Summary
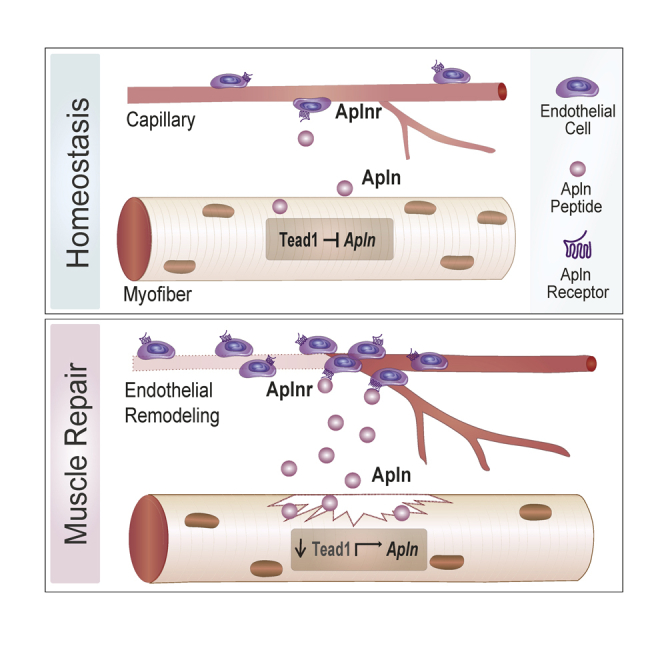
Apelin (Apln) is a myokine that regulates skeletal muscle plasticity and metabolism and declines during aging. Through a yeast one-hybrid transcription factor binding screen, we identified the TEA domain transcription factor 1 (Tead1) as a novel regulator of the Apln promoter. Single-cell analysis of regenerating muscle revealed that the apelin receptor (Aplnr) is enriched in endothelial cells, whereas Tead1 is enriched in myogenic cells. Knock-down of Tead1 stimulates Apln secretion from muscle cells in vitro and myofiber-specific overexpression of Tead1 suppresses Apln secretion in vivo. Apln secretion via Tead1 knock-down in muscle cells stimulates endothelial cell expansion via endothelial Aplnr. In vivo, Apln peptide supplementation enhances endothelial cell expansion while Tead1 muscle overexpression delays endothelial remodeling following muscle injury. Our work describes a novel paracrine crosstalk in which Apln secretion is controlled by Tead1 in myogenic cells and influences endothelial remodeling during muscle repair.
Subject areas: Molecular biology, Molecular mechanism of gene regulation, Cell biology
Graphical abstract
Highlights
-
•
A transcription factor binding screen identified that Tead1 binds the Apln promoter
-
•
Tead1 represses the secretion of the Apln peptide from myogenic cells
-
•
Apln stimulates endothelial cell remodeling during muscle regeneration
-
•
Tead1-mediated regulation of Apln in myogenic cells controls endothelial cell remodeling
Molecular biology; Molecular mechanism of gene regulation; Cell biology
Introduction
Skeletal muscle is a plastic tissue with intrinsic capacity for structural and functional adaptations in response to nutrition, physical activity, and various physiological needs of the body. In particular, muscle physiology adapts to different types of exercise training but also declines in muscle-wasting conditions which can arise from genetic monogenic mutations causing muscular dystrophies, chronic diseases such as cancer or COPD, aging, traumatic or sports injuries, or simply inactivity during immobilization or prolonged bed rest (Egerman and Glass, 2014; Sartori et al., 2021). At the cellular level, the ability to contract is tightly controlled by the number of contractile proteins and the bioenergetic capacity of myofibers, but also by local cellular interactions with motoneurons and blood vessels as well as endocrine signals from the rest of the body. In addition, skeletal muscle can be repaired when damaged during pathological conditions owing to a tissue-resident population of stem cells called satellite cells (Ancel et al., 2021; Fuchs and Blau, 2020; Yin et al., 2013).
Secretion of myokines by myofibers and paracrine communication within skeletal muscle has emerged as an active mechanism through which muscle physiology and repair are coordinated through cross-talk with satellite cells, immune cells, fibro-adipogenic progenitors, and endothelial cells which regulate angiogenesis and remodel the vasculature to ensure efficient oxygen and nutrient supply to muscle fibers (Chazaud, 2020; Lazure et al., 2020; Mashinchian et al., 2018; Whitham and Febbraio, 2016). Exercise training is well described to remodel the vascularization of skeletal muscle via mechanisms involving paracrine secretion of the vascular endothelial growth factor (VEGF) by myofibers, which stimulates endothelial cells and regulates angiogenesis (Bloor, 2005; Gorski and De Bock, 2019). The remodeling of blood vessels during muscle repair is also a topic of active investigation as the spatiotemporal coordination of angiogenesis and myogenesis is key to rebuilding functional vascularized myofibers during tissue repair following muscle injury. Muscle stem cells have been shown to attract endothelial cells by secreting local VEGF in the niche (Verma et al., 2018). Macrophages invading muscle after tissue injury were also recently described to modulate endothelial cell remodeling via the secretion of lactate (Zhang et al., 2020). Conversely, vascular remodeling during muscle repair supports muscle stem cell expansion and commitment to myogenesis (Christov et al., 2007). Beyond the identification of these first signals, the understanding of the complex inter-cellular crosstalk between muscle fibers, muscle stem cells, and endothelial cells requires further investigation.
Apelin (Apln) is a small peptide regulating paracrine and endocrine communication in adipose tissue and the cardiovascular system. Apln is produced in skeletal muscle in response to exercise and contraction and has recently emerged as a modulator of muscle physiology and as a therapeutic target for muscle and metabolic diseases (Besse-Patin et al., 2014; Castan-laurell et al., 2012; Kadoglou et al., 2012; Rai et al., 2017). In humans, APLN is produced as a 77 amino acid pre-pro peptide, which is cleaved by endopeptidases, and then converted into the bioactive peptides APLN-36, APLN-17, and APLN-13 (Japp and Newby, 2008; Nyimanu et al., 2019; Tatemoto et al., 1998). APLN signals by binding to the G protein-coupled Apln receptor (APLNR, angiotensin receptor-like 1) and intra-cellular coupling to Gαi. Downstream to this receptor complex, APLN signaling diverges on several signaling pathways such as phosphoinositol 3-kinase (Pl3K), phospholipase C (PLC), AMPK, and mitogenic extracellular signal-regulated kinase (ERK) in skeletal muscle (Besse-Patin et al., 2014; Chapman et al., 2014; Szokodi et al., 2002). Systemic Apln levels and local production by muscle decline with aging and an exogenous administration of the Apln peptide ameliorates age-associated pathologies such as cardiac hypertrophy, insulin resistance, and sarcopenia (Attane et al., 2012; Rai et al., 2017; Vinel et al., 2018, 2019).
The transcriptional regulation of the Apln promoter directly influences its circulating levels, and HIF and USF have been identified as transcription factors regulating Apln transcription based on targeted hypotheses (Han et al., 2008; Wang et al., 2006). It remains, however, unclear which network of transcription factors coordinates Apln expression in skeletal muscle and how the endogenous regulation of Apln expression influences paracrine communication. In this study, we performed a systematic evaluation of transcription factors regulating the Apln promoter using a yeast one-hybrid (Y1H) screen with a library containing 745 mammalian transcription factors (Gubelmann et al., 2013). Through this approach, we identified TEA domain family member 1 (Tead1) as a regulator of Apln expression in myofibers in vitro and in vivo. Finally, single-cell RNAseq analysis of healthy and regenerating muscle identified endothelial cells as the major cell type expressing Aplnr. Based on this observation, we demonstrate that Apln treatment or modulation of Apln production by Tead1 in myofibers drives endothelial remodeling and angiogenesis during muscle repair through paracrine signaling.
Results
Tead1 interacts with the apelin gene promoter in myogenic cells
To gain insight into the transcriptional mechanisms of Apln regulation, we investigated cis-regulatory elements of the Apelin precursor gene (Apln) and its regulatory transcription factors. Classic methods to discover TF-DNA interactions, such as gel shift assay and reporter assay combined with promoter deletion do not easily scale to a high-throughput evaluation of TF-DNA interactions. Hence, we used a high throughput yeast one-hybrid (Y1H) assay, which allows us to probe interactions between TFs and DNA sequences of interest at a large scale (Gubelmann et al., 2013). To identify a putative promoter region for Apln, we examined the epigenetic regulatory signatures and sequence conservation of its 5′ upstream region in the WashU Epigenome Browser (Figure S1A). The conservation score between 20 mammalian species described in (Miller et al., 2007; Zhou and Wang, 2012) is high within the −400 bp region upstream of the Apln transcriptional start site (TSS). Similarly, this region is enriched in H3K9ac and H3K4me3 histone marks and EP300 binding based on public ChIP-seq analyses (Gates et al., 2017; Liang et al., 2004), suggesting that it may contain the core Apln promoter. To determine the transcriptional activity of the Apln promoter region, we performed dual reporter gene assays in C2C12 myoblast cells (Figure S1B). −200/-1 bp and −400/-1 bp fragments of the Apln promoter showed the highest transcriptional activity, confirming that this proximal promoter contains the core activating elements for Apln transcription.
We then performed Y1H screens of different Apln promoter regions. Yeast lines containing the respective Apln promoter fragments were each mated with a library of yeast strains, each containing a prey vector that encodes one out of 745 mouse transcription factors that are fused to the Gal4 activation domain. Positive TF-Apln promoter fragment interactions were then identified based on the ability of the respective diploid yeast to grow on a selective plate (Figure 1A and (Gubelmann et al., 2013)). Among the positive Y1H interactions with the −200/-1 bp Apln promoter region, we identified six TFs (Tead1, Zic3, Barx1, Zfp319, Gcm2, and Zdhhc9) out of the 745 candidates in the TF ORF library (Figures 1B and 1C). Binding of the same six TFs was detected for the −400/-1 bp Apln promoter (Figures S1C and S1D), and longer promoter fragments did not result in further binding of additional TFs (data not shown).
