Significance
Biofilms are an important mode of bacterial growth where surface-attached cells enjoy survival advantages provided by the extracellular matrix composed of secreted, diffusible biopolymers. An important question is how diffusible matrix production can be stable under the evolutionary pressure of exploitation. Here, we show that some matrix proteins in Vibrio cholerae biofilms can indeed be exploited by nonproducers, but the exploitation happens within a quantifiable spatial range around the producer cell clusters. An ecological model considering the length scales of exploitation and cell group structure reveals the stable conditions for diffusible matrix production that are consistent with those in natural environments. Our study provides concepts and tools for studying public goods sharing in spatially structured bacterial biofilms.
Keywords: biofilm, matrix, adhesion protein, public goods dilemma, social evolution
Abstract
Biofilm formation is an important and ubiquitous mode of growth among bacteria. Central to the evolutionary advantage of biofilm formation is cell–cell and cell–surface adhesion achieved by a variety of factors, some of which are diffusible compounds that may operate as classical public goods—factors that are costly to produce but may benefit other cells. An outstanding question is how diffusible matrix production, in general, can be stable over evolutionary timescales. In this work, using Vibrio cholerae as a model, we show that shared diffusible biofilm matrix proteins are indeed susceptible to cheater exploitation and that the evolutionary stability of producing these matrix components fundamentally depends on biofilm spatial structure, intrinsic sharing mechanisms of these components, and flow conditions in the environment. We further show that exploitation of diffusible adhesion proteins is localized within a well-defined spatial range around cell clusters that produce them. Based on this exploitation range and the spatial distribution of cell clusters, we constructed a model of costly diffusible matrix production and related these length scales to the relatedness coefficient in social evolution theory. Our results show that production of diffusible biofilm matrix components is evolutionarily stable under conditions consistent with natural biofilm habitats and host environments. We expect the mechanisms revealed in this study to be relevant to other secreted factors that operate as cooperative public goods in bacterial communities and the concept of exploitation range and the associated analysis tools to be generally applicable.
Biofilms are an ancient and ubiquitous form of bacterial life in which cells form surface-attached groups embedded in an extracellular matrix (1). Biofilm-dwelling bacteria gain survival advantages from the matrix, including resistance to chemical and biological threats such as antibiotic exposure, host immune system defenses, and invasion and predation by other species (2–6). Biofilm formation also contributes to the pathogenicity of many infectious bacterial species and thus makes an outsized impact on human health (7–10). The evolutionary advantage of biofilm formation is often tied to cell–cell and cell–surface adhesion, through which biofilm-forming species gain strength in numbers, resistance to physical disturbance, and physical association with favorable surfaces (11, 12). Indeed, in the native habitats of many biofilm-formers, nutrient distribution is heterogeneous and often localized around solid substrates (13–15). Among the components of the extracellular matrix, exopolysaccharides and accessory proteins have been suggested to play dominant roles in biofilm adhesion to both biotic and abiotic surfaces in different species (3).
In order to function properly, the adhesive molecules associated with biofilm matrix must be secreted outside of cells. This raises an important question: Is the production of adhesive molecules exploitable, and if so, what conditions allow for their evolutionary stability? In many contexts, secreted beneficial substances that diffuse away from producing cells can be scavenged by exploitative mutants (often termed cheaters), leading to a public goods dilemma (16, 17) in which cooperative behaviors partially or completely break down because producing cells, which pay a metabolic cost, are outcompeted by exploitative mutants (17–20). The diffusive nature of secreted public goods necessarily leads to a characteristic length scale over which they can be shared or exploited (21–24). Previous literature has introduced the idea that secretion of public goods can be stabilized against exploitation if this length scale is smaller than or similar to the scale over which clonemates are clustered together, allowing cooperative cells to preferentially help each other relative to the population as a whole (25–29). This principle is directly analogous to the classical statement of social evolution theory that cooperation can be favored by natural selection when the benefits of receiving cooperative help, weighted by the relatedness coefficient among givers and receivers of cooperation, exceed the costs of cooperation (30).
Several studies have explored the competitive dynamics of partially shared matrix components in colonies on agar or pellicles at the air–liquid interface (31–35), which can serve as excellent experimental platforms; but the continuous film of bacteria formed in these cases does not always correspond to scenarios in nature where isolated cell clusters are separated from each other in space. In many cases, biofilm-forming bacteria form cell clusters due to constraints on movement or mother-daughter cell adhesion, and these clusters may be sparsely or densely distributed in space depending on environmental conditions (36). The physical size of these clonal clusters and the distance between them therefore represent two independent length scales. In principle, these two length scales should be compared with the diffusive length scale of the public goods to determine the physical conditions that stabilize cooperative product secretion against cheating.
The biofilm-producing pathogen and marine microbe Vibrio cholerae offers an excellent model for studies that may provide a close coupling of theory and quantitative empirical testing on distance-dependent cooperative behavior. V. cholerae produces a suite of matrix components—including vibrio polysaccharide (VPS) and the proteins RbmA, RbmC, and Bap1—that have been extensively characterized. VPS and RbmA are shared in a restricted fashion within secreting cell lineage groups and thus are minimally susceptible to exploitation by nonproducing cells (4, 37–40); this pattern has also been observed for major matrix components in Pseudomonas aeruginosa and Bacillus subtilis (33, 34, 41, 42). On the other hand, RbmC and Bap1, which are particularly important for adhesion of cell clusters to underlying surfaces, can diffuse far away from producing cells (37, 43). Here, we establish that these two matrix proteins are exploitable by nonproducing cells, creating an associated public goods dilemma. Competition assays between producers and nonproducers of RbmC and Bap1 in static culture and with flow reveal the conditions under which populations of adhesion protein producers can resist invasion by cheating cells. We further show that exploitation of these matrix components takes place within a quantifiable spatial range, which depends critically on rates of diffusion and advection. As a result, sparse distribution of cell groups and environmental flow that limits the spatial range of shared products work jointly to limit exploitation of the diffusible matrix. This resolution links well to the ecology of V. cholerae biofilms in natural habitats and can be straightforwardly related to fundamental social evolution theory (30).
Results
RbmC and Bap1 Are Exploitable Public Goods in V. cholerae Biofilms.
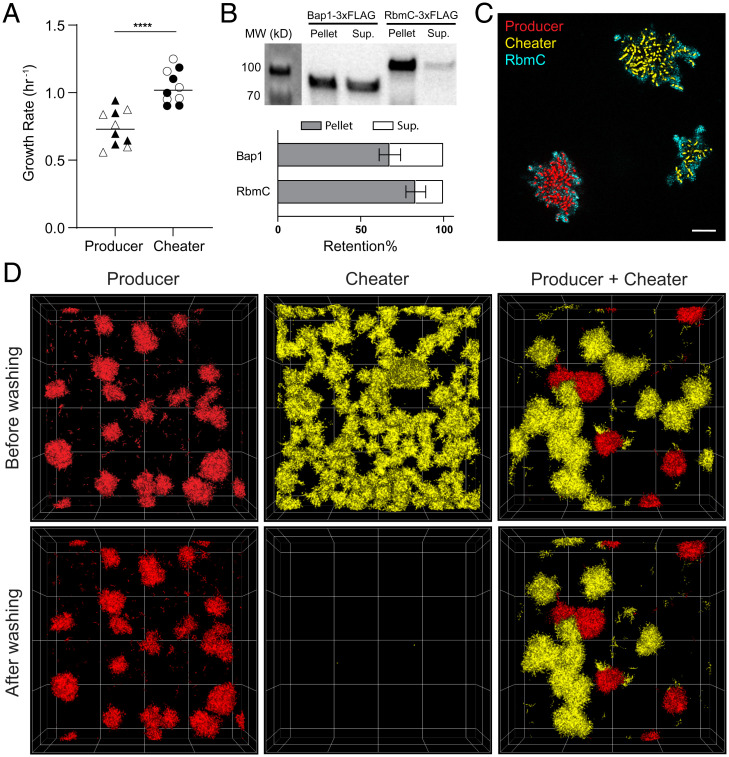
To study competition between diffusible matrix (RbmC and Bap1) producers and potential cheaters in V. cholerae biofilms, we used a strain background (vpvCW240R) locked in a high level of cyclic diguanylate that constitutively up-regulates biofilm production machinery (44). Using a constitutive biofilm-producing background allowed us to perform biofilm culturing experiments more readily while avoiding confounding factors from biofilm gene regulation, but our results can be obtained with a wild-type V. cholerae background as well (SI Appendix, Fig. S1). We generated an isogenic mutant with double deletions of rbmC and bap1 as the cheating strain because of their functional redundancy in conferring surface adhesion (37, 43, 45). The producer and cheater each expressed a different fluorescent protein to distinguish them from each other (SI Appendix, Fig. S2). We first examined whether there is a fitness cost for producing RbmC and Bap1 by comparing producer and cheater growth rates in monoculture. As shown in Fig. 1A, the growth rate of the producer was significantly lower than that of the cheater, demonstrating a substantial cost of producing these matrix components.
