Abstract
Objective
Military servicemen deployed to war zones are at increased risk of developing posttraumatic stress disorder (PTSD) and successful adaptation to stress is important. Epigenetic alterations in response to trauma have been identified as mechanism of adaptation and may therefore predict deployment-related PTSD symptoms. To date, human studies of epigenetic marks for traumatic stress have been largely constrained by short-term analyses of one or two time points.
Method
This study in a prospective Dutch military cohort (N = 125) examined longitudinal changes of DNA methylation profiles before, as well as one and six months after deployment-related combat exposure in relation to the development of PTSD symptoms over a period of up to five years after deployment. We investigated the predictive value of specific methylation changes for immediate and delayed-onset PTSD symptoms and recovery. This epigenetic prediction was compared to polygenic risk score predictions obtained from the currently available largest genome-wide association study of PTSD.
Results
A total of fourteen genomic regions were identified in which PTSD symptom levels were associated with methylation changes over time (pre-deployment, one, and six months post-deployment). Of these regions, four were significant determinants of longitudinal development of PTSD symptoms. In addition, we observed that, together with risk level during deployment (operating inside or outside the military base) and physical childhood trauma, post-deployment decreases in methylation at a genomic region in EP300/miRNA1281 was associated with a delayed onset of PTSD compared to a resilient profile. Polygenic risk, in contrast, was related to PTSD onset within six months after deployment but was not associated with long term outcomes.
Conclusion
The present study suggests predictive utility of changes in DNA methylation for the subsequent development of PTSD symptoms and showed that the currently available measure of polygenic risk is primarily related to non-delayed disease onset.
Keywords: DNA methylation, EP300, Epigenetics, miRNA1281, Posttraumatic stress disorder, Prediction
Highlights
-
•
First study of methylation changes over a deployment period using three time-points.
-
•
Post-deployment methylation decreases in EP300 were associated with delayed onset PTSD.
-
•
Genetic risk was exclusively related to a non-delayed PTSD onset.
1. Introduction
Epigenetic modifications in response to trauma or severe stress may be a critical factor in risk or resilience to stress-related disorders. They reflect the complex interplay between environment and genes, and could therefore be one of the mechanisms in the pathway between trauma and the development of posttraumatic stress disorder (PTSD). This interplay is particularly relevant for military populations, as they regularly encounter stressful events during deployment and show a high burden of PTSD following deployment [1,2,3]. One of the best characterized mechanisms of epigenetic regulation is DNA methylation, and mounting evidence from animal models and human clinical studies suggests that changes to DNA methylation resulting from trauma are associated with PTSD (reviewed in: [4,5,6]). Candidate gene as well as epigenome-wide studies have highlighted genes involved in the immune system and HPA axis that are involved in PTSD [5]. However, human studies have been largely constrained by relatively short-term analyses of post-trauma symptoms, generally up to six to twelve months after deployment.
Moreover, genome-wide epigenetic studies in PTSD that use longitudinal data are scarce. The largest study on methylation changes so far suggests the implication of immune-related genes in the human leukocyte antigen region, HEXDC, and MAD1L1, a gene previously associated with PTSD [7]. A genome-wide DNA methylation study of our group in a Dutch military sample pinpointed novel genomic regions where decreases in blood DNA methylation across a period of exposure to combat trauma were related to increasing levels of PTSD symptoms over a six-month period. Targeted analyses of these findings replicated the observed association at the genomic regions in ZFP57, RNF39, and HIST1H2APS2 in an independent prospective military cohort of US marines [8]. ZFP57 methylation was also shown to reverse following successful PTSD treatment, which provides further support for the association of decreased methylation of ZFP57 to symptoms of PTSD [9].
There are good reasons to investigate the relation between DNA methylation changes in more intervals around the trauma exposure and development of PTSD symptoms in the short term and longer follow up. Firstly, it has the potential to capture the dynamics of DNA methylation changes during deployment and immediately after return for the identification of genes and genetic pathways that are related to PTSD and response to trauma. Secondly, it enables study of how these dynamic changes are related to short and longer term outcomes. A third reason is the potential for the prediction of PTSD, since there are currently no clear biological measures that can be used to screen individuals for an increased vulnerability to develop PTSD symptoms after deployment. Studies by others and our group show that PTSD can develop with a latency of months to several years, as demonstrated by identification of a delayed onset PTSD developmental trajectory in addition to resilient and recovery trajectories [1,10,11]. Routine screening for PTSD usually discontinues after one or two years post-deployment. Identification of biological markers reflecting vulnerability for delayed onset PTSD may therefore have an important role for prevention and early intervention. Another relevant question is how such epigenetic changes compare to genetic prediction for PTSD. In the past years substantial progress has been made to illuminate the role of genes in PTSD susceptibility leading to genome wide significant identification of risk genes [12]. The question remains, however, how these risk genes are related to longer term PTSD outcomes.
The current study is, to our knowledge, the first to investigate longitudinal changes of DNA methylation profiles across a period of combat exposure using three time points (pre-deployment, one month- and six months post-deployment) in relation to the development of PTSD symptoms in a cohort of deployed military servicemen. In order to assess the predictive value of methylation patterns for the development of PTSD symptoms over time, we identified genetic regions where methylation changes are related to changes in PTSD symptoms and used these to predict developmental trajectories over a five-year follow-up period. Because of the higher clinical relevance of identifying a predictive biomarker for the development of PTSD symptoms before PTSD symptomatology is present and the limitation that methylation changes can only be determined after deployment, the focus in these analyses was on predicting delayed onset of PTSD symptoms years after deployment. Identification of such a biomarker for late-onset PTSD may be very useful for targeted screening and early intervention. Finally, we compared predictions based on methylation changes to that of polygenic risk scores (PRS), a measure for one’s genetic liability to PTSD.
2. Materials and methods
2.1. Discovery data set
2.1.1. Participants
Samples are from a subset of participants from the Prospective Research in Stress-related Military Operations (PRISMO study). PRISMO is a large prospective cohort study on the development and biological underpinnings of stress-related mental health symptoms in Dutch military personnel deployed to Afghanistan for at least four months between 2005 and 2008 [13]. The current study draws on peripheral blood samples from 125 PRISMO participants obtained one month before deployment and one and six months after the deployment period, and survey data obtained at six different time points spread out over five years (Fig. 1). No blood samples were available for the one year-, two year-, and five year follow-up measurement. A subset of PRISMO study participants was pre-selected based on two criteria: 1) available DNA, and 2) prioritization of participants who developed PTSD at any of the time points.
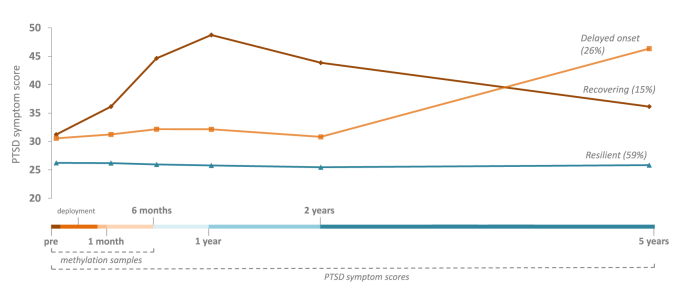
Fig. 1.
Schematic illustration of the overall study design and the latent developmental trajectories of self-reported posttraumatic stress symptoms as measured by the Self-Report Inventory for PTSD (SRIP) over the study’s time period.
Note: Latent developmental trajectories and symptom scores are based on the full PRISMO sample as described in Ref. [1]; trajectory membership percentages are based on the sample of the present study; PTSD = posttraumatic stress disorder; methylation samples were available at the first three measurement points, PTSD symptom scores were available at all measurement points (pre-deployment up to five years post-deployment).
