Abstract
Plant argonautes (AGOs) play important roles in the defense responses against viruses. The expression of Nicotiana benthamiana AGO5 gene (NbAGO5) is highly induced by Bamboo mosaic virus (BaMV) infection; however, the underlying mechanisms remain elusive. In this study, we have analyzed the potential promoter activities of NbAGO5 and its interactions with viral proteins by using a 2,000 bp fragment, designated as PN1, upstream to the translation initiation of NbAGO5. PN1 and seven serial 5′-deletion mutants (PN2–PN8) were fused with a β-glucuronidase (GUS) reporter and introduced into the N. benthamiana genome by Agrobacterium-mediated transformation for further characterization. It was found that PN4-GUS transgenic plants were able to drive strong GUS expression in the whole plant. In the virus infection tests, the GUS activity was strongly induced in PN4-GUS transgenic plants after being challenged with potexviruses. Infiltration of the transgenic plants individually with BaMV coat protein (CP) or triple gene block protein 1 (TGBp1) revealed that only TGBp1 was crucial for inducing the NbAGO5 promoter. To identify the factors responsible for controlling the activity of the NbAGO5 promoter, we employed yeast one-hybrid screening on a transcription factor cDNA library. The result showed that NbNAC42 and NbZFP3 could directly bind the 704 bp promoter regions of NbAGO5. By using overexpressing and virus-induced gene silencing techniques, we found that NbNAC42 and NbZFP3 regulated and downregulated, respectively, the expression of the NbAGO5 gene. Upon virus infection, NbNAC42 played an important role in regulating the expression of NbAGO5. Together, these results provide new insights into the modulation of the defense mechanism of N. benthamiana against viruses. This virus inducible promoter could be an ideal candidate to drive the target gene expression that could improve the anti-virus abilities of crops in the future.
Keywords: argonaute, AGO5, BaMV, TGBp1, potexvirus, NAC, zinc finger protein (ZFP)
Introduction
Nicotiana benthamiana can be infected by diverse plant viruses, making it a popular model host in many aspects of plant virology (Goodin et al., 2008). A variety of host factors in N. benthamiana involved in the infection processes of viruses have been identified in previous studies (Huang et al., 2012, 2017b, 2019, 2021). However, the defense machineries of N. benthamiana against viruses remain relatively unexplored. Among the few well-studied virus resistance mechanisms, it has been shown that plant viruses may induce a potent and specific antiviral RNA silencing host response in which argonautes (AGOs) play a central role (Carbonell and Carrington, 2015). Antiviral AGOs may associate with virus-derived small RNAs (vsiRNAs) to repress the expression of the cognate viral RNAs or DNAs, or with endogenous small RNAs (sRNAs) to regulate host gene expression and promote antiviral defense (Baulcombe, 1999; Carbonell and Carrington, 2015; Csorba et al., 2015; Alazem et al., 2017). The gene silencing mechanism, mainly through the interactions of AGOs with sRNAs, plays an important role in the regulation of gene expression in plants (Meister, 2013), and is an integral part of the essential innate immunity to fight against both RNA and DNA viruses in plants.
Both mono- and dicotyledonous plants encode several AGO proteins that can be categorized into three distinct clades: AGO 1/5/10, AGO 2/3/7, and AGO 4/6/8/9 based on phylogenetic and functional relationships (Vaucheret, 2008; Fang and Qi, 2016). Among them, AGO 1/5/10 clade is mainly involved in anti-viral activities. Several virus-resistant AGO members have been identified in N. benthamiana. NbAGO1 has been reported to be required for resistance to Tomato ring spot virus (ToRSV) (Ghoshal and Sanfacon, 2014), Tomato bushy stunt virus (TBSV), and Cymbidium ringspot virus (CymRSV) (Gursinsky et al., 2015), Turnip crinkle virus (TCV) (Ludman et al., 2017), and Bamboo mosaic virus (BaMV) (Huang et al., 2019); NbAGO2 has been shown to play a role in the resistance against TBSV, Tobacco mosaic virus (TMV) (Diao et al., 2019), Potato virus X (PVX), Turnip mosaic virus (TuMV), and TCV (Fatyol et al., 2016; Alazem et al., 2017); NbAGO4 is involved in PVX resistance (Bhattacharjee et al., 2009); NbAGO5 is also able to bind Cucumber mosaic virus (CMV) vsiRNAs (Takeda et al., 2008), indicating a role in antiviral defense. It has also been shown that AGO5 is induced by PVX and cooperates with AGO2 to restrict viral infection (Brosseau and Moffett, 2015); NbAGO7 is related to resistance against Foxtail mosaic virus (FoMV), Sunn-hemp mosaic virus (SHMV), and TBSV (Odokonyero et al., 2017). The presence of specialized proteins known as viral suppressors of RNA silencing (VSRs) in viral genomes is one of the strongest indications of the significance of RNA silencing as an antiviral response (Csorba et al., 2015). The successful infection of a given host by a virus generally indicates that the virus has overcome the RNA silencing mechanisms of the host, possibly via the action of its VSR. Most VSRs are multifunctional proteins with a secondary role of VSR activity because many viral proteins are involved in the replication, movement, and/or encapsidation of viruses. VSRs have different modes of action, including sequestration of sRNAs and proteins necessary for the onset of RNA silencing and the propagation of signals. VSRs may also exert their functions by shifting the balances among different defense-related host factors. Previous studies have shown that NbAGO10 could facilitate the accumulation of viruses mainly because P28 (TGBp1) in BaMV promoted the expression of NbAGO10, which competes with NbAGO1 for vsiRNA and leads to a decrease in the antiviral ability of AGO1 (Huang et al., 2019). Thus, specific AGOs may be recruited in the defense responses against different viruses. However, the underlying mechanism for the modulation of specific AGO gene expression by specific viral factors awaits further exploration.
In plants, the crosstalk among phytohormones is crucial in the responses to multiple biotic and abiotic stimuli, including the infection of viruses. It has been shown that the promoters of almost all AGO-encoding genes contain motifs involved in the binding by salicylic acid (SA) or abscisic acid (ABA)-responsive transcription factors (TFs) (Alazem et al., 2019). During virus infection in plants, several AGO-encoding genes are upregulated (Bai et al., 2012; Alazem et al., 2017; Paudel et al., 2018; Diao et al., 2019; Sheng et al., 2019). In addition to phytohormones, plants have evolved a number of molecular mechanisms for dealing with stress, including the augmentation or reduction of the effects of TFs on specific target genes. In the responses to abiotic and biotic stressors, plant TFs may operate as nodes in a regulatory network. Many members of diverse families of TFs, including MYB (Yang and Klessig, 1996; Yang et al., 2020), WRKY (Park et al., 2006; Chen et al., 2013), bZIP (Gaguancela et al., 2016; Gayral et al., 2020), AP2/ERF (Huang et al., 2016), and NAC (Ren et al., 2000; Selth et al., 2005; Donze et al., 2014; Huang et al., 2017a; Sun et al., 2019), have been demonstrated to be involved in the responses to viral stimuli. In addition, TFs with “zinc finger” domains (zinc finger protein, ZFP) known for their finger-like structure and the ability to bind Zn+2 ions have also been implicated in response to viral infections (Guo et al., 2004; Huh et al., 2011). However, it remains to be explored to what extent phytohormones and TFs affect the expression and function of AGOs during virus infection.
In Arabidopsis thaliana, AGO5 is normally expressed mostly in flowers and other reproductive tissues, but the AGO5 gene is systemically activated in leaves after the infection of PVX or Plantago asiatica mosaic virus, implicating that AGO5 plays an important role in limiting the spread of systemic viruses (Brosseau and Moffett, 2015). In orchids, the AGO5 family proteins play an important part in the defense against the Cymbidium mosaic virus (CymMV) and the Odontoglossum ringspot virus (ORSV) (Kuo et al., 2021). Interestingly, we have found that NbAGO5 is strongly induced by BaMV infection in N. benthamiana, which provides a feasible system to elucidate the underlying mechanisms involved in antiviral activities. Thus, the objective of this study was to identify the factors which trigger the expression of NbAGO5, by investigating the NbAGO5 promoter activities in transgenic plants. In addition, we also aimed at identifying the TFs that affect the expression of NbAGO5 promoter by screening the Arabidopsis TFs cDNA library using yeast one-hybrid assay.
Materials and Methods
Plant Cultivation Conditions
Unless otherwise stated, all N. benthamiana plants were grown in a growth room maintained at 26 ± 1°C under a photoperiod of 16 h-light/8 h-dark.
Mechanical Inoculation of Bamboo Mosaic Virus
The fourth leaves from the bottom to the top along the stem of seedlings of the 30-day-old N. benthamiana were mechanically inoculated with 0.5 μg of the BaMV severe strain (BaMV-S) virion (Lin et al., 1994). The relative expression of Argonautes mRNAs in inoculated leaves was determined using real-time quantitative reverse transcription PCR (qRT-PCR), with actin serving as an internal control.
Construction of Viral Infectious Clones and Inoculation Assay
The infectious constructs, based on the plasmid backbone pKn (Prasanth et al., 2011), of the viruses used in this study included pKB (Liou et al., 2014) for BaMV, pKP (Huang et al., 2012) for PVX, pKF (Huang et al., 2020) for FoMV, pKCy1 (Huang et al., 2020) for CymMV, and pKT for TMV. For the construction of pKT, the cDNA of TMV was reverse transcribed with primer KpnI_TMV-R (Supplementary Table 1) from TMV RNA that was purified from TMV_Taiwan isolates. After amplification with primer TMV_5′-F (Supplementary Table 1), the PCR products were cloned into pCass (Ding et al., 1995) at the appropriate restriction sites to generate pCT, and then sub-cloned into SbfI and KpnI restriction sites of pKn to generate pKT construct.
For the construction of CMV infectious clones, the cDNAs of CMV_NT9 RNA1, RNA2, and RNA3 were reverse transcribed with primers, SacI_CMV1a_R, HpaI-KpnI_CMV2a_R, and SacI_CMV3a_R, respectively (Supplementary Table 1). After amplification with forward primers for CMV RNA1, RNA2, and RNA3, BamHI/SacI_CMV1a, BamHI/HpaI-KpnI_CMV2a, and BamHI/SacI_CMV3a, respectively, the PCR products were cloned into pEpyon-GFP (Huang et al., 2017b) at the appropriate restriction sites to generate constructs, pECMV1, pECMV2, and pECMV3.