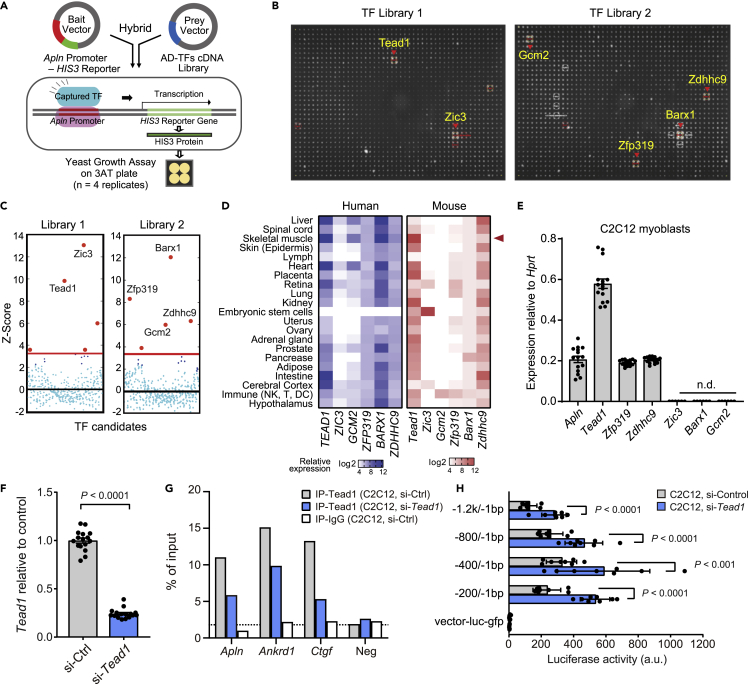
Figure 1.
A transcription factor screen identifies Tead1 as a direct regulator of apelin transcription
(A) Overview of the yeast one-hybrid (Y1H) assay to screen 745 transcription factors for their ability to interact with the Apelin (Apln) promoter. Binding of a transcription factor is readout via expression of the HIS3 reporter which enables yeast growth on a selective 3AT-containing medium plate.
(B) Y1H screen results using the −200/-1 bp fragment of the mouse Apln promoter as bait and 745 mouse TFs as prey (n = 4 replicates per tested TF, hence the formation of a quadrant in case of positive interaction).
(C) Z-score-normalized Y1H spot intensities for all 745 TFs. Six TFs with Z-scores above the background threshold (red line) are noted here and in (B).
(D) Relative mRNA expression of the six TF candidates in bulk mRNA profiling of human and mouse tissues. Microarray data for humans (left) from the GeneAtlas UI33A and mouse (right) from the GeneAtlas MOE430 of the bioGPS gene annotation portal. n = 2 replicates per tissue. Expression data are normalized and presented in a log2-scaled heatmap by species.
(E) mRNA expression of Apln and the six TFs in C2C12 myoblasts measured by RT-qPCR relative to Hprt. n.d., not detected. Mean ± SE of mean (SEM) of n = 16 replicates.
(F) mRNA expression of Tead1 in C2C12 myoblasts transfected with scrambled control or Tead1 targeted siRNAs for 3 d n = 16 replicates.
(G) ChIP-qPCR assay of Tead1 testing for binding to known target promoters (Ctgf, Ankrd1), Apln promoter (−177/-77 bp), or a random negative control in C2C12 myoblasts treated with either scrambled control or Tead1-targeted siRNA for 3 days. ChIP was performed with Tead1 or IgG control antibodies and qPCR was normalized to IP input. n = 1 biological replicate.
(H) Luciferase activity of five Apln promoter fragments transfected into C2C12 myoblasts at D3 with scrambled or Tead1 siRNA. Vector control contains no Apln promoter. Mean ± SEM of n = 8 biological replicates. p values are reported from two-tailed, unpaired t-tests between siRNA conditions.
To prioritize the relevance of the candidate Apln promoter binding-TFs to skeletal muscle physiology, we compared the mRNA expression of the six TFs in mouse and human tissues using datasets in the bioGPS portal (Su et al., 2004; Wu et al., 2013). Tead1, Zfp319, Zdhhc9, and Barx1 expressions were all detectable in skeletal muscle from both mice and humans (Figure 1D). Tead1, Zfp319, and Zdhhc9 were also expressed in C12C12 myoblasts (Figure 1E) and were selected for functional validation through siRNA knock-down. Knock-down of Zdhhc9 did not influence Apln expression in C2C12 cells and knock-down of Zfp319 reduced Apln mRNA but did not influence Apln peptide secretion (Figures S2A–S2F). Given that Zdhhc9 and Zfp319 are not expressed in myogenic cells from single-cells and nucleus RNAseq dataset (Figure S4A), we tested whether Tead1 binds and regulates the Apln promoter in myogenic cells. In chromatin immunoprecipitation (ChIP) experiments, we found that Tead1 specifically binds to the proximal Apln promoter (Figure 1G), as well as regulatory regions of the previously reported TEAD1 target genes Ankrd1 and Ctgf (Stein et al., 2015). The binding of Tead1 to the Apln promoter was reduced upon siRNA knockdown of Tead1 (Figures 1F and 1G). We examined publically available Tead1 ChIP-seq analysis of mouse heart tissue (Akerberg et al., 2019) and observed Tead1 binding enrichment associated close to the proximal promoter region of Apln. This agrees with our ChIP-qPCR data demonstrating Tead1 binding on Apln promoter region using the primer sets −123/-23 bp from the TSS. Motif enrichment analysis did not detect the canonical Tead family MCAT binding motif (CATTCC) in the -1 kb/-1 bp but identified significant enrichment of CATT and ATTC Tead motifs in multiple regions within the −500/-1 bp of Apln. Functionally, the siRNA knockdown of Tead1 increased the activity of an Apln promoter-luciferase reporter (Figure 1H), highlighting that Tead1 represses the Apln promoter in muscle cells as also previously reported in other cell types (Kim et al., 2015).
Tead1 suppresses apelin secretion in muscle
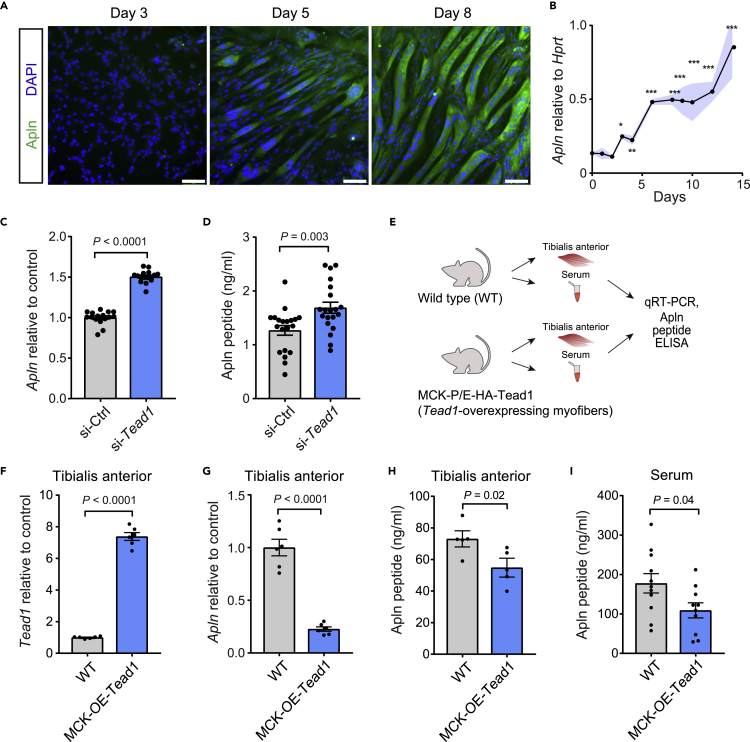
The Apln peptide is a myokine previously documented to be produced by skeletal muscle during contraction (Vinel et al., 2018). Apln is also expressed in myogenic cells (Latroche et al., 2017) (Vinel et al., 2018), and induced at the transcript and protein levels during myogenic differentiation (Figures 2A and 2B). To evaluate if Tead1 regulates the endogenous Apln promoter, we performed Tead1 knockdown by siRNA in muscle cells. The Apln transcript and peptide levels were significantly increased by 50 and 24%, respectively, when Tead1 was knocked down (Figures 2C, 2D, and 1F). The repression of Apln by Tead1 in myofibers was then tested in vivo by analyzing Apln levels in mice overexpressing Tead1 specifically in mature myofibers under the control of the muscle creatine kinase (MCK) promoter (Figures 2E–2I) (Southard et al., 2016; Tsika et al., 2008). Consistent with the in vitro data, myofiber-specific Tead1 overexpression resulted in lower levels of Apln mRNA and peptide in skeletal muscle (Figures 2F–2H). Overexpression of Tead1 in myofibers also reduced systemic Apln levels in serum (Figure 2I), demonstrating that the regulation of the Apln gene by Tead1 in muscle directly influences systemic levels of Apln in the circulation and that skeletal muscle is a direct contributor to circulating Apln levels.
Figure 2.
Apelin is repressed by Tead1 in muscle cells in vitro and in vivo
(A) Immunostaining of Apln protein during C2C12 myotube differentiation. Scale bars, 100 μm.
(B) Quantification of Apln mRNA by RT-qPCR during C2C12 myotube differentiation relative to Hprt using 2–dCt method. n = 4 cell culture replicates per time point.
(C and D) Quantification of apelin mRNA by RT-qPCR (C) or apelin peptide in supernatant by ELISA (D) in C2C12 myoblasts transfected with scrambled control or Tead1 targeted siRNAs for 3days n = 16-20 replicates per condition.
