SUMMARY
Several influenza antibodies with broad group 2-neutralization have recently been isolated. Here, we analyze the structure, class, and binding of one of these antibodies from an H7N9 vaccine trial, 315–19-1D12. The cryo-EM structure of 315–19-1D12 Fab in complex with the hemagglutinin (HA) trimer revealed the antibody to recognize the helix A region of the HA stem, at the supersite of vulnerability recognized by group 1-specific and by cross-group neutralizing antibodies. 315–19-1D12 derived from HV1–2 and KV2–28 genes and appeared to form a new antibody class. Bioinformatic analysis indicated its group 2-neutralization specificity to be a consequence of four key residue positions. We specifically tested the impact of the group 1-specific N33 glycan, which decreased but did not abolish group 2-binding of 315–19-1D12. Overall, this study highlights the recognition of a broad group 2-neutralizing antibody, revealing unexpected diversity in neutralization specificity for antibodies that recognize the HA-stem supersite.
Keywords: antibody class, hemagglutinin, influenza, neutralizing antibodies, stem supersite
Graphical Abstract
eTOC Blurb
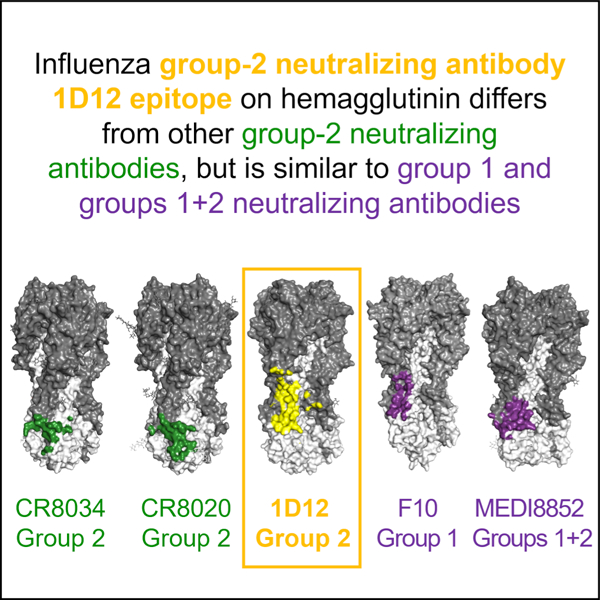
Influenza hemagglutinin stem-targeting antibodies can neutralize group 1 and/or 2 viruses. Cheung et al. isolate a broad group 2-neutralizing antibody, 1D12, from an H7N9-vaccine trial. Its epitope overlaps better with group 1 and cross-group epitopes than with previously described group 2 epitopes, revealing unexpected differences in neutralization specificity versus epitope.
INTRODUCTION
Influenza A and B viruses cause seasonal flu epidemics and are severe threats to public health, resulting in significant disease burden in terms of morbidity, complications, hospitalizations, and deaths, as well as the potential for flu pandemics (reviewed in Wu and Wilson, 2017). For decades, influenza A subtypes H1N1 and H3N2 have been the major strains circulating in humans in annual flu epidemics. However, over the last 20 years, sporadic cross-overs of divergent influenza viruses have occurred with what appears to be increasing frequency; these often involving the group 1-H5N1 strain, which can be highly lethal, but generally does not transmit between humans (Ferguson et al., 2005; Subbarao et al., 1998; reviewed in Neumann et al., 2009; Webster et al., 1992). In 2013, the first human infection with the group 2-H7N9 avian influenza virus was reported in China (Li and Chen, 2020). Since then, the subtype H7 avian influenza viruses have rapidly evolved to be highly pathogenic, causing an increasing number of outbreaks among poultry and humans (Su et al., 2017). As most humans are immunologically naïve to subtype H7 viruses, the H7N9 influenza is considered potentially the most serious future pandemic threat (Su et al., 2017). Moreover, none of the strains used in annual flu vaccination are known to induce protective responses against H7 viruses. Thus, there is an urgent need to develop more effective vaccines and antiviral drugs to combat this disease.
Hemagglutinin (HA) is the major glycoprotein on the influenza surface and principal target of neutralizing antibodies. HA is synthesized as a monomer that assembles into a trimeric precursor, which upon cleavage by host proteases, forms the fusion-competent mature trimer comprised of disulfide-linked HA1 and HA2 subunits (Gething et al., 1980; Verhoeyen et al., 1980; Ward and Dopheide, 1981; Wilson et al., 1981). The trimeric HA consists of a membrane-distal and antigenically variable globular head domain and a more conserved elongated membrane-proximal stem domain dominated by helices. The globular head is made up of HA1 and contains the receptor-binding site (RBS) for viral attachment as well as vestigial esterase subdomain. The stem domain comprises HA2 (F fusion subdomain) and the N- and C-terminal segments of HA1 (F′ fusion subdomain) (Rosenthal et al., 1998; Russell et al., 2004) and is responsible for mediating membrane fusion (Carr and Kim, 1993; Weis et al., 1988; Wilson et al., 1981). In general, neutralizing antibodies against HA are divided into two categories: head-targeting and stem-targeting (Corti et al., 2017; Ekiert and Wilson, 2012). Although head-targeting antibodies are generally more potent, they tend to have restricted recognition, even within a single subtype due to sequence diversity of the head (Cheung et al., 2020; Krause et al., 2011; Nogales et al., 2018; Qiu et al., 2020). In contrast, stem antibodies, which bind the relatively conserved HA stem, generally have considerably greater neutralization breadth and some even have extensive heterosubtypic activities (Corti et al., 2011; De Jong et al., 2020; Dreyfus et al., 2012; Joyce et al., 2016; Kallewaard et al., 2016; Wu and Wilson, 2017). A stem supersite of vulnerability involving the helix A region is recognized by pan group 1-neutralizing and cross-group neutralizing antibodies (Corti et al., 2011; Ekiert et al., 2009; Joyce et al., 2016; McCarthy et al., 2018; Pan et al., 2014; Sui et al., 2009); however a group 2-specific glycan at position 38 impedes recognition of this site, and pan-group 2-neutralizing antibodies identified thus far bind to a distinct region on HA that is substantially closer to the membrane (Ekiert et al., 2011; Friesen et al., 2014).
In the VRC 315 clinical trial (ClincialTrials.gov identifier NCT02206464), healthy, H7N9-naïve adults vaccinated with H7N9 show a transient expansion of memory B cells specific for conserved epitopes on H7 HA with little observed adaptation (Andrews et al., 2019). One of these antibodies, 315–19-1D12 (abbreviated as 1D12) binds group 2 HAs, including H3, H7 and H10, and neutralizes potently many group 2 strains, including a wide spectrum of H3 viruses spanning from 1968–2014, but not group 1 viruses (Creanga et al., 2021). To gain insight into the broad group 2-recognition by this antibody, we determined the cryogenic electron microscopy (cryo-EM) structure of 1D12 in complex with HA trimer and analyzed the revealed epitope relative to other broad influenza-neutralizing antibodies as well as its class and the residue responsible for its neutralization specificity. These studies revealed 1D12 to target the helix A region of the HA stem, an epitope differing from previously described human pan-group 2-neutralizing antibodies, CR8020 (Friesen et al., 2014) and CR8043 (Ekiert et al., 2011) but overlapping with the epitopes of broad cross-group neutralizing antibodies and of group 1-specific neutralizing antibodies such as CR6261 and F10 (Ekiert et al., 2009; Sui et al., 2009). Overall, this study highlights a broad group 2-neutralizing antibody recognizing an atypical group 2-binding site and provides insight into the relationship between recognized epitope and neutralization specificity.
RESULTS
Isolation of the broad group 2 influenza neutralizing antibody 1D12 from a human donor receiving the H7N9 experimental vaccine
We isolated antibody 1D12 from a human donor primed with DNA plasmid (VRC-FLUDNA071–00-VP) that encoded for A/Anhui/1/2013 and a monovalent influenza subunit virion A/Shanghai/02/2013 (SH13) H7N9 vaccine (MIV) manufactured by Sanofi Pasteur Inc (Swiftwater, PA), followed by an H7N9 MIV boost 16 weeks later (Andrews et al., 2019). PBMCs were collected from the donor two weeks after the boost and sorted using the H3 (A/Texas/50/2012) and H7 (SH13) HA probes. 1D12 antibody was isolated from a B cell showing cross-reactivity to both H3 and H7 HA trimers (Figure S1A). Recombinant 1D12 was observed to bind multiple group 2 HAs, including H3, H7, and H10, but no HA from group 1 viruses except H9 (Figures S1B and S1C). The neutralization profile of 1D12 was similar to that of 315–27-1C08 from the same clinical trial and that of CR8020 (Friesen et al., 2014), as well as that of 315–09-1B12 except that the latter also neutralized group 1-H5N1 strains (Andrews et al., 2017; Creanga et al., 2021).
Structural analysis of 1D12 antibody in complex with H7 A/Shanghai/02/2013 (SH13) HA
To elucidate the molecular basis for the broad group 2 reactivity of 1D12, we determined a structure of H7 SH13 HA in complex with 1D12 Fab at 2.76 Å resolution by cryo-EM (Figures 1A and S2, Table S1). Structural analysis revealed 1D12 to bind the HA stem, with 35.3% of the contact surface on HA1 and 64.7% on HA2 (Table S2). Binding involved complementarity-determining regions (CDRs) H1, H3, L1, and L3 (Figure 1A, inset). The heavy and light chains contributed 544.1 Å2 and 600.1 Å2, respectively, to the overall buried surface area (BSA) of 1144.2 Å2 (Figure 1B). The heavy chain framework 1 (FR1) residues Thr28 and Phe29, as well as CDR H1 residues Gly31 and Tyr32 interacted with Lys39HA1 and Gln42 HA1 of helix A (Figure 1C). CDR H3 residues Lys95, Tyr100e, and Tyr100h formed hydrophobic interactions and hydrogen bonds with the HA2 residues Asp19HA2, Gln42HA2, Asp46HA2, and Thr49HA2, which are highly conserved within the group 2 viruses (Figure 1D). The light chain CDR L1 residues Arg24, Ser27a, Leu27c, His27d, and Leu27e interacted with loops of F fusion subdomain through extensive hydrogen bonds (Figure 1E); and Arg93 of CDR L3 also formed hydrogen bonds with Ile56HA2 of helix A and Asn291HA1 (Figure 1F). A summary of these interactions between 1D12 and H7 SH13 HA were annotated on the heavy and light chain sequences in Figure 1G. Overall, 1D12 utilized both heavy and light chain CDRs to bind a relatively conserved region of HA stem, away from the membrane-proximal base.
Figure 1. Cryo-EM structure of 1D12 Fab in complex with H7 SH13 HA trimer at 2.8 Å resolution.
