomes. Plant AGOs participate in posttranscriptional gene silencing (PTGS) through endonucleolytic cleavage or translational repression, in transcriptional gene silencing through RNA-directed DNA methylation (RdDM), and in other emerging functions (Carbonell, 2017; Carbonell and Carrington, 2015; Fang and Qi, 2016). Current knowledge of plant AGOs has been derived mainly from angiosperms, especially Oryza sativa and Arabidopsis thaliana (Fang and Qi, 2016). Multiple AGOs tend to be present in each species, such as 10 in Arabidopsis, representing diversification of RNAi pathways (Carbonell, 2017; Fang and Qi, 2016). In Arabidopsis, AGO1 is the major RNAi effector that associates with nearly all microRNAs (miRNAs) as well as small interfering RNAs (siRNAs) from some endogenous loci, transgenes or viruses and, together with its associated sRNAs, leads to PTGS by cleaving target transcripts or causing inhibition of translation (Baumberger and Baulcombe, 2005; Borges and Martienssen, 2015; Garcia-Ruiz et al., 2015; Morel et al., 2002; Qi et al., 2005; Rogers and Chen, 2013; Vaucheret et al., 2004; Wang et al., 2011). AGO2 is well known for its anti-viral functions, but also mediates the activities of a few miRNAs (Harvey et al., 2011; Jaubert et al., 2011; Schuck et al., 2013; Zhang et al., 2011). AGO2 also facilitates DNA double-stranded break repair (Gao et al., 2014). AtAGO4, 6 and 9 associate with endogenous 24-nucleotide (nt) siRNAs derived from transposable elements (TEs) and lead to RdDM of homologous TEs to ensure genome stability (Duan et al., 2015; Olmedo-Monfil et al., 2010; Qi et al., 2006; Zheng et al., 2007; Zilberman et al., 2003). The sRNAs bound by AtAGO8 have not been determined, but AtAGO8 belongs to the same clade as AtAGO4, 6 and 9 and, like the other AGOs in this clade, controls early megaspore formation (Hernández-Lagana et al., 2016; Olmedo-Monfil et al., 2010). AtAGO5 promotes megagametogenesis and anti-viral responses through unknown sRNA partners (Brosseau and Moffett, 2015; Tucker et al., 2012), and associates with miR156 to regulate flowering (Borges et al., 2011; Roussin-Léveillée et al., 2020). Some AGOs, such as AGO7 and AGO10, associate with, and act through, specific miRNAs. AtAGO10 competes with AGO1 for miR165/166 (Zhu et al., 2011) and channels it for degradation (Yu et al., 2021). Through this molecular activity, AGO10 impacts developmental processes such as shoot apical meristem (SAM) maintenance and axillary meristem development (Liu et al., 2009; Zhang et al., 2020; Zhou et al., 2015; Zhu et al., 2011). AtAGO7 associates with miR390 and triggers the production of trans-acting siR-NAs from TAS3 transcripts, targets of miR390 (Adenot et al., 2006; Allen et al., 2005; Fahlgren et al., 2006; Howell et al., 2007; Montgomery et al., 2008).
With the sequencing of plant genomes and the profiling of transcriptomes, more and more plant AGO genes have been discovered. For example, there are six AGO genes in the moss Physcomitrella patens (Arif et al., 2013), 13 in Citrus sinensis (Sabbione et al., 2019), 15 in Solanum lycopersicum (Bai et al., 2012), 17 in maize (Qian et al., 2011) and 19 in rice (Kapoor et al., 2008). AGO proteins in angiosperms have diversified into three major clades according to their phylogenetic relationships: AGO1/5/10, AGO4/6/8/9 and AGO2/3/7, which are named after AGO1-10 from A. thaliana. The origin of these three clades can be traced back to the early common ancestor of land plants, and the divergence occurred early in land plant evolution (Fang and Qi, 2016; Singh et al., 2015; Vaucheret, 2008; You et al., 2017). Some researchers suggest that AGO5 forms an individual clade based on phylogenetic topology (Rodríguez-Leal et al., 2016; Singh et al., 2015). Moreover, the linage-specific AGO18 was found in maize and rice (Kapoor et al., 2008; Qian et al., 2011). Recent studies found that an extra AGO clade existed in land plants: AGO-like, whereas it seems to be lost in angiosperms (You et al., 2017). Although the evolutionary framework of the plant AGO family has been established, the current phylogenetic classification system relies heavily on a few species, which limited a clear understanding of the evolutionary origin and phylogenetic relationships of plant AGOs. Given the presence of a large number of AGOs in any angiosperm species, and with more AGOs being discovered from many species, including basal plant lineages, a more accurate and complete phylogenetic system is urgently needed to further classify the AGO family.
In order to further understand the conservation and diversification of plant AGOs, we sought to build a comprehensive phylogeny of plant AGOs. Here, we mined genomic and/or transcriptomic data from 244 green plants and identified 2958 AGO proteins, with which phylogenetic analyses were performed. Based on the phylogeny, we explored the origin and divergence of the plant AGO family. The results showed that there are four major AGO clades in land plants, including AGO1/5/10, AGO2/3/7, AGO4/6/8/9 and AGO-like. The divergence of those AGO clades could be traced back to charophytes before the emergence of land plants. In addition, we also provide a more complete phylogenetic architecture of angiosperm AGOs than before. Compared with other land plants, the AGOs in angiosperms are further classified to eight clades: AGO2, AGO7, AGO6, AGO4, AGO1, AGO10a, AGO10b and AGO5. In the AGO4 clade, we found that there are novel sub-clades in eudicots, which do not have orthologs in Arabidopsis. We also reclassified the homologs of AGO10 into AGO10a and AGO10b. Finally, we discuss the possible evolutionary relationships of the AGO protein family in the green lineage.
RESULTS
Identification of AGO proteins from plant genomes and/or transcriptomes
In order to comprehensively identify AGO proteins in plants, we selected 245 plant species from different lineages, including chlorophytic algae, charophytes, bryophytes, ferns, lycophytes, gymnosperms and angiosperms (Table S1), the taxonomy of which is shown in Figure S1. Most of the species (238/244) used in our analysis have sequenced genomes. To gain a better understanding of the evolutionary history of AGO proteins in the green lineage, we also identified AGO protein sequences from six charophytes in the OneKP transcriptome database (https://db.cngb.org/onekp/). In all, 2958 AGO candidate protein sequences were retrieved from 244 plant species by performing homology searches and domain predictions (Table S2). The copy number of AGO proteins varies greatly among the different plant lineages, ranging on average from 3 copies in chlorophytic algae, 5.8 in bryophytes, 6.8 in gymnosperms, to 13.1 in angiosperms (Table 1). These data indicate a trend of gradual expansion of the AGO family during evolution. Among the angiosperms examined, Poaceae (Gramineae) plants have the largest AGO family, such as 19 in rice, 17 in maize and 15 in sorghum, with an average of 18.48 members (Table 1).
Table 1.
The number of AGO proteins in green plants
| Taxonomy | Number of species | Number of AGO | Average number of AGO per species |
|---|---|---|---|
| Chlorophytes | 3 | 9 | 3.00 |
| Charophytes | 13 | 48 | 3.69 |
| Bryophytes | 5 | 29 | 5.80 |
| Ferns | 2 | 12 | 6.00 |
| Lycophytes | 1 | 7 | 7.00 |
| Gymnosperms | 5 | 34 | 6.80 |
| Angiosperms | 215 | 2819 | 13.11 |
| Poaceae | 40 | 739 | 18.48 |
AGO, Argonaute.
