Abstract
Microglia have emerged as important players in brain aging and pathology. To understand how genetic risk for neurological and psychiatric disorders is related to microglial function, large transcriptome studies are essential. Here, we describe the transcriptome analysis of 255 primary human microglia samples isolated at autopsy from multiple brain regions of 100 human subjects. We performed systematic analyses to investigate various aspects of microglial heterogeneities, including brain region and aging. We mapped expression and splicing quantitative trait loci and showed that many neurological disease susceptibility loci are mediated through gene expression or splicing in microglia. Fine-mapping of these loci nominated candidate causal variants that are within microglia-specific enhancers, finding associations with microglia expression of USP6NL for Alzheimer’s disease and P2RY12 for Parkinson’s disease. We have built the most comprehensive catalog to date of genetic effects on the microglia transcriptome and propose candidate functional variants in neurological and psychiatric disorders.
Introduction
Microglia, the myeloid immune cells of the brain, are a cell type of compelling interest in the pathogenesis of several brain disorders1–3. Microglia play critical roles in inflammatory responses, regulation of brain homeostasis, neurodevelopment, and neurogenesis. Microglia are highly dynamic cells that are strongly influenced by different environmental signals which result in distinct phenotypes and functions across brain regions4–9. In addition, microglial functions vary across different ages, disease pathologies, and between sexes10–16. For decades, changes in microglial density, morphology, and transcriptional state have been observed in postmortem brain tissue of patients with neurological and psychiatric disorders17–21. This was initially suggested to reflect a response of the immune system to underlying disease processes. However, recent evidence from genome-wide association studies (GWAS) and other follow-up analyses has suggested that a proportion of the genetic risk of neurological and psychiatric diseases acts through myeloid cells22–25. As the myeloid cells of the nervous system, microglia may therefore play a causal role in disease.
To better understand this potential causal role of microglia in brain pathology and to identify microglia-related targets for treatment in the long-term, there is a critical need to identify which gene(s) are influenced by disease-associated genetic risk variants in microglia. This is a complicated task, as most of the common variants that have been identified are located outside protein-coding regions. These variants influence gene expression through complex regulatory mechanisms, such as altering enhancer activity, which often affect a gene beyond the nearest gene. Applying a combination of genetic and transcriptomic analyses on the same samples of a set of different donors, one can elucidate which gene is under influence of which genetic variant, by calling quantitative trait loci (QTLs). Investigations of QTLs in microglia have been limited by the availability of microglial samples from the number of subjects required to perform well-powered genomic analyses. Recently, Young et al. constructed expression QTLs (eQTLs) in primary human microglia (n = 93 individuals/samples), and detected 401 eQTLs, some of which colocalized with AD loci, including BIN126. However, the microglial transcriptome is highly heterogeneous compared to other cell types27, so larger sample sizes are needed to further identify statistically significant eQTLs. In addition, it is becoming increasingly clear that genetic risk can also be mediated through mRNA splicing28,29. For instance, the CD33 locus in AD influences CD33 splicing, resulting in isoforms with different biological and likely pathological functions29.
In the present study, we describe the Microglia Genomic Atlas (MiGA), a genetic and transcriptomic resource comprised of 255 primary human microglia samples isolated ex vivo from four different brain regions of 100 human subjects with neurodegenerative, neurological, or neuropsychiatric disorders, as well as unaffected controls (Fig. 1). We performed systematic analyses to investigate sources of microglial heterogeneity, including brain region, age, and sex. We further performed expression and splicing QTL analyses in each region and performed a meta-analysis across the four regions to increase our discovery power. We then performed colocalization and used fine-mapping and microglia-specific epigenomic data to prioritize genes and variants that influence neurological disease susceptibility through gene expression and splicing in microglia. With this approach, we have built the most comprehensive resource to date of cis-genetic effects on the microglial transcriptome and propose underlying molecular mechanisms of potentially causal functional variants in several brain disorders.
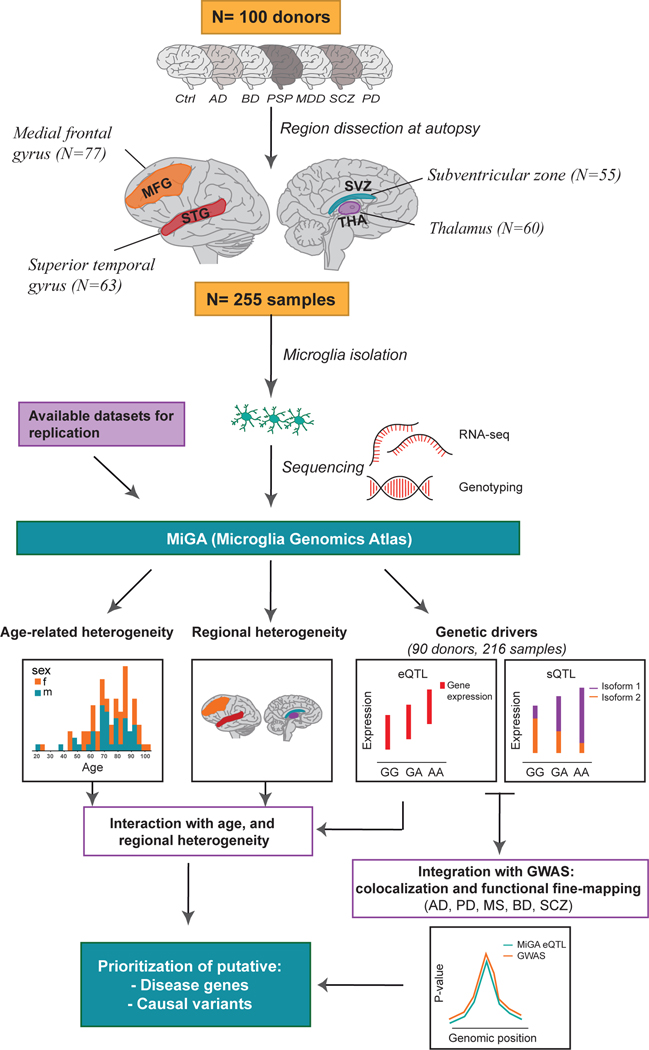
Figure 1. Overview of the Microglia Genomic Atlas (MiGA).
Primary human CD11b+ microglia were isolated at autopsy from 100 donors with neurological and psychiatric diseases, as well as unaffected subjects (controls) generating a total of 255 samples from four brain regions: medial frontal gyrus (MFG), superior temporal gyrus (STG), subventricular zone (SVZ) and thalamus (THA). Samples were isolated from two brain banks: the Netherlands Brain Bank (NBB) and the Neuropathology Brain Bank and Research CoRE at Mount Sinai Hospital. RNA was isolated and sequenced. Genome-wide genotyping was performed using DNA isolated from all donors. The following analysis were performed with the MiGA dataset: (i) age-related analysis; (ii) regional heterogeneity analysis by looking at the differentially expressed genes among the brain regions; (iii) expression quantitative trait loci (eQTL) analysis; (iv) splicing quantitative trait loci (sQTL) analysis; (v) colocalization and functional fine-mapping integrating the eQTL results with the most recent GWAS from five diseases: Alzheimer’s disease (AD), Parkinson’s disease (PD), Multiple sclerosis (MS), Bipolar disorder (BD) and Schizophrenia (SCZ).
Results
Biological factors driving the microglia transcriptome
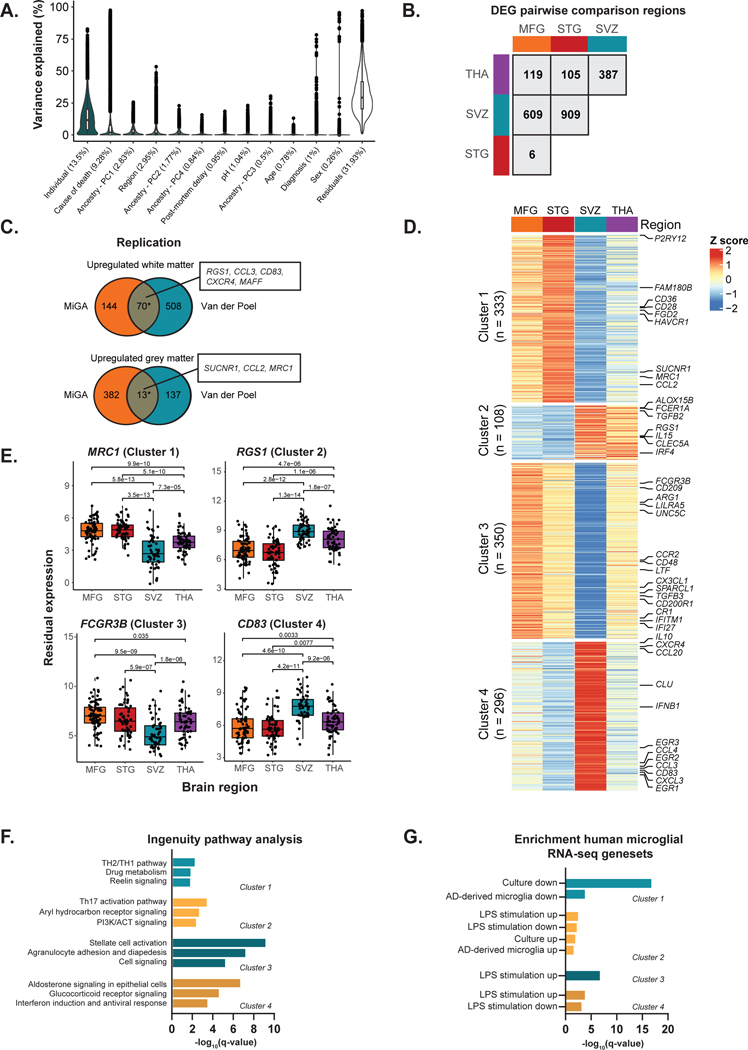
We isolated microglia cells from different brain regions of 115 donors. Details of the donors and quality controls are described in the Supplementary Note, Supplementary Fig. 1–4 and Supplementary Table 1. We explored the variation of a wide range of biological factors in driving the human microglia transcriptome before and after controlling for technical confounders (Supplementary Fig. 5 and 6). Using principal component analysis (PCA), we observed no clear separation by any factor after regressing out the technical factors, with the exception of age (Supplementary Fig. 7). Sex explained little variance (Fig. 2a), and we observed no differentially expressed genes between males and females (Supplementary Table 2). Using a linear mixed model to estimate the variance explained for a set of factors per gene30, we found that donor identity explained the most variance per gene (mean 13.5%) (Fig. 2a). Brain region explained comparatively little variance overall (mean 2.95%), but we identified a subset of genes that were strongly variable between regions. We performed pairwise comparisons of differential gene expression between each pair of regions, accounting for shared donors in a linear mixed model31 (Fig. 2b, Supplementary Tables 3–8). The largest number of differentially expressed genes (FDR < 0.05; |log2 fold change| > 1) were between the subventricular zone (SVZ) and the two cortical regions (609 in medial frontal gyrus (MFG), 909 in superior temporal gyrus (STG)), whereas comparing STG to MFG found the fewest (6 genes). We compared our findings in a published dataset of white and grey matter microglia5 and found small, but significant overlaps with our MFG vs SVZ comparison (upregulated OR = 18.4; P < 1 × 10−16, downregulated OR = 4.83; P = 9 × 10−6; Fisher’s exact test; Fig. 2c).
Figure 2. Regional heterogeneity analysis.
A) Distributions of variance explained per gene for the non-technical factors. Mean variance explained by each factor is in brackets. Data are presented as a percentage (%) of total variance explained. Box plots show median, box spans first to third quartiles, and whiskers extends 1.5 times the interquartile range (IQR) from the box. B) Number of differentially expressed genes by pairwise region comparison (FDR < 0.05 and | logFC | >1). C) Replication analysis between the medial frontal gyrus (MFG) vs subventricular zone (SVZ) differentially expressed genes and a published dataset of microglia samples isolated from white (n = 5) and grey matter (n = 11) of controls5. Asterisk indicates significant enrichment by a one-sided Fisher’s exact test (upregulated OR = 18.4; P < 1 × 10−16, downregulated OR = 4.83; P = 9 × 10−6). Selected genes in overlap are highlighted. D) Heatmap of K-means clustering of 1,087 differentially expressed genes from the pairwise comparison. K-means was performed on z-scored values of the median per region of voom transformed expression. The colors represent row scaled z-score levels: red and blue indicate high and low relative region expression, respectively. n refers to the number of differentially expressed genes in each cluster. E) Examples of differentially expressed genes in each region. P-values (two-sided) are FDR-adjusted from the linear mixed model of the differential expression analysis. Box plots show median, box spans first to third quartiles, and whiskers extend 1.5 times the interquartile range (IQR) from the box. F) Functional Enrichment Analysis of each K-means cluster using Ingenuity Pathway Analysis (IPA). Only significantly enriched terms shown (q-value < 0.05). G) Enrichment analysis with curated human microglia RNA-seq gene sets33,34 using one-sided Fisher’s exact test at Bonferroni adjusted P < 0.05.
We then performed k-means clustering32 of the genes found in the pairwise comparisons. We identified k = 4 as the optimal clustering partitioning after minimizing the total within-cluster sum of squares (WSS) (Fig. 2d). Cluster 1 contained genes that were upregulated in the cortical regions compared to subcortical regions, such as P2RY12, CD36, and MRC1, and was enriched in genes that were downregulated in AD brain33 and in response to in vitro culture34. Cluster 2 contained genes that were downregulated in the cortex compared to the subcortical brain (e.g. FCER1A, IL15, RGS1). Cluster 3 contained genes specifically downregulated in the SVZ (e.g. CX3CL1, CCR2, FCGR3B) and cluster 4 contained genes upregulated in SVZ (e.g. IL10, CLU, CD83), compared to the other three regions (Fig. 2e). We found that genes implicated in inflammatory processes were highly expressed in cluster 2 (Fig. 2f), whereas genes related to homeostatic functions of microglial cells were mainly present in cluster 1. Cluster 4 included genes that were involved in biological functions related to hormonal signaling and interferon response (Fig. 2f). Analysis of upstream regulators of the four clusters using Ingenuity Pathway Analysis (IPA) was inconclusive (Supplementary Table 9). We overlapped the region-specific genes with gene sets altered after stimulation with lipopolysaccharide (LPS) or interferon-gamma (IFNγ; generated in-house), following in vitro culture34, and in microglia derived from AD patient brains compared to controls33. Cluster 1 genes were enriched in genes that were downregulated in AD brain and in response to in vitro culture. Cluster 2 genes were significantly enriched for genes upregulated following in vitro culture34 and in AD-derived microglia33. Cluster 2, 3 and 4 genes showed enrichment with LPS responsive genes in both directions (Fig. 2g, Supplementary Table S9).
