Abstract
Background
The roots of Panax ginseng contain two types of tetracyclic triterpenoid saponins, namely, protopanaxadiol (PPD)-type saponins and protopanaxatiol (PPT)-type saponins. In P. ginseng, the protopanaxadiol 6-hydroxylase (PPT synthase) enzyme catalyses protopanaxatriol (PPT) production from protopanaxadiol (PPD). In this study, we constructed homozygous mutant lines of ginseng by CRISPR/Cas9-mediated mutagenesis of the PPT synthase gene and obtained the mutant ginseng root lines having complete depletion of the PPT-type ginsenosides.
Methods
Two sgRNAs (single guide RNAs) were designed for target mutations in the exon sequences of the two PPT synthase genes (both PPTa and PPTg sequences) with the CRISPR/Cas9 system. Transgenic ginseng roots were generated through Agrobacterium-mediated transformation. The mutant lines were screened by ginsenoside analysis and DNA sequencing.
Result
Ginsenoside analysis revealed the complete depletion of PPT-type ginsenosides in three putative mutant lines (Cr4, Cr7, and Cr14). The reduction of PPT-type ginsenosides in mutant lines led to increased accumulation of PPD-type ginsenosides. The gene editing in the selected mutant lines was confirmed by targeted deep sequencing.
Conclusion
We have established the genome editing protocol by CRISPR/Cas9 system in P. ginseng and demonstrated the mutated roots producing only PPD-type ginsenosides by depleting PPT-type ginsenosides. Because the pharmacological activity of PPD-group ginsenosides is significantly different from that of PPT-group ginsenosides, the new type of ginseng mutant producing only PPD-group ginsenosides may have new pharmacological characteristics compared to wild-type ginseng. This is the first report to generate target-induced mutations for the modification of saponin biosynthesis in Panax species using CRISPR–Cas9 system.
Keywords: CRISPR/Cas9 system, construction of mutant, genetic transformation, ginsenoside, protopanaxadiol 6-hydroxylase, saponins, sgRNA, triterpene
Abbreviations: CYP, cytochrome P450; qPCR, quantitative real-time RT-PCR; RNAi, RNA interference; RT-PCR, reverse transcription-PCR; CRISPR/Cas9, clustered regularly interspaced short palindromic repeats associated with nuclease Cas9; sgRNA, single guide RNA
Graphical abstract
We generated new mutant ginseng root lines by changing the ginsenoside saponin production using the CRISPR/Cas9 knock-out system. Ginsenoside analysis revealed the complete or partial depletion of PPT-type ginsenosides in mutant lines. Furthermore, the reduction of PPT-type ginsenosides in mutant lines led to increased accumulation of PPD-type saponins. The new type of ginseng mutants producing only PPD-group ginsenosides may have different pharmacological characteristics compared to wild-type ginseng and new sources for ginseng breeding.
1. Introduction
Triterpenoid saponins are present in many higher plants and exhibit a wide range of saponin structures, depending on their aglycone structures and sugar moieties. Certain triterpenoid saponins also have commercial value and are utilized as drugs and medicines [[1], [2], [3]]. The roots of ginseng (Panax ginseng Meyer) contain pharmacologically active compounds, among which ginsenoside saponins are the primary compounds responsible for the pharmacological activities [2,3]. Ginsenosides are classified as protopanaxadiol (PPD)- or protopanaxatriol (PPT)-type saponins based on their aglycone structures. PPD-type ginsenosides have glycosidic bonds at the C-3 and C-20 hydroxyl groups of the PPD aglycone. PPT-group ginsenosides have glycosidic bonds at the C-6 and C-20 hydroxyl groups of the PPT aglycone. The PPD-type ginsenosides include the ginsenosides Rb1, Rc, Rd, Rg3, and Rh2. The PPT-type ginsenosides include the ginsenosides Re, Rf, Rg1, Rg2, and Rh1.
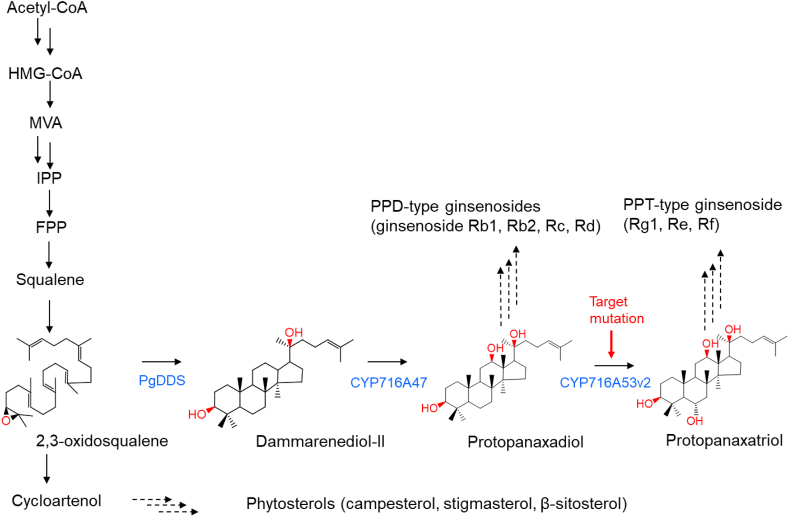
The first step in ginsenoside saponin biosynthesis begins with the enzyme activity of squalene synthase producing squalene, which is converted to 2,3-oxidosqualene by 2,3-oxidosqualene synthase. Dammarenediol-II enzyme catalyses the conversion of 2,3-oxidosqualene into dammarenediol-II, a unique triterpene structure found in Panax species [4]. Dammarenediol 12-hydroxylase (CYP716A47) catalyses the production of PPD from dammarenediol-II [5]. The production of PPT from PPD is catalysed by protopanaxadiol 6-hydroxylase (CYP716A53v2) [6]. The two compounds (PPD and PPT) are basic aglycones of dammarene-type ginsenoside saponins and are subsequently glycosylated by various types of glycosyltransferases (Fig. 1) [7].
Fig. 1.
Biosynthetic pathway for ginsenosides in P. ginseng. Squalene epoxidase converts squalene to 2,3-oxidosqualene, which is subsequently converted to a triterpene aglycone (dammarenediol-II) by dammarenediol synthase. Dammarenediol-II undergoes oxidation and glycosylation and is finally converted to PPD- and PPT-group ginsenosides.
Ginsenosides have cancer-preventing activities against various tumours [8]. Although the anticancer activities of ginsenosides are related to the number and position of hydroxyl groups in ginsenoside aglycones, the number and kinds of sugar molecules bounded aglycones, and stereoselectivity [9], PPD-group ginsenosides have exhibited more powerful anticancer activity than PPT-group ginsenosides [[10], [11], [12], [13]]. The ginsenosides Rg3 and Rh2, which belong to the PPD-group ginsenosides, are known to have strong anticancer activities [14]. PPD aglycone, a hydrolyzed sapogenin belonging to the PPD-group of ginsenosides, has the most potent anticancer properties [15]. Thus, ginseng plants with increased production of PPD-group ginsenosides may exhibit particular medicinal properties, such as anticancer activities.
