Abstract
Soybean is one of the important food, feed, and biofuel crops in the world. Soybean genome modification by genetic transformation has been carried out for trait improvement for more than 4 decades. However, compared to other major crops such as rice, soybean is still recalcitrant to genetic transformation, and transgenic soybean production has been hampered by limitations such as low transformation efficiency and genotype specificity, and prolonged and tedious protocols. The primary goal in soybean transformation over the last decade is to achieve high efficiency and genotype flexibility. Soybean transformation has been improved by modifying tissue culture conditions such as selection of explant types, adjustment of culture medium components and choice of selection reagents, as well as better understanding the transformation mechanisms of specific approaches such as Agrobacterium infection. Transgenesis-based breeding of soybean varieties with new traits is now possible by development of improved protocols. In this review, we summarize the developments in soybean genetic transformation to date, especially focusing on the progress made using Agrobacterium-mediated methods and biolistic methods over the past decade. We also discuss current challenges and future directions.
Keywords: soybean transformation, transformation efficiency, genotype, Agrobacterium, biolistic method, genome editing
Introduction
Soybean [Glycine max (L.) Merrill] is a legume crop belonging to the family of Leguminosae, a subfamily of Papilionoideae. Soybean is grown worldwide and is one of the most important crop plants for its high seed oil and protein content, and for its capability to fix atmospheric nitrogen by symbioses with soil-borne microorganisms. Recent studies on high-quality reference genome sequencing of a United States variety, Williams82 (Schmutz et al., 2010), a Japanese variety, Enrei (Shimomura et al., 2015), a Chinese cultivar, Zhonghuang13, and a wild soybean, W05 (Shen et al., 2018; Xie et al., 2019) have estimated that there exist a total of 46,430 protein-coding genes in soybean, 70% more than that in Arabidopsis. Soybean is an ancient polyploidy (palaeopolyploid) plant with a highly duplicated genome. Nearly 75% of the genes are present in multiple copies, representing a threefold redundancy due to its long evolutionary history (Schmutz et al., 2010). Some repetitive sequence families may be species-specific (Morgante et al., 1997). Several other databases have been developed, including an expressed sequence tag (EST) database, full-length cDNAs and cDNA microarrays (Stacey et al., 2004; Umezawa et al., 2008), and a haplotype map (GmHapMap) (Torkamaneh et al., 2019). These resources provide a wide range of opportunities for soybean improvement by marker-assisted breeding and with transgenic and genome editing approaches, and for understanding gene function through various forward and reverse genetic approaches. Most of these approaches are reliant on high-throughput transformation systems.
Genetic transformations allow for various genes of interest to be introduced and expressed in cells of living organisms, which can also overcome barriers of sexual incompatibility. Soybean genetic transformation was originally developed in late 1980s. The first fertile transgenic soybeans were produced by either regeneration of cotyledonary nodes infected with Agrobacterium tumefaciens (Hinchee et al., 1988) or by particle bombardment using meristems of immature soybean seeds (Mccabe et al., 1988). The development of soybean transgenic methods before 2013 has previously been extensively reviewed (Homrich et al., 2012; Yamada et al., 2012; Lee et al., 2013; Mariashibu et al., 2013). Soybean improvements using these transformation methods have been continued over the last 30 years. Since the first transgenic herbicide-resistant soybean product was commercialized in the mid 1990s, soybean has become one of the most important crops improved using modern biotechnology and one of the major commercially grown transgenic plants around the world. Genetically modified (GM) soybean, especially the GM Roundup Ready soybean resistant to glyphosate herbicides, has been grown in many countries including the United States, Argentina, and Brazil (Pagano and Miransari, 2016), which has made it a leading biotech crop. This soybean variety allows for growers to spray herbicides to kill any weeds in the field while not killing the soybean crop1. It was reported that about 105 million hectares of GM soybean was grown in 2017, and that about 272 million metric tons of seeds were produced, which accounted for 80% of all soybean production in the world (Voora et al., 2020). Genetic engineering has been conducted to enhance the protein quality of soybean by altering biosynthetic feedback pathways that increase lysine and sulfur-containing amino acids (Falco et al., 1995). Many types of GM soybeans have improved traits such as increased oleic acid content, decreased linolenic acid content, delayed flowering time, modified plant architecture and increased yield (Yamada et al., 2012). With increasing soybean demands around the world, especially from China, developing GM soybean varieties with high quality and yield is the main task for soybean researchers and breeders. Recently, genome editing (GE) technologies, especially the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated (Cas) technology, have been used for studying soybean genetics and commercial trait development (reviewed in Xu et al., 2020). However, using genome editing technologies on plants has been heavily dependent on efficient transformation systems and regeneration of plants containing edited events (Ran et al., 2017; Gao, 2021). Therefore, an efficient and genotype-flexible transformation system is key to realizing soybean improvement using these new technologies. Unfortunately, soybean remains recalcitrant to routine transformations compared to other major cultivated crops such as rice (Chen et al., 2018b). Low transformation frequency and genotype inflexibility are major hurdles that limit soybean transgenesis and breeding. In this review, we will summarize the major achievements that have been made in this field since 2013, and describe current best methods used for achieving stable and transient transformations in soybean. We also describe the remaining challenges that need to be addressed.
Current Transformation Methods Developed for Soybean
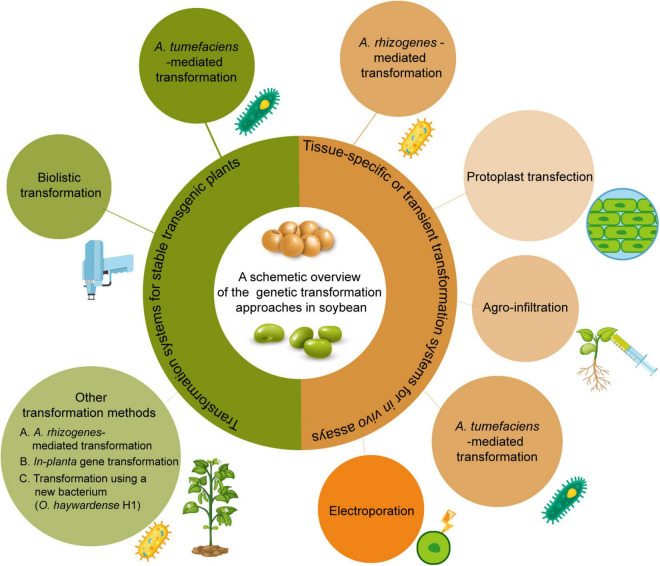
Various transformation methods have been developed for soybean. Here, we will summarize each transformation method and its ability to produce either stable transgenic plants or transient events used for soybean research (Figure 1).
FIGURE 1.
Current available genetic transformation methods for soybean. The left part with greenish color shows transformation methods to produce stable whole plants. The right part with yellowish color shows transformation methods for transient assay.
Tissue-Specific or Transient Transformation Systems for in vivo Assays
Transient assays are used for a variety of studies including the functional genomics of in vivo gene expression and subcellular gene localization, and determination of genome editing efficiency. For soybean, Agrobacterium rhizogenes-mediated transformation, protoplast transfection, Agro-infiltration, and electroporation have been developed. Agrobacterium transformation and protoplast transfection are frequently performed for transient assays.
Agrobacterium rhizogenes-Mediated Transformation
Agrobacterium rhizogenes-mediated transformation leads to development of a hairy-root phenotype. This method relies on co-transfer of T-DNAs from the Ri plasmid and a binary vector containing a gene of interest into the plant genome (Christey, 2001; Broothaerts et al., 2005). Large numbers of transgenic hairy-roots can be obtained in the absence of exogenous plant growth regulators (Collier et al., 2005), and each represents an independent transformation event (Kereszt et al., 2007). The relatively short timeframe (approximately 6–8 weeks) for recovering transformants is a major advantage for screening genes and promoters or expressing foreign genes in a stably transformed plant as a bioreactor (Cho et al., 2000; Bahramnejad et al., 2019). This method is also used for studying functional genomics in soybean roots. This approach has been used to characterize promoters (Hernandez-Garcia et al., 2009; Li et al., 2014), propagation of nematodes (Cho et al., 2000), symbiotic interactions (Hayashi et al., 2012), pathogenic interactions (Li et al., 2010), gene silencing by RNA interference (RNAi) (Subramanian et al., 2004), and recently for measuring genome editing activity (Du et al., 2016; Cheng et al., 2021). Recently, a reporter gene AtMyb75, encoding an R2R3 type MYB transcription factor, was ectopically expressed in hairy roots-mediated by A. rhizogenes and induced purple/red colored anthocyanin accumulation in soybean hairy roots. This is a convenient, non-destructive, low cost, directly visual selection of transgenic hairy roots (Fan et al., 2020). Several efficient transformation protocols have been developed for studying functional genomics and root biology (Kereszt et al., 2007; Kuma et al., 2015; Chen et al., 2018a,c; Fan et al., 2020; Song et al., 2021).
Protoplast Transfection
The first genetic transformation of soybean protoplasts was achieved by electroporation by Lin et al. (1987). Dhir et al. (1992) was the first to report the transformation of immature cotyledon-derived protoplasts and regeneration of transgenic plants from calli derived from electroporation-transfected protoplasts. Protoplasts could be a good explant for transformation if an efficient regeneration system is established, especially since a large number of protoplasts can be transfected at a time and many forms of genetic materials such as DNA, RNA, and protein can be delivered. Unfortunately, protoplast transfection has not yet been conducted for soybean transgenic plant production. The main challenge is achieving protoplast regeneration, which has yet to be reported in soybean. Protoplast-based transfection has been mainly conducted to evaluate gene functions (Yi et al., 2010; Faria et al., 2011; Kidokoro et al., 2015; Xiong et al., 2019), screen promoters (Sultana et al., 2019), and validate vectors for GE (Sun et al., 2015; Demorest et al., 2016; Do et al., 2019; Patil et al., 2022). Recently, Wu and Hanzawa (2018) developed a method to isolate protoplasts from leaves of soybean seedlings and established a PEG-mediated transfection method that can achieve high transfection efficiency compared to other transient assays.
Agro-Infiltration
Agrobacteria can be infiltrated into the intercellular space of plant tissues to permit the delivery of genes from different organisms into plant genomes (Grimsley et al., 1986). Ever since this method was successfully established for soybean (King et al., 2015), it has been used for virus-induced gene silencing (VIGS) (Kim et al., 2016) and expression of hairpin RNA for RNAi against two-spot spider mites (Dubey et al., 2017).
Agrobacterium tumefaciens-Mediated Transformation
Except for stable transformation, A. tumefaciens is used to carry out transformation for soybean transient assay. Kun et al. (2017) established an Agrobacterium-mediated transient system using calli induced from hypocotyl explants. It has been successfully used in many specific assays including Western blot and Co-IP assay for protein analysis. The system is genotype-flexible and cost-saving. However, it takes a couple of months to complete the assay.
Electroporation
Electroporation is a technique that utilizes a high intensity electric pulse to create transient pores in the cell membrane, thereby facilitating the uptake of macromolecules such as DNA. Christou et al. (1988) conducted electroporation to deliver constructs into soybean calli and showed stable integration of genes but did not succeed in regenerating plants. Later, Chowrira et al. (1995) reported on electroporation of intact nodal meristems which avoided the soybean tissue culture process completely, but no transgenic plants have been recovered.
Transformation Systems for Stable Transgenic Plants
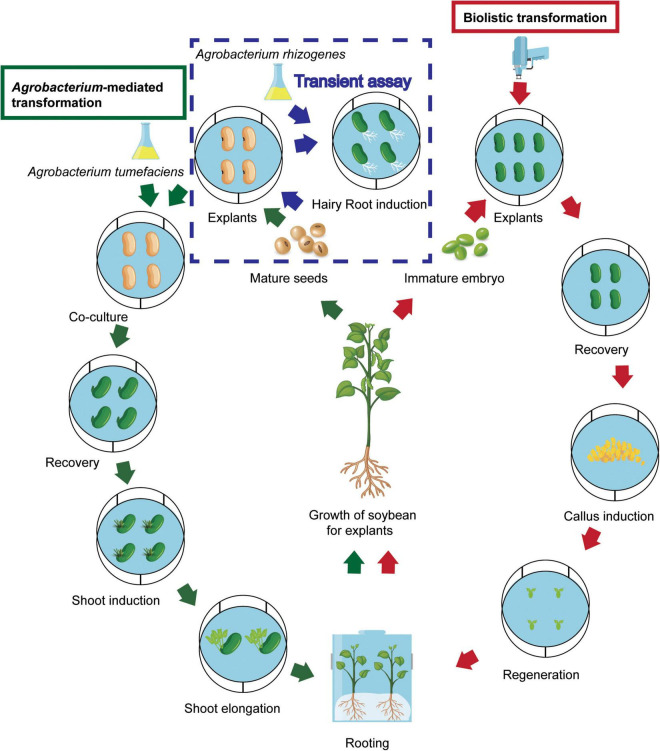
Agrobacterium-mediated transformation and biolistic methods, and in planta transformation and protoplast transfection methods have been applied for generation of transgenic soybean plants. Among these methods, the A. tumefaciens-mediated and biolistic methods are the two major platforms for stable soybean transformation. The general transformation procedure of both methods is shown in Figure 2. The other methods mentioned above are used less because of relatively low efficiency and the specific technique and equipment required in these methods.
FIGURE 2.
General procedure of Agrobacterium-mediated and biolistic soybean transformation. The route following the green arrows is the Agrobacterium tumefaciens-mediated transformation procedure. The route following the red arrows is the biolistic transformation procedure. The route following the blue arrows in the frame with broken blue line shows Agrobacterium rhizogenes-mediated transformation for transient assay.
A. tumefaciens-Mediated Transformation
A. tumefaciens-mediated transformation of soybean was first initiated using cotyledonary nodes by Hinchee et al. (1988). Since the system was established based on regeneration of mature or immature seed explants, the simplicity and relatively high TF of the method have made it a favorite method for soybean. Relatively high efficient Agrobacterium-mediated transformation protocol has been gradually developed through improving factors such as using an appropriate Agrobacterium strain, a good explant, culture media with adequate antioxidant chemicals and combinations of appropriate plant growth regulators for a specific soybean genotype (reviewed in Yamada et al., 2012; Lee et al., 2013; Li et al., 2017; Table 1). Key elements of the progress are summarized in a later section. Main advantages of Agrobacterium transformation include relatively high ratio of single-copy gene insertion, relative simplicity of the transformation procedure, and low cost (Hwang et al., 2017). However, there is a limitation in delivery of genetic material. It delivers DNA plasmids but cannot deliver DNA fragments, RNAs, or proteins.
TABLE 1.
Progress of soybean stable genetic transformation approaches for whole transgenic plants.
| Method | Explant | Genotype | Selectable marker/agent | Physical treatment | Specific chemicals in medium | Agro-strain | Available TF (%) | References |
| Agrobacterium | Immature cotyledon | PI283332 and Peking | NptII/G418 | Wounding | \ | EHA101 and LBA4404 | \ | Parrott et al., 1989 |
| Jack | Hph/Hygromycin B | Wounding | AS | EHA105 | 0.03 | Yan et al., 2000 | ||
| Jack, Williams, Ina, Macon, Dwight, and Rend | Hph/Hygromycin B | Wounding; orientation of explant (downward of the adaxial side) | AS | KYRT1 | 1.3 (1.1–1.7) (Jack) | Ko et al., 2003 | ||
| Mature cotyledonary node | Delmar, Maple Presto, and Peking | NptII/Kanamycin | \ | \ | A208 | \ | Hinchee et al., 1988 | |
| 28 genotypes | NptII/Kanamycin | Wounding and sonication | AS | KYRT1 | 1–2 | Meurer et al., 1998 | ||
| A3237 | Bar/Glufosinate | Wounding | AS, glutamine, and asparagine | EHA101 and EHA105 | 0.9 | Zhang et al., 1999 | ||
| 12 genotypes | NptII/Kanamycin | Wounding | \ | A281, C58, ACH5, and EHA105 | 0.4 (one genotype) | Donaldson and Simmonds, 2000 | ||
| Bert | Bar/Glufosinate | Wounding | AS and L-cysteine | AGL1 | 2.1 | Olhoft and Somers, 2001 | ||
| 12 genotypes | Bar/Glufosinate, Hph/Hygromycin B, and NptII/Kanamycin | Wounding | AS, D-cysteine, and other thiol compounds | AGL1, LBA4404, GV3101, EHA105, and EHA101 | \ | Olhoft et al., 2001 | ||
| 12 genotypes | Bar/Glufosinate | Wounding | AS, L-cysteine, DTT, asparagine, and glutamine | EHA101 | 2–6.3 (glufosinate) 0–2.9 (bialaphos) | Paz et al., 2004 | ||
| Williams82 | Bar/Glufosinate | Wounding | AS and L-cysteine | EHA101 | 5.9 | Zeng et al., 2004 | ||
| 5 genotypes (Chinese soybean) | Hph/Hygromycin B | Wounding | AS, Silwet L-77, and L-cysteine, asparagine, and L-pyroglutamic acid | EHA105 | 3.8–11.7 | Liu et al., 2008 | ||
| Kariyutaka | Bar/Glufosinate or Basta | Wounding (micro brush) | Silwet L-77 | EHA105 | 4.4 | Yamada et al., 2010 | ||
| PK416, JS90-41, Hara Soy, Co1, and Co2 | Hph/Hygromycin B | Sonication and vacuum infiltration, wounding (hypodermic needle) | AS, DTT, L-cysteine, and sodium thiosulfate (STS) | LBA4404, EHA101, and EHA105 | 13.3–18.6 | Arun et al., 2015 | ||
| JS-335 | Bar/Glufosinate | Sonication and vacuum infiltration | AS, DTT, L-cysteine, and STS | EHA105 | 12.6 (10.5–16.2, J8335-bar); | Hada et al., 2018 | ||
| Jack and Zhonghuang 10 | G2Epsps/Glyphosate | Sonication | Silwet L-77, AS, DTT, L-cysteine, and Na2S2O3 | Ag10 | 2.9–5.7 | Guo et al., 2015 | ||
| Jidou17 | NptII/Kanamycin | Sonication | DDT, L-cysteine, sodium thiosulfate, and α-Aminooxyacetic acid | EHA105 | \ | Zhang et al., 2016 | ||
| 7 genotypes | Bar/Glufosinate | Wounding | α-lipoic acid (α-LA), DTT, L-cysteine, AgNO3, glutamine, and asparagine | EHA101 | 14.7 | Yang J. et al., 2016 | ||
| Half-seed | Bert | Hph/Hygromycin B | Wounding | AS, DTT, L-cysteine, and STS | LBA4404 and EHA105 | 16.4 (9.4–26.2 LBA4404); 14 (9.4–26.2 EHA105) | Olhoft et al., 2003 | |
| Thorne, Williams, Williams79, and Williams82 | Bar/Glufosinate | Wounding | AS, L-cysteine, and DTT | EHA101 | 3.8 (1.4–8.7) | Paz et al., 2006 | ||
| 7 genotypes | Hph/Hygromycin B | Wounding (multi-needle) | AS, DTT, L-cysteine, and STS | LBA4404 | \ | Zhang and Xue, 2019 | ||
| 5 US (Williams82) and 5 Chinese genotypes | Bar/Glufosinate | Wounding | AS, DTT, and L-cysteine | EHA105 | 0–6.71 | Jia et al., 2015 | ||
| 7 genotypes | Bar/Glufosinate | Wounding | L-cysteine and DTT | EHA105 | 0.5 (0–0.9) | Sato et al., 2007 | ||
| 7 genotypes | Hph/Hygromycin B | Wounding (multi-needle) | AS, DTT, L-cysteine, and STS | LBA4404 | \ | Zhang and Xue, 2019 | ||
| 5 US (Williams82) and 5 Chinese genotypes | Bar/Glufosinate | Wounding | AS, DTT, and L-cysteine | EHA105 | 0–6.71 | Jia et al., 2015 | ||
| 7 genotypes | Bar/Glufosinate | Wounding | L-cysteine and DTT | EHA105 | 0.5 (0–0.9) | Sato et al., 2007 | ||
| DS97–12 | Hph/Hygromycin B | Sonication and vacuum infiltration | Polyamine (spermidine, spermine, and putrescine) | EHA105 | 29.3 | Arun et al., 2016 | ||
| Williams82 | Bar/Glufosinate | Wounding | AS, L-cysteine, and DDT | EHA101 | 1.0–3.5 (35s or NOS promoter) | Testroet et al., 2017 | ||
| 8 genotypes | Bar/Glufosinate | Wounding | AS, DDT, STS, L-cysteine, AgNO3, L-asparagine, L-pyroglutamic acid, and L-ascorbic acid | EHA101 | 7.3–10.0 | Li et al., 2017 | ||
| Jack, Williams82, Zigongdongdou, and Heihe27 | Bar/Glufosinate | Wounding | DTT, AS, L-asparagine, and L-glutamine | EHA101 | 7.6 (2.6–11.1) | Chen et al., 2018b | ||
| DS-9712 | NptII/Kanamycin | Sonication and vacuum infiltration | AS and L-cysteine | EHA105 | 14.51 | Hada et al., 2018 | ||
| PUSA 9712 | Bar/Basta | \ | SNP | EHA101 | 34.6 | Karthik et al., 2020 | ||
| Maverick and 20 proprietary elite lines | Pat/Glufosinate | Wounding | L-asparagine and L-pyroglutamic acid | EHA101 and EHA105 | 18.7 (12.1–23.0) | Pareddy et al., 2020 | ||
| whole cotyledonary node | ZhongHuang13 | NptII/Kanamycin | Wounding | L-cysteine | EHA105 | 23.1 | Zhang et al., 2014 | |
| Calluses induced from either cot-node | 5 genotypes | Bar/Glufosinate | \ | AS, DTT, L-cysteine, and STS | AGL1 | 1.3 (0.3–4.3) | Hong et al., 2007 | |
| Hypocotyls | Heinong44 | NptII/Kanamycin | \ | AS, L-cysteine, DTT, AgNO3, and STS | EHA105 | 9.3 | Wang and Xu, 2008 | |
| Embryogenic cell suspension | Chapman | Hph/Hygromycin B | Sonication | AS | EHA105 | \ | Trick and Finer, 1998 | |
| Embryogenic axes | P29T50, P33T50,93Y21, DM118, and 98C21 | SpcN/Spectinomycin | Sonication | AS and DDT | Ochrobactrum haywardense H1 | 35 | Cho et al., 2022 | |
| Biolistic method | Immature embryo axis | Williams82 and Mandarin Ottawa | NptII/Kanamycin | Electrical, arc-discharge gun/Gold particles | Plasmid DNA | Mccabe et al., 1988 | ||
| Williams82 | NptII/Kanamycin | PDS 1000/Tungsten | Plasmid DNA | Sato et al., 1993 | ||||
| Somatic embryogenic suspension | Fayette | Hph/Hygromycin B | DuPont Biolistics TM Particle Delivery System (Model BPG)/Tungsten particles | Plasmid DNA | 0.4 | Finer and Mcmullen, 1991 | ||
| Fayette | Npt II/G418 | PDS 1000/Tungsten particles | Plasmid DNA | Four plants per bombarded flask | Sato et al., 1993 | |||
| Fayette | Hph/Hygromycin B | PDS 1000/Tungsten particles | Multiple plasmid DNA | \(co-transformation) | Hadi et al., 1996 | |||
| \ | Hph/Hygromycin B | PDS 1000/Tungsten particles | Plasmid DNA | \(protocol) | Finer and Larkin, 2008 | |||
| \ | Hph/Hygromycin B | PDS 1000/Tungsten particles | Plasmid DNA | \(protocol) | Finer, 2016 | |||
| 93B86 | Hph/Hygromycin B and Als/Chlorsulfuron | PDS 1000/Gold particles | Plasmid DNA and DNA fragment | \(targeted insertion) | Li et al., 2015 | |||
| Mature embryo axis | BR-16, Doko RC, BR-91, and Conquista | AHAS/Imazapyr | HPHMAS/Tungsten | \ | Plasmid DNA | 0.1–7.8 | Aragão et al., 2000 | |
| BR-16, BR-91, Celeste, Conquista, Doko RC, Nina, Indiana, and Itaipu | AHAS/Imazapyr | PDS1000/Tungsten | Plasmid DNA | ≤0.2 (protocol) | Rech et al., 2008 | |||
| Conquista | AHAS/Imazapyr | HPHMAS/Tungsten | \ | DNA fragments | 0.8 | Vianna et al., 2011 | ||
| INCASoy-36 | Cp4epsps/Glyphosate | PDS 1000/Tungsten | Plasmid DNA | 6 | Soto et al., 2017 | |||
| Immature embryo | Maverick | Hph/Hygromycin B, DSM2/Glufosinate | Cold treatment and plasmolysis | Plasmid DNA | 2–5.5 (hph) and 1–2.7 (DSM2) | Chennareddy et al., 2018 |
(1) HPHMAS: The high-pressure helium-driven microparticle acceleration system. (2) \ means not available. (3) Protocol means the reference is a published protocol.
Biolistic Transformation
Biolistic transformation, known as gene gun or particle bombardment, delivers small tungsten or gold particles coated with desired genes to target plant cells (Christou et al., 1988). Since an electrical-discharge gene gun was first used in soybean to regenerate a fertile transgenic plant (Mccabe et al., 1988), gene delivery to meristematic soybean cells by particle bombardment has been considered to be more genotype-flexible for transfer of foreign DNA into soybean (Homrich et al., 2012). Recently, embryogenic callus based biolistic method becomes more popular due to its relatively higher efficiency compared to other explants and its directly delivering way which meets the need for genome editing using RNA and RNPs editing reagents for recovery of DNA-free edited events. In comparison to the A. tumefaciens-mediated method, the biolistic method offers benefits with their capacity to transform organelles and deliver RNA, proteins, nanoparticles, dyes, and complexes to cells (Klein et al., 1987; Liang et al., 2017). The drawback is mainly high transgene copy and relatively high cost, and its application has been restricted in limited soybean genotypes because of unavailable meristematic explants. Compared to plasmid bombardment, utilization of specific constructs including linear minimal expression cassettes (MECs) in biolistic transformation enables the production of plants carrying much simpler patterns of transgene integration, which has been confirmed in plants such as wheat (Ismagul et al., 2018). The major progress in soybean biolistic transformation is presented in a later section and summarized in Table 1.