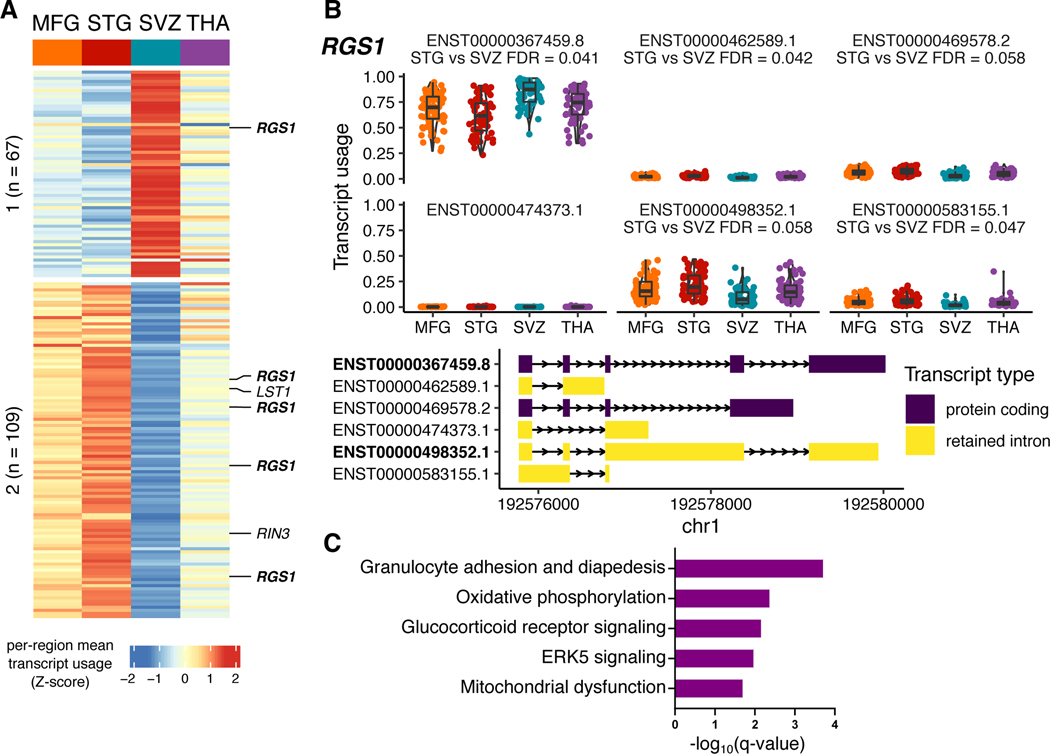
We examined changes in splicing between microglia regions using a differential transcript usage (DTU) framework. 176 transcripts in 132 genes had evidence of DTU (log odds ratio > 1; empirical FDR < 0.1), with a majority of transcripts coming from comparisons with the SVZ (Extended Data Fig. 1a). 31 DTU genes were also differentially expressed between pairs of regions (OR = 5.47, P = 2.9 × 10−12; Fisher exact test). RGS1 is an example of a gene with a shift in the ratio of the two most abundant isoforms in the SVZ compared to the other regions (Extended Data Fig. 1b). The regional DTU gene set includes genes involved in mitochondrial functions, glucocorticoid receptor signaling pathways, and host defense against infections (Extended Data Fig. 1c), pathways also observed in the regional expression analysis.
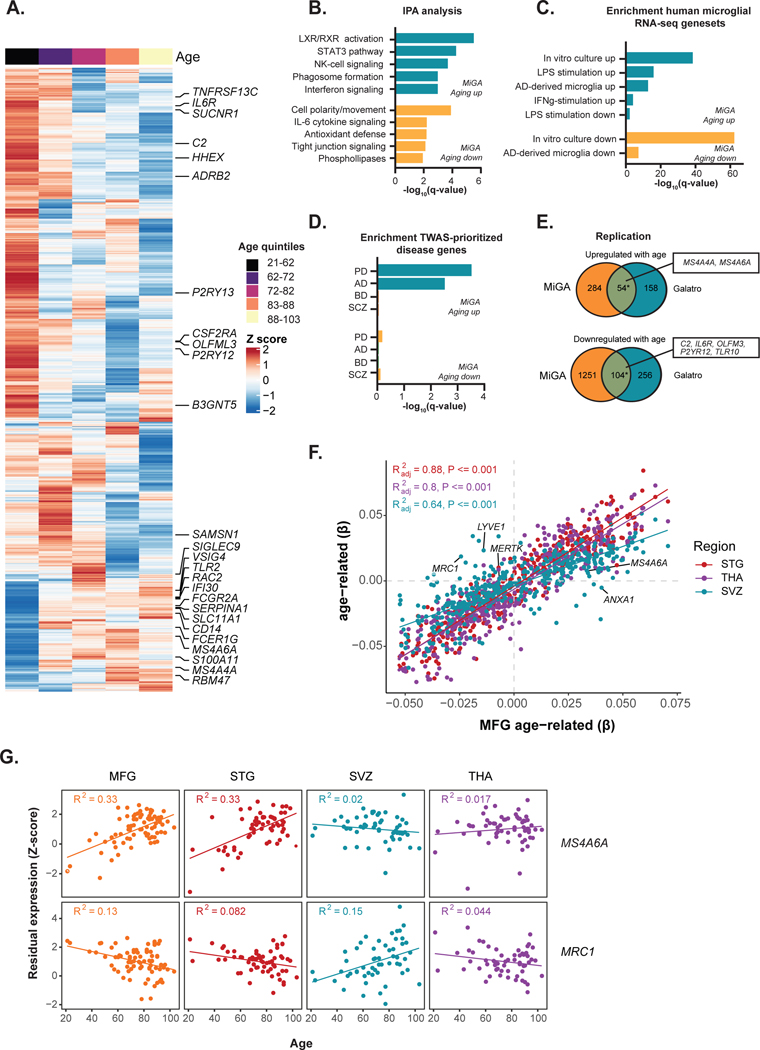
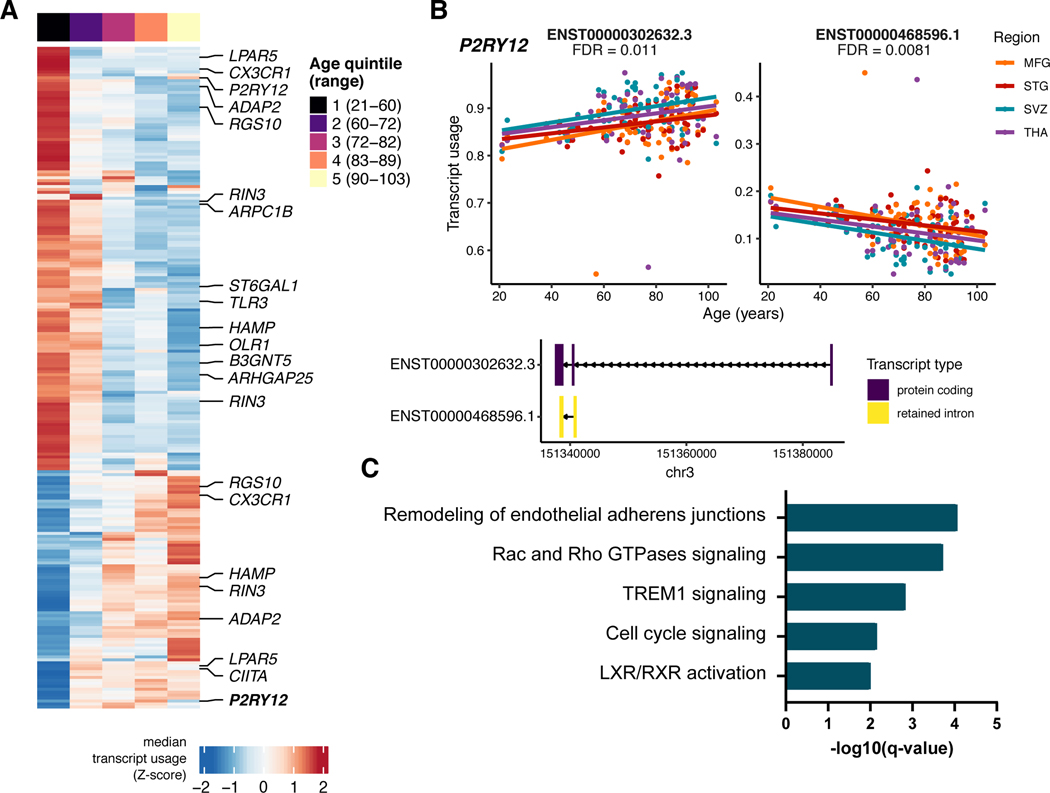
We explored the effect of diagnosis on the microglia transcriptome and detected 24 genes, such as MCF2 and AIDH3B1, differentially expressed in the dementia group compared to controls (FDR < 0.05; Supplementary Table 10). No significant gene expression changes were found for PD, MDD and BD/SCZ (Supplementary Tables 11–13). To assess the effect of aging on the microglial transcriptome, we fitted a linear mixed model accounting for shared donors across all four regions. We observed 1,693 genes (338 up, 1,355 down at FDR < 0.05) associated with the chronological age of subjects (Fig. 3A, Supplementary Table 14). Similarly, we found 225 transcripts from 150 genes exhibiting DTU with age (FDR < 0.1) (Extended Data Fig. 2a), where the balance between a long and short isoform shifts over age (Extended Data Fig. 2b). 36 of these DTU genes also showed an association with age at the gene expression level (OR = 3.47, P = 7 × 10−8, Fisher Exact test). Genes upregulated in aging were significantly enriched for several Gene Ontology (GO) biological processes including lipid metabolism, immune responses such as Natural Killer (NK) cell and interferon signaling, and phagosome formation (Fig. 3b). The downregulated genes were significantly enriched for cell motility, polarity, IL-6 cytokine signaling (Fig. 3b), and for genes also downregulated following in vitro culture34 and in AD-derived microglia33 (Fig. 3c). The genes associated with aging DTU were enriched in similar functions (Extended Data Fig. 2c).
Figure 3. Age-related analysis.
A) Heatmap of 1,693 genes associated with age (FDR < 0.05). Each gene (row) plotted as a Z-score of median expression averaged first by donor (across multiple regions) and then by age quintiles with 20 donors each. Genes are ordered by Ward’s hierarchical clustering. B) Ingenuity Pathway Analysis of age-related genes. Only significantly enriched terms shown (q-value < 0.05). C) Enrichment analysis with curated human microglia RNA-seq gene sets33,34 using a one sided Fisher’s exact test at Bonferroni-adjusted P < 0.05. D) Genes associated with age show overrepresentation for TWAS prioritized genes for Alzheimer’s (AD) or Parkinson’s disease (PD), but not for genes in Schizophrenia or Bipolar disorder. P-value based on one-sided Fisher’s exact test. E) Replication analysis with an independent dataset of human microglia samples, 49 healthy controls with ages between 31 and 102 years old16. Asterisk indicates significant enrichment by a one-sided Fisher’s exact test (upregulated genes OR = 23.4; P < 1 × 10−16, downregulated genes OR = 5.97; P < 1 × 10−16). Selected overlapping genes are highlighted. F) Scatter plot showing the size correlation of the age-related genes by brain region. Only the genes that are significant (FDR < 0.05) in at least one region are shown. The x-axis shows the beta values in the MFG and the y-axis shows the betas in other brain regions (STG, SVZ and THA). G) Scatter plot showing the Z-score transformed residual expression for selected genes (MRC1 and MS4A6A) by age and brain region.
We next used gene sets prioritized by transcriptome-wide association study (TWAS) in different diseases35–38 (Supplementary Table 15). The upregulated genes in chronological aging showed overrepresentation for genes in AD (e.g. MS4A6A, FCER1G, and CR1) or PD (e.g. BST1, PTPN22, and TNFSF13) GWAS loci, but not for genes in schizophrenia (SCZ) or bipolar disorder (BD) (Fig. 3d). We replicated our findings using an external microglia aging dataset from the parietal cortex16, and from peripheral blood39 (Supplementary Fig. 8). The number of genes that overlapped between the datasets was small, but significant (upregulated genes OR = 23.4; P < 1 × 10−16, downregulated genes OR = 5.97; P < 1 × 10−16; Fisher’s Exact test; Fig. 3e).
It is not known whether the impact of aging on the microglial transcriptome is uniform throughout the human brain. Although most genes showed concordant effect size and direction across regions (Fig. 3f), 91 genes demonstrated age-region relationships after fitting an interaction term model (FDR < 0.05) (Supplementary Fig. 9, Supplementary Table 16). 35 genes (e.g., MRC1, CD24) changed specifically in SVZ and not in other regions (adj. R2 > 3* interquartile range (IQR)) (Fig. 3g; Supplementary Fig. 9). Together, our results indicate that the microglial phenotype ages in a generally uniform manner across brain regions, with a distinct aging trajectory observed in a minority of genes.