For the construction of transient expression clones of BaMV-CP and -TGBp1, the corresponding coding regions were amplified by PCR using pKB as the template with the primers, XbaI_BaCP-F and SacI_BaCP-R for BaMV-CP, XbaI_BaTGBp1-F and SacI_BaTGBp1-R for BaMV-TGBp1 (Supplementary Table 1), and cloned into pEpyon-GFP at the BamHI and EcoRI restriction sites to generate pEBaCP and pEBaTGBp1 constructs, respectively. The infectious constructs and transient expression plasmids were introduced into Agrobacterium tumefaciens strain GV3850 individually by electroporation. The third, fourth, and fifth leaves from the bottom to the top along the stem of seedlings of the 30-day-old N. benthamiana were infiltrated with A. tumefaciens harboring the above constructs as described previously (Huang et al., 2012).
Real-Time Quantitative Reverse Transcription
Total RNA was extracted from leaf tissues using TriPure Isolation Reagent (Roche Life Science, Mannheim, Germany) according to the manufacturer’s instructions. First-strand cDNAs were synthesized using GoScript Reverse Transcriptase (Promega, Madison, WI, United States) with the oligo dT(18) primer. The qRT-PCR analyzes of NbAGO5 or TFs mRNA levels were performed with a TOptical Gradient 96 Real-Time PCR Thermal Cycler (Biometra, Göttingen, Germany) in reactions containing 3 μl of two-fold-diluted cDNAs as templates specific primers (Supplementary Table 1), and the KAPA SYBR® FAST qPCR master mix (Kapa Biosystems, Wilmington, MA, United States). The internal control was actin, and the experiments were performed in triplicate.
Isolation of NbAGO5 Promoter Sequence
Primers were designed based on the 2 kb region upstream to the ATG start codon of NbAGO5 gene in N. benthamiana genome (Nbv6.1trP59647) downloaded from the QUT database1. The putative NbAGO5 promoter region was amplified by PCR from N. benthamiana genomic DNA with PNP1 primers (Supplementary Table 1). The amplicon containing the 2,000 bp putative promoter region, designated PN1, of the NbAGO5 gene was cloned and sequenced.
In silico Analysis of NbAGO5 Promoter Sequence
To predict the potential interacting transcription factors, the cis-regulatory elements of NbAGO5 promoter were analyzed using PlantCARE web server2 (Lescot et al., 2002).
Construction of Promoter: β-Glucuronidase Fusion and Deletion Vectors
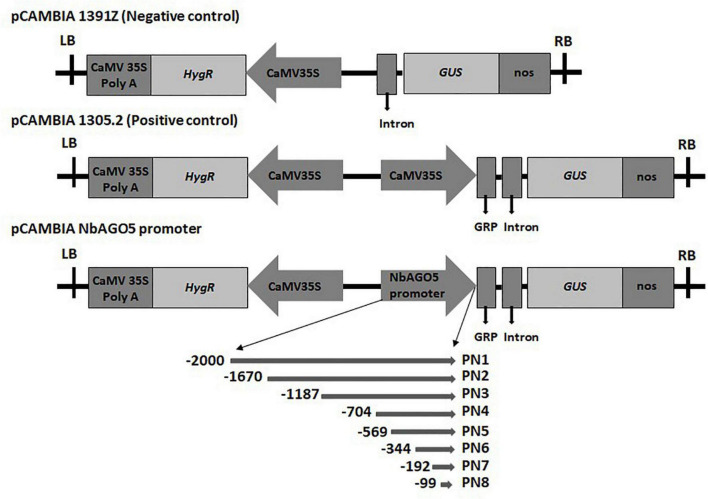
The initial division of the putative “full-length” promoter region was based on the presence of two restriction sites, HindIII and XbaI, which allows for a convenient sub-cloning process. To further fine-map the functional promoter region, PN2 and PN5 were further divided into shorter fragments based on the design of proper primer sequences for PCR amplification. Five serial 5′-deletion fragments (ranging from nucleotide positions −1,187, −704, −344, −192, and −99 to position −1, with the translation initiation site designated as “+1”; Figure 1) were amplified by PCR from the PN1 region with specific oligonucleotide primers (Supplementary Table 1). For the generation of NbAGO5:GUS constructs, the PCR products were subsequently constructed into the pCAMBIA1305.2 vector (Marker Gene Technologies, Ipswich, England) at the XbaI or HindIII/NcoI restriction sites. The resulting constructs were confirmed by sequencing and used for N. benthamiana stable transformation.
FIGURE 1.
Schematic diagrams of the truncated fragments of different deleted versions fused with the GUS reporter gene. The pCAMBIA1391Z vector acts as a negative control that does not have a promoter to drive GUS. The pCAMBIA1301 vector was used as a positive control, and the pCAMBIA NbAGO5 promoter constructs with different lengths of NbAGO5 promoter was created by replacing the 35S promoter (A). LB, left border; CaMV 35S PolyA, cauliflower mosaic virus 35S terminator; HygR, hygromycin resistance gene; 35S promoter, cauliflower mosaic virus 35S promoter; GRP, glycine-rich signal peptide sequence; GUS, β-glucuronidase reporter gene; Nos, nopaline synthase terminator; RB, right border.
Culture and Stable Transformation of Nicotiana benthamiana
The NbAGO5:GUS plasmids were introduced into A. tumefaciens strain Agro 3,850 individually by electroporation (Kotnik, 2013). The A. tumefaciens-mediated transformation procedures were followed as previously described (Horsch et al., 1985). Regeneration of the transformants was performed using the method as described (van Tunen et al., 1989). The T0 positive transformants were screened by hygromycin resistance (Clough and Bent, 1998). The homozygous transgenic lines of T3 generation were chosen for subsequent investigations via segregation ratio analysis.
Hormone Treatments in Transgenic Nicotiana benthamiana
Seeds of transgenic plants were germinated and grown in liquid MS medium at 20°C under a 16/8 h photoperiod in 6-well plates. When seedlings were 6 days old, the liquid MS medium was replaced with fresh MS containing ABA (10 μM), SA (50 μM), Methyl jasmonate (MeJA) (50 μM), or mock (0.5% EtOH in ddH2O) (Alazem et al., 2019). Two days after treatment, 10 seedlings from each line were infiltrated with the above viral infectious constructs, and the leaves were collected at 2 days post infiltration (dpi) and analyzed for GUS activities.
Analysis of Virus-Induced Gene Expression in Transgenic Nicotiana benthamiana
For analyzing stress responses caused by viruses, A. tumefaciens cells harboring infectious clones of potexviruses (BaMV, PVX, FoMV, and CymMV) were infiltrated (hereafter referred to as agroinfiltrated) into transgenic plants as follows. The A. tumefaciens cultures were collected by centrifugation and resuspended in infiltration buffer (10 mM MES buffer, pH 5.5, and 10 mM MgCl2), adjusted to OD600 = 0.5, and infiltrated into the leaves of each test plant through a needleless syringe. The third, fourth, and fifth leaves, from the bottom to the top of the 30-day-old N. benthamiana seedlings, were agroinfiltrated. The leaves were harvested at 3 dpi and subjected to a GUS activity assay.
Histochemical β-Glucuronidase Staining and β-Glucuronidase Activity Quantification
Total proteins from infiltrated plants were extracted and quantified using the method described previously (Bradford, 1976). Histochemical GUS staining and GUS fluorometric analyzes were performed by following standard methods (Jefferson et al., 1987).
Yeast One-Hybrid Screening at Arabidopsis Transcription Factors Library
To identify the TFs that could interact with the NbAGO5 promoter, a systematic screening of the Y1H TFs library, which contains approximately 1,350 Arabidopsis TFs (Mitsuda et al., 2010) was undertaken in yeast strain YM4271.
For Y1H assays, the prey protein was fused to the activation domain (AD) of the yeast Gal4 transcription factor. The positive interaction between the prey protein and the bait DNA sequence (putative AGO5 promoter region) caused HIS3 reporter gene expression in Saccharomyces cerevisiae. Preparation of yeast competent cell, yeast transformation, and selection of interaction were carried out according to the user manual of the manufacturer (Invitrogen, Thermo Fisher, Taipei, Taiwan). The bait and prey-designated plasmids were combined as indicated and were transformed simultaneously into a Y187 cell and plated on an SD/-Leu-Ura for selection of successfully co-transformed cells. Three colonies were selected at random, dissolved in water, and plated on an SD/-Leu-Ura-His for the selection of a specific interaction.
For the construction of bait plasmid pHISi-PN4, the 704 bp promoter region of NbAGO5 (PN4) was amplified from the genomic DNA of N. benthamiana by using appropriate primers (Supplementary Table 1) with the restriction endonuclease sites of EcoRI and SacI, cloned in the vector pHisi to give pHisi–PN4, and integrated into the yeast genome Y187. The Y1H assay was carried out as described previously (Mitsuda et al., 2010). In the screenings of Arabidopsis TFs library, the degree of positive interaction between a prey TF and the bait sequence was scored between 0 and 3 in each screening according to the yeast growth status under selective media so that each TF has its respective interaction strength.
Corresponding Transcription Factors in Nicotiana benthamiana Transcriptome
The above Y1H screening generated candidate A. thaliana TFs with high binding affinity to the putative promoter region of NbAGO5. To identify the corresponding TFs in N. benthamiana genome, the sequences of the candidate A. thaliana TFs were used as the query to search for the corresponding TF homologs in N. benthamiana transcriptome database.3 Following the identification of the corresponding TFs in N. benthamiana, oligonucleotide primers were designed (Supplementary Table 1) to amplify the respective N. benthamiana TFs by PCR. The PCR products were then cloned into pGADT7 (Mitsuda et al., 2010), pEpyon, and pTRV2 vector (Ratcliff et al., 2001) at EcoRI and BamHI restriction sites for further Y1H, over-expression, and virus-induced gene silencing (VIGS) analyzes, respectively.
Yeast One-Hybrid Assay
To verify the interactions between the putative AGO5 promoter region and candidate N. benthamiana TFs or BaMV encoded proteins, yeast one-hybrid assays were performed. The coding regions of BaMV-CP and -TGBp1 and candidate N. benthamiana TFs were amplified by PCR with specific primer pairs (Supplementary Table 1) and cloned into the respective restriction sites of pGADT7 vector to give the prey constructs pGADT7-CP, pGADT7-TGBp1, pGADT7-NbNAC42, and pGADT7-NbZFP3. The plasmids and pHisi–PN4 were transformed into the yeast genome Y187. The transformants were grown on SD minimal medium lacking Leu, Ura, and His but containing 20 mM 3-AT. The cell concentration was adjusted to OD600 = 1 and serial dilutions were spotted on plates and grown for 3 days at 28°C.
Transient Expression and Virus-Induced Gene Silencing of Candidate Transcription Factors in Nicotiana benthamiana
The coding regions of candidate TFs were amplified with specific primers (Supplementary Table 1) and cloned into transient expression vector pEpyon. The pEpyon-based constructs of candidate TFs were introduced into A. tumefaciens strain GV3850 individually by electroporation. The A. tumefaciens cultures were collected by centrifugation and resuspended in infiltration buffer (10 mM MES buffer, pH 5.5 and 10 mM MgCl2), adjusted to OD600 = 0.5, and infiltrated into the leaves of each test plant through needleless syringes. The leaves were harvested at 3 dpi and subjected to GUS activity assay and western blot as described previously (Huang et al., 2019).