Other Stable Transformation Methods
A. rhizogenes-Mediated Transformation
Transgenic plants can also be produced by regeneration of hairy roots transformed with A. rhizogenes. Success of stable transformation has been reported in many plant species (Christey, 2001). In soybean, stable soybean transgenic plants were produced from hairy roots using primary-node explants infected by a disarmed A. rhizogenes strain SHA17 (Olhoft et al., 2007) and the several reports of targeted mutation events using genome editing also have been obtained from hairy roots through A. rhizogenes-mediated transformation (Curtin et al., 2011; Haun et al., 2014; Demorest et al., 2016). However, genotype inflexibility has been the main hurdle for using the method in soybean.
In-Planta Gene Transformation
This is an alternative method in which Agrobacterium is used to infect explants, but it does not involve in vitro culture and regeneration of plant cells or tissues (Kalbande and Patil, 2016), thereby reducing time and labor cost, and, most importantly, avoiding somaclonal variation occurrence during in vitro culture-mediated genetic transformation and regeneration. In soybean, an Agrobacterium suspension is directly injected into the ovary (Liu et al., 2009), axillary meristematic region of germinated seedling (Chee et al., 1989), or stigma in which exogenous DNA was introduced into cells via the “pollen-tube-pathway” (Hu and Wang, 1999). Transgenic events could be obtained from progeny seeds. Liu et al. (2009) reported the transfer of a minimal linear marker-free and vector-free smGFP cassette into soybean by pollen tube-mediated gene transfer. Mangena (2019) summarized the progress made in in planta transformation and formulated a simple protocol using in planta Agrobacterium injection of seedlings. Although this could be a tissue culture bypass method and attempts for new ways are made from time to time, its efficiency has been very low and it is often not repeatable. This method has not been widely used.
Transformation Using a New Bacterium
Recently a novel bacterium, Ochrobactrum haywardense H1 (Oh H1), was discovered and it is capable of efficient plant transformation (Cho et al., 2022). Ochrobactrum is able to host for Agrobacterium-derived vir and T-DNA and helps to deliver transgenes in soybean. Oh H1-8 generated high-quality transgenic events by single-copy, plasmid backbone-free insertion at frequencies higher than those of Agrobacterium strains. It achieved high transformation efficiency in several soybean genotypes, which can be up to 35%. The application of the new bacterium-mediated transformation in soybean needs to be evaluated further.
Progress Made to Improve Soybean Transformation Over the Last Decade
Since 2010, increasing the transformation frequency (TF) has been the main focus for soybean transformation improvement. Several major factors affecting soybean TF based on Agrobacterium-mediated transformation have been identified, and progress has been made in establishing a high-throughput transformation system (Zhang et al., 2014; Arun et al., 2015, 2016; Yang X. F. et al., 2016; Li et al., 2017; Chen et al., 2018b; Karthik et al., 2020; Pareddy et al., 2020). Some confirmed positive elements in Agrobacterium-mediated transformation protocols have also been applied for enhancing soybean biolistic transformation (Table 1).
Agrobacterium-Mediated Transformation
Soybean transgenic plant production still relies on Agrobacterium-mediated transformation (Figure 2 and Table 1). Recently, high TFs of over 10% have been obtained in more and more soybean genotypes using improved protocols (Zhang et al., 2014; Arun et al., 2015, 2016; Yang X. F. et al., 2016; Li et al., 2017; Chen et al., 2018b; Karthik et al., 2020; Pareddy et al., 2020). The enhancement of TF is based on changes in several factors, including explant, selectable marker, and culture medium composition such as antioxidants, of these protocols (Table 1).
Adjustment of Infection Method and Improving Regeneration
Reducing the explant tissue browning and necrosis caused by Agrobacterium enhances construct delivery and regeneration of transformed cells. Changing the ways for preparation of Agrobacterium infection solutions and co-cultivation media, and modifying infection methods can achieve this goal and eventually increase transformation efficiency. Addition of antioxidants such as dithiothreitol (DTT) in infection solutions and extending co-cultivation time to 5 days achieved an infection efficiency of more than 96% and, hence, increased TF (Li et al., 2017). Infection solutions prepared with a two-round overnight culture of Agrobacterium using AB minimal media in second round culture significantly increased transformation frequency in comparison with the culture using normal YEP medium (Pareddy et al., 2020). It was also found to be beneficial to A. tumefaciens infection when the co-cultivation temperature for soybean transformation was set to 23°C under dim light (Yang X. F. et al., 2016). The same group also demonstrated to alleviate explant necrosis and significantly improve the transformation efficiency when antioxidants alone such as α-lipoic acid (α-LA, 0.12 mM) and silver nitrate (AgNO3, 20 μM), or combinations of antioxidants such as L-cysteine (1 mM) + DTT (3.3 mM) + AgNO3 (20 μM), and L-cysteine (1 mM) + DTT (3.3 mM), were added in the solid co-cultivation medium. For improving regeneration, it was found that adding 6-benzylaminopurine (BAP) in a germinating medium could significantly increase regeneration efficiency, which led to enhancement of TF; the optimal BAP concentration for shoot formation was 0.5 mg/L (Zhang et al., 2014). More examples are presented in Table 1.
Genotype Effect and Explant Choice
In the tissue culture-based transformation process, the composition of culture media and susceptibility of selected explants to Agrobacterium influence soybean transgenic frequency. A highly efficient in vitro culturing system and regeneration of cells susceptible to Agrobacterium are prerequisites for a reliable transformation protocol. Until now, the TF for most tested genotypes of soybean has remained quite low at a level mostly below 5% when conducting Agrobacterium-mediated transformation [summarized in Yamada et al. (2012); Jia et al. (2015), and Li et al. (2017); Table 1]. Since 2000, many research groups have used model soybean varieties such as Jack, Bert, and Williams serials and other specific genotypes because of their amenability to transformation (Olhoft and Somers, 2001; Olhoft et al., 2001, 2003; Paz et al., 2004, 2006; Zeng et al., 2004; Luth et al., 2015). Recently, soybean transformations with high TFs have been reported using specific genotypes. For example, it was claimed 23.1% with Zhonghuang13 (Zhang et al., 2014) and an average of 14% TF for a local Indian genotype, DS-9712 (Hada et al., 2018). Improvement based on Agrobacterium-mediated soybean transformation has been made to expand target genotypes from conventional model varieties to many elite varieties (Ko et al., 2003; Yi and Yu, 2006; Sato et al., 2007; Song et al., 2013; Arun et al., 2015; Pareddy et al., 2020). For example, over 5% TF for more than 10 varieties was achieved with a robust protocol (Pareddy et al., 2020).
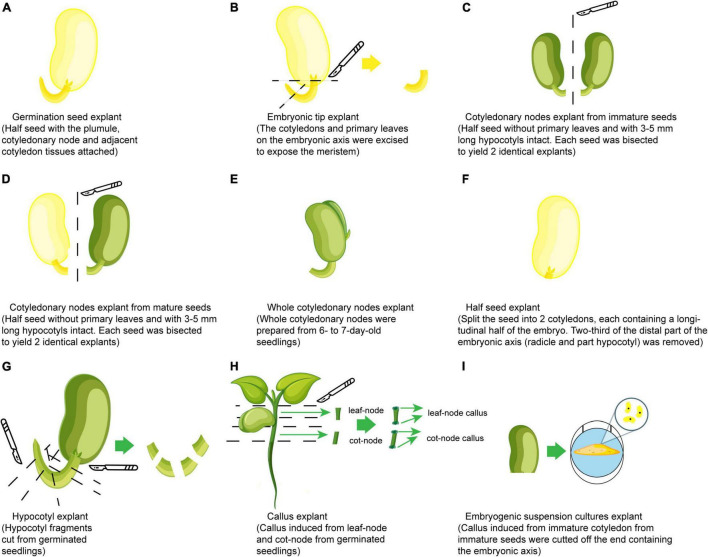
Since Hinchee et al. (1988) obtained transgenic events, the cotyledonary node of mature seeds has been the most favorite explant used for Agrobacterium-mediated soybean transformation using many other explants such as embryonic tips and calli (Figure 3). Cotyledonary node regions have axillary meristems at the junction between cotyledon and hypocotyl, which can proliferate and regenerate by the formation of multiple adventitious shoots on a culture medium containing cytokinin. Successful transformation has been achieved using similar organogenesis from various explants, which include germination seeds (Chee et al., 1989), embryonic shoot tips (Martinell et al., 2002; Liu et al., 2004), cotyledonary nodes from immature seeds (Parrott et al., 1989; Yan et al., 2000; Ko et al., 2003), cotyledonary nodes from mature seeds (Meurer et al., 1998; Zhang et al., 1999; Donaldson and Simmonds, 2000; Olhoft and Somers, 2001; Olhoft et al., 2001; Paz et al., 2004; Zeng et al., 2004; Liu et al., 2008), half-seeds (Paz et al., 2006; Pareddy et al., 2020), whole cotyledonary nodes (Zhang et al., 2014) and hypocotyls (Dan and Reichert, 1998; Liu et al., 2004; Wang and Xu, 2008), and other explants with different regeneration procedures such as calli induced from geminated seedlings (Hong et al., 2007) and embryogenic suspension cultures (Trick and Finer, 1998). However, successful and repeatable production of transgenic soybean via Agrobacterium-mediated transformation has mainly been based on protocols with explants containing cotyledonary nodes from young seedlings and imbibed mature seeds (Zhang et al., 1999; Olhoft et al., 2003; Paz et al., 2006). Recently, half-seeds have gradually become the trend for explants since (Paz et al., 2006) their first use, because half-seed explants possess advantages to have more nutrition supply for shoot regeneration compared to cotyledonary nodes and to be prepared within a short time (less than 1 day) due to using imbibed seeds, which reduces the period of total regeneration and labor cost. Based on descriptions of explants in several reports (Paz et al., 2006; Pareddy et al., 2020), half-seed, whole cotyledon, and split seed explants can now be put under the same category of half-seed explants. Obtaining TFs of over 10% for soybean with half-seed explants have been demonstrated in many reports (Zhang et al., 2014; Arun et al., 2016; Li et al., 2017; Chen et al., 2018b; Hada et al., 2018) (Table 1). The highest TF of 34.6% has been obtained using these explants together with nitric oxide treatment in a co-cultivation medium in the protocol made by Karthik et al. (2020). Some specific explant treatments such as sonication in combination with vacuum infiltration, sonication in combination with surfactant, or just sonication (Mariashibu et al., 2013; Arun et al., 2015; Guo et al., 2015; Zhang et al., 2016; Hada et al., 2018), and pre-wounding with a multi-needle consisting of 30 thin fibers (Xue et al., 2006) or a micro-brush (Yamada et al., 2010) were also used before Agrobacterium infection to increase infection rate and TFs, because these treatments facilitate the penetration of Agrobacterium into plant tissues and increase the contact between plant cells and the bacterium, and stimulate the infection ability of the bacterium, which leads to T-DNA transfer into plant cells.
FIGURE 3.
Types of explants used for soybean transformation. (A) Germination seed. (B) Embryonic shoot tips. (C) Cotyledonary nodes from immature seeds. (D) Cotyledonary nodes from mature seeds (yellow cotyledon means use cotyledon directly from mature seed; the green one means use mature cotyledon after germination under light). (E) Whole cotyledonary node. (F) Half-seed. (G) Hypocotyl. (H) Callus induced from geminated seedlings. (I) Embryogenic suspension cultures.
Addition of Antioxidants in Medium
Antioxidants, in general, are known to reduce pathogen-induced programed cell death (Mittler et al., 1999). These include inhibitors of polyphenol oxidases (PPOs) and peroxidases (PODs) through the action of their thiol group, such as compounds L-cysteine, DTT, and sodium thiosulfate. They are commonly used to reduce enzymatic browning in food processing caused by deposition of tannins (Nicolas et al., 1994; Ghidelli et al., 2014). Polyvinylpyrrolidone (PVP), DTT, L-cysteine, glutathione, α-LA, L-ascorbic acid, and citric acid have been confirmed to decrease tissue necrosis of explants used for Agrobacterium-mediated transformation (Barampuram and Zhang, 2011). Either one or more than 2 of the chemicals have been used in soybean transformation (Olhoft and Somers, 2001; Olhoft et al., 2003; Paz et al., 2004; Yi and Yu, 2006; Liu et al., 2008). L-cysteine and DTT have been frequently used in soybean transformation since its first use by Olhoft et al. (2001). Reports clearly showed that there was less browning on the cut and damaged surfaces of the hypocotyl, cotyledon node region, and on the cotyledon of explants, which increased the TF of stable transformations (Olhoft et al., 2001, 2003; Paz et al., 2004; Li et al., 2017). A high average TF of 12.7% resulted from the combination of L-cysteine and DTT, which was significantly greater than that of either L-cysteine or DTT alone (Olhoft et al., 2003). The positive effect has been continuously confirmed in recent reports (Table 1). Another type of antioxidants is a group of sulfur-containing compounds involved in several multienzyme complexes such as α-LA. These include pyruvate dehydrogenase, α-ketoglutarte dehydrogenase, branched-chain ketoacid dehydrogenase, and glycine decarboxylase (Dan et al., 2009). Adding the antioxidant α-LA in a co-cultivation medium could increase transient GUS expression and increased the percentage of shoot induction (Yang X. F. et al., 2016). In this report, 0.12 mM α-LA was found to be the most useful for alleviating browning and necrosis. Other antioxidants conventionally used in plant tissue culture, such as ascorbic acid, PVP, and citric acid, may promote soybean transformation efficiency, but their roles have not yet been made clear (Li et al., 2017). Plant hormone-like antioxidants such as sodium nitroprusside (SNP), a nitric oxide (NO) donor, play varied roles in growth and development of plants. Nitric oxide is involved in cell metabolism and morphogenesis and acts as a signaling molecule in response to various biotic and abiotic stresses (Verma et al., 2020), and can alleviate abiotic stress threat in plants reacting quickly with ROS. SNP significantly enhanced regeneration and development rate of soybean plants (Karthik et al., 2019); addition of SNP also significantly increased soybean TF by up to 34.6% with the half-seed method (Karthik et al., 2020).
Addition of Other Chemicals in Culture Medium
Except for antibiotics, chemicals related to host defense response, ethylene inhibitors, surfactants, demethylating reagents, polyamines, and antagonist α-aminooxyacetic acid (AOA) are proved to have a positive effect on improving TF. L-glutamine and L-asparagine are types of chemicals that weaken host defense responses. It has been reported that the addition of L-glutamine into a culture medium alone or in combination with a cold shock pretreatment could enhance Agrobacterium transformation efficiency (Zhang et al., 2013). Although the mechanism is still not clear, L-glutamine could play a role in lessening host defense responses by attenuating the expression of certain pathogenesis-related genes (PRs), and potentially improve the efficiency of Agrobacterium-mediated plant transformation (Zhang et al., 2013, 2014). It was demonstrated that TF was significantly increased in soybean when additional L-glutamine or L-asparagine alone, or both of them were added in all culture media (Chen et al., 2018b). The TF was 8.8 ± 1.5 (L-glutamine), 5.9 ± 2.1% (L-asparagine), 11 ± 0 (both), and 3.5 ± 2.4% (without any one of them). Ethylene inhibitors such as AgNO3 have a positive effect on transformation. It has been reported that Ag+ interferes with the binding of ethylene receptor sites and helps reduce ethylene production by promotion of polyamine biosynthesis (Roustan et al., 1990). The main function of AgNO3 is to eliminate the potential danger to plant cells and tissues in liverwort caused by ethylene (Beyer, 1979). It has already been confirmed to promote somatic embryo production and shoot regeneration in wheat and maize (Carvalho et al., 1997; Fernandez et al., 1999). This effect has been proved to improve soybean TF (Olhoft et al., 2004; Li et al., 2017). A nearly 10% TF with genotype Heilong44 was reported when BAP and AgNO3 were added into a culture medium (Wang and Xu, 2008). Surfactants such as SilwetL-77 and pluronic acid F68 also increase TF, which initially showed to enhance T-DNA delivery in wheat Agrobacterium-mediated transformation when added into an inoculation medium (Cheng et al., 1997). This was also confirmed in soybean transformation. It was reported that adding SilwetL-77 to an infection medium coupled with hygromycin-based selection strategies led to transformation efficiencies ranging from 3.8 to 11.7% in Chinese soybean varieties (Liu et al., 2008). SilwetL-77 has been frequently used to increase soybean TF (Yamada et al., 2010; Guo et al., 2015). Surfactants may enhance T-DNA delivery by aiding A. tumefaciens attachment and/or by elimination of certain substances that inhibit A. tumefaciens attachment (Opabode, 2006). Polyamines enhance plant cell differentiation, induce totipotency, and increase cell division (Rakesh et al., 2021). Addition of polyamines in the plant transformation process leads to vir gene induction and T-DNA transfer, and increases transformation efficiency (Kumar and Rajam, 2005). As high as 29.3% TF in soybean has been achieved by addition of spermidine, spermine, and putrescine in a culture medium compared with its counterparts (14.6%) and with respective plant growth regulator (PGR) alone (Arun et al., 2016). Demethylating reagents commonly applied in epigenetic research such as 5-azacytidine (5-Azac), significantly improve the transient transfection efficiency and transgene expression level in low-efficiency genotypes. Treatment with 5-Azac improved the shoot regeneration efficiency in low-efficiency genotypes during the process of Agrobacterium-mediated soybean transformation. This indicates that lower methylation level in transgenes contributed to enhance shoot regeneration in Agrobacterium-mediated soybean transformation (Zhao et al., 2019b). Antagonist AOA relieves the structural membrane barriers of Agrobacterium entering cells, hinders the perception of intercellular signal transmission, and thus effectively alleviates defense responses and increases the susceptibility of cells to Agrobacterium infection. Combined use of AOA and sonication treatments (novel method) greatly improved T-DNA delivery efficiency in soybean (Zhang et al., 2015, 2016).
Refining Selection Agents
The most frequently used selectable markers in both the somatic embryogenesis- and organogenesis-based soybean transformation methods are genes conferring resistance to herbicides or antibiotics so as to reduce escape rate significantly. The selectable markers include bar and pat genes conferring resistance to phosphinothricin, the active ingredient in BASTA and bialaphos herbicides (Zhang et al., 1999; Olhoft and Somers, 2001; Olhoft et al., 2001; Paz et al., 2004; Testroet et al., 2017; Pareddy et al., 2020), EPSPS (5-enolpyruvylshikimate-3-phosphate synthase) genes conferring resistance to the herbicide glyphosate (Martinell et al., 1999; Clemente et al., 2000; Yao, 2001; Guo et al., 2015; Xiao et al., 2019), and the nptII gene conferring resistance to the antibiotics kanamycin (Homrich et al., 2012) and hph or hpt II (hygromycin phosphotransferase) genes conferring resistance to hygromycin B (Yan et al., 2000; Ko et al., 2003; Olhoft et al., 2003; Liu et al., 2008). Recently, Hph and Bar or Pat have been proven to be the most favorite selectable markers (Table 1). An average transformation frequency as high as 29.3% was achieved with the half-seed (Arun et al., 2016) and 13.3–18.6% with cotyledonary node explant (Arun et al., 2015) employing hygromycin B selection, which is better than or comparable with that by Olhoft et al. (2003). Glufosinate has also been used as a selection agent based on the bar or the pat gene and initially had less than 10% TF in soybean transformations involving the half-seed (Paz et al., 2006) and embryo tip (Dang and Wei, 2007) explants. Recently TFs of over 10% have been obtained with cotyledonary nodes (Hada et al., 2014; Yang X. F. et al., 2016) and half-seeds (Li et al., 2017; Chen et al., 2018b; Pareddy et al., 2020). A 34.6% TF was reported using a protocol with addition of sodium nitroprusside (SNP) when Basta was sprayed for selection (Karthik et al., 2020). Since an epsps/glyphosate selection based protocol is established (Martinell et al., 1999), glyphosate has gradually been incorporated into transformation as a selectable agent and has shown its beneficial side for high stringency. In order to quickly and efficiently screen glyphosate-tolerant events, a rapid and convenient spotting method was established for screening regenerated glyphosate-tolerant T0 plantlets (Guo et al., 2020a). In this report, an optimized Agrobacterium-mediated soybean transformation system with rapid and effective selection of transformed cells was developed, with TFs ranging from 2.9 to 5.6%. Especially, 96% regenerated T0 plantlets showed clear tolerance to glyphosate and their transgenic nature were confirmed by molecular analysis. Spectinomycin was also used as a selective agent to obtain transgenic soybean when the aminoglycoside-3′′-adenylyltransferase gene (aadA) was used as a selectable marker (Martinell et al., 2002). The spectinomycin selection protocol demonstrated higher frequency of transformation, a shorter period of time needed to complete each protocol, and lower cost compared with the glyphosate selective protocol. Soybean transformation using a GFP as a detection marker was also reported, and transgenic plants could be identified at an early stage, although the frequency was not high (2.5%) (Yang S. et al., 2019). Combined with a normal selectable marker and a selection agent, GmFAST (fluorescence-accumulating seed technology) has recently been developed to identify homologous transgenic seeds. It is a marker composed of a soybean seed-specific promoter coupled to the OLE1-GFP gene, which encodes a GFP fusion of the oil-body membrane protein OLEOSIN1 of Arabidopsis thaliana and is a time-saving and efficient method to produce homologous transgenic events (Iwabuchi et al., 2020).
Generally, the efficiency of Agrobacterium-mediated transformation in soybean has been enhanced by improving both Agrobacterium infection and explant regeneration. Addition of antioxidants such as DDT, L-cysteine, and NO in a co-cultivation medium and some infection-assisting specific chemicals including surfactants and AgNO3, and some regeneration-promoting elements such as polyamines (Table 1), plays an important role in the improvement. All of the measures have facilitated Agrobacterium to transform the meristematic region of soybean explants. Recently, a specific Agrobacterium-mediated protocol was reported, which conducted bombardment to make wounding and reduce the lab work time to only 2 days in the transformation process and kept the rest time to grow T0 plants in a glasshouse (Paes de Melo et al., 2020). Transgenic events were screened using a swab to spread glufosinate solution on leaves of putative events and the TF reached nearly 10%. This method avoided many tissue culture steps and may be a cost-saving protocol.
Biolistic Transformation
Since Mccabe et al. (1988) reported the first transgenic soybean plant using the biolistic method, many reports of soybean biolistic transformation have been published and the development of this method in the first 25 years has been reviewed by Homrich et al. (2012); Lee et al. (2013), Mariashibu et al. (2013), and Mangena et al. (2017). Initially, meristems of soybean tissues as the target tissue were used for bombardment such as embryonic axes of immature and mature seeds (Sato et al., 1993; Aragão et al., 2000; Rech et al., 2008; Soto et al., 2017). In later studies, somatic embryos (Finer and Mcmullen, 1991; Finer and Larkin, 2008; Finer, 2016) were the most frequently used explants for biolistic transformation. However, chimeric transgenic plants were produced because of multiple cell layers (L1, L2, and L3) in the original apical meristem of soybean (Christou, 1990; Christou and Mccabe, 1992). Fortunately, using secondary somatic embryos and new selective markers such as EPSPS has eliminated transgenic chimeras (Sato et al., 1993; Martinell et al., 1999). Somatic embryo regeneration and proliferation were initiated either on semi-solid media (Parrott et al., 1989) or liquid suspension culture media (Finer and Nagasawa, 1988). Co-transformation of multiple plasmids or multiple gene inserts in same constructs with selectable markers has been achieved (Hadi et al., 1996; Li et al., 2015). Since 2010, factors such as explant type, abiotic stress treatment, selectable marker, and tissue culture method have been the main focus to improve biolistic transformation TF, and reliable protocols for the biolistic method with embryogenesis-based explant have been developed to produce a reasonable number of transgenic plants (Table 1). For example, a TF of up to 6% was achieved with cp4epsps as selectable marker when embryonic axes of mature seeds of the INCASoy-36 Cuban cultivar were bombarded (Soto et al., 2017). Chennareddy et al. (2018) combined an immature half-seed explant with an intact embryonic axis, cold and plasmolysis pre-treatment, and a specific somatic embryogenic callus regeneration medium in their protocol. They achieved 5% TF with HPH/hygromycin selection and 2.7% with DSM2/glufosinate selection. A selection system using NPTII/G418 was developed for a biolistic-transformed embryogenic callus rather than the most used HPH/hygromycin system and similar TF in comparison with the HPH system was obtained (Itaya et al., 2018). The current status is that soybean biolistic transformation still relies on an embryogenic callus, since it is the prerequisite for establishing a robust transformation system for a specific genotype. Selection for the amenability of an embryogenic callus induced from local elite varieties (genotypes) is the main focus (Joyner et al., 2010; Abbasi et al., 2016; Islam et al., 2017; Raza et al., 2020). An improved biolistic soybean transformation protocol was published using an embryogenic callus induced from an immature cotyledon explant (Finer, 2016), which is a robust one and can produce quite a lot of transgenic plants within 6–9 months.
Recent Applications of Soybean Transformation for Trait Improvement
Transgenic technology has been used to improve soybean agronomic traits, which include yield component, grain quality, and biotic and abiotic stress tolerance, and economic traits such as oil and biofuel quality, and specific chemical content in seed for human health, and other traits. Trait improvements through forward and reverse genetic approaches in the last 5 years are summarized in Table 2; i.e., downregulation of the pyruvate dehydrogenase kinase gene GmPDHK through RNAi made an average of 42.2% protein content in seeds of transgenic plants, which is significantly increased compared with the non-transgenic control (Jones et al., 2020). Soybean seeds with linolenic acid content in excess of 50% of the total oil have been generated by increasing the expression of the FAD3 gene, which encodes the enzyme that converts linoleic acid to linolenic acid (Yeom et al., 2020). Overexpressing the GmmiR156b (Squamosa promoter-binding protein-like, SPL) gene in soybean and transgenic plants produced significantly increased numbers of long branches, nodes, and pods that exhibited increased 100-seed weight, resulting in a 46–63% increase in yield per plant and no significant impact on plant height in a growth room or under field conditions (Sun et al., 2019). Stable GmMYB14-overexpressing (GmMYB14-OE) transgenic soybean plants demonstrate semi-dwarfism and a compact plant architecture associated with decreased cell size, causing decreased plant height, internode length, leaf area, leaf petiole length, and leaf petiole angle, and improved yield in high density and drought tolerance under field conditions (Chen et al., 2021b). Salt-tolerant transgenic soybean and its applications in field are summarized in a review (Cao et al., 2018). Resistance to soybean cyst nematode (SCN; Heterodera glycines) in stably transformed soybean plants is enhanced by downregulation of the HgY25 and HgPrp17 genes, which are related to reproduction and fitness (Tian et al., 2019). Overexpression of PAC1 and GmKR3, a TIR–NBS–LRR-type R gene, can increase multiple virus resistance in transgenic soybean and, thus, provide an efficient control strategy against RNA viruses such as SMV, BCMV, WMV, and BPMV (Xun et al., 2019). Overexpression of GmDR1 [Glycine max disease resistance 1 (Glyma.10G094800)] led to enhanced resistance not only against F. virguliforme but also against spider mites (Tetranychus urticae, Koch), soybean aphids (Aphis glycines, Matsumura), and SCN (Ngaki et al., 2021). Many types of herbicide-resistant transgenic soybean, such as glyphosate-resistant, dicamba-, and 2,4-D-resistant, are grown widely in the United States (Nandula, 2019). Transgenic soybean plays an important role in soybean production worldwide now, and transgenic soybean covers 50% of the global transgenic crop area, occupying 94.1 million ha (Nandula, 2019). Therefore, a better soybean transformation system is the base for soybean improvement through transgenic technology.
