Summary
Background
Tumor heterogeneity of human colorectal cancer (CRC)-initiating cells (CRCICs) in cancer tissues often represents aggressive features of cancer progression. For high-resolution examination of CRCICs, we performed single-cell whole-exome sequencing (scWES) and bulk cell targeted exome sequencing (TES) of CRCICs to investigate stemness-specific somatic alterations or clonal evolution.
Methods
Single cells of three subpopulations of CRCICs (CD133+CD44+, CD133−CD44+, and CD133+CD44− cells), CRC cells (CRCCs), and control cells from one CRC tissue were sorted for scWES. Then, we set up a mutation panel from scWES data and TES was used to validate mutation distribution and clonal evolution in additional 96 samples (20 patients) those were also sorted into the same three groups of CRCICs and CRCCs. The knock-down experiments were used to analyze stemness-related mutant genes. Neoantigens of these mutant genes and their MHC binding affinity were also analyzed.
Findings
Clonal evolution analysis of scWES and TES showed that the CD133+CD44− CRCICs were the likely origin of CRC before evolving into other groups of CRCICs/CRCCs. We revealed that AHNAK2, PLIN4, HLA-B, ALK, CCDC92 and ALMS1 genes were specifically mutated in CRCICs followed by the validation of their functions. Furthermore, four predicted neoantigens of AHNAK2 were identified and validated, which might have applications in immunotherapy for CRC patients.
Interpretation
All the integrative analyses above revealed clonal evolution of CRC and new markers for CRCICs and demonstrate the important roles of CRCICs in tumorigenesis and progression of CRCs.
Funding
A full list of funding bodies that contributed to this study can be found in the Acknowledgements section.
Keywords: Colorectal cancer, Cancer initiating cells, Single-cell sequencing, Mutation, Clonal evolution
Abbreviations: CRCICs, colorectal cancer initiating cells; CRCCs or E+ cells, common colorectal cancer cells with EpCAM+ but lacking stem-cell markers; E/133+44+ cells, EpCAM+CD133+CD44+ CRCICs; E/133+ cells, EpCAM+CD133+CD44− CRCICs; E/44+ cells, EpCAM+CD133−CD44+ CRCICs; scWES, single-cell whole-exome sequencing; TES, targeted exome sequencing
Research in context.
Evidence before this study
Colorectal cancer (CRC) is one of the most prevalent cancers, which mortality worldwide among cancers is high (9.2%). Although several NGS-based genome studies of colorectal cancer (CRC) were reported, the integrated mechanisms by which cell-to-cell heterogeneity promotes tumorigenesis as well as the originating evolution of CRC has not yet been fully elucidated. In particular, emerging data indicate that both CD133 and CD44 are putative CRCIC markers; cells with these two cell surface markers could induce xenograft tumors in immune-deficient mice. However, the exact mechanism of the tumorigenicity of CRCICs of CD133 and CD44 subpopulations remains unclear.
Added value of this study
Our study performed scWES of CRCCs and three subpopulations of CRCICs, comprising EpCAM+CD133+CD44+ cells, EpCAM+CD133+CD44− cells, and EpCAM+CD133−CD44+ cells. When compared to common CRC cells, the integrated genetic and evolutionary mechanisms of these three types of CRCICs was identified. Our data by clinical samples of test sets and validation sets and cell-line experiments demonstrate that part of EpCAM+CD133+CD44− CRCICs that displayed the least number of mutations were the origin of CRC, and then evolved into other groups of CRCICs and CRCCs. Finally, we found and functional validated 6 genes were stem-specifically mutated and 33 genes were prone to stem-mutated in both scWES data and target sequencing data. Moreover, several neoantigens were identified and validated, especially neoantigen KLDLKVPKA (Chr14: 105410531A>G, S3753P) with high MHC-I binding affinity in AHNAK2 gene that was shared in 6/21 CRC patients.
Implications of all the available evidence
This study reported the clonal evolution of CRCICs, and it also be the report of mutational features, including neoantigens of CRCICs, which could provide a foundation for further precision treatment of CRC.
Alt-text: Unlabelled box
Introduction
Colorectal cancer (CRC) remains one of the leading causes of cancer incidence (9.2%) and mortality (9.8%) worldwide.1 Increasing studies have indicated that CRC tumors are highly hetero-cellular, and that their increased hetero-cellularity-based heterogeneity is associated with poorer survival. However, the integrated mechanisms by which cell-to-cell heterogeneity promotes tumorigenesis in CRC have not yet been fully elucidated.2
Over the past two decades, CRC has been characterized as a disease of significant overgrowth of colorectal epithelial cells, which can be identified by cell surface marker EpCAM.3, 4, 5 As such, most attempts to treat CRC have focused on inhibiting proliferation of colorectal epithelial cells. Advances in methodological approaches for the investigation of cancer genetics and genomics have led to the identification of a variety of molecular targets or signaling pathways that are altered during the development and progression of CRC including KRAS, WNT, TP53, PI3K, and DNA-mismatch-repair pathway genes.6 However, the targeted therapeutics developed for these molecules show only modest efficacy.7 Thus, to treat CRC more effectively, there is a need to understand the genetic features and clonal evolution of diverse tumor cell populations in CRC.
Cancer cells are thought to be stochastic and originate from the accumulation of mutations in normal cells. In CRC, a subset of cells referred to as CRC-initiating cells (CRCICs), also called cancer stem cells, have been characterized as having self-renewal ability and the potential to give rise to other differentiated progenies.3,4 Although recent studies have reported that the frequency of CRCICs is low, such that they represent only a minor subset of CRC cells, they are thought to have significant roles in tumorigenesis, tumor relapse, and treatment resistance.8 More importantly, these CRCICs are more frequently enriched in advanced and aggressive tumors, indicating that CRCICs may represent ideal targets for cancer therapy. Emerging data indicate that CD133 and CD44 are putative CRCIC markers,3,4,8 and cells with these two markers could induce xenograft tumors in immune-deficient mice.9,10 Specifically, proliferation speed and ability to form colorectal spheres/tumors were significantly higher for CD133+CD44+ cells compared with CD133+CD44− and CD133−CD44+ cells.4,10,11 Thus, these markers can help to discern the molecular sub-types and clonal-evolution of three distinct groups of CRCICs: the EpCAM+CD133+CD44+ (E/133+44+) cells, EpCAM+CD133+CD44− (E/133+) cells and EpCAM+CD133−CD44+ (E/44+) cells. These markers could also be used to comparison of the genetic differences between CRCICs and common colorectal cancer cells (CRCCs) (EpCAM+CD133−CD44− cells, or E+ cells). These exact mechanisms of the tumorigenicity of CRCICs remains unclear. Further insight into the genetic characteristics of CRCICs may enable deeper understanding of the pathogenesis of CRC and provide new therapeutic targets. A more complete understanding and an integrated view of the genetic properties and functions of various subpopulations of CRCICs related to CD133 and/or CD44 are urgently required.
In the present study, we performed single-cell whole-exome sequencing (scWES) of CRCCs (EpCAM+ cells) and three above mentioned subpopulations of CRCICs (E/133+44+ cells, E/133+ cells, E/44+ cells), to identify the integrated genetic and evolutionary mechanisms of CRCICs among various subpopulations. The results were then validated by functional analysis and targeted exome sequencing (TES) for an additional 20 CRC patients. Our findings may provide novel potential CRCIC biomarkers and targets in targeted therapies and immunotherapy for CRC patients in the future.
Methods
Patients and samples collection
The patient in scWES study was a typical CRC patient, who was 76-year-old Chinese female with adenocarcinoma of the transverse colorectal that close to the splenic flexure. Another 20 CRC patients were participated in exome sequencing of target genes. By the time we collected the samples of each patient, they didn't receive any treatments. Samples collection was that: after surgery, part of living normal-colon epithelial tissue and CRC tissue were obtained. We performed frozen sections and H/E staining for diagnosis of CRC and normal control. When pathologists confirmed, each left fresh sample was divided into two parts, one was as tissue samples and directly cryopreserved, and one was dissociated by 1mg/ml collagenase IV (C5138, Sigma-Aldrich, Merck KGaA) at 37°C for 75 min. Single-cell suspensions were obtained and then treated with red blood cell lysis buffer (B541001, Sangon), and then were suspended in physiological saline and 3% bovine serum albumin (BSA, E661003, Sangon) for FACS analysis (BD FACSAriaTM III, BD). 1 × 106 cells were incubated with BV421-conjugated CD326 (563180, BD, USA), PE-conjugated CD133 (130-080-801, Miltenyi Biotec, Germany), APC-conjugated CD44 (559942, BD) for 30 min at room temperature in the dark. Isotypic IgG and unstained cells were used as negative controls. After sorting, we obtained EpCAM+CD133+CD44+ (E/133+44+), EpCAM+CD133−CD44+ (E/44+), EpCAM+CD133+CD44−(E/133+), EpCAM+CD133−CD44− (E+) subpopulations. In scWES part, each cell was captured by micromanipulator system (MP‑285; Sutter Instruments) and transferred into a thin-wall PCR tube (N8010180, Applied Biosystems, USA) for next step. In exome sequencing of target genes, each subpopulation was collected and directly cryopreserved.
