Abstract
Type 2 diabetes is the most prevalent endocrine disease in the world, and recently the gut microbiota have become a potential target for its management. Recent studies have illustrated that this disease may predispose individuals to certain microbiome compositions, and treatments like metformin have been shown to change gut microbiota and their associated metabolic pathways. However, given the limitations and side effects associated with pharmaceuticals currently being used for therapy of diabetes, there is a significant need for alternative treatments. In this study, we investigated the effects of a root extract from Rhodiola rosea in a Leptin receptor knockout (db/db) mouse model of type 2 diabetes. Our previous work showed that Rhodiola rosea had anti-inflammatory and gut microbiome-modulating properties, while extending lifespan in several animal models. In this study, treatment with Rhodiola rosea improved fasting blood glucose levels, altered the response to exogenous insulin, and decreased circulating lipopolysaccharide and hepatic C-reactive protein transcript levels. We hypothesize that these changes may in part reflect the modulation of the microbiota, resulting in improved gut barrier integrity and decreasing the translocation of inflammatory biomolecules into the bloodstream. These findings indicate that Rhodiola rosea is an attractive candidate for further research in the management of type 2 diabetes.
Subject terms: Biomarkers, Biomarkers, Endocrinology, Metabolic disorders
Introduction
Type 2 diabetes (T2D) is a metabolic disease that currently ranks as one of the largest concerns of global public health, affecting an estimated 476 million people worldwide1. With both genetic and environmental factors contributing to this complex disease, diabetes is a leading cause of mortality in many countries and globally impacts life expectancy in both developed and developing nations1,2. One of the hallmarks of the disease is hyperglycemia resulting from defects in insulin secretion, insulin action, or both3. Of the three forms of clinical diabetes, the vast majority (about 90%) of patients have type 2 diabetes (T2D), which is characterized by insulin resistance3. Whereas there are many environmental and behavioral factors that modulate genetic susceptibility to this form of diabetes, the prevalence of T2D has risen steadily in recent decades4. Due to rising health costs associated with the increasing incidence and prevalence of diabetes worldwide5,6, identifying and evaluating safe and cost-effective therapeutic interventions in the management of T2D is of increasing importance. Current treatment algorithms for T2D include lifestyle changes, as well as oral and parenteral drugs7. However, many current treatments have significant limitations or side effects that can impact this large patient population. Although metformin is first-line treatment for T2D, it has side effects including nausea and diarrhea and is contraindicated in patients predisposed to lactic acidosis7,8. Insulin secretagogues such as sulfonylureas and meglitinides tend to lose their efficacy over time due to beta cell failure9,10, while alpha-glucosidase inhibitors, thiazolidinediones, and dipeptidyl peptidase-4 inhibitors have unique mechanisms of action, but adverse effects such as weight gain, heart failure, and gastrointestinal issues have led to poor compliance with therapy11–14. Thus, there is a need for new therapeutic interventions in T2D that are safe and effective.
There is significant evidence that inflammation plays a central role in the pathogenesis of T2D through two distinct pathways15. Obesity, specifically visceral adiposity, causes systemic inflammation through infiltration of adipose tissue by macrophages16,17 and production of pro-inflammatory cytokines such as TNF-α, IL-1β, IL-6, and CCL218,19, which act as antagonists of insulin signaling20. Recent studies also suggest that differences in the human gut microbiome between normal and diabetic subjects21 are linked to systemic inflammation through altered gut integrity22,23, increased circulating gram-negative bacteria and endotoxin22,24, as well as lipopolysaccharide (LPS)-induced inflammatory cytokine secretion through TLR-4 signaling24,25. Evidence in humans also suggests that certain gut microbiome compositions may predispose individuals to conditions such as obesity and diabetes26,27. As crucial components that moderate host health and physiology, the gut microbiome can contribute to inflammation, alterations of intestinal linings, dyslipidemia, and a wide variety of other changes23. However, studies also indicate that modulation of the gut microbiome can decrease LPS-induced systemic inflammation in a mouse T2D model, thereby counteracting these changes25. Together, these observations suggest that targeting inflammation through the microbiome could be a novel approach to treating T2D17,28.
Plant-derived therapeutics such as cinnamon and curcumin extracts have been shown to have potential anti-diabetic properties, although their efficacy has not been evaluated in randomized clinical trials29,30. The adaptogenic plant Rhodiola rosea is used as a medicinal in traditional medical practices worldwide and has been shown to have anti-inflammatory and gut microbiome-modulating properties31–33. As an adaptogen, R. rosea represents an important category of pharmacological substances that are known to aid the body in resisting a wide variety of stressors (i.e. biological, chemical, physical, etc.) to maintain homeostasis and stabilize physiological processes that may be disrupted32. Rhodiola rosea extract appears to be safe in human studies34–36, which makes it an attractive candidate for the treatment of T2D. Rhodiola rosea extended lifespan in several animal models including worms, snails, and flies37–40. When tested on a fly model deficient in the insulin receptor substrate chico, R. rosea still extended lifespan but decreased expression levels of Drosophila insulin-like peptide (dILP) 2, 3, and 5 in wild-type flies39, suggesting complex effects on the insulin signaling pathway. However, the effects of this plant extract on an animal model of diabetes have not yet been evaluated.
Here, we investigated the effects of a root extract from R. rosea in a Leptin receptor knockout (db/db) mouse model of T2D. Leptin is a key adipokine responsible for maintenance of energy homeostasis and body mass, whereas mice lacking the Leptin receptor display hyperphagia and consequently develop obesity, visceral adiposity, hyperglycemia, and hyperinsulinemia41–43. In other mouse models, deficiencies in Leptin as well as beta-cell dysfunction have been linked to T2D, suggesting that this adipokine plays a key role in the pathogenesis of diabetes44. Like human diabetics, db/db mice show augmented expression of the differentiation marker Aldh1a3 and reduced nuclear expression of the transcription factor Nkxx 6.1, and exhibit hyperglycemia compared to age-matched congenic non-db/db mice45. These observations make the db/db mouse model suitable for the goals of this study.