TABLE 2.
Summary of transgenic approaches for soybean trait improvement and functional genomics in the last 5 years.
| Target traits | Transgene | Source of gene | Delivery method | Effect on trait or function | Genotype | References |
| Seed components and quality | ||||||
| Seed protein and amino acid | Zmδ-zeins and Zmγ-zein | Z. mays | A. tumefaciens | Increase 27% the methionine content | Williams82 | Kim and Krishnan, 2019 |
| Zmβ-zein | Z. mays | A. tumefaciens | Increase 15% the methionine content | Jack | Guo et al., 2020b | |
| GmPDHK | G. max | Biolistic method | Increase average 42.2% protein content | Jack | Jones et al., 2020 | |
| Glyma.10G38760a | G. max | A. tumefaciens | Increase sulfur amino acid content | Maverick | Kim et al., 2020 | |
| Oil | GmFAD2-1B | G. max | A. tumefaciens | Increase oleic acid content | Williams82 | Yang J. et al., 2018 |
| GmSDP1-1 | G. max | A. tumefaciens | Increase oil content | Kariyutak | Kanai et al., 2019 | |
| PfFAD3-1 | P. fendleri | A. tumefaciens | Increase α-linolenic acid production | Kwangankong | Yeom et al., 2020 | |
| GmOLEO1 | G. max | A. tumefaciens | Increase 10.6% seed oil content and enriched smaller OBs | Williams82 | Zhang et al., 2019a | |
| GmWRI1b | G. max | A. tumefaciens | Increases total seed oil production | \ | Guo et al., 2020c | |
| Glyma.13G30950 | G. max | A. tumefaciens | Increase seed pods and oil production | Kariyutaka | Iwabuchi et al., 2020 | |
| GmDGAT2A | G. max | A. tumefaciens | Increase oil production and α-linoleic acid content | P03 | Jing et al., 2021 | |
| GmZF392 | G. max | A. tumefaciens | Increase seed oil accumulation | Jack | Lu et al., 2021 | |
| GmWRI1a | G. max | A. tumefaciens | Increase seed oil content | Dongnong50 | Wang Z. et al., 2022 | |
| AhDGAT3 | A. hypogaea | A. tumefaciens | Increase oleic acid and total fatty acid | Jack | Xu et al., 2022 | |
| Bioreactor | rhBMP2 | H. sapiens | Biolistic method | Result in production of bone morphogenetic protein BMP2 | BRS16 | Queiroz et al., 2019 |
| The lunasin gene | G. max | A. tumefaciens | Result in production of bioactive lunasin peptide | \ | Hao et al., 2020 | |
| The hIFN-γ gene | H. sapiens | A. tumefaciens | Result in production of human IFN-γ protein | Williams | Mehrizadeh et al., 2021 | |
| Phytate content | GmIPK2 | G. max | A. tumefaciens | Result in production of low phytate | Pusa-16 | Punjabi et al., 2018 |
| GmMIPS1 | G. max | A. tumefaciens | Regulate phytate biosynthesis | DS-9712 | Kumar et al., 2019 | |
| EcMappA | E. coli | A. tumefaciens | Result in production of a thermostable phytase | Wandou-28 | Zhao et al., 2019c | |
| Specific chemical compounds | ZmGB1 | Z. mays | A. tumefaciens | Increase glycinebetaine content | A5403, A4922, A3469, and A3244 | Castiglioni et al., 2018 |
| GmCHI1A | G. max | A. tumefaciens | Increase seed isoflavones | DT2008 | Nguyen et al., 2020 | |
| GmMATE1 | G. max | A. tumefaciens | Increase seed isoflavones | C08 and W05 | Ng et al., 2021 | |
|
GmMYB176 and GmbZIP5 |
G. max | A. rhizogenes | Increase seed isoflavones | Harosoy63 | Anguraj Vadivel et al., 2021 | |
| Agronomic traits | ||||||
| Seed yield and plant biomass | GmPT7 | G. max | A. tumefaciens | Increase symbiotic N2 fixation and yield | HN66 | Chen et al., 2019b |
| GmmiR156b | G. max | A. tumefaciens | Improve the shoot architecture and yield | Williams82 | Sun et al., 2019 | |
| psNTP9 | P. sativum | A. tumefaciens | Increase soybean yield | Williams82 | Veerappa et al., 2019 | |
| GmWRI1b | G. max | A. tumefaciens | Improve plant architecture and associated yield parameters, and increases total seed oil production | \ | Guo et al., 2020c | |
| HaHB4 | H. annuus | A. tumefaciens | Enhance drought tolerance with yield reduced | Williams82 | Ribichich et al., 2020 | |
| GmMYB14 | G. max | A. tumefaciens | Enhance high-density yield and drought tolerance | Tianlong1 | Chen et al., 2021b | |
| ZmSOC1 | Z. mays | A. tumefaciens | Increase soybean yield | Jack | Han et al., 2021 | |
| GmFULa | G. max | A. tumefaciens | Increase soybean yield | Zigongdongdou | Yue et al., 2021 | |
| GmHSP17.9 | G. max | A. rhizogenes | Increase nodule number, nodule fresh weight, and seed yield | Williams82 | Yang et al., 2022 | |
| Plant architecture | GmIDL2a and GmIDL4a | G. max | A. rhizogenes | Increase the lateral roots densities of the primary roots | XIAOLIDOU | Liu C. et al., 2018 |
| GmmiR156b | G. max | A. tumefaciens | Improve the shoot architecture and yield | Williams82 | Sun et al., 2019 | |
| GmYUC2a | G. max | A. rhizogenes | Delay nodule development and a reduced number of nodules | Williams82 | Wang et al., 2019c | |
| GmGASA32 | G. max | A. rhizogenes | Increase plant height | Williams82 | Chen et al., 2020b | |
| GmWRI1b | G. max | A. tumefaciens | Improve plant architecture and associated yield parameters, and increases total seed oil production | \ | Guo et al., 2020c | |
| Glyma.13G30950 | G. max | A. tumefaciens | Increase seed pods and oil production | Kariyutaka | Iwabuchi et al., 2020 | |
| GmPIF4b | G. max var. Bragg | A. tumefaciens | Affect plant morphology and accelerating reproductive phase transitions | Bragg | Arya et al., 2021 | |
| AtBIC1 | A. thaliana | A. tumefaciens | Increase plant height | Kwangankong | Cho et al., 2021 | |
| GmDIR27 | G. max | A. tumefaciens | Increase pod dehiscence | Williams82 | Ma X. et al., 2021 | |
| GA2ox8A and GA2ox8B | G. max | A. tumefaciens | Decrease trailing growth and shoot length | W05 | Wang et al., 2021d | |
| GmGAMYB | G. max | A. tumefaciens | Promote flowering and increase plant height | DongNong50 | Yang et al., 2021 | |
| GmBICs | G. max | A. tumefaciens | Increase stem elongation | TianLong1 | Mu et al., 2022 | |
| Iron, nitrogen, and phosphorus use efficiency | GmbHLH57 and GmbHLH300 | G. max | A. rhizogenes | Enhance Fe uptake and increase the Fe content in plants | Williams82 | Li et al., 2018 |
| GmPT7 | G. max | A. tumefaciens | Enhance symbiotic N2 fixation and yield | HN66 | Chen et al., 2019b | |
| GmWRI1s | G. max | A. rhizogenes | Increase nodule numbers | Tianlong1 | Chen et al., 2020a | |
| GmPAP12 | G. max | A. rhizogenes | Increase nodule numbers | Williams82 | Wang et al., 2020d | |
| GmAAP6a | G. max | A. tumefaciens | Enhance tolerance to low nitrogen and improve seed nitrogen status | Tianlong1 | Liu et al., 2020 | |
| GmMDH12 | G. max | A. tumefaciens | Decrease nodule size and mediates malate synthesis | YC03-3 | Zhu et al., 2021 | |
| GmNMHC5 | G. max | A. tumefaciens | Increase nodulation | Jack | Wang W. et al., 2022 | |
| GmNINs | G. max | A. rhizogenes | Decrease nodule numbers | Williams82 and Huachun6 | Fu et al., 2022 | |
| GmD27c | G. max | A. rhizogenes | Increase nodule numbers | Tianlong1 | Rehman et al., 2022 | |
| GmSPX8 | G. max | A. rhizogenes | Increase nodule number, nodule fresh weight, and nitrogenase activity | Zhonghuang15 | Xing et al., 2022 | |
| GmHSP17.9 | G. max | A. rhizogenes | Increase nodule number, nodule fresh weight, and seed yield | Williams82 | Yang et al., 2022 | |
| EsPHT1;4 | E. salsugineum | A. tumefaciens | Increase tolerance to low phosphorus stress | YD22 | Yang et al., 2020b | |
| GmETO1 | G. max | A. tumefaciens | Enhance Pi deficiency tolerance | NN94156 and Bogao | Zhang H. et al., 2020 | |
| Flowering time | GmFT1a and GmFT2a/5a | G. max | A. tumefaciens | GmFT1a and GmFT2a/5a have opposite roles in controlling flowering | Zigongdongdou and Heihe27 | Liu W. et al., 2018 |
| GmFT2b | G. max | A. tumefaciens | Promote flowering | Jack | Chen et al., 2020c | |
| GmGAMYB | G. max | A. tumefaciens | Promote flowering and increase of plant height | DongNong50 | Yang et al., 2021 | |
| E1 (Glyma06g23026) | G. max | A. tumefaciens | Promote flowering | Zigongdongdou | Liu et al., 2022a | |
| Abiotic and biotic traits | ||||||
| Nematode resistance | HgY25 | H. glycines | Biolistic method | Enhance resistance to soybean cyst nematodes | Jack | Tian et al., 2019 |
| BtCry14Ab | B. thuringiensis | Biolistic method | Enhance resistance to soybean cyst nematodes | Jack | Kahn et al., 2021 | |
| GmSYP31A | G. max | A. tumefaciens | Enhance resistance to soybean cyst nematodes | Williams82 | Wang et al., 2021b | |
| Hg-rps23, Hg-snb1, and Hg-cpn1 | H. glycines | A. tumefaciens | Enhance resistance to soybean cyst nematodes | Williams82 | Zhang et al., 2022c | |
| Insect resistance | BtCry8-like gene | B. thuringiensis | A. tumefaciens | Result in resistance to Holotrichia parallela | Jinong28 | Qin et al., 2019 |
| BtCry1Ia5 | B. thuringiensis | A. tumefaciens | Result in resistance to Spodoptera littoralis | Giza21 and Giza111 | Moghaieb et al., 2019 | |
| Virus resistance | The coat protein gene of MYMIV | Mung bean yellow mosaic India virus (MYMIV) | A. tumefaciens | Result in resistance to yellow mosaic viruses | JS335 | Kumari et al., 2018 |
| SMV P3 cistron fragment (2,529–2,830 nt) | SMV SC3 | A. tumefaciens | Enhance resistance to multiple Potyvirus strains and isolates | Shennong9 and Williams82 | Yang X. et al., 2018 | |
| GmeIF4E | G. max | A. tumefaciens | Result in resistance to multiple potyvirids | Tianlong1 | Gao et al., 2020 | |
| The AC2 gene | MYMIV | A. tumefaciens | Enhance MYMIV resistance | JS335 | Ramesh et al., 2019 | |
| GmKR3 | G. max | A. tumefaciens | Result in resistance to multiple viruses | Jack | Xun et al., 2019 | |
| The protein kinase PBS1 | TuMV | A. tumefaciens | Enhance potyvirus resistance | Williams82 | Pottinger et al., 2020 | |
| GmVma12 | G. max | A. tumefaciens | Enhance SMV resistance | Tianlong1 | Luan et al., 2020 | |
| GmST1 | G. max | A. tumefaciens | Enhance resistance to soybean mosaic virus strains G2 and G3 |
Dongnong93−046 | Zhao et al., 2021 | |
| GmNF-YC4-2 | G. max | A. tumefaciens | Result in broad disease resistance for bacterial, viral, and fungal infections |
Williams82 | O’Conner et al., 2021 | |
| Fungal disease resistance | hrpZm | P. syringae | A. tumefaciens | Enhance tolerance to Phytophthora root and stem rot caused by P. sojae | Williams82 and Shennong9 | Du et al., 2018 |
| AtPSS1 | A. thaliana | A. tumefaciens | Result inresistance to F. virguliforme | Williams82 | Wang et al., 2018a | |
| GmCHI1A | G. max | A. rhizogenes | Result in resistance to P. sojae | Williams82 (carrying Rps 1k) | Zhou et al., 2018 | |
| GmPI4L | G. max | A. tumefaciens | Result in resistance to P. sojae | Dongnong50 | Chen et al., 2019c | |
| Hrf2 | X. oryzaepv. oryzicola | A. tumefaciens | Result in resistance to P. sojae | Shennong9 | Niu et al., 2019 | |
| GmSnRK1.1 | G. max | A. tumefaciens | Result in resistance to P. sojae | Suinong10 | Wang et al., 2019a | |
| GmC4H1 | G. max | A. rhizogenes | Result in resistance to P. sojae | Conrad | Yan et al., 2019 | |
| TaOXO | T. aestivum | A. tumefaciens | Enhance resistance to sclerotinia stem rot | Willams82 | Yang X. et al., 2019 | |
| GmMYB29A2 | G. max | A. rhizogenes | Result in resistance to P. sojae | Harosoy, H63, Williams, and W82 | Jahan et al., 2020 | |
| NmDef02 | N. megalosiphon | Biolistic method | Enhance resistance to soybean rust and anthracnose | DT-84 | Soto et al., 2020 | |
| CmCH1 | C. minitans | A. tumefaciens | Enhanced resistance to Sclerotiniasclerotiorum | Williams82 | Yang et al., 2020c | |
| GmDR1 | G. max | A. tumefaciens | Result in broad spectrum immunity against fungal disease | Williams82 | Ngaki et al., 2021 | |
| AtFOLT1 | A. thaliana | A. tumefaciens | Enhance resistance to broad-spectrum disease | Williams82 | Kambakam et al., 2021 | |
| NLR gene | G. max | O. haywardense | Result in broad-spectrum resistance to P. sojae | 93Y21 | Wang et al., 2021c | |
| GmTNL16 | G. max | A. rhizogenes | Enhance resistance to P. sojae | Williams | Li et al., 2022 | |
| GmNAC1 | G. max | A. tumefaciens | Enhance resistance to P. sojae | Tianlong1 and Suinong10 | Yu et al., 2022 | |
| Herbicide tolerance | G10-EPSPS | D. radiodurans | A. tumefaciens | Result in glyphosate tolerance | Zhongdou32 | Xiao et al., 2019 |
| G2-EPSPS and G10-EPSPS | P. fluorescens G2 | A. tumefaciens | Result in glyphosate tolerance | Jack | Guo et al., 2020a | |
| Cytochrome P450 geneP450-N-Z1 | C. dactylon | A. tumefaciens | Result in multiple herbicides tolerance | Tianlong1 | Zheng et al., 2022 | |
| Drought tolerance | PgTIP1 | P. ginseng | A. tumefaciens | Enhance both salt and drought tolerance | Hybrid strain 4076 | An et al., 2018 |
| GmPIP2;9 | G. max | A. tumefaciens | Increase drought tolerance | Williams82 | Lu et al., 2018 | |
| AtABF3 | A. thaliana | A. tumefaciens | Enhance drought tolerance | Kwangankong | Kim et al., 2018 | |
| GmWRKY12 | G. max | A. rhizogenes | Increase drought and salt tolerance | Williams82 | Shi et al., 2018 | |
| GmBIN2 | G. max | A. rhizogenes | Enhance tolerance to salt and drought | Dongnong50 | Wang et al., 2018b | |
| GmBiP | G. max | A. tumefaciens | Enhance drought tolerance | Conquista | Coutinho et al., 2019 | |
| AtYUCCA6 | A. thaliana | A. tumefaciens | Enhance drought tolerance | Kwangankong | Park et al., 2019 | |
| GmWRKY54 | G. max | A. rhizogenes | Enhance drought tolerance | Williams82 | Wei W. et al., 2019 | |
| FvC5SD | F. velutipes | A. tumefaciens | Enhance drought stress tolerance | Shennong9 | Zhang et al., 2019b | |
| GmNFYA5 | G. max | A. rhizogenes | Enhance drought tolerance | Williams82 | Ma et al., 2020 | |
| AtNCED3 | A. thaliana | A. tumefaciens | Enhance drought tolerance | BRS184 | Molinari et al., 2020 | |
| GmDREB2 | G. max | A. tumefaciens | Enhance drought tolerance | DT84 | Pham et al., 2020 | |
| AtΔKinase | A. thaliana | A. tumefaciens | Increase tolerance to water deficit stress | Williams82 | Shanmugam et al., 2020 | |
| HaHB4 | H. annuus | A. tumefaciens | Enhance drought tolerance with yield reduced | Williams82 | Ribichich et al., 2020 | |
| GmNAC8 | G. max | A. tumefaciens | Enhance drought tolerance | Tianlong1 | Yang et al., 2020a | |
| GmbZIP2 | G. max | A. tumefaciens | Enhance tolerance to salt, drought, or cold condition | Williams82 | Yang et al., 2020d | |
| GmbZIP15 | G. max | A. tumefaciens | Decrease tolerance to drought and salt tolerance | C03-3 | Zhang M. et al., 2020 | |
| Gmgma-miR398c | G. max | A. rhizogenes | Negatively regulate drought tolerance | Williams82 | Zhou et al., 2020 | |
| GmNTF2B-1 | G. max | A. rhizogenes | Enhance drought tolerance | Williams82 | Chen et al., 2021a | |
| GmMYB14 | G. max | A. tumefaciens | Enhance high-density yield and drought tolerance | Tianlong1 | Chen et al., 2021b | |
| GmTGA15 | G. max | A. rhizogenes | Enhance drought tolerance | Williams82 | Chen et al., 2021c | |
| GmPI-PLC7 | G. max | A. rhizogenes | Increase drought and salt tolerance | Williams82 | Chen et al., 2021d | |
| GmCIPK2 | G. max | A. tumefaciens | Enhance drought tolerance | Williams82 | Xu et al., 2021 | |
| GsPOD40 | G. max | A. tumefaciens | Enhance drought tolerance | PI342618B/DTP and Tianlong1 | Aleem et al., 2022 | |
| GmDREB1 | G. max | A. tumefaciens | Enhance drought tolerance | P3 | Chen et al., 2022 | |
| sHSP26 | G. max | A. tumefaciens | Enhance drought tolerance | Jinong18 | Liu et al., 2022b | |
| GmDREB2 | G. max | A. tumefaciens | Enhance drought tolerance | BRS283 | Marinho et al., 2022 | |
| GmEF8 | G. max | A. rhizogenes | Enhance drought and heat tolerance | Williams82 | Zhang et al., 2022a | |
| Salt and other stress tolerance | PgTIP1 | P. ginseng | A. tumefaciens | Enhance both salt and drought tolerance | Hybrid strain 4076 | An et al., 2018 |
| ZmGB1 | Z. mays | A. tumefaciens | Enhance tolerance to abiotic stress | \ | Castiglioni et al., 2018 | |
| GmWRKY12 | G. max | A. rhizogenes | Increase drought and salt tolerance | Williams82 | Shi et al., 2018 | |
| AtXTH31 | A. thaliana | A. tumefaciens | Enhance tolerance to flooding stress | Maverick | Song et al., 2018 | |
| GmBIN2 | G. max | A. rhizogenes | Enhance tolerance to salt and drought | Dongnong50 | Wang et al., 2018b | |
| MsWRKY11 | M. sativa (alfalfa) | A. tumefaciens | Enhance salt tolerance | Dongnong50 | Wang et al., 2018c | |
| GmHsp90A2 | G. max | A. tumefaciens | Increase tolerance to heat stress | Qihuang22 | Huang et al., 2019 | |
| AtAVP1 and AtNHX1 | A. thaliana | A. tumefaciens | Increase salt tolerance | DT26 | Nguyen N. T. et al., 2019 | |
| GmDREB-6 | G. max | A. tumefaciens | Enhance salt tolerance | DT84 | Nguyen Q. H. et al., 2019 | |
| GsCLC-c2 | G. soja | A. tumefaciens | Enhance salt tolerance | N23674 | Wei P. et al., 2019 | |
| GmERF135 | G. max | A. rhizogenes | Enhance salt tolerance | Tiefeng8 | Zhao et al., 2019a | |
| GmCDF1 | G. max | A. rhizogenes | Negatively regulate salt tolerance | Kefeng1 and Nannong1138–2 | Zhang et al., 2019c | |
| GmSAP16 | G. max | A. tumefaciens | Enhance drought and salt tolerance | Williams82 | Zhang et al., 2019d | |
| J | G. max | A. tumefaciens | Increase salt tolerance | Huaxia3 | Cheng et al., 2020 | |
| GsSnRK1 | G. soja | A. tumefaciens | Increase salt and alkaline stresses tolerance | Dongnong50 | Feng et al., 2020 | |
| GmMYB68 | G. max | A. tumefaciens | Increase salt and alkaline stresses tolerance | Williams82 | He et al., 2020 | |
| Gs5PTase8 | G. soja | A. rhizogenes | Enhance salt tolerance | Mengjin1 and Union | Jia et al., 2020 | |
| GsAAE3 | G. soja | A. rhizogenes | Increase tolerance to Cd and Al stresses | BW69 | Xian et al., 2020 | |
| GmbZIP2 | G. max | A. tumefaciens | Enhance tolerance to salt, drought, or cold condition | Williams82 | Yang et al., 2020d | |
| GsJAZ2 | G. soja (G07256) | Biolistic method | Enhance tolerance to alkaline stress | HF55 | Zhao et al., 2020 | |
| GmbZIP15 | G. max | A. tumefaciens | Decrease tolerance to drought and salt tolerance | C03-3 | Zhang M. et al., 2020 | |
| GmPI-PLC7 | G. max | A. rhizogenes | Increase drought and salt tolerance | Williams82 | Chen et al., 2021d | |
| AgGlpF | A. glaucus | A. tumefaciens | Enhance salt tolerance | Williams82 | Li et al., 2021a | |
| GmNAC06 | G. max | A. rhizogenes | Enhance salt tolerance | Williams82 | Li et al., 2021b | |
| GsCLC-c2 | G. soja | A. tumefaciens | Enhance Cl–/salt tolerance | BB52 | Liu et al., 2021 | |
| GsBET11a | G. soja | A. tumefaciens | Enhance salt tolerance | G07256 and Dongnong50 | Sun X. et al., 2021 | |
| GmNHX5 | G. max | A. rhizogenes | Enhance salt tolerance | Jidou-7 | Sun T. et al., 2021 | |
| GmAKT1 | G. max | A. rhizogenes | Enhance salt tolerance | Dongnong50 | Wang et al., 2021e | |
| GmbHLH3 | G. max | A. rhizogenes | Enhance Cl–/salt tolerance | N23674 | Liu et al., 2022c | |
| GmEF8 | G. max | A. rhizogenes | Enhance drought and heat tolerance | Williams82 | Zhang et al., 2022a |
\ means not available.
Challenges and Future Directions in Soybean Transformation
Although much effort has been made to improve the transformation systems for soybean, there are some challenges such as genotype flexibility, low transformation frequency, time to time chimerism in T0 transgenic plants, and availability of a system for new breeding technologies such as genome editing.