PTSD symptoms were assessed at six different time points (pre-deployment, one month-, six month-, one year-, two year- and five year post-deployment) using the Self-Report Inventory for PTSD (SRIP), a questionnaire with good internal consistency, discriminant validity and concurrent validity with other PTSD measures [14,15]. As recommended in the literature, a cut-off score of 38 was used to indicate substantial PTSD symptoms [15]. No subjects scored above cut-off pre-deployment. At the follow-up measurements at one and six months post-deployment, respectively, 29 subjects and 30 subjects scored above cut-off. Three trajectories of posttraumatic stress symptoms (resilient, recovering, and delayed onset, see Fig. 1) from pre-deployment up to five year post-deployment were previously identified in a latent growth mixture model using the full PRISMO sample (N = 960), as described in Ref. [1]. The model included a group with a low and stable PTSD trajectory (i.e. resilient), a group that had a moderate level of PTSD symptoms that increased heavily in the last time period (i.e. delayed onset), and a group that had increasing symptoms in the first year after deployment and then showed a recovery process (i.e. recovering) (Fig. 1). Of note are the elevated symptom levels pre-deployment in the delayed onset and recovering trajectory.
Exposure to traumatic stress during deployment was measured with a 19-item deployment experience checklist, the Deployment Experience Scale (DES), which covered a range of potentially traumatic experiences that can occur during deployment [16]. Childhood trauma was assessed using the Dutch version of the Early Trauma Inventory Self Report-Short Form (ETISR-SF) [17]. Demographics and other characteristics of the participants are described in Table 1. Participants received financial compensation for participation. Written informed consent was obtained from all participants in accordance with procedures approved by the Institutional Review Board of the University Medical Center Utrecht.
Table 1.
Demographics and clinical characteristics of the full PRISMO sample and the different PTSD developmental trajectories.
| All (N = 125) | Resilient (N = 74) | Recovering (N = 19) | Delayed onset (N = 32) | p-value | |
|---|---|---|---|---|---|
| Gender (%) | |||||
| Male | 92.8 | 94.6 | 89.5 | 90.6 | 0.559 |
| Female | 7.2 | 5.4 | 10.5 | 9.4 | |
| Age (SD) | 27.3 (8.9) | 27.1 (8.7) | 26.6 (8.5) | 28.2 (9.7) | 0.797 |
| Educational level (%) † | |||||
| Low | 40.0 | 38.4 | 41.2 | 43.3 | 0.622 |
| Moderate | 52.5 | 56.2 | 52.9 | 43.3 | |
| High | 7.5 | 5.5 | 5.9 | 13.3 | |
| Rank (%) | |||||
| Private | 44.0 | 44.6 | 47.4 | 40.6 | 0.897 |
| Corporal | 23.2 | 24.3 | 26.3 | 18.8 | |
| Non-commissioned officer | 24.8 | 24.3 | 15.8 | 31.3 | |
| Staff officer | 8.0 | 6.8 | 10.5 | 9.4 | |
| Previous deployment(s) (% yes) | 46.2 | 45.1 | 44.4 | 50.0 | 0.899 |
| Function (%) | |||||
| Inside the military base | 21.6 | 17.1 | 17.6 | 34.5 | 0.045 |
| Outside the military base | 67.2 | 75.7 | 70.6 | 44.8 | |
| Both inside and outside the military base | 11.2 | 7.1 | 11.8 | 20.7 | |
| Deployment year (%) | |||||
| 2005 or 2006 | 13.6 | 13.5 | 15.8 | 12.5 | 0.936 |
| 2007 or 2008 | 86.4 | 86.5 | 84.2 | 87.5 | |
| New deployment(s) (% yes) | 22.1 | 22.0 | 22.2 | 22.2 | 1.000 |
| Deployment stressor score (SD) ‡ | 6.4 (3.6) | 6.5 (3.5) | 7.7 (3.0) | 5.5 (3.9) | 0.142 |
| Childhood trauma score (SD) ¶ | 3.9 (3.4) | 3.6 (2.9) | 4.1 (3.5) | 4.8 (4.3) | 0.270 |
Note: data are % or mean (SD). Differences in descriptive characteristics between participants in the different trajectories were tested with one-way ANOVA (continuous) or Fisher’s Exact (categorical). † Education (International Standard Classification of Education levels): Low = primary and lower secondary education; Moderate = upper secondary, postsecondary non-tertiary, and short cycle tertiary education; High = bachelor, master, and doctoral education; ‡ Deployment stressor score measured with the Deployment Experience Scale; ¶ Childhood trauma score measured with the Early Trauma Inventory Self Report-Short Form; SD = standard deviation.
2.1.2. DNA isolation genotyping and methylation quantification
For genotyping, DNA was isolated from whole blood obtained via venipuncture using standard protocol. DNA concentration and quality were examined using Nanodrop (Thermo Fisher Scientific, MA, USA).
Genotyping was conducted using Illumina Human OmniExpress 24 v1.1. DNA for the methylation assay was quantified fluorescently prior to bisulfite conversion (Zymo Research, CA, USA). Genome-wide DNA methylation was interrogated using the Infinium Methylation EPIC BeadChip (Illumina, Inc., CA, USA). Batches were minimized by putting the three time points of one participant on the same array and equally distributing PTSD status over the arrays. Also, batches were minimized using information from the control probes as implemented in the functional normalization procedure of Meffil [18]. The dataset was preprocessed in R version 3.3.3 with the meffil package [18], using functional normalization [19]. There were no samples with fewer than three beads in 20% of the probes. Single nucleotide polymorphism (SNP) profile included on the array matched their genetic identity. Nine samples had to be removed, five because of failed hybridization as indicated by outliers (3 SD from the methylation mean), one outside the predefined boundaries of the control probe, one for gender mismatch, and two for gender estimate outlier. 1152 probes with a detection p-value greater than 0.01 were removed. Non-specific probes and those with SNPs in the probe sequence were removed [20,21]. After quality control, 864,528 CpGs in 361 samples and 133 different individuals were left for further analysis. The level of DNA methylation is expressed as a ‘beta’ value ranging from 0 (no cytosine methylation) to 1 (complete cytosine methylation). Analyses were performed using M-values (log2 ratio of beta values) [22].
2.2. Replication data set
Replication of the identified DMRs was sought in the Marine Resiliency Study (MRS) [23]. MRS is a large prospective PTSD study with a longitudinal follow-up in a cohort of 2599 marines deployed to either Iraq or Afghanistan. Measurements were obtained approximately one month pre-deployment and one week, three months and six months post-deployment. PTSD symptoms were measured using a structured diagnostic interview and the Clinician Administered PTSD Scale (CAPS) [24]. Peripheral blood samples were collected pre-deployment and three and six months post-deployment. A subset of 128 men was selected for DNA methylation analysis, with a mean age at baseline of 22 years. The participants showed no PTSD diagnoses (CAPS ≤ 25) pre-deployment. In the follow-up measurements at three months and six months post-deployment, respectively 51 and 36 participants were diagnosed with PTSD. The institutional review boards of the University of California San Diego, VA San Diego Research Service, and Naval Health Research Center approved the study. Written informed consent was obtained from all participants.
Genome-wide DNA methylation levels were assessed in DNA extracted from whole blood using the Infinium HumanMethylation450 array. Baseline and follow-up samples were positioned differently between studies. Methylation level βs were calculated using the Minfi package and normalized to correct for type-I and II probe design bias using the BMIQ procedure implemented in watermelon. Batch and plate effects were removed using COMBAT. Relative proportions of cell compositions were estimated to account for cellular heterogeneity in blood-derived samples using the Minfi package.
2.3. Polygenic risk scores
PRS of PTSD were calculated for each subject based on the Psychiatric Genomics Consortium PTSD (PGC-PTSD) Freeze 2 European ancestry GWAS [12], using PRSice version 2.2.11.b [25], but with the PRISMO samples being left out of the GWAS meta-analysis. We selected SNPs associated at the optimal p-value threshold of 0.45 or lower.