Genome editing is a powerful tool for increasing plant yields, improving food quality, enhancing crop disease resistance and developing new cultivars to meet market demands [16]. Since the CRISPR-Cas9 system was successfully adopted to edit plant genomes in 2013, numerous efforts have attempted to utilize it to transform living organisms. The CRISPR-Cas9 system constitutes a revolutionary tool for functional gene analysis, and it is also a promising approach for the development of new traits of interest in plants [17]. Recently, the CRISPR/Cas9 method has been rapidly developed as a useful tool for targeted genetic modification in various plant species. However, no report has been published for induction of mutant using CRISPR/Cas9 method in Panax species.
The two CYP genes (CYP716A47 and CYP716A53v2) play critical roles in the production of basic aglycones (PPD and PPT, respectively) in P. ginseng. We postulated that knocking out the CYP716A53v2 gene may clearly alter the composition of ginsenosides because all PPT-type ginsenosides are derived from the activity of the PPT synthase enzyme. In this report, we describe the first successful knockout of the PPT synthase gene by targeted mutagenesis using the CRISPR-Cas9 system in P. ginseng. We designed sgRNAs for targeted mutation of the PPT synthase gene. Transgenic ginseng root lines were generated via Agrobacterium-mediated transformation with the CRISPR-Cas9 system vector. Saponin analysis demonstrated that PPT-type ginsenosides were completely reduced or depleted in the roots by mutation of the PPT synthase gene in P. ginseng. This article is the first report for the CRISPR/Cas9-mediated mutation in P. ginseng.
2. Materials and methods
2.1. Design of sgRNAs in exon sequences of the CYP716A53v2 gene
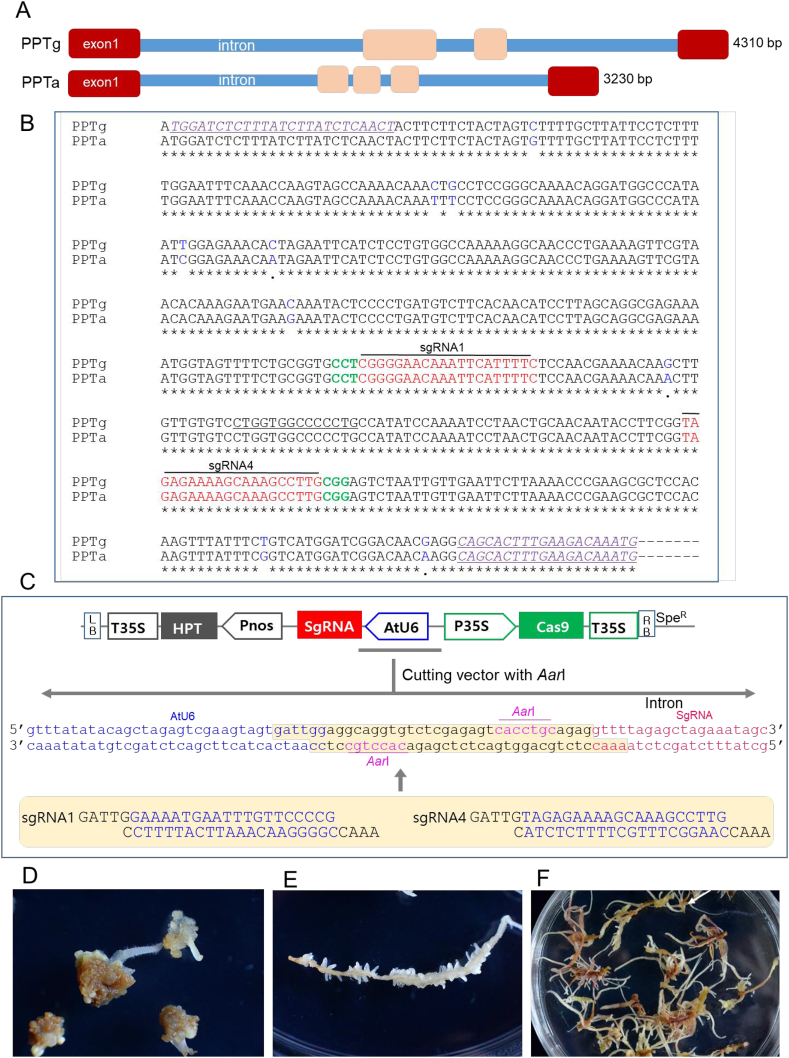
P. ginseng plants contain two types of sequences encoding for PPT synthase gene. The sequence of CYP716A53v2 gene, characterized as PPT synthase gene (protopanaxadiol 6-hydroxylase), was registered in GenBank (accession number, JX036031). A putative paralog gene was registered as a partial sequence (GenBank accession numbers, GAAG01029440.1 and GAAG01029441.1). In the Ginseng Genome Database accessed at http://ginsengdb.snu.ac.kr/, two genomic sequences for PPT synthase genes (Fig. S1) were retrieved from the genomic nucleotide sequence of Pg_scaffold1770 and Pg_scaffold0325. The genomic sequence for the CYP716A53v2 gene was named PPTg and another sequence for a paralog was named PPTa (Fig. S1). sgRNAs (sgRNA1 and sgRNA4) were designed using the common nucleotide region of both PPTa and PPTg sequences in 473-bp exon 1 fragment (Fig. 2B). Two target sites for sgRNAs (named sgRNA1 and sgRNA4) were designed using the two sequences (Fig. 2B). These target sequences were selected by first identifying the NGG protospacer adjacent motif (PAM) sequence required for Streptococcus pyogenes Cas9 and then capturing the 20 nucleotides immediately upstream of the PAM sequence for use as the spacer in the sgRNAs.
Fig. 2.
Selection of target sites in the PPT synthase gene sequence, CRISPR/Cas9 binary vector construction, and ginseng transformation. (A) Intron and exon structure of PPTg and PPTg sequences. The first exon regions (exon 1) with 473 bp containing start codon in both PPTg and PPTg were highly homologous. (B) Locations of two target sites (sgRNA1 and sgRNA4) for CRISPR/Cas9-mediated target mutagenesis in the N-terminal site of PPTa and PPTg exon sequences. sgRNA1 (designed using the reverse-frame sequence) and sgRNA4 are marked in red, PAM sequences are marked in green, and primers used for sequencing are underlined and purple. Nine blue letters are single nucleotide polymorphisms between the PPTa and PPTg sequences. (C) Schematic representation of the CRISPR/Cas9 binary vector used for ginseng transformation. Arabidopsis thaliana promoters and terminators drive the expression of gRNA1 (AtU6–26p and AtU6–26t) and gRNA4 (AtU6–29p and AtU6–29t). The caulifower mosaic virus promoter (CaMV 35S) drives the expression of the Cas9 gene. The two sgRNAs were inserted into vectors using AarI enzyme sites. (D) Formation of new adventitious roots on the surfaces of hygromycin-resistant callus derived from root segments on medium with 2 mg/l IBA and 20 mg/l hygromycin. (E) Proliferation of an excised single root by induction of lateral root formation on medium with 2 mg/l IBA. (F) Proliferation of adventitious root masses from a single root for further analysis.