Evolutionary origin of AGO proteins in land plants
A previous study showed that AGO proteins can be divided into four clades in land plants (You et al., 2017). To investigate the evolutionary origin of AGO proteins in land plants, we used dataset I, which includes 187 AGO proteins from 33 plants in the green lineage, to reconstruct an unrooted AGO phylogenetic tree. The species in dataset I encompassed more non-angiosperm species than those used in the You et al. study. The result showed that there exist five major AGO clades in plants, which are Chlorophytic AGO, AGO1/5/10, AGO-like, AGO4/6/8/9 and AGO2/3/7 (Figure 1). Except for the Chlorophytic AGO clade, other clades were mainly composed of land plant AGOs. The topology of the phylogenetic tree clearly separated the chlorophytes from the other clades, which suggests that all land plant AGOs originated from AGOs in a common ancestor. To find out whether the divergence of land plant AGOs occurred in the common ancestor of eukaryotes, we added some fungal and animal AGOs on the basis of dataset I to rebuild a phylogenetic tree. The results showed that AGOs in land plants are clearly distinct from those in animals and fungi, which suggests that the divergence of land plant AGOs occurred after the emergence of plants (Figure S2). Each of the AGO1/5/10, AGO4/6/8/9 and AGO2/3/7 clades included not only land plants but also charophytes (Figure 1), which indicates that the divergence of land plant AGOs may have occurred as early as in the common ancestor of charophytes and land plants. Moreover, not all charophytic algae lineages were found in the three clades. Only AGOs from Zygnematophyceae were present, while Zygnematophyceae is considered to be the most closely related to land plants (Cheng et al., 2019). The results suggested that the diversity of AGOs in charophytes during the later stage of evolution from hydrobiontic algae to land plants is similar to that in land plants, and implies that the three land plant AGO clades emerged in the common ancestor of Zygnematophyceae and land plants. Although the above results were robustly supported by the maximum likelihood (ML) tree constructed by the IQ-TREE2 tool (Figure S3), the support for the charophytic node in the tree constructed by RAxML was weak. Due to a lack of more charophytic data to be included for further analyses, we cannot rule out the possible origin of diversified AGO clades in an ancestral land plant.
Figure 1.
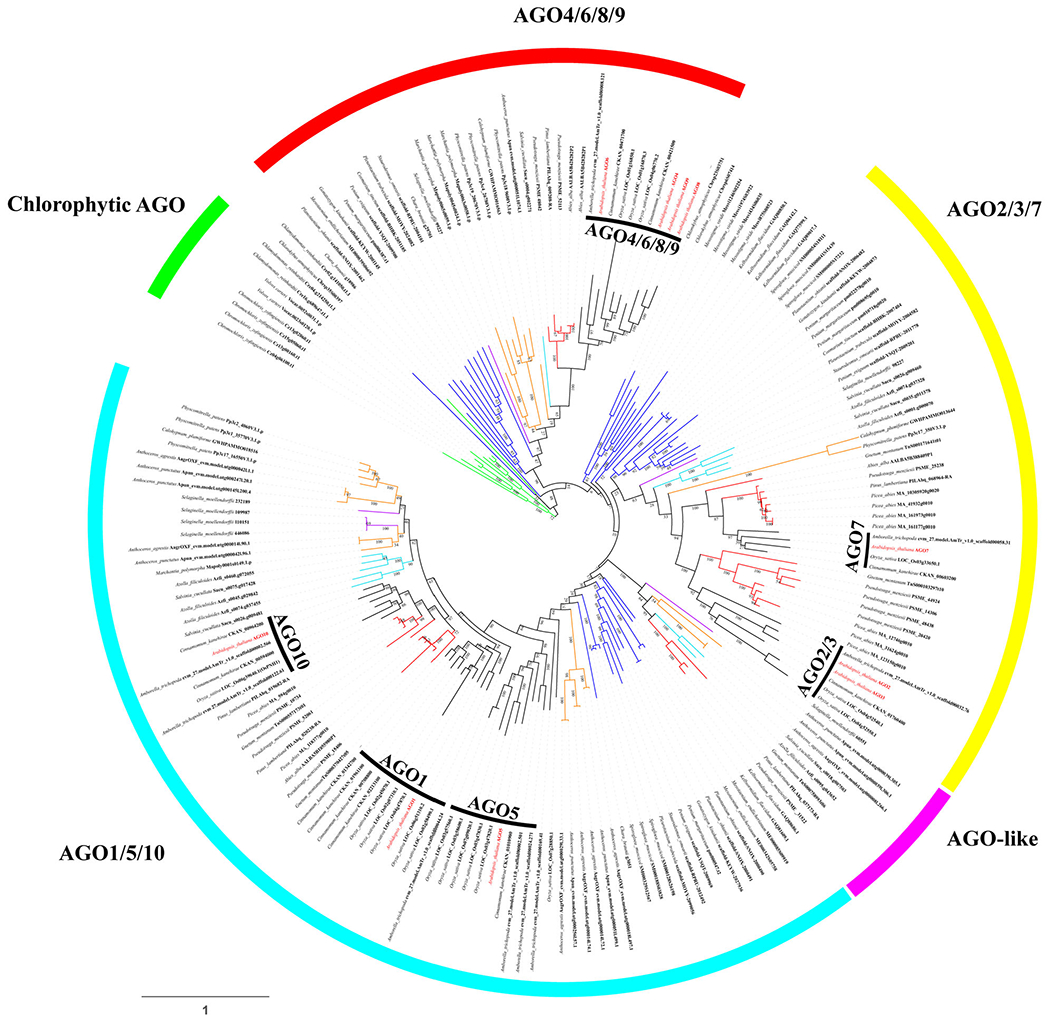
A phylogenetic tree of Argonaute (AGO) proteins in green plants. The tree was constructed using dataset I implemented in RAxML. Dataset I includes 187 AGO proteins from 3 chlorophytes, green; 13 charophytes, blue; 5 bryophytes, orange; 1 lycophyte, purple, 2 ferns, light blue; 5 gymnosperms, red; 4 angiosperms, black. The sequences from chlorophytes (Chlamydomonas reinhardtii, Volvox carteri and Chromochloris zofingiensis) are clustered and named Chlorophytic AGO. The AGO1/5/10, AGO2/3/7, AGO4/6/8/9 clades were named after the 10 Arabidopsis AGOs, and the AGO-like clade was named according to You et al. (2017). AGOs from charophytes, bryophytes, lycophytes, ferns, gymnosperms and angiosperms are present in all clades except for the AGO-like clade. The inner circle (black bars) is identified AGO subclade. The numbers next to the nodes are bootstrap support values (from 0 to 100).
Among the four land plant AGO clades, all except for AGO-like contain AGO proteins in angiosperms. The AGO-like clade only consists of AGOs in hornworts, lycophytes, ferns and gymnosperms, which is consistent with the previous finding that the AGO-like clade is present in early-diverging land plants and lost in angiosperms (You et al., 2017). The angiosperm AGO4/6/8/9 clade is monophyletic (Figure 1), suggesting that the emergence of AGO4/6/8/9 occurred in the common ancestor of angiosperms. In the AGO2/3/7 clade, the divergence of AGO2/3 and AGO7 was observed in seed plants (gymnosperms and angiosperms), but only one copy exists in earlier-diverging plants, which suggests that the diversification into AGO2/3 and AGO7 occurred in the ancestor of seed plants. In the AGO1/5/10 clade, the topology of the AGO1 and AGO10 branches is irreproducible in RAxML-inferred (Figure 1) and IQ-TREE-inferred ML trees (Figure S3). The IQ-TREE-inferred tree shows that AGO1 and AGO10 both have counterparts in gymnosperms (Figure S3), which suggests that the divergence of AGO1 and AGO10 may have occurred in early seed plants. However, this result is in conflict with the RAxML-inferred gene tree (Figure 1). Furthermore, AGO5 can only be detected in angiosperms. Therefore, we assume that AGO5 is an angiosperm-specific AGO.
Phylogenetic classification of the AGO family in angiosperms
In order to further elucidate the phylogenetic relationship of the AGO family in angiosperms, we reconstructed a phylogenetic tree with dataset II, which consists of 2377 AGO proteins from 190 angiosperm species. The phylogenetic analysis revealed that angiosperm AGOs clustered into eight major groups: AGO2, AGO7, AGO4, AGO6, AGO10a, AGO1, AGO5 and AGO10b (Figures 2 and S4). AGO2/3/7 can be further divided into AGO2/3 and AGO7. Due to the fact that AGO2 and AGO3 are lineage-specific pairs within Brassicaceae (Figure S5a), AGO2/3 was denoted as AGO2. It can be concluded that most plants only have one gene corresponding to Arabidopsis AGO2 and 3. AGO1/5/10 can be divided into AGO1, AGO5 and AGO10. And AGO10 can be further divided into two clades, which we hereby name AGO10a and AGO10b. AGO4/6/8/9 can be divided into AGO4/8/9 and AGO6. AGO4, AGO8 and AGO9 are also lineage-specific paralogs within Brassicaceae (Figure S5b), so AGO4/8/9 was denoted as AGO4. The topological structure of the above eight branches is similar to that of species tree, suggesting that these clades originated independently. At the same time, the subsequent evolution is dynamic, accompanied by duplication and lost events (Singh et al., 2015).
Figure 2.