2.4. Statistical analysis
An overview of the statistical analyses can be found in Table 2. For the methylation analysis, independent surrogate variables (ISVA) were calculated as implemented in Meffil to adjust for technical batch effects. Cell type composition was estimated using the Houseman algorithm [26]. Inspection of the potential confounding was performed using the surrogate variables and their correlation to known confounders (genetic ancestry, cell type composition, age, smoking, and gender). Optimal fit was obtained based on qq-plotting to avoid type I error inflation. In the optimal model, three ISVA’s were included that effectively accounted for technical batches (see Appendix Fig. A1) alongside five cell types, age, smoking, gender, and two genetic principal components. To identify differentially methylated positions (DMPs), longitudinal analyses were conducted using DNA methylation levels (one month pre-deployment (T0) and one (T1) and six months (T2) post-deployment) as the outcome and SRIP scores (T0, T1 and T2) as a determinant in a mixed model. Baseline SRIP score (T0), the time variable, an interaction term between time and SRIP scores, and the known confounders were included in the model. The interaction term was included to assess whether the association between methylation level and PTSD scores significantly changed over time. A random intercept was used to account for the variance between participants. The QQ-plot of the expected p-values versus the observed values and a lambda of 0.989 (see Appendix Fig. A2) indicated absence of type-I error inflation and no artificial differences between groups. False discovery rate p-values were calculated according to the Benjamini-Hochberg method (p < 0.05). The assumptions of the linear regression mixed models were evaluated by inspecting the distribution of residuals for the identified loci. Differentially methylated regions (DMRs) were calculated based on the p-values for each methylation locus using the DMRcate package [27]. A DMR consists of a strongly associated locus (p < 0.0001) and several other significantly associated loci within the proximity of 1000 base pairs. The furthest loci define the borders (start and stop location) of a DMR.
Table 2.
Overview statistical analyses.
| Analysis | Variables | Statistical model | Cohort/data set |
|---|---|---|---|
| 1. Discovery DMPs | Outcome: DNA methylation levels (T0, T1, T2) | Mixed model | PRISMO |
| Predictor: PTSD score (T0, T1, T2) | |||
| 2. Identification DMRs and PRS as determinants | Outcome: PTSD score (T0, T1, T2) | Mixed model | PRISMO |
| Predictor: | |||
| a. DMR methylation levels (T0, T1, T2) | |||
| b. PRS | |||
| 3. Replication DMRs | Outcome: PTSD score (T0, T1, T2) | Mixed model | MRS |
| Predictor: DMR methylation levels (T0, T1, T2) | |||
| 4. Correlations DMRs, PRS, and PTSD symptoms | a. DMR change score (T0-T1 or T1-T2) x PTSD score (T0 to T5) |
Pearson’s correlation | PRISMO |
| b. DMR change score x PRS | |||
| c. PRS x PTSD score (T0 to T5) | |||
| 5. Association DMRs and PRS with PTSD trajectories | Outcome: PTSD trajectory | Multinomial logistic regression | PRISMO (imputed) |
| Predictor: | |||
| a. DMR change score | |||
| b. PRS | |||
| 6. Prediction model delayed onset PTSD | Outcome: delayed PTSD trajectory (vs. resilient trajectory) | Stepwise backward logistic regression model | PRISMO (imputed) |
| Predictors: DMR change score, variables in Table 1 |
Note: DMP = differentially methylation position; PTSD = posttraumatic stress disorder; PRS = polygenic risk score.
Standardized methylation levels (z-values) at T0, T1, and T2 of identified DMRs of the first set of analyses were then used as determinants of longitudinal development of PTSD symptom scores over the three time points in a mixed model. Goodness of fit of the models was determined using loglikelihood-ratio-tests. The optimal fitting models included a random intercept and random slope, and assumed a quadratic development over time. Genetic ancestry (two genetic principal components), cell type composition (five cell types), age, smoking, and gender were used as covariates. To assess methylation scores as determinant for PTSD symptom development, a model with and without interaction terms between time and methylation scores and time2 and methylation scores were compared on goodness of fit using a loglikelihood-ratio-test. A p-value < 0.05 was considered statistically significant. A similar procedure was followed to assess PRS as a determinant of PTSD symptom scores over time in a separate model; age and gender were used as covariates in the mixed model, and goodness of fit was compared between the models with and without interactions between time and PRS.
To assess the robustness of the identified DMRs in an independent dataset, the methylation scores of the DMRs were tested as determinants of longitudinal development of PTSD symptom scores in the MRS data set using the same mixed model analysis with a quadratic term for time and a random intercept and random slope. The included covariates in the models were five cell types, three ancestry-related principal components (based on previous MRS analyses), age, and smoking. Goodness of fit was compared between the models with and without interactions between the time variables and methylation scores.
For follow up analysis of the significant DMRs in the discovery and validation analysis, DNA methylation levels at each time point were adjusted for cell type composition by computing the residuals in a linear regression model, and corresponding methylation change scores were calculated (T0-T1, T1-T2) where a positive change score indicated a decrease in methylation level, and a negative change score indicated an increase in methylation level. To assess whether DMR change scores were correlated to PTSD scores at specific time points, Pearson’s correlations between the methylation change scores and PTSD symptom scores at six time points (pre-deployment till five-year post-deployment) were calculated. In addition, correlations between PRS and PTSD symptom scores, and PRS and methylation change scores were calculated. Standardized methylation change scores and PRS were then assessed as determinants for PTSD developmental trajectories over the five-year follow-up period using separate multinomial logistic regression models in SPSS using an imputed data set (see section 2.5). To assess the potential utility of DNA methylation change scores and PRS as biological markers for a late onset of PTSD, the DMR methylation change scores and PRS that were significantly associated with the delayed PTSD trajectory were tested in a stepwise backward logistic regression model to predict a delayed onset PTSD versus a resilient profile using SAS version 9.4. Several demographic and psychological factors that are known from the literature to possibly relate to changes in post-traumatic stress symptoms (for a review [28]: were included in the full model, and can be found in Table 1. All variables measured on a continuous scale were standardized using z-scores. Variables with a p-value > 0.10 were eliminated step-by-step. Firth correction was applied for bias-reduction of the maximum likelihood estimates. Long-term follow-up data on PTSD symptom scores and trajectories were not yet available in the MRS data set and therefore replication of the association with delayed-onset PTSD symptoms could not be obtained.
2.5. Imputation of missing data
Missing values in the PRISMO dataset were assumed to be missing at random, and were managed using data imputation (see Appendix Table A.1). As multiple imputation is not suitable for a stepwise selection approach, single Bayesian stochastic regression imputation using fifty iterations was performed in SPSS. The imputation model included all the predictor variables (and covariates) that were used in the multinomial logistic regression analyses, as well as the outcome variable (PTSD symptom trajectory).
3. Results
3.1. Genome-wide DNA methylation and polygenic risk in relation to PTSD symptom development
Analyses identified fourteen DMRs in which PTSD symptom levels were associated with changes in DNA methylation (see Appendix Table A.2). Of these DMRs, four were significant determinants of longitudinal development of PTSD symptom scores in the optimized mixed models (DMR1, DMR2, DMR6, and DMR7; Table 3). DMR1 was located in or near the transcription start sites of the TUBA3FP pseudogene and P2RX6 gene, DMR 2 in or near the EP300 and miRNA1281 genes, and DMR6 in or near the IMPA1 gene. DMR7 was not located in or near any transcription start sites. The direction of effect was negative for all loci in DMR1, DMR2, and DMR7, indicating that decreased DNA methylation levels at the DMRs were associated with increased PTSD symptom scores over time. The effect was positive for DMR6, indicating that increased methylation was associated with increased PTSD symptom scores over time. PRS was not a significant determinant of longitudinal PTSD symptom scores (p = 0.446).
Table 3.