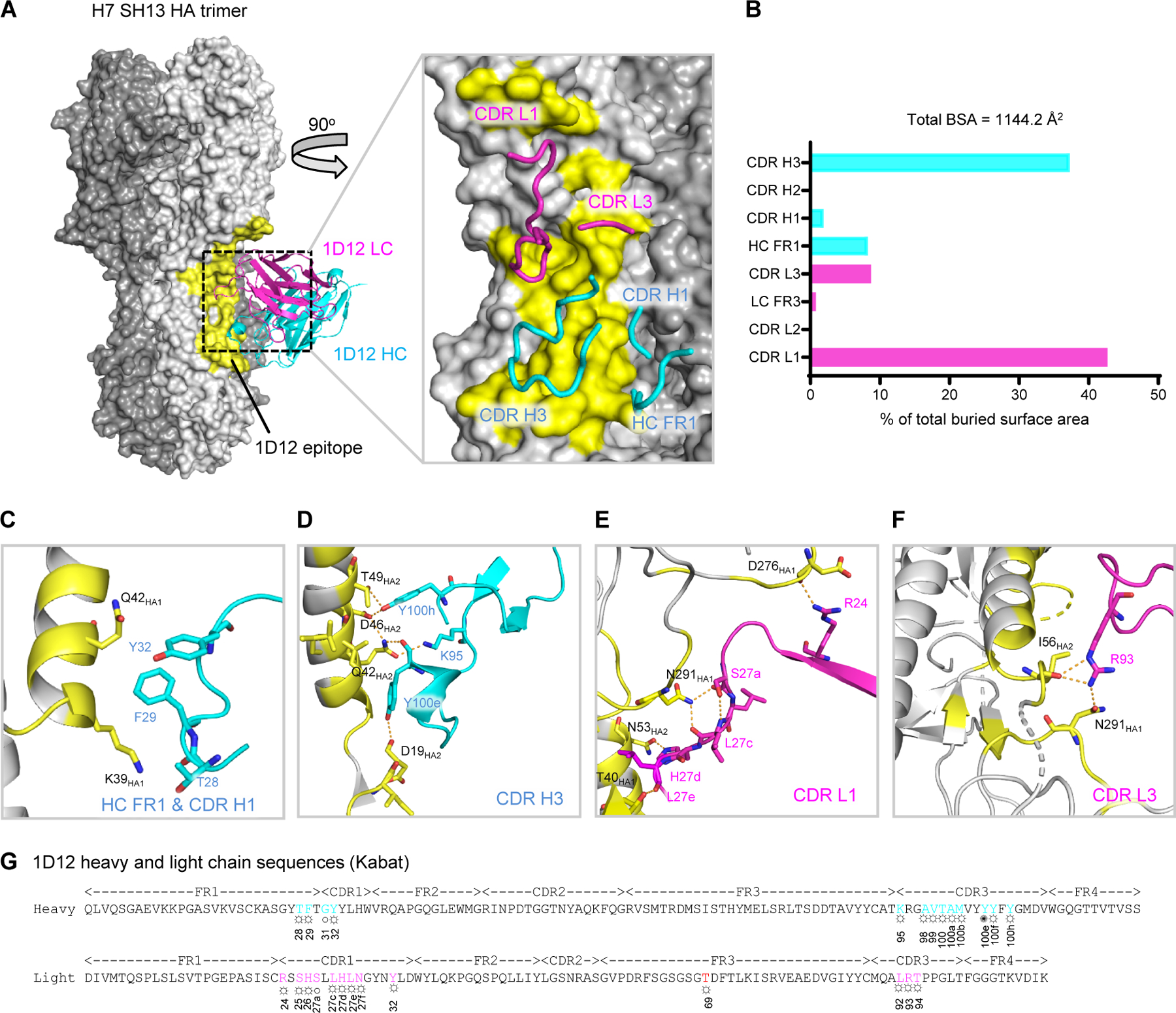
(A) Overall structure of 1D12 Fab bound to the H7 SH13 HA trimer. Only one antibody is shown for clarity. The HA trimer is shown in gray with varied shades for the protomers. The 1D12 epitope is colored yellow; heavy and light chain variable domains colored cyan and magenta, respectively. Zoom-in view highlights interactions of CDR and FR loops with HA. (B) Percentage of total buried surface area on SH13 HA contributed by the FRs and CDRs of 1D12 Fab. Buried surface areas were calculated with PISA (https://www.ebi.ac.uk/pdbe/pisa/). (C-F) CDR interactions show in ribbons with interacting residues in stick representation, colored as in (A). (G) 1D12 sequence annotated with Kabat numbering scheme. Heavy and light residues in contact with HA are colored cyan and magenta. ○ denotes main-chain interaction, ☼ denotes side-chain interaction and ✺ denotes both main- and side-chain interactions. See also Figures S1 and S2, and Tables S1 and S2.
1D12 shares an overlapping epitope with cross-group broadly neutralizing antibodies
To delineate the 1D12 epitope versus other HA-recognizing antibodies, we analyzed surface areas of interaction of 1D12 as well as of all antibody-HA complexes in the PDB and calculated pairwise epitope overlap. Despite binding predominantly group 2 HAs (Figure S1, B and C), the 1D12 epitope was distinct from those of the two other known group 2-specific human antibodies, CR8020 and CR8043 (Ekiert et al., 2011; Friesen et al., 2014). Owing to the conserved N-linked glycan at Asn38HA1 in most group 2 HAs (Ekiert et al., 2011), binding of CR8020 and CR8043 is confined primarily to a portion of the fusion peptide and the edge of the membrane proximal β-sheet (Figure 2B) (Ekiert et al., 2011; Friesen et al., 2014). However, the 1D12 epitope shifted laterally, with the Asn38HA1 glycan at one edge to have favorable interactions with 1D12, enabling it to bind higher up on the stem. As a result, 1D12 shared less than 25% overlapping epitope with CR8020 and CR8043 (Figure 2A,B). By contrast, 1D12 epitope exhibited a much higher percentage of overlap (ranging from 54.5%-69.7%) with cross-group neutralizing antibodies, such as 31.a.83, 39.29, 16.a.26 and 16.g.07, and group 1-specific antibodies, like CR6261 and F10, which bind a supersite of vulnerability encompassing helix A in the middle portion of the HA stem (Figure 2C).
Figure 2. Similar HA binding region observed in 1D12 and cross-group broadly neutralizing antibodies.
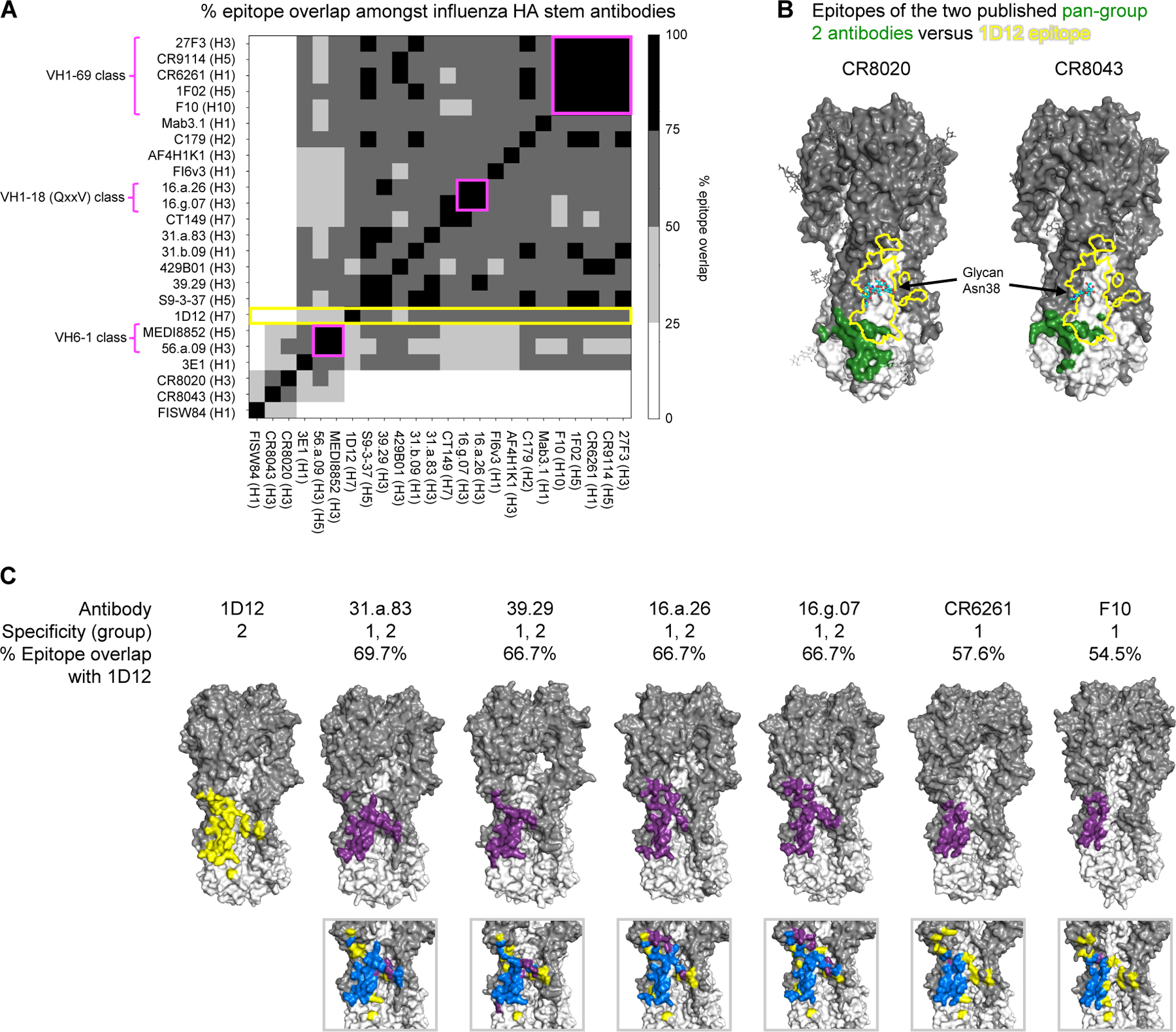
(A) Comprehensive analysis of pairwise % epitope overlap amongst influenza HA stem antibodies. % overlap with the epitope of 1D12 is indicated by yellow. % epitope overlap amongst known antibody classes are indicated by magenta. (B) Epitopes of pan-group 2 antibodies CR8020 and CR8043. The HAs of the CR8020 and CR8043 complexes are depicted as surface representation, with HA1 in dark gray and HA2 in white. The epitopes (colored forest-green) are located near the base of HA and have little overlap with that of 1D12 (yellow outline). Glycan at Asn38HA1 is shown in cyan for carbon atoms. (C) Epitopes of HA stem antibodies that are most similar to that of 1D12 correspond to those of groups 1 and 2 broadly neutralizing and group 1-specific neutralizing antibodies. Epitope of 1D12 is indicated in yellow. Epitopes of all other antibodies are indicated in purple. Overlap of epitopes are indicated in blue.