DNA extraction for tissue sequencing analysis
QIAamp DNA mini kit (51304, Qiagen, Germany) was used for genomic DNA extraction from the fresh frozen tissue sample according to manufacture protocols. The DNA sample quality and integrity were controlled by A260/A280 ratio and agarose gel electrophoresis. The concentration of genomic DNA was measured by Nanodrop 2000 (Thermo, USA) and Qubit 3.0 (Life Technologies, USA).
Whole genome amplification (multiple displacement amplification, MDA) of lysed single cells and lysed subpopulation cells
Lysed single cells: Whole genome amplification was performed on single cells using MDA-2 (REPLI-g Single Cell Kit) (150345, Qiagen).12 A reaction of a total 50 ul volume was incubated at 30°C for 3 h and inactivate the DNA polymerase at 65°C for 3 min.
Lysed subpopulation cells: Whole genome amplification for lysed subpopulation cells was named MDA-1 (REPLI-g UltraFast Mini kit) (150035, Qiagen).12 A reaction of a total 20 ul volume was incubated at 30°C for 1.5h and inactivate the DNA polymerase at 65°C for 3 min.
Amplified DNAs were directly used or stored at -20°C. The Qubit 3.0 (Life Tech.) was used to measure the concentration of MDA-1 and MDA-2 products. The quality and genome-coverage for PCR were checked by five housekeeping genes located on different chromosomes. Only the products successfully amplified by at least four genes were chosen for exome capture. PCR primers used were as follows: 2p (Forward [F] primer-5’-GTCTTTAGCTGCTGAGGAAATG-3’, Reverse [R] primer-5’-AGCAGAATTCTGCACATGACG-3’), 3p (F-5’-ATTTATTGCAAACTCCCTAATATCA-3’, R-5’-CCTCCATTGGCATGAAGTCT-3’), 4p (F-5’-AACTGAATGGCAGTGAAAACA-3’, R-5’-CCCTAGCCTGTCATTGCTG-3’), 5p (F-5’-GGGTAAGATCCAGAGCCACA-3’, R-5’-CCTCATTCCTTCTCGAAGCA-3’), β-actin (F-5’-GCCAACTTGTCCTTACCCAGAG-3’, R-5’-GCCAGGAACTCCCCAATAAGC-3’).
Exome capture, library preparation, sequencing and quality control (QC) of the sequencing raw data
scWES: Exome capture was used 96 rxn xGen® Exome Research Panel v1.0 (IDT, USA). Library was constructed by KAPA Hyper Prep Kits (96 Rxn) (KAPA Biosystem, USA).
Exome sequencing of target genes: To get the target gene regions, we designed probes on the website of Agilent (https://earray.chem.agilent.com/suredesign/index.htm?sessiontimeout=true) about 258 genes of scWES data according the design description. The qualified genomic DNA was fragmented by Covaris technology with resultant library fragments 180-280 bp, and then adapters were ligated to both ends of the fragments. Extracted DNA was then amplified by ligation-mediated PCR (LM-PCR), purified, and hybridized to the probe for enrichment, non-hybridized fragments were then washed out. Both non-captured and captured LM-PCR products were subjected to real-time PCR to estimate the magnitude of enrichment. And then the libraries were sequenced on Illumina Hiseq 4000 with PE150. The sequencing kit was Illumina Hiseq SBS Kit 300 cycles (Illumina, USA). The Illumina library adapters and unreliable low-quality read ends were trimmed or dumped from the raw sequencing data using cutadapt13 and in-house QC program implemented in C. Raw sequencing data of patient for scWES were deposited in the SRA database, with project number SRP098870.
Sequence alignment and processing
Valid sequencing data was mapped to the reference human genome (UCSC hg19) by Burrows-Wheeler Aligner (BWA)14 in order to get the original mapping results stored in BAM format. Then SAMtools, Picard (http://broadinstitute.github.io/picard/) and GATK (Genome Anlysis Toolkit) (http://www.broadinstitute.org/gatk/) were used to sort BAM files and do duplicate marking, local realignment, and base quality recalibration to generate final BAM file for computation of the sequence coverage and depth.
For the tumor samples with the matched normal tissues, the somatic SNVs and InDels were detected by GATK. ANNOVAR15 was performed to do annotation for VCF (Variant Call Format), including the databases such as dbSNP (Sherry et al., 2001), the 1000 Genomes Project,16 Exome Aggregation Consortium (ExAC: http://exac.broadinstitute.org/), SIFT,17 Polyphen,18 and so on.
Somatic point-mutation detection
The GATK was used to detect single nucleotide variants (SNVs). All the samples VCF files were merged, and then variants detected in the tumor single cells were filtered out to eliminate germline mutations using matched normal tissue samples and the normal single cells. A candidate somatic mutation was called if the following criteria were met: Single nucleotide variants and indels were annotated using Annovar 9 (version 2013 Nov20, Aug 23rd) (we downloaded databases dbSNP build 138, polyphen and avsift using the Annovar perl scripts); Filtered low-quality mutations (QUAL<100, ANNOVA; Mutant cell < 2) and also fileted relative to the 1000 genomes (> 0.01) and Repeat (have comments); Retained the CDS region; The minimum distance of SNPs are larger than 5 bp. A non-negative matrix factorization algorithm (https://www.mathworks.com/help/stats/nonnegative-matrix-factorization.html) was used for decomposing somatic SNV spectra.
Single cell classification
Tissue mutation was detected from sequence read density using the Whole Exome Sequencing (WES) variations method: MuTect (http://www.broadinstitute.org/cancer/cga/mutect/). Clustering analysis based on the WES SNVs was applied to classify single cells. We determined that the SNVs are classified under the default parameters. The Euclidean distance between pairs of SNVs of single cells was calculated by:
Where A and B were from different single cells. ‘i’ is the index for the SNV. Based on the above Euclidean distances, the bionjs cluster method implemented in the R package named ape (https://cran.r-project.org/web/packages/ape/index.html) was applied to create clusters of single cells to infer the single cell status.
Phylogenetic tree construction
For the identification of significantly mutated genes (SMG, we also named driver genes) we used the high confidence somatic SNVs and InDels as input for Genome MuSiC19 of SMG test. This test assessed multiple methods of calculating summarized P-values, including a convolution test (CT), a Fisher's combined P-value test (FCPT), and the likelihood ratio test (LRT), using a partially simulated data set. The result of the SMG test is a compilation of the P-values for each test and each gene under the null hypothesis that the number of mutations seen in the gene is in accordance with those seen in the background. False discovery rates are also reported for each test and each gene.
Using OncoNEM (oncogenetic nested effects model),20 we built the phylogenetic tree based on the driver of functional (missense, stopgain, stoploss) mutations (Panel of 20 patients used all the mutations). OncoNEM is an automated method for reconstructing clonal lineage trees from somatic single nucleotide variants (SSNVs) of multiple single tumor cells that exploits the nested structure of mutation patterns of related cells. OncoNEM probabilistically accounts for genotyping errors and tests for unobserved subpopulations, addressing both challenges described above. It simultaneously clusters cells with similar mutation patterns into subpopulations and infers relationships and genotypes of observed and unobserved subpopulations, yielding results that are more accurate than those of previous methods. OncoNEM inference is a three-step process of initial search, testing for unobserved clones and clustering. Step 1. Initial search: building a cell tree. Step 2. Refinement: testing for unobserved clones. Step 3. Refinement: clustering cells into clones.
Sanger sequencing for validation of somatic mutations
A part of somatic mutations was validated by Sanger sequencing. The PCR primers for each mutation were designed by PrimerSelect in DNASTAR Lasergene. v7.1 and initially used to amplify the source MDA-amplified DNA from 170 mutated single cells and 24 no-mutation single cells. 10 ng template DNA from each sample was used for per PCR reaction. Each PCR product was cloned into a pUCm-T Vector (B522211, Sangon) and 10-15 clones were sequenced on ABI PRISM 3730xl Genetic Analyzer. The sequences were assembled and analyzed using DNASTAR Lasergene. v7.1.