List of differentially methylated regions (DMRs) that were significant determinants of longitudinal development of PTSD symptom score.
| Chromosomal position of DMR | Genes | Probes | P-value (Stouffer method) | P-value in determinant analysis | |
|---|---|---|---|---|---|
| DMR1 | chr22: 21368603, 21368765 |
TUBA3FP P2RX6 |
cg06912512 cg19789653 cg09481857 cg21014483 cg01038149 |
8.84 E−05 | 0.002 |
| DMR2 | chr22: 41487073, 41487283 | EP300 miR1281 | cg00500400 cg08131204 |
1.18 E−06 | 0.016 |
| DMR6 | chr8 82598501, 82598664 |
IMPA1 | cg05798523 cg23402311 cg03588978 cg04364718 cg12093930 |
1.87 E−06 | 0.033 |
| DMR7 | chr8: 144973617, 144973638 | - | cg26529963 cg03000485 |
9.19 E−05 | 0.040 |
Note: the column on determinant analysis provides the P-values for the analyses in which methylation levels were tested as determinants of longitudinal development of PTSD symptom scores; PTSD = posttraumatic stress disorder; DMR = differentially methylated region.
3.2. Replication of DMRs related to PTSD symptom development
Replication failed for the identified DMRs in the independent MRS data set. Longitudinal changes in DNA methylation were not significantly associated with longitudinal changes in PTSD symptom scores at DMR1 (p = 0.873), DMR2 (p = 0.725), DMR6 (p = 0.085), and DMR7 (p = 0.535).
3.3. Correlation DNA methylation, polygenic risk, and PTSD symptoms
Of the four DMRs at which changes in methylation levels were significant determinants of changes in PTSD symptom scores (section 3.1), the methylation change between T1 (one month post-deployment) and T2 (six months post-deployment) at DMR1 was positively correlated with PTSD symptoms at T1 (one month post-deployment; r = 0.348, p = 0.001), T2 (six months post-deployment; r = 0.244, p = 0.020) and T6 (five years post-deployment; r = 0.281, p = 0.027). Methylation change between T1 and T2 at DMR2 was positively correlated with PTSD symptoms at T2 (r = 0.219, p = 0.038) and T6 (five years post-deployment; r = 0.326, p = 0.010). DMR6 methylation change between T1 and T2 was negatively correlated with PTSD symptoms at T1. Methylation changes in DMR7 were not correlated with PTSD symptoms, nor were methylation changes between T0 (pre-deployment) and T1 at any of the DMRs. PRS was only correlated with PTSD symptoms at T2 (r = 0.218, p = 0.032). There were no correlations between PRS and DMR methylation changes.
3.4. Association methylation changes and polygenic risk with PTSD trajectories
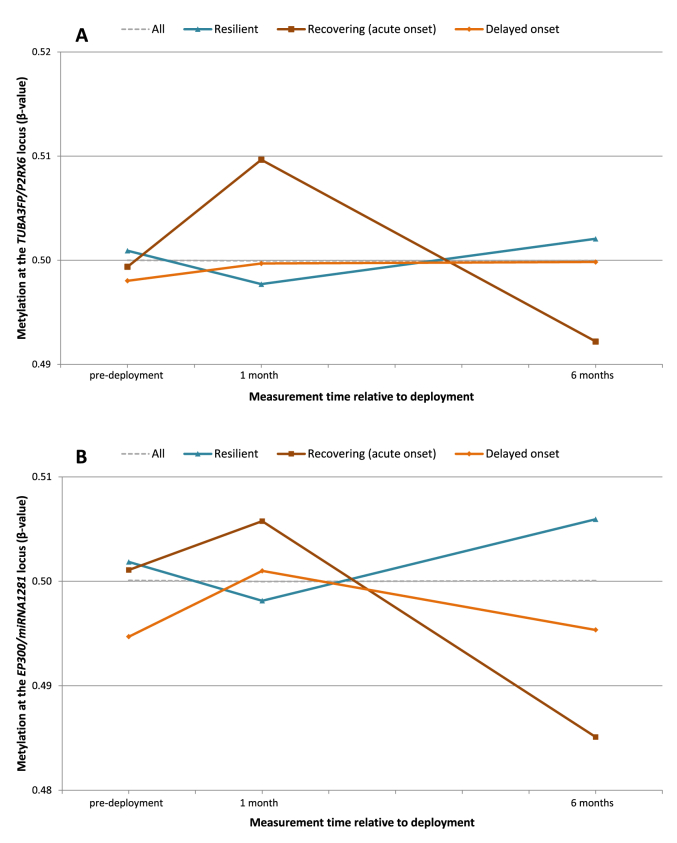
DNA methylation change between T1 and T2 at DMR1 was significantly associated with PTSD trajectory (loglikelihood-ratio-test: p = 0.010). The multinomial logistic regression model indicated an association between DMR1 methylation change between T1 and T2 and a recovering PTSD developmental trajectory compared to a resilient trajectory (OR = 2.37, p = 0.008), and a delayed trajectory compared to a recovering trajectory (OR = 0.497, p = 0.042). DMR2 methylation change between T1 and T2 was also associated with PTSD trajectory (loglikelihood-ratio-test: p = 0.001). The model indicated an association between DMR2 methylation change between T1 and T2 and a recovering trajectory compared to a resilient trajectory (OR = 1.65, p = 0.010), and a delayed onset trajectory compared to a resilient trajectory (OR = 2.73, p = 0.001). Methylation changes of DMR6 and DMR7 were not associated with PTSD developmental trajectory, nor were methylation changes between T0 and T1 in DMR1 and DMR2. PRS was not significantly associated with PTSD trajectory. Full results of the logistic regression models can be found in Appendix Table A.3. Mean methylation levels for DMR1 and DMR2 on each time point for the full sample and the different trajectories can be found in Fig. 2.
Fig. 2.
Mean methylation (β-value) corrected for cell type composition at the (a) DMR1 TUBA3FP/P2RX6 and (b) DMR2 EP300/miRNA1281 locus on each time point for the complete sample (dotted line) and separated for posttraumatic stress disorder symptom trajectories (colored lines).
3.5. Prediction model delayed onset PTSD
As only DMR2 methylation change score between T1 and T2 was associated with a delayed onset trajectory in the previous analysis, DMR2 T1-T2 methylation change (among other factors) was included in the full prediction model for a delayed onset PTSD trajectory (N = 106). The final prediction model included three variables. The first variable was one’s thread level during deployment (function inside the military base vs. outside the base: OR = 4.11, p = 0.009; function both inside and outside the base vs. outside the base: OR = 6.77, p = 0.010), the second variable was physical childhood trauma (OR = 1.96, p = 0.006), and the third variable was DMR2 T1-T2 methylation change score (OR = 1.74, p = 0.029). The model had an area under the curve (AUC) of 0.79.
4. Discussion
In the present study in a Dutch military cohort, we investigated longitudinal changes in blood DNA methylation profiles across a period of combat exposure in relation to the development of posttraumatic stress symptoms, and studied the predictive value of these methylation changes for the delayed onset of PTSD over a follow-up period of five years. Methylation of four genomic regions served as significant determinants of the longitudinal development of PTSD symptoms. Furthermore, we found that increases in methylation between one month post-deployment and six months post-deployment within the P2RX6 gene were associated with a delayed onset PTSD trajectory compared to a recovering trajectory, while post-deployment decreases in methylation within EP300/miRNA1281 were associated with a delayed onset PTSD trajectory compared to a resilient profile. Our findings provide preliminary evidence for the predictive utility of DNA methylation for the late onset development of PTSD symptoms.
Evidence was found for an association of DNA methylation changes with PTSD symptom levels for DMRs that were located in or near the transcription start sites of the Purinergic Receptor P2X 6 (P2RX6) gene, E1A binding protein p300 (EP300) and microRNA 1281 (miRNA1281) genes, and the Inositol Monophosphatase 1 (IMPA1) gene. P2RX6 belongs to the family of P2X receptors, which are ligand-gated ion channels. Interestingly, the related functional gene cluster (a group of functionally highly related genes) includes RYR2 and CACNA1C [29], previously implicated in PTSD pathogenesis [30,31]. Moreover, P2RX6 expression is suggested to be associated with anxiety behavior, schizophrenia, and alcohol and drug dependence [29,32,33].