Genotype Flexibility
Like in other recalcitrant plant species, genotype inflexibility has been an obstacle that restricted the scope of soybean transformation. The ideal soybean transformation target material for trait improvement would be any elite variety with excellent agronomic characteristics. However, most reliable transformations are still based on specific genotypes although genotypes amenable to transformation have expanded to some preferred genotypes. For example, in the early stage, successful Agrobacterium-mediated transformation occurred in several genotypes and their derivatives such as Williams, Williams79, and Williams82 (Paz et al., 2004, 2006). High-efficiency Agrobacterium-mediated transformation is only achieved in a limited number of elite lines (Zhang et al., 2014; Arun et al., 2015, 2016; Yang J. et al., 2016; Li et al., 2017; Chen et al., 2018b). High-efficiency genotypes possess greater susceptibility to Agrobacterium infection, which has been confirmed in many reports (Jia et al., 2015; Yang J. et al., 2016; Yang X. F. et al., 2016; Zhao et al., 2019b). The competency of cotyledons of seeds to Agrobacterium infection and the ability to regenerate plants are key factors. These may be determined by cell defense response, including attachment of A. tumefaciens to plant cells, plant signals sensed by A. tumefaciens, regulating vir gene expression, T-DNA/virulence protein transport or initial contact of A. tumefaciens to plants and cytoplasmic trafficking, and nuclear import of T-DNA and effector proteins (Hwang et al., 2017). An important step to enhance the transformation efficiency of recalcitrant genotypes is to improve the genotypes’ susceptibility to Agrobacterium infection. Many commonly used treatments to increase transformation efficiency such as heat shock, cold shock, antioxidants, and hypoxia may act by suppression of cellular response to Agrobacterium infection (Zhang et al., 2013). Combinations of various positive factors discovered or developed recently have promoted Agrobacterium-mediated soybean transformation to extend genotype scope (Table 1). For example, transgenic events have been obtained from 19 out of 20 genotypes based on an improved protocol (Pareddy et al., 2020) and 7 out of 8 genotypes (Zhao et al., 2019b). One important progress in these reports is that over 5% of TFs were obtained in nearly half of these genotypes. The second factor that affects genotype flexibility is the regeneration ability of donor genotypes, which restricts TFs for both Agrobacterium-mediated and biolistic transformations. Increasing the amenability of many soybean genotypes to regenerate may be conducted by either adding some specific chemicals in the culture medium described above or using plant regeneration factors or regeneration booster genes. Significant progress has been made to improve transformations from various tissue types using plant regeneration factors such as maize (Zea mays) morphogenic genes, Baby boom (BBM) and Wuschel2 (WUS2) genes in maize plant (Lowe et al., 2016), and plant growth regulators such as GROWTH-REGULATING FACTORS (GRF) genes used in monocot and dicot species including soybean (Gordon-Kamm et al., 2019; Debernardi et al., 2020; Kong et al., 2020). Use of these genes significantly increased transformation frequency and reduced genotype obstacle for transformation, providing a good solution for genotype-inflexibility bottleneck in transformation of crops including soybean. For example, introducing AtGRF5 and GRF5 orthologs into soybean cells could improve regeneration and, hence, increase transformation TFs significantly (Kong et al., 2020). GRFs can also enhance shoot organogenesis and callus regeneration, which has been confirmed in dicots including sugar beet, canola, and sunflower. Meanwhile, somatic embryogenesis can be promoted using some genes introduced into explants in soybean, such as soybean orthologs of the Arabidopsis (A. thaliana) MADS box genes AGAMOUS-Like15 (GmAGL15) and GmAGL18, which can also expand soybean genotypes suitable for transformation, especially for biolistic transformation (Zheng and Perry, 2014). Transformation bypass tissue culture such as in planta transformation is an alternative way to overcome genotype inflexibility in soybean (Liu et al., 2009; Mangena, 2019). Nanotechnology-based transformation can also be employed to overcome host range limitation including genotype inflexibility, and can simplify delivery way using pollen channel, and highly increase efficiency (Wang and Zhao, 2019). By integration of multiple-omics technologies, genes related to transformation efficiency should be discovered for increasing transformation efficiency. Use of the novel bacterium O. haywardense H1 may also increase the genotype scope for transformation, since it was claimed to be less genotype sensitive when it was used for soybean transformation (Cho et al., 2022).
Low Transformation Frequency
The average TF for varieties (genotypes) reported is lower than 5%, although improvements have been made by modifying the main factors described above (Table 1). Since the biolistic method tends to use an embryogenic callus as explant because of less chimerism compare to an embryo axis, TFs for biolistic transformation are dependent on the success of embryogenic callus induction for a specific genotype. Therefore, the main focus to improve TFs is to select genotypes that are amenable to embryogenic callus induction, or to stimulate a genotype to produce an embryogenic callus. As described above, the regeneration booster provides a new way to induce an embryogenic callus without genotype limitation, which has been confirmed in monocot plants (Lowe et al., 2018; Gordon-Kamm et al., 2019; Debernardi et al., 2020). Enhancement of TFs for soybean Agrobacterium-mediated transformation is mainly achieved by improving regeneration rates of explants and increasing the susceptibility of explants to Agrobacterium. Half-seed explants have been the major choice, because these explants could provide more nutrition and less damage than cotyledonary nodes (Table 1). Continuously modifying MS-based culture medium composition (Murashige and Skoog, 1962), especially by addition of chemicals discovered through the study of omics, has played a big role in TF improvement, and has been summarized in the section above. Combinations of many factors have promoted the TFs of soybean transformation (Table 1). More efforts should be made to increase the average TFs close to that of other major crops. Again, the morphogenic genes including GRFs described above may play an important role in enhancing soybean transformation frequency.
Chimerism in T0 Transgenic Plant
Chimerism in legume transformation is fairly common, which causes non-transmission of transgenes to subsequent generations either completely or at a lower ratio expected by Mendelian genetics. Therefore, minimizing chimerism in transgenic plants is required to obtain transmission of transgenes to the T1 generation. In soybean, Agrobacterium-mediated transformation of cotyledonary nodes by organogenesis has been extensively conducted for transgenic production in research and commercial product development (Barwale et al., 1986; Homrich et al., 2012; Yamada et al., 2012; Lee et al., 2013; Mariashibu et al., 2013; Mangena et al., 2017). Plant regeneration by organogenesis with an explant containing an embryo axis may be the main cause, since shoots regenerated from soybean shoot tips were derived from 3 superimposed cellular layers (L1, L2, and L3) in the original apical meristem (Christou, 1990; Christou and Mccabe, 1992). Transformed cells existed primarily in the L1 and L2 layers but not in the L3 layer of the apical meristems of regenerated shoots, indicating possible escape in the regenerated shoots during transformation, and this chimerism has been confirmed (Mccabe et al., 1988; Sato et al., 1993). Currently, the chimerism in transgenic soybean is still a major concern in the research community, and inheritance study has been always an important part in transformation protocol development (Pareddy et al., 2020). Improvement for reducing escapes or chimeric rate has been made when strict select stringency was used, especially some new selectable markers/reagents such as AHAS/imazapyr (Aragão et al., 2000; Rech et al., 2008), EPSPS/glyphosate (Martinell et al., 1999; Guo et al., 2015, 2020a; Soto et al., 2017), and AADA/spectinomycin (Martinell et al., 2002). Meanwhile, the modified protocols made use of specific explants, such as somatic embryogenic calli, to reduce the chance of infection with cells at the late development stage, and combined proper selection of chemical agents with high stringency to decrease escape rate dramatically, which led to more than 90% T0 transgenic plants transmitting their transgenes into T1 generation (Soto et al., 2017; Chennareddy et al., 2018; Guo et al., 2020a). Therefore, transgenic soybean with chimeric issues may due to insufficient selection that existed in various protocols.
Development of Transformation Method for New Breeding Technology
Genome editing is the recent advancement in genome engineering, which has revolutionized crop research and plant breeding. GE, through site-specific nucleases (SSNs), can precisely make changes in targeted genome sequence sites by disruption including insertion and deletion, base changes, sequence replacement, and sequence insertion. SSNs include zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and CRISPR/CAS. GE is a fast-developing technology that will potentially play an important role in genomics study and will create opportunities for rapid development of elite cultivars with desired traits. The development of soybean GE has been reviewed in Xu et al. (2020). Recent GE applications in soybean for trait improvement have been summarized in Table 3. For example, function analysis of photo period-related genes such as LHY homologs, J and E1, and tof 16 (Time of Flowering 16) using GE technology showed that more than 80% accessions in low latitude harbor the mutations of tof16 and J, which suggests that loss of functions of Tof16 and J is the major genetic basis of soybean adaptation into tropics. Therefore, maturity and yield traits can be quantitatively improved by modulating the genetic complexity of various alleles of LHY homologs, J, and E1 (Bu et al., 2021; Dong et al., 2021). The findings uncover the adaptation trajectory of soybean from its temperate origin to the tropics. Knockout of GmJAG1, which controls the number of seeds per pod (NSPP), increases by over 8% the yield of a Chinese variety, Huachun 6 (Cai et al., 2021). GmMs1 KO events in soybean were created, which showed male sterility phenotype (Jiang et al., 2021; Nadeem et al., 2021). SCN-resistant mechanisms such as t-SNAREs binding Rhg1 α-SNAP (Dong et al., 2020) and WI12Rhg1 interacting with DELLAs (Dong and Hudson, 2022) were found using GE as a tool. Targeted chromosome cleavage by CRISPR/Cas9 can conceivably induce rearrangements and, thus, emergence of new resistance gene paralogs. CRISPR/Cas9-mediated chromosome rearrangements in nucleotide-binding-site-leucine-rich-repeat (NBS-LRR) gene families of soybean produced a new disease-resistant gene (Nagy et al., 2021). Raffinose family oligosaccharides (RFOs) are major soluble carbohydrates in soybean seeds that cannot be digested by humans and other monogastric animals. Double mutation events, knockouts in two soybean galactinol synthase (GOLS) genes, GmGOLS1A and its homeolog GmGOLS1B, showed a reduction in the total RFO content of soybean seeds from 64.7 to 41.95 mg/g dry weight, a 35.2% decrease (Le et al., 2020). This product improved the soybean nutrition quality. Two transcription systems were also tested in soybean including the single transcriptional unit (STU), SpCas9 and sgRNA are driven by only one promoter, and in the conventional system, the two-component transcriptional unit (TCTU), SpCas9, is under the control of a pol II promoter, and sgRNAs are under the control of a pol III promoter. The results showed that the STU is more efficient (Carrijo et al., 2021). Cpf1 (Cas12a) systems have also been established in soybean for GE (Duan et al., 2021; Kim and Choi, 2021). Meanwhile, different GE systems for soybean have been established using specific editing reagent delivery methods developed for soybean transformation, which produce transgene-free GE events either with the biolistic method (Adachi et al., 2021) and selectable marker-free GT systems by O. haywardense H1-8-mediated delivery (Kumar et al., 2022), or by organ-specific editing using an egg cell-specific promoter (Zheng et al., 2020). All these GE studies on soybean demonstrate that the ability to conduct genome editing directly depends on plant transformation technologies, since recovery of stable events with the target gene edited is normally based on available transformation systems including editing reagent delivery and edited event regeneration. GE has the potential to avoid many regulatory issues regarding transgenics if specific editing reagents are used. Based on the CRISPR/CAS system, gRNA in the form of in vitro-synthesized RNA molecule, together with Cas9 as DNA construct, can be stably integrated into the host genome and constitutively expressed, which might lead to a transgenic event for a GE event. This issue can be resolved by introduction of editing tools without genomic integration or transient expression. Transgene-free or DNA-free edited events in many crops can now be obtained either by delivering the RNA form of sgRNA and Cas9 or Cas9 protein (RNP) using the biolistic method, or by protoplast transfection (Chen et al., 2019a; Xu et al., 2020; Gao, 2021; Kim and Choi, 2021). Transgene-free events can also be recovered with the Agrobacterium-mediated method without selection (Liang et al., 2017). However, genotype flexibility limitation is a major issue for soybean GE in the biolistic method, and low TF for some elite varieties is the main hurdle in the Agrobacterium-mediated method.
TABLE 3.
List of soybean genes edited for functional genetics study and trait improvement using genome editing technology.
| Trait | Gene/targeting location | GE platform | Delivery method | Edited events | Editing outcomes | References |
| Yield | ||||||
| Plant architecture | GmLHY1a (Glyma.16G017400), GmLHY1b (Glyma.07G048500), GmLHY2a (Glyma.19G260900), and GmLHY2b (Glyma.03G261800) | CRISPR/Cas9 | A. tumefaciens | Whole plant | Knockout (multiplex) | Cheng et al., 2019 |
| GmSPL9a (Glyma.02G177500), GmSPL9b (Glyma.09G113800), GmSPL9c (Glyma.03G143100), and GmSPL9d (Glyma.19G146000) | CRISPR/Cas9 | A. tumefaciens | Whole plant | Knockout (multiplex) | Bao et al., 2019 | |
| GmAP1a (Glyma.16G091300), GmAP1b (Glyma.08G269800), GmAP1c (Glyma.01G064200), and GmAP1d (Glyma.02G121600) | CRISPR/Cas9 | A. tumefaciens | Whole plant | Knockout (multiplex) | Chen et al., 2020d | |
| Seed weight and organ size | GmPPD1 (Glyma.10G244400) and GmPPD2 (Glyma.20G150000) | CRISPR/Cas9 | A. tumefaciens | Whole plant | Knockout (multiplex) | Kanazashi et al., 2018 |
| GmSWEET10a (Glyma.15G049200) and GmSWEET10b (Glyma.08G183500) | CRISPR/Cas9 | A. tumefaciens | Whole plant | Knockout (multiplex) | Wang et al., 2020c | |
| GmKIX8-1 (Glyma.17G112800) | CRISPR/Cas9 | A. tumefaciens | Whole plant | Knockout | Nguyen et al., 2021 | |
| Seed number | GmJAG1 (Glyma.20G25000) and GmJAG2 (Glyma.10G42020) | CRISPR/Cas9 | A. tumefaciens | Whole plant | Knockout (multiplex) | Cai et al., 2021 |
| Photoperiod | GmFT2a (Glyma.16G26660) | CRISPR/Cas9 | A. tumefaciens | Whole plant | Knockout | Cai et al., 2018 |
| GmE1 (Glyma.06G207800) | CRISPR/Cas9 | A. tumefaciens | Whole plant | Knockout | Han et al., 2019 | |
| GmFT2a (Glyma.16G26660) and GmFT5a (Glyma.16G04830) | CRISPR/Cas9 | A. tumefaciens | Whole plant | Knockout (multiplex) | Cai et al., 2020b | |
| GmFT2a (Glyma.16G150700) and GmFT4 (Glyma.08G363100) | BE base editor | A. tumefaciens | Whole plant | Base editing | Cai et al., 2020a | |
| GmFT2b (Glyma.16G26690) | CRISPR/Cas9 | A. tumefaciens | Whole plant | Knockout | Chen et al., 2020c | |
| GmAP1a (Glyma.16G091300), GmAP1b (Glyma.08G269800), GmAP1c (Glyma.01G064200), and GmAP1d (Glyma.02G121600) | CRISPR/Cas9 | A. tumefaciens | Whole plant | Knockout (multiplex) | Chen et al., 2020d | |
| GmPRR37 (Glyma.12G073900) | CRISPR/Cas9 | A. tumefaciens | Whole plant | Knockout | Wang et al., 2020b | |
| GmLUX1 (Glyma.12G060200) and GmLUX2 (Glyma.11G136600) | CRISPR/Cas9 | A. tumefaciens | Whole plant | Knockout (multiplex) | Bu et al., 2021 | |
| GmLNK2a (Glyma.04G141400), GmLNK2b (Glyma.11G154700), GmLNK2c (Glyma.13G199300), and GmLNK2d (Glyma.15G237600) | CRISPR/Cas9 | A. tumefaciens | Whole plant | Knockout (multiplex) | Li et al., 2021c | |
| Nutrition and quality | ||||||
| Storage protein | Glyma.20G148400, Glyma.20G146200, Glyma.10G246300, Glyma.20G148200, Glyma.10G037100, Glyma.03G163500, Glyma.19G164900, Glyma.13G123500, and Glyma.19G164800 | CRISPR/Cas9 | A. rhizogenes | Hairy root | Knockout (multiplex) | Li et al., 2019 |
| Seed oil | GmFAD2-1A (Glyma.10G278000) and GmFAD2-1B (Glyma.20G111000) | TALENs | A. rhizogenes and disarmed A. rhizogenes | Hairy root and whole plant | Knockout (multiplex) | Haun et al., 2014 |
| GmFAD2-2 | CRISPR/Cas9 | A. tumefaciens | Whole plant | Knockout | al Amin et al., 2019 | |
| GmFAD2-1A (Glyma.10G278000) | ZFNs | Biolistic method | Whole plant | Knock in (NHEJ) | Bonawitz et al., 2019 | |
| GmFAD2-1A (Glyma.10G278000) and GmFAD2-1B (Glyma.20G111000) | CRISPR/Cas9 | A. tumefaciens | Whole plant | Knockout (multiplex) | Do et al., 2019 | |
| GmGOLS1A (Glyma.03G222000) and GmGOLS1B (Glyma.19G219100) | CRISPR/Cas9 | A. tumefaciens | Whole plant | Knockout (multiplex) | Le et al., 2020 | |
| GmFAD2–1A (Glyma.10G278000) and GmFAD2–2A (Glyma.19G147300) | CRISPR/Cas9 | A. tumefaciens | Whole plant | Knockout (multiplex) | Wu et al., 2020 | |
| GmFATB1a (Glyma.05G012300) and GmFATB1b (Glyma.17G012400) | CRISPR/Cas9 | A. tumefaciens | Whole plant | Knockout (multiplex) | Ma J. et al., 2021 | |
| Glyma.15G117700 | CRISPR/Cas9 | A. tumefaciens | Whole plant | Knockout | Qu et al., 2021 | |
| Bean flavor-free soybean | GmLox1 (Glyma.13G347600), GmLox2 (Glyma.13G347500), and GmLox3 (Glyma.15G026300) | CRISPR/Cas9 | A. tumefaciens | Whole plant | Knockout (multiplex) | Wang et al., 2020a |
| Disease resistance | ||||||
| Cyst nematode resistance | GmSyn12 (Glyma.12G194800), GmSyn13 (Glyma.13G307600), GmSyn16 (Glyma.16G154200), and GmSyn02 (Glyma.02G072900) | CRISPR/Cas9 | A. rhizogenes | Hairy root | Knockout (multiplex) | Dong et al., 2020 |
| Rps1 families (Glyma.03G034400, Glyma.03G0034800, Glyma.03G039200, Glyma.03G039500, Glyma.03G037100, Glyma.03G037300, Glyma.03G037400, Glyma.03G037400, Glyma.03G037000, Glyma.03G034500, Glyma.03G039300, Glyma.03G045700, Glyma.03G043600, Glyma.03G045300, Glyma.03G043000, Glyma.03G043500, Glyma.03G044000, Glyma.03G043200, Glyma.03G045000, Glyma.03G046500, Glyma.03G047000, Glyma.03G043900) and Rpp1L families (Glyma.18G281700, Glyma.18G281600, Glyma.18G281500, and Glyma.18G280300) | CRISPR/Cas9 | A. tumefaciens | Whole plant | Knockout (multiplex) | Nagy et al., 2021 | |
| Rhg1-locus (Glyma.18G02270), DELLA18 (Glyma.18G040000), and DELLA11 (Glyma.11G216500) | CRISPR/Cas9 | A. rhizogenes | Hairy root | Knockout (multiplex) | Dong and Hudson, 2022 | |
| Insect resistance | GmUGT (Glyma.07G110300) | CRISPR/Cas9 | A. tumefaciens | Whole plant | Knockout | Zhang et al., 2022b |
| Abiotic stress tolerance | ||||||
| Drought tolerance | GmLHY1a (Glyma.16G017400), GmLHY1b (Glyma.07G048500), GmLHY2a (Glyma.19G260900), and GmLHY2b (Glyma.03G261800) | CRISPR/Cas9 | A. tumefaciens | Whole plant | Knockout (multiplex) | Wang et al., 2021a |
| Salt tolerance | GmNAC06 (Glyma06G21020) | CRISPR/Cas9 | A. rhizogenes | Hairy root | Knockout | Li et al., 2021b |
| Nitrogen fixation | ||||||
| GmNSP1a (Glyma.07G039400) and GmNSP1b (Glyma.16G008200) | CRISPR/Cas9 | A. tumefaciens | Whole plant | Knockout (multiplex) | He et al., 2021 | |
| Herbicide resistance | GmALS1 (Glyma.04G37270.1), GmALS2 (Glyma.06G17790.1), GmALS3 (Glyma.13G31470.1), and GmALS4 (Glyma.15G07860.1) | CRISPR/Cas9 | Biolistic method | Whole plant | Knockin (HDR) | Li et al., 2015 |
| Root nodulation | GmRIC1 (Glyma.13G292300), GmRIC2 (Glyma.06G284100), GmRDN1-1 (Glyma.02G279600), GmRDN1-2 (Glyma.14G035100), and GmRDN1-3 (Glyma.20G040500) | CRISPR/Cas9 | A. tumefaciens | Whole plant | Knockout (multiplex) | Bai et al., 2020 |
| GmSPL9d (Glyma.19G146000) and GmmiR156 | CRISPR/Cas9 | A. rhizogenes | Hairy root | Knockout (multiplex) | Yun et al., 2022 | |
| Allergy reduction | Gly m Bd 28K (Glyma.U020300) and Gly m Bd 30K (Glyma.08G116300) | CRISPR/Cas9 | A. tumefaciens | Whole plant | Knockout (multiplex) | Sugano et al., 2020 |
| GE platform adoption in soybean | GmDCL1a (Glyma.03G42290), GmDCL1b (Glyma.19G45060), GmDCL4a (Glyma.17G11240), GmDCL4b (Glyma.13G22450), GmRDR6a (Glyma.04G07150), GmRDR6b (Glyma.06G07250), and GmHEN1a (Glyma.08G08650) | ZFNs | A. rhizogenes | Hairy root | Knockout (multiplex) | Curtin et al., 2011 |
| GmDCL4a (Glyma.17G11240) and GmDCL4b (Glyma.13G22450) | ZFNs | A. rhizogenes | Hairy root | Knockout | Sander et al., 2011 | |
| Bar transgene, GmFEI1 (Glyma.01G35390), GmFEI2 (Glyma.09G34940), and GmSHR | CRISPR/Cas9 | A. rhizogenes | Hairy root | Knockout (multiplex) | Cai et al., 2015 | |
| GmGS (Glyma.18G04660 and Glyma.18G041100) and GmCHI20 (Glyma.20G38560 and Glyma.20G241500) | CRISPR/Cas9 | A. rhizogenes | Hairy root | Knockout | Michno et al., 2015 | |
| GFP transgene, Glyma07g14530, 01gDDM1 (Glyma.11G38150), 11gDDM1 (Glyma.11G07220), Glyma04g36150, Glyma06g18790, miR1509, and miR1514 | CRISPR/Cas9 | A. rhizogenes | Hairy root | Knockout | Jacobs et al., 2015 | |
| Glyma.06G14180, Glyma.08G02290, and Glyma.12G37050 | CRISPR/Cas9 | A. rhizogenes | Hairy root | Knockout (multiplex) | Sun et al., 2015 | |
| GmPDS11 (Glyma.11G253000) and GmPDS18 (Glyma.18G003900) | TALENs | A. tumefaciens | Whole plant | Knockout | Du et al., 2016 | |
| FAD2-1A (Glyma.10G42470) and FAD2-1B (Glyma.20G24530) | CRISPR/AsCpf1 or LpCpf1 | Protoplast transfection | Protoplast | Knockout (RNP) | Kim et al., 2017 | |
| GmIPK1 (Glyma.14G072200) and GmIPK2 (Glyma.12G240900) (STU and TCTU system*) | CRISPR/Cas9 | A. rhizogenes | Hairy root | Knockout (multiplex) | Carrijo et al., 2021 | |
| Glyma.15G249000 and Glyma.13G259100 | CRISPR/Cas9 | A. rhizogenes | Hairy root | Knockout (multiplex) | Luo et al., 2021 | |
| GmPDS11g (Glyma.11g253000) and GmPDS18g (Glyma.18g003900) | CRISPR/Cas9 | A. tumefaciens | Whole plant | Knockout (multiplex) | Lu and Tian, 2022 | |
| Transgene-free edited events | Target sites DD38 and DD51 | CRISPR/Cas9 | O. haywardense H1-8 | Targeted insertion | Kumar et al., 2022 | |
| Gly m Bd 30K (Glyma.08G116300) | CRISPR/Cas9 | Biolistic method | Whole plant | Knockout | Adachi et al., 2021 | |
| Egg cell promoter driving Cas9 | GmAGO7a (Glyma.01G053100) and GmAGO7b (Glyma.02G111600) | CRISPR/Cas9 | A. rhizogenes andA. tumefaciens | Hairy root and whole plant | Knockout (multiplex) | Zheng et al., 2020 |
| Targeted deletions of DNA fragments | GmFT2a (Glyma.16G26660) and GmFT5a (Glyma.16G04830) | CRISPR/Cas9 | A. tumefaciens | Whole plant | Knockout (4.5 kb in GmFT2a) | Cai et al., 2018 |
| Growth of soybean trichomes | GmCPR5 (Glyma.06G145800) | CRISPR/Cas9 | Biolistic method | Whole plant | Knockout | Campbell et al., 2019 |
| Fertility | GmMs1 (Glyma.13G114200) | CRISPR/Cas9 | Biolistic method | Whole plant | Knockout | Nadeem et al., 2021 |
| GmMs1 (Glyma.13G114200) | CRISPR/Cas9 | Biolistic method | Whole plant | Knockout | Jiang et al., 2021 | |
| miRNA pathway and small RNA processing | GmDCL1a (Glyma.03G42290), GmDCL1b (Glyma.19G45060), GmDCL4a (Glyma.17G11240), GmDCL4b (Glyma.13G22450), GmRDR6a (Glyma.04G07150), GmRDR6b (Glyma.06G07250), GmHEN1a (Glyma.08G08650), and GFP transgene | ZFNs | A. rhizogenes | Hairy root | Knockout | Curtin et al., 2011 |
| GmDRB2a (Glyma.12G075700), GmDRB2b (Glyma.11G145900), GmDCL3a (Glyma.04G057400), GmHEN1a (Glyma.08G081600), and GmHEN1b (Glyma.05G126600) | CRISPR/Cas9 | A. rhizogenes | Hairy root | Knockout (multiplex) | Curtin et al., 2018 | |
| GmDCL2a (Glyma.09G025400), GmDCL2b (Glyma.09G025300), and GmDCL3a (Glyma.04G057400) | TALENs | Disarmed A. rhizogenes | Whole plant | Knockout (multiplex) | Curtin et al., 2018 | |
| Sucrose export related embryo development | GmSWEET15a (Glyma.05G126600) and GmSWEET15b (Glyma.05G1266000) | CRISPR/Cas9 | A. tumefaciens | Whole plant | Knockout (multiplex) | Wang et al., 2019b |
| Circadian rhythmicity | GmLCLa1 (Glyma.16G01980), GmLCLa2 (Glyma.07G05410), GmLCLb1 (Glyma.03G42260), and GmLCLb2 (Glyma.19G45030) | CRISPR/Cas9 | A. tumefaciens | Whole plant | Knockout (multiplex) | Wang et al., 2020e |
| Soybean knockout library | 70 sgRNAs to target 102 genes | CRISPR/Cas9 | A. tumefaciens (pooled) | Whole plant | Knockout (multiplex) | Bai et al., 2020 |
*STU, single transcriptional unit; SpCas9 and sgRNA are driven by only one promoter; and the two-component transcriptional unit (TCTU) in the conventional system, and SpCas9 and sgRNA are under the control of different promoters.