(E–I) Analysis of Tead1 and in adult mice over-expression Tead1 in skeletal muscle myofibers under the muscle creatine kinase (MCK) promoter (MCK-OE-Tead1 mice), compared to WT C57BL6 controls. (F and G) Tead1 mRNA (F) and Apln mRNA (G) expression levels were measured by RT-qPCR and normalized to Hprt in tibialis anterior (TA) muscles. n = 6 mice per condition. (H and I) Apln peptide concentration measured by ELISA in TA muscles (H) or serum (I). n = 5 mice per condition for TA; n = 11 mice per condition for serum. All data are presented as mean ± SEM, and p values are reported from two-tailed, unpaired t-tests between conditions. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Tead1-Apln-Aplnr expression patterns suggest paracrine signaling to endothelial cells
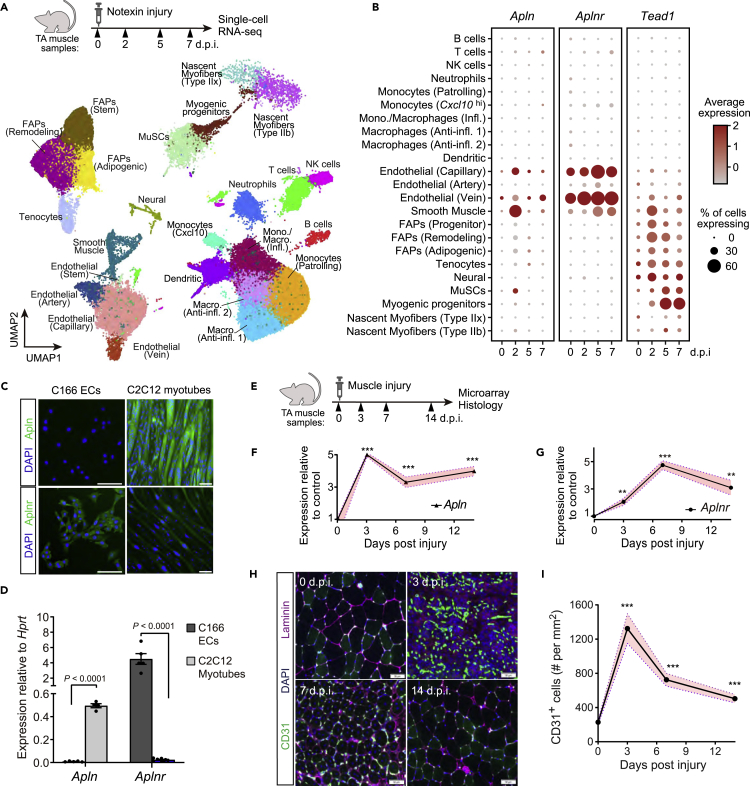
The recovery of skeletal muscle following tissue damage is essential to maintain muscle mass and strength and relies on coordinated expansion and differentiation of myogenic and non-myogenic cell types (Bentzinger et al., 2013). Our previous work demonstrating that Apln promotes muscle regeneration (Vinel et al., 2018) prompted us to examine how Tead1 and Apln signaling crosstalk in the niche during muscle repair by analyzing a notexin-induced muscle regeneration single-cell RNA sequencing dataset previously reported (De Micheli et al., 2020). In this dataset analyzing mononucleated cells, Apln expression in mature multinucleated myofibers cannot be captured as Apln is induced during terminal phases of myogenic differentiation after myogenic fusion and maturation (Figures 2A and 2B). Out of the mononucleated niche cells analyzed, low levels of Apln expression were detected in endothelial and smooth muscle cells (Figures 3A and 3B). Conversely, Tead1 was weakly expressed in most non-immune cell clusters, including in the myogenic progenitor cell (Myod1+) cluster, compared to all non-myogenic cells at 7 days post-injury (d.p.i.; false discovery-adjusted p = 2.2 × 10−164). To understand how Apln secretion signals during muscle regeneration, we analyzed which recipient cells from the muscle stem cell niche express the apelin receptor at 0-7 days.p.i. in this dataset (Figures 3A, 3B, and S3). Aplnr expression was detected in the capillary (Kdr+ Pecam1+) and vein (Vwf+ Pecam1+) endothelium cell clusters at most time points. At 7 days.p.i., Aplnr was enriched in all endothelial cells relative to all non-endothelial cells (false discovery-adjusted p = 0.038). To confirm the specificity of Apln and Aplnr expression in the niche and identify simple cell culture models to study apelin signaling across the niche, we analyzed the expression of Apln and Aplnr mRNA and protein in endothelial cells and myotubes in vitro. As expected from the single-cell RNAseq, Aplnr is specifically expressed by mouse C166 yolk sac-derived endothelial cells (Wang et al., 1996), while Apln is specifically expressed by C2C12 myotubes (Figures 3C and 3D). Given that Aplnr is highly enriched in endothelial cells, we hypothesized that Apln may regulate endothelial cell behavior and angiogenesis following muscle injury. To evaluate this possibility, the dynamics of Apln production and endothelial cell remodeling were measured in an in vivo model of muscle regeneration (Figures 3E–3G). Transient activation of Apln expression during muscle regeneration directly mirrored the dynamics of CD31/Pecam1+ endothelial cells remodeling, with both Apln expression and CD31+ endothelial cells peaking in the initial phases of muscle repair at 3 days.p.i. (Figures 3F, 3H and 3I). Thus, Aplnr is enriched in endothelial cells and the timing of Apln production correlates with endothelial remodeling during tissue repair.
Figure 3.
Apln, Aplnr, and Tead1 expression dynamics in regenerating skeletal muscle
(A and B) Single-cell RNA-sequencing analysis of a notexin injury response in TA muscle adult mice. TA muscle samples from 0, 2, 5, and 7 days post-injury (d.p.i.) with n = 2-3 mice per time-point were analyzed from De Micheli et al. (2020) (A) UMAP projection of scRNAseq data demonstrating cell-type annotations of clusters using markers shown in Figure S3.
(B) Dot plots showing expression of Apln, Aplnr, and Tead1 by cell-type cluster and time-point. Dot size shows the frequency of cells expressing non-zero transcript level. Dot color shows average expression level.
(C and D) In vitro expression of Apln and Aplnr protein by immunofluorescence (C) and mRNA by qRT-PCR (D) in C166 endothelial cells (ECs) and C2C12 myotubes differentiated for 8days n = 5 for C166 ECs; n = 4 for C2C12 myotubes. Scale bar, 100 μm.
(E–I) Regeneration of TA muscles of adult WT mice after IM injection of glycerol analyzed at 0, 3, 7, and 14 days.p.i. by gene expression microarray and immunohistology.
(E) Experimental overview. (F and G) Apln and Aplnr mRNA levels from transcriptomic analyses, normalized and presented as fold-change relative to mean of 0 days.p.i. Data are mean ± SEM of n = 5 (0, 14 days.p.i.) and n = 6 (3, 7 days.p.i.) mice. (H) Representative images of CD31 and Laminin immunostaining in regenerating muscle samples at 0, 3, 7, and 14 days.p.i. Scale bar, 50 μm. (I) Quantification of CD31+ endothelial cells per cross-sectional area.
Data are mean ± SEM of n = 5 TA muscles. In (F–G) and (I), p values are reported by two-tailed unpaired t-test compared to 0 days.p.i.; with ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Apelin stimulates endothelial remodeling during muscle regeneration
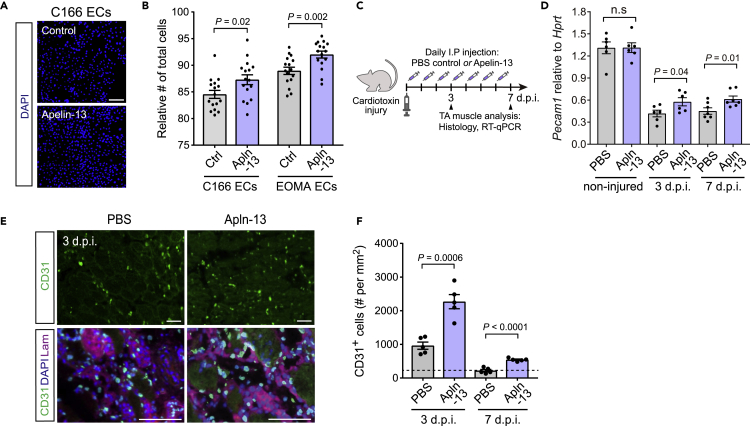
To examine if Apln directly regulates endothelial cell remodeling, we treated C166 ECs as well as the mouse EOMA hemangioendothelioma EC line with the recombinant bioactive Apln-13 peptide for 5 days and observed increased endothelial cell expansion with both EC lines (Figures 4A and 4B) (Obeso et al., 1990). We next asked if Apln signaling influences angiogenesis during muscle regeneration in vivo. Based on the prior report of reduced Apln production during aging (Vinel et al., 2018), we examined the effect of daily Apln-13 administration at 0.5 μmol/kg/day in aged mice following cardiotoxin-induced muscle injury (Figure 4C). Apln-13 treatment increased the abundance of CD31+ endothelial cells by IHC and elevated Pecam1 expression by bulk muscle RT-qPCR at both 3 and 7 days.p.i. compared to vehicle-treated controls without changing Pecam1 during homeostasis (Figures 4D–4F). These data indicate that short-term Apln supplementation induces the proliferation of endothelial cells and acts as a pro-angiogenic factor exclusively during the early stage of muscle regeneration without affecting the intact endothelium.
Figure 4.
Apln promotes endothelial cell remodeling in vitro and in vivo
(A and B) EC expansion of C166 and EOMA lines treated for 5days with the recombinant Apln-13 peptide at 10μM.
(A) Representative images of DAPI staining of C166 ECs. Scale bar, 100 μm.
(B) Quantification of the total number of C166 and EOMA cells treated with Apln compared to the control medium. (n = 16 cell culture replicates per group).
(C–F) Daily Apln-13 administration at 0.5 μmol/kg/day for 7days in aged mice following cardiotoxin-induced injury in TA muscle. (D) Expression of Pecam1 mRNA by RT-qPCR in whole TA muscles at 3 and 7 days.p.i. and non-injured TA muscles (n = six to seven TA muscles per group). (E) Representative images of CD31 and Laminin immunostaining in CTX-injured TA muscles at 3 days.p.i. Scale bar, 50 μm. (F) Quantification of CD31+ endothelial cells at 3 and 7 days.p.i. in regenerating regions (n = 5 TA muscles per group).
Dashed line indicates the basal level of CD31+ cell per mm2 in non-injured TA muscles (from Figure 3I). In (B), (D) and (I), p values are reported by two-tailed unpaired t-test compared to 0 days.p.i.; n.s represents p > 0.05.