Results
Rhodiola rosea alters glucose homeostasis in diabetic db/db mice
To assess the effects of Rhodiola rosea on a mouse model of T2D, we treated a cohort of 6 week-old leptin receptor-knockout (db/db) mice with an extract of R. rosea that was verified for quality based on validated biomarkers (see “Methods”), administered daily at a dose of 25 mg/kg by oral gavage for 4 weeks. The timeline of the study is depicted in Fig. 1. Control mice received gavage with water. We chose oral gavage as the route of admistration over adding the extract to food to allow more precise control of the amount of R. rosea administered. Following completion of treatment at 10 weeks of age, both R. rosea-treated and control db/db mice exhibited fasting hyperglycemia, with fasting blood glucose levels at 11 weeks of age significantly lower in R. rosea-treated mice than in control mice (Fig. 2). When subjected to a parenteral glucose challenge (glucose tolerance test, GTT) administered at the conclusion of treatment at week 10, there was no further elevation in blood glucose in either group, perhaps due to the extremely high fasting blood glucose levels in these mice, with a subsequent modest decrease in glucose levels over 120 min that might reflect induction of an endogenous insulin response (Fig. 3).
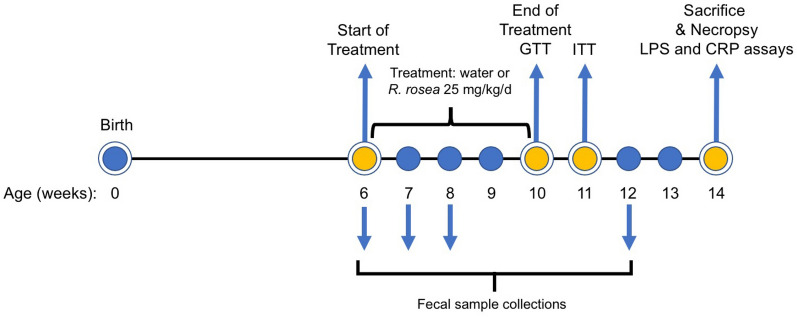
Figure 1.
Timeline and experimental design. Cohorts of db/db mice were treated beginning at 6 weeks of age with R. rosea extract (25 mg/kg daily by oral gavage) or with water for a total of 4 weeks. A glucose tolerance test (GTT) was administered at the end of treatment (week 10), and an insulin tolerance test (ITT) administered 1 week later (week 11). Fecal samples were collected for microbiome analysis before treatment at week 6, and subsequently at weeks 7, 8 and 12.
Figure 2.
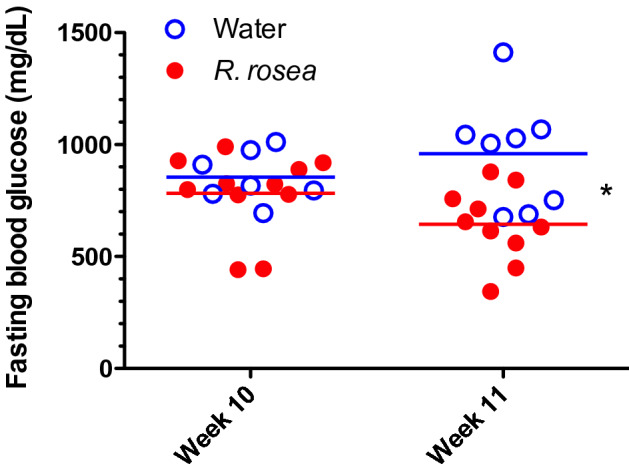
Rhodiola rosea improves fasting blood glucose in db/db mice. Folowing a 4 h fast, blood glucose levels were determined at the end of treatment (week 10) and 1 week later. The difference in mean blood glucose level at week 11 was significant (*P = 0.0303, repeated measures ANOVA). n = 8 control, n = 11 R. rosea-treated; one control sample from week 10 and one R-rosea-treated sample from week 11 did not give interpretable results and were omitted.
Figure 3.
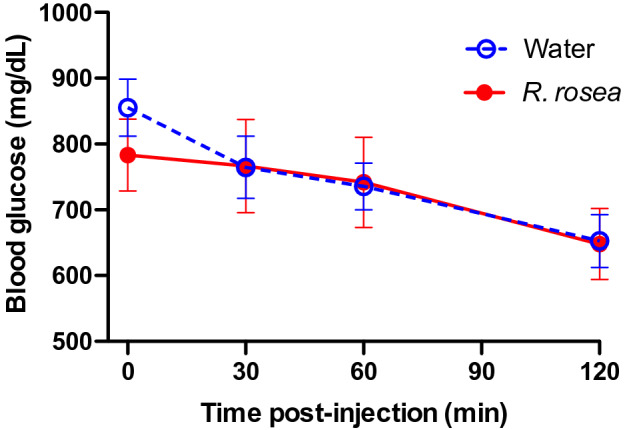
Lack of effect of Rhodiola rosea on glucose tolerance in db/db mice. Cohorts of mice (n = 7 control, n = 11 R. rosea-treated) at the end of the treatment period (week 10) were fasted for 4 h and then challenged with 0.5 g/kg glucose by intraperitoneal injection, followed by determination of blood glucose levels at 30, 60, and 120 min post-injection. None of the differences between control and R. rosea-treated mice were significant (one-way repeated measures ANOVA).
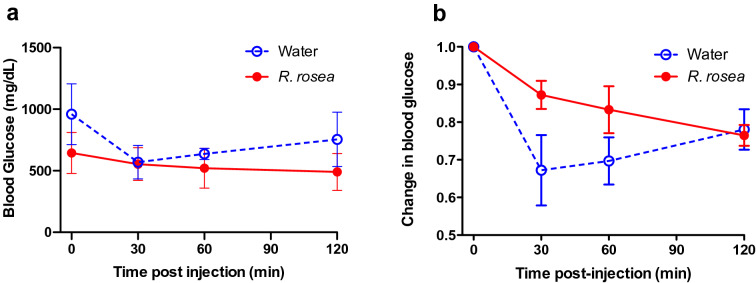
Following a week of recovery, we tested the response of the two treatment cohorts to an exogenous insulin challenge (insulin tolerance test, ITT) at 11 weeks of age, 1 week after supplementation had ended (Fig. 4). Rhodiola rosea-treated mice showed a continuous decline in blood glucose levels over the 2 h period following insulin administration, with mean glucose values below those of water-treated mice at every time point (Fig. 4a). By contrast, the control mice exhibited an initial steeper decline in blood glucose in response to insulin (~ 33% decrease at 30 m; Fig. 4b) but a subsequent increase in blood glucose levels over the following 90 min. These results suggest that R. rosea treatment alters the response to insulin in diabetic db/db mice in a complex manner, resulting in a more prolonged response to insulin.
Figure 4.