Fig. 1.
RbmC and Bap1 are exploitable public goods in V. cholerae biofilms. (A) Growth rates of the producer strain and the cheater strain (Δbap1ΔrbmC) from n = 10 independent replicates (****P < 0.0001, Mann–Whitney U test). Strains expressing mScarlet-I and mNeonGreen are shown by filled and empty markers, respectively. Horizontal bars inducate the mean values. (B) Top: Western blot showing Bap1 and RbmC in the biofilm (Pellet) and in the cell-free supernatant (Sup.). Bottom: Quantification of the fraction of protein retained in biofilm. Error represents the SE from n = 3 independent replicates. (C) A representative confocal image 7 µm above the substratum of a coculture of producer (red) expressing 3×FLAG-tagged RbmC and cheater (yellow), stained with Cy3-conjugated anti-FLAG antibodies (cyan) to show the spatial distribution of RbmC. (Scale bar, 10 µm.) (D) Monoculture of producer (red), monoculture of cheater (yellow), and coculture of producer and cheater before and after washing. The producer constitutively expressed mScartlet-I, and the cheater constitutively expressed mNeonGreen. Image size: 222 × 222 × 60 µm.
To document the degree of retention versus diffusive sharing of RbmC and Bap1, we performed a quantitative Western assay and found that 83% of RbmC and 68% of Bap1 were retained within producer biofilms, with the remaining protein detected in the surrounding liquid medium (Fig. 1B). Recapitulating prior literature, this result confirms that a substantial fraction of RbmC and Bap1 molecules can leave the cell clusters that produce them (37, 43). Next, to confirm that the molecules diffusing into the medium can be exploited by the cheaters, we tagged the C terminus of RbmC or Bap1 with a 3×FLAG epitope and followed their spatial distributions in cocultured biofilms using in situ immunostaining. The staining results show that significant amounts of RbmC and Bap1 were relocated from producer to cheater biofilm clusters (Fig. 1C and SI Appendix, Fig. S3).
To determine whether exogenously acquired Bap1 and RbmC are functional in adhering cheater cell clusters to the substratum, we grew producer and cheater biofilms separately and in coculture in 96-well plates. After growing biofilms statically for 16 h, we compared the biofilm biomass remaining on the glass substratum after a washing step that removed any weakly adherent cell clusters (Materials and Methods, Static Competition Assay with End-point Flow Disturbance) (Fig. 1D). We found that in monocultures, almost all producer cell clusters remained surface-bound after washing, whereas nearly zero biomass was found for cheater biofilms after the disturbance, consistent with the absence of surface adhesion in the cheater strain. In contrast, in the coculture of producer and cheater, some of the cheater cell groups were able to resist the strong fluid flow during the washing step. Coupling this observation with the immunostaining results, we inferred that adhesion can be partially restored in cheater biofilms by taking advantage of the adhesion proteins produced by the cohabitating producers. Overall, these results confirm that the shared matrix proteins RbmC and Bap1 in V. cholerae biofilms are cooperative products that draw a significant cost to produce and are susceptible to cheater exploitation.
Exploitation Range and Community Spatial Structure Jointly Determine the Outcome of Competition between Producers and Cheaters.
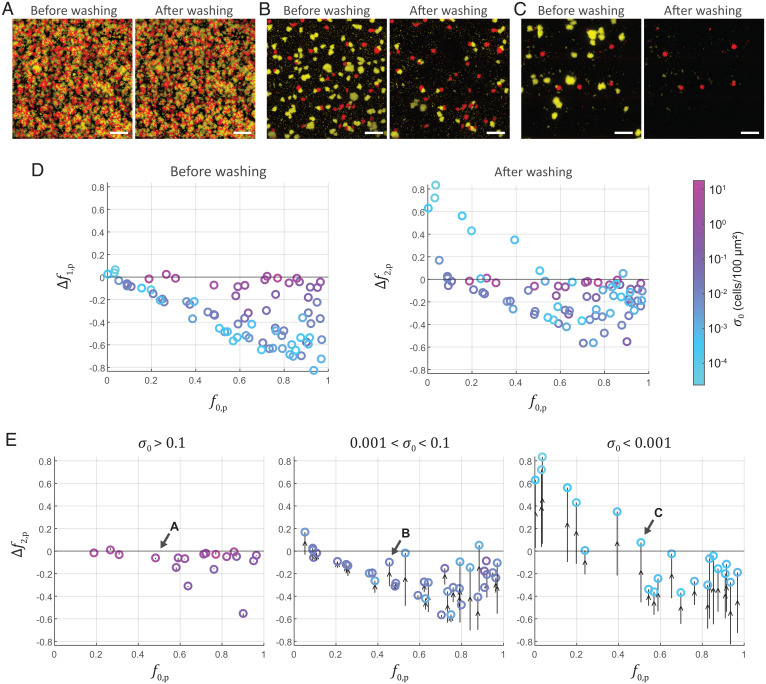
Because RbmC and Bap1 are costly to produce but exploitable by cheating strains, it is possible that nonproducing cells could outcompete producing cells in coculture. To test if this is true, we first competed producer against cheater in a static environment. The initial biomass of both strains was quantified by the inoculation number densities ( and for producer and cheater, respectively; is the total inoculation number density). After 16 h of growth, the total biomass in three-dimensional (3D) space was quantified both before and after washing (Materials and Methods, Microscopy, Biomass Quantification, and Growth Rate Measurement; SI Appendix, Fig. S4). The competition assay was initiated at different initial frequencies of the producer for a range of total inoculation number density . Representative images and quantitative outcomes of experiments are shown in Fig. 2A–D, where and are the frequency changes of the producer compared to before and after washing, respectively.
Fig. 2.
Competition between adhesion protein producer and cheater strains in a static environment. (A–C) Producer (red) and cheater (yellow) biofilms grown in a static environment before and after washing with different inoculation number densities corresponding to data labeled in E. (Scale bars, 100 µm.) (D) Adhesion protein producers in competition with cheaters before and after washing. (E) Adhesion protein producers in competition with cheaters in different ranges (color-coded) in a static environment. Each data marker denotes the frequency change in producer after washing (), and the corresponding frequency change before washing () is connected to it by an arrow ( → ). Arrows labeled with A, B, and C indicate data points corresponding to images in (A), (B), and (C), respectively.
We found that before washing, cheaters outcompeted producers at all initial frequencies, and the fitness of the producer decreased as the inoculation density decreased (Fig. 2D). This is because in a static environment, both producers and cheaters compete for the same nutrient pool, and the cheater has a substantial advantage in growth rate (Fig. 1A); also, importantly, the benefit of shared adhesion protein production to biofilm cells does not manifest in a disturbance-free environment. The decrease in producer fitness with decreasing is due to finite resources in the static environment: At high , both strains can only undergo a short period of growth before reaching the environment’s carrying capacity, limiting the total change in relative abundance of the two strains that can occur. A classic resource-limited competition model reproduced the competition results in a static environment (SI Appendix, Fig. S5).