1D12 does not belong to any known influenza antibody class
To determine whether 1D12 belongs to a previously identified influenza antibody class, i.e., sharing the same recognition mode and similar B cell ontogeny as other influenza antibodies (Kwong and Mascola, 2012; Zhou et al., 2013), we performed informatic and experimental analyses to define its antibody class (Figure 3A). 1D12 was derived from IGHV1–2 and IGLV2–28 germline genes (Figures 3B and S3A). Although the mature 1D12 antibody exhibited low somatic hypermutation (<4% in both heavy and light chains) (Figure S3, A and B), the inferred germline antibody bound none of the group 1 or group 2 HAs tested (Figure S3C). We searched PubMed (https://pubmed.ncbi.nlm.nih.gov) and IEDB (Vita et al., 2019) for influenza A antibodies that used these same germline genes and identified sequences for five other IGHV1–2-encoded and five other IGLV2–28-encoded influenza antibodies. Six of the ten antibodies (CH65, K03.12, 02–1G08, 02–2B07, FLD194 and 3C11) were excluded because they targeted the HA head instead of stem, and thus have a different mode of recognition and could not be of the same class. Further, of the four remaining antibodies, 315–27-1C08 and 315–04-1B12 shared the same IGHV and IGLV genes and belonged to the same class so 315–27-1C08 was used as a representative for both, leaving a total of three antibodies to examine in greater detail (Figure 3B).
Figure 3. Antibody 1D12 appears to be a unique class.
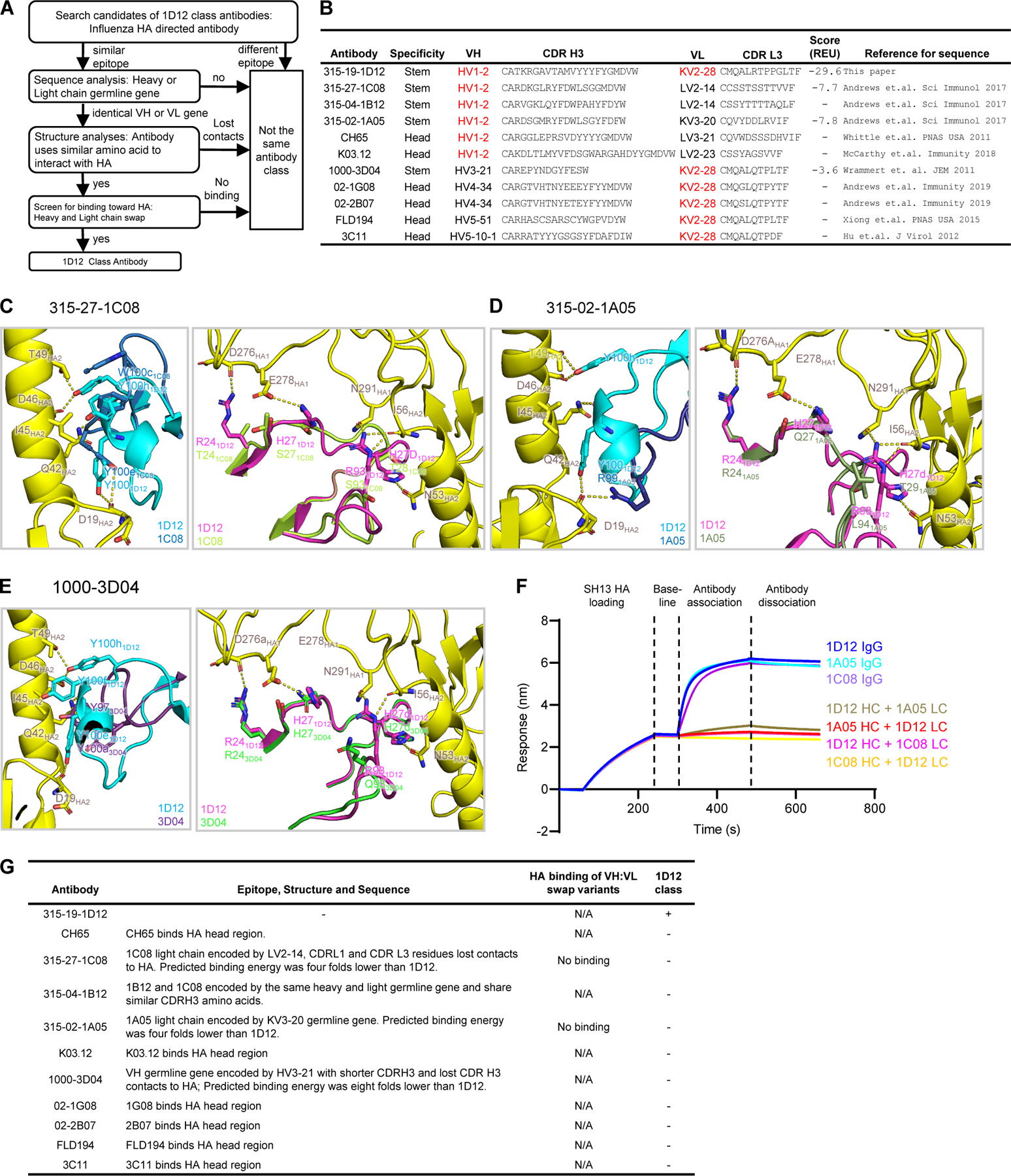
(A) Workflow for determination of 1D12 antibody as a new influenza antibody class. (B) Classification of influenza antibodies encoded by either HV1–2 or KV2–28 from published papers and this paper. (C) - (E) Superposition of 1D12-HA complex with modeled HA complex structures of 315–27-1C08, 315–02-1A05 and 1000–3D04 showing their predicted binding interactions at CDR H3 (left panel of each set) and CDR L1 (right panel of each set). Interacting residues are shown in stick representation. (F) HA binding analysis of antibodies 1D12, 315–02-1A05, and 315–27-1C08, and their heavy and light chain-swapped variants by biolayer interferometry using Octet RED.(G) Summary of classification of 1D12 class antibody. See also Figure S3.
We modeled the heavy and light chains of 315–27-1C08 (Figure 3C), 315–02-1A05 (Figure 3D), and 1000–3D04 (Figure 3E) to analyze whether these antibodies could have the same interactions with HA as 1D12. Even though both 315–27-1C08 and 315–02-1A05 are encoded by the HV1–2 germline gene, modeling indicated contact residues of 1D12 CDRs H3 and L3 to not be conserved in 315–27-1C08 and 315–02-1A05, indicating a 4-fold decrease in the predicted binding energy compared to 1D12 (Figure 3B). Although both 1000–3D04 and 1D12 used the same KV2–28 gene, and therefore light chain contacts to HA are mostly preserved, 1000–3D04 used a completely different heavy chain germline gene (HV3–21), where its shorter CDR H3 length would likely abolish contacts to HA. More importantly, the predicted binding energy score was 8-fold lower than 1D12, indicating 1000–3D04 may use a different binding mode to interact HA (Figure 3B). To verify our bioinformatic analysis, we swapped the light chains between 315–27-1C08 (LV2–14), 315–02-1A05 (LV2–14) and 315–19-1D12 (LV2–28) and observed a complete loss of binding for HA for all swapped variants, indicating that these different heavy and light chains failed to yield functional antibodies, and therefore both 315–27-1C08 and 315–02-1A05 were unlikely to be in the same class as 1D12 (Figure 3F). We also applied informatic analyses to known influenza antibodies and none appeared to belong to the same class as 1D12 (Figure S3D). These results suggest antibody 1D12 to comprise the first member of a newly identified antibody class.
Conserved glycan at Asn33 on HA1 of group 1 HAs limits 1D12 specificity
As described above, 1D12 shared <25% overlapping epitope with two other group 2-specific antibodies CR8020, and CR8043 (Figure 2A). Despite the very similar footprint with the cross-group broadly neutralizing antibodies, 1D12 failed to neutralize group 1 viruses (Creanga et al., 2021), and it could not bind HA of group 1 viruses except H9 (Figure S1B-C). To explain this phenomenon, we modeled the 1D12 Fab binding to the H1N1 CA09 HA (PDB: 4M4Y) (Hong et al., 2013) and H5N1 VN04 HA (PDB: 4MHH) (Zhu et al., 2013), and we found that the presence of the glycan at Asn33HA1, which is structurally distinct from Asn38HA1 and conserved in many group 1 viruses, could hinder the HA interaction with 1D12 (Figures 4A-D, Table S3). This finding is consistent with the observation that 1D12 could bind to H9N2 HK98 HA (PDB: 1JSD) (Ha et al., 2002), which lacks glycan at position 33 (Figure 4D, Table S3). Indeed, as we performed mutagenesis to introduce a glycosylation sequon in wildtype H7 SH13, H3 Vic11, and H9 HK99 HAs at position 33, the mutants showed decreased affinity toward 1D12, relative to their wildtype counterparts (Figure 4E). These results suggest that, although the glycan at Asn33 contributes to the group-2 specificity of 1D12, it is not the only determining factor.
Figure 4. Glycan at Asn33 is present in all HA1 of group 1 except H9 and hinders 1D12 binding.