Genetic regulatory effects in microglia
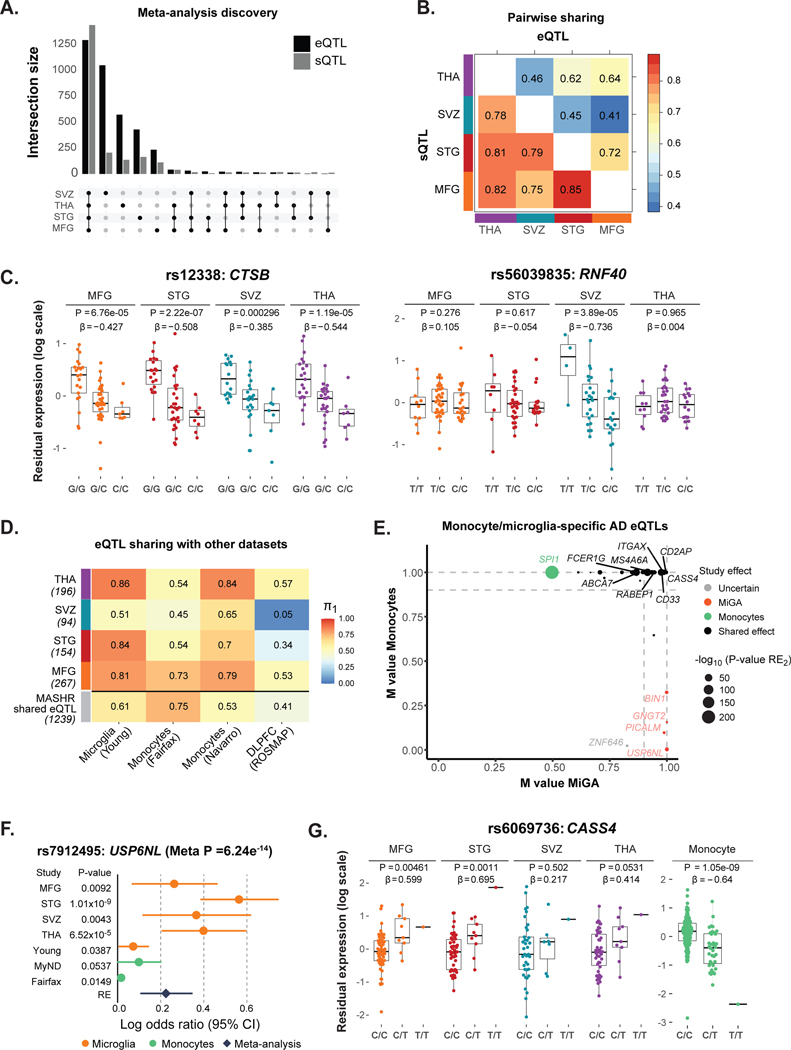
We performed cis-eQTL and cis-sQTL analyses in primary human microglia from four different brain regions. After QC, 216 samples from 90 individuals of European ancestry were used for the analysis (Supplementary Fig. 10). In the region-specific analysis, we observed between 67 and 199 genes with a cis-eQTL (eGenes) and 253 to 426 genes with a cis-sQTL (sGenes) per region (FDR < 0.05; Supplementary Table 17, Supplementary Fig. 11). cis-QTL discovery was highly correlated with the sample size for each region (Spearman’s ρ = 0.8 for eQTLs, and 1 for sQTLs), contributing to the low number of eQTLs detected in the region by region analysis. We therefore performed a meta-analysis across all four regions using the multivariate adaptive shrinkage (mashR)40 (v0.2–11) method to increase power and to assess shared QTLs between regions. In total, we identified 3,611 eGenes and 4,614 sGenes, at a local false sign rate (lfsr) < 0.05 in at least one region (Fig. 4a, Supplementary Tables 18–19).
Figure 4. Genetic regulatory effects in microglia.
A) Number of genes with a cis-eQTL (eGenes) and cis-sQTL (sGenes) at local false sign rate (lfsr) < 0.05, from a meta-analysis across all four brain regions using mashR. B) Pairwise shared eQTLs (upper triangle) and sQTLs (lower triangle) across the four brain regions. Numbers represent the proportion of significant effects (lfsr < 0.05) that are shared in magnitude (i.e. effect estimates that are in the same direction and within a factor of 2 in size). C) Examples of shared (CTSB gene, rs12338) and region-specific effect (RNF40, rs56039835). eQTL boxplots with residual gene expression (PEER adjusted) per individual stratified by genotype. The eQTL nominal P-value and effect size from the linear regression model are listed on top. Box plots show median, box spans first to third quartiles, and whiskers extend 1.5 times the interquartile range (IQR) from the box. D) Replication of MiGA eQTLs effects (region-by-region analysis q-value < 0.10 and mashR results at lfsr < 0.05) compared to four external eQTL datasets: microglia26, monocytes41,42, and dorsolateral prefrontal cortex (ROSMAP)43. The proportion of replication is measured by Storey’s π1. E) Meta-analysis results for colocalized eGenes in AD loci. The m-values represent the posterior probability that the effect exists in each study (i.e. MiGA for microglia, or Navarro and Fairfax for monocytes), as reported by METASOFT. Small m-values (< 0.1) suggest that the gene and the SNP do not have an association in the study; large m-values (> 0.9) suggest that the gene and the SNP have a strong association. Otherwise, the prediction is uncertain. The x-axis shows the maximum m-values among the four brain regions in MiGA, and the y-axis shows maximum m-values for monocytes41,42. Colors indicate cell type: orange for the genes with strong effect in MiGA only, green for monocytes and black for the shared effects between microglia and monocytes. F) Example of microglia-specific eQTL found only in MiGA. The gene USP6NL (rs7912495) has significant effect in all four brain regions from MiGA but does not have an effect in other datasets (MiGA dataset includes MFG, STG, SVZ and THA: N = 216 samples; Young: N = 93 samples; MyND: N = 180 samples; Fairfax: N = 300 samples). Error bars indicate the log odds ratio (95% confidence interval). G) Example of discordant eQTL effects for CASS4 (rs6069736) between microglia and monocytes. The nominal P-values are from the linear regression model in the region-by-region eQTL analysis. Box plots show median, box spans first to third quartiles, and whiskers extend 1.5 times the interquartile range (IQR) from the box.
We observed a high degree of eQTL sharing (effect estimates that are in the same direction and are similar sizes, within a factor of 2) between MFG and STG (72%), as expected, given that these two cortical regions have similar gene expression patterns (Fig. 4b, upper triangle). Microglia from SVZ exhibited lower pairwise sharing of eQTLs with other regions, with the lowest sharing by magnitude observed between SVZ and MFG (41%), consistent with observed transcriptomic differences between these two regions. For sQTLs, we found overall higher regional sharing effects compared to eQTLs, but still following the same trends as for eQTLs (Fig. 4b, lower triangle). In addition, while the majority of the eQTLs were shared across regions, we identified 1,791 (49.6%) eQTLs with a stronger effect in one region than in any other (lfsr < 0.05 and > 2-fold effect size in one region compared to others). Microglia from the SVZ had the most region-specific effects with 1,045, most likely because the transcriptomic profile of this region is most distinct (Supplementary Table 20). We include examples of shared and region-specific eQTLs (Fig. 4c).
To assess eQTL reproducibility and cell-type specificity, we compared MiGA eQTLs with four other external eQTL datasets, including microglia26, monocytes41,42, and bulk brain dorsolateral prefrontal cortex (DLPFC)43 using the q-value π1 metric44. We found that eQTL sharing was both cell type- and region-dependent (Fig. 4d), with the highest sharing between MiGA and the Young et al. microglia (π1 = 0.81–0.86), but with a lower sharing in the SVZ (π1 = 0.51). Sharing with monocyte eQTLs was generally slightly lower than with microglia and sharing with bulk DLPFC eQTLs was lowest. Together, these results highlight shared genetic regulation between microglia and monocytes, which is only partly captured in whole-tissue brain data22.
We performed a cross-study eQTL meta-analysis (MiGA, Young26, MyND42, and Fairfax41) using METASOFT45) to assess the sharing of effects between distinct cell types. We focused on genes associated with AD23,46 through METASOFT’s m-value, which is the posterior probability that the effect exists in a particular study. The comparison of m-values between microglia (MiGA) and monocytes (MyND42, and Fairfax41) showed that a large number of eQTLs in AD loci have shared effects between these two cell types, for example, MS4A6A, RABEP1, CD33, FCER1G, and ABCA7. However, there were eQTLs with specific microglial effects that were absent in monocytes (e.g., BIN1, PICALM, USP6NL and GNGT2; Fig. 4e). The USP6NL gene is an example of an eQTL with a strong effect in MiGA but not in monocytes or Young et al. (Fig. 4f). Generally, directions of effect between monocytes and microglia were concordant (Supplementary Fig. 12), with the exception of CASS4. eQTLs for CASS4 are significant in both MiGA and monocytes (MyND) but with opposite directions of effect (Fig. 4g), suggesting that the causal variant is located in a complex regulatory element where both enhancing and repressing mechanisms are at play.
Genetic effects in microglia mediate neurological disease
We next explored whether disease-associated genetic variants may potentially act through microglia eQTL or sQTL using the coloc R package47 (v3.2–1) and publicly available GWAS summary statistics for AD23,46,48,49, PD50, SCZ51, BD35, and Multiple Sclerosis (MS)52.
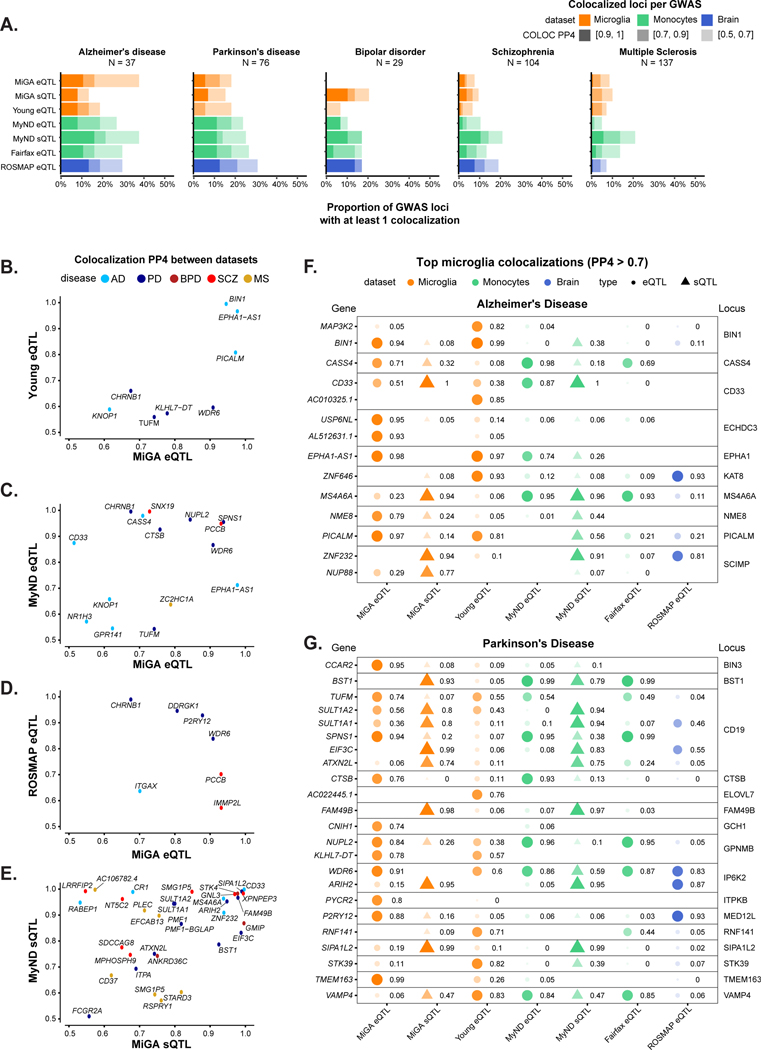
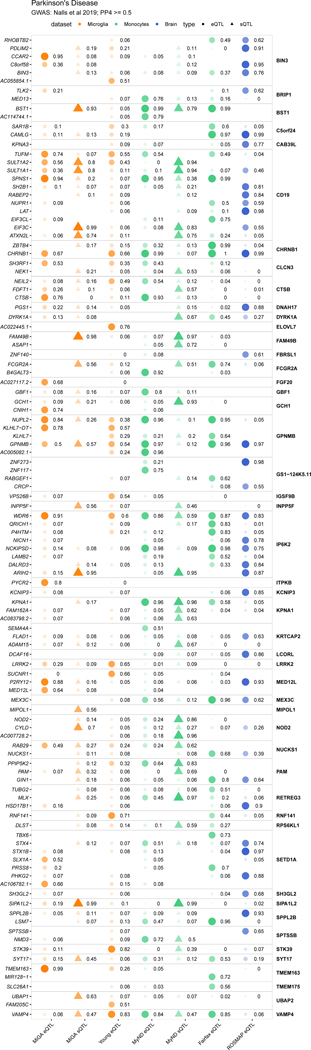
We compared our MiGA QTLs to the same set of published microglia, monocytes, and bulk brain tissue QTLs as before. AD and PD had the highest number of colocalizing loci in each QTL dataset, compared to the other diseases (Fig. 5a, Supplementary Table 21), with 10–30% of loci containing at least one colocalized gene, depending on the stringency of the H4 posterior probability (PP4), with lower proportions observed in BD, SCZ and MS.
Figure 5. Summary of colocalization analyses.
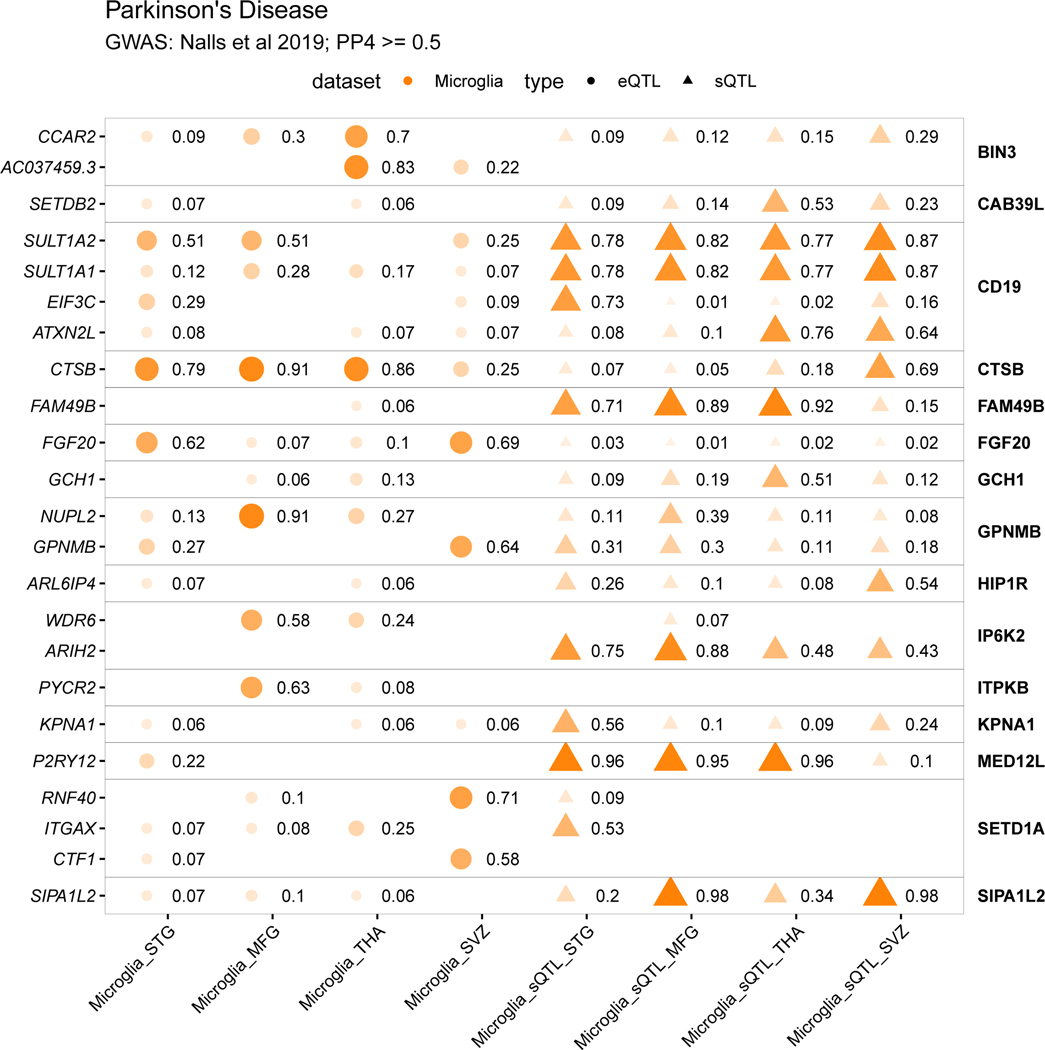
A) The proportion of GWAS loci that have at least one colocalized gene in each QTL dataset. Fill opacity used to represent numbers of loci at different stringency levels for colocalization posterior probability H4 (PP4): 0.5–0.7 (lightest), 0.7–0.9 (medium); 0.9–1 (darkest). Bars are colored by the cell or tissue type of the QTLs: microglia (orange), monocytes (green), or dorsolateral prefrontal cortex (DLPFC; blue). N refers to the number of GWAS loci. B-E) Pairwise comparison of the coloc PP4 for genes between QTL datasets. Points are colored by disease. B), C), D) compare the MiGA microglia eQTLs to the Young et al. microglia eQTLs, MyND monocyte eQTLs, and ROSMAP DLPFC eQTLs, respectively. E) Comparison of the MiGA splicing QTLs and the MyND monocyte splicing QTLs. F) All genes in the AD GWAS that have a PP4 > 0.7 in one of the three microglia QTL datasets. Shape opacity and size scaled to the magnitude of PP4. Circles represent colocalizations with expression QTLs and triangles represent those with splicing QTLs. G) The same for Parkinson’s Disease.