In the competition data after washing, producer fitness was systematically increased compared to the competition before washing, suggesting that cheater biomass is preferentially removed compared to producer (Fig. 2D). Importantly, this pattern shifted with and corresponded to negative frequency-dependent selection for RbmC and Bap1 production at : At low initial coverage, the producer was positively selected at low initial frequency. This is evident in Fig. 2E where the competition data are sorted based on inoculation number density . We expect that at even lower inoculation number densities, the stable point will move to (i.e., the producer will always win regardless of the initial frequency); obtaining sufficient statistics at such low is, however, experimentally difficult due to limits on field-of-view acquisition with sufficient detail to make accurate measurements. We also note that the magnitude of , the fitness difference in producer before and after washing, increased with decreasing (Fig. 2E), hinting at the importance of the distance between producer and cheater clusters for the effect of shear disturbance on population dynamics.
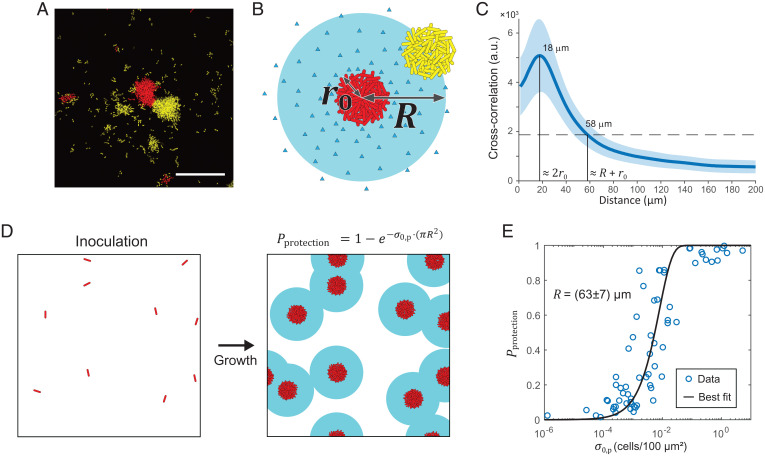
To understand the mechanism underlying the sharing dynamics in the competition assay, we analyzed how exploitation of diffusible matrix adhesion proteins takes place in a spatial context. We observed that at low , cheater biomass was distributed predominantly around isolated producer clusters after washing (Fig. 3A). Based on this observation, we hypothesized that there exists a finite spatial range in which cheater cell clusters are protected against fluid disturbance by RbmC and Bap1 secreted by the producer (Fig. 3B). We defined the radius of this range as the exploitation radius , which we extracted by calculating the cross-correlation between the producer and cheater biomass distributions in the cocultures after washing (Fig. 3C). Conceptually, the farther away a cheater cluster is from a producer cluster, the less likely that it persists on the substrate after washing; this leads to a decaying likelihood of finding cheater biomass around producers with increasing distance. By assuming an exponentially decaying cross-correlation, we arrived at an exploitation radius value of 49 ± 11 µm (Fig. 3C and SI Appendix, Fig. S6; Materials and Methods, Quantification of Exploitation Range and Protection Probability). Note that this value is significantly larger than the physical size of the producer cell cluster, ( µm). This value agrees with the visual inspection of cheater distribution in Fig. 3A. Using this method, we also confirmed the partial redundancy of RbmC and Bap1 in providing adhesion between surfaces and cell clusters, as both single mutants gave a similar exploitation radius (SI Appendix, Fig. S7).
Fig. 3.
A spatial exploitation model quantitively explains competition dynamics. (A) Representative confocal image of a coculture of producer (red) and cheater (yellow). The maximum intensity projection image shows the distribution of cheater biomass around isolated producer clusters after washing. (Scale bar, 50 µm.) (B) Schematic of exploitation radius provided by diffusion of adhesion proteins (blue triangles) from a producer cluster (red cluster). A cheater cluster at the edge of the exploitation range is shown as a yellow cluster. is the physical radius of the producer cluster. (C) Exploitation radius determined as µm by analyzing the cross-correlation between the distribution of producer and cheater biomasses. Error represents the SE from n = 4 independent replicates. The peak at 18 µm corresponds to . The horizontal dashed line indicating of the maximum cross-correlation intersects at 58 µm, corresponding to . (D) Schematic of the spatial model quantifying the protection probability conferred by randomly distributed producer clusters. Each inoculated individual cell contributes to an area of protection around it, and all producer clusters collectively yield , where is the inoculation number density of the producer. (E) Experimentally measured plotted versus . Fitting the data with the equation in D yields µm with 95% confidence bounds. a.u., arbitrary units.
Next, to cross-check the value of exploitation radius and to show how this concept can quantitatively explain competition dynamics, we constructed a spatial model to interpret the dependence of competition outcome on inoculation number density and initial producer frequency (Materials and Methods, Spatial Exploitation Model Based on Randomly Distributed Disks). Briefly, we assumed that each producer cell cluster originates from a randomly distributed founder cell, inoculated at a number density (Fig. 3D). As such, cheaters within a distance from any producer cluster remain adherent after washing; otherwise, they are removed. In this setting, the probability of a cheater cluster falling into the exploitation radius of any producer cluster and being protected from flow disturbance is related to the total exploitable area provided collectively by all of the producers in the system. To calculate the latter, we treated each producer cell cluster as contributing a disk of area of protection (Fig. 3D). Note that these imaginary disks can overlap with each other because there is no constraint on the distance between inoculated producer cells. The relation between , , and can be derived from probability theory: (Fig. 3D; Materials and Methods). Importantly, can be easily extracted from the biomass data before and after washing by identifying as the portion of cheater biomass remaining adherent after washing (SI Appendix, Fig. S4). Fitting versus from experimental data with the theoretical relation yields 63 ± 7 µm, where the error corresponds to 95% confidence bounds (Fig. 3E). This value is slightly higher than the value extracted above (49 ± 11 µm), but the agreement between the two estimates—one from direct quantification of biomass distribution and the other derived from the spatial exploitation model—is remarkable. The two methods serve as a consistency test for each other and corroborate our hypothesis that exploitation of matrix proteins can take place beyond the physical size of the clonal cluster but only within a finite, well-defined length scale. A two-part model combining the spatial exploitation model with the structureless competition model can qualitatively reproduce all the experimental data in the 96-well plate (SI Appendix, Fig. S5).
Flow Shifts the Competition between Producer and Cheater by Modulating the Exploitation Range of Shared Adhesion Proteins.
Thus far, we have studied the population dynamics of RbmC/Bap1-producing and cheating strains in static liquid environments; however, in natural contexts of V. cholerae and many other microbes, including on marine detritus (so-called marine snow) (46), biofilms grow under continuous fluid flow. Indeed, flow has been shown previously to alleviate the potential for exploitation in case of secreted digestive enzymes (27) in biofilms growing on chitin.To understand how a continuous flow might shift the competition between diffusible matrix producers and cheaters, we conducted competition assays in microfluidic channels with a flow rate of 1 µL/min, corresponding to an average flow speed of 694 µm/s (Materials and Methods, Competition Assay under Flow). This flow speed falls into the range of the measured speed of sinking marine snow particles, the natural context of (non–host-associated) V. cholerae and many other marine bacteria (47, 48).