To transiently knockdown the expression of the candidate TFs, the VIGS technique (Ratcliff et al., 2001) was employed. The target sequences of candidate TFs were amplified from N. benthamiana genomic DNA with the oligonucleotide primers (Supplementary Table 1), which were designed based on the sequences of specific TFs downloaded from the SGN database (Solanaceae Genomics Network4). The PCR products were subsequently constructed into the pTRV2 vector at the corresponding restriction sites. For knockdown experiments, the pTRV1- and pTRV2-based constructs were electroporated into the A. tumefaciens strain C58C1 and infiltrated into test plants as described previously (Huang et al., 2012).
Statistical Analysis
All inoculation and Y1H experiments were performed with at least three biological replicates. The data were analyzed using a basic statistic tool in EXCEL 2016 for Windows 10 (Microsoft), including ANOVA. Statistical differences between means of groups were determined by Student’s t-test, with P-value ≤ 0.05; ≤0.01, or ≤0.001 as statistically significant or highly significant (denoted by “*”; “**” or “***”), respectively.
Results
NbAGOs mRNA Expression Profile After Bamboo Mosaic Virus Infection
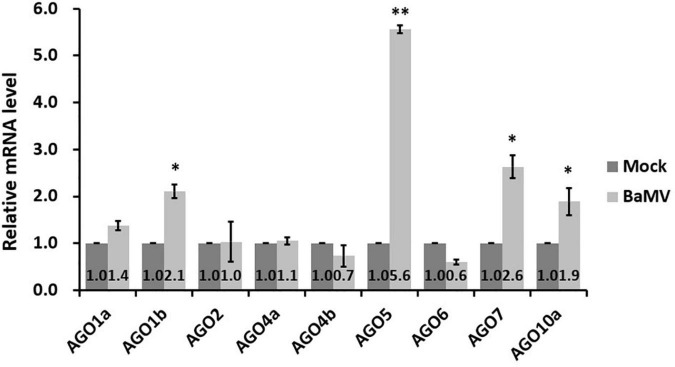
The NbAGOs corresponding to the ten AtAGOs, including isoforms, have been identified in the N. benthamiana genome. There are seven members of NbAGOs: namely NbAGO1, NbAGO2, NbAGO4, NbAGO5, NbAGO6, NbAGO7, and NbAGO10. Two isoforms were found for NbAGO1, NbAGO4, and NbAGO10. To analyze the mRNA expression profiles of different NbAGOs in response to the BaMV infection, total RNAs from mock- and BaMV-infected N. benthamiana leaves were extracted at 5 dpi and the mRNA levels of NbAGOs were quantified by qRT-PCR. The result revealed that NbAGO5 mRNA levels were 5.6-fold higher in BaMV-inoculated leaves than that in mock-inoculated leaves at 5 dpi (Figure 2), which was the largest increase of all NbAGOs. In addition, the expression levels of NbAGO1b, NbAGO7, and NbAGO10a were also significantly higher than those in plants inoculated with an empty vector. In contrast, the expression level of NbAGO10b was not detectable in leaves of mock-inoculated or BaMV-inoculated N. benthamiana and was not shown in Figure 2. Thus, NbAGO5 and its putative promoter region were chosen as the main subjects of the following analyzes.
FIGURE 2.
Realtime RT-PCR analyzes of Argonaute genes in Bamboo mosaic virus (BaMV)-inoculated Nicotiana benthamiana. Nicotiana benthamiana leaves were mechanically inoculated with 0.5 μg of BaMV virions and then collected at 5 dpi for analysis. Real-time quantitative reverse transcription PCR (qRT-PCR) was used to determine the relative expression of Argonautes mRNAs in inoculated leaves, with actin serving as an internal control. Each column represents the average of at least three replicates ± standard error (SE). Statistical significance was analyzed using two-tailed student’s t-tests; *p < 0.05; **P < 0.01.
Isolation and Sequence Analysis of the NbAGO5 Promoter
Initially, the putative NbAGO5 promoter region was amplified and cloned from genomic DNA using specific primer pairs designed for the 2,000-nucleotide fragment upstream to the start codon of NbAGO5 gene, designated as PN1. Following sequence verification, the putative NbAGO5 promoter region was used as the query to search against the PlantCARE database for the presence of possible cis-acting elements for transcription. The positions and descriptions of the identified cis-acting elements are shown in Supplementary Table 2. The results revealed that the 2,000 bp promoter sequence contains a number of TATA-box and CAAT-box core cis-acting elements. Some cis-acting elements for the perception of multiple environmental stimuli are also present, including eight types of light-responsive elements (G-box, I-box, MRE, TCT-motif, AE-box, GT1-motif, Sp1, and TCT-motif), 3 types of abscisic acid-responsive elements (ABRE, ABRE3a, and ABRE4), one of ethylene-responsive element (ERE), one of SA responsive element (TCA-element), two gibberellin-responsive elements (GARE-motif and P-box), one known element required for meristem expression (CAT-box), a cis-acting regulatory element essential for the anaerobic induction (ARE), and some elements (MYB) that have different responses in growth or environmental stress. Thus, the putative NbAGO5 promoter region has the potential to perceive the stimuli associated with growth, development, phytohormones and biotic or abiotic stresses in plants.
Mapping of the Core Region of NbAGO5 Promoter in Transgenic Nicotiana benthamiana Through 5′ Serial Deletions
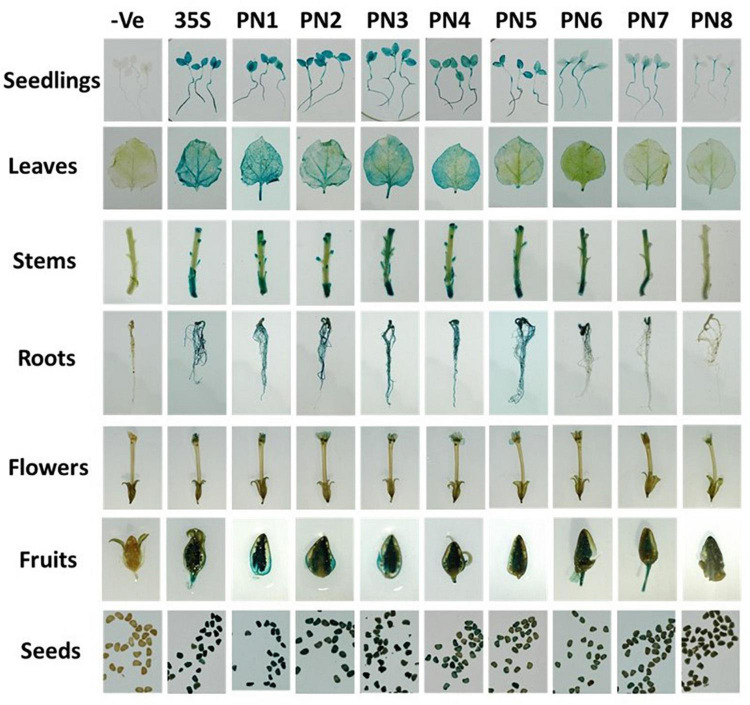
To map the core region of NbAGO5 promoter, 5′-serial deletions of PN1 fragment were prepared and fused with the GUS gene to produce reporter constructs used for generating transgenic N. benthamiana. Five to ten independent transgenic lines from each of the constructs (PN1–PN8, with CaMV35S promoter as a control) were screened in T1 generation by histochemical GUS staining. The homozygous lines in T3 generation were chosen for further analysis. The results of GUS staining showed that fragments PN1 to PN8 were all capable of driving GUS expression, but with obvious difference in their promoter activities in transgenic N. benthamiana (Figure 3). To analyze the expression profiles of transgenic N. benthamiana harboring GUS reporter gene driven by PN1 to PN8 or the CaMV 35S promoter under normal conditions, samples from different tissues, including the radicle of seeds from 10-day-old seedlings, roots, stems, and leaves of 70-day-old plants, flowers, fruits, and seeds from 90-day-old plants were subjected to GUS histochemical staining (Figure 3). GUS expressions driven by PN7 and PN8 were the lowest among all tissues. High GUS expression was detected in almost all tissues of plants harboring fragments PN1–PN6, including the seedlings, leaves, cotyledon, stem, petals, sepals, stigma, and seed. It is worth noting that, in leaves, PN4 seemed to possess the minimal length required for efficient promoter activity for driving GUS expression in transgenic plants, and thus might represent the core promoter region of NbAGO5 gene.
FIGURE 3.
Histochemical GUS staining in tissues of T3 transgenic N. benthamiana. Plant organs from homozygous T3 generation transgenic lines were incubated in GUS staining solution for 24 h at 37°C. Then the samples were observed and photographed after decolorization.
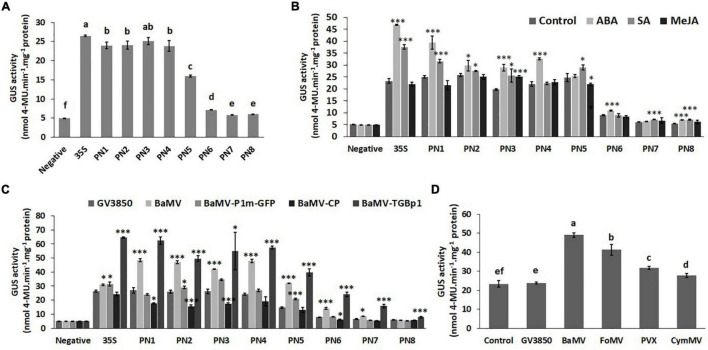
To further quantify the promoter strengths of PN1–PN8 under normal conditions and to verify the core region of the NbAGO5 promoter, fluorometric GUS assays were performed with leaves of transgenic N. benthamiana harboring different constructs. The result (Figure 4A) revealed that the promoter activities remained high with deletions up to position −704 (PN4), but significantly decreased for PN5–PN8, with PN7 and PN8 showing the lowest activities. The sharpest decrease was observed between PN5 and PN7, as compared to those of PN1–PN4, suggesting that the core NbAGO5 promoter region may reside between positions −704 and −192, relative to the initiation codon, whereas the region between −191 to −1 may still exhibit the basal level of promoter activity. Among them, the GUS expression driven by PN6 was obviously higher than those of PN7 and PN8. These results were in accordance with the result of the identification of cis-acting elements within PN1 (Supplementary Table 2), which showed that positions −191 to -1 bp do not contain TATA-box, whereas the region between −704 and -192 bp contains multiple cis-acting elements, such as TATA-box (Supplementary Table 2). Together, these results indicated that PN4 contains the core NbAGO5 promoter region sufficient to drive high-level expression in transgenic N. benthamiana under normal conditions.