Cell culture and FACS for validation of evolution analysis
CRC cell lines SW620 (cat. Number: TCHu 101), HCT116 (cat. Number: TCHu 99) and HT29 (cat. Number: TCHu 103) were chosen for validation of CD133+CD44−-originating evolution. SW620, HCT116 and HT29 cell lines were purchased from the Cell Bank of the Chinese Academy of Sciences, Shanghai, China and passaged for less than 4 months. We authenticated the source of cell lines by short tandem repeat analysis (Genetic Testing Biotechnology Corporation, Suzhou, China) and no cross-contaminated cell lines were found. Besides, regularly mycoplasma testing cell cultures of these three cell lines to ensure absence of mycoplasma by using Mycoplasma PCR Detection Kit (C0301S, Beyotime, Shanghai, China). SW620 cultured in L-15 medium (11415-064, Gibco, Thermo, USA), and HCT116 and HT29 cultured in McCoy's 5A medium (16600-082, Gibco) plus NaHCO3, and all mediums were added 500 ng/mL penicillin-streptomycin (15140-122, Gibco) and 10% fetal bovine serum (10099-141, Gibco), and incubated at 37°C in 5% CO2/95% air. Each cultured-cells were digested by trypsin enzyme (3197, Gibco) and resuspended in phosphate buffered saline (PBS, 14190, Gibco), and every 1 × 106 cells were incubated with BV421-conjugated CD326 (563180, BD, USA), PE-conjugated CD133 (130-080-801, Miltenyi Biotec, Germany), APC-conjugated CD44 (559942, BD) for 30 min at room temperature in the dark. Isotypic IgG and unstained cells were used as negative controls. After sorting, we obtained E/133+44+, E/44+, E/133+, E+ subpopulations of each cell lines. We back tested each subpopulation to ensure purity. Then each subpopulation was cultured and passaged. The subpopulation of E+ cells was for adherent culture, while E/133+44+, E/44+, E/133+ were for suspension culture with sphere-formation medium that included DMEM/F15 (11995, Gibco), B27 (17504-044, Gibco), epidermal growth factor (PHG0311L, Gibco), basic fibroblast growth factor (PHG0264, Gibco), insulin (12585-014, Gibco), transferrin (T8158-100MG, Sigma, USA) and bovine serum albumin (H1130, Solarbio, China). Subculture of each subpopulation was analyzed again by FACS (BD FACSAriaTM III, BD) with the same markers.
Lentivirus preparation and infection
HEK293T cells (632180, Clontech, USA) were co-transfected with target gene constructs and lentivirus-packing plasmids (Prre[#12251], pREV[#12253], and pVSVG[#8454], Addgene, USA) by lipofectamine 2000 (11668-027, Invitrogen, USA). And packing the lentivirus vector (pLVX-shRNA1 [QYV0022, Quallityard Biotech, Beijing, China] or GV493 [Genechem, Shanghai, China]) plasmid as a negative control (shNC). Three shRNAs have tested for one gene. The shRNAs with the best knock-down efficiency were then selected. The medium was changed after 16 h. The lentivirus-containing supernatant was collected 48 h later and used for infection in the presence of 10 ug/ml Polybrene (C0351, Beyotime, Shanghai, China). In the end, we seeded HCT116 or SW620 cells in 6-well (140675, Thermo) at the density of 0.5 × 104 cells, incubated the cells with the virus for 48-72 h, and gently aspirated the media from the cells. When the cells in the un-transduced well (0 µl lentivirus, above) were dying and transgene expression in a polyclonal population might drop, FACS was used to choose target cells. Then continued infection the target until the appropriate of fluorescence intensity.
Sphere formation
Knock-down cells and negative control cells (shNC cells) were suspended on 6-well ultra-low attachment plates (3471, Corning, USA) for about 8 days or 14 days at a density of 500 cells/well in sphere-formation medium described above in a CO2 incubator. Photographs of each well with all sphere cells were obtained by Olympus IX83 microscope (including panoramic scanning system) (Olympus, Japan). Then the number of all sphere cells were counted.
FACS for the cell proportion changes of knock-down cells
1 × 106 knock-down cells were incubated with BV421-conjugated CD326 (563180, BD, USA), PE-conjugated CD133 (130-080-801, Miltenyi Biotec, Germany), APC-conjugated CD44 (559942, BD) for 30 min at room temperature in the dark. Isotypic IgG and unstained cells were used as negative controls. We set gates for EpCAM+CD133+CD44+ (E/133+44+), EpCAM+CD133−CD44+ (E/44+), EpCAM+CD133+CD44−(E/133+) and EpCAM+CD133−CD44− (E+), and then calculated the proportions of each subpopulation.
Neoantigens predict and analysis
HLA typing was performed using Polysolver.21 All samples have developed a specialized genotyping algorithm for the HLA locus that is based on read alignments. Missense mutations were used to generate a list of peptides ranging 9-11 amino acids in length with the mutated residues represented in each position. Prediction for binding affinity of every mutant peptide and its corresponding wild-type peptide to the patient's germline HLA alleles was performed using the NetMHCpan (v3.0).22 Candidate neoantigens were identified as those with a predicted mutant peptide binding affinity of < 500 nM.
AlphaFold23 v2.2 was used to predict the 3D variant structure of antigens, including neoantigens and their wild type. The monomer model was selected for training model, the maximum template release date was 2020/05/14 and full genetic database was chosen for preset MSA database. No JAX model evaluations were applied for reducing inference time. All 5 predictions were generated for each antigen. We kept the model for each antigen with highest predicted local distance difference test (pLDDT), and all chosen models had pLDDT > 70.
MHC binding affinity of predicted neoantigens
Peptides listed in Table 1 were synthesized (HYBIO, Shenzhen, China). Peptides were loaded at 1mM onto the QuickSwitch Quant HLA-A*02:01 Tetramers-PE (TB-7300-K1; MBL International; Japan). Generation of new specificity tetramer using peptide exchange and quantification of peptide exchange according to manufacturer's instructions.
Table 1.
Neoantigens of the stem-related mutant genes found in both scWES data and targeted sequencing data.
 |
# Psc: Patient with scWES; Px: x mean the patient number of the 20 patients with targeted sequencing data.
&% Peptide exchange: The results from the QuickSwitchTM Quant Tetramer Kit (MBL, Japan), which is based on the capacity of MHC class I molecules to exchange peptides. Higher exchange mean the peptide had higher binding with MHC class I.
⁎ means the changed amino acid.
Statistics
All scWES and TES data were statitics by R. All function experiments were repeated at least three times, and the data were obtained at least in triplicate. Student t-test was used. Data are presented as mean ± SD. * P < 0.05; ** P < 0.01.
Ethics approval and consent to participate
Patients’ consents, all sample collections and patient recruitments followed institutional review board protocols from Hangzhou Cancer Hospital. The approval number is HZCH-2016-03.
Consent for publication
Written informed consent for publication was obtained from all participants.
Role of the funding source
We thank grants for study design, data collection, all materials (including cell lines), sequencing, data analyses, interpretation and writing of report.
Results
Heterogeneous CRCICs in CRC patients
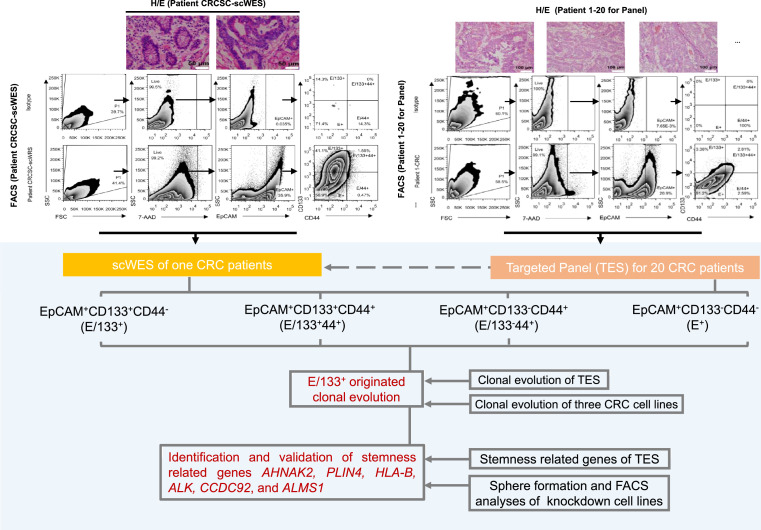
A colorectal cancer tissue from one CRC patient (patient CRCIC-scWES, Psc) (Figure 1) was collected followed by the sorting of CRCCs (EpCAM+ cells) and three subpopulations of CRCICs including EpCAM+CD133+CD44+ (E/133+44+) cells, EpCAM+CD133+CD44− (E/133+) cells, and EpCAM+CD133−CD44+ (E/44+) cells by fluorescence-activated cell sorting (FACS). All individual cells were used for single-cell whole-exome sequencing (scWES) to characterize CRCICs.
Figure 1.