The Tead1-Apelin axis regulates endothelial remodeling during muscle regeneration
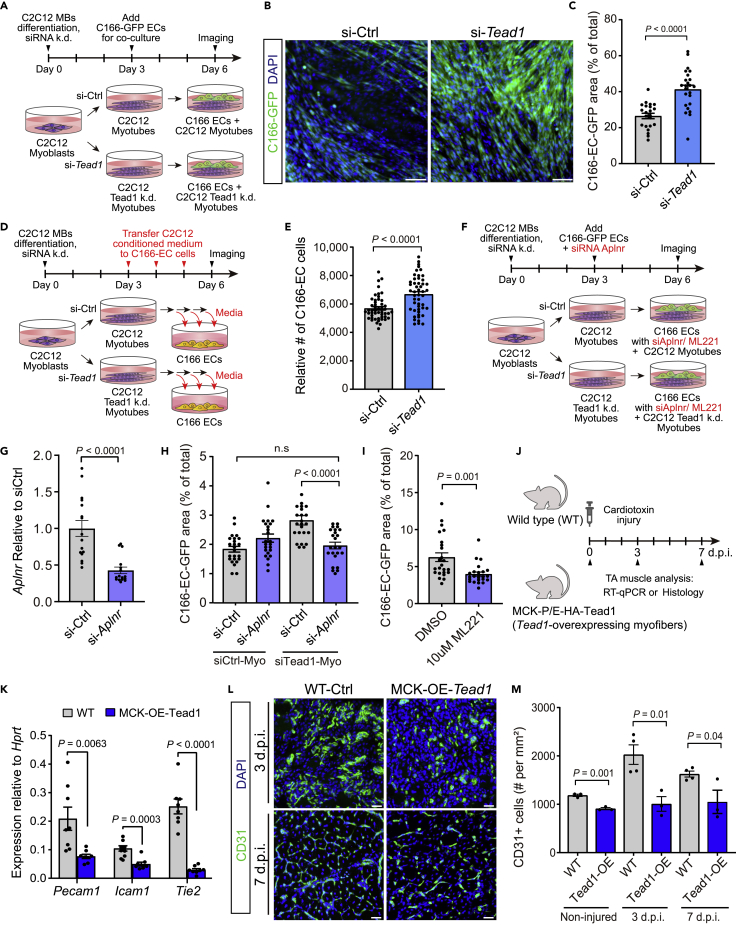
Given the prior findings, we hypothesized that the paracrine communication between myogenic cells and endothelial cells via Apln may be modulated by myogenic Tead1. We tested this in a co-culture system with GFP-expressing C166 ECs cultured directly on pre-differentiated C2C12 myotubes, with or without Tead1 depletion by siRNA (Figure 5A). After 3days of co-culture, EC growth reflected by total GFP+ cell area was enhanced in co-cultures on si-Tead1 C2C12 myotubes compared to C2C12 myotubes (Figures 5B and 5C). To test if this co-culture effect arises from direct interactions or secreted factors, we assayed C166 EC expansion in the presence of transferred conditioned media from C2C12 myotube mono-cultures starting at 3 days of differentiation and continued daily (Figure 5D). In this system, we found that the conditioned medium from si-Tead1 treated C2C12 myotubes enhanced C166 EC cell growth compared to the conditioned medium from control C2C12 cultures, suggesting that Tead1-regulated secreted factor(s) mediate the myogenic-endothelial cell communication effect (Figure 5E). We next tested if this cell communication occurs through Aplnr by using siRNA knock-down (kd) of Aplnr in ECs or cotreating co-culture system with the Aplnr antagonist ML221 (Maloney et al., 2010). The paracrine effect on EC induced by kd of Tead1 in myotubes was attenuated in the presence of Aplnr kd in ECs or ML221, demonstrating that Tead1 regulates myogenic-endothelial crosstalk via Apln-Aplnr signaling (Figures 5F–5I).
Figure 5.
Myogenic Tead1 inhibits myogenic-endothelial cross-talk
(A-C) Co-culture of GFP expressing C166 ECs with myotubes derived from C2C12 myoblasts transfected with scrambled control or Tead1 targeted siRNAs for 3days
(B) Representative images of C166-GFP EC and C2C12 co-cultures at 6days, with DAPI counter-stain. Scale bar, 100 μm.
(C) Quantification of the total GFP + cell area relative to the total image area. (n = 24 cell culture replicates per group).
(D and E) Culture of C166 ECs in presence of conditioned medium harvested from myotubes derived from C2C12 myoblasts transfected with scrambled control or Tead1 targeted siRNAs. Conditioned media were collected from myotube cultures and applied daily to C166 EC cultures for 3 days.
(E) Quantification of the number of C166 ECs per well in conditioned medium from control and Tead1 siRNA treated C2C12 myotubes. (n = 49 wells cell culture replicates per group).
(F-I) Co-culture of GFP expressing C166 ECs with myotubes derived from C2C12 myoblasts transfected with scrambled control or Tead1 targeted siRNAs in presence of Aplnr siRNAs or Aplnr inhibitor (ML221).
(G) Aplnr expression after the transfection of siRNAs targeted Aplnr compared to si-scrambled control (n = 16 per group).
(H) Quantification of the total GFP + cell area relative to total image area after siAplnr k/d (n = 24 per group).
(I) Quantification of the total GFP + cell area relative to total image area after 10μM ML221 (Aplnr inhibitor) compared to DMSO control (n = 24 per group).
(J–M) Analysis of MCK-Tead1 overexpressing mice following cardiotoxin-induced injury in TA muscle compared to WT controls. (K) Expression of Pecam1, Icam1, and Tek (Tie2) mRNA by RT-qPCR in tibialis anterior muscles isolated from adult myofiber-specific Tead1-overexpressing mice (MCK-OE-Tead1) compared to WT C57BL6 controls. (n = 4 mice per group). (L) Representative images of CD31 immunostaining in CTX-injured TA muscles at 3, 7 days.p.i. Scale bar, 50 μm. (M) Quantification of CD31+ endothelial cells at non-injured, 3 and 7 days.p.i. in regenerating regions (n = three to four TA muscles per group).
All graphs are reported as mean ± SEM and p values are reported from two-tailed, unpaired t-test between the conditions.
Finally, we examined MCK-OE-Tead1 mice to test if Tead1-overexpressing myofibers, which reduces Apln secretion (Figures 2E–2I), influence endothelial phenotypes in vivo after muscle injury (Figures 5J–5M). In MCK-OE-Tead1 transgenic muscles, the basal levels of mRNA expression of the canonical endothelial genes Pecam1, Icam1, and Tek (also known as Tie2) and the number of CD31+ ECs by IHC were decreased (Figures 5K–5M). In addition, the expansion of ECs after muscle injury was reduced at 3 and 7 days.p.i in the MCK-OE-Tead1 mice (Figures 5L and 5M). This observation demonstrates that Tead1 acts by suppressing pro-angiogenic paracrine secretion from myofibers, and establishes the Tead1-Apln regulation in myofibers as a contributing mechanism to myogenic-endothelial cross-talk during muscle regeneration.
Discussion
Serum and muscle Apln decline during aging in humans and low Apln levels are associated with loss of muscle mass and strength in older people (Vinel et al., 2018). Pre-clinical studies have shown the therapeutic potential of Apln supplementation as Apln-13 injection in aged mice could increase muscle strength and physical performance and boost regeneration after muscle injury (Vinel et al., 2018). These proof-of-concept observations bear promising therapeutic potential, but the applicability in humans is limited by the short half-life of the Apln peptide and the fact that it is not orally bioavailable. Stabilized Apln peptides and synthetic small molecule agonists of the Apln receptor constitute possible alternatives but understanding the endogenous regulatory pathways that modulate Apln production endogenously will allow the developing parallel strategies to modify local production specifically in target tissues.
In this study, we used a screen of 745 mammalian transcription factors to identify novel transcriptional regulators of Apln expression. Out of four confirmed screening hits, we characterize Tead1 as a bona fide regulator of Apln expression in myogenic cells using functional assays in vitro and in vivo. Tead1 has been shown to regulate the expression of several skeletal muscle-specific genes (Joshi et al., 2017; Qiu et al., 2011). Myofiber-specific overexpression of Tead1 induces a switch to a slow muscle contractile phenotype in vivo (Tsika et al., 2008), and induces hyperplasia of muscle stem cells (Southard et al., 2016). Although Tead1 generally activates transcription by recruiting coactivators such as Yap-Taz (Huh et al., 2019; Stein et al., 2015), repressive actions of Tead1 have been previously reported via co-repressor recruitment and coactivator squelching (Kim et al., 2015). In this study, we demonstrated that Tead1 binds to the Apln promoter in myotubes and knock-down of Tead1 is sufficient to boost Apln transcription and secretion, demonstrating that an endogenous repressive tone limits Apln secretion in muscle cells. Conversely, mice with myofiber-specific overexpression of Tead1 have reduced muscle and serum Apln concentrations, highlighting that the regulation of Apln by Tead1 is also at play in vivo and that skeletal muscle is a dominant contributor to systemic Apln levels.
To understand how apelin signals across the multiple cell types efficiently maintain and repair skeletal muscle, we analyzed the Tead1-Apln-Aplnr regulatory network using recent single-cell RNAseq datasets of the muscle stem cell niche. Consistent with previous reports (Latroche et al., 2017; Vinel et al., 2018), Apln was secreted by myogenic cells and myofibers and to some extent in endothelial and smooth muscle populations. Although Aplnr mRNA was not detected in myogenic cells by scRNAseq as the low expression of this GPCR cannot be captured with the sensitivity of scRNAseq, Apln can signal in an autocrine fashion in myogenic cells as these muscle stem cells express the Aplnr protein at sufficient levels to drive myogenic commitment (Latroche et al., 2017; Vinel et al., 2018). Consistent with these observations, Apln production increases in vivo after muscle injury, and systemic Apln injection further stimulates muscle stem cell amplification and myofiber repair after a muscle injury (Vinel et al., 2018). Our scRNAseq analysis also detected very strong enrichment of the Aplnr mRNA in various endothelial cell populations during muscle regeneration, highlighting that endothelial cells are active receiving cells for Apln signals in the niche.