We then compared different QTL datasets to find shared evidence of colocalization at the level of individual genes within a GWAS locus (Fig. 5b–e). The sharing between our microglia and previously published microglia26 was low (Fig. 5b), with only a few known loci in AD and PD (BIN1, PICALM, CHRNB1), presumably due to lower power in the Young et al. data compared to our multi-tissue meta-analysis. Overall, 11% of MiGA eQTL colocalizations could be reproduced in the Young et al. data, and 15% of the Young et al. colocalizations could be found in the MiGA data, at a relaxed PP4 > 0.5, whereas sharing between the two monocyte datasets41,42 was 17–24% with the same parameters (Supplementary Fig. 13). Substantially lower sharing (18–21%) was observed between the MiGA eQTLs and those of our lab’s recent monocyte dataset (Fig. 5c; Supplementary Fig. 13) than between the respective splicing QTLs (23–53%) (Fig. 5e; Supplementary Fig. 13). This suggests that splicing QTLs are less cell type-specific, presumably due to the association with distinct types of regulatory elements.
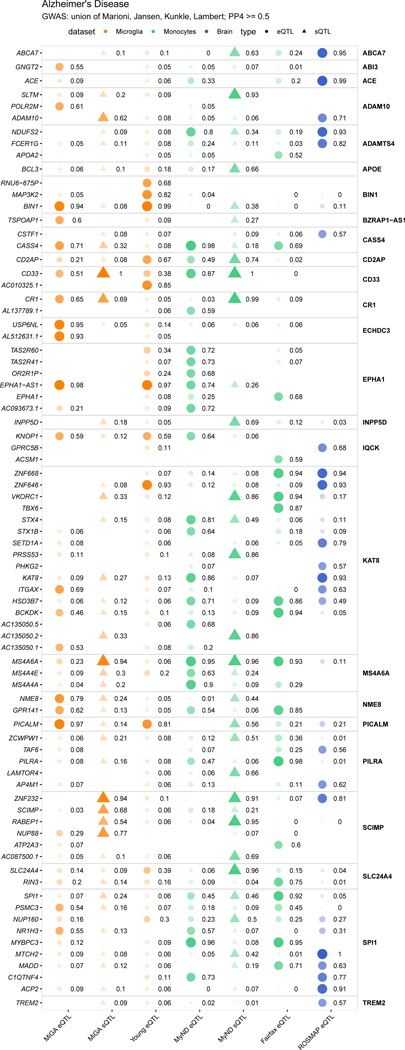
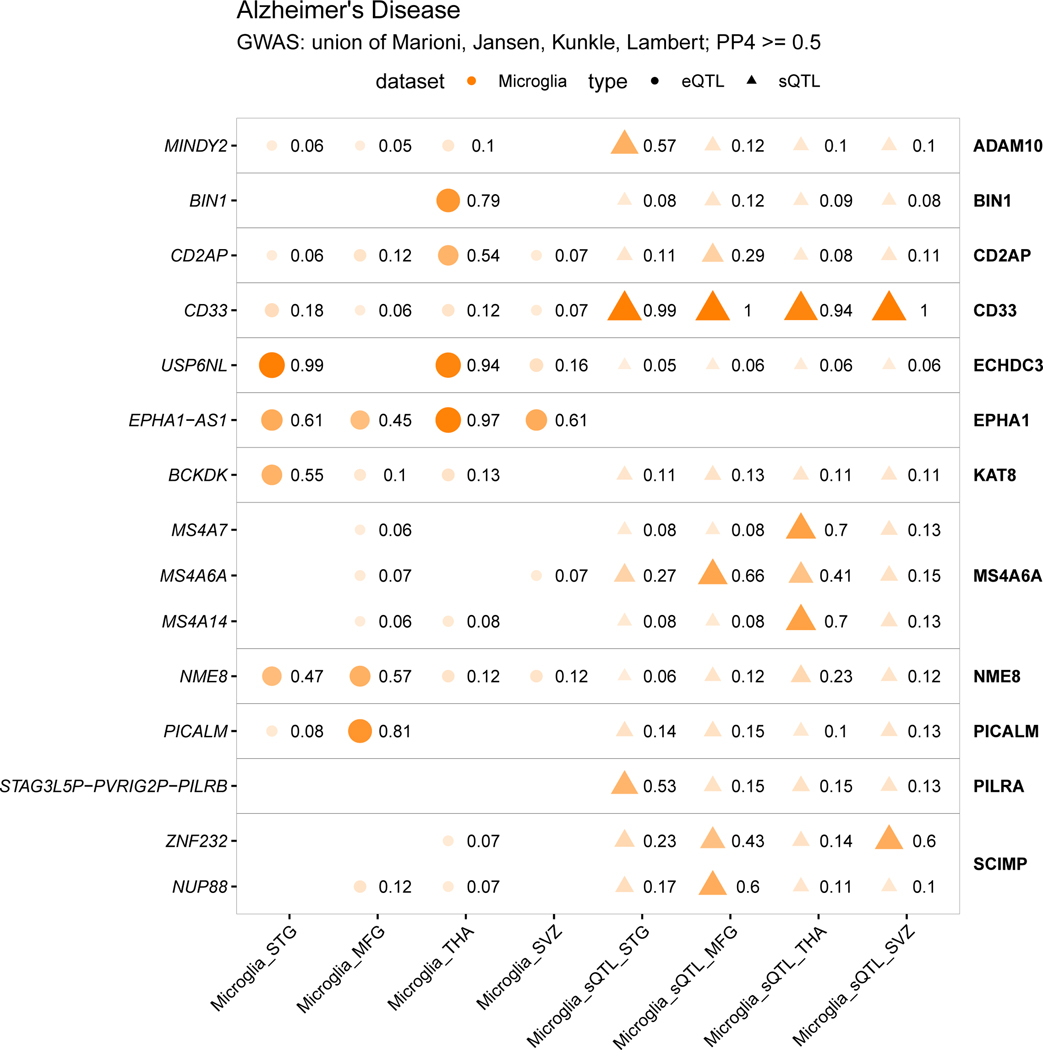
We present colocalizations in AD (Fig. 5f) and PD (Fig. 5g) in each QTL dataset. We emphasize microglia by including only genes that colocalize with one of the three microglia QTL datasets at PP4 > 0.7. In each disease there are genes that appear to be microglia-specific (BIN1, PYCR2), shared between microglia and monocytes (CASS4, CTSB), and shared between microglia and brain (ZNF646, P2RY12). We also observe multiple splicing QTLs, some previously reported (CD33, FAM49B), and some unreported (MS4A6A, BST1). We present full plots for all colocalizations in each disease in the supplementary materials (Extended Data Fig. 3–6; Supplementary Fig. 14–19).
Neurological disease loci regulate microglia gene expression
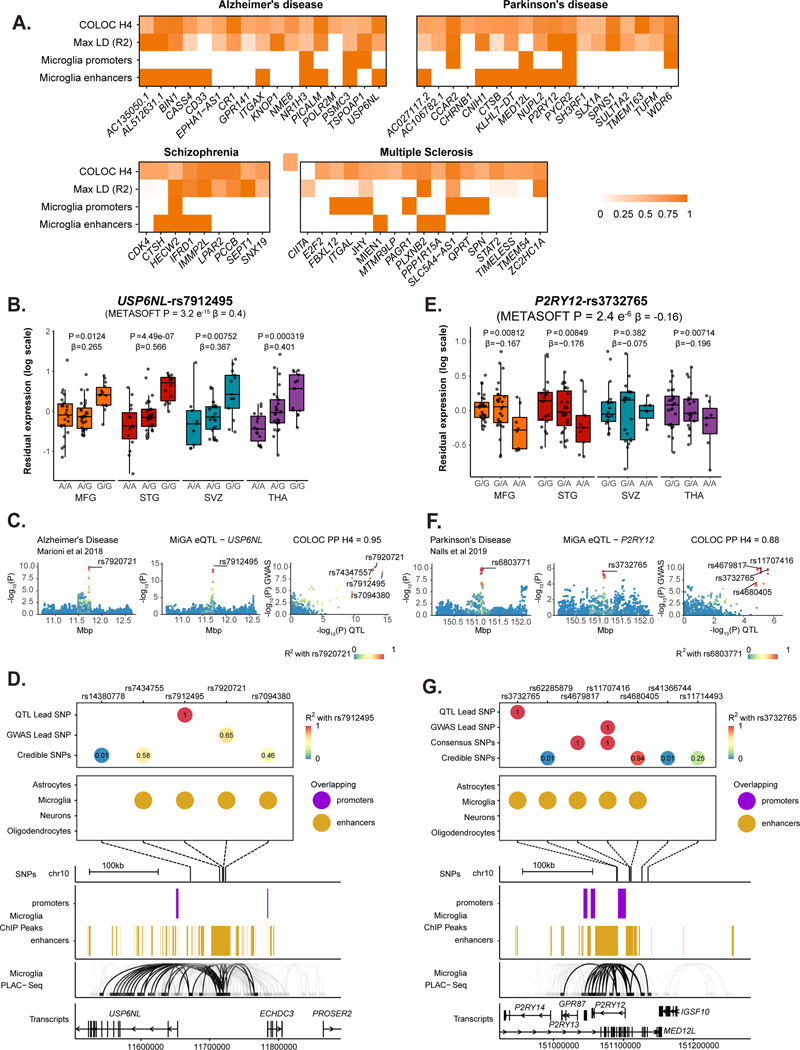
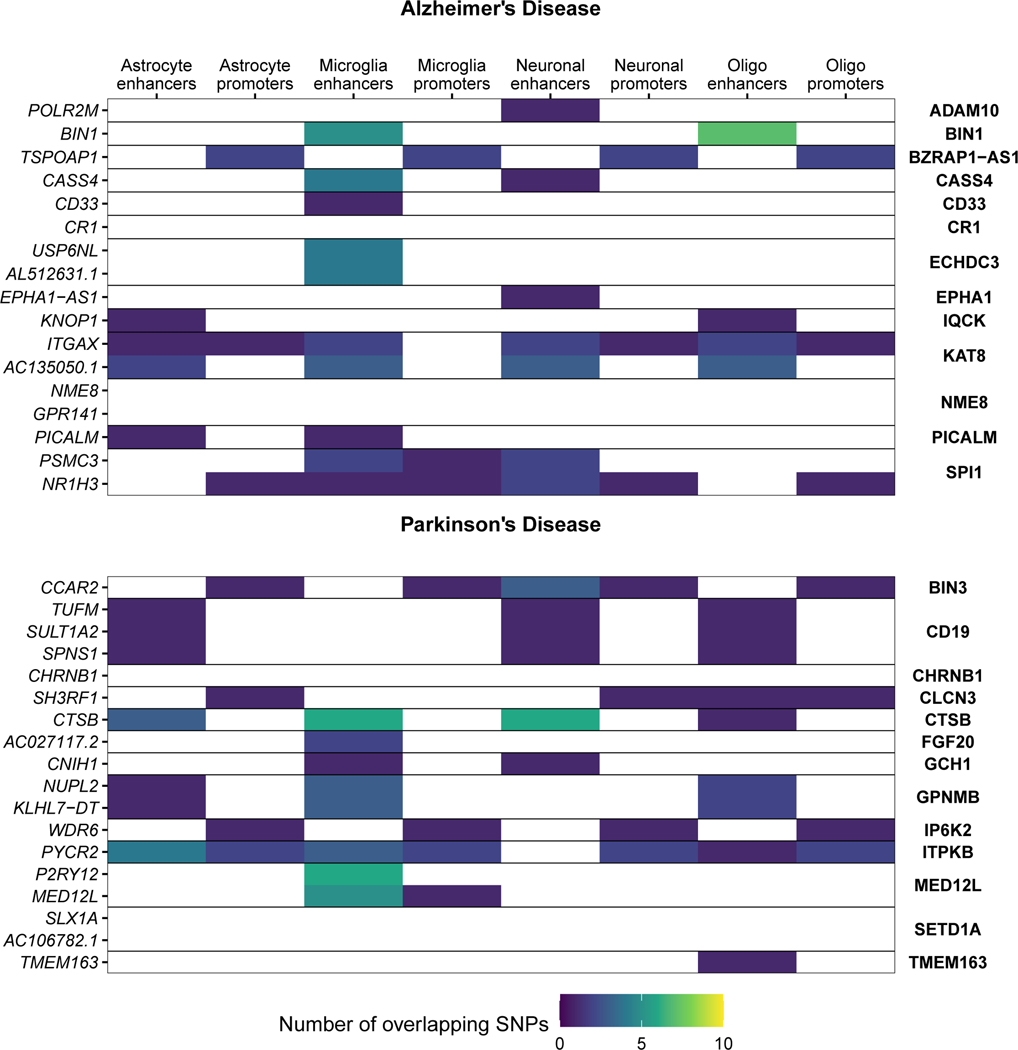
We next examined whether the microglia eQTLs that colocalized with disease GWAS loci were due to genetic variation within microglia-specific regulatory regions. As further outlined in the Supplementary Note. We found that 10 out of 17 genes colocalizing in AD, 8 out of 18 in PD, 4 out of 9 in SCZ, and 3 out of 17 in MS include SNPs that overlap with microglial enhancers (Fig. 6a; Extended Data Fig. 7; Supplementary Fig. 20). This approach allowed us to prioritize disease loci that likely act on disease risk by modulating gene expression specifically in microglia. Here we discuss two examples.