Overview of the study. Fresh colorectal tissues were obtained after surgery, diagnosed with hematoxylin and eosin (H/E) staining, and digested with collagenase IV. Single-cell suspensions were used for FACS analysis. Three groups of CRC stem cells (CRCICs; E/133+44+, E/44+, E/133+) and one group of CRC cells (CRCCs, E+) were sorted. All four groups of single cells from one CRC patient were isolated manually with a micromanipulator for scWES. After bioinformatics analysis, stem-cell-specific clonal evolution and somatic mutations, including stem-cell-specific mutations, were found. The Samples from 20 CRC patients were sorted into the same three groups of CRCICs and one group of CRCCs. All mutations obtained by scWES were made into targeted panel and targeted sequencing was performed in samples from 20 patients to validate the results of scWES. E/133+ originated clonal evolution obtained by scWES data was validated by targeted sequencing data and CRC cell lines. Stemness related genes, AHNAK2, PLIN4, HLA-B, ALK, CCDC92, and ALMS1, obtained by scWES data were validated by targeted sequencing data and functional analyses (sphere formation and FASC analyses) of knockdown cell lines.
By isolating live, single cells representing CRCICs and CRCCs, we discovered several features of the tumor cell subpopulations. Of the isolated 208 single cells (48 E/133+44+ cells, 64 E/133+ cells, 33 E/44+ cells, and 63 E+ cells), the genomes of 146 single cells (45 E/133+44+ cells, 50 E/133+ cells, 23 E/44+ cells, and 28 E+ cells) were used successfully for multiple displacement amplification (MDA). Notably, less than half of the CRCCs achieved MDA (28/63, 45%) compared to the other three subpopulations of CRCICs (94%, 78%, and 70%, respectively). Moreover, the genomes of single white blood cells were also amplified by the MDA method as a control, and all of them (37/37, 100%) were successfully amplified and sequenced by scWES. After filtering, single cells with poor data quality (see Methods) were eliminated, and scWES data for 101 CRC single cells (36 E/133+44+ cells, 41 E/133+ cells, 12 E/44+ cells, and 12 E+ cells) and 36 white blood cells were analyzed and compared (Supplementary Table 1). WES data for normal colorectal epithelial tissue and CRC tissue samples were also examined (Supplementary Table 1).
Mutation signatures revealed in CRC single cells
Based on these data, we observed a wide spectrum of mutations across individual CRC cells, leveraging a depth range of 136-449X and 20X mean coverage of 88.3% (Supplementary Table 1). To discern high-risk mutations, we first removed germline single-nucleotide polymorphisms (SNPs) by comparison with WES data for normal colorectal epithelial tissue, and then applied ANNOVAR for variant annotations. The scWES data were filtered for low-quality mutations (QUAL<100, ANNOVAR; mutant cell < 2) by using the 1000 genomes and ExAC database. Then the somatic mutations of scWES data were obtained (Supplementary Table 2).
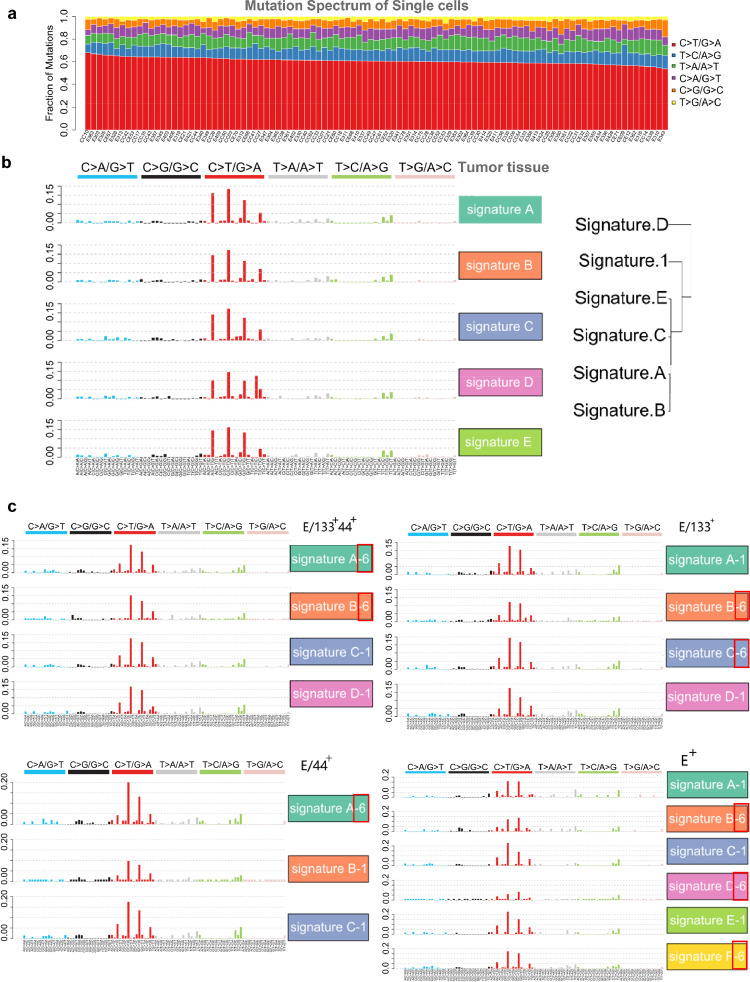
We found somatic mutations of scWES data showed a preference mutation spectrum for C/G to T/A, which was similar to the previous reports of the mutation pattern seen in CRC and other cancers (Figure 2a).2,24 Decomposing somatic SNV spectra with a non-negative matrix factorization algorithm revealed five signatures of the tumor tissue (signature A, B, C, D and E), which were all similar to signature 1 (age signature) of the 30 most common cancer signatures, as seen in the pan-cancer analysis (http://cancer.sanger.ac.uk/cosmic/signatures) (Figure 2b).2 This also replicates a similar result seen in previous reports on other two colorectal cancers.25 Interestingly, a decomposition of mutation spectra of each group cells (three CRCIC groups and CRCCs) with the same algorithm showed similarity to cancer signature 1 and 6, which is associated with aging and defective DNA mismatch repair, respectively (Figure 2c). As such, these data demonstrate CRC similarity to signature 6, indicating that novel mutation signatures of colorectal cancer can be uncovered at the single-cell level.
Figure 2.
Mutation Spectra and Mutation Signatures of Tumor, CRCICs and CRCCs from Patient for scWES, which are Based on the Somatic SNV of Tissue WES Data and scWES Data. a. Mutation spectra of all single cells. b. Signature A. B. C. D and E of the tumor tissue are similar to signature 1 of most cancer samples from pan-cancer analysis. c. Mutation signatures of CRCICs and CRCCs from patient for scWES, which are based on the somatic SNV of scWES data. Signature A. B. C. D and E are similar to signature 1 (named ‘-1’) or 6 (named ‘-6’ and red frame, which is not identified in Tissue WES data) of most cancer samples from pan-cancer analysis.
