Abstract
A total of 137 soilborne and plant-associated bacterial strains belonging to different Pseudomonas species were tested for their ability to synthesize N-acyl-homoserine lactones (NAHL). Fifty-four strains synthesized NAHL. Interestingly, NAHL production appears to be more common among plant-associated than among soilborne Pseudomonas spp. Indeed, 40% of the analyzed Pseudomonas syringae strains produced NAHL which were identified most often as the short-chain NAHL, N-hexanoyl-l-homoserine lactone, N-(3-oxo-hexanoyl)-homoserine lactone, and N-(3-oxo-octanoyl)-l-homoserine lactone (no absolute correlation between genomospecies of P. syringae and their ability to produce NAHL could be found). Six strains of fluorescent pseudomonads, belonging to the species P. chlororaphis, P. fluorescens, and P. putida, isolated from the plant rhizosphere produced different types of NAHL. In contrast, none of the strains isolated from soil samples were shown to produce NAHL. The gene encoding the NAHL synthase in P. syringae pv. maculicola was isolated by complementation of an NAHL-deficient Chromobacterium mutant. Sequence analysis revealed the existence of a luxI homologue that we named psmI. This gene is sufficient to confer NAHL synthesis upon its bacterial host and has strong homology to psyI and ahlI, two genes involved in NAHL production in P. syringae pv. tabaci and P. syringae pv. syringae, respectively. We identified another open reading frame that we termed psmR, transcribed convergently in relation to psmI and partly overlapping psmI; this gene encodes a putative LuxR regulatory protein. This gene organization, with luxI and luxR homologues facing each other and overlapping, has been found so far only in the enteric bacteria Erwinia and Pantoea and in the related species P. syringae pv. tabaci.
In most habitats, microbes compete to ensure their survival and multiplication. In the plant and soil environments, the mechanisms that allow a bacterium to outcompete other microbes are diverse (for reviews, see references 6, 8, and 47). Some of these traits are expressed constitutively, i.e., at any moment in the life of the microbial cell. Others are expressed only at times most favorable to allow an efficient biological effect. The regulation of these processes may depend on environmental parameters or alterations sensed by the microbes (see, for example, references 25 and 34; for a review, see reference 5), such as changes in microbial cell density. Indeed, microbes have evolved regulatory systems allowing gene expression only when the microbial cell density is appropriate. Such a phenomenon, which couples the microbial cell density to the expression of the relevant biological traits, is known as quorum sensing (QS) (for reviews, see references 20, 24, 41, and 46).
The mechanism underlying QS has been described for several microbes, including gram-negative bacteria, gram-positive bacteria, and streptomycetes (26). It involves the synthesis of low-molecular-weight molecules that diffuse in and out of the bacterial cell. As the bacterial population density increases, the amount of signal molecules synthesized and, consequently, their concentration in the environment increase. Once a critical concentration is reached, the signal molecules can be bound by a ligand protein, which acts as a transcription regulator in the microbial cell, allowing, upon binding, the expression of QS-regulated genes (for reviews, see references 20, 24, 41, and 46).
In gram-negative bacteria, the QS signal molecules are almost exclusively N-acyl-homoserine lactones (NAHL). Light emission by the fish symbiont Photobacterium fischeri (also known as Vibrio fischeri) was the first biological function known to be regulated in a QS-dependent fashion (13). In this bacterium, the NAHL-derived mediator was identified as N-(3-oxo-hexanoyl)-homoserine lactone (OHHL) (14), the synthesis of which relies on the activity of the NAHL synthase LuxI (encoded at the luxI locus). OHHL can be bound by LuxR (encoded by the luxR gene), which in turn is converted, upon binding, to a functional transcription regulator (for a review, see reference 46). This regulator attaches to a 20-nucleotide, inverted-repeated sequence located upstream of the operon encoding the proteins responsible for luminescence (16), allowing its transcription. This palindrome sequence is known as the lux operator or the lux box. It has been found in the promoter regions of several QS-regulated genes (see, for example, references 1, 2, and 19). Interestingly, the first gene of the lux operon is luxI. Thus, the activation of the lux operon stimulates the production of the protein responsible for the production of OHHL (44).
Several bacterial traits are known to be regulated by LuxI- and LuxR-like proteins as a function of cell density (41). Indeed, luxI and luxR sequences have been detected in various gram-negative bacteria, and the production of NAHL signal molecules is widespread (4). These characteristics apply to plant-associated bacteria whether they are beneficial or deleterious for plant growth and health (37). Among the systems described so far, the conjugal transfer of the Ti plasmid of Agrobacterium tumefaciens depends on the presence of the relevant opine(s) in the environment as well as on a QS regulatory mechanism (18, 39). Similarly, the production of macerating exoenzymes (40) or that of the antibiotic carbapenem by Erwinia carotovora strains is regulated in a cell-density-dependent fashion (33; for a review, see reference 46). All the above functions involve the production of different NAHL, which differ from one species to the other or one strain to the other but whose structures are closely related.
Fluorescent pseudomonads have also developed QS-regulated synthesis of secondary metabolites implicated in antagonistic activities against plant pathogens, such as phenazines and pyoverdines (36, 45, 51). Because of the role of QS in the regulation of important physiological processes in plant-bacterium associations and because NAHL production in plant-associated Pseudomonas species has been investigated only in a small number of studies (4, 12), we decided to conduct a broad survey of NAHL production among pseudomonads isolated from plant tissues, the plant rhizosphere, or the bulk soil. NAHL production was not uncommon among plant-associated pseudomonads but was not detected in soil isolates. Among the plant-associated bacteria, members of the pathogenic Pseudomonas syringae and related species often produced NAHL. We therefore decided to characterize the genetic organization of the NAHL-dependent regulatory system in a representative isolate, P. syringae pv. maculicola strain CFBP 10912–9. Genes involved in QS regulation were isolated and analyzed. They appear to be distantly related members of the luxR-luxI gene family, with an unusual organization characterized by luxR and luxI facing and overlapping each other.
(Part of this work was presented at the 9th International Congress on Molecular Plant-Microbe Interactions, Amsterdam, The Netherlands, July 1999.)
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Pseudomonas strains and their origins and characteristics are listed in Tables 1, 2 and 3. The cytochrome c oxidase-positive strains of fluorescent pseudomonads are a representative subset of the diversity of a larger collection of strains (n = 340) isolated from two bulk soils and the rhizosphere of two plant species cultivated in these two soils (28). All bacterial strains were different, even though they belong to the same genomospecies and the same pathovar. Three complementary NAHL biosensors were used: A. tumefaciens strain NT1(pDCI41E33) (43), Chromobacterium violaceum strain CVO26 (32), and Escherichia coli strain JM109(pSB401) (50). Plasmid pDCI41E33 from A. tumefaciens harbors traR but not traI and contains a traG::lacZ fusion. CVO26 is a mini-Tn5-generated mutant of C. violaceum strain ATCC 31532 with all genes involved in violacein production and mutated copies of two regulatory genes, making violacein production dependent upon exogenous NAHL. Plasmid pSB401 harbors the luxR gene and the lux operon of P. fischeri with a deleted luxI region, making light emission dependent on the presence of exogenous NAHL. Other E. coli strains used were DH5α (42) and DB82(pRK2013) (11).
TABLE 1.