FIGURE 4.
Promoter activity of NbAGO5 under normal condition, hormone treatments, and challenge with the virus in Transgenic N. benthamiana. (A) The fluorometric GUS assays were performed with leaves of transgenic N. benthamiana harboring different constructs under normal conditions and (B) hormone treatments. (C) The GUS activities in the plants infiltrated with A. tumefaciens harboring infectious construct of BaMV or expression vector for BaMV-CP or BaMV-TGBp1 were analyzed at 3 dpi. (D) PN4 transgenic N. benthamiana plants were agro-infiltrated with infectious constructs of various potexviruses, including BaMV, PVX, FoMV, and CymMV. The GUS activities were then analyzed at 3 dpi. Each column represents the mean of at least three replicates ± standard error (SE). Statistical significance was analyzed using two-tailed student’s t-tests; *p < 0.05; **p < 0.01; ***P < 0.001. The different lowercase letters above the bars indicate statistically significant differences among the means based on two-way ANOVA followed by Duncan’s multiple range test (P < 0.05).
Hormone Treatments and Challenges With the Virus in Transgenic Nicotiana benthamiana
Since the analysis of cis-acting elements in the putative NbAGO5 promoter region revealed the presence of several hormone-responsive motifs, we further tested whether the treatment of phytohormones could modulate the expression of the GUS reporter gene in transgenic plants. The result revealed that not all transgenic plant seedlings improved their GUS expression following hormone treatments (Figure 4B). ABA and SA may be important factors in affecting the GUS expression driven by PN1-PN3 regions, but only ABA was effective in PN4 plants (Figure 4B).
In order to test the influence of BaMV on the activity of NbAGO5 promoter, transgenic N. benthamiana was infiltrated with A. tumefaciens harboring infectious construct of BaMV, or expression vector for BaMV-CP or BaMV-TGBp1. The GUS activities in the infiltrated plants were then analyzed at 3 dpi. The results of quantitative analyzes of GUS activity showed that higher NbAGO5 promoter activity levels were observed when PN4 transgenic N. benthamiana was inoculated with BaMV and BaMV-TGBp1 at 3 dpi, whereas the expression of BaMV-CP did not alter NbAGO5 promoter activity (Figure 4C). The results indicated that BaMV-TGBp1 may be the viral factor for the induction of high-level NbAGO5 gene expression.
For testing the effect of infections by other potexviruses, PN4 transgenic N. benthamiana plants were agro-infiltrated with infectious constructs of various potexviruses, including BaMV, PVX, FoMV, and CymMV, and the GUS activities were analyzed at 3 dpi. The result showed that GUS activities in transgenic N. benthamiana were induced by the infection of all potexviruses tested, with various activities in the order of BaMV > FoMV > PVX > CymMV (Figure 4D). The result suggested that NbAGO5 may participate in the defense against potexviruses.
The above quantitative analyses of NbAGO5 promoter activity in transgenic N. benthamiana PN4 line demonstrated that it was higher after potexvirus infection.
Yeast One-Hybrid Screening of the Transcription Factor Interacting With the NbAGO5 Promoter
Yeast One-Hybrid (Y1H) screening was used to identify possible trans-activators using a 704 bp region (PN4, positions -704 to -1 upstream of the translational initiate site ATG) of the putative NbAGO5 promoter as the prey. The bait plasmid pHISi-PN4 was used for Y1H screening in the library of around 1,350 Arabidopsis thaliana TFs (Mitsuda et al., 2010). The pHISi-PN4 bait plasmid was transformed into a Y187 cell and plated on SD/-Leu-Ura for the selection of successfully co-transformed cells, which could be used for screenings of Arabidopsis TFs for positive interactions with the PN4 region by the addition of 75, 100, and 125 mM 3-AT. A total of 31 positive TFs, including 1, 11, and 19 with strong, weak, and very weak interactions, respectively, were obtained from this screening. We classified these candidate TFs and used them as the queries to search in N. benthamiana genome database by BLAST (Altschul et al., 1990) for positive ones, excluding very weak interactions (Table 1). The result revealed five TFs, including NAC, B3, C2H2ZnF, FAR1_related, and Trihelix classified as transcription factors in A. thaliana genome. Through homology searches in N. benthamiana database using BLAST, 8 homologous genes (NAC21/22, NAC42, NAC68, NAC94, ZFP1, ZFP3, Far1-related sequence 4 isoform 3, and Trihelix transcription factor GT-2) were identified in the N. benthamiana genome. To further verify the functions in antiviral defense, primers were designed based on the above 8 homologous genes (Supplementary Table 1), and the expression of these genes was analyzed by qRT-PCR with specific primers in healthy and BaMV-infected N. benthamiana leaves. The result showed that only NbNAC42 and NbZFP3 could be detected and isolated from health and BaMV-infected leaves of N. benthamiana (Supplementary Figure 1). Thus NbNAC42 and NbZFP3 were chosen as the main subjects for the following analyzes.
TABLE 1.
Transcription factors binding to NbAGO5 promoter by screening of yeast one-hybrid libraries.
| Screening of Y1H libraries | Isolated TF |
TF classification | Interaction strength | Homology gene in N. benthamiana transcriptome | |
| Locus | Name | ||||
| Arabidopsis | AT3G12910 | NAC | NAC | Strong | NAC42, NAC68, NAC94 |
| AT3G49610 | B3 | B3 | Weak | No | |
| AT5G04390 | C2H2ZnF | C2H2ZnF | Weak | ZFP1, ZFP3 | |
| AT3G10470 | C2H2ZnF | C2H2ZnF | Weak | ZFP1, ZFP3 | |
| AT2G37430 | ZAT11 | C2H2ZnF | Weak | ZFP1 | |
| AT2G28200 | C2H2ZnF | C2H2ZnF | Weak | ZFP1, ZFP3 | |
| AT3G46070 | C2H2ZnF | C2H2ZnF | Weak | ZFP1 | |
| AT2G28710 | C2H2ZnF | C2H2ZnF | Weak | ZFP1 | |
| AT3G19580 | ZF2 | C2H2ZnF | Weak | ZFP1 | |
| AT3G22170 | CPD45 | FAR1_related | Weak | Far1-related sequence 4 isoform 3 | |
| AT3G12977 | NAC1L | NAC | Weak | NAC21/22 | |
| AT2G35640 | Homeodomain-like superfamily protein | Trihelix | Weak | Trihelix transcription factor GT-2 | |
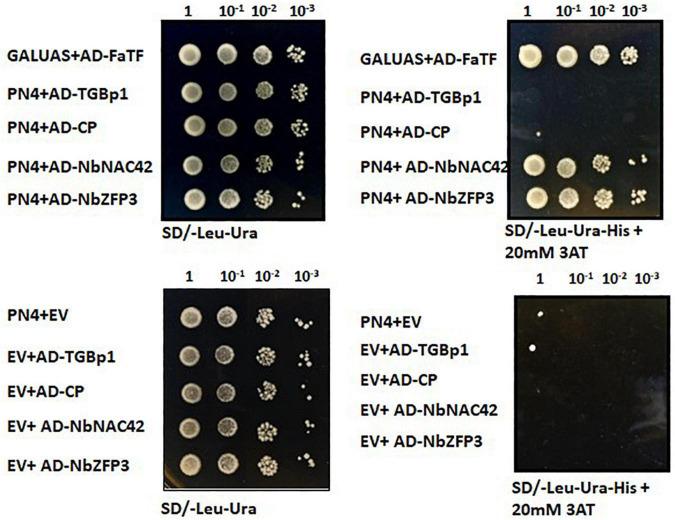
To further analyze the functions of NbNAC42 and NbZFP3 protein in BaMV infection, Y1H experiments were performed using the PN4 fragment of putative NbAGO5 promoter as the bait. The result revealed that both NbNAC42 and NbZFP3 proteins could interact strongly with the PN4 region of the putative NbAGO5 promoter, but BaMV-CP and BaMV-TGBp1 did not (Figure 5), suggesting NbNAC42 and NbZFP3 proteins may play a central role in regulating the expression of NbAGO5. In contrast to the result in Figure 4, which showed that BaMV-TGBp1 could induce a high-level gene expression of NbAGO5, the result of this Y1H experiment indicated that the up-regulation of NbAGO5 expression may be an indirect induction effect of BaMV-TGBp1, possibly through the regulation of NbNAC42 and NbZFP3 proteins, and not caused by the direct interaction between BaMV-TGBp1 and the PN4 region of the putative NbAGO5 promoter.
FIGURE 5.
Yeast one-hybrid assay for the interaction between NbAGO5 promoter (704 bp) and transcription factors or BaMV proteins. Yeast strain Y187 expressing PN4 (prey) was transformed with pGADT7-CP, pGADT7-TGBp1, pGADT7-NbNAC42, or pGADT7-NbZFP3 (bait). Transformed yeast cells were selected on SD minimal medium without Leu, Ura, and His but containing 20 mM 3-AT. Empty prey and bait vectors were used as negative controls. Coexpression of GALUAS along with FaTF was used as the positive control.
Effects of NbNAC42 and NbZFP3 on the Expression of NbAGO5 in Nicotiana benthamiana
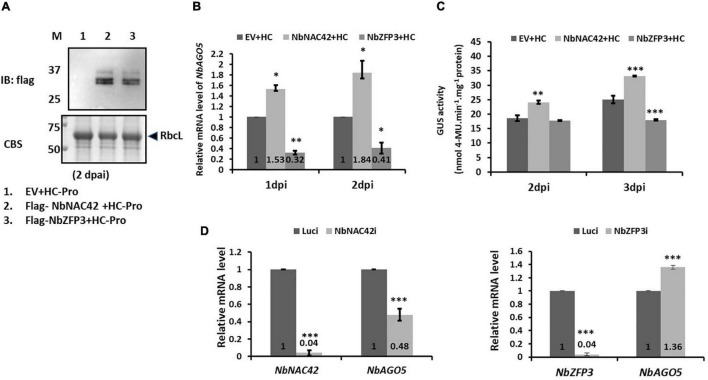
To verify the effect of NbNAC42 and NbZFP3 on the regulation of NbAGO5 expression, transient over-expression, and silencing analyzes were performed. For transient expression, the coding regions of NbNAC42 and NbZFP3 genes were amplified, with Flag-tag, and cloned into pEpyon to generate pEpyon-NbNAC42 and pEpyon-NbZFP3. Following the verification of the constructs, the third, fourth, and fifth leaves (from the bottom to the top along the stem of seedlings) of the 30-day-old N. benthamiana were infiltrated with A. tumefaciens GV3850 cells harboring pEpyon-NbNAC42 and pEpyon-NbZFP3. Leaves were sampled for analysis at 1 and 2 dpi. The expression of Flag-tagged NbNAC42 and NbZFP3 was confirmed by immunoblot assay using an anti-Flag antibody (Figure 6A). The expression level of NbAGO5 was then analyzed by qRT-PCR. The result revealed that mRNA levels of NbAGO5 at 1 and 2 dpi were 1.53- and 1.84-fold higher, respectively, in leaves of plants transiently over-expressing NbNAC42 than those of the plants infiltrated with control constructs. In contrast, expression of NbAGO5 was 0.32- and 0.41-fold lower at 1 and 2 dpi, respectively, in plants transiently expressing NbZFP3 than those in plants infiltrated with the control construct (Figure 6B). Similar trends were observed in transgenic plants harboring PN4-GUS reporter constructs. The GUS expression level at 3 dpi was 1.32-fold higher in plants transiently overexpressing NbNAC42, but 0.71-fold lower in those transiently expressing NbZFP3, as compared to the GUS expression level in plants agro-infiltrated with empty vector (EV) (Figure 6C).