EP300 (also referred to as p300) encodes a histone acetyltransferase that regulates transcription via chromatin remodeling [34]. It also acts as a scaffold for transcription factors to activate gene transcription [35]. Modifications in chromatin structure have been widely implicated in memory and cognition, and more specifically in contextual fear memory [36]. EP300 is suggested to be required for newly acquired and reactivated fear memories in the amygdala, as inhibition of EP300 impairs fear memory consolidation, reconsolidation and synaptic plasticity in the lateral amygdala in rodents [37]. As it has often been proposed that in PTSD the traumatic memory has been over-consolidated and reconsolidated, these findings suggest a mediating role for EP300 in the development of PTSD symptoms. However, in the present study DNA methylation was assessed in blood, and due to likely tissue specific differences, it is difficult to infer causality from these findings. Less is known about the miRNA1281 in relation to PTSD development. One microRNA expression study found a downregulation in miRNA1281 expression in combat veterans with PTSD compared to combat-exposed controls, but this result has not yet been replicated in other expression studies [38].
IMPA1 encodes a modulator of intracellular signal transduction, and is proposed as a physiologically relevant target for lithium administered to bipolar disorder patients [39,40,41]. So far, no direct associations with PTSD have been reported. However, IMPA1 is a putative target of microRNA 135, a regulatory element in serotonergic activity associated with stress-related neuropsychiatric disorders [42].
Both CpGs in the EP300/miRNA1281 locus were involved in methylation-quantitative trait loci (mQTL) based on the mQTL Database [43]. All indicated SNPs were located in intergenic sequences on chromosome 5 and 10, and their clinical relevance is unknown. However, the clustered SNPs on chromosome 5 were proximate to the ADAMTS16 gene, a gene previously indicated in functional impairment in psychiatric disorders [44]. For the other loci, no clustered SNPs were identified. Based on the iMethyl database [45], the P2RX6 locus included an expression quantitative trait methylation (eQTM) pointing to an association with transcription of the phosphatase-coding gene PPM1F. This gene plays a broad role in both the stress response and serotonergic signaling, and is suggested to moderate the association between PTSD and cortical thickness [46,47]. The EP300/miRNA1281 locus included an eQTM pointing to the protease-coding gene DES11. How this specific correlations between methylation level and gene expression could relate to PTSD development in unclear.
We attempted to replicate our identified loci in an independent prospective military cohort, but the associations between methylation of the P2RX6, EP300/miRNA1281, and IMPA1 loci and PTSD symptoms up to six months after deployment were not significant in this replication dataset. Moreover, this study identifies different loci compared to previous methylome wide PTSD studies. This may indicate false-positive findings but may also be related to vast differences in methodology and populations between studies. Several confounders are at play such as genetic ancestry, personality, culture, environmental and combat exposures, and nutrition. Specific to the replication data set, differences in PTSD assessment (self-report vs. clinical diagnoses) and the use of different arrays (850 k vs. 450 k) might explain any discrepancy between the results of the separate data sets. However, we also did not find overlap in our results and the results reported by Ref. [8]; despite our samples were drawn from the same cohort of military personnel. Putative reasons for that are the fact that overlap is only partial, and more importantly, changes over three time points pose a very different concept of DNA methylation changes. In the current analysis only the most versatile loci are identified.
Unique to this study is that besides the association between DNA methylation changes and PTSD symptom development shortly after deployment, we also studied the relationship between DNA methylation changes and longer term PTSD outcomes using PTSD symptom trajectories. Our study suggests predictive utility of DNA methylation at EP300/miRNA1281 for a delayed onset of PTSD symptoms between two and five years after the original trauma exposure. Most research addresses prevalence rates and risk factors for acute development of PTSD symptoms after trauma exposure, and thereby overlooks those who seem well initially but develop symptoms later in time. A challenge is to identify who is most at risk for developing symptomatology after the acute phase, and target follow-up screening and monitoring accordingly [48]. In our prediction model we observed that, together with thread level during deployment and physical childhood trauma, decreases in methylation within EP300/miRNA1281 between one month and six months post-deployment were most strongly associated with the development of a delayed onset of PTSD symptoms. Although the results need to be interpreted with caution given the low number of participants represented in the delayed onset trajectory, this study suggests possible utility of DNA methylation measures for screening trauma-exposed individuals for an increased risk to develop a delayed onset of PTSD. This could be useful for prevention and early intervention in this group. Increased methylation changes at the P2RX6 locus between one month and six months post-deployment also predicted a delayed onset trajectory but was only able to distinct that from a recovering trajectory. Although the clinical utility of this finding is lower, it does suggest that individuals with an acute and delayed onset of PTSD differ in epigenetic response after deployment. Long-term follow-up data on symptom scores were not available in the replication dataset and therefore we were not able to validate these findings in an independent cohort.
It is noteworthy that not the initial change in methylation level (pre-deployment to one month post-deployment) but the change after deployment was associated with PTSD trajectory. Compared to pre-deployment, the trajectories with substantial PTSD symptom development (either acute or delayed) show an initial increase in methylation level, whereas methylation levels drop in the resilient trajectory. However, these initial methylation responses at P2RX6 and EP300/miRNA1281 were not significantly different between trajectories. This suggests that, at least for the identified genomic regions, the initial epigenetic response following trauma is similar for both PTSD cases and controls, while they differ in their ability to reverse their methylation levels during the aftermath of trauma, and that such reversal is protective for PTSD.
We also compared the association between PTSD symptoms and epigenetic measures with the association between PTSD symptoms and individual genetic liability to PTSD (PRS). PRS showed a significant correlation with acute onset PTSD within six months but had no relation with later onset (in contrast to the DNA methylation prediction). This suggests that discovery of the genetic risk is driven by onset soon after deployment, and that different (genetic) processes may underlie delayed onset. It also points to the possibility that sampling practice within the PGC in other PTSD developmental trajectories lead to underrepresentation of delayed onset PTSD. In this study, DNA methylation performed better as predictor of PTSD compared to genetic risk. The findings open a new perspective on the role of genetic risk to PTSD and the potential role of DNA methylation. Despite the documented relation between genetic vulnerability and epigenetic regulation, our data underscore the vast differences between the two and in this small dataset no common outcomes were present. Further studies that for instance investigate genetic risk for late onset or interrogate the relationship between PRS and DNA methylation are of interest to unravel the complex interplay between genetic predisposition and environmental exposures that impact on transcriptional regulation.
The current study possessed several strengths, including the prospective sampling, the use of an unbiased epigenome-wide approach, methylation measures at three consecutive time points, and a follow-up period of five years. Nonetheless, the study’s results should be interpreted in the context of its limitations. The size of our sample was relatively small, and predominantly consisted of European males. It may therefore be difficult to extrapolate the findings to other samples and populations. In addition, DNA methylation was assessed in blood, and due to tissue specificity, it is difficult to infer causality from these findings. Regarding the analyses using the PTSD trajectories, the elevated pre-deployment scores of the delayed onset and recovering trajectory may limit the extent to which the analyses can truly predict the development of PTSD symptoms, as individuals in these trajectories already experienced a certain amount of PTSD symptoms before deployment. Furthermore, the use of backward regression might have introduced additional bias and therefore limit the generalizability of the findings from the prediction model. Also, the reported effect sizes are small. Due to the explorative nature of the analyses we did not adjust for the multiple prediction analysis. Adjustment for multiple testing would render the effects non-significant, and is obviously a point of concern. Finally, as in most DNA methylation studies, the actual DNA methylation differences driving the associations were small. Although these differences are likely to be biologically relevant for transcript length [49] and count [50], the field is only in the early stages of understanding the complexity of transcriptional regulation [51].
Overall this study provided new insights into the potential relationships between epigenetic alterations at different time periods and the development of PTSD symptoms up to five years post-deployment. In addition, a possible epigenetic mark was identified with the potential to contribute to successful classification of individuals with increased risk for developing a delayed onset of PTSD symptomatology. Finally, the results suggest that the current genetic background is most strongly related to acute disease onset, and that different processes may be at play in those individuals that develop PTSD after years of delay.