The injured muscle stem cell niche is a hypoxic environment and the efficient coupling of myogenesis and angiogenesis is required to rebuild functional myofibers with adequate vascularization for oxygen and nutrient supply (Barnouin et al., 2017; Drouin et al., 2019; Duscha et al., 1999; Latroche et al., 2015; Luque et al., 1995). In particular, muscle stem cells and their myogenic progeny attract endothelial cells and orchestrate re-vascularization during tissue repair (Christov et al., 2007; Latroche et al., 2015, 2017). Apln has been previously reported to regulate angiogenesis and vascular formation during development and adult physio-pathology in tissues like the heart or the retina (Kidoya and Takakura, 2012; Wu et al., 2017), and was identified as a mediator of the cross-talk between myogenic and endothelial cells in skeletal muscle using the loss of function approaches (Latroche et al., 2017). Our experiments with the recombinant Apln peptide further link Apln signaling to endothelial remodeling in muscle, and open translational opportunities by proving that Apln-mediated activation of angiogenesis can be further enhanced beyond its endogenous tone through therapeutic activation of Apln signaling in vivo. Importantly, cellular co-culture, conditioned medium transfer, myofiber-specific Tead1 gain or loss of function, and AplnR inhibition in ECs established the directionality of the Apln-mediated myogenic-angiogenic crosstalk, demonstrating that Tead1 regulation of Apln in myogenic cells directly stimulates endothelial cell expansion through Aplnr. Considering that capillarization in skeletal muscle positively correlates with myofiber size and function (Barnouin et al., 2017; Takahashi et al., 2002), our results support a model where the regulation of endothelial cell remodeling and angiogenesis by Apln contributes to the reported benefits of Apln treatment on muscle mass and muscle strength (Vinel et al., 2018).
Collectively, our experiments establish a novel regulatory pathway controlling Apln secretion where Tead1 transcriptionally controls Apln expression in myofibers to regulate endothelial remodeling via paracrine communication. This regulation highlights that the beneficial effects of Apln during muscle regeneration are mediated by inter-cellular paracrine communication across the niche and further strengthen the importance of myogenesis-angiogenesis coupling during muscle repair. The therapeutic applications of recombinant Apln and Aplnr agonists are actively investigated in cardiovascular and metabolic diseases (Maloney et al., 2010; Narayanan et al., 2016; Read et al., 2016). In addition to the clinical studies on heart failure, pulmonary disease, and type 2 diabetes (Brame, NCT02129309; Cheriyan, NCT02150694; Novartis, NCT02696967; Gourdy, NCT02724566), our results suggest that Apln agonists could plausibly be used to prevent skeletal muscle diseases by enhancing myogenic-angiogenic signaling (Brame, 2014; Cheriyan, 2015; Novartis Pharmaceuticals, 2020).
Limitations of the study
While our study established that endothelial cells are the cells with the highest expression of Aplnr mRNA in skeletal muscle using scRNAseq, the Aplnr is a GPCR, which can still induce intra-cellular signaling at low expression levels and may also act in other cell types. For example, low but significant expression of the Aplnr protein was measured by FACS in MuSCs and the Apln peptide can directly regulate myogenic cell differentiation in vitro (Vinel et al., 2018), suggesting that Apln can enhance muscle regeneration via endothelial remodeling but also through a direct effect in myogenic cells. Similarly, we have studied the regulation of Apln by Tead1 in myofibers which are the cells with strongest Tead1 expression in skeletal muscle, but Tead1 is also expressed in smooth muscle cells, pericytes, neural cells, and FAPs (Figure S4). Thus, other cell types of the niche may also contribute to the regulation of Apln. Finally, while we detected binding of Tead1 to the Apln promoter and direct repressive roles of Tead1 have been described on other promoters (Kim et al., 2015), we have not characterized the full epigenetic mechanisms of repression and cannot fully exclude that Tead1 may also repress Apln indirectly by activating a transcriptional repressor.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Myosin Heavy Chain antibody, clone A4.1025 | Millipore | Cat#05–716; RRID:AB_309930 |
| Anti-Tead1 (TEF-1 Pure) | BD Biosciences | Cat# 610922; RRID:AB_398237 |
| Anti-IgG | Millipore | Cat# Magna 0014 |
| Anti-APJ receptor antibody | Abcam | Cat# ab214369 |
| Anti-Apelin antibody | Abcam | Cat# ab125213 RRID:AB_10999708 |
| Anti-Laminin antibody | LS Bio | Cat# LS-C96142 RRID:AB_2033342 |
| Anti-CD31 (Pecam1) antibody | BD Biosciences | Cat# 557355 RRID:AB_396660 |
| Critical commercial assays | ||
| One-Glo Firefly Luciferase Detection Kit | Promega | Cat# E8120 |
| Nano-Glo Dual-Luciferase Detection Kit | Promega | Cat# N1620 |
| Glo-lysis Buffer | Promega | Cat# E2661 |
| Lipofectamine RNAiMax | Thermofisher | Cat# S-006-100 |
| Apelin EIA kit | Phoenix pharmaceuticals | Cat# EK-057-23 |
| Gibson assembly kit | NEB | Cat# E5510 |
| FastLane cell multiplex NR kit | Qiagen | Cat# 216713 |
| miRNeasy Mini Kit | Qiagen | Cat# 217004 |
| Covaris truChIP chromatin shearing kit | Covaris | Cat# SKU:500465 |
| Magna ChIP A/G kit | Millipore/Sigma | Cat#17–10085 |
| Minielute DNA purification kit | Qiagen | Cat# 28004 |
| Maxima Sybr Green/ROX master mix | Thermo Scientific | Cat# K0221 |
| Chemicals, peptides, and recombinant proteins | ||
| Recombinant Phy Apelin-13 | Bachem | Cat# U-01260 |
| Cardiotoxin | Laxotan | Cat# L8102 |
| Notexin | Laxotan | Cat# L8104 |
| Deposited data | ||
| GSE1133 | bioGPS | GeneAtlas UI33A (gcrma) for human samples |
| GSE1133 | bioGPS | GeneAtlas MOE430 (gcrma) for mouse tissues |
| GSE45577 | GEO | (Lukjanenko et al., 2013) https://doi.org/10.1371/journal.pone.0071084 |
| GSE143435, GSE143437, GSE159500 | GEO | (McKellar et al., 2021) https://doi.org/10.1101/2020.12.01.407460 |
| Experimental models: Cell lines | ||
| C2C12 myoblast cell line | ATCC | Cat# CRL1772 RRID:CVCL_0188 |
| C166 endothelial cell line | ATCC | Cat# CRL2581 RRID:CVCL_6581 |
| EOMA endothelial cell line | ATCC | Cat# CRL2586 RRID:CVCL_3507 |
| C166GFP endothelial cell line | ATCC | Cat# CRL2583 RRID:CVCL_6582 |
| Experimental models: Organisms/strains | ||
| C57BL/6J – 24month old | Jackson | Cat# 000664 RRID:IMSR_JAX:000664 |
| Mck-Tead1 OE mice | (Southard et al., 2016) | |
| Oligonucleotides | ||
| si-RNA scramble pool | Dharmacon | Cat# D-001810-10–50 |
| si-Tead1 pool | Dharmacon | Cat# L-048419-01–0005 |
| si-Zdhhc9 pool | Dharmacon | Cat# L-058018-01–0005 |
| si-Zfp319 pool | Dharmacon | Cat# L-059731-01–0005 |
| Apln taqman probe | Life Technologies | Mm00443562_m1 |
| Hprt taqman probe | Life Technologies | Mm00446968_m1 |
| Tead1 taqman probe | Life Technologies | Mm00493507_m1 |
| Barx1 taqman probe | Life Technologies | Mm01353100_m1 |
| Zic3 taqman probe | Life Technologies | Mm00494362_m1 |
| Zdhhc9 taqman probe | Life Technologies | Mm00552609_m1 |
| Gcm2 taqman probe | Life Technologies | Mm00492312_m1 |
| Pecam1 taqman probe | Life Technologies | Mm01242576_m1 |
| Icam 1 taqman probe | Life Technologies | Mm00516023_m1 |
| Tek (Tie2) taqman probe | Life Technologies | Mm00443243_m1 |
| Recombinant DNA | ||
| N/A | ||
| Software and algorithms | ||
| SnapGene | Snapgene | N/A |
| metaXpress | Molecular Devices | N/A |
| VS-ASW FL software | Nikon | N/A |
| MetaXpress software | Molecular Devices | N/A |
| NIS-Elements | Nikon | N/A |
| GraphPad Prism Software | GraphPad | N/A |
| ApE | ApE | version 7–9 |
| Transcription factor-DNA interaction detection in yeast (TIDY) | (Hens et al., 2011) |
https://updeplasrv1.epfl.ch/software/ https://doi.org/10.1038/nmeth.1763 |
| Other | ||
| Seurat | (Butler et al., 2018) | version3.1.5 |
| Harmony | (Korsunsky et al., 2019) | version 1.0 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled where possible by the lead contact, Jerome Feige (jerome.feige@rd.nestle.com).
Materials availability
All stable reagents generated in this study are available from the lead contact without restriction.
Experimental model and subject details
Cell culture and differentiation
C2C12 cells were harvested in a growth medium formulated with 20% of heat-inactivated Fetal Bovine Serum (FBS) and 1% penicillin and streptomycin (Pen/Strep) in DMEM with or without glutamax supplemented (Blau et al., 1985; Yaffe and Saxel, 1977). For the myotube differentiation, 5% horse serum was added to DMEM with 1%Pen/Strep, and media was changed daily. For the endothelial cell growth assay, we used C166 (ATCC) and EOMA (ATCC) endothelial cells from ATCC and cultured them in DMEM supplemented with 20% FBS and 1% Pen/Strep (Obeso et al., 1990; Wang et al., 1996).