FIGURE 6.
Effects of overexpression and silencing of NbNAC42 and NbZFP3 on NbAGO5 levels in N. benthamiana. (A) Immunoblot analysis using anti-Flag antibody confirmed the expression of Flag-tagged NbNAC42 and NbZFP3. Total protein extracts were prepared from N. benthamiana leaves at 2 days post-agroinfiltration (dpai) and underwent SDS-PAGE, followed by Coomassie blue staining (CBS), anti-Flag immunoblotting. M, prestained protein markers on the left with molecular mass (in kDa). RbcL, RuBisCO large subunit is the loading control. (B) The expression level of NbAGO5 was analyzed by qRT-PCR. (C) The effects of NbNAC42 and NbZFP3 on PN4 activity. The GUS expression level of PN4 transgenic N. benthamiana plants transiently expressed NbNAC42 and NbZFP3 by Agrobacterium-mediated expression was analyzed at 2 and 3 dpi. (D) The transcript level of NbAGO5 in N. benthamiana transiently knock-down the expression of NbNAC42 and NbZFP3. Statistical significance was analyzed using two-tailed student’s t-tests; *p < 0.05; **p < 0.01; ***P < 0.001.
For gene silencing analysis, the TRV-based VIGS system (Ratcliff et al., 2001) was used to transiently knockdown the expression of NbNAC42 and NbZFP3 in N. benthamiana. NbNAC42 and NbZFP3 mRNA levels in leaves infiltrated with TRV2-NbNAC42 and TRV2-NbZFP3 were both decreased to 0.04-fold of those of the negative control (Luci, leaves infiltrated with TVR2-Luc) at 10 dpi (Figure 6D). The result of the qRT-PCR revealed that the mRNA level of NbAGO5 was decreased to 0.48-fold in NbNAC42-knockdown plants as compared to that of the negative control, but increased to 1.36-fold of that of the negative control when NbZFP3-knockdown plants (Figure 6D). The results of over-expression and silencing analyzes further supported the hypothesis that NbAGO5 expression was positively and negatively associated with NbNAC42 and NbZFP3, respectively.
Responses of NbNAC42 and NbZFP3 on Challenges of Various Viruses in Nicotiana benthamiana
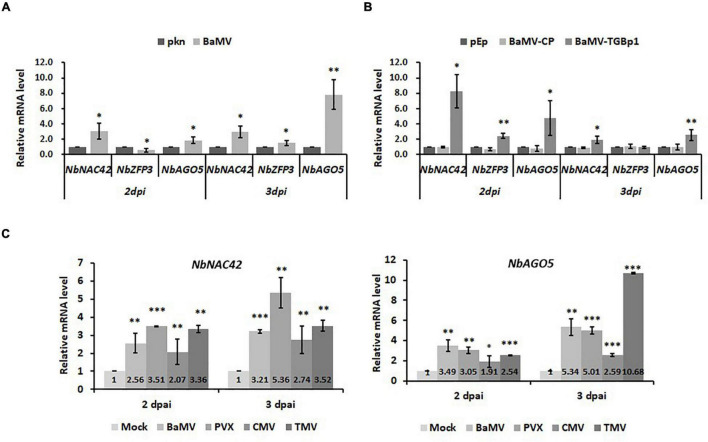
To verify the effects of NbNAC42 and NbZFP3 on the expression of NbAGO5 in N. benthamiana infected by different viruses, the following experiments were performed. Nicotiana benthamiana plants were inoculated with BaMV, PVX, CMV, or TMV, or infiltrated with the constructs, pEpyon-BaMV-TGBp1, and pEpyon-BaMV-CP, for transient over-expression of BaMV-TGBp1 and -CP, respectively. Leaf samples were collected at 2 and 3 dpi and analyzed by qRT-PCR with specific primers (Supplementary Table 1). The result revealed that the mRNA expression levels of NbNAC42 and NbAGO5 were significantly increased following the challenge of viruses or the expression of BaMV-TGBp1 at both 2 and 3 dpi as compared to those in the control plants (Figure 7B). This result was similar in accordance with those observed in previous experiments (Figure 6B), in which the transient over-expression of NbNAC42 in N. benthamiana leaves led to a significant increase in the relative mRNA level of NbAGO5. Thus, NbNAC42 seems to play an important role in inducing NbAGO5 expression. In contrast, the relative mRNA level of NbZFP3 was much less than that of NbNAC42. The expression of NbZFP3 in BaMV-inoculated plants was even reduced at 2 dpi (Figure 7A) but increased in the same plants at 3 dpi and in those expressing BaMV-TGBp1 (Figure 7B). This observation indicated that even if NbZFP3 may negatively regulate the expression of NbAGO5 (Figure 6D), the expression level and speed of NbZFP3 might not be sufficient to antagonize the positive effect of NbNAC42 on NbAGO5 expression in N. benthamiana. In addition, it was also found that the mRNA level of NbNAC42 and NbAGO5 were significantly increased in response to the infections of BaMV, PVX, CMV, and TMV at 2 and 3 dpi as compared to those in control groups (Figure 7C). The result suggested that the NbNAC42 may serve as the sensor of the antiviral surveillance system in N. benthamiana by interacting with viral factors (such as BaMV-TGBp1), and subsequently activating the expression of NbAGO5 which in turn participates in the RNA silencing mechanism in the defense against different viruses.
FIGURE 7.
Expression of NbNAC42 and NbZFP3 in response to challenges of various viruses in N. benthamiana. The expression level of NbAGO5, NbNAC42, and NbZFP3 were analyzed by qRT-PCR in the plants infiltrated with A. tumefaciens harboring infectious construct of BaMV (A), or expression vectors for BaMV-CP or BaMV-TGBp1 (B). (C) The mRNA level of NbNAC42 and NbAGO5 were analyzed by qRT-PCR in the plants infiltrated with A. tumefaciens harboring infectious constructs of BaMV, PVX, CMV, and TMV at 2 and 3 dpi as compared to those in control groups. Relative mRNA accumulation levels of three genes were individually shown above; numbers the compared to controls (pkn or pEpyon). Data are means ± SE from at least three independent experiments. Statistical significance was analyzed using two-tailed student’s t-tests; *p < 0.05; **p < 0.01; ***P < 0.001.
Discussion
Plant AGO proteins are guide-dependent nucleases involved in important functions, including developmental processes and stress responses against biological or abiotic stresses (Carbonell, 2017). For defense against plant viruses, it is known that different AGO proteins may be recruited in a virus-specific manner. However, the mechanisms for the specific induction of unique AGO proteins have not been explored extensively. In this study, we have attempted to elucidate the chain of command, at least in part, for the defense against specific viruses in N. benthamiana. The result revealed that BaMV encoded protein, TGBp1, may trigger the defense response by inducing the expression of NbNAC42, which in turn enhanced the expression of NbAGO5 and the subsequent antiviral defense through the RNA silencing mechanism. To counteract the host defense system, BaMV-TGBp1 may have evolved the ability to induce the expression of NbZFP3, which suppressed the expression of NbAGO5, as depicted in the proposed model shown in Figure 8. However, the level of NbZFP3 enhancement might not be sufficient to suppress the RNA silencing mechanism of the host against the invading virus.
FIGURE 8.
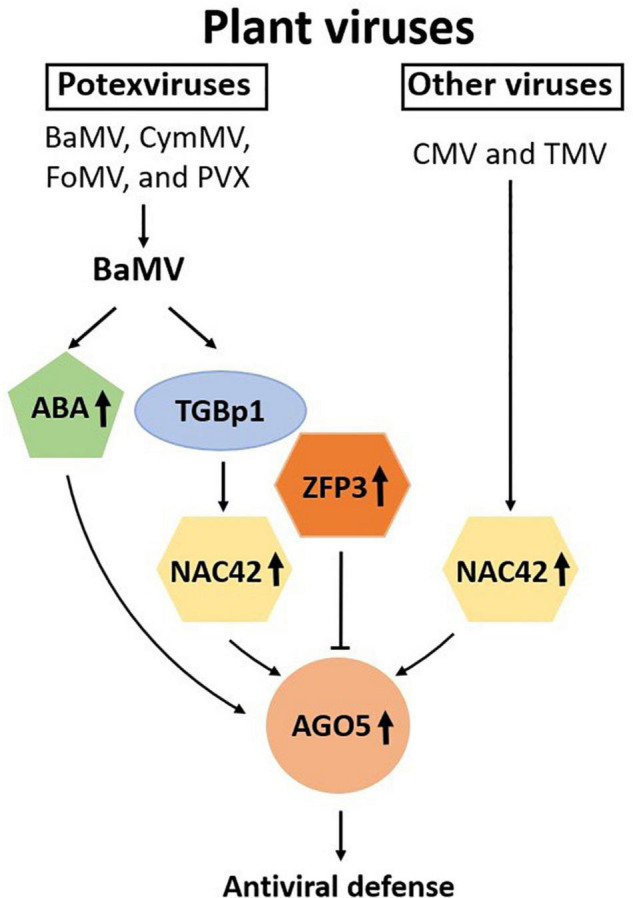
Schematic representation of the involvement of NAC and ZFP transcription factors and ABA in the regulation of NbAGO5 expression in response to virus infection. Plant argonautes (AGOs) play an important role in antiviral defense. The expression of NbAGO5 gene is strongly induced after virus infection in N. benthamiana. NbNAC42 and NbZFP3 positively and negatively regulate the expression of NbAGO5, respectively. The plant hormone ABA, whose biosynthesis has been shown to be activated by BaMV infection (Alazem et al., 2014), is also involved in the induction of NbAGO5 promoter activity. TGBp1, a BaMV-encoded protein, may regulate NbAGO5 expression and plant defense response through activation of the NbNAC42 expression.