Mutation characteristics of CRCIC single cells
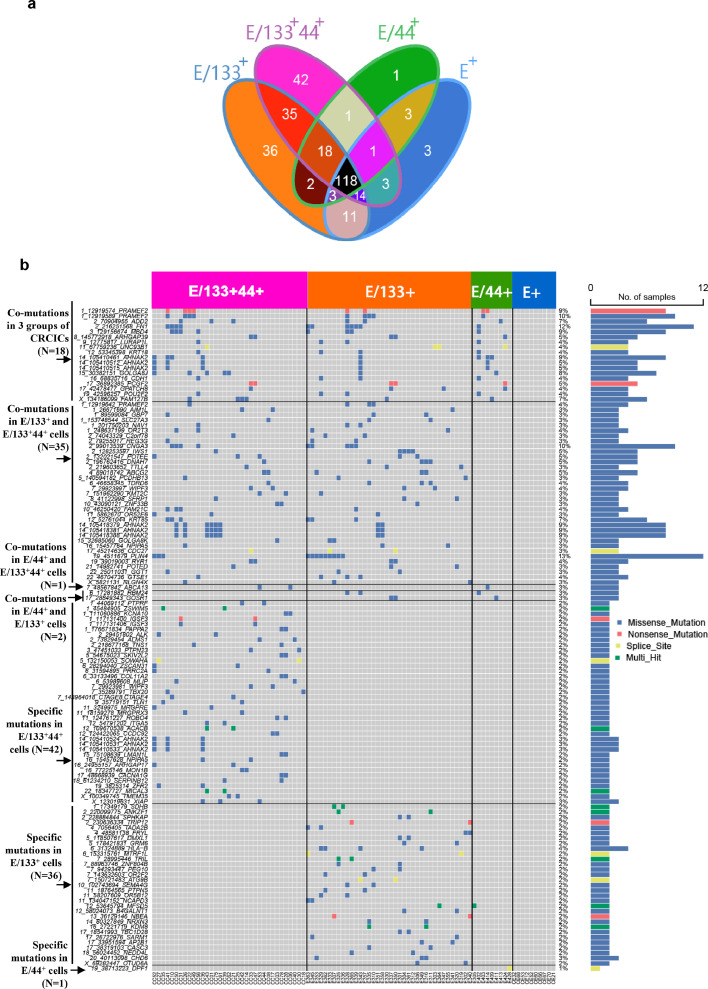
In total, we identified 291 somatic mutational sites (Supplementary Table 2a) in individual CRCICs and CRCCs by GATK (Genome Anlysis Toolkit). Some of these mutations was validated by Sanger sequencing (Supplementary Table 3). The distribution of somatic mutations of each groups including three CRCICs groups and CRCCs, were shown in Figure 3a. Interestingly, CRCICs and CRCCs displayed different mutation characteristics, and a total of 118 somatic mutational sites existed in both CRCICs and CRCCs. Another 135 mutations had low mutation frequency (4.87% [2/41] to 16.7% [2/12]) and were specifically mutated in CRCICs. Mutations specifically mutated in each type of CRCICs, or mutated in both or three types of CRCICs were also clustered in Figure 3b. Among the 135 mutations, 18 mutations including CDH1, ARHGAP39, LURAP1L, PCGF2 and MBD4, were shared by all three subpopulations of CRCICs, and 113 mutations were specifically mutated in CD133-positive CRCICs. We then analyzed the related pathways of these specific mutant genes in CRCICs and determined whether they were related to stemness. The results showed that many of them had been previously reported to be correlated with stemness or cancer stem cells (Supplementary Table 4). We successfully constructed 11 knock-down cell lines with genes co-mutated in three groups of CRCICs on HCT116 and SW620 cell lines to verify their functions, the information of their shRNA and knock down efficiency shown in Supplementary Table 5, and found that most of these genes are indeed related to stemness (Supplementary Figure 1). Sphere formation and FACS analysis showed that CDH1, ARHGAP39, LURAP1L, PCGF2 and MBD4 were positively associated with stemness (Supplementary Figure 1a, b) and knocking down each of them could increase the proportion of CD133+ and/or CD44+ cells (Supplementary Figure 1c, d).
Figure 3.
CRCICs-specific SNVs of scWES data. a. Somatic mutations of all four groups (three groups of CRCICs and CRCCs) were showed clearly in Venn diagram. Each combination in Venn diagram was marked with one color background. b. The mutational landscape of corresponding mutations that specific for CRCICs.
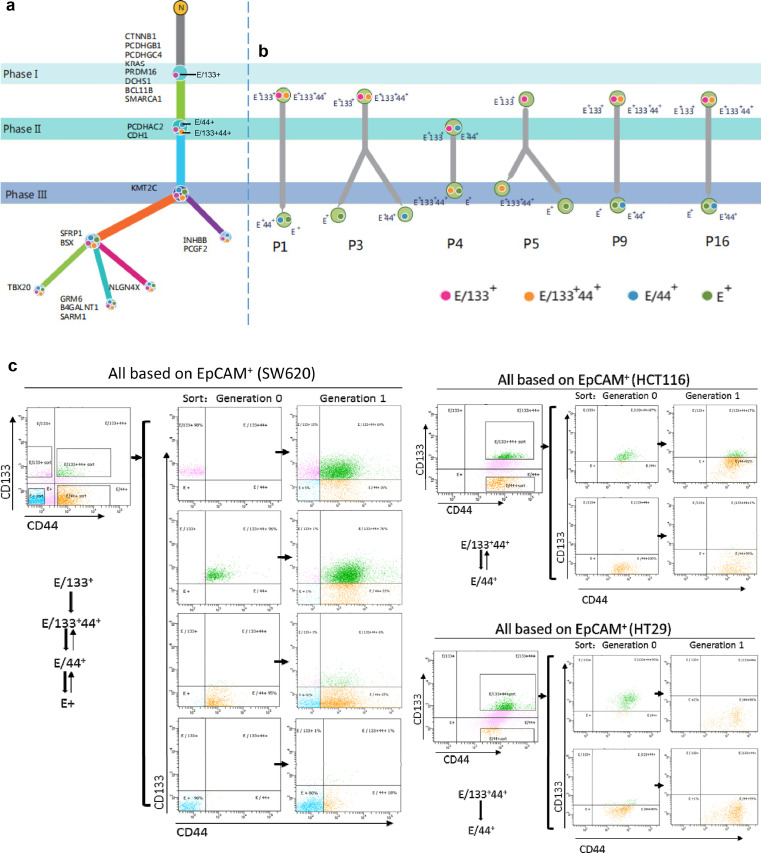
CRCICs, especially E/133+cells, were origins of clonal evolution of CRC
For scWES data, we then used the OncoNEM algorithm to obtain two clonal evolution branches (Figure 4a). All clonal evolution branches predicted that CRCs originated from E/133+ CRCICs, which were mutated at the first phase and had the fewest mutations. E/133+44+ and E/44+ cells were mutated at the second phase, and E+ cells branched last and were mutated at the third phase.
Figure 4.
Clonal evolution of CRCICs and CRCCs of scWES and TES data, and its validation of CRC cell lines. a. Clonal evolution of the scWES data. We used the OncoNEM method to analyze the clonal evolution of all single cells. The three phases are marked as I, II, III, showing the evolution of each step with specific gene mutations. E/133+ cells were marked as solid pink circles, while E/133+44+ cells were solid yellow circles, E/44+ cells were solid blue circles, and E+ cells were solid green circles. E/133+ cells are the original CRC cells. b. The targeted sequencing panel that included all mutant genes of scWES data were used to analyze the clonal evolution of other six patients. The cancer cells of these six patients were origin from cancer initiating cells (E/133+44+ or E/133+). The E/133+44+, E/44+, E/133+ and E+ cells of each patient were sorted by FACS. The three phases are marked as I, II, III, showing the evolution of each step with specific gene mutations. E/133+ cells are the original CRC cells. c. E/133+44+, E/44+, E/133+, and E+ cells from cell lines SW620, HCT116, and HT29 were sorted by FACS and then cultured. After one generation, E/133+ cells could grow into E/133+44+, E/44+, and E+ cells.
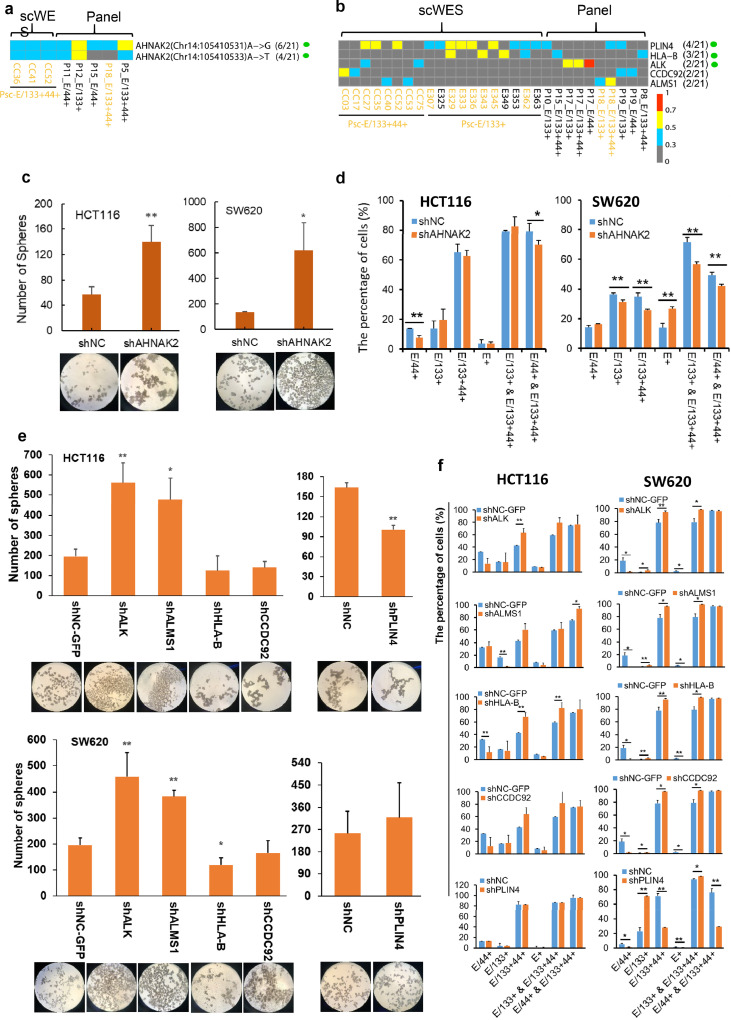
To validate the E/133+ origin of CRC evolution, the same three subpopulations of CRCICs, CRCCs and control cells in another 20 patients (96 samples in total) were sorted by FACS (Figure 1, the upper right panel) and subjected to targeted exome-seq (TES) using the panel of all-mutated genes identified in scWES data (Supplementary Table 6a, b, c). We also observed that CD133 markers were highly frequent in cancer originating cells (Figure 4b and Supplementary Figure 2). Cancer cells derived from 6 of 20 CRC patients were origin from E/133+ (Figure 4b, P1, P3, P4, P5, P9, P16).