Mouse experiments and muscle injury models
All animal experiments were approved by The Cornell University Institutional Animal Care and Use Committee (IACUC) or the cantonal authorities of Vaud, Switzerland under license VD2620. 25 μL of 10 uM cardiotoxin (Laxotan, Cat#L8102), 10 μL of 10 μg/mL Notexin (Laxotan, Cat#L8104), or 25 μL of 50% glycerol was injected to tibialis anterior (TA) muscle through intermuscular injections under isoflurane induced anethesia. Mice were sacrificed at indicated time points with cervical dislocation (US) or CO2 (CH). After TAs were harvested, the TA muscles were embedded in OCT, and frozen in liquid nitrogen-cooled isopentane for histological analysis. Conclusions from Apln treatment in aged mice during muscle regeneration were derived from re-use of existing samples previously generated and described in (Vinel et al., 2018). Briefly, Apln-13 was administered by daily i.p. injection at 0.5 μmol/kg/day in 24 month old mice following cardiotoxin-induced injury in TA muscle.
Method details
Dual reporter assay
C2C12 cells were co-transfected with the apelin promoter plasmid fused with the nano-luciferase encoding gene and a plasmid with the constitutively active cytomegalovirus (CMV) promoter conjugated with firefly luciferase. Stop&Glo reagents were purchased from Promega (Cat# E8120, N1620, E2661). Three days after transfection, cells were washed with PBS and lysed. After 15 min of incubation, 65 μL of the lysate (supernatant) were mixed with ONE GLO EX firefly luciferase detection reagent and measured luminescence with the integration time of 1 min. Subsequently, 65 μL of NanoDLR STOP&GLO was added and luminescence was measured after mixed on an orbital shaker. The activity of NanoLuc was normalized based on the constitutively active firefly luciferase activity.
Yeast one-hybrid screen
A large-scale library of TF Open Reading Frame (ORF) clones (768 mouse TFs) was created as described previously (Gubelmann et al., 2013). For bait construction, four different sizes of the Apln promoter region were selected as described in the promoter characterization section. Those promoter fragments were inserted into a yeast-compatible pMW2 vector containing the HIS3 gene with the Gibson assembly kit (NEB) after which bait-HIS3 reporter yeast lines were generated, as described previously (Gubelmann et al., 2013). Yeast lines that showed minimal background reporter expression were then selected for mating with the compatible mouse TF ORF yeast library whereby each interaction is tested in quadruplicate, also as described (Gubelmann et al., 2013). After one week of incubation, positive TF-DNA interactions were then identified in semi-automated fashion based on growth on a selective yeast plate containing 3-amino-1,2,4-triazole (3AT) using “transcription factor-DNA interaction detection in yeast (TIDY)” software (Hens et al., 2011). In short, TIDY calculates the intensity values of each quadrant (i.e. four replicates of the same, tested protein-DNA interaction) and groups these into 10 clusters. Among these clusters, the highest intensity value of the largest cluster, which most likely represents the bulk of negative interactions, was used as the background threshold. Then, the TF yeast quadrants whose intensity values are above at least 20% of the background value are selected as positive hits.
Cell knock-down experiments
C2C12 myoblasts were harvested without pen/strep 24 h prior to transfection and the cells were split and distributed at 1000 per well in 96-well plate. When the cells were in the suspension, the complex of transfection reagent RNAiMax (Thermofisher) with the siRNAs was directly added to the cells. The siRNAs were synthesized by Dharmacon (Cat# L-048419-01–0005 for si-Tead1, L-058018-01–0005 for Zdhhc9, L-059731-01–0005 for Zfp319), and we used scrambled siRNAs for the negative control (Cat#D-001810-10–50). After 3 days of incubation, mRNA or protein were extracted and the following experiments were performed.
Chromatin immunoprecipitation (ChIP) qPCR assay
Cells were treated with siRNAs as described in Knock-down experiments and harvested until 100% confluency. Cells were fixed with 1% formaldehyde, and nuclei were prepared with Covaris truChIP Chromatin Shearing Kit (Cat# SKU:500465) by following the manual for “high cell” from the producer. The extracted chromatin was sonicated for 90 s using Covaris E220 (5% duty cycle and intensity 4). Immunoprecipitation was performed using Magna ChIP A/G kit from Millipore Sigma (Cat# 17–10085) and 10 μg of chromatin was incubated with 1 μg of anti-Tead1 (Cat# 610922 BD Biosciences) or IgG (Cat# Magna 0014) which were pre-incubated with 20 μL of magnetic beads overnight at 4°C. Followed by low salt, high salt, Lici, and TE washing steps with immunoprecipitation using a magnetic rack as described in the manual of Magna ChIP kit, DNA was eluted, reverse-crosslinked at 65°C overnight, and purified using Minielute PCR column purification (Qiagen, Cat#28004). For qPCR, Maxima Sybr Green/ROX qPCR master mix (Thermo scientific, Cat# K0221) was used and performed on ViiA seven Real-Time PCR system (Life Technologies). Ankrd1, Ctgf, Apln, Neg primers were used for qPCR detection as Table S1.
Real-Time qPCR
Total RNA was extracted with the miRNeasy mini kit (Qiagen, Cat# 217004) or Fastlane cell multiplex kit (Qiagen, Cat# 216713). TaqMan probes and the FastLane Cell Multiplex NR kit (Qiagen) were used to measure RNA levels. Following real time qPCR, the expression level of target gene mRNA was analyzed with a ddCT algorithm and normalized to a reference gene, Hprt. The TaqMan probes used are listed in the key resources table.
Protein extraction
After TA muscle is harvested, samples were weighed and calculated the lysis buffer as 10 μL per mg of tissue. The lysis buffer was formulated with 50mM Tris-HCl (pH 7.5), 1 mM EDTA, 1 mM EGFA, 0.27 M Sucrose, 1% Triton X-100, 20mM Glycerol-2-phosphate disodium, 50 mM NaF, and 5 mM Na4P2O7 with a protease inhibitor cocktail (Roche, Cat# 4693159001). The samples were homogenized by polytron and incubated on ice with the lysis buffer for 30 min. The protein was obtained by transferring the supernatant after spinning down the samples with a centrifuge at 3500 g with 4°C for 5 min and the concentration of protein was further determined by bicinchoninic acid assay (Peirce, Cat #23225).
Elisa detection of the apelin peptide
Following protein extraction, the apelin peptide was measured with the EIA kit (Phoenix pharmaceuticals, Cat# EK-057-23). 5 mg of protein from samples in 50 μL of lysis buffer were distributed on the immunoplate, which was pre-coated with secondary antibody and 25 μL of biotinylated peptide. After 2 h of incubation, the biotinylated peptide was catalyzed by streptavidin-horseradish peroxidase solution for 1 h. TMB substrate solution was added for 1 h at room temperature. Then, the reaction was terminated with 2N HCl. The concentration of apelin peptide was detected by absorbance at 450 nm and then quantified based on a standard curve.
Gene expression analysis with single-cell RNA sequencing data
Previously reported single-cell RNA sequencing data was prepared as described in McKellar et al. (2021) (McKellar et al., 2021). Briefly, raw reads were aligned to the mm10 reference genome using Cellranger version 3.1.0. Count matrices generated by Cellranger were analyzed using Seurat, version3.1.5 (Butler et al., 2018). Cells with fewer than 1000 transcripts detected or greater than 30% of transcripts mapping to mitochondrial genes were removed from the analysis. Batch correction was performed using harmony, version 1.0 (Korsunsky et al., 2019). After batch correction, clustering was performed via Seurat (FindClusters) using default parameters. Each cluster was labeled based on canonical gene expression. To minimize batch effects in gene expression values, but retain the clustering resolution enabled through the large-scale resource, only samples from Gene Expression Omnibus accession numbers GSE143435, GSE143437, and GSE159500 were subset out and used for this analysis. After quality filtering and subsetting, 67,985 cells were used.
Histology of endothelial cell infiltration in muscle tissue
Mouse muscle samples were dissected, embedded in OCT, and frozen in liquid nitrogen-cooled isopentane. Samples were then sectioned at 10 μm on a cryostat and fixed with 4% paraformaldehyde for 15 min at room temperature. After permeabilization with cold 100% methanol for 6 min, blocking was performed with 4% BSA for 3 h. Following blocking, primary antibodies, rat anti-mouse CD31 (BD Biosciences, Cat# 557355), chicken anti-mouse laminin (LS Bio, Cat# LS-C96142) were incubated at 4°C overnight at 1:500. Secondary antibodies with Hoechst (goat anti-rat A488 1/2000, goat anti-chicken-A647 1/200, Hoechst 1/5000) were incubated for 1 h at room temperature. Subsequently, the stained sections were mounted and imaged using a 10X objective on an Olympus VS120 fluorescence slide scanner and quantified in three randomly selected injured areas of each sample using the VS-ASV 2.8 software. The total number of endothelial cells was quantified as the total number of CD31+/DAPI + nuclei per area analyzed.
Quantification and statistical analysis
Statistical significance (P-value) for qPCR, luciferase activity, and cell quantification data were assessed using Student’s t-test (2 groups) or two-way ANOVA (multiple groups) in the software GraphPad Prism seven to 9. The exact sample number of each figure is reported in the figure legends.
Acknowledgments
This study was funded by Nestlé, EPFL Institutional Support, SNSF grant 310030_182655, SNSF Doc. Mobility grant_P1ELP3_187970, and the National Institutes of Health award R01AG058630. We also thank Joris Michaud, Eugenia Migliavacca, Cedric Gobet, Mathieu Membrez, Omid Mashinchian, Gabrielle Dammone, Tanja Sonntag, Caterina Collodet, Guillaume Jacot for technical support and helpful discussions.
Author contributions
U.L designed and performed experiments and analyzed results. B.D., B.D.C, and J.N.F. designed and supervised the study. P.S., S.K., J.R, M.D., C.L. performed experiments and generated critical experimental tools. D.W.M. performed single-cell RNAseq data analysis. U.L., B.D.C, and J.N.F. interpreted the results and wrote the article.
Declaration of interests
U.L., P.S., S.K., M.D. and J.N.F. are or were employees of the Société des Produits Nestlé S.A.
Inclusion and diversity
We worked to ensure sex balance in the selection of non-human subjects and to ensure diversity in the selection of cell lines. One or more of the authors of this article self-identifies as an under-represented ethnic minority in science. Although citing references scientifically relevant for this work, we also actively worked to promote gender balance in our reference list.
Published: July 15, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104589.