Previously, we have found that an AGO5 family protein of Phalaenopsis aphrodite, PaAGO5b, is the only AGO proteins highly induced following the infection of Cymbidium mosaic virus and Odontoglossum ringspot virus, and we also demonstrated that PaAGO5b plays an important role in the defense against these viruses. However, the mechanism for the elicitation of PaAGO5b expression remains to be investigated. Here, among the N. benthamiana AGO family proteins tested, NbAGO5 was also found to be highly induced in N. benthamiana infected by BaMV. We have characterized the putative promoter of NbAGO5 to understand its expression patterns in response to virus infection and identified two TFs, namely NbNAC42 and NbZFP3, for the modulation of NbAGO5 expression. The core promoter region of NbAGO5 was mapped to segment PN4, a 704 bp segment (nucleotide positions -704 to −1, upstream of the translational initiate codon of the NbAGO5 gene) which was sufficient to drive high-level expression under the induction of virus challenges. We have found that PN4 is the key region required for response to virus infections. In A. thaliana, AtAGO5 has been found to be expressed in meristems (Tucker et al., 2012) and affect the megagametogenesis process (Mi et al., 2008; Tucker et al., 2012). In N. benthamiana, GUS histochemical staining analysis revealed that GUS was highly expressed in both flowers and seeds of transgenic plants (Figure 3), suggesting the involvement of NbAGO5 during the stages of pollination and reproduction. This observation is in accordance with the finding that the putative NbAGO5 promoter region contains a CAT-BOX element that has a cis-acting regulatory function related to meristem expression.
In addition, two types of NAC protein binding sites with the core CACG and CATGTG elements (Tran et al., 2004; He et al., 2005) were identified in the putative NbAGO5 promoter region. We also confirmed in Y1H experiments that NbNAC42 has a strong interaction with the putative NbAGO5 promoter region. NAC-TFs are named after three functional NAC domain-containing genes: no apical meristem (NAM) (Souer et al., 1996), Arabidopsis thaliana Transcription Activator Factor 1/2 (ATAF1/2), and cup-shaped cotyledon (CUC) (Aida et al., 1997). NAC proteins are divided into two structural regions: one N-terminal DNA-binding domain of 151–159 amino acids and C-terminal transcriptional regulatory region that are highly divergent (Aida et al., 1997; Kikuchi et al., 2000; Jensen et al., 2010). Comparative genomic and gene functional analyzes were used to divide NAC proteins into six major orthologous groups (Groups I–VI) using 2,106 non-redundant sequences from 24 different green plant species (Pereira-Santana et al., 2015). NbNAC42 is classified as Group IV. The members of NAC group IV perform a variety of functions, such as the regulation of responses in drought, salinity (Tak et al., 2017), longevity (Wu et al., 2012), and resistance against the virus (Thirumalaikumar et al., 2018), as demonstrated for JUNGBRUNNEN1 (JUB1 or NAC042). Although many NACs have been characterized as having anti-viral ability (Ren et al., 2000; Selth et al., 2005; Donze et al., 2014; Huang et al., 2017a; Sun et al., 2019), the activation mechanisms and the downstream targets for NAC proteins remain largely unknown. The results of this study revealed that the expression of NbNAC42 could be enhanced by BaMV-TGBp1, and that NbNAC42 achieves the antiviral effect by up-regulating the expression of NbAGO5, which is involved in the RNA silencing-mediated antiviral defense system.
Zinc finger proteins have been shown to be regulators of abiotic stress responses in plants (Han et al., 2020) and divided into subgroups based on the order of Cys and His residues in their secondary structures, such as Cys2/His2-type (C2H2), C3H, C3HC4, C2HC5, C4HC3, C2HC, C4, C6, and C8 (Miller et al., 1985; Klug, 2010; Han et al., 2014). NbZFP3 is classified as C2H2-type, which could directly target genes involved in hormone signal transduction (Kodaira et al., 2011) and regulate the responses to stresses of salt (Huang et al., 2007; Ma et al., 2016), osmotic pressure (Sun et al., 2010; Han et al., 2019), reactive oxygen species (Rizhsky et al., 2004; Vogel et al., 2005; Sun et al., 2010; Liu et al., 2017), and cold (Vogel et al., 2005; Kim et al., 2016) stress. C2H2-type ZFPs have the Ethylene-responsive element-binding factor-associated amphiphilic repression (EAR) motif [with the conserved amino acid sequence of L/FDLN L/F(x)P] in the C-terminal region (Ohta et al., 2001). The EAR motif has been shown to lower the transcriptional activity of the reporter gene and other TFs (Kazan, 2006). NbZFP3 has an EAR motif starting at amino acid position 312, which specifically binds to the A(G/C)T repeat sequences (Sakamoto et al., 2004) in their target promoters. The negative correlation between the expression of NbAGO5 and NbZFP3 could be due to the binding of NbZFP3 to the NbAGO5 promoter through the EAR motif, which in turn repressed the expression of NbAGO5. This may serve as the strategy of the invading virus to suppress the defense system of the host. However, it should be noted that the induction level of NbZFP3 by the infection of BaMV or the expression of TGBp1 might not be high enough to counteract the enhancement effect on NbAGO5 expression by NbNAC42, as shown in Figure 7B.
It is worth noting that the JA-responsive transcription factor, JAMYB, in rice has been shown to activate the expression of AGO18, a core RNA silencing component, by directly binding to its promoter, thus enhancing the antiviral defense (Yang et al., 2020). It was shown that the activation of JAMYB expression is mainly by the CP of the rice stripe virus, which stimulates the accumulation of JA. In comparison, the results of our work showed that the AGO5 promoter (PN4) contains an MYB binding site; however, there appeared to be no interaction with MYB-type TFs in the screenings of the Arabidopsis TFs library (Table 1). Furthermore, infecting N. benthamiana with BaMV or CMV increased ABA levels and activated the SA and ABA pathways, reversing the antagonistic relationship between these two pathways (Alazem et al., 2014). Therefore, this means that the AGO family with antiviral ability is diversified in the mechanisms of initiating antiviral defense. This also implies that, as plants evolve, different defense mechanisms could be activated in response to different virus threats. In Figure 7C, the result revealed that potexviruses may trigger the defense response by inducing the expression of NbNAC42. For BaMV infections, we showed that BaMV- TGBp1 may serve as an activator for NbNAC42 expression (Figure 7B). We also tested the effect of other viruses on the expression of NbNAC42, such as CMV and TMV (Figure 7C). Thus, the expression of NbNAC42 may be sensitive to various virus infections, and in turn, may enhance the expression of NbAGO5 to defend against viruses.
In addition to the contribution to the understanding of antiviral defense mechanisms, the virus-inducible NbAGO5 promoter may have practical applications in plant biotechnology. Currently, the commonly used promoters for the expression of foreign genes in plants are constituent promoters, such as the CaMV35S promoter and the maize ubiquitin promoter. These constituent promoters are usually active in almost all tissues and developmental stages of plants, which might result in excessive energy loss and possible morphological and physiological dysfunction. In contrast, inducible or tissue-specific promoters may regulate the expression of target genes in particular conditions or tissues, which is more energy conservative and easier for the maintenance of normal physiological functions of plants. In this study, we have demonstrated that the NbAGO5 promoter could be positively and negatively regulated by NbNAC42 and NbZFP3, respectively, in response to virus infections or hormone treatment (such as ABA, Figure 4B). Thus, this viral stress-inducible promoter would be an ideal candidate for the controlled overexpression of foreign proteins for academic or industrial purposes.
Overall, this study demonstrated the involvement of NbAGO5 in the defense against specific viruses in N. benthamiana, and revealed the underlying mechanism for the modulation of NbAGO5 expression by two antagonizing TFs, NbNAC42, and NbZFP3, in response to specific virus infections or the presence of specific viral protein, such as BaMV-TGBp1. These findings provided further insights into the chain of command in the antiviral defense system in plants and may be further applied in the fields of crop improvement or plant biotechnology.
Data Availability Statement
The original contributions presented in this study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
Y-DK and KV performed the isolation and functional analysis of the NbAGO5 promoter. C-MY and NM performed the yeast one hybrid screening analysis. Y-DK and Y-WH revealed the underlying mechanism for the modulation of NbAGO5 expression by two antagonizing TFs in the defense against viruses. Y-DK drafted the manuscript. C-CH, C-MY, N-SL, and Y-HH helped to draft the article structure of the manuscript. Y-HH supervised the entire study and organized the participation of each lab participant. All authors read and approved the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Fumie Tobe for Y1H screening. Chin-Wei Lee for her assistance in the construction of the plasmids; Meng-Jhe Luo and Chu-I Sun for their assistance in qRT-PCR analysis.