2.2. In vitro assay to test sgRNA cleavage efficiency
All steps for the in vitro sgRNA cleavage assay were performed according to the manufacturer's instructions provided in the Guide-it Complete sgRNA Screening System (Takara Clontech, Japan) with several modifications, as follows: the cleavage reaction mixture consisted of 100 ng of cleavage template, 20 ng of synthesized sgRNA, and 250 ng of Cas9 nuclease in 10 μl of 1x Cas9 reaction buffer with 1x BSA. Also, the primers used for amplifying cleavage templates were forward primer 5′- CAA AAT GAA TTT GTT CCC CGA GG3′, reverse primer 5′- CTG CAA CGC TGA GTA ACC AA-3’. The efficiency of Cas9-mediated cleavage was measured by agarose gel electrophoresis.
2.3. Vector construction
A pHAtC binary vector (accession number KU213971) carrying the Cas9 gene (driven by the CaMV 35S promoter), gRNA (driven by the AtU6–29p promoter), and HPT (driven by the nos promoter) scaffolds was used (Fig. 1C). To generate the construct expressing sgRNA1, we designed two oligomers: the forward (5′-GAT TGG AAA ATG AAT TTG TTC CCC G-3′) oligomer and the reverse (5′-AAA CCG GGG AAC AAA TTC ATT TTC C C-3′) oligomer. To generate the construct expressing sgRNA4, we used two oligomer pairs: the forward (5′-GAT TGT AGA GAA AAG CAA AGC CTT G-3′) and oligomer and the reverse (5′-AAA CCA AGG CTT TGC TTT TCT CTA C-3′) oligomer. Two oligo strands together were annealed in equal molar concentrations (100 μM) with the following conditions: initial denaturation at 95 °C for 10 min, decreasing 1 °C per cycle up to 60 °C, incubation at 60 °C for 10 min, decreasing 1 °C per cycle up to 20 °C, and holding at 4 °C for stabilization. These annealed oligomers were ligated to the pHAtC vector digested to AarI and transformed into DH5α-competent Escherichia coli cells on LB plates containing 50 mg/ml spectinomycin. The construct was sequenced and subsequently transformed into A. tumefaciens GV3101 cells using standard molecular biology techniques.
2.4. Construction of transgenic P. ginseng root lines
Stratified seeds (selected pure line, line 5) of Korean ginseng were harvested from a research field of the Department of Herbal Crop Research, National Institute of Horticultural and Herbal Science (Chungcheongbuk-do, Korea), RDA. Zygotic embryos were induced by transferring embryos onto 1/2 MS [18] medium with 5 mg/L GA3. To induce adventitious roots, the root parts of a single plantlet germinated from a zygotic embryo were pieced into approximately 10-mm segments and transferred onto MS medium lacking NH4NO3 supplemented with 2 mg/l IBA to induce new adventitious roots.
Transgenic ginseng plants were generated via Agrobacterium tumefaciens-mediated genetic transformation using segments of adventitious roots. Root segments measuring approximately 10 mm in length were dipped in Agrobacterium solution supplemented with 50 μg/l acetosyringone for 10 min and then blotted onto filter paper. After drying of the remnant bacteria on the surfaces of root segments, these root segments were transferred onto MS medium lacking NH4NO3 supplemented with 2 mg/L IBA and co-cultivated with Agrobacterium for three days. Thereafter, the root segments were transferred onto the same medium with additional supplementation with 500 mg/L cefotaxime to remove Agrobacterium. After two weeks, the root segments were transferred onto the same MS medium lacking NH4NO3 supplemented with 2 mg/l IBA, 500 mg/L cefotaxime, and 20 mg/L hygromycin to induce hygromycin-resistant adventitious roots. When new adventitious roots were formed on hygromycin-resistant calli in the presence of IBA, each actively growing adventitious root was cut and transferred onto new medium supplemented with 2 mg/l IBA and 20 mg/L hygromycin. When the hygromycin-resistant root segments were grown to root masses, each of the root clusters was regarded as an independent transgenic line.
2.5. Ginsenoside analysis by UPLC
The ginseng adventitious roots were dried in a ventilated oven at 50 °C for approximately 1 day. Milled powders (100 mg) were soaked in 100% methanol in Eppendorf tubes and sonicated at 20 kHz at a range of processing temperatures (25–40 °C). The supernatants were collected by centrifugation and filtered with a SepPak C-18 cartridge (Waters, Milford, MA, USA).
An ACQUITY ultra-performance liquid chromatography (UPLC) (Waters Corporation, Milford, MA, USA) equipped with a Phtodiode Array (PDA) detector was used for ginsenoside analysis. UPLC separation was performed using an ACQUITY UPLC BEH C18 column (50 mm × 2.1 mm, 1.7 μm). The column oven temperature was set at 40 °C, and the flow rate was maintained at 0.4 mL/min.
For ginsenoside analysis, a binary gradient method was used to vary the mobile phase composition over time using the same protocol as described by [19]. The mobile phase solvents A and B consisted of water and acetonitrile, respectively. The mobile phase (delivered at 0.5 mL/min) comprised Solvent A (water) and Solvent B (acetonitrile). A binary gradient elution was performed for ginsenoside analysis: initial 15% B from 0 to 0.5 min, linear gradient 15-45% B from 0.5 min to 8 min, linear gradient 45-100% B from 8 min to 10 min, and a final return to 15% B, which was maintained until 2 min to balance the column. The total run time was 18 min, and the sample injection volume was 2 μL. The standard compounds of ginsenosides (Rb1, Rb2, Rc, Rd, Re, Rf, and Rg1) for UPLC analysis were purchased from Sigma-Aldrich Co.
For single ion mode (SIM) analysis to detect PPT-type ginsenosides in ginseng mutant roots, a liquid chromatography ion trap time-of-flight mass spectrometry (LC-IT-TOF-MS) system (Shimadzu, Kyoto, Japan) equipped with an APCI source was used. Chromatographic separation was performed on a YMC-Pack Pro C18 RS column (5 μm, 2.0 × 150 mm, YMC, Japan) at 40 °C. The other analytical methods were followed the protocol by Park et al [24].
2.6. Targeted deep sequencing
Genomic DNA (gDNA) was extracted from adventitious root lines using a DNeasy Plant Mini Kit (Qiagen, Germany) according to the supplier's instructions. The gDNAs were used to perform target amplification Deep sequencing and Sanger sequencing. The primers used for the first PCR were 5′- TGG ATC TCT TTA TCT CAT CTC AAC T-3′ for forward and 5′- CAT TTG TCT TCA AAG TGC TG-3′ for reverse. For the adaptor PCR, forward (5′-ACA TCC TTA GCA GGC GAG AA-3′) and reverse primers (5′-CCG ATC CAT GAC CGA AAT AA-3′) were used. Amplifications were performed using Phusion polymerase (New England Biolabs, Ipswich, MA). Amplified PCR products were sequenced using the Illumina MiSeq platform. Mutations induced by sgRNAs were calculated based on the presence of insertions or deletions around the Cas9 RNA-guided endonuclease cleavage site (3 bp upstream of PAM). The ratio of the number of mutant reads to the total number of target reads was used as the editing efficiency. The targeted deep sequencing data were analysed using a Cas analyser [20].