Names, origins, and NAHL-producing capacities of strains of cytochrome c oxidase-negative species of fluorescent pseudomonadsa
| Genomospeciesb | Pathovar | Strainc | Host plant | NAHL synthesized |
|---|---|---|---|---|
| P. syringae | syringae | CFBP 1392 | Syringa vulgaris | |
| syringae | Meyer | Phaseolus vulgaris | HHL, OHHL | |
| aptata | CFBP 1617 | Beta vulgaris | ||
| lapsa | CFBP 1731 | Triticum aestivum | ||
| papulans | CFBP 1754 | Malus sylvestris | ||
| pisi | CFBP 2105 | Pisum sativum | ||
| CFBP 10267 | Pisum sativum | |||
| atrofaciens | CFBP 2213 | Triticum aestivum | ||
| aceris | CFBP 2339 | Acer sp. | OHHL, OOHL | |
| panici | CFBP 2345 | Panicum sp. | ||
| dysoxylis | CFBP 2356 | Dysoxylum spectabile | ||
| japonica | CFBP 2896 | Hordeum vulgare | ||
| P. savastanoi | phaseolicola | CFBP 1390 | Phaseolus vulgaris | |
| ulmi | CFBP 1407 | Ulmus sp. | HHL, OHHL, OOHL | |
| mori | CFBP 1642 | Morus alba | ||
| lachrymans | CFBP 1644t | Cucumis sativus | HHL, OHHL, OOHL | |
| savastanoi | CFBP 1670 | Olea europaea | HHL, OHHL | |
| CFBP 62 | Olea europaea | HHL, OHHL | ||
| CFBP 1020 | Olea europaea | HHL, OHHL | ||
| CFBP 2088 | Nerium oleander | HHL, OHHL | ||
| CFBP T12-1 | Nerium oleander | HHL, OHHL | ||
| CFBP 2093 | Fraxinus excelsior | HHL, OHHL | ||
| CFBP 2167 | Fraxinus excelsior | HHL, OHHL | ||
| CFBP 2169 | Fraxinus excelsior | HHL, OHHL | ||
| CFBP 2172 | Fraxinus excelsior | HHL, OHHL | ||
| CFBP 2174 | Fraxinus excelsior | HHL, OHHL | ||
| CFBP T36-10 | Fraxinus excelsior | HHL, OHHL | ||
| CFBP 2094 | Fraxinus excelsior | HHL, OHHL | ||
| CFBP 2176 | Fraxinus excelsior | HHL, OHHL | ||
| sesami | CFBP 1671 | Sesamum indicum | ||
| tabaci | CFBP 2106 | Sesamum indicum | HHL, OHHL | |
| morsprunorum | CFBP 2116t | Prunus cerasus | ||
| glycinea | CFBP 2214 | Glycinea max | ||
| CFBP 1645 | Glycinea javanica | |||
| ciccaronei | CFBP 2342 | Ceratonia siliqua | HHL, OHHL | |
| eriobotryae | CFBP 2343 | Eriobotrya japonica | HHL, OHHL | |
| mellea | CFBP 2344 | Nicotiana tabacum | HHL, OHHL, OOHL | |
| aesculi | CFBP 2894 | Aesculus indica | HHL, OHHL, OOHL | |
| hibisci | CFBP 2895 | Hibiscus japonica | HHL, OHHL, OOHL | |
| myricae | CFBP 2897 | Myrica rubra | HHL, OHHL | |
| CFBP 11005 | Myrica rubra | HHL, OHHL | ||
| photiniae | CFBP 2899 | Photinia glabra | HHL, OHHL | |
| CFBP 11033 | Photinia glabra | HHL, OHHL, OOHL | ||
| dencropanacis | CFBP 3226 | Dendropanax trifidus | OHHL | |
| amygdali | CFBP 3340 | Prunus amydalus | ||
| CFBP 10788 | Prunus amydalus | |||
| ficuserectae | CFBP 3224 | Ficus erecta | HHL, OHHL | |
| meliae | CFBP 3225 | Melia azedarach | HHL, OHHL | |
| P. tomato | persicae | CFBP 1573 | Prunus persicae | |
| CFBP 1067 | Prunus persicae | |||
| anthirrini | CFBP 1620 | Antirrhinum majus | OHHL | |
| maculicola | CFBP 1657 | Brassica oleracea | OHHL, OOHL | |
| CFBP 11056 | Brassica oleracea | HHL, OHHL, OOHL | ||
| CFBP 1637 | Raphanus sativus | |||
| CFBP 11079-5 | Raphanus sativus | OHHL, OOHL | ||
| CFBP 10912-9 | Raphanus sativus | OHHL, OOHL | ||
| viburni | CFBP 1702 | Viburnum sp. | OHHL, OOHL | |
| berberidis | CFBP 1727 | Berberidis sp. | ||
| CFBP 3208 | Berberidis gagnepainii | |||
| apii | CFBP 2103 | Apium graveolens | OHHL, OOHL | |
| tomato | CFBP 2212 | Lycopersicon esculentum | OHHL | |
| CFBP 8486 | Lycopersicon esculentum | |||
| CFBP 10199 | Lycopersicon esculentum | |||
| CFBP 10208 | Lycopersicon esculentum | |||
| delphinii | CFBP 2215 | Delphinium sp. | HHL, OHHL | |
| passiflorae | CFBP 2346 | Passiflora edulis | OHHL, OOHL | |
| morsprunorum | CFBP 2351 | Prunus domestica | ||
| lachrymans | CFBP 2440 | Cucumis sativus | ||
| philadelphi | CFBP 2898 | Philadelphus coronarius | ||
| ribicola | CFBP 10971t | Ribes aureum | HHL, OHHL | |
| primulae | CFBP 11007t | Primula sp. | BHL, HHL | |
| P. porri | porri | CFBP 1908 | Allium porrum | |
| CFBP 2395 | Allium porrum | |||
| garcae | CFBP 1634 | Coffea arabica | ||
| striafaciens | CFBP 1674 | Avena sativa | ||
| CFBP 1686t | Avena sativa | |||
| coronofaciens | CFBP 2216 | Avena sativa | ||
| atropurpurea | CFBP 2340 | Lolium multiforum | ||
| oryzae | CFBP 3228 | Oryza sativa | ||
| zizaniae | CFBP 11040 | Zizania aquatica | ||
| P. avellanae | avellanae | CFBP 10963 | Corylus avellana | |
| theae | CFBP 2353 | Thea sinensis | ||
| P. helianthi | helianthi | CFBP 2067 | Helianthus annuus | OHHL, HHL |
| tagetis | CFBP 1694 | Tagetes erecta | OHHL, HHL | |
| P. tremae | tremae | CFBP 3229 | Trema orientalis | |
| P. cannabina | cannabina | CFBP 2341 | Cannabis sativa | |
| CFBP 1619 | Cannabis sativa | |||
| P. viridiflava | viridiflava | CFBP 2107 | Phaseolus sp. | |
| primulae | CFBP 1660 | Primula sp. | ||
| ribicola | CFBP 2348 | Ribes aureum |
Strains were grown in both ABG medium and KBm.
CFBP, Collection Française des Bactéries Phytopathogènes (French Collection of Plant-Pathogenic Bacteria), Institut National de la Recherche Agromomique, Angers, France. The bacteria assayed were all unique strains.
TABLE 2.