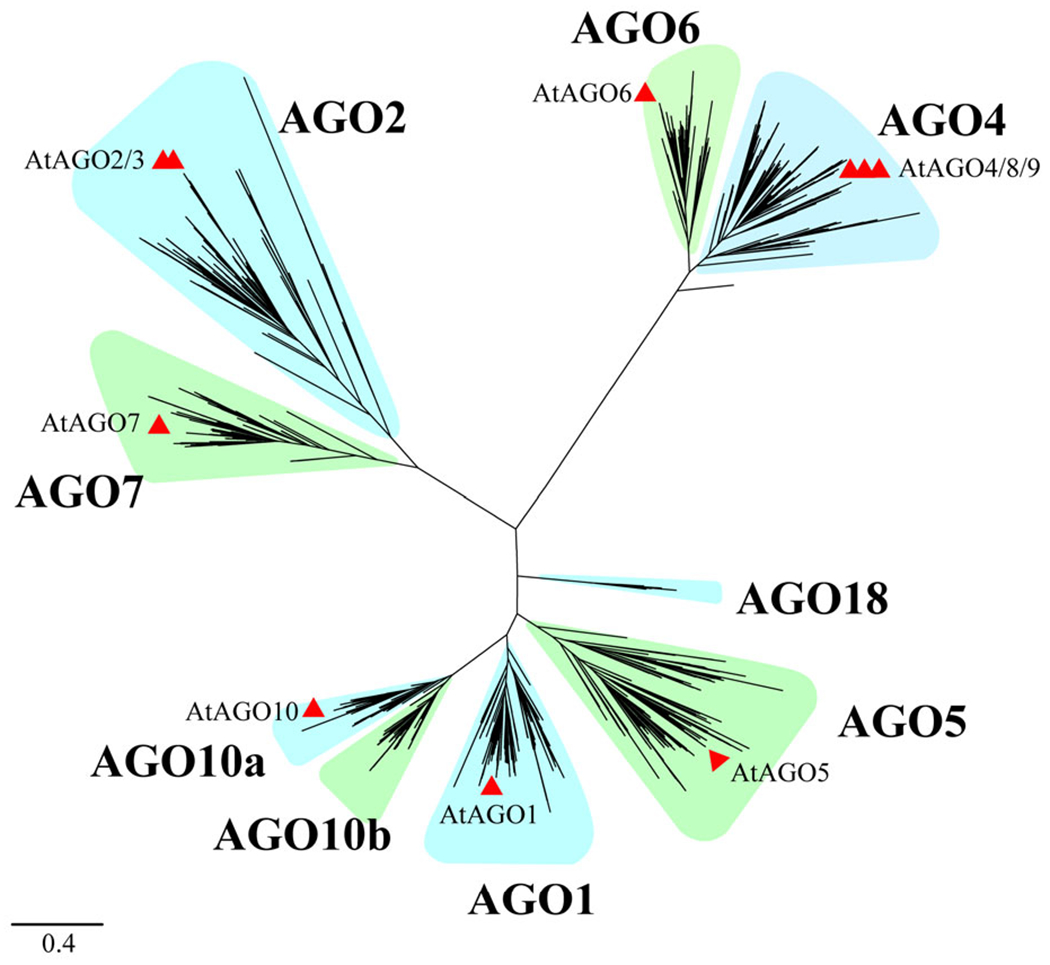
Phylogenetic classification of Argonautes (AGOs) in angiosperms. The topology shows that AGOs in angiosperms can be clearly classified into eight sub-groups: AGO2, AGO7, AGO4, AGO6, AGO10a, AGO1, AGO5 and AGO10b. The Poaceae-specific AGO18 clade is closer to AGO1/5/10 clade.
In the AGO4/6/8/9 clade, AGO6 and AGO4 shared a common basal angiosperm as their root, but other angiosperm lineages such as eudicots, monocots and magnoliidae can be found in both AGO6 and AGO4 branches (Figure S6). This indicates that the divergence between AGO6 and AGO4 occurred after the emergence of angiosperms but at least before the dicots/monocots/magnoliidae divergence. Previous studies have shown that the AGO4 clade has a considerable number of copies in some species (Bai et al., 2012; Kapoor et al., 2008; Sabbione et al., 2019), presumably due to large-scale gene duplication events. Our results showed that AGO4 formed three sub-branches in eudicots: Eudicots-Group I (AGO4-I); Eudicots-Group II (AGO4-II); and Eudicots-Group III (AGO4-III; Figure 3). Through further analysis of the three sub-branches, it was found that AGO4 of all Brassicaceae plants including Arabidopsis were clustered in the AGO4-I branch. The differentiation between AGO4 and AGO8/9 is due to gene duplication in the order Brassicales, while the differentiation between AGO8 and AGO9 is due to gene duplication in the Brassicaceae family (Figure S5b).
Figure 3.
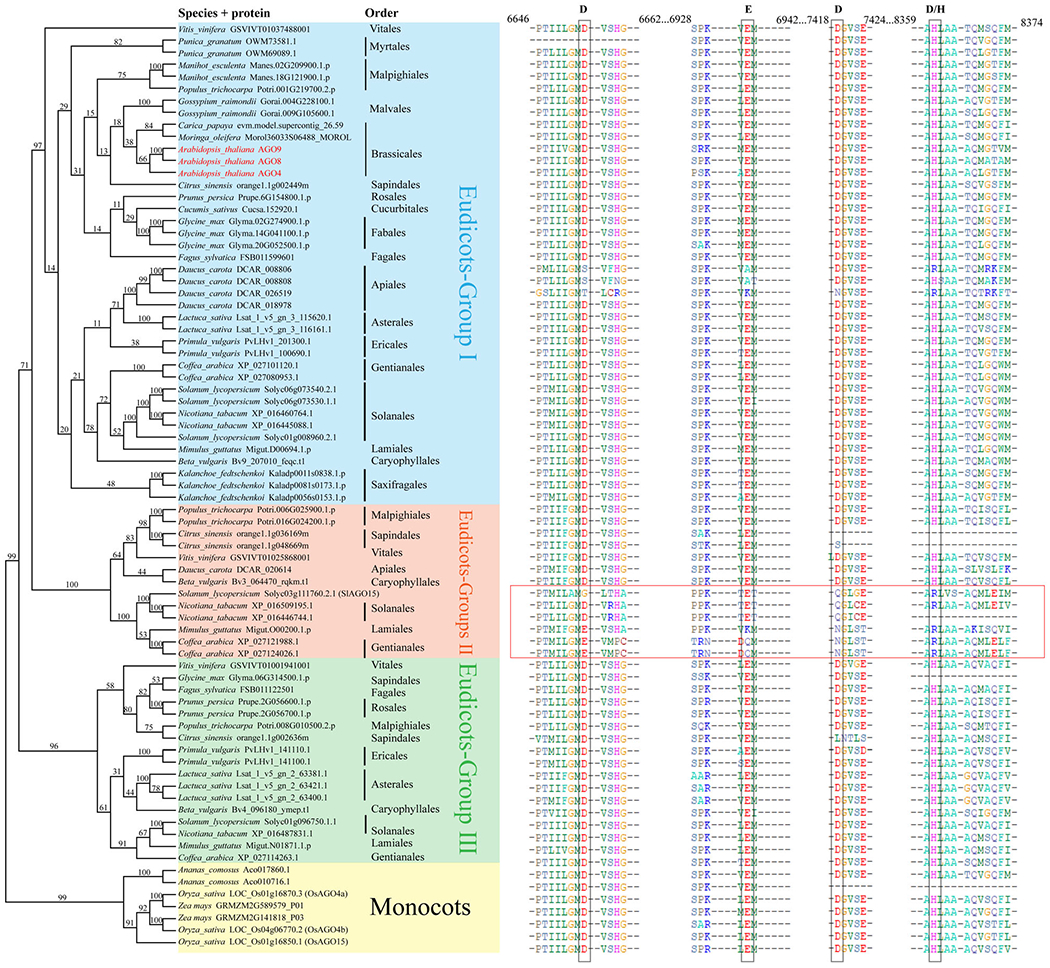
Phylogenetic relationship and multiple sequence alignment of the DEDD/H motif in the AGO4 clade. To aid presentation, some representative species from each order were included. The topology shows that AGO4 in eudicots can be clearly classified into three sub-groups: Eudicots-Group I; Eudicots-Group II; and Eudicots-Group III. Residues in the catalytic tetrad DEDD/H (Asp-Glu-Asp-Asp/His) motif are indicated by black rectangles, and AGO-II members that do not possess a canonical ’DEDH/D’ motif are indicated with the red rectangle. The numbers at the top of the sequence alignment are the positions of amino acids. The numbers next to the nodes are bootstrap support values (from 0 to 100). AGO, Argonaute.