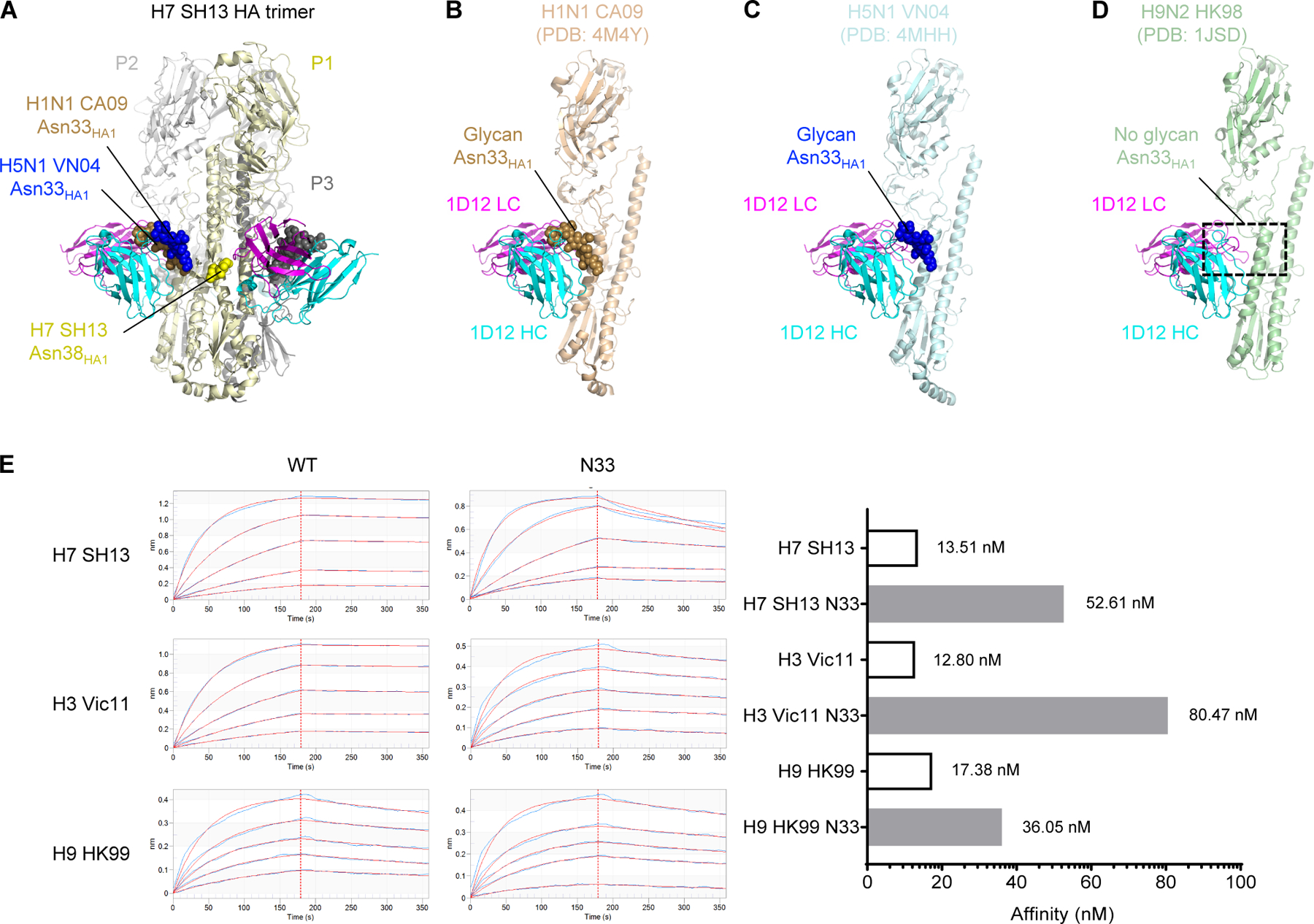
(A) A trimer of the H7 SH13 structure is shown with 1D12. The glycans at positions N33 and N38 of protomer 1 (P1, light yellow) are shown as spheres for this trimer and aligned trimers from H1N1 and H5N1. (B-D) Modelling of the 1D12 Fab onto the H1N1 CA09 HA (PDB: 4M4Y) (B), H5N1 VN04 HA (PDB: 4MHH) (C), and H9N2 HK98 (PDB: 1JSD) (D). The protein of protomer P2 is shown with the glycans from protomer P1. The presence of glycan at HA1 Asn33 in H1 CA09 and H5 VN04 resulted in clashes with the 1D12 Fab. Absence of glycan at HA1 Asn33 in H9 HK98 allowed 1D12 binding without clashes. (E) Octet affinity measurements of 1D12 Fab binding to wild-type group 2 (H3 and H7) as well as group 1 (H9) HAs with and without glycan Asn33 introduced. The KD values were shown on the right. Hemagglutinin analyte concentrations used 2-fold dilutions starting at 100 nM and ending with 6.25 nM.
In silico analysis explains 1D12’s group 2 specificity
The broad group 2-neutralizing capability of antibody 1D12 was surprising in light of its recognition of an epitope that overlapped primarily with antibodies that had group 1-neutralizing or cross-group-neutralizing capabilities. To provide insight into the influenza group 2-specificity of 1D12, we first identified an extended set of 46 epitope positions using buried surface area and sequence analysis, which encompasses all 39 residues determined to be part of the 1D12 epitope (Table S3). Sequence analysis of 55 influenza HAs (Data S1) revealed that the neutralization resistant and sensitive set of strains chemically diverged in the amino acid composition for 20 positions (colored amino acids in Figure 5A). Additionally, we developed an in silico pipeline to estimate binding energies between HA trimers and 1D12, as a surrogate for neutralization. The pipeline includes three steps: (i) modeling of diverse HA trimers from diverse influenza subtypes, (ii) complex formation of HA trimer and 1D12 antibody, and (iii) in silico binding energy calculation using Rosetta Interface Analyzer (Stranges and Kuhlman, 2013). Notably, in silico binding energies accurately distinguished between HA strains that were neutralization resistant or neutralization sensitive (P-value = 3.11e-07, classification accuracy = 89%, ROC AUC = 0.87) (Figure 5B, Figure S5 A). Analysis of position-specific in silico energies suggested multiple positions to contribute to the overall energetic differences between sensitive and resistant strains, as opposed to one single key position (Figure S4).
Figure 5. Resistance and sensitivity of neutralization by 1D12 appears to relate to a collection of sequence changes, not to a single amino acid change in the epitope.
(A) Sequence analysis of epitope residues by chemical properties and corresponding buried surface area. Chemically different residues between the two sets are colored. (B) In silico binding energy distributions for neutralization resistant and sensitive HAs to 1D12. Mann-Whitney test P-value = 3.11e-07. (C) Binding energy distributions for resistant (top) and sensitive (bottom) HAs to 1D12 after introducing consensus sensitive amino acid (top) or consensus resistant amino acid into WT HA sequences. Mean values for each distribution are depicted by red circles. Epitope positions with highest expected mutational effected are highlighted by black dashed boxes and assessed statistically using Mann-Whitney test (****: P-value < 0.0001, ***: P-value < 0.001, ns: not significant). (D) HA-1D12 binding energy distributions after introduction of consensus resistant (cyan) or consensus sensitive (salmon) amino acids in key epitope residues. Single, double, triple and quadruple mutants are highlighted using black, grey, brown and blue dashed boxes, respectively. (E) Key epitope residues are depicted on HA trimer by red spheres. 1D12 light (magenta) and heavy (cyan) chains are shown in surface representation. See also Figures S4 and S5, and Data S1 and S2.
Next, we used the in silico pipeline to further estimate the effects that the 20 chemically-divergent residue positions have on the 1D12 binding energies. Hence, we mutated HA sequences representing neutralization resistant strains at each of the 20 residue positions to the consensus sensitive amino acid of that position, and vice versa all neutralization sensitive HA sequences to the consensus resistant amino acid of that position (colored residues in Figure 5A). Four epitope residue positions showed the expected outcome of increasing the binding strength (lower binding energies) for the neutralization resistant HAs and decreasing the binding strength for the neutralization sensitive HAs (Figure 5C). In particular, residue position 33 – a N-glycosylation site within the neutralization resistant set – along with residue positions 278, 366, and 377 had expected effects. We also assessed if single-, double-, triple-, or quadruple-mutation combinations of the aforementioned residues are sufficient to reverse the binding energies so that the resistant set has higher binding strength to 1D12 than the sensitive set of HAs. Selected double mutants, such as mutations at 33–278 and 33–366, and triple as well as quadruple mutants showed reverse binding strength as expected (Figure 5D). Single mutations, however, did not seem to be sufficient to reverse the binding strength of HAs to 1D12 (Figure 5D). Interestingly, two of the mutations (366 and 377) were located in Helix A of HA2, while two positions are in HA1 (Figure 5E). Overall, these results indicate the group 2-specificity of antibody 1D12 to result from its recognition of a collection of epitope residues, rather than a specific residue, such as the glycan at Asn38HA1, which is a central player in the group 2-specificity of known pan-group 2-neutralizing antibodies.
To validate in silico predictions, we performed binding studies introducing resistant mutations at the four identified epitope positions (33, 278, 366, 377) into two sensitive strains, in particular mutations that should revert the binding (Data S2). We selected strains most likely to knock out binding from the in-silico predictions which resulted in H3N2_Texas_77 and H3N2_Shanghai_87 (Figure S5B). H3N2_Shanghai_87 wildtype did not show any binding to 1D12 antibody at any of the 4 concentrations tested nor to the 640-C1 or MEDI8852 positive control antibodies and was therefore not considered for analysis, since it most likely did not express into well-formed HA trimers. H3N2_Texas_77 wildtype did show high binding to 1D12 at different concentrations, as well as to the positive controls. The in silico energy was then correlated to the experimental binding. Favorable in silico energies were correlated with an increase in experimental binding as predicted (Figure S5C). At best, we found a good correlation R=0.63 with significant P-value of 0.015 at a 10ug/ml 1D12 antibody concentration, at worst R=0.56 and P-value 0.038 at a 0.6ug/ml 1D12 antibody, with similar correlation values at other concentrations.
DISCUSSION
The isolation and characterization of antibodies targeting the stem region of the influenza virus HA have provided valuable insights to the development of therapeutics and vaccines. Over the last few decades, a substantial number of stem-targeting antibodies neutralizing heterosubtypic group 1 influenza viruses, or both group 1 and 2 viruses, have been identified (Corti et al., 2010; Corti et al., 2011; Dreyfus et al., 2013; Ekiert et al., 2009; Ekiert et al., 2011; Friesen et al., 2014; Kashyap et al., 2008; Sui et al., 2009; Throsby et al., 2008; Wrammert et al., 2011). However, heterosubtypic group 2-specific neutralizing antibodies are far less common. So far, only two human group 2-specific neutralizing antibodies (CR8020 and CR8043) have been reported (Ekiert et al., 2011; Friesen et al., 2014).
In this study, we isolated a broad group 2-neutralizing antibody 1D12 from a human vaccinee who received a DNA prime and protein boost of an experimental H7N9 vaccine. Compared to the two previously isolated group 2 antibodies CR8020 and CR8043, which derive from HV1–3 and HV1–18 germline genes respectively, 1D12 is encoded by the HV1–2 gene (Figure 3). HV1–2 is a frequent germline gene for encoding HIV CD4-binding site neutralizing antibodies (Navis et al., 2014), but its usage is much rarer in influenza antibodies. Structural analysis of 1D12 indicated that this antibody bound at a higher position on HA stem at helix A (Figure 1), a site that is different from the epitopes of the other known group 2-specific antibodies CR8020 and CR8043, but instead overlaps considerably with that of the group 1-specific and cross-group antibodies (Figure 2). This binding site has been traditionally considered to be inaccessible to group 2-specific antibodies owing to steric hindrance of the conserved glycan at Asn38HA1 in group 2 HA (Tan et al., 2014). Unexpectedly, we found that a human group 2 influenza-neutralizing antibody can indeed accommodate a glycan at position 38HA1 for binding at the typical cross-group neutralizing site (Figure 1).