Names, origins, and NAHL-producing capacities of strains of cytochrome c oxidase-positive species of fluorescent pseudomonadsa
| Species | Biovar | Strainb | Origin | Host plantc | NAHL synthesized |
|---|---|---|---|---|---|
| P. chlororaphis | DTR133 | Rhizosphere | Lycopersicon esculentum | HHL, OHHL | |
| P. fluorescens | II | DLRp214 | Rhizoplane | Linum usitatissimum | |
| CLRp812 | Rhizoplane | Linum usitatissimum | |||
| CLE513 | Root tissue | Linum usitatissimum | |||
| CS611 | Bulk soil | NA | |||
| CTR1015 | Rhizosphere | Lycopersicon esculentum | |||
| CTR212 | Rhizosphere | Lycopersicon esculentum | |||
| C7R12 | Rhizosphere | Linum usitatissimum | |||
| III | CS613 | Bulk soil | |||
| IV | DLR426 | Rhizosphere | Linum usitatissimum | ||
| DTR335 | Rhizosphere | Lycopersicon esculentum | Unidentified | ||
| DLE411J | Root tissue | Linum usitatissimum | |||
| VI | CLR711 | Rhizosphere | Linum usitatissimum | HHL | |
| CTRp112 | Rhizoplane | Lycopersicon esculentum | |||
| Undetermined | CFBP 2129 | Water | NA | ||
| GRA-3 | Rhizosphere | ND | |||
| CFBP 11393 | Rhizosphere | Lycopersicon esculentum | OOHL, OHL, DHL | ||
| CFBP 11363 | Rhizosphere | Zea mays | OOHL, OHL, DHL | ||
| CFBP 11350 | Rhizosphere | Zea mays | |||
| CFBP 11388 | Rhizosphere | Lycopersicon esculentum | |||
| CFBP 11345 | Rhizosphere | Zea mays | |||
| CFBP 11273 | Rhizosphere | Glycine max | |||
| CFBP 11346 | Rhizosphere | Zea mays | |||
| CFBP 11369 | Rhizosphere | Zea mays | |||
| CFBP 11387 | Rhizosphere | Lycopersicon esculentum | |||
| CFBP 11385 | Rhizosphere | Lycopersicon esculentum | |||
| CFBP 11366 | Rhizosphere | Zea mays | |||
| CFBP 11394 | Rhizosphere | Lycopersicon esculentum | |||
| Intermediate species (P. fluorescens and P. putida) | Undetermined | DS824 | Bulk soil | NA | |
| DS821 | Bulk soil | NA | |||
| P. putida | A | DTRp621 | Rhizoplane | Lycopersicon esculentum | |
| DLR223 | Rhizosphere | Linum usitatissimum | |||
| DLR228 | Rhizosphere | Linum usitatissimum | |||
| DS1026 | Bulk soil | NA | |||
| DLE3216 | Root tissue | Linum usitatissimum | |||
| DS131 | Bulk soil | NA | |||
| CS111 | Bulk soil | NA | |||
| CS413 | Bulk soil | NA | |||
| CS714 | Bulk soil | NA | |||
| Undetermined | CFBP 2066 | Bulk soil | NA | ||
| N5F5 | Rhizosphere | Beta vulgaris | OHHL, OOHL, ODHL, one unidentified | ||
| P. cichorii | CFBP 2101 | Plant | Cichorium endivia |
Strains were grown in both ABG medium and KBm.
See Table 1, footnote c. Strains other than CFBP strains were from our laboratories. The bacteria assayed were all unique strains.
NA, not available; ND, not determined.
TABLE 3.
Names, origins, and NAHL-producing capacities of strains of nonfluorescent species of Pseudomonasa
| Species | Strainb | Originc | Host plant | NAHL synthesized |
|---|---|---|---|---|
| P. stutzeri | CFBP 2443 | ND | ND | |
| P. corrugata | 8-1 | Plant | Lycopersicon esculentum | HHL, OHHL, OHL |
| 82.23.6 | Plant | Lycopersicon esculentum | HHL, OHHL, OHL | |
| 632.2 | Plant | Lycopersicon esculentum | HHL, OHHL, OHL | |
| 83.83.4 | Plant | Lycopersicon esculentum | HHL, OHHL, OHL |
Strains were grown in both ABG medium and KBm.
See Table 1, footnote c. Strains other than the CFBP strain were from our laboratories. The bacteria assayed were all unique strains.
ND, not determined.
Pseudomonas strains were grown at 25°C, Chromobacterium and Agrobacterium strains were grown at 28 to 29°C, and E. coli strains were grown at 37°C. The media used were modified Luria-Bertani medium (LBm) (49); modified King B medium (KBm) (23); and ABM or ABG medium, which consisted of AB minimal medium (7) supplemented with mannitol or glucose at 0.5% (wt/vol), respectively. When required, antibiotics were incorporated into the media at the following concentrations: ampicillin (100 μg/ml), carbenicillin (100 μg/ml), rifampin (150 μg/ml), tetracycline (10 μg/ml), and kanamycin (100 μg/ml); however, E. coli strain DB82(pRK2013) was grown in the presence of 50 μg of kanamycin/ml. 5-Bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) was used at 40 μg/ml when necessary.
Chemicals.
QS systems involve the production of several NAHL with closely related structures. We designate these molecules as a function of the number of carbon atoms of the lipid moiety and as a function of the substitution at position 3 of the fatty acid chains. The molecules are designated as follows: HSL, N-acyl-l-homoserine lactone; BHL, N-butanoyl-l-homoserine lactone (or C4-HSL); HHL, N-hexanoyl-l-homoserine lactone (or C6-HSL); OHL, N-octanoyl-l-homoserine lactone (or C8-HSL); DHL, N-decanoyl-l-homoserine lactone (or C10-HSL); OBHL, N-(3-oxo-butanoyl)-l-homoserine lactone (or 3-oxo-C4-HSL); OHHL, N-(3-oxo-hexanoyl)-l-homoserine lactone (or 3-oxo-C6-HSL); OOHL, N-(3-oxo-octanoyl)-l-homoserine lactone (or 3-oxo-C8-HSL); ODHL, N-(3-oxo-decanoyl)-l-homoserine lactone (or 3-oxo-C10 HSL); and OdDHL, N-(3-oxo-dodecanoyl)-l-homoserine lactone (or 3-oxo-C12-HSL). Authentic NAHL samples were kindly provided by Paul Williams (University of Nottingham).
NAHL extraction.
For rapid determination of NAHL production in liquid medium, 1-ml samples of the strains were cultured in KBm. Strains were grown to stationary phase and centrifuged for 10 min at ca. 10,000 × g and 4°C. A 250-μl sample of the native culture supernatant was used to load wells in plate assays as described below. Strains identified as positive were further tested on ABG medium and retested on KBm. The spent culture supernatant was extracted as follows. Bacteria were grown in 5 ml of ABG medium or KBm to stationary phase. The media were centrifuged for 10 min at 7,500 × g and 4°C. The supernatant was extracted twice with 100% ethyl acetate to yield 10 ml of extract. The extract was dried over anhydrous sodium sulfate, filtered, and evaporated to dryness in a rotary evaporator at room temperature. The dried extract was resuspended in 600 μl of 100% ethyl acetate, evaporated again in a rotary evaporator at room temperature, and resuspended in 50 μl of 100% ethyl acetate. The final volume of the extract was therefore 1/100 that of the original culture medium.
NAHL detection and characterization.
Individual strains were screened for the production of NAHL by two different assays. First, strains were screened on solid medium in a streak plate assay as described by Piper et al. (38) using the NAHL biosensors C. violaceum CVO26 and E. coli JM109(pSB401) on LBm and A. tumefaciens NT1(pDCI41E33) on ABM supplemented with 40 μg of X-Gal/ml. This assay was used to survey NAHL production by growing bacteria. Second, the presence of NAHL in native culture supernatants was assayed by the plate assay of McClean et al. (32) using the same three biosensors. Assay plates were incubated for 24 h for Chromobacterium and E. coli and for up to 48 h for Agrobacterium. NAHL produced by strains positive in any of the above-mentioned screens were further characterized by thin-layer chromatography (TLC) essentially as described by Shaw et al. (43) using the Agrobacterium sensor or by McClean et al. (32) using the Chromobacterium sensor. The concentrated extract was spotted onto a C18 reverse-phase TLC plate developed with a methanol-water solvent mixture. After elution, the plate was overlaid with soft agar containing bacterial indicators; this procedure generated a chromogenic compound upon sensing of a trace amount of one or more NAHL.
Modifications to the previously published protocols were as follows. TLC plates were C18 reverse-phase TLC plates (Silicagel; 60 Å, 20 by 20 cm, 0.2-mm thick). Elution buffers were methanol-water (60/40, vol/vol) for general use and methanol-water (70/30, vol/vol) for improved resolution of NAHL with acyl chains longer than eight carbon atoms. Characterization of the NAHL was based on the evaluation of the Rfs and shapes of the spots and on the differential responses of the sensor strains (including “reverse detection” by Chromobacterium [32]). An NAHL multistandard was always spotted on the TLC plates before migration of and along with the analyzed samples. The distributions of NAHL producers in different classes, defined according to their origins and to their relationships with the plant (soilborne isolates, nonpathogenic isolates from roots or leaves, and pathogenic isolates), were compared pairwise to a theoretical even distribution by a chi-square test.
DNA manipulations and sequencing.