Figure 6. Enhancer-promoter interaction data links GWAS variants to microglia-specific regulatory regions.
A) Overview of fine-mapping and epigenomic overlap analyses for all eQTL genes with PP4 > 0.5 in each disease. Max linkage disequilibrium (LD) refers to the highest LD coefficient between the lead eQTL SNP and any of the lead GWAS SNP or fine-mapped SNPs. Microglia enhancers and promoters refer to whether any of the SNPs for that eGene overlap a microglia enhancer or promoter, as defined by Nott et al56. B-D) Analysis of the USP6NL gene. B) USP6NL expression is associated with the rs7912495 genotype in all four microglia regions. The nominal P-value from the linear regression model in the region-by-region eQTL analysis is indicated on top of the boxplots. The beta and P-value from the meta-analysis are also indicated on top of the Figure. Box plots show median, box spans first to third quartiles, and whiskers extend 1.5 times the interquartile range (IQR) from the box. C) The meta-analyzed USP6NL eQTL colocalizes with the ECHDC3 Alzheimer’s disease risk locus (PP4 = 0.95). D) Fine-mapping of the ECHDC3 locus and combining with the lead QTL and lead GWAS SNPs. SNPs are colored by the LD with the lead QTL SNP. 4 out of 5 of the SNPs overlap a microglia-specific enhancer element as defined by ChIP-seq. Genomic plots (hg19) of the fine-mapped SNPs and the epigenomic data from microglia ChIP-seq, and PLAC-seq junctions. Junctions that overlap the fine-mapped SNPs are emphasized. E-G) Analysis of the P2RY12 gene. E) P2RY12 expression is associated with the rs3732765 genotype. The nominal P-value from the linear regression model in the region-by-region eQTL analysis is indicated on top of the boxplots. The beta and P-value from the meta-analysis are also indicated on top of the Figure. Box plots show median, box spans first to third quartiles, and whiskers extends 1.5 times the interquartile range (IQR) from the box. F) The P2RY12 eQTL colocalizes with the MED12L Parkinson’s Disease locus. G) Fine-mapping of the MED12L locus discovers SNPs that in strong LD with the eQTL lead SNP that overlap microglia-specific enhancer regions. Genomic plots show that the microglia enhancer connects with the P2RY12 promoter.
The ECHDC3 locus has been associated with AD risk in several GWAS23,46,49. The lead SNP rs7920721 sits in an intergenic region that separates two genes, ECHDC3 and USP6NL. Previous analyses have prioritized ECHDC3, as it is upregulated in AD post-mortem brains53,54, and an eQTL for ECHDC3 was seen in whole blood55, though it did not colocalize with the GWAS SNP23.
USP6NL harbors an expression QTL observed in all four microglial regions, with the lead QTL SNP rs7912495-G increasing USP6NL expression (Fig. 6b). The meta-analyzed eQTL colocalizes with the ECHDC3 locus in all 4 AD GWAS used in this study, with the highest PP4 (0.95) seen in 46 (Fig. 6c; Extended Data Fig. 3–4). No colocalization was observed in any other QTL dataset, though we note that USP6NL is expressed five-fold higher in microglia than in monocytes (MiGA median TPM = 15.77; MyND42 monocyte median TPM = 3.13). Fine-mapping of the ECHDC3 locus suggested three additional SNPs as well as the lead GWAS SNP (rs7920721) and lead QTL SNP (rs7912495). The GWAS lead SNP and the QTL lead SNP are in moderate LD (r2 = 0.65), as are two of the three fine-mapped SNPs (Fig. 6d; Supplementary Table 22). Of the five SNPs of interest, 4 of them overlap a microglia-specific enhancer. Using proximity ligation-assisted ChIP-seq (PLAC-seq) data56, we observed that the overlapping microglia enhancer region has extensive long-range connections to regions overlapping the USP6NL promoter and gene body. Notably, there was no colocalization of the upstream ECHDC3 gene in any tested cell type, suggesting that USP6NL is the AD risk gene at this locus. The lead QTL SNP rs7912495-G increases AD risk (β = −0.0492; P = 6.8×10−10; 46) and we propose that it achieves this through upregulating USP6NL expression in microglia. Transcription factor binding motif analysis was inconclusive, with three of the tested SNPs rs143807787, rs74347557, and rs7912495 predicted to disrupt multiple motifs in different directions (Supplementary Table 23).
The MED12L locus was identified in the latest PD GWAS50. The lead SNP rs11707416 sits within a large intron of the MED12L gene, which overlaps with several smaller genes, one of which is P2RY12. A previous study prioritized P2RY12 at this locus due to an overlap with eQTLs in blood and brain57.
P2RY12 is an eQTL (lead SNP rs3732765) identified in the METASOFT meta-analysis, with the lead QTL SNP rs3732765-A decreasing P2RY12 expression (Fig. 6e). The eQTL colocalizes with the PD GWAS MED12L locus (PP4 = 0.88; Fig. 6f). Colocalization was also observed with a P2RY12 eQTL in the dorsolateral prefrontal cortex (PP4 = 0.93; Fig. 5d; Extended Data Fig. 5). Fine-mapping revealed that the lead GWAS SNP rs11707416 was suggested as a causal SNP by multiple fine-mapping tools (a consensus SNP) and is in perfect LD (r2 = 1) with the lead QTL SNP rs3732765 (Fig. 6g). In addition, there are 4 other SNPs prioritized by fine-mapping, 2 of which were in perfect or very high LD with the lead QTL SNP. Of the 7 total SNPs in the set, 5 overlapped a microglia-specific enhancer region on either side of the P2RY12 promoter. PLAC-seq56 revealed long-range connections between the enhancer and the P2RY12 promoter but not to MED12L (Fig. 6g). No colocalization was observed with any MED12L QTL. Altogether this suggests that P2RY12 is the causal gene at the locus. The lead QTL SNP rs3732765-A decreases PD risk (β = −0.06; P = 2.4×10−10; 58), and we propose that it acts through downregulating P2RY12 expression in microglia. Effects on transcription factor binding were predicted for rs11707416, rs41366744, rs4680405, and rs62285879, again for multiple motifs (Supplementary Table 23).
Splicing QTLs identify additional disease-associated loci
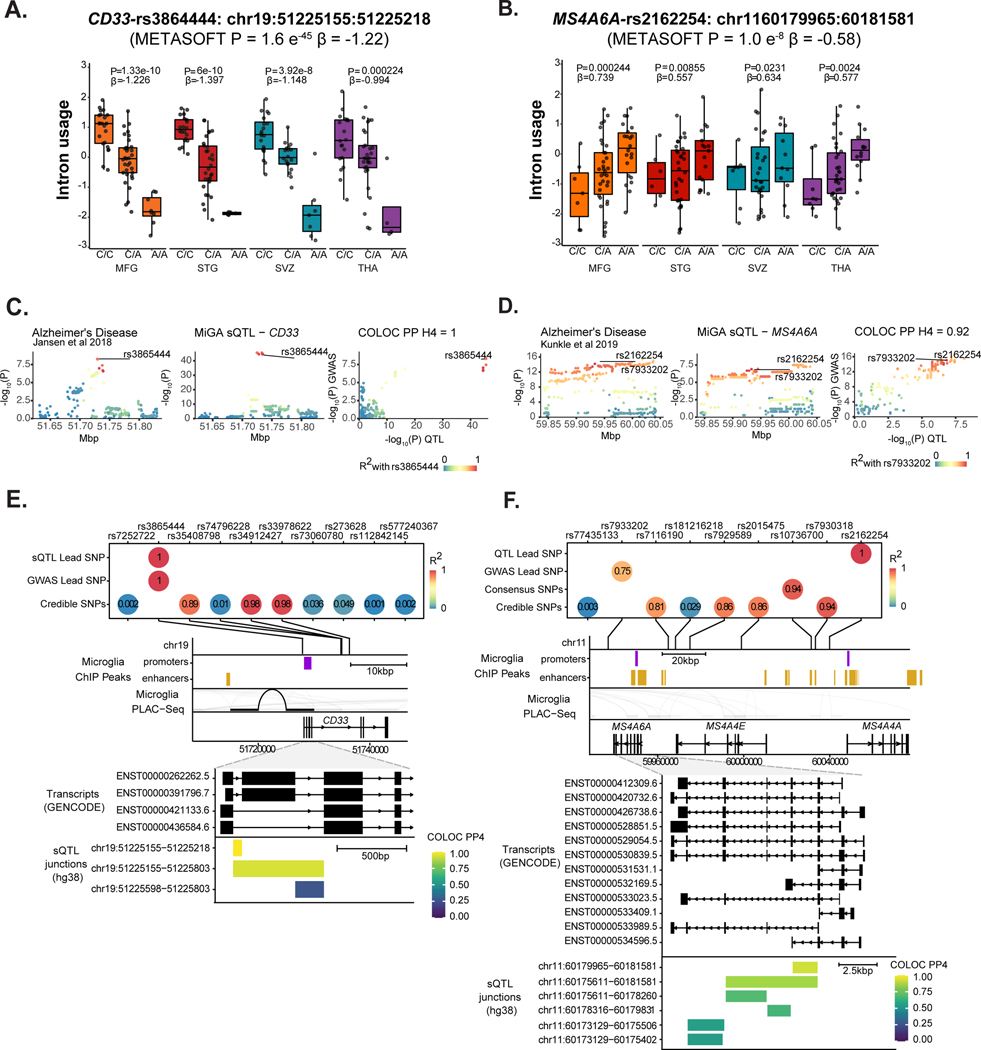
We repeated our colocalization and fine-mapping analyses with sQTLs across the different diseases. Overall we found 81 splicing junctions in 31 genes with a colocalized sQTL at PP4 > 0.7 with 26 GWAS loci (Supplementary Table 21). We highlight two examples of sQTLs associated with Alzheimer’s Disease and identify key challenges ahead for the interpretation of such events. The CD33 risk locus has been implicated in AD susceptibility59. Previous analyses in peripheral monocytes found association between lead GWAS SNP rs3865444 and the inclusion of CD33 exon 259. In MiGA, we also found a strong colocalization with an sQTL associating the same SNP rs3865444-A with reduced intron usage of intron 1, corresponding to reduced inclusion of exon 2 (Fig. 7a–e). Another sQTL was identified in MS4A6A. The MS4A gene cluster is a gene-dense region spanning 600kb, containing 12 genes. We observed colocalization with eQTLs and sQTLs in MS4A6A, as well as eQTLs in MS4A4A and MS4A4E (Fig. 5f). In MiGA, we observed colocalization solely with sQTLs in MS4A6A (Fig. 7f–j). We overlaid all sQTL junctions that colocalized with the AD risk locus and found that the strongest colocalization signals highlighted a cluster of introns in the middle of the gene, with the 5’ intron in the cluster having the strongest colocalization. Notably, 2 transcripts containing this intron have a premature polyadenylation site. rs2162254-A is associated with an increased usage of this intron, which may result in increased production of the shorter isoforms, which could have a downstream consequence on MS4A6A protein function.
Figure 7. Splicing QTLs in CD33 and MS4A6A colocalize with Alzheimer’s Disease risk loci.
A-E) Analysis of CD33. A) Intron usage in CD33 is associated with rs3865444 in all four microglia regions. The nominal P-value from the linear regression model in the region-by-region sQTL analysis is indicated on top of the boxplots. The beta and P-value from the meta-analysis are also indicated on top of the Figure. Box plots show median, box spans first to third quartiles, and whiskers extends 1.5 times the interquartile range (IQR) from the box. B) The meta-analyzed sQTL colocalizes with an AD risk locus, PP4 =1. C) The lead GWAS SNP and lead sQTL SNP are the same causal variant rs3865444. Three other SNPs prioritized by fine-mapping are in high LD. D) The lead SNP falls near the CD33 promoter overlapping a PLAC-seq junction. E) Overlaying CD33 protein-coding transcripts (GENCODE v30) with the sQTL introns, colored by strength of colocalization probability (PP4). Introns with highest PP4 all connect CD33 exon 2 splicing to the AD risk locus. F-J) Analysis of MS4A6A. F) MS4A6A intron usage is associated with rs2162254 across all four regions. Box plots show median, box spans first to third quartiles, and whiskers extend 1.5 times the interquartile range (IQR) from the box. G) The sQTL locus strongly colocalizes with an AD risk locus. H) The lead QTL and lead GWAS SNPs are in moderate LD (R2 = 0.75), as are multiple SNPs prioritized by fine-mapping. I) The MS4A locus contains multiple genes and putative enhancer regions. No SNP overlaps PLAC-seq peaks. J) All protein-coding MS4A6A transcripts (GENCODE v30) with sQTL introns overlaid, coloured by COLOC PP4. A complex cluster of introns all colocalize with the AD risk locus.
Discussion
Here we present the Microglia Genomic Atlas (MiGA), a comprehensive genetic and transcriptomic resource comprised of primary human microglia samples across multiple disease pathologies. We demonstrate that transcriptional heterogeneity in human microglia varies between brain regions and across aging. We generated a catalog of eQTLs and sQTLs in microglia and thereby validated and extended the list of disease genes and putative causal variants underlying risk for neurodegenerative and psychiatric diseases.
