ABSTRACT
Mechanisms of evolution and evolution of antibiotic resistance are both fundamental and world health problems. Stress-induced mutagenesis defines mechanisms of mutagenesis upregulated by stress responses, which drive adaptation when cells are maladapted to their environments—when stressed. Work in mutagenesis induced by antibiotics had produced tantalizing clues but not coherent mechanisms. We review recent advances in antibiotic-induced mutagenesis that integrate how reactive oxygen species (ROS), the SOS and general stress responses, and multichromosome cells orchestrate a stress response-induced switch from high-fidelity to mutagenic repair of DNA breaks. Moreover, while sibling cells stay stable, a mutable “gambler” cell subpopulation is induced by differentially generated ROS, which signal the general stress response. We discuss other evolvable subpopulations and consider diverse evolution-promoting molecules as potential targets for drugs to slow evolution of antibiotic resistance, cross-resistance, and immune evasion. An FDA-approved drug exemplifies “stealth” evolution-slowing drugs that avoid selecting resistance to themselves or antibiotics.
KEYWORDS: antibiotic resistance, antibiotics, cell subpopulations, evolvability, evolution, stress-induced mutagenesis, antievolvability drugs, drug resistance evolution
INTRODUCTION
Evolution drives real-world problems in infectious disease and biology, from dangerous new viral variants (1) to antibiotic resistance (2). An estimated 1.27 million deaths worldwide resulted from antibiotic-resistant infections in 2019 (3). The World Health Organization (WHO) has issued a call to action against evolution of antibiotic resistance in priority pathogens (4). Antibiotic resistance occurs either by transfer of resistance genes from one to another bacterium (reviewed in reference 5), or by de novo mutations that confer resistance. Although horizontal gene transfer (HGT) is important in many clinical circumstances (6, 7), for some specific widespread pathogens and widely used antibiotics, de novo mutations cause clinically relevant resistance.
De novo mutations can cause antibiotic resistance in various ways: they can alter the target protein and prevent antibiotic binding (8), or upregulate efflux pumps (9) or enzymes that degrade antibiotics (10, 11), reducing effective antibiotic concentrations. Mutations are the primary source of resistance of enterobacterial nosocomial infections in hospitals (12). In the World Health Organization (WHO) list of priority antibiotic-resistant pathogens (4), several acquire resistance by mutagenesis. These include Helicobacter pylori resistance to tetracycline (13), Mycobacterium tuberculosis resistance to isoniazid (14), and carbapenem resistance in Enterobacteriaceae (15). Mutagenesis is the main route to resistance to widely used fluoroquinolones (16) and the “last-chance” antibiotic daptomycin (17) and underlies chromosomally mediated colistin resistance (18). Moreover, even plasmid-borne β-lactamases, shared by HGT, require mutagenesis to confer resistance to newer-generation β-lactam antibiotics, which are then rendered ineffective (10). Identification of the mutagenic mechanisms that promote antibiotic resistance could allow new strategies to combat the now critical problem (4). The escape of pathogens from our immune defenses and drugs is a problem in the molecular, systems-biological, and populational mechanisms of evolution.
Mutations (including all de novo genomic changes) drive evolution, and our paradigm for both is changing. Mutagenesis and evolution are being recognized as dynamic, regulated processes with molecular mechanisms that can be both understood and, potentially, inhibited clinically (19–25). This view is necessitated by understanding of stress-induced mutagenesis: molecular mechanisms of mutagenesis that are upregulated by stress responses. The existence of stress-induced mutagenesis mechanisms implies that mutation rates, and the ability to evolve, increase preferentially when cells are poorly adapted to their environment, when stressed (reviewed in references 23, 25, and 26).
Stress-induced mutagenesis departs from ideas established before knowledge of the molecular basis of genes. Mutations were assumed to occur randomly both in time and in genomic space and constantly and gradually (27). Luria and Delbrück defined a mathematical relationship between the birth of mutations and cell divisions that occurred before exposure to a killing environment of lytic bacteriophage, in which Escherichia coli phage-resistant mutants were selected and then quantified (28). Because they used a killing selection for mutants, they saw only mutants already present and failed to detect any possible stress-induced mutagenesis.
Discovery of the bacterial SOS DNA damage response (29–32) led Harrison Echols to propose that stress sensing could, via the SOS response, promote genetic instability, and “inducible evolution” (33). The SOS response upregulates DNA damage tolerance and repair and instigates mutagenesis, prophage induction, and inhibition of cell division (reviewed in reference 34). Others argued, however, that SOS mutagenesis is an unavoidable by-product of repairing DNA damage, that nonmutagenic DNA repair could not evolve (e.g., see references 35 and 36), and that cells must repair DNA damage to survive.
The possibility of stress-inducible evolution (33) was difficult to consider until discoveries that mismatch repair, which corrects DNA replication errors, could be downregulated, increasing the mutation rate without assisting DNA repair (as shown in references 37, to ,40 and reviewed in references 23, 25, and 26), and that the general stress response was required for transposon movement (41, 42) and other mutagenesis under stress (43) and not for concurrent repair (44, 45). Many different stress responses are now documented to upregulate mechanisms of mutagenesis, including mutagenesis unrelated to the SOS response (e.g., see references 46, to ,52). These various mechanisms promote aneuploidy (53, 54) (in eukaryotes), base substitutions and indels (insertions or deletions of one or a few base pairs) (reviewed here, and see references 25 and 26), transpositions (41, 42), and copy number alterations (CNAs) and other genome rearrangements (49, 50, 55). Additionally, reactive oxygen modifies transposase accuracy directly, without stress responses, and so, similarly, causes stress-induced transposon mutagenesis during oxidative stress (56).
In this review, we discuss current understanding of mechanisms of stress-induced mutagenesis. We examine mutagenesis induced by antibiotics and its promotion of antibiotic resistance and cross-resistance to antibiotics not yet encountered. We discuss two mechanisms of starvation stress-induced mutagenesis (21) that also underlie quinolone-induced mutagenesis (19, 57): two kinds of mutagenic repair of DNA breaks. Quinolones induce a switch from accurate to mutagenic modes of DNA break repair, using the general stress and SOS responses, which link mutagenesis to times of stress (57). The mutations are focused in hot spots near sites of DNA breakage (58) and occur in a transiently differentiated mutable “gambler” cell subpopulation (57). We examine how each of these departures from “random” mutagenesis can promote evolution. “Persisters” are subpopulations of transiently nongrowing or slowly growing cells that survive antibiotics temporarily, without a resistance mutation (reviewed in references 59 and 60) and cause relapse of infections by resuming growth after antibiotic clearance (59). We consider the possible relevance of persisters and other evolvable cell subpopulations to gambler cells and suggest criteria for choosing evolution-promoting molecules as potential targets for new drugs to slow evolution of antibiotic resistance and immune evasion, without selecting resistance to themselves or antibiotics. We call these “stealth” evolution inhibitors. In addition, we explore possible useful next steps.
STRESS-INDUCED MUTAGENESIS IN BACTERIAL EVOLUTION
Most of this review is focused on mutagenesis and evolution in the laboratory. Stress-inducible mutagenesis, however, appears to contribute meaningfully to natural bacterial evolution. The large majority (more than 80%) of 787 E. coli natural isolates from diverse environments worldwide display stress-induced mutagenesis in a laboratory setting, showing that they possess the capability (46). Moreover, their ability to do so is correlated with the ecological niche of the isolates, suggesting that stress-inducible mutagenesis is selected (46). Selection of stress-induced mutagenic abilities is also supported by mathematical modeling. Bacterial populations capable of stress-induced mutagenesis showed improved fitness in changing environments (61–63). Separately, in whole-genome sequences of wild E. coli isolates, “mutational signatures” of the sequence differences between their genomes were dominated by specific base substitutions and indels that characterize mutagenesis that depends on the general (sigma-S, or σS) stress response (64). The data imply that most natural variation arose by σS-dependent (stress-induced) mutagenesis (64). Moreover, the multiple molecular mechanisms of stress-induced mutagenesis discovered in bacteria have predicted mechanisms at work in evolution of cancers (as reviewed in reference 25 and see references 65, to ,67). Widespread evidence from bacteria to humans (25 [and see reference 68]) implies that much of the mutagenesis underlying evolution is stress induced.
THE GENERAL STRESS RESPONSE SWITCH TO MUTAGENIC BREAK REPAIR
The general or starvation stress response in E. coli promotes at least two mechanisms of mutagenesis, both of which switch the otherwise accurate mechanism of DNA double-strand break (DSB) repair to mutagenic repair routes (Fig. 1). Both are activated by the general stress response, which occurs via production of the σS transcriptional activator (43–45) encoded by the rpoS gene (69). Bacterial sigma factors, including σS, are interchangeable subunits of RNA polymerase (RNAP), which when plugged into RNAP, direct RNAP and transcription to some genes and away from others. The σS regulon, reviewed by Battesti et al. (69), is upregulated in response to starvation, cold shock, osmotic shock, acid shock, oxidative stress, and antibiotics (24, 70) and protects cellular hardware from damage during those stresses.
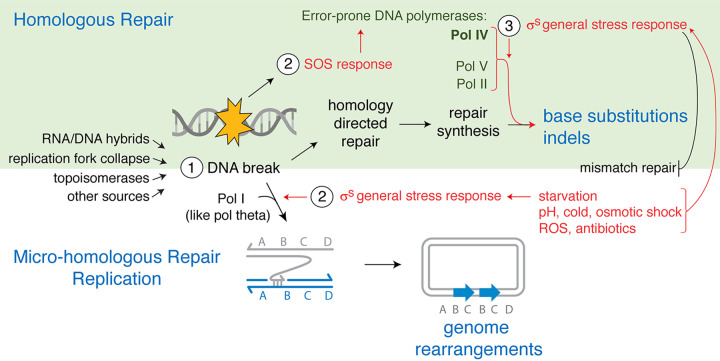
FIG 1.
Temporal regulation of mutagenesis by stress responses in E. coli mutagenic break repair. (Step 1) DNA double-strand breaks (DSBs) are generated by various processes and can then be repaired by homologous or microhomologous repair mechanisms. (Top) During homology-directed DSB repair (HR) (reviewed in references 74 and 75), ssDNA exposed at the DSB ends base pairs with complementary sequence in a sister chromosome, promoting repair DNA synthesis. (Step 2) DSBs also induce the SOS response, which transcriptionally upregulates the error-prone DNA polymerases (Pols) IV, V, and II (82); however, repair remains accurate unless another stressor induces the general stress (σS) response (44, 45). (Step 3) The σS response induces two kinds of switches to mutagenic DSB repair. In cells that are also SOS induced, the σS response, by unknown means, allows the use of, or persistence of errors made by, the error-prone DNA Pols in repair, causing base substitutions (45, 84) and indels (43, 44, 83, 166, 167). σS also downregulates mismatch repair (37–40, 88), which allows errors in DNA synthesis to persist. (Bottom) Less frequently, microhomologous MBR of a DSB occurs. It is SOS independent and requires (step 2) the σS response and DNA Pol I for template switching to regions containing microhomology (49, 50) (few complementary bases). The repair replication creates genome rearrangements. A duplicated chromosome segment is shown (blue arrows). Parallel lines represent base-paired DNA strands, and half arrowheads represent the 3′ DNA ends.