Different from Brassicaceae, some dicotyledonous plants, such as Solanales (such as S. lycopersicum), Vitales (such as Vitis vinifera) and Malpighiales (such as Populus trichocarpa), contain all three AGO4 branches; others, such as Fabales (e.g. Glycine max), possess two branches (Figure 3). However, Cucurbitales and Brassicales only harbor AGO4-I (Figure 3). Arabidopsis has no homologs in AGO4-II or AGO4-III (Figure 3), which indicates that these two branches may have some currently unknown functions. Some AGO-II members do not possess a canonical ‘DEDH/D’ catalytic tetrad in the PIWI domain (Figure 3), which suggests that these AGOs may have different biochemical properties.
OsPNH1 has been considered as a homolog of AtAGO10 in rice, and the two genes have similar functions in the maintenance of the SAM (Moussian et al., 1998; Nishimura et al., 2002). However, our phylogenetic analysis shows that AtAGO10 and OsPNH1 are located in different clades, AGO10a and AGO10b. AGO10a and AGO10b are widely distributed in angiosperms, including basal angiosperms, eudicots, monocots and even magnoliids (Figure 4). Interestingly, no AGOs from the Poales species included in this analysis were found in the AGO10a clade, and Brassicaceae AGOs did not appear in the AGO10b clade, suggesting that AGO10a and AGO10b were lost in Poales and Brassicaceae, respectively. Different from the loss of AGO10b in Brassicaceae, two sister families of Brassicaceae, Moringaceae (such as Moringa oleifera) and Caricaceae (such as Carica papaya), both contain AGO10a and AGO10b proteins. A similar situation was found in the sister family of Brassicaceae, Bromeliaceae (for example, Ananas comosus), which also has both AGO10a and AGO10b. The above results suggest that a family-level loss of AGO10a and AGO10b occurred.
Figure 4.
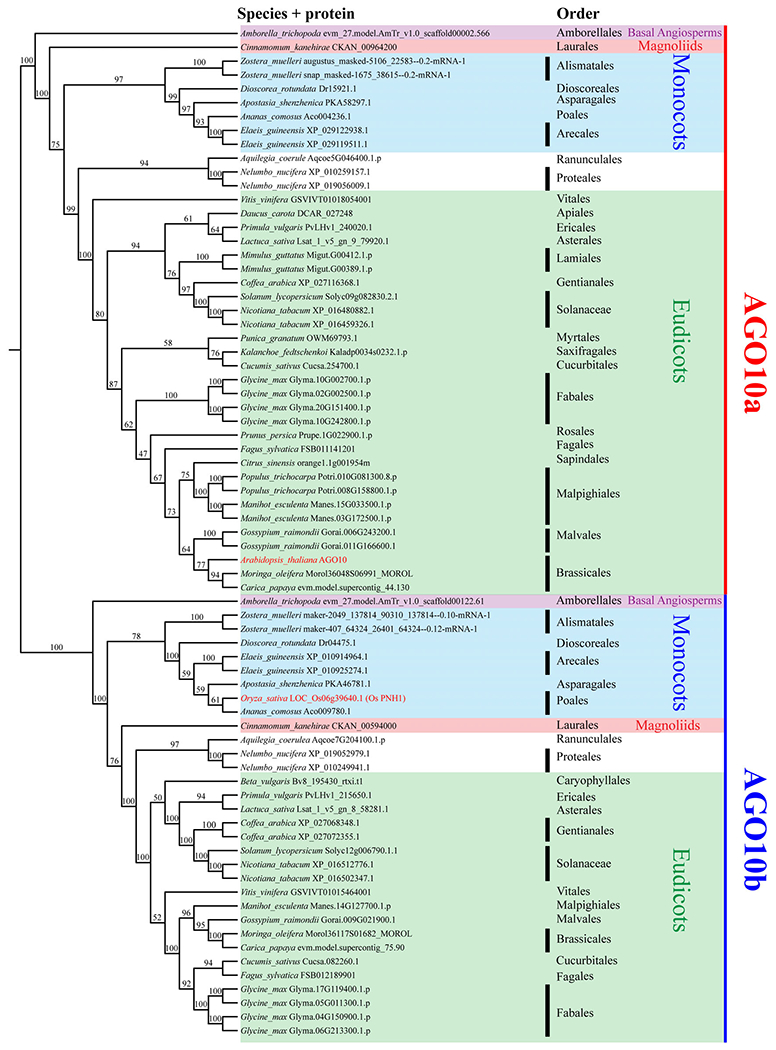
Phylogenetic relationship within the AGO10 clade. To aid presentation, only selected species were included to represent each order. The topology shows that AGO10 in eudicots can be clearly classified into two clades, AGO10a and AGO10b. Each of them includes all major lineages in angiosperms: basal angiosperms, purple; monocots, blue; magnoliids, red; eudicots, green. Most commercial crops contain members of both clades, such as Glycine max, Vitis vinifera and Gossypium raimondii, etc. However, no Arabidopsis AGO10 is found in AGO10b and no Oryza AGO10 is in AGO10a. The numbers next to the nodes are bootstrap support values (from 0 to 100). AGO, Argonaute.
Phylogenetic classification of the AGO family in Poaceae
The family Poaceae contains major crops in the world. Understanding RNAi-mediated mechanisms requires a deeper understanding of the AGO family in Poaceae plants, which is also a prerequisite for crop improvement via RNAi. Here, 739 AGO proteins from 40 Poaceae species were used to investigate the evolution of Poaceae AGOs. Poaceae AGOs were placed in eight clades: AGO2, AGO7, AGO1, AGO5, AGO10a, AGO18, AGO4, AGO6, and the Poaceae-specific clade AGO18. Except for AGO18, the distribution of AGOs in Poaceae was consistent with that in angiosperms (Figure S7). Expansion was observed in AGO4, AGO5, AGO18 and AGO1 branches, resulting in a large number of AGO copies in Poaceae species. According to the topology, AGO1 in Poaceae experienced two expansion events. AGO1 expanded in monocots for the first time, resulting in the formation of two subclades: Monocots-Group I and Monocots-Group II (Figure 5). The result of this expansion can be found in some lineages, such as Poales, Arecales, Asparagales and Dioscoreales. The divergence between OsAGO1ab and OsAGO1cd is the result of this monocot-specific expansion (Figure 5). Subsequently, AGO1 expanded in Poaceae for the second time, resulting in more than one copy of AGO1 in each group (Figure 5). In addition, inside of Poaceae, AGO4 and AGO5 formed three sub-branches (Figure S7a,e). And AGO2, AGO10b and AGO18 formed a specific sub-branch (Figure S7g,h).
Figure 5.
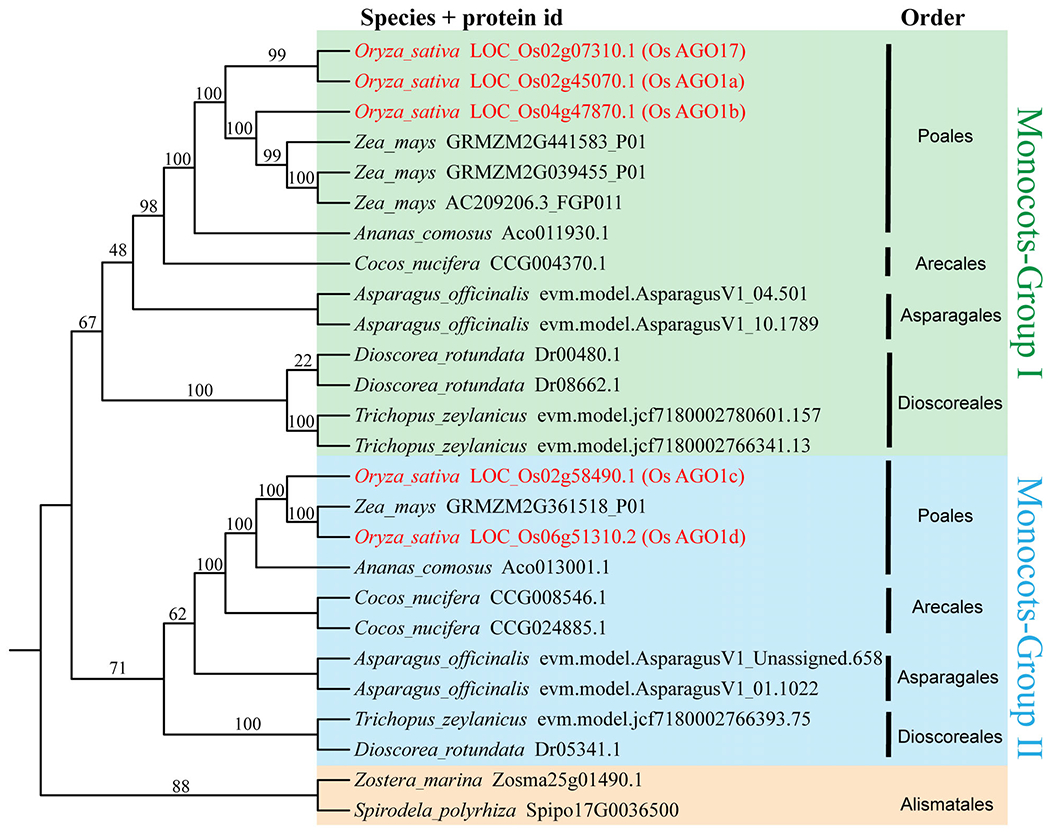
Phylogenetic relationship of monocot Argonautes (AGOs) in the AGO1 clade. To aid presentation, only selected species were included to represent each order. There was an expansion of AGO1 in monocots and these proteins are classified into two clades: Monocots-Group I; and Monocots-Group II. Poales, Arecales, Asparagales and Dioscoreales all have AGO1 members in the two clades. However, an early monocotyledonous order, Alismatales, does not have two groups. The numbers next to the nodes are bootstrap support values (from 0 to 100).