Altered insulin tolerance in Rhodiola rosea-treated db/db mice. Insulin tolerance test (ITT) of db/db mice. ITT was conducted at 11 weeks of age. Following a 4 h fast, mice were injected intraperitoneally with 0.75 IU/kg insulin. Blood was sampled pre-injection and at 30, 60 and 120 minues post-injection and glucose levels determined. The difference between the two data sets is significant (P < 0.05, one-way repeated measures ANOVA; n = 7 for water and n = 11 for R. rosea-treated).
Weight loss does not account for the effect of Rhodiola rosea on db/db mice
To assess whether changes in glucose homeostasis of R. rosea-supplemented mice were due to a decrease in obesity, we measured the body mass of the mice on a weekly basis during the study, beginning at the time of treatment initiation at week 6 (Fig. 5). The two cohorts had very similar mean weight before starting treatment, as expected. Treatment with R. rosea was associated with a transient decrease in weight, followed by a recovery in weight gain over time after week 8. Although the mean body weight of R. rosea-treated db/db mice was lower than control water-treated mice in weeks 8 through 14 of the study, these differences were not statistically significant.
Figure 5.
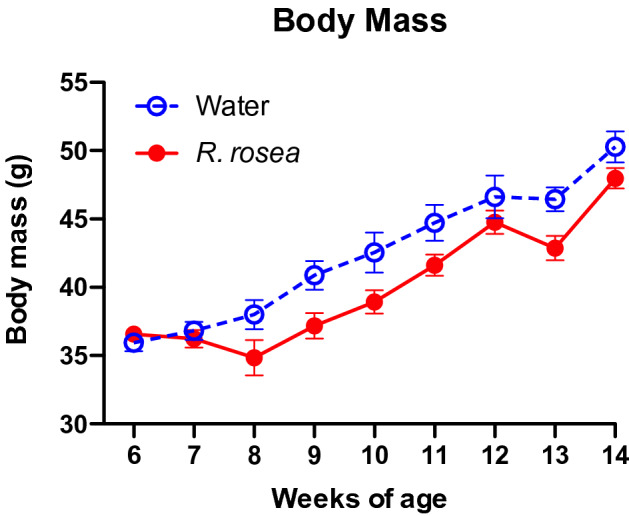
Effect of Rhodiola rosea treatment on body weight of db/db mice. Mice in each cohort (n = 8 water-treated, n = 11 R. rosea-treated) were weighed weekly starting just before initiation of treatment (week 6). None of the paired differences in weight were significant (one-way repeated measures ANOVA).
Rhodiola rosea modestly modulates the fecal microbiome of db/db mice
To assess the effect of R. rosea on the fecal microbiome of db/db mice, we collected serial stool samples before treatment (week 6) and at weeks 7, 8 and 12 (Fig. 1) from females of the two cohorts and analyzed the composition of the microbial community of the feces with 16S rRNA amplicon next-generation sequencing. The microbiome of both cohorts were dominated by Bacteroidetes and Firmicutes, and most of the reads could be designated as amplicon sequence variants (ASVs) within four major taxonomic assignments: the families Rikenellaceae and S24-7 (Bacteroidetes), and the family Lachnospiraceae and genus Lactobacillus (Firmicutes). Compared to the treatment, time of sample collection accounted for the most variation in the microbiota composition of water and R. rosea-treated mice (Supplementary Fig. 1). When the microbiota composition of the two cohorts was subjected to Bray–Curtis analysis46, which does not account for phylogenetic relationships, there was a significant effect of R. rosea treatment on the fecal microbiome of db/db mice (Fig. 6 and Table 1). The only taxa that varied with time and treatment were reads assigned to the Desulfovibrionales, clustered at the order level, which were more abundant in R. rosea-treated mice than control mice (Supplementary Fig. 2). While a previous study demonstrated reduction in Desulfovibrionales abundance in the intestinal microbiota of BALB/c mice following treatment with salidroside47, a glycoside found in R. rosea, the difference in Desulfovibrionales abundance was observed before R. rosea was administered, suggesting the changes might be attributed to differences in microbiota composition before the experiment began (e.g. early cage effects) rather than to R. rosea treatment. Together, these findings identify modest but significant variation in the composition of the mouse fecal microbiota associated with R. rosea treatment.
Figure 6.
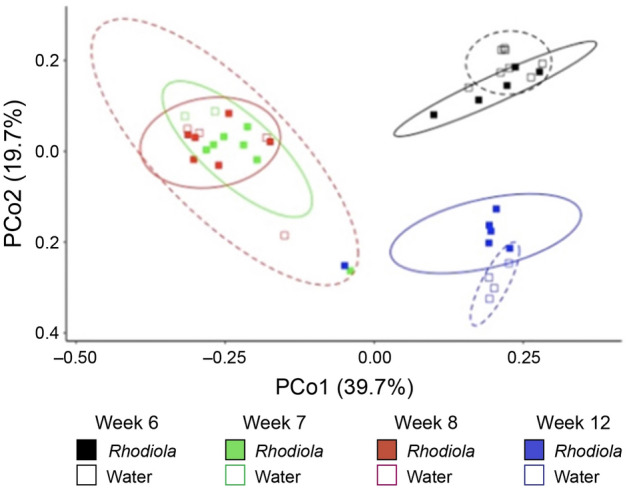
Effects of Rhodiola rosea on the fecal microbiome of db/db mice. Bray–Curtis principal coordinate ordination of the data from Supplemental Fig. 1. The corresponding PERMANOVA (Table 1) identifies significant differences in microbiota composition with treatment and time. The number of fecal samples from different mice analyzed is n = 5 (R. rosea week 6), n = 7 (R. rosea week 7), n = 6 (R. rosea week 8), n = 6 (R. rosea week 12), n = 8 (water week 6), n = 2 (water week 7), n = 4 (water week 8), and n = 4 (water week 12).
Table 1.