Mutagenic DNA break repair (MBR) in E. coli, studied during starvation, can occur during DSB repair by homologous recombination (44, 45, 58, 71–73), causing base substitutions and indels (25, 26) (Fig. 1, top). MBR requires three simultaneous events to occur (44, 45) (Fig. 1, top). First, a DSB must occur (Fig. 1, step 1) and be repaired by homology-directed repair (HR) (Fig. 1, top). DSB repair in E. coli uses the RecBCD homology-directed repair mechanism and occurs similarly in other bacteria (74, 75). DSBs are bound and “resected” by RecBCD, a DSB end-specific exonuclease that exposes single-stranded DNA (ssDNA) and then loads RecA onto the ssDNA end for repair (76). The RecA-DNA complex also signals DNA damage, which activates the SOS response (34, 77) (Fig. 1, top, step 2). The ssDNA-RecA complex displaces a DNA strand of similar sequence, usually in a sister chromosome, base pairs with the complementary sequence (reviewed in references 74, 75, and 78), and then initiates repair synthesis using the high-fidelity replicative DNA polymerase III (Pol III) (79). DSB repair and MBR require the RuvABC Holliday junction-resolvase complex (72–75) and RecA and RecBCD (44, 45, 71).
Second, the SOS response (Fig. 1, top, step 2) must be induced for homology-directed MBR (80), and this occurs following DNA breakage in about 25% of cells with a single reparable DSB (81). The SOS response transcriptionally upregulates about 40 genes in E. coli, including error-prone DNA Pols IV, V, and II (Fig. 1, top) (reviewed in references 34 and 82), all of which promote components of stress-induced mutagenesis. Pol IV is required for all homology-directed MBR and promotes formation of indels and base substitutions (44, 45, 83–85). The Pol IV 10-fold upregulation by the SOS response accounts for the SOS role in stress-induced MBR (86). Despite the 10-fold upregulation and efficient homology-directed DSB repair (44, 45), repair remains high fidelity and nonmutagenic unless a third event occurs: activation of the σS general stress response (Fig. 1, top, step 3) (43–45).
The σS response is an “AND gate” for MBR, for which at least two responses must occur simultaneously (SOS AND σS). That is, the cell must sense at least two different stressors before committing to mutagenesis: DNA damage and the σS inducer. The σS-inducing stressor most studied is starvation (21). Because a constitutive σS response allows MBR in the absence of any stress (44, 45), the stress response is required, but stress is not. The σS response, by unknown means, licenses the use of the SOS-upregulated DNA Pols in DSB repair and/or allows their errors to persist and become mutations (Fig. 1, top, base substitutions and indels). σS upregulates Pol IV about 2-fold (87), downregulates mismatch repair (38, 88), and may downregulate high-fidelity replicative DNA Pol III (about 2-fold reduction in mRNA) (89). Pol III competes with Pol IV in DSB repair-associated replication in cells (90) and biochemically at model strand displacement loops (D-loops) (91). Any of these effects could underlie the role of σS in stress-induced MBR.
Importantly, Pol IV and mutagenesis are not needed for efficient DSB repair (44, 45), which works as well (45) or better (44) in its absence, thus refuting arguments that high-fidelity DNA repair cannot evolve (35, 36). It did, but E. coli cells do not use it under σS-inducing stress.
σS also promotes a “microhomologous” mechanism of DSB repair, which causes genome rearrangements (43) (Fig. 1, bottom), including copy number alterations (CNAs) and other rearrangements (49, 50, 55). Microhomologous MBR does not require an SOS response (83). Microhomologous MBR might occur in starving cells that lack a sister chromosome template for homology-directed repair or those not undergoing the SOS response upon DNA breakage. Microhomologous MBR (Fig. 1, bottom) occurs via a microhomology-mediated break-induced replication (MMBIR) mechanism (44, 50, 92, 93) and is reviewed elsewhere (25, 92).
MUTATION HOT SPOTS AND CLUSTERS
MBR causes mutations near DSBs, implicating the tracts of DSB repair synthesis as the MBR sites (44, 58). When a site-specific DSB is delivered to the chromosome of starving cells, σS- and Pol IV-dependent mutations occur maximally within the first kilobase pair on either side of the break site and fall off logarithmically to about 60 kb on either side of it, with a long tail of low-level Pol IV-dependent mutations up to a megabase pair away (58), presumably in the tracts of DSB repair synthesis. The mutations occur in clusters, with the probability of a mutation being higher at sites near another mutation (94). Mutation clusters can promote “concerted evolution” that requires multiple simultaneous mutations within a gene or linked genes to allow function. There is no evidence of increased DNA breakage during MBR in starving cells (93) compared with nonstressed cells (81, 95), only of the σS-dependent increase in the mutagenicity of repair (44, 45). Thus, how and where spontaneous DNA breakage occurs might shape mutational landscapes and genome evolution. The first detailed maps of spontaneous DNA breakage in proliferating (mostly unstressed) E. coli cells show hot spots (96). Detailed maps of genomic mutations under various stress conditions would be invaluable for testing the prediction that these bacterial “fragile sites” (96) are mutable genomic regions.
PROTEIN NETWORKS WITH STRESS RESPONSES AS HUBS
In a screen for MBR-defective mutants, our lab identified a network of more than 93 diverse proteins that promote MBR (21). A small number of these were most of the previously known MBR proteins: stress response activators, proteins that perform DSB repair, and error-prone DNA polymerases. However, most of the network proteins are highly diverse and were not obvious candidates for roles in mutagenesis. For example, the largest single category of network genes functions in the electron transfer chain (ETC). Functional tests showed that more than half of the 93 proteins promote mutagenesis, acting upstream of (i.e., before) activation of the three key stress response regulators in MBR: σS (31 proteins), σE (44 proteins), and the SOS response (6 proteins). For example, if the σS response is activated constitutively, the proteins required for its activation are no longer needed for MBR (21). Thus, most of the network proteins promote MBR by sensing stress and transducing the signals that activate the stress responses required to switch to error-prone DSB repair (21). Activation of stress responses appears to be the most important criterion for the E. coli decision to allow mutagenesis, having the largest allotment of genes. Moreover, the stress-response activators are nonredundant network hubs (21).
QUINOLONE ANTIBIOTICS
In the rest of this article, we focus mostly on the very widely used fluoroquinolone antibiotics and how they induce mutagenesis to fluoroquinolone resistance and cross-resistance to other antibiotics, and we compare these findings with data on mutagenesis induced by other antibiotics, about which less is known.
Quinolone antibiotics bind and inhibit bacterial type II topoisomerases (topos) while they are in the act of relieving DNA supercoils (97), which result from unwinding of DNA during DNA replication and transcription. Type II topos bind DNA and cleave both strands, creating a DSB, and attach covalently to each 5′-end strand (98). The broken DNA allows another duplex to pass through, and is then religated, releasing the topo. The DSBs undo supercoils or allow decatenation of linked sister chromosomes following DNA replication (99). Quinolones bind type II topos after DNA breakage and before the religation step and so leave the DNA broken (97). The commonest route to resistance clinically (12) and in the lab (10, 100) is by de novo mutations that either alter the topo so that the drug no longer binds or cause upregulation of efflux pumps that export the drug.
QUINOLONES INDUCE MUTAGENESIS AND ANTIBIOTIC CROSS-RESISTANCE
Ciprofloxacin (cipro) is the most commonly used fluoroquinolone (16, 101). “Subinhibitory” concentrations of cipro (below the minimal inhibitory concentration, or MIC) occur in ecosystems and during antibiotic therapies at the beginning, the end, and when doses are missed. Subinhibitory cipro can both induce and select cipro-resistant mutants (19, 100), making quantification of cipro induction of mutagenesis challenging. That fluoroquinolones induce mutagenesis was shown by exposing cells to another fluoroquinolone, norfloxacin, and then selecting and quantifying mutants resistant to antibiotics not yet encountered (102), antibiotic “cross-resistant” mutants. The norfloxacin-induced mutagenesis required reactive oxygen species (ROS) (102), which are induced by the antibiotic and also underlie its antibiotic (killing) activity (103). Yet, how the ROS might promote mutagenesis was unclear; direct oxidation of DNA bases seemed easy to imagine.
We found that cells grown in subinhibitory cipro at the “minimum antibiotic concentration (MAC),” at which the final CFU are 10% of identical drug-free cultures, induce mutations that confer resistance to two different antibiotics (57). Rifampin-resistant mutants carry base substitutions in the (essential) rpoB gene (104), and ampicillin-resistant mutants carry any loss-of-function mutation in the ampD gene (57, 105). These are induced about 30-fold and 15-fold, respectively, by MAC cipro (57). The cipro-induced rifampin- and ampicillin-resistant mutants have a slight growth disadvantage in MAC cipro and so are not selected by the cipro. Rather, bona fide induction of mutagenesis occurs (57). Base substitutions and indels are generated along with larger genomic rearrangements, including deletions in the ampD gene (57). In Fig. 1 (bottom), the microhomologous rearrangement pathway is shown and requires the σS and not the SOS response (83), and it might account for the cipro-induced larger deletions (92). The starvation stress-induced MBR mechanism (Fig. 1, top) provided a useful entry into how cipro induces mutagenesis (57).
Cipro-induced mutagenesis is MBR.
Cipro-induced mutagenesis occurs via the stress-induced MBR pathway, requiring proteins of DSB repair, the stress-response regulators for the SOS and σS responses, and the error-prone DNA polymerases IV, V, and II (57) (Fig. 1, top). Supporting an MBR mechanism, cipro-induced mutagenesis is blocked by an induced DSB end-specific binding protein, Gam of phage Mu (57), demonstrating a role for DSBs. Cipro-induced mutagenesis also requires ROS and is blocked by ROS-quenching agents and an inhibitor of Fenton chemistry (57), which generates ROS. Cipro-induced DSBs, quantified as foci of GamGFP (a fusion of the phage Mu Gam protein to green fluorescent protein) (95), are unaffected by ROS-reducing agents (57), indicating that the role of ROS in MBR is not generating the DNA breaks. As described below, ROS promote mutagenesis by activating the σS general stress response.
Mutable gambler cell subpopulation via general stress response.
Antibiotics, including fluoroquinolones, induced the SOS (106–108) and general stress responses (24, 70) in studies that used bulk cell measurements. At the single-cell level, an interesting transient “differentiation” is seen. Flow cytometry and microscopy with ROS stain and fluorescence reporter genes for an active SOS (81, 109) or σS response (21) revealed the cascade of events outlined in Fig. 2 (57).