DISCUSSION
This study examined the phylogenetic classification and characteristics of thousands of AGOs from hundreds of plant species and enriched our understanding of green plant AGOs, especially angiosperm AGOs. The phylogenetic insight helps further reveal the molecular and biological functions of various AGO proteins (Figure 6).
Figure 6.
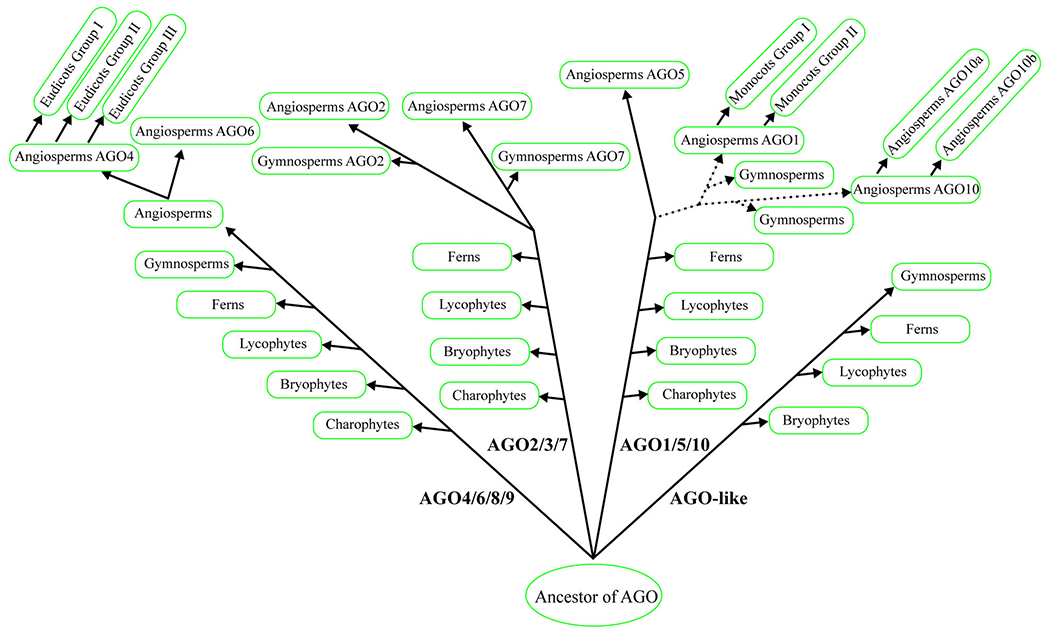
A proposed evolutionary model of Argonautes (AGOs) in plants. The model is based on the phylogeny of AGOs and the cladogram of green plant species. The origin of plant AGOs can traced back to chlorophytes. During the evolutionary process from hydrobiontic plants to land plants, AGOs in the green lineage diverged into four major groups. In seed plants, the AGO family underwent further expansion, especially in angiosperms. In total, eight clades were formed in angiosperms and this was accompanied by independent loss/gain in different lineages. The dotted lines describe the relationship between AGO1 and AGO10, because the divergence of AGO1/AGO10 is unclear.
Phylogenetic relationship of plant AGOs
As genome-based AGO protein identification may miss some proteins due to the quality of genome assembly and the accuracy of genome annotation, we included many species in each phylogenetic group to reduce the impact of potentially missed proteins. Our search for AGO proteins shows that the AGO family is widely distributed in the green lineage, and its copy number varies among different species (Table 1). AGO can be detected in unicellular algae, which implies that the RNAi pathway was established in the common ancestor of green plants (Molnar et al., 2007), but the presence of only one clade of AGO in unicellular algae suggests that AGOs may not have diversified to possess complex functions in these organisms as in higher plants. During evolution, the AGO family subsequently expanded and formed four major clades in land plants. In addition, AGOs from Zygnematophyceaes clustered together with land plant AGOs, which suggested that the divergence of land plant AGOs predated the emergence of land plants, perhaps in the common ancestor of charophytic algae and land plants. How charophytic algae evolved to conquer land is still under active research (Nishiyama et al., 2018; Rensing, 2018). Our results show that Zygnematophyceae species have a diversified AGO family similar to land plants. On the other hand, AGO-mediated regulation of genes and genomes impacts a multitude of biological processes and may have helped plants adapt to the land environment. Thus, the finding that the diversification of the AGO family prior to the emergence of land plants provides new insights into land plant evolution.
The AGO family has considerable diversity in angiosperms, which may be related to the diversification and function of key developmental processes (Cibrian-Jaramillo and Martienssen, 2009). Several previous studies have focused on the evolution of the AGO family in angiosperms (Rodríguez-Leal et al., 2016; Singh et al., 2015). Here, we performed a more comprehensive analysis by including more AGO proteins from a large number of angiosperm plants. Based on our analysis, we propose a hypothetical model for the evolution of AGO proteins in green plants. A common ancestor of AGO exists in early plant ancestors. Subsequently, multi-protein families of AGOs formed during the evolution from hydrobiontic plants to land plants. Four major AGO clades are found in land plants, each of which evolved independently. Notably, gene loss occurred in the AGO-like clade so that it is only present in some land plants. Except for the AGO-like clade, the expansion of the AGO family in the other three clades occurred in parallel in angiosperms, which leads to eight major branches. Among them, the divergence of AGO2/3 occurred prior to the emergence of angiosperms and after the fern-seed plant split; the divergence of AGO4 and AGO6 was the latest, after the emergence of angiosperms, which explains the high degree of sequence conservation (Rodríguez-Leal et al., 2016). In addition, within the AGO4 and AGO1 branches, a large expansion occurred in dicots and monocots, resulting in the formation of three and two subclades, respectively. Besides, there are many lineage-specific duplication and loss events at different taxonomic levels. Those events may contribute to functional diversity of RNAi (Carbonell, 2017), and may pertain to lineage- or species-specific functions.
Insights into AGO evolution and functional diversification in angiosperms
To understand the evolutionary implications of the phylogenetic architecture of the AGO family, it is best to crossreference the phylogenetic relationships of AGOs with function.
The expansion in the AGO family leads to functional diversification (Fang and Qi, 2016). Generally, AGOs in the same clade are functionally similar and specific. The AGO7 clade is involved in the biogenesis of ta-siRNAs by specifically binding miR390, and this is conserved in Arabidopsis (Montgomery et al., 2008), maize (Douglas et al., 2010) and rice (Nagasaki et al., 2007). Another example is the AGO4 clade, in which members bind endogenous 24-nt siRNAs and direct DNA methylation (Duan et al., 2015; Olmedo-Monfil et al., 2010; Qi et al., 2006; Wu et al., 2010; Zilberman et al., 2003). Furthermore, different members of the same clade may have functional specialization and/or redundancy. The four AGO1 homologs (OsAGO1a, OsAGO1b, OsAGO1c and OsAGO1d) in rice show preference and exclusion for given miRNAs (Wu et al., 2009), and OsAGO1a/b and OsAGO1c/d also show different abilities in antiviral defense (Wu et al., 2015). However, AtAGO3, which is not located in the same branch as AGO4, can also bind 24-nt sRNAs and partially supplement the function of AGO4 (Zhang et al., 2016). Therefore, the functions of AGOs are complex and cannot be simply classified according to the phylogenetic relationship.