Regional and age-related differences in microglial density, morphology, gene, and protein expression have previously been described for both animals and humans5,6,9,60,61. Our analyses suggest some pathways that may be involved in regulating the regional heterogeneity, such as reelin, interferon and glucocorticoid signaling pathways. In addition, we found age-related changes in genes involved in a wide range of inflammatory responses, in line with previous results in aging in microglia16,26,62 and peripheral blood39. Of interest also are a downregulation of C2, P2RY12, and P2RY13, key players in microglia-neuron interactions63,64, as well as genes related to age-related disorders: MS4A4A, MS4A6A, BST1 and P2RY12. Our pathway analyses identified immune-related pathways that may be of relevance for the mechanisms of microglial aging, including STAT-3 and IL-6 signaling, as well as LXR/RXR activation, which has emerged as a key player in regulating cholesterol homeostasis and inflammation in the brain with a potential role in neurodegenerative disorders65–67. Based on previous studies in humans and mice6,8, we expected to find region-specific patterns of age-related changes in microglia. MS4A6A, a gene related to AD risk22,68,69, was one of the genes that showed a region-specific effect of age68,70. By mapping both expression and splicing QTLs in human microglia we have created a resource that has informed our own genetic studies and will be useful for the genetics and neuroscience community. We have identified specific disease colocalizations that may not be captured in monocytes or bulk brain tissue, like BIN1, USP6NL, and PICALM in AD, P2RY12 in PD, PLXNB2 in MS, and IFRD1 in SCZ. We also found colocalizations with opposing effects, such as CASS4. Disease-associated eQTLs results were partly shared between MiGA and the microglia eQTL study by Young et al26. Differences between the studies in age, diagnoses of the included donors, studied brain region, recruitment of tissue (surgical versus autopsy), postmortem delay, and sample size (93 individuals/samples versus 90 individuals/216 samples) have likely contributed to a lack of sharing of part of the hits. By mapping sQTLs we have shown that the known AD risk association with CD33 exon 2 splicing is also present in microglia, and added disease associations that may act through splicing, such as MS4A6A in AD, SIPA1L2 and FAM49B in PD, IRF3 in SCZ, STK4 and GMIP in BD, and CD37 and EFCAB13 in MS. Interpretation of these sQTLs will be improved with the generation of long-read RNA-seq in microglia to identify novel transcripts.
We have performed comprehensive fine-mapping of GWAS loci in five diseases through an ensemble of four different methods and microglia-specific epigenomic datasets to identify credible sets of putative causal variants. This approach allowed us to identify candidate functional variants in multiple disease susceptibility loci that modulate microglia-specific enhancer activity and regulate causal gene expression, which in turn likely modify disease risk by altering the function of microglia (or other myeloid cells) in the brain. In Alzheimer’s disease, we propose USP6NL to be the causal gene in the ECHDC3 locus, due to both a convincing colocalization with AD GWAS and eQTL and the overlap of fine-mapped putative causal SNPs within a defined microglia enhancer which connects with the USP6NL promoter. USP6NL, a GTPase-activating protein involved in control of endocytosis, adds to a growing list of genes (BIN1, PICALM, RABEP1, RIN3, and SORL1) that implicate the dysfunction of the myeloid endolysosomal system in AD37,71.
In Parkinson’s disease, we propose P2RY12 in the MED12L locus through a similar mechanism. P2RY12 is a particularly interesting gene due to the increasing body of literature on its importance for the functioning of microglia72, as well as the proposed link between PD and purinergic signaling73. P2RY12 is one of the P2Y metabotropic G-protein-coupled purinergic receptors, which is highly expressed in microglia in comparison to other brain and myeloid cell types. P2RY12 expression is lost upon microglia activation74, culture34 and in our analyses we have shown that expression is decreased with aging. P2RY12 has been shown to play a role in microglia migration, activation, and neuronal activity64,75. Further validation work is required to test whether the enhancers we prioritize with fine-mapping regulate these genes specifically in microglia.
We recognize several limitations to the current study. First, our sample size is still small in comparison to monocyte and brain datasets22,41–43. We increased power by combining the four regions in a meta-analysis, with the caveat of not adjusting for shared donors, which will have increased our false discovery rate. Another limitation is a variety of known and unknown pre- and post-mortem factors that have an impact on the microglial transcriptome, as shown by our variance partition analyses, that we could not control for in our analyses. There are several methodological differences (recruitment of tissue, studied brain region, postmortem delay, pH, age, diagnosis, medication use) that could interfere with the interpretation of comparisons between MiGA and other microglial datasets5,9,42. We sorted the microglial cells with CD11b+ beads. This marker is not restricted to microglia and may capture small fractions of other myeloid cells. Besides neuroinflammation, hypoxia, and long postmortem intervals, technical artifacts (enzymatic digestion, temperature changes, sorting) may cause microglial activation. We could not control for all these potential confounders, even though these factors could contribute to gene expression changes76,77. Furthermore, our ability to detect additional disease-associated eQTLs may be obscured due to the use of bulk RNA-sequencing data. Future work with large numbers of single-cell RNA-seq profiles from many individuals creates opportunities for mapping eQTLs across microglial subpopulations27, although single-cell data is in general sparse and noisy, which may result in reduced power compared to bulk RNA-seq78. Lastly, many eQTLs are conditional and only revealed after specific stimuli that change the activation state of specific cell types. Thus, mapping response-eQTLs after stimulation of specific-stimuli in primary microglia may reveal additional associations that may provide further mechanistic insights into the disease-associated variants41,78,79.
In summary, we have performed a comprehensive assessment of the transcriptomic landscape of human microglia from multiple brain regions. We have generated an atlas of genetic effects on the human microglia transcriptome, which allowed us to identify potential causal genes and variants underlying risk for neurodegenerative and psychiatric diseases. Our findings represent mechanistic hypotheses that can now be tested with further experimental work at both the level of individual variants and the candidate genes.
Methods
Human brain tissue
Post-mortem brain samples were obtained from the Netherlands Brain Bank (NBB)80 and the Neuropathology Brain Bank and Research CoRE at Mount Sinai Hospital. The permission to collect human brain material was obtained from the Ethical Committee of the VU University Medical Center, Amsterdam, The Netherlands, and the Mount Sinai Institutional Review Board (IRB). Informed consent for autopsy, the use of brain tissue and accompanied clinical information for research purposes was obtained per donor ante-mortem. All autopsies were performed with written consent from the legal next-of-kin. The study was performed under IRB-approved guidance and regulations to keep all patient information strictly de-identified. All research conformed to the principles of the Helsinki Declaration. Neuropathological assessments have been performed by the NBB (see Code availability). In total, 100 donors were included with a mean age of 73.6 years (range 21 – 103 years) and 58 donors were female. Detailed information per donor, including tissue type, age, sex, postmortem interval, pH of cerebrospinal fluid, cause of death, diagnosis, use of medication and neuropathological information is provided in Supplementary Table 1. Participants did not receive compensation.
Microglial isolation and RNA sequencing
Microglia were isolated from four regions, including medial frontal gyrus (MFG; 77 samples), superior temporal gyrus (STG; 63 samples), thalamus (THA; 60 samples), subventricular zone (SVZ; 55 samples). Microglia were isolated as described before in detail60,81,82 and in the Supplementary Note. Microglia were stored in RLT buffer + 1% 2-Mercaptoethanol or lysed in TRIzol reagent (Invitrogen, USA). RNA was isolated using RNeasy Mini kit (Qiagen) adding the DNase I optional step or as described in detail before81. Library preparation was performed at Genewiz using the Ultra-low input system which uses Poly-A selection. SMART-Seq v4 Ultra Low Input RNA Kit was used for library construction using 100 ng of RNA. The libraries were sequenced as 150 bp on fragments with an average read depth of 29 million (ranging from 14–82M) read pairs on the Illumina HiSeq 2500. RNA-seq data was processed using the RAPiD pipeline83. RAPiD aligns samples to the hg38 genome build using STAR84 (v2.7.2a) using the GENCODE v30 transcriptome reference and calculates quality control metrics using Picard85. RNA-seq quality control was performed applying filters to remove samples: 1) samples with less than 10M reads aligned from STAR; 2) samples with more than 20% of the reads aligned to ribosomal regions; 3) samples with less than 10% of the reads mapping to coding regions; 4) samples from brain regions with fewer than 20 donors. Estimated transcript abundance was obtained using RSEM86 (v1.3.1) and transcripts were summed to the gene level with tximport87. Genes with more than 1 read count per million (CPM) in 30% of the samples were kept for downstream analysis. Gene level read counts were normalized as transcripts per million mapped reads (TPM) to adjust for sequencing library size differences.
DNA isolation and genotyping
Genomic DNA was extracted from medial frontal gyrus, superior temporal gyrus, thalamus, or cerebellum using the Qiagen DNeasy Blood & Tissue Kit and followed the manufacturer’s instructions. Details are described in the Supplementary Note. DNA quality and concentration was assessed using a Nanodrop. Samples were genotyped using the Illumina Infinium Global Screening Array (GSA), which contains a genome-wide backbone of 642,824 common variants plus custom disease SNP content (~ 60,000 SNPs).
External Datasets
We downloaded genome-wide association study (GWAS) and genome-wide association study by proxy (GWAX) summary statistics for the following diseases: Alzheimer’s disease (AD)23,46,48,49, Parkinson’s disease (PD)58, Schizophrenia (SCZ)51, Bipolar Disorder (BD)35, and Multiple Sclerosis (MS)52. For each GWAS we downloaded the full summary statistics and a list of genome-wide significant loci, as defined separately by each study. Missing fields in the nominal statistics were dealt with as follows: standard error was calculated from the effect size and P-value; minor allele frequency was taken from the European samples from 1000 Genomes88; SNP coordinates or RS ID were matched using Ensembl (release 99). We took the lists of top genome-wide associated loci from the supplementary materials from each study. For the PD GWAS we removed any loci that did not pass the final quality control filtering according to the “Failed final filtering and QC” column. To avoid double-counting in colocalization, if multiple GWAS loci overlapped (within 1 megabase), we retained the locus with the lowest P-value. Due to the complex LD structure within both regions89,90, loci overlapping the human MHC/HLA region (hg19 chr6:28,477,797–33,448,354) or the MAPT H1/H2 haplotype region (hg19 chr17:43,628,944–44,571,603) were removed. When conditionally independent loci were listed, only the primary association was kept due to lack of conditional summary statistics. Loci from the four AD GWAS were given consensus names using the most recent GWAS as a guide. This resulted in the following locus numbers for each disease: 37 for AD, 71 for PD, 104 for SCZ, 29 for BD, and 137 for MS.
Expression quantitative trait loci (eQTL) full summary statistics were downloaded for microglia26, monocytes41,42 and dorsolateral prefrontal cortex43. All summary statistics were coordinate sorted and indexed with tabix91. Epigenomic data from purified human microglia, neurons, astrocytes, oligodendrocytes56 were downloaded through the echolocatoR package92. To replicate and validate our findings we downloaded processed lists of differentially expressed genes from previous microglia studies5,16,26.
Sources of transcriptomic variation
To understand major sources of variation in the gene expression data at the sample level, we used PCA and linear regression to measure the effect of the following experimental confounders on gene expression variance: sex, age, post mortem interval (PMI), pH, and technical covariates estimated by Picard (Supplementary Fig. 5). We then applied variancePartition (v1.17.7), which uses a linear mixed model to attribute a percentage of variation in expression based on selected covariates on each gene30. As highly correlated covariates cannot be included in the model, we selected covariates that were not very strongly correlated to run the variancePartition analysis (Supplementary Fig. 6b). Gene counts were normalized using trimmed means of M-values (TMM) values calculated from edgeR93 and voom transformed94, which is a method that estimates the mean-variance relationship of the log-counts as input to variancePartition. The technical covariates included in the analysis were % mRNA bases (Picard), mean insert size (Picard), % ribosomal bases (Picard), % read alignment (Picard), and sequencing lane. The biological covariates were donor ID, donor age, sex, brain region, cause of death, sample pH, main diagnosis, post-mortem interval (PMI) in minutes, and the first 4 genotyping ancestry MDS values (C1-C4).
Differential Expression Analysis
Differential expression analysis was performed between the brain regions using the R package Differential expression for repeated measures (DREAM) from VariancePartition31. DREAM uses a linear model to increase power and decrease false positives for RNA-seq datasets with repeated measurements. For the analysis, inputs included the count matrix and the covariate file. These data were normalized using the function voomWithDreamWeights that also performs voom transformation. Since one donor can have different brain regions, we modeled the individual as a random effect and added selected covariates to adjust for possible technical and biological confounders. The final model accounted for sex, donor ID, age, region, cause of death, the first 4 ancestry MDS values (C1–4), % mRNA bases, median insert size, and % ribosomal bases. P-values were then adjusted for multiple testing correction using the Benjamini-Hochberg False Discovery Rate (FDR) correction. For all the Differential Expression Analysis, donor ID and cause of death covariates were modeled as random effects and the others covariates modeled as fixed effects (see Code availability session for GitHub repository page with code). Details about the differential age-by-region, sex-related and diagnosis analysis are described in the Supplementary Note.
Pathway and Gene Set Enrichment analysis
Pathway analysis:
we performed canonical pathway analyses in the Ingenuity Pathway Analysis (IPA) software independently using the following input gene sets: upregulated DEGs aging (n = 338 genes), downregulated DEGs aging (n = 1,355 genes), and clusters of gene sets for specific brain regions; cluster 1 (n = 333 genes upregulated in MFG and STG), cluster 2 (n = 108 genes upregulated in SVZ and THA), cluster 3 (n = 350 genes downregulated in SVZ), and cluster 4 (n = 296 genes upregulated in SVZ) at FDR < 0.05. In addition, we analyzed the canonical pathways associated with splicing in the regional differential transcript usage (DTU) gene set (n = 132) and aging DTU gene set (n = 150) at FDR < 0.01 in IPA. We show the top 10 enriched significant pathways in Supplementary Table 9 and 15. Three to five out of the 10 significant enriched pathways, specifically related to microglia function, with at least four genes that overlap are described in the main text. Additionally, to identify upstream transcriptional regulators that may explain the observed gene expression changes across the different regional clusters we used the IPA upstream regulator analysis. We show the top 20 upstream molecules in Supplementary Table 9.
Gene set enrichment analysis:
to test specific pathways we used curated gene sets and tested statistical enrichment using Fisher exact test at FDR < 0.05 for the following curated gene lists: (1) Human Alzheimer disease (HAM) curated lists: 53 upregulated and 22 downregulated genes from Srinivasan et al. 202033 (2) Cultured microglia curated lists: raw counts were extracted from Gosselin et al. 201734. The Bioconductor package DESeq295 was employed to determine differential gene expression between ex vivo and microglia samples cultured for 7 days; 3,674 upregulated and 4,121 downregulated genes were detected and used in further analyses. (3) IFN-y stimulated microglia curated gene list: 74 upregulated and 6 downregulated genes were detected following the methods as described below in IFNy and LPS stimulated microglia (4) LPS stimulated microglia curated genes list: 472 upregulated and 316 downregulated genes were detected following the methods as described below in IFN-y and LPS stimulated microglia. (5) Aging in human peripheral blood curated gene list: 600 upregulated and 897 downregulated genes from Peters et al. 201539. (6) Microglia-specific curated list: 249 genes from Patir et al. 201996. Additionally, we included specific disease-related lists based on the latest TWAS results: (10) Alzheimer’s disease curated gene list: 36 genes from Raj et al. 201837 (11) Parkinson’s disease curated gene list: 77 genes from Li et al. 201938 (12) Schizophrenia curated gene list: 43 genes from Gusev et al. 201836 (13) Bipolar disorder curated gene list: 16 genes from TWAS results from latest BD GWAS35.