Supplemental information
Data and code availability
All data or any additional information required to reanalyze the data reported in this paper are available from the lead contact upon request. Transcriptomic datasets used in this study are available in the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) under the specific references below:
References
- Akerberg B.N., Gu F., VanDusen N.J., Zhang X., Dong R., Li K., Zhang B., Zhou B., Sethi I., Ma Q., et al. A reference map of murine cardiac transcription factor chromatin occupancy identifies dynamic and conserved enhancers. Nat. Commun. 2019;10:4907. doi: 10.1038/s41467-019-12812-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ancel S., Stuelsatz P., Feige J.N. Muscle stem cell quiescence: controlling stemness by staying asleep. Trends Cell Biol. 2021;31:556–568. doi: 10.1016/j.tcb.2021.02.006. [DOI] [PubMed] [Google Scholar]
- Attane C., Foussal C., Le Gonidec S., Benani A., Daviaud D., Wanecq E., Guzman-Ruiz R., Dray C., Bezaire V., Rancoule C., et al. Apelin treatment increases complete fatty acid oxidation, mitochondrial oxidative capacity, and biogenesis in muscle of insulin-resistant mice. Diabetes. 2012;61:310–320. doi: 10.2337/db11-0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnouin Y., McPhee J.S., Butler-Browne G., Bosutti A., De Vito G., Jones D.A., Narici M., Behin A., Hogrel J.-Y., Degens H. Coupling between skeletal muscle fiber size and capillarization is maintained during healthy aging. J. Cachexia Sarcopenia Muscle. 2017;8:647–659. doi: 10.1002/jcsm.12194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentzinger C.F., Wang Y.X., Dumont N.A., Rudnicki M.A. Cellular dynamics in the muscle satellite cell niche. EMBO Rep. 2013;14:1062–1072. doi: 10.1038/embor.2013.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besse-Patin A., Montastier E., Vinel C., Castan-Laurell I., Louche K., Dray C., Daviaud D., Mir L., Marques M.-A., Thalamas C., et al. Effect of endurance training on skeletal muscle myokine expression in obese men: identification of apelin as a novel myokine. Int. J. Obes. 2014;38:707–713. doi: 10.1038/ijo.2013.158. [DOI] [PubMed] [Google Scholar]
- Blau H.M., Pavlath G.K., Hardeman E.C., Chiu C.-P., Silberstein L., Webster S.G., Miller S.C., Webster C. Plasticity of the differentiated state. Science. 1985;230:758–766. doi: 10.1126/science.2414846. [DOI] [PubMed] [Google Scholar]
- Bloor C.M. Angiogenesis during exercise and training. Angiogenesis. 2005;8:263–271. doi: 10.1007/s10456-005-9013-x. [DOI] [PubMed] [Google Scholar]
- Brame A.N. Local haemodynamic effects of Apelin agonists and antagonists in man in vivo. 2014. clinicaltrials.gov
- Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castan-laurell I., Dray C., Knauf C., Kunduzova O., Valet P. Apelin, a promising target for type 2 diabetes treatment? Trends Endocrinol. Metab. 2012;23:234–241. doi: 10.1016/j.tem.2012.02.005. [DOI] [PubMed] [Google Scholar]
- Chapman N.A., Dupré D.J., Rainey J.K. The apelin receptor: physiology, pathology, cell signalling, and ligand modulation of a peptide-activated class A GPCR. Biochem. Cell Biol. Biochim. Biol. Cell. 2014;92:431–440. doi: 10.1139/bcb-2014-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chazaud B. Inflammation and skeletal muscle regeneration: leave it to the macrophages! Trends Immunol. 2020;41:481–492. doi: 10.1016/j.it.2020.04.006. [DOI] [PubMed] [Google Scholar]
- Cheriyan J. Haemodynamic effects of apelin agonists and antagonists in man in COPD with raised pulmonary artery pressures. 2015. clinicaltrials.gov NCT02129309.
- Christov C., Chrétien F., Abou-Khalil R., Bassez G., Vallet G., Authier F.-J., Bassaglia Y., Shinin V., Tajbakhsh S., Chazaud B., et al. Muscle satellite cells and endothelial cells: close neighbors and privileged partners. Mol. Biol. Cell. 2007;18:1397–1409. doi: 10.1091/mbc.e06-08-0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Micheli A.J., Laurilliard E.J., Heinke C.L., Ravichandran H., Fraczek P., Soueid-Baumgarten S., De Vlaminck I., Elemento O., Cosgrove B.D. Single-cell analysis of the muscle stem cell hierarchy identifies heterotypic communication signals involved in skeletal muscle regeneration. Cell Rep. 2020;30:3583–3595.e5. doi: 10.1016/j.celrep.2020.02.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouin G., Couture V., Lauzon M.-A., Balg F., Faucheux N., Grenier G. Muscle injury-induced hypoxia alters the proliferation and differentiation potentials of muscle resident stromal cells. Skelet. Muscle. 2019;9:18. doi: 10.1186/s13395-019-0202-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duscha B.D., Kraus W.E., Keteyian S.J., Sullivan M.J., Green H.J., Schachat F.H., Pippen A.M., Brawner C.A., Blank J.M., Annex B.H. Capillary density of skeletal muscle: a contributing mechanism for exercise intolerance in class II–III chronic heart failure independent of other peripheral alterations. J. Am. Coll. Cardiol. 1999;33:1956–1963. doi: 10.1016/S0735-1097(99)00101-1. [DOI] [PubMed] [Google Scholar]
- Egerman M.A., Glass D.J. Signaling pathways controlling skeletal muscle mass. Crit. Rev. Biochem. Mol. Biol. 2014;49:59–68. doi: 10.3109/10409238.2013.857291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs E., Blau H.M. Tissue stem cells: architects of their niches. Cell Stem Cell. 2020;27:532–556. doi: 10.1016/j.stem.2020.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gates L.A., Shi J., Rohira A.D., Feng Q., Zhu B., Bedford M.T., Sagum C.A., Jung S.Y., Qin J., Tsai M.-J., et al. Acetylation on histone H3 lysine 9 mediates a switch from transcription initiation to elongation. J. Biol. Chem. 2017;292:14456–14472. doi: 10.1074/jbc.M117.802074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski T., De Bock K. Metabolic regulation of exercise-induced angiogenesis. Vasc. Biol. 2019;1:H1–H8. doi: 10.1530/VB-19-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubelmann C., Waszak S.M., Isakova A., Holcombe W., Hens K., Iagovitina A., Feuz J., Raghav S.K., Simicevic J., Deplancke B. A yeast one-hybrid and microfluidics-based pipeline to map mammalian gene regulatory networks. Mol. Syst. Biol. 2013;9:682. doi: 10.1038/msb.2013.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S., Wang G., Qi X., Lee H.M., Englander E.W., Greeley G.H. A possible role for hypoxia-induced apelin expression in enteric cell proliferation. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2008;294:R1832–R1839. doi: 10.1152/ajpregu.00083.2008. [DOI] [PubMed] [Google Scholar]
- Hens K., Feuz J.-D., Isakova A., Iagovitina A., Massouras A., Bryois J., Callaerts P., Celniker S.E., Deplancke B. Automated protein-DNA interaction screening of Drosophila regulatory elements. Nat. Methods. 2011;8:1065–1070. doi: 10.1038/nmeth.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh H.D., Kim D.H., Jeong H.-S., Park H.W. Regulation of TEAD transcription factors in cancer biology. Cells. 2019;8 doi: 10.3390/cells8060600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Japp A.G., Newby D.E. The apelin–APJ system in heart failure: pathophysiologic relevance and therapeutic potential. Biochem. Pharmacol. 2008;75:1882–1892. doi: 10.1016/j.bcp.2007.12.015. [DOI] [PubMed] [Google Scholar]
- Joshi S., Davidson G., Le Gras S., Watanabe S., Braun T., Mengus G., Davidson I. TEAD transcription factors are required for normal primary myoblast differentiation in vitro and muscle regeneration in vivo. PLoS Genet. 2017;13:e1006600. doi: 10.1371/journal.pgen.1006600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoglou N.P.E., Vrabas I.S., Kapelouzou A., Lampropoulos S., Sailer N., Kostakis A., Liapis C.D., Angelopoulou N. The impact of aerobic exercise training on novel adipokines, apelin and ghrelin, in patients with type 2 diabetes. Med. Sci. Monit. 2012;18:CR290–CR295. doi: 10.12659/MSM.882734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidoya H., Takakura N. Biology of the apelin-APJ axis in vascular formation. J. Biochem. 2012;152:125–131. doi: 10.1093/jb/mvs071. [DOI] [PubMed] [Google Scholar]
- Kim M., Kim T., Johnson R.L., Lim D.-S. Transcriptional Co-repressor function of the hippo pathway transducers YAP and TAZ. Cell Rep. 2015;11:270–282. doi: 10.1016/j.celrep.2015.03.015. [DOI] [PubMed] [Google Scholar]
- Korsunsky I., Millard N., Fan J., Slowikowski K., Zhang F., Wei K., Baglaenko Y., Brenner M., Loh P., Raychaudhuri S. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods. 2019;16:1289–1296. doi: 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latroche C., Gitiaux C., Chrétien F., Desguerre I., Mounier R., Chazaud B. Skeletal muscle microvasculature: a highly dynamic lifeline. Physiology. 2015;30:417–427. doi: 10.1152/physiol.00026.2015. [DOI] [PubMed] [Google Scholar]