Financial and competing interests disclosure
The recruitment and assessments in the PRISMO study were funded by the Dutch Ministry of Defence, The Netherlands. The Marine Resilience Study was funded by the Department of Veterans Affairs Health Service Research, United States project SDR09-0128, the Marine Corps, United States, the Navy Bureau of Medicine and Surgery, United States, and the National Institutes of Health, United States (R01MH093500 and R01MH108826). B.P.F. Rutten was funded by a VIDI award (no. 91718336) from the Netherlands Scientific Organization, The Netherlands. The funders had no role in the design and reporting of the study. The authors declare no conflict of interest.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We like to thank all PRISMO and MRS participants for their commitment to the study. We would also like to thank Danny Nispeling for the bioinformatics support.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cpnec.2020.100018.
Contributor Information
Sija J. van der Wal, Email: s.j.vanderwal@umcutrecht.nl.
Adam X. Maihofer, Email: amaihofer@ucsd.edu.
Christiaan H. Vinkers, Email: c.vinkers@amsterdamumc.nl.
Alicia K. Smith, Email: aksmit3@emory.edu.
Caroline M. Nievergelt, Email: cnievergelt@ucsd.edu.
Dawayland O. Cobb, Email: dawayland.cobb@emory.edu.
Monica Uddin, Email: monica43@usf.edu.
Dewleen G. Baker, Email: dgbaker@health.ucsd.edu.
Nicolaas P.A. Zuithoff, Email: n.p.a.zuithoff@umcutrecht.nl.
Bart P.F. Rutten, Email: b.rutten@maastrichtuniversity.nl.
Eric Vermetten, Email: h.g.j.m.vermetten@lumc.nl.
Elbert Geuze, Email: s.g.geuze@umcutrecht.nl.
Marco P. Boks, Email: m.p.m.boks@umcutrecht.nl.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Eekhout I., Reijnen A., Vermetten E., Geuze E. Post-traumatic stress symptoms 5 years after military deployment to Afghanistan: an observational cohort study. The Lancet Psychiatry. 2016;3:58–64. doi: 10.1016/S2215-0366(15)00368-5. [DOI] [PubMed] [Google Scholar]
- 2.Marmar C.R., Schlenger W., Henn-Haase C., Qian M., Purchia E., Li M., Corry N., Williams C.S., Ho C.L., Horesh D., Karstoft K.I., Shalev A., Kulka R.A. Course of posttraumatic stress disorder 40 years after the Vietnamwar findings from the national Vietnam veterans longitudinal study. JAMA Psychiatry. 2015;72:875–881. doi: 10.1001/jamapsychiatry.2015.0803. [DOI] [PubMed] [Google Scholar]
- 3.Stevelink S.A.M., Jones M., Hull L., Pernet D., Maccrimmon S., Goodwin L., Macmanus D., Murphy D., Jones N., Greenberg N., Rona R.J., Fear N.T., Wessely S. Mental health outcomes at the end of the British involvement in the Iraq and Afghanistan conflicts: a cohort study. Br. J. Psychiatry. 2018;213:690–697. doi: 10.1192/bjp.2018.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daskalakis N.P., Rijal C.M., King C., Huckins L.M., Ressler K.J. Recent genetics and epigenetics approaches to PTSD. Curr. Psychiatr. Rep. 2018;20 doi: 10.1007/s11920-018-0898-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morrison F.G., Miller M.W., Logue M.W., Assef M., Wolf E.J. DNA methylation correlates of PTSD: recent findings and technical challenges. Prog. Neuro-Psychopharmacol. Biol. Psychiatry. 2019;90:223–234. doi: 10.1016/j.pnpbp.2018.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zannas A.S., Provençal N., Binder E.B. Epigenetics of posttraumatic stress disorder: current evidence, challenges, and future directions. Biol. Psychiatr. 2015;78:327–335. doi: 10.1016/j.biopsych.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 7.Snijders C., Maihofer A.X., Ratanatharathorn A., Baker D.G., Boks M.P., Geuze E., Jain S., Kessler R.C., Pishva E., Risbrough V.B., Stein M.B., Ursano R.J., Vermetten E., Vinkers C.H., Smith A.K., Uddin M., Rutten B.P.F., Nievergelt C.M. Longitudinal epigenome-wide association studies of three male military cohorts reveal multiple CpG sites associated with post-traumatic stress disorder. Clin. Epigenet. 2020;12:1–13. doi: 10.1186/s13148-019-0798-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rutten B.P.F., Vermetten E., Vinkers C.H., Ursini G., Daskalakis N.P., Pishva E., de Nijs L., Houtepen L.C., Eijssen L., Jaffe A.E., Kenis G., Viechtbauer W., van den Hove D., Schraut K.G., Lesch K.-P., Kleinman J.E., Hyde T.M., Weinberger D.R., Schalkwyk L., Lunnon K., Mill J., Cohen H., Yehuda R., Baker D.G., Maihofer A.X., Nievergelt C.M., Geuze E., Boks M.P.M. Longitudinal analyses of the DNA methylome in deployed military servicemen identify susceptibility loci for post-traumatic stress disorder. Mol. Psychiatr. 2018;23:1145–1156. doi: 10.1038/mp.2017.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vinkers C.H., Geuze E., van Rooij S.J.H., Kennis M., Schür R.R., Nispeling D.M., Smith A.K., Nievergelt C.M., Uddin M., Rutten B.P.F., Vermetten E., Boks M.P. Successful treatment of post-traumatic stress disorder reverses DNA methylation marks. Mol. Psychiatr. 2019 doi: 10.1038/s41380-019-0549-3. [DOI] [PubMed] [Google Scholar]
- 10.Karstoft K.I., Armour C., Elklit A., Solomon Z. Long-term trajectories of posttraumatic stress disorder in veterans: the role of social resources. J. Clin. Psychiatr. 2013;74:1163–1168. doi: 10.4088/JCP.13.m08482. [DOI] [PubMed] [Google Scholar]
- 11.Porter B., Bonanno G.A., Frasco M.A., Dursa E.K., Boyko E.J. Prospective post-traumatic stress disorder symptom trajectories in active duty and separated military personnel. J. Psychiatr. Res. 2017;89:55–64. doi: 10.1016/j.jpsychires.2017.01.016. [DOI] [PubMed] [Google Scholar]
- 12.Nievergelt C.M., Maihofer A.X., Klengel T., Atkinson E.G., Chen C.Y., Choi K.W., Coleman J.R.I., Dalvie S., Duncan L.E., Gelernter J., Levey D.F., Logue M.W., Polimanti R., Provost A.C., Ratanatharathorn A., Stein M.B., Torres K., Aiello A.E., Almli L.M., Amstadter A.B., Andersen S.B., Andreassen O.A., Arbisi P.A., Ashley-Koch A.E., Austin S.B., Avdibegovic E., Babić D., Bækvad-Hansen M., Baker D.G., Beckham J.C., Bierut L.J., Bisson J.I., Boks M.P., Bolger E.A., Børglum A.D., Bradley B., Brashear M., Breen G., Bryant R.A., Bustamante A.C., Bybjerg-Grauholm J., Calabrese J.R., Caldas- de- Almeida J.M., Dale A.M., Daly M.J., Daskalakis N.P., Deckert J., Delahanty D.L., Dennis M.F., Disner S.G., Domschke K., Dzubur-Kulenovic A., Erbes C.R., Evans A., Farrer L.A., Feeny N.C., Flory J.D., Forbes D., Franz C.E., Galea S., Garrett M.E., Gelaye B., Geuze E., Gillespie C., Uka A.G., Gordon S.D., Guffanti G., Hammamieh R., Harnal S., Hauser M.A., Heath A.C., Hemmings S.M.J., Hougaard D.M., Jakovljevic M., Jett M., Johnson E.O., Jones I., Jovanovic T., Qin X.J., Junglen A.G., Karstoft K.I., Kaufman M.L., Kessler R.C., Khan A., Kimbrel N.A., King A.P., Koen N., Kranzler H.R., Kremen W.S., Lawford B.R., Lebois L.A.M., Lewis C.E., Linnstaedt S.D., Lori A., Lugonja B., Luykx J.J., Lyons M.J., Maples-Keller J., Marmar C., Martin A.R., Martin N.G., Maurer D., Mavissakalian M.R., McFarlane A., McGlinchey R.E., McLaughlin K.A., McLean S.A., McLeay