Results of PERMANOVA on Bray–Curtis distances of microbiota data.
| Degrees of freedom | Sum of squares | Mean squares | f | R2 | P value | |
|---|---|---|---|---|---|---|
| Treatment | 1 | 0.2 | 0.2 | 3.64 | 0.04 | 0.02 |
| Time point | 3 | 2.89 | 0.96 | 17.69 | 0.55 | 0 |
| Interaction | 3 | 0.35 | 0.12 | 2.14 | 0.07 | 0.02 |
| Residuals | 34 | 1.85 | 0.05 | NA | 0.35 | NA |
| Total | 41 | 5.29 | NA | NA | 1 | NA |
Rhodiola rosea decreases circulating lipopolysaccharide levels and a marker of inflammation
To determine whether the changes of the fecal microbiota associated with R. rosea treatment had any physiological consequences, we determined the lipopolysaccharide (LPS) content from serum samples from the two cohorts via a limulus amoebocyte lysate (LAL) assay. Treatment with R. rosea decreased the LPS levels in the serum by almost 50% (Fig. 7a). Whereas circulating LPS (derived from gram-negative bacteria) triggers production of inflammatory cytokines by tissue macrophages and other cells, to further observe inflammation levels exhibited by the db/db mice, we determined if R. rosea treatment was associated with changes in C-reactive protein (CRP), an inflammatory marker exclusively produced by hepatocytes. Similar to LPS, R. rosea treatment decreased hepatic CRP transcript levels by about 40% (Fig. 7b).
Figure 7.
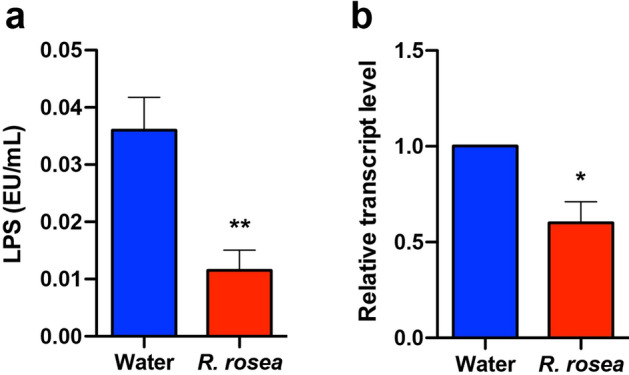
Rhodiola rosea treatment reduces circulating LPS and hepatocyte CRP expression. At 14 weeks of age, mice were sacrificed and serum and liver tissue obtained. (a) Serum LPS levels, measured by the LAL assay (n = 8 for water-treated and n = 6 for R. rosea-treated mice). **P < 0.01, Mann–Whitney U test. (b) Relative expression of CRP transcripts in mouse liver, calculated by the ΔΔCt method (n = 6 for water-treated and n = 5 for R. rosea-treated mice). *P < 0.05, one sample Wilcoxon test.
Discussion
The goal of this study was to evaluate the impact of R. rosea, a medicinal plant of emerging interest and possible therapeutic value, on the phenotype and the fecal microbiota of the db/db mouse model of T2D. Although this study shares a common format with a number of published microbiota-disease interaction articles48–51, it is one of the few studies that evaluates the impact of a botanical extract on the fecal microbiota throughout the progression of T2D. The format of our study was correlative and associative but the results may serve as the basis for future mechanism-based studies. We elected to use db/db mice for this study to test whether R. rosea can improve glucose homeostasis in a T2D model that is similar to severe and advanced human T2D in terms of visceral adiposity and insulin resistance, without resorting to a high-fat diet or other dietary manipulations. The use of this severe model may have prevented us from observing beneficial effects of R. rosea that might be relevant to less severe phenotypes of diabetes (i.e., pre-diabetes).
Daily treatment with R. rosea for 1 month significantly lowered the fasting blood glucose level in db/db mice at 11 weeks of age (Fig. 2). There was no effect of R. rosea on the response to a parenteral glucose load (Fig. 3), which may be a reflection of the substantial baseline hyperglycemia observed in both cohorts (Fig. 3 and reference 45). By contrast, R. rosea treatment significantly altered the response of db/db mice to exogenous insulin in a complex fashion, manifested as a decrease in acute blood glucose lowering but a sustained hypoglycemic response that persisted over 2 h (Fig. 4). These effects were not a consequence of decreased food intake and reduction in obesity (Fig. 5). Taken together, these results suggest that administration of R. rosea may favorably modulate the T2D phenotype, either by improving the function of insulin-responsive tissues in db/db mice or ameliorating the exhaustion of pancreatic beta cells that is observed in this model45.
Multiple studies have demonstrated major changes in the gut microbiota during the development of obesity52 and T2D in humans, including significantly reduced proportions of phylum Firmicutes and class Clostridia, in addition to compositional changes in the microbiota27,53,54. In addition, the presence of sulfate-reducing bacterial species from the Desulfovibrionales order has been associated with the pathology of T2D21. db/db mice display elevated levels of gut bacteria from the S24-7 family compared to wild-type mice55 and we observed a similar trend, with S24-7 bacteria dominating the composition of the fecal microbiome in both cohorts at all time points (Supplementary Fig. 1). While Bray–Curtis analysis demonstrated significant effects of R. rosea treatment on the db/db fecal microbiome (Fig. 6), our previous studies suggest that R. rosea does not modulate the gut microbiome through direct anti-microbial activity, as the plant extract did not suppress growth when tested on bacteria isolated from the Drosophila gut33.
As a consequence of changes in the local microbiome, gut integrity and intestinal permeability may be lowered through multiple mechanisms19,22,25, leading to endotoxemia and chronic inflammation16 that is postulated to contribute to the pathogenesis of T2D16–19,26,27. Mice with obesity induced by diet25 or genetic mutation (db/db)56 also have impaired gut integrity and increases in circulating LPS and inflammatory cytokines. Importantly, we observed significantly decreased levels of circulating LPS and decreased hepatic CRP transcripts (Fig. 7) in the R. rosea-treated cohort, providing a potential functional connection between modulation of the gut microbiome, inflammation, and glucose homeostasis by R. rosea. It is noteworthy that the changes in these inflammatory biomarkers persisted a month following cessation of R. rosea treatment, suggesting a long-term effect of exposure to Rhodiola on the pathophysiology of T2D in db/db mice.
In conclusion, while previous studies have illustrated that R. rosea has anti-inflammatory and gut microbiome-modulating properties and can extend lifespan in several animal models, the present study demonstrates that short-term exposure to R. rosea has beneficial effects on glucose homeostasis in the Leptin receptor knockout (db/db) mouse model of severe T2D, and suggests a possible mechanism of action. While only a modest modulation of the fecal microbiome was observed in this study, we hypothesize that these changes may have improved the integrity of the gut barrier, leading to decreased systemic inflammation. Rhodiola rosea is a good candidate for further investigation as a potential treatment for T2D, but further mechanistic studies in mice, and ultimately human clinical trials, are indicated.
Methods
All methods were performed in accordance with applicable guidelines and regulations.