3. Results
3.1. Design of sgRNAs in exon sequences of the CYP716A53v2 gene
It has been suggested that P. ginseng is allotetraploid (2n = 48) [21]. Thus, the PPT synthase gene might be expected to be existed in at least duplicated form. The CYP716A53v2 gene (GenBank accession number, JX036031) has been functionally characterized as a PPT synthase gene (protopanaxadiol 6-hydroxylase) [6]. Another homologous gene (paralog) was registered as a partial sequence (GenBank accession numbers, GAAG01029440.1 and GAAG01029441.1) that has 98% homology to the 5′ region of the CYP716A53v2 sequence, including the ATG start codon. Using the Ginseng Genome Database http://ginsengdb.snu.ac.kr/ [22], two genomic sequences both for CYP716A53v2 gene and a paralog were retrieved (Fig. S1). The genomic sequence for CYP716A53v2 gene had 4310 bp long constituted with four exon sites (Fig. 2A). Other genomic sequences for a paralog gene had 3230 bp long with five exons (Fig. 2A). To distinguish the sequence of CYP716A53v2 gene and a paralog, the former sequence was named to PPTg and the later was named to PPTa. The two sequences had high homologous exon region (exon 1) at 473-bp long including start codon (Fig. 2B) having 9 single nucleotide polymorphism (SNP) sites (Fig. 2B).
We designed two sgRNAs (sgRNA1 and sgRNA4) from the first exon sequence of both PPTa and PPTg sequences (Fig. 2B). The two different target sites, sgRNA1 and sgRNA4, had 100% identity in both PPTg and PPTa sequences, and were designed for the knockout of the genes in P. ginseng (Fig. 2B). These target sites precede an NGG protospacer adjacent motif (PAM) sequence required for Streptococcus pyogenes Cas9 (Fig. 2B). Potential off-target sites of these two sgRNA1 and sgRNA4 were searched for using CRISPR-OFFinder [23], and no potential off-target sites were detected.
An in vitro cleavage assay was conducted to exclude inferior sgRNAs. When the gRNAs, Cas9, and PCR fragments containing the target sequence were mixed and incubated, we observed cleavage of the target DNAs in two sgRNA constructs (Fig. S2). Finally, we decided to use sgRNA1 and sgRNA4 for targeted mutagenesis in ginseng.
3.2. Vector construction and production of transgenic ginseng root lines
Two sgRNAs (sgRNA1 and sgRNA4) were cloned into a pHAtC binary vector (accession number KU213971) carrying the Cas9 gene (driven by the CaMV 35S promoter), sgRNA (driven by the AtU6–29p promoter), and HPT (driven by the nos promoter) scaffolds (Fig. 2C). To improve codelivery, both the sgRNA and Cas9 were subcloned into one expression vector (Fig. 2C).
Transgenic ginseng roots were generated via A. tumefaciens-mediated genetic transformation. Genetic transformation of ginseng was started using in vitro-maintained adventitious roots induced from a single plant to minimize the genetic variation of the wild-type control. Root segments were co-cultivated with Agrobacterium for three days and then transferred onto MS medium lacking NH4NO3 supplemented with 2 mg/L IBA and 500 mg/L cefotaxime, both to remove Agrobacterium and to induce adventitious roots. After two weeks, the root segments were transferred onto the same medium supplemented with 2 mg/l IBA, 500 mg/L cefotaxime, and 20 mg/L hygromycin to induce the formation of hygromycin-resistant adventitious roots. The initial root segments were turned to callus masses during the culture, and new adventitious roots were induced on the surfaces of hygromycin-resistant callus in the presence of IBA (Fig. 2D). Each actively growing adventitious root grown on medium with hygromycin was cut and transferred onto new medium supplemented with 2 mg/l IBA and 20 mg/L hygromycin. If the excised roots were resistant to hygromycin, they produced numerous lateral roots (Fig. 2E) and subsequently grew into root masses, which are regarded as independent transgenic lines (Fig. 2F).
3.3. Selection of mutant lines by ginsenoside analysis in putative transgenic ginseng root lines
We expected that mutation in PPTg and/or PPTa sequences induced by the CRISPR/Cas9 system may result in a reduction and/or complete lack of PPT-type ginsenosides in transgenic lines. For screening of mutant lines, we adopted HPLC analysis to find the reduction of chromatogram for PPT-group ginsenosides (Rg1, Re, and Rf) in mutant lines compared to those of wild-type roots. HPLC analysis revealed that three major PPT-group ginsenosides (Rg1, Re, and Rf) and four major PPD-group ginsenosides (Rb1, Rb2, Rc, and Rd) were clearly detected in the wild-type ginseng roots in comparison with standard ginsenosides (Fig. S2). Some lines (such as the Cr7 line induced by target mutation using sgRNA1) showed no peaks for ginsenoside Rg1, Re, and Rf, indicating complete depletion of PPT-group ginsenosides (Fig. S2A). Conversely, some transgenic lines (such as the Cr3 line induced by target mutation using sgRNA4) showed partial reduction of the three PPT-group ginsenosides (Fig. S2B). Interestingly, the reduction of PPT-group ginsenosides (Rg1, Re, and Rf) in the mutant lines resulted in enhanced peak height in PPD-group ginsenosides (Rb1, Rc, Rb2, and Rd) (Fig. S2B).
Mutant lines, among the transgenic lines were screened for the complete depletion or reduction of PPT-type ginsenosides by HPLC chromatogram analysis. Among the total 94 independent root lines induced from the target mutation of the sgRNA1 site, 4 lines showed a partial reduction in PPT-group ginsenosides compared to wild-type roots, and 3 lines showed a complete reduction. Among the total 73 independent root lines induced from the target mutation of the sgRNA4 site, 3 lines showed a partial reduction in PPT-group ginsenosides compared to those of wild-type roots, and 1 line showed a complete reduction. Transformation efficiency calculated by the reduction of PPT-group ginsenosides revealed that among the total analysed transgenic lines using the vector for targeting sgRNA1, 7.4% of lines showed the mutant phenotype, and among the transgenic lines using the vector for targeting sgRNA4, 5.4% of lines showed the mutant phenotype.