Despite the potential lack of a one-to-one correlation between phylogenetic position and function, our studies provide insights that may guide future investigations of AGO function, particularly in non-model organisms. A basic understanding of the function of Arabidopsis AGO4 in RdDM was established; however, we found that there are three branches in AGO4 in dicots, while AGO4 and its paralogs in the AGO4 clade in Arabidopsis are only in AGO4-I (Figure 3). Whether AGO4-II and AGO4-III are also involved in RdDM remains to be investigated. This also raises the notion that studying AGOs only in Arabidopsis or a few model species is not sufficient. In addition, some AGO4-II members have different amino acids in canonical ‘DEDH/D’ catalytic tetrad (Figure 3). Solarium tuberosum AGO15 (StAGO15) is the closest homolog to S. lycopersicum Solyc03g11760, which belongs to AGO4-II (Figure 3). Its catalytic site has been replaced by G-E-Q-R, but it is unknown whether this affects its slicer activity (Liao et al., 2020). Its expression is elevated upon pathogen infection (Liao et al., 2020).
Previous phylogenetic studies were not able to distinguish AGO10a and AGO10b when using only Arabidopsis and rice as the representatives of dicots and monocots. Meanwhile, current research about AGO10 has mainly focused on AGO10a, with little being known about AGO10b. Here we could differentiate these two branches by including more data. In S. lycopersicum, SlAGO10a and SlAGO10b are expressed strongly under heat and salt stress conditions, respectively (Bai et al., 2012). AtAGO10 competes for miR165/166 with AGO1 to prevent miR165/166 from loading into AGO1 (Zhu et al., 2011). Whether AGO10b also has a similar function needs further investigation. Future studies should focus on species having both AGO10a and AGO10b genes to identify the functional differences between them.
EXPERIMENTAL PROCEDURES
Data sources and acquisition of sequences
A total 244 plant species were screened for AGOs. Among them, 238 have sequenced genomes and the genomic sequences were retrieved from public databases (Table S1), with the primary sources being NCBI (https://www.ncbi.nlm.nih.gov/), Phytozome v12.0 (https://phytozome.jgi.doe.gov/pz/portal.html) and Gigadb (www.gigadb.org). Six charophytic transcriptomes were collected from the OneKp database (https://db.cngb.org/onekp/) in order to obtain a larger representation of charophytes. The animal and fungal AGOs were reported (Swarts et al., 2014).
The putative AGOs were identified through hmmsearch. First, protein sequences of AGOs in A. thaliana, O. sativa, A. trichopoda, Selaginella moellendorffii and P. patens retrieved from Phytozome v12.0 were used as query sequences in hmmsearch with e-value <1-e5 to identify AGO homologs in other plants. Second, domain search is further performed on the output results by using the Simple Modular Architecture Research Tool (SMART) in normal mode (Letunic and Bork, 2018). Only sequences with PAZ and PIWI domains were retained. Finally, we removed sequences that have obvious errors. According to the database Phytozome, some genes contain multiple transcript isoforms due to alternative splicing. Thus, we chose the representative protein for the gene if it was annotated in Phytozome and, if not, the longest protein was selected. In total, 2958 AGO proteins were collected for further analysis.
Multiple sequence alignment (MSA) and phylogenetic analysis
The MSA of the above identified protein sequences was performed by mafft v7.455 using the parameters ‘L-INS-I’ (Katoh and Standley, 2013). As the poor quality in MSA due to variable lengths of numerous AGOs from different species may lead to wrong phylogenetic inferences, regions that do not encode the functional domains of AGOs were removed to improve the quality of alignment. Then, the aligned protein sequences were used to identify the corresponding coding nucleotide sequences, and the nucleotide alignment was used for subsequent phylogenetic analyses.
To explore the phylogenetic relationship of different species, we divided the alignment into three parts based on taxonomy and constructed phylogenetic trees separately: Dataset I, 29 non-angiosperms + 4 angiosperms; Dataset II, 190 angiosperms (containing 15 Poaceae species); Dataset III, 42 Poales including 40 Poaceae plants, 1 Cyperaceae plant and 1 Bromeliaceae plant + A. thaliana (Details in Table S2). The ML tree was constructed by RAxML-HPC tools with the parameter ‘-f a -m GTRGAMMAI’ and 1000 bootstrap replicates (Stamatakis, 2014). Because support in the phylogenetic tree of dataset I was low, we also reconstructed a phylogenetic tree using IQ-TREE2 with the parameter ‘-m MFP --epsilon 0.00001’ and 2000 ultra bootstrap replicates (Minh et al., 2020). The final tree was drawn by figtree, and proteins in each tree were labeled with the species names + protein name (tree.bio.ed.ac.uk/software/figtree).
Supplementary Material
Figure S2. Phylogenetic tree of plant AGOs constructed with AGOs from dataset I, six animals and seven fungi.
Figure S3. Phylogenetic tree of non-flowering plant AGOs constructed with dataset I using IQ-TREE2. The green branches indicate chlorophytes; blue, charophytes; orange, bryophytes; purple, lycophyte; light blue, fern; red, gymnosperms; black, angiosperms.
Figure S1. Phylogeny of green plants showing the species included in this study. Phylogeny was adapted from The Taxonomy Database in NCBI (www.ncbi.nlm.nih.gov/taxonomy/). Phylogeny of green plants is divided into two groups: (a) phylogeny of non-flowering plants; (b) phylogeny of flowering plants.
Figure S5. Phylogenetic tree of the AGO family in Brassicales. (a) AGO2/3 in Brassicales; (b) AGO4/8/9 in Brassicales.
Figure S6. A brief phylogenetic tree showing the AGO4/6 clade in angiosperms. AGO6 and AGO4 shared a common basal angiosperm origin, such that eudicots, monocots and magnoliidae can be found in both AGO6 and AGO4 branches.
Figure S7. Phylogenetic tree of Poaceae AGOs constructed with dataset III. The AGOs clustered into eight major groups. (a) AGO4; (b) AGO6; (c) AGO2; (d) AGO7; (e) AGO5; (f) AGO1; (g)AGO10b; (h) AGO18. Numbers at the nodes are bootstrap support values (from 0 to 100). The red stars indicate lineage-specific gene expansion events. The colored lines indicate out groups – red, Arabidopsis thaliana; blue, Ananas comosus (Bromeliaceae, Poales); and green, Kobresia littledalei (Cyperaceae, Poales). The black lines represent various Poaceae species.
Figure S4. Phylogenetic tree of flowering plant AGOs constructed with 2377 AGO protein sequences from dataset II. The AGOs clustered into eight major groups. A: AGO4; B: AGO6; C: AGO2; D: AGO7; E: AGO5; F: AGO1; G: AGO10a; H: AGO10b. Numbers at the nodes are bootstrap support values (from 0 to 100).
Table S1. Green plants, sources of sequence and genome version information used in this study
Table S2. Classification of AGOs used in this study
ACKNOWLEDGEMENTS
This project is supported by the Shenzhen Fundamental Research fund (20200812151848001, JCYJ20170818092637786), National Natural Science Foundation of China (89200138), Guangdong Innovation Research Team Fund (2014ZT05S078).
Footnotes
CONFLICT OF INTEREST
The authors declare no competing interests.
SUPPORTING INFORMATION
Additional Supporting Information may be found in the online version of this article.
DATA AVAILABILITY STATEMENT
All raw sequencing data were downloaded from public database. The detailed info could be found in Table S1.