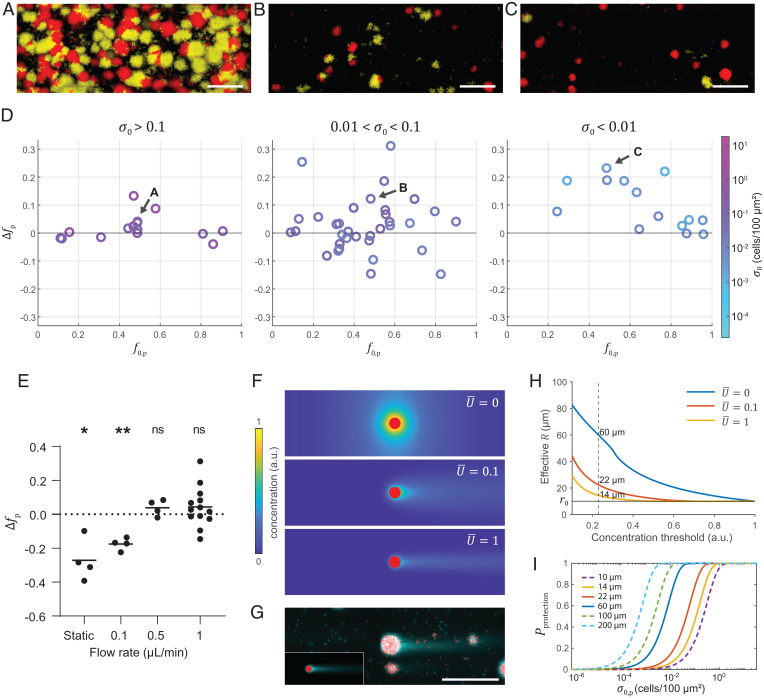
The presence of flow dramatically changed the population dynamics relative to static cocultures: Competition was generally neutral at medium to high inoculation density (), indicating that RbmC/Bap1-producer and cheater strains can coexist under this condition (Fig. 4A–D). In contrast, at , producers outcompeted cheaters regardless of initial population composition. Compared to a static environment, continuous flow generally enhanced the fitness of the RbmC/Bap1-producer against cheating strains (Fig. 2E). For example, in the range and , an increase in the flow rate from 0 to 1 µL/min increased the fitness of the producer, an effect that saturated beyond 0.5 µL/min under our experimental conditions (Fig. 4E).
Fig. 4.
Flow shifts the competition between producer and cheater by modulating the exploitation range of the adhesion protein. (A–C) Representative maximum intensity projection images of producer (red) and cheater (yellow) biofilms grown in a microfluidic channel with different corresponding to data labeled in D. (Scale bars, 100 µm.) (D) Producers in competition with cheaters at different in a flow environment (flow rate = 1 µL/min). Arrows labeled with A, B, and C indicate data points corresponding to images in (A), (B), and (C), respectively. (E) Dependence of on flow rate. Data labeled static were obtained from the static competition assay in 96-well plate. Data were taken at and . One sample t test was used to examine if is statistically different from 0 (*P < 0.05 and **P < 0.01; n = 4, 4, 4, and 13). (F) Simulated depth-averaged adhesion protein concentration in a flow chamber at different average flow speeds . The concentration is normalized by the value at the edge of the producer cluster at . The red disk corresponds to a producer cluster of radius µm. (G) Distribution of adhesion protein Bap1 (cyan) around producer clusters (red) at the surface of the flow chamber shown by Cy3-conjugated anti-FLAG antibody staining. The corresponding simulation result is shown in the inset (Lower left). (Scale bar, 100 µm.) (H) Effective as a function of concentration threshold for protection at different flow rates in simulations. The horizontal line indicates the edge of the producer cluster at . A threshold concentration of 0.23 (vertical dashed line) yields 60, 22, and 14 µm for = 0, 0.1, and 1, respectively. (I) versus calculated for different values. a.u., arbitrary units; ns, not significant.
To understand mechanistically how fluid flow shifts the competition between RbmC/Bap1 producers and cheaters, we performed numerical modeling of the fluid flow and adhesion protein concentration around a producer cluster attached to the surface at different average flow speeds ( in simulations corresponds to 1 µL/min in experiments). The producer cluster is modeled as a surface-attached hemisphere and secretes matrix molecules at a constant rate from the biofilm–fluid interface (Materials and Methods, Flow and Adhesion Protein Simulation in a Microfluidic Environment). The simulation results show that compared to the purely diffusive case, the adhesion protein concentration was drastically reduced around the producer cluster due to advection at a finite flow speed (Fig. 4F). Furthermore, the advective transport resulted in a comet-tail profile of the protein concentration at the glass surface along the wake of the fluid flow, which we confirmed in the immunostaining experiment performed under the same condition (Fig. 4G).
To explain the difference in fitness of the producers in the static and flow environments, we again applied the concept of exploitation range. Assuming a threshold concentration above which cheater biofilms receive sufficient adhesion proteins to resist flow, the corresponding effective around producer cell clusters can be derived based on the simulated concentration profiles of RbmC/Bap1 at different flow speeds (Fig. 4F). As increases, the effective reduces sharply: For example, setting an arbitrary threshold, as shown in Fig. 4H, yields 60, 22, and 14 µm for = 0, 0.1, and 1, respectively. The ensuing , which is dependent on , decreases dramatically with decreasing as shown in Fig. 4I. Our simulation results thus show that flow shrinks the effective exploitation radius conferred by an individual producer cluster (27), which in turn reduces and positively shifts the competitive dynamics toward a regime that increasingly favors RbmC/Bap1-producing cells. The dependence of on flow therefore qualitatively accounts for the dependence of fitness on flow (Fig. 4E). Combined with the distance between producer cell clusters driven by sparse single cells colonizing the surface, flow limits the spatial range of sharable matrix components and averts exploitation of sharable matrix adhesion proteins.
Discussion
Cell–cell attachment and cell–surface adhesion are often critical to the evolutionary advantage of biofilm formation. The extracellular matrix plays an important role in controlling biofilm organization and adhesion (37–39, 45, 49–54), and understanding the evolutionary dynamics of its production remains an active area of work in this research domain. The biofilm matrix often contains components that vary in their diffusion properties and potential for exploitation by cells that do not produce them. In this study, we explored the population dynamics of diffusible matrix protein production in V. cholerae biofilms. We carried out competition assays under static and flow conditions, both of which are frequently encountered by V. cholerae in native habitats and in hosts (47, 48, 55). We found that the matrix components RbmC and Bap1, as diffusible public goods, can be exploited and confer resistance to physical disturbance to clusters of nonproducing cells within a finite spatial range around producer cell clusters. This exploitation range depends on the diffusion–advection condition of the environment, where a continuous flow effectively shrinks the exploitation range and consequently suppresses cheater exploitation. In their marine and freshwater reservoirs, V. cholerae cells often attach to floating food particles and experience shear flows (47, 48). During infection, V. cholerae biofilms are primarily localized on the tips of intestinal villi and therefore experience peristaltic flow in the gut (55). The flow speed and the associated shear stress used in this study fall within the range measured in these environments (56). The population densities of V. cholerae cells in natural ecosystems have been measured as ranging from 102 to 105 cells per liter (57), corresponding to an average distance between cells on the order of millimeters in 3D space. Thus, our results suggest that the combination of flow and low cell density drives the competition dynamics in favor of RbmC and Bap1 producers and leads to the evolutionary stability of these diffusible matrix proteins in V. cholerae.
It is interesting to contrast our results with those for the other major matrix protein in V. cholerae biofilms, RbmA, which (together with VPS) is responsible for high-density cell–cell packing within biofilm cell clusters. RbmA is secreted and shared in a limited fashion within cell lineage groups producing it, conferring protection from exploitation with little dependence on the distance between clusters of producing versus nonproducing cells (4). We envision that the different sharing dynamics of RbmA, RbmC, and Bap1 are constrained by their function as components of the biofilm matrix. RbmA, which holds mother-daughter cell lineages together, must be sequestered and stay in close proximity to producing cells to perform this function. RbmC and Bap1, on the other hand, must travel a distance away from producing cells to confer adhesion between cell groups and underlying surfaces, making them inherently sharable and exploitable; the exact biochemical and biophysical mechanisms underlying this difference await future research. As a consequence, different matrix components—even those produced by cells of one species within the same biofilm—can have different environmental constraints and population structures as conditions for their evolutionary stability.