S., Mehta D., Milberg W.P., Miller M.W., Morey R.A., Morris C.P., Mors O., Mortensen P.B., Neale B.M., Nelson E.C., Nordentoft M., Norman S.B., O’Donnell M., Orcutt H.K., Panizzon M.S., Peters E.S., Peterson A.L., Peverill M., Pietrzak R.H., Polusny M.A., Rice J.P., Ripke S., Risbrough V.B., Roberts A.L., Rothbaum A.O., Rothbaum B.O., Roy-Byrne P., Ruggiero K., Rung A., Rutten B.P.F., Saccone N.L., Sanchez S.E., Schijven D., Seedat S., Seligowski A.V., Seng J.S., Sheerin C.M., Silove D., Smith A.K., Smoller J.W., Sponheim S.R., Stein D.J., Stevens J.S., Sumner J.A., Teicher M.H., Thompson W.K., Trapido E., Uddin M., Ursano R.J., van den Heuvel L.L., Van Hooff M., Vermetten E., Vinkers C.H., Voisey J., Wang Y., Wang Z., Werge T., Williams M.A., Williamson D.E., Winternitz S., Wolf C., Wolf E.J., Wolff J.D., Yehuda R., Young R.M.D., Young K.A., Zhao H., Zoellner L.A., Liberzon I., Ressler K.J., Haas M., Koenen K.C. International meta-analysis of PTSD genome-wide association studies identifies sex- and ancestry-specific genetic risk loci. Nat. Commun. 2019;10:1–16. doi: 10.1038/s41467-019-12576-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van der Wal S.J., Gorter R., Reijnen A., Geuze E., Vermetten E. Cohort profile: the prospective research in stress-related military Operations (PRISMO) study in the Dutch armed forces. BMJ Open. 2019;9:1–10. doi: 10.1136/bmjopen-2018-026670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hovens J.E., Bramsen I., Van Der Ploeg H.M. Self-rating inventory for posttraumatic stress disorder: review of the psychometric properties of a new brief Dutch screening instrument. Percept. Mot. Skills. 2002;94:996–1008. doi: 10.2466/pms.2002.94.3.996. [DOI] [PubMed] [Google Scholar]
- 15.Hovens J.E., van der Ploeg H.M., Bramsen I., Klaarenbeek M.T., Schreuder J.N., Rivero V.V. The development of the self-rating inventory for posttraumatic stress disorder. Acta Psychiatr. Scand. 1994;90:172–183. doi: 10.1111/j.1600-0447.1994.tb01574.x. [DOI] [PubMed] [Google Scholar]
- 16.Reijnen A., Rademaker A.R., Vermetten E., Geuze E. Prevalence of mental health symptoms in Dutch military personnel returning from deployment to Afghanistan: a 2-year longitudinal analysis. Eur. Psychiatr. 2015;30:341–346. doi: 10.1016/j.eurpsy.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 17.Bremner J.D., Bolus R., Mayer E.A. Psychometric properties of the early trauma inventory-self report. J. Nerv. Ment. Dis. 2007;195:211–218. doi: 10.1097/01.nmd.0000243824.84651.6c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Min J.L., Hemani G., Smith G.D., Relton C., Suderman M. Meffil: efficient normalization and analysis of very large DNA methylation datasets. Bioinformatics. 2018;34:3983–3989. doi: 10.1093/bioinformatics/bty476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fortin J.-P., Labbe A., Lemire M., Zanke B.W., Hudson T.J., Fertig E.J., Greenwood C.M., Hansen K.D. Functional normalization of 450k methylation array data improves replication in large cancer studies. Genome Biol. 2014;15:503. doi: 10.1186/s13059-014-0503-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Y., Lemire M., Choufani S., Butcher D.T., Grafodatskaya D., Zanke B.W., Gallinger S., Hudson T.J., Weksberg R. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics. 2013;8:203–209. doi: 10.4161/epi.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pidsley R., Zotenko E., Peters T.J., Lawrence M.G., Risbridger G.P., Molloy P., Van Djik S., Muhlhausler B., Stirzaker C., Clark S.J. Critical evaluation of the Illumina MethylationEPIC BeadChip microarray for whole-genome DNA methylation profiling. Genome Biol. 2016;17:1–17. doi: 10.1186/s13059-016-1066-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Du P., Zhang X., Huang C., Jafari N., Kibbe W.A., Hou L., Lin S.M. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinf. 2010;11:587. doi: 10.1186/1471-2105-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baker D.G., Nash W.P., Litz B.T., Geyer M.A., Risbrough V.B., Nievergelt C.M., O’Connor D.T., Larson G.E., Schork N.J., Vasterling J.J., Hammer P.S., Webb-Murphy J.A. Predictors of risk and resilience for posttraumatic stress disorder among ground combat marines: methods of the marine resiliency study. Prev. Chronic Dis. 2012;9:1–11. doi: 10.5888/pcd9.110134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blake D.D., Weathers F.W., Nagy L.M., Kaloupek D.G., Gusman F.D., Charney D.S., Keane T.M. The development of a clinician-administered PTSD scale. J. Trauma Stress. 1995;8:75–90. doi: 10.1007/BF02105408. [DOI] [PubMed] [Google Scholar]
- 25.Choi S.W., O’Reilly P.F. PRSice-2: polygenic Risk Score software for biobank-scale data. GigaScience. 2019;8:1–6. doi: 10.1093/gigascience/giz082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Houseman E.A., Kile M.L., Christiani D.C., Ince T.A., Kelsey K.T., Marsit C.J. Reference-free deconvolution of DNA methylation data and mediation by cell composition effects. BMC Bioinf. 2016;17:1–15. doi: 10.1186/s12859-016-1140-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peters T.J., Buckley M.J., Statham A.L., Pidsley R., Samaras K., V Lord R., Clark S.J., Molloy P.L. De novo identification of differentially methylated regions in the human genome. Epigenet. Chromatin. 2015;8:1–16. doi: 10.1186/1756-8935-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xue C., Ge Y., Tang B., Liu Y., Kang P., Wang M., Zhang L. A meta-analysis of risk factors for combat-related PTSD among military personnel and veterans. PLoS One. 2015;10:1–21. doi: 10.1371/journal.pone.0120270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bassett A.S., Lowther C., Merico D., Costain G., Chow E.W.C., van Amelsvoort T., McDonald-McGinn D., Gur R.E., Swillen A., Van den Bree M., Murphy K., Gothelf D., Bearden C.E., Eliez S., Kates W., Philip N., Sashi V., Campbell L., Vorstman J., Cubells J., Repetto G.M., Simon T., Boot E., Heung T., Evers R., Vingerhoets C., van Duin E., Zackai E., Vergaelen E., Devriendt K., Vermeesch J.R., Owen M., Murphy C., Michaelovosky E., Kushan L., Schneider M., Fremont W., Busa T., Hooper S., McCabe K., Duijff S., Isaev K., Pellecchia G., Wei J., Gazzellone M.J., Scherer S.W., Emanuel B.S., Guo T., Morrow B.E., Marshall C.R. Rare genome-wide copy number variation and expression of schizophrenia in 22q11.2 deletion syndrome. Am. J. Psychiatr. 2017;174:1054–1063. doi: 10.1176/appi.ajp.2017.16121417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krzyzewska I.M., Ensink J.B.M., Nawijn L., Mul A.N., Koch S.B., Venema A., Shankar V., Frijling J.L., Veltman D.J., Lindauer R.J.L., Olff M., Mannens M.M.A.M., van Zuiden M., Henneman P. Genetic variant in CACNA1C is associated with PTSD in traumatized police officers. Eur. J. Hum. Genet. 