Conclusion
As summarized above, development of soybean transformation protocols, which pose genotype-flexibility and relatively high efficiency and can easily be adapted in any laboratory, is still a main task for researchers. Reducing biological restrictions such as genotype dependence or tissue-specific and method restrictions will eventually lead to transformation automation and versatile and high throughput, which will facilitate the application of next-generation breeding technologies such as genome editing for soybean improvement. These goals may be achieved with fast progress in fundamental research to unravel basic biological process and genetic background, especially when more regeneration regulators such as morphogenic genes are identified. Transgenic soybean in which various genes can be manipulated will accelerate the validation of gene function in the context of complex gene networks at different plant developmental stages, which will accelerate the understanding of the mechanism of soybean cell regeneration, and it is beneficial for us to modify transformation protocols. New technologies like nanoparticle delivery also bring us hope to break through these barriers as well as the transformation bypass method.
Author Contributions
HX and YG collected the materials. HX drew the figures and tables. YR wrote the manuscript. LQ and YR designed the article structure. All authors contributed to the article and approved the submitted version.
Conflict of Interest
HX and YR were employed by Tianjin Genovo Biotechnology Co., Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
This study was supported by the National Transgenic Major Program of China (2016ZX08004001) and the Agricultural Science and Technology Innovation Program (ASTIP) of Chinese Academy of Agricultural Sciences.
Footnotes
References
- Abbasi Z., Hooshyar S., Bagherieh-Najjar M. B. (2016). Improvement of callus production and shoot regeneration using various organs of soybean (Glycine max L. Merr) by response surface methodology. Vitro Cell. Dev. Pl. 52 537–545. 10.1007/s11627-016-9778-1 [DOI] [Google Scholar]
- Adachi K., Hirose A., Kanazashi Y., Hibara M., Hirata T., Mikami M., et al. (2021). Site-directed mutagenesis by biolistic transformation efficiently generates inheritable mutations in a targeted locus in soybean somatic embryos and transgene-free descendants in the T1 generation. Transgenic Res. 30 77–89. 10.1007/s11248-020-00229-4 [DOI] [PubMed] [Google Scholar]
- al Amin N., Ahmad N., Wu N., Pu X., Ma T., Du Y., et al. (2019). CRISPR-Cas9 mediated targeted disruption of FAD2–2 microsomal omega-6 desaturase in soybean (Glycine max.L). BMC Biotechnol. 19:9. 10.1186/s12896-019-0501-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleem M., Riaz A., Raza Q., Aleem M., Aslam M., Kong K., et al. (2022). Genome-wide characterization and functional analysis of class III peroxidase gene family in soybean reveal regulatory roles of GsPOD40 in drought tolerance. Genomics 114 45–60. 10.1016/j.ygeno.2021.11.016 [DOI] [PubMed] [Google Scholar]
- An J., Cheng C., Hu Z., Chen H., Cai W., Yu B. (2018). The Panax ginseng PgTIP1 gene confers enhanced salt and drought tolerance to transgenic soybean plants by maintaining homeostasis of water, salt ions and ROS. Environ. Exp. Bot. 155 45–55. 10.1016/j.envexpbot.2018.06.025 [DOI] [Google Scholar]
- Anguraj Vadivel A. K., McDowell T., Renaud J. B., Dhaubhadel S. (2021). A combinatorial action of GmMYB176 and GmbZIP5 controls isoflavonoid biosynthesis in soybean (Glycine max). Commun. Biol. 4:356. 10.1038/s42003-021-01889-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragão F. J. L., Sarokin L., Vianna G. R., Rech E. L. (2000). Selection of transgenic meristematic cells utilizing a herbicidal molecule results in the recovery of fertile transgenic soybean [Glycine max (L.) Merril] plants at a high frequency. Theor. Appl. Genet. 101 1–6. 10.1007/s001220051441 [DOI] [Google Scholar]
- Arun M., Chinnathambi A., Subramanyam K., Karthik S., Sivanandhan G., Theboral J., et al. (2016). Involvement of exogenous polyamines enhances regeneration and Agrobacterium-mediated genetic transformation in half-seeds of soybean. 3 Biotech 6:148. 10.1007/s13205-016-0448-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arun M., Subramanyam K., Mariashibu T. S., Theboral J., Shivanandhan G., Manickavasagam M., et al. (2015). Application of sonication in combination with vacuum infiltration enhances the Agrobacterium-mediated genetic transformation in Indian soybean cultivars. Appl. Biochem. Biotechnol. 175 2266–2287. 10.1007/s12010-014-1360-x [DOI] [PubMed] [Google Scholar]
- Arya H., Singh M. B., Bhalla P. L. (2021). Overexpression of PIF4 affects plant morphology and accelerates reproductive phase transitions in soybean. Food Energy Secur. 10:e291. 10.1002/fes3.291 [DOI] [Google Scholar]
- Bahramnejad B., Naji M., Bose R., Jha S. (2019). A critical review on use of Agrobacterium rhizogenes and their associated binary vectors for plant transformation. Biotech. Adv. 37:107405. 10.1016/j.biotechadv.2019.06.004 [DOI] [PubMed] [Google Scholar]
- Bai M., Yuan J., Kuang H., Gong P., Li S., Zhang Z., et al. (2020). Generation of a multiplex mutagenesis population via pooled CRISPR-Cas9 in soya bean. Plant Biotechnol. J. 18 721–731. 10.1111/pbi.13239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao A., Chen H., Chen L., Chen S., Hao Q., Guo W., et al. (2019). CRISPR/Cas9-mediated targeted mutagenesis of GmSPL9 genes alters plant architecture in soybean. BMC Plant Biol. 19:131. 10.1186/s12870-019-1746-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barampuram S., Zhang Z. J. (2011). “Recent advances in plant transformation,” in Plant Chromosome Engineering. Methods in Molecular Biology (Methods and Protocols, ed. Birchler J. A. (Totowa, NJ: Humana Press; ), 1–35. [DOI] [PubMed] [Google Scholar]
- Barwale U. B., Kerns H. R., Widholm Jack M. (1986). Plant regeneration from callus cultures of several soybean genotypes via embryogenesis and organogenesis. Planta 167 473–481. 10.1007/BF00391223 [DOI] [PubMed] [Google Scholar]
- Beyer E. (1979). Effect of silver ion, carbon dioxide, and oxygen on ethylene action and metabolism. Plant Physiol. 63 169–173. 10.1104/pp.63.1.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonawitz N. D., Ainley W. M., Itaya A., Chennareddy S. R., Cicak T., Effinger K., et al. (2019). Zinc finger nuclease-mediated targeting of multiple transgenes to an endogenous soybean genomic locus via non-homologous end joining. Plant Biotechnol. J. 17 750–761. 10.1111/pbi.13012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broothaerts W., Mitchell H. J., Weir B., Kaines S., Smith L. M., Yang W., et al. (2005). Gene transfer to plants by diverse species of bacteria. Nature 433 629–633. 10.1038/nature03309 [DOI] [PubMed] [Google Scholar]
- Bu T., Lu S., Wang K., Dong L., Li S., Xie Q., et al. (2021). A critical role of the soybean evening complex in the control of photoperiod sensitivity and adaptation. Proc. Natl. Acad. Sci. U. S. A. 118:e2010241118. 10.1073/pnas.2010241118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y., Chen L., Liu X., Guo C., Sun S., Wu C., et al. (2018). CRISPR/Cas9-mediated targeted mutagenesis of GmFT2a delays flowering time in soya bean. Plant Biotechnol. J. 16 176–185. 10.1111/pbi.12758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y., Chen L., Liu X., Sun S., Wu C., Jiang B., et al. (2015). CRISPR/Cas9-mediated genome editing in soybean hairy roots. PLoS One 10:e0136064. 10.1371/journal.pone.0136064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y., Wang L., Chen L., Wu T., Liu L., Sun S., et al. (2020b). Mutagenesis of GmFT2a and GmFT5a mediated by CRISPR/Cas9 contributes for expanding the regional adaptability of soybean. Plant Biotechnol. J. 18 298–309. 10.1111/pbi.13199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y., Chen L., Zhang Y., Yuan S., Su Q., Sun S., et al. (2020a). Target base editing in soybean using a modified CRISPR/Cas9 system. Plant Biotechnol. J. 18 1996–1998. 10.1111/pbi.13386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Z., Xian P., Cheng Y., Ma Q., Lian T., Nian H., et al. (2021). CRISPR/Cas9-mediated gene editing of GmJAGGED1 increased yield in the low latitude soybean variety Huachun 6. Plant Biotechnol. J. 19 1898–1900. 10.1111/pbi.13673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell B. W., Hoyle J. W., Bucciarelli B., Stec A. O., Samac D. A., Parrott W. A., et al. (2019). Functional analysis and development of a CRISPR/Cas9 allelic series for a CPR5 ortholog necessary for proper growth of soybean trichomes. Sci. Rep. 9:14757. 10.1038/s41598-019-51240-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D., Li Y., Liu B., Kong F., Tran L. P. (2018). Adaptive mechanisms of soybean grown on salt−affected soils. Land Degrad. Dev. 29 1054–1064. 10.1002/ldr.2754 [DOI] [Google Scholar]
- Carrijo J., Illa-Berenguer E., LaFayette P., Torres N., Aragao F. J. L., Parrott W., et al. (2021). Two efficient CRISPR/Cas9 systems for gene editing in soybean. Transgenic Res. 30 239–249. 10.1007/s11248-021-00246-x [DOI] [PubMed] [Google Scholar]
- Carvalho C. H. S., Bohorova N., Bordallo P. N., Abreu L. L., Valicente F. H., Bressan W., et al. (1997). Type II callus production and plant regeneration in tropical maize genotypes. Plant Cell Rep. 17 73–76. 10.1007/s002990050355 [DOI] [PubMed] [Google Scholar]
- Castiglioni P., Bell E., Lund A., Rosenberg A. F., Galligan M., Hinchey B. S., et al. (2018). Identification of GB1, a gene whose constitutive overexpression increases glycinebetaine content in maize and soybean. Plant Direct 2:e00040. 10.1002/pld3.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chee P. P., Fober K. A., Slightom J. L. (1989). Transformation of soybean (Glycine max) by infecting germinating seeds with Agrobacterium tumefaciens. Plant Physiol. 91 1212–1218. 10.1104/pp.91.3.1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K., Liu W., Li X., Li H. (2020b). Overexpression of GmGASA32 promoted soybean height by interacting with GmCDC25. Plant Signal. Behav. 16:1855017. 10.1080/15592324.2020.1855017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B., Zhang G., Li P., Yang J., Guo L., Benning C., et al. (2020a). Multiple GmWRI1s are redundantly involved in seed filling and nodulation by regulating plastidic glycolysis, lipid biosynthesis and hormone signalling in soybean (Glycine max). Plant Biotechnol. J. 18 155–171. 10.1111/pbi.13183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Cai Y., Qu M., Wang L., Sun H., Jiang B., et al. (2020c). Soybean adaption to high-latitude regions is associated with natural variations of GmFT2b, an ortholog of FLOWERING LOCUS T. Plant Cell Environ. 43 934–944. 10.1111/pce.13695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Nan H., Kong L., Yue L., Yang H., Zhao Q., et al. (2020d). Soybean AP1 homologs control flowering time and plant height. J. Integr. Agr. 62 1868–1879. 10.1111/jipb.12988 [DOI] [PubMed] [Google Scholar]
- Chen L., Yang H., Fang Y., Guo W., Chen H., Zhang X., et al. (2021b). Overexpression of GmMYB14 improves high-density yield and drought tolerance of soybean through regulating plant architecture mediated by the brassinosteroid pathway. Plant Biotechnol. J. 19 702–716. 10.1111/pbi.13496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K., Su C., Tang W., Zhou Y., Xu Z., Chen J., et al. (2021a). Nuclear transport factor GmNTF2B-1 enhances soybean drought tolerance by interacting with oxidoreductase GmOXR17 to reduce reactive oxygen species content. Plant J. 107 740–759. 10.1111/tpj.15319 [DOI] [PubMed] [Google Scholar]
- Chen Z., Fang X., Yuan X., Zhang Y., Li H., Zhou Y., et al. (2021c). Overexpression of transcription factor GmTGA15 enhances drought tolerance in transgenic soybean hairy roots and Arabidopsis plants. Agronomy 11 170. 10.3390/agronomy11010170 [DOI] [Google Scholar]
- Chen Z. F., Ru J. N., Sun G. Z., Du Y., Chen J., Zhou Y. B., et al. (2021d). Genomic-wide analysis of the PLC family and detection of GmPI-PLC7 responses to drought and salt stresses in soybean. Front. Plant Sci. 12:631470. 10.3389/fpls.2021.631470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K., Tang W., Zhou Y., Chen J., Xu Z., Ma R., et al. (2022). AP2/ERF transcription factor GmDREB1 confers drought tolerance in transgenic soybean by interacting with GmERFs. Plant Physiol. Biochem. 170 287–295. 10.1016/j.plaphy.2021.12.014 [DOI] [PubMed] [Google Scholar]
- Chen K., Wang Y., Zhang R., Zhang H., Gao C. (2019a). CRISPR/Cas genome editing and precision plant breeding in agriculture. Ann. Rev. Plant Biol. 70 667–697. 10.1146/annurev-arplant-050718-100049 [DOI] [PubMed] [Google Scholar]
- Chen L., Qin L., Zhou L., Li X., Chen Z., Sun L., et al. (2019b). A nodule-localized phosphate transporter GmPT7 plays an important role in enhancing symbiotic N2 fixation and yield in soybean. New Phytol. 221 2013–2025. 10.1111/nph.15541 [DOI] [PubMed] [Google Scholar]
- Chen X., Fang X., Zhang Y., Wang X., Zhang C., Yan X., et al. (2019c). Overexpression of a soybean 4-coumaric acid: coenzyme A ligase (GmPI4L) enhances resistance to Phytophthora sojae in soybean. Front. Plant Sci. 46:304–313. 10.1071/FP18111 [DOI] [PubMed] [Google Scholar]
- Chen L., Cai Y., Liu X., Yao W., Guo C., Sun S., et al. (2018b). Improvement of soybean Agrobacterium-mediated transformation efficiency by adding glutamine and asparagine into the culture media. Int. J. Mol. Sci. 19:3039. 10.3390/ijms19103039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Cai Y., Liu X., Guo C., Sun S., Wu C., et al. (2018a). Soybean hairy roots produced in vitro by Agrobacterium rhizogenes-mediated transformation. Crop J. 6 162–171. 10.1016/j.cj.2017.08.006 [DOI] [Google Scholar]
- Chen L., Chunyu Z., Mingxia F., Wenjuan M., Meiming C., Fengchun C., et al. (2018c). GmIDL2a and GmIDL4a, encoding the inflorescence deficient in abscission-like protein, are involved in soybean cell wall degradation during lateral root emergence. Int. J. Mol. Sci. 19:2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng M., Fry J. E., Pang S., Zhou H., Hironaka C. M., Duncan D. R., et al. (1997). Genetic transformation of wheat mediated by Agrobacterium tumefaciens. Plant Physiol. 115 971–980. 10.1104/pp.115.3.971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Q., Dong L., Su T., Li T., Gan Z., Nan H., et al. (2019). CRISPR/Cas9-mediated targeted mutagenesis of GmLHY genes alters plant height and internode length in soybean. BMC Plant Biol. 19:562. 10.1186/s12870-019-2145-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Q., Gan Z., Wang Y., Lu S., Hou Z., Li H., et al. (2020). The soybean gene J contributes to salt stress tolerance by up-regulating salt-responsive genes. Front. Plant Sci. 11:272. 10.3389/fpls.2020.00272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Wang X., Cao L., Ji J., Liu T., Duan K. (2021). Highly efficient Agrobacterium rhizogenes-mediated hairy root transformation for gene functional and gene editing analysis in soybean. Plant Methods 17:73. 10.1186/s13007-021-00778-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chennareddy S., Cicak T., Mall T., Effinger K., Sardesai N., Pareddy D., et al. (2018). Improved direct transformation via particle bombardment of split-immature embryo explants in soybean (Glycine max). Plant Cell Tiss. Org. 135 23–35. 10.1007/s11240-018-1440-7 [DOI] [Google Scholar]
- Cho H. J., Farrand S. K., Noel G. R., Widholm J. M. (2000). High-efficiency induction of soybean hairy roots and propagation of the soybean cyst nematode. Planta 210 195–204. 10.1007/PL00008126 [DOI] [PubMed] [Google Scholar]
- Cho H. J., Moy Y., Rudnick N. A., Klein T. M., Yin J., Bolar J., et al. (2022). Development of an efficient marker-free soybean transformation method using the novel bacterium Ochrobactrum haywardense H1. Plant Biotechnol. J. 20 977–990. 10.1111/pbi.13777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H. S., Lee Y. J., Kim H. J., Park M.-Y., Yeom W. W., Song J. H., et al. (2021). Overexpression of Arabidopsis thaliana blue-light inhibitor of cryptochromes 1 gene alters plant architecture in soybean. Plant Biotechnol. Rep. 15 459–469. 10.1007/s11816-021-00693-2 [DOI] [Google Scholar]
- Chowrira G. M., Akella V., Lurquin P. F. (1995). Electroporation-mediated gene transfer into intact nodal meristemsin planta. Mol. Biotech. 3 17–23. 10.1007/BF02821331 [DOI] [PubMed] [Google Scholar]
- Christey M. C. (2001). Use of ri-mediated transformation for production of transgenic plants. In Vitro Cell. Dev. Pl. 37 687–700. 10.1007/s11627-001-0120-0 [DOI] [Google Scholar]
- Christou P. (1990). Morphological description of transgenic soybean chimeras created by the delivery, integration and expression of foreign DNA using electric discharge particle acceleration. Ann. Bot. 66 379–386. 10.1093/oxfordjournals.aob.a088039 [DOI] [Google Scholar]
- Christou P., McCabe D., Swain W. (1988). Stable transformation of soybean callus by DNA-coated gold particles. Plant Physiol. 87 671–674. 10.1104/pp.87.3.671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christou P., Mccabe D. E. (1992). Prediction of germ-line transformation events in chimeric Ro transgenic soybean plantlets using tissue-specific expression patterns. Plant J. 2 283–290. 10.1111/j.1365-313X.1992.00283.x [DOI] [Google Scholar]
- Clemente T. E., LaVallee B. J., Howe A. R., Conner-Ward D., Rozman R. J., Hunter P. E., et al. (2000). Progeny analysis of glyphosate selected transgenic soybeans derived from Agrobacterium-Mediated transformation. Crop Sci. 40 797–803. 10.2135/cropsci2000.403797x [DOI] [Google Scholar]
- Collier R., Fuchs B., Walter N., Kevin Lutke W., Taylor C. G. (2005). Ex vitro composite plants: an inexpensive, rapid method for root biology. Plant J. 43 449–457. 10.1111/j.1365-313X.2005.02454.x [DOI] [PubMed] [Google Scholar]
- Coutinho F. S., Dos Santos D. S., Lima L. L., Vital C. E., Santos L. A., Pimenta M. R., et al. (2019). Mechanism of the drought tolerance of a transgenic soybean overexpressing the molecular chaperone BiP. Physiol. Mol. Biol. Plants 25 457–472. 10.1007/s12298-019-00643-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtin S. J., Xiong Y., Michno J. M., Campbell B. W., Stec A. O., Cermak T., et al. (2018). CRISPR/Cas9 and TALENs generate heritable mutations for genes involved in small RNA processing of Glycine max and Medicago truncatula. Plant Biotechnol. J. 16 1125–1137. 10.1111/pbi.12857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtin S. J., Zhang F., Sander J. D., Haun W. J., Starker C., Baltes N. J., et al. (2011). Targeted mutagenesis of duplicated genes in soybean with zinc-finger nucleases. Plant Physiol. 156 466–473. 10.1104/pp.111.172981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan Y., Armstrong C. L., Dong J., Feng X., Fry J. E., Keithly G. E., et al. (2009). Lipoic acid—an unique plant transformation enhancer. In Vitro Cell. Dev. Biol. Plant 45 630–638. 10.1007/s11627-009-9227-5 [DOI] [Google Scholar]
- Dan Y., Reichert N. A. (1998). Organogenic regeneration of soybean from hypocotyl explants. In Vitro Cell. Dev. Pl. 34 14–21. 10.1007/BF02823117 [DOI] [Google Scholar]
- Dang W., Wei Z. M. (2007). An optimized Agrobacterium-mediated transformation for soybean for expression of binary insect resistance genes. Plant Sci. 173 381–389. 10.1016/j.plantsci.2007.06.010 [DOI] [Google Scholar]
- Debernardi J. M., Tricoli D. M., Ercoli M. F., Hayta S., Ronald P., Palatnik J. F., et al. (2020). A GRF-GIF chimeric protein improves the regeneration efficiency of transgenic plants. Nat. Biotech. 38 1274–1279. 10.1038/s41587-020-0703-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demorest Z. L., Coffman A., Baltes N. J., Stoddard T. J., Clasen B. M., Luo S., et al. (2016). Direct stacking of sequence-specific nuclease-induced mutations to produce high oleic and low linolenic soybean oil. BMC Plant Biol. 16:225. 10.1186/s12870-016-0906-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhir S. K., Dhir S., Savka M. A., Belanger F., Kriz A. L., Farrand S. K., et al. (1992). Regeneration of transgenic soybean (Glycine max) plants from electroporated protoplasts. Plant Physiol. 99 81–88. 10.1104/pp.99.1.81 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Do P. T., Nguyen C. X., Bui H. T., Tran L. T. N., Stacey G., Gillman J. D., et al. (2019). Demonstration of highly efficient dual gRNA CRISPR/Cas9 editing of the homeologous GmFAD2-1A and GmFAD2-1B genes to yield a high oleic, low linoleic and alpha-linolenic acid phenotype in soybean. BMC Plant Biol. 19:311. 10.1186/s12870-019-1906-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson P. A., Simmonds D. H. (2000). Susceptibility to Agrobacterium tumefaciens and cotyledonary node transformation in short-season soybean. Plant Cell Rep. 19 478–484. 10.1007/s002990050759 [DOI] [PubMed] [Google Scholar]
- Dong J., Hudson M. E. (2022). WI12Rhg1 interacts with DELLAs and mediates soybean cyst nematode resistance through hormone pathways. Plant Biotechnol. J. 20 283–296. 10.1111/pbi.13709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J., Zielinski R. E., Hudson M. E. (2020). t-SNAREs bind the Rhg1 α-SNAP and mediate soybean cyst nematode resistance. Plant J. 104 318–331. 10.1111/tpj.14923 [DOI] [PubMed] [Google Scholar]
- Dong L., Fang C., Cheng Q., Su T., Kou K., Kong L., et al. (2021). Genetic basis and adaptation trajectory of soybean from its temperate origin to tropics. Nat. Commun. 12:5445. 10.1038/s41467-021-25800-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H., Zeng X., Zhao M., Cui X., Wang Q., Yang H., et al. (2016). Efficient targeted mutagenesis in soybean by TALENs and CRISPR/Cas9. J. Biotech. 217 90–97. 10.1016/j.jbiotec.2015.11.005 [DOI] [PubMed] [Google Scholar]
- Du Q., Yang X., Zhang J., Zhong X., Kim K. S., Yang J., et al. (2018). Over-expression of the Pseudomonas syringae harpin-encoding gene hrpZm confers enhanced tolerance to Phytophthora root and stem rot in transgenic soybean. Transgenic Res. 27 277–288. 10.1007/s11248-018-0071-4 [DOI] [PubMed] [Google Scholar]
- Duan K., Cheng Y., Ji J., Wang C., Wei Y., Wang Y. (2021). Large chromosomal segment deletions by CRISPR/LbCpf1-mediated multiplex gene editing in soybean. J. Integr. Plant Biol. 63 1620–1631. 10.1111/jipb.13158 [DOI] [PubMed] [Google Scholar]
- Dubey V. K., Lee U. G., Kwon D. H., Lee S. H. (2017). Agroinfiltration-based expression of hairpin RNA in soybean plants for RNA interference against Tetranychus urticae. Pestic. Biochem. Physiol. 142 53–58. 10.1016/j.pestbp.2017.01.004 [DOI] [PubMed] [Google Scholar]
- Falco S. C., Guida T., Locke M., Mauvais J., Sanders C., Ward R. T., et al. (1995). Transgenic canola and soybean seeds with increased lysine. Biotechnology 13 577–582. 10.1038/nbt0695-577 [DOI] [PubMed] [Google Scholar]
- Fan Y., Wang X., Li H., Liu S., Jin L., Lyu Y., et al. (2020). Anthocyanin, a novel and user-friendly reporter for convenient, non-destructive, low cost, directly visual selection of transgenic hairy roots in the study of rhizobia-legume symbiosis. Plant Methods 16:94. 10.1186/s13007-020-00638-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria J. A. Q. A., Reis P. A. B., Reis M. T. B., Rosado G. L., Pinheiro G. L., Mendes G. C., et al. (2011). The NAC domain-containing protein, GmNAC6, is a downstream component of the ER stress- and osmotic stress-induced NRP-mediated cell-death signaling pathway. BMC Plant Biol. 11:129. 10.1186/1471-2229-11-129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X., Feng P., Yu H., Yu X., Sun Q., Liu S., et al. (2020). GsSnRK1 interplays with transcription factor GsERF7 from wild soybean to regulate soybean stress resistance. Plant Cell Environ. 43 1192–1211. 10.1111/pce.13726 [DOI] [PubMed] [Google Scholar]