Our results have a straightforward correspondence with social evolution theory. Hamilton’s rule provides the canonical condition under which cooperation is favored by selection: The fitness benefit of receiving cooperative help, weighted by the relatedness coefficient that quantifies the correlation between recipient genotype and cooperative actor genotype (58–60), must exceed the cost of the cooperative behavior (18). Putting this principle into the context of our experiments, the cost of RbmC and Bap1 production is set by their regulation (here, nearly constitutive) and nutrient supply conditions, and the benefit is set by the extent to which shear stress is applied to biofilms (SI Appendix, Fig. S8). The relatedness coefficient—taking note again that this effectively measures the extent to which RbmC/Bap1-producing cells benefit each other relative to the total population composition (28, 58, 60–62)—is controlled by the range over which RbmC and Bap1 diffuse and the spatial distribution of producer and cheater cell clusters (26, 28, 29, 58, 62, 63). In many reported cases, the clustering of clonal cell lineages within mixed biofilm communities can be sufficient to stabilize cooperation against cheating (25–29). Here, however, the spatial range of RbmC/Bap1 sharing was larger than the size of clonal cell clusters due to the leakage of these proteins from groups of producing cells, and the associated public goods dilemma could not be resolved by clonal group clustering alone. Instead, the solution is rooted in the combination of biophysical mechanisms of protein retention and diffusion, community spatial structure, and environmental perturbations, all captured in the spatial exploitation model (Fig. 3).
The key parameter in our model, the exploitation radius R, can additionally depend on many physiological factors including the production rate of the matrix, duration of biofilm growth, aggregation of adhesion proteins, uptake of adhesion protein by cheater matrix, and biofilm dispersal (64). Competition for binding to the exopolysaccharides and for surface adsorption from other biomacromolecules in the environment may also reduce (65). In addition, although we have assumed to be constant in our spatial model of , does appear to vary and drop to a much smaller value when the inoculation number density goes above 10−1 cells/100 µm2 (SI Appendix, Fig. S9). This can be understood as follows: At a high total inoculation number density, the size of each producer cluster is smaller due to nutrient limitation, and the concentration of secreted proteins around each producer cluster is consequently reduced. Additionally, the elevated incorporation of adhesion proteins into the surrounding cheater clusters further constrains the exploitation range. In the limit of a confluent biofilm layer, the system reduces to the well-studied case of a densely packed bacterial colony (21, 23). Another approximation in our model is that the density of producer clusters is taken as the inoculation number density of producer cells, . In practice, we observed a modest increase in the number of producer clusters at the end of the experiment due to cell dispersal and recolonization (SI Appendix, Fig. S10). To account for these details, a multiscale model that considers these additional factors should be implemented, which we leave for future studies. Nevertheless, our simplified spatial model already provides a good quantitative description of the underlying dynamics of cooperative matrix protein production, especially in the low seeding density regime most relevant to the natural conditions of V. cholerae. While the concept of exploitation range has been discussed in relation to public goods production in bacterial communities (26–29, 66–69), direct quantitation of the exploitation radius and a close comparison between model and experiment, as we achieved here, represent important steps forward. Our study thus provides both a conceptual guideline and technical tool set for future studies on how to quantify public goods sharing in relation to population structure in the biofilm context.
Finally, previous work has shown that in the cases of extracellular enzymes (27, 70, 71), siderophores (23, 72), and autoinducers (73–75) as diffusive public goods, clonal segregation and efficient consumption of public goods (or their enzymatic products and complexes) through high local cell density and a high uptake rate can minimize the potential for exploitation in the context of continuous films (21, 26, 27). In the context of spatially discrete clonal clusters, we expect the evolutionary stability of these public goods to be determined by the interplay between the exploitation range and the spatial structure as shown here.
Materials and Methods
Methods for protein quantification, the two-part competition model, and the statistical analysis are further described in SI Appendix, Materials and Methods.
Strains and Media.
All V. cholerae strains used in this study were derivatives of the wild-type Vibrio cholerae O1 biovar El Tor strain C6706 and are listed in SI Appendix, Table S1. All strains harbor a missense mutation in the vpvC gene (vpvCW240R) that elevates intracellular cyclic diguanylate (c-di-GMP) level and are robust biofilm formers (44). This allowed us to focus on the ecological consequences of adhesion protein production in V. cholerae biofilms rather than behaviors involving gene regulation. Additional mutations were genetically engineered using the natural transformation method or through the suicide vector pKAS32 (76, 77). All strains were grown overnight in lysogeny broth (LB) at 37 °C with shaking. Competition assays were performed in M9 medium and supplemented with 2 mM MgSO4, 100 μM CaCl2, 0.5% glucose, and 0.1 mg/mL bovine serum albumin (BSA). BSA was required for blocking nonspecific surface adsorption in the immunostaining experiments; to be consistent, we included BSA in all competition assays as well.
Static Competition Assay with End-point Flow Disturbance.
V. cholerae strains carrying different fluorescent proteins were streaked onto an LB agar plate and grown overnight at 37 °C. Single colonies with a rugose morphology were picked and grown overnight at 37 °C with glass beads. The LB culture was then vortexed vigorously before back-diluting 30-fold in M9 medium and grown for 3 to 4 h with shaking and with glass beads. After another vortexing, the inoculant was washed with fresh M9 medium without glucose three times. The inoculant was then bead bashed using a Digital Disrupter Genie with small glass beads (acid-washed, 425 to 500 μm; Sigma-Aldrich). This procedure ensured that large cell clusters formed in culture were broken apart to allow more accurate measurement of optical density at 600 nm(OD600). The inoculants of the two competing strains were then mixed at different ratios and OD600, and 100 µL of the mixture was added to wells of 96-well plates with a #1.5 coverslip bottom (MatTek). The cells were allowed 1 h to attach, after which the wells were washed twice with fresh M9 medium. The wells were subsequently filled with 100 µL M9 growth medium with glucose and 0.1 mg/mL BSA and imaged by a confocal fluorescence microscope (see below) to quantify the inoculation number density for both strains. Inoculation number densities, instead of OD600, were used to quantify due to the asymmetry in inoculation efficiency between producer and cheater (SI Appendix, Fig. S11). The cultures were then grown for 16 to 18 h at 30 °C before biomass quantification. After growth, one set of biomass quantification for both strains was done by fluorescence confocal microscopy before a flow disturbance to the culture was introduced by washing the well vigorously twice with fresh medium, after which another set of biomass quantification was done. Immunostaining of adhesion proteins was done using producer strains expressing C-terminal 3×FLAG-tagged RbmC or Bap1 cocultured with the cheater under the same growth conditions for competition assays, except that the growth medium contained Cy3-conjugated anti-FLAG antibodies (2 µg/mL; Sigma-Aldrich).
Competition Assay under Flow.
The inoculants of the competition strains were prepared in the same way as described in the static competition assay. The inoculants were then introduced into microfluidic channels (channel dimensions: 1 cm in length, 400 µm in width, and 60 µm in height) through the outlet without inoculating the inlet. This ensured that no biofilms were grown in the zero-flow zone directly beneath the inlet. The cells were allowed 2 h to attach, after which sterile inlet and outlet polytetrafluoroethylene tubing was connected to the microfluidic chamber, and the M9 growth medium was flown through the channel at flow rates controlled by a syringe pump. The flow rates used in this study ranged from 0.1 to 1 µL/min, corresponding to average flow speeds from 69.4 to 694 µm/s and shear rates from 7 to 70 s−1. The microfluidic channels were transferred to a confocal fluorescence microscope and imaged to quantify the inoculation number density for both strains. The cultures under flow were then grown for 16 to 18 h at 30 °C before biomass quantification. After growth, one set of biomass quantification for both strains was done by fluorescence microscopy.
Microscopy, Biomass Quantification, and Growth Rate Measurement.
Fluorescence confocal microscopy was conducted using a Yokogawa W1 confocal scanner unit connected to a Nikon Ti2-E inverted microscope with a Perfect Focus System. Cells constitutively expressing SCFP3A, mNeonGreen, and mScarlet-I cytosolically were excited by lasers at 445 nm, 488 nm, and 561 nm, respectively, and the fluorescent signal was recorded by an sCMOS camera (Photometrics Prime BSI) after the corresponding filters. Confocal images were taken using a 60× water immersion objective (numerical aperture = 1.2) at 2 × 2–pixel binning, and the resulting pixel size was 217 nm. All images presented are raw images rendered with Nikon NIS-Elements. Producers are pseudocolored red, cheaters are pseudocolored yellow, and the immunosignals are pseudocolored cyan throughout the text unless indicated otherwise.