All molecular techniques, such as DNA extraction and restriction analysis, were performed using standard protocols (42). Routine cloning at various sites of the ColE1-derived vector pUC19 (or related vectors) was done with E. coli strain DH5α as a recipient strain. A genomic DNA bank was obtained from 10912–9 by cloning the dephosphorylated restriction products of a partial Sau3AI digestion of genomic DNA from this strain at the BamHI site of the broad-host-range cosmid vector pCP13/B (10) according to standard protocols (42). When necessary, cosmid clones were transferred to gram-negative recipients by triparental mating with DB82(pRK2013) (11).
Sequencing was performed in part at the Institut des Sciences Végétales with an Applied Biosystems 370 sequencer and in part at Eurogentec (Herstal, Belgium). Sequences were assembled with Sequencer software (GeneCodes, Ann Harbor, Mich.). Nucleotide and amino acid sequence comparisons were made using the BLAST protocol available online at the National Center for Biotechnology Information website. Multiple alignments were performed using the Pileup subroutine of the GCG package (version 10; GCG Inc., Madison, Wis.).
Mutagenesis of the psmI gene was performed by introducing a gentamicin resistance cassette obtained from pmGm (35) as an SmaI fragment at the SspI site of psmI, interrupting the open reading frame (ORF) at position 85. To evaluate homoserine lactone production, the wild-type psmI or psmR locus and the psmI::Gm construct were cloned into the broad-host-range vector pBBR1MCS-3 (27), which was further transferred to various gram-negative bacterial hosts by triparental mating using DB82(pRK2013) (11) as a helper.
Nucleotide sequence accession number.
Sequences determined here have been deposited in GenBank under accession number AF234628.
RESULTS
NAHL production is more common among plant-associated than among soilborne Pseudomonas spp.
Using the protocols described in Materials and Methods, strains which induced the production of violacein (or which inhibited it) in the C. violaceum sensor, strains which activated the biosynthesis of beta-galactosidase in A. tumefaciens NT1(pDCI41E33), and strains which induced light emission in E. coli JM109(pSB401) remained NAHL producers. The others were regarded as nonproducing strains under our experimental conditions. The results of this analysis and the identification of the synthesized NAHL molecules are shown in Tables 1, 2 and 3 and in Fig. 1. As the characterization of the NAHL molecules is based on the examination of the shapes and Rfs of the spots and on the differential responses of the sensors (with respect to standard reference samples), the identification results must be interpreted with care.
FIG. 1.
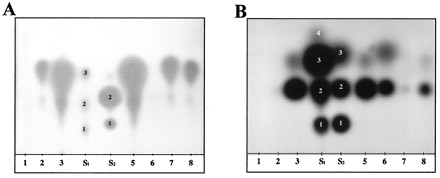
Production of NAHL by various isolates of Pseudomonas. Supernatant extracts were obtained as indicated in Materials and Methods from strains of the P. tomato genomospecies: 1, CFBP 1573; 2, CFBP 1620; 3, CFBP 1702; 5, CFBP 2215; 6, CFBP 11007t; 7, CFBP 10912–9; and 8, CFBP 2212. S1 was a mixture of keto-NAHL (OBHL, 4; OHHL, 3; OOHL, 2; and ODHL, 1) used as a migration standard; S2, another migration standard, was a mixture of reduced NAHL (BHL, 3; HHL, 2; and OOHL, 1). (A) The sensor strain was Agrobacterium NT1(pDCI41E33) (43), which exhibits a high level of sensitivity toward reduced NAHL (concentration of standards according to Shaw et al. [43]). (B) The sensor strain was Chromobacterium CVO26 (32), which senses preferentially very-short-chain (C6 and C8) keto-NAHL (concentration of standards according to McClean et al. [32]).
Among the 90 pathogenic strains of oxidase-negative fluorescent pseudomonads, 44 were identified as NAHL producers. Within this group, plant-pathogenic bacteria belonging to P. syringae and related species are listed by genomospecies (Table 1). These are taxonomic subdivisions which can be regarded de facto as species; indeed, strains belonging to the same genomospecies show a DNA-DNA relatedness of at least 66% (21, 22), whereas relatedness between strains belonging to different genospecies is lower than 59%. The ratio of NAHL producers varies from one genomospecies to another. Thus, among the strains of the genomospecies P. syringae, only 2 out of 12 produced OHHL and HHL; none of the strains of the genomospecies P. porri, P. avellanae, P. tremae, P. cannabina, and P. viridiflava synthesized NAHL (under our experimental conditions). In contrast, NAHL-producing strains were predominant in three genomospecies. In the P. savastanoi genomospecies, 28 of 35 strains and, most notably, all 13 strains of P. savastanoi pv. savastanoi produced NAHL, while 12 of 23 strains of the genomospecies P. tomato produced NAHL. Finally, the two strains defining the P. helianthi genomospecies also produced NAHL.
Among the 42 oxidase-positive Pseudomonas strains, 27 were isolated from the rhizosphere and rhizoplane, 10 were from bulk soil, 4 were from plant tissues, and 1 was from water (Table 2). Within this group, only 6 of 42 strains produced NAHL. These six strains were isolated from the rhizosphere of four different plants. In contrast to their oxidase-negative counterparts, from which they are taxonomically distinct, oxidase-positive pseudomonads synthesized a variety of NAHL molecules, such as HHL, OHHL, OOHL, DHL, and ODHL. P. putida strain P5F5 also produced an unidentified NAHL, the migration of which was related to that of OdDHL but was not strictly identical (data not shown).
Overall, the data revealed that out of 137 different strains analyzed, 54 (40%) produced NAHL. Remarkably, NAHL-producing strains were found only among bacteria isolated from plants; none of the soilborne strains tested was able to synthesize these molecules. More precisely, the NAHL-producing strains were more abundant among the pathogenic strains (49%) than among the rhizosphere strains (28%) and the soilborne strains (0%). Statistical analysis of these data indicated that the frequencies of NAHL-producing strains differed significantly according to their origins and to their relationships with the plant (χ2 = 11.53, P < 0.05).
Cloning of the genes responsible for NAHL production in P. syringae pv. maculicola.
The above results indicate that NAHL production is not uncommon among plant-associated Pseudomonas species and especially among strains of P. syringae. However, little is known about the biological traits regulated via QS in P. syringae. To identify the NAHL biosynthetic pathway and the relevant regulated biological functions, we attempted to identify the genes involved in the synthesis of these compounds by a P. syringae strain. The strain chosen for this study was P. syringae pv. maculicola strain CFBP 10912–9 (P. tomato genomospecies) (Table 1), essentially because it produces large amounts of OHHL and OOHL.
To isolate the genes responsible for NAHL production, a genomic cosmid library of 10912–9 DNA was conjugated en masse by triparental mating into a rifampin-resistant derivative of the biosensor CVO26. Screening of ca. 5,000 transconjugants selected as being resistant to tetracycline and rifampin was used to identify the genes encoding the production of NAHL; this procedure yielded 20 clones that restored violacein production in the biosensor. The recombinant plasmids in the clones, all with inserts ranging from 20 to 35 kb, contained one or more similarly sized EcoRI and XhoI fragments and could be organized in two classes according to their restriction patterns. A representative of each class, termed pMES-A and pMES-B, was retained for further studies. The two cosmids were transferred into the non-NAHL-producing P. syringae pv. persicae strain CFBP 1573 (Table 1). Both conferred to that strain the ability to produce NAHL. Furthermore, TLC profiles of concentrated culture supernatants from CFBP 1573(pMES-A) and 10912–9 were indistinguishable (Fig. 2). The region of overlap between the two cosmid classes was mapped, subcloned into the ColE1-based vector pUC19, and introduced into E. coli DH5α, which does not produce any detectable NAHL. Two pUC19-derived clones, containing a ca. 2-kb XhoI fragment (pMEX-A) and a 4.5-kb PstI fragment (pMEP-A), conferred to that strain the ability to produce the same NAHL signal molecules as the parent strain, P. syringae pv. maculicola strain CFBP 10912–9 (Fig. 2), even in the absence of isopropyl-β-d-thiogalactopyranoside (IPTG). Interestingly, the original cosmid clones, pMES-A and pMES-B, did not confer to the E. coli strain the ability to produce NAHL. Similarly, two pUC19-derived clones containing the 2-kb XhoI fragment and the 4.5-kb PstI fragment in the orientations opposite those in pMEX-A and pMEP-A, respectively, did not induce the production of NAHL in DH5α.