- Fernandez S., Michaux-Ferrière N., Coumans M. F. (1999). The embryogenic response of immature embryo cultures of durum wheat (Triticum durum Desf.): histology and improvement by AgNO3. Plant Growth Regul. 28 147–155. 10.1023/A:1006142504577 [DOI] [Google Scholar]
- Finer J. J. (2016). Generation of transgenic soybean (Glycine max) via particle bombardment of embryogenic cultures. Curr. Protoc. Plant Bio. 1 592–603. 10.1002/cppb.20039 [DOI] [PubMed] [Google Scholar]
- Finer J. J., Larkin K. M. (2008). “Genetic transformation of soybean using particle bombardment and SAAT approaches,” in Handbook of New Technologies For Genetic Improvement of Legumes, ed. PB K. (Boca Raton, USA: CRC Press/Taylor and Francis Group; ), 103–123. [Google Scholar]
- Finer J. J., Mcmullen M. D. (1991). Transformation of soybean via particle bombardment of embryogenic suspension culture tissue. In Vitro Cell. Dev. Pl. 27 175–182. 10.2307/4292952 [DOI] [Google Scholar]
- Finer J. J., Nagasawa A. (1988). Development of an embryogenic suspension culture of soybean (Glycine max Merrill.). Plant Cell Tiss. Org. 15 125–136. 10.1007/BF00035754 [DOI] [Google Scholar]
- Fu M., Sun J., Li X., Guan Y., Xie F. (2022). Asymmetric redundancy of soybean Nodule Inception (NIN) genes in root nodule symbiosis. Plant Physiol. 188 477–489. 10.1093/plphys/kiab473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C. (2021). Genome engineering for crop improvement and future agriculture. Cell 184 1621–1635. 10.1016/j.cell.2021.01.005 [DOI] [PubMed] [Google Scholar]
- Gao L., Luo J., Ding X., Wang T., Hu T., Song P., et al. (2020). Soybean RNA interference lines silenced for eIF4E show broad potyvirus resistance. Mol. Plant Pathol. 21 303–317. 10.1111/mpp.12897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghidelli C., Mateos M., Rojas-Argudo C., Pérez-Gago M. B. (2014). Effect of antioxidants on enzymatic browning of eggplant extract and fresh-cut tissue. J. Food Process. Pres. 38 1501–1510. 10.1111/jfpp.12109 [DOI] [Google Scholar]
- Gordon-Kamm W., Sardesai N., Arling M., Lowe K., Hoerster G., Betts S., et al. (2019). Using morphogenic genes to improve recovery and regeneration of transgenic plants. Plants 8 38. 10.3390/plants8020038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimsley N., Hohn B., Hohn T., Walden R. (1986). “Agroinfection,” an alternative route for viral infection of plants by using the Ti plasmid. Proc. Natl. Acad. Sci. U. S. A. 83 3282–3286. 10.1073/pnas.83.10.3282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B., Guo Y., Hong H., Jin L., Zhang L., Chang R. Z., et al. (2015). Co-expression of G2-EPSPS and glyphosate acetyltransferase GAT genes conferring high tolerance to glyphosate in soybean. Front. Plant Sci. 6:847. 10.3389/fpls.2015.00847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B. F., Hong H. L., Han J. N., Zhang L. J., Qiu L. J. (2020a). Development and identification of glyphosate-tolerant transgenic soybean via direct selection with glyphosate. J. Integr. Agr. 19 1186–1196. 10.1016/S2095-3119(19)62747-4 [DOI] [Google Scholar]
- Guo C., Liu X., Chen L., Cai Y., Yao W., Yuan S., et al. (2020b). Elevated methionine content in soybean seed by overexpressing maize β-zein protein. Oil Crop Sci. 5 11–16. 10.1016/j.ocsci.2020.03.004 [DOI] [Google Scholar]
- Guo W., Chen L., Chen H., Yang H., You Q., Bao A., et al. (2020c). Overexpression of GmWRI1b in soybean stably improves plant architecture and associated yield parameters, and increases total seed oil production under field conditions. Plant Biotechnol. J. 18 1639–1641. 10.1111/pbi.13324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hada A., Gupta A. K., Jeevaraj T., Manickavasagam M., Ganapathi A., Jolly M., et al. (2014). Developing rapid and reliable regeneration system using cotyledonary-node method in Indian soybean genotypes (Glycine max L.). Int. J. Inno.Res. Sci. 3 12678–12686. [Google Scholar]
- Hada A., Krishnan V., Mohamed Jaabir M. S., Kumari A., Jolly M., Praveen S., et al. (2018). Improved Agrobacterium tumefaciens-mediated transformation of soybean [Glycine max (L.) Merr.] following optimization of culture conditions and mechanical techniques. In Vitro Cell. Dev. Pl. 54 672–688. 10.1007/s11627-018-9944-8 [DOI] [Google Scholar]
- Hadi M. Z., McMullen M. D., Finer J. J. (1996). Transformation of 12 different plasmids into soybean via particle bombardment. Plant Cell Rep. 15 500–505. 10.1007/BF00232982 [DOI] [PubMed] [Google Scholar]
- Han J., Guo B., Guo Y., Zhang B., Wang X., Qiu L. J. (2019). Creation of early flowering germplasm of soybean by CRISPR/Cas9 technology. Front. Plant Sci. 10:1446. 10.3389/fpls.2019.01446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X., Wang D., Song G. Q. (2021). Expression of a maize SOC1 gene enhances soybean yield potential through modulating plant growth and flowering. Sci. Rep. 11:12758. 10.1038/s41598-021-92215-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y., Fan X., Guo H., Yao Y., Ren G., Lv X., et al. (2020). Overexpression of the bioactive lunasin peptide in soybean and evaluation of its anti-inflammatory and anti-cancer activities invitro. J. Biosci. Bioeng. 129 395–404. 10.1016/j.jbiosc.2019.11.001 [DOI] [PubMed] [Google Scholar]
- Haun W., Coffman A., Clasen B. M., Demorest Z. L., Lowy A., Ray E., et al. (2014). Improved soybean oil quality by targeted mutagenesis of the fatty acid desaturase 2 gene family. Plant Biotechnol. J. 12 934–940. 10.1111/pbi.12201 [DOI] [PubMed] [Google Scholar]
- Hayashi M., Saeki Y., Haga M., Harada K., Kouchi H., Umehara Y. (2012). Rj (rj) genes involved in nitrogen-fixing root nodule formation in soybean. Breeding Sci. 61 544–553. 10.1270/jsbbs.61.544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C., Gao H., Wang H., Guo Y., He M., Peng Y., et al. (2021). GSK3-mediated stress signaling inhibits legume–rhizobium symbiosis by phosphorylating GmNSP1 in soybean. Mol. Plant 14 488–502. 10.1016/j.molp.2020.12.015 [DOI] [PubMed] [Google Scholar]
- He Y., Dong Y., Yang X., Guo D., Qian X., Yan F., et al. (2020). Functional activation of a novel R2R3-MYB protein gene, GmMYB68, confers salt-alkali resistance in soybean (Glycine max L.). Genome 63 13–26. 10.1139/gen-2018-0132 [DOI] [PubMed] [Google Scholar]
- Hernandez-Garcia C. M., Martinelli A. P., Bouchard R. A., Finer J. J. (2009). A soybean (Glycine max) polyubiquitin promoter gives strong constitutive expression in transgenic soybean. Plant Cell Rep. 28 837–849. 10.1007/s00299-009-0681-7 [DOI] [PubMed] [Google Scholar]
- Hinchee M. A. W., Connor-Ward D. V., Newell C. A., Mcdonnell R. E., Sato S. J., Gasser C. S., et al. (1988). Production of transgenic soybean plants using Agrobacterium-mediated DNA transfer. Biotechnology 6 915–922. 10.1038/nbt0888-915 [DOI] [Google Scholar]
- Homrich M. S., Wiebke-Strohm B., Weber R. L., Bodanese-Zanettini M. H. (2012). Soybean genetic transformation: a valuable tool for the functional study of genes and the production of agronomically improved plants. Genet. Mol. Biol. 35 998–1010. 10.1590/s1415-47572012000600015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong H., Zhang H., Olhoft P., Hill S., Wiley H., Toren E., et al. (2007). Organogenic callus as the target for plant regeneration and transformation via Agrobacterium in soybean (Glycine max (L.) Merr.). In Vitro Cell. Dev. Pl. 43 558–568. 10.1007/s11627-007-9066-1 [DOI] [Google Scholar]
- Hu C. Y., Wang L. (1999). In planta soybean transformation technologies developed in China: procedure, confirmation and field performance. In Vitro Cell. Dev. Pl. 35 417–420. 10.2307/4293277 [DOI] [Google Scholar]
- Huang Y., Xuan H., Yang C., Guo N., Wang H., Zhao J., et al. (2019). GmHsp90A2 is involved in soybean heat stress as a positive regulator. Plant Sci. 285 26–33. 10.1016/j.plantsci.2019.04.016 [DOI] [PubMed] [Google Scholar]
- Hwang H. H., Yu M., Lai E. M. (2017). Agrobacterium-mediated plant transformation: biology and applications. Arabidopsis Book 15:e0186. 10.1199/tab.0186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam N., Islam T., Hossain M., Bhattacharjee B., Hossain M., Islam S. M. S. (2017). Embryogenic callus induction and efficient plant regeneration in three varieties of soybean (Glycine max). Plant Tiss. Cul. Biotechnol. 27 41–50. 10.3329/ptcb.v27i1.35011 [DOI] [Google Scholar]
- Ismagul A., Yang N., Maltseva E., Iskakova G., Mazonka I., Skiba Y., et al. (2018). A biolistic method for high-throughput production of transgenic wheat plants with single gene insertions. BMC Plant Biol. 18:135. 10.1186/s12870-018-1326-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itaya A., Zheng S., Simmonds D. (2018). Establishment of neomycin phosphotransferase II (nptII) selection for transformation of soybean somatic embryogenic cultures. In Vitro Cell. Dev. Pl. 54 184–194. 10.1007/s11627-017-9875-9 [DOI] [Google Scholar]
- Iwabuchi K., Shimada T. L., Yamada T., Hara-Nishimura I. (2020). A space-saving visual screening method. Glycine max FAST, for generating transgenic soybean. Plant Signal. Behav. 15:1722911. 10.1080/15592324.2020.1722911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs T. B., LaFayette P. R., Schmitz R. J., Parrott W. A. (2015). Targeted genome modifications in soybean with CRISPR/Cas9. BMC Biotechnol. 15:16. 10.1186/s12896-015-0131-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahan M. A., Harris B., Lowery M., Infante A. M., Percifield R. J., Kovinich N. (2020). Glyceollin transcription factor GmMYB29A2 regulates soybean resistance to Phytophthora sojae. Plant Physiol. 183 530–546. 10.1104/pp.19.01293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Q., Sun S., Kong D., Song J., Wu L., Yan Z., et al. (2020). Ectopic expression of Gs5PTase8, a soybean inositol polyphosphate 5-phosphatase, enhances salt tolerance in plants. Int. J. Mol. Sci. 21 1023. 10.3390/ijms21031023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y., Yao X., Zhao M., Zhao Q., Du Y., Yu C., et al. (2015). Comparison of soybean transformation efficiency and plant factors affecting transformation during the Agrobacterium infection process. Int. J. Mol. Sci. 16 18522–18543. 10.3390/ijms160818522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang B., Chen L., Yang C., Wu T., Yuan S., Wu C., et al. (2021). The cloning and CRISPR/Cas9-mediated mutagenesis of a male sterility gene MS1 of soybean. Plant Biotechnol. J. 19 1098–1100. 10.1111/pbi.13601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing G., Tang D., Yao Y., Su Y., Shen Y., Bai Y., et al. (2021). Seed specifically over-expressing DGAT2A enhances oil and linoleic acid contents in soybean seeds. Biochem. Bioph. Res. Co. 568 143–150. 10.1016/j.bbrc.2021.06.087 [DOI] [PubMed] [Google Scholar]
- Jones S. I., Hunt M. R., Vodkin L. O. (2020). An embryo lethal transgenic line manifests global expression changes and elevated protein/oil ratios in heterozygous soybean plants. PLoS One 15:e0233721. 10.1371/journal.pone.0233721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyner E. Y., Boykin L. S., Lodhi M. A. (2010). Callus induction and organogenesis in soybean [Glycine max (L.) Merr.] cv. pyramid from mature cotyledons and embryos. Open Plant Sci. J. 4 18–21. 10.2174/1874294701004010018 [DOI] [Google Scholar]
- Kahn T. W., Duck N. B., McCarville M. T., Schouten L. C., Schweri K., Zaitseva J., et al. (2021). A Bacillus thuringiensis Cry protein controls soybean cyst nematode in transgenic soybean plants. Nat. Commun. 12:3380. 10.1038/s41467-021-23743-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalbande B. B., Patil A. S. (2016). Plant tissue culture independent Agrobacterium tumefaciens mediated In-planta transformation strategy for upland cotton (Gossypium hirsutum). J. Genet. Eng. Biotechnol. 14 9–18. 10.1016/j.jgeb.2016.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambakam S., Ngaki M. N., Sahu B. B., Kandel D. R., Singh P., Sumit R., et al. (2021). Arabidopsis non-host resistance PSS30 gene enhances broad-spectrum disease resistance in the soybean cultivar Williams 82. Plant J. 107 1432–1446. 10.1111/tpj.15392 [DOI] [PubMed] [Google Scholar]
- Kanai M., Yamada T., Hayashi M., Mano S., Nishimura M. (2019). Soybean (Glycine max L.) triacylglycerol lipase GmSDP1 regulates the quality and quantity of seed oil. Sci. Rep. 9:8924. 10.1038/s41598-019-45331-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanazashi Y., Hirose A., Takahashi I., Mikami M., Endo M., Hirose S., et al. (2018). Simultaneous site-directed mutagenesis of duplicated loci in soybean using a single guide RNA. Plant Cell Rep. 37 553–563. 10.1007/s00299-018-2251-3 [DOI] [PubMed] [Google Scholar]
- Karthik S., Pavan G., Manickavasagam M. (2020). Nitric oxide donor regulates Agrobacterium-mediated genetic transformation efficiency in soybean [Glycine max (L.) Merrill]. Plant Cell Tiss. Org. 141 655–660. 10.1007/s11240-020-01808-3 [DOI] [Google Scholar]
- Karthik S., Tuteja N., Ganapathi A., Manickavasagam M. (2019). Pea p68, a DEAD-box helicase, enhances salt tolerance in marker-free transgenic plants of soybean [Glycine max (L.) Merrill]. 3 Biotech 9:10. 10.1007/s13205-018-1553-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kereszt A., Li D., Indrasumunar A., Nguyen C. D., Nontachaiyapoom S., Kinkema M., et al. (2007). Agrobacterium rhizogenes-mediated transformation of soybean to study root biology. Nat. Protoc. 2 948–952. 10.1038/nprot.2007.141 [DOI] [PubMed] [Google Scholar]
- Kidokoro S., Watanabe K., Ohori T., Moriwaki T., Maruyama K., Mizoi J., et al. (2015). Soybean DREB1/CBF-type transcription factors function in heat and drought as well as cold stress-responsive gene expression. Plant J. 81 505–518. 10.1111/tpj.12746 [DOI] [PubMed] [Google Scholar]
- Kim H., Choi J. (2021). A robust and practical CRISPR/crRNA screening system for soybean cultivar editing using LbCpf1 ribonucleoproteins. Plant Cell Rep. 40 1059–1070. 10.1007/s00299-020-02597-x [DOI] [PubMed] [Google Scholar]
- Kim H., Kim S. T., Ryu J., Kang B. C., Kim J. S., Kim S. G. (2017). CRISPR/Cpf1-mediated DNA-free plant genome editing. Nat. Commun. 8:14406. 10.1038/ncomms14406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. J., Cho H. S., Pak J. H., Kwon T., Lee J. H., Kim D. H., et al. (2018). Confirmation of drought tolerance of ectopically expressed AtABF3 gene in soybean. Mol. Cells 41 413–422. 10.14348/molcells.2018.2254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. H., Lim S., Kang Y. J., Yoon M. Y., Nam M., Jun T. H., et al. (2016). Optimization of a virus-induced gene silencing system with soybean yellow common mosaic virus for gene function studies in soybeans. Plant Pathol. J. 32 112–122. 10.5423/PPJ.OA.04.2015.0063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W.-S., Krishnan H. B. (2019). Impact of co-expression of maize 11 and 18 kDa δ-zeins and 27 kDa γ-zein in transgenic soybeans on protein body structure and sulfur amino acid content. Plant Sci. 280 340–347. 10.1016/j.plantsci.2018.12.016 [DOI] [PubMed] [Google Scholar]
- Kim W.-S., Sun-Hyung J., Oehrle N. W., Jez J. M., Krishnan H. B. (2020). Overexpression of ATP sulfurylase improves the sulfur amino acid content, enhances the accumulation of Bowman–Birk protease inhibitor and suppresses the accumulation of the β-subunit of β-conglycinin in soybean seeds. Sci. Rep. 10:14989. 10.1038/s41598-020-72134-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- King J. L., Finer J. J., McHale L. K. (2015). Development and optimization of agroinfiltration for soybean. Plant Cell Rep. 34 133–140. 10.1007/s00299-014-1694-4 [DOI] [PubMed] [Google Scholar]
- Klein T. M., Wolf E. D., Wu R., Sanford J. C. (1987). High-velocity microprojectiles for delivering nucleic acids into living cells. Nature 327 70–73. 10.1038/327070a0 [DOI] [PubMed] [Google Scholar]
- Ko T. S., Lee S., Krasnyanski S., Korban S. S. (2003). Two critical factors are required for efficient transformation of multiple soybean cultivars: agrobacterium strain and orientation of immature cotyledonary explant. Theor. Appl. Genet. 107 439–447. 10.1007/s00122-003-1264-6 [DOI] [PubMed] [Google Scholar]
- Kong J., Martin-Ortigosa S., Finer J., Orchard N., Gunadi A., Batts L. A., et al. (2020). Overexpression of the transcription factor GROWTH-REGULATING FACTOR5 improves transformation of dicot and monocot Species. Front. Plant Sci. 11:572319. 10.3389/fpls.2020.572319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuma K. M., Lopes-Caitar V. S., Romero C. C. T., Silva S. M. H., Kuwahara M. K., Carvalho M. C. C. G., et al. (2015). A high efficient protocol for soybean root transformation by Agrobacterium rhizogenes and most stable reference genes for RT-qPCR analysis. Plant Cell Rep. 34 1987–2000. 10.1007/s00299-015-1845-2 [DOI] [PubMed] [Google Scholar]
- Kumar A., Kumar V., Krishnan V., Hada A., Marathe A. C. P. (2019). Seed targeted RNAi-mediated silencing of GmMIPS1 limits phytate accumulation and improves mineral bioavailability in soybean. Sci. Rep. 9:7744. 10.1038/s41598-019-44255-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Liu Z.-B., Sanyour-Doyel N., Lenderts B., Worden A., Anand A., et al. (2022). Efficient gene targeting in soybean using Ochrobactrum haywardense-mediated delivery of a marker-free donor template. Plant Physiol. 00 1–10. 10.1093/plphys/kiac075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S. V., Rajam M. (2005). Polyamines enhance Agrobacterium tumefaciens vir gene induction and T-DNA transfer. Plant Sci. 168 475–480. 10.1016/j.plantsci.2004.09.018 [DOI] [Google Scholar]
- Kumari A., Hada A., Subramanyam K., Theboral J., Misra S., Ganapathi A., et al. (2018). RNAi-mediated resistance to yellow mosaic viruses in soybean targeting coat protein gene. Acta Physiol. Plant 40:32. 10.1007/s11738-018-2608-9 [DOI] [Google Scholar]
- Kun X., Zhang X.-M., Fan C.-M., Chen F.-L., Zhu J.-L., Zhang S.-L., et al. (2017). A callus transformation system for gene functional studies in soybean. J. Integr. Agr. 16 1913–1922. 10.1016/S2095-3119(16)61621-0 [DOI] [Google Scholar]
- Le H., Nguyen N. H., Ta D. T., Le T. N. T., Bui T. P., Le N. T., et al. (2020). CRISPR/Cas9-mediated knockout of galactinol synthase-encoding genes reduces raffinose family oligosaccharide levels in soybean seeds. Front. Plant Sci. 11:612942. 10.3389/fpls.2020.612942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Park S.-Y., Zhang Z. J. (2013). “An overview of genetic transformation of soybean,” in A Comprehensive Survey of International Soybean Research - Genetics, Physiology, Agronomy and Nitrogen Relationships, ed. Board J. E. (London, UK: IntechOpen; ). [Google Scholar]
- Li C., Nguyen V., Liu J., Fu W., Chen C., Yu K., et al. (2019). Mutagenesis of seed storage protein genes in soybean using CRISPR/Cas9. BMC Res. Notes 12:176. 10.1186/s13104-019-4207-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Zhang H., Wang X., Liao H. (2014). A comparison study of Agrobacterium-mediated transformation methods for root-specific promoter analysis in soybean. Plant Cell Rep. 33 1921–1932. 10.1007/s00299-014-1669-5 [DOI] [PubMed] [Google Scholar]
- Li F., Ni H., Yan W., Xie Y., Liu X., Tan X., et al. (2021a). Overexpression of an aquaporin protein from Aspergillus glaucus confers salt tolerance in transgenic soybean. Transgenic Res. 30 727–737. 10.1007/s11248-021-00280-9 [DOI] [PubMed] [Google Scholar]
- Li M., Chen R., Jiang Q., Sun X., Zhang H., Hu Z. (2021b). GmNAC06, a NAC domain transcription factor enhances salt stress tolerance in soybean. Plant Mol. Biol. 105 333–345. 10.1007/s11103-020-01091-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Cheng Q., Gan Z., Hou Z., Zhang Y., Li Y., et al. (2021c). Multiplex CRISPR/Cas9-mediated knockout of soybean LNK2 advances flowering time. Crop J. 9 767–776. 10.1016/j.cj.2020.09.005 [DOI] [Google Scholar]
- Li J., Todd T. C., Oakley T. R., Lee J., Trick H. N. (2010). Host-derived suppression of nematode reproductive and fitness genes decreases fecundity of Heterodera glycines Ichinohe. Planta 232 775–785. 10.1007/s00425-010-1209-7 [DOI] [PubMed] [Google Scholar]
- Li L., Gao W., Peng Q., Zhou B., Kong Q., Ying Y., et al. (2018). Two soybean bHLH factors regulate response to iron deficiency. J. Integr. Plant Biol. 60 608–622. 10.1111/jipb.12651 [DOI] [PubMed] [Google Scholar]
- Li S., Cong Y., Liu Y., Wang T., Shuai Q., Chen N., et al. (2017). Optimization of Agrobacterium-mediated transformation in soybean. Front. Plant Sci. 8:246. 10.3389/fpls.2017.00246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Liu Z. B., Xing A., Moon B. P., Koellhoffer J. P., Huang L., et al. (2015). Cas9-guide RNA directed genome editing in soybean. Plant Physiol. 169 960–970. 10.1104/pp.15.00783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Sushuang D., Huidong X., Xingxing F., Ruidong S., Jinming Z., et al. (2022). A novel TIR-NBS-LRR gene regulates immune response to Phytophthora root rot in soybean. Crop J. 10.1016/j.cj.2022.03.003 [DOI] [Google Scholar]
- Liang Z., Chen K., Li T., Zhang Y., Wang Y., Zhao Q., et al. (2017). Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nat. Commun. 8:14261. 10.1038/ncomms14261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W., Odell J. T., Schreiner R. M. (1987). Soybean protoplast culture and direct gene uptake and expression by cultured soybean protoplasts. Plant Physiol. 84 856–861. 10.1104/pp.84.3.856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Zhang C., Fan M., Ma W., Chen M., Cai F., et al. (2018). GmIDL2a and GmIDL4a, encoding the inflorescence deficient in abscission-like protein, are involved in soybean cell wall degradation during lateral root emergence. Int. J. Mol. Sci. 19:2262. 10.3390/ijms19082262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Jiang B., Ma L., Zhang S., Zhai H., Xu X., et al. (2018). Functional diversification of Flowering Locus T homologs in soybean: GmFT1a and GmFT2a/5a have opposite roles in controlling flowering and maturation. New Phytol. 217 1335–1345. 10.1111/nph.14884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H. K., Yang C., Wei Z. M. (2004). Efficient Agrobacterium tumefaciens-mediated transformation of soybeans using an embryonic tip regeneration system. Planta 219 1042–1049. 10.1007/s00425-004-1310-x [DOI] [PubMed] [Google Scholar]
- Liu J., Su Q., An L., Yang A. (2009). Transfer of a minimal linear marker-free and vector-free smGFP cassette into soybean via ovary-drip transformation. Biotechnol. Lett. 31 295–303. 10.1007/s10529-008-9851-x [DOI] [PubMed] [Google Scholar]
- Liu L. F., Le G., Zhang L. X., Cai Y. P., Song W. W., Han T. F. (2022a). Co-silencing E1 and its homologs in an extremely late-maturing soybean cultivar confers super-early maturity and adaptation to high-latitude short-season regions. J. Integr. Agr. 21 326–335. 10.1016/S2095-3119(20)63391-3 [DOI] [Google Scholar]
- Liu S. Y., Liu J. F., Zhang Y. Z., Jiang Y. S., Hu S. W., Qu J., et al. (2022b). Cloning of the soybean sHSP26 gene and analysis of its drought resistance. Phyton 97 1465–1482. 10.32604/phyton.2022.018836 [DOI] [Google Scholar]
- Liu X., Pi B., Du Z., Yang T., Gu M., Sun S., et al. (2022c). The transcription factor GmbHLH3 confers Cl–/salt tolerance to soybean by upregulating GmCLC1 expression for maintenance of anion homeostasis. Environ. Exp. Bot. 194:104755. 10.1016/j.envexpbot.2021.104755 [DOI] [Google Scholar]
- Liu S., Wang D., Mei Y., Xia T., Xu W., Zhang Y., et al. (2020). Overexpression of GmAAP6a enhances tolerance to low nitrogen and improves seed nitrogen status by optimizing amino acid partitioning in soybean. Plant Biotechnol. 18 1749–1762. 10.1111/pbi.13338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S. J., Wei Z. M., Huang J. Q. (2008). The effect of co-cultivation and selection parameters on Agrobacterium-mediated transformation of Chinese soybean varieties. Plant Cell Rep. 27 489–498. 10.1007/s00299-007-0475-8 [DOI] [PubMed] [Google Scholar]