Recent studies have demonstrated that influenza broadly neutralizing antibodies can evolve and develop from either group 1- or group 2-preferring germline precursors in humans (Wu et al., 2020). The two different types of precursors undergo distinct evolutionary pathways to acquire somatic hypermutations, leading to increased affinity and breadth (Wu et al., 2020). Because the 1D12 epitope overlapped considerably with that of the cross-group neutralizing antibodies, 1D12 could potentially evolve into a cross-group neutralizing antibody, given a sufficient level of somatic hypermutations being accumulated. Mutagenesis studies introducing glycan at Asn33HA1 in group 2 strains showed reduced antibody binding (Figure 4), suggesting that this glycan served only as part of the molecular barrier for 1D12 to bind group 1 HA. Because 1D12 light chain contacted extensively with the vestigial esterase region and the lower part of the HA head, accounting for approximately 24% of total BSA, we reasoned that the inability of 1D12 to bind group 1 HA could also result from the collective loss of interactions with these residues that are highly variable between groups 1 and 2 HAs (Figure 5).
In summary, our findings revealed that 1D12, despite being a group 2-specific antibody, could accommodate the glycan at Asn38HA1 and bound at the supersite recognized by the group 1 and cross-group antibodies, which is located in the middle of the HA stem instead of at the base as the other two known group 2 antibodies with available structures (Ekiert et al., 2011; Friesen et al., 2014). The structural characterization of 1D12-HA complex here provides insight into the relationship between recognized HA epitope and specificity of influenza neutralization and adds to the accumulating understanding by the field needed to develop vaccines that afford broad protection from diverse influenza strains.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Peter D. Kwong (pdkwong@nih.gov).
Materials availability
Plasmids generated in this study will be available upon request.
Data and code availability
Cryo-EM electron density map of 315–19-1D12 in complex with H7 SH13 HA trimer has been deposited in the Electron Microscopy Data Bank (EMDB) under accession number EMD-21961, and the fitted atomic coordinates have been deposited in the Protein Data Bank (PDB; www.pdb.org) under accession code 6WXL. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines
Human embryonic kidney (HEK)-derived 293T cells were obtained from the American Type Culture Collection (ATCC), and maintained in complete Dulbecco’s Modified Eagle Medium (herein referred to as cDMEM) containing high glucose Dulbecco’s Modified Eagle Medium (DMEM, Thermo Fisher), 1X Penicillin-Streptomycin (Pen Strep, Thermo Fisher) and 10% fetal bovine serum (FBS, Gemini Bio Products) at 37°C. Expi293F cells, obtained from Thermo Fisher, were maintained in Expi293 Expression Medium (Thermo Fisher), at 37oC with shaking at 120 rpm.
METHOD DETAILS
Flow cytometry and single-cell sorting
Cryopreserved peripheral blood mononuclear cells from blood collected before vaccination or two weeks after boost from trial volunteer were stained with the anti-human mAbs CD3, CD56, CD14, CD27, and CD38 from BioLegend; IgG and IgM were from BD Biosciences; and CD19 was from Beckman Coulter. HA probes were expressed, biotinylated, and labeled with fluorochromes, as described previously (Whittle et al., 2014). Briefly, HA ectodomain was C-terminally fused to the T4 fibritin trimerization motif, AviTag sequence GLNDIFEAQKIEWHE, and a hexahistidine affinity tag and expressed in Expi293 cells grown in Expi239 medium (Invitrogen) by transient transfection. Cells were grown for 5 days, and the culture supernatant was clarified by centrifugation. The protein was purified by Ni Sepharose Fast Flow resin (GE Healthcare) and then passed through a Superdex 200 16/60 column in 10 mM Tris (pH 8.0). The fractions containing trimeric HA were pooled and concentrated. The protein was biotinylated using biotin protein ligase (Avidity), following manufacturer’s recommendation, and then exchanged into PBS. Biotinylated protein was then incubated with fluorochrome-labeled streptavidin (ThermoFisher) at a 1:4 molar ratio. Aqua dead cell stain was added for live/dead discrimination (Thermo Fisher Scientific). Stained samples were run on an LSR II (BD Biosciences), and data were analyzed using FlowJo (TreeStar). To single-cell sort, HA-specific B cells were stained as above, and CD19+ IgG+ HA+ B cells were single cell–sorted into 96-well plates using a FACSAria II (BD Biosciences). Addition of multiple HA probes and index sorting were used to determine the binding of each sorted B cell to HA of multiple subtypes simultaneously.
Single-cell Ig amplification and cloning
Reverse transcription was performed on sorted cells, and multiplexed polymerase chain reaction (PCR) was used to amplify Ig heavy and light chain genes, as described previously (Doria-Rose et al., 2016; Tiller et al., 2008). Briefly, cDNA was directly made from cells sorted into dry 96-well plates using Superscript III Reverse Transcriptase (ThermoFisher), dNTPs (ThermoFisher), random hexamers (GeneLink), IGEPAL (Sigma-Aldrich) and RNAseOUT (ThermoFisher). Immunoglobulin heavy and light chain (kappa and lambda) genes were then PCR amplified separately with two rounds of nested PCR, 50 cycles each, using DreamTaq Mastermix (ThermoFisher) and previously published primer pools and cycle conditions (Doria-Rose et al., 2016). The paired heavy and light chain Ig sequences were obtained from an average of 70% of single cells. PCR products were sequenced by Beckman Coulter or Genewiz and analyzed using IMGT (Brochet et al., 2008). Heavy and light variable region sequences were synthesized and cloned by GenScript into pVRC8400 vectors, containing human IgG1, kappa, or lambda constant region and a human cytomegalovirus promoter.
Preparation of HA trimers
All HA constructs being studied were fused to a thrombin cleavage sequence, T4 fibritin trimerization motif, and a hexahistidine affinity tag at their C-termini. DNA sequences encoding H7 SH13, H3 VIC11, and H9 HK99 were synthesized by GenScript. Asn33 mutation was introduced into H7 SH13, H3 VIC11, and H9 HK99 HA constructs by GeneImmune Biotechnology LLC. DNA sequences of H7 SH13, H3 VIC11, and H9 HK99 wildtype and mutant HA trimers were cloned into a pVRC8400 expression plasmid as previously described (Whittle et al., 2014). Expi293 cells were diluted to 1.2 × 106 cells/ml and transfected with 1,000 μg/liter of HA expression plasmids using Turbo293 transfection reagent. At day six, the media was clarified by centrifugation at 10,000 × g and filtered, concentrated, and loaded on Complete His-Tag Resin (Roche) by gravity flow. The resin was washed with three column volumes of PBS with 50 mM imidazole (Roche) and the target protein was subsequently eluted in three column volumes of PBS with 300 mM imidazole. The eluted protein was concentrated and loaded on a Superdex 200 16/60 size exclusion column (GE Healthcare). Fractions corresponding to SH13 HA trimer were collected, pooled and concentrated.
Antibody production and Fab preparation
To produce the 315–19-1D12 antibody, Expi293 cells were transfected with pVRC8400 plasmids encoding Ig heavy and light chain pairs with ExpiFectamine (Thermo Fisher Scientific). The cells were grown at 33oC for five days and IgG purified from the cell supernatant using Sepharose Protein A (Pierce). The purified IgG protein was cleaved by LysC enzyme (1:4000 w/w) (Roche) at 37°C overnight for Fab preparation. The enzymatic digestion reaction was stopped by adding in protease inhibitor (Roche). The cleavage mixture was subsequently passed through a Protein A column to separate the Fc fragments from the Fab. The Fab collected in the flow-through was further purified by a Superdex 200 16/60 column in a buffer containing 5 mM HEPES pH 7.5 and 150 mM NaCl.
Cryo-electron microscopy structural analyses
The H7 SH13 HA trimer was incubated with molar excess of antibody Fab fragments of 315–191-D12 and 2.3 μl of the complex at 2 mg/ml concentration was deposited on a C-flat 1.2/1.3 carbon grid (protochip.com). An FEI Vitrobot Mark IV was used for vitrification with a wait time of 30 seconds, blot time of 3 seconds, blot force of 1 and humidity of 100%. Leginon (Suloway et al., 2005) was used for automated data collection on a Titan Krios microscope was using a Gatan K3 direct detection device. Exposures were collected in movie mode for 2 seconds with the total dose of 49.9 e–/Å2. Pre-processing was performed with Appion (Lander et al., 2009; Voss et al., 2010); frames were aligned and dose-weighted with MotionCor2 (Zheng et al., 2017), ctf was estimated with CTFFind4 (Rohou and Grigorieff, 2015; Zhang, 2016), DoG Picker (Lander et al., 2009; Voss et al., 2010) was used for particle picking, RELION (Scheres, 2012) was then used for particle extraction. CryoSPARC 2.15 (Punjani et al., 2017) was subsequently used for the remaining steps of 2D classifications, ab initio 3D reconstruction, and nonuniform 3D refinement. The initial reconstruction was performed using C1 symmetry, confirming 3 Fab molecules per trimer, C3 symmetry was then applied for the final refinement. Model building through coot was followed by simulated annealing and real space refinement in Phenix (Adams et al., 2004) and then iteratively improved with manual fitting of the coordinates in Coot (Emsley and Cowtan, 2004). Geometry and map fitting were assessed through Molprobity (Davis et al., 2004) and EMRinger (Barad et al., 2015). PyMOL (www.pymol.org) and ChimeraX (Pettersen et al., 2021) were used to generate figures.
Calculation of pairwise epitope overlap
To determine the epitope residues, the surface area of each hemagglutinin residue buried by the antibody of each antibody is calculated based on published antibody-HA complex structure using NACCESS (http://www.bioinf.manchester.ac.uk/naccess/). A residue is considered an epitope residue if it has >5Å2 of such buried surface area. The epitope overlap between two antibodies defined as (# of overlapping epitope residues between 2 antibodies) / (maximum number of epitope residues for the 2 antibodies).
Homology modeling of antibody
Antibody sequences were threaded on H7 SH13 HA-1D12 Fab complex structure (PDB: 6WXL) using software NEST (Koehl and Delarue, 1995). The models were energy minimized using YASARA (Krieger and Vriend, 2015) to give optimized structure models. Rosetta package, InterfaceAnalyzer, was applied to predict binding energy between antibody and influenza A virus HA (Stranges and Kuhlman, 2013). The values of dG_separated reported by Interface Analyzer were used in Figure 3B.
Germline reversion
The inferred germline-reverted antibody was constructed by reverting the V (HV1–2*06 and KV2–28*01) and J (HJ6*02 and KJ4*01) amino acids to germline versions. Amino acids on CDR 3 region within the range of germline gene were also reverted to germline amino acids.
HA binding assay
Meso Scale Discovery (MSD) 384 well Streptavidin coated SECTOR® Imager 2400 Reader Plates were blocked with 5% MSD Blocker A for 30–60 minutes, then washed six times with the wash buffer (PBS+0.05% Tween). The plates were then coated with recombinant biotinylated HA protein for one hour and washed. mAbs were diluted in 1% MSD Blocker A to 1μg/ml, serially diluted 3-fold, and added to the coated plates. Sera was diluted 1:100 in 1% MSD Blocker A and serially diluted 3-fold before adding to coated plates. After one hour incubation, plates were washed and incubated with SULFO-TAG conjugated anti-human IgG for one hour. After washing, the plates were read using 1X MSD Read Buffer using a MSD SECTOR® Imager 2400. Binding curves were plotted and the area under the curve (AUC) was determined using Prism 7.