FIG. 2.
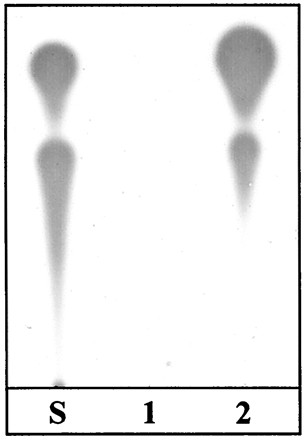
Production of NAHL by the cloned psmI gene. Concentrated extracts of bacterial spent supernatants were obtained and analyzed as indicated in Materials and Methods using the Agrobacterium sensor strain NT1(pDCI41E33) (43). Extracts were obtained from P. tomato pv. persicae CFBP 1573 (lane 1) and CFBP 1573(pMES-A) (lane 2). A result identical to that shown in lane 2 was obtained using wild-type P. syringae pv. maculicola strain CFBP 10912–9 (not shown). S, migration standard consisting of OHHL (top) and OOHL (middle).
Sequence analysis and identification of psmI and psmR
The DNA sequence of the 2-kb XhoI/XhoI fragment and that of its adjacent regions were determined. DNA sequence analysis revealed the presence of three ORFs organized as shown in Fig. 3. One of the three ORFs, which we named psmI, could encode a 244-amino-acid protein with an estimated mass of 27.16 kDa. This sequence is closely related to those of two luxI gene homologues, ahlI (12) and psyI (P. M. Oger et al., unpublished data [GenBank accession number AF266600]), found in other members of the P. syringae genomospecies (Fig. 4). The product of this ORF is only distantly related to the other LuxI proteins. The psmI ORF is preceded by sequences with reasonable matches to consensus ribosome binding site (RBS) sequences and −10 and −35 promoter elements (Fig. 3). In addition, we were able to identify a sequence closely related to the lux box consensus sequence at positions −76 to −56 upstream of the psmI start codon, indicating that this ORF could be regulated by a QS-dependent regulator. Interestingly, this regulatory box lies between the best matches to −35 and −10 sequences and overlaps the putative −10 sequence (Fig. 3). Since psmI is the only ORF on the 2-kb XhoI fragment of pMEX-A whose product shows homology to NAHL synthases, its expression must be responsible for NAHL synthesis in DH5α(pMEX-A). Interestingly, the XhoI site used for the cloning lies within the psmI ORF. Therefore, the truncated psmI ORF of pMEX-A still encodes a functional or partially functional NAHL synthase.
FIG. 3.
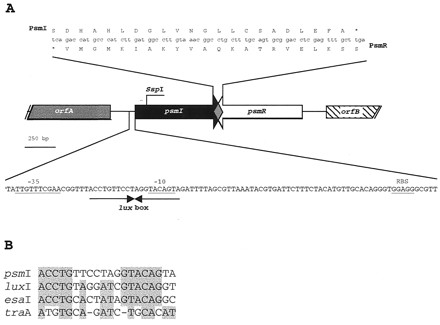
(A) Genetic organization of the psmI-psmR locus from P. syringae pv. maculicola strain CFBP 10912–9. In the diagram in the center of panel A, the putative identified ORFs are represented by arrows indicating the direction of transcription. The upper diagram shows a detail of the overlap between the psmI and psmR genes. The lower diagram shows a detail of the promoter region of the psmI gene. The sequences of the putative regulatory elements (−35, −10, and RBS) are underlined. The position of the putative lux box is indicated by the convergent arrows. The gentamicin cassette used to disrupt the psmI gene was inserted at the SspI site shown above the psmI ORF. (B) Sequence alignment of lux boxes from the psmI gene of P. syringae pv. maculicola, the luxI gene of P. fischeri, the esaI gene of P. stewartii, and the traA gene of A. tumefaciens.
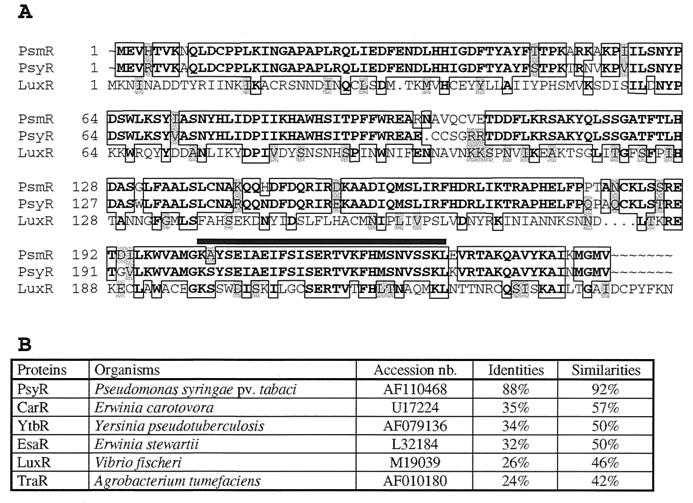
FIG. 4.
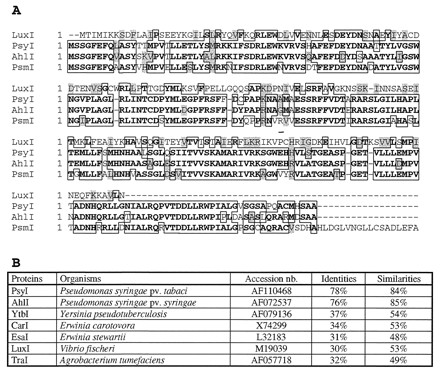
Relatedness of the PsmI protein to other NAHL synthases. (A) Protein sequence alignment of PsmI with PsyI, AhlI, and LuxI, NAHL synthases from P. syringae pv. tabaci, P. syringae pv. syringae, and P. fischeri, respectively. Bold letters and boxes indicate identity; grey shading indicates similarity. (B) Similarity and identity with other LuxI-like proteins.
The second complete ORF, which we called psmR, was found downstream of psmI (Fig. 3). It could encode a protein of 247 amino acids with a mass of 28.34 kDa. The deduced protein sequence exhibits a helix-turn-helix motif, suggesting that it can bind nucleic acid. In agreement with this information, the psmR gene product exhibits homology with the LuxR family of regulatory proteins (Fig. 5), especially with PsyR from P. syringae pv. tabaci (Oger et al., unpublished data; AF266600). As shown in Fig. 3, the psmI and psmR genes are transcribed convergently, and the 3′ ends of their coding regions overlap. Determination of the GC contents of both psmI and psmR yielded values (ca. 55 and 52%, respectively) slightly below those reported for other P. tomato genes (58%).
FIG. 5.
Relatedness of the PsmR protein to other LuxR-type regulators. (A) Protein sequence alignment of PsmR with PsyR and LuxR from P. syringae pv. tabaci and P. fischeri, respectively. Bold letters and boxes indicate identity; grey shading indicates similarity. The black bar indicates the putative DNA-binding domain. (B) Similarity and identity with other LuxR-like proteins.
The third ORF, which we termed orfA, is located ca. 218 bp upstream of the putative start codon of psmI and is most probably transcribed divergently from it. It is closely related (85% identity) to the orf1 gene, identified upstream of the luxI homologue psyI in P. syringae pv. tabaci. The putative products of orfA and orf1 have weak homologies (32 and 54%, respectively) to the C-terminal ends of members of the histidinol-phosphate aminotransferase family and therefore could have a similar enzymatic function. However, the N-terminal ends of the characterized sequences of the putative proteins OrfA and Orf1 do not share homology with aminotransferases. No lux regulatory sequences could be identified upstream of orfA or psmR, suggesting that these genes may not be regulated by QS.
Analysis of psmI by complementation and mutational analysis.