Mouse strains and handling
BKS.Cg-Dock7m+/+ Leprdb/J and C57BL/6J mice were obtained from the Jackson Laboratory (Bar Harbor, ME). Leprdb/J mice were bred to generate homozygous Leprdb/Leprdb (db/db) pups which were used to model T2D. During the study, mice were group-housed by sex in microisolator cages with filter tops (Techniplast) on a ventilated rack, provided with contact wood chip bedding (autoclaved Envigo Teklad corncob, 1/8 in.) and were allowed ad libitum access to food (Purina rodent chow #5001) and RO water. The animal room was maintained in 12 h light/dark cycles (0630 on/1830 off) at a temperature of 72° F ± 2° and humidity of 50% ± 5%. Enrichment was provided as two Nestlets (6 g, from Ancare) per cage. The animal facility was a SPF barrier facility; the health of the mice was monitored twice weekly by staff, testing for pathogens was done by monitoring exhaust dust and sentinels exposed to dirty bedding. Surveillance pathogen testing included Sendai virus, MHV, MPV, PVM, Reo-3, Mycoplasma, TMEV, LCMV, MVM, MNV, MKPV, EDIM, Ectromelia, pinworms, fur mites, Helicobacter, C. bovis, and S. muris.
Mice were weaned when they were 3 weeks old, and entered into the study at 6 weeks of age. At that point, mice were weighed, and mice of the same size (36 ± 1.5 g) randomly assigned to control (water) or experimental (R. rosea) treatment, with equal numbers of males and females per group. A cohort size of n = 9 was predicted to give 90% power to detect a decrease in fasting blood glucose from 800 to 600 mg/dL given an estimated standard deviation of 150 at a significance of α = 0.05 (one-sample t-test). At the end of the study, mice were humanely euthanized by a AVMA-approved method (CO2 asphyxiation using a gradual-fill method followed by cervical dislocation). The study was approved by the Instituitional Animal Care and Use Committee (IACUC) of UCI (Protocol #AUP-16-52). All experiments were performed in accordance with guidelines from UCI IACUC, U.S. Dept. of Agriculture APHIS, and ARRIVE version 2.0 (https://arriveguidelines.org).
Study timeline
The study timeline is summarized in Fig. 1. Mice were administered either water (control) or 25 mg/kg R. rosea extract (treatment) by oral gavage daily for the 4 weeks of treatment, initiated when the mice were 6 weeks old and continued until the mice were 10 weeks old. This dose was selected based on our own preliminary dose-finding work and previously published studies where R. rosea was evaluated in rodent diabetes models57,58. The quality of the R. rosea extract was verified by HPLC that showed the extract contained 1.3% salidroside and 3.9% rosavins, consistent with a high-quality extract (data on file). Fecal samples were collected at 6 weeks of age (prior to treatment), 7 weeks of age, 8 weeks of age and 12 weeks of age. The glucose tolerance test (GTT) was performed at week 10 and insulin tolerance test (ITT) was performed at week 11. Mice were sacrificed and subjected to necropsy at week 14.
Glucose tolerance test (GTT) and insulin tolerance test (ITT)
Given the importance of in vivo mouse models for studying the pathogenesis of T2D and various treatment interventions, several methods have been developed to investigate glucose tolerance, as well as the secretion and action of insulin in these models59. To ascertain potential differences between treatment and control groups with regards to glucose homeostasis, both glucose and insulin tolerance tests were performed. Four hours before the assays were performed, the food was removed to induce a state of fasting. Mice were placed briefly in an immobilization device without anesthesia, and blood samples (~ 2 µL) obtained from the mouse tail vein punctured by a 21 Ga needle. The first drop of blood was wiped away, and the second drop used to record values. Samples were blinded before determination of blood glucose levels using an AlphaTrak2 Blood Glucose Monitoring System. Blood glucose values were recorded 30, 60, and 120 min after intraperitoneal injection of either 0.5 g/kg glucose (5 μL per gram body weight of a 10% solution) or 0.75 IU/kg (3 μL per gram body weight of 0.25 IU/mL insulin stock). The number of mice utilized was 11 for the R. rosea-treated cohort and 7 water-treated control mice. Data were plotted using GraphPad Prism v8, statistical analysis was done in SAS using one-way repeated measures ANOVA.
Body mass assay
Mice from both treatment cohorts (n = 8 for water-treated, n = 11 for R. rosea-treated) were weighed weekly from weeks 6–14. Data from male and female mice in each cohort were pooled and plotted using GraphPad Prism v8. Statistical analysis was performed in SAS using one-way repeated measures ANOVA.
Microbiome analysis
Fecal samples from female mice were collected at 6 weeks of age (prior to treatment), 7 weeks of age, 8 weeks of age, and 12 weeks of age (Fig. 1). DNA was extracted using the Zymobiomics Mini DNA kit. 16S rRNA amplicon PCR was performed, targeting the V4-V5region using the EMP primers 515F (barcoded) and 926R60. The samples were prepared into a library that was sequenced at the UC Irvine Genomics High Throughput Facility on an Illumina MiSeq, using paired-end 300 bp v3 sequencing chemistry. The raw sequence data were imported into QIIME2 (qiime2.org)61,62 and demultiplexed60. This bioinformatics platform has recently been rewritten and reengineered for the next generation of microbiome sequencing, facilitating taxanomic and phylogenetic analyses. While dozens of software packages written in various programming languages are often needed for comprehensive analyses of this type of sequencing data, QIIME2 allows for “sequence quality checking, denoising, taxonomic classification, alignment, and phylogenetic tree building”—allowing for seamless analysis, description, and quantification of microbial communities60.
The sequences were assigned a taxonomic classification using the q2-feature-classifier63 on a GreenGenes database downloaded in July 2020 from the QIIME2 website (qiime2.org)64. A total of 6,352,455 read pairs passed Illumina quality filters. However, the quality scores of the reverse reads were generally lower than the forward reads, and no read pairs passed DADA2 quality filter steps65. Therefore, we analyzed the data only using the forward reads, which had higher average quality scores than the reverse reads. Forward reads that passed quality filtering and denoising by default DADA2 parameters in QIIME2 were rarefied to 11,390 reads per sample and beta-diversity distance metrics were calculated using QIIME246,66,67 (Supplementary Fig. 3). As part of calculating the Unifrac beta-diversity metrics we constructed a phylogenetic tree with fasttree268 based on mafft alignment69. Permutational multivariate analysis of variance (PERMANOVA)70 and analysis of microbial communities (ANCOM)71 analyses were performed in R.