Affinity measurement
Affinity expressed as dissociation constant (KD) value was determined by biolayer interferometry (BLI) using an Octet RED instrument (ForteBio, Inc.). Histidine-tagged HAs (wildtype or mutants) were loaded onto NTA sensors for 180 s. For the measurement of kON, the association of 1D12 Fab was measured for 180 s by exposing the sensors to six concentrations (100 nM, 50 nM, 25 nM, 12.5 nM, 6.25 nM, 0 nM) of Fabs in PBS. For the measurement of kOFF, the dissociation of 1D12 Fab was measured for 180 s. Experiments were performed at 30°C. Data analysis and curve fitting were carried out using the Octet Data Analysis Software 9.0 (fortéBio). Experimental data were fitted with the binding equations describing a 1:1 interaction. Global analysis of the data sets assuming reversible binding (full dissociation) were carried out using nonlinear least-squares fitting allowing a single set of binding parameters to be obtained simultaneously for all of the concentrations used in each experiment.
In silico protein-protein interface analysis
In silico pipeline was developed to approximate protein-protein interface binding energies to correlate with the neutralization outcome. First, homology models of HA molecules of the diverse influenza subtypes were generated using YASARA software (Stranges and Kuhlman, 2013), where the experimentally determined H7 SH13 HA-1D12 Fab cryo-EM structure served as a template. The modelled HA structure was then superimposed onto the cryo-EM complex structure and minimized using YASARA. Subsequently, Rosetta’s InterfaceAnalyzer (https://new.rosettacommons.org/docs/latest/application_documentation/analysis/interface-analyzer) was used to determine the binding energies between the modelled HA structure and the 1D12 antibody.
An extended epitope set of 46 positions was determined by taking the union of positions with strictly positive BSA values across all virus strains. This extended epitope definition was chosen to cover all energy values present in Rosetta’s InterfaceAnalyzer outputs.
Sequence analysis was performed by first aligning HA sequences with the 2HMG sequence as reference using MAFFT. For logo plots, residues were clustered per position into 6 chemical categories {GSTYC} (Polar), {QN} (Neutral), {KRH} (Basic), {DE} (Acidic), {AVLIPWFM} (Hydrophobic) and {O} where O represents N-glycosylated residues with the pattern NxS or NxT with x≠P.
A residue was colored grey at a given position if the similarity between the frequency distribution for the sensitive set P and the resistant set Q was lower than 0.5, as measured by the Jensen-Shannon distance JSD. , where m is the is the pointwise mean of P and Q, and D is the Kullback-Leibler divergence with x being a chemical category.
Experimental validation of in silico method
We performed binding studies introducing resistant mutations at the four identified epitope positions (33, 278, 366, 377) into two sensitive strains, in particular mutations that should revert the binding.
Two strains with the highest average in-silico delta energies ∆Ej across the 4 key positions with the constraint that these strains should also contain the most frequent and representative residue from the sensitive set as shown in figure 5A were first selected.
In other words, we selected strains most likely to knock out binding in-silico with residues Q33, S278, L366 and N377. This resulted in H3N2_Texas_77 and H3N2_ Shanghai_87. The resistant mutations introduced were also the most representative of the full resistant set (glycan33N_E35T, 278D, 366Q and 377T).
For H3N2_Texas_1977, a ∆Binding was calculated by ∆Binding = Bindmut − BindWT where Bindmut corresponds to the ELISA reading of a given mutant and BindWT corresponds to the HA_A_H3N2_Texas_1_1977_DS2–6R ELISA reading. A mutant was not considered in the analysis if the MEDI8852 control reading was below the average negative controls (Medium and blank in Data S2, average = 0.103). A correlation was calculated for each individual 1D12 antibody concentration. P-values were calculated using the python library statsmodels.api.OLS.
96-well expression and ELISA assessment
Wells with 100 μL of log-phase HEK 293T cells at 2.5 x 105/ml density in RealFect Media (ABI Scientific) were inoculated in a 96-well cell culture microplate (Corning Scientific), and allowed to grow for 24 hours at 37°C, 5% CO2. Immediately before transfection, 40 μL of media was removed from each well. 250 ng of plasmid DNA encoding His-tagged HA protein (GenScript synthesized) in 10 μL of Opti-MEM I Reduced Serum Medium (Invitrogen) was mixed with 0.75 μL of TrueFect Max transfection reagent (United Biosystems) in 10μL of Opti-MEM I Reduced Serum Medium at RT for 15 minutes, then added into a cell well in 96-well cell culture microplate. One day post transfection, 30 μL per well of CelBooster Cell Growth Enhancer (ABI Scientific) was added. Four days post transfection, the supernatant in the cell well was harvested, and analyzed in a 96-well ELISA format. Briefly, 30 μL of expression supernatant and 70 μL of PBS per well were incubated in Ni-coated plate (Thermo Scientific) at RT for 2 hrs. Captured HA proteins were detected by incubating with anti-HA primary antibodies at 10 μg/ml concentration at RT for 60 minutes, followed with HRP-conjugate (Jackson ImmunoResearch Labs) at RT for 30 minutes. After final washing, the reaction signal was developed by addition of BioFX-TMB (SurModics) at RT for 10 minutes. The reaction was stopped by the addition of 0.5N H2SO4. The signal was measured at 450nm on a micro plate reader (SpectraMax Plus, Molecular Devices).
QUANTIFICATION AND STATISTICAL ANALYSIS
Cryo-EM data were processed and analyzed using CryoSPARC. Cryo-EM structural statistics were analyzed with Phenix and Molprobity. BLI data were analyzed with ForteBio Data Analysis 9.0 software supplied by the instrument manufacturer. B cell sorting data were analyzed with GraphPad Prism. Statistical details of experiments are described in Method Details or figure legends. The in silico energy distribution in Figures 5B, C, D was analyzed with the Mann-Whitney test with n_sensitive=26 and n_resistant=29. Additional details are described in Figure 5 legend. Experimental validation of in silico results in Figure S5C was analyzed as detailed above in the section of Experimental validation of in silico method.
Supplementary Material
Data S1. Sequences of influenza hemagglutinin used in the in silico 1D12 binding analysis, related to Figure 5
Data S2. Antigenic assessment of influenza HA mutants with 96-well ELISA screening, related to Figure 5
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER | |
|---|---|---|---|
| Antibodies | |||
| 315–19-1D12 | This paper | N/A | |
| Bacterial and virus strains | |||
| None | |||
| Biological Samples | |||
| PBMC of donor 19 (VRC 315 trial) | (Andrews et al., 2017) | N/A | |
| Chemicals, peptides, and recombinant proteins | |||
| 2’-(4-Methylumbelliferyl)-alpha-D-N-aceylneuramainic acid sodium salt hydrate | Sigma | Cat# 8639 | |
| AbBooster medium | ABI scientific | Cat# PB2668 | |
| Anti-total IgG-alkaline peroxidase antibody | Southern Biotechnologies | Cat# OB2041–04 | |
| BirA | Avidity | Cat# BirA | |
| Bovine serum albumin | Sigma | Cat# A2058 | |
| Cell culture lysis buffer | Promega | Cat# E1531 | |
| COmplete His-Tag Resin | Roche | Cat# 05893801001 | |
| DreamTaq Mastermix | ThermoFisher | Cat# K1082 | |
| Endoproteinase LysC | Roche | Cat# 11058533103 | |
| Endoglycosidase H | New England Biolabs | Cat# P0702L | |
| Fugene6 | Promega | Cat# E2691 | |
| HEPES | Life Technologies | Cat# 15630–080 | |
| IGEPAL CA-630 | Sigma-Aldrich | Cat# I3021 | |
| IgG elution buffer | Thermo Scientific | Cat# 21009 | |
| Luciferase assay reagent | Promega | Cat# E1500 | |
| Ni-NTA Capture Biosensors | fortéBIO | Cat# 18–5103 | |
| p-nitrophenyl phosphate | Sigma | Cat# 487663 | |
| Protease inhibitor cocktail tablets | Roche | Cat# 37378900 | |
| Random Hexamers | GeneLink | Cat# 26–4000-03 | |
| Recombinant H1 CA09 protein | Protein Sciences | Cat# NR-44074 | |
| Recombinant Protein A sepharose | GE Healthcare | Cat# 17–1279-03 | |
| RNAseOUT | ThermoFisher | Cat# 10777019 | |
| Sodium chloride | Quality Biological, Inc | Cat# 351–036-101 | |
| Streptavidin PE | ThermoFisher | Cat# SA10041 | |
| Superscript III Reverse Transcriptase | ThermoFisher | Cat# 18080093 | |
| Thrombin | Millipore | Cat# 2964423 | |
| Turbo293 transfection reagent | SPEED BioSystem | Cat# PXX1002 | |
| Critical commercial assays | |||
| DNA gene synthesis and cloning | GenScript Biotech Corporation | N/A | |
| Site-directed mutagenesis | GeneImmune Biotechnology LLC | N/A | |
| Deposited data | |||
| Cryo-EM data and structure of SH13 HA in complex with 1D12 Fab | This paper | PD: 6WXL; EMDB: EMD-21961 | |
| Experimental models: Cell lines | |||
| Human: Expi293F cells | Thermo Fisher | Cat# A14527; RRID: CVCL_D615 | |
| Human: FreeStyle 293-F cells | Thermo Fisher | Cat# R79007 | |
| Human HEK 293T/17 cells | ATCC | Cat# CRL-11268 | |
| Recombinant DNA | |||
| H3N2_A/Victoria/361/2011 hemagglutinin wild-type plasmid | NIH/VRC | N/A | |
| H3N2_A/Victoria/361/2011 hemagglutinin plasmid with Asn33 introduced | NIH/VRC | N/A | |
| H7N9 A/Shanghai/2/2013 hemagglutinin wild-type plasmid | NIH/VRC | N/A | |
| H7N9 A/Shanghai/2/2013 hemagglutinin plasmid with Asn33 introduced | NIH/VRC | N/A | |
| H9N2 A/Hong Kong/1073/99 Hong Kong hemagglutinin wild-type plasmid | NIH/VRC | N/A | |
| H9N2 A/Hong Kong/1073/99 Hong Kong hemagglutinin plasmid with Asn33 introduced | NIH/VRC | N/A | |
| pVRC8400–1D12-heavy-chain | NIH/VRC | N/A | |
| pVRC8400–1D12-light-chain | NIH/VRC | N/A | |
| Software and algorithms | |||
| Chimera | (Pettersen et al., 2004) | https://www.cgl.ucsf.edu/chimera/ | |
| Coot | (Emsley and Cowtan, 2004) | https://sbgrid.org/software/ | |
| CryoSPARC 2.15 | (Punjani et al., 2017) | https://cryosparc.com | |
| EMRinger | (Barad et al., 2015) | http://emringer.com/ | |
| GCTF | (Zhang, 2016) | https://www.mrc-lmb.cam.ac.uk/kzhang/Gctf/ | |
| ForteBio Data Analysis 9.0 software | ForteBio, Inc. | N/A | |
| IMGT | (Lefranc et al., 2015) | http://www.imgt.org | |
| Leginon | (Suloway et al., 2005) | https://sbgrid.org/software/titles/leginon | |
| MolProbity | (Williams et al., 2018) | http://molprobity.biochem.duke.edu | |
| NACCESS | http://www.bioinf.manchester.ac.uk/naccess/ | N/A | |
| NEST | (Koehl and Delarue, 1995) | N/A | |
| PDBePISA | (Krissinel and Henrick, 2007) | http://www.ebi.ac.uk/pdbe/pisa/ | |
| Phenix | (Adams et al., 2004) | https://sbgrid.org/software/ | |
| PRISM | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ | |
| PyMOL | Schrödinger | https://pymol.org | |
| Relion | (Scheres, 2012) | https://www3.mrc-lmb.cam.ac.uk/relion/index.php/Main_Page | |
| Rosetta | (Lyskov et al., 2013; Weitzner et al., 2017) | https://rosie.graylab.jhu.edu | |
| YASARA | (Krieger and Vriend, 2015) | http://www.yasara.org | |
HIGHLIGHTS.