To quantify the inoculation number density before growth, an image at the surface was taken for each wavelength at the center of each well or microfluidic channel. To quantify the biomass after growth, a 3D tiled image with a 3-µm step size in z was taken at three to five evenly distributed positions within the same area for each inoculation number density. The total height of the 3D image was chosen to include all biomass of the surface-attached biofilm and was 60 to 72 µm in the static competition assay and 60 µm for the competition assay under flow.
For accurate segmentation and quantification of the biomass, we used a modified Bernsen’s local thresholding method, which is less sensitive to depth-dependent intensity variations and contrast reduction (due to pinhole crosstalk and scattering) in the 3D confocal image. Briefly, each two-dimensional(2D) image slice first went through a constant background subtraction and denoising, and the image contrast was enhanced by subtracting the image by its Gaussian blur. To determine whether a pixel belonged to cells or background, if the contrast in the local neighborhood around a given pixel was higher than the contrast threshold (50 in all data set), the pixel was set to belong to cells if its value was higher than 1.5 times the median value of the neighborhood; otherwise, a pixel was set to belong to cells if its value was higher than an intensity threshold determined based on the brightness of the image. Examples of segmented images are shown in SI Appendix, Fig. S4. The total area of cells in each 2D image was converted to a number density using the average cell sizes, measured separately at low cell densities for different fluorescence channels. The number densities in each 2D slice were then added to obtain the total 3D biomass.
Growth rates of producer and cheater biofilms were measured by time-course imaging of biofilm growing in a 96-well plate. Confocal images were taken every 2 h for 40 h, and biomass was quantified as described above at each time point. The effect of confocal imaging was confirmed to be nonsignificant to biofilm growth under the conditions used in the experiment. We extracted growth rates as slopes of fitting lines to the growth curves in semilog plots in their exponential phase. Carrying capacities used in the competition model were measured as the maximum biomass in the stationary phase.
Quantification of Exploitation Range and Protection Probability.
Cross-correlations between producer and cheater biomass distributions were used to quantify the exploitation radius . We modeled the cheater biomass distribution as being exponentially decaying around a producer cell cluster of radius with the decaying constant equal to ; this yielded an exploitation radius . We found that the cross-correlation between the producer and cheater images gives adequate information to extract information on and (SI Appendix, Fig. S6). To obtain cross-correlations from experimental images, inoculant mixtures of producer strain with OD600 = 0.001 and cheater strain with OD600 = 0.1 were used for the static competition assay with isolated producer clusters. Maximum intensity projections of confocal images for both strains after end-point flow disturbance were thresholded and used for cross-correlation calculations. The field of view of each image was ∼1.8 mm × 1.8 mm. The cross-correlation between producer and cheater was calculated as
where and are the binary images for producer and cheater, and are the row and column indices of the image. was then radially averaged and converted to real spatial units. The error in Fig. 3C was calculated as the SE from the n = 4 replicates. For a negative control of the correlated biomass distribution, we compared the normalized cross-correlation between producer and cheater biofilms with that between producer and producer biofilms (SI Appendix, Fig. S6). The normalized cross-correlation is defined as
where and are the mean values of and , respectively.
Spatial Exploitation Model Based on Randomly Distributed Disks.
Assuming each producer cluster contributes a circular disk of protection with area , the overall protection probability conferred by producer biofilms to cheater biofilms in an area of interest can be modeled by the coverage problem of randomly distributed circular disks (78): Let there be interpenetrable disks with area distributed randomly in an area of interest . The uncovered probability of has a mean value . In the limit of large and , the uncovered probability approaches . Therefore, for producer clusters with number density and exploitation radius , the protection probability reads .
Flow and Adhesion Protein Simulation in a Microfluidic Environment.
To gain insight into the effect of different fluid flow speeds on the transport of molecules released by the producer, we solved the governing equations of the fluid flow and a coupled scalar transport equation to model the molecular distribution around a producer. We assumed the producer cluster was shaped like a solid hemisphere adhered to the bottom surface of a rectangular channel and was subjected to fluid flow, as shown in SI Appendix, Fig. S12. The fluid flow is governed by the Navier-Stokes and continuity equations, and the molecular transport is described by the coupled advection-diffusion equation, which are respectively given by:
| [1] |
| [2] |
| [3] |
In these equations, is the fluid velocity vector, is the pressure, and is the concentration of molecules released by the producer. Eqs. 1–3 are nondimensionalized using a length scale µm, which is the average diameter of a producer cluster, as found in experiments. The timescale used is the advective timescale , where is the dimensional average inflow velocity. The concentration is normalized using an arbitrary reference concentration of adhesion .
There are two important nondimensional parameters that describe the equations. The Reynolds number is defined as the ratio of the inertial to the viscous forces, where is the kinematic viscosity of water. Our experiments were typically in the range , meaning viscous effects dominated the dynamics. At these Re, the left-hand side of Eq. 1 can be neglected, reducing to the Stokes equation. The Péclet number, , is defined as the ratio of the diffusive timescale to the advective timescale; is the diffusion coefficient of the adhesion molecules based on the Stokes-Einstein equation and their estimated size. suggests fluid advection has a larger role in transporting the molecules than diffusive transport. We varied the Péclet number in the range , in agreement with variable experimental conditions of µm/s.
We simulated fluid flow through a channel of dimension as shown in SI Appendix, Fig. S12. The solid hemisphere modeling the producer cluster was positioned at . In the experiments, the aspect ratio of the channel was , where . This large aspect ratio made simulations computationally expensive, so we instead used a channel of dimensions in the simulation, which preserved the important cross-stream dimension.
A no-slip boundary condition was imposed for the top and bottom surfaces of the domain ( at ). A Dirichlet boundary condition was applied at the inlet ( at ), where is the average nondimensional inflow velocity into the channel scaled by , and corresponds to an average velocity of 694 µm/s. An open/outflow boundary condition was applied to the outlet at . Along the material walls at , we imposed a free-slip or no-penetration boundary condition. The boundary condition for the concentration was no-flux on all material walls, except for the surface of the solid hemisphere, for which we used a time-varying Dirichlet boundary condition of the form
| [4] |
Here, molecules/µm3/s is the total production rate of the molecules from the surface of the producer cluster (43), and quantifies the fraction of molecules retained in the producer cluster based on experimentally measured values. Eq. 4 suggests that there is a constant rate of production of molecules at the surface of the hemisphere.
We numerically solved the governing Eqs. 1–4 using the spectral element fluid solver NEK5000 (79). NEK5000 has been used to study a broad range of fluid flow problems (80), including complicated chaotic fluid flows and reaction-advection-diffusion systems (81–83). Our approach uses a semi-implicit operator-splitting scheme, which is second order accurate in time and exponentially convergent in space. Our domain consisted of 392 hexahedral spectral elements, and we used a 17th-order Lagrangian interpolant polynomial for spatial discretization, which we found was sufficient to resolve both the fluid flow and the intricate features of the molecular distribution around the producer.
To resolve the small Re of the flow, we evolved Eqs. 1 and 2 using an extremely small time step of . We simulated the flow field for steps and ensured that the flow field reached a steady state. We then stopped the evolution of the fluid flow and used the steady velocity profile to solve the advection-diffusion equation (Eq. 3). Since we did not evolve the flow field, we used larger time steps (typically ) to evolve the concentration field for a total of 3 min (SI Appendix, Fig. S13).