Moreover, we also sorted the same groups of CRCICs and CRCCs from three CRC cell lines SW620, HCT116, and HT29, and then cultured them to observe their evolutions. After one generation, the subculture in each cell line showed different cellular components (Figure 4c). In SW620 cell lines, from generation 0 to 1, the isolated E133+ cells (approximately 98% of total cells) developed to 15% of E/133+ cells, 64% of E/133+44+ cells, 16% of E/44+ cells and 5% of E+ cells. Thus, E/133+ cells could develop into E/133+44+, E/44+, and E+ cells. Nearly all of the isolated E/133+44+ cells (96%) of SW620 cell lines developed to E/133+44+ cells (76%), E/44+ cells (22%). Nearly all of the isolated E/44+ cells (95%) developed to E/133+44+ cells (6%), E/44+ cells (67%) and E+ cells (26%). Nearly all of the isolated E+ cells developed to E+ cells (80%) and E/44+ cells (18%). Similar phenomenon was also found in HCT116 and HT29 cell lines. Thus, the other types of cells were very difficult to develop into E/133+ cells. Mutual conversions between E/133+44+ and E/44+ cells or between E/44+ and E+ cells were also observed (Figure 4c). These results supported E/133+ as the origin of CRCICs evolution.
Functional analysis of stemness-related genes identified by scWES data and TES data
In the 20 patients with TES sequencing (Supplementary Table 6, 7), twenty-five percent (5/20) of them harbored the same stem-cell-specific mutations of AHNAK2 as those found in the scWES data (Chr14_105410531 and Chr14_105410533) (Figure 5a). Furthermore, 45% (9/20) of the patients harbored at least one mutation of stem-cell-specific mutated genes AHNAK2, PLIN4, HLA-B, ALK, CCDC92, and ALMS1 (Figure 5b). In previous researches (Supplementary Table 4), the AHNAK2, HLA-B and ALMS1 were related to normal stem cells (embryonic stem cells,26 mesenchymal stem cells27 and neural stem cells,28 respectively). The mRNA transcripts of ALK not only expressed in neural stem cells,29 but also expressed in non-small cell lung cancer and could promote cancer stem cells-like properties.30 While no report of the relationships between stemness and CCDC92 or PLIN4, here we using knock-down experiments to assess the stemness characteristics of co-mutant genes AHNAK2, ALK, ALMS1, HLA-B, CCDC92 and PLIN4. Down-regulated expressions of AHNAK2, ALK and ALMS1 increased the ability of sphere formation in both cell lines HCT116 and SW620 (Figure 5c-f), showing their cancer stem properties. Moreover, cell lines of shALK and shALMS1 with increased sphere numbers also showed significantly increased the proportion of CD133+ cells (Figure 5e, f; E/133+& E/133+44+ set of columns). Significantly increased sphere numbers of shAHNAK2 cells caused increased numbers of E/133+ cells in HCT116 cell lines, while it is puzzling why it caused obvious decreased numbers of CD44+ CRCIC cells (Figure 5c, d). An increased percentage of CD133+ cells were observed in cells of shHLA-B, shCCDC92 and shPLIN4, all of which without obvious changes in sphere formation (Figure 5e, f). Exception of the above-mentioned six genes, in all 20 CRC patients, we found another 33 genes, including FN1, that tended to mutate in cancer initiating cells (Supplementary Figure 3).
Figure 5.
The same CRCICs mutational sites or mutated genes found in scWES data and targeted sequencing data, and the knock down expression analysis of these mutated genes. a. The same CRCICs mutational sites found in both data. 6/21 and 4/21 means the corresponding patients with the mutation vs. total 21 patients. b. The same CRCICs mutant genes found in both data. Px means the serial number of each CRC patient for targeted sequencing data. Psc means the patient for scWES data. Green solid circle means that the subcellular location of the gene is at the cell surface. ‘x’ in (x/21) means how many patients has this mutation, while “21” in (x/21) means there are totally 21 patients including the patient for scWES and the 20 patients for validation. KRAS-mutant cells are marked with yellow fonts. a, b. Genes with variant allele frequency (VAF) of ‘≥ 0.7 to 1’, ‘≥ 0.5 to < 0.7’, ‘≥ 0.3 to < 0.5’, ‘0 to <0.3’ were labeled with squares of red, yellow, blue, and grew, respectively. c. Sphere formation of AHNAK2 knock-down HCT116 and SW620 cells. Magnification: 40 X. d. Compared with shNC cells, the percentage changes of each CRCIC or CRCC group in AHNAK2 knock-down HCT116 and SW620 cells by FACS assays. e. Sphere formation of other five genes knock-down HCT116 and SW620 cells. Magnification: 40 X. f. Compared with shNC cells, the percentage changes of each CRCIC or CRCC group in other five genes knock-down HCT116 and SW620 cells by FACS assays, n=3. Student t-test was used. Data are presented as mean ± SD. * P < 0.05; ** P < 0.01. shNC and shNC-GFP cells are negative control cells transfected with lentivirus-packing plasmids.
Potential therapeutic targets
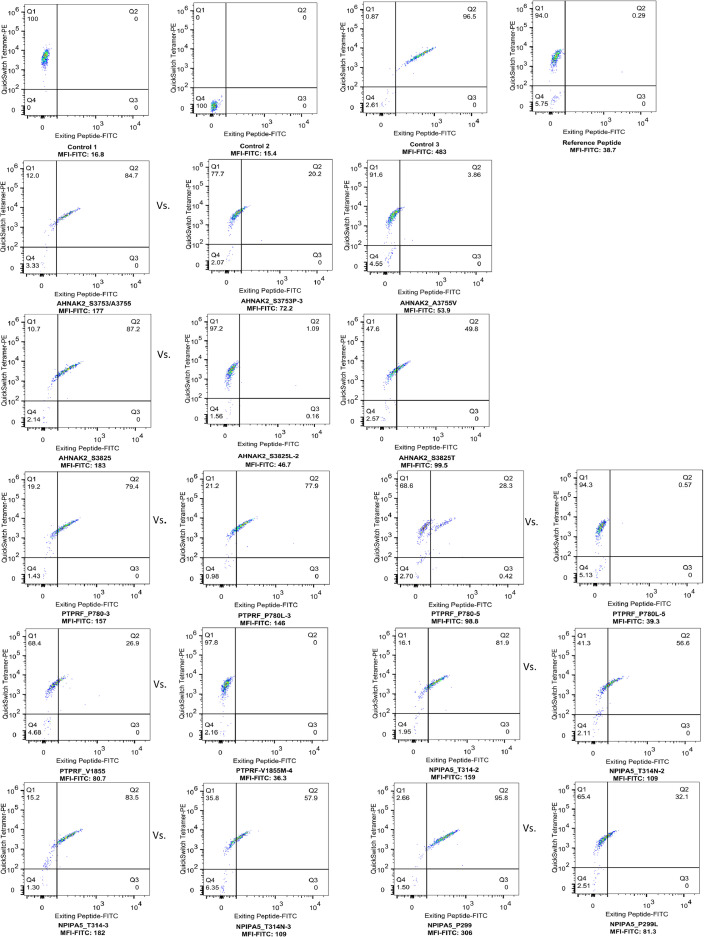
We analyzed neoantigens of all mutations of scWES data and TES data. All neoantigens had predicted mutant peptide binding affinity of < 500 nM, and were lower than that of their corresponding wild peptides (Supplementary Table 8a, b). Interestingly, in 6 of stem-cell-specific genes mentioned above, or 33 of likely stem-cell-specific genes, 30 genes were located at the plasma membrane or extracellularly (green solid circles in Figure 5a, b and Supplementary Figure 3), including AHNAK2, PLIN4, HLA-B, and ALK, suggesting that these genes may have potential as prognostic markers for CRCICs and also as targets for antibody-mediated immunotherapies. Of them, 10 neoantigens were predicted from the cell-surface-located AHNAK2, PTPRF, and NPIPA5 genes and these neoantigens had obvious changes of 3D structure (Table 1). We synthesized these neoantigen peptides and the corresponding wild peptides and assessed their MHC binding affinity using a QuickSwitchTM Quant Tetramer Kit (MBL, Japan). The results showed that neoantigen peptide KLDLKVPKA (Chr14: 105410531A>G, S3753P) of the AHNAK2 gene, which was found in 6 patients in both the scWES and targeted exome-seq data, had high peptide exchange (88.16% vs. 66.01% for the wild peptide), indicating higher binding affinity with MHC class I. Similarly, other initiating cell-specific neoantigen peptides in the AHNAK2, PTPRF, and NPIPA5 genes, which were present in the scWES or TES data, also showed high peptide exchange (Table 1 and Figure 6). These neoantigens might be useful for CD8-mediated immunotherapy in CRC, which was worthy to be investigated in the future.