Limulus amebocyte lysate assay of serum LPS
Serum samples were obtained at week 14 (Fig. 1) from n = 6 R. rosea-treated mice and n = 9 control water-treated mice. Samples were blinded, diluted 40-fold, and LPS levels determined using the Pierce™ Chromogenic Endotoxin Quant Kit according to the manufacturer’s recommended protocol. Data were plotted and analyzed using GraphPad Prism v8.
CRP transcript assay
Mice were sacrificed at week 14 (Fig. 1) and RNA extracted from liver tissue using Trizol. A total of 5 samples were collected from R. rosea-treated mice and 6 samples from control water-treated mice. Samples were blinded and the RNA extract was treated with DNA-free™ DNA Removal Kit from Invitrogen to remove any contaminating DNA. A cDNA library was generated with the iScript™ cDNA Synthesis Kit from Bio-Rad. qPCR was performed on a BioRadMJ Mini Personal Thermal Cycler with iQ SYBR Green Supermix. The PCR amplification program consisted of an initial denaturation set at 94 °C for 3 min, followed by 40 three-step cycles at 94 °C for 10 s, 60 °C at 30 s and at 72 °C for 45 s. The ΔΔCt Method was used to quantify the relative expression of genes of interest46. The CRP (gene of interest) and GAPDH (reference gene) primers were derived from PrimerBank (Supplementary Table 1). Data were plotted and analyzed using GraphPad Prism v8.
Ethical statement
All experiments involving laboratory mice were carried out in compliance with ARRIVE guidelines 2.0 and with the approval of the UCI Institutional Animal Care and Use Committee (UCI IACUC; Animal Welfare Assurance # A3416-01) under the auspices of protocol AUP-16-52 (Van Etten laboratory; approval date 13 December 2016).
Supplementary Information
Acknowledgements
We would like to acknowledge the UCI Microbiome Center for granting a Pilot Award, providing consulting on study design and supporting the generation of the microbiome amplicon sequence data. We would like to thank Ms. Claudia Weihe, Dr. Jennifer Martiny and Dr. Katrine Whiteson from the UCI Microbiome Center. We thank the UCI School of Medicine-Pharmaceutical Scienes Collaborative Research Funds and Mr. and Mrs. John P. and Lois C. Wareham for providing funding for this work.
Author contributions
M.J. and R.A.V. designed the study, oversaw the experiments, and wrote and revised the manuscript; J.M.C. and R.C. analyzed microbiome data and participated in writing the results and discussion; J.G.J.A., D.K., and C.D.N. performed the mouse and biochemical experiments; N.J., P.T., and Z.-Y.L. maintained the mouse colonies and participated in the mouse experiments; all authors read and approved the final version of the manuscript.
Data availability
16S rRNA metagenomic data are deposited in the NCBI Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra), accession # PRJNA848938. CRP transcript data are deposited in DRYAD (https://datadryad.org), 10.7280/D1FX2D. Other data in this manuscript are freely available to qualified researchers by contacting the lead author at mjafari@hs.uci.edu.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Jasmin Grace Juanson Arabit, Robert Courville and Dara Kiani.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-14241-7.
References
- 1.Lin X, et al. Global, regional, and national burden and trend of diabetes in 195 countries and territories: An analysis from 1990 to 2025. Sci. Rep. 2020;10:14790. doi: 10.1038/s41598-020-71908-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mambiya M, et al. The play of genes and non-genetic factors on type 2 diabetes. Front. Public Health. 2019;7:349. doi: 10.3389/fpubh.2019.00349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Association AD. Diagnosis and classification of diabetes mellitus. Diabetes Care. 2013;36(Suppl 1):S67–S74. doi: 10.2337/dc13-S067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murea M, Ma L, Freedman BI. Genetic and environmental factors associated with type 2 diabetes and diabetic vascular complications. Rev. Diabet. Stud. 2012;9:6–22. doi: 10.1900/RDS.2012.9.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smyth S, Heron A. Diabetes and obesity: The twin epidemics. Nat. Med. 2006;12:75–80. doi: 10.1038/nm0106-75. [DOI] [PubMed] [Google Scholar]
- 6.Association AD. Economic costs of diabetes in the U.S. in 2017. Diabetes Care. 2018;41:917–928. doi: 10.2337/dci18-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marín-Peñalver JJ, Martín-Timón I, Sevillano-Collantes C, Del Cañizo-Gómez FJ. Update on the treatment of type 2 diabetes mellitus. World J. Diabetes. 2016;7:354–395. doi: 10.4239/wjd.v7.i17.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee H, Ko G. Effect of metformin on metabolic improvement and gut microbiota. Appl. Environ. Microbiol. 2014;80:5935–5943. doi: 10.1128/AEM.01357-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takahashi A, et al. Sulfonylurea and glinide reduce insulin content, functional expression of K(ATP) channels, and accelerate apoptotic beta-cell death in the chronic phase. Diabetes Res. Clin. Pract. 2007;77:343–350. doi: 10.1016/j.diabres.2006.12.021. [DOI] [PubMed] [Google Scholar]
- 10.Maedler K, et al. Sulfonylurea induced beta-cell apoptosis in cultured human islets. J. Clin. Endocrinol. Metab. 2005;90:501–506. doi: 10.1210/jc.2004-0699. [DOI] [PubMed] [Google Scholar]
- 11.Chiasson JL, et al. Acarbose treatment and the risk of cardiovascular disease and hypertension in patients with impaired glucose tolerance: The STOP-NIDDM trial. JAMA. 2003;290:486–494. doi: 10.1001/jama.290.4.486. [DOI] [PubMed] [Google Scholar]
- 12.Abe M, Okada K, Soma M. Antidiabetic agents in patients with chronic kidney disease and end-stage renal disease on dialysis: Metabolism and clinical practice. Curr. Drug Metab. 2011;12:57–69. doi: 10.2174/138920011794520053. [DOI] [PubMed] [Google Scholar]
- 13.Bogacka I, Xie H, Bray GA, Smith SR. The effect of pioglitazone on peroxisome proliferator-activated receptor-gamma target genes related to lipid storage in vivo. Diabetes Care. 2004;27:1660–1667. doi: 10.2337/diacare.27.7.1660. [DOI] [PubMed] [Google Scholar]