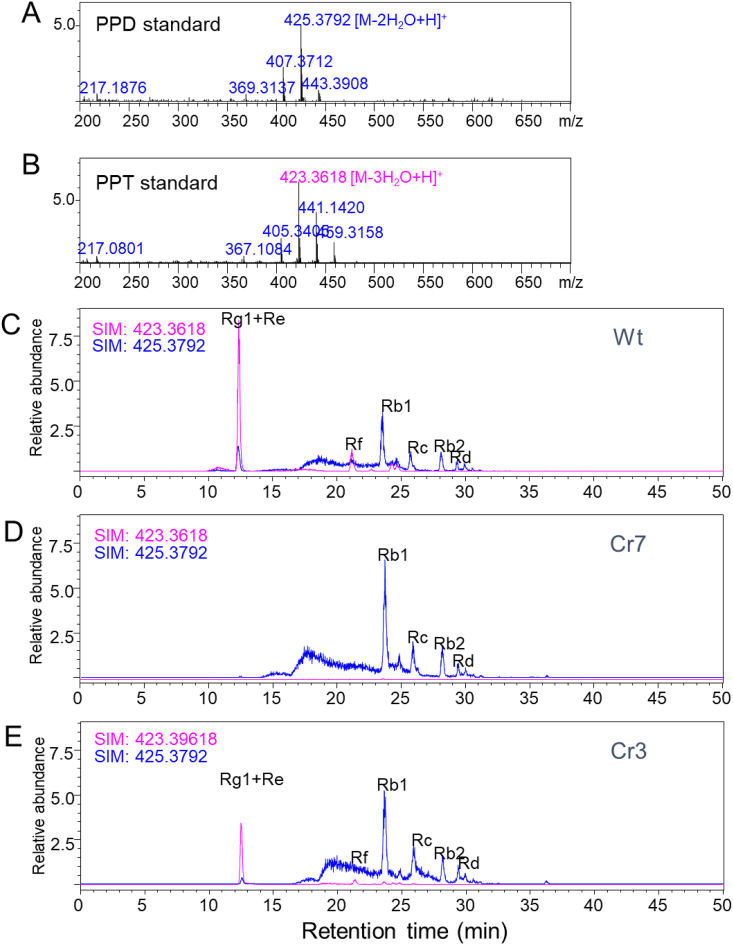
It has not been determined whether PPT-type ginsenosides are completely depleted in some mutant lines, although a clear reduction in PPT-type ginsenosides was observed by HPLC analysis. Ginsenosides were analysed again by selected ion mode (SIM), which enables the mass spectrometer to detect specific aglycone structure with very high sensitivity. The PPT-type ginsenosides share the common MS fragmentation patterns of PPT aglycone (405 [M-4H2O + H]+, 423 [M-3H2O + H] +, 441 [M-2H2O + H] +, and 459 [M-H2O + H] +) as seen in standard PPT aglycone (Fig. 3A). All the PPD-type ginsenosides share the common MS fragmentation patterns of PPD aglycone (407 [M-3H2O + H]+, 425 [M-2H2O + H] +, 443 [M-H2O + H] +) as seen in standard PPD aglycone (Fig. 3B). We analysed the PPT-type and PPD-type ginsenoside signals by SIM mode analysis using a single ion fragment of m/z 423.3618 and m/z 425.3792 in both wild-type and mutant lines using an LC-IT-TOF-MS system, respectively. The chromatogram of wild-type roots showed clear peaks for PPT-type ginsenosides (Rg1, Re, and Rf) by SIM (m/z 423.3999) analysis (Fig. 3C). However, Cr7 mutant roots did not contain any signal at the retention times of ginsenosides Rg1, Re, and Rf (Fig. 3D). In Cr3 roots, the PPT signal at the retention times of ginsenosides Rg1, Re, and Rf was partially reduced compared to wild-type roots (Fig. 3E).
Fig. 3.
SIM mode analysis of PPT-type ginsenosides in wild-type and mutant lines using an LC-IT-TOF-MS system. (A-B) MS spectrum of protopanaxatiol (A) and protopanaxatiol (B) standard aglycone. (C) SIM chromatograms of ginsenoside extracts from wild-type roots. (D-E) SIM chromatograms of ginsenosides of two putative mutant lines (Cr3 and Cr7).
3.4. Ginsenoside analysis in the five selected mutant lines
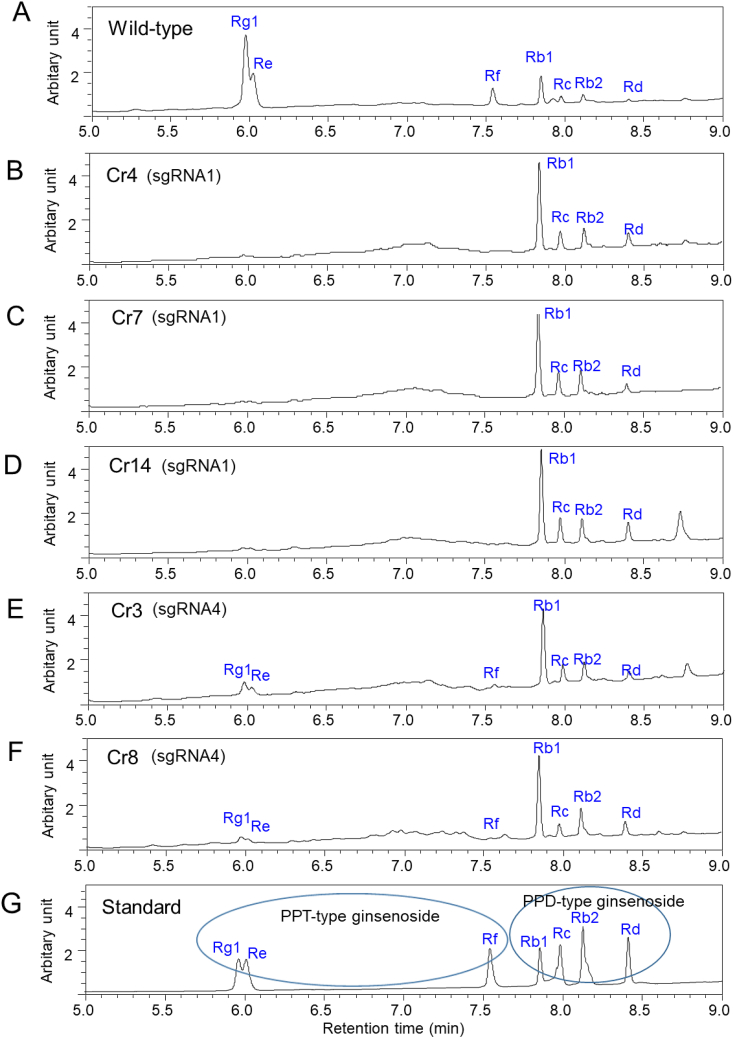
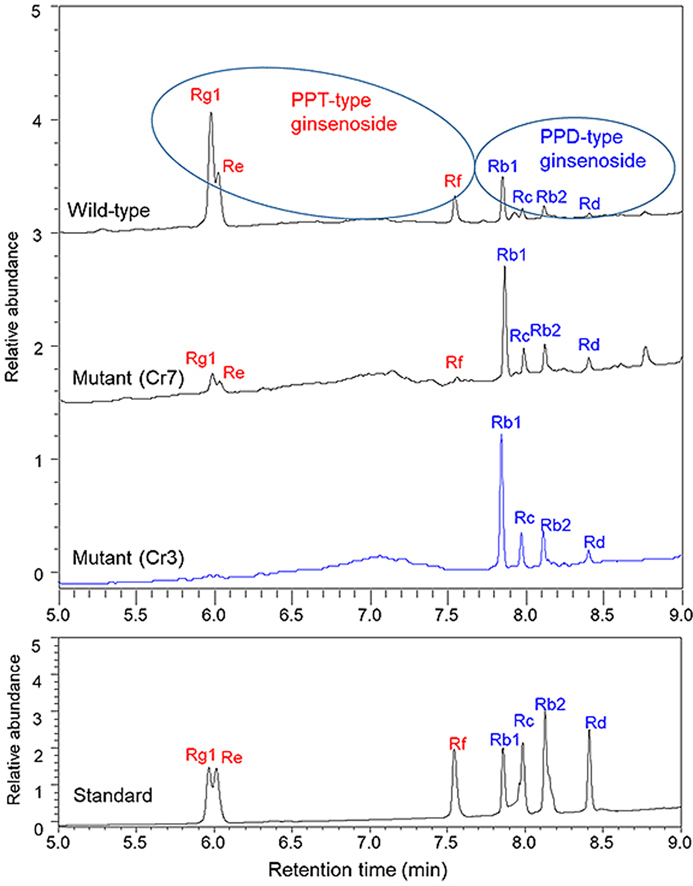
We finally selected three homozygous mutant lines (Cr4, Cr7, and Cr14) and two heterozygous mutant lines (Cr3 and Cr8) after sequencing the target mutation site. HPLC analysis revealed that ginsenoside Rg1 and Re were detected in major amounts in wild-type roots (Fig. 4A, Table 1). In contrast, all three PPT-group ginsenosides (Rg1, Re, and Rf) were not detected at the retention times as seen in ginsenoside standards (Fig. 4G) in all the three homozygous mutant lines (Cr4, Cr7, and Cr14) induced by the sgRNA1 target mutation (Fig. 4B–D). Instead, all the PPD-group ginsenosides (Rb1, Rb2, Rc, and Rd) in the three mutant lines (Cr4, Cr7, and Cr14) were strongly increased compared to the wild-type line (Fig. 4B–D). Two heterozygous mutant lines (Cr3 and Cr8) induced by sgRNA4 target mutations showed chromatogram peaks for PPT-group ginsenosides (Rg1, Re, and Rf) that were partially reduced (Fig. 4E and F) compared to those of wild-type roots (Fig. 4A). Interestingly, the reduction of PPT-group ginsenosides (Rg1, Re, and Rf) in all five mutant lines resulted in enhanced peak height for PPD-group ginsenosides (Rb1, Rc, Rb2, and Rd) (Fig. 4B–F). The contents of each PPD- and PPT-type ginsenoside and the total ginsenoside are shown in Table 1. PPT-type ginsenoside accumulation was completely inhibited in three homozygous mutant lines (Cr4, Cr7, and Cr14) and strongly reduced in two heterozygous mutant lines (Cr3 and Cr8). Instead, the accumulation of PPD-group ginsenosides (Rb1, Rc, Rb2, and Rd) was clearly enhanced in all mutant lines (Table 1). Wild-type roots contained a total of 6.68 mg/g DW ginsenosides. The total amount of ginsenosides in the mutant lines was not significantly reduced compared to that of wild-type roots (Table 1).