FIG 2.
Pathway to and potential intervention points in formation of mutable gambler cells. cipro-induced DSBs activate the SOS response, which slows aerobic respiration (140). We suggest that increased autoxidation of reduced ubiquinone leads to (as observed in reference 57) a cell subpopulation with high levels of reactive oxygen species (ROS). ROS activate σS by upregulating transcription of sRNAs DsrA and ArcZ (57), which, with the Hfq RNA chaperone, increase translation of rpoS (σS) mRNA in the cells with high ROS (57). This σS-high “gambler” cell subpopulation allows mutagenic DNA break repair (MBR) (Fig. 1, top) and produces antibiotic cross-resistant mutants induced by cipro (57). The gambler subpopulation is transiently mutable (57). The SOS response also upregulates the SulA inhibitor of cell division, which promotes formation of multichromosome cells (168), which facilitate cipro-induced MBR (57). Potential antievolvability drug targets (depicted as “–| target,” in which “–|” indicates inhibition) have been identified by the discovery of the various steps of gambler cell formation and the action illustrated. The FDA-approved human drug edaravone inhibits gambler cell formation by quenching ROS (57), which does not select antievolvability drug resistance. Previous and proposed drugs to target the SOS response (155–157), DSB repair (158), any other DNA repair (154) and/or the error-prone DNA polymerases do reduce fitness in the antibiotic and so select resistance.
Cipro-induced DNA breaks were visible as GamGFP foci (95) in essentially all cells, as was the SOS response (57) (Fig. 2). The SOS response and DSBs are unaffected by ROS quenching, indicating that the DSBs and SOS response occur independently of ROS (57). Intriguingly, the sequences of the cipro-induced mutations do not show the ROS-mediated mutation signature of 8-oxo-dG (oxidized guanine: G·C→T·A and A·T→C·G) (110), reinforcing the conclusion that the ROS role in mutagenesis is not via oxidized (damaged) DNA (57).
Surprisingly, ROS-dyed and σS-active cells composed only a roughly 20% cell subpopulation. The same cells display first ROS and then σS activity. The ROS induce the σS response, in that quenching the ROS prevented σS induction (Fig. 2) (57). As indicated in Fig. 2, ROS induce transcription of two small RNAs (sRNAs), ArcZ and DsrA, which with their RNA chaperone Hfq (57) increase translation of rpoS mRNA (69) (Fig. 2). Removal of any of these components prevented σS induction and mutagenesis (57) (Fig. 2). Additionally, the ROS ceased to be needed for mutagenesis if the σS response was artificially upregulated (57). Thus, the major role of ROS in cipro-induced mutagenesis is inducing the σS response, which subpopulation cells do by upregulating the two sRNAs (Fig. 2). This is unlike ROS roles in starvation stress-induced MBR (111), DNA damage, and antibiotic activity (112, 113).
The σS-active cells, enriched by fluorescence-activated cell sorting (FACS), generated most of the cipro-induced mutants—more than 400 times more than arose from the σS-inactive cells (57). Thus, ROS activation of σS creates a mutable “gambler” cell subpopulation. The gamblers “experiment” with mutagenesis, which might lead to either adaptation or loss of fitness, while most of the cells take no similar risk (57) (Fig. 2). This might be a “bet-hedging” strategy (114, 115), in which some members of a population are more likely to succeed by adaptation to a stressful environment and others stay the course, and so benefit if the environment reverts to its prestress state.
Gamblers are transient in that the mutants they generate do not retain a σS-activated phenotype (57). Whereas low mutation rates under stressful conditions can limit adaptability (62, 63), populations of constitutively hypermutating cells show reduced long-term fitness (116). The transient gambler subpopulation appears to be an intermediate in which short-term increases of mutability may promote adaptation under stress without harming long-term fitness once adaptation occurs.
In one mathematical model, the apparent mutability of gamblers might have resulted from differential cell death (117); however, death rates were equal in gamblers and nongamblers, ruling out this hypothesis (57). This apparent discrepancy might reflect the model’s assumptions of no antibiotic-induced increase in chromosomes per cell (discussed below) and that cells in a population are equally mutable (117). The gambler subpopulation shows neither to be the case (57).
Many antibiotics activate σS (24 [reviewed in reference 23]), suggesting that MBR may be a conserved response for survival of antibiotics (70). Whether other antibiotics or σS inducers differentiate transient mutable gambler cell subpopulations and MBR remains to be determined. In starving colonies, cell subpopulations have been observed that have stress responses activated (118) and might be gamblers.
EVOLVABLE CELL SUBPOPULATIONS, PERSISTERS, AND METABOLISM
Transient cell subpopulations provide alternative physiological options that allow survival, with or without mutation(s) or horizontal gene acquisition. Bacillus subtilis activates both starvation stress-induced mutagenesis and a subpopulation “competent” for DNA uptake with the same ComK transcriptional activator (119). The mutants are not enriched among transformants (52), suggesting alternative responses to the same stress. Plasmid transmission (HGT) and loss may also affect antibiotic resistance, with high conjugation frequency (10−3) promoting plasmid transmission and the resistance conferred and high loss (10−3) causing plasmid eradication and loss of resistance (120).
Present at about 10−5 of the population, persisters survive β-lactams (121, 122), fluoroquinolones (121–124), and aminoglycosides (121, 122) and underlie much of antibiotic treatment failure (reviewed in references 60 and 125). Persisters can form stochastically (126) or be induced via stress responses, including SOS (123) and σS (127), similarly to gamblers (57). Persistence resembles antibiotic “tolerance” (reviewed in reference 60), a physiological state in which whole populations survive even higher antibiotic levels (60) without a mutation.
Whereas persisters are found under high doses (60), gamblers and mutagenesis are maximal with “subinhibitory” antibiotics (19, 24, 57, 102) (10% survival). Gamblers might become or harbor future persisters or could be an alternative or unrelated program.
Reduced energy metabolism characterizes gamblers and allows persisters to withstand most antibiotics. For example, tricarboxylic acid (TCA) enzyme promoter activity varies in proliferating cells, and those with low TCA gene transcription become persisters (121). Some antibiotics kill regardless of metabolic activity, but are toxic at high doses. Zheng et al. (122) combined these with antibiotics that attack metabolically active cells to eradicate persisters (122). The metabolism-dependent antibiotics kill most of the cells, while rare persisters succumb to the metabolism-insensitive killers at lower, nontoxic doses (122).
Metabolism can affect resistance directly. Mutations that reduce TCA cycle activity cause resistance to some antibiotics (128) and appear in many clinically relevant pathogens (128). Similarly, the electron transfer component ubiquinone (UQ) acts early in gambler cell differentiation, upstream of reactive oxygen (57) (Fig. 2), which may result from ubiquinone autooxidation (129, 130) (discussed in the next section). Even “taking the chance” that some cells become mutable begins only when metabolism is threatened. Matic and colleagues suggest that energy metabolism is a key universal sensor for many stresses, including antibiotics (70).
In growing cells, spontaneous mutations, seen as foci of MutL-GFP mismatch-interacting protein (131), occurred mostly in subpopulations with stress responses activated (132), detected with fluorescence reporter genes. Cells with high SOS, RpoH/σH heat shock protein (protein stress), or OxyR oxidative stress response activity showed more MutL-GFP foci than those without stress response induction, linking spontaneous mutability to stress and stress responses (132). Furthermore, the cells with increased mutations also showed increased translation errors (132), suggesting a vicious cycle of mutations fueling poor proteostasis, which because proteins make DNA, feeds back to increased mutability. Rarely examined, translation errors might often accompany mutagenesis.
Do efflux pumps induce gamblers?
Surprisingly, like gamblers, subpopulations with high activity of the AcrAB-TolC efflux pump (133, 134) show reduced levels of MutS mismatch repair protein and an increased mutation rate (135). Deletion of acrB blocked pump activity and MutS reduction. We hypothesize that increased efflux activity reduces the effective within-cell drug concentration to subinhibitory levels that both aid survival and induce mutagenesis (57, 102): a one-two punch against the antibiotic. The NorA efflux pump promotes both immediate survival and the evolution of cipro resistance in Staphylococcus aureus (136), and its chemical inhibition by reserpine reduced cipro resistance, supporting the hypothesis that preventing cipro efflux can raise the in-cell drug concentration (136) to beyond the subinhibitory level at which mutagenesis is induced (57, 102).
Most evolutionary interpretations (35, 36) and models (117) assume homogeneous populations. The variability of metabolic rates, prevalence of drug-induced and spontaneous mutations and persisters in metabolically depressed subpopulations, and gambler promotion of adaptability (55, 57, 135, 137–139) indicate that those models could be improved by incorporating heterogeneity. Evolvable cell subpopulations may be the rule, not the exception.
Why some cells and not others? First steps.
How mutable subpopulations begin can reveal circumstances that necessitate acceleration of evolution. For gamblers, the ROS-high cell subpopulation (and mutagenesis) (Fig. 2) are induced by an SOS response in all cells (Fig. 2), which is dependent on UbiD synthesis of ubiquinone (UQ), a component of the electron transfer chain (ETC). UQ was not needed if σS was artificially upregulated, indicating that subpopulation induction is its sole role in MBR. The SOS response could promote ROS by its suppression of aerobic respiration (140), which causes autoxidation of reduced quinols, leading to ROS (129, 130). There might be heterogeneity in the SOS slowing of the ETC and a respiration threshold below which autoxidation of ubiquinone occurs. ETC activity might vary between cells, or, alternatively, heterogeneous production of an SOS-regulated protein(s) might cause only some cells to slow the ETC. SOS-induced TisB and DinQ disrupt membrane potential (124, 141) and so are candidates for an ETC inhibitor. The distribution of their induction among cells is unknown (142). Coupling mutagenesis to the ETC (21, 57) highlights ATP production in basic sensing of stress.
Multiple chromosomes and mutagenesis.
E. coli cells grown in fluoroquinolones form long, multichromosome cell “filaments” (143). The SOS-induced inhibitor of cell division SulA (144) blocks polymerization of the microtubule-like cell division (FtsZ) ring, causing more chromosomes per single long cell “filament” (145, 146) (Fig. 2, bottom). SulA (144) promotes mutability in both starved (80) and cipro-treated cells, both per cell and per chromosome (57). The mutability might reflect a requirement for complementation of deleterious mutations long enough to make an adaptive mutation. Recombination between chromosomes might be the advantage (57, 143) for generating the mutants by MBR and/or for losing or buffering deleterious alleles. Mathematical modeling indicates that multichromosome cells survive increased mutation rates better than nonfilaments (57). Cipro-induced mutability shows both heterogeneous mutability between cells and promotion of mutations in multichromosome cells (Fig. 2), both evolution accelerators as predicted by modeling (57, 63).
In a striking similarity to gamblers (57), multiple chromosomes promoted persister formation directly (147). Cells with more than 1 chromosome survived quinolones better than single-chromosome cells separated by FACS (147). Moreover, the survival was RecA and RecB dependent, supporting the need for recombinational repair and/or for an SOS response to survive cipro-induced DSBs (19).