- Liu X., Liu F., Zhang L., Cheng C., Wei P., Yu B. (2021). GsCLC-c2 from wild soybean confers chloride/salt tolerance to transgenic Arabidopsis and soybean composite plants by regulating anion homeostasis. Physiol. Plantarum 172 1867–1879. 10.1111/ppl.13396 [DOI] [PubMed] [Google Scholar]
- Lowe K., La Rota M., Hoerster G., Hastings C., Wang N., Chamberlin M., et al. (2018). Rapid genotype “independent” Zea mays L. (maize) transformation via direct somatic embryogenesis. In Vitro Cell. Dev. Pl. 54 240–252. 10.1007/s11627-018-9905-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe K., Wu E., Wang N., Hoerster G., Hastings C., Cho M. J., et al. (2016). Morphogenic regulators Baby boom and Wuschel improve monocot transformation. Plant Cell 28 1998–2015. 10.1105/tpc.16.00124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L., Dong C., Liu R., Zhou B., Wang C., Shou H. (2018). Roles of soybean plasma membrane intrinsic protein GmPIP2;9 in drought tolerance and seed development. Front. Plant Sci. 9:530. 10.3389/fpls.2018.00530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L., Wei W., Li Q.-T., Bian X.-H., Lu X., Hu Y., et al. (2021). A transcriptional regulatory module controls lipid accumulation in soybean. New Phytol. 231 661–678. 10.1111/nph.17401 [DOI] [PubMed] [Google Scholar]
- Lu Q. S. M., Tian L. (2022). An efficient and specific CRISPR-Cas9 genome editing system targeting soybean phytoene desaturase genes. BMC Biotechnol. 22:7. 10.1186/s12896-022-00737-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan H., Liao W., Song Y., Niu H., Zhi H. (2020). Transgenic plant generated by RNAi-mediated knocking down of soybean Vma12 and soybean mosaic virus resistance evaluation. AMB Express 10 1–10. 10.1186/s13568-020-00997-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Na R., Nowak J. S., Qiu Y., Lu Q. S., Yang C., et al. (2021). Development of a Csy4-processed guide RNA delivery system with soybean-infecting virus ALSV for genome editing. BMC Plant Biol. 21:419. 10.1186/s12870-021-03138-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luth D., Warnberg K., Wang K. (2015). “Soybean [Glycine max (L.) Merr.],” in Agrobacterium Protocols. Methods in Molecular Biology, ed. Wang K. (New York, NY: Springer; ), 275–284. [DOI] [PubMed] [Google Scholar]
- Ma J., Sun S., Whelan J., Shou H. (2021). CRISPR/Cas9-mediated knockout of GmFATB1 significantly reduced the amount of saturated fatty acids in soybean seeds. Int. J. Mol. Sci. 22:3877. 10.3390/ijms22083877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Xu W., Liu T., Chen R., Zhu H., Zhang H., et al. (2021). Functional characterization of soybean (Glycine max) DIRIGENT genes reveals an important role of GmDIR27 in the regulation of pod dehiscence. Genomics 113 979–990. 10.1016/j.ygeno.2020.10.033 [DOI] [PubMed] [Google Scholar]
- Ma X. J., Yu T. F., Li X. H., Cao X. Y., Ma J., Chen J., et al. (2020). Overexpression of GmNFYA5 confers drought tolerance to transgenic Arabidopsis and soybean plants. BMC Plant Biol. 20:123. 10.1186/s12870-020-02337-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangena P. (2019). A simplified in-planta genetic transformation in soybean. Res. J. Biotech. 14 117–125. [Google Scholar]
- Mangena P., Mokwala P., Nikolova R. V. (2017). “Challenges of in vitro and in vivo Agrobacterium-mediated genetic transformation in soybean,” in Soybean - The Basis of Yield, Biomass and Productivity, ed. Kasai M. (London: IntechOpen; ), 75–94. [Google Scholar]
- Mariashibu T. S., Subramanyam K., Arun M., Mayavan S., Rajesh M., Theboral J., et al. (2013). Vacuum infiltration enhances the Agrobacterium-mediated genetic transformation in Indian soybean cultivars. Acta Physiol. Plant 35 41–54. 10.1007/s11738-012-1046-3 [DOI] [PubMed] [Google Scholar]
- Marinho J. P., Pagliarini R. F., Molinari M. D. C., Marcolino-Gomes J., Caranhoto A. L. H., Marin S. R. R., et al. (2022). Overexpression of full-length and partial DREB2A enhances soybean drought tolerance. Agronomy Sci. Biotech. 8 1–21. 10.33158/ASB.r141.v8.2022 [DOI] [Google Scholar]
- Martinell B. J., Julson L. A., Hinchee M. A. W., Connor-Ward D., McCabe D., Emler C. (1999). Efficiency Soybean Transformation Protocol. Patent Version No.: US5914451. St. Louis, Mo: U.S. Patent and Trademark Office, Monsanto Company. [Google Scholar]
- Martinell B. J., Julson L. S., Emler C. A., Huang Y., Mccabe D. E., Williams E. J. (2002). Soybean Agrobacterium Transformation Method. Patent Version No.: US6384301. St. Louis, MO: U.S. Patent and Trademark Office, Monsanto Company. [Google Scholar]
- Mccabe D. E., Swain W. F., Martinell B. J., Christou P. (1988). Stable transformation of soybean (Glycine Max) by particle acceleration. Nat. Biotech. 6 923–926. 10.1038/nbt0888-923 [DOI] [Google Scholar]
- Mehrizadeh V., Dorani E., Mohammadi S. A., Ghareyazie B. (2021). Expression of recombinant human IFN-γ protein in soybean (Glycine max L.). Plant Cell Tiss. Org. 146 127–136. 10.1007/s11240-021-02052-z [DOI] [Google Scholar]
- Meurer C. A., Dinkins R. D., Collins G. B. (1998). Factors affecting soybean cotyledonary node transformation. Plant Cell Rep. 18 180–186. 10.1007/s002990050553 [DOI] [PubMed] [Google Scholar]
- Michno J. M., Wang X., Liu J., Curtin S. J., Kono T. J., Stupar R. M. (2015). CRISPR/Cas mutagenesis of soybean and Medicago truncatula using a new web-tool and a modified Cas9 enzyme. GM Crops Food 6 243–252. 10.1080/21645698.2015.1106063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R., Herr E. H., Orvar B. L., van Camp W., Willekens H., Inzé D., et al. (1999). Transgenic tobacco plants with reduced capability to detoxify reactive oxygen intermediates are hyperresponsive to pathogen infection. Proc. Natl. Acad. Sci. U. S. A. 96 14165–14170. 10.1073/pnas.96.24.14165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moghaieb R. E. A., Khashaba E. H., Abdel Elazim A. M. (2019). The toxicity of Cry1Ia5 transgenic soybean plants against Spodoptera littoralis. J. Plant Prot. Res. 59 185–191. 10.24425/jppr.2019.129286 [DOI] [Google Scholar]
- Molinari M. D. C., Fuganti-Pagliarini R., Marin S. R. R., Ferreira L. C., Barbosa D. A., Marcolino-Gomes J., et al. (2020). Overexpression of AtNCED3 gene improved drought tolerance in soybean in greenhouse and field conditions. Genet. Mol. Biol. 43:e20190292. 10.1590/1678-4685-gmb-2019-0292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgante M., Jurman I., Shi L., Zhu T., Keim P., Rafalski J. A. (1997). The STR120 satellite DNA of soybean: organization, evolution and chromosomal specificity. Chromosome Res. 5 363–373. 10.1023/a:1018492208247 [DOI] [PubMed] [Google Scholar]
- Mu R., Lyu X., Ji R., Liu J., Zhao T., Li H., et al. (2022). GmBICs modulate low blue light-induced stem elongation in soybean. Front. Plant Sci. 13:803122. 10.3389/fpls.2022.803122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T., Skoog F. (1962). A Revised Medium for Rapid Growth and Bio Assays with Tobacco Tissue Cultures. Physiol. Plant. 15 473–497. 10.1111/j.1399-3054.1962.tb08052.x [DOI] [Google Scholar]
- Nadeem M., Chen A., Hong H., Li D., Li J., Zhao D., et al. (2021). GmMs1 encodes a kinesin-like protein essential for male fertility in soybean (Glycine max L.). J. Integr. Plant Biol. 63 1054–1064. 10.1111/jipb.13110 [DOI] [PubMed] [Google Scholar]
- Nagy E. D., Stevens J. L., Yu N., Hubmeier C. S., LaFaver N., Gillespie M., et al. (2021). Novel disease resistance gene paralogs created by CRISPR/Cas9 in soy. Plant Cell Rep. 40 1047–1058. 10.1007/s00299-021-02678-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandula V. K. (2019). Herbicide resistance traits in maize and soybean: current status and future outlook. Plants 8:377. 10.3390/plants8090337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng M.-S., Ku Y.-S., Yung W.-S., Cheng S.-S., Man C.-K., Yang L., et al. (2021). MATE-type proteins are responsible for isoflavone transportation and accumulation in soybean seeds. Int. J. Mol. Sci. 22:12017. 10.3390/ijms222112017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngaki M. N., Sahoo D. K., Wang B., Bhattacharyya M. K. (2021). Overexpression of a plasma membrane protein generated broad-spectrum immunity in soybean. Plant Biotechnol. J. 19 502–516. 10.1111/pbi.13479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen C., Paddock K., Zhang Z., Stacey M. (2021). GmKIX8-1 regulates organ size in soybean and is the causative gene for the major seed weight QTL qSw17-1. New Phytol. 229 920–934. 10.1111/nph.16928 [DOI] [PubMed] [Google Scholar]
- Nguyen H. Q., Le T. H. T., Nguyen T. N. L., Nguyen T. G., Sy D. T., Tu Q. T., et al. (2020). Overexpressing GmCHI1A increases the isoflavone content of transgenic soybean (Glycine max (L.) Merr.) seeds. In Vitro Cell. Dev. Pl. 56 842–850. 10.1007/s11627-020-10076-x [DOI] [Google Scholar]
- Nguyen N. T., Vu H. T., Nguyen T. T., Nguyen L.-A. T., Nguyen M.-C. D., Hoang K. L., et al. (2019). Co-expression of Arabidopsis AtAVP1 and AtNHX1 to improve salt tolerance in soybean. Crop Sci. 59 1133–1143. 10.2135/cropsci2018.10.0640 [DOI] [Google Scholar]
- Nguyen Q. H., Vu L. T. K., Nguyen L. T. N., Pham N. T. T., Nguyen Y. T. H., Le S. V., et al. (2019). Overexpression of the GmDREB6 gene enhances proline accumulation and salt tolerance in genetically modified soybean plants. Sci. Rep. 9:19663. 10.1038/s41598-019-55895-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas J. J., Richard-Forget F. C., Goupy P. M., Amiot M. J., Aubert S. Y. (1994). Enzymatic browning reactions in apple and apple products. Criti. Rev. Food Sci. 34 109–157. 10.1080/10408399409527653 [DOI] [PubMed] [Google Scholar]
- Niu L., Yang J., Zhang J., He H., Xing G., Zhao Q., et al. (2019). Introduction of the harpinXooc-encoding gene hrf2 in soybean enhances resistance against the oomycete pathogen Phytophthora sojae. Transgenic Res. 28 257–266. 10.1007/s11248-019-00119-4 [DOI] [PubMed] [Google Scholar]
- O’Conner S., Zheng W., Qi M., Kandel Y., Fuller R., Whitham S. A., et al. (2021). GmNF-YC4-2 increases protein, exhibits broad disease resistance and expedites maturity in soybean. Int. J. Mol. Sci. 22:3586. 10.3390/ijms22073586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olhoft P. M., Bernal L. M., Grist L. B., Hill D. S., Mankin S. L., Shen Y., et al. (2007). A novel Agrobacterium rhizogenes-mediated transformation method of soybean [Glycine max (L.) Merrill] using primary-node explants from seedlings. In Vitro Cell. Dev. Pl. 43 536–549. 10.1007/s11627-007-9050-9 [DOI] [Google Scholar]
- Olhoft P. M., Flagel L. E., Donovan C. M., Somers D. A. (2003). Efficient soybean transformation using hygromycin B selection in the cotyledonary-node method. Planta 216 723–735. 10.1007/s00425-002-0922-2 [DOI] [PubMed] [Google Scholar]
- Olhoft P. M., Flagel L. E., Somers D. A. (2004). T-DNA locus structure in a large population of soybean plants transformed using the Agrobacterium-mediated cotyledonary-node method. Plant Biotechnol. J. 2 289–300. 10.1111/j.1467-7652.2004.00070.x [DOI] [PubMed] [Google Scholar]
- Olhoft P. O., Lin K. L., Galbraith J. G., Nielsen N. N., Somers D. S. (2001). The role of thiol compounds in increasing Agrobacterium-mediated transformation of soybean cotyledonary-node cells. Plant Cell Rep. 20 731–737. 10.1007/s002990100388 [DOI] [Google Scholar]
- Olhoft P. O., Somers D. S. (2001). L-Cysteine increases Agrobacterium-mediated T-DNA delivery into soybean cotyledonary-node cells. Plant Cell Rep. 20 706–711. 10.1007/s002990100379 [DOI] [Google Scholar]
- Opabode J. T. (2006). Agrobacterium-mediated transformation of plants: emerging factors that influence efficiency. Biotech. Mol. Bio. Rev. 1 12–20. [Google Scholar]
- Paes de Melo B., Lourenço-Tessutti I., Morgante C., Santos N., Pinheiro L., Lins C., et al. (2020). Soybean embryonic axis transformation: combining biolistic and Agrobacterium-mediated protocols to overcome typical complications of in vitro plant regeneration. Front. Plant Sci. 11:1228. 10.3389/fpls.2020.01228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano M. C., Miransari M. (2016). “The importance of soybean production worldwide,” in Abiotic and Biotic Stresses in Soybean Production, ed. Miransari M. (San Diego: Academic Press; ), 1–26. [Google Scholar]
- Pareddy D., Chennareddy S., Anthony G., Sardesai N., Mall T., Minnicks T., et al. (2020). Improved soybean transformation for efficient and high throughput transgenic production. Transgenic Res. 29 267–281. 10.1007/s11248-020-00198-8 [DOI] [PubMed] [Google Scholar]
- Park J. S., Kim H. J., Cho H. S., Jung H. W., Cha J. Y., Yun D. J., et al. (2019). Overexpression of AtYUCCA6 in soybean crop results in reduced ROS production and increased drought tolerance. Plant Biotechnol. Rep. 13 161–168. 10.1007/s11816-019-00527-2 [DOI] [Google Scholar]
- Parrott W. A., Williams E. G., Hildebrand D. F., Collins G. B. (1989). Effect of genotype on somatic embryogenesis from immature cotyledons of soybean. Plant Cell Tiss. Org. 16 15–21. 10.1007/BF00044068 [DOI] [Google Scholar]
- Patil G. B., Stupar R. M., Zhang F. (2022). “Protoplast Isolation, transfection, and gene editing for soybean (Glycine max),” in Proc. Natl. Acad. Sci, eds Wang K., Zhang F. (New York, NY: Springer US; ), 173–186. 10.1007/978-1-0716-2164-6_13 [DOI] [PubMed] [Google Scholar]
- Paz M. M., Martinez J. C., Kalvig A. B., Fonger T. M., Wang K. (2006). Improved cotyledonary node method using an alternative explant derived from mature seed for efficient Agrobacterium-mediated soybean transformation. Plant Cell Rep. 25 206–213. 10.1007/s00299-005-0048-7 [DOI] [PubMed] [Google Scholar]
- Paz M. M., Shou H., Guo Z., Zhang Z., Banerjee A. K., Wang K. (2004). Assessment of conditions affecting Agrobacterium-mediated soybean transformation using the cotyledonary node explant. Euphytica 136 167–179. 10.1023/B:EUPH.0000030669.75809.dc [DOI] [Google Scholar]
- Pham T. T. N., Nguyen H. Q., Nguyen T. N. L., Dao X. T., Sy D. T., Le V. S., et al. (2020). Overexpression of the GmDREB2 gene increases proline accumulation and tolerance to drought stress in soybean plant. Australian J. Crop Sci. 14 495–503. 10.21475/ajcs.20.14.03.p2173 [DOI] [Google Scholar]
- Pottinger S. E., Bak A., Margets A., Helm M., Tang L., Casteel C., et al. (2020). Optimizing the PBS1 decoy system to confer resistance to potyvirus infection in Arabidopsis and soybean. Mol. Plant Microbe Interact. 33 932–944. 10.1094/MPMI-07-19-0190-R [DOI] [PubMed] [Google Scholar]
- Punjabi M., Bharadvaja N., Jolly M., Dahuja A., Sachdev A. (2018). Development and evaluation of low phytic acid soybean by siRNA triggered seed specific silencing of inositol polyphosphate 6-/3-/5-kinase gene. Front. Plant Sci. 9:804. 10.3389/fpls.2018.00804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin D., Liu X. Y., Miceli C., Zhang Q., Wang P. W. (2019). Soybean plants expressing the Bacillus thuringiensis cry8-like gene show resistance to Holotrichia parallela. BMC Biotechnol. 19:66. 10.1186/s12896-019-0563-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu S., Jiao Y., Abraham L., Wang P. (2021). Correlation analysis of new soybean [Glycine max (L.) Merr] gene Gm15G117700 with oleic acid. Phyton 90 1177–1192. 10.32604/phyton.2021.015206 [DOI] [Google Scholar]
- Queiroz L. N., Maldaner F. R., Mendes E. A., Sousa A. R., D’Allastta R. C., Mendonca G., et al. (2019). Evaluation of lettuce chloroplast and soybean cotyledon as platforms for production of functional bone morphogenetic protein 2. Transgenic Res. 28 213–224. 10.1007/s11248-019-00116-7 [DOI] [PubMed] [Google Scholar]
- Rakesh B., Sudheer W. N., Nagella P. (2021). Role of polyamines in plant tissue culture: an overview. Plant Cell Tiss. Org. 145 487–506. 10.1007/s11240-021-02029-y [DOI] [Google Scholar]
- Ramesh S. V., Shivakumar M., Praveen S., Chouhan B. S., Chand S. (2019). Expression of short hairpin RNA (shRNA) targeting AC2 gene of Mungbean yellow mosaic India virus (MYMIV) reduces the viral titre in soybean. 3 Biotech 9:334. 10.1007/s13205-019-1865-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran Y., Liang Z., Gao C. (2017). Current and future editing reagent delivery systems for plant genome editing. Sci. China Life Sci. 60 490–495. 10.1007/s11427-017-9022-1 [DOI] [PubMed] [Google Scholar]
- Raza G., Singh M., Bhalla P. (2020). Somatic embryogenesis and plant regeneration from commercial soybean cultivars. Plants 9:38. 10.3390/plants9010038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rech E. L., Vianna G. R., Aragao F. J. (2008). High-efficiency transformation by biolistics of soybean, common bean and cotton transgenic plants. Nat. Protoc. 3 410–418. 10.1038/nprot.2008.9 [DOI] [PubMed] [Google Scholar]
- Rehman N. U., Abbas F., Imran M., Alam I., Imran M., Ullah I., et al. (2022). Genome wide analysis of DWARF27 genes in soybean and functional characterization of GmD27c reveals eminent role of strigolactones in rhizobia interaction and nodulation in Glycine max. Mol. Bio. Rep. [Epub ahead of print]. 10.1007/s11033-022-07127-4 [DOI] [PubMed] [Google Scholar]
- Ribichich K. F., Chiozza M., Avalos-Britez S., Cabello J. V., Arce A. L., Watson G., et al. (2020). Successful field performance in warm and dry environments of soybean expressing the sunflower transcription factor HB4. J. Exp. Bot. 71 3142–3156. 10.1093/jxb/eraa064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roustan J. P., Latche A., Fallot J. (1990). Inhibition of ethylene production and stimulation of carrot somatic embryogenesis by salicylic acid. Biol. Plant. 32 273–276. 10.1007/BF02886947 [DOI] [Google Scholar]
- Sander J. D., Dahlborg E. J., Goodwin M. J., Cade L., Zhang F., Cifuentes D., et al. (2011). Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA). Nat. Methods 8 67–69. 10.1038/nmeth.1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato H., Yamada T., Kita Y., Ishimoto M., Kitamura K. (2007). Production of transgenic plants and their early seed set in Japanese soybean variety, Kariyutaka. Plant Biotechnol. 24 533–536. 10.5511/plantbiotechnology.24.533 [DOI] [Google Scholar]
- Sato S., Newell C., Kolacz K., Tredo L., Finer J., Hinchee M. (1993). Stable transformation via particle bombardment in two different soybean regeneration systems. Plant Cell Rep. 12 408–413. 10.1007/BF00234702 [DOI] [PubMed] [Google Scholar]
- Schmutz J., Cannon S. B., Schlueter J., Ma J., Mitros T., Nelson W., et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463 178–183. 10.1038/nature08670 [DOI] [PubMed] [Google Scholar]
- Shanmugam S., Zhao S., Nandy S., Srivastava V., Khodakovskaya M. (2020). Modification of soybean growth and abiotic stress tolerance by expression of truncated ERECTA protein from Arabidopsis thaliana. PLoS One 15:e0233383. 10.1371/journal.pone.0233383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Liu J., Geng H., Zhang J., Liu Y., Zhang H., et al. (2018). De novo assembly of a Chinese soybean genome. Sci. China Life Sci. 61 871–884. 10.1007/s11427-018-9360-0 [DOI] [PubMed] [Google Scholar]
- Shi W. Y., Du Y. T., Ma J., Min D. H., Jin L. G., Chen J., et al. (2018). The WRKY transcription factor GmWRKY12 confers drought and salt tolerance in soybean. Int. J. Mol. Sci. 19:4087. 10.3390/ijms19124087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura M., Kanamori H., Komatsu S., Namiki N., Mukai Y., Kurita K., et al. (2015). The Glycine max cv. enrei genome for improvement of Japanese soybean cultivars. Int. J. Genomics 2015: 358127. 10.1155/2015/358127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Toth K., Montes-Luz B., Stacey G. (2021). Soybean hairy root transformation: a rapid and highly efficient method. Curr. Protoc. 1:e195. 10.1002/cpz1.195 [DOI] [PubMed] [Google Scholar]
- Song L., Valliyodan B., Prince S., Wan J., Nguyen H. T. (2018). Characterization of the XTH gene family: new insight to the roles in soybean flooding tolerance. Int. J. Mol. Sci. 19:2705. 10.3390/ijms19092705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z. Y., Tian J.l, Fu W. Z., Li L., Lu L. H., Zhou L., et al. (2013). Screening Chinese soybean genotypes for Agrobacterium-mediated genetic transformation suitability. J. Zhejiang Univ. Sci. B 14 289–298. 10.1631/jzus.B1200278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto N., Delgado C., Hernández Y., Rosabal Y., Ferreira A., Pujol M., et al. (2017). Efficient particle bombardment-mediated transformation of Cuban soybean (INCASoy-36) using glyphosate as a selective agent. Plant Cell Tiss. Org. 128 187–196. 10.1007/s11240-016-1099-x [DOI] [Google Scholar]
- Soto N., Hernandez Y., Delgado C., Rosabal Y., Ortiz R., Valencia L., et al. (2020). Field resistance to Phakopsora pachyrhizi and Colletotrichum truncatum of transgenic soybean expressing the NmDef02 plant defensin gene. Front. Plant Sci. 11:562. 10.3389/fpls.2020.00562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey G., Vodkin L., Parrott W. A., Shoemaker R. C. (2004). National science foundation-sponsored workshop report. draft plan for soybean genomics. Plant Physiol. 135 59–70. 10.1104/pp.103.037903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian S., Hu X., Lu G., Odelland J. T., Yu O. (2004). The promoters of two isoflavone synthase genes respond differentially to nodulation and defense signals in transgenic soybean roots. Plant Mol. Biol. 54 623–639. 10.1023/B:PLAN.0000040814.28507.35 [DOI] [PubMed] [Google Scholar]
- Sugano S., Hirose A., Kanazashi Y., Adachi K., Hibara M., Itoh T., et al. (2020). Simultaneous induction of mutant alleles of two allergenic genes in soybean by using site-directed mutagenesis. BMC Plant Biol. 20:513. 10.1186/s12870-020-02708-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultana M. S., Frazier T. P., Millwood R. J., Lenaghan S. C., Stewart C. N. (2019). Development and validation of a novel and robust cell culture system in soybean (Glycine max (L.) Merr.) for promoter screening. Plant Cell Rep. 38 1329–1345. 10.1007/s00299-019-02455-5 [DOI] [PubMed] [Google Scholar]
- Sun T., Ma N., Wang C., Fan H., Wang M., Zhang J., et al. (2021). A golgi-localized sodium/hydrogen exchanger positively regulates salt tolerance by maintaining higher K+/Na+ ratio in soybean. Front. Plant Sci. 12:638340. 10.3389/fpls.2021.638340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Cai X., Yin K., Gu L., Shen Y., Hu B., et al. (2021). Wild soybean SNARE proteins BET1s mediate the subcellular localization of the cytoplasmic receptor-like kinases CRCK1s to modulate salt stress responses. Plant J. 105 771–785. 10.1111/tpj.15072 [DOI] [PubMed] [Google Scholar]
- Sun X., Hu Z., Chen R., Jiang Q., Song G., Zhang H., et al. (2015). Targeted mutagenesis in soybean using the CRISPR-Cas9 system. Sci. Rep. 5:10342. 10.1038/srep10342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z., Su C., Yun J., Jiang Q., Wang L., Wang Y., et al. (2019). Genetic improvement of the shoot architecture and yield in soya bean plants via the manipulation of GmmiR156b. Plant Biotechnol. J. 17 50–62. 10.1111/pbi.12946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testroet A., Lee K., Luth D., Wang K. (2017). Comparison of transformation frequency using the bar gene regulated by the CaMV 35S or NOS promoter in Agrobacterium-mediated soybean (Glycine max L.) transformation. In Vitro Cell. Dev. Pl. 53 188–199. 10.1007/s11627-017-9810-0 [DOI] [Google Scholar]
- Tian B., Li J., Vodkin L. O., Todd T. C., Finer J. J., Trick H. N. (2019). Host-derived gene silencing of parasite fitness genes improves resistance to soybean cyst nematodes in stable transgenic soybean. Theor. Appl. Genet. 132 2651–2662. 10.1007/s00122-019-03379-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torkamaneh D., Laroche J., Valliyodan B., O’Donoughue L., Cober E., Rajcan I., et al. (2019). Soybean haplotype map (GmHapMap): a universal resource for soybean translational and functional genomics. BioRxiv [preprint] 10.1101/534578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trick H. N., Finer J. J. (1998). Sonication-assisted Agrobacterium-mediated transformation of soybean [Glycine max (L.) Merrill] embryogenic suspension culture tissue. Plant Cell Rep. 17 482–488. 10.1007/s002990050429 [DOI] [PubMed] [Google Scholar]