- 14.Rosenwasser RF, Sultan S, Sutton D, Choksi R, Epstein BJ. SGLT-2 inhibitors and their potential in the treatment of diabetes. Diabetes Metab. Syndr. Obes. 2013;6:453–467. doi: 10.2147/DMSO.S34416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gregor MF, Hotamisligil GS. Inflammatory mechanisms in obesity. Annu. Rev. Immunol. 2011;29:415–445. doi: 10.1146/annurev-immunol-031210-101322. [DOI] [PubMed] [Google Scholar]
- 16.Sweet MJ, Hume DA. Endotoxin signal transduction in macrophages. J. Leukoc. Biol. 1996;60:8–26. doi: 10.1002/jlb.60.1.8. [DOI] [PubMed] [Google Scholar]
- 17.Xu H, et al. Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J. Clin. Investig. 2003;112:1821–1830. doi: 10.1172/JCI200319451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shoelson SE, Lee J, Goldfine AB. Inflammation and insulin resistance. J. Clin. Investig. 2006;116:1793–1801. doi: 10.1172/JCI29069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berg AH, Scherer PE. Adipose tissue, inflammation, and cardiovascular disease. Circ. Res. 2005;96:939–949. doi: 10.1161/01.RES.0000163635.62927.34. [DOI] [PubMed] [Google Scholar]
- 20.Peraldi P, Hotamisligil GS, Buurman WA, White MF, Spiegelman BM. Tumor necrosis factor (TNF)-alpha inhibits insulin signaling through stimulation of the p55 TNF receptor and activation of sphingomyelinase. J. Biol. Chem. 1996;271:13018–13022. doi: 10.1074/jbc.271.22.13018. [DOI] [PubMed] [Google Scholar]
- 21.Qin J, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 22.Cani PD, et al. Changes in gut microbiota control inflammation in obese mice through a mechanism involving GLP-2-driven improvement of gut permeability. Gut. 2009;58:1091–1103. doi: 10.1136/gut.2008.165886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kirschman LJ, Milligan-Myhre KC. The costs of living together: Immune responses to the microbiota and chronic gut inflammation. Appl. Environ. Microbiol. 2019;85:e02147-18. doi: 10.1128/AEM.02147-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cani PD, et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. 2007;56:1761–1772. doi: 10.2337/db06-1491. [DOI] [PubMed] [Google Scholar]
- 25.Cani PD, et al. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes. 2008;57:1470–1481. doi: 10.2337/db07-1403. [DOI] [PubMed] [Google Scholar]
- 26.Musso G, Gambino R, Cassader M. Interactions between gut microbiota and host metabolism predisposing to obesity and diabetes. Annu. Rev. Med. 2011;62:361–380. doi: 10.1146/annurev-med-012510-175505. [DOI] [PubMed] [Google Scholar]
- 27.Kalliomäki M, Collado MC, Salminen S, Isolauri E. Early differences in fecal microbiota composition in children may predict overweight. Am. J. Clin. Nutr. 2008;87:534–538. doi: 10.1093/ajcn/87.3.534. [DOI] [PubMed] [Google Scholar]
- 28.Fantuzzi G. Adipose tissue, adipokines, and inflammation. J. Allergy Clin. Immunol. 2005;115:911–919. doi: 10.1016/j.jaci.2005.02.023. [DOI] [PubMed] [Google Scholar]
- 29.Chuengsamarn S, Rattanamongkolgul S, Luechapudiporn R, Phisalaphong C, Jirawatnotai S. Curcumin extract for prevention of type 2 diabetes. Diabetes Care. 2012;35:2121–2127. doi: 10.2337/dc12-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kirkham S, Akilen R, Sharma S, Tsiami A. The potential of cinnamon to reduce blood glucose levels in patients with type 2 diabetes and insulin resistance. Diabetes Obes. Metab. 2009;11:1100–1113. doi: 10.1111/j.1463-1326.2009.01094.x. [DOI] [PubMed] [Google Scholar]
- 31.Lee Y, et al. Anti-Inflammatory and Neuroprotective Effects of Constituents Isolated from Rhodiola rosea. Evid. Based Complement. Alternat. Med. 2013;2013:514049. doi: 10.1155/2013/514049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bawa AS, Khanum F. Anti-inflammatory activity of Rhodiola rosea—"a second-generation adaptogen". Phytother. Res. 2009;23:1099–1102. doi: 10.1002/ptr.2749. [DOI] [PubMed] [Google Scholar]
- 33.Labachyan KE, Kiani D, Sevrioukov EA, Schriner SE, Jafari M. The impact of Rhodiola rosea on the gut microbial community of Drosophila melanogaster. Gut Pathog. 2018;10:12. doi: 10.1186/s13099-018-0239-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Darbinyan V, et al. Clinical trial of Rhodiola rosea L. extract SHR-5 in the treatment of mild to moderate depression. Nord. J. Psychiatry. 2007;61:343–348. doi: 10.1080/08039480701643290. [DOI] [PubMed] [Google Scholar]
- 35.Olsson EM, von Schéele B, Panossian AG. A randomised, double-blind, placebo-controlled, parallel-group study of the standardised extract shr-5 of the roots of Rhodiola rosea in the treatment of subjects with stress-related fatigue. Planta Med. 2009;75:105–112. doi: 10.1055/s-0028-1088346. [DOI] [PubMed] [Google Scholar]
- 36.Shevtsov VA, et al. A randomized trial of two different doses of a SHR-5 Rhodiola rosea extract versus placebo and control of capacity for mental work. Phytomed. Int. J. Phytother. Phytopharmacol. 2003;10:95–105. doi: 10.1078/094471103321659780. [DOI] [PubMed] [Google Scholar]
- 37.Chen C, et al. Rhodiola rosea extends lifespan and improves stress tolerance in silkworm, Bombyx mori. Biogerontology. 2016;17:373–381. doi: 10.1007/s10522-015-9622-8. [DOI] [PubMed] [Google Scholar]
- 38.Wiegant FA, et al. Plant adaptogens increase lifespan and stress resistance in C. elegans. Biogerontology. 2009;10:27–42. doi: 10.1007/s10522-008-9151-9. [DOI] [PubMed] [Google Scholar]
- 39.Schriner SE, et al. Extension of Drosophila lifespan by Rhodiola rosea through a mechanism independent from dietary restriction. PLoS ONE. 2013;8:e63886. doi: 10.1371/journal.pone.0063886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boon-Niermeijer EK, van den Berg A, Wikman G, Wiegant FA. Phyto-adaptogens protect against environmental stress-induced death of embryos from the freshwater snail Lymnaea stagnalis. Phytomed. Int. J. Phytother. Phytopharmacol. 2000;7:389–399. doi: 10.1016/S0944-7113(00)80060-4. [DOI] [PubMed] [Google Scholar]
- 41.Tartaglia LA, et al. Identification and expression cloning of a leptin receptor, OB-R. Cell. 1995;83:1263–1271. doi: 10.1016/0092-8674(95)90151-5. [DOI] [PubMed] [Google Scholar]