Supplementary Material
Acknowledgments
We thank Andrew Sharo, Sara Siwiecki, and Carissa Chan for their help in the initial experiments. We thank Xin Huang, Japinder Nijjer, and Qiuting Zhang for helpful discussions. Alexandre Persat, Kevin Foster, Benjamin Wucher, and James Winans provided helpful feedback on the manuscript. S.M. thanks Paul Fischer, Som Dutta, and the Nek5000 user group for helpful discussions regarding numerical computations. Numerical computations were performed using the resources of the Minnesota Supercomputing Institute at the University of Minnesota. J.Y. holds a Career Award at the Scientific Interface from the Burroughs Wellcome Fund. C.D.N. is supported by the Simons Foundation Award No. 826672. J.T. holds a Career Award at the Scientific Interface from the Burroughs Wellcome Fund. J.-S.B.T. is a Damon Runyon Fellow supported by the Damon Runyon Cancer Research Foundation (Grant No. DRG-2446-21). Research reported in this publication was supported by the National Institute of General Medical Sciences of the NIH under Award No. DP2GM146253 (awarded to J.Y.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2123469119/-/DCSupplemental.
Data Availability
All data are included in the article and/or SI Appendix. The original data can be accessed through doi:10.5061/dryad.rfj6q57d9.
References
- 1.Flemming H.-C., et al. , Biofilms: An emergent form of bacterial life. Nat. Rev. Microbiol. 14, 563–575 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Stewart P. S., Costerton J. W., Antibiotic resistance of bacteria in biofilms. Lancet 358, 135–138 (2001). [DOI] [PubMed] [Google Scholar]
- 3.Persat A., et al. , The mechanical world of bacteria. Cell 161, 988–997 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nadell C. D., Drescher K., Wingreen N. S., Bassler B. L., Extracellular matrix structure governs invasion resistance in bacterial biofilms. ISME J. 9, 1700–1709 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wucher B. R., Elsayed M., Adelman J. S., Kadouri D. E., Nadell C. D., Bacterial predation transforms the landscape and community assembly of biofilms. Curr. Biol. 31, 2643–2651.e3 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vidakovic L., Singh P. K., Hartmann R., Nadell C. D., Drescher K., Dynamic biofilm architecture confers individual and collective mechanisms of viral protection. Nat. Microbiol. 3, 26–31 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hall-Stoodley L., Costerton J. W., Stoodley P., Bacterial biofilms: From the natural environment to infectious diseases. Nat. Rev. Microbiol. 2, 95–108 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Barrasso K., et al. , Impact of a human gut microbe on Vibrio cholerae host colonization through biofilm enhancement. eLife 11:e73010 (2022). [DOI] [PMC free article] [PubMed]
- 9.Foster T. J., Geoghegan J. A., Ganesh V. K., Höök M., Adhesion, invasion and evasion: The many functions of the surface proteins of Staphylococcus aureus. Nat. Rev. Microbiol. 12, 49–62 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tamayo R., Patimalla B., Camilli A., Growth in a biofilm induces a hyperinfectious phenotype in Vibrio cholerae. Infect. Immun. 78, 3560–3569 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang Z., Nero T., Mukherjee S., Olson R., Yan J., Searching for the secret of stickiness: How biofilms adhere to surfaces. Front. Microbiol. 12, 686793 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nadell C. D., Drescher K., Foster K. R., Spatial structure, cooperation and competition in biofilms. Nat. Rev. Microbiol. 14, 589–600 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Cordero O. X., Datta M. S., Microbial interactions and community assembly at microscales. Curr. Opin. Microbiol. 31, 227–234 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Datta M. S., Sliwerska E., Gore J., Polz M. F., Cordero O. X., Microbial interactions lead to rapid micro-scale successions on model marine particles. Nat. Commun. 7, 11965 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ebrahimi A., Schwartzman J., Cordero O. X., Cooperation and spatial self-organization determine rate and efficiency of particulate organic matter degradation in marine bacteria. Proc. Natl. Acad. Sci. U.S.A. 116, 23309–23316 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stocker R., Seymour J. R., Ecology and physics of bacterial chemotaxis in the ocean. Microbiol. Mol. Biol. Rev. 76, 792–812 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.West S. A., Cooper G. A., Ghoul M. B., Griffin A. S., Ten recent insights for our understanding of cooperation. Nat. Ecol. Evol. 5, 419–430 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamilton W. D., The genetical evolution of social behaviour. I. J. Theor. Biol. 7, 1–16 (1964). [DOI] [PubMed] [Google Scholar]
- 19.Hardin G., The tragedy of the commons. The population problem has no technical solution; it requires a fundamental extension in morality. Science 162, 1243–1248 (1968). [PubMed] [Google Scholar]
- 20.West S. A., Diggle S. P., Buckling A., Gardner A., Griffin A. S., The social lives of microbes. Annu. Rev. Ecol. Evol. Syst. 38, 53–77 (2007). [Google Scholar]
- 21.Dal Co A., van Vliet S., Kiviet D. J., Schlegel S., Ackermann M., Short-range interactions govern the dynamics and functions of microbial communities. Nat. Ecol. Evol. 4, 366–375 (2020). [DOI] [PubMed] [Google Scholar]
- 22.Van Vliet S., Hauert C., Fridberg K., Ackermann M., Dal Co A., Global dynamics of microbial communities emerge from local interaction rules. PLoS Comput. Biol. 18, e1009877 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Julou T., et al. , Cell-cell contacts confine public goods diffusion inside Pseudomonas aeruginosa clonal microcolonies. Proc. Natl. Acad. Sci. U.S.A. 110, 12577–12582 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Allen B., Gore J., Nowak M. A., Spatial dilemmas of diffusible public goods. eLife 2, e01169 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nadell C. D., Foster K. R., Xavier J. B., Emergence of spatial structure in cell groups and the evolution of cooperation. PLOS Comput. Biol. 6, e1000716 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nadell C. D., et al. , Cutting through the complexity of cell collectives. Proc. Biol. Sci. 280, 20122770 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Drescher K., Nadell C. D., Stone H. A., Wingreen N. S., Bassler B. L., Solutions to the public goods dilemma in bacterial biofilms. Curr. Biol. 24, 50–55 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Driscoll W. W., Pepper J. W., Theory for the evolution of diffusible external goods. Evolution 64, 2682–2687 (2010). [DOI] [PubMed] [Google Scholar]
- 29.Allison S. D., Cheaters, diffusion and nutrients constrain decomposition by microbial enzymes in spatially structured environments. Ecol. Lett. 8, 626–635 (2005). [Google Scholar]
- 30.Hamilton W. D., The genetical evolution of social behaviour. II. J. Theor. Biol. 7, 17–52 (1964). [DOI] [PubMed] [Google Scholar]
- 31.Rainey P. B., Rainey K., Evolution of cooperation and conflict in experimental bacterial populations. Nature 425, 72–74 (2003). [DOI] [PubMed] [Google Scholar]
- 32.van Gestel J., Weissing F. J., Kuipers O. P., Kovács A. T., Density of founder cells affects spatial pattern formation and cooperation in Bacillus subtilis biofilms. ISME J. 8, 2069–2079 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Irie Y., et al. , The Pseudomonas aeruginosa PSL polysaccharide is a social but noncheatable trait in biofilms. MBio 8, e00374-e17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Otto S. B., et al. , Privatization of biofilm matrix in structurally heterogeneous biofilms. mSystems 5, e00425–e20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dragoš A., et al. , Collapse of genetic division of labour and evolution of autonomy in pellicle biofilms. Nat. Microbiol. 3, 1451–1460 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Rossy T., Nadell C. D., Persat A., Cellular advective-diffusion drives the emergence of bacterial surface colonization patterns and heterogeneity. Nat. Commun. 