Figure 6.
MHC binding affinity of initiating-cell specific neoantigen peptides of AHNAK2, PTPRF and NPIPA5 gene and their corresponding wild peptides. Control 1 is the beads captured QuickSwitchTM Tetramer-PE for adjusting compensation. Control 2 is the beads that have not captured any tetramer-PE and no Exiting Peptide-FITC. The low MFI (mean fluorescence intensity) FITC15.4 corresponds to 100% peptide exchange. Control 3 is the beads that have captured Tetramer-PE and Exiting Peptide-FITC. MFI FITC483 corresponds to 0% peptide exchange. The reference peptide with MFI FITC 38.7 serves as a positive control for peptide exchange of the tetramer. The higher MFI FITC in neoantigens vs. that in corresponding wild type peptides meaning the higher frequency of peptide exchange and higher MHC binding affinity of neoantigens. The sequence of each peptide and the percentage of peptide exchange was shown in Table 1.
Discussion
Colorectal cancer is thought to occur when the mutation burden accumulated in the colon crypt exceeds a certain threshold, leading to clonal expansion and eventual tumor transformation. Olpe et al.31 found KDM6A and KRAS mutations were associated with crypt fission and the crypt diffusion process may cause accommodation of the additional crypts to a threshold beyond which polyp growth may occur. With the development of gene detection technology, more and more studies are exploring the clonal expansion of colorectal cancer. By using tissue WES of primary CRC tumors and matched lung metastasis (LM), Zhang et al.32 found primary and LM were founded by the same clone. However, Dang et al.33 found metastasis-seeding clones of four CRC patients were not identified in any primary region. Besides, with the development of single-cell sequencing technology, the nature and extent of intratumor diversification were investigated,34 and CRC cells exhibited a wide variety of mutations and carried several times more somatic mutations than normal colorectal cells. In general, the mechanism of clonal diffusion of colorectal cancer is being explored, and it has also been found that the occurrence of colorectal cancers may derive from the intestinal cells that acquire stem cell properties following malignant transformation (called colorectal cancer stem cells, or initiating cells, CRCICs).35 CRCs is prone to recurrence and metastasis, in part because of the persistence of CRCICs. CRCICs are known to undergo asymmetric division and to have chemo-resistant characteristics. Identification of CICs in cancer growth is gradually attracting research attention.36,37 In this study, using scWES and TES data, we revealed the genetic basis of CRCICs. The observed origination of CRCCs from CRCICs, or even from CD133+ CRCICs, provides genetic evidence to support the hypothesis8,38 of CRCICs’ or even CD133+ CRCICs’ origin. More attention to E/133+ cells and better control of this cluster may be beneficial with respect to prognosis in CRC.
In addition, we used targeted sequencing and functional analyses to validate stemness related mutant genes. AHNAK2, PLIN4, HLA-B, ALK, CCDC92, and ALMS1 all showed stemness characteristics. Previously, a few reports showed part of these genes were associated with cancer: AHNAK2 (AHNAK nucleoprotein 2) was found as a novel prognostic marker and oncogenic protein for clear cell renal cell carcinoma.39 PLIN4 (perilipin 4), HLA-B (major histocompatibility complex, class I, B) and ALMS1 (Alstom syndrome protein 1) was mutated in gastric carcinoma,40 acute myelogenous leukemia41 and chronic lymphocytic leukemia,42 respectively. ALK (anaplastic lymphoma receptor tyrosine kinase) is a biomarker in lung cancer and ALK variation was an important factor in acquired resistance.43,44 No reports were about CCDC92 (coiled-coil domain containing 92) and cancer. Here, these genes provided some evidence that they are correlated with CRC and CRCICs, especially CD133+ CRCICs. They may represent possible prognostic markers for CRCICs.
Currently, identification of CICs in cancer growth has inspired the design of novel treatment strategies to overcome treatment resistance by targeting both CICs and non-CIC tumor cells.36,37 Several reagent targeting cancer stem cells were developed, such as cancer stemness kinase inhibitor amcasertib (BBI-503), cancer stemness inhibitor napabucasin (BBI-608).45 Adoptive cell therapy with tumor infiltrating lymphocytes is a new weapon for cancer precision immunotherapy.46,47 It is all known that obtaining unique neoantigen repertoire of each patient is one of the challenges in translating neoantigen-targeted therapies.46 Identification of shared mutated targets and obtaining shared immunogenetic neoantigens among patients would facilitate the therapies that could be more broadly applied to CRC patients. Here we identified one shared neoantigen KLDLKVPKA (Chr14: 105410531A>G, S3753P) with high MHC-I binding affinity in AHNAK2 gene, which shared in the stem cells of 6/21 (28.57%) CRC patients in our data. We believe this epitope will be useful for immunotherapy of CRC. Similarly, we found several neoantigens of other genes that specific in scWES data or bulk cell TES data, meaning we can also pay attention to discover specific neoantigens for the precise treatment of each patient.
Under the technical conditions, we will try to further develop some explorations in the future. Firstly, this study used knock-down experiments which only preliminarily find the relationship between these genes and CRCICs. The relationships and mechanisms between some mutated genes or mutations and CRCICs are still unclear. For example, we don't know why knocking down the expression of AHNAK2 genes significantly increased the number of spheroids, but not the percentage of CD133+ cells. Perhaps the future use of point mutation-related functional validation can better uncover their relationships. In addition, in the new antigen part, only the affinity of MHC-I has been verified, and there is still a lack of subsequent direct correlation experiments of T cells, which cannot provide more evidence for cell therapy. This is also where we need to improve in the future.
Totally, this is a report of CRCICs from scWES aspect, which revealed more information about CRCICs, and this is a study revealed clonal evolution and mutational features, including neoantigens of CRCICs combing the data of scWES and bulk cell targeted sequencing. These results may provide a foundation for further research in CRC.
Contributors
X.Zhang, L.Yang, S.Wu, and T. Enver designed the study. X.Zhang, W.Lei, and L.Yang analyzed the data and performed statistical analyses. X.Zhang, Q.Hou and M.Huang performed experiments. R.Zhou confirmed the status of samples. X.Zhang, L.Yang, W.Lei, S.Wu and T. Enver interpreted and discussed the data with all authors. X. Zhang, W. Lei and L. Yang and R. Zhou verified the underlying data. X.Zhang, L.Yang, W.Lei and S.Wu wrote the manuscript. All authors read and approved this manuscript.
Data sharing statement
Raw sequencing data of patient for scWES were deposited in the SRA database, with project number SRP098870.
Declaration of interests
There are none to declare.
Acknowledgements
This work was supported by the grants from Hangzhou agricultural and social development research initiative design project (No. 20172016A04), Sanming Project of Medicine in Shenzhen (No. SZSM201612063), Zhejiang Health Science and Technology Project (No.2018KY195), Hangzhou Science and Technology Development Program (No. 20150733Q63). The work of Wanjun Lei was supported by the grant No. 20172016A04.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2022.104125.
Contributor Information
Tariq Enver, Email: t.enver@ucl.ac.uk.
Shixiu Wu, Email: wushixiu@medmail.com.cn.