- 42.Bray GA, York DA. Hypothalamic and genetic obesity in experimental animals: An autonomic and endocrine hypothesis. Physiol. Rev. 1979;59:719–809. doi: 10.1152/physrev.1979.59.3.719. [DOI] [PubMed] [Google Scholar]
- 43.Kobayashi K, et al. The db/db mouse, a model for diabetic dyslipidemia: Molecular characterization and effects of Western diet feeding. Metab. Clin. Exp. 2000;49:22–31. doi: 10.1016/S0026-0495(00)90588-2. [DOI] [PubMed] [Google Scholar]
- 44.Chen WS, et al. Leptin deficiency and beta-cell dysfunction underlie type 2 diabetes in compound Akt knockout mice. Mol. Cell. Biol. 2009;29:3151–3162. doi: 10.1128/MCB.01792-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Burke SJ, et al. db/db mice exhibit features of human type 2 diabetes that are not present in weight-matched C57BL/6J mice fed a western diet. J. Diabetes Res. 2017;2017:8503754. doi: 10.1155/2017/8503754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sorensen TA. A method of establishing groups of equal amplitude in plant sociology based on similarity of species content and its application to analyses of the vegetation on Danish commons. Biol. Skar. 1948;5:1–34. [Google Scholar]
- 47.Yuan Y, Wu X, Zhang X, Hong Y, Yan H. Ameliorative effect of salidroside from Rhodiola rosea L. on the gut microbiota subject to furan-induced liver injury in a mouse model. Food Chem. Toxicol. 2019;125:333–340. doi: 10.1016/j.fct.2019.01.007. [DOI] [PubMed] [Google Scholar]
- 48.Membrez M, et al. Gut microbiota modulation with norfloxacin and ampicillin enhances glucose tolerance in mice. FASEB J. 2008;22:2416–2426. doi: 10.1096/fj.07-102723. [DOI] [PubMed] [Google Scholar]
- 49.Chou CJ, Membrez M, Blancher F. Gut decontamination with norfloxacin and ampicillin enhances insulin sensitivity in mice. Nestle Nutr. Workshop Ser. Pediatr. Program. 2008;62:127–137. doi: 10.1159/000146256. [DOI] [PubMed] [Google Scholar]
- 50.Geurts L, et al. Altered gut microbiota and endocannabinoid system tone in obese and diabetic leptin-resistant mice: Impact on apelin regulation in adipose tissue. Front. Microbiol. 2011;2:149. doi: 10.3389/fmicb.2011.00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu W, et al. Grape seed proanthocyanidin extract ameliorates inflammation and adiposity by modulating gut microbiota in high-fat diet mice. Mol. Nutr. Food Res. 2017;61:1601082. doi: 10.1002/mnfr.201601082. [DOI] [PubMed] [Google Scholar]
- 52.Jumpertz R, et al. Energy-balance studies reveal associations between gut microbes, caloric load, and nutrient absorption in humans. Am. J. Clin. Nutr. 2011;94:58–65. doi: 10.3945/ajcn.110.010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu X, et al. Molecular characterisation of the faecal microbiota in patients with type II diabetes. Curr. Microbiol. 2010;61:69–78. doi: 10.1007/s00284-010-9582-9. [DOI] [PubMed] [Google Scholar]
- 54.Larsen N, et al. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS ONE. 2010;5:e9085. doi: 10.1371/journal.pone.0009085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu F, et al. Abnormal gut microbiota composition contributes to the development of type 2 diabetes mellitus in db/db mice. Aging. 2019;11:10454–10467. doi: 10.18632/aging.102469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brun P, et al. Increased intestinal permeability in obese mice: New evidence in the pathogenesis of nonalcoholic steatohepatitis. Am. J. Physiol. Gastrointest. Liver Physiol. 2007;292:G518–525. doi: 10.1152/ajpgi.00024.2006. [DOI] [PubMed] [Google Scholar]
- 57.Déciga-Campos M, et al. Antihyperalgesic activity of Rhodiola rosea in a diabetic rat model. Drug Dev. Res. 2016;77:29–36. doi: 10.1002/ddr.21289. [DOI] [PubMed] [Google Scholar]
- 58.Niu CS, Chen LJ, Niu HS. Antihyperglycemic action of rhodiola-aqeous extract in type1-like diabetic rats. BMC Complement. Altern. Med. 2014;14:20. doi: 10.1186/1472-6882-14-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Beguinot F, Nigro C. Measurement of glucose homeostasis in vivo: Glucose and insulin tolerance tests. Methods Mol. Biol. 2012;933:219–228. doi: 10.1007/978-1-62703-068-7_14. [DOI] [PubMed] [Google Scholar]
- 60.Hall M, Beiko RG. 16S rRNA gene analysis with QIIME2. Methods Mol. Biol. 2018;1849:113–129. doi: 10.1007/978-1-4939-8728-3_8. [DOI] [PubMed] [Google Scholar]
- 61.Bolyen E, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Whelan FJ, Rossi L, Stearns JC, Surette MG. Culture and molecular profiling of the respiratory tract microbiota. Methods Mol. Biol. 2018;1849:49–61. doi: 10.1007/978-1-4939-8728-3_4. [DOI] [PubMed] [Google Scholar]
- 63.Bokulich NA, et al. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2's q2-feature-classifier plugin. Microbiome. 2018;6:90. doi: 10.1186/s40168-018-0470-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McDonald D, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Callahan BJ, et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lozupone CA, Hamady M, Kelley ST, Knight R. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 2007;73:1576–1585. doi: 10.1128/AEM.01996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lozupone C, Knight R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Price MN, Dehal PS, Arkin AP. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Oksanen, J. et al. vegan: Community Ecology Package, version 2.5-7, https://CRAN.R-project.org/package=vegan (2020).
- 71.Mandal S, et al. Analysis of composition of microbiomes: A novel method for studying microbial composition. Microb. Ecol. Health Dis. 2015;26:27663. doi: 10.3402/mehd.v26.27663. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
16S rRNA metagenomic data are deposited in the NCBI Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra), accession # PRJNA848938. CRP transcript data are deposited in DRYAD (https://datadryad.org), 10.7280/D1FX2D. Other data in this manuscript are freely available to qualified researchers by contacting the lead author at mjafari@hs.uci.edu.