TARGETS FOR ANTIEVOLVABILITY DRUGS
Mechanisms of stress-induced mutagenesis promise to reveal possible targets for “antievolvability” drugs to slow evolution of antibiotic resistance, cross-resistance, and immune evasion (19, 21, 22, 25) for better clinical outcomes (19–22, 24, 25). With identification of mechanisms and molecules that promote evolvability, it may be appropriate to consider criteria for choosing antievolvability drug targets. One approach could be to target essentially any protein required for stress-induced mutagenesis (or other evolution), preferably in various bacteria and antibiotics. A more deliberate “stealth” approach could focus, we suggest, on targets the loss of which causes no immediate fitness decrease—as for example, the loss of DNA repair proteins does—so that resistance to the antievolvability drug will not be selected directly. In this light, some previously proposed (Mfd) or targeted (RecA, SOS) evolvability-promoting molecules include nonstealthy targets that select resistance.
Targeting proteins also needed for antibiotic survival.
Mfd is an RNAP translocase that functions in transcription-coupled nucleotide excision repair (NER) (TCR), among other roles. Mfd promotes mutagenesis (93, 148–151) to drug resistance (93, 150) and was suggested as a possible target for antievolvability drugs because it acts in diverse bacteria, including in the E. coli MBR mechanism (93). In E. coli MBR, Mfd promotes DNA DSBs at some genomic sites (93) in a pathway that also required RNA-DNA hybrids and the σE membrane protein stress response regulator (93, 152). An enzyme-induced DSB delivered near the mutation reporter gene substituted for all of these components, indicating their roles in generation of the spontaneous DSBs (93, 152). Mfd was postulated to promote DSB formation and mutagenesis by stabilizing RNA-DNA hybrid “R-loops” in DNA, which can prime DNA synthesis/replication that creates a DSB when it encounters a single-stranded nick in the DNA (93). σE-dependent transcription was postulated to generate the RNA in the R-loops (93, 152).
Mfd also promotes mutagenesis in Bacillus subtilis (148–150, 153), Pseudomonas aeruginosa, and Salmonella enterica serovar Typhimurium, including within host cells (150). In B. subtilis, ComK-dependent (52) stress-induced mutagenesis requires Mfd and UvrA, a nucleotide excision repair (NER) protein that works in TCR and global (Mfd-independent) NER, implying that mutagenesis occurred dependently on TCR (148, 149); possible molecular mechanisms have not been defined. In another B. subtilis mutagenesis assay (150), Mfd-mediated mutagenesis also required UvrA, transcription, and Mfd interaction with an RNA polymerase subunit, implicating TCR (150). The authors suggest targeting Mfd or its interaction with the RNA polymerase to inhibit mutagenesis and antibiotic resistance (154).
“Stealth” targeting of network hubs not needed for immediate antibiotic survival.
Another possible approach to targeting evolution makes use of a functional network analysis of evolution-promoting mechanisms. Functional network analysis of starvation stress-induced MBR showed the stress response activators to be key nonredundant hubs in the MBR network (21), making them attractive targets (21), inhibition of which might collapse the entire network. These hubs include σS and SOS response activators (21). Current inhibitors of the SOS response target RecA (activator), LexA (155–157) (repressor), and RecB/AddAB (158) (activators). Inhibition of these proteins increases bacterial killing (by blocking DNA repair) and so may be expected to select strongly for resistance (19): a nonstealth approach. We suggest aiming new drugs, instead, at evolution-promoting hubs that have little effect on survival of antibiotics. In the pathogen Candida albicans, HSP90 inhibitors (159) and the natural product beauvericin (160) prevent the evolution of resistance to antifungal drugs without altering killing. Resistance to beauvericin and similar evolvability inhibitors is, therefore, unlikely to be selected directly.
As a proof of concept, the ROS-reducing FDA-approved drug edaravone inhibits formation of cipro-induced gambler cells and mutagenesis (57) (Fig. 2). Edaravone is used for amyotrophic lateral sclerosis (ALS) and cerebral infarction (161). Edaravone did not change cipro induction of DSBs, the SOS response, or antibiotic killing (Fig. 2) (57), any of which could select edaravone- and cipro-resistant cells. Antioxidants, including edaravone, given either with antibiotics or alone, were beneficial in mouse infection models (162–164). The authors studied immune-modulating effects and not antibiotic resistance, a potential factor in their effectiveness. In humans, edaravone reduced septic shock and mortality in septic peritonitis patients receiving standard care (including antibiotics) (165). This might result partly from slowed evolution of antibiotic resistance; however, data on resistance were not reported. Edaravone is a promising proof of concept for stealth drugs that decrease evolvability without selection for antibiotic resistance (or edaravone resistance) because it reduces mutagenesis, not survival (57). Other possible hub-related targets for antievolvability drugs in cipro-induced MBR are noted in Fig. 2 (not meant to be an exhaustive list). Future analyses of evolution-promoting functional networks may reveal other promising targets as network hubs.
Antievolvability drugs might be used as adjuvants to traditional antibiotics, to extend their utility by slowing development of resistance (19, 22, 25, 57). Alternatively, as monotherapies, the slowing of pathogen evolution might tilt their evolutionary races against our immune defenses in favor of the immune system and allow clearance of infections without reducing the beneficial diversity of our microbiota with antibiotics (21, 22, 57).
CONCLUSIONS
Mutagenesis mechanisms upregulated by stress responses promote mutagenesis preferentially when cells are maladapted to their environment and speed adaption in stressful and changing environments (models shown in references 62 and 63). In stress-induced mutagenic break repair, upregulation of mutagenesis does not aid cell survival of the DNA breaks (44, 45)—but may be induced because it accelerates adaptation (62, 63) when cells are stressed and struggling to survive. Gambler cells depart further from random mutagenesis, honing regulation further by limiting the risks of mutagenesis to part of a population that experiments with new genotypes, while other parts “hedge” the population’s “bets” by remaining stable. An antimutagenic FDA-approved drug, edaravone, quenches ROS and prevents formation of the gambler cell subpopulation in the laboratory (57) (Fig. 2): an example of “stealth drugging” evolution without selecting resistance directly. Edaravone might or might not be optimal for clinical inhibition of antibiotic resistance. There are likely to be more, as-yet-undescribed regulatory steps in quinolone-induced mutagenesis and other antibiotic-induced evolvability mechanisms, some of which might afford attractive potential drug targets. Understanding the details of the mechanisms of mutagenesis is likely to expand options for combating the evolution of pathogens and antibiotic resistance.
ACKNOWLEDGMENTS
We thank Devon Fitzgerald and Christophe Herman for comments on the manuscript.
The work was supported by National Institutes of Health (NIH) grant R35-GM122598 (S.M.R.), NIH Director’s Pioneer Award DP1-AG072751 (S.M.R.), NIH R01-CA250905 (S.M.R. and Kyle M. Miller), NIH R01-GM106373 (P.J.H.), and a grant from the W. M. Keck Foundation (S.M.R. and K.M. Miller).
Footnotes
This article is a direct contribution from Susan M. Rosenberg, a Fellow of the American Academy of Microbiology, who arranged for and secured reviews by Ivan Matic, Institut Cochin, and Jue D. Wang, University of Wisconsin—Madison.
Contributor Information
Susan M. Rosenberg, Email: smr@bcm.edu.
Leah E. Cowen, University of Toronto
REFERENCES
- 1.Anonymous . 2021. COVID evolution and the Webb telescope—the week in infographics. Nature doi: 10.1038/d41586-021-03694-x. [DOI] [PubMed] [Google Scholar]
- 2.Ventola CL. 2015. The antibiotic resistance crisis: part 1: causes and threats. P T 40:277–283. [PMC free article] [PubMed] [Google Scholar]
- 3.Murray CJL, Ikuta KS, Sharara F, Swetschinski L, Robles Aguilar G, Gray A, Han C, Bisignano C, Rao P, Wool E, Johnson SC, Browne AJ, Chipeta MG, Fell F, Hackett S, Haines-Woodhouse G, Kashef Hamadani BH, Kumaran EAP, McManigal B, Agarwal R, Akech S, Albertson S, Amuasi J, Andrews J, Aravkin A, Ashley E, Bailey F, Baker S, Basnyat B, Bekker A, Bender R, Bethou A, Bielicki J, Boonkasidecha S, Bukosia J, Carvalheiro C, Castañeda-Orjuela C, Chansamouth V, Chaurasia S, Chiurchiù S, Chowdhury F, Cook AJ, Cooper B, Cressey TR, Criollo-Mora E, Cunningham M, Darboe S, Day NPJ, De Luca M, Dokova K, et al. 2022. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399:629–655. doi: 10.1016/S0140-6736(21)02724-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Govindaraj Vaithinathan A, Vanitha A. 2018. WHO global priority pathogens list on antibiotic resistance: an urgent need for action to integrate One Health data. Perspect Public Health 138:87–88. doi: 10.1177/1757913917743881. [DOI] [PubMed] [Google Scholar]
- 5.Thomas CM, Nielsen KM. 2005. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat Rev Microbiol 3:711–721. doi: 10.1038/nrmicro1234. [DOI] [PubMed] [Google Scholar]
- 6.Peterson E, Kaur P. 2018. Antibiotic resistance mechanisms in bacteria: relationships between resistance determinants of antibiotic producers, environmental bacteria, and clinical pathogens. Front Microbiol 9:2928. doi: 10.3389/fmicb.2018.02928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lerminiaux NA, Cameron ADS. 2019. Horizontal transfer of antibiotic resistance genes in clinical environments. Can J Microbiol 65:34–44. doi: 10.1139/cjm-2018-0275. [DOI] [PubMed] [Google Scholar]