To confirm the involvement of psmI in NAHL synthesis, we recloned the 4.5-kb PstI fragment in the broad-host-range vector pBBR1MCS-3 and transferred it by triparental mating to P. fluorescens recipient strain 1855.344, which does not produce any NAHL (4; unpublished results). Strain 1855.344 harboring the cloned 4.5-kb PstI fragment now produced the same NAHL as wild-type strain 10912–9. A cassette encoding gentamycin resistance was cloned at the SspI site of the 4.5-kb PstI fragment to disrupt psmI (Fig. 3). The resulting construction was also transferred to 1855.344. The resulting transconjugants did not produce any NAHL (data not shown).
DISCUSSION
The strain survey presented in the first part of this study indicates that NAHL production is not uncommon among plant-associated Pseudomonas strains. Several conclusions can be drawn. For instance, among oxidase-negative Pseudomonas strains, no clear correlation between taxonomic position and NAHL production is evident, with the exception of the 8 strains of the P. porri genomospecies (which did not appear to produce NAHL) and the 13 P. savastanoi strains (which all produced HHL and OHHL). During the course of this work, two surveys of NAHL production by gram-negative strains were published (4, 12). A few strains of the P. syringae group were examined for NAHL production. Some discrepancies between those studies and ours can be observed, e.g., for two P. savastanoi strains (CFBP 2088 and CFBP 2093). While Cha et al. (4) did not observe NAHL production for these strains, we report here that they do produce the NAHL signal molecules HHL and OHHL. This difference most likely results from the different growth conditions used in the two studies. However, this difference emphasizes that NAHL production among the strains analyzed by us and by others might have an even wider distribution. Strains which did not produce any NAHL under our experimental conditions might indeed produce such signal molecules under other growth conditions, e.g., in a more favorable or in an “inducing” environment, as reported for the opine-dependent NAHL synthesis involved in the regulation of Ti plasmid transfer in Agrobacterium (for a review, see reference 18).
As indicated above, most of the analyzed Pseudomonas strains produced short-chain NAHL (eight or fewer carbon atoms, e.g., HHL, OHHL, and OOHL). This feature is not due to a technical bias, since two of the biosensors are able to detect long-chain NAHL (Chromobacterium [reverse staining] and Agrobacterium) (32, 43). Indeed, one of the soil P. putida strains was found to produce ODHL and possibly OdDHL, two “long-chain” NAHL. The generalized production of HHL and OHHL by P. syringae strains is interesting and might reflect a common origin for the different QS systems in these strains.
Interestingly, our results go beyond those previously published (4, 12), as they indicate that NAHL production seems more common among pseudomonads closely associated with plants than among their soilborne counterparts. Indeed, the percentage of NAHL producers decreased from 49% among plant-pathogenic bacteria to 28 and 0% among nonpathogenic bacteria associated with the plant and soilborne bacteria, respectively. This finding suggests that the closer the relationship of the bacteria with the host plant, the higher the probability that it produces NAHL. That NAHL production appears to be more common among isolates closely associated with plant tissues may be related to the wealth of available carbon sources (31) in this highly competitive environment. This fact is consistent with the detection of QS-regulated functions in other plant-associated, plant-symbiont, or plant-pathogenic bacteria (for reviews, see references 20, 37, 41, and 46) or in other microbial hosts of rich ecotopes (e.g., Photobacterium, Shigella, and so forth). Alternatively, our results could reveal the limited ability of our experimental conditions to detect NAHL production in soilborne pseudomonads. If this is true, then the observed variation does not correlate with the mere presence of luxI and luxR loci but reflects differences in terms of regulation of these loci (e.g., additional regulatory levels, requirements for unknown inducers, and so forth). However, the statistical analysis demonstrates that these putative differences discriminating soilborne, plant-associated, and plant-pathogenic pseudomonads are statistically and biologically significant, as they correlate with the ecology of the bacteria.
The wide occurrence of NAHL producers among pseudomonads, especially among strains of P. syringae and related species, stands in contrast with the lack of information on the function(s) regulated in a QS-dependent way in these bacteria. A first step toward the elucidation and understanding of the QS-regulated functions in P. syringae strains involves the isolation of the relevant genes. Cloning and sequencing of the region responsible for NAHL production in P. syringae pv. maculicola led to the identification of two genes, psmI and psmR. To our knowledge, this is the first report of the presence of a luxR homologue in P. syringae. Sequence analysis of these genes revealed their high degree of homology to members of the luxI and luxR gene families, especially with the psyI and psyR genes from P. syringae pv. tabaci (Oger et al., unpublished results) and with the ahlI gene from P. syringae pv. syringae (12), respectively.
The psmI gene is likely to code for an NAHL synthase. Several lines of evidence support this hypothesis. First, it has strong homology to other genes encoding such enzymatic activities (Fig. 4). Second, the cloned gene conferred NAHL production upon nonproducing hosts (e.g., P. syringae pv. persicae, P. fluorescens, E. coli, and the two NAHL reporter strains tested). Third, bacteria hosting psmI exhibited a production pattern analogous to that of the P. syringae pv. maculicola strain. Finally, a mutated psmI gene does not confer NAHL production upon the bacterial host anymore. These features clearly indicate that the psmI gene is the necessary and sufficient genetic determinant accounting for the production of all NAHL signal molecules in P. syringae pv. maculicola CFBP 10912–9.
NAHL production in DH5α was observed only with clones harboring the luxI gene inserted into a pUC19 plasmid (e.g., pMEX-A) and under the control of the lac promoter and not with the full-size cosmid clones harboring both the psmI and the psmR genes, although the psmI gene was expressed in the P. syringae pv. persicae background. This result may have been due to the organization or sequence of the promoter regions of P. syringae pv. maculicola genes that are not recognized by the E. coli transcription machinery, although the psmI gene is preceded by reasonable matches to consensus −35 and −10 sequences. This observation has also been reported for several nonenteric bacterial species, such as P. fluorescens, P. syringae pv. tabaci, and A. tumefaciens. For example, the expression of Agrobacterium virulence genes in E. coli requires the presence of the alpha subunit of the RNA polymerase from Agrobacterium (29). Our data also suggest that PsmR could act as a repressor which prevents the expression of psmI. In agreement with this hypothesis, a palindromic, lux box-like sequence has been detected within the promoter region of psmI, just upstream of the putative −10 sequence and overlapping it. Interestingly, this lux box is located at a position similar to that found in P. stewartii for a system involving the LuxR-like repressor protein EsaR (3). This finding is also consistent with the observation that in other systems which involve activator proteins, the regulatory lux sequences are located upstream of the proposed −35 elements (1, 16, 52). Recent results obtained by Luo and Farrand (30) confirmed that the activity of the LuxR-type regulator TraR, although an intrinsic property of the molecule, is also strongly affected by the positioning of the lux box (17).
Sequence data revealed a gene organization with two genes (psmI and psmR) facing each other and slightly overlapping. This organization is only the third example reported so far for bacteria. The first two were described for Yersinia (48) and Erwinia (Pantoea) (2). Although not demonstrated for P. syringae pv. maculicola or for any of the systems with convergently transcribed luxR and luxI genes, this organization may play an additional role in the regulation of the expression of the two genes, as the elevated transcription of one of the two may impair the expression of the other.
In some organisms, e.g., A. tumefaciens and P. fischeri, the genes regulated in a QS-dependent fashion are located downstream of the traI and luxI genes, respectively, and are coordinately regulated with these genes. In P. syringae pv. maculicola, this appears not to be the case. In this respect, the gene organization of psmI and psmR is similar to that in the enteric bacterium Erwinia (Pantoea), in which QS-regulated genes are not linked to the regulatory loci. The major difference between the organization of QS systems is of interest. In the systems in which a luxI homologue is the first gene of the LuxR-regulated operon, QS regulates only a single function, i.e., conjugal transfer for A. tumefaciens and bioluminescence for P. fischeri. In the systems in which a luxI homologue is not associated with a QS-regulated function, QS is most often involved in a complex regulatory scheme that controls the expression of more than one operon or function. This information suggests that QS may also regulate more than one trait in P. syringae pv. maculicola. Whether the functions regulated in a QS-dependent fashion in P. syringae pathovars are important for plant-microbe associations remains to be determined. However, with respect to previously published data, a possible correlation between NAHL synthesis and pathogenicity (2, 40), siderophore biogenesis (45), swarming (15), or biofilm formation (9) may be proposed.