REFERENCES
- Adenot X, Elmayan T, Lauressergues D, Boutet S, Bouche N, Gasciolli V et al. (2006) DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Current Biology, 16, 927–932. [DOI] [PubMed] [Google Scholar]
- Allen E, Xie Z, Gustafson AM & Carrington JC (2005) MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell, 121,207–221. [DOI] [PubMed] [Google Scholar]
- Arif MA, Frank W & Khraiwesh B (2013) Role of RNA interference (RNAi) in the moss physcomitrella patens. International Journal of Molecular Sciences, 14, 1516–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai M, Yang GS, Chen WT, Mao ZC, Kang HX, Chen GH et al. (2012) Genome-wide identification of Dicer-like, Argonaute and RNA-dependent RNA polymerase gene families and their expression analyses in response to viral infection and abiotic stresses in Solanum lycopersicum. Gene, 501, 52–62. [DOI] [PubMed] [Google Scholar]
- Baumberger N & Baulcombe DC (2005) Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proceedings of the National Academy of Sciences of the United States of America, 102, 11928–11933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges F & Martienssen RA (2015) The expanding world of small RNAs in plants. Nature Reviews Molecular Cell Biology, 16, 727–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges F, Pereira PA, Slotkin RK, Martienssen RA & Becker JD (2011) MicroRNA activity in the Arabidopsis male germline. Journal of Experimental Botany, 62, 1611–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosseau C & Moffett P (2015) Functional and genetic analysis identify a role for Arabidopsis ARGONAUTE5 in antiviral RNA silencing. The Plant Cell, 27, 1742–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonell A (2017) Plant ARGONAUTEs: features, functions, and unknowns. Methods in Molecular Biology, 1640,1–21. [DOI] [PubMed] [Google Scholar]
- Carbonell A & Carrington JC (2015) Antiviral roles of plant ARGO-NAUTES. Current Opinion in Plant Biology, 27,111–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S, Xian W, Fu Y, Marin B, Keller J, Wu T et al. (2019) Genomes of subaerial zygnematophyceae provide insights into Land plant evolution. Cell, 179, 1057–1067.e14. [DOI] [PubMed] [Google Scholar]
- Cibrian-Jaramillo A & Martienssen RA (2009) Darwin’s “Abominable Mystery”: the role of RNA interference in the evolution of flowering plants. Cold Spring Harbor Symposia on Quantitative Biology, 74, 267–273. [DOI] [PubMed] [Google Scholar]
- Douglas RN, Wiley D, Sarkar A, Springer N, Timmermans MC & Scanlon MJ (2010) Ragged seedling2 encodes an ARGONAUTE7-like protein required for mediolateral expansion, but not dorsiventrality, of maize leaves. The Plant Cell, 22, 1441–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan CG, Zhang HM, Tang K, Zhu XH, Qian WQ, Hou YJ et al. (2015) Specific but interdependent functions for Arabidopsis AGO4 and AGO6 in RNA-directed DNA methylation. EMBO Journal, 34, 581–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren N, Montgomery TA, Howell MD, Allen E, Dvorak SK, Alexander AL et al. (2006) Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Current Biology, 16, 939–944. [DOI] [PubMed] [Google Scholar]
- Fang X & Qi Y (2016) RNAi in plants: an Argonaute-Centered view. The Plant Cell, 28, 272–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank F, Hauver J, Sonenberg N & Nagar B (2012) Arabidopsis Argonaute MID domains use their nucleotide specificity loop to sort small RNAs. EMBO Journal, 31, 3588–3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M, Wei W, Li M-M, Wu Y-S, Ba Z, Jin K-X et al. (2014) Ago2 facilitates Rad51 recruitment and DNA double-strand break repair by homologous recombination. Cell Research, 24, 532–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Ruiz H, Carbonell A, Hoyer JS, Fahlgren N, Gilbert KB, Takeda A et al. (2015) Roles and programming of Arabidopsis ARGONAUTE proteins during Turnip mosaic virus infection. PLoS Path, 11, e1004755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S, Jin L, Huang Y, Zhang F & Kay MA (2012) Slicing-independent RISC activation requires the argonaute PAZ domain. Current Biology, 22, 1536–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey JJW, Lewsey MG, Kanu P, Jack W, Susanne H, Carr JP et al. (2011) An antiviral defense role of AGO2 in plants. PLoS One, 6, e14639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Lagana E, Rodríguez-Leal D, Lúa J & Vielle-Calzada JP (2016) A multigenic network of ARGONAUTE4 clade members controls early megaspore formation in Arabidopsis. Genetics, 204, 1045–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell MD, Fahlgren N, Chapman EJ, Cumbie JS, Sullivan CM, Givan SA et al. (2007) Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA- and tasiRNA-directed targeting. The Plant Cell, 19, 926–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G & Simard MJ (2008) Argonaute proteins: key players in RNA silencing. Nature Reviews Molecular Cell Biology, 9, 22–32. [DOI] [PubMed] [Google Scholar]
- Jaubert M, Bhattacharjee S, Mello AF, Perry KL & Moffett P (2011) ARGONAUTE2 mediates RNA-silencing antiviral defenses against Potato virus X in Arabidopsis. Plant Physiology, 156, 1556–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor M, Arora R, Lama T, Nijhawan A, Khurana JP, Tyagi AK et al. (2008) Genome-wide identification, organization and phylogenetic analysis of Dicer-like, Argonaute and RNA-dependent RNA Polymerase gene families and their expression analysis during reproductive development and stress in rice. BMC Genomics, 9, 451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K & Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution, 30, 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I & Bork P (2018) 20 years of the SMART protein domain annotation resource. Nucleic Acids Research, 46, D493–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Z, Hoden KP, Singh RK & Dixelius C (2020) Genome-wide identification of Argonautes in Solanaceae with emphasis on potato. Scientific Reports, 10, 20577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingel A, Simon B, Izaurralde E & Sattler M (2004) Nucleic acid 3′-end recognition by the Argonaute2 PAZ domain. Nature Structural & Molecular Biology, 11, 576–577. [DOI] [PubMed] [Google Scholar]
- Liu JD, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ et al. (2004) Argonaute2 is the catalytic engine of mammalian RNAi. Science, 305, 1437–1441. [DOI] [PubMed] [Google Scholar]
- Liu Q, Yao X, Pi L, Wang H, Cui X & Huang H (2009) The ARGONAUTE10 gene modulates shoot apical meristem maintenance and establishment of leaf polarity by repressing miR165/166 in Arabidopsis. The Plant Journal, 58, 27–40. [DOI] [PubMed] [Google Scholar]
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A et al. (2020) IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Molecular Biology and Evolution, 37, 1530–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar A, Schwach F, Studholme DJ, Thuenemann EC & Baulcombe DC (2007) MiRNAs control gene expression in the single-cell alga Chlamydomonas reinhardtii. Nature, 447, 1126–1129. [DOI] [PubMed] [Google Scholar]
- Montgomery TA, Howell MD, Cuperus JT, Li DW, Hansen JE, Alexander AL et al. (2008) Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell, 133, 128–141. [DOI] [PubMed] [Google Scholar]
- Morel JB, Godon C, Mourrain P, Beclin C, Boutet S, Feuerbach F et al. (2002) Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. The Plant Cell, 14, 629–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussian B, Schoof H, Haecker A, Jurgens G & Laux T (1998) Role of the ZWILLE gene in the regulation of central shoot meristem cell fate during Arabidopsis embryogenesis. EMBO Journal, 17, 1799–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasaki H, Itoh J-I, Hayashi K, Hibara K-I, Satoh-Nagasawa N, Nosaka M et al. (2007) The small interfering RNA production pathway is required for shoot meristem initiation in rice. Proceedings of the National Academy of Sciences of the United States of America, 104, 14867–14871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura A, Ito M, Kamiya N, Sato Y & Matsuoka M (2002) OsPNH1 regulates leaf development and maintenance of the shoot apical meristem in rice. The Plant Journal, 30, 189–201. [DOI] [PubMed] [Google Scholar]
- Nishiyama T, Sakayama H, de Vries J, Buschmann H, Saint-Marcoux D, Ullrich KK et al. (2018) The chara genome: secondary complexity and implications for plant terrestrialization. Cell, 174(2), 448–464.e24. [DOI] [PubMed] [Google Scholar]