2018;26:247–257. doi: 10.1038/s41431-017-0059-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu X., Betzenhauser M.J., Reiken S., Meli A.C., Xie W., Chen B.X., Arancio O., Marks A.R. Role of leaky neuronal ryanodine receptors in stress-induced cognitive dysfunction. Cell. 2012;150:1055–1067. doi: 10.1161/CIRCULATIONAHA.110.956839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sarro E.C., Sullivan R.M., Barr G. Unpredictable neonatal stress enhances adult anxiety and alters amygdala gene expression related to serotonin and GABA. Neuroscience. 2014;258:147–161. doi: 10.1016/j.neuroscience.2013.10.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wetherill L., Lai D., Johnson E.C., Anokhin A., Bauer L., Bucholz K.K., Dick D.M., Hariri A.R., Hesselbrock V., Kamarajan C., Kramer J., Kuperman S., Meyers J.L., Nurnberger J.I., Schuckit M., Scott D.M., Taylor R.E., Tischfield J., Porjesz B., Goate A.M., Edenberg H.J., Foroud T., Bogdan R., Agrawal A. Genome-wide association study identifies loci associated with liability to alcohol and drug dependence that is associated with variability in reward-related ventral striatum activity in African- and European-Americans. Gene Brain Behav. 2019;18:1–16. doi: 10.1111/gbb.12580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jin Q., Yu L.R., Wang L., Zhang Z., Kasper L.H., Lee J.E., Wang C., Brindle P.K., Dent S.Y.R., Ge K. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 2011;30:249–262. doi: 10.1038/emboj.2010.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chan H.M., La Thangue N.B. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J. Cell Sci. 2001;114:2363–2373. doi: 10.1242/jcs.114.13.2363. [DOI] [PubMed] [Google Scholar]
- 36.Roth T.L., Sweatt J.D. Regulation of chromatin structure in memory formation. Curr. Opin. Neurobiol. 2009;19:336–342. doi: 10.1038/jid.2014.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maddox S.A., Watts C.S., Schafe G.E. p300/CBP histone acetyltransferase activity is required for newly acquired and reactivated fear memories in the lateral amygdala. Learn. Mem. 2013;20:109–119. doi: 10.1101/lm.029157.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou J., Nagarkatti P., Zhong Y., Ginsberg J.P., Singh N.P., Zhang J., Nagarkatti M. Dysregulation in microRNA expression is associated with alterations in immune functions in combat veterans with post-traumatic stress disorder. PLoS One. 2014;9 doi: 10.1371/journal.pone.0094075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berridge M.J., Downes C.P., Hanley M.R. Neural and developmental actions of lithium: a unifying hypothesis. Cell. 1989;59:411–419. doi: 10.1016/0092-8674(89)90026-3. [DOI] [PubMed] [Google Scholar]
- 40.Cryns K., Shamir A., Van Acker N., Levi I., Daneels G., Goris I., Bouwknecht J.A., Andries L., Kass S., Agam G., Belmaker H., Bersudsky Y., Steckler T., Moechars D. IMPA1 is essential for embryonic development and lithium-like pilocarpine sensitivity. Neuropsychopharmacology. 2008;33:674–684. doi: 10.1038/sj.npp.1301431. [DOI] [PubMed] [Google Scholar]
- 41.Damri O., Sade Y., Toker L., Bersudsky Y., Belmaker R.H., Agam G., Azab A.N. Molecular effects of lithium are partially mimicked by inositol-monophosphatase (IMPA)1 knockout mice in a brain region-dependent manner. Eur. Neuropsychopharmacol. 2015;25:425–434. doi: 10.1016/j.euroneuro.2014.06.012. [DOI] [PubMed] [Google Scholar]
- 42.Issler O., Haramati S., Paul E.D., Maeno H., Navon I., Zwang R., Gil S., Mayberg H.S., Dunlop B.W., Menke A., Awatramani R., Binder E.B., Deneris E.S., Lowry C.A., Chen A. MicroRNA 135 is essential for chronic stress resiliency, antidepressant efficacy, and intact serotonergic activity. Neuron. 2014;83:344–360. doi: 10.1016/j.neuron.2014.05.042. [DOI] [PubMed] [Google Scholar]
- 43.Gaunt T.R., Shihab H.A., Hemani G., Min J.L., Woodward G., Lyttleton O., Zheng J., Duggirala A., McArdle W.L., Ho K., Ring S.M., Evans D.M., Davey Smith G., Relton C.L. Systematic identification of genetic influences on methylation across the human life course. Genome Biol. 2016;17:61. doi: 10.1186/s13059-016-0926-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mcgrath L.M., Cornelis M.C., Lee P.H., Robinson E.B., Duncan L.E., Barnett J.H., Huang J., Gerber G., Sklar P., Sullivan P., Perlis R.H., Smoller J.W. Genetic predictors of risk and resilience in psychiatric disorders: a cross-disorder genome-wide association study of functional impairment in major depressive disorder, bipolar disorder, and schizophrenia. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2013;162:779–788. doi: 10.1002/ajmg.b.32190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hachiya T., Furukawa R., Shiwa Y., Ohmomo H., Ono K., Katsuoka F., Nagasaki M., Yasuda J., Fuse N., Kinoshita K., Yamamoto M., Tanno K., Satoh M., Endo R., Sasaki M., Sakata K., Kobayashi S., Ogasawara K., Hitomi J., Sobue K., Shimizu A. Genome-wide identification of inter-individually variable DNA methylation sites improves the efficacy of epigenetic association studies. npj Genomic Med. 2017;2:1–13. doi: 10.1038/s41525-017-0016-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sullivan D.R., Morrison F.G., Wolf E.J., Logue M.W., Fortier C.B., Salat D.H., Fonda J.R., Stone A., Schichman S., Milberg W., McGlinchey R., Miller M.W. The PPM1F gene moderates the association between PTSD and cortical thickness. J. Affect. Disord. 2019;259:201–209. doi: 10.1016/j.jad.2019.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wingo A.P., Velasco E., Florido A., Lori A., Choi D.C., Jovanovic T., Ressler K.J., Andero R. Expression of the PPM1F gene is regulated by stress and associated with anxiety and depression. Biol. Psychiatr. 2018;83:284–295. doi: 10.1016/0191-8869(88)90077-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Forbes D., Alkemade N., Nickerson A., Bryant R.A., Creamer M., Silove D., McFarlane A.C., Van Hoof M., Phelps A.J., Rees S., Steele Z., ODonnell M. Prediction of late-onset psychiatric disorder in survivors of severe injury: findings of a Latent Transition Analysis. J. Clin. Psychiatr. 2016;77:807–812. doi: 10.4088/JCP.15m09854. [DOI] [PubMed] [Google Scholar]
- 49.Leenen F.A.D., Vernocchi S., Hunewald O.E., Schmitz S., Molitor A.M., Muller C.P., Turner J.D. Where does transcription start? 5’-RACE adapted to next-generation sequencing. Nucleic Acids Res. 2016;44:262802645. doi: 10.1093/nar/gkv1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Eijk K.R., de Jong S., Boks M.P.M., Langeveld T., Colas F., Veldink J.H., de Kovel C.G.F., Janson E., Strengman E., Langfelder P., Kahn R.S., van den Berg L.H., Horvath S., Ophoff R.A. Genetic analysis of DNA methylation and gene expression levels in whole blood of healthy human subjects. BMC Genom. 2012;13:636. doi: 10.1186/1471-2164-13-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rajarajan P., Akbarian S. Use of the epigenetic toolbox to contextualize common variants associated with schizophrenia risk. Dialogues Clin. Neurosci. 2019;21:407–416. doi: 10.31887/DCNS.2019.21.4/sakbarian. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.