ACKNOWLEDGMENTS
This work was made possible by an E.U. grant (BIO4 CT96–181) to Y.D. and G.S. M.E. was supported by the same grant, and E.G. was supported by a grant from Académie d'Agriculture de France.
We thank Paul Williams and Andrea Hardman (Nottingham, United Kingdom) for NAHL samples and René Bally and Marie-Louise Bouillant (Lyon, France), Stephen K. Farrand (Champaign-Urbana, Ill.), and Louis Gardan (Angers, France) for helpful discussions and communication of unpublished material.
Footnotes
This paper is dedicated to the memory of Gordon Stewart.
REFERENCES
- 1.Baldwin T O, Devine J H, Heckel R C, Lin J W, Shadel G S. The complete nucleotide sequence of the lux regulon of Vibrio fischeri and the luxABN region of Photobacterium leiognathi and the mechanism of control of bacterial bioluminescence. J Biolumin Chemilumin. 1989;4:326–341. doi: 10.1002/bio.1170040145. [DOI] [PubMed] [Google Scholar]
- 2.Beck von Bodman S, Farrand S K. Capsular polysaccharide biosynthesis and pathogenicity in Erwinia stewartii require induction by an N-acylhomoserine lactone autoinducer. J Bacteriol. 1995;177:5000–5008. doi: 10.1128/jb.177.17.5000-5008.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beck von Bodman S, Majerczak D R, Coplin D L. A negative regulator mediates quorum-sensing control of exopolysaccharide production in Pantoea stewartii subsp. stewartii. Proc Natl Acad Sci USA. 1998;95:7687–7692. doi: 10.1073/pnas.95.13.7687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cha C, Gao P, Chen Y C, Shaw P D, Farrand S K. Production of acyl-homoserine lactone quorum-sensing signals by gram-negative plant-associated bacteria. Mol Plant Microbe Interact. 1998;11:1119–1129. doi: 10.1094/MPMI.1998.11.11.1119. [DOI] [PubMed] [Google Scholar]
- 5.Chang C, Stewart R C. The two-component system. Regulation of diverse signaling pathways in prokaryotes and eukaryotes. Plant Physiol. 1998;117:723–731. doi: 10.1104/pp.117.3.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chet I, Ordenlich A, Shapira R, Oppenheim A. Mechanisms of biocontrol of soil-borne plant pathogens by Rhizobacteria. In: Keister D L, Cregan P B, editors. The rhizosphere and plant growth. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1991. pp. 229–236. [Google Scholar]
- 7.Chilton M-D, Currier T C, Farrand S K, Bendich A J, Gordon M P, Nester E W. Agrobacterium tumefaciens and PS8 bacteriophage DNA not detected in crown gall tumor DNA. Proc Natl Acad Sci USA. 1974;71:3672–3676. doi: 10.1073/pnas.71.9.3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Curl E A, Truelove B. Microbial interactions. Adv Ser Agric Sci. 1986;15:140–166. [Google Scholar]
- 9.Davies D G, Parsek M R, Pearson J P, Iglewski B H, Costerton J W, Greenberg E P. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science. 1998;280:295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- 10.Dessaux Y, Tempé J, Farrand S K. Genetic analysis of mannityl opine catabolism in octopine-type Agrobacterium tumefaciens strain 15955. Mol Gen Genet. 1987;208:301–308. doi: 10.1007/BF00330457. [DOI] [PubMed] [Google Scholar]
- 11.Ditta G, Stanfield S, Corbin D, Helinski D R. Broad host range DNA cloning system for Gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci USA. 1980;77:7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dumenyo C K, Mukherjee A, Chunn W, Chatterjee A R. Genetic and physiological evidence for the production of N-acyl homoserine lactones by Pseudomonas syringae pv. syringae and other fluorescent plant pathogenic Pseudomonas species. Eur J Plant Pathol. 1998;104:569–582. [Google Scholar]
- 13.Eberhard A. Inhibition and activation of bacterial luciferase synthesis. J Bacteriol. 1972;109:1101–1105. doi: 10.1128/jb.109.3.1101-1105.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eberhard A, Burlingame A L, Eberhard C, Kenyon G L, Nealson K H, Oppenheimer N J. Structural identification of autoinducer of Photobacterium fischeri luciferase. Biochemistry. 1981;20:2444–2449. doi: 10.1021/bi00512a013. [DOI] [PubMed] [Google Scholar]
- 15.Eberl L, Winson M K, Sternberg C, Stewart G S, Christiansen G, Chhabra S R, Bycroft B, Williams P, Molin S, Givskov M. Involvement of N-acyl-l-homoserine lactone autoinducers in controlling the multicellular behaviour of Serratia liquefaciens. Mol Microbiol. 1996;20:127–136. doi: 10.1111/j.1365-2958.1996.tb02495.x. [DOI] [PubMed] [Google Scholar]
- 16.Egland K A, Greenberg E P. Quorum sensing in Vibrio fischeri: elements of the luxI promoter. Mol Microbiol. 1999;31:1197–1204. doi: 10.1046/j.1365-2958.1999.01261.x. [DOI] [PubMed] [Google Scholar]
- 17.Egland K A, Greenberg E P. Conversion of the Vibrio fischeri transcriptional activator LuxR to a repressor. J Bacteriol. 2000;182:805–811. doi: 10.1128/jb.182.3.805-811.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farrand S K. Conjugal plasmids and their transfer. In: Spaink H P, Kondorosi A, Hooykaas P J J, editors. The Rhizobiaceae. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. pp. 199–233. [Google Scholar]
- 19.Fuqua C, Winans S C. Conserved cis-acting promoter elements are required for density-dependent transcription of Agrobacterium tumefaciens conjugal transfer genes. J Bacteriol. 1996;178:435–440. doi: 10.1128/jb.178.2.435-440.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuqua C, Greenberg E P. Self perception in bacteria: quorum sensing with acylated homoserine lactones. Curr Opin Microbiol. 1998;1:183–189. doi: 10.1016/s1369-5274(98)80009-x. [DOI] [PubMed] [Google Scholar]
- 21.Gardan L, Bollet C, Abu Ghorrah M, Grimont F, Grimont P A D. DNA relatedness among pathovar strains of Pseudomonas syringae pv. savastanoi Janse (1982) and proposal of Pseudomonas savastanoi sp. nov. Int J Syst Bacteriol. 1992;42:606–612. [Google Scholar]
- 22.Gardan L, Shafik H, Belouin S, Broch R, Grimont F, Grimont P A D. DNA relatedness among the pathovars of Pseudomonas syringae and description of Pseudomonas tremae sp. nov. and Pseudomonas cannabina sp. nov. (ex Sutic and Dowson 1959) Int J Syst Bacteriol. 1999;49:469–478. doi: 10.1099/00207713-49-2-469. [DOI] [PubMed] [Google Scholar]
- 23.Glickmann E, Gardan L, Jacquet S, Hussain S, Elasri M, Petit A, Dessaux Y. Auxin production is a common feature of most pathovars of Pseudomonas syringae. Mol Plant Microbe Interact. 1998;11:156–162. doi: 10.1094/MPMI.1998.11.2.156. [DOI] [PubMed] [Google Scholar]
- 24.Gray K M. Intercellular communication and group behavior in bacteria. Trends Microbiol. 1997;5:184–188. doi: 10.1016/S0966-842X(97)01002-0. [DOI] [PubMed] [Google Scholar]
- 25.Joshi H M, Tabita F R. A global two component signal transduction system that integrates the control of photosynthesis, carbon dioxide assimilation, and nitrogen fixation. Proc Natl Acad Sci USA. 1996;25:14515–14520. doi: 10.1073/pnas.93.25.14515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kleerebezem M, Quadri L E, Kuipers O P, de Vos W M. Quorum-sensing by peptide pheromones and two-component signal-transduction systems in Gram-positive bacteria. Mol Microbiol. 1997;24:895–904. doi: 10.1046/j.1365-2958.1997.4251782.x. [DOI] [PubMed] [Google Scholar]