Genotype Quality Control and Imputation
Samples were genotyped using the Illumina Infinium Global Screening Array (GSA) plus a custom disease SNP content (~ 60,000 SNPs) for a total of 760,329 common variants. To select high-quality data, we applied an initial genotyping quality control using bcftools (v1.9) and vcftools (v0.1.15), keeping SNPs with call rate > 95%, minor allele frequency (MAF) > 5%, Hardy-Weinberg equilibrium (HWE) P-value > 1 × 10−6, and sample call rate > 95%.
Duplicated and up to third-degree related samples were removed based on pairwise kinship coefficients estimated using KING97 (Supplementary Fig. 10a). DNA samples were matched to the RNA-seq data to confirm the same donor origin using the MBV tool from QTLtools98 (Supplementary Fig. 10b) and sex mismatching samples were removed by comparing DNA inferred sex from PLINK to RNA gene expression of the UTY and XIST genes (Supplementary Fig. 10c). This resulted in 593,748 genotyped variants passing all QC steps in 98 donors, of which 90 donors were of European ancestry. Genetic ancestry of samples was confirmed by principal components analysis using the PLINK program99 and MDS (multidimensional scaling) values of study subjects were compared to those of 1000 Genome Project samples (Phase 3) (Supplementary Fig. 10d).
Genotype imputation was performed for those 90 donors through the Michigan Imputation Server v1.4.1 (Minimac 4)100 using the 1000 Genomes (Phase 3) v5 (GRCh37) European panel and Eagle v2.4 phasing101 in quality control and imputation mode with rsq filter set to 0.3. Following imputation, variants were lifted over to the GRCh38 reference to match the RNA-seq data using Picard liftoverVCF and the “b37ToHg38.over.chain.gz” liftover chain file. Finally, we applied another round of variant quality controls, removing indels and multi-allelic SNPs, and keeping only variants with MAF > 5% and Hardy-Weinberg P-value >1×10−6. After imputation, liftover, and QC, a total of 5,803,004 variants were included in downstream analyses. These variants were additionally annotated using dbSNP (All_20180418.vcf.gz) and snpEff v4.3i102.
Quantitative Trait Loci mapping
To perform expression QTL (eQTL) mapping, we followed the latest pipeline created by the GTEX consortium103. We completed a separate normalization and filtering method to previous analyses. Gene expression matrices were created from the RSEM output using tximport87. Matrices were then converted to GCT format, TMM normalized, filtered for lowly expressed genes, removing any gene with less than 0.1 TPM in 20% of samples and at least 6 counts in 20% of samples. Each gene was then inverse-normal transformed across samples. After filtering, we tested a total of 18,430 genes. Then, PEER104 factors were calculated to estimate hidden confounders within our expression data. We created a combined covariate matrix that included the PEER factors and the first 4 genotyping ancestry MDS values as input to the analysis. We tested numbers of PEER factors from 0 to 20 and found that between 5 and 10 factors produced the largest number of eGenes in each region (Supplementary Fig. 11).
To test for cis-eQTLs, linear regression was performed using the tensorQTL105 (v1.0.2) cis_nominal mode for each SNP-gene pair using a 1 megabase window within the transcription start site (TSS) of a gene. To test for association between gene expression and the top variant in cis we used tensorQTL cis permutation pass per gene with 1000 permutations. To identify eGenes, we performed q-value correction of the permutation P-values for the top association per gene44 at a threshold of 0.05.
We performed splicing quantitative trait loci (sQTL) analysis using the splice junction read counts generated by regtools106(v0.5.1). Junctions were clustered using Leafcutter107(v0.2.8), specifying for each junction in a cluster a maximum length of 100kb. Following the GTEx pipeline, introns without read counts in at least 50% of samples or with fewer than 10 read counts in at least 10% of samples were removed. Introns with insufficient variability across samples were removed. Filtered counts were then quantile normalized using prepare_phenotype_table.py from Leafcutter, merged, and converted to BED format, using the coordinates from the middle of the intron cluster. We created a combined covariate matrix that included the PEER factors and the first 4 genotyping ancestry MDS values as input to the analysis. We mapped sQTLs with between 0 and 20 PEER factors as covariates in our QTL model and determined 5 to be optimal in MFG, STG and THA. 0 PEER factors were used for SVZ (Supplementary Fig. 11).
To test for cis sQTLs, linear regression was performed using the tensorQTL nominal pass for each SNP-junction pair using a 100kb window from the center of each intron cluster. Although junctions were initially grouped together into clusters, we tested each SNP-junction pair separately, which is the standard approach103,107. To test for association between intronic ratio and the top variant in cis we used tensorQTL permutation pass, grouping junctions by their cluster using --grp option. To identify significant clusters, we performed q-value correction using a threshold of 0.05.
We estimated pairwise replication (π1) of cis-eQTLs with the external eQTL datasets using the q-value R package44. Briefly, this involves taking the SNP-gene pairs that are significant in our microglia data at q-value < 0.1 and extracting the unadjusted P-values for the matched SNP-gene pairs in the external dataset.
Meta-analysis of microglia QTLs
METASOFT
Meta-analysis of the four microglia brain regions (MFG, STG, THA and SVZ), along with monocytes (MyND and Fairfax) and dorsolateral prefrontal cortex (ROSMAP) was performed using METASOFT45 (v2.0.1). Effect sizes and standard errors of each SNP-Gene pair were used as input. We carried out a random effects meta-analysis using their RE2 model, optimized to detect associations under heterogeneity.
mashr: Multivariate Adaptive Shrinkage in R
To estimate and compare the genetic effects in gene expression and splicing proportions across different brain regions, we performed a Multivariate Adaptive Shrinkage (MASH) through the R package mashR40. MASH employs an empirical Bayes method to estimate patterns of similarity among conditions and improve the accuracy of effect estimates.
Following the pipeline applied by GTEx Consortium (see Code availability) we used as input, the nominal associations (P-values, betas, and standard errors) from eQTL and sQTL (gene-SNP pair for eQTL or junction-SNP pair for sQTL) for each region. Then, we selected the strongest associations after computing a sparse factorization matrix of the z-scores using the Sparse Factor Analysis (SFA) software with K=5 factors. Secondly, we computed data-driven covariance matrices priors by applying the Extreme Deconvolution method and computed the canonical covariance matrices, including the identity matrix, and matrices representing condition-specific effects. Next, using the entire dataset, we computed the maximum-likelihood estimates of the weights for each combination and learned how each pattern-effect size combination occurs in the data. Finally, we computed the posterior statistics using the fitted MASH model from the previous step. This step creates the tables with posterior means and local false sign rate (lfsr), a measure analogous to FDR, that accounts for effect size and standard error rather than only P-values108. This approach improves effect size estimates and allows for more quantitative assessments of effect-size heterogeneity compared to simple region-specific assessments40.
Statistics and reproducibility
We generated genotyping and RNA-seq, including bulk and single-cell data from human CD11b+ microglia as described in the Supplementary Note. The statistical tests were performed and indicated in the figure legends or outlined in the Methods. The age range of the 100 donors is between 21 and 103 years old; 58 of them were female. All analyses were adjusted for age, gender, and other covariates. In total, 59 out of 314 samples were excluded due to insufficient RNA-seq quality or insufficient sample size by brain region. The investigators were not blinded for group allocation (diagnosis, sex, age etc.) during data analysis since adjustment for these factors was necessary for the data analyses. Supplementary Figure 1 shows a flowchart of quality control, and all measures applied are available Online and in the Methods section. Further information on statistics and reproducibility is available in the corresponding sections of Methods and in the Reporting Summary.
Data availability
Raw and processed RNA-seq and genotype data sets are deposited in the National Institute on Aging Genetics of Alzheimer’s Disease Data Storage Site (NIAGADS at https://dss.niagads.org/datasets/ng00105/; Accession number: NG00105.v1). The user will need to log into NIAGADS Data Access Request (DAR) to start an application. Instructions to download the dataset can be found at https://www.niagads.org/data/request/data-request-instructions. All differential expression, gene lists, and fine-mapping results are present as supplementary tables. The GWAS fine-mapping results are available from the echoLocatoR Shiny application at https://rajlab.shinyapps.io/Fine_Mapping_Shiny. Full nominal and permuted eQTL and sQTL summary statistics per brain region are available from Zenodo at https://doi.org/10.5281/zenodo.4118605 (eQTL) and https://doi.org/10.5281/zenodo.4118403 (sQTL). Results for eQTL and sQTL meta-analysis (mashR and METASOFT) and colocalization (COLOC) are available from Zenodo at https://doi.org/10.5281/zenodo.4118676.
Code availability
All the code used to perform the analysis is available at https://github.com/RajLabMSSM/MiGA_public_release. To perform expression QTL (eQTL) mapping, we followed the latest pipeline created by the GTEx consortium103 (https://github.com/broadinstitute/gtex-pipeline). To estimate and compare the genetic effects in gene expression and splicing proportions across different brain regions, we used the mashR pipeline40 (https://stephenslab.github.io/gtexresults/gtex.html). Tools used for genotyping quality control or specific R packages are described in the Methods session and Supplementary Note.
Extended Data
Extended Data Fig. 1. Regional heterogeneity analysis for transcript usage.
A) Heatmap of relative transcript usage between regions using all 176 transcripts from pairwise comparisons of differential transcript usage (DTU; empirical FDR < 0.1), plotted as row-scaled z-scores of mean transcript usage per region; red and blue indicates high and low relative transcript usage, respectively. Transcripts form 2 k-means clusters, n refers to the number of transcripts in each cluster. Core microglia genes from Patir et al. highlighted. B) Transcript usage plots for the gene RGS1. The two most abundant transcripts are bolded. The DTU signal is driven by a reduction of the intron retention transcript ENST00000498352.1 and a corresponding increase in the protein-coding transcript ENST00000367459.8 in the SVZ compared to the other regions. Boxplots show the median with the first and third quartiles of the distribution. C) Functional Enrichment Analysis of all 132 genes with regional DTU using Ingenuity Pathway Analysis (IPA). Significantly enriched terms shown (q-value < 0.05).
Extended Data Fig. 2. Age-related analysis for transcript usage.
A) Heatmap of the 225 transcripts associated with age (empirical FDR < 0.1). Each row plotted as Z-score of median expression averaged first by donor (across multiple regions) and then by age quintiles with 20 donors each. Transcripts are ordered by Ward’s hierarchical clustering. Core microglia genes from Patir et al. highlighted. B) Example transcript usage for P2RY12. The association is caused by an increase in the long protein-coding transcript ENST00000302632.3 and a corresponding decrease in the short intron retention transcript ENST00000468596.1 during aging. C) Functional Enrichment Analysis of all 150 genes with DTU in aging using Ingenuity Pathway Analysis (IPA). Only significantly enriched terms shown (q-value < 0.05).
Extended Data Fig. 3. Full colocalization results in Alzheimer’s Disease.
Colocalization PP4 displayed for each GWAS locus (right text) and gene (left text) for each QTL dataset. An empty value means no QTL was present for testing for that gene in that dataset.
Extended Data Fig. 4. Colocalization results for each regional microglia dataset in Alzheimer’s Disease.
Colocalization PP4 displayed for each GWAS locus (right text) and gene (left text) for each QTL dataset. An empty value means no QTL was present for testing for that gene in that dataset.
Extended Data Fig. 5. Full colocalization results in Parkinson’s Disease.
Colocalization PP4 displayed for each GWAS locus (right text) and gene (left text) for each QTL dataset. An empty value means no QTL was present for testing for that gene in that dataset.
Extended Data Fig. 6. Colocalization results for each regional microglia dataset in Parkinson’s Disease.
Colocalization PP4 displayed for each GWAS locus (right text) and gene (left text) for each QTL dataset. An empty value means no QTL was present for testing for that gene in that dataset.
Extended Data Fig. 7. Overlap of colocalized microglia eQTLs with epigenomic features in AD and PD.
Cell-type specific promoters and enhancers56 were overlapped with SNP sets for each colocalizing microglia QTL - GWAS locus. SNP sets consisted of the lead GWAS SNP, the lead QTL SNP and any fine-mapped consensus or credible SNPs. Results are summarized here by the number of SNPs in the set that overlap with a particular feature type.
Supplementary Material
Acknowledgments
We thank members of the Raj and de Witte labs for their feedback on the manuscript. The authors thank the teams of the Netherlands Brain Bank and the Mount Sinai Neuropathology Brain Bank and Research CoRE for their services. We thank the study participants for their generous gifts of brain donation. The microglia were isolated through the efforts of a large team and we would like to thank Manja Litjens, Roland D. van Dijk, Alba Fernández-Andreu, Paul R. Ormel, Hans C. van Mierlo, Y. He, Stephanie Gumbs, Miriam E van Strien, Saskia Burm, Vanessa Donega, and Elly M. Hol for all their contributions to this effort. The authors thank Michael Chao for his assistance with genotyping QC. This work was supported in part through the computational and data resources and staff expertise provided by Scientific Computing at the Icahn School of Medicine at Mount Sinai. Research reported in this paper was supported by the Office of Research Infrastructure of the National Institutes of Health under award number S10OD026880. T.R. is supported by grants from the US National Institutes of Health (NIH) NIA R21-AG063130, NIA R01-AG054005, NIA U01-AG068880, NIA RF1-AG065926, NIA R56-AG055824, and NINDS R01-NS116006. G.S. was supported through ZonMw and the foundation “De Drie Lichten” in the Netherlands. E.N. was supported by Ramon Areces fellowship. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Footnotes
Competing interests statement
The authors declare no competing interests.