Structure of 1D12 in complex with HA at 2.8 Å reveals an epitope in the HA stem
1D12 epitope differs from that of other known broad group 2-neutralizing antibodies
1D12 derives from IGHV1–2 and IGKV2–28 and forms another antibody class
HA residues 33, 278, 366, and 377 delineate neutralization specificity of 1D12
ACKNOWLDEGEMENTS
We thank J. Stuckey for assistance with figures, and members of the Structural Biology Section and Structural Bioinformatics Core, Vaccine Research Center, for discussions and comments on the manuscript. Support for this work was provided by the Intramural Research Program of the Vaccine Research Center, National Institute of Allergy and Infectious Diseases. Some of this work was performed at the Simons Electron Microscopy Center (SEMC) and National Resource for Automated Molecular Microscopy located at the New York Structural Biology Center, supported by grants from the Simons Foundation (SF349247), NYSTAR, and the NIH National Institute of General Medical Sciences (GM103310).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS: The authors declare no competing interests.
REFERENCES
- Adams PD, Gopal K, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Pai RK, Read RJ, Romo TD, et al. (2004). Recent developments in the PHENIX software for automated crystallographic structure determination. J Synchrotron Radiat 11, 53–55. [DOI] [PubMed] [Google Scholar]
- Andrews SF, Chambers MJ, Schramm CA, Plyler J, Raab JE, Kanekiyo M, Gillespie RA, Ransier A, Darko S, Hu J, et al. (2019). Activation Dynamics and Immunoglobulin Evolution of Pre-existing and Newly Generated Human Memory B cell Responses to Influenza Hemagglutinin. Immunity 51, 398–410. [DOI] [PubMed] [Google Scholar]
- Andrews SF, Joyce MG, Chambers MJ, Gillespie RA, Kanekiyo M, Leung K, Yang ES, Tsybovsky Y, Wheatley AK, Crank MC, et al. (2017). Preferential induction of cross-group influenza A hemagglutinin stem-specific memory B cells after H7N9 immunization in humans. Sci Immunol 2, eaan2676. [DOI] [PubMed] [Google Scholar]
- Barad BA, Echols N, Wang RY, Cheng Y, DiMaio F, Adams PD, and Fraser JS (2015). EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy. Nat Methods 12, 943–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brochet X, Lefranc MP, and Giudicelli V (2008). IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res 36, W503–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr CM, and Kim PS (1993). A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell 73, 823–832. [DOI] [PubMed] [Google Scholar]
- Cheung CS, Fruehwirth A, Paparoditis PCG, Shen CH, Foglierini M, Joyce MG, Leung K, Piccoli L, Rawi R, Silacci-Fregni C, et al. (2020). Identification and Structure of a Multidonor Class of Head-Directed Influenza-Neutralizing Antibodies Reveal the Mechanism for Its Recurrent Elicitation. Cell Rep 32, 108088. [DOI] [PubMed] [Google Scholar]
- Corti D, Cameroni E, Guarino B, Kallewaard NL, Zhu Q, and Lanzavecchia A (2017). Tackling influenza with broadly neutralizing antibodies. Curr Opin Virol 24, 60–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti D, Suguitan AL Jr., Pinna D, Silacci C, Fernandez-Rodriguez BM, Vanzetta F, Santos C, Luke CJ, Torres-Velez FJ, Temperton NJ, et al. (2010). Heterosubtypic neutralizing antibodies are produced by individuals immunized with a seasonal influenza vaccine. J Clin Invest 120, 1663–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti D, Voss J, Gamblin SJ, Codoni G, Macagno A, Jarrossay D, Vachieri SG, Pinna D, Minola A, Vanzetta F, et al. (2011). A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 333, 850–856. [DOI] [PubMed] [Google Scholar]
- Creanga A, Gillespie RA, Fisher BE, Andrews SF, Lederhofer J, Yap C, Hatch L, Stephens T, Tsybovsky Y, Crank MC, et al. (2021). A comprehensive influenza reporter virus panel for high-throughput deep profiling of neutralizing antibodies. Nature Communications 12, 1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis IW, Murray LW, Richardson JS, and Richardson DC (2004). MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res 32, W615–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jong NMC, Aartse A, Van Gils MJ, and Eggink D (2020). Development of broadly reactive influenza vaccines by targeting the conserved regions of the hemagglutinin stem and head domains. Expert Rev Vaccines 19, 563–577. [DOI] [PubMed] [Google Scholar]
- Doria-Rose NA, Bhiman JN, Roark RS, Schramm CA, Gorman J, Chuang GY, Pancera M, Cale EM, Ernandes MJ, Louder MK, et al. (2016). New Member of the V1V2-Directed CAP256-VRC26 Lineage That Shows Increased Breadth and Exceptional Potency. J Virol 90, 76–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfus C, Ekiert DC, and Wilson IA (2013). Structure of a classical broadly neutralizing stem antibody in complex with a pandemic H2 influenza virus hemagglutinin. J Virol 87, 7149–7154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfus C, Laursen NS, Kwaks T, Zuijdgeest D, Khayat R, Ekiert DC, Lee JH, Metlagel Z, Bujny MV, Jongeneelen M, et al. (2012). Highly conserved protective epitopes on influenza B viruses. Science 337, 1343–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekiert DC, Bhabha G, Elsliger MA, Friesen RH, Jongeneelen M, Throsby M, Goudsmit J, and Wilson IA (2009). Antibody recognition of a highly conserved influenza virus epitope. Science 324, 246–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekiert DC, Friesen RH, Bhabha G, Kwaks T, Jongeneelen M, Yu W, Ophorst C, Cox F, Korse HJ, Brandenburg B, et al. (2011). A highly conserved neutralizing epitope on group 2 influenza A viruses. Science 333, 843–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekiert DC, and Wilson IA (2012). Broadly neutralizing antibodies against influenza virus and prospects for universal therapies. Curr Opin Virol 2, 134–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, and Cowtan K (2004). Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- Ferguson NM, Cummings DA, Cauchemez S, Fraser C, Riley S, Meeyai A, Iamsirithaworn S, and Burke DS (2005). Strategies for containing an emerging influenza pandemic in Southeast Asia. Nature 437, 209–214. [DOI] [PubMed] [Google Scholar]
- Friesen RH, Lee PS, Stoop EJ, Hoffman RM, Ekiert DC, Bhabha G, Yu W, Juraszek J, Koudstaal W, Jongeneelen M, et al. (2014). A common solution to group 2 influenza virus neutralization. Proc Natl Acad Sci U S A 111, 445–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gething MJ, Bye J, Skehel J, and Waterfield M (1980). Cloning and DNA sequence of double-stranded copies of haemagglutinin genes from H2 and H3 strains elucidates antigenic shift and drift in human influenza virus. Nature 287, 301–306. [DOI] [PubMed] [Google Scholar]
- Ha Y, Stevens DJ, Skehel JJ, and Wiley DC (2002). H5 avian and H9 swine influenza virus haemagglutinin structures: possible origin of influenza subtypes. Embo j 21, 865–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong M, Lee PS, Hoffman RM, Zhu X, Krause JC, Laursen NS, Yoon SI, Song L, Tussey L, Crowe JE Jr., et al. (2013). Antibody recognition of the pandemic H1N1 Influenza virus hemagglutinin receptor binding site. J Virol 87, 12471–12480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Voss J, Zhang G, Buchy P, Zuo T, Wang L, Wang F, Zhou F, Wang G, Tsai C, et al. (2012). A human antibody recognizing a conserved epitope of H5 hemagglutinin broadly neutralizes highly pathogenic avian influenza H5N1 viruses. J Virol 86, 2978–2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce MG, Wheatley AK, Thomas PV, Chuang GY, Soto C, Bailer RT, Druz A, Georgiev IS, Gillespie RA, Kanekiyo M, et al. (2016). Vaccine-Induced Antibodies that Neutralize Group 1 and Group 2 Influenza A Viruses. Cell 166, 609–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallewaard NL, Corti D, Collins PJ, Neu U, McAuliffe JM, Benjamin E, Wachter-Rosati L, Palmer-Hill FJ, Yuan AQ, Walker PA, et al. (2016). Structure and Function Analysis of an Antibody Recognizing All Influenza A Subtypes. Cell 166, 596–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashyap AK, Steel J, Oner AF, Dillon MA, Swale RE, Wall KM, Perry KJ, Faynboym A, Ilhan M, Horowitz M, et al. (2008). Combinatorial antibody libraries from survivors of the Turkish H5N1 avian influenza outbreak reveal virus neutralization strategies. Proc Natl Acad Sci U S A 105, 5986–5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehl P, and Delarue M (1995). A self consistent mean field approach to simultaneous gap closure and side-chain positioning in homology modelling. Nat Struct Biol 2, 163–170. [DOI] [PubMed] [Google Scholar]
- Krause JC, Tsibane T, Tumpey TM, Huffman CJ, Basler CF, and Crowe JE Jr. (2011). A broadly neutralizing human monoclonal antibody that recognizes a conserved, novel epitope on the globular head of the influenza H1N1 virus hemagglutinin. J Virol 85, 1090510908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger E, and Vriend G (2015). New ways to boost molecular dynamics simulations. J Comput Chem 36, 996–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, and Henrick K (2007). Inference of macromolecular assemblies from crystalline state. J Mol Biol 372, 774–97. [DOI] [PubMed] [Google Scholar]