- 27.Kovach M E, Elzer P H, Hill D S, Robertson G T, Farris M A, Roop II R M, Peterson K M. Four new derivatives of the broad host range cloning vector, pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 28.Latour X, Corberand T, Laguerre G, Allard F, Lemanceau P. The composition of fluorescent pseudomonad populations associated with roots is influenced by plant and soil type. Appl Environ Microbiol. 1996;62:2449–2556. doi: 10.1128/aem.62.7.2449-2456.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lohrke S M, Nechaev S, Yang H, Severinov K, Jin S J. Transcriptional activation of Agrobacterium tumefaciens virulence gene promoters in Escherichia coli requires the A. tumefaciens RpoA gene, encoding the alpha subunit of RNA polymerase. J Bacteriol. 1999;181:4533–4539. doi: 10.1128/jb.181.15.4533-4539.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo Z-Q, Farrand S K. Signal-dependent DNA binding and functional domains of the quorum-sensing activator TraR as identified by repressor activity. Proc Natl Acad Sci USA. 1999;96:9009–9014. doi: 10.1073/pnas.96.16.9009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lynch J M, Whipps J M. Substrate flow in the rhizosphere. In: Keister D L, Cregan P B, editors. The rhizosphere and plant growth. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1991. pp. 15–24. [Google Scholar]
- 32.McClean K H, Winson M K, Fish L, Taylor A, Chhabra S R, Camara M, Daykin M, Lamb J H, Swift S, Bycroft B W, Stewart G S A B, Williams P. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology. 1997;143:3703–3711. doi: 10.1099/00221287-143-12-3703. [DOI] [PubMed] [Google Scholar]
- 33.McGowan S, Sebaihia M, Jones S, Yu B, Bainton N, Chan P F, Bycroft B, Stewart G S, Williams P, Salmond G P. Carbapenem antibiotic production in Erwinia carotovora is regulated by CarR, a homologue of the LuxR transcriptional activator. Microbiology. 1995;141:541–550. doi: 10.1099/13500872-141-3-541. [DOI] [PubMed] [Google Scholar]
- 34.Mills S D, Jasalavich C A, Cooksey D A. A two-component regulatory system required for copper-inducible expression of the copper resistance operon of Pseudomonas syringae. J Bacteriol. 1993;6:1656–1664. doi: 10.1128/jb.175.6.1656-1664.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murillo J, Shen H, Gerhold D, Sharma A, Cooksey D A, Keen N T. Characterization of pPT23B, the plasmid involved in syringolide production by Pseudomonas syringae pv. tomato PT23. Plasmid. 1994;31:275–287. doi: 10.1006/plas.1994.1029. [DOI] [PubMed] [Google Scholar]
- 36.Pierson L S, III, Keppenne V D, Wood D W. Phenazine antibiotic biosynthesis in Pseudomonas aureofaciens 30–84 is regulated by PhzR in response to cell density. J Bacteriol. 1994;176:3966–3974. doi: 10.1128/jb.176.13.3966-3974.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pierson L S, III, Wood D W, Pierson E A. Homoserine lactone-mediated gene regulation in plant-associated bacteria. Annu Rev Phytopathol. 1998;36:207–225. doi: 10.1146/annurev.phyto.36.1.207. [DOI] [PubMed] [Google Scholar]
- 38.Piper K R, Beck von Bodman S, Farrand S K. Conjugation factor of Agrobacterium tumefaciens regulates Ti plasmid transfer by autoinduction. Nature. 1993;362:448–450. doi: 10.1038/362448a0. [DOI] [PubMed] [Google Scholar]
- 39.Piper K R, Beck von Bodman S, Hwang I, Farrand S K. Hierarchical gene regulatory systems arising from fortuitous gene associations: controlling quorum sensing by the opine regulon in Agrobacterium. Mol Microbiol. 1999;32:1077–1089. doi: 10.1046/j.1365-2958.1999.01422.x. [DOI] [PubMed] [Google Scholar]
- 40.Pirhonen M, Flego D, Heikinheimo R, Palva E T. A small diffusible signal molecule is responsible for the global control of virulence and exoenzyme production in the plant pathogen Erwinia carotovora. EMBO J. 1993;12:2467–2476. doi: 10.1002/j.1460-2075.1993.tb05901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robson N D, Cox A R J, McGowan S J, Bycroft B W, Salmond G P C. Bacterial N-acyl-homoserine-lactone-dependent signalling and its potential biotechnological application. Trends Biotechnol. 1997;15:458–464. doi: 10.1016/S0167-7799(97)01102-5. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 43.Shaw P D, Gao P, Daly S L, Cha C, Cronan J E, Jr, Rinehart K L, Farrand S K. Detecting and characterizing N-acyl-homoserine lactone signal molecules by thin-layer chromatography. Proc Natl Acad Sci USA. 1997;94:6036–6041. doi: 10.1073/pnas.94.12.6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sitnikov D M, Schineller J B, Baldwin T O. Transcriptional regulation of bioluminescence genes from Vibrio fischeri. Mol Microbiol. 1995;17:801–812. doi: 10.1111/j.1365-2958.1995.mmi_17050801.x. [DOI] [PubMed] [Google Scholar]
- 45.Stintzi A, Evans K, Meyer J-M, Poole K. Quorum sensing and siderophore biosynthesis in Pseudomonas aeruginosa: lasR/lasI mutants exhibit reduced pyoverdine biosynthesis. FEMS Microbiol Lett. 1998;166:341–345. doi: 10.1111/j.1574-6968.1998.tb13910.x. [DOI] [PubMed] [Google Scholar]
- 46.Swift S, Bainton N J, Winson M K. Gram-negative bacterial communication by N-acyl homoserine lactones: a universal language? Trends Microbiol. 1994;2:193–198. doi: 10.1016/0966-842x(94)90110-q. [DOI] [PubMed] [Google Scholar]
- 47.Thomashow L S, Weller D M. Role of antibiotics and siderophores in biocontrol of take-all disease of wheat. In: Keister D L, Cregan P B, editors. The rhizosphere and plant growth. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1991. pp. 245–251. [Google Scholar]
- 48.Throup J P, Camara M, Briggs G S, Winson M K, Chhabra S R, Bycroft B W, Williams P, Stewart G S. Characterisation of the yenI/yenR locus from Yersinia enterocolitica mediating the synthesis of two N-acylhomoserine lactone signal molecules. Mol Microbiol. 1995;17:345–356. doi: 10.1111/j.1365-2958.1995.mmi_17020345.x. [DOI] [PubMed] [Google Scholar]
- 49.Vaudequin-Dransart V, Petit A, Poncet C, Ponsonnet C, Nesme X, Jones J B, Bouzar H, Chilton W S, Dessaux Y. Novel Ti plasmids in Agrobacterium strains isolated from fig tree and chrysanthemum tumors and their opinelike molecules. Mol Plant Microbe Interact. 1995;8:311–321. doi: 10.1094/mpmi-8-0311. [DOI] [PubMed] [Google Scholar]
- 50.Winson M K, Swift S, Fish L, Throup J P J P, Jørgensen F, Chhabra S R, Bycroft B W, Williams P, Stewart G S A B. Construction of luxCDABE-based plasmid sensors for investigating N-acyl homoserine lactone-mediated quorum sensing. FEMS Microbiol Lett. 1998;163:185–192. doi: 10.1111/j.1574-6968.1998.tb13044.x. [DOI] [PubMed] [Google Scholar]
- 51.Wood D W, Gong F, Daykin M M, Williams P, Pierson L S., III N-Acyl-homoserine lactone-mediated regulation of phenazine gene expression by Pseudomonas aureofaciens 30–84 in the wheat rhizosphere. J Bacteriol. 1997;179:7663–7670. doi: 10.1128/jb.179.24.7663-7670.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu J, Winans S C. Autoinducer binding by the quorum-sensing regulator TraR increases affinity for target promoters in vitro and decreases TraR turnover rates in whole cells. Proc Natl Acad Sci USA. 1999;96:4832–4837. doi: 10.1073/pnas.96.9.4832. [DOI] [PMC free article] [PubMed] [Google Scholar]