References
- 1.Priller J. & Prinz M. Targeting microglia in brain disorders. Science 365, 32–33 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Ransohoff RM & El Khoury J. Microglia in health and disease. Cold Spring Harb. Perspect. Biol 8, a020560 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prinz M, Jung S. & Priller J. Microglia biology: One century of evolving concepts. Cell 179, 292–311 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Tan Y-L, Yuan Y. & Tian L. Microglial regional heterogeneity and its role in the brain. Mol. Psychiatry 25, 351–367 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van der Poel M. et al. Transcriptional profiling of human microglia reveals grey-white matter heterogeneity and multiple sclerosis-associated changes. Nat. Commun 10, 1139 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grabert K. et al. Microglial brain region-dependent diversity and selective regional sensitivities to aging. Nat. Neurosci 19, 504–516 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Biase LM et al. Local cues establish and maintain region-specific phenotypes of basal ganglia microglia. Neuron 95, 341–356.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soreq L. et al. Major shifts in glial regional identity are a transcriptional hallmark of human brain aging. Cell Rep. 18, 557–570 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Masuda T. et al. Spatial and temporal heterogeneity of mouse and human microglia at single-cell resolution. Nature 566, 388–392 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Olah M. et al. A transcriptomic atlas of aged human microglia. Nat. Commun 9, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mathys H. et al. Single-cell transcriptomic analysis of Alzheimer’s disease. Nature 570, 332–337 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mrdjen D. et al. High-dimensional single-cell mapping of central nervous system immune cells reveals distinct myeloid subsets in health, aging, and disease. Immunity 48, 599 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Hammond TR et al. Single-cell RNA sequencing of microglia throughout the mouse lifespan and in the injured brain reveals complex cell-state changes. Immunity 50, 253–271.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keren-Shaul H. et al. A unique microglia type associated with restricting development of Alzheimer’s disease. Cell 169, 1276–1290.e17 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Masuda T. et al. Author Correction: Spatial and temporal heterogeneity of mouse and human microglia at single-cell resolution. Nature 568, E4 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Galatro TF et al. Transcriptomic analysis of purified human cortical microglia reveals age-associated changes. Nat. Neurosci 20, 1162–1171 (2017). [DOI] [PubMed] [Google Scholar]
- 17.McGeer PL et al. Microglia in degenerative neurological disease. Glia 7, 84–92 (1993). [DOI] [PubMed] [Google Scholar]
- 18.Kreutzberg GW Microglia: a sensor for pathological events in the CNS. Trends Neurosci. 19, 312–318 (1996). [DOI] [PubMed] [Google Scholar]
- 19.Trépanier MO, Hopperton KE, Mizrahi R, Mechawar N. & Bazinet RP Postmortem evidence of cerebral inflammation in schizophrenia: a systematic review. Mol. Psychiatry 21, 1009–1026 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hopperton KE, Mohammad D, Trépanier MO, Giuliano V. & Bazinet RP Markers of microglia in post-mortem brain samples from patients with Alzheimer’s disease: a systematic review. Mol. Psychiatry 23, 177–198 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parikshak NN et al. Genome-wide changes in lncRNA, splicing, and regional gene expression patterns in autism. Nature 540, 423–427 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raj T. et al. Polarization of the effects of autoimmune and neurodegenerative risk alleles in leukocytes. Science 344, 519–523 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kunkle BW et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat. Genet 51, 414–430 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCauley ME & Baloh RH Inflammation in ALS/FTD pathogenesis. Acta Neuropathol. 137, 715–730 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gandal MJ et al. Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science 362, eaat8127 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Young Adam. M. H. et al. A map of transcriptional heterogeneity and regulatory variation in human microglia. Nat. Genetics 53, 861–868 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Masuda T, Sankowski R, Staszewski O. & Prinz M. Microglia heterogeneity in the single-cell era. Cell Rep. 30, 1271–1281 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Li YI et al. RNA splicing is a primary link between genetic variation and disease. Science 352, 600–604 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raj T. et al. CD33: increased inclusion of exon 2 implicates the Ig V-set domain in Alzheimer’s disease susceptibility. Hum. Mol. Genet 23, 2729–2736 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoffman GE & Schadt EE variancePartition: interpreting drivers of variation in complex gene expression studies. BMC Bioinformatics 17, 483 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoffman GE & Roussos P. dream: Powerful differential expression analysis for repeated measures designs. Bioinformatics (2020) doi: 10.1093/bioinformatics/btaa687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hartigan JA & Wong MA Algorithm AS 136: A K-means clustering algorithm. J. R. Stat. Soc. Ser. C. Appl. Stat 28, 100 (1979). [Google Scholar]
- 33.Srinivasan K. et al. Alzheimer’s patient microglia exhibit enhanced aging and unique transcriptional activation. Cell Rep. 31, 107843 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gosselin D. et al. An environment-dependent transcriptional network specifies human microglia identity. Science 356, eaal3222 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stahl EA et al. Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat. Genet 51, 793–803 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gusev A. et al. Transcriptome-wide association study of schizophrenia and chromatin activity yields mechanistic disease insights. Nat. Genet 50, 538–548 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raj T. et al. Integrative transcriptome analyses of the aging brain implicate altered splicing in Alzheimer’s disease susceptibility. Nat. Genet 50, 1584–1592 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li YI, Wong G, Humphrey J. & Raj T. Prioritizing Parkinson’s disease genes using population-scale transcriptomic data. Nat. Commun 10, 994 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peters MJ et al. The transcriptional landscape of age in human peripheral blood. Nat. Commun 6, 8570 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Urbut SM, Wang G, Carbonetto P. & Stephens M. Flexible statistical methods for estimating and testing effects in genomic studies with multiple conditions. Nat. Genet 51, 187–195 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fairfax BP et al. Innate immune activity conditions the effect of regulatory variants upon monocyte gene expression. Science 343, 1246949 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Navarro E. et al. Dysregulation of mitochondrial and proteolysosomal genes in Parkinson’s disease myeloid cells. Nat Aging 1, 850–863 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ng B. et al. An xQTL map integrates the genetic architecture of the human brain’s transcriptome and epigenome. Nat. Neurosci 20, 1418–1426 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Storey JD The positive false discovery rate: a Bayesian interpretation and the q-value. Ann. Stat 31, 2013–2035 (2003). [Google Scholar]
- 45.Han B. & Eskin E. Random-effects model aimed at discovering associations in meta-analysis of genome-wide association studies. Am. J. Hum. Genet 88, 586–598 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marioni RE et al. GWAS on family history of Alzheimer’s disease. Transl. Psychiatry 8, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Giambartolomei C. et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 10, e1004383 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lambert J-C et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat. Genet 45, 1452–1458 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jansen IE et al. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat. Genet 51, 404–413 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nalls MA et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet Neurol. 18, 1091–1102 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.International Multiple Sclerosis Genetics Consortium. Multiple sclerosis genomic map implicates peripheral immune cells and microglia in susceptibility. Science 365, eaav7188 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang B. et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell 153, 707–720 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Desikan RS et al. Polygenic overlap between C-reactive protein, plasma lipids, and Alzheimer disease. Circulation 131, 2061–2069 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhernakova DV et al. Identification of context-dependent expression quantitative trait loci in whole blood. Nat. Genet 49, 139–145 (2017). [DOI] [PubMed] [Google Scholar]
- 56.Nott A. et al. Brain cell type-specific enhancer-promoter interactome maps and disease-risk association. Science 366, 1134–1139 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grenn FP et al. The Parkinson’s disease genome-wide association study locus browser. Mov. Disord 35, 2056–2067 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nalls MA et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet Neurol. 18, 1091–1102 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hollingworth P. et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat. Genet 43, 429–435 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Böttcher C. et al. Human microglia regional heterogeneity and phenotypes determined by multiplexed single-cell mass cytometry. Nat. Neurosci 22, 78–90 (2019). [DOI] [PubMed] [Google Scholar]
- 61.Mittelbronn M, Dietz K, Schluesener HJ & Meyermann R. Local distribution of microglia in the normal adult human central nervous system differs by up to one order of magnitude. Acta Neuropathol. 101, 249–255 (2001). [DOI] [PubMed] [Google Scholar]
- 62.Olah M. et al. Identification of a microglia phenotype supportive of remyelination. Glia 60, 306–321 (2012). [DOI] [PubMed] [Google Scholar]
- 63.Stevens B. et al. The classical complement cascade mediates CNS synapse elimination. Cell 131, 1164–1178 (2007). [DOI] [PubMed] [Google Scholar]
- 64.Badimon A. et al. Negative feedback control of neuronal activity by microglia. Nature 586, 417–423 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Savage JC et al. Nuclear receptors license phagocytosis by trem2+ myeloid cells in mouse models of Alzheimer’s disease. J. Neurosci 35, 6532–6543 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Courtney R. & Landreth GE LXR regulation of brain cholesterol: From development to disease. Trends Endocrinol. Metab. 27, 404–414 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kao Y-C, Ho P-C, Tu Y-K, Jou I-M & Tsai K-J Lipids and Alzheimer’s disease. Int. J. Mol. Sci 21, 1505 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Proitsi P. et al. Alzheimer’s disease susceptibility variants in the MS4A6A gene are associated with altered levels of MS4A6A expression in blood. Neurobiol. Aging 35, 279–290 (2014). [DOI] [PubMed] [Google Scholar]
- 69.Huang K-L et al. A common haplotype lowers PU.1 expression in myeloid cells and delays onset of Alzheimer’s disease. Nat. Neurosci 20, 1052–1061 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Deming Y. et al. The MS4A gene cluster is a key modulator of soluble TREM2 and Alzheimer’s disease risk. Sci. Transl. Med 11, eaau2291 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Novikova G. et al. Integration of Alzheimer’s disease genetics and myeloid genomics identifies disease risk regulatory elements and genes. Nat. Commun 12, 1610 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mildner A, Huang H, Radke J, Stenzel W. & Priller J. P2Y12 receptor is expressed on human microglia under physiological conditions throughout development and is sensitive to neuroinflammatory diseases. Glia 65, 375–387 (2017). [DOI] [PubMed] [Google Scholar]
- 73.Tóth A, Antal Z, Bereczki D. & Sperlágh B. Purinergic signalling in Parkinson’s disease: A multi-target system to combat neurodegeneration. Neurochem. Res 44, 2413–2422 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.van Wageningen TA et al. Regulation of microglial TMEM119 and P2RY12 immunoreactivity in multiple sclerosis white and grey matter lesions is dependent on their inflammatory environment. Acta Neuropathol. Commun 7, 206 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Haynes SE et al. The P2Y12 receptor regulates microglial activation by extracellular nucleotides. Nat. Neurosci 9, 1512–1519 (2006). [DOI] [PubMed] [Google Scholar]
- 76.Marsh SE et al. Single cell sequencing reveals glial specific responses to tissue processing & enzymatic dissociation in mice and humans. bioRxiv (2020) doi: 10.1101/2020.12.03.408542. [DOI] [Google Scholar]
- 77.Mattei D. et al. Enzymatic dissociation induces transcriptional and proteotype bias in brain cell populations. Int. J. Mol. Sci 21, 7944 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lee MN et al. Common genetic variants modulate pathogen-sensing responses in human dendritic cells. Science 343, 1246980 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ramdhani S. et al. Tensor decomposition of stimulated monocyte and macrophage gene expression profiles identifies neurodegenerative disease-specific trans-eQTLs. PLoS Genet. 16, e1008549 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.de Lange GM, Rademaker M, Boks MP & Palmen SJMC Brain donation in psychiatry: results of a Dutch prospective donor program among psychiatric cohort participants. BMC Psychiatry 17, 347 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Melief J. et al. Characterizing primary human microglia: A comparative study with myeloid subsets and culture models. Glia 64, 1857–1868 (2016). [DOI] [PubMed] [Google Scholar]
- 82.Sneeboer MAM et al. Microglia in post-mortem brain tissue of patients with bipolar disorder are not immune activated. Transl. Psychiatry 9, 153 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shah H. PgmNr 1856: RAPiD—an agile and dependable RNA-Seq framework. (ASHG, 2015). [Google Scholar]
- 84.Dobin A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Picard. http://broadinstitute.github.io/picard/.
- 86.Li B. & Dewey CN RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Soneson C, Love MI & Robinson MD Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res. 4, 1521 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.1000 Genomes Project Consortium et al. A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Boettger LM, Handsaker RE, Zody MC & McCarroll SA Structural haplotypes and recent evolution of the human 17q21.31 region. Nat. Genet 44, 881–885 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Allcock RJN et al. The MHC haplotype project: a resource for HLA-linked association studies. Tissue Antigens 59, 520–521 (2002). [DOI] [PubMed] [Google Scholar]
- 91.Li H. Tabix: fast retrieval of sequence features from generic TAB-delimited files. Bioinformatics 27, 718–719 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schilder BM, Humphrey J. & Raj T. echolocatoR: an automated end-to-end statistical and functional genomic fine-mapping pipeline. Bioinformatics (2021) doi: 10.1093/bioinformatics/btab658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Robinson MD, McCarthy DJ & Smyth GK edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Law CW, Chen Y, Shi W. & Smyth GK voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15, R29 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Love MI, Huber W. & Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Patir A, Shih B, McColl BW & Freeman TC A core transcriptional signature of human microglia: Derivation and utility in describing region-dependent alterations associated with Alzheimer’s disease. Glia 67, 1240–1253 (2019). [DOI] [PubMed] [Google Scholar]
- 97.Manichaikul A. et al. Robust relationship inference in genome-wide association studies. Bioinformatics 26, 2867–2873 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fort A. et al. MBV: a method to solve sample mislabeling and detect technical bias in large combined genotype and sequencing assay datasets. Bioinformatics 33, 1895–1897 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chang CC et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4, 7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Das S. et al. Next-generation genotype imputation service and methods. Nat. Genet 48, 1284–1287 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Loh P-R et al. Reference-based phasing using the Haplotype Reference Consortium panel. Nat. Genet 48, 1443–1448 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cingolani P. et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6, 80–92 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.The GTEx Consortium. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 369, 1318–1330 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Stegle O, Parts L, Piipari M, Winn J. & Durbin R. Using probabilistic estimation of expression residuals (PEER) to obtain increased power and interpretability of gene expression analyses. Nat. Protoc 7, 500–507 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Taylor-Weiner A. et al. Scaling computational genomics to millions of individuals with GPUs. Genome Biol. 20, 228 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cotto KC et al. RegTools: Integrated analysis of genomic and transcriptomic data for the discovery of splicing variants in cancer. bioRxiv (2018) doi: 10.1101/436634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Li YI et al. Annotation-free quantification of RNA splicing using LeafCutter. Nat. Genet 50, 151–158 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Stephens M. False discovery rates: a new deal. Biostatistics 18, 275–294 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw and processed RNA-seq and genotype data sets are deposited in the National Institute on Aging Genetics of Alzheimer’s Disease Data Storage Site (NIAGADS at https://dss.niagads.org/datasets/ng00105/; Accession number: NG00105.v1). The user will need to log into NIAGADS Data Access Request (DAR) to start an application. Instructions to download the dataset can be found at https://www.niagads.org/data/request/data-request-instructions. All differential expression, gene lists, and fine-mapping results are present as supplementary tables. The GWAS fine-mapping results are available from the echoLocatoR Shiny application at https://rajlab.shinyapps.io/Fine_Mapping_Shiny. Full nominal and permuted eQTL and sQTL summary statistics per brain region are available from Zenodo at https://doi.org/10.5281/zenodo.4118605 (eQTL) and https://doi.org/10.5281/zenodo.4118403 (sQTL). Results for eQTL and sQTL meta-analysis (mashR and METASOFT) and colocalization (COLOC) are available from Zenodo at https://doi.org/10.5281/zenodo.4118676.