Footnotes
Funding
This work was financially supported by the Ministry of Science and Technology, Taiwan (MOST-110-2313-B-005-019) and the Advanced Plant Biotechnology Center from The Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.924482/full#supplementary-material
References
- Aida M., Ishida T., Fukaki H., Fujisawa H., Tasaka M. (1997). Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell 9 841–857. 10.1105/tpc.9.6.841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alazem M., He M. H., Moffett P., Lin N. S. (2017). Abscisic acid induces resistance against bamboo mosaic virus through argonaute2 and 3. Plant Physiol. 174 339–355. 10.1104/pp.16.00015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alazem M., Kim K. H., Lin N. S. (2019). Effects of abscisic acid and salicylic acid on gene expression in the antiviral RNA silencing pathway in arabidopsis. Int. J. Mol. 20:2538. 10.3390/ijms20102538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alazem M., Lin K. Y., Lin N. S. (2014). The abscisic acid pathway has multifaceted effects on the accumulation of Bamboo mosaic virus. Mol. Plant Microbe Interact. 27 177–189. 10.1094/MPMI-08-13-0216-R [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Bai M., Yang G. S., Chen W. T., Mao Z. C., Kang H. X., Chen G. H., et al. (2012). Genome-wide identification of Dicer-like, Argonaute and RNA-dependent RNA polymerase gene families and their expression analyses in response to viral infection and abiotic stresses in Solanum lycopersicum. Gene 501 52–62. 10.1016/j.gene.2012.02.009 [DOI] [PubMed] [Google Scholar]
- Baulcombe D. (1999). Viruses and gene silencing in plants. Arch. Virol. Suppl. 15 189–201. 10.1007/978-3-7091-6425-9_14 [DOI] [PubMed] [Google Scholar]
- Bhattacharjee S., Zamora A., Azhar M. T., Sacco M. A., Lambert L. H., Moffett P. (2009). Virus resistance induced by NB-LRR proteins involves Argonaute4-dependent translational control. Plant J. 58 940–951. 10.1111/j.1365-313X.2009.03832.x [DOI] [PubMed] [Google Scholar]
- Bradford M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72 248–254. 10.1006/abio.1976.9999 [DOI] [PubMed] [Google Scholar]
- Brosseau C., Moffett P. (2015). Functional and genetic analysis identify a role for arabidopsis ARGONAUTE5 in antiviral RNA silencing. Plant Cell 27 1742–1754. 10.1105/tpc.15.00264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonell A. (2017). Plant ARGONAUTEs: features, Functions, and Unknowns. Methods Mol. Biol. 1640 1–21. 10.1007/978-1-4939-7165-7_1 [DOI] [PubMed] [Google Scholar]
- Carbonell A., Carrington J. C. (2015). Antiviral roles of plant ARGONAUTES. Curr. Opin. Plant Biol. 27 111–117. 10.1016/j.pbi.2015.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Zhang L., Li D., Wang F., Yu D. (2013). WRKY8 transcription factor functions in the TMV-cg defense response by mediating both abscisic acid and ethylene signaling in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 110 E1963–E1971. 10.1073/pnas.1221347110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Csorba T., Kontra L., Burgyan J. (2015). viral silencing suppressors: tools forged to fine-tune host-pathogen coexistence. Virology 47 85–103. [DOI] [PubMed] [Google Scholar]
- Diao P., Zhang Q., Sun H., Ma W., Cao A., Yu R., et al. (2019). miR403a and SA are involved in NbAGO2 mediated antiviral defenses against TMV Infection in Nicotiana benthamiana. Genes 10:526. 10.3390/genes10070526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S. W., Rathjen J. P., Li W. X., Swanson R., Healy H., Symons R. H. (1995). Efficient infection from cDNA clones of cucumber mosaic cucumovirus RNAs in a new plasmid vector. J. Gen. Virol. 76(Pt 2), 459–464. 10.1099/0022-1317-76-2-459 [DOI] [PubMed] [Google Scholar]
- Donze T., Qu F., Twigg P., Morris T. J. (2014). Turnip crinkle virus coat protein inhibits the basal immune response to virus invasion in Arabidopsis by binding to the NAC transcription factor TIP. Virology 449 207–214. 10.1016/j.virol.2013.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X., Qi Y. (2016). RNAi in Plants: an argonaute-centered view. Plant Cell 28 272–285. 10.1105/tpc.15.00920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatyol K., Ludman M., Burgyan J. (2016). Functional dissection of a plant Argonaute. Nucleic Acids Res. 44 1384–1397. 10.1093/nar/gkv1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaguancela O. A., Zuniga L. P., Arias A. V., Halterman D., Flores F. J., Johansen I. E., et al. (2016). The IRE1/bZIP60 pathway and bax inhibitor 1 suppress systemic accumulation of potyviruses and potexviruses in arabidopsis and nicotiana benthamiana plants. Mol. Plant Microbe Interact. 29 750–766. 10.1094/MPMI-07-16-0147-R [DOI] [PubMed] [Google Scholar]
- Gayral M., Arias Gaguancela O., Vasquez E., Herath V., Flores F. J., Dickman M. B., et al. (2020). Multiple ER-to-nucleus stress signaling pathways are activated during Plantago asiatica mosaic virus and Turnip mosaic virus infection in Arabidopsis thaliana. Plant J. 103 1233–1245. 10.1111/tpj.14798 [DOI] [PubMed] [Google Scholar]
- Ghoshal B., Sanfacon H. (2014). Temperature-dependent symptom recovery in Nicotiana benthamiana plants infected with tomato ringspot virus is associated with reduced translation of viral RNA2 and requires ARGONAUTE 1. Virology 45 188–197. 10.1016/j.virol.2014.03.026 [DOI] [PubMed] [Google Scholar]
- Goodin M. M., Zaitlin D., Naidu R. A., Lommel S. A. (2008). Nicotiana benthamiana: its history and future as a model for plant-pathogen interactions. Mol. Plant Microbe Interact. 21 1015–1026. 10.1094/Mpmi-21-8-1015 [DOI] [PubMed] [Google Scholar]
- Guo X., Carroll J. W., Macdonald M. R., Goff S. P., Gao G. (2004). The zinc finger antiviral protein directly binds to specific viral mRNAs through the CCCH zinc finger motifs. J. Virol. 78 12781–12787. 10.1128/JVI.78.23.12781-12787.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gursinsky T., Pirovano W., Gambino G., Friedrich S., Behrens S. E., Pantaleo V. (2015). Homeologs of the nicotiana benthamiana antiviral ARGONAUTE1 show different susceptibilities to microRNA168-Mediated Control. Plant Physiol. 168 938–952. 10.1104/pp.15.00070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han G., Lu C., Guo J., Qiao Z., Sui N., Qiu N., et al. (2020). C2H2 zinc finger proteins: master regulators of abiotic stress responses in plants. Front. Plant Sci. 11:115. 10.3389/fpls.2020.00115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han G., Wang M., Yuan F., Sui N., Song J., Wang B. (2014). The CCCH zinc finger protein gene AtZFP1 improves salt resistance in Arabidopsis thaliana. Plant Mol. Biol. 86 237–253. 10.1007/s11103-014-0226-5 [DOI] [PubMed] [Google Scholar]
- Han G., Yuan F., Guo J., Zhang Y., Sui N., Wang B. (2019). AtSIZ1 improves salt tolerance by maintaining ionic homeostasis and osmotic balance in Arabidopsis. Plant Sci. 285 55–67. 10.1016/j.plantsci.2019.05.002 [DOI] [PubMed] [Google Scholar]
- He X. J., Mu R. L., Cao W. H., Zhang Z. G., Zhang J. S., Chen S. Y. (2005). AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J. 44 903–916. 10.1111/j.1365-313X.2005.02575.x [DOI] [PubMed] [Google Scholar]
- Horsch R. B., Rogers S. G., Fraley R. T. (1985). Transgenic plants. Cold Spring Harb. Symp. Quant. Biol. 50 433–437. 10.1101/sqb.1985.050.01.054 [DOI] [PubMed] [Google Scholar]
- Huang J., Yang X., Wang M. M., Tang H. J., Ding L. Y., Shen Y., et al. (2007). A novel rice C2H2-type zinc finger protein lacking DLN-box/EAR-motif plays a role in salt tolerance. Biochim. Biophys. Acta 1769 220–227. 10.1016/j.bbaexp.2007.02.006 [DOI] [PubMed] [Google Scholar]
- Huang Y. W., Hu C. C., Tsai C. H., Lin N. S., Hsu Y. H. (2017b). Chloroplast Hsp70 isoform is required for age-dependent tissue preference of bamboo mosaic virus in mature nicotiana benthamiana leaves. Mol. Plant Microbe Interact. 30 631–645. 10.1094/MPMI-01-17-0012-R [DOI] [PubMed] [Google Scholar]
- Huang Y., Li T., Xu Z. S., Wang F., Xiong A. S. (2017a). Six NAC transcription factors involved in response to TYLCV infection in resistant and susceptible tomato cultivars. Plant Physiol. Biochem. 120 61–74. 10.1016/j.plaphy.2017.09.020 [DOI] [PubMed] [Google Scholar]
- Huang Y., Zhang B. L., Sun S., Xing G. M., Wang F., Li M. Y., et al. (2016). AP2/ERF transcription factors involved in response to tomato yellow leaf curly virus in tomato. Plant Genome 9 1–15. 10.3835/plantgenome2015.09.0082 [DOI] [PubMed] [Google Scholar]
- Huang Y. W., Chang C. Y., Hsu Y. H. (2020). Virus-Induced gene silencing in poaceae using a foxtail mosaic virus vector. Methods Mol. Biol. 2172 15–25. 10.1007/978-1-0716-0751-0_2 [DOI] [PubMed] [Google Scholar]
- Huang Y. W., Hu C. C., Liou M. R., Chang B. Y., Tsai C. H., Meng M., et al. (2012). Hsp90 interacts specifically with viral RNA and differentially regulates replication initiation of Bamboo mosaic virus and associated satellite RNA. PLoS Pathog. 8:e1002726. 10.1371/journal.ppat.1002726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. W., Hu C. C., Tsai C. H., Lin N. S., Hsu Y. H. (2019). Nicotiana benthamiana Argonaute10 plays a pro-viral role in Bamboo mosaic virus infection. New Phytol. 224 804–817. 10.1111/nph.16048 [DOI] [PubMed] [Google Scholar]
- Huang Y. W., Sun C. I., Hu C. C., Tsai C. H., Meng M., Lin N. S., et al. (2021). NbPsbO1 interacts specifically with the Bamboo Mosaic Virus (BaMV) subgenomic RNA (sgRNA) promoter and is required for efficient BaMV sgRNA transcription. J. Virol. 95:e0083121. 10.1128/JVI.00831-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh S. U., Kim M. J., Ham B. K., Paek K. H. (2011). A zinc finger protein Tsip1 controls Cucumber mosaic virus infection by interacting with the replication complex on vacuolar membranes of the tobacco plant. New Phytol. 191 746–762. 10.1111/j.1469-8137.2011.03717.x [DOI] [PubMed] [Google Scholar]
- Jefferson R. A., Kavanagh T. A., Bevan M. W. (1987). GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6 3901–3907. 10.1002/j.1460-2075.1987.tb02730.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen M. K., Kjaersgaard T., Nielsen M. M., Galberg P., Petersen K., O’Shea C., et al. (2010). The Arabidopsis thaliana NAC transcription factor family: structure-function relationships and determinants of ANAC019 stress signalling. Biochem. J. 426 183–196. 10.1042/BJ20091234 [DOI] [PubMed] [Google Scholar]
- Kazan K. (2006). Negative regulation of defence and stress genes by EAR-motif-containing repressors. Trends Plant Sci. 11 109–112. 10.1016/j.tplants.2006.01.004 [DOI] [PubMed] [Google Scholar]
- Kikuchi K., Ueguchi-Tanaka M., Yoshida K. T., Nagato Y., Matsusoka M., Hirano H. Y. (2000). Molecular analysis of the NAC gene family in rice. Mol. Gen. Genet. 262 1047–1051. 10.1007/pl00008647 [DOI] [PubMed] [Google Scholar]