Fig. 4.
HPLC analysis of CRISPR/Cas9-mediated mutant lines of ginseng roots. (A) Ginsenoside chromatograms of extracts from wild-type roots. (B-D) Ginsenoside chromatograms of three mutant lines (Cr4, Cr7, and Cr14) using sgRNA1 of P. ginseng. (E-F) Ginsenoside chromatograms of two mutant lines (Cr3 and Cr8) using sgRNA4. (G) HPLC chromatogram of ginsenoside standards.
Table 1.
Change of PPT- and PPD-type ginsenoside concentration in CRISPR/Cas9-mediated mutant lines
| Line | Ginsenoside concentration (mg/g) |
Total | ||||||
|---|---|---|---|---|---|---|---|---|
| PPT-type |
PPD-type |
|||||||
| Rg1 | Re | Rf | Rb1 | Rc | Rb2 | Rd | ||
| Wild-type | 2.84 ± 0.13 | 1.13 ± 0.13 | 0.52 ± 0.04 | 1.53 ± 0.14 | 0.20 ± 0.05 | 0.31 ± 0.02 | 0.15 ± 0.01 | 6.68 ± 0.52 |
| Cr4 (sgRNA1) | - | - | - | 4.62 ± 0.12 | 0.42 ± 0.02 | 0.95 ± 0.03 | 0.37 ± 0.03 | 6.36 ± 0.20 |
| Cr7 (sgRNA1) | - | - | - | 4.43 ± 0.17 | 0.36 ± 0.01 | 0.96 ± 0.02 | 0.36 ± 0.03 | 6.11 ± 0.23 |
| Cr14 (sgRNA1) | - | - | - | 4.58 ± 0.21 | 0.44 ± 0.02 | 0.78 ± 0.04 | 0.48 ± 0.02 | 6.24 ± 0.29 |
| Cr3 (sgRNA4) | 0.38 ± 0.02 | 0.22 ± 0.03 | - | 3.76 ± 0.19 | 0.34 ± 0.03 | 0.92 ± 0.13 | 0.33 ± 0.02 | 5.45 ± 0.42 |
| Cr8 (sgRNA4) | 0.15 ± 0.03 | 0.09 ± 0.01 | 0.14 ± 0.01 | 3.67 ± 0.31 | 0.23 ± 0.04 | 1.06 ± 0.07 | 0.44 ± 0.01 | 5.78 ± 0.48 |
3.5. Deep sequencing of sgRNA regions and confirmation of indels in PPTa and PPTg sequences
For evaluation of the mutation events at the loci of the target site of sgRNAs, targeted Deep sequencing for the three mutant lines (Cr4, Cr7, and Cr14 induced by sgRNA1) and Sanger sequencing for one mutant line (Cr3 induced by sgRNA4) analysis was conducted.
In the PPTg and PPTa sequences, a one SNP in the flanking region including the target sgRNA1 region and two SNPs in the flanking region including the target sgRNA4 region were detected as shown in Fig. 2B. Deep sequencing analysis was performed to differentiate the mutation events in PPTa and PPTg sequences in the flanking region including the target sgRNA1 region in three mutant lines (Cr4, Cr7, and Cr14) induced by sgRNA1 including wild-type sequences (Fig. 5A). As expected, all the mutations were located in the vicinity of the PAM target of the Cas9-induced cleavage site (Fig. 5). Mutations occurred in sgRNA1 region of PPTg and PPTa sequences in all alleles (Fig. 5). In the Cr4 mutant line, a 12-bp deletion and a 1-bp insertion occurred in the PPTa sequence, and a 1-bp deletion and a 1-bp insertion occurred in the PPTg sequence (Fig. 5A). In the Cr7 mutant line, only a 1-bp deletion occurred in all the PPTa and PPTg sequences (Fig. 5 A). In the Cr14 mutant line, an 8-bp deletion occurred in two alleles of the PPTg sequence, and a 3-bp deletion occurred in two alleles of the PPTa sequence (Fig. 5A). Sanger sequencing analysis was performed to differentiate the mutation events in PPTa and PPTg sequences in the flanking region including the target sgRNA4 region in Cr3 mutant line induced by sgRNA4 including wild-type sequences (Fig. 5B). As expected, a mutation was located in the vicinity of the PAM target of the Cas9-induced cleavage site (Fig. 5B). Cr3 mutant line induced by targeted sgRNA4 showed 7 bp deletion in PPTg sequence but no indel in PPTa sequence (Fig. 5B).
Fig. 5.