- Olmedo-Monfil V, Duran-Figueroa N, Arteaga-Vazquez M, Demesa-Arevalo E, Autran D, Grimanelli D et al. (2010) Control of female gamete formation by a small RNA pathway in Arabidopsis. Nature, 464, 628–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JS, Roe SM & Barford D (2005) Structural insights into mRNA recognition from a PIWI domain-siRNA guide complex. Nature, 434, 663–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Denli AM & Hannon GJ (2005) Biochemical specialization within Arabidopsis RNA silencing pathways. Molecular Cell, 19, 421–428. [DOI] [PubMed] [Google Scholar]
- Qi Y, He X, Wang XJ, Kohany O, Jurka J & Hannon GJ (2006) Distinct catalytic and non-catalytic roles of ARGONAUTE4 in RNA-directed DNA methylation. Nature, 443, 1008–1012. [DOI] [PubMed] [Google Scholar]
- Qian Y, Cheng Y, Cheng X, Jiang H, Zhu S & Cheng B (2011) Identification and characterization of Dicer-like, Argonaute and RNA-dependent RNA polymerase gene families in maize. Plant Cell Reports, 30, 1347–1363. [DOI] [PubMed] [Google Scholar]
- Rensing SA (2018) Great moments in evolution: the conquest of land by plants. Current Opinion in Plant Biology, 42, 49–54. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Leal D, Castillo-Cobian A, Rodriguez-Arevalo I & Vielle-Calzada JP (2016) A Primary sequence analysis of the ARGONAUTE protein family in plants. Frontiersin Plant Science, 7, 1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers K & Chen X (2013) Biogenesis, turnover, and mode of action of plant microRNAs. The Plant Cell, 25, 2383–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussin-Léveillée C, Silva-Martins G & Moffett P (2020) ARGONAUTE5 represses age-dependent induction of flowering through physical and functional interaction with miR156 in Arabidopsis. Plant and Cell Physiology, 61, 957–966. [DOI] [PubMed] [Google Scholar]
- Sabbione A, Daurelio L, Vegetti A, Talón M, Tadeo F & Dotto M (2019) Genome-wide analysis of AGO, DCL and RDR gene families reveals RNA-directed DNA methylation is involved in fruit abscission in Citrus sinensis. BMC Plant Biology, 19, 401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck J, Gursinsky T, Pantaleo V, Burgyan J & Behrens SE (2013) AGO/RISC-mediated antiviral RNA silencing in a plant in vitro system. Nucleic Acids Research, 41, 5090–5103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RK, Gase K, Baldwin IT & Pandey SP (2015) Molecular evolution and diversification of the Argonaute family of proteins in plants. BMC Plant Biology, 15, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JJ, Liu JD, Tolia NH, Schneiderman J, Smith SK, Martienssen RA et al. (2003) The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Natural Structural Biology, 10, 1026–1032. [DOI] [PubMed] [Google Scholar]
- Song JJ, Smith SK, Hannon GJ & Joshua-Tor L (2004) Crystal structure of Argonaute and its implications for RISC slicer activity. Science, 305, 1434–1437. [DOI] [PubMed] [Google Scholar]
- Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics, 30, 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarts DC, Makarova K, Wang YL, Nakanishi K, Ketting RF, Koonin EV et al. (2014) The evolutionary journey of Argonaute proteins. Nature Structural & Molecular Biology, 21, 743–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker MR, Okada T, Hu Y, Scholefield A, Taylor JM & Koltunow AM (2012) Somatic small RNA pathways promote the mitotic events of megagametogenesis during female reproductive development in Arabidopsis. Development, 139, 1399–1404. [DOI] [PubMed] [Google Scholar]
- Vaucheret H (2008) Plant ARGONAUTES. Trends in Plant Science, 13, 350–358. [DOI] [PubMed] [Google Scholar]
- Vaucheret H, Vazquez F, Crété P & Bartel DP (2004) The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes & Development, 18, 1187–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XB, Jovel J, Udomporn P, Wang Y, Wu Q, Li WX et al. (2011) The 21-nucleotide, but not 22-nucleotide, viral secondary small interfering RNAs direct potent antiviral defense by two cooperative argonautes in Arabidopsis thaliana. The Plant Cell, 23, 1625–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Yang Z, Wang YU, Zheng L, Ye R, Ji Y et al. (2015) Viralinducible Argonaute18 confers broad-spectrum virus resistance in rice by sequestering a host microRNA. eLife, 4, e05733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Zhang Q, Zhou H, Ni F, Wu X & Qi Y (2009) Rice microRNA effector complexes and targets. The Plant Cell, 21, 3421–3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Zhou H, Zhang Q, Zhang J, Ni F, Liu C et al. (2010) DNA methylation mediated by a microRNA pathway. Molecular Cell, 38, 465–475. [DOI] [PubMed] [Google Scholar]
- Yan KS, Yan S, Farooq A, Han A, Zeng L & Zhou MM (2003) Structure and conserved RNA binding of the PAZ domain. Nature, 426, 468–474. [DOI] [PubMed] [Google Scholar]
- You CJ, Cui J, Wang H, Qi XP, Kuo LY, Ma H et al. (2017) Conservation and divergence of small RNA pathways and microRNAs in land plants. Genome Biology, 18, 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YU, Ji L, Le BH, Zhai J, Chen J, Luscher E et al. (2021) Correction: ARGONAUTE10 promotes the degradation of miR165/6 through the SDN1 and SDN2 exonucleases in Arabidopsis. PLoS Biology, 19, e3001120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Fan LS, Le BH, Ye PY, Mo BX & Chen XM (2020) Regulation of ARGONAUTE10 expression enables temporal and spatial precision in axillary meristem initiation in Arabidopsis. Developmental Cell, 55(5), 603–616.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XM, Zhao HW, Gao S, Wang WC, Katiyar-Agarwal S, Huang HD et al. (2011) Arabidopsis Argonaute 2 regulates innate immunity via miRNA393*-mediated silencing of a golgi-localized SNARE gene, MEMB12. Molecular Cell, 42, 356–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Liu X, Guo X, Wang XJ & Zhang X (2016) Arabidopsis AGO3 predominantly recruits 24-nt small RNAs to regulate epigenetic silencing. Nat Plants, 2, 16049. [DOI] [PubMed] [Google Scholar]
- Zheng X, Zhu J, Kapoor A & Zhu JK (2007) Role of Arabidopsis AGO6 in siRNA accumulation, DNA methylation and transcriptional gene silencing. EMBO Journal, 26, 1691–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Honda M, Zhu H, Zhang Z, Guo X, Li T et al. (2015) Spatiotemporal sequestration of miR165/166 by Arabidopsis Argonaute10 promotes shoot apical meristem maintenance. Cell Reports, 10, 1819–1827. [DOI] [PubMed] [Google Scholar]
- Zhu HL, Hu FQ, Wang RH, Zhou X, Sze SH, Liou LW et al. (2011) Arabidopsis Argonaute10 specifically sequesters miR166/165 to regulate shoot apical meristem development. Cell, 145, 242–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D, Cao X & Jacobsen SE (2003) ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science, 299, 716–719. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S2. Phylogenetic tree of plant AGOs constructed with AGOs from dataset I, six animals and seven fungi.
Figure S3. Phylogenetic tree of non-flowering plant AGOs constructed with dataset I using IQ-TREE2. The green branches indicate chlorophytes; blue, charophytes; orange, bryophytes; purple, lycophyte; light blue, fern; red, gymnosperms; black, angiosperms.
Figure S1. Phylogeny of green plants showing the species included in this study. Phylogeny was adapted from The Taxonomy Database in NCBI (www.ncbi.nlm.nih.gov/taxonomy/). Phylogeny of green plants is divided into two groups: (a) phylogeny of non-flowering plants; (b) phylogeny of flowering plants.
Figure S5. Phylogenetic tree of the AGO family in Brassicales. (a) AGO2/3 in Brassicales; (b) AGO4/8/9 in Brassicales.
Figure S6. A brief phylogenetic tree showing the AGO4/6 clade in angiosperms. AGO6 and AGO4 shared a common basal angiosperm origin, such that eudicots, monocots and magnoliidae can be found in both AGO6 and AGO4 branches.
Figure S7. Phylogenetic tree of Poaceae AGOs constructed with dataset III. The AGOs clustered into eight major groups. (a) AGO4; (b) AGO6; (c) AGO2; (d) AGO7; (e) AGO5; (f) AGO1; (g)AGO10b; (h) AGO18. Numbers at the nodes are bootstrap support values (from 0 to 100). The red stars indicate lineage-specific gene expansion events. The colored lines indicate out groups – red, Arabidopsis thaliana; blue, Ananas comosus (Bromeliaceae, Poales); and green, Kobresia littledalei (Cyperaceae, Poales). The black lines represent various Poaceae species.
Figure S4. Phylogenetic tree of flowering plant AGOs constructed with 2377 AGO protein sequences from dataset II. The AGOs clustered into eight major groups. A: AGO4; B: AGO6; C: AGO2; D: AGO7; E: AGO5; F: AGO1; G: AGO10a; H: AGO10b. Numbers at the nodes are bootstrap support values (from 0 to 100).
Table S1. Green plants, sources of sequence and genome version information used in this study
Table S2. Classification of AGOs used in this study
Data Availability Statement
All raw sequencing data were downloaded from public database. The detailed info could be found in Table S1.