Appendix. Supplementary materials
References
- 1.Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:41. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dalerba P, Dylla SJ, Park I-K, et al. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci USA. 2007;104:10158–10163. doi: 10.1073/pnas.0703478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haraguchi N, Ohkuma M, Sakashita H, et al. CD133+CD44+ population efficiently enriches colon cancer initiating cells. Ann Surg Oncol. 2008;15:2927–2933. doi: 10.1245/s10434-008-0074-0. [DOI] [PubMed] [Google Scholar]
- 5.Munz M, Baeuerle P, Gires O. The emerging role of EpCAM in cancer and stem cell signaling. Cancer Res. 2009;69:5627–5629. doi: 10.1158/0008-5472.CAN-09-0654. [DOI] [PubMed] [Google Scholar]
- 6.Muzny DM, Bainbridge MN, Chang K, et al. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murciano-Goroff Y, Betof Warner A, Wolchok JD. The future of cancer immunotherapy: microenvironment-targeting combinations. Cell Res. 2020;30:507–519. doi: 10.1038/s41422-020-0337-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ricci-Vitiani L, Lombardi DG, Pilozzi E, et al. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 9.Du L, Wang H, He L, et al. CD44 is of functional importance for colorectal cancer stem cells. Clin Cancer Res Off J Am Assoc Cancer Res. 2008;14:6751–6760. doi: 10.1158/1078-0432.CCR-08-1034. [DOI] [PubMed] [Google Scholar]
- 10.Wang C, Xie J, Guo J, Manning HC, Gore JC, Guo N. Evaluation of CD44 and CD133 as cancer stem cell markers for colorectal cancer. Oncol Rep. 2012;28:1301–1308. doi: 10.3892/or.2012.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen K, Pan F, Jiang H, et al. Highly enriched CD133+CD44+ stem-like cells with CD133+CD44high metastatic subset in HCT116 colon cancer cells. Clin Exp Metastasis. 2011;28:751–763. doi: 10.1007/s10585-011-9407-7. [DOI] [PubMed] [Google Scholar]
- 12.Hou Y, Wu K, Shi X, et al. Comparison of variations detection between whole-genome amplification methods used in single-cell resequencing. GigaScience. 2015;4:37. doi: 10.1186/s13742-015-0068-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetJournal. 2011;17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 14.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinforma Oxf Engl. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.1000 Genomes Project Consortium. Abecasis GR, Auton A, Brooks LD, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet. 2013;Chapter 7:Unit 7:20. doi: 10.1002/0471142905.hg0720s76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dees ND, Zhang Q, Kandoth C, et al. MuSiC: identifying mutational significance in cancer genomes. Genome Res. 2012;22:1589–1598. doi: 10.1101/gr.134635.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ross EM, Markowetz F. OncoNEM: inferring tumor evolution from single-cell sequencing data. Genome Biol. 2016;17:69. doi: 10.1186/s13059-016-0929-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shukla SA, Rooney MS, Rajasagi M, et al. Comprehensive analysis of cancer-associated somatic mutations in class I HLA genes. Nat Biotechnol. 2015;33:1152–1158. doi: 10.1038/nbt.3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nielsen M, Andreatta M. NetMHCpan-3.0; improved prediction of binding to MHC class I molecules integrating information from multiple receptor and peptide length datasets. Genome Med. 2016;8:33. doi: 10.1186/s13073-016-0288-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tunyasuvunakool K, Adler J, Wu Z, et al. Highly accurate protein structure prediction for the human proteome. Nature. 2021;596:590–596. doi: 10.1038/s41586-021-03828-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Helleday T, Eshtad S, Nik-Zainal S. Mechanisms underlying mutational signatures in human cancers. Nat Rev Genet. 2014;15:585–598. doi: 10.1038/nrg3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu H, Zhang X-Y, Hu Z, et al. Evolution and heterogeneity of non-hereditary colorectal cancer revealed by single-cell exome sequencing. Oncogene. 2017;36:2857–2867. doi: 10.1038/onc.2016.438. [DOI] [PubMed] [Google Scholar]
- 26.Rigbolt KTG, Prokhorova TA, Akimov V, et al. System-wide temporal characterization of the proteome and phosphoproteome of human embryonic stem cell differentiation. Sci Signal. 2011;4:rs3. doi: 10.1126/scisignal.2001570. [DOI] [PubMed] [Google Scholar]
- 27.Isa A, Nehlin JO, Sabir HJ, et al. Impaired cell surface expression of HLA-B antigens on mesenchymal stem cells and muscle cell progenitors. PloS One. 2010;5:e10900. doi: 10.1371/journal.pone.0010900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang C-P, Li X, Wu Y, et al. Comprehensive integrative analyses identify GLT8D1 and CSNK2B as schizophrenia risk genes. Nat Commun. 2018;9:838. doi: 10.1038/s41467-018-03247-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Furuta M, Shiraishi T, Okamoto H, Mineta T, Tabuchi K, Shiwa M. Identification of pleiotrophin in conditioned medium secreted from neural stem cells by SELDI-TOF and SELDI-tandem mass spectrometry. Brain Res Dev Brain Res. 2004;152:189–197. doi: 10.1016/j.devbrainres.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 30.Guo F, Liu X, Qing Q, et al. EML4-ALK induces epithelial-mesenchymal transition consistent with cancer stem cell properties in H1299 non-small cell lung cancer cells. Biochem Biophys Res Commun. 2015;459:398–404. doi: 10.1016/j.bbrc.2015.02.114. [DOI] [PubMed] [Google Scholar]
- 31.Olpe C, Khamis D, Chukanova M, et al. A diffusion-like process accommodates new crypts during clonal expansion in human colonic epithelium. Gastroenterology. 2021;161:548–559.e23. doi: 10.1053/j.gastro.2021.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang N, Di J, Wang Z, Gao P, Jiang B, Su X. Genomic profiling of colorectal cancer with isolated lung metastasis. Cancer Cell Int. 2020;20:281. doi: 10.1186/s12935-020-01373-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dang HX, Krasnick BA, White BS, et al. The clonal evolution of metastatic colorectal cancer. Sci Adv. 2020;6:eaay9691. doi: 10.1126/sciadv.aay9691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roerink SF, Sasaki N, Lee-Six H, et al. Intra-tumour diversification in colorectal cancer at the single-cell level. Nature. 2018;556:457–462. doi: 10.1038/s41586-018-0024-3. [DOI] [PubMed] [Google Scholar]
- 35.Testa U, Pelosi E, Castelli G. Colorectal cancer: genetic abnormalities, tumor progression, tumor heterogeneity, clonal evolution and tumor-initiating cells. Med Sci Basel Switz. 2018;6:E31. doi: 10.3390/medsci6020031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pützer BM, Solanki M, Herchenröder O. Advances in cancer stem cell targeting: how to strike the evil at its root. Adv Drug Deliv Rev. 2017;120:89–107. doi: 10.1016/j.addr.2017.07.013. [DOI] [PubMed] [Google Scholar]
- 37.Marquardt S, Solanki M, Spitschak A, Vera J, Pützer BM. Emerging functional markers for cancer stem cell-based therapies: understanding signaling networks for targeting metastasis. Semin Cancer Biol. 2018;53:90–109. doi: 10.1016/j.semcancer.2018.06.006. [DOI] [PubMed] [Google Scholar]
- 38.O'Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 39.Wang M, Li X, Zhang J, et al. AHNAK2 is a novel prognostic marker and oncogenic protein for clear cell renal cell carcinoma. Theranostics. 2017;7:1100–1113. doi: 10.7150/thno.18198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang J, Huang JY, Chen YN, et al. Whole genome and transcriptome sequencing of matched primary and peritoneal metastatic gastric carcinoma. Sci Rep. 2015;5:13750. doi: 10.1038/srep13750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Planelles D, Balas A, Gil C, Muñoz C, Rodríguez-Cebriá M, Vicario JL. Somatic mutation in the HLA-B gene of a patient with acute myelogenous leukaemia. HLA. 2016;88:35–37. doi: 10.1111/tan.12830. [DOI] [PubMed] [Google Scholar]
- 42.Rajasagi M, Shukla SA, Fritsch EF, et al. Systematic identification of personal tumor-specific neoantigens in chronic lymphocytic leukemia. Blood. 2014;124:453–462. doi: 10.1182/blood-2014-04-567933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo J, Guo L, Sun L, et al. Capture-based ultra-deep sequencing in plasma ctDNA reveals the resistance mechanism of ALK inhibitors in a patient with advanced ALK-positive NSCLC. Cancer Biol Ther. 2018;19:359–363. doi: 10.1080/15384047.2018.1433496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin JJ, Zhu VW, Yoda S, et al. Impact of EML4-ALK variant on resistance mechanisms and clinical outcomes in ALK-positive lung cancer. J Clin Oncol Off J Am Soc Clin Oncol. 2018;36:1199–1206. doi: 10.1200/JCO.2017.76.2294. JCO2017762294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sonbol MB, Ahn DH, Bekaii-Saab T. Therapeutic targeting strategies of cancer stem cells in gastrointestinal malignancies. Biomedicines. 2019;7:17. doi: 10.3390/biomedicines7010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lo W, Parkhurst M, Robbins PF, et al. Immunologic recognition of a shared p53 mutated neoantigen in a patient with metastatic colorectal cancer. Cancer Immunol Res. 2019;7:534–543. doi: 10.1158/2326-6066.CIR-18-0686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kula T, Dezfulian MH, Wang CI, et al. T-Scan: a genome-wide method for the systematic discovery of T cell epitopes. Cell. 2019;178:1016–1028.e13. doi: 10.1016/j.cell.2019.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.