Genotyping of targeted induced mutant lines of ginseng roots induced by CRISPR/Cas9. (A) The figure presents mutations in individual alleles in Cr4, Cr7, and Cr14 mutants induced by sgRNA1 aligned to two corresponding wild-type fragments (PPTa and PPTg), as determined by Deep DNA sequencing. Target sequences of the sgRNA1 site are bold and in blue, and the protospacer adjacent motif (PAM) is underlined. Deleted nucleotides are shown with red hyphens, and inserted nucleotides are shown in red bold. The net change in length is noted to the right of each sequence (insertion and deletion). SNPs between PPTg and PPTa sequences are marked in dark red or green. (B) The figure presents mutations in individual alleles in Cr3 mutant induced by sgRNA4 aligned to two corresponding wild-type fragments (PPTa and PPTg), as determined by Sanger sequencing.
Deep sequencing analysis revealed that the sequenced reads showed 100% indel frequency in all 98,146 total reads in the Cr4 mutant line (Table S1). In the Cr7 mutant line, 100% of sequenced reads (a total of 101,867 reads) also showed indel frequency. In the Cr14 mutant line, 99.5% of reads showed indel frequency. In contrast, sequencing of off-target sites (sgRNA4 region) showed no indel frequency (near 0%) in all three mutant lines (Cr4, Cr7, and Cr14).
4. Discussion
4.1. Selection of putative mutant lines by ginsenoside saponin analysis
PPT synthase (CYP716A53v2, protopanaxadiol 6-hydroxylase) is the key enzyme for the biosynthesis of PPT-type ginsenosides in P. ginseng [6]. In this work, we applied the CRISPR/Cas9 system to induce the target mutation of the PPT synthase gene in P. ginseng. In situ hybridization and genomic in situ hybridization analysis have indicated that P. ginseng is allotetraploid [21,24]. Two types of sequences for PPT synthase gene was registered in NCBI database. Only one gene (CYP716A53v2 gene) was characterized as PPT synthase, but another gene is registered as partial fragments in NCBI database and remained to be functionally characterized. In Ginseng Genome Database, http://ginsengdb.snu.ac.kr/ [22], two genomic sequences for the two PPT synthase genes (named as PPTg and PPTa sequences) were obtained. Although the two mRNA sequences had 98% identity, genomic sequences were highly different because PPTg genomic sequences had 4310-bp long constituted with 4 exon sites and PPTa genomic sequence had 3230-bp long with 5 exons. This result indicates that P. ginseng plants may contain at least two homologous sequences coding for PPT synthase with different exon and intron structure.
The target sites of sgRNAs (sgRNA1 and sgRNA4) were selected as the regions that showed 100% identity in both PPTg and PPTa sequences. We constructed two vectors for the CRISPR/Cas9 system using two sgRNAs (sgRNA1 and sgRNA4). Transgenic root lines were obtained by Agrobacterium-mediated genetic transformation of ginseng.
4.2. Screening of mutant lines by ginsenoside analysis
We expected that disruption of the PPT synthase gene induced by CRISPR/Cas9-mediated targeted mutation might result in reduced accumulation and/or complete depletion of PPT-type ginsenosides, which would enable us to identify the mutant by analysis of ginsenoside chromatograms using HPLC. A total of 167 independent lines using the two vectors targeting sgRNA1 or sgRNA4 were generated, and the ginsenoside profiles were analysed to select the putative mutant lines. We observed that PPT-type ginsenosides were completely or partially reduced in several transgenic lines compared with wild-type roots. This result reveals that mutation of the PPT synthase gene clearly resulted in a change in PPT-type ginsenoside accumulation. The SIM mode of LC-MS-MS analysis is suitable for the detection of trace components when the mass spectra of the components are known. SIM analysis employing the target ion (m/z 423) of the protopanaxatriol aglycone of PPT-type ginsenosides using an LC-IT-TOF-MS system revealed that PPT-type ginsenosides were completely depleted in some mutant lines (Cr4, Cr7, and Cr14). From this result, we postulated that the complete depletion of PPT-type ginsenosides might be attributable to the mutation of all alleles of PPT synthase genes in tetraploid ginseng chromosomes. Thus, screening ginsenoside profiles by LC analysis is a very efficient way to select homozygous mutant lines.
4.3. Identification of mutant DNA sequences
The two PPTg and PPTa sequences had 9 SNP regions on exon sequences of 473-bp long including start codon. Thus, PCR primers to detect the mutation site of PPTg and PPTa sequences were designed to be able to identify both sequences of PPTg and PPTa, including the SNP recognition site. Genomic sequence analysis of target sgRNA regions using Deep and Sanger sequencing was performed in mutant lines that showed the reduction or complete lack of PPT-type ginsenosides. Three lines (Cr4, Cr7, and Cr14) showing complete lack of PPT-type ginsenosides were suggested to have mutations in all 4 alleles. Deep sequencing in the three homozygous mutants (Cr4, Cr7, and Cr14) revealed that nearly all the sequences (more than 99.5% of reads) showed the mutation in the target sgRNA1 site. The mutation occurs in the sgRNA1 target site of both the PPTa and the PPTg sequences. Sequencing of the off-target site (sgRNA4 target region) in Cr4, Cr7, and Cr14 mutants revealed that the mutation rate was nearly zero in all the three mutant lines (Table S1). This result indicates that target-induced mutation by CRISPR/Cas9 system was highly specific for the target sgRNA site in P. ginseng.
4.4. Depletion of PPT-group ginsenosides resulted in the enhanced accumulation of PPD-group ginsenosides
Interestingly, the reduction or depletion of PPT-group ginsenosides in mutant lines resulted in the increased accumulation of PPD-group ginsenosides (Rb1, Rb2, Rc, and Rd). Thus, the total amount of ginsenosides was not clearly changed in mutant lines compared to wild-type roots. This result indicates that total triterpene flux towards ginsenoside biosynthesis may not be affected by blocking PPT-type ginsenoside biosynthesis. We suggest that PPD- and PPT-type ginsenoside production may be competing pathways that exhibit see-saw effects depending on the amount of common intermediate triterpene (dammarenediol-II). We previously reported that the reduced production of PPT-type ginsenosides by RNA interference to knock down the CYP716A53v2 gene in P. ginseng resulted in increased accumulation of PPD-type ginsenosides [25]. Compared to knock down system by RNAi, which showed the partial reduction of PPT-type ginsenosides in transgenic ginseng plants, CRISPR/Cas9 system is very effective to complete depletion of PPT-type ginsenosides in P. ginseng because the target genes are completely knockout.
It has been reported that the biological activities of PPD-group ginsenosides are different from those of PPT-group ginsenosides, and PPD group ginsenosides are known to have higher anticancer activities than PPT-group ginsenosides [[10], [11], [12], [13]]. The mutant ginseng roots producing only PPD-group ginsenosides induced by the CRISPR/Cas9 system may exhibit altered biological properties and enhanced anticancer activities. In the future, we are going to regenerate the plants from these mutant root lines of P. ginseng induced by CRISPR/Cas9 system and transferred to soil to set the seeds. This article is the first report for constructing mutant lines using CRISPR/Cas9 system in Panax species. Construction of mutated ginseng plants via the CRISPR/Cas9 system might have high value for innovative new breeding.
Declaration of competing interest
The authors declare that they have no conflicts of interest.