- 8.Tran TT, Panesso D, Mishra NN, Mileykovskaya E, Guan Z, Munita JM, Reyes J, Diaz L, Weinstock GM, Murray BE, Shamoo Y, Dowhan W, Bayer AS, Arias CA. 2013. Daptomycin-resistant Enterococcus faecalis diverts the antibiotic molecule from the division septum and remodels cell membrane phospholipids. mBio 4:e00281-13. doi: 10.1128/mBio.00281-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Piddock LJ. 2006. Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin Microbiol Rev 19:382–402. doi: 10.1128/CMR.19.2.382-402.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blair JM, Webber MA, Baylay AJ, Ogbolu DO, Piddock LJ. 2015. Molecular mechanisms of antibiotic resistance. Nat Rev Microbiol 13:42–51. doi: 10.1038/nrmicro3380. [DOI] [PubMed] [Google Scholar]
- 11.Livermore DM. 2008. Defining an extended-spectrum beta-lactamase. Clin Microbiol Infect 14(Suppl 1):3–10. doi: 10.1111/j.1469-0691.2007.01857.x. [DOI] [PubMed] [Google Scholar]
- 12.Mehrad B, Clark NM, Zhanel GG, Lynch JP, III, 2015. Antimicrobial resistance in hospital-acquired Gram-negative bacterial infections. Chest 147:1413–1421. doi: 10.1378/chest.14-2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nishizawa T, Suzuki H. 2014. Mechanisms of Helicobacter pylori antibiotic resistance and molecular testing. Front Mol Biosci 1:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gillespie SH. 2002. Evolution of drug resistance in Mycobacterium tuberculosis: clinical and molecular perspective. Antimicrob Agents Chemother 46:267–274. doi: 10.1128/AAC.46.2.267-274.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guh AY, Bulens SN, Mu Y, Jacob JT, Reno J, Scott J, Wilson LE, Vaeth E, Lynfield R, Shaw KM, Vagnone PM, Bamberg WM, Janelle SJ, Dumyati G, Concannon C, Beldavs Z, Cunningham M, Cassidy PM, Phipps EC, Kenslow N, Travis T, Lonsway D, Rasheed JK, Limbago BM, Kallen AJ. 2015. Epidemiology of carbapenem-resistant Enterobacteriaceae in 7 US communities, 2012–2013. JAMA 314:1479–1487. doi: 10.1001/jama.2015.12480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacoby GA. 2005. Mechanisms of resistance to quinolones. Clin Infect Dis 41(Suppl 2):S120–S126. doi: 10.1086/428052. [DOI] [PubMed] [Google Scholar]
- 17.Palmer KL, Daniel A, Hardy C, Silverman J, Gilmore MS. 2011. Genetic basis for daptomycin resistance in enterococci. Antimicrob Agents Chemother 55:3345–3356. doi: 10.1128/AAC.00207-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cannatelli A, Giani T, D'Andrea MM, Di Pilato V, Arena F, Conte V, Tryfinopoulou K, Vatopoulos A, Rossolini GM, COLGRIT Study Group . 2014. MgrB inactivation is a common mechanism of colistin resistance in KPC-producing Klebsiella pneumoniae of clinical origin. Antimicrob Agents Chemother 58:5696–5703. doi: 10.1128/AAC.03110-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cirz RT, Chin JK, Andes DR, de Crecy-Lagard V, Craig WA, Romesberg FE. 2005. Inhibition of mutation and combating the evolution of antibiotic resistance. PLoS Biol 3:e176. doi: 10.1371/journal.pbio.0030176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith PA, Romesberg FE. 2007. Combating bacteria and drug resistance by inhibiting mechanisms of persistence and adaptation. Nat Chem Biol 3:549–556. doi: 10.1038/nchembio.2007.27. [DOI] [PubMed] [Google Scholar]
- 21.Al Mamun AA, Lombardo MJ, Shee C, Lisewski AM, Gonzalez C, Lin D, Nehring RB, Saint-Ruf C, Gibson JL, Frisch RL, Lichtarge O, Hastings PJ, Rosenberg SM. 2012. Identity and function of a large gene network underlying mutagenic repair of DNA breaks. Science 338:1344–1348. doi: 10.1126/science.1226683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosenberg SM, Queitsch C. 2014. Medicine. Combating evolution to fight disease. Science 343:1088–1089. doi: 10.1126/science.1247472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blazquez J, Rodriguez-Beltran J, Matic I. 2018. Antibiotic-induced genetic variation: how it arises and how it can be prevented. Annu Rev Microbiol 72:209–230. doi: 10.1146/annurev-micro-090817-062139. [DOI] [PubMed] [Google Scholar]
- 24.Gutierrez A, Laureti L, Crussard S, Abida H, Rodriguez-Rojas A, Blazquez J, Baharoglu Z, Mazel D, Darfeuille F, Vogel J, Matic I. 2013. β-Lactam antibiotics promote bacterial mutagenesis via an RpoS-mediated reduction in replication fidelity. Nat Commun 4:1610. doi: 10.1038/ncomms2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fitzgerald DM, Hastings PJ, Rosenberg SM. 2017. Stress-induced mutagenesis: implications in cancer and drug resistance. Annu Rev Cancer Biol 1:119–140. doi: 10.1146/annurev-cancerbio-050216-121919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fitzgerald DM, Rosenberg SM. 2019. What is mutation? A chapter in the series: how microbes “jeopardize” the modern synthesis. PLoS Genet 15:e1007995. doi: 10.1371/journal.pgen.1007995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mayr E. 1982. The growth of biological thought: diversity, evolution, and inheritance. Belknap Press, Cambridge, MA. [Google Scholar]
- 28.Luria SE, Delbruck M. 1943. Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28:491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Witkin EM, George DL. 1973. Ultraviolet mutagenesis in polA and UvrA polA derivatives of Escherichia coli B-R: evidence for an inducible error-prone repair system. Genetics 73(Suppl 73):91–100. [PubMed] [Google Scholar]
- 30.Witkin EM. 1967. The radiation sensitivity of Escherichia coli B: a hypothesis relating filament formation and prophage induction. Proc Natl Acad Sci USA 57:1275–1279. doi: 10.1073/pnas.57.5.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Radman M. 1975. SOS repair hypothesis: phenomenology of an inducible DNA repair which is accompanied by mutagenesis. Basic Life Sci 5A:355–367. doi: 10.1007/978-1-4684-2895-7_48. [DOI] [PubMed] [Google Scholar]
- 32.Radman M. 1974. Phenomenology of an inducible mutagenic DNA repair pathway in Escherichia coli: SOS repair hypothesis, p 128–142. In Prakash L, Sherman F, Miller MW, Lawrence CW, Tabor HW (ed), Molecular and environmental aspects of mutagenesis. Charles C. Thomas, Springfield, IL. [Google Scholar]
- 33.Echols H. 1981. SOS functions, cancer and inducible evolution. Cell 25:1–2. doi: 10.1016/0092-8674(81)90223-3. [DOI] [PubMed] [Google Scholar]
- 34.Chatterjee N, Walker GC. 2017. Mechanisms of DNA damage, repair, and mutagenesis. Environ Mol Mutagen 58:235–263. doi: 10.1002/em.22087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lynch M. 2010. Evolution of the mutation rate. Trends Genet 26:345–352. doi: 10.1016/j.tig.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andersson DI, Hughes D. 2014. Microbiological effects of sublethal levels of antibiotics. Nat Rev Microbiol 12:465–478. doi: 10.1038/nrmicro3270. [DOI] [PubMed] [Google Scholar]
- 37.Longerich S, Galloway AM, Harris RS, Wong C, Rosenberg SM. 1995. Adaptive mutation sequences reproduced by mismatch repair deficiency. Proc Natl Acad Sci USA 92:12017–12020. doi: 10.1073/pnas.92.26.12017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsui HC, Feng G, Winkler ME. 1997. Negative regulation of mutS and mutH repair gene expression by the Hfq and RpoS global regulators of Escherichia coli K-12. J Bacteriol 179:7476–7487. doi: 10.1128/jb.179.23.7476-7487.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harris RS, Feng G, Ross KJ, Sidhu R, Thulin C, Longerich S, Szigety SK, Winkler ME, Rosenberg SM. 1997. Mismatch repair protein MutL becomes limiting during stationary-phase mutation. Genes Dev 11:2426–2437. doi: 10.1101/gad.11.18.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brégeon D, Matic I, Radman M, Taddei F. 1999. Inefficient mismatch repair: genetic defects and down regulation. J Genet 78:21–28. doi: 10.1007/BF02994699. [DOI] [Google Scholar]
- 41.Maenhaut-Michel G, Shapiro JA. 1994. The roles of starvation and selective substrates in the emergence of araB-lacZ fusion clones. EMBO J 13:5229–5239. doi: 10.1002/j.1460-2075.1994.tb06854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Twiss E, Coros AM, Tavakoli NP, Derbyshire KM. 2005. Transposition is modulated by a diverse set of host factors in Escherichia coli and is stimulated by nutritional stress. Mol Microbiol 57:1593–1607. doi: 10.1111/j.1365-2958.2005.04794.x. [DOI] [PubMed] [Google Scholar]
- 43.Lombardo MJ, Aponyi I, Rosenberg SM. 2004. General stress response regulator RpoS in adaptive mutation and amplification in Escherichia coli. Genetics 166:669–680. doi: 10.1534/genetics.166.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ponder RG, Fonville NC, Rosenberg SM. 2005. A switch from high-fidelity to error-prone DNA double-strand break repair underlies stress-induced mutation. Mol Cell 19:791–804. doi: 10.1016/j.molcel.2005.07.025. [DOI] [PubMed] [Google Scholar]
- 45.Shee C, Gibson JL, Darrow MC, Gonzalez C, Rosenberg SM. 2011. Impact of a stress-inducible switch to mutagenic repair of DNA breaks on mutation in Escherichia coli. Proc Natl Acad Sci USA 108:13659–13664. doi: 10.1073/pnas.1104681108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bjedov I, Tenaillon O, Gerard B, Souza V, Denamur E, Radman M, Taddei F, Matic I. 2003. Stress-induced mutagenesis in bacteria. Science 300:1404–1409. doi: 10.1126/science.1082240. [DOI] [PubMed] [Google Scholar]
- 47.Mihaylova VT, Bindra RS, Yuan J, Campisi D, Narayanan L, Jensen R, Giordano F, Johnson RS, Rockwell S, Glazer PM. 2003. Decreased expression of the DNA mismatch repair gene Mlh1 under hypoxic stress in mammalian cells. Mol Cell Biol 23:3265–3273. doi: 10.1128/MCB.23.9.3265-3273.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bindra RS, Gibson SL, Meng A, Westermark U, Jasin M, Pierce AJ, Bristow RG, Classon MK, Glazer PM. 2005. Hypoxia-induced down-regulation of BRCA1 expression by E2Fs. Cancer Res 65:11597–11604. doi: 10.1158/0008-5472.CAN-05-2119. [DOI] [PubMed] [Google Scholar]
- 49.Hastings PJ, Bull HJ, Klump JR, Rosenberg SM. 2000. Adaptive amplification: an inducible chromosomal instability mechanism. Cell 103:723–731. doi: 10.1016/s0092-8674(00)00176-8. [DOI] [PubMed] [Google Scholar]