- Umezawa T., Sakurai T., Totoki Y., Toyoda A., Seki M., Ishiwata A., et al. (2008). Sequencing and analysis of approximately 40,000 soybean cDNA clones from a full-length-enriched cDNA library. DNA Res. 15 333–346. 10.1093/dnares/dsn024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veerappa R., Slocum R. D., Siegenthaler A., Wang J., Clark G., Roux S. J. (2019). Ectopic expression of a pea apyrase enhances root system architecture and drought survival in Arabidopsis and soybean. Plant Cell Environ. 42 337–353. 10.1111/pce.13425 [DOI] [PubMed] [Google Scholar]
- Verma N., Tiwari S., Singh V. P., Prasad S. M. (2020). Nitric oxide in plants: an ancient molecule with new tasks. Plant Growth Regul. 90 1–13. 10.1007/s10725-019-00543-w [DOI] [Google Scholar]
- Vianna G. R., Aragao F. J., Rech E. L. (2011). A minimal DNA cassette as a vector for genetic transformation of soybean (Glycine max). Genet. Mol. Biol. 10 382–390. 10.4238/vol10-1gmr1058 [DOI] [PubMed] [Google Scholar]
- Voora V., Larrea C., Bermudez S. (2020). “Global market report: soybeans,” in International Institute for Sustainable Development, ed. Baliño S. (Winnipeg, Manitoba: International Institute for Sustainable Development; ). [Google Scholar]
- Wang B., Sumit R., Sahu B. B., Ngaki M. N., Srivastava S. K., Yang Y., et al. (2018a). Arabidopsis novel glycine-rich plasma membrane PSS1 protein enhances disease resistance in transgenic soybean plants. Plant Physiol. 176 865–878. 10.1104/pp.16.01982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. S., Chen Q. S., Xin D. W., Qi Z. M., Zhang C., Li S. N., et al. (2018b). Overexpression of GmBIN2, a soybean glycogen synthase kinase 3 gene, enhances tolerance to salt and drought in transgenic Arabidopsis and soybean hairy roots. J. Integr. Agr. 17 1959–1971. 10.1016/S2095-3119(17)61863-X [DOI] [Google Scholar]
- Wang Y., Jiang L., Chen J., Tao L., An Y., Cai H., et al. (2018c). Overexpression of the alfalfa WRKY11 gene enhances salt tolerance in soybean. PLoS One 13:e0192382. 10.1371/journal.pone.0192382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Xu Y. (2008). Hypocotyl-based Agrobacterium-mediated transformation of soybean (Glycine max) and application for RNA interference. Plant Cell Rep. 27 1177–1184. 10.1007/s00299-008-0535-8 [DOI] [PubMed] [Google Scholar]
- Wang Y., Yang Z., Kong Y., Li X., Li W., Du H., et al. (2020d). GmPAP12 is required for nodule development and nitrogen fixation under phosphorus starvation in soybean. Front. Plant Sci. 11:450. 10.3389/fpls.2020.00450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Liu S., Wang J., Yokosho K., Zhou B., Yu Y.-C., et al. (2020c). Simultaneous changes in seed size, oil content, and protein content driven by selection of SWEET homologues during soybean domestication. Nat. Sci. Rev. 7 1776–1786. 10.1093/nsr/nwaa110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Sun S., Wu T., Liu L., Sun X., Cai Y., et al. (2020b). Natural variation and CRISPR/Cas9-mediated mutation in GmPRR37 affect photoperiodic flowering and contribute to regional adaptation of soybean. Plant Biotechnol. J. 18 1869–1881. 10.1111/pbi.13346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Kuang H., Zhang Z., Yang Y., Yan L., Zhang M., et al. (2020a). Generation of seed lipoxygenase-free soybean using CRISPR-Cas9. Crop J. 8 432–439. 10.1016/j.cj.2019.08.008 [DOI] [Google Scholar]
- Wang Y., Yuan L., Su T., Wang Q., Gao Y., Zhang S., et al. (2020e). Light- and temperature-entrainable circadian clock in soybean development. Plant Cell Environ. 43 637–648. 10.1111/pce.13678 [DOI] [PubMed] [Google Scholar]
- Wang X., Li M.-W., Wong F.-L., Luk C.-Y., Chung C. Y.-L., Yung W.-S., et al. (2021d). Increased copy number of gibberellin 2-oxidase 8 genes reduced trailing growth and shoot length during soybean domestication. Plant J. 107 1739–1755. 10.1111/tpj.15414 [DOI] [PubMed] [Google Scholar]
- Wang R., Deng M., Yang C., Yu Q., Zhang L., Zhu Q., et al. (2021b). A Qa-SNARE complex contributes to soybean cyst nematode resistance via regulation of mitochondria-mediated cell death. J. Exp. Bot. 72 7145–7162. 10.1093/jxb/erab301 [DOI] [PubMed] [Google Scholar]
- Wang W., Chen L., Fengler K., Bolar J., Llaca V., Wang X., et al. (2021c). A giant NLR gene confers broad-spectrum resistance to Phytophthora sojae in soybean. Nat. Commun. 12:6263. 10.1038/s41467-021-26554-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Zhao J., Fang Q., Chang X., Sun M., Li W., et al. (2021e). GmAKT1 is involved in K+ uptake and Na+/K+ homeostasis in Arabidopsis and soybean plants. Plant Sci. 304 110736. 10.1016/j.plantsci.2020.110736 [DOI] [PubMed] [Google Scholar]
- Wang K., Bu T., Cheng Q., Dong L., Su T., Chen Z., et al. (2021a). Two homologous LHY pairs negatively control soybean drought tolerance by repressing the abscisic acid responses. New Phytol. 229 2660–2675. 10.1111/nph.17019 [DOI] [PubMed] [Google Scholar]
- Wang Y., Yang W., Zuo Y., Zhu L., Hastwell A. H., Chen L., et al. (2019c). GmYUC2a mediates auxin biosynthesis during root development and nodulation in soybean. J. Exp. Bot. 70 3165–3176. 10.1093/jxb/erz144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Wang H., He S., Meng F., Zhang C., Fan S., et al. (2019a). GmSnRK1.1, a sucrose non-fermenting-1(SNF1)-related protein kinase, promotes soybean resistance to Phytophthora sojae. Front. Plant Sci. 10:996. 10.3389/fpls.2019.00996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Yokosho K., Guo R., Whelan J., Ruan Y. L., Ma J. F., et al. (2019b). The soybean sugar transporter GmSWEET15 mediates sucrose export from endosperm to early embryo. Plant Physiol. 180 2133–2141. 10.1104/pp.19.00641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Zhao F.-J. (2019). Engineering crops without genome integration using nanotechnology. Trends Plant Sci. 24 574–577. 10.1016/j.tplants.2019.05.004 [DOI] [PubMed] [Google Scholar]
- Wang W., Wang Z., Hou W., Chen L., Jiang B., Ma W., et al. (2022). GmNMHC5 may promote nodulation via interaction with GmGAI in soybean. Crop J. 10 273–279. 10.1016/j.cj.2021.03.019 [DOI] [Google Scholar]
- Wang Z., Wang Y., Shang P., Yang C., Yang M., Huang J., et al. (2022). Overexpression of soybean GmWRI1a stably increases the seed oil content in soybean. Int. J. Mol. Sci. 23:5084. 10.3390/ijms23095084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei P., Che B., Shen L., Cui Y., Wu S., Cheng C., et al. (2019). Identification and functional characterization of the chloride channel gene, GsCLC-c2 from wild soybean. BMC Plant Biol. 19:121. 10.1186/s12870-019-1732-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W., Liang D. W., Bian X. H., Shen M., Xiao J. H., Zhang W. K., et al. (2019). GmWRKY54 improves drought tolerance through activating genes in abscisic acid and Ca2+ signaling pathways in transgenic soybean. Plant J. 100 384–398. 10.1111/tpj.14449 [DOI] [PubMed] [Google Scholar]
- Wu F., Hanzawa Y. (2018). A simple method for isolation of soybean protoplasts and application to transient gene expression analyses. J. Vis. Exp. 131:e57258. 10.3791/57258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N., Lu Q., Wang P., Zhang Q., Zhang J., Qu J., et al. (2020). Construction and analysis of GmFAD2-1A and GmFAD2-2A soybean fatty acid desaturase mutants based on CRISPR/Cas9 technology. Int. J. Mol. Sci. 21:1104. 10.3390/ijms21031104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xian P., Cai Z., Cheng Y., Lin R., Lian T., Ma Q., et al. (2020). Wild soybean oxalyl-CoA synthetase degrades oxalate and affects the tolerance to cadmium and aluminum stresses. Int. J. Mol. Sci. 21:8869. 10.3390/ijms21228869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao P. Y., Liu Y., Cao Y. P. (2019). Overexpression of G10-EPSPS in soybean provides high glyphosate tolerance. J. Integr. Agr. 18 1851–1858. 10.1016/S2095-3119(18)62124-0 [DOI] [Google Scholar]
- Xie M., Chung C. Y., Li M. W., Wong F. L., Wang X., Liu A., et al. (2019). A reference-grade wild soybean genome. Nat. Commun. 10 1–12. 10.1038/s41467-019-09142-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing X., Du H., Yang Z., Li X., Kong Y., Li W., et al. (2022). GmSPX8, a nodule-localized regulator confers nodule development and nitrogen fixation under phosphorus starvation in soybean. BMC Plant Biol. 22:161. 10.1186/s12870-022-03556-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L., Li C., Li H., Lyu X., Zhao T., Liu J., et al. (2019). A transient expression system in soybean mesophyll protoplasts reveals the formation of cytoplasmic GmCRY1 photobody-like structures. Sci. China Life Sci. 62 1070–1077. 10.1007/s11427-018-9496-5 [DOI] [PubMed] [Google Scholar]
- Xu H., Zhang L., Zhang K., Ran Y. (2020). Progresses, challenges, and prospects of genome editing in soybean (Glycine max). Front. Plant Sci. 11:571138. 10.3389/fpls.2020.571138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M., Li H., Liu Z.-N., Wang X.-H., Xu P., Dai S.-J., et al. (2021). The soybean CBL-interacting protein kinase, GmCIPK2, positively regulates drought tolerance and ABA signaling. Plant Physiol. Biochem. 167 980–989. 10.1016/j.plaphy.2021.09.026 [DOI] [PubMed] [Google Scholar]
- Xu Y., Yan F., Zong Y., Li J., Gao H., Liu Y., et al. (2022). Proteomic and lipidomics analyses of high fatty acid AhDGAT3 transgenic soybean reveals the key lipase gene associated with the lipid internal mechanism. Genome 65 153–164. 10.1139/gen-2021-0043 [DOI] [PubMed] [Google Scholar]
- Xue R. G., Xie H. F., Zhang B. (2006). A multi-needle-assisted transformation of soybean cotyledonary node cells. Biotechnol. Lett. 28 1551–1557. 10.1007/s10529-006-9123-6 [DOI] [PubMed] [Google Scholar]
- Xun H., Yang X., He H., Wang M., Guo P., Wang Y., et al. (2019). Over-expression of GmKR3, a TIR-NBS-LRR type R gene, confers resistance to multiple viruses in soybean. Plant Mol. Biol. 99 95–111. 10.1007/s11103-018-0804-z [DOI] [PubMed] [Google Scholar]
- Yamada T., Takagi K., Ishimoto M. (2012). Recent advances in soybean transformation and their application to molecular breeding and genomic analysis. Breed. Sci. 61 480–494. 10.1270/jsbbs.61.480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T., Watanabe S., Arai M., Harada K., Kitamura K. (2010). Cotyledonary node pre-wounding with a micro-brush increased frequency of Agrobacterium-mediated transformation in soybean. Plant Biotechnol. 27 217–220. 10.5511/plantbiotechnology.27.217 [DOI] [Google Scholar]
- Yan B., Srinivasa Reddy M. S., Collins G. B., Dinkins R. D. (2000). Agrobacterium tumefaciens- mediated transformation of soybean [Glycine max (L.) Merrill.] using immature zygotic cotyledon explants. Plant Cell Rep. 19 1090–1097. 10.1007/s002990000236 [DOI] [PubMed] [Google Scholar]
- Yan Q., Si J., Cui X., Peng H., Dou D. (2019). The soybean cinnamate 4-hydroxylase gene GmC4H1 contributes positively to plant defense via increasing lignin content. Plant Growth Regul. 88 139–149. 10.1007/s10725-019-00494-2 [DOI] [Google Scholar]
- Yang S., Feng Y., Zhao Y., Bai J., Wang J. (2020b). Overexpression of a Eutrema salsugineum phosphate transporter gene EsPHT1;4 enhances tolerance to low phosphorus stress in soybean. Biotechnol. Lett. 42 2425–2439. 10.1007/s10529-020-02968-0 [DOI] [PubMed] [Google Scholar]
- Yang X., Yang J., Li H., Niu L., Xing G., Zhang Y., et al. (2020c). Overexpression of the chitinase gene CmCH1 from Coniothyrium minitans renders enhanced resistance to Sclerotinia sclerotiorum in soybean. Transgenic Res. 29 187–198. 10.1007/s11248-020-00190-2 [DOI] [PubMed] [Google Scholar]
- Yang Y., Yu T.-F., Ma J., Chen J., Zhou Y.-B., Chen M., et al. (2020d). The soybean bZIP transcription factor gene GmbZIP2 confers drought and salt resistance in transgenic plants . Int. J. Mol. Sci. 21:670. 10.3390/ijms21020670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Huang Y., Lv W., Zhang Y., Bhat J. A., Kong J., et al. (2020a). GmNAC8 acts as a positive regulator in soybean drought stress. Plant Sci. 293:110442. 10.1016/j.plantsci.2020.110442 [DOI] [PubMed] [Google Scholar]
- Yang J., Xing G., Niu L., He H., Guo D., Du Q., et al. (2018). Improved oil quality in transgenic soybean seeds by RNAi-mediated knockdown of GmFAD2-1B. Transgenic Res. 27 155–166. 10.1007/s11248-018-0063-4 [DOI] [PubMed] [Google Scholar]
- Yang X., Niu L., Zhang W., Yang J., Xing G., He H., et al. (2018). RNAi-mediated SMV P3 cistron silencing confers significantly enhanced resistance to multiple Potyvirus strains and isolates in transgenic soybean. Plant Cell Rep. 37 103–114. 10.1007/s00299-017-2186-0 [DOI] [PubMed] [Google Scholar]
- Yang J., Xing G., Qian D. U., Sui L., Guo D., Niu L., et al. (2016). Effects of different soybean genotypes on the transformation efficiency of soybean and analysis of the T-DNA insertions in the soybean genome. Soybean Sci. 35 562–567. 10.11861/j.issn.1000-9841.2016.04.0562 [DOI] [Google Scholar]
- Yang X. F., Yu X. Q., Zhou Z., Ma W. J., Tang G. X. (2016). A high-efficiency Agrobacterium tumefaciens mediated transformation system using cotyledonary node as explants in soybean (Glycine max L.). Acta Physiol. Plant. 38:60. 10.1007/s11738-016-2081-2 [DOI] [Google Scholar]
- Yang S., Yanfeng H., Cheng Z., Rice J., Miao L., Ma J., et al. (2019). An efficient Agrobacterium-mediated soybean transformation method using green fluorescent protein as a selectable marker. Plant Signal. Behav. 14 1–7. 10.1080/15592324.2019.1612682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Yang J., Wang Y., He H., Niu L., Guo D., et al. (2019). Enhanced resistance to sclerotinia stem rot in transgenic soybean that overexpresses a wheat oxalate oxidase. Transgenic Res. 28 103–114. 10.1007/s11248-018-0106-x [DOI] [PubMed] [Google Scholar]
- Yang X., Li X., Shan J., Li Y., Zhang Y., Wang Y., et al. (2021). Overexpression of GmGAMYB accelerate the transition to flowering and increases plant height in soybean. Front. Plant Sci. 12:667242. 10.3389/fpls.2021.667242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Du H., Xing X., Li W., Kong Y., Li X., et al. (2022). A small heat shock protein, GmHSP17.9, from nodule confers symbiotic nitrogen fixation and seed yield in soybean. Plant Biotechnol. J. 20 103–115. 10.1111/pbi.13698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao S. (2001). Optimization of Agrobacterium-mediated Genetic Transformation of Soybean Using Glufosinate as a Selective Agent. Ph.D. thesis. Baton Rouge, LA: Louisiana State University. https://digitalcommons.lsu.edu/gradschool_disstheses/329 [Google Scholar]
- Yeom W., Kim H., Lee K.-R., Cho H., Kim J.-Y., Jung H. W., et al. (2020). Increased production of α-linolenic acid in soybean seeds by overexpression of lesquerella FAD3-1. Front. Plant Sci. 10:1812. 10.3389/fpls.2019.01812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi J., Derynck M. R., Li X., Telmer P., Marsolais F., Dhaubhadel S. (2010). A single-repeat MYB transcription factor, GmMYB176, regulates CHS8 gene expression and affects isoflavonoid biosynthesis in soybean. Plant J. 62 1019–1034. 10.1111/j.1365-313X.2010.04214.x [DOI] [PubMed] [Google Scholar]
- Yi X., Yu D. (2006). Transformation of multiple soybean cultivars by infecting cotyledonary-node with Agrobacterium tumefaciens. Afr. J. Biotech. 5 1989–1993. 10.5897/AJB2006.000-5100 [DOI] [Google Scholar]
- Yu G., Zou J., Wang J., Zhu R., Qi Z., Jiang H., et al. (2022). A soybean NAC homolog contributes to resistance to Phytophthora sojae mediated by dirigent proteins. Crop J. 10 332–341. 10.1016/j.cj.2021.08.009 [DOI] [Google Scholar]
- Yue Y., Sun S., Li J., Yu H., Wu H., Sun B., et al. (2021). GmFULa improves soybean yield by enhancing carbon assimilation without altering flowering time or maturity. Plant Cell Rep. 40 1875–1888. 10.1007/s00299-021-02752-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun J., Sun Z., Jiang Q., Wang Y., Wang C., Luo Y., et al. (2022). The miR156b-GmSPL9d module modulates nodulation by targeting multiple core nodulation genes in soybean. New Phytol. 233 1881–1899. 10.1111/nph.17899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng P., Vadnais D. A., Zhang Z., Polacco J. C. (2004). Refined glufosinate selection in Agrobacterium-mediated transformation of soybean [Glycine max (L.) Merrill]. Plant Cell Rep. 22 478–482. 10.1007/s00299-003-0712-8 [DOI] [PubMed] [Google Scholar]
- Zhang B., Xue R. G. (2019). Refined factors in multi-needle-assisted transformation of soybean. Asian J. Biotechnol. Genet. Eng. 2 1–7. [Google Scholar]
- Zhang D., Zhang H., Hu Z., Chu S., Yu K., Lv L., et al. (2019a). Artificial selection on GmOLEO1 contributes to the increase in seed oil during soybean domestication. PLoS Genet. 15:e1008267. 10.1371/journal.pgen.1008267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Li T., Wang Y., Zhang Y., Dong Y. S. (2019b). FvC5SD overexpression enhances drought tolerance in soybean by reactive oxygen species scavenging and modulating stress-responsive gene expression. Plant Cell Rep. 38 1039–1051. 10.1007/s00299-019-02424-y [DOI] [PubMed] [Google Scholar]
- Zhang W., Liao X., Cui Y., Ma W., Zhang X., Du H., et al. (2019c). A cation diffusion facilitator, GmCDF1, negatively regulates salt tolerance in soybean. PLoS Genet. 15:e1007798. 10.1371/journal.pgen.1007798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. Z., Zheng W. J., Cao X. Y., Cui X. Y., Zhao S. P., Yu T. F., et al. (2019d). Genomic analysis of stress associated proteins in soybean and the role of GmSAP16 in abiotic stress responses in Arabidopsis and soybean. Front. Plant Sci. 10:1453. 10.3389/fpls.2019.01453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Chen C., Ge H., Liu J., Luo Y., Liu K., et al. (2014). Efficient soybean regeneration and Agrobacterium-mediated transformation using a whole cotyledonary node as an explant. Biotech. App. Biochem. 61 620–625. 10.1002/bab.1207 [DOI] [PubMed] [Google Scholar]
- Zhang H., Yang Y., Sun C., Liu X., Lv L., Hu Z., et al. (2020). Up-regulating GmETO1 improves phosphorus uptake and use efficiency by promoting root growth in soybean. Plant Cell Environ. 43 2080–2094. 10.1111/pce.13816 [DOI] [PubMed] [Google Scholar]
- Zhang M., Liu Y., Cai H., Guo M., Chai M., She Z., et al. (2020). The bZIP transcription factor GmbZIP15 negatively regulates salt- and drought-stress responses in soybean. Int. J. Mol. Sci. 21:7778. 10.3390/ijms21207778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zhao Q., Zhang J., Niu L., Yang J., Liu X., et al. (2022c). Enhanced resistance to soybean cyst nematode in transgenic soybean via host-induced silencing of vital Heterodera glycines genes. Transgenic Res. 31 239–248. 10.1007/s11248-022-00298-7 [DOI] [PubMed] [Google Scholar]
- Zhang H.-Y., Hou Z.-H., Zhang Y., Li Z.-Y., Chen J., Zhou Y.-B., et al. (2022a). A soybean EF-Tu family protein GmEF8, an interactor of GmCBL1, enhances drought and heat tolerance in transgenic Arabidopsis and soybean. Int. J. Biol. Macromol. 205 462–472. 10.1016/j.ijbiomac.2022.01.165 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Guo W., Chen L., Shen X., Yang H., Fang Y., et al. (2022b). CRISPR/Cas9-mediated targeted mutagenesis of GmUGT enhanced soybean resistance against leaf-chewing insects through flavonoids biosynthesis. Front. Plant Sci. 13:802716. 10.3389/fpls.2022.802716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W. J., Dewey R. E., Boss W., Phillippy B. Q., Qu R. (2013). Enhanced Agrobacterium-mediated transformation efficiencies in monocot cells is associated with attenuated defense responses. Plant Mol. Biol. 81 273–286. 10.1007/s11103-012-9997-8 [DOI] [PubMed] [Google Scholar]
- Zhang Y. M., Liu Z. H., Yang R. J., Li G. L., Guo X. L., Zhang H. N., et al. (2016). Improvement of soybean transformation via Agrobacterium tumefaciens methods involving α-aminooxyacetic acid and sonication treatments enlightened by gene expression profile analysis. Plant Cell Rep. 35 1259–1271. 10.1007/s00299-016-1958-2 [DOI] [PubMed] [Google Scholar]
- Zhang Y.-M., Zhang H.-M., Liu Z.-H., Guo X.-L., Li H.-C., Li G.-L., et al. (2015). Inhibition of isoflavone biosynthesis enhanced T-DNA delivery in soybean by improving plant–Agrobacterium tumefaciens interaction. Plant Cell Tiss. Org. 121 183–193. 10.1007/s11240-014-0693-z [DOI] [Google Scholar]
- Zhang Z., Xing A., Staswick P., Clemente T. E. (1999). The use of glufosinate as a selective agent in Agrobacterium-mediated transformation of soybean. Plant Cell Tiss. Org. 56 37–46. 10.1023/A:1006298622969 [DOI] [Google Scholar]
- Zhao C., Pan X., Yu Y., Zhu Y., Kong F., Sun X., et al. (2020). Overexpression of a TIFY family gene, GsJAZ2, exhibits enhanced tolerance to alkaline stress in soybean. Mol. Breed. 40:33. 10.1007/s11032-020-01113-z [DOI] [Google Scholar]
- Zhao Q., Du Y., Wang H., Rogers H. J., Yu C., Liu W., et al. (2019b). 5-Azacytidine promotes shoot regeneration during Agrobacterium-mediated soybean transformation. Plant Physiol. Biochem. 141 40–50. 10.1016/j.plaphy.2019.05.014 [DOI] [PubMed] [Google Scholar]
- Zhao M. J., Yin L. J., Ma J., Zheng J. C., Wang Y. X., Lan J. H., et al. (2019a). The roles of GmERF135 in improving salt tolerance and decreasing ABA sensitivity in soybean. Front. Plant Sci. 10:940. 10.3389/fpls.2019.00940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Zhu L., Lin C., Shen Z., Xu C. (2019c). Transgenic soybean expressing a thermostable phytase as substitution for feed additive phytase. Sci. Rep. 9:14390. 10.1038/s41598-019-51033-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Jing Y., Luo Z., Gao S., Teng W., Zhan Y., et al. (2021). GmST1, which encodes a sulfotransferase, confers resistance to soybean mosaic virus strains G2 and G3. Plant Cell Environ. 44 2777–2792. 10.1111/pce.14066 [DOI] [PubMed] [Google Scholar]
- Zheng N., Li T., Dittman J., Su J., Li R., Gassmann W., et al. (2020). CRISPR/Cas9-based gene editing using egg cell-specific promoters in Arabidopsis and soybean. Front. Plant Sci. 11:800. 10.3389/fpls.2020.00800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q., Perry S. E. (2014). Alterations in the transcriptome of soybean in response to enhanced somatic embryogenesis promoted by orthologs of Agamous-like15 and Agamous-like18. Plant Physiol. 164 1365–1377. 10.1104/pp.113.234062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng T., Yu X., Sun Y., Zhang Q., Zhang X., Tang M., et al. (2022). Expression of a cytochrome P450 gene from bermuda grass Cynodon dactylon in soybean confers tolerance to multiple herbicides. Plants 11:949. 10.3390/plants11070949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Huang J. L., Zhang X. L., Zhu L. M., Wang X. F., Guo N., et al. (2018). Overexpression of chalcone isomerase (CHI) increases resistance against Phytophthora sojae in soybean. J. Plant Biol. 61 309–319. 10.1007/s12374-018-0017-7 [DOI] [Google Scholar]
- Zhou Y., Liu W., Li X., Sun D., Xu K., Feng C., et al. (2020). Integration of sRNA, degradome, transcriptome analysis and functional investigation reveals gma-miR398c negatively regulates drought tolerance via GmCSDs and GmCCS in transgenic Arabidopsis and soybean. BMC Plant Biol. 20:190. 10.1186/s12870-020-02370-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S., Chen Z., Xie B., Guo Q., Chen M., Liang C., et al. (2021). A phosphate starvation responsive malate dehydrogenase, GmMDH12 mediates malate synthesis and nodule size in soybean (Glycine max). Environ. Exp. Bot. 189:104560. 10.1016/j.envexpbot.2021.104560 [DOI] [Google Scholar]