Acknowledgments
This work was supported by grants from the Next-Generation BioGreen 21 Program (PJ01344401) of the Rural Development Administration, and by R&D Program for Forest Science Technology (Project No. 2021339A00-2123-CD02) provided by Korea Forest Service (Korea Forestry Promotion Institute), Republic of Korea.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jgr.2021.06.004.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Hostettmann K.A., Marston A. Cambridge University Press; Cambridge: United Kingdom: 1995. Saponins. Chemistry and pharmacology of natural products. [Google Scholar]
- 2.Vogler B.K., Pittler M.H., Ernst E. The efficacy of ginseng. A systematic review of randomised clinical trials. Eur J Clin Pharmacol. 1999;55:567–575. doi: 10.1007/s002280050674. [DOI] [PubMed] [Google Scholar]
- 3.Shibata S. Chemistry and cancer preventing activities of ginseng saponins and some related triterpenoid compounds. J Korean Med Sci. 2001;16:S28–S37. doi: 10.3346/jkms.2001.16.S.S28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Han J.Y., Kwon Y.S., Yang D.C., Jung Y.R., Choi Y.E. Expression and RNA interference-induced silencing of the dammarenediol synthase gene in Panax ginseng. Plant Cell Physiol. 2006;47:1653–1662. doi: 10.1093/pcp/pcl032. [DOI] [PubMed] [Google Scholar]
- 5.Han J.Y., Kim H.J., Kwon Y.S., Choi Y.E. The Cyt P450 enzyme CYP716A47 catalyzes the formation of protopanaxadiol from dammarenediol-II during ginsenoside biosynthesis in Panax ginseng. Plant Cell Physiol. 2011;52:2062–2073. doi: 10.1093/pcp/pcr150. [DOI] [PubMed] [Google Scholar]
- 6.Han J.Y., Hwang H.S., Choi S.W., Kim H.J., Choi Y.E. Cytochrome P450 CYP716A53v2 catalyzes the formation of protopanaxatriol from protopanaxadiol during ginsenoside biosynthesis in Panax ginseng. Plant Cell Physiol. 2012;53:1535–1545. doi: 10.1093/pcp/pcs106. [DOI] [PubMed] [Google Scholar]
- 7.Yan X., Fan Y., Wei W., Wang P., Liu Q., Wei Y., Zhang L., Zhao G., Yue J., Zhou Z. Production of bioactive ginsenoside compound K in metabolically engineered yeast. Cell Res. 2014;24:770–773. doi: 10.1038/cr.2014.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mao Q., Zhang P.H., Wang Q., Li S.L. Ginsenoside F2 induces apoptosis in humor gastric carcinoma cells through reactive oxygen species-mitochondria pathway and modulation of ASK-1/JNK signaling cascade in vitro and in vivo. Phytomedicine. 2014;21(4):515–522. doi: 10.1016/j.phymed.2013.10.013. [DOI] [PubMed] [Google Scholar]
- 9.Ahuja A., Kim J.H., Yi Y.S., Cho J.Y. Functional role of ginseng-derived compounds in cancer. J Ginseng Res. 2018;42(3):248–254. doi: 10.1016/j.jgr.2017.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jin J., Shahi S., Kang H.K., van Veen H.W., Fan T.-P. Metabolites of ginsenosides as novel BCRP inhibitors. Biochem Biophys Res Commun. 2006;345(4):1308–1314. doi: 10.1016/j.bbrc.2006.04.152. [DOI] [PubMed] [Google Scholar]
- 11.Shim S.H., Baek K.-H., Kim Y.S. Inhibition of human 20S proteasome by ginsenosides from Panax ginseng. Bull Korean Chem Soc. 2009;30:1385–1387. [Google Scholar]
- 12.Lee J.I., Ha Y.W., Choi T.W., et al. Cellular uptake of ginsenosides in Korean white ginseng and red ginseng and their apoptotic activities in human breast cancer cells. Planta Med. 2011;77(2):133–140. doi: 10.1055/s-0030-1250160. [DOI] [PubMed] [Google Scholar]
- 13.Chen X.J., Zhang X.J., Shui Y.M., Wan J.B., Gao J.L. Anticancer activities of protopanaxadiol- and protopanaxatriol-type ginsenosides and their metabolites. Evid-Based Complement Altern Med. 2016;2016:1–19. doi: 10.1155/2016/5738694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Helms S. Cancer prevention and therapeutics: Panax ginseng. Altern Med Rev. 2004;9:259–274. [PubMed] [Google Scholar]
- 15.Wang M., Li H., Liu W., Cao H., Hu X., Gao X., Xu F., Li Z., Hua H., Li D. Dammarane-type leads panaxadiol and protopanaxadiol for drug discovery: biological activity and structural modification. Eur J Med Chem. 2020;189:112087. doi: 10.1016/j.ejmech.2020.112087. [DOI] [PubMed] [Google Scholar]
- 16.Begemann M.B., Gray B.N., January E., Gordon G.C., He Y., Liu H., Wu X., Brutnell T.P., Mockler T.C., Oufattole M. Precise insertion and guided editing of higher plant genomes using Cpf1 CRISPR nucleases. Sci Rep. 2017;7:11606. doi: 10.1038/s41598-017-11760-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petolino J.F., Srivastava V., Daniell H. Editing Plant Genomes: a new era of crop improvement. Plant Biotechnol J. 2016;14:435–436. doi: 10.1111/pbi.12542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murashige T., Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue. Physiol Plant. 1962;15:473–497. [Google Scholar]
- 19.Kim J.Y., Adhikari P.B., Ahn C.H., Kim D.H., Kim Y.C., Han J.Y., et al. High frequency somatic embryogenesis and plant regeneration of interspecific ginseng hybrid between Panax ginseng and Panax quinquefolius. J Ginseng Res. 2019;43:38–48. doi: 10.1016/j.jgr.2017.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park J., Lim K., Kim J.S., Bae S. Cas-analyzer: an online tool for assessing genome editing results using NGS data. Bioinformatics. 2017;33:286–288. doi: 10.1093/bioinformatics/btw561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choi H.I., Waminal N.E., Park H.M., Kim N.H., Choi B.S., Park M., Choi D., Lim Y.P., Kwon S.J., Park B.S., et al. Major repeat components covering one-third of the ginseng (Panax ginseng C.A. Meyer) genome and evidence for allotetraploidy. Plant J. 2014;77:906–916. doi: 10.1111/tpj.12441. [DOI] [PubMed] [Google Scholar]
- 22.Jayakodi M., Choi B.S., Lee S.C., Kim N.H., Park J.Y., Jang W., et al. Ginseng Genome Database: an open-access platform for genomics of Panax ginseng. BMC Plant Biology. 2018;18:62. doi: 10.1186/s12870-018-1282-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bae S., Park J., Kim J.S. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 2014;30:1473–1475. doi: 10.1093/bioinformatics/btu048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi H.W., Koo D.H., Bang K.H., Paek K.Y., Seong N.S., Bang J.W. FISH and GISH analysis of the genomic relationships among Panax species. Genes Genom. 2009;31:99–105. [Google Scholar]
- 25.Park S.B., Chun J.H., Ban Y.W., Han J.Y., Choi Y.E. Alteration of Panax ginseng saponin composition by overexpression and RNA interference of the protopanaxadiol 6-hydroxylase gene (CYP716A53v2) J Ginseng Res. 2016;40:47–54. doi: 10.1016/j.jgr.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.