- 50.Slack A, Thornton PC, Magner DB, Rosenberg SM, Hastings PJ. 2006. On the mechanism of gene amplification induced under stress in Escherichia coli. PLoS Genet 2:e48. doi: 10.1371/journal.pgen.0020048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sung HM, Yeamans G, Ross CA, Yasbin RE. 2003. Roles of YqjH and YqjW, homologs of the Escherichia coli UmuC/DinB or Y superfamily of DNA polymerases, in stationary-phase mutagenesis and UV-induced mutagenesis of Bacillus subtilis. J Bacteriol 185:2153–2160. doi: 10.1128/JB.185.7.2153-2160.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martin HA, Kidman AA, Socea J, Vallin C, Pedraza-Reyes M, Robleto EA. 2020. The Bacillus subtilis K-state promotes stationary-phase mutagenesis via oxidative damage. Genes (Basel) 11:190. doi: 10.3390/genes11020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsai HJ, Nelliat AR, Choudhury MI, Kucharavy A, Bradford WD, Cook ME, Kim J, Mair DB, Sun SX, Schatz MC, Li R. 2019. Hypo-osmotic-like stress underlies general cellular defects of aneuploidy. Nature 570:117–121. doi: 10.1038/s41586-019-1187-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sheltzer JM, Torres EM, Dunham MJ, Amon A. 2012. Transcriptional consequences of aneuploidy. Proc Natl Acad Sci USA 109:12644–12649. doi: 10.1073/pnas.1209227109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lin D, Gibson IB, Moore JM, Thornton PC, Leal SM, Hastings PJ. 2011. Global chromosomal structural instability in a subpopulation of starving Escherichia coli cells. PLoS Genet 7:e1002223. doi: 10.1371/journal.pgen.1002223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Robbins JB, Smith D, Belfort M. 2011. Redox-responsive zinc finger fidelity switch in homing endonuclease and intron promiscuity in oxidative stress. Curr Biol 21:243–248. doi: 10.1016/j.cub.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pribis JP, Garcia-Villada L, Zhai Y, Lewin-Epstein O, Wang AZ, Liu J, Xia J, Mei Q, Fitzgerald DM, Bos J, Austin RH, Herman C, Bates D, Hadany L, Hastings PJ, Rosenberg SM. 2019. Gamblers: an antibiotic-induced evolvable cell subpopulation differentiated by reactive-oxygen-induced general stress response. Mol Cell 74:785–800.e7. doi: 10.1016/j.molcel.2019.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shee C, Gibson JL, Rosenberg SM. 2012. Two mechanisms produce mutation hotspots at DNA breaks in Escherichia coli. Cell Rep 2:714–721. doi: 10.1016/j.celrep.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lewis K. 2010. Persister cells. Annu Rev Microbiol 64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 60.Balaban NQ, Helaine S, Lewis K, Ackermann M, Aldridge B, Andersson DI, Brynildsen MP, Bumann D, Camilli A, Collins JJ, Dehio C, Fortune S, Ghigo JM, Hardt WD, Harms A, Heinemann M, Hung DT, Jenal U, Levin BR, Michiels J, Storz G, Tan MW, Tenson T, Van Melderen L, Zinkernagel A. 2019. Definitions and guidelines for research on antibiotic persistence. Nat Rev Microbiol 17:441–448. doi: 10.1038/s41579-019-0196-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tenaillon O, Denamur E, Matic I. 2004. Evolutionary significance of stress-induced mutagenesis in bacteria. Trends Microbiol 12:264–270. doi: 10.1016/j.tim.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 62.Ram Y, Hadany L. 2012. The evolution of stress-induced hypermutation in asexual populations. Evolution 66:2315–2328. doi: 10.1111/j.1558-5646.2012.01576.x. [DOI] [PubMed] [Google Scholar]
- 63.Ram Y, Hadany L. 2014. Stress-induced mutagenesis and complex adaptation. Proc Biol Sci 281:20141025. doi: 10.1098/rspb.2014.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Maharjan R, Ferenci T. 2015. Mutational signatures indicative of environmental stress in bacteria. Mol Biol Evol 32:380–391. doi: 10.1093/molbev/msu306. [DOI] [PubMed] [Google Scholar]
- 65.Pal D, Pertot A, Shirole NH, Yao Z, Anaparthy N, Garvin T, Cox H, Chang K, Rollins F, Kendall J, Edwards L, Singh VA, Stone GC, Schatz MC, Hicks J, Hannon GJ, Sordella R. 2017. TGF-beta reduces DNA ds-break repair mechanisms to heighten genetic diversity and adaptability of CD44+/CD24- cancer cells. eLife 6:e21615. doi: 10.7554/eLife.21615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cipponi A, Goode DL, Bedo J, McCabe MJ, Pajic M, Croucher DR, Rajal AG, Junankar SR, Saunders DN, Lobachevsky P, Papenfuss AT, Nessem D, Nobis M, Warren SC, Timpson P, Cowley M, Vargas AC, Qiu MR, Generali DG, Keerthikumar S, Nguyen U, Corcoran NM, Long GV, Blay JY, Thomas DM. 2020. MTOR signaling orchestrates stress-induced mutagenesis, facilitating adaptive evolution in cancer. Science 368:1127–1131. doi: 10.1126/science.aau8768. [DOI] [PubMed] [Google Scholar]
- 67.Russo M, Crisafulli G, Sogari A, Reilly NM, Arena S, Lamba S, Bartolini A, Amodio V, Magri A, Novara L, Sarotto I, Nagel ZD, Piett CG, Amatu A, Sartore-Bianchi A, Siena S, Bertotti A, Trusolino L, Corigliano M, Gherardi M, Lagomarsino MC, Di Nicolantonio F, Bardelli A. 2019. Adaptive mutability of colorectal cancers in response to targeted therapies. Science 366:1473–1480. doi: 10.1126/science.aav4474. [DOI] [PubMed] [Google Scholar]
- 68.Melamed D, Nov Y, Malik A, Yakass MB, Bolotin E, Shemer R, Hiadzi EK, Skorecki KL, Livnat A. 2022. De novo mutation rates at the single-mutation resolution in a human HBB gene region associated with adaptation and genetic disease. Genome Res 32:488–498. doi: 10.1101/gr.276103.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Battesti A, Majdalani N, Gottesman S. 2011. The RpoS-mediated general stress response in Escherichia coli. Annu Rev Microbiol 65:189–213. doi: 10.1146/annurev-micro-090110-102946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mathieu A, Fleurier S, Frenoy A, Dairou J, Bredeche MF, Sanchez-Vizuete P, Song X, Matic I. 2016. Discovery and function of a general core hormetic stress response in E. coli induced by sublethal concentrations of antibiotics. Cell Rep 17:46–57. doi: 10.1016/j.celrep.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 71.Harris RS, Longerich S, Rosenberg SM. 1994. Recombination in adaptive mutation. Science 264:258–260. doi: 10.1126/science.8146657. [DOI] [PubMed] [Google Scholar]
- 72.Harris RS, Ross KJ, Rosenberg SM. 1996. Opposing roles of the Holliday junction processing systems of Escherichia coli in recombination-dependent adaptive mutation. Genetics 142:681–691. doi: 10.1093/genetics/142.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Foster PL, Trimarchi JM, Maurer RA. 1996. Two enzymes, both of which process recombination intermediates, have opposite effects on adaptive mutation in Escherichia coli. Genetics 142:25–37. doi: 10.1093/genetics/142.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dillingham MS, Kowalczykowski SC. 2008. RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol Mol Biol Rev 72:642–671. doi: 10.1128/MMBR.00020-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kuzminov A. 2011. Homologous recombination-experimental systems, analysis, and significance. EcoSal Plus 4. doi: 10.1128/ecosalplus.7.2.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Churchill JJ, Anderson DG, Kowalczykowski SC. 1999. The RecBC enzyme loads RecA protein onto ssDNA asymmetrically and independently of chi, resulting in constitutive recombination activation. Genes Dev 13:901–911. doi: 10.1101/gad.13.7.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McPartland A, Green L, Echols H. 1980. Control of recA gene RNA in E. coli: regulatory and signal genes. Cell 20:731–737. doi: 10.1016/0092-8674(80)90319-0. [DOI] [PubMed] [Google Scholar]
- 78.Wiktor J, Gynna AH, Leroy P, Larsson J, Coceano G, Testa I, Elf J. 2021. RecA finds homologous DNA by reduced dimensionality search. Nature 597:426–429. doi: 10.1038/s41586-021-03877-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Motamedi MR, Szigety SK, Rosenberg SM. 1999. Double-strand-break repair recombination in Escherichia coli: physical evidence for a DNA replication mechanism in vivo. Genes Dev 13:2889–2903. doi: 10.1101/gad.13.21.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McKenzie GJ, Harris RS, Lee PL, Rosenberg SM. 2000. The SOS response regulates adaptive mutation. Proc Natl Acad Sci USA 97:6646–6651. doi: 10.1073/pnas.120161797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pennington JM, Rosenberg SM. 2007. Spontaneous DNA breakage in single living Escherichia coli cells. Nat Genet 39:797–802. doi: 10.1038/ng2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nohmi T. 2006. Environmental stress and lesion-bypass DNA polymerases. Annu Rev Microbiol 60:231–253. doi: 10.1146/annurev.micro.60.080805.142238. [DOI] [PubMed] [Google Scholar]
- 83.McKenzie GJ, Lee PL, Lombardo MJ, Hastings PJ, Rosenberg SM. 2001. SOS mutator DNA polymerase IV functions in adaptive mutation and not adaptive amplification. Mol Cell 7:571–579. doi: 10.1016/S1097-2765(01)00204-0. [DOI] [PubMed] [Google Scholar]
- 84.Petrosino JF, Galhardo RS, Morales LD, Rosenberg SM. 2009. Stress-induced beta-lactam antibiotic resistance mutation and sequences of stationary-phase mutations in the Escherichia coli chromosome. J Bacteriol 191:5881–5889. doi: 10.1128/JB.00732-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bull HJ, Lombardo MJ, Rosenberg SM. 2001. Stationary-phase mutation in the bacterial chromosome: recombination protein and DNA polymerase IV dependence. Proc Natl Acad Sci USA 98:8334–8341. doi: 10.1073/pnas.151009798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Galhardo RS, Do R, Yamada M, Friedberg EC, Hastings PJ, Nohmi T, Rosenberg SM. 2009. DinB upregulation is the sole role of the SOS response in stress-induced mutagenesis in Escherichia coli. Genetics 182:55–68. doi: 10.1534/genetics.109.100735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Layton JC, Foster PL. 2003. Error-prone DNA polymerase IV is controlled by the stress-response sigma factor, RpoS, in Escherichia coli. Mol Microbiol 50:549–561. doi: 10.1046/j.1365-2958.2003.03704.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen J, Gottesman S. 2017. Hfq links translation repression to stress-induced mutagenesis in E. coli. Genes Dev 31:1382–1395. doi: 10.1101/gad.302547.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dong T, Schellhorn HE. 2009. Control of RpoS in global gene expression of Escherichia coli in minimal media. Mol Genet Genomics 281:19–33. doi: 10.1007/s00438-008-0389-3. [DOI] [PubMed] [Google Scholar]