- Kim Y. H., Kim M. D., Park S. C., Jeong J. C., Kwak S. S., Lee H. S. J. P. B. (2016). Transgenic potato plants expressing the cold-inducible transcription factor SCOF-1 display enhanced tolerance to freezing stress. Plant Breed. 135 513–518. [Google Scholar]
- Klug A. (2010). The discovery of zinc fingers and their applications in gene regulation and genome manipulation. Annu. Rev. Biochem. 79 213–231. 10.1146/annurev-biochem-010909-095056 [DOI] [PubMed] [Google Scholar]
- Kodaira K. S., Qin F., Tran L. S., Maruyama K., Kidokoro S., Fujita Y., et al. (2011). Arabidopsis Cys2/His2 zinc-finger proteins AZF1 and AZF2 negatively regulate abscisic acid-repressive and auxin-inducible genes under abiotic stress conditions. Plant Physiol. 157 742–756. 10.1104/pp.111.182683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotnik T. (2013). Lightning-triggered electroporation and electrofusion as possible contributors to natural horizontal gene transfer. Phys. Life Rev. 10 351–370. 10.1016/j.plrev.2013.05.001 [DOI] [PubMed] [Google Scholar]
- Kuo S. Y., Hu C. C., Huang Y. W., Lee C. W., Luo M. J., Tu C. W., et al. (2021). Argonaute 5 family proteins play crucial roles in the defence against Cymbidium mosaic virus and Odontoglossum ringspot virus in Phalaenopsis aphrodite subsp. formosana. Mol. Plant Pathol. 22 627–643. 10.1111/mpp.13049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescot M., Dehais P., Thijs G., Marchal K., Moreau Y., Van de Peer Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30 325–327. 10.1093/nar/30.1.325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin N. S., Lin B. Y., Lo N. W., Hu C. C., Chow T. Y., Hsu Y. H. (1994). Nucleotide sequence of the genomic RNA of bamboo mosaic potexvirus. J. Gen. Virol. 75(Pt 9), 2513–2518. 10.1099/0022-1317-75-9-2513 [DOI] [PubMed] [Google Scholar]
- Liou M. R., Huang Y. W., Hu C. C., Lin N. S., Hsu Y. H. (2014). A dual gene-silencing vector system for monocot and dicot plants. Plant Biotechnol. J. 12 330–343. 10.1111/pbi.12140 [DOI] [PubMed] [Google Scholar]
- Liu D., Yang L., Luo M., Wu Q., Liu S., Liu Y. (2017). Molecular cloning and characterization of PtrZPT2-1, a ZPT2 family gene encoding a Cys2/His2-type zinc finger protein from trifoliate orange (Poncirus trifoliata (L.) Raf.) that enhances plant tolerance to multiple abiotic stresses. Plant Sci. 263 66–78. 10.1016/j.plantsci.2017.07.012 [DOI] [PubMed] [Google Scholar]
- Ludman M., Burgyan J., Fatyol K. (2017). Crispr/Cas9 mediated inactivation of argonaute 2 reveals its differential involvement in antiviral responses. Sci. Rep. 7:1010. 10.1038/s41598-017-01050-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Liang W., Gu P., Huang Z. (2016). Salt tolerance function of the novel C2H2-type zinc finger protein TaZNF in wheat. Plant Physiol. Biochem. 106 129–140. 10.1016/j.plaphy.2016.04.033 [DOI] [PubMed] [Google Scholar]
- Meister G. (2013). Argonaute proteins: functional insights and emerging roles. Nat. Rev. Genet. 14 447–459. 10.1038/nrg3462 [DOI] [PubMed] [Google Scholar]
- Mi S., Cai T., Hu Y., Chen Y., Hodges E., Ni F., et al. (2008). Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5’ terminal nucleotide. Cell 133 116–127. 10.1016/j.cell.2008.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J., McLachlan A. D., Klug A. (1985). Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 4 1609–1614. 10.1002/j.1460-2075.1985.tb03825.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda N., Ikeda M., Takada S., Takiguchi Y., Kondou Y., Yoshizumi T., et al. (2010). Efficient yeast one-/two-hybrid screening using a library composed only of transcription factors in Arabidopsis thaliana. Plant Cell Physiol. 51 2145–2151. 10.1093/pcp/pcq161 [DOI] [PubMed] [Google Scholar]
- Odokonyero D., Mendoza M. R., Moffett P., Scholthof H. B. (2017). Tobacco Rattle Virus (TRV)-mediated silencing of nicotiana benthamiana ARGONAUTES (NbAGOs) reveals new antiviral candidates and dominant effects of TRV-NbAGO1. Phytopathology 107 977–987. 10.1094/PHYTO-02-17-0049-R [DOI] [PubMed] [Google Scholar]
- Ohta M., Matsui K., Hiratsu K., Shinshi H., Ohme-Takagi M. (2001). Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13 1959–1968. 10.1105/tpc.010127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C. J., Shin Y. C., Lee B. J., Kim K. J., Kim J. K., Paek K. H. (2006). A hot pepper gene encoding WRKY transcription factor is induced during hypersensitive response to Tobacco mosaic virus and Xanthomonas campestris. Planta 223 168–179. 10.1007/s00425-005-0067-1 [DOI] [PubMed] [Google Scholar]
- Paudel D. B., Ghoshal B., Jossey S., Ludman M., Fatyol K., Sanfacon H. (2018). Expression and antiviral function of ARGONAUTE 2 in Nicotiana benthamiana plants infected with two isolates of tomato ringspot virus with varying degrees of virulence. Virology 524 127–139. 10.1016/j.virol.2018.08.016 [DOI] [PubMed] [Google Scholar]
- Pereira-Santana A., Alcaraz L. D., Castano E., Sanchez-Calderon L., Sanchez-Teyer F., Rodriguez-Zapata L. (2015). Comparative Genomics of NAC Transcriptional Factors in Angiosperms: implications for the adaptation and diversification of flowering plants. PLoS One 10:e0141866. 10.1371/journal.pone.0141866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth K. R., Huang Y. W., Liou M. R., Wang R. Y., Hu C. C., Tsai C. H., et al. (2011). Glyceraldehyde 3-phosphate dehydrogenase negatively regulates the replication of Bamboo mosaic virus and its associated satellite RNA. J. Virol. 85 8829–8840. 10.1128/JVI.00556-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliff F., Martin-Hernandez A. M., Baulcombe D. C. (2001). Technical advance. tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J. 25 237–245. 10.1046/j.0960-7412.2000.00942.x [DOI] [PubMed] [Google Scholar]
- Ren T., Qu F., Morris T. J. (2000). HRT gene function requires interaction between a NAC protein and viral capsid protein to confer resistance to turnip crinkle virus. Plant Cell 12 1917–1926. 10.1105/tpc.12.10.1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizhsky L., Davletova S., Liang H., Mittler R. (2004). The zinc finger protein Zat12 is required for cytosolic ascorbate peroxidase 1 expression during oxidative stress in Arabidopsis. J. Biol. Chem. 279 11736–11743. 10.1074/jbc.M313350200 [DOI] [PubMed] [Google Scholar]
- Sakamoto H., Maruyama K., Sakuma Y., Meshi T., Iwabuchi M., Shinozaki K., et al. (2004). Arabidopsis Cys2/His2-type zinc-finger proteins function as transcription repressors under drought, cold, and high-salinity stress conditions. Plant Physiol. 136 2734–2746. 10.1104/pp.104.046599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selth L. A., Dogra S. C., Rasheed M. S., Healy H., Randles J. W., Rezaian M. A. (2005). A NAC domain protein interacts with tomato leaf curl virus replication accessory protein and enhances viral replication. Plant Cell 17 311–325. 10.1105/tpc.104.027235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng Y., Yang L., Li C., Wang Y., Guo H. (2019). Transcriptomic changes in Nicotiana tabacum leaves during mosaic virus infection. 3 Biotech 9:220. 10.1007/s13205-019-1740-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souer E., van Houwelingen A., Kloos D., Mol J., Koes R. (1996). The no apical meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 85 159–170. 10.1016/s0092-8674(00)81093-4 [DOI] [PubMed] [Google Scholar]
- Sun D., Zhang X., Zhang Q., Ji X., Jia Y., Wang H., et al. (2019). Comparative transcriptome profiling uncovers a Lilium regale NAC transcription factor, LrNAC35, contributing to defence response against cucumber mosaic virus and tobacco mosaic virus. Mol. Plant Pathol. 20 1662–1681. 10.1111/mpp.12868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S. J., Guo S. Q., Yang X., Bao Y. M., Tang H. J., Sun H., et al. (2010). Functional analysis of a novel Cys2/His2-type zinc finger protein involved in salt tolerance in rice. J. Exp. Bot. 61 2807–2818. 10.1093/jxb/erq120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tak H., Negi S., Ganapathi T. R. (2017). Banana NAC transcription factor MusaNAC042 is positively associated with drought and salinity tolerance. Protoplasma 254 803–816. 10.1007/s00709-016-0991-x [DOI] [PubMed] [Google Scholar]
- Takeda A., Iwasaki S., Watanabe T., Utsumi M., Watanabe Y. (2008). The mechanism selecting the guide strand from small RNA duplexes is different among argonaute proteins. Plant Cell Physiol. 49 493–500. 10.1093/pcp/pcn043 [DOI] [PubMed] [Google Scholar]
- Thirumalaikumar V. P., Devkar V., Mehterov N., Ali S., Ozgur R., Turkan I., et al. (2018). NAC transcription factor JUNGBRUNNEN1 enhances drought tolerance in tomato. Plant Biotechnol. J. 16 354–366. 10.1111/pbi.12776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran L. S., Nakashima K., Sakuma Y., Simpson S. D., Fujita Y., Maruyama K., et al. (2004). Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16 2481–2498. 10.1105/tpc.104.022699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker M. R., Okada T., Hu Y., Scholefield A., Taylor J. M., Koltunow A. M. (2012). Somatic small RNA pathways promote the mitotic events of megagametogenesis during female reproductive development in Arabidopsis. Development 139 1399–1404. 10.1242/dev.075390 [DOI] [PubMed] [Google Scholar]
- van Tunen A. J., Hartman S. A., Mur L. A., Mol J. N. (1989). Regulation of chalcone flavanone isomerase (CHI) gene expression inPetunia hybrida: the use of alternative promoters in corolla, anthers and pollen. Plant Mol. Biol. 12 539–551. 10.1007/BF00036968 [DOI] [PubMed] [Google Scholar]
- Vaucheret H. (2008). Plant ARGONAUTES. Trends Plant Sci. 13 350–358. 10.1016/j.tplants.2008.04.007 [DOI] [PubMed] [Google Scholar]
- Vogel J. T., Zarka D. G., Van Buskirk H. A., Fowler S. G., Thomashow M. F. (2005). Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J. 41 195–211. 10.1111/j.1365-313X.2004.02288.x [DOI] [PubMed] [Google Scholar]
- Wu A., Allu A. D., Garapati P., Siddiqui H., Dortay H., Zanor M. I., et al. (2012). JUNGBRUNNEN1, a reactive oxygen species-responsive NAC transcription factor, regulates longevity in Arabidopsis. Plant Cell 24 482–506. 10.1105/tpc.111.090894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Klessig D. F. (1996). Isolation and characterization of a tobacco mosaic virus-inducible myb oncogene homolog from tobacco. Proc. Natl. Acad. Sci.U.S.A. 93 14972–14977. 10.1073/pnas.93.25.14972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Huang Y., Yang J., Yao S., Zhao K., Wang D. (2020). Jasmonate signaling enhances RNA silencing and antiviral defense in rice. Cell Host Microbe 28 89–103. 10.1016/j.chom.2020.05.001 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in this study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.