10, 2471 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berk V., et al. , Molecular architecture and assembly principles of Vibrio cholerae biofilms. Science 337, 236–239 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fong J. C. N., Syed K. A., Klose K. E., Yildiz F. H., Role of Vibrio polysaccharide (vps) genes in VPS production, biofilm formation and Vibrio cholerae pathogenesis. Microbiology (Reading) 156, 2757–2769 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fong J. C., et al. , Structural dynamics of RbmA governs plasticity of Vibrio cholerae biofilms. eLife 6, e26163 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nadell C. D., Bassler B. L., A fitness trade-off between local competition and dispersal in Vibrio cholerae biofilms. Proc. Natl. Acad. Sci. U.S.A. 108, 14181–14185 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nadell C. D., Ricaurte D., Yan J., Drescher K., Bassler B. L., Flow environment and matrix structure interact to determine spatial competition in Pseudomonas aeruginosa biofilms. eLife 6, e21855 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Madsen J. S., et al. , Facultative control of matrix production optimizes competitive fitness in Pseudomonas aeruginosa PA14 biofilm models. Appl. Environ. Microbiol. 81, 8414–8426 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Absalon C., Van Dellen K., Watnick P. I., A communal bacterial adhesin anchors biofilm and bystander cells to surfaces. PLoS Pathog. 7, e1002210 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beyhan S., Yildiz F. H., Smooth to rugose phase variation in Vibrio cholerae can be mediated by a single nucleotide change that targets c-di-GMP signalling pathway. Mol. Microbiol. 63, 995–1007 (2007). [DOI] [PubMed] [Google Scholar]
- 45.Yan J., Sharo A. G., Stone H. A., Wingreen N. S., Bassler B. L., Vibrio cholerae biofilm growth program and architecture revealed by single-cell live imaging. Proc. Natl. Acad. Sci. U.S.A. 113, E5337–E5343 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alcolombri U., et al. , Sinking enhances the degradation of organic particles by marine bacteria. Nat. Geosci. 14, 775–780 (2021). [Google Scholar]
- 47.Diercks A.-R., Asper V. L., In situ settling speeds of marine snow aggregates below the mixed layer: Black Sea and Gulf of Mexico. Deep Sea Res. Part I Oceanogr. Res. Pap. 44, 385–398 (1997). [Google Scholar]
- 48.Nowald N., et al. , “In-situ sinking speed measurements of marine snow aggregates acquired with a settling chamber mounted to the Cherokee ROV” in OCEANS 2009-EUROPE, (Institute of Electrical and Electronics Engineers, 2009), pp. 1–6. [Google Scholar]
- 49.Drescher K., et al. , Architectural transitions in Vibrio cholerae biofilms at single-cell resolution. Proc. Natl. Acad. Sci. U.S.A. 113, E2066–E2072 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hartmann R., et al. , Emergence of three-dimensional order and structure in growing biofilms. Nat. Phys. 15, 251–256 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jeckel H., et al. , Multispecies phase diagram reveals biophysical principles of bacterial biofilm architectures. bioRxiv [Preprint] (2021). https://www.biorxiv.org/content/10.1101/2021.08.06.455416v1. Accessed 15 September 2021.
- 52.Beroz F., et al. , Verticalization of bacterial biofilms. Nat. Phys. 14, 954–960 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Q., et al. , Morphogenesis and cell ordering in confined bacterial biofilms. Proc. Natl. Acad. Sci. U.S.A. 118, e2107107118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nijjer J., et al. , Mechanical forces drive a reorientation cascade leading to biofilm self-patterning. Nat. Commun. 12, 6632 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gallego-Hernandez A. L., et al. , Upregulation of virulence genes promotes Vibrio cholerae biofilm hyperinfectivity. Proc. Natl. Acad. Sci. U.S.A. 117, 11010–11017 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kulthong K., et al. , Transcriptome comparisons of in vitro intestinal epithelia grown under static and microfluidic gut-on-chip conditions with in vivo human epithelia. Sci. Rep. 11, 3234 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kirchberger P. C., et al. , Culture-independent tracking of Vibrio cholerae lineages reveals complex spatiotemporal dynamics in a natural population. Environ. Microbiol. 22, 4244–4256 (2020). [DOI] [PubMed] [Google Scholar]
- 58.Frank S. A., The Foundations of Social Evolution (Princeton University Press, 1998). [Google Scholar]
- 59.Gardner A., The price equation. Curr. Biol. 18, R198–R202 (2008). [DOI] [PubMed] [Google Scholar]
- 60.Pepper J. W., Relatedness in trait group models of social evolution. J. Theor. Biol. 206, 355–368 (2000). [DOI] [PubMed] [Google Scholar]
- 61.Damore J. A., Gore J., Understanding microbial cooperation. J. Theor. Biol. 299, 31–41 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Queller D. C., Genetic relatedness in viscous populations. Evol. Ecol. 8, 70–73 (1994). [Google Scholar]
- 63.Brockhurst M. A., Habets M. G. J. L., Libberton B., Buckling A., Gardner A., Ecological drivers of the evolution of public-goods cooperation in bacteria. Ecology 91, 334–340 (2010). [DOI] [PubMed] [Google Scholar]
- 64.Rumbaugh K. P., Sauer K., Biofilm dispersion. Nat. Rev. Microbiol. 18, 571–586 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Callow J. A., Callow M. E., Trends in the development of environmentally friendly fouling-resistant marine coatings. Nat. Commun. 2, 244 (2011). [DOI] [PubMed] [Google Scholar]
- 66.Koschwanez J. H., Foster K. R., Murray A. W., Sucrose utilization in budding yeast as a model for the origin of undifferentiated multicellularity. PLoS Biol. 9, e1001122 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mitri S., Xavier J. B., Foster K. R., Social evolution in multispecies biofilms. Proc. Natl. Acad. Sci. U.S.A. 108 (suppl. 2), 10839–10846 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Estrela S., Brown S. P., Community interactions and spatial structure shape selection on antibiotic resistant lineages. PLoS Comput. Biol. 14, e1006179 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Van Dyken J. D., Müller M. J. I., Mack K. M. L., Desai M. M., Spatial population expansion promotes the evolution of cooperation in an experimental Prisoner’s Dilemma. Curr. Biol. 23, 919–923 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Frost I., et al. , Cooperation, competition and antibiotic resistance in bacterial colonies. ISME J. 12, 1582–1593 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sharma A., Wood K. B., Spatial segregation and cooperation in radially expanding microbial colonies under antibiotic stress. ISME J. 15, 3019–3033 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Griffin A. S., West S. A., Buckling A., Cooperation and competition in pathogenic bacteria. Nature 430, 1024–1027 (2004). [DOI] [PubMed] [Google Scholar]
- 73.Rumbaugh K. P., et al. , Quorum sensing and the social evolution of bacterial virulence. Curr. Biol. 19, 341–345 (2009). [DOI] [PubMed] [Google Scholar]
- 74.van Gestel J., et al. , Short-range quorum sensing controls horizontal gene transfer at micron scale in bacterial communities. Nat. Commun. 12, 2324 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Darch S. E., et al. , Spatial determinants of quorum signaling in a Pseudomonas aeruginosa infection model. Proc. Natl. Acad. Sci. U.S.A. 115, 4779–4784 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dalia A. B., McDonough E., Camilli A., Multiplex genome editing by natural transformation. Proc. Natl. Acad. Sci. U.S.A. 111, 8937–8942 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Skorupski K., Taylor R. K., Positive selection vectors for allelic exchange. Gene 169, 47–52 (1996). [DOI] [PubMed] [Google Scholar]
- 78.Hall P., Introduction to the Theory of Coverage Processes (Wiley, 1988). [Google Scholar]
- 79.nek5000 website. https://nek5000.mcs.anl.gov/ (2008). Accessed 15 June 2021.
- 80.Fischer P. F., An overlapping Schwarz method for spectral element solution of the incompressible Navier–Stokes equations. J. Comput. Phys. 133, 84–101 (1997). [Google Scholar]
- 81.Mukherjee S., Paul M. R., Velocity and geometry of propagating fronts in complex convective flow fields. Phys. Rev. E 99, 012213 (2019). [DOI] [PubMed] [Google Scholar]
- 82.Mukherjee S., Front Propagation and Feedback in Convective Flow Fields (Virginia Polytechnic Institute and State University, 2020). [Google Scholar]
- 83.Paul M. R., Chiam K.-H., Cross M. C., Fischer P. F., Greenside H. S., Pattern formation and dynamics in Rayleigh–Bénard convection: Numerical simulations of experimentally realistic geometries. Physica D 184, 114–126 (2003). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are included in the article and/or SI Appendix. The original data can be accessed through doi:10.5061/dryad.rfj6q57d9.