- Latroche C., Weiss-Gayet M., Muller L., Gitiaux C., Leblanc P., Liot S., Ben-Larbi S., Abou-Khalil R., Verger N., Bardot P., et al. Coupling between myogenesis and angiogenesis during skeletal muscle regeneration is stimulated by restorative macrophages. Stem Cell Rep. 2017;9:2018–2033. doi: 10.1016/j.stemcr.2017.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazure F., Blackburn D.M., Corchado A.H., Sahinyan K., Karam N., Sharanek A., Nguyen D., Lepper C., Najafabadi H.S., Perkins T.J., et al. Myf6/MRF4 is a myogenic niche regulator required for the maintenance of the muscle stem cell pool. EMBO Rep. 2020;21:e49499. doi: 10.15252/embr.201949499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G., Lin J.C.Y., Wei V., Yoo C., Cheng J.C., Nguyen C.T., Weisenberger D.J., Egger G., Takai D., Gonzales F.A., et al. Distinct localization of histone H3 acetylation and H3-K4 methylation to the transcription start sites in the human genome. Proc. Natl. Acad. Sci. U S A. 2004;101:7357–7362. doi: 10.1073/pnas.0401866101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukjanenko L., Brachat S., Pierrel E., Lach-Trifilieff E., Feige J.N. Genomic profiling reveals that transient adipogenic activation is a hallmark of mouse models of skeletal muscle regeneration. PLoS One. 2013;8:e71084. doi: 10.1371/journal.pone.0071084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luque E., Peña J., Martin P., Jimena I., Vaamonde R. Capillary supply during development of individual regenerating muscle fibers. Anat. Histol. Embryol. 1995;24:87–89. doi: 10.1111/j.1439-0264.1995.tb00016.x. [DOI] [PubMed] [Google Scholar]
- Maloney P.R., Khan P., Hedrick M., Gosalia P., Milewski M., Li L., Roth G.P., Sergienko E., Suyama E., Sugarman E., et al. Probe Reports from the NIH Molecular Libraries Program. National Center for Biotechnology Information; 2010. Functional antagonists of the Apelin (APJ) receptor. [PubMed] [Google Scholar]
- Mashinchian O., Pisconti A., Le Moal E., Bentzinger C.F. In: Current Topics in Developmental Biology. Sassoon D., editor. Academic Press; 2018. Chapter two - the muscle stem cell niche in health and disease; pp. 23–65. [DOI] [PubMed] [Google Scholar]
- McKellar D.W., Walter L.D., Song L.T., Mantri M., Wang M.F.Z., De Vlaminck I., Cosgrove B.D. Large-scale integration of single-cell transcriptomic data captures transitional progenitor states in mouse skeletal muscle regeneration. Commun. Biol. 2021;4:1280. doi: 10.1038/s42003-021-02810-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller W., Rosenbloom K., Hardison R.C., Hou M., Taylor J., Raney B., Burhans R., King D.C., Baertsch R., Blankenberg D., et al. 28-Way vertebrate alignment and conservation track in the UCSC Genome Browser. Genome Res. 2007;17:1797–1808. doi: 10.1101/gr.6761107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan S., Maitra R., Deschamps J.R., Bortoff K., Thomas J.B., Zhang Y., Warner K., Vasukuttan V., Decker A., Runyon S.P. Discovery of a novel small molecule agonist scaffold for the APJ receptor. Bioorg. Med. Chem. 2016;24:3758–3770. doi: 10.1016/j.bmc.2016.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novartis Pharmaceuticals A randomized, subject and investigator-blind, placebo-controlled study of CLR325 in chronic stable heart failure patients. 2020. clinicaltrials.gov
- Nyimanu D., Kay R.G., Sulentic P., Kuc R.E., Ambery P., Jermutus L., Reimann F., Gribble F.M., Cheriyan J., Maguire J.J., et al. Development and validation of an LC-MS/MS method for detection and quantification of in vivo derived metabolites of [Pyr 1 ]apelin-13 in humans. Sci. Rep. 2019;9:19934. doi: 10.1038/s41598-019-56157-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obeso J., Weber J., Auerbach R. A hemangioendothelioma-derived cell line: its use as a model for the study of endothelial cell biology. Lab. Invest. J. Tech. Methods Pathol. 1990;63:259–269. [PubMed] [Google Scholar]
- Qiu H., Wang F., Liu C., Xu X., Liu B. TEAD1-dependent expression of the FoxO3a gene in mouse skeletal muscle. BMC Mol. Biol. 2011;12:1. doi: 10.1186/1471-2199-12-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai R., Ghosh A.K., Eren M., Mackie A.R., Levine D.C., Kim S.-Y., Cedernaes J., Ramirez V., Procissi D., Smith L.H., et al. Downregulation of the apelinergic Axis Accelerates aging, whereas its systemic restoration improves the mammalian healthspan. Cell Rep. 2017;21:1471–1480. doi: 10.1016/j.celrep.2017.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read C., Fitzpatrick C.M., Yang P., Kuc R.E., Maguire J.J., Glen R.C., Foster R.E., Davenport A.P. Cardiac action of the first G protein biased small molecule apelin agonist. Biochem. Pharmacol. 2016;116:63–72. doi: 10.1016/j.bcp.2016.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori R., Romanello V., Sandri M. Mechanisms of muscle atrophy and hypertrophy: implications in health and disease. Nat. Commun. 2021;12:330. doi: 10.1038/s41467-020-20123-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southard S., Kim J.-R., Low S., Tsika R.W., Lepper C. Myofiber-specific TEAD1 overexpression drives satellite cell hyperplasia and counters pathological effects of dystrophin deficiency. Elife. 2016;5:e15461. doi: 10.7554/eLife.15461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein C., Bardet A.F., Roma G., Bergling S., Clay I., Ruchti A., Agarinis C., Schmelzle T., Bouwmeester T., Schübeler D., et al. YAP1 exerts its transcriptional control via TEAD-mediated activation of enhancers. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1005465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su A.I., Wiltshire T., Batalov S., Lapp H., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. U S A. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szokodi I., Tavi P., Földes G., Voutilainen-Myllylä S., Ilves M., Tokola H., Pikkarainen S., Piuhola J., Rysä J., Tóth M., et al. Apelin, the novel endogenous ligand of the orphan receptor APJ, regulates cardiac contractility. Circ. Res. 2002;91:434–440. doi: 10.1161/01.res.0000033522.37861.69. [DOI] [PubMed] [Google Scholar]
- Takahashi A., Kureishi Y., Yang J., Luo Z., Guo K., Mukhopadhyay D., Ivashchenko Y., Branellec D., Walsh K. Myogenic akt signaling regulates blood vessel recruitment during myofiber growth. Mol. Cell Biol. 2002;22:4803–4814. doi: 10.1128/MCB.22.13.4803-4814.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatemoto K., Hosoya M., Habata Y., Fujii R., Kakegawa T., Zou M.X., Kawamata Y., Fukusumi S., Hinuma S., Kitada C., et al. Isolation and characterization of a novel endogenous peptide ligand for the human APJ receptor. Biochem. Biophys. Res. Commun. 1998;251:471–476. doi: 10.1006/bbrc.1998.9489. [DOI] [PubMed] [Google Scholar]
- Tsika R.W., Schramm C., Simmer G., Fitzsimons D.P., Moss R.L., Ji J. Overexpression of TEAD-1 in transgenic mouse striated muscles produces a slower skeletal muscle contractile phenotype. J. Biol. Chem. 2008;283:36154–36167. doi: 10.1074/jbc.M807461200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma M., Asakura Y., Murakonda B.S.R., Pengo T., Latroche C., Chazaud B., McLoon L.K., Asakura A. Muscle satellite cell cross-talk with a vascular niche maintains quiescence via VEGF and notch signaling. Cell Stem Cell. 2018;23:530–543.e9. doi: 10.1016/j.stem.2018.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinel C., Lukjanenko L., Batut A., Deleruyelle S., Pradère J.-P., Le Gonidec S., Dortignac A., Geoffre N., Pereira O., Karaz S., et al. The exerkine apelin reverses age-associated sarcopenia. Nat. Med. 2018;24:1360–1371. doi: 10.1038/s41591-018-0131-6. [DOI] [PubMed] [Google Scholar]
- Vinel C., Schanstra J.P., Boizard F., Péreira O., Auriau J., Dortignac A., Breuil B., Feuillet G., Nkuipou-Kenfack E., Zürbig P., et al. Apelin affects the mouse aging urinary peptidome with minimal effects on kidney. Sci. Rep. 2019;9:10647. doi: 10.1038/s41598-019-47109-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.J., Greer P., Auerbach R. Isolation and propagation of yolk-sac-derived endothelial cells from a hypervascular transgenic mouse expressing a gain-of-function fps/fes proto-oncogene. In Vitro Cell. Dev. Biol. Anim. 1996;32:292–299. doi: 10.1007/BF02723062. [DOI] [PubMed] [Google Scholar]
- Wang G., Qi X., Wei W., Englander E.W., Greeley G.H. Characterization of the 5’-regulatory regions of the rat and human apelin genes and regulation of breast apelin by USF. FASEB J. 2006;20:2639–2641. doi: 10.1096/fj.06-6315fje. [DOI] [PubMed] [Google Scholar]
- Whitham M., Febbraio M.A. The ever-expanding myokinome: discovery challenges and therapeutic implications. Nat. Rev. Drug Discov. 2016;15:719–729. doi: 10.1038/nrd.2016.153. [DOI] [PubMed] [Google Scholar]
- Wu C., Macleod I., Su A.I. BioGPS and MyGene.info: organizing online, gene-centric information. Nucleic Acids Res. 2013;41:D561–D565. doi: 10.1093/nar/gks1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Chen L., Li L. Apelin/APJ system: a novel promising therapy target for pathological angiogenesis. Clin. Chim. Acta. 2017;466:78–84. doi: 10.1016/j.cca.2016.12.023. [DOI] [PubMed] [Google Scholar]
- Yaffe D., Saxel O. Serial passaging and differentiation of myogenic cells isolated from dystrophic mouse muscle. Nature. 1977;270:725–727. doi: 10.1038/270725a0. [DOI] [PubMed] [Google Scholar]
- Yin H., Price F., Rudnicki M.A. Satellite cells and the muscle stem cell niche. Physiol. Rev. 2013;93:23–67. doi: 10.1152/physrev.00043.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Muri J., Fitzgerald G., Gorski T., Gianni-Barrera R., Masschelein E., D’Hulst G., Gilardoni P., Turiel G., Fan Z., et al. Endothelial lactate controls muscle regeneration from ischemia by inducing M2-like macrophage polarization. Cell Metab. 2020;31:1136–1153.e7. doi: 10.1016/j.cmet.2020.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Wang T. Using the Wash U Epigenome Browser to examine genome-wide sequencing data. Curr. Protoc. Bioinformatics. 2012;10 doi: 10.1002/0471250953.bi1010s40. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data or any additional information required to reanalyze the data reported in this paper are available from the lead contact upon request. Transcriptomic datasets used in this study are available in the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) under the specific references below: