Abstract
35S rRNA transcripts include a 5′-external transcribed spacer followed by rRNAs of the small and large ribosomal subunits. Their processing yields massive precursors that include dozens of assembly factor proteins. In Saccharomyces cerevisiae, nucleolar assembly factors form 2 coaxial layers/volumes around ribosomal DNA. Most of these factors are cyclically recruited from a latent state to an operative state, and are extensively conserved. The layers match, at least approximately, known subcompartments found in higher eukaryotic cells. ∼80% of assembly factors are essential. The number of copies of these assembly factors is comparable to the number of nascent transcripts. Moreover, they exhibit “isoelectric balance,” with RNA-binding candidate “nucleator” assembly factors being notably basic. The physical properties of pre-small subunit and pre-large subunit assembly factors are similar, as are their 19 motif signatures detected by hierarchical clustering, unlike motif signatures of the 5′-external transcribed spacer rRNP. Additionally, many assembly factors lack shared motifs. Taken together with the progression of rRNP composition during subunit maturation, and the realization that the ribosomal DNA cable is initially bathed in a subunit-nonspecific assembly factor reservoir/microenvironment, we propose a “3-step subdomain assembly model”: Step (1): predominantly basic assembly factors sequentially nucleate sites along nascent rRNA; Step (2): the resulting rRNPs recruit numerous less basic assembly factors along with notably basic ribosomal proteins; Step (3): rRNPs in nearby subdomains consolidate. Cleavages of rRNA then promote release of rRNPs to the nucleoplasm, likely facilitated by the persistence of assembly factors that were already associated with nucleolar precursors.
Keywords: nucleolus, ribosomal DNA, ribosome assembly factors, coaxial cable, ribosome biogenesis, chromosome XII, S. cerevisiae
Introduction
Many varieties of mRNA are produced throughout the nucleoplasm. In contrast, rRNA—that accounts for 60% of RNA synthesis in yeast—is transcribed in the nucleolar crescent (Warner 1999). As a step toward understanding issues relevant to all gene expression, we here focus on the successive steps of rRNA packaging that occur in the nucleolus of Saccharomyces cerevisiae. It is striking that packaging yields highly complex seemingly oversized precursor particles that ultimately give rise to ribosomal subunits, that are significantly smaller.
In S. cerevisiae, there are ∼150 head-to-tail repeats of ribosomal DNA (rDNA) along the long arm of chromosome XII, which reaches from the spindle pole body across the nucleoplasm into and through the nucleolus (Albert et al. 2013). Electron microscopic spreads and biochemical studies show that their rRNA transcripts are packaged cotranscriptionally by protein assembly factors (AFs). Most of the ∼200 AFs localize to the nucleolus and are thought to act during transcription, while others function in the nucleoplasm or after export to the cytoplasm (Panse and Johnson 2010; Baßler and Hurt 2019; Klinge and Woolford 2019). Considering that ∼80% of yeast genes are not required for mitotic growth, an extraordinarily high fraction (78%) of genes encoding AFs are essential (Giaever et al. 2002). Although the organization of precursors of both small and large subunits (SSU, LSU) is surely dynamic, several presumably low-energy assembly intermediates have been retrieved and imaged by cryo-EM.
Each rRNA transcript has a tripartite organization, including an external transcribed spacer (5′-ETS, 0.7 kb), the “pre-40S” segment (1.8 kb) that includes the SSU RNA (18S), and the segment (6.6 kb) that includes the LSU RNAs (5.8S, 25S) (Fig. 1a). The nascent rRNA transcript is cleaved between the 5′-ETS sequence and the pre-40S sequence, between the pre-40S and pre-LSU sequences, and just before the 3′ terminus. Intriguingly, there is no evidence that the 5′-ETS rRNP segment is present in the chromatin-rich nucleoplasm or in the cytoplasm. This may signify that these components do not leave the nucleolar crescent or that they are quickly returned to the nucleolus. The 5′-ETS rRNA itself is efficiently degraded (Osheim et al. 2004; Talkish et al. 2016; Baßler and Hurt 2019; Klinge and Woolford 2019; Turowski et al. 2020; Black and Johnson 2021; Lau et al. 2021).
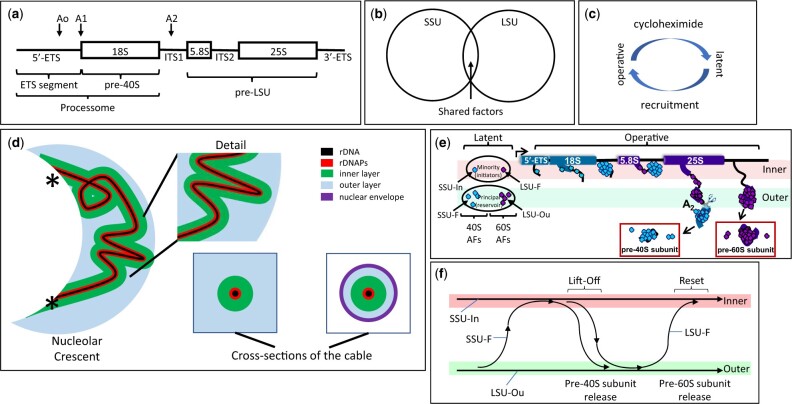
Fig. 1.
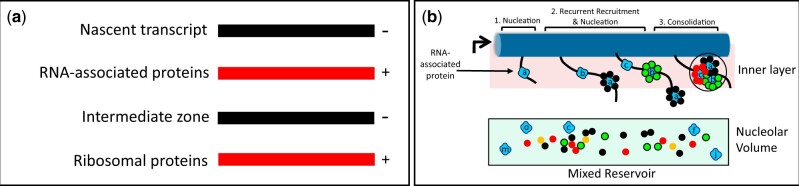
rRNA Processing in the Nucleolus. a) rDNA organization. A single tripartite rDNA unit, indicating sites of rRNA cleavage and the segments that give rise to the rRNAs of the 5′-ETS, pre-40S and pre-LSU. The “processome” is also known as the “90S preribosome.” ETS: external transcribed spacer. The term “tripartite” has also been used by some authors to specify the 18S, 5.8S, and 25S segments of rRNA. b) Subunit specialization of AFs. The Venn diagram indicates the largely distinct repertoires of AFs that contribute to the biogenesis of each type of subunit. The overlap includes several DExD/H proteins (Dbp3, Has1, Prp43), snoRNP proteins, subunits of RNA polymerase A and Rrp5 that binds sequences on either side of ITS1 (internal transcribed spacer). c) Activity cycle of AFs. Each AF is thought to cycle between a latent state (when not associated with rRNPs) and an operative state. Recruitment to the operative state requires production of new copies of ribosomal proteins and is correspondingly blocked by cycloheximide. d) Coaxial organization of the nucleolus when subunits are being produced. From left-to-right: Overview of the nucleolar crescent showing the rDNA segment (black) colocalizes with rDNA-associated proteins (rDNAPs) (red) and is enclosed by an “inner” layer of AFs (for SSU assembly, green). The remaining volume of the nucleolus is largely occupied by AFs that are engaged in assembling LSUs (blue). The points at which chromosome XII enters and leaves the crescent are indicated by (*). Two cross-sections of the coaxial cable are indicated in the lower right, either in cycling cells (left) or after metaphase arrest (right). At metaphase, when the nucleolus become elongated in the mother cell, cross-sections show the layered organization of the cable and the close apposition of the surrounding nuclear envelope (NE, purple). Note: We propose that the contour of rDNA weaves repeatedly throughout the nucleolus. See Tartakoff et al. (2021) for further detail. e) Single cycle of rRNP formation. The diagram illustrates the proposed loading of subunit-specific AFs onto rRNA transcripts during a single cycle of transcription (see Table 1 and Supplementary Table 1 for nomenclature). At the extreme left, the AFs are in their latent form, not having encountered nascent rRNA. The “principal groups” of AFs (SSU-F, LSU-Ou) are in the outer layer/volume of the coaxial structure, but a limited number (“minority group”) of AFs (SSU-In, LSU-F) are in the inner layer. As transcription begins, the SSU-In group binds the nascent transcript, followed by the SSU-F group that is recruited from the outer layer. When the 18S rRNA sequences have been transcribed and the initial downstream sequences appear, they bind LSU-F AFs and the nascent transcript extends/transfers to the outer layer where the SSU precursor is released (due to cleavage) and the LSU-Ou AFs bind. When transcription has almost completed, a site near the 3′ terminus is cleaved, thereby releasing the LSU precursor. Although not indicated, addition of ribosomal proteins occurs in conjunction with AFs. f) Flux of AFs during a single cycle of transcription. This diagram summarizes the changes of localization of AFs (lift-off) indicated in (e) as well as their return (reset) when transcription is complete, as described in Tartakoff et al. (2021).
Given the large number of AFs associated with each precursor, and judging from cryo-EM, AFs appear to be polyvalent. Some AFs govern the insertion of specific ribosomal proteins, while others covalently modify rRNA, possibly to exert quality control (Cole et al. 2009; Liang et al. 2009; Panse and Johnson 2010; Baßler and Hurt 2019; Klinge and Woolford 2019). AFs that are DExD/H-box proteins, GTPases and ATPases likely adjust the progressive organization of immature particles as ribosomal proteins are added.
In the large majority of cases, individual AFs are required to produce either SSU or LSU, but not both (Fig. 1b) (Musters et al. 1989; Woolford and Baserga 2013). This could signify that SSU AFs have common biochemical properties that distinguish them from LSU AFs. Alternatively, - given the known mobility of AFs within the nucleus (Phair and Misteli 2000; Chen and Huang 2001; Tartakoff et al. 2021) - the subunit specificity of the two groups of biochemically similar AFs could reflect their being recruited to specific binding sites of each type of precursor.
Our recent studies of S. cerevisiae in which rDNA has been “linearized” concluded that rDNA and its most closely associated proteins (rDNAPs) (the axis) are normally folded inside the nucleolus as a curvilinear cable. Since we observed that the localization of many AFs changed when subunit assembly stopped, we reasoned that each AF normally cycles between a “latent” state (when not associated with immature subunits) and an “operative” state (when incorporated into immature subunits) (Fig. 1c). AFs in the latent state may remain extensively associated with each other. When subunits are not being made, we found that the large majority of latent AFs avoid the axis itself, fill the surrounding volume of the nucleolus, and do not intermix with chromatin. This observation gave first indications that—when transcription does occur—latent AFs are recruited to nascent rRNA from a surrounding reservoir (Tartakoff et al. 2021).
In cells that are making subunits, we found that the axis is the central element of a coaxial cable and is ensheathed by 2 layers/volumes of AFs. Judging from the distribution of specific AFs and rRNA sequences, the inner layer is engaged in assembling SSUs while the outer layer—that extends as far as the surface of the nucleolus—assembles LSUs (Fig. 1d and Table 1). These layers are roughly equivalent to the DFC and GC, as described below. In accord with this information, we found that subunit-specific AFs fall into 4 groups (Table 1; Supplementary Table 1) [and Fig S6 in Tartakoff et al. (2021)]. The groups of AFs are distinguished by their localizations when subunits are being produced, as compared with their localizations when subunits are not being produced. The abbreviations used for the four groups are explained in the legend of Table 1.
Table 1.
Subsets of nucleolar assembly factors.
| Subset | Operative distribution | Latent distribution | Suggested rolea | |
|---|---|---|---|---|
| SSU-F | Principal subsets | Inner layer | Outer layer | SSU knob formation |
| LSU-Ou | Outer layer | Outer layer | LSU knob formationb | |
| SSU-In | Minority subsets | Inner layer | Inner layer | Axis adherence |
| LSU-F | Outer layer | Inner layer | Axis adherence | |
| shared AFs | N.A. | Inner layer | Inner layer | Unknowna |
| snoRNP proteins | N.A. | Inner layer | Inner layer | RNA methylation, Ψ formation |
Based on our previous studies (Tartakoff et al. 2021), subunit-specific AFs can be divided into 4 groups according to their localizations both before and after interruption of subunit assembly. As indicated in Fig. 1f, many AFs are thought to transit between the inner and outer layers during each cycle of transcription. Two types of AFs function in production of SSU, the SSU-F and SSU-In. The localization of the SSU-F proteins is facultative—they move from the inner to the outer layer/volume when subunit production stops. The SSU-In AFs, in contrast, remain along the inner layer. There are two corresponding types of LSU AFs, the LSU-F and LSU-Ou AFs. The LSU-F AFs localize to the inner layer only when subunits are not being produced. The LSU-Ou AFs localize to the outer layer regardless of whether subunits are being made.
For the names of each subset of proteins (see Tartakoff et al. 2021).
At least three DExH/D proteins are in this category: Dbp3, Has1, Prp43. The RNA-binding protein, Rrp5, is also involved in production of both subunits.
See Osheim et al. (2004) for images of the knobs that form at the 5' ends of nascent transcripts.
These observations gave rise to a model whereby “minority” subsets of AFs localize along the rDNA axis when latent, bind nascent rRNA, and then recruit numerous AFs (“the principal group”) from the surrounding reservoir/volume, along with ribosomal proteins (Fig. 1e). As part of this process, nascent rRNP intermediates translocate (“lift-off”) from the inner layer to the outer layer/volume of the coaxial cable (Fig. 1f). It seems likely that the coaxial structure is universal, and that it is most readily detected in yeast since we can linearize rDNA and since ITS1 cleavage occurs during transcription, thereby separating SSU precursors from LSU precursors. In higher eukaryotes, this cleavage occurs after chain termination (Osheim et al. 2004).
Classic studies have often focused on the nucleolus and ribosome biogenesis in animal cells (Hadjiolov 1985; Scheer and Weisenberger 1994; Grummt 2003; Raska et al. 2006; Zemp and Kutay 2007; Hernandez-Verdun et al. 2010; Pederson 2011; Farley et al. 2015; Yao et al. 2019). In most higher eukaryotic cells 3 nucleolar subcompartments have been detected (initially by electron microscopy): the fibrillar center (FC), the dense fibrillar component (DFC), and the granular component (GC). These subcompartments are thought to constitute distinct protein phases whose coherence depends on interactions among intrinsically disordered protein domains (Feric et al. 2016; Turoverov et al. 2019; Lafontaine et al. 2021). Additional studies indicate that transcription occurs along the interface between the FC and the DFC (or in the FC) and that transcripts then travel centrifugally through the DFC and the GC (Scheer and Hock 1999; Raska et al. 2006; Hernandez-Verdun et al. 2010; Lamaye et al. 2011; James et al. 2014; Farley et al. 2015; Yao et al. 2019).
The identification of human homologs of yeast AFs has made possible many comparative studies (Hinsby et al. 2006; Tafforeau et al. 2013; Badertscher et al. 2015; Aubert et al. 2018; Bohnsack and Bohnsack 2019; Stenstrom et al. 2020; Singh et al. 2021). Our present focus is on the yeast nucleolar AFs for which human homologs can be identified (Supplementary Table 2). Supplementary Table 1 also identifies AF homologs that have been implicated in disease.
The nucleolus defines the microenvironment in which rDNA transcription and maturation of nascent subunit precursors occur. The spatial separation of the nucleolus from the chromatin-filled nucleoplasm might be of value for (a) causing a corresponding local increase in the concentrations of nucleolar components, (b) Juxtaposing domains to power thermodynamically downhill events as cargoes transfer between them (“vectorial 2-phase partitioning”) (Tartakoff et al. 2021), or (c) Providing a mechanism to exclude outsiders.
Explanation (a) seems unlikely to be determinative since AF concentrations would decrease only ∼3× if nucleolar proteins were intermixed with chromatin in yeast. Explanation (b) can account for translocation of assembly intermediates between the inner and outer layers of the coaxial nucleolar structure (Tartakoff et al. 2021). It may also be relevant to transfer of assembly intermediates from the nucleolus to the nucleoplasm, as we discuss below. Explanation (c) is relevant since a variety of nucleoplasmic proteins appear to be completely absent from the nucleolus, judging both from classic cytological studies of higher eukaryotes and from more recent studies of yeast (Tartakoff et al. 2021). Their absence could be critical. For example, free histones could sequester key acidic AFs, repress rDNA expression and possibly promote rDNA recombination (Torres-Rosell et al. 2007). According to this contrarian view, the value of having a nucleolus is that it insulates early stages of ribosome biogenesis from the nucleoplasm.
Abundance and Physical Properties of Assembly Factors
Copy number of individual assembly factors
To appreciate the complexity of the nucleolar microenvironment in S. cerevisiae, we tabulated the copy number/cell of subsets of AFs, based on a genome-wide compilation of values from several laboratories (Ho et al. 2018) (Fig. 2 and Table 2; Supplementary Fig. 1). The copy-number of nucleolar AFs belonging to each of the four subunit-specific groups is relatively uniform, with a mean of ∼ 5,400 copies/cell. AFs that are found primarily in the nucleoplasm (8 AFs) or cytoplasm (13 AFs) also have similar mean values (5,369 and 4,807). Transcription of a single copy of rDNA in yeast engages ∼50 copies of RNA polymerase A (French et al. 2003; Turowski et al. 2020). If only 80 out of the total of 150 rDNA repeats are active (Dammann et al. 1993), there will be 80 × 50 = 4,000 nascent transcripts, each of which will eventually associate with each AF. Hence the number of copies of the subunit-specific AFs are of the same order of magnitude as rRNA transcripts. Modest alterations of the titer or activity of AFs therefore could affect ribosome production. Moreover, when a yeast cell is making subunits there should be no large pool of latent AFs.
Fig. 2.
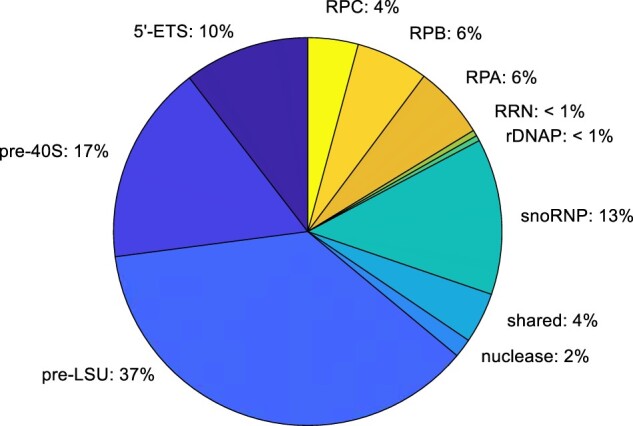
Copy number of assembly factors and other relevant proteins per cell. Integration of the number of copies of each subset of proteins. Some categories, including the subunit-specific AFs, are underestimates since the only AFs that are included are those for which there are homologs in man. This pie chart cannot be directly compared with charts based on isolated fractions (Andersen et al. 2005), since those charts illustrate the total number of different kinds of proteins that are present in each category, rather than the aggregate number of molecules present in each category. We estimate that there are ∼1,080,000 subunit-specific AFs in each cell (200 × 5,400 copies). This number is about twice that estimated for histones. Since the volume of a cell with a diameter of 5 µg is ∼6.5 µ 10−11 ml, the yeast nucleus makes up about 7% of the cell volume (Jorgensen et al. 2007), and the volume of the nucleolus is about one-third the volume of the nucleus, if one takes 100 kDa as the average MW of AFs, the total concentration of AFs in the nucleolus would reach ∼ 100 mg/ml. Each AF therefore would have a concentration of ∼ 0.5 mg/ml.
Table 2.
Copy number for groups of assembly factors.
| Subset | Average copy number per cell × number of different proteins = Total number of copies | ||
|---|---|---|---|
| 5′-ETS | 5,056 ± 2,269 | 23 | 116,288 |
| pre-40S | 4,525 ± 2,295 | 43 | 194,575 |
| pre-LSU | 6,045 ± 2,786 | 69 | 417,105 |
| Nuclease | 3,497 ± 1,808 | 5 | 17,485 |
| Shareda | 11,797 ± 4,840 | 4 | 47,188 |
| snoRNP | 20,730 ± 9,334 | 8 | 165,840 |
| rDNAPb | 1,705 ± 965 | 6 | 10,230 |
| RRN | 794 ± 789 | 7 | 5,558 |
| RPAc | 9,651 ± 4,297 | 7 | 67,557 |
| RPBc | 9,580 ± 4,371 | 7 | 67,060 |
| RPCc | 6,677 ± 2,485 | 7 | 46,739 |
| Histones | 51,652 ± 40,471 | 8 | 413,216 |
Tabulation of the average copy numbers of proteins of interest.
See Table 1.
The rDNAP proteins are Csm1, Fob1, Lrs4, Tof2, Top1, and Top2.
The five subunits that are shared among polymerases are not included in this tabulation.
As indicated in Table 2, there are ∼12,000 copies/cell of each of 3 DExD/H-box ATPases that contribute to both types of subunit (Dbp3, Has1, Prp43) as well as for Rrp5 that binds rRNA segments on both sides of ITS1 (Rocak et al. 2005; Linder and Jankowsky 2011; Lebaron et al. 2013). snoRNP proteins are more abundant than other AFs (∼21,000 copies/cell), perhaps since rRNA undergoes dozens of snoRNP-dependent modifications that depend on many guide snoRNAs, of which there are a total of ∼27,000 copies/cell (Pircher et al. 2014; Sharma and Lafontaine 2015; Taoka et al. 2016). The relatively high values for snoRNP proteins presumably account for their often having been used as nucleolar markers for cytological studies.
The protein composition of the nucleolus has previously been approximated by analyzing subcellular fractions recovered from cell lines, by immunolabeling, and by bioinformatic means (Andersen et al. 2005; Coute et al. 2006; Staub et al. 2006; Ahmad et al. 2009; Patel et al. 2010; Beck et al. 2011; Stenstrom et al. 2020). The approach that we have taken focuses on proteins that are known to be required to make ribosomes and is free from any losses or redistribution that might occur during isolation.
We find that regulators of rDNA transcription (Rrn proteins) are present at only ∼5 copies/rDNA repeat and that each of the rDNAPs (rDNA-associated proteins) is slightly more abundant at ∼12 copies/repeat (Supplementary Fig. 1). Since the levels of these proteins are so modest, a small reduction of their availability might have a large impact on overall rRNA transcription. Limitation of their availability might also affect the balance of inactive vs active rDNA loci (Granneman and Baserga 2004; McStay 2006; Hamperl et al. 2013).
Physical properties of assembly factors
We characterized AFs that associate with each rRNA domain (5′-ETS, pre-40S, pre-LSU). The comparisons are based on
1D plots of isoelectric point (IEP), predicted disorder and molecular weight (Fig. 3a;Supplementary Fig. 2). The plots include both yeast AFs and their homologs in man.
2D plots that compare IEP and MW (Fig. 3, b and c). These subgroups are distinct from those identified in Table 1. In the 2D plots the AFs fall into four quadrants, as described below. Again, both yeast and human AFs are included.
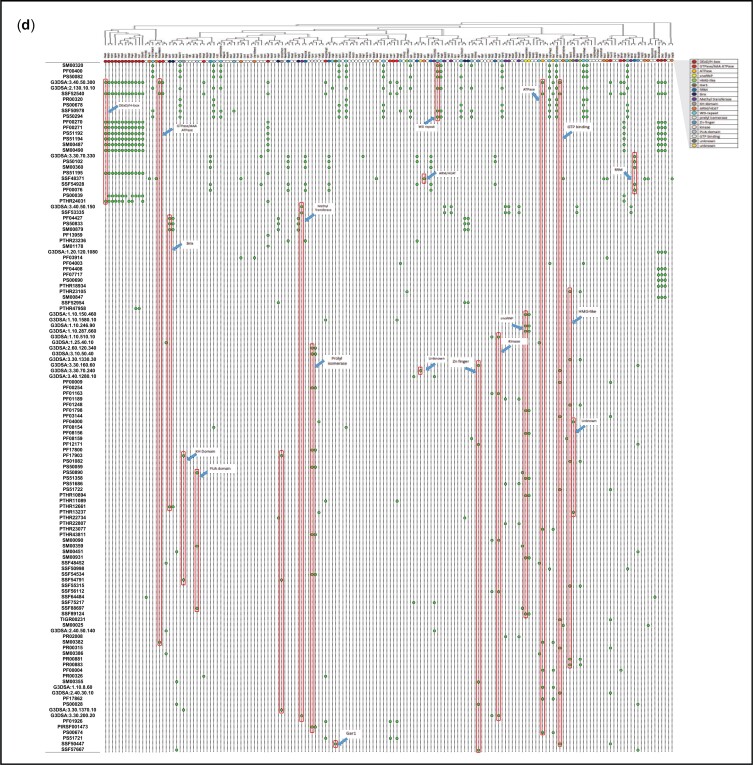
Hierarchical clustering of motif signatures (Fig. 3d). For this purpose, all motifs listed by the Saccharomyces Genome Database were included if they are present in more than a single AF.
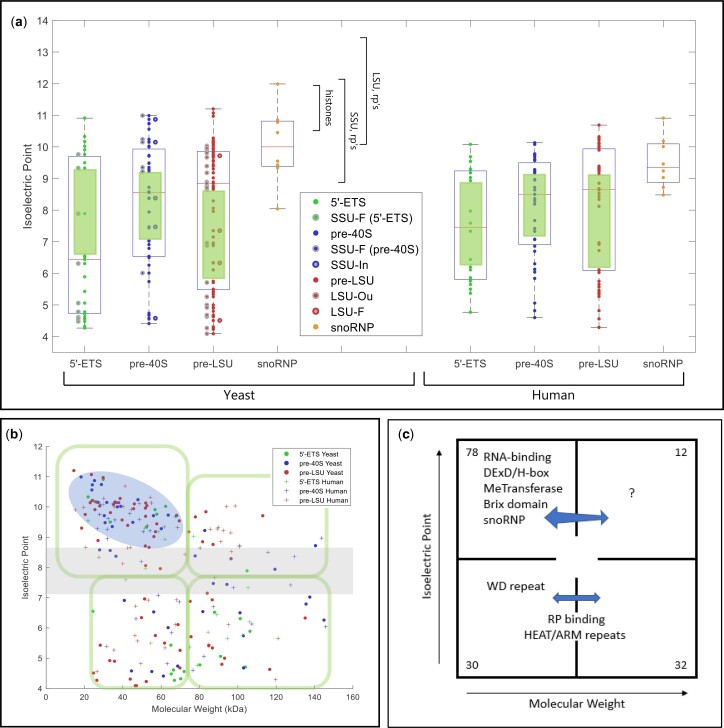
Fig. 3.
Physical parameters and motifs of AFs. a) IEP of yeast and human AFs. AFs were grouped to illustrate the characteristics of those that associate with the 5′-ETS, the pre-40S segment, or the pre-LSU segment (see Fig. 1a). The central column of values includes all relevant AFs listed in Supplementary Table 2. For the 5′-ETS, the points to the left are SSU-F. For the pre-40S, the points to the left are SSU-F and the points to the right are SSU-In. For the pre-LSU, the points to the left are LSU-Ou and the points to the right are LSU-F. Human AFs are subgrouped according to the information for their yeast homologs. The gap/thinning in the IEP plot around neutral pH is marked by green boxes. Supplementary Table 4 lists the extreme values of physical parameters for AFs. Outlier AFs for each subset were calculated based on the IQR rule (see Materials and Methods). Human AFs are subgrouped according to the information for their yeast homologs. The IEPs of yeast histones and yeast ribosomal proteins are indicated by brackets. A subset of ribosomal proteins is much more acidic (RPS0A, RPS0B, RPS12, RPS21A, RPS21B, RPL5, RPL22A, RPL22B, RPP0, RPP1A, RPP1B, RPP2A, RPP2B). Many (perhaps all) of these are added after export. b) Nucleolar AF 2D plot. Molecular weights and IEPs of yeast and human AFs associated with each of the three domains (5′-ETS, pre-40S, pre-LSU). The 4 quadrants are discussed in the text. The pale blue oblong encloses the most obvious of the subgroupings. The gray horizonal band highlights the pH neutral zone in which there are few AFs. The plots in Fig. 3, a and b display the intrinsic properties of AFs; however, the characteristics of nascent rRNPs also depend on 1) covalent modifications of AFs and RNA (ubiquitylation, methylation, phosphorylation, etc.); 2) the length of the progressively elongating rRNA itself; and 3) the growing complement of ribosomal proteins. The stoichiometry of most of these post-translational modifications is unknown. c) Diagrammatic summary of enrichment of yeast AFs in distinct quadrants. The 4 quadrants identified in (b) are indicated schematically, along with the names of groups of AFs that are enriched in distinct quadrants. The numbers in the corners indicate the total number of yeast AFs in each quadrant defined by the green enclosures in (b). Blue arrows are positioned asymmetrically to indicate quadrant bias. RP, ribosomal proteins. d) Hierarchical clustering of yeast AFs. AFs were clustered by amino acid sequence and annotated by reference to motifs that occur in at least 2 AFs in the Saccharomyces Genome Database. Subunits of RNA polymerase A are included, as well as AFs that localize primarily to the nucleoplasm or cytoplasm. The colored ovals along the top correspond to the prototypic motif signatures in Supplementary Table 5. The open/white ovals designate AFs that lack motif signatures as well as the subunits of RNA polymerase A. Motifs are listed along the vertical axis on the left. The phylogenetic tree of yeast proteins was generated based on the multiple sequence alignments by Clustal Omega (Sievers and Higgins 2018). The protein domains shared by 2 or more than 2 genes were included in our study to generate the 2D map.
Fig. 3.
(Continued)
In the 1D plots, each of the 3 parameters shows wide dispersion and the values for the 3 domains in yeast overlap extensively: IEP's range from pH 4 to 11, predicted disorder ranges from <10% to 80%, and molecular weights range from ∼10 to 280 kDa. Interestingly, the plots of IEP for each of the 3 domains indicate that the distribution is bimodal: a concentration of AFs is isoelectric near pH 9–10.5, and a broader peak has IEPs from pH 4 to 6, with thinning at neutral pH values (green rectangles). Figure 3a (right) shows the overall similarity for human AFs.
The 2D plots identify subgroups that are differentially enriched in the quadrants. The most densely populated grouping (basic proteins of modest size) is encircled and the neutral pH zone—with only few AFs—is in gray. The differential enrichments are best appreciated in the overlay plots of Supplementary Fig. 3 and in Supplementary Table 3 that focus on the distributions of functionally/structurally defined subsets of AFs. Table 3 summarizes the composition of the three domains, showing that the 5′-ETS domain is quite distinct from the pre-40S and pre-LSU groups, that resemble each other. This resemblance is consistent with a model in which the majority of these AFs (the “principal group”) constitute a shared reservoir from which individual AFs can be recruited to either subunit. Supplementary Table 4 lists the extreme values of the physical properties of single AFs.
Table 3.
Assembly factors of successive domains of nascent transcripts. 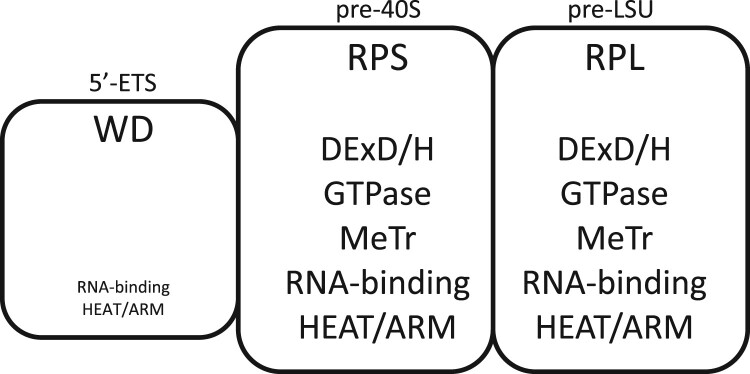
| Featurea | 5′-ETS segment (0–0.7 kb) | pre-40S segment (0.7–2.5 kb) | pre-LSU (2.5–9.1 kb) |
|---|---|---|---|
| Ribosomal proteins | None | Progressively increasing | Progressively increasing |
| WD repeats | 12 (52 %) | 1 (2 %) | 6 (9 %) |
| DExD/H motifs | 0b (0 %) | 7 (16 %) | 7 (10 %) |
| GTPases | 0 | 1 | 2 |
| Methyl transferases | 0 | 3 | 5 |
| Brix motifs | 1 | 0 | 5 |
| HEAT or ARM motifs | 1 | 5 | 6 |
| RRM, pumilio, KH | 1 | 6 | 7 |
| Number of AFs | 23 | 43 | 69 |
The properties of AFs associated with each segment are indicated.
Entries are based on Woolford and Baserga (2013) and Chaker-Margot et al. (2015). snoRNP proteins are not included in these tabulations.
See Supplementary Table 5 for the names of AFs with characteristic motifs. Additionally, Utp6, Utp10, and Utp20 can be described as having α-solenoid motifs and further AFs with related motifs are Noc2, Noc4/Utp19, and Sda1 (Dlakic and Tollervey 2004).
The DExD/H protein, Dbp4, may be present (Soltanieh et al. 2015).
The distributions in the 2D plots are as follows:
Upper left: enriched in proteins of modest size that interact with RNA: AFs with RRMs, KH domains, DExD/H-box proteins, methyl transferases, snoRNP proteins, and Brix domain proteins. The mean IEP for this entire group is pH 9.2 ± 0.6. About half of the proteins in this quadrant are known to bind RNA. A lesser number of basic RNA-binding proteins is in the upper right-hand quadrant.
Lower left: enriched in WD repeat proteins. The β-barrel structures of WD proteins generally form scaffolds for multiprotein complexes (Makarova et al. 2005; Smith 2008). In the 5′-ETS domain, they are thought to constitute extensively cross-linked structures (Barandun et al. 2018; Cheng et al. 2020). Nucleolar WD proteins are slightly acidic (mean IEP = pH 6.6) and relatively small (mean = 75 kDa).
Lower left and lower right: these quadrants include the majority of proteins with HEAT repeat motifs and tend to be neutral (mean IEP = pH 7.0). Although not all large, these proteins range in size up to 267 kDa, consistent with their fulfilling a spatially distributed organizational role (Dlakic and Tollervey 2004; Dez et al. 2007). The corresponding figures for proteins with structurally related ARM repeats are pH 7.8 and 118 kDa. The lower quadrants also include relatively acidic AFs that contact ribosomal proteins in assembly intermediates (Bms1, Erb1, Mak16, Mpp10, Rrp1, Utp3, etc.: mean IEP = pH 5.0 ± 0.8) (Kornprobst et al. 2016; Barandun et al. 2017; Sanghai et al. 2018).
Upper right: this quadrant (large basic proteins) is mixed, both with regard to domain specificity and putative functions.
Physical properties of assembly factors are conserved through evolution
To learn whether the physical parameters (charge, predicted disorder, molecular weight) of yeast AFs are biologically significant, one can inquire whether they are conserved in man. As judged from box-and-whisker plots of the human homologs, the dispersion of their values is comparable to that for yeast AFs (Fig. 3a). The diagonal plots in Supplementary Fig. 4 compare values for individual AFs and document extensive conservation.
Separation of the nucleolus from chromatin
What characteristics of the nucleolus and chromatin—apart from the defining presence of rDNA and its paucity of nucleosomes (Merz et al. 2008; Hamperl et al. 2013; Panday and Grove 2017)—cause these compartments not to intermix? Is this an example of condensate formation and phase separation, as discussed for the subcompartments themselves (Berry et al. 2015; Hult et al. 2017; Caragine et al. 2019; Lafontaine et al. 2021). Although controversial, condensates are thought also to be characteristic of sites of transcription by polymerase II (Rieder et al. 2012; Cho et al. 2018; McSwiggen et al. 2019; Narlikar et al. 2021; Sharp et al. 2022). An alternative hypothesis to account for the lack of intermixing is that - unless endowed with special (unspecified) characteristics - high concentrations of only few proteins are thermodynamically compatible with chromatin.
Additional relevant parameters include (1) high concentrations of AFs; (2) mutual-coherence among AFs; and (3) surface barriers. These same features are potentially relevant to separation of domains within chromatin (McKeown and Shaw 2009; Korolev, Allahverdi, et al. 2012; Korolev, Fan, et al. 2012; Strom et al. 2017; Shin et al. 2018; Gibson et al. 2019; Erdel et al. 2020; Hansen et al. 2021; Rippe 2022). Any impact of high concentrations of AFs and their mutual coherence could be strongly accentuated by cooperativity due to the adjacency of rDNA repeats.
High protein concentrations
The concentration of each AF in the nucleolus is ∼0.5 mg/ml (Fig. 2 legend). This concentration is of the same order of magnitude as the concentration of selected proteins that is required for formation of a separate phase in physiological salt solution (Asherie 2004; Hyman et al. 2014; Feric et al. 2016; You et al. 2020).
Mutual-coherence
Considering the close-packed arrangements of AFs in the assembly intermediates that have been imaged by cryo-EM, it seems plausible that AFs interact even when they are latent. Indeed, many AFs have been purified as subcomplexes or modules in the absence of rRNA (UTP complexes, proteins associated with Mpp10, Noc4, etc.) (Gallagher et al. 2004; Krogan et al. 2004; Hinsby et al. 2006; Merl et al. 2010; Woolford and Baserga 2013; Wada et al. 2014; Vincent et al. 2018). Moreover, interactions among many AFs have been documented in 2-hybrid assays (Baßler et al. 2017; Vincent et al. 2018). The tendency of AFs to form coherent units is also evident in the “mininucleoli” that accumulate after cell division in animal cells before rRNA synthesis resumes (Dimario 2004; Hernandez-Verdun 2011). Possibly similar accumulations are found in yeast that express rRNA from high-copy plasmids (Oakes et al. 1998).
Observations of yeast arrested at metaphase are also indicative of coherence. In this situation—when rRNA synthesis and processing continue—the nuclear envelope spans the bud neck and nucleolar markers and rDNA remain in the mother cell where they abut on the bud neck. Nevertheless, chromatin is ductile and oscillates across the bud neck (Palmer et al. 1989; Rai et al. 2017; Tartakoff et al. 2021). Thus, the nucleolar mass behaves as expected for an extensively cross-linked unit. It remains an open question as to whether the nucleolus includes any internal organizational matrix (Fath et al. 2000; Hirai et al. 2013).
Surface barriers
Several human AFs localize to the periphery of the nucleolus (Van Hooser et al. 2005; Booth et al. 2016; Stenstrom et al. 2020). Furthermore, one protein has been identified that lines the surface of the nucleolus in Xenopus oocytes (Voltmer-Irsch et al. 2007). In yeast, no protein has yet been detected at the interface between the nucleolus and chromatin.
A secluded environment for nascent rRNPs—the viral analogy
The presence of the many AFs in nascent rRNPs—as well as the surrounding reservoir of latent AFs—could limit or buffer their interactions with nucleoplasmic proteins. By analogy, replication of viral genomes and assembly of nucleocapsids often occur in compositionally distinct microenvironments (“factories”)—either in the nucleus or cytoplasm—that are not surrounded by membranes (Okano et al. 1999; Novoa et al. 2005; Erickson et al. 2012; Strang et al. 2012; McSwiggen et al. 2019; Charman and Weitzman 2020; Snijder et al. 2020; Iarovaia et al. 2021). There is no comprehensive information on the makeup of these factories; however, as for nucleolus, we propose that these zones restrict encroachment by surrounding host factors. This parallel is dramatically emphasized by the observation that when one RNA segment of the influenza genome is absent from viral particles, its place is taken by rRNAs (Noda et al. 2018). Furthermore, infection by many viruses alters the appearance of the nucleolus and/or relocalizes AFs, e.g., (Matthews et al. 2011; Salvetti and Greco 2014). Interestingly, a variety of proteins implicated in replication of yeast viral particles depend on host cell AFs (Mak5, Mak16, etc.) (Wickner 1986).
Translocation of rRNPs between domains
In its simplest form, the progress of rRNP maturation involves passage from the inner layer that surrounds the rDNA axis to the outer layer/volume, subsequent translocation into the chromatin-domain, and export to the cytoplasm. As we have previously discussed (Tartakoff et al. 2021), transfer of nascent rRNPs from the inner layer of the coaxial structure to the outer layer can be attributed to thermodynamic properties of the associated AFs since they are most stable in the outer layer, judging from the distribution of their latent forms. In other words, when they are recruited to nascent rRNA in the inner layer they are in a metastable condition. The potential energy that is characteristic of this condition could power transfer of nascent rRNPs to the outer layer, providing an example of vectorial 2-phase partitioning (Supplementary Fig. 5).
Subsequent translocation into the chromatin domain could be closely related. Indeed, this event seems analogous to transfer between solutions of distinct polymers that constitute “aqueous 2-phase systems.” Such systems have often been used for purification of corresponding macromolecular cargoes that are initially present in one of the phases and transfer to the other. Immature ribosomal subunits, in this sense, can be considered cargoes that are “purified” by being transferred (Walter 1994; Johansson et al. 1998; Filho 2004; Asenjo and Andrews 2011; Monterroso et al. 2016).
In particular, we previously noticed that the outer layer/volume includes multiple AFs that localize primarily to the chromatin domain and are recovered in LSU subunit precursors (Tartakoff et al. 2021). Especially, if these AFs enter the outer layer/volume and line the surface of rRNP particles in that compartment, this “coating” could overcome any energy barrier that limits their diffusion into the chromatin domain— Supplementary Fig. 5. Deeper understanding of the physico-chemical properties of chromatin are, however, required to pursue this hypothesis (Filho 2004; Korolev, Allahverdi, et al. 2012; Korolev, Fan, et al. 2012; Hult et al. 2017; Strickfaden 2021).
There has been discussion of whether changes of the protein/RNA ratio of rRNPs account for their progressive transfer to the chromatin domain (Riback et al. 2020). Since the data underlying this suggestion come from experiments in which bacterial subunits are mixed with single recombinant nucleolar proteins, it is difficult to evaluate their in vivo relevance. Further in situ cryo-EM tomographic studies have described successive stages of maturation of intermediates as they enter the chromatin domain (Erdmann et al. 2021).
Functional Subsets of Assembly Factors
Motif signatures of assembly factors
Hierarchical clustering identifies nineteen prototypic motif signatures that are shared by two or more AFs (Fig. 3d and Supplementary Table 5). These signatures include 1–10 motifs and correspond to GTPases, ATPases, methyl transferases, structural proteins (ARM, HEAT, and WD repeats), RNA-binding proteins, subsets of snoRNP proteins, prolyl isomerases, kinases, etc. Most signatures are found among both SSU and LSU AFs, and no AF has more than a single signature (among those listed). Many AFs that share a signature belong to distinct clades, implying that their primary sequences are not closely related to each other. The clustering analysis also calls attention to “variant” motif signatures for several types of AFs.
One-third of the AFs in the clustering diagram (51 proteins) lack motif signatures that are shared with other AFs (Supplementary Table 5, legend). The IEP values of these AFs have a bimodal distribution, their sizes are comparable to those of other AFs (10–100 kDa), and their levels of predicted disorder are modest (51% ± 20% vs 37% ±15%). Approximately two-thirds of these proteins are essential for haploid growth and ∼ 2/3 have homologs in man. They do not correspond to the “complexes with undetermined function” that have previously been described (Hinsby et al. 2006). The exceptional yeast AFs are found associated with each of the 3 domains of rRNA (14%, 30%, 56% for the 5′-ETS, pre-40S, and pre-LSU domains). Nine have motifs implicated in binding RNA and 7 have motifs that resemble single ribosomal proteins, some of which are known to function as placeholders (Espinar-Marchena et al. 2017).
The assembly factors of nascent subunits during their maturation
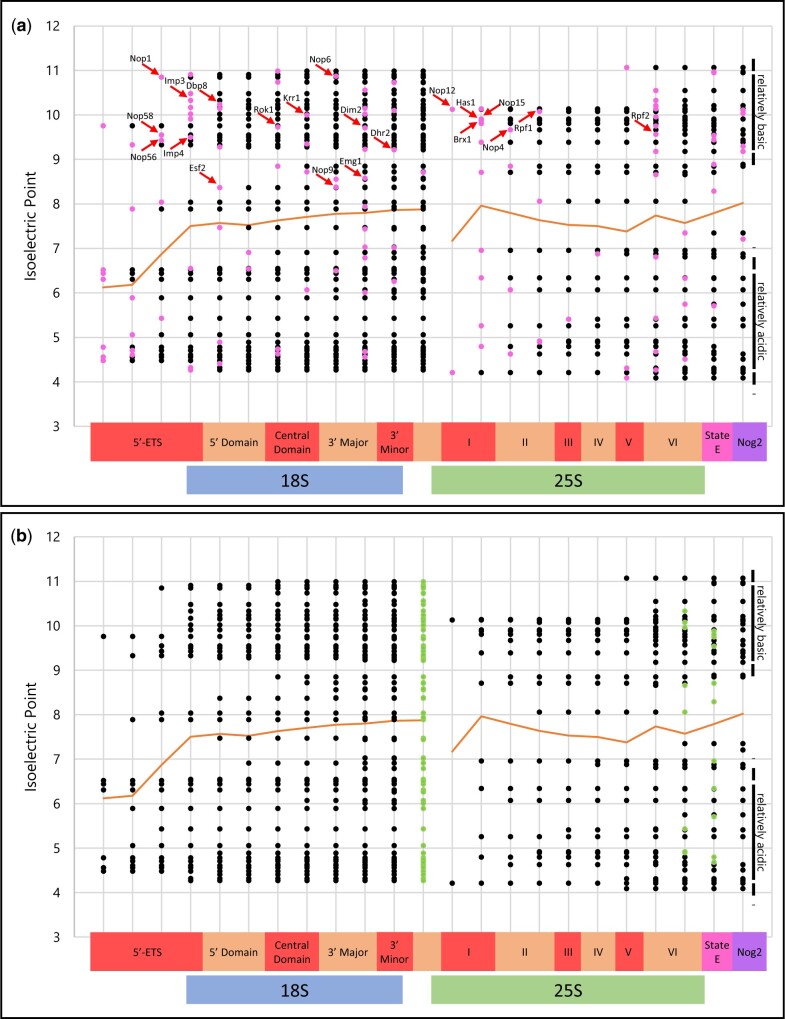
The progressive contributions of AFs to the isoelectric properties of rRNPs during transcript elongation are illustrated in the “progressive plots” of Fig. 4, a and b, based on the information in (Klinge and Woolford 2019) that estimates the timing of addition of 109 AFs along the rRNA segment from the 5′-ETS through the 3′-ETS. Since nascent rRNA transcripts in growing yeast are usually cleaved in ITS1 (Osheim et al. 2004), in these graphs the data points “restart” after ITS1. These “progressive plots” are highlighted to indicate the timing of addition (pink) and removal (green) of individual AFs.
Fig. 4.
The AF complement of nascent subunits during their maturation—“progressive plots.” Individual AFs are positioned according to a recent summary (Klinge and Woolford 2019). The continuous near-horizonal line indicates the mean IEP of all AFs that have been added. (a) Additions of AFs in each step are shown in pink. (b) Removals of AFs for each subsequent (n + 1) step are in green. rRNA domains are indicated along the horizontal axis at the bottom. Note the relative thinning of the isoelectric distributions at neutral pH values (also seen in Fig. 3a). State E and the Nog2 particle are described (Wu et al. 2016; Kater et al. 2017). The arrows indicate proteins that have classic RNA-binding motifs, as enumerated in Supplementary Table 6. The H/ACA snoRNP proteins are not included in these figures because they are absent from (Klinge and Woolford 2019). They associate with the central domain of the pre-40S segment, according to experiments with 3′-truncated transcripts expressed in vivo (Chaker-Margot et al. 2015; Zhang et al. 2016).
For both subunits, addition of AFs is incremental. For SSU precursors, there is no indication of release of AFs until transcription reaches the end of the 3′-minor domain (Zhang et al. 2016). Correspondingly, AFs begin to be released from LSU precursors approximately when rRNA undergoes cleavage near the 3′ terminus and intermediates enter the nucleoplasm (Panse and Johnson 2010; Gamalinda et al. 2014; Nerurkar et al. 2015; Kater et al. 2017).
The progressive plot representation of the AF complement of the nascent rRNPs exhibits an unanticipated degree of bimodal “isoelectric balance,” with some AFs being significantly more basic than others and only few AFs being isoelectric near neutral pH. As shown in Fig. 3a, this bimodality is also found among human AFs. Moreover—at least in yeast—after the relatively acidic 5′-ETS segment, cumulative near-neutrality of the mean values of the associated AFs is characteristic of the remaining length of the rRNA transcript.
The progressive plot in Fig. 4a shows that—with few exceptions—addition of basic AFs that are known to bind RNA (arrows) is accompanied by addition of more acidic AFs. We develop below the hypothesis that selected basic RNA-binding AFs are responsible for a succession of nucleation events during transcript elongation.
The 5′-ETS domain as a unique element
The 5′-ETS segment does not acquire ribosomal proteins and is ultimately removed, rather than arriving in the cytoplasm. It therefore is of interest that this segment lacks DexD/H-box proteins, GTPases, methyltransferases, and kinases (Table 3) (Kornprobst et al. 2016; Barandun et al. 2017; Baßler et al. 2017; Sun et al. 2017; Hunziker et al. 2019). If spatial rearrangements within assembly intermediates are needed to accommodate the arrival of new ribosomal proteins, such enzyme activities could be important only in the segments that do include ribosomal proteins, thereby accounting for their absence from the 5′-ETS rRNP.
As is well-known, the 5′-ETS domain is unusually enriched in WD-repeat proteins (Barandun et al. 2017; Cheng et al. 2020). The 5′-ETS rRNP helps position U3 snoRNA and contributes to reorganization of downstream segments (Musters et al. 1990; Beltrame and Tollervey 1995; Kornprobst et al. 2016; Hunziker et al. 2019). Its massive and apparently cross-linked structure, as well as its relatively low IEP, could also be integral to the thermodynamically favorable translocation of rRNP intermediates from the inner to the outer layer of the coaxial structure (lift-off) during transcription (Fig. 1, e and f). The human 5′-ETS is about 4× as long as the yeast 5′-ETS, but 75% of its length is dispensable (Singh et al. 2021).
A 3-step Subdomain Assembly Model
On the basis of the observations summarized above, we propose a “3-step subdomain folding model” of production of nascent subunits that divides the process into steps that recur within the 5′-ETS domain, as well as the pre-40S and pre-LSU domains. These steps are nucleation, recruitment, and consolidation (Fig. 5, a and b). This model is distinct from suggestions that AFs are uniformly distributed along rRNA or that rRNP formation is initiated at only a pair of sites, one corresponding to each subunit. Key evidence that multiple rRNPs can form separately within the 5′-ETS domain, as well as the pre-40S and pre-LSU domains comes from the in vivo observation that when single subdomains or 3′-truncated rRNAs are expressed in vivo they recruit distinct groups of AFs. Moreover, as described below, subdomain folding is detected by cryo-EM examination. It is conventional to distinguish 4 subdomains within the pre-40S segment and 6 subdomains within the pre-LSU segment (Perez-Fernandez et al. 2007; Chaker-Margot et al. 2015; Zhang et al. 2016; Chen et al. 2017; Hunziker et al. 2019).
Fig. 5.
Distributions of AFs—models. a) Isoelectric balance. The nascent rRNA recruits basic proteins to its successive domains. Such proteins then attract less basic AFs. These AFs, in turn, preferentially bind basic ribosomal proteins, while other ribosomal proteins bind directly to rRNA. The layered model incorporates these observations, suggesting that they reflect a widespread tendency. There is however no reason to expect that intermediates are organized as concentric spheres. b) Sequential nucleation, recruitment and consolidation of AFs from a shared reservoir. From left-to-right: initiator RNA-associated proteins (blue, a, b, c, etc.) bind successive sites along transcripts, latent AFs (multicolored) are recruited to the initiators from the shared reservoir throughout the nucleolus, the resulting rRNPs then consolidate into composite units resembling those of Fig. 1e. This sequence of events is depicted in association with the inner layer, as is appropriate for pre-40S intermediates. In this diagram, all AFs that bind a given initiator have the same shape and color. This is not intended to imply that they are chemically identical. In the reservoir, some AFs are monomers, while others (possibly the majority) are associated with other AFs.
Nucleation
As each subdomain of rRNA is transcribed, we propose that “packaging signals” emerge along their length and bind nucleator AFs to form primary rRNP complexes. At least a subset of these AFs (the SSU-In, LSU-F subsets) already reside in the inner layer, judging from our earlier studies. Nucleation events can be physically independent of each other, considering the classic observation that transcription of 5′-ETS/pre-40S and 5.8S/25S segments of rRNA from a pair of plasmids is sufficient to sustain growth, i.e. no continuity is required between the 3′ end of the pre-40S segment and the 5′ end of the 5.8S segment (Liang and Fournier 1997). Independent nucleation also provides a simple basis for understanding that many lesions that impair assembly of SSUs allow production of LSUs to continue (and vice versa). Nucleation sites may include those that can be linked to RNA polymerase A (Turowski et al. 2020; Azouzi et al. 2021) and/or sites that associate with the DExD/H-box protein, Prp43 (Bohnsack et al. 2009). Genesis of ribosomal subunits seems analogous to encapsidation of the genomes of many RNA viruses in which “packaging signals” interact with notably basic domains of viral capsid proteins, which in turn interact with more externally disposed proteins (e.g. Masters 2019; Wulan et al. 2015; Lakdawala et al. 2016; Labaronne et al. 2016; Kaddis Maldonado and Parent 2016).
Candidate nucleator AFs are among those that have been recovered when 3′-truncated transcripts (or single domains) of SSU (or LSU) rRNA are expressed in vivo and used to retrieve AFs. Supplementary Table 6 lists such AFs that have motifs that are known to interact with RNA, as well as the snoRNP proteins that are retrieved. The mean IEP of this entire group is pH 8.8 ± 1.9 and the members of this group with IEP > pH 8.4 are marked with arrows in Fig. 4a to indicate the timing of their addition to nascent rRNPs. Critically, formation of SSU terminal knobs is inhibited upon depletion of either of two of these proteins (Dbp4, Imp3) that bind RNA and were previously implicated in early steps of SSU assembly (Supplementary Table 1), while production of LSUs continues (Osheim et al. 2004; Soltanieh et al. 2015).
Recruitment
Primary rRNP complexes recruit latent AFs from the shared reservoir, many of which are less basic. In doing so, the nucleators exhibit polyvalency, being able to bind rRNA and other AFs. In fact, for each rRNA subdomain, the total number of AFs retrieved along with 3′-truncated transcripts significantly exceeds the number of candidate nucleators that, by definition, are basic and bind RNA. Thus, for the successive SSU RNA domains that associate with 3′ truncated transcripts (including snoRNP proteins), the ratios of total AFs-to-candidate nucleators are 23/6 (5′-ETS), 9/3 (5′-domain), 10/7 (central domain), 4/3 (3′-major domain), and 18/5 (3′-minor domain). For LSU domains, the ratios are 20/5 (domain I), 8/2 (domain II), 2/1 (domain III), 2/1 (domain IV), 9/2 (domain V), and 5/1 (domain VI) (Chaker-Margot et al. 2015; Zhang et al. 2016; Chen et al. 2017; Hunziker et al. 2019). The resulting composite rRNPs may in fact account for the irregularities seen along elongating nascent rRNA (smaller than the terminal knobs) (e.g. Osheim et al. 2004).
In sum, as schematized in Fig. 5, a and b, we propose that the sequential interactions between rRNA, nucleator largely basic AFs, additional less-basic AFs, and ribosomal proteins are favored by charge alternation. There is, however, no reason to think that there is strict conformity to a layered or concentric structure.
Experimental identification of interactions between AFs might also be informative in this regard. The many pair-wise interactions that have been reported presumably concern latent AFs since the protocols used did not ensure association with rRNA. Interestingly, in addition to SSU-SSU and LSU-LSU interactions, these studies document many heterotypic interactions between SSU and LSU AFs (Hinsby et al. 2006; Baßler et al. 2017; Vincent et al. 2018). This is unlike the homotypic interactions among AFs seen in extracts of cells in which rRNA transcription has been inhibited (Merl et al. 2010). Relatively acidic secondary AFs—along with rRNA itself—could also be well-suited to promote integration of basic ribosomal proteins.
When stable rRNP domains have formed, they are expected to be visible in cryo-EM images. It therefore is of interest that such images show that the 5′-ETS domain and relatively 3′ rRNP domain of SSU precursors can be visible in particles that lack structured intervening domains (Chaker-Margot et al. 2015; Zhang et al. 2016; Cheng et al. 2019). Observations of what appears to be nonuniform domain folding have also been made for LSU intermediates (Kater et al. 2017). In both cases, it seems likely that nucleation does occur sequentially following the 5′ to 3′ order, but that the intervening domains are conformationally immature in the rRNPs that have been imaged.
Consolidation
When 2 or more nascent rRNP units that are destined to contribute to the same subunit have formed—and may already contact each other—they consolidate to form larger assemblies, ultimately giving rise to rRNP knobs (Osheim et al. 2004). These events likely are cooperative. Consolidation could be part of the large-scale structural rearrangements that have been detected by cryo-EM (Leidig et al. 2014; Wu et al. 2016; Kater et al. 2017; Poll et al. 2017; Cheng et al. 2019; Aquino et al. 2021) and could include the restructuring of domains that appeared to be delayed in their assembly, as mentioned above.
One key observation supports the hypothesis that consolidation occurs in cis, i.e. that it requires the coordinated folding of colinear domains. This evidence again comes from experiments in which a pair of plasmids was used to drive production of subunits. In these experiments, 5.8S and 25S rRNA sequences can function when they are expressed by the same plasmid but not if they are in trans, i.e. when one plasmid codes for the of 5′-ETS/pre-40S/5.8S segment and the other codes for the 25S segment (Liang and Fournier 1997). An example of a transcriptional unit that can function in trans is the 5S rRNP. Thus, in yeast, the 5S DNA sequences are positioned between the rDNA repeats. 5S rRNA itself—once transcribed by RNA polymerase III—is secondarily incorporated into the LSU precursor.
In addition to formulating this “3-step subdomain folding model,” we have discussed the importance of the nucleolar microenvironment, the isoelectric balance that is characteristic of rRNP formation, and the contribution of vectorial 2-phase partitioning for translocation of rRNPs. The observation that AFs form polyvalent closely interlocking structures in nascent rRNPs likely accounts for why the large majority of these proteins are essential. We suspect that the principles that become evident in this nearly macroscopic example of gene expression can serve as a guide for understanding many examples of transcription by RNA polymerase II.
Evolutionary Considerations
Many AF homologs are thought to have existed in LECA (the last eukaryotic common ancestor) (Staub et al. 2004; Ebersberger et al. 2014; Bohnsack and Bohnsack 2019). These homologs include representatives of each of the 4 subunit-specific groups of AFs, snoRNP proteins, and the AFs that contribute to both types of subunit (Supplementary Table 7). The coaxial organization of the nucleolus and the sequential reorganization of nascent rRNPs therefore could have an ancient origin. AF homologs are, however, poorly represented in LUCA (the last universal cellular ancestor) and in the Archaea (Staub et al. 2004; Bower-Phipps et al. 2012; Feng et al. 2013; Ebersberger et al. 2014). Interestingly, despite the small size of the 5′-ETS segment of rDNA in Escherichia coli and despite the absence of contiguous rDNA repeats, Miller spreads from E. coli nevertheless appear very similar to those of yeast and higher organisms (Gotta et al. 1991; Davis and Williamson 2017).
Data availability
The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables.
Supplemental material is available at GENETICS online.
Supplementary Material
Acknowledgments
The authors thank the NIH for R01GM089872 (AT), R01HL126626 and HL141423 (GHM), and P30CA43703-12 and thank the Visconsi family for support. The authors also thank Dalya Khalife and Jingwei Li for database scouring.
This investigation was a shared virtual research effort. All members of the group collected information on subsets of AFs. They also had more specific responsibilities, as enumerated below.
Contributor Information
Samantha Lin, Pathology Department and The Cell Biology Program, Case Western Reserve University, Cleveland, OH 44106, USA.
Suchita Rajan, Pathology Department and The Cell Biology Program, Case Western Reserve University, Cleveland, OH 44106, USA.
Sofia Lemberg, Pathology Department and The Cell Biology Program, Case Western Reserve University, Cleveland, OH 44106, USA.
Mark Altawil, Pathology Department and The Cell Biology Program, Case Western Reserve University, Cleveland, OH 44106, USA.
Katherine Anderson, Pathology Department and The Cell Biology Program, Case Western Reserve University, Cleveland, OH 44106, USA.
Ruth Bryant, Pathology Department and The Cell Biology Program, Case Western Reserve University, Cleveland, OH 44106, USA.
Sebastian Cappeta, Pathology Department and The Cell Biology Program, Case Western Reserve University, Cleveland, OH 44106, USA.
Brandon Chin, Pathology Department and The Cell Biology Program, Case Western Reserve University, Cleveland, OH 44106, USA.
Isabella Hamdan, Pathology Department and The Cell Biology Program, Case Western Reserve University, Cleveland, OH 44106, USA.
Annelise Hamer, Pathology Department and The Cell Biology Program, Case Western Reserve University, Cleveland, OH 44106, USA.
Rachel Hyzny, Pathology Department and The Cell Biology Program, Case Western Reserve University, Cleveland, OH 44106, USA.
Andrew Karp, Pathology Department and The Cell Biology Program, Case Western Reserve University, Cleveland, OH 44106, USA.
Daniel Lee, Pathology Department and The Cell Biology Program, Case Western Reserve University, Cleveland, OH 44106, USA.
Alexandria Lim, Pathology Department and The Cell Biology Program, Case Western Reserve University, Cleveland, OH 44106, USA.
Medha Nayak, Pathology Department and The Cell Biology Program, Case Western Reserve University, Cleveland, OH 44106, USA.
Vishnu Palaniappan, Pathology Department and The Cell Biology Program, Case Western Reserve University, Cleveland, OH 44106, USA.
Soomin Park, Pathology Department and The Cell Biology Program, Case Western Reserve University, Cleveland, OH 44106, USA.
Sarika Satishkumar, Pathology Department and The Cell Biology Program, Case Western Reserve University, Cleveland, OH 44106, USA.
Anika Seth, Pathology Department and The Cell Biology Program, Case Western Reserve University, Cleveland, OH 44106, USA.
Uva Sri Dasari, Pathology Department and The Cell Biology Program, Case Western Reserve University, Cleveland, OH 44106, USA.
Emili Toppari, Pathology Department and The Cell Biology Program, Case Western Reserve University, Cleveland, OH 44106, USA.
Ayush Vyas, Pathology Department and The Cell Biology Program, Case Western Reserve University, Cleveland, OH 44106, USA.
Julianne Walker, Pathology Department and The Cell Biology Program, Case Western Reserve University, Cleveland, OH 44106, USA.
Evan Weston, Pathology Department and The Cell Biology Program, Case Western Reserve University, Cleveland, OH 44106, USA.
Atif Zafar, Pathology Department and The Cell Biology Program, Case Western Reserve University, Cleveland, OH 44106, USA.
Cecelia Zielke, Pathology Department and The Cell Biology Program, Case Western Reserve University, Cleveland, OH 44106, USA.
Ganapati H Mahabeleshwar, Pathology Department and The Cell Biology Program, Case Western Reserve University, Cleveland, OH 44106, USA.
Alan M Tartakoff, Pathology Department and The Cell Biology Program, Case Western Reserve University, Cleveland, OH 44106, USA.
Conflicts of interest
None declared.
| Name | Task |
|---|---|
| Lin, Samantha | Taught the group regarding MATLAB, analyzed physical properties, formulated many figures, wrote an extensive draft of the manuscript. |
| Altawil, Mark | Disease relations of AFs |
| Anderson, Katherine | Comparison of disorder algorithms |
| Bryant, Ruth | Human chromosome map |
| Cappeta, Sebastian | Yeast chromosome map |
| Chin, Brandon | Disease relations of AFs |
| Hamdan, Isabella | Linear organization of AFs |
| Hamer, Annelise | Linear organization of AFs |
| Hyzny, Rachel | Klyuveromyces chromosome map |
| Karp, Andrew | Linear organization of AFs |
| Lee, Daniel | Analysis of peptide motifs, hierarchical clustering |
| Lemberg, Sofia | Group coordinator, formulated the cumulative plots |
| Lim, Alexandria | Linear organization of AFs |
| Mahabaleshwar, Ganapati H. | Funded the clustering analysis |
| Nayak, Medha | Identification of NLSs |
| Palaniappan, Vishnu | Chromosome maps |
| Park, Soomin | Evaluation of protein abundance |
| Rajan, Suchita | Group coordinator, identification of NLSs |
| Satishkumar, Sarika | Investigation of physical properties of subsets of AFs |
| Seth, Anika | Coordination, data collection |
| Sri Dasari, Uva | Linear organization of AFs |
| Toppari, Emili | Identification of peptide motifs |
| Vyas, Ayush | Investigation of physical properties of subsets of AFs |
| Walker, Julianne | Modeling of transcription, physical properties of AFs. |
| Weston, Evan | Evaluation of protein abundance. |
| Zafer, Atif | Hierarchical clustering |
| Zielke, Cecelia | Disease linkage |
| Tartakoff, Alan M. | Conceived of the project, coordinated sessions, writing. |
Literature cited
- Ahmad Y, Boisvert F-M, Gregor P, Cobley A, Lamond AI.. NOPdb: nucleolar proteome database–2008 update. Nucleic Acids Res. 2009;37(Database Issue):D181–D184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert B, Mathon J, Shukla A, Saad H, Normand C, Léger-Silvestre I, Villa D, Kamgoue A, Mozziconacci J, Wong H, et al. Systematic characterization of the conformation and dynamics of budding yeast chromosome XII. J Cell Biol. 2013;202(2):201–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen JS, Lam YW, Leung AKL, Ong S-E, Lyon CE, Lamond AI, Mann M.. Nucleolar proteome dynamics. Nature. 2005;433(7021):77–83. [DOI] [PubMed] [Google Scholar]
- Aquino GRR, Hackert P, Krogh N, Pan K-T, Jaafar M, Henras AK, Nielsen H, Urlaub H, Bohnsack KE, Bohnsack MT, et al. The RNA helicase Dbp7 promotes domain V/VI compaction and stabilization of inter-domain interactions during early 60S assembly. Nat Commun. 2021;12(1):6152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asenjo JA, Andrews BA.. Aqueous two-phase systems for protein separation: a perspective. J Chromatogr A. 2011;1218(49):8826–8835. [DOI] [PubMed] [Google Scholar]
- Asherie N. Protein crystallization and phase diagrams. Methods. 2004;34(3):266–272. [DOI] [PubMed] [Google Scholar]
- Aubert M, O’Donohue M-F, Lebaron S, Gleizes P-E.. Pre-ribosomal RNA processing in human cells: from mechanisms to congenital diseases. Biomolecules. 2018;8(4):123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azouzi C, Jaafar M, Dez C, Abou Merhi R, Lesne A, Henras AK, Gadal O.. Coupling between production of ribosomal RNA and maturation: just at the beginning. Front Mol Biosci. 2021;8:778778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badertscher L, Wild T, Montellese C, Alexander LT, Bammert L, Sarazova M, Stebler M, Csucs G, Mayer TU, Zamboni N, et al. Genome-wide RNAi screening identifies protein modules required for 40S subunit synthesis in human cells. Cell Rep. 2015;13(12):2879–2891. [DOI] [PubMed] [Google Scholar]
- Barandun J, Chaker-Margot M, Hunziker M, Molloy KR, Chait BT, Klinge S.. The complete structure of the small-subunit processome. Nat Struct Mol Biol. 2017;24(11):944–953. [DOI] [PubMed] [Google Scholar]
- Barandun J, Hunziker M, Klinge S.. Assembly and structure of the SSU processome-a nucleolar precursor of the small ribosomal subunit. Curr Opin Struct Biol. 2018;49:85–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baßler J, Ahmed YL, Kallas M, Kornprobst M, Calviño FR, Gnädig M, Thoms M, Stier G, Ismail S, Kharde S, et al. Interaction network of the ribosome assembly machinery from a eukaryotic thermophile. Protein Sci. 2017;26(2):327–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baßler J, Hurt E.. Eukaryotic ribosome assembly. Annu Rev Biochem. 2019;88:281–306. [DOI] [PubMed] [Google Scholar]
- Beck M, Schmidt A, Malmstroem J, Claassen M, Ori A, Szymborska A, Herzog F, Rinner O, Ellenberg J, Aebersold R, et al. The quantitative proteome of a human cell line. Mol Syst Biol. 2011;7:549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrame M, Tollervey D.. Base pairing between U3 and the pre-ribosomal RNA is required for 18S rRNA synthesis. EMBO J. 1995;14(17):4350–4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry J, Weber SC, Vaidya N, Haataja M, Brangwynne CP.. RNA transcription modulates phase transition-driven nuclear body assembly. Proc Natl Acad Sci U S A. 2015;112(38):e5237–e5245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black JJ, Johnson AW.. Genetics animates structure: leveraging genetic interactions to study the dynamics of ribosome biogenesis. Curr Genet. 2021;67(5):729–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnsack KE, Bohnsack MT.. Uncovering the assembly pathway of human ribosomes and its emerging links to disease. EMBO J. 2019;38(13):e100278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnsack MT, Martin R, Granneman S, Ruprecht M, Schleiff E, Tollervey D.. Prp43 bound at different sites on the pre-rRNA performs distinct functions in ribosome synthesis. Mol Cell. 2009;36(4):583–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth DG, Beckett AJ, Molina O, Samejima I, Masumoto H, Kouprina N, Larionov V, Prior IA, Earnshaw WC.. 3D-CLEM reveals that a major portion of mitotic chromosomes is not chromatin. Mol Cell. 2016;64(4):790–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower-Phipps KR, Taylor DW, Wang H-W, Baserga SJ.. The box C/D sRNP dimeric architecture is conserved across domain Archaea. RNA. 2012;18(8):1527–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caragine CM, Haley SC, Zidovska A.. Nucleolar dynamics and interactions with nucleoplasm in living cells. Elife. 2019;8:e47533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaker-Margot M, Hunziker M, Barandun J, Dill BD, Klinge S.. Stage-specific assembly events of the 6-MDa small-subunit processome initiate eukaryotic ribosome biogenesis. Nat Struct Mol Biol. 2015;22(11):920–923. [DOI] [PubMed] [Google Scholar]
- Charman M, Weitzman MD.. Replication compartments of DNA viruses in the nucleus: location, location, location. Viruses. 2020;12(2):151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Huang S.. Nucleolar components involved in ribosome biogenesis cycle between the nucleolus and nucleoplasm in interphase cells. J Cell Biol. 2001;153(1):169–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Xie Z, Yang F, Ye K.. Stepwise assembly of the earliest precursors of large ribosomal subunits in yeast. Nucleic Acids Res. 2017;45(11):6837–6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Baßler J, Fischer P, Lau B, Kellner N, Kunze R, Griesel S, Kallas M, Berninghausen O, Strauss D, et al. Thermophile 90S pre-ribosome structures reveal the reverse order of co-transcriptional 18S rRNA subdomain integration. Mol Cell. 2019;75(6):1256–1269.e7. [DOI] [PubMed] [Google Scholar]
- Cheng J, Lau B, La Venuta G, Ameismeier M, Berninghausen O, Hurt E, Beckmann R.. 90S pre-ribosome transformation into the primordial 40S subunit. Science. 2020;369(6510):1470–1476. [DOI] [PubMed] [Google Scholar]
- Cho W-K, Spille J-H, Hecht M, Lee C, Li C, Grube V, Cisse II.. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science. 2018;361(6400):412–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole SE, LaRiviere FJ, Merrikh CN, Moore MJ.. A convergence of rRNA and mRNA quality control pathways revealed by mechanistic analysis of nonfunctional rRNA decay. Mol Cell. 2009;34(4):440–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coute Y, Burgess J, Diaz J, Chichester C, Lisacek, F, Greco A, Sanchez J. Deciphering the human nucleolar proteome. Mass Spectrom Rev. 2006;25(2):215–234. [DOI] [PubMed] [Google Scholar]
- Dammann R, Lucchini R, Koller T, Sogo JM.. Chromatin structures and transcription of rDNA in yeast Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21(10):2331–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JH, Williamson JR.. Structure and dynamics of bacterial ribosome biogenesis. Philos Trans R Soc Lond B Biol Sci. 2017;372(1716):20160181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dez C, Dlakic M, Tollervey D.. Roles of the HEAT repeat proteins Utp10 and Utp20 in 40S ribosome maturation. RNA. 2007;13(9):1516–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimario PJ. Cell and molecular biology of nucleolar assembly and disassembly. Int Rev Cytol. 2004;239:99–178. [DOI] [PubMed] [Google Scholar]
- Dlakic M, Tollervey D.. The Noc proteins involved in ribosome synthesis and export contain divergent HEAT repeats. RNA. 2004;10(3):351–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersberger I, Simm S, Leisegang MS, Schmitzberger P, Mirus O, von Haeseler A, Bohnsack MT, Schleiff E.. The evolution of the ribosome biogenesis pathway from a yeast perspective. Nucleic Acids Res. 2014;42(3):1509–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdel F, Rademacher A, Vlijm R, Tünnermann J, Frank L, Weinmann R, Schweigert E, Yserentant K, Hummert J, Bauer C, et al. Mouse heterochromatin adopts digital compaction states without showing hallmarks of HP1-driven liquid-liquid phase separation. Mol Cell. 2020;78(2):236–249.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann PS, Hou Z, Klumpe S, Khavnekar S, Beck F, Wilfling F, Plitzko JM, Baumeister W.. In situ cryo-electron tomography reveals gradient organization of ribosome biogenesis in intact nucleoli. Nat Commun. 2021;12(1):5364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson KD, Bouchet-Marquis C, Heiser K, Szomolanyi-Tsuda E, Mishra R, Lamothe B, Hoenger A, Garcea RL.. Virion assembly factories in the nucleus of polyomavirus-infected cells. PLoS Pathog. 2012;8(4): e1002630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinar-Marchena FJ, Babiano R, Cruz J.. Placeholder factors in ribosome biogenesis: please, pave my way. Microb Cell. 2017;4(5):144–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farley KI, Surovtseva Y, Merkel J, Baserga SJ.. Determinants of mammalian nucleolar architecture. Chromosoma. 2015;124(3):323–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fath S, Milkereit P, Podtelejnikov AV, Bischler N, Schultz P, Bier M, Mann M, Tschochner H.. Association of yeast RNA polymerase I with a nucleolar substructure active in rRNA synthesis and processing. J Cell Biol. 2000;149(3):575–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng JM, Tian HF, Wen JF.. Origin and evolution of the eukaryotic SSU processome revealed by a comprehensive genomic analysis and implications for the origin of the nucleolus. Genome Biol Evol. 2013;5(12):2255–2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feric M, Vaidya N, Harmon TS, Mitrea DM, Zhu L, Richardson TM, Kriwacki RW, Pappu RV, Brangwynne CP.. Coexisting liquid phases underlie nucleolar subcompartments. Cell. 2016;165(7):1686–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filho PMR. Thermodynamic modeling of the partitioning of biomolecules in aqueous two-phase systems using a modified Flory–Huggins equation. Process Biochem. 2004;39:2075–2083. [Google Scholar]
- French SL, Osheim YN, Cioci F, Nomura M, Beyer AL.. In exponentially growing Saccharomyces cerevisiae cells, rRNA synthesis is determined by the summed RNA polymerase I loading rate rather than by the number of active genes. Mol Cell Biol. 2003;23(5):1558–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher JEG, Dunbar DA, Granneman S, Mitchell BM, Osheim Y, Beyer AL, Baserga SJ.. RNA polymerase I transcription and pre-rRNA processing are linked by specific SSU processome components. Genes Dev. 2004;18(20):2506–2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamalinda M, Ohmayer U, Jakovljevic J, Kumcuoglu B, Woolford J, Mbom B, Lin L, Woolford JL.. A hierarchical model for assembly of eukaryotic 60S ribosomal subunit domains. Genes Dev. 2014;28(2):198–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Véronneau S, Dow S, Lucau-Danila A, Anderson K, André B, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418(6896):387–391. [DOI] [PubMed] [Google Scholar]
- Gibson BA, Doolittle LK, Schneider MWG, Jensen LE, Gamarra N, Henry L, Gerlich DW, Redding S, Rosen MK.. Organization of chromatin by intrinsic and regulated phase separation. Cell. 2019;179(2):470–484.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotta SL, Miller OL Jr, French SL.. rRNA transcription rate in Escherichia coli. J Bacteriol. 1991;173(20):6647–6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granneman S, Baserga SJ.. Ribosome biogenesis: of knobs and RNA processing. Exp Cell Res. 2004;296(1):43–50. [DOI] [PubMed] [Google Scholar]
- Grummt I. Life on a planet of its own: regulation of RNA polymerase I transcription in the nucleolus. Genes Dev. 2003;17(14):1691–1702. [DOI] [PubMed] [Google Scholar]
- Hadjiolov AA. The Nucleolus and Ribosome Biogenesis. Berlin (Germany: ): Springer Verlag; 1985. [Google Scholar]
- Hamperl S, Wittner M, Babl V, Perez-Fernandez J, Tschochner H, Griesenbeck J.. Chromatin states at ribosomal DNA loci. Biochim Biophys Acta. 2013;1829(3–4):405–417. [DOI] [PubMed] [Google Scholar]
- Hansen JC, Maeshima K, Hendzel MJ.. The solid and liquid states of chromatin. Epigenetics Chromatin. 2021;14(1):50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Verdun D, Roussel P, Thiry M, Sirri V, Lafontaine DLJ.. The nucleolus: structure/function relationship in RNA metabolism. Wiley Interdiscip Rev RNA. 2010;1(3):415–431. [DOI] [PubMed] [Google Scholar]
- Hernandez-Verdun D. Assembly and disassembly of the nucleolus during the cell cycle. Nucleus. 2011;2(3):189–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinsby AM, Kiemer L, Karlberg EO, Lage K, Fausbøll A, Juncker AS, Andersen JS, Mann M, Brunak S.. A wiring of the human nucleolus. Mol Cell. 2006;22(2):285–295. [DOI] [PubMed] [Google Scholar]
- Hirai Y, Louvet E, Oda T, Kumeta M, Watanabe Y, Horigome T, Takeyasu K.. Nucleolar scaffold protein, WDR46, determines the granular compartmental localization of nucleolin and DDX21. Genes Cells. 2013;18(9):780–797. [DOI] [PubMed] [Google Scholar]
- Ho B, Baryshnikova A, Brown GW.. Unification of protein abundance datasets yields a quantitative Saccharomyces cerevisiae proteome. Cell Syst. 2018;6(2):192–205.e3. [DOI] [PubMed] [Google Scholar]
- Hult C, Adalsteinsson D, Vasquez P, Lawrimore J, Bennett M, York A, Cook D, Yeh E, Forest M, Bloom K. Enrichment of dynamic chromosomal crosslinks drive phase separation of the nucleolus. Nucleic Acids Res. 2017;45(19):11159–11173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunziker M, Barandun J, Buzovetsky O, Steckler C, Molina H, Klinge S.. Conformational switches control early maturation of the eukaryotic small ribosomal subunit. Elife. 2019;8:e45185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman AA, Weber CA, Julicher F.. Liquid-liquid phase separation in biology. Annu Rev Cell Dev Biol. 2014;30:39–58. [DOI] [PubMed] [Google Scholar]
- Iarovaia OV, Ioudinkova ES, Velichko AK, Razin SV.. Manipulation of cellular processes via nucleolus hijaking in the course of viral infection in mammals. Cells. 2021;10(7):1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James A, Wang Y, Raje H, Rosby R, DiMario P.. Nucleolar stress with and without p53. Nucleus. 2014;5(5):402–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson HO, Karlström G, Tjerneld F, Haynes CA.. Driving forces for phase separation and partitioning in aqueous two-phase systems. J Chromatogr B Biomed Sci Appl. 1998;711(1–2):3–17. [DOI] [PubMed] [Google Scholar]
- Jorgensen P, Edgington NP, Schneider BL, Rupes I, Tyers M, Futcher B.. The size of the nucleus increases as yeast cells grow. Mol Biol Cell. 2007;18(9):3523–3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaddis Maldonado RJ, Parent LJ.. Orchestrating the selection and packaging of genomic RNA by retroviruses: an Ensemble of viral and host factors. Viruses. 2016;8(9):257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kater L, Thoms M, Barrio-Garcia C, Cheng J, Ismail S, Ahmed YL, Bange G, Kressler D, Berninghausen O, Sinning I, et al. Visualizing the assembly pathway of nucleolar pre-60S ribosomes. Cell. 2017;171(7):1599–1610.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinge S, Woolford JL Jr.. Ribosome assembly coming into focus. Nat Rev Mol Cell Biol. 2019;20(2):116–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornprobst M, Turk M, Kellner N, Cheng J, Flemming D, Koš-Braun I, Koš M, Thoms M, Berninghausen O, Beckmann R, et al. Architecture of the 90S pre-ribosome: a structural view on the birth of the eukaryotic ribosome. Cell. 2016;166(2):380–393. [DOI] [PubMed] [Google Scholar]
- Korolev N, Allahverdi A, Lyubartsev AP, Nordenskiöld L.. The polyelectrolyte properties of chromatin. Soft Matter. 2012;8(36):9322. [Google Scholar]
- Korolev N, Fan Y, Lyubartsev AP, Nordenskiöld L.. Modelling chromatin structure and dynamics: status and prospects. Curr Opin Struct Biol. 2012;22(2):151–159. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Peng W-T, Cagney G, Robinson MD, Haw R, Zhong G, Guo X, Zhang X, Canadien V, Richards DP, et al. High-definition macromolecular composition of yeast RNA-processing complexes. Mol Cell. 2004;13(2):225–239. [DOI] [PubMed] [Google Scholar]
- Labaronne A, Swale C, Monod A, Schoehn G, Crépin T, Ruigrok R.. Binding of RNA by the nucleoproteins of Influenza Viruses A and B. Viruses. 2016;8(9):247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine DLJ, Riback JA, Bascetin R, Brangwynne CP.. The nucleolus as a multiphase liquid condensate. Nat Rev Mol Cell Biol. 2021;22(3):165–182. [DOI] [PubMed] [Google Scholar]
- Lakdawala SS, Fodor E, Subbarao K.. Moving on out: transport and packaging of influenza viral RNA into virions. Annu Rev Virol. 2016;3(1):411–427. [DOI] [PubMed] [Google Scholar]
- Lamaye F, Galliot S, Alibardi L, Lafontaine DLJ, Thiry M.. Nucleolar structure across evolution: the transition between bi- and tri-compartmentalized nucleoli lies within the class Reptilia. J Struct Biol. 2011;174(2):352–359. [DOI] [PubMed] [Google Scholar]
- Lau B, Cheng J, Flemming D, La Venuta G, Berninghausen O, Beckmann R, Hurt E.. Structure of the maturing 90S pre-ribosome in association with the RNA exosome. Mol Cell. 2021;81(2):293–303.e4. [DOI] [PubMed] [Google Scholar]
- Lebaron S, Segerstolpe A, French SL, Dudnakova T, de Lima Alves F, Granneman S, Rappsilber J, Beyer AL, Wieslander L, Tollervey D, et al. Rrp5 binding at multiple sites coordinates pre-rRNA processing and assembly. Mol Cell. 2013;52(5):707–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidig C, Thoms M, Holdermann I, Bradatsch B, Berninghausen O, Bange G, Sinning I, Hurt E, Beckmann R.. 60S ribosome biogenesis requires rotation of the 5S ribonucleoprotein particle. Nat Commun. 2014;5:3491. [DOI] [PubMed] [Google Scholar]
- Liang WQ, Fournier MJ.. Synthesis of functional eukaryotic ribosomal RNAs in trans: development of a novel in vivo rDNA system for dissecting ribosome biogenesis. Proc Natl Acad Sci U S A. 1997;94(7):2864–2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang XH, Liu Q, Fournier MJ.. Loss of rRNA modifications in the decoding center of the ribosome impairs translation and strongly delays pre-rRNA processing. RNA. 2009;15(9):1716–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder P, Jankowsky E.. From unwinding to clamping - the DEAD box RNA helicase family. Nat Rev Mol Cell Biol. 2011;12(8):505–516. [DOI] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Mekhedov SL, Mirkin BG, Koonin EV.. Ancestral paralogs and pseudoparalogs and their role in the emergence of the eukaryotic cell. Nucleic Acids Res. 2005;33(14):4626–4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters PS. Coronavirus genomic RNA packaging. Virology. 2019;537:198–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews D, Emmott E, Hiscox J.. Viruses and the nucleolus. In: M Olson, editor. The Nucleolus. Berlin (Germany: ): Springer; 2011. p. 321–345. [Google Scholar]
- McKeown PC, Shaw PJ.. Chromatin: linking structure and function in the nucleolus. Chromosoma. 2009;118(1):11–23. [DOI] [PubMed] [Google Scholar]
- McStay B. Nucleolar dominance: a model for rRNA gene silencing. Genes Dev. 2006;20(10):1207–1214. [DOI] [PubMed] [Google Scholar]
- McSwiggen DT, Hansen AS, Teves SS, Marie-Nelly H, Hao Y, Heckert AB, Umemoto KK, Dugast-Darzacq C, Tjian R, Darzacq X, et al. Evidence for DNA-mediated nuclear compartmentalization distinct from phase separation. Elife. 2019;8:e47098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSwiggen DT, Mir M, Darzacq X, Tjian R.. Evaluating phase separation in live cells: diagnosis, caveats, and functional consequences. Genes Dev. 2019;33(23–24):1619–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merl J, Jakob S, Ridinger K, Hierlmeier T, Deutzmann R, Milkereit P, Tschochner H.. Analysis of ribosome biogenesis factor-modules in yeast cells depleted from pre-ribosomes. Nucleic Acids Res. 2010;38(9):3068–3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merz K, Hondele M, Goetze H, Gmelch K, Stoeckl U, Griesenbeck J.. Actively transcribed rRNA genes in S. cerevisiae are organized in a specialized chromatin associated with the high-mobility group protein Hmo1 and are largely devoid of histone molecules. Genes Dev. 2008;22(9):1190–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monterroso B, Zorrilla S, Sobrinos-Sanguino M, Keating CD, Rivas G.. Microenvironments created by liquid-liquid phase transition control the dynamic distribution of bacterial division FtsZ protein. Sci Rep. 2016;6:35140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musters W, Boon K, van der Sande CA, van Heerikhuizen H, Planta RJ.. Functional analysis of transcribed spacers of yeast ribosomal DNA. EMBO J. 1990;9(12):3989–3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musters W, Venema J, van der Linden G, van Heerikhuizen H, Klootwijk J, Planta RJ.. A system for the analysis of yeast ribosomal DNA mutations. Mol Cell Biol. 1989;9(2):551–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narlikar GJ, Myong S, Larson D, Maeshima K, Francis N, Rippe K, Sabari B, Strader L, Tjian R.. Is transcriptional regulation just going through a phase? Mol Cell. 2021;81(8):1579–1585. [DOI] [PubMed] [Google Scholar]
- Nerurkar P, Altvater M, Gerhardy S, Schütz S, Fischer U, Weirich C, Panse VG.. Eukaryotic ribosome assembly and nuclear export. Int Rev Cell Mol Biol. 2015;319:107–140. [DOI] [PubMed] [Google Scholar]
- Noda T, Murakami S, Nakatsu S, Imai H, Muramoto Y, Shindo K, Sagara H, Kawaoka Y.. Importance of the 1 + 7 configuration of ribonucleoprotein complexes for influenza A virus genome packaging. Nat Commun. 2018;9(1):54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novoa RR, Calderita G, Arranz R, Fontana J, Granzow H, Risco C.. Virus factories: associations of cell organelles for viral replication and morphogenesis. Biol Cell. 2005;97(2):147–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes M, Aris JP, Brockenbrough JS, Wai H, Vu L, Nomura M.. Mutational analysis of the structure and localization of the nucleolus in the yeast Saccharomyces cerevisiae. J Cell Biol. 1998;143(1):23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano K, Mikhailov VS, Maeda S.. Colocalization of baculovirus IE-1 and two DNA-binding proteins, DBP and LEF-3, to viral replication factories. J Virol. 1999;73(1):110–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osheim YN, French SL, Keck KM, Champion EA, Spasov K, Dragon F, Baserga SJ, Beyer AL.. Pre-18S ribosomal RNA is structurally compacted into the SSU processome prior to being cleaved from nascent transcripts in Saccharomyces cerevisiae. Mol Cell. 2004;16(6):943–954. [DOI] [PubMed] [Google Scholar]
- Palmer RE, Koval M, Koshland D.. The dynamics of chromosome movement in the budding yeast Saccharomyces cerevisiae. J Cell Biol. 1989;109(6 Pt 2):3355–3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panday A, Grove A.. Yeast HMO1: linker histone reinvented. Microbiol Mol Biol Rev. 2017;81(1): [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panse VG, Johnson AW.. Maturation of eukaryotic ribosomes: acquisition of functionality. Trends Biochem Sci. 2010;35(5):260–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel AK, Olson D, Tikoo SK.. Proteomic analysis of bovine nucleolus. Genomics Proteomics Bioinformatics. 2010;8(3):145–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T. The nucleolus. Cold Spring Harb Perspect Biol. 2011;3(3) a000638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Fernandez J, Roman A, De Las Rivas J, Bustelo X, Dosil M.. The 90S preribosome is a multimodular structure that is assembled through a hierarchical mechanism. Mol Cell Biol. 2007;27(15):5414–5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phair RD, Misteli T.. High mobility of proteins in the mammalian cell nucleus. Nature. 2000;404(6778):604–609. [DOI] [PubMed] [Google Scholar]
- Pircher A, Bakowska-Zywicka K, Schneider L, Zywicki M, Polacek N.. An mRNA-derived noncoding RNA targets and regulates the ribosome. Mol Cell. 2014;54(1):147–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poll G, Muller C, Bodden M, Teubl F, Eichner N, Lehmann G, Griesenbeck J, Tschochner H, Milkereit P.. Structural transitions during large ribosomal subunit maturation analyzed by tethered nuclease structure probing in S. cerevisiae. PLoS One. 2017;12(7):e0179405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai U, Najm F, Tartakoff AM.. Nucleolar asymmetry and the importance of septin integrity upon cell cycle arrest. PLoS One. 2017;12(3): e0174306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raska I, Shaw PJ, Cmarko D.. Structure and function of the nucleolus in the spotlight. Curr Opin Cell Biol. 2006;18(3):325–334. [DOI] [PubMed] [Google Scholar]
- Riback JA, Zhu L, Ferrolino MC, Tolbert M, Mitrea DM, Sanders DW, Wei M-T, Kriwacki RW, Brangwynne CP.. Composition-dependent thermodynamics of intracellular phase separation. Nature. 2020;581(7807):209–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder D, Trajanoski Z, McNally JG.. Transcription factories. Front Genet. 2012;3:221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippe K. Liquid-liquid phase separation in chromatin. Cold Spring Harb Perspect Biol. 2022;14(2):a040683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocak S, Emery B, Tanner NK, Linder P.. Characterization of the ATPase and unwinding activities of the yeast DEAD-box protein Has1p and the analysis of the roles of the conserved motifs. Nucleic Acids Res. 2005;33(3):999–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvetti A, Greco A.. Viruses and the nucleolus: the fatal attraction. Biochim Biophys Acta. 2014;1842(6):840–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanghai ZA, Miller L, Molloy KR, Barandun J, Hunziker M, Chaker-Margot M, Wang J, Chait BT, Klinge S.. Modular assembly of the nucleolar pre-60S ribosomal subunit. Nature. 2018;556(7699):126–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer U, Hock R.. Structure and function of the nucleolus. Curr Opin Cell Biol. 1999;11(3):385–390. [DOI] [PubMed] [Google Scholar]
- Scheer U, Weisenberger D.. The nucleolus. Curr Opin Cell Biol. 1994;6(3):354–359. [DOI] [PubMed] [Google Scholar]
- Sharma S, Lafontaine DLJ.. ‘View from a bridge’: a new perspective on eukaryotic rRNA base modification. Trends Biochem Sci. 2015;40(10):560–575. [DOI] [PubMed] [Google Scholar]
- Sharp PA, Chakraborty AK, Henninger JE, Young RA.. RNA in formation and regulation of transcriptional condensates. RNA. 2022;28(1):52–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y, Chang Y-C, Lee DSW, Berry J, Sanders DW, Ronceray P, Wingreen NS, Haataja M, Brangwynne CP.. Liquid nuclear condensates mechanically sense and restructure the genome. Cell. 2018;175(6):1481–1491.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Higgins DG.. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 2018;27(1):135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S, Vanden Broeck A, Miller L, Chaker-Margot M, Klinge S.. Nucleolar maturation of the human small subunit processome. Science. 2021;373(6560): eabj5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TF. Diversity of WD-repeat proteins. Subcell Biochem. 2008;48:20–30. [DOI] [PubMed] [Google Scholar]
- Snijder EJ, Limpens RWAL, de Wilde AH, de Jong AWM, Zevenhoven-Dobbe JC, Maier HJ, Faas FFGA, Koster AJ, Bárcena M.. A unifying structural and functional model of the coronavirus replication organelle: tracking down RNA synthesis. PLoS Biol. 2020;18(6):e3000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltanieh S, Osheim YN, Spasov K, Trahan C, Beyer AL, Dragon F.. DEAD-box RNA helicase Dbp4 is required for small-subunit processome formation and function. Mol Cell Biol. 2015;35(5):816–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staub E, Fiziev P, Rosenthal A, Hinzmann B.. Insights into the evolution of the nucleolus by an analysis of its protein domain repertoire. Bioessays. 2004;26(5):567–581. [DOI] [PubMed] [Google Scholar]
- Staub E, Mackowiak S, Vingron M.. An inventory of yeast proteins associated with nucleolar and ribosomal components. Genome Biol. 2006;7(10):R98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenstrom L, Mahdessian D, Gnann C, Cesnik A, Ouyang W, Leonetti M, Uhlen M, Cuylen-Haering S, Thul P, Lundberg E.. Mapping the nucleolar proteome reveals a spatiotemporal organization related to intrinsic protein disorder. Mol Syst Biol. 2020;16(8):e9469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strang BL, Boulant S, Chang L, Knipe DM, Kirchhausen T, Coen DM.. Human cytomegalovirus UL44 concentrates at the periphery of replication compartments, the site of viral DNA synthesis. J Virol. 2012;86(4):2089–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickfaden H. Reflections on the organization and the physical state of chromatin in eukaryotic cells. Genome. 2021;64(4):311–325. [DOI] [PubMed] [Google Scholar]
- Strom AR, Emelyanov AV, Mir M, Fyodorov DV, Darzacq X, Karpen GH.. Phase separation drives heterochromatin domain formation. Nature. 2017;547(7662):241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Zhu X, Qi J, An W, Lan P, Tan D, Chen R, Wang B, Zheng S, Zhang C, et al. Molecular architecture of the 90S small subunit pre-ribosome. Elife. 2017;6: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tafforeau L, Zorbas C, Langhendries J-L, Mullineux S-T, Stamatopoulou V, Mullier R, Wacheul L, Lafontaine DLJ.. The complexity of human ribosome biogenesis revealed by systematic nucleolar screening of pre-rRNA processing factors. Mol Cell. 2013;51(4):539–551. [DOI] [PubMed] [Google Scholar]
- Talkish J, Biedka S, Jakovljevic J, Zhang J, Tang L, Strahler JR, Andrews PC, Maddock JR, Woolford JL.. Disruption of ribosome assembly in yeast blocks cotranscriptional pre-rRNA processing and affects the global hierarchy of ribosome biogenesis. RNA. 2016;22(6):852–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taoka M, Nobe Y, Yamaki Y, Yamauchi Y, Ishikawa H, Takahashi N, Nakayama H, Isobe T.. The complete chemical structure of Saccharomyces cerevisiae rRNA: partial pseudouridylation of U2345 in 25S rRNA by snoRNA snR9. Nucleic Acids Res. 2016;44(18):8951–8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartakoff AM, Chen L, Raghavachari S, Gitiforooz D, Dhinakaran A, Ni C-L, Pasadyn C, Mahabeleshwar GH, Pasadyn V, Woolford JL, et al. The nucleolus as a polarized coaxial cable in which the rDNA axis is surrounded by dynamic subunit-specific phases. Curr Biol. 2021;31(12):2507–2519.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Rosell J, Sunjevaric I, De Piccoli G, Sacher M, Eckert-Boulet N, Reid R, Jentsch S, Rothstein R, Aragón L, Lisby M, et al. The Smc5-Smc6 complex and SUMO modification of Rad52 regulates recombinational repair at the ribosomal gene locus. Nat Cell Biol. 2007;9(8):923–931. [DOI] [PubMed] [Google Scholar]
- Turoverov KK, Kuznetsova IM, Fonin AV, Darling AL, Zaslavsky BY, Uversky VN.. Stochasticity of biological soft matter: emerging concepts in intrinsically disordered proteins and biological phase separation. Trends Biochem Sci. 2019;44(8):716–728. [DOI] [PubMed] [Google Scholar]
- Turowski TW, Petfalski E, Goddard BD, French SL, Helwak A, Tollervey D.. Nascent transcript folding plays a major role in determining RNA polymerase elongation rates. Mol Cell. 2020;79(3):488–503.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hooser AA, Yuh P, Heald R.. The perichromosomal layer. Chromosoma. 2005;114(6):377–388. [DOI] [PubMed] [Google Scholar]
- Vincent NG, Charette JM, Baserga SJ.. The SSU processome interactome in Saccharomyces cerevisiae reveals novel protein subcomplexes. RNA. 2018;24(1):77–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voltmer-Irsch S, Kneissel S, Adenot PG, Schmidt-Zachmann MS.. Regulatory mechanisms governing the oocyte-specific synthesis of the karyoskeletal protein NO145. J Cell Sci. 2007;120(Pt 8):1412–1422. [DOI] [PubMed] [Google Scholar]
- Wada K, Sato M, Araki N, Kumeta M, Hirai Y, Takeyasu K, Furukawa K, Horigome T.. Dynamics of WD-repeat containing proteins in SSU processome components. Biochem Cell Biol. 2014;92(3):191–199. [DOI] [PubMed] [Google Scholar]
- Walter HJG. Aqueous two-phase systems. In: Methods of Enzymology. New York: Academic Press; 1994. [Google Scholar]
- Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24(11):437–440. [DOI] [PubMed] [Google Scholar]
- Wickner RB. Double-stranded RNA replication in yeast: the killer system. Annu Rev Biochem. 1986;55:373–395. [DOI] [PubMed] [Google Scholar]
- Woolford JL Jr, Baserga SJ.. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics. 2013;195(3):643–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Tutuncuoglu B, Yan K, Brown H, Zhang Y, Tan D, Gamalinda M, Yuan Y, Li Z, Jakovljevic J, et al. Diverse roles of assembly factors revealed by structures of late nuclear pre-60S ribosomes. Nature. 2016;534(7605):133–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulan WN, Heydet D, Walker EJ, Gahan ME, Ghildyal R.. Nucleocytoplasmic transport of nucleocapsid proteins of enveloped RNA viruses. Front Microbiol. 2015;6:553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao R-W, Xu G, Wang Y, Shan L, Luan P-F, Wang Y, Wu M, Yang L-Z, Xing Y-H, Yang L, et al. Nascent pre-rRNA sorting via phase separation drives the assembly of dense fibrillar components in the human nucleolus. Mol Cell. 2019;76(5):767–783.e11. [DOI] [PubMed] [Google Scholar]
- You K, Huang Q, Yu C, Shen B, Sevilla C, Shi M, Hermjakob H, Chen Y, Li T.. PhaSepDB: a database of liquid-liquid phase separation related proteins. Nucleic Acids Res. 2020;48(D1): D354–D359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemp I, Kutay U.. Nuclear export and cytoplasmic maturation of ribosomal subunits. FEBS Lett. 2007;581(15):2783–2793. [DOI] [PubMed] [Google Scholar]
- Zhang L, Wu C, Cai G, Chen S, Ye K.. Stepwise and dynamic assembly of the earliest precursors of small ribosomal subunits in yeast. Genes Dev. 2016;30(6):718–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables.
Supplemental material is available at GENETICS online.