- 90.Hastings PJ, Hersh MN, Thornton PC, Fonville NC, Slack A, Frisch RL, Ray MP, Harris RS, Leal SM, Rosenberg SM. 2010. Competition of Escherichia coli DNA polymerases I, II and III with DNA Pol IV in stressed cells. PLoS One 5:e10862. doi: 10.1371/journal.pone.0010862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pomerantz RT, Kurth I, Goodman MF, O'Donnell ME. 2013. Preferential D-loop extension by a translesion DNA polymerase underlies error-prone recombination. Nat Struct Mol Biol 20:748–755. doi: 10.1038/nsmb.2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hastings PJ, Ira G, Lupski JR. 2009. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet 5:e1000327. doi: 10.1371/journal.pgen.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wimberly H, Shee C, Thornton PC, Sivaramakrishnan P, Rosenberg SM, Hastings PJ. 2013. R-loops and nicks initiate DNA breakage and genome instability in non-growing Escherichia coli. Nat Commun 4:2115. doi: 10.1038/ncomms3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bull HJ, McKenzie GJ, Hastings PJ, Rosenberg SM. 2000. Evidence that stationary-phase hypermutation in the Escherichia coli chromosome is promoted by recombination. Genetics 154:1427–1437. doi: 10.1093/genetics/154.4.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shee C, Cox BD, Gu F, Luengas EM, Joshi MC, Chiu LY, Magnan D, Halliday JA, Frisch RL, Gibson JL, Nehring RB, Do HG, Hernandez M, Li L, Herman C, Hastings PJ, Bates D, Harris RS, Miller KM, Rosenberg SM. 2013. Engineered proteins detect spontaneous DNA breakage in human and bacterial cells. eLife 2:e01222. doi: 10.7554/eLife.01222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mei Q, Fitzgerald DM, Liu J, Xia J, Pribis JP, Zhai Y, Nehring RB, Paiano J, Li H, Nussenzweig A, Hastings PJ, Rosenberg SM. 2021. Two mechanisms of chromosome fragility at replication-termination sites in bacteria. Sci Adv 7:eabe2846. doi: 10.1126/sciadv.abe2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Anderson VE, Osheroff N. 2001. Type II topoisomerases as targets for quinolone antibacterials: turning Dr. Jekyll into Mr. Hyde. Curr Pharm Des 7:337–353. doi: 10.2174/1381612013398013. [DOI] [PubMed] [Google Scholar]
- 98.Wang JC. 1998. Moving one DNA double helix through another by a type II DNA topoisomerase: the story of a simple molecular machine. Q Rev Biophys 31:107–144. doi: 10.1017/s0033583598003424. [DOI] [PubMed] [Google Scholar]
- 99.Cannan WJ, Pederson DS. 2016. Mechanisms and consequences of double-strand DNA break formation in chromatin. J Cell Physiol 231:3–14. doi: 10.1002/jcp.25048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang Q, Lambert G, Liao D, Kim H, Robin K, Tung CK, Pourmand N, Austin RH. 2011. Acceleration of emergence of bacterial antibiotic resistance in connected microenvironments. Science 333:1764–1767. doi: 10.1126/science.1208747. [DOI] [PubMed] [Google Scholar]
- 101.Hicks LA, Bartoces MG, Roberts RM, Suda KJ, Hunkler RJ, Taylor TH, Jr, Schrag SJ. 2015. US outpatient antibiotic prescribing variation according to geography, patient population, and provider specialty in 2011. Clin Infect Dis 60:1308–1316. doi: 10.1093/cid/civ076. [DOI] [PubMed] [Google Scholar]
- 102.Kohanski MA, DePristo MA, Collins JJ. 2010. Sublethal antibiotic treatment leads to multidrug resistance via radical-induced mutagenesis. Mol Cell 37:311–320. doi: 10.1016/j.molcel.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. 2007. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 104.Reynolds MG. 2000. Compensatory evolution in rifampin-resistant Escherichia coli. Genetics 156:1471–1481. doi: 10.1093/genetics/156.4.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Petrosino JF, Pendleton AR, Weiner JH, Rosenberg SM. 2002. Chromosomal system for studying AmpC-mediated beta-lactam resistance mutation in Escherichia coli. Antimicrob Agents Chemother 46:1535–1539. doi: 10.1128/AAC.46.5.1535-1539.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Phillips I, Culebras E, Moreno F, Baquero F. 1987. Induction of the SOS response by new 4-quinolones. J Antimicrob Chemother 20:631–638. doi: 10.1093/jac/20.5.631. [DOI] [PubMed] [Google Scholar]
- 107.Miller C, Thomsen LE, Gaggero C, Mosseri R, Ingmer H, Cohen SN. 2004. SOS response induction by beta-lactams and bacterial defense against antibiotic lethality. Science 305:1629–1631. doi: 10.1126/science.1101630. [DOI] [PubMed] [Google Scholar]
- 108.Maiques E, Ubeda C, Campoy S, Salvador N, Lasa I, Novick RP, Barbe J, Penades JR. 2006. β-Lactam antibiotics induce the SOS response and horizontal transfer of virulence factors in Staphylococcus aureus. J Bacteriol 188:2726–2729. doi: 10.1128/JB.188.7.2726-2729.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nehring RB, Gu F, Lin HY, Gibson JL, Blythe MJ, Wilson R, Bravo Nunez MA, Hastings PJ, Louis EJ, Frisch RL, Hu JC, Rosenberg SM. 2016. An ultra-dense library resource for rapid deconvolution of mutations that cause phenotypes in Escherichia coli. Nucleic Acids Res 44:e41. doi: 10.1093/nar/gkv1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Schaaper RM, Dunn RL. 1987. Escherichia coli mutT mutator effect during in vitro DNA synthesis. Enhanced A.G replicational errors. J Biol Chem 262:16267–16270. doi: 10.1016/S0021-9258(18)49248-4. [DOI] [PubMed] [Google Scholar]
- 111.Moore JM, Correa R, Rosenberg SM, Hastings PJ. 2017. Persistent damaged bases in DNA allow mutagenic break repair in Escherichia coli. PLoS Genet 13:e1006733. doi: 10.1371/journal.pgen.1006733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dwyer DJ, Collins JJ, Walker GC. 2015. Unraveling the physiological complexities of antibiotic lethality. Annu Rev Pharmacol Toxicol 55:313–332. doi: 10.1146/annurev-pharmtox-010814-124712. [DOI] [PubMed] [Google Scholar]
- 113.Foti JJ, Devadoss B, Winkler JA, Collins JJ, Walker GC. 2012. Oxidation of the guanine nucleotide pool underlies cell death by bactericidal antibiotics. Science 336:315–319. doi: 10.1126/science.1219192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Veening J-W, Smits WK, Kuipers OP. 2008. Bistability, epigenetics, and bet-hedging in bacteria. Annu Rev Microbiol 62:193–210. doi: 10.1146/annurev.micro.62.081307.163002. [DOI] [PubMed] [Google Scholar]
- 115.Norman TM, Lord ND, Paulsson J, Losick R. 2015. Stochastic switching of cell fate in microbes. Annu Rev Microbiol 69:381–403. doi: 10.1146/annurev-micro-091213-112852. [DOI] [PubMed] [Google Scholar]
- 116.Denamur E, Matic I. 2006. Evolution of mutation rates in bacteria. Mol Microbiol 60:820–827. doi: 10.1111/j.1365-2958.2006.05150.x. [DOI] [PubMed] [Google Scholar]
- 117.Frenoy A, Bonhoeffer S. 2018. Death and population dynamics affect mutation rate estimates and evolvability under stress in bacteria. PLoS Biol 16:e2005056. doi: 10.1371/journal.pbio.2005056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Saint-Ruf C, Garfa-Traore M, Collin V, Cordier C, Franceschi C, Matic I. 2014. Massive diversification in aging colonies of Escherichia coli. J Bacteriol 196:3059–3073. doi: 10.1128/JB.01421-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sung HM, Yasbin RE. 2002. Adaptive, or stationary-phase, mutagenesis, a component of bacterial differentiation in Bacillus subtilis. J Bacteriol 184:5641–5653. doi: 10.1128/JB.184.20.5641-5653.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Campos M, San Millan A, Sempere JM, Lanza VF, Coque TM, Llorens C, Baquero F. 2020. Simulating the influence of conjugative-plasmid kinetic values on the multilevel dynamics of antimicrobial resistance in a membrane computing model. Antimicrob Agents Chemother 64:e00593-20. doi: 10.1128/AAC.00593-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zalis EA, Nuxoll AS, Manuse S, Clair G, Radlinski LC, Conlon BP, Adkins J, Lewis K. 2019. Stochastic variation in expression of the tricarboxylic acid cycle produces persister cells. mBio 10:e01930-19. doi: 10.1128/mBio.01930-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zheng EJ, Stokes JM, Collins JJ. 2020. Eradicating bacterial persisters with combinations of strongly and weakly metabolism-dependent antibiotics. Cell Chem Biol 27:1544–1552.e3. doi: 10.1016/j.chembiol.2020.08.015. [DOI] [PubMed] [Google Scholar]
- 123.Dorr T, Lewis K, Vulic M. 2009. SOS response induces persistence to fluoroquinolones in Escherichia coli. PLoS Genet 5:e1000760. doi: 10.1371/journal.pgen.1000760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Dorr T, Vulic M, Lewis K. 2010. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol 8:e1000317. doi: 10.1371/journal.pbio.1000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lewis K. 2012. Persister cells: molecular mechanisms related to antibiotic tolerance. Handb Exp Pharmacol 2012:121–133. doi: 10.1007/978-3-642-28951-4_8:121-33. [DOI] [PubMed] [Google Scholar]
- 126.Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. 2004. Bacterial persistence as a phenotypic switch. Science 305:1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 127.Radzikowski JL, Vedelaar S, Siegel D, Ortega AD, Schmidt A, Heinemann M. 2016. Bacterial persistence is an active sigmaS stress response to metabolic flux limitation. Mol Syst Biol 12:882. doi: 10.15252/msb.20166998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lopatkin AJ, Bening SC, Manson AL, Stokes JM, Kohanski MA, Badran AH, Earl AM, Cheney NJ, Yang JH, Collins JJ. 2021. Clinically relevant mutations in core metabolic genes confer antibiotic resistance. Science 371:eaba0862. doi: 10.1126/science.aba0862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Skulachev VP. 1998. Uncoupling: new approaches to an old problem of bioenergetics. Biochim Biophys Acta 1363:100–124. doi: 10.1016/S0005-2728(97)00091-1. [DOI] [PubMed] [Google Scholar]
- 130.Gonzalez-Flecha B, Demple B. 1995. Metabolic sources of hydrogen peroxide in aerobically growing Escherichia coli. J Biol Chem 270:13681–13687. doi: 10.1074/jbc.270.23.13681. [DOI] [PubMed] [Google Scholar]
- 131.Elez M, Murray AW, Bi LJ, Zhang XE, Matic I, Radman M. 2010. Seeing mutations in living cells. Curr Biol 20:1432–1437. doi: 10.1016/j.cub.2010.06.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Woo AC, Faure L, Dapa T, Matic I. 2018. Heterogeneity of spontaneous DNA replication errors in single isogenic Escherichia coli cells. Sci Adv 4:eaat1608. doi: 10.1126/sciadv.aat1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Pu Y, Zhao Z, Li Y, Zou J, Ma Q, Zhao Y, Ke Y, Zhu Y, Chen H, Baker MAB, Ge H, Sun Y, Xie XS, Bai F. 2016. Enhanced efflux activity facilitates drug tolerance in dormant bacterial cells. Mol Cell 62:284–294. doi: 10.1016/j.molcel.2016.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bergmiller T, Andersson AMC, Tomasek K, Balleza E, Kiviet DJ, Hauschild R, Tkacik G, Guet CC. 2017. Biased partitioning of the multidrug efflux pump AcrAB-TolC underlies long-lived phenotypic heterogeneity. Science 356:311–315. doi: 10.1126/science.aaf4762. [DOI] [PubMed] [Google Scholar]
- 135.El Meouche I, Dunlop MJ. 2018. Heterogeneity in efflux pump expression predisposes antibiotic-resistant cells to mutation. Science 362:686–690. doi: 10.1126/science.aar7981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Papkou A, Hedge J, Kapel N, Young B, MacLean RC. 2020. Efflux pump activity potentiates the evolution of antibiotic resistance across S. aureus isolates. Nat Commun 11:3970. doi: 10.1038/s41467-020-17735-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Torkelson J, Harris RS, Lombardo MJ, Nagendran J, Thulin C, Rosenberg SM. 1997. Genome-wide hypermutation in a subpopulation of stationary-phase cells underlies recombination-dependent adaptive mutation. EMBO J 16:3303–3311. doi: 10.1093/emboj/16.11.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Gonzalez C, Hadany L, Ponder RG, Price M, Hastings PJ, Rosenberg SM. 2008. Mutability and importance of a hypermutable cell subpopulation that produces stress-induced mutants in Escherichia coli. PLoS Genet 4:e1000208. doi: 10.1371/journal.pgen.1000208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Matic I. 2019. Mutation rate heterogeneity increases odds of survival in unpredictable environments. Mol Cell 75:421–425. doi: 10.1016/j.molcel.2019.06.029. [DOI] [PubMed] [Google Scholar]
- 140.Swenson PA, Schenley RL. 1974. Respiration, growth and viability of repair-deficient mutants of Escherichia coli after ultraviolet irradiation. Int J Radiat Biol Relat Stud Phys Chem Med 25:51–60. doi: 10.1080/09553007414550051. [DOI] [PubMed] [Google Scholar]
- 141.Weel-Sneve R, Kristiansen KI, Odsbu I, Dalhus B, Booth J, Rognes T, Skarstad K, Bjoras M. 2013. Single transmembrane peptide DinQ modulates membrane-dependent activities. PLoS Genet 9:e1003260. doi: 10.1371/journal.pgen.1003260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Courcelle J, Khodursky A, Peter B, Brown PO, Hanawalt PC. 2001. Comparative gene expression profiles following UV exposure in wild-type and SOS-deficient Escherichia coli. Genetics 158:41–64. doi: 10.1093/genetics/158.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Bos J, Zhang Q, Vyawahare S, Rogers E, Rosenberg SM, Austin RH. 2015. Emergence of antibiotic resistance from multinucleated bacterial filaments. Proc Natl Acad Sci USA 112:178–183. doi: 10.1073/pnas.1420702111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Huisman O, D'Ari R. 1981. An inducible DNA replication-cell division coupling mechanism in E. coli. Nature 290:797–799. doi: 10.1038/290797a0. [DOI] [PubMed] [Google Scholar]
- 145.Bi E, Lutkenhaus J. 1993. Cell division inhibitors SulA and MinCD prevent formation of the FtsZ ring. J Bacteriol 175:1118–1125. doi: 10.1128/jb.175.4.1118-1125.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Scheffers D, Driessen AJ. 2001. The polymerization mechanism of the bacterial cell division protein FtsZ. FEBS Lett 506:6–10. doi: 10.1016/s0014-5793(01)02855-1. [DOI] [PubMed] [Google Scholar]
- 147.Murawski AM, Brynildsen MP. 2021. Ploidy is an important determinant of fluoroquinolone persister survival. Curr Biol 31:2039–2050 e7. doi: 10.1016/j.cub.2021.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ross C, Pybus C, Pedraza-Reyes M, Sung HM, Yasbin RE, Robleto E. 2006. Novel role of mfd: effects on stationary-phase mutagenesis in Bacillus subtilis. J Bacteriol 188:7512–7520. doi: 10.1128/JB.00980-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Gomez-Marroquin M, Martin HA, Pepper A, Girard ME, Kidman AA, Vallin C, Yasbin RE, Pedraza-Reyes M, Robleto EA. 2016. Stationary-phase mutagenesis in stressed Bacillus subtilis cells operates by Mfd-dependent mutagenic pathways. Genes (Basel) 7:33. doi: 10.3390/genes7070033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ragheb MN, Thomason MK, Hsu C, Nugent P, Gage J, Samadpour AN, Kariisa A, Merrikh CN, Miller SI, Sherman DR, Merrikh H. 2019. Inhibiting the evolution of antibiotic resistance. Mol Cell 73:157–165.e5. doi: 10.1016/j.molcel.2018.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Portman JR, Brouwer GM, Bollins J, Savery NJ, Strick TR. 2021. Cotranscriptional R-loop formation by Mfd involves topological partitioning of DNA. Proc Natl Acad Sci USA 118. doi: 10.1073/pnas.2019630118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Gibson JL, Lombardo MJ, Thornton PC, Hu KH, Galhardo RS, Beadle B, Habib A, Magner DB, Frost LS, Herman C, Hastings PJ, Rosenberg SM. 2010. The sigma(E) stress response is required for stress-induced mutation and amplification in Escherichia coli. Mol Microbiol 77:415–430. doi: 10.1111/j.1365-2958.2010.07213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Million-Weaver S, Samadpour AN, Moreno-Habel DA, Nugent P, Brittnacher MJ, Weiss E, Hayden HS, Miller SI, Liachko I, Merrikh H. 2015. An underlying mechanism for the increased mutagenesis of lagging-strand genes in Bacillus subtilis. Proc Natl Acad Sci USA 112:E1096–E1105. doi: 10.1073/pnas.1416651112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Merrikh H, Kohli RM. 2020. Targeting evolution to inhibit antibiotic resistance. FEBS J 287:4341–4353. doi: 10.1111/febs.15370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Bunnell BE, Escobar JF, Bair KL, Sutton MD, Crane JK. 2017. Zinc blocks SOS-induced antibiotic resistance via inhibition of RecA in Escherichia coli. PLoS One 12:e0178303. doi: 10.1371/journal.pone.0178303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Mo CY, Culyba MJ, Selwood T, Kubiak JM, Hostetler ZM, Jurewicz AJ, Keller PM, Pope AJ, Quinn A, Schneck J, Widdowson KL, Kohli RM. 2018. Inhibitors of LexA autoproteolysis and the bacterial SOS response discovered by an academic-industry partnership. ACS Infect Dis 4:349–359. doi: 10.1021/acsinfecdis.7b00122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Alam MK, Alhhazmi A, DeCoteau JF, Luo Y, Geyer CR. 2016. RecA inhibitors potentiate antibiotic activity and block evolution of antibiotic resistance. Cell Chem Biol 23:381–391. doi: 10.1016/j.chembiol.2016.02.010. [DOI] [PubMed] [Google Scholar]
- 158.Amundsen SK, Spicer T, Karabulut AC, Londono LM, Eberhart C, Fernandez Vega V, Bannister TD, Hodder P, Smith GR. 2012. Small-molecule inhibitors of bacterial AddAB and RecBCD helicase-nuclease DNA repair enzymes. ACS Chem Biol 7:879–891. doi: 10.1021/cb300018x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Cowen LE, Lindquist S. 2005. Hsp90 potentiates the rapid evolution of new traits: drug resistance in diverse fungi. Science 309:2185–2189. doi: 10.1126/science.1118370. [DOI] [PubMed] [Google Scholar]
- 160.Shekhar-Guturja T, Gunaherath GM, Wijeratne EM, Lambert JP, Averette AF, Lee SC, Kim T, Bahn YS, Tripodi F, Ammar R, Dohl K, Niewola-Staszkowska K, Schmitt L, Loewith RJ, Roth FP, Sanglard D, Andes D, Nislow C, Coccetti P, Gingras AC, Heitman J, Gunatilaka AA, Cowen LE. 2016. Dual action antifungal small molecule modulates multidrug efflux and TOR signaling. Nat Chem Biol 12:867–875. doi: 10.1038/nchembio.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Watanabe K, Tanaka M, Yuki S, Hirai M, Yamamoto Y. 2018. How is edaravone effective against acute ischemic stroke and amyotrophic lateral sclerosis? J Clin Biochem Nutr 62:20–38. doi: 10.3164/jcbn.17-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Brynildsen MP, Winkler JA, Spina CS, MacDonald IC, Collins JJ. 2013. Potentiating antibacterial activity by predictably enhancing endogenous microbial ROS production. Nat Biotechnol 31:160–165. doi: 10.1038/nbt.2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Galvao AM, Wanderley MS, Silva RA, Filho CA, Melo-Junior MR, Silva LA, Streck EL, Dornelas de Andrade AF, Souza Maia MB, Barbosa de Castro CM. 2014. Intratracheal co-administration of antioxidants and ceftriaxone reduces pulmonary injury and mortality rate in an experimental model of sepsis. Respirology 19:1080–1087. doi: 10.1111/resp.12363. [DOI] [PubMed] [Google Scholar]
- 164.Li Z, Ma QQ, Yan Y, Xu FD, Zhang XY, Zhou WQ, Feng ZC. 2016. Edaravone attenuates hippocampal damage in an infant mouse model of pneumococcal meningitis by reducing HMGB1 and iNOS expression via the Nrf2/HO-1 pathway. Acta Pharmacol Sin 37:1298–1306. doi: 10.1038/aps.2016.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Elbaradey GF, Elshmaa NS, Hodeib H. 2016. Role of edaravone in management of septic peritonitis. J Anaesthesiol Clin Pharmacol 32:465–469. doi: 10.4103/0970-9185.194770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Rosenberg SM, Longerich S, Gee P, Harris RS. 1994. Adaptive mutation by deletions in small mononucleotide repeats. Science 265:405–407. doi: 10.1126/science.8023163. [DOI] [PubMed] [Google Scholar]
- 167.Foster PL, Trimarchi JM. 1994. Adaptive reversion of a frameshift mutation in Escherichia coli by simple base deletions in homopolymeric runs. Science 265:407–409. doi: 10.1126/science.8023164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Huisman O, D'Ari R, Gottesman S. 1984. Cell-division control in Escherichia coli: specific induction of the SOS function SfiA protein is sufficient to block septation. Proc Natl Acad Sci USA 81:4490–4494. doi: 10.1073/pnas.81.14.4490. [DOI] [PMC free article] [PubMed] [Google Scholar]