- Kwong PD, and Mascola JR (2012). Human antibodies that neutralize HIV-1: identification, structures, and B cell ontogenies. Immunity 37, 412–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander GC, Stagg SM, Voss NR, Cheng A, Fellmann D, Pulokas J, Yoshioka C, Irving C, Mulder A, Lau PW, et al. (2009). Appion: an integrated, database-driven pipeline to facilitate EM image processing. J Struct Biol 166, 95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefranc MP, Giudicelli V, Duroux P, Jabado-Michaloud J, Folch G, Aouinti S, Carillon E, Duvergey H, Houles A, Paysan-Lafosse T, et al. (2015). IMGT(R), the international ImMunoGeneTics information system(R) 25 years on. Nucleic Acids Res 43, D413–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, and Chen H (2020). H7N9 Influenza Virus in China. Cold Spring Harb Perspect Med [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyskov S, Chou FC, Conchuir SO, Der BS, Drew K, Kuroda D, Xu J, Weitzner BD, Renfrew PD, Sripakdeevong P, et al. (2013). Serverification of molecular modeling applications: the Rosetta Online Server that Includes Everyone (ROSIE). PLoS One 8, e63906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy KR, Watanabe A, Kuraoka M, Do KT, McGee CE, Sempowski GD, Kepler TB, Schmidt AG, Kelsoe G, and Harrison SC (2018). Memory B Cells that Cross-React with Group 1 and Group 2 Influenza A Viruses Are Abundant in Adult Human Repertoires. Immunity 48, 174–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navis M, Tran K, Bale S, Phad GE, Guenaga J, Wilson R, Soldemo M, McKee K, Sundling C, Mascola J, et al. (2014). HIV-1 receptor binding site-directed antibodies using a VH1–2 gene segment orthologue are activated by Env trimer immunization. PLoS Pathog 10, e1004337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann G, Noda T, and Kawaoka Y (2009). Emergence and pandemic potential of swineorigin H1N1 influenza virus. Nature 459, 931–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales A, Piepenbrink MS, Wang J, Ortega S, Basu M, Fucile CF, Treanor JJ, Rosenberg AF, Zand MS, Keefer MC, et al. (2018). A Highly Potent and Broadly Neutralizing H1 Influenza-Specific Human Monoclonal Antibody. Sci Rep 8, 4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Sasaki T, Kubota-Koketsu R, Inoue Y, Yasugi M, Yamashita A, Ramadhany R, Arai Y, Du A, Boonsathorn N, et al. (2014). Human monoclonal antibodies derived from a patient infected with 2009 pandemic influenza A virus broadly cross-neutralize group 1 influenza viruses. Biochem Biophys Res Commun 450, 42–48. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–12. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Meng EC, Couch GS, Croll TI, Morris JH, and Ferrin TE (2021). UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci 30, 70–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punjani A, Rubinstein JL, Fleet DJ, and Brubaker MA (2017). cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods 14, 290–296. [DOI] [PubMed] [Google Scholar]
- Qiu Y, Stegalkina S, Zhang J, Boudanova E, Park A, Zhou Y, Prabakaran P, Pougatcheva S, Ustyugova IV, Vogel TU, et al. (2020). Mapping of a Novel H3-Specific Broadly Neutralizing Monoclonal Antibody Targeting the Hemagglutinin Globular Head Isolated from an Elite Influenza Virus-Immunized Donor Exhibiting Serological Breadth. J Virol 94, e01035–01019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohou A, and Grigorieff N (2015). CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol 192, 216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal PB, Zhang X, Formanowski F, Fitz W, Wong CH, Meier-Ewert H, Skehel JJ, and Wiley DC (1998). Structure of the haemagglutinin-esterase-fusion glycoprotein of influenza C virus. Nature 396, 92–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell RJ, Gamblin SJ, Haire LF, Stevens DJ, Xiao B, Ha Y, and Skehel JJ (2004). H1 and H7 influenza haemagglutinin structures extend a structural classification of haemagglutinin subtypes. Virology 325, 287–296. [DOI] [PubMed] [Google Scholar]
- Scheres SH (2012). RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 180, 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranges PB, and Kuhlman B (2013). A comparison of successful and failed protein interface designs highlights the challenges of designing buried hydrogen bonds. Protein Sci 22, 74–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S, Gu M, Liu D, Cui J, Gao GF, Zhou J, and Liu X (2017). Epidemiology, Evolution, and Pathogenesis of H7N9 Influenza Viruses in Five Epidemic Waves since 2013 in China. Trends Microbiol 25, 713–728. [DOI] [PubMed] [Google Scholar]
- Subbarao K, Klimov A, Katz J, Regnery H, Lim W, Hall H, Perdue M, Swayne D, Bender C, Huang J, et al. (1998). Characterization of an avian influenza A (H5N1) virus isolated from a child with a fatal respiratory illness. Science 279, 393–396. [DOI] [PubMed] [Google Scholar]
- Sui J, Hwang WC, Perez S, Wei G, Aird D, Chen LM, Santelli E, Stec B, Cadwell G, Ali M, et al. (2009). Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol 16, 265–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, and Carragher B (2005). Automated molecular microscopy: the new Leginon system. J Struct Biol 151, 41–60. [DOI] [PubMed] [Google Scholar]
- Tan GS, Lee PS, Hoffman RM, Mazel-Sanchez B, Krammer F, Leon PE, Ward AB, Wilson IA, and Palese P (2014). Characterization of a broadly neutralizing monoclonal antibody that targets the fusion domain of group 2 influenza A virus hemagglutinin. J Virol 88, 13580–13592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Throsby M, van den Brink E, Jongeneelen M, Poon LL, Alard P, Cornelissen L, Bakker A, Cox F, van Deventer E, Guan Y, et al. (2008). Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS One 3, e3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller T, Meffre E, Yurasov S, Tsuiji M, Nussenzweig MC, and Wardemann H (2008). Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J Immunol Methods 329, 112–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhoeyen M, Fang R, Jou WM, Devos R, Huylebroeck D, Saman E, and Fiers W (1980). Antigenic drift between the haemagglutinin of the Hong Kong influenza strains A/Aichi/2/68 and A/Victoria/3/75. Nature 286, 771–776. [DOI] [PubMed] [Google Scholar]
- Vita R, Mahajan S, Overton JA, Dhanda SK, Martini S, Cantrell JR, Wheeler DK, Sette A, and Peters B (2019). The Immune Epitope Database (IEDB): 2018 update. Nucleic Acids Res 47, D339–D343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss NR, Lyumkis D, Cheng A, Lau PW, Mulder A, Lander GC, Brignole EJ, Fellmann D, Irving C, Jacovetty EL, et al. (2010). A toolbox for ab initio 3-D reconstructions in single-particle electron microscopy. J Struct Biol 169, 389–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward CW, and Dopheide TA (1981). Amino acid sequence and oligosaccharide distribution of the haemagglutinin from an early Hong Kong influenza virus variant A/Aichi/2/68 (X-31). Biochem J 193, 953–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster RG, Bean WJ, Gorman OT, Chambers TM, and Kawaoka Y (1992). Evolution and ecology of influenza A viruses. Microbiol Rev 56, 152–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis W, Brown JH, Cusack S, Paulson JC, Skehel JJ, and Wiley DC (1988). Structure of the influenza virus haemagglutinin complexed with its receptor, sialic acid. Nature 333, 426–431. [DOI] [PubMed] [Google Scholar]
- Weitzner BD, Jeliazkov JR, Lyskov S, Marze N, Kuroda D, Frick R, Adolf-Bryfogle J, Biswas N, Dunbrack RL Jr., and Gray JJ (2017). Modeling and docking of antibody structures with Rosetta. Nat Protoc 12, 401–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle JR, Wheatley AK, Wu L, Lingwood D, Kanekiyo M, Ma SS, Narpala SR, Yassine HM, Frank GM, Yewdell JW, et al. (2014). Flow cytometry reveals that H5N1 vaccination elicits cross-reactive stem-directed antibodies from multiple Ig heavy-chain lineages. J Virol 88, 4047–4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle JR, Zhang R, Khurana S, King LR, Manischewitz J, Golding H, Dormitzer PR, Haynes BF, Walter EB, Moody MA, et al. (2011). Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc Natl Acad Sci U S A 108, 14216–14221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CJ, Headd JJ, Moriarty NW, Prisant MG, Videau LL, Deis LN, Verma V, Keedy DA, Hintze BJ, Chen VB, et al. (2018). MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci 27, 293–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson IA, Skehel JJ, and Wiley DC (1981). Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature 289, 366–373. [DOI] [PubMed] [Google Scholar]
- Wrammert J, Koutsonanos D, Li GM, Edupuganti S, Sui J, Morrissey M, McCausland M, Skountzou I, Hornig M, Lipkin WI, et al. (2011). Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J Exp Med 208, 181–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu NC, Andrews SF, Raab JE, O’Connell S, Schramm CA, Ding X, Chambers MJ, Leung K, Wang L, Zhang Y, et al. (2020). Convergent Evolution in Breadth of Two V(H)6–1-Encoded Influenza Antibody Clonotypes from a Single Donor. Cell Host Microbe 28, 434–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu NC, and Wilson IA (2017). A Perspective on the Structural and Functional Constraints for Immune Evasion: Insights from Influenza Virus. J Mol Biol 429, 2694–2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X, Corti D, Liu J, Pinna D, Foglierini M, Calder LJ, Martin SR, Lin YP, Walker PA, Collins PJ, et al. (2015). Structures of complexes formed by H5 influenza hemagglutinin with a potent broadly neutralizing human monoclonal antibody. Proc Natl Acad Sci U S A 112, 9430–9435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K (2016). Gctf: Real-time CTF determination and correction. J Struct Biol 193, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, and Agard DA (2017). MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Zhu J, Wu X, Moquin S, Zhang B, Acharya P, Georgiev IS, Altae-Tran HR, Chuang GY, Joyce MG, et al. (2013). Multidonor analysis reveals structural elements, genetic determinants, and maturation pathway for HIV-1 neutralization by VRC01-class antibodies. Immunity 39, 245–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Guo YH, Jiang T, Wang YD, Chan KH, Li XF, Yu W, McBride R, Paulson JC, Yuen KY, et al. (2013). A unique and conserved neutralization epitope in H5N1 influenza viruses identified by an antibody against the A/Goose/Guangdong/1/96 hemagglutinin. J Virol 87, 12619–12635. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Sequences of influenza hemagglutinin used in the in silico 1D12 binding analysis, related to Figure 5
Data S2. Antigenic assessment of influenza HA mutants with 96-well ELISA screening, related to Figure 5
Data Availability Statement
Cryo-EM electron density map of 315–19-1D12 in complex with H7 SH13 HA trimer has been deposited in the Electron Microscopy Data Bank (EMDB) under accession number EMD-21961, and the fitted atomic coordinates have been deposited in the Protein Data Bank (PDB; www.pdb.